-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaCoordination of Flower Maturation by a Regulatory Circuit of Three MicroRNAs
The development of multicellular organisms relies on interconnected genetic programs that control progression through their life cycle. MicroRNAs (miRNAs) and transcription factors (TFs) play key roles in such regulatory circuits. Here, we describe how three evolutionary conserved miRNA-TF pairs interact to form multiple checkpoints during reproductive development of Arabidopsis thaliana. Genetic, cellular, and physiological experiments show that miR159 - and miR319-regulated MYB and TCP transcription factors pattern the expression of miR167 family members and their ARF6/8 targets. Coordinated action of these miRNA-TF pairs is crucial for the execution of consecutive hormone-dependent transitions during flower maturation. Cross-regulation includes both cis - and trans-regulatory interactions between these miRNAs and their targets. Our observations reveal how different miRNA-TF pairs can be organized into modules that coordinate successive steps in the plant life cycle.
Published in the journal: . PLoS Genet 9(3): e32767. doi:10.1371/journal.pgen.1003374
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003374Summary
The development of multicellular organisms relies on interconnected genetic programs that control progression through their life cycle. MicroRNAs (miRNAs) and transcription factors (TFs) play key roles in such regulatory circuits. Here, we describe how three evolutionary conserved miRNA-TF pairs interact to form multiple checkpoints during reproductive development of Arabidopsis thaliana. Genetic, cellular, and physiological experiments show that miR159 - and miR319-regulated MYB and TCP transcription factors pattern the expression of miR167 family members and their ARF6/8 targets. Coordinated action of these miRNA-TF pairs is crucial for the execution of consecutive hormone-dependent transitions during flower maturation. Cross-regulation includes both cis - and trans-regulatory interactions between these miRNAs and their targets. Our observations reveal how different miRNA-TF pairs can be organized into modules that coordinate successive steps in the plant life cycle.
Introduction
In mammals, reproductive organs are already formed in the embryo, with gametogenesis being completed after birth. By contrast, plant reproductive development is initiated only after the transition from the juvenile to the adult phase, with the production of flowers that harbor the reproductive organs. Despite these differences, both kingdoms tightly control the progression from initiation to maturation of reproductive organs and the acquisition of reproductive competence. The progression relies on successive genetic programs that govern a delicate balance of cell division and differentiation, organ growth and maturation, as well as cell death. Coordination of these events in plants makes use of a host of interacting hormones, including cytokinins, auxin, gibberellins (GAs), jasmonate (JA), ethylene and brassinosteroids. Hormone action is often mediated by dedicated transcription factors (TFs) such as the auxin response factors (ARFs), several of which are microRNA (miRNA) regulated. As an example, expression of the paralogous master regulators ARF6 and ARF8 (“ARF6/8”) of Arabidopsis thaliana are fine-tuned in specific floral organs by miR167 [1]. Increased miR167 levels or reduced ARF6/8 function both result in underdeveloped floral organs and impaired male and female fertility [1]–[4]. ARF6/8 activity is also sufficient for driving growth of floral organs, as shown with plants in which miR167 function is blocked [1], [5]. ARF6/8 regulate the expression of auxin homeostatic genes and help to set the boundaries of cytokinin-dependent meristematic activity, in addition to modulating JA biosynthesis [2], [4], [6]. Therefore, some of the defects in flowers with reduced ARF6/8 levels resemble symptoms of auxin, GA and JA deficiency and cytokinin overproduction [7]–[14]. Further complexity comes from miR167 having several isoforms that differ in their expression patterns and in their ability to downregulate their ARF6/8 targets [1].
Defects seen in arf6/8 mutant flowers are reminiscent of ones observed when the function of two other miRNAs, miR159 and miR319, is compromised. Such flowers suffer from retarded development of organs in the three outer whorls, the sepals, petals and anthers. The targets of these miRNAs are MYB and TCP transcription factors [15]–[18]. miR159-mediated restriction of MYB33 and MYB65 activity to anthers is necessary for normal floral organ growth and fertility [16], [19], [20]. MiR159 levels are positively regulated by GA [15], and at least in rice, this is also true for miR319 expression [21]. MiR319 is encoded by three genomic loci with overlapping expression patterns in A. thaliana. Transcription of MIR319A coincides with MIR319C at the base of all floral organs, in stamen filaments and petals, while MIR319B is restricted to stamens and the abscission zone of sepals [22], [23]. Reduction of miR319 activity results in smaller flowers with short petals, strap-like petals and underdeveloped stamens; in extreme cases, petals and stamens are lost [5], [23].
Here, we report how these three evolutionary conserved miRNA-TF pairs (miRNA-TF nodes hereafter) are organized into a regulatory network that is responsible for several checkpoints in floral organ maturation. MiR159 and miR319 regulate directly interacting targets, which in turn control the expression of MIR167 family members during this process. Restriction of MIR167A expression allows the progression from meristematic, cytokinin-dependent programs to auxin-dependent organogenesis, culminating in GA - and JA-dependent maturation. We propose that the convergence of unrelated miRNA targets onto common downstream targets is an important feature of miRNA networks in plant development.
Results
Floral defects caused by altered miR159, miR167, and miR319 activities
Flowers of Pro35S:MIM159 and Pro35S:MIM319 transgenic plants, in which miR159 and miR319 activities are impaired by constitutive expression of target mimics (Figure S1), have defects reminiscent of those found in plants with reduced ARF6/8 activity [5]. We therefore compared in detail the consequences of reducing ARF6/8, miR159 and miR319 function during reproductive development. Similar to plants with reduced ARF6/8 function (arf6-2 arf8-3 double mutants, Pro35S:MIR167A and Pro35S:MIR167C plants), the maturation of sepals, petals and anthers was delayed to various degrees in Pro35S:MIM159 and Pro35S:MIM319 plants. In addition, the flower stems, or pedicels, were shorter, and the angles of flowers relative to the main stem were more acute (Figure 1A and Figure S2). Stamens did not completely elongate in any of the genotypes. The strongest defects were seen in arf6 arf8, Pro35S:MIR167A and Pro35S:MIM319 plants, which never shed pollen (Figure 1A). Thus, stamen filament elongation seemed to be more sensitive to loss of ARF6/8 activity than anther dehiscence.
Fig. 1. Effects of miR159, miR167, and miR319 on flower morphology. 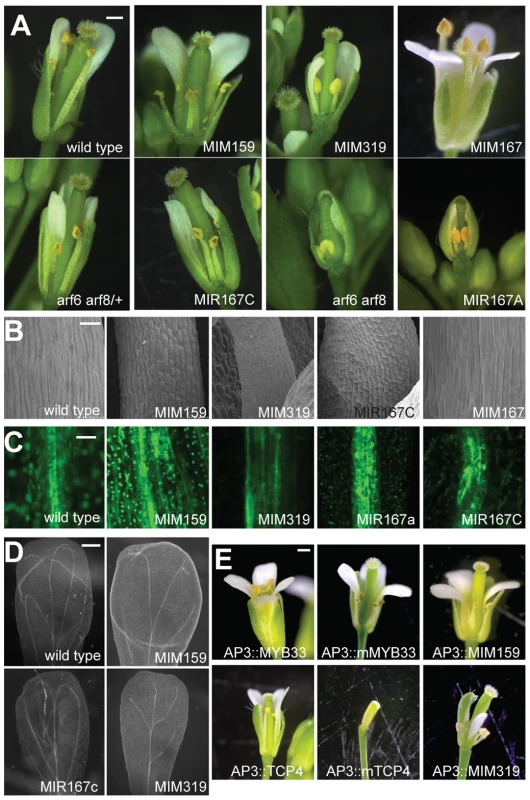
(A) Entire flowers, with some petals and sepals removed to reveal interior organs. Petals and stamens are underdeveloped in Pro35S:MIM159 (‘MIM159’), Pro35S:MIM319 (‘MIM319’), Pro35S:MIR167A (‘MIR167A’), Pro35S:MIR167C (‘MIR167C‘) plants and arf6 arf8 mutants, all of which have reduced ARF6/8 activity. Pro35S:MIM167 (‘MIM167’) plants, which have increased ARF6/8 activity, show an opposite phenotype of organ overgrowth, especially in anther filaments. (B) Epidermis of stamen filaments. (C) Reduced ARF6/8 activity in different transgenic backgrounds leads to expanded expression of procambial marker Q0990. (D) Effects of reduced ARF6/8 activity on vasculature pattern in mature petals (dark field view of cleared organs). (E) Entire flowers of plants in which miR159 and miR319 targets are specifically overexpressed or their functions attenuated in petals and stamens. Scale bars indicate 1 mm (A, E), 30 µm (B), 10 µm (C), 200 µm (D). See also Figure S2. Smaller organ size was likely due to reduced cell size, as seen by scanning electron microscopy of stamen filaments (Figure 1B). In addition to epidermal defects, vascular development in stamen filaments appeared to be arrested at the procambium stage, since the expression of the procambial marker Q0990 [24] was expanded in plants with diminished miR159, miR319 and ARF6/8 activities (Figure 1C). Petal vasculature was also compromised, lacking secondary loops in Pro35S:MIM319 plants, or having only two instead of four loops in Pro35S:MIM159 and Pro35S:MIR167c plants (Figure 1D).
