-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaPolycomb Group Gene Regulates Rice () Seed Development and Grain Filling via a Mechanism Distinct from
Cereal endosperm represents 60% of the calories consumed by human beings worldwide. In addition, cereals also serve as the primary feedstock for livestock. However, the regulatory mechanism of cereal endosperm and seed development is largely unknown. Polycomb complex has been shown to play a key role in the regulation of endosperm development in Arabidopsis, but its role in cereal endosperm development remains obscure. Additionally, the enzyme activities of the polycomb complexes have not been demonstrated in plants. Here we purified the rice OsFIE2-polycomb complex using tandem affinity purification and demonstrated its specific H3 methyltransferase activity. We found that the OsFIE2 gene product was responsible for H3K27me3 production specifically in vivo. Genetic studies showed that a reduction of OsFIE2 expression led to smaller seeds, partially filled seeds, and partial loss of seed dormancy. Gene expression and proteomics analyses found that the starch synthesis rate limiting step enzyme and multiple storage proteins are down-regulated in OsFIE2 reduction lines. Genome wide ChIP–Seq data analysis shows that H3K27me3 is associated with many genes in the young seeds. The H3K27me3 modification and gene expression in a key helix-loop-helix transcription factor is shown to be regulated by OsFIE2. Our results suggest that OsFIE2-polycomb complex positively regulates rice endosperm development and grain filling via a mechanism highly different from that in Arabidopsis.
Published in the journal: . PLoS Genet 9(3): e32767. doi:10.1371/journal.pgen.1003322
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003322Summary
Cereal endosperm represents 60% of the calories consumed by human beings worldwide. In addition, cereals also serve as the primary feedstock for livestock. However, the regulatory mechanism of cereal endosperm and seed development is largely unknown. Polycomb complex has been shown to play a key role in the regulation of endosperm development in Arabidopsis, but its role in cereal endosperm development remains obscure. Additionally, the enzyme activities of the polycomb complexes have not been demonstrated in plants. Here we purified the rice OsFIE2-polycomb complex using tandem affinity purification and demonstrated its specific H3 methyltransferase activity. We found that the OsFIE2 gene product was responsible for H3K27me3 production specifically in vivo. Genetic studies showed that a reduction of OsFIE2 expression led to smaller seeds, partially filled seeds, and partial loss of seed dormancy. Gene expression and proteomics analyses found that the starch synthesis rate limiting step enzyme and multiple storage proteins are down-regulated in OsFIE2 reduction lines. Genome wide ChIP–Seq data analysis shows that H3K27me3 is associated with many genes in the young seeds. The H3K27me3 modification and gene expression in a key helix-loop-helix transcription factor is shown to be regulated by OsFIE2. Our results suggest that OsFIE2-polycomb complex positively regulates rice endosperm development and grain filling via a mechanism highly different from that in Arabidopsis.
Introduction
Rice (Oryza sativa) serves as the staple food for over half of the world's population. The main body of rice grain is endosperm, which is consumed as food. Endosperm stores energy primarily in the form of starch, storage proteins, and lipids. As in other angiosperms, rice seed development requires initiation signals from double fertilization. During double fertilization, one sperm cell fuses with the egg cell to develop into the diploid embryo while the other sperm cell fuses with the central cell of the female gametophyte to develop into the triploid endosperm. Before fertilization, premature divisions of the egg and the central cells are suppressed. Substantial progress has been made in understanding the role of the polycomb group (PcG) genes in the repression of central cell division before fertilization in Arabidopsis.
Polycomb group (PcG) genes, first discovered in Drosophila melanogaster, play an important role in maintaining the repressed state of homeotic (HOX) genes by posttranslational modifications of histones [1]. PcG genes also control a large number of genes regulating many cellular functions and developmental pathways, such as cell proliferation, stem cell maintenance, imprinting and cancer [2], [3]. PcG proteins form three principle types of multiprotein complexes, Polycomb Repressive Complex 1 (PRC1), Polycomb Repressive Complex 2 (PRC2) and Pho RC. PcG complexes have been identified in different organisms, including Drosophila melanogaster and Arabidopsis thaliana [3]–[9]. In Drosophila melanogaster, PRC2 complex consists of E (Z), ESC, Su (Z) 12 and p55 [10].
The identification and characterization of PRC2 complex genes in plants indicated remarkable structural and functional conservation of the PRC2 complexes between plants and animals. Genetic and molecular studies suggested the presence of three PRC2-like complexes in Arabidopsis thaliana: the Fertilization Independent Seed (FIS), Embryonic Flower (EMF) and Vernalization (VRN) complexes [3], [8], [9], [11]–[17]. The EMF complex, which probably contains CLF/SWN, EMF2, FIE and MSI1, promotes vegetative development by repressing the transcription of flowering activators such as Flowering Locus T (FT) and Agamous-like 19 (AGL19) [18]–[20]. The VRN complex is involved in cold-induced epigenetic silencing of Arabidopsis Flowering locus C (FLC). The VRN complex is purified biochemically by tandem affinity purification method [3]. It includes VRN2, SWINGER (E (Z) homolog), FIE (ESC homolog), MSI1 (p55 homolog), VRN5, VIN3, and VEL1 [3], [4], [13], [14], [21], [22]. The VRN complex associates with PHD-finger proteins for better H3K27me3 deposition. Mathematical modeling showed that a polycomb-based switch underlying quantitative epigenetic memory [23].
Mutations of FIS complex genes (MEA, FIS2, FIE and MSI1) result in the formation of multinucleate central cell that develops to the point of cellularization in the absence of double fertilization. The partially developed seed like structures eventually atrophy. Multinucleate central cell that develops to an endosperm was capable of nourishing an embryo [24]. Mutations in Arabidopsis FIS genes also show an interesting phenotype after fertilization, including endosperm over-proliferation, embryo arrest and abortion. In FIS mutants, endosperm nuclei continue to divide even after the wild type endosperm stopped to replicate and develops large balloon like seeds with delayed cellularization in the endosperm. Gradually, the endosperm collapses and the seed aborts. It is not clear whether the endosperm and embryo phenotypes are both directly due to the FIS gene mutation or whether one is the primary defect and the other is a downstream event [25].
Mutations in Arabidopsis FIS genes also show parent-of-origin effect on seed development [5]–[8], [26]. Every seed that inherits a maternal mutant FIS allele aborts regardless of the presence of a wild type paternal allele [5], [27]–[30]. MEADEA (FIS1) and FIS2 are imprinted genes and expressed maternally throughout endosperm development in Arabidopsis [5], [31]–[33]. FIE (FIS3) maternal allele is expressed in the early endosperm [12], [16]. A recent study using the FIE mutant showed that the PRC2 complex is essential for the transition from embryonic phase to the seedling stage [34]. The FIE mutant show delayed germination. 40% of the homozygous FIE mutants failed to germinate after 20 days while the wildtype germinated within 2 days. In addition, polycomb group proteins are required to couple seed coat initiation to fertilization [35].
Cereal crops, such as Maize, barley and Rice, have multiple homologs of PRC2 core complex genes. Two homologs of ESC are identified in maize, ZmFie1 and ZmFie2. The ZmFie1 is expressed only in endosperm and imprinted during endosperm development, whereas ZmFie2 is expressed in the egg cell and more intensively in the central cell [36]–[38]. Maize has three E (Z) homologs: MEZ1, MEZ2, and MEZ3 [39], [40]. Four polycomb gene homologs are identified in barley, HvFIE, HvE(Z), HvSu(Z)12a and HvSu(Z)12b [41]. Genome wide analysis studies indicated the presence of multiple homologs of ESC, E (Z), and Su (Z) 12 in Rice genome [42]. Rice has two ESC homologs (OsFIE1 (Os08g04290) and OsFIE2 (Os08g04270)), two E (Z) homologs (OsSET1 (Os03g19480) and OsCLF (Os06g16390)) and two Su (Z) 12 homologs (OsEMF2a (Os04g08034) and OsEMF2b (Os09g13630)) [42]–[45].
Interestingly, not all the FIS complex functions identified in Arabidopsis are conserved in other plants [42], [46]. Both imprinted genes MEA and FIS2 are involved in the central cell repression and endosperm development in Arabidopsis. But rice, maize and barley genomes do not have MEA and FIS2 gene orthologs [42], [46], [47]. In addition, all the reported rice polycomb genes are widely expressed in different tissues including endosperm except that OsFIE1 is imprinted and the maternal allele is expressed specifically in endosperm [42], [45]. Mutations in Arabidopsis FIS genes show autonomous endosperm development whereas T-DNA insertion line in OsFIE1 gene did not result in autonomous central cell proliferation in rice [42], suggesting that even though OsFIE1 is imprinted in rice endosperm, it is not involved in the repression of central cell proliferation. Knockdown of ZmFIE1 and ZmFIE2 genes in maize also produced no autonomous central cell proliferation [46]. In sexual Hieracium pilosella, RNAi lines of HFIE failed to show the autonomous central cell proliferation [48]. But down-regulation of HFIE in sexual Hieracium resulted in seed abortion after fertilization. Similarly, specific HFIE down regulation in the apomictic Hieracium resulted in embryo abortion and defective endosperm, indicating that HFIE gene is important for the development of viable seed in both sexual and apomictic Hieracium [48]. The presence of homologs of Arabidopsis polycomb genes in rice, maize and barley genomes indicates the conservation of polycomb genes between monocot and dicot plants. Whereas, the lack of some important seed specific FIS genes (MEA and FIS2) in rice, maize and barley genomes suggests that there might be different functional regulatory mechanisms involved in the embryo and endosperm development in monocots.
Despite the extensive genetic studies on polycomb complex genes during the seed development in plants, biochemical characterization of the polycomb complex is still very limited except that the VRN complex has been purified via tandem affinity purification in Arabidopsis. The enzyme activities of the PcG complex have not been demonstrated in vitro for plants. To elucidate the biological function of the polycomb complex and its molecular mechanism in the regulation of gene expression, we purified the polycomb complex, identified the components of the complex via mass spectrometry analysis, and detected the enzyme activity of the complex by in vitro assay. In addition, we generated RNAi and over expression lines of OsFIE2. The phenotype of the mutant lines clearly demonstrated an essential role of the complex in the regulation of endosperm development, grain filling and seed dormancy. The regulatory mechanisms are different from those in Arabidopsis. Transcription and proteomic analysis of selected nutrient metabolic pathway genes showed that many of them are subject to OsFIE2 regulation, directly or indirectly. Finally, the H3K27me3 binding sites in young endosperm were identified using ChIP-Seq approach. Our results suggest that the polycomb group genes control seed development and grain filling by regulating a large number of genes.
Results
Tandem affinity purification of OsFIE2-interacting proteins
The FIE proteins belong to a family of WD-repeat proteins that promote protein-protein interaction in various multiprotein complexes. They are necessary for the viable seed formation in both Arabidopsis and Hieracium. To identify the proteins associated with rice OsFIE2, the tandem affinity purification (TAP) approach was used [3], [49]–[57]. The TAP tag used in this study contains protein A, the TEV recognition site and the calmodulin-binding protein (CBP) domain [49]–[51]. The final N-Terminal TAP-Tagged transgenic rice plants and cell suspension cultures expressing the TAP-Tagged OsFIE2 were generated. The expression of TAP-tagged OsFIE2 protein was verified by Western blots in multiple transgenic lines (Figure 1A-i). A cell culture system established from the most highly expressed transgenic line 04A was selected for future studies.
Fig. 1. Tandem affinity purification of OsFIE2 complex. 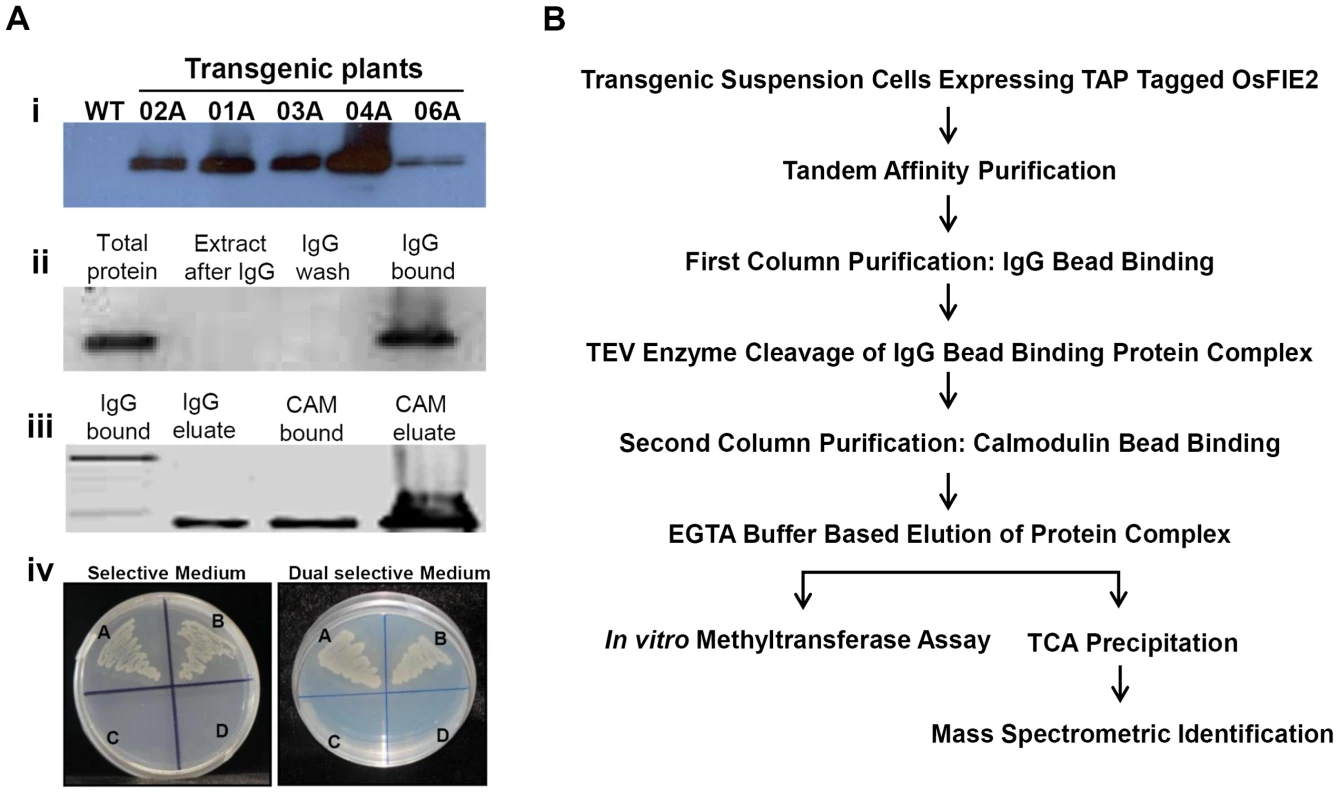
(A) Western blot analyses and E.coli two hybrid assay. (i) TAP tagged OsFIE2 expression in different transgenic lines. The transgenic lines are indicated on the top. Antibodies specific for the protein A domain of the TAP tag were used for Western blots shown in panels i and ii. (ii) Western blot analysis of the samples collected in the first column purification phase. (iii) Western blot analysis of the samples collected in the second column purification phase. CBP domain was detected by using biotinylated CAM [51]. (iv) E. coli two hybrid assay. A: pBT-LGF2+pTRG-Gal II - Positive control. B: Recombinant pBT-OsSET1+ Recombinant pTRG-OsFIE2. C: Recombinant pBT-OsSET1+ Empty pTRG - Negative control. D: Empty pBT+Recombinant pTRG-OsFIE2 - Negative control. Co-transformations were performed with above mentioned pair wise plasmid combinations. Positive interactions were identified by screening co-transformed cells on selective medium containing 3-Amino-1, 2, 4-triazole (3-AT). Positive interactions were further confirmed by screening on dual selective medium containing 3-AT and streptomycin. (B) Schematic representation of different steps used in tandem affinity purification of OsFIE2 complex. Different steps invloved in the TAP purification method are depicted in Figure 1B. In the first step of the tandem purification, protein A domain of OsFIE2 TAP tag was bound to the IgG sepharose beads. The TAP tagged proteins enrichment to the IgG beads were revealed by Western blot analysis (Figure 1A-ii). The IgG bound proteins were eluted by digestion with AcTEV protease. The AcTEV cut eluate was incubated with CAM agarose beads. The protein enrichment by the CAM beads was detected by Western blot analysis (Figure 1A-iii). Finally, OsFIE2-TAP complex proteins were eluted with buffer containing EGTA. The eluate was TCA precipitated and processed for mass spectrometry analyses after trypsin digestion.
