-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaLong Interspersed Element–1 (LINE-1): Passenger or Driver in Human Neoplasms?
LINE-1 (L1) retrotransposons make up a significant portion of human genomes, with an estimated 500,000 copies per genome. Like other retrotransposons, L1 retrotransposons propagate through RNA sequences that are reverse transcribed into DNA sequences, which are integrated into new genomic loci. L1 somatic insertions have the potential to disrupt the transcriptome by inserting into or nearby genes. By mutating genes and playing a role in epigenetic dysregulation, L1 transposons may contribute to tumorigenesis. Studies of the “mobilome” have lagged behind other tumor characterizations at the sequence, transcript, and epigenetic levels. Here, we consider evidence that L1 retrotransposons may sometimes drive human tumorigenesis.
Published in the journal: . PLoS Genet 9(3): e32767. doi:10.1371/journal.pgen.1003402
Category: Review
doi: https://doi.org/10.1371/journal.pgen.1003402Summary
LINE-1 (L1) retrotransposons make up a significant portion of human genomes, with an estimated 500,000 copies per genome. Like other retrotransposons, L1 retrotransposons propagate through RNA sequences that are reverse transcribed into DNA sequences, which are integrated into new genomic loci. L1 somatic insertions have the potential to disrupt the transcriptome by inserting into or nearby genes. By mutating genes and playing a role in epigenetic dysregulation, L1 transposons may contribute to tumorigenesis. Studies of the “mobilome” have lagged behind other tumor characterizations at the sequence, transcript, and epigenetic levels. Here, we consider evidence that L1 retrotransposons may sometimes drive human tumorigenesis.
Introduction to LINE-1 (L1) Retrotransposons
Repetitive sequences collectively make up greater than half of the human genome and are subdivided into two principal types (Figure 1) [1], [2]. The first is the tandem repeat, or satellite, in which each repeat unit is immediately adjacent to others. Tandem repeat sequences are formed in situ by replication or recombination events [3]. The second type consists of interspersed repeats, which are repeated sequences that are distributed throughout the genome rather than occurring in tandem [4]. Interspersed repeat sequences are derived from transposable elements or mobile DNAs, further described as either DNA or RNA transposons, depending on the mechanism of their spread. While DNA transposons use a “cut-and-paste” mechanism, RNA transposons use a “copy-and-paste” mode of moving in genomes. RNA transposons use an RNA intermediate and are also referred to as retrotransposons, retroposons, or retroelements; we will use the term retrotransposons in this review. The only active mobile DNAs in modern-day humans are the autonomous L1 retrotransposon and non-protein-coding (nonautonomous) sequences its machinery mobilizes.
Fig. 1. Repetitive sequences in the human genome. 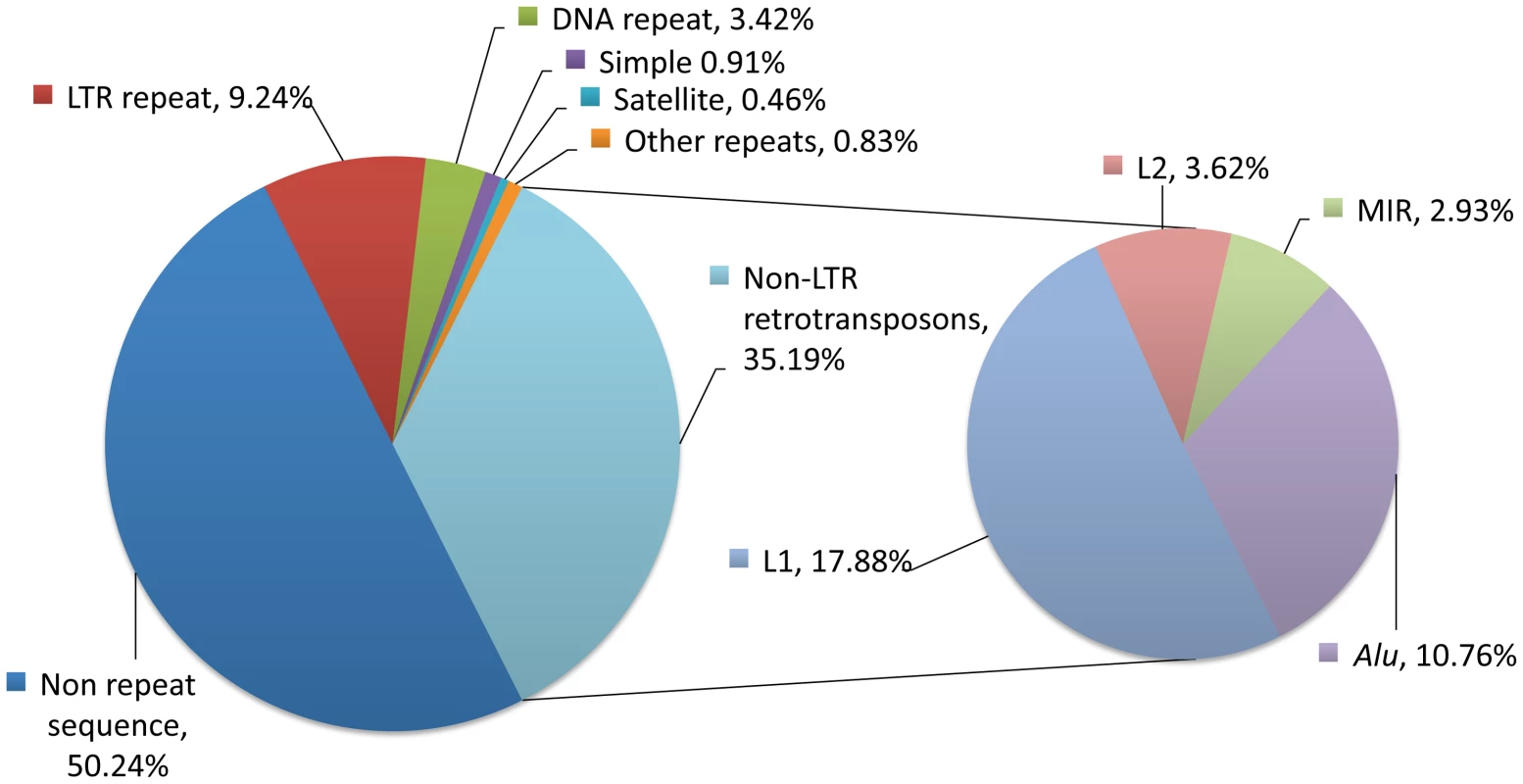
