-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaGene Expression and Stress Response Mediated by the Epigenetic Regulation of a Transposable Element Small RNA
The epigenetic activity of transposable elements (TEs) can influence the regulation of genes; though, this regulation is confined to the genes, promoters, and enhancers that neighbor the TE. This local cis regulation of genes therefore limits the influence of the TE's epigenetic regulation on the genome. TE activity is suppressed by small RNAs, which also inhibit viruses and regulate the expression of genes. The production of TE heterochromatin-associated endogenous small interfering RNAs (siRNAs) in the reference plant Arabidopsis thaliana is mechanistically distinct from gene-regulating small RNAs, such as microRNAs or trans-acting siRNAs (tasiRNAs). Previous research identified a TE small RNA that potentially regulates the UBP1b mRNA, which encodes an RNA–binding protein involved in stress granule formation. We demonstrate that this siRNA, siRNA854, is under the same trans-generational epigenetic control as the Athila family LTR retrotransposons from which it is produced. The epigenetic activation of Athila elements results in a shift in small RNA processing pathways, and new 21–22 nucleotide versions of Athila siRNAs are produced by protein components normally not responsible for processing TE siRNAs. This processing results in siRNA854's incorporation into ARGONAUTE1 protein complexes in a similar fashion to gene-regulating tasiRNAs. We have used reporter transgenes to demonstrate that the UPB1b 3′ untranslated region directly responds to the epigenetic status of Athila TEs and the accumulation of siRNA854. The regulation of the UPB1b 3′ untranslated region occurs both on the post-transcriptional and translational levels when Athila TEs are epigenetically activated, and this regulation results in the phenocopy of the ubp1b mutant stress-sensitive phenotype. This demonstrates that a TE's epigenetic activity can modulate the host organism's stress response. In addition, the ability of this TE siRNA to regulate a gene's expression in trans blurs the lines between TE and gene-regulating small RNAs.
Published in the journal: . PLoS Genet 8(2): e32767. doi:10.1371/journal.pgen.1002474
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002474Summary
The epigenetic activity of transposable elements (TEs) can influence the regulation of genes; though, this regulation is confined to the genes, promoters, and enhancers that neighbor the TE. This local cis regulation of genes therefore limits the influence of the TE's epigenetic regulation on the genome. TE activity is suppressed by small RNAs, which also inhibit viruses and regulate the expression of genes. The production of TE heterochromatin-associated endogenous small interfering RNAs (siRNAs) in the reference plant Arabidopsis thaliana is mechanistically distinct from gene-regulating small RNAs, such as microRNAs or trans-acting siRNAs (tasiRNAs). Previous research identified a TE small RNA that potentially regulates the UBP1b mRNA, which encodes an RNA–binding protein involved in stress granule formation. We demonstrate that this siRNA, siRNA854, is under the same trans-generational epigenetic control as the Athila family LTR retrotransposons from which it is produced. The epigenetic activation of Athila elements results in a shift in small RNA processing pathways, and new 21–22 nucleotide versions of Athila siRNAs are produced by protein components normally not responsible for processing TE siRNAs. This processing results in siRNA854's incorporation into ARGONAUTE1 protein complexes in a similar fashion to gene-regulating tasiRNAs. We have used reporter transgenes to demonstrate that the UPB1b 3′ untranslated region directly responds to the epigenetic status of Athila TEs and the accumulation of siRNA854. The regulation of the UPB1b 3′ untranslated region occurs both on the post-transcriptional and translational levels when Athila TEs are epigenetically activated, and this regulation results in the phenocopy of the ubp1b mutant stress-sensitive phenotype. This demonstrates that a TE's epigenetic activity can modulate the host organism's stress response. In addition, the ability of this TE siRNA to regulate a gene's expression in trans blurs the lines between TE and gene-regulating small RNAs.
Introduction
Transposable elements (TEs) are mobile fragments of DNA that can accumulate and occupy large fractions of a genome, including over 45% of the human genome [1]. When active, TEs have the potential to create mutations by inserting into genes or generating breaks in DNA. To suppress the mutagenic potential of TEs, the eukaryotic genome has evolved defense mechanisms to inhibit TE proliferation, which are distinct from the developmental regulation of genes [2]. TEs are targeted for epigenetic repression mediated by the overlapping signals of cytosine DNA methylation, repressive histone tail modifications, and remodeling of chromatin into transcriptionally recalcitrant condensed heterochromatin (reviewed in [3]). Gene regulation can be influenced by the epigenetic regulation of TEs; however, this only occurs due to the proximity of a preexisting or newly transposed TE to a gene. This regulation of genes by neighboring TEs in cis can be due to multiple mechanisms, including interruption of a regulatory element, or by local spreading of repressive chromatin modifications such as DNA or histone methylation, resulting in position-effect variegation and potentially the formation of heritable epialleles [4]–[5].
TEs are major producers of small RNAs that act to maintain the TE in an epigenetically silenced state. In plants, and perhaps in animals, heterochromatin modifications are targeted by the activity of small RNAs. For example, in the mouse TE-derived piwi-interacting RNAs (piRNAs) guide DNA methylation to TEs [6]. In the reference plant Arabidopsis thaliana, the cycle of RNA-directed DNA methylation (RdDM) is initiated by the plant-specific RNA Polymerase IV (PolIV), which produces a non-protein coding transcript that is converted into double stranded RNA (dsRNA) by the activity of RNA-dependent RNA Polymerase 2 (RDR2)(reviewed in [7]). Dicer-like 3 (DCL3) cleaves this TE dsRNA into small interfering RNAs (siRNAs) of 24 nucleotides (nt) in length, which are incorporated into either Argonaute 4 (AGO4), AGO6, or potentially AGO9 [8]. These siRNA-loaded Argonaute proteins act to maintain the heterochromatic state of TEs by targeting them for DNA and histone tail methylation.
Athila LTR retrotransposons are the largest family of TEs in Arabidopsis, occupying over 2.7% of the genome [9]. Athila elements are transcriptionally silenced, and silencing is dependent on symmetrical CG DNA methylation. When DNA methylation is removed, either in a DNMT1-homolog maintenance of DNA methylation 1 (met1) mutant, or in a swi/snf family chromatin remodeling protein decrease in DNA methylation 1 (ddm1) mutant, transcriptional activation occurs [10]–[11]. Athila retrotransposons are also transcriptionally activated in the vegetative nucleus of wild-type (wt) pollen grains [12]. In all of these examples heterochromatin modifications and condensation are lost, and global activation of TEs occurs [4], [12]–[13].
Upon global activation of TEs, there are widespread shifts in the accumulation of small RNAs derived from TE transcripts. Transcriptional activation of most silenced TEs results in the loss of their corresponding siRNAs [12]–[14]. However, some retrotransposon families, including Athila, are unusual in the fact that they produce more siRNAs when epigenetically active than when epigenetically silenced [12], [15]–[16]. The Athila siRNAs that increase in abundance are primarily 21–22 nt in length and are produced from the non-protein coding region downstream of the gag and pol ORFs of the consensus Athila element. The abundance and specific location of these siRNAs generates islands of 21–22 nt siRNAs in the genome when epigenetic silencing of Athila is lost [12].
In Arabidopsis, as well as in animals, the production of small RNAs and subsequent targeting of TEs is distinct from the production of gene-regulating small RNAs (reviewed in [17]). The first examples of a TE piRNA or siRNA regulating a genic mRNA in trans were only recently discovered in Drosophila and mouse [18]–[19]. In addition, recently an example of a plant viral siRNA was shown to regulate a gene [20]. However, these examples represent exceptions to the general rule of separation between TE/viral and gene-regulating small RNAs [21]. For example, there is a clear distinction between the biogenesis mechanism and target of TE siRNAs and microRNAs. MicroRNAs act in plants and animals to regulate gene expression on the post-transcriptional or translational level. In Arabidopsis, DCL1 produces 21 nt microRNAs from stem-loop precursor transcripts generated by RNA polymerase II (PolII), and these microRNAs are loaded primarily into AGO1. Thus, microRNAs are not amplified by an RNA-dependent RNA polymerase, and only one or two single small RNA species accumulate from the microRNA locus. In contrast, most plant siRNAs are the products of RNA-dependent RNA polymerases, and cleavage of these long dsRNAs produces clusters of siRNAs from each locus. However, the notion that only microRNAs regulate genes, while endogenous siRNAs do not, is incorrect, as some inverted repeat-derived siRNAs act to regulate genes, [22] and other siRNAs act to regulate genes through the trans-acting siRNA (tasiRNA) pathway in Arabidopsis. This pathway begins with the cleavage of a non-protein coding transcript by the microRNA-loaded AGO1 or AGO7, which initiates the DCL4 - and RDR6-dependent phased production of siRNAs (reviewed in [23]). These siRNAs are loaded into AGO1 and regulate gene expression similar to a microRNA. DCL4, RDR6 and AGO1, as well as DCL2, also act on viral transcripts in the virus-induced gene silencing (VIGS) pathway to initiate and amplify the 21–22 nt siRNA signal that participates in the post-transcriptional degradation of the viral mRNAs, as well as to transport these siRNAs to unaffected regions of the plant to mount a systemic resistance to the spread of the virus [24]–[28]. Therefore, the Arabidopsis AGO1 protein is highly versatile, as it is involved in the microRNA, tasiRNA and VIGS pathways. It is currently unknown if, how or why AGO1 distinguishes between a gene-regulating tasiRNA and a VIGS siRNA involved in viral transcript processing, as both are generated using the same DCL4 and RDR6 machinery.
Arteaga-Vázquez et al demonstrated that 12 elements of the Athila6 subfamily each encode a small RNA, for which they predicted and provided indirect evidence targets a genic transcript for translational repression [29]. They predicted that this small RNA was generated as a microRNA from a stem-loop precursor transcript and determined that it was processed by the microRNA machinery DCL1, HEN1 and HYL1. Additionally, they predicted that this microRNA, which they named microRNA854, targets the 3′ untranslated region (UTR) of the UPB1b gene, a homolog of the mammalian TIA-1 that encodes an RNA binding protein involved in the formation of stress granules [30]–[31]. They observed that 21 nt microRNA854 accumulates in wt vegetative tissues and found that the microRNA854-targeted UPB1b 3′UTR inhibits translation in wt plants when the 3′UTR is added to a reporter transcript. Lastly, Arteaga-Vázquez et al provided evidence that microRNA854 is highly conserved from plants to mammals.
We were unable to detect the accumulation of 21 nt microRNA854 in wt seedling, root, leaf and inflorescence tissues. Due to the failure to meet multiple criteria in order to validate this small RNA as a microRNA [32], including the biogenesis pathway of this small RNA (see below), we have renamed this small RNA siRNA854 to avoid confusion. We have directly demonstrated that the TE-derived siRNA854 regulates in trans the transcript of the UBP1b gene. We show that the accumulation of siRNA854 is under the same trans-generational epigenetic regulation and inheritance patterns to which Athila TEs are subject. Upon Athila6 epigenetic activation, siRNA854 production is shifted from a 24 nt TE siRNA dependent on PolIV and RDR2, to 21–22 nt siRNAs that are dependent upon DCL2, DCL4 and RDR6 and are incorporated into AGO1. We demonstrate that UBP1b regulation is altered only when Athila6 is epigenetically activated, resulting in the phenocopy of the stress-sensitive upb1b mutant phenotype.
Results
Epigenetic activation of Athila6 results in production of Athila siRNAs and siRNA854
To determine when siRNA854 accumulates, we interrogated publicly available deep sequencing small RNA libraries [33] and found that in the plant body of wild-type Columbia ecotype plants (wt Col), 21 nt siRNA854 does not accumulate. Only one read of 21 nt siRNA854 was detected in over 6.6 million genome-matched reads of wt Col leaf and inflorescence small RNAs combined (Table 1). However, when epigenetic repression of Athila6 is lost, 21 nt siRNA854 levels increase. Table 1 shows that, compared to the extremely low levels in wt Col inflorescence and leaf tissue, siRNA854 accumulates in met1 and ddm1 mutant inflorescences. Increased siRNA854 levels were also detected in pollen of wt Col plants, albeit to a lower level of 21 nt siRNA854 reads per million than in met1 or ddm1 mutants.
