-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaA Negative Feedback Loop That Limits the Ectopic Activation of a Cell Type–Specific Sporulation Sigma Factor of
Two highly similar RNA polymerase sigma subunits, σF and σG, govern the early and late phases of forespore-specific gene expression during spore differentiation in Bacillus subtilis. σF drives synthesis of σG but the latter only becomes active once engulfment of the forespore by the mother cell is completed, its levels rising quickly due to a positive feedback loop. The mechanisms that prevent premature or ectopic activation of σG while discriminating between σF and σG in the forespore are not fully comprehended. Here, we report that the substitution of an asparagine by a glutamic acid at position 45 of σG (N45E) strongly reduced binding by a previously characterized anti-sigma factor, CsfB (also known as Gin), in vitro, and increased the activity of σG in vivo. The N45E mutation caused the appearance of a sub-population of pre-divisional cells with strong activity of σG. CsfB is normally produced in the forespore, under σF control, but sigGN45E mutant cells also expressed csfB and did so in a σG-dependent manner, autonomously from σF. Thus, a negative feedback loop involving CsfB counteracts the positive feedback loop resulting from ectopic σG activity. N45 is invariant in the homologous position of σG orthologues, whereas its functional equivalent in σF proteins, E39, is highly conserved. While CsfB does not bind to wild-type σF, a E39N substitution in σF resulted in efficient binding of CsfB to σF. Moreover, under certain conditions, the E39N alteration strongly restrains the activity of σF in vivo, in a csfB-dependent manner, and the efficiency of sporulation. Therefore, a single amino residue, N45/E39, is sufficient for the ability of CsfB to discriminate between the two forespore-specific sigma factors in B. subtilis.
Published in the journal: . PLoS Genet 7(9): e32767. doi:10.1371/journal.pgen.1002220
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002220Summary
Two highly similar RNA polymerase sigma subunits, σF and σG, govern the early and late phases of forespore-specific gene expression during spore differentiation in Bacillus subtilis. σF drives synthesis of σG but the latter only becomes active once engulfment of the forespore by the mother cell is completed, its levels rising quickly due to a positive feedback loop. The mechanisms that prevent premature or ectopic activation of σG while discriminating between σF and σG in the forespore are not fully comprehended. Here, we report that the substitution of an asparagine by a glutamic acid at position 45 of σG (N45E) strongly reduced binding by a previously characterized anti-sigma factor, CsfB (also known as Gin), in vitro, and increased the activity of σG in vivo. The N45E mutation caused the appearance of a sub-population of pre-divisional cells with strong activity of σG. CsfB is normally produced in the forespore, under σF control, but sigGN45E mutant cells also expressed csfB and did so in a σG-dependent manner, autonomously from σF. Thus, a negative feedback loop involving CsfB counteracts the positive feedback loop resulting from ectopic σG activity. N45 is invariant in the homologous position of σG orthologues, whereas its functional equivalent in σF proteins, E39, is highly conserved. While CsfB does not bind to wild-type σF, a E39N substitution in σF resulted in efficient binding of CsfB to σF. Moreover, under certain conditions, the E39N alteration strongly restrains the activity of σF in vivo, in a csfB-dependent manner, and the efficiency of sporulation. Therefore, a single amino residue, N45/E39, is sufficient for the ability of CsfB to discriminate between the two forespore-specific sigma factors in B. subtilis.
Introduction
When cells of Bacillus subtilis enter stationary phase and face severe nutrient depletion, they may embark into a developmental pathway that results in the production of a dormant, highly resistant endospore [1]. Sporulation involves the asymmetric division of the rod-shape cell into a smaller forespore, the future spore, and a larger mother cell. Soon after asymmetric cell division, the mother cell engulfs the forespore, eventually releasing it as a free protoplast within its cytoplasm. Following engulfment completion, the forespore becomes encased in a series of protective layers after which it is released into the environment through lysis of the mother cell [1]. Underlying the differentiation process are mother cell - and forespore-specific programs of gene expression administered by a cascade of cell type-specific RNA polymerase sigma factors. σF and σE govern the initial stages in development in the forespore and in the mother cell, respectively. At late stages of development, σF is replaced by σG (Figure 1A), whereas σK replaces σE. The sporulation-specific sigma factors are produced prior to their period of activity, and maintained inactive until the successful conclusion of key morphological events during development. Both σF and σE are synthesized in the predivisional cell. Proper septation is a prerequisite for the activation of σF in the forespore and soon after a signaling pathway initiated by σF leads to the activation of σE in the mother cell. Likewise, synthesis of σG and σK is initially driven by σF and σE, respectively. However, σE-dependent gene expression is required for the activation of σG following engulfment completion and when active, σG initiates a signaling pathway that causes the activation of σK ([1]–[3] see also below). The double responsiveness of the cell-type specific σ factors to proper morphogenesis and to intercompartmental signaling pathways effectively links the forespore and mother cell programs of gene expression and keeps gene expression in close register with the course of morphogenesis. Importantly, proper timing of sigma factor activation is essential for the fidelity of the developmental process [reviewed by [1]–[3]].