About half of our DNA bears homology to known classes of repeats (left chart). The largest class of repeats is the non-LTR retrotransposons, which consists mostly of LINE-1 (L1), L2, MIRs, and Alu elements (right chart). L2 and MIR sequences are not currently active, but subsets of L1 (17.88%), Alu (10.76%), and SVA sequences (not shown, 0.1%) are currently mobile in human genomes and are sources of genetic polymorphisms. Proportions were determined using a RepeatMasker (version rm-20110920, default settings, RepBase sequence database version 16.08) analysis of the Human February 2009 (GRCh37/hg19) assembly. LTR, long terminal repeat retrotransposons; L1, long interspersed element–1; L2, long interspersed element–2; MIR, mammalian wide interspersed repeat; Alu, a short interspersed element named for the AluI restriction enzyme; SVA, a composite retrotransposon consisting of short interspersed repeat (SINE-R), variable number tandem repeat (VNTR), and Alu like sequence segments. The L1 life cycle entails three steps (Figure 2, red box). The first step is transcription of a genomic L1 into RNA, which is mediated by RNA polymerase II from an internal L1 promoter. Transcription from an internal antisense L1 promoter may occur concurrently. In the second step, the RNA is translated into two L1-encoded proteins: ORF1p, an RNA-binding protein, and ORF2p, a protein with reverse transcriptase and endonuclease activities. These proteins associate with the L1 transcript, and the resulting ribonucleoprotein (RNP) complexes are then transferred to the nucleus. The third step is termed target-primed reverse transcription (TPRT). In the course of TPRT, ORF2p cleaves the target DNA (often at a 5′-TTTTAA-3′ consensus sequence) and uses the 3′ hydroxyl group to prime the reverse transcription reaction. Synthesis of the second strand and resolution of the structure are poorly understood. Because the L1 life cycle generates DNA breaks, cell host proteins that mediate DNA repair are likely involved.
Fig. 2. DNA methylation and related mechanisms inhibit LINE-1 (L1) expression, and hypomethylation of DNA allows the L1 retrotransposon “life cycle” to proceed. 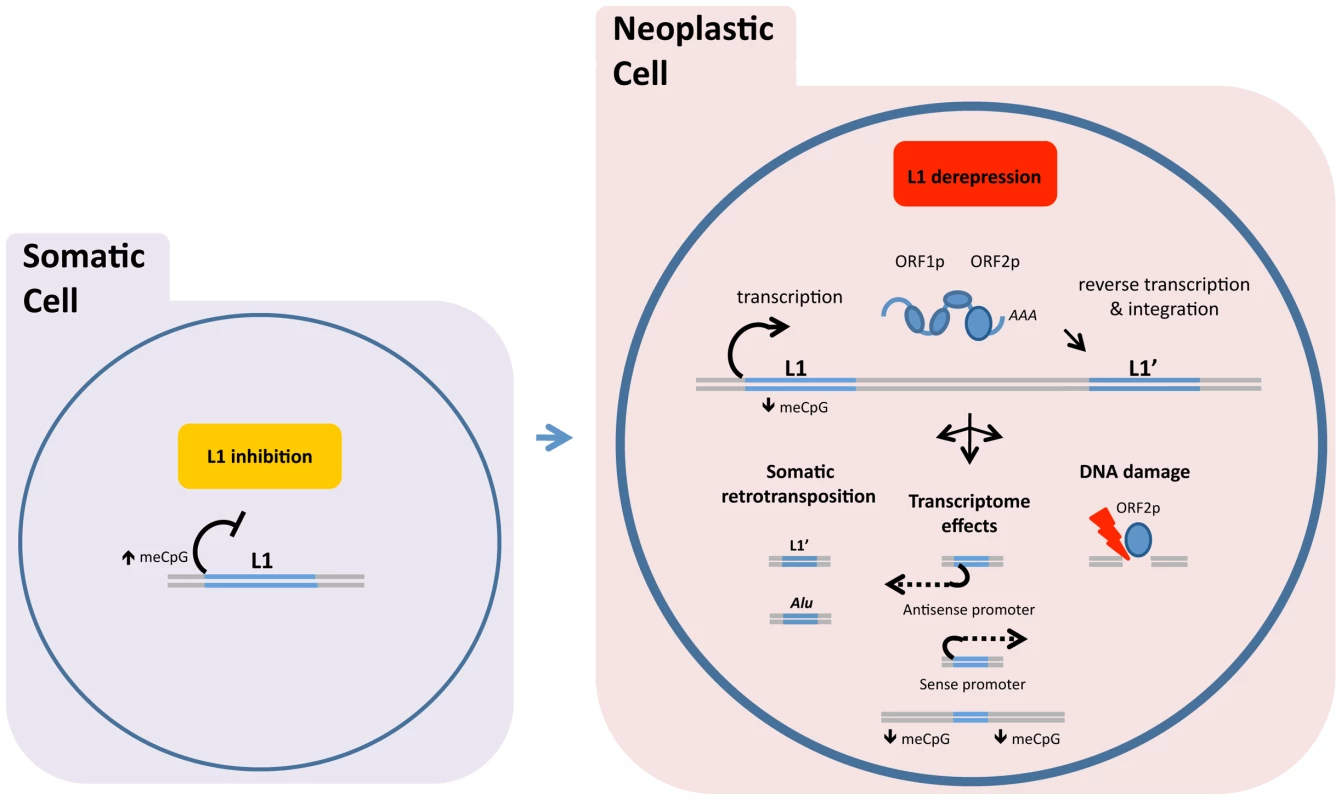
In normal somatic cells, DNA methylation and related mechanisms inhibit LINE-1 (L1) expression (left image). In neoplastic cells, hypomethylation of DNA allows the L1 retrotransposon “life cycle” to proceed (right image). Retrotransposition is shown in a simplified schematic under the red box as (from left to right) transcription, assembly of ORF1p and ORF2p with L1 RNA, and insertion of a new L1 sequence (L1′). Related tumor effects are conceptually shown as (i) somatic retrotransposition of L1 and nonautonomous repeat elements, such as Alu repeats; (ii) transcriptional changes induced by L1-encoded promoters (in antisense and sense) or impacts on area methylation; and (iii) L1 ORF2p-generated DNA breaks. ASP, L1 antisense promoter. In the human genome, the majority of our estimated 500,000 L1 copies are (1) present on both homologous chromosomes, (2) truncated at the 5′ end (mean length, 0.9 kb), and (3) incapable of encoding ORF1p and ORF2p and transposing. A relatively small number are potentially active, full-length L1 elements (approximately 6 kb long) with intact coding sequences for ORF1p and ORF2p [5]. Full-length L1 insertions largely reflect the activity of the Ta1 subfamily of human-specific L1. Functional human-specific L1 insertions passed in the germline have deleterious effects on fitness and are hence under negative selection but continue to be a source of genetic diversity.
In this review, we discuss inhibition of L1 retrotransposition in normal somatic cells and activation of L1 in cancer cells. We also consider possible causal roles of L1 in tumorigenesis, discussing ways in which it may influence regulation of host genes or genomic stability apart from the canonical transposition pathway.
