-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaMutations in or near the Transmembrane Domain Alter PMEL Amyloid Formation from Functional to Pathogenic
PMEL is a pigment cell-specific protein that forms physiological amyloid fibrils upon which melanins ultimately deposit in the lumen of the pigment organelle, the melanosome. Whereas hypomorphic PMEL mutations in several species result in a mild pigment dilution that is inherited in a recessive manner, PMEL alleles found in the Dominant white (DW) chicken and Silver horse (HoSi)—which bear mutations that alter the PMEL transmembrane domain (TMD) and that are thus outside the amyloid core—are associated with a striking loss of pigmentation that is inherited in a dominant fashion. Here we show that the DW and HoSi mutations alter PMEL TMD oligomerization and/or association with membranes, with consequent formation of aberrantly packed fibrils. The aberrant fibrils are associated with a loss of pigmentation in cultured melanocytes, suggesting that they inhibit melanin production and/or melanosome integrity. A secondary mutation in the Smoky chicken, which reverts the dominant DW phenotype, prevents the accumulation of PMEL in fibrillogenic compartments and thus averts DW–associated pigment loss; a secondary mutation found in the Dun chicken likely dampens a HoSi–like dominant mutation in a similar manner. We propose that the DW and HoSi mutations alter the normally benign amyloid to a pathogenic form that antagonizes melanosome function, and that the secondary mutations found in the Smoky and Dun chickens revert or dampen pathogenicity by functioning as null alleles, thus preventing the formation of aberrant fibrils. We speculate that PMEL mutations can model the conversion between physiological and pathological amyloid.
Published in the journal: . PLoS Genet 7(9): e32767. doi:10.1371/journal.pgen.1002286
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002286Summary
PMEL is a pigment cell-specific protein that forms physiological amyloid fibrils upon which melanins ultimately deposit in the lumen of the pigment organelle, the melanosome. Whereas hypomorphic PMEL mutations in several species result in a mild pigment dilution that is inherited in a recessive manner, PMEL alleles found in the Dominant white (DW) chicken and Silver horse (HoSi)—which bear mutations that alter the PMEL transmembrane domain (TMD) and that are thus outside the amyloid core—are associated with a striking loss of pigmentation that is inherited in a dominant fashion. Here we show that the DW and HoSi mutations alter PMEL TMD oligomerization and/or association with membranes, with consequent formation of aberrantly packed fibrils. The aberrant fibrils are associated with a loss of pigmentation in cultured melanocytes, suggesting that they inhibit melanin production and/or melanosome integrity. A secondary mutation in the Smoky chicken, which reverts the dominant DW phenotype, prevents the accumulation of PMEL in fibrillogenic compartments and thus averts DW–associated pigment loss; a secondary mutation found in the Dun chicken likely dampens a HoSi–like dominant mutation in a similar manner. We propose that the DW and HoSi mutations alter the normally benign amyloid to a pathogenic form that antagonizes melanosome function, and that the secondary mutations found in the Smoky and Dun chickens revert or dampen pathogenicity by functioning as null alleles, thus preventing the formation of aberrant fibrils. We speculate that PMEL mutations can model the conversion between physiological and pathological amyloid.
Introduction
Amyloid fibrils are polymers of single proteins that oligomerize and assemble into a characteristic fibrillar structure with a cross-beta sheet backbone [1], [2]. Amyloid formation is typically associated with pathologies, such as the Aβ aggregates in Alzheimer Disease or prion aggregates in inherited or acquired spongiform encephalopathies. However, the amyloid fold has also been exploited for functional means in prokaryotes, lower eukaryotes and even in mammals [2]–[6]. The structural and biogenetic features that distinguish functional from pathological amyloid are not well understood. Discerning these features might lead to novel therapies for amyloid diseases.
A potential model for distinguishing functional from pathological amyloids is the pigment cell-specific integral membrane glycoprotein, PMEL (also called gp100, Pmel17, or Silver) [7], [8]. Functional amyloid fibrillar sheets, composed largely of lumenal proteolytic fragments of PMEL, form the structural foundation of eumelanosomes, which are membrane-bound, pigment cell-specific lysosome-related organelles within which black and brown melanin pigments are synthesized and stored [9], [10]. The fibrils begin to form in association with intralumenal membrane vesicles (ILVs) within multivesicular melanosome precursors [11], [12], to which PMEL is selectively delivered during biosynthetic transport [13], [14]. Either in the trans Golgi network [15] or in association with the ILVs or with membrane domains destined for the ILVs [16], PMEL is cleaved by two site-specific proteases to liberate a lumenal fragment called Mα [13], [14], [16]–[18] (see Figure 1). Mα then undergoes ordered oligomerization into protofibrils that are detergent insoluble [17], detectable by electron microscopy (EM) [11], [13], [14], [17], and reactive with amyloidogenic dyes [19]. The Mα fibrils are further matured by proteolytic processing [20]–[23] and assemble into sheets [11], [24], upon which melanins deposit as they are synthesized during melanosome maturation [10], [24]. In vitro, denatured recombinant Mα fragments that are diluted into non-denaturing buffers rapidly assemble into fibrils that are classified as amyloid by a number of biophysical measures [19], [23], [25]. The physiological function of the fibrils is not entirely clear, but they likely serve to condense melanin intermediates to facilitate their detoxification, polymerization, and/or intercellular or intracellular transfer [8]. This function seems to be important for optimal pigmentation, as animal models with apparently hypomorphic mutations in PMEL show varying levels of hypopigmentation, ranging from modest in the silver mouse [26] to more pronounced in Merle dogs [27] and fading vision zebrafish [28]. In the accompanying paper by Hellström et al. [29], we show that a complete loss of PMEL expression in the Pmel -/- mouse also presents with a modest pigment dilution.
Fig. 1. Schematic diagram of PMEL proteolytic processing. 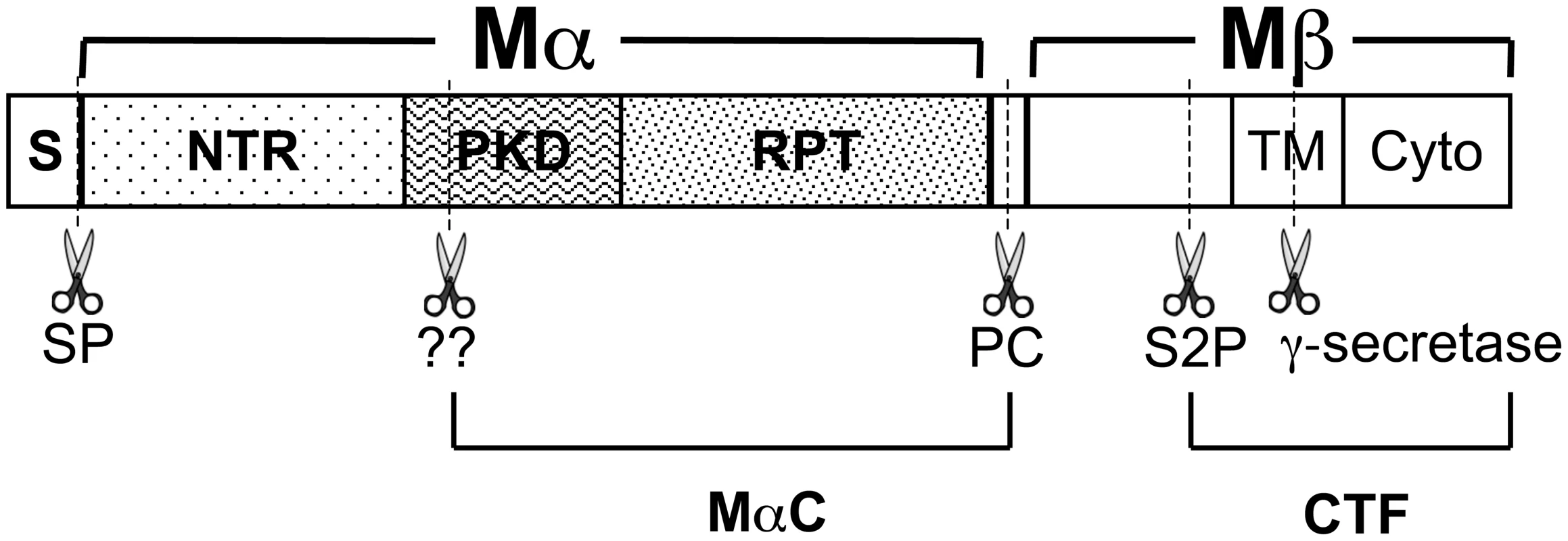
Diagram of PMEL domain structure, relevant proteolytic fragments and proteolytic cleavage sites (dashed lines and scissors). Within the primary sequence are indicated the signal peptide (S), N-terminal region (NTR), polycystic kidney disease repeat domain (PKD), repeat domain (RPT), transmembrane domain (TM) and cytoplasmic domain (Cyto). The signal peptide is removed from the N-terminus of PMEL in the ER by signal peptidase (SP). PMEL is cleaved by a proprotein convertase (PC) to produce Mα and Mβ fragments, which are linked by disulfide bonds (not shown). Subsequent cleavage by a site 2 protease (S2P) at a site proximal to the lumenal side of the TM produces a C-terminal fragment (CTF) and liberates Mα. As yet unidentified enzymes (marked “??”) further cleave Mα at as yet unknown sites to produce MαC. MαC fragments and a fragment of the PKD domain have been shown to accumulate in fibrillar stage II melanosomes [23]. Compared to the more modest effects of the PMEL mutations described above, genetic models in the chicken and horse show that in some cases, PMEL mutations can result in severe hypopigmentation. In the chicken, a nine bp in-frame insertion within the coding region for the PMEL transmembrane domain (TMD; TMinsWAP) is associated with the nearly complete loss of feather eumelanin in Dominant White (DW) chickens [30]. The pigment loss in DW chickens is associated with poor melanocyte survival in culture and with melanocyte depletion along the feather shaft in vivo [31], both exaggerations of the increased cell cycle length [32] and slow, progressive melanocyte loss [33] observed in homozygous mice bearing the recessive, hypomorphic PMEL silver mutation. However, unlike mouse silver, the DW allele is associated with impaired melanosome maturation and with melanosome loss within epidermal melanocytes [34], [35]. Moreover, the DW allele differs from mouse silver and other PMEL hypomorphs in that it confers hypopigmentation in a dominant fashion, suggesting that the DW PMEL product must either inhibit endogenous PMEL function in a dominant-negative manner or confer some gain-of-function that is detrimental to melanocyte function and/or health. Interestingly, a concomitant deletion within the conserved lumenal Polycystic Kidney Disease-1 homology (PKD) domain in the Smoky chicken (PKDΔLVVT) reverts the dominant phenotype of the TMinsWAP DW mutation, resulting in a much more modest hypopigmentation that is inherited in a recessive manner [30]. In Silver horses (HoSi), a dominant missense mutation in PMEL results in a transversion that substitutes a cysteine for the second of three consecutive arginine residues immediately following the PMEL TMD (TMR625C), causing a dilution of black pigment that is most noticeable in the mane and tail of the animal [36], [37]. An orthologously identical TMR625C mutation in the Dun chicken PMEL allele is accompanied by an additional gene deletion (TMΔ5) that eliminates five residues from the PMEL TMD [30]. However, while Dun is likewise dominant, it confers a more modest hypopigmentation, suggesting either species-specific variation in the TMR625C PMEL phenotype or that the TMΔ5 secondary mutation might partially dampen the pigmentation defect that the TMR625C mutation confers individually. Why the mutations in the DW chicken and Silver horse present a dominant phenotype and how they might be reverted by the additional deletions in the Smoky and (perhaps) Dun chickens is not understood.
