-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaA Regulatory Network for Coordinated Flower Maturation
For self-pollinating plants to reproduce, male and female organ development must be coordinated as flowers mature. The Arabidopsis transcription factors AUXIN RESPONSE FACTOR 6 (ARF6) and ARF8 regulate this complex process by promoting petal expansion, stamen filament elongation, anther dehiscence, and gynoecium maturation, thereby ensuring that pollen released from the anthers is deposited on the stigma of a receptive gynoecium. ARF6 and ARF8 induce jasmonate production, which in turn triggers expression of MYB21 and MYB24, encoding R2R3 MYB transcription factors that promote petal and stamen growth. To understand the dynamics of this flower maturation regulatory network, we have characterized morphological, chemical, and global gene expression phenotypes of arf, myb, and jasmonate pathway mutant flowers. We found that MYB21 and MYB24 promoted not only petal and stamen development but also gynoecium growth. As well as regulating reproductive competence, both the ARF and MYB factors promoted nectary development or function and volatile sesquiterpene production, which may attract insect pollinators and/or repel pathogens. Mutants lacking jasmonate synthesis or response had decreased MYB21 expression and stamen and petal growth at the stage when flowers normally open, but had increased MYB21 expression in petals of older flowers, resulting in renewed and persistent petal expansion at later stages. Both auxin response and jasmonate synthesis promoted positive feedbacks that may ensure rapid petal and stamen growth as flowers open. MYB21 also fed back negatively on expression of jasmonate biosynthesis pathway genes to decrease flower jasmonate level, which correlated with termination of growth after flowers have opened. These dynamic feedbacks may promote timely, coordinated, and transient growth of flower organs.
Published in the journal: . PLoS Genet 8(2): e32767. doi:10.1371/journal.pgen.1002506
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002506Summary
For self-pollinating plants to reproduce, male and female organ development must be coordinated as flowers mature. The Arabidopsis transcription factors AUXIN RESPONSE FACTOR 6 (ARF6) and ARF8 regulate this complex process by promoting petal expansion, stamen filament elongation, anther dehiscence, and gynoecium maturation, thereby ensuring that pollen released from the anthers is deposited on the stigma of a receptive gynoecium. ARF6 and ARF8 induce jasmonate production, which in turn triggers expression of MYB21 and MYB24, encoding R2R3 MYB transcription factors that promote petal and stamen growth. To understand the dynamics of this flower maturation regulatory network, we have characterized morphological, chemical, and global gene expression phenotypes of arf, myb, and jasmonate pathway mutant flowers. We found that MYB21 and MYB24 promoted not only petal and stamen development but also gynoecium growth. As well as regulating reproductive competence, both the ARF and MYB factors promoted nectary development or function and volatile sesquiterpene production, which may attract insect pollinators and/or repel pathogens. Mutants lacking jasmonate synthesis or response had decreased MYB21 expression and stamen and petal growth at the stage when flowers normally open, but had increased MYB21 expression in petals of older flowers, resulting in renewed and persistent petal expansion at later stages. Both auxin response and jasmonate synthesis promoted positive feedbacks that may ensure rapid petal and stamen growth as flowers open. MYB21 also fed back negatively on expression of jasmonate biosynthesis pathway genes to decrease flower jasmonate level, which correlated with termination of growth after flowers have opened. These dynamic feedbacks may promote timely, coordinated, and transient growth of flower organs.
Introduction
In typical angiosperms, late in flower development, sepals open to expose the inner organs; the petals, stamen filaments, and style elongate; the anthers dehisce to release pollen; and the stigma and transmitting tract mature so as to permit pollen germination and pollen tube growth. These events often occur quite quickly, and are transient, so that flowers open and pollinate, but then stop growing. Effective reproduction therefore requires accurate coordination of these events. Variation in spatial arrangement and timing of maturation of different organs may affect the pollination mode and the mating system. In plants with self-pollinating flowers such as Arabidopsis thaliana, stamens and gynoecium grow to about the same length and mature synchronously, allowing efficient self-fertilization [1]. In outcrossing plants, differential growth of stamens and style or staggered timing of anther and gynoecium maturation can instead promote cross-pollination.
The Arabidopsis transcription factors AUXIN RESPONSE FACTOR 6 (ARF6/At1g30330) and ARF8/At5g37020 act partially redundantly to promote late stages in petal, stamen and gynoecium development. arf6 arf8 double null mutant flowers arrest at flower stage 12 as closed buds with short petals, short stamen filaments, undehisced anthers, and immature gynoecia with short stigmatic papillae and poor support of pollen tube growth, and are largely male - and female-sterile [2]–[4]. arf6 and arf8 single mutants and sesquimutants (homozygous for one mutation and heterozygous for the other) have delayed stamen filament elongation and decreased fecundity. ARF6 and ARF8 are each expressed in multiple flower tissues including sepals, petals, stamen filaments, style, transmitting tract, ovule funiculi, and nectaries [2], [4]. ARF6 and ARF8 thus act in several organs to promote the transition from closed buds to mature fertile flowers, and to ensure coordinated development of male and female organs, leading to efficient self-fertilization.
arf6-2 arf8-3 flowers have very low jasmonic acid (JA) levels and decreased expression of several jasmonate biosynthesis genes, and exogenous methyl jasmonate (MeJA) rescued the petal elongation and anther dehiscence defects, but not the stamen elongation defect or gynoecium arrest, of arf6 arf8 flowers [2], [3]. Mutants affected in jasmonate synthesis or signaling similarly have delayed stamen growth and indehiscent anthers [5]–[8]. Similarly to stamens, petals of jasmonate pathway mutants have been reported to have delayed growth [6]. However, in contrast, other groups have reported that petals of jasmonate pathway mutants are larger than those of wild-type flowers [5], [8], [9]. Jasmonates can inhibit petal expansion by activating alternative splicing of a bHLH31/BPE/At1g59640 transcript [9], [10]. arf8 mutants also had enlarged petals, suggesting that ARF8 and BPE act in a common pathway [11]. These results indicate that ARF6 and ARF8 trigger anther dehiscence by promoting jasmonate production, can promote or inhibit petal growth through jasmonate-dependent pathways, and regulate other aspects of flower maturation independently of jasmonate.
The role of jasmonate in stamen development was investigated in more detail by examining MeJA-induced global gene expression changes in the stamens of jasmonate-deficient opr3 mutant plants [8], [12]. Two closely related R2R3 MYB transcription factor genes, MYB21/At3g27810 and MYB24/At5g40350 [13], were rapidly induced by jasmonate. myb21 mutants had short stamen filaments and petals, and myb21 myb24 double mutants had indehiscent anthers. These phenotypes were not rescued by exogenous JA or MeJA application, indicating that MYB21 and MYB24 act downstream of jasmonate signaling to promote stamen and petal development [12], [14]. Gibberellin-deficient mutants also have delayed stamen development, decreased JA level, and decreased expression of MYB21, MYB24, and a third closely related gene, MYB57/At3g01530 [14]. A fourth closely related gene, MYB108/At3g06490, also contributes to stamen development partially redundantly with MYB24 [15]. MYB57 and MYB108 are also induced by jasmonate. MYB108 has also been isolated as BOTRYTIS OVERSENSITIVE 1 (BOS1), and is required for JA-mediated biotic and abiotic stress responses [16]. MYB21 and MYB24 can activate transcription, and overexpression of MYB21 or MYB24 caused defects in flower development [17]–[20]. Other genes encoding members of this clade, MYB2, MYB62, MYB78, MYB112 and MYB116, were not appreciably expressed in flowers [21].
To understand how these components interact to regulate flower maturation, we have analyzed the relative timing of flower organ growth in arf, myb, and jasmonate pathway mutants, and compared expression of MYB and jasmonate pathway genes in wild-type and mutant flowers. These analyses suggest a hierarchical regulatory pathway that triggers flower maturation, and also reveal contrasting effects of jasmonate on petal growth at different developmental stages. Analyses of global gene expression patterns in wild-type, myb21 myb24, and arf6 arf8 flowers reveal that the flower maturation network controls putative chemical attractant functions of flowers, and that both positive and negative feedback loops control auxin and jasmonate responses during flower maturation.
Results
MYB Genes Are Expressed Downstream of ARF6 and ARF8 in Multiple Flower Organs
Before characterizing mutant phenotypes, we examined expression of MYB genes in wild-type, arf6-2 arf8-3, and jasmonate pathway mutant flowers. In wild-type flowers, MYB21 and MYB24 were first expressed at stages 11–12 shortly before flower opening, whereas in arf6-2 arf8-3 flowers MYB21 and MYB24 mRNAs were almost undetectable (Figure 1A; Figure S1A, S1C) [2]. Conversely, ARF6 and ARF8 mRNA levels were normal in myb21-5 myb24-5 flowers (Figure 1C, Table S3). MYB21 and MYB24 were also underexpressed in jasmonate-deficient aos-2 mutant inflorescence apices and in jasmonate-resistant coi1-1 apices (Figure 1B). Both MYB21 and MYB24 baseline expression levels were lower in arf6-2 arf8-3 inflorescences than in aos-2 or coi1-1 inflorescences (Figure 1B). Exogenous methyl jasmonate induced MYB21 and MYB24 genes in arf6-2 arf8-3 and aos-2 mutant inflorescence apices, but not in coi1-1 apices (Figure 1B) [12], [14], [15]. P35S:ARF6 plants that overexpress ARF6 did not have increased MYB21 mRNA level (Figure S1A); and PARF6:mARF6 plants expressing an ARF6 transgene that is immune to regulation by miR167, and which have an expanded ARF6 expression domain in the ovules [4], did not have a similarly expanded MYB21 expression domain (Figure 2F, 2G). These results suggest that ARF6 and ARF8 induce these MYB genes indirectly, at least partly by increasing jasmonate levels. Exogenous MeJA only partially restored MYB21 and MYB24 expression and stamen and petal growth in arf6-2 arf8-3 flowers (Figure 1B) [2], raising the possibility that ARF6 and ARF8 may also regulate MYB21 and MYB24 by additional jasmonate-independent mechanisms.