Inhibition of L1 Retrotransposition in Normal Germline and Somatic Cells
L1 is regulated by distinct pathways in different cell contexts. In the male germline, L1 is inhibited via an elaborate system involving Piwi-interacting RNAs (piRNAs) that ultimately methylates genomic L1 sequences. This depends on methylation regulator DNMT3L [6] and PIWIL4 (also known as MIWI2) [7], as well as PIWI proteins involved in piRNA production. In embryonic stem cells, which can be used to model chromatin regulation in preimplantation-stage embryos, inherited L1 methylation is maintained by DNA methyltransferases DNMT1, DNMT3A, and DNMT3B [8]. In embryonal carcinoma cell lines, which are sometimes used for the same purpose, newly retrotransposed L1 sequences are silenced by histone alterations, including deacetylation of H4 and dimethylation of H3K9 [9].
In addition to the host factors mentioned above, other proteins have been implicated in L1 repression in various somatic tissues. These include methyl CpG binding protein 2 (MECP2) [10], lymphoid-specific helicase (HELLS) [11]–[13], a retinoblastoma protein-containing complex (RB1) [14], the 3′ repair exonuclease 1 (TREX1) [15], excision repair cross complementing 1 (ERCC) [16], and apolipoprotein B mRNA editing enzyme, catalytic proteins APOBEC3A, APOBEC3B, and APOBEC1 [17], [18]. In aggregate, these and other proteins seem to prevent L1 expression or somatic retrotransposition in all normal tissues except for the developing brain [19], [20]. However, the relative importance of each of these proteins to transposon silencing in the breadth of human cell types remains to be fully characterized.
L1 Retrotransposition in Neoplasms
Somatic L1 insertions have the inherent potential to drive tumorigenesis by activating oncogenes or inactivating tumor suppressor genes. However, somatic L1-related rearrangements that have driven tumorigenesis have rarely been discovered in humans. Before 2010, only two such cases had been reported. The first case involved a gene-activating rearrangement at the MYC oncogene locus in breast ductal adenocarcinoma [21]. The second case involved a newly integrated L1 sequence that inactivated the APC tumor suppressor gene in colon cancer cells. This insertion had sequence features characteristic of an L1 retrotransposition [22]. The scarcity of somatic L1 insertions that have been found to drive tumorigenesis may be owed in part to the limitations of traditional molecular assays, which have not lent themselves to identifying de novo L1 insertions.
The introduction of targeted next-generation sequencing and analysis methods in recent years has led to additional reports of somatic L1 retrotransposition in colon cancer [23], [24], as well as in lung [25], prostate [24], and ovarian [24] carcinomas. In one study, researchers targeted insertion sites for sequencing and found nine tumor-specific somatic L1 insertions in six of 20 primary lung carcinomas tested [25]. In another study, Lee and colleagues [24] surveyed whole genome sequences from a variety of tumors included in The Cancer Genome Atlas project and discovered 183 L1 insertions in colorectal, prostatic, and ovarian carcinomas. The number of insertions per tumor ranged from an average of four in several ovarian carcinoma specimens to 106 in a single colon carcinoma specimen. All of the insertions were severely truncated at the 5′ end, perhaps reflecting inhibition of TPRT by robust somatic cell mechanisms.
