-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaPositive and Negative Regulation of Gli Activity by Kif7 in the Zebrafish Embryo
Loss of function mutations of Kif7, the vertebrate orthologue of the Drosophila Hh pathway component Costal2, cause defects in the limbs and neural tubes of mice, attributable to ectopic expression of Hh target genes. While this implies a functional conservation of Cos2 and Kif7 between flies and vertebrates, the association of Kif7 with the primary cilium, an organelle absent from most Drosophila cells, suggests their mechanisms of action may have diverged. Here, using mutant alleles induced by Zinc Finger Nuclease-mediated targeted mutagenesis, we show that in zebrafish, Kif7 acts principally to suppress the activity of the Gli1 transcription factor. Notably, we find that endogenous Kif7 protein accumulates not only in the primary cilium, as previously observed in mammalian cells, but also in cytoplasmic puncta that disperse in response to Hh pathway activation. Moreover, we show that Drosophila Costal2 can substitute for Kif7, suggesting a conserved mode of action of the two proteins. We show that Kif7 interacts with both Gli1 and Gli2a and suggest that it functions to sequester Gli proteins in the cytoplasm, in a manner analogous to the regulation of Ci by Cos2 in Drosophila. We also show that zebrafish Kif7 potentiates Gli2a activity by promoting its dissociation from the Suppressor of Fused (Sufu) protein and present evidence that it mediates a Smo dependent modification of the full length form of Gli2a. Surprisingly, the function of Kif7 in the zebrafish embryo appears restricted principally to mesodermal derivatives, its inactivation having little effect on neural tube patterning, even when Sufu protein levels are depleted. Remarkably, zebrafish lacking all Kif7 function are viable, in contrast to the peri-natal lethality of mouse kif7 mutants but similar to some Acrocallosal or Joubert syndrome patients who are homozygous for loss of function KIF7 alleles.
Published in the journal: . PLoS Genet 9(12): e32767. doi:10.1371/journal.pgen.1003955
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003955Summary
Loss of function mutations of Kif7, the vertebrate orthologue of the Drosophila Hh pathway component Costal2, cause defects in the limbs and neural tubes of mice, attributable to ectopic expression of Hh target genes. While this implies a functional conservation of Cos2 and Kif7 between flies and vertebrates, the association of Kif7 with the primary cilium, an organelle absent from most Drosophila cells, suggests their mechanisms of action may have diverged. Here, using mutant alleles induced by Zinc Finger Nuclease-mediated targeted mutagenesis, we show that in zebrafish, Kif7 acts principally to suppress the activity of the Gli1 transcription factor. Notably, we find that endogenous Kif7 protein accumulates not only in the primary cilium, as previously observed in mammalian cells, but also in cytoplasmic puncta that disperse in response to Hh pathway activation. Moreover, we show that Drosophila Costal2 can substitute for Kif7, suggesting a conserved mode of action of the two proteins. We show that Kif7 interacts with both Gli1 and Gli2a and suggest that it functions to sequester Gli proteins in the cytoplasm, in a manner analogous to the regulation of Ci by Cos2 in Drosophila. We also show that zebrafish Kif7 potentiates Gli2a activity by promoting its dissociation from the Suppressor of Fused (Sufu) protein and present evidence that it mediates a Smo dependent modification of the full length form of Gli2a. Surprisingly, the function of Kif7 in the zebrafish embryo appears restricted principally to mesodermal derivatives, its inactivation having little effect on neural tube patterning, even when Sufu protein levels are depleted. Remarkably, zebrafish lacking all Kif7 function are viable, in contrast to the peri-natal lethality of mouse kif7 mutants but similar to some Acrocallosal or Joubert syndrome patients who are homozygous for loss of function KIF7 alleles.
Introduction
Hedgehog (Hh) proteins play a fundamental role in animal development, controlling cell type specification, proliferation and survival in a variety of contexts through a signaling pathway, the core components of which are shared across species [reviewed in 1], [2], [3]. Hh signaling also functions post-embryonically, regulating tissue homeostasis [4], metabolism [5] and physiological processes [6], while aberrant pathway activity underlies the etiology of a variety of cancers [7], [8].
The kinesin family protein Costal2 (Cos2) is a central component of the intracellular Hedgehog signaling complex in Drosophila [9]. Cos2 physically interacts with the Gli family protein Cubitus interruptus (Ci), restraining the transcriptional activating activity of its full length form both by anchoring it in the cytoplasm as well as by recruiting the serine-threonine kinases that prime it for processing into a truncated transcriptional repressor [10], [11], [12]. Consistent with these effects, loss of Cos2 activity results in the ectopic activation of Hh target genes, both in embryos and imaginal discs [13], [14]. In addition to its negative regulatory role, Cos2 has also been shown to potentiate Hh pathway activity by promoting the dissociation of Ci from Suppressor of Fused (Sufu) [15], another negative pathway regulator that acts to inhibit nuclear import of Ci [10], [16].
The closest vertebrate homologue of Cos2 is Kif7 [17], mouse mutants of which similarly exhibit evidence both of gain and loss of Hh pathway activity [18], [19], [20], implying a conservation of Cos2/Kif7 function between insects and vertebrates. Like Cos2, Kif7 promotes the processing of Gli transcription factors to their repressor forms both in mouse and human cells [18], [19], [20], [21] a function that, in part, can explain the Hh gain of function phenotypes in the limbs and in the neural tube of kif7 mutants. One significant difference between Kif7 and Cos2 however, is the association of the former with the Primary Cilium, an organelle that is absent from most Drosophila cells but of central importance for Hh signaling in vertebrates [22], [23]. Tagged forms of Kif7 have been shown to localize to the primary cilium tip in response to Hh pathway activation when expressed in cultured mammalian cells [19], [20]. Similarly, translocation of Gli proteins to the primary cilium tip is also induced by Hh signaling [24], a process that is proposed to be required for their activation through dissociation from Suppressor of Fused (Sufu) [25], [26]. Paradoxically, given its role as a negative regulator of the pathway, loss of Kif7 function abrogates the Hh induced translocation of Gli to the primary cilium tip [19]. Whilst such an effect can be reconciled with the partial attenuation of pathway activation observed in the neural tube of kif7 mutant embryos [18], [19], [20], how the localization of Kif7 to the primary cilium relates to its repressive function remains unclear.
The first evidence of a conserved function for Kif7 in vertebrate Hh signaling was based on morpholino mediated transient knock-down experiments in zebrafish, [17]. Morphant kif7embryos exhibit rather subtle defects in cell fate specification principally in the myotome that contrasts with the more robust de-repression of the Hh response seen in Drosophila cos2 mutants. While this could reflect a divergence in Kif7 function between species, it might also be attributable to the transient nature of the morpholino mediated knock-down. To explore the role of zebrafish Kif7 further, we have generated loss of function alleles by zinc finger nuclease (ZFN) mediated targeted mutagenesis [27], [28], [29] and used these to dissect the role of Kif7 in modulating the activity of the Gli transcription factors. In addition, using an antibody raised against the zebrafish protein, we have analyzed the levels and sub-cellular distribution of endogenous Kif7 in the presence and absence of Hh pathway activation. Our findings imply a previously unrecognized role for Kif7 in sequestering Gli1 in the cytoplasm, a function analogous to that of Drosophila Cos2, and suggest that Kif7 functions in the primary cilium principally to potentiate Gli2 activity.
Results
Generation of mutant alleles of zebrafish kif7 using zinc finger nucleases
To analyze the function of zebrafish Kif7, we generated stable germ line transmissible mutant alleles of kif7 using zinc finger nuclease (ZFN) mediated targeted mutagenesis [27], [28]. A budding yeast-based system identified sequences in the kif7coding region that are potentially amenable to targeted mutagenesis. Targeting this sequence in the 3rd coding exon had the potential to create mutations in a region conserved with the mammalian Kif7 protein at cds nt729–765 (A729TCCAAATTCCATTTTGTGGACCTGGCAGGATCAGAG765) (Fig. 1A). Embryos injected with capped RNA encoding a ZFN pair selected for recognition of this sequence were found to carry a variety of deletions or insertions at the target site (data not shown) confirming the efficacy of this approach. Accordingly, we grew up adults from injected embryos and screened their progeny for transmission of kif7 lesions. Three individuals transmitting deletion mutations were recovered and two alleles that cause frame-shifts resulting in premature termination codons (Table 1) predicted to yield proteins truncated in the motor domain (Fig. 1B) were selected for further analysis.
Fig. 1. Targeted mutation of the zebrafish kif7 gene. 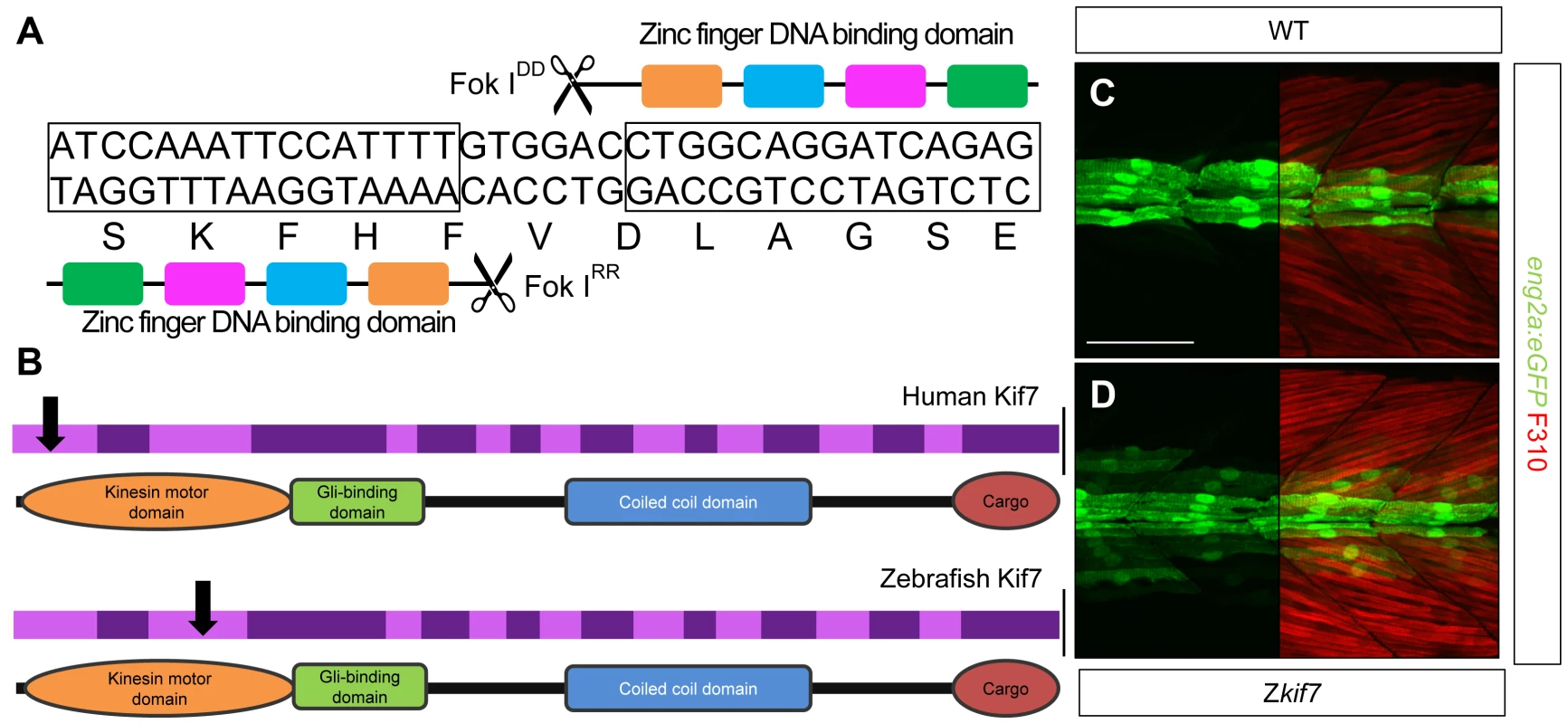
(A) Schematic representation of the nucleotide sequence in exon 3 of the zebrafish kif7 gene targeted by the Zinc finger nucleases. (B) Schematic representation of the human and zebrafish Kif7 coding sequences showing conserved exonic structure (dark shading) corresponding to different protein domains (drawn to scale). The black arrows indicate the approximate position of homozygous viable mutations found in some human patients and of the induced lesions in the zebrafish gene. (C,D) Expression of eng2a:gfp reporter gene in muscle fibers in the tail somites of wild-type (C) and kif7 homozygous (D) embryos at 2.5 dpf. Low level ectopic expression of the reporter is detected in fibers surrounding the muscle pioneers in the kif7 mutants. Merged images showing fast twitch muscle fibers stained with mAb F310 (red) are shown in the right-hand panels. Scale bar: 50 µm. Tab. 1. Alleles and translational products of zebrafish <i>kif7</i> mutants generated with CompoZ ZFN. 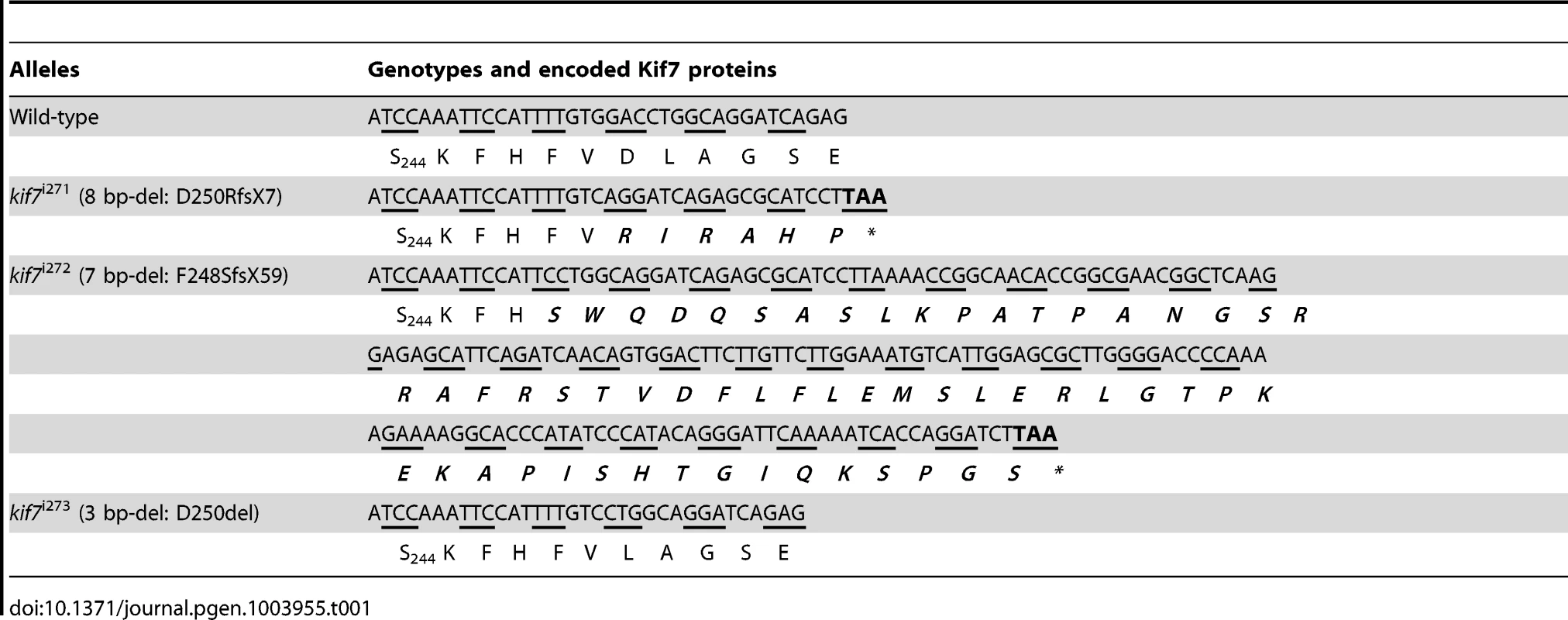
Loss of zygotic kif7 function causes a mild Hh pathway de-repression phenotype
Animals homozygous or trans-heterozygous for the kif7 mutant alleles predicted to encode truncated proteins, completed embryogenesis and showed no defects in the specification of slow-twitch muscle fibres, muscle pioneers (MPs) or medial fast fibres (MFFs) as visualized by Prox1 and Eng2a expression respectively (data not shown), sensitive read-outs of Hh activity in the zebrafish embryo [30]; nor did they display any other manifestations of aberrant Hh pathway activity at 24 hpf. By 2.5 dpf, however, some mutant larvae exhibited ectopic expression of the eng2a:eGFP reporter gene in the myotome (Fig. 1C,D). Nevertheless, the homozygous and trans-heterozygous fish were fully viable and grew into phenotypically normal adults. This contrasts with the peri-natal lethality of mice homozygous for kif7 loss of function alleles [18], [19], [20], but mirrors the finding that some Joubert syndrome patients are homozygous for mutated KIF7 alleles that cause severe truncation of the protein ([31]; see Fig. 1B).