Fig. 1. Segregation of σF and σG activities and mutagenesis of sigG. 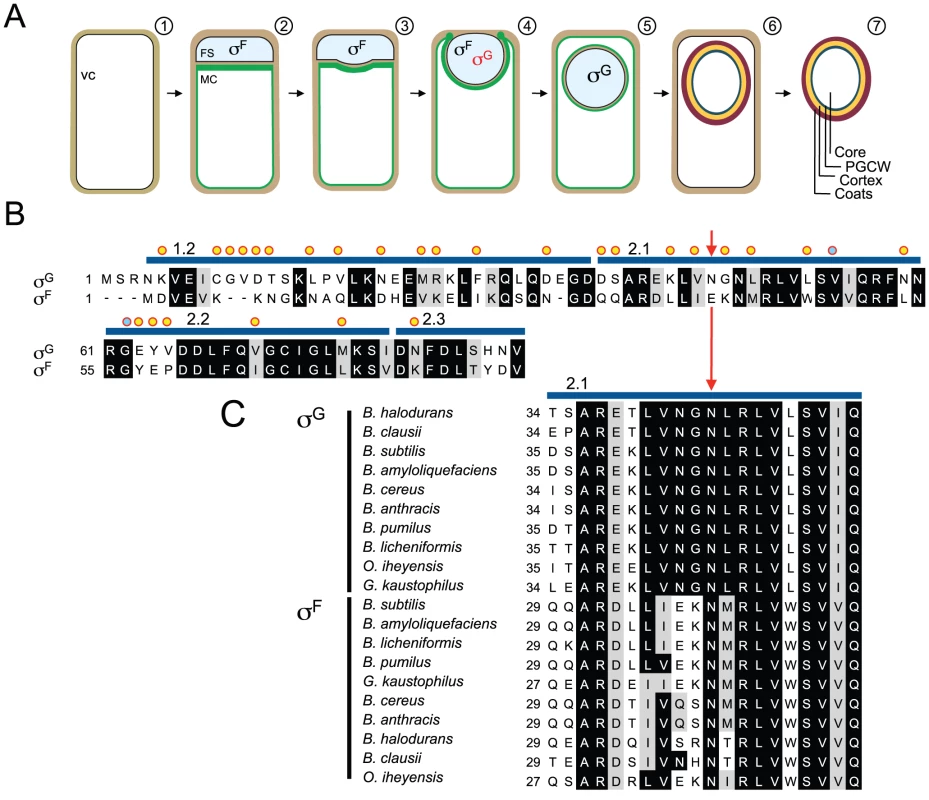
Panel A shows the main stages in sporulation and the temporal windows of activity of σF and σG in the forespore. Shown is (1) a pre-divisional cell, (2) a cell that has completed asymmetric division forming a forespore (FS) and a mother cell compartment (MC), (3, 4) a cell during and (5) after engulfment completion, (6) the following assembly of the spore protective layers and (7) release of the mature spore upon mother cell lysis. The mother cell membranes are colored in green. σF is activated in the forespore soon after polar division, and transcribes the sigG gene, but σG only becomes active following engulfment completion. Red and black denote inactive and active σG, respectively. Panel B shows an alignment between the N-terminal regions of σG and σF from B. subtilis. Residues in σG that were subject to mutagenesis are indicated by yellow circles or, for N45, by a red arrow. The blue lines above the sequence delimit conserved regions 1.2 through 2.3. Residues previously implicated in binding of SpoIIAB to σF are marked with a blue circle. Panel C shows the alignment of region 2.1 of B. subtilis σG (highlighting residue N45 with a red arrow) with the same region of σG and σF from other, selected, Bacillus species. The alignments (B and C) were prepared using ClustalW (http://www.ch.embnet.org/software/ClustalW.html). Black and grey backgrounds highlight identical or conserved residues, respectively. This study addresses the mechanisms involved in the regulation of the activity of σG. Productive transcription of the sigG gene (coding for σG) is controlled by σF [4], [5]. However, sigG is not transcribed as soon as σF becomes active. The delay appears to result from an as yet poorly understood dependency of sigG transcription upon the activity of σE in the mother cell [6], [7]. σG can be detected in the forespore towards the end of the engulfment sequence, but its window of activity begins only after engulfment completion. Activity of σG requires the assembly of a novel type of secretion system formed by eight mother cell proteins (AA through AH) coded for by the σE-controlled spoIIIA operon, and by the forespore-specific, σF-controlled protein SpoIIQ [8]–[14], with the assistance of the membrane protein translocase SpoIIIJ [8],[15]–[18]. The SpoIIIA-SpoIIQ complex spans the intermembrane space that separates the forespore and the mother cell establishing a direct connection between the cytoplasm of the two cells [8], [10], [14], [19]. Recent work has lead to the concept that the channel acts as a feeding tube, maintaining the potential for macromolecular synthesis when the forespore becomes isolated from the external medium [9]. This model brings the important implication that the activation of σG in engulfed forespores does not necessarily involve counteracting a specific inhibitor or inhibitors of σG. However, once active, σG recognizes its own promoter, creating a positive feedback loop that causes its levels to increase rapidly [4], [5]. This autoregulatory effect implies the tight regulation of σG activation so that its normal timing and cell specificity are both observed, and raises questions regarding the mechanisms that prevent activation of the positive feedback in the forespore prior to engulfment completion, or in non-sporulating cells.