Tab. 1. Frequency of 21 nt siRNA854 in SBS small RNA libraries. 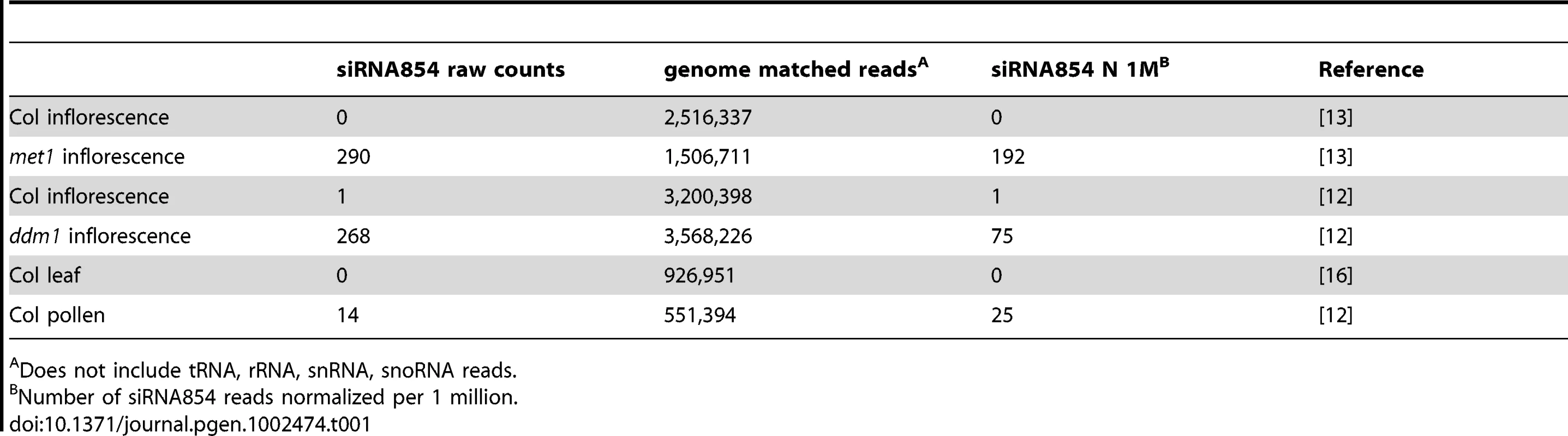
Does not include tRNA, rRNA, snRNA, snoRNA reads. In the plant body, retrotransposons such as Athila6 are tightly epigenetically suppressed by heritable symmetric DNA methylation and RdDM [3]. In each case of 21 nt siRNA854 accumulation (met1, ddm1 and pollen) loss of TE epigenetic silencing is known to occur [11]–[12], [34]–[35]. To determine if the Athila6 retrotransposon is specifically activated in these mutants and pollen, we performed real-time quantitative RT-PCR (qRT-PCR) and found that in met1 and ddm1 mutants and wt pollen, Athila6 transcript accumulation is significantly increased compared to wt Col whole seedlings, leaf and inflorescence tissue (Figure 1A). We used qRT-PCR to measure Athila6 expression (Figure 1), as well as a separate Athila6 primer set specific to the microRNA stem-loop structure previously proposed (Figure S1) [29]. Both primer sets provided similar data, showing that neither the proposed stem-loop nor flanking Athila6 region transcripts accumulate in wt Col seedlings, leaves or inflorescences, while both regions are expressed in ddm1and met1mutants. In addition, Athila6 transcripts accumulate in wt Col pollen (Figure 1A). Compared relatively, pollen has considerably less Athila6 transcript accumulation than either ddm1 or met1 mutants, perhaps due to the fact that pollen TE reactivation only occurs in the pollen vegetative nucleus, one of three nuclei expressing mRNA in mature pollen [12].
Fig. 1. Expression of the Athila6 retrotransposon leads to accumulation of Athila 21–22 nt siRNAs, including siRNA854. 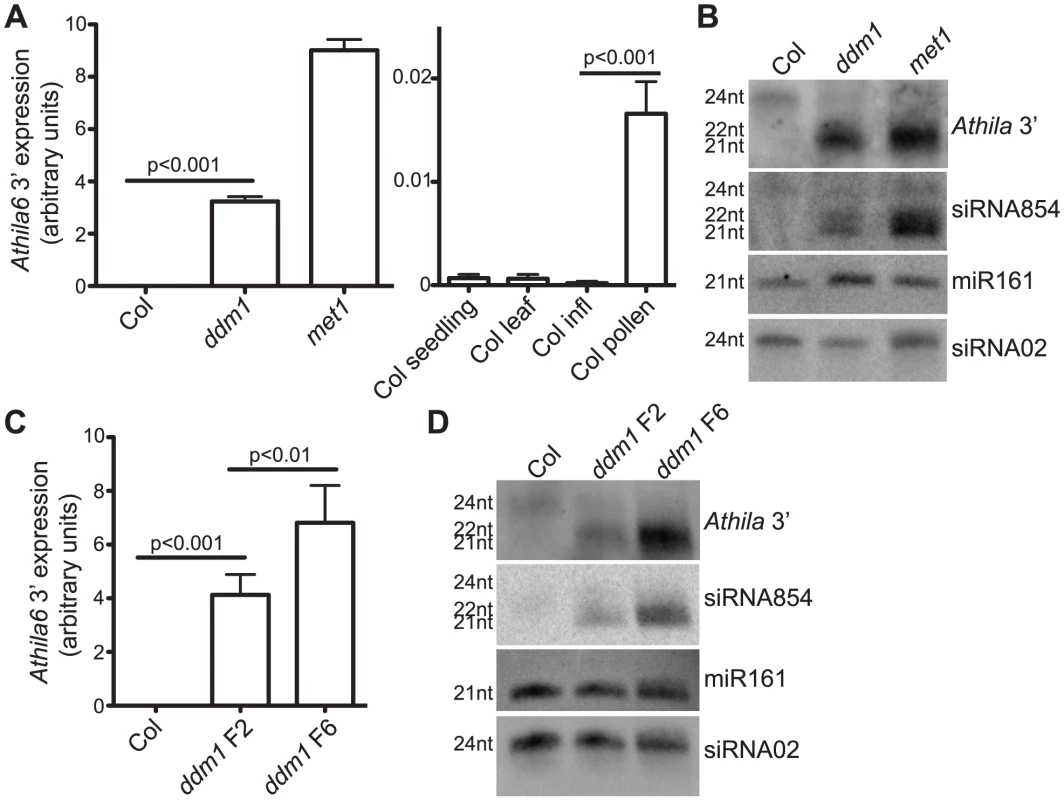
(A) qRT-PCR of the Athila6 retrotransposon, using primers specific for a location 3′ of the gag/pol protein coding region. Expression in wt Col inflorescences (infl) is activated in ddm1 and met1 mutants (left). Expression is also significantly activated (although to a lesser degree) in Col pollen compared to Col whole seedling, leaf and infl. Athila6 transcript accumulation increases >80-fold in wt pollen compared to wt Col infl. Athila6 transcript accumulation increases >24,000-fold in ddm1 mutants compared to wt Col infl expression. (B) Northern blot detecting siRNA854 and the flanking 3′ region of Athila6 (Athila6 3′) in ddm1 and met1 mutant infl. 21–22 nt Athila6 siRNAs and siRNA854 only accumulate when the retrotransposon is transcriptionally activated. The DNA oligonucleotide probe used to detect siRNA854 is 21 nt in length, and is shown in Table S1. (C) qRT-PCR of the Athila6 retrotransposon region 3′ of gag/pol, demonstrating that higher retrotransposon expression levels accumulate when ddm1 is maintained as a homozygote over several generations. (D) Small RNA Northern blot detecting siRNA854 and the Athila6 3′ region. Increased levels of Athila6 21–22 nt siRNAs and siRNA854 correspond to samples with higher transcript levels. For Northern blots in parts B and D, microRNA161 (miR161) and a heterochromatic-region 24 nt siRNA (siRNA02) are shown as loading controls. To examine siRNA854 accumulation in more detail, we performed small RNA Northern blots and found in wt Col and ddm1 and met1 inflorescences, a 24 nt version containing the siRNA854 sequence accumulates, while 21–22 nt versions of this sequence accumulate only in ddm1 and met1 (Figure 1B), as well as in pollen (Table 1). We then probed this Northern blot with a 300 bp siRNA854-flanking region of Athila6 (Athila6 3′ probe) and found that this region also produces other 24 and 21–22 nt siRNAs at levels comparable to those of siRNA854. These results demonstrate that the production of 21–22 nt siRNAs from this entire region is under the same epigenetic regulation as siRNA854, and combined with the results of deep sequencing of small RNAs from ddm1, met1 and pollen [12]–[13], demonstrates that siRNA854 is one member of a larger region of siRNA production. Our data refutes previous data that characterized a specific microRNA produced from this region of the Athila6 retrotransposon [29].
The phenotype of ddm1 mutant plants becomes more severe in progressive generations. Second generation homozygotes for the recessive ddm1-2 allele (ddm1 F2) display little to no morphological phenotype, while after propagation as a homozygote for four additional generations (ddm1 F6), leaf size and infertility phenotypes are severe [36]. Figure 1C shows that increasing transcript accumulation of the Athila6 retrotransposon is associated with the progression of ddm1 from the F2 to F6 generation. To determine if the different transcript levels of Athila6 directly correlate with the accumulation of 21–22 nt siRNA854 and flanking 21–22 nt Athila6 3′ siRNAs, we examined siRNA854 accumulation by Northern blot in ddm1 F2 and F6 individuals. F6 ddm1 individuals produce increased levels of siRNA854 and other Athila6 3′ siRNAs compared to F2 generation ddm1 individuals (Figure 1D). These data, together with the transcript accumulation and siRNA accumulation in met1 and pollen (Figure 1A and 1B, Table 1), suggests that the epigenetic activation and level of Athila6 steady-state transcripts directly and positively correlates with the accumulation level of Athila6 21–22 nt siRNAs, including siRNA854.
siRNA854 biogenesis is atypical for a retrotransposon siRNA and requires RDR6, DCL2, DCL4, and AGO1
To determine the biogenesis mechanism responsible for producing the 21–22 nt versions of Athila siRNAs and siRNA854, we first screened four tissues of wt Col and ddm1 mutant plants and determined that Athila6 21–22 nt siRNAs are not detected in wt Col seedlings, roots, leaves or inflorescences (Figure 2A). Athila6 21–22 nt siRNAs are most easily detectable in ddm1 inflorescence tissue, while leaf and seedling tissues have lower relative levels, and the siRNAs are undetectable in roots (Figure 2A). In wt Col, 24 nt Athila6 siRNAs weakly accumulate in the root and inflorescence (Figure 2A), and these inflorescence 24 nt siRNAs are dependent on the PolIV component NRPD1A, RDR2, and the small RNA-modifying protein HEN1 (Figure 2B). HEN1 is responsible for the accumulation of both microRNAs and siRNAs [37], while the requirement of NRPD1A and RDR2 demonstrates that, like other known 24 nt TE siRNAs, Athila6 24 nt siRNAs are generated by the RdDM pathway which is responsible for maintaining epigenetically silenced regions of the genome [21], [38]. The 24 nt siRNA854 accumulation in wt Col inflorescences is not dependent on RDR6, the PolV component NRDE1, or the microRNA processing DCL1 (Figure 2B). Contrary to previously published results, these data demonstrate that there is no siRNA854 version in wt Col inflorescences that is dependent on the microRNA machinery.
Fig. 2. Biogenesis of Athila6 siRNAs and siRNA854. 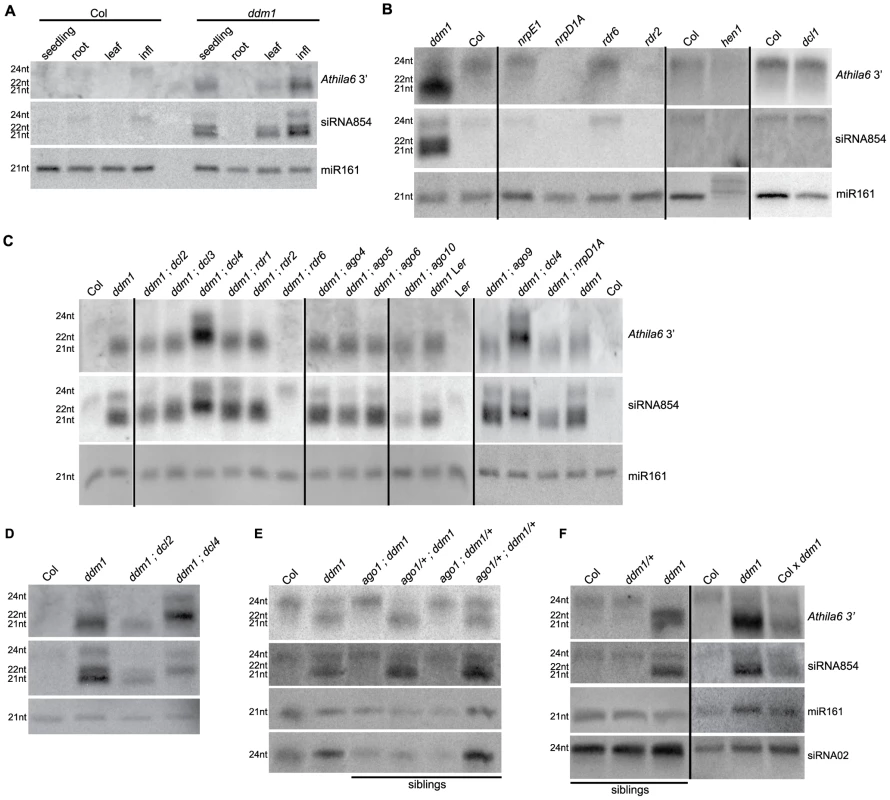
All parts correspond to Northern blots detecting siRNA854 and a flanking region of Athila6 3′ of the gag/pol protein coding region. (A) In wt Col, only the 24 nt versions of siRNA854 and Athila6 siRNAs accumulate, while in ddm1 seedling, leaf and infl, 21 nt and 22 nt versions of siRNA854 and flanking Athila6 siRNAs accumulate. (B) Biogenesis pathway of the 24 nt Athila6 siRNAs. hen1, rdr2 and nrpD1A mutants fail to accumulate Athila6 24 nt siRNAs. (C) Biogenesis pathway of the 21–22 nt Athila6 siRNAs that accumulate upon transcriptional activation. ddm1 double mutants were generated for mutants involved in known siRNA production pathways. 21–22 nt Athila6 3′ siRNAs and siRNA854 fail to accumulate in ddm1;rdr6 double mutants, while the 21 nt siRNAs shift to 22 nt in ddm1;dcl4 double mutants. (D) Higher resolution Northern showing that the 21 nt version of siRNA854 and Athila6 3′ siRNAs are absent in ddm1;dcl4 double mutants, while the 22 nt version is absent in ddm1;dcl2 double mutants. (E) The accumulation of Athila6 3′ siRNAs in a ddm1;ago1 segregating family produced from ddm1 homozygote P1 plants. AGO1 is necessary for the accumulation of 21–22 nt siRNA854 and Athila6 3′ siRNAs from both ddm1 homozygotes and ddm1 heterozygotes (ddm1/+). (F) 21–22 nt Athila6 3′ siRNAs and siRNA854 accumulate in ddm1 heterozygotes produced by crossing wt to an individual homozygous for the recessive ddm1-2 allele (Col x ddm1). ddm1/+ heterozygous plants produced from parents that have not been homozygous for ddm1 for at least 6 generations do not accumulate Athila6 21–22 nt siRNAs and are shown as a control (ddm1/+ in segregating family). To determine the small RNA biogenesis pathway responsible for producing 21–22 nt siRNA854 and Athila6 siRNAs when Athila is epigenetically activated, we generated 12 double mutant combinations with ddm1, using mutants for genes with known roles in the various Arabidopsis small RNA biogenesis pathways, such as different AGO, DCL and RDR genes. Double mutant inflorescences were used to assay the accumulation of siRNA854 (Figure 2C). As some of these double mutants are in Col x Landsberg erecta (Ler) mixed genetic backgrounds, as a control we confirmed that Ler ddm1 mutants also accumulate Athila6 siRNAs, while wt Ler does not. We found that the RdDM pathway responsible for producing Athila6 24 nt siRNAs involving NRPD1A and RDR2 does not generate the 21 nt or 22 nt siRNA854. This represents a change in siRNA biogenesis pathways for Athila siRNAs, as their epigenetic reactivation results in a new set of proteins responsible for the 21–22 nt siRNA production. We determined that RDR6 function is required for both 21 nt and 22 nt siRNA854 accumulation, as in ddm1;rdr6 double mutants neither of these siRNAs accumulate (Figure 2C), while RDR6 is not responsible for the 24 nt version of these siRNAs (Figure 2B). RDR6's involvement suggests that an Athila6 transcript is copied into dsRNA, which is required for 21–22 nt siRNA production. We also found that in ddm1;dcl4 double mutants, the 21 nt siRNA854 is absent, while increased levels of the 22 nt and 24 nt versions are detected. There are well-described hierarchical relationships among DCL2, DCL3, and DCL4. When DCL4 is not present to generate 21 nt siRNAs, DCL2 primarily substitutes for this function and generates 22 nt siRNAs, while DCL3 substitutes for DCL4 to a lesser degree and produces 24 nt siRNAs [39]. Conversely, ddm1;dcl2 double mutants lose the 22 nt version of Athila6 siRNAs, including siRNA854 (Figure 2D), demonstrating that the 21 nt and 22 nt versions of siRNA854 that accumulate in ddm1 mutants are generated by the activity of DCL4 and DCL2, respectively. The production of 21 nt or 22 nt siRNA854 in either ddm1;dcl2 or ddm1;dcl4 double mutants suggests that the processing by DCL4 and DCL2 proteins occurs after RDR6 converts the Athila6 transcript into dsRNA. In addition, the proteins responsible for the biogenesis of the 24 nt version of siRNA854 and, separately, for the 21–22 nt version are identical to those responsible for generating the corresponding sizes of the flanking Athila6 siRNAs, further indicating that siRNA854 is not solely or specifically cleaved from this region.