The complex proteins were identified using LC-MS/MS mass spectrometry analyses with the protein identification criteria of minimal two peptides as we reported previously [58]–[60]. The OsFIE2 complex includes OsFIE2 (homolog of ESC), OsCLF (homolog of E (Z)), OsSET1 (homolog of E (Z)), and OsEMF2b (homolog of Su (Z) 12) (Table 1). The histone H4 protein was identified in this study (Table 1). It is reported that Drosophila histone H4 strongly binds to polycomb protein p55 [61]. The RbAp48/46, mammalian polycomb protein, also binds to the histone H4 [62], [63]. Whether H4 is a stable component of the rice OsFIE2 complex remains to be further verified although it is possible. Two other proteins identified in our purification were elongation factor 1-alpha and heat shock cognate 70 kDa protein 2. These two proteins had been identified in an affinity purification using the same TAP system in rice and were considered as nonspecific interaction [53]. We also identified the heat shock proteins during the purification of another protein using the same TAP system (Mujahid and Peng, unpublished results). Therefore, we concluded that these two proteins were not the interactive proteins of the OsFIE2-TAP complex.
Tab. 1. 2D-LC-MS/MS analysis of OsFIE2 associated complex proteins identified by Tandem affinity purification. 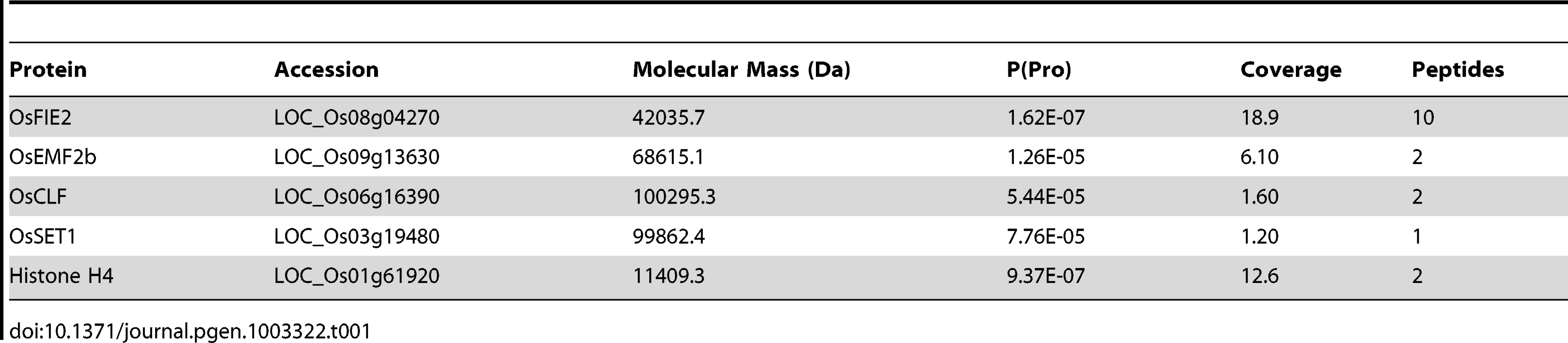
OsFIE2 interacts with OsSET1 in Escherichia coli two-hybrid system
To understand the structure of the OsFIE2-PRC2 complex, we carried E. coli two-hybrid assays for pairwise interactions of the four polycomb subunits. E. coli two-hybrid assay has some advantages over yeast two-hybrid such as E. coli grows much faster than yeast and it can be transformed with higher efficiency so larger number of interactions can be rapidly screened. In addition, E. coli two-hybrid assay reduces the chance that the host harbors a eukaryotic homologue of one of the interacting protein partners [64]. Our results showed that OsFIE2 interacted with OsSET1 strongly (Figure 1A-iv). However, no other interactions were detected using the E. coli two-hybrid system. The results support that OsSET1 is a component of the OsFIE2 complex and suggest that post translational modification(s), other cellular component(s) such as long non-coding RNAs, or the nuclear environment is required to reveal other protein-protein interactions within the OsFIE2 complex.
In vitro activity of TAP purified OsFIE2 complex
Mammalian and Drosophila PRC2 complexes are shown to methylate H3K27 (histone H3 at lysine 27) and, to a lesser extent, H3K9 both in vivo and in vitro [65]–[68]. The substrates include H3K27, H3K27me, H3K27me2, and H3K9me. Although VRN complexes have been purified in Arabidopsis, no enzyme activities are reported in vitro. We carried out in vitro histone methyltransferase assay using the TAP purified OsFIE2 protein complex as enzyme, chicken core histones as substrate, and S-adenosyl-[methyl-3H]-l-methionine as methyl donor. The TAP purified OsFIE2-PRC2 complex demonstrated strong methyltransferase activity against histone H3 only (Figure 2A), indicating that our purified OsFIE2-PRC2 complex is a functional complex with H3 specific enzyme activity.
Fig. 2. In vitro histone methyl transferase assay and immunoblot analyses of H3 modifications in transgenic lines. 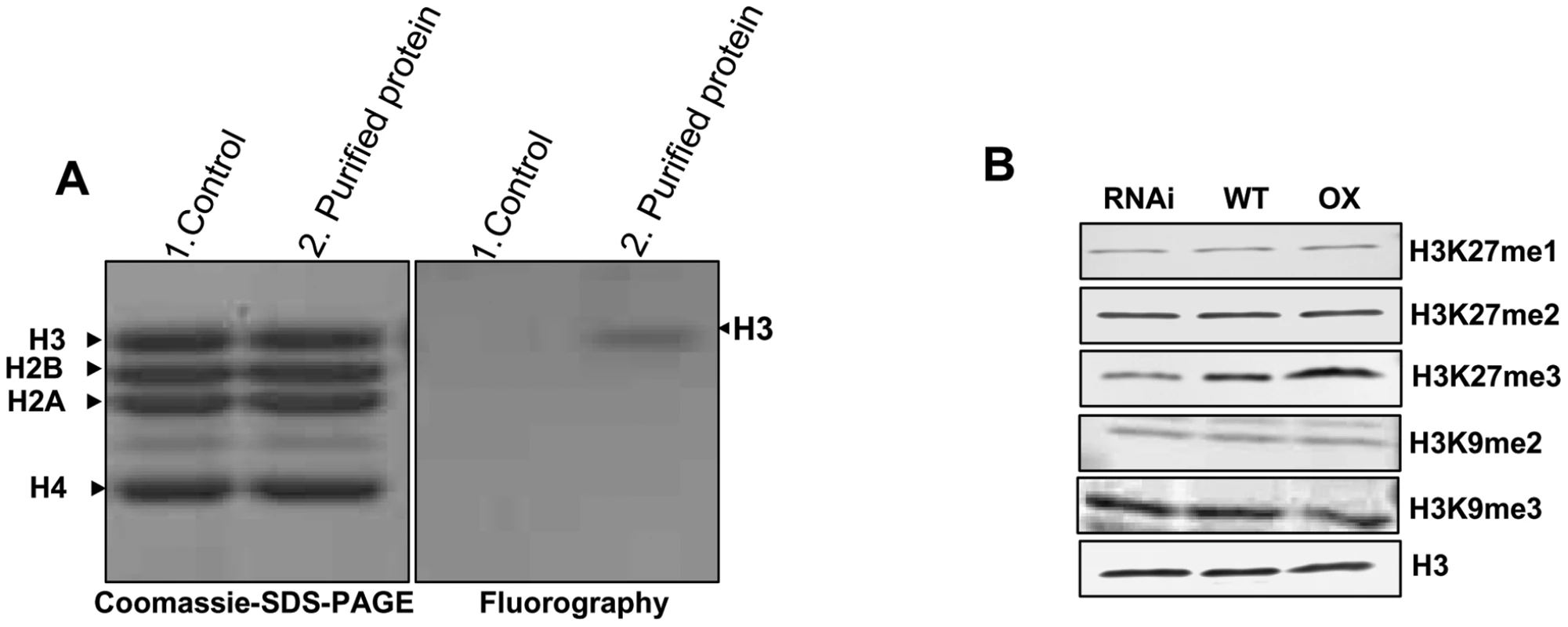
(A) In vitro histone methyltransferase assay of TAP purified OsFIE2-PRC2 complex: Chicken core histones were incubated with S-adenosyl-[methyl-3H]-l-methionine and affinity purified OsFIE2-PRC2 complex proteins. In the control reaction, TAP elution buffer was used instead of purified OsFIE2-PRC2 complex proteins. Methylated [3H] histones were resolved by 15% SDS-PAGE, stained with coomassie (Left side) and visualized by fluorography (Right side). (B) H3 methylation status in OsFIE2-RNAi line NO3, OsFIE2-overexpression line 04A and wild type plants. Equal amounts of protein samples were loaded for Western blot analyses. The specific antibodies used are indicated on the right and the protein sample sources are shown on the top. Western blot signals were normalized with the unmodified H3 band as control using the PDQUEST software. To further illustrate the enzyme specificity of the OsFIE2-PRC2 complex, we examined the H3K27 modification state in the OsFIE2 overexpression line and the OsFIE2 RNAi lines. Our results showed that H3K27me3 was reduced to about 54% in the RNAi line NO3 (Figure 2b). Meanwhile, H3K27me3 level had increased to about 152% in the overexpression line 04A compared with the wild-type. In contrast, the content of H3K27me and H3K27me2 had no detectable change, suggesting that OsFIE2 PRC2 complex specifically catalyzed the conversion of H3K27me2 to H3K27me3. We also examined H3K9me2 and H3K9me3 levels in OsFIE2 overexpression and RNAi lines (Figure 2B). The results indicated that H3K9 methylation state was not affected by OsFIE2 overexpression or reduction, suggesting that H3K9me and H3K9me2 are not the primary substrates of the OsFIE2-PRC2 complex at genome level compared with H3K27me3. However, we can not rule out the possibility of the gene specific effect of OsFIE2 PRC on H3K9me and H3K9me2 in the rice genome. To further validate the results, we tested other RNAi and overexpression lines. Similar results and conclusions were obtained (Figure S1).
Generation of OsFIE2-RNAi and overexpression transgenic rice plants
To study the biological role of OsFIE2 in rice, we made OsFIE2-RNAi constructs and obtained twenty independent RNAi lines using Agrobacterium mediated transformation. Six independent RNAi transgenic lines (No3, No8, No9, No10, No13 and No15) with different OsFIE2 expression levels as revealed by quantitative real time PCR (Figure 3A) were selected for further analysis. No 8 and No 9 had less than 20% of the expression level of the wild-type. No 3 and No 10 had about 40% to 50% expression levels and No15 and No 13 had about 70% to 90% expression levels. We categorized Lines No8 and No9 as severe RNAi lines and the others as weak RNAi lines. The vegetative development of the weak RNAi plants was the same as the wildtype.