Fig. 1. Gene expression and jasmonate production in wild-type and mutant flowers. 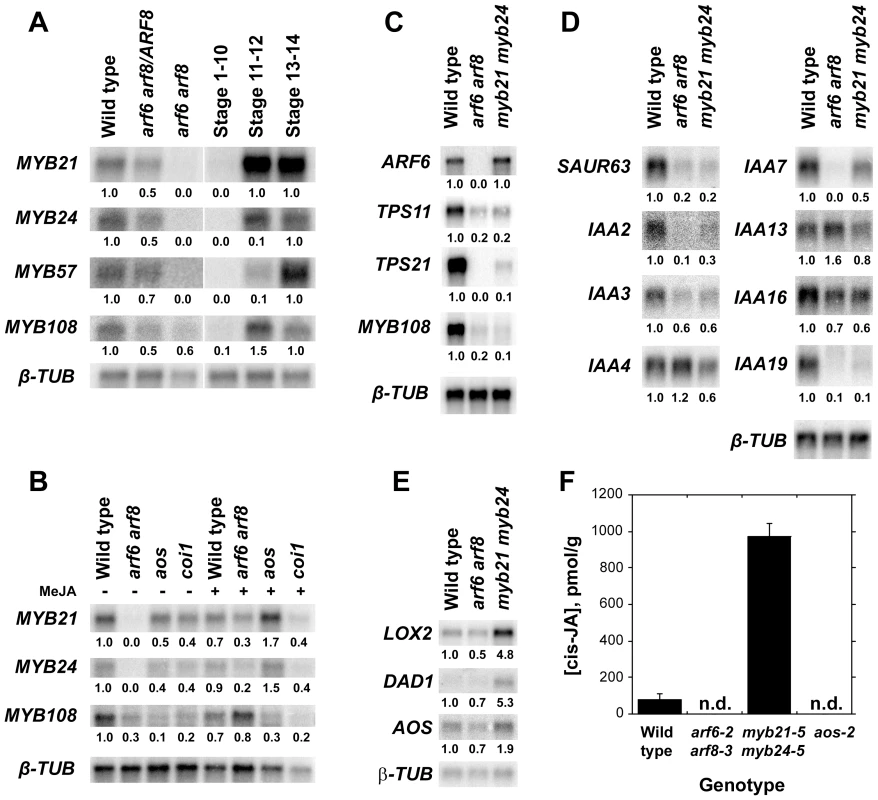
(A–B) RNA gel blot hybridization using MYB21, MYB24, MYB57 and MYB108 probes. (A) RNA from wild-type, arf6-2 arf8-3/ARF8 and arf6-2 arf8-3 inflorescences (left panel), and wild-type stage 1–10, stage 11–12 and stage 13–14 flowers (right panel). (B) RNA from untreated (left panel) or MeJA treated (right panel) wild-type, arf6-2 arf8-3, aos-2 and coi1-1 inflorescences. (C) RNA gel blot hybridization using ARF6, TPS11, TPS21, MYB108 and SAUR63 probes. RNA from wild-type, arf6-2 arf8-3 and myb21-5 myb24-5 inflorescences. (D) RNA gel blot hybridization using SAUR63, IAA2, IAA3, IAA4, IAA7, IAA13, IAA16 and IAA19 probes. RNA from wild-type, arf6-2 arf8-3 and myb21-5 myb24-5 stage 12-13 flowers. (E) RNA gel blot hybridization using LOX2, DAD1, and AOS probes. PolyA+ RNA from wild-type, arf6-2 arf8-3 and myb21-5 myb24-5 stage 12–13 flowers. In A–E, numbers beneath each band indicate measured signal level relative to the β-TUBULIN control. (F) cis-JA concentrations in wild-type, arf6-2 arf8-3, myb21-5 myb24-5, and aos-2 stage 12-13 flowers. Data are means of two measurements ± SD. n.d., not detected. Fig. 2. Expression patterns of MYB21 and MYB24. 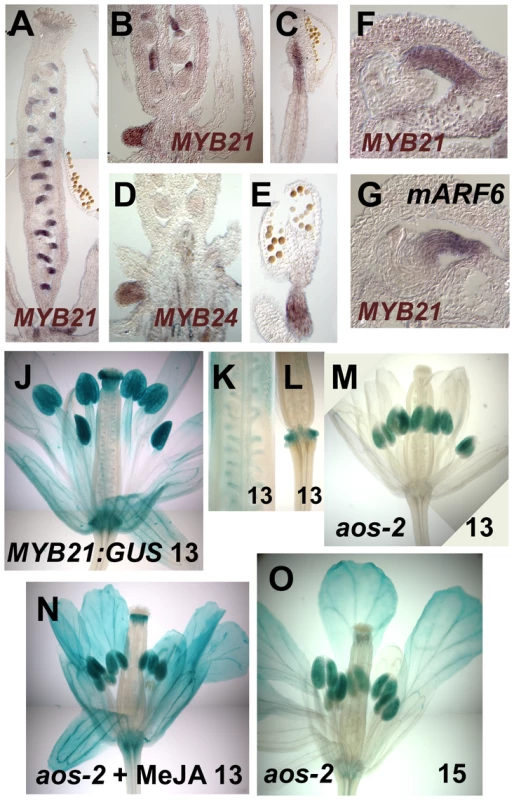
(A–C) In situ hybridization with a MYB21 antisense probe in stage 12 wild-type gynoecia (A,B) or stamen filament (C). (D,E) In situ hybridization with a MYB24 antisense probe in stage 12 wild-type nectary (D) and stament filament (E). (F) MYB21 in situ hybridization in a wild-type ovule. (G) MYB21 in situ hybridization in a mARF6 ovule. (J–O) X-Gluc staining of PMYB21:MYB21:GUS flowers. (J) Stage 13 wild-type whole flower. (K) Gynoecium showing ovule funiculi. (L) Gynoecium base showing nectary. (M–O) aos-2 PMYB21:MYB21:GUS flowers at stage 13 (M), (N) MeJA-treated stage 13, (O) Stage 15 untreated. By in situ hybridization and using transgenic plants carrying a PMYB21:MYB21:GUS protein fusion reporter, we detected MYB21 expression in sepals, petals, the apical part of stamen filaments, the style, ovule funiculi, and nectaries of stage 13 and 14 flowers (Figure 2A–2C, 2F, 2J–2L; Figure S1D). In the aos-2 jasmonate-deficient background, PMYB21:MYB21:GUS expression was decreased in these organs, but was restored by exogenous methyl jasmonate (Figure 2M, 2N). MYB24 and PMYB24:MYB24:GUS were likewise expressed in stamen filaments, style, and nectaries of stage 13 and 14 flowers, but not in ovule funiculi (Figure 2D, 2E; Figure S1E). Available microarray expression data are consistent with expression of both MYB21 and MYB24 in sepals, petals, stamens and carpels [21]. Expression of PMYB21:MYB21:GUS and PMYB24:MYB24:GUS in anthers or pollen (Figure 2, Figure S1) is likely an artifact of our fusion constructs, because in situ hybridizations revealed stamen filament but not anther expression (Figure 2C, 2E) [14]; microarray data from dry or germinated pollen revealed no expression of MYB21 or MYB24 [22]; and X-Gluc staining was present in anthers of arf6-2 arf8-3 PMYB21:MYB21:GUS plants although arf6-2 arf8-3 flowers lacked detectable MYB21 transcript (Figure 1A, 1B; Figure S1A–S1C).
MYB21 and MYB24 Promote Petal and Stamen Development
To determine timing of stamen and petal growth in flowers of different genotypes, we measured organ lengths of flowers along a developmental series from closed buds to open flowers (Figure 3) [23]. Wild-type gynoecia elongated at a fairly constant rate through these stages, so that gynoecium length provided an internal reference for developmental stage. In addition, in independent experiments we measured organ lengths of flowers at defined positions on the inflorescence relative to the position of the first open flower in wild-type plants (Table S1). In wild-type Arabidopsis flowers, sepals stopped growing at stage 12, shortly before flowers opened [1], [23]. Petals and stamens grew slowly through early stages, but grew much more rapidly at stage 12 and stage 13, when the flowers opened (Figure 3A, 3I, 3J). Wild-type flowers generally self-pollinated as they opened. Just after this stage, petals and stamens stopped elongating, and about two days thereafter they began to senesce [1], [24].
Fig. 3. Inflorescence apices and flower phenotypes of myb21, myb24, myb108, and aos mutants. 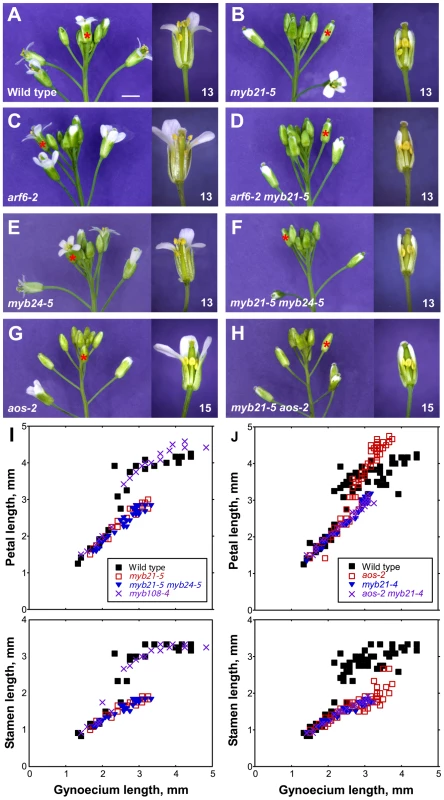
(A–H) Photographs of inflorescences (left panels) and individual flowers (right panels) of indicated genotypes. Asterisks indicate the position of the first open flower (stage 13) in the inflorescences shown, or the corresponding flower based upon bud size and position compared to a wild-type inflorescence. Individual flowers shown in the right panels are the first open flower (stage 13, A–F) or the fourth open flower (stage 15, G–H). Some sepals and petals have been removed to show inner organs. Scale bar: left panels, 3 mm, right panels, 1 mm. (I, J) Scatter plots showing petal and stamen lengths relative to gynoecium length of individual flowers of indicated genotypes. In I, data from a single experiment are shown. In J, measurements from two experiments were combined. Figure S2 shows similar data for additional genotypes. arf6-2 and arf8-3 single mutants had delayed petal and stamen growth compared to wild type, but at a slightly later stage arf6-2 and arf8-3 mutant petals and stamens did reach wild-type lengths relative to gynoecium length (Figure 3C, Figure S2A). Although arf8-3 mutants have been reported to have longer and wider petals than wild type [11], under our growth conditions petals of arf6-2 and arf8-3 flowers appeared wider but were not longer than wild-type petals. arf6-2 arf8-3 double mutant flowers arrested with short stamens, petals, and gynoecia (Figure S2A) [2], [3].
We recovered the presumed null mutations myb21-4 and myb21-5, each of which has a stop codon in the MYB21 coding sequence, in a screen for arf6-2 enhancers (Figure S3); and we used available T-DNA insertion alleles in MYB24 (Materials and Methods, Figure S4, Table S2). arf6-2 myb21-4 and arf6-2 myb21-5 plants had flower buds with small unreflexed petals and short stamens, and set seed only when manually pollinated (Figure 3D, Table S1). The myb21-5 mutation also enhanced arf8-3 phenotypes, but did not affect organ lengths of the more severely affected arf6-2 arf8-3 flowers (Table S1), indicating that MYB21 can be placed in the same genetic pathway as ARF6 and ARF8.
Similarly to other myb21 mutants [12], [14], [20], myb21-4 and myb21-5 single mutant flowers had short petals, short stamens with reduced epidermal cell length, and delayed flower opening and anther dehiscence (Figure 3B, 3I, Figure S2C, Figure S5A–S5C, Table S1). The myb21-4 and myb21-5 mutants had stronger phenotypes than the myb21-2 T-DNA insertion allele, which is in an intron and makes some full-length transcript (Table S1, Figure S4) [12]. Flowers of myb24-2 and myb24-5 single mutant plants appeared normal (Table S1, Figure 3E). Flowers of myb21-5 myb24-5 double mutants grew similarly to myb21-5 flowers up to stage 13 (Figure 3B, 3F, 3I; Table S1). However, whereas myb21-5 flowers sometimes opened, myb21-5 myb24-5 flower buds remained closed (Figure 3B, 3F). Moreover, myb21-5 myb24-5 anthers failed to release pollen until after the flowers started to senesce, and treatment with exogenous MeJA failed to accelerate pollen release (Table S1).
ARF6, ARF8, and MYB21 Promote Gynoecium Growth
As well as acting in petals and stamens, MYB21 and MYB24 are expressed in the gynoecium, suggesting that they may regulate aspects of gynoecium development or function. Gynoecia of wild-type, arf6-2, and arf8-3 flowers grew to at least 4 mm long even if unpollinated (Figure S2A). Gynoecia of arf6-2 arf8-3, myb21 and myb21 myb24 flowers were shorter than wild-type gynoecia, and arrested at about 3 mm long (Table S1; Figure 3I, 3J; Figure S2A). This phenotype was largely attributable to decreased valve lengths in the mutants (Figure S6F). myb21 mutations also decreased stigma lengths, although this effect was only statistically significant for both tested myb21 alleles in myb24-5 or arf6-2/+ arf8-3 genetic backgrounds (Figure S6A–S6D, S6G). In the arf6-2/+ arf8-3 genetic background, myb21 mutations also decreased the proportion of ovules that were fertilized by wild-type pollen, from about 78% in arf6-2/ARF6 arf8-3 ovules, to just 35–40% in arf6-2/ARF6 arf8-3 myb21-4 or arf6-2/ARF6 arf8-3 myb21-5 ovules. In many poorly fertilized gynoecia, pollen tubes only entered the apical part of the transmitting tract. These stigma and fertilization phenotypes were similar to, although less severe than, those observed for arf6-2 arf8-3 plants (Figure S6E) [4].
MYB21 Can Promote Petal Growth Independently of Jasmonate Response
Flowers of jasmonate-deficient (aos-2) or -insensitive (coi1-1) mutants had short stamens and indehiscent anthers similar to those of myb21 myb24 mutants (Figure 3G, 3J; Figure S2C; Table S1). Similarly, at the time of wild-type flower opening (staged according to gynoecium length), aos-2 and coi1-1 flowers had delayed petal growth just as myb21 and myb21 myb24 flowers did, indicating that jasmonates promote petal growth at stage 12 (Figure 3G, 3J; Figure S2C). However, at stages 14–15 after pollination has normally occurred in wild-type flowers, petals of aos-2 and coi1-1 flowers continued to grow, so as to become larger than wild-type petals (Figure 3G, 3J; Figure S2C). Mutant flowers also senesced later than wild-type flowers, possibly accounting in part for the prolonged growth phase of these petals.