To determine which Argonaute protein(s) are responsible for siRNA854 accumulation, we tested ddm1 double mutants with ago1, ago4, ago5, ago6 and ago10. ddm1 double mutants with ago4, ago5, and ago6 did not result in loss of siRNA854, and ago10 double mutants displayed only reduced accumulation (Figure 2C). In the ddm1;ago1 double mutant both the 21 nt and 22 nt versions of siRNA854 fail to accumulate (Figure 2E), demonstrating that AGO1 is essential for 21–22 nt siRNA854 accumulation. The requirement of RDR6, DCL2, DCL4 and AGO1 suggests that either the known VIGS pathway of post-transcriptional degradation of viral RNAs, or the tasiRNA pathway is responsible for Athila6 21–22 nt siRNA biogenesis.
While generating the ddm1 double mutant plants, we encountered an unusual pattern of inheritance of Athila6 siRNAs, which stems from the atypical genetic inheritance of ddm1 mutants. For example, both the ddm1 mutant phenotype and Athila6 expression become more severe over increasing generations (Figure 1C) [36]. In addition, ddm1/+ heterozygote plants produced by crossing a plant homozygous for the ddm1-2 recessive allele to wt Col inherit epigenetically decondensed and transcriptionally uncontrolled chromatin from the ddm1 parent, which is not fully restored in the ddm1/+ heterozygote [40]–[42]. This mutant chromatin in a ddm1/+ heterozygous individual continues to express TEs [42]. In Figure 2F we demonstrate that a ddm1/+ heterozygote produced from a ddm1 homozygous parent (Col x ddm1 in Figure 2F) still produces 21–22 nt siRNA854 and Athila6 3′ siRNAs, although to a considerably lower level than the ddm1 homozygote. This is in contrast to a ddm1/+ heterozygote that was not the progeny of a homozygous parent, but has been backcrossed to wt Col for six generations while being maintained as a heterozygote. In this ddm1/+ heterozygote (ddm1/+ in segregating family, Figure 2F) the amount of mutant chromatin inherited from the ddm1 homozygous parent has been diluted away by segregation in each generation of crossing to wt Col, demonstrating that there is an effect of the parent's genotype on the production of Athila6 siRNAs in ddm1/+ heterozygous plants. The requirement of AGO1 for the production of 21–22 nt siRNA854 in ddm1/+ heterozygotes was demonstrated using an F2 family segregating for ago1 and ddm1, which was produced from a ddm1 homozygous P1 individual (Figure 2E). In ago1 mutants that are ddm1/+ heterozygotes (ago1;ddm1/+), neither 21 nor 22 nt versions of siRNA854 accumulate, while they do in the corresponding ago1/+;ddm1/+ double heterozygote siblings. These data demonstrate that AGO1 is necessary for siRNA854 accumulation in ddm1 mutants and in the progeny of ddm1 homozygotes.
siRNA854 accumulation represses reporter gene transcripts with the UBP1b 3′UTR
The 21 nt version of siRNA854 was previously predicted to target the 3′UTR of the UBP1b gene in four locations using modified criteria that allows for non-canonical or ‘wobble’ G:U base pairing [29]. G:U base pairing has been demonstrated to be tolerated in microRNA target sites, even within the critical first 7 base pairs (bp) or ‘seed’ pairing region [43]. However, the targeting of the 3′UTR by small RNAs was not previously shown directly, and complementarity of siRNA854 to the UBP1b 3′UTR relies heavily on non-canonical base pairing and lacks a strong 7 bp seed-pairing region (shown in Figure S2). To directly test if the 21 nt siRNA854 sequence has the ability to target the UBP1b 3′UTR, we took advantage of the fact that wt Col inflorescences do not accumulate 21–22 nt siRNA854 (Table 1, Figure 1, Figure 2). To directly examine the role of the siRNA854 sequence, we constructed plants constitutively expressing a GUS reporter gene with the GUS mRNA fused to the UBP1b 3′UTR and transformed these plants with artificial microRNA (amiRNA) constructs expressing the siRNA854 sequence, or an unrelated sequence as a control, from the Arabidopsis microRNA319a stem-loop transcript [44]. Quantitative assays to detect GUS activity, as well as qualitative plant staining, demonstrate that the plants with the control amiRNA have high levels of GUS activity, while plants expressing the siRNA854 sequence from a microRNA stem-loop display significantly lower levels of GUS activity (Figure 3A). These data demonstrate that although the alignment of siRNA854 to the UBP1b 3′UTR lacks a strong seed pairing region and relies on G:U base pairing, the 21 and/or 22 nt siRNA854 sequence can target the UBP1b 3′UTR resulting in decreased reporter protein accumulation.
Fig. 3. Accumulation of 21–22 nt siRNA854 negatively regulates transgene transcripts with the UBP1b 3′ UTR. 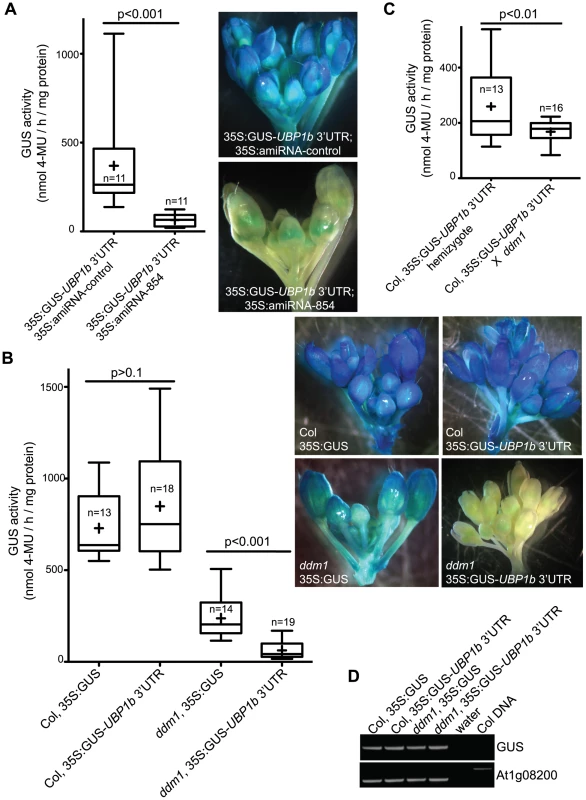
(A) Plants homozygous for a transgene constitutively expressing the GUS reporter protein fused to the UBP1b 3′ UTR (35S:GUS-UBP1b 3′UTR) were transformed with an artificial microRNA (amiRNA) with the siRNA854 sequence (35S:amiRNA-854), or a second control sequence that does not target UBP1b (35S-amiRNA-control). GUS activity was monitored using a quantitative assay (left) and inflorescence staining (right). Plants expressing the siRNA854 sequence as an artificial microRNA show significantly reduced GUS levels. (B) Col and ddm1 plants homozygous for a constitutively expressed GUS transgene (35S:GUS) or the same reporter transgene with the UBP1b 3′ UTR from part A. As in A, GUS activity was monitored using a quantitative assay (left) and plant staining (right). Wt Col plants show no differential regulation between the two transgene variations, while in the ddm1 mutant background the GUS activity of the UBP1b 3′ UTR transgene is significantly less than the control 35S:GUS transgene. (C) A 35S:GUS-UBP1b 3′UTR transgene in the wt Col background was crossed to a ddm1 homozygote, and the GUS activity was measured in the F1 plant. GUS activity of the same hemizygous transgene in the wt Col background is shown as a control. (D) RT-PCR of the same transgenic individuals from part B. The GUS activity differences observed in part B are not reflected in transgene transcript levels, demonstrating that this regulation is not due to post-transcriptional mRNA degradation. In A, B and C, the box plot whiskers represent the minimum and maximum of the dataset, the top and bottom of the box are the 75th and 25th percentile (respectively), the middle line is the median, and + is the mean. The number of individuals assayed (n) is shown in or near the box. The developmental expression profiles of UBP1b and the six Athila6 elements on the Affymetrix ATH1 gene expression microarray are negatively correlated, with UPB1b expressed highly in all wt tissues except mature pollen, specifically where Athila6 activation occurs (Figure S3A). To determine if the increased levels of endogenous 21–22 nt siRNA854 observed when Athila6 is epigenetically activated can regulate the UBP1b 3′UTR, we transformed both wt Col and ddm1 plants with either the GUS-UBP1b 3′UTR transgene from Figure 3A, or a control transgene without the UBP1b 3′UTR. We assayed GUS activity in plants homozygous for the transgenes and found that in wt Col, the presence of the UBP1b 3′UTR did not affect GUS activity (Figure 3B). In contrast, when this same analysis was previously published, the same GUS-UPB1b 3′UTR transgene in a wt Col plant resulted in little to no GUS protein production in leaves and inflorescences [29]. Our data, which demonstrate no inhibition of the UBP1b 3′UTR in wt Col leaves and inflorescences, are supported by the fact that there is no 21/22 nt siRNA854 detected in leaves or inflorescence by either Northern blot (Figure 1, Figure 2A) or small RNA deep sequencing (1 read in a combined 6.6 million)(Table 1).
In ddm1 mutants, the GUS-UBP1b 3′UTR and GUS control (no 3′UTR) transgenes both display reduced GUS activity (Figure 3B). It remains enigmatic why the constitutively expressed GUS transgene without a 3′UTR has reduced expression in ddm1 compared to wt Col. However, the presence of the UBP1b 3′UTR resulted in a significant reduction of GUS activity compared to the no 3′UTR control in ddm1 (Figure 3B). To make sure that position effects of these transgene insertions were not the cause of this differential regulation, we crossed a wt Col plant with the UBP1b 3′UTR transgene that displayed high GUS activity to a ddm1 homozygote, as the resulting heterozygote will have siRNA854 accumulation (Figure 2F). The GUS activity in this ddm1 heterozygote is significantly reduced compared to both the wt Col homozygous GUS transgene parent and to wt Col plants heterozygous for the same GUS transgene (Figure 3C). Therefore, the regulation of the UBP1b 3′UTR is sensitive to the levels of siRNA854, with either the production of this sequence as an amiRNA, or accumulation of this sequence as an siRNA in ddm1 resulting in repression of GUS activity. We determined that all of the transgenes in either wt Col or ddm1 from Figure 3B have GUS transcripts that accumulate to similar levels (Figure 3D), indicating that the regulation of these transgene transcripts is not due to post-transcriptional degradation of the GUS RNA, but is likely rather due to the inhibition of translation of these mRNAs.
