-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaLoss of Tgif Function Causes Holoprosencephaly by Disrupting the Shh Signaling Pathway
Holoprosencephaly (HPE) is a severe human genetic disease affecting craniofacial development, with an incidence of up to 1/250 human conceptions and 1.3 per 10,000 live births. Mutations in the Sonic Hedgehog (SHH) gene result in HPE in humans and mice, and the Shh pathway is targeted by other mutations that cause HPE. However, at least 12 loci are associated with HPE in humans, suggesting that defects in other pathways contribute to this disease. Although the TGIF1 (TG-interacting factor) gene maps to the HPE4 locus, and heterozygous loss of function TGIF1 mutations are associated with HPE, mouse models have not yet explained how loss of Tgif1 causes HPE. Using a conditional Tgif1 allele, we show that mouse embryos lacking both Tgif1 and the related Tgif2 have HPE-like phenotypes reminiscent of Shh null embryos. Eye and nasal field separation is defective, and forebrain patterning is disrupted in embryos lacking both Tgifs. Early anterior patterning is relatively normal, but expression of Shh is reduced in the forebrain, and Gli3 expression is up-regulated throughout the neural tube. Gli3 acts primarily as an antagonist of Shh function, and the introduction of a heterozygous Gli3 mutation into embryos lacking both Tgif genes partially rescues Shh signaling, nasal field separation, and HPE. Tgif1 and Tgif2 are transcriptional repressors that limit Transforming Growth Factor β/Nodal signaling, and we show that reducing Nodal signaling in embryos lacking both Tgifs reduces the severity of HPE and partially restores the output of Shh signaling. Together, these results support a model in which Tgif function limits Nodal signaling to maintain the appropriate output of the Shh pathway in the forebrain. These data show for the first time that Tgif1 mutation in mouse contributes to HPE pathogenesis and provide evidence that this is due to disruption of the Shh pathway.
Published in the journal: . PLoS Genet 8(2): e32767. doi:10.1371/journal.pgen.1002524
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002524Summary
Holoprosencephaly (HPE) is a severe human genetic disease affecting craniofacial development, with an incidence of up to 1/250 human conceptions and 1.3 per 10,000 live births. Mutations in the Sonic Hedgehog (SHH) gene result in HPE in humans and mice, and the Shh pathway is targeted by other mutations that cause HPE. However, at least 12 loci are associated with HPE in humans, suggesting that defects in other pathways contribute to this disease. Although the TGIF1 (TG-interacting factor) gene maps to the HPE4 locus, and heterozygous loss of function TGIF1 mutations are associated with HPE, mouse models have not yet explained how loss of Tgif1 causes HPE. Using a conditional Tgif1 allele, we show that mouse embryos lacking both Tgif1 and the related Tgif2 have HPE-like phenotypes reminiscent of Shh null embryos. Eye and nasal field separation is defective, and forebrain patterning is disrupted in embryos lacking both Tgifs. Early anterior patterning is relatively normal, but expression of Shh is reduced in the forebrain, and Gli3 expression is up-regulated throughout the neural tube. Gli3 acts primarily as an antagonist of Shh function, and the introduction of a heterozygous Gli3 mutation into embryos lacking both Tgif genes partially rescues Shh signaling, nasal field separation, and HPE. Tgif1 and Tgif2 are transcriptional repressors that limit Transforming Growth Factor β/Nodal signaling, and we show that reducing Nodal signaling in embryos lacking both Tgifs reduces the severity of HPE and partially restores the output of Shh signaling. Together, these results support a model in which Tgif function limits Nodal signaling to maintain the appropriate output of the Shh pathway in the forebrain. These data show for the first time that Tgif1 mutation in mouse contributes to HPE pathogenesis and provide evidence that this is due to disruption of the Shh pathway.
Introduction
Holoprosencephaly (HPE) is a prevalent human disorder affecting forebrain and craniofacial development, with an incidence of up to 1∶250 during embryogenesis, and a high frequency of intrauterine lethality [1], [2]. Recent estimates of the frequency of HPE live births are as high as 1.3 per 10,000 [3], and many children born with severe HPE phenotypes die soon after birth [4], [5]. The primary defect in HPE is a failure of ventral forebrain development with concomitant defects in midline facial structures [6], [7]. In its most severe form (alobar HPE) the forebrain fails to divide, resulting in a single brain ventricle. Less devastating forms of HPE allow near or complete separation of left and right hemispheres [8], [9]. At least 12 genetic loci have been implicated in HPE by mapping of the minimal chromosomal regions deleted in affected families [10]–[12]. Perhaps the best studied HPE gene, Sonic hedgehog (SHH), maps to the HPE3 locus [13]. In humans heterozygous SHH loss of function mutations account for 17% of familial HPE and 3.7% of sporadic cases [13]–[15], suggesting a loss of function haploinsufficient phenotype [16], [17]. The genes encoding the transcription factors TGIF1, Six3 and Zic2 have been identified as the affected genes at other HPE loci [18]–[20]. Interestingly, recent work has shown that Six3 specifically activates expression of Shh in the forebrain, and in mice Shh and Six3 mutations synergize to cause HPE, further emphasizing the importance of the Shh pathway [21], [22].
To establish forebrain dorsoventral patterning, the proper output of the Shh signaling pathway is essential in prechordal plate (PrCP), a primitive streak-derived axial tissue. In mouse embryos at 7.75 dpc, Shh expression is seen in the PrCP underlying the forebrain precursor tissue. Shh expression in the PrCP is essential for activating Shh expression in the overlying ventral diencephalon tissue by 9.0 dpc, where Shh specifies ventral identity [1], [23]. Gli3, a zinc-finger transcription factor that primarily acts as a repressor of Shh signaling, has been shown to play a crucial role in forebrain dorsoventral patterning. In the developing neural tissue, Gli3 is expressed in a gradient that is higher dorsally, and Gli3 homozygous null embryos have a forebrain with dorsally expanded ventral tissue, that lacks dorsal identity [24]–[26]. It has been shown that the proper balance between Gli3 and the ventralizing Shh is critical during forebrain patterning [25], [27]. The lack of ventral identity seen in Shh null embryos is partially rescued when the dose of Gli3 is reduced genetically, suggesting that the mutual antagonism of these two factors is critical for forebrain dorso-ventral patterning. However, since the forebrain develops relatively normally in the absence of both Shh and Gli3, there must be additional pathways that specify telencephalon development, which likely depend on Foxg1 and FGF signaling [28]. Disruption of FGF signaling in the anterior by deletion of the Fgfr1 and Fgfr2 genes results in defective ventral telencephalon development, without disruption of the Shh signaling pathway [29].
TGIF1 (Thymine/Guanine-Interacting Factor) is a homeodomain protein, which binds directly to DNA via a thymine/guanine-containing consensus site, or interacts with Transforming Growth Factor (TGF) β-activated Smad proteins [30], [31]. In response to binding of a TGFβ family ligand to its receptors, the receptor complex phosphorylates and activates specific receptor Smad (R-Smad) proteins: Smad2 or Smad3 in the case of TGFβ, Nodal and Activin [32], [33]. Activated R-Smads complex with the co-Smad, Smad4, translocate to the nucleus and activate target gene expression via direct binding to DNA, or by interactions with other sequence specific DNA binding proteins [33]. Once recruited to DNA, a Smad complex activates transcription in part through interactions with general coactivators, such as p300/CBP [33]. The presence of specific Smad transcriptional corepressors, such as TGIF1, limits the transcriptional response by competing with coactivators and by recruiting general corepressor complexes to the Smads [31], [34]. The more recently identified TGIF2 is homologous to TGIF1 and functions similarly. TGIF2 interacts directly with DNA, or with TGFβ activated Smads and represses gene expression via the mSin3/HDAC complex, but unlike TGIF1, it does not interact with CtBP corepressors [35]–[37]. Thus overall Tgif function (TGIF1 and TGIF2) limits the magnitude of the transcriptional response to TGFβ family ligands. In addition to regulating TGFβ signaling, TGIF1 can also repress gene expression via the RXR retinoid receptor [30], [38], [39].
The TGIF1 gene lies within the minimal HPE4 locus, and TGIF1 sequences were shown to be absent from individuals affected with HPE [20]. In addition to the more common deletions of TGIF1, single amino acid miss-sense mutations have been identified, some of which reduce transcriptional repression by TGIF1 [20], [40]–[42]. Heterozygous loss of TGIF1 causes HPE in humans, suggesting a haploinsufficient phenotype [20]. While there is no evidence for mutations in the human TGIF2 gene being associated with HPE, it is clearly possible that these two related proteins share overlapping functions during embryogenesis [42]. In mice, loss of Tgif1 does not have severe phenotypic consequences, at least in a mixed strain background [38], [43]–45. In a more pure C57BL/6 strain background placental defects and reduced viability are associated with loss of Tgif1, and an intragenic mutation in Tgif1 that may result in expression of a truncated polypeptide caused some anterior defects [46], [47]. As with Tgif1, Tgif2 null mice are normal on a mixed strain background, but the combination of both mutations results in early embryonic lethality with gastrulation defects in all embryos that are homozygous null for both genes [48]. Genetically reducing Nodal signaling in these embryos reduces the severity of the gastrulation defects, consistent with an inhibitory role for Tgifs in the TGFβ/Nodal pathway. While this demonstrates an essential role for TGIF function early in embryogenesis, the function of Tgifs after gastrulation is less well understood.
Here, we investigated the role of Tgif1 and Tgif2 during forebrain development. We demonstrate that loss of Tgif function is indeed important in HPE pathogenesis, and that Tgif1 and Tgif2 play overlapping essential roles during ventral forebrain development by regulating Shh signaling. Conditional loss of function of Tgif1 in the background of a Tgif2 null mutation causes HPE. Furthermore, we show that the HPE phenotype is partially rescued when the dose of Gli3 is reduced. Additionally, we show that reducing Nodal signaling reduces the severity of the HPE phenotype, and partially restores the output of the Shh pathway. This provides the first evidence that Tgifs are required for proper Shh signaling during ventral forebrain development, and verifies that TGIF1 is a bona fide HPE gene.
Results
Loss of Tgif1 and Tgif2 causes HPE
We have previously shown that loss of both Tgif1 and Tgif2 results in a failure of gastrulation [48]. Conditional deletion of Tgif1 in the epiblast, using a loxP flanked Tgif1 allele [45] and the Sox2-Cre transgene, which is expressed in the epiblast after 5.5 dpc [49], in the background of a Tgif2 null mutation allows these embryos (which we refer to as cdKO, for conditional double knock-out) to complete gastrulation. However, most cdKO embryos do not survive past 11.0 dpc, have left-right asymmetry defects, and have severe anterior defects. Scanning electron microscopy (SEM) analysis of frontal forebrain structure revealed that the ventral lips of the cephalic folds are fused in cdKO embryos at 8.25 dpc, as seen in Shh null embryos (asterisks, Figure 1A). It has been shown previously that the normally separated cephalic neural tube is fused in mouse mutants with HPE, including Shh null embryos [50], [51]. Additional SEM analysis at later stages shows that the midbrain neural tube fails to close in cdKO embryos even at 9.25 dpc (Figure 1B). Since human TGIF1 mutations are associated with HPE, we next analyzed the forebrain morphology of control and cdKO embryos to determine whether there was additional morphological evidence to suggest that cdKO embryos have HPE. Whole-mount morphology of the cdKO forebrain at 9.0 dpc showed that overall forebrain size and morphology were relatively normal compared to the control. H&E staining showed that neuroepithelium and surface ectoderm were present, but that the neuroepithelium is thinner and lacks any indication of ventral morphology of the control (Figure 1C). By 10.0 dpc the cdKO forebrain was clearly abnormal, and was significantly smaller than the control (Figure 1D). Further analysis of forebrain structure by H&E staining showed that ventral forebrain morphology was defective, and that cdKO embryos appeared to have a single thickened layer of surface ectoderm in the ventral forebrain, suggesting that the nasal field has not separated by 10.0 dpc (Figure 1D). Since classic HPE phenotypes, such as cyclopia, are more apparent after 11.0 dpc, we analyzed a large number of embryos at 12.5 dpc in an attempt to identify any cdKO embryos that survive to this stage. Although the most of the cdKO embryos die by 11.0 dpc, we were able to identify two cdKO embryos that had survived to 12.5 dpc. For this analysis, we dissected a total of 117 embryos at 12.5 dpc, 76 (65%) of which appeared normal, 39 (33%) were in the process of being resorbed, and only two were doubly homozygous null for Tgif1 and Tgif2. Both cdKO embryos showed cyclopia, and one of the two had developed a proboscis, similar to that in an equivalent stage Shh null embryo (Figure 1E). H&E staining of coronal sections through the brain tissue clearly showed that only one nasal epithelium structure was present in the proboscis tissue of both cdKO and Shh null embryos (Figure 1E, i), and that only one eye field was present in cdKO and Shh null embryos (Figure 1, ii). Thus, the morphological abnormalities in the cdKO forebrain appeared to be quite similar to those seen in Shh null embryos, suggesting that cdKO embryos exhibit a classic form of HPE.
