-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaTay Bridge Is a Negative Regulator of EGFR Signalling and Interacts with Erk and Mkp3 in the Wing
The regulation of Extracellular regulated kinase (Erk) activity is a key aspect of signalling by pathways activated by extracellular ligands acting through tyrosine kinase transmembrane receptors. In this process, participate proteins with kinase activity that phosphorylate and activate Erk, as well as different phosphatases that inactivate Erk by de-phosphorylation. The state of Erk phosphorylation affects not only its activity, but also its subcellular localization, defining the repertoire of Erk target proteins, and consequently, the cellular response to Erk. In this work, we characterise Tay bridge as a novel component of the EGFR/Erk signalling pathway. Tay bridge is a large nuclear protein with a domain of homology with human AUTS2, and was previously identified due to the neuronal phenotypes displayed by loss-of-function mutations. We show that Tay bridge antagonizes EGFR signalling in the Drosophila melanogaster wing disc and other tissues, and that the protein interacts with both Erk and Mkp3. We suggest that Tay bridge constitutes a novel element involved in the regulation of Erk activity, acting as a nuclear docking for Erk that retains this protein in an inactive form in the nucleus.
Published in the journal: . PLoS Genet 9(12): e32767. doi:10.1371/journal.pgen.1003982
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003982Summary
The regulation of Extracellular regulated kinase (Erk) activity is a key aspect of signalling by pathways activated by extracellular ligands acting through tyrosine kinase transmembrane receptors. In this process, participate proteins with kinase activity that phosphorylate and activate Erk, as well as different phosphatases that inactivate Erk by de-phosphorylation. The state of Erk phosphorylation affects not only its activity, but also its subcellular localization, defining the repertoire of Erk target proteins, and consequently, the cellular response to Erk. In this work, we characterise Tay bridge as a novel component of the EGFR/Erk signalling pathway. Tay bridge is a large nuclear protein with a domain of homology with human AUTS2, and was previously identified due to the neuronal phenotypes displayed by loss-of-function mutations. We show that Tay bridge antagonizes EGFR signalling in the Drosophila melanogaster wing disc and other tissues, and that the protein interacts with both Erk and Mkp3. We suggest that Tay bridge constitutes a novel element involved in the regulation of Erk activity, acting as a nuclear docking for Erk that retains this protein in an inactive form in the nucleus.
Introduction
The Epidermal Growth Factor Receptor (EGFR) signalling pathway is a conserved module that plays multiple roles during development and tissue homeostasis in eukaryotic organisms [1]–[3]. The best-characterized functions of the pathway involve the EGFR downstream proteins Sos, Ras, Raf, Mek and Erk, the MAPK that is encoded by rolled in Drosophila melanogaster [4]. The activity of these core components is required in multiple developmental contexts, influencing cell proliferation, migration, apoptosis, epithelial integrity and cell fate acquisition [1], [5]. A key node in the regulation of EGFR signalling occurs at the level of Erk phosphorylation and de-phosphorylation by Mek and dual-specificity phosphatases, respectively [6]–[8]. In general, upon activation by Mek, the Erk serine/threonine kinase is transported into the nucleus, where it can phosphorylate specific transcription factors, regulating their activity and consequently gene expression. Erk is de-phosphorylated and inactivated by dual-specificity phosphatases, which promote Erk accumulation in an inactive state in the cytoplasm [2], [9].
The nucleus-cytoplasm compartmentalization of Erk is also regulated by several proteins acting as scaffolds, which influence the kinetics of Erk activation by favouring its association with upstream components, or that target Erk to different substrates by regulating its subcellular localization [10]–[11]. Thus, Kinase suppressor of Ras (Ksr) and MEK partner 1 (MP-1) facilitate the phosphorylation of Erk by Mek [11]–[16], whereas β-arrestin and Sef (Similar Expression to FGF genes) serve as scaffolds directing Erk activity toward different subcellular localizations and sets of target proteins [17]–[18]. In fact, because Erk lacks nuclear localization or export sequences, it appears that its subcellular compartmentalization is mostly determined by binding to scaffolds, anchors and substrates [8], [10], [19]. In the absence of active export, Erk tends to accumulate inside the nucleus, and it has been suggested that imported Erk binds to nuclear anchoring proteins that difficult its free diffusion to the cytoplasm [6].
The EGFR signalling system has been extensively characterised in Drosophila, an organism that has been instrumental to identify the intricacies of signalling regulation in vivo [1], [20]–[22]. Furthermore, the exquisite sensitivity of several developmental processes to variations in levels of EGFR signalling has driven the search and identification of many components of the pathway through genetic screens, expression profiling and cell culture experiments [22]–[25]. The wing disc, the epithelial tissue that gives rise to the adult wing and part of the thorax, is particularly sensitive to changes in the levels of EGFR signalling [26]–[27]. The function of EGFR in this tissue is required for cell proliferation and viability [28], for the specification of the wing disc and its territorial subdivision [26], [29]–[32], and also in cell fate choices affecting sensory organs and veins [33]–[34]. In this last process, the function of the pathway is needed to promote the formation of the veins, longitudinal stripes of cells that differentiate a cuticle thicker and more pigmented than the cuticle of inter-vein cells [35]–[36].
We conducted a gain-of-function screen aimed to identify genes regulating wing vein differentiation, expecting that some of these genes would encode novel components of the signalling pathways driving the formation of these structures [37]. In this screen, we identified a P-UAS insertion in the gene tay bridge (tay) that in combination with a vein-specific Gal4 driver causes the elimination of the longitudinal veins, a phenotype reminiscent of loss of EGFR activity in the developing veins [27], [37]. Tay encodes a large protein of 2486 amino acids expressed predominantly in the central nervous system [38]. Mutant tay flies present a constriction in the protocerebral bridge, and display reduced walking speed, reduced sensitivity to the effects of alcohol and defective compensation of rotatory stimuli during walking [38]–[39]. The Carboxi-terminal part of Drosophila Tay presents homology with mammalian AUTS2, a neuronal nuclear protein that is related to autism [40]–[41], mental retardation [42], [43], Attention Deficit Hyperactivity Disorder [44], and alcohol drinking behaviour [39]. Auts2 expression is maximal in maturating neurons and declines as these cells become mature, suggesting that its function is required for neuronal differentiation [41], [45].
Here we report a genetic and developmental analysis of tay in the wing disc, and show that the function of Tay here is primarily related to the regulation of EGFR signalling. Thus, excess and loss of tay results in opposite phenotypes of loss - and extra veins, respectively, that are caused by changes in the levels of Erk activity. In addition, Tay level of expression modifies the phenotypic outcomes of altered EGFR signalling. We identify molecular interactions between Tay and Erk that might underline both the effects of Tay on Erk phosphorylation and the effects of Erk on Tay nuclear accumulation. All together, our results suggest that Tay is a novel component of the EGFR/Erk signalling pathway that regulates the nucleus/cytoplasm distribution of Erk.
Results
The phenotypes of EP-866/Gal4 combinations are due to the over-expression of tay
EP-866 is a P-GS element inserted in the first intron of tay, and was selected in a gain-of-function screen designed to identify genes that, when over-expressed, affect the differentiation of the wing veins [37]. The combination of EP-866 with a variety of Gal4 lines reduces the size of the wing and causes the partial loss of longitudinal veins (Fig. 1A–D; Fig. S1H–J). The most extreme phenotypes are observed in combinations of EP-866 with Gal4 drivers expressed in the entire wing blade and hinge (nub-Gal4/EP-866; Fig. 1B). A weaker version of this phenotype is detected in combinations with a Gal4 driver expressed only in the central region of the wing blade (salEPv-Gal4/EP-866; Fig. 1C). The reduction in wing size and loss of veins occurs in a compartment-specific manner, as they are also observed in combinations with the hh-Gal4 and ap-Gal4 drivers (Fig. S1J and data not shown). In all cases, the drastic reduction in wing size is associated with a reduction of cell proliferation, and not to the induction of cell death. Thus, wing discs of combinations between EP-866 and Gal4 drivers show a very low number of mitotic cells and no activation of Caspase3 (Fig. S1A–G′). When the gene affected by the EP-866 insertion is over-expressed during pupal development, the size of the wing is normal, but the veins fail to differentiate (Fig. 1D). EP-866/Gal4 combinations also display phenotypes in other adult structures, including fusion of tarsal joints in the legs (dll-Gal4/EP-866; Fig. S1A, C), a significant reduction in the size of the eye (ey-Gal4/EP-866; data not shown) and loss of sensory organs in the thorax (ap-Gal4/EP-866; Fig. S1B, D). The strength of the EP-866/Gal4 phenotype increases with the number of copies of both the Gal4 and the EP-866 insertion (Fig. S1K–M).
Fig. 1. Genetic characterization of the EP-866 insertion and expression analysis of tay. 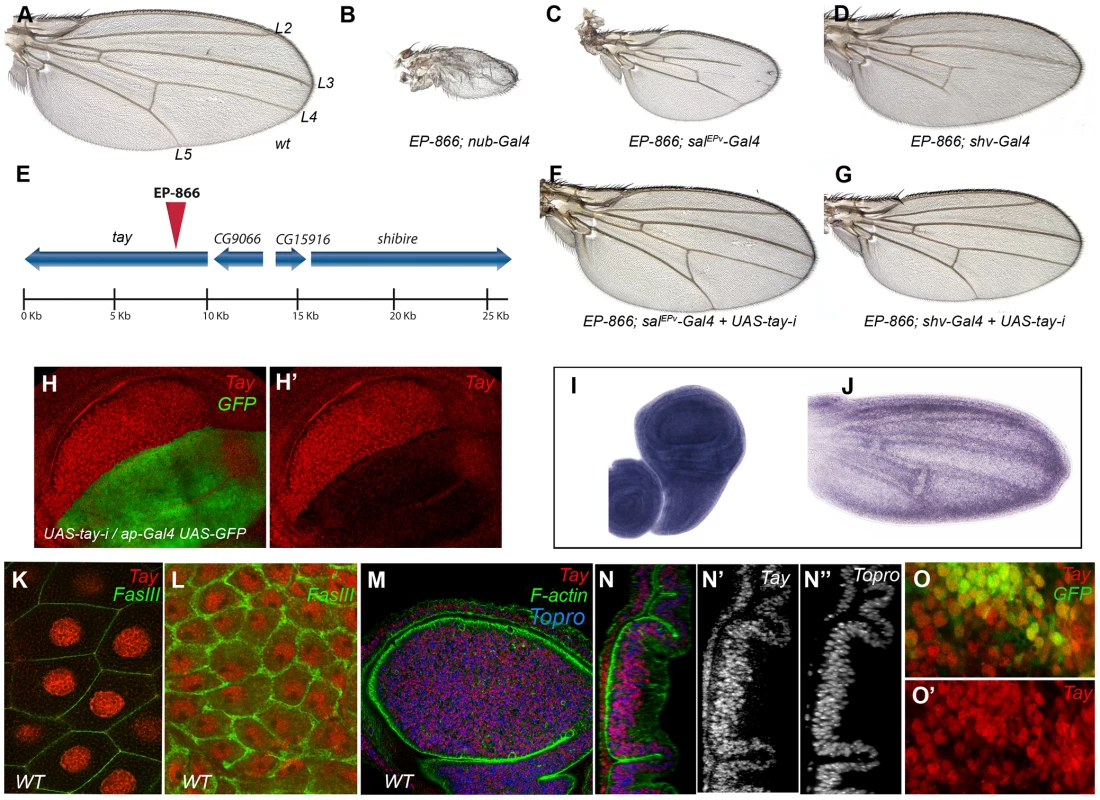
(A) Wild type wing (wt) indicating the position of the longitudinal veins L2–L5. (B–D) Combinations of EP-866 with Gal4 drivers expressed in the entire wing blade and hinge (EP-866/+; nub-Gal4/+; B), in the central region of the wing blade (EP-866; salEPv-Gal4/+; C) and in the pupal wing veins (EP-866/+; shv-Gal4/+; D). (E) Schematic representation of the genomic region around the insertion site of EP-866, showing the position and orientation of tay, CG9066, CG15916 and shibire coding regions. (F–G) Rescue of the EP-866/Gal4 phenotype by expression of RNA interference directed against tay (UAS-tay-i). In both cases, EP-866; salEPv-Gal4/+; UAS-tay-i/+ (F) and EP-866/+; shv-Gal4/UAS-tay-i (G) the phenotype is rescued (compare F with C and G with D). (H–H′) Expression of Tay (red) and GFP (green) in ap-Gal4 UAS-GFP/+; UAS-tay-i/+ third instar wing discs. The red channel (H′) shows the strong reduction in Tay protein levels. (I–J) in situ hybridization with a tay probe in a late third instar wing disc (I) and in a pupal wing 28 h after puparium formation (J). (K–L) High magnification pictures of a salivary gland (K) and the peripodial membrane of the wing disc (L) showing the accumulation of Tay (red channel) in the cell nucleus. The expression of FasIII (green) shows the contour of the cells. (M–N″) Expression of Tay (red), F-actin (green) and To-Pro (blue) in a third instar wing disc. (N–N″) Orthogonal section of the disc showed in M showing the individual channels for Tay (N′) and To-Pro (N″). (O–O′) Expression of Tay (red) in EP-866; salEPv-Gal4/+; UAS-GFP/+ discs, showing nuclear localization of Tay in cells over-expressing the protein. The expression of GFP is in green (O). The most likely candidate to cause the over-expression phenotype of EP-866/Gal4 combinations is the gene tay (Fig. 1E). Nonetheless, the genes CG15916 (5 Kb) and shibire (7 Kb) are close to the EP-866 insertion, and adjacent to tay is located CG9066, which is oriented in the 3′ to 5′ direction of transcription regarding the UAS sequences of the P-GS insertion. We know that tay, CG15916 and shi are over-expressed when EP-866 is combined with the salEPv-Gal4 [37]. However, the phenotypes of wing size reduction and loss of veins observed in EP-866/salEPv-Gal4 and EP-866/shv-Gal4 flies are suppressed when we introduced a UAS-tay-RNAi construct in these combinations (Fig. 1F–G, compare with 1C and D, respectively). In addition, the over-expression of Tay results in identical phenotypes of variable vein loss and wing size reduction (see below), indicating that tay causes the over-expression phenotypes of EP-866/Gal4 combinations.