In addition to the increased levels of siRNA854 in ddm1 mutants, siRNA854 also accumulates in wt Col pollen (Table 1). To determine if endogenous siRNA854 in pollen is able to regulate the UBP1b 3′UTR, we performed similar transgene reporter assays as above in wt Col pollen. We used a pollen vegetative cell promoter to drive GFP and added one of three different 3′UTRs to these reporter transgenes. GFP fluorescence was quantitatively measured by subtracting the fluorescence of segregating pollen grains that did not inherit the transgene from the fluorescence of pollen grains that did inherit the transgene. With no 3′UTR, transgene protein accumulates, and a moderate level of fluorescence is observed (Figure 4). When the wt UBP1b 3′UTR is added to this transgene, significantly less fluorescence is observed, likely due to the accumulation of siRNA854 in wt Col pollen. To test if the binding of siRNA854 was specifically responsible for this regulation, we generated a version of the UBP1b 3′UTR that lacks all four of the siRNA854 predicted target sites, resulting in a shorter 3′UTR (shown in Figure S2). This deleted 3′UTR transgene (DEL transgene) resulted in significantly increased fluorescence compared to the wt UBP1b 3′UTR (Figure 4). We also produced a UBP1b 3′UTR variation of the same length as the wt UBP1b 3′UTR, in which each of the perfectly complementary base pairs in all four of the siRNA854 predicted target sites have been switched to bases that do not show complementarity (or G:U pairing) with siRNA854 (Figure S2). Pollen grains with the base-modified 3′UTR (MOD transgene) on the GFP transcript display significantly increased fluorescence compared to the wt UBP1b 3′UTR (Figure 4), demonstrating that these bases are necessary for the regulation of the UBP1b 3′UTR. Pollen from both the MOD and DEL 3′UTR transgenes display fluorescence levels even higher than the control lacking a 3′UTR, likely due to the ability of the UBP1b 3′UTR, when not targeted by small RNAs, to stabilize transcripts or promote their translation. Lastly, we transformed the GFP transgene with and without the wt UBP1b 3′UTR into rdr6 mutants. We observed that the expression of Lat52:GFP (no 3′UTR) in rdr6 mutant pollen is higher than that of the same transgene in wt Col pollen (Figure 4). This difference is likely due to the role of RDR6 in post-transcriptional silencing of transgenes [45]. We speculate that the wt Col Lat52:GFP transgene is subject to a certain low amount of post-transcriptional regulation mediated by RDR6. When this transgene is present in rdr6 mutant plants, this post-transcriptional regulation does not occur, resulting in higher expression of the transgene compared to wt Col. Interestingly, we did not observe a reduction in pollen fluorescence for the Lat52:GFP-UBP1b 3′UTR transgene in rdr6 compared to the no-3′UTR control transgene in rdr6 (Figure 4), demonstrating that RDR6 is necessary for the targeting of the UBP1b 3′UTR in pollen. The combined data from Figure 3 and Figure 4 demonstrate that the RDR6-dependent accumulation of siRNA854 and base pairing with the UBP1b 3′UTR target sites are required for the inhibitory regulation of UBP1b 3′UTR reporter genes.
Fig. 4. Accumulation of 21–22 nt siRNA854 in wt pollen regulates transgene transcripts with the UBP1b 3′UTR. 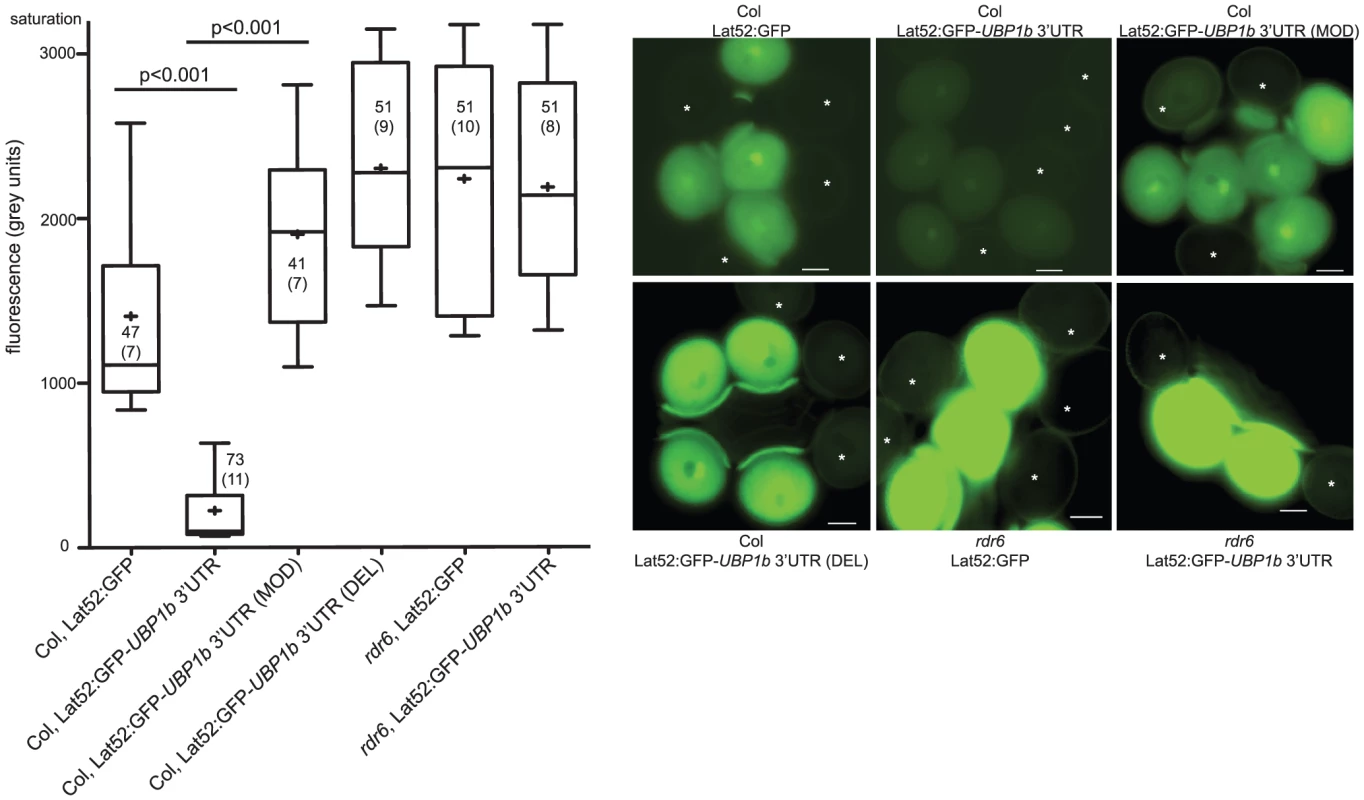
GFP transgenes with pollen-specific expression were used to assay the activity of endogenous siRNA854 in pollen. Background corrected GFP fluorescence levels are shown on the left, while representative pollen images are on the right. On each image, an asterisk marks non-fluorescent pollen grains that did not inherit the transgene from the hemizygous parent that were used for background correction. The addition of the UBP1b 3′UTR to a GFP transgene results in reduced fluorescence. Abrogation of the perfectly complementary base pairing in the predicted siRNA854 target sites of the UBP1b 3′UTR (MOD), or removal of these target sites altogether (DEL), alleviates this repression. The repression of the UBP1b 3′ UTR in pollen is lost in rdr6 mutant plants. Box plot whiskers represent the 90th and 10th percentile of the dataset, the top and bottom of the box are the 75th and 25th percentile (respectively), the middle line is the median, and + is the mean. Number of pollen grains measured and the number of transgenic individuals they came from (in parentheses) is shown in or near the box. Scale bars = 10 µm. siRNA854 represses endogenous UBP1b in pollen
We aimed to determine if siRNA854 has a regulatory effect on the endogenous UBP1b gene or transcript. To aid our characterization of UBP1b we isolated two mutant upb1b alleles, which are in the Ws background. UBP1b insertion alleles result in a lack of polyadenylated transcripts, although un-spliced and non-polyadenylated transcripts are still produced (Figure S4).
First, we wondered if the sequence similarity between the 21, 22 or 24 nt versions of siRNA854 was directing DNA methylation to the UBP1b 3′UTR through the RdDM pathway, as a possible mechanism of epiallele production. We have determined that the DNA methylation status of the UBP1b 3′UTR is not altered in ddm1 inflorescences relative to wt Col (Figure S5A). Next, we utilized microarray data and RT-PCR to analyze UPB1b transcript levels, and we found they accumulate to the highest levels in inflorescence tissue, intermediate levels in seedling and leaf tissues, and either to extremely low levels or not at all in wt Col pollen (Figure S3). We measured UBP1b transcript accumulation in ddm1 mutants at two developmental time points: inflorescence buds and mature pollen. In inflorescence tissue, the transcript accumulation of UPB1b is not significantly altered in ddm1 F2 or ddm1 F6 inflorescences (Figure 5A). Therefore, qRT-PCR expression analysis and DNA methylation analysis both demonstrate that in inflorescence tissue there is no transcriptional or post-transcriptional effect of siRNA854 on UBP1b transcript accumulation. We continued to assay UBP1b in inflorescence tissue of wt Col and ddm1 supposing that the regulation may be on the translational level, as was observed for the inflorescences of the GUS-UBP1b 3′UTR transgene transcript in Figure 3. We assayed two known microRNA-induced alterations to transcripts associated with translational regulation (reviewed in [46]). We determined that in ddm1 inflorescence tissue the polyA tail length of UBP1b is unaffected, and uncapped transcript does not accumulate (Figure S5B and S5C). Without the availability of a specific antibody to assay endogenous UBP1b protein accumulation, we can provide no direct evidence that endogenous UPB1b transcripts are regulated by the elevated siRNA854 levels that accumulate in ddm1 inflorescences.
Fig. 5. Tissue-specific regulation of UBP1b by siRNA854. 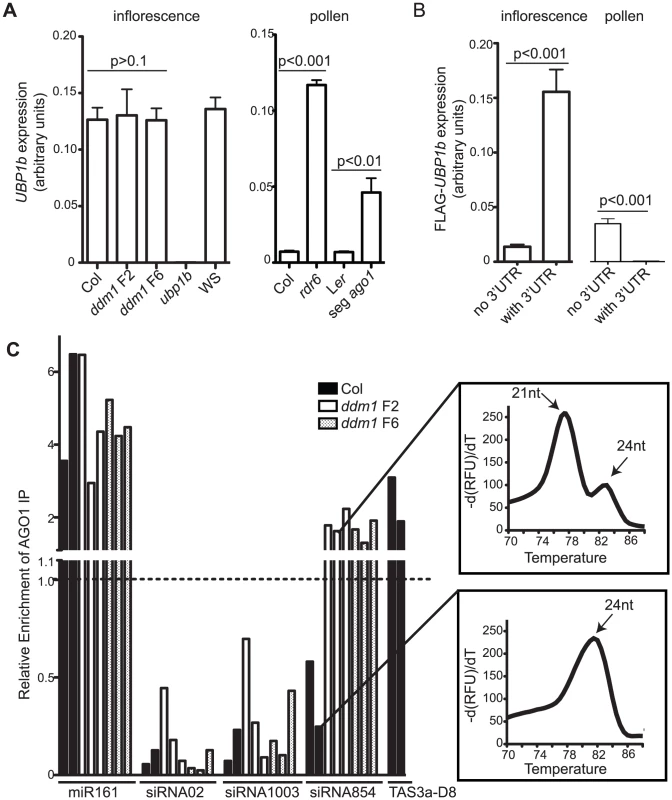
(A) qRT-PCR of UBP1b in inflorescence tissue and mature pollen. In inflorescence tissue, UBP1b transcript accumulation is not affected in ddm1 mutants. In wt Col pollen, UBP1b expression significantly increases in rdr6 mutants and in a pool of pollen that is segregating 1∶1 ago1 mutant pollen (seg ago1). (B) qRT-PCR of FLAG-tagged UBP1b transgenes with and without the UBP1b 3′UTR and under control of their native promoters. In inflorescence tissue, the addition of the UBP1b 3′UTR results in increased transcript levels. In pollen, the UBP1b promoter is active, and addition of the UBP1b 3′UTR results in significantly decreased levels of mRNA. (C) qRT-PCR of small RNAs from AGO1-IP biological replicates demonstrating that in the plant body of ddm1 mutants siRNA854 is enriched in AGO1 protein complexes, while it is not in wt Col. Relative enrichment values over 1.0 indicate AGO1-enrichment, whereas relative enrichment values under 1.0 indicate no enrichment. Relative enrichment was calculated based on amplification of the input sample for each IP. MiR161 and TAS3a-D8 are shown as positive controls while siRNA02 and siRNA1003 are 24 nt siRNA negative controls not bound by AGO1. qRT-PCR melting curves for siRNA854 amplification products from the AGO1-IP demonstrate that the non-enriched siRNA854 in wt Col is the larger 24 nt version (higher melting temperature), while the AGO1-enriched siRNA854 in ddm1 is the 21–22 nt version (lower melting temperature). In contrast to inflorescence tissue, the transcript accumulation of UBP1b in pollen is regulated at the post-transcriptional level by siRNA854. In wt Col pollen, the UBP1b transcript does not accumulate (Figure S3). To determine if this is a consequence of the increased levels of siRNA854 in pollen, or if the UBP1b promoter is simply not active in mature pollen, we performed qRT-PCR in rdr6 mutant plants as well as from plants heterozygous for ago1. Plants homozygous for the recessive ago1-11 allele do not produce pollen, so we used an ago1-11/+ heterozygote that produces pollen segregating 1∶1 wt and mutant for ago1. In both rdr6 pollen and ago1 segregating pollen there is a significant increase in UBP1b transcript accumulation, with rdr6 having a >16-fold increase (Figure 5A). This demonstrates that the UBP1b promoter is active in pollen, but the transcripts are subject to post-transcriptional degradation. Attempts at identifying siRNA854-induced cleavage sites in the UPB1b 3′UTR from inflorescence and pollen were inconclusive (data not shown), likely due to the high rate of non-small RNA-induced processing and degradation of the endogenous UBP1b 3′UTR detected in whole genome degradome analysis [47]–[48].