Fig. 3. Analysis of OsFIE2-RNAi plants. 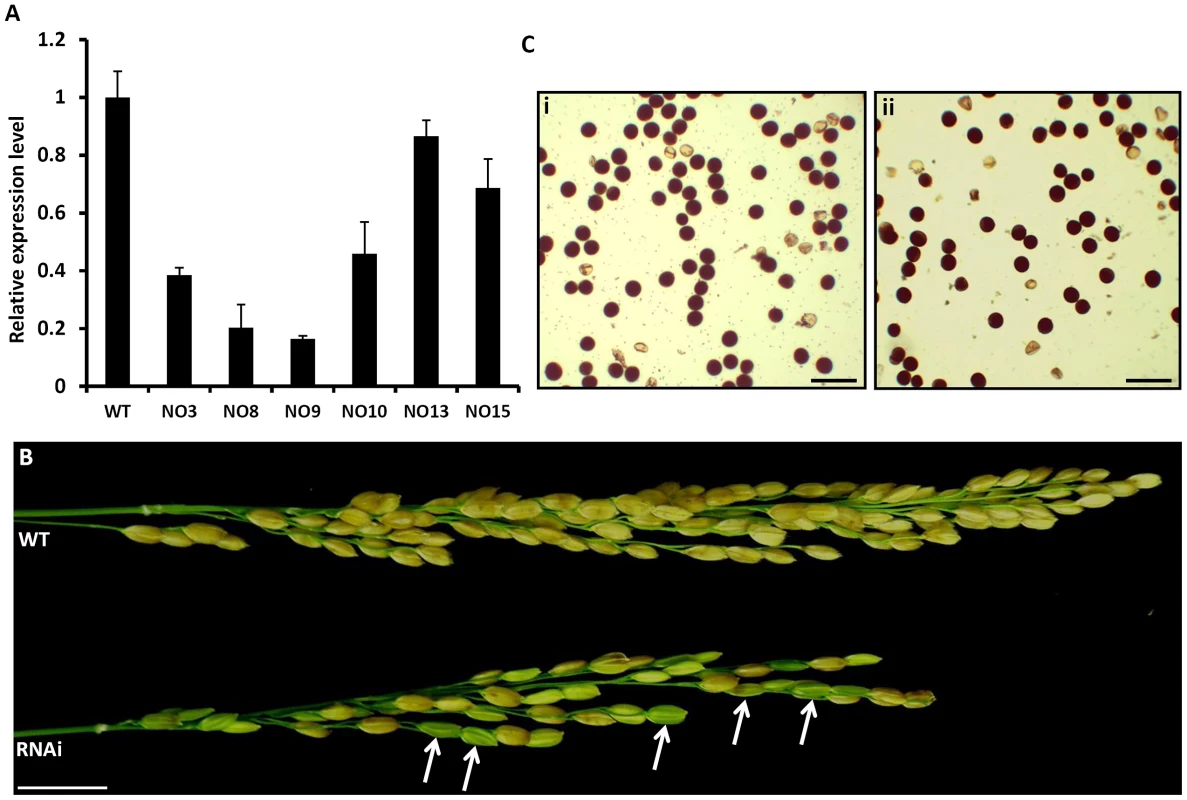
(A) Quantitative real-time PCR analysis of OsFIE2 gene expression in different RNAi lines and wild type. Rice Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as internal control. Same amount of cDNA template was used in each sample. Primers are listed in Table S3. (B) Comparison of mature panicles of OsFIE2-RNAi line NO3 (bottom) and wild type (top). Panicles of RNAi plants contain more partially filled seeds and less filled seeds when compared to wild type panicles. Partially filled or unfilled seeds are marked with arrow in panicle of OsFIE2-RNAi plant. (C) Images of pollen viability tests. Pollen grains are stained with I2-KI solution. Viable pollen grains are stained dark and nonviable pollen grains are stained light yellow. (i) Image of wild-type pollen. (ii) Image of OsFIE2-RNAi line NO3 pollen. The scale bars = 2 cm in (B) and 100 µm in (C). Phenotype evaluation of seed development in OsFIE2-RNAi T1, the endosperm was at T2 generation, and generations following revealed substantial differences compared with the wild-type. The ratio of mature and fully filled seeds was substantially reduced in RNAi lines (Figure 3B and Table 2). Fully filled mature seeds were produced in a ratio ranging from 66% to 81% in the weak RNAi lines, while the wild-type was about 88%. Interestingly, per thousand seed weight of the fully filled seeds in the weak RNAi lines was smaller in all the tested lines (Figure 4E, 4F and Table 2). The rest of the seeds were partially filled or not filled (Figure 4B). For the partially filled seeds, we found that some of them lost dormancy with seed germination before the seed mature (Figure 4D). We carried out a series of sectioning to examine the seed development in both wildtype and the RNAi lines. Defective shrunken endosperm tissue was observed in the partially filled seeds of RNAi lines (Figure 4H, 4J and K). In addition, we observed big embryos in the partially filled seeds without clear germination, which might be due to partial germination or uncontrolled embryo development (Figure 4J and 4K). Overall, the embryo sizes in the partially filled seeds increased to different degrees. Interestingly, the Arabidopsis fie mutants showed substantial delay in germination in contrast to loss of dormancy [34].
Fig. 4. Phenotypes of OsFIE2-RNAi seeds. 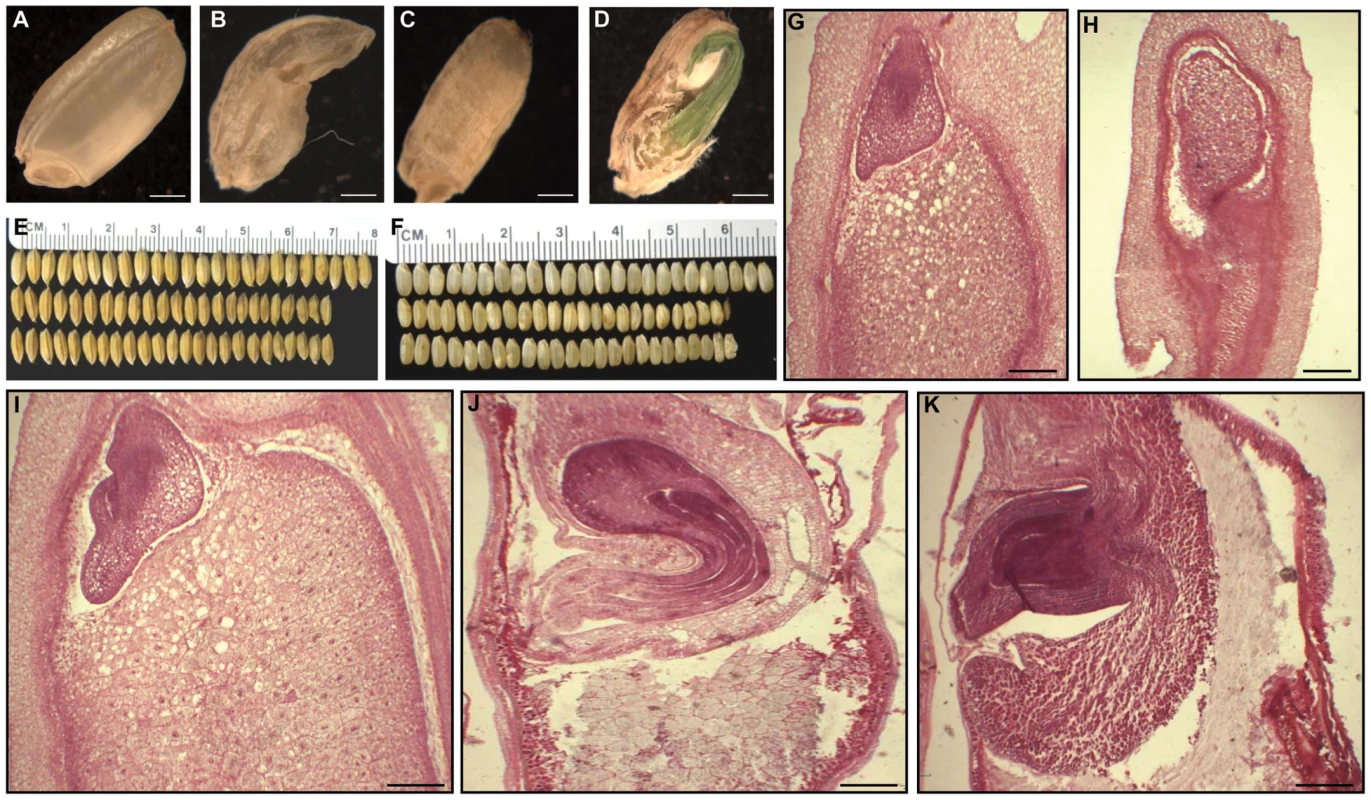
(A) Mature wild type seed with well-developed starchy endosperm. (B–D) Phenotype severity levels of partially filled seeds of OsFIE2-RNAi plant. (B) Severe partially filled seed phenotype of OsFIE2-RNAi plant with defective shrunken endosperm due to lack of proper starch filling. (C) Partially filled seed of OsFIE2-RNAi plants with partially filled shrunken endosperm. (D) Partially filled and lack of dormancy seed of the OsFIE2-RNAi plant. (E) Comparison of mature seeds of OsFIE2-RNAi line NO3 (bottom two rows) and wild-type (top row). (F) Comparison of mature seeds (without husk) of OsFIE2-RNAi line NO3 (bottom two rows) and wild-type (top row). Seed size of the OsFIE2-RNAi seeds is substantially smaller compared to the wild type seeds. (G,H) Images of longitudinal sections of OsFIE2-RNAi and wild-type early stage seeds (6 days After Fertilization). (G) Wild type early seed with well-developed starchy endosperm and viable embryo. (H) Image of partially filled early seed of OsFIE2-RNAi plants. Please compare the size ratio of embryo to seed in this seed with wild-type seeds (G). (I–K) Images of longitudinal sections of OsFIE2-RNAi and wild-type mature seeds. (I) Wild type mature seed with well-developed starchy endosperm and viable embryo. (J,K) Images of partially filled mature seeds of OsFIE2-RNAi plants with partially filled shrunken defective endosperm and large embryo. The scale bars = 1 mm in (A) to (D) and 100 µm in (G–K). Tab. 2. Agronomic traits of T1 generation OsFIE2-RNAi transgenic lines. 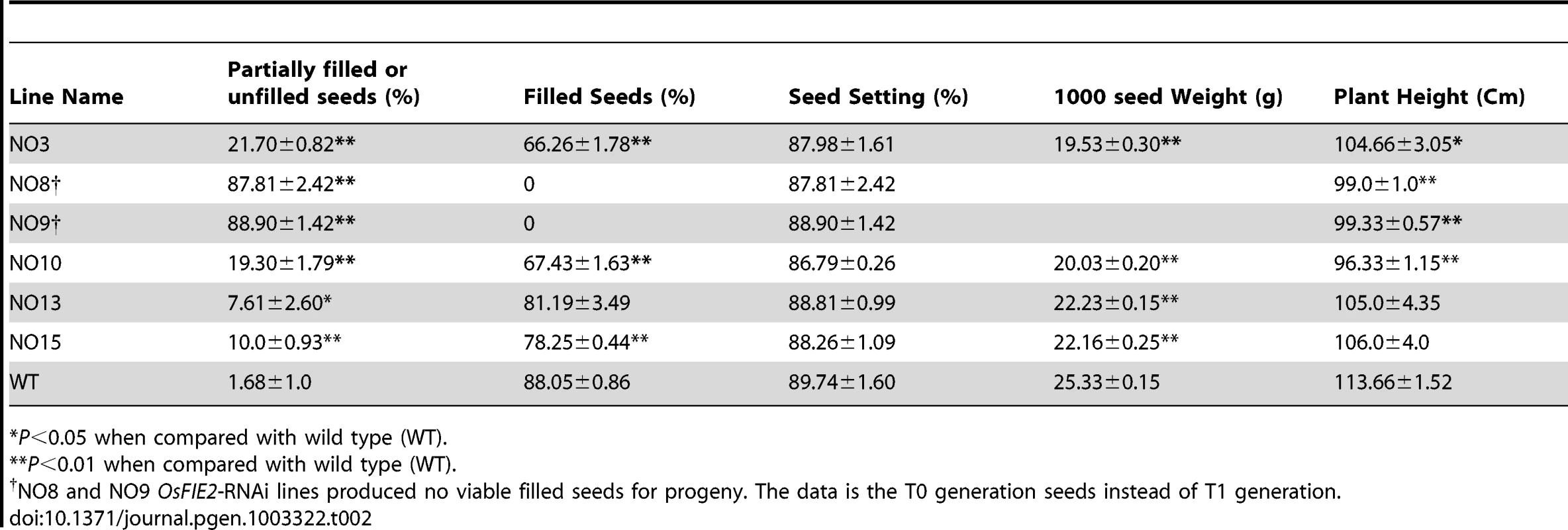
P<0.05 when compared with wild type (WT). The T0 plants of the two severe OsFIE2-RNAi lines (No8 and No9) did not produce viable seeds. We failed to obtain the reproductive progeny of the two transgenic lines although we successfully maintained the plants by asexually amplifying new tillers. To verify the phenotype observed in the severe lines, we carried out a second batch of transformation and generated another 18 independent lines. One line displayed severe phenotype but also failed to produce viable seeds. Since all the three severe RNAi lines failed to produce viable progeny, we could not verify if the observed phenotype is inheritable meiotically. Therefore, the phenotypes of the severe lines are not discussed further in this manuscript. The small seeds and partially filled seed phenotype were repeatable in the weak RNAi lines generated in the second batch (data not shown). To examine pollen quality, we carried out pollen viability test using iodine-potassium iodide staining method (Figure 3C). We found that the viable pollen ratio was about 87% in wild-type and about 86% in weak RNAi lines, suggesting that the pollen grains developed normally.
Since autonomous endosperm development was reported in Arabidopsis FIS/FIE mutants, we emasculated anthers before anthesis in over 100 spikelets in the RNAi severe and weak lines, respectively. After examination with microscopy and the naked eye, no sign of autonomous endosperm development was observed in the absence of fertilization event. Meanwhile, we examined the expression of the OsFIE2 gene and other polycomb group genes in anthers with reverse transcription PCR to study if these genes subject to imprinting regulation as in Arabidopsis. The OsFIE2 gene was highly expressed while OsFIE1 gene expression was not detected in anthers (Figure 5A). We also examined the expression of other OsFIE2-PRC2 complex genes and OsFIE1 in other plant tissues, including leaf, sheath, stem nodes, young and mature flowers, panicles, and endosperm tissues in three different stages. All the OsFIE2 complex genes were highly expressed in these tested tissues except that OsCLF might have minor quantity changes in five days old endosperm tissues (Figure 5B). In contrast, OsFIE1 was only detected in five days and ten days old endosperm tissues. Luo et al. (2011) [45] carried out a genome wide analysis of imprinted genes in rice endosperm. They found that only OsFIE1 but not other polycomb genes were imprinted in rice endosperm. Given that OsFIE2 was highly expressed in anther but not OsFIE1, our results were consistent with their report (Luo et al. 2011) [45] that OsFIE1 was probably paternal imprinted but not OsFIE2. The dominant negative nature of RNAi line prevented us to further test if OsFIE2 is imprinted by reciprocal crosses.
