-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaEthylene Promotes Hypocotyl Growth and HY5 Degradation by Enhancing the Movement of COP1 to the Nucleus in the Light
In the dark, etiolated seedlings display a long hypocotyl, the growth of which is rapidly inhibited when the seedlings are exposed to light. In contrast, the phytohormone ethylene prevents hypocotyl elongation in the dark but enhances its growth in the light. However, the mechanism by which light and ethylene signalling oppositely affect this process at the protein level is unclear. Here, we report that ethylene enhances the movement of CONSTITUTIVE PHOTOMORPHOGENESIS 1 (COP1) to the nucleus where it mediates the degradation of LONG HYPOCOTYL 5 (HY5), contributing to hypocotyl growth in the light. Our results indicate that HY5 is required for ethylene-promoted hypocotyl growth in the light, but not in the dark. Using genetic and biochemical analyses, we found that HY5 functions downstream of ETHYLENE INSENSITIVE 3 (EIN3) for ethylene-promoted hypocotyl growth. Furthermore, the upstream regulation of HY5 stability by ethylene is COP1-dependent, and COP1 is genetically located downstream of EIN3, indicating that the COP1-HY5 complex integrates light and ethylene signalling downstream of EIN3. Importantly, the ethylene precursor 1-aminocyclopropane-1-carboxylate (ACC) enriched the nuclear localisation of COP1; however, this effect was dependent on EIN3 only in the presence of light, strongly suggesting that ethylene promotes the effects of light on the movement of COP1 from the cytoplasm to the nucleus. Thus, our investigation demonstrates that the COP1-HY5 complex is a novel integrator that plays an essential role in ethylene-promoted hypocotyl growth in the light.
Published in the journal: . PLoS Genet 9(12): e32767. doi:10.1371/journal.pgen.1004025
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004025Summary
In the dark, etiolated seedlings display a long hypocotyl, the growth of which is rapidly inhibited when the seedlings are exposed to light. In contrast, the phytohormone ethylene prevents hypocotyl elongation in the dark but enhances its growth in the light. However, the mechanism by which light and ethylene signalling oppositely affect this process at the protein level is unclear. Here, we report that ethylene enhances the movement of CONSTITUTIVE PHOTOMORPHOGENESIS 1 (COP1) to the nucleus where it mediates the degradation of LONG HYPOCOTYL 5 (HY5), contributing to hypocotyl growth in the light. Our results indicate that HY5 is required for ethylene-promoted hypocotyl growth in the light, but not in the dark. Using genetic and biochemical analyses, we found that HY5 functions downstream of ETHYLENE INSENSITIVE 3 (EIN3) for ethylene-promoted hypocotyl growth. Furthermore, the upstream regulation of HY5 stability by ethylene is COP1-dependent, and COP1 is genetically located downstream of EIN3, indicating that the COP1-HY5 complex integrates light and ethylene signalling downstream of EIN3. Importantly, the ethylene precursor 1-aminocyclopropane-1-carboxylate (ACC) enriched the nuclear localisation of COP1; however, this effect was dependent on EIN3 only in the presence of light, strongly suggesting that ethylene promotes the effects of light on the movement of COP1 from the cytoplasm to the nucleus. Thus, our investigation demonstrates that the COP1-HY5 complex is a novel integrator that plays an essential role in ethylene-promoted hypocotyl growth in the light.
Introduction
The phytohormone ethylene plays significant roles in many developmental processes and stress responses in plants. Molecular and genetic analyses have revealed a linear signalling pathway, which is initiated by ethylene perception at the endoplasmic reticulum membrane, resulting in transcriptional regulation in the nucleus [1]–[3]. The ethylene receptors ETHYLENE RESPONSE 1 (ETR1), ETHYLENE RESPONSE SENSOR 1 (ERS1), ETR2, ERS2, and ETHYLENE INSENSITIVE 4 (EIN4) are members of a family of two-component His protein kinase receptors that negatively affect ethylene signaling [4], [5]. In the absence of ethylene, these receptors directly suppress the ethylene response by interacting with a Raf-like mitogen-activated protein kinase kinase kinase family protein, CONSTITUTIVE TRIPLE RESPONSE 1 (CTR1) [6]–[8]. This negative regulator interacts with and directly phosphorylates the cytosolic C-terminal domain of EIN2 in Arabidopsis [9]. EIN2 is a central positive regulator of ethylene signalling that is localised to the endoplasmic reticulum membrane through its N-terminal domain [10]. The phosphorylation of EIN2 by CTR1 prevents EIN2 from signalling in the absence of ethylene, whereas the inhibition of CTR1 upon ethylene perception is a signal for cleavage and translocation of EIN2 from the cytoplasm to the nucleus [9], [11]–[13]. The transcription factors EIN3 and EIN3-LIKE 1 (EIL1) act downstream of EIN2, which is required for the ethylene-induced stabilisation of EIN3/EIL1 [14], [15]. EIN3 further activates the expression of ethylene-responsive genes in different physiological processes [15]–[21].
When emerging from the soil, newly etiolated Arabidopsis thaliana seedlings display long hypocotyls, apical hooks, and closed cotyledons. Exposure to light inhibits hypocotyl growth and promotes the greening and expansion of the cotyledons and leaves [22]. During this processes, light modulates multiple hormonal pathways, including those involving gibberellins, abscisic acid, auxin, brassinosteroids, cytokinins, and ethylene, to regulate these developmental changes [23]–[32]. Increasing evidence suggests that gibberellins and cytokinins regulate the accumulation of the light signalling component LONG HYPOCOTYL 5 (HY5), a basic leucine zipper (bZIP) transcription factor that acts downstream of the light signal and positively regulates the transcription of light-induced genes [33], [34]. Light promotes the accumulation of HY5 protein by inhibiting the accumulation of CONSTITUTIVELY PHOTOMORPHOGENIC 1 (COP1) in the nucleus [35], [36]. Furthermore, HY5 reportedly promotes photomorphogenesis, in part by modulating auxin, cytokinin, and gibberellins signalling [32], , revealing the integration of light and phytohormone signalling. In addition, a recent study reported that EIN3/EIL1 cooperate with phytochrome interacting factor 1 (PIF1) and COP1 to optimise the de-etiolation of Arabidopsis seedlings and demonstrated that ethylene plays a key role in the establishment of green seedlings upon exposure to light [19]. Importantly, ethylene also inhibits Arabidopsis hypocotyl elongation in the dark, whereas ethylene and its precursor 1-aminocyclopropane-1-carboxylate (ACC) increase hypocotyl elongation in light-grown Arabidopsis seedlings [24], [39], [40], and EIN3 transcriptionally activates two contrasting pathways: the PIF3-dependent growth-promoting pathway and an ethylene response factor 1 (ERF1)-mediated growth inhibiting pathway, to fine-tune ethylene-promoted hypocotyl growth in the light [39]. Moreover, ethylene regulates the biosynthesis, transport, and distribution of IAA during light-mediated hypocotyl growth, dependent on the effect of COP1 on gene transcription downstream of EIN3 [29], [40]. These findings indicate the regulation of seedling growth by ethylene-light interactions; however, it remains unclear how the ethylene and light signalling pathways are integrated at the protein level to achieve the conserved regulation of plant hypocotyl growth.
In this study, we found that ethylene enhances the movement of COP1 to the nucleus to degrade HY5 in the light, revealing that the COP1-HY5 complex is a novel integrator of light-ethylene interactions during hypocotyl growth. Our data show that ethylene regulates hypocotyl growth by reducing HY5 protein levels. More importantly, ethylene-promoted hypocotyl elongation was inhibited in the absence of COP1 function, and COP1 was found to be required for HY5 stability. Further, ACC affected the EIN3-dependent shuttling of COP1 between the nucleus and cytoplasm. These results reveal that the light-regulated COP1-HY5 cascade is involved in ethylene-promoted hypocotyl growth.
Results
HY5 mediates ethylene-promoted hypocotyl elongation in the light
Ethylene confers opposing effects on hypocotyl growth that are dependent on the conditions under which plants are grown (light or dark). Ethylene and its precursor ACC can stimulate hypocotyl growth in the light, but inhibit hypocotyl elongation in etiolated seedlings. HY5 is an important positive regulator of photomorphogenesis; the hy5 mutant displays a long hypocotyl when grown in the light, but a normal hypocotyl when grown in the dark [41]. A recent report revealed that HY5 binds to the promoters of cell elongation-related genes and recruits PKL/EPP1 through their physical interaction to regulate hypocotyl growth [42]. Interestingly, although treatment with 10 µM ACC diminished hypocotyl growth in dark-grown Col-0 and hy5 plants, ACC did not significantly promote hypocotyl growth in light-grown hy5 seedlings (Figure 1A–D). This result was confirmed in hy5-215 (Col-0 background) and hy5-ks50 (Ws background, Figure S1). In addition, the root of hy5 mutants responded normally to ethylene because ACC inhibited the growth of root as the same of Col-0 in the light or in the dark (Figure 1A and 1C, Figure S1A). These results indicate that HY5 plays an essential role in ethylene-promoted hypocotyl growth. As a key factor in photomorphogenic development, HY5 represents a convergence point for light and multiple hormone signalling pathways [29]; combined with our results, these observations suggest that HY5 participates in ethylene-promoted hypocotyl elongation in the light but not in the dark.