Three negative regulators of σG are known, the LonA protease, and the anti-sigma factors SpoIIAB and CsfB [12], [20]–[22]. LonA, an ATP-dependent serine protease, acts mainly to prevent inappropriate activity of σG under culture conditions in which sporulation is not favored [22]–[24]. During sporulation LonA may only be active in the mother cell, because its forced expression in the forespore strongly interferes with sporulation [23], [24]. Genetic and biochemical experiments have shown that SpoIIAB, the anti-sigma factor that maintains σF inactive prior to the asymmetric division of sporulating cells, also binds to σG [12], [24]–[26]. However, while SpoIIAB contributes to the inactivity of σG under non-sporulation conditions and in the mother cell during sporulation it does not play a critical role in the negative regulation of σG in the forespore [8], [21], [24]). A third negative regulator of σG is CsfB (also known as Gin), a novel type of Zn2+ anti-sigma factor [20], [27], [28]. CsfB combines two properties expected for a factor capable of inhibiting σG prior to engulfment completion: specificity for σG (unlike SpoIIAB, CsfB does not binds to σF) and its early presence in the forespore compartment [20], [28], [29]. However, although one group initially proposed that CsfB had a key role in the negative regulation of σG in the pre-engulfed forespore [20], other groups did not observe massive premature activation of σG in the forespore upon deletion of the csfB gene [8], [27].
While the auto regulatory nature of σG seems to justify the existence of multiple negative regulators, none of the known regulators per se, seems to have a decisive role in preventing activation of the σG positive feedback loop. Because σF and σG are very similar proteins, we reasoned that the residues in which the two proteins differ could hold the key to their differential regulation. We changed all the residues within conserved regions 1.2 through the beginning of region 2.3 of σG in which it differs from σF to the residue found in this latter protein. We report the identification of a mutation (N45E) that reduces binding of CsfB to σG in vivo and in vitro. The mutation also results in the appearance of a population of stationary phase cells in which σG becomes active. We show that σG drives expression of csfB in these cells, setting-up a negative feedback loop that limits its activation across the population.
We further show the importance of N45 in σG and its equivalent in σF (E39), in the different responsiveness of the two forespore-specific sigma factors to CsfB. While unable to bind to wild type σF, CsfB interacts with a form of σF in which E39 is replaced by an N residue, found in the corresponding position of σG (N45). Importantly, we show that the E39N substitution can strongly inhibit the forespore-specific activity of σF and the efficiency of sporulation. Thus, a single amino acid residue allows CsfB to discriminate between the two highly similar forespore-specific sigma factors. This property is likely to be widespread, because N45 is invariant in Bacillus orthologues of σG, while with a single exception N is excluded from the equivalent position in the σF proteins of the same species.
Results
A mutation in conserved region 2.2 that increases the activity of σG
Since σF is active in the forespore in a temporal window when σG is kept inactive (Figure 1A), we reasoned that we would be able to find one or more substitutions that would render σG prematurely active. We initiated this study by changing most of the residues within regions 1.2 and 2.1 through the beginning of region 2.3 of σG that differed from σF to the amino acid found at the equivalent position in this latter protein (Figure 1B). The mutations were generated in vitro and transferred by congression to the sigG locus (see the Materials and Methods section). We then screened for mutants exhibiting elevated levels of σG -directed gene expression under non-sporulation conditions (during growth in LB) as these conditions previously led to the identification of two negative regulators of σG [21], [22]. This is possible because active σG utilizes its own promoter, leading to the establishment of a positive auto regulatory loop that reinforces its activity [5]. We found a single substitution at codon 45 of the sigG gene, an asparagine to a glutamic acid (henceforth N45E) that increased the activity of σG in vivo, as monitored using a fusion of the σG-responsive sspE promoter to lacZ [5]. The sspE gene codes for an abundant small acid-soluble protein required for the efficient return of spores to vegetative growth, and that is normally expressed in the forespore when σG becomes active [30]–[32]. The N45E mutation stimulated PsspE-lacZ transcription in colonies of cells growing on solid medium as well as in cells growing in liquid medium, where β-galactosidase activity was 2 fold higher in N45E mutant cells than in wild type cells (Figure 2A and 2B). On liquid medium, the activity of σGN45E was higher when the cells entered stationary phase (Figure 2B). The augmented expression of PsspE-lacZ could be due to increased activity of σG or alternatively to the titration by σGN45E of a negative regulator of σF, which at least under some conditions is also able to direct transcription from the sspE promoter [5]. To test the model that σGN45E could titrate an inhibitor of σF, we first examined the effect of two additional point mutations, F91A and Y94A, in region 2.3 of σG (see Figure S1A). These residues are presumed to play a role in promoter melting (reviewed by [33]), and alanine substitutions at these positions, while allowing the accumulation of σG, inactivate the sigma factor (Text S1 and Figure S1). Importantly, the N45E-stimulated expression of PsspE-lacZ was abolished in a N45E/F91A/Y94A triple mutant (data not shown). This finding established that the N45E stimulated transcription of PsspE-lacZ was dependent on σG itself. None of the other sigG mutations screened increased expression of PsspE-lacZ, as illustrated by the sigGV44I mutant, bearing a valine to isoleucine substitution at codon 44 (V44I) (Figure 2A and 2B).