Loss of zygotic and maternally derived Kif7 activity causes strong de-repression of Hh signaling in mesodermal derivatives
We surmised that the lack of disruption of Hh pathway activity in the absence of zygotic kif7 function could be due to maternally derived kif7 mRNA present in newly fertilized eggs. To test this inference, we crossed kif7 homozygous females to kif7 homozygous males. The resulting maternal and zygotic (MZ) kif7 mutant embryos were devoid of full length Kif7 protein detectable by Western blot analysis (Fig. 2E) and exhibited a significant expansion of eng2a:eGFP reporter gene expression in the myotome at 30 hpf (Fig. 2A,B). Consistent with the de-repression of the Hh pathway implied by this effect, a reporter gene for ptch2, a direct target of the Hh pathway, was ectopically expressed throughout the myotomal compartment of the somites (Fig. 2F–I). MZkif7 embryos also showed an increase in the number of Prox1+ve slow-twitch muscle cells, the specification of which is Hh–dependent (Fig. 2 C,D; see also Fig. 7I); however, the ectopic expression of Eng, revealed both by the eng2a:GFP reporter (Fig. 2B) and by 4D9 mAb staining (Fig. 2D) was restricted to fast-twitch muscle fibers, indicating that loss of Kif7 activity is not sufficient for the maximal pathway activation required for MP induction [30]. Inhibition of Smo activity in MZkif7 embryos (via mutation or treatment with the Smo antagonist cyclopamine) had little effect on the ectopic expression of ptch2 (data not shown) or Eng (Fig. 3C) consistent with Kif7 acting downstream of Smo to suppress transcriptional activation of Hh targets by the Gli transcription factors in the myotome. In both instances, however, there was a loss of MPs, indicating that removal of Kif7 is not sufficient for maximal pathway activation in the absence of Smo (Fig. 3B,C). Hh signaling is also required for definitive haematopoiesis in the zebrafish embryo [32] and inhibition of Smo activity by cyclopamine treatment blocks the formation of haematopoietic stem cells (HSCs) as revealed by the loss of c-myb expression in the dorsal aorta (Fig. 3E). Complete elimination of Kif7 activity reversed this effect (Fig. 3F) indicating that Kif7 also acts in the non-myogenic mesoderm to modulate Hh pathway activity.
Fig. 2. Absence of Kif7 protein and de-repression of Hh target genes in MZkif7 embryos. 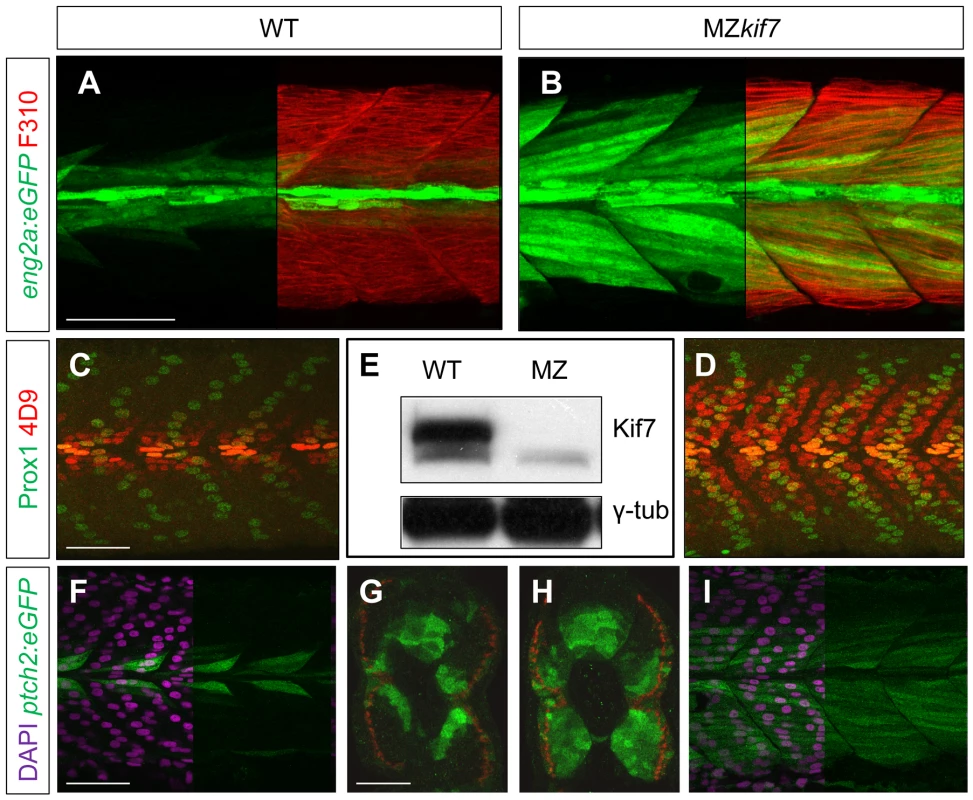
(A,B) Lateral images of 30 hpf wild-type (A) and MZkif7 mutant (B) embryos expressing eng2a:eGFP (green); note dramatic expansion of eng2a:eGFP expression within the fast-twitch fibers revealed by F310 staining (red) in the merged image (right panel). (C,D) Parasagittal optical sections of wild-type (C) and MZkif7 mutant (D) 30 hpf embryos stained with anti-Prox1 (green) and mAb4D9 (red) (E) Western blot of wild-type (WT) and MZkif7 mutant (MZ) embryo extracts probed with polyclonal rabbit anti-Kif7 antiserum showing complete loss of Kif7 protein (upper band) from MZkif7 embryos. Loading control: γ-tubulin (γ-tub). (F,I) Parasagittal and (G,H) transverse optical sections of 30 hpf wild-type (F,G) and MZkif7 (H,I) embryos expressing a ptch2:eGFP transgene (green) showing the expansion of the ptch2 expression domain in the myotome and neural tube in the absence of Kif7. Nuclei are revealed in left half (of panels F,I) by DAPI staining (purple). The edge of the myotome is shown by mAbF9 (in G,H) marking the superficial slow-twitch muscle fibers. Scale bar: 50 µm. Fig. 3. Kif7 acts downstream of Smo to control Hh target genes. 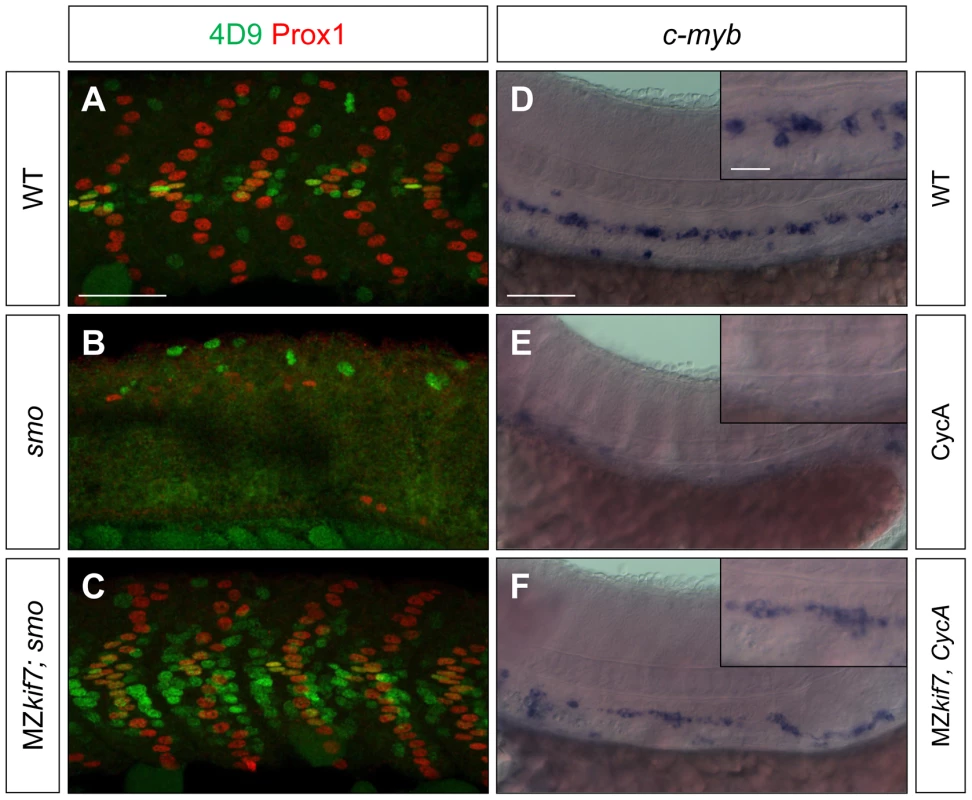
(A,B,C) Parasagittal optical sections of 30 hpf wild-type (A), smo mutant (B) and smo;MZkif7 (C) double mutant embryos stained with mAb4D9 (green) and Prox1 (red). Scale bar 50 µm. (D,E,F) Lateral view at the level of the yolk extension of 36 hpf wild-type (D), cyclopamine exposed (E) and MZkif7; cyclopamine (CycA) exposed (F) hybridized with a probe for c-myb RNA, marking hematopoietic stem cells in the ventral floor of the dorsal aorta. Scale bar 50 µm (detail in insets; scale bar 10 µm). Fig. 4. Loss of Sufu further enhances Gli activity in MZkif7 mutants. 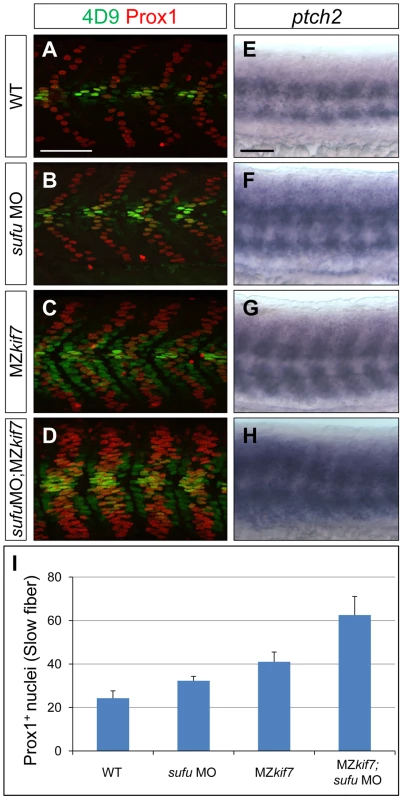
(A–D) Parasagittal optical sections of 30 hpf WT (A,B) or MZkif7(C,D) embryos stained with anti-Prox1 (red) and 4D9 (green) antibodies; morpholino mediated knock-down of sufu results in significant increase in Prox1 expressing slow twitch muscle cells in WT (B, quantified in I) and a further increase in MZkif7(D) mutants; Scale bar: 50 µm. (E–H) Lateral view of 30 hpf WT (A) or MZkif7(C) embryos hybridized with an antisense probe for ptch2 mRNA; MZkif7 embryo shows an expansion of ptch2 expression; embryos of similar stage and genotype injected with sufu MO result in an enhancement of the levels of ptch2 staining in WT (F) and in MZkif7 (H) embryos; Scale bar: 50 µm. (I) Quantification of Prox1+ slow twitch muscle fiber nuclei per hemisegment in WT, sufu MO injected, MZkif7 and MZkif7 injected with sufu MO. Error bars represent standard deviation obtained from 16–20 hemisegments from >6 embryos. Note the significant increase in MZkif7 compared to WT and a further enhancement in slow fibers by removal of Sufu in MZkif7 mutants. Loss of Kif7 has previously been reported to disrupt left-right (L-R) asymmetry reflecting a presumed role in ciliogensis in Kupfer's vesicle [33]. We analyzed the establishment of L-R asymmetry in MZkif7 embryos using lefty2 expression [34] as an assay and found no example of situs inversus (Fig. S1). We conclude that the reported defect is likely an off-target effect of the morpholino.