Given the severity of their pigmentation phenotype, the association with loss of melanosome integrity and melanocyte viability in vivo and in cell culture, and the dominant nature of the mutations, we hypothesized that the TMinsWAP and TMR625C mutations of PMEL in the DW chicken and the Silver horse, respectively, alter the cellular or biophysical properties of PMEL to ultimately convert functional amyloid into a pathological form. Here, by recapitulating these mutations in the context of human PMEL (hPMEL), we provide evidence to support this hypothesis. Moreover, we show that the secondary PKDΔLVVT and TMΔ5 mutations in the Smoky and Dun chicken PMEL orthologues represent null or partial null alleles that revert the pathological effects of the TMinsWAP and TMR625C mutations on pigmentation. Finally, we show that changes in or near the TMD of an integral membrane amyloidogenic protein can influence the oligomerization of a distal lumenal fragment into functional amyloid. We discuss these findings with regard to their potential implications for the formation of functional vs. pathological amyloid.
Results
Mutations found in DW and HoSi PMEL alter the oligomerization potential of the human PMEL transmembrane domain
Human PMEL (hPMEL) forms pre-amyloid oligomers that are stabilized by disulfide bonds [13], and a recent study found that hPMEL formed fewer disulfide-bonded oligomers when coexpressed with chicken PMEL bearing the DW-associated TMinsWAP mutation [38]. However, it is not known whether the result in that study reflected a direct effect of the TMinsWAP mutation or an inability of hPMEL to form the appropriate disulfide bonds with chicken PMEL. To specifically test whether and how mutations in or near the PMEL TMD influence PMEL oligomerization, we introduced mutations analogous to those found in the DW chicken (TMinsWAP) and Silver horse (TMR625C) in the context of hPMEL (Figure 2A). Upon expression of these mutants or wild-type full-length hPMEL in HeLa cells, we found that the TMD mutants were equally effective as wild-type hPMEL at forming disulfide bonded oligomers, as detected by non-reducing SDS-PAGE and immunoblotting (Figure S1A). These data suggest that the PMEL mutations associated with the DW chicken and HoSi do not affect the formation of disulfide-bonded PMEL oligomers when presented within the context of hPMEL. The previously observed reduction in disulfide-bonded oligomers by hPMEL upon coexpression of chicken DW PMEL [38] thus likely reflects the inability of these cross-species PMEL isoforms to form oligomers, which is supported by our inability to co-immunoprecipitate wild-type mouse PMEL with wild-type hPMEL (data not shown).
Fig. 2. PMEL TMD mutations cause increased TMD–mediated dimerization. 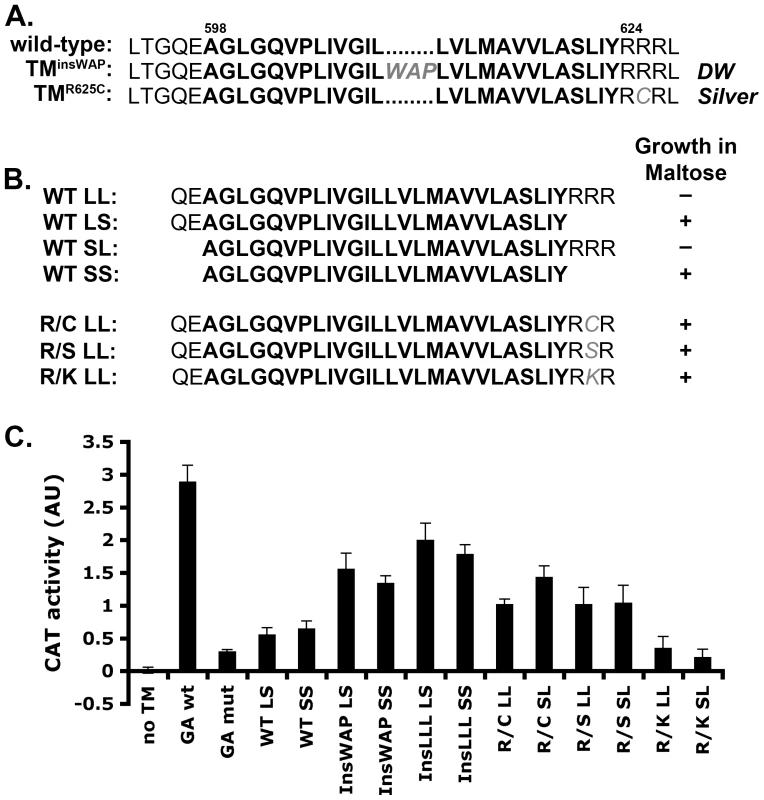
A. Wild-type human PMEL (hPMEL) TMD sequence (bold) and surrounding amino acids. In the context of hPMEL, homologous mutations to those found in the DW chicken (TMinsWAP) and Silver horse (TMR625C) are shown in gray. B. Left, TMD sequences (bold) used to make the ToxR-TMD-MBP chimeras with (L) or without (S) the N-terminal boundary residues QE and C-terminal boundary residues RRR. Right, MBP-deficient bacteria transformed with the indicated TMD-containing chimera were plated on medium with maltose as the only source of carbon; shown is whether colonies grew (+) or not (-). Growth indicates proper insertion of the chimera into the bacterial membrane. C. Representative histogram (from at least 3 similar experiments) of CAT activity, representing TMD-mediated dimerization of the chimera, as measured spectrophotometrically and normalized to protein concentration. GA, Glycophorin A TM domain. Bars, standard deviation. While oligomerization of the hPMEL lumenal domain is stabilized by disulfide bonds, it is not known whether the TMD itself has oligomeric properties that might otherwise influence lumenal domain interactions. To determine whether the hPMEL TMD can oligomerize on its own and whether the TMinsWAP and TMR625C mutations influence this property, we turned to the widely used TOXCAT assay [39]. In this assay, a chimeric protein consisting of the transcription factor ToxR, the TMD of interest, and maltose binding protein (MBP) is expressed in MBP-deficient bacteria. Proper insertion of the chimeric protein into the plasma membrane confers growth in maltose as the only carbon source, and oligomerization mediated by the TMD activates ToxR and stimulates ToxR-dependent transcription of the chloramphenicol acetyl transferase (CAT) gene. Upon expression of ToxR-TMD-MBP chimeras containing the PMEL TMD with or without the natural border residues at either end (Figure 2B), only chimeras lacking the C-terminal border residues, RRR (SS, LS), conferred growth in maltose — regardless of the presence of the N-terminal border residues, QE — despite equivalent expression of all chimeras as determined by immunoblotting (Figure S1B). This suggested that the C-terminal residues interfered with insertion of the chimera into the plasma membrane with the proper orientation. Interestingly, altering the second arginine in the C-terminal border sequence to either Cys (R/C; as in the TMR625C mutant), Ser (R/S), or Lys (R/K) allowed the chimeric protein to insert properly into the membrane, as evidenced by maltose complementation (Figure 2B). This indicates that the TMR625C substitution associated with HoSi PMEL alters the properties of the PMEL TMD.
For those chimeras that confer growth in maltose, we then tested their ability to dimerize by measuring CAT activity using a spectrophotometric assay (Figure 2C) [40]. As a positive control we used a chimera containing the strongly dimerizing TMD of Glycophorin A (GA wt), and as a negative control we used either the vector without a TMD (no TM) or a chimera containing the non-dimerizing Glycophorin A G83I mutant TMD (GA mut) [39], [41]. The wild-type PMEL TMD, lacking (SS) or containing (LS) the N-terminal border residues, conferred similar CAT activity as the negative controls. In contrast, the TMinsWAP (InsWAP) and TMR625C (R/C) mutations conferred substantial CAT activity. These data indicate that whereas the wild-type hPMEL TMD is not capable of oligomerization, both the DW-associated TMD insertion and the HoSi-associated R/C transversion facilitate TMD dimerization. Insertion of three leucines (InsLLL) in place of the WAP insertion conferred an even slightly higher CAT activity, suggesting that the increased dimerization mediated by the TMinsWAP mutation is likely due to the increase in TMD length. To test whether the R/C mutation reflected a specific property of the cysteine (such as disulfide bond formation), we tested whether replacement of the same arginine residue by serine (R/S) or lysine (R/K) affected CAT activity. Interestingly, whereas both of these mutants conferred proper insertion into the plasma membrane as indicated by growth in maltose, the R/S mutant, but not R/K, conferred CAT activity similar to that by the R/C mutant. This suggests that the dimerization conferred by the R/C mutant reflects decreased repulsion between adjacent basic RRR (or RKR) motifs.
DW – and HoSi–associated TMD mutations do not affect PMEL trafficking to premelanosomal compartments or proteolytic activation
We next tested whether the altered TMD properties associated with the TMinsWAP and TMR625C mutations influenced PMEL trafficking or processing. hPMEL without (wild-type) or with the TMinsWAP or TMR625C mutations was expressed ectopically in non-melanocytic HeLa cells by transient transfection. As previously shown, wild-type hPMEL expressed in these cells is enriched at steady state within late endosomes and lysosomes [13] (Figure 3Aa–c). Neither the TMinsWAP nor the TMR625C mutation affected this steady state localization, as shown by the predominant labeling for these mutants by immunofluorescence microscopy (IFM) on the interior of structures labeled by the late endosome/lysosome marker, LAMP1 (Figure 3Ad–i). These data suggest that the TMD mutations in dominant PMEL mutants do not affect hPMEL trafficking, supporting a similar earlier conclusion for chicken DW PMEL [38].