To determine if the UBP1b 3′UTR is specifically responsible for the differential UBP1b accumulation in inflorescence and pollen, we generated two transgenes with the native UBP1b promoter and coding region, with and without the 3′UTR. This transgene also has a 5′ FLAG epitope tag to distinguish it from the endogenous UBP1b. We found that the presence of the UBP1b 3′UTR in inflorescence tissue increases the transcript accumulation levels >11-fold, presumably by stabilizing this transcript (Figure 5B). In wt Col pollen the UBP1b promoter is active, and without the 3′UTR this transcript accumulates to levels 4-fold less compared to inflorescence tissue. However, in contrast to inflorescence tissue, the addition of the UBP1b 3′UTR resulted in >73-fold reduced transcript accumulation in wt pollen. Together, these data demonstrate that in wt Col pollen the presence of the UBP1b 3′UTR causes a decrease in UBP1b transcript accumulation.
The mature pollen grain contains two sperm cells with highly condensed chromatin imbedded into the larger vegetative cell, which displays a chromatin-decondensed vegetative nucleus. Communication between the vegetative cell and imbedded sperm cells has been previously hypothesized to occur (reviewed in [49]). To determine in which cell post-transcriptional silencing in the pollen grain is taking place, we aimed to decipher where in the mature pollen grain the repression of the UBP1b 3′UTR is occurring. Since we demonstrated that both RDR6 and AGO1 are necessary for the repression of the endogenous UBP1b transcript in the mature pollen grain (Figure 5A), we examined the localization of the RDR6 and AGO1 proteins by fusing them to GFP and expressing them from their native promoters. We found that both of these proteins localize to the nucleus and cytoplasm of the pollen vegetative cell and are not detectable in sperm cells (Figure S6), in agreement with mined microarray data from purified sperm cells [50]. The pollen vegetative cell is also the location of epigenetic TE reactivation [12], suggesting that the activation of Athila6, cleavage into siRNAs and potentially the repression of the UBP1b 3′UTR are all occurring in this cell.
Since a functional AGO1 protein is required for the accumulation of siRNA854 (Figure 2E), and UBP1b transcript levels increase in segregating mutant ago1 pollen (Figure 5A), we aimed to determine if siRNA854 is incorporated into AGO1 protein complexes. We performed an immunoprecipitation (IP) of AGO1 complexes using a commercially available AGO1 antibody and purified the incorporated small RNA. To verify the specificity of the AGO1 antibody, we demonstrated that this antibody does not detect any other proteins by first performing a Western blot on protein extracts from Col, Ler, and ago1-11 inflorescences. We found that the AGO1 antibody yields no cross-reactive bands (Figure S7A). Additionally, we used Western blot analysis to confirm the success of the IP by both detecting the presence of AGO1 protein in the IP Input extract and AGO1 IP samples, and observing the absence of AGO1 protein in the no antibody IP control (Figure S7B). After the IP, we purified the AGO1-bound small RNAs and used qRT-PCR to assay levels of siRNA854, a known AGO1-incorporated microRNA (miR161), a known AGO1-incorporated tasiRNA (TAS3a-D8), and two 24 nt heterochromatic siRNAs not present in AGO1 complexes (siRNA02 and siRNA1003) [51]–[52]. We found no AGO1-IP enrichment of siRNA854 or the control siRNAs, siRNA02 and siRNA1003, in the wt Col plant body, while we did detect enrichment of the control microRNA miR161 and control tasiRNA TAS3a-D8 (Figure 5C). In contrast, in ddm1 F2 and F6 plants we found enrichment of siRNA854 in AGO1. Analysis of the melting curves generated from the products of the qRT-PCR demonstrate that the background levels of siRNA854 from wt Col plants are the larger 24 nt version (which have higher melting temperatures) compared to the AGO1-enriched 21–22 nt siRNA854 from ddm1 plants (lower melting temperatures)(Figure 5C). The level of enrichment of siRNA854 in AGO1 complexes in ddm1 mutants is not as high as miR161, but is more similar to the level of enrichment of the tasiRNA TAS3a-D8 in wt Col (Figure 5C), likely due the fact that both siRNA854 and TAS3a-D8 are single siRNAs from transcripts that generate multiple siRNAs through the activity of RDR6 and DCL4. In addition, we detected no difference in enrichment levels between ddm1 F2 and ddm1 F6 plants. However, since there are higher levels of siRNA854 in the ddm1 F6 plants (Figure 1D) and input (non-IP) sample that was used for normalization, there are likely more AGO1 protein complexes interacting with siRNA854 in F6 ddm1 plants compared to the F2 generation. These data demonstrate that only when epigenetically activated, the Athila6-generated 21–22 nt siRNA854 is incorporated into AGO1, and this complex is responsible for the regulation of the UBP1b transcript.
Altered stress-regulation in ddm1 mutants
TIA-1 has a known role in the sensing and response to cellular stress, and mutant cells unable to form stress granules display increased sensitivity to stress [53]–[55]. We have experimentally determined that the germination and growth of ubp1b mutant plants are also sensitive to both ionic (+NaCl) and osmotic (+mannitol) stress conditions compared to its corresponding wt background Ws, particularly when grown on 100 mM NaCl or 300 mM mannitol (Figure 6A). In addition, wt Ws itself is more sensitive to these stresses than wt Col, as at higher NaCl or mannitol concentrations, wt Col survives but wt Ws does not. Since ddm1 plants have increased levels of siRNA854 (Table 1, Figure 1), and siRNA854 can target the UBP1b 3′UTR (Figure 3, Figure 4), we tested ddm1 seedlings to determine if they display a similar stress sensitivity as upb1b mutant plants. Plants that have been homozygous ddm1 for six generations (ddm1 F6) are significantly more sensitive than the corresponding wt Col for both ionic and osmotic stress, while ddm1 F2 is only sensitive to ionic stress (Figure 6A). Similar to ubp1b mutants, ddm1 mutant plants are sensitive to ionic and osmotic stress conditions. These data, combined with our demonstrated regulation of UBP1b levels by siRNA854 (Figure 5), suggest that the ddm1 stress sensitivity acts directly through epigenetic activation of Athila6 and production of siRNA854, which results in the post-transcriptional and/or translational repression of UBP1b.
Fig. 6. UBP1b protein localization and stress sensitivity of ddm1. 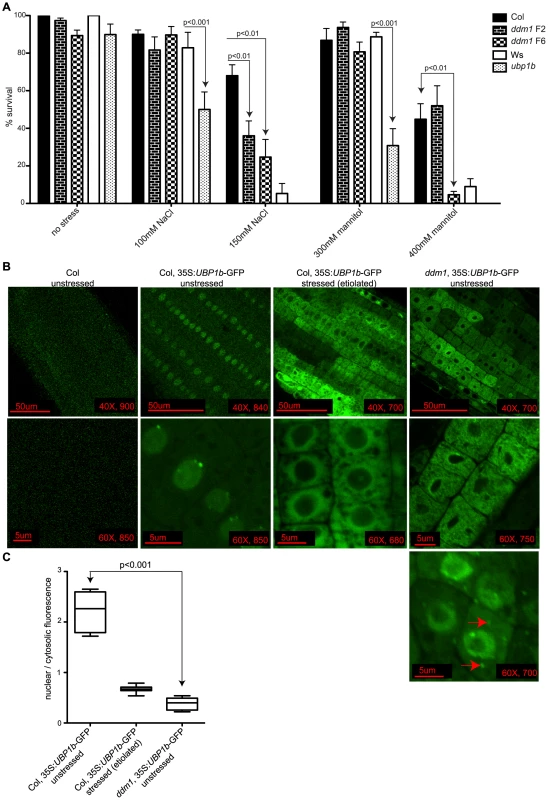
(A) Survival of plants grown under conditions of ionic and osmotic stress. Seeds were spotted on plates containing no stressor, increasing levels of NaCl or increasing levels of mannitol. After 11 days the number of surviving plants were counted. Wt plants of the Ws background are more sensitive to the stressors tested than wt Col. Compared to the corresponding wt Ws control, ubp1b mutants are more sensitive to both ionic (100 mM NaCl) and osmotic (300 mM mannitol) stresses. No upb1b seedlings survived under stress conditions of 150 mM NaCl or 400 mM mannitol. ddm1 F6 generation plants are also sensitive to both ionic (150 mM NaCl) and osmotic (400 mM mannitol) stresses compared to wt Col. (B) Imaging of a constitutively expressed UBP1b protein fused to GFP (without its native 3′ UTR) in the root cell elongation zone. In unstressed wt Col seedling roots the UBP1b-GFP protein is localized to the nucleus, where specific peri-nuclear bright foci are observed. Growth of the same plants under stress conditions (due to etiolation) results in the relocalization of the UBP1b-GFP fusion protein from the nucleus into the cytosol. In unstressed ddm1 mutants, the UBP1b-GFP protein accumulates in the cytosol, while occasional cells display distinct cytoplasmic foci (arrows). For each image, scale bars, magnification, and exposure time (in ms) are shown. (C) Measurements of fluorescence intensity were taken from the lines in B, and a ratio of nuclear to cytosolic fluorescence was calculated. The localization of UBP1b-GFP fluorescence is significantly different in unstressed wt Col compared to unstressed ddm1. The mammalian homolog of UBP1b is TIA-1, an RNA binding protein localized to the nucleus that moves into the cytoplasm and aggregates into stress granules upon induction of stresses such as treatment with arsenite, glucose deprivation, and viral infection [56]–[58]. To visualize the sub-cellular localization of the UBP1b protein in response to cellular stress in Arabidopsis, as well as to determine the influence of ddm1 on this process, we expressed an siRNA854-resistant version of UBP1b (without its native 3′UTR) fused to GFP, under constitutive expression. In wt Col seedling roots this protein is localized to the nucleus, with distinct bright peri-nuclear foci observed (Figure 6B). However, when transformed into ddm1 plants, this same transgene displayed cytosolic fluorescence (Figure 6B). We aimed to induce stress in both the wt Col and ddm1 UBP1b-GFP lines; however, the ionic and osmotic conditions from Figure 6A inhibited growth in the ddm1 plants. Therefore, we experimentally determined that germination and growth in the dark (etiolation) would generate a UPB1b protein stress response, without killing the plant. We germinated and grew the wt Col UBP1b-GFP plants under etiolation conditions and observed a shift in the sub-cellular localization of UBP1b-GFP, as fluorescence accumulated around the periphery of the nucleus and in the cytoplasm (Figure 6B). This change in sub-cellular distribution of UBP1b-GFP under a condition of stress (etiolation) is analogous to the movement of TIA-1 out of the nucleus under stress conditions [59]. In etiolated ddm1 plants, the UBP1b-GFP fluorescence pattern is the same as in non-stressed ddm1 plants (data not shown). We quantified these fluorescence patterns by measuring the amount of nuclear vs. cytoplasmic fluorescence and found a statistically significant difference between the localization of UBP1b-GFP in unstressed wt Col compared to unstressed ddm1 (Figure 6C). The unstressed ddm1 fluorescence pattern resembles the stressed wt Col roots (Figure 6B and 6C). Additionally, in a very small number of ddm1 cells (roughly 1/1000), we observe a second accumulation pattern that displays distinct cytoplasmic foci reminiscent of mammalian stress granules (red arrows, Figure 6B), as well as fluorescence at the periphery of the nucleus. Together, these data suggest that the translocation of the UBP1b protein in ddm1 mutant cells is a response to an intracellular stress as a result of the ddm1 mutation, perhaps due to altered gene expression in ddm1 mutants, or due to the loss of DNA methylation, loss of repressive histone modifications, and activation of TEs [4], [60].
Discussion
The epigenetic control of Athila6 and siRNA854
SiRNA854 is a gene-regulating endogenous siRNA that is produced from the Athila6 family of LTR retrotransposons, and its accumulation is strictly dependent upon the transcriptional epigenetic activation of the Athila6 element. Athila elements, as well as nearly all other types of TEs in wt Col Arabidopsis, are normally transcriptionally silenced and are associated with DNA methylation and 24 nt siRNAs involved in maintaining the transcriptionally silenced state [7], [13]. The 21–22 nt versions of siRNA854 are only produced upon Athila6 transcriptional activation, such as in ddm1 and met1 mutants, or in the vegetative cell of wt pollen. Like Athila6 activity itself, siRNA854 production displays two unusual epigenetic trans-generational inheritance patterns. First, siRNA854 is produced in a ddm1/+ heterozygote that was generated from a ddm1 homozygote, and the pathway of this accumulation remains dependent on AGO1 in the ddm1/+ heterozygote. Second, there is a positive correlation between the increasing levels of Athila6 mRNA and accumulation of Athila6 21–22 nt siRNAs (including siRNA854) as ddm1 mutants are propagated from the F2 to the F6 generation. The progressively increasing transcript levels in ddm1 F2 to F6 generations could be due to increased rates of transcription, perhaps due to progressive loss of heterochromatin control, such as Athila DNA methylation, each generation. The positive correlation in Athila mRNA and siRNA levels suggests that the Athila6 transcript is the limiting factor in siRNA854 production, and that at least some Athila6 mRNA transcripts can accumulate although the 3′ region of Athila6 is degraded into siRNAs and amplified using a RNA-dependent RNA polymerase. This correlation could potentially be due to two different subsets of elements that increase in expression from the ddm1 F2 to F6 generation, one subset that is cleaved into siRNAs and one subset that is not. Alternatively, Athila6 transcripts may be converted into siRNAs at a very slow rate, allowing time for the transcripts to accumulate before degradation.