Gli2a processing is delayed in the absence of Kif7
Cos2/Kif7 has been implicated in the control of Gli protein processing both in flies and mammals; in the case of Drosophila, at least, this is mediated through the recruitment of the kinases that prime the Gli–family protein, Ci, for proteasomal cleavage [12]. Previously we showed that Gli2a is processed in a Hh dependent manner in zebrafish embryos, the ratio of full length (FL) to the truncated repressor (R) form of Gli2a being markedly increased in ptch1;ptch2 double mutant embryos and substantially decreased in embryos treated with cyclopamine [35]. We investigated the role of Kif7 in Gli2a processing using Western blot analysis to compare the levels of FL and R forms in MZkif7 embryos with those in wild-type embryos in which the Hh pathway was fully activated (by injection of Shh mRNA) or fully repressed (by exposure to cyclopamine). At 22 hpf, whereas the FL∶R ratio was significantly increased in the Shh mRNA injected embryos and reduced in embryos exposed to cyclopamine, the ratio did not differ significantly between MZkif7 and wild-type embryos, although the levels of FL protein appeared less sensitive to cyclopamine treatment in the MZkif7 embryos (Fig. 4A,B). In younger (18 hpf) embryos, by contrast, we observed a consistent increase in the FL∶R ratio in MZkif7 embryos, though not as great as in Shh mRNA injected embryos (Fig. 4A,C). At this stage, the FL form of Gli2a appears as a doublet, the slower mobility form of which is sensitive to cyclopamine treatment and therefore presumably Smo dependent. Whereas in Shh mRNA injected embryos, it is this putative Smo dependent form that accumulates at the expense of the truncated R form, in MZkif7 it is the cyclopamine resistant form that accumulates (see Fig. 4A). This suggests that Kif7 is required for a Smo-dependent modification of full length Gli2a as well as for its efficient cleavage to the R form. Notably, the increased FL∶R ratio in MZkif7 embryos is effectively suppressed by cyclopamine treatment (Fig. 4A,C), suggesting that Kif7 may potentiate Gli2a cleavage by opposing Smo activity.
Fig. 5. Regulation of Gli processing and activity by Kif7. 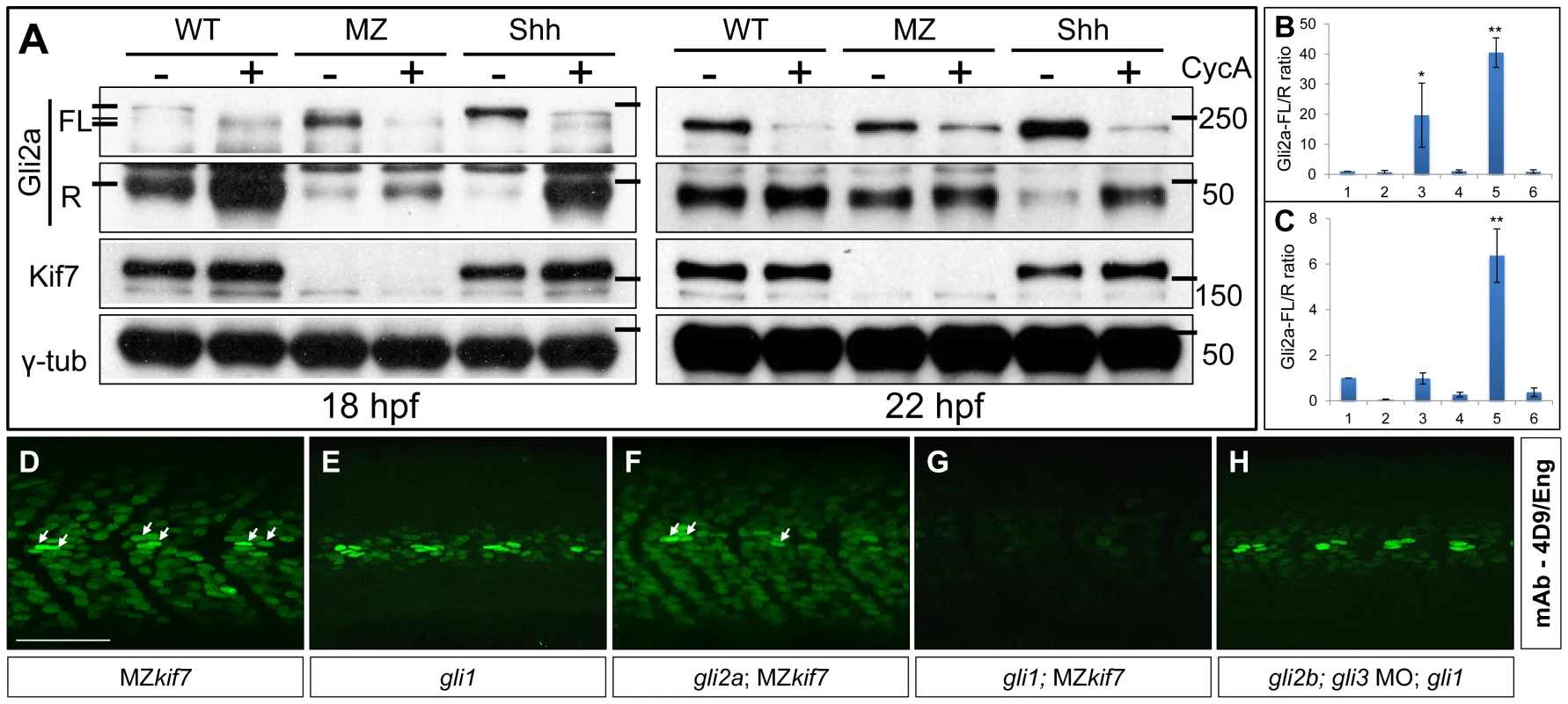
(A) Western blot analysis showing Gli2a processing in wild-type (WT), MZkif7 (MZ) and Shh mRNA injected (Shh) embryos at 18 hpf (left panel) and at 22 hpf (right panel) compared to the same set treated with cyclopamine (CycA). Full-length (FL) and repressor (R) forms of Gli2a are indicated. The lower panels shows the same blot re-probed with Kif7 and γ-tubulin (loading control) antibodies (B,C) Quantification of the Gli2a FL∶R ratio normalized to WT at 18 hpf (B) and 22 hpf (C). Bar graphs numbered 1–6 represent lanes on the blots in (A) from left to right. Error bars represent standard deviation obtained from three independent Western blots including those shown in (A). Single asterisk: P<0.02; double asterisk: P<0.001. (D–H) Parasagittal optical sections of 30 hpf embryos of different genotypes showing expression of Eng-expressing muscle cells as revealed by mAb4D9 (green). The MZkif7 phenotype (D) is largely unaffected by removal of Gli2a (F), other than the loss of the some MP cells (arrows); by contrast, Eng expression is nearly eliminated in the MZkif7;gli1 double mutant embryo (G), despite Gli1 being dispensable for Eng expression in the presence of Kif7 function (E). Knockdown of gli2b and gli3 activity using morpholinos in gli1 mutants (H) has no effect on Eng expression in the myotome. Scale bar: 50 µm. Kif7 functions principally to restrain Gli1 activity
Previous studies have shown that Gli1 and Gli2a act redundantly to mediate Hh activity in the myotome [30]; embryos homozygous for loss of function alleles of gli1 (also known as detour (dtr): [36]) or gli2a [37] show normal specification of Hh-dependent muscle cell types, whereas in the absence of both genes, all Hh-dependent muscle cell types fail to form [30], [37]. By contrast, simultaneous morpholino mediated knockdown of gli3 and of the gli2a paralogue gli2b had no discernible effect on Eng expression either in wild-type or gli1 mutant embryos (data not shown and Fig. 4G). It follows that Gli1 and Gli2a are the principal mediators of Hh signaling in the myotome and that either protein can respond to Hh activity to elicit the full range of cellular responses to the signal. To investigate the role of Kif7 in regulating Gli activity, we generated MZkif7 embryos homozygous for either gli1 or gli2a loss of function mutations. MZkif7;gli2a double mutants exhibited little modification of the ectopic Eng expression seen in MZkif7 mutants alone, except for a slight reduction in the number of MP cells in each somite (Fig. 4E). By contrast, in MZkif7; gli1 double mutants, Eng+ve cells were barely discernible in the myotome (Fig. 4F). These findings imply that Kif7 acts principally to restrain Gli1 activity.
Endogenous Kif7 interacts with Gli1 protein in the developing embryo
The genetic data imply that Kif7 may physically interact with Gli1. Since no antibody specific for zebrafish Gli1 is available, we tested this inference by expressing a GFP tagged form of Gli1 in wild-type embryos. Injection of mRNA encoding a similarly modified form of Gli1 tagged with mCherry resulted in ectopic activation of a ptch2:eGFP reporter gene (Fig. 5A,B), confirming that the tag does not disrupt the function of the protein. Immunoprecipitation of the eGFP-Gli1 from injected wild-type embryos revealed an interaction with both Kif7 and the negative Hh pathway regulator Sufu (Fig. 5C). The levels of Kif7 pulled down with eGFP-Gli1 were increased in embryos treated with cyclopamine (Fig. 5D), implying that the interaction is sensitive to Smo activation. Consistent with this, co-injection of Shh mRNA with the eGFP-Gli1 mRNA resulted in a decrease in the levels of Kif7 relative to Gli1 (Fig. 5D). A similar trend was also seen for the interaction between Sufu and Gli1 (Fig. 5E).