Since petals and stamens were particularly sensitive to depletion of miR159 and miR319, we investigated the specific functions of two of their main targets, MYB33 and TCP4 [16], [17], [19], [20], [25], by expressing miRNA-non-targetable versions (mMYB33 and mTCP4) under the control of the petal - and stamen-specific APETALA3 (AP3) promoter [26]. We grew those plants along with plants expressing MIM159 and MIM319 decoys under the same AP3 promoter for comparison (Figure 1E). While mis-expression of the unmodified versions had little, if any, phenotypic effects, the non-targetable forms led to underdeveloped petals and anthers in ProAP3:mMYB33 flowers, and elimination of petals and anthers in ProAP3:mTCP4 plants (Figure 1E) [23]. Flowers expressing the miRNA insensitive form of MYB33 faithfully resembled the developmental impairments found in petals and stamens of ProAP3:MIM159 plants (Figure 1E). Nevertheless, flowers from ProAP3:mTCP4 presented more severe developmental defects than ProAP3:MIM319 flowers, in line with previous reports [23]. The phenotypic differences might be explained by a crosstalk between different miR319 TCP targets, or, as suggested in [23], by TCP4 movement or feed-forward regulation.
A sequence of hormone-related events regulated by coordinated miRNA action
Previous studies that have examined the role of different hormones during flower maturation suggested a sequential pattern for hormone action in the three outer whorls of the flower, especially in petals and stamens. Early in flower development, cytokinin-dependent maintenance of meristematic activity is delimited by the action of auxins [2]. Auxins subsequently trigger GA - and JA-based programs required for organ elongation and maturation during floral development [4], [27], [28].
A major consequence of ARF6/8 deficiency, and therefore reduced auxin signaling, is the expanded expression of positive meristem regulators involved in cytokinin action, such as the class I KNOX genes SHOOTMERISTEMLESS (STM) and BREVIPEDICELLUS (BP) [2]. The phenotypic alterations described above, along with premature flower bud opening, might thus be attributed to the expansion of cytokinin-related meristematic activity and reduced auxin action [12], [13], [29], [30]. As similar developmental defects were found in plants deficient in miR159 and miR319 activity, we asked whether their absence was also affecting the sequence of hormone-related events leading to flower maturation. We analyzed STM and BP activity in Pro35S:MIM159 and Pro35S:MIM319 inflorescences, and in MIR167C overexpressors, which have reduced ARF6/8 expression. In all three backgrounds, STM and BP expression were increased (Figure 2A). Suppression of the bp-1 mutant phenotype by Pro35S:MIM319 indicated that increased expression of other KNOXI genes could bypass the BP requirement (Figure 2B, 2C). Increased KNOXI activity might also explain the abnormal angle of petiole growth in mutants affected in ARF6/8, miR159 and miR319 activities. Additional defects caused by increased KNOXI activity are in vasculature development and stamen elongation and maturation, which are linked to reduced auxin transport [2], [10]. Accordingly, STM and BP upregulation was paralleled by reduced expression of the PIN1 auxin transporter gene in Pro35S:MIM159, Pro35S:MIR319 and Pro35S:MIR167C lines (Figure 2A). These results showed that miR159, miR319, just like ARF6/8, were negative regulators of the expression of class I KNOX genes, thereby confining cytokinin-dependent meristematic activity.
Fig. 2. Effects of miRNA mis-regulation on hormone pathways important for flower formation and maturation. 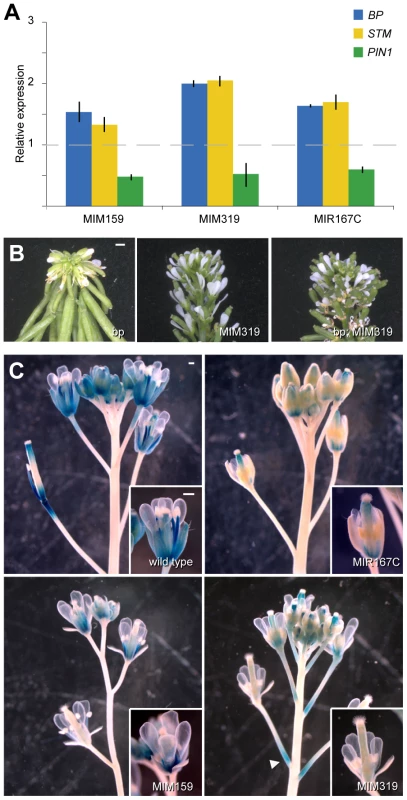
(A) Expression of genes involved in cytokinin-signaling (BP and STM) and auxin transport (PIN1) in flowers, as monitored by real-time RT-PCR. Error bars indicate range of two biological and two technical replicates. Measurements in mutants were normalized to values from wild-type inflorescences (dashed line). (B) Pro35S:MIM319 suppresses the downward-bent pedicels in bp-1 mutants (Ler background). (C) Misregulation of miR159, miR167 or miR319 reduces ProLOX2:GUS reporter activity, which is most active in sepals and stamens of wild-type flowers. Arrowhead points to ectopic GUS activity at the base of Pro35S:MIM319 pedicels. Scale bars indicate 1 mm. In addition to demarcating cytokinin action, ARF6/8 also contribute to flower development in later stages of maturation. ARF6/8 activate the expression of genes encoding two major JA biosynthetic enzymes, LOX2 and DAD1 [2], [4], a role that they share with the miR319 target TCP4 [31]. JA is important for the latest stages of floral organ development, including final stamen elongation, positioning of the anthers at the stigma at the time of dehiscence, and pollen maturation. Hence, without JA, plants are infertile [32]–[34]. In addition, JA modulates petal growth and vascular development [35], [36]. LOX2 is not essential for fertility [37], but the LOX2 promoter is active in stamens and sepals, where its expression increases from stages 12 to 15 of flower development [38], [39]. Consistent with a role of miR159, miR167 and miR319 in JA regulation, LOX2 promoter activity was reduced in sepals of Pro35S:MIM319, Pro35S:MIM159 and Pro35S:MIR167c plants (Figure 2C), with ectopic activation at the base of Pro35S:MIM319 pedicels, where MIR319B is normally expressed (Figure 2C, Figure S4C).
Taken together, the phenotypic and physiological resemblance of plants with a reduction in ARF6/8 or miR159/miR319 activities supports links between miR167, miR159 and miR319 in growth and hormone-dependent maturation of sepals, petals and anthers. The coordinated roles of the three miRNAs and their targets initially enable organogenesis by confining cytokinin-dependent meristematic activity. Later on, they contribute to the JA-dependent maturation of floral organs.
Mediation of miR159 and miR319 effects by miR167
We next assayed ARF6/8 expression in Pro35S:MIM159 and Pro35S:MIM319 plants in order to determine how ARF6/8 are regulated by miR159 and miR319. In wild type, both genes are expressed in the vasculature of floral organs, including in sepals, in petals, in stamen filaments, and in the precursor of the transmitting tract, the medial ridge of the carpel [1]. Expression of ARF6/8 was slightly reduced in Pro35S:MIM159 and Pro35S:MIM319 flowers, especially in the stamen filament vasculature (Figure 3A). In addition, we found that the expression of the ProARF8:ARF8-GUS reporter was specifically absent from stamens when miR159 and miR319 activities were reduced (Figure 3B).
