-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaAstakine 2—the Dark Knight Linking Melatonin to Circadian Regulation in Crustaceans
Daily, circadian rhythms influence essentially all living organisms and affect many physiological processes from sleep and nutrition to immunity. This ability to respond to environmental daily rhythms has been conserved along evolution, and it is found among species from bacteria to mammals. The hematopoietic process of the crayfish Pacifastacus leniusculus is under circadian control and is tightly regulated by astakines, a new family of cytokines sharing a prokineticin (PROK) domain. The expression of AST1 and AST2 are light-dependent, and this suggests an evolutionarily conserved function for PROK domain proteins in mediating circadian rhythms. Vertebrate PROKs are transmitters of circadian rhythms of the suprachiasmatic nucleus (SCN) in the brain of mammals, but the mechanism by which they function is unknown. Here we demonstrate that high AST2 expression is induced by melatonin in the brain. We identify RACK1 as a binding protein of AST2 and further provide evidence that a complex between AST2 and RACK1 functions as a negative-feedback regulator of the circadian clock. By DNA mobility shift assay, we showed that the AST2-RACK1 complex will interfere with the binding between BMAL1 and CLK and inhibit the E-box binding activity of the complex BMAL1-CLK. Finally, we demonstrate by gene knockdown that AST2 is necessary for melatonin-induced inhibition of the complex formation between BMAL1 and CLK during the dark period. In summary, we provide evidence that melatonin regulates AST2 expression and thereby affects the core clock of the crustacean brain. This process may be very important in all animals that have AST2 molecules, i.e. spiders, ticks, crustaceans, scorpions, several insect groups such as Hymenoptera, Hemiptera, and Blattodea, but not Diptera and Coleoptera. Our findings further reveal an ancient evolutionary role for the prokineticin superfamily protein that links melatonin to direct regulation of the core clock gene feedback loops.
Published in the journal: . PLoS Genet 9(3): e32767. doi:10.1371/journal.pgen.1003361
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003361Summary
Daily, circadian rhythms influence essentially all living organisms and affect many physiological processes from sleep and nutrition to immunity. This ability to respond to environmental daily rhythms has been conserved along evolution, and it is found among species from bacteria to mammals. The hematopoietic process of the crayfish Pacifastacus leniusculus is under circadian control and is tightly regulated by astakines, a new family of cytokines sharing a prokineticin (PROK) domain. The expression of AST1 and AST2 are light-dependent, and this suggests an evolutionarily conserved function for PROK domain proteins in mediating circadian rhythms. Vertebrate PROKs are transmitters of circadian rhythms of the suprachiasmatic nucleus (SCN) in the brain of mammals, but the mechanism by which they function is unknown. Here we demonstrate that high AST2 expression is induced by melatonin in the brain. We identify RACK1 as a binding protein of AST2 and further provide evidence that a complex between AST2 and RACK1 functions as a negative-feedback regulator of the circadian clock. By DNA mobility shift assay, we showed that the AST2-RACK1 complex will interfere with the binding between BMAL1 and CLK and inhibit the E-box binding activity of the complex BMAL1-CLK. Finally, we demonstrate by gene knockdown that AST2 is necessary for melatonin-induced inhibition of the complex formation between BMAL1 and CLK during the dark period. In summary, we provide evidence that melatonin regulates AST2 expression and thereby affects the core clock of the crustacean brain. This process may be very important in all animals that have AST2 molecules, i.e. spiders, ticks, crustaceans, scorpions, several insect groups such as Hymenoptera, Hemiptera, and Blattodea, but not Diptera and Coleoptera. Our findings further reveal an ancient evolutionary role for the prokineticin superfamily protein that links melatonin to direct regulation of the core clock gene feedback loops.
Introduction
The physiology and behavior of most organisms are regulated according to daily environmental changes in a circadian manner. Circadian rhythms are often monitored by assaying behavioral and/or molecular fluctuations [1]. Clock genes and hormones are core regulators or pacemakers in circadian regulation, which reside in the suprachiasmatic nucleus (SCN) in mammals and the brain visual center in insects [2], [3]. In Drosophila, the proteins encoded by clock genes form two distinct heterodimers: Period (PER) and Timeless (TIM) form one heterodimer, and Clock (CLK) and Cycle (CYC) form the other [4]. In mammals, the homologues of the CLK and PER proteins have the same name, while the homologues of TIM and CYC are called cryptochrome (CRY) and brain and muscle aryl hydrocarbon receptor nuclear translocator (ARNT)-like 1 (BMAL1) respectively [5]. The PER-TIM heterodimer functions as an autoregulatory negative feedback loop that causes a decrease in transcriptional activation by CLK and CYC [6]. Conversely, the CLK-CYC heterodimer directly interacts with the upstream E-boxes (CACGTG) of the PER and TIM genes to activate their transcription [7]. These clock genes are detected in several neural and non-neural tissues, suggesting that these feedback loops are not restricted to neurons [8]. RACK1 was recently identified as an inhibitor of mammalian CLK-BMAL1 activity by recruiting PKCα during the negative feedback phase of the cycle [9].
As described above, the SCN serves as the “master clock” in the mammalian brain [10]. The SCN is reset on a circadian basis by light input from the retina during the day and by melatonin secretion from the pineal gland at night [11], [12]. Several animal studies have documented that melatonin is mainly synthesized and released during darkness, while the melatonin level is low in the presence of light [13]. This hormone is formed not only in the pineal gland but also in the photoreceptive structures of both vertebrates and invertebrates [14]. The melatonin synthesis pattern is known to reflect both daily rhythms and changes in photoperiod, demonstrating that melatonin has major roles in regulating both circadian and seasonal rhythms [15]. Several circadian rhythms, such as the rest/activity cycle, core body temperature, neuronal electrical activity and locomotor activity, are driven by melatonin [16], [17], [18], [19]. However, melatonin does not directly induce immediate changes in clock gene mRNA expression in the rat SCN, suggesting that the phase-shifting effect of melatonin on the SCN molecular loop implicates a post-translational rather than a transcriptional effect [15]. Little is known about the molecular mechanism by which melatonin drives the clock gene feedback loops in the SCN. In addition, the nocturnal increase in the circadian secretion of the cytokine IL-2 and its immunobiological activity are under melatonin control during the dark period [20]. There is a link between the rest/activity cycle and the immune system via melatonin-cytokine interactions [21].
Neuronal tissues that contain circadian clocks are present in the brain of the crayfish Cherax destructor and Procambarus clarkii, the latter of which is highly related to Pacifastacus leniusculus. These species have photoreceptor cells not only within their eyes, but also in extraretinal parts of the brain. In Cherax this area is located in the anterior median brain soma cluster 6 [22], and the axons from these cells terminate in an area within the protocerebral bridge [23]. It is clear that these brain photoreceptors are sufficient to maintain the circadian locomotor rhythm in these animals [23]. These areas have also recently been shown to be immunoreactive for proteins encoded by the clock genes CLK, PER and TIM [24]. Several studies imply a conserved role of melatonin as an important transducer of circadian information in invertebrates and vertebrates, and melatonin production has been demonstrated in the eyestalks of several crustacean species [25], [26]. The presence of MT2 melatonin receptors have not been conclusively shown in crayfish, but indirect evidences for such a receptor were presented by Mendoza-Vargas et al [27] since the application of the MT2 receptor selective agonist 8-M-PDOT or antagonist DH97 had a significant effect on how melatonin affected the retinal photoreceptors [27].
In mammals, the secretion of prokineticin 2 (PROK2) by the SCN is implicated in the regulation of the sleep/wake cycle and locomotor activity and is known to suppress the function of melatonin [28]. The transcription of PROK2 is activated by the CLK-BMAL1 heterodimer and light [29], [30].
Crayfish astakines are invertebrate homologues of the prokineticins (PROK), and there are two forms in the crayfish P. leniusculus: astakine 1 (AST1, AAX14635) and astakine 2 (AST2, EF568370) [31]. These astakines have been identified in several invertebrates, and similar to PROK2, the expression profiles of crayfish AST1 and AST2 follow a daily rhythm [32]. In contrast to AST1, the rhythmic expression of AST2 is at its maximum level in the dark phase [32]. Although the patterns of melatonin and AST2 release are circadian-regulated in crayfish [32], [33], nothing is known about their role in regulating rhythmic oscillations. In this study, we provide evidence that melatonin regulates the circadian rhythm and that this regulation is mediated by AST2 during the dark phase. Our results imply that AST2 acts as novel negative feedback regulator of CLK-BMAL1 activity.
Results
Melatonin induces the expression of AST2
In a recent report, we showed that AST2 mRNA is expressed in the hematopoietic tissue (HPT) of P. leniusculus at higher levels at night than during the day, the time when melatonin is also at a high level [32], [33]. Thus, we hypothesized that there may be a physiological link between these two molecules. We examined the expression of AST1 and AST2 in HPT cells incubated with melatonin in vitro (Figure 1A–1B) and in HPT, hemocytes and brain injected with melatonin in vivo (Figure 1C–1H). Melatonin incubation or injection clearly affected AST2 expression (Figure 1B, 1D, 1F and 1H), whereas AST1 expression was unaffected by this treatment (Figure 1A, 1C, 1E and 1G). These results were also confirmed at the level of translation using an ELISA assay (Figure 2A–2C), in which melatonin treatment resulted in higher levels of the AST2 protein. When cell lysates from the brain and HPT were analyzed by western blot at 9 : 00 and 20 : 00, respectively, the level of AST2 was clearly higher at night than during the day (Figure 2D).