Fig. 3. TMD mutations do not affect PMEL trafficking, maturation, or processing. 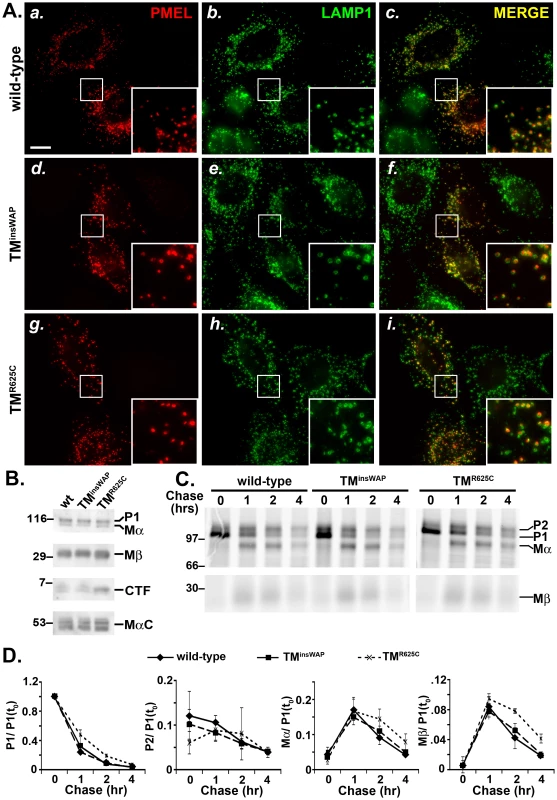
HeLa cells were transiently transfected with low (A and C–D) or high (B) DNA levels of pCI plasmids encoding wild-type, TMinsWAP or TMR625C hPMEL, and processed for analysis 48 hrs post-transfection. A. IFM analysis. HeLa cells were fixed and co-stained with antibodies against hPMEL (red; panels a, d and g) and LAMP1 (green; panels b, e, and h). Right panels (c, f, i) show the merged images, and insets show a 4X magnification of the boxed regions. Notice the similar pattern of labeling of wild-type (a–c), TMinsWAP (d–f) and TMR625C hPMEL within LAMP1-labelled late endosomes/lysosomes. Bar, 20 µm. B. Immunoblot analysis. Transfected HeLa cells were lysed and fractionated into detergent soluble and insoluble fractions. Detergent soluble fractions (top three panels) were probed with antibodies to either the hPMEL N-terminus to detect P1 and Mα (top), or the C-terminus to detect Mβ and CTF (middle panels). Detergent insoluble, fibril-enriched fractions were probed with HMB45 to detect the PMEL-derived MαC fragments (lower panel). Left, molecular weight markers; right, relevant bands are indicated. C. Metabolic pulse chase/ immunoprecipitation analysis. Transfected HeLa cells were metabolically labeled with 35S methionine/ cysteine, chased for the indicated times, and lysed in Triton X-100. PMEL was immunoprecipitated from detergent soluble fractions using an antibody to the C-terminus. Left, molecular weight markers; right, relevant bands are indicated. D. Quantification of abundance of relevant PMEL fragments from at least three pulse chase experiments as shown in C.; the band intensity of each fragment was normalized to that of P1 at time zero [P1(t0)]. Bars, standard error. In order to liberate the amyloidogenic Mα fragment, PMEL undergoes regulated proteolytic cleavage in the lumenal domain by a proprotein convertase (PC) and an as yet unidentified site 2 protease (S2P) [17], [18] (Figure 1). Mα is then further processed by as yet unidentified proteases during fibril maturation [12], [21]–[23]. The products of these cleavages are detected and semi-quantified by immunoblot analysis of detergent-soluble lysates from HeLa cells transfected with wild-type hPMEL; the characteristic Mα and Mβ fragments result from PC cleavage of full-length hPMEL [17], the C-terminal fragment (CTF) results from S2P cleavage of Mβ [18], and the MαC fragments that are enriched in detergent-insoluble fibrils result from further proteolysis of Mα [21] (Figure 1). Similar levels of all fragments are present in lysates from cells expressing TMinsWAP or TMR625C hPMEL, with the exception of a slight increase in the levels of the CTF relative to Mβ in cells expressing the TMR625C mutant (Figure 3B). Moreover, whereas wild-type and TMinsWAP hPMEL often form two species of CTF (see doublet in Figure 3B), TMR625C hPMEL forms predominantly a single species. These data suggest that TMinsWAP and TMR625C hPMEL are effectively cleaved to fibrillogenic fragments by the PC and by proteases within late endosomal compartments but that S2P cleavage is favored at one of two sites in TMR625C hPMEL and the ensuing CTF is likely more stable than the wild-type CTF.
To test whether processing kinetics are altered by the TMD mutations, we analyzed PMEL maturation in transfected HeLa cells by metabolic pulse/ chase analysis of wild-type, TMinsWAP, or TMR625C hPMEL immunoprecipitated from detergent-soluble lysates. As shown in Figure 3C, both the TMinsWAP and TMR625C mutant hPMEL were matured to the Golgi-processed P2 form, cleaved to the Mα/ Mβ forms, and disappeared from detergent-soluble lysates with roughly wild-type kinetics (Figure 3D). Moreover, for all hPMEL variants, Mα was secreted into the medium with similar kinetics and efficiency, and CTF was generated with similar kinetics (data not shown).
Altogether, these results indicate that the TMD mutations found in the DW chicken and the Silver horse affect neither the delivery of PMEL to late endocytic compartments nor its ability to be processed to amyloidogenic Mα and MαC fragments.
hPMEL with TMinsWAP and TMR625C mutations is fibrillogenic within endosomes
PMEL fibril formation is often inferred from the detection of detergent-insoluble MαC fragments by immunoblotting, but non-fibrillar mutants of PMEL that form disorganized aggregates — such as hPMEL lacking a PC cleavage site — can also generate similar fragments [12], [17]. To test whether the TMinsWAP and TMR625C variants are capable of supporting fibril formation, HeLa cells transiently expressing these variants or wild-type hPMEL were analyzed by standard electron microscopy (EM). As shown in Figure 4, stage II-melanosome-like compartments with fibrillar arrays were detected in cells expressing any of the hPMEL variants, but not in cells expressing empty vector (Figure S2). As for cells expressing wild-type hPMEL, fibrils in cells expressing TMinsWAP or TMR625C were detected within organelles that often contained internal membrane vesicles or sheets, but not in secretory or early endocytic compartments. Immunoelectron microscopy (IEM) analysis using immunogold labeling of ultrathin cryosections further showed that all of the hPMEL variants were incorporated into the fibrils (data not shown, but see below for incorporation of variants into melanosome fibrils in melanocytic cells). These data indicate that the TMinsWAP and TMR625C mutations do not impair the intrinsic ability of hPMEL to form fibrils within endosomal compartments.
Fig. 4. PMEL TMD mutants can form fibrils when overexpressed in non-pigmented cells. 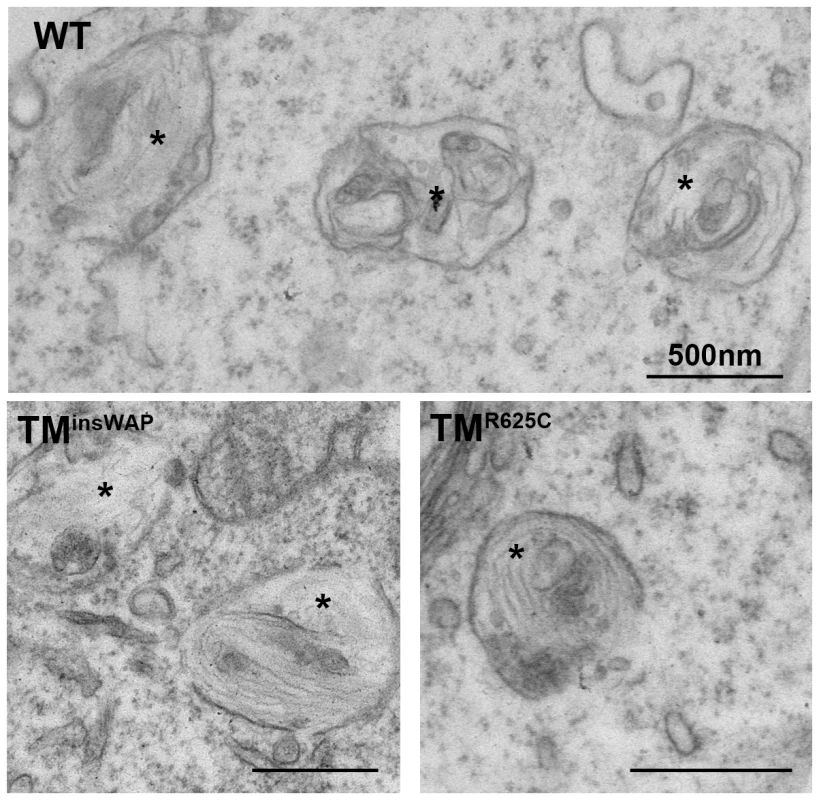
HeLa cells transiently overexpressing wild-type, TMinsWAP or TMR625C hPMEL were fixed and embedded in epon resin for conventional electron microscopy analysis. Note that elongated fibrils in the lumen of multivesicular endocytic compartments (labeled with asterisks) are observed in cells expressing all three variants of hPMEL. Bars, 500 nm. TMinsWAP and TMR625C mutations induce altered morphology and melanization of hPMEL fibrils in pigmented cells
Unlike in non-pigment cells, in which fibril maturation is inefficient and occurs within late endosomes [13], PMEL protofibrils in pigment cells mature efficiently into sheets [11] that accumulate in stage II melanosomes and that then serve as sites of melanin deposition during melanosome maturation [24]. To determine whether the TMinsWAP and TMR625C mutations influence fibril maturation or downstream pigmentation, we analyzed the behavior of hPMEL with or without these mutations expressed transiently or stably in pigment cells. In order to distinguish the transgene from the endogenous mouse PMEL, we expressed the hPMEL variants in “wild-type” mouse melanocyte cell lines, and exploited antibodies (NKI-beteb and HMB-50) that only detect hPMEL. By IFM, wild-type, TMinsWAP and TMR625C hPMEL each localized to punctate structures that partially overlapped with late endocytic compartments marked by LAMP2 and that did not overlap with mature pigmented eumelanosomes (Figure 5 and Figure S3). These data suggest that the wild-type, TMinsWAP and TMR625C hPMEL variants localize similarly in melanocytes. Endogenous mouse PMEL does not significantly co-localize with late endocytic markers (not shown) and thus the partial co-localization of ectopic hPMEL with LAMP2 might reflect less efficient delivery of this isoform to early stage melanosomes, likely due to the low expression levels attained by infection (see below) and the inability of hPMEL to interact with the mouse isoform as detected by coimmunoprecipitation (data not shown).