Fig. 6. Functional tagged Gli1 associates with Kif7 and Sufu in a Hh dependent manner. 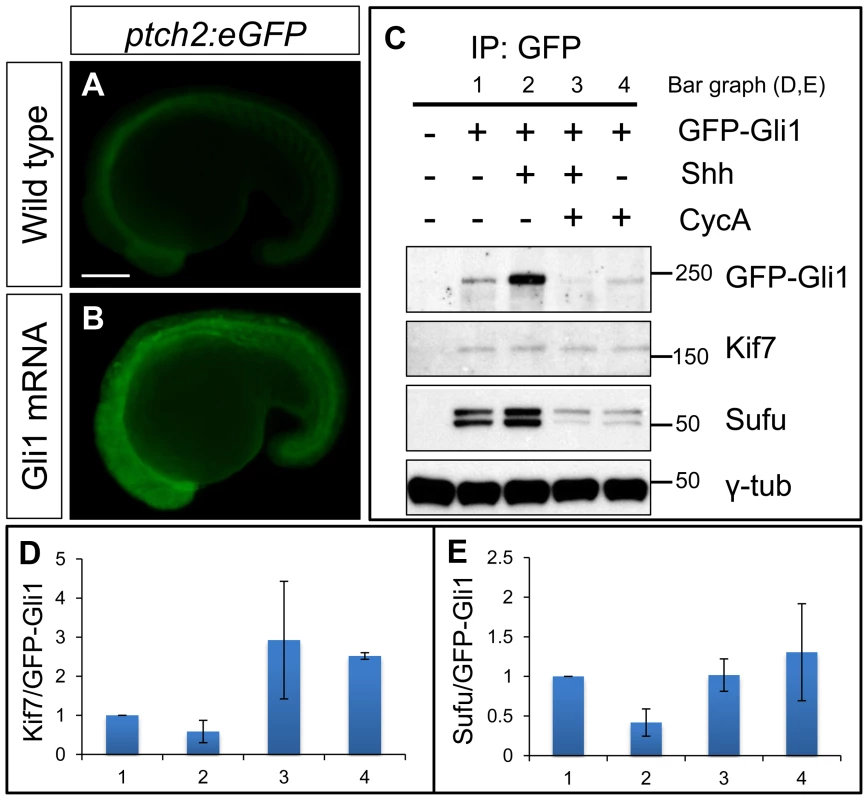
(A,B) Lateral views of 20ss ptc2:eGFP embryos uninjected (A) and injected with mCherry-Gli1 mRNA (B) showing the ectopic activation of ptch2:eGFP reporter. Scale bar: 150 µm. (C) Western blot analysis of anti-GFP immune-precipitates from uninjected embryos or embryos injected with a combination of GFP-Gli1 and/or Shh mRNA and/or exposed to cyclopamine (CycA); note the stabilization of GFP-Gli1 in the presence of ectopic Shh and a reduced association between Kif7 and Gli1 (see D for quantification); inhibition of Hh pathway activity by cyclopamine reverses this effect and further enhances the association. The inhibitory association of Sufu and Gli1 is also reduced in response to pathway activation by Shh mRNA injection (see E for quantification). (D) The ratio of Kif7:GFP-Gli1 from experiments described in panel (C); WT (1), Shh mRNA injected (2), CycA exposed and Shh mRNA injected (3), and CycA exposed embryos; showing reduced Kif7-Gli1 association upon pathway activation and increased association when the pathway is inhibited. Error bars represent standard deviation obtained from three independent biological replicates. (E) The ratio of Sufu:GFP-Gli1 from experiments described in panel (C); WT (1), Shh mRNA injected (2), CycA exposed and Shh mRNA injected (3), and CycA exposed embryos; showing reduced Sufu-Gli1 association upon pathway activation and restoration of this association upon pathway inhibition. Error bars represent standard deviation obtained from three independent biological replicates. Kif7 potentiates Gli2a activity by promoting its dissociation from Sufu
In Drosophila, Cos2 exerts a positive effect on Ci activity by promoting its dissociation from Sufu [15] and in mammalian cells dissociation of Gli3 from Sufu has been shown to be of key importance for maximal pathway activation [25], [26]. We immunoprecipitated Gli2a from wild-type and MZkif7 embryos and used Western blotting to analyze its interaction with Sufu and Kif7. As expected, both Sufu and Kif7 proteins co-precipitated with Gli2a from wild-type embryos, whereas only Sufu could be detected in the immunoprecipitates from MZkif7 embryos (Fig. 6A). Notably, the levels of Sufu were increased approximately 2-fold relative to wild-type in the MZkif7 embryos, consistent with a decrease in dissociation of the two proteins in the absence of Kif7 function. Similarly, the levels of Gli2a that co-immunoprecipitated with Sufu from MZkif7 embryos were increased (2-fold) relative to wild-type controls (Fig. 6B); by contrast, activation of the pathway upstream of Kif7 through Shh mRNA injection, had no significant effect on the levels of Gli2a pulled down with Sufu, whilst a modest increase was seen in embryos treated with cyclopamine (Fig. 6B).
Fig. 7. Kif7 modulates Gli2a localization and its association with Sufu. 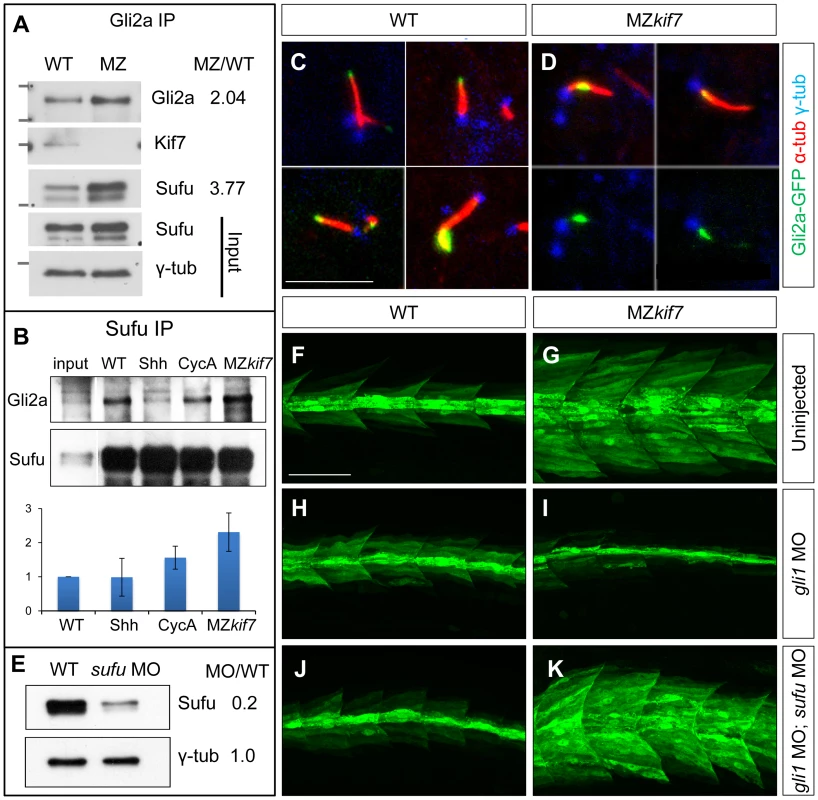
(A) Western blot analysis of anti-Gli2a immune-precipitates of WT and MZkif7 (MZ) embryos, showing association of Kif7 with Gli2a in wild-type embryos and increased association of Sufu with Gli2a in MZkif7 embryos, as indicated by the ratio of the normalized intensities of the MZ to WT signals. (B) Western blot analysis of anti-Sufu immuno-precipitates of WT, Shh RNA injected (Shh), cyclopamine (cycA) exposed and MZkif7 (MZ) embryos; the Gli2aFL:Sufu ratios normalized to wild-type are shown below each lane. Error bars represent standard deviation obtained from three independent biological replicates. Note the increase in levels of Gli2aFL that co-precipitates with Sufu in MZkif7 embryos. (C,D) Localization of a functional eGFP tagged Gli2a protein (green) to primary cilia in wild-type (C) and MZkif7 (D) embryos. The axonemes of the primary cilia are marked by acetylated α-tubulin (red) and the basal bodies by γ-tubulin (blue) staining. In wild-type, Gli2a localizes to the tip of the cilia whereas in MZkif7, localization is restricted to the base of the cilia; the lower two panels in (D) are the same images as in the upper panels but with the red channel removed to show the juxtaposed GFP-Gli2a and γ-tubulin signals more clearly. Scale bar: 5 µm. (E) Western blot of wild-type (WT) and sufu morpholino-injected (sufu MO) embryo extracts probed with anti-Sufu and anti γ-tubulin, showing significant depletion of Sufu levels in the morphants relative to wild-type (MO/WT). (F–K) Parasagittal optical sections of 30 hpf wild-type (WT) or MZkif7 embryos showing the effect on eng2a:eGFP expression of morpholino mediated knockdown of gli1 (H,I) or gli1 and sufu (J,K). Depletion of gli1 in WT embryos (H) has no discernible effect, whereas it causes a drastic suppression of the ectopic expression in MZkif7 (I). This suppression is abrogated by simultaneous removal of Sufu and Gli1 from MZkif7 embryos (K). Scale bar: 50 µm. Dissociation of the Gli proteins from Sufu has been postulated to occur at the tip of the primary cilium [26]; in line with our findings, we found that the Gli2a-GFP fusion protein, which localizes to the tip of the primary cilium in wild-type embryos in response to Hh activity [38], was consistently located close to the base of the axoneme of primary cilia of myotomal cells in MZkif7 mutant embryos (Fig. 6C,D,S2).
Together, these data suggest a role for Kif7 in promoting the activation of full length Gli2a through its transport to the tip of primary cilium and dissociation from the Sufu protein. To test this inference, we used two previously characterized antisense morpholino oligonucleotides [30], [36] to reduce Sufu and Gli1 activity simultaneously in MZkif7 mutant embryos. Such embryos showed an 80% reduction in Sufu protein levels (Fig. 6E) and a significant restoration of the MZkif7 phenotype, with ectopic eng2a:eGFP expression throughout the fast-twitch fibers (cf. Fig. 6I & K). It follows that Kif7 not only potentiates Gli2a activity by promoting its dissociation from Sufu but also restrains it in a manner independent of its processing. Notably, knock-down of Sufu in MZkif7 mutant embryos resulted in a substantial increase in the number of slow twitch fibers and MP cells, implying a greater degree of Hh pathway de-repression than occurs in embryos lacking only Sufu or Kif7 (Fig. 7A–D,I). Consistent with this phenotype, these embryos showed increased expansion of the ptch2 expression domain (Fig. 7E–H). This implies that Sufu also attenuates Gli1 activity and that Kif7 acts in concert with Sufu to restrain Gli1 activity in the myotome.
Differing requirements for Kif7 in Hh pathway activity in the neurectoderm and the mesoderm
Shh plays a major role in patterning the neural tube and the de-repression of the pathway caused by ptch mutations results in the ectopic expression of Hh target genes both in mouse [39], [40] and fish embryos [41], [42](Fig. 8C,G,K). In mouse embryos homozygous for loss of function kif7 alleles, there is a modest expansion of the expression domains of ptch1 and of the ventral neural tube markers nkx2.2 and olig2 [18], [19], [20] consistent with a partial de-repression of Hh pathway activity. In zebrafish MZkif7 embryos, by contrast, no changes in the neural tube expression domains of fkd4, nkx2.2a or olig2 could be detected (Fig. 8B,F,J). Notably, however, we found that olig2 expression could be partially uncoupled from its dependence on Smo activity by complete elimination of Kif7 activity (Fig. 8M,N). Taken together, these findings imply that the role of Kif7 in regulating Hh pathway activity is dispensable in the neural tube in zebrafish.
Fig. 8. Hh target gene regulation in the neural tube is largely independent of Kif7 and Sufu function. 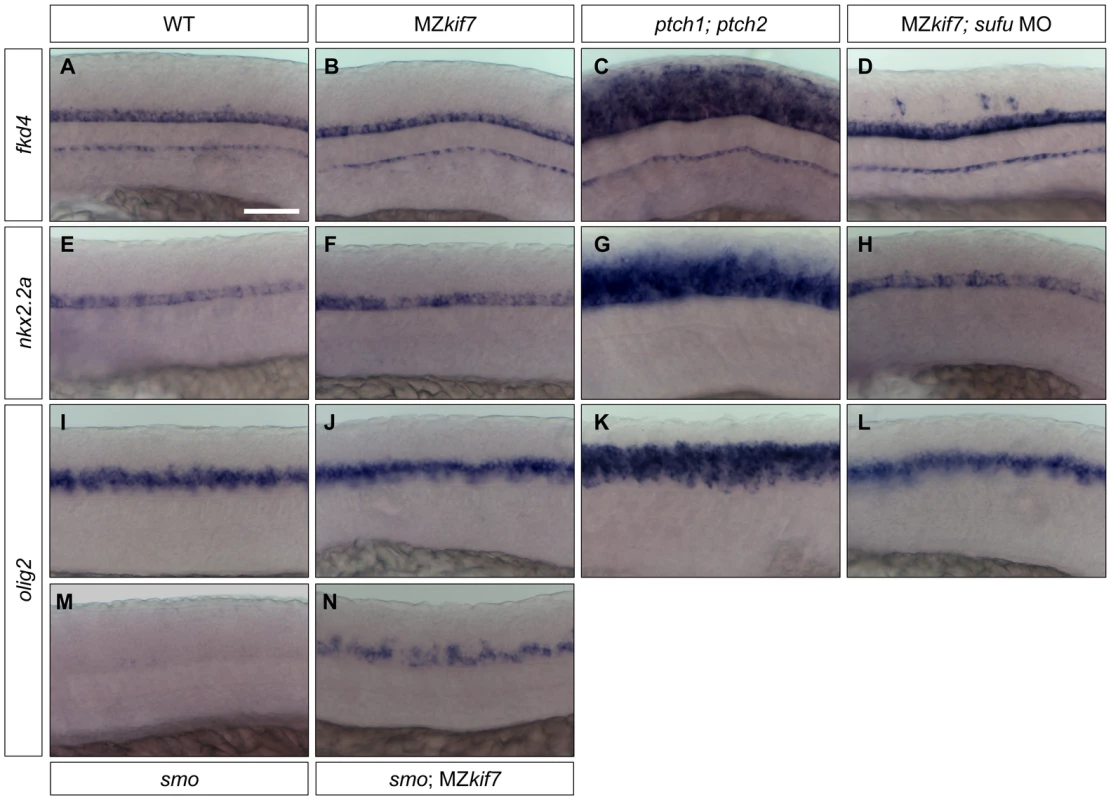
(A–L) Lateral view at the level of yolk extension of 30 hpf WT (A,E,I), MZkif7 (B,F,J), ptch1;ptch2 double mutants (C,G,K) and sufu MO injected MZkif7 mutants (D,H,L) hybridized with antisense probes for fkd4 (A–D), nkx2.2a (E–H) or olig2 (I–L); Note the ventrally restricted domains of fkd4, olig2 and nkx2.2a typical of WT embryos are dramatically expanded in ptch1;ptch2 double mutant embryos (C,G,K) but are unaffected in the absence of Kif7 function. sufu MO injected MZkif7 embryos show wild-type patterns of nkx2.2a or olig2 but ectopically expression fkd4 in scattered cells. (M,N) expression of olig2 is completely lost from the neural tube in smo mutant embryos (M) but is partially restored by removal of kif7 function (N). Scale bar: 50 µm. Given that Sufu knock down significantly enhances the MZkif7 phenotype in the myotome, we investigated whether Sufu activity might account for the lack of pathway de-repression in the neural tube in MZkif7 mutants. Surprisingly, knock down of Sufu function in MZkif7 embryos had no effect on the expression domains of either nkx2.2a or olig2 (Fig. 8H,L); however, ectopic expression of fkd was consistently observed in the occasional cell scattered throughout the neural tube (Fig. 8D).
