-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaActivin Signaling Targeted by Insulin/dFOXO Regulates Aging and Muscle Proteostasis in
Reduced insulin/IGF signaling increases lifespan in many animals. To understand how insulin/IGF mediates lifespan in Drosophila, we performed chromatin immunoprecipitation-sequencing analysis with the insulin/IGF regulated transcription factor dFOXO in long-lived insulin/IGF signaling genotypes. Dawdle, an Activin ligand, is bound and repressed by dFOXO when reduced insulin/IGF extends lifespan. Reduced Activin signaling improves performance and protein homeostasis in muscles of aged flies. Activin signaling through the Smad binding element inhibits the transcription of Autophagy-specific gene 8a (Atg8a) within muscle, a factor controlling the rate of autophagy. Expression of Atg8a within muscle is sufficient to increase lifespan. These data reveal how insulin signaling can regulate aging through control of Activin signaling that in turn controls autophagy, representing a potentially conserved molecular basis for longevity assurance. While reduced Activin within muscle autonomously retards functional aging of this tissue, these effects in muscle also reduce secretion of insulin-like peptides at a distance from the brain. Reduced insulin secretion from the brain may subsequently reinforce longevity assurance through decreased systemic insulin/IGF signaling.
Published in the journal: . PLoS Genet 9(11): e32767. doi:10.1371/journal.pgen.1003941
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003941Summary
Reduced insulin/IGF signaling increases lifespan in many animals. To understand how insulin/IGF mediates lifespan in Drosophila, we performed chromatin immunoprecipitation-sequencing analysis with the insulin/IGF regulated transcription factor dFOXO in long-lived insulin/IGF signaling genotypes. Dawdle, an Activin ligand, is bound and repressed by dFOXO when reduced insulin/IGF extends lifespan. Reduced Activin signaling improves performance and protein homeostasis in muscles of aged flies. Activin signaling through the Smad binding element inhibits the transcription of Autophagy-specific gene 8a (Atg8a) within muscle, a factor controlling the rate of autophagy. Expression of Atg8a within muscle is sufficient to increase lifespan. These data reveal how insulin signaling can regulate aging through control of Activin signaling that in turn controls autophagy, representing a potentially conserved molecular basis for longevity assurance. While reduced Activin within muscle autonomously retards functional aging of this tissue, these effects in muscle also reduce secretion of insulin-like peptides at a distance from the brain. Reduced insulin secretion from the brain may subsequently reinforce longevity assurance through decreased systemic insulin/IGF signaling.
Introduction
Reduced insulin/IGF-1 signaling increases the lifespan of nematodes, flies and rodents [1], [2]. In Caenorhabditis elegans, mutants in insulin-like receptor daf-2 live twice as long as wild type [3], [4]. Mutation of insulin receptor InR and insulin receptor substrate (chico) increase adult lifespan in the fruit fly Drosophila melanogaster [5], [6]. It is reported that mice with mutation at the IGF-1 receptor (Igf1r) extend lifespan [7], as do mutants of the insulin receptor substrate (Irs2) [8] and of the insulin receptor within adipose tissues [9].
Genetic evidence places the forkhead transcription factor FOXO as the downstream effector of insulin/IGF-1 signaling [3], [10], [11], [12]. Activated insulin/IGF-1 signaling enhances the phosphorylation of FOXO, which is sequestered in the cytoplasm. Conversely, reduced insulin results in FOXO nuclear translocation, which thus promotes or represses the transcription of FOXO target genes [11] (Figure S1A). In C. elegans lifespan extension of daf-2 and age-1(PI3 kinase) mutants requires daf-16, a FOXO homolog in worms [3]. Recent work likewise shows that FOXO is required for insulin-mediated lifespan extension in Drosophila [13], [14]. FOXO also appears to function in human aging where independent studies found polymorphisms of FoxO3A to associate with exceptional longevity [15], [16]. Insulin signaling through its control of FOXO is a potentially conserved system to regulate aging but despite this emerging consensus, the proximal targets of insulin/FOXO signaling that orchestrate these mechanisms of longevity assurance are essentially unknown.
One aspect that is clear is that insulin/FOXO signaling operates both nonautonomously and autonomously to control Drosophila aging. Systemically reducing insulin signaling by mutations of the insulin receptor (InR) and insulin receptor substrate (chico) slows the decline in cardiac performance of aging flies while a similar outcome is produced by overexpressing FOXO and PTEN just within cardiomyocytes [17]. Likewise, overexpressing FOXO in muscle maintains muscle protein homeostasis and delays muscle function decline with age while dFOXO expressed in muscles extends lifespan [18], as does expression of dFOXO only from fat body [19], [20]. dFOXO expressed from fat body reduces secretion of systemic insulin-like peptides (DILP2 and DILP5), which are produced predominantly in the brain. dFOXO of Drosophila fat body modulates lifespan by inducing fat body dilp6 transcription, which in turn suppresses neuronal DILP secretion [21]. These findings suggest that insulin/FOXO signaling within some organs controls both the systemic level of circulating DILPs while systemic DILPs regulate somatic maintenance of insulin sensitive tissues. Identifying the FOXO target genes and somatic maintenance pathways in such tissues will elucidate how reduced insulin/IGF-1 assures longevity.
Genome-wide studies with C. elegans have been used to probe how daf-16 controls lifespan in response to insulin signaling. Microarray analyses have identified many mRNA that are affected directly or indirectly by daf-16, and reducing some of these genes by RNA interference (RNAi) increases longevity [22], [23]. Chromatin immunoprecipitation (ChIP) and DNA adenine methyltransferase identification (DamID) have been used to identify the direct targets of DAF-16 to clarify which pathways are proximally responsible for the impact of daf-16 upon aging [24], [25]. Oh et al. [24] thus described 103 genes to be direct targets of daf-16 in the long-lived daf-2(e1370) mutant, and three out of 33 tested target genes were found to increase lifespan when gene expressions were reduced by RNAi: lin-2, egl-10 and sca-1.
In Drosophila, work to date has identified binding targets of dFOXO primarily from wild type adults. Alic et al identified 1423 dFOXO binding sites in wildtype adult female Drosophila using a ChIP-on-chip approach [26]. Among 1755 unique genes that are less than 1 kb away from these dFOXO binding sites, about 365 genes are transcriptionally regulated by dFOXO. These targets could potentially modulate many insulin related phenotypes including growth, reproduction, and metabolism, as well as aging.
Here we aim to understand how reduced insulin/IGF-1 signaling extends Drosophila lifespan by identifying genes transcriptionally regulated by dFOXO in long-lived insulin signaling mutants. We conducted ChIP analysis with a dFOXO antibody followed by Illumina high-throughput sequencing from chico heterozygous mutants, which are long-lived and normal sized, and from adult flies with ablated insulin producing cells (IPCs), which are also long-lived [27]. dFOXO was seen to bind at promoters of 273 genes common to these genotypes, thus providing a candidate set of potential factors in control of aging. Pathways enriched within this set include those for G-proteins, Wnt and Transforming growth factor-beta (TGF-β). We subsequently focused on TGF-β signaling because dFOXO binds in the promoter and represses dawdle (daw), an Activin-like ligand in TGF-β superfamily. In genetic trials, reducing daw or its downstream transcription factor Smox increases lifespan, preserves muscle function and reduces poly-ubiquitinated protein accumulation. The muscle specific benefits of activated dFOXO are mediated through the control of autophagy by Smox, which we find to bind and transcriptionally repress Atg8a/LC3, a reported longevity assurance gene of Drosophila [28]. Expressing Atg8a in muscle was also sufficient to increase lifespan. Furthermore, reducing daw in muscle decreased DILP2 peptide secretion from the brain while peripheral insulin/IGF signaling was correspondingly reduced. Our results suggest that insulin/IGF signaling controls lifespan in part through dFOXO-mediated repression of muscle Activin signaling and its downstream functions including muscle autophagy, muscle proteostasis and subsequent remote control of systemic insulin/IGF signaling.
Results
Activin-like ligand dawdle is a direct transcriptional target of dFOXO that regulates longevity
To understand how dFOXO extends Drosophila lifespan we sequenced promoters derived from chromatin-immunoprecipitation with antibody against dFOXO in two genotypes of long-lived flies with reduced insulin signaling. Heterozygotes of chico1 live 36% longer than co-segregating wildtype sibs [13], [29]. Unlike many mutants of the insulin-signaling pathway, chico+/− adults have normal development time, body size and fecundity. Aging is likewise retarded by partially ablating adult IPCs by inducing apoptosis with a cell specific inducible driver (Dilp2-GeneSwitch-gal4>UAS-reaper) [27]. We conducted ChIP-Seq analysis from 15-day old female adults from both genetic manipulations. This revealed dFOXO to bind at 1331 and 763 promoter regions (Figure S1B), from chico and IPCs ablated flies respectively, corresponding to 2042 and 1012 candidate genes (Figure 1A and Table S7).