Gynoecia and valves of aos-2 and coi1-1 mutant flowers grew slightly less than those of unfertilized wild-type flowers, but more than those of myb21 or myb21 myb24 flowers (Figure 3J, Figure S2C, Figure S6F). Stigmas of aos-2 and coi1-1 flowers were as long as those of wild-type flowers, and aos-2 and coi1-1 gynoecia supported full fertilization after being pollinated manually (Figure S6G). Thus, myb21 mutations had stronger effects on both petal and gynoecium growth than did aos-2 or coi1-1 mutations. The weaker phenotypes of aos-2 and coi1-1 than myb21 and myb21 myb24 mutants appears inconsistent with the hierarchical model in which jasmonates induce MYB genes which in turn cause petal expansion. These results might have arisen if the aos-2 and coi1-1 mutants each retain some jasmonate response. However, we detected no cis-JA in aos-2 flowers (Figure 1F), and the coi1-1 mutation is a null mutation in the only known JA-Ile receptor. Moreover, flowers of aos-2 coi1-1 double mutant plants had enlarged petals and delayed senescence as did flowers of either single mutant (data not shown), suggesting that aos-2 and coi1-1 mutations each eliminated jasmonate response in flowers.
We therefore explored in more detail how the jasmonate pathway affects MYB21 expression. In wild-type flowers, MYB21 expression was high at stage 12, and then decreased at stages 13 and 14 (Figure 4, Table S3). Whereas at stage 12 aos-2 and coi1-1 flowers had lower expression of MYB21 than did wild-type flowers, at stage 14 they had higher expression (Figure 4). Similarly, aos-2 PMYB21:MYB21:GUS plants had reduced X-Gluc staining at stage 13, but had X-Gluc staining in petals at stage 15 (Figure 2M, 2O). Thus, in both wild-type and jasmonate pathway mutant plants, petal growth correlated with MYB21 expression. Moreover, petals of myb21-4 aos-2, myb21-5 aos-2, and coi1-1 myb21-4 double mutant flowers failed to enlarge at late stages, and flower buds of these double mutants never opened (Table S1; Figure 3H, 3J; Figure S2C). Thus, MYB21 is active and promotes petal elongation in stage 14 aos-2 and coi1-1 flowers.
Fig. 4. Expression of MYB21, MYB24, and jasmonate pathway genes in wild-type and mutant flowers at stages 12, 13, and 14. 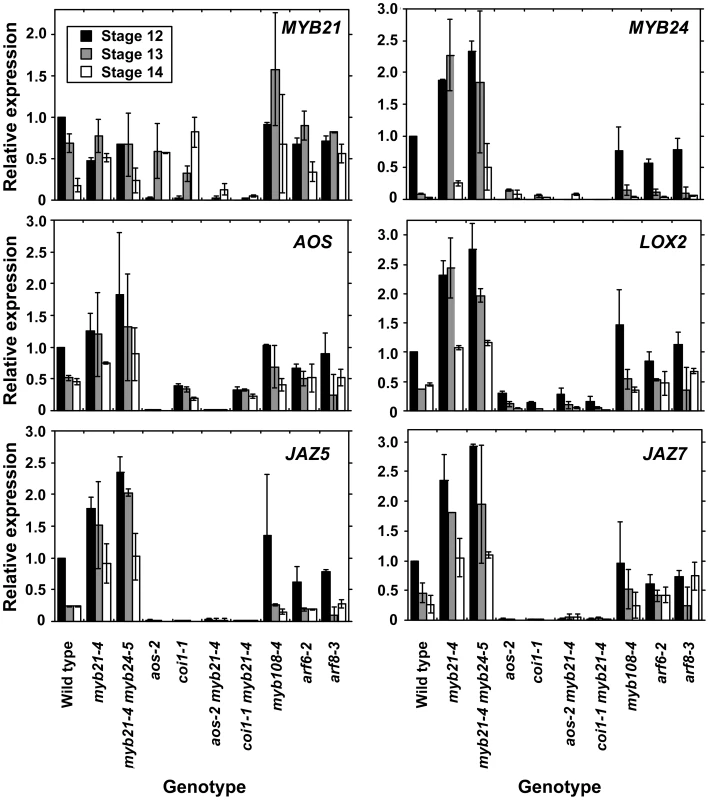
Gene expression was measured by quantitative RT-PCR. Shown are means of two biological replicates each having three technical replicates (± SD). Within each biological replicate, expression levels were normalized to expression in wild-type stage 12 flowers. Global Gene Expression Changes in Maturation-Deficient Flowers
These analyses revealed that starting at flower stage 12, ARF6 and ARF8 promote MYB21 and MYB24 expression in multiple flower organs largely by increasing jasmonate levels. MYB21 and MYB24 in turn promote petal and stamen filament growth, anther dehiscence, and gynoecium growth and maturation, with MYB21 having a predominant role. To explore gene expression patterns underlying this regulatory hierarchy, we used Affymetrix ATH1 gene chip arrays to monitor global gene expression in wild-type, arf6-2 arf8-3 and myb21-5 myb24-5 closed buds (stage 12 flowers) and newly open flowers (stage 13). Expression data for each array probe set were compared statistically between genotypes, and in addition a two-fold expression ratio cutoff was applied to remove genes with statistically significant but small relative differences in expression level (Figure 5, Table S3). We focussed our analyses on gene expression changes at stage 12, when flowers of both double mutants have similar morphology to wild-type flowers. Stage 13 data are presented for reference (Table S3), but presumably include many indirect effects caused by developmental arrest of mutant flowers at stage 12. As most array probe sets correspond uniquely to a single gene, in the following analyses we refer to probe sets as “genes.”
Fig. 5. Global analyses of gene expression in arf6-2 arf8-3 and myb21-5 myb24-5 stage 12 flowers. 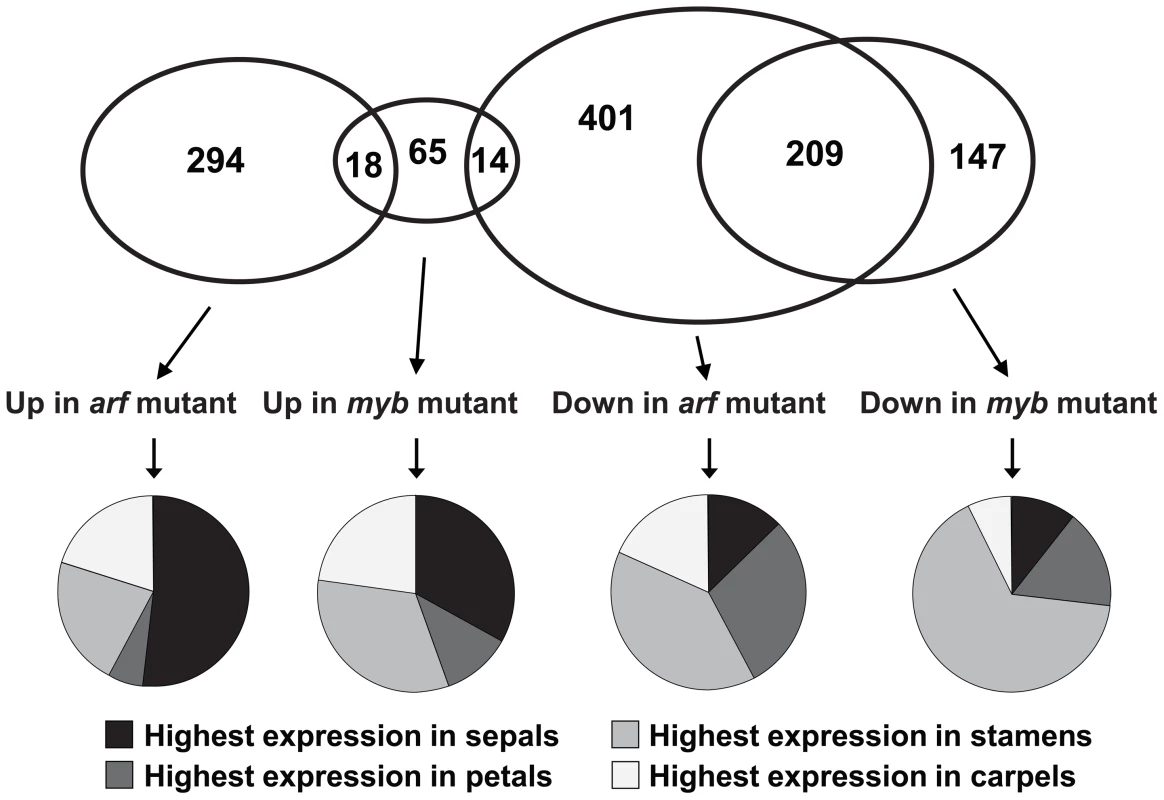
Venn diagram indicates numbers of genes with higher or lower expression in mutant compared to wild-type flowers, based on a t-test (P<0.05) and a two-fold ratio of expression values. Pie charts indicate the proportion of genes in each expression class having highest expression in sepals, petals, stamens, or carpels of wild-type stage 12 flowers [21]. Table S3 lists these genes and provides details of their expression levels. At flower stage 12, 624 genes were expressed at a lower level in arf6-2 arf8-3 flowers than in wild-type flowers, and 312 genes were expressed at a higher level (Figure 5). In myb21-5 myb24-5 stage 12 flowers, 356 genes were underexpressed and 97 were overexpressed relative to wild-type flowers. Of the genes underexpressed in arf6-2 arf8-3 flowers, 33% (209/624) were also underexpressed in myb21-5 myb24-5 flowers, and 2% (14/624) were overexpressed. Of the genes overexpressed in arf6-2 arf8-3 flowers, 6% (18/312) were also overexpressed in myb21-5 myb24-5 flowers, and none was underexpressed. Thus, the myb mutations affected a greater proportion of genes that were underexpressed in arf6-2 arf8-3 flowers than of genes that were overexpressed in arf6-2 arf8-3 flowers. As MYB21 and MYB24 can activate genes [17], [18], the 209 genes underexpressed in both myb21-5 myb24-5 and arf6-2 arf8-3 flowers may include genes that the MYB proteins activate. Independent RNA blot hybridization and qRT-PCR experiments confirmed expression characteristics deduced from the array data for about 15 genes of interest (Figure 1, Figure 4, Figure S1C).
To discern patterns in the gene expression data, we compared our data to global gene expression datasets generated by other workers (Table S3). Gibberellins, acting in part through derepression of DELLA protein activity, also promote late stages of petal, stamen, and gynoecium development [14], [25]–[28]. We compared our gene expression results to a list of genes that were over - or under-expressed in ga1-3 gibberellin-deficient mutant flowers [29]. 28% (172/624) of genes that were underexpressed in arf6-2 arf8-3 flowers were also underexpressed in ga1-3 flowers, and just 1.3% (8/624) were overexpressed in ga1-3 flowers (Table S3). Similarly, 25% (77/312) of genes that were overexpressed in arf6-2 arf8-3 flowers were also overexpressed in ga1-3 flowers, and just 4.5% (14/312) were underexpressed in ga1-3 flowers. Thus, ARF6 and ARF8 and gibberellin induce and repress an overlapping set of downstream responses in flowers, in most cases in the same direction.
We used data on gene expression in wild-type stage 12 sepals, petals, stamens and carpels [21] to determine in which organs each gene affected in arf6-2 arf8-3 or myb21-5 myb24-5 flowers was expressed (Figure 5, Table S3). Although most of the affected genes were expressed in multiple flower organs, to identify trends in the data it proved convenient to bin genes according to the organ in which they had highest expression in wild-type stage 12 flowers. Of the genes that were underexpressed in arf6-2 arf8-3 flowers, substantial numbers were most highly expressed in sepals (79/624, 13%), petals (185/624, 30%), stamens (246/624, 39%) or carpels (114/624, 18%) of wild-type flowers. In contrast, of the genes that were overexpressed in arf6-2 arf8-3 flowers, over half (161/312, 52%) were most highly expressed in sepals of wild-type flowers, whereas just 22% (69/312) were most highly expressed in wild-type stamens. In myb21-5 myb24-5 flowers, 66% (234/356) of underexpressed genes had highest expression in stamens of wild-type flowers. Of the genes that were overexpressed in the myb21-5 myb24-5 flowers, an equal number had highest expression in wild-type sepals as in wild-type stamens (32/97 in each case).
Positive Feedbacks on Auxin Response and MYB Function
Among the genes with decreased expression in arf6-2 arf8-3 flowers were several known auxin-inducible genes including IAA1, SHY2/IAA3, IAA6, AXR2/IAA7, IAA17, IAA19, SAUR9 (SMALL AUXIN UP RNA9), SAUR23, SAUR25, SAUR27, SAUR35, SAUR42, SAUR62-SAUR68, and SAUR70 (Table S3). Many of these have auxin response elements in their presumed promoters and are good candidates to be direct targets of ARF6 and ARF8 [30]. Although the hierarchical regulatory model does not predict that MYB21 or MYB24 should affect expression of direct ARF targets, several of these IAA and SAUR genes (IAA6, IAA19, SAUR9, 25, 35, 64, 66, 67, and 68) were also underexpressed in stage 12 myb21-5 myb24-5 flowers (Table S3). RNA gel blot hybridization experiments confirmed that IAA19 and SAUR63 were underexpressed in myb21-5 myb24-5 flowers, and that in addition IAA2, SHY2/IAA3 and AXR2/IAA7 were more modestly underexpressed in myb21-5 myb24-5 flowers (Figure 1D). These results suggest that MYB21 and MYB24 participate in positive feedback loops that promote ARF activity.