Fig. 5. Wild-type and TMD mutant hPMEL variants localize similarly when expressed in mouse melanocytes. 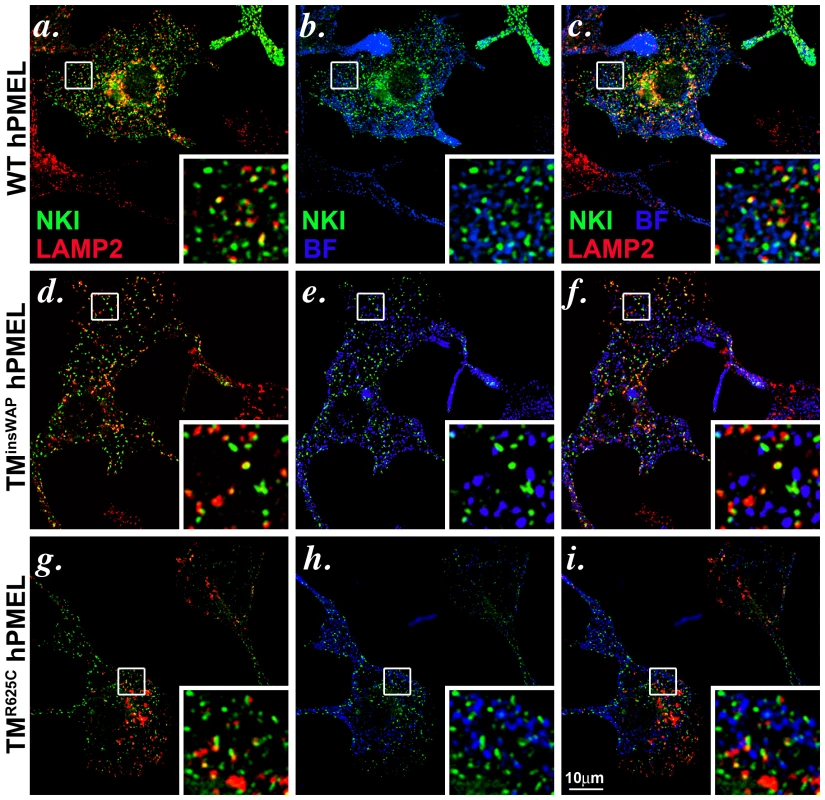
Immortalized melan-Ink4a mouse melanocytes that stably expressed wild-type (a–c), TMinsWAP (d–f) or TMR625C (g–i) variants of hPMEL were fixed and analyzed by IFM relative to the lysosomal marker, LAMP2 (red), and to pigment granules visualized by bright field microscopy (BF; pseudocolored blue). Ectopic hPMEL was detected using NKI-Beteb (green), an antibody that recognizes human PMEL, but not murine PMEL. Shown are overlaps of PMEL relative to LAMP2 (a, d, g) or melanosomes (b, e, h) alone or together (c, f, i). Insets show a 4X magnification of the boxed areas. Note that both mutants show a similar pattern of staining and co-localization as wild-type hPMEL. Individual labels and the original bright field images are shown in Figure S3. We next tested whether expression of the TMinsWAP and TMR625C hPMEL variants affect the morphology or degree of pigmentation of individual melanosomes. In mouse melan-Ink4a melanocyte stable transfectants that expressed very low levels of the dominant TMinsWAP and TMR625C hPMEL variants (∼10–20% of endogenous mouse PMEL as assessed by immunoblotting), no changes in overall pigmentation were observed relative to cells expressing wild type hPMEL (bright field images in Figure S3, with melanosomes pseudocolored blue in the insets), but this is likely due to the low expression levels (see below). Nevertheless, by IEM of ultrathin cryosections from each of the stable cell lines, thin and disorganized immature protofibrils (arrowheads) that were densely immunogold labeled with anti-hPMEL antibodies were observed within Stage I melanosomes (Figure 6a–c). However, upon maturation of the protofibrils into elongated fibrillar sheets, wild-type and mutant hPMEL showed strikingly different characteristics (compare Figure 6d to 6e–f). In cells expressing wild-type hPMEL, only very sparse immunogold labeling was observed on the parallel sheets of organized fibrils, and most of the organelles with thickened fibrillar sheets, corresponding to Stage III or IV melanosomes, were densely pigmented (Figure 6d) as has been previously reported [24]. This is consistent with the notion that although a fraction of hPMEL is delivered to the late endocytic pathway in mouse melanocytes, sufficient protein is properly trafficked to melanosomes to form fibrils. The lack of labeling on the pigmented fibrils likely reflects epitope sequestration as pigmentation proceeds [13], [14], [42]. In contrast to cells expressing wild-type hPMEL, compartments with tightly packed fibrils that were densely immunogold labeled for hPMEL were easily observed in cells expressing the TMR625C and especially the TMinsWAP variants (asterisks, Figure 6e–f; more images can be found in Figure S4). The sheets seemed unusually tightly packed with no space between the fibrils (note the spacing between fibrils in organelles that lack labeling for the transgene, indicated by brackets, and the loss of spacing in organelles that are labeled for the TMinsWAP and TMR625C transgenes). The organelles that were immunogold labeled largely showed no overt pigmentation, suggesting that either the fibrils were no longer capable of binding to melanins or that they inhibited melanin production. In some cases, particularly in cells expressing the TMR625C variant, labeling could be observed on the non-pigmented periphery of melanized melanosomes (Figure S4h and i).
Fig. 6. PMEL TMD mutants generate fibrils with aberrant morphology. 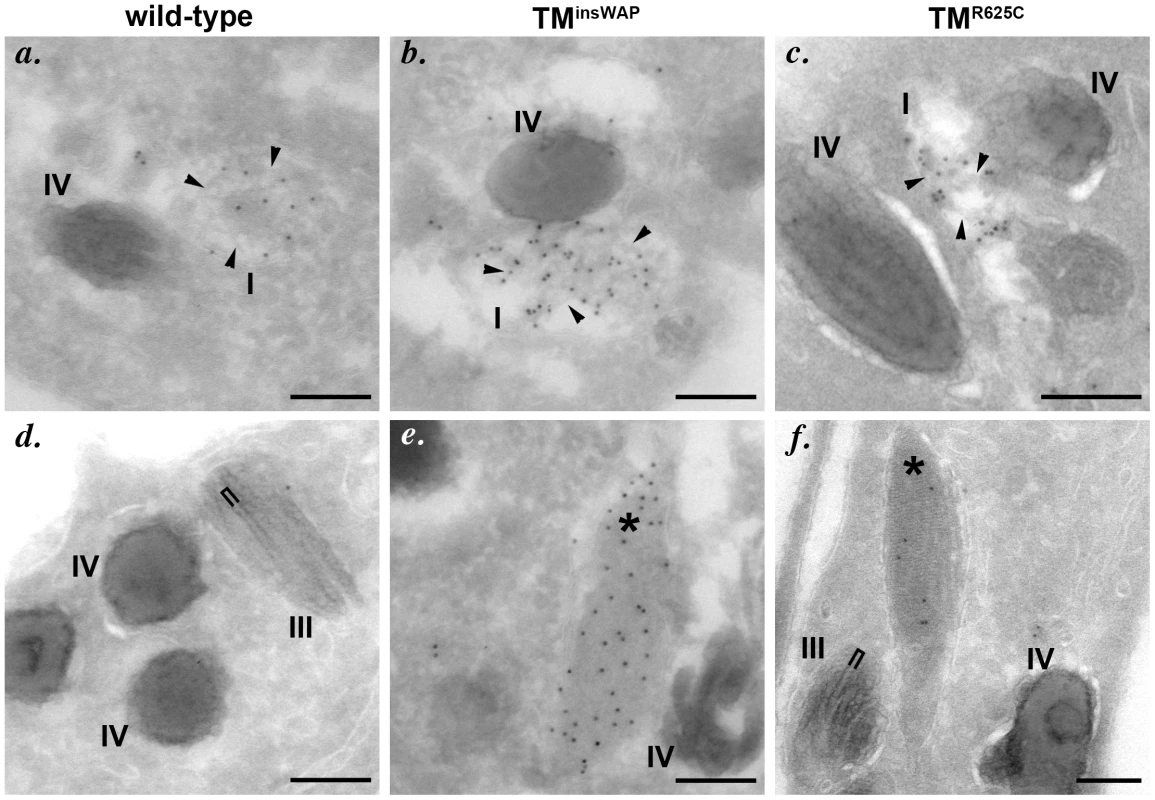
Immortalized melan-Ink4a melanocytes that stably express wild-type (a, d), TMinsWAP (b, e) or TMR625C (c, f) variants of hPMEL were fixed and processed for cryoimmunoelectron microscopy. Ultrathin cryosections were immunogold labeled for hPMEL using the hPMEL-specific NKI-beteb antibody and 10 nm protein A gold. Note that dense immunolabeling is observed on short immature fibrils (arrowheads) within stage I melanosomes from cells expressing all three hPMEL variants (upper panels, a–c), but that mature, elongated fibrils are immunogold labeled only in compartments (asterisks) within cells expressing the TMD mutants (lower panels, d–f). Also note the densely packed profile of the immunogold-labeled mature fibrils in cells expressing TMD mutant hPMEL as compared to the thick pigmented fibrils that lack labeling, which are found within stage III melanosomes in cells expressing wild-type hPMEL and which show some spacing between neighboring fibrils (bracket). Bars, 200 nm. To better test whether the variant hPMEL isoforms affected overall pigmentation, we transiently expressed them in melan-mu:MuHA, a highly pigmented mouse melanocyte cell line that is less likely than melan-Ink4a to de-differentiate (our unpublished observations), using recombinant retroviruses that coexpress EGFP. The highest hPMEL expressers were enriched by cell sorting for high EGFP expression. By immunoblotting, transgene expression was substantially higher than in the stable transfectants but still less than endogenous mouse PMEL expression (data not shown). Cells were then processed for standard EM analysis. Whereas cells expressing wild-type hPMEL were normally pigmented as compared to cells expressing empty vector (not shown) and harbored few stage II (unpigmented) melanosomes, cells expressing the TMR625C and especially the TMinsWAP variants were hypopigmented, harbored fewer pigmented melanosomes and were enriched in early stage melanosomes (Figure 7A–7B). Quantification showed that cells expressing TMinsWAP hPMEL showed an increase in non-pigmented Stage II (p = 0.006) with a concomitant loss of pigmented Stage III melanosomes (p = 0.014) as compared to wild-type hPMEL (Figure 7B, 7C); cells expressing TMR625C hPMEL likewise showed a decrease in Stage IV melanosomes (p = 0.027) with a concomitant increase in Stage II (p = 0.018) and Stage I melanosomes (p = 0.014). Importantly, mutant PMEL-expressing cells with the highest increase in early stage melanosomes consistently showed the most marked decrease in pigmented organelles, suggesting a defect in melanosome maturation or pigment production that might be associated with higher expression levels of mutant PMEL. Moreover, quantification of the number of pigmented (Stage III and IV) organelles per unit area was decreased most strikingly in cells expressing TMinsWAP (p = 0.002; Figure 7D). In cells expressing TMR625C there was also a tendency towards a decrease in the number of pigmented organelles (see Figure 7A–7C), but it was not significantly different from cells expressing wild-type PMEL (Figure 7D), likely due to the high variability encountered in these cells (see Figure 7B). These results suggest that even after short periods of time and with modest expression levels, the fibrils formed by the TMinsWAP—and to a lesser degree the TMR625C variant hPMEL—impair pigmentation.