Fig. 1. AST2 expression is induced by melatonin treatment in vitro and in vivo. 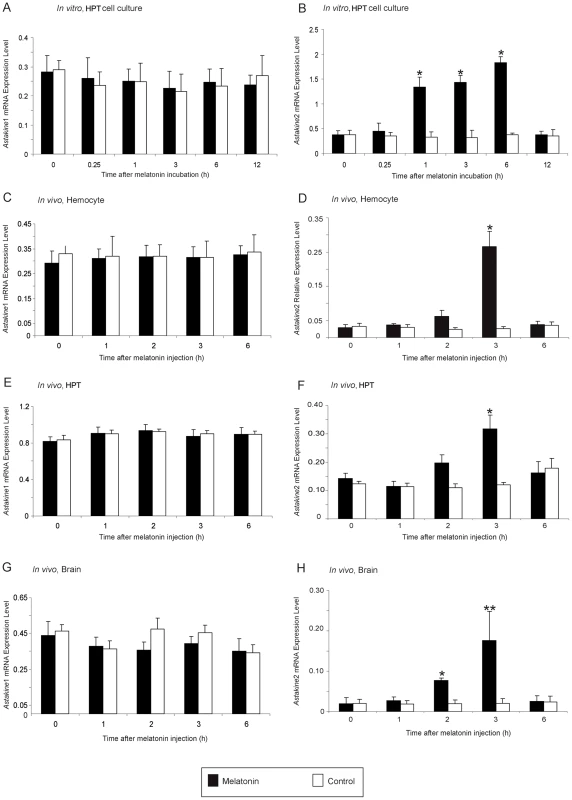
A) Relative expression of AST1 mRNA estimated by qPCR, after incubation with melatonin in cultured HPT cells in vitro at daytime. Black bars = melatonin (3 µM), white bars = control. B) Relative expression of AST2 mRNA estimated by qPCR, after incubation with melatonin in cultured HPT cells in vitro at daytime. Black bars = melatonin (3 µM), white bars = control. C) Relative expression of AST1 mRNA in hemocytes estimated by qPCR, after injection of melatonin in live crayfish. Black bars = melatonin (4.3 nmol/g), white bars = control. D) Relative expression of AST2 mRNA in hemocytes estimated by qPCR, after injection of melatonin in live crayfish at daytime. Black bars = melatonin (4.3 nmol/g), white bars = control. E) Relative expression of AST1 mRNA in HPT estimated by qPCR, after injection of melatonin in live crayfish at daytime. Black bars = melatonin (4.3 nmol/g), white bars = control. F) Relative expression of AST2 mRNA in HPT estimated by qPCR, after injection of melatonin in live crayfish at daytime. Black bars = melatonin (4.3 nmol/g), white bars = control. G) Relative expression of AST1 mRNA in the brain estimated by qPCR, after injection of melatonin in live crayfish at daytime. Black bars = melatonin (4.3 nmol/g), white bars = control. H) Relative expression of AST2 mRNA in the brain estimated by qPCR, after injection of melatonin in live crayfish at daytime. Black bars = melatonin (4.3 nmol/g), white bars = control. Expression of the 40S ribosomal protein was used as an internal control. Error bars indicate standard deviation (SD) from three replicates and the experiment has been repeated three times with similar results. The asterisks indicate significant differences (*P<0.05, **P<0.01); one-way ANOVA with Duncan's new multiple-range test and the Tukey test. Fig. 2. Melatonin induces higher AST2 protein levels in vitro and in vivo. 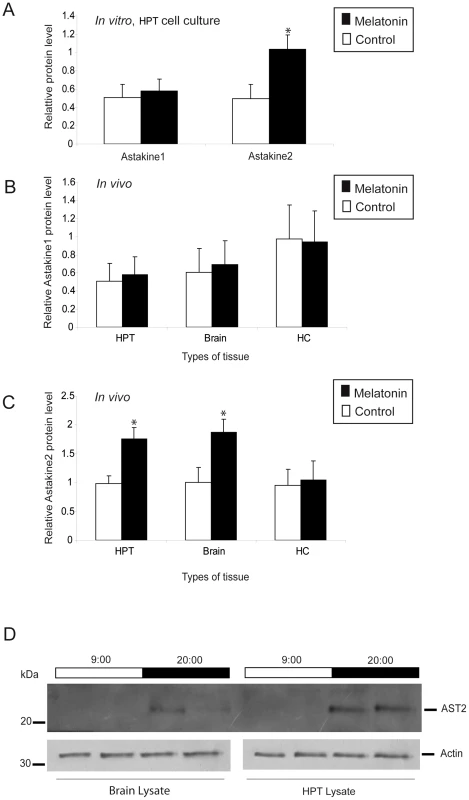
A) Relative levels of AST1 or AST2 protein in cultured HPT cells in vitro as estimated by ELISA, after incubation with melatonin. Black bars = melatonin (3 µM), white bars = control. The level of β-actin was used as an internal control. B) Relative levels of AST1 in the HPT, brain and hemocytes in live crayfish as estimated by ELISA, after injection of melatonin. Black bars = melatonin (4.3 nmol/g), white bars = control. The level of β-actin was used as an internal control. C) Relative levels of AST2 in the HPT, brain and hemocytes in live crayfish as estimated by ELISA, after injection of melatonin. Black bars = melatonin (4.3 nmol/g), white bars = control. The level of β-actin was used as an internal control. The asterisks indicate significant differences (*P<0.05); one-way ANOVA with Duncan's new multiple-range test and the Tukey test. Results are representative of three independent experiments. Error bars indicate SD from three replicates and the experiment has been repeated three times with similar results. D) Western blot analysis of AST2 in the brain and HPT at 9:00 and 20:00, using an antibody against AST2. The expression level of actin was used as an internal control. Light and dark periods are indicated on the top of the blot. AST2 binds to PlRACK1
In contrast to AST1, which is a secreted protein, AST2 is mainly an intracellular protein (Figure S1). To identify any protein that interacts with AST2 in HPT cells, recombinant AST2 (GST-AST2) was used as a bait to bind to proteins in HPT and hemocyte lysates by far overlay or pull-down assays (Figure 3A–3B). After excluding the background proteins found in the control, a protein with high similarity to Penaeus monodon RACK1 was identified as an AST2-binding protein by mass spectra analysis. The Mascot search resulted in significant hits where P. monodon RACK1 had the highest significant protein score 781 (Table S1). This protein was the significant hit detected in both the far overlay and the GST pull-down assay.
Fig. 3. AST2 interacts with PlRACK1. 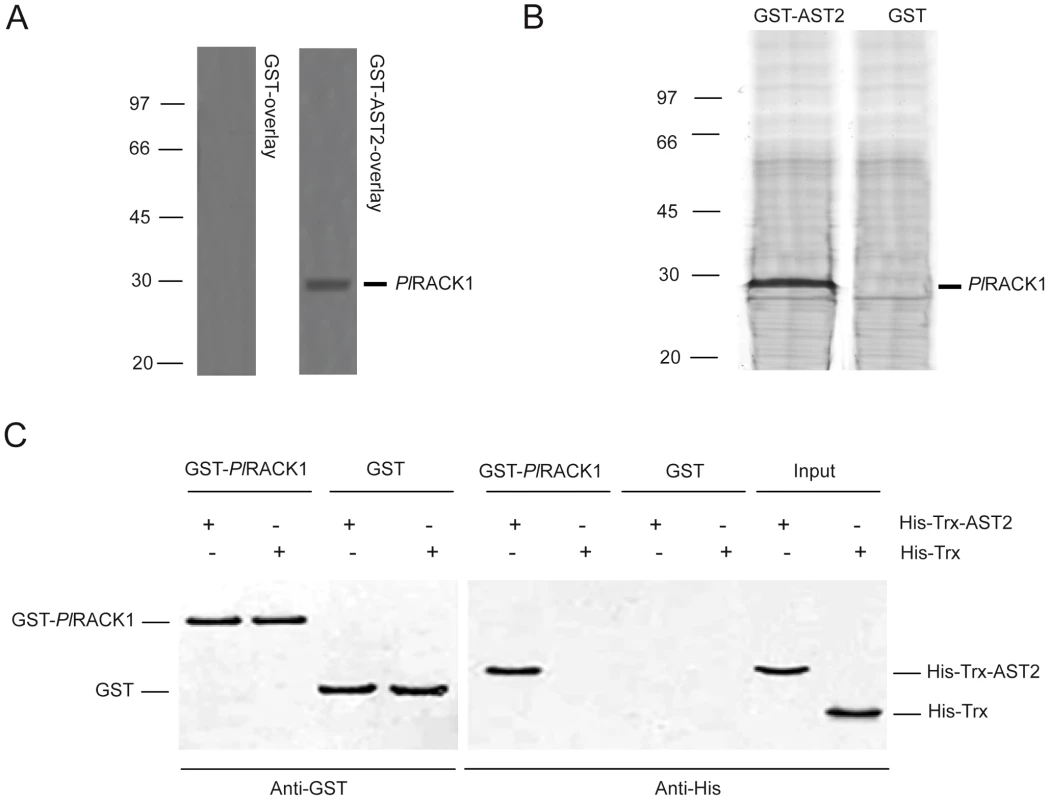
A) PlRACK1 was identified as an AST2 binding protein using a far western assay with recombinant GST-AST2 or GST as a control. A HPT protein extract was subjected to SDS-PAGE, electroblotted to a PVDF membrane and overlayed with either GST (left) or GST-AST2 (right) alone and binding was detected using a GST antibody. B) The binding between recombinant AST2 with PlRACK1 was confirmed by a GST pull-down assay using GST-AST2 as the binding protein and proteins in a HPT cell lysate as bait. GST was used as a control in both assays. C) GST pull-down of His-Trx-AST2 by GST-PlRACK1. The bound proteins were analyzed by 12.5% SDS-PAGE. Bands corresponding to GST and GST-PlRACK1 were detected with an anti-GST antibody (lanes 1–4). The eluted material was also examined for the presence of AST2 with an anti-His antibody (lanes 5–8). Lanes 9 and 10 contain purified His-Trx-AST2 and HisTrx, respectively. By designing degenerate primers from the obtained amino acid sequences, a complete RACK1 cDNA was identified by RACE technique from P. leniusculus (PlRACK1). The open reading frame of PlRACK1 contains 957 bp (JQ686053), encoding 318 amino acids with a calculated molecular mass of 35.7 kDa (predicted MW using ProtParam program), and its deduced amino acid sequence was found to consist of seven conserved WD40 repeat binding sites (Figure S2). PlRACK1 mRNA was highly expressed in different crayfish tissues (Figure S3). To confirm the interaction between PlRACK1 and AST2, recombinant GST-PlRACK1 was produced and purified with a predicted molecular mass of 62 kDa (Figure S5A). The GST-PlRACK1 was used in an in vitro binding assay with a His-Trx-AST2 fusion protein (predicted mass of 33 kDa). The His-Trx and GST proteins were also expressed and purified to serve as control proteins (Figure S5B: the predicted mass of His-Trx and GST proteins are 20 and 26 kDa, respectively). As shown in Figure 3C, western blot analysis revealed that the His-Trx-AST2 protein was pulled down by GST - PlRACK1, but not by GST alone. These data indicate that there is an interaction between PlRACK1 and AST2.
PlBMAL1 forms a complex with PlRACK1
The finding that PlRACK1 is a binding partner of the melatonin-induced protein AST2 led us to examine whether PlRACK1 associates with BMAL1 to thereby act as a regulator of circadian rhythm, as has recently been reported in mice [9]. BMAL1 cDNA was amplified from P. leniusculus (PlBMAL1, JQ670886) with an open reading frame of 1,995 bp that encodes a 664 amino acid protein with a predicted molecular mass of 73.64 kDa. One close sequence to PlBMAL1 was that of BMAL1 from Drosophila melanogaster (identity = 55%). Domain homology analysis using SMART revealed that the deduced amino acid sequence was highly conserved in its HLH (helix-loop-helix) DNA-binding domain and the protein dimerization PAS and PAC regions (Figure S4A). The expression of PlBMAL1 was highest in hemocytes, followed by testis, nerve, intestine and brain (Figure S4B). An interaction between PlBMAL1 and PlRACK1 was demonstrated using a GST pull-down experiment with recombinant GST-PlRACK1 (predicted mass of 62 kDa) and His-Trx-PlBMAL1 (predicted mass of 94 kDa as shown in Figure S6A–S6B). This in vitro GST pull-down assay showed an apparent binding between these two recombinant proteins (Figure 4A). The far western blotting using PlRACK1 without GST tag (predicted mass of 36 kDa) as a prey and GST - PlBMAL1 (predicted mass of 100 kDa as shown in Figure S6C) as bait, also confirmed this interaction (Figure 4B). The results show that the GST-PlBMAL1 was detected at the place where the PlRACK1 was located, whereas the GST control protein could not be detected. Binding of PlRACK1 to AST2 as well as to PlBMAL1 was detected using far western after SDS-PAGE at reducing condition. The crystal structure of human RACK1, with high homology to PlRACK1, is known as a sevenfold ß-propeller structure [34]. A comparison of the structure of human RACK1 with yeast RACK1 (that lacks most of the cysteines present in animal RACK1's) show a high similarity in the overall structure, indicating that the cysteines are of minor importance for the main structure characteristics of this group of proteins, which may explain retained binding properties in the far western assay.