Fig. 5. Expression profile analyses of polycomb genes in different rice tissues. 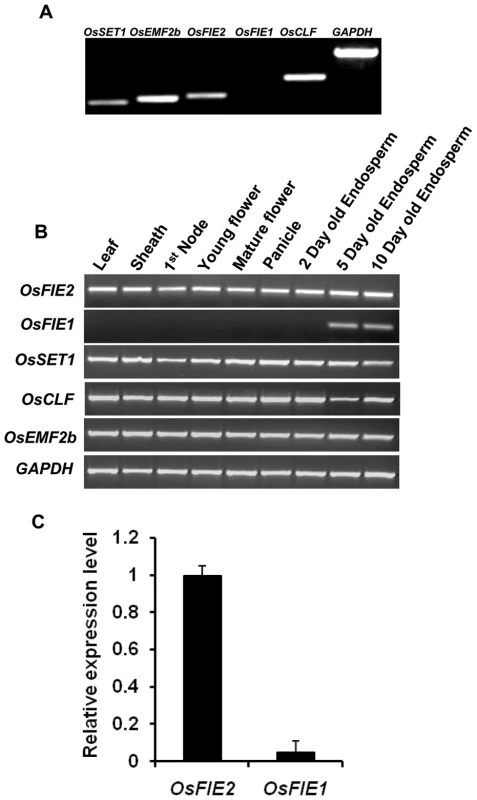
GAPDH was used as internal control. Same amount of cDNA template was used in each sample. Primers are listed in Table S3. (A) Reverse Transcription-PCR analysis of the expression of the rice polycomb genes in anther tissue. 28 cycles were used. (B) Reverse-Transcription-PCR analysis of the expression of rice polycomb genes in different rice tissues. (C) Comparison of OsFIE2 and OsFIE1 expressions in 5-days old rice endosperm by quantitative real-time PCR. Surprisingly, the endosperm specific OsFIE1 mutant had no endosperm phenotype [42], but the endosperm unspecific gene OsFIE2 RNAi lines all had reduced seed size and partially filled phenotype. We compared OsFIE1 and OsFIE2 expression level in endosperm with real-time PCR (Figure 5C). The results showed that the OsFIE1 expression level was only about 4% of the expression level of OsFIE2 in endosperm. The OsFIE1 and OsFIE2 proteins share 72% of overall amino acid sequence identities. If the N-terminal 82 amino acid stretch specific for OsFIE1 is excluded, the two proteins share about 85% amino acid sequence identities. If OsFIE1 and OsFIE2 are functionally redundant, our results would imply that OsFIE1 level is too low to replace the function of OsFIE2 because a reduction of OsFIE2 expression to 20% could not produce viable progeny but the OsFIE1 expression is only 4% of the OsFIE2gene. In contrast, OsFIE2 level is high enough to replace OsFIE1 function. Therefore, mutation on OsFIE1 gene may not show phenotype, but mutation on the OsFIE2 gene will show phenotype. Due to the high homology between OsFIE2 and OsFIE1, we tested if the OsFIE1 expression was also reduced in the OsFIE2 RNAi lines. Our results showed that OsFIE1 expression was not affected in the OsFIE2 RNAi lines (Figure S3).
We also generated multiple OsFIE2 overexpression transgenic lines. We found no obvious phenotype in all the transgenic plants and the endosperm and embryo development were normal as wild type (data not shown).
OsFIE2 regulates the production of storage proteins and storage starch in the rice endosperm
Given that the mature seeds of the RNAi lines are much smaller than the wild-type and many seeds are only partially filled, we examined the expression levels of selected starch biosynthesis pathway genes in the endosperm using quantitative real-time PCR. The healthy looking developing seeds of weak RNAi lines in the grain filling stage were selected for the experiment. The genes tested included ADP-glucose pyrophosphorylase small subunit 1 (AGPS1)-AK073146, AGPS2a-AK071826, AGPS2b-AK103906, AGPL1-AK100910, AGPL2-AK071497 and AGPL3-AK069296. Among them, the expression of AGPS2b reduced to 9% compared with the wild type (Figure 6). Other genes had no substantial change. Our results indicated that the AGPS2b gene, encoding the rate limiting step enzyme in the starch biosynthesis pathway [69] is subjected to the regulation of OsFIE2 either directly or indirectly.
Fig. 6. Quantitative real-time PCR analyses of gene expression in the storage protein and starch synthesis pathways in the OsFIE2 RNAi and wild-type endosperm. 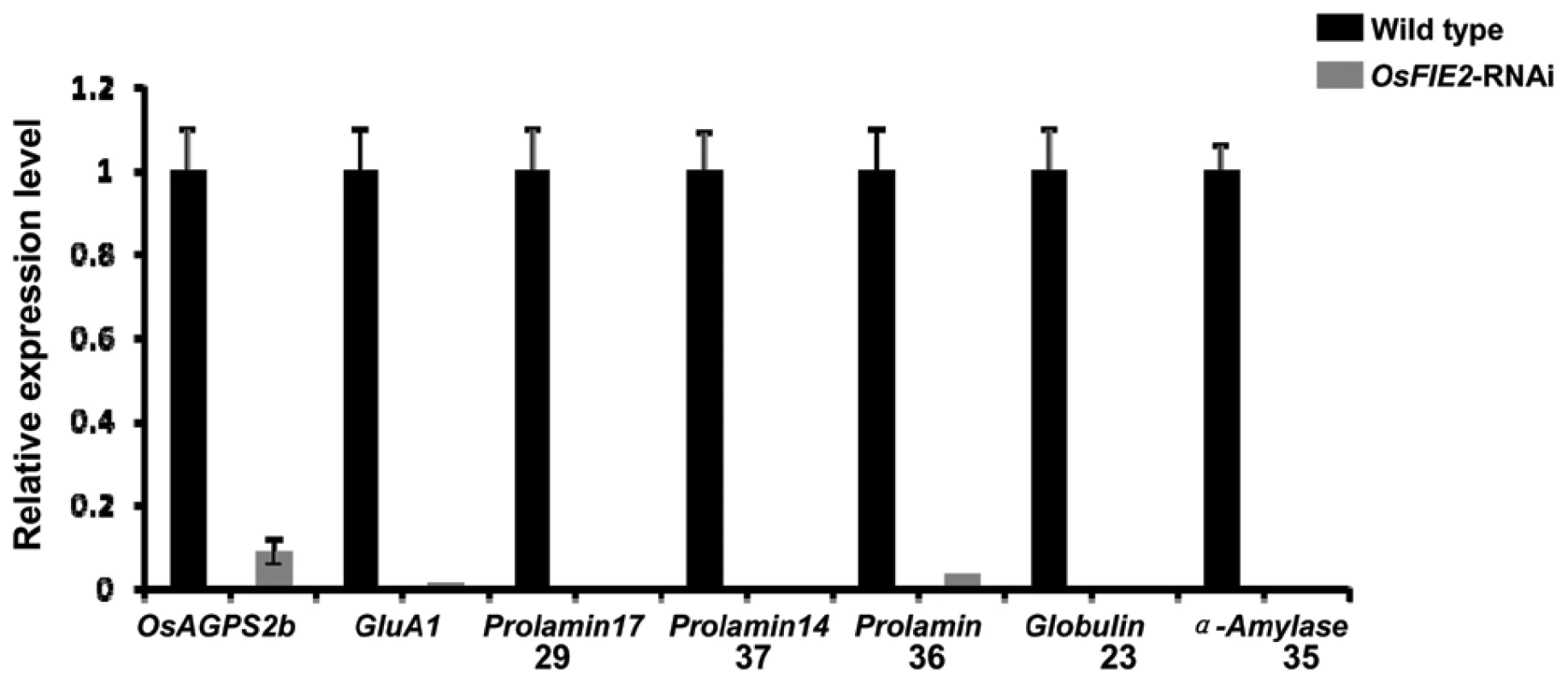
We used GAPDH as internal control and same amount of cDNA template was used in each sample. Numbers 29, 37, 36, 23, 35 indicates the genes encoding proteins corresponding to 2-DE gel spot numbers shown in Figure 7 and Table 3. During the seed filling stage, massive amounts of storage proteins are accumulated in cereal endosperm [70], [71]. To investigate the possible role of OsFIE2 in the regulation of storage protein accumulation, we examined the storage protein content in the endosperm of fully filled mature seeds in the weak RNAi lines NO 3, NO 10, and NO15 using two-dimensional (2-DE) gel analysis. We isolated total proteins from the mature seeds of both OsFIE2-RNAi and wild-type seeds using phenol extraction method as we reported [72]. After 2-DE gel separation and PDQUEST analysis, differentially regulated protein spots were robotically excised and analyzed using MALDI TOF-TOF mass spectrometry. Proteins with reduced accumulation in the RNAi seeds included prolamin, globulin, alpha-amylase and seed allergenic proteins (Figure 7 and Table 3). The results were similar in all three tested RNAi lines. Real-time PCR analysis demonstrated the expression of some genes also reduced at the mRNA level (Figure 6), suggesting a regulation of these genes at the transcriptional level. We also found that the expression of Glutelin A1 (Os01g55690) reduced in RNAi seeds compared with the wild type (Figure 6).
Fig. 7. 2-DE gel images of rice seed proteins from weak OsFIE2-RNAi and wild-type plants. 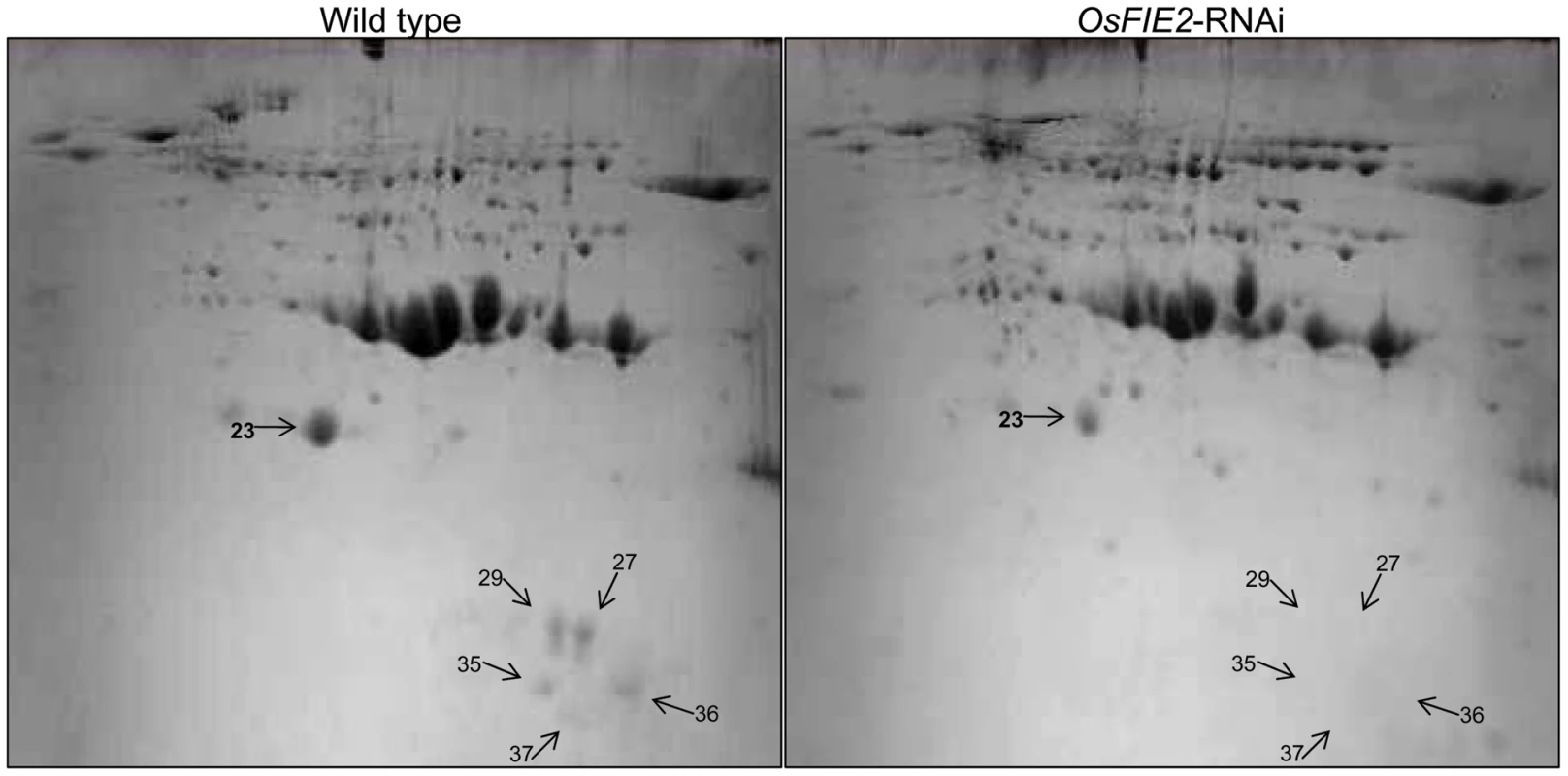
Proteins were extracted from fully filled mature rice seeds, separated by 2-DE , and visualized by coomassie blue staining. Differentially expressed protein spots were marked with arrows and numbers. Tab. 3. Differentially regulated proteins identified in 2D-gel images of <i>OsFIE2</i> RNAi and wild-type seed proteins. 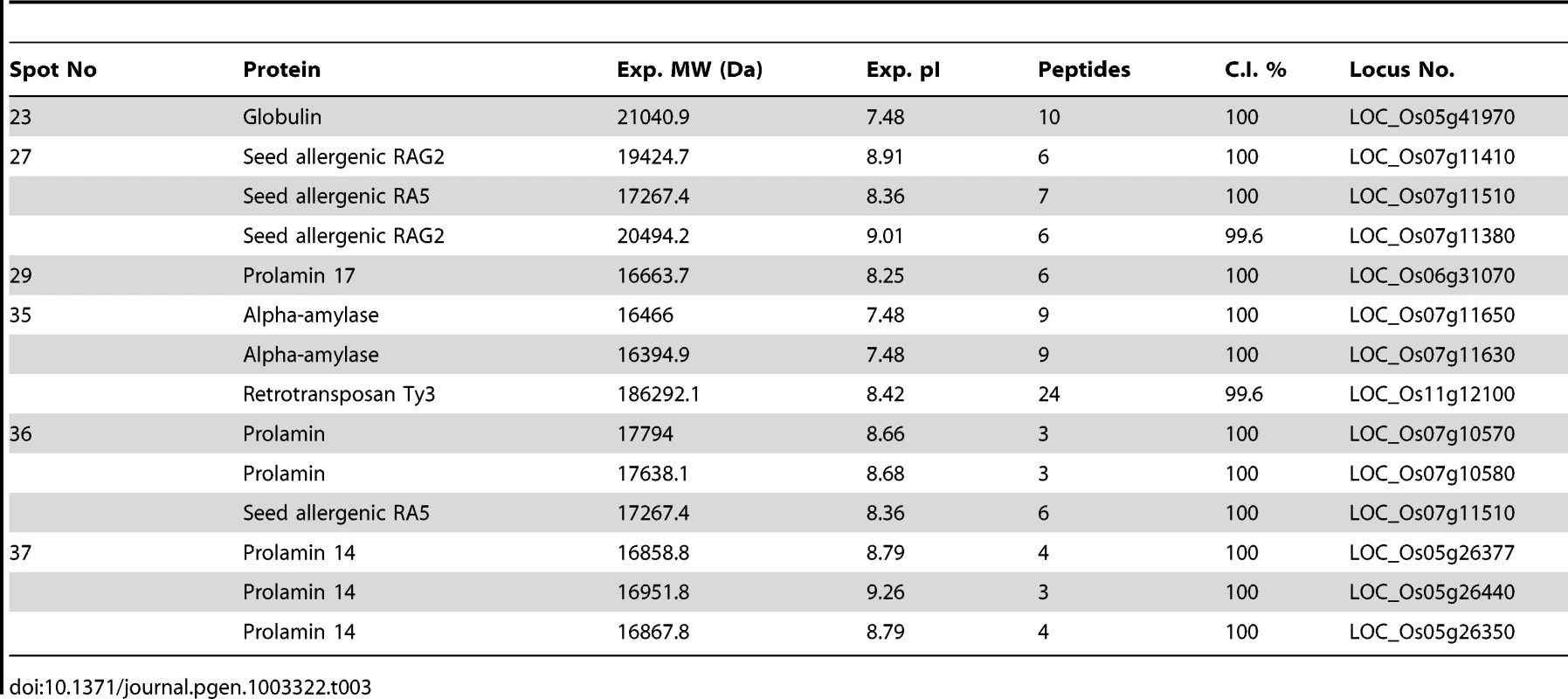
Genome-wide identification of H3K27me3 binding sites
Our studies above indicated that OsFIE2 played a critical role in regulating endosperm development and grain filling and the OsFIE2-PRC2 complex specifically catalyzes the methylation of H3K27me2 to H3K27me3. We hypothesized that some key regulatory genes controlling storage starch and protein synthesis in endosperm must be subjected to the regulation of H3K27me3 either directly or indirectly. We have carried out a genome wide ChIP-Seq experiment using 6 to 7 days post pollination endosperm tissues as materials to compare four algorithms used for ChIP-Seq data analysis [73]. The endosperm ChIP-Seq results were verified by ChIP-PCR using 28 selected genes [73] and the antibody used for the experiment (antibodies against H3K27me3, produced by Millipore) is the antibody with 100% specificity for H3K27me3 [74]. Given that the role of H3K27me3 is primary for gene repression and 6 to 7 days post pollination is in the stage before starch and storage protein synthesis, we expected that H3K27me3 should be still associated with genes regulating storage nutrient synthesis in this stage. Therefore, we decided to use the published data set to search for genes regulated by H3K27me3 modification in endosperm. Given that the antibody might bind to non specific sites in the mutant background due to reduced H3K27me3 [34], the results obtained in wild-type background should be the best one could get.
