-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
Reklama–Independent Phenotypic Switching in and a Dual Role for Wor1 in Regulating Switching and Filamentation
Phenotypic switching allows for rapid transitions between alternative cell states and is important in pathogenic fungi for colonization and infection of different host niches. In Candida albicans, the white-opaque phenotypic switch plays a central role in regulating the program of sexual mating as well as interactions with the mammalian host. White-opaque switching is controlled by genes encoded at the MTL (mating-type-like) locus that ensures that only a or α cells can switch from the white state to the mating-competent opaque state, while a/α cells are refractory to switching. Here, we show that the related pathogen C. tropicalis undergoes white-opaque switching in all three cell types (a, α, and a/α), and thus switching is independent of MTL control. We also demonstrate that C. tropicalis white cells are themselves mating-competent, albeit at a lower efficiency than opaque cells. Transcriptional profiling of C. tropicalis white and opaque cells reveals significant overlap between switch-regulated genes in MTL homozygous and MTL heterozygous cells, although twice as many genes are white-opaque regulated in a/α cells as in a cells. In C. albicans, the transcription factor Wor1 is the master regulator of the white-opaque switch, and we show that Wor1 also regulates switching in C. tropicalis; deletion of WOR1 locks a, α, and a/α cells in the white state, while WOR1 overexpression induces these cells to adopt the opaque state. Furthermore, we show that WOR1 overexpression promotes both filamentous growth and biofilm formation in C. tropicalis, independent of the white-opaque switch. These results demonstrate an expanded role for C. tropicalis Wor1, including the regulation of processes necessary for infection of the mammalian host. We discuss these findings in light of the ancestral role of Wor1 as a transcriptional regulator of the transition between yeast form and filamentous growth.
Published in the journal: . PLoS Genet 9(3): e32767. doi:10.1371/journal.pgen.1003369
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003369Summary
Phenotypic switching allows for rapid transitions between alternative cell states and is important in pathogenic fungi for colonization and infection of different host niches. In Candida albicans, the white-opaque phenotypic switch plays a central role in regulating the program of sexual mating as well as interactions with the mammalian host. White-opaque switching is controlled by genes encoded at the MTL (mating-type-like) locus that ensures that only a or α cells can switch from the white state to the mating-competent opaque state, while a/α cells are refractory to switching. Here, we show that the related pathogen C. tropicalis undergoes white-opaque switching in all three cell types (a, α, and a/α), and thus switching is independent of MTL control. We also demonstrate that C. tropicalis white cells are themselves mating-competent, albeit at a lower efficiency than opaque cells. Transcriptional profiling of C. tropicalis white and opaque cells reveals significant overlap between switch-regulated genes in MTL homozygous and MTL heterozygous cells, although twice as many genes are white-opaque regulated in a/α cells as in a cells. In C. albicans, the transcription factor Wor1 is the master regulator of the white-opaque switch, and we show that Wor1 also regulates switching in C. tropicalis; deletion of WOR1 locks a, α, and a/α cells in the white state, while WOR1 overexpression induces these cells to adopt the opaque state. Furthermore, we show that WOR1 overexpression promotes both filamentous growth and biofilm formation in C. tropicalis, independent of the white-opaque switch. These results demonstrate an expanded role for C. tropicalis Wor1, including the regulation of processes necessary for infection of the mammalian host. We discuss these findings in light of the ancestral role of Wor1 as a transcriptional regulator of the transition between yeast form and filamentous growth.
Introduction
The incidence of opportunistic fungal infections has increased in recent years as a result of immunosuppressive diseases such as AIDS, as well as the use of immunosuppressive drugs in modern medical practices [1]. Candida species are typically harmless commensals of humans but are also important fungal pathogens, responsible for both systemic and mucosal opportunistic infections [2]. Most clinically relevant species belong to the Candida clade of hemiascomycete yeasts, which diverged from the model yeast Saccharomyces cerevisiae 300–600 million years ago [3], [4]. Three Candida clade pathogens, Candida albicans, Candida dubliniensis, and Candida tropicalis, have been shown to undergo an epigenetic switch between distinct ‘white’ and ‘opaque’ states [5]–[8], and this phenotypic switch plays a crucial role in modulating behavior. The white-opaque switch has been extensively studied in C. albicans, where the two states differ in metabolic preferences [9], environmental responses [10]–[14], interactions with host immune cells [15], [16], and the ability to undergo sexual reproduction [17], [18].
C. albicans and C. dubliniensis are closely related species that can undergo productive mating with one another [7]. While C. albicans represents the most commonly isolated Candida species in the clinic, C. dubliniensis is rarely found in infections, which may reflect the more limited ability of this species to undergo filamentation [2], [19]. C. tropicalis is also a prevalent human pathogen, particularly in individuals with neutropenia or hematologic malignancies [2], and shows a similar overall genome structure to that of C. albicans, including synteny at the mating-type-like (MTL) locus [20]. C. albicans and C. tropicalis both contain more than 6,000 protein-coding genes, and analysis of the 5,254 orthologs shared between the two species indicates an average protein sequence identity of ∼70% [20]. However, relatively little is known about the biology of C. tropicalis compared to that of the model species C. albicans, including the factors that promote pathogenesis in the mammalian host.
In all three Candida species, white and opaque forms are distinguished by differences in cell shape, colony morphology, and gene expression profiles. White cells are generally round and give rise to smooth, shiny colonies, while opaque cells are elongated and produce duller, darker colonies [5], [6]. The transcriptional profiles of white and opaque forms are also significantly different, with one-sixth of the transcriptome regulated by the switch in C. albicans [9], [21], [22]. Furthermore, white and opaque forms exhibit differences in the propensity to undergo filamentous growth; C. albicans white cells are induced to filament in response to multiple environmental stimuli while opaque cells do not generally undergo filamentation [23], [24].
The white-opaque switch plays a particularly prominent role in regulating mating, as only cells in the opaque state undergo efficient conjugation [5], [18]. Switching is regulated by WOR1 such that loss of this gene prevents formation of the opaque state while, in C. albicans, WOR1 overexpression drives cells into the opaque state [5], [25]–[27]. Genes encoded at the MTL locus control white-opaque switching in C. albicans; only a or α cells switch to the opaque state as a complex between MTLa1 and MTLα2 proteins blocks a/α cell switching due to repression of WOR1 [18], [21], [26]. An analysis of 220 clinical isolates of C. albicans further demonstrated that only MTL homozygous (a/a or α/α) strains could form stable opaque cells at a detectable frequency [17]. MTL regulation ensures that opaque formation only occurs in cells that have the potential to undergo mating, which can take place between MTLa and MTLα cells or via same-sex mating of these cell types [18], [28].
In addition to Wor1, three other transcription factors, Czf1, Wor2, and Efg1, regulate the C. albicans white-opaque switch via a network of positive and negative feedback loops [29]. While Wor1 is the master regulator of the opaque state, Czf1 and Wor2 also play positive roles in promoting formation of opaque cells, while Efg1 antagonizes opaque formation and promotes switching to the white state [29]. Surprisingly, the mechanism regulating white-opaque switching in C. tropicalis appears distinct from that in C. albicans; while switching is dependent on Wor1 in both species, the three associated network transcription factors are not white-opaque regulated in C. tropicalis [5]. It is therefore likely that significant differences exist between the mechanisms regulating phenotypic switching in C. albicans and C. tropicalis [5].
The white-opaque switch is thought to have evolved relatively recently in the Candida clade, probably just prior to the divergence of C. albicans/C. dubliniensis and C. tropicalis [5], [8]. Although this phenotypic switch is limited to within the Candida clade, the master transcriptional regulator Wor1 is conserved across the ascomycete lineage [30]. In S. cerevisiae, the Wor1 homolog Mit1 acts as part of a transcriptional network to regulate pseudohyphal formation [31], while in the more distantly related ascomycete Histoplasma capsulatum, the Wor1 homolog Ryp1 is a master regulator of the transition between yeast and mycelial forms [32]. It has therefore been proposed that the ancestral role of Wor1 was the transcriptional regulation of morphological changes, including the control of filamentous growth [31].
In this study, we investigated the white-opaque transition in C. tropicalis and uncovered several features that further distinguish it from the analogous switch in C. albicans. We first demonstrate that a stable white-opaque switch can occur in C. tropicalis a/α cells, indicating that the a1/α2 complex does not repress switching in this species. Transcriptional profiling reveals that genes regulated by the white-opaque switch in a/α cells show a significant overlap with those regulated by the switch in a or α cell types. However, many white-opaque regulated genes are unique to MTL heterozygous or MTL homozygous cells, indicating that the MTL configuration significantly impacts gene expression in the two phenotypic states. Despite these transcriptional differences, the phenotypic switch in all three cell types (a, α, and a/α) is dependent on the master regulator, Wor1; deletion of Wor1 blocked white-to-opaque switching while overexpression of Wor1 forced cells into the opaque state. We further show that Wor1 promotes filamentous growth in C. tropicalis, as elevated WOR1 expression led to a significant increase in both filamentous growth and biofilm formation. These studies provide new insights into the evolution of Wor1 homologs as conserved regulators of filamentation and epigenetic switching.
Results
Comparative Analysis of Mating in C. tropicalis White and Opaque Cells
A key feature of the C. albicans white-opaque switch is that a/α cells are locked in the white state due to repression of WOR1 transcription by the a1/α2 heterodimer. A direct consequence of this control is that a/α mating products formed between opaque a and α cells typically form white colonies [18]. Rare opaque colonies (<5%) were observed in C. albicans mating products, but in each case these colonies had undergone loss of a1 or α2, thereby relieving repression of WOR1 and allowing propagation of the opaque state [18]. In C. tropicalis, the white-opaque switch also regulates mating, although the difference in mating efficiency between white and opaque forms is less striking than that in C. albicans. Whereas C. albicans opaque cells mate a million times more efficiently than white cells [18], mating of C. tropicalis opaque cells is only about a hundred times greater than that of white cells [5].
These observations led us to hypothesize that C. tropicalis white cells may be capable of appreciable mating frequencies, even without switching to the opaque state. Close inspection of C. tropicalis a/α mating products revealed that colonies generated by mating white a and α cells were distinct from those formed by mating opaque cells. This is illustrated in Figure 1A, where colonies formed from mating between white a and α cells exhibited a shiny, smooth appearance, while colonies formed by products of mating between opaque cells were consistently duller and darker. Examination of the cells from these colonies revealed that the products of white×white mating were round, resembling classical white cells, while the products of opaque × opaque mating were elongated, reminiscent of opaque cells (Figure 1B). PCR analysis of the mating products confirmed that a1 and α2 were still present in the products formed from mating white or opaque cells (Figure 1C). The a1 and α2 loci were also sequenced and shown to exactly match the published gene sequences [20], indicating that these genes were not mutated in the C. tropicalis strains used for these experiments. Together, these results indicate that the C. tropicalis white-opaque switch is not under strict regulation by the MTL locus. Thus, unlike C. albicans, mating of C. tropicalis opaque a and α cells (and formation of the a1/α2 complex) does not force cells back to the white form. Instead, C. tropicalis a/α mating products inherit the phenotype of their parental cells and can even stably propagate in the opaque form.