Fig. 1. Analysis of the HPE phenotype in cdKO embryos. 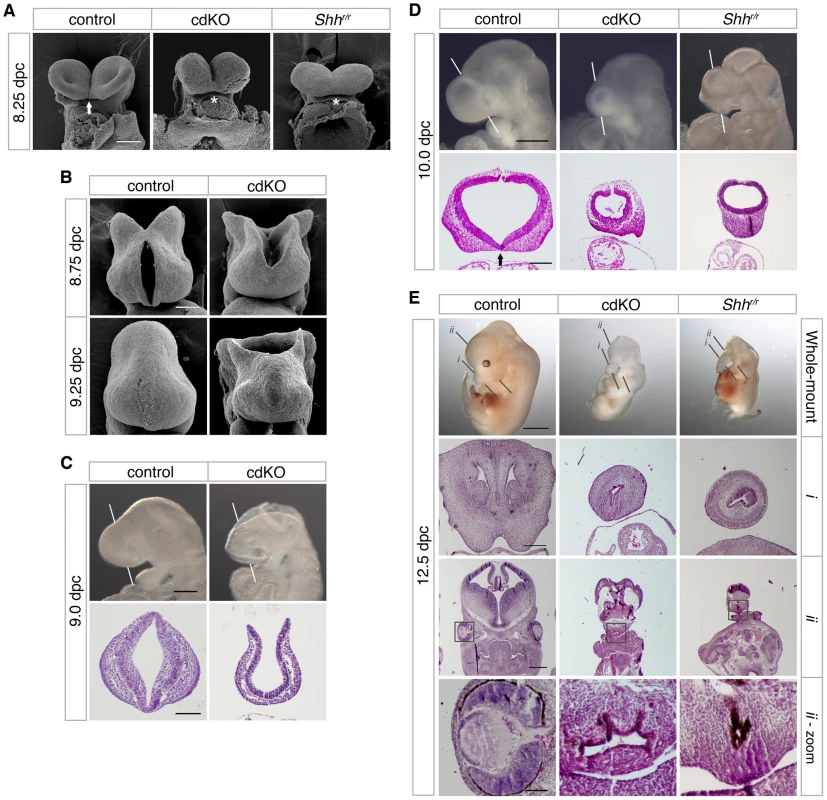
(A) Scanning electron microscopy (SEM) images of the frontal anterior view of embryos at 8.25 dpc, from Tgif1;Tgif2 conditional double intercrosses with epiblast specific deletion of the conditional Tgif1 allele (referred to as cdKO), Shh mutant intercrosses and a stage matched control are shown. The genotype of the control embryos is not indicated as they are representative of normal embryos from these crosses. The arrow indicates the separation of ventral lips of the cephalic folds in the control, that is defective in the cdKO and Shh null (marked by asterisks). Note, the conditional Shh null allele is referred to as ‘r’, for recombined. (B) SEM images of the frontal view of the forebrain of control and cdKO embryos at 8.75 and 9.25 dpc are shown. (C and D) Whole-mount images and hematoxylin and eosin (H&E) stained coronal section of fixed and paraffin-embedded control and cdKO embryos at 9.0 (C) and control, cdKO and Shhr/r at 10.0 dpc (D). The white lines indicate the plane of the coronal sections through the forebrain vesicle. Embryos are representative of at least 3 analyzed. In D, the division of the nasal field by the neuroepithelium is marked by an arrow. Note the continuous thickened layer of surface ectoderm in the mutants. (E) Whole mount images and H&E stained sections of fixed and paraffin-embedded control, cdKO and Shh null embryos at 12.5 dpc are shown. The two planes of section are indicated in the upper panels, and a magnified view of the eye is shown at the bottom (boxed region in section ii). Only two cdKO embryos were identified at this stage. Scale bars: 100 µm in A and B; 250 µm for whole-mount and 100 µm for section in C; 500 µm for whole-mount and 200 µm for sections in D; 2 mm for whole-mount, 250 µm for i, 500 µm for ii and 100 µm for ii-zoom in E. Anterior patterning in cdKO embryos
The defects in forebrain structure led us to test whether anterior patterning is defective in cdKO embryos. The expression of Six3, a transcription factor that activates the Shh gene in ventral forebrain [21], [22], was seen in forebrain in both control and cdKO embryos (Figure 2A). Foxg1, a transcription factor that is required for proper forebrain patterning [52], is expressed in approximately the appropriate pattern in cdKO embryos (Figure 2A). Although there was no major change in the expression pattern, the expression levels of Six3 and Foxg1 were slightly increased in cdKO embryos. In addition, the expression of Fgf8 was clearly increased in the cdKO forebrain, but was still present in approximately the same region as in control embryos (Figure 2A). Consistent with these observations, Fgf8 has been shown to be a FoxH1/Smad2 target gene in the anterior, so may be up-regulated in the absence of Tgifs due to derepression of Smad dependent transcription [53]. Hesx1, which is a highly specific marker for ventral diencephalon [54], shows the appropriate expression pattern in the cdKO ventral diencephalon tissue at 9.0 dpc, suggesting that the midline of the ventral diencephalon is formed in cdKO embryos (Figure 2B). Emx2, a transcription factor that is required for dorsal forebrain patterning [55], was slightly decreased, but was present in a similar domain as in the control (Figure 2B). We next analyzed prospective forebrain tissue in younger embryos. At 7.25 dpc Hesx1 was expressed in the anterior of both control and cdKO embryos (Figure 2C). We have shown previously that the forebrain marker, Otx2, was expressed in cdKO embryos at 7.5 dpc [48], and Six3 was also expressed in the prospective forebrain tissue of cdKO embryos at early head fold (EHF) stage (Figure 2C). Taken together these results suggest that forebrain tissue is for the most part correctly patterned in cdKO embryos.
Fig. 2. Analysis of anterior patterning in cdKO embryos. 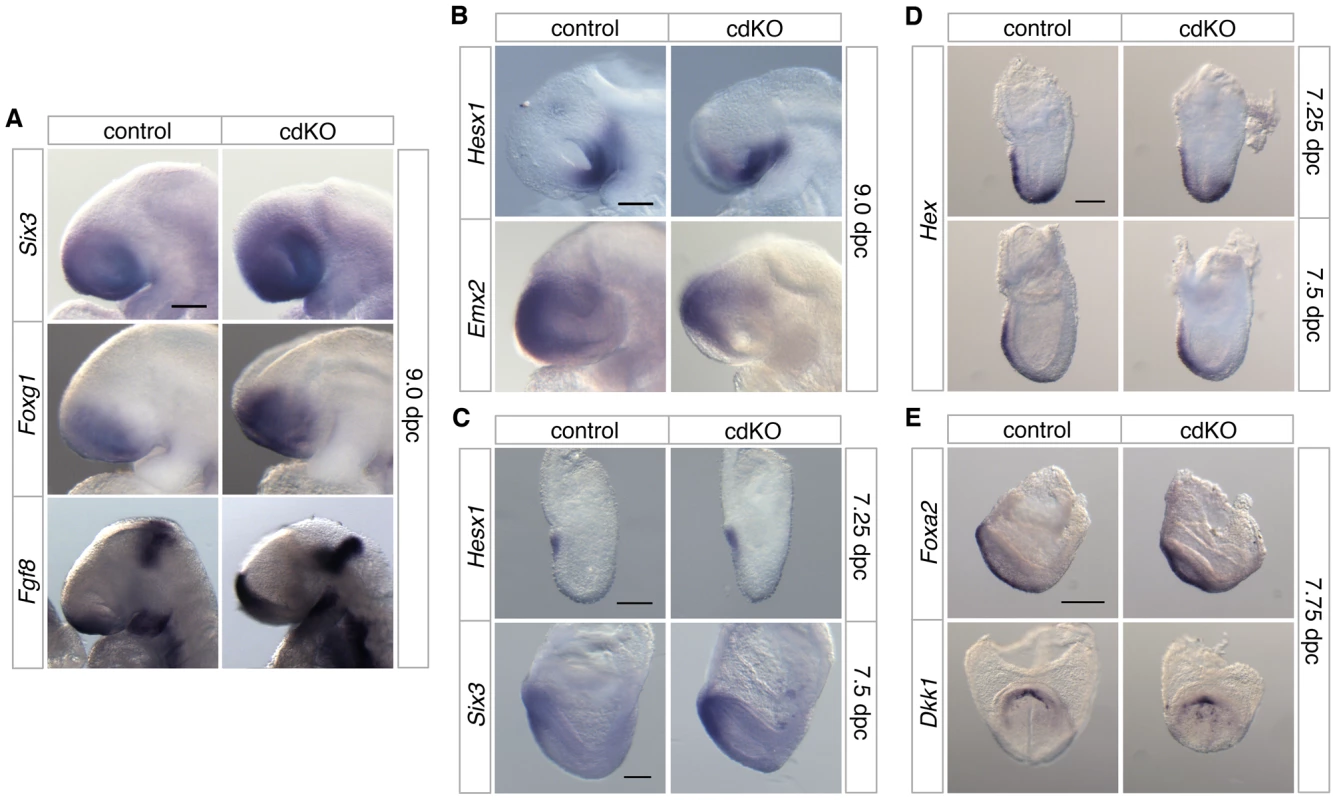
Stage matched control and cdKO embryos were analyzed by in situ hybridization with anti-sense probes for Six3, Foxg1 and Fgf8 at 9.0 dpc (A), Hesx1 and Emx2 at 9.0 dpc (B) and Hesx1 and Six3 at 7.25 and 7.5 dpc respectively (C). Stage matched control and cdKO embryos were analyzed at the indicated stages by in situ hybridization for Hex (D), and Foxa2 and Dkk1 (E). Images shown are representative of at least 3 embryos each. Scale bars: 125 µm in A, B, C and D; 250 µm in E. In the mouse, forebrain induction and patterning is mediated by primitive streak-derived anterior midline tissue, which includes anterior definitive endoderm (ADE) and PrCP [23], [56]. At 7.25 dpc the expression of Hex, a transcription factor that is essential for endoderm development [57], was seen in both control and cdKO embryos in anterior visceral endoderm and also in the ADE migrating out of the primitive streak at this stage (Figure 2D). By 7.5 dpc, Hex expression in anteriorly migrated ADE tissue was present, and did not appear to be significantly different between control and cdKO embryos (Figure 2D). A member of the Forkhead transcription factor family, Foxa2, which is normally expressed in axial tissue [58], was expressed in midline tissue of cdKO embryos at the EHF stage (Figure 2E). The PrCP can be identified by expression of Gsc and Dkk1 at late head fold (LHF) stage and at 8.0 dpc [56], [59]. Appropriate expression of both Gsc and Dkk1 was seen in cdKO embryos (Figure 2E and [48]), suggesting that the PrCP is present in the absence of Tgifs. This analysis suggests that anterior structures are initially patterned relatively normally in cdKO embryos.
Shh signaling is defective in cdKO embryos
While there are clearly some phenotypic differences, such as the failure of the midbrain to close in cdKO embryos, the similarities between cdKO and Shh null embryos raised the possibility that HPE in cdKO embryos may be due to defects in the Shh signaling pathway. At 9.5 dpc, Shh was expressed throughout the neural tube in the floor plate, including the midline of the ventral diencephalon of control embryos (Figure 3A). However, Shh expression was clearly reduced in the ventral diencephalon of cdKO embryos. Similarly, in cdKO embryos Shh expression was reduced in the anterior midline at 8.25 dpc (Figure 3B). By 8.75 dpc Shh expression was present in the ventral forebrain in the control, whereas expression was clearly reduced in the cdKO ventral forebrain tissue (Figure 3B). Transverse sections showed that Shh expression is present but is reduced in the midline tissue including the PrCP (arrows, Figure 3B), and that Shh expression is not detected in the ventral forebrain (Figure 3B). We next analyzed the expression pattern of Shh signaling components at 9.0 dpc. Ptch1 encodes a 12 transmembrane Shh receptor, and Gli1, a transcription factor that mediates Shh signaling [60]. Both genes are direct downstream targets of Shh signaling and are normally expressed strongly in the ventral diencephalon. In cdKO embryos the expression of Gli1 was clearly reduced primarily in the ventral forebrain, while expression was more normal throughout the neural tube up to the forebrain-midbrain boundary (Figure 3C). Ptch1 expression was more similar between cdKO and control embryos, although there was a slight decrease in expression in the anterior in cdKO embryos (brackets, Figure 3C). Together, these results suggest that forebrain patterning is relatively normal, but that the Shh signaling pathway is defective specifically in the ventral forebrain and PrCP. Thus it appears that Tgif function may be required for normal Shh signaling in anterior tissues.