Fig. 4. PlRACK1 can bind to PlBMAL1. 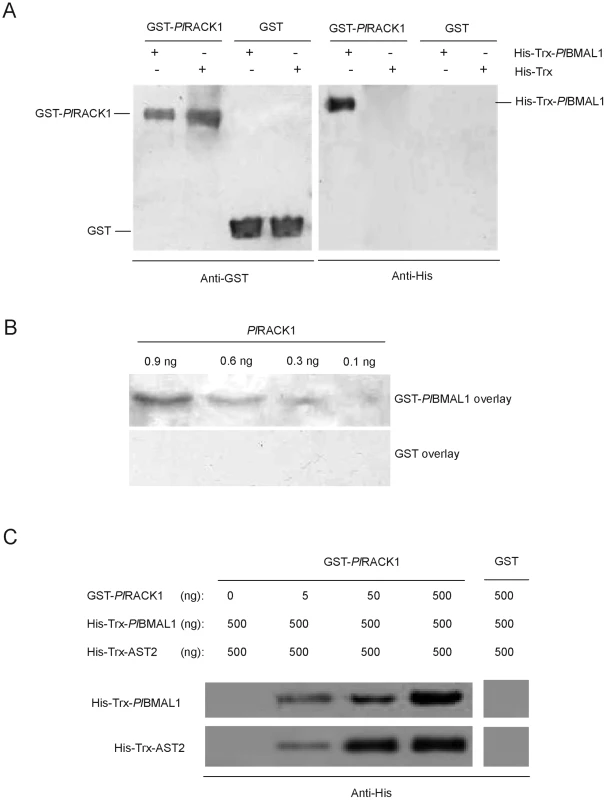
A) In vitro GST pull-down assay of His-Trx-PlBMAL1 by GST-PlRACK1. The elution fractions of the GST-PlRACK1 pull-down assay were examined by western blot analysis using anti-GST and anti-His antibodies. Lanes 1 and 5: elution fraction of GST-PlRACK1 pull-down of HisTrx-PlBMAL1; Lanes 2 and 6: GST-PlRACK1 pull-down of His-Trx; Lanes 3 and 7: GST pull-down of HisTrx-PlBMAL1; Lanes 4 and 8; GST pull-down of His-Trx. B) The protein-protein interaction of PlRACK1 and PlBMAL1 was analyzed by far western blotting. C) Binding of AST2 and PlBMAL1 to GST-PlRACK1. GST-PlRACK1 (0, 5, 50 or 500 ng) was bound to Glutathione Sepharose beads and incubated with 500 ng His-Trx-PlBMAL1 and 500 ng His-Trx-AST2. Bound proteins were eluted and immunoblotted for PlBMAL1 and AST2 using an anti-His antibody and for PlRACK1 with an anti-GST antibody. Interaction of AST2 with PlRACK1-PlBMAL1 in vitro
To investigate if and how AST2 regulates an oscillatory mechanism, we examined whether AST2 interacts with the PlBMAL1-PlRACK1 complex. This interaction is mediated by AST2 binding to PlRACK1, since we could show that AST2 is not able to bind to BMAL1 alone (Figure S7). The results from GST pull-down experiments showed that recombinant His-Trx-AST2 strongly associated with the recombinant GST-PlRACK1/His-Trx-PlBMAL1 complex, and the binding of His-Trx-PlBMAL1 and His-Trx-AST2 was dependent on the concentration of GST-PlRACK1 (Figure 4C). These results suggest that AST2 plays an essential role as a stabilizer of the PlBMAL1-PlRACK1 complex, thereby suppressing the CLK-BMAL1 interaction during the dark period.
AST2 associates to form an AST2-PlRACK1-PlBMAL1 complex during the dark period in vivo
The amount of AST2 protein varies in a circadian manner and is highest during early night in the brain and HPT, while PlBMAL1 and PlRACK1 protein levels do not change significantly according to daily rhythms (Figure 5A–5B). Because our in vitro experiments indicated an interaction between AST2 and PlRACK1 and between PlRACK1 and PlBMAL1, we confirmed these two interactions with in vivo experiments. To study the in vivo interactions between these three proteins during the light and dark periods, we performed immunoprecipitation experiments of brain and HPT extracts. A high-molecular-weight complex was present at night and disappeared during the day, as shown in Figure 5C and 5D. This complex was abundant in the brain; it was also found at very low levels in a few HPT samples (Figure 5C). Reducing SDS-PAGE revealed that AST2, PlBMAL1 and PlRACK1 were components of this approximately 200 kDa complex (Figure 5C) (the native mass of these three proteins are 14, 74 and 36 kDa, respectively). During the light period, this high molecular weight complex could not be detected; instead, a smaller complex of AST2-PlRACK1 was identified (Figure 5D).
Fig. 5. PlRACK1, PlBMAL1, and AST2 form an approximately 200-kDa protein complex during the dark period. 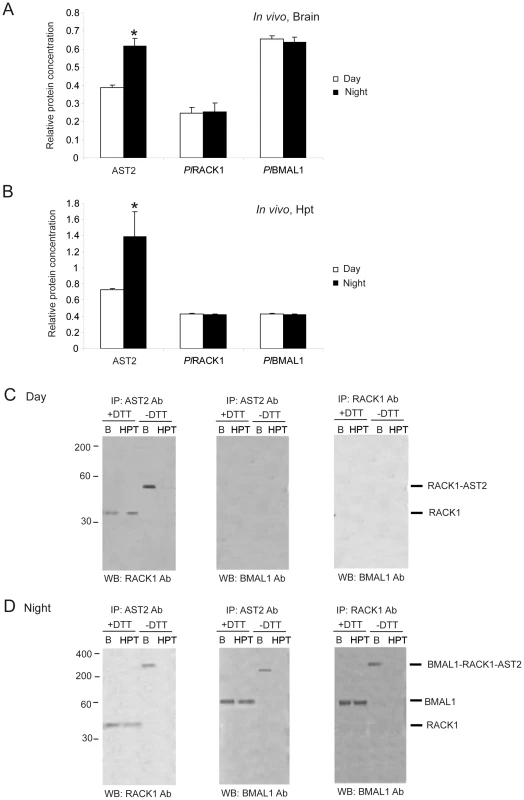
A) Relative amounts of AST2, PlRACK1 and PlBMAL1 proteins in brain extracts as determined by ELISA at 3 h after light turned off or on. B) Relative amounts of AST2, PlRACK1 and PlBMAL1 proteins in HPT extracts as determined by ELISA at 3 h after light turned off or on. The asterisks indicate significant differences (*P<0.05); one-way ANOVA with Duncan's new multiple-range test and the Tukey test. Results are representative of three independent experiments. Error bars indicate SD from three replicates and the experiment has been repeated three times with similar results. C) Immunoprecipitation (IP) of brain extract using antibodies against RACK1, BMAL1 and AST2 revealed that a PlRACK1-AST2 complex was present in the brain (B) during the day. D) Immunoprecipitation (IP) of brain extract using antibodies against RACK1, BMAL1 and AST2 revealed the presence of a high-molecular-weight complex (approximately 200 kDa) composed of all three proteins in the brain (B) at night. C–D) The immunoprecipitated complex was analyzed by western blotting (WB) using another antibody. “B” and “HPT” represent brain and hematopoietic tissue, respectively. The antibodies used for immunoprecipitation (IP) and detection (WB) is indicated at the top and bottom of the blots, respectively, and +DTT and −DTT represent reducing and non-reducing conditions, respectively. Molecular masses are indicated at the left. The CLK protein is detected as a component of the 400-kDa complex
To determine whether CLK (JQ670885) forms a complex with BMAL1, crayfish brain lysates were prepared, immunoprecipitated with antibodies against BMAL1 and probed for CLK. The CLK protein with a molecular mass of 95 kDa clearly coimmunoprecipitated with endogenous PlBMAL1 (Figure 6A), indicating that CLK is one protein component of this large approximately 400 kDa complex. Confirmation of complex formation was accomplished by switching the antibodies used for immunoprecipitation and for western blot detection, respectively (Figure 6A). We then monitored the CLK protein levels in the brain during the day and night by western blotting. As shown in Figure 6B, the CLK protein level oscillated in a circadian rhythm, and its expression peaked in the light period (Figure 6F). Furthermore, the approximately 400 kDa CLK-PlBMAL1 complex was barely detectable at night and reached maximal levels in the morning (Figure 6B and 6C). More interestingly, the lowest level of the CLK-PlBMAL1 complex in brain lysates coincided with the time at which the AST2-PlRACK1-PlBMAL1 complex was at its highest level (Figure 6B and 6D). Thus, these data indicate that AST2 is an important regulator of circadian rhythm by interfering with formation of the CLK-PlBMAL1 complex.
Fig. 6. A 400-kDa CLK-PlBMAL1 complex is present in the light in the crayfish brain. 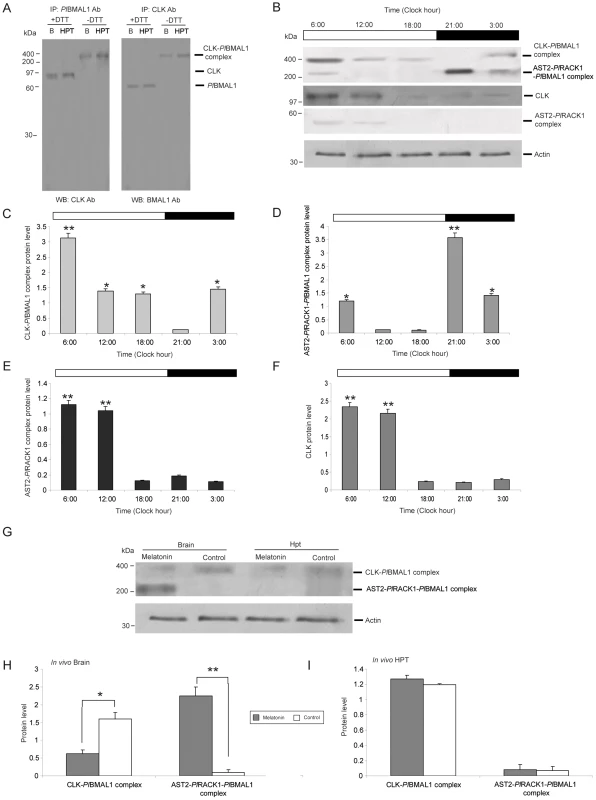
A) Immunoprecipitation (IP) of endogenous CLOCK (CLK) and PlBMAL1 in the brain (B) and HPT. Total proteins were extracted from the brain and HPT and were immunoprecipitated (IP) with the antibodies (Ab) indicated on the top of the blots, followed by western blot (WB) detection with antibodies against CLK or BMAL1 as shown at the bottoms of the blots. Reducing and non-reducing conditions of the samples are indicated by +DTT or −DTT, respectively. B) The levels of the CLK-PlBMAL1 and AST2-PlRACK1-PlBMAL1 protein complexes were analyzed by SDS-PAGE under non-reducing conditions (sample without DTT) followed by western blotting using an antibody against BMAL1. The AST2-PlRACK1 heterodimer and CLK were detected by western blotting using antibodies against AST2 and CLK, respectively. Time points were taken at 03:00, 06:00, 12:00, 18:00, and 21:00 (n = 4). An actin protein was used as an internal control. The horizontal band at the top of the histogram indicates the light condition (white = light, black = dark). C) Relative amounts of CLK-PlBMAL1 protein complex in brain extracts at different time points (n = 3) as determined by western blotting. D) Relative amounts of AST2-PlRACK1-PlBMAL1 protein complex in brain extracts at different time points (n = 3) as determined by western blotting. E) Relative amounts of AST2-PlRACK1 protein complex in brain extracts at different time points (n = 3) as determined by western blotting. F) Relative amounts of CLK protein in brain extracts at different time points (n = 3) as determined by western blotting. Average protein level in Graphs C–F was quantitated using Quantity One. The asterisks indicate significant differences (*P<0.05, **P<0.01); one-way ANOVA with Duncan's new multiple-range test and the Tukey test. Results are representative of three independent experiments. Error bars indicate SD from three replicates and the experiment has been repeated three times with similar results. G) Melatonin injection inhibited the formation of the CLK-PlBMAL1 complex; this inhibition is mediated by the AST2-PlRACK1-PlBMAL1 complex. The levels of the CLK-PlBMAL1 and AST2-PlRACK1-PlBMAL1 protein complexes were analyzed by SDS-PAGE under non-reducing conditions (sample without DTT) followed by western blotting using an antibody against BMAL1. The level of β-actin was used as an internal control. H) Relative levels of CLK-PlBMAL1 and AST2-PlRACK1-PlBMAL1 complexes in vivo, in the brain of crayfish after injection of melatonin and then brain extracts were analyzed by western blotting using an antibody against BMAL1. Grey bars = melatonin (4.3 nmol/g), white bars = control injection (PBS). I) Relative levels of CLK-PlBMAL1 and AST2-PlRACK1-PlBMAL1 complexes in vivo, in the HPT of crayfish after injection of melatonin and then HPT extracts were analyzed by western blotting using an antibody against BMAL1. Grey bars = melatonin (4.3 nmol/g), white bars = control injection (PBS). The level of β-actin was used as an internal control. Asterisks indicate significant differences (*P<0.05, **P<0.01). Quantity One analysis was used to quantify the intensity of protein bands. Graphs (H and I) represent the quantification of each complex formation, using Quantity one. Results are representative of three independent experiments. Statistical significance: *P<0.05, **P<0.01 using Student's paired t-test (error bars indicate SD from nine replicates). The effect of melatonin injection during the light period
Because we detected a clear stimulatory effect of melatonin on the expression of AST2 (Figure 2C), we decided to further confirm this putative role of AST2 by determining the presence, in vivo, of the approximately 400 kDa CLK-PlBMAL1 and approximately 200 kDa AST2-PlRACK1-PlBMAL1 complexes, respectively, after melatonin injection during the light period. Six hours after melatonin injection, the approximately 200 kDa complex was detected in the crayfish brains, but not in the HPT (Figure 6G–6I). Conversely, the amount of the approximately 400 kDa complex containing CLK-PlBMAL1 was reduced post-melatonin injection (Figure 6G and 6H). These results indicate that either secretion of natural melatonin at night or melatonin treatment causes an increase in the AST2 protein level and thereby increases the amount of the approximately 200 kDa complex.