Fig. 2. Identification of mutants with increased activity of σG. 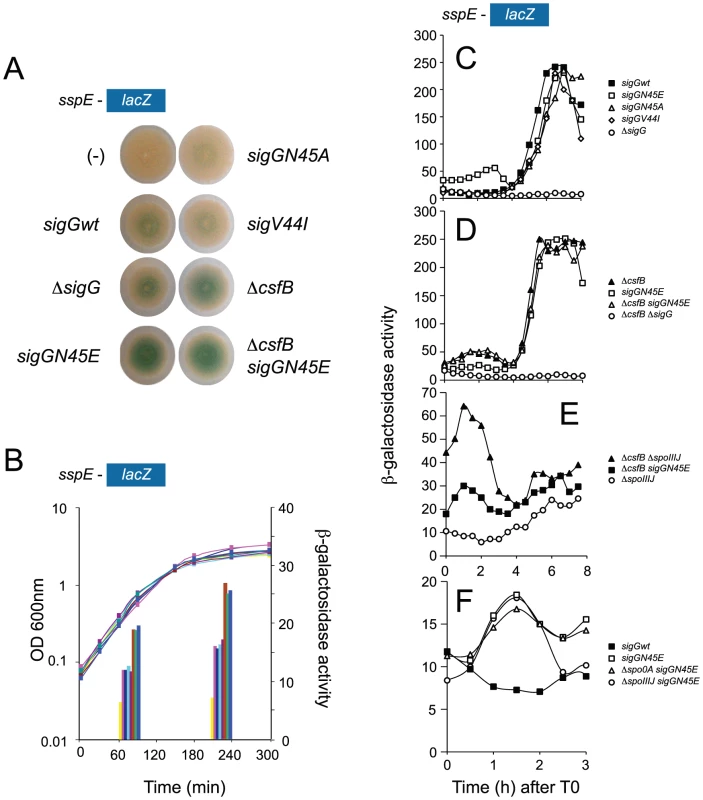
Panel A illustrates a colony screening for enhanced β-galactosidase production from the σG-controlled PsspE-lacZ reporter fusion on LB plates containing X-Gal. Panel B shows the quantitative analysis of enzyme activity for the same strains shown in A, on liquid LB cultures at mid-log and at the onset of stationary phase. The following strains are shown (Table S1 shows the complete genotype of all strains): wild type background (pink in panel B), sigGN45E (brown in panel B), sigGN45A (light blue in panel B), sigGV44I (purple in panel B), ΔcsfB (green in panel B), and ΔcsfB sigGN45E (blue in panel B). Controls for background levels are the wild type MB24 bearing no reporter fusion (“−” symbol in panel A, yellow bars in panel B), and a strain carrying the PsspE-lacZ fusion in conjunction with a sigG deletion mutation (dark blue in panel B). In panels C, D, E, and F the activity of wild type σG or its mutant forms σGN45A, σGN45E, or σGV44I was examined during sporulation in DSM medium in a wild type background or in strains carrying mutations in genes known to control the initiation of sporulation or to influence the activity of wild type σG. All strains used in this analysis carry the σG-controlled PsspE-lacZ fusion. Panel C, shows the activity profile for wild type σG, and for σGN45E, σGN45A, or σGV44I in an otherwise wild type background. Residual expression of PsspE-lacZ in a ΔsigG mutant is included for reference. Panel D compares the activity of wild type σG in a ΔcsfB mutant to the activity of σGN45E in a wild type background or in a ΔcsfB mutant. Expression of PsspE-lacZ in a ΔsigG ΔcsfB mutant is also represented. Panel E compares the activity of wild type σG in a ΔspoIIIJ mutant and a ΔspoIIIJ ΔcsfB double mutant to that of σGN45E in a ΔspoIIIJ mutant. Panel F shows the activity σGN45E in a wild type background, or in the absence of sigF or spo0A (AH6626, open triangles). The activity of wild type σG in a wild type background is shown for reference. The various strains were grown in sporulation medium (DSM) and sampled at hourly intervals after T0 (denoting the end of the logarithmic phase of growth). β-galactosidase activity is shown in Miller Units. In all cases, the various forms of σG are produced from the wt or sigG mutant alleles present at the sigG locus. Panels C-F show the results of representative experiments, which in all cases were conducted independently at least three times. We hypothesized that the N45 residue was a contact site for a putative inhibitor of σG, which was eliminated by the N45E substitution. As a test of this idea we replaced the asparagine residue by an alanine (henceforth N45A), a substitution expected to remove any positive contribution of the wild type amino acid side chain to a presumed interaction while maintaining protein structure [34]. Unexpectedly, the N45A substitution did not increase σG-directed transcription on colonies of cells growing on LB medium nor on liquid medium cultures (Figure 2A and 2B). This observation suggests that the side chain of N45 may not be essential for a direct interaction of σG with an inhibitory factor. One alternative possibility is that N45E interferes with the binding of a putative inhibitor to σG.