Drosophila Cos2 can substitute for Kif7 in the zebrafish embryo
Sequence comparison has revealed significant structural conservation between Drosophila Cos2 and zebrafish Kif7 [17]. To investigate the extent to which function is also conserved, we exploited the ability to produce pure populations of MZkif7 embryos to assay the rescuing activity of in vitro synthesized mRNA encoding eGFP tagged forms of Cos2 and Kif7. Consistent with previous reports of mRNA mediated rescue of the kif7 morphant phenotype [21], injection of zebrafish kif7 mRNA into MZkif7 embryos resulted in substantial suppression of the ectopic eng2a:eGFP expression (Fig. 9A–D,G and data not shown). Remarkably, a significant degree of rescue was also seen following injection of Drosophila cos2 mRNA (Fig. 9E,F). Confocal imaging of injected embryos revealed that the exogenous Kif7 protein localized to the tips of primary cilia (Fig. 9H,J) as previously reported [35]. By contrast, the tagged Cos2 protein was found exclusively in the cytoplasm, with no evidence of localization to the primary cilia (Fig. 9I,K). Immunoprecipitation of Gli2a from embryos injected with Cos2 mRNA revealed an interaction between the two proteins (Fig. 9L), consistent with the notion that Cos2 restrains the activity of Gli2a as well as Gli1 by retaining it in the cytoplasm.
Fig. 9. Drosophila Cos2 rescues Kif7 function and binds endogenous Gli2a. 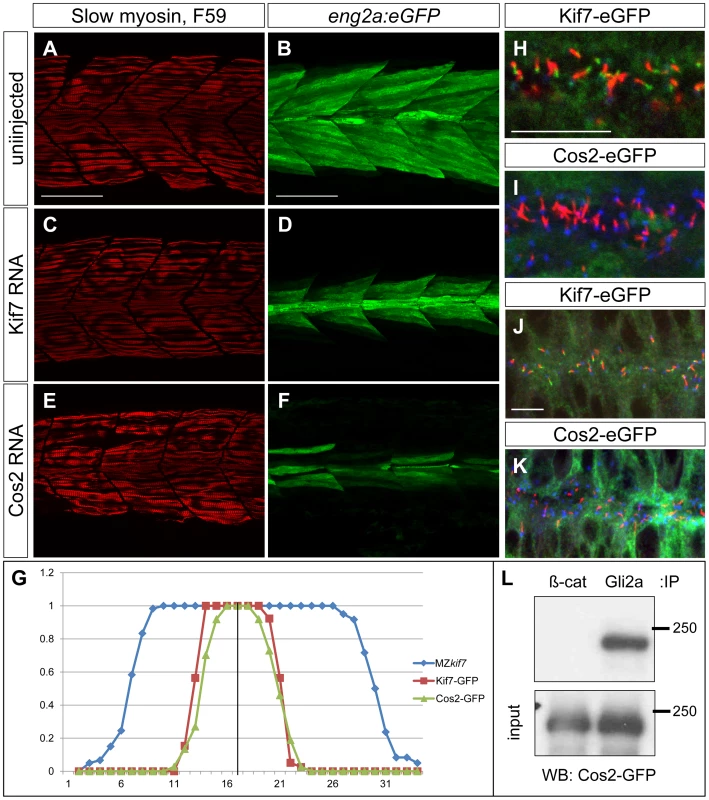
(A–F) Parasagittal optical sections of 30 hpf MZkif7;eng2a:eGFP embryos showing slow twitch muscle fibers revealed by mAbF59 in red (A,C,E) and eng2a expression in green (B,D,F); control (A,B) embryo exhibits the typical expansion of GFP expression which is suppressed in embryos injected with zebrafish kif7 mRNA (C,D) or Drosophila cos2 mRNA (E,F). Scale bar: 50 µm. (G) Quantification of the eng2a:eGFP expression at 30 hpf in MZkif7 and in MZkif7 injected with mRNA encoding zebrafish Kif7 or Drosophila Cos2. Individual fibers (MFFs) were counted at the level of the yolk extension in 33–36 hemisegments from >10 embryos for each sample. Optical sections showing localization of zebrafish Kif7-eGFP (H,J) or Drosophila Cos2-GFP (I,K) in the otic vesicle (H,I) or neural tube (J,K) of 20ss embryo injected with mRNA encoding the tagged proteins: eGFP (green), α-acetylated tubulin (red) and γ-tubulin (red in H,J and blue in I,K). Scale bar 10 µm. (L) Western blot analysis of anti-β-catenin or anti-Gli2a immune precipitates from 20ss embryos injected with Cos2-eGFP RNA probed with anti-GFP revealing co-precipitation of the tagged Cos2 with endogenous Gli2a. The stability and subcellular localization of Kif7 is regulated by Hh activity
Using a rabbit polyclonal antibody raised against the C-terminal domain of zebrafish Kif7 (see Materials and Methods) we analyzed the sub-cellular distribution of the endogenous protein in fixed embryos. In wild-type embryos, Kif7 was found to accumulate in large puncta within the cytoplasm of cells in the neural tube, otic vesicle and somites; in addition, we found that it localized to the primary cilia of some cells, especially in the otic vesicle (Fig. 10A,D,G and data not shown). A similar distribution was observed in smo mutant embryos (Fig. 10B,E,H). By contrast, we found that the cytoplasmic puncta are completely absent in ptch1;ptch2 double mutants, the protein localizing exclusively to the tips of primary cilia (Fig. 10C,F,I). We used spot segmentation and brightness measurement to quantify Kif7 protein levels in the otic vesicles (see Materials & Methods); this revealed an approximately two fold reduction in fluorescence intensity in ptch1;ptch2 double mutants compared to smo mutants or wild-type siblings (Fig. 10J) suggesting that Hh pathway activity may induce increased turnover and/or dispersion of the Kif7 protein. Consistent with the former possibility, Western blot analysis of total extracts from embryos injected with Shh mRNA showed a similar two fold reduction in Kif7 levels relative to wild-type, an effect that could be reversed by exposure of the injected embryos to cyclopamine (Fig. 10K,L).
Fig. 10. Kif7 protein localization is modulated by Hh pathway activity. 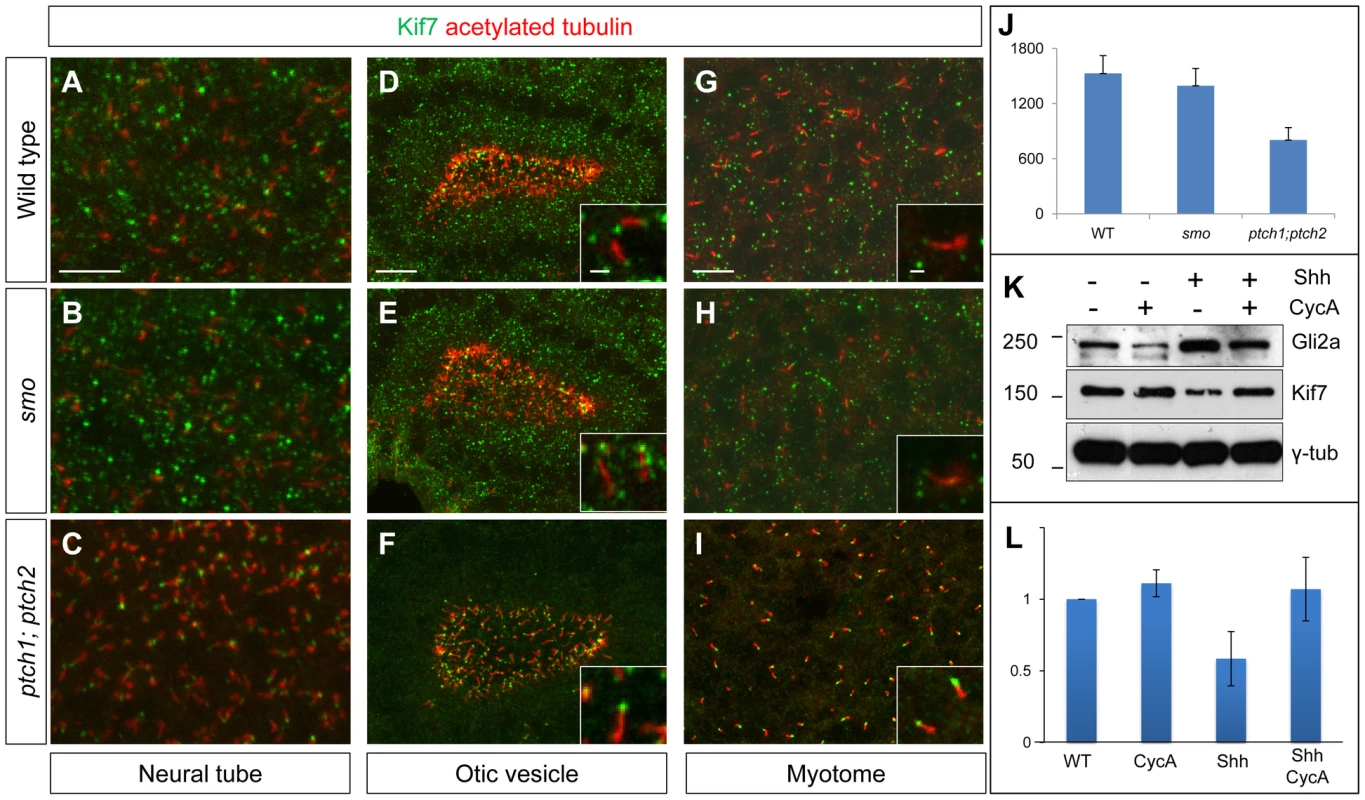
(A–C) Parasagittal optical sections of the neural tube in 20ss embryos, showing the distribution of the endogenous Kif7 protein. In wild-type (A) and smo (B) embryos, Kif7 accumulates in puncta throughout the cytoplasm as well as in the primary cilium. In ptch1; ptch2 double mutant embryos (C) by contrast, the cytoplasmic puncta are completely absent with Kif7 remaining only at the tips of the cilia. Scale bar: 10 µm. (D–I) similar distributions of Kif7 are seen in the otic vesicle (D–F) and the myotome (G–I) of wild-type and mutant embryos. Insets show a magnified view of a part of each image; note that Kif7 accumulates at the tips of some primary cilia in wild-type (D,G), smo mutant (E,H) but at elevated levels in all cilia in ptch1;ptch2 double mutant (F,I) embryos. Scale bars: 10 µm; Inset scale bar: 1 µm. (J) Quantification of fluorescence intensity of Kif7 from the otic vesicle at 20ss from wild-type (WT), smo mutants and ptch1;2 double mutants, revealing a decrease in Kif7 levels detected by immunofluorescence. Error bars represent standard deviation in spot intensity in pre-processed confocal stacks. (K) Western blot analysis of endogenous Gli2a and Kif7 protein from wild-type (WT), cyclopamine exposed wild-type (WT), Shh RNA injected (Shh) and cyclopamine treated Shh RNA injected (Shh CycA) wild-type embryos exposed to cyclopamine. Note the increase in full-length Gli2a levels following Shh overexpression. Relative levels of Kif7 protein normalized to wild-type are indicated in (L). Discussion
Through ZFN mediated targeted mutagenesis, we have confirmed a role for Kif7 in Hh pathway regulation in the zebrafish, previously inferred from transient knockdown experiments using morpholino antisense oligonucleotides [17]. Notably, we find that maternal expression of Kif7 is sufficient to support normal Hh pathway activity during embryogenesis, underlining the limitations of forward genetic screens for zygotic lethals in identifying genes with developmental functions.
Previous studies in mouse have revealed a role for mammalian Kif7 in promoting the processing of the Gli3 protein to its repressor form [18], [19], [20], analogous to the role of Cos2 in promoting cleavage of Ci in Drosophila. In cultured mammalian cells, tagged forms of Kif7 were found to localize to the base of primary cilia, translocating to their tips in response to Hh pathway activation [19], [20]. This led to the suggestion that Kif7 acts to localize Gli proteins to the base of the cilium, a site enriched in proteasomes, thereby promoting their proteolytic cleavage [20]. In this view, translocation of Kif7 to the cilia tips in response to Hh would abrogate Gli processing, leading to the activation of Hh target gene expression. Notably, however, we find that GFP tagged Gli2a remains close to the base of the primary cilium in MZkif7 mutants; moreover, the processing of endogenous Gli2a appears to be delayed rather than disrupted in the absence of Kif7 function. Thus although at 18 hpf the levels of the R form in MZ kif7 embryos are reduced compared to wild-type, by 22 hpf the Gli2a FL∶R ratio is similar to that in wild-type embryos. Moreover, our genetic data indicate that Gli2a activity is largely dispensable for the ectopic activation of the Hh target genes in MZkif7 mutant embryos, whereas loss of Gli1 activity effectively suppresses their expression. This suggests that Gli1 is the major target of Kif7 repression; consistent with this, we find that GFP tagged Gli1 protein interacts with endogenous Kif7, an interaction that is attenuated by the activation of the Hh pathway. These findings contrast with the evidence from mouse studies implicating Gli2 as the principal Kif7 target [20]; notably, however, the activity of mammalian Gli1 expressed in transgenic Drosophila can be suppressed by endogenous Cos2 function [43]. We note that ectopic Gli1 activity has also been shown to underlie the Hh gain of function phenotypes of the zebrafish igu and oval mutants [44], [45], [46], in both of which the primary cilium is disrupted [38], [44], [47]. How this relates to our proposed role for Kif7 requires further investigation.