Malone et al. (2011) [73] compared four Algorithms for ChIP-Seq data analysis and concluded that the overall biological conclusion of the results obtained by the four algorithms is the same. Since the FindPeak algorithm combines excellent sensitivity with accuracy compared with the other three Algorithms [73], the findPeak results are discussed here. The FindPeak program analyses found that 76 endosperm specific genes, including DNA binding proteins, and 97 nutrition metabolic pathway genes were enriched by antibodies against H3K27me3 in the endosperm ChIP-Seq experiment (Table S1). The total list of enriched peaks is shown in Table S2. The selected examples of the peak profiles are shown in Figure S2. Examples of enriched endosperm specific genes include: MYB protein, putative (LOC_Os02g09670); helix-loop-helix DNA-binding domain containing protein (LOC_Os04g35010); basic region leucine zipper domain containing protein (LOC_Os09g34880); RNA recognition motif containing protein (LOC_Os09g14550); trehalose-6-phosphate synthase (LOC_Os09g25890); OsIAA29-Auxin-responsive Aux/IAA gene family member (LOC_Os11g11430); etc. The ChIP enriched nutrition metabolic pathway genes include: seed specific protein Bn15D1B (LOC_Os04g50970); starch synthase (LOC_Os06g04200); globulin 2 (LOC_Os11g34780); glutelin (LOC_Os02g15090); seed maturation protein PM23 (LOC_Os03g41080); starch binding domain containing protein (LOC_Os05g37450); etc.
OsFIE2 regulates the histone modification and expression of a helix-loop-helix DNA–binding gene
Our ChIP-Seq results showed that the helix-loop-helix DNA-binding domain containing protein (LOC_Os04g35010) is enriched in the ChIP-Seq experiment for H3K27me3 modification (Figure S2 and Table S1). The gene's Arabidopsis ortholog RGE1 (AT1G49770) is expressed in the endosperm surrounding region which directly surrounds the developing embryo. It exerts its effect non autonomously in the developing embryo [75], [76]. Loss of RGE1 function results in small embryos. Mutant seedlings are extremely sensitive to desiccation due to the abnormal cuticle. Since we observed large embryo phenotype in our RNAi lines, we checked the expression of the rice LOC_Os4g35010 gene in both wild-type and the RNAi lines in developing seeds. We found that this gene was over expressed in the RNAi lines compared with wild-type as shown in Figure 8. Further ChIP-PCR experiment using antibodies for H3K27me3 showed that the H3K27me3 modification was substantially enriched in this locus in the wild-type but the enrichment substantially reduced in the RNAi lines. Our results confirmed the ChIP-Seq result and suggest that this gene's H3K27me3 modification level and expression are regulated by OsFIE2 expression.
Fig. 8. Reverse Transcription–PCR and ChIP PCR analysis of helix-loop-helix DNA binding domain containing gene in wild-type and OsFIE2-RNAi plants. 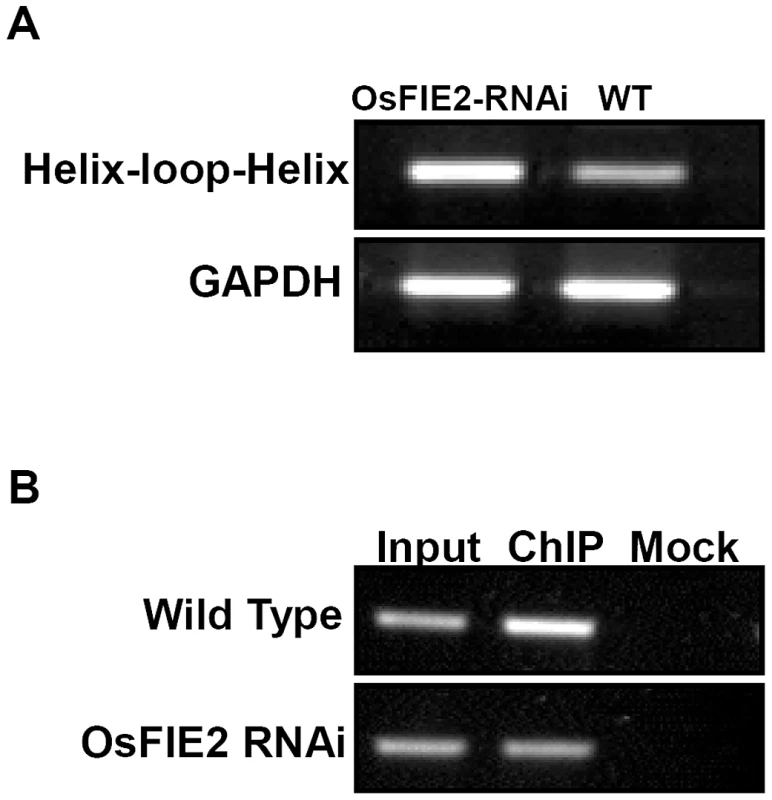
(A) Reverse Transcription-PCR analysis of the expression of helix-loop-helix DNA binding domain containing gene in endosperm tissue. GAPDH was used as internal control. Same amount of cDNA template was used in each sample. Primers are listed in Table S3. (B) ChIP PCR analysis of helix-loop-helix DNA binding domain containing gene in wild type and OsFIE2-RNAi plants. Chromatin is isolated from endosperm tissue of wild type and OsFIE2-RNAi plants. Antibodies for H3K27me3 were used for chromatin immunoprecipitation (ChIP). Input: DNA sample extracted from the chromatin before ChIP enrichment. ChIP: DNA sample extracted from H3K27me3 ChIP enriched chromatin. Mock: DNA sample that went through the immunoprecipitation procedure without antibody. Discussion
OsFIE2 forms a multi-protein complex specifically catalyzing the production of H3K27me3
The role of polycomb group genes in plant development has been extensively studied in Arabidopsis. The polycomb complex that regulates vernalization process has been purified in Arabidopsis using Tandem affinity tag approach and the seven putative subunits of the complex have been identified following LC-MS/MS analysis [3], [14]. However, no enzyme activities had been tested in vitro for the PRC2 complexes in plants. Thus, the mechanism of PRC2 complex action has not been demonstrated in plants. Given the unique and essential function of PRC2 complexes in cereal crops, purifying and characterizing the complex in cereals is particularly important. In this report, we purified the OsFIE2-PRC2 protein complex in rice and demonstrated the Histone methyltransferase activity in vitro.
We find that OsFIE2 (the ESC homologue) forms a stable protein complex with polycomb proteins OsCLF (the E (Z) homologue), OsSET1 (the E (Z) homologue), and OsEMF2b (the Su (Z) 12 homologue). Our E. coli two hybrid studies show that OsFIE2 may directly interact with OsSET1. The OsFIE2-PRC2 complex genes are well expressed in young and mature endosperm tissues in rice. Along with the PRC2 core components, the histone H4 protein was co-purified with the OsFIE2 protein. Interaction of histone H4 with polycomb proteins was reported in mammals and Drosophila. Mammalian ortholog of p55, RbAp 48/46, directly binds to histone H4 [62], [63]. GST pull-down assay revealed that Drosophila histone H4 strongly and specifically binds to p55 [61]. Therefore, H4 may directly interact with OsFIE2-PRC2 complex although further experiments are required to confirm the interaction. There are four p55 homologous genes in the rice genome [46]. But we did not detect the p55 homologous proteins in purified complex although we successfully detected methyltransferase activity with the purified complex. One possibility is that the quantity of p55 homologous protein is very low in the complex. The mass spectrometer failed to detect it. Alternatively, the polycomb complex in rice has components different from those in other organisms. Nevertheless, it is clear that the rice OsFIE2-PRC2 complex is conserved in overall structure compared with the Drosophila PRC2 complex. Our E. coli two hybrid studies show that OsFIE2 may directly interact with OsSET1. Other interactions were not detected in E. coli two hybrid assays among the components, suggesting that posttranslational modifications or additional components may be involved in complex formation. It is interesting to note that non coding RNAs play a key role in the polycomb complex [77], [78]. But it is still unknown if the RNAs play any critical structure role in the complex. Our expression studies show that all the OsFIE2-PRC2 complex genes are well expressed in both young and mature endosperm tissues of rice.
In Mammals and Drosophila, PRC2 complexes were shown to methylate histone H3 at lysine 27 and, to a lesser extent, H3K9 both in vivo and in vitro [65]–[68]. In Arabidopsis vernalization mutant vrn2, encoding a subunit of PRC2 complex, both methylation marks at H3K9 and H3K27 were lost [22], [79]. In two other mutants vrn1 and vin3, the H3K9me2 mark is missing. Vin3 has also been shown to be a component of the PRC2 complex with VRN2 [3], suggesting a role of the PRC2 complex in regulating H3K9 methylation. Therefore, detecting the methyltransferase activity and revealing the substrate specificity of the OsFIE2-PRC2 complex is important for understanding the molecular mechanism of PRC2 complex function in plants. Our studies with the overexpression and RNAi lines demonstrated that manipulation of the OsFIE2 gene expression level had no effect on the cellular level of H3K9me2, H3K9me3, H3K27me and H3K27me2 but resulted in a substantial change in the cellular level of H3K27me3, suggesting that OsFIE2-PRC2 complex is primary for regulating the formation of H3K27me3 at genome level. However, we can not rule out the possibility of the gene specific effect of OsFIE2 PRC on other modifications in the rice genome. The results suggest that the function of the rice OsFIE2-PRC2 complex is different from the Arabidopsis VRN2-PRC2 complex, the later contained more subunits and may have H3K9 methyltransferase activity [3], [22], [79]. The OsFIE2-PRC2 complex is also different from the Arabidopsis endosperm development related MEA-FIE complex because the orthologs of the Arabidopsis MEDEA and FIS2 were not present in the rice genome.
OsFIE2 regulation model in rice seed and endosperm development is different from that in Arabidopsis
The polycomb gene FIE is well studied in Arabidopsis and is required to repress the endosperm development in the absence of fertilization [6], [7], [13], [15], [16]. Loss-of-function mutations of genes in the MEA-FIS - FIE-MSI1 complex can form autonomous diploid endosperm in the absence of fertilization [6], [7]. Mutation in MSI1 gene exhibits very strong phenotype of autonomous endosperm development [8], [26]. It is shown that MEDEA represses expression of Arabidopsis MADS-box gene PHERES1 during the seed development [8], [9]. Interestingly, MEA gene itself is one of the target genes of the FIS complex, indicating a self-imprinting mechanism in Arabidopsis [32], [33], [80].
We found that emasculated OsFIE2-RNAi florets did not show autonomous endosperm development. Similarly, no autonomous endosperm development was observed in rice T-DNA mutants of OsFIE1 and OsEMF2b [42]. The maize RNAi plants of ZmFIE1 and ZmFIE2 also produced no autonomous endosperm in the absence of fertilization [46]. Further, RNAi lines of HFIE in sexual Hieracium pilosella, failed to show the autonomous central cell proliferation [48]. These results, together, suggested that the function of FIE gene is not to suppress endosperm development. The function of the FIE containing PRC2 complex in repressing autonomous endosperm development in Arabidopsis is not conserved among plants.
We found that down regulation of OsFIE2 gene in RNAi plants resulted in the production of small seeds and partially filled seeds (Figure 3 and Figure 4). Our results suggested that the OsFIE2 positively regulated endosperm development in rice instead of repressing endosperm development as predicted using Arabidopsis as the model. We found that some of the partially filled seeds germinated before maturation. Cross sectioning of the un-germinated partially filled seeds found that the embryos were bigger than the wild-type. It is still unknown if the large embryo was due to partial germination or other developmental defect. In addition, it is also unknown if the early germination is directly regulated by polycomb complex or it is a secondary effect due to partial filling or others.
Interestingly, the Arabidopsis FIE mutant showed delayed seed germination [34]. While the wild-type seeds germinated within 2 days, approximately 40% of the homozygous FIE mutants stayed dormant for the course of the entire experiment, which lasted 20 days. Therefore, the FIE containing polycomb complex in rice and Arabidopsis have distinct roles. While the Arabidopsis complex suppress endosperm development in the early stage and promote seed germination after maturation, the rice OsFIE2 complex, in contrast, is required for seed and endosperm development and suppress early germination of the seeds either directly or indirectly.
Interestingly, no phenotype was reported in mutants of the endosperm specific gene OsFIE1, a gene sharing the highest homology with OsFIE2 in the rice genome. Given the high homology of these two genes, these two genes might be functionally redundant. Analysis of the gene expression profile indicates that the expression level of OsFIE2 is about 25 times higher than that of the OsFIE1 gene in the endosperm. If these two genes are truly redundant in function, OsFIE1 mutant will display no phenotype because OsFIE2 can replace OsFIE1 function. In contrast, OsFIE2 mutant will display phenotype because the expression level of OsFIE1 is too low to replace the function of OsFIE2. Indeed, no phenotype was reported for OsFIE1 mutant but striking phenotypes were observed in our OsFIE2 RNAi lines.