The fact that the somatic insertions reported by Lee and colleagues were skewed toward genes that are commonly downregulated or mutated in the tumors that they studied suggests that L1 insertions may contribute to cancer formation. In order to determine whether L1 actually does so, it would help to identify specific loci that are recurrently disrupted by L1 insertions. Ongoing work to profile L1 positions in additional tumor types, metastatic samples, and samples of disease relapse after therapy may lead to the identification of such loci.
L1 Expression in Human Neoplasms
Whereas normal adult tissues do not express L1 ORF1p [26], [27], selected human neoplasms do express both L1 RNA and proteins. Several epithelial neoplasms, including renal, ovarian, lung, and prostate carcinomas, express L1 RNA at detectable levels [28]. This L1 derepression can be associated with poor clinical features. For example, in pancreatic ductal adenocarcinomas, higher levels of L1 RNA correlate with higher grade lesions and poorer clinical outcomes [29]. Similarly, in high-grade breast carcinomas, higher levels of nuclear L1 ORF1p protein, as determined by immunohistochemistry, correlate with poorer clinical outcomes [26].
In addition to these definitive cases of L1 expression, there have been other cases indicating L1 derepression. For example, L1 promoter hypomethylation has been reported in multiple myeloma [30], chronic myeloid leukemia (CML) [31], and chronic lymphocytic leukemia, suggesting that L1 might be transcribed in these cancers [32]. Of note, CML cases with L1 promoter CpG dinucleotide hypomethylation tend to be aggressive neoplasms leading to poor prognosis, although whether L1 is causally related to the aggressiveness is not known [31].
L1 RNA expression, ORF1p expression, and L1 methylation status are more readily detected than ORF2p expression using current reagents. While it is plausible that ORF2p expression also occurs in cases exhibiting the first three features, it is possible that examples of decoupled regulation will be described. ORF2p expression is a major mechanism, in addition to retrotransposition, through which L1 may impact the genome. Cell-culture-based studies convincingly show that ORF2p expression can be mutagenic, inducing not only mutations associated with canonical retrotransposition events but also DNA breaks [33]–[35] and large genomic deletions [36]. ORF2p has also recently been implicated in recurrent DNA translocations that occur in conjunction with other DNA-binding proteins that may target its endonuclease activity within the genome [37].
L1 and Antitumor Effects of Reverse Transcriptase (RT) Inhibition
Several nucleoside analogues have been shown in biochemical analyses to inhibit the RT activity of ORF2p and, in cell-culture-based retrotransposition assays, to reduce the number of L1 retrotransposition events [38], [39]. Nucleoside analogues function by becoming incorporated into growing DNA strands, where they act as chain terminators. In contrast, other types of RT inhibitors, known as nonnucleoside RT inhibitors (NNRTIs), function by binding to a “pocket” of the RT enzyme, and may be more restricted in their scope of activity. NNRTIs are much less potent than nucleoside analogues, or do not affect ORF2p activity at all in retrotransposition assays. For example, efavirenz, an NNRTI antiretroviral drug, affects ORF2p activity only at very high concentrations, and nevirapine, also an NNRTI antiretroviral drug, has negligible impact on ORF2p activity.
In cell-culture-based cancer models, both nucleoside RT inhibitors and NNRTIs have been shown to promote senescence and differentiation, and reduce invasive growth. For example, the nucleoside analog abacavir inhibits the proliferation and migration of a prostate cancer cell line [40], and the NNRTIs efavirenz and nevirapine inhibit the growth of malignant melanoma and prostate cancer cell lines in a dose-dependent manner [41]. Nevirapine also inhibits the growth of teratocarcinoma, colon carcinoma, lung carcinoma, and acute myeloid leukemia cell lines in a dose-dependent manner [42]. Whether L1 RT activity is relevant to these observations is hotly debated. Some or all of these drug effects may be mediated through pathways other than inhibition of L1 RT.
Impacts of L1 on Transcription Initiation in Cancer
In some cases, transcription is initiated within an L1 to form a chimeric mRNA that contains L1 sequence (sense or antisense) and downstream exons of a host gene. In fact, in normal mouse tissues, transcription is initiated frequently within L1 sequence, as Faulkner et al. determined by sequencing the 5′-most nucleotides of RNAs from both normal and neoplastic tissues, using cap-analysis gene expression tag technology (CAGE) [43]. Transcription initiation often occurs within 5′ L1 sequence, which contains both sense and antisense promoter activities. The CAGE analysis also revealed highly specific patterns of transcriptional activity from L1 across samples. In human cancers, aberrant expression of chimeric transcripts may play important roles in tumorigenesis. Investigators have noted transcript variants initiated by L1 antisense promoters in bladder carcinoma [44], chronic myeloid leukemia (CML) [31], esophageal adenocarcinoma [45], and breast carcinoma [46].
L1 transcription initiation can be drug induced. For example, Weber et al. showed that transcription of the L1-cMet sequence, which originates within the MET proto-oncogene locus, is specifically induced in CML cells by the demethylating agents azacytidine and decitabine [47]. Their work further indicated that L1-cMet functions as a potential tumor suppressor by interfering with expression of the cMet transcript, ultimately leading to decreased cMet signaling. The fact that L1 derepression is associated with tumor suppression in some cases and cancer progression in others, as mentioned earlier, makes us cognizant that the roles of L1 derepression in cancer are complex.