Tay is a large nuclear protein related to human AUTS2
tay encodes a protein of 2486 amino-acids which most remarkable characteristic is a 30% of identity in the 1764–2019 amino acid region with a 486–782 stretch of the 1295 amino acid long human protein AUTS2 (Autism Susceptibility Candidate 2) (see below). The expression of tay occurs ubiquitously in all imaginal discs (Fig. 1I and data not shown), although we can also observe higher levels of expression in cells adjacent to the veins during pupal development (Fig. 1J). Tay is also expressed at other developmental stages, and during embryonic development its mRNA and protein are detected prominently in the central nervous system (Fig. S3E–G and data not shown). To visualize the accumulation of the Tay protein, we generated a specific polyclonal antibody (Fig. S2B), and found that the protein is present in the nucleus of all imaginal discs and salivary gland cells (Fig. 1K–N). The accumulation of Tay is very much reduced or lost in dorsal wing compartments expressing a tay RNA interference (Fig. 1H–H′). We also confirmed the specificity of this antibody by staining cells homozygous for a tay deficiency, where we found that the signal is completely lost (Fig. S2C–C′). The subcellular localization of the protein in wing discs over-expressing Tay is mostly nuclear, although some cytoplasmic staining is detected at higher level of over-expression (Fig. 1O–O′). These observations suggest that the adult phenotypes associated to Tay over-expression are caused by the accumulation of Tay at higher than normal levels in the nuclei of imaginal cells that normally express the gene. Interestingly, we also detected Tay in the cytoplasm of a subset of motoneurons in the central nervous system (CM and JFdC, data not shown), indicating that the protein subcellular localization is regulated in a cell-type specific manner.
Loss of tay function causes excess of vein differentiation
To identify the normal requirement of Tay during wing development, we reduced the levels of tay mRNA by expressing its RNA interference (tay-RNAi) in different domains of the wing disc. When tay-RNAi is expressed in the wing blade (638-Gal4/UAS-tay-i) the wings are reduced in size (32% smaller than wild type wings without changes in cellular size), display ectopic veins and show some defects in the most distal region of the wing margin (Fig. 2B). These phenotypes are caused by the reduction of tay, because they are enhanced in a genetic background with only one copy of the gene (Fig. 2C–D; 638-Gal4/+; Df(1)tay/UAS-tay-i). To generate stronger loss-of-function conditions, we made two small deficiencies by transposition (EP-866Rev34 and EP-866Rev40; see Fig. S2A), and a deficiency that eliminates tay and the adjacent gene CG16952 (Df(1)tay; Fig. S2A). These alleles are embryonic lethal in homozygous flies, and consequently they were analysed in mitotic recombination clones. The results obtained in Df(1)tay, EP-866rev40 and EP-866rev34 clones were identical, with cells deficient for tay forming clones that differentiate ectopic veins in inter-vein territories (Fig. 2E–G and data not shown). Interestingly, only a fraction of the mutant cells in each clone differentiate as ectopic veins of normal thickness (Fig. 2E–G). These phenotypes were very similar to those observed in wings expressing the tay-RNAi (compare with Fig. 2A–D).
Fig. 2. Loss-of-function analysis of tay. 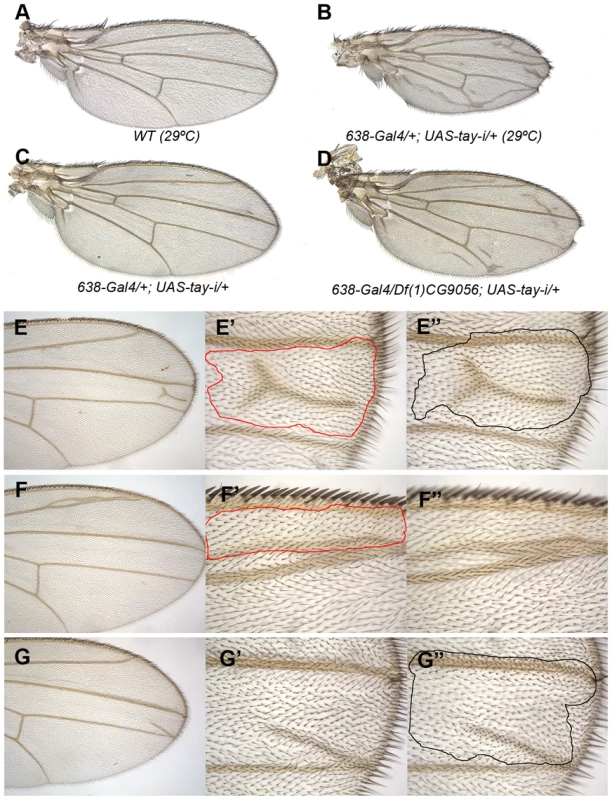
(A) Wild type wing grown at 29°C. (B) Wing size reduction, extra veins and loss of wing margin in 638-Gal4/+; UAS-tay-i/+ wings at 29°C. (C–D) The weak phenotype of 638-Gal4/+; UAS-tay-i/+ wings at 25°C (C) is increased in flies heterozygous for a tay deficiency (638-Gal4/Df(1)tay; UAS-tay-i/+; D). (E–G) Examples of mitotic recombination clones in flies of f36a Df(1)tay FRT18A/FRT18A UbiGFP; hsFLP/+ genotype. In E, example of a dorso-ventral clone located between the L3 and L4 veins (E′ and E″ are higher magnifications of the dorsal and ventral wing surfaces, respectively). In F example of a dorsal clone located anterior to the L2 vein. This clone (F′) differentiates vein cells and induces vein differentiation in the ventral wing surface (F″). In G example of a ventral clone located between the L3 and L4 veins that differentiate an ectopic vein in the ventral surface (G″) and induces vein differentiation in the dorsal surface (G′). Genetic interactions between Tay and upstream components of the EGFR pathway
The over-expression of tay in the wing imaginal disc prevents vein differentiation, macrochaetae formation and wing growth. Conversely, loss of tay function causes the formation of veins in inter-vein regions. These phenotypes are reminiscent to those caused by alterations in the levels of EGFR signalling, because loss of EGFR function impedes vein differentiation, and the increase in EGFR activity causes the formation of extra veins [27], [46]. To study the possible interactions between Tay and EGFR signalling, we made genetic combinations in which tay gain or loss of expression conditions were introduced in genetic backgrounds with modified EGFR activity. We find that the reduction of tay expression enhances the extra-vein phenotype caused by increased EGFR signalling. Thus, knock-down of tay enhances vein differentiation in RasV12 (Fig. 3A–C) and ectopic rhomboid (Fig. 3D–F) backgrounds. These observations suggest that Tay function is necessary either to attenuate EGFR signalling or to reduce the response to particular levels of EGFR signalling. Compatible with these possibilities, Tay over-expression enhances the loss-of-vein phenotype caused by reduced activity of the pathway, for example in a situation when the expression of EGFR is reduced (Fig. 3G–I). Interestingly, the reduction of tay expression does not modify the complete loss of vein phenotype caused by strong reductions in EGFR signalling (Fig. 3J–L), indicating that Tay function is mostly required to modulate the levels of EGFR signalling once the pathway has been activated.
Fig. 3. Genetic interactions between Tay and EGFR signalling. 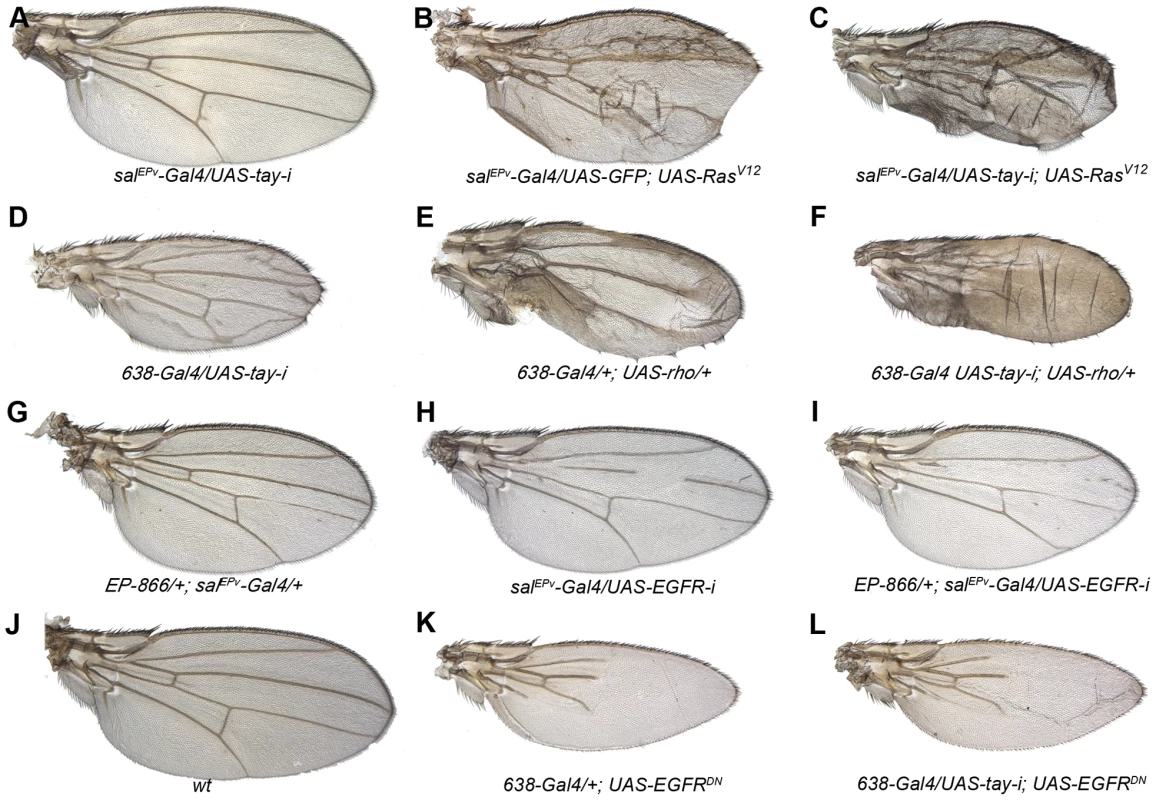
(A–C) The reduction of Tay (salEPv-Gal4/UAS-tay-i; A) increases the phenotype of ectopic veins caused by RasV12 over-expression (salEPv-Gal4/UAS-GFP; UAS-RasV12/+; B) in salEPv-Gal4/UAS-tay-i; UAS-RasV12/+ (C).). (D–F) Interaction of tay and rhomboid. The reduction of Tay (638-Gal4/UAS-tay-i; D) and the increase in rhomboid expression (638-Gal4/+; UAS-rho/+; E) cause ectopic veins, and this phenotype is strongly enhanced in the double combination (638-Gal4/UAS-tay-i; UAS-rho/+; F). (G–I) Interaction of Tay and EGFR. The expression of Tay (EP-866/+; salEPv-Gal4/+; G) enhances the phenotype of EGFR loss (salEPv-Gal4/UAS-EGFRi; H) in EP-866; salEPv-Gal4/+; UAS- EGFRi/+ flies (I). Interaction of Tay and EGFR. The reduction of tay expression does not modify the strong loss of veins phenotype caused by the expression of a dominant negative form of EGFR (638-Gal4/+; UAS-EGFRDN/+; K) in 638-Gal4/UAS-tay-i; UAS-EGFRDN/+ flies (L). A wild type wing is shown in J. Tay behaves as a negative regulator of the EGFR pathway
To analyse whether changes in the expression of tay directly affect EGFR signalling, we monitored the levels of di-Phosphorylated Erk (dP-Erk) and the expression of the EGFR transcriptional targets Delta and argos in tay over-expression conditions. The accumulation of dP-Erk in wild type wing discs is maximal in the developing L3 and L4 longitudinal veins and in the marginal veins [34]; Fig. 4B). dP-Erk accumulation is strongly reduced in these territories when Tay is over-expressed in the wing blade (Fig. 4F, compare with 4B). The expression of Delta (Dl), which is regulated by EGFR signalling during imaginal development [47], is maximal in the veins L3, L4 and L5 and in the marginal veins in wild type wing discs (Fig. 4C). Over-expression of tay in the central region of the wing blade causes a reduction of Dl expression in the veins L3 and L4 (Fig. 4G, compare with 4C). The vein L5 is not affected, because it is located outside the domain of salEPv-Gal4 expression (Fig. 4F–G). Therefore, this vein serves as an internal control in these experiments. We also observed changes in the transcription of argos, which expression is also regulated by the EGFR pathway and is maximal in the veins L3, L4 and L5 and in the marginal veins in wild type wing discs [48]; Fig. 4D). Over-expression of tay reduces argos-LacZ expression (Fig. 4H, compare with 4D). In all cases, the changes in Erk phosphorylation and in Dl/argos gene expression caused by Tay over-expression were consistently stronger than the loss of vein phenotype observed in the corresponding adult wings, as these wings still differentiate some stretches of the L3 and L4 veins (Fig. 4E).