Although we found that endogenous Kif7 protein localizes to the primary cilia in the developing zebrafish embryo, we also detected significant accumulation of endogenous Kif7 protein outside the primary cilium, in cytoplasmic puncta. These puncta represent the major site of Kif7 accumulation in unstimulated cells, but disappear upon pathway activation. We surmise that this cytoplasmic pool of Kif7 plays a central role in the negative regulation of Gli proteins, an interpretation supported by our finding that Cos2, which functions independently of primary cilia in Drosophila and does not localize to primary cilia when expressed in zebrafish embryos, can nevertheless effect a substantial rescue of the MZkif7 phenotype. In this view, the Hh mediated dispersal of cytoplasmic Kif7 would facilitate pathway activation by releasing Gli proteins from their cytoplasmic sequestration, similar to the regulation of Cos2 by Hh in Drosophila [48]. Consistent with this, we find the interaction between Gli1 and Kif7 to be abrogated by Shh overexpression.
Translocation of Gli proteins to the tip of the primary cilium has been implicated in their activation by dissociation from Sufu [25], [26] and in line with the block in Gli2a translocation seen in MZkif7 embryos, we find a concomitant increase in the association of Gli2a with Sufu, similar to that recently reported in keratinocytes of mouse Kif7 mutants [49]. This suggests that the principal role of Kif7 in the primary cilium is to mediate Gli protein activation by promoting its dissociation from Sufu. Consistent with this, ectopic Hh target gene activation is significantly enhanced by depletion of Sufu protein in MZkif7 mutant embryos. Such a role mirrors the promotion of dissociation of Sufu from Ci by Cos2 in Drosophila, an effect mediated by hyperactivation of the Fused serine threonine kinase in response to Cos2 dimerization [15]. In mouse, activity of the Fused orthologue Stk36 has been shown to be dispensable for Hh signaling [50] though in zebrafish, morpholino mediated knockdown experiment suggest that its involvement in the pathway has been conserved [30], [33]. Whether or not Stk36 is mediates the positive regulatory activity of Kif7 in zebrafish remains to be determined. Interestingly, we also observed accumulation of a faster mobility form of the FL Gli2a protein in MZkif7 embryos, similar to previous observations of Gli2 in kif7 mutant mouse keratinocytes [49]. Moreover, this form accumulates at the expense of a slower migrating form that we found to be cyclopamine sensitive, suggesting that Kif7 is also required for a Smo-dependent modification of the FL form, a modification that may contribute to Gli2a activation.
It is remarkable that even in the total absence of Kif7 function, zebrafish can complete embryogenesis and survive to become fertile adults. This stands in contrast to the peri-natal lethality caused by loss of function alleles of kif7 in the mouse [18], [19], [20] but interestingly, mirrors the finding that some Acrocallosal and Joubert syndrome patients are homozygous for loss of function alleles of the human KIF7 gene [21], [31]. A further notable interspecies difference is the lack of requirement for Kif7 in the patterning of the neural tube in zebrafish. Despite this apparent dispensability, the Hh dependent regulation of Kif7 protein sub-cellular localization occurs within the neural tube and evidence of its activity can be detected in the absence of Smo function. We surmised that Kif7 might function redundantly with Sufu to repress Gli activity in the neural tube; however, despite causing a significant enhancement of the myotomal phenotype in MZkif7 mutants, morpholino mediated depletion of Sufu protein caused only sporadic ectopic expression of fkd4, a marker of floorplate, that by contrast is expressed throughout the neural tube in the absence of Ptch function. In these respects, the regulation of Gli activity by Hh signaling seems to differ significantly between mammals and zebrafish; in mouse the requirement for Sufu is absolute, its elimination causing almost total de-repression of pathway activity with consequent widespread defects in neural tube patterning similar to those caused by lack of Ptch1 function [51], [52]. Although Sufu is effectively dispensable in Drosophila [53], it seems surprising that Gli activity can be restrained in the absence of both Sufu and Kif7 in the zebrafish neural tube; one caveat to this conclusion, however, is that the morpholino mediated depletion of Sufu is not complete. Nevertheless, it seems clear that some other component(s) must be responsible for the suppression of Gli activity. In the absence of Kif27 [17], [33] or any other close homologue of Kif7 from the zebrafish genome, the identity of such components remains a mystery.
Materials and Methods
Ethics statement
The research described in this paper uses the zebrafish as an alternative to mammalian experimental models. Adult zebrafish were raised and maintained under internationally accepted conditions. All experimental procedures were performed in compliance with and approved by the A*STAR Biological Resource Centre Institutional Animal Care and Use Committee (IACUC Project #110638). Most experimentation and analysis was restricted to the first five days post fertilization (dpf). Homozygous mutant fish were regularly monitored and any showing signs of distress were humanely euthanized following accepted protocols.
Zebrafish strains and husbandry
Adult fish were maintained on a 14 hour light/10 hour dark cycle at 28°C in the AVA (Singapore) certificated IMCB Zebrafish Facility. Previously described zebrafish strains used were: ptch1hu1602, ptch2tj222 [41]; iguts294 [46]; Tg(eng2a:eGFP)i233 [54]; gli1 (dtrts269) [55]; gli2a (yotty119) [56] gli2ai276 [37]; smob641 [57].
DNA expression constructs and zebrafish transgenic lines
Ptch2:HAeGFP: Homologous recombination in bacteria [58] was used to insert eGFP-FRT-AMP-FRT in frame after the first codon of ptch2 in BAC CH211-226H23. An HA tag was inserted in frame at the N-terminus of eGFP, while amplifying the targeting cassette. The FRT-Amp-FRT was excised using arabinose induction and a fragment containing 8 kb upstream of the insertion along with eGFP was cloned into pMiniTol2 using homologous recombination for gap repair [58]. The targeting cassettes and plasmids used have been described previously [54]. More than 10 stable transgenic lines were generated with this construct and one of the brightest of these (allele designation i271) selected for further experiments.
pCS2-mCherry-Gli1, pCS2-eGFP-Gli1: A Vn-Gli1-pCS2+ plasmid [54] was digested with SmaI and AgeI (to release the Vn fragment); eGFP and mCherry were amplified with oligos containing the SmaI and AgeI and cloned into the same site of Vn-Gli1-pCS2+ plasmid.
pCS2-Kif7-eGFP, pCS2-Cos2-eGFP: Zebrafish Kif7 and Drosophila Cos2 [10] were amplified using BamHI and SmaI adaptors and cloned into the same sites of a eGFP-pCS2+ plasmid (eGFP cloned between SmaI, XbaI sites)
Generation, selection and genotyping of kif7 mutant alleles
Plasmids encoding Zinc-finger nucleases (ZFN) specific for the zebrafish kif7 (set 3) gene were purchased from Sigma (see accompanying data sheet). Capped polyadenylated RNA from each plasmid was produced by in vitro transcription and a range of doses was injected into one-cell stage zebrafish embryos. Embryos injected with approximately 600 pg had around 30% rate of deformity at 24 hpf (hours post-fertilization). Genomic DNA prepared from non-deformed 24 hpf embryos injected with this dose was used as a PCR template to analyze potential somatic mutations. Roche Titanium 454 amplicon sequencing showed that 2.5% (6.5% is by 8 of 127 colony PCR and sequencing) of the amplicon molecules had insertions or deletions at the target site. G0 adults derived from embryos injected with ZFN capped RNA were in-crossed and their progenies (G1) individually genotyped by PCR using the forward primer (Kif7 exF2 : 5′CGAGGTGCTGAGTCTCTTAGAGT) and reverse primer (Kif7 exR1 : 5′TGAATCCCTGTATGGGATATGGGT) followed by Sanger sequencing.
Founders transmitting two different alleles (kif7Δ8, an 8-bp del: GGACCTGG kif7Δ7, a 7-bp del: TTGTGGA) were selected and used to establish stable mutant lines. The full nucleic acid sequences of mutated alleles together with their allele designations are shown in Table 1).
In situ hybridization and immunofluorescence
Standard in situ hybridization (ISH) was performed with anti-DIG alkaline phosphatase and chromogenic substrate NBT/BCIP as previously described [59]. RNA probes were prepared from templates as previously described: nkx2.2 [60], olig2 [61], ptch2 (formerly ptc1) [62], prdm1a [63].
Whole-mount antibody staining was performed as previously described [35], [64] at the following dilutions: mAb 4D9 (anti-Engrailed; DHSB) at 1∶50–1∶200; mAb F310 (1∶50; DHSB); mAb F59 (1∶50; DHSB); rabbit anti-Prox1 (1∶2000); rabbit anti-γ-tubulin (1∶500; Sigma); mouse anti-γ-tubulin (1∶500; Sigma); mouse anti-acetylated α-tubulin (1∶800; Sigma); rabbit anti-Kif7 (1∶500); chick anti-GFP (1∶400; Abcam); rabbit anti-GFP-488 (1∶750; Invitrogen); rabbit anti-DSred (1∶200; Clontech). The secondary antibodies were: Alexa488-conjugated goat anti-mouse or rabbit, Alexa546-conjugated goat anti-mouse and Alexa568-conjugated goat anti-rabbit, Alexa633-conjugated goat anti-secondary antibodies (1∶1000, Invitrogen). Bright field microscopy images were acquired with an AxioCam HRc mounted on a Zeiss AXIO Imager M2, Olympus DP70 on MVX10 or Leica DFC300 FX mounted on MZ16FA. Fluorescent specimens were imaged using the 60× or 100× oil immersion objective on an Olympus Fluoview 1000 confocal microscope. Images were acquired using Olympus FV10-ASW software.
Synthetic RNA, DNA and morpholino for injection
Capped synthetic mRNAs for the following genes was synthesized using the SP6 mMessage mMachine Kit (Ambion), the plasmids were linearized with restriction enzymes as indicated: pCS2-GFP-Kif7 (NotI) [17]; pCS2-Cos2-GFP (NotI) [65]; pCS2-KIF7-GFP (NotI). BAC Gli2a-GFP was used as previously described [38]. Morpholino oligonucleotides were obtained from Gene Tools (USA) and injected into newly fertilized embryos. The sequences of those targeting Gli2a and Sufu antisense were as described [30]. The sequence of the Gli1 morpholino was as described by [36].
Cyclopamine treatment of embryos
Cyclopamine treatment followed a standard method with immersion in 40 µM cyclopamine (Toronto Research Chemicals) from 50% epiboly as previously described [30].
Generation of antibodies
Sufu: The full-length zebrafish Sufu coding sequence [30] was cloned into the BamHI-EcoR1 sites of pGEX-6p-1 (Amersham). The resulting GST-zSufu fusion protein was expressed in E.coli strain BL21 and purified by SDS-PAGE and electro-elution. After dialysis in PBS, the antigen was injected into mice. Splenocytes were extracted from immunoreactive animals and fused with melanoma cells to generate hybridomas. Of 904 hybridoma clones screened, 50 were positive by ELISA and one of these, 2A10, tested positive by western blot of zebrafish embryo extracts. Polyclonal rabbit anti-zebrafish Sufu antibody was generated by Absea Biotechnology Ltd. (China) using the same pGEX-zSufu expression construct.
Kif7: Polyclonal rabbit anti-zKif7 antibody was generated by Strategic Diagnostics Inc. (USA) using a peptide corresponding to residues 1231–1330 of the zebrafish Kif7 protein sequence [17].
Fluorescence image analysis
(1) Image pre-processing
Acquired image stacks were reconstructed and the voxel size normalized using linear interpolation such that the scales of voxels were isotropic, i.e. each voxel is 0.2×0.2×0.2 µm. The images were smoothened by a Gaussian kernel to reduce noise and enhanced based on the histogram. A Laplacian kernel was applied to enhance the boundaries between them.
(2) Spot segmentation and brightness measurement
To formalize the spot segmentation problem, the pre-processed image stacks at each time point were defined on a subset of three-dimensional space . We used to represent the image intensity of the spots after the pre-processing. The intensity function of the image stack was normalized such that . The local maximum in was detected as spot centers, denoted by seeds. The spots were then segmented by Evolving Generalized Voronoi Diagrams approach [66], [67]. The segmented regions of represent spot segments, represented by for , where L is the number of detected spots. Each forms connected regions in . A simple statement defines the connected region:
-
Connected region: A set of points form a connected region if and , a path connecting and such that .