Additional ARF and MYB genes were underexpressed in mutant flowers, and might also constitute positive feedbacks if they share targets with ARF6 and ARF8 or MYB21. The ARF16 (At4g30080) gene encodes an Auxin Response Factor that is phylogenetically distant from ARF6 and ARF8, and regulates root cap differentiation together with its closest paralog ARF10 [31], [32]. ARF16 was underexpressed in arf6-2 arf8-3 flowers but had normal expression level in myb21-5 myb24-5 flowers. arf10-3 arf16-2 flowers, as well as P35S:MIR160c flowers overexpressing a microRNA that targets ARF10 and ARF16 [31], had delayed stamen and petal growth, similarly to arf6-2 or arf8-3 single mutant flowers (Figure S2A, S2B). These results suggest that ARF10 and ARF16 act downstream of ARF6 and ARF8 to amplify stamen and petal growth at stage 12.
Analogously, MYB57 and MYB108, closely related genes to MYB21 and MYB24, were underexpressed in arf6-2 arf8-3 and myb21-5 myb24-5 flowers (Figure 1A, 1B; Table S3). A PMYB57:MYB57:GUS reporter was expressed in stamen filaments of opened wild-type flowers (Figure S1F). A PMYB108:GUS reporter was expressed in sepals and stamen filaments, particularly in the vasculature of these organs, and in the style (Figure S1G). The myb57-1 mutant (Figure S3) had no obvious floral phenotypes (data not shown), although myb57-1 can enhance a myb21 mutation [14]. Flowers of myb108 mutants (Figure S3) had normal organ lengths at stages 12 and 13, but had slightly delayed anther dehiscence (Table S1; Figure 3I; Figure S5E, S5F) [15]. In addition, myb108-4 petals continued to grow after wild-type petals had stopped expanding, resulting in slightly longer petals at stage 14 (Figure 3I, Figure S5D–S5F). Similarly to the jasmonate pathway mutants, stage 14 myb108-4 flowers had elevated MYB21 expression, and myb21-5 myb108-4 flowers had small petals (Figure 4, Table S1). Thus, increased MYB21 expression may also cause persistent petal growth in myb108 mutants.
ARF6 and ARF8 Regulate Nectary Development
ARF6, ARF8, MYB21 and MYB24 are each expressed in nectaries. A previous study identified 270 genes that were preferentially expressed in nectaries [33]. Of these, 18 were underexpressed in arf6-2 arf8-3 only, 6 were underexpressed in myb21-5 myb24-5 only, and 14 were underexpressed in both mutants (Table S3). In contrast, just 5 of the nectary-enriched genes were overexpressed in either mutant. Among the underexpressed genes were CRABSCLAW (CRC/At1g69180), which is required for nectary formation [34]; YABBY5 (At2g26580) encoding a protein closely related to CRC; CWIV4 (At2g36190) encoding a cell wall invertase required for nectary sink strength and nectar production [35]; SWEET9 (At2g39060) encoding a nectary-specific glucose transporter [36]; and JMT (At1g19640) encoding S-adenosyl-L-methionine:jasmonic acid carboxyl methyltransferase, which makes the volatile compound methyl jasmonate [37]. Each of these genes was underexpressed in both arf6-2 arf8-3 and myb21-5 myb24-5 flowers, except for CRC which was underexpressed in arf6-2 arf8-3 flowers only. Consistent with these gene expression changes, nectaries in arf6-2 arf8-3 flowers were very small and only apparent by light microscopy in a fraction of flowers (Figure 6A, 6B). Nectaries in coi1-1, arf6-2 and arf8-3 single mutants and in myb21-5 myb24-5 double mutant flowers appeared normal (Figure 6C, 6D; data not shown). These morphological and gene expression results indicate that ARF6 and ARF8 affect nectary growth and function, and that MYB21 and MYB24 affect nectary gene expression but not nectary formation.
Fig. 6. Phenotypes related to insect attraction. 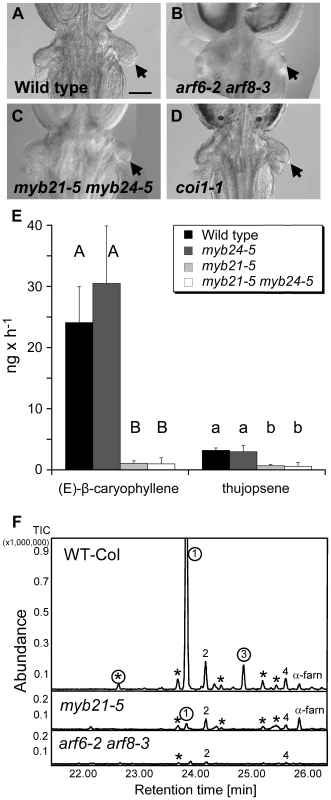
(A–D) Base of gynoecia of indicated genotypes. Arrows indicate nectaries. Scale bar, 0.1 mm. (E) Comparative quantitative analyses of floral volatile sesquiterpene emissions from wild-type, myb21-5, myb24-5, and myb21-5 myb24-5 mutants. Emitted compounds were collected for 7 h from 40 detached inflorescences by a closed-loop stripping procedure. Emission was determined in ng h−1 per 40 inflorescences. Values are averages and standard deviations of three independent collections. Only emissions of (E)-β-caryophyllene, the product of TPS21, and thujopsene, the product of TPS11, are shown. Different letters indicate significant differences in emissions of each compound between genotypes ( p≤0.001). (F) GC-MS analyses of sesquiterpene hydrocarbons collected via SPME from 20 inflorescences of wild-type, myb21-5 and arf6-2 arf8-3 mutants. Peaks marked with circles represent sesquiterpenes produced by the terpene synthase TPS21. Compounds not labeled with circles are products of the terpene synthase TPS11, with the exception of α-farnesene (α-farn). 1, (E)-β-caryophyllene; 2, thujopsene; 3, α-humulene; 4, β-chamigrene. Peaks marked with asterisks are other sesquiterpene products of TPS11 or TPS21 [43]. MYB21 Promotes Production of Volatile Sesquiterpenes
As flowers open, they emit volatile compounds, which may attract insect pollinators or predators, or may have a role in pathogen defense [38]–[41]. The Arabidopsis terpene synthase genes TPS11 (At5g44630) and TPS21 (At5g23960) synthesize a mixture of volatile sesquiterpenes emitted from flowers [42], [43]. Both genes were highly expressed in wild-type carpels, and TPS11 was also expressed in nectaries [33], [43]. Both TPS11 and TPS21 were underexpressed in arf6-2 arf8-3 and myb21-5 myb24-5 flowers (Figure 1C, Table S3). Consistent with these patterns, arf6-2 arf8-3 flowers emitted dramatically less sesquiterpenes produced by both TPS11 and TPS21 (Figure 6F). Similarly, myb21-5 flowers had strongly reduced emission of sesquiterpenes produced by TPS21 (e.g. (E)-β-caryophyllene, α-humulene), and partially reduced levels of volatile sesquiterpenes produced by TPS11 (e.g. thujopsene, β-chamigrene) (Figure 6E, 6F). These effects are consistent with the gene expression patterns, as TPS11 expression was reduced in myb21-5 myb24-5 flowers by less than was TPS21 expression (Table S3, Figure 1C). The myb24-5 mutation did not affect emission of volatile sesquiterpenes, either by itself or in combination with myb21-5 (Figure 6E). (E)-β-caryophyllene and thujopsene emissions were also reduced in flowers of the opr3 jasmonate-deficient mutant (Figure S7).
MYB21 and MYB24 Mediate Secondary Jasmonate Responses in Stamens
In a gene chip array dataset of gene expression in stamens of jasmonate-deficient opr3 mutant stage 12 flowers treated with exogenous methyl jasmonate (MeJA), 31 genes were induced by at least 2-fold after 30 minutes of MeJA treatment, 179 additional genes were first induced after 2 hours, and 393 more genes were first induced after 8 hours [12]. MYB21 and MYB24 were themselves induced at the two hour time point in this dataset. Consistent with their reduced jasmonate production, arf6-2 arf8-3 flowers underexpressed many of these MeJA-responsive genes, with the greatest proportional effect on the earliest MeJA-responsive genes. Thus, about 45% (14/31) of the genes induced by MeJA in stamens within 30 minutes were underexpressed in arf6-2 arf8-3 flowers (Table S3).
In the myb21-5 myb24-5 flowers, none of the early MeJA-inducible genes was underexpressed, and the proportion of MeJA-responsive genes affected was highest among those induced by MeJA at 8 hours. Of 86 late (8 hour) MeJA-inducible genes underexpressed in arf6-2 arf8-3 flowers in our experiment, 50 (58%) were also underexpressed in myb21-5 myb24-5 flowers, indicating that MYB21 and MYB24 mediate a large portion of late responses to jasmonate in flowers (Table S3).
MYB21 Decreases Jasmonate Levels through a Negative Feedback Loop
Strikingly, 13 of the 14 genes that were underexpressed in arf6-2 arf8-3 flowers but overexpressed in myb21-5 myb24-5 flowers were MeJA-induced in stamens (Table S3). Using a less stringent 1.3-fold expression ratio cutoff, 71 genes were underexpressed in arf6-2 arf8-3 flowers and overexpressed in myb21-5 myb24-5 flowers, and 44 of these were MeJA-induced in stamens (13 of these at the earliest 0.5 h time point) (Table S4). Among these genes were MYC2 (At1g32640), which binds to jasmonate-inducible promoters to mediate induction [44]; seven JAZ genes encoding negative regulators of jasmonate response [45]; and several genes encoding known or putative enzymes in the jasmonate biosynthesis pathway. These included LOX2 (At3g45140) and LOX4 (At1g72520) encoding lipoxygenases involved in generating the fatty acid precursor [46], [47]; AOS (At5g42650) encoding allene oxide synthase [48]; OPR3 (At2g06050) encoding 12-oxophytodienoate reductase [8]; and 4CL11 (At5g38120) and 4CL9/OPCL1 (At1g20510), encoding 4-coumarate CoA ligases [49] (Table S4). RNA blot hybridization with polyA+ mRNA and qRT-PCR experiments confirmed increased expression of LOX2 and AOS in myb21-4 myb24-5 and myb21-5 myb24-5 flowers (Figure 1E, Figure 4). The phospholipase DAD1 (At2g44810) was expressed at a low level in all samples in the gene chip array experiment, but was also seen to be overexpressed in myb21-5 myb24-5 flowers by RNA blot hybridization (Figure 1E). Consistent with their increased expression of jasmonate biosynthetic genes, stage 12–13 myb21-5 myb24-5 flowers had about 12-fold higher level of cis-JA than did wild-type flowers (Figure 1F). The AOS, LOX2, JAZ5, and JAZ7 genes were also overexpressed in myb21-4 single mutant flowers, to the same degree as in myb21-4 myb24-5 double mutant flowers (Figure 4). These data indicate that MYB21 acts within a negative feedback loop that regulates expression of multiple JA biosynthetic genes.
As mentioned above, the nectary-expressed JMT (At1g19640) gene whose product makes methyl jasmonate was underexpressed in both arf6-2 arf8-3 and myb21-5 myb24-5 flowers. However, the At3g11480 gene encoding a JMT-related protein was overexpressed in myb21-5 myb24-5 flowers, suggesting that At3g11480 rather than JMT/At1g19640 might produce MeJA as part of the MYB-regulated negative feedback loop. JAR1 (At2g46370), encoding an enzyme that synthesizes the active JA-Ile, did not show statistically different expression between wild-type and mutant flowers. jar1 plants are male-fertile, suggesting that another enzyme produces JA-Ile in flowers [50]. The most closely related Arabidopsis gene to JAR1 is GH3-10/DFL2 (At4g03400), which had normal expression in both mutants at stage 12, but was underexpressed in both mutants at stage 13 (data not shown).