Fig. 4. Effects of Tay on EGFR signalling. 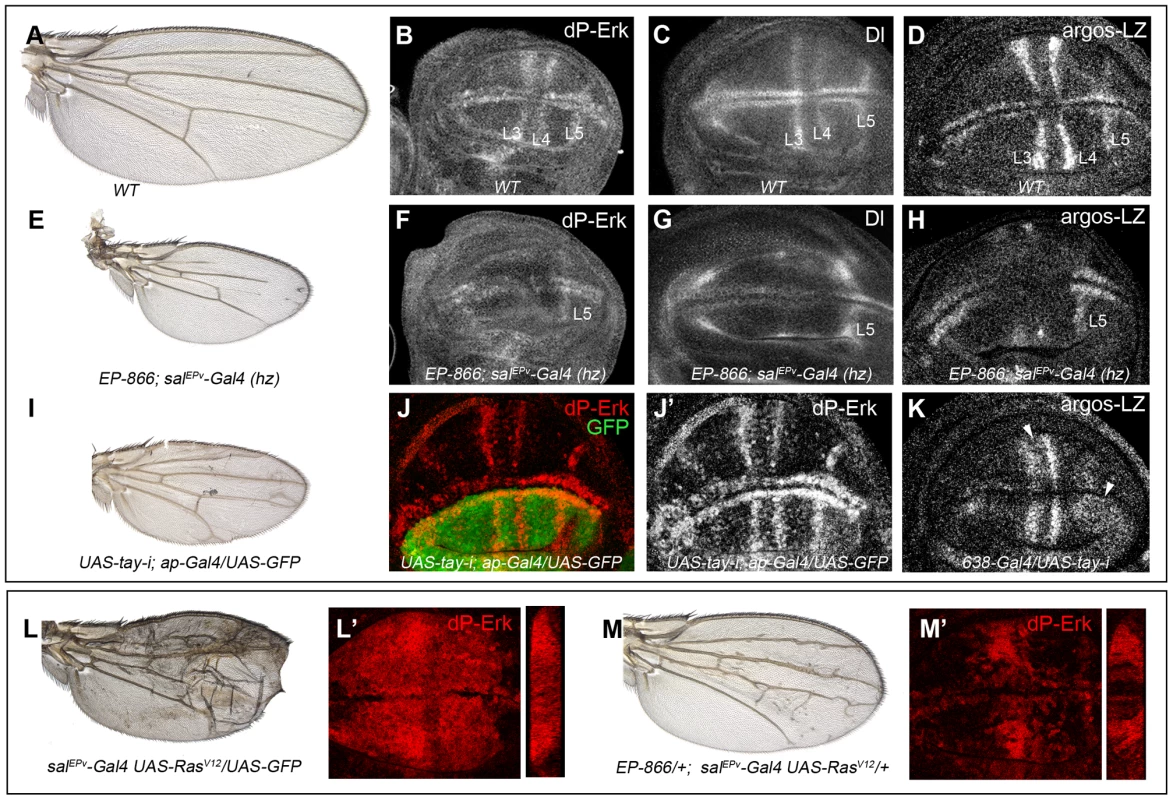
(A–D) Wild type wing (A) and late third instar wing discs showing the expression of dP-Erk (B), Delta (Dl, C) and argos-lacZ (D). (E–H) EP-866; salEPv-Gal4/salEPv-Gal4 wing (E) showing reduced size and partial loss of the L2, L3 and L4 veins. Late third instar wing disc of the same genotype showing dP-Erk (F), Delta (G) and argos-lacZ (H) expression. Only the expression of dP-Erk, Delta and argos-lacZ in the L5 vein, which is outside the domain of salEPv-Gal4, is present. (I–J′) Wing of UAS-tay-i; ap-Gal4/UAS-GFP genotype (I) and corresponding third instar wing disc showing the expression of dP-ERK (J–J′). Note the difference in expression levels between dorsal (labelled in green in J) and ventral inter vein cells. J′ corresponds to the red channel of J. (K) Expression of argos-lacZ in wing disc of UAS-tay-i/638-Gal4 genotype. Note that the reduction of tay expression in the entire wing blade produces ectopic expression of argos-lacZ, mainly in the L3/L4 intervein and around the L5 vein (white arrowheads). (L–L′) Phenotype of RasV12 over-expression (salEPv-Gal4 UAS-RasV12/UAS-GFP; L), and accumulation of dP-Erk (red; L′) in salEPv-Gal4 UAS-RasV12/UAS-GFP wing disc. (M–M′) The expression of Tay reduces the phenotype of RasV12 wings in EP-866/+; salEPv-Gal4/UAS-RasV12 flies (M), and also reduces the accumulation of dP-Erk (red; M′) in EP-866/+; salEPv-Gal4/UAS-RasV12 wing discs. Tangential sections through the length of the wing epithelium are shown to the right of L′ and M′. We also checked the effects of loss of Tay in the accumulation of dP-Erk. For this experiment we expressed tay-RNAi in the dorsal compartment of the wing (ap-Gal4/UAS-tay-i). In these discs the ventral compartment serves as an internal control. We observed that the reduction of tay expression increases dP-Erk accumulation in dorsal compartments compared with the ventral ones (Fig. 4J–J′). In addition, the expression of tay-RNAi in the entire wing blade (638-Gal4/UAS-tay-i) causes ectopic argos-lacZ expression (Fig. 4K, compare with 4D). Finally, we check whether excess of Tay can modulate dP-Erk accumulation under strong conditions of constitutive pathway activation. We find that Tay over-expression reduces the levels of dP-Erk induced by RasV12 in the central region of the wing disc (Fig. 4L′, M′), and also the phenotype of ectopic veins caused by RasV12 (Fig. 4L, M), suggesting that the negative effect of Tay on the activity of the EGFR pathway occurs downstream of Ras activation and affects the accumulation of dP-Erk. The effects of Tay loss and gain on dP-Erk accumulation were also detected in other imaginal discs, such as the eye disc (not shown) and the leg disc (Fig. S3C–D′), and also in embryos mutant for tay (Fig. S3A–B′), suggesting that Tay functions as a general modulator of Erk phosphorylation.
Genetic interactions between Tay, Erk and Mkp3
The preferential nuclear localization of Tay and its effects on EGFR signalling and Erk phosphorylation prompted us to study the interactions between Tay and EGFR pathway components which subcellular localization shifts between the nucleus and the cytoplasm. We focussed this analysis on Erk and its specific phosphatase Mkp3. These proteins can interact with each other in the cytoplasm, where Mkp3 retains ERK and prevents its phosphorylation, and also in the nucleus, where Mkp3 de-phosphorylates and inactivates Erk [8], [49]. In addition, the phenotypes caused by the loss of Erk or Mkp3 are very similar to those cause by tay over-expression or loss of function, respectively. To study the genetic interactions between Tay and Erk we over-expressed wild type Erk or its mutant form sevenmaker (Erksem), which bears a single amino acid substitution preventing Erk interactions with Mkp3 [50]–[51]. The use of Erksem allows the analysis of Erk over-expression conditions in the absence of its interaction with Mkp3. The formation of ectopic veins caused by a reduction in Tay levels is only weakly increased when the normal form of Erk is over-expressed (Fig. 5D–F). In contrast, loss of tay in a background of Erksem over-expression causes a strong increase in the differentiation of extra-vein tissue (Fig. 5H), compared with loss of only tay (Fig. 5D) or with Erksem over-expression (Fig. 5G). Interestingly, Tay over-expression reduces, but does not suppress, the ectopic veins caused by Erksem (Fig. 5A–B). These results suggest that Erksem is much more effective when Tay levels are reduced, and, conversely, that Tay is less effective antagonizing Erk when this protein cannot interact with Mkp3.
Fig. 5. Genetic interactions between tay, Mkp3 and Erk. 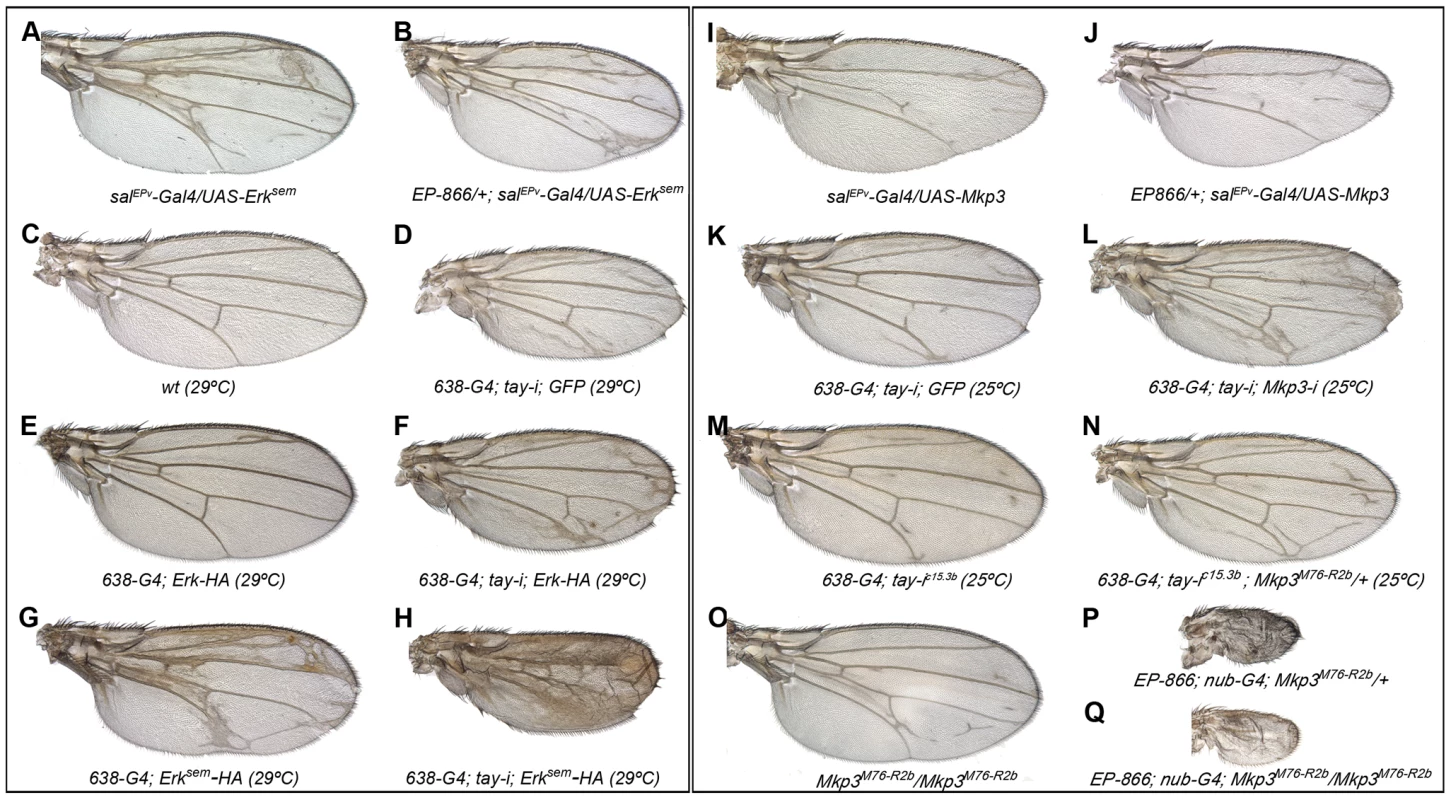
(A–H) Genetic interactions between tay and Erk. (A–B) The over-expression of Tay does not supress the phenotype of ectopic veins caused by the expression of Erksem (salPEv-Gal4/UAS-Erksem; A) in EP-866/+; salEPv-Gal4/UAS-Erksem flies (B). (C–D) Control wild type wings grown at 29°C (C) and 638-Gal4/UAS-tay-i; UAS-GFP/+ wing showing ectopic veins in inter-vein regions and reduced size (D). (E–F) The ectopic vein phenotype of 638-Gal4/+; UAS-Erk-HA/+ flies (E) is enhanced when the expression of tay is reduced in 638-Gal4/UAS-tay-i; UAS-Erk-HA/+ flies (F). (G–H) The ectopic vein phenotype of 638-Gal4/+; UAS-Erksem-HA/+ wings (G) is strongly increased when the expression of tay is reduced in 638-Gal4/UAS-tay-i; UAS-Erksem-HA/+ flies (H). (I-P) Genetic interactions between tay and Mkp3. (I–J) The loss of veins phenotype of salEPv-Gal4/UAS-Mkp3 wings (I) is not modified when Tay is over-expressed in EP-866/+; salEPv-Gal4/UAS-Mkp3 wings (J). (K–L) The ectopic veins phenotype of 638-Gal4/UAS-tay-i; UAS-GFP/+ wings (K) is increased when the expression of Mkp3 is also reduced in 638-Gal4/UAS-tay-i; UAS-Mkp3-i/+ flies (L). (M–N) The phenotype of ectopic veins of 638-Gal4/+; UAS-tay-i/+ wings (M) is increased in heterozygous Mkp3 flies (638-Gal4/UAS-tay-i; Mkp3M76-R2b/+; N). (O–P) The phenotype of loss of veins and strong wing size reduction produced by the over-expression of Tay in the entire wing blade and hinge (EP-866; nub-Gal4/+; Mkp3M76-R2b/+; P) is not modified in homozygous Mkp3 background (EP-866; nub-Gal4/+; Mkp3M76-R2b/Mkp3M76-R2b; Q). Control Mkp3M76-R2b homozygous wings are shown in panel O, and control heterozygous Mkp3M76-R2b/+ wings are indistinguishable from wild type. In the case of Mkp3, the loss of veins caused by its over-expression (Fig. 5I) is not modified by loss (not shown) or excess of tay (Fig. 5J), confirming that Tay levels are not relevant upon a strong loss of Erk activation. In contrast, the formation of extra veins observed in tay loss-of-function conditions (Fig. 5K, M) depends on the gene dosage of Mkp3, becoming stronger in Mkp3M76-R2b heterozygous flies (Fig. 5N, compare with M) or upon expression of Mkp3-RNAi (Fig. 5L, compare with K). One possible explanation for these interactions is that Tay participates in the regulation of Erk inactivation, perhaps by promoting its de-phosphorylation. This possibility is compatible with the strong reduction of Erk phosphorylation caused by Tay over-expression, and implies that Tay over-expression phenotypes should be dependent on the presence and activity of Erk phosphatases such as Mkp3. However, we notice that the phenotype of Tay over-expression is not modified in Mkp3 null mutant backgrounds (Fig. 5O–Q). Thus, although we cannot exclude a role of Mkp3 in Tay function, this result indicates that the effects of Tay over-expression are not mediated exclusively by the activity of Mkp3.
Effects of Tay in the activation of ERK
Next, we wanted to visualize the activation of Erk in genetic backgrounds where the level of Erk and Tay expression is changed and the activity of the EGFR pathway is increased. To this end, we made tagged forms of Tay (Tay-Flag), Erk (Erk-HA) and Erksem (Erksem-HA) and studied the accumulation of dP-Erk in wing discs of different genotypes. The expression of Erk-HA and Erksem-HA causes very weak (Erk-HA; Fig. 5E) or moderate (Erksem-HA; Fig. 5G and 6I) extra veins. In none of these over-expression backgrounds we were able to detect changes in the pattern or level of dP-Erk accumulation (Fig. 6A and 6E). The reduction of Erk phosphorylation caused by Tay over-expression (Fig. 4) is still observed when either Erk-HA (Fig. 6B) or Erksem-HA (Fig. 6F) is expressed in combination with Tay. The strong activation of the pathway caused by RasV12 is also observed in backgrounds of Erk-HA or Erksem-HA expression (Fig. 6C and G, respectively). The introduction of Tay in these backgrounds causes a moderate reduction in dP-Erk accumulation (Fig. 6D and H, compare with 6C and G), although the resulting phenotype of ectopic vein differentiation is not reduced (Fig. 6K–L). From these observations we conclude that Tay is still effective in promoting the de-phosphorylation of Erk under conditions of Erk and Erksem over-expression, but less so in backgrounds of strong pathway activation.