AST2 is required for delayed CLK-BMAL1 formation
To further test whether AST2 is crucial for the interference of CLK-BMAL1 heterodimer formation, AST2 mRNA expression was partially knocked down by RNAi, to ca 50% of the control. As shown in Figure 7A and 7B, AST2 expression could be partly decreased in the brain by injection of dsRNA. A reduction of the approximately 200 kDa complex occurred during the night as a result of this AST2 deficiency when compared to injection of the dsGFP control (Figure 7C and 7E). In contrast to the control group, the formation of the ≈400 kDa complex in AST2-silenced animals occurred at 21 : 00, as shown in Figure 7C and 7D. Thus, this experiment further confirms that AST2 is required to interfere with the formation of the CLK-PlBMAL1 heterodimer, and that high AST2 levels at night cause lower amounts of CLK-PlBMAL1 heterodimer formation during the dark period (Figure 7C–7G).
Fig. 7. AST2 is required to regulate the CLK-PlBMAL1 heterodimer formation. 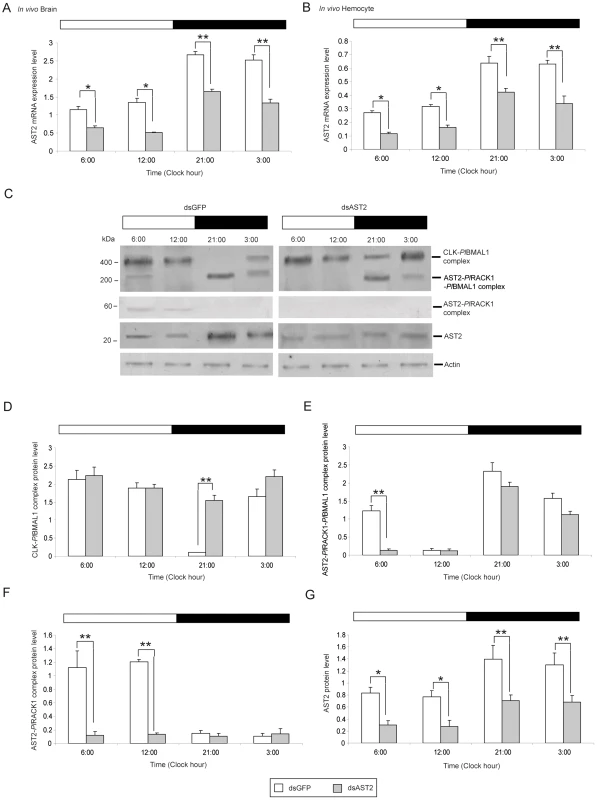
A) The relative expression levels of AST2 in the brain at 03:00, 06:00, 12:00, and 21:00 (n = 9) after partial knock down of AST2 (grey) in the brain by dsRNA injection and detection by qPCR; dsGFP injection was used as a control (white). B) The relative expression levels of AST2 in hemocytes at 03:00, 06:00, 12:00, and 21:00 (n = 9) after partial knock down of AST2 (grey) by dsRNA injection and detection by qPCR; dsGFP injection was used as a control (white). Graph A and B represent AST2 mRNA levels, using qPCR. Results are representative of three independent experiments. Error bars indicate SD of nine replicates. The asterisks indicate significant differences (*P<0.05, **P<0.01); Student's paired t-test. C) The effect of dsAST2 on complex formation in the brain in vivo was examined by western blotting of brain extracts and detection as follows: for the CLK-PlBMAL1 complex an anti-BMAL1 antibody, for the AST2- PlRACK1-PlBMAL1 complex an anti-BMAL1 antibody, and for the AST2-PlRACK1 an anti- AST2 antibody was used. The horizontal band at the top of the histogram indicates the light condition (white = light, black = dark). D) Relative amounts of CLK-PlBMAL1 protein complex in the brain in vivo was determined at different time points (n = 9) by western blotting of brain extracts, using an antibody against BMAL1. Grey bars = dsAST2, white bars = dsGFP. E) Relative amounts of AST2-PlRACK1-PlBMAL1 protein complex in the brain in vivo was determined at different time points (n = 9) by western blotting of brain extracts, using an antibody against BMAL1. Grey bars = dsAST2, white bars = dsGFP. F) Relative amounts of AST2-PlRACK1 protein complex in the brain in vivo was determined at different time points (n = 9) by western blotting of brain extracts, using an antibody against RACK1. Grey bars = dsAST2, white bars = dsGFP. G) Relative amounts of AST2 protein in the brain in vivo was determined at different time points (n = 3) as determined by western blotting of brain extracts, using an antibody against AST2. Grey bars = dsAST2, white bars = dsGFP. Quantity One analysis was used to quantify the intensity of protein bands from three independent experiments and results are presented in graphs D to G. Statistical significance: *P<0.05, **P<0.01 using Student's paired t-test (error bars indicate SD from nine replicates). AST2 gene knockdown induces CLK/BMAL1 E-box binding in the dark
Since knockdown of AST2 resulted in more CLK-PlBMAL1 complex formation during the dark period, we decided to test whether AST2 also had an effect on CLK-PlBMAL1 E-box binding. We assayed the CLK-PlBMAL1 E-box binding by a DNA-protein binding assay. As expected, we could demonstrate CLK-PlBMAL1 specific E-box binding during the night in AST2 silenced animals, whereas the control group (dsGFP) did not show any E-box binding in the dark (Figure 8A–8B). A super shift of this E-box binding was induced after incubation with anti-CLK antibodies (Figure 8A), and controls using mutated or unlabelled E-box oligonucleotides did not show any binding (Figure 8B and Figure S8A–S8B). In contrast, melatonin injection during the day (which leads to enhanced levels of AST2, Figure 2A) reduced the DNA binding activity of CLK-PlBMAL1 compared to other groups (Figure S8B). The complex formation of CLK-PlBMAL1 or AST2-PlRACK1-PlBMAL1 was followed by western blotting after knockdown of AST2 expression by dsRNA or increasing AST2 by addition of melatonin (Figure 8C–8E). These results reveal that AST2 is an important regulator of the E-box binding activity of the CLK-PlBMAL1 complex.
Fig. 8. AST2 knockdown enhanced the DNA binding activity of CLK/PlBMAL1. 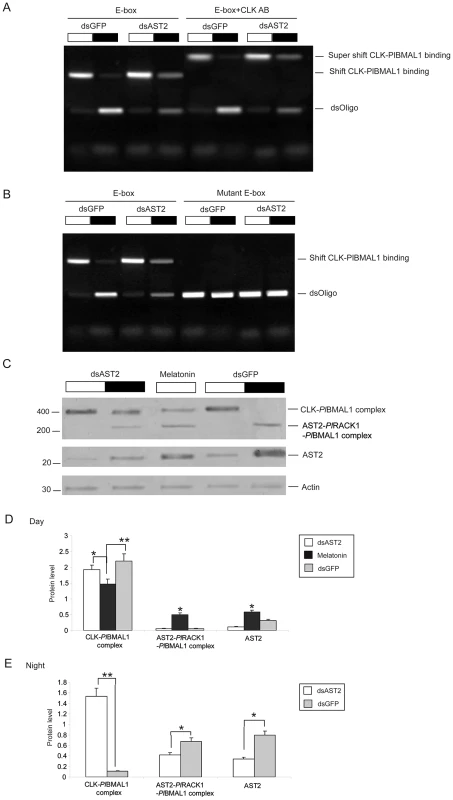
A) Crayfish received dsGFP or dsAST2 treatments, then crude brain lysates were harvested during day and night, and were used to study E-box binding with fluorophore labeled oligonucleotides in an EMSA assay. As a control the brain lysates were incubated with an antibody against CLK, to show a supershift. The E-box binding activity of CLK-PlBMAL1 is indicated on the right side of the gel. B) The effect of AST2 knock down on CLK-PlBMAL1 E-box binding was analyzed by EMSA. A mutant E-box was used as control. C) Western blot showing the effect of AST2 knock down or melatonin treatment on the protein levels of CLK/PlBMAL1, AST2-PlRACK1-PlBMAL1 complexes (upper panel), and AST2 level (middle panel). Actin was used as an internal control (lower panel). The horizontal band on top of this histogram indicates day (white) or night (black). D) Relative amounts of CLK-PlBMAL1 and AST2-PlRACK1-PlBMAL1 protein complexes in brain extracts isolated during the day (n = 9) as determined by western blotting, using an antibody against BMAL1. The AST2 protein was also detected by western blotting, using an antibody against AST2. White bars = dsAST2, black bars = melatonin, grey bars = dsGFP. E) Relative amounts of CLK-PlBMAL1 and AST2-PlRACK1-PlBMAL1 protein complexes in brain extracts isolated during the night (n = 9) as determined by western blotting, using an antibody against BMAL1. The AST2 protein was also detected by western blotting, using an antibody against AST2. White bars = dsAST2, and grey bars = dsGFP. In three independent experiments, proteins were visualized by western blotting and quantitated using the Quantity One software. These results are shown in graph D and E. Error bars indicate SD from nine replicates. The asterisks indicate significant differences (*P<0.05, **P<0.01); one-way ANOVA with Duncan's new multiple-range test and the Tukey test. Discussion
Various proteins are regulated by the circadian rhythm, and there are a growing number of examples of circadian regulation of stem cell activities, such as the proliferation and recruitment of hematopoietic progenitor cells [33], [35]. We have recently shown that the hematopoietic cytokine AST1 is regulated by light and that this regulation has an impact on hemocyte synthesis [26]. AST1 contains a prokineticin domain (pfam06607) and shares similarities with vertebrate PROK2, which is regulated by light at the transcriptional level by core clock genes [22]. Apart from functioning in circadian clocks, vertebrate PROKs have documented roles in the regulation of intestinal muscle contractility, neurogenesis, pain perception, food uptake, and appetite regulation [36], [37]. In similarity with invertebrate astakines the vertebrate PROKs also play roles in hematopoiesis and proinflammatory immune responses. Therefore, the structural and functional similarities between astakines and the vertebrate PROKs point to an ancient similar role for these proteins [38].
The oscillation in PROK2 transcription in mice occurs in the SCN, and it acts as an output molecule, signaling the rhythm to other cells and tissues and thereby changing circadian behavior [39]. Signal transduction from vertebrate PROK's are mediated by two closely related G-protein coupled receptors, and the AVIT motif has been shown to be crucial to this receptor binding and the activity of the PROK's [40]. An analysis of the gene structure for human PROK2 reveals that exon 2–4 share similarities to crayfish AST2, while the first exon encodes the signal peptide and the AVIT motif [41], [42]. Thus, all invertebrate astakines found so far, differ from the vertebrates PROKs in their N-terminal and lack of the AVIT-motif [25] a fact that indicates different receptor binding properties for this invertebrate group of PROK domain containing proteins. Similar to PROK2, AST1 is a secreted molecule with circadian expression, and these data indicate a conserved role for this domain throughout evolution, from arthropods to humans [25], [26]. In this study, we report a more direct role for another crayfish prokineticin, namely AST2, as an intracellular regulator of the heterodimeric transcription factors CLK and PlBMAL1. The expression of AST2 does not exhibit a light-dependent pattern that is similar to that of AST1 or PROK2 in mice; instead, we detected high levels of AST2 expression in the HPT and brain during the dark period when melatonin levels are high. We also showed that melatonin administration clearly increased the expression of AST2 at both the mRNA and protein levels.