SiRNA854 biogenesis
Most Arabidopsis TEs lose siRNA production when epigenetically activated. There is an unknown factor that causes some TEs, such as Athila, to produce siRNAs even when transcriptionally active. In contrast to some TE families such as ATGP1, which simply retains 24 nt siRNAs when activated, Athila is one of only very few element families identified to date that produces 21–22 nt siRNAs when epigenetically activated and expressed [12], [15]–[16]. The production of 21–22 nt siRNAs represents a shift in small RNA biogenesis pathways from PolIV-dependent 24 nt siRNAs processed by the RdDM pathway, to a post-transcriptional silencing pathway that presumably uses PolII-derived transcripts and involves DCL2, DCL4, RDR6 and AGO1, with at least siRNA854 eventually incorporated into AGO1. These DCL, RDR and AGO proteins act in both the tasiRNA and VIGS pathways, and Athila processing shows hallmarks of both. For example, the VIGS pathway likely acts on Athila6 transcripts, as Athila has evolved from an LTR retrovirus, and, due to the conservation of the envelope protein coding domain, the Athila4 subfamily has even been classified as an Arabidopsis endogenous retrovirus [61]. Athila6 siRNAs may be produced via the VIGS pathway; however, siRNA854's ability to regulate UBP1b in trans is functionally more similar to the tasiRNA pathway. Therefore, we defer classifying Athila6 21–22 nt siRNAs as either tasiRNAs or VIGS siRNAs.
The classification of Athila6 21–22 nt siRNAs as either tasiRNAs or VIGS siRNAs perhaps can be resolved once the initiation of their production is understood. We currently have three models for how the initiation of Athila6 21–22 nt siRNAs may occur. First, the secondary structure of the Athila6 transcript, particularly in the region of siRNA production, may fold back into hairpin-like structures, producing a substrate for DCL4 cleavage. Second, overlapping sense and antisense Athila6 transcripts may result in the formation of a dsRNA trigger, as Athila elements accumulate in nested arrays of elements near the centromere that favor the production of read-through transcripts (reviewed in [62]). A pathway of natural antisense transcript siRNA (nat-siRNA) production exists in Arabidopsis; however, PolIV and RDR2 are required for this pathway [63]–[64], and we have experimentally determined that these proteins are not required for Athila6 21–22 nt siRNA accumulation. A third proposed mechanism of 21–22 nt Athila6 siRNA initiation may be similar to tasiRNA initiation and the initiation of islands of 21–22 nt siRNA accumulation in maize and rice. MicroRNA(s) may initiate the cleavage of an Athila6 transcript, causing the production of RDR6 - and DCL4-dependent siRNAs. In rice, the production of 21 nt phased siRNAs is dependent first on microRNA cleavage, and then on OsDCL4 for production, and these siRNAs preferentially accumulate in male reproductive organs [65]. One microRNA that shows potential seed region complementarity to Athila6 is the 22 nt microRNA845b; however, Athila siRNAs are produced on either side of the predicted Athila6 cleavage site, and our experiments to date provide no evidence that microRNA845b is required for Athila6 21–22 nt siRNA biogenesis (data not shown). In addition to acting downstream of siRNA854 production, it is currently unknown if AGO1 acts upstream of DCL4 and RDR6 to initiate Athila6 transcript cleavage, but it is likely that a 22 nt siRNA initiates the RDR6-dependent amplification of Athila6 siRNAs [66]. If initiated by a tasiRNA-like mechanism, tasiRNA-like phasing should be detected in the Athila6 siRNA production. We have not detected any such phasing of Athila6 siRNAs (data not shown), but this analysis is complicated by the 12 nearly identical Athila6 elements that carry siRNA854, and dozens more Athila elements that are cleaved into siRNAs at the same time. If each element that produces 21–22 nt siRNAs is correctly phased, but not in the same register as each other, our analysis would detect no phasing. Therefore, although we have identified AGO1, RDR6 and DCL4 as necessary for the accumulation of siRNA854, the trigger for Athila6 siRNA initiation remains to be elucidated.
The regulation of UBP1b by siRNA854
We used the UBP1b 3′UTR in reporter assays to demonstrate that whenever we observe the accumulation of the 21–22 nt siRNA854 sequence (in ddm1 mutants, wt pollen, or expressed as an amiRNA), we observe decreased reporter protein accumulation. Expression of the siRNA854 sequence as an amiRNA in the vegetative tissue of wt plants demonstrates that the potentially complicating secondary effects occurring from loss of heterochromatin control in ddm1 mutant plants and wt pollen are not responsible for repression of the UPB1b 3′UTR. Both the siRNA854-amiRNA and endogenous siRNA854 are able to inhibit protein production from reporter transcripts bearing the UBP1b 3′UTR, and in pollen this regulation is dependent on the siRNA854 target sites in the 3′UTR, as well as on RDR6.
We have also demonstrated that the endogenous UBP1b transcript is regulated by siRNA854. In inflorescence tissue, this regulation is likely on the translational level, and this result is supported by the translational regulation of the GUS-UBP1b 3′UTR transcript in inflorescences. In contrast, in mature pollen we detect post-transcriptional regulation of the endogenous UBP1b transcript by siRNA854. This pollen post-transcriptional regulation of the endogenous UPB1b transcript is under the control of RDR6 and AGO1, suggesting that the accumulation of siRNA854 is necessary for this regulation. The basis of the switch from translational control in inflorescence tissue to post-transcriptional control in pollen remains puzzling. One possibility is that the four predicted target sites for siRNA854 in the UBP1b 3′UTR are not equally available to base pair in inflorescence tissue compared to pollen. Therefore, in pollen the interaction of siRNA854 with one target site may cause transcript cleavage, while in inflorescence tissue the interaction with a different target site may lead to translational inhibition. Lastly, the observation of 21–22 nt siRNA854 still present in ddm1 heterozygotes produced from ddm1 homozygous parents suggests that there may be a trans-generational epigenetic component to the regulation of UBP1b, as UBP1b may continue to respond to Athila activity even when the plant is no longer homozygous for ddm1. This trans-generational regulation was observed with the GUS-UBP1b 3′UTR transgene in an individual heterozygous for the recessive ddm1-2 allele, due to the inheritance of mutant chromatin from the parental plant.
Athila and stress
Under the stress condition of etiolation, the UBP1b-GFP protein traffics from the nucleus and accumulates in the cytoplasm. In unstressed ddm1 mutants, this siRNA854-resistant form of UBP1b is also located in the cytoplasm, suggesting that some aspect of the ddm1 mutation triggers this stress-sensing change in protein location, independent of siRNA854. It is currently unknown which characteristic(s) of the ddm1 mutation triggers this response, as ddm1 mutants display aberrant control of genic epialleles, global TE activation, TE mobilization, and general chromatin decondensation [4], [35], [60], [67]. Conversely, ddm1 mutant seedlings show a phenotype similarly sensitive to ionic and osmotic stress as upb1b mutants. Several studies have shown that TEs are reactivated during stress conditions [68]–[69]; however, in this case TEs are regulating the stress-responsive pathway. Taken together, these data suggest that an antagonism exists between the UBP1b-induced stress response, which is activated in ddm1 mutants, and Athila6, which inhibits this response by targeting UBP1b through siRNA854. This antagonism may also exist in animal cells, as some DNA viruses generate microRNAs that specifically target the UBP1b homologue TIA-1 mRNA [70], while other RNA viruses specifically target stress granule proteins for proteolysis [71], presumably for the same reason that Athila targets UBP1b. Since TIA-1 is known to repress the activity of some animal viruses and retrotransposons through the formation of stress granules [72]–[73], we speculate that the same is true for Athila. Therefore, we envision a three-layered host repression of Athila activity. First, transcriptional regulation dependent on DNA methylation epigenetically silences Athila. Second, when transcriptionally active, Athila mRNA accumulation is inhibited by the post-transcriptional regulation mediated by the tasiRNA/VIGS siRNA pathway components DCL2, DCL4, RDR6 and AGO1. Third, we speculate that Athila transcripts may be translationally inhibited due to their sequestration in stress granules, targeted by the UBP1b protein. Transcripts in stress granules are not degraded, but are not translated due to their separation from active ribosome complexes (reviewed in [54]). Akin to a virus encoding a suppressor of gene silencing [20], Athila may encode siRNA854 to inhibit UBP1b protein formation and interfere with the function of this translational-level repression.
TE regulation of genes
AGO1 is known to mediate gene regulation via siRNAs in the tasiRNA pathway [74]–[75]. We have demonstrated that an siRNA which is not part of one of the four known tasiRNA producing loci (TAS1-4), but rather part of an epigenetically regulated TE, is able to act on genic transcripts in trans in a similar fashion to a tasiRNA. We think the key aspect of this regulation is the incorporation of siRNA854 into AGO1. AGO1 is the main Argonaute protein responsible for gene regulation in Arabidopsis (reviewed in [76]). This protein is likely unable to distinguish between an siRNA generated from a transcriptionally reactivated TE and one generated from a tasiRNA precursor transcript, at least in the case of siRNA854. Sequencing from AGO1 immunoprecipitations has demonstrated a higher than expected level of siRNAs [52], [77], providing evidence that AGO1 is likely regulating both genic transcripts using microRNAs, as well as viral, TE or other repeat transcripts via siRNAs and post-transcriptional silencing. Further investigation is required to understand if and how AGO1 protein complexes determine which siRNAs should target genic mRNAs in trans and which should not. Therefore, the possibility currently exists that siRNA854 does not act alone, and the genome-wide regulation of many transcripts is altered by TE or viral siRNAs loaded into AGO1. It is an intriguing possibility, since both TE epigenetic activation and viral infection lead to a series of still unexplained changes in gene regulation and phenotype. In fact, one longstanding enigmatic viral symptom of the Cucumber mosaic virus was recently found to be caused by a viral satellite siRNA targeting a host gene in trans [20]. In animals, many viruses encode microRNAs that target cellular genes to generate a favorable cellular environment [78]. In order to gain this same advantage, plant TEs may carry sequences that do not require a microRNA stem-loop structure, but utilize a different mechanism by co-opting the tasiRNA/VIGS siRNA biogenesis machinery to regulate a diverse set of cellular transcripts.
Materials and Methods
Plant material
The mutant alleles used in this study are listed in Table S1. All mutants are in the Col background except ago1 (Ler), ago10 (Ler), hen1-1 (Ler), ubp1b (FLAG_298B04)(Ws), upb1b (FLAG_071F09)(Ws), and ddm1 Ler. Plants were grown under standard long-day growth chamber conditions. Etiolated and stress-test plants were stratified for 3 days at 4°C and grown for 11 days on 1/2X MS media+Gamborg's vitamins with supplemented sucrose in 16 hours of light per day, with the exception of etiolated seedlings, which were grown without light. For the stress-test analysis, the number of plants surviving after 11 days was counted. Fifty seedlings of each genotype for each condition were grown, and the analysis was replicated three or more times.
qRT–PCR
Total RNA was extracted using TRIzol reagent (Invitrogen) or the RNeasy Plant Kit (Qiagen). Total RNA was DNAseI treated and reverse transcribed using an oligo-dT primer and SuperScript III Reverse Transcriptase (Invitrogen). qRT-PCR was performed with iQ SYBR Green Supermix (BioRad Laboratories) using 3 technical replicates each of 3 or more biological replicates. qRT-PCR primers are shown in Table S1. qPCR reactions were annealed at 60°C unless otherwise noted. Since most standard qRT-PCR control genes are not highly expressed in pollen, the relative expression values for all experiments were calculated based on the expression of the experimentally validated control gene At1g08200. qPCR was performed on a CFX96 thermocycler and the results analyzed on the CFX Manager Software package (BioRad Laboratories). Relative expression was calculated using the ‘delta-delta method’ formula 2−[ΔCP sample−ΔCP control], where 2 represents perfect PCR efficiency. Statistical significance was calculated using unpaired T-tests.
Small RNA Northern blots
Total RNA was extracted using TRIzol reagent (Invitrogen), and small RNA was enriched by polyethylene glycol precipitation. The quantity of small RNA loaded in each lane ranged from 16–60 µg between blots, though the same amount of RNA was loaded per lane on each blot for comparison between samples. We accounted for the equal loading and sizes of small RNAs by re-probing our Northern blots with a known 21 nt microRNA (miR161) and/or a known 24 nt siRNA (siRNA02). In addition, our small RNA Northern blot analysis is supported by independent small RNA deep sequencing data [33]. Gel electrophoresis, blotting and cross-linking were performed as in Pall et al. [79]. Probes for siRNA854, miR161, and siRNA02 were generated by 5′ labeling DNA oligonucleotides with P32-ATP, whereas the probe for Athila 3′ was generated by randomly degrading a P32-UTP labeled in vitro transcribed RNA as in [80]. Sequences of DNA oligonucleotides and primers for generating the in vitro transcription template are listed in Table S1.
Transgene construction and analysis
The 35S:amiRNA-siRNA854 transgene was generated by cloning the sequence 5′GATGAGGATAGGGAGGAGGAG into the microRNA319a stem-loop transcript as in [44]. This transcript was sub-cloned into the 35S promoter binary plasmid pB2GW7. The wt version of the UBP1b 3′UTR was amplified from the wt Col genome, and the 35S:GUS-UBP1b 3′UTR transgene was produced as in [29]. GUS staining was performed as in [81]. For GUS protein activity quantification, protein was quantified using the DC assay (BioRad Laboratories), and 1 mg was used to assay the cleavage of MUG into fluorescent 4-MU as in [82]–[83]. Fluorescence was measured in 96-well format with a Tecan-SpectraFluor Plus microplate reader, and the specific activity of GUS in each sample was calculated as nmol of 4-MU formed per hour per mg of protein (nmol 4-MU/h/mg). RT-PCR of these lines was performed with oligo-dT primed cDNA for 28 cycles of PCR using primers listed in Table S1.