Fig. 1. ChIP-Seq to identify dFOXO direct target genes and lifespan screen for 23 selected candidates. 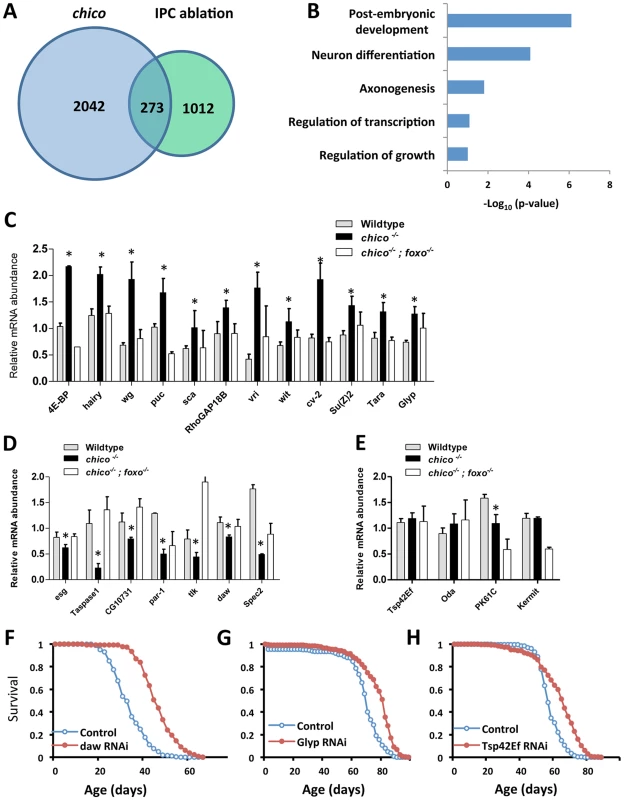
(A) Venn diagram to show dFOXO target genes identified in ChIP-Seq analysis. 15-day-old female insulin mutants (chico −/+ and IPC ablation) were used in ChIP-Seq experiments. dFOXO was enriched at promoters of 273 genes common to these genotypes. (B) Pathway analysis for 273 dFOXO targets, determined by DAVID functional classification. (C–E) Expression analysis of 23 selected dFOXO target genes indicates dFOXO acts as both activator and repressor. Asterisk indicates significant difference between chico−/− and wildtype (p<0.05); three biological replicates per genotype. (F–H) Lifespan analysis for three dFOXO target genes (daw, Glyp and Tsp42Ef) (Log-rank test, p<0.0001). Ubiquitous GeneSwitch (GS)-Gal4 drivers, Tub-GS-Gal4 or Tub-GS-dicer2-Gal4 (with UAS-dicer2 to enhance the knockdown) were used in lifespan screen (lifetable statistics summarized in Table S1). We identified 273 genes common to both longevity-assurance genotypes (Figure 1A). Biological functions defined by Gene Ontology (david.abcc.ncifcrf.gov) in this overlapping set include development, growth and neuron differentiation (Figure 1B). Pathway analysis (david.abcc.ncifcrf.gov) revealed enrichment in Wnt and TGF-β signaling (Figure S1C). Corresponding to previous work, we also found significant binding of dFOXO at puckered (puc) in both longevity assurance genotypes (Table S1). In the JNK signaling pathway, puc is a negative regulator of JUN kinase basket (bsk), and mutation of puc extends Drosophila lifespan [30].
To determine how candidate dFOXO targets affect longevity we selected 23 genes for further analyses (Figures 1C–1E, Table S1) based on their placement in recognized signaling pathways or because they showed a strong dFOXO binding. dFOXO binding at the promoter of these candidates was verified by ChIP followed with gene specific qPCR. In this analysis dFOXO was significantly enriched at all candidate targets in both insulin mutants when compared to wildtype (Figures S1E–S1F).
To measure the impact of insulin/IGF-1 on candidate transcription we quantified mRNA in adults of wildtype (WT), chico null mutant (chico −/−) and chico; foxo double mutant (chico −/−; foxo −/−) (Figures 1C–1E). Transcripts of 12 genes were up-regulated in chico −/− relative to wildtype but not in chico −/−; foxo −/−, indicating that dFOXO induces these genes. The expression of seven genes was repressed in chico −/− relative to wildtype but not in chico −/−; foxo −/−, suggesting that dFOXO directly represses these genes. Four genes were not differentially expressed despite their enriched dFOXO binding in the insulin mutants; activated dFOXO may be required but not sufficient to control the expression of these genes. Thus, dFOXO can function as both a transcriptional activator and repressor [26], but this factor may also become poised at genes upon reduced insulin signaling and not affect transcriptional changes until the required co-factors are induced by other signals.
To determine if candidate dFOXO targets contribute to aging regulation we measured lifespan and age-specific mortality when each was reduced by RNAi or over-expressed from transgenes. Cohorts of control and mis-expression genotypes were coisogenic; misexpression was induced only in adults via GeneSwitch (GS)-Gal4 driving either UAS-RNAi or UAS-transgene. The effect of RNAi on lifespan was assessed for all 23 candidates. Knockdown of three genes (daw, Glyp and Tsp42Ef) extended lifespan by consistently reducing age-specific mortality (Figures 1F–1H, Figure S2 and Table S1), while knockdown of 14 genes shortened lifespan (Table S1). Among the candidates whose transcriptions were positively regulated by dFOXO, seven transgenic lines were available to test the effect of overexpression on lifespan; two cases had no effect on lifespan while five cases reduced survival (Table S2).
Among the observed longevity assurance genes, daw-RNAi induced by two independent ubiquitous GeneSwitch drivers respectively extended lifespan 12% to 35% (mean lifespan) by consistently reducing mortality rate (Table S1, S3). Dawdle is one of two Drosophila Activin-like ligands [31], [32], belonging to the Transforming Growth Factor-β (TGF-β) protein superfamily. To date, daw is reported to function in axon guidance [31], [32], cell proliferation and larval brain development [33]. Our results indicate that daw acts as a downstream target of dFOXO to modulate lifespan, suggesting that the Activin branch of TGF - β signaling may participate in control of aging.
Activin signaling within muscle regulates Drosophila lifespan
Drosophila has two TGF-β ligand subfamilies: bone morphogenetic proteins (BMP) (ligands: Dpp, Gbb and Scw) and Activin (ligands: Daw and Act-β). These ligands signal through subfamily-specific Type I receptors (Tkv and Sax for BMP, Babo for Activin) and shared Type II receptors (Punt, Wit). BMP-like ligands and Activin-like ligands activate distinct downstream signaling cascades leading respectively to phosphorylation of the Smad transcription factors Mad and Smox (Figure 2A) [34].
Fig. 2. Reducing Activin signaling, but not BMP signaling prolongs lifespan in Drosophila. 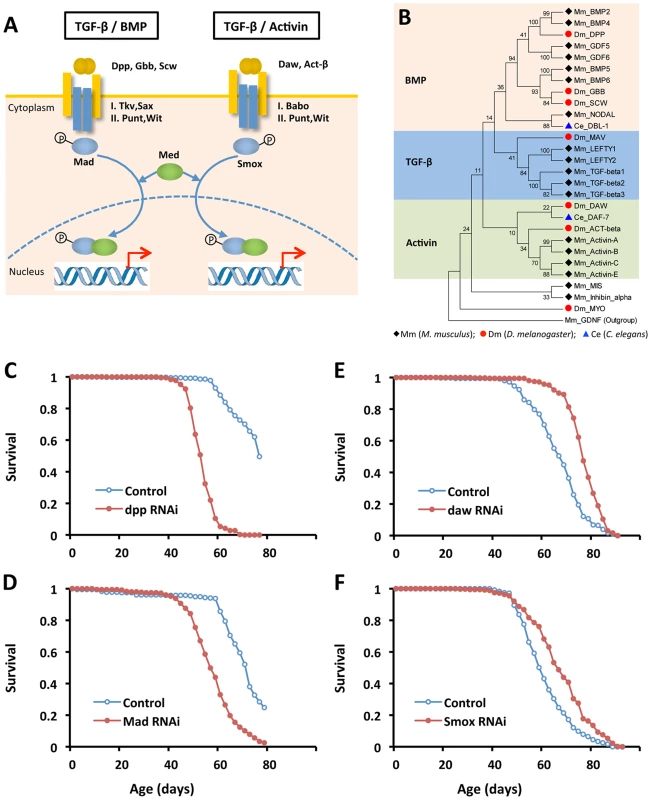
(A) Schematic showing distinct TGF-β pathways in Drosophila: BMP and Activin. (B) Phylogenetic analysis of TGF-β ligands from worm, fly and mouse. Ligand sequences were retrieved from Flybase, Wormbase and Genebank, respectively. The phylogeny was constructed using MEGA 5.0. (C–F) Lifespan analysis of TGF-β pathways in Drosophila using ubiquitous GeneSwitch (GS)-Gal4 drivers, Tub-GS-Gal4 or da-GS-gal4 (Log-rank test, p<0.0001). See Table S3 for survival analysis. Since daw-RNAi increases lifespan, we determined whether other elements of either TGF-β pathway could likewise control aging (Figures 2C–2F and Figure S3). RNAi for Smox, the Activin associated Smad transcription factor, extended lifespan 10% (Figure 2F). RNAi for Activin receptor babo and the Activin-like ligand Act-β did not affect survival. Repressing the BMP branch of TGF-β signaling via RNAi for dpp, gbb, Mad and Tkv consistently reduced survival (Table S3). Ubiquitously overexpressing genes in either Activin or BMP subfamily shortened lifespan (data not shown), as did RNAi for co-Smad (Med), the shared Type-II receptor (Punt and Wit) and two other TGF - β ligands (Myo and Mav) (Table S3).
The TGF-β signaling pathways of Drosophila are homologous to C. elegans TGF-β/dauer and Sma/Mab. Recent reports clarify that the TGF-β/dauer pathway can regulate somatic aging, while the Sma/Mab pathway appears to modulate reproductive aging [35], [36]. We performed a phylogenetic analysis on TGF-β ligands of C. elegans, Drosophila and mouse (Figure 2B). Similar to previous published phylogenetic analysis [37], [38], we found that the TGF-β/dauer ligand of C. elegans, DAF-7, is closely related to Activin-like ligands in Drosophila (Daw and Activin-β) and mouse (Activin-A, B, C and E), while the Sma/Mab ligand in C. elegans, DBL-1, is similar to BMP-like ligands in Drosophila and mouse. Together these results suggest that Activin may be a conserved longevity pathway.