Fig. 3. miR167-dependent regulation of ARF6/8 by miR159 and miR319. 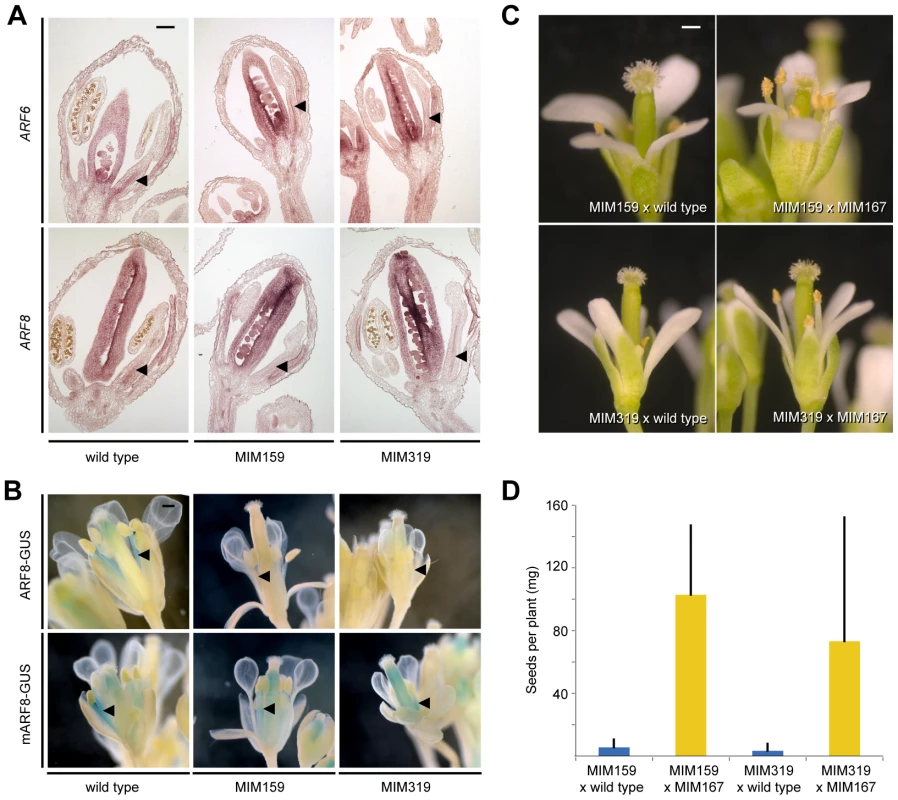
(A) ARF6/8 RNA expression in stamen filaments (triangles). (B) ARF8 reporter activity in stamen filaments (triangles). (C) Partial suppression of floral organ defects triggered by Pro35S:MIM159 and Pro35S:MIM319 upon reduction of miR167 activity. (D) Suppression of fertility defects caused by Pro35S:MIM159 (p<<0.001) and Pro35S:MIM319 (p<0.001). Error bars indicate standard deviation for at least 10 individuals from two independent crosses. Scale bars indicate 100 µm (A), 1 mm (B, C). To determine whether miR159 and miR319 targets are likely to regulate ARF6/8 directly or through miR167, we compared the response of ProARF8:ARF8-GUS and its miRNA insensitive form, ProARF8:mARF8-GUS, to changes in miR159 and miR319 activity (Figure 3B). While the expression of ProARF8:ARF8-GUS was altered in Pro35S:MIM159 and Pro35S:MIM319 plants, that of ProARF8:mARF8-GUS was not, indicating that miR159 and miR319 targets regulate ARF8 by increasing miR167 activity (Figure 3B). This hypothesis was further supported by the finding that petal and stamen development as well as fertility in Pro35S:MIM159 and Pro35S:MIM319 flowers were substantially restored when we reduced miR167 activity with the Pro35S:MIM167 construct (Figure 3C, 3D). These results placed miR159 and miR319 upstream of miR167. Plant miRNAs can regulate the expression of their targets through mRNA cleavage and/or translational inhibition [5], [40]. The observed difference between the effects of changes in miR167 expression on ARF6/8 mRNA and protein indicated that miR167 acts through both mRNA cleavage and translational inhibition.
Simultaneous sequestration of both miR159 and miR319 resulted in additive phenotypic and molecular effects (Figure 4A, 4B, 4D), suggesting that the miR159 - and miR319-regulated MYB and TCP transcription factors regulate shared target genes. We found that miR159 targets affected anther and petal development even when miR319 targets were suppressed in jaw-D plants (Figure 4C), indicating that MYBs and TCPs work in parallel to trigger miR167-mediated ARF6/8 regulation. Since TFs that regulate the same gene often form higher-order heteromeric complexes [41], we hypothesized that this might be the case for miR159-targetd MYBs and miR319-targeted TCPs. Consistent with this idea, the expression patterns of MYB33 and TCP4 in the affected floral organs overlap (Figure S3) and the proteins can interact in yeast and plant assays (Figure 4E, 4F). This implies that the two miRNA pathways converge on common downstream targets by modulating the composition of regulatory TF complexes.
Fig. 4. Interaction of miR159 and miR319 targets. 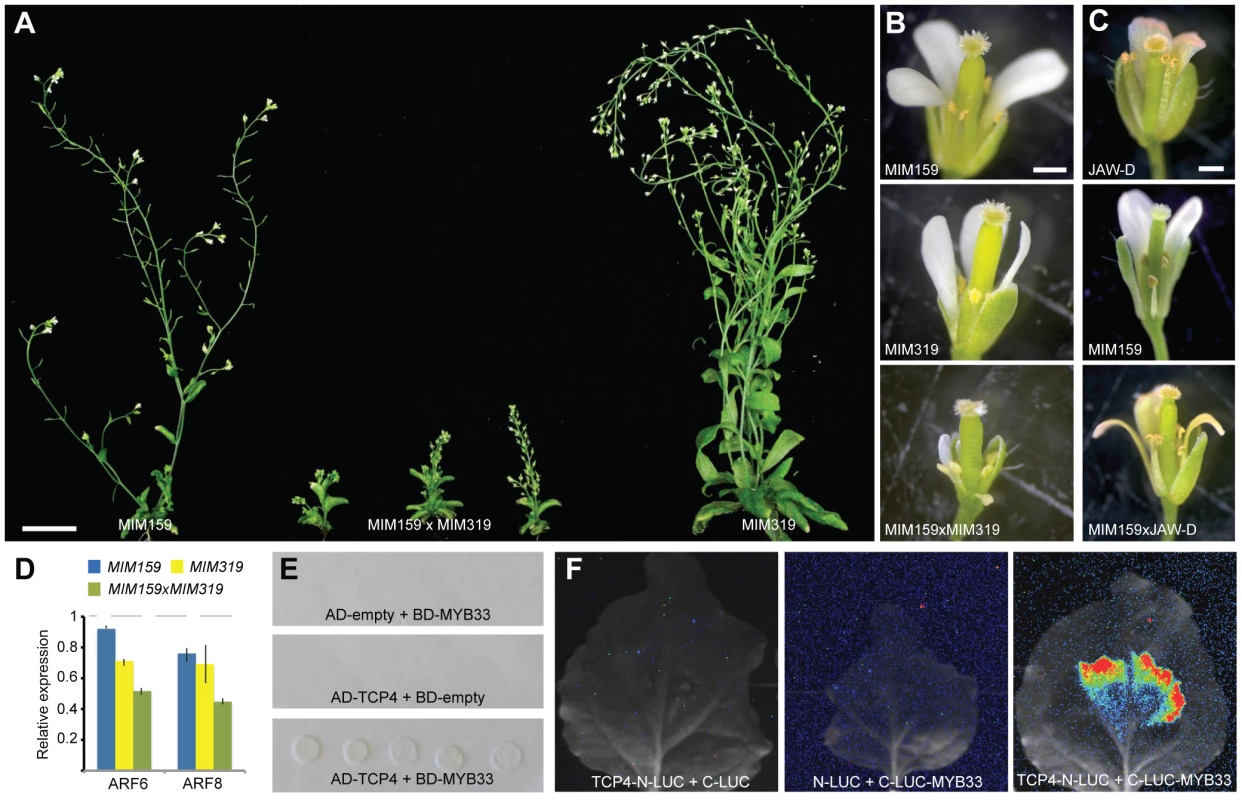
(A) Comparison of 45-day old plants. (B) Close-up of flowers in single and double Pro35S:MIM159 and Pro35S:MIM319 expressing plants. (C) Altered floral development as consequence of reduced miR159 activity and/or miR319 overexpression. (D) ARF6/8 expression levels in single and double Pro35S:MIM159 and Pro35S:MIM319 plants as monitored by real-time RT-PCR. Error bars indicate range of two biological and two technical replicates. Measurements in mutants were normalized to values from wild-type inflorescences (dashed lines). (E) Interaction of miR159 target MYB33 and miR319 target TCP4 in yeast-two-hybrid system. (F) Interaction of MYB33 and TCP4 assayed by firefly luciferase complementation in N. benthamiana. Luciferase activity is shown in false color, with highest levels red and lowest levels blue. Scale bars indicate 1 mm (B,C). Differential regulation of MIR167 genes by miR159 and miR319 targets
The divergent promoter activities of MIR167 family members are paralleled by their different abilities to downregulate ARF6/8, both of which indicate subfunctionalization [1]. It appears likely that the different MIRNA167 genes fine-tune ARF6/8 expression within different organs. To better understand miR159 - and miR319-dependent regulation of MIR167 genes, we assayed the transcriptional activity of MIR167A, MIR167B and MIR167C in Pro35S:MIM159 and Pro35S:MIM319 plants. We also analyzed Pro35S:MIR167C plants deficient in ARF6/8 function, to address whether altered reporter expression was a direct consequence of the absence of miR159 and miR319, or a downstream consequence of disrupting development by reducing ARF6/8 levels.