Fig. 3. Defective Shh signaling in the forebrain of cdKO embryos. 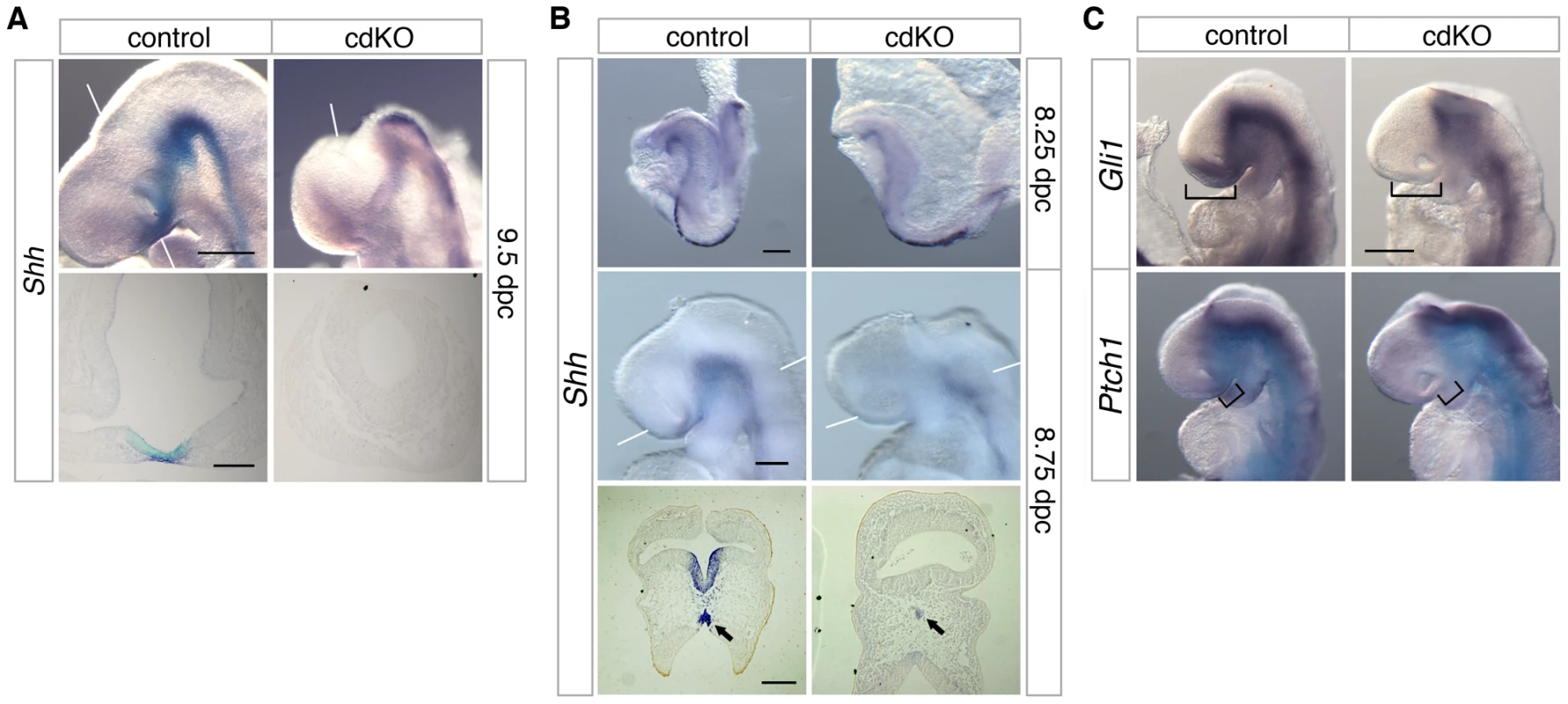
(A and B) Stage matched control and cdKO embryos at the indicated ages were analyzed by in situ hybridization for Shh. Whole mount and images of coronal sections through the forebrain vesicle of paraffin-embedded control and cdKO embryos at 9.5 dpc (A) and transverse sections through ventral forebrain and neural tube at 8.75 dpc (B) are shown. The arrows in B indicate the Shh expression in midline tissue. (C) Stage matched control and cdKO embryos at 9.0 dpc were analyzed by in situ hybridization for Gli1 and Ptch1. Brackets in C indicate the expression domain that is reduced in the cdKO. White lines indicate the plane of sections. Images shown are representative of at least 3 embryos. Scale bars: 250 µm for whole-mount and 100 µm for sections in A; 125 µm for whole-mount and 100 µm for sections in B; 250 µm in C. Shh signaling is rescued by a reduction in Gli3 levels
The transcription factor, Gli3, acts as a potent repressor of the Shh signaling pathway. In the absence of Shh, it has been shown that there is some increase in Gli3 expression [24], and the HPE phenotype in Shh null embryos is partially rescued when Gli3 gene dosage is reduced, suggesting that the proper balance of dorsalizing and ventralizing signals is critical during forebrain development [27], [61]. We, therefore, analyzed the expression level of Gli3 in control and cdKO embryos. Strikingly, Gli3 expression was clearly increased throughout the neural tube including the forebrain in cdKO embryos (Figure 4A). We also performed WISH for Gli3 in Shh null embryos and compared the level of Gli3 expression with cdKO embryos. Surprisingly, Gli3 expression was higher in cdKO embryos than in Shh null embryos (Figure 4A), suggesting that there may be an additional Tgif-mediated mechanism, distinct from the reduction in Shh expression, that regulates Gli3 expression.
Fig. 4. Rescue of Shh signaling by a reduction in Gli3 levels. 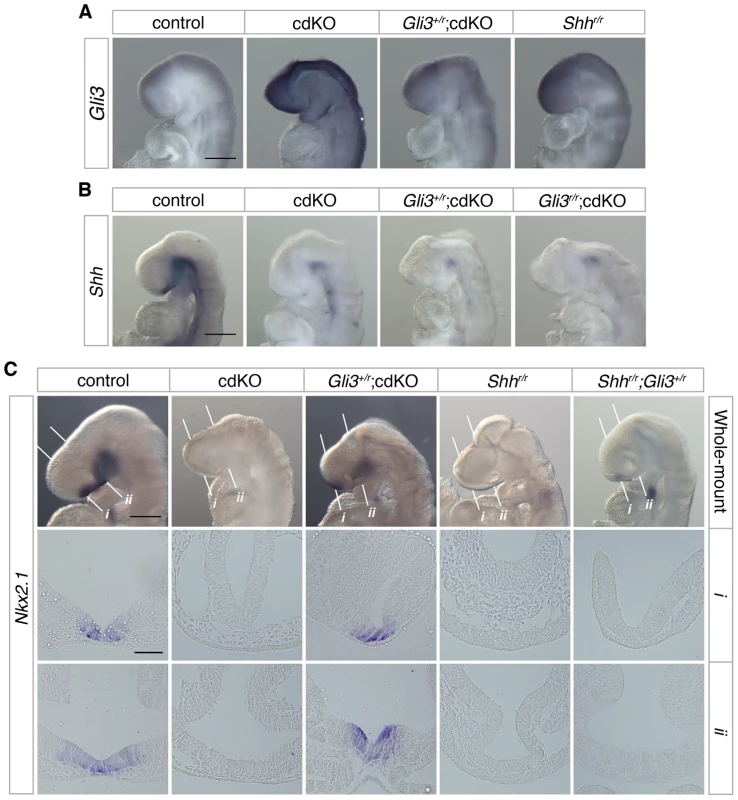
(A) Stage matched control, cdKO, Gli3+/r;cdKO and Shhr/r embryos at 9.0 dpc were analyzed by in situ hybridization for Gli3. (B) Stage matched control, cdKO, Gli3+/r;cdKO and Gli3r/r;cdKO embryos at 9.0 dpc were analyzed by in situ hybridization for Shh. (C) Stage matched control, cdKO, Gli3+/r;cdKO, Shhr/r and Gli3+/r;Shhr/r embryos at 9.0 dpc were analyzed by in situ hybridization for Nkx2.1. Whole mount and coronal sections through the rostral (i) and caudal (ii) forebrain are shown. The white lines indicate the planes of the sections. Embryos shown are representative of at least 3. Scale bars: 250 µm for whole-mount and 50 µm for sections. To determine whether the increased level of Gli3 contributes to defective Shh signaling in the absence of Tgif function, we performed a genetic rescue experiment by introducing a Gli3 mutant allele into the cdKO background. The Gli3 allele has exon 8 flanked by loxP sites such that Cre-mediated recombination creates a null allele [62], which is referred to here as Gli3r. In Gli3+/r;cdKO embryos, Gli3 expression was significantly reduced, to below the expression level seen in Shh null embryos (Figure 4A). In contrast, Shh expression was not restored in cdKO embryos that were heterozygous for Gli3, or in cdKO embryos that were homozygous null for the Gli3 gene (Gli3r/r;cdKO), suggesting that the reduction in Shh expression is at least partially independent of Gli3 activity in cdKO embryos (Figure 4B). We then analyzed the expression of Nkx2.1, a downstream target gene of Shh signaling in the forebrain 1,23, in control and a series of mutant embryos. At 9.0 dpc, the expression of Nkx2.1 was seen in the ventral diencephalon in control embryos, whereas, Nkx2.1 expression was not detected in cdKO or Shh null embryos (Figure 4C). In Gli3+/r;cdKO embryos, Nkx2.1 expression was clearly restored while Nkx2.1 expression in the ventral diencephalon was not rescued in Gli3+/r;Shhr/r embryos (Figure 4C). These results suggest that a reduction in the excess Gli3 expression partially restores the output of the Shh signaling pathway in cdKO embryos, without affecting Shh expression itself.
Reduced Gli3 levels rescue cdKO ventral forebrain morphology
Initial observation of Gli3 heterozygous cdKO embryos suggests that there may be some phenotypic rescue of the cdKO phenotype. Instead of the round forebrain morphology seen in cdKO embryos, a more structured forebrain vesicle was observed in Gli3+/r;cdKO embryos at 10.0 dpc (Figure 5A). To further determine the degree of phenotypic rescue, we H&E stained coronal sections through the forebrain vesicle of control, cdKO and Gli3 heterozygous cdKO embryos. Gli3+/r;cdKO embryos clearly had a more organized forebrain neuroepithelium morphology, and the neuroepithelium appeared to have initiated division of the nasal placode (arrows, Figure 5A), suggesting that the altered balance between Gli3 and Shh expression in cdKO embryos does contribute to the HPE phenotype. In addition, SEM analysis of Gli3 heterozygous cdKO embryos at 8.25 dpc shows a partial rescue of the forebrain structure, such that the Gli3 heterozygous forebrain appears to be less disorganized than the cdKO, and the ventral lips of the cephalic folds appear to be partially separated in the Gli3+/r;cdKO (arrows, Figure 5B). Thus, it appears that reducing Gli3 levels results in some rescue of the cdKO phenotype. To address this further, we tested for changes in proliferation and examined forebrain patterning.