L1 insertions may also influence transcription initiation from the canonical transcriptional start site of a gene locus. Estecio et al. found evidence of this by noting an increased number of L1 insertions near selectively hypomethylated transcriptional start sites of single-copy genes in cancer cell lines [48]. Thus, epigenetic dysregulation of L1 sequences may promote transcription of nearby oncogenes.
Conclusions
There is growing evidence that L1 expression and even complete retrotransposition occur in selected human cancers, suggesting that L1 may drive tumorigenesis. Even if L1 does not drive tumorigenesis through new insertions or ORF2p genetic damage, L1 sequences are such prevalent genomic passengers that they are likely to contribute to gene expression during tumor development, and affect responses of tumors to treatment. New reagents, experimental approaches, and informatics tools are being developed to provide more complete pictures of L1-mediated roles in human neoplasms.
Zdroje
1. Smit A, Hubley R, Green P (1996–2010) RepeatMasker Open. http://www.repeatmasker.org.
2. de KoningAP, GuW, CastoeTA, BatzerMA, PollockDD (2011) Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet 7: e1002384 doi:10.1371/journal.pgen.1002384..
3. GemayelR, VincesMD, LegendreM, VerstrepenKJ (2010) Variable tandem repeats accelerate evolution of coding and regulatory sequences. Annual Review of Genetics 44 : 445–477.
4. SingerMF (1982) SINEs and LINEs: highly repeated short and long interspersed sequences in mammalian genomes. Cell 28 : 433–434.
5. BoissinotS, DavisJ, EntezamA, PetrovD, FuranoAV (2006) Fitness cost of LINE-1 (L1) activity in humans. Proc Natl Acad Sci U S A 103 : 9590–9594.
6. Bourc'hisD, BestorTH (2004) Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature 431 : 96–99.
7. CarmellMA, GirardA, van de KantHJ, Bourc'hisD, BestorTH, et al. (2007) MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Developmental Cell 12 : 503–514.
8. LiangG, ChanMF, TomigaharaY, TsaiYC, GonzalesFA, et al. (2002) Cooperativity between DNA methyltransferases in the maintenance methylation of repetitive elements. Molecular and Cellular Biology 22 : 480–491.
9. Garcia-PerezJL, MorellM, ScheysJO, KulpaDA, MorellS, et al. (2010) Epigenetic silencing of engineered L1 retrotransposition events in human embryonic carcinoma cells. Nature 466 : 769–773.
10. MuotriAR, MarchettoMC, CoufalNG, OefnerR, YeoG, et al. (2010) L1 retrotransposition in neurons is modulated by MeCP2. Nature 468 : 443–446.
11. HuangJ, FanT, YanQ, ZhuH, FoxS, et al. (2004) Lsh, an epigenetic guardian of repetitive elements. Nucleic Acids Research 32 : 5019–5028.
12. SunLQ, LeeDW, ZhangQ, XiaoW, RaabeEH, et al. (2004) Growth retardation and premature aging phenotypes in mice with disruption of the SNF2-like gene, PASG. Genes & Development 18 : 1035–1046.
13. FanT, SchmidtmannA, XiS, BrionesV, ZhuH, et al. (2008) DNA hypomethylation caused by Lsh deletion promotes erythroleukemia development. Epigenetics: Official Journal of the DNA Methylation Society 3 : 134–142.
14. Montoya-DurangoDE, LiuY, TenengI, KalbfleischT, LacyME, et al. (2009) Epigenetic control of mammalian LINE-1 retrotransposon by retinoblastoma proteins. Mutation Research 665 : 20–28.
15. StetsonDB, KoJS, HeidmannT, MedzhitovR (2008) Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell 134 : 587–598.
16. GasiorSL, Roy-EngelAM, DeiningerPL (2008) ERCC1/XPF limits L1 retrotransposition. DNA Repair 7 : 983–989.
17. BogerdHP, WiegandHL, HulmeAE, Garcia-PerezJL, O'SheaKS, et al. (2006) Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc Natl Acad Sci U S A 103 : 8780–8785.
18. IkedaT, Abd El GalilKH, TokunagaK, MaedaK, SataT, et al. (2011) Intrinsic restriction activity by apolipoprotein B mRNA editing enzyme APOBEC1 against the mobility of autonomous retrotransposons. Nucleic Acids Research 39 : 5538–5554.
19. MuotriAR, ChuVT, MarchettoMC, DengW, MoranJV, et al. (2005) Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature 435 : 903–910.
20. BaillieJK, BarnettMW, UptonKR, GerhardtDJ, RichmondTA, et al. (2011) Somatic retrotransposition alters the genetic landscape of the human brain. Nature 479 : 534–537.
21. MorseB, RothergPG, SouthVJ, SpandorferJM, AstrinSM (1988) Insertional mutagenesis of the myc locus by a LINE-1 sequence in a human breast carcinoma. Nature 333 : 87–90.
22. MikiY, NishishoI, HoriiA, MiyoshiY, UtsunomiyaJ, et al. (1992) Disruption of the APC gene by a retrotransposal insertion of L1 sequence in a colon cancer. Cancer Research 52 : 643–645.
23. SolyomS, EwingAD, RahrmannEP, DoucetT, NelsonHH, et al. (2012) Extensive somatic L1 retrotransposition in colorectal tumors. Genome Res 22 : 2328–2338.