A GUS reporter under control of the MIR167A promoter is expressed in anther procambium, sepal vasculature and ovaries; activity of the MIR167B promoter has been detected in ovaries and at the base of flower pedicels; and the MIR167C promoter is mainly active in stamen filaments and petals [1]. As expected, in Pro35S:MIM159 and Pro35S:MIM319 flowers, ProMIR167A:GUS was ectopically expressed in tissues with abnormal development, including sepal vasculature, petals and stamen filaments, while MIR167C overexpression had no effect (Figure 5A and Figure S4A, S4B). In addition, similarly to the LOX2 promoter in Pro35S:MIM319 flowers (Figure 2C), the MIR167A promoter was ectopically activated at the base of pedicels, in contrast to the repression of the MIR167B promoter in this region (Figure 5A and Figure S4A, S4C). Likewise, reduced ARF6/8 function in Pro35S:MIM159 and Pro35S:MIM319 plants or in MIR167C overexpressors caused the MIR167C promoter to be less active in stamen filaments (Figure 5A). These results point to miR159 and miR319 as coordinating the expression pattern of MIR167 family members in petals, sepals and stamen.
Fig. 5. miR159- and miR319-dependent expression of miR167 in inflorescences. 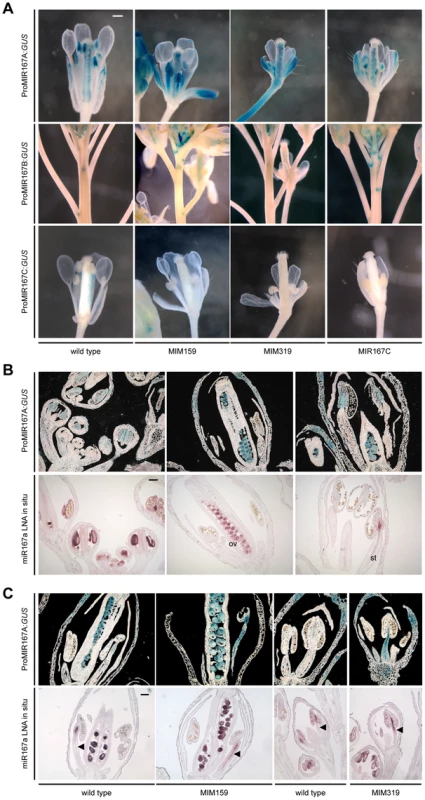
(A) Promoter activity of MIR167 family members. Some sepals and petals have been removed to reveal interior organs. (B) Comparison of MIR167A promoter activity (darkfield images of sections, top rows) and mature miR167a expression in wild-type flowers. ov, ovule; st, stamen. (C) Comparison of MIR167A promoter activity and mature miR167a expression in transgenic plants, in which miR167a becomes ectopically activated in stamen filaments (triangles). Scale bars indicate 1 mm (A), 100 µm (B, C). See also Figure S4. MIR167A promoter activity correlated with mature miRNA levels, as shown by in situ hybridization with an LNA probe (Figure 5B). MIR167A was initially expressed in emerging stamen, becoming confined to the procambium of stamen filaments from stage 7 on, and its levels were increased in Pro35S:MIM159 and Pro35S:MIM319 flowers (Figure 5C). The expression pattern of miR167a in ovaries, which was unaffected by changes in miR159 and miR319, resembled its promoter activity as well (Figure 5B, 5C) [1], [42]. Together, these results suggested that the effects of miR159 and miR319 are mainly mediated by MIR167A. The MIR167A promoter has two predicted TCP binding sites (Table S1). Mutating either of two potential TCP sites rendered the ProMIR167A:GUS reporter unresponsive to increased TCP levels in Pro35S:MIM319 plants (Figure 6A, 6B, and Figure S5A). We tested whether MIR167A regulation by TCP4 is direct by chromatin immunoprecipitation (ChIP) using anti-GFP antibodies and extracts from ProTCP4:TCP4-GFP Pro35S:MIM319 inflorescences. Quantitative PCR revealed specific enrichment overlapping one of the two potential TCP binding motifs (Figure 6C, Table S1). The A. thaliana genome encodes 24 TCPs, including several in the miR319-regulated TCP4 clade, and many can homo - and heterodimerize (Figure S5B) [43]–[46]. It is therefore conceivable that one of the TCP consensus motifs is bound by a TCP other than TCP4.
Fig. 6. Direct regulation of MIR167A by TCP4. 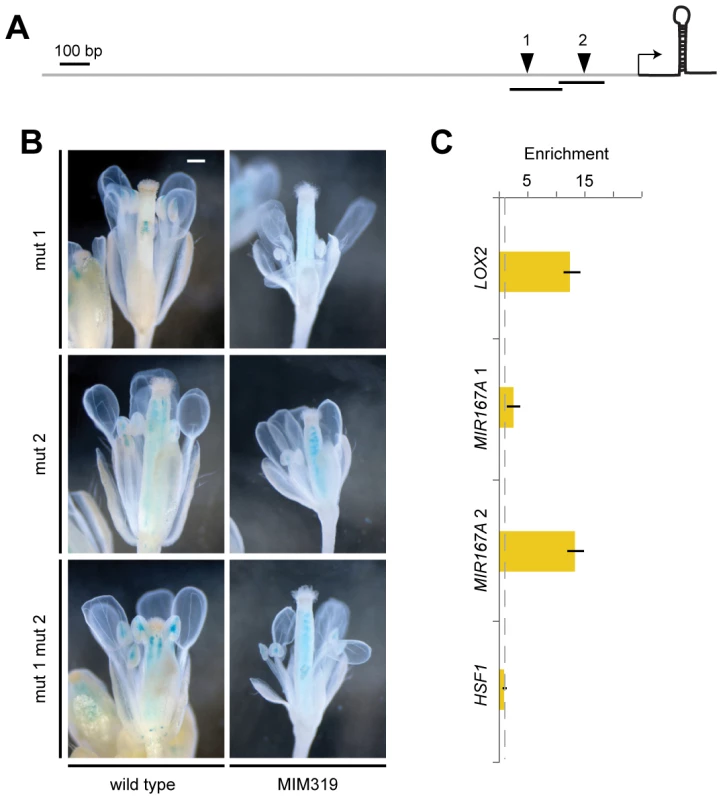
(A) Diagram of MIR167A promoter with potential TCP binding sites (inverted triangles) that were mutated (1: 5′-G[G/t]TCCC-3′; 2: 5′-GGA[C/a]CA-3′; lower case indicates mutant). Transcription start site is from [70]; transcribed region is not to scale. (B) Reporter gene assay with ProMIR167A:GUS. In contrast to the wild-type promoter (Figure 4A), the mutant promoter variants do not respond to a change in miR319 activity. Scale bar indicates 1 mm. (C) TCP4-GFP ChIP with material from ProTCP4:TCP4-GFP Pro35S:MIM319 inflorescences, normalized against empty vector control (dotted line). Amplicons 1 and 2 are indicated as lines in (A); LOX2 served as positive, HSF1 as negative control. Error bars indicate range of two biological and two technical replicates. See also Tables S1, S2 and Figure S5. Discussion
We have analyzed how three miRNA-target nodes interact to control consecutive checkpoints during floral organ maturation in the three outer whorls. First, we have discovered that the miR159-MYB and miR319-TCP nodes can independently regulate the miR167-ARF node. Second, we conclude that the convergent downstream effects of miR159 and miR319 are at least partially due to direct interaction of their MYB and TCP transcription factor targets. The finding of a link between these three miRNA-TF nodes reinforces the observation that miRNAs in plants are disproportionately often involved in auxin signaling [47] and that they can be organized into miRNA networks [48].