Fig. 6. Effects of Tay over-expression in the activation of Erk. 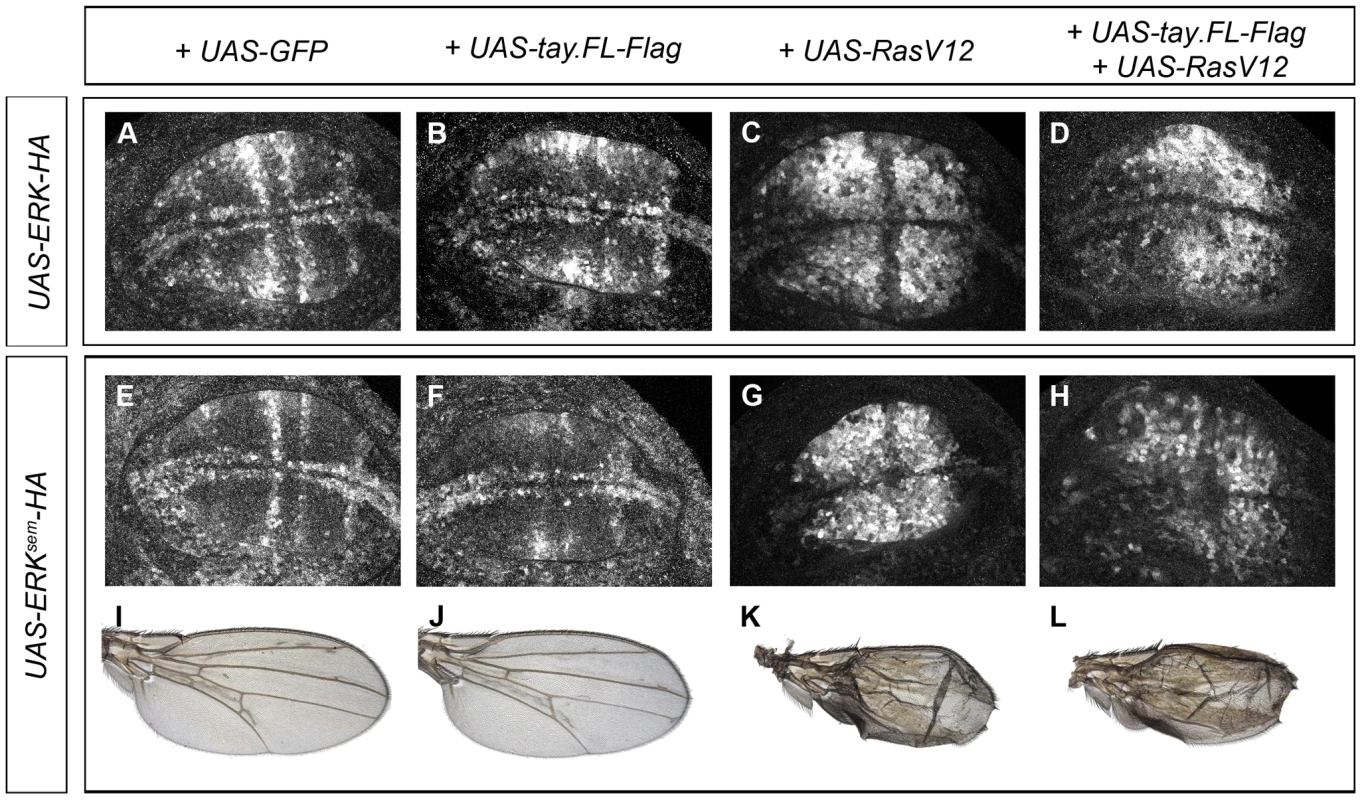
(A–D) Accumulation of dP-Erk in late third instar wing discs over-expressing Erk-HA. (A) Control salEPv-Gal4/UAS-Erk-HA; UAS-GFP/+ wing disc. (B) salEPv-Gal4/UAS-Erk-HA; UAS-tay.FL-Flag/+ wing disc. (C) salEPv-Gal4/UAS-Erk-HA; UAS-RasV12/+ wing disc. (D) salEPv-Gal4/UAS-Erk-HA; UAS-RasV12/UAS-tay.FL-Flag wing disc. (E–F) Accumulation of dP-Erk in late third instar wing discs over-expressing Erksem-HA. (E) Control salEPv-Gal4/UAS-Erksem-HA; UAS-GFP/+ wing disc. (F) salEPv-Gal4/UAS-Erksem-HA; UAS-tay.FL-Flag/+ wing disc. (G) salEPv-Gal4/UAS-Erksem-HA; UAS-RasV12/+ wing disc. (H) salEPv-Gal4/UAS-Erksem-HA; UAS-RasV12/UAS-tay.FL-Flag wing disc. (I–L) Wings of salEPv-Gal4/UAS-Erksem-HA; UAS-GFP/+ (I), salEPv-Gal4/UAS-Erksem-HA; UAS-tay.FL-Flag/+ (J), salEPv-Gal4/UAS-Erksem-HA; UAS-RasV12/+ (K) and salEPv-Gal4/UAS-Erksem-HA; UAS-RasV12/UAS-tay.FL-Flag (L) genotypes. Effects of Tay in the subcellular localization of Erk, Erksem and Mkp3
The subcellular localization of Mkp3 and Erk is dynamic, shifting between the nucleus and the cytoplasm [8], [49]. We wanted to analyse whether Tay influences the accumulation of these proteins in wing imaginal cells in over-expression conditions. First, we confirmed that Mkp3-Myc is preferentially localised in the cytoplasm (Fig. S4A–A′″), and that both Erk-HA and Erksem-HA are detected in the nucleus and in the cytoplasm, with Erk-HA distributed at higher levels in the cytoplasm (Fig. 7A and E, respectively and Fig. S4C–C″ and E–E′″). The co-expression of Mkp3-Myc and Tay-Flag does not modify the preferential cytoplasmic (Mkp3) or nuclear (Tay) accumulation of these proteins (Fig. S4B–B′″). The co-expression of Mkp3-Myc and Erk-HA results in a clear cytoplasmic retention of Erk-HA (Fig. 7B, compare with A). In contrast, Mkp3-Myc does not modify the homogeneous nucleus-cytoplasm distribution of Erksem-HA (Fig. 7F, compare with E). Neither the localization of Tay-Flag or Erk-HA changes when both are co-expressed in the same cells of the central region of the wing disc (Fig. 7C and Fig. S4D–D′″). In addition, the expression of RasV12 does not affect the localization of Erk-HA, which is still localised in the nucleus and cytoplasm (Fig. 7D, compare with A, and Fig. S5A–A″).
Fig. 7. Subcellular localization of Erk in over-expression conditions. 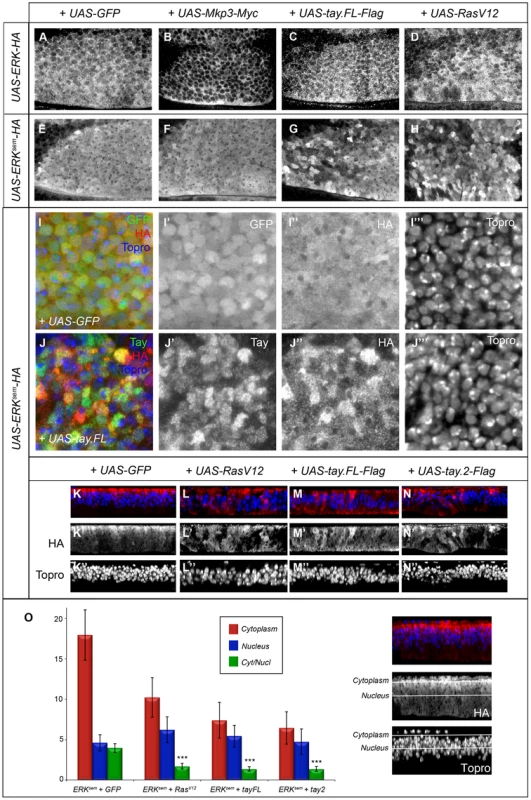
(A–B′) Subcellular localization of Erk (HA) in the anterior-dorsal region of wing discs of the following genotypes: salEPv-Gal4/UAS-Erk-HA; UAS-GFP/+ (A); salEPv-Gal4/UAS-Erk-HA; UAS-Mkp3-Myc/+ (B); salEPv-Gal4/UAS-Erk-HA; UAS-tay.FL-Flag/+ (C) and salEPv-Gal4/UAS-Erk-HA; UAS-RasV12/+ (D). Note the cytoplasmic retention of Erk-HA in the presence of Mkp3-Myc (B) compared to the control (A). (E–H) Subcellular localization of Erksem (HA) in wing imaginal discs of the following genotypes: salEPv-Gal4/UAS-Erksem-HA; UAS-GFP/+ (E); salEPv-Gal4/+; UAS-Erksem-HA/UAS-Mkp3-Myc (F); salEPv-Gal4/UAS-Erksem-HA; UAS-tay.FL-Flag/+ (G) and salEPv-Gal4/UAS-Erksem-HA; UAS-RasV12/+ (D). Note the similar appearance of Erksem accumulation in the presence of Tay.FL-Flag (G) and Rasv12 (H). (I–J) Higher magnification pictures of wing discs showing the expression of Erksem-HA (I″ and J″), Tay (J′) and To-pro (I′″ and J′″). The composite images are shown in panels I and J. In all cases Erksem-HA was over-expressed in the central region of the wing disc in larvae of the salEPv-Gal4/UAS-Erksem-HA; UAS-GFP/+ (I–I′″) and salEPv-Gal4/UAS-Erksem-HA; UAS-tay.FL-Flag/+ genotype (J–J′″). Note the more heterogeneous appearance of Erksem-HA in presence of Tay.FL (J–J′″) compare to controls (I–I′″). (K–N) Transverse sections through the wing disc showing the expression of Erksem-HA (red) and To-pro (blue) in wing discs of salEPv-Gal4/UAS-Erksem-HA; UAS-GFP/+ (K–K″), salEPv-Gal4/UAS-Erksem-HA; UAS-rasV12/+ (L–L″), salEPv-Gal4/UAS-Erksem-HA; UAS-tay.FL-Flag/+ (M-M″) and salEPv-Gal4/UAS-Erksem-HA; UAS-tay.2-Flag/+ (N–N″). (O) Average values and standard deviation of Erksem signal intensity in 6 sections of 6 discs in each genotype (n = 36) taken from the apical (cytoplasm; red columns and see white line in the picture to the right) or medial (nucleus; blue columns and see white line in the picture to the right) level of the epithelium. The Cytoplasm/Nucleus ratio (green columns) was calculated as the average ratio of 36 pair of measures. In the three genotypes analysed the Cytoplasm/Nucleus ratio is significantly lower that control (p>0.005, t-student). In contrast, both Erksem-HA and Tay-Flag display a heterogeneous distribution when co-expressed (Fig. 7G, J–J″ and Fig. S4F–F′″). We took higher magnification pictures of sections taken from the most anterior region of the salEPv-Gal4 domain of expression, because in these cells the level of over-expression are lower and Tay retains its nuclear localization (Fig. S7). We observed that the nuclear level of Erksem-HA and Tay in each cell are not correlated (r2 = 0.09; n = 60). A similar heterogeneous distribution of ERKsem was observed in a RasV12 background (Fig. 7H and Fig. S5C–C″), and also when both Tay-Flag and Erksem-HA were co-expressed in a Rasv12 background (Fig. S5D–D″). We do not understand the molecular bases for these changes in Erksem and Tay accumulation in the presence of each other or upon strong pathway activation, but they might be related to a dynamic regulation of protein turnover when Tay and Erk are co-expressed at higher levels. To get a quantitative view of Erksem nuclear-cytoplasmic localization, we took serial sections of the wing disc, quantified the levels of Erksem in the cytoplasm (apical in the epithelium; Fig. 7O) and nucleus (medial in the epithelium; Fig. 7O), and calculated the average cytoplasm/nucleus ratio of Erksem signal in different genetic backgrounds (Fig. 7K–O). These measures show that Erksem is mostly localised apically in the cell (cytoplasm), and that both the presence of Rasv12 (Fig. 7K) or Tay-Flag (Fig. 7L) strongly reduce the amount of cytoplasmic Erksem and weakly increase the level of nuclear Erksem (Fig. 7N). In this manner, the expression of either RasV12 or Tay changes Erksem localization in a similar manner, but although both Tay and RasV12 reduce the cytoplasm/nucleus ratio of Erksem accumulation, Erk activation, as visualised by the presence of dP-Erk (see Fig. 6), only occurs in RasV12 conditions.
Tay binds both ERK and MKP3
We next considered the possibility that Tay might be directly interacting with Erk or Mkp3 in co-immunoprecipitation and pull-down experiments. Co-immunoprecipitation experiments were carried out from protein extracts obtained from embryos expressing combinations of Tay.FL-Flag, Mkp3-Myc, Erksem-HA and Erk-HA (see Fig. S2D–E). Tay.FL-Flag was never detected in western blots, perhaps because the size of the protein prevents its transference to the membrane. However, when Tay-Flag is co-expressed with Mkp3-Myc or Erksem-HA, we detected co-immunoprecipitation when the IP was made using anti-Flag and the western blot revealed using anti-Myc (Fig. 8A, line T+M from IP lanes) or anti-HA (Fig. 8B, line T+E from IP lanes). In protein extracts from embryos expressing only Mkp3-Myc or Erksem-HA and IP with anti-Flag, we never detected Myc or HA (Fig. 8A–B, lines M and E, from IP lanes, respectively). The interaction between Tay and Erk and between Tay and Mkp3 might be direct, because they were also observed in pull-down experiments using in vitro translated Tay incubated with Erk-GST and Mkp3-GST fusion proteins (Fig. 8C).