Fig. 1. Products of C. tropicalis white×white and opaque×opaque mating maintain parental phenotypes. 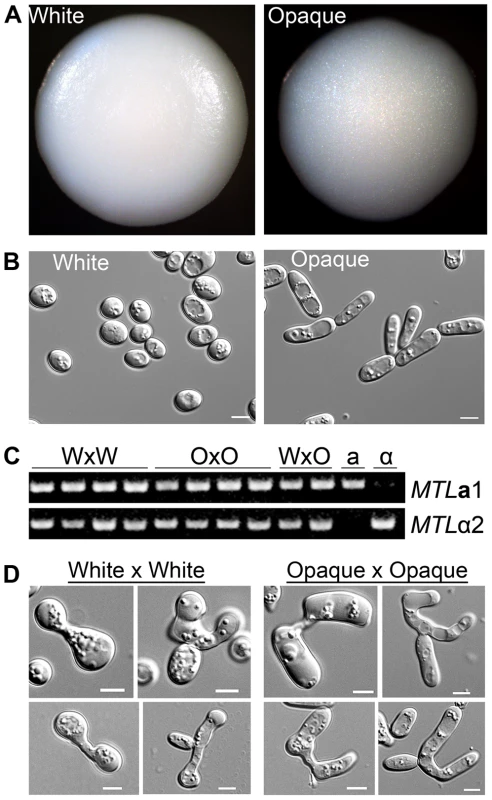
(A) Colony morphology of mating products. Left, product of mating C. tropicalis white a (CAY3376) with white α (CAY3391) cells. Right, product of mating opaque a (CAY3378) with opaque α (CAY3392) cells. Colonies were grown on SCD medium lacking histidine and arginine for 3 days at room temperature to select for mating products. (B) Cell morphology of cells taken from the corresponding colonies. Scale bars = 5 µm. (C) PCR to verify the presence of MTLa1 and MTLα2 genes in white×white (WxW) mating products, opaque×opaque (OxO) mating products, white×opaque (WxO) mating products, and in the parental a and α strains. (D) Left, pictures of zygotes analyzed from white×white mating experiments and, right, pictures of zygotes analyzed from opaque×opaque mating experiments. Scale bars = 5 µm. These experiments reveal another important aspect of C. tropicalis biology, as they establish that white cells can mate without switching to the opaque form. Our observation that the products of white and opaque cell mating are distinguishable, and that white×white mating generates white mating products, indicates that C. tropicalis white cells can directly undergo conjugation with one another. In support of the fact that both white and opaque cells can mate, we also observed structural differences in zygotes formed between white cells and those formed between opaque cells. Zygotes formed from white cells were generally smaller and contained rounder cells than zygotes formed from opaque cells (Figure 1D). We conclude that C. tropicalis white cells are competent for mating, albeit at a lower efficiency than that of opaque cells.
To complete this analysis, we also examined the products of mating in crosses between white and opaque cells. Interestingly, the products of these mixed crosses were predominantly opaque (data not shown). Since Wor1 expression is essential for induction of the opaque form, we surmise that Wor1 protein from the parental opaque cell is present at sufficient levels in the zygote to result in stable propagation of the opaque state in these mating products.
Phenotypic Switching in C. tropicalis a/α Cells
A previous study by Xie et al. indicated that a/α cells of C. tropicalis were able to form opaque cells based on similar cell morphologies to a/a and α/α opaque cell types [8]. To address whether a/α cells could directly switch to the opaque state, cells were grown on medium containing N-acetylglucosamine, as this was previously shown to induce C. tropicalis switching [8]. Rare colony switching was observed with darker sectors forming at the edge of a/α colonies, similar to that in conventional white-to-opaque sectoring of a and α colonies (Figure 2A). Cells from the a/α sectors were elongated rather than spherical, a characteristic feature of opaque cells [5]. Additionally, white a/α cells had a smooth surface while opaque a/α cells had an uneven or pimpled surface, and thus resembled conventional white and opaque cells (Figure S1A and [33]). Once formed, diploid opaque a/α cells were stably maintained in the opaque state, similar to tetraploid opaque a/α mating products (Figure 1A and data not shown). The frequency of white-to-opaque switching in a/a and a/α cells varied widely from experiment to experiment (0–7% switching), but the difference in switching between strains was not statistically significant.
Fig. 2. C. tropicalis a/α cells undergo a white-opaque phenotypic switch related to that in a and α cells. 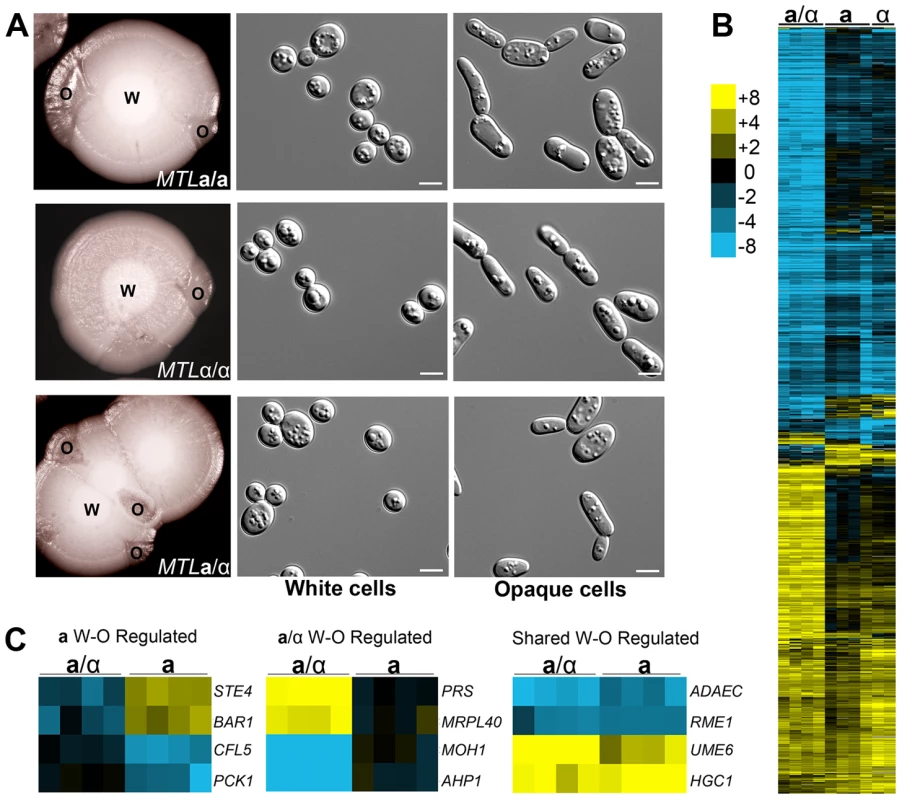
(A) White (W) colonies sectoring to opaque (O) in a (CAY1503), α (CAY1505), and a/α (CAY1511) strains and the corresponding cell morphologies observed from these colonies. Cells were grown in SCD at 37°C for 5 days and plated to Lee's + N-acetylglucosamine at room temperature for 10 days. Scale bars = 5 µm. (B) Opaque-white gene expression profiles of a/α, a, and α cells. cDNA prepared from white and opaque states of CAY1511 (a/α), CAY1504 (a), and CAY1505 (α) in independent experiments was hybridized against a universal reference. Opaque cell gene expression was divided by that in white cells and filtered for genes with a fold-change greater than 4 and clustered by Average Linkage Clustering. Genes with elevated expression in white cells are shown in blue and genes upregulated in opaque cells are shown in yellow. (C) Examples of genes whose expression was white-opaque regulated in a cells (left), a/α cells (middle), or in both a and a/α cells (right). As the a1/α2 complex acts to repress white-to-opaque switching in C. albicans, we also examined whether the a1 and α2 genes were expressed in C. tropicalis opaque a/α cells. Reverse transcription (RT)-PCR was performed on each cell type and revealed that a1 and α2 genes were actively expressed in a/α cells regardless of phenotype (Figure S1B). This result indicates that the white-to-opaque switch in C. tropicalis appears independent of a1 and α2 function.
We further examined 8 additional C. tropicalis clinical a/α strains for their ability to undergo the white-opaque switch. Opaque formation was observed in 2 of these strains, indicating that other a/α strains are able to undergo the white-opaque switch (Table S1). The presence of the a1 and α2 genes in switching strains was confirmed by PCR. These genes were sequenced from opaque cells derived from one of the three clinical a/α strains and also found to match the published sequence (data not shown). Our results therefore establish that C. tropicalis white-opaque switching occurs independent of MTL control and that multiple a/α strains can switch to a stable opaque state.
Transcriptional Profiling of C. tropicalis White and Opaque States
To ascertain the relationship between genes regulated by the white-opaque switch in MTL heterozygous a/α cells with those in MTL homozygous a/a or α/α cells, transcriptional profiling was performed on cells from both morphological states in each cell type (Figure 2B). SAM (Statistical Analysis of Microarrays) was used to determine gene expression changes that were significant for white-opaque regulated genes in a and a/α cells (see Materials and Methods and Tables S5 and S6). Expression profiles of a/α white and opaque forms showed significant overlap with the corresponding white - and opaque-specific genes in a cells (Figure 2B). In particular, of the 120 genes significantly upregulated in white a cells, 73 of these genes were also upregulated in white a/α cells, while of the 129 genes upregulated in opaque a cells, 22 of these genes were also upregulated in opaque a/α cells (Figure S2). These results establish that C. tropicalis a/α cells undergo a white-opaque switch related to that in MTL homozygous cells [5], and that there is significant overlap between white-opaque genes regulated in MTL homozygous and heterozygous cell types (p<1e-150 and p<1e-20 for white - and opaque-specific genes, respectively).
Despite significant overlap between white-opaque regulated genes in the different cell types, a large number of switch-regulated genes were unique to either MTL heterozygous or MTL homozygous cells. This was particularly striking in a/α cells, where the expression of 549 genes was significantly different between white and opaque forms, while only 249 genes were white-opaque regulated in a cells (Figure 2B, 2C and Figure S2). Many of the additional white-opaque regulated genes in a/α cells were involved in translation, biosynthetic processes, or gene expression based on GO term analysis (Text S1 and Figure S3). This indicates that the switch in a/α cells regulates additional cellular processes to those in a or α cells, and these differences could have important implications for the role of the switch in MTL heterozygous strains.
Conversely, white and opaque a and α cells also differentially expressed genes that were not white-opaque regulated in a/α cells. For example, the mating-related genes STE4 and BAR1 were expressed at higher levels in opaque MTL homozygous cells than in white cells, but were not differentially expressed between white and opaque states in MTL heterozygous cells (Figure 2C). Taken together, these results indicate that MTL homozygous and MTL heterozygous cells share related white-opaque expression profiles, but that the genes regulated by this switch are also influenced by the configuration of the MTL locus.