Fig. 1. HY5 is required for ethylene-promoted hypocotyl growth in the light. 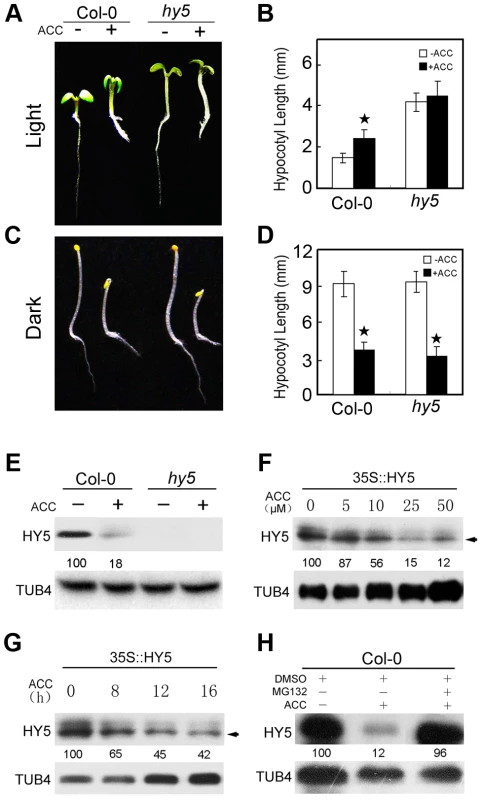
(A, C) Morphological observations and (B, D) statistical analyses of hypocotyl length. Images were taken after 5 days of incubation in MS medium supplemented with or without 10 µM ACC. The data indicate the mean values plus the standard deviation (SD) from three independent experiments with approximately 30 seedlings. P-values (ACC treatment vs. non-treatment) were calculated by a two-tailed Student's t-test assuming equal variances (*P<0.05). (E) HY5 accumulation in Col-0 and hy5. Protein extracts were prepared from 5-day-old seedlings after treatment with or without 25 µM ACC for 15 h. (F) The effects of different ACC concentrations on HY5 stability in 35S::HY5 seedlings. (G) A time course of HY5 stability in response to ACC treatment in 35S::HY5 seedlings. Five-day-old seedlings were treated either with the indicated concentrations of ACC treatment for 16 h or with 25 µM ACC for the indicated times. The arrow in (F) and (G) indicates endogenous HY5, whereas the upper band corresponds to transgenic HY5. (H) Effect of the 26S proteolysis inhibitor MG132 on HY5 protein accumulation in Col-0 under normal growth conditions. Five-day-old seedlings were treated with 25 µM ACC plus 0.1% DMSO or 5 µM MG132 for 15 h. Immunoblotting was performed with anti-HY5 and -TUB4 antibodies. The TUB4 signals confirm equal protein loading. Numbers indicate the relative protein levels of HY5. To confirm the function of HY5 in regulating hypocotyl growth, we generated a hybrid of the ethylene overproducing mutant eto2 [43] and transgenic plants constitutively expressing HY5 (35S::HY5) [44]. As shown in Figure S2A and S2B, eto2 displayed longer hypocotyls than Col-0 plants, while the hypocotyl length of eto2×35S::HY5 was intermediate between that of eto2 and Col-0. The long hypocotyl caused by endogenous ethylene overproduction was diminished by the constitutive expression of HY5 (Figure S2), indicating that HY5 negatively regulates ethylene-promoted hypocotyl growth in the light.
Ethylene suppresses HY5 accumulation in a dose - and time-dependent manner
To determine how ethylene modulates HY5 accumulation, we first examined whether ethylene transcriptionally activates HY5 expression. The expression levels of HY5 and its homologue HYH were similar in Col-0, eto, ein2, and ein3-1 under normal growth conditions (Figure S3), suggesting that ethylene does not transcriptionally regulate HY5 expression. We next detected HY5 protein accumulation in Col-0 and hy5 mutant plants. There was no band detected in hy5 mutant while ACC greatly reduced HY5 protein accumulation in Col-0 (Figure 1E), suggesting that ethylene mediates an increase in HY5 instability at the post-transcriptional level to regulate hypocotyl growth.
To further understand how ethylene suppresses HY5 accumulation, we analysed the stability of HY5 in the transgenic line 35S::HY5. Because ACC induces hypocotyl growth in a dose-dependent manner [24], we first examined the induction of HY5 degradation by ethylene using different concentrations of ACC. Treatment with 5, 10, or 25 µM ACC for 16 h slightly, greatly, and significantly suppressed HY5 accumulation in 35S::HY5, respectively. When the applied ACC concentration increased, HY5 protein accumulation gradually decreased (Figure 1F). Furthermore, we observed that treatment with ACC for 8 h resulted in obvious degradation of HY5; when the treatment was increased to 16 h, the HY5 protein level was further reduced (Figure 1G), suggesting that ethylene significantly suppresses HY5 stability in a dose - and time-dependent manner. We also found that treatment with the 26S protein degradation inhibitor MG132 prevented the decrease in HY5 protein levels caused by ACC treatment (Figure 1H), suggesting that ethylene regulates HY5 stability via 26S proteasome-mediated degradation.
HY5 acts downstream of EIN3 in ethylene-promoted hypocotyl elongation
To further study the mechanism of ethylene-promoted hypocotyl elongation in the light, we examined the hypocotyl lengths of ethylene signalling mutants. Similar to eto2, the constitutive ethylene response mutant ctr1-1 displayed longer hypocotyls than Col-0 or the ethylene-insensitive mutants etr1-1, ein2, ein3-1, and ein3-1 eil1. Unlike Col-0, the hypocotyl lengths of etr1-1, ein2, ein3-1, and ein3-1 eil1 did not increase following treatment with 10 µM ACC (Figure 2A and 2B). An analysis of HY5 expression in these mutants by Western blotting produced results that were consistent with those of our hypocotyl elongation assays (Figure 2C). Namely, HY5 accumulation was reduced in eto2 and ctr1-1 compared to Col-0 in the absence of ACC, and ACC treatment enhanced the decrease in HY5 protein, especially in ctr1-1. However, in etr1-1, ein2, ein3-1, and ein3-1 eil1, only slight differences from Col-0 were observed, and ACC did not promote HY5 degradation (Figure 2C). Consistent with the hypocotyl lengths shown in Figure 2A and 2B, these results indicate that HY5 acts downstream of ethylene signalling to regulate hypocotyl growth.
Fig. 2. Ethylene signalling positively promotes hypocotyl growth and HY5 degradation. 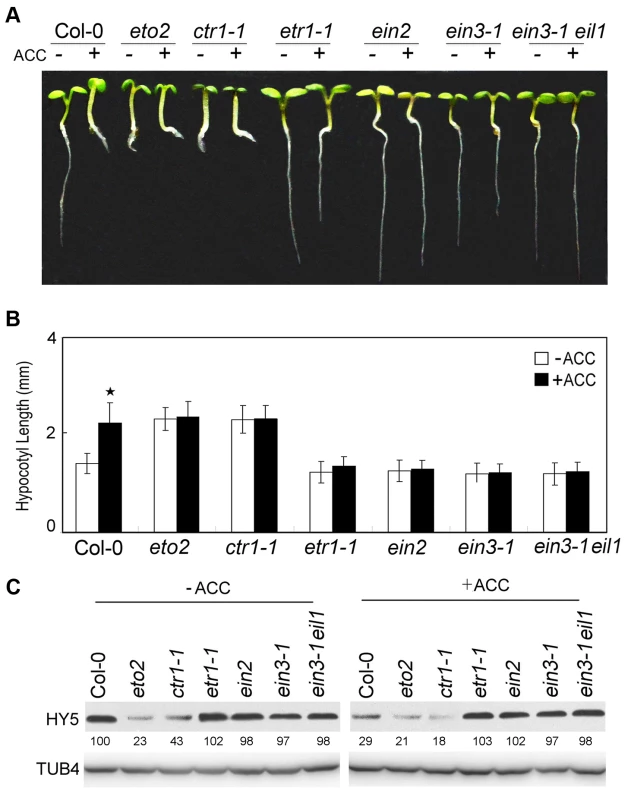
(A) Morphological observations and (B) statistical analyses of hypocotyl length after 5 days of incubation in MS medium supplemented with or without 10 µM ACC. The data indicate the mean values plus the SD from three independent experiments with approximately 30 seedlings. P-values (ACC treatment vs. non-treatment) were determined with a two-tailed Student's t-test assuming equal variances (*P<0.05). (C) HY5 accumulation after treatment with or without ACC treatment. Protein extracts were prepared from 5-day-old seedlings after treatment with or without 25 µM ACC for 15 h. Immunoblotting was performed with anti-HY5 and -TUB4 antibodies. The TUB4 signals indicate equal protein loading. Numbers indicate the relative protein levels of HY5. We next analysed the genetic relationship between HY5 and ethylene signalling. The hypocotyl lengths in the etr1-1 hy5, ein2 hy5, and ein3-1 hy5 double mutants were obviously longer than that in Col-0 and similar to that in hy5 (Figure 3A and 3B). Taken together with the finding that ACC did not promote the degradation of HY5 in ein3-1 (Figure 2C), our data indicate that HY5 acts downstream of EIN3 to mediate ethylene-promoted hypocotyl growth.
Fig. 3. HY5 operates downstream of EIN3 to modulate ethylene-promoted hypocotyl growth. 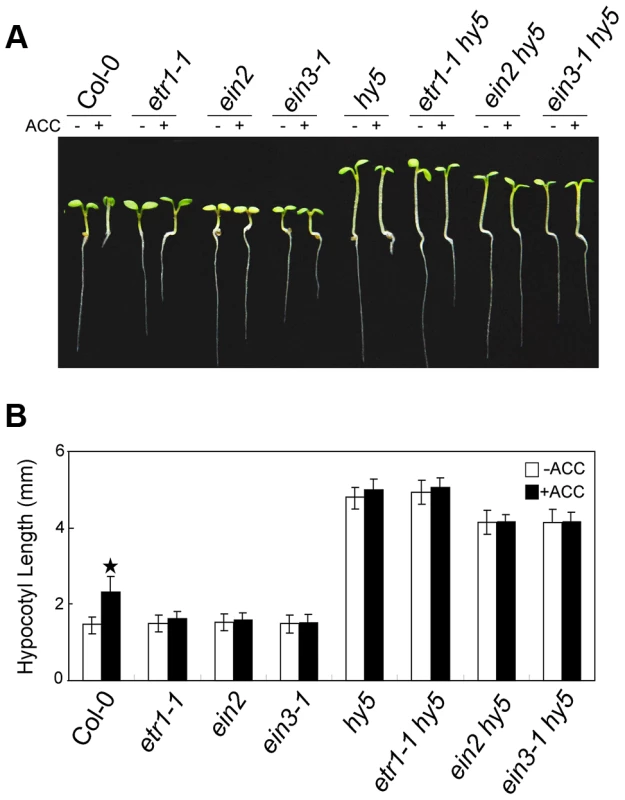
(A) Morphological observations and (B) statistical analyses of hypocotyl length after 5 days of incubation in MS medium supplemented with or without 10 µM ACC. The data indicate the mean values plus SD from three independent experiments with approximately 30 seedlings. P-values (ACC treatment vs. non-treatment) were determined with a two-tailed Student's t-test assuming equal variances (*P<0.05). COP1 mediates ethylene-promoted HY5 degradation
COP1 is an E3 ligase that negatively modulates HY5 activity via protein-protein interactions [33], [36]. COP1 has three structural domains that are critical for its molecular association with HY5: an N-terminal RING-finger domain followed by a predicted coiled-coil domain and C-terminal WD-40 repeats [33]. For the regulation of HY5 stability by ethylene through 26S proteasome-mediated degradation, we hypothesised that COP1 may also function in this process. Therefore, we measured the ethylene response in Arabidopsis cop1 mutants (cop1-4 and cop1-6, both in Col-0 background) and transgenic lines. It has previously evidenced that the cop1-4 mutation alters the Gln-283 CAA codon to a UAA stop codon, resulting in a truncated COP1 protein containing only the N-terminal 282 amino acids, and the cop1-6 mutation changes the splicing junction “AG” at the 3′ end of intron 4 to “GG”, generating a five-amino acid insertion in the COP1 protein [45]. Interestingly, those cop1 mutants that reduced activity of COP1 displayed shorter hypocotyls than Col-0, and ACC failed to promote hypocotyl elongation in cop1-4 or cop1-6, whereas the full-length COP1 overexpressor (GUS-COP1) displayed longer hypocotyls than Col-0 (Figure 4A and 4B). These results indicate that COP1 positively regulates ethylene-promoted hypocotyl growth.