24. LeeE, IskowR, YangL, GokcumenO, HaseleyP, et al. (2012) Landscape of somatic retrotransposition in human cancers. Science 337 : 967–971.
25. IskowRC, McCabeMT, MillsRE, ToreneS, PittardWS, et al. (2010) Natural mutagenesis of human genomes by endogenous retrotransposons. Cell 141 : 1253–1261.
26. HarrisCR, NormartR, YangQ, StevensonE, HafftyBG, et al. (2010) Association of nuclear localization of a long interspersed nuclear element-1 protein in breast tumors with poor prognostic outcomes. Genes Cancer 1 : 115–124.
27. SuY, DaviesS, DavisM, LuH, GillerR, et al. (2007) Expression of LINE-1 p40 protein in pediatric malignant germ cell tumors and its association with clinicopathological parameters: a report from the Children's Oncology Group. Cancer Lett 247 : 204–212.
28. BelancioVP, Roy-EngelAM, PochampallyRR, DeiningerP (2010) Somatic expression of LINE-1 elements in human tissues. Nucleic Acids Research 38 : 3909–3922.
29. TingDT, LipsonD, PaulS, BranniganBW, AkhavanfardS, et al. (2011) Aberrant overexpression of satellite repeats in pancreatic and other epithelial cancers. Science 331 : 593–596.
30. BollatiV, FabrisS, PegoraroV, RonchettiD, MoscaL, et al. (2009) Differential repetitive DNA methylation in multiple myeloma molecular subgroups. Carcinogenesis 30 : 1330–1335.
31. Roman-GomezJ, Jimenez-VelascoA, AgirreX, CervantesF, SanchezJ, et al. (2005) Promoter hypomethylation of the LINE-1 retrotransposable elements activates sense/antisense transcription and marks the progression of chronic myeloid leukemia. Oncogene 24 : 7213–7223.
32. FabrisS, BollatiV, AgnelliL, MorabitoF, MottaV, et al. (2011) Biological and clinical relevance of quantitative global methylation of repetitive DNA sequences in chronic lymphocytic leukemia. Epigenetics 6 : 188–194.
33. BelancioVP, Roy-EngelAM, DeiningerPL (2010) All y'all need to know 'bout retroelements in cancer. Seminars in Cancer Biology 20 : 200–210.
34. WallaceNA, BelancioVP, DeiningerPL (2008) L1 mobile element expression causes multiple types of toxicity. Gene 419 : 75–81.
35. GasiorSL, WakemanTP, XuB, DeiningerPL (2006) The human LINE-1 retrotransposon creates DNA double-strand breaks. Journal of Molecular Biology 357 : 1383–1393.
36. SymerDE, ConnellyC, SzakST, CaputoEM, CostGJ, et al. (2002) Human l1 retrotransposition is associated with genetic instability in vivo. Cell 110 : 327–338.
37. LinC, YangL, TanasaB, HuttK, JuBG, et al. (2009) Nuclear receptor-induced chromosomal proximity and DNA breaks underlie specific translocations in cancer. Cell 139 : 1069–1083.
38. DaiL, HuangQ, BoekeJD (2011) Effect of reverse transcriptase inhibitors on LINE-1 and Ty1 reverse transcriptase activities and on LINE-1 retrotransposition. BMC Biochemistry 12 : 18.
39. JonesRB, GarrisonKE, WongJC, DuanEH, NixonDF, et al. (2008) Nucleoside analogue reverse transcriptase inhibitors differentially inhibit human LINE-1 retrotransposition. PLoS One 3: e1547 doi:10.1371/journal.pone.0001547..
40. CarliniF, RidolfiB, MolinariA, ParisiC, BozzutoG, et al. (2010) The reverse transcription inhibitor abacavir shows anticancer activity in prostate cancer cell lines. PloS One 5: e14221 doi:10.1371/journal.pone.0014221..
41. SciamannaI, LandriscinaM, PittoggiC, QuirinoM, MearelliC, et al. (2005) Inhibition of endogenous reverse transcriptase antagonizes human tumor growth. Oncogene 24 : 3923–3931.
42. MangiacasaleR, PittoggiC, SciamannaI, CaredduA, MatteiE, et al. (2003) Exposure of normal and transformed cells to nevirapine, a reverse transcriptase inhibitor, reduces cell growth and promotes differentiation. Oncogene 22 : 2750–2761.
43. FaulknerGJ, KimuraY, DaubCO, WaniS, PlessyC, et al. (2009) The regulated retrotransposon transcriptome of mammalian cells. Nature Genetics 41 : 563–571.
44. WolffEM, ByunHM, HanHF, SharmaS, NicholsPW, et al. (2010) Hypomethylation of a LINE-1 promoter activates an alternate transcript of the MET oncogene in bladders with cancer. PLoS Genet 6: e1000917 doi:10.1371/journal.pgen.1000917..
45. LinL, WangZ, PrescottMS, van DekkenH, ThomasDG, et al. (2006) Multiple forms of genetic instability within a 2-Mb chromosomal segment of 3q26.3-q27 are associated with development of esophageal adenocarcinoma. Genes, Chromosomes & Cancer 45 : 319–331.
46. CruickshanksHA, TufarelliC (2009) Isolation of cancer-specific chimeric transcripts induced by hypomethylation of the LINE-1 antisense promoter. Genomics 94 : 397–406.
47. WeberB, KimhiS, HowardG, EdenA, LykoF (2010) Demethylation of a LINE-1 antisense promoter in the cMet locus impairs Met signalling through induction of illegitimate transcription. Oncogene 29 : 5775–5784.