The miR159-miR167-miR319 circuit acts in sepals, petals and anthers to modulate the activity of ARF6/8, which control a large number of floral genes [4]. MiR159 and miR319 dampen the expression of their TF targets, which can otherwise lead to miR167 misexpression both individually and cooperatively through engaging in common protein complexes. In addition to regulation by miR159 and miR319 targets, there is cross-regulation among MIR167 genes, one example being repression of MIR167C in stamen filaments by miR167a. All these inputs are required to avoid miR167 misexpression. Moreover, the interactions can be complex, with miR319 contributing to activation of MIR167B and repression of MIR167A at the base of pedicels.
MiR159 and miR319 regulation enable ARF6/8 to play a central role in setting the cytokinin-auxin differentiation boundary by delimiting the expression of KNOXI genes. Inhibition of KNOXI expression leads to repression of cytokinin-based programs, which allows cells to exit the undifferentiated state [2], [12], [13]. MiR159 - and miR319-dependent ARF6/8 function contributes to later aspects of development by promoting JA synthesis in sepals through the induction of LOX2 expression and in petals and anthers through the regulation of DAD1 expression [2]. In addition, LOX2 is directly controlled by miR319-targeted TCP transcription factors at the base of pedicels independently of ARF6/8 levels. The tissue-specific differences in JA regulation by miR319 deserve further investigation.
Petal and anther development are particularly sensitive to perturbations in the miR159-miR167-miR319 network. Hormone action in the development of these floral organs starts with the maintenance of meristematic activity linked to KNOXI-dependent cytokinin biosynthesis and signal transduction (Figure 7). Auxin action is a prerequisite for the initiation of flower organ primordia [9], [49]–[51]. Later on, miR159 - and miR319-dependent ARF6/8 activities control a checkpoint for a transition that requires inhibition of KNOXI genes, which in turn regulates auxin transport and GA signaling (Figure 7) [2], [7], [52], [53]. Both processes are essential for the elongation and maturation of petals and anthers as well as for JA biosynthesis, which contributes to the last steps of the maturation of both organs (Figure 7) [2], [4], [7], [8], [10], [11], [32], [33], [35], [54]. Auxin action, mediated by ARF5/MP and ARF6/8, also promotes cambium development [4], [55], [56], and we propose that progression of vascular development which appears to follow a similar sequence of signaling events as floral organ maturation (Figure 7) [36], [57], is mediated by the miR159-miR167-miR319 network as well.
Fig. 7. Summary of interactions described in this work driving developmental progression of petals and anthers. 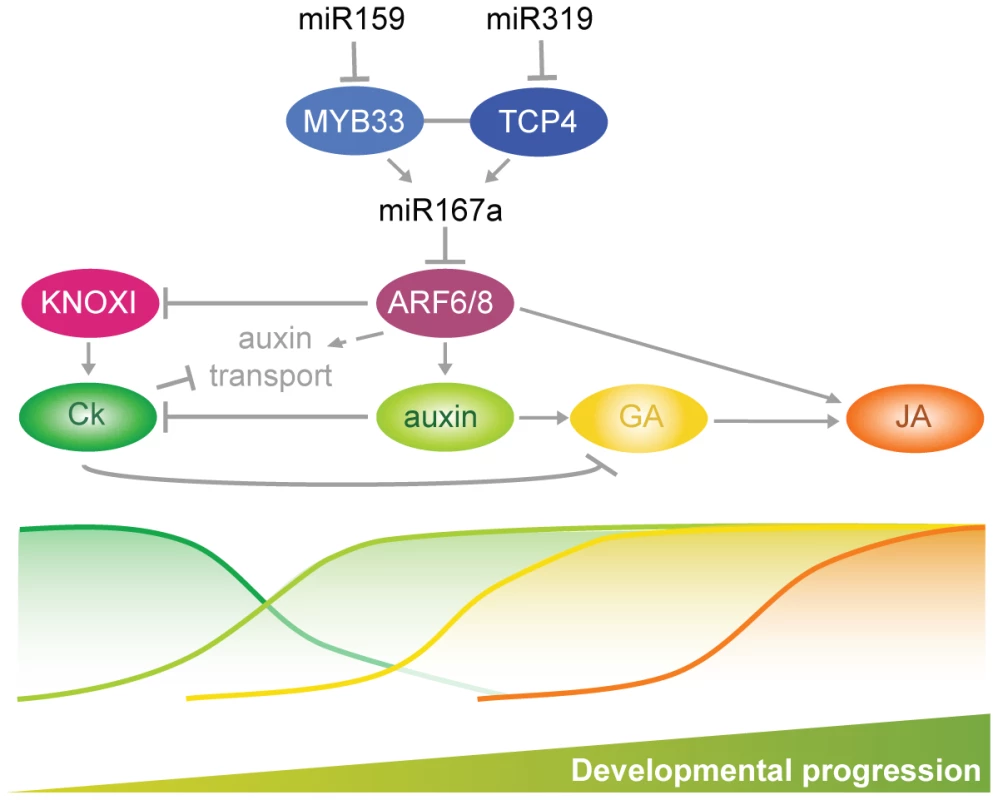
Sepals, petals and stamens, which make up the three outer whorls of floral organs, have key roles in evolutionary transitions between different plant mating strategies. The most common change is from obligate cross-pollination (allogamy) to self-fertilization (autogamy), which is found in up to half of all flowering plants, including many domesticated species [58]. The window of time during which the stigma is exserted beyond the outcrossing-protective sepals and petals before the stamens elongate, is a central determinant of outcrossing opportunities [59], [60]. Since the miR159-167-miR319 network is crucial for the development of sepals, petals and stamens, modulation of its activity might provide a regulatory entry point for different reproductive strategies.
Materials and Methods
Plant material
Plants were grown on soil in long days (16 h light/8 hours dark) under a mixture of cool and warm white fluorescent light at 23°C and 65% humidity. ProMIR167A:GUS, ProMIR167B:GUS, ProMIR167c:GUS ProARF8:ARF8-GUS, ProARF8:mARF8-GUS and afr6-2 arf8-3 [1], [4], artificial miRNA target mimics [5], ProLOX2:GUS and ProTCP4:TCP4-GFP [31], the Q0990 marker [24], and bp-1 mutants [61] have been described.
Transgenic plants
MIR167A and MIR167C fragments were PCR amplified from Col-0 genomic DNA, and placed behind the constitutive CaMV 35S promoter (Pro35S) [62]. Unmodified and modified MYB33 and TCP4 coding sequences [17] were placed behind the AP3 promoter [26]. The ProMIR319B:GUS reporter included a 2,662 bp genomic DNA fragment that begins 182 bp upstream of the first nucleotide of pre-miR319b. The MIR167A promoter was according to [1]. See Table S2 for oligonucleotide primers. Constructs were introduced into Col-0 plants by Agrobacterium tumefaciens-mediated transformation [63]. Their names can be found in Table S3.
RNA analyses
Total RNA was extracted from 30-day old inflorescences of T1 or F1 plants using TRIzol Reagent (Invitrogen) with two biological replicates using tissue pooled from 10 to 15 plants each. After reverse transcription with the RevertAid First Strand cDNA Synthesis Kit (Fermentas) of 1 µg of total RNA that had been treated with DNase I (Fermentas). PCR was carried out in presence of SYBR Green (Invitrogen) and monitored in real time with the Opticon Continuous Fluorescence Detection System (MJR/BioRad). For in situ hybridization [64] to detect ARF6/8, we used sections of inflorescences from 30-day old plants, with sense and antisense probes as described [1]. For LNA in situ hybridization [65] to detect miR167, 10% polyvinylalcohol was added to the colorimetric reaction buffer to increase the sensitivity of the assay.
