-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaVariants Induce Differential Protection to Viruses in : A Phenotypic and Phylogenomic Analysis
Wolbachia are intracellular bacterial symbionts that are able to protect various insect hosts from viral infections. This tripartite interaction was initially described in Drosophila melanogaster carrying wMel, its natural Wolbachia strain. wMel has been shown to be genetically polymorphic and there has been a recent change in variant frequencies in natural populations. We have compared the antiviral protection conferred by different wMel variants, their titres and influence on host longevity, in a genetically identical D. melanogaster host. The phenotypes cluster the variants into two groups — wMelCS-like and wMel-like. wMelCS-like variants give stronger protection against Drosophila C virus and Flock House virus, reach higher titres and often shorten the host lifespan. We have sequenced and assembled the genomes of these Wolbachia, and shown that the two phenotypic groups are two monophyletic groups. We have also analysed a virulent and over-replicating variant, wMelPop, which protects D. melanogaster even better than the closely related wMelCS. We have found that a ∼21 kb region of the genome, encoding eight genes, is amplified seven times in wMelPop and may be the cause of its phenotypes. Our results indicate that the more protective wMelCS-like variants, which sometimes have a cost, were replaced by the less protective but more benign wMel-like variants. This has resulted in a recent reduction in virus resistance in D. melanogaster in natural populations worldwide. Our work helps to understand the natural variation in wMel and its evolutionary dynamics, and inform the use of Wolbachia in arthropod-borne disease control.
Published in the journal: . PLoS Genet 9(12): e32767. doi:10.1371/journal.pgen.1003896
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003896Summary
Wolbachia are intracellular bacterial symbionts that are able to protect various insect hosts from viral infections. This tripartite interaction was initially described in Drosophila melanogaster carrying wMel, its natural Wolbachia strain. wMel has been shown to be genetically polymorphic and there has been a recent change in variant frequencies in natural populations. We have compared the antiviral protection conferred by different wMel variants, their titres and influence on host longevity, in a genetically identical D. melanogaster host. The phenotypes cluster the variants into two groups — wMelCS-like and wMel-like. wMelCS-like variants give stronger protection against Drosophila C virus and Flock House virus, reach higher titres and often shorten the host lifespan. We have sequenced and assembled the genomes of these Wolbachia, and shown that the two phenotypic groups are two monophyletic groups. We have also analysed a virulent and over-replicating variant, wMelPop, which protects D. melanogaster even better than the closely related wMelCS. We have found that a ∼21 kb region of the genome, encoding eight genes, is amplified seven times in wMelPop and may be the cause of its phenotypes. Our results indicate that the more protective wMelCS-like variants, which sometimes have a cost, were replaced by the less protective but more benign wMel-like variants. This has resulted in a recent reduction in virus resistance in D. melanogaster in natural populations worldwide. Our work helps to understand the natural variation in wMel and its evolutionary dynamics, and inform the use of Wolbachia in arthropod-borne disease control.
Introduction
Many arthropods are infected by bacterial secondary (facultative) symbionts [1]. These are vertically transmitted bacteria that are not essential for the host to survive or reproduce, but nonetheless can have important effects on their host's biology. The fitness of these secondary symbionts is directly linked to their host's fitness; their transmission through successive generations is dependent on the breeding success of their hosts. This close association and dependence is predicted to favour the evolution of mutualism [2]. Nonetheless, the presence of replicating bacteria in the host is bound to have a cost. This fitness cost and imperfect vertical transmission would theoretically lead to elimination of vertically transmitted symbionts from host populations [3], [4]. Specific phenotypes associated with secondary symbionts explain their maintenance. Some secondary symbionts are parasites and manipulate their host reproductive biology [3], [5]. Others are mutualists and confer a fitness advantage to their hosts (e.g. resistance to environmental stress or pathogens) [6]. Genetic variability of the symbiont may impact all these associated phenotypes. Therefore, understanding this genotypic and phenotypic variability is essential to understand facultative symbionts population genetics.
In recent years it has become clear that symbionts can modulate the interactions between hosts and parasites in many taxa [6]–[20]. Insects are no exception to this pattern, and secondary symbionts can play a key role in protecting their hosts against infection or parasitism [6], [7], [9]–[12], [14], [17], [19]. Protection to pathogens may be the fitness advantage that enables these bacteria to invade insect populations. For example, the recent spread of Spiroplasma in North American populations of Drosophila neotestacea may be a consequence of the protection to nematode parasites conferred by these bacteria [10]. Also, the bacterium Hamiltonella defensa increases in frequency in aphid cage populations in the presence of parasitoid wasps, to which it provides protection, but decreases in the absence of it [21]. Presence of a protective symbiont can, therefore, be treated as an heritable, albeit non-Mendelian, condition-dependent beneficial genetic change [6].
The intracellular α-proteobacteria Wolbachia protects Drosophila melanogaster against viral infections [12], [14]. Wolbachia are estimated to infect 40% of arthropod species [22] and are, therefore, some of the most common intracellular bacteria known. Its success may be related to its anti-viral protective effect on natural hosts [12], [14], [23], [24], although this protection is not always observed [23], [25], [26]. Other mechanisms, many involving manipulation of the host reproduction, can also maintain Wolbachia in natural populations [27], [28]. The most common manipulation is cytoplasmic incompatibility (CI), which renders the crosses between Wolbachia infected males and uninfected females sterile or with low viability, giving a relative fitness advantage to infected females. Nonetheless, even when Wolbachia can cause CI, a beneficial effect, like protection to viruses, may contribute to the invasion of a new host [29].
Wolbachia-conferred protection against viruses is of particular interest because of potential applications in vector-borne disease control. Mosquitoes infected with Wolbachia can be more resistant to human arboviruses [24], [25], [30]–[33] and other human pathogens [30], [34]–[36]. A large effort is being made to use Aedes aegypti mosquitoes trans-infected with Wolbachia variants from D. melanogaster in limiting dengue virus transmission [30], [31]. Pilot releases of these trans-infected mosquitoes have already been conducted successfully [37] and intervention in dengue endemic areas is planned [38].
There can be a great deal of genetic variation in how symbionts modulate host-pathogen interactions. Different D. simulans lines infected with different Wolbachia strains, for instance, show variation in the protection to viruses [23]. The protection ranges from nearly complete to none, and the combinations showing higher protection have higher levels of the endosymbiont [23]. While these Wolbachia strains are distantly related, other studies have found variation within populations of closely related symbionts. For example, H. defensa protects aphids from parasitoid wasps only when it carries a lysogenic bacteriophage [39]. Understanding this genetic variation among symbionts may explain the frequency of different variants in natural populations and give insight into the mechanisms underlying the interactions.
In natural populations of D. melanogaster there has been a recent replacement of Wolbachia variants [40]–[42]. Wolbachia is present in most natural populations of D. melanogaster, although with variable frequencies of infection [41]–[50]. Only a single strain, wMel, is known to infect D. melanogaster, but several closely related genotypes of this strain - wMel, wMel2, wMel3, wMelCS and wMelCS2 - were defined on the basis of polymorphic genetic markers [40]. The frequencies of these genotypes in isolates from natural populations of D. melanogaster have changed during the 20th century. Early isolates have a high proportion of wMelCS type, while the wMel genotype is predominant in late 20th century isolates [40]. This wMel genotype replacement was supported by the analysis of Wolbachia genotype-associated mitochondrial DNA haplotypes [41]. More recently the genomes of 179 different wMel variants and 290 associated and non-associated mitochondria were assembled [42]. Their analysis showed that all wMel variants come from a single infection event and the most recent ancestor of all wMel and mitochondria dates to about 8,000 years ago [42]. The low genetic diversity and excessive rare variants in wMel, in the well sampled North American population of the Drosophila Genetic Reference Panel [51], are consistent with a recent sweep of wMel variants [42]. However, the wMelCS and wMel types diverged several thousand years ago [42] and the sweep is incomplete, since there are still wMelCS variants in natural populations [41], [42], [48], [50].
Phenotypic differences associated with different wMel variants could explain why their frequencies have changed. CI, despite being weak in D. melanogaster, has been shown to vary in level in flies harbouring different wMel genotypes [52], [53]. However, the contribution of host or symbiont genetic variation to these differences is not resolved in these studies. Overall clear phenotypic differences between natural wMel variants are not known.
A wMel variant that clearly induces a particular phenotype is wMelPop [54]. This variant was isolated from a laboratory stock and it is pathogenic: it overproliferates and shortens host lifespan [54]–[56]. In terms of genetic markers wMelPop is indistinguishable from wMelCS [57], however, no wMelCS variant with a similar phenotype has been isolated from the wild. Both wMelPop and wMel genotype have been introduced into Ae. aegypti as a strategy to block dengue [30], [31] and they protect differently from viral infection [31], [32]. Because of the potential field application and the pathogenicity of wMelPop it is also important to understand in more detail the phenotypic and genomic differences between wMelPop and other variants.
Here we compare the antiviral protection conferred by different wMel variants, in genetically identical D. melanogaster hosts. We show that wMelCS-like confer greater antiviral protection than wMel-like variants, but have higher bacterial densities and can reduce the survival of the flies. Through the assembly of their genomes and phylogenetic analysis we reconstruct the relationship of the strains. We also investigate in detail the phenotypic differences between the closely related wMelCS and wMelPop and propose a genomic basis for them. This analysis strengthens the notion that susceptibility to infectious disease can rapidly evolve due to changes in symbionts found in the host population.
Results
Phylogenomic Analysis of wMel Variants
To address the question of how genetic variability within the Wolbachia wMel strain affects resistance to viruses, we analysed the five genotypes described by Riegler et al. on the basis of a small number of genetic markers [40]. For the genotypes wMel and wMel3 we used one D. melanogaster line, while for wMel2, wMelCS and wMelCS2 we used two lines for each genotype (Table 1). We will refer to each wMel originating from a unique D. melanogaster line as a variant. In order to determine the phylogeny of these variants we sequenced and assembled their genomes and associated mitochondria. We sequenced 75 bp paired-end libraries and mapped the reads to the wMel reference genome (GenBank ID: AE017196) and to the mitochondrial genome in D. melanogaster Release 5 genome sequence (chrU:5288528–5305749). This mapping strategy was previously used to assemble and analyse the genomes of 179 Wolbachia and 290 mitochondria [42]. We produced a phylogenetic tree of the wMel variants together with the 179 Wolbachia genomes described in Richardson et al. [42] (Figure 1 and Figure S1).
Fig. 1. Phylogeny of wMel variants. 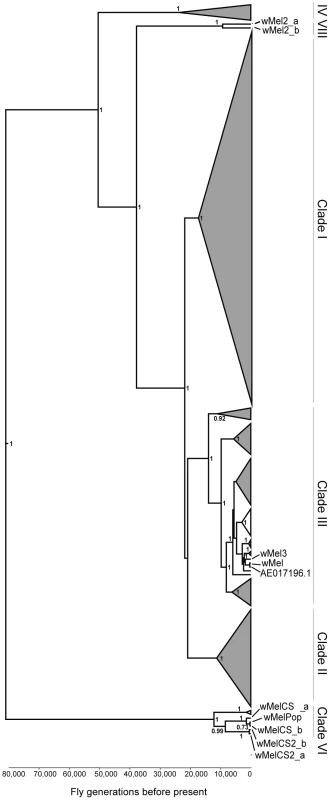
Phylogenomic tree was reconstructed using the concatenated sequences of complete Wolbachia and mitochondrial genomes. The length of the branches reflects the estimated number of Drosophila generations, which was calibrated using the mitochondrial mutation rate. The node labels show posterior supports >0.5. The clades are named after Richardson et al. [42]. Tab. 1. wMel variants used. 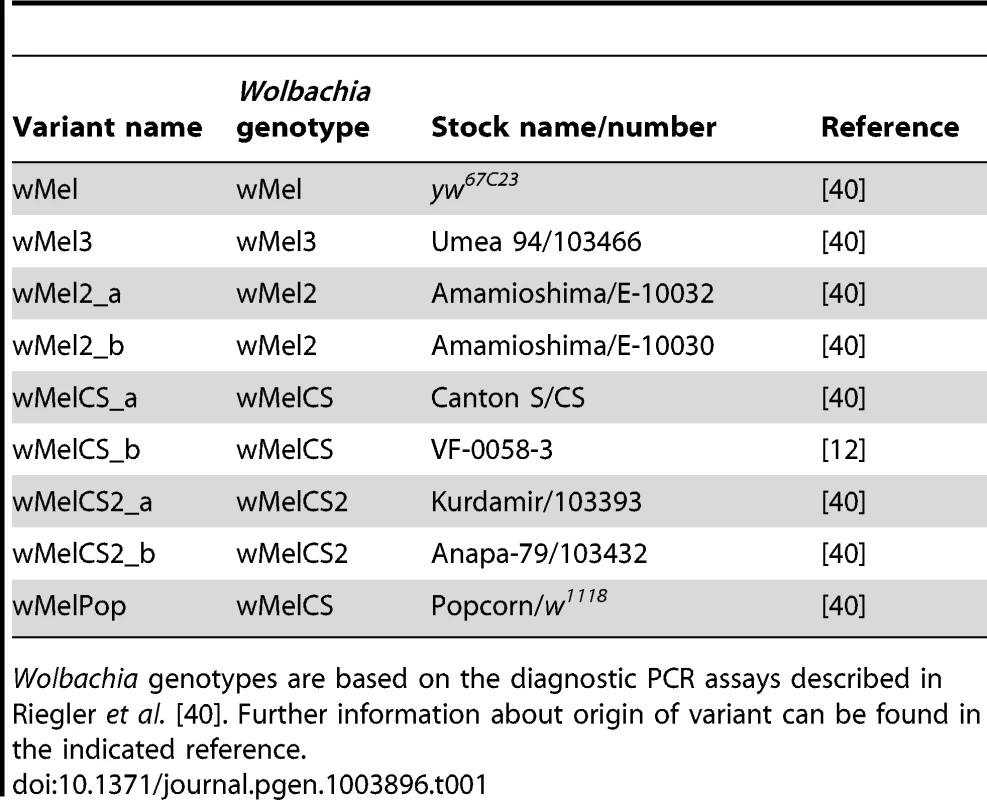
Wolbachia genotypes are based on the diagnostic PCR assays described in Riegler et al. [40]. Further information about origin of variant can be found in the indicated reference. The wMel variant genome clusters together with the reference genome AE017196 in clade III. The only differences we found between them were five positions with an ambiguous call for the wMel nucleotide. This indicates a good quality of the sequencing and assembly, since the reference genome was sequenced from this variant [58]. wMel3 is also assigned to clade III and is the most closely related, out of all the genomes in the phylogenetic tree, to wMel and AE017196. This wMel3 variant is the only known variant with this genotype. The original D. melanogaster stock that had wMel3 was probably related to the laboratory stock used for Wolbachia wMel sequencing. The only genomic marker from Riegler et al. that distinguishes wMel3 from wMel is the absence of the IS5 (ISWip1) WD0516/7 [40]. This seems to be a consequence of a very recent excision of this mobile element in the wMel3 variant since it is present in the closely related wMel variant and in Mel2_a and wMel2_b. Sanger sequencing of this region shows that this would be a precise excision of the transposon (data not shown).
wMel2_a and wMel2_b variants form the new major clade VIII. We estimate that the most recent common ancestor of this clade and clade III dates to 37,537 fly generations before present. The original flies carrying these two variants were captured in the Amami-oshima islands in Japan [40] and eight other lines with wMel2 genotypes have origins in China, Thailand, Philippines and India [41]. Therefore, clade VIII may be exclusive to Asian D. melanogaster populations.
wMelCS_a, wMelCS_b, wMelCS2_a and wMelCS2_b variants belong to clade VI and are relatively closely related. As expected from the genomic markers, wMelCS_a and wMelCS_b are more similar to each other than to wMelCS2_a and wMelCS2_b. Our data confirms that wMelCS-like variants belong to clade VI, as predicted by Richardson et al., based on ISWip1 in silico mapping [42].