Fig. 4. COP1 is required for ethylene-promoted hypocotyl growth. 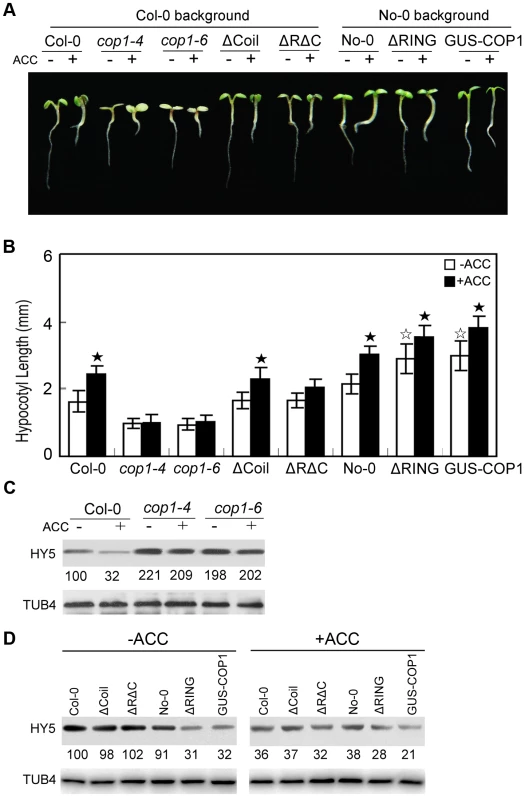
(A) Morphological observations and (B) statistical analyses of hypocotyl length after 5 days of incubation in MS medium supplemented with or without 10 µM ACC. The data indicate the mean values plus the SD from three independent experiments with approximately 30 seedlings. P-values (★: ACC treatment vs. non-treatment, ☆: transgenic lines vs. wild type) were determined with a two-tailed Student's t-test assuming equal variances (*P<0.05). (C, D) HY5 accumulation with or without ACC treatment. Protein extracts were prepared from 5-day-old seedlings after treatment with or without 25 µM ACC for 15 h. Immunoblotting was performed using anti-HY5 and -TUB4 antibodies. The TUB4 signals indicate equal protein loading. Numbers indicate the relative protein levels of HY5. The RING-finger domain, coiled-coil domain, and WD-40 repeats of COP1 are essential for its interaction with HY5 [33], [46]. Accordingly, a transgenic line overexpressing COP1 without the RING-finger domain (ΔRING, No-0 background) showed significantly longer hypocotyls than the wild type, whereas a transgenic line overexpressing COP1 without the coiled-coil domain (ΔCoil, Col-0 background) displayed a similar hypocotyl length to the wild type in the absence of ACC under white light (Figure 4A and 4B). Distinctive to the cop1 mutant, ACC significantly enhanced hypocotyl growth in the ΔRING and ΔCoil transgenic lines, similar to the effect of ACC on their corresponding controls (Figure 4A and 4B), suggesting the redundant function of the RING-finger and coiled-coil domains in hypocotyl growth. Furthermore, transgenic line overexpressing COP1 without the RING-finger and coiled-coil domains (ΔRΔC, Col-0 background) did not show obvious hypocotyl elongation compared to Col-0 in the absence of ACC under white light, and ACC did not significantly improve hypocotyl growth in the ΔRΔC line (Figure 4A and 4B). Thus the above observations indicate that mutation of protein-protein interaction domains of COP1 alters ethylene responsive hypocotyl elongation.
To elucidate the mechanism through which ethylene suppresses HY5 accumulation, we analysed the HY5 protein levels in the cop1 mutants and transgenic lines. As expected, HY5 accumulation was detected in cop1-4 and cop1-6, while ACC-induced HY5 degradation was inhibited in cop1-4 and cop1-6 (Figure 4C), revealing the essential role of COP1 in ethylene-promoted hypocotyl growth and HY5 stability. Moreover, the abundance of HY5 in these transgenic lines was correlated with the observed extent of photomorphogenic development. When grown under the same white light conditions, GUS-COP1 and ΔRING had reduced while ΔCoil and ΔRΔC showed similar levels of HY5 protein relative to wild type, and ACC treatment enhanced the degradation of HY5 in these transgenic lines (Figure 4D), indicating that the ACC-induced suppression of HY5 is mediated by the E3 ligase COP1, dependent on the interaction of COP1 with HY5.
Because HY5 is not the unique targets of COP1 [47]–[49], then we further questioned whether the role of HY5 in ethylene-promoted growth is specifically targeted by the interaction with COP1. To address this query, we performed assays of ethylene responsiveness using transgenic lines of the deletion of N-terminal 77 amino acids of HY5 (HY5-ΔN77) in Col-0 or hy5 background, which has been evidenced that this fragment is essential for the interaction of HY5 with COP1, and HY5-ΔN77 transgene is being driven by the CaMV35S promoter [33]. Transcriptional detections with quantitative real-time PCR (qPCR) showed that the expression of truncated HY5 (deletion of the N-terminal 77 amino acids) or full-length HY5 was overexpressed in individual transgenic lines (Figure S4). Further observations showed that the transgenic lines of HY5-ΔN77 displayed shortened-hypocotyl in Col-0 background, but the HY5-ΔN77 partially complemented the hy5 long hypocotyl phenotype. More importantly, ACC did not significantly promote the elongation of hypocotyl in HY5-ΔN77/Col-0 and HY5-ΔN77/hy5 (Figure 5). These results indicate that the COP1-HY5 interaction is required for the degradation of HY5 by ethylene, and subsequent hypocotyl growth.
Fig. 5. COP1-HY5 interaction is required for HY5-mediated hypocotyl growth. 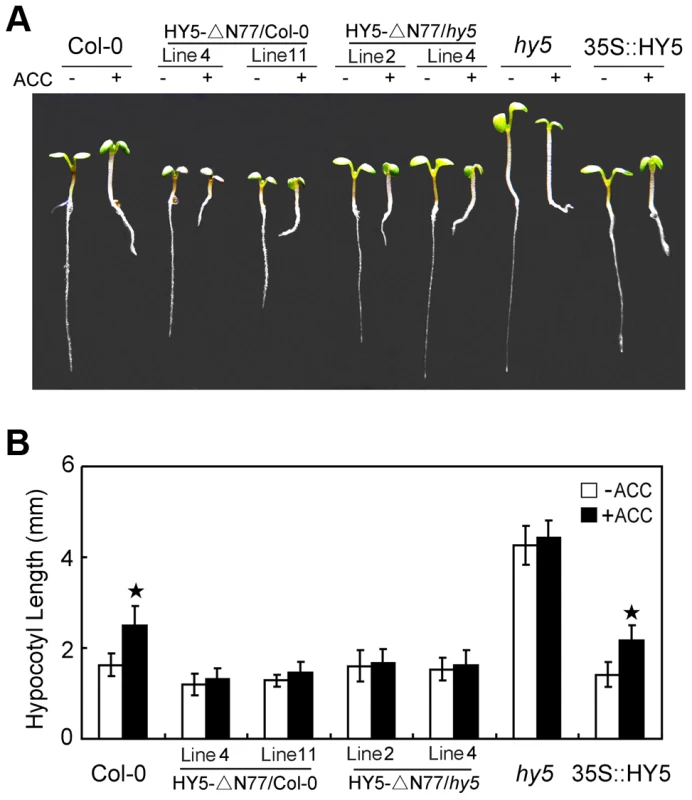
(A) Morphological observations and (B) statistical analyses of hypocotyl length in transgenic lines of HY5-ΔN77 driven by the CaMV35S promoter. The images were taken after 5 days of incubation in MS medium supplemented with or without 10 µM ACC. The data indicate the mean values plus the SD from three independent experiments with approximately 30 seedlings. P-values (ACC treatment vs. non-treatment) were determined with a two-tailed Student's t-test assuming equal variances (*P<0.05). We then generated eto2 cop1-4 and ctr1 cop1-4 double mutants to determine whether ethylene-mediated hypocotyl elongation is COP1-dependent. Both of the mutants displayed hypocotyl lengths that were similar to those of cop1-4 and were not affected by ACC treatment (Figure S5). To investigate the relationship between COP1 and ethylene signalling, we evaluated the hybrids EIN3ox×cop1-4 and ein3-1×GUS-COP1. Both EIN3ox and GUS-COP1 displayed longer hypocotyls than the controls, indicating that EIN3 and COP1 positively regulate ethylene-promoted hypocotyl growth. However, EIN3ox×cop1-4, which lacked COP1, displayed a short hypocotyl phenotype that was similar to that of cop1-4, and ein3-1×GUS-COP1, which lacked EIN3, showed the same long hypocotyl phenotype as GUS-COP1 (Figure 6A and 6B), indicating that COP1 might be genetically downstream of EIN3. This conclusion was supported by Western blotting analyses: as shown in Figure 6C, twice as much HY5 protein accumulated in EIN3ox×cop1-4 as in cop1-4, and this accumulation was not affected by ACC treatment. Correspondingly, the HY5 level in ein3-1×GUS-COP1 was dramatically reduced compared to that in the controls (Figure 6C), consistent with the function of HY5, COP1 likely acts downstream of EIN3 in ethylene-promoted hypocotyl elongation. Thus, our results demonstrate that the ethylene-enhanced degradation of HY5 is mediated by COP1 and that the COP1-HY5 complex acts downstream of EIN3 to mediate ethylene-promoted hypocotyl growth.