Fig. 7. Reduced melanosome pigmentation in melanocytes expressing PMEL TMD mutants. 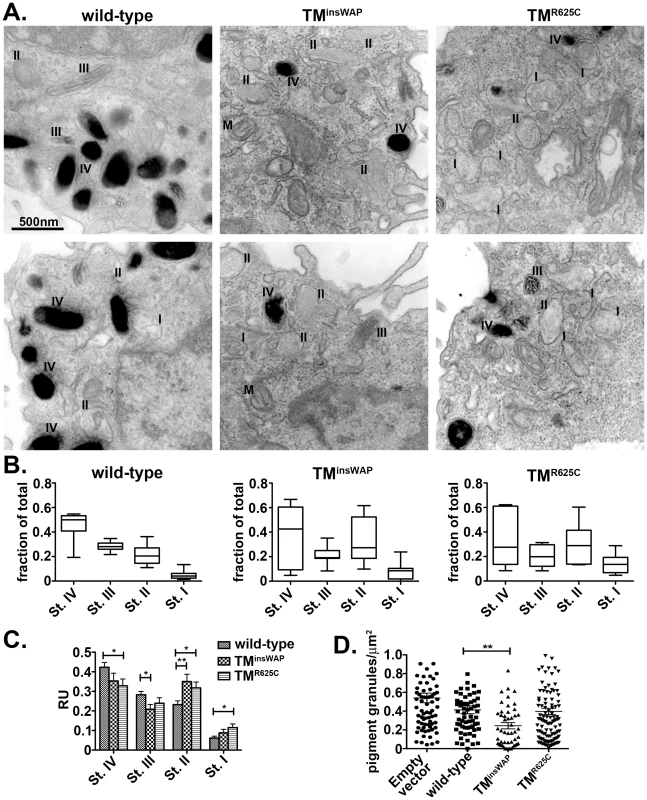
Immortalized melanMu:MuHA pigmented melanocytes were transiently infected with retroviruses that encode wild-type, TMinsWAP or TMR625C hPMEL-and co-expressed EGFP. Four days post-infection, cells expressing high levels of hPMEL transgene were selected by flow cytometric sorting for high EGFP expression, then fixed, embedded in epon resin, and processed for conventional thin section EM. A. Cells expressing wild-type hPMEL (left panels) show predominantly pigmented stage III and IV (III, IV) melanosomes and few unpigmented Stage I and II melanosomes (I, II). By contrast, cells expressing either TMinsWAP (middle panels) or TMR625C (right panels) hPMEL variants harbor many fewer Stage IV melanosomes and many more non-pigmented, Stage I-II melanosomes. M, mitochondria. B. The total number of melanosomes of each stage in 6–7 whole cells from each set of samples was quantified relative to the total number of melanosomes per cell. The mean, median, maximum (max) and minimum (min) values, and 25th and 75th quartile values for distance moved in each experimental set are shown. C. Quantification of the number of each stage melanosome divided by the number of melanosomes in that field; at least 60 fields of equal size/magnification were analyzed, containing more than 400 melanosomes of different maturation stages. *, p <0.05; **, p <0.01 as determined by ANOVA and Student's t-test. D. Quantification of the number of pigmented granules (Stage III-IV) per µm2 of cell area in cells expressing wild-type, TMinsWAP or TMR625C hPMEL variants or cells transduced with virus that did not co-express hPMEL. Each measurement is shown individually; median values are indicated. **, p<0.01 as determined by ANOVA and Student's t-test. Together, these results suggest that the mutations found in the dominant DW chicken PMEL and Silver horse influence the assembly of the fibrils into sheets, creating a tightly packed structure that may be inaccessible to pigment and/or that inhibits melanin biosynthesis.
The Smoky chicken mutation prevents formation of aberrant PMEL fibrils
The Smoky chicken is a recessive revertant of the DW allele, reflecting a second site mutation that results in deletion of four residues from the PKD domain (PKDΔLVVT) (Figure 8A) [30]. In homozygous form, the Smoky allele imparts modest pigment dilution in the feathers—similar to the modest pigment dilution observed in the silver mouse [26], the Pmel -/- mouse [29] and the fading vision zebrafish [28]—as compared to the dramatic loss of pigment imparted by the dominant DW allele. To investigate how the PKDΔLVVT mutation might reverse the DW phenotype, we created hPMEL variants with either the PKDΔLVVT deletion alone or together with the WAP insertion (PKDΔLVVT-TMinsWAP) as found in the Smoky chicken PMEL allele. The variants were expressed in HeLa cells, and their maturation, proteolytic processing, and trafficking were assessed by metabolic labeling/pulse chase and immunoprecipitation analysis and by IFM. As shown in Figure 8B, introduction of the PKDΔLVVT deletion, either by itself or in combination with the TMinsWAP insertion, impaired PMEL exit from the endoplasmic reticulum (ER) as shown by a decreased maturation into the fully glycosylated, post-Golgi P2 form. Furthermore, the PKDΔLVVT and PKDΔLVVT-TMinsWAP hPMEL forms were not efficiently processed by proprotein convertase cleavage into Mα and Mβ fragments. These data are reminiscent of the effects of deletion of the entire PKD domain [16], [22] and suggest that the PKDΔLVVT deletion impairs PMEL maturation.
Fig. 8. The PKDΔLLVT secondary mutation found in the Smoky chicken PMEL prevents delivery of TMinsWAP mutant hPMEL to fibrillogenic compartments. 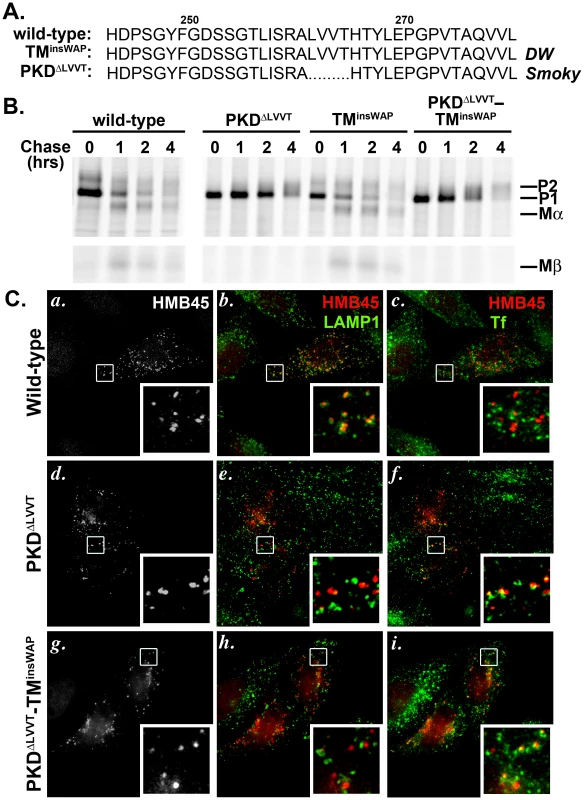
A. Sequence of PKD domain residues 245–280 in wild-type and mutant hPMEL. The PMEL gene in the Smoky chicken harbors an in-frame deletion that eliminates four amino acids from the PKD domain, corresponding to LVVT in hPMEL (PKDΔLLVT), in addition to the TMinsWAP mutation. B. Metabolic labeling/pulse chase analysis of HeLa cells transiently transfected with wild-type, PKDΔLVVT, TMinsWAP or PKDΔLVVT-TMinsWAP variants of hPMEL. Cells were labeled, chased and Triton X-100-soluble cell lysates were immunoprecipitated with an antibody to the hPMEL C-terminus as in Figure 3C. Note that variants bearing the PKDΔLVVT deletion, regardless of the presence of the TMinsWAP insertion, do not mature efficiently to P2 and fail to accumulate Mα and Mβ fragments. C. IFM analysis of HeLa cells transiently expressing wild-type (a–c), PKDΔLVVT (d–f) or PKDΔLVVT-TMinsWAP (g–i) variants of hPMEL. Cells were fixed after exposure for 30 min at 37°C to fluorophore-conjugated transferrin (Tf) to label early sorting and recycling endosomes, then stained with an antibody that recognizes only mature PMEL (HMB45) and an anti-LAMP1 antibody. Left panels (a, d, g) show PMEL labeling only; middle panels (b, e, h) show overlap of LAMP1 (green) and HMB45-labeled PMEL (pseudocolored red); and right panels (c, f, i) show overlap of internalized Tf (green) and PMEL (red). Insets show a 4X magnification of the boxed areas. Note that wild-type PMEL overlaps with LAMP1 but not internalized Tf, whereas PKDΔLLVT overlaps partially with Tf but not LAMP1. Although the pulse chase data suggested that the PKDΔLVVT mutation impairs PMEL exit from the ER, a fraction of the Golgi-modified P2 form did accumulate over time. To determine whether this fraction of “mature” PKDΔLVVT PMEL is properly trafficked to late endosomal compartments, we analyzed its localization in HeLa cells by IFM using an antibody (HMB45) that only recognizes Golgi-modified PMEL [12], [20]. Unlike wild-type hPMEL, this “mature” form of PKDΔLVVT or PKDΔLVVT-TMinsWAP hPMEL did not localize significantly to late endosomal compartments marked by LAMP1 (Figure 8C, central panels). Rather, it partially overlapped with endocytic recycling compartments labeled by internalized transferrin (Figure 8C, right panels), much like PMEL lacking the entire PKD domain [16]. This indicates that the PKDΔLVVT deletion impairs not only PMEL maturation in the early biosynthetic pathway but also its selective incorporation into ILVs for trafficking to late endosomal compartments, thus precluding access to proprotein convertase cleavage [16]. Consistent with the requirement for sorting to ILVs and proprotein convertase cleavage for fibril formation, MαC fragments are not detected in detergent-insoluble fractions of cells expressing the PKDΔLVVT or PKDΔLVVT-TMinsWAP mutants (Figure S5). We conclude that the PKDΔLVVT mutation reverts the dominant phenotype of the TMinsWAP mutation by impairing access of PMEL to fibrillogenic compartments and blocking aberrant amyloid fibril formation.
The results described above suggest that primary PMEL mutations that inhibit pigmentation in a dominant fashion can be reverted by second site mutations that prevent the aberrant PMEL variants from accumulating in fibrillogenic compartments. The dominant but milder Dun chicken PMEL allele contains both the orthologous TMR625C mutation found in the HoSi allele and an additional deletion of 5 amino acids within the TMD (TMΔ5; Figure S6A). We therefore predicted that the TMΔ5 mutation, like the Smoky-associated PKDΔLVVT mutation, would impair PMEL accumulation within fibrillogenic endosomal compartments. Consistent with this prediction, when expressed in HeLa cells, introduction of the TMΔ5 mutation in the context of hPMEL decreases the expression level of all mature (post-ER) PMEL species, including the fibrillar MαC forms (Figure S6B), even when corrected for mRNA expression (data not shown). This likely reflects enhanced ER-associated degradation due to poor folding or membrane incorporation, as the material that does exit the ER appears to be processed with normal kinetics (Figure S6C-S6D). These data suggest that while the TMΔ5 mutation does not prevent delivery of PMEL to fibrillogenic compartments (Figure S6E), it can impair the efficiency with which aberrant PMEL fibrils accumulate within such compartments. Given that the primary effect of the TMΔ5 mutation is on ER exit, it is highly likely to have similar effects in the context of the TMR625C mutation, as in Dun chickens, which does not affect ER exit (Figure 3).
Discussion
Unlike most commonly known forms of amyloid, which are thought to provoke pathologic processes, PMEL is an example of a benign and functional amyloid. Here we show how mutations in the PMEL TMD are associated with an aberrant amyloid fibril biogenetic pathway, altering the normally physiological amyloid to produce a pathological form that impairs pigmentation within melanocytes. Epidermal melanocytes from animals harboring these mutations are depleted of melanosomes [34] and have decreased viability in vitro and perhaps in vivo [31], suggesting that the formation of these aberrant fibrils impairs melanosome integrity and may be toxic to the pigment cell. Although the TMD does not form part of the amyloid core, mutations in this domain influence TMD oligomeric properties that reverberate distally on the association between the amyloidogenic domains of PMEL, as evidenced by an abnormal packing of the mutant PMEL fibrils. We also show that secondary mutations found in animals in which the pigment dilution associated with the primary pathogenic TMD mutations are dampened or reverted prevent the accumulation of these PMEL isoforms in fibrillogenic compartments, thus mimicking a PMEL knockout. This finding indicates that it is less detrimental to express no fibrils at all than to express aberrant fibrils that inhibit pigmentation and might be toxic to the melanocyte.