In leaves, jasmonate induces genes encoding enzymes in the jasmonate biosynthesis pathway, indicating that a positive feedback loop amplifies jasmonate synthesis [7], [8], [47], [51]–[54]. In qRT-PCR assays, stage 12, 13, and 14 aos-2 and coi1-1 flowers had lower levels of AOS and LOX2 than did wild-type flowers (Figure 4), confirming that such a positive feedback loop operates in flowers. To explore how the MYB21-mediated negative feedback and the COI1-mediated positive feedback interact, we assessed expression of these genes in aos-2 myb21-4 and coi1-1 myb21-4 double mutant flowers. In flowers of both double mutants, AOS and LOX2 levels were as low as in aos-2 or coi1-1 mutant flowers. These results indicate that COI1 is required to activate jasmonate biosynthesis in myb21-4 flowers, and suggest that MYB21 acts by inhibiting the COI1-mediated positive feedback loop in jasmonate biosynthesis. AOS, LOX2, JAZ5, and JAZ7 were also underexpressed in arf6-2 arf8-3 myb21-4 triple mutant flowers (Figure S1C), indicating that jasmonate overproduction in myb21 mutant flowers also depends on ARF6 and ARF8.
Discussion
Functions of the Flower Maturation Regulatory Network
The phenotypic and gene expression analyses presented here show that, in addition to previously described petal, stamen, and gynoecium growth and maturation [2], [3], the ARF6 and ARF8 regulatory network promotes nectary development and floral scent production. This regulatory network should promote reproduction by both self-pollination and outcrossing. Thus, coordination of timing of stamen filament elongation, pollen release, stigma growth, and style and transmitting tract support of pollen tube growth ensures efficient self-fertilization; whereas coordination of petal growth, nectary development, and sesquiterpene production with stamen and gynoecium development would attract pollinators to flowers when they are reproductively competent. Although Arabidopsis self-pollinates efficiently, outcrossing by insect pollination has been observed in field populations [55], [56]. Terpene formation coordinated with gynoecium development also helps to protect reproductive organs against invasion by microbial pathogens (M. Huang, A. M. Sanchez-Moreiras, C. Abel, J. Gershenzon, and D. Tholl, unpublished results).
ARF6 and ARF8 activate jasmonate biosynthesis, which in turn activates MYB21 and MYB24. Genes underexpressed in arf6-2 arf8-3 and myb21-5 myb24-5 flowers may promote aspects of flower maturation deficient in both mutants. Such genes include MYB108, which promotes anther dehiscence; several SAUR genes that promote organ elongation (K. Chae, C. G. Isaacs, P. H. Reeves, G. S. Maloney, G. K. Muday, and J. W. Reed, unpublished results); and the TPS11 and TPS21 genes required for sesquiterpene production. Genes affected in arf6-2 arf8-3 but not myb21-5 myb24-5 flowers must act upstream of MYB21 or mediate MYB21-independent functions. These include ARF16, which contributes to petal and stamen elongation; several genes involved in nectary formation or function; and three closely related bHLH transcription factors, HALF-FILLED(HAF)/bHLH075, BRASSINOSTEROID ENHANCED EXPRESSION1 (BEE1)/bHLH044 and BEE3/bHLH050, which act redundantly to promote transmitting tract differentiation (Table S3) [57], [58]. Other genes identified in this dataset may allow further dissection of general and organ-specific aspects of flower maturation, such as stylar factors that promote stigma growth non-cell-autonomously and/or potentiate pollen tube growth [22].
Sepal growth normally ceases at stage 12 when petal and stamen filament growth accelerates, and sepals of mutant flowers appeared outwardly normal. Nevertheless, 240 genes having preferential expression in wild-type sepals had altered expression in arf6-2 arf8-3 flowers, and about two thirds (161/240) of these were overexpressed. In contrast, most affected genes with preferential expression in wild-type petals, stamens, or gynoecia were underexpressed in arf6-2 arf8-3 flowers (91%, 78%, and 64%, respectively). Internal organs in the mutant flowers might have decreased sink strength, which might induce gene expression changes in sepals indirectly, or might cause internal organs to resemble sepals physiologically and express higher levels of “sepal” genes.
Dynamic Interactions among Hormone Response Pathways during Flower Maturation
Three mobile hormone signals - auxin, gibberellin, and jasmonate - regulate flower maturation, and this network incorporates both signal amplification and feedback mechanisms (Figure 7). Auxin can activate ARF6 and ARF8 activity by destabilizing Aux/IAA transcriptional repressor proteins, and both msg2/iaa19 gain-of-function mutants and yucca2 yucca6 mutants deficient in auxin biosynthesis have delayed stamen development [23], [59]–[64]. These results indicate that auxin indeed promotes wild-type flower maturation. Temperature stress, shade light and the circadian rhythm can each regulate auxin levels and/or response [65]–[71], and these environmental factors might thereby regulate flower growth according to light or temperature conditions, or ensure appropriate diurnal timing of flower opening and pollination.
Fig. 7. Genetic model of Arabidopsis flower maturation. 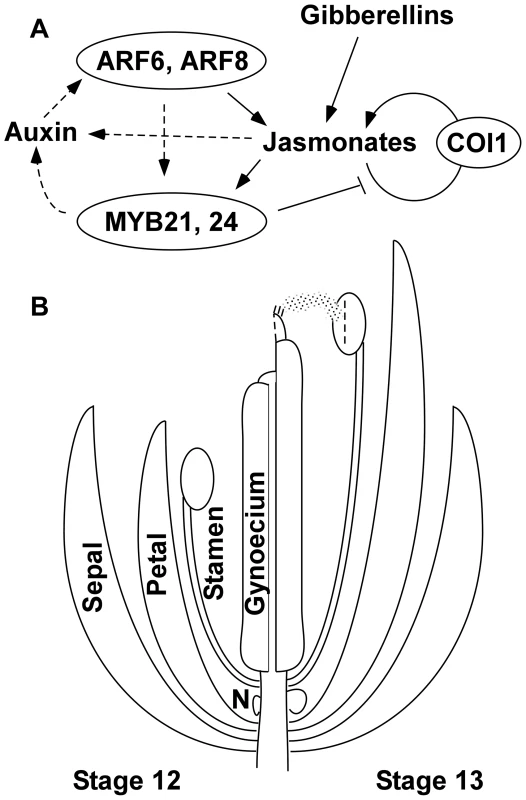
(A) Diagram of principal regulatory pathways. Arrows indicate regulatory events established in this work or by previous studies. Both gibberellins and ARF6 and ARF8 auxin response factors promote jasmonate biosynthesis at flower stage 12. Auxin presumably enables ARF activity, and this may also be regulated by the circadian rhythm. Jasmonates in turn activate expression of genes for jasmonate biosynthesis, in a positive feedback loop requiring the JA-Ile receptor COI1. The underexpression of potential direct ARF6- and ARF8-targets in myb21 myb24 flowers suggests that MYB21 and MYB24 may also participate in an additional positive feedback loop that promotes ARF6 and ARF8 activity, possibly through effects on auxin level (shown as dashed arrows). MYB21 represses jasmonate biosynthesis, and after the flower has opened (stage 13 and later), this negative feedback arrests flower maturation functions. In the absence of jasmonate signaling, ARF6 and ARF8 also contribute to MYB21 expression in late-stage petals. (B) Illustration of flower developmental events regulated by the network between flower stage 12 (left) and stage 13 (right). The network induces downstream effectors that promote multiple events including petal and stamen filament elongation (regulated by ARF16 and by SAUR proteins), anther dehiscence (regulated by MYB108), volatile compound production (by TPS11 and TPS21 terpene synthases), nectary growth and development (regulated by CRC), and gynoecium growth and maturation. These and other effector genes may be activated directly or indirectly by MYB21 and MYB24, or by ARF6 and ARF8 independently of the MYB proteins. N, nectary. Similarly to arf6 arf8 mutants, gibberellin-deficient mutants have arrested petal, stamen, and gynoecium development, are deficient in jasmonate production, and are both male - and female-sterile [14], [26], [28]. Although the two pathways had overlapping effects on gene expression, based on our gene chip expression data, arf6-2 arf8-3 flowers had normal gibberellin biosynthetic gene expression levels, and known auxin biosynthetic genes did not appear in the gibberellin-responsive gene lists. Thus, the two pathways may be integrated through shared downstream targets rather than acting hierarchically. Auxins and gibberellins also each regulate hypocotyl elongation and fruit growth, by both hierarchical and parallel mechanisms [72]–[75].
ARF6 and ARF8 and gibberellins each activate jasmonate biosynthesis. ARF6 and ARF8 may do this in part through TCP4 (At3g15030), which was underexpressed in arf6-2 arf8-3 flowers and activates developmental expression of LOX2 [76]. JA-Ile in turn activates a positive feedback loop of jasmonate synthesis by causing COI1-dependent turnover of JAZ transcriptional repressor proteins, which then (at least in leaves) releases the bHLH proteins MYC2, MYC3, and MYC4 to activate transcription of jasmonate biosynthesis genes as well as MYC2 itself [77]–[79]. Jasmonate synthesis has been postulated to occur in stamen filaments, based on the expression pattern of DAD1 [6], [80]. However, other genes can act redundantly with DAD1 during wound-induced jasmonate production [81], and other jasmonate biosynthetic genes were expressed in multiple flower organs (Table S3) [21], [47], suggesting that jasmonates are synthesized broadly throughout the flower. If synthesis were first triggered in stamen filaments, the positive feedback of jasmonate synthesis and movement of MeJA or another jasmonate pathway compound might amplify jasmonate production throughout the flower, thereby causing a coordinated burst of stamen and petal growth and emission of floral scents.
Jasmonates induce MYB21 and MYB24, and MYB21 and MYB24 then activate secondary gene expression responses to jasmonate leading to petal and stamen filament elongation and anther dehiscence. MYB21 and MYB24 are also required for expression of several known primary auxin-responsive genes. This finding suggests that MYB21 and MYB24 also affect ARF6 and ARF8 activity, and that a portion of the myb21 myb24 flower phenotypes may be caused by decreased ARF activity.
MYB21 also induces a negative feedback on jasmonate biosynthesis. Jasmonate overproduction in myb21 flowers requires the COI1-dependent positive feedback pathway that activates jasmonate biosynthesis genes, suggesting that MYB21 acts on a component of this pathway. JAZ genes encoding repressors of jasmonate response are themselves jasmonate-inducible, and the MYB proteins might amplify this negative feedback loop if they activate JAZ gene expression. However, the increased rather than decreased expression of JAZ and other primary jasmonate responsive genes in myb21-5 myb24-5 flowers suggests that other proteins such as MYC2 are sufficient to activate primary jasmonate response. Alternatively, as suggested by the recent discovery that MYB21 and MYB24 proteins can interact with JAZ1, JAZ8, JAZ10, and JAZ11 proteins [20], MYB21 might stabilize JAZ proteins by interfering with their COI1-mediated turnover. This negative feedback pathway may also act in flowers of the jar1-1 mutant deficient in the enzyme that makes active JA-Ile, which similarly had elevated jasmonic acid levels [82].
In wild-type flowers, jasmonic acid levels increase at stages 11–12 just before flowers open, and then decrease at stages 13–14, when flower organs stop growing [2]. Mathematical modeling suggests that after wounding of leaves, positive feedback increases the amplitude of jasmonate synthesis, whereas negative feedback mediated by the JAZ proteins determines the duration of the jasmonate pulse [83]. In flowers, the linked positive and negative feedback loops regulating jasmonate production and auxin response provide a plausible mechanism for inducing coordinated rapid increase in petal and stamen growth at stage 12, followed by a quick cessation of growth after stage 13 once the flower has opened and pollen has been released. MYB21 and MYB24 are not expressed in wounded leaves, and recruitment of the MYB factors into the feedback mechanisms may be an evolutionary innovation that has contributed to the adaptation of this network to regulate flower opening.
The prolonged growth seen in petals of stage 13–14 jasmonate pathway mutant flowers arises from jasmonate-independent MYB21 expression. As arf6 arf8 flowers do not express MYB21, an ARF-dependent but jasmonate-independent mechanism can apparently activate MYB21. This or a similar pathway apparently also acts in stage 14 arf8 and myb108 mutant flowers [11]. BIGPETAL (BPE)/bHLH31 (At1g59640) is activated in petals by jasmonate-induced alternative splicing and represses petal growth [9], [10], [12], [58], and it will be interesting to test whether it represses MYB21.
Action of this Network in Other Angiosperms
In tobacco and petunia, putative orthologs of MYB21 and MYB24 regulate both floral scent production and flower opening [84]–[86]. The network described here may thus provide a useful context to understand flower maturation in other angiosperms, and the roles of genes responsible for natural variation in flower morphology [87]. For example, variation in the expression level of the tomato Style2.1 gene determines the extent of style growth, which in turn affects whether the plant self-pollinates or outcrosses [88]. An Arabidopsis homolog of Style2.1, PRE1/bHLH136/BNQ1 (At5g39860), is underexpressed in arf6-2 arf8-3 flowers and may contribute to Arabidopsis flower organ elongation [89], [90].