Mating of C. tropicalis a/α Cell Types
A key role of the white-opaque switch in Candida species is the regulation of sexual reproduction [5], [18]. Given that C. tropicalis a/α cells undergo the white-to-opaque switch, we tested whether these cell types undergo productive mating. We found that a/α cells could not mate with other a/α cells, and that same-sex mating of a or α cells was also not observed (data not shown), in agreement with previous studies [5]. However, low levels of mating were obtained between a/α cells and either a or α cells as partners. The frequency of this a/α cell mating was 103 to 105-fold lower than conventional mating between a and α cells (Figure 3 and Figure S4). Mating could be detected using both white and opaque a/α cells, and although a/α cell mating was higher in the opaque state, this difference was not significant.
Fig. 3. The white-opaque switch regulates C. tropicalis mating in a, α, and a/α cells. 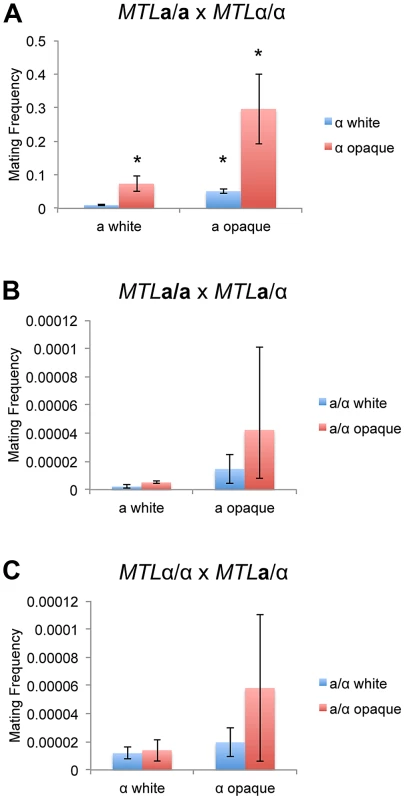
Mating frequency of (A) a x α (CAY1503 x CAY1505), (B) a x a/α (CAY1503 x CAY1511), and (C) α x a/α (CAY1505 x CAY1513) white and opaque cells. Mating experiments were performed on Spider medium for 3 days at room temperature and plated to selective media to quantify mating frequency (see Materials and Methods). Error bars indicate SEM for 3 independent experiments. * = significantly different from white×white crosses, p<0.01. C. albicans a/α cells from the SC5314 background do not undergo mating with other cell types ([18] and data not shown). However, overexpression of WOR1 in these cell types can override a/α control, forcing them to adopt the opaque state and promoting low frequency mating with a or α cells [25]. These studies indicated that productive mating was likely due to loss of MTL genes, allowing C. albicans a/α cells to mate as a or α cells [25]. We similarly suggest that low level mating of C. tropicalis a/α cells is due to MTL instability, as loss of MTL genes was frequently detected in the mating products from a/α crosses (Text S1, Figure S5, Table S11). In addition, genetic recombination at the MTL was observed in a subset of mating products, indicating that homozygosis of the MTL could promote a/α mating (Figure S5). However, regardless of the mechanism involved, mating of a/α cells occurred at a very low frequency compared to conventional mating between a and α cells.
Transcriptional Regulation of C. tropicalis Phenotypic Switching
WOR1 is the master transcriptional regulator of the white-opaque switch in both C. tropicalis [5] and C. albicans [25]–[27], and its expression is therefore critical for opaque cell formation. To further investigate the role of WOR1, we constructed Δwor1/Δwor1 and WOR1 overexpression strains in C. tropicalis a, α, and a/α backgrounds. Initially, a pACT1-WOR1 construct was used to overexpress WOR1. However, unlike in C. albicans, this construct failed to induce switching to opaque in C. tropicalis (data not shown and [8]). Instead, an alternative promoter, TDH3, was utilized for WOR1 expression and found to be sufficient to induce opaque formation in C. tropicalis a, α, and a/α cells (Figure 4A). Thus, WOR1 expression is both necessary and sufficient for formation of the opaque state in C. tropicalis.
Fig. 4. WOR1 is the master regulator of the white-opaque switch in C. tropicalis. 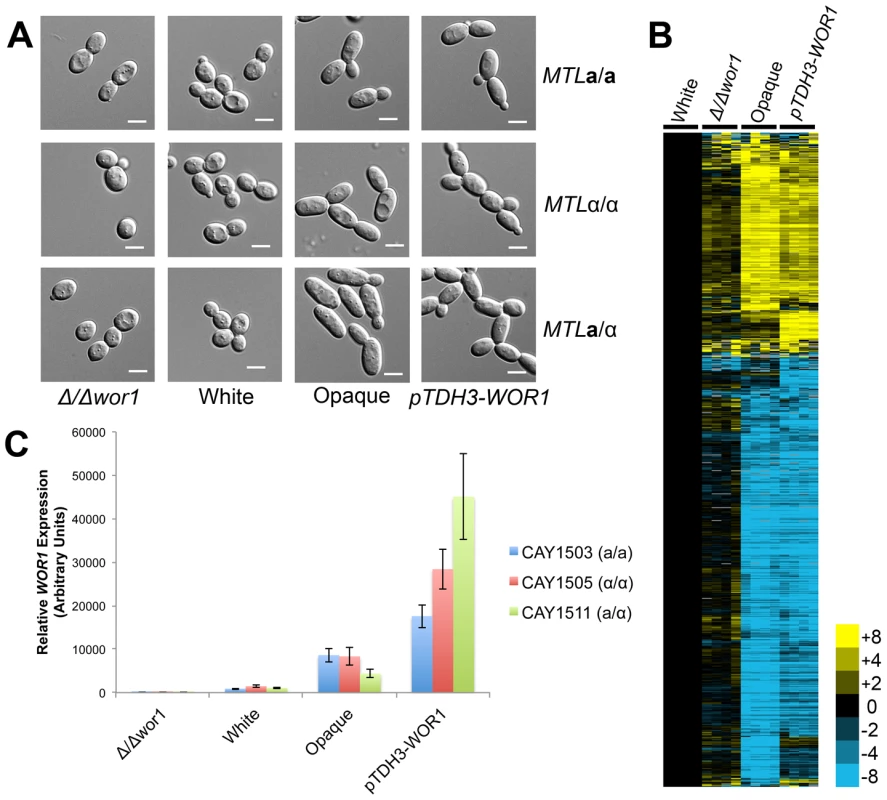
(A) Cell morphologies of Δwor1, white, opaque, and pTDH3-WOR1 (WOR1-overexpressing) strains. Cells were grown in Spider medium at room temperature to 0.8–1.0 OD600. Scale bars = 5 µm. (B) Gene expression in a/α white cells (CAY1511), Δwor1 cells (CAY4043), opaque cells (CAY4048), and pTDH3-WOR1 cells (CAY4045), relative to white cells (CAY1511). Expression profiles for each state were divided by white expression values and filtered for those genes with an expression change greater than 4-fold in 4 or more experiments. (C) WOR1 expression in Δwor1, white, opaque, and pTDH3-WOR1 strains derived from a, α, and a/α cells. Expression levels measured by qRT-PCR. Error bars indicate SEM for replicate experiments from 3 different biological replicates. Previously, we showed that wor1 mutant a or α cells do not undergo detectable mating in C. tropicalis [5]. Mating assays were also performed on WOR1 overexpression strains and these strains exhibited increased mating between a and α cells (∼70% mating) relative to wild-type opaque strains (∼15% mating, Figure S4). This result further establishes the key role of Wor1 in directing expression of genes necessary for efficient conjugation.
Transcriptional profiling was also performed on wor1 mutant and WOR1 overexpression a/α strains, and profiles compared to wild-type white and opaque cells (Figure 4B). Consistent with the key role of WOR1 in directing the white-opaque switch, the expression profile of Δwor1 strains was similar to that of wild-type white cells, while strains overexpressing WOR1 had an expression profile similar to that of wild-type opaque cells (Figure 4B). However, although the global expression patterns were similar, many genes were differentially expressed between Δwor1 strains and white cells (Tables S7 and S8) and between WOR1 overexpression strains and opaque cells (Tables S9 and S10). Expression of WOR1 was also compared by RT-PCR and was found to have the following relative transcription levels: WOR1 overexpresser > wild-type opaque > wild-type white > wor1 mutant (Figure 4C). WOR1 expression was ∼2–8-fold higher in WOR1 overexpression strains relative to opaque strains, ∼4–10-fold higher in opaque cells relative to white cells, and ∼1000-fold higher in white cells relative to wor1 mutants. The difference in expression levels between white cells and wor1 mutants is consistent with low-level expression of this gene in the white state. We also note that WOR1 expression levels were in line with the relative mating efficiencies of these strains (Figure S4).
In C. albicans, the white-opaque switch is regulated by Wor1 together with three other factors, Wor2, Czf1, and Efg1, as part of an interacting transcriptional circuit [29]. The EFG1 gene is missing from the C. tropicalis genome although a gene encoding a related APSES transcription factor, EFH1, is present [20]. Previous studies failed to observe differential expression of WOR2, CZF1, or EFH1 between C. tropicalis white and opaque a cells [5]. We examined the expression of these genes in the larger white-opaque data set present in a/α cells, and between wor1 mutant and WOR1 overexpression a/α strains. CZF1 and EFH1 did not show significant expression differences between these profiles, while WOR2 expression was decreased in opaque cells, the opposite of that expected based on its expression in C. albicans. These results are therefore consistent with transcriptional control of the white-opaque switch being divergent between C. tropicalis and C. albicans (outside of the conserved role of Wor1).
We also identified several other transcription factors regulated by the C. tropicalis white-opaque switch. Among these factors, UME6, a gene known to have a role in regulation of filamentous growth in C. albicans, had consistently higher expression in C. tropicalis opaque cells. Conversely, upregulated genes in white cells included RME1 and NDT802, genes with homologs involved in regulating meiosis in S. cerevisiae, as well as ADAEC, a gene that is also white-specific in C. albicans. Further studies are now required to determine if any of these transcription factors play an active role in regulating the white-opaque switch in C. tropicalis.
Wor1 Regulates Filamentation and Biofilm Formation in C. tropicalis
In several diverse fungal species, Wor1 homologs do not mediate white-opaque phenotypic switching but still act as transcriptional regulators of cellular morphogenesis. This is evident in S. cerevisiae and H. capsulatum, where Wor1 homologs regulate the transition from budding yeast forms to filamentous forms [31], [32]. Colony morphologies of C. tropicalis strains were compared on several media, and it was found that WOR1 overexpression strains were markedly more wrinkled than wild-type white or opaque cells when grown on Spider medium (Figure 5A). Spider medium is a low nutrient medium that also induces filamentation in C. albicans [34]. Examination of cells from the wrinkled colonies confirmed that Wor1 overexpression strains were significantly more filamentous than other cell types due to a higher percentage of both hyphal and pseudohyphal cells (Figure 5B). In addition, filamentation generally increased as WOR1 gene expression levels increased. Thus, for a/α cells, filamentation increased from wor1 mutants (4.4%) to white cells (10.7%) to opaque cells (23.9%) to WOR1 overexpressing cells (32.9%, Figure 5B). These results indicate that filamentation correlates with WOR1 expression in C. tropicalis, and that Wor1 regulates filamentous growth independent of its regulation of the white-opaque switch.