Activity of σGN45E during sporulation
We next studied the effect of the N45E substitution on the activity of σG during sporulation in liquid Difco sporulation medium (DSM). In this system, sporulation is induced by exhaustion of key nutrients, and its onset defined as the point at which a culture enters stationary phase. None of the sigG mutants that we screened in LB medium caused a Spo− phenotype (data not shown), but we looked at PsspE-lacZ transcription during sporulation as the mutations could alter the normal activity profile of σG. In wild type cells, expression of PsspE-lacZ was sharply induced 4 hours after the onset of stationary phase and reached maximum levels around hour 6 (Figure 2C). In keeping with the link between the activity of σG and engulfment completion, induction of PsspE-lacZ expression at hour 4 coincided with forespore engulfment in most cells of the population, as judged by FM4-64 staining (not shown). In N45E cells PsspE-lacZ expression followed a bi-modal pattern, with an early period that peaked 2 hours after the onset of stationary phase and a second, starting at hour 4, superimposable to the window of σG activity seen for wild type cells (Figure 2C). The activity profile of σG and σGN45E paralleled the accumulation of the proteins, as assessed by immunobloting with an anti-σG antibody [17]. Both σG and σGN45E accumulated to maximum levels at hour 4 of sporulation in consonance with the main period of PsspE-lacZ expression, following engulfment completion (Figure S2C). However, σGN45E begun to accumulate earlier than the wild type protein, soon after the onset of stationary phase in DSM, which correlates with the first period of PsspE-lacZ expression in the N45E mutant (Figure S2C).
Only the second period of PsspE-lacZ expression was seen for the V44I and the N45A mutants (Figure 2C), consistent with the observation that these mutations did not enhance expression of the reporter fusion in our initial screen (see above). Also consistent with the conclusion of our initial screen that the increased expression of the PsspE-lacZ reporter was not indirectly caused by titration of a negative regulator of σF (see above), the N45E mutation did not increase expression of a lacZ fusion to the promoter for a gene, spoIIQ, controlled by σF (spoIIQ-lacZ, [13]) (Text S1 and Figure S2A). In addition, the first period of σGN45E activity was still observed independently of sigF, coding for σF, which normally drives transcription of sigG in the forespore (Figure 2F; see also below). While the sspE promoter can also be utilized by σF [8], [27], it is clear that the first period of σGN45E activity is σF-independent. This first period also occurred in cells with deletion mutations of the spoIIIJ (Figure 2E) and spoIIIA loci (not shown), which are required for σG activity following engulffment completion. In fact, the first peak of σGN45E activity was seen even in cells of a spo0A deletion mutant ([35], [36], and references therein), which codes for the master regulatory protein governing entry into sporulation and without which the asymmetric division that produces the forespore compartment does not takes place [37](Figure 2F).
Altogether, these results show that the effect of the N45E substitution on PsspE-lacZ transcription during stationary phase in sporulation medium was dependent on and mediated by σG. The results also show that the second peak of σGN45E activity remained dependent on the normal control mechanisms that govern σG production and activation during sporulation.
σGN45E is activated in a population of stationary phase cells
The results described in the preceding section could be explained if the N45E mutant segregated two distinct cellular populations, one with a normal pattern of σG activity, the other activating σG independently of sporulation. To test this possibility, the activity of σGN45E was localized during stationary phase in DSM, using a PsspE-cfp transcriptional fusion [10]. Note that under our experimental conditions, asymmetric division was completed in most of the cell population between hours 2 and 3 after entry into stationary phase (as assayed by staining with the membrane dye FM4-64), and engulfment was completed around hour 4 (above). In agreement with previous results, expression of PsspE-cfp in wild type cells was only detected in the forespore at hour 4 after the onset of stationary phase, and only in cells in which the forespore had been engulfed by the mother cell (Figure 3 and Table 1). Note that no fluorescence was detected in cells of a sigG deletion mutant (Figure 3), confirming that the detected expression of the fluorescent reporter relied on σG. In the N45E mutant, however, around 1% of the cells scored between hour 0 and 2 after the onset of stationary phase showed strong whole-cell fluorescence (Figure 3 and Table 1). These cells had no morphological signs of sporulation, i.e., they did not show asymmetric septa or engulfing membranes as assessed by FM4-64 staining. Consistent with the absence of asymmetric septation, we found that these cells did not show Pspo0A-yfp expression (not shown) and time-lapse microscopy experiments revealed that they eventually lysed (Figure S3). A second, larger population of N45E cells consisted of organisms that resembled the wild type in that they begun to display forespore-specific cfp fluorescence at hour 3 (Figure 3; Table 1). These cells did not show premature, whole-cell expression of PsspE-cfp. The results show that the first period of σG activity in the N45E mutant can be accounted for by a sub-population of cells that enter stationary phase and that do not enter in sporulation.