Interactions among TMDs are known to influence multisubunit complex assembly and function in vivo [43]–[48]. Here, we show that whereas the TMD of hPMEL normally does not promote oligomerization, introduction of either the TMinsWAP or the TMR625C mutations found in the DW chicken or Silver horse PMEL orthologues results in substantial oligomerization potential. Oligomerization by the PMEL TMD was similarly enhanced by insertion of three leucine residues in place of the DW-associated WAP insertion, suggesting that the effect reflected increased TMD length rather than specific amino acid side chain interactions. Although the observed increase in dimerization by the TMR625C could not be directly compared to the wild-type TMD with the extended cytosolic domain because the latter did not insert properly into the E. coli plasma membrane, a similar degree of oligomerization was observed upon alteration of R625 to serine as with the HoSi-associated cysteine, but not to lysine, all of which supported proper membrane insertion. This suggests that increased oligomerization mediated by the TMR625C mutation reflects removal of a positive charge from the TMD boundary, decreasing the electrostatic repulsion between neighboring PMEL molecules by the membrane proximal arginine triplet. Interestingly, the TMR625C mutation was associated with greater CTF stability. In addition, whereas we often observed a CTF doublet for both wild-type and TMinsWAP PMEL, reflecting the two possible S2P sites [18], we always detected a single TMR625C CTF species. The altered TMD mediated oligomerization of this mutant might thus result in either a greater accessibility of one site over the other or aberrant partitioning of PMEL to membrane subdomains that preferentially harbor a site-specific enzyme, akin to what has been proposed to occur between α - and β-secretases in the cleavage of APP to produce pathologic Aβ [49].
How increased TMD-mediated dimerization might influence PMEL folding, assembly, and fibril formation is not yet clear. A previous study found that the DW chicken PMEL associated with membrane microdomains to a similar degree as wild-type hPMEL [38], suggesting that the TMinsWAP mutation does not alter membrane partitioning. The same study suggested that maturation and proteolytic processing of DW chicken PMEL was not substantially different from that of wild-type hPMEL. Consistently, we find that neither the TMinsWAP nor the TMR625C mutation affect hPMEL biosynthetic trafficking, proteolytic maturation, delivery to ILVs within endosomes, or the initial stages of protofibril formation. While we could not detect a previously described effect of the TMinsWAP mutation (or of the TMR625C mutation) in reducing disulfide bond-mediated dimerization of the PMEL lumenal domain [38]—which likely reflected more a lack of heteromeric interactions between chicken and human PMEL than an effect of the TMD mutation itself—it is highly likely that the induced TMD interactions impact the orientation and proximity of PMEL dimers that form early in PMEL biosynthesis [13], [38]. Although the induced conformational changes are likely subtle and do not impact recognition by the ER quality control system, biosynthetic trafficking, or the ability to form fibrils, they do appear to have downstream effects on the assembly of fibrils into sheets and/or in the packing of the sheets. One potential explanation for these effects is that non-amyloidogenic domains of PMEL dimers that protrude from the fibrils and regulate the packing of fibrils into higher order assemblies might be positioned differently. An alternative explanation is that oligomerization via the PMEL TMD might increase the kinetics of higher order fibril assembly. Either effect might result in more tightly packed fibrils within early stage melanosomes.
How might the TMinsWAP or TMR625C PMEL variants impair melanogenesis and melanosome integrity? If indeed increased TMD oligomerization translates a conformational change to the lumenal domain to alter either the mode or kinetics of fibril polymerization into sheets, several mechanisms could be envisioned. Both altered conformation or kinetics—either by physical blockade through tighter packing or by overly rapid kinetics of sheet assembly—would potentially preclude the delivery of melanogenic enzymes, such as tyrosinase, to the lumen of the maturing melanosome [50]. This would in turn have the effect of concentrating the formation of oxidative melanin intermediates at the limiting membrane of the maturing melanosome and subjecting the limiting membrane to oxidative attack, potentially damaging the integrity of the organelle. This would explain both the loss of melanization (this study and refs. [31], [34]), despite the presence of a potentially active tyrosinase [34], [35], and the decrease in melanosome numbers [34], [35]. Release of melanosomal contents might then impact cell viability [31]. Alternatively, it is known that melanosomes are highly enriched in divalent cations [51]–[53], and PMEL has been suggested to sequester calcium [54]; alterations to PMEL fibril packing might reduce its ability to sequester divalent cations, with potential harmful effects on melanosomes by further oxidative damage. Potential negative effects on copper-dependent tyrosinase activity within melanosomes might result from a similar loss of copper sequestration [55]. A third possibility is that the altered conformation of the fibrils—which very likely are a variant form of amyloid—makes them inherently toxic. For example, Aβ amyloid has been shown to insert into and disrupt lipid bilayers [56]; a similar property of the TMinsWAP and TMR625C PMEL amyloid fibrils could potentially disrupt the melanosome membrane directly, leading to a loss of melanosome integrity and consequent loss of pigmentation. Finally, it is possible that pigmentation and melanosome viability are disrupted by an intermediate in fibril or sheet assembly that might persist due to a decrease in kinetics or that might be produced only by the variants. None of these possibilities are mutually exclusive, and it is possible that a combination of effects inhibits pigmentation.
It has not been understood why the phenotype of the Smoky chicken, with a PKDΔLVVT mutation in addition to the DW-associated TMinsWAP mutation, restores substantial pigmentation relative to the parental DW chicken [30]. We show here that the secondary PKDΔLVVT mutation prevents the accumulation of PMEL in fibrillogenic compartments, likely explaining the decreased pigment dilution observed in Smoky vs. DW chickens. The PKDΔLVVT mutation largely impairs PMEL maturation through the early biosynthetic pathway, causing retention in the ER. Moreover, the small fraction of PMEL that exits the ER is not selectively targeted to multivesicular endosomes and rather accumulates in early endosomal recycling compartments. This correlates with a lack of proprotein convertase cleavage into Mα and Mβ fragments and decreased accumulation of PMEL fragments in detergent insoluble, fibril-enriched fractions. The behavior of this mutant form of hPMEL is similar to that of hPMEL in which the entire PKD domain is deleted [16], supporting a critical role for the PKD domain in targeting PMEL to fibrillogenic compartments and perhaps directly in fibrillogenesis [23]. The results indicate that the Smoky allele is functionally a PMEL null allele that counteracts the DW mutant's pathogenic effects on pigmentation in a recessive manner by preventing the formation of aberrant fibrils. Thus, Smoky chickens show a slight pigment dilution similar to that observed in PMEL knockout mice [29] or in the hypomorphic silver mouse [26], [33] rather than a dramatic loss of eumelanin pigment as observed in the DW chicken and Silver horse. Since melanocytes in the DW chicken show decreased viability [31] and melanosome integrity [34], our studies further suggest that the formation of tightly packed TMinsWAP fibrils may be toxic to pigment cells and that it is therefore less detrimental to the cell to have no PMEL fibrils at all.
The reversion of the dominant DW phenotype by the PKDΔLVVT mutation in Smoky chickens suggests that a general mechanism for averting PMEL amyloid pathology is to prevent access of the aberrant amyloidogenic protein to compartments within which amyloid formation occurs. The Dun chicken appears to be another example of such a mechanism. Whereas the Dun chicken PMEL allele contains a mutation orthologous to that of the dominant TMR625C mutation in the toxic Silver horse PMEL, it also has a secondary deletion of 5 amino acids in the TMD (TMΔ5) [30]. Introduction of this secondary mutation into hPMEL impairs ER exit, trafficking through the plasma membrane (data not shown), and accumulation of all mature PMEL species at steady state, suggesting inefficient PMEL folding and a greater propensity for degradation. Thus, while the mechanism is different from that of the secondary PKDΔLVVT mutation in Smoky chickens, the overall effect of the TMΔ5 mutation in Dun chickens might be similar — a reduction of aberrant PMEL accumulation in fibrillogenic compartments. We therefore liken this secondary mutation to a revertant of the Silver horse phenotype. We speculate that mutations in other toxic/ pathological forms of amyloidogenic proteins that prevent appropriate accumulation of the amyloidogenic species within amyloidogenic compartments will be associated with protection from disease.
Materials and Methods
Antibodies and reagents
The mouse monoclonal antibodies used, their targets and sources were as follows: HMB45 and NKI-Beteb to PMEL were from Lab Vision (Freemont, CA); TA99 to TYRP1 was from American Type Culture Collection (Manassas, VA); H4A3 to LAMP1 was from Developmental Studies Hybridoma Bank (University of Iowa, Iowa City, IA). The rabbit polyclonal antibodies used, their targets and sources were αPep13h to the C-terminal peptide of hPMEL [13], αPmel-N to the N-terminal peptide of hPMEL [17], α-LAMP1 from Affinity BioReagents (Golden, CO), and α-MBP from New England Biolabs (Beverly, MA). Rat α-LAMP2 was from Developmental Studies Hybridoma Bank. Unless otherwise specified, chemicals were obtained from Sigma-Aldrich (St. Lois, MO). Tissue culture reagents were from Invitrogen (Carlsbad, CA). FuGENE-6 and hygromycin were from Roche Diagnostics (Indianapolis, IN).
DNA constructs and cloning
Wild-type hPMEL (long form) in pCI has been described [13] and was used as a template for site directed mutagenesis via PCR to generate the TMinsWAP, TMR625C, PKDΔLVVT, PKDΔLVVT-TMinsWAP, TMΔ5 mutations; the primers used are indicated in Table S1. Wild-type and mutant hPMEL XhoI-NotI inserts from pCI were subcloned into pBMN-IRES-Hygro (a gift from R. Scheller, Genentech, San Francisco, CA) or pBMN-IRES-EGFP retroviral vectors for stable or transient infection, respectively. For the TOXCAT assays, the pccKan expression vector was used. The pccGpA WT and pccGpA G83I mutant derivatives of pccKan, encoding wild-type or mutant glycophorin A, as well as the MBP deficient Escherichia coli (malE-) have been described [39]. PMEL wild-type or mutant TMDs were PCR-amplified from their pCI templates and cloned into the NheI-BamHI sites of pccKan. All plasmid inserts were verified by DNA sequencing.
Maltose complementation and TOXCAT
malE- bacteria were transformed with pccKan or its derivatives encoding the ToxR-TMD-MBP chimeras. Ampicillin resistant colonies were selected, grown to mid-log phase in Luria Broth, streaked over amp-M9 agar plates with either glucose (positive control) or maltose as the only source of carbon [39], and incubated for 2–4 days at 37°C. CAT activity of clones that grew in maltose was measured using a spectrophotometric assay [40]. Briefly, the pellet from 1 ml of OD600 = 0.75 culture was resuspended in 300 µl of sonication buffer (25 mM Tris pH 7.8 2 mM EDTA), sonicated, and centrifuged 30 min at 4°C, 13000 x g to obtain cell free extracts. 10 µl of lysate was combined with 230 µl of CAT reaction buffer (100 mM Tris pH 7.8, 0.4 mg/ml DTNB, 0.1 M AcetylCoA) and the A412 nm was recorded every minute for 5 min to obtain the background. Then, 10 µl of 2.5 mM chloramphenicol was added to initiate the CAT reaction and the A412 nm was recorded every minute for 30 min to determine the CAT activity according to the method by Shaw [40]. The change in A412 nm was linear throughout the experiment.