Variants of the genotypes wMel, wMel2 and wMel3 are more closely related to each other than to variants of the wMelCS and wMelCS2 genotypes. We estimate that the most recent common ancestor of all these variants dates back to 80,000 fly generations before present and corresponds to the most recent common ancestor of all wMel variants.
The laboratory variant wMelPop is indistinguishable from wMelCS, based on genomic markers [57]. We have also sequenced and assembled its genome and found it to be closely related to wMelCS_b (Figure 1 and Figure S1).
wMel Variants Provide Differing Levels of Protection to Viruses
To compare the phenotypic effects of the wMel variants, we replaced the first, second and third chromosome of Drosophila lines carrying these variants with chromosomes of the DrosDel w1118 isogenic line [59], using balancer chromosomes. All lines were cleaned of possible chronic viral infections, as previously described [12], [60]. The microbiota associated with these lines, as well as the control Wolbachia-free DrosDel w1118 isogenic line (w1118 iso), are expected to be diverse and, presumably, eliminated by the virus cleaning procedure. To homogenize the microbiota associated with these lines, surface sterilized embryos of each line were raised in fly food containing an inoculum of Drosophila-associated microbiota from a reference stock.
We tested the mortality after Drosophila C virus (DCV) infection in the lines harbouring different Wolbachia wMel variants (Figure 2A). DCV is a non-enveloped, positive sense single-stranded RNA virus of the Dicistroviridae family, that is a natural pathogen of D. melanogaster [60], [61]. It has been shown before that Wolbachia gives strong resistance to this virus [12], [14]. All lines with Wolbachia survive the DCV challenge better than the w1118 iso line without Wolbachia, demonstrating that all these wMel variants confer protection to DCV. We have analysed the survival data of these infected lines with a Cox proportional hazard mixed effect model [62]. This method determines the Cox hazard ratio for each line, which in this experiment is a measure of the risk of death of DCV-infected flies from each Wolbachia line relative to the risk of death of DCV-infected flies from the Wolbachia-free line (Figure 2B). A Tukey's test on Cox hazard ratios allows the comparison between all the lines and shows that the Wolbachia variants segregate into two groups (Figure 2B). The Cox hazard ratios of the wMelCS-like lines (wMelCS_a, wMelCS_b, wMelCS2_a and wMelCS2_b) are not significantly different from each other but are lower and significantly different from wMel-like lines (wMel, wMel2_a, wMel2_b, and wMel3). Therefore, variants of clade VI (wMelCS-like) confer higher protection to DCV infection than variants of clades III and VIII (wMel-like). There are still some statistically significant differences in survival between lines of the wMel-like group (Figure 2B). However, the statistical analysis does not allow a clear subdivision. In the timeframe of this experiment there is no significant difference in survival between the lines pricked with buffer only (Figure 2E). Therefore, we conclude that the differences in survival upon viral challenge are due to variability in protection to viruses.
Fig. 2. Wolbachia wMel variants confer different protection to Drosophila C Virus. 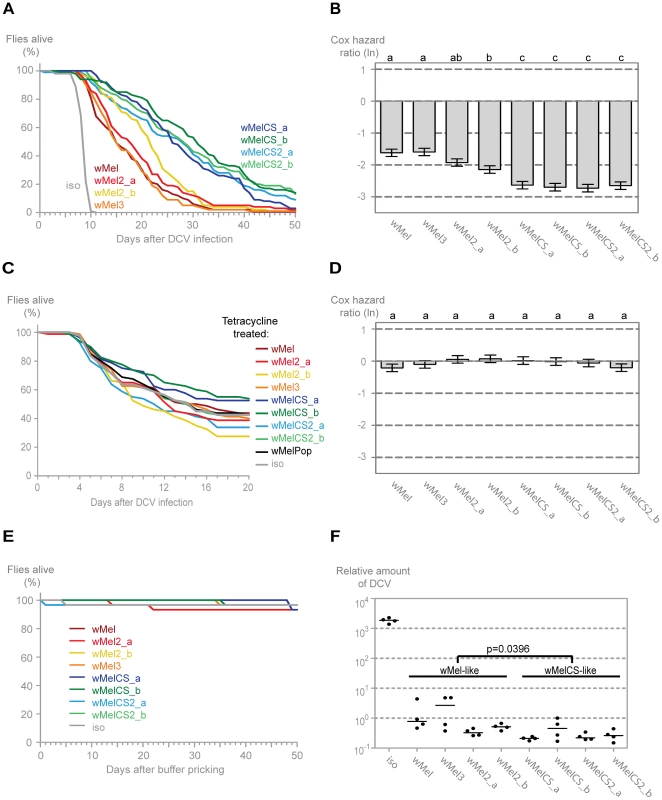
(A) One hundred males of each wMel variant line and w1118 iso were pricked with DCV (107.5 TCID50/ml) and survival was followed daily. Two more replicates were performed with similar results. (B) Cox hazard ratio of each wMel variant line compared to w1118 iso when infected with DCV (107.5 TCID50/ml). The natural logarithm of the Cox hazard ratio is shown. Error bars represent standard error. Letters refer to compact letter display of Tukey's test of all pairwise comparisons. Analysis is based on three independent replicates (doses: 107 TCID50/ml, 107.5 TCID50/ml and 109 TCID50/ml), each with 100 flies per line, with 10 flies per vial. w1118 iso is assigned to group “d” in the compact letter display of Tukey's test (not shown). (C) Eighty males of each tetracycline treated line, derived from the wMel variants lines and w1118 iso, were pricked with DCV (105.5 TCID50/ml) and survival was followed daily. Two more replicates were performed with similar results. (D) Cox hazard ratio of each tetracycline treated line, derived from the wMel variants lines, compared to w1118 iso tetracycline treated line, when infected with DCV. Analysis is based on three independent replicates, one with 80 flies per line (105.5 TCID50/ml) and two with 100 flies per line (one at 107 TCID50/ml and one at 107.5 TCID50/ml), with 10 flies per vial. w1118 iso tetracycline treated line is assigned to group “a” in the compact letter display of Tukey's test (not shown). (E) Thirty males of each wMel variant line and w1118 iso were pricked with buffer and survival was followed daily. (F) 3–6 day old males of each wMel variant line and w1118 iso were pricked with DCV (107.5 TCID50/ml) and collected 3 days later for RNA extraction and RT-qPCR. Relative amount of DCV was calculated using host Rpl32 gene expression as a reference and values are relative to median of wMelCS_b samples. Each point represents a replicate (ten males per replicate, four replicates per Drosophila line), and lines are medians of the replicates. DCV loads are two-fold higher in wMel-like variants than in wMelCS-like variants (linear mixed-effect model, p = 0.0396). Wolbachia and mitochondria are both maternally inherited. Consequently, introducing Wolbachia variants into the same host genetic background implies co-inheritance of the mitochondria associated with them. To determine the influence mitochondria may have on the survival upon the viral infections we cured all the lines of Wolbachia by treating them with tetracycline and we re-homogenized the microbiota in the newly established lines. We have performed this infection with the same dose of DCV as for the Wolbachia lines and also with a lower dose in order to better reveal potential differences between lines (all Wolbachia-free lines are more susceptible to viral infection) (Figure 2C and 2D). We did not observe any statistically significant difference in survival between the tetracycline-treated lines after DCV infection. A direct comparison between wMel and wMelCS-like derived lines also showed no significant difference (Tukey's test on the mixed effects Cox model fit of the survival data, p = 0.953). We conclude that the genetic variability in the mitochondria is not separating these lines regarding the susceptibility to DCV and the original segregation is due to Wolbachia variation. However, we cannot formally exclude the possibility of a Wolbachia-mitochondria genetic interaction.
To determine if the differences in survival between the two groups were due to differences in viral titres, we assessed the viral load in infected flies using real-time quantitative reverse transcription PCR (qRT-PCR) with DCV-specific primers [63]. We assayed titres at 3 days post-infection, since at this point there is already extensive viral replication but it is not yet at its maximum and there is still no lethality associated with infection [12]. All wMel variants confer resistance to DCV, having on average 5000-fold less virus than control (Figure 2F). The comparison of the virus titres between the wMel-like and wMelCS-like groups shows a significant two-fold difference (linear mixed-effect model, p = 0.040). The Drosophila lines with wMelCS-like variants have lower viral titres, in agreement with better survival after DCV infection.
To assess if the wMel variants show differential protection against other viruses we analysed their interaction with Flock House virus (FHV). This is also a non-enveloped positive sense single-stranded RNA virus. However, it belongs to the Nodaviridae family and it is not a natural pathogen of D. melanogaster [64], [65]. We have shown before that Wolbachia protect Drosophila against FHV infections not by limiting the pathogen burden but by increasing survival under similar pathogen load; that is by increasing tolerance to this virus [12], [66]. Consistently with the DCV results we observe that all variants give protection to FHV (Figure 3A). Moreover, the variants split into the same two wMel and wMelCS-like groups, with the latter conferring greater protection (Tukey's test on the mixed effects Cox model fit of the survival data (Figure 3B)). There are, again, some statistically significant differences within the wMel-like group but not between the same variants that show differences in survival after DCV infection.
Fig. 3. Wolbachia wMel variants confer different protection to Flock house virus. 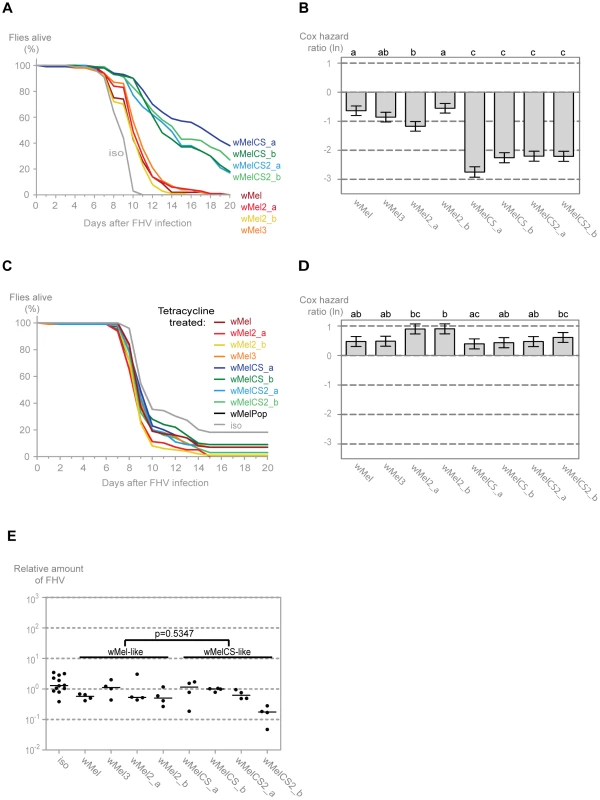
(A) One hundred males of each wMel variant line and w1118 iso were pricked with FHV (109 TCID50/ml) and survival was followed daily. One more replicate was performed with similar results. (B) Cox hazard ratio of each wMel variant line compared to w1118 iso when infected with FHV (109 TCID50/ml). The natural logarithm of the Cox hazard ratio is shown. Error bars represent standard error. Letters refer to compact letter display of Tukey's test of all pairwise comparisons. Analysis is based on two independent replicates, one with 100 flies per line and one with 50 flies per line, with 10 flies per vial. w1118 iso is assigned to group “d” in the compact letter display of Tukey's test (not shown). (C) One hundred males of each tetracycline treated line, derived from the wMel variants lines and w1118 iso, were pricked with FHV (108 TCID50/ml) and survival was followed daily. (D) Cox hazard ratio of each tetracycline treated line, derived from the wMel variants lines, compared to w1118 iso tetracycline treated line, when infected with FHV (108 TCID50/ml). Analysis is based on one replicate with 100 flies per line, 10 flies per vial. w1118 iso tetracycline treated line is assigned to group “a” in the compact letter display of Tukey's test (not shown). (E) 3–6 day old males of each wMel variant line and w1118 iso were pricked with FHV (109 TCID50/ml) and collected 3 days later for RNA extraction and RT-qPCR. Relative amount of FHV was calculated using host Rpl32 mRNA as a reference and values are relative to median of wMelCS_b samples. Each point represents a replicate (ten males per replicate, four replicates per Drosophila line), and lines are medians of the replicates. FHV loads are not significantly different between wMel and wMelCS-like variants (linear mixed-effect model, p = 0.5347). The survival of the tetracycline treated lines upon infection with a lower dose of FHV showed similar results to the DCV challenge (Figure 3C). Although there are some statistical differences between lines, there is no clear segregation between wMel and wMelCS-like derived lines (Figure 3D) and a direct comparison between these groups showed no significant difference (Tukey's test on the mixed effects Cox model fit of the survival data, p = 0.153). This shows that the difference in survival to FHV infection in the non-tetracycline treated lines is not solely due to differences in mitochondria. The differences that we can still observe between lines may be a consequence of differences between mitochondria or due to incomplete isogenization or homogenization of the microbiota in these lines.
To test if there was also a difference in FHV titres between the two groups of wMel variants we measured the levels of this virus three days after infection by qRT-PCR. The comparison of the virus titres between the wMel-like and wMelCS-like groups shows no statistically significant difference (Figure 3E, linear mixed-effect model, p = 0.535). Therefore the differences in survival between the two groups are due to differences in tolerance to FHV, not resistance.
The above results show that all tested wMel variants confer protection to DCV and FHV. There is a differential protection that separates the wMel variants into two groups. The wMelCS-like group lines, compared with the wMel-like lines, have a better survival upon infection with both viruses, higher resistance to DCV, and higher tolerance to FHV.
Wolbachia Densities and Host Lifespan Are wMel Variant Dependent
In order to characterize better the differences between wMel variants and understand the basis of the differential protection, we analysed the titres of Wolbachia in the different lines. We determined by qPCR the levels of Wolbachia genomes relative to host genomes in males of two age groups: 3–4 and 6–7-day-old. (Figure 4A). We observe that, for each variant, the titre of Wolbachia is very similar between the two age groups and that there is no tendency for higher or lower titres at these two time points. However, lines with different wMel variants vary in Wolbachia titres and can, once more, be separated into wMelCS and wMel-like groups. A pairwise Wilcoxon rank sum test shows that wMelCS-like variants titres are not significantly different between them but different when compared to wMel-like variants. wMel-like variants show some differences between themselves but with no clear sub-groups. The median Wolbachia titre of wMelCS-like lines is 2.55 times higher than wMel-like lines. These results show that wMel titres are, at least partially, controlled by the symbiont genotype.