Fig. 5. Rescued ventral forebrain structure in Gli3 mutant cdKO embryos. 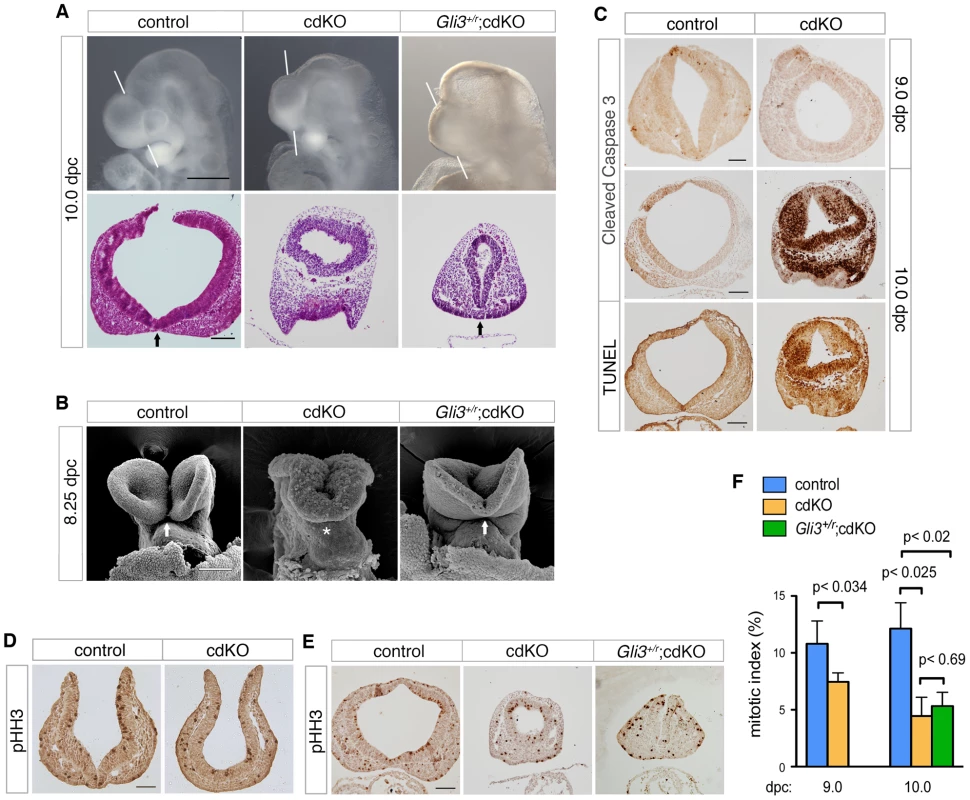
(A) Whole-mount images and H&E stained coronal sections through the forebrain vesicle of control, cdKO and Gli3+/r;cdKO embryos at 10.0 dpc are shown. The white lines indicate the plane of coronal sections. Arrows indicate the division of the nasal field by the neuroepithelium. (B) SEM images of frontal anterior view of control, cdKO and Gli3+/r;cdKO are shown at 8.25 dpc. The arrows indicate the separation of the ventral lips of the cephalic folds in the control, and the partial rescue of this morphology in the Gli3+/r;cdKO, compared to the complete failure in the cdKO (asterisk). (C) Coronal sections of control and cdKO embryos at 9.0 and 10.0 dpc were analyzed by IHC with antibodies for cleaved caspase 3, or by TUNEL at 10.0 dpc. (D) Coronal sections of control and cdKO embryos at 9.0 dpc were analyzed by IHC with antibodies for Histone H3, phosphorylated on serine 10 (pHH3). (E) Coronal sections of control, cdKO and Gli3+/r;cdKO embryos were analyzed by IHC for pHH3 (F) The mitotic index of the forebrain neuroepithelium of control and cdKO embryos at 9.0 or 10.0 dpc, and of Gli3+/r;cdKO at 10.0 dpc was calculated for each section as the percentage of pHH3-stained nuclei. This data is from four control and five cdKO embryos at 9.0 dpc, and three embryos each at 10.0 dpc. Average+s.d. is shown, with the statistical significance as calculated by Student's t-test. Embryos are representative of at least 3 analyzed, unless otherwise noted. Scale bars: 50 µm for sections of 9.0 dpc embryos; 100 µm for sections from 10.0 dpc embryos. Since the anterior of the cdKO is clearly reduced in size by 10.0 dpc, we tested whether the apparent morphological rescue by Gli3 heterozygosity might be due to a restoration of proliferation. Antibody staining for cleaved caspase 3, which is a marker of apoptotic cells, identified very few apoptotic cells in either control or cdKO forebrain at 9.0 dpc (Figure 5C). Although the cdKO embryos were still alive at 10.0 dpc, cells that were positive for cleaved caspase were present throughout the cdKO forebrain neuroepithelium, but were rarely seen in the control (Figure 5C). Consistent with this, TUNEL analysis showed increased apoptosis in the cdKO forebrain at 10.0 dpc (Figure 5C). To determine whether proliferation is reduced in cdKO embryos, we stained multiple coronal sections of control and cdKO forebrains at 9.0 and 10.0 dpc with an antibody to Histone H3, phosphorylated on serine 10 (pHH3), which is a marker for cells in late G2 and mitosis. Mitotic cells were seen throughout neuroepithelium for both control and cdKO at 9.0 dpc (Figure 5D). Quantification of the proportion of mitotic cells in the neuroepithelium showed that there was a significant reduction in proliferation at 9.0 dpc, that was more pronounced by 10.0 dpc (Figure 5E and 5F). These results suggest that cdKO embryos have proliferation defects in the forebrain neuroepithelium, and that the reduced proliferation is seen prior to any increase in apoptosis. We next tested whether the apparent rescue of forebrain morphology in Gli3+/r;cdKO embryos was accompanied by a restoration of normal levels of proliferation. However, in Gli3+/r;cdKO embryos, proliferation levels were not different from the cdKO at 10.0 dpc (Figure 5E and 5F). This suggests that the phenotypic rescue in Gli3+/r;cdKO embryos is independent of changes in proliferation, and that the morphological defects in the cdKO are not solely due to reduced proliferation.
To further characterize ventral structure, we analyzed the expression pattern of Pax7, a nasal field marker, as well as the eye field marker, Pax2 [61]. Normally by 10.0 dpc, the nasal field is well separated as evidenced by the position of the ventral neuroepithelium clearly separating the facial field (see Figure 1D, for example). In Shh null embryos, Pax7 expression is present in a single central region suggesting that the nasal field is not fully separated, whereas when the dose of Gli3 is reduced in Shh null embryos Pax7 expression becomes separated to the two nasal fields [61]. In cdKO embryos, Pax7 expression was observed as a single continuous band, suggesting that nasal field separation is defective (Figure 6A). In Gli3+/r;cdKO embryos, Pax7 expression was clearly well separated and was more similar to that seen in controls, suggesting that the nasal field separation defect is partially rescued in Gli3 heterozygous cdKO embryos (Figure 6A). Similarly, Pax2 expression was reduced and was seen as a single continuous band in cdKO embryos, suggesting that eye field separation is defective (Figure 6B). In Gli3+/r;cdKO embryos, the Pax2 expression level was increased, and appeared as less of a continuous band with distinct eye fields on both sides of the forebrain (Figure 6B). These results suggest that the increase in Gli3 expression, and the altered balance between Gli3 and Shh contribute to the HPE phenotype seen in cdKO embryos resulting in a disruption of the separation of facial primordia.
Fig. 6. Defective separation of facial features. 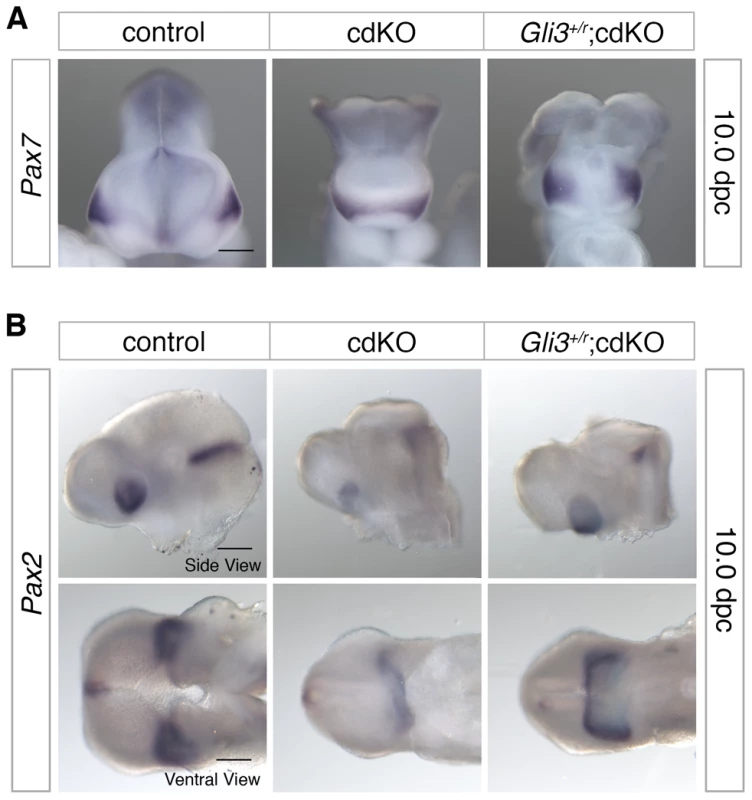
(A) Frontal forebrain images of stage matched control, cdKO and Gli3+/r;cdKO embryos analyzed by in situ hybridization for Pax7. (B) Side and ventral views of embryos analyzed for Pax2 expression are shown. The Gli3+/r;cdKO embryos shown in A and B are representative of 7 and 4 embryos respectively, other images are representative of at least 3. Scale bars: 250 µm for Pax2 and Pax7 side view, and 200 µm for Pax2 ventral view. Nodal dependence of forebrain development in the cdKO embryos
The TGFβ/Nodal signaling pathway has been linked to HPE pathogenesis. For example, HPE has been reported in mouse mutants that result in reduced TGFβ/Nodal signaling, such as Nodal;Smad2 double heterozygotes [63]. Since mutations in these genes result in a reduction in the output of TGFβ/Nodal signaling, rather than the expected increase in cdKO embryos, we generated mice that are heterozygous for both Nodal and Smad2 genes for comparison to our cdKOs. The Smad2 null allele is referred to here as ‘r’ and the Nodal null allele as ‘z’ (see Materials and Methods for a full explanation). Of 41 Nodal;Smad2 double heterozygotes analyzed between 10.5 and 12.5 dpc only one had HPE, although an additional 15 of the 41 double heterozygotes had anterior truncations or a severe growth delay. The Nodal;Smad2 double heterozygous embryo with HPE had a proboscis and a partial failure to separate the eyes, but was significantly larger than cdKO and Shh null embryos (Figure S1). H&E staining of sections through the nasal structure showed a single nasal epithelium that appears structurally similar to that of cdKO and Shh null embryos (Figure S1, i). H&E staining of sections through the eye field showed that a laterally elongated, large optic structure containing two distinct eyes had begun to form, while cdKO and Shh null embryos had only one small pigmented eye field vesicle (Figure S1, ii). Thus, in contrast to the cdKO embryos, it appears that in embryos with reduced Nodal pathway activity HPE is relatively rare.
Our previous analysis of Tgif1;Tgif2 double null mutants showed that Tgifs limit Nodal signaling [48]. To test whether the HPE phenotypes in cdKO embryos were due to increased Nodal signaling, we generated cdKO embryos that carry a Nodal heterozygous mutation. Initial examination of the Nodal heterozygous cdKO embryos suggests that there may be some rescue of the HPE phenotype (Figure 7A). From 317 embryos dissected at 10.0 dpc we identified 38 Nodal heterozygous cdKO embryos, representing 12% of the total, which compares well to the expected 12.5% from these crosses. Other than two severely delayed embryos, and a small proportion (less than 10%) that had severe anterior truncations, the Nodal heterozygous cdKO embryos could be divided into two main phenotypic classes. Around one quarter of the total showed a partial rescue of the cdKO phenotype, such that the forebrain vesicle was better organized and larger in size compared to the cdKO (Figure 7A). Additionally, it appears that there is some improvement in the morphogenesis of the ventral neuroepithelium in these embryos (arrowhead, Figure 7A). The other major phenotype, seen in almost two thirds of Nodal heterozygous cdKO embryos was a reduction in the forebrain. Nodal+/z;cdKO embryos with a reduced forebrain also had a highly disorganized neuroepithelium (Figure 7A). These results suggest that the HPE phenotype seen in cdKO embryos can be at least partially rescued by Nodal heterozygosity, consistent with the defects being due to increased activity of the Nodal/Smad pathway.