48. EstecioMR, GallegosJ, VallotC, CastoroRJ, ChungW, et al. (2010) Genome architecture marked by retrotransposons modulates predisposition to DNA methylation in cancer. Genome Research 20 : 1369–1382.
Štítky
Genetika Reprodukční medicína
Článek Ubiquitous Polygenicity of Human Complex Traits: Genome-Wide Analysis of 49 Traits in KoreansČlánek Alternative Splicing and Subfunctionalization Generates Functional Diversity in Fungal ProteomesČlánek RFX Transcription Factor DAF-19 Regulates 5-HT and Innate Immune Responses to Pathogenic Bacteria inČlánek Surveillance-Activated Defenses Block the ROS–Induced Mitochondrial Unfolded Protein ResponseČlánek Deficiency Reduces Adipose OXPHOS Capacity and Triggers Inflammation and Insulin Resistance in Mice
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 3
-
Všechny články tohoto čísla
- Power and Predictive Accuracy of Polygenic Risk Scores
- Rare Copy Number Variants Are a Common Cause of Short Stature
- Coordination of Flower Maturation by a Regulatory Circuit of Three MicroRNAs
- Ubiquitous Polygenicity of Human Complex Traits: Genome-Wide Analysis of 49 Traits in Koreans
- Genomic Evidence for Island Population Conversion Resolves Conflicting Theories of Polar Bear Evolution
- Mechanistic Insight into the Pathology of Polyalanine Expansion Disorders Revealed by a Mouse Model for X Linked Hypopituitarism
- Genome-Wide Association Study and Gene Expression Analysis Identifies as a Predictor of Response to Etanercept Therapy in Rheumatoid Arthritis
- Problem Solved: An Interview with Sir Edwin Southern
- Long Interspersed Element–1 (LINE-1): Passenger or Driver in Human Neoplasms?
- Mouse HFM1/Mer3 Is Required for Crossover Formation and Complete Synapsis of Homologous Chromosomes during Meiosis
- Alternative Splicing and Subfunctionalization Generates Functional Diversity in Fungal Proteomes
- A WRKY Transcription Factor Recruits the SYG1-Like Protein SHB1 to Activate Gene Expression and Seed Cavity Enlargement
- Microhomology-Mediated Mechanisms Underlie Non-Recurrent Disease-Causing Microdeletions of the Gene or Its Regulatory Domain
- Ancient Evolutionary Trade-Offs between Yeast Ploidy States
- Differential Evolutionary Fate of an Ancestral Primate Endogenous Retrovirus Envelope Gene, the EnvV , Captured for a Function in Placentation
- A Feed-Forward Loop Coupling Extracellular BMP Transport and Morphogenesis in Wing
- The Tomato Yellow Leaf Curl Virus Resistance Genes and Are Allelic and Code for DFDGD-Class RNA–Dependent RNA Polymerases
- The U-Box E3 Ubiquitin Ligase TUD1 Functions with a Heterotrimeric G α Subunit to Regulate Brassinosteroid-Mediated Growth in Rice
- Role of the DSC1 Channel in Regulating Neuronal Excitability in : Extending Nervous System Stability under Stress
- –Independent Phenotypic Switching in and a Dual Role for Wor1 in Regulating Switching and Filamentation
- Pax6 Regulates Gene Expression in the Vertebrate Lens through miR-204
- Blood-Informative Transcripts Define Nine Common Axes of Peripheral Blood Gene Expression
- Genetic Architecture of Skin and Eye Color in an African-European Admixed Population
- Fine Characterisation of a Recombination Hotspot at the Locus and Resolution of the Paradoxical Excess of Duplications over Deletions in the General Population
- Estrogen Mediated-Activation of miR-191/425 Cluster Modulates Tumorigenicity of Breast Cancer Cells Depending on Estrogen Receptor Status
- Complex Patterns of Genomic Admixture within Southern Africa
- Yap- and Cdc42-Dependent Nephrogenesis and Morphogenesis during Mouse Kidney Development
- Molecular Networks of Human Muscle Adaptation to Exercise and Age
- Alp/Enigma Family Proteins Cooperate in Z-Disc Formation and Myofibril Assembly
- Polycomb Group Gene Regulates Rice () Seed Development and Grain Filling via a Mechanism Distinct from
- RFX Transcription Factor DAF-19 Regulates 5-HT and Innate Immune Responses to Pathogenic Bacteria in
- Distinct Molecular Strategies for Hox-Mediated Limb Suppression in : From Cooperativity to Dispensability/Antagonism in TALE Partnership
- A Natural Polymorphism in rDNA Replication Origins Links Origin Activation with Calorie Restriction and Lifespan
- TDP2–Dependent Non-Homologous End-Joining Protects against Topoisomerase II–Induced DNA Breaks and Genome Instability in Cells and
- Recurrent Rearrangement during Adaptive Evolution in an Interspecific Yeast Hybrid Suggests a Model for Rapid Introgression
- Genome-Wide Association Study in Mutation Carriers Identifies Novel Loci Associated with Breast and Ovarian Cancer Risk
- Coincident Resection at Both Ends of Random, γ–Induced Double-Strand Breaks Requires MRX (MRN), Sae2 (Ctp1), and Mre11-Nuclease
- Identification of a -Specific Modifier Locus at 6p24 Related to Breast Cancer Risk
- A Novel Function for the Hox Gene in the Male Accessory Gland Regulates the Long-Term Female Post-Mating Response in
- Tdp2: A Means to Fixing the Ends
- A Novel Role for the RNA–Binding Protein FXR1P in Myoblasts Cell-Cycle Progression by Modulating mRNA Stability