Fig. 8. Tay interacts with Erk and Mkp3. 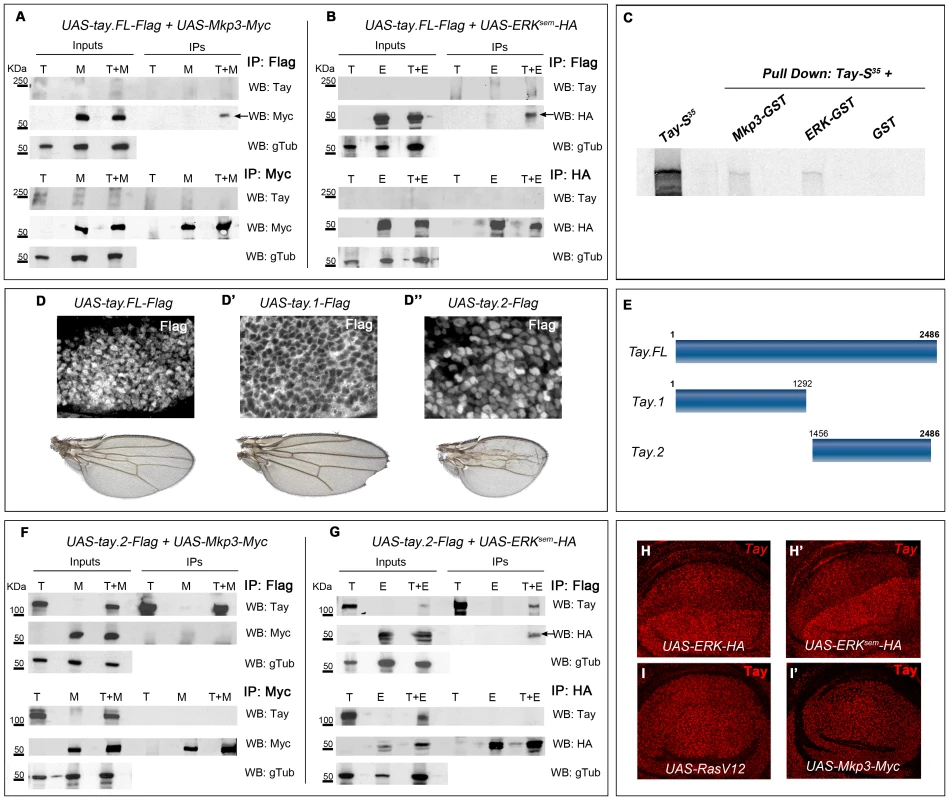
(A–B) Interactions between Tay and Mkp3 (A) and between Tay and Erk (B). Embryo extracts were immunoprecipitated with anti-Flag (upper lines in A and B), anti-Myc (lower lines in A) and anti-HA (lower lines in B). Immunoprecipitates (IPs) and lysates (Inputs) were immunoblotted with anti-Myc (detecting Mkp3-Myc protein), anti-HA (detecting Erksem-HA protein) and anti-Tay (we could not detect neither endogenous Tay nor Tay-Flag protein). In control IPs carried out with da-Gal4/UAS-tay.FL-Flag (T) and da-Gal4/UAS-Mkp3-Myc (M) embryos in A, and da-Gal4/UAS-tay.FL-Flag (T) and da-Gal4/UAS-Erksem-HA (E) embryos in B, we did not detect any Tay, Mkp3 or Erk proteins. In Co-IPs experiments carried out with da-Gal4/UAS-tay.FL-Flag; UAS-Mkp3-Myc/+ embryos (T+M) in A and da-Gal4/UAS-tay.FL-Flag; UAS-Erksem-HA/+ (T+E) embryos in B, we detected Mkp3-Myc protein in A and Erksem-HA protein in B when the extracts were immunoprecipitated with anti-Flag (black arrows). (C) Pull down assay showing the interactions Tay-Mkp3 and Tay-Erk. Tay protein was in vitro translated and radiolabelled with MetS35. Tay was detected when it was incubated with Mkp3-GST protein and Erk-GST protein, but was not detected in control pulldowns with GST. (D–D″) Subcellular localization of Tay (Flag, white in D–D″) in wing imaginal discs of salEPv-Gal4/UAS-tay.FL-Flag (D), salEPv-Gal4/UAS-tay.1-Flag (D′) and salEPv-Gal4/UAS-tay.2-Flag (D″) and their corresponding wings. (E) Schematic representation of Tay (Tay.FL) and the N-terminal (Tay.1) and C-terminal (Tay.2) forms. (F–G) Interactions between Tay.2 and Mkp3 (F) and Tay.2 and Erk (G). Embryo extracts were immunoprecipitated with anti-Flag (upper lines in F and G), anti-Myc (lower lines in F) and anti-HA (lower lines in G). Immunoprecipitates (IPs) and lysates (Inputs) were immunoblotted with anti-Myc (detecting Mkp3-Myc protein), anti-HA (detecting Erksem-HA protein) and anti-Tay (detecting Tay.2-Flag protein). In control IPs carried out with da-Gal4/UAS-tay.2-Flag (T) and da-Gal4/UAS-Mkp3-Myc (M) embryos in F, and with da-Gal4/UAS-tay.2-Flag (T) and da-Gal4/UAS-Erksem-HA (E) embryos in G, we did not detect any Tay.2, Mkp3 or Erk protein. In Co-IPs experiments carried out with da-Gal4/UAS-tay.2-Flag; UAS-Mkp3-Myc/+ embryos (T+M) in F, we did not detect Mkp3-Myc protein when the extracts were immunoprecipitated with anti-Flag. In Co-IPs experiments carried out with da-Gal4/UAS-tay.2-Flag; UAS-Erksem-HA/+ embryos (T+E) in G, we detected Erksem-HA protein when the extracts were immunoprecipitated with anti-Flag (black arrow). (H–I′) Accumulation of Tay in wing discs over-expressing Erk, Erksem, RasV12 and Mkp3. The over-expression is limited to the dorsal compartment of ap-Gal4/UAS-Erk-HA (H) and ap-Gal4/UAS-Erksem-HA (H′) discs, or to the central domain of the wing blade in salEPv-Gal4/UAS-RasV12 (I) and salEPv-Gal4/UAS-Mkp3-Myc (I′) discs. Note the increased levels of Tay in dorsal compartments over-expressing Erk-HA and Erksem-HA (H–H′). To identify the region of Tay involved in these interactions, we made several truncated forms of the protein (Fig. 8E and data not shown), and expressed them in the wing disc. We found that the 1292 amino acid N-terminal fragment of Tay (Tay.1) is located exclusively in the cytoplasm (Fig. 8D′), and its over-expression does not affect the differentiation of veins (Fig. 8D′). In contrast, the 1030 amino acid C-terminal fragment (Tay.2) is accumulated preferentially in the nucleus (Fig. 8D″), similar to the full-length Tay-Flag protein (Fig. 8D). Interestingly, the expression of Tay.2 consistently results in stronger phenotypes of vein loss and reduced wing size than those caused by the over-expression of the full-length protein (Fig. 8D″ compare with D). This C-terminal fragment includes the domain of homology detected between Tay and human AUTS2. The distribution of Erksem is not modified in the presence of the N-terminal portion of Tay (data not shown). In contrast, Tay.2 results in the same changes in the cytoplasm/nucleus ratio of Erksem accumulation as Tay.FL (Fig. 7N–O).
The C-terminal 1030 amino acid Tay fragment (Tay.2) contains all the information necessary to regulate the subcellular localization of the protein, and also all the domains necessary to reproduce the effects of the full-length protein (see above). We repeated the immunoprecipitation experiments using this fragment, and found that Tay.2 retains its interaction with Erksem (Fig. 8G, line T+E, IP lanes), but loses its ability to interact with Mkp3 (Fig. 8F, line T+M, IP lanes). The failure of Tay.2 to interact with Mkp3 might increase the titration of ERK by Tay.2, explaining why Tay.2 interferes with EGFR signalling more efficiently than the full-length protein.
We also found that the levels of Tay accumulation in the nucleus are much higher than normal in cells over-expressing Erk or Erksem (Fig. 8H–H′ and Fig. S7A–D). As Erk or Erksem over-expression do not change the expression of tay (not show), these observations indicate that Erk increases the stability of Tay in the nucleus. This effect is independent of EGFR signalling, as neither RasV12 nor Mkp3 over-expression modified the accumulation of endogenous (Fig. 8I–I′) or over-expressed Tay (Fig. S6A–B″). We conclude from these data that Tay can interact with Erk in the nucleus and that Erk protects Tay from degradation.
The expression of human AUTS2 in the wing disc causes the formation of ectopic veins
Drosophila Tay and human AUTS2 are very different proteins in sequence and length, but they share a small 250 amino acid stretch with significant homology (Fig. 9A). Our deletion analysis of Tay indicates that this region is included in the smaller fragment of Tay that we found has biological activity and nuclear localization (C.M. and J.F.dC., unpublished results). We wanted to check whether AUTS2 expressed in flies was able to reproduce some of the effects observed in Tay over-expression conditions. A Flag-tagged form of AUTS2 expressed in the wing disc is localised exclusively in the nuclei (Fig. 9B–C), the same as Tay. Interestingly, the expression of AUTS2 in the wing leads to a phenotype of ectopic vein formation reminiscent to the consequence of Tay loss (Fig. 9D). The extra veins that develop in AUTS2 over-expression conditions depend on EGFR signalling, because they are eliminated when the expression of Erk is reduced (Fig. 9E–F). AUTS2 also enhances the formation of extra veins caused by the expression of Erksem (Fig. 9H–I), and causes an increase in the levels of activated Erk (ap-Gal4/UAS-hAUTS2-Flag; Fig. 9J–J′). These data suggest that AUTS2 is able to interact with some, but not all, targets of Tay, and raise the possibility that AUTS2 normal function in humans is related to the regulation of the Erk signalling pathway, albeit in an opposite manner as Tay.
Fig. 9. Subcelullar localization and phenotypes of human AUTS2 in flies. 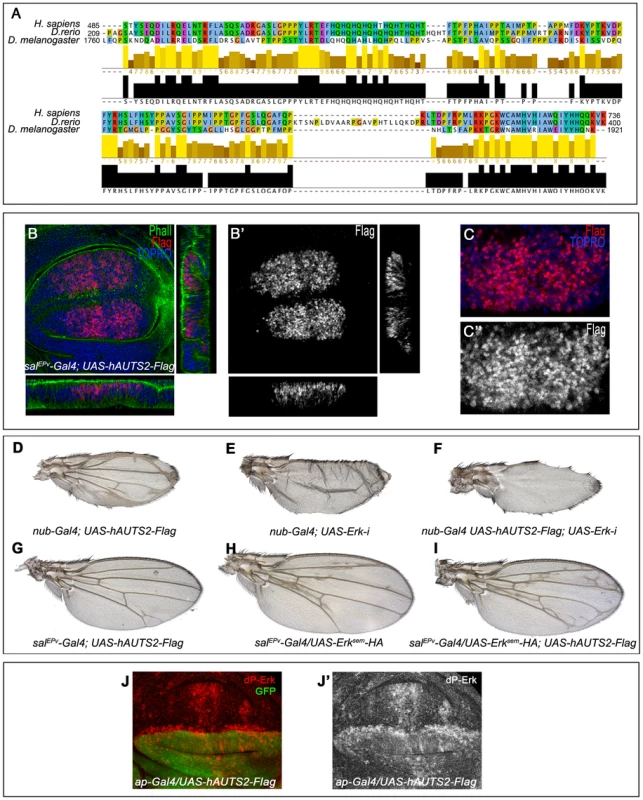
(A) Sequence conservation between human AUTS2 (H. sapiens; amino acids 485–736), Zebra fish AUTS2 (D. renio; amino acids 209–400) and Tay (D. melanogaster; amino acids 1760–1921). The colour code indicates the physicochemical properties of each amino acid (Jalview software) and the bars below each amino acid the identity in two or three species. (B–B′) Subcellular localization of AUTS2 (Flag red in B, white in B′) in wing discs of salEPv-Gal4/+; UAS-hAUTS2-Flag/+. Below and to the right are the corresponding horizontal and transversal sections of the disc. The expression of F-actin is in green and To-Pro in blue. B′ is the red channel of B showing the accumulation of AUTS2. (C–C′) Higher magnification of the dorsal region of a salEPv-Gal4/+; UAS-hAUTS2-Flag/+ disc, showing the expression of AUTS2 (red in C, white in C″) and To-Pro (blue). The single red channel is shown in C′. (D–I) The phenotype of ectopic veins produced by the expression of AUTS2 in the entire wing blade and hinge (nub-Gal4/UAS-hAUTS2-Flag; D) is abolished when the expression of AUTS2 in accompanied with a reduction of Erk expression in nub-Gal4/UAS-hAUTS2-Flag; UAS-Erk-i/+ flies (F), producing a loss of vein phenotype very similar to nub-Gal4/UAS-Erk-i flies (E). (G–I) Expression of AUTS2 in salEPv-Gal4/UAS-hAUTS2-Flag flies (G) and expression of Erksem in salEPv-Gal4/UAS-Erksem-HA flies (H) causes weak phenotypes of ectopic veins that are enhanced when AUTS2 and Erksem are expressed together in salEPv-Gal4/UAS-Erksem-HA; UAS-hAUTS2-Flag flies (I). (J–J′) Third instar wing disc showing the expression of dP-Erk (red in J, white in J′) in ap-Gal4/UAS-hAUTS2-Flag wing discs. Note the difference in expression levels between dorsal (labelled in green in J) and ventral cells. J′ corresponds to the red channel of J. Discussion
Signalling by Erk in response to growth factors regulates growth, differentiation and survival of cells in a variety of developmental contexts [7], [52]–[53]. The extent and level of Erk activation relies on its phosphorylation state, which in turns regulates Erk subcellular localization and interactions with downstream effectors and other proteins [7], [10], [54]–[55]. Erk activation is transient, and failures in the mechanisms responsible for its inactivation can drive developmental defects and oncogenic transformations [10]. In this work we identified Tay as a novel nuclear component that interacts with Erk and is involved in the maintenance of appropriate levels of Erk activity.