Fig. 7. Effects of Nodal heterozygosity of the cdKO phenotype. 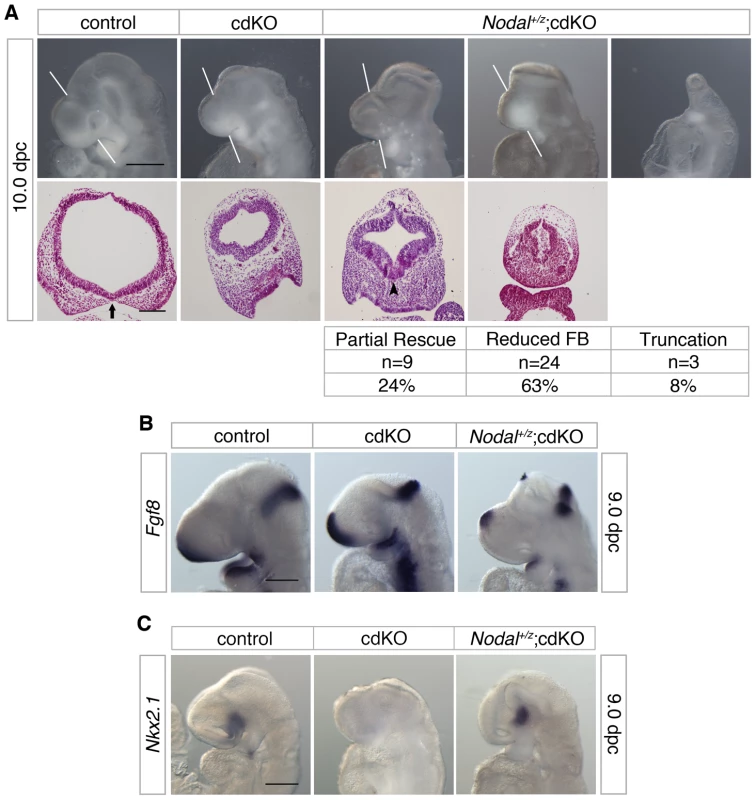
(A) Whole-mount images and H&E stained coronal sections through the forebrain vesicle of control, cdKO and Nodal+/z;cdKO embryos at 10.0 dpc are shown. The white lines indicate the plane of coronal sections. Three Nodal+/z;cdKO embryos are shown that are representative of the three classes of phenotype seen. The numbers of Nodal+/z;cdKO embryos analyzed at 10.0 dpc (from a total of 317 embryos) are shown below for each class of phenotype, together with the percentage of the Nodal+/z;cdKO embryos with each phenotype: Partial rescue of the HPE phenotype; Reduced forebrain (FB); and severe truncation. Two additional embryos were too severely delayed to be classified. Note the improved ventral neuroepithelium morphogenesis in the left hand Nodal+/z;cdKO embryo (arrowhead). The separation of the facial field by the neuroepithelium in the control is indicated by an arrow. (B) Control, cdKO and Nodal+/z;cdKO embryos at 9.0 dpc were analyzed for Fgf8 expression, and for Nkx2.1 expression in (C). Embryos in B and C are representative of at least three each. Scale bars: 250 µm for whole-mount and 100 µm for sections in A; 250 µm in B and C. To confirm that the Nodal heterozygous mutation was reducing expression of Smad2 target genes, we analyzed expression of Fgf8, which is a direct Smad2/FoxH1 target [53]. As shown earlier, Fgf8 expression is increased in cdKO embryos (Figure 2A), whereas, Fgf8 expression was significantly reduced in the forebrain of Nodal+/z;cdKO embryos, consistent with a reduction in Nodal signaling to Smad2 (Figure 7B). In order to determine whether reducing Nodal signaling in cdKO embryos could affect the output of the Shh signaling pathway, we analyzed the expression level of Nkx2.1, a target of Shh signaling in the forebrain at 9.0 dpc. Strikingly, Nkx2.1 expression was restored in the ventral forebrain of Nodal+/z;cdKO while Nkx2.1 expression was clearly reduced in cdKO embryos (Figure 7C). Taken together, these results suggest that Nodal signaling plays a role in regulating Shh signaling during forebrain development, and that unchecked Nodal signaling in the absence of Tgifs is responsible, at least partially, for disrupting Shh signaling in cdKO embryos.
Tgifs coordinate Nodal and Gli3 signaling to regulate Fgf8 expression
Fgf8 plays a role in coordinating multiple patterning centers during forebrain development [64], [65]. In the telencephalon, Fgf8 is a target of TGFβ/Nodal signaling, and is also negatively regulated by Gli3, a potent inhibitory factor of Shh signaling, during early forebrain development [24]. Analysis of Fgf8 expression in Shh null embryos at 8.5 dpc showed that Fgf8 was expressed in the ventral forebrain (Figure 8A). However, consistent with previous work [64], Fgf8 expression was reduced in the telencephalon of Shh null embryos at 8.5 dpc and effectively absent by 9.0 dpc (Figure 8A). In contrast to the reduction of Fgf8 expression in the Shh null embryos, the cdKO forebrain at 9.0 dpc showed increased expression of Fgf8, most likely due to increased Nodal signaling (Figure 2A and Figure 7B). Interestingly, however, analysis at 9.5 dpc revealed that Fgf8 expression was not maintained in cdKO embryos, while Fgf8 expression was clearly restored in Gli3+/r;cdKO embryos (Figure 8B). This result suggests that, by 9.5 dpc, Fgf8 expression is no longer maintained by Nodal signaling and that the excess Gli3 in the cdKO limits Fgf8 expression. We next analyzed the expression pattern of Foxg1, a target of Fgf8 signaling at 9.5 dpc. Foxg1 expression was increased in the cdKO forebrain tissue at 9.0 dpc consistent with the increased expression of Fgf8 (see Figure 2A). At 9.5 dpc, Foxg1 expression in the telencephalon was clearly reduced in the cdKO, whereas, the level of Foxg1 expression was restored to levels similar to that in controls in Gli3+/r;cdKO embryos (Figure 8C). Analysis at 10.0 dpc also revealed that Foxg1 expression was reduced in the neuroepithelium, but was partially restored in Gli3+/r;cdKO embryos. The expression of Foxg1 in the optic vesicle was reduced and was seen as a continuous band in the cdKO (Figure 8C). Although in Gli3+/r;cdKO embryos Foxg1 expression was lower than in controls in the optic vesicle, the expression domains were clearly better separated than in cdKO embryos, providing further evidence for a partial rescue of eye field separation (arrowheads, Figure 8C). Taken together, these results suggest that, at 9.0 dpc Fgf8 expression is dependent on TGFβ/Nodal signaling, whereas, by 9.5 dpc the effect of TGFβ/Nodal signaling decreases and repression of Fgf8 by Gli3 becomes more pronounced.
Fig. 8. Analysis of Fgf8 expression. 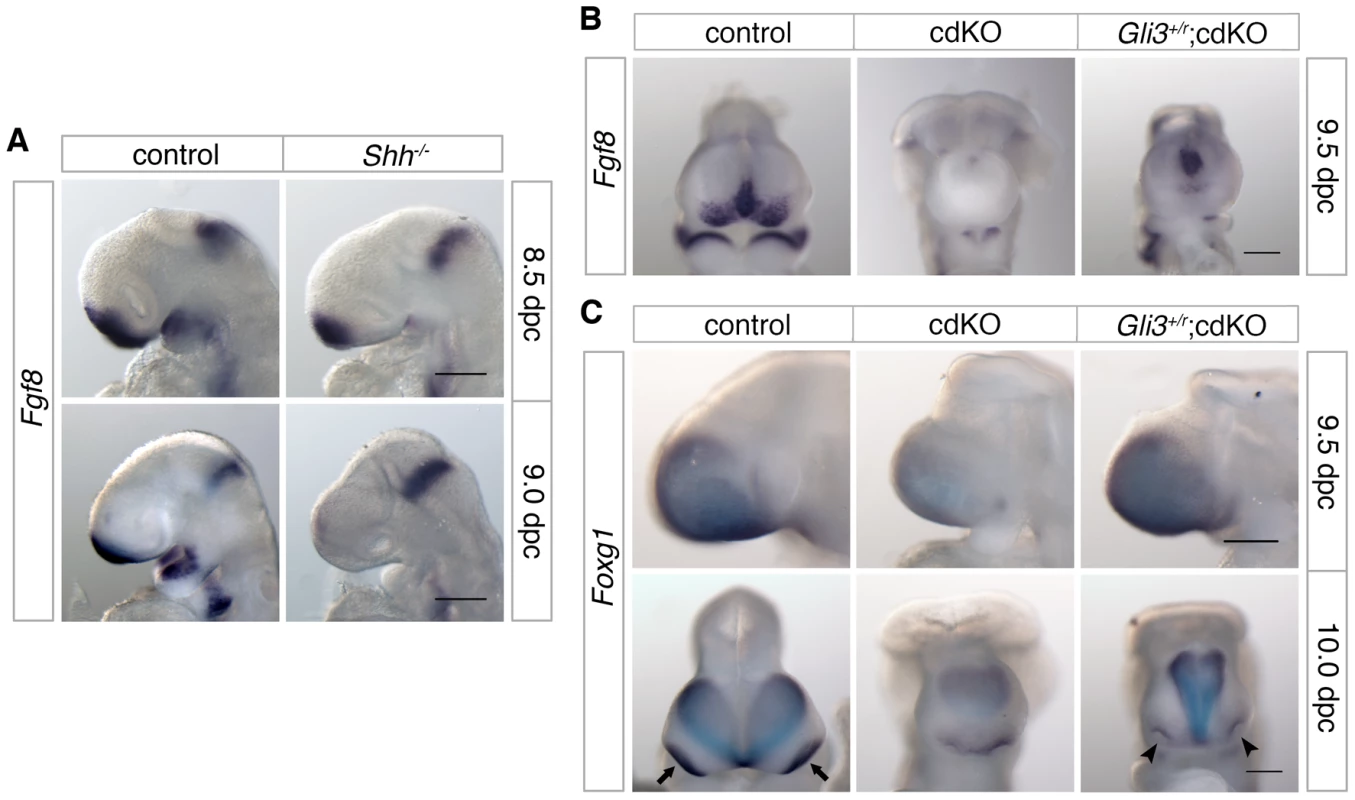
(A) Control and Shh null embryos were analyzed for Fgf8 expression at 8.5 and 9.0 dpc. (B) Control, cdKO and Gli3+/r;cdKO embryos were analyzed for Fgf8 expression at 9.5 dpc, and in (C) for Foxg1 expression at 9.5 and 10.0 dpc. Embryos are representative of at least three of each genotype at each stage and 5 each for panel B. Arrows indicate the eye field expression of Foxg1, and show the partial rescue of eye field separation in the Gli3+/r;cdKO embryo (arrowheads). Scale bar: 180 µm at 8.5 dpc and 250 µm at 9.0 dpc in A; 250 µm in B, C and D. Discussion
Of the 12 genetic loci associated with HPE in humans, the best characterized (SHH, SIX3 and ZIC2) are all linked to the Shh pathway. In contrast, while mutations in the TGIF1 gene, which encodes a corepressor for TGFβ/Nodal signaling, are associated with HPE pathogenesis, the underlying role of Tgif function in forebrain development has remained unclear. We now demonstrate that all embryos with a conditional epiblast-specific double knock-out of Tgif1 and Tgif2 exhibit early HPE-like phenotypes that are reminiscent of those seen in Shh null embryos. Our results provide strong evidence that a major function of Tgifs in the forebrain is to maintain the proper balance between Shh and its antagonist, Gli3, by limiting Nodal signaling. These results resolve the conundrum of how Tgif function is associated with HPE, and identify novel points of coordination between the Shh, Nodal and FGF signaling pathways during anterior development (Figure 9).
Fig. 9. Model for the role of Tgifs in signaling during forebrain development. 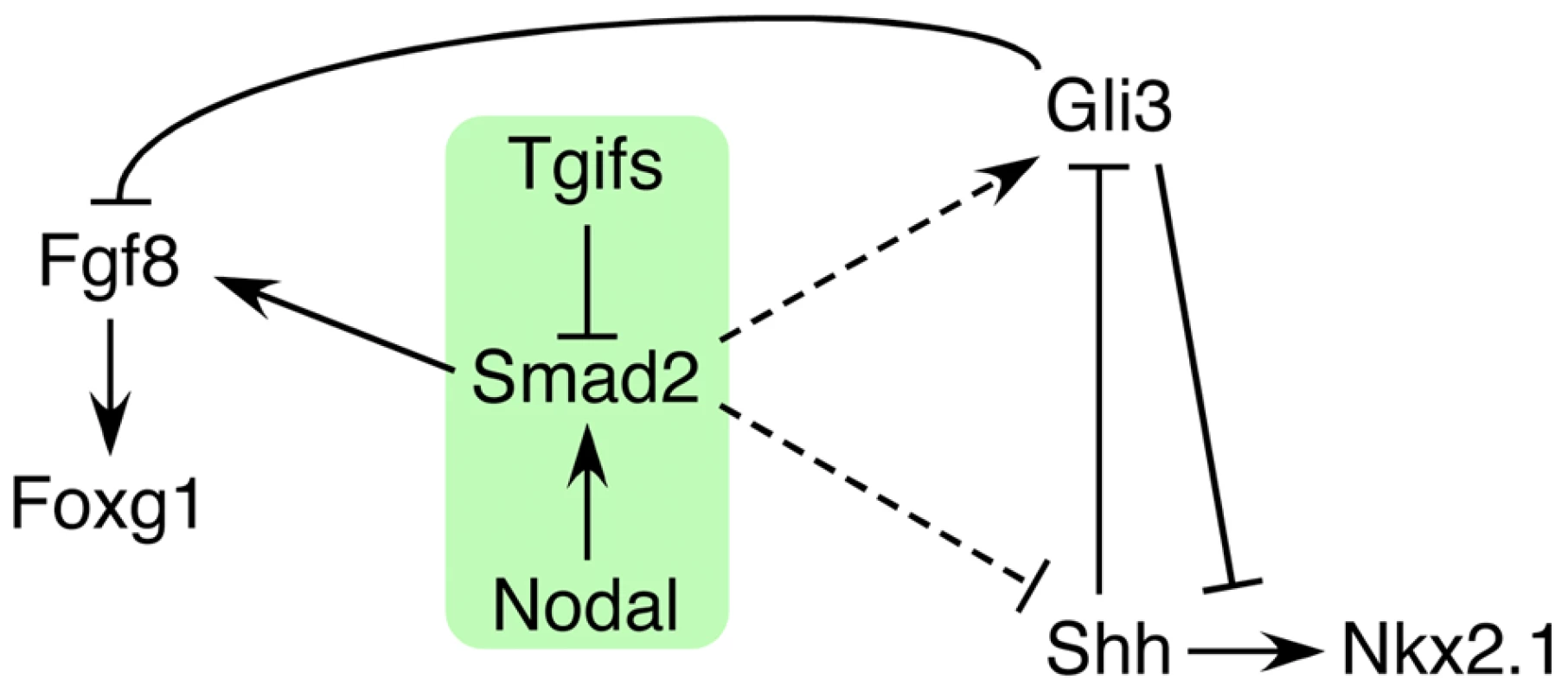
A tentative model is shown that describes the data presented here. Briefly, Tgifs limit Smad2 transcriptional activity, which is required for activation of Fgf8 expression. Tgif regulation of the Nodal-Smad2 pathway is required for the correct balance between Gli3 and Shh activity in the Shh pathway. Dashed lines indicate that the links from the Nodal-Smad2 pathway to Shh signaling components may not be direct, and that the regulation may be of both Shh and Gli3, or may occur primarily via one of them. SHH, SIX3, ZIC2 and TGIF1, are the four genes that are most commonly screened as a part of the genetic evaluation of human HPE patients [66]. Mice homozygous for a Shh null allele exhibit defects in midline facial features including cyclopia and proboscis that are typically seen in severe cases of human HPE, suggesting that SHH mutations do contribute to HPE in humans [50]. Recent work showed that the transcription factor Six3 is directly linked to Shh signaling by acting as a transcriptional activator of the Shh gene, specifically in the ventral forebrain [21], [22]. ZIC2, encodes a zinc-finger containing transcription factor, that has been shown to be important for forebrain patterning and Shh signaling [67], [68]. Thus, the best characterized HPE mutations appear to target the Shh signaling pathway. In contrast, the role in HPE pathogenesis of mutations in TGIF1, which encodes a corepressor for TGFβ/Nodal signaling, has long remained unclear. Loss of function mutations in the Tgif1 gene in mice have no severe phenotypes in a mixed strain background, although an intragenic mutation in Tgif1, which may create a hypomorphic allele, has been shown to cause anterior defects in a strain specific manner [47]. However, HPE phenotypes have not been seen in Tgif1 or Tgif2 mutants, and these analyses have not yet shed light on any potential role in HPE pathogenesis.