Materials and Methods
Plant Material and Isolation of arf6-2 Enhancer Mutants
All genotypes were in the Columbia ecotype of Arabidopsis thaliana. arf6-2 and arf8-3 mutants were previously described [2]. The myb21-4 and myb21-5 mutants were isolated from an EMS mutagenesis screen for enhancers of the arf6-2 mutant. arf6-2 seeds were treated with 0.2% EMS for 16 hours, and 10,000 M2 plants derived from approximately 5000 M1 parents were screened for reduced fecundity. In addition to the myb21 mutations described here, we isolated three new arf8 alleles in this screen (Table S2). arf6-2 myb21 plants from the screen were back-crossed once to arf6-2, and then crossed twice to wild type prior to further analysis. Backcrosses indicated that the myb21 phenotype was caused by a recessive mutation at a single genetic locus. To map the mutations, arf6-2 enhancer mutants were crossed to an arf6-2 line that had been introgressed into the Landsberg erecta ecotype. A bulked-segregant analysis approach using 29 markers evenly distributed over the genome was used to establish a preliminary map position [91], and the map position was then refined using closely linked SSLP, CAPS and dCAPS markers (Figure S3).
T-DNA insertion mutations in MYB21, MYB24, MYB57, MYB108, and AOS from the SALK Genomic Analysis Laboratory were provided by the Arabidopsis Biological Resource Centre [92]. Homozygous mutants were identified within segregating T3 and T4 populations. Details on these mutants, and PCR primers used to identify mutant alleles, are provided in Figure S4, Table S2, and Table S5. Double and triple mutants were identified in the F2 progeny of crosses from the respective single or double mutant parents. Most genotypes were fertile when manually self-pollinated. However, myb21 myb24, myb21 aos-2 and myb21-5 arf6-2 arf8-3 plants were maintained as myb21/+ myb24, myb21 aos-2/+ and myb21 arf8-3 arf6-2/+ stocks. coi1-1 seeds were provided by John Turner (University of East Anglia, Norwich, UK), and PLAT52:GUS seed were provided by Mark Johnson (Brown University, Providence, RI). ams seeds (SALK_152147) [93] were provided by Hong Ma (Pennsylvania State University, College Station, PA).
Phenotypic Analyses
To measure flower organs across a developmental series, flower buds were dissected, and flower organs were placed on an agar plate and measured using a camera lucida attachment on a dissecting microscope. For measurements of floral organ lengths and timing of anther dehiscence in Table S1, the first open flower of wild-type plants was designated as flower 1 (stage 13) [1]. For genotypes in which flower opening was impaired, equivalent stage flowers were identified based upon bud size and position on the inflorescence stem. Scanning electron microscopy was carried out as previously described [2]. Fertilization frequencies were assessed by X-Gluc staining 24 hours after pollination with the pollen-specific reporter line PLAT52:GUS [94]. In these assays, 87% or more of wild-type, ams male-sterile, myb21-5, myb21 myb24, and myb21 myb24 myb108 ovules were fertilized, as judged by strong X-Gluc staining in ovules in which a pollen tube had ruptured.
Transgenic Plants
To make GUS reporter constructs, promoter and genomic sequences lacking the endogenous stop codon were cloned into the Gateway pENTR/D-TOPO vector (Invitrogen Life Technologies, Carlsbad, CA). The upstream region (2266 bp) and first exon of MYB21 were amplified by PCR using the primers MYB21 PF and MYB21 R4 (Table S5). The introns and second and third exons were amplified using the primers MYB21 F4 and MYB21 R7. These two PCR products were cloned separately into pENTR/D-TOPO, and then the promoter and first exon of MYB21 were excised and ligated into the construct containing the 3′end of the MYB21 gene using NotI and PstI. The upstream region (2207 bp) and first exon of MYB24 was amplified by PCR using the primers MYB24 PF and MYB24 R3. The entire predicted coding region of MYB24 was amplified using the primers MYB24 F2 and MYB24 R2. These two PCR products were cloned separately into pENTR/D-TOPO, and then the promoter and first exon were excised and ligated into the construct containing the 3′end of MYB24 gene using NotI and PstI. The MYB57 upstream region (2414 bp) and predicted coding region were amplified using the primers MYB57 PF and MYB57 R2 and cloned into pENTR/D-TOPO. For MYB108, only the promoter was used in the GUS reporter construct. The upstream region of MYB108 (2090 bp) was amplified using the primers MYB108 PF and MYB108 R2 and cloned into pENTR/D-TOPO. The promoter and genomic sequences were fused to the GUS reporter by recombining entry clones into the destination vector pGWB3 [95] using LR clonase (Invitrogen). Transformation of Arabidopsis plants and histochemical staining were performed as described previously [96], [97]. The PMYB21:MYB21:GUS and PMYB24:MYB24:GUS constructs partially rescued the phenotype of myb21-2 myb24-2 flowers, indicating that they retained some MYB21 and MYB24 activity.
For P35S:MYB21 and P35S:Green fluorescent protein(GFP):MYB21 constructs, a genomic MYB21 fragment was amplified using the primers MYB21 GWF and MYB21 GWR* (Table S5), and cloned into the Gateway pENTR/D-TOPO vector. For P35S:MYB21, the entry clone was recombined into pB2GW7 [98]. For P35S:GFP:MYB21 the entry clone was recombined into pGWB6 [95]. Transgenic T1 P35S:MYB21 and P35S:GFP:MYB21 plants showed a range of phenotypes, including narrow leaves, dwarfism, and floral defects, similar to those previously described [19], [20]. For our analyses, we used weaker lines that had less severe phenotypes.
Hormone Treatments
For gene expression analyses, plants were sprayed with 1 mM MeJA (Bedoukian Research, Inc, Danbury, CT) or 10 µM IAA (Sigma) in 1% methanol 0.05% Tween-20, or with solvent alone, and were harvested after two hours (MeJA) or the specified time periods (IAA). To restore fertility to JA-deficient plants and to assess the effect of jasmonate on aos-2 PMYB21:MYB21:GUS plants, flowers were sprayed with 1 mM MeJA daily for 4 days.
Gene Expression Analyses
Flowers or whole inflorescences were frozen in liquid N2 and total RNA was isolated using Trizol reagent (Invitrogen Life Technologies, Carlsbad, CA) or with RNeasy plant mini kits (Qiagen). Poly (A+) RNA was extracted from 50 µg of total RNA using oligo(dT)25 Dynabeads according to manufacturers' instructions (Dynal A.S., Oslo, Norway). RNA gel blot hybridizations were performed as described [99]. Probes were created by PCR using genomic DNA or Peking-Yale cDNA clones (Arabidopsis Biological Resource Center) as template using primer pairs listed in Table S5.
For real-time quantitative RT-PCR analyses, total RNA was extracted from stage 12, 13 and 14 flowers in the morning between 2 and 4 hours after subjective dawn. cDNA was synthesized using the Reverse Transcription System (Promega A3500) with random primers according to the manufacturer's instructions. 0.1 µg of total RNA was used for the 20 µl volume reaction and incubated for 1 hr at 42°C. The RT reaction mixture was diluted 10-fold and 1 µl was used as a template in 10 µl PCR reactions using the Applied Biosystems real-time PCR systems in standard mode with SYBR Green Master Mix (Applied Biosystems) following the manufacturer's protocol. The primers used for qRT-PCR analysis are listed in Table S5. Reactions were performed in triplicate and the products were checked by melting curve analysis. Transcript levels were normalized to the level of reference transcript UBQ10.
For Affymetrix gene chip gene expression analyses, RNA was isolated from stage 12 (largest closed buds) and stage 13 (first open flowers) harvested in the morning between 2 and 4 hours after subjective dawn. Three biological replicates were performed. Probe synthesis and gene chip hybridizations were performed by the UNC-CH Functional Genomics Core Facility. Total RNA (1000 ng) was used to synthesize cDNA followed by aRNA. The MessageAmp II-Biotin Enhanced Kit (Ambion) was used to generate biotinylated aRNA from the cDNA reaction. The aRNA was then fragmented in fragmentation buffer from the Ambion kit at 94°C for 35 minutes before the chip hybridization. Fragmented aRNA (15 µg) was then added to a hybridization cocktail (0.05 µg/µl fragmented aRNA, 50 pM control oligonucleotide B2, BioB, BioC, BioD and cre hybridization controls, 0.1 mg/ml herring sperm DNA, 0.5 mg/ml acetylated BSA, 100 mM MES, 1 M [Na+], 20 mM EDTA, 0.01% Tween 20). aRNA (10 µg) was used for hybridization in a volume of 200 µl per slide. ATH1 arrays [100] (Affymetrix, Santa Clara, CA) were hybridized for 16 hours at 45°C in the GeneChip Hybridization Oven 640 (Affymetrix). The arrays were washed and stained with R-phycoerythrin streptavidin in the GeneChip Fluidics Station 450 (Affymetrix) using wash protocol EukGE-WS2v4, and arrays were scanned with the GeneChip Scanner 3000 7G Plus with autoloader. Affymetrix MAS 5.0 GeneChip Operating Software was used for washing, scanning and basic analysis. Sample quality was assessed by examination of 3′ to 5′ intensity ratios of selected genes. Data were analyzed using Genespring GX 10.0.1 software. Raw data were background corrected and normalized using the RMA algorithm with no baseline correction. Means for each gene over the three biological replicates were calculated, and statistical differences between wild-type and mutant expression levels assessed by t-test without multiple testing correction. Genes reported in Table S3 are those with P<0.05 and having 2-fold or greater expression level difference from the corresponding wild-type sample. Gene chip hybridization data have been deposited in the NCBI GEO database (http://www.ncbi.nlm.nih.gov/geo/) with accession number GSE32193.
In Situ Hybridization
In situ hybridizations were carried out as previously described [4]. MYB21 and MYB24 probes were PCR amplified from genomic DNA using primers that spanned the last exon (MYB21 ins-HindIII F, MYB21 ins-BamHI R; MYB24 ins-HindIII F, MYB24 ins-BamHI R) (Table S5). PCR products were then cloned into the pGEM-T vector (Promega). MYB21 and MYB24 sense probes produced no signal in wild-type flowers.
Jasmonic Acid and Volatile Sesquiterpene Collection and Analysis from Flowers
Jasmonic acid was measured as described [101] from stage 12–13 flowers collected in the morning and frozen in liquid nitrogen. To measure sesquiterpenes, volatile compounds were collected in 1 L bell jars with 40 detached inflorescences placed in a small glass beaker filled with tap water, under controlled temperature and light conditions (22°C, 150 µmol m−2 s−1 PAR). Emitted volatile compounds were collected for 7 h on 5 mg Charcoal filter traps (Gränicher and Quartero, Daumazan, France) in a closed-loop stripping procedure [102] and then eluted from the traps with 40 µl CH2Cl2 containing 1-bromodecane (20 ng/µl) as a standard. Sample analysis and quantification of terpenes was performed by gas chromatography–mass spectrometry (GC-MS) on a Shimadzu QP 2010s GC-MS instrument as described previously [103]. Separation was performed on a (5%-phenyl)-methylpolysiloxane (DB5) column (Restek, 30 m×0.25 mm i.d.×0.25 m –thickness). Helium was the carrier gas (flow rate 1.4 ml min−1), a splitless injection (injection volume 1 µl) was used, and a temperature gradient of 5°C/min from 40°C (2 min hold) to 220°C was applied. Compounds were identified by comparison of retention times and mass spectra with those of authentic standards. Trapping and GC-MS analysis of volatiles from flowers of opr3 and Wassilewskija wild type were performed as described in [43]. Statistical significance of differences in volatile emission was determined with SAS9.1 (SAS Institute Inc., Cary, NC, USA) using student's t-test or ANOVA with Tukey post-hoc test.
For an alternative fast sampling and analysis of volatile compounds, 20 inflorescences were placed in a 20 ml screw cap glass vial containing 4 ml of water. Inflorescences were incubated in the sealed vial for 2 h under the conditions described above. Volatile compounds were then trapped by solid phase microextraction (SPME) for 30 min at 40°C and injected into the GC by thermal desorption using an automated SPME sampling device (Combi-PAL, CTC Analytics, Zwingen, Switzerland).
Gene Accession Numbers
Arabidopsis Genome Initiative locus identifiers for the genes studied in this article are as follows: AOS (At5g42650); ARF6 (At1g30330); ARF8 (At5g37020); COI1 (At2g39440); IAA2 (At 3g23030); IAA3 (At1g04240); IAA4 (At5g43700); IAA7 (At3g23050); IAA13 (At2g33310); IAA16 (At3g04730); IAA19 (At3g15540); MYB21 (At3g27810); MYB24 (At5g40350); MYB57 (At3g01530); MYB108 (At3g06490); OPR3 (At2g06050); SAUR63 (At1g29440); AtTPS11 (At5g44630); AtTPS21 (At5g23960).