Histology
Inflorescences were fixed in 90% acetone, and GUS activity was assayed as described [66]. For ARF8-GUS reporters, ten-fold lower concentrations of ferri - and ferrocyanate were used to increase sensitivity. Gus pictures are representative of three different experiments that included at least 10 samples each. Petals were sequentially cleared with chloral hydrate and ethanol/glycerol/lactic acid (3∶1∶1).
Chromatin immunoprecipitation
About 150 inflorescences were collected and fixed in two biological replicates, and immunoprecipitation was performed as described [67], with 3 µl of anti-rabbit GFP antibody (ab290; Abcam). Twenty µl eluate from Minelute columns (Qiagen) was diluted 1∶5 and used as template for two technical replicates of real-time PCR. Enrichment was calculated by comparing amplification in post-binding and input fraction and normalized to enrichment in samples from empty vector plants. We used a DNA fragment located within the 1 Kb upstream region of the Hsf1 gene (at4g17750) and lacking any putative TCP binding site as negative control.
Protein–protein interaction
Interaction in the yeast two-hybrid system was assayed on selective medium (Leu−, Trp−, His−) supplemented with 60 mM 3-amino 1,2,4-triazole (3-AT). The assay was repeated four times. For the luciferase complementation assay [69], fusions of non-targetable forms of MYB33, TCP2 and TCP4 were expressed from the 35S promoter. Agrobacterium tumefaciens cultures with the different constructs at OD600 = 0.3 each were mixed in equal ratios with a P19 silencing suppressor culture at OD600 = 0.1. Leaves were imaged three days after inoculation. Details for the constructs can be found in Table S3.
Supporting Information
Zdroje
1. WuMF, TianQ, ReedJW (2006) Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development 133 : 4211–4218.
2. TabataR, IkezakiM, FujibeT, AidaM, TianCE, et al. (2010) Arabidopsis AUXIN RESPONSE FACTOR6 and 8 regulate jasmonic acid biosynthesis and floral organ development via repression of class 1 KNOX genes. Plant Cell Physiol 51 : 164–175.
3. RuP, XuL, MaH, HuangH (2006) Plant fertility defects induced by the enhanced expression of microRNA167. Cell Res 16 : 457–465.
4. NagpalP, EllisCM, WeberH, PloenseSE, BarkawiLS, et al. (2005) Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development 132 : 4107–4118.
5. TodescoM, Rubio-SomozaI, Paz-AresJ, WeigelD (2010) A collection of target mimics for comprehensive analysis of microRNA function in Arabidopsis thaliana. PLoS Genet 6: e1001031 doi:10.1371/journal.pgen.1001031.
6. ReevesPH, EllisCM, PloenseSE, WuMF, YadavV, et al. (2012) A regulatory network for coordinated flower maturation. PLoS Genet 8: e1002506 doi:10.1371/journal.pgen.1002506.
7. WenCK, ChangC (2002) Arabidopsis RGL1 encodes a negative regulator of gibberellin responses. Plant Cell 14 : 87–100.
8. GriffithsJ, MuraseK, RieuI, ZentellaR, ZhangZL, et al. (2006) Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell 18 : 3399–3414.
9. ChengY, DaiX, ZhaoY (2006) Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev 20 : 1790–1799.
10. FengXL, NiWM, ElgeS, Mueller-RoeberB, XuZH, et al. (2006) Auxin flow in anther filaments is critical for pollen grain development through regulating pollen mitosis. Plant Mol Biol 61 : 215–226.
11. CecchettiV, AltamuraMM, FalascaG, CostantinoP, CardarelliM (2008) Auxin regulates Arabidopsis anther dehiscence, pollen maturation, and filament elongation. Plant Cell 20 : 1760–1774.
12. ScofieldS, DewitteW, MurrayJA (2007) The KNOX gene SHOOT MERISTEMLESS is required for the development of reproductive meristematic tissues in Arabidopsis. Plant J 50 : 767–781.
13. OriN, EshedY, ChuckG, BowmanJL, HakeS (2000) Mechanisms that control KNOX gene expression in the Arabidopsis shoot. Development 127 : 5523–5532.
14. PautotV, DockxJ, HamantO, KronenbergerJ, GrandjeanO, et al. (2001) KNAT2: evidence for a link between knotted-like genes and carpel development. Plant Cell 13 : 1719–1734.
15. AchardP, HerrA, BaulcombeDC, HarberdNP (2004) Modulation of floral development by a gibberellin-regulated microRNA. Development 131 : 3357–3365.
16. MillarAA, GublerF (2005) The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Plant Cell 17 : 705–721.
17. PalatnikJF, AllenE, WuX, SchommerC, SchwabR, et al. (2003) Control of leaf morphogenesis by microRNAs. Nature 425 : 257–263.
18. RhoadesMW, ReinhartBJ, LimLP, BurgeCB, BartelB, et al. (2002) Prediction of plant microRNA targets. Cell 110 : 513–520.
19. AllenRS, LiJ, StahleMI, DubroueA, GublerF, et al. (2007) Genetic analysis reveals functional redundancy and the major target genes of the Arabidopsis miR159 family. Proc Natl Acad Sci USA 104 : 16371–16376.
20. AllenRS, LiJ, Alonso-PeralMM, WhiteRG, GublerF, et al. (2010) MicroR159 regulation of most conserved targets in Arabidopsis has negligible phenotypic effects. Silence 1 : 18.
21. LiuQ, ZhangYC, WangCY, LuoYC, HuangQJ, et al. (2009) Expression analysis of phytohormone-regulated microRNAs in rice, implying their regulation roles in plant hormone signaling. FEBS Lett 583 : 723–728.
22. WarthmannN, DasS, LanzC, WeigelD (2008) Comparative analysis of the MIR319a microRNA locus in Arabidopsis and related Brassicaceae. Mol Biol Evol 25 : 892–902.
23. NagA, KingS, JackT (2009) miR319a targeting of TCP4 is critical for petal growth and development in Arabidopsis. Proc Natl Acad Sci USA 106 : 22534–22539.
24. SawchukMG, HeadP, DonnerTJ, ScarpellaE (2007) Time-lapse imaging of Arabidopsis leaf development shows dynamic patterns of procambium formation. New Phytol 176 : 560–571.
25. PalatnikJF, WollmannH, SchommerC, SchwabR, BoisbouvierJ, et al. (2007) Sequence and expression differences underlie functional specialization of Arabidopsis microRNAs miR159 and miR319. Dev Cell 13 : 115–125.
26. JackT, FoxGL, MeyerowitzEM (1994) Arabidopsis homeotic gene APETALA3 ectopic expression: transcriptional and posttranscriptional regulation determine floral organ identity. Cell 76 : 703–716.
27. WolbangCM, ChandlerPM, SmithJJ, RossJJ (2004) Auxin from the developing inflorescence is required for the biosynthesis of active gibberellins in barley stems. Plant Physiol 134 : 769–776.
28. ChengH, SongS, XiaoL, SooHM, ChengZ, et al. (2009) Gibberellin acts through jasmonate to control the expression of MYB21, MYB24, and MYB57 to promote stamen filament growth in Arabidopsis. PLoS Genet 5: e1000440 doi:10.1371/journal.pgen.1000440.
29. LiXG, SuYH, ZhaoXY, LiW, GaoXQ, et al. (2010) Cytokinin overproduction-caused alteration of flower development is partially mediated by CUC2 and CUC3 in Arabidopsis. Gene 450 : 109–120.
30. BartrinaI, OttoE, StrnadM, WernerT, SchmüllingT (2011) Cytokinin regulates the activity of reproductive meristems, flower organ size, ovule formation, and, thus, seed yield in Arabidopsis thaliana. Plant Cell tpc.110.079079.
31. SchommerC, PalatnikJF, AggarwalP, ChetelatA, CubasP, et al. (2008) Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol 6: e230 doi:10.1371/journal.pbio.0060230.
32. FeysB, BenedettiCE, PenfoldCN, TurnerJG (1994) Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 6 : 751–759.
33. StintziA, BrowseJ (2000) The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc Natl Acad Sci USA 97 : 10625–10630.
34. CaldelariD, WangG, FarmerEE, DongX (2010) Arabidopsis lox3 lox4 double mutants are male sterile and defective in global proliferative arrest. Plant Mol Biol 75 : 25–33.
35. BrioudesF, JolyC, SzécsiJ, VaraudE, LerouxJ, et al. (2009) Jasmonate controls late development stages of petal growth in Arabidopsis thaliana. Plant J 60 : 1070–1080.