The average brightness of each spot is then calculated based on Eq. (1),
where the denominator is the volume of spots.Western blot analysis
Embryos were de-chorionated, de-yolked, and homogenized manually in ice cold PBS without Ca2+ and Mg2+ in the presence of complete protease inhibitor cocktail (Roche). The embryo pellet was lysed in RIPA buffer (50 mM Tris.HCl, pH 8.0/150 mM NaCl/1%NP-40/0.5% Na.Deoxycholate/0.1%SDS/protease inhibitor cocktail/1 mM PMSF). Samples were microcentrifuged for 10 min at 4°C, loading buffer (62.6 mM Tris HCl, pH 6.8; 2% SDS; 0.01% bromophenol blue; 10% glycerol; 100 mM DTT) was added to the supernatant and the equivalent of 30 embryos run on each lane of a 7.5% acrylamide denaturing gel at 30 mA for 120 mins, and electroblotted onto Immobilon-P polyvinylidene fluoride (PVDF) membrane (Millipore). PVDF strips were blocked in 5% milk powder PBS 0.1%Tween20 for 1 hr, and incubated with rabbit anti-zebrafish Gli2a (1∶5000) [54], rabbit anti-zebrafish Kif7 (1∶5000), mouse anti-Sufu (1∶100) for 1 hr at room temperature. After washing, primary antibody was detected with ECL HRP-conjugated anti-rabbit lgG (1∶50,000) and anti-mouse IgG (1∶50,000). Chemiluminescent Substrate was SuperSignal West Femto (Pierce). The loading amount of protein extract among specimens was evaluated by gamma-Tubulin level with either rabbit or mouse anti-gamma Tubulin (1∶5000; Sigma). Signal quantification was performed using Adobe Photoshop or Image J software.
Immunoprecipitation
Embryos were de-chorionated using pronase (2 mg/ml, Sigma) and rinsed with egg water and then PBS with 2× protease inhibitor (PI; Roche). Embryos were de-yolked by yellow tip on ice and rinsed in ice-cold PBS/2× PI. The embryos were centrifuged at 200×g for 5 mins. The pellet was stored at −80C degree. For 1000× embryos, 1 ml of triton buffer (20 mM Tris-HCl, pH 8.0/137 mM NaCl/10% Glycerol/1%Triton X100/2 mM EDTA)/2×PI/10 mM PMSF is added to extract the proteins. Tubes were left horizontally on ice for 30 mins or longer. The lysates were spun at 10,000×g for 15 mins. The suspension was transferred into a new 2 ml eppendorf. Forty µl of lysate was taken out as the input control. In 1 ml of lysate, 150 µl of magnetic Dyna-bead (Invitrogen) was added to pre-clear for 3 hours at 4°C degree. After removing the pre-clear beads, 10 µg of rabbit primary Ab was added into lysate. For eGFP immunoprecipitations we added the lysate to 20 µl of GFP-Trap beads (Chromotek) and other steps remained unchanged. The binding between rAb and the specific protein was facilitated by shaking at 4°C overnight. The 120 µl of dynabead protein A was added in 1 ml of lysate and incubated at 4°C degree for no more than 1 hour to pull down the complex of rAb-target protein. The beads were washed with 1.5 ml of lysis buffer/2×PI per tube 4 to 6 times. During the last time of washing, the beads were transferred to a new tube. Elution was performed by adding 40 µl of 2× SDS NuPage LDS/100 mM DTT (Invitrogen) per tube and by heating at 95°C for 5 mins.
Supporting Information
Zdroje
1. HooperJE, ScottMP (2005) Communicating with Hedgehogs. Nat Rev Mol Cell Biol 6 : 306–317.
2. InghamPW, PlaczekM (2006) Orchestrating ontogenesis: variations on a theme by sonic hedgehog. Nat Rev Genet 7 : 841–850.
3. InghamPW, NakanoY, SegerC (2011) Mechanisms and functions of Hedgehog signalling across the metazoa. Nat Rev Genet 12 : 393–406.
4. ShinK, LeeJ, GuoN, KimJ, LimA, et al. (2011) Hedgehog/Wnt feedback supports regenerative proliferation of epithelial stem cells in bladder. Nature 472 : 110–114.
5. TeperinoR, AmannS, BayerM, McGeeSL, LoipetzbergerA, et al. (2012) Hedgehog partial agonism drives Warburg-like metabolism in muscle and brown fat. Cell 151 : 414–426.
6. BabcockDT, ShiS, JoJ, ShawM, GutsteinHB, et al. (2011) Hedgehog signaling regulates nociceptive sensitization. Curr Biol 21 : 1525–1533.
7. TaipaleJ, BeachyPA (2001) The Hedgehog and Wnt signalling pathways in cancer. Nature 411 : 349–354.
8. TheunissenJW, de SauvageFJ (2009) Paracrine Hedgehog signaling in cancer. Cancer Res 69 : 6007–6010.
9. RobbinsDJ, NybakkenKE, KobayashiR, SissonJC, BishopJM, et al. (1997) Hedgehog elicits signal transduction by means of a large complex containing the kinesin-related protein costal2. Cell 90 : 225–234.
10. WangG, AmanaiK, WangB, JiangJ (2000) Interactions with Costal2 and suppressor of fused regulate nuclear translocation and activity of cubitus interruptus. Genes Dev 14 : 2893–2905.
11. LumL, ZhangC, OhS, MannRK, von KesslerDP, et al. (2003) Hedgehog signal transduction via Smoothened association with a cytoplasmic complex scaffolded by the atypical kinesin, Costal-2. Mol Cell 12 : 1261–1274.
12. ZhangW, ZhaoY, TongC, WangG, WangB, et al. (2005) Hedgehog-regulated Costal2-kinase complexes control phosphorylation and proteolytic processing of Cubitus interruptus. Dev Cell 8 : 267–278.
13. CapdevilaJ, EstradaMP, Sanchez-HerreroE, GuerreroI (1994) The Drosophila segment polarity gene patched interacts with decapentaplegic in wing development. EMBO J 13 : 71–82.
14. ForbesAJ, NakanoY, TaylorAM, InghamPW (1993) Genetic analysis of hedgehog signalling in the Drosophila embryo. Dev Suppl 115–124.
15. RanieriN, RuelL, GalletA, RaisinS, TherondPP (2012) Distinct phosphorylations on kinesin costal-2 mediate differential hedgehog signaling strength. Dev Cell 22 : 279–294.
16. MonnierV, DussillolF, AlvesG, Lamour-IsnardC, PlessisA (1998) Suppressor of fused links fused and Cubitus interruptus on the hedgehog signalling pathway. Curr Biol 8 : 583–586.
17. TaySY, InghamPW, RoyS (2005) A homologue of the Drosophila kinesin-like protein Costal2 regulates Hedgehog signal transduction in the vertebrate embryo. Development 132 : 625–634.
18. CheungHO, ZhangX, RibeiroA, MoR, MakinoS, et al. (2009) The kinesin protein Kif7 is a critical regulator of Gli transcription factors in mammalian hedgehog signaling. Sci Signal 2: ra29.
19. Endoh-YamagamiS, EvangelistaM, WilsonD, WenX, TheunissenJW, et al. (2009) The mammalian Cos2 homolog Kif7 plays an essential role in modulating Hh signal transduction during development. Curr Biol 19 : 1320–1326.
20. LiemKFJr, HeM, OcbinaPJ, AndersonKV (2009) Mouse Kif7/Costal2 is a cilia-associated protein that regulates Sonic hedgehog signaling. Proc Natl Acad Sci U S A 106 : 13377–13382.
21. PutouxA, ThomasS, CoeneKL, DavisEE, AlanayY, et al. (2011) KIF7 mutations cause fetal hydrolethalus and acrocallosal syndromes. Nat Genet 43 : 601–606.
22. GoetzSC, AndersonKV (2010) The primary cilium: a signalling centre during vertebrate development. Nat Rev Genet 11 : 331–344.
23. HuangfuD, AndersonKV (2005) Cilia and Hedgehog responsiveness in the mouse. Proc Natl Acad Sci U S A 102 : 11325–11330.
24. HaycraftCJ, BanizsB, Aydin-SonY, ZhangQ, MichaudEJ, et al. (2005) Gli2 and Gli3 localize to cilia and require the intraflagellar transport protein polaris for processing and function. PLoS Genet 1: e53.
25. HumkeEW, DornKV, MilenkovicL, ScottMP, RohatgiR (2010) The output of Hedgehog signaling is controlled by the dynamic association between Suppressor of Fused and the Gli proteins. Genes Dev 24 : 670–682.
26. TukachinskyH, LopezLV, SalicA (2010) A mechanism for vertebrate Hedgehog signaling: recruitment to cilia and dissociation of SuFu-Gli protein complexes. J Cell Biol 191 : 415–428.
27. DoyonY, McCammonJM, MillerJC, FarajiF, NgoC, et al. (2008) Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat Biotechnol 26 : 702–708.
28. FoleyJE, YehJR, MaederML, ReyonD, SanderJD, et al. (2009) Rapid mutation of endogenous zebrafish genes using zinc finger nucleases made by Oligomerized Pool ENgineering (OPEN). PLoS One 4: e4348.
29. MengX, NoyesMB, ZhuLJ, LawsonND, WolfeSA (2008) Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat Biotechnol 26 : 695–701.
30. WolffC, RoyS, InghamPW (2003) Multiple muscle cell identities induced by distinct levels and timing of hedgehog activity in the zebrafish embryo. Curr Biol 13 : 1169–1181.
31. DafingerC, LiebauMC, ElsayedSM, HellenbroichY, BoltshauserE, et al. (2011) Mutations in KIF7 link Joubert syndrome with Sonic Hedgehog signaling and microtubule dynamics. J Clin Invest 121 : 2662–2667.
32. GeringM, PatientR (2005) Hedgehog signaling is required for adult blood stem cell formation in zebrafish embryos. Dev Cell 8 : 389–400.
33. WilsonCW, NguyenCT, ChenMH, YangJH, GacayanR, et al. (2009) Fused has evolved divergent roles in vertebrate Hedgehog signalling and motile ciliogenesis. Nature 459 : 98–102.
34. BisgroveBW, EssnerJJ, YostHJ (1999) Regulation of midline development by antagonism of lefty and nodal signaling. Development 126 : 3253–3262.
35. BenJ, ElworthyS, NgAS, van EedenF, InghamPW (2011) Targeted mutation of the talpid3 gene in zebrafish reveals its conserved requirement for ciliogenesis and Hedgehog signalling across the vertebrates. Development 138 : 4969–4978.
36. KarlstromRO, TyurinaOV, KawakamiA, NishiokaN, TalbotWS, et al. (2003) Genetic analysis of zebrafish gli1 and gli2 reveals divergent requirements for gli genes in vertebrate development. Development 130 : 1549–1564.
37. WangX, ZhaoZ, MullerJ, IyuA, KhngAJ, et al. (2013) Targeted inactivation and identification of targets of the Gli2a transcription factor in the Zebrafish. Biology Open doi: 10.1242/bio.20136262
38. KimHR, RichardsonJ, van EedenF, InghamPW (2010) Gli2a protein localization reveals a role for Iguana/DZIP1 in primary ciliogenesis and a dependence of Hedgehog signal transduction on primary cilia in the zebrafish. BMC Biol 8 : 65.
39. GoodrichLV, MilenkovicL, HigginsKM, ScottMP (1997) Altered neural cell fates and medulloblastoma in mouse patched mutants. Science 277 : 1109–1113.
40. MotoyamaJ, MilenkovicL, IwamaM, ShikataY, ScottMP, et al. (2003) Differential requirement for Gli2 and Gli3 in ventral neural cell fate specification. Dev Biol 259 : 150–161.
41. KoudijsMJ, den BroederMJ, GrootE, van EedenFJ (2008) Genetic analysis of the two zebrafish patched homologues identifies novel roles for the hedgehog signaling pathway. BMC Dev Biol 8 : 15.
42. KoudijsMJ, den BroederMJ, KeijserA, WienholdsE, HouwingS, et al. (2005) The zebrafish mutants dre, uki, and lep encode negative regulators of the hedgehog signaling pathway. PLoS Genet 1: e19.
43. MarksSA, KalderonD (2011) Regulation of mammalian Gli proteins by Costal 2 and PKA in Drosophila reveals Hedgehog pathway conservation. Development 138 : 2533–2542.
44. HuangPS, CooperAF (2009) Dampened Hedgehog signaling but normal Wnt signaling in zebrafish without cilia. Development 136 : 3089–3098.
45. SekimizuK, NishiokaN, SasakiH, TakedaH, KarlstromRO, et al. (2004) The zebrafish iguana locus encodes Dzip1, a novel zinc-finger protein required for proper regulation of Hedgehog signaling. Development 131 : 2521–2532.
46. WolffC, RoyS, LewisKE, SchauerteH, Joerg-RauchG, et al. (2004) iguana encodes a novel zinc-finger protein with coiled-coil domains essential for Hedgehog signal transduction in the zebrafish embryo. Genes Dev 18 : 1565–1576.
47. TaySY, YuX, WongKN, PanseP, NgCP, et al. (2010) The iguana/DZIP1 protein is a novel component of the ciliogenic pathway essential for axonemal biogenesis. Dev Dyn 239 : 527–534.
48. RuelL, RodriguezR, GalletA, Lavenant-StacciniL, TherondPP (2003) Stability and association of Smoothened, Costal2 and Fused with Cubitus interruptus are regulated by Hedgehog. Nat Cell Biol 5 : 907–913.
49. LiZJ, NieuwenhuisE, NienW, ZhangX, ZhangJ, et al. (2012) Kif7 regulates Gli2 through Sufu-dependent and -independent functions during skin development and tumorigenesis. Development 139 : 4152–4161.