Fig. 6. COP1 acts downstream of EIN3 to modulate ethylene-promoted hypocotyl growth. 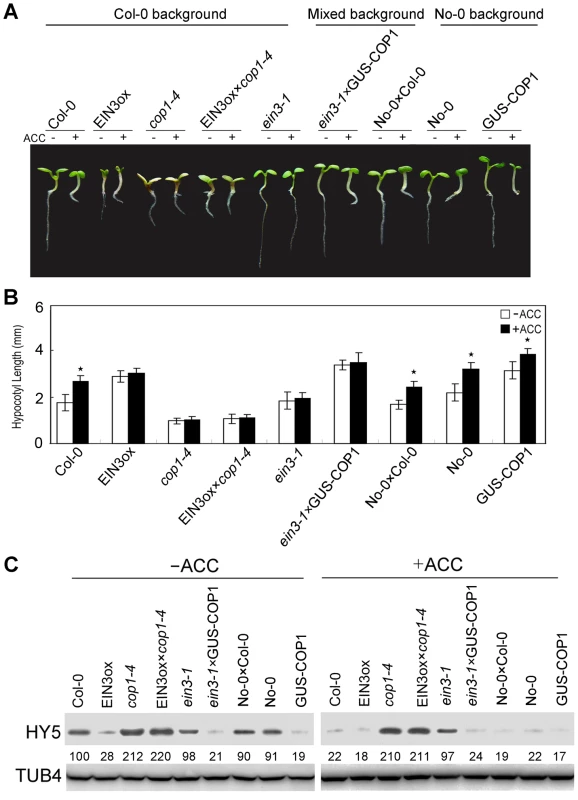
(A) Morphological observations and (B) statistical analyses of hypocotyl length after 5 days of incubation in MS medium supplemented with or without 10 µM ACC. The data indicate the mean values plus the SD from three independent experiments with approximately 30 seedlings. P-values (ACC treatment vs. non-treatment) were determined with a two-tailed Student's t-test assuming equal variances (*P<0.05). (C) HY5 accumulation with or without ACC treatment. Protein extracts were prepared from 5-day-old seedlings after treatment with or without 25 µM ACC for 15 h. Immunoblotting was performed with anti-HY5 and -TUB4 antibodies. The TUB4 signals indicate equal protein loading. Because the ein3-1 and GUS-COP1 were produced in different backgrounds, an F1 hybrid of No-0×Col-0 was used as the control. The numbers below the gel indicate the protein levels of HY5. To further confirm the ethylene responsiveness of the physical interaction between COP1 and HY5, we examined the expression of the ethylene-regulated genes ERF1 [16], ESE1 [21], and CHIB [50] in hy5. Our data show that the expression of ERF1, ESE1, and CHIB was inhibited in cop1-4, but increased in hy5 (Figure S6), indicating the participation of COP1-HY5 not only in ethylene-promoted hypocotyl growth, but also in ethylene-regulated gene expression.
Interestingly, we did not observe an obvious difference in root length between Col-0 and the ethylene signalling mutants in the absence of ACC. In particular, root growth in the mutants eto2 and ctr1-1 was constitutively inhibited. However, ACC significantly diminished root growth in Col-0, but not in ein2 or the ein3 eil1 double mutant, and it partially inhibited root growth in etr1-1 and ein3-1 (Figure S7A), demonstrating the essential role of ethylene in the regulation of root growth. However, the root length in the hy5 or cop1 mutants and the transgenic lines expressing truncated versions of COP1 was reduced by treatment with ACC (Figure S7B), indicating that the COP-HY5 complex mainly takes part in ethylene-regulated hypocotyl growth rather than root growth.
Ethylene enhances COP1 nuclear localisation
COP1 activity has been correlated with its movement between the cytoplasm and nucleus [35]. COP1 is excluded from the nucleus in the light, whereas in the dark or shade it accumulates in the nucleus and directs its targets, including HY5, to the proteasomal degradation machinery [36], [47]–[49], [51], [52]. This nucleocytoplasmic partitioning of COP1 is required to promote HY5 protein accumulation [53], [54]. To determine whether ethylene influences COP1 localisation to the nucleus, we quantified the nuclear and cytoplasmic localisation of COP1 in a transgenic line that constitutively expresses a GUS-COP1 fusion (GUS-COP1). GUS was mainly localised to the nucleus in the dark, but it translocated to the cytosol upon light irradiation in the absence of ACC (Figure 7A and 7B), consistent with previous data [35]. Importantly, ACC treatment did not induce obvious changes in GUS localisation in the dark, whereas under continuous white light for 24 h, ACC treatment enhanced the nuclear localisation of GUS-COP1 (Figure 7A), suggesting that ACC affects the distribution of COP1 and hypocotyl growth in the light rather than in the dark. Further, we used 4-day-old seedlings grown in long days (16/8 h), and then the plants were shifted to continuous white light for 36–48 h to conduct experiments. The level of GUS staining in the nucleus did not change following treatment with ACC under continuous white light (Figure S8). Moreover, under an 8-h/16-h dark/light cycle approximately 18% of the hypocotyl cells exhibited nucleus-enriched GUS staining in the absence of ACC; however, after addition of ACC under the same growth conditions, the proportion of cells with nucleus-enriched GUS staining increased to 58% (Figure S9A–D). These results indicate that ACC reversed the localisation of COP1 in the light and further enhanced HY5 degradation.
Fig. 7. The ethylene precursor ACC improves COP1 nucleus-enriched localisation in the light. 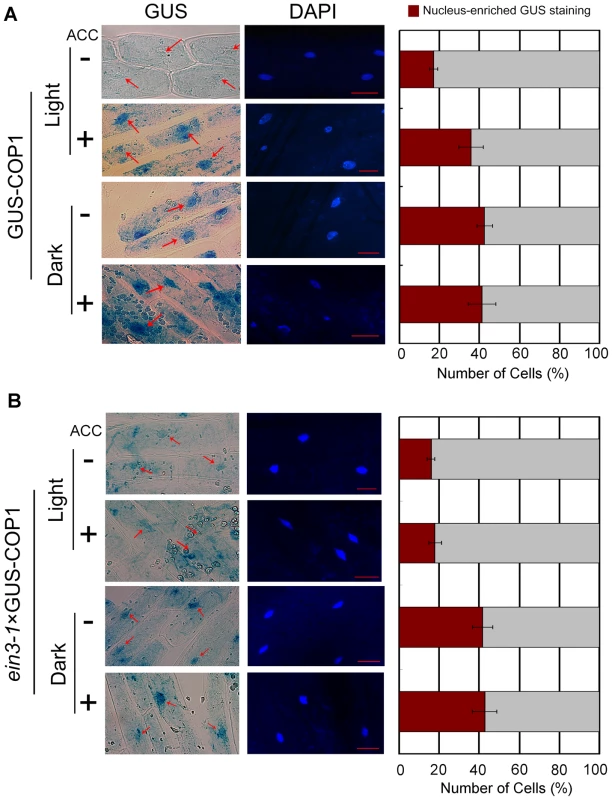
Images (left panel) and statistical summaries (right panel) of GUS-COP1 localisation in the hypocotyl in Col-0 (A) and ein3-1 (B) under different growth conditions. The degree of nuclear enrichment of GUS staining is shown as the percentage of cells with nucleus-enriched GUS relative to the total number of GUS-stained hypocotyl cells. At least 100 cells were counted for each sample. GUS-COP1 transgenic seedlings were first grown on MS for 5 days and then grown in the dark (labelled “Dark”) or under continuous white light (50 µmol/m2s; labelled “Light”) for another 24 h with or without 25 µM ACC. The cell nuclei were stained with 0.5 µg/mL DAPI. Scale bar: 5 µm. Because COP1 is a downstream component of EIN3, we examined whether ethylene modulates COP1 localisation via the action of EIN3. To address this issue, we constructed a hybrid of GUS-COP transgenic line using the ein3-1 mutant to detect GUS-COP1 shuttling between the cytoplasm and nucleus and grew the hybrid under continuous white light for 24 h. Our results indicate that the loss of function of EIN3 in the ein3×GUS-COP1 hybrid disabled the shuttling of COP1 to the nucleus promoted by ACC. In comparison, the shuttling of COP1 in ein3×GUS-COP1 was controlled by darkness (Figure 7B). These results demonstrate that ethylene modulates COP1 localisation via the action of EIN3.
To directly demonstrate that ACC promotes the shuttling of COP1 between the nucleus and cytoplasm, we performed cell fractionation experiments using 5-day-old Col-0 plants grown under an 8-h/16-h dark/light cycle. To exclude the influence of de novo protein production and degradation during the process, we supplemented with cycloheximide (CHX) and MG132 with or without ACC in the light for 15 h. Our results showed that COP1 was mainly localized in the cytoplasm in the light without ACC treatment, while it shuttled to the nucleus in the presence of ACC (Figure 8A), evidencing that ACC enriches the nuclear distribution of COP1 in the light. Therefore our data suggest that ACC promotes COP1 translocation to the nucleus, resulting in the subsequent degradation of HY5 via 26S proteolysis. This experiment was also performed using ein2 and ein3-1 plants. In accordance with the localisation of GUS-COP1 in the ein3-1 background, COP1 was only weakly detected in the nucleus with or without ACC (Figure 8B), demonstrating that ethylene-regulated COP1 shuttling occurs downstream of EIN3.
Fig. 8. Biochemical detection of COP1 in the cytoplasm and nucleus. 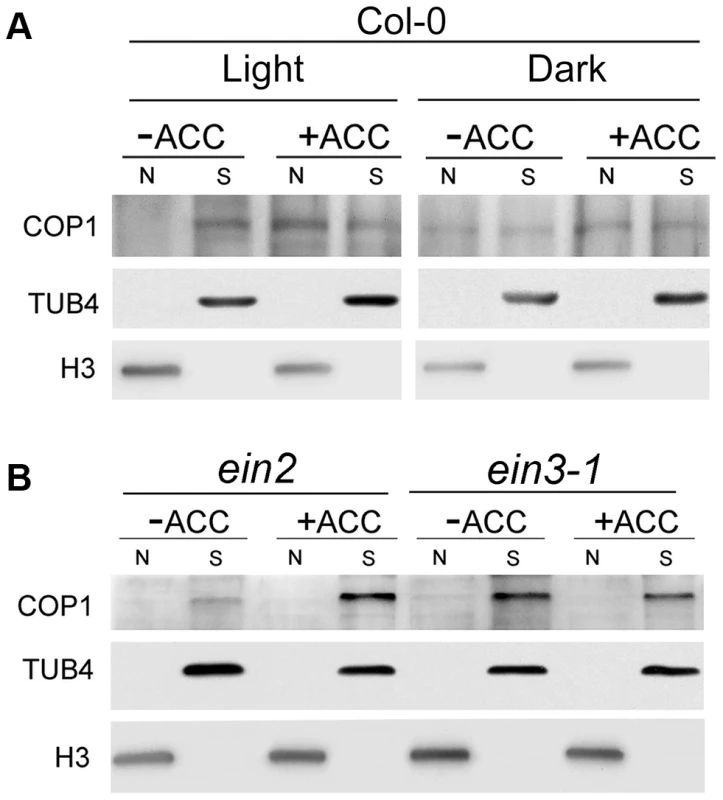
Seedlings of five-day-old (A) Col-0 and (B) ein2 and ein3-1 Arabidopsis plants grown under long-day conditions (16-h light/8-h dark) were treated with 50 µM CHX and 5 µM MG132, supplemented with or without 25 µM ACC for 15 h, and then the nuclear proteins were extracted as described in the Materials and methods (N: nuclear protein, S: soluble fraction, cytoplasmic protein). Immunoblotting was performed using anti-COP1, -histone 3 (H3), and -TUB4 antibodies. The TUB4 and H3 signals confirm equal protein loading. All of the seedlings were illuminated with white light (50 µmol/m2s). Discussion
To understand the interactions between environmental cues and phytohormones, it is critical to elucidate the coordination of ethylene and light signalling during seedling hypocotyl growth. A recent study showed that EIN3 transcriptionally activates the expression of PIF3 and ERF1 to integrate these pathways [39], revealing the transcriptional regulation of a light regulator by ethylene. The bZIP transcriptional regulator HY5 has been shown to be a key factor in hypocotyl growth through its interaction with COP1 as a photomorphogenic repressor [33], [44]. In the present report, we demonstrated that the light regulators COP1 and HY5 are essential for ethylene-promoted hypocotyl growth in the light but not in the dark, and that these proteins act downstream of the ethylene signalling pathway component EIN3. Importantly, the ethylene-dependent activity of EIN3 enhanced the shuttling of COP1 between the nucleus and cytoplasm to promote HY5 degradation. Therefore, to our knowledge, this study presents the first evidence that the COP1-HY5 complex integrates ethylene-promoted hypocotyl growth in the light, increasing our understanding of the mechanism by which light suppresses (while ethylene promotes) hypocotyl elongation in the light.
Ethylene suppresses hypocotyl growth through the ethylene-induced gene ERF1 whereas it promotes hypocotyl elongation downstream of EIN3 via PIF3 in the light [39]. This is consistent with the finding that PIF3 and HY5 independently regulate PHYB-mediated hypocotyl elongation inhibition [55]. Moreover, regulation of the biosynthesis, transport, and distribution of IAA by ethylene was observed during light-mediated hypocotyl growth, further confirming the transcriptional regulation of hypocotyl growth by ethylene [29], [40]. In addition, increasing evidence indicates that the components of the ethylene signalling pathway play critical roles in plant growth and development [56]–[58]. In the present investigation, we demonstrated that ethylene-promoted hypocotyl growth in the light, but not in the dark, is mediated by the photomorphogenic factor HY5. The hypothesis that HY5 functions downstream of the ethylene signalling pathway is further supported by biochemical assays demonstrating that, under normal growth conditions, HY5 protein levels were not altered in loss-of-function ethylene signalling mutants but were significantly depressed in the ctr1-1 background. Moreover, the addition of ACC greatly reduced HY5 accumulation in eto2, ctr1-1, and transgenic EIN3ox plants but not in etr1-1, ein2, ein3, and ein3-1 eil1 plants. Therefore, the results of our genetic and phenotypic analyses and biochemical assays suggest that HY5 represents a novel negative regulator that functions downstream of the ethylene signalling component EIN3 and participates in ethylene-promoted hypocotyl growth.
The direct interaction between COP1 and CSN1 reportedly stimulates the nuclear localisation of COP1 [59], and the plant CSN-interacting F-box protein COP9 INTERACTING F-BOX KELCH 1 contributes to the control of hypocotyl elongation [60]. Indeed, several components of the photomorphogenic response, including CSN3, CSN6A, and CSN6B, interact with EIN2. It has been reported that EIN2 probably functions to direct the activity of the CSN through ENHANCED ETHYLENE RESPONSE 5, which is part of the ethylene response pathway [61], suggesting that photomorphogenic factors are involved in ethylene signalling. Importantly, we found that the EIN3-dependent nuclear localisation of COP1 could be enhanced by ethylene in the light but not in the dark, suggesting that ethylene affects the shuttling of COP1 to the nucleus. The cop1 mutant has been shown to display a photomorphogenic response in the dark but a short hypocotyl in the light [35]. In the present study, exogenous ACC and endogenous ethylene stimulated hypocotyl growth, but this stimulation was not observed in cop mutant plants. These results suggest that ethylene-mediated hypocotyl growth is COP1-dependent, consistent with the finding that COP1 is involved in the ethylene-promoted photomorphogenic response [19], [40]. We found that cop1 mutants have shorter hypocotyls, while eto2 and ctr1 display longer hypocotyls, in the light. The short hypocotyl lengths of the eto2 cop1-4 and ctr1 cop1-4 double mutants, however, were similar to that of the cop1-4 mutant, and this phenotype was unaffected by the addition of ACC. Furthermore, genetic analyses and biochemical assays showed that the hypocotyl length and HY5 stability in the hybrids EIN3ox×cop1-4 and ein3-1×GUS-COP1 were consistent with the function of COP1, indicating that COP1 acts downstream of the ethylene signalling component EIN3, similar to the manner in which HY5 is regulated by EIN3 during hypocotyl growth.
Interestingly, we also found that ACC enhanced the movement of COP1 to the nucleus in root cells upon light illumination (Figure S9E and S9F), suggesting the general regulation of the movement of COP1 in the light by ethylene. However, root elongation in hy5, cop1, and Col-0 seedlings was not significantly affected by ACC treatment, suggesting that the effect of ACC/ethylene on root growth is independent of COP1-HY5. Moreover, the length of the root in the ethylene signaling mutants was similar to that in Col-0 in the absence of ACC treatment. After ACC treatment, root growth was completely abolished in Col-0, but it was similar in terms of length in ein2 and ein3-1 eil1 with the control, implying that ethylene is essential for root growth. Reflecting the redundancy of EIN3 and EIL1, root growth in ein3-1 and the gain-of-function mutant etr1-1 was partially inhibited, compared to the corresponding controls. This observation suggests that the regulatory role of ethylene in hypocotyl growth is different from that in root elongation, further supporting the regulation of HY5 in hypocotyl growth rather than in root growth. Thus, the involvement of HY5 in the ethylene response might be an organ-specific effect.
Importantly, the data from our phenotypic and biochemical analyses indicate that a loss of function of CTR1 constitutively improves hypocotyl growth by decreasing the level of HY5, while the hypocotyl length in ctr1 is similar to that in ACC-treated Col-0, but shorter than that in hy5, suggesting that other factors coordinate with ethylene to affect HY5 protein levels. This hypothesis is supported by the observation that ACC moderately induces but does not significantly improve growth of the hypocotyl in the hy5 mutant, consistent with previous observations [40], indicating that ethylene coordinates with other phytohormones, including auxin, to affect hypocotyl growth [62], [63].
The finding that ethylene promotes hypocotyl elongation in the light by increasing nuclear-localised COP1, resulting in a decreased level of HY5, combined with the observation that the effect of ethylene on HY5 protein levels was abolished in etr1-1, ein2, ein3-1, and ein3/eil1, indicates that the effect of ethylene on hypocotyl growth requires transcriptional regulation by EIN3. This hypothesis is supported by the following observations. EIN3 overexpression decreased the HY5 protein level in a Col-0 background but not in a cop1-4 background, whereas COP1 overexpression reduced the HY5 level irrespective of EIN3. Moreover, the loss of function of EIN3 abolished the movement of COP1 in the light, suggesting that EIN3 is required for the COP1-mediated decrease in HY5. Therefore, based on the present data and previous reports showing that ethylene promotes EIN3/EIL1 accumulation whereas high EIN3/EIL1 levels are detrimental to plant growth and development [14], [15], we propose that COP1-HY5 functions as a regulatory module in ethylene-promoted hypocotyl growth. In the absence of ethylene, COP1 accumulates mainly in the cytoplasm in the light, releasing the suppression of COP1 on HY5. In the presence of ethylene, this effect is reversed, even though in the light the nucleus is enriched with COP1, resulting in the degradation of HY5. Ethylene production is negatively regulated by HY5 [64], [65]; decreases in HY5 accumulation enhance the synthesis of ethylene. Thus, in the present study, we found that the COP1-HY5 complex acts as an integrator downstream of EIN3 to fine-tune the regulation of hypocotyl growth by light and ethylene. Although previous data [19], [39], [40] and our findings suggest that cotyledon greening and hypocotyl growth are COP1-related, multiple factors downstream of COP1 may participate in these growth processes. Additional genetic and biochemical tests should be performed to better understand this integration.
Materials and Methods
Plant materials and growth conditions
All Arabidopsis thaliana mutants and transgenic lines were generated in the Columbia (Col-0) background with the exception of overexpression line of GUS-COP1, overexpressing COP1 without the RING-finger domain (ΔRING, Nossen, No-0 background) and hy5-ks50 (Wassilewskija, Ws background). Homozygous eto1-1 (CS3072), eto2 (CS8059), eto3 (CS8060), etr1-1 (CS237), ctr1-1 (CS8057), ein2 (CS8844), ein3-1 (CS8052), and hy5 (SALK_096651) lines were obtained from the Arabidopsis Biological Resource Center (Columbus, OH). The sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: COP1 (At2g32950), HY5 (At5g11260), HYH (At3g17609), ETR1 (Ag1g66340), CTR1 (At5g03730), EIN2 (At5g03280), EIN3 (At3g20770), EIL1 (At2g27050), ERF1 (AT3G23240), ESE1 (AT3G23220), and CHIB (AT3G12500). The mutants or transgenic plants hy5-215 and hy5-ks50 [41]; cop1-4 and cop1-6 [45]; ein3-1 eil1, EIN3ox, and EIN3ox×cop1-4 [19]; 35S::HY5 [44]; ΔRING, ΔCoil, and ΔRΔC [46]; HY5-ΔN77/Col-0 and HY5-ΔN77/hy5 driven by the CaMV35S promoter [33] were generated previously. Seeds were surface-sterilised and sown on Murashige and Skoog (MS) medium containing 3% sucrose and 0.5% Phytagel. The plates were chilled at 4°C in the dark for 3 days and then moved to 22°C under a 16-h white light (50 µmol/m2s)/8-h dark cycle. All of the chemicals used were obtained from Sigma-Aldrich (St. Louis, MO).
RNA extraction, reverse transcription, and quantitative real-time PCR (qPCR)
Total RNA was extracted from 5-day-old seedlings using TRIzol reagent (Invitrogen, Carlsbad, CA) and treated with RNase free-DNase I (Promega, Madison, WI) before the latter procedures were performed. Five micrograms of total RNA were reverse-transcribed to cDNA with M-MLV reverse transcriptase (Promega) according to the manufacturer's instructions. Gene expression was measured by qPCR analysis (SYBR Premix; Takara Bio Inc., Shiga, Japan). All amplification reactions were performed in 96-well optical reaction plates with 45 cycles of denaturation for 15 s at 95°C, annealing for 20 s at 56°C, and extension for 45 s at 72°C. The expression levels were normalised to that of TUB4. The primers used for qPCR are listed in Table S1. Each qPCR was repeated independently three times with the same expression pattern.
Hypocotyl measurements
For the measurement of hypocotyl length, surface-sterilised seeds were deposited on plates containing MS medium with or without ACC. The plant materials and growth conditions were described in the previous section. At least 50 seedlings were measured from digital images of 5-day-old seedlings using ImageJ software.
Western blot analysis
Proteins were extracted from approximately 50 seedlings treated with or without ACC in 100 µL of extraction buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 1 mM EDTA, 10 mM NaF, 25 mM β-glycerophosphate, 2 mM sodium orthovanadate, 10% glycerol, 0.1% Tween 20, and 1 mM PMSF). After centrifugation, 10 µg of the supernatant was separated via SDS-PAGE and Western blotting was performed as described previously [36] using anti-HY5 antibodies (1∶500). TUB4 (1∶5,000) was used as a loading control.
The relative protein levels of HY5 were calculated using Image Gauge V3.12 (Fujifilm, Tokyo, Japan). The values were normalised to 100 in Col-0 without ACC treatment based on the TUB4 signal as an internal loading control.
Genetic manipulation
35S::HY5 and eto2 plants were crossed, and putative homozygous F2 progeny were screened for kanamycin resistance during seed germination. Kanamycin-resistant homozygous eto2 seedlings were identified as described previously [66]. Non-segregating lines of the progeny of the eto2 homozygous kanamycin-resistant plants were screened to identify homozygous hybrids of eto2×35S::HY5.
The double mutants ein2 hy5, ein3-1 hy5, and etr1-1 hy5 were generated by crossing the recessive ethylene mutant ein2, ein3-1, or etr1-1 with hy5. The F2 progeny of the crosses were grown in the dark for 4 days with 10 µM ACC to isolate ethylene-insensitive individuals that were homozygous for ein2, ein3-1, or etr1-1. The isolated individuals were subsequently screened for the presence of long hypocotyls, and the homozygous hy5 mutations were confirmed by PCR-based genotyping.
The double mutants eto2 cop1-4 and ctr1-1 cop1-4 were generated by crossing the ethylene mutant eto2, or ctr1-1 with cop1-4. The F2 progeny from the crosses were grown in the dark for 4 days to isolate homozygous cop1-4 lines by the phenotype. Individuals were subsequently screened by PCR-based genotyping, and the PCR products were digested with AvaI (New England Biolabs, Ipswich, MA) for eto2 or Tsp509I (New England Biolabs) for ctr1-1 to identify homozygous double mutant lines. The primer information is summarised in Table S1.
ein3-1 and GUS-COP1 plants were crossed, and the resulting putative homozygous plants (F2 progeny) were screened for kanamycin resistance during seed germination. After the identification of non-segregating progeny of the ein3-1 homozygous plants, seedlings were screened to identify homozygous hybrids of ein3-1×GUS-COP1.
Subcellular localisation of GUS-COP1
Transgenic GUS-COP1 seeds were germinated and grown on MS medium for 5 days. After treatment with or without 25 µM ACC for 24 h in the dark, continuous light, or light-dark (16 h/8 h) conditions, the seedlings were fixed with cold acetone and then soaked in GUS-DAPI buffer (0.5 µg/mL DAPI, 0.5 mg/mL 5-bromo-4-chloro-3-indolyl-β-D-glucuronic acid, 5 mM ferricyanide and ferrocyanide, 10 mM EDTA, and 0.1% [v/v] Tween 20 in 100 mM sodium phosphate, pH 7.0) and left in the dark at 37°C overnight. The tissues were cleared in 70% (v/v) ethanol and mounted on glass slides. To quantify the nuclear-cytoplasmic localisation of GUS-COP1, we calculated the percentages of cells that exhibited nucleus-enriched GUS staining. The cells in which GUS-COP1 staining was stronger in the nucleus than in the cytoplasm were scored as ‘nuclear,’ and the ratio of these cells to the total number of cells that displayed GUS staining, including both nucleus-enriched cells and cells displaying evenly distributed GUS staining between the nucleus and cytoplasm, was calculated. At least 10 seedlings with 20 cells per seedling were analysed for each GUS fusion under both the MS and ACC conditions.
Nuclear protein extraction
Nuclear proteins were extracted at 4°C using a CelLytic PN Extraction Kit (Sigma-Aldrich) as described previously [67] with minor modifications. After the samples grown on MS medium for 5-days were treated with 50 µM CHX and 5 µM MG132 (to exclude de novo protein production and degradation during the process), supplemented with or without 25 µM ACC, in the light for 15 h, the seedlings (3 g) were ground to a fine powder in liquid nitrogen using a precooled mortar and pestle. Next, 15 mL of 1× NIB were mixed with the samples. The suspension was passed through a 100-µm filter mesh into 50-mL tubes. The organelles in the tubes, including the nuclei, were pelleted by centrifugation at 1,200 g for 10 min, and the supernatant, including the cytoplasmic fraction, was collected and analysed as the soluble fraction. The pellet was completely resuspended in 1 mL of 1× NIBA (NIB buffer containing a protease inhibitor cocktail), and the organelle membranes were lysed by adding 10% Triton X-100 to a final concentration of 0.3%. To produce a semi-pure nuclear preparation, the lysates were applied to a 0.8-mL cushion of 1.5 M sucrose in 1× NIBA in 1.5-mL tubes. After centrifugation at 12,000 g for 10 min, the upper phase and sucrose cushion were removed, and the pellet was washed twice with NIBA buffer. The nuclear pellet was resuspended in 50 µL of nuclear extraction buffer and vortexed for 30 min. The insoluble material was removed by centrifugation at 12,000 g for 10 min. The final nuclear protein fraction was transferred to a new tube and stored at −80°C.
Supporting Information
Zdroje
1. WangKL, LiH, EckerJR (2002) Ethylene biosynthesis and signaling networks. Plant Cell 14 Suppl: S131–151.
2. GuoH, EckerJR (2004) The ethylene signaling pathway: new insights. Curr Opin Plant Biol 7 : 40–49.
3. StepanovaAN, AlonsoJM (2009) Ethylene signaling and response: where different regulatory modules meet. Curr Opin Plant Biol 12 : 548–555.
4. HuaJ, MeyerowitzEM (1998) Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell 94 : 261–271.
5. HuaJ, SakaiH, NourizadehS, ChenQG, BleeckerAB, et al. (1998) EIN4 and ERS2 are members of the putative ethylene receptor gene family in Arabidopsis. Plant Cell 10 : 1321–1332.
6. KieberJJ, RothenbergM, RomanG, FeldmannKA, EckerJR (1993) CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell 72 : 427–441.
7. ClarkKL, LarsenPB, WangX, ChangC (1998) Association of the Arabidopsis CTR1 Raf-like kinase with the ETR1 and ERS ethylene receptors. Proc Natl Acad Sci U S A 95 : 5401–5406.
8. HuangY, LiH, HutchisonCE, LaskeyJ, KieberJJ (2003) Biochemical and functional analysis of CTR1, a protein kinase that negatively regulates ethylene signaling in Arabidopsis. Plant J 33 : 221–233.
9. JuC, YoonGM, ShemanskyJM, LinDY, YingZI, et al. (2012) CTR1 phosphorylates the central regulator EIN2 to control ethylene hormone signaling from the ER membrane to the nucleus in Arabidopsis. Proc Natl Acad Sci U S A 109 : 19486–19491.
10. BissonMM, BleckmannA, AllekotteS, GrothG (2009) EIN2, the central regulator of ethylene signalling, is localized at the ER membrane where it interacts with the ethylene receptor ETR1. Biochem J 424 : 1–6.
11. JiY, GuoH (2013) From endoplasmic reticulum (ER) to nucleus: EIN2 bridges the gap in ethylene signaling. Mol Plant 6 : 11–14.
12. QiaoH, ShenZ, HuangSS, SchmitzRJ, UrichMA, et al. (2012) Processing and subcellular trafficking of ER-tethered EIN2 control response to ethylene gas. Science 338 : 390–393.
13. WenX, ZhangC, JiY, ZhaoQ, HeW, et al. (2012) Activation of ethylene signaling is mediated by nuclear translocation of the cleaved EIN2 carboxyl terminus. Cell Res 22 : 1613–1616.
14. GuoH, EckerJR (2003) Plant responses to ethylene gas are mediated by SCF(EBF1/EBF2)-dependent proteolysis of EIN3 transcription factor. Cell 115 : 667–677.