The modified and deleted versions of the UBP1b 3′UTR were synthesized by IDT. The Lat52 promoter-driven GFP-UPB1b 3′UTR transgene was constructed by cloning the either the wt UBP1b 3′UTR, Modified UBP1b 3′UTR or Deleted 3′UTR version into the SacI site at the end of the mGFP coding sequence of the binary plasmid pMDC107, and then by adding the Lat52 promoter to the KpnI site upstream of mGFP in these clones. GFP fluorescence quantification was performed on a Nikon C2 confocal microscope with the NIS-Elements software suite (Nikon Corporation). GFP quantification was performed with the same microscope settings (exposure time, laser power) on the same day. Subtraction of the fluorescence of pollen grains that did not inherit the GFP transgene from the same hemizygous plant negated the background pollen auto-fluorescence.
The FLAG-UBP1b transgene was constructed by adding the FLAG epitope sequence to the 5′ end of the UBP1b CDS as in [51]. This FLAG-UBP1b fragment was amplified and cloned into pENTR/D-TOPO (Invitrogen). The UBP1b promoter and 5′UTR were inserted 5′ to the FLAG tag, and the UBP1b 3′UTR was inserted 3′ of the UBP1b coding region by In-Fusion Recombination (Clontech). Subsequent constructs were recombined into pBGW by Gateway LR Recombination (Invitrogen). Specific qRT-PCR primer sets detecting the FLAG-UBP1b transgene are shown in Table S1. Attempts at identifying the FLAG-UBP1b protein using a FLAG-epitope antibody were repeatedly unsuccessful.
The 35S:UBP1b-GFP transgene was generated by cloning the UBP1b coding region into the binary plasmid pK7FWG2. Seedlings were grown on 1/2X MS media for 11 days before their roots were analyzed by confocal microscopy. The ratio of nuclear to cytosolic fluorescence was calculated by using the NIS-Elements software (Nikon Corporation) by manually defining the average fluorescence touching an analysis line transecting the nucleus and cytosol of an individual cell. The ratios of 25 cells were examined per condition.
All Arabidopsis transformations were performed using Agrobacterium strain GV3101 and standard laboratory techniques. Statistical significance was calculated using unpaired T-tests.
AGO1 immunoprecipitation and small RNA analysis
The AGO1 protein was immunoprecipitated as follows using a commercially available polyclonal AGO1 antibody (Agrisera AB) specific to the unique N-terminal peptide of AGO1, which has been demonstrated to lack cross-reactivity with various over-expressed AGO proteins [84]. Inflorescence tissue was ground with liquid nitrogen and homogenized in 2 ml extraction buffer (100 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, and 5 mM DTT) containing 1 tablet/10 mL protease inhibitor cocktail (Roche) per gram of tissue. In a standard immunoprecipitation reaction, Arabidopsis protein extract was pre-cleared by incubation with 10 µl of goat anti-rabbit magnetic beads (NEB). Pre-cleared extracts were then incubated overnight with goat anti-rabbit magnetic beads pre-incubated with 1 µg α-AGO1. All washes were performed with extraction buffer. Immunoprecipitated, mock-immunoprecipitated and input sample RNA was isolated using TRIzol (Invitrogen). 125 ng of each RNA sample was subjected to polyA tailing, cDNA synthesis, and qRT-PCR according to the QuantiMir product specifications (System Biosciences Incorporated). The PCR was annealed at 61.5°C and performed on 2–3 biological replicate immunoprecipitations for each genotype tested, each one having 3 technical qPCR replicates. Each qRT-PCR IP C(t) value was normalized to the amplification of its own input sample, using the ‘delta-delta method’ formula 2−[ΔCP IP−ΔCP Input], where 2 represents perfect PCR efficiency.
Supporting Information
Zdroje
1. LanderESLintonLMBirrenBNusbaumCZodyMC 2001 Initial sequencing and analysis of the human genome. Nature 409 860 921 doi:10.1038/35057062
2. RoudierFCOAhmedIBerardCBESarazinAMary-HuardT 2011 Integrative epigenomic mapping defines four main chromatin states in Arabidopsis. EMBO J 30 1928 1938 doi:doi:10.1038/emboj.2011.103
3. SlotkinRKMartienssenR 2007 Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet 8 272 285 doi:10.1038/nrg2072
4. LippmanZGendrelA-VBlackMVaughnMWDedhiaN 2004 Role of transposable elements in heterochromatin and epigenetic control. Nature 430 471 476 doi:10.1038/nature02651
5. SunF-LHaynesKSimpsonCLLeeSDCollinsL 2004 cis-Acting determinants of heterochromatin formation on Drosophila melanogaster chromosome four. Mol Cell Biol 24 8210 8220 doi:10.1128/MCB.24.18.8210-8220.2004
6. AravinAASachidanandamRBourc'hisDSchaeferCPezicD 2008 A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell 31 785 799 doi:10.1016/j.molcel.2008.09.003
7. LawJAJacobsenSE 2010 Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet doi:10.1038/nrg2719
8. HaveckerERWallbridgeLMHardcastleTJBushMSKellyKA 2010 The Arabidopsis RNA-directed DNA methylation argonautes functionally diverge based on their expression and interaction with target loci. Plant Cell 22 321 334 doi:10.1105/tpc.109.072199
9. PereiraV 2004 Insertion bias and purifying selection of retrotransposons in the Arabidopsis thaliana genome. Genome Biol 5 R79 doi:10.1186/gb-2004-5-10-r79
10. WrightDAVoytasDF 1998 Potential retroviruses in plants: Tat1 is related to a group of Arabidopsis thaliana Ty3/gypsy retrotransposons that encode envelope-like proteins. Genetics 149 703 715
11. SteimerAAmedeoPAfsarKFranszPMittelsten ScheidO 2000 Endogenous targets of transcriptional gene silencing in Arabidopsis. Plant Cell 12 1165 1178
12. SlotkinRKVaughnMBorgesFTanurdzicMBeckerJD 2009 Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 136 461 472 doi:10.1016/j.cell.2008.12.038
13. ListerRO'MalleyRTonti-FilippiniJGregoryBBerryC 2008 Highly Integrated Single-Base Resolution Maps of the Epigenome in Arabidopsis. Cell 133 523 536 doi:10.1016/j.cell.2008.03.029
14. LippmanZMayBYordanCSingerTMartienssenR 2003 Distinct mechanisms determine transposon inheritance and methylation via small interfering RNA and histone modification. PLoS Biol 1 e67 doi:10.1371/journal.pbio.0000067
15. MirouzeMReindersJBucherENishimuraTSchneebergerK 2009 Selective epigenetic control of retrotransposition in Arabidopsis. Nature 461 427 430 doi:10.1038/nature08328
16. TanurdzicMVaughnMWJiangHLeeT-JSlotkinRK 2008 Epigenomic consequences of immortalized plant cell suspension culture. PLoS Biol 6 e302 doi:10.1371/journal.pbio.0060302
17. CzechBHannonGJ 2011 Small RNA sorting: matchmaking for Argonautes. Nat Rev Genet 12 19 31 doi:10.1038/nrg2916
18. RougetCPapinCBoureuxAMeunierA-CFrancoB 2010 Maternal mRNA deadenylation and decay by the piRNA pathway in the early Drosophila embryo. Nature 467 1128 1132 doi:doi:10.1038/nature09465
19. WatanabeTTomizawaS-IMitsuyaKTotokiYYamamotoY 2011 Role for piRNAs and noncoding RNA in de novo DNA methylation of the imprinted mouse Rasgrf1 locus. Science 332 848 852 doi:10.1126/science.1203919
20. SmithNAEamensALWangM-B 2011 Viral small interfering RNAs target host genes to mediate disease symptoms in plants. PLoS Pathog 7 e1002022 doi:10.1371/journal.ppat.1002022
21. XieZJohansenLKGustafsonAMKasschauKDLellisAD 2004 Genetic and functional diversification of small RNA pathways in plants. PLoS Biol 2 e104 doi:10.1371/journal.pbio.0020104
22. DunoyerPBrosnanCASchottGWangYJayF 2010 An endogenous, systemic RNAi pathway in plants. EMBO J 29 1699 1712 doi:10.1038/emboj.2010.65
23. ChapmanEJCarringtonJC 2007 Specialization and evolution of endogenous small RNA pathways. Nat Rev Genet 8 884 896 doi:10.1038/nrg2179
24. DunoyerPHimberCVoinnetO 2005 DICER-LIKE 4 is required for RNA interference and produces the 21-nucleotide small interfering RNA component of the plant cell-to-cell silencing signal. Nat Genet 37 1356 1360 doi:10.1038/ng1675
25. SchwachFVaistijFEJonesLBaulcombeDC 2005 An RNA-dependent RNA polymerase prevents meristem invasion by potato virus X and is required for the activity but not the production of a systemic silencing signal. Plant Physiol 138 1842 1852 doi:10.1104/pp.105.063537
26. DelerisAGallego-BartolomeJBaoJKasschauKDCarringtonJC 2006 Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science 313 68 71 doi:10.1126/science.1128214
27. Garcia-RuizHTakedaAChapmanEJSullivanCMFahlgrenN 2010 Arabidopsis RNA-dependent RNA polymerases and dicer-like proteins in antiviral defense and small interfering RNA biogenesis during Turnip Mosaic Virus infection. Plant Cell 22 481 496 doi:10.1105/tpc.109.073056
28. DunoyerPSchottGHimberCMeyerDTakedaA 2010 Small RNA duplexes function as mobile silencing signals between plant cells. Science 328 912 916 doi:10.1126/science.1185880
29. Arteaga-VázquezMCaballero-PérezJVielle-CalzadaJ-P 2006 A family of microRNAs present in plants and animals. Plant Cell 18 3355 3369 doi:10.1105/tpc.106.044420
30. KedershaNLGuptaMLiWMillerIAndersonP 1999 RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J Cell Biol 147 1431 1442
31. WeberCNoverLFauthM 2008 Plant stress granules and mRNA processing bodies are distinct from heat stress granules. Plant J 56 517 530 doi:10.1111/j.1365-313X.2008.03623.x
32. MeyersBCAxtellMJBartelBBartelDPBaulcombeD 2008 Criteria for annotation of plant MicroRNAs. Plant Cell 20 3186 3190 doi:10.1105/tpc.108.064311
33. NakanoMNobutaKVemarajuKTejSSSkogenJW 2006 Plant MPSS databases: signature-based transcriptional resources for analyses of mRNA and small RNA. Nucleic Acids Res 34 D731 5 doi:10.1093/nar/gkj077
34. JohnsonLCaoXJacobsenS 2002 Interplay between two epigenetic marks. DNA methylation and histone H3 lysine 9 methylation. Curr Biol 12 1360 1367
35. HirochikaHOkamotoHKakutaniT 2000 Silencing of retrotransposons in arabidopsis and reactivation by the ddm1 mutation. Plant Cell 12 357 369
36. KakutaniTJeddelohJAFlowersSKMunakataKRichardsEJ 1996 Developmental abnormalities and epimutations associated with DNA hypomethylation mutations. Proc Natl Acad Sci USA 93 12406 12411
37. BoutetSVazquezFLiuJBéclinCFagardM 2003 Arabidopsis HEN1: a genetic link between endogenous miRNA controlling development and siRNA controlling transgene silencing and virus resistance. Curr Biol 13 843 848
38. KannoTHuettelBMetteMFAufsatzWJaligotE 2005 Atypical RNA polymerase subunits required for RNA-directed DNA methylation. Nat Genet 37 761 765 doi:10.1038/ng1580
39. GasciolliVMalloryACBartelDPVaucheretH 2005 Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr Biol 15 1494 1500 doi:10.1016/j.cub.2005.07.024
40. VongsAKakutaniTMartienssenRARichardsEJ 1993 Arabidopsis thaliana DNA methylation mutants. Science 260 1926 1928
41. KakutaniTMunakataKRichardsEJHirochikaH 1999 Meiotically and mitotically stable inheritance of DNA hypomethylation induced by ddm1 mutation of Arabidopsis thaliana. Genetics 151 831 838
42. TeixeiraFKHerediaFSarazinARoudierFBoccaraM 2009 A role for RNAi in the selective correction of DNA methylation defects. Science 323 1600 1604 doi:10.1126/science.1165313
43. DidianoDHobertO 2006 Perfect seed pairing is not a generally reliable predictor for miRNA-target interactions. Nat Struct Mol Biol 13 849 851 doi:10.1038/nsmb1138
44. SchwabROssowskiSRiesterMWarthmannNWeigelD 2006 Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18 1121 1133 doi:10.1105/tpc.105.039834
45. LuoZChenZ 2007 Improperly terminated, unpolyadenylated mRNA of sense transgenes is targeted by RDR6-mediated RNA silencing in Arabidopsis. Plant Cell 19 943 958 doi:10.1105/tpc.106.045724
46. LiuJ 2008 Control of protein synthesis and mRNA degradation by microRNAs. Curr Opin Cell Biol 20 214 221 Available: http://www.sciencedirect.com/science/article/pii/S095506740800015X. Accessed 30 Aug. 2011
47. Addo-QuayeCEshooTWBartelDPAxtellMJ 2008 Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr Biol 18 758 762 doi:10.1016/j.cub.2008.04.042
48. GermanMAPillayMJeongD-HHetawalALuoS 2008 Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat Biotechnol 26 941 946 doi:10.1038/nbt1417
49. McCueADCrestiMFeijóJASlotkinRK 2011 Cytoplasmic connection of sperm cells to the pollen vegetative cell nucleus: potential roles of the male germ unit revisited. J Exp Bot 62 1621 1631 doi:10.1093/jxb/err032
50. BorgesFGomesGGardnerRMorenoNMcCormickS 2008 Comparative transcriptomics of Arabidopsis sperm cells. Plant Physiol 148 1168 1181 doi:10.1104/pp.108.125229
51. BaumbergerNBaulcombeDC 2005 Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc Natl Acad Sci USA 102 11928 11933 doi:10.1073/pnas.0505461102
52. WangHZhangXLiuJKibaTWooJ 2011 Deep sequencing of small RNAs specifically associated with Arabidopsis AGO1 and AGO4 uncovers new AGO functions. Plant J 67 292 304 doi:10.1111/j.1365-313X.2011.04594.x
53. PiecykMWaxSBeckARKedershaNGuptaM 2000 TIA-1 is a translational silencer that selectively regulates the expression of TNF-alpha. EMBO J 19 4154 4163 doi:10.1093/emboj/19.15.4154
54. Eisinger-MathasonTSKAndradeJGroehlerALClarkDEMuratore-SchroederTL 2008 Codependent functions of RSK2 and the apoptosis-promoting factor TIA-1 in stress granule assembly and cell survival. Mol Cell 31 722 736 doi:10.1016/j.molcel.2008.06.025
55. ZouTYangXPanDHuangJSahinM 2011 SMN deficiency reduces cellular ability to form stress granules, sensitizing cells to stress. Cell Mol Neurobiol 31 541 550 doi:10.1007/s10571-011-9647-8
56. GilksNKedershaNAyodeleMShenLStoecklinG 2004 Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol Biol Cell 15 5383 5398 doi:10.1091/mbc.E04-08-0715
57. BuchanJRParkerR 2009 Eukaryotic stress granules: the ins and outs of translation. Mol Cell 36 932 941 doi:10.1016/j.molcel.2009.11.020
58. BeckhamCJParkerR 2008 P bodies, stress granules, and viral life cycles. Cell Host Microbe 3 206 212 doi:10.1016/j.chom.2008.03.004
59. KedershaNChoMRLiWYaconoPWChenS 2000 Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J Cell Biol 151 1257 1268
60. GendrelA-VLippmanZYordanCColotVMartienssenRA 2002 Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science 297 1871 1873 doi:10.1126/science.1074950
61. WrightDAVoytasDF 2002 Athila4 of Arabidopsis and Calypso of soybean define a lineage of endogenous plant retroviruses. Genome Res 12 122 131 doi:10.1101/gr.196001
62. SlotkinRK 2010 The epigenetic control of the Athila family of retrotransposons in Arabidopsis. Epigenetics 5 483 490
63. BorsaniOZhuJVersluesPESunkarRZhuJ-K 2005 Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 123 1279 1291 doi:10.1016/j.cell.2005.11.035
64. RonMAlandete-SaezMEshed-WilliamsLFletcherJCMcCormickS 2010 Proper regulation of a sperm-specific cis-nat-siRNA is essential for double fertilization in Arabidopsis. Genes Dev 24 1010 1021 doi:10.1101/gad.1882810