To understand how Activin regulates Drosophila aging we determined which tissues produced this control. daw mRNA is highly expressed in muscle and fat body, a tissue with both liver and adipose-like activities (Figure 3A). Smox protein is more widely distributed (Figure 3B). To assess the role of Activin in muscle and fat body we knocked down daw, Smox and babo with tissue-specific drivers. Lifespan was extended by inactivating each of these genes in muscle, but not in fat body (Figures 3C–3H, Figure S4 and Table S4). Since daw is a dFOXO downstream target that is down-regulated in chico mutants (Figure 1D), we determined whether reduced insulin/IGF-1 signaling modulates Activin within muscle. chico mutants showed reduced daw mRNA sampled from thorax (containing mostly flight muscle), and this effect was reversed in chico; foxo double mutants (Figure 3I). Furthermore, Smox protein was less phosophorylated in chico mutants (Figure 3J). Insulin/IGF-1 signaling, through dFOXO, thus appears to modulate muscle Activin signaling, which in turn is sufficient to regulate longevity.
Fig. 3. Inactivation of genes in Activin signaling (daw, Smox and babo) in muscle, but not in fat body extended lifespan. 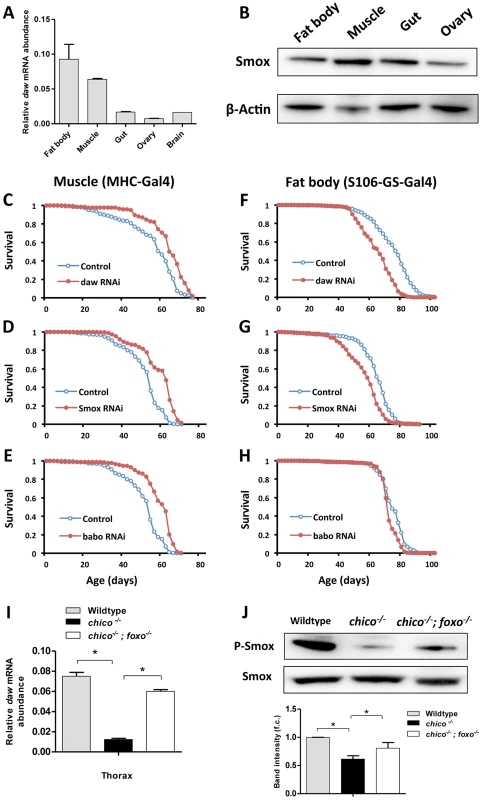
(A) Tissue-specific gene expression pattern of daw. (B) Tissue-specific distribution of transcription factor Smox using 7-day-old Oregon R females. (C–E) Lifespan analysis of Activin signaling using muscle-specific Gal4 driver (MHC-Gal4). Lifespan was extended by inactivating Activin genes (daw, Smox and babo) in muscle (Log-rank test, p<0.0001). (F–H) Lifespan analysis of Activin signaling using adult fat body-specific Gal4 driver (S106-GS-Gal4). Fat body-specific inactivation of Activin genes (daw and Smox) shortens lifespan (Log-rank test, p<0.0001). See Table S4 for survival analysis. (I, J) mRNA expression of daw and phosphorylation of Smox are down-regulated by chico mutation and rescued by mutation of dFOXO. Muscle and fat body were dissected from 7-day-old female wildtype, chico−/− and chico;foxo double mutants. Band intensity was quantified using Bio-Rad Image Lab software. The average band intensity from four independent experiments is shown. Asterisk indicates significant difference between treatment and control (p<0.05). Muscle Activin signaling regulates proteostasis and autophagy
Muscle performance in many animals declines in parallel to the accumulation of misfolded protein aggregates [39]. Insulin/IGF-1 signaling in Drosophila may affect this process since over-expressing dFOXO in Drosophila muscle slows the aggregate accumulation and promotes macroautophagy [18]. Here we determine whether dFOXO mediates its effects on muscle proteostasis and function through its control of Activin.
Experimentally reducing Activin prevents the decline of muscle function with age. Flight activity typically declines in aging flies [18], as it does in our wildtype control (Figures 4A–4C). RNAi against the Activin factors daw, Smox and babo each retarded this decline (Figures 4A–4C). Likewise, the ability to climb at advanced ages was preserved in daw RNAi flies relative to wildtype (Figure S6D). Progression of these composite movement traits was associated with changes in protein aggregates within muscle. Aggregates visualized with Poly-Ubiquitin FK2 antibody increase with age in wildtype muscle, but this change was significantly delayed by muscle specific RNAi against daw, Smox or babo (Figure 4D, F).
Fig. 4. Activin signaling regulates muscle aging and proteostasis. 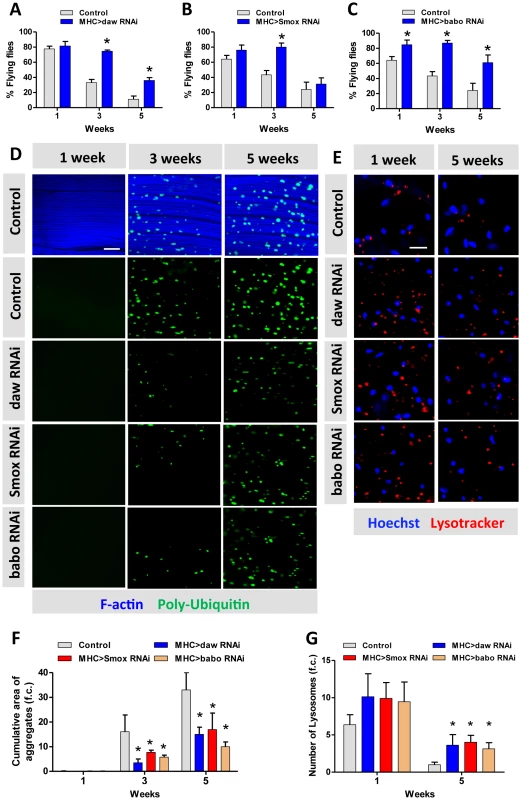
(A–C) Decline of flight with age is delayed in daw, Smox and babo RNAi flies. 40 females were scored for each genotype at each time point. Flying ability was measured at one week, three weeks and five weeks. (D) Poly-Ubiquitin-positive protein aggregates are reduced at old age in daw, Smox and babo RNAi flies. Aggregates were visualized with Poly-Ubiquitin FK2 antibody at one week, three weeks and five weeks. Scale bar: 20 µm. (E) RNAi for daw, Smox and babo preserves the decline of lysotracker-positive organelles (lysosomes). Scale bar: 20 µm. (F) Quantification of the cumulative area of protein aggregates for Figure 4D (n = 20). (G) Quantification of the number of lysotracker-positive stain for Figure 4E (n = 10). Asterisk indicates significant difference between treatment and control (p<0.05). Macroautophagy modulates protein aggregate accumulation [40]. We used two markers of lysosome/autophagy activity, lysotracker and cherry-tagged-Atg8a (homolog of LC3), to determine if Activin regulates muscle proteostasis through macroautophagy. The intensity of lysosome markers decreased with age in wildtype flight muscle, but was maintained in aged muscle expressing RNAi for daw, Smox, or babo (Figure 4E, quantified in Figure 4G). We likewise observed more autophagosomes in flight muscle with inactivated TGF - β/Activin signaling (via RNAi for daw, Smox, or babo) (Figure 5A). In contrast, constitutively activated Activin signaling (via overexpressing babo-Act) reduced the number of autophagosomes (Figure 5A, 5B). Since Activin signaling is transcriptionally regulated by dFOXO via daw, these results may explain reported associations between reduced insulin signaling and elevated autophagy [18]. Reduced insulin signaling represses Activin, which in turn releases repression of autophagy and thereby reduces accumulation of protein aggregates.
Fig. 5. Activin regulates muscle autophagy. 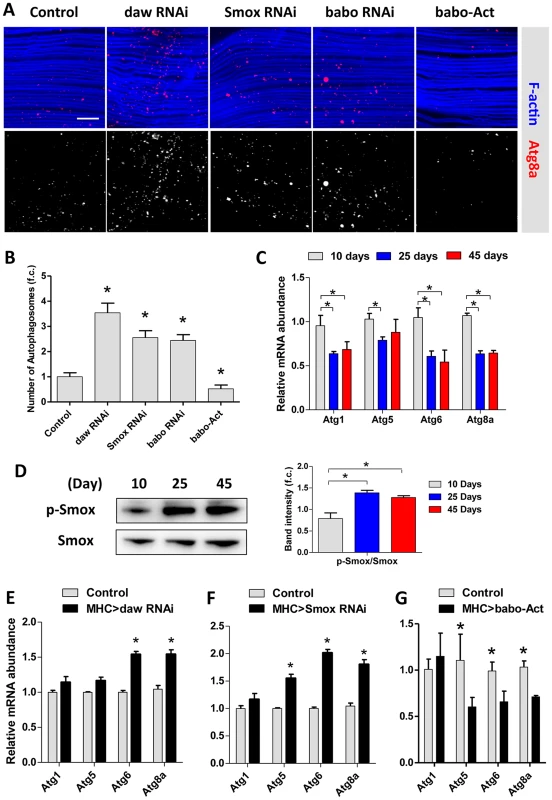
(A) Autophagosomes indicated using an Atg8a-Cherry reporter in Activin RNAi flies or babo over-expressing (babo-Act) flies. 3-day old females. (B) Quantification of autophagosomes for Figure 5A (n = 20). (C) mRNA expression of autophagy genes (Atg1, Atg5, Atg6 and Atg8a) in aging muscle, at 10 days, 25 days and 45 days. (D) Phosphorylation of Smox in muscle increases with age. The average band intensity from three independent experiments was quantified using Image Lab software. (E) Inactivation of daw in muscle up-regulates autophagy gene expression. (F) RNAi against Smox in muscle up-regulates autophagy gene expression. (G) Constitutive activation of babo (babo-Act) in muscle inhibits autophagy gene expression. Asterisk indicates significant difference between treatment and control (p<0.05). Activin signaling represses autophagy via transcriptional regulation on Atg8a
Drosophila encode 18 autophagy genes [41]. Many of these are less expressed in aged flies (Figure 5C) [18]. Since reduced Smox mRNA produces elevated autophagy in aged muscle, we studied the phosphorylation of this transcription factor in old flies. Smox phosphorylation is increased in aging muscle (Figure 5D), suggesting that Activin may be a negative regulator of Atg gene expression. Indeed, Atg6 and Atg8a mRNA were increased when daw and Smox were reduced in muscle (Figure 5E, 5F), while mRNA of Atg5, Atg6 and Atg8a were reduced by over-expressing constitutively active form of the babo receptor (Figure 5G).