Fig. 4. Wolbachia densities and Drosophila longevity are variant dependent. 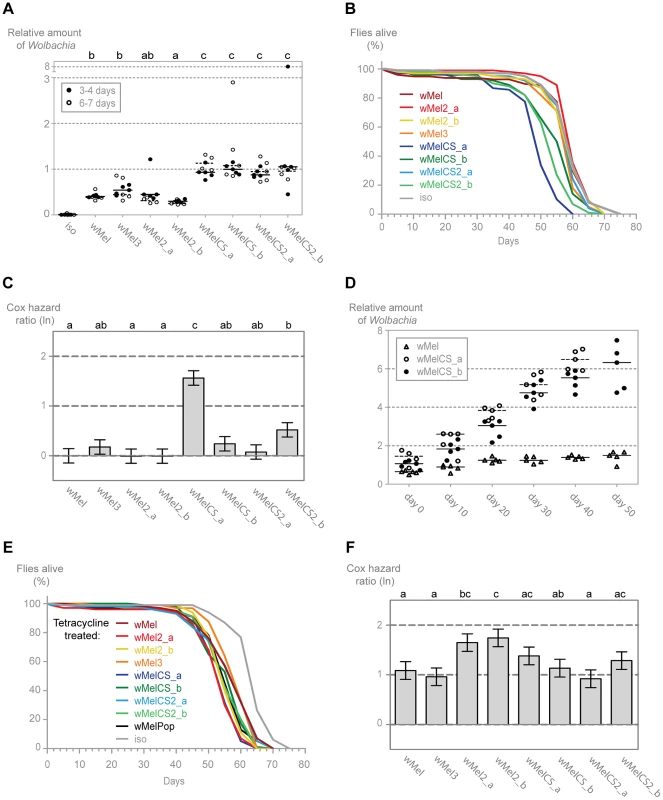
(A) 3–4 and 6–7 day old males of each wMel variant line and w1118 iso were collected for DNA extraction and qPCR. Relative amount of Wolbachia genomic DNA was calculated using host Rpl32 as a reference gene and values are relative to median of 3–4 days-old wMelCS_b samples. Each point represents a replicate (ten males per replicate, four replicates per Drosophila line), and lines are medians of the replicates. The compact letters display of pairwise Wilcoxon rank sum tests between variants is shown on the top. (B) The survival of one hundred males of each wMel variant line and w1118 iso was checked every five days. One more replicate was performed with similar results. (C) Cox hazard ratio of each wMel variant line compared to w1118 iso. The natural logarithm of the Cox hazard ratio is shown. Error bars represent standard error. Letters refer to compact letter display of Tukey's test of all pairwise comparisons. Analysis is based on two independent replicates, each with 100 flies per line, with 10 flies per vial. w1118 iso is assigned to group “a” in the compact letter display of Tukey's test (not shown). (D) Males of wMel, wMelCS_a and wMelCS_b lines were collected for DNA extraction and qPCR every 10 days. Day 0 corresponds to 3–6 days-old flies, wMelCS_a were collected up to 40 days and wMel and wMelCS_b up to 50 days. There are no further time points due to high mortality. Each point represents a replicate (ten males per replicate, five replicates per time point), and lines are medians of the replicates. Relative amount of Wolbachia genomic DNA was calculated using host Rpl32 as a reference gene and values are relative to median of samples of wMelCS_b at day zero. (E) The survival of one hundred males of each tetracycline treated line derived from the wMel variants lines and w1118 iso was checked every five days. The experiment was repeated once with similar results. (F) Cox hazard ratio of each tetracycline treated line, derived from the wMel variants lines, compared to w1118 iso tetracycline treated line. Analysis is based on two independent replicates, each with 100 flies per line, with 10 flies per vial. w1118 iso tetracycline treated line is assigned to group “d” in the compact letter display of Tukey's test (not shown). To determine if these differences in Wolbachia titres have any long-term effect on the D. melanogaster, we followed the long-term survival of these lines in the absence of any viral challenge (Figures 4B and 4C). The three lines with the shortest average lifespan are all infected with wMelCS-like variants. Of these, the wMelCS_a line has a significantly greater mortality rate compared to all other variants and w1118 iso, and wMelCS2_b has a statistically significant greater mortality rate than w1118 iso and three of the wMel-like lines. Despite its shorter mean lifespan, when analysed as proportional hazards the wMelCS_b line is not significantly different from the control. Furthermore, when wMelCS and wMel-like group survivals are directly compared the difference is not significant (Tukey's test on the mixed effects Cox model fit of the survival data, p = 0.073). Therefore we can only state that some wMelCS-like variants have a deleterious effect on longevity. Nonetheless, these results exclude the hypothesis that Drosophila lines with wMel-like Wolbachia succumb to viral infection faster due to a deleterious effect of the variants they are harbouring.
Prompted by these lifespan shortening effects, we investigated how the Wolbachia titres of wMelCS_a, wMelCS_b and wMel change through the host life (Figure 4D). We observe that in these three variants Wolbachia levels increase with Drosophila age (this was not evident in the data set of Figure 4A due to the small interval between the two age groups analysed). Based on the comparison of linear and log-linear models, a linear growth explains better these increases than an exponential one (Table S1). The titres at eclosion are not significantly different between the three variants in a multiple linear regression analysis (intercept wMel: 0.722; intercept difference between wMelCS_a and wMel: 0.416, p = 0.090; intercept difference between wMelCS_b and wMel: 0.222, p = 0.328). However, while there is a significant increase in wMel titres with the host age (slope wMel: 0.017, p = 0.003), the wMelCS_a and wMelCS_b growth rate is 6.5–8 times faster than wMel (slope difference between wMelCS_a and wMel: 0.115, p<0.001; slope difference between wMelCS_b and wMel: 0.092, p<0.001). wMelCS_a also has a twenty percent faster growth than wMelCS_b (slope difference between wMelCS_a and wMelCS_b: 0.024, p = 0.010). These results show that the two tested wMelCS-like variants have a higher growth rate than the wMel-like variant tested. Moreover, wMelCS_a, the variant that shortens host lifespan, has the highest growth rate.
Finally, we analysed the lifespan of the tetracycline treated lines in order to assess the mitochondria contribution to the differences seen in the wMel variants lines survival (Figure 4E and 4F). Although we see small differences between the lines they do not match the differences seen in the wMel variant lines (e.g. the wMelCS_a line treated with tetracycline does not have the shortest lifespan) (Figure 4F). There is no difference in survival of wMel and wMelCS-group derived lines (Tukey's test on the mixed effects Cox model fit of the survival data, p = 0.615). We do observe, however, a statistically significant difference between these groups and w1118 iso derived line (p<0.001 for wMel-group vs w1118 iso derived lines, p<0.001 for wMelCS-group vs w1118 iso derived lines). The w1118 iso line was subjected to the same tetracycline treatment and the difference in survival may be due to variation in mitochondria (see [67]).
Since the wMelCS-like variants have higher titres of Wolbachia and better protection to viruses we tested the correlation between Wolbachia titres and the survival upon viral infections, viral titres and long-term survival (data in Table S2). We found significant correlations between Wolbachia titres and survival upon DCV and FHV infection (Pearson's product moment correlation, p = 0.034 and p = 0.002 with Bonferroni correction, respectively), but not with the other phenotypes.
Given the recurrent phenotypic differences between the wMel-like group and the wMelCS-like group, we tested if, overall, our data led to the clustering of wMel variants into these two groups. To do this we analysed data of survival to viral infection, viral titres upon infections, long-term survival and Wolbachia titres together (Figure 5 and Table S2). A cluster analysis of the scaled values, based on Euclidian distances, shows that the wMel variants phenotypes cluster them into a wMel and a wMelCS-like group. This phenotypic clustering (Figure 5) has a phylogenetic basis (Figure 1) and the two groups correspond to the basal clade VI (wMelCS-like) and to variants of the more closely related clades III and VIII (wMel-like).
Fig. 5. Phenotype-based cluster analysis of wMel variants. 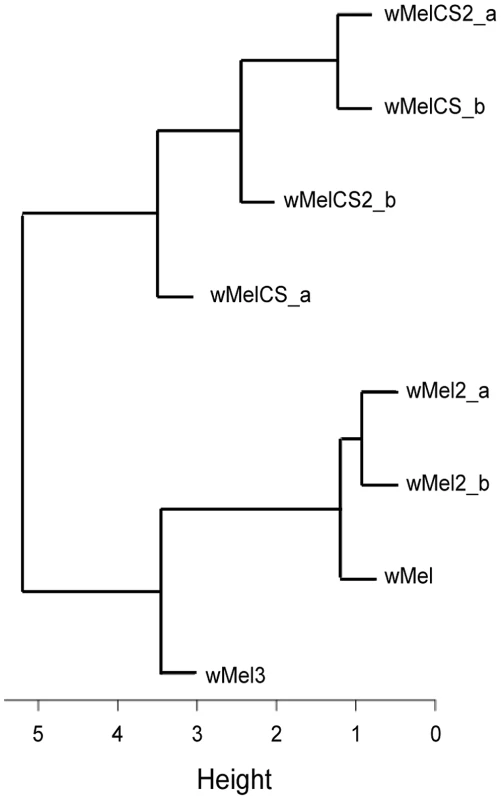
Cluster diagram of the wMel variants based on the Euclidian distance of the scaled values of Cox hazard ratios of long-term survival, survival to FVH and DCV infections, FHV and DCV titres upon infection, and Wolbachia titres (Data in Table S2). Wolbachia wMelPop Provides the Strongest Resistance against Viruses
The life shortening wMelPop Wolbachia strain is known to over-proliferate in its native D. melanogaster host [54] and it has been shown to confer protection to DCV [14]. Importantly, this variant has been transferred to Aedes aegypti where it also limits infection by several viruses, like dengue and Chikungunya, and the malaria parasite Plasmodium gallinaceum [30], [68]. wMelPop is indistinguishable from wMelCS based on genomic markers [57], therefore we made a detailed comparison between wMelPop and wMelCS_b in the same conditions as for the other wMel variants. However, this set of experiments was performed with 1–2-day-old flies to minimize the variability due to different Wolbachia levels within the wMelPop sample or the wMelPop deleterious effect.
Upon challenge with DCV, young flies carrying wMelPop have 235-times lower viral loads than the flies with wMelCS_b (over 3000-fold less than w1118 iso) (Figure 6A, Mann-Whitney test, p<0.001). wMelPop also has much lower titres of FHV three days post infection when compared with wMelCS_b (Figure 6B, Mann-Whitney test, p = 0.007). In most of the wMelPop samples FHV titres were below the limit of detection of the qRT-PCR. Therefore the difference between the medians of wMelCS_b and wMelPop is not quantifiable but it is over ten thousand fold (over one million-fold when compared with w1118 iso). These results show that wMelPop gives stronger resistance to viruses than the closely related wMelCS_b. This data also demonstrates that Wolbachia can confer strong resistance to FHV.
Fig. 6. wMelPop confers the strongest antiviral protection. 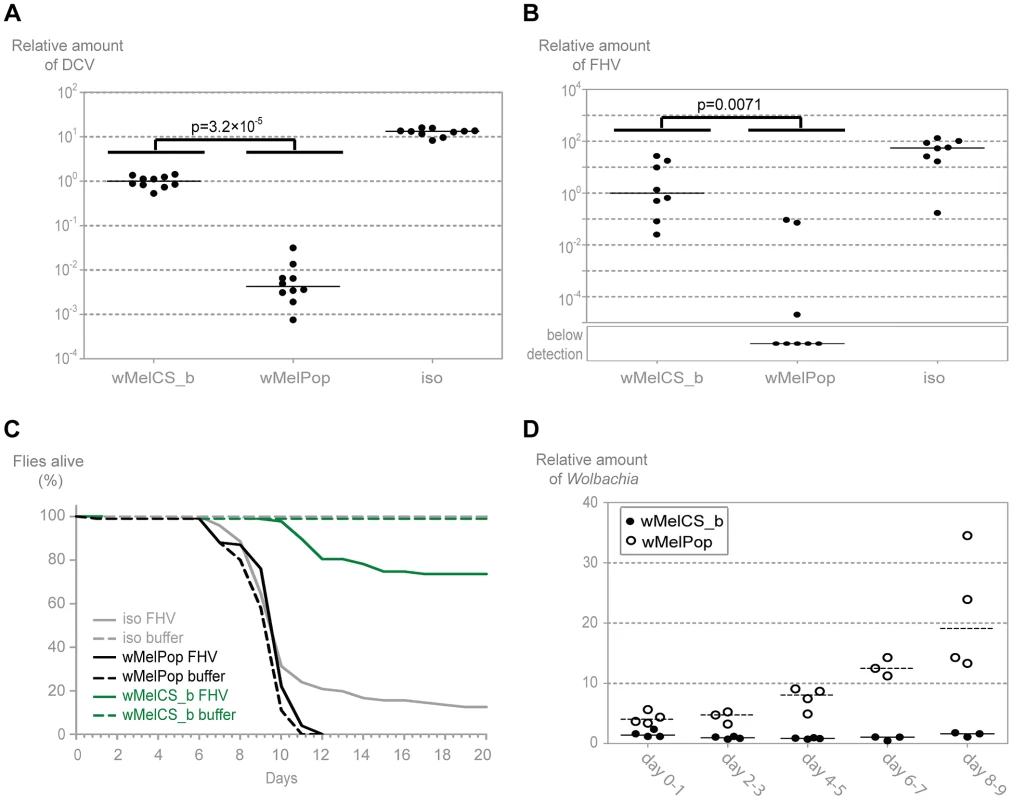
(A) 1–2 day old males of the lines wMelPop, wMelCS_b, and w1118 iso were pricked with DCV (109 TCID50/ml) and collected 3 days later for RNA extraction and RT-qPCR. Relative amount of DCV was calculated using host Rpl32 expression as a reference and values are relative to median of wMelCS_b samples. Each point represents a replicate (ten males per replicate, ten replicates per Drosophila line), and lines are medians of the replicates. DCV titres are 235 times lower in wMelPop line than in wMelCS_b line (Mann-Whitney test, p = 3.2×10−5). (B) 1–2 day old males of the lines wMelCS_b, wMelPop, and w1118 iso were pricked with FHV (107 TCID50/ml) and collected 3 days later for RNA extraction and RT-qPCR. Relative amount of FHV was calculated using host Rpl32 expression as a reference and values are relative to median of wMelCS_b samples. Each point represents a replicate (ten males per replicate, eight replicates per Drosophila line), and lines are medians of the replicates. FHV titres are lower in wMelPop line than in wMelCS_b line (Mann-Whitney test, p = 0.007). (C) One hundred 1–2 day old males of the lines wMelCS_b, wMelPop, and w1118 iso were pricked with FHV (107 TCID50/ml) or buffer, and the survival was followed daily. (D) Males of wMelCS_b and wMelPop lines were collected for DNA extraction and qPCR every 2 days. Each point represents a replicate (ten males per replicate, three to four replicates per time point), and lines are medians of the replicates. Relative amount of Wolbachia genomic DNA was calculated using host Rpl32 as a reference gene and values are relative to median of samples of wMelCS_b at day 2–3. We tested wMelPop protection to viral infection in terms of survival upon infection with FHV (Figure 6C). Contrary to wMelCS_b, the presence of wMelPop does not increase survival of FHV infected flies (Tukey's test on the mixed effects Cox model fit, wMelCS_b versus w1118 iso lines infected with FHV, p<0.001; wMelPop versus w1118 iso lines infected with FHV, p = 0.229). This is due to a very strong pathogenic effect of wMelPop, even in the absence of FHV; at 25°C all flies are dead by day 12 (wMelPop versus w1118 iso lines not infected with FHV, p<0.001). This pathogenic effect at 25°C has been reported before [54]–[56] but seems stronger in our experiment. Nonetheless, FHV does not cause any mortality in the wMelPop line (wMelPop line infected and not infected with FHV, p = 0.816), which is consistent with the strong resistance we observed. Therefore, although wMelPop confers strong resistance to FHV, it does not increase lifespan of an FHV infected host because it is very deleterious by itself.