Fig. 5. C. tropicalis WOR1 expression regulates filamentous growth. 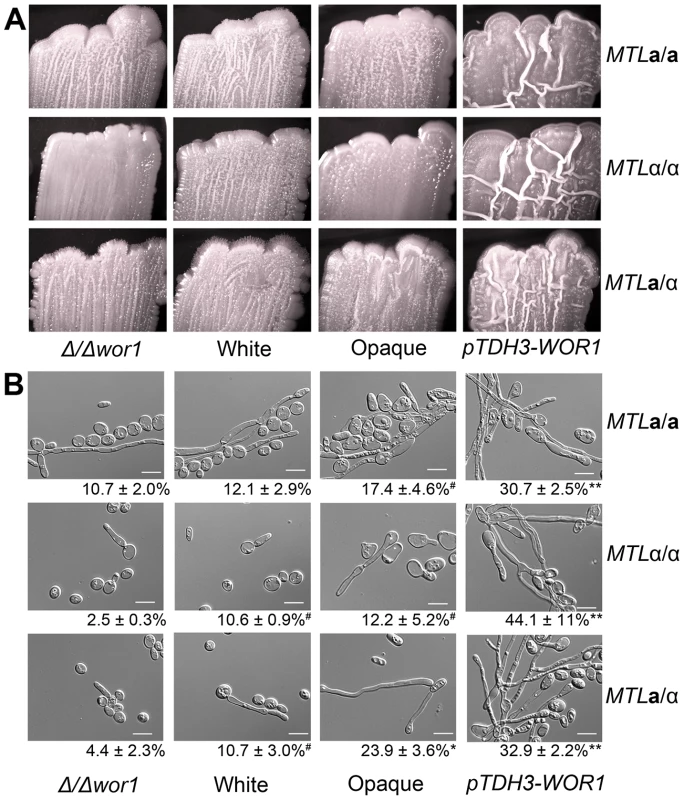
(A) Wild-type white, wild-type opaque, αwor1 mutant, and WOR1-overexpression (pTDH3-WOR1) strains were grown on Spider medium at 37°C for 7 days and photographed. (B) Cells from corresponding patches in (A). The percentage of filamentous cells is shown below each image. Scale bar = 10 µm. ** = significantly different from opaque, white, and Δwor1 strains, p<0.001. * = significantly different from white and Δwor1, p<0.001. # = significantly different from Δwor1, p<0.05. Filamentous growth is directly associated with biofilm formation in C. albicans. Biofilms are surface-associated communities of cells and often involve both yeast and filamentous cells in stable, complex structures that form on biotic and abiotic surfaces [35], [36]. Furthermore, C. albicans biofilms are responsible for the seeding of serious bloodstream infections and are associated with antifungal drug resistance [35], [36]. We therefore examined whether C. tropicalis wor1 mutant cells, white cells, opaque cells, or cells overexpressing WOR1 displayed differences in their ability to form biofilms using an adherence to plastic assay. White cells, opaque cells, and wor1 mutants generally performed poorly in these adherence assays, regardless of MTL configuration (Figure 6A). In contrast, cells overexpressing WOR1 generated robust biofilms on the polystyrene plates, as noted by visual inspection as well as quantification of the biofilm by optical density (Figure 6A). Increased biofilm formation was also demonstrated by an elevated level of staining with crystal violet, which indicates extracellular matrix production (Figure 6B), and by increased XTT reduction, indicating that there were significantly more adherent, viable cells in the WOR1 overexpressing biofilms (Figure 6C). Many cells within the WOR1-overexpressing biofilms were filamentous (22–59%), whereas comparatively few cells were filamentous in white, opaque, or Δwor1 cells (0–12%, Figure 6D).
Fig. 6. Biofilm formation is induced by WOR1 overexpression in C. tropicalis. 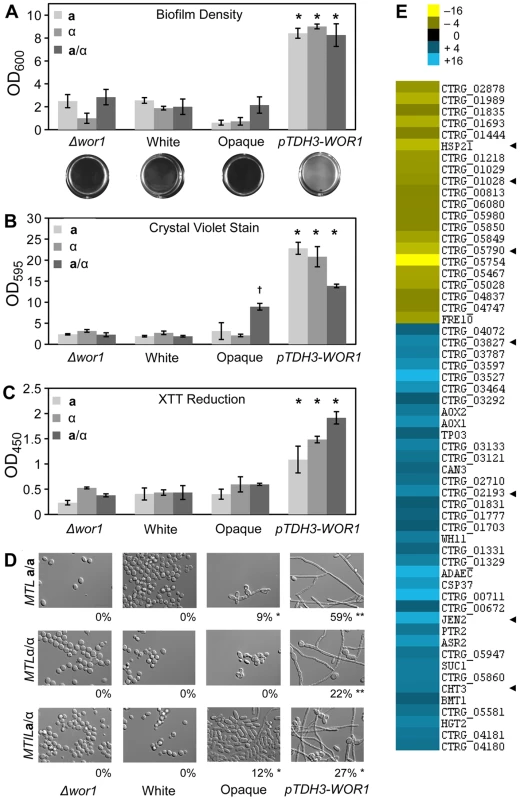
(A) Quantification of cells adhering to a 12-well polystyrene dish following a 48-hour incubation in Lee's medium. Representative images of wells appear below the corresponding strains. Data from three experimental replicates, error bars indicate SD. * = p<0.001 when compared to Δwor1, white, and opaque strains. (B, C) Quantification of biofilm production by colorometric assays. (B) Beta-glucan content was determined by crystal violet staining. (C) Cellular viability was measured by formazan formation upon XTT reduction. Each biofilm experiment includes three or more replicates, error bars indicate S.D. * = p<0.001 when compared to Δwor1, white, and opaque strains. † = p<0.001 when compared to Δwor1, white, and pTDH3-WOR1 strains. (D) Morphology of adherent cells taken from (A). Percentage of filamentous cells indicated below image. Scale bars = 20 µm. Statistical significance was determined using Mann-Whitney pair-wise tests. ** = significantly different from opaque, white, and αwor1 strains, p<0.001. * = significantly different from white and αwor1 strains, p<0.001. (E) Genes induced in MTLa/a WOR1 overexpressing cells under biofilm conditions. Heat map shows relative expression changes between pTDH3-WOR1 (CAY3853) and opaque (CAY3378) strains in the adherence to plastic biofilm assay. Data set was filtered for genes with a fold-change greater than 4 and clustered by Average Linkage Clustering. Data shows average of two independent biological replicates. Arrowheads highlight C. albicans homologs that are biofilm regulated. Transcriptional profiling was performed under biofilm culture conditions on WOR1 overexpression strains and compared to gene expression in wild-type opaque strains. This revealed that 58 genes were differentially expressed (>4-fold) between these strains (Figure 6E). Gene expression changes included CHT3, which encodes a chitinase whose expression is repressed in C. albicans hyphal cells [37], and which was also repressed in the C. tropicalis WOR1 overexpression strain. Conversely, JEN2 was upregulated in C. tropicalis WOR1-mediated biofilms, and encodes a dicarboxylic acid transporter whose expression is also upregulated in C. albicans biofilms [38]. In addition, several C. tropicalis white-specific genes (e.g., ADAEC and WH11) were further downregulated in the WOR1-overexpressing cells relative to opaque cells (Figure 6E). However, no genes with defined roles in filament regulation were obtained from these profiling studies, although many of the differentially regulated genes currently have no known function.
Together, these results demonstrate that Wor1 promotes filamentous growth and biofilm formation in C. tropicalis and these functions are independent of the white-opaque switch. The role of Wor1 in this species is therefore analogous to that of several distantly related Wor1 homologs in the fungal lineage. The increase in filamentous growth is presumably a major reason for the increased adhesion and biofilm formation in the C. tropicalis Wor1-overexpression strains.
Contribution of MTL Configuration to C. tropicalis Fitness
We next determined if the configuration of the MTL locus contributes to overall fitness in C. tropicalis. In particular, we investigated the possibility that a/α cells exhibit increased fitness over MTL homozygous cells. We first analyzed 150 natural C. tropicalis isolates (Table S1) to determine their MTL configuration and found that 119 isolates (∼80%) were a/α strains, while 23 (∼15%) were a strains and 8 (∼5%) were α strains. A similar analysis performed by Xie et al. showed an even higher prevalence of a/α strains (145/150 isolates) in the natural population [8]. Thus, similar to the case in C. albicans [17], [39], a/α genotypes are the predominant cell types in natural C. tropicalis isolates.
We speculated that the predominance of a/α cells could result from increased fitness of these cell types relative to a or αcells. Thus, the ability to switch as an a/α cell could allow cells to retain their optimal fitness yet also be competent to undergo the white-opaque switch. The fitness of C. tropicalis a, α, and a/α strains was addressed using an in vivo model of murine candidiasis. Competition experiments were performed between a/α and a cells, or between a/α and α cells, using strains carrying different auxotrophic markers. Neutropenia was induced prior to infection with C. tropicalis cells as this increases the fungal load associated with systemic disease [40]. Mixtures of strains were injected into the tail veins of neutropenic mice and cells recovered from the brain and kidneys 72 hours following infection and characterized for cell type.
Regardless of the combination of auxotrophic markers used, a/α cells consistently colonized host organs at a higher fungal burden than a or α cells in competition assays (Figure 7). This difference was statistically significant for fungal colonization of the brain, where approximately 70% of the recovered cells were a/α cell types. These experiments indicate that a/α cells are fitter than a or α cells in this in vivo model, as they outcompeted MTL homozygous cell types during systemic infection. We propose that phenotypic switching in a/α cells allows these cells to have the potential to adopt both white and opaque forms while still maintaining their fitness advantage over a or α strains.