Drosophila Smox protein is homologous to vertebrate Smad2 and Smad3 transcription factors. Human Smad3 protein recognizes the consensus sequence GTCTAGAC [42], although a single copy of the Smad box (GTCT) is also reported to support Smad3 binding at the MH1 domain [43]. We searched the promoter regions of Atg8a and identified at least two adjacent Smad boxes located within Atg8a (Figure 6A). ChIP-PCR with affinity-purified Smox antibody showed that Smox binds to the promoter region of Atg8a (Figure 6B), but not Atg1 and Atg6 (Figure 6C). In contrast to Smox, dFOXO does not bind to the promoter of Atg8a (Figure S6E). This is unlike mammalian FoxO3 which induces autophagy by directly binding to the promoters of LC3b, Gabarapl1, and Atg12l in C2C12 myotubes [44]. Consistent with our model for negative regulation on Activin signaling by activated dFOXO, chico −/− inhibits Smox binding at Atg8a (Figure 6D).
Fig. 6. Activin signaling represses autophagy via transcriptional regulation on Atg8a. 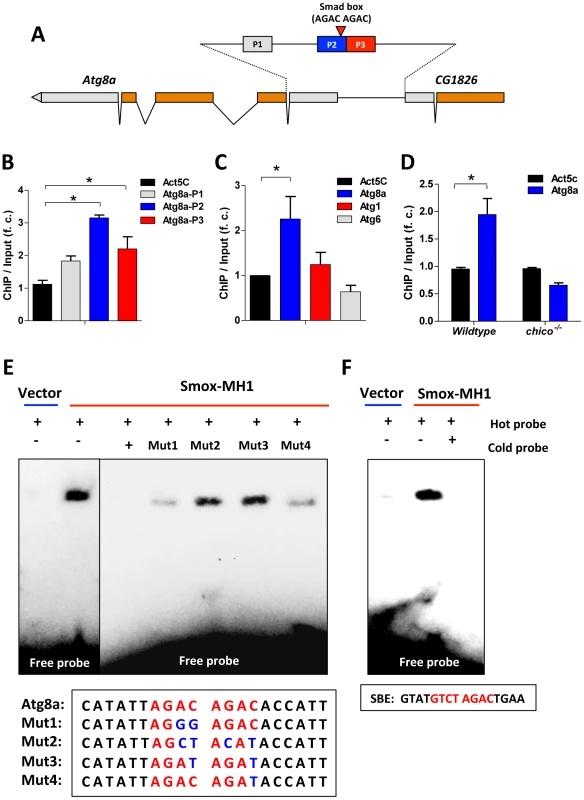
(A) Schematic of Atg8a genomic region (Smad box and ChIP-PCR target regions (P1–P3) are shown). Gray bar represents UTR and orange bar represents exon. (B) ChIP-PCR shows Smox binds to the promoter of Atg8a with binding enrichment calculated as the fold change of ChIP DNA vs. input DNA. The binding to the coding region of Actin gene (Act5C) was used as a negative control. (C) Smox binds to the promoter of Atg8a, but not Atg1 and Atg6. The primers targeting the promoter regions containing putative Smad box in Atg8a, Atg1 and Atg6 were used in ChIP-PCR. (D) The binding of Smox to Atg8a promoter is abolished by chico mutation. Asterisk indicates significant difference between treatment and control (p<0.05). (E). EMSA analysis reveals that recombinant Smox protein (MH1-DNA binding domain) binds to Smad binding element (AGAC AGAC) located in Atg8a promoter. Biotin-labeled Atg8a oligonucleotide probe (5′- CATATTAGAC AGACACCATT -3′) and its mutated forms are labeled with biotin. Halo-tagged Smox-MH1 DNA binding domain (amino acids 1–140) are expressed in E. coli and purified before used in EMSA analysis. (F) Recombinant Smox protein can also bind to mammalian SBE (GTATGTCT AGACTGAA). An electrophoretic mobility shift assay (EMSA) confirms that Smox binds directly within Atg8a promoter. We expressed and purified a recombinant protein of the Smox-MH1 DNA binding domain (amino acids 1–140) and measured its interaction with biotin-labeled Atg8a oligonucleotide probes containing Smad box (5′-AGAC AGAC-3′). Smox-MH1 strongly bound to the Atg8a probe, and this interaction was blocked by addition of unlabeled wildtype cold probes (Figure 6E). To define the required sequences of this Smad box (AGAC) we competed labeled wildtype probe with mutated cold probes. Unlabeled cold probes with mutations in both Smad boxes (Mut2 and Mut3) did not compete with the wildtype binding, but cold probe mutants for only a single only Smad box (Mut1 and Mut4) retained some competitive ability (Figure 6E). Furthermore, in vitro expressed Smox-MH1 also binds to the oligonucleotide probe for the vertebrate Smad binding element (5′-GTCT AGAC-3′) (Figure 6F). Together these data identify an invertebrate Smad binding element (AGAC AGAC) in the promoter region of the autophagy gene Atg8a. This Smad binding element contains a direct repeat of two Smad boxes (AGAC). Upon activation, Smox, the Drosophila homologue of Smad2/3, binds to the Smad box located within the promoter of Atg8a. Activin signaling represses autophagy via direct transcriptional regulation on the key autophagy gene Atg8a.
To test whether increasing Atg8a expression within muscle is sufficient to promote lifespan, we over-expressing Atg8a using a muscle-specific driver (MHC-Gal4). Lifespan was modestly but significantly increased, suggesting that Atg8a gene is a specific instance of a longevity assurance genes that that functions through muscle downstream of Activin signaling (Figure 7A, Table S4). To further examine whether Atg8a is required for Activin-mediated lifespan extension, we silenced both Atg8a and daw using muscle-specific RNAi (Figure 7B, Figure S7, Table S5). Lifespan extension when daw RNAi was expressed in muscle was rescued when Atg8a was simultaneously reduced by RNAi in this tissue, indicating Activin regulates longevity through muscle Atg8a.
Fig. 7. Muscle Activin signaling regulates longevity through Atg8a and remotely controls brain insulin secretion. 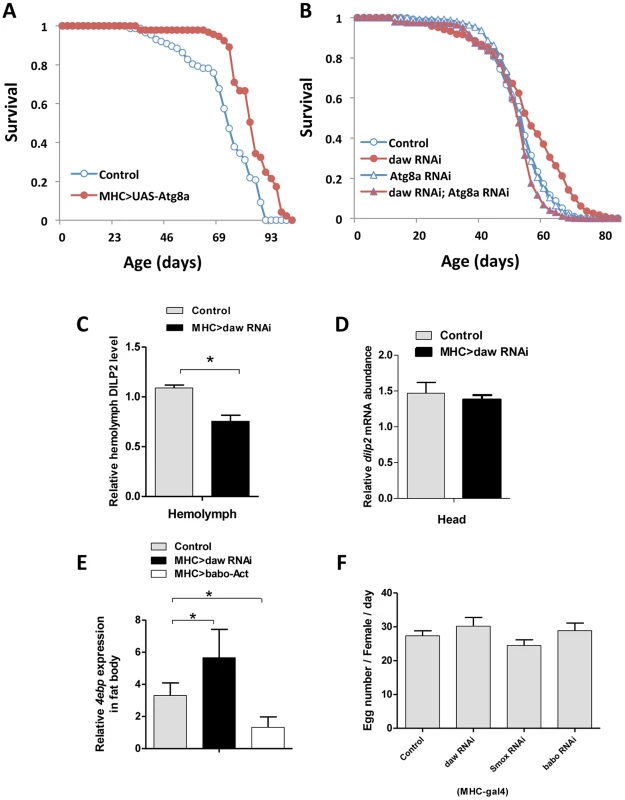
(A) Lifespan analysis of muscle-specific Atg8a overexpression. (B) Genetic epistasis between daw and Atg8a in muscle (MHC-Gal4). Simultaneous expression of RNAi for daw and Atg8a blocks the longevity benefit of daw RNAi alone (while Atg8a RNAi alone does not affect survival). See Table S5, S6 for survival analysis. (C, D) Muscle-specific daw RNAi reduces circulating DILP2 level, but has no effects on dilp2 mRNA expression in the head. (E) 4ebp mRNA expression in fat body is regulated by muscle Activin signaling. 4ebp mRNA is elevated in fat body when daw is reduced in muscle, while it is repressed when muscle babo is induced. (F) Female fecundity is not affected by reducing muscle Activin signaling. Asterisk indicates significant difference between treatment and control (p<0.05). Muscle Activin signaling remotely controls brain insulin secretion
Previous studies from C. elegans recognize cross-talk between insulin/IGF-1 and TGF-β pathways [36], [45]. DAF-16 is nuclear localized in many mutants of the TGF-β/dauer pathway [36]. To determine if muscle Activin signaling affects Drosophila lifespan through systemic insulin/IGF-1signaling we measured circulating insulin-like peptides in adults with tissue specific daw-RNAi. Knockdown of daw in muscle reduced the level of hemolymph DILP2 (Figure 7C), while dilp2 mRNA remained constant (Figure 7D), suggesting the daw specifically modulates DILP2 secretion. In contrast, knockdown of daw in fat body increased the level of circulating DILP2 (Figure S6F). These contrasting tissue associated changes correspond to the observed effects upon lifespan when daw is reduced in each tissue (Figure 3C–3H). Notably, dilp2 mRNA is reduced in other tissue-limited genetic manipulations that extend lifespan [19], [46], and knockout of the dilp2 locus is sufficient to extend lifespan [47]. We now see that reducing muscle Activin signaling via daw RNAi also remotely controls DILP2 secretion from the brain. This is sufficient to decrease systemic insulin signaling because insulin/FOXO sensitive 4ebp mRNA is elevated in peripheral tissues (e.g. fat body) (Figure 7E).