Given the strong anti-viral resistance and pathogenic effect we observe with wMelPop at 25°C, we decided to measure how Wolbachia titres change with age in flies infected with wMelPop and wMelCS_b (Figure 6D). wMelPop growth is better explained by an exponential model of growth than a linear model (Table S1) with an estimated doubling time of 3.4 days. Wolbachia titres and growth rate are significantly higher in wMelPop (log-linear model, intercept difference between wMelPop and wMelCS_b: 0.904, p<0.001; slope difference: 0.224, p<0.001). At the day of our viral infection (1–2 days) wMelPop titres are 3 to 5 times higher than wMelCS_b titres.
Once again we observe that the wMel variant with higher titres gives stronger protection to viruses. In wMelPop the exponential growth leads to a much stronger resistance to DCV and FHV but severely reduces the host lifespan.
Genetic Basis of the Phenotypic Differences between wMel Variants
Having identified phenotypic differences between wMel variants we asked what their genetic bases were. To answer that used the information from the sequence analysis and their assembled genomes. From the multiple alignments we extracted variant sites that were different between all wMel-like and all wMelCS-like variants, in order to focus on common differences. We detected 108 single nucleotide polymorphisms (SNPs) between these two groups of variants, a tandem duplication and seven insertion-deletion polymorphisms (indels) (Tables 2, S3 and S4). 83 of the SNPs map to annotated wMel genes [58], of which 59 are non-synonymous substitutions. The 55 genes that differ in these 59 SNPs encode proteins with a wide variety of functions, based on predicted conserved domains (Table 2). This set contains a high number of genes coding ankyrin-repeat containing (ANK) proteins: WD0073, WD0292, WD0514, WD0636, WD0754, and WD0766.
Tab. 2. Non-synonymous SNPs between wMel-like and wMelCS-like Wolbachia variants. 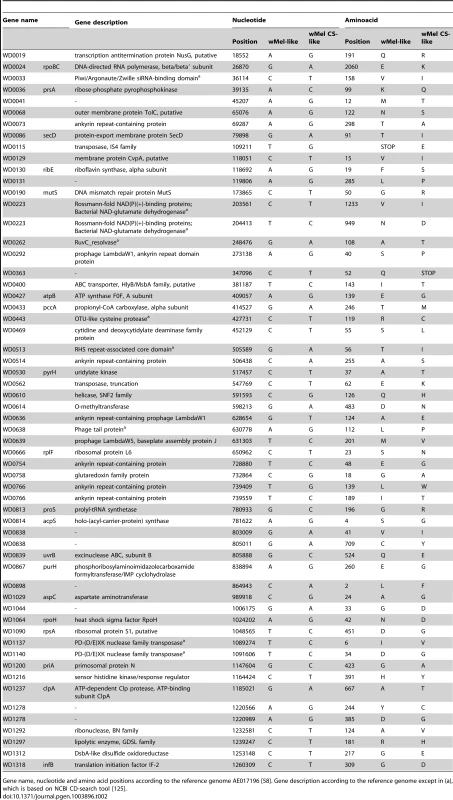
Gene name, nucleotide and amino acid positions according to the reference genome AE017196 [58]. Gene description according to the reference genome except in (a), which is based on NCBI CD-search tool [125]. In order to understand the basis of the strong phenotypic differences between the closely related wMelCS_b and wMelPop variants we have investigated the differences between their genomes. Previous studies have not identified any genetic differences between wMelCS and wMelPop [57], [69]. From the genome sequence analysis we found only two SNPs unique to wMelPop, and six positions where there was an ambiguous call for the wMelPop nucleotide. We Sanger sequenced these regions in wMelCS_b and wMelPop, and found that only two synonymous SNPs were true differences between these variants (position 943,443, G>A, unique to wMelPop; position 858,287, T>C, unique to wMelCS_b). In our analysis of split sequencing reads, there were no indel polymorphisms unique to wMelPop that met our filtering criteria. Therefore we cannot identify any SNPs or small indels that could be clearly related to the phenotypic differences.
To identify other possible differences between wMelPop and wMelCS_b we analysed copy number variation in their genomes. We mapped the sequence reads to the wMel reference genome and examined variation in the depth of coverage. In wMelPop there is a large increase in read depth in a ∼21 kB region. Using the mean shift approach implemented in CNVnator [70] we estimated that this region has been amplified approximately five times (Figure 7A and Figure S2; t test: p<10−20; breakpoints: 486,601–507,800). Due to the extensive amplification, probable association with the over-proliferative phenotype, and containing eight predicted genes, we call it the Octomom region.
Fig. 7. Genomic region amplified in wMelPop. 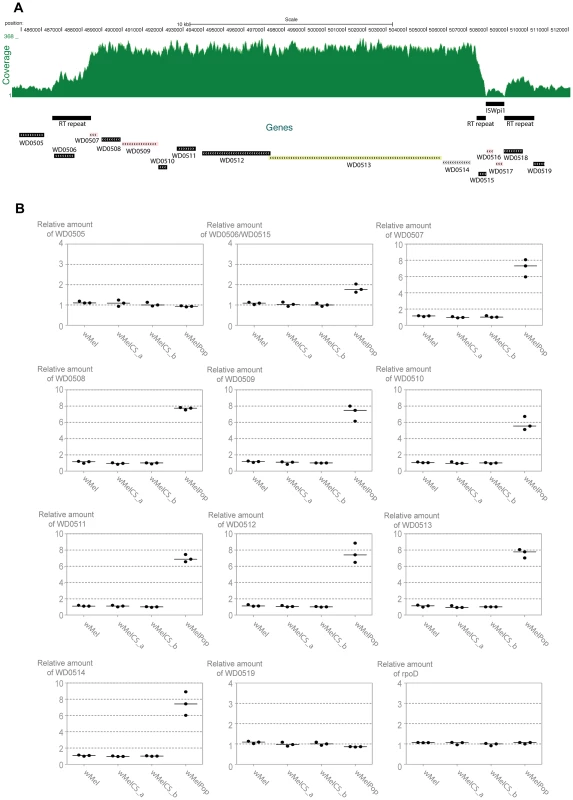
(A) Depth of coverage of sequence reads of wMelPop mapped to wMel reference genome (GenBank: AE017196) in region 484,564 to 512,000. Nucleotide positions, predicted genes, RT repeats and the ISWpi1 element in this region of wMel are shown. The 5′ RT repeat extends from 486,532 to 488,449 (1912 bp). The 3′ RT repeat in wMel extends from 507,470 to 510,325 but is split in two parts due to the insertion of an ISWpi1 (IS5) transposon from 507,928 to 508,848. This ISWpi1 is not present in wMelPop or the closely related wMelCS_b. Figure modified from USCS genome browser (http://genome.ucsc.edu/) [121], [122]. (B) Relative amounts of genomic copy number of Octomom genes (WD0506–14), genes adjacent to Octomom region (WD0505 and WD0519) and control gene rpoD in wMel, wMelCS_a, wMelCS_b and wMelPop were calculated using wsp as a reference gene. Values are relative to median of wMelCS_b samples. Each point represents a replicate (ten males per replicate, three replicates per Drosophila line) and lines are medians of the replicates. There are two repeated regions, with the same orientation, flanking the Octomom region (RT repeat in Figure 7A). The 5′ repeat region contains WD0506, which is annotated as a pseudogene in the reference genome (GenBank: AE017196 [58]), but it may encode a 329 aa protein with a reverse transcriptase (RT) with group II intron origin domain. In the wMel reference genome the 3′ repeat region is split in two parts due to the insertion of an ISWpi1 (IS5) transposon (Figure 7A) (ISWpi1 is repeated 13 times in the wMel genome [58], [71]). This ISWpi1 insertion, however, is absent in the wMelCS-like variants, including wMelPop. In fact presence/absence of this insertion is one of the genomic markers used to distinguish wMel variants (IS5 WD0516/7) [40]. Accordingly, in the coverage plot (Figure 7A) that there is no coverage at the interface between the ISWpi1 and the RT repeat regions in wMelPop. Therefore, this region in wMelPop is 100% identical to the 5′ RT repeat (confirmed, in the region of ISWpi1 insertion, by Sanger sequencing, data not shown). Two other 100% identical RT repeats occur in the genome, at positions 243,822–245,739 and 584,482-582,565 and a smaller 718 bp sequence at positions 633,948–634,665, also 100% identical in its length.
The amplified region in wMelPop contains eight predicted genes between the RT repeats, WD0507–WD0514 (Figure 7A, Table S5). WD0507–11 encode proteins potentially involved in DNA replication, repair, recombination, transposition or transcription. The genes WD0512–14 have previously been shown to be an operon [72]. WD0513 protein has an Rhs domain and WD0514 encodes a ANK repeat protein, but the function of any of the three proteins encoded in this operon is unknown.
The Octomom region was first noticed because of its presence in the strain wMel but absence in many other Wolbachia strains [72]. It has since been found that there are homologues of WD0512–14 in wPip [73], [74] and of WD0514 in several strains of Wolbachia supergroup A [75]. We find orthologues of all the genes of the Octomom region, including the RT repeat, in the genome of wPip (GenBank: AM999887.1 [76]). In wPip WD0507–10 orthologues have conserved synteny with wMel. We also find WD0507–509 homologue syntenic blocks in the prophages WOVitA1 of wVitA (GenBank: HQ906662.1 [77]) and WOVitB1 of wVitB (GenBank: HQ906666.1 [77])) and in wAlbB (GenBank: CAGB01000117.1).
WD0512–3 and their wPip homologue are also an interesting example of a horizontal gene transfer between Wolbachia and mosquitoes [73], [74], [78]. Previously, their homologues have only been found in Culicidae (Aedes, Anopheles and Culex). We have also found homologues of WD0513 in the recently sequenced genome of Daphnia pulex (GenBank: EFX66732.1 [79]). DAPPUDRAFT_229333 and DAPPUDRAFT_300516 are 35% and 32% identical to this protein, respectively.
To confirm the depth of coverage results we performed qPCR to determine relative genomic copy numbers of the genes immediately adjacent and inside the Octomom region in wMel, wMelCS_a, wMelCS_b and wMelPop (Figure 7B). All genes tested showed the same relative amount in wMel, wMelCS_a, wMelCS_b. The genes immediately outside the Octomom region WD0505 and WD0519, as well as two other control genes located elsewhere in the genome, rpoD and gmk, show the same copy number in wMelPop and in the other wMel variants (between 0.86 and 1.09 relative to wMelCS_b) (Figure 7B and data not shown). In contrast, in wMelPop the eight genes inside Octomom, WD0507–14, have estimated copy numbers between 5.54 and 7.78 times the levels of wMelCS_b (with a median of 7.42 times). These results confirm the extensive amplification detected by the depth of coverage analysis and show a 7-fold amplification of this region.
The results for WD0506/WD0515 (the qPCR primers amplify both) show 1.77 fold difference between wMelPop and wMelCS_b (Figure 7B). There are 4 identical copies of the amplified region in wMel, wMelCS_a and wMelCS_b (in the 4 full RT repeats). If this region were also amplified 7 times in wMelPop we would expect 2.75 more copies in wMelPop than in wMelCS_b. The fold difference between the wMelPop and wMelCS_b is lower than expected but shows an amplification of this region and indicates that in wMelPop there are 3 more copies of this gene.
The only other large duplication in the wMel variants detected using CNVnator were in wMel2_a and wMel2_b, where a large region corresponding to the phage WO-B has been duplicated (Figure S2; t tests: p<0.001; breakpoints in both: 569,001–634,000). This is a stable duplication since the most recent common ancestor of these lines dates to an estimated 9,252 host generations ago (Figure 1). Independent WO-B prophage amplifications have been shown before in Wolbachia strains; it is present in two copies in wRi [80] and five copies in wPip [76].
Discussion
We have found that genetically closely related variants of Wolbachia from D. melanogaster vary in the degree to which they protect their hosts against viral infection. The Wolbachia variants that provide the greatest protection have higher titres and often shorten the lifespan of their hosts (Table S6). Previous work has shown that in natural populations these highly protective wMelCS-like variants were recently largely replaced by less protective wMel-like variants. The genome sequences of strains conferring different levels of protection have allowed us both to reconstruct the evolution of antiviral protection and identify candidate genes that may affect it.
Phylogeny and Genomics of wMel Variants
Large-scale genome sequencing of wMel variants from natural populations of D. melanogaster has previously identified two major monophyletic groups of Wolbachia [42]. We were working with a set of Wolbachia variants that had been identified using a small number of genetic markers, so we sequenced the genomes of these variants and their associated mitochondria in order to determine where they fall on the phylogeny. We found that these variants belong to both major monophyletic groups, which diverged approximately 80,000 fly generations before present. This date corresponds to the most common recent ancestor of all wMel variants in D. melanogaster.
We found that there are striking differences in the degree to which the strains from the different phylogenetic clades protect flies against viruses, with the less common wMelCS-like clade providing the stronger protection and having higher Wolbachia densities. This phylogenetic basis for the phenotypic differences confirms that genetic differences between wMel variants are responsible for the variation in symbiont titres and resistance to viruses. Therefore, we identified the common genetic differences between all the wMelCS-like variants and all the wMel-like variants. We found eight indels and 108 SNPs that differ between them, with polymorphisms in the coding sequence of 58 proteins. This number is still too high in order to speculate on possible individual contributions to the phenotypic differences. Future experimental work will help to further reduce this finite number of candidate differences.
We have also compared the genome of the pathogenic variant wMelPop with the closely related wMelCS_b. The wMelPop genome has only two unique differences from the other strains – a single synonymous SNP and a 7-fold amplification of a ∼21 kb region which we named Octomom. The Octomom amplification is therefore the most probable cause of wMelPop pathogenicity and increased protection against viruses. In bacterial genomes copy number variation is very common and mostly involves unequal recombination between two direct sequence repeats, amplifying the region in between [81]. The Octomom region in wMelCS is flanked by two identical 1912b direct repeats, which may provide the origin for the initial duplication in wMelPop. In bacteria and viruses gene amplifications have been shown to increase growth or virulence [81]–[85]. In the future it will be important to show functional data linking this amplification and the pathogenic phenotype of wMelPop.