36. SehrEM, AgustiJ, LehnerR, FarmerEE, SchwarzM, et al. (2010) Analysis of secondary growth in the Arabidopsis shoot reveals a positive role of jasmonate signalling in cambium formation. Plant J 63 : 811–822.
37. BellE, CreelmanRA, MulletJE (1995) A chloroplast lipoxygenase is required for wound-induced jasmonic acid accumulation in Arabidopsis. Proc Natl Acad Sci USA 92 : 8675–8679.
38. JensenAB, RaventosD, MundyJ (2002) Fusion genetic analysis of jasmonate-signalling mutants in Arabidopsis. Plant J 29 : 595–606.
39. SchmidM, DavisonTS, HenzSR, PapeUJ, DemarM, et al. (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37 : 501–506.
40. BrodersenP, Sakvarelidze-AchardL, Bruun-RasmussenM, DunoyerP, YamamotoYY, et al. (2008) Widespread translational inhibition by plant miRNAs and siRNAs. Science 320 : 1185–1190.
41. SmaczniakC, ImminkRG, MuinoJM, BlanvillainR, BusscherM, et al. (2012) Characterization of MADS-domain transcription factor complexes in Arabidopsis flower development. Proc Natl Acad Sci U S A 109 : 1560–1565.
42. ValocziA, VarallyayE, KauppinenS, BurgyanJ, HaveldaZ (2006) Spatio-temporal accumulation of microRNAs is highly coordinated in developing plant tissues. Plant J 47 : 140–151.
43. CubasP, LauterN, DoebleyJ, CoenE (1999) The TCP domain: a motif found in proteins regulating plant growth and development. Plant J 18 : 215–222.
44. KosugiS, OhashiY (2002) DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J 30 : 337–348.
45. AggarwalP, Das GuptaM, JosephAP, ChatterjeeN, SrinivasanN, et al. (2010) Identification of specific DNA binding residues in the TCP family of transcription factors in Arabidopsis. Plant Cell 22 : 1174–1189.
46. Martin-TrilloM, CubasP (2010) TCP genes: a family snapshot ten years later. Trends Plant Sci 15 : 31–39.
47. Rubio-SomozaI, CuperusJT, WeigelD, CarringtonJC (2009) Regulation and functional specialization of small RNA-target nodes during plant development. Curr Opin Plant Biol 12 : 622–627.
48. Rubio-SomozaI, WeigelD (2011) MicroRNA networks and developmental plasticity in plants. Trends Plant Sci 16 : 258–264.
49. TaoY, FerrerJL, LjungK, PojerF, HongF, et al. (2008) Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133 : 164–176.
50. StepanovaAN, Robertson-HoytJ, YunJ, BenaventeLM, XieDY, et al. (2008) TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133 : 177–191.
51. ZhaoZ, AndersenSU, LjungK, DolezalK, MiotkA, et al. (2010) Hormonal control of the shoot stem-cell niche. Nature 465 : 1089–1092.
52. JasinskiS, PiazzaP, CraftJ, HayA, WoolleyL, et al. (2005) KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr Biol 15 : 1560–1565.
53. FleishonS, ShaniE, OriN, WeissD (2011) Negative reciprocal interactions between gibberellin and cytokinin in tomato. New Phytol 190 : 609–617.
54. PengJ (2009) Gibberellin and jasmonate crosstalk during stamen development. J Integr Plant Biol 51 : 1064–1070.
55. HardtkeCS, BerlethT (1998) The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J 17 : 1405–1411.
56. DonnerTJ, SherrI, ScarpellaE (2010) Auxin signal transduction in Arabidopsis vein formation. Plant Signal Behav 5 : 70–72.
57. DettmerJ, EloA, HelariuttaY (2009) Hormone interactions during vascular development. Plant Mol Biol 69 : 347–360.
58. ChenKY, CongB, WingR, VrebalovJ, TanksleySD (2007) Changes in regulation of a transcription factor lead to autogamy in cultivated tomatoes. Science 318 : 643–645.
59. PengP, ChanSW, ShahGA, JacobsenSE (2006) Plant genetics: increased outcrossing in hothead mutants. Nature 443: E8 discussion E8-9.
60. TantikanjanaT, NasrallahJB (2012) Non-cell-autonomous regulation of crucifer self-incompatibility by Auxin Response Factor ARF3. Proc Natl Acad Sci U S A 109 : 19468–19473.
61. VenglatSP, DumonceauxT, RozwadowskiK, ParnellL, BabicV, et al. (2002) The homeobox gene BREVIPEDICELLUS is a key regulator of inflorescence architecture in Arabidopsis. Proc Natl Acad Sci USA 99 : 4730–4735.
62. OdellJT, NagyF, ChuaN-H (1985) Identification of DNA-sequences required for activity of the cauliflower mosaic virus-35S promoter. Nature 313 : 810–812.
63. Weigel D, Glazebrook J (2002) Arabidopsis: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. 354 p.
64. WollmannH, MicaE, TodescoM, LongJA, WeigelD (2010) On reconciling the interactions between APETALA2, miR172 and AGAMOUS with the ABC model of flower development. Development 137 : 3633–3642.
65. WangJW, CzechB, WeigelD (2009) miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138 : 738–749.
66. BlázquezMA, SoowalLN, LeeI, WeigelD (1997) LEAFY expression and flower initiation in Arabidopsis. Development 124 : 3835–3844.
67. Gomez-MeñaC, de FolterS, CostaMM, AngenentGC, SablowskiR (2005) Transcriptional program controlled by the floral homeotic gene AGAMOUS during early organogenesis. Development 132 : 429–438.
68. MathieuJ, YantLJ, MürdterF, KüttnerF, SchmidM (2009) Repression of flowering by the miR172 target SMZ. PLoS Biol 7: e1000148 doi:10.1371/journal.pbio.1000148.
69. ChenH, ZouY, ShangY, LinH, WangY, et al. (2008) Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol 146 : 368–376.
70. XieZ, AllenE, FahlgrenN, CalamarA, GivanSA, et al. (2005) Expression of Arabidopsis MIRNA genes. Plant Physiol 138 : 2145–2154.
Štítky
Genetika Reprodukční medicína
Článek Ubiquitous Polygenicity of Human Complex Traits: Genome-Wide Analysis of 49 Traits in KoreansČlánek Alternative Splicing and Subfunctionalization Generates Functional Diversity in Fungal ProteomesČlánek RFX Transcription Factor DAF-19 Regulates 5-HT and Innate Immune Responses to Pathogenic Bacteria inČlánek Surveillance-Activated Defenses Block the ROS–Induced Mitochondrial Unfolded Protein ResponseČlánek Deficiency Reduces Adipose OXPHOS Capacity and Triggers Inflammation and Insulin Resistance in Mice
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 3
-
Všechny články tohoto čísla
- Power and Predictive Accuracy of Polygenic Risk Scores
- Rare Copy Number Variants Are a Common Cause of Short Stature
- Coordination of Flower Maturation by a Regulatory Circuit of Three MicroRNAs
- Ubiquitous Polygenicity of Human Complex Traits: Genome-Wide Analysis of 49 Traits in Koreans
- Genomic Evidence for Island Population Conversion Resolves Conflicting Theories of Polar Bear Evolution
- Mechanistic Insight into the Pathology of Polyalanine Expansion Disorders Revealed by a Mouse Model for X Linked Hypopituitarism
- Genome-Wide Association Study and Gene Expression Analysis Identifies as a Predictor of Response to Etanercept Therapy in Rheumatoid Arthritis
- Problem Solved: An Interview with Sir Edwin Southern
- Long Interspersed Element–1 (LINE-1): Passenger or Driver in Human Neoplasms?