Fig. 7. Heterozygosity at the MTL provides a fitness advantage in vivo. 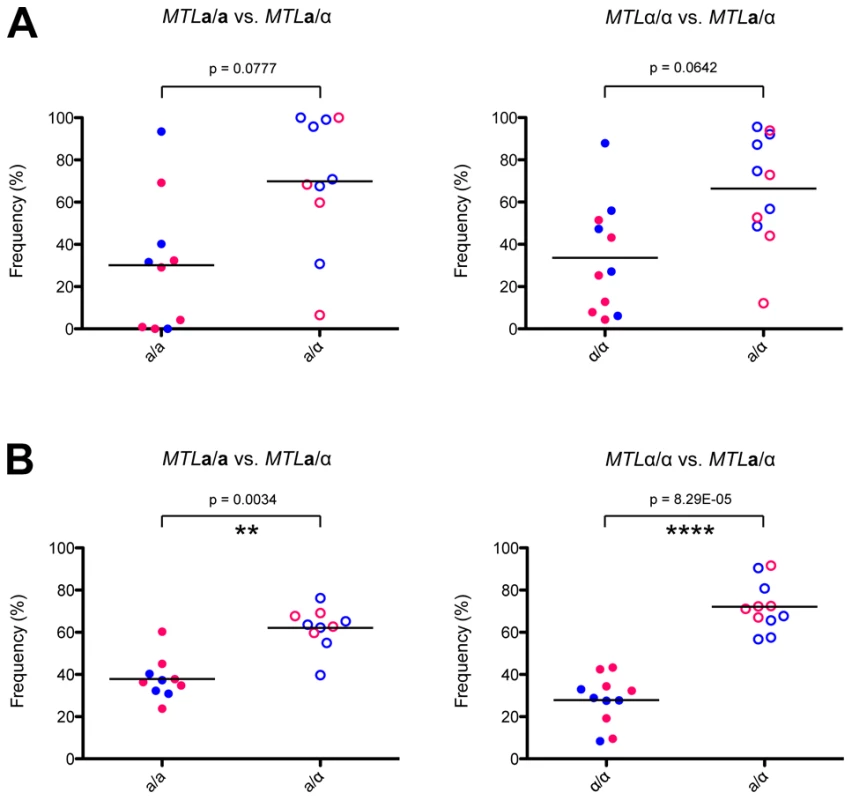
MTL homozygous and heterozygous C. tropicalis cells were mixed 1∶1 and injected systemically via the mouse tail vein. After 72 hours, C. tropicalis cells were recovered from the kidney (A) and the brain (B), and strain genotypes identified by plating to auxotrophic media. In both organs, a/α cells (CAY1511 or CAY1513) showed increased fitness relative to a (CAY1502 or CAY1503) orα (CAY1505 or CAY1509) cells, and this difference was statistically significant in the brain. ** = p<0.01, **** = p<0.0001. Blue circles indicate strains were His-, red circles indicate strains were Arg-. Discussion
In this study, we show that a Wor1-regulated phenotypic switch operates in C. tropicalis a, α, and a/α cell types and is therefore independent of MTL control. Even in strains expressing intact a1 and α2 genes, white cells could stably switch to opaque cells, indicating that MTL genes do not block switching. The regulation of the white-opaque switch in C. tropicalis therefore deviates from that in C. albicans, where the action of a1/α2 prohibits efficient opaque formation in a/α cells [18]. The discovery that C. tropicalis a/α cells can undergo stable formation of the opaque state raises new questions as to the role(s) of the switch in vivo, as well as the transcriptional regulation of the switch in this species. We note that Xie et al. also recently observed a white-opaque-like morphological switch in two C. tropicalis a/α strains [8], and although the integrity of a1 and α2 genes was not reported, it is likely that these strains were also capable of MTL-independent white-opaque switching.
C. tropicalis Mating and the White-Opaque Switch
Despite its apparent independence from MTL control, the C. tropicalis white-opaque switch still regulates sexual mating in this species. Mating between C. tropicalis a and α opaque cells occurs approximately 100 times more efficiently than that between white cells. It was not known, however, if mating of white cells required that cells first switch to the opaque state prior to mating, or if white cells were themselves mating competent. Analysis of C. tropicalis white×white and opaque×opaque mating products addressed this question, as they revealed that mating products inherited the phenotypic state of the parental cells (i.e. mating between white cells generated white mating products while those between opaque cells generated opaque mating products). This simple observation established two important facts about phenotypic switching and mating in C. tropicalis. First, it demonstrated that the C. tropicalis white-opaque switch is independent of MTL status, as a/α cells formed by mating would be expected to be in the white state, regardless of the phenotype of the parental cells. Indeed, this is what has been observed in C. albicans, where the products of mating are white or, if opaque, have undergone concomitant loss of MTL genes [18]. Second, it showed that C. tropicalis white cells are themselves mating competent; if they had switched to opaque prior to mating then the products of mating would also have been opaque. Thus, C. tropicalis white and opaque cells are both mating competent, although the efficiency of mating is higher in the opaque state than in the white state.
The discovery that C. tropicalis white cells are mating competent is interesting given that mating is completely abolished in mutant cells lacking WOR1. Thus, although C. tropicalis white cells mate inefficiently, their mating frequency is still more than 1000-fold higher than that of wor1 mutants [5]. This indicates that significant WOR1 expression occurs in C. tropicalis white cells (supported by array and qPCR data), and that this expression is sufficient for basal induction of genes necessary for conjugation. In opaque cells, Wor1 levels are further increased over white cells (4–10 fold) and higher mating efficiencies are observed in cells with the opaque phenotype. Candidate mating genes include STE4 and BAR1, both of which are elevated in the opaque state and are known to regulate mating in diverse fungal species [41]–[43]. Mating efficiency was further increased in Wor1 overexpression strains compared to wild-type opaque cells. These results demonstrate that mating efficiency can be separated from the white-opaque switch per se and, at least to a first approximation, C. tropicalis mating correlates directly with Wor1 expression levels.
Mating was also observed between C. tropicalis a/α cells and either a or α partners. The efficiency of a/α cell mating was very low (∼10−6) regardless of whether white or opaque cells were used, and was approximately 5 orders of magnitude lower than conventional mating between opaque a and α cells. In fact, mating of a/α cells was 1000-fold lower than that between white a and α cells. Furthermore, the majority of a/α mating products had lost MTL genes or had undergone recombination at the MTL, and therefore had presumably mated as a or α cell types. Aberrant mating of a/α cells has also been described in C. albicans cells overexpressing WOR1 and was also attributed to rare loss of MTL genes [25]. Moreover, it is known that C. albicans a/α cells undergo occasional mating upon loss of a1 or α2 genes, as a1/α2 represses the expression of haploid-specific genes necessary for mating [18], [21].
Profiling of C. tropicalis Phenotypic States
Transcriptional profiling of the three different genotypes (a, α, and a/α) in C. tropicalis revealed that there was significant overlap between white-opaque regulated genes in each cell type. This is to be expected given that Wor1 regulates the switch irrespective of MTL configuration. The key role of this transcription factor in determining the phenotypic state was further illustrated by comparing the profiles of white cells, opaque cells, wor1 mutants, and WOR1 overexpression strains. The profiles of white cells and wor1 mutant cells were similar, as were those of opaque cells and WOR1 overexpressing cells. In addition, wor1 mutant strains did not undergo switching to opaque, while WOR1 overexpression locked cells in the opaque state. Together, these results establish Wor1 as the master regulator of the C. tropicalis white-opaque switch in a/α cells, similar to its role in a and α cells.
Surprisingly, significant differences in white-opaque regulated genes were noted between MTL homozygous and MTL heterozygous strains. For example, STE4 and BAR1 were expressed at elevated levels in opaque a or α cells, but not in opaque a/α cells. This may reflect the fact that the white-opaque switch regulates mating between a and α cells, but does not significantly promote mating in a/α cells. In addition, ∼400 switch-regulated genes were unique to a/α cells and were not observed in a or α cell types. The gene set unique to opaque a/α cells included a significant association with translation, biosynthetic processes, and gene expression control. A direct implication of these transcriptional differences is that behavioral differences (including effects on host interactions and virulence) may occur between white and opaque cells from MTL homozygous and MTL heterozygous cell types, and additional experiments will now test this possibility.
Evolution and Function of Phenotypic Switching in Candida Species
The white-opaque switch affects virtually every aspect of Candida biology, from mating to interactions with host immune cells to pathogenesis [44]–[47]. MTL regulation of the switch in C. albicans ensures that only those cells that are capable of mating undergo the switch to the mating-competent form [18]. The discovery that C. tropicalis can form opaque cells regardless of MTL status suggests that the switch may have originally evolved to regulate processes other than sexual mating. Furthermore, the majority of natural C. tropicalis and C. albicans isolates are a/α strains (80–95%); thus, while most C. albicans strains are unable to switch, it appears that the majority of C. tropicalis isolates are competent for switching to the opaque form.
Many of the genes controlled by the white-opaque switch in C. albicans are metabolism genes, perhaps reflective of the fact that white and opaque cells colonize different host niches [9], [21]. C. albicans white cells exhibit greater virulence in models of systemic infection, while opaque cells are more efficient at colonization of the skin and are unstable at 37°C, rapidly switching back to the white form [48], [49]. In contrast to C. albicans, C. tropicalis opaque cells are stable at 37°C [5] and may colonize host niches that are refractory to C. albicans opaque cells. In addition, we found that C. tropicalis a/α cells exhibited increased fitness relative to a or α strains in a neutropenic model of candidiasis. Thus, the ability of a/α cells to switch phenotypes may be beneficial to C. tropicalis as it allows cells to propagate with optimal fitness, but still have the potential to form opaque cells and thereby adapt to different host niches.
Alternatively, the white-opaque switch could have evolved to regulate mating in the ancestor to C. tropicalis and C. albicans, and MTL control of the switch subsequently evolved in C. albicans as a means of restricting switching to MTL homozygous strains. If the major role of the white-opaque switch is to regulate mating, then fine-tuning of the switching mechanism could have been beneficial to help prevent futile switching to the opaque state in a/α cells that are sterile. Certainly, the regulation of mating by the white-opaque switch is stricter in C. albicans than in C. tropicalis; white cells of C. tropicalis undergo appreciable mating frequencies while C. albicans white cells do not.
Finally, it is formally possible that MTL control of the white-opaque switch was present in the ancestor to C. albicans and C. tropicalis, but has since been lost in C. tropicalis. Experiments will now be required to characterize C. tropicalis white and opaque states in vivo, and to determine if they exhibit different preferences for host colonization and pathogenesis as has been documented for C. albicans.
A Dual Role for Wor1 in Regulating Phenotypic Switching and Filamentation in C. tropicalis
An unexpected outcome of our analysis of white and opaque cells was the discovery of a link between C. tropicalis Wor1 and filamentous growth. Overexpression of WOR1 in C. tropicalis a, α, or a/α cells resulted in increased filamentation relative to wild-type white or opaque cells. C. tropicalis Wor1 is therefore a key regulator of filamentation, and this control can be separated from regulation of the white-opaque switch. The role of Wor1 was most clearly manifested in biofilm assays, where increased expression of WOR1 led to enhanced biofilm formation. This is presumably a direct result of the increase in filamentous growth, as filamentation and biofilm formation are inter-related processes in multiple Candida species [35], [36], [50]. Regulation of filamentation by the white-opaque switch has also been noted in C. albicans, although here white cells have been shown to be more conducive to undergoing filamentation than opaque cells [23], [24].