The functions of the genes in the Octomom region are unknown. The WD0506–WD0511 proteins have predicted domains that are related to interactions with nucleic acids and could have a role in DNA replication, transcription or repair. Therefore, the amplification of these genes could have a direct effect on the replication of Wolbachia. WD0512–14 have homology to proteins or protein domains of eukaryotes and are, consequently, candidate effector proteins of Wolbachia. Hypothetically they could mediate the pathogenicity through interaction with the host.
Key adaptive traits of bacterial pathogens and symbionts are often controlled by genes that are frequently gained and lost through evolution, which are collectively known as the ‘accessory genome’. The Octomom region appears to fit this pattern as it is partially or totally absent in several Wolbachia strains [72], [86], [87]. WD0512–3 homologues have also been suggested to be amplified in wSim [86], although they are not present in its unassembled genome sequence [87]. Homologues of some Octomom genes in other strains have been described [73]–[75], [86] and here we identify more in WOVitA1, WOVitB1, and wAlbB. Moreover, we detect orthologues of all the Octomom genes in wPip, although not as one syntenic block. As the number of sequenced genomes of different Wolbachia strains increases it will be interesting to understand the evolutionary history of this region. In particular if there is horizontal gene transfer between strains, as suggested before [72], [74]. This would be compatible with our finding of some of these genes in prophage regions of WOVitA1 and WOVitB1.
The horizontal transfer of these genes may also occur between Wolbachia and their insect hosts, as two of the genes in the Octomom region, WD0512–3, are homologous to genes previously identified only in Culicidae mosquitoes [73], [74], [78]. The direction of the horizontal gene transfer between mosquitoes and Wolbachia is not clear [73], [74], [78]. In mosquitoes these homologues constitute a family of proteins termed salivary gland surface proteins (SGSs). There is evidence that Ae. aegypti aaSGS1 is a receptor for malaria sporozoite in salivary glands [78] and An. gambiae Sgs4 and Sgs5 are components of the saliva [88]. We have identified two other homologues of WD0513 in the crustacean Daphnia pulex. The number of sequenced crustaceans genomes is very low so we do not know how prevalent these genes are in crustaceans. However, the absence of homologues in any other sequenced insect opens the possibility that there was also horizontal gene transfer between Daphnia/Crustaceans and either mosquitoes or Wolbachia.
Phenotypes Associated with wMel Variants
Symbionts could protect their hosts against infection either by limiting pathogen titres (resistance) or by reducing the harmful effects of those pathogens (tolerance) [66]. We have previously reported that Wolbachia provides tolerance to FHV and resistance to DCV [12]. In this study we found similar FHV titres in lines with wMelCS-like and wMel-like variants, despite the former having far lower mortality rates. This indicates that natural wMel variants differ in how they modulate tolerance to FHV infection rather than resistance (although wMelPop confers strong resistance to FHV, see below). On the other hand, the levels of DCV change between the two groups, with wMelCS-like variants having a two-fold reduction in DCV titres when compared with wMel-like variants. This difference is small, especially when compared to the 5,000-fold reduction in titres in relation to the control without Wolbachia, but is reflected in a substantial change in survival. Therefore, it is possible that there is also a tolerance component in the variants differential protection to DCV. However, with our data we cannot distinguish between these hypotheses since it is possible that even a small change in viral titre is sufficient to explain the better survival (see also discussion in [89]). Nonetheless, induced tolerance to DCV has been shown before for the Wolbachia strain wRi in D. simulans [23]. Therefore, the interaction of Wolbachia with different viruses may always have components of resistance and tolerance modulation.
The more protective wMelCS-like variants reach 2.5 higher titres than wMel-like variants in the first days after adult eclosion, and then continue to proliferate during the lifespan of their host. These results show that in D. melanogaster the control of Wolbachia levels is also dependent on the endosymbiont genotype. It has been shown before that the host genotype and Wolbachia strain can influence Wolbachia titres [56], [90]–[95]. In Leptopilina heterotoma each strain's titre is even independent of the presence of the other strains [93]. Different strains of Wolbachia also reach different levels in D. simulans, although the host nuclear genetic background has not been controlled in this study [23]. Our results show that these differences are also seen between Wolbachia variants that are very closely related to each other (their most recent common ancestor is estimated to date to only 8,000 years or 80,000 fly generations before the present).
The positive correlation between Wolbachia titres and protection against viral infection suggests that this may be the cause of the greater protection provided by the wMelCS-like variants. It is important to note that the strains are not phylogenetically independent, so the association between protection to viruses and titres might have arisen independently in the ancestors of the wMel-like and wMelCS-like groups. However, this seems unlikely, as a density effect has been previously reported in Wolbachia-mediated antiviral protection. Wolbachia-host combinations with higher titres of Wolbachia show higher protection [23], [94], [96] and decreasing levels of Wolbachia with antibiotic treatment lowers protection [94], [97]. Correlations between titres and other Wolbachia-associated phenotypes have been shown before (e.g. with cytoplasmic incompatibility) [95], [98]–[103]. Therefore, the simplest hypothesis is that the differential protection to viruses of the wMel variants is a consequence of their titres.
The localization of the protective symbionts and the pathogens could be an important factor to understand their interaction [23]. Wolbachia, DCV and FHV have been shown to infect several tissues of D. melanogaster [104]–[108]. Although the information on localizations is not necessarily exhaustive there are some tissues of overlap between Wolbachia and the two viruses where the interaction could occur. It will be important in the future to determine the tissue distribution of the different Wolbachia variants and how it contributes to the overall differences in titres. It will also be interesting to know if Wolbachia titres increase with host age is uniform between all the tissues. It has been previously shown that some Wolbachia strains grow at different rates in heads and ovaries [56].
We found that some of the most protective wMel variants reduce the survival of their hosts, suggesting that there may be a trade-off between symbiont-mediated protection and other components of fitness. This cost could be either due to the metabolic cost of their replication or damage caused by their presence. The difference between the wMel-like and wMelCS-like strains was less clear-cut for this trait. We observed that two wMelCS-like lines had significantly greater mortality rates. A third line infected with wMelCS_b has previously been shown to have a shorter lifespan than the control [12], although this is not significant in this report. The fourth wMelCS-like line did not show any detectable effect on lifespan. This variation could be due to the cost of Wolbachia infection being difficult to assess in normal laboratory conditions. This reduction in longevity may not directly affect the fitness of flies in the wild since probably not many flies live up to this late age and their fertility would be very low. However, the assay can be interpreted as a proxy for fitness costs associated with the Wolbachia variants, which are expressed in other unknown ways in the wild.
The phenotypes of the line carrying the laboratory variant wMelPop are consistent with the differences between natural variants. Our results are in agreement with previous reports that wMelPop can reach high titres and shorten lifespan [54]–[56], as well as to give strong protection to viruses [31], [32]. Here we directly compare this variant with wMelCS_b, its closest related variant, in their natural host. wMelPop is the variant that reaches higher levels in D. melanogaster, gives the strongest resistance to viruses, and most severely shortens the host lifespan at 25°C. The pathogenic effect was described before at 25°C [54]–[56]. Yet, this phenotype at 25°C seems to be stronger in our experimental conditions. This is probably related with the wMelPop exponential growth that we detect. We also observed that flies with wMelPop have very strong resistance to DCV and FHV. The strong resistance to FHV induced by wMelPop may indicate that there is no qualitative difference between the interference of Wolbachia with DCV and FHV. Again, it may be only a question of different degrees of resistance and tolerance to different viruses.
Evolution and Dynamics of Wolbachia in Populations
Analysis of Drosophila lines collected from the early 20th century to the present has indicated that natural selection has driven a recent and fast replacement of wMelCS-like variants by wMel-like variants and their associated mitochondria [40], [41]. A more recent phylogenomic analysis of Wolbachia and mitochondria is consistent with a wMel-like global replacement, although it indicates that this event is not complete and started before the 20th century [42]. Overall, it is clear that there was a relatively recent and rapid replacement of wMelCS with wMel-like variants at a worldwide level. Therefore, our results indicate that this has resulted in a recent and rapid decline in the level of anti-viral protection that Wolbachia provides D. melanogaster in the wild. Consequently, we can conclude that the driving force for this change in wMel frequencies was not an increase in viral protection. On the other hand, the wMelCS-like variants that have higher titres and can have a cost, have been replaced with variants with lower titres and, most probably, lower cost to their hosts.
Our data suggests that the balance between benefit (protection to viruses) and cost may have shifted recently, resulting in selection favouring lower levels of protection. In the simplest scenario, the rate at which this replacement has occurred would allow us to easily estimate the net benefit that the low protection strain has had. There are however several complexities that could affect the dynamics of this replacement. First, if the viruses are predominantly transmitted within D. melanogaster populations rather than among different fly species, then the spread of a low protection strain might increase the viral prevalence [29]. This might make the fitness of the low protection strain negatively frequency dependant, potentially stably maintaining both strains in the population. Second, the difference in the density of the high and low protection variants might affect other aspects of Wolbachia's fitness, such as its vertical transmission efficiency or the strength of cytoplasmic incompatibility [95], [103], [109]. These parameters can be experimentally measured and their effects explored with simple extensions to standard models.
In order to block transmission of dengue, the wMel and wMelPop variants were recently introduced into the mosquito Ae. aegypti [30], [31]. Our work in D. melanogaster is in agreement with the mosquito data showing that wMelPop confers both a higher protection to viruses and a higher fitness cost when compared to wMel [31], [32], [110]. The deployment of these Wolbachia infected mosquitoes in the field has to take in consideration the trade-off between fitness costs which make it difficult to invade a population and protection to dengue. Our analysis indicates that wMelCS-like variants have an intermediate phenotype in terms of benefit and cost, and could be considered as an alternative.
Our data also indicate that if there is a strong selection for a mosquito-Wolbachia combination with lower fitness costs, this might result in lower protection to viruses. The dynamics of this selection may influence the success of this strategy to control dengue infection. In addition to the replacement of wMelCS-like variants with wMel-like variants in D. melanogaster, rapid evolution of Wolbachia has been observed in natural populations of D. simulans, resulting in an increase in fertility of Wolbachia infected flies [111]. Finally, if the Octomom region amplification is the basis of wMelPop higher titres and protection to viruses, it could have important consequences on its long-term maintenance in mosquito populations. Duplications in bacterial genomes can be very unstable due to homologous recombination [81]. If loss of the duplication is frequent in a wMelPop infected mosquito population, a rapid selection of a variant with low replication and low protection to viruses may be expected.
The differences in protection to viral infection with wMel variants demonstrate that in order to understand Wolbachia protection to viruses in D. melanogaster one has to consider not only presence or absence of Wolbachia but also the genetic variability of the symbiont. Our results provide another example of how bacterial symbionts can cause rapid evolution in natural populations and control important traits. Furthermore, they illustrate how the ease with which genomes can be sequenced can provide clues to the molecular basis of these traits.
Materials and Methods
Fly Strains and Husbandry
D. melanogaster lines with Wolbachia are described in Table 1. Lines with Wolbachia variants described in Riegler et al. [40] were kindly provided by Markus Riegler and Scott O'Neill. wMelCS_b source and DrosDel w1118 isogenic background were described elsewhere [12], [59]. wMel variants were introduced in the DrosDel w1118 iso isogenic background by chromosomes replacement using a first and third double balancer line and a second chromosome balancer line. The crosses were performed with Wolbachia-infected females, ensuring endosymbiont transmission through the germline. The fourth chromosome was not isogenized. All the Wolbachia genotypes were confirmed by PCR, as described in Riegler et al. [40] (data not shown).
The lines were cleaned of possible chronic viral infections as described elsewhere [12], [60].
In order to homogenize the gut microbiota, embryos from each line were sterilized with 2% sodium hypochlorite, followed by 70% ethanol and washed with sterile water. Embryos were placed in new food vials and 150 µl of a bacterial inoculum from a reference stock was added. The inoculum was produced by mixing 5 ml of sterile water with 2 g of food from 10 days old vials containing VF-0058–3 flies [12], and filtering it to remove eggs and larvae.
Tetracycline-treated lines were cleaned of Wolbachia infection by raising them for two generations in ready-mix dried food (Philip Harris) with 0.05 mg/ml of tetracycline hydrochloride (Sigma). Experiments were performed on lines that were raised without antibiotics for at least 6 generations.
Drosophila lines were maintained on standard cornmeal diet at a constant temperature of 25°C. We focused the analysis on males in the assumption that Wolbachia levels would be more stable in these. Wolbachia is present in ovaries and the sizes of these vary greatly with mating status and physiology of the female.
Long-Term Survival Analysis
To measure the lifespan of different fly lines, 10 flies were placed per vial (without yeast) per replicate, at 25°C. Vials were checked for survival and changed every 5 days.
The analysis of survival data was performed with the Cox proportional hazard mixed effect model. Fixed effects include genotype and repeat of the experiment while replicate vials within the same experiment were considered as a random effect. This method accounts for variation between vials of the same line in the same experiment and variation between replicates of the experiment. Model fitting was done using the coxme package in R [112]. Tukey's test was applied for pairwise comparisons of Cox hazard ratios between all wMel variants and DrosDel w1118 iso.
Virus Production and Infection
Viruses were produced and titrated as in Teixeira et al. [12], with minor changes. DCV was titrated in Schneider's Line 2 (SL-2), while FHV was titrated in Schneider Drosophila line 2 (DL2).
For viral infections CO2 anesthetized flies were pricked in the thorax. The 0.15 mm diameter needles used for infection (Austerlitz Insect Pins) were dipped into a virus solution diluted to the desired concentration in 50 mM Tris-HCl, pH 7.5. After the infection flies were kept in vials without yeast, 10 flies per vial. DCV infected flies were maintained at 18°C, while FHV infected flies were maintained at 25°C. Vials were checked for survival daily and changed every 5 days. Unless otherwise stated, infection was performed on 3 to 6 days-old flies.
Survival analysis was done as above.
RNA Extractions and cDNA Synthesis
For each sample 10 flies were pooled and homogenized with a plastic pestle in 1 ml of Trizol Reagent (Invitrogen). RNA was extracted according to manufacturer's protocol and re-suspended in 50 µl of DEPC-treated water (Ambion). RNA concentrations were determined using NanoDrop ND-1000 Spectrophotometer. cDNA was prepared from 1 µg of total RNA using Random Primers and M-MLV Reverse Transcriptase (both Promega). Primers were allowed to bind to the template RNA for 5 min at 70°C and the reaction proceeded to 25°C for 10 min, 37°C for 60 min and 80°C for 10 min.
DNA Extractions
For Wolbachia relative quantification, ten flies were used per replicate. DNA was extracted according to DrosDel protocol (http://www.drosdel.org.uk/molecular_methods.php) [59]. For wMel Octomom genes relative quantification, total DNA was extracted from replicates of ten flies using a standard phenol-chloroform protocol. The DNA concentrations were checked with NanoDrop ND-1000 Spectrophotometer.