It is well established that the neurohormone melatonin has an important function in the SCN in regulating the feedback loop of the core clock genes and also functions as an output signal that has an effect on peripheral cells [43]. Melatonin is mainly produced within the eyestalks of crayfish, and its production is elevated during the dark period [27]. Melatonin is known to influence numerous physiological processes in crayfish [18], [33], [44], although the molecular mechanism by which melatonin exerts its effect has not been fully clarified. However, AST2 is an intracellular protein that is present in several crustaceans and insect species [25]. Here, for the first time, we identify a direct role for AST2 in the regulation of the circadian clock and show that the protein acts as a link between melatonin and the heterodimeric transcriptional activator CLK-PlBMAL1. Intracellular PlRACK1 was identified as an AST2-binding protein, and in the brain, the binding of PlBMAL1 to this complex mediated a decrease in the formation of the CLK-PlBMAL1 heterodimer. This process may be very important in all animals that have AST2 molecules i.e., spiders, ticks, crustaceans, scorpions, several insect groups such as Hymenoptera, Hemiptera, Blattodea but not Diptera and Coleoptera. If any vertebrate PROK is performing the same function still remains to be demonstrated.
Recently, RACK1 and protein kinase Cα (PKCα) were shown to interact with BMAL1 in a circadian manner, resulting in suppressed CLK-BMAL1 activity [9]. This result is consistent with our observation of the PlRACK1-PlBMAL1 interaction in the crayfish brain. At night, we detected an approximately 200 kDa complex in the crayfish brain and found that the components of this complex consisted of AST2, PlRACK1 and PlBMAL1. Conversely, this complex disappeared during the light period due to lower levels of AST2 during the day. Thus, our results show that AST2 acts as a regulator of the CLK-PlBMAL1 complex by binding to PlRACK1-PlBMAL1 in an oscillatory manner that is induced by changes in the level of melatonin.
Interestingly, enhancement of the approximately 400 kDa complex consisting of PlBMAL1 and CLK was detected when the level of the approximately 200 kDa complex was decreased either during the light period or by knockdown of AST2 expression. A similar high-molecular-weight complex of PlBMAL1 and CLK (approximately 400 kDa) was also recently detected in mice [9]. The formation of the approximately 400 kDa crayfish complex is dependent on the expression of CLK. The expression of mammalian CLK is not rhythmic [45], [46], while zebrafish CLK shows robust rhythmic expression, as was the case for crayfish CLK in our study [47], [48], [49]. As with zebrafish and Drosophila, the CLK of crayfish is directly regulated by light [50], [51]. Unlike those in mammals, the protein products of PER and TIM (or CRY in mammals) in Drosophila act to inhibit the transcriptional activity of the CLK-CYC complex (CLK-BMAL1) by interacting with CLK or a CLK-containing complex during the night but not during most of the day [52]. In our experiments, the AST2-PlRACK1-PlBMAL1 complex (approximately 200 kDa) competes for binding to PlBMAL1 to interfere with the formation of the CLK-PlBMAL1 heterodimer during the dark period. Further it is clear from our DNA mobility shift assay that AST2 can inhibit the transcriptional activation activity of CLK-PlBMAL1. This 200 kDa AST2-containing complex directly interacts with different proteins and is clearly induced by melatonin. Therefore, our findings reveal an ancient evolutionary role for a prokineticin superfamily protein that links melatonin to direct regulation of the core clock gene feedback loops, as indicated in the model (Figure 9).
Fig. 9. Molecular model of the circadian regulation by CLK, PlBMAL1, PlRACK1, and AST2. 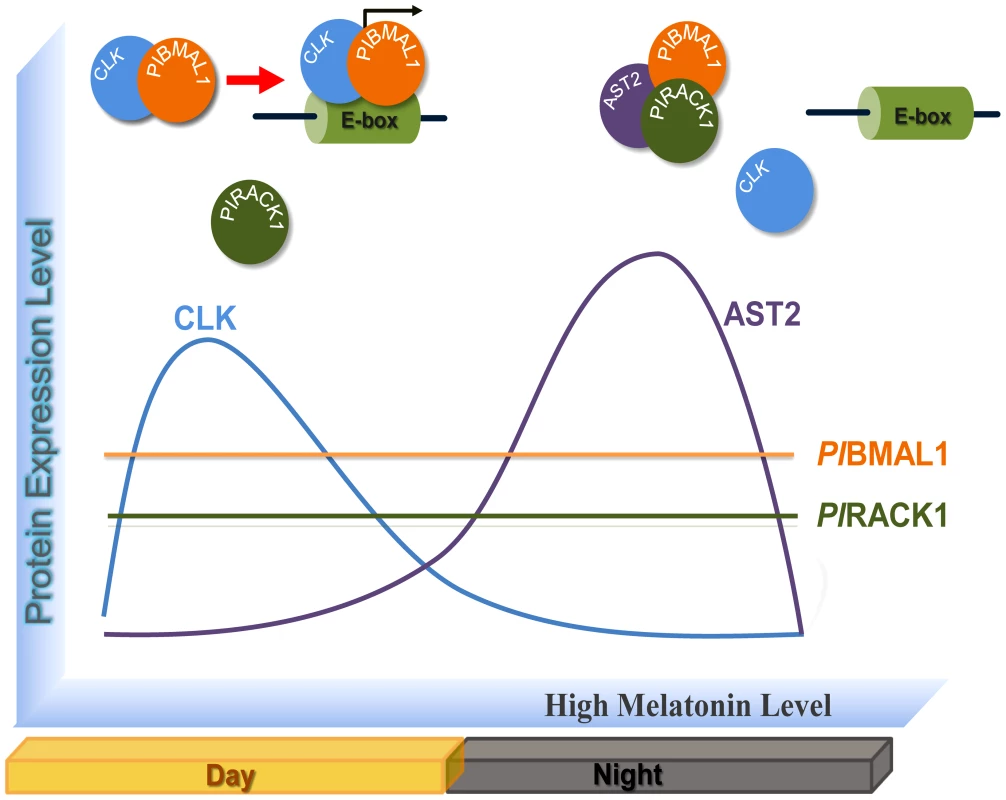
The protein level of CLK is enhanced during the light period, and a CLK-PlBMAL1 complex is formed that acts as a transcriptional activator. During the dark period melatonin secretion causes an upregulation of AST2 and this high AST2 level results in the formation of a complex between PlBMAL1 and PlRACK1. Then AST2 binds to PlRACK 1 in the PlBMAL-PlRACK1 complex and forms the AST2-PlRACK1-PlBMAL1 complex and this interferes with the formation and activity of the CLK-PlBMAL1 heterodimer. Materials and Methods
Crayfish
Healthy intermolt freshwater crayfish (P. leniusculus) were obtained from Lake Hjälmaren, Sweden and were maintained in aerated tap water at 10°C.
Cloning and sequence analysis of full-length P. leniusculus RACK1 (PlRACK1) and PlBMAL1 cDNAs
Total RNA was isolated from crayfish brains using TRIzol LS (Invitrogen) followed by RNase-free DNase I (Ambion) treatment. cDNA synthesis was accomplished with an oligo (dT) primer and ThermoScript RT-PCR (Invitrogen) according to the manufacturer's protocol. A partial sequence of PlRACK1 was amplified using primers that were designed based on the shrimp (P. monodon) PmRACK1 sequence (GenBank accession no. EF569136), whereas PlBMAL1 primers were based on the cDNA sequences from several organisms (listed in Table S2). To obtain the full-length cDNAs of the PlRACK1 and PlBMAL1 genes, 5′ and 3′ rapid amplification of cDNA ends (RACE) technology was performed using the SMARTer RACE cDNA Amplification Kit (Clontech) according to the manufacturer's instructions. The complete PlRACK1 cDNA sequence was amplified using gene-specific primers for PlRACK1 (Table S2) and the SMART Universal Primer A. The PCR products were then cloned into the TOPO TA cloning vector and sequenced. The deduced amino acid sequence of the PlRACK1 gene was searched against the GenBank database with the BLAST program (http://www.ncbi.nlm.nih.gov/BLAST/). Sequence alignment of PlRACK1 was generated by ClustalW, and the protein domains were analyzed with the SMART program (http://smart.embl-heidelberg.de/).
Tissue distribution analysis
Total RNAs were isolated from different crayfish tissues using TRIzol reagent (Invitrogen) followed by RNase-free DNase I (Ambion) treatment. For reverse transcription, 1 µg of total RNA was reverse transcribed to produce cDNA using ThermoScrip RT-PCR (Invitrogen). To amplify and visualize the cDNAs of PlRACK1 and PlBMAL1, two sets of primers were used: the forward and reverse primers listed in Table S2. The PCR cycles were as follows: 1 cycle (95°C for 2 min); 28 cycles (95°C, 30 s; 55°C, 30 s; 72°C, 40 s) followed by an extension step (72°C for 5 min). The transcription of the 40S ribosomal protein was used as an internal control. All PCR products were analyzed on 1% GelRed-stained agarose gels.
Crayfish HPT cell culture and maintenance
The hematopoietic tissue was dissected, washed with CPBS (crayfish phosphate buffered saline (pH 6.8), 10 mM Na2HPO4, 10 mM KH2PO4, 150 mM NaCl, 10 µM CaCl2, and 10 µM MnCl2,) and incubated in 600 µl of 0.1% collagenase (types I and IV) (Sigma) in CPBS at room temperature for 45 min to separate the HPT cells. The separated cells were washed twice with CPBS by centrifugation at 800×g for 5 min at room temperature. The cell pellet was re-suspended in modified L-15 medium, and cells were then seeded at a density of 2.5×106 cells/150 µl in 96-well plates. The HPT cells were supplemented with partially purified plasma as a source of AST1 after 1 h of attachment at room temperature. The culture plates were incubated at 16°C, and 1/3 of the medium was changed at 48 h intervals.
Melatonin treatment
For the in vivo experiments, 100 µl of a melatonin solution (Sigma) was dissolved in ethanol and diluted in crayfish saline (to a final ethanol concentration of 1%). This solution was injected into the base of a walking leg such that the amount of injected melatonin was 4.3 nmol/g fresh weight. Control crayfish received 100 µl of saline with 1% ethanol. The hemolymph, brain, and HPT of these crayfish were collected at 0, 1, 2, 3, and 6 h post-injection for the extraction of total RNA, whereas samples for total protein extraction were dissected at 3 h post-injection. All injections were performed during the day.
For the in vitro experiments, HPT cell cultures were prepared and incubated at 16°C for 12 h. The medium was then replaced with 150 µl of L-15 medium containing 5 µl of the melatonin solution at final concentration of 0.43 nmol/well (3 µM) (the control group was supplemented with 5 µl of saline with 1% ethanol) and 5 µl of crude AST1, followed by incubation for 0, 0.25, 1, 3, 6, and 12 h at 20°C. Thereafter, the cells were harvested at each time point for extraction of total RNA and protein. The transcription and translation levels of AST1 and AST2 in vitro and in vivo were detected by quantitative RT-PCR and ELISA, respectively.
Sampling times
The light in the aquarium room was turned on and off at 4 : 00 and 20 : 00, respectively. Brains were dissected at 06 : 00, 12 : 00, 16 : 00, 21 : 00, and 03 : 00 from three crayfish at each sampling time, which were kept in normal light/dark conditions. The experiments were repeated at least twice. Complex formation and CLK expression were determined by western blotting using antibodies against BMAL1 and CLK, respectively. An anti-actin antibody (Santa Cruz Biotechnology) was used as an internal control.
Generation of dsRNA
A T7 promoter sequence was incorporated into gene-specific primers for AST2 and GFP (italic letters) at their 5′ ends (AST2 RNAi-F, 5′ - TAA TAC GAC TCA CTA TAG GGT CCA CGC CTC TGA GTC TTT T-3′; AST2 RNAi-R, 5′ - TAA TAC GAC TCA CTA TAG GGA TGC CCA GAG TGT TGT CCT C-3′ and GFP 63+, 5′ - TAA TAC GAC TCA CTA TAG GGC GAC GTA AAC GGC CAC AAG T-3′; GFP 719-, 5′ - TAA TAC GAC TCA CTA TAG GGT TCT TGT ACA GCT CGT CCA TG-3′). These primers were used to amplify PCR products as templates for dsRNA synthesis. A GFP transcript was amplified with the pd2EGFP-1 vector (Clontech) as a template and was used as a control. The amplified products were then purified using a GenElute Gel extraction kit (Sigma) followed by in vitro transcription using a MegaScript kit (Ambion), and the dsRNA was purified with TRIzol LS (Invitrogen).
dsRNA in vivo
Small intermolt crayfish (15±2 g of fresh weight) were divided into two groups with three crayfish in each group (n = 3). The first and second groups were injected with 300 µg of GFP control dsRNA and 300 µg of dsAST2, respectively, at the base of the fourth walking leg. Twenty-four hours after the first dsRNA injection, four drops of crayfish hemolymph were bled for total RNA isolation to test the efficiency of the RNAi. Then, dsRNA was injected a second time into both groups as described above. Twenty-four hours after the second dsRNA injection, the brains were dissected at 06 : 00, 12 : 00, 21 : 00, and 03 : 00 to determine complex formation by western blotting.