The observation that C. tropicalis Wor1 induces filamentation shows interesting parallels with the role of Wor1 homologs in more distantly related ascomycete species. In S. cerevisiae and H. capsulatum, the Wor1 homologs Mit1 and Ryp1, respectively, are both master regulators of filamentation [31], [32]. It has therefore been proposed that the ancestral function of Wor1/Mit1/Ryp1 was to control morphological transitions such as that between yeast and filamentous forms [31]. The discovery that C. tropicalis Wor1 promotes filamentation suggests that it has retained the ancestral function of Wor1 in this species.
In contrast to filamentous growth, the white-opaque switch evolved only recently in the ascomycete lineage, probably immediately prior to the divergence of C. tropicalis and C. albicans [5], [8], [51]. We therefore propose that C. tropicalis Wor1 has retained both the ancestral role of the Wor1 family of transcription factors (regulation of filamentous growth), as well as adopting control over the more recently evolved white-opaque switch. In contrast, overexpression of C. albicans Wor1 did not promote biofilm formation under the conditions tested (data not shown) and can even inhibit filamentous growth [8]. Further studies on C. tropicalis Wor1 will therefore shed light on its role as a key regulator of both morphological switches: the recently evolved white-opaque switch, as well as the ancestral program of filamentous growth.
Materials and Methods
Ethics Statement
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals as defined by the National Institutes of Health (PHS Assurance #A3284-01). Animal protocols were reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) of Brown University. All animals were housed in a centralized and AAALAC-accredited research animal facility that is fully staffed with trained husbandry, technical, and veterinary personnel.
Media
Yeast extract peptone dextrose medium (YPD), synthetic complete dextrose medium (SCD), and Spider medium were made as described previously [34], [52]. YPD plates containing 200 µg/ml nourseothricin (NAT) were used for selection of strains that were resistant to nourseothricin (Werner Bioagents, Jena, Germany) as previously described [53]. Lee's media containing 12.5 g/L N-acetylglucosamine (Alpha Aesar) was used for switching assays, and Lee's media containing 1.25% glucose was used for biofilm assays. [12], [54]. Lee's media was supplemented with 0.004% histidine when used in experiments with -His strains.
Plasmids and Strains
C. tropicalis strains used in this study are listed in Tables S1 and S2. Gene deletions were constructed by using the SAT1 flipper strategy as described [55]. Strains were transformed with 1–4 µg of DNA by using a modified electroporation protocol [5]. To delete WOR1, HIS1, ARG4, MTLa2, and MTLα1 genes using the SAT1-flipper method, oligonucleotides were used to amplify ∼900 bp of the 5′ and 3′ homologous flanks of each gene. The resulting PCR products were digested with restriction enzymes noted in Table S3 and cloned into the plasmid pSFS2A [55]. The resulting plasmids were digested with restriction enzymes as noted in Table S4 to liberate cassettes containing the 5′ and 3′ gene flanks as well as the SAT1 selectable marker and used for transformation. Correct genomic integration of transformant colonies was confirmed by PCR of the 5′ and 3′ junctions (for oligonucleotides, see Table S3). The SAT1 marker was recycled by growing transformants on maltose media at room temperature or 30°C and subsequent replica patching to both YPD and YPD+NAT plates. Alternatively, the SAT1 marker was recycled by growing cells in liquid YEP + 2% maltose at room temperature for ∼2 days and selected by plating to YPD+ low NAT (10 µg/mL), as previously described [55]. The transformation process was repeated to delete the remaining copy of the gene, and loss of the ORF was confirmed by PCR. To overexpress WOR1, fusion PCRs were performed to create a pTDH3-WOR1 construct, which was cloned into pSFS2A. The plasmid was linearized in the TDH3 promoter with SmaI and transformed. Correct genomic integration was confirmed by PCR. To insert a SAT1 marker next to the MTL, PCR was performed to amplify a sequence immediately upstream of the MTL, which was cloned into pSFS2A. The plasmid was linearized within this sequence with EcoRI and transformed into C. tropicalis. Correct genomic integration next to the MTLa or MTLα locus was confirmed by PCR.
Mating Assays
Quantitative mating assays between C. tropicalis strains were performed as described previously [5], [18]. In brief, C. tropicalis cells were taken from Spider plates that had been grown at room temperature for 1–2 days and resuspended in water. Approximately 1×107 cells of each strain were mixed and pipetted onto 0.8-µm pore-size nitrocellulose filters and grown on the surface of Spider medium for 1–3 days at room temperature. Cells were collected from the filters and plated at different dilutions onto His − Arg − media to select for mating products and onto His − and Arg − plates to monitor each parent population. The limiting parent was used to calculate mating frequencies as follows: mating efficiency = conjugants/(limiting parent + conjugants) = the greater of (Arg − His−)/Arg − or (Arg − His−)/His−. Statistical significance was determined using a Student's T-test.
Phenotypic Switching Assays
White phase cells were inoculated into liquid SCD medium and incubated at room temperature overnight. Cells from this culture were diluted to 0.1 OD600 and incubated at 37°C for 5 days. Cultures were diluted in water and plated onto Spider medium or Lee's medium containing N-acetylglucosamine at a concentration of ∼100 colonies per plate. Colonies were examined for opaque sectors after growth at room temperature for 7–10 days.
RT–PCR
RNA was isolated from cells grown in Spider liquid at 0.8–1.0 OD600 using the Ribopure-Yeast Kit (Ambion). RNA was treated with Turbo DNaseI (Ambion), and 2 µg of RNA used for subsequent cDNA generation using the GoScript enzyme (Promega). qRT-PCR was then performed by using the gene specific primers listed in Table S3 with the SYBR Green Kit (Applied Biosystems) and ran on an Applied Biosystems 7300 Real-Time PCR System.
Microarrays
Cells were harvested after being grown to OD600 1.0–1.2 in Spider medium at room temperature. Cells were collected, flash frozen, and stored at −80°C. For microarrays performed on biofilms, cells were collected after the two-day incubation detailed in the adherence assay. Total RNA was extracted from cell pellets using the RiboPure-Yeast Kit protocol (Ambion). RNA was treated with Turbo DNaseI (Ambion) to eliminate DNA contamination and re-extracted with phenol/chloroform. Aminoallyl-labeled cDNA synthesis and hybridization to custom Agilent C. tropicalis microarrays was previously described by Porman et al. [5]. Arrays were scanned on a GenePix 4000 scanner (Axon Instruments), data quantified using GENEPIX PRO version 3.0 and normalized using Goulphar (http://transcriptome.ens.fr/goulphar). Data analysis was performed as previously described [5]. GO term analysis was facilitated by CGD (http://candidagenome.org) and Princeton University's Generic GO Term Mapper (http://go.princeton.edu/cgi-bin/GOTermMapper). Array data is available from GEO (accession numbers GSE40179, GSE42517 and GSE43267).
Microscopy
Digital images of colonies were collected using a Zeiss Stemi 2000-C microscope equipped with an Infinity 2 digital camera and Infinity Analyzer software (Lumenera Corperation, Ottawa, Canada). Differential interference contrast (DIC) of cells were captured using a Zeiss Inverted Microscope (Axio Observer. Z1) fitted with an AxioCam HR. Images were processed with AxioVision Rel. 4.8 (Zeiss, Germany).
For electron micrographs, cells were resuspended in water and attached to poly-L-lysine coated-coverslips. Samples were fixed with 2.5% (w/v) glutaraldehyde in 0.1 M Na-cacodylate buffer, pH 7.4 at 4°C, and washed with 0.1 M Na-cacodylate buffer, pH 7.4. Samples were then treated with 1% aqueous osmium tetroxide in 0.1 M Na-Cacodylate buffer, pH 7.4, at 25°C for 90 minutes, and washed with 0.1 M Na-Cacodylate buffer, pH 7.4. Cells were gradually dehydrated using a gradient ethanol series, dried in a critical point dryer, and coated with 20 nm gold palladium (60∶40) in an Emitech K550 sputter coater. Images were captured with a Hitachi S-2700 scanning electronic microscope with Quartz PCI software.
To quantify filamentation from colonies grown on Spider medium, cells were removed from the center of patches and counted for the fraction of filamentous cells. At least 4 fields of view and 400 cells were counted for each data point. Statistical significance was determined using a Student's T-test.
Adherence Assays
Cultures were inoculated in 3 ml of Spider medium then incubated at 25°C overnight. 2 ODs of cells were spun down and resuspended in 1 ml of Lee's + Glucose medium and transferred to a well of a 12-well polystyrene plate. Plates were incubated at 25°C for 1–2 days without shaking, then decanted. Each well was washed 3 times with 1 ml of water to remove non-adherent cells. Plates were imaged using a Chemidoc XRS+ with Image Lab software (Bio-Rad). Adherent cells were scraped off the plastic surface and resuspended in 1 ml of water and optical density was determined. For significant differences between data sets, each was tested for normal distribution. A one-way ANOVA was performed on OD600 results. Nine representative microscope fields were counted for each condition to determine the fraction of filamentous cells. Statistical significance was determined using a Mann-Whitney pair-wise test due to non-parametric datasets.
Crystal Violet Staining
Samples were prepared similarly to the adherence assays. Following the 3 washes, 12-well plates were decanted and left to dry for 45 minutes, and subsequently stained with 385 µL of 0.4% aqueous crystal violet per well for 45 minutes. Each well was washed 3 times with 1 mL of water, then destained with 700 µL of 95% ethanol. Finally, 100 µL of each destain solution was transferred to a 96-well plate and diluted 10-fold. Optical density was read at 595 nm using a BioTek Synergy HT plate reader and statistics were performed similarly to the adherence assays.
XTT Reduction Assay
Samples were prepared similarly to the adherence assays. Following the 3 washes, the plates were decanted, then 315 µL of 1 mg/mL XTT (2,3-bis-(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide) and 35 µL of 360 µg/mL phenazine methosulfate were added to each well. The plates were incubated at room temperature for 20 minutes, then 100 µL of solution from each well was transferred to a 96-well plate and diluted. Optical density was read at 450 nm using a BioTek Synergy HT plate reader and statistics were performed similarly to the adherence assays.
Analysis of C. tropicalis Fitness In Vivo
Female BALB/c mice (18–20 g, Charles River Laboratories) were made neutropenic by intraperitoneal injection of 200 µg anti-Gr-1 mAb (clone: RB6-8C5, BioXCell, West Lebanon, NH) one day prior to infection with C. tropicalis. C. tropicalis strains were grown overnight in Spider medium at 30°C, diluted to 0.2 OD600 in fresh Spider medium and grown at 30°C to log phase. Cells were collected and washed three times in sterile phosphate-buffered saline (PBS). Mice were infected with a mixture of two C. tropicalis strains in a 50∶50 ratio with a total inoculum of ∼1.0×106 colony forming units (CFUs) by injection into the tail vein. Each C. tropicalis mixture included one isolate auxotrophic for histidine (His) and one isolate auxotrophic for arginine (Arg) biosynthesis. Dilutions of the inoculum were plated onto SCD medium lacking either histidine or arginine to confirm initial cell concentrations. Mice were euthanized 72 hours after infection and kidney and brains isolated. Organs were homogenized through a 70 µm filter and dilutions of the organ suspensions plated onto SCD medium lacking His or Arg. CFUs were counted on each plate to quantify the relative abundance of each strain. Mice where no cells were recovered after infection were excluded from analyses. Statistical significance was determined using a Student's T-test.