Tay is a nuclear protein that antagonize EGFR signalling
We have addressed the requirements and function of tay mostly in the wing disc, a convenient developmental system to analyse the contribution of signalling pathways to the regulation of organ size and pattern formation [56]. Tay was previously described as a protein that regulates locomotion and other neural aspects [38]–[39]. We have observed that changes in the level of EGFR signalling in the nervous system also cause locomotion defects (Molnar and de Celis, in preparation), which is indicative of a role of Tay in the regulation of EGFR signalling also in the nervous system. In the context of wing development and vein differentiation, the loss of tay results in the differentiation of extra veins in inter-vein territories. This phenotype is very similar to those obtained in conditions of excess of EGFR signalling, suggesting that Tay negatively regulates the activity or the response to this pathway. In addition, loss of tay also causes a reduction in the size of the wing blade, a phenotype that is not expected in a situation of excess of EGFR/ERK activity. This last result suggests that Tay might also have functions independent of its role in the regulation of EGFR signalling. The consequences of gain of Tay expression mostly indicate that the role of Tay is related to the modulation of EGFR signalling. Thus, excess of Tay expression in different imaginal discs results in phenotypes that can be attributed to loss of EGFR signalling, such as loss of veins and bristles [33], wing size reduction and failures in tarsal joint formation [57] and ommatidial differentiation (data not shown).
We further explore the relationships between Tay and EGFR signalling in genetic combinations in which the activity of the pathway is altered in backgrounds with modified levels of Tay expression. In all cases, we observed synergistic interactions between loss of tay and excess of EGFR, and between excess of tay and loss of EGFR activity. Furthermore, we notice that the extra veins differentiating in tay mutants require EGFR function, suggesting that Tay modulates EGFR signalling during vein formation. All together, the results of genetic combinations indicate that cells with lower levels of Tay become more sensitive to an increase in EGFR signalling, and that Tay over-expression prevents cells to acquire the level of EGFR signalling required for vein formation.
The negative effect of Tay on EGFR signalling is more directly visualised by considering the effects of Tay in Erk phosphorylation and in the expression of the EGFR/Erk targets genes Dl and argos. Thus, Tay over-expression strongly suppresses Erk phosphorylation and prevents the expression of Dl and argos in the developing veins. Conversely, in loss of tay conditions we detect an increase in the levels of phosphorylated Erk, which is accompanied by a moderate ectopic expression of argos. The extra-vein phenotype of loss of tay is not as extreme as the massive vein differentiation that occurs upon strong and constitutive activation of the EGFR pathway. In fact, tay mutant wings differentiate a similar pattern of extra veins as moderate increases in EGFR signalling caused by, for example, mutations in the Mkp3 gene [58]. This suggest us that Tay primary function is to prevent increases in EGFR/Erk signalling in places where the pathway must be active but only at low levels. Thus, high levels of EGFR activity and dP-Erk accumulation are restricted to the presumptive veins in wild type third instar wing discs, but the pathway is also active at lower levels in the inter-veins, where it promotes cell proliferation and survival [28]. In tay or Mkp3 mutant backgrounds, a fraction of these cells initiates the vein differentiation program, escaping the negative feed-back loops that maintain low dP-Erk levels and entering the positive feed-back loops that normally operate in vein territories through the regulation of rhomboid expression [59]. In this model, Tay would participate in a mechanism that favours Erk de-phosphorylation and its nuclear retention in an inactive form. This mechanism of Tay action is compatible with the effects of its over-expression, which essentially cause a failure to accumulate dP-Erk in vein territories, and consequently a loss of vein differentiation.
Tay interacts with Erk and Mkp3
Signalling by Erk proteins in the nucleus is in part regulated by the rate of Erk nucleus/cytoplasm shuttling [60]. In the nucleus, signal termination involves Erk de-phosphorylation by nuclear phosphatases and also its sequestration away from cytoplasmic Erk kinases [61]. Because Erk does not contain nuclear localization nor export sequences, its subcellular localization relies on proteins acting as anchors [8]. We observed direct interactions between Tay and Erk and between Tay and Mkp3, and these interactions were also detected in immunoprecipitation experiments from embryo protein extracts. These data suggests that Tay could form part of protein complexes including both Erk and Mkp3 in the nucleus.
A direct interaction between Tay and Erk is also compatible with several observations regarding Tay stability and Erk subcellular localization. First, Erk and Erksem increase the accumulation of Tay in the nucleus, and do so independently of EGFR signalling, as neither RasV12 nor Mkp3 over-expression modified Tay accumulation. Second, Tay over-expression prevents the accumulation of dP-Erk, whereas loss of Tay has the converse effect. Finally, Tay over-expression modifies Erksem subcellular localization, increasing the nucleus/cytoplasm ratio of Erksem accumulation. In this regard, it is worth noting that the expression of RasV12 has the same effects on Erksem subcellular localization as the over-expression of Tay, as both Tay and RasV12 increase the nuclear/cytoplasm ratio of Erksem accumulation. We notice that the effects of Tay on Erk localization are only manifest when we used the Erksem form. Because we also see that Erksem is not retained in the cytoplasm by Mkp3, we reason that Erksem, liberated of cytoplasmic anchorage by Mkp3, is more sensitive to pathway activation and to the presence of other anchoring proteins, and that Tay might play this role in the nucleus.
We also observed a direct interaction between Tay and Mkp3. Mkp3 is a dual-specificity phosphatase that is predominantly localised in the cytoplasm, but it shuttles between the nucleus and cytoplasm and could play a role in translocating inactive Erk from the nucleus to the cytoplasm [8]. It is possible that Tay could promote the nuclear function of Mkp3, but in addition, Tay should also act independently of Mkp3 to promote Erk inactivation and retention, because Tay is able to down-regulate Erk activity in Mkp3 mutant backgrounds.
Most of the Tay interacting region with Erk is localised to the C-terminal part of Tay, a 1000 amino acid long region that includes the domain of homology between Tay and human AUTS2. This fragment of Tay fails to interact with Mkp3, and is even more efficient than the full-length protein in its effects on Erk subcellular localization and in its antagonism on Erk signalling. Intriguingly, AUTS2 expressed in the wing disc also interferes with EGFR signalling, but it does so in an opposite manner to Tay or to the Tay C-terminal domain. We cannot extract many conclusions from the consequences of AUTS2 expression in the wing disc, but speculate that this protein retains some of its interactions with Drosophila Erk that might protect this protein from inactivation by nuclear phosphatases. Similarly, the effects of AUTS2 on Drosophila EGFR signalling are compatible with a role for this protein in the regulation of Erk activity in humans, and that this effects might underline the effects of zebrafish, murine and human mutations in the onset of neurological disorders.
From the analysis in the wing disc we conclude that Tay interacts with Erk in the nucleus, affecting its phosphorylation and promoting its nuclear retention. In this context, it is interesting to note that the free diffusion of human ERK2 is impeded within the nucleus, and that this limitation in mobility increases after ERK2 stimulation [6]. This has lead to postulate that ERK2 retention in the nucleus involves high-affinity interactions with unidentified low-mobility sites that are constitutively expressed [6]. We suggest that Tay could play such a role in vivo, acting as a nuclear anchor for Erk that facilitates its inactivation by nuclear phosphatases and its retention in an inactive state.
Materials and Methods
Genetic strains
We used the Mkp3 allele Mkp3M76-R2b [58], and the deficiencies EP-866rev34, EP-866rev40 and Df(1)tay (see below). We used the following Gal4 lines: shv3kpn-Gal4 [62], 638-Gal4, nub-Gal4, salEPv-Gal4 [63], ap-Gal4, hh-Gal4, bs-Gal4, 1348-Gal4, dll-Gal4, eye-Gal4 and da-Gal4 [64]. We also used the UAS lines: UAS-RasV12 [65], UAS-Rafact [66], UAS-Erksem [67], UAS-Erk-HA [55], UAS-Erksem-HA [55], UAS-rhomboid [47], UAS-EGFR, UAS-EGFRDN [68] and UAS-GFP [69] and the P-GS lines EP-M76 and EP-866 [37]. We generated the following UAS lines: UAS-tay-i, UAS-tay.FL-Flag, UAS-tay.1-Flag, UAS-tay.2-Flag, UAS-hAUTS2-Flag, and UAS-Mkp3-Myc. We also used the RNA interference lines UAS-Mkp3-i (23911), UAS-tay-i (29021) and UAS-rolled-i (35641) from the VDRC Stock Center, and the lines UAS-EGFR-i (10079R-2) and UAS-ras-i (9375R-1) from NIG-Fly.
Generation of tay alleles
Df(1)tay: We used the insertions e03798 and d06351 [70], which are separated by 15 Kb of DNA including tay and part of CG16952. Flipase (FLP)-induced recombination was induced by a daily 1 h heat shock at 37°C to the progeny of e03798/d06351; hsFLP/+ females and FM7 males. Ten putative e03798-d06351/FM7 offspring females were individually crossed to FM7 males, and after 3 days, were used to extract genomic DNA to determinate by PCR the existence of FLP recombination. The position of the flanking insertions e03798 and d06351 and the extent of the tay deficiency are described in Suppl. Fig. S2A.
EP-866rev40 and EP-866rev34: We used Δ2–3 as a source of transposase to mobilize the EP-866 P-GS element. Males carrying both EP-866 and Δ2–3 were crossed with N55e11/FM7c females. The offspring EP-866 males with white phenotype were selected to make individual stocks. A complementation test was done to analyse the behaviour of these new alleles.
Molecular identification of the EP-866rev40 and EP-866rev34 deficiencies
Fifty wild type (control) and homozygous EP-866rev40 and EP-866rev34 embryos were used to extract genomic DNA to identify by PCR the genomic region excised by the mobilization of the P-GS. We used the following primers: 5′GCCGTGGAAATGGACTCTG3′ and 5′TTGCTGCTGCTGGTGAAAT3′. The size of the amplified fragments was 3629pb in wild type embryos, 2373pb in EP-866rev34 embryos and 932pb in EP-866rev40 embryos. The size of the generated deficiencies was confirmed by sequencing the PCR fragments sub-cloned in the pGEM-T-Easy vector (Promega) confirming an excision of 1276pb in EP-866rev34 and of 2717pb in EP-866rev40.
Generation of FLP recombination clones
Homozygous Df(1)tay, EP-866rev40 and EP-866rev34 clones were induced in larvae of the following genotypes: Df(1)tay f36a FRT18A/FRT18A UbiGFP; hsFLP/+; EP-866rev40 f36a FRT18A/FRT18A UbiGFP; hsFLP/+ and EP-866rev34 f36a FRT18A/FRT18A UbiGFP; hsFLP/+, respectively. Homozygous tay mutant cells were recognized in the adult wing by the cellular marker forked (f) and in the wing disc by the absence of GFP.
Generation of tay and Mkp3 constructs
UAS-tay-i
The EST LD22609 (DGRC) was used as a template to amplify a 700pb tay fragment using the following primers: 5′GCGCTCTAGAGCGGCAGCGATGGGCACAGTA3′ and 5′GCGCTCTAGAGCATGCGTAGCAGCAGGCGGCGGATAA3′. The amplified fragment was digested with the restriction enzyme XbaI (underlined sequence in the primers) and cloned into the pWIZ vector [71], previously digested with AvrII. The resulting plasmid was digested with NheI to clone the tay PCR fragment digested with XbaI. The orientation of both XbaI fragments cloned into pWIZ was checked to confirm an inverted position.
Tay-Flag constructs
In order to generate epitope tagged Tay proteins, the cDNA clone LD22609 was used as a template to amplify tay fragments using the following primers: Tay.FL 5′CACCATGGACACATCAAATGCCAGCGC3′ and 5′TCGACTGGGCGCCACCGATG3′; Tay.1 5′CACCATGGACACATCAAATGCCAGCGC3′ and 5′TGGAGGCAGGTATGACCCGTG3′ and Tay.2 5′CACCATGTCGCAGAATCAGCCAATGGTT3′ and 5′TCGACTGGGCGCCACCGATG3′. These PCR products were directionally subcloned into pENTR/D-TOPO (Invitrogen). For generating the C-terminal-Flag-tagged fusion proteins, we used the LR Clonase II reaction with Tay-pENTR/D-TOPO clones and the pTWF (3XFlag-tag at the C-terminal) vector for expression in vivo in Gal4-expressing cells following the instructions from Invitrogen.
Mkp3-Myc construct
The cDNA clone SD06439 (DGCR) was used as a template to amplify the Mkp3 cDNA using the following primers: 5′CACCATGCCAGAAACGGAGCACGAG3′ and 5′TTTAAGACCCGTGTCCGACGG3′. This PCR product was directionally cloned into pENTR/D-TOPO (Invitrogen). For generating the C-terminal-Myc-tagged fusion proteins, we used the LR Clonase II reaction with Mkp3-pENTR/D-TOPO and the pTWM (6XMyc-tag at the C-terminal) vector for expression in vivo in Gal4-expressing cells following the instructions from Invitrogen.
UAS-hAUTS2-Flag
The Full Length clone IRAKp961I02133Q (imaGenes) was used as a template to amplify the human AUTS2 cDNA using the following primers: 5′CACCATGGATGGCCCGACGCGGGGC3′ and 5′TCGGGCCTCGATATCCTTCAG3′. These PCR products were directionally cloned into pENTR/D-TOPO (Invitrogen). For generating the C-terminal-Flag-tagged fusion proteins, we used the LR Clonase II reaction with hAUTS2-pENTR/D-TOPO and the pTWF (3XFlag-tag at the C-terminal) vector for expression in vivo in Gal4-expressing cells following the instructions from Invitrogen.
Generation of Tay antiserum
Protein expression and purification
Fusion protein containing amino acids 1756–2049 of Tay was generated using LD22609 as template and the following primer pair: 5′GCGCGGATCCTCTGCAGAATCGCTTTTTCAG3′ and 5′GCGCATGAATTCCTCACACTGCGGTTCCAATATGACT3′ containing BamHI and EcoRI restriction sites respectively (underlined sequence). The amplified fragment was digested with the restriction enzymes BamHI and EcoRI, cloned in the BamHI-EcoRI site of the Gluthatione-S-Transferase (GST) gene fusion pGEX-2T (Promega) vector and transformed in E. coli BL21 (DE3). Selected clones were verified by sequencing.
Antibody generation
The GST-Tay1756–2049 protein was expressed in E. coli BL21 (DE3) and purified using Glutathione Sepharose 4B (GE Healthcare). The anti-Tay antibody was prepared by immunizing rats and guinea pigs with purified GST-Tay1756–2049 following conventional procedures.