Plasmid constructs
PlRACK1 was amplified using the synthesized forward primer RACK1_F1 and reverse primer RACK1_R1 (Table S2) by PCR. The conditions for the PCR reactions were essentially identical to those described in the manufacturer's protocol for the vector. All constructs were sequence-verified. The PCR product was cloned into the pGEX-4T-1 bacterial expression vector at the BamHI and SalI cleavage sites.
PlBMAL1 was amplified from crayfish testis cDNA by PCR using 1 unit of Paq5000 DNA polymerase (Stratagene) and a pair of specific primers listed in Table S2. EcoRI and XhoI sites were engineered into the primers to facilitate subsequent cloning. PlBMAL1 was further subcloned into the pGEX-4T-1 and pET-32a vectors. The recombinant plasmids containing the PlRACK1 and PlBMAL1 genes were confirmed by DNA sequencing.
Bacterial protein expression and purification
The full-length cDNA encoding PlRACK1was cloned into the pGEX-4T-1 bacterial expression vector to express a glutathione S-transferase (GST)-fused PlRACK1 protein in the Escherichia coli strain BL21(DE3). Protein expression was induced by adding IPTG to a final concentration of 1 mM at 25°C for 6 h. The cells were harvested by centrifugation followed by suspension in lysis buffer (50 mM NaH2PO4 (pH 8.0), 300 mM NaCl and 2% Triton X-100) and lysis by sonication. Inclusion body pellets were solubilized in denaturing buffer (50 mM Tris (pH 8.0), 8 M guanidine (GdnHCl) and 10 mM DTT) followed by centrifugation (13,000 rpm for 15 min). The protein was refolded in refolding buffer (55 mM Tris-HCl, 0.44 M L-arginine, 21 mM NaCl, 0.88 mM KCl and 10 mM DTT). The refolded GST-PlRACK1protein was dialyzed in phosphate-buffered saline (PBS, pH 7.4) and applied to Glutathione Sepharose 4B resin (Amersham Biosciences, Inc.) for 30 min at room temperature. The beads were washed with ice-cold PBS followed by incubation in reducing buffer (50 mM Tris-HCl and 20 mM reduced glutathione, pH 8.0) for 30 min at room temperature. After centrifugation at 1,000×g for 5 min, the supernatant was collected and analyzed by 12.5% SDS-PAGE. The protein concentration was measured, and the proteins were stored at −80°C. The full-length sequence of PlBMAL1 was ligated into EcoRI/XhoI-linearized pGEX-4T-1 and pET-32a vectors to create a plasmid that expresses GST - and His-Trx fusion proteins, respectively. The recombinant GST-PlBMAL1 protein was expressed and purified in a similar manner as the GST-PlRACK1 protein. The His-Trx-PlBMAL1 recombinant protein was purified using a Ni-NTA column. His-Trx-AST2 was expressed and purified as described in earlier publications [25]. Briefly, His-Trx tagged AST2 was expressed in E. coli BL21 (DE3) and induced by the addition of 0.02 mM IPTG. The cells were resuspended in lysis buffer (50 mM NaH2PO4, pH 8.0, 300 mM NaCl and 2% Triton X-100) and purified using HisPur Cobalt Resin (Thermo Scientific). The protein products of the His-Trx tag expression vector pET-32a and the expression vector pGEX-4T-1, which are used for the incorporation of N-terminal thioredoxin and GST tags, respectively, were expressed in E. coli BL21(DE3) cells. The thioredoxin and GST proteins were purified with HisPur Cobalt or Glutathione Sepharose resins, respectively, as control proteins. All samples were analyzed by 12% SDS-PAGE.
In vitro GST pull-down assay
To identify putative AST2-binding proteins, total protein was extracted from crayfish brains in buffer containing 50 mM Tris-HCl (pH 7.2), 100 mM NaCl, 10% glycerol, 1 mM PMSF, 10 µM MG132 and Complete Mini Protease Inhibitor tablets according to the manufacturer's instructions (Roche Diagnostics). Cell debris was removed by centrifugation at 10,000×g for 10 min. Recombinant GST–AST2 was expressed in E. coli and purified using glutathione Sepharose 4B resin according to standard protocols. GST–AST2 or GST (3–4 µg) was incubated with 800 µg of crude brain total protein extract at 4°C for 1 h unless otherwise specified. Glutathione beads were recovered by brief centrifugation and washed three times with 1 ml of washing buffer (50 mM Tris-HCl (pH 7.2), 100 mM NaCl, 10% glycerol and 0.1% Tween 20). GST was used as a control protein in this experiment. Pull-down mixtures were separated by SDS–PAGE, and after excluding the background proteins found in the control, the remaining band of about 30 kDa was sequenced by mass spectrometry after trypsin cleavage. Identification of the protein was based on raw MS/MS data, and ions search using the Mascot program (http://www.matrixscience.com).
The interaction between GST-PlRACK1and HisTrx-AST2 was examined by incubating purified GST-PlRACK1or the GST control with Glutathione Sepharose 4B resin (50 µl a 50% bed slurry) at room temperature for 1 h. The beads were washed three times with PBS, purified HisTrx-AST2 or the His-Trx control (5 µg) was added, and incubated for another 2 h at room temperature in PBS buffer. After washing with PBS, the precipitated proteins were eluted with SDS-sample buffer.
To determine the formation of a PlRACK1-PlBMAL1 complex, 5 µg purified His-Trx-PlBMAL1 protein was incubated with 5 µg purified GST-PlRACK1 protein bound to Glutathione Sepharose 4B beads. The beads were then washed five times, and the remaining proteins were eluted with SDS-sample buffer.
To explore whether AST2 can bind to PlRACK1 together with PlBMAL1, purified His-Trx-PlBMAL1 (500 ng) and His-Trx-AST2 (500 ng) were incubated with Glutathione Sepharose 4B-bound GST-PlRACK1 (0, 5, 50, 500 ng) in PBS buffer at 25°C for 2 h. The protein-bead complexes were washed, and the bound proteins were eluted with SDS-sample buffer. All protein samples were resolved by SDS-PAGE and were analyzed by western blot.
Blot overlay assays
In addition to an in vitro GST pull-down assay, a far western overlay assay was used to identify any putative AST2 binding protein. The protein was extracted from crayfish HPT and then subjected to 12% SDS-PAGE, transferred to PDVF membranes, and blocked with 10% skim milk in TBST buffer (10 mM Tris-HCl (pH 7.5), 150 mM NaCl and 0.05% Tween 20) at room temperature. After three washes with TBST, the membranes were incubated with 25 mM GST-AST2 or GST alone in 10 ml TBST buffer at room temperature. The blots were washed twice and incubated for 1 h with 1∶2,000 dilution of anti GST antibody and then washed twice, incubated for 1 h with 1∶1,000 dilution of anti mouse antibody for 1 h, and washed three times before detection with an ECL western blotting reagent kit. The control reaction was incubated with GST instead of GST-AST2.
Purified GST-PlRACK1 protein was cleaved by incubation with thrombin (GE Healthcare, 1 µl thrombin per 100 µg recombinant protein) at 22°C overnight to remove the GST fusion tag. Free PlRACK1was eluted with PBS from the Glutathione Sepharose 4B beads. PlRACK1 was run on 12.5% SDS-PAGE, transferred to PDVF membranes and incubated at 4°C overnight with purified GST-PlBMAL1 or GST alone (7 µg) in 10 ml TBST buffer. The blot was washed three times (10 min each) in TBST containing 0.05% Tween 20. Protein interactions were detected by western blotting using an anti-GST antibody. Blot overlay assays were also used to confirm the binding of AST2. Preparation of total crayfish brain homogenates and gel overlays with recombinant GST-AST2 (far western blots) were performed as described above.
Protein extraction
Whole brains were dissected, and HPT cells were isolated, rinsed three times with PBS and prepared for western analysis, immunoprecipitation and ELISA. Cell extracts for western blotting and immunoprecipitation were prepared by suspending the PBS-washed cells in NP-40 lysis buffer (50 mM Tris (pH 8.0), 150 mM NaCl, and 1.0% NP-40) with protease inhibitor cocktail tablets (Roche). Following incubation on ice for 30 min, non-extractable material was removed by centrifugation at 13,000 rpm for 10 min at 4°C. Cleared supernatants were used for immunoprecipitation, whereas whole homogenized cells were mixed with 1X SDS loading buffer with and without DTT, followed by western analysis. Cell extracts analyzed directly by ELISA were homogenized in bicarbonate/carbonate coating buffer (pH 9.6).
Western blot analysis
Protein samples were separated by 12.5% SDS-PAGE and transferred to PVDF membranes (Bio-Rad, America) in electroblotting buffer (25 mM Tris, 190 mM glycine, 20% methanol) for 120 min. After rinsing with TBST buffer, the membranes were immersed in blocking buffer (5% skim milk powder in TBST) at 4°C, overnight, followed by incubation with primary antibody for 1 h (anti-GST or anti-His diluted in 5% skim milk in TBST). Subsequently, the membranes were incubated in HRP-conjugated rabbit anti-mouse IgG (GE Healthcare) for 1 h and visualized by the enhanced chemiluminescence detection system (Amersham Biosciences). The X-ray images from western blotting were analyzed according to the Quantity One user guide. Briefly, the western blotting films were scanned and saved as digital images as a quantity-one file (or .1sc) by GS-800 calibrated imaging densitometer. These quantity-one files were used for protein analysis. Protein bands in all scanned images (.1sc file) were quantified and the values of the band intensities were calculated by using the Volume and Match tools (Quantity One software version 4.6.0; Bio-Rad Laboratories). These values were used to perform statistical analysis as described in the “Statistical analysis” part below.
Immunoprecipitation
Immunoprecipitations were performed by first incubating cell lysates from brain or HPT with the indicated primary antibodies overnight at 4°C. Immune complexes were then precipitated by incubating reactions for an additional 2 h at 4°C with either Sepharose-conjugated protein A or agarose-conjugated protein G. The immunoprecipitates were washed four times with 1% Triton X-100 lysis buffer and were resuspended in SDS-PAGE sample buffer for western blotting.
Transcription analysis
The transcript levels of AST1 and AST2 were detected by quantitative RT-PCR using QuantiTect SYBR green PCR kit (QIAGEN). The relative expression was normalized to the expression of the mRNA encoding the crayfish ribosomal protein gene (R40s) for each sample. The primers used are shown in Table S2. The qPCR reactions contained 5 µl of 1∶10 diluted cDNA template, 1xQuantiTect SYBR Green PCR master mix (QIAGEN) and 5 µM forward and reverse primers in a 25 µl reaction volume. The following amplification profile was used: 95°C for 15 min, followed by 45 cycles of 94°C for 15 s, 58°C for 30 s, and 72°C for 30 s. All qPCR reactions were performed in triplicates. The hemocytes, HPTs and brains from a least three crayfish were used for each time point. The statistical comparisons were performed as shown below (Statistical analysis).
ELISA
Cell extracts, containing protein at 20 µg/ml were coated in wells and incubated for 2 h at room temperature, followed by incubation with blocking solution (1% BSA in PBS). Following three careful washes with PBS, the different proteins were detected with the corresponding primary antibody (1∶1000) followed by HRP-conjugated secondary antibodies (1∶3000). After the addition of 100 µl tetramethylbenzidine (TMB) substrate (Sigma) and incubation for 20 min, the reaction was terminated by 100 µl of stop solution (0.5 M sulfuric acid) and the absorbance of the resulting color was measured at 450 nm. All samples were performed in triplicates and each experiment was repeated three times.