Supporting Information
Zdroje
1. SmythDRBowmanJLMeyerowitzEM 1990 Early flower development in Arabidopsis. Plant Cell 1 37 52
2. NagpalPEllisCMWeberHPloenseSEBarkawiLS 2005 Auxin Response Factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development 132 4107 4118
3. TabataRIkezakiMFujibeTAidaMTianCE 2010 Arabidopsis AUXIN RESPONSE FACTOR6 and 8 regulate jasmonic acid biosynthesis and floral organ development via repression of class 1 KNOX genes. Plant Cell Physiol 51 164 175
4. WuM-FTianQReedJW 2006 Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression and regulates both female and male reproduction. Development 133 4211 4218
5. FeysBBenedettiCEPenfoldCNTurnerJG 1994 Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 6 751 759
6. IshiguroSKawai-OdaAUedaJNishidaIOkadaK 2001 The DEFECTIVE IN ANTHER DEHISCENCE1 gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell 13 2191 2209
7. SandersPMLeePYBiesgenCBooneJDBealsTP 2000 The Arabidopsis DELAYED DEHISCENCE1 gene encodes an enzyme in the jasmonic acid synthesis pathway. Plant Cell 12 1041 1061
8. StintziABrowseJ 2000 The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc Natl Acad Sci (USA) 97 10625 10630
9. BrioudesFJolyCSzecsiJVaraudELerouxJ 2009 Jasmonate controls late development stages of petal growth in Arabidopsis thaliana. Plant J 60 1070 1080
10. SzecsiJJolyCBordjiKVaraudECockJM 2006 BIGPETALp, a bHLH transcription factor is involved in the control of Arabidopsis petal size. Embo J 25 3912 3920
11. VaraudEBrioudesFSzecsiJLerouxJBrownS 2011 AUXIN RESPONSE FACTOR8 Regulates Arabidopsis Petal Growth by Interacting with the bHLH Transcription Factor BIGPETALp. Plant Cell 23 973 983
12. MandaokarAThinesBShinBMarkus LangeBChoiG 2006 Transcriptional regulators of stamen development in Arabidopsis identified by transcriptional profiling. The Plant Journal 46 984 1008
13. StrackeRWerberMWeisshaarB 2001 The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol 4 447 456
14. ChengHSongSXiaoLSooHMChengZ 2009 Gibberellin acts through jasmonate to control the expression of MYB21, MYB24, and MYB57 to promote stamen filament growth in Arabidopsis. PLoS Genet 5 e1000440 doi:10.1371/journal.pgen.1000440
15. MandaokarABrowseJ 2009 MYB108 acts together with MYB24 to regulate jasmonate-mediated stamen maturation in Arabidopsis. Plant Physiol 149 851 862
16. MengisteTChenXSalmeronJDietrichR 2003 The BOTRYTIS SUSCEPTIBLE1 gene encodes an R2R3MYB transcription factor protein that is required for biotic and abiotic stress responses in Arabidopsis. Plant Cell 15 2551 2565
17. YangXYLiJGPeiMGuHChenZL 2007 Over-expression of a flower-specific transcription factor gene AtMYB24 causes aberrant anther development. Plant Cell Reports 26 219 228
18. LiJYangXWangYLiXGaoZ 2006 Two groups of MYB transcription factors share a motif which enhances trans-activation activity. Biochem Biophys Res Commun 341 1155 1163
19. ShinBChoiGYiHYangSChoI 2002 AtMYB21, a gene encoding a flower-specific transcription factor, is regulated by COP1. Plant J 30 23 32
20. SongSQiTHuangHRenQWuD 2011 The Jasmonate-ZIM Domain Proteins Interact with the R2R3-MYB Transcription Factors MYB21 and MYB24 to Affect Jasmonate-Regulated Stamen Development in Arabidopsis. Plant Cell 23 1000 1013
21. SchmidMDavisonTSHenzSRPapeUJDemarM 2005 A gene expression map of Arabidopsis thaliana development. Nat Genet 37 501 506
22. QinYLeydonARManzielloAPandeyRMountD 2009 Penetration of the stigma and style elicits a novel transcriptome in pollen tubes, pointing to genes critical for growth in a pistil. PLoS Genet 5 e1000621 doi:10.1371/journal.pgen.1000621
23. TashiroSTianCEWatahikiMKYamamotoKT 2009 Changes in growth kinetics of stamen filaments cause inefficient pollination in massugu2, an auxin insensitive, dominant mutant of Arabidopsis thaliana. Physiol Plant 137 175 187
24. Carbonell-BejeranoPUrbezCCarbonellJGranellAPerez-AmadorMA 2010 A fertilization-independent developmental program triggers partial fruit development and senescence processes in pistils of Arabidopsis. Plant Physiol 154 163 172
25. HuJMitchumMGBarnabyNAyeleBTOgawaM 2008 Potential sites of bioactive gibberellin production during reproductive growth in Arabidopsis. Plant Cell 20 320 336
26. ChengHQinLLeeSFuXRichardsDE 2004 Gibberellin regulates Arabidopsis floral development via suppression of DELLA protein function. Development 131 1055 1064
27. KoornneefMvan der VeenJH 1980 Induction and analysis of gibberellin sensitive mutants in Arabidopsis thaliana (L.) Heynh. Theoretical and Applied Genetics 58 257 263
28. GotoNPharisRP 1999 Role of gibberellins in the development of floral organs of the gibberellin-deficient mutant, ga1-1, of Arabidopsis thaliana. Canadian Journal of Botany 77 944 954
29. CaoDChengHWuWSooHMPengJ 2006 Gibberellin mobilizes distinct DELLA-dependent transcriptomes to regulate seed germination and floral development in Arabidopsis. Plant Physiol 142 509 525
30. HagenGGuilfoyleT 2002 Auxin-responsive gene expression: genes, promoters, and regulatory factors. Plant Mol Biol 49 373 385
31. WangJWWangLJMaoYBCaiWJXueHW 2005 Control of Root Cap Formation by MicroRNA-Targeted Auxin Response Factors in Arabidopsis. Plant Cell 17 2204 2216
32. RemingtonDLVisionTJGuilfoyleTJReedJW 2004 Contrasting modes of diversification in the Aux/IAA and ARF gene families. Plant Physiology 135 1738 1752
33. KramBWXuWWCarterCJ 2009 Uncovering the Arabidopsis thaliana nectary transcriptome: investigation of differential gene expression in floral nectariferous tissues. BMC Plant Biol 9 92
34. BowmanJLSmythDR 1999 CRABS CLAW, a gene that regulates carpel and nectary development in Arabidopsis, encodes a novel protein with zinc finger and helix-loop-helix domains. Development 126 2387 2396
35. RuhlmannJMKramBWCarterCJ 2010 CELL WALL INVERTASE 4 is required for nectar production in Arabidopsis. J Exp Bot 61 395 404
36. ChenLQHouBHLalondeSTakanagaHHartungML 2010 Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 468 527 532
37. SeoHSSongJTCheongJJLeeYHLeeYW 2001 Jasmonic acid carboxyl methyltransferase: a key enzyme for jasmonate-regulated plant responses. Proc Natl Acad Sci U S A 98 4788 4793
38. PicherskyEGershenzonJ 2002 The formation and function of plant volatiles: perfumes for pollinator attraction and defense. Curr Opin Plant Biol 5 237 243
39. DeansSGWWatermanPG 1993 Biological activity of volatile oils. HayRKMWatermanPG Volatile Oil Crops: Their Biology, Biochemistry and Production Essex, England Longman Scientific and Technical 97 111
40. ChenFThollDD'AureaJCFarooqAPicherskyE 2003 Biosynthesis and emission of terpenoid volatiles from Arabidopsis flowers. Plant Cell 15 481 494
41. DudarevaNPicherskyEGershenzonJ 2004 Biochemistry of plant volatiles. Plant Physiol 135 1893 1902
42. AubourgSLecharnyABohlmannJ 2002 Genomic analysis of the terpenoid synthase ( AtTPS) gene family of Arabidopsis thaliana. Mol Genet Genomics 267 730 745
43. ThollDChenFPetriJGershenzonJPicherskyE 2005 Two sesquiterpene synthases are responsible for the complex mixture of sesquiterpenes emitted from Arabidopsis flowers. Plant J 42 757 771
44. LorenzoOChicoJMSanchez-SerranoJJSolanoR 2004 JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16 1938 1950
45. ChiniAFonsecaSFernandezGAdieBChicoJM 2007 The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448 666 671
46. CaldelariDWangGFarmerEEDongX 2011 Arabidopsis lox3 lox4 double mutants are male sterile and defective in global proliferative arrest. Plant Mol Biol 75 25 33
47. JensenABRaventosDMundyJ 2002 Fusion genetic analysis of jasmonate-signalling mutants in Arabidopsis. Plant J 29 595 606
48. KubigsteltigILaudertDWeilerEW 1999 Structure and regulation of the Arabidopsis thaliana allene oxide synthase gene. Planta 208 463 471
49. KooAJChungHSKobayashiYHoweGA 2006 Identification of a peroxisomal acyl-activating enzyme involved in the biosynthesis of jasmonic acid in Arabidopsis. J Biol Chem 281 33511 33520
50. StaswickPETiryakiI 2004 The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. Plant Cell 16 2117 2127
51. BellEMulletJE 1993 Characterization of an Arabidopsis lipoxygenase gene responsive to methyl jasmonate and wounding. Plant Physiol 103 1133 1137
52. LaudertDWeilerEW 1998 Allene oxide synthase: a major control point in Arabidopsis thaliana octadecanoid signalling. Plant J 15 675 684
53. DevotoAEllisCMagusinAChangHSChilcottC 2005 Expression profiling reveals COI1 to be a key regulator of genes involved in wound - and methyl jasmonate-induced secondary metabolism, defence, and hormone interactions. Plant Mol Biol 58 497 513
54. SasakiYAsamizuEShibataDNakamuraYKanekoT 2001 Monitoring of methyl jasmonate-responsive genes in Arabidopsis by cDNA macroarray: self-activation of jasmonic acid biosynthesis and crosstalk with other phytohormone signaling pathways. DNA Research 8 153 161
55. HoffmannMHBremerMSchneiderKBurgerFStolleEMortizG 2003 Flower Visitors in a Natural Population of Arabidopsis thaliana. Plant Biol (Stuttg) 5 491 494
56. TanYYXuHHTaoWJHoffmannMHWangXF 2005 Transgenic GFP as a molecular marker for approaches to quantify pollination mechanism and gene flow in Arabidopsis thaliana. Plant Biol (Stuttg) 7 405 410
57. CrawfordBCYanofskyMF 2011 HALF FILLED promotes reproductive tract development and fertilization efficiency in Arabidopsis thaliana. Development 138 2999 3009
58. FriedrichsenDMNemhauserJMuramitsuTMaloofJNAlonsoJ 2002 Three redundant brassinosteroid early response genes encode putative bHLH transcription factors required for normal growth. Genetics 162 1445 1456
59. UlmasovTHagenGGuilfoyleTJ 1999 Activation and repression of transcription by auxin-response factors. Proc Natl Acad Sci USA 96 5844 5849
60. TiwariSBHagenGGuilfoyleTJ 2003 The roles of Auxin Response Factor domains in auxin-responsive transcription. Plant Cell 15 533 543
61. GrayWKepinskiSRouseDLeyserOEstelleM 2001 Auxin regulates SCFTIR1-dependent degradation of AUX/IAA proteins. Nature 414 271 276
62. TatematsuKKumagaiSMutoHSatoAWatahikiMK 2004 MASSUGU2 encodes Aux/IAA19, an auxin-regulated protein that functions together with the transcriptional activator NPH4/ARF7 to regulate differential growth responses of hypocotyl and formation of lateral roots in Arabidopsis thaliana. Plant Cell 16 379 393
63. ZhaoYChristensenSKFankhauserCCashmanJRCohenJD 2001 A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291 306 309
64. ChengYDaiXZhaoY 2006 Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev 20 1790 1799
65. SakataTOshinoTMiuraSTomabechiMTsunagaY 2010 Auxins reverse plant male sterility caused by high temperatures. Proc Natl Acad Sci U S A 107 8569 8574
66. TaoYFerrerJLLjungKPojerFHongF 2008 Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133 164 176
67. WeinigC 2002 Phytochrome photoreceptors mediate plasticity to light quality in flowers of the Brassicaceae. American Journal of Botany 89 230 235
68. CovingtonMFHarmerSL 2007 The circadian clock regulates auxin signaling and responses in Arabidopsis. PLoS Biol 5 e222 doi:10.1371/journal.pbio.0050222
69. JouveLGasparTKeversCGreppinHDegli AgostiR 1999 Involvement of indole-3-acetic acid in the circadian growth of the first internode of Arabidopsis. Planta 209 136 142