Fig. 3. Localization of σG activity. 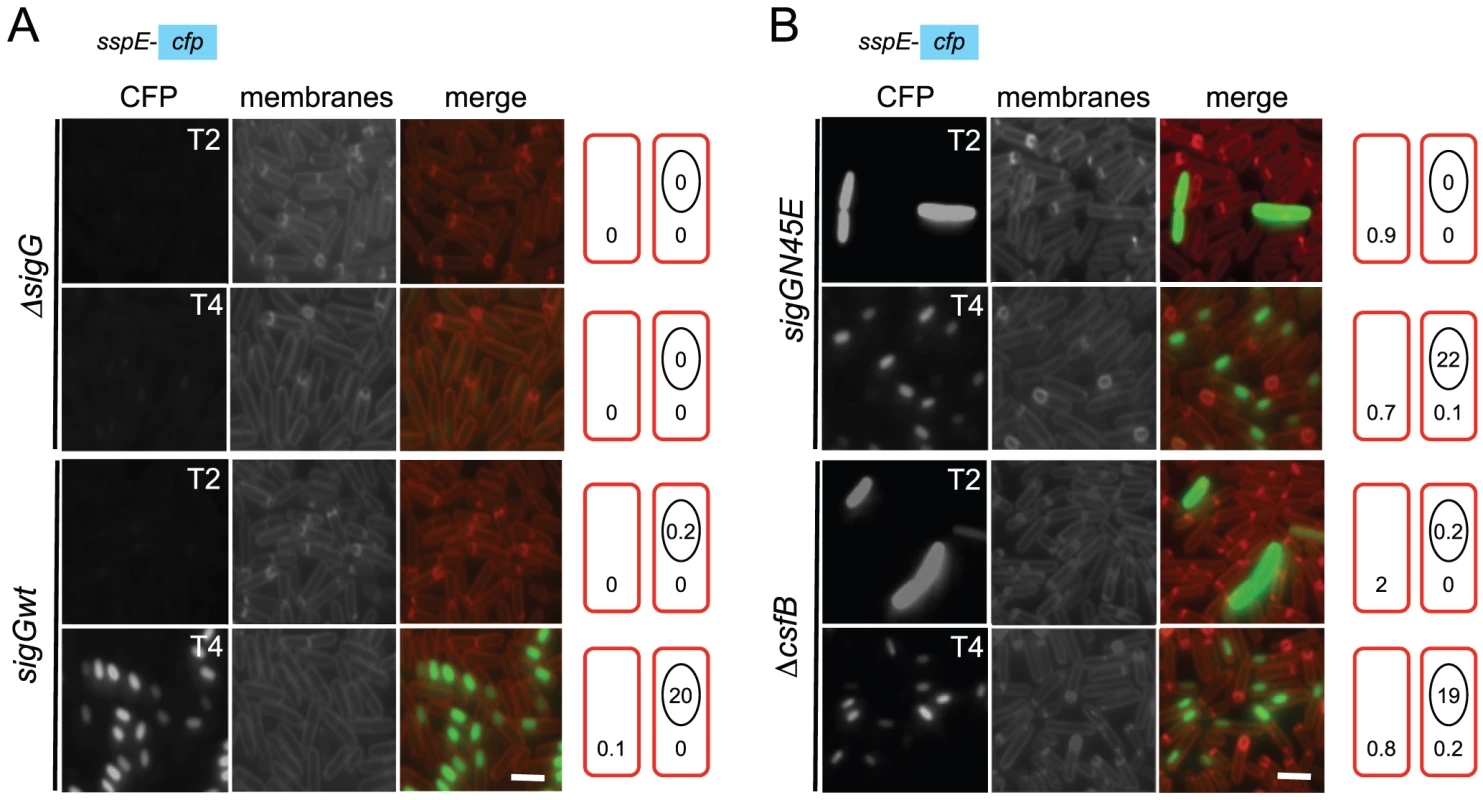
The activity of σG was monitored by fluorescence microscopy 2 (T2) and 4 (T4) hours after the onset of stationary phase in sporulation medium (DSM), for the following panel of strains (A and B), all of which carrying a fusion of the σG-controlled PsspE promoter to cfp at the non-essential yycR locus: the wild-type, a ΔsigG mutant, the sigGN45E mutant, and the ΔcsfB mutant. The membranes were visualized with the lypophylic membrane dye FM4-64 which is unable to reach the forespore membranes following engulfment completion, and thus serves as a reporter of the engulfment status of the forespore. Fluorescence from PsspE-cfp was false colored in green. Scale bars, 1 mm. The cartoons represent the classes of cells showing CFP accumulation at the indicated times and the numbers, the percentage of cells with CFP fluorescence in the indicated compartment. Tab. 1. Localization of CFP expressed from the σG-controlled PsspE promoter. 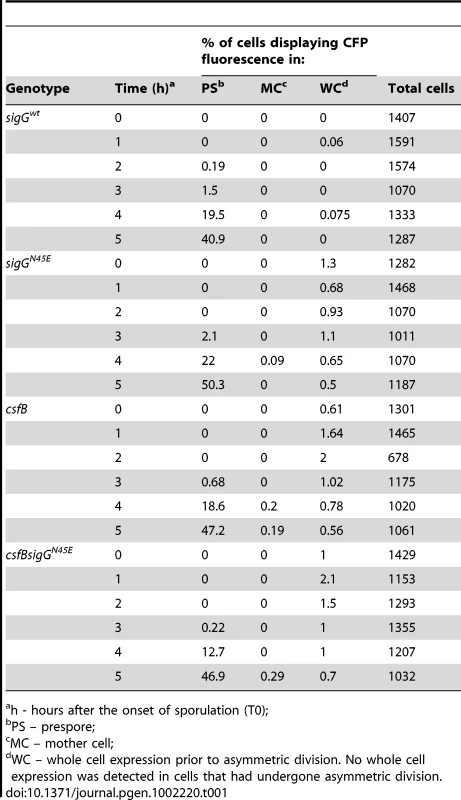
h - hours after the onset of sporulation (T0); csfB and the sigGN45E allele are epistatic
We then focused our attention in the mechanism of activation of σGN45E in post-exponential phase cells. We considered the possibility that the N45E substitution made σG less responsive to the SpoIIAB anti-σG factor, which binds to and contributes to the negative regulation of σG in non-sporulating cells [8], [12], [24]. However, we found the activity of σGN45E to remain sensitive to SpoIIAB in vivo (Text S1 and Figure S2B). While the possibility that the N45E substitution made σG refractory to SpoIIAB seemed discarded, the profile of σGN45E activity, in particular the first period of activity detected in stationary phase DSM cultures, was reminiscent of the effect reported for a mutation in csfB, which codes for the CsfB anti-σG factor [8], [20], [27]. For this reason, we examined the contribution of a csfB deletion mutation to the effect of the sigGN45E allele on σG-directed gene expression. On LB medium supplemented with X-Gal, the double mutant exhibited levels of β-galactosidase activity similar to the single sigGN45E or csfB mutants (Figure 2A and 2B). In DSM the double mutant showed the bi-modal temporal pattern of PsspE-lacZ expression seen for the csfB or sigGN45E single mutants, but with β-galactosidase levels during the first period of expression higher than those of the sigGN45E mutant (Figure 2D). There was no detectable effect of the mutations alone or in combination, on the second period of PsspE-lacZ activity (Figure 2D). When examined by fluorescence microscopy, the sigGN45E/csfB double mutant resembled the single mutants: about 1% of the cells displayed early whole-cell fluorescence (between hours 0 and 2 of sporulation) whereas most of the population showed CFP fluorescence in the forespore following engulfment completion (Figure 3 and Table 1). Presumably, the fraction of pre-divisional cells with a strong whole-cell CFP signal corresponds to the β-galactosidase producing cells during the first hours of sporulation (Figure 2C–2F). In conclusion, sigGN45E cells phenocopied the csfB mutant and the sigGN45E/csfB double mutant did not differ significantly from either single mutant. These findings suggest that the csfB and sigGN45E alleles exert their effect on σG by acting on the same pathway.