Supporting Information
Zdroje
1. BrownGD, DenningDW, LevitzSM (2012) Tackling human fungal infections. Science 336 : 647.
2. PfallerMA, DiekemaDJ (2007) Epidemiology of invasive candidiasis: a persistent public health problem. Clin Microbiol Rev 20 : 133–163.
3. HedgesSB, BlairJE, VenturiML, ShoeJL (2004) A molecular timescale of eukaryote evolution and the rise of complex multicellular life. BMC Evol Biol 4 : 2.
4. PesoleG, LottiM, AlberghinaL, SacconeC (1995) Evolutionary origin of nonuniversal CUGSer codon in some Candida species as inferred from a molecular phylogeny. Genetics 141 : 903–907.
5. PormanAM, AlbyK, HirakawaMP, BennettRJ (2011) Discovery of a phenotypic switch regulating sexual mating in the opportunistic fungal pathogen Candida tropicalis. Proc Natl Acad Sci U S A 108 : 21158–21163.
6. SlutskyB, StaebellM, AndersonJ, RisenL, PfallerM, et al. (1987) “White-opaque transition”: a second high-frequency switching system in Candida albicans. J Bacteriol 169 : 189–197.
7. PujolC, DanielsKJ, LockhartSR, SrikanthaT, RadkeJB, et al. (2004) The closely related species Candida albicans and Candida dubliniensis can mate. Eukaryot Cell 3 : 1015–1027.
8. XieJ, DuH, GuanG, TongY, KourkoumpetisTK, et al. (2012) N-acetylglucosamine induces white-to-opaque switching and mating in Candida tropicalis, providing new insights into adaptation and fungal sexual evolution. Eukaryot Cell 11 : 773–782.
9. LanCY, NewportG, MurilloLA, JonesT, SchererS, et al. (2002) Metabolic specialization associated with phenotypic switching in Candida albicans. Proc Natl Acad Sci U S A 99 : 14907–14912.
10. AlbyK, BennettRJ (2009) Stress-induced phenotypic switching in Candida albicans. Mol Biol Cell 20 : 3178–3191.
11. HuangG, SrikanthaT, SahniN, YiS, SollDR (2009) CO(2) regulates white-to-opaque switching in Candida albicans. Curr Biol 19 : 330–334.
12. HuangG, YiS, SahniN, DanielsKJ, SrikanthaT, et al. (2010) N-acetylglucosamine induces white to opaque switching, a mating prerequisite in Candida albicans. PLoS Pathog 6: e1000806 doi:10.1371/journal.ppat.1000806.
13. KolotilaMP, DiamondRD (1990) Effects of neutrophils and in vitro oxidants on survival and phenotypic switching of Candida albicans WO-1. Infect Immun 58 : 1174–1179.
14. Ramirez-ZavalaB, ReussO, ParkYN, OhlsenK, MorschhauserJ (2008) Environmental induction of white-opaque switching in Candida albicans. PLoS Pathog 4: e1000089 doi:10.1371/journal.ppat.1000089.
15. GeigerJ, WesselsD, LockhartSR, SollDR (2004) Release of a potent polymorphonuclear leukocyte chemoattractant is regulated by white-opaque switching in Candida albicans. Infect Immun 72 : 667–677.
16. LohseMB, JohnsonAD (2008) Differential phagocytosis of white versus opaque Candida albicans by Drosophila and mouse phagocytes. PLoS ONE 3: e1473 doi:10.1371/journal.pone.0001473.
17. LockhartSR, PujolC, DanielsKJ, MillerMG, JohnsonAD, et al. (2002) In Candida albicans, white-opaque switchers are homozygous for mating type. Genetics 162 : 737–745.
18. MillerMG, JohnsonAD (2002) White-opaque switching in Candida albicans is controlled by mating-type locus homeodomain proteins and allows efficient mating. Cell 110 : 293–302.
19. StokesC, MoranGP, SpieringMJ, ColeGT, ColemanDC, et al. (2007) Lower filamentation rates of Candida dubliniensis contribute to its lower virulence in comparison with Candida albicans. Fungal Genet Biol 44 : 920–931.
20. ButlerG, RasmussenMD, LinMF, SantosMA, SakthikumarS, et al. (2009) Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 459 : 657–662.
21. TsongAE, MillerMG, RaisnerRM, JohnsonAD (2003) Evolution of a combinatorial transcriptional circuit: a case study in yeasts. Cell 115 : 389–399.
22. TuchBB, MitrovichQM, HomannOR, HerndayAD, MonighettiCK, et al. (2010) The transcriptomes of two heritable cell types illuminate the circuit governing their differentiation. PLoS Genet 6: e1001070 doi:10.1371/journal.pgen.1001070.
23. AndersonJ, CundiffL, SchnarsB, GaoMX, MackenzieI, et al. (1989) Hypha formation in the white-opaque transition of Candida albicans. Infect Immun 57 : 458–467.
24. ErnstJF (2000) Transcription factors in Candida albicans - environmental control of morphogenesis. Microbiology 146 (Pt 8) 1763–1774.
25. HuangG, WangH, ChouS, NieX, ChenJ, et al. (2006) Bistable expression of WOR1, a master regulator of white-opaque switching in Candida albicans. Proc Natl Acad Sci U S A 103 : 12813–12818.
26. SrikanthaT, BornemanAR, DanielsKJ, PujolC, WuW, et al. (2006) TOS9 regulates white-opaque switching in Candida albicans. Eukaryot Cell 5 : 1674–1687.
27. ZordanRE, GalgoczyDJ, JohnsonAD (2006) Epigenetic properties of white-opaque switching in Candida albicans are based on a self-sustaining transcriptional feedback loop. Proc Natl Acad Sci U S A 103 : 12807–12812.
28. AlbyK, SchaeferD, BennettRJ (2009) Homothallic and heterothallic mating in the opportunistic pathogen Candida albicans. Nature 460 : 890–893.
29. ZordanRE, MillerMG, GalgoczyDJ, TuchBB, JohnsonAD (2007) Interlocking Transcriptional Feedback Loops Control White-Opaque Switching in Candida albicans. PLoS Biol 5: e256 doi:10.1371/journal.pbio.0050256.
30. LohseMB, ZordanRE, CainCW, JohnsonAD (2010) Distinct class of DNA-binding domains is exemplified by a master regulator of phenotypic switching in Candida albicans. Proc Natl Acad Sci U S A 107 : 14105–14110.
31. CainCW, LohseMB, HomannOR, SilA, JohnsonAD (2012) A conserved transcriptional regulator governs fungal morphology in widely diverged species. Genetics 190 : 511–521.
32. NguyenVQ, SilA (2008) Temperature-induced switch to the pathogenic yeast form of Histoplasma capsulatum requires Ryp1, a conserved transcriptional regulator. Proc Natl Acad Sci U S A 105 : 4880–4885.
33. AndersonJM, SollDR (1987) Unique phenotype of opaque cells in the white-opaque transition of Candida albicans. J Bacteriol 169 : 5579–5588.
34. LiuH, KohlerJ, FinkGR (1994) Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science 266 : 1723–1726.
35. FinkelJS, MitchellAP (2011) Genetic control of Candida albicans biofilm development. Nat Rev Microbiol 9 : 109–118.
36. NobileCJ, MitchellAP (2006) Genetics and genomics of Candida albicans biofilm formation. Cell Microbiol 8 : 1382–1391.
37. McCreathKJ, SpechtCA, RobbinsPW (1995) Molecular cloning and characterization of chitinase genes from Candida albicans. Proc Natl Acad Sci U S A 92 : 2544–2548.
38. NettJE, LepakAJ, MarchilloK, AndesDR (2009) Time course global gene expression analysis of an in vivo Candida biofilm. J Infect Dis 200 : 307–313.
39. LegrandM, LephartP, ForcheA, MuellerFM, WalshT, et al. (2004) Homozygosity at the MTL locus in clinical strains of Candida albicans: karyotypic rearrangements and tetraploid formation. Mol Microbiol 52 : 1451–1462.
40. FromtlingRA, AbruzzoGK, GiltinanDM (1987) Candida tropicalis infection in normal, diabetic, and neutropenic mice. J Clin Microbiol 25 : 1416–1420.
41. DignardD, AndreD, WhitewayM (2008) Heterotrimeric G-protein subunit function in Candida albicans: both the alpha and beta subunits of the pheromone response G protein are required for mating. Eukaryot Cell 7 : 1591–1599.
42. JonesSKJr, BennettRJ (2011) Fungal mating pheromones: choreographing the dating game. Fungal Genet Biol 48 : 668–676.
43. LengelerKB, DavidsonRC, D'SouzaC, HarashimaT, ShenWC, et al. (2000) Signal transduction cascades regulating fungal development and virulence. Microbiol Mol Biol Rev 64 : 746–785.
44. AlbyK, BennettRJ (2010) Sexual reproduction in the Candida clade: cryptic cycles, diverse mechanisms, and alternative functions. Cell Mol Life Sci
45. LohseMB, JohnsonAD (2009) White-opaque switching in Candida albicans. Curr Opin Microbiol 12 : 650–654.
46. MorschhauserJ (2010) Regulation of white-opaque switching in Candida albicans. Med Microbiol Immunol 199 : 165–172.
47. SollDR (2009) Why does Candida albicans switch? FEMS Yeast Res 9 : 973–989.
48. KvaalC, LachkeSA, SrikanthaT, DanielsK, McCoyJ, et al. (1999) Misexpression of the opaque-phase-specific gene PEP1 (SAP1) in the white phase of Candida albicans confers increased virulence in a mouse model of cutaneous infection. Infect Immun 67 : 6652–6662.
49. KvaalCA, SrikanthaT, SollDR (1997) Misexpression of the white-phase-specific gene WH11 in the opaque phase of Candida albicans affects switching and virulence. Infect Immun 65 : 4468–4475.
50. LaffeySF, ButlerG (2005) Phenotype switching affects biofilm formation by Candida parapsilosis. Microbiology 151 : 1073–1081.
51. TuchBB, GalgoczyDJ, HerndayAD, LiH, JohnsonAD (2008) The evolution of combinatorial gene regulation in fungi. PLoS Biol 6: e38 doi:10.1371/journal.pbio.0060038.
52. Guthrie C, Fink GR (1991) Guide to Yeast Genetics and Molecular Biology. San Diego: Academic Press.
53. ReussO, VikA, KolterR, MorschhauserJ (2004) The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene 341 : 119–127.