Real-Time Quantitative PCR
The real-time qPCR reactions were carried out in 7900HT Fast Real-Time PCR System (Applied Biosystems) or CFX384 Real-Time PCR Detection System (BioRad). For each reaction in 384-well plate (Applied Biosystems or BioRad) we used 6 µl of iQ SYBR Green supermix (Bio Rad), 0,5 µl of each primer solution at 3,6 µM and 5 µl of diluted DNA. Each plate contained three technical replicates of every sample for each set of primers. Primers used are described in Table S7.
The thermal cycling protocol for the amplification of Wolbachia genes was as follows: initial 50°C for 2 min, denaturation for 10 min at 95°C followed by 40 cycles of 30 s at 95°C, 1 min at 59°C and 30 s s at 72°C. Amplification of DCV and FHV was performed using the same conditions, except an annealing temperature of 56°C. Melting curves were analysed to confirm specificity of amplified products. We obtained Ct values for manual threshold of 10 using the program SDS 2.4 or with Bio-Rad CFX Manager with default threshold settings.
Relative amounts were calculated by the Pfaffl Method [113] using Drosophila Rpl32 as a reference gene for wsp and viruses and wsp as a reference for Wolbachia Octomom genes.
Kruskal-Wallis rank sum test (kruskal.test in R) was performed on Wolbachia and viruses quantification data to detect differences within all the lines. Pairwise comparison between all variants was performed with Wilcoxon rank sum test with Holm correction (pairwise.wilcox.test in R). Direct comparison between wMel-like and wMelCS-like variants was performed with a linear mixed-effects model fit by maximizing the restricted log-likelihood on the log of the values (lme in R). Time course analysis of Wolbachia titres was performed with a linear model fit (lm in R).
Cluster Analysis and Correlations
The data in Table S2 was used for the cluster analysis of wMel variants (hclust in R). In each column the mean was subtracted from the data, for centering, and the result divided by the standard deviation, for scaling. Complete linkage hierarchical clustering was performed on Euclidian distances between wMel variants.
Correlations were calculated using Pearson's product moment correlation (cor.test in R).
Sequencing and Genome Assembly
The genome assembly of the wMel variants was done with the invaluable help of Casey Bergman (University of Manchester).
For each fly line, 20 females were anaesthetized under CO2 and washed in 50% bleach solution for 3 min. Females were then briefly washed in distilled water and dissected under a microscope. The two ovaries of the 20 females were pooled for DNA extraction. DNA was extracted using the Gentra Puregene DNA Purification kit according to the manufacturer's protocol, including an RNase A treatment. Yields of purified DNA ranged between 1.1 and 4.2 µg. Library preparation and sequencing were performed at the Eastern Sequence and Informatics Hub (Cambridge, UK). 75 bp paired-end libraries were prepared with an insert size of 300 bp and sequenced in one lane of HiSeq2000 (Illumina). Base calling was performed using the Offline Basecaller (version 1.9.3) from Illumina, and demultiplexing was handled by bespoke Eastern Sequence and Informatics Hub software. The reads are submitted to the Sequence Read Archive (accession number: ERP002662).
Forward and reverse fastq sequences were mapped individually to single database containing a mitochondrial reference sequence extracted from the D. melanogaster Release 5 genome sequence (chrU:5288528–5305749) and the D. melanogaster Wolbachia endosymbiont reference genome (GenBank ID: AE017196) and converted to paired end alignments using BWA version 0.5.9-r16 [114]. BWA output was converted to SAM format and reads mapping to the mitochondria or Wolbachia reference sequences were extracted and sorted using SAMtools version 0.1.18. Sorted BAM files were used for variant base calling followed by a standard SAMtools version 0.1.16 pileup pipeline [115]. Individual strain consensus fastq sequences were generated using pileup2fq.pl with minimum and maximum read depths set to 10 and 100, respectively, and converted to fasta format using seqtk (https://github.com/lh3/seqtk). Individual reference-based fasta consensus sequence files were merged into multiple alignments from http://bergman.smith.man.ac.uk/data/wolbachia/DGRP_DPGP_Wolbachia_v1.tgz [42]. Alignment columns that had an N in any strain (which can represent either a fully ambiguous character or a deletion relative to the reference) were then removed.
Fasta file of assembled sequences of Wolbachia variants and associated mitochondria are in Dataset S1 and S2, respectively. Tables of variants for these Wolbachia and mitochondria together with data from Wolbachia-carrying strains described in Richardson et al. [42] are in Dataset S3 and S4, respectively.
Phylogenetic Analysis
We produced a dated evolutionary history of Wolbachia using BEAST v1.7.2 [116]. The Wolbachia and Drosophila mitochondrial phylogenies have been shown to be fully congruent [42], so they share the same evolutionary history. We therefore concatenated the Wolbachia variants alignments with their respective host Drosophila mitochondrial alignments, removing all indels. We included the Wolbachia reference strain AE017196, even though no host Drosophila mitochondrial alignment exists, after checking that its inclusion made no qualitative difference to either the dates or topology. This alignment was then partitioned into eight different groups representing different categories of sites; first and second codon positions, third codon positions, noncoding RNA genes and intergenic sites (for both the Wolbachia and Drosophila mitochondria). Each partition had their own HKY+Γ model of evolution [117], [118] but linked to the same dated phylogeny and constant population size coalescent tree prior. In order to calibrate the molecular dating, we assigned a prior lognormal distribution of rate based on the Drosophila mutation rate [42], [119] to third codon positions of the Drosophila mitochondria, sites that are less likely to be under purifying selection. Rates at all other site classes were given a prior of uniform distribution between 0 and 1, whereas priors on all other parameters were given default values as specified in BEAUti v1.7.2 [116].
Genetic Polymorphism and Predicted Genes Analyses
For single nucleotide polymorphism analysis a multiple alignment was built, with only the sequences of the wMel variants analysed in this report, and alignment columns that had an N in any strain were removed. Variant sites were then extracted and mapped back to reference coordinates using custom R and PERL scripts (Dataset S5). Variants that differ between all wMel-like and all wMelCS-like variants were identified and mapped to predicted genes or non-coding regions with Galaxy [120]. Identification of synonymous or non-synonymous substitutions was performed with custom Python scripts.
To identify duplications and deletions that have led to copy number variation (CNVs), we examined depth of sequence coverage across the Wolbachia genome. To do this we partitioned the genome into non-overlapping 200 bp bins and used the mean shift approach implemented in CNVnator [70] to infer differences in copy number and identify break-points. The variants wMel, wMel3 and wMelCS2_a were not analysed as they had highly variable coverage. Analysis of regions containing duplications was aided by UCSC Genome Browser http://genome.ucsc.edu/ [121], [122].
We also used the program Pindel [123] to search for ‘split reads’, which map to two different positions in the Wolbachia genome. To reduce artefacts we only retained structural variants where at least one strain had 10 or more supporting reads and where at least one strain had no supporting reads. As we know the phylogeny of these strains, we expect most true structural variants to be present in monophyletic clades (i.e. they have only arisen once). Out of 18 variants detected, 17 fulfilled this criterion, suggesting that our methods are robust.
Predicted protein domain analysis was based on the reference genome [58] or using NCBI CD-search [124]).
Supporting Information
Zdroje
1. MoranN, McCutcheonJ, NakabachiA (2008) Genomics and Evolution of Heritable Bacterial Symbionts. Annu Rev Genet 42 : 165–190 doi:10.1146/annurev.genet.41.110306.130119
2. AxelrodR, HamiltonWD (1981) The evolution of cooperation. Science 211 : 1390–1396.
3. TurelliM (1994) Evolution of Incompatibility-Inducing Microbes and Their Hosts. Evolution 48 : 1500–1513.
4. HoffmannAA, HercusM, DagherH (1998) Population dynamics of the Wolbachia infection causing cytoplasmic incompatibility in Drosophila melanogaster. Genetics 148 : 221–231.
5. EngelstaedterJ, HurstGDD (2009) The Ecology and Evolution of Microbes that Manipulate Host Reproduction. Annu Rev Ecol Evol S 40 : 127–149 doi:10.1146/annurev.ecolsys.110308.120206
6. JaenikeJ (2012) Population genetics of beneficial heritable symbionts. Trends Ecol Evol 27 : 226–232 doi:10.1016/j.tree.2011.10.005
7. HaineER (2008) Symbiont-mediated protection. Proc Biol Sci 275 : 353–361 doi:10.1098/rspb.2007.1211
8. LittmanDR, PamerEG (2011) Role of the commensal microbiota in normal and pathogenic host immune responses. Cell Host Microbe 10 : 311–323 doi:10.1016/j.chom.2011.10.004
9. OliverKM, RussellJA, MoranNA, HunterMS (2003) Facultative bacterial symbionts in aphids confer resistance to parasitic wasps. Proc Natl Acad Sci USA 100 : 1803–1807 doi:10.1073/pnas.0335320100
10. JaenikeJ, UncklessR, CockburnSN, BoelioLM, PerlmanSJ (2010) Adaptation via symbiosis: recent spread of a Drosophila defensive symbiont. Science 329 : 212–215 doi:10.1126/science.1188235
11. XieJ, VilchezI, MateosM (2010) Spiroplasma bacteria enhance survival of Drosophila hydei attacked by the parasitic wasp Leptopilina heterotoma. PLoS ONE 5: e12149 doi:10.1371/journal.pone.0012149
12. TeixeiraL, FerreiraA, AshburnerM (2008) The bacterial symbiont Wolbachia induces resistance to RNA viral infections in Drosophila melanogaster. PLoS Biol 6: e1000002 doi:10.1371/journal.pbio.1000002
13. Gil-TurnesMS, HayME, FenicalW (1989) Symbiotic marine bacteria chemically defend crustacean embryos from a pathogenic fungus. Science 246 : 116–118.
14. HedgesLM, BrownlieJC, O'NeillSL, JohnsonKN (2008) Wolbachia and Virus Protection in Insects. Science 322 : 702–702 doi:10.1126/science.1162418
15. BartonES, WhiteDW, CathelynJS, Brett-McClellanKA, EngleM, et al. (2007) Herpesvirus latency confers symbiotic protection from bacterial infection. Nature 447 : 326–329 doi:10.1038/nature05762
16. GrivelJC, ItoY, FagàG, SantoroF, ShaheenF, et al. (2001) Suppression of CCR5 - but not CXCR4-tropic HIV-1 in lymphoid tissue by human herpesvirus 6. Nat Med 7 : 1232–1235 doi:10.1038/nm1101-1232
17. ScarboroughCL, FerrariJ, GodfrayHCJ (2005) Aphid protected from pathogen by endosymbiont. Science 310 : 1781 doi:10.1126/science.1120180
18. KaltenpothM, GöttlerW, HerznerG, StrohmE (2005) Symbiotic bacteria protect wasp larvae from fungal infestation. Curr Biol 15 : 475–479 doi:10.1016/j.cub.2004.12.084
19. WeissBL, WangJ, AksoyS (2011) Tsetse immune system maturation requires the presence of obligate symbionts in larvae. PLoS Biol 9: e1000619 doi:10.1371/journal.pbio.1000619
20. CirimotichCM, DongY, ClaytonAM, SandifordSL, Souza-NetoJA, et al. (2011) Natural microbe-mediated refractoriness to Plasmodium infection in Anopheles gambiae. Science 332 : 855–858 doi:10.1126/science.1201618
21. OliverKM, CamposJ, MoranNA, HunterMS (2008) Population dynamics of defensive symbionts in aphids. Proc Biol Sci 275 : 293–299 doi:10.1098/rspb.2007.1192
22. ZugR, HammersteinP (2012) Still a host of hosts for Wolbachia: analysis of recent data suggests that 40% of terrestrial arthropod species are infected. PLoS ONE 7: e38544 doi:10.1371/journal.pone.0038544
23. OsborneSE, LeongYS, O'NeillSL, JohnsonKN (2009) Variation in antiviral protection mediated by different Wolbachia strains in Drosophila simulans. PLoS Pathog 5: e1000656 doi:10.1371/journal.ppat.1000656
24. GlaserRL, MeolaMA (2010) The native Wolbachia endosymbionts of Drosophila melanogaster and Culex quinquefasciatus increase host resistance to West Nile virus infection. PLoS ONE 5: e11977 doi:10.1371/journal.pone.0011977
25. BianG, XuY, LuP, XieY, XiZ (2010) The endosymbiotic bacterium Wolbachia induces resistance to dengue virus in Aedes aegypti. PLoS Pathog 6: e1000833 doi:10.1371/journal.ppat.1000833
26. LongdonB, FabianDK, HurstGD, JigginsFM (2012) Male-killing Wolbachia do not protect Drosophila bifasciata against viral infection. BMC Microbiol 12 Suppl 1: S8 doi:10.1186/1471-2180-12-S1-S8
27. StouthamerR, BreeuwerJA, HurstGD (1999) Wolbachia pipientis: microbial manipulator of arthropod reproduction. Annu Rev Microbiol 53 : 71–102 doi:10.1146/annurev.micro.53.1.71
28. WerrenJH, BaldoL, ClarkME (2008) Wolbachia: master manipulators of invertebrate biology. Nat Rev Microbiol 6 : 741–751 doi:10.1038/nrmicro1969
29. FentonA, JohnsonKN, BrownlieJC, HurstGDD (2011) Solving the WolbachiaParadox: Modeling the Tripartite Interaction between Host, Wolbachia, and a Natural Enemy. Am Nat 178 : 333–342 doi:10.1086/661247
30. MoreiraLA, Iturbe-OrmaetxeI, JefferyJA, LuG, PykeAT, et al. (2009) A Wolbachia symbiont in Aedes aegypti limits infection with dengue, Chikungunya, and Plasmodium. Cell 139 : 1268–1278 doi:10.1016/j.cell.2009.11.042
31. WalkerT, JohnsonPH, MoreiraLA, Iturbe-OrmaetxeI, FrentiuFD, et al. (2011) The wMel Wolbachia strain blocks dengue and invades caged Aedes aegypti populations. Nature 476 : 450–453 doi:doi:10.1038/nature10355
32. van den HurkAF, Hall-MendelinS, PykeAT, FrentiuFD, McElroyK, et al. (2012) Impact of Wolbachia on Infection with Chikungunya and Yellow Fever Viruses in the Mosquito Vector Aedes aegypti. PLoS Neglected Tropical Diseases 6: e1892 doi:10.1371/journal.pntd.0001892
33. BlagroveMSC, Arias-GoetaC, FaillouxA-B, SinkinsSP (2012) Wolbachia strain wMel induces cytoplasmic incompatibility and blocks dengue transmission in Aedes albopictus. Proc Natl Acad Sci USA 109 : 255–260 doi:10.1073/pnas.1112021108
34. KambrisZ, CookPE, PhucHK, SinkinsSP (2009) Immune activation by life-shortening Wolbachia and reduced filarial competence in mosquitoes. Science 326 : 134–136 doi:10.1126/science.1177531
35. HughesGL, KogaR, XueP, FukatsuT, RasgonJL (2011) Wolbachia infections are virulent and inhibit the human malaria parasite Plasmodium falciparum in anopheles gambiae. PLoS Pathog 7: e1002043 doi:10.1371/journal.ppat.1002043
36. KambrisZ, BlagboroughAM, PintoSB, BlagroveMSC, GodfrayHCJ, et al. (2010) Wolbachia stimulates immune gene expression and inhibits plasmodium development in Anopheles gambiae. PLoS Pathog 6: e1001143 doi:10.1371/journal.ppat.1001143
37. HoffmannAA, MontgomeryBL, PopoviciJ, Iturbe-OrmaetxeI, JohnsonPH, et al. (2011) Successful establishment of Wolbachia in Aedes populations to suppress dengue transmission. Nature 476 : 454–457 doi:10.1038/nature10356
38. Iturbe-OrmaetxeI, WalkerT, O NeillSL (2011) Wolbachia and the biological control of mosquito-borne disease. EMBO Rep 12 : 508–518 doi:10.1038/embor.2011.84
39. OliverKM, DegnanPH, HunterMS, MoranNA (2009) Bacteriophages encode factors required for protection in a symbiotic mutualism. Science 325 : 992–994 doi:10.1126/science.1174463
40. RieglerM, SidhuM, MillerWJ, O'NeillSL (2005) Evidence for a Global Wolbachia Replacement in Drosophila melanogaster. Current Biology 15 : 1428–1433 doi:10.1016/j.cub.2005.06.069
41. NunesMDS, NolteV, SchlöttererC (2008) Nonrandom Wolbachia infection status of Drosophila melanogaster strains with different mtDNA haplotypes. Mol Biol Evol 25 : 2493–2498 doi:10.1093/molbev/msn199
42. RichardsonMF, WeinertLA, WelchJJ, LinheiroRS, MagwireMM, et al. (2012) Population Genomics of the Wolbachia Endosymbiont in Drosophila melanogaster. PLoS Genet 8: e1003129 doi:10.1371/journal.pgen.1003129
43. HoldenPR, JonesP, BrookfieldJF (1993) Evidence for a Wolbachia symbiont in Drosophila melanogaster. Genet Res 62 : 23–29.