50. MerchantM, EvangelistaM, LuohSM, FrantzGD, ChalasaniS, et al. (2005) Loss of the serine/threonine kinase fused results in postnatal growth defects and lethality due to progressive hydrocephalus. Mol Cell Biol 25 : 7054–7068.
51. CooperAF, YuKP, BruecknerM, BraileyLL, JohnsonL, et al. (2005) Cardiac and CNS defects in a mouse with targeted disruption of suppressor of fused. Development 132 : 4407–4417.
52. SvardJ, Heby-HenricsonK, Persson-LekM, RozellB, LauthM, et al. (2006) Genetic elimination of Suppressor of fused reveals an essential repressor function in the mammalian Hedgehog signaling pathway. Dev Cell 10 : 187–197.
53. PreatT (1992) Characterization of Suppressor of fused, a complete suppressor of the fused segment polarity gene of Drosophila melanogaster. Genetics 132 : 725–736.
54. MauryaAK, TanH, SourenM, WangX, WittbrodtJ, et al. (2011) Integration of Hedgehog and BMP signalling by the engrailed2a gene in the zebrafish myotome. Development 138 : 755–765.
55. KarlstromRO, TroweT, KlostermannS, BaierH, BrandM, et al. (1996) Zebrafish mutations affecting retinotectal axon pathfinding. Development 123 : 427–438.
56. van EedenFJ, GranatoM, SchachU, BrandM, Furutani-SeikiM, et al. (1996) Mutations affecting somite formation and patterning in the zebrafish, Danio rerio. Development 123 : 153–164.
57. VargaZM, AmoresA, LewisKE, YanYL, PostlethwaitJH, et al. (2001) Zebrafish smoothened functions in ventral neural tube specification and axon tract formation. Development 128 : 3497–3509.
58. LeeEC, YuD, Martinez de VelascoJ, TessarolloL, SwingDA, et al. (2001) A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics 73 : 56–65.
59. OxtobyE, JowettT (1993) Cloning of the zebrafish krox-20 gene (krx-20) and its expression during hindbrain development. Nucleic Acids Res 21 : 1087–1095.
60. BarthKA, WilsonSW (1995) Expression of zebrafish nk2.2 is influenced by sonic hedgehog/vertebrate hedgehog-1 and demarcates a zone of neuronal differentiation in the embryonic forebrain. Development 121 : 1755–1768.
61. ParkHC, MehtaA, RichardsonJS, AppelB (2002) olig2 is required for zebrafish primary motor neuron and oligodendrocyte development. Dev Biol 248 : 356–368.
62. ConcordetJP, LewisKE, MooreJW, GoodrichLV, JohnsonRL, et al. (1996) Spatial regulation of a zebrafish patched homologue reflects the roles of sonic hedgehog and protein kinase A in neural tube and somite patterning. Development 122 : 2835–2846.
63. BaxendaleS, DavisonC, MuxworthyC, WolffC, InghamPW, et al. (2004) The B-cell maturation factor Blimp-1 specifies vertebrate slow-twitch muscle fiber identity in response to Hedgehog signaling. Nat Genet 36 : 88–93.
64. ElworthyS, HargraveM, KnightR, MebusK, InghamPW (2008) Expression of multiple slow myosin heavy chain genes reveals a diversity of zebrafish slow twitch muscle fibres with differing requirements for Hedgehog and Prdm1 activity. Development 135 : 2115–2126.
65. LiuY, CaoX, JiangJ, JiaJ (2007) Fused-Costal2 protein complex regulates Hedgehog-induced Smo phosphorylation and cell-surface accumulation. Genes Dev 21 : 1949–1963.
66. YuW, LeeHK, HariharanS, BuW, AhmedS (2009) Quantitative neurite outgrowth measurement based on image segmentation with topological dependence. Cytometry A 75 : 289–297.
67. YuW, LeeHK, HariharanS, BuW, AhmedS (2010) Evolving generalized Voronoi diagrams for accurate cellular image segmentation. Cytometry Part A 77A: 379–386.
Štítky
Genetika Reprodukční medicína
Článek Interaction between and during Mammalian Jaw Patterning and in the Pathogenesis of SyngnathiaČlánek Clustering of Tissue-Specific Sub-TADs Accompanies the Regulation of Genes in Developing LimbsČlánek Transcription Factor Occupancy Can Mediate Active Turnover of DNA Methylation at Regulatory RegionsČlánek Tay Bridge Is a Negative Regulator of EGFR Signalling and Interacts with Erk and Mkp3 in the Wing
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 12
-
Všechny články tohoto čísla
- Stressing the Importance of CHOP in Liver Cancer
- The AmAZI1ng Roles of Centriolar Satellites during Development
- Flies Get a Head Start on Meiosis
- Recommendations from Jane Gitschier's Bookshelf
- And Baby Makes Three: Genomic Imprinting in Plant Embryos
- Bugs in Transition: The Dynamic World of in Insects
- Defining the Role of ATP Hydrolysis in Mitotic Segregation of Bacterial Plasmids
- Synaptonemal Complex Components Promote Centromere Pairing in Pre-meiotic Germ Cells
- Cohesinopathies of a Feather Flock Together
- Genetic Recombination Is Targeted towards Gene Promoter Regions in Dogs
- Parathyroid-Specific Deletion of Unravels a Novel Calcineurin-Dependent FGF23 Signaling Pathway That Regulates PTH Secretion
- MAN1B1 Deficiency: An Unexpected CDG-II
- Phosphate Flow between Hybrid Histidine Kinases CheA and CheS Controls Cyst Formation
- Basolateral Mg Extrusion via CNNM4 Mediates Transcellular Mg Transport across Epithelia: A Mouse Model
- Truncation of Unsilences Paternal and Ameliorates Behavioral Defects in the Angelman Syndrome Mouse Model
- Autozygome Sequencing Expands the Horizon of Human Knockout Research and Provides Novel Insights into Human Phenotypic Variation
- Huntington's Disease Induced Cardiac Amyloidosis Is Reversed by Modulating Protein Folding and Oxidative Stress Pathways in the Heart
- Low Frequency Variants, Collapsed Based on Biological Knowledge, Uncover Complexity of Population Stratification in 1000 Genomes Project Data
- Targeted Ablation of and in Retinal Progenitor Cells Mimics Leber Congenital Amaurosis
- Genomic Imprinting in the Embryo Is Partly Regulated by PRC2
- Binary Cell Fate Decisions and Fate Transformation in the Larval Eye
- The Stress-Regulated Transcription Factor CHOP Promotes Hepatic Inflammatory Gene Expression, Fibrosis, and Oncogenesis
- A Global RNAi Screen Identifies a Key Role of Ceramide Phosphoethanolamine for Glial Ensheathment of Axons
- Functional Analysis of the Interdependence between DNA Uptake Sequence and Its Cognate ComP Receptor during Natural Transformation in Species
- Cross-Modulation of Homeostatic Responses to Temperature, Oxygen and Carbon Dioxide in
- Alcohol-Induced Histone Acetylation Reveals a Gene Network Involved in Alcohol Tolerance
- Molecular Characterization of Host-Specific Biofilm Formation in a Vertebrate Gut Symbiont
- CRIS—A Novel cAMP-Binding Protein Controlling Spermiogenesis and the Development of Flagellar Bending
- Dual Regulation of the Mitotic Exit Network (MEN) by PP2A-Cdc55 Phosphatase
- Expanding the Marine Virosphere Using Metagenomics
- Detection of Slipped-DNAs at the Trinucleotide Repeats of the Myotonic Dystrophy Type I Disease Locus in Patient Tissues
- Interaction between and during Mammalian Jaw Patterning and in the Pathogenesis of Syngnathia
- Mutations in the UQCC1-Interacting Protein, UQCC2, Cause Human Complex III Deficiency Associated with Perturbed Cytochrome Protein Expression
- Reactivation of Chromosomally Integrated Human Herpesvirus-6 by Telomeric Circle Formation
- Anoxia-Reoxygenation Regulates Mitochondrial Dynamics through the Hypoxia Response Pathway, SKN-1/Nrf, and Stomatin-Like Protein STL-1/SLP-2
- The Midline Protein Regulates Axon Guidance by Blocking the Reiteration of Neuroblast Rows within the Drosophila Ventral Nerve Cord
- Tomato Yield Heterosis Is Triggered by a Dosage Sensitivity of the Florigen Pathway That Fine-Tunes Shoot Architecture
- Selection on Plant Male Function Genes Identifies Candidates for Reproductive Isolation of Yellow Monkeyflowers
- Role of Tomato Lipoxygenase D in Wound-Induced Jasmonate Biosynthesis and Plant Immunity to Insect Herbivores
- Meiotic Cohesin SMC1β Provides Prophase I Centromeric Cohesion and Is Required for Multiple Synapsis-Associated Functions
- Identification of Sphingolipid Metabolites That Induce Obesity via Misregulation of Appetite, Caloric Intake and Fat Storage in
- Genome-Wide Screen Reveals Replication Pathway for Quasi-Palindrome Fragility Dependent on Homologous Recombination
- Histone Methylation Restrains the Expression of Subtype-Specific Genes during Terminal Neuronal Differentiation in
- A Novel Intergenic ETnII-β Insertion Mutation Causes Multiple Malformations in Mice
- The NuRD Chromatin-Remodeling Enzyme CHD4 Promotes Embryonic Vascular Integrity by Transcriptionally Regulating Extracellular Matrix Proteolysis
- A Domesticated Transposase Interacts with Heterochromatin and Catalyzes Reproducible DNA Elimination in
- Acute Versus Chronic Loss of Mammalian Results in Distinct Ciliary Phenotypes
- MBD3 Localizes at Promoters, Gene Bodies and Enhancers of Active Genes
- Positive and Negative Regulation of Gli Activity by Kif7 in the Zebrafish Embryo
- A Hereditary Spastic Paraplegia Mouse Model Supports a Role of ZFYVE26/SPASTIZIN for the Endolysosomal System
- The CCR4-NOT Complex Mediates Deadenylation and Degradation of Stem Cell mRNAs and Promotes Planarian Stem Cell Differentiation
- Reconstructing Native American Migrations from Whole-Genome and Whole-Exome Data
- Contributions of Protein-Coding and Regulatory Change to Adaptive Molecular Evolution in Murid Rodents
- Comprehensive Analysis of Transcriptome Variation Uncovers Known and Novel Driver Events in T-Cell Acute Lymphoblastic Leukemia
- A -Acting Protein Effect Causes Severe Eye Malformation in the Mouse
- Clustering of Tissue-Specific Sub-TADs Accompanies the Regulation of Genes in Developing Limbs
- Germline Progenitors Escape the Widespread Phenomenon of Homolog Pairing during Development
- Transcription Factor Occupancy Can Mediate Active Turnover of DNA Methylation at Regulatory Regions
- Somatic mtDNA Mutation Spectra in the Aging Human Putamen
- ESCRT-I Mediates FLS2 Endosomal Sorting and Plant Immunity
- Ethylene Promotes Hypocotyl Growth and HY5 Degradation by Enhancing the Movement of COP1 to the Nucleus in the Light
- The PAF Complex and Prf1/Rtf1 Delineate Distinct Cdk9-Dependent Pathways Regulating Transcription Elongation in Fission Yeast
- Dual Regulation of Gene Expression Mediated by Extended MAPK Activation and Salicylic Acid Contributes to Robust Innate Immunity in
- Quantifying Missing Heritability at Known GWAS Loci
- Smc5/6-Mms21 Prevents and Eliminates Inappropriate Recombination Intermediates in Meiosis
- Smc5/6 Coordinates Formation and Resolution of Joint Molecules with Chromosome Morphology to Ensure Meiotic Divisions
- Tay Bridge Is a Negative Regulator of EGFR Signalling and Interacts with Erk and Mkp3 in the Wing
- Meiotic Crossover Control by Concerted Action of Rad51-Dmc1 in Homolog Template Bias and Robust Homeostatic Regulation
- Active Transport and Diffusion Barriers Restrict Joubert Syndrome-Associated ARL13B/ARL-13 to an Inv-like Ciliary Membrane Subdomain
- An Regulatory Circuit Modulates /Wnt Signaling and Determines the Size of the Midbrain Dopaminergic Progenitor Pool
- Variants Induce Differential Protection to Viruses in : A Phenotypic and Phylogenomic Analysis
- Base Pairing Interaction between 5′- and 3′-UTRs Controls mRNA Translation in
- Evidence That Masking of Synapsis Imperfections Counterbalances Quality Control to Promote Efficient Meiosis
- Insulin/IGF-Regulated Size Scaling of Neuroendocrine Cells Expressing the bHLH Transcription Factor in
- Sumoylated NHR-25/NR5A Regulates Cell Fate during Vulval Development
- TATN-1 Mutations Reveal a Novel Role for Tyrosine as a Metabolic Signal That Influences Developmental Decisions and Longevity in
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The NuRD Chromatin-Remodeling Enzyme CHD4 Promotes Embryonic Vascular Integrity by Transcriptionally Regulating Extracellular Matrix Proteolysis
- Mutations in the UQCC1-Interacting Protein, UQCC2, Cause Human Complex III Deficiency Associated with Perturbed Cytochrome Protein Expression
- The Midline Protein Regulates Axon Guidance by Blocking the Reiteration of Neuroblast Rows within the Drosophila Ventral Nerve Cord
- Tomato Yield Heterosis Is Triggered by a Dosage Sensitivity of the Florigen Pathway That Fine-Tunes Shoot Architecture
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání




