-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaSmc5/6-Mms21 Prevents and Eliminates Inappropriate Recombination Intermediates in Meiosis
Repairing broken chromosomes via joint molecule (JM) intermediates is hazardous and therefore strictly controlled in most organisms. Also in budding yeast meiosis, where production of enough crossovers via JMs is imperative, only a subset of DNA breaks are repaired via JMs, closely regulated by the ZMM pathway. The other breaks are repaired to non-crossovers, avoiding JM formation, through pathways that require the BLM/Sgs1 helicase. “Rogue” JMs that escape the ZMM pathway and BLM/Sgs1 are eliminated before metaphase by resolvases like Mus81-Mms4 to prevent chromosome nondisjunction. Here, we report the requirement of Smc5/6-Mms21 for antagonizing rogue JMs via two mechanisms; destabilizing early intermediates and resolving JMs. Elimination of the Mms21 SUMO E3-ligase domain leads to transient JM accumulation, depending on Mus81-Mms4 for resolution. Absence of Smc6 leads to persistent rogue JMs accumulation, preventing chromatin separation. We propose that the Smc5/6-Mms21 complex antagonizes toxic JMs by coordinating helicases and resolvases at D-Loops and HJs, respectively.
Published in the journal: . PLoS Genet 9(12): e32767. doi:10.1371/journal.pgen.1004067
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004067Summary
Repairing broken chromosomes via joint molecule (JM) intermediates is hazardous and therefore strictly controlled in most organisms. Also in budding yeast meiosis, where production of enough crossovers via JMs is imperative, only a subset of DNA breaks are repaired via JMs, closely regulated by the ZMM pathway. The other breaks are repaired to non-crossovers, avoiding JM formation, through pathways that require the BLM/Sgs1 helicase. “Rogue” JMs that escape the ZMM pathway and BLM/Sgs1 are eliminated before metaphase by resolvases like Mus81-Mms4 to prevent chromosome nondisjunction. Here, we report the requirement of Smc5/6-Mms21 for antagonizing rogue JMs via two mechanisms; destabilizing early intermediates and resolving JMs. Elimination of the Mms21 SUMO E3-ligase domain leads to transient JM accumulation, depending on Mus81-Mms4 for resolution. Absence of Smc6 leads to persistent rogue JMs accumulation, preventing chromatin separation. We propose that the Smc5/6-Mms21 complex antagonizes toxic JMs by coordinating helicases and resolvases at D-Loops and HJs, respectively.
Introduction
Sexual reproduction in eukaryotes relies on the generation of haploid gametes from diploid somatic cells by a process called meiosis. Meiosis achieves the required reduction in ploidy by executing a single round of DNA replication followed by two consecutive rounds of chromosome segregation. The correct segregation of homologous chromosomes depends on the formation of chiasmata, which are crossovers (COs) held in place by distal sister chromatid cohesion [1], [2]. Crossovers, and in many organisms also the identification and pairing of homologous chromosomes, require the repair of programmed meiotic DNA double-strand breaks (DSBs) in prophase I at sites that have completed pre-meiotic DNA replication [3]. As a consequence of the repair of DSBs, stable recombination intermediates called double Holliday Junctions (dHJs) can arise, and can be resolved to generate crossovers (COs) or non-crossovers (NCOs).
Sophisticated mechanisms controlling CO numbers and distribution ensure that each bivalent (two paired homologous chromosomes) receives at least its obligate CO. In budding yeast, the ZMM pathway (an acronym for the involved proteins Zip1-4, Mer3, Msh4/5 [4]) is a key part of this control. It guides a subset of DSBs to become allelic COs between homologs by allowing these breaks to form dHJs specifically resolved to COs depending on Exo1-Mlh1/3 [5]. A sophisticated machinery, including the Synaptonemal Complex (SC), regulates the progression of recombination intermediates in the ZMM pathway. The SC is a tripartite proteinaceous structure connecting bivalents along their whole length at a distance of 100 nm in the pachytene stage of meiosis. After completion of synapsis, Polo-like kinase Cdc5 activation triggers the resolution of dHJs shortly before cells become committed to enter the first meiotic division [6], [7].
Non-ZMM DSBs are not destined to become COs and follow another main route, involving fast and minimal-risk repair by Synthesis Dependent Strand Annealing (SDSA). In SDSA the invasion of one broken DNA terminus into an intact template allows DNA repair synthesis beyond the break. Importantly, interaction remains limited by the rapid displacement of the invading strand. This pathway does not produce COs but may nevertheless facilitate homolog recognition. Recently, the RecQ helicase BLM/Sgs1 [8] has been shown to be a central player in both primary pathways of meiotic recombination [5], [9]–[12]. In the SDSA pathway, Sgs1 promotes strand displacement thereby preventing stabilization of the invasion. In the ZMM pathway, Sgs1 delays repair until the formation of stable Single End Invasion (SEI) and dHJ intermediates is appropriate, presumably after the cell has accumulated some information about the correctness of the invaded target. In budding yeast, the ZMM and SDSA pathways together accomplish the large majority of meiotic recombination events [5].
If recombination intermediates escape the two pathways described above, unregulated, mitotic like Joint Molecules (JMs) can arise, such as dHJs not associated with the proper ZMM machinery. We will refer to these intermediates as “rogue”, in the sense that they are “unprincipled, unreliable and with potentially destructive properties” which could result in non-allelic COs after escaping the two canonical, safe pathways. Although these rogue intermediates constitute only a minor fraction of recombination events in wild type meiosis, they can block chromosome segregation if unresolved. Resolution of such JMs is mediated by the overlapping activity of the three HJ resolvases Mus81-Mms4, Slx1-Slx4, and Yen1 [5], [7], [9]. Cdc5/Plk1 also controls the activation of Mus81-Mms4 and Slx1-Slx4.
dHJs represent potential danger for their inability to elicit a DNA damage checkpoint response [10]. Due to their stability, they may cause chromosome nondisjunction or even block segregation if not resolved. Conversely, resolution of HJ intermediates between non-allelic positions can result in deletions or translocations, even in dicentric chromosomes. This explains attempts of the cell to avoid the formation of such stable recombination intermediates right away, mediated by the action of helicases such as Srs2 and BLM/Sgs1 that can destabilize Rad51 filaments and nascent invasions [13]–[17]. Even if dHJs formed, the cell may be able to unwind them conservatively through the action of Sgs1-Rmi1-Top3 [18], [19], a process termed dHJ dissolution. Ultimately, once the cell has passed the DNA damage checkpoint and is committed for division, resolvases will be given priority for timely removal of linking Holliday Junctions (HJs) to avoid chromosome nondisjunction events. While a key set of DNA metabolizing activities have recently been described, important questions concerning the chromosomal context remain unanswered. Coordination of local recruitment, regulation and orientation of anti-JM helicases and resolvases in the in vivo context remain completely obscure to date.
In this study, we provide evidence that the Smc5/6-Mms21 complex mediates such functions. SMC complexes are evolutionarily conserved from gram-negative bacteria to mammals, serving critical functions in chromosome metabolism, thereby helping to preserve chromosomal and genomic integrity. Eukaryotes have three distinct, essential ring - shaped SMC complexes at their disposal; the cohesin complex (Smc1-Smc3), linking sister chromatids until the metaphase/anaphase transition, condensin (Smc2-Smc4), thought to regulate higher order chromosome structure and finally the Smc5/6-Mms21 complex, involved in recombinational DNA repair.
All SMC ring complexes exhibit the ability to associate with DNA and the hinge region of several SMC complexes, including condensin, has been shown to bind to DNA [20]. At least for cohesin and condensin there is direct evidence that DNA is topologically entrapped inside the ring [21]–[24].
While this has not yet been tested for the Smc5-Smc6 complex, it was shown that both Smc5 and Smc6 individually bind stably to DNA with a strong preference for single stranded DNA [25], [26]. Taking into consideration the high similarity in structure and size to the other SMCs, it is not unlikely that Smc5/6-Mms21 also encloses DNA strands topologically.
The SMC proteins consist of two extended domains that fold back on themselves at their central hinge region into an anti-parallel coiled-coil structure. This brings the two terminal Walker A/B motifs in close proximity to form ABC-like ATPases. Two SMC proteins connect via their hinge regions, while the kleisin subunit closes the ring by linking the terminal SMC ATPase heads. Different SMC complexes contain characteristic non-SMC subunits that form integral components of the functional complex.
In budding and fission yeast the Smc5/6-Mms21 complex comprises six Non-SMC Elements (NSEs), Nse1 to Nse6, of which Nse4 is the kleisin (Figure 1A). Beside Smc5 and 6, at least Nse1 to Nse4 are conserved from yeast to man [27]–[29].
Fig. 1. Smc6, but not Mms21 SUMO ligase activity is required for chromosome segregation after meiotic recombination. 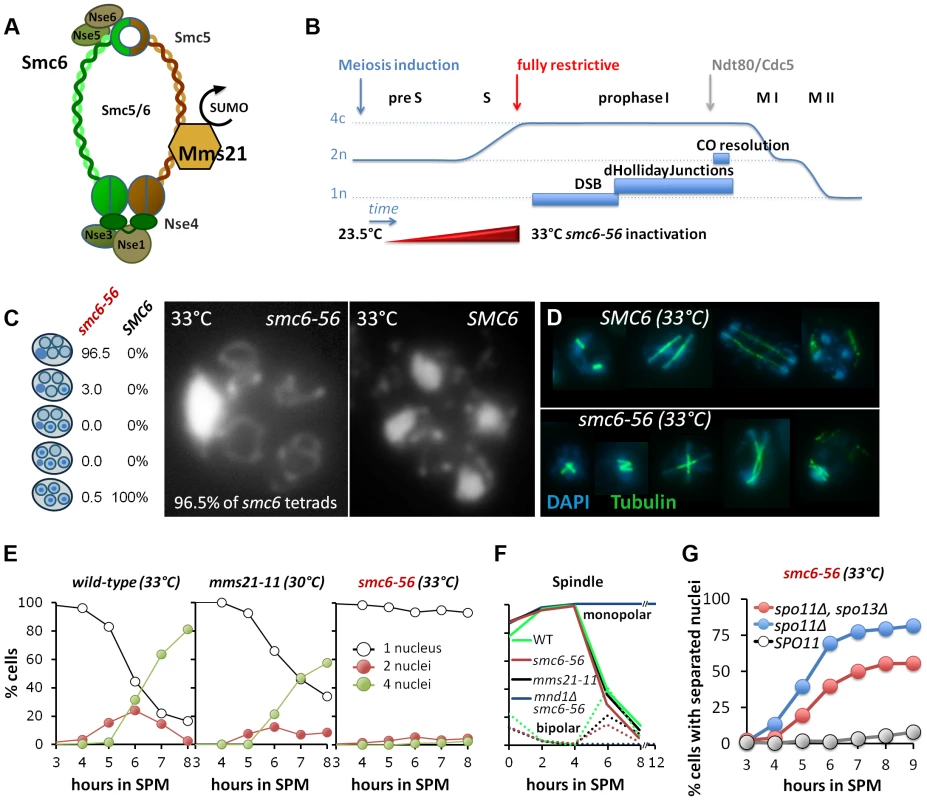
(A) Schematic representation of the Smc5/6-Mms21 subunits. (B) Schematic of the temperature shift experiments. To avoid mitosis and premeiotic S-phase a temperature shift from 23.5 to 33°C was carried out in increments upon induction of meiosis (see experimental procedures) reaching 33°C by 2.5 hours in SPM when cells are past bulk DNA replication. This scheme eliminates Smc6 at the point when the earliest DSBs become detectable and interference with mitosis and pre-meiotic S-phase is little. (C) DNA staining with DAPI after spore formation. The right micrograph shows the presence of 4 nuclei and mitochondrial DNA in a wild type tetrad. The left micrograph shows the nuclear DNA outside the four spores, pressed against the ascus wall, while only the mitochondrial DNA had segregated into the spores. Left table: Percent of asci with 0,1,2,3,4 spores containing nuclear DNA (n = 200 tetrads). (D) Spindle staining by anti-tubulin. The upper panel shows examples of spindle morphologies post anaphase I in wild type. The lower panel shows aberrant spindles of the smc6-56 mutant, consistent with physical impediment of DNA separation. (E) Meiotic nuclear divisions are blocked in smc6-56 mutants at 33°C. Left panel: wild type (33°C), middle: mms21-11 (30°C), right: smc6-56 (33°C). Empty black circles: cells containing one DAPI stained nucleus (1n), filled red circles: cells with 2 nuclei (2n), filled green circles: cells with 4 nuclei (4n). (F) Meiotic progression is normal in smc6-56 (33°C): Meiotic progression was followed by spindle morphology by tubulin labeling in an in situ staining procedure. Monopolar and bi-polar spindles were plotted separately against hours in SPM. n = 200 cells per experiment, continuous lines: monopolar spindles, dotted lines: bipolar spindles. Green: Wild type (30°C), black: mms21-11 (30°C), red: smc6-56 (33°C), blue: mnd1Δ smc6-56 (33°C). (G) spo11Δ suppresses the chromatin separation defect of smc6-56. Cells containing at least 2 separated nuclei plotted against the time in sporulation. Grey filled circles: smc6-56 SPO11, blue filled circles: smc6-56 spo11Δ, red filled circles: smc6-56 spo11Δ spo13Δ. A unique feature of the Smc5/6-Mms21 complex, compared to cohesin and condensin, is its SUMO E3 ligase subunit Mms21/Nse2 [27], conferring the ability to post-translationally modify target proteins. The SUMOylation substrates of Mms21 remain poorly defined to date, however, candidates include Smc5, Scc1, the kleisin subunit of cohesin, as well as telomeric proteins [30],[31].
Mms21 is stably bound through an extensive N-terminal coiled-coil interface to the coiled coil domain of Smc5 [32], [33] while the E3 SPL RING structure that recruits the SUMO E2 ligase Ubc9 resides at its C-terminus [32]. Mms21 is an essential subunit of the Smc5/6-Mms21 complex in Saccharomyces cerevisiae and the mere disruption of its interaction with Smc5 is lethal [32], [34]. Notably, elimination of Mms21's SUMO E3 ligase activity alone is not lethal, but sensitizes the cell to genotoxic agents [27].
Yeast cells mutated in the Smc5/6-Mms21 complex become hypersensitive to genotoxic agents including hydroxyurea, MMS, ionizing irradiation and UV [27], [35], [36], and DNA regions vulnerable to homologous recombination (HR) like rDNA and telomeres are particularly affected [37]. Accordingly, Smc5/6 was also found to accumulate at such sites prone to recombinogenic damage [38], [39]. Accumulation of X-shaped DNA intermediates upon challenge and aberrant processing of stalled replication forks have been reported in Smc5/6 mutants [40] and roles for the Smc5/6-Mms21 complex in multiple repair pathways, including homologous recombination, have been suggested [41]. Complete elimination of functional Smc5/6-Mms21 complex from vegetative cells leads to heterogeneous defects, including cells arresting in metaphase, chromosome missegregation and eventually lethality [42].
In meiosis, phenotypes of Smc5/6 mutants in S. pombe and S. cerevisiae represent exacerbated manifestations of those observed during mitosis, including catastrophic failures in meiotic divisions [43], [44]. In a synapsis mutant (zip1Δ) of S. cerevisiae, the homologous chromosomes tended to become more attached to each other and chromosomal entanglements seemed to increase. These defects were partly caused during premeiotic S-phase, as they were not fully dependent on initiating meiotic recombination [44]. Another study in S. pombe found accumulation of X-shaped DNA molecules in nse6 mutants, originating from meiotic recombination [43].
Here we study the role of Smc6 in meiotic recombination by eliminating Smc6 after premeiotic S-phase is largely complete. This leads to an accumulation of unresolved JMs at meiotic recombination hotspots and a corresponding uniform block in nuclear divisions. We observe strong overlaps of Smc6-chromatin binding with that of Sgs1 at a substantial number of meiotic DSB-hotspots. In chromosome spreads, Smc5/6 foci co-localize with Rad51/Dmc1 foci side by side. We further show that the E3 ligase deficient mms21-11 allele restores dHJ and CO formation and improves spore viability in the robust ZMM mutant zip3Δ, implying an important role of Mms21 in the prevention of dHJ formation in this background. Resolution of the JMs in the mms21-11 mutant depends on the Mus81-Mms4 resolvase. The Smc5/6-Mms21 complex is also required for HJ resolvase activity responsible for eliminating rogue HJ intermediates. A dramatic accumulation of unresolved JMs and a pronounced reduction in COs and NCOs was observed in the smc6-56 sgs1 double mutant, in which all meiotic recombination follows an aberrant non-ZMM, non-SDSA pathway, while COs form at near normal levels in the smc6-56 SGS1. We conclude that Smc5/6-Mms21 collaborates with helicases and resolvases to both prevent and eliminate JMs that arise outside the ZMM recombination pathway.
Results
Smc6, but not Mms21 SUMO ligase activity is required for chromosome segregation after meiotic recombination
In order to characterize the role of the Smc5/6-Mms21 complex in the context of meiotic recombination, we utilized two previously described mutants with distinctive phenotypes. smc6-56 is a temperature sensitive allele, carrying three missense mutations in the N-terminus proximal coiled-coil region (A287V, H379R, I421T) conferring lethality at restrictive temperature [45]. The mms21-11 allele terminates after Thr183 and lacks the C-terminal SPL-RING domain, thus depriving Mms21 of its SUMO E3 ligase activity by abolishing its interaction with the SUMO E2 enzyme Ubc9 [27], [46].
To distinguish the role of the Smc5/6-Mms21 complex in meiotic DSB repair from that of mitosis and premeiotic S-phase we inactivated the smc6-56 allele by shifting the temperature of the synchronized cultures gradually to restrictive conditions at 33°C (Figure 1B). Cells are allowed to exit mitosis and undergo most of meiotic DNA replication under permissive or semi-permissive conditions (Figure S1A, B). Fully restrictive conditions were applied from 2.5 hrs post induction of meiosis, when most cells are in late S-phase and the earliest meiotic DSBs become detectable [47]. The constitutive mms21-11 mutant was analyzed at 30°C.
Under these conditions, smc6-56 causes neither a delay in meiotic progression and spore formation, nor does it produce evident defects in meiotic DSB formation, repair, or chromosome axis architecture. Chromosome synapsis is flawless with normal timing (Figure S1C), meiosis I and meiosis II spindles form with normal kinetics, and a mnd1Δ smc6-56 double mutant arrests indistinguishably from mnd1Δ which prevents DSB repair at the strand invasion step [48]–[50] in prophase1 indicating a functional DNA damage checkpoint (Figure 1F), suggesting efficient DSB-turnover in smc6-56 comparable to wild type.
However, 92–99% of cells with inactivated Smc6 failed to segregate the chromatin during both meiotic divisions and thus produced a single nucleus outside of four empty spores (Figure 1C, E). Unable to separate the chromatin, anaphase I and II spindles ultimately collapse, resulting in aberrant spindle morphologies from Meiosis II onwards (Figure 1D). The timing of spore formation and the number of cells forming spores was as in wild-type, however, the number of aberrant asci containing one, two or three spores was increased at the expense of complete tetrads (21–27%, less than half of wild type). In 88–95% of the tetrads, all four spores were empty and only 0.5–4.5% of tetrads had DNA in all four spores. Most empty spores ultimately collapse and partially lyse by 24 hours. Notably, those few tetrads that managed to receive DNA in all 4 spores and survive zymolyase digestion show a fairly high spore viability (53.75%, n = 80 spores/20 tetrads).
To test whether the observed meiotic catastrophe depended on meiotic recombination, DSB formation was abolished by introducing the spo11Δ mutation in the smc6-56 background. Indeed, meiotic divisions were largely restored with 80% of the cells completing at least one division and 60% of the cells finishing both meiotic divisions (Figure 1G). To demonstrate that also the sister chromatids can separate, the triple mutant with spo13Δ was analyzed. We found that almost 60% of the cells underwent the single division with normal kinetics (Figure 1G). Thus, the applied regime of conditional inactivation of Smc6 separates most of the defects related to mitosis and meiotic S-phase from those in meiotic recombination. We conclude that the Smc5/6-Mms21 complex is essential for allowing chromosome disjunction upon initiation of meiotic recombination.
In contrast to the smc6-56 mutant, mms21-11 showed almost normal kinetics of meiotic divisions, forming tetra-nucleated cells with only a slight delay (Figure 1E). Synapsis and sporulation are efficient (Figure S1C) and the resulting tetrads exhibited high spore viability (88.75%; n = 400 spores of 100 tetrads). Only a small fraction of tetrads (3–3.25%) were missing DNA in one or more spores. Consequently, the SUMO E3 ligase activity of the Smc5/6-Mms21 complex is not essential to prevent massive chromosome non-disjunction.
The Smc5/6-Mms21 complex binds early to meiotic chromatin and associates with sites of DSB repair
To characterize the binding of Smc5/6-Mms21 to meiotic chromatin in the context of meiotic recombination we used the epitope tagged SMC6-myc13 construct and prepared meiotic nuclear spreads from synchronized cultures at various time points for cytology. Smc6-myc13 does not show any obvious defects and co-localizes with Smc5-HA3 on chromosome spreads, suggesting it is a valid representation of the complex (Figure S2A).
Smc6-myc13 localizes in individual foci which appear early in meiosis at approximately the same time as Rec8 (Figure S2B) and soon accumulate to considerable numbers until earliest prophase 1 (Figure S3). We used antibody-staining against the synapsis specific Zip1 protein for staging (from isolated Zip1 foci up to full SCs). In nuclei engaged in meiotic recombination (Zip1 positive) an average of 118±18 (n = 21 nuclei scored) Smc6 foci were scored and this number did not change significantly in different stages of prophase 1. After prophase 1, the intensity of foci decreased slightly, but numbers remained high until Metaphase II/Anaphase II transition (Figure 2C, S3). A particular enrichment was observed for the rDNA region on chromosome 12, which remains unsynapsed in the course of meiotic recombination (Figure 2A white arrows, S3).
Fig. 2. The Smc5/6-Mms21 complex binds early to meiotic chromatin and associates with sites of DSB repair. 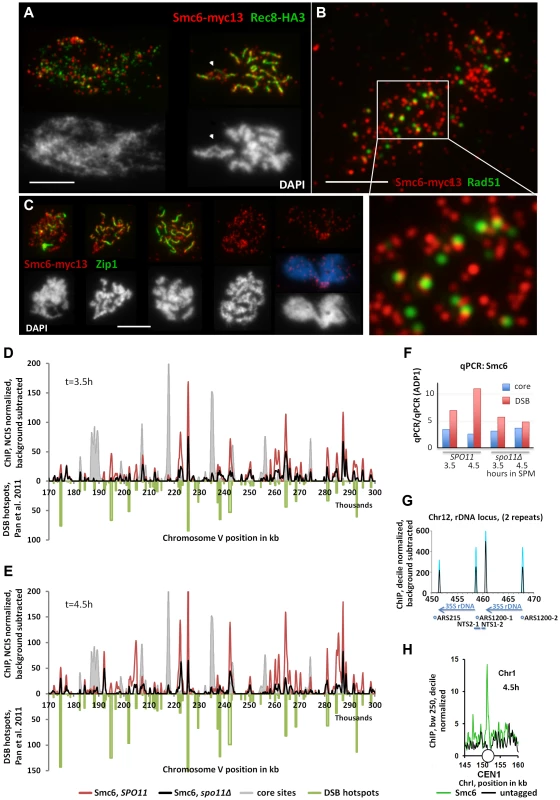
(A) Co-immuno labeling for Smc6-myc13 and cohesin subunit Rec8-HA3 of a spread, leptotene nucleus and pachytene nucleus. Red: Smc6-myc13, Green: Rec8-HA, white bar: 5 µm. (B) Co-immuno labeling for Smc6-myc13 and recombinase Rad51 of a spread meiotic prophase I nucleus. Red: Smc6-myc13, Green: Rad51, white bar: 5 µm. White rectangle indicates position of magnified sub region. (C) Co-immuno labeling for Smc6-myc13 and synapsis specific Zip1 protein of several meiotic stages. (Figure S3 displays a complete series of double stained nuclei, staged according to Zip1 and DNA morphology): Red: Smc6-myc13, Green: Zip1, White (and blue) figures below the immunostained nuclei represent the corresponding nuclear DNA stained with DAPI. White bars: 5 µm. Stages correspond to (from left to right): Early zygotene, late zygotene, pachytene, diplotene, late anaphase I. (D) Smc6-myc13 (t = 3.5 hours in SPM) localizes at DSB sites. ChIP-seq signals on a 130 kb region of chromosome V are shown after smoothing (bandwidth: 500 bp), NCIS normalization and background subtraction as described in Materials and Methods. Red: Smc6-myc13, black: Smc6-myc13, spo11Δ, filled grey profile: Mer2-HAint. (t = 4h) to illustrate core site signals. DSB hotspots defined in [53] were plotted in green on the negative scale to indicate their positions and relative intensities. The diagram on the right shows the results of qPCR at three positions on chromosome III from the same experiment: a DSB site (ca. at 211k, YCR047C), a core site (ca. at 219k) and a cold spot (ca. at 136, ADP1). To correct for possible differences in the efficiencies of the IPs across the different strains, the enrichment of core and DSB qPCR signals relative to the ADP1 signal per ChIP is plotted for the indicated genotypes and time points. Core/ADP1 shown in blue, DSB/ADP1 in red. (E) Same as (D) but Smc6-myc13 analyzed at t = 4.5 hours in SPM. (F) qPCR on chromosome III from the same experiment as in (D,E), a DSB site (ca. at 211k, YCR047C), a core site (ca. at 219k) and a cold spot (ca. at 136, ADP1). To correct for possible differences in the efficiencies of the IPs across the different strains, the enrichment of core and DSB qPCR signals relative to the ADP1 signal is plotted for the indicated genotypes and time points, Core/ADP1 shown in blue, DSB/ADP1 in red. (G) Representation of the rDNA locus on chromosome XII as provided by the Saccharomyces Genome Database (SGD) showing two (of approximately 200) rDNA repeats. Profiles for Smc6-myc13 at 4.5 hours in SPM are shown (blue: WT, black: spo11Δ; smoothed at bandwidth 250bp, decile normalized and background subtracted (minus untagged)). Sharp Smc6 signals flank the 35S rDNA transcriptional unit independent of Spo11. Positions of replication origins (ARS), nontranscribed spacers (NTS) and RDN37 repeats (blue arrows) are indicated below. (H) Centromeres display a Smc6 signal. As in (G) but without background subtraction for a small region around CEN I (white circle). Green: Smc6-myc13, t = 4.5, black: untagged. The preferred binding sites of Smc6 in wild-type meiosis differ markedly from those of the related cohesin complex, which binds specifically to chromosome regions that constitute the chromosome axis upon condensation [1], [51], [52]. On meiotic chromatin, foci of Smc6-myc13 and Rec8-HA3 (a tagged version of the meiotic kleisin-subunit of cohesin) generally exclude each other (Figure 2A, S2B). When chromosomes condense, Smc6 signal often protrudes from the Rec8-axes, suggesting preferential binding of the Smc5/6-Mms21 complex to DNA regions not associated with the axis (Figure 2A).
In order to determine the chromosomal binding sites of Smc5/6-Mms21 complex, ChIP-Seq (Chromatin-Immuno Precipitation followed by next generation sequencing) of Smc6-myc13 was performed. Synchronous meiotic cultures were crosslinked with formaldehyde, subjected to ChIP and the precipitated DNA deep sequenced on an Illumina platform.
Figure 2D, E show a 130 kb representative region of Chromosome V with about 55 medium to weak DSB hotspots mapped previously [53]. Figure 2D, E demonstrate that the majority of Smc6 peaks localize precisely to these hotspots. In the example shown, the 3.5 hour Smc6 peaks fall into 45 (82%) hotspots, while 49 (89%) hotspots coincide with a 4.5 hour Smc6 peak. The vast majority of peaks are precisely on top of the hotspots (up to 80% genome wide), however, we note that the intensities of the co-localizing Smc6 peaks are often not proportional to the intensity of the break site, resulting in the relatively low genome wide Pearson correlations despite precise co-localization.
To address whether Smc6 localization depended on the formation of DSBs by Spo11, the experiment was repeated in a spo11Δ mutant. A genome wide reduction of Smc6 at DSB sites was observed, however, surprisingly, Smc6 still accumulated at the majority of hotspots (Figure 2D, E, S4). The genome wide ChIP experiments were accompanied by qChip at a DSB hotspot (YCR047, 211k), a core site (219k) and a cold region (ADP1, 136k) all on chromosome III (see Figure S4). qChIP confirmed the results on the corresponding positions of the ChIP-Seq profiles (Figure 2F). The ChIP-Seq experiments were repeated confirming the reported results (not shown). The qPCR of the biological repeat confirmed the Spo11 improved enrichment of Smc6 at the hotspot precisely (Figure S4C). Not all binding sites of Smc6 are DSB-sites. For instance, sharp Smc6-peaks mark all the centromeres (Figure 2H, S4A,B). Furthermore, confirming our cytological observation of abundant rDNA localization of Smc6 foci, Spo11 independent Smc6 signals flank the 35S rDNA transcriptional units (Figure 2G). Binding of Smc6 to the rDNA region is in agreement with the crucial anti-recombination role that Smc5/6-Mms21 plays at this repetitive DNA locus [37]. For the remaining Smc6 signal we often observe overlaps with meiotic chromosome axis sites as defined by Mer2 and Hop1 [51]. Such axis specific enrichment for Smc6-myc13 is, however, rather low (Figure 2, S4). Similar conclusions have been drawn in the study of Copsey and coworkers (accompanying manuscript) [54], although more prominent binding of Smc5 at core sites is found than in our study. The quantitative discrepancies in the recovery for Smc5 and Smc6 may reflect different biological properties of the complex. For instance, it was reported for the related cohesin complex, that different cohesin populations exist regarding stability of chromatin binding [55]. Further, we observed previously that the choice of a tag can influence the balance between core and DSB site residency [51], [56]. Differences can also arise due to the different subunits analyzed, the different platforms and resolutions used and the highly stringent background subtraction that we use. Importantly, both studies observe signals consistent with a role of Smc5/6 at sites of meiotic recombination.
Due to observed recruitment of Smc6 to DSB hotspots, we asked whether Rad51, the eukaryotic strand exchange protein which assembles along the resected DSB-ends and facilitates the strand invasion step in HR repair, co-localizes with Smc6-myc13. The number of Smc6-myc13 foci far exceeded (about 4-fold) the Rad51 foci, consistent with DSB independent loading of Smc6 to chromatin. Strikingly, we found almost no on-top co-localization of Smc6 with Rad51 (Figure 2B). Counting foci on 6 meiotic nuclei confirmed this impression. Only 0–10% of the Rad51 foci directly co-localized with Smc6 (on average 6.5±4%). However, a total of 85±4% (n = 6 nuclei scored) Rad51 foci were in a side-by-side configuration with one or two Smc6 foci, even on nuclei with strongly spread chromatin (Figure 2B). A similar relation of Smc6-myc13 with the ZMM-DSB marker protein Zip4-myc9 (Figure S2C) was observed.
In summary, we identify a precise and sensitive association of Smc6 with meiotic recombination hotspots and enrichment of Smc6 to DSB hotspots upon actual DSB formation. The frequent side-by-side co-localization of Smc6 and Rad51 recombinase foci is indicative of a primarily spatial separation with juxtaposed positioning of the two complexes.
The Smc5/6-Mms21 complex is required to prevent the accumulation of toxic Joint Molecules
Unresolved Holliday Junctions (HJs) stably connect chromosomes that had engaged in homologous recombination and thus may result in chromosome nondisjunction. A sufficiently large number of such unresolved recombination intermediates prevent nuclear divisions resulting in mitotic or meiotic catastrophe [7], [10], [12]. Since previous studies reported an accumulation of repair intermediates in mutants of the Smc5/6-Mms21 complex [37], [57], [58], we asked whether an accumulation of unresolved recombination intermediates in the smc6-56 mutant might explain the Spo11 dependent failure in nuclear divisions.
Meiotic recombination was followed in a physical assay at the URA3-arg4 locus under conditions that preserve HJ intermediates [59]. This Southern based assay allows quantification of the key intermediates of meiotic recombination: DSBs, dHJ intermediates, as well as crossover and non-crossover recombination products due to restriction polymorphisms between the parental chromosomes (Figure 3A). Using this recombination reporter system, wild type and mutant strains were analyzed under restrictive conditions for smc6-56 in a time course experiment. Joint molecule levels were measured by probing against ARG4 DNA on genomic XmnI restriction-fragments. No differences were noted in the transient appearance of DSBs.
Fig. 3. The Smc5/6-Mms21 complex is required to prevent the accumulation of toxic Joint Molecules. 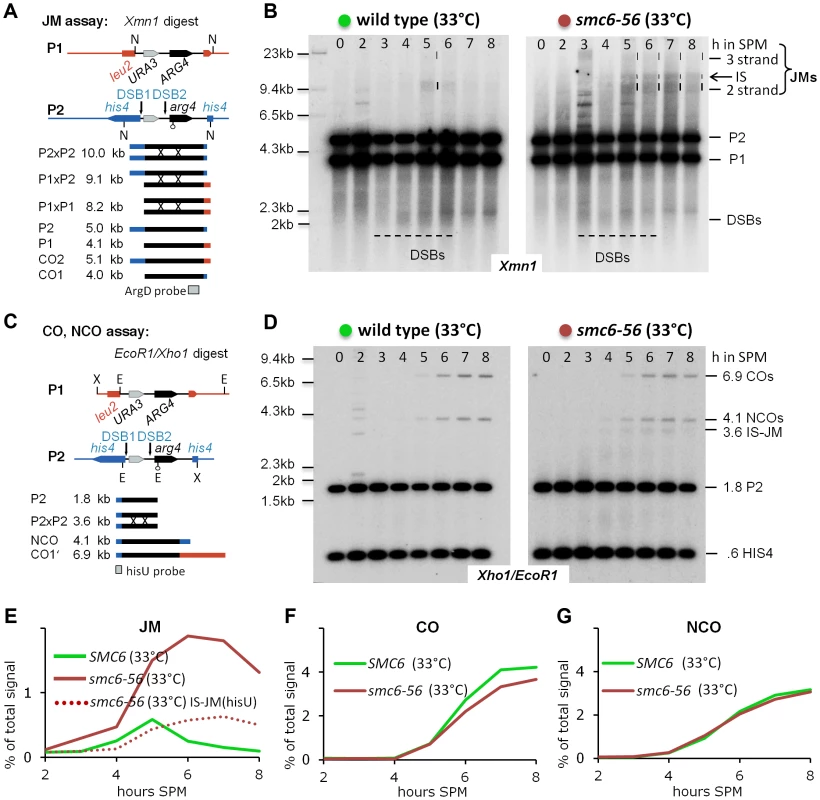
(A) Schematic representation of the genetic loci, restriction sites and the probe used to demonstrate joint molecule formation (modified from [6]). (B) Joint molecules accumulate and persist in smc6-56. Southern blotting of samples taken from synchronous meiotic time courses at restrictive temperature with DNA extracted and digested with Xmn1 under conditions preserving JMs and using the probe indicated in (A). Lane number indicates hours in SPM. Size markers are provided to the left of the blots while the identity of the labeled species is indicated to the right. The bracket indicates the region where various JMs (2 strand (IH and IS), 3 strand or 4 strand JMs) migrate to. P1, P2: parental fragments. Dashed lines highlight the regions on the blot where DSB signals or JM signals appear. Left Panel: Wild type, right panel: smc6-56. (C) Schematic representation of the genetic loci, restriction sites and the probe used to detect and quantify IS-JMs, COs and NCOs (modified from [6]). (D) As in (B) from the same time course experiment but genomic DNA digested with Xho1 and EcoR1 and probe corresponding to (C). Note: hisU probe detects only one parental fragment (P2). (E) JMs accumulate and persist in smc6-56 at restrictive temperature. Total JM signals were quantified, subtracted from background and plotted as % of total signal as a function of time in SPM. Green: Wild type (33°C), red: smc6-56 (33°C), red dotted line: Inter sister JM from the blot shown in (D). (F) Near normal levels of CO in smc6-56: Representation as in (E) but quantification of CO product. (G) Normal levels of NCO in smc6-56: Representation as in (E) but quantification of NCO product. However, while joint molecules appear transiently in wild type with low steady state levels and a clear peak at 5 hours when cells are in the pachytene stage of meiosis, the smc6-56 mutant accumulated high molecular weight recombination intermediates that failed to be resolved (Figure 3B). The observed persistent joint molecules in smc6-56 amount to more than twice that of the JM peak in wild type, exceeding the amount of stable joint molecules reported to block chromosome segregation (for example in the mms4-mn yen1Δ double mutant [9]). In addition, while joint molecules rarely form between sister chromatids in wild type (IS:IH≤0.2), in the smc6-56 mutant a substantial amount of inter sister JMs (IS-mom:IH 0.32–0.38, by two different assays; XmnI and XhoI/EcoRI) contribute to the overall persistent JMs (Figure 3B, D). Using a HIS4 fragment to probe for COs and NCOs after a different digest (XhoI/EcoRI) revealed that the levels of both these recombination products were not significantly altered in smc6-56 (Figure 3C, D, F, G). Consequently, the sum of COs, NCOs and JM-intermediates in the mutant exceeds corresponding numbers in the wild type, uncovering a role of Smc5/6-Mms21 in early destabilization of intermediates to prevent stable JM formation.
In summary, the Smc5/6-Mms21 complex is required to prevent the accumulation of aberrant, unresolved recombination intermediates. The results further suggest that most of the stabilized JMs arise from recombination events not resulting in CO or NCO products in wild type, such as inter sister SDSA or dissolution of rogue dHJs. In particular, this suggests that the CO specific ZMM-pathway is not affected by the defect in smc6-56.
Mms21 SUMO E3 ligase activity prevents tight homolog associations in zip3Δ mutants
If the smc6-56 mutant confers persistent joint molecules to the cell that impede nuclear divisions, then there are two feasible explanations as to why the mms21-11 mutant does not. Either mms21-11 represents a plain Smc5/6-Mms21 hypomorph in meiosis with remaining activity at a level such that formation of aberrant joint molecules is negligible. Alternatively, mms21-11 may represent a separation of function mutant of the complex in which prevention of aberrant JM formation and the resolution of such by the Smc5/6-Mms21 complex had become separated. If the latter is true, it should be possible to detect inappropriate joint molecule formation as well as their removal in mms21-11.
To test if the mms21-11 mutant indeed fails to prevent the formation of additional and atypical joint molecules in considerable amounts, we assayed on surface spread meiotic nuclei for the presence of excess axial associations in the background of the ZMM mutant zip3Δ. In mutants of the ZMM pathway chromosome synapsis is defective, the repair of ZMM-breaks is inhibited through activity of the Sgs1 helicase [11], and dHJ and CO formation are impaired while Non-ZMM breaks are repaired by SDSA [4]. Axial associations as first described in the zip1Δ mutant are the cytological manifestation of stable recombination intermediates between the homologs, approximating dHJs and COs numbers in wild type [60]. In the SK1 strain background, zip3Δ confers one of the most severe ZMM phenotypes, exhibiting a strong reduction in JMs and a robust prophase I arrest [4].
Accordingly, at 5 hrs in SPM, when in wild type cells most homologs are synapsed, univalents almost bare of axial associations and recognizable pairing dominate the zip3Δ mutant phenotype (Figure 4A). 63% of nuclei have no more than 2 chromosomes per nucleus paired or connected by axial associations (n = 100; Figure 4B). In contrast, pairing and axial associations are frequent in the zip3Δ mms21-11 double mutant, with 5–6 chromosome pairs on average in one experiment and as many as 7 in a biological repeat (n = 100; Figure 4A, B). A similar improvement in pairing can be seen for zip3Δ smc6-56, but not for mnd1Δ mutants, which are defective in the strand invasion step (Figure S5A) [48], [49].
Fig. 4. Mms21 SUMO E3 ligase activity antagonizes inappropriate Joint Molecule formation in zip3Δ and in mms4-mn mutants. 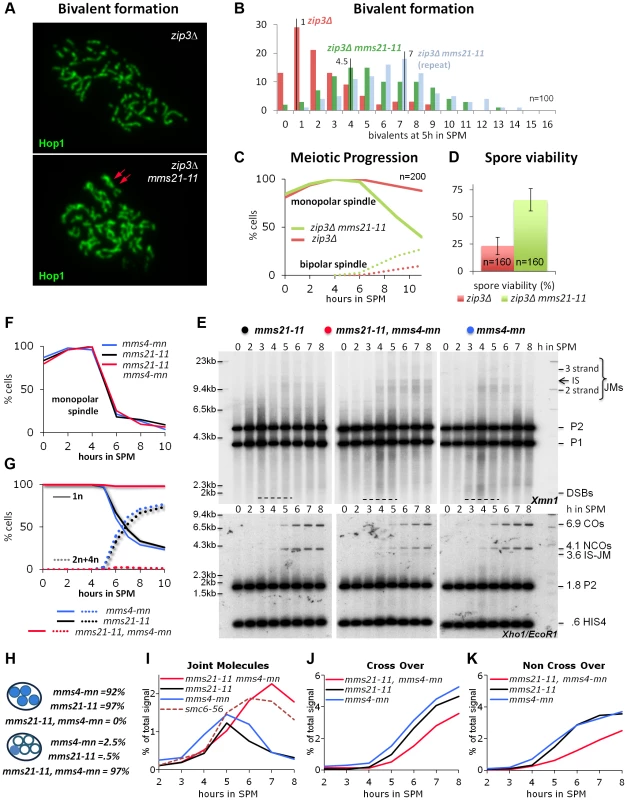
(A) mms21-11 restores bivalent formation and axial associations in zip3Δ mutants. Shown are representative nuclei after chromosome spreading and immunolabeling with Hop1 to visualize chromosome axes. Top panel: zip3Δ. Bottom panel: zip3Δ mms21-11 double mutant. Red arrows point at a mms21-11 dependent bivalent with apparently two axial associations. Several such bivalents emerge in this nucleus. (B) Quantification of bivalent formation: Spread nuclei were classified according to the number of bivalents present. The discrete density of the distribution is plotted (incidences against class). n = 100 nuclei were classified per experiment. Red: zip3Δ, green and blue: two biological repeats of zip3Δ mms21-11. (C) mms21-11 improves meiotic progression in zip3Δ: Meiotic progression was followed via spindle morphology. Monopolar and bi-polar spindles were plotted against hours in SPM. n = 200 cells per experiment, continuous lines: monopolar spindles, dotted lines: bipolar spindles, red: zip3Δ, green: zip3Δ mms21-11. (D) mms21-11 improves spore viability in zip3Δ: Spore viability was assayed by tetrad dissection in n = 40 tetrads per mutant, red: zip3Δ, green: zip3Δ mms21-11. (E) JMs accumulate in mms21-11 that require the Mus81-Mms4 resolvase for resolution: Same JM, CO and NCO assay as described in Figure 3 (A through D). Southern blotting of samples from synchronous meiotic time courses at 30°C. Upper panels: Xmn1 digest, JM detection. Left: mms21-11, Middle: mms21-11 mms4-mn, Right: mms4-mn. Lower panels: Xho1/EcoR1 digest, CO/NCO detection. Left: mms21-11, Middle: mms21-11 mms4-mn, Right: mms4-mn. (F) Meiotic progression is normal in all mms21-11 and mms4-mn mutants: Meiotic spindles were labeled. Monopolar spindles were plotted against hours in SPM. n = 200 cells per experiment, continuous lines: monopolar spindles. Red: mms4-mn mms21-11, black: mms21-11, blue: mms4-mn. (G) Meiotic nuclear divisions are blocked in mms4-mn mms21-11 double mutants. Continuous lines: cells containing one DAPI stained nucleus (1n), dotted lines: cells with 2 or 4 nuclei/DAPI-bodies (2n+4n). Red: mms4-mn mms21-11, black: mms21-11, blue: mms4-mn. (H) Segregation of DNA into spores: Numbers represent the percentage of tetrads with either DNA in all 4 spores (upper 3 lines), or all chromosomes outside the spores (lower 3 lines). (I) JMs accumulate and persist in mms4-mn mms21-11 double mutants. Total JM signals were quantified, subtracted from background and plotted as % of total signal as a function of time in SPM. Red: mms4-mn mms21-11, black: mms21-11, blue: mms4-mn, red dashed line: smc6-56 (33°C). (J) Reduced levels of CO in mms4-mn mms21-11 double mutants: Representation as in (I) but quantification of CO product. (K) Reduced levels of NCO in mms4-mn mms21-11 double mutants: Representation as in (I) but quantification of NCO product. In addition to the increased number of axial associations, mms21-11 ameliorated the pachytene arrest of zip3Δ. Following the spindle morphology as a marker of meiotic progression, we find 88% of zip3Δ cells at 11 hrs still in prophase I, whereas 60% of the zip3Δ mms21-11 cells had at the same time already progressed beyond prophase I (n = 200; Figure 4C). Additionally, sporulation of zip3Δ was improved from 12.75% to 22.5% in the double mutant at 24 hours (n = 400). Thus, ZMM DSBs of zip3Δ are turned over in the mms21-11 background into intermediates that do not elicit a DNA damage checkpoint response. The suppression of the prophase I arrest is not due to a checkpoint defect because the resulting tetrads of the double mutant exhibit a greatly improved spore viability of 65.63%, from 23.13% in zip3Δ (n = 160 spores; Figure 4D). The opposite would be expected for a checkpoint failure. Impairment of an early anti-recombinogenic function and inappropriate transformation of DSBs to JMs in mutants of the Smc5/6-Mms21 complex is further supported by the observation that neither absence of the three resolvases Mus81-Mms4, Slx1-Slx4 and Yen1, responsible for the removal of unregulated JMs [5], [9], nor the absence of all four meiosis relevant resolvases, Mlh1/3-Exo1, Mus81-Mms4, Slx1-Slx4 and Yen1 which account for ≥90% of the meiotic resolution activity [5] can improve chromosome pairing in zip3Δ (Figure S5B,C in comparison to nse4-mn and sgs1-mn, meiosis specific null alleles of the Smc5/6 kleisin subunit and the Sgs1/BLM helicase, respectively).
These results indicate that the SUMO E3 ligase deficient mms21-11 allele fails to antagonize dHJ-formation during DSB repair in the zip3Δ mutant, thereby improving bivalent formation, facilitating meiotic progression and ultimately greatly improving spore viability. This is similar to the effect of sgs1-mn (zip3Δ sgs1-mn: 71.25%, n = 160 spores). While restoration of COs in zip3Δ mms21-11 remains to be confirmed by physical analysis, we conclude that Mms21 as part of the Smc5/6 complex is specifically required for an early, antagonistic function of the complex in meiotic recombination, most likely by preventing the formation of illegitimate joint molecules, a role previously reported for the helicase BLM/Sgs1 [10]–[12], [61].
Lack of the Mms21 SUMO E3 ligase leads to rogue HJ intermediates, depending on Mus81-Mms4 for resolution
Three resolvases with overlapping function are responsible for eliminating rogue HJ intermediates in budding yeast: Mus81-Mms4, Slx1-Slx4, and Yen1. JMs arising outside the ZMM pathway depend on these three partially redundant resolvases for resolution, with the XPF family nuclease Mus81-Mms4 showing the biggest contribution [5], [9]. If, as the Zip3 results suggest, considerable amounts of rogue joint molecules form also in the mms21-11 single mutant, these joint molecules must apparently be efficiently removed prior to nuclear divisions as implied by functional chromatin segregation and high spore viability in mms21-11. The most important “rogue JM resolvase” Mus81-Mms4 is the most likely candidate to carry out this vital role. If mms21-11 represented a separation of function mutant of the Smc5/6-Mms21 complex, unable to prevent aberrant JMs, additional inactivation of Mus81-Mms4 should block nuclear divisions, thereby recreating the full smc6-56 mutant phenotype.
To test this hypothesis Mus81-Mms4 function was eliminated in the mms21-11 background using a characterized meiotic null allele of MMS4 (mms4-mn) [10]. Indeed, in the mms21-11 mms4-mn double mutant meiotic segregation of chromosomes is blocked, resulting in meiotic catastrophe, while only rarely a cell of the single mutants shows this defect (Figure 4G,H). Chromosome synapsis and meiotic progression were not different from wild type for the single mutants or the double mutant (Figure 4F). As predicted by these results, persistent JMs accumulated in the mms21-11 mms4-mn double mutant at levels comparable to those found in the smc6-56 mutant while the JMs of both single mutants were successfully resolved (Figure 4E,I).
In wild type meiosis, most COs are formed in the ZMM pathway from dHJs resolved via Exo1-Mlh1/3, while the NCOs arise from non-ZMM DSBs repaired through SDSA [5], [9]. In the mms21-11 mutant, a fraction of both COs and NCOs become dependent on MMS4 as their formation is reduced in mms21-11 mms4-mn (Figure 4J,K) but not in MMS21 mms4-mn. Thus, these recombination products form in mms21-11 from the resolution of rogue dHJs by Mus81-Mms4, implying that a significant fraction of ZMM and SDSA breaks in mms21-11 switch to a rogue JM fate.
In summary, the evidence indicates that the Mms21 SUMO E3 ligase is required to prevent the formation of inappropriate HJ intermediates. However, in contrast to the smc6-56 mutant, the aberrant JMs can still be removed in mms21-11, but require for this the activity of the Mus81-Mms4 resolvase.
The Smc5/6-Mms21 complex is required for the function of the “rogue JM resolvases”
Despite the failure to resolve a considerable amount of JMs (Figure 3B–D), the levels and kinetics of meiotic recombination products in the smc6-56 mutant are only marginally affected. Consequently, the Smc5/6-Mms21 complex is not essential for the resolution of all JMs in budding yeast meiosis. However, as the mms21-11 mutant does generate aberrant HJ intermediates that need Mus81-Mms4 for resolution, the Smc5/6-Mms21 complex could be required exclusively for the removal of the ZMM-independent “rogue JMs”.
To specifically address the ability of the smc6-56 mutant to remove ZMM-independent JMs, JM resolution and product formation was analyzed in the background of an SGS1 meiotic null allele (sgs1-mn). In sgs1-mn mutants, nearly all ZMM recombination intermediates adopt a rogue JM fate and, consequently, nearly all recombination products, namely COs and NCOs, depend on the “rogue JM resolvases” Mus81-Mms4, Slx1-Slx4, and Yen1 [5], [9]. If the Smc5/6-Mms21 complex were critically required for the removal of rogue JMs via these resolvases, the smc6-56 mutant should strongly inhibit both JM resolution and product formation in the sgs1-mn background.
As shown in Figure 5A–E, the sgs1-mn smc6-56 double mutant does indeed dramatically accumulate JMs, accompanied by a substantial loss in recombination products consistent with a failure of the rogue JM resolvases in non-ZMM JM removal. In contrast, the sgs1-mn single mutant efficiently resolves JMs to CO and NCO products (Figure 5A–E), as expected [5], [9], [10]. Similarly to the smc6-56 single mutant, nuclear divisions are completely blocked in the double mutant, while meiotic progression based on spindle morphology is unaffected in single and double mutants (Figure 5F,G).
Fig. 5. The Smc5/6-Mms21 complex is required for the function of the “rogue JM resolvases”. 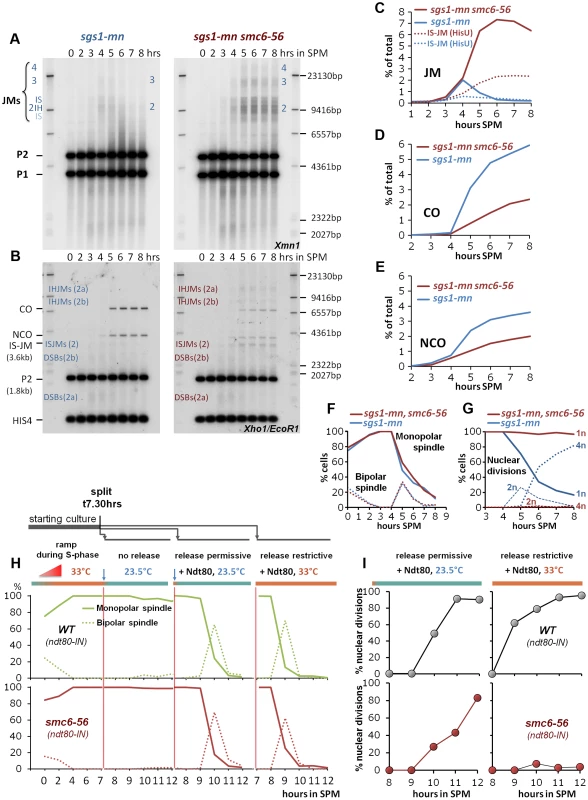
(A) Rogue JMs forming in sgs1-mn require Smc6 for their resolution: Same JM assay as described in Figure 3 (A, B). Southern blotting for JMs from synchronous meiotic time courses at 33°C. Xmn1 digest, JM detection. Left: sgs1-mn, Right: sgs1-mn smc6-56. (B) CO and NCO products from sgs1-mn are severely repressed in the absence of Smc6: Same CO and NCO assay as described in Figure 3 (C, D). Southern blotting of samples from synchronous meiotic time courses at 33°C. Xho1/EcoR1 digests, IS-JM/CO/NCO detection. Left: sgs1-mn, Right: sgs1-mn smc6-56. (C) Total JM signals were quantified, subtracted from background and plotted as % of total signal as a function of time in SPM. Continuous lines: JM from (A). Dotted lines: Inter sister JM from (B). Blue: sgs1-mn, red: sgs1-mn smc6-56. (D) CO signals from (B) were quantified, subtracted from background and plotted as % of total signal as a function of time in SPM. Blue: sgs1-mn, red: sgs1-mn smc6-56. (E) NCO signals from (B) were quantified, subtracted from background and plotted as % of total signal as a function of time in SPM. Blue: sgs1-mn, red: sgs1-mn smc6-56. (F) Meiotic progression is unaffected in sgs1-mn smc6-56: Meiotic progression was followed by spindle labeling. n = 200 cells per experiment, continuous lines: monopolar spindles, dotted lines: bipolar spindles. Blue: sgs1-mn, red: sgs1-mn smc6-56. (G) Meiotic nuclear divisions are blocked in sgs1-mn smc6-56 double mutants. Continuous lines: cells containing one DAPI stained nucleus (1n), dashed lines: cells with 2 nuclei, dotted lines: cells with 4 nuclei (4n). Blue: sgs1-mn, red: sgs1-mn smc6-56. (H, I) Shift to permissive temperature during ndt80 release restores nuclear divisions in smc6-56. (H) Upper panels, green: Wild type, lower panels, red: smc6-56. Continuous lines: Monopolar spindles. Dotted lines: bipolar spindles. From left to right: 1st panel: Time course experiment under restrictive conditions in the ndt80-IN strain background in the absence of inducer (estradiol). 2nd panel: shift to permissive temperature (23.5°C), without release (− estradiol). 3rd panel: release (+ estradiol) into permissive temperature (causes short and synchronous burst of bipolar spindles). 4th panel: release (+ estradiol) into restrictive temperature (causes short and synchronous burst of bipolar spindles). (I) % Multi nuclear cells (2n+4n) are plotted against hours in SPM. Upper panels (grey filled circles): SMC6 ndt80-IN, lower panels (red filled circles): smc6-56 ndt80-IN. Left panels: ndt80 release (+ estradiol) into permissive conditions restores divisions in the smc6-56 background. Right panel: release (+ estradiol) into restrictive temperature maintains the block of division. In the sgs1-mn background where nearly all DSBs adopt a rogue fate, the amount of persistent JMs in the sgs1-mn smc6-56 mutant is 3 to 4-fold elevated compared to the smc6-56 single mutant, reaching a high level of 6–7% unresolved JMs. The failure to resolve JMs in the sgs1-mn smc6-56 double mutant resulted in corresponding depletion of 60% of the sgs1-mn COs and 45% of the sgs1-mn NCOs (Figure 5C–E). We conclude that the Smc5/6-Mms21 complex is specifically required for the resolution of the unregulated, “rogue” JMs and that the resolution activity lost due to smc6-56 equals the effect of loss of at least the most active rogue JM resolvase Mus81-Mms4 [9], [10].
To investigate whether JMs might become terminally inaccessible to resolvases due to an early defect in smc6-56, cells were allowed for 7 h 30 min to complete prophase I until the arrest in late pachytene of ndt80 under restrictive temperature. Cells were then released into permissive temperature to supply functional Smc6 for JM resolution. The release was mediated by addition of estradiol using an estradiol inducible NDT80 allele (ndt80-IN). Ndt80 expression induces pachytene exit and Cdc5 expression which in turn activates the resolvases for resolution of dHJs [6], [7], [62]. If JMs had derailed and become unresolvable early, the late addition of Smc6 should not be able to support nuclear divisions.
The key result of the experiment is shown in Figure 5I, namely that providing Smc6 after the ndt80 arrest restores nuclear divisions to wild type levels, with only a slight delay, required to resolve the accumulated JMs. Notably, all the controls were as expected, that is cells efficiently resumed meiotic progression upon estradiol addition independent of the temperature (Figure 5H). Also, SMC6 performed nuclear divisions independent of the restrictive conditions, while of course successful nuclear divisions in smc6-56 depended on the termination of restrictive conditions (Figure 5H,I). We conclude that it is sufficient to provide Smc6 function after the ndt80 arrest point to ensure chromosome segregation. The converse experiment, in which cells process DSBs under permissive conditions but are released under restrictive conditions from the ndt80 arrest, shows that cells block (Figure S6) consistent with a critical role of Smc5/6-Mms21 complex in mediating the function of rogue JM resolvases beyond only Mus81-Mms4.
In summary, we conclude that Smc5/6-Mms21 promotes the function of the “rogue JM resolvases” at the time of resolution - and thus rather directly, and that the integrity and accessibility of the aberrantly formed JMs is not affected.
Smc5/6-Mms21 supports the function of the anti-recombinogenic helicase BLM/Sgs1
The biological function of the Mms21 SUMO E3 ligase domain is presumably mediated through regulation of downstream factors. The most obvious candidates for destabilizing early intermediates are helicases that can unwind recombinogenic structures. The helicase reported to interact with the Smc5/6-Mms21 complex is Mph1, an anti-recombinogenic, FANCM like helicase, but unlike Sgs1 or Mms21 SUMO ligase activity, Mph1 is dispensable for coping with the bulk of induced lesions [63], [64]. However, absence of Mph1 activity partially alleviates defects in Mms21 SUMO E3 ligase mutants, suggesting dysregulation of Mph1 activity [63], [64]. The same study also found that this suppression depends on the activity of Sgs1, the sgs1Δ mutant being epistatic. These data suggest that Mph1 activity becomes toxic in the absence of Smc5/6-Mms21 and may hamper Sgs1 mediated repair. It is therefore possible that the Mms21 SUMO E3 ligase mediates its anti-JM formation activity by coordinating the two helicases, Sgs1 and Mph1. However, observations in a previous study in which mms21-11 and sgs1Δ conferred a synthetic growth defect in the double mutant led to the notion that Sgs1 and Smc5/6-Mms21 may not work together [64].
In this study a striking overlap between the biological functions of Mms21 and Sgs1 is apparent. Similarly to Sgs1, Mms21 prevents the formation of rogue JMs and is required for SDSA and the repair block of early ZMM breaks, although the defect of mms21-11 is clearly less pronounced than that of an sgs1 mutant. This similarity in behavior of mms21-11 and sgs1 mutants is also observed in studies on mitotic cells [40]. In vegetative growth, we found very similar synthetic interactions for mms21-11 as for sgs1Δ and mph1Δ (Figure S7B, C; and [65]), including reduced growth for the mms21-11 sgs1Δ double mutant. For the synthetic interaction of mms21-11 and sgs1-mn in meiosis, we observed a moderate chromosome segregation defect in the double mutant. Specifically, 60% of tetrads were missing DNA in at least one spore and 39% of the tetrads (n = 200) were devoid of DNA in all four spores (Figure 6A).
Fig. 6. The Smc5/6-Mms21 complex supports the anti-recombinogenic function of Sgs1. 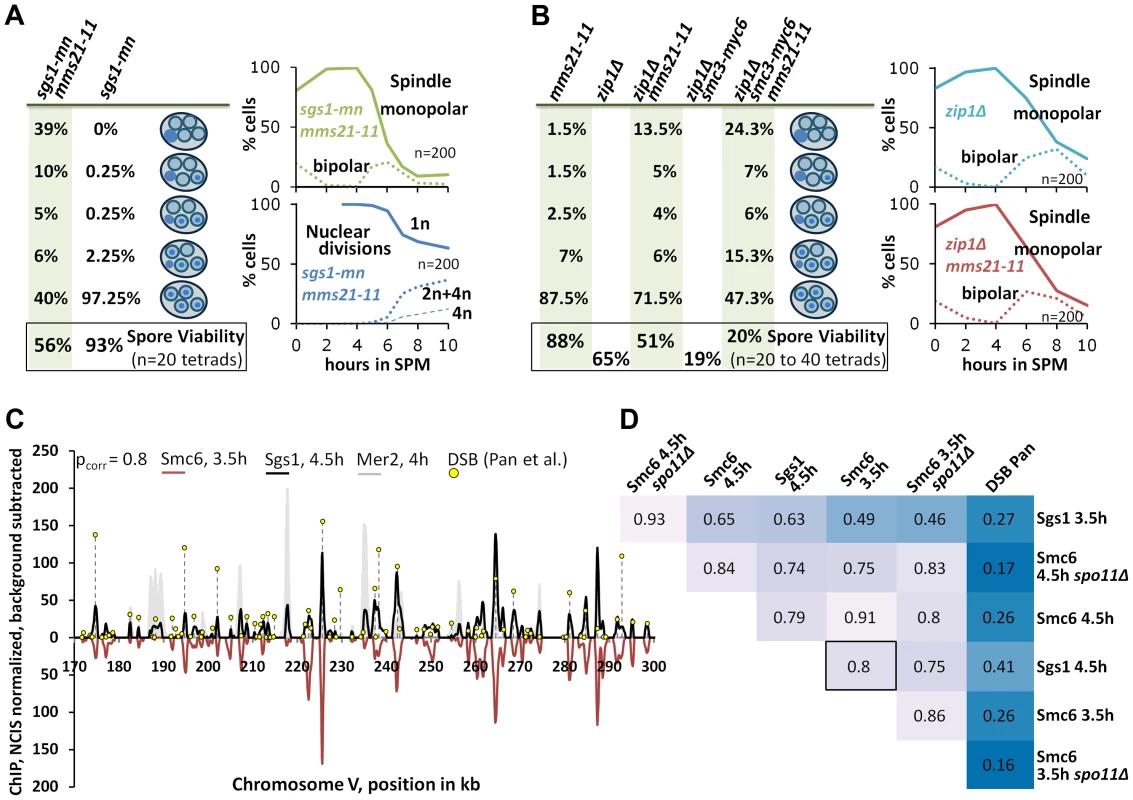
(A) Lack of the Mms21 SUMO ligase mildly compromises chromatin separation in the sgs1-mn background. Left column on green background: From top down: Percent of tetrads with 0, 1,2,3,4 spores containing nuclear DNA. Bottom: Spore viability from tetrad dissection. Right upper panel: Meiotic progression of sgs1-mn mms21-11 was followed by spindle labeling. n = 200 cells, continuous line: monopolar spindles, dotted line: bipolar spindles. Right lower panel: Continuous line: cells containing one DAPI stained nucleus (1n), dotted lines: cells with 2 or 4 nuclei (2n+4n), dashed lines: cells with 4 nuclei (4n). (B) As in (A). The Mms21 SUMO ligase guarantees complete chromatin separation in the zip1Δ mutant indicating that the Mms21 SUMO ligase is required for full Smc5/6-Mms21 resolvase activity. Top to bottom: Percent of tetrads with 0,1,2,3,4 spores containing nuclear DNA. Number at the bottom: Spore viability from tetrad dissection given as percentage. Genotypes indicated above each column. Right upper panel: Meiotic progression of zip1Δ was followed by spindle labeling. n = 200 cells, continuous line: monopolar spindles, dotted line: bipolar spindles. Right lower panel: Meiotic progression of zip1Δ mms21-11 was followed by spindle labeling. n = 200 cells, continuous line: monopolar spindles, dotted line: bipolar spindles. (C; and D) Chromosomal interaction sites and intensities are highly similar between Sgs1 and Smc6. DNA-interaction sites for Sgs1-myc18 (t = 4.5 hours in SPM) and Smc6-myc13 (t = 3.5 hours in SPM) on a 130 kb region of chromosome V. ChIP-seq signals are shown after smoothing (bandwidth: 500 bp), NCIS normalization and background subtraction as described in Materials and Methods. Black: Sgs1. Smc6 signals were plotted in red on the negative scale to facilitate comparison. The obvious Sgs1/Smc6 symmetry in the example region is corroborated genome-wide by the high Pearson correlation (pcorr = 0.8) over more than 6000 peaks per profile. Yellow lollipops: DSB positions as defined in [53] (corresponding to ∼7000 DSB-hotspots across the genome). These positions map overwhelmingly often precisely at the ChIP-seq peaks. The filled grey profile represents Mer2-HAint. (t = 4h) to illustrate core site signals. Note that smaller Sgs1 signals, and to a lesser extent also Smc6 signals also overlap with core sites, as defined by Mer2. (D) Pearson correlation coefficient matrix between the peaks of all profiles shown in this work. Matching peaks (after smoothing to a bandwidth of 300 bp and subtracting the smoothed untagged control) were identified (based on exceeding a certain threshold, set here to Q = 0.7) and their intensities compared by Pearson correlation. The coloring should facilitate comparisons: dark blue represents low correlations whereas lighter shades indicate increased correlation. Intensities between DSBs and ChIP-seq profiles do not match (pcorr<.3), whereas intensities of peaks between Sgs1 and Smc6 profiles usually highly (frequently pcorr>.75). Since Mms21 is an essential subunit of the Smc5/6-Mms21 complex and because the Smc5/6 complex itself is targeted by Mms21 for SUMOylation [27], [28] we tested whether the mms21-11 allele might confer a mild defect in the Smc5/6-Mms21 rogue JM resolvase activity which could explain a synthetic interaction between mms21-11 and sgs1. Since the JMs of ZMM mutants also depend on the rogue JM resolvases for their resolution [9], we tested whether mms21-11 is permissive for DNA segregation in a zip1Δ mutant meiosis. zip1Δ is the most permissive ZMM mutant (presumably providing no substantial Sgs1 mediated ZMM-DSB repair block), forming JMs readily from its DSBs and consequently exhibiting a very short prophase 1 delay [4]. Accordingly, while DSB turnover and meiotic progression in the zip3Δ mutant could be enhanced by mms21-11, the already swift exit of zip1Δ from prophase 1 in SK1 is not improved further by mms21-11 (Figure 6B). However, chromatin segregation is notably affected in the zip1Δ mms21-11 double mutant with 28.5% of the tetrads missing DNA in at least one spore and 13.5% of the tetrads being completely devoid of DNA in all four spores (n = 200). Consistent with a reduced number of crossovers, spore viability is decreased from 65% of zip1Δ to 51.25% in the double mutant (n = 80 spores of 20 tetrads). Combination with a hypomorphic smc3-myc6 allele, which decreases the viability of zip1Δ to 20.0% (n = 160 spores of 40 tetrads), even exaggerates this segregation defect to 52.7% of the tetrads lacking DNA in at least one spore and 24.3% of the tetrads (n = 300) devoid of DNA in all spores in the resulting zip1Δ smc3-myc6 mms21-11 triple mutant. We conclude that mms21-11 indeed mediates a defect to the rogue JM resolvase function of Smc5/6-Mms21.
Given that already low amounts of unresolved JMs completely block nuclear divisions and that sgs1-mn imposes a rogue JM fate on basically all recombination events [9], the defect in the rogue JM resolvase function of mms21-11, although relevant, must be rather weak.
Finally, we tested by ChIP-Seq if the localization pattern of the epitope tagged Sgs1-myc18 would lend support to the idea of cooperation between Sgs1 and the Smc5/6-Mms21 complex. Indeed, 70% of the 1000 strongest Sgs1-myc18 peaks map precisely to sites of meiotic DSB formation, as identified by Pan and coworkers [53] at near single nucleotide resolution where they overlap nearly perfectly with peaks of the Smc6 profiles (Figure 6C). However, the intensity of hotspot-matching Sgs1 peaks is not proportional to the activity of corresponding hotspots but proportional to the corresponding Smc6 peaks. Thus the correlation coefficients between the peaks of Sgs1 profiles and their matching DSB sites is low (cor = .27 [Sgs1, 3.5h], cor = .41 [Sgs1, 4.5h], Figure 6D). Similarly, Smc6 peaks don't correlate well with DSB intensities (cor = .26 [Smc6, 3.5h and Smc6, 4.5h]) despite localizing precisely at hotspots (Figure 6D) for most peaks. This suggests that the proteins are not recruited proportionally to the DSB activity. In contrast, peak intensities match very well between Sgs1 and Smc6 (cor = ∼.8 for different profile comparisons, Figure 6D). Smc6 and Sgs1 also match on many non-DSB positions including prominent peaks at the centromeres (figure 2G, S4, S7A). Sgs1, and to a lesser extent Smc6 also bind to core sites, but in general these signals are much smaller than their signals at DSB sites. These findings show that Smc6 and Sgs1 populate largely the same chromosomal target sites at high resolution and suggest that they are accumulated by some common characteristics at these sites that is different from actual hotspot activity.
Discussion
The Smc5/6-Mms21 complex antagonizes unregulated JMs
Broken chromosomes pose a considerable threat for cells; unrepaired, they cause potentially lethal loss of genetic information, but their repair is equally risky. Inappropriate recombination intermediates and outcomes are inevitable if repair is not carefully controlled. JMs typically arise as HR intermediates with both ends of the DSB lesion engaged with one or more repair templates. Having eliminated the primary lesion and any major stretches of single stranded DNA, JMs do not trigger a DNA damage checkpoint response. Thus, if JMs arise between different chromosomes they mediate dangerous, stable connections due to reciprocal base pairing and catenation which will cause chromosome non-disjunction if not removed in time before anaphase. JMs generated at non-allelic positions are particularly dangerous because their resolution can lead to deletions or translocations and thus need to be avoided in the first place. In many organisms, HR via JMs is strongly down regulated for much of their life cycle, the exception being meiosis where the generation of crossovers via JMs is imperative. Mechanisms and factors that mediate surveillance of such dangerous intermediates or the coordination of the known JM-antagonists are not well known or understood to date.
In meiosis, cells are forced to generate high numbers of crossovers and consequently evolved a specialized recombination pathway on top of the mitotic machinery to do so safely. In budding yeast, the conserved ZMM recombination pathway was first described to perform this task [4], [66], [67]. It involves a sophisticated machinery which works on a subset of DSBs (ZMM-DSBs) to generate appropriate amounts of stable JMs to achieve at least one obligatory CO per bivalent, and it also ensures that COs are formed between the appropriate partners. The ZMM-dHJs are transformed into COs at pachytene exit by a dedicated ZMM resolution machinery, dependent on Exo1-Mlh1/3 [5]. In contrast, non-ZMM DSBs are repaired fast and safely by SDSA to yield NCOs and are thought to support homology search. Recently, BLM/Sgs1 helicase was identified as being central to SDSA mediated NCO formation, as well as in preserving early ZMM recombination intermediates [10]–[12], [61]. Unregulated or “rogue” JMs that arise from non-ZMM DSBs by escaping destabilization through Sgs1, or that escape the ZMM pathway, are resolved by a group of rogue JM resolvases: Mus81-Mms4, Slx1-Slx4 and Yen1 [5], [9].
Here we demonstrate that the Smc5/6-Mms21 complex specifically binds to sites of meiotic DSBs and plays a dual role in homologous recombination as an antagonist of unregulated JMs and HJ intermediates. It does so by promoting anti-JM formation activity through its Mms21 SUMO E3 ligase and, if inappropriate JMs have already formed, by mediating their resolution through rogue JM resolvases. Similar conclusions have been drawn in an independent study (accompanying manuscript) [54]. In particular, that study showed directly that Exo1-Mlh1/3 dependent resolution of ZMM-dHJs does not require Smc5/6-Mms21.
Phenotypes associated with defects in these functions include the recombination dependent failure to separate chromatin during meiosis upon Smc6 inactivation and the concomitant accumulation of persistent JMs that also include a considerable fraction of unresolved IS-JMs and some three-strand JMs indicative of aberrant JM formation. This phenotype resembles the behavior of sgs1-mn mms4-mn mutants [10], [12] in which most meiotic recombination events give rise to “rogue” non-ZMM JMs and block nuclear divisions, because they depend on the inactivated “rogue JM resolvase” Mus81-Mms4 for resolution. In analogy, this implies that the Smc5/6-Mms21 complex mediates both functions in the management of unregulated JMs. It supports the prevention of rogue JMs as does Blm/Sgs1 and mediates their efficient removal through the rogue JM resolvases. Early defects of Smc5/6-Mms21 mutants were also noted in the accompanying manuscript by Copsey and coworkers [54], who observed an increase in IS JMs, as well as an increase of Zip3 foci indicating compensation for derailed ZMM intermediates, and high levels of unresolved JMs.
This view is corroborated by the role of the SUMO E3 ligase domain of Mms21. mms21-11, which lacks this domain, largely separates the two functions of the complex. This allele is defective in the anti rogue JM formation activity of the complex but only mildly affects Smc5/6-Mms21 resolution function. In the mms21-11 mutant, a significant amount of additional JMs are formed and in the background of the meiotic null allele mms4-mn, unresolved JMs accumulate comparable to the smc6-56 mutant (Figure 3B,E, 4E,I). Thus, the SUMO E3 ligase domain of Mms21 is required for the anti rogue JM formation activity of the complex but is largely dispensable for the resolution activity.
On the other hand, inactivation of Smc6 severely compromised JM resolution and recombination product formation in an sgs1-mn mutant background (Figure 5A–E, see also accompanying manuscript for a mutant in another Smc5/6-Mms21 subunit, Nse4 [54]), where JM resolution is almost fully dependent on the rogue JM resolvases [5], [9]. The resolution deficiency in sgs1-mn smc6-56 equals or exceeds that seen in sgs1-mn mms4-mn [9], [10], thus resolution activity equivalent to at least the Mus81-Mms4 resolvase must have been lost (identical conclusion in the accompanying study [54]). Providing Smc5/6-Mms21 function after the Ndt80-IN pachytene arrest proves sufficient to ensure meiotic chromosome segregation. This indicates that the lack of early (destabilizing) function produced no irreversible damage and can be compensated for by the late (resolution) function. This also suggests that late recruitment of the complex to promote resolution is possible. In this way, the Smc5/6-Mms21 complex supports meiotic recombination pathway choice and safeguards the turnover of non-ZMM intermediates, particularly the otherwise unregulated non-ZMM JMs (Figure 7A). These observations imply that the Smc5/6-Mms21 complex recognizes recombinogenic lesions and locally mediates antagonistic activities. How might the Smc5/6-Mms21 complex mediate this activity?
Fig. 7. Model of how Smc5/6-Mms21 may antagonize inappropriate JMs in meiotic recombination. 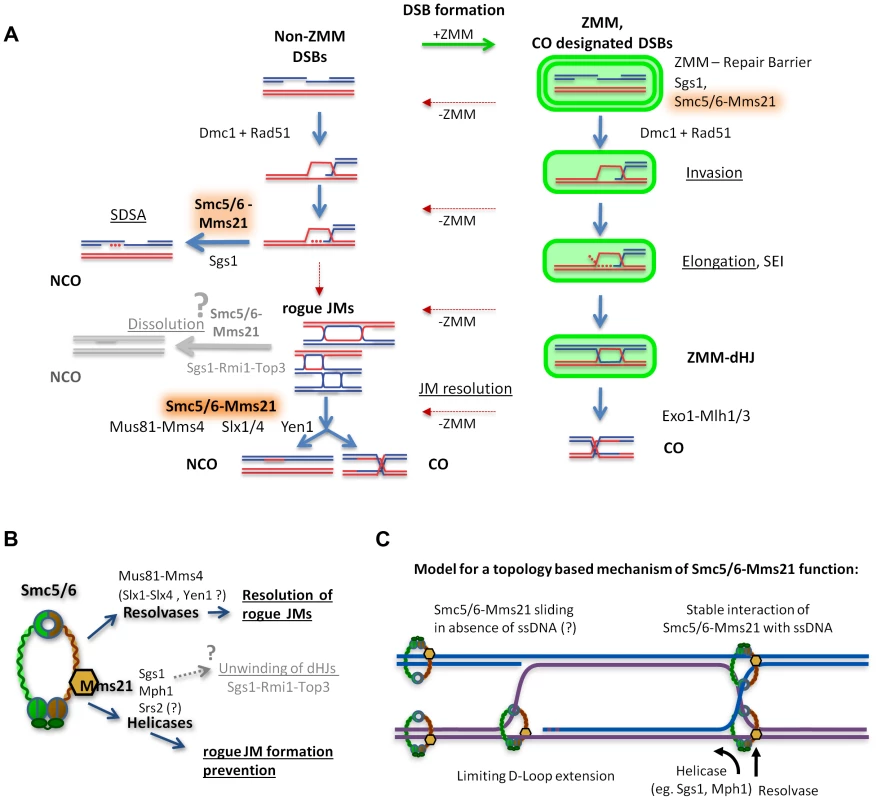
(A) A representation of homologous recombination and intermediate pathway choice in the course of budding yeast meiosis is shown, along with the points of function for Smc5/6-Mms21 in antagonizing inappropriate JMs. Meiotic DSBs can engage for two alternative fates: non-ZMM DSBs and ZMM-DSBs (highlighted in green). Regular pathway progression is marked with blue arrows and aberrant deviations are depicted in red. By default, stable strand invasions are disassembled for all breaks through the action of Sgs1 helicase and Smc5/6-Mms21. Non-ZMM DSBs are consequently repaired by SDSA into non-crossovers. In contrast, ZMM-DSBs are in addition also prevented from being repaired at all until licensed in the ZMM pathway for progression to stable invasions, here referred to as “ZMM-Repair Barrier”. When a ZMM-DSB is allowed to form stable SEIs and dHJs, the ZMM-pathway protects that DSB from the action of Sgs1 and Smc5/6-Mms21 and marks the resulting dHJ for CO specific resolution by Exo1-Mlh1/3. Intermediates that deviate from their supposed repair pathways by inappropriate intermediate stabilization and absence of a ZMM label result in “rogue” JMs that depend on Smc5/6-Mms21 for resolution by the rogue JM resolvase Mus81-Mms4 (and probably also Slx1-Slx4 and Yen1). (B) Our data implicates the Smc5/6-Mms21 complex in at least two independent mechanisms for rogue JM avoidance: On one hand prevention of JM formation through Mms21 SUMO E3 mediated regulation of anti-recombinogenic helicases, and on the other hand, the promotion of resolution of JMs by rogue JM resolvases. We also assume that through regulation of helicases, the Smc5/6-Mms21 complex may very likely also be involved in regulating dHJ dissolution by Sgs1-Rmi1-Top3. (C) Smc5/6-Mms21 may survey the DNA for displaced ssDNA at D-loops or HJs by topologically entrapping dsDNA as a sliding SMC ring and binding stably to ssDNA (e.g. at the hinge or any other part of the ring) upon encounter. Stable binding to such ssDNA will constrain D-loop extension and HJ branch migration and label sites with a ssDNA/dsDNA interface where appropriate action of anti-recombinogenic helicases (like Sgs1 or Mph1) and resolvases will be required. The Smc5/6-Mms21 complex affects the activity of anti-JM factors
The Smc5/6-Mms21 complex binds to DSB sites, suggesting it might locally recruit and/or orchestrate the function of anti-recombinogenic helicases and of resolvases. The absence of the complex in meiosis renders the activity of Sgs1 insufficient for normal intermediate destabilization and protection and strongly impairs the function of the rogue JM resolvases, supporting the above model. Notably, the defect of smc6-56 and mms21-11 mutants on rogue JM prevention is weaker than that of sgs1-mn leaving the ZMM pathway largely operative and thus CO formation unaffected by Smc6 inactivation. The phenotype of the SUMO E3 domain deletion suggests that Smc5/6-Mms21 mediates its anti-rogue JM formation activity through SUMOylation dependent regulation and coordination of anti-recombinogenic helicase activity at the site of the lesion. Sgs1 and Mph1 are the most likely targets. Interestingly, it has been reported that the FANCM-like helicase Mph1 becomes toxic in the absence of the Mms21 ligase activity [63]. The observed auto-SUMOylation of the complex could also be critical in this function for recruiting or sequestering regulatory targets [27]. In addition, since DNA damage induced SUMOylation of Sgs1 has been reported [40], Sgs1 may represent a direct substrate for post translational regulation by Smc5/6-Mms21. However, it was also reported that Mms21 is not essential for this SUMOylation and that different SUMO E3 ligases may provide redundancy [40], [68]. Consistent with a regulatory role of the Smc5/6-Mms21 complex for anti-recombinogenic helicase function, we found that the recruitment of Sgs1 to meiotic DSBs does require neither Mms21 SUMO E3 ligase activity nor an intact complex since Sgs1-myc18 enrichment at DSB hotspots comparable to wild-type is detected by qChIP in both mms21-11 and smc6-56 mutants (Figure S8A,B).
Beyond its supportive role in the prevention of rogue JMs, we find a profound defect of smc6-56 in resolution of rogue JMs in meiosis, implying a direct function of Smc5/6-Mms21 in promoting rogue JM resolvase activity. There are three rogue JM resolvases identified in S. cerevisiae, Mus81-Mms4, Slx1-Slx4, and Yen1.
Mus81-Mms4 is the one resolvase which facilitates the removal of the bulk of rogue JMs arising from homologous recombination [5], [9]. Comparing our results to published data, we estimate that in the smc6-56 mutant resolution activity equivalent to at least the Mus81-Mms4 resolvase must have been lost [9], [10]. There is no evidence that Mus81-Mms4 is subject to SUMOylation or direct interaction with Smc5/6. However, the complex may stabilize the HJ and present the substrate DNA to Mus81-Mms4, or it may mediate processing steps preceding resolution. With the Smc5/6-Mms21 complex, we could identify the second known factor critical for full activity of the Mus81-Mms4 resolvase after the identification of its activating kinase Cdc5 [7].
In contrast to Mus81-Mms4, direct interaction of the Slx1-Slx4 resolvase with the Smc5/6-Mms21 binding partner Rtt107 was observed, although it is unclear whether Rtt107 binds Slx1-Slx4 and Smc5/6-Mms21 alternatively or simultaneously [38]. Slx4 is also a SUMO substrate and interacts with Rad1-Rad10, a ssDNA nuclease involved in DNA processing during repair [69], [70]. Therefore, direct regulation of Slx1-Slx4 and Slx4-Rad1-Rad10 via Smc5/6-Mms21 is possible. However, during meiosis, Slx1-Slx4 only plays a minor role for rogue JM resolution, and also Rad1 did not appear to be required for JM resolution [5], [9]. Yen1 is the third rogue JM resolvase in budding yeast. Biochemical studies indicate that HJs are its natural substrate without the need for prior processing [71]. Yen1 becomes active around the onset of metaphase 2 and will resolve most rogue JMs still present, even in the absence of Mus81-Mms4 and Slx1-Slx4 [5], [7], [9] but not at elevated JM levels as in an sgs1 background. smc6-56 may represent a strong hypomorph for one, two or all rogue JM resolvases, however, the strong JM accumulation seen in sgs1-mn smc6-56 suggests that more than just the function of Mus81-Mms4 is affected.
Intriguingly, we find preloading of Smc6 to break sites. This could be a reasonable safety measure when anticipating the programmed generation of hundreds of DSBs. It was reported recently that Rtt107 promotes recruitment of Smc5/6-Mms21 specifically to the HO site upon DSB formation [38]. However, as Rtt107 is neither essential (like Smc5/6) nor required for Smc5/6-Mms21 function, Rtt107 may rather enhance the recruitment of Smc5/6-Mms21 to specific loci in anticipation of DNA damage. Meiotic DSB-hotspots are located in promoters and nuclease sensitive DNA regions [72]–[74] and are frequently associated with chromatin features such as H3K4me3 [75] that may directly, or via Rtt107, allow for local enrichment of Smc5/6-Mms21 to protect vulnerable DNA regions.
A topological model for the function of the Smc5/6-Mms21 complex
Smc5/6-Mms21 represents a ring shaped SMC complex, highly similar to its closely related brethren, cohesin and condensin [28] for which topological mechanisms of function were successfully demonstrated [23], [76]. While evidence for topological binding of the Smc5/6-Mms21 complex to DNA is still missing to date, its overall high similarity to cohesin and condensin and also previously proposed DNA damage independent loading through the cohesin loaders Scc2-Scc4 to chromatin [39], make a topological component in the function of Smc5/6-Mms21 likely.
If the Smc5/6-Mms21 complex would serve solely as a platform for JM antagonizing factors and their regulators, it would appear inconceivable why the SMC ring should have been maintained throughout evolution. Instead, it is far more likely that a preceding topological function of the evolutionary ancestor SMC complex fulfilled a function rudimentarily similar to Smc5/6-Mms21, which was ultimately enhanced in the course of evolution. In addition, preemptive loading of Smc5/6-Mms21 to DNA would appear considerably more effective in preventing dangerous lesions if the Smc5/6-Mms21 complex were not statically bound but rather could slide along longer regions of DNA, as proposed for cohesin [77].
How could the Smc5/6-Mms21 complex mediate its function in antagonizing JMs through an underlying topological association with DNA? We propose the following model to explain the previously observed functions for the Smc5/6-Mms21 complex.
The Smc5/6-Mms21 ring, preemptive of DNA damage, is loaded onto dsDNA, entrapping (in contrast to cohesin) only one dsDNA molecule. Such topological loading onto a single dsDNA molecule and the previously reported strong ssDNA binding properties of Smc5 and Smc6 [25], [26] are sufficient to counteract and stabilize recombination intermediates, and can serve to direct the activity of anti-recombinogenic helicases as well as resolvases at (and to) the according site, namely, the junction between the HR partners (Figure 7C, S9A).
In such a model, Smc5/6-Mms21 could slide freely along the intact dsDNA until encountering the ssDNA/dsDNA interface of an occurring lesion. Such an interface would be present at a progressing D-Loop, or at the HJ of the mature intermediate. Stable binding of the Smc5/6-Mms21 ring at such ssDNA junction sites may limit the spatial freedom for subsequent D-Loop extension, reduce the chance for second end capture, and impede further branch migration of HJs towards detrimental dHJ extension (as such expanding branch migration would require to overcome one obstructing strand of the HJ topologically) (Figure 7C, S9A). We also believe that this model could be extended to mitosis and be applied to recombinogenic structures such as cruciform DNA structures arising from replication fork regression.
By these means, the Smc5/6-Mms21 complex could survey for recombinogenic lesions, stabilize them through binding and, ultimately, employ counteractive measures by means of helicases and resolvases. As a corollary, this model postulates that Smc5/6-Mms21 mediates its function from outside the lesion, with involvement from only the intact donor DNA molecule being sufficient, to ultimately associate with the very borders of the recombinogenic lesion. Our observations that Smc5/6-Mms21 specifically enriches to sites of DSB formation after break formation but, nonetheless, fails to reveal significant on-top co-localization with the recombinosome marker Rad51, are consistent with this supposition.
In addition, it is to date unknown how anti-recombinogenic helicases like Sgs1-Rmi1-Top3 or Mph1 are directed to mediate their function in the context of a lesion. Association of a helicase to the wrong strand of the recombination junction will, instead of disassembly, result in extension. This problem is also eminent in the function of the Sgs1-Rmi1-Top3 dHJ dissolvase. While hetero-duplex DNA may provide information about the relative position of the parent molecules, it cannot account for directing the dissolution of JMs between perfectly identical sister-chromatids. Since in our model Smc5/6-Mms21 would inherently mark the parent associated “outsides” of a lesion, it may thus provide the lesion with a polarity and direct the anti-recombinogenic helicases for acting in the appropriate orientation (Figure S9B).
By these means, the Smc5/6-Mms21 complex could stably mark recombination intermediates and thus orchestrate the activity of anti-recombinogenic helicases and resolvases at the very site where JM antagonizing effectors are needed. While downstream effectors of the complex may vary to suit the specific needs of the individual organism or cell type, we postulate that the underlying JM restraining activity of Smc5/6-Mms21 complex is conserved throughout organisms.
Materials and Methods
Yeast strains
All strains used in this study are derivatives of SK1. Detailed genotypes are provided in Table S1. Strains were constructed by crossing or LiAc transformation using standard procedures. The URA3-arg4 recombination reporter was described in [47]. In the meiotic null mutants mms4-mn, sgs1-mn and nse4-mn the respective promoters are replaced by a CLB2 promoter fragment [78] and the estrogen inducible ndt80-IN system has been described [79].
Growth conditions and synchronized sporulation
Yeast strains were grown at 30°C in supplemented YPD (1% Difco yeast extract, 2% Difco peptone, 2% dextrose, 75 mg/L Ade, 75 mg/L Ura, 75 mg/L Trp) with exception of the smc6-56 mutants which were grown at 23.5°C permissive temperature. All manipulation of strains followed standard procedures. For synchronous sporulation in liquid culture, strains were grown at 30°C overnight in SPS pre-sporulation media (0.5% Difco yeast extract, 1% Difco peptone, 0.17% Difco yeast nitrogen base w/o AA&AS, 1% potassium acetate, 0.5% ammonium sulphate, 0.05 M potassium-biphthalate, pH 5.5) to an OD660 of 1.1–1.3 (4×107 cells/ml). Meiosis was induced by a subsequent wash and transfer of the cells into pre-warmed supplemented SPM (1% potassium acetate, 0.001% PPG2000, 4 mg/L Ura, 4 mg/L Trp, 4 mg/L His, 4 mg/L Arg, 6 mg/L Leu) at equal volume. Maximum aeration was provided for efficient meiosis. For synchronous meiosis, smc6-56 mutants were allowed to exit mitosis and proceed through most of pre-meiotic S-phase under (semi-) permissive conditions with the following temperature regime applied (after pre-growth at 23.5°C): At 0 hr SPM, upshift to 26°C, at 1 hr15 min to 28°C, at 1 hr40 min to 30°C, at 2 hr20 min to 33°C. Every temperature shift is translated to the culture media within 10 minutes. We find that smc6-56 is just able to perform nuclear divisions if undergoing meiosis at 30°C as long as not burdened by any additional number of rogue JMs (data not shown). Therefore, we define 28–30°C as semi-permissive temperature for smc6-56 in meiosis. ndt80-IN expression and exit from ndt80-IN pachytene arrest was induced by the addition of β-estradiol (ED, 5 mM stock in EtOH, stored at −20°C) to a final concentration of 1 µM at 7 hr30 min SPM, at which point approximately 80–90% of possible JMs had already formed [9]. The separation of early and late functions of Smc5/6-Mms21 in the respective experiments was performed using smc6-56 and the ndt80-IN system. Elimination of the early, pre pachytene-exit function and providing Smc5/6-Mms21 only after pachytene: Normal S-phase upshift to restrictive 33°C and arrest in pachytene followed at 7 hr30 min by addition of ED for Ndt80 induction and linear downshift to 23.5°C over 25 minutes. Elimination of the late, post pachytene function of Smc5/6-Mms21: Cells were transferred into SPM at 23.5°C and allowed to arrest in pachytene. The temperature was raised to 25.5°C from 6 hr45 min to 7 hrs, to 33°C until 7 hr20 min, and ED for ndt80-IN release was added at 7 hr30 min.
Physical recombination intermediate assays
Preparation of genomic DNA and Southern blotting of one dimensional 0.6% agarose gels for physical detection of recombination intermediates was performed as described in detail [47], [80], [81] using the CTAB/CoHex/Mg2+ procedure which stabilizes HJ intermediates. Relevant sets of JM experiments were performed in parallel for comparability. For radioactive Southern hybridization, the ArgD probe was used for detection of both maternal and paternal URA3-arg4EcPal/URA3-ARG4 loci and the HisU probe for allele specific detection of the maternal his4::URA3-arg4EcPal locus. Both probes are described [47]. As size marker, lambda HindIII digested DNA was used and probed against. Storage phosphor screens were used for signal detection and scanned with the Bio-Rad Molecular Imager FX. Quantification of signals was performed using Fuji Image Gauge Ver4.0. Frequencies are given as a percentage of total DNA signal.
Cytology
Nuclear divisions were followed on ethanol fixed and DAPI stained whole cells. For spindle staining on whole yeast cells, 1 ml aliquots of culture were fixed in 3.2% formaldehyde overnight at 4°C. Cells were washed and cell walls digested at 37°C using 100 µg Zymolyase 20T (SEIKAGAKU #120491), 70 mM DTT and 1 M sorbitol, in 200 µl. Cells were mounted on poly-L-lysine coated slides and fixed for 3 minutes in ice cold methanol and 10 seconds in ice cold acetone. Cells were blocked and immunostained in 0.5% BSA fraction V and 0.2% gelatin in 1×PBS. Microtubuli were detected using rat anti-tubulin-alpha (Serotec MCA78, 1∶200) and FITC-conjugated rabbit anti-rat (Sigma F1763, 1∶100) antibodies. Vectashield with DAPI (Vector Laboratories H-1200) was used to stain the DNA and stabilize the label. Chromosome surface spreads and immunostainings were performed as described [82]. For antibodies used in this study see Supplemental Information, Table S2. Evaluation of synapsis followed the criteria used in [83]. Chromosome pairing in zip3Δ mutant surface spread nuclei was evaluated by counting pairs of Hop1 axes of roughly equal length and in close proximity to each other, strictly parallel to each other, or connected via axial associations, or engaged in (pseudo-) synapsis. Images were taken on a Zeiss Axioskop fluorescence microscope with a Photometrics CH250 CCD camera using IPLab Spectrum with magnification and exposure times constant.
Flow cytometry
Flow cytometric quantification of cellular DNA content was performed with a BD Biosciences FACSCanto on ethanol fixed cells in the presence of 20 µg/ml propidium iodide in 50 mM Tris pH 7.5. Prior measurement, cells were treated overnight with 2 mg/ml RNaseA in 50 mM Tris 15 mM NaCl pH 7.5 at 40°C and one hour with 350 µg Proteinase K at 50°C in 500 µl volume. After brief sonication, propidium iodide was added 15 minutes before performing measurements. 5000 events were counted for each sample. Analysis was performed with Treestar FlowJo v10.
ChIP-seq
ChIP from meiotic cultures was performed as described by Panizza and coworkers [51]. In brief, 4×109 cells were collected per time point and incubated with para-formaldehyde (1% final concentration) for 15 minutes at 25°C. Cross-linking was stopped by addition of glycine to 131 mM. 4×109 cells were divided into 8 aliquots and separately opened using a multibead shocker (YASUI-KIKAI, Osaka) at 2,500 rpm, 15 cycles of 30 sec on and 30 sec off, at 4°C. Extracts were sonicated to an average of 200 bp by a Covaris S2 instrument. After removing the cell debris, the supernatant was used for the chromatin immunoprecipitation. For each sample, 50 µl Dynabeads Pan mouse IgG (Invitrogen) were incubated with the 9E11 anti-myc (mouse) antibody for 6–15 hr at 4°C. The precipitated DNA was used as a template for quantitative real-time PCR using GoTaq qPCR Master Mix for SYBR assay (Promega) (primer sequences: DSB1: CCGCAGAAGCCAACAAACGG, CTTTCGGTGGAACCTCGACC; DSB2: CGTGCCAGATTGAATTTTGA, GAATGGCCTTGGTAGCAAAT; DSB3: ACTTCCAACTGCAGGACGAC, ATCTGGCGGATGAACTTGAG; DSB4: ACGAACAGAGTCCCGAACCT, GCGGTTAATTCGATGGAAAG; CORE1: TGGATGGCAACTGAAGGAGC, TGGAATACCTATGAGTTGACTGC; ADP1: GGTGATGATTGCTCTCTGCC, CGTCACAATTGATCCCTCCC).
For genome-wide analysis (ChIP-seq), one-tenth of the ChIP DNA was analyzed by real-time PCR (qChIP) while the remaining DNA was concentrated by precipitation in ethanol. For the preparation of libraries for Illumina sequencing, we strictly followed the protocols provided by Illumina (Illumina ChIP-seq DNA sample prep protocol). Briefly, DNA-ends were repaired to convert overhangs to phosphorylated blunt ends with T4 DNA polymerase, E. coli DNA Pol I (Klenow fragment), and T4 polynucleotide kinase (PNK). An ‘A’ nucleotide was added to the 3′ end of the blunt ended fragments using Klenow fragment (3′ to 5′ exo minus). This prepared the DNA fragments for ligation to the adapters (Truseq Adaptor1-5) which have a single ‘T’ base overhang at their 3′ ends. After ligation, excess adaptors were removed by selecting a certain size range from 200–500 bp with QIAquick Gel Extraction Kit (Qiagen). Purified templates were PCR amplified with 15–18 cycles by KAPA-HiFi Hot Start PCR Kit (KAPA biosystems). Before hybridization to the flow cell, the amount and the size of the DNA library was controlled to be at least 1 µg/ul (for an average fragment size of 300 bp).
Processing of ChIP-seq data
Sequencing was performed at the CSF NGS Unit (csf.ac.at) with Illumina HiSeq 2000 resulting in 50 bp single-end reads in multiplex (Illumina TruSeq adapters 1–5). 2.3–8.6M reads per sample could be aligned with high confidence to the S. cerevisiae strain S288C, genome version R64-1-1 (20110203; http://downloads.yeastgenome.org/sequence/S288C_reference/genome_releases/). We used NextGenMap (http://cibiv.github.com/NextGenMap) for fast and sensitive alignment allowing up to 5000 hits per sequence. Read depth per position was generated in Java, dividing each hit by the number of genome-wide matches in case of multi-mapping reads.
All subsequent analyses were performed using R (version 2.15.2). After summing up the read depth of both strands, gaps were filled up with zeros followed by smoothing with ksmooth (Nadaraja-Watson kernel) with bandwidths as indicated and a resolution of 10 bp. Samples were normalized relative to their negative controls using NCIS [84]. This method estimates the fraction of background signal in each sample. After normalization, the untagged control sample was subtracted from each profile for background removal. The profiles remained robust against changes between different negative controls.
Peaks were defined as being flanked on both sides by valleys with a minimum depth. The maximal distance between matching peaks from different profiles was set to be equal to the smoothing bandwidth. Peak heights were subsequently compared by pair-wise Pearson correlation (R, cor). The significance of the number of overlapping peaks between profiles was assessed by a hyper-geometric random model. For matching peaks with DSB hotspots, peaks were required to map precisely between the borders of the DSB hotspots. The significance of the number of peaks matching to hotspot regions was assessed by a binomial random model. Here hotspots are defined according to [53] as groups of Spo11-oligo 5′-ends mapping less than 50 bp from each other. To account for the differences in genome versions, the raw data taken from [53] (GEO accession GSE26449) were re-aligned to the same genome version as the other samples, R64-1-1. Mapped sequence reads and generated profiles for this study are provided at the GEO repository, accession number GSE51977.
Supporting Information
Zdroje
1. KleinF, MahrP, GalovaM, BuonomoSB, MichaelisC, et al. (1999) A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell 98 : 91–103.
2. BuonomoSB, ClyneRK, FuchsJ, LoidlJ, UhlmannF, et al. (2000) Disjunction of homologous chromosomes in meiosis I depends on proteolytic cleavage of the meiotic cohesin Rec8 by separin. Cell 103 : 387–398.
3. BordeV, GoldmanAS, LichtenM (2000) Direct coupling between meiotic DNA replication and recombination initiation. Science 290 : 806–809.
4. BornerGV, KlecknerN, HunterN (2004) Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell 117 : 29–45.
5. ZakharyevichK, TangS, MaY, HunterN (2012) Delineation of joint molecule resolution pathways in meiosis identifies a crossover-specific resolvase. Cell 149 : 334–347.
6. SourirajanA, LichtenM (2008) Polo-like kinase Cdc5 drives exit from pachytene during budding yeast meiosis. Genes Dev 22 : 2627–2632.
7. MatosJ, BlancoMG, MaslenS, SkehelJM, WestSC (2011) Regulatory control of the resolution of DNA recombination intermediates during meiosis and mitosis. Cell 147 : 158–172.
8. BennettRJ, SharpJA, WangJC (1998) Purification and characterization of the Sgs1 DNA helicase activity of Saccharomyces cerevisiae. J Biol Chem 273 : 9644–9650.
9. De MuytA, JessopL, KolarE, SourirajanA, ChenJ, et al. (2012) BLM helicase ortholog Sgs1 is a central regulator of meiotic recombination intermediate metabolism. Mol Cell 46 : 43–53.
10. JessopL, LichtenM (2008) Mus81/Mms4 endonuclease and Sgs1 helicase collaborate to ensure proper recombination intermediate metabolism during meiosis. Mol Cell 31 : 313–323.
11. JessopL, RockmillB, RoederGS, LichtenM (2006) Meiotic chromosome synapsis-promoting proteins antagonize the anti-crossover activity of Sgs1. PLoS Genet 2: e155.
12. OhS, LaoJ, TaylorA, SmithG, HunterN (2008) RecQ helicase, Sgs1, and XPF family endonuclease, Mus81-Mms4, resolve aberrant joint molecules during meiotic recombination. Mol Cell 31 : 324–336.
13. FabreF, ChanA, HeyerWD, GangloffS (2002) Alternate pathways involving Sgs1/Top3, Mus81/Mms4, and Srs2 prevent formation of toxic recombination intermediates from single-stranded gaps created by DNA replication. Proc Natl Acad Sci U S A 99 : 16887–16892.
14. LiuJ, RenaultL, VeauteX, FabreF, StahlbergH, et al. (2011) Rad51 paralogues Rad55-Rad57 balance the antirecombinase Srs2 in Rad51 filament formation. Nature 479 : 245–248.
15. AntonyE, TomkoE, XiaoQ, KrejciL, LohmanT, et al. (2009) Srs2 disassembles Rad51 filaments by a protein-protein interaction triggering ATP turnover and dissociation of Rad51 from DNA. Mol Cell 35 : 105–115.
16. SugawaraN, GoldfarbT, StudamireB, AlaniE, HaberJE (2004) Heteroduplex rejection during single-strand annealing requires Sgs1 helicase and mismatch repair proteins Msh2 and Msh6 but not Pms1. Proc Natl Acad Sci U S A 101 : 9315–9320.
17. ChenCF, BrillSJ (2010) An essential DNA strand-exchange activity is conserved in the divergent N-termini of BLM orthologs. EMBO J 29 : 1713–1725.
18. WuL, HicksonID (2003) The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature 426 : 870–874.
19. DayaniY, SimchenG, LichtenM (2011) Meiotic recombination intermediates are resolved with minimal crossover formation during return-to-growth, an analogue of the mitotic cell cycle. PLoS Genet 7: e1002083.
20. GrieseJJ, WitteG, HopfnerKP (2010) Structure and DNA binding activity of the mouse condensin hinge domain highlight common and diverse features of SMC proteins. Nucleic Acids Res 38 : 3454–3465.
21. HaeringCH, LoweJ, HochwagenA, NasmythK (2002) Molecular architecture of SMC proteins and the yeast cohesin complex. Mol Cell 9 : 773–788.
22. HiranoM, HiranoT (2006) Opening closed arms: long-distance activation of SMC ATPase by hinge-DNA interactions. Mol Cell 21 : 175–186.
23. CuylenS, MetzJ, HaeringCH (2011) Condensin structures chromosomal DNA through topological links. Nat Struct Mol Biol 18 : 894–901.
24. IvanovD, NasmythK (2005) A topological interaction between cohesin rings and a circular minichromosome. Cell 122 : 849–860.
25. RoyMA, SiddiquiN, D'AmoursD (2011) Dynamic and selective DNA-binding activity of Smc5, a core component of the Smc5-Smc6 complex. Cell Cycle 10 : 690–700.
26. RoyMA, D'AmoursD (2011) DNA-binding properties of Smc6, a core component of the Smc5-6 DNA repair complex. Biochem Biophys Res Commun 416 : 80–85.
27. ZhaoX, BlobelG (2005) From The Cover: A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc Natl Acad Sci U S A 102 : 4777–4782.
28. DuanX, YangY, ChenYH, ArenzJ, RangiGK, et al. (2009) Architecture of the Smc5/6 complex of Saccharomyces cerevisiae reveals a unique interaction between the Nse5-6 subcomplex and the hinge regions of Smc5 and Smc6. J Biol Chem 284 : 8507–8515.
29. De PiccoliG, Torres-RosellJ, AragonL (2009) The unnamed complex: what do we know about Smc5-Smc6? Chromosome Res 17 : 251–263.
30. McAleenanA, Cordon-PreciadoV, Clemente-BlancoA, LiuIC, SenN, et al. (2012) SUMOylation of the alpha-kleisin subunit of cohesin is required for DNA damage-induced cohesion. Curr Biol : CB 22 : 1564–1575.
31. PottsPR (2009) The Yin and Yang of the MMS21-SMC5/6 SUMO ligase complex in homologous recombination. DNA Repair (Amst) 8 : 499–506.
32. DuanX, SarangiP, LiuX, RangiGK, ZhaoX, et al. (2009) Structural and functional insights into the roles of the Mms21 subunit of the Smc5/6 complex. Mol Cell 35 : 657–668.
33. TaylorEM, CopseyAC, HudsonJJ, VidotS, LehmannAR (2008) Identification of the proteins, including MAGEG1, that make up the human SMC5-6 protein complex. Mol Cell Biol 28 : 1197–1206.
34. McDonaldWH, PavlovaY, YatesJR3rd, BoddyMN (2003) Novel essential DNA repair proteins Nse1 and Nse2 are subunits of the fission yeast Smc5-Smc6 complex. J Biol Chem 278 : 45460–45467.
35. De PiccoliG, Cortes-LedesmaF, IraG, Torres-RosellJ, UhleS, et al. (2006) Smc5-Smc6 mediate DNA double-strand-break repair by promoting sister-chromatid recombination. Nat Cell Biol 8 : 1032–1034.
36. CostGJ, CozzarelliNR (2006) Smc5p promotes faithful chromosome transmission and DNA repair in Saccharomyces cerevisiae. Genetics 172 : 2185–2200.
37. Torres-RosellJ, MachinF, FarmerS, JarmuzA, EydmannT, et al. (2005) SMC5 and SMC6 genes are required for the segregation of repetitive chromosome regions. Nat Cell Biol 7 : 412–419.
38. LeungGP, LeeL, SchmidtTI, ShirahigeK, KoborMS (2011) Rtt107 is required for recruitment of the SMC5/6 complex to DNA double strand breaks. J Biol Chem 286 : 26250–26257.
39. LindroosHB, StromL, ItohT, KatouY, ShirahigeK, et al. (2006) Chromosomal association of the Smc5/6 complex reveals that it functions in differently regulated pathways. Mol Cell 22 : 755–767.
40. BranzeiD, SollierJ, LiberiG, ZhaoX, MaedaD, et al. (2006) Ubc9 - and Mms21-mediated sumoylation counteracts recombinogenic events at damaged replication forks. Cell 127 : 509–522.
41. MurrayJ, CarrA (2008) Smc5/6: a link between DNA repair and unidirectional replication? Nat Rev Mol Cell Biol 9 : 177–182.
42. HarveySH, SheedyDM, CuddihyAR, O'ConnellMJ (2004) Coordination of DNA damage responses via the Smc5/Smc6 complex. Mol Cell Biol 24 : 662–674.
43. Wehrkamp-RichterS, HyppaRW, PruddenJ, SmithGR, BoddyMN (2012) Meiotic DNA joint molecule resolution depends on Nse5-Nse6 of the Smc5-Smc6 holocomplex. Nucleic Acids Res 40 : 9633–9646.
44. FarmerS, San-SegundoPA, AragonL (2011) The Smc5-Smc6 complex is required to remove chromosome junctions in meiosis. PloS One 6: e20948.
45. OnodaF, TakedaM, SekiM, MaedaD, TajimaJ, et al. (2004) SMC6 is required for MMS-induced interchromosomal and sister chromatid recombinations in Saccharomyces cerevisiae. DNA Repair (Amst) 3 : 429–439.
46. DuanX, YeH (2009) Purification, crystallization and preliminary X-ray crystallographic studies of the complex between Smc5 and the SUMO E3 ligase Mms21. Acta Crystallogr Sect F Struct Biol Cryst Commun 65 : 849–852.
47. AllersT, LichtenM (2001) Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106 : 47–57.
48. PezzaR, VoloshinO, VanevskiF, Camerini-OteroR (2007) Hop2/Mnd1 acts on two critical steps in Dmc1-promoted homologous pairing. Genes Dev 21 : 1758–1766.
49. ZierhutC, BerlingerM, RuppC, ShinoharaA, KleinF (2004) Mnd1 is required for meiotic interhomolog repair. Curr Biol 14 : 752–762.
50. GertonJL, DeRisiJL (2002) Mnd1p: an evolutionarily conserved protein required for meiotic recombination. Proc Natl Acad Sci U S A 99 : 6895–6900.
51. PanizzaS, MendozaMA, BerlingerM, HuangL, NicolasA, et al. (2011) Spo11-accessory proteins link double-strand break sites to the chromosome axis in early meiotic recombination. Cell 146 : 372–383.
52. GlynnEF, MegeePC, YuHG, MistrotC, UnalE, et al. (2004) Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae. PLoS Biol 2: E259.
53. PanJ, SasakiM, KniewelR, MurakamiH, BlitzblauHG, et al. (2011) A hierarchical combination of factors shapes the genome-wide topography of yeast meiotic recombination initiation. Cell 144 : 719–731.
54. CopseyA, TangS, JordanP, BlitzblauH, NewcombeS, et al. (2013) Smc5/6 coordinates formation and resolution of joint molecules with chromosome morphology to ensure meiotic divisions. PLoS Genet 6: e1001028.
55. GerlichD, KochB, DupeuxF, PetersJM, EllenbergJ (2006) Live-cell imaging reveals a stable cohesin-chromatin interaction after but not before DNA replication. Curr Biol 16 : 1571–1578.
56. CarballoJA, PanizzaS, SerrentinoME, JohnsonAL, GeymonatM, et al. (2013) Budding yeast ATM/ATR control meiotic double-strand break (DSB) levels by down-regulating Rec114, an essential component of the DSB-machinery. PLoS Genet 9: e1003545.
57. AmpatzidouE, IrmischA, O'ConnellMJ, MurrayJM (2006) Smc5/6 is required for repair at collapsed replication forks. Mol Cell Biol 26 : 9387–9401.
58. ChavezA, GeorgeV, AgrawalV, JohnsonFB (2010) Sumoylation and the structural maintenance of chromosomes (Smc) 5/6 complex slow senescence through recombination intermediate resolution. J Biol Chem 285 : 11922–11930.
59. AllersT, LichtenM (2001) Intermediates of yeast meiotic recombination contain heteroduplex DNA. Mol Cell 8 : 225–231.
60. ChuaPR, RoederGS (1998) Zip2, a meiosis-specific protein required for the initiation of chromosome synapsis. Cell 93 : 349–359.
61. OhS, LaoJ, HwangP, TaylorA, SmithG, et al. (2007) BLM ortholog, Sgs1, prevents aberrant crossing-over by suppressing formation of multichromatid joint molecules. Cell 130 : 259–272.
62. ClyneRK, KatisVL, JessopL, BenjaminKR, HerskowitzI, et al. (2003) Polo-like kinase Cdc5 promotes chiasmata formation and cosegregation of sister centromeres at meiosis I. Nat Cell Biol 5 : 480–485.
63. ChavezA, AgrawalV, JohnsonFB (2011) Homologous recombination-dependent rescue of deficiency in the structural maintenance of chromosomes (Smc) 5/6 complex. J Biol Chem 286 : 5119–5125.
64. ChenY, ChoiK, SzakalB, ArenzJ, DuanX, et al. (2009) Interplay between the Smc5/6 complex and the Mph1 helicase in recombinational repair. Proc Natl Acad Sci U S A 106 : 21252–21257.
65. St OngeRP, ManiR, OhJ, ProctorM, FungE, et al. (2007) Systematic pathway analysis using high-resolution fitness profiling of combinatorial gene deletions. Nature Genet 39 : 199–206.
66. LynnA, SoucekR, BornerGV (2007) ZMM proteins during meiosis: crossover artists at work. Chromosome Res 15 : 591–605.
67. AgarwalS, RoederG (2000) Zip3 provides a link between recombination enzymes and synaptonemal complex proteins. Cell 102 : 245–255.
68. LuCY, TsaiCH, BrillSJ, TengSC (2010) Sumoylation of the BLM ortholog, Sgs1, promotes telomere-telomere recombination in budding yeast. Nucleic Acids Res 38 : 488–498.
69. CremonaCA, SarangiP, YangY, HangLE, RahmanS, et al. (2012) Extensive DNA damage-induced sumoylation contributes to replication and repair and acts in addition to the mec1 checkpoint. Mol Cell 45 : 422–432.
70. FlottS, AlabertC, TohGW, TothR, SugawaraN, et al. (2007) Phosphorylation of Slx4 by Mec1 and Tel1 regulates the single-strand annealing mode of DNA repair in budding yeast. Mol Cell Biol 27 : 6433–6445.
71. IpS, RassU, BlancoM, FlynnH, SkehelJ, et al. (2008) Identification of Holliday junction resolvases from humans and yeast. Nature 456 : 357–361.
72. WuTC, LichtenM (1994) Meiosis-induced double-strand break sites determined by yeast chromatin structure. Science 263 : 515–518.
73. WuTC, LichtenM (1995) Factors that affect the location and frequency of meiosis-induced double-strand breaks in Saccharomyces cerevisiae. Genetics 140 : 55–66.
74. OhtaK, ShibataT, NicolasA (1994) Changes in chromatin structure at recombination initiation sites during yeast meiosis. EMBO J 13 : 5754–5763.
75. BordeV, RobineN, LinW, BonfilsS, GeliV, et al. (2009) Histone H3 lysine 4 trimethylation marks meiotic recombination initiation sites. EMBO J 28 : 99–111.
76. HaeringC, FarcasA, ArumugamP, MetsonJ, NasmythK (2008) The cohesin ring concatenates sister DNA molecules. Nature 454 : 297–301.
77. LengronneA, KatouY, MoriS, YokobayashiS, KellyGP, et al. (2004) Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature 430 : 573–578.
78. LeeBH, AmonA (2003) Role of Polo-like kinase CDC5 in programming meiosis I chromosome segregation. Science 300 : 482–486.
79. CarlileT, AmonA (2008) Meiosis I is established through division-specific translational control of a cyclin. Cell 133 : 280–291.
80. OhSD, JessopL, LaoJP, AllersT, LichtenM, et al. (2009) Stabilization and electrophoretic analysis of meiotic recombination intermediates in Saccharomyces cerevisiae. Methods Mol Biol 557 : 209–234.
81. AllersT, LichtenM (2000) A method for preparing genomic DNA that restrains branch migration of Holliday junctions. Nucleic Acids Res 28: e6.
82. NairzK, KleinF (1997) mre11S–a yeast mutation that blocks double-strand-break processing and permits nonhomologous synapsis in meiosis. Genes Dev 11 : 2272–2290.
83. KlutsteinM, XaverM, ShemeshR, ZenvirthD, KleinF, et al. (2009) Separation of roles of Zip1 in meiosis revealed in heterozygous mutants of Saccharomyces cerevisiae. Mol Genet Genomics 282 : 453–462.
84. LiangK, KelesS (2012) Normalization of ChIP-seq data with control. BMC bioinformatics 13 : 199.
Štítky
Genetika Reprodukční medicína
Článek Interaction between and during Mammalian Jaw Patterning and in the Pathogenesis of SyngnathiaČlánek Clustering of Tissue-Specific Sub-TADs Accompanies the Regulation of Genes in Developing LimbsČlánek Transcription Factor Occupancy Can Mediate Active Turnover of DNA Methylation at Regulatory RegionsČlánek Tay Bridge Is a Negative Regulator of EGFR Signalling and Interacts with Erk and Mkp3 in the Wing
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 12
-
Všechny články tohoto čísla
- Stressing the Importance of CHOP in Liver Cancer
- The AmAZI1ng Roles of Centriolar Satellites during Development
- Flies Get a Head Start on Meiosis
- Recommendations from Jane Gitschier's Bookshelf
- And Baby Makes Three: Genomic Imprinting in Plant Embryos
- Bugs in Transition: The Dynamic World of in Insects
- Defining the Role of ATP Hydrolysis in Mitotic Segregation of Bacterial Plasmids
- Synaptonemal Complex Components Promote Centromere Pairing in Pre-meiotic Germ Cells
- Cohesinopathies of a Feather Flock Together
- Genetic Recombination Is Targeted towards Gene Promoter Regions in Dogs
- Parathyroid-Specific Deletion of Unravels a Novel Calcineurin-Dependent FGF23 Signaling Pathway That Regulates PTH Secretion
- MAN1B1 Deficiency: An Unexpected CDG-II
- Phosphate Flow between Hybrid Histidine Kinases CheA and CheS Controls Cyst Formation
- Basolateral Mg Extrusion via CNNM4 Mediates Transcellular Mg Transport across Epithelia: A Mouse Model
- Truncation of Unsilences Paternal and Ameliorates Behavioral Defects in the Angelman Syndrome Mouse Model
- Autozygome Sequencing Expands the Horizon of Human Knockout Research and Provides Novel Insights into Human Phenotypic Variation
- Huntington's Disease Induced Cardiac Amyloidosis Is Reversed by Modulating Protein Folding and Oxidative Stress Pathways in the Heart
- Low Frequency Variants, Collapsed Based on Biological Knowledge, Uncover Complexity of Population Stratification in 1000 Genomes Project Data
- Targeted Ablation of and in Retinal Progenitor Cells Mimics Leber Congenital Amaurosis
- Genomic Imprinting in the Embryo Is Partly Regulated by PRC2
- Binary Cell Fate Decisions and Fate Transformation in the Larval Eye
- The Stress-Regulated Transcription Factor CHOP Promotes Hepatic Inflammatory Gene Expression, Fibrosis, and Oncogenesis
- A Global RNAi Screen Identifies a Key Role of Ceramide Phosphoethanolamine for Glial Ensheathment of Axons
- Functional Analysis of the Interdependence between DNA Uptake Sequence and Its Cognate ComP Receptor during Natural Transformation in Species
- Cross-Modulation of Homeostatic Responses to Temperature, Oxygen and Carbon Dioxide in
- Alcohol-Induced Histone Acetylation Reveals a Gene Network Involved in Alcohol Tolerance
- Molecular Characterization of Host-Specific Biofilm Formation in a Vertebrate Gut Symbiont
- CRIS—A Novel cAMP-Binding Protein Controlling Spermiogenesis and the Development of Flagellar Bending
- Dual Regulation of the Mitotic Exit Network (MEN) by PP2A-Cdc55 Phosphatase
- Expanding the Marine Virosphere Using Metagenomics
- Detection of Slipped-DNAs at the Trinucleotide Repeats of the Myotonic Dystrophy Type I Disease Locus in Patient Tissues
- Interaction between and during Mammalian Jaw Patterning and in the Pathogenesis of Syngnathia
- Mutations in the UQCC1-Interacting Protein, UQCC2, Cause Human Complex III Deficiency Associated with Perturbed Cytochrome Protein Expression
- Reactivation of Chromosomally Integrated Human Herpesvirus-6 by Telomeric Circle Formation
- Anoxia-Reoxygenation Regulates Mitochondrial Dynamics through the Hypoxia Response Pathway, SKN-1/Nrf, and Stomatin-Like Protein STL-1/SLP-2
- The Midline Protein Regulates Axon Guidance by Blocking the Reiteration of Neuroblast Rows within the Drosophila Ventral Nerve Cord
- Tomato Yield Heterosis Is Triggered by a Dosage Sensitivity of the Florigen Pathway That Fine-Tunes Shoot Architecture
- Selection on Plant Male Function Genes Identifies Candidates for Reproductive Isolation of Yellow Monkeyflowers
- Role of Tomato Lipoxygenase D in Wound-Induced Jasmonate Biosynthesis and Plant Immunity to Insect Herbivores
- Meiotic Cohesin SMC1β Provides Prophase I Centromeric Cohesion and Is Required for Multiple Synapsis-Associated Functions
- Identification of Sphingolipid Metabolites That Induce Obesity via Misregulation of Appetite, Caloric Intake and Fat Storage in
- Genome-Wide Screen Reveals Replication Pathway for Quasi-Palindrome Fragility Dependent on Homologous Recombination
- Histone Methylation Restrains the Expression of Subtype-Specific Genes during Terminal Neuronal Differentiation in
- A Novel Intergenic ETnII-β Insertion Mutation Causes Multiple Malformations in Mice
- The NuRD Chromatin-Remodeling Enzyme CHD4 Promotes Embryonic Vascular Integrity by Transcriptionally Regulating Extracellular Matrix Proteolysis
- A Domesticated Transposase Interacts with Heterochromatin and Catalyzes Reproducible DNA Elimination in
- Acute Versus Chronic Loss of Mammalian Results in Distinct Ciliary Phenotypes
- MBD3 Localizes at Promoters, Gene Bodies and Enhancers of Active Genes
- Positive and Negative Regulation of Gli Activity by Kif7 in the Zebrafish Embryo
- A Hereditary Spastic Paraplegia Mouse Model Supports a Role of ZFYVE26/SPASTIZIN for the Endolysosomal System
- The CCR4-NOT Complex Mediates Deadenylation and Degradation of Stem Cell mRNAs and Promotes Planarian Stem Cell Differentiation
- Reconstructing Native American Migrations from Whole-Genome and Whole-Exome Data
- Contributions of Protein-Coding and Regulatory Change to Adaptive Molecular Evolution in Murid Rodents
- Comprehensive Analysis of Transcriptome Variation Uncovers Known and Novel Driver Events in T-Cell Acute Lymphoblastic Leukemia
- A -Acting Protein Effect Causes Severe Eye Malformation in the Mouse
- Clustering of Tissue-Specific Sub-TADs Accompanies the Regulation of Genes in Developing Limbs
- Germline Progenitors Escape the Widespread Phenomenon of Homolog Pairing during Development
- Transcription Factor Occupancy Can Mediate Active Turnover of DNA Methylation at Regulatory Regions
- Somatic mtDNA Mutation Spectra in the Aging Human Putamen
- ESCRT-I Mediates FLS2 Endosomal Sorting and Plant Immunity
- Ethylene Promotes Hypocotyl Growth and HY5 Degradation by Enhancing the Movement of COP1 to the Nucleus in the Light
- The PAF Complex and Prf1/Rtf1 Delineate Distinct Cdk9-Dependent Pathways Regulating Transcription Elongation in Fission Yeast
- Dual Regulation of Gene Expression Mediated by Extended MAPK Activation and Salicylic Acid Contributes to Robust Innate Immunity in
- Quantifying Missing Heritability at Known GWAS Loci
- Smc5/6-Mms21 Prevents and Eliminates Inappropriate Recombination Intermediates in Meiosis
- Smc5/6 Coordinates Formation and Resolution of Joint Molecules with Chromosome Morphology to Ensure Meiotic Divisions
- Tay Bridge Is a Negative Regulator of EGFR Signalling and Interacts with Erk and Mkp3 in the Wing
- Meiotic Crossover Control by Concerted Action of Rad51-Dmc1 in Homolog Template Bias and Robust Homeostatic Regulation
- Active Transport and Diffusion Barriers Restrict Joubert Syndrome-Associated ARL13B/ARL-13 to an Inv-like Ciliary Membrane Subdomain
- An Regulatory Circuit Modulates /Wnt Signaling and Determines the Size of the Midbrain Dopaminergic Progenitor Pool
- Variants Induce Differential Protection to Viruses in : A Phenotypic and Phylogenomic Analysis
- Base Pairing Interaction between 5′- and 3′-UTRs Controls mRNA Translation in
- Evidence That Masking of Synapsis Imperfections Counterbalances Quality Control to Promote Efficient Meiosis
- Insulin/IGF-Regulated Size Scaling of Neuroendocrine Cells Expressing the bHLH Transcription Factor in
- Sumoylated NHR-25/NR5A Regulates Cell Fate during Vulval Development
- TATN-1 Mutations Reveal a Novel Role for Tyrosine as a Metabolic Signal That Influences Developmental Decisions and Longevity in
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The NuRD Chromatin-Remodeling Enzyme CHD4 Promotes Embryonic Vascular Integrity by Transcriptionally Regulating Extracellular Matrix Proteolysis
- Mutations in the UQCC1-Interacting Protein, UQCC2, Cause Human Complex III Deficiency Associated with Perturbed Cytochrome Protein Expression
- The Midline Protein Regulates Axon Guidance by Blocking the Reiteration of Neuroblast Rows within the Drosophila Ventral Nerve Cord
- Tomato Yield Heterosis Is Triggered by a Dosage Sensitivity of the Florigen Pathway That Fine-Tunes Shoot Architecture
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