OsFIE2 regulation in gene expression in seeds
Our real-time PCR results showed that the starch synthesis rate limiting step enzyme (glucose pyrophosphorylase) subunit OsAGPS2b was down regulated in the OsFIE2 RNAi lines. Meanwhile, proteomics analysis revealed that the expression of prolamin, globulin, alpha-amylase, and seed allergenic proteins are reduced in OsFIE2 RNAi lines (Figure 6, Figure 7, and Table 3). These results suggest that the partially filled and smaller seeds were probably due to a reduction of starch and storage protein synthesis.
To understand how the polycomb group genes regulate gene expression in rice endosperm, we analyzed our prior published rice endosperm ChIP-Seq data set. The ChIP-Seq data has been well verified by ChIP-PCR experiments with twenty eight genes [73]. We found that a large number of endosperm specific genes and storage nutrient metabolic pathway genes were subjected to the regulation of H3K27me3 in endosperm (Table S1 and Table S2). However, most genes shown to be down regulated by real-time PCR in the OsFIE2 RNAi lines (Figure 6) are not the direct targets of the H3K27me3, suggesting a complicated gene expression regulation network in endosperm. Seed development is a complex quantitative trait. If the starch and protein synthesis genes were directly regulated by polycomb complex, it would act as a simple Mendelian trait. Therefore, it is understandable that many of the starch and storage protein genes are not the direct targets of OsFIE2 complex. In addition, there are multiple polycomb complexes in plants. It is also possible that many ChIP-Seq identified loci are not the direct target of OsFIE2 complex but the targets of other PRC2 complexes.
Arabidopsis helix-loop-helix DNA binding domain containing gene RGE1 (AT1G49770) is specifically expressed in the endosperm surrounding region which directly surrounds the developing embryo. It exerts its effect non autonomously in the developing embryo. Loss of RGE1 function results in small embryos. We found that the expression of rice RGE1 gene was increased and the modification of H3K27me3 at the locus was reduced in the RNAi lines, suggesting that it is a potential target subject to OsFIE2 regulation. Interestingly, while a mutation of the RGE1 gene leads to small embryo in Arabidopsis, we observed large embryo in the RNAi lines showing high expression of the rice RGE1 gene.
The MADS box gene OsMADS6 (LOC_Os02g45770) is highly expressed both in flower and endosperm. Prior studies have shown that OsMADS6 is subjected to the regulation of H3K27me3 [71] and our ChIP-seq data analysis results verified that OsMADS6 was enriched by antibodies for H3K27me3 (Table S1 and S2). OsMADS6 controls grain filling by regulating the gene encoding OsAGPS1 (ADP-glucose pyrophosphorylase small subunit1, AK073146), a subunit of the ADP-glucose pyrophosphorylase [71]. Meanwhile, we found that the expression of another subunit gene OsAGPS2b of the glucose pyrophosphorylase enzyme was down regulated in the OsFIE2 RNAi lines. Further, a starch synthase and several storage protein genes were shown to be enriched in the ChIP-Seq experiment. Our results suggest that the OsFIE2 polycomb complex controls endosperm development and grain filling by regulating multiple levels of targets including transcription regulators as well as metabolic pathway genes. The large number of genes subject to the regulation of H3K27me3 in endosperm is consistent with the critical role of OsFIE2 gene in endosperm development and grain filling and suggests a highly sophisticated regulatory network.
Materials and Methods
Plant materials and growth conditions
Rice (Oryza sativa, cultivar Nipponbare) growth conditions were similar to our previous report [81]. All rice plants were grown in the greenhouse of the Department of Biochemistry and Molecular Biology, Mississippi State University, MS, USA. Wild-type Nipponbare (rice subspecies japonica) was used as control.
Generation of TAP-tagged OsFIE2 overexpression transgenic plants and cell suspension cultures
The OsFIE2 full length cDNA clone (AK111761, Jo23058f21) was obtained from National Institute of Agrobiological Sciences, Ibaraki, Japan. The cDNA was amplified using primers 61TAPF and 61TAPR and cloned into pENTR™/D-TOPO (Invitrogen) entry vector. The cDNA clone in the entry vector was transferred to final UbI-NTAP-1300 destination vector [51] by using a single LR clonase recombination reaction (Invitrogen). The final N-Terminal TAP-tagged OsFIE2 construct was transformed to Agrobacterium strain EH105A by electroporation. Rice transformation was performed as described Wu et al. (2003) [82]. Transformed resistant callus was screened using hygromycin. Transgenic plants were regenerated from the resistant callus. Mature seeds from the T1 transgenic plants with high expression of TAP-tagged OsFIE2 were used to induce transgenic callus. Cell suspension cultures were generated from transgenic callus and maintained in suspension medium (3.2 g/liter Gamborg B5 basal medium (Phytotechnology Laboratories™), 0.5 g/liter MES, 20 g/liter sucrose, 2 mg/liter 2, 4-Dichloro acetic acid, 2 g/liter N-Z-AmineA, PH 5.7, adjusted with 1 M KOH) at 25°C in darkness by constant shaking (150 rpm) on a gyratory shaker.
Generation of OsFIE2-RNAi transgenic lines
A portion of coding sequence fragment was amplified using primer set 61RNAiF and 61RNAiR from OsFIE2 cDNA clone (AK111761, Jo23058f21) and cloned into pENTR™/D-TOPO (Invitrogen) entry vector. The amplified fragment in the entry vector was transferred to final pANDA vector [83] by using a single LR clonase recombination reaction (Invitrogen). The final OsFIE2-RNAi construct was transformed to Agrobacterium strain EH105A by electroporation. Rice transformation was performed as described Wu et al. (2003) [82]. Transformed resistant callus was screened by using hygromycin. Transgenic plants were regenerated from the resistant callus.
Tandem affinity purification
Exponentially growing cell suspension cultures (30 g) were harvested 3 days after subculturing and ground in liquid nitrogen. Protein extracts were prepared in two volumes of extraction buffer (20 mM Tris-Hcl, pH 8.0, 150 mM NaCl, 0.1% IGEPAL (Sigma), 2.5 mM EDTA, 2 mM benzamidine, 10 mM β-mercaptoethanol, 20 mM NaF, 2 mM phenyl methanesulfonylfluoride (PMSF), 1% Protease cocktail (Sigma), 10 µM leupeptin (Sigma), and 10 µM 3,4-dichloroisocoumarin (Sigma)). The suspension was homogenized with the help of Polytron PTA 20 TS homogenizer for 2 min on ice and filtered through two layers of miracloth. The soluble protein fraction was collected by centrifugation twice at 30,000 g for 20 min at 4°C. Affinity purifications were performed as described by Rohila et al. (2006) and Van Leene et al. (2007) [53], [54] with some modifications. The protein extract was incubated with 400 µl IgG sepharose beads (GE Healthcare) for 1 h at 4°C. The IgG beads were loaded on to a polyprep chromatography column (Bio-Rad Laboratories, CA, USA) and washed with 10 ml of extraction buffer lacking protease inhibitors and 10 ml of TEV (Tobacco etch virus) cleavage reaction buffer (10 mM Tris–HCl, pH 8.0, 150 mM NaCl, 0.1% IGEPAL, 0.5 mM EDTA, 1 mM DTT). Bound proteins were eluted by digestion with 150 U of Ac TEV enzyme in TEV cleavage reaction buffer containing 1 µM E-64 protease inhibitor for 1 h at 16°C. IgG-eluted fraction was adjusted to 2 mM Cacl2 and diluted with 3 volumes of calmodulin binding buffer (10 mM Tris–HCl pH 8.0, 150 mM NaCl, 1 mM Mg-acetate, 1 mM imidazole, 2 mM CaCl2, 0.1% IGEPAL, 10 mM β-mercaptoethanol) and the fraction was incubated with 400 µl of calmodulin-agarose beads (Stratagene, CA) for 1 h at 4°C. The calmodulin-agarose beads were loaded on to a polyprep chromatography column and washed with 10 ml of calmodulin binding buffer. The protein complexes were eluted with 2 ml of elution buffer (10 mM Tris–HCl pH 8.0, 150 mM NaCl, 1 mM Mg-acetate, 1 mM imidazole, 25 mM EGTA, 0.1% IGEPAL, 10 mM β-mercaptoethanol) and proteins were precipitated with TCA. The protein pellet was washed with cold acetone.
Mass spectrometric analysis of TAP purified complex proteins
Tandem affinity purified complex proteins were trypsin digested and identified using LC-MS/MS mass spectrometry analysis as described by Chitteti et al. (2008); Tan et al. (2007); Tan et al. (2010) [58], [59], [60].
Western blot analysis
Protein extracts were prepared according to the TAP protocol. Protein samples were collected in different stages of the TAP purification and separated on a 12% SDS-polyacrylamide gel and transferred on to PVDF (Millipore) membrane. In the first step of purification, presence of TAP-tagged protein was identified by using peroxidase anti peroxidase conjugate (PAP, Sigma) antibody, which is specific to protein A domain of the TAP tag, as previously described Rivas et al. (2002) [56]. In the second step of purification, CBP domain was detected by using biotinylated CAM as previously described Rohila et al. (2004) [51].
To check the methylation status, protein extracts from OsFIE2-RNAi, OsFIE2-overexpression and wild type leaf tissues were separated by 15% SDS-PAGE gel and transferred to immobilon membrane. Immunoblots were performed using antibodies for H3K27me1, H3K27me2, H3K27me3, H3K9me2, H3K9me3 and unmodified H3 (Table S4) by following the standard Western blot procedure [59]. Equal amounts of protein samples were loaded for Western blot analysis. Western blot signals were normalized using unmodified H3 band intensity as a control and quantified with the help of PDQUEST software.
In vitro histone methyltransferase assay
Procedures for histone methyltranferase assay were adapted from the reported method Li et al. (2002) [84]. Briefly the assay was carried out by incubating TAP purified OsFIE2 protein complex, 10 µg of chicken core histones (Upstate) and 2 µCi of S-adenosyl-[methyl-3H]-l-methionine (GE Healthcare) in 50 µl of reaction buffer (50 mM Tris–HCl, pH 8.5, 20 mM KCl, 10 mM MgCl2, 1 mM CaCl2, 10 mM 2-mercaptoethanol, 1 mM dithiothreitol, and 250 mM sucrose) for 2 h at 30°C. The reaction was stopped by addition of SDS loading buffer and boiling at 100°C for 10 minutes. The proteins were separated by 15% SDS-PAGE gel and visualized by coomassie staining and fluorography.
Protein extraction and two-dimensional polyacrylamide gel electrophoresis
Total seed proteins were isolated from the mature seeds of both OsFIE2-RNAi and wild type plants. Mature seeds were ground into fine powder and protein extractions were performed using phenol extraction method as described Chitteti and Peng (2007) and Li et al. (2008) [72], [85]. Eight hundred micrograms of seed proteins were separated by using a standard 2-DE gel electrophoresis as described by Chitteti and Peng (2007) [85]. After PDQUEST analysis, the spots of interest were robotically excised from 2-DE gels by proteome works spot cutter (BioRad). In-gel digestion and MALD TOF-TOF mass spectrometry was performed as described by Chitteti and Peng (2007) [85].
RNA isolation and RT–PCR analysis
Total RNA extraction from different tissues of rice plants was performed by using Trizol (Invitrogen) according to the manufacturer's instructions. Three micrograms of total RNA was used for the cDNA synthesis by using Moloney murine leukemia virus (M-MLV) Reverse Transcriptase (Invitrogen). Rice Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (Loc_Os04g40950) was used as an internal control. PCR products were analyzed by 1% agarose gel electrophoresis. Primers used in the experiment are listed in Table S3.
Microscopy
Rice developing grains were fixed in FAA (3.7% formaldehyde, 5% acetic acid, 50% ethanol) at 4°C for overnight. Samples were dehydrated through graded ethanol series. After infiltration with xylene, samples were embedded in paraffin (Sigma-Aldrich), sectioned at 10 µm, and stained with 0.1% Eosin Y. Sections were observed and photographed using a bright-field microscope (Nikon).
Quantitative real-time PCR analysis
Real-time quantitative PCR analysis was performed with LightCycler 480 Real-Time PCR System (Roche Applied Science), using LightCycler 480 SYBR Green I Master. The relative quantification of transcript levels was performed by using 2−ΔΔC T Method [86]. The rice GAPDH (Loc_Os04g40950) gene was used as an internal control. Primers used in the experiment are listed in the Table S3.
E. coli two-hybrid assay
E. coli two hybrid assay was carried out according to the instructions given by manufacturers (Agilent Technologies). Recombinant bait (pBT) and target (pTRG) vectors were co-transformed into BacterioMatch II competent cells and positive interactions were screened by using selective and dual selective medium [64].
Pollen viability assay
To examine the pollen viability of OsFIE2-RNAi and wild-type plants, anthers were collected from well developed spikelets just before anthesis. Mature pollen grains were stained with 1% iodine in 3% potassium iodide solution (KI-I2) [87].Viable pollen grains were stained black and nonviable pollen grains were stained light yellow. The viable pollen grains were examined and counted under microscope.
Chromatin extraction and chromatin immunoprecipitation
The chromatin was extracted from endosperm following the protocol of Gendrel et al. (2005) [88] with minor modifications as reported recently Malone et al. (2011) [73]. Chromatin immunoprecipitation (ChIP) experiments were performed as reported Malone et al. (2011); Gendrel et al. (2005) [73], [88] using H3K27me3 antibody (Millipore) and protein A agarose/Salmon Sperm DNA.
Library preparation and solexa sequencing
The library preparation and Solexa Sequencing were the same as reported Malone et al. (2011) [73]. Input and ChIP samples were processed following Illumina's protocol from the ChIP DNA Sample Prep Kit. Briefly, 10 ng input and ChIP enriched DNA was subjected to end repair, addition of “A” bases to 3′ ends, ligation of adapters, agarose gel size selection for fragments with average size about 186 bp, and PCR amplification to produce a DNA library of adapter-modified fragments. DNA sequencing was carried out using the Illumina/Solexa Genome Analyzer sequencing system at a concentration of 2 to 4 pM. Cluster amplification, linearization, blocking and sequencing primer reagents were provided in the Solexa Cluster Amplification kits and were used according to the manufacturer's specifications.
Mapping the short reads to the genome and identifying peaks
Mapping the short reads is the same as reported Malone et al. (2011) [73]. The generated short reads were mapped onto the genome using SeqMap [89] allowing up to two mismatches between the short read and genome. The Illumina reads were aligned to TIGR version 6 of the rice genome [90]. The alignments were output in ELAND format. Only reads which mapped uniquely to the genome were retained. FindPeaks [91] was used to identify peaks with the mapped reads and the parameters used were the same as reported [73]. All the ChIP-Seq data were deposit to NCBI's Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) with the deposition number GSE27048 for genome-wide maps of chromatin state in rice endosperm.