15. AnF, ZhaoQ, JiY, LiW, JiangZ, et al. (2010) Ethylene-induced stabilization of ETHYLENE INSENSITIVE3 and EIN3-LIKE1 is mediated by proteasomal degradation of EIN3 binding F-box 1 and 2 that requires EIN2 in Arabidopsis. Plant Cell 22 : 2384–2401.
16. SolanoR, StepanovaA, ChaoQ, EckerJR (1998) Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev 12 : 3703–3714.
17. KonishiM, YanagisawaS (2008) Ethylene signaling in Arabidopsis involves feedback regulation via the elaborate control of EBF2 expression by EIN3. Plant J 55 : 821–831.
18. ChenH, XueL, ChintamananiS, GermainH, LinH, et al. (2009) ETHYLENE INSENSITIVE3 and ETHYLENE INSENSITIVE3-LIKE1 repress SALICYLIC ACID INDUCTION DEFICIENT2 expression to negatively regulate plant innate immunity in Arabidopsis. Plant Cell 21 : 2527–2540.
19. ZhongS, ZhaoM, ShiT, ShiH, AnF, et al. (2009) EIN3/EIL1 cooperate with PIF1 to prevent photo-oxidation and to promote greening of Arabidopsis seedlings. Proc Natl Acad Sci U S A 106 : 21431–21436.
20. BoutrotF, SegonzacC, ChangKN, QiaoH, EckerJR, et al. (2010) Direct transcriptional control of the Arabidopsis immune receptor FLS2 by the ethylene-dependent transcription factors EIN3 and EIL1. Proc Natl Acad Sci U S A 107 : 14502–14507.
21. ZhangL, LiZ, QuanR, LiG, WangR, et al. (2011) An AP2 domain-containing gene, ESE1, targeted by the ethylene signaling component EIN3 is important for the salt response in Arabidopsis. Plant Physiol 157 : 854–865.
22. von ArnimA, DengXW (1996) Light control of seedling development. Annu Rev Plant Physiol Plant Mol Biol 47 : 215–243.
23. CaryAJ, LiuW, HowellSH (1995) Cytokinin action is coupled to ethylene in its effects on the inhibition of root and hypocotyl elongation in Arabidopsis thaliana seedlings. Plant Physiol 107 : 1075–1082.
24. SmalleJ, HaegmanM, KurepaJ, Van MontaguM, StraetenDV (1997) Ethylene can stimulate Arabidopsis hypocotyl elongation in the light. Proc Natl Acad Sci U S A 94 : 2756–2761.
25. LiH, JohnsonP, StepanovaA, AlonsoJM, EckerJR (2004) Convergence of signaling pathways in the control of differential cell growth in Arabidopsis. Dev Cell 7 : 193–204.
26. ChenH, ZhangJ, NeffMM, HongSW, ZhangH, et al. (2008) Integration of light and abscisic acid signaling during seed germination and early seedling development. Proc Natl Acad Sci U S A 105 : 4495–4500.
27. de LucasM, DaviereJM, Rodriguez-FalconM, PontinM, Iglesias-PedrazJM, et al. (2008) A molecular framework for light and gibberellin control of cell elongation. Nature 451 : 480–484.
28. FengS, MartinezC, GusmaroliG, WangY, ZhouJ, et al. (2008) Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451 : 475–479.
29. LauOS, DengXW (2010) Plant hormone signaling lightens up: integrators of light and hormones. Curr Opin Plant Biol 13 : 571–577.
30. BaiMY, ShangJX, OhE, FanM, BaiY, et al. (2012) Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nat Cell Biol 14 : 810–817.
31. WangQ, ZhuZ, OzkardeshK, LinC (2013) Phytochromes and phytohormones: the shrinking degree of separation. Mol Plant 6 : 5–7.
32. CluisCP, MouchelCF, HardtkeCS (2004) The Arabidopsis transcription factor HY5 integrates light and hormone signaling pathways. Plant J 38 : 332–347.
33. AngLH, ChattopadhyayS, WeiN, OyamaT, OkadaK, et al. (1998) Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol Cell 1 : 213–222.
34. ChattopadhyayS, AngLH, PuenteP, DengXW, WeiN (1998) Arabidopsis bZIP protein HY5 directly interacts with light-responsive promoters in mediating light control of gene expression. Plant Cell 10 : 673–683.
35. von ArnimAG, DengXW (1994) Light inactivation of Arabidopsis photomorphogenic repressor COP1 involves a cell-specific regulation of its nucleocytoplasmic partitioning. Cell 79 : 1035–1045.
36. OsterlundMT, HardtkeCS, WeiN, DengXW (2000) Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405 : 462–466.
37. VandenbusscheF, HabricotY, CondiffAS, MaldineyR, Van der StraetenD, et al. (2007) HY5 is a point of convergence between cryptochrome and cytokinin signalling pathways in Arabidopsis thaliana. Plant J 49 : 428–441.
38. AlabadiD, Gallego-BartolomeJ, OrlandoL, Garcia-CarcelL, RubioV, et al. (2008) Gibberellins modulate light signaling pathways to prevent Arabidopsis seedling de-etiolation in darkness. Plant J 53 : 324–335.
39. ZhongS, ShiH, XueC, WangL, XiY, et al. (2012) A molecular framework of light-controlled phytohormone action in Arabidopsis. Curr Biol 22 : 1530–1535.
40. LiangX, WangH, MaoL, HuY, DongT, et al. (2012) Involvement of COP1 in ethylene - and light-regulated hypocotyl elongation. Planta 236 : 1791–1802.
41. OyamaT, ShimuraY, OkadaK (1997) The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev 11 : 2983–2995.
42. JingY, ZhangD, WangX, TangW, WangW, et al. (2013) Arabidopsis chromatin remodeling factor PICKLE interacts with transcription factor HY5 to regulate hypocotyl cell elongation. Plant Cell 25 : 242–256.
43. ChaeHS, FaureF, KieberJJ (2003) The eto1, eto2, and eto3 mutations and cytokinin treatment increase ethylene biosynthesis in Arabidopsis by increasing the stability of ACS protein. Plant Cell 15 : 545–559.
44. LeeJ, HeK, StolcV, LeeH, FigueroaP, et al. (2007) Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19 : 731–749.
45. McNellisTW, von ArnimAG, ArakiT, KomedaY, MiseraS, et al. (1994) Genetic and molecular analysis of an allelic series of cop1 mutants suggests functional roles for the multiple protein domains. Plant Cell 6 : 487–500.
46. ToriiKU, McNellisTW, DengXW (1998) Functional dissection of Arabidopsis COP1 reveals specific roles of its three structural modules in light control of seedling development. EMBO J 17 : 5577–5587.
47. SeoHS, YangJY, IshikawaM, BolleC, BallesterosML, et al. (2003) LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature 423 : 995–999.
48. YangJ, LinR, SullivanJ, HoeckerU, LiuBX, et al. (2005) Light regulates COP1-mediated degradation of HFR1, a transcription factor essential for light signaling in Arabidopsis. Plant Cell 17 : 804–821.
49. JangIC, YangJY, SeoHS, ChuaNH (2005) HFR1 is targeted by COP1 E3 ligase for post-translational proteolysis during phytochrome A signaling. Genes Dev 19 : 593–602.
50. SamacDA, HironakaCM, YallalyPE, ShahDM (1990) Isolation and Characterization of the Genes Encoding Basic and Acidic Chitinase in Arabidopsis thaliana. Plant Physiol 93 : 907–914.
51. PacinM, LegrisM, CasalJJ (2013) COP1 re-accumulates in the nucleus under shade. Plant J 75 : 631–641.
52. SaijoY, SullivanJA, WangH, YangJ, ShenY, et al. (2003) The COP1-SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Genes Dev 17 : 2642–2647.
53. SubramanianC, KimBH, LyssenkoNN, XuX, JohnsonCH, et al. (2004) The Arabidopsis repressor of light signaling, COP1, is regulated by nuclear exclusion: mutational analysis by bioluminescence resonance energy transfer. Proc Natl Acad Sci U S A 101 : 6798–6802.
54. YiC, DengXW (2005) COP1-from plant photomorphogenesis to mammalian tumorigenesis. Trends Cell Biol 15 : 618–625.
55. ShinJ, ParkE, ChoiG (2007) PIF3 regulates anthocyanin biosynthesis in an HY5-dependent manner with both factors directly binding anthocyanin biosynthetic gene promoters in Arabidopsis. Plant J 49 : 981–994.
56. ZhangZ, HuangR (2010) Enhanced tolerance to freezing in tobacco and tomato overexpressing transcription factor TERF2/LeERF2 is modulated by ethylene biosynthesis. Plant Mol Biol 73 : 241–249.
57. AchardP, ChengH, De GrauweL, DecatJ, SchouttetenH, et al. (2006) Integration of plant responses to environmentally activated phytohormonal signals. Science 311 : 91–94.
58. AlonsoJM, HirayamaT, RomanG, NourizadehS, EckerJR (1999) EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284 : 2148–2152.
59. WangX, LiW, PiquerasR, CaoK, DengXW, et al. (2009) Regulation of COP1 nuclear localization by the COP9 signalosome via direct interaction with CSN1. Plant J 58 : 655–667.
60. FranciosiniA, LombardiB, IafrateS, PecceV, MeleG, et al. (2013) The Arabidopsis COP9 SIGNALOSOME INTERACTING F-BOX KELCH 1 protein forms an SCF ubiquitin ligase and regulates hypocotyl elongation. Mol Plant 6 : 1616–1629.
61. ChristiansMJ, RoblesLM, ZellerSM, LarsenPB (2008) The eer5 mutation, which affects a novel proteasome-related subunit, indicates a prominent role for the COP9 signalosome in resetting the ethylene-signaling pathway in Arabidopsis. Plant J 55 : 467–477.
62. SunJ, QiL, LiY, ChuJ, LiC (2012) PIF4-mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating Arabidopsis hypocotyl growth. PLoS Genet 8: e1002594.
63. ChapmanEJ, GreenhamK, CastillejoC, SartorR, BialyA, et al. (2012) Hypocotyl transcriptome reveals auxin regulation of growth-promoting genes through GA-dependent and -independent pathways. PLoS One 7: e36210.
64. LiZ, HuangR (2011) The reciprocal regulation of abscisic acid and ethylene biosyntheses. Plant Signal Behav 6 : 1647–1650.