70. MichaelTPBretonGHazenSPPriestHMocklerTC 2008 A morning-specific phytohormone gene expression program underlying rhythmic plant growth. PLoS Biol 6 e225 doi:10.1371/journal.pbio.0060225
71. AranaMVMarin-de la RosaNMaloofJNBlazquezMAAlabadiD 2011 Circadian oscillation of gibberellin signaling in Arabidopsis. Proc Natl Acad Sci U S A 108 9292 9297
72. Vivian-SmithAKoltunowAM 1999 Genetic analysis of growth-regulator-induced parthenocarpy in Arabidopsis. Plant Physiol 121 437 451
73. DorceyEUrbezCBlazquezMACarbonellJPerez-AmadorMA 2009 Fertilization-dependent auxin response in ovules triggers fruit development through the modulation of gibberellin metabolism in Arabidopsis. Plant Journal 58 318 332
74. DaviesP 2004 The plant hormones: their nature, occurrence, and functions. DaviesP Plant hormones: Biosynthesis, Signal Transduction, Action Kluwer Academic Publishers 1 15
75. NozueKHarmerSLMaloofJN 2011 Genomic analysis of circadian clock-, light-, and growth-correlated genes reveals PHYTOCHROME-INTERACTING FACTOR5 as a modulator of auxin signaling in Arabidopsis. Plant Physiol 156 357 372
76. SchommerCPalatnikJFAggarwalPChetelatACubasP 2008 Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol 6 e230 doi:10.1371/journal.pbio.0060230
77. Fernandez-CalvoPChiniAFernandez-BarberoGChicoJMGimenez-IbanezS 2011 The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23 701 715
78. NiuYFigueroaPBrowseJ 2011 Characterization of JAZ-interacting bHLH transcription factors that regulate jasmonate responses in Arabidopsis. J Exp Bot 62 2143 2154
79. QiTSongSRenQWuDHuangH 2011 The Jasmonate-ZIM-Domain Proteins Interact with the WD-Repeat/bHLH/MYB Complexes to Regulate Jasmonate-Mediated Anthocyanin Accumulation and Trichome Initiation in Arabidopsis thaliana. Plant Cell 23 1795 1814
80. ItoTNgKHLimTSYuHMeyerowitzEM 2007 The homeotic protein AGAMOUS controls late stamen development by regulating a jasmonate biosynthetic gene in Arabidopsis. Plant Cell 19 3516 3529
81. EllingerDStinglNKubigsteltigIIBalsTJuengerM 2010 DONGLE and DEFECTIVE IN ANTHER DEHISCENCE1 lipases are not essential for wound - and pathogen-induced jasmonate biosynthesis: redundant lipases contribute to jasmonate formation. Plant Physiol 153 114 127
82. SuzaWPStaswickPE 2008 The role of JAR1 in Jasmonoyl-L: -isoleucine production during Arabidopsis wound response. Planta 227 1221 1232
83. BanerjeeSBoseI 2011 Transient pulse formation in jasmonate signaling pathway. J Theor Biol 273 188 196
84. ColquhounTASchwietermanMLWeddeAESchimmelBCMarciniakDM 2011 EOBII Controls Flower Opening by Functioning as a General Transcriptomic Switch. Plant Physiol 156 974 984
85. LiuGRenGGuirgisAThornburgRW 2009 The MYB305 transcription factor regulates expression of nectarin genes in the ornamental tobacco floral nectary. Plant Cell 21 2672 2687
86. Spitzer-RimonBMarhevkaEBarkaiOMartonIEdelbaumO 2010 EOBII, a gene encoding a flower-specific regulator of phenylpropanoid volatiles' biosynthesis in petunia. Plant Cell 22 1961 1976
87. SicardALenhardM 2011 The selfing syndrome: a model for studying the genetic and evolutionary basis of morphological adaptation in plants. Ann Bot 107 1433 1443
88. ChenKYCongBWingRVrebalovJTanksleySD 2007 Changes in regulation of a transcription factor lead to autogamy in cultivated tomatoes. Science 318 643 645
89. LeeSYangKYKimYMParkSYKimSY 2006 Overexpression of PRE1 and its homologous genes activates Gibberellin-dependent responses in Arabidopsis thaliana. Plant Cell Physiol 47 591 600
90. MaraCDHuangTIrishVF 2010 The Arabidopsis floral homeotic proteins APETALA3 and PISTILLATA negatively regulate the BANQUO genes implicated in light signaling. Plant Cell 22 690 702
91. MichelmoreRWParanIKesseliRV 1991 Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci U S A 88 9828 9832
92. AlonsoJMStepanovaANLeisseTJKimCJChenH 2003 Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653 657
93. XuJYangCYuanZZhangDGondweMY 2010 The ABORTED MICROSPORES regulatory network is required for postmeiotic male reproductive development in Arabidopsis thaliana. Plant Cell 22 91 107
94. JohnsonMAvon BesserKZhouQSmithEAuxG 2004 Arabidopsis hapless mutations define essential gametophytic functions. Genetics 168 971 982
95. NakagawaTKuroseTHinoTTanakaKKawamukaiM 2007 Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104 34 41
96. TianQUhlirNJReedJW 2002 Arabidopsis SHY2/IAA3 inhibits auxin-regulated gene expression. Plant Cell 14 301 319
97. EllisCMNagpalPYoungJCHagenGGuilfoyleTJ 2005 AUXIN RESPONSE FACTOR1 and AUXIN RESPONSE FACTOR2 regulate senescence and floral organ abscission in Arabidopsis thaliana. Development 132 4563 4574
98. KarimiMInzeDDepickerA 2002 GATEWAY™ vectors for Agrobacterium-mediated plant transformation. Trends in Plant Science 17 193 195
99. NagpalPWalkerLYoungJSonawalaATimpteC 2000 AXR2 encodes a member of the Aux/IAA protein family. Plant Physiol 123 563 573
100. RedmanJCHaasBJTanimotoGTownCD 2004 Development and evaluation of an Arabidopsis whole genome Affymetrix probe array. Plant J 38 545 561
101. MuellerMJMene-SaffraneLGrunCKargKFarmerEE 2006 Oxylipin analysis methods. Plant J 45 472 489
102. DonathJBolandW 1995 Biosynthesis of acyclic homoterpenes: enzyme selectivity and absolute configuration of the nerolidol precursor. Phytochemistry 39 785 790
103. LeeSBadieyanSBevanDRHerdeMGatzC 2010 Herbivore-induced and floral homoterpene volatiles are biosynthesized by a single P450 enzyme (CYP82G1) in Arabidopsis. Proc Natl Acad Sci U S A 107 21205 21210
Štítky
Genetika Reprodukční medicína
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 2
-
Všechny články tohoto čísla
- Upsetting the Dogma: Germline Selection in Human Males
- A Strong Deletion Bias in Nonallelic Gene Conversion
- Positive Selection for New Disease Mutations in the Human Germline: Evidence from the Heritable Cancer Syndrome Multiple Endocrine Neoplasia Type 2B
- Genome-Wide Association Study in East Asians Identifies Novel Susceptibility Loci for Breast Cancer
- Mixed Effects Modeling of Proliferation Rates in Cell-Based Models: Consequence for Pharmacogenomics and Cancer
- Reduction of NADPH-Oxidase Activity Ameliorates the Cardiovascular Phenotype in a Mouse Model of Williams-Beuren Syndrome
- Genome-Wide Association Study Identifies Chromosome 10q24.32 Variants Associated with Arsenic Metabolism and Toxicity Phenotypes in Bangladesh
- Structural Basis of Transcriptional Gene Silencing Mediated by MOM1
- Genomic Restructuring in the Tasmanian Devil Facial Tumour: Chromosome Painting and Gene Mapping Provide Clues to Evolution of a Transmissible Tumour
- Genome-Wide Association Study Identifies Novel Loci Associated with Circulating Phospho- and Sphingolipid Concentrations
- Contrasting Properties of Gene-Specific Regulatory, Coding, and Copy Number Mutations in : Frequency, Effects, and Dominance
- The Origin and Nature of Tightly Clustered Deletions in Precursor B-Cell Acute Lymphoblastic Leukemia Support a Model of Multiclonal Evolution
- Ultrafast Evolution and Loss of CRISPRs Following a Host Shift in a Novel Wildlife Pathogen,
- Phosphorylation of Chromosome Core Components May Serve as Axis Marks for the Status of Chromosomal Events during Mammalian Meiosis
- Psoriasis Patients Are Enriched for Genetic Variants That Protect against HIV-1 Disease
- A Pathogenic Mechanism in Huntington's Disease Involves Small CAG-Repeated RNAs with Neurotoxic Activity
- The Mitochondrial Chaperone Protein TRAP1 Mitigates α-Synuclein Toxicity
- Homeobox Genes Critically Regulate Embryo Implantation by Controlling Paracrine Signaling between Uterine Stroma and Epithelium
- Developmental Transcriptional Networks Are Required to Maintain Neuronal Subtype Identity in the Mature Nervous System
- Down-Regulating Sphingolipid Synthesis Increases Yeast Lifespan
- Gene Expression and Stress Response Mediated by the Epigenetic Regulation of a Transposable Element Small RNA
- Loss of Tgif Function Causes Holoprosencephaly by Disrupting the Shh Signaling Pathway
- Sequestration of Highly Expressed mRNAs in Cytoplasmic Granules, P-Bodies, and Stress Granules Enhances Cell Viability
- Discovery of a Modified Tetrapolar Sexual Cycle in and the Evolution of in the Species Complex
- The Role of Glypicans in Wnt Inhibitory Factor-1 Activity and the Structural Basis of Wif1's Effects on Wnt and Hedgehog Signaling
- Nondisjunction of a Single Chromosome Leads to Breakage and Activation of DNA Damage Checkpoint in G2
- A Regulatory Network for Coordinated Flower Maturation
- Coexpression Network Analysis in Abdominal and Gluteal Adipose Tissue Reveals Regulatory Genetic Loci for Metabolic Syndrome and Related Phenotypes
- Diced Triplets Expose Neurons to RISC
- The Williams-Beuren Syndrome—A Window into Genetic Variants Leading to the Development of Cardiovascular Disease
- The Empirical Power of Rare Variant Association Methods: Results from Sanger Sequencing in 1,998 Individuals
- Systematic Detection of Epistatic Interactions Based on Allele Pair Frequencies
- Familial Identification: Population Structure and Relationship Distinguishability
- Raf1 Is a DCAF for the Rik1 DDB1-Like Protein and Has Separable Roles in siRNA Generation and Chromatin Modification
- Loss of Function of the Cik1/Kar3 Motor Complex Results in Chromosomes with Syntelic Attachment That Are Sensed by the Tension Checkpoint
- Computational Prediction and Molecular Characterization of an Oomycete Effector and the Cognate Resistance Gene
- The Dynamics and Prognostic Potential of DNA Methylation Changes at Stem Cell Gene Loci in Women's Cancer
- GTPase Activity and Neuronal Toxicity of Parkinson's Disease–Associated LRRK2 Is Regulated by ArfGAP1
- Evaluation of the Role of Functional Constraints on the Integrity of an Ultraconserved Region in the Genus
- Neurophysiological Defects and Neuronal Gene Deregulation in Mutants
- Genetic and Functional Analyses of Mutations Suggest a Multiple Hit Model of Autism Spectrum Disorders
- Negative Supercoiling Creates Single-Stranded Patches of DNA That Are Substrates for AID–Mediated Mutagenesis
- Rewiring of PDZ Domain-Ligand Interaction Network Contributed to Eukaryotic Evolution
- The Eph Receptor Activates NCK and N-WASP, and Inhibits Ena/VASP to Regulate Growth Cone Dynamics during Axon Guidance
- Repression of a Potassium Channel by Nuclear Hormone Receptor and TGF-β Signaling Modulates Insulin Signaling in
- The Retrohoming of Linear Group II Intron RNAs in Occurs by Both DNA Ligase 4–Dependent and –Independent Mechanisms
- Cell Lineage Analysis of the Mammalian Female Germline
- Association of a Functional Variant in the Wnt Co-Receptor with Early Onset Ileal Crohn's Disease
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Gene Expression and Stress Response Mediated by the Epigenetic Regulation of a Transposable Element Small RNA
- Contrasting Properties of Gene-Specific Regulatory, Coding, and Copy Number Mutations in : Frequency, Effects, and Dominance
- Homeobox Genes Critically Regulate Embryo Implantation by Controlling Paracrine Signaling between Uterine Stroma and Epithelium
- Nondisjunction of a Single Chromosome Leads to Breakage and Activation of DNA Damage Checkpoint in G2
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