Antibodies
Polyclonal antibodies against recombinant crayfish AST1 and AST2 were raised in rabbit as have been described earlier [31]. A goat polyclonal antibody to human BMAL1 (N-5: SC-8550) and goat polyclonal antibody to mouse CLK (S-19: SC-6927) were obtained from Santa Cruz Biotechnology. A goat polyclonal antibody to RACK1 (GNB2L1: Orb22484) was obtained from Biorbyt. A goat polyclonal antibody against actin (C-11: SC-1615) was used as internal control. The specificity of each antibody was tested by western blots, and all were found to be specific for each protein.
Electrophoretic mobility shift assay (EMSA)
Mobility shift assays were performed according to reference [53]. Briefly, a synthetic fluorophore labeled oligonucleotide probe including an E-box element (CACGTG) was used as a substrate in this assay. To generate an E-box containing double-stranded DNA fragment, the two fluorophore labeled E-box oligonucleotides described below were annealed. A 30 µL EMSA reaction mixture contained ∼100 mM KCl, 50 ng of crude crayfish brain lysates, 1 µg poly (dI-dC), and 10 fmol of labeled probe. For control 10 fmol of unlabeled competitor oligonucleotide or mutant E-box oligonucleotide were incubated as control. After incubation for 1 hour on ice, antibodies against CLK protein was added and incubated another 20 minutes on ice. Protein-DNA complexes were resolved by 5% polyacrylamide gel electrophoresis. Specific DNA-protein and antibody-supershifted complexes were observed as retarded bands in the gel. E-box (5′-TTT AGT GAA AAG CCG CCG CTC ACG TGG CGA ACT GCG TGA CTT G-3′ and 5′-TTT CAA GTC ACG CAG TTC GCC ACG TGA GCG GCG GCT TTT CAC T-3′) and E-box mutant (5′-TTT AGT GAA AAG CCG CCG CTC AGC TGG CGA ACT GCG TGA CTT G-3′ and 5′-TTT CAA GTC ACG CAG TTC GCC AGC TGA GCG GCG GCT TTT CAC T-3′) sequences were used in the gel shift analysis (the E-box, and E-box mutant sequence is underlined).
Statistical analysis
All statistical comparisons were examined by one-way analysis of variance, followed by Duncan's new multiple-range test and the Tukey test. When two treatments were compared, a Student's paired t-test was used. Differences were considered statistically significant at P<0.05 and/or P<0.01. The results are expressed as the mean ± standard deviation (SD).
Supporting Information
Zdroje
1. RosbashM, AlladaR, DembinskaM, GuoWQ, LeM, et al. (1996) A Drosophila circadian clock. Cold Spring Harb Symp Quant Biol 61 : 265–278.
2. WeaverDR (1998) The suprachiasmatic nucleus: a 25-year retrospective. J Biol Rhythms 13 : 100–112.
3. Helfrich-ForsterC, StenglM, HombergU (1998) Organization of the circadian system in insects. Chronobiol Int 15 : 567–594.
4. StoneEF, FultonBO, AyresJS, PhamLN, ZiauddinJ, et al. (2012) The circadian clock protein timeless regulates phagocytosis of bacteria in Drosophila. PLoS Pathog 8: e1002445 doi:10.1371/journal.ppat.1002445
5. AlladaR, EmeryP, TakahashiJS, RosbashM (2001) Stopping time: the genetics of fly and mouse circadian clocks. Annu Rev Neurosci 24 : 1091–1119.
6. LowreyPL, TakahashiJS (2004) Mammalian circadian biology: elucidating genome-wide levels of temporal organization. Annu Rev Genomics Hum Genet 5 : 407–441.
7. WangGK, OusleyA, DarlingtonTK, ChenD, ChenY, et al. (2001) Regulation of the cycling of timeless (tim) RNA. J Neurobiol 47 : 161–175.
8. KingDP, ZhaoY, SangoramAM, WilsbacherLD, TanakaM, et al. (1997) Positional cloning of the mouse circadian clock gene. Cell 89 : 641–653.
9. RoblesMS, BoyaultC, KnuttiD, PadmanabhanK, WeitzCJ (2010) Identification of RACK1 and protein kinase Calpha as integral components of the mammalian circadian clock. Science 327 : 463–466.
10. SaperCB, ScammellTE, LuJ (2005) Hypothalamic regulation of sleep and circadian rhythms. Nature 437 : 1257–1263.
11. JohnsonRF, MooreRY, MorinLP (1988) Loss of entrainment and anatomical plasticity after lesions of the hamster retinohypothalamic tract. Brain Res 460 : 297–313.
12. CassoneVM, ChesworthMJ, ArmstrongSM (1986) Entrainment of rat circadian rhythms by daily injection of melatonin depends upon the hypothalamic suprachiasmatic nuclei. Physiol Behav 36 : 1111–1121.
13. MaestroniGJ, ContiA, PierpaoliW (1986) Role of the pineal gland in immunity. Circadian synthesis and release of melatonin modulates the antibody response and antagonizes the immunosuppressive effect of corticosterone. J Neuroimmunol 13 : 19–30.
14. HardelandR, PoeggelerB (2003) Non-vertebrate melatonin. J Pineal Res 34 : 233–241.
15. PoirelVJ, BoggioV, DardenteH, PevetP, Masson-PevetM, et al. (2003) Contrary to other non-photic cues, acute melatonin injection does not induce immediate changes of clock gene mRNA expression in the rat suprachiasmatic nuclei. Neuroscience 120 : 745–755.
16. KrauchiK, Wirz-JusticeA (2001) Circadian clues to sleep onset mechanisms. Neuropsychopharmacology 25: S92–96.
17. SharmaVK, SingaravelM, SubbarajR, ChandrashekaranMK (1999) Timely administration of melatonin accelerates reentrainment to phase-shifted light-dark cycles in the field mouse Mus booduga. Chronobiol Int 16 : 163–170.
18. ShibataS, CassoneVM, MooreRY (1989) Effects of melatonin on neuronal activity in the rat suprachiasmatic nucleus in vitro. Neurosci Lett 97 : 140–144.
19. DijkDJ, CajochenC (1997) Melatonin and the circadian regulation of sleep initiation, consolidation, structure, and the sleep EEG. J Biol Rhythms 12 : 627–635.
20. LissoniP, RovelliF, BrivioF, BrivioO, FumagalliL (1998) Circadian secretions of IL-2, IL-12, IL-6 and IL-10 in relation to the light/dark rhythm of the pineal hormone melatonin in healthy humans. Nat Immun 16 : 1–5.
21. PetrovskyN (2001) Towards a unified model of neuroendocrine-immune interaction. Immunol Cell Biol 79 : 350–357.
22. SandemanD, SandemanR, DerbyC, SchmidtM (1992) Morphology of the brain of crayfish, crabs and spiny lobsters: a common nomenclature for homologous structures. Biol Bull 183 : 304–326.
23. SullivanJM, GencoMC, MarlowED, BentonJL, BeltzBS, et al. (2009) Brain photoreceptor pathways contributing to circadian rhythmicity in crayfish. Chronobiol Int 26 : 1136–1168.
24. Escamilla-ChimalEG, Velazquez-AmadoRM, FiordelisioT, Fanjul-MolesML (2010) Putative pacemakers of crayfish show clock proteins interlocked with circadian oscillations. J Exp Biol 213 : 3723–3733.
25. WithyachumnarnkulB, AjpruS, RachawongS, Pongsa-AsawapaiboonA, SumridthongA (1999) Sexual dimorphism in N-acetyltransferase and melatonin levels in the giant freshwater prawn Macrobrachium rosenbergii de Man. J Pineal Res 26 : 174–177.
26. TildenAR, AltJ, BrummerK, GrothR, HerwigK, et al. (2001) Influence of photoperiod on N-acetyltransferase activity and melatonin in the fiddler crab Uca pugilator. Gen Comp Endocrinol 122 : 233–237.
27. Mendoza-VargasL, Solis-ChagoyanH, Benitez-KingG, Fuentes-PardoB (2009) MT2-like melatonin receptor modulates amplitude receptor potential in visual cells of crayfish during a 24-hour cycle. Comp Biochem Physiol A Mol Integr Physiol 154 : 486–492.
28. ChengMY, BullockCM, LiC, LeeAG, BermakJC, et al. (2002) Prokineticin 2 transmits the behavioural circadian rhythm of the suprachiasmatic nucleus. Nature 417 : 405–410.
29. PittendrighCS (1993) Temporal organization: reflections of a Darwinian clock-watcher. Annu Rev Physiol 55 : 16–54.
30. WilsbacherLD, YamazakiS, HerzogED, SongEJ, RadcliffeLA, et al. (2002) Photic and circadian expression of luciferase in mPeriod1-luc transgenic mice invivo. Proc Natl Acad Sci U S A 99 : 489–494.
31. LinX, NovotnyM, SöderhällK, SöderhällI (2010) Ancient cytokines, the role of astakines as hematopoietic growth factors. J Biol Chem 285 : 28577–28586.
32. WatthanasurorotA, SöderhällK, JiravanichpaisalP, SöderhällI (2011) An ancient cytokine, astakine, mediates circadian regulation of invertebrate hematopoiesis. Cell Mol Life Sci 68 : 315–323.
33. TildenAR, BrauchR, BallR, JanzeAM, GhaffariAH, et al. (2003) Modulatory effects of melatonin on behavior, hemolymph metabolites, and neurotransmitter release in crayfish. Brain Res 992 : 252–262.
34. Ruiz CarrilloD, ChandrasekaranR, NilssonM, CornvikT, LiewCW, et al. (2012) Structure of human Rack1 protein at a resolution of 2.45 A. Acta Crystallogr Sect F Struct Biol Cryst Commun 68 : 867–872.
35. Mendez-FerrerS, ChowA, MeradM, FrenettePS (2009) Circadian rhythms influence hematopoietic stem cells. Curr Opin Hematol 16 : 235–242.
36. MonnierJ, SamsonM (2008) Cytokine properties of prokineticins. FEBS J 275 : 4014–4021.
37. NegriL, LattanziR, GianniniE, CanestrelliM, NicotraA, et al. (2009) Bv8/Prokineticins and their Receptors A New Pronociceptive System. Int Rev Neurobiol 85 : 145–157.
38. LinX, SöderhällI (2011) Crustacean hematopoiesis and the astakine cytokines. Blood 117 : 6417–6424.
39. ProsserHM, BradleyA, CheshamJE, EblingFJ, HastingsMH, et al. (2007) Prokineticin receptor 2 (Prokr2) is essential for the regulation of circadian behavior by the suprachiasmatic nuclei. Proc Natl Acad Sci U S A 104 : 648–653.
40. NegriL, LattanziR, GianniniE, ColucciMA, MignognaG, et al. (2005) Biological activities of Bv8 analogues. Br J Pharmacol 146 : 625–632.
41. ChenJ, KueiC, SuttonS, WilsonS, YuJ, et al. (2005) Identification and pharmacological characterization of prokineticin 2 beta as a selective ligand for prokineticin receptor 1. Mol Pharmacol 67 : 2070–2076.
42. UrayamaK, GuiliniC, MessaddeqN, HuK, SteenmanM, et al. (2007) The prokineticin receptor-1 (GPR73) promotes cardiomyocyte survival and angiogenesis. FASEB J 21 : 2980–2993.
43. HardelandR, CardinaliDP, SrinivasanV, SpenceDW, BrownGM, et al. (2011) Melatonin–a pleiotropic, orchestrating regulator molecule. Prog Neurobiol 93 : 350–384.
44. VerdeMA, Barriga-MontoyaC, Fuentes-PardoB (2007) Pigment dispersing hormone generates a circadian response to light in the crayfish, Procambarus clarkii. Comp Biochem Physiol A Mol Integr Physiol 147 : 983–992.
45. ShearmanLP, SriramS, WeaverDR, MaywoodES, ChavesI, et al. (2000) Interacting molecular loops in the mammalian circadian clock. Science 288 : 1013–1019.