- Association Mapping and the Genomic Consequences of Selection in Sunflower
- Histone Deacetylase 2 (HDAC2) Regulates Chromosome Segregation and Kinetochore Function via H4K16 Deacetylation during Oocyte Maturation in Mouse
- A Novel Mutation in the Upstream Open Reading Frame of the Gene Causes a MEN4 Phenotype
- Ataxin1L Is a Regulator of HSC Function Highlighting the Utility of Cross-Tissue Comparisons for Gene Discovery
- Human Spermatogenic Failure Purges Deleterious Mutation Load from the Autosomes and Both Sex Chromosomes, including the Gene
- A Conserved Upstream Motif Orchestrates Autonomous, Germline-Enriched Expression of piRNAs
- Statistical Analysis Reveals Co-Expression Patterns of Many Pairs of Genes in Yeast Are Jointly Regulated by Interacting Loci
- Matefin/SUN-1 Phosphorylation Is Part of a Surveillance Mechanism to Coordinate Chromosome Synapsis and Recombination with Meiotic Progression and Chromosome Movement
- A Role for the Malignant Brain Tumour (MBT) Domain Protein LIN-61 in DNA Double-Strand Break Repair by Homologous Recombination
- The Population and Evolutionary Dynamics of Phage and Bacteria with CRISPR–Mediated Immunity
- Long Noncoding RNA MALAT1 Controls Cell Cycle Progression by Regulating the Expression of Oncogenic Transcription Factor B-MYB
- Surveillance-Activated Defenses Block the ROS–Induced Mitochondrial Unfolded Protein Response
- DNA Topoisomerase III Localizes to Centromeres and Affects Centromeric CENP-A Levels in Fission Yeast
- Genome-Wide Control of RNA Polymerase II Activity by Cohesin
- Divergent Selection Drives Genetic Differentiation in an R2R3-MYB Transcription Factor That Contributes to Incipient Speciation in
- NODULE INCEPTION Directly Targets Subunit Genes to Regulate Essential Processes of Root Nodule Development in
- Spreading of a Prion Domain from Cell-to-Cell by Vesicular Transport in
- Deficiency in Origin Licensing Proteins Impairs Cilia Formation: Implications for the Aetiology of Meier-Gorlin Syndrome
- Deficiency Reduces Adipose OXPHOS Capacity and Triggers Inflammation and Insulin Resistance in Mice
- The Conserved SKN-1/Nrf2 Stress Response Pathway Regulates Synaptic Function in
- Functional Genomic Analysis of the Regulatory Network in
- Astakine 2—the Dark Knight Linking Melatonin to Circadian Regulation in Crustaceans
- CRL2 E3-Ligase Regulates Proliferation and Progression through Meiosis in the Germline
- Both the Caspase CSP-1 and a Caspase-Independent Pathway Promote Programmed Cell Death in Parallel to the Canonical Pathway for Apoptosis in
- PRMT4 Is a Novel Coactivator of c-Myb-Dependent Transcription in Haematopoietic Cell Lines
- A Copy Number Variant at the Locus Likely Confers Risk for Canine Squamous Cell Carcinoma of the Digit
- Evidence of Gene–Environment Interactions between Common Breast Cancer Susceptibility Loci and Established Environmental Risk Factors
- HIV Infection Disrupts the Sympatric Host–Pathogen Relationship in Human Tuberculosis
- Trans-Ethnic Fine-Mapping of Lipid Loci Identifies Population-Specific Signals and Allelic Heterogeneity That Increases the Trait Variance Explained
- A Gene Transfer Agent and a Dynamic Repertoire of Secretion Systems Hold the Keys to the Explosive Radiation of the Emerging Pathogen
- The Role of ATM in the Deficiency in Nonhomologous End-Joining near Telomeres in a Human Cancer Cell Line
- Dynamic Circadian Protein–Protein Interaction Networks Predict Temporal Organization of Cellular Functions
- Nuclear Myosin 1c Facilitates the Chromatin Modifications Required to Activate rRNA Gene Transcription and Cell Cycle Progression
- Robust Prediction of Expression Differences among Human Individuals Using Only Genotype Information
- A Single Cohesin Complex Performs Mitotic and Meiotic Functions in the Protist
- The Role of the Arabidopsis Exosome in siRNA–Independent Silencing of Heterochromatic Loci
- Elevated Expression of the Integrin-Associated Protein PINCH Suppresses the Defects of Muscle Hypercontraction Mutants
- Twist1 Controls a Cell-Specification Switch Governing Cell Fate Decisions within the Cardiac Neural Crest
- Genome-Wide Testing of Putative Functional Exonic Variants in Relationship with Breast and Prostate Cancer Risk in a Multiethnic Population
- Heteroduplex DNA Position Defines the Roles of the Sgs1, Srs2, and Mph1 Helicases in Promoting Distinct Recombination Outcomes
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Fine Characterisation of a Recombination Hotspot at the Locus and Resolution of the Paradoxical Excess of Duplications over Deletions in the General Population
- Molecular Networks of Human Muscle Adaptation to Exercise and Age
- Recurrent Rearrangement during Adaptive Evolution in an Interspecific Yeast Hybrid Suggests a Model for Rapid Introgression
- Genome-Wide Association Study and Gene Expression Analysis Identifies as a Predictor of Response to Etanercept Therapy in Rheumatoid Arthritis
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