Tgif2, a closely related Tgif1 paralog present in mouse and human, shares conserved functions with Tgif1 [69]. Both Tgif1 and Tgif2 show ubiquitous expression in the embryo proper from at least 6.0 dpc, consistent with the possibility of overlapping function during early development. As with Tgif1 mutations, mice that carry a homozygous Tgif2 mutation do not show appreciable phenotypes in a mixed strain background. Mice with both Tgif1 and Tgif2 mutations, with at least one wild-type allele of either Tgif1 or Tgif2, are also viable and fertile in a mixed strain background [48]. In contrast, embryos with homozygous mutation of both Tgif1 and Tgif2 fail to gastrulate, providing strong evidence that Tgif1 and Tgif2 perform essential overlapping functions during embryogenesis. Thus, although there is no evidence suggesting that human TGIF2 is associated with HPE [42], it is possible at least in mice, that both proteins share overlapping functions in anterior development. We generated embryos with Sox2-Cre mediated conditional deletion of Tgif1 in the background of a Tgif2 null, which allows the resulting embryos to undergo gastrulation successfully. At 10.0 dpc, the cdKO embryos have an HPE-like forebrain and neuroepithelium morphology, and the expression patterns of Pax2 and Pax7 suggest that separation of midline facial features is defective. Moreover, SEM analysis shows that separation of the ventral lips of the cephalic neural fold is defective, consistent with the failure to divide midline facial features. These phenotypes are typical of early HPE mouse mutants such as Shh null embryos, clearly demonstrating that Tgif1 and Tgif2 share redundant functions and together are essential players in normal forebrain development. Although the majority of cdKO embryos fail to survive past 11.0 dpc, from an analysis of 117 embryos where approximately 30 were expected to be cdKO, we were able to identify two embryos lacking both Tgif1 and Tgif2 at 12.5 dpc, which had presumably survived to this point due to a slight delay in recombination of the conditional Tgif1 allele. Interestingly, these two embryos also showed remarkable similarity to Shh null embryos at the same stage. Specifically, one had a proboscis and both had cyclopia, further reinforcing the idea that the early phenotypes analyzed in detail here are clear precursors of later HPE. While the fact that relatively few embryos survive past 11.0 dpc limits our ability to analyze later HPE phenotypes in detail, those cdKO embryos that do survive to 12.5 dpc have classic HPE phenotypes. Despite the similarity of the HPE-like phenotypes, it should be noted that there are some differences between our cdKO and Shh null embryos. Such differences include the failure of the midbrain neural tube to close, which is not seen in Shh nulls, and the fact that the majority of cdKO embryos die by 11.0 dpc, whereas most Shh null embryos survive to late gestation. These differences aside, this work provides the first clear evidence from mouse models for a role for loss of Tgif function in HPE pathogenesis.
Our data suggest that Tgif function is required for appropriate Shh signaling during forebrain development. In cdKO embryos, Shh expression is present but reduced in the PrCP, and is undetectable in the neuroepithelium, suggesting that Shh is transcriptionally activated but that its expression is not properly maintained. In addition to the defective Shh expression in the forebrain, the expression of downstream targets of Shh signaling is significantly reduced in the forebrain. Expression of Gli3, which encodes a repressor for the Shh signaling pathway in the forebrain, is up-regulated in Shh null embryos, and the HPE phenotype of Shh null embryos is partially rescued when the genetic dose of Gli3 is reduced [27], [61]. Similarly, cdKO embryos showed an increased level of Gli3 expression in the forebrain. Intriguingly, the increase in Gli3 expression in cdKO was clearly higher than in Shh null embryos, suggesting that there is an additional, Shh-independent, Tgif-dependent mechanism that regulates Gli3 gene expression. In cdKO embryos with a reduced dose of Gli3, there was a phenotypic rescue in the morphology of the forebrain neuroepithelium and also of the craniofacial features. Additionally, Nkx2.1 expression was restored in the ventral diencephalon of cdKO embryos carrying a Gli3 heterozygous mutation, while, in agreement with previous work, there was no rescue of Nkx2.1 expression in the diencephalon of Shh null embryos with a Gli3 heterozygous mutation [61]. This suggests that some level of Shh expression is required for Nkx2.1 expression, and also suggests that sufficient Shh expression is present to activate Nkx2.1 in the ventral diencephalon of cdKO embryos. However, it should be noted that Shh expression was not rescued in the ventral forebrain of Gli3 mutant cdKO embryos. Although many mutations that cause HPE may do so by affecting the Shh pathway, and specifically the balance between Shh and Gli3, it is worth pointing out that Gli3 heterozygosity does not rescue all mouse models of HPE. For example, the phenotype of Fgfr1;Fgfr2 double mutant embryos is not rescued by Gli3 mutation, suggesting that there is some specificity to the rescue by Gli3 mutations [29]. Taken together, these data provide strong evidence that Tgifs play a critical role in regulating Shh signaling during forebrain development, and that the loss of Tgif-mediated regulation of the Shh pathway is important for HPE pathogenesis.
Studies in humans and mice have implicated both the retinoic acid and TGFβ/Nodal pathways in HPE pathogenesis. Retinoic acid mediated teratogenesis in humans is known to contribute to CNS anomalies such as hydrocephalus, and in a few rare cases, HPE, and in mice in utero administration of retinoic acid to pregnant females on gestational day 7 leads to embryos with severe craniofacial phenotypes including HPE [70], [71]. However, mutations in genes associated with retinoic acid signaling have not been identified in HPE patients. Mutations that likely reduce the output of the TGFβ/Nodal pathway have been found in human patients with HPE or laterality defects. Mutations in TDGF1 (also referred to as CRIPTO), an EGF-CFC family member that acts as a co-factor for the NODAL ligand, and in the gene encoding the forkhead transcription factor FOXH1 (also known as FAST1), which complexes with SMAD2 and SMAD4 to mediate TGFβ/NODAL signaling, have been identified [72], [73]. However, these mutations are found very rarely in HPE, and in general are not complete loss of function alleles. Studies in Tdgf1 null and Foxh1 null embryos show that these genes are required for the activity of the early organizing centers during gastrulation [74], [75]. In Tdgf1 null embryos, marker analysis shows that expression of organizer genes including Brachyury, Cerl1 and Lhx1 is defective. Similarly in Foxh1 null embryos, expression of organizer genes such as Foxa2 and Goosecoid, is reduced, and analysis of forebrain markers such as Six3, Hesx1 and Fgf8 shows that the forebrain tissue is significantly reduced, exhibiting a mild anterior truncation phenotype [75]. It has also been suggested that Nodal;Smad2 double heterozygous mutations can result in HPE, again indicating that a reduction in TGFβ/Nodal signaling is important in HPE pathogenesis [1]. However, the morphology of these embryos suggests that in most cases forebrain tissue is reduced or missing, rather than exhibiting a clear HPE phenotype as seen in Shh null embryos, for example. Thus it appears that, at least in mice, a reduction in the TGFβ/Nodal signaling pathway primarily results in defective early organizing centers, leading to phenotypes such as a small or truncated forebrain. In contrast, in our cdKO embryos, marker analysis shows that the organizing centers are formed, and that the forebrain does not show an anterior truncation phenotype. In addition, the forebrain morphology shows an HPE phenotype that is similar in many respects to that seen in Shh null embryos, and forebrain markers show relatively normal expression patterns, suggesting that the forebrain is reasonably formed in cdKO embryos. Our own analysis of embryos that are heterozygous for both Smad2 and Nodal is in agreement with the idea that HPE is relatively rare in this genetic combination – only one out of 41 double heterozygotes analyzed at 10.5–12.5 dpc had HPE, with an additional 15 showing severe growth delays or anterior truncations. Additionally, it is interesting to note that the comparison of cdKO, Shh null and Smad2/Nodal double heterozygous embryos with HPE at 12.5 dpc suggests that, at least superficially, the Shh null and cdKO are more similar to each other than to the Smad2/Nodal double heterozygote. Thus the loss of Tgif1 and Tgif2 causes a classic HPE phenotype, rather than the predominance of anterior truncations that are seen in embryos with reduced activity of the TGFβ/Nodal pathway.
Our results, together with evidence from mouse mutants with reduced Nodal activity, support a model in which decreased Nodal signaling primarily results in a truncation of anterior tissues, whereas increased Nodal signaling (as in our cdKO embryos) causes classic HPE phenotypes. One alternate interpretation of this difference between the HPE phenotype in cdKO embryos and other TGFβ/Nodal mouse mutants is that the effects of loss of Tgif function are independent of TGFβ/Nodal signaling during forebrain development. However, we have shown that embryos that are homozygous null for both Tgif1 and Tgif2 fail gastrulation, and that the gastrulation defect is dependent on increased TGFβ/Nodal signaling. Similarly, left-right asymmetry defects in cdKO embryos can be partially rescued by reducing the dose of Nodal [48]. Here we show that at 9.0 dpc, Fgf8 expression is increased in the cdKO, consistent with the derepression of a Smad/Foxh1 target gene [53]. Importantly, this excess Fgf8 expression is reduced in the Nodal heterozygote. Reducing the dose of Nodal also results in a partial rescue of the HPE phenotypes in a proportion of cdKO embryos. Most of the remaining Nodal heterozygous cdKO embryos have a mild anterior truncation, which might indicate that there are additional Nodal and Tgif specific phenotypes, but could also reflect the effect of mutating multiple components of the Nodal pathway. However, with the restoration of Nkx2.1 expression in the Nodal heterozygous cdKO forebrain, this is clearly consistent with a model in which Tgifs limit Nodal signaling and that the absence of this restraint causes disruption of the Shh pathway and HPE. It should, however, be noted that we have not yet exhaustively analyzed the Shh signaling pathway in Nodal heterozygous cdKO embryos, and it will clearly be of interest in the future to determine precisely how Nodal heterozygosity rescues Nkx2.1 expression and forebrain morphology. One attractive candidate for the Nodal target would be the Gli3 gene, given its striking upregulation in the cdKO. However, this remains to be tested and potential effects of other pathways, such as FGF signaling, that specify forebrain patterning should also be considered. On balance, it is reasonable at this point to suggest that the HPE phenotype seen in cdKO embryos is dependent on excessive TGFβ/Nodal signaling due to the loss of Tgif-mediated repression, and that disruption of the Shh pathway makes a major contribution to the phenotype.