Unlike the consistent response of 4eBP in longevity mutants with reduced insulin, some but not all insulin/IGF pathway mutants have reduced fecundity. Here we found normal fecundity in females with reduced muscle Activin signaling (Figure 7F), suggesting the effects of muscle Activin on lifespan are not mediated through trade-off between longevity and reproduction.
Discussion
Insulin/IGF-1 signaling modulates longevity in many animals. Genetic analysis in C. elegans and Drosophila shows that insulin/IGF-1 signaling requires the DAF-16/FOXO transcription factor to extend lifespan, while in humans several polymorphisms of FoxO3A are associated with exceptional longevity [15], [16]. Although many downstream effectors of FOXO have been identified through genome-wide studies [22], [24], [25], [26], the targets of FOXO responsible for longevity assurance upon reduced insulin signaling are largely unknown [24]. Here we found 273 genes targeted by Drosophila FOXO using ChIP-Seq with two long-lived insulin mutant genotypes. We focused on daw, an Activin ligand, which is transcriptionally repressed by FOXO upon reduced insulin/IGF signaling. Inactivation of daw and of its downstream signaling partners babo and Smox extend lifespan. These results are reminiscent of observations from C. elegans where reduced TGF-β/dauer signaling extends longevity [36]. Notably, the lifespan extension of TGF-β/dauer mutants (e.g. daf-7 (e1372) mutants) can be suppressed by daf-16 mutants, suggesting that TGF-β signaling intersects with the insulin/IGF-1 pathway for longevity in C. elegans [36]. In our phylogenetic analysis, DAF-7, Daw and mammalian Activin-like proteins share common ancestry. Activin signaling, in response to insulin/IGF-1, may thus represent a taxonomically conserved longevity assurance pathway.
Longevity benefits of reduced Activin (TGF-β/dauer) in C. elegans were resolved only when the matricide or ‘bagging’ (due to progeny hatching within the mother) was prevented by treating daf-7(e1372) mutants with 5-fluorodeoxyuridine (FUdR) to block progeny development [36]. This approach made it possible to distinguish the role of Activin in somatic aging from the previously recognized influence of BMP (Sma/Mab signaling) upon reproductive aging in C. elegans [35], [48]. Activin, of course, is a somatically expressed regulatory hormone of mammalian menstrual cycles that induces follicle-stimulating hormone (FSH) in the pituitary gland. In young females, FSH is suppressed within a cycle when maturing follicles secrete the related TGF-β hormone Inhibin [49]. In mammalian reproductive aging, the effect of Activin in the pituitary becomes unopposed as the stock of primary follicles declines, thus inducing elevated production of FSH. We now find that reduced Activin but not BMP signaling favors somatic persistence in Drosophila. These parallels between reproductive and somatic aging among invertebrate models and humans suggest that unopposed Activin signaling is pro-aging while favoring reproduction.
Reduced insulin/IGF signaling extends lifespan through interacting autonomous and non-autonomous actions. Reducing IIS in some distal tissues has been shown to slow aging because this reduces insulin secretion from a few neurons: reducing IIS by increasing dFOXO in fat body or muscle extends Drosophila fly lifespan while decreasing IPC production of systemically secreted DILP2 [18], [19]. Here we identify Activin as a direct, downstream target of insulin/dFOXO signaling within muscles that has the capacity to non-autonomously regulate lifespan. Knockdown of Activin in muscle but not in fat body is sufficient to prolong lifespan. RNAi for muscle Activin signaling led to decreased circulating DILP2 and increased peripheral insulin signaling. Muscle is thus proposed to produce a signaling factor, a myokine, which impacts organism-wide aging and metabolism [18], [50], [51] (Figure S8).
Aging muscle may produce different myokine-like signals in response to their physiological state. Aged muscles degenerate in many ways including changes in composition, mitochondria, regenerative potential and within-cell protein homeostasis [52]. Protein homeostasis is normally maintained, at least in part, by autophagy [40], [53]. Loss of macroautophagy and chaperone-mediated autophagy with age will accelerate the accumulation of damaged proteins [54]. Expression of Atg8a in Drosophila CNS is reported to extend lifespan by 56% [28], while recent studies find elevated autophagy in long-lived mutants including those of the insulin/IGF-1 signaling pathway [18], [55], [56]. Our results now show that insulin/IGF signaling can regulate autophagy through its control of Activin via dFOXO. Poly-ubiquitinated proteins accumulate in aging Drosophila while lysosome activity and macroautophagy decline. Muscle performance with age (flight, climbing) was preserved by inactivating Activin within this tissue. This genetic treatment also reduced the accumulation of protein aggregates. These effects are mediated by blocking the transcription factor Smox, which otherwise represses Atg8a. Smox directly regulates Atg8a through its conserved Smad binding motif (AGAC AGAC). These results, however, contrast with an observation where TGF-β1 promotes autophagy in mouse mesangial cells [57].
Insulin/IGF-1 signaling is a widely conserved longevity assurance pathway. Our data indicate that reduced insulin/IGF-1 retards aging at least in part through its FOXO-mediated control of Activin. Furthermore, affecting Activin only in muscle is sufficient to slow its functional decline as well as to extend lifespan. Autophagy within aging muscle controls these outcomes, and we now find that Activin directly regulates autophagy through Smox-mediated repression of Atg8a. If extrapolated to mammals, pharmaceutical manipulations of Activin may reduce age-dependent muscle pathology associated with impaired autophagy, and potentially increase healthy and total lifespan through beneficial signaling derived from such preserved tissue.
Materials and Methods
Fly husbandry and stocks
Flies were reared and maintained at 25°C, 40% relative humidity and 12-hour light/dark. Adults were maintained upon agar-based diet with cornmeal (0.8%), sugar (10%), and yeast (8% unless otherwise noted). RU486 (mifepristone, Sigma, St. Louis, MO, USA) used to activate GeneSwitch-Gal4 was dissolved in ethanol to a concentration of 200 µM and added to the food.
Fly stocks included: MHC-Gal4 [18], Tub-GS-Gal4 [58]; da-GS-Gal4 [59]; S106-GS-Gal4 [58]; dilp2-GS-Gal4 (provided by H. Jasper); dilp2-GS-Gal4,UAS-dicer2 (provided by S. Helfand). RNAi lines for dFOXO target genes and TGF-β pathway are from Bloomington stock center (TRiP line) and Vienna Drosophila RNAi Center (see the supplemental Table for the stock number). UAS-lines are: UAS-hairy [60], UAS-puc [61], UAS-RhoGAP18B [62], UAS-vri [63], UAS-wit [64], UAS-cv-2 [65], UAS-babo-Act (also known as UAS-babo*1A2) [66], UAS-Atg8-Cherry [67]. MHC-Gal4 is a constitutive muscle-specific driver, with expression restricted to the thoracic region and legs (data not shown).
Chico mutants were made in our lab as describe previously [13], [29]: y1; cn1; ry506 (wildtype), y1; cn1 chico1/cn1; ry506 (chico−/+), y1; cn1 chico1/cn1 chico1; ry506 (chico−/−), y1; cn1 chico1/cn1 chico1; foxo21 ry506 (chico−/−, foxo−/−). Adult on-set IPC ablation flies were made by crossing Dilp2-GS-Gal4 to UAS-rpr and inducing the cell death in IPC cells by feeding flies with RU486 for 15 days.
ChIP-Seq, ChIP-PCR and data analysis
Two insulin mutants were used in ChIP-Seq experiments, chico −/+ and IPC ablation. Chromatin immunoprecipitation (ChIP) was performed according to previously published methods with modification [68], [69], [70]. About 200–250 adult females (∼200 mg) at the age of 15-day-old were pooled for each ChIP sample. Two biological replicates were prepared for each genotype. Flies were homogenized and cross-linked in 1× PBS containing 1% formaldehyde. The fly lysate were sonicated using a Branson 450 sonicator to break down the chromatin into a pool of DNA fragment with average size of 500 bp. Immunoprecipitation was performed using Dynal protean A beads (Invitrogen, Grand Island, NY, USA) and affinity purified anti-dFOXO antibody made in our laboratory. Following the wash with LiCl and TE buffer, the DNA-protein complex was eluted from the Dynal beads and reverse cross-linked. After Proteinase K digestion, dFOXO-bound DNA fragments were purified and diluted in Tris-HCl buffer. About 20 ng of ChIP DNA (dFOXO-bound DNA) and input DNA (DNA sample before the immunoprecipitation) were used in library preparation following the methods described in [71]. The libraries were then size-selected (150 bp-350 bp) and purified by agarose gel, and subjected to the Illumina Genome Analyzer IIx Sequencer (Illumina, San Diego, CA, USA).