Supporting Information
Zdroje
1. SchuettengruberB, ChourroutD, VervoortM, LeblancB, CavalliG (2007) Genome regulation by polycomb and trithorax proteins. Cell 128 : 735–745.
2. SchwartzYB, PirrottaV (2008) Polycomb complexes and epigenetic states. Curr Opin Cell Biol 20 : 266–273.
3. De LuciaF, CrevillenP, JonesAM, GrebT, DeanC (2008) A PHD-polycomb repressive complex 2 triggers the epigenetic silencing of FLC during vernalization. Proc Natl Acad Sci U S A 105 : 16831–16836.
4. GoodrichJ, PuangsomleeP, MartinM, LongD, MeyerowitzEM, et al. (1997) A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature 386 : 44–51.
5. GrossniklausU, Vielle-CalzadaJP, HoeppnerMA, GaglianoWB (1998) Maternal control of embryogenesis by MEDEA, a polycomb group gene in Arabidopsis. Science 280 : 446–450.
6. OhadN, YadegariR, MargossianL, HannonM, MichaeliD, et al. (1999) Mutations in FIE, a WD polycomb group gene, allow endosperm development without fertilization. Plant Cell 11 : 407–416.
7. LuoM, BilodeauP, KoltunowA, DennisES, PeacockWJ, et al. (1999) Genes controlling fertilization-independent seed development in Arabidopsis thaliana. Proc Natl Acad Sci U S A 96 : 296–301.
8. KohlerC, HennigL, BouveretR, GheyselinckJ, GrossniklausU, et al. (2003a) Arabidopsis MSI1 is a component of the MEA/FIE Polycomb group complex and required for seed development. EMBO J 22 : 4804–4814.
9. KohlerC, HennigL, SpillaneC, PienS, GruissemW, et al. (2003b) The Polycomb-group protein MEDEA regulates seed development by controlling expression of the MADS-box gene PHERES1. Genes Dev 17 : 1540–1553.
10. HuhJH, BauerMJ, HsiehTF, FischerRL (2008) Cellular programming of plant gene imprinting. Cell 132 : 735–744.
11. ChanvivattanaY, BishoppA, SchubertD, StockC, MoonYH, et al. (2004) Interaction of Polycomb-group proteins controlling flowering in Arabidopsis. Development 131 : 5263–5276.
12. LuoM, BilodeauP, DennisES, PeacockWJ, ChaudhuryA (2000) Expression and parent-of-origin effects for FIS2, MEA, and FIE in the endosperm and embryo of developing Arabidopsis seeds. Proc Natl Acad Sci U S A 97 : 10637–10642.
13. KatzA, OlivaM, MosqunaA, HakimO, OhadN (2004) FIE and CURLY LEAF polycomb proteins interact in the regulation of homeobox gene expression during sporophyte development. Plant J 37 : 707–719.
14. WoodCC, RobertsonM, TannerG, PeacockWJ, DennisES, et al. (2006) The Arabidopsis thaliana vernalization response requires a polycomb-like protein complex that also includes VERNALIZATION INSENSITIVE 3. Proc Natl Acad Sci U S A 103 : 14631–14636.
15. SpillaneC, MacDougallC, StockC, KohlerC, Vielle-CalzadaJP, et al. (2000) Interaction of the Arabidopsis polycomb group proteins FIE and MEA mediates their common phenotypes. Curr Biol 10 : 1535–1538.
16. YadegariR, KinoshitaT, LotanO, CohenG, KatzA, et al. (2000) Mutations in the FIE and MEA genes that encode interacting polycomb proteins cause parent-of-origin effects on seed development by distinct mechanisms. Plant Cell 12 : 2367–2382.
17. WangD, TysonMD, JacksonSS, YadegariR (2006) Partially redundant functions of two SET-domain polycomb-group proteins in controlling initiation of seed development in Arabidopsis. Proc Natl Acad Sci U S A 103 : 13244–13249.
18. YoshidaN, YanaiY, ChenL, KatoY, HiratsukaJ, et al. (2001) EMBRYONIC FLOWER2, a novel polycomb group protein homolog, mediates shoot development and flowering in Arabidopsis. Plant Cell 13 : 2471–2481.
19. JiangH, WongWH (2008) SeqMap: mapping massive amount of oligonucleotides to the genome. Bioinformatics 24 : 2395–2396.
20. SchonrockN, BouveretR, LeroyO, BorghiL, KohlerC, et al. (2006) Polycomb-group proteins repress the floral activator AGL19 in the FLC-independent vernalization pathway. Genes Dev 20 : 1667–1678.
21. GendallAR, LevyYY, WilsonA, DeanC (2001) The VERNALIZATION 2 gene mediates the epigenetic regulation of vernalization in Arabidopsis. Cell 107 : 525–535.
22. SungS, AmasinoRM (2004) Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature 427 : 159–164.
23. AngelA, SongJ, DeanC, HowardM (2011) A Polycomb-based switch underlying quantitative epigenetic memory. Nature 476 : 105–108.
24. NowackMK, ShirzadiR, DissmeyerN, DolfA, EndlE, et al. (2007) Bypassing genomic imprinting allows seed development. Nature 447 : 312–315.
25. GehringM, ChoiY, FischerRL (2004) Imprinting and seed development. Plant Cell 16 Suppl: S203–213.
26. GuittonAE, PageDR, ChambrierP, LionnetC, FaureJE, et al. (2004) Identification of new members of Fertilisation Independent Seed Polycomb Group pathway involved in the control of seed development in Arabidopsis thaliana. Development 131 : 2971–2981.
27. ChaudhuryAM, MingL, MillerC, CraigS, DennisES, et al. (1997) Fertilization-independent seed development in Arabidopsis thaliana. Proc Natl Acad Sci U S A 94 : 4223–4228.
28. KiyosueT, OhadN, YadegariR, HannonM, DinnenyJ, et al. (1999) Control of fertilization-independent endosperm development by the MEDEA polycomb gene in Arabidopsis. Proc Natl Acad Sci U S A 96 : 4186–4191.
29. XiaoW, GehringM, ChoiY, MargossianL, PuH, et al. (2003) Imprinting of the MEA Polycomb gene is controlled by antagonism between MET1 methyltransferase and DME glycosylase. Dev Cell 5 : 891–901.
30. LeroyO, HennigL, BreuningerH, LauxT, KohlerC (2007) Polycomb group proteins function in the female gametophyte to determine seed development in plants. Development 134 : 3639–3648.
31. KinoshitaT, YadegariR, HaradaJJ, GoldbergRB, FischerRL (1999) Imprinting of the MEDEA polycomb gene in the Arabidopsis endosperm. Plant Cell 11 : 1945–1952.
32. JullienPE, KatzA, OlivaM, OhadN, BergerF (2006a) Polycomb group complexes self-regulate imprinting of the Polycomb group gene MEDEA in Arabidopsis. Curr Biol 16 : 486–492.
33. JullienPE, KinoshitaT, OhadN, BergerF (2006b) Maintenance of DNA methylation during the Arabidopsis life cycle is essential for parental imprinting. Plant Cell 18 : 1360–1372.
34. BouyerD, RoudierF, HeeseM, AndersenED, GeyD, et al. (2011) Polycomb repressive complex 2 controls the embryo-to-seedling phase transition. PLoS Genet 7: e1002014 doi:10.1371/journal.pgen.1002014.
35. RoszakP, KohlerC (2011) Polycomb group proteins are required to couple seed coat initiation to fertilization. Proc Natl Acad Sci U S A 108 : 20826–20831.
36. DanilevskayaON, HermonP, HantkeS, MuszynskiMG, KolliparaK, et al. (2003) Duplicated fie genes in maize: expression pattern and imprinting suggest distinct functions. Plant Cell 15 : 425–438.
37. Gutierrez-MarcosJF, CostaLM, Dal PraM, ScholtenS, KranzE, et al. (2006) Epigenetic asymmetry of imprinted genes in plant gametes. Nat Genet 38 : 876–878.
38. HermonP, SrilunchangKO, ZouJ, DresselhausT, DanilevskayaON (2007) Activation of the imprinted Polycomb Group Fie1 gene in maize endosperm requires demethylation of the maternal allele. Plant Mol Biol 64 : 387–395.
39. SpringerNM, DanilevskayaON, HermonP, HelentjarisTG, PhillipsRL, et al. (2002) Sequence relationships, conserved domains, and expression patterns for maize homologs of the polycomb group genes E(z), esc, and E(Pc). Plant Physiol 128 : 1332–1345.
40. HaunWJ, Laoueille-DupratS, O'Connell MJ, SpillaneC, GrossniklausU, et al. (2007) Genomic imprinting, methylation and molecular evolution of maize Enhancer of zeste (Mez) homologs. Plant J 49 : 325–337.
41. KapazoglouA, TondelliA, PapaefthimiouD, AmpatzidouH, FranciaE, et al. (2010) Epigenetic chromatin modifiers in barley: IV. The study of barley polycomb group (PcG) genes during seed development and in response to external ABA. BMC Plant Biol 10 : 73.
42. LuoM, PlattenD, ChaudhuryA, PeacockWJ, DennisES (2009) Expression, imprinting, and evolution of rice homologs of the polycomb group genes. Mol Plant 2 : 711–723.
43. LiangYK, WangY, ZhangY, LiSG, LuXC, et al. (2003) OsSET1, a novel SET-domain-containing gene from rice. J Exp Bot 54 : 1995–1996.
44. ThakurJK, MalikMR, BhattV, ReddyMK, SoporySK, et al. (2003) A POLYCOMB group gene of rice (Oryza sativa L. subspecies indica), OsiEZ1, codes for a nuclear-localized protein expressed preferentially in young seedlings and during reproductive development. Gene 314 : 1–13.
45. LuoM, TaylorJM, SpriggsA, ZhangH, WuX, et al. (2011) A genome-wide survey of imprinted genes in rice seeds reveals imprinting primarily occurs in the endosperm. PLoS Genet 7: e1002125 doi:10.1371/journal.pgen.1002125.
46. RodriguesJC, LuoM, BergerF, KoltunowAM (2010) Polycomb group gene function in sexual and asexual seed development in angiosperms. Sex Plant Reprod 23 : 123–133.
47. SpillaneC, SchmidKJ, Laoueille-DupratS, PienS, Escobar-RestrepoJM, et al. (2007) Positive darwinian selection at the imprinted MEDEA locus in plants. Nature 448 : 349–352.
48. RodriguesJC, TuckerMR, JohnsonSD, HrmovaM, KoltunowAM (2008) Sexual and apomictic seed formation in Hieracium requires the plant polycomb-group gene FERTILIZATION INDEPENDENT ENDOSPERM. Plant Cell 20 : 2372–2386.
49. RigautG, ShevchenkoA, RutzB, WilmM, MannM, et al. (1999) A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol 17 : 1030–1032.
50. PuigO, CasparyF, RigautG, RutzB, BouveretE, et al. (2001) The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24 : 218–229.
51. RohilaJS, ChenM, CernyR, FrommME (2004) Improved tandem affinity purification tag and methods for isolation of protein heterocomplexes from plants. Plant J 38 : 172–181.
52. RubioV, ShenY, SaijoY, LiuY, GusmaroliG, et al. (2005) An alternative tandem affinity purification strategy applied to Arabidopsis protein complex isolation. Plant J 41 : 767–778.
53. RohilaJS, ChenM, ChenS, ChenJ, CernyR, et al. (2006) Protein-protein interactions of tandem affinity purification-tagged protein kinases in rice. Plant J 46 : 1–13.
54. Van LeeneJ, StalsH, EeckhoutD, PersiauG, Van De SlijkeE, et al. (2007) A tandem affinity purification-based technology platform to study the cell cycle interactome in Arabidopsis thaliana. Mol Cell Proteomics 6 : 1226–1238.
55. BrownAP, AffleckV, FawcettT, SlabasAR (2006) Tandem affinity purification tagging of fatty acid biosynthetic enzymes in Synechocystis sp. PCC6803 and Arabidopsis thaliana. J Exp Bot 57 : 1563–1571.
56. RivasS, RomeisT, JonesJD (2002) The Cf-9 disease resistance protein is present in an approximately 420-kilodalton heteromultimeric membrane-associated complex at one molecule per complex. Plant Cell 14 : 689–702.
57. AbeM, FujiwaraM, KurotaniK, YokoiS, ShimamotoK (2008) Identification of dynamin as an interactor of rice GIGANTEA by tandem affinity purification (TAP). Plant Cell Physiol 49 : 420–432.
58. ChittetiBR, TanF, MujahidH, MageeBG, BridgesSM, et al. (2008) Comparative analysis of proteome differential regulation during cell dedifferentiation in Arabidopsis. Proteomics 8 : 4303–4316.
59. TanF, LiG, ChittetiBR, PengZ (2007) Proteome and phosphoproteome analysis of chromatin associated proteins in rice (Oryza sativa). Proteomics 7 : 4511–4527.
60. TanF, ZhangK, MujahidH, VermaDP, PengZ (2010) Differential histone modification and protein expression associated with cell wall removal and regeneration in rice (Oryza sativa). J Proteome Res 10 : 551–563.
61. TieF, StrattonCA, KurzhalsRL, HartePJ (2007) The N terminus of Drosophila ESC binds directly to histone H3 and is required for E(Z)-dependent trimethylation of H3 lysine 27. Mol Cell Biol 27 : 2014–2026.
62. VerreaultA, KaufmanPD, KobayashiR, StillmanB (1998) Nucleosomal DNA regulates the core-histone-binding subunit of the human Hat1 acetyltransferase. Curr Biol 8 : 96–108.
63. VermaakD, WadePA, JonesPL, ShiYB, WolffeAP (1999) Functional analysis of the SIN3-histone deacetylase RPD3-RbAp48-histone H4 connection in the Xenopus oocyte. Mol Cell Biol 19 : 5847–5860.
64. JoungJK, RammEI, PaboCO (2000) A bacterial two-hybrid selection system for studying protein-DNA and protein-protein interactions. Proc Natl Acad Sci U S A 97 : 7382–7387.
65. CaoR, WangL, WangH, XiaL, Erdjument-BromageH, et al. (2002) Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298 : 1039–1043.
66. CzerminB, MelfiR, McCabeD, SeitzV, ImhofA, et al. (2002) Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111 : 185–196.
67. KuzmichevA, NishiokaK, Erdjument-BromageH, TempstP, ReinbergD (2002) Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev 16 : 2893–2905.
68. MullerJ, HartCM, FrancisNJ, VargasML, SenguptaA, et al. (2002) Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 111 : 197–208.