Immunohistochemistry
We used the rabbit antibodies: anti-phospho-Histone3, anti-activated Cas3 and anti-diphosphorylated ERK1&2 (Cell Signalling). We also use the mouse monoclonal antibodies: anti-c-Myc 9E10 (Santa Cruz Biotechnology), anti-HA 12CA5 (Sigma), anti-FlagM2 (Sigma), anti-βGal (Promega), and anti-FasIII, anti-Dl and anti-Arm from the Hybridoma Bank at University of Iowa (Iowa City, IA). Alexa Fluor secondary antibodies (used at 1∶200 dilution) were from Invitrogen. To stain the nuclei we used To-Pro and to stain F-actin we used Alexa Fluor Phalloidin, from Invitrogen. Imaginal wing discs were dissected, fixed, and stained as described in [72]. Confocal images were taken in a LSM510 confocal microscope (Zeiss). In situ hybridization with the tay probe were carried out as described [72]. We used the cDNA LD22609 as template to synthesize the tay probe. The quantification of Erksem nuclear and cytoplasmic staining was carried out in Z-sections taken from 6 proximo-distal planes of 6 discs of each genotype along the length of the epithelium with the program ImageJ.
Pull-down assays
The fusion proteins Mkp3-GST and Erk-GST and the GST protein (negative control) were expressed in E. coli BL21 (DE3), using the constructs pGEX2TK-DMkp3 and pGEX4T1-DErk [73] and the vector pGEX2T, respectively, and were purified using Glutathione Sepharose 4B (GE Healthcare). The complete Tay protein was generated from the cDNA LD22609 using the TNT T7 Coupled Reticulocyte Lysate System (Promega) and radiolabeled with S35-Met. The pull-down assay was performed incubating over-night at 4°C the same amount of GST or GST fusion proteins bound to Glutathione Sepharose4B with in vitro translated Tay. After centrifugation and washes the proteins were resolved by 6% SDS/PAGE and the existence of pull-down proteins was analysed by autoradiography. The pulldown experiments were repeated five times with the same results.
Western blot and immunoprecipitation
Embryos lysate
Embryos were collected 24 hours after egg laying, washed in a 0.7% NaCl + 0.04% Triton X-100 solution, de-corionated in 50% bleach and frozen at −80°C. Embryos were homogenised in immunoprecipitation (IP) buffer (40 mM Tris-HCl, 200 mM NaCl, 1% NP-40, 0.1% SDS, 0.5%DOC, 6 mM EDTA, 6 mM EGTA, 100 µM PMSF and complete EDTA-free protease inhibitor cocktail tablet (Roche)) and incubated 2 h at 4°C. Lysates were clarified by centrifugation. Protein concentration in these lysates was determined using the Lowry-Peterson protocol.
Immunoprecipitation
50 µg and 2 mg of total protein from each lysate were used to assess protein expression levels (input) and for the immunoprecipitation reactions, respectively. The immunoprecipitation reactions were performed by incubating the lysates with 1 mg/ml BSA, 10% Glycerol and specific antibodies for Mkp3-Myc (15 µl of anti-Myc agarose, Santa Cruz Biotechnology), for Erksem-HA (15 µl of anti-HA agarose, Santa Cruz Biotechnology) and for Tay.FL-Flag and Tay.2-Flag (15 µl of anti-FlagM2 agarose, Sigma) at 4°C for 12–16 h. Erksem is a mutated form of Erk that shows less sensitivity to Mkp3 [50].
Immunoblotting
Embryo lysates or immunoprecipitated complexes were resolved by 7% SDS-PAGE and proteins were transferred to nitrocellulose membranes using a semi-dry blotting apparatus (BioRad). Erksem and Mkp3 proteins were detected by incubating with anti-HA (HA 12CA5, Hybridome Bank) and anti-Myc (c-Myc 9E10, Santa Cruz Biotechnology) monoclonal antibodies, respectively. Tay proteins were detected by incubating with anti-Tay guinea pig serum. Immunoblots were developed using IR680 and 800 labelled antibodies (Li-Cor) with the Odyssey Infrared Imaging System (Li-Cor). The immunoprecipitation experiments for Tay.FL and Mkp3 were repeated 7 times, for Tay.FL and Erksem 5 times, for Tay.2 and Mkp3 5 times and for Tay.2 and Erksem 4 times.
Supporting Information
Zdroje
1. ShiloBZ (2003) Signaling by the Drosophila epidermal growth factor receptor pathway during development. Exp Cell Res 284 : 140–149.
2. ShaulYD, SegerR (2007) The MEK/ERK cascade: from signaling specificity to diverse functions. Biochimica et Biophysica Acta 1773 : 1213–1226.
3. AvrahamR, YardenY (2011) Feedback regulation of EGFR signalling: decision making by early and delayed loops. Nat Rev Mol Cell Biol 12 : 104–117.
4. BiggsWH, ZavitzKH, DicksonB, van der StratenA, BrunnerD, et al. (1994) The Drosophila rolled locus encodes a MAP kinase required in the sevenless signal transduction pathway. EMBO J 13 : 1628–1635.
5. DoroquezDB, RebayI (2006) Signal integration during development: mechanisms of EGFR and Notch pathway function and cross-talk. Critical Reviews in Biochemistry and Molecular Biology 41 : 339–385.
6. CostaM, MarchiM, CardarelliF, RoyA, BeltramF, et al. (2006) Dynamic regulation of ERK2 nuclear translocation and mobility in living cells. Journal of Cell Science 119 : 4952–4963.
7. von KriegsheimA, BaiocchiA, BirtwistleM, SumptonD, BienvenutW, et al. (2009) Cell fate decisions are specified by the dynamic ERK interactome. Nature cell biology 11 : 1458–1464.
8. CauntC, ArmstrongS, RiversC, NormanM, McArdleC (2008) Spatiotemporal regulation of ERK2 by dual specificity Phosphatases. JOURNAL OF BIOLOGICAL CHEMISTRY 283 : 26612–26623.
9. ZehoraiE, YaoZ, PlotnikovA, SegerR (2010) The subcellular localization of MEK and ERK—A novel nuclear translocation signal (NTS) paves a way to the nucleus. Molecular and Cellular Endocrinology 314 : 213–220.
10. KolchW (2005) Coordinating ERK/MAPK signaling through scaffolds and inhibitors. Nature Reviews Mol Cel BIol 6 : 827–837.
11. TeisD, WunderlichW, HuberLA (2002) Localization of the MP1-MAPK scaffold complex to endosomes is mediated by p14 and required for signal transduction. Dev Cell 3 : 803–814.
12. TherrienM, ChangHC, SolomonNM, KarimFD, WassarmanDA, et al. (1995) KSR, a novel protein kinase required for RAS signal transduction. Cell 83 : 879–888.
13. TherrienM, MichaudNR, RubinGM, MorrisonDK (1996) KSR modulates signal propagation within the MAPK cascade. Genes Dev 10 : 2684–2695.
14. KornfeldK, HomDB, HorvitzHR (1995) The ksr-1 gene encodes a novel protein kinase involved in Ras-mediated signaling in C. elegans. Cell 83 : 903–913.
15. SchaefferHJ, CatlingAD, EblenST, CollierLS, KraussA, et al. (1998) MP1: a MEK binding partner that enhances enzymatic activation of the MAP kinase cascade. Science 281 : 1668–1671.
16. NguyenA, BurackWR, StockJL, KortumR, ChaikaO, et al. (2002) Kinase Suppressor of Ras (KSR) ss a scaffold which facilitates mitogen-activated protein kinase activation in vivo. Mol Cel Biol 22 : 3035–3045.
17. LuttrellLM, RoudabushFL, ChoyEW, MillerWE, FieldME, et al. (2001) Activation and targeting of extracellular signal-regulated kinases by β-arrestin scaffolds. PNAS 98 : 2449–2454.
18. ToriiS, KusakabeM, YamamotoT, MaekawaM, NishidaE (2004) Sef is a spatial regulator for Ras/MAP kinase signaling. Dev Cell 7 : 33–44.
19. BhattacharyyaR, RemenyiA, YehB, LimW (2006) Domains, motifs, and scaffolds: the role of modular interactions in the evolution and wiring of cell signaling circuits. Annu Rev Biochem 75 : 655–680.
20. PerrimonN (1994) Signalling pathways initiated by receptor protein tyrosine kinases in Drosophila. Curr Opin Cell Biol 6 : 260–266.
21. PerrimonN, McMahonAP (1999) Negative feedback mechanisms and their roles during pattern formation. Cell 97 : 13–16.
22. FriedmanA, PerrimonN (2006) High-throughput approaches to dissecting MAPK signaling pathways. Methods 40 : 262–271.
23. KarimFD, ChangHC, TherrienM, WassarmanDA, LavertyT, et al. (1996) A Screen for Genes That Function Downstream of Ras1 During Drosophila Eye Development. Genetics 143 : 315–329.
24. HuangAM, RubinGM (2000) A Misexpression Screen Identifies Genes That Can Modulate RAS1 Pathway Signaling in Drosophila melanogaster. Genetics 156 : 1219–1230.
25. ButcharJ, CainD, ManivannanS, McCueA, BonannoL, et al. (2012) New negative feedback regulators of EGFR signaling in Drosophila. Genetics 191 : 1213–1226.
26. WangSH, SimcoxA, CampbellG (2000) Dual role for Drosophila epidermal growth factor receptor signalling in early wing disc development. Genes Dev 14 : 2271–2276.
27. SotillosS, de CelisJF (2005) Interactions between the Notch, EGFR, and decapentaplegic signaling pathways regulate vein differentiation during Drosophila pupal wing development. Dev Dyn 232 : 738–752.
28. Diaz-BenjumeaF, Garcia-BellidoA (1990) Behavior of cells mutant for an EGF receptor homologue of Drosophila in genetics mosaics. Proc Roy Soc Lond Biol Sci 36–44.
29. KubotaK, GotoS, EtoK, HayashiS (2000) EGF receptor attenuates Dpp signaling and helps to distinguish the wing and leg cell fates in Drosophila. Development 127 : 3769–3776.
30. PallaviSK, ShashidharaLS (2003) Egfr/Ras pathway mediates interactions between peripodial and disc proper cells in Drosophila wing discs. Development 130 : 4931–4941.
31. ZeccaM, StruhlG (2002) Control of growth and patterning of the Drosophila wing imaginal disc by EGFR-mediated signaling. Development 129 : 1369–1376.
32. ZeccaM, StruhlG (2002) Subdivision of the Drosophila wing imaginal disc by EGFR-mediated signaling. Development 129 : 1357–1368.
33. CuliJ, Martin-BlancoE, ModolellJ (2001) The EGF receptor and N signalling pathways act antagonistically in Drosophila mesothorax bristle patterning. Development 128 : 299–308.
34. GabayL, SegerR, ShiloB-Z (1997) In Situ Activation Pattern of Drosophila EGF Receptor Pathway During Development. Science 277 : 1103–1106.
35. BierE (2000) Drawing lines in the Drosophila wing: initiation of wing vein development. Curr Opin Genet Dev 10 : 393–398.
36. de CelisJF (2003) Pattern formation in the Drosophila wing: the development of the veins. BioEssays 25 : 443–451.
37. MolnarC, Lopez-VareaA, HernandezR, de CelisJF (2006) A gain-of-function screen identifying genes required for vein formation in the Drosophila melanogaster wing. Genetics 174 : 1635–1659.
38. PoeckB, TriphanT, NeuserK, StraussR (2008) Locomotor control by the central complex in Drosophila—An analysis of the tay bridge mutant. Developmental Neurobiology 68 : 1046–1058.
39. SchumannG, CoinbL, LourdusamyaA, CharoenP, et al. (2011) Genome-wide association and genetic functional studies identify autism susceptibility candidate 2 gene (AUTS2) in the regulation of alcohol consumption . PNAS 108 : 7119–7124.
40. SultanaR, YuCE, YuJ, MunsonJ, ChenD, et al. (2002) Identification of a novel gene on chromosome 7q11.2 interrupted by a translocation breakpoint in a pair of autistic twins. Genomics 80 : 129–134.
41. BedogniF, HodgeR, NelsonB, FrederickE, ShibaN, et al. (2010) Autism susceptibility candidate 2 (Auts2) encodes a nuclear protein expressed in developing brain regions implicated in autism neuropathology. Gene Expr Patterns 10 : 1255–1266.
42. KalscheuerVM, FitzPatrickD, TommerupN, BuggeM, NiebuhrE, et al. (2007) Mutations in autism susceptibility candidate 2 (AUTS2) in patients with mental retardation. Hum Genet 121 : 501–509.
43. BeundersG, VoorhoeveE, GolzioC, PardoLM, RosenfeldJA, et al. (2013) Exonic deletions in AUTS2 cause a syndromic form of intellectual disability and suggest a critical role for the C terminus. American journal of human genetics 92 : 210–220.
44. EliaJ, GaiX, XieHM, PerinJC, GeigerE, et al. (2010) Rare structural variants found in attention-deficit hyperactivity disorder are preferentially associated with neurodevelopmental genes. Mol Psychiatry 15 : 637–646.
45. OksenbergN, StevisonL, WallJD, AhituvN (2013) Function and Regulation of AUTS2, a Gene Implicated in Autism and Human Evolution. PLOS Genetics 9: e1003221.
46. SturtevantMA, RoarkM, BierE (1993) The Drosophila rhomboid gene mediates the localized formation of wing veins and interacts genetically with components of the EGF-R signaling pathway. Genes Dev 7 : 961–973.
47. de CelisJF, BrayS, Garcia-BellidoA (1997) Notch signalling regulates veinlet expression and establishes boundaries between veins and interveins in the Drosophila wing. Development 124 : 1919–1928.
48. GolemboM, SchweitzerR, FreemanM, ShiloBZ (1996) argos transcription is induced by the Drosophila EGF receptor pathway to form an inhibitory feedback loop. Development 122 : 223–230.
49. KarlssonM, MathersJ, DickinsonRJ, MandlM, KeyseSM (2004) Both nuclear-cytoplasmic shuttling of the dual specificity phosphatase MKP-3 and its ability to anchor MAP Kinase in the cytoplasm are mediated by a conserved nuclear export signal. The Journal of Biological Chemistry 279 : 41882–41891.