To map the dFOXO binding sites, we pooled the raw reads (about 20 million reads per sample) from two replicates into one data file and aligned it to Drosophila reference genome using Bowtie short read aligner [72]. About 70% of raw reads have at least one alignment. The enrichment of dFOXO binding between ChIP DNA and input DNA was determined using peak calling package PeakSeq [73]. Enriched regions with FDR of 0.01 were selected. Target genes, which were detected 5 kb away from the center of the binding sites, were also obtained. The ChIP-Seq raw data are archived at NCBI GEO with Accession # GSE44686.
For ChIP-PCR analysis, the binding enrichment was calculated as the fold change of ChIP DNA versus input DNA. The binding to the coding region of Actin gene (Act5C) and sry genomic region were used as negative controls.
Pathway and motif analysis
The DAVID functional classification tool was used for pathway and molecular function analysis on the dFOXO target genes [74]. Genomic sequence near the dFOXO binding region (∼200 bp) was downloaded from the Flybase (http://flybase.org/) and de novo motif analysis was performed using MEME Suite [75].
Quantitative RT–PCR
Total RNA was extracted from 10 whole flies or from tissue of 15 flies in Trizol reagent (Invitrogen, Grand Island, NY, USA). DNase-treated total RNA was quantified with a NanoDrop ND-1000. About 50–100 ng of total RNA was used for quantification with SuperScript One-Step RT-PCR reagent (Invitrogen, Grand Island, NY, USA) and measured on an ABI prism 7300 Sequence Detection System (Applied Biosystems, Carlsbad, CA, USA). Three biological replicates were used for each experimental treatment. mRNA abundance of each gene was normalized relative to ribosomal protein L32 (RpL32, also known as rp49) by the method of comparative CT. Primer sequences are shown in Table S8.
Phylogenetic analysis
Full length TGF-β ligands from worm, fly and mouse were aligned in ClustalW. From the alignments, a phylogenetic tree was constructed using MEGA 5.0 [76], according to the neighbor-joining method with a bootstrap test calculated with 2000 replicates and a poisson correction model. Mouse Glial cell line-derived neurotrophic factor (GDNF) was used as the out-group.
Demography and survival analysis
Two to three-day-old female adult flies were collected with light CO2 anesthesia and pooled in 1 L demography cages at a density of 100 to 125 flies per cages. Three independent cages were initiated per genotype. Food vials with media containing vehicle only or RU486 were changed every two days, at which time dead flies were removed and recorded. Survival analysis was conducted with JMP statistical software with data from replicate cages combined. Survival distributions were compared by the Log-Rank test. Cox proportional hazard survival analysis was used to assess how reduced daw and Atg8a interacted to affect mortality.
Flying and climbing assays
Flying and climbing assays were scored as described in [18]. In the flying assay, flies were released at the top of a 250 ml cylinder (about 30 cm long). The number of flies that didn't fall straight to the bottom of the cylinder was recorded. A total of 40 females were scored for each genotype.
In the climbing assay (also known as negative geotaxis assay), flies were first tapped down to the bottom of a standard (empty) food vial, and the percentage of flies that climbed up 8 cm within 20 seconds was recorded. A total of 80 females (10 flies per vial) were scored for each genotype.
Immunostaining and imaging
Antibodies for immunostaining included: anti-polyubiquitin FK2 (1∶200) (Assay Designs/Enzo Life Sciences, Farmingdale, NY, USA), and anti-rabbit IgG-DyLight 488 (1∶300) (Jackson ImmunoResearch, West Grove, PA, USA). F-actin was visualized by Alexa Fluor 488-conjugated Phalloidin (Invitrogen, Grand Island, NY, USA). Lysosome was monitored by LysoTracker Red DND-99 at the concentration of 100 nM (Invitrogen, Grand Island, NY, USA). DNA was stained with Hoechst 33342 (1 µg/ml) (Invitrogen, Grand Island, NY, USA). Samples were processed as described in [18], and imaged with a Leica SP2 laser scanning confocal microscope. To quantify the area of protein aggregates and the number of lysotracker or Atg8a-positive dots, grayscale images were converted to binary images (halftone or black & white) with a grayscale cutoff of 20 pixels using ImageJ software [77]. The number/area of positive immunostaining was measured with the “Analyze Particles” function.
Smox antibody and Western blot
Smox polyclonal antibody was generated against the peptide sequence (DSIVDYPLDNHTHQ) corresponding to amino acids 143–156 (Covance, Dedham, MA, USA) and affinity purified (Thermo/Pierce, Waltham, MA, USA) (specificity documented in Figure S5). Phospho-Smad2 antibody was from Cell Signaling Technology (#3108) (Danvers, MA, USA). Thorax tissue from ten female adults was homogenized in RIPA buffer (Thermo/Pierce, Waltham, MA, USA) with protease inhibitor cocktail (Sigma, St. Louis, MO, USA). Supernatant was incubated with SDS loading buffer (Invitrogen, Grand Island, NY, USA) at 70°C for 10 min. About 30 µg of denatured protein was separated on 10% SDS-polyacrylamide precast gels (Invitrogen Grand Island, NY, USA) and transferred to nitrocellulose membranes. Following incubation with primary and secondary antibodies, the blots were visualized with Pierce ECL Western Blotting Substrate (Thermo Fisher Scientific, Waltham, MA, USA). Band intensity was quantified with Image Lab software (Bio-Rad, Hercules, CA, USA).
Smox protein production and Electrophoretic Mobility Shift Assays (EMSA)
cDNA for Smox-MH1 DNA binding domain (1–420 nt) was cloned into pFN29K-His6HaloTag protein expression vector (Promega, Madison, WI, USA). After expression in E. coli, recombinant proteins were purified using HaloTag purification kit (Promega, Madison, WI, USA). Empty vector was used as a negative control.
Biotin-labeled DNA probes were generated using 3′-end biotin labeling (Fisher/Thermo, Waltham, MA, USA). The binding reactions were carried out in a 10 µl of assay mixture containing 10 mM Tris·HCl (pH 7.5), 150 mM KCl, 5 mM MgCl2, 10 ng/µL poly(dI-dC), ∼50 ng labeled probe and 20 µg purified recombinant protein. After incubation at room temperature for 20 min, the mixtures were electrophoresed on 0.8% agarose gels in 0.5× Tris/borate/EDTA buffer. Biotin-labeled DNAs were transferred to a positive-charged nylon membrane (Invitrogen, Grand Island, NY, USA) and detected using GelShift Chemiluminescent EMSA (Active Motif, Carlsbad, CA, USA).
Female fecundity
10-day-old mated female flies were maintained on standard food (2% yeast) for five days at three females per vial and 8–10 vials per group. Flies were passed daily to new vials over five days and eggs were counted daily.
Enzyme Immunoassay (EIA) for hemolymph DILP2
We followed our recently reported EIA assay to measure hemolymph DILP2 [21]. Briefly, about 0.5 µL of hemolymph was collected by decapitation of 15 female flies. Hemolymph was then incubated overnight in a 96-well EIA/RIA plate (Corning Incorporated, Corning, NY, USA) at room temperature. Anti-DILP2 antibody (gift from P. Leopold) was used at 1∶2500 dilution. After the incubation with a HRP-conjugated secondary antibody (1∶2500), hemolymph samples were treated with TMB solution (3,3′,5,5′-teramethylbenzidine; American Qualex antibodies, San Clemente, CA);absorbance was recorded at 450 nm upon a plate reader.
Statistical analysis
Data are presented as mean ± SEM from three independent biological replicates, unless otherwise noted. Statistical significances were evaluated by t-test and one-way ANOVA analyses using GraphPad Prism Software.
Supporting Information
Zdroje
1. TatarM, BartkeA, AntebiA (2003) The endocrine regulation of aging by insulin-like signals. Science 299 : 1346–1351.
2. KenyonC (2010) The genetics of ageing. Nature 464 : 504–512.
3. KenyonC, ChangJ, GenschE, RudnerA, TabtiangR (1993) A C. elegans mutant that lives twice as long as wild type. Nature 366 : 461–464.
4. KimuraKD, TissenbaumHA, LiuY, RuvkunG (1997) daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science 277 : 942–946.
5. ClancyDJ, GemsD, HarshmanLG, OldhamS, StockerH, et al. (2001) Extension of life-span by loss of CHICO, a Drosophila insulin receptor substrate protein. Science 292 : 104–106.
6. TatarM, KopelmanA, EpsteinD, TuMP, YinCM, et al. (2001) A mutant Drosophila insulin receptor homolog that extends life-span and impairs neuroendocrine function. Science 292 : 107–110.
7. HolzenbergerM, DupontJ, DucosB, LeneuveP, GeloenA, et al. (2003) IGF-1 receptor regulates lifespan and resistance to oxidative stress in mice. Nature 421 : 182–187.
8. TaguchiA, WartschowLM, WhiteMF (2007) Brain IRS2 signaling coordinates life span and nutrient homeostasis. Science 317 : 369–372.
9. BluherM, KahnBB, KahnCR (2003) Extended longevity in mice lacking the insulin receptor in adipose tissue. Science 299 : 572–574.