65. SongXLiPZhaiJZhouMMaL 2011 Roles of DCL4 and DCL3b in Rice Phased Small RNA Biogenesis. Plant J in press.
66. CuperusJTCarbonellAFahlgrenNGarcia-RuizHBurkeRT 2010 Unique functionality of 22-nt miRNAs in triggering RDR6-dependent siRNA biogenesis from target transcripts in Arabidopsis. Nat Struct Mol Biol 17 997 1003 doi:10.1038/nsmb.1866
67. MiuraAYonebayashiSWatanabeKToyamaTShimadaH 2001 Mobilization of transposons by a mutation abolishing full DNA methylation in Arabidopsis. Nature 411 212 214 doi:10.1038/35075612
68. GrandbastienM-A 2004 Stress activation and genomic impact of plant retrotransposons. J Soc Biol 198 425 432
69. BeauregardACurcioMJBelfortM 2008 The take and give between retrotransposable elements and their hosts. Annu Rev Genet 42 587 617 doi:10.1146/annurev.genet.42.110807.091549
70. AparicioOCarneroEAbadXRazquinNGuruceagaE 2010 Adenovirus VA RNA-derived miRNAs target cellular genes involved in cell growth, gene expression and DNA repair. Nucleic Acids Res 38 750 763 doi:10.1093/nar/gkp1028
71. WhiteJPCardenasAMMarissenWELloydRE 2007 Inhibition of cytoplasmic mRNA stress granule formation by a viral proteinase. Cell Host Microbe 2 295 305 doi:10.1016/j.chom.2007.08.006
72. EsclatineATaddeoBRoizmanB 2004 Herpes simplex virus 1 induces cytoplasmic accumulation of TIA-1/TIAR and both synthesis and cytoplasmic accumulation of tristetraprolin, two cellular proteins that bind and destabilize AU-rich RNAs. J Virol 78 8582 8592 doi:10.1128/JVI.78.16.8582-8592.2004
73. GoodierJLZhangLVetterMRKazazianHH 2007 LINE-1 ORF1 protein localizes in stress granules with other RNA-binding proteins, including components of RNA interference RNA-induced silencing complex. Mol Cell Biol 27 6469 6483 doi:10.1128/MCB.00332-07
74. VazquezFVaucheretHRajagopalanRLepersCGasciolliV 2004 Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol Cell 16 69 79 doi:10.1016/j.molcel.2004.09.028
75. MontgomeryTAYooSJFahlgrenNGilbertSDHowellMD 2008 AGO1-miR173 complex initiates phased siRNA formation in plants. Proc Natl Acad Sci USA 105 20055 20062 doi:10.1073/pnas.0810241105
76. HutvágnerGSimardMJ 2008 Argonaute proteins: key players in RNA silencing. Nature Reviews Molecular Cell Biology 9 22 32 doi:10.1038/nrm2321
77. MiSCaiTHuYChenYHodgesE 2008 Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell 133 116 127 doi:10.1016/j.cell.2008.02.034
78. GrundhoffASullivanCS 2011 Virus-encoded microRNAs. Virology 411 325 343 doi:10.1016/j.virol.2011.01.002
79. PallGSCodony-ServatCByrneJRitchieLHamiltonA 2007 Carbodiimide-mediated cross-linking of RNA to nylon membranes improves the detection of siRNA, miRNA and piRNA by northern blot. Nucleic Acids Res 35 e60 doi:10.1093/nar/gkm112
80. SlotkinRKFreelingMLischD 2005 Heritable transposon silencing initiated by a naturally occurring transposon inverted duplication. Nat Genet 37 641 644 doi:10.1038/ng1576
81. SundaresanVSpringerPVolpeTHawardSJonesJD 1995 Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev 9 1797 1810
82. JeffersonRA 1987 Assaying Chimeric Genes in Plants: The GUS Gene Fusion System. Plant Molecular Biology Reporter 5 387 405
83. TwellDYamaguchiJMcCormickS 1990 Pollen-specific gene expression in transgenic plants: coordinate regulation of two different tomato gene promoters during microsporogenesis. Development 109 705 713
84. BaumbergerNTsaiC-HLieMHaveckerEBaulcombeDC 2007 The Polerovirus silencing suppressor P0 targets ARGONAUTE proteins for degradation. Curr Biol 17 1609 1614 doi:10.1016/j.cub.2007.08.039
85. KidnerCAMartienssenRA 2004 Spatially restricted microRNA directs leaf polarity through ARGONAUTE1. Nature 428 81 84 doi:10.1038/nature02366
Štítky
Genetika Reprodukční medicína
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 2
-
Všechny články tohoto čísla
- Upsetting the Dogma: Germline Selection in Human Males
- A Strong Deletion Bias in Nonallelic Gene Conversion
- Positive Selection for New Disease Mutations in the Human Germline: Evidence from the Heritable Cancer Syndrome Multiple Endocrine Neoplasia Type 2B
- Genome-Wide Association Study in East Asians Identifies Novel Susceptibility Loci for Breast Cancer
- Mixed Effects Modeling of Proliferation Rates in Cell-Based Models: Consequence for Pharmacogenomics and Cancer
- Reduction of NADPH-Oxidase Activity Ameliorates the Cardiovascular Phenotype in a Mouse Model of Williams-Beuren Syndrome
- Genome-Wide Association Study Identifies Chromosome 10q24.32 Variants Associated with Arsenic Metabolism and Toxicity Phenotypes in Bangladesh
- Structural Basis of Transcriptional Gene Silencing Mediated by MOM1
- Genomic Restructuring in the Tasmanian Devil Facial Tumour: Chromosome Painting and Gene Mapping Provide Clues to Evolution of a Transmissible Tumour
- Genome-Wide Association Study Identifies Novel Loci Associated with Circulating Phospho- and Sphingolipid Concentrations
- Contrasting Properties of Gene-Specific Regulatory, Coding, and Copy Number Mutations in : Frequency, Effects, and Dominance
- The Origin and Nature of Tightly Clustered Deletions in Precursor B-Cell Acute Lymphoblastic Leukemia Support a Model of Multiclonal Evolution
- Ultrafast Evolution and Loss of CRISPRs Following a Host Shift in a Novel Wildlife Pathogen,
- Phosphorylation of Chromosome Core Components May Serve as Axis Marks for the Status of Chromosomal Events during Mammalian Meiosis
- Psoriasis Patients Are Enriched for Genetic Variants That Protect against HIV-1 Disease
- A Pathogenic Mechanism in Huntington's Disease Involves Small CAG-Repeated RNAs with Neurotoxic Activity
- The Mitochondrial Chaperone Protein TRAP1 Mitigates α-Synuclein Toxicity
- Homeobox Genes Critically Regulate Embryo Implantation by Controlling Paracrine Signaling between Uterine Stroma and Epithelium
- Developmental Transcriptional Networks Are Required to Maintain Neuronal Subtype Identity in the Mature Nervous System
- Down-Regulating Sphingolipid Synthesis Increases Yeast Lifespan
- Gene Expression and Stress Response Mediated by the Epigenetic Regulation of a Transposable Element Small RNA
- Loss of Tgif Function Causes Holoprosencephaly by Disrupting the Shh Signaling Pathway
- Sequestration of Highly Expressed mRNAs in Cytoplasmic Granules, P-Bodies, and Stress Granules Enhances Cell Viability
- Discovery of a Modified Tetrapolar Sexual Cycle in and the Evolution of in the Species Complex
- The Role of Glypicans in Wnt Inhibitory Factor-1 Activity and the Structural Basis of Wif1's Effects on Wnt and Hedgehog Signaling
- Nondisjunction of a Single Chromosome Leads to Breakage and Activation of DNA Damage Checkpoint in G2
- A Regulatory Network for Coordinated Flower Maturation
- Coexpression Network Analysis in Abdominal and Gluteal Adipose Tissue Reveals Regulatory Genetic Loci for Metabolic Syndrome and Related Phenotypes
- Diced Triplets Expose Neurons to RISC
- The Williams-Beuren Syndrome—A Window into Genetic Variants Leading to the Development of Cardiovascular Disease
- The Empirical Power of Rare Variant Association Methods: Results from Sanger Sequencing in 1,998 Individuals
- Systematic Detection of Epistatic Interactions Based on Allele Pair Frequencies
- Familial Identification: Population Structure and Relationship Distinguishability
- Raf1 Is a DCAF for the Rik1 DDB1-Like Protein and Has Separable Roles in siRNA Generation and Chromatin Modification
- Loss of Function of the Cik1/Kar3 Motor Complex Results in Chromosomes with Syntelic Attachment That Are Sensed by the Tension Checkpoint
- Computational Prediction and Molecular Characterization of an Oomycete Effector and the Cognate Resistance Gene
- The Dynamics and Prognostic Potential of DNA Methylation Changes at Stem Cell Gene Loci in Women's Cancer
- GTPase Activity and Neuronal Toxicity of Parkinson's Disease–Associated LRRK2 Is Regulated by ArfGAP1
- Evaluation of the Role of Functional Constraints on the Integrity of an Ultraconserved Region in the Genus
- Neurophysiological Defects and Neuronal Gene Deregulation in Mutants
- Genetic and Functional Analyses of Mutations Suggest a Multiple Hit Model of Autism Spectrum Disorders
- Negative Supercoiling Creates Single-Stranded Patches of DNA That Are Substrates for AID–Mediated Mutagenesis
- Rewiring of PDZ Domain-Ligand Interaction Network Contributed to Eukaryotic Evolution
- The Eph Receptor Activates NCK and N-WASP, and Inhibits Ena/VASP to Regulate Growth Cone Dynamics during Axon Guidance
- Repression of a Potassium Channel by Nuclear Hormone Receptor and TGF-β Signaling Modulates Insulin Signaling in
- The Retrohoming of Linear Group II Intron RNAs in Occurs by Both DNA Ligase 4–Dependent and –Independent Mechanisms
- Cell Lineage Analysis of the Mammalian Female Germline
- Association of a Functional Variant in the Wnt Co-Receptor with Early Onset Ileal Crohn's Disease
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Gene Expression and Stress Response Mediated by the Epigenetic Regulation of a Transposable Element Small RNA
- Contrasting Properties of Gene-Specific Regulatory, Coding, and Copy Number Mutations in : Frequency, Effects, and Dominance
- Homeobox Genes Critically Regulate Embryo Implantation by Controlling Paracrine Signaling between Uterine Stroma and Epithelium
- Nondisjunction of a Single Chromosome Leads to Breakage and Activation of DNA Damage Checkpoint in G2
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