54. BedellGW, SollDR (1979) Effects of low concentrations of zinc on the growth and dimorphism of Candida albicans: evidence for zinc-resistant and -sensitive pathways for mycelium formation. Infect Immun 26 : 348–354.
55. ParkYN, MorschhauserJ (2005) Tetracycline-inducible gene expression and gene deletion in Candida albicans. Eukaryot Cell 4 : 1328–1342.
Štítky
Genetika Reprodukční medicína
Článek Ubiquitous Polygenicity of Human Complex Traits: Genome-Wide Analysis of 49 Traits in KoreansČlánek Alternative Splicing and Subfunctionalization Generates Functional Diversity in Fungal ProteomesČlánek RFX Transcription Factor DAF-19 Regulates 5-HT and Innate Immune Responses to Pathogenic Bacteria inČlánek Surveillance-Activated Defenses Block the ROS–Induced Mitochondrial Unfolded Protein ResponseČlánek Deficiency Reduces Adipose OXPHOS Capacity and Triggers Inflammation and Insulin Resistance in Mice
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 3
-
Všechny články tohoto čísla
- Power and Predictive Accuracy of Polygenic Risk Scores
- Rare Copy Number Variants Are a Common Cause of Short Stature
- Coordination of Flower Maturation by a Regulatory Circuit of Three MicroRNAs
- Ubiquitous Polygenicity of Human Complex Traits: Genome-Wide Analysis of 49 Traits in Koreans
- Genomic Evidence for Island Population Conversion Resolves Conflicting Theories of Polar Bear Evolution
- Mechanistic Insight into the Pathology of Polyalanine Expansion Disorders Revealed by a Mouse Model for X Linked Hypopituitarism
- Genome-Wide Association Study and Gene Expression Analysis Identifies as a Predictor of Response to Etanercept Therapy in Rheumatoid Arthritis
- Problem Solved: An Interview with Sir Edwin Southern
- Long Interspersed Element–1 (LINE-1): Passenger or Driver in Human Neoplasms?
- Mouse HFM1/Mer3 Is Required for Crossover Formation and Complete Synapsis of Homologous Chromosomes during Meiosis
- Alternative Splicing and Subfunctionalization Generates Functional Diversity in Fungal Proteomes
- A WRKY Transcription Factor Recruits the SYG1-Like Protein SHB1 to Activate Gene Expression and Seed Cavity Enlargement
- Microhomology-Mediated Mechanisms Underlie Non-Recurrent Disease-Causing Microdeletions of the Gene or Its Regulatory Domain
- Ancient Evolutionary Trade-Offs between Yeast Ploidy States
- Differential Evolutionary Fate of an Ancestral Primate Endogenous Retrovirus Envelope Gene, the EnvV , Captured for a Function in Placentation
- A Feed-Forward Loop Coupling Extracellular BMP Transport and Morphogenesis in Wing
- The Tomato Yellow Leaf Curl Virus Resistance Genes and Are Allelic and Code for DFDGD-Class RNA–Dependent RNA Polymerases
- The U-Box E3 Ubiquitin Ligase TUD1 Functions with a Heterotrimeric G α Subunit to Regulate Brassinosteroid-Mediated Growth in Rice
- Role of the DSC1 Channel in Regulating Neuronal Excitability in : Extending Nervous System Stability under Stress
- –Independent Phenotypic Switching in and a Dual Role for Wor1 in Regulating Switching and Filamentation
- Pax6 Regulates Gene Expression in the Vertebrate Lens through miR-204
- Blood-Informative Transcripts Define Nine Common Axes of Peripheral Blood Gene Expression
- Genetic Architecture of Skin and Eye Color in an African-European Admixed Population
- Fine Characterisation of a Recombination Hotspot at the Locus and Resolution of the Paradoxical Excess of Duplications over Deletions in the General Population
- Estrogen Mediated-Activation of miR-191/425 Cluster Modulates Tumorigenicity of Breast Cancer Cells Depending on Estrogen Receptor Status
- Complex Patterns of Genomic Admixture within Southern Africa
- Yap- and Cdc42-Dependent Nephrogenesis and Morphogenesis during Mouse Kidney Development
- Molecular Networks of Human Muscle Adaptation to Exercise and Age
- Alp/Enigma Family Proteins Cooperate in Z-Disc Formation and Myofibril Assembly
- Polycomb Group Gene Regulates Rice () Seed Development and Grain Filling via a Mechanism Distinct from
- RFX Transcription Factor DAF-19 Regulates 5-HT and Innate Immune Responses to Pathogenic Bacteria in
- Distinct Molecular Strategies for Hox-Mediated Limb Suppression in : From Cooperativity to Dispensability/Antagonism in TALE Partnership
- A Natural Polymorphism in rDNA Replication Origins Links Origin Activation with Calorie Restriction and Lifespan
- TDP2–Dependent Non-Homologous End-Joining Protects against Topoisomerase II–Induced DNA Breaks and Genome Instability in Cells and
- Recurrent Rearrangement during Adaptive Evolution in an Interspecific Yeast Hybrid Suggests a Model for Rapid Introgression
- Genome-Wide Association Study in Mutation Carriers Identifies Novel Loci Associated with Breast and Ovarian Cancer Risk
- Coincident Resection at Both Ends of Random, γ–Induced Double-Strand Breaks Requires MRX (MRN), Sae2 (Ctp1), and Mre11-Nuclease
- Identification of a -Specific Modifier Locus at 6p24 Related to Breast Cancer Risk
- A Novel Function for the Hox Gene in the Male Accessory Gland Regulates the Long-Term Female Post-Mating Response in
- Tdp2: A Means to Fixing the Ends
- A Novel Role for the RNA–Binding Protein FXR1P in Myoblasts Cell-Cycle Progression by Modulating mRNA Stability
- Association Mapping and the Genomic Consequences of Selection in Sunflower
- Histone Deacetylase 2 (HDAC2) Regulates Chromosome Segregation and Kinetochore Function via H4K16 Deacetylation during Oocyte Maturation in Mouse
- A Novel Mutation in the Upstream Open Reading Frame of the Gene Causes a MEN4 Phenotype
- Ataxin1L Is a Regulator of HSC Function Highlighting the Utility of Cross-Tissue Comparisons for Gene Discovery
- Human Spermatogenic Failure Purges Deleterious Mutation Load from the Autosomes and Both Sex Chromosomes, including the Gene
- A Conserved Upstream Motif Orchestrates Autonomous, Germline-Enriched Expression of piRNAs
- Statistical Analysis Reveals Co-Expression Patterns of Many Pairs of Genes in Yeast Are Jointly Regulated by Interacting Loci
- Matefin/SUN-1 Phosphorylation Is Part of a Surveillance Mechanism to Coordinate Chromosome Synapsis and Recombination with Meiotic Progression and Chromosome Movement
- A Role for the Malignant Brain Tumour (MBT) Domain Protein LIN-61 in DNA Double-Strand Break Repair by Homologous Recombination
- The Population and Evolutionary Dynamics of Phage and Bacteria with CRISPR–Mediated Immunity
- Long Noncoding RNA MALAT1 Controls Cell Cycle Progression by Regulating the Expression of Oncogenic Transcription Factor B-MYB
- Surveillance-Activated Defenses Block the ROS–Induced Mitochondrial Unfolded Protein Response
- DNA Topoisomerase III Localizes to Centromeres and Affects Centromeric CENP-A Levels in Fission Yeast
- Genome-Wide Control of RNA Polymerase II Activity by Cohesin
- Divergent Selection Drives Genetic Differentiation in an R2R3-MYB Transcription Factor That Contributes to Incipient Speciation in
- NODULE INCEPTION Directly Targets Subunit Genes to Regulate Essential Processes of Root Nodule Development in
- Spreading of a Prion Domain from Cell-to-Cell by Vesicular Transport in
- Deficiency in Origin Licensing Proteins Impairs Cilia Formation: Implications for the Aetiology of Meier-Gorlin Syndrome
- Deficiency Reduces Adipose OXPHOS Capacity and Triggers Inflammation and Insulin Resistance in Mice
- The Conserved SKN-1/Nrf2 Stress Response Pathway Regulates Synaptic Function in
- Functional Genomic Analysis of the Regulatory Network in
- Astakine 2—the Dark Knight Linking Melatonin to Circadian Regulation in Crustaceans
- CRL2 E3-Ligase Regulates Proliferation and Progression through Meiosis in the Germline
- Both the Caspase CSP-1 and a Caspase-Independent Pathway Promote Programmed Cell Death in Parallel to the Canonical Pathway for Apoptosis in
- PRMT4 Is a Novel Coactivator of c-Myb-Dependent Transcription in Haematopoietic Cell Lines
- A Copy Number Variant at the Locus Likely Confers Risk for Canine Squamous Cell Carcinoma of the Digit
- Evidence of Gene–Environment Interactions between Common Breast Cancer Susceptibility Loci and Established Environmental Risk Factors
- HIV Infection Disrupts the Sympatric Host–Pathogen Relationship in Human Tuberculosis
- Trans-Ethnic Fine-Mapping of Lipid Loci Identifies Population-Specific Signals and Allelic Heterogeneity That Increases the Trait Variance Explained
- A Gene Transfer Agent and a Dynamic Repertoire of Secretion Systems Hold the Keys to the Explosive Radiation of the Emerging Pathogen
- The Role of ATM in the Deficiency in Nonhomologous End-Joining near Telomeres in a Human Cancer Cell Line
- Dynamic Circadian Protein–Protein Interaction Networks Predict Temporal Organization of Cellular Functions
- Nuclear Myosin 1c Facilitates the Chromatin Modifications Required to Activate rRNA Gene Transcription and Cell Cycle Progression
- Robust Prediction of Expression Differences among Human Individuals Using Only Genotype Information
- A Single Cohesin Complex Performs Mitotic and Meiotic Functions in the Protist
- The Role of the Arabidopsis Exosome in siRNA–Independent Silencing of Heterochromatic Loci
- Elevated Expression of the Integrin-Associated Protein PINCH Suppresses the Defects of Muscle Hypercontraction Mutants
- Twist1 Controls a Cell-Specification Switch Governing Cell Fate Decisions within the Cardiac Neural Crest
- Genome-Wide Testing of Putative Functional Exonic Variants in Relationship with Breast and Prostate Cancer Risk in a Multiethnic Population
- Heteroduplex DNA Position Defines the Roles of the Sgs1, Srs2, and Mph1 Helicases in Promoting Distinct Recombination Outcomes
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Fine Characterisation of a Recombination Hotspot at the Locus and Resolution of the Paradoxical Excess of Duplications over Deletions in the General Population
- Molecular Networks of Human Muscle Adaptation to Exercise and Age
- Recurrent Rearrangement during Adaptive Evolution in an Interspecific Yeast Hybrid Suggests a Model for Rapid Introgression
- Genome-Wide Association Study and Gene Expression Analysis Identifies as a Predictor of Response to Etanercept Therapy in Rheumatoid Arthritis
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