- Mouse HFM1/Mer3 Is Required for Crossover Formation and Complete Synapsis of Homologous Chromosomes during Meiosis
- Alternative Splicing and Subfunctionalization Generates Functional Diversity in Fungal Proteomes
- A WRKY Transcription Factor Recruits the SYG1-Like Protein SHB1 to Activate Gene Expression and Seed Cavity Enlargement
- Microhomology-Mediated Mechanisms Underlie Non-Recurrent Disease-Causing Microdeletions of the Gene or Its Regulatory Domain
- Ancient Evolutionary Trade-Offs between Yeast Ploidy States
- Differential Evolutionary Fate of an Ancestral Primate Endogenous Retrovirus Envelope Gene, the EnvV , Captured for a Function in Placentation
- A Feed-Forward Loop Coupling Extracellular BMP Transport and Morphogenesis in Wing
- The Tomato Yellow Leaf Curl Virus Resistance Genes and Are Allelic and Code for DFDGD-Class RNA–Dependent RNA Polymerases
- The U-Box E3 Ubiquitin Ligase TUD1 Functions with a Heterotrimeric G α Subunit to Regulate Brassinosteroid-Mediated Growth in Rice
- Role of the DSC1 Channel in Regulating Neuronal Excitability in : Extending Nervous System Stability under Stress
- –Independent Phenotypic Switching in and a Dual Role for Wor1 in Regulating Switching and Filamentation
- Pax6 Regulates Gene Expression in the Vertebrate Lens through miR-204
- Blood-Informative Transcripts Define Nine Common Axes of Peripheral Blood Gene Expression
- Genetic Architecture of Skin and Eye Color in an African-European Admixed Population
- Fine Characterisation of a Recombination Hotspot at the Locus and Resolution of the Paradoxical Excess of Duplications over Deletions in the General Population
- Estrogen Mediated-Activation of miR-191/425 Cluster Modulates Tumorigenicity of Breast Cancer Cells Depending on Estrogen Receptor Status
- Complex Patterns of Genomic Admixture within Southern Africa
- Yap- and Cdc42-Dependent Nephrogenesis and Morphogenesis during Mouse Kidney Development
- Molecular Networks of Human Muscle Adaptation to Exercise and Age
- Alp/Enigma Family Proteins Cooperate in Z-Disc Formation and Myofibril Assembly
- Polycomb Group Gene Regulates Rice () Seed Development and Grain Filling via a Mechanism Distinct from
- RFX Transcription Factor DAF-19 Regulates 5-HT and Innate Immune Responses to Pathogenic Bacteria in
- Distinct Molecular Strategies for Hox-Mediated Limb Suppression in : From Cooperativity to Dispensability/Antagonism in TALE Partnership
- A Natural Polymorphism in rDNA Replication Origins Links Origin Activation with Calorie Restriction and Lifespan
- TDP2–Dependent Non-Homologous End-Joining Protects against Topoisomerase II–Induced DNA Breaks and Genome Instability in Cells and
- Recurrent Rearrangement during Adaptive Evolution in an Interspecific Yeast Hybrid Suggests a Model for Rapid Introgression
- Genome-Wide Association Study in Mutation Carriers Identifies Novel Loci Associated with Breast and Ovarian Cancer Risk
- Coincident Resection at Both Ends of Random, γ–Induced Double-Strand Breaks Requires MRX (MRN), Sae2 (Ctp1), and Mre11-Nuclease
- Identification of a -Specific Modifier Locus at 6p24 Related to Breast Cancer Risk
- A Novel Function for the Hox Gene in the Male Accessory Gland Regulates the Long-Term Female Post-Mating Response in
- Tdp2: A Means to Fixing the Ends
- A Novel Role for the RNA–Binding Protein FXR1P in Myoblasts Cell-Cycle Progression by Modulating mRNA Stability
- Association Mapping and the Genomic Consequences of Selection in Sunflower
- Histone Deacetylase 2 (HDAC2) Regulates Chromosome Segregation and Kinetochore Function via H4K16 Deacetylation during Oocyte Maturation in Mouse
- A Novel Mutation in the Upstream Open Reading Frame of the Gene Causes a MEN4 Phenotype
- Ataxin1L Is a Regulator of HSC Function Highlighting the Utility of Cross-Tissue Comparisons for Gene Discovery
- Human Spermatogenic Failure Purges Deleterious Mutation Load from the Autosomes and Both Sex Chromosomes, including the Gene
- A Conserved Upstream Motif Orchestrates Autonomous, Germline-Enriched Expression of piRNAs
- Statistical Analysis Reveals Co-Expression Patterns of Many Pairs of Genes in Yeast Are Jointly Regulated by Interacting Loci
- Matefin/SUN-1 Phosphorylation Is Part of a Surveillance Mechanism to Coordinate Chromosome Synapsis and Recombination with Meiotic Progression and Chromosome Movement
- A Role for the Malignant Brain Tumour (MBT) Domain Protein LIN-61 in DNA Double-Strand Break Repair by Homologous Recombination
- The Population and Evolutionary Dynamics of Phage and Bacteria with CRISPR–Mediated Immunity
- Long Noncoding RNA MALAT1 Controls Cell Cycle Progression by Regulating the Expression of Oncogenic Transcription Factor B-MYB
- Surveillance-Activated Defenses Block the ROS–Induced Mitochondrial Unfolded Protein Response
- DNA Topoisomerase III Localizes to Centromeres and Affects Centromeric CENP-A Levels in Fission Yeast
- Genome-Wide Control of RNA Polymerase II Activity by Cohesin
- Divergent Selection Drives Genetic Differentiation in an R2R3-MYB Transcription Factor That Contributes to Incipient Speciation in
- NODULE INCEPTION Directly Targets Subunit Genes to Regulate Essential Processes of Root Nodule Development in
- Spreading of a Prion Domain from Cell-to-Cell by Vesicular Transport in
- Deficiency in Origin Licensing Proteins Impairs Cilia Formation: Implications for the Aetiology of Meier-Gorlin Syndrome
- Deficiency Reduces Adipose OXPHOS Capacity and Triggers Inflammation and Insulin Resistance in Mice
- The Conserved SKN-1/Nrf2 Stress Response Pathway Regulates Synaptic Function in
- Functional Genomic Analysis of the Regulatory Network in
- Astakine 2—the Dark Knight Linking Melatonin to Circadian Regulation in Crustaceans
- CRL2 E3-Ligase Regulates Proliferation and Progression through Meiosis in the Germline
- Both the Caspase CSP-1 and a Caspase-Independent Pathway Promote Programmed Cell Death in Parallel to the Canonical Pathway for Apoptosis in
- PRMT4 Is a Novel Coactivator of c-Myb-Dependent Transcription in Haematopoietic Cell Lines
- A Copy Number Variant at the Locus Likely Confers Risk for Canine Squamous Cell Carcinoma of the Digit
- Evidence of Gene–Environment Interactions between Common Breast Cancer Susceptibility Loci and Established Environmental Risk Factors
- HIV Infection Disrupts the Sympatric Host–Pathogen Relationship in Human Tuberculosis
- Trans-Ethnic Fine-Mapping of Lipid Loci Identifies Population-Specific Signals and Allelic Heterogeneity That Increases the Trait Variance Explained
- A Gene Transfer Agent and a Dynamic Repertoire of Secretion Systems Hold the Keys to the Explosive Radiation of the Emerging Pathogen
- The Role of ATM in the Deficiency in Nonhomologous End-Joining near Telomeres in a Human Cancer Cell Line
- Dynamic Circadian Protein–Protein Interaction Networks Predict Temporal Organization of Cellular Functions
- Nuclear Myosin 1c Facilitates the Chromatin Modifications Required to Activate rRNA Gene Transcription and Cell Cycle Progression
- Robust Prediction of Expression Differences among Human Individuals Using Only Genotype Information
- A Single Cohesin Complex Performs Mitotic and Meiotic Functions in the Protist
- The Role of the Arabidopsis Exosome in siRNA–Independent Silencing of Heterochromatic Loci
- Elevated Expression of the Integrin-Associated Protein PINCH Suppresses the Defects of Muscle Hypercontraction Mutants
- Twist1 Controls a Cell-Specification Switch Governing Cell Fate Decisions within the Cardiac Neural Crest
- Genome-Wide Testing of Putative Functional Exonic Variants in Relationship with Breast and Prostate Cancer Risk in a Multiethnic Population
- Heteroduplex DNA Position Defines the Roles of the Sgs1, Srs2, and Mph1 Helicases in Promoting Distinct Recombination Outcomes
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Fine Characterisation of a Recombination Hotspot at the Locus and Resolution of the Paradoxical Excess of Duplications over Deletions in the General Population
- Molecular Networks of Human Muscle Adaptation to Exercise and Age
- Recurrent Rearrangement during Adaptive Evolution in an Interspecific Yeast Hybrid Suggests a Model for Rapid Introgression
- Genome-Wide Association Study and Gene Expression Analysis Identifies as a Predictor of Response to Etanercept Therapy in Rheumatoid Arthritis
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