The N45E substitution reduces binding of CsfB to σG
The idea that both csfB and the sigGN45E allele act on the same pathway suggested to us that the N45E substitution could interfere with binding of CsfB to the mutant form of σG. In earlier work, CsfB and σG were found to directly interact in a yeast two-hybrid assay, and the first 71 residues of σG to be required for the CsfB-dependent inhibition of σG in vivo [20]. We used a similar approach to investigate whether σGN45E was less efficiently bound by CsfB. σG, σGN45A, σGN45E or CsfB were translationally fused to the C-terminus of the Gal4 DNA binding (BD) and activation domains (AD), and the various fusion proteins expressed in different combinations in yeast cells and checked for their ability to interact in vivo, as assessed by the expression of a lacZ gene preceded by a Gal4-responsive element. As shown in Figure 4A and 4B), CsfB interacts efficiently with σG and only slightly less well with σGN45A. In contrast, CsfB interacts only weakly with σGN45E.
Fig. 4. Interactions of SpoIIAB and CsfB with σG, σGN45A, and σGN45E. 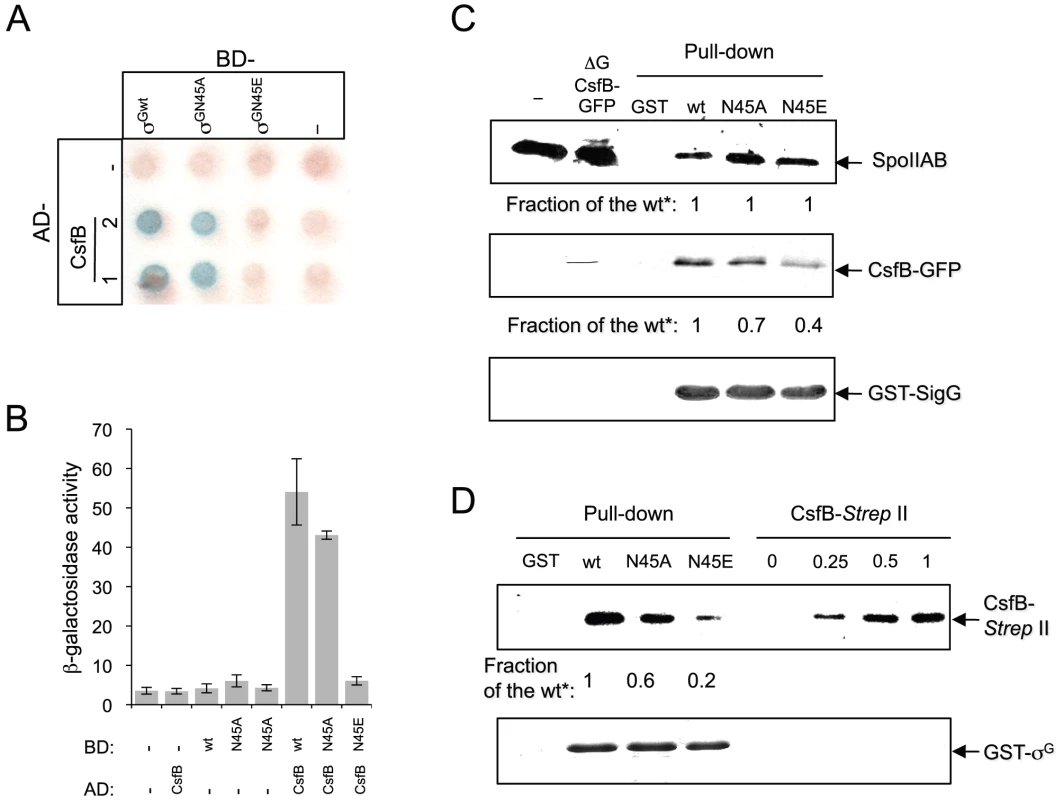
Panels A and B: colony lift (A) and quantitative assay on liquid medium (B) for the detection of β-galactosidase activity in yeast strains expressing fusions of CsfB to the GAL4 activation domain (AD; two colonies are shown) and fusions of σG, σGN45A or σGN45E to the GAL4 binding domain (BD), as indicated. Assays in which the BD and AD were expressed from empty vectors were used as negative controls (“−”). β-galactosidase activity was detected with X-Gal after a reaction time of 30 min (A), or with the ONPG substrate (B) and expressed in Miller units. The data represented in panel B are the average of three independent experiments. Panel C shows the results of pull-down with GST (lane GST), GST-σG, GST-σGN45A, or GST-σGN45E fusion proteins (as indicated above the panel) immobilized on glutathione agarose beads. Cell extracts from a B. subtilis strain producing a CsfB-GFP fusion were prepared from DSM cultures 2 hours into stationary phase, and incubated with the various beads preparations. Bound proteins were visualized, following elution, by immunoblotting with anti-SpoIIAB and anti-GFP antibodies. The extract used in the assay (ΔG CsfB-GFP) as well as an extract prepared from the wild type MB24 strain (control for the GFP antibody) at hour 2 of sporulation in DSM, were also directly loaded on the same gel. Panel D: GST pull-down assays with purified CsfB-Strep II tag. GST and the various GST-σG fusions (wt, N45A, and N45E, as indicated) were bound to glutathione beads and incubated with purified CsfB-Strep II (100 nM). Bound proteins were detected, following elution, with an anti-Strep II tag antibody. In the lanes denoted “CsfB-Strep II” the indicated amounts of purified protein (in ng) were analyzed by immunoblot as a control for the intensity of the signal in the pull-down experiments. In panels C and D, “Fraction of the wt*” refers to the binding ratio between of GST-σG, GST-σGN45A, or GST-σGN45E to SpoIIAB or to CsfB (C), or CsfB-strep II tag (D). The numbers were determined through densitometric analysis of the blots, and are averages of three independent experiments. We then used affinity chromatography to further investigate the interaction between CsfB