46. OishiK, SakamotoK, OkadaT, NagaseT, IshidaN (1998) Antiphase circadian expression between BMAL1 and period homologue mRNA in the suprachiasmatic nucleus and peripheral tissues of rats. Biochem Biophys Res Commun 253 : 199–203.
47. IshikawaT, HirayamaJ, KobayashiY, TodoT (2002) Zebrafish CRY represses transcription mediated by CLOCK-BMAL heterodimer without inhibiting its binding to DNA. Genes Cells 7 : 1073–1086.
48. BaeK, LeeC, SidoteD, ChuangKY, EderyI (1998) Circadian regulation of a Drosophila homolog of the mammalian Clock gene: PER and TIM function as positive regulators. Mol Cell Biol 18 : 6142–6151.
49. GlossopNR, LyonsLC, HardinPE (1999) Interlocked feedback loops within the Drosophila circadian oscillator. Science 286 : 766–768.
50. GiebultowiczJM, StanewskyR, HallJC, HegeDM (2000) Transplanted Drosophila excretory tubules maintain circadian clock cycling out of phase with the host. Curr Biol 10 : 107–110.
51. YamazakiS, NumanoR, AbeM, HidaA, TakahashiR, et al. (2000) Resetting central and peripheral circadian oscillators in transgenic rats. Science 288 : 682–685.
52. LeeC, BaeK, EderyI (1998) The Drosophila CLOCK protein undergoes daily rhythms in abundance, phosphorylation, and interactions with the PER-TIM complex. Neuron 21 : 857–867.
53. LeeC, BaeK, EderyI (1999) PER and TIM inhibit the DNA binding activity of a Drosophila CLOCK-CYC/dBMAL1 heterodimer without disrupting formation of the heterodimer: a basis for circadian transcription. Mol Cell Biol 19 : 5316–5325.
Štítky
Genetika Reprodukční medicína
Článek Ubiquitous Polygenicity of Human Complex Traits: Genome-Wide Analysis of 49 Traits in KoreansČlánek Alternative Splicing and Subfunctionalization Generates Functional Diversity in Fungal ProteomesČlánek RFX Transcription Factor DAF-19 Regulates 5-HT and Innate Immune Responses to Pathogenic Bacteria inČlánek Surveillance-Activated Defenses Block the ROS–Induced Mitochondrial Unfolded Protein ResponseČlánek Deficiency Reduces Adipose OXPHOS Capacity and Triggers Inflammation and Insulin Resistance in Mice
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 3
-
Všechny články tohoto čísla
- Power and Predictive Accuracy of Polygenic Risk Scores
- Rare Copy Number Variants Are a Common Cause of Short Stature
- Coordination of Flower Maturation by a Regulatory Circuit of Three MicroRNAs
- Ubiquitous Polygenicity of Human Complex Traits: Genome-Wide Analysis of 49 Traits in Koreans
- Genomic Evidence for Island Population Conversion Resolves Conflicting Theories of Polar Bear Evolution
- Mechanistic Insight into the Pathology of Polyalanine Expansion Disorders Revealed by a Mouse Model for X Linked Hypopituitarism
- Genome-Wide Association Study and Gene Expression Analysis Identifies as a Predictor of Response to Etanercept Therapy in Rheumatoid Arthritis
- Problem Solved: An Interview with Sir Edwin Southern
- Long Interspersed Element–1 (LINE-1): Passenger or Driver in Human Neoplasms?
- Mouse HFM1/Mer3 Is Required for Crossover Formation and Complete Synapsis of Homologous Chromosomes during Meiosis
- Alternative Splicing and Subfunctionalization Generates Functional Diversity in Fungal Proteomes
- A WRKY Transcription Factor Recruits the SYG1-Like Protein SHB1 to Activate Gene Expression and Seed Cavity Enlargement
- Microhomology-Mediated Mechanisms Underlie Non-Recurrent Disease-Causing Microdeletions of the Gene or Its Regulatory Domain
- Ancient Evolutionary Trade-Offs between Yeast Ploidy States
- Differential Evolutionary Fate of an Ancestral Primate Endogenous Retrovirus Envelope Gene, the EnvV , Captured for a Function in Placentation
- A Feed-Forward Loop Coupling Extracellular BMP Transport and Morphogenesis in Wing
- The Tomato Yellow Leaf Curl Virus Resistance Genes and Are Allelic and Code for DFDGD-Class RNA–Dependent RNA Polymerases
- The U-Box E3 Ubiquitin Ligase TUD1 Functions with a Heterotrimeric G α Subunit to Regulate Brassinosteroid-Mediated Growth in Rice
- Role of the DSC1 Channel in Regulating Neuronal Excitability in : Extending Nervous System Stability under Stress
- –Independent Phenotypic Switching in and a Dual Role for Wor1 in Regulating Switching and Filamentation
- Pax6 Regulates Gene Expression in the Vertebrate Lens through miR-204
- Blood-Informative Transcripts Define Nine Common Axes of Peripheral Blood Gene Expression
- Genetic Architecture of Skin and Eye Color in an African-European Admixed Population
- Fine Characterisation of a Recombination Hotspot at the Locus and Resolution of the Paradoxical Excess of Duplications over Deletions in the General Population
- Estrogen Mediated-Activation of miR-191/425 Cluster Modulates Tumorigenicity of Breast Cancer Cells Depending on Estrogen Receptor Status
- Complex Patterns of Genomic Admixture within Southern Africa
- Yap- and Cdc42-Dependent Nephrogenesis and Morphogenesis during Mouse Kidney Development
- Molecular Networks of Human Muscle Adaptation to Exercise and Age
- Alp/Enigma Family Proteins Cooperate in Z-Disc Formation and Myofibril Assembly
- Polycomb Group Gene Regulates Rice () Seed Development and Grain Filling via a Mechanism Distinct from
- RFX Transcription Factor DAF-19 Regulates 5-HT and Innate Immune Responses to Pathogenic Bacteria in
- Distinct Molecular Strategies for Hox-Mediated Limb Suppression in : From Cooperativity to Dispensability/Antagonism in TALE Partnership
- A Natural Polymorphism in rDNA Replication Origins Links Origin Activation with Calorie Restriction and Lifespan
- TDP2–Dependent Non-Homologous End-Joining Protects against Topoisomerase II–Induced DNA Breaks and Genome Instability in Cells and
- Recurrent Rearrangement during Adaptive Evolution in an Interspecific Yeast Hybrid Suggests a Model for Rapid Introgression
- Genome-Wide Association Study in Mutation Carriers Identifies Novel Loci Associated with Breast and Ovarian Cancer Risk
- Coincident Resection at Both Ends of Random, γ–Induced Double-Strand Breaks Requires MRX (MRN), Sae2 (Ctp1), and Mre11-Nuclease
- Identification of a -Specific Modifier Locus at 6p24 Related to Breast Cancer Risk
- A Novel Function for the Hox Gene in the Male Accessory Gland Regulates the Long-Term Female Post-Mating Response in
- Tdp2: A Means to Fixing the Ends
- A Novel Role for the RNA–Binding Protein FXR1P in Myoblasts Cell-Cycle Progression by Modulating mRNA Stability
- Association Mapping and the Genomic Consequences of Selection in Sunflower
- Histone Deacetylase 2 (HDAC2) Regulates Chromosome Segregation and Kinetochore Function via H4K16 Deacetylation during Oocyte Maturation in Mouse
- A Novel Mutation in the Upstream Open Reading Frame of the Gene Causes a MEN4 Phenotype
- Ataxin1L Is a Regulator of HSC Function Highlighting the Utility of Cross-Tissue Comparisons for Gene Discovery
- Human Spermatogenic Failure Purges Deleterious Mutation Load from the Autosomes and Both Sex Chromosomes, including the Gene
- A Conserved Upstream Motif Orchestrates Autonomous, Germline-Enriched Expression of piRNAs
- Statistical Analysis Reveals Co-Expression Patterns of Many Pairs of Genes in Yeast Are Jointly Regulated by Interacting Loci
- Matefin/SUN-1 Phosphorylation Is Part of a Surveillance Mechanism to Coordinate Chromosome Synapsis and Recombination with Meiotic Progression and Chromosome Movement
- A Role for the Malignant Brain Tumour (MBT) Domain Protein LIN-61 in DNA Double-Strand Break Repair by Homologous Recombination
- The Population and Evolutionary Dynamics of Phage and Bacteria with CRISPR–Mediated Immunity
- Long Noncoding RNA MALAT1 Controls Cell Cycle Progression by Regulating the Expression of Oncogenic Transcription Factor B-MYB
- Surveillance-Activated Defenses Block the ROS–Induced Mitochondrial Unfolded Protein Response
- DNA Topoisomerase III Localizes to Centromeres and Affects Centromeric CENP-A Levels in Fission Yeast
- Genome-Wide Control of RNA Polymerase II Activity by Cohesin
- Divergent Selection Drives Genetic Differentiation in an R2R3-MYB Transcription Factor That Contributes to Incipient Speciation in
- NODULE INCEPTION Directly Targets Subunit Genes to Regulate Essential Processes of Root Nodule Development in
- Spreading of a Prion Domain from Cell-to-Cell by Vesicular Transport in
- Deficiency in Origin Licensing Proteins Impairs Cilia Formation: Implications for the Aetiology of Meier-Gorlin Syndrome
- Deficiency Reduces Adipose OXPHOS Capacity and Triggers Inflammation and Insulin Resistance in Mice
- The Conserved SKN-1/Nrf2 Stress Response Pathway Regulates Synaptic Function in
- Functional Genomic Analysis of the Regulatory Network in
- Astakine 2—the Dark Knight Linking Melatonin to Circadian Regulation in Crustaceans
- CRL2 E3-Ligase Regulates Proliferation and Progression through Meiosis in the Germline
- Both the Caspase CSP-1 and a Caspase-Independent Pathway Promote Programmed Cell Death in Parallel to the Canonical Pathway for Apoptosis in
- PRMT4 Is a Novel Coactivator of c-Myb-Dependent Transcription in Haematopoietic Cell Lines
- A Copy Number Variant at the Locus Likely Confers Risk for Canine Squamous Cell Carcinoma of the Digit
- Evidence of Gene–Environment Interactions between Common Breast Cancer Susceptibility Loci and Established Environmental Risk Factors
- HIV Infection Disrupts the Sympatric Host–Pathogen Relationship in Human Tuberculosis
- Trans-Ethnic Fine-Mapping of Lipid Loci Identifies Population-Specific Signals and Allelic Heterogeneity That Increases the Trait Variance Explained
- A Gene Transfer Agent and a Dynamic Repertoire of Secretion Systems Hold the Keys to the Explosive Radiation of the Emerging Pathogen
- The Role of ATM in the Deficiency in Nonhomologous End-Joining near Telomeres in a Human Cancer Cell Line
- Dynamic Circadian Protein–Protein Interaction Networks Predict Temporal Organization of Cellular Functions
- Nuclear Myosin 1c Facilitates the Chromatin Modifications Required to Activate rRNA Gene Transcription and Cell Cycle Progression
- Robust Prediction of Expression Differences among Human Individuals Using Only Genotype Information
- A Single Cohesin Complex Performs Mitotic and Meiotic Functions in the Protist
- The Role of the Arabidopsis Exosome in siRNA–Independent Silencing of Heterochromatic Loci
- Elevated Expression of the Integrin-Associated Protein PINCH Suppresses the Defects of Muscle Hypercontraction Mutants
- Twist1 Controls a Cell-Specification Switch Governing Cell Fate Decisions within the Cardiac Neural Crest
- Genome-Wide Testing of Putative Functional Exonic Variants in Relationship with Breast and Prostate Cancer Risk in a Multiethnic Population
- Heteroduplex DNA Position Defines the Roles of the Sgs1, Srs2, and Mph1 Helicases in Promoting Distinct Recombination Outcomes
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Fine Characterisation of a Recombination Hotspot at the Locus and Resolution of the Paradoxical Excess of Duplications over Deletions in the General Population
- Molecular Networks of Human Muscle Adaptation to Exercise and Age
- Recurrent Rearrangement during Adaptive Evolution in an Interspecific Yeast Hybrid Suggests a Model for Rapid Introgression
- Genome-Wide Association Study and Gene Expression Analysis Identifies as a Predictor of Response to Etanercept Therapy in Rheumatoid Arthritis
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