The increased Fgf8 expression seen at 9.0 dpc in cdKO embryos is consistent with an increase in Nodal signaling, and is in fact reduced in the Nodal heterozygote. However, this also appears to be somewhat at odds with the increased Gli3 expression seen in cdKO embryos, since Gli3 represses Fgf8 expression in the anterior. However, by 9.5 dpc, we show that Fgf8 expression in the cdKO telencephalon is essentially lost, consistent with increased repression by Gli3. It is likely that by this stage the effect of Nodal signaling is diminishing, even in the cdKO, and so the excess Gli3 predominates. In support of this, Gli3 heterozygosity restores some Fgf8 expression and restores expression of Foxg1, which is a downstream target of FGF signals in the anterior [65]. Analysis of Fgf8 expression in Shh null embryos reveals that expression is already lost by 9.0 dpc, while at this stage in the cdKO it is increased. However, as with the Gli3 heterozygous cdKO at 9.5 dpc, the loss of Fgf8 expression in Shh null embryos can be rescued by Gli3 heterozygosity [24], [61]. Thus the loss of Fgf8 expression in the anterior may contribute to the HPE phenotypes seen in both Shh null and cdKO embryos, and the difference in timing of the loss of expression may also be in part responsible for some of the differences between these two models. Given that loss of Fgf8 expression is common to the Shh null and cdKO HPE models, it is tempting to speculate that in the small proportion of Smad2/Nodal double heterozygous mutants with the HPE phenotype is in part due to a failure to fully activate Fgf8 expression.
Taken together, our data suggest a model in which Tgifs limit the activity of the Nodal-Smad2 pathway, which is required for full activation of Smad/Foxh1 targets, such as Fgf8 (Figure 9). In addition we provide evidence that regulation of Nodal signaling by Tgifs is required to maintain the appropriate balance between Shh and Gli3 levels in the forebrain. However, it should be noted that we do not yet know whether this occurs via direct regulation of Gli3 or Shh expression (dashed lines in Figure 9), or whether the regulation is less direct. An additional possibility is that at least some of the regulation of the Shh pathway by Tgifs is independent of Nodal/Smad2. For example, Gli3 might be a direct target of Tgif repression, although the rescue of Nkx2.1 expression in the Nodal heterozygotes is consistent with a Nodal dependent regulation of the Shh pathway. In summary, this work provides the first clear evidence for a role for loss of Tgif function in HPE pathogenesis, and suggests that Tgifs regulate Shh signaling pathway activity. We propose that Tgif function limits Gli3 expression, and that by a mechanism that is independent of changes in Gli3 levels, Tgifs are required for full Shh expression in the PrCP and neuroepithelium. Thus, the Tgifs have significant contributions to HPE pathogenesis by functioning as key regulators of Shh signaling during forebrain development, most likely by limiting Nodal signaling.
Materials and Methods
Ethics statement
All animal procedures were approved by the Animal Care and Use Committee of the University of Virginia, which is fully accredited by the AAALAC.
Mice and DNA analysis
The loxP flanked Tgif allele [45], Tgif2 null [48], loxP flanked Gli3 allele [62], Nodal mutants [76], loxP flanked Smad2 allele [77], and the Sox2-Cre line [49] have been described previously. Conditional Shh mice were obtained from Jackson labs (stock 4293; [78]). The Gli3, Shh and Smad2 alleles each contain loxP flanked exons, which when recombined result in null alleles, and are referred to here as ‘r’ for recombined (null). The Nodal null allele is referred to as ‘z’, for an introduced lacZ reporter. All mouse lines were maintained on a mixed C57BL/6J×129Sv/J background. Genomic DNA for PCR genotype analysis was purified from ear punch, at post-natal day 21 (P21), or yolk sac (7.0–10.0 dpc) by HotShot [79].
In situ hybridization
Whole-mount in situ hybridization was performed on 7.5–10.0 dpc embryos with digoxigenin-labeled riboprobes, as described [80]. Stained embryos were processed for sectioning and histology as described [58]. All images are representative of at least three embryos analyzed.
Histology, immunohistochemistry (IHC), and whole-mount analysis
Embryos were fixed overnight in 4% paraformaldehyde at 4°C, dehydrated through an ethanol series (70%, 90%, 95%, 100% ×2 for 30 minutes each), incubated in xylene twice for 60 minutes and 1∶1 xylene/paraffin for 60 minutes at 60°C, then embedded in paraffin wax, and sectioned at 7 µm. For Hematoxylin and Eosin (H&E) histological analysis, sections were de-paraffinized with xylene and stained with H&E. Multiple sections per embryo were incubated with primary antibodies for pHH3 or active caspase 3 as described [48]. For IHC, antibody staining was detected using Vectastain ABC (Vector Laboratories) and developed with Impact DAB (Vector Laboratories). For H&E and IHC images were captured using an Olympus BX51 microscope and either an Olympus SZX12 or DP70 digital camera, and manipulated in Adobe Photoshop. Images of 7.0–10.0 dpc embryos were captured using a Leica MZ16 stereomicroscope and QImaging 5.0 RTV digital camera.
Scanning electron microscopy
Embryos were fixed overnight in 4% paraformaldehyde at 4°C, and then fixed with osmium tetraoxide for 30 min and dehydrated through an ethanol series (40%, 60%, 80% and 100% ×2 for 15 minutes each). Dehydrated samples were further processed in an Autosamdri-815 (Tousimis Research Corporation) and were gold coated by using a SCD005 Sputter Coater (Bal-Tec). Images were captured using a JSM-6400 Scanning Electron Microscope (JEOL).
Supporting Information
Zdroje
1. GengXOliverG 2009 Pathogenesis of holoprosencephaly. J Clin Invest 119 1403 1413
2. MuenkeMBeachyPA 2001 Holoprosencephaly. C.R.SA.L.BW.S.SD.VB.C The metabolic and molecular bases of inherited disease: McGraw-Hill 6203 6230
3. LeonciniEBaranelloGOrioliIMAnnerenGBakkerM 2008 Frequency of holoprosencephaly in the International Clearinghouse Birth Defects Surveillance Systems: searching for population variations. Birth Defects Res A Clin Mol Teratol 82 585 591
4. MatsunagaEShiotaK 1977 Holoprosencephaly in human embryos: epidemiologic studies of 150 cases. Teratology 16 261 272
5. RoachEDemyerWConneallyPMPalmerCMerrittAD 1975 Holoprosencephaly: birth data, genetic and demographic analyses of 30 families. Birth Defects Orig Artic Ser 11 294 313
6. RubensteinJLBeachyPA 1998 Patterning of the embryonic forebrain. Curr Opin Neurobiol 8 18 26
7. GoldenJA 1998 Holoprosencephaly: a defect in brain patterning. J Neuropathol Exp Neurol 57 991 999
8. OlsenCLHughesJPYoungbloodLGSharpe-StimacM 1997 Epidemiology of holoprosencephaly and phenotypic characteristics of affected children: New York State, 1984–1989. Am J Med Genet 73 217 226
9. CroenLAShawGMLammerEJ 1996 Holoprosencephaly: epidemiologic and clinical characteristics of a California population. Am J Med Genet 64 465 472
10. RoesslerEMuenkeM 1998 Holoprosencephaly: a paradigm for the complex genetics of brain development. J Inherit Metab Dis 21 481 497
11. WallisDEMuenkeM 1999 Molecular Mechanisms of Holoprosencephaly. Mol Genet Metab 68 126 138
12. MuenkeMBeachyPA 2000 Genetics of ventral forebrain development and holoprosencephaly. Curr Opin Genet Dev 10 262 269
13. RoesslerEBelloniEGaudenzKJayPBertaP 1996 Mutations in the human Sonic Hedgehog gene cause holoprosencephaly. Nat Genet 14 357 360
14. RoesslerEBelloniEGaudenzKVargasFSchererSW 1997 Mutations in the C-terminal domain of Sonic Hedgehog cause holoprosencephaly. Hum Mol Genet 6 1847 1853
15. NanniLMingJEBocianMSteinhausKBianchiDW 1999 The mutational spectrum of the sonic hedgehog gene in holoprosencephaly: SHH mutations cause a significant proportion of autosomal dominant holoprosencephaly. Hum Mol Genet 8 2479 2488
16. RoesslerEWardDEGaudenzKBelloniESchererSW 1997 Cytogenetic rearrangements involving the loss of the Sonic Hedgehog gene at 7q36 cause holoprosencephaly. Hum Genet 100 172 181
17. GurrieriFTraskBJvan den EnghGKraussCMSchinzelA 1993 Physical mapping of the holoprosencephaly critical region on chromosome 7q36. Nat Genet 3 247 251
18. WallisDERoesslerEHehrUNanniLWiltshireT 1999 Mutations in the homeodomain of the human SIX3 gene cause holoprosencephaly. Nat Genet 22 196 198
19. BrownSAWarburtonDBrownLYYuCYRoederER 1998 Holoprosencephaly due to mutations in ZIC2, a homologue of Drosophila odd-paired. Nat Genet 20 180 183
20. GrippKWWottonDEdwardsMCRoesslerEAdesL 2000 Mutations in TGIF cause holoprosencephaly and link NODAL signalling to human neural axis determination. Nat Genet 25 205 208
21. GengXSpeirsCLagutinOInbalALiuW 2008 Haploinsufficiency of Six3 fails to activate Sonic hedgehog expression in the ventral forebrain and causes holoprosencephaly. Dev Cell 15 236 247
22. JeongYLeskowFCEl-JaickKRoesslerEMuenkeM 2008 Regulation of a remote Shh forebrain enhancer by the Six3 homeoprotein. Nat Genet 40 1348 1353
23. ShimamuraKRubensteinJL 1997 Inductive interactions direct early regionalization of the mouse forebrain. Development 124 2709 2718
24. AotoKNishimuraTEtoKMotoyamaJ 2002 Mouse GLI3 regulates Fgf8 expression and apoptosis in the developing neural tube, face, and limb bud. Dev Biol 251 320 332
25. FuccilloMJoynerALFishellG 2006 Morphogen to mitogen: the multiple roles of hedgehog signalling in vertebrate neural development. Nat Rev Neurosci 7 772 783
26. ToleSRagsdaleCWGroveEA 2000 Dorsoventral patterning of the telencephalon is disrupted in the mouse mutant extra-toes(J). Dev Biol 217 254 265
27. RalluMMacholdRGaianoNCorbinJGMcMahonAP 2002 Dorsoventral patterning is established in the telencephalon of mutants lacking both Gli3 and Hedgehog signaling. Development 129 4963 4974
28. HebertJMFishellG 2008 The genetics of early telencephalon patterning: some assembly required. Nat Rev Neurosci 9 678 685
29. GutinGFernandesMPalazzoloLPaekHYuK 2006 FGF signalling generates ventral telencephalic cells independently of SHH. Development 133 2937 2946
30. BertolinoEReimundBWildt-PerinicDClercR 1995 A novel homeobox protein which recognizes a TGT core and functionally interferes with a retinoid-responsive motif. J Biol Chem 270 31178 31188
31. WottonDLoRSLeeSMassagueJ 1999 A Smad transcriptional corepressor. Cell 97 29 39
32. HeldinC-HMiyazonoKten DijkeP 1997 TGF-ß signalling from cell membrane to nucleus through SMAD proteins. Nature 390 465 471
33. MassagueJSeoaneJWottonD 2005 Smad transcription factors. Genes Dev 19 2783 2810
34. WottonDMassagueJ 2001 Smad transcriptional corepressors in TGF beta family signaling. Curr Top Microbiol Immunol 254 145 164
35. ImotoIPimkhaokhamAWatanabeTSaito-OharaFSoedaE 2000 Amplification and overexpression of TGIF2, a novel homeobox gene of the TALE superclass, in ovarian cancer cell lines. Biochem Biophys Res Commun 276 264 270
36. MelhuishTAGalloCMWottonD 2001 TGIF2 interacts with histone deacetylase 1 and represses transcription. J Biol Chem 276 32109 32114
37. MelhuishTAWottonD 2006 The Tgif2 gene contains a retained intron within the coding sequence. BMC Mol Biol 7 2
38. BartholinLPowersSEMelhuishTALasseSWeinsteinM 2006 TGIF inhibits retinoid signaling. Mol Cell Biol 26 990 1001
39. MelhuishTAChungDDBjerkeGAWottonD 2010 Tgif1 represses apolipoprotein gene expression in liver. J Cell Biochem 111 380 390
40. AguilellaCDubourgCAttia-SobolJVigneronJBlayauM 2003 Molecular screening of the TGIF gene in holoprosencephaly: identification of two novel mutations. Hum Genet 112 131 134
41. ChenCPChernSRDuSHWangW 2002 Molecular diagnosis of a novel heterozygous 268C→T (R90C) mutation in TGIF gene in a fetus with holoprosencephaly and premaxillary agenesis. Prenat Diagn 22 5 7