Cell culture and transgene expression
HeLa cells were grown as described previously [13] and transiently transfected with 0.1 µg DNA/ 3-cm dish for low or 7.5 µg DNA/ 10-cm dish for high transgene expression using FuGENE-6 according to the manufacturer's instructions. HeLa cells were analyzed either 48 hrs (immunofluorescence microscopy, metabolic labeling/pulse chase and immunoblotting) or 72–96 hrs (electron microscopy) post-transfection. The immortalized mouse melanocyte cell lines melan-Ink4a [57] and melan-mu:MuHA rescued cells (MuHA; [58]) were grown as described previously [57] and stably or transiently infected with the retroviral vectors described above. Stable melan-Ink4a transductants were selected with 200–400 µg hygromycin B and processed for immunofluorescence (IFM) or immunoelectron microscopy (IEM). Transiently infected MuHA cells were sorted (University of Pennsylvania Cell Sorting Core Facility, Philadelphia, PA) for high EGFP expression 96 hrs post-infection and processed for conventional electron microscopy (EM).
Immunofluorescence microscopy
Cells were fixed with 2% formaldehyde, incubated with primary and fluorochrome-conjugated secondary antibodies as described previously [13] and analyzed on a DM IRBE microscope (Leica Microsystems, Wetzlar, Germany). Digital images were captured with an Orca camera (Hamamatsu, Bridgewater, NJ) and deconvolved and manipulated with OpenLab software (Improvision, Lexington, MA). Insets were magnified using Adobe Photoshop (Adobe Systems, Mountain View, CA). For recycling endosome labeling by continuous transferrin (Tf) uptake, HeLa cells were starved in serum-free media containing 0.5% BSA 15 mM HEPES for 30 min at 37°C, incubated with 7.5 µg/ml Alexafluor488-conjugated human Tf (Molecular Probes, Eugene, OR) diluted in starvation media for an additional 30 min at 37°C, fixed, and processed for IFM as indicated above.
Electron microscopy
For conventional electron microscopy, cells were fixed in 2% glutaraldehyde 4% paraformaldehyde, dehydrated and embedded in epon resin. Ultrathin sections were contrasted with 2% uranyl acetate and analyzed by transmission electron microscopy. For immunoelectron microscopy (IEM), cells were fixed with 2% paraformaldehyde 0.5% glutaraldehyde and ultrathin frozen sections were single - or double-immunogold labeled as described previously [14], [59] using Protein A gold conjugates. For quantification of numbers of Stage I-IV melanosomes, at least 50 random fields of each cell type (>400 compartments) were analyzed.
Immunoblotting, metabolic labeling/pulse chase, and immunoprecipitation analyses
For immunoblotting, cells were harvested with 5 mM EDTA in PBS, washed with 30 mM NEM in PBS, and frozen. Thawed cells were lysed with TX-100 lysis buffer as described previously [60] in the presence of protease inhibitors and NEM and fractionated into detergent soluble and insoluble fractions by centrifugation. For metabolic labeling, cells were harvested with trypsin-EDTA, starved for 30 min in methionine/cysteine free media, labeled with 35S methionine-cysteine for 30 min and chased for the indicated periods of time. Cells were then lysed as indicated above and detergent soluble fractions were immunoprecipitated with antibodies directed to the C-terminus of hPMEL, followed by SDS-PAGE fractionation and analysis with a STORM PhosphorImager and ImageQuant software (GE Healthcare, Buckinghamshire, United Kingdom).
Supporting Information
Zdroje
1. ChitiFDobsonCM 2006 Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem 75 333 366
2. GreenwaldJRiekR 2010 Biology of amyloid: structure, function, and regulation. Structure 18 1244 1260
3. FowlerDMKoulovAVBalchWEKellyJW 2007 Functional amyloid--from bacteria to humans. Trends Biochem Sci 32 217 224
4. MajiSKPerrinMHSawayaMRJessbergerSVadodariaK 2009 Functional amyloids as natural storage of peptide hormones in pituitary secretory granules. Science 325 328 332
5. WicknerRBShewmakerFKryndushkinDEdskesHK 2008 Protein inheritance (prions) based on parallel in-register beta-sheet amyloid structures. Bioessays 30 955 964
6. HammerNDWangXMcGuffieBAChapmanMR 2008 Amyloids: friend or foe? J Alzheimers Dis 13 407 419
7. TheosACTruschelSTRaposoGMarksMS 2005 The Silver locus product Pmel17/ gp100/ Silv/ ME20: Controversial in name and in function. Pigment Cell Res 18 322 336
8. WattBRaposoGMarksMS 2010 Pmel17: An amyloid determinant of organelle structure. BucciantiniM Functional Amyloid Aggregation Trivandrum, Kerala, India Research Signpost
9. HearingVJ 2005 Biogenesis of pigment granules: a sensitive way to regulate melanocyte function. J Dermatol Sci 37 3 14
10. RaposoGMarksMS 2007 Melanosomes - dark organelles enlighten endosomal membrane transport. Nat Rev Mol Cell Biol 8 786 798
11. HurbainIGeertsWJCBoudierTMarcoSVerkleijA 2008 Electron tomography of early melanosomes: implications for melanogenesis and the generation of fibrillar amyloid sheets. Proc Natl Acad Sci USA 105 19726 19731
12. HarperDCTheosACHermanKETenzaDRaposoG 2008 Premelanosome amyloid-like fibrils are composed of only golgi-processed forms of pmel17 that have been proteolytically processed in endosomes. J Biol Chem 283 2307 2322
13. BersonJFHarperDTenzaDRaposoGMarksMS 2001 Pmel17 initiates premelanosome morphogenesis within multivesicular bodies. Mol Biol Cell 12 3451 3464
14. RaposoGTenzaDMurphyDMBersonJFMarksMS 2001 Distinct protein sorting and localization to premelanosomes, melanosomes, and lysosomes in pigmented melanocytic cells. J Cell Biol 152 809 823
15. LeonhardtRMVigneronNRahnerCCresswellP 2011 Proprotein convertases process Pmel17 during secretion. J Biol Chem 286 9321 9337
16. TheosACTruschelSTTenzaDHurbainIHarperDC 2006 A lumenal domain-dependent pathway for sorting to intralumenal vesicles of multivesicular endosomes involved in organelle morphogenesis. Dev Cell 10 343 354
17. BersonJFTheosACHarperDCTenzaDRaposoG 2003 Proprotein convertase cleavage liberates a fibrillogenic fragment of a resident glycoprotein to initiate melanosome biogenesis. J Cell Biol 161 521 533
18. KummerMPMaruyamaHHuelsmannCBachesSWeggenS 2009 Formation of Pmel17 amyloid is regulated by juxtamembrane metalloproteinase cleavage, and the resulting C-terminal fragment is a substrate for gamma-secretase. J Biol Chem 284 2296 2306
19. FowlerDMKoulovAVAlory-JostCMarksMSBalchWE 2006 Functional amyloid formation within mammalian tissue. PLoS Biol 4 e6 doi:10.1371/journal.pbio.0040006
20. ChiamentiAMVellaFBonettiFPeaMFerrariS 1996 Anti-melanoma monoclonal antibody HMB-45 on enhanced chemiluminescence-western blotting recognizes a 30-35 kDa melanosome-associated sialated glycoprotein. Melanoma Res 6 291 298
21. KushimotoTBasrurVValenciaJMatsunagaJVieiraWD 2001 A model for melanosome biogenesis based on the purification and analysis of early melanosomes. Proc Natl Acad Sci USA 98 10698 10703
22. HoashiTMullerJVieiraWDRouzaudFKikuchiK 2006 The repeat domain of the melanosomal matrix protein Pmel17/gp100 is required for the formation of organellar fibers. J Biol Chem 281 21198 22208
23. WattBvan NielGFowlerDMHurbainILukKC 2009 N-terminal domains elicit formation of functional Pmel17 amyloid fibrils. J Biol Chem 284 35543 35555
24. SeijiMFitzpatrickTMSimpsonRTBirbeckMSC 1963 Chemical composition and terminology of specialized organelles (melanosomes and melanin granules) in mammalian melanocytes. Nature 197 1082 1084
25. McGlincheyRPShewmakerFMcPhiePMonterrosoBThurberK 2009 The repeat domain of the melanosome fibril protein Pmel17 forms the amyloid core promoting melanin synthesis. Proc Natl Acad Sci USA 106 13731 13736
26. DunnLCThigpenLW 1930 The silver mouse: a recessive color variation. J Heredity 21 495 498
27. ClarkLAWahlJMReesCAMurphyKE 2006 Retrotransposon insertion in SILV is responsible for merle patterning of the domestic dog. Proc Natl Acad Sci USA 103 1376 1381
28. SchonthalerHBLampertJMvon LintigJSchwarzHGeislerR 2005 A mutation in the silver gene leads to defects in melanosome biogenesis and alterations in the visual system in the zebrafish mutant fading vision. Dev Biol 284 421 436
29. HellströmARWattBFardSSTenzaDMannströmP 2011 Inactivation of the Pmel gene alters melanosome shape but has only a subtle effect on visible pigmentation. PLoS Genet 7 e1002285 doi:10.1371/journal.pgen.1002285
30. KerjeSSharmaPGunnarssonUKimHBagchiS 2004 The Dominant white, Dun and Smoky color variants in chicken are associated with insertion deletion polymorphisms in the PMEL17 gene. Genetics 168 1507 1518
31. HamiltonH 1940 A study of the physiological properties of melanophores with special reference to their role in feather coloration. Anat Rec 78 525 548
32. SpanakisELaminaPBennettDC 1992 Effects of the developmental colour mutations silver and recessive spotting on proliferation of diploid and immortal mouse melanocytes in culture. Development 114 675 680
33. QuevedoWCFleischmannRDDyckmanJ 1981 Premature loss of melanocytes from hair follicles of light (Blt) and silver (si) mice. SeijiM Phenotypic expression in pigment cells Tokyo Tokyo Univ Press 177 184
34. BrumbaughJA 1971 The ultrastructural effects of the I and S loci upon black-red melanin differentiation in the fowl. Dev Biol 24 392 412
35. BrumbaughJALeeKW 1975 The gene action and function of two dopa oxidase positive melanocyte mutants of the fowl. Genetics 81 333 347
36. BrunbergEAnderssonLCothranGSandbergKMikkoS 2006 A missense mutation in PMEL17 is associated with the Silver coat color in the horse. BMC Genet 7 46
37. ReissmannMBierwolfJBrockmannGA 2007 Two SNPs in the SILV gene are associated with silver coat colour in ponies. Anim Genet 38 1 6
38. KuliawatRSantambrogioL 2009 A mutation within the transmembrane domain of melanosomal protein Silver (Pmel17) changes lumenal fragment interactions. Eur J Cell Biol 88 653 667
39. RussWPEngelmanDM 1999 TOXCAT: a measure of transmembrane helix association in a biological membrane. Proc Natl Acad Sci USA 96 863 868
40. ShawWV 1975 Chloramphenicol acetyltransferase from chloramphenicol-resistant bacteria. Methods Enzymol 43 737 755