69. OhdanT, FranciscoPBJr, SawadaT, HiroseT, TeraoT, et al. (2005) Expression profiling of genes involved in starch synthesis in sink and source organs of rice. J Exp Bot 56 : 3229–3244.
70. GirouxMJ, BoyerC, FeixG, HannahLC (1994) Coordinated Transcriptional Regulation of Storage Product Genes in the Maize Endosperm. Plant Physiol 106 : 713–722.
71. SheKC, KusanoH, KoizumiK, YamakawaH, HakataM, et al. (2010) A novel factor FLOURY ENDOSPERM2 is involved in regulation of rice grain size and starch quality. Plant Cell 22 : 3280–3294.
72. LiG, NallamilliBR, TanF, PengZ (2008) Removal of high-abundance proteins for nuclear subproteome studies in rice (Oryza sativa) endosperm. Electrophoresis 29 : 604–617.
73. MaloneBM, TanF, BridgesSM, PengZ (2011) Comparison of four ChIP-Seq analytical algorithms using rice endosperm H3K27 trimethylation profiling data. PLoS ONE 6: e25260 doi:10.1371/journal.pone.0025260.
74. EgelhoferTA, MinodaA, KlugmanS, LeeK, Kolasinska-ZwierzP, et al. (2011) An assessment of histone-modification antibody quality. Nat Struct Mol Biol 18 : 91–93.
75. KondouY, NakazawaM, KawashimaM, IchikawaT, YoshizumiT, et al. (2008) Retarded growth of EMBRYO1, a new basic helix-loop-helix protein, expression in endosperm to control embryo growth. Plant Physiol 147 : 1924–1935.
76. YangS, JohnstonN, TalidehE, MitchellS, JeffreeC, et al. (2008) The endosperm-specific ZHOUPI gene of Arabidopsis thaliana regulates endosperm breakdown and embryonic epidermal development. Development 135 : 3501–3509.
77. SwiezewskiS, LiuF, MagusinA, DeanC (2009) Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature 462 : 799–802.
78. HeoJB, SungS (2011) Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 331 : 76–79.
79. BastowR, MylneJS, ListerC, LippmanZ, MartienssenRA, et al. (2004) Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 427 : 164–167.
80. GehringM, HuhJH, HsiehTF, PentermanJ, ChoiY, et al. (2006) DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell 124 : 495–506.
81. ZhangJ, NallamilliBR, MujahidH, PengZ (2010) OsMADS6 plays an essential role in endosperm nutrient accumulation and is subject to epigenetic regulation in rice (Oryza sativa). Plant J 64 : 604–617.
82. WuC, LiX, YuanW, ChenG, KilianA, et al. (2003) Development of enhancer trap lines for functional analysis of the rice genome. Plant J 35 : 418–427.
83. MikiD, ShimamotoK (2004) Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant Cell Physiol 45 : 490–495.
84. LiJ, MoazedD, GygiSP (2002) Association of the histone methyltransferase Set2 with RNA polymerase II plays a role in transcription elongation. J Biol Chem 277 : 49383–49388.
85. ChittetiBR, PengZ (2007) Proteome and phosphoproteome dynamic change during cell dedifferentiation in Arabidopsis. Proteomics 7 : 1473–1500.
86. LivakKJ, SchmittgenTD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25 : 402–408.
87. ChhunT, AyaK, AsanoK, YamamotoE, MorinakaY, et al. (2007) Gibberellin regulates pollen viability and pollen tube growth in rice. Plant Cell 19 : 3876–3888.
88. GendrelAV, LippmanZ, MartienssenR, ColotV (2005) Profiling histone modification patterns in plants using genomic tiling microarrays. Nat Methods 2 : 213–218.
89. JiangH, WongWH (2008) SeqMap: mapping massive amount of oligonucleotides to the genome. Bioinformatics 24 : 2395–2396.
90. OuyangS, ZhuW, HamiltonJ, LinH, CampbellM, et al. (2007) The TIGR Rice Genome Annotation Resource: improvements and new features. Nucleic Acids Res 35: D883–887.
91. FejesAP, RobertsonG, BilenkyM, VarholR, BainbridgeM, et al. (2008) FindPeaks 3.1: a tool for identifying areas of enrichment from massively parallel short-read sequencing technology. Bioinformatics 24 : 1729–1730.
Štítky
Genetika Reprodukční medicína
Článek Ubiquitous Polygenicity of Human Complex Traits: Genome-Wide Analysis of 49 Traits in KoreansČlánek Alternative Splicing and Subfunctionalization Generates Functional Diversity in Fungal ProteomesČlánek RFX Transcription Factor DAF-19 Regulates 5-HT and Innate Immune Responses to Pathogenic Bacteria inČlánek Surveillance-Activated Defenses Block the ROS–Induced Mitochondrial Unfolded Protein ResponseČlánek Deficiency Reduces Adipose OXPHOS Capacity and Triggers Inflammation and Insulin Resistance in Mice
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 3
-
Všechny články tohoto čísla
- Power and Predictive Accuracy of Polygenic Risk Scores
- Rare Copy Number Variants Are a Common Cause of Short Stature
- Coordination of Flower Maturation by a Regulatory Circuit of Three MicroRNAs
- Ubiquitous Polygenicity of Human Complex Traits: Genome-Wide Analysis of 49 Traits in Koreans
- Genomic Evidence for Island Population Conversion Resolves Conflicting Theories of Polar Bear Evolution
- Mechanistic Insight into the Pathology of Polyalanine Expansion Disorders Revealed by a Mouse Model for X Linked Hypopituitarism
- Genome-Wide Association Study and Gene Expression Analysis Identifies as a Predictor of Response to Etanercept Therapy in Rheumatoid Arthritis
- Problem Solved: An Interview with Sir Edwin Southern
- Long Interspersed Element–1 (LINE-1): Passenger or Driver in Human Neoplasms?
- Mouse HFM1/Mer3 Is Required for Crossover Formation and Complete Synapsis of Homologous Chromosomes during Meiosis
- Alternative Splicing and Subfunctionalization Generates Functional Diversity in Fungal Proteomes
- A WRKY Transcription Factor Recruits the SYG1-Like Protein SHB1 to Activate Gene Expression and Seed Cavity Enlargement
- Microhomology-Mediated Mechanisms Underlie Non-Recurrent Disease-Causing Microdeletions of the Gene or Its Regulatory Domain
- Ancient Evolutionary Trade-Offs between Yeast Ploidy States
- Differential Evolutionary Fate of an Ancestral Primate Endogenous Retrovirus Envelope Gene, the EnvV , Captured for a Function in Placentation
- A Feed-Forward Loop Coupling Extracellular BMP Transport and Morphogenesis in Wing
- The Tomato Yellow Leaf Curl Virus Resistance Genes and Are Allelic and Code for DFDGD-Class RNA–Dependent RNA Polymerases
- The U-Box E3 Ubiquitin Ligase TUD1 Functions with a Heterotrimeric G α Subunit to Regulate Brassinosteroid-Mediated Growth in Rice
- Role of the DSC1 Channel in Regulating Neuronal Excitability in : Extending Nervous System Stability under Stress
- –Independent Phenotypic Switching in and a Dual Role for Wor1 in Regulating Switching and Filamentation
- Pax6 Regulates Gene Expression in the Vertebrate Lens through miR-204
- Blood-Informative Transcripts Define Nine Common Axes of Peripheral Blood Gene Expression
- Genetic Architecture of Skin and Eye Color in an African-European Admixed Population
- Fine Characterisation of a Recombination Hotspot at the Locus and Resolution of the Paradoxical Excess of Duplications over Deletions in the General Population
- Estrogen Mediated-Activation of miR-191/425 Cluster Modulates Tumorigenicity of Breast Cancer Cells Depending on Estrogen Receptor Status
- Complex Patterns of Genomic Admixture within Southern Africa
- Yap- and Cdc42-Dependent Nephrogenesis and Morphogenesis during Mouse Kidney Development
- Molecular Networks of Human Muscle Adaptation to Exercise and Age
- Alp/Enigma Family Proteins Cooperate in Z-Disc Formation and Myofibril Assembly
- Polycomb Group Gene Regulates Rice () Seed Development and Grain Filling via a Mechanism Distinct from
- RFX Transcription Factor DAF-19 Regulates 5-HT and Innate Immune Responses to Pathogenic Bacteria in
- Distinct Molecular Strategies for Hox-Mediated Limb Suppression in : From Cooperativity to Dispensability/Antagonism in TALE Partnership
- A Natural Polymorphism in rDNA Replication Origins Links Origin Activation with Calorie Restriction and Lifespan
- TDP2–Dependent Non-Homologous End-Joining Protects against Topoisomerase II–Induced DNA Breaks and Genome Instability in Cells and
- Recurrent Rearrangement during Adaptive Evolution in an Interspecific Yeast Hybrid Suggests a Model for Rapid Introgression
- Genome-Wide Association Study in Mutation Carriers Identifies Novel Loci Associated with Breast and Ovarian Cancer Risk
- Coincident Resection at Both Ends of Random, γ–Induced Double-Strand Breaks Requires MRX (MRN), Sae2 (Ctp1), and Mre11-Nuclease
- Identification of a -Specific Modifier Locus at 6p24 Related to Breast Cancer Risk
- A Novel Function for the Hox Gene in the Male Accessory Gland Regulates the Long-Term Female Post-Mating Response in
- Tdp2: A Means to Fixing the Ends
- A Novel Role for the RNA–Binding Protein FXR1P in Myoblasts Cell-Cycle Progression by Modulating mRNA Stability
- Association Mapping and the Genomic Consequences of Selection in Sunflower
- Histone Deacetylase 2 (HDAC2) Regulates Chromosome Segregation and Kinetochore Function via H4K16 Deacetylation during Oocyte Maturation in Mouse
- A Novel Mutation in the Upstream Open Reading Frame of the Gene Causes a MEN4 Phenotype
- Ataxin1L Is a Regulator of HSC Function Highlighting the Utility of Cross-Tissue Comparisons for Gene Discovery
- Human Spermatogenic Failure Purges Deleterious Mutation Load from the Autosomes and Both Sex Chromosomes, including the Gene
- A Conserved Upstream Motif Orchestrates Autonomous, Germline-Enriched Expression of piRNAs
- Statistical Analysis Reveals Co-Expression Patterns of Many Pairs of Genes in Yeast Are Jointly Regulated by Interacting Loci
- Matefin/SUN-1 Phosphorylation Is Part of a Surveillance Mechanism to Coordinate Chromosome Synapsis and Recombination with Meiotic Progression and Chromosome Movement
- A Role for the Malignant Brain Tumour (MBT) Domain Protein LIN-61 in DNA Double-Strand Break Repair by Homologous Recombination
- The Population and Evolutionary Dynamics of Phage and Bacteria with CRISPR–Mediated Immunity
- Long Noncoding RNA MALAT1 Controls Cell Cycle Progression by Regulating the Expression of Oncogenic Transcription Factor B-MYB
- Surveillance-Activated Defenses Block the ROS–Induced Mitochondrial Unfolded Protein Response
- DNA Topoisomerase III Localizes to Centromeres and Affects Centromeric CENP-A Levels in Fission Yeast
- Genome-Wide Control of RNA Polymerase II Activity by Cohesin
- Divergent Selection Drives Genetic Differentiation in an R2R3-MYB Transcription Factor That Contributes to Incipient Speciation in
- NODULE INCEPTION Directly Targets Subunit Genes to Regulate Essential Processes of Root Nodule Development in
- Spreading of a Prion Domain from Cell-to-Cell by Vesicular Transport in
- Deficiency in Origin Licensing Proteins Impairs Cilia Formation: Implications for the Aetiology of Meier-Gorlin Syndrome
- Deficiency Reduces Adipose OXPHOS Capacity and Triggers Inflammation and Insulin Resistance in Mice
- The Conserved SKN-1/Nrf2 Stress Response Pathway Regulates Synaptic Function in
- Functional Genomic Analysis of the Regulatory Network in
- Astakine 2—the Dark Knight Linking Melatonin to Circadian Regulation in Crustaceans
- CRL2 E3-Ligase Regulates Proliferation and Progression through Meiosis in the Germline
- Both the Caspase CSP-1 and a Caspase-Independent Pathway Promote Programmed Cell Death in Parallel to the Canonical Pathway for Apoptosis in
- PRMT4 Is a Novel Coactivator of c-Myb-Dependent Transcription in Haematopoietic Cell Lines
- A Copy Number Variant at the Locus Likely Confers Risk for Canine Squamous Cell Carcinoma of the Digit
- Evidence of Gene–Environment Interactions between Common Breast Cancer Susceptibility Loci and Established Environmental Risk Factors
- HIV Infection Disrupts the Sympatric Host–Pathogen Relationship in Human Tuberculosis
- Trans-Ethnic Fine-Mapping of Lipid Loci Identifies Population-Specific Signals and Allelic Heterogeneity That Increases the Trait Variance Explained
- A Gene Transfer Agent and a Dynamic Repertoire of Secretion Systems Hold the Keys to the Explosive Radiation of the Emerging Pathogen
- The Role of ATM in the Deficiency in Nonhomologous End-Joining near Telomeres in a Human Cancer Cell Line
- Dynamic Circadian Protein–Protein Interaction Networks Predict Temporal Organization of Cellular Functions
- Nuclear Myosin 1c Facilitates the Chromatin Modifications Required to Activate rRNA Gene Transcription and Cell Cycle Progression
- Robust Prediction of Expression Differences among Human Individuals Using Only Genotype Information
- A Single Cohesin Complex Performs Mitotic and Meiotic Functions in the Protist
- The Role of the Arabidopsis Exosome in siRNA–Independent Silencing of Heterochromatic Loci
- Elevated Expression of the Integrin-Associated Protein PINCH Suppresses the Defects of Muscle Hypercontraction Mutants
- Twist1 Controls a Cell-Specification Switch Governing Cell Fate Decisions within the Cardiac Neural Crest
- Genome-Wide Testing of Putative Functional Exonic Variants in Relationship with Breast and Prostate Cancer Risk in a Multiethnic Population
- Heteroduplex DNA Position Defines the Roles of the Sgs1, Srs2, and Mph1 Helicases in Promoting Distinct Recombination Outcomes
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Fine Characterisation of a Recombination Hotspot at the Locus and Resolution of the Paradoxical Excess of Duplications over Deletions in the General Population
- Molecular Networks of Human Muscle Adaptation to Exercise and Age
- Recurrent Rearrangement during Adaptive Evolution in an Interspecific Yeast Hybrid Suggests a Model for Rapid Introgression
- Genome-Wide Association Study and Gene Expression Analysis Identifies as a Predictor of Response to Etanercept Therapy in Rheumatoid Arthritis
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