44. MateosM, CastrezanaSJ, NankivellBJ, EstesAM, MarkowTA, et al. (2006) Heritable endosymbionts of Drosophila. Genetics 174 : 363–376 doi:10.1534/genetics.106.058818
45. SolignacM, VautrinD, RoussetF (1994) Widespread Occurrence of the Proteobacteria Wolbachia and Partial Cytoplasmic Incompatibility in Drosophila melanogaster. Cr Acad Sci Iii-Vie 317 : 461–470.
46. HoffmannAA (1988) Partial cytoplasmic incompatibility between two Australian populations of Drosophila melanogaster. Entomol Exp Appl 48 : 61–67 doi:10.1111/j.1570-7458.1988.tb02299.x
47. HoffmannAA, ClancyDJ, MertonE (1994) Cytoplasmic incompatibility in Australian populations of Drosophila melanogaster. Genetics 136 : 993–999.
48. IlinskyYY, ZakharovIK (2007) The endosymbiont Wolbachia in Eurasian populations of Drosophila melanogaster. Russ J Genet+ 43 : 748–756 doi:10.1134/S102279540707006X
49. VerspoorRL, HaddrillPR (2011) Genetic Diversity, Population Structure and Wolbachia Infection Status in a Worldwide Sample of Drosophila melanogaster and D. simulans Populations. PLoS ONE 6: e26318 doi:10.1371/journal.pone.0026318
50. IlinskyY (2013) Coevolution of Drosophila melanogaster mtDNA and Wolbachia Genotypes. PLoS ONE 8: e54373 doi:10.1371/journal.pone.0054373
51. MackayTFC, RichardsS, StoneEA, BarbadillaA, AyrolesJF, et al. (2012) The Drosophila melanogaster Genetic Reference Panel. Nature 482 : 173–178 doi:10.1038/nature10811
52. VenetiZ, ClarkME, ZabalouS, KarrTL, SavakisC, et al. (2003) Cytoplasmic incompatibility and sperm cyst infection in different Drosophila-Wolbachia associations. Genetics 164 : 545–552.
53. IlinskyYY, ZakharovIK (2011) Cytoplasmic incompatibility in Drosophila melanogaster is caused by different Wolbachia genotypes. Russ J Genet Appl Res 1 : 458–462 doi:10.1134/S2079059711020031
54. MinKT, BenzerS (1997) Wolbachia, normally a symbiont of Drosophila, can be virulent, causing degeneration and early death. Proc Natl Acad Sci USA 94 : 10792–10796.
55. ReynoldsKT, ThomsonLJ, HoffmannAA (2003) The effects of host age, host nuclear background and temperature on phenotypic effects of the virulent Wolbachia strain popcorn in Drosophila melanogaster. Genetics 164 : 1027–1034.
56. McGrawEA, MerrittDJ, DrollerJN, O'NeillSL (2002) Wolbachia density and virulence attenuation after transfer into a novel host. Proc Natl Acad Sci USA 99 : 2918–2923 doi:10.1073/pnas.052466499
57. RieglerM, Iturbe-OrmaetxeI, WoolfitM, MillerWJ, O'NeillSL (2012) Tandem repeat markers as novel diagnostic tools for high resolution fingerprinting of Wolbachia. BMC Microbiol 12 Suppl 1: S12 doi:10.1186/1471-2180-12-S1-S12
58. WuM, SunLV, VamathevanJ, RieglerM, DeboyR, et al. (2004) Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: a streamlined genome overrun by mobile genetic elements. PLoS Biol 2: E69 doi:10.1371/journal.pbio.0020069
59. RyderE, BlowsF, AshburnerM, Bautista-LlacerR, CoulsonD, et al. (2004) The DrosDel collection: a set of P-element insertions for generating custom chromosomal aberrations in Drosophila melanogaster. Genetics 167 : 797–813 doi:10.1534/genetics.104.026658
60. Brun G, Plus N (1978) The viruses of Drosophila melanogaster. Ashburner M, Wright T, editors. Genetics and Biology of Drosophila. New York: Academic Press. pp. 625–702
61. JohnsonKN, ChristianPD (1998) The novel genome organization of the insect picorna-like virus Drosophila C virus suggests this virus belongs to a previously undescribed virus family. J Gen Virol 79 : 191–203.
62. CoxDR (1972) Regression models and life-tables. Journal of the Royal Statistical Society Series B (Methodological) 34 : 187–220.
63. DeddoucheS, MattN, BuddA, MuellerS, KempC, et al. (2008) The DExD/H-box helicase Dicer-2 mediates the induction of antiviral activity in Drosophila. Nat Immunol 9 : 1425–1432 doi:10.1038/ni.1664
64. Ball LA, Johnson KN (1998) Nodaviruses of Insects. Ball LA, Miller LK, editors. The insect viruses. New York: Plenum Press. pp. 225–268
65. DearingSC, ScottiPD, WigleyPJ, DhanaSD (1980) A small RNA virus isolated from the grass grub, Costelytra zealandica (Coleoptera, Scarabaeidae). New Zeal J Zool 7 : 267–269.
66. SchneiderDS, AyresJS (2008) Two ways to survive infection: what resistance and tolerance can teach us about treating infectious diseases. Nat Rev Immunol 8 : 889–895 doi:10.1038/nri2432
67. ClancyDJ (2008) Variation in mitochondrial genotype has substantial lifespan effects which may be modulated by nuclear background. Aging Cell 7 : 795–804 doi:10.1111/j.1474-9726.2008.00428.x
68. McMenimanCJ, LaneRV, CassBN, FongAWC, SidhuM, et al. (2009) Stable Introduction of a Life-Shortening Wolbachia Infection into the Mosquito Aedes aegypti. Science 323 : 141–144 doi:10.1126/science.1165326
69. Iturbe-OrmaetxeI, WoolfitM, RancèsE, DuplouyA, O'NeillSL (2011) A simple protocol to obtain highly pure Wolbachia endosymbiont DNA for genome sequencing. J Microbiol Methods 84 : 134–136 doi:10.1016/j.mimet.2010.10.019
70. AbyzovA, UrbanAE, SnyderM, GersteinM (2011) CNVnator: an approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res 21 : 974–984 doi:10.1101/gr.114876.110
71. CordauxR, PichonS, LingA, PerezP, DelaunayC, et al. (2008) Intense Transpositional Activity of Insertion Sequences in an Ancient Obligate Endosymbiont. Mol Biol Evol 25 : 1889–1896 doi:10.1093/molbev/msn134
72. Iturbe-OrmaetxeI, BurkeGR, RieglerM, O'NeillSL (2005) Distribution, expression, and motif variability of ankyrin domain genes in Wolbachia pipientis. J Bacteriol 187 : 5136–5145 doi:10.1128/JB.187.15.5136-5145.2005
73. KlassonL, KambrisZ, CookPE, WalkerT, SinkinsSP (2009) Horizontal gene transfer between Wolbachia and the mosquito Aedes aegypti. BMC Genomics 10 : 33 doi:10.1186/1471-2164-10-33
74. WoolfitM, Iturbe-OrmaetxeI, McGrawEA, O'NeillSL (2009) An ancient horizontal gene transfer between mosquito and the endosymbiotic bacterium Wolbachia pipientis. Mol Biol Evol 26 : 367–374 doi:10.1093/molbev/msn253
75. SioziosS, IoannidisP, KlassonL, AnderssonSGE, BraigHR, et al. (2013) The Diversity and Evolution of Wolbachia Ankyrin Repeat Domain Genes. PLoS ONE 8: e55390 doi:10.1371/journal.pone.0055390
76. KlassonL, WalkerT, SebaihiaM, SandersMJ, QuailMA, et al. (2008) Genome evolution of Wolbachia strain wPip from the Culex pipiens group. Mol Biol Evol 25 : 1877–1887 doi:10.1093/molbev/msn133
77. KentBN, SalichosL, GibbonsJG, RokasA, NewtonILG, et al. (2011) Complete bacteriophage transfer in a bacterial endosymbiont (Wolbachia) determined by targeted genome capture. Genome Biol Evol 3 : 209–218 doi:10.1093/gbe/evr007
78. KorochkinaS, BarreauC, PradelG, JefferyE, LiJ, et al. (2006) A mosquito-specific protein family includes candidate receptors for malaria sporozoite invasion of salivary glands. Cell Microbiol 8 : 163–175 doi:10.1111/j.1462-5822.2005.00611.x
79. ColbourneJK, PfrenderME, GilbertD, ThomasWK, TuckerA, et al. (2011) The ecoresponsive genome of Daphnia pulex. Science 331 : 555–561 doi:10.1126/science.1197761
80. KlassonL, WestbergJ, SapountzisP, NäslundK, LutnaesY, et al. (2009) The mosaic genome structure of the Wolbachia wRi strain infecting Drosophila simulans. Proc Natl Acad Sci USA 106 : 5725–5730 doi:10.1073/pnas.0810753106
81. AnderssonDI, HughesD (2009) Gene amplification and adaptive evolution in bacteria. Annu Rev Ecol Evol S 43 : 167–195 doi:10.1146/annurev-genet-102108-134805
82. MekalanosJJ (1983) Duplication and amplification of toxin genes in Vibrio cholerae. Cell 35 : 253–263.
83. KrollJS, LoyndsBM, MoxonER (1991) The Haemophilus influenzae capsulation gene cluster: a compound transposon. Mol Microbiol 5 : 1549–1560.
84. MavinguiP, LaeremansT, FloresM, RomeroD, Martínez-RomeroE, et al. (1998) Genes essential for nod factor production and nodulation are located on a symbiotic amplicon (AMPRtrCFN299pc60) in Rhizobium tropici. J Bacteriol 180 : 2866–2874.
85. EldeNC, ChildSJ, EickbushMT, KitzmanJO, RogersKS, et al. (2012) Poxviruses deploy genomic accordions to adapt rapidly against host antiviral defenses. Cell 150 : 831–841 doi:10.1016/j.cell.2012.05.049
86. IshmaelN, Dunning HotoppJC, IoannidisP, BiberS, SakamotoJ, et al. (2009) Extensive genomic diversity of closely related Wolbachia strains. Microbiology (Reading, Engl) 155 : 2211–2222 doi:10.1099/mic.0.027581-0
87. SalzbergSL, Dunning HotoppJC, DelcherAL, PopM, SmithDR, et al. (2005) Serendipitous discovery of Wolbachia genomes in multiple Drosophila species. Genome Biol 6: R23 doi:10.1186/gb-2005-6-3-r23
88. KingJG, VernickKD, HillyerJF (2011) Members of the Salivary Gland Surface Protein (SGS) Family Are Major Immunogenic Components of Mosquito Saliva. Journal of Biological Chemistry 286 : 40824–40834 doi:10.1074/jbc.M111.280552
89. AyresJS, SchneiderDS (2008) A signaling protease required for melanization in Drosophila affects resistance and tolerance of infections. PLoS Biol 6 : 2764–2773 doi:10.1371/journal.pbio.0060305
90. VenetiZ, ClarkME, ZabalouS, KarrTL, SavakisC, et al. (2003) Cytoplasmic incompatibility and sperm cyst infection in different Drosophila-Wolbachia associations. Genetics 164 : 545–552.
91. KondoN, ShimadaM, FukatsuT (2005) Infection density of Wolbachia endosymbiont affected by co-infection and host genotype. Biol Lett 1 : 488–491 doi:10.1098/rsbl.2005.0340
92. MoutonL, HenriH, CharifD, BoulétreauM, VavreF (2007) Interaction between host genotype and environmental conditions affects bacterial density in Wolbachia symbiosis. Biol Lett 3 : 210–213 doi:10.1098/rsbl.2006.0590
93. MoutonL, HenriH, BouletreauM, VavreF (2003) Strain-specific regulation of intracellular Wolbachia density in multiply infected insects. Mol Ecol 12 : 3459–3465.
94. LuP, BianG, PanX, XiZ (2012) Wolbachia Induces Density-Dependent Inhibition to Dengue Virus in Mosquito Cells. PLoS Neglected Tropical Diseases 6: e1754 doi:10.1371/journal.pntd.0001754
95. JaenikeJ (2009) Coupled population dynamics of endosymbionts within and between hosts. Oikos 118 : 353–362.
96. FrentiuFD, RobinsonJ, YoungPR, McGrawEA, O'NeillSL (2010) Wolbachia-mediated resistance to dengue virus infection and death at the cellular level. PLoS ONE 5: e13398 doi:10.1371/journal.pone.0013398
97. OsborneSE, Iturbe-OrmaetxeI, BrownlieJC, O'NeillSL, JohnsonKN (2012) Antiviral protection and the importance of Wolbachia density and tissue tropism in Drosophila. Appl Environ Microbiol 78 : 6922–6929 doi:10.1128/AEM.01727-12
98. BressacC, RoussetF (1993) The reproductive incompatibility system in Drosophila simulans: DAPI-staining analysis of the Wolbachia symbionts in sperm cysts. J Invertebr Pathol 61 : 226–230 doi:10.1006/jipa.1993.1044
99. BreeuwerJA, WerrenJH (1993) Cytoplasmic incompatibility and bacterial density in Nasonia vitripennis. Genetics 135 : 565–574.