41. LemmonMATreutleinHRAdamsPDBrüngerATEngelmanDM 1994 A dimerization motif for transmembrane alpha-helices. Nat Struct Biol 1 157 163
42. DonatienPDOrlowSJ 1995 Interaction of melanosomal proteins with melanin. Eur J Biochem 232 159 164
43. CossonPBonifacinoJS 1992 Role of transmembrane domain interactions in the assembly of class II MHC molecules. Science 258 659 662
44. MendrolaJMBergerMBKingMCLemmonMA 2002 The single transmembrane domains of ErbB receptors self-associate in cell membranes. J Biol Chem 277 4704 4712
45. MunterL-MVoigtPHarmeierAKadenDGottschalkKE 2007 GxxxG motifs within the amyloid precursor protein transmembrane sequence are critical for the etiology of Aβ42. EMBO J 26 1702 1712
46. NgDPDeberCM 2010 Modulation of the oligomerization of myelin proteolipid protein by transmembrane helix interaction motifs. Biochemistry 49 6896 6902
47. RutledgeTCossonPManoliosNBonifacinoJSKlausnerRD 1992 Transmembrane helical interactions: zeta chain dimerization and functional association with the T cell antigen receptor. Embo J 11 3245 3254
48. TannerKGKyteJ 1999 Dimerization of the extracellular domain of the receptor for epidermal growth factor containing the membrane-spanning segment in response to treatment with epidermal growth factor. J Biol Chem 274 35985 35990
49. EhehaltRKellerPHaassCThieleCSimonsK 2003 Amyloidogenic processing of the Alzheimer β-amyloid precursor protein depends on lipid rafts. J Cell Biol 160 113 123
50. TheosACTenzaDMartinaJAHurbainIPedenAA 2005 Functions of AP-3 and AP-1 in tyrosinase sorting from endosomes to melanosomes. Mol Biol Cell 16 5356 5372
51. SamuelsonDASmithPUlshaferRJHendricksDGWhitleyRD 1993 X-ray microanalysis of ocular melanin in pigs maintained on normal and low zinc diets. Exp Eye Res 56 63 70
52. SzekeresL 1975 Fine structure and X-ray microanalysis of melanosomes in pigmented nevi and melanomas. Arch Derm Forsch 252 297 304
53. ZarebaMSzewczykGSarnaTHongLSimonJD 2006 Effects of photodegradation on the physical and antioxidant properties of melanosomes isolated from retinal pigment epithelium. Photochem Photobiol 82 1024 1029
54. ChakrabortyAKPlattJTKimKKKwonBSBennettDC 1996 Polymerization of 5,6-dihydroxyindole-2-carboxylic acid to melanin by the pmel 17/silver locus protein. Eur J Biochem 236 180 188
55. SettySRGTenzaDSviderskayaEVBennettDCRaposoG 2008 Cell-specific ATP7A transport sustains copper-dependent tyrosinase activity in melanosomes. Nature 454 1142 1146
56. GorbenkoGPKinnunenPK 2006 The role of lipid-protein interactions in amyloid-type protein fibril formation. Chem Phys Lipids 141 72 82
57. SviderskayaEVHillSPEvans-WhippTJChinLOrlowSJ 2002 p16Ink4a in melanocyte senescence and differentiation. J Natl Cancer Inst 94 446 454
58. SettySRGTenzaDTruschelSTChouESviderskayaEV 2007 BLOC-1 is required for cargo-specific sorting from vacuolar early endosomes toward lysosome-related organelles. Mol Biol Cell 18 768 780
59. RaposoGKleijmeerMJPosthumaGSlotJWGeuzeHJ 1997 Immunogold labeling of ultrathin cryosections: application in immunology. HerzenbergLAWeirDHerzenbergLABlackwellC Handbook of Exp Immunol Cambridge, MA Blackwell Science, Inc 1 11 5th ed.
60. BersonJFFrankDWCalvoPABielerBMMarksMS 2000 A common temperature-sensitive allelic form of human tyrosinase is retained in the endoplasmic reticulum at the nonpermissive temperature. J Biol Chem 275 12281 12289
Štítky
Genetika Reprodukční medicína
Článek Produces a Long Noncoding RNA and Has General Functions in the Germline, Stem Cells, and BrainČlánek Inference of Relationships in Population Data Using Identity-by-Descent and Identity-by-StateČlánek A Genome-Wide Meta-Analysis of Six Type 1 Diabetes Cohorts Identifies Multiple Associated LociČlánek Retrotransposon-Induced Heterochromatin Spreading in the Mouse Revealed by Insertional PolymorphismsČlánek Essential Roles of BCCIP in Mouse Embryonic Development and Structural Stability of ChromosomesČlánek The Receptor Tyrosine Kinase Alk Controls Neurofibromin Functions in Drosophila Growth and LearningČlánek PCNA Ubiquitination Is Important, But Not Essential for Translesion DNA Synthesis in Mammalian CellsČlánek Genome-Wide Association Study Identifies Four Loci Associated with Eruption of Permanent Teeth
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2011 Číslo 9
-
Všechny články tohoto čísla
- Temporal Trends in Results Availability from Genome-Wide Association Studies
- Association of eGFR-Related Loci Identified by GWAS with Incident CKD and ESRD
- Genetic Variants at Chromosomes 2q35, 5p12, 6q25.1, 10q26.13, and 16q12.1 Influence the Risk of Breast Cancer in Men
- Large-Scale Gene-Centric Analysis Identifies Novel Variants for Coronary Artery Disease
- Genetic Association for Renal Traits among Participants of African Ancestry Reveals New Loci for Renal Function
- Transcriptome Kinetics Is Governed by a Genome-Wide Coupling of mRNA Production and Degradation: A Role for RNA Pol II
- Conserved Regulation of p53 Network Dosage by MicroRNA–125b Occurs through Evolving miRNA–Target Gene Pairs
- Heterozygous Mutations of Are Associated with an Increased Risk of Isolated Metopic Craniosynostosis in Humans and Mice
- Study of FoxA Pioneer Factor at Silent Genes Reveals Rfx-Repressed Enhancer at and a Potential Indicator of Esophageal Adenocarcinoma Development
- Cholesterol Metabolism Is Required for Intracellular Hedgehog Signal Transduction
- Produces a Long Noncoding RNA and Has General Functions in the Germline, Stem Cells, and Brain
- Age-Dependent Recombination Rates in Human Pedigrees
- Sequence Conservation and Functional Constraint on Intergenic Spacers in Reduced Genomes of the Obligate Symbiont
- Sex Chromosome Mosaicism and Hybrid Speciation among Tiger Swallowtail Butterflies
- A Negative Feedback Loop That Limits the Ectopic Activation of a Cell Type–Specific Sporulation Sigma Factor of
- Phased Whole-Genome Genetic Risk in a Family Quartet Using a Major Allele Reference Sequence
- Mutations in or near the Transmembrane Domain Alter PMEL Amyloid Formation from Functional to Pathogenic
- Inactivation of Alters Melanosome Shape But Has Only a Subtle Effect on Visible Pigmentation
- Novel Interactions between Actin and the Proteasome Revealed by Complex Haploinsufficiency
- Germline Genetic Variants Disturbing the /LIN28 Double-Negative Feedback Loop Alter Breast Cancer Susceptibility
- Separation of Recombination and SOS Response in RecA Suggests LexA Interaction Sites
- Inference of Relationships in Population Data Using Identity-by-Descent and Identity-by-State
- Misregulation of Scm3p/HJURP Causes Chromosome Instability in and Human Cells
- A Noncoding Point Mutation of Causes Multiple Developmental Malformations and Obesity in Twirler Mice
- The Evolutionarily Conserved Longevity Determinants HCF-1 and SIR-2.1/SIRT1 Collaborate to Regulate DAF-16/FOXO
- A Genome-Wide Metabolic QTL Analysis in Europeans Implicates Two Loci Shaped by Recent Positive Selection
- Bacterial Communities of Diverse Species: Ecological Context of a Host–Microbe Model System
- A Genome-Wide Meta-Analysis of Six Type 1 Diabetes Cohorts Identifies Multiple Associated Loci
- Elongator Complex Influences Telomeric Gene Silencing and DNA Damage Response by Its Role in Wobble Uridine tRNA Modification
- Elevated Proteasome Capacity Extends Replicative Lifespan in
- Genome-Wide Analysis of Heteroduplex DNA in Mismatch Repair–Deficient Yeast Cells Reveals Novel Properties of Meiotic Recombination Pathways
- MicroRNA Predictors of Longevity in
- An Iterative Genetic and Dynamical Modelling Approach Identifies Novel Features of the Gene Regulatory Network Underlying Melanocyte Development
- Atypical AT Skew in Firmicute Genomes Results from Selection and Not from Mutation
- Retrotransposon-Induced Heterochromatin Spreading in the Mouse Revealed by Insertional Polymorphisms
- Genomic Analysis of QTLs and Genes Altering Natural Variation in Stochastic Noise
- The Abnormal Phenotypes of Cartilage and Bone in Calcium-Sensing Receptor Deficient Mice Are Dependent on the Actions of Calcium, Phosphorus, and PTH
- Cell Type–Specific Transcriptome Analysis Reveals a Major Role for and miR-200b in Mouse Inner Ear Morphogenesis
- Essential Roles of BCCIP in Mouse Embryonic Development and Structural Stability of Chromosomes
- IAP1-Mediated Ubiquitylation Controls Activation of the Initiator Caspase DRONC Independent of Protein Degradation
- VANG-1 and PRKL-1 Cooperate to Negatively Regulate Neurite Formation in
- The Receptor Tyrosine Kinase Alk Controls Neurofibromin Functions in Drosophila Growth and Learning
- Comparative and Functional Genomics of PD630 for Biofuels Development
- Identification of Type 1 Diabetes–Associated DNA Methylation Variable Positions That Precede Disease Diagnosis
- PCNA Ubiquitination Is Important, But Not Essential for Translesion DNA Synthesis in Mammalian Cells
- Genetic Effects at Pleiotropic Loci Are Context-Dependent with Consequences for the Maintenance of Genetic Variation in Populations
- Genome-Wide Association Study Identifies Four Loci Associated with Eruption of Permanent Teeth
- Bmp and Nodal Independently Regulate Expression to Maintain Unilateral Nodal Activity during Left-Right Axis Specification in Zebrafish
- Inter-Allelic Prion Propagation Reveals Conformational Relationships among a Multitude of [] Strains
- Emergence and Modular Evolution of a Novel Motility Machinery in Bacteria
- Histone Methyltransferase MET-2 Shields the Male X Chromosome from Checkpoint Machinery and Mediates Meiotic Sex Chromosome Inactivation
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Retrotransposon-Induced Heterochromatin Spreading in the Mouse Revealed by Insertional Polymorphisms
- The Evolutionarily Conserved Longevity Determinants HCF-1 and SIR-2.1/SIRT1 Collaborate to Regulate DAF-16/FOXO
- Genome-Wide Analysis of Heteroduplex DNA in Mismatch Repair–Deficient Yeast Cells Reveals Novel Properties of Meiotic Recombination Pathways
- Association of eGFR-Related Loci Identified by GWAS with Incident CKD and ESRD
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