42. El-JaickKBPowersSEBartholinLMyersKRHahnJ 2007 Functional analysis of mutations in TGIF associated with holoprosencephaly. Mol Genet Metab 90 97 111
43. JinJZGuSMcKinneyPDingJ 2006 Expression and functional analysis of Tgif during mouse midline development. Dev Dyn 235 547 553
44. MarLHoodlessPA 2006 Embryonic fibroblasts from mice lacking Tgif were defective in cell cycling. Mol Cell Biol 26 4302 4310
45. ShenJWalshCA 2005 Targeted disruption of Tgif, the mouse ortholog of a human holoprosencephaly gene, does not result in holoprosencephaly in mice. Mol Cell Biol 25 3639 3647
46. BartholinLMelhuishTAPowersSEGoddard-LeonSTreilleuxI 2008 Maternal Tgif is required for vascularization of the embryonic placenta. Dev Biol 319 285 297
47. KuangCXiaoYYangLChenQWangZ 2006 Intragenic deletion of Tgif causes defectsin brain development. Hum Mol Genet 15 3508 3519
48. PowersSETaniguchiKYenWMelhuishTAShenJ 2010 Tgif1 and Tgif2 regulate Nodal signaling and are required for gastrulation. Development 137 249 259
49. HayashiSLewisPPevnyLMcMahonAP 2002 Efficient gene modulation in mouse epiblast using a Sox2Cre transgenic mouse strain. Mech Dev 119 Suppl 1 S97 S101
50. ChiangCLitingtungYLeeEYoungKECordenJL 1996 Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature 383 407 413
51. MaYErknerAGongRYaoSTaipaleJ 2002 Hedgehog-mediated patterning of the mammalian embryo requires transporter-like function of dispatched. Cell 111 63 75
52. MartynogaBMorrisonHPriceDJMasonJO 2005 Foxg1 is required for specification of ventral telencephalon and region-specific regulation of dorsal telencephalic precursor proliferation and apoptosis. Dev Biol 283 113 127
53. SilvestriCNarimatsuMvon BothILiuYTanNB 2008 Genome-wide identification of Smad/Foxh1 targets reveals a role for Foxh1 in retinoic acid regulation and forebrain development. Dev Cell 14 411 423
54. ThomasPBeddingtonR 1996 Anterior primitive endoderm may be responsible for patterning the anterior neural plate in the mouse embryo. Curr Biol 6 1487 1496
55. Fukuchi-ShimogoriTGroveEA 2003 Emx2 patterns the neocortex by regulating FGF positional signaling. Nat Neurosci 6 825 831
56. LewisSLTamPP 2006 Definitive endoderm of the mouse embryo: formation, cell fates, and morphogenetic function. Dev Dyn 235 2315 2329
57. Martinez BarberaJPClementsMThomasPRodriguezTMeloyD 2000 The homeobox gene Hex is required in definitive endodermal tissues for normal forebrain, liver and thyroid formation. Development 127 2433 2445
58. SasakiHHoganBL 1993 Differential expression of multiple fork head related genes during gastrulation and axial pattern formation in the mouse embryo. Development 118 47 59
59. LewisSLKhooPLAndrea De YoungRBildsoeHWakamiyaM 2007 Genetic interaction of Gsc and Dkk1 in head morphogenesis of the mouse. Mech Dev 124 157 165
60. InghamPWMcMahonAP 2001 Hedgehog signaling in animal development: paradigms and principles. Genes Dev 15 3059 3087
61. AotoKShikataYImaiHMatsumaruDTokunagaT 2009 Mouse Shh is required for prechordal plate maintenance during brain and craniofacial morphogenesis. Dev Biol 327 106 120
62. BlaessSStephenDJoynerAL 2008 Gli3 coordinates three-dimensional patterning and growth of the tectum and cerebellum by integrating Shh and Fgf8 signaling. Development 135 2093 2103
63. NomuraMLiE 1998 Smad2 role in mesoderm formation, left-right patterning and craniofacial development. Nature 393 786 790
64. OhkuboYChiangCRubensteinJL 2002 Coordinate regulation and synergistic actions of BMP4, SHH and FGF8 in the rostral prosencephalon regulate morphogenesis of the telencephalic and optic vesicles. Neuroscience 111 1 17
65. StormEEGarelSBorelloUHebertJMMartinezS 2006 Dose-dependent functions of Fgf8 in regulating telencephalic patterning centers. Development 133 1831 1844
66. SolomonBDMercierSVelezJIPineda-AlvarezDEWyllieA 2010 Analysis of genotype-phenotype correlations in human holoprosencephaly. Am J Med Genet C Semin Med Genet 154C 133 141
67. SanekNATaylorAANyholmMKGrinblatY 2009 Zebrafish zic2a patterns the forebrain through modulation of Hedgehog-activated gene expression. Development 136 3791 3800
68. WarrNPowles-GloverNChappellARobsonJNorrisD 2008 Zic2-associated holoprosencephaly is caused by a transient defect in the organizer region during gastrulation. Hum Mol Genet 17 2986 2996
69. HymanCABartholinLNewfeldSJWottonD 2003 Drosophila TGIF proteins are transcriptional activators. Mol Cell Biol 23 9262 9274
70. LammerEJChenDTHoarRMAgnishNDBenkePJ 1985 Retinoic acid embryopathy. N Engl J Med 313 837 841
71. SulikKKDehartDBRogersJMChernoffN 1995 Teratogenicity of low doses of all-trans retinoic acid in presomite mouse embryos. Teratology 51 398 403
72. De La CruzJMBamfordRNBurdineRDRoesslerEBarkovichAJ 2002 A loss-of-function mutation in the CFC domain of TDGF1 is associated with human forebrain defects. Hum Genet 110 422 428
73. RoesslerEOuspenskaiaMVKarkeraJDVelezJIKantipongA 2008 Reduced NODAL signaling strength via mutation of several pathway members including FOXH1 is linked to human heart defects and holoprosencephaly. Am J Hum Genet 83 18 29
74. DingJYangLYanYTChenADesaiN 1998 Cripto is required for correct orientation of the anterior-posterior axis in the mouse embryo. Nature 395 702 707
75. HoodlessPAPyeMChazaudCLabbeEAttisanoL 2001 FoxH1 (Fast) functions to specify the anterior primitive streak in the mouse. Genes Dev 15 1257 1271
76. CollignonJVarletIRobertsonEJ 1996 Relationship between asymmetric nodal expression and the direction of embryonic turning. Nature 381 155 158
77. JuWOgawaAHeyerJNierhofDYuL 2006 Deletion of Smad2 in mouse liver reveals novel functions in hepatocyte growth and differentiation. Mol Cell Biol 26 654 667
78. LewisPMDunnMPMcMahonJALoganMMartinJF 2001 Cholesterol modification of sonic hedgehog is required for long-range signaling activity and effective modulation of signaling by Ptc1. Cell 105 599 612
79. TruettGEHeegerPMynattRLTruettAAWalkerJA 2000 Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT). Biotechniques 29 52 54
80. WilkinsonDG 1992 In situ hybridization: a practical approach. 75 83
Štítky
Genetika Reprodukční medicína
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 2
-
Všechny články tohoto čísla
- Upsetting the Dogma: Germline Selection in Human Males
- A Strong Deletion Bias in Nonallelic Gene Conversion
- Positive Selection for New Disease Mutations in the Human Germline: Evidence from the Heritable Cancer Syndrome Multiple Endocrine Neoplasia Type 2B
- Genome-Wide Association Study in East Asians Identifies Novel Susceptibility Loci for Breast Cancer
- Mixed Effects Modeling of Proliferation Rates in Cell-Based Models: Consequence for Pharmacogenomics and Cancer
- Reduction of NADPH-Oxidase Activity Ameliorates the Cardiovascular Phenotype in a Mouse Model of Williams-Beuren Syndrome
- Genome-Wide Association Study Identifies Chromosome 10q24.32 Variants Associated with Arsenic Metabolism and Toxicity Phenotypes in Bangladesh
- Structural Basis of Transcriptional Gene Silencing Mediated by MOM1
- Genomic Restructuring in the Tasmanian Devil Facial Tumour: Chromosome Painting and Gene Mapping Provide Clues to Evolution of a Transmissible Tumour
- Genome-Wide Association Study Identifies Novel Loci Associated with Circulating Phospho- and Sphingolipid Concentrations
- Contrasting Properties of Gene-Specific Regulatory, Coding, and Copy Number Mutations in : Frequency, Effects, and Dominance
- The Origin and Nature of Tightly Clustered Deletions in Precursor B-Cell Acute Lymphoblastic Leukemia Support a Model of Multiclonal Evolution
- Ultrafast Evolution and Loss of CRISPRs Following a Host Shift in a Novel Wildlife Pathogen,
- Phosphorylation of Chromosome Core Components May Serve as Axis Marks for the Status of Chromosomal Events during Mammalian Meiosis
- Psoriasis Patients Are Enriched for Genetic Variants That Protect against HIV-1 Disease
- A Pathogenic Mechanism in Huntington's Disease Involves Small CAG-Repeated RNAs with Neurotoxic Activity
- The Mitochondrial Chaperone Protein TRAP1 Mitigates α-Synuclein Toxicity
- Homeobox Genes Critically Regulate Embryo Implantation by Controlling Paracrine Signaling between Uterine Stroma and Epithelium
- Developmental Transcriptional Networks Are Required to Maintain Neuronal Subtype Identity in the Mature Nervous System
- Down-Regulating Sphingolipid Synthesis Increases Yeast Lifespan
- Gene Expression and Stress Response Mediated by the Epigenetic Regulation of a Transposable Element Small RNA
- Loss of Tgif Function Causes Holoprosencephaly by Disrupting the Shh Signaling Pathway
- Sequestration of Highly Expressed mRNAs in Cytoplasmic Granules, P-Bodies, and Stress Granules Enhances Cell Viability
- Discovery of a Modified Tetrapolar Sexual Cycle in and the Evolution of in the Species Complex
- The Role of Glypicans in Wnt Inhibitory Factor-1 Activity and the Structural Basis of Wif1's Effects on Wnt and Hedgehog Signaling
- Nondisjunction of a Single Chromosome Leads to Breakage and Activation of DNA Damage Checkpoint in G2
- A Regulatory Network for Coordinated Flower Maturation
- Coexpression Network Analysis in Abdominal and Gluteal Adipose Tissue Reveals Regulatory Genetic Loci for Metabolic Syndrome and Related Phenotypes
- Diced Triplets Expose Neurons to RISC
- The Williams-Beuren Syndrome—A Window into Genetic Variants Leading to the Development of Cardiovascular Disease
- The Empirical Power of Rare Variant Association Methods: Results from Sanger Sequencing in 1,998 Individuals
- Systematic Detection of Epistatic Interactions Based on Allele Pair Frequencies
- Familial Identification: Population Structure and Relationship Distinguishability
- Raf1 Is a DCAF for the Rik1 DDB1-Like Protein and Has Separable Roles in siRNA Generation and Chromatin Modification
- Loss of Function of the Cik1/Kar3 Motor Complex Results in Chromosomes with Syntelic Attachment That Are Sensed by the Tension Checkpoint
- Computational Prediction and Molecular Characterization of an Oomycete Effector and the Cognate Resistance Gene
- The Dynamics and Prognostic Potential of DNA Methylation Changes at Stem Cell Gene Loci in Women's Cancer
- GTPase Activity and Neuronal Toxicity of Parkinson's Disease–Associated LRRK2 Is Regulated by ArfGAP1
- Evaluation of the Role of Functional Constraints on the Integrity of an Ultraconserved Region in the Genus
- Neurophysiological Defects and Neuronal Gene Deregulation in Mutants
- Genetic and Functional Analyses of Mutations Suggest a Multiple Hit Model of Autism Spectrum Disorders
- Negative Supercoiling Creates Single-Stranded Patches of DNA That Are Substrates for AID–Mediated Mutagenesis
- Rewiring of PDZ Domain-Ligand Interaction Network Contributed to Eukaryotic Evolution
- The Eph Receptor Activates NCK and N-WASP, and Inhibits Ena/VASP to Regulate Growth Cone Dynamics during Axon Guidance
- Repression of a Potassium Channel by Nuclear Hormone Receptor and TGF-β Signaling Modulates Insulin Signaling in
- The Retrohoming of Linear Group II Intron RNAs in Occurs by Both DNA Ligase 4–Dependent and –Independent Mechanisms
- Cell Lineage Analysis of the Mammalian Female Germline
- Association of a Functional Variant in the Wnt Co-Receptor with Early Onset Ileal Crohn's Disease
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Gene Expression and Stress Response Mediated by the Epigenetic Regulation of a Transposable Element Small RNA
- Contrasting Properties of Gene-Specific Regulatory, Coding, and Copy Number Mutations in : Frequency, Effects, and Dominance
- Homeobox Genes Critically Regulate Embryo Implantation by Controlling Paracrine Signaling between Uterine Stroma and Epithelium
- Nondisjunction of a Single Chromosome Leads to Breakage and Activation of DNA Damage Checkpoint in G2
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