65. LiZ, ZhangL, YuY, QuanR, ZhangZ, et al. (2011) The ethylene response factor AtERF11 that is transcriptionally modulated by the bZIP transcription factor HY5 is a crucial repressor for ethylene biosynthesis in Arabidopsis. Plant J 68 : 88–99.
66. De GrauweL, ChaerleL, DugardeynJ, DecatJ, RieuI, et al. (2008) Reduced gibberellin response affects ethylene biosynthesis and responsiveness in the Arabidopsis gai eto2-1 double mutant. New Phytol 177 : 128–141.
67. JangIC, HenriquesR, SeoHS, NagataniA, ChuaNH (2010) Arabidopsis PHYTOCHROME INTERACTING FACTOR proteins promote phytochrome B polyubiquitination by COP1 E3 ligase in the nucleus. Plant Cell 22 : 2370–2383.
Štítky
Genetika Reprodukční medicína
Článek Interaction between and during Mammalian Jaw Patterning and in the Pathogenesis of SyngnathiaČlánek Clustering of Tissue-Specific Sub-TADs Accompanies the Regulation of Genes in Developing LimbsČlánek Transcription Factor Occupancy Can Mediate Active Turnover of DNA Methylation at Regulatory RegionsČlánek Tay Bridge Is a Negative Regulator of EGFR Signalling and Interacts with Erk and Mkp3 in the Wing
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 12
-
Všechny články tohoto čísla
- Stressing the Importance of CHOP in Liver Cancer
- The AmAZI1ng Roles of Centriolar Satellites during Development
- Flies Get a Head Start on Meiosis
- Recommendations from Jane Gitschier's Bookshelf
- And Baby Makes Three: Genomic Imprinting in Plant Embryos
- Bugs in Transition: The Dynamic World of in Insects
- Defining the Role of ATP Hydrolysis in Mitotic Segregation of Bacterial Plasmids
- Synaptonemal Complex Components Promote Centromere Pairing in Pre-meiotic Germ Cells
- Cohesinopathies of a Feather Flock Together
- Genetic Recombination Is Targeted towards Gene Promoter Regions in Dogs
- Parathyroid-Specific Deletion of Unravels a Novel Calcineurin-Dependent FGF23 Signaling Pathway That Regulates PTH Secretion
- MAN1B1 Deficiency: An Unexpected CDG-II
- Phosphate Flow between Hybrid Histidine Kinases CheA and CheS Controls Cyst Formation
- Basolateral Mg Extrusion via CNNM4 Mediates Transcellular Mg Transport across Epithelia: A Mouse Model
- Truncation of Unsilences Paternal and Ameliorates Behavioral Defects in the Angelman Syndrome Mouse Model
- Autozygome Sequencing Expands the Horizon of Human Knockout Research and Provides Novel Insights into Human Phenotypic Variation
- Huntington's Disease Induced Cardiac Amyloidosis Is Reversed by Modulating Protein Folding and Oxidative Stress Pathways in the Heart
- Low Frequency Variants, Collapsed Based on Biological Knowledge, Uncover Complexity of Population Stratification in 1000 Genomes Project Data
- Targeted Ablation of and in Retinal Progenitor Cells Mimics Leber Congenital Amaurosis
- Genomic Imprinting in the Embryo Is Partly Regulated by PRC2
- Binary Cell Fate Decisions and Fate Transformation in the Larval Eye
- The Stress-Regulated Transcription Factor CHOP Promotes Hepatic Inflammatory Gene Expression, Fibrosis, and Oncogenesis
- A Global RNAi Screen Identifies a Key Role of Ceramide Phosphoethanolamine for Glial Ensheathment of Axons
- Functional Analysis of the Interdependence between DNA Uptake Sequence and Its Cognate ComP Receptor during Natural Transformation in Species
- Cross-Modulation of Homeostatic Responses to Temperature, Oxygen and Carbon Dioxide in
- Alcohol-Induced Histone Acetylation Reveals a Gene Network Involved in Alcohol Tolerance
- Molecular Characterization of Host-Specific Biofilm Formation in a Vertebrate Gut Symbiont
- CRIS—A Novel cAMP-Binding Protein Controlling Spermiogenesis and the Development of Flagellar Bending
- Dual Regulation of the Mitotic Exit Network (MEN) by PP2A-Cdc55 Phosphatase
- Expanding the Marine Virosphere Using Metagenomics
- Detection of Slipped-DNAs at the Trinucleotide Repeats of the Myotonic Dystrophy Type I Disease Locus in Patient Tissues
- Interaction between and during Mammalian Jaw Patterning and in the Pathogenesis of Syngnathia
- Mutations in the UQCC1-Interacting Protein, UQCC2, Cause Human Complex III Deficiency Associated with Perturbed Cytochrome Protein Expression
- Reactivation of Chromosomally Integrated Human Herpesvirus-6 by Telomeric Circle Formation
- Anoxia-Reoxygenation Regulates Mitochondrial Dynamics through the Hypoxia Response Pathway, SKN-1/Nrf, and Stomatin-Like Protein STL-1/SLP-2
- The Midline Protein Regulates Axon Guidance by Blocking the Reiteration of Neuroblast Rows within the Drosophila Ventral Nerve Cord
- Tomato Yield Heterosis Is Triggered by a Dosage Sensitivity of the Florigen Pathway That Fine-Tunes Shoot Architecture
- Selection on Plant Male Function Genes Identifies Candidates for Reproductive Isolation of Yellow Monkeyflowers
- Role of Tomato Lipoxygenase D in Wound-Induced Jasmonate Biosynthesis and Plant Immunity to Insect Herbivores
- Meiotic Cohesin SMC1β Provides Prophase I Centromeric Cohesion and Is Required for Multiple Synapsis-Associated Functions
- Identification of Sphingolipid Metabolites That Induce Obesity via Misregulation of Appetite, Caloric Intake and Fat Storage in
- Genome-Wide Screen Reveals Replication Pathway for Quasi-Palindrome Fragility Dependent on Homologous Recombination
- Histone Methylation Restrains the Expression of Subtype-Specific Genes during Terminal Neuronal Differentiation in
- A Novel Intergenic ETnII-β Insertion Mutation Causes Multiple Malformations in Mice
- The NuRD Chromatin-Remodeling Enzyme CHD4 Promotes Embryonic Vascular Integrity by Transcriptionally Regulating Extracellular Matrix Proteolysis
- A Domesticated Transposase Interacts with Heterochromatin and Catalyzes Reproducible DNA Elimination in
- Acute Versus Chronic Loss of Mammalian Results in Distinct Ciliary Phenotypes
- MBD3 Localizes at Promoters, Gene Bodies and Enhancers of Active Genes
- Positive and Negative Regulation of Gli Activity by Kif7 in the Zebrafish Embryo
- A Hereditary Spastic Paraplegia Mouse Model Supports a Role of ZFYVE26/SPASTIZIN for the Endolysosomal System
- The CCR4-NOT Complex Mediates Deadenylation and Degradation of Stem Cell mRNAs and Promotes Planarian Stem Cell Differentiation
- Reconstructing Native American Migrations from Whole-Genome and Whole-Exome Data
- Contributions of Protein-Coding and Regulatory Change to Adaptive Molecular Evolution in Murid Rodents
- Comprehensive Analysis of Transcriptome Variation Uncovers Known and Novel Driver Events in T-Cell Acute Lymphoblastic Leukemia
- A -Acting Protein Effect Causes Severe Eye Malformation in the Mouse
- Clustering of Tissue-Specific Sub-TADs Accompanies the Regulation of Genes in Developing Limbs
- Germline Progenitors Escape the Widespread Phenomenon of Homolog Pairing during Development
- Transcription Factor Occupancy Can Mediate Active Turnover of DNA Methylation at Regulatory Regions
- Somatic mtDNA Mutation Spectra in the Aging Human Putamen
- ESCRT-I Mediates FLS2 Endosomal Sorting and Plant Immunity
- Ethylene Promotes Hypocotyl Growth and HY5 Degradation by Enhancing the Movement of COP1 to the Nucleus in the Light
- The PAF Complex and Prf1/Rtf1 Delineate Distinct Cdk9-Dependent Pathways Regulating Transcription Elongation in Fission Yeast
- Dual Regulation of Gene Expression Mediated by Extended MAPK Activation and Salicylic Acid Contributes to Robust Innate Immunity in
- Quantifying Missing Heritability at Known GWAS Loci
- Smc5/6-Mms21 Prevents and Eliminates Inappropriate Recombination Intermediates in Meiosis
- Smc5/6 Coordinates Formation and Resolution of Joint Molecules with Chromosome Morphology to Ensure Meiotic Divisions
- Tay Bridge Is a Negative Regulator of EGFR Signalling and Interacts with Erk and Mkp3 in the Wing
- Meiotic Crossover Control by Concerted Action of Rad51-Dmc1 in Homolog Template Bias and Robust Homeostatic Regulation
- Active Transport and Diffusion Barriers Restrict Joubert Syndrome-Associated ARL13B/ARL-13 to an Inv-like Ciliary Membrane Subdomain
- An Regulatory Circuit Modulates /Wnt Signaling and Determines the Size of the Midbrain Dopaminergic Progenitor Pool
- Variants Induce Differential Protection to Viruses in : A Phenotypic and Phylogenomic Analysis
- Base Pairing Interaction between 5′- and 3′-UTRs Controls mRNA Translation in
- Evidence That Masking of Synapsis Imperfections Counterbalances Quality Control to Promote Efficient Meiosis
- Insulin/IGF-Regulated Size Scaling of Neuroendocrine Cells Expressing the bHLH Transcription Factor in
- Sumoylated NHR-25/NR5A Regulates Cell Fate during Vulval Development
- TATN-1 Mutations Reveal a Novel Role for Tyrosine as a Metabolic Signal That Influences Developmental Decisions and Longevity in
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The NuRD Chromatin-Remodeling Enzyme CHD4 Promotes Embryonic Vascular Integrity by Transcriptionally Regulating Extracellular Matrix Proteolysis
- Mutations in the UQCC1-Interacting Protein, UQCC2, Cause Human Complex III Deficiency Associated with Perturbed Cytochrome Protein Expression
- The Midline Protein Regulates Axon Guidance by Blocking the Reiteration of Neuroblast Rows within the Drosophila Ventral Nerve Cord
- Tomato Yield Heterosis Is Triggered by a Dosage Sensitivity of the Florigen Pathway That Fine-Tunes Shoot Architecture
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