10. OggS, ParadisS, GottliebS, PattersonGI, LeeL, et al. (1997) The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature 389 : 994–999.
11. PuigO, MarrMT, RuhfML, TjianR (2003) Control of cell number by Drosophila FOXO: downstream and feedback regulation of the insulin receptor pathway. Genes Dev 17 : 2006–2020.
12. JungerMA, RintelenF, StockerH, WassermanJD, VeghM, et al. (2003) The Drosophila forkhead transcription factor FOXO mediates the reduction in cell number associated with reduced insulin signaling. J Biol 2 : 20.
13. YamamotoR, TatarM (2011) Insulin receptor substrate chico acts with the transcription factor FOXO to extend Drosophila lifespan. Aging Cell 10 : 729–732.
14. SlackC, GiannakouME, FoleyA, GossM, PartridgeL (2011) dFOXO-independent effects of reduced insulin-like signaling in Drosophila. Aging Cell 10 : 735–748.
15. FlachsbartF, CaliebeA, KleindorpR, BlancheH, von Eller-EbersteinH, et al. (2009) Association of FOXO3A variation with human longevity confirmed in German centenarians. Proc Natl Acad Sci U S A 106 : 2700–2705.
16. WillcoxBJ, DonlonTA, HeQ, ChenR, GroveJS, et al. (2008) FOXO3A genotype is strongly associated with human longevity. Proc Natl Acad Sci U S A 105 : 13987–13992.
17. WessellsRJ, FitzgeraldE, CypserJR, TatarM, BodmerR (2004) Insulin regulation of heart function in aging fruit flies. Nat Genet 36 : 1275–1281.
18. DemontisF, PerrimonN (2010) FOXO/4E-BP signaling in Drosophila muscles regulates organism-wide proteostasis during aging. Cell 143 : 813–825.
19. HwangboDS, GershmanB, TuMP, PalmerM, TatarM (2004) Drosophila dFOXO controls lifespan and regulates insulin signalling in brain and fat body. Nature 429 : 562–566.
20. GiannakouME, GossM, JungerMA, HafenE, LeeversSJ, et al. (2004) Long-lived Drosophila with overexpressed dFOXO in adult fat body. Science 305 : 361.
21. BaiH, KangP, TatarM (2012) Drosophila insulin-like peptide-6 (dilp6) expression from fat body extends lifespan and represses secretion of Drosophila insulin-like peptide-2 from the brain. Aging Cell 11 : 978–985.
22. MurphyCT, McCarrollSA, BargmannCI, FraserA, KamathRS, et al. (2003) Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans.. Nature 424 : 277–283.
23. LeeSS, KennedyS, TolonenAC, RuvkunG (2003) DAF-16 target genes that control C. elegans life-span and metabolism. Science 300 : 644–647.
24. OhSW, MukhopadhyayA, DixitBL, RahaT, GreenMR, et al. (2006) Identification of direct DAF-16 targets controlling longevity, metabolism and diapause by chromatin immunoprecipitation. Nat Genet 38 : 251–257.
25. SchusterE, McElweeJJ, TulletJM, DoonanR, MatthijssensF, et al. (2010) DamID in C. elegans reveals longevity-associated targets of DAF-16/FoxO. Mol Syst Biol 6 : 399.
26. AlicN, AndrewsTD, GiannakouME, PapatheodorouI, SlackC, et al. (2011) Genome-wide dFOXO targets and topology of the transcriptomic response to stress and insulin signalling. Mol Syst Biol 7 : 502.
27. HaseltonA, SharminE, SchraderJ, SahM, PoonP, et al. (2010) Partial ablation of adult Drosophila insulin-producing neurons modulates glucose homeostasis and extends life span without insulin resistance. Cell Cycle 9 : 3063–3071.
28. SimonsenA, CummingRC, BrechA, IsaksonP, SchubertDR, et al. (2008) Promoting basal levels of autophagy in the nervous system enhances longevity and oxidant resistance in adult Drosophila. Autophagy 4 : 176–184.
29. TuMP, EpsteinD, TatarM (2002) The demography of slow aging in male and female Drosophila mutant for the insulin-receptor substrate homologue chico. Aging Cell 1 : 75–80.
30. WangMC, BohmannD, JasperH (2005) JNK extends life span and limits growth by antagonizing cellular and organism-wide responses to insulin signaling. Cell 121 : 115–125.
31. ParkerL, EllisJE, NguyenMQ, AroraK (2006) The divergent TGF-β ligand Dawdle utilizes an activin pathway to influence axon guidance in Drosophila. Development 133 : 4981–4991.
32. SerpeM, O'ConnorMB (2006) The metalloprotease tolloid-related and its TGF-β-like substrate Dawdle regulate Drosophila motoneuron axon guidance. Development 133 : 4969–4979.
33. ZhuCC, BooneJQ, JensenPA, HannaS, PodemskiL, et al. (2008) Drosophila Activin - and the Activin-like product Dawdle function redundantly to regulate proliferation in the larval brain. Development 135 : 513–521.
34. Jensen PA (2012) Regulation of Insect Development by TGF-β Signaling. In: Gilbert LI, editor. Insect Molecular Biology and Biochemistry. Academic Press. pp. 450–479.
35. LuoS, ShawWM, AshrafJ, MurphyCT (2009) TGF-β Sma/Mab signaling mutations uncouple reproductive aging from somatic aging. PLoS Genet 5: e1000789.
36. ShawWM, LuoS, LandisJ, AshrafJ, MurphyCT (2007) The C. elegans TGF-β Dauer pathway regulates longevity via insulin signaling. Curr Biol 17 : 1635–1645.
37. SchmiererB, HillCS (2007) TGF-β-SMAD signal transduction: molecular specificity and functional flexibility. Nat Rev Mol Cell Biol 8 : 970–982.
38. KahlemP, NewfeldSJ (2009) Informatics approaches to understanding TGF-β pathway regulation. Development 136 : 3729–3740.
39. BrignullHR, MorleyJF, MorimotoRI (2007) The stress of misfolded proteins: C. elegans models for neurodegenerative disease and aging. Adv Exp Med Biol 594 : 167–189.
40. RubinszteinDC (2006) The roles of intracellular protein-degradation pathways in neurodegeneration. Nature 443 : 780–786.
41. ChangYY, NeufeldTP (2010) Autophagy takes flight in Drosophila. FEBS Lett 584 : 1342–1349.
42. ZawelL, DaiJL, BuckhaultsP, ZhouS, KinzlerKW, et al. (1998) Human Smad3 and Smad4 are sequence-specific transcription activators. Mol Cell 1 : 611–617.
43. ShiY, WangYF, JayaramanL, YangH, MassagueJ, et al. (1998) Crystal structure of a Smad MH1 domain bound to DNA: insights on DNA binding in TGF-β signaling. Cell 94 : 585–594.
44. ZhaoJ, BraultJJ, SchildA, CaoP, SandriM, et al. (2007) FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab 6 : 472–483.
45. NarasimhanSD, YenK, BansalA, KwonES, PadmanabhanS, et al. (2011) PDP-1 links the TGF-β and IIS pathways to regulate longevity, development, and metabolism. PLoS Genet 7: e1001377.
46. LeeKS, KwonOY, LeeJH, KwonK, MinKJ, et al. (2008) Drosophila short neuropeptide F signalling regulates growth by ERK-mediated insulin signalling. Nat Cell Biol 10 : 468–475.
47. GronkeS, ClarkeDF, BroughtonS, AndrewsTD, PartridgeL (2010) Molecular evolution and functional characterization of Drosophila insulin-like peptides. PLoS Genet 6: e1000857.
48. LuoS, KleemannGA, AshrafJM, ShawWM, MurphyCT (2010) TGF-β and insulin signaling regulate reproductive aging via oocyte and germline quality maintenance. Cell 143 : 299–312.
49. de KretserDM, HedgerMP, LovelandKL, PhillipsDJ (2002) Inhibins, activins and follistatin in reproduction. Hum Reprod Update 8 : 529–541.
50. KatewaSD, DemontisF, KolipinskiM, HubbardA, GillMS, et al. (2012) Intramyocellular fatty-acid metabolism plays a critical role in mediating responses to dietary restriction in Drosophila melanogaster. Cell Metab 16 : 97–103.
51. BostromP, WuJ, JedrychowskiMP, KordeA, YeL, et al. (2012) A PGC1-α-dependent myokine that drives brown-fat-like development of white fat and thermogenesis. Nature 481 : 463–468.
52. NairKS (2005) Aging muscle. Am J Clin Nutr 81 : 953–963.
53. MizushimaN, KlionskyDJ (2007) Protein turnover via autophagy: implications for metabolism. Annu Rev Nutr 27 : 19–40.
54. CuervoAM, BergaminiE, BrunkUT, DrogeW, FfrenchM, et al. (2005) Autophagy and aging: the importance of maintaining “clean” cells. Autophagy 1 : 131–140.
55. HansenM, ChandraA, MiticLL, OnkenB, DriscollM, et al. (2008) A role for autophagy in the extension of lifespan by dietary restriction in C. elegans. PLoS Genet 4: e24.
56. MelendezA, TalloczyZ, SeamanM, EskelinenEL, HallDH, et al. (2003) Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science 301 : 1387–1391.
57. DingY, KimJK, KimSI, NaHJ, JunSY, et al. (2010) TGF-β1 protects against mesangial cell apoptosis via induction of autophagy. J Biol Chem 285 : 37909–37919.
58. RomanG, EndoK, ZongL, DavisRL (2001) P[Switch], a system for spatial and temporal control of gene expression in Drosophila melanogaster. Proc Natl Acad Sci U S A 98 : 12602–12607.