100. BourtzisK, NirgianakiA, MarkakisG, SavakisC (1996) Wolbachia infection and cytoplasmic incompatibility in Drosophila species. Genetics 144 : 1063–1073.
101. BordensteinSR, MarshallML, FryAJ, KimU, WernegreenJJ (2006) The tripartite associations between bacteriophage, Wolbachia, and arthropods. PLoS Pathog 2: e43 doi:10.1371/journal.ppat.0020043
102. BoyleL, O'NeillS, RobertsonH, KarrT (1993) Interspecific and intraspecific horizontal transfer of Wolbachia in Drosophila. Science 260 : 1796–1799 doi:10.1126/science.8511587
103. UncklessR, BoelioL, HerrenJ, JaenikeJ (2009) Wolbachia as populations within individual insects: causes and consequences of density variation in natural populations. Proc Biol Sci 276 : 2805–2811 doi:10.1098/rspb.2009.0287
104. ClarkME, AndersonCL, CandeJ, KarrTL (2005) Widespread prevalence of Wolbachia in laboratory stocks and the implications for Drosophila research. Genetics 170 : 1667–1675 doi:10.1534/genetics.104.038901
105. Lautié-HarivelN, Thomas-OrillardM (1990) Location of Drosophila C virus target organs in Drosophila host population by an immunofluorescence technique. Biol Cell 69 : 35–39.
106. DostertC, JouanguyE, IrvingP, TroxlerL, Galiana-ArnouxD, et al. (2005) The Jak-STAT signaling pathway is required but not sufficient for the antiviral response of Drosophila. Nat Immunol 6 : 946–953 doi:10.1038/ni1237
107. Galiana-ArnouxD, DostertC, SchneemannA, HoffmannJA, ImlerJ-L (2006) Essential function in vivo for Dicer-2 in host defense against RNA viruses in Drosophila. Nat Immunol 7 : 590–597 doi:10.1038/ni1335
108. EleftherianosI, WonS, ChtarbanovaS, SquibanB, OcorrK, et al. (2011) ATP-sensitive potassium channel (KATP)-dependent regulation of cardiotropic viral infections. Proc Natl Acad Sci USA 108 : 12024–12029 doi:10.1073/pnas.1108926108
109. HoffmannAA, TurelliM, HarshmanLG (1990) Factors affecting the distribution of cytoplasmic incompatibility in Drosophila simulans. Genetics 126 : 933–948.
110. HussainM, LuG, TorresS, EdmondsJH, KayBH, et al. (2013) Effect of Wolbachia on replication of West Nile virus in a mosquito cell line and adult mosquitoes. J Virol 87 : 851–858 doi:10.1128/JVI.01837-12
111. WeeksAR, TurelliM, HarcombeWR, ReynoldsKT, HoffmannAA (2007) From parasite to mutualist: rapid evolution of Wolbachia in natural populations of Drosophila. PLoS Biol 5: e114 doi:10.1371/journal.pbio.0050114
112. Team RC (2012) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. http://www.R-project.org/.
113. PfafflMichael W (2001) A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res 29: e45.
114. LiH, DurbinR (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25 : 1754–1760 doi:10.1093/bioinformatics/btp324
115. LiH, HandsakerB, WysokerA, FennellT, RuanJ, et al. (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25 : 2078–2079 doi:10.1093/bioinformatics/btp352
116. DrummondAJ, SuchardMA, XieD, RambautA (2012) Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29 : 1969–1973 doi:10.1093/molbev/mss075
117. HasegawaM, KishinoH, YanoT (1985) Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol 22 : 160–174.
118. ShapiroB, RambautA, DrummondAJ (2006) Choosing appropriate substitution models for the phylogenetic analysis of protein-coding sequences. Mol Biol Evol 23 : 7–9 doi:10.1093/molbev/msj021
119. Haag-LiautardC, CoffeyN, HouleD, LynchM, CharlesworthB, et al. (2008) Direct estimation of the mitochondrial DNA mutation rate in Drosophila melanogaster. PLoS Biol 6: e204 doi:10.1371/journal.pbio.0060204
120. GiardineB, RiemerC, HardisonRC, BurhansR, ElnitskiL, et al. (2005) Galaxy: a platform for interactive large-scale genome analysis. Genome Res 15 : 1451–1455 doi:10.1101/gr.4086505
121. SchneiderKL, PollardKS, BaertschR, PohlA, LoweTM (2006) The UCSC Archaeal Genome Browser. Nucleic Acids Res 34: D407–D410 doi:10.1093/nar/gkj134
122. KarolchikD, BaertschR, DiekhansM, FureyTS, HinrichsA, et al. (2003) The UCSC Genome Browser Database. Nucleic Acids Res 31 : 51–54.
123. YeK, SchulzMH, LongQ, ApweilerR, NingZ (2009) Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics 25 : 2865–2871 doi:10.1093/bioinformatics/btp394
124. Marchler-BauerA, LuS, AndersonJB, ChitsazF, DerbyshireMK, et al. (2010) CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res 39: D225–D229 doi:10.1093/nar/gkq1189
125. Marchler-BauerA, ZhengC, ChitsazF, DerbyshireMK, GeerLY, et al. (2013) CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Res 41: D348–D352 doi:10.1093/nar/gks1243
126. BerryB, DeddoucheS, KirschnerD, ImlerJ-L, AntoniewskiC (2009) Viral suppressors of RNA silencing hinder exogenous and endogenous small RNA pathways in Drosophila. PLoS ONE 4: e5866 doi:10.1371/journal.pone.0005866
Štítky
Genetika Reprodukční medicína
Článek Interaction between and during Mammalian Jaw Patterning and in the Pathogenesis of SyngnathiaČlánek Clustering of Tissue-Specific Sub-TADs Accompanies the Regulation of Genes in Developing LimbsČlánek Transcription Factor Occupancy Can Mediate Active Turnover of DNA Methylation at Regulatory Regions
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 12
-
Všechny články tohoto čísla
- Stressing the Importance of CHOP in Liver Cancer
- The AmAZI1ng Roles of Centriolar Satellites during Development
- Flies Get a Head Start on Meiosis
- Recommendations from Jane Gitschier's Bookshelf
- And Baby Makes Three: Genomic Imprinting in Plant Embryos
- Bugs in Transition: The Dynamic World of in Insects
- Defining the Role of ATP Hydrolysis in Mitotic Segregation of Bacterial Plasmids
- Synaptonemal Complex Components Promote Centromere Pairing in Pre-meiotic Germ Cells
- Cohesinopathies of a Feather Flock Together
- Genetic Recombination Is Targeted towards Gene Promoter Regions in Dogs
- Parathyroid-Specific Deletion of Unravels a Novel Calcineurin-Dependent FGF23 Signaling Pathway That Regulates PTH Secretion
- MAN1B1 Deficiency: An Unexpected CDG-II
- Phosphate Flow between Hybrid Histidine Kinases CheA and CheS Controls Cyst Formation
- Basolateral Mg Extrusion via CNNM4 Mediates Transcellular Mg Transport across Epithelia: A Mouse Model
- Truncation of Unsilences Paternal and Ameliorates Behavioral Defects in the Angelman Syndrome Mouse Model
- Autozygome Sequencing Expands the Horizon of Human Knockout Research and Provides Novel Insights into Human Phenotypic Variation
- Huntington's Disease Induced Cardiac Amyloidosis Is Reversed by Modulating Protein Folding and Oxidative Stress Pathways in the Heart
- Low Frequency Variants, Collapsed Based on Biological Knowledge, Uncover Complexity of Population Stratification in 1000 Genomes Project Data
- Targeted Ablation of and in Retinal Progenitor Cells Mimics Leber Congenital Amaurosis
- Genomic Imprinting in the Embryo Is Partly Regulated by PRC2
- Binary Cell Fate Decisions and Fate Transformation in the Larval Eye
- The Stress-Regulated Transcription Factor CHOP Promotes Hepatic Inflammatory Gene Expression, Fibrosis, and Oncogenesis
- A Global RNAi Screen Identifies a Key Role of Ceramide Phosphoethanolamine for Glial Ensheathment of Axons
- Functional Analysis of the Interdependence between DNA Uptake Sequence and Its Cognate ComP Receptor during Natural Transformation in Species
- Cross-Modulation of Homeostatic Responses to Temperature, Oxygen and Carbon Dioxide in
- Alcohol-Induced Histone Acetylation Reveals a Gene Network Involved in Alcohol Tolerance
- Molecular Characterization of Host-Specific Biofilm Formation in a Vertebrate Gut Symbiont
- CRIS—A Novel cAMP-Binding Protein Controlling Spermiogenesis and the Development of Flagellar Bending
- Dual Regulation of the Mitotic Exit Network (MEN) by PP2A-Cdc55 Phosphatase
- Expanding the Marine Virosphere Using Metagenomics
- Detection of Slipped-DNAs at the Trinucleotide Repeats of the Myotonic Dystrophy Type I Disease Locus in Patient Tissues
- Interaction between and during Mammalian Jaw Patterning and in the Pathogenesis of Syngnathia
- Mutations in the UQCC1-Interacting Protein, UQCC2, Cause Human Complex III Deficiency Associated with Perturbed Cytochrome Protein Expression
- Reactivation of Chromosomally Integrated Human Herpesvirus-6 by Telomeric Circle Formation
- Anoxia-Reoxygenation Regulates Mitochondrial Dynamics through the Hypoxia Response Pathway, SKN-1/Nrf, and Stomatin-Like Protein STL-1/SLP-2
- The Midline Protein Regulates Axon Guidance by Blocking the Reiteration of Neuroblast Rows within the Drosophila Ventral Nerve Cord
- Tomato Yield Heterosis Is Triggered by a Dosage Sensitivity of the Florigen Pathway That Fine-Tunes Shoot Architecture
- Selection on Plant Male Function Genes Identifies Candidates for Reproductive Isolation of Yellow Monkeyflowers
- Role of Tomato Lipoxygenase D in Wound-Induced Jasmonate Biosynthesis and Plant Immunity to Insect Herbivores
- Meiotic Cohesin SMC1β Provides Prophase I Centromeric Cohesion and Is Required for Multiple Synapsis-Associated Functions
- Identification of Sphingolipid Metabolites That Induce Obesity via Misregulation of Appetite, Caloric Intake and Fat Storage in
- Genome-Wide Screen Reveals Replication Pathway for Quasi-Palindrome Fragility Dependent on Homologous Recombination
- Histone Methylation Restrains the Expression of Subtype-Specific Genes during Terminal Neuronal Differentiation in
- A Novel Intergenic ETnII-β Insertion Mutation Causes Multiple Malformations in Mice
- The NuRD Chromatin-Remodeling Enzyme CHD4 Promotes Embryonic Vascular Integrity by Transcriptionally Regulating Extracellular Matrix Proteolysis
- A Domesticated Transposase Interacts with Heterochromatin and Catalyzes Reproducible DNA Elimination in
- Acute Versus Chronic Loss of Mammalian Results in Distinct Ciliary Phenotypes
- MBD3 Localizes at Promoters, Gene Bodies and Enhancers of Active Genes
- Positive and Negative Regulation of Gli Activity by Kif7 in the Zebrafish Embryo
- A Hereditary Spastic Paraplegia Mouse Model Supports a Role of ZFYVE26/SPASTIZIN for the Endolysosomal System
- The CCR4-NOT Complex Mediates Deadenylation and Degradation of Stem Cell mRNAs and Promotes Planarian Stem Cell Differentiation
- Reconstructing Native American Migrations from Whole-Genome and Whole-Exome Data
- Contributions of Protein-Coding and Regulatory Change to Adaptive Molecular Evolution in Murid Rodents
- Comprehensive Analysis of Transcriptome Variation Uncovers Known and Novel Driver Events in T-Cell Acute Lymphoblastic Leukemia
- A -Acting Protein Effect Causes Severe Eye Malformation in the Mouse
- Clustering of Tissue-Specific Sub-TADs Accompanies the Regulation of Genes in Developing Limbs
- Germline Progenitors Escape the Widespread Phenomenon of Homolog Pairing during Development
- Transcription Factor Occupancy Can Mediate Active Turnover of DNA Methylation at Regulatory Regions
- Somatic mtDNA Mutation Spectra in the Aging Human Putamen
- ESCRT-I Mediates FLS2 Endosomal Sorting and Plant Immunity
- Ethylene Promotes Hypocotyl Growth and HY5 Degradation by Enhancing the Movement of COP1 to the Nucleus in the Light
- The PAF Complex and Prf1/Rtf1 Delineate Distinct Cdk9-Dependent Pathways Regulating Transcription Elongation in Fission Yeast
- Dual Regulation of Gene Expression Mediated by Extended MAPK Activation and Salicylic Acid Contributes to Robust Innate Immunity in
- Quantifying Missing Heritability at Known GWAS Loci
- Smc5/6-Mms21 Prevents and Eliminates Inappropriate Recombination Intermediates in Meiosis
- Smc5/6 Coordinates Formation and Resolution of Joint Molecules with Chromosome Morphology to Ensure Meiotic Divisions
- Tay Bridge Is a Negative Regulator of EGFR Signalling and Interacts with Erk and Mkp3 in the Wing
- Meiotic Crossover Control by Concerted Action of Rad51-Dmc1 in Homolog Template Bias and Robust Homeostatic Regulation
- Active Transport and Diffusion Barriers Restrict Joubert Syndrome-Associated ARL13B/ARL-13 to an Inv-like Ciliary Membrane Subdomain
- An Regulatory Circuit Modulates /Wnt Signaling and Determines the Size of the Midbrain Dopaminergic Progenitor Pool
- Variants Induce Differential Protection to Viruses in : A Phenotypic and Phylogenomic Analysis
- Base Pairing Interaction between 5′- and 3′-UTRs Controls mRNA Translation in
- Evidence That Masking of Synapsis Imperfections Counterbalances Quality Control to Promote Efficient Meiosis
- Insulin/IGF-Regulated Size Scaling of Neuroendocrine Cells Expressing the bHLH Transcription Factor in
- Sumoylated NHR-25/NR5A Regulates Cell Fate during Vulval Development
- TATN-1 Mutations Reveal a Novel Role for Tyrosine as a Metabolic Signal That Influences Developmental Decisions and Longevity in
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The NuRD Chromatin-Remodeling Enzyme CHD4 Promotes Embryonic Vascular Integrity by Transcriptionally Regulating Extracellular Matrix Proteolysis
- Mutations in the UQCC1-Interacting Protein, UQCC2, Cause Human Complex III Deficiency Associated with Perturbed Cytochrome Protein Expression
- The Midline Protein Regulates Axon Guidance by Blocking the Reiteration of Neuroblast Rows within the Drosophila Ventral Nerve Cord
- Tomato Yield Heterosis Is Triggered by a Dosage Sensitivity of the Florigen Pathway That Fine-Tunes Shoot Architecture
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