50. BottCM, ThorneycroftSG, MarshallCJ (1994) The sevenmaker gain-of-function mutation in p42 MAP kinase leads to enhanced signalling and reduced sensitivity to dual specificity phosphatase action. FEBS Lett 352 : 201–205.
51. ChuY, SolskiPA, Khosravi-FarR, DerCJ, KellyK (1996) The mitogen-activated protein kinase phosphatases PAC1, MKP-1, and MKP-2 have unique substrate specificities and reduced activity in vivo toward the ERK2 sevenmaker mutation. J Biol Chem 271 : 6497–6501.
52. RouxPP, BlenisJ (2004) ERK and p38 MAPK-activated protein kinases: a family of protein kinases with diverse biological functions. Microbiol Mol Biol Rev 68 : 320–344.
53. ChuderlandD, SegerR (2005) Protein–Protein interactions in the regulation of the extracellular signal regulated kinase. Molecular Biothechnology 29 : 57–74.
54. BurackWR, ShawAS (2000) Signal transduction: hanging on a scaffold. Curr Opin Cell Biol 12 : 211–216.
55. KimM, LeeJ, KohH, LeeSH, JangC, et al. (2006) Inhibition of ERK-MAP kinase signaling by RSK during Drosophila development. EMBO J 25 : 3056–3067.
56. Molnar C, Resnik-Docampo M, Organista MF, Martín M, Hevia CF, et al.. (2011) Signalling Pathways in Development and Human Disease: A Drosophila Wing Perspective. In: Plaseska-Karanfilska DD, editor. Human Genetic Diseases. Rijeka: InTech.
57. GalindoMI, BishopSA, GreigS, CousoJP (2002) Leg patterning driven by proximal-distal interactions and EGFR signaling. Science 297 : 256–259.
58. Ruiz-GómezA, López-VareaA, MolnarC, de la Calle-MustienesE, Ruiz-GómezM, et al. (2005) Conserved cross-interactions between Ras/MAPK signalling and the dual-specificity phosphatase MKP3. Dev Dyn 232 : 695–708.
59. Martin-BlancoE, RochF, NollE, BaonzaA, DuffyJB, et al. (1999) A temporal switch in DER signaling controls the specification and differentiation of veins and interveins in the Drosophila wing. Development 126 : 5739–5747.
60. AndoR, MizunoH, MiyawakiA (2004) Regulated fast nucleocytoplasmic shuttling observed by reversible protein highlighting. Science 306 : 1370–1373.
61. VolmatV, CampsM, ArkinstallS, PouyssegurJ, LenormandP (2001) The nucleus, a site for signal termination by sequestration and inactivation of p42/p44 MAP kinases. Journal of Cell Science 114 : 3433–3443.
62. SotillosS, de CelisJF (2006) Regulation of decapentaplegic expression during Drosophila wing veins pupal development. Mechanisms of development 123 : 241–251.
63. CruzC, GlavicA, CasadoM, de CelisJF (2009) A gain of function screen identifying genes required for growth and pattern formation of the Drosophila melanogaster wing. Genetics 183 : 1005–10026.
64. CallejaM, MorenoE, PelazS, MorataG (1996) Visualization of gene expression in living adult Drosophila. Science 274 : 252–255.
65. LeeT, FeigL, MontellDJ (1996) Two distinct roles for Ras in a developmentally regulated cell migration. Development 122 : 409–418.
66. BrandAH, PerrimonN (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118 : 401–415.
67. BrunnerD, OellersN, SzabadJ, BiggsWH, ZipurskySL, et al. (1994) A gain-of-function mutation in Drosophila MAP kinase activates multiple receptor tyrosine kinase signalling pathaways. Cell 875–888.
68. BuffE, CarmenaA, GisselbrechtS, JimenezF, MichelsonAM (1998) Signalling by the Drosophila epidermal growth factor receptor is required for the specification and diversification of embryonic muscle progenitors. Development 125 : 2075–2086.
69. ItoK, AwanoW, SuzukiK, HiromiY, YamamotoD (1997) The Drosophila mushroom body is a quadruple structure of clonal units each of which contains a virtually identical set of neurones and glial cells. Development 124 : 761–771.
70. ThibaultST, SingerMA, MiyazakiWY, MilashB, DompeNA, et al. (2004) A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat Genet 36 : 283–287.
71. LeeYS, CarthewRW (2003) Making a better RNAi vector for Drosophila: use of intron spacers. Methods 30 : 322–329.
72. de CelisJF (1997) Expression and function of decapentaplegic and thick veins in the differentiation of the veins in the Drosophila wing. Development 124 : 1007–1018.
73. KimSH, KwonHB, KimYS, RyuJH, KimKS, et al. (2002) Isolation and characterization of a Drosophila homologue of mitogen-activated protein kinase phosphatase-3 which has a high substrate specificity towards extracellular-signal-regulated kinase. Biochem J 361 : 143–151.
Štítky
Genetika Reprodukční medicína
Článek Interaction between and during Mammalian Jaw Patterning and in the Pathogenesis of SyngnathiaČlánek Clustering of Tissue-Specific Sub-TADs Accompanies the Regulation of Genes in Developing LimbsČlánek Transcription Factor Occupancy Can Mediate Active Turnover of DNA Methylation at Regulatory Regions
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 12
-
Všechny články tohoto čísla
- Stressing the Importance of CHOP in Liver Cancer
- The AmAZI1ng Roles of Centriolar Satellites during Development
- Flies Get a Head Start on Meiosis
- Recommendations from Jane Gitschier's Bookshelf
- And Baby Makes Three: Genomic Imprinting in Plant Embryos
- Bugs in Transition: The Dynamic World of in Insects
- Defining the Role of ATP Hydrolysis in Mitotic Segregation of Bacterial Plasmids
- Synaptonemal Complex Components Promote Centromere Pairing in Pre-meiotic Germ Cells
- Cohesinopathies of a Feather Flock Together
- Genetic Recombination Is Targeted towards Gene Promoter Regions in Dogs
- Parathyroid-Specific Deletion of Unravels a Novel Calcineurin-Dependent FGF23 Signaling Pathway That Regulates PTH Secretion
- MAN1B1 Deficiency: An Unexpected CDG-II
- Phosphate Flow between Hybrid Histidine Kinases CheA and CheS Controls Cyst Formation
- Basolateral Mg Extrusion via CNNM4 Mediates Transcellular Mg Transport across Epithelia: A Mouse Model
- Truncation of Unsilences Paternal and Ameliorates Behavioral Defects in the Angelman Syndrome Mouse Model
- Autozygome Sequencing Expands the Horizon of Human Knockout Research and Provides Novel Insights into Human Phenotypic Variation
- Huntington's Disease Induced Cardiac Amyloidosis Is Reversed by Modulating Protein Folding and Oxidative Stress Pathways in the Heart
- Low Frequency Variants, Collapsed Based on Biological Knowledge, Uncover Complexity of Population Stratification in 1000 Genomes Project Data
- Targeted Ablation of and in Retinal Progenitor Cells Mimics Leber Congenital Amaurosis
- Genomic Imprinting in the Embryo Is Partly Regulated by PRC2
- Binary Cell Fate Decisions and Fate Transformation in the Larval Eye
- The Stress-Regulated Transcription Factor CHOP Promotes Hepatic Inflammatory Gene Expression, Fibrosis, and Oncogenesis
- A Global RNAi Screen Identifies a Key Role of Ceramide Phosphoethanolamine for Glial Ensheathment of Axons
- Functional Analysis of the Interdependence between DNA Uptake Sequence and Its Cognate ComP Receptor during Natural Transformation in Species
- Cross-Modulation of Homeostatic Responses to Temperature, Oxygen and Carbon Dioxide in
- Alcohol-Induced Histone Acetylation Reveals a Gene Network Involved in Alcohol Tolerance
- Molecular Characterization of Host-Specific Biofilm Formation in a Vertebrate Gut Symbiont
- CRIS—A Novel cAMP-Binding Protein Controlling Spermiogenesis and the Development of Flagellar Bending
- Dual Regulation of the Mitotic Exit Network (MEN) by PP2A-Cdc55 Phosphatase
- Expanding the Marine Virosphere Using Metagenomics
- Detection of Slipped-DNAs at the Trinucleotide Repeats of the Myotonic Dystrophy Type I Disease Locus in Patient Tissues
- Interaction between and during Mammalian Jaw Patterning and in the Pathogenesis of Syngnathia
- Mutations in the UQCC1-Interacting Protein, UQCC2, Cause Human Complex III Deficiency Associated with Perturbed Cytochrome Protein Expression
- Reactivation of Chromosomally Integrated Human Herpesvirus-6 by Telomeric Circle Formation
- Anoxia-Reoxygenation Regulates Mitochondrial Dynamics through the Hypoxia Response Pathway, SKN-1/Nrf, and Stomatin-Like Protein STL-1/SLP-2
- The Midline Protein Regulates Axon Guidance by Blocking the Reiteration of Neuroblast Rows within the Drosophila Ventral Nerve Cord
- Tomato Yield Heterosis Is Triggered by a Dosage Sensitivity of the Florigen Pathway That Fine-Tunes Shoot Architecture
- Selection on Plant Male Function Genes Identifies Candidates for Reproductive Isolation of Yellow Monkeyflowers
- Role of Tomato Lipoxygenase D in Wound-Induced Jasmonate Biosynthesis and Plant Immunity to Insect Herbivores
- Meiotic Cohesin SMC1β Provides Prophase I Centromeric Cohesion and Is Required for Multiple Synapsis-Associated Functions
- Identification of Sphingolipid Metabolites That Induce Obesity via Misregulation of Appetite, Caloric Intake and Fat Storage in
- Genome-Wide Screen Reveals Replication Pathway for Quasi-Palindrome Fragility Dependent on Homologous Recombination
- Histone Methylation Restrains the Expression of Subtype-Specific Genes during Terminal Neuronal Differentiation in
- A Novel Intergenic ETnII-β Insertion Mutation Causes Multiple Malformations in Mice
- The NuRD Chromatin-Remodeling Enzyme CHD4 Promotes Embryonic Vascular Integrity by Transcriptionally Regulating Extracellular Matrix Proteolysis
- A Domesticated Transposase Interacts with Heterochromatin and Catalyzes Reproducible DNA Elimination in
- Acute Versus Chronic Loss of Mammalian Results in Distinct Ciliary Phenotypes
- MBD3 Localizes at Promoters, Gene Bodies and Enhancers of Active Genes
- Positive and Negative Regulation of Gli Activity by Kif7 in the Zebrafish Embryo
- A Hereditary Spastic Paraplegia Mouse Model Supports a Role of ZFYVE26/SPASTIZIN for the Endolysosomal System
- The CCR4-NOT Complex Mediates Deadenylation and Degradation of Stem Cell mRNAs and Promotes Planarian Stem Cell Differentiation
- Reconstructing Native American Migrations from Whole-Genome and Whole-Exome Data
- Contributions of Protein-Coding and Regulatory Change to Adaptive Molecular Evolution in Murid Rodents
- Comprehensive Analysis of Transcriptome Variation Uncovers Known and Novel Driver Events in T-Cell Acute Lymphoblastic Leukemia
- A -Acting Protein Effect Causes Severe Eye Malformation in the Mouse
- Clustering of Tissue-Specific Sub-TADs Accompanies the Regulation of Genes in Developing Limbs
- Germline Progenitors Escape the Widespread Phenomenon of Homolog Pairing during Development
- Transcription Factor Occupancy Can Mediate Active Turnover of DNA Methylation at Regulatory Regions
- Somatic mtDNA Mutation Spectra in the Aging Human Putamen
- ESCRT-I Mediates FLS2 Endosomal Sorting and Plant Immunity
- Ethylene Promotes Hypocotyl Growth and HY5 Degradation by Enhancing the Movement of COP1 to the Nucleus in the Light
- The PAF Complex and Prf1/Rtf1 Delineate Distinct Cdk9-Dependent Pathways Regulating Transcription Elongation in Fission Yeast
- Dual Regulation of Gene Expression Mediated by Extended MAPK Activation and Salicylic Acid Contributes to Robust Innate Immunity in
- Quantifying Missing Heritability at Known GWAS Loci
- Smc5/6-Mms21 Prevents and Eliminates Inappropriate Recombination Intermediates in Meiosis
- Smc5/6 Coordinates Formation and Resolution of Joint Molecules with Chromosome Morphology to Ensure Meiotic Divisions
- Tay Bridge Is a Negative Regulator of EGFR Signalling and Interacts with Erk and Mkp3 in the Wing
- Meiotic Crossover Control by Concerted Action of Rad51-Dmc1 in Homolog Template Bias and Robust Homeostatic Regulation
- Active Transport and Diffusion Barriers Restrict Joubert Syndrome-Associated ARL13B/ARL-13 to an Inv-like Ciliary Membrane Subdomain
- An Regulatory Circuit Modulates /Wnt Signaling and Determines the Size of the Midbrain Dopaminergic Progenitor Pool
- Variants Induce Differential Protection to Viruses in : A Phenotypic and Phylogenomic Analysis
- Base Pairing Interaction between 5′- and 3′-UTRs Controls mRNA Translation in
- Evidence That Masking of Synapsis Imperfections Counterbalances Quality Control to Promote Efficient Meiosis
- Insulin/IGF-Regulated Size Scaling of Neuroendocrine Cells Expressing the bHLH Transcription Factor in
- Sumoylated NHR-25/NR5A Regulates Cell Fate during Vulval Development
- TATN-1 Mutations Reveal a Novel Role for Tyrosine as a Metabolic Signal That Influences Developmental Decisions and Longevity in
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The NuRD Chromatin-Remodeling Enzyme CHD4 Promotes Embryonic Vascular Integrity by Transcriptionally Regulating Extracellular Matrix Proteolysis
- Mutations in the UQCC1-Interacting Protein, UQCC2, Cause Human Complex III Deficiency Associated with Perturbed Cytochrome Protein Expression
- The Midline Protein Regulates Axon Guidance by Blocking the Reiteration of Neuroblast Rows within the Drosophila Ventral Nerve Cord
- Tomato Yield Heterosis Is Triggered by a Dosage Sensitivity of the Florigen Pathway That Fine-Tunes Shoot Architecture
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