59. TricoireH, BattistiV, TrannoyS, LasbleizC, PretAM, et al. (2009) The steroid hormone receptor EcR finely modulates Drosophila lifespan during adulthood in a sex-specific manner. Mech Ageing Dev 130 : 547–552.
60. JenningsBH, WainwrightSM, Ish-HorowiczD (2008) Differential in vivo requirements for oligomerization during Groucho-mediated repression. EMBO Rep 9 : 76–83.
61. ZeitlingerJ, BohmannD (1999) Thorax closure in Drosophila: involvement of Fos and the JNK pathway. Development 126 : 3947–3956.
62. RothenfluhA, ThrelkeldRJ, BaintonRJ, TsaiLT, LasekAW, et al. (2006) Distinct behavioral responses to ethanol are regulated by alternate RhoGAP18B isoforms. Cell 127 : 199–211.
63. SzuplewskiS, KottlerB, TerracolR (2003) The Drosophila bZIP transcription factor Vrille is involved in hair and cell growth. Development 130 : 3651–3662.
64. MarquesG, BaoH, HaerryTE, ShimellMJ, DuchekP, et al. (2002) The Drosophila BMP type II receptor Wishful Thinking regulates neuromuscular synapse morphology and function. Neuron 33 : 529–543.
65. SerpeM, UmulisD, RalstonA, ChenJ, OlsonDJ, et al. (2008) The BMP-binding protein Crossveinless 2 is a short-range, concentration-dependent, biphasic modulator of BMP signaling in Drosophila. Dev Cell 14 : 940–953.
66. JensenPA, ZhengX, LeeT, O'ConnorMB (2009) The Drosophila Activin-like ligand Dawdle signals preferentially through one isoform of the Type-I receptor Baboon. Mech Dev 126 : 950–957.
67. NezisIP, LamarkT, VelentzasAD, RustenTE, BjorkoyG, et al. (2009) Cell death during Drosophila melanogaster early oogenesis is mediated through autophagy. Autophagy 5 : 298–302.
68. LeeTI, JohnstoneSE, YoungRA (2006) Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat Protoc 1 : 729–748.
69. SandmannT, JakobsenJS, FurlongEE (2006) ChIP-on-chip protocol for genome-wide analysis of transcription factor binding in Drosophila melanogaster embryos. Nat Protoc 1 : 2839–2855.
70. TelemanAA, HietakangasV, SayadianAC, CohenSM (2008) Nutritional control of protein biosynthetic capacity by insulin via Myc in Drosophila. Cell Metab 7 : 21–32.
71. QuailMA, KozarewaI, SmithF, ScallyA, StephensPJ, et al. (2008) A large genome center's improvements to the Illumina sequencing system. Nat Methods 5 : 1005–1010.
72. LangmeadB, TrapnellC, PopM, SalzbergSL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25.
73. RozowskyJ, EuskirchenG, AuerbachRK, ZhangZD, GibsonT, et al. (2009) PeakSeq enables systematic scoring of ChIP-seq experiments relative to controls. Nat Biotechnol 27 : 66–75.
74. Huang daW, ShermanBT, LempickiRA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4 : 44–57.
75. BaileyTL, BodenM, BuskeFA, FrithM, GrantCE, et al. (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37: W202–208.
76. TamuraK, PetersonD, PetersonN, StecherG, NeiM, et al. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28 : 2731–2739.
77. SchneiderCA, RasbandWS, EliceiriKW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9 : 671–675.
78. SlaidinaM, DelanoueR, GronkeS, PartridgeL, LeopoldP (2009) A Drosophila insulin-like peptide promotes growth during nonfeeding states. Dev Cell 17 : 874–884.
Štítky
Genetika Reprodukční medicína
Článek Ribosome Synthesis and MAPK Activity Modulate Ionizing Radiation-Induced Germ Cell Apoptosis inČlánek Fission Yeast Shelterin Regulates DNA Polymerases and Rad3 Kinase to Limit Telomere Extension
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 11
-
Všechny články tohoto čísla
- Molecular Recognition by a Polymorphic Cell Surface Receptor Governs Cooperative Behaviors in Bacteria
- The Light Skin Allele of in South Asians and Europeans Shares Identity by Descent
- Ribosome Synthesis and MAPK Activity Modulate Ionizing Radiation-Induced Germ Cell Apoptosis in
- Retrotransposon Silencing During Embryogenesis: Cuts in LINE
- Roles of XRCC2, RAD51B and RAD51D in RAD51-Independent SSA Recombination
- Parallel Evolution of Chordate Regulatory Code for Development
- A Genetic Approach to the Recruitment of PRC2 at the Locus
- Deletion of the Murine Cytochrome P450 Locus by Fused BAC-Mediated Recombination Identifies a Role for in the Pulmonary Vascular Response to Hypoxia
- Elevated Mutagenesis Does Not Explain the Increased Frequency of Antibiotic Resistant Mutants in Starved Aging Colonies
- Deletion of an X-Inactivation Boundary Disrupts Adjacent Gene Silencing
- Interplay between Active Chromatin Marks and RNA-Directed DNA Methylation in
- Recombinogenic Conditions Influence Partner Choice in Spontaneous Mitotic Recombination
- Crosstalk between NSL Histone Acetyltransferase and MLL/SET Complexes: NSL Complex Functions in Promoting Histone H3K4 Di-Methylation Activity by MLL/SET Complexes
- A New Role for the GARP Complex in MicroRNA-Mediated Gene Regulation
- RNAi-Dependent and Independent Control of LINE1 Accumulation and Mobility in Mouse Embryonic Stem Cells
- Loss of DNMT1o Disrupts Imprinted X Chromosome Inactivation and Accentuates Placental Defects in Females
- Inhibition of the Smc5/6 Complex during Meiosis Perturbs Joint Molecule Formation and Resolution without Significantly Changing Crossover or Non-crossover Levels
- Disruption of Lipid Metabolism Genes Causes Tissue Overgrowth Associated with Altered Developmental Signaling
- Translation Initiation Factors eIF3 and HCR1 Control Translation Termination and Stop Codon Read-Through in Yeast Cells
- Recruitment of TREX to the Transcription Machinery by Its Direct Binding to the Phospho-CTD of RNA Polymerase II
- MYB97, MYB101 and MYB120 Function as Male Factors That Control Pollen Tube-Synergid Interaction in Fertilization
- Oct4 Is Required ∼E7.5 for Proliferation in the Primitive Streak
- Contrasted Patterns of Crossover and Non-crossover at Meiotic Recombination Hotspots
- Transposable Prophage Mu Is Organized as a Stable Chromosomal Domain of
- Ash1l Methylates Lys36 of Histone H3 Independently of Transcriptional Elongation to Counteract Polycomb Silencing
- Fine-Mapping the Genetic Association of the Major Histocompatibility Complex in Multiple Sclerosis: HLA and Non-HLA Effects
- Genomic Mechanisms Accounting for the Adaptation to Parasitism in Nematode-Trapping Fungi
- Decoding a Signature-Based Model of Transcription Cofactor Recruitment Dictated by Cardinal Cis-Regulatory Elements in Proximal Promoter Regions
- Removal of Misincorporated Ribonucleotides from Prokaryotic Genomes: An Unexpected Role for Nucleotide Excision Repair
- Fission Yeast Shelterin Regulates DNA Polymerases and Rad3 Kinase to Limit Telomere Extension
- Activin Signaling Targeted by Insulin/dFOXO Regulates Aging and Muscle Proteostasis in
- Activin-Like Kinase 2 Functions in Peri-implantation Uterine Signaling in Mice and Humans
- Demographic Divergence History of Pied Flycatcher and Collared Flycatcher Inferred from Whole-Genome Re-sequencing Data
- Recurrent Tissue-Specific mtDNA Mutations Are Common in Humans
- The Histone Variant His2Av is Required for Adult Stem Cell Maintenance in the Testis
- The Maternal-to-Zygotic Transition Targets Actin to Promote Robustness during Morphogenesis
- Reconstructing the Population Genetic History of the Caribbean
- and Are Required for Growth under Iron-Limiting Conditions
- Whole Genome, Whole Population Sequencing Reveals That Loss of Signaling Networks Is the Major Adaptive Strategy in a Constant Environment
- Neuron-Specific Feeding RNAi in and Its Use in a Screen for Essential Genes Required for GABA Neuron Function
- RNA∶DNA Hybrids Initiate Quasi-Palindrome-Associated Mutations in Highly Transcribed Yeast DNA
- Mouse BAZ1A (ACF1) Is Dispensable for Double-Strand Break Repair but Is Essential for Averting Improper Gene Expression during Spermatogenesis
- Genetic and Functional Studies Implicate Synaptic Overgrowth and Ring Gland cAMP/PKA Signaling Defects in the Neurofibromatosis-1 Growth Deficiency
- DUX4 Binding to Retroelements Creates Promoters That Are Active in FSHD Muscle and Testis
- Pathways-Driven Sparse Regression Identifies Pathways and Genes Associated with High-Density Lipoprotein Cholesterol in Two Asian Cohorts
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- and Are Required for Growth under Iron-Limiting Conditions
- Genetic and Functional Studies Implicate Synaptic Overgrowth and Ring Gland cAMP/PKA Signaling Defects in the Neurofibromatosis-1 Growth Deficiency
- The Light Skin Allele of in South Asians and Europeans Shares Identity by Descent
- RNA∶DNA Hybrids Initiate Quasi-Palindrome-Associated Mutations in Highly Transcribed Yeast DNA
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