and the different forms of σG. Whole cell extracts were prepared from cultures of a B. subtilis strain producing a functional CsfB-GFP fusion, 2 hours after the onset of sporulation, when σF is active and CsfB is known to accumulate [29]. The extracts were incubated with GST-σGwt, GST-σGN45A, GST-σGN45E or GST alone bound to glutathione agarose beads. Bound proteins were eluted and identified by immnunoblot with an anti-GFP antibody (see Materials and Methods). These experiments showed that CsfB was pulled down efficiently by immobilized GST - σGwt but not by GST itself (Figure 4C). GST-σGN45A pulled down CsfB-GFP less efficiently that the wild type (the efficiency was 0.7× of the wild type) but importantly, for σGN45E the efficiency of the pull down was about 0.4× of the wild type (Figure 4C; note that the numbers in the panel represent averages for three independent experiments). We also note that in these assays SpoIIAB was pulled down by all forms of GST-σG with similar efficiency (Figure 4C), suggesting that the N45A or N45E substitutions do not significantly affect binding of SpoIIAB to σG, and in line with the results of the in vivo activity experiments in which σGN45E was still susceptible to SpoIIAB (see above; Figure S2A). To discard the possibility that the reduced retention of CsfB by σGN45E was caused by increased binding of a competing protein present in the B. subtilis extracts, the assay was repeated using a CsfB-Strep II-tagged protein overproduced and purified from E. coli. CsfB-Strep II was soluble when overproduced in a minimal medium only in the presence of Zn2+, or in LB, which contains high levels of Zn2+ (Text S1 and Figure S4). The CsfB-Strep II protein purified from LB medium had bound Zn2+ (metal to protein ratio of 1∶1), as determined by atomic absorption spectroscopy. We incubated purified CsfB with GST or the various GST-σG forms immobilized on glutathione beads. After washing, CsfB was detected in the eluates by immunoblot with an anti-Strep II tag antibody. The CsfB protein was retained by GST-σG and by GST-σGN45A (∼0.6× the efficiency of the wild type), and to a lower level (∼0.2× of the wild type) by GST-σGN45E (Figure 4D). In these experiments, the signal in the pull down could be matched to that of a dilution of purified CsfB-Strep II (Figure 4D). Although the differences between σG/σGN45A and σGN45E were more pronounced in the yeast two-hybrid experiments, both this assay and the pull-downs are in general agreement. Together, the results show that N45E is the substitution with the greatest impact on binding of CsfB to σG.
A negative feedback loop that limits the ectopic activation of σG
csfB was first identified as a gene under the control of σF, and hence transcribed in the forespore soon after asymmetric septation [29]. Yet, in our hands, the main effect of a csfB deletion on the activity of σG activity was manifested in predivisional cells, i.e., before the activation of σF (Figure 3). Previously, Chary et al., (2007) have speculated that there is a basal level of σF-directed transcription during vegetative growth. However, our results suggest that the increased activity of σGN45E, which as we show is at least partially resistant to CsfB, was dependent solely on σG (see above). Therefore, and although the expression of csfB in the forespore is not thought to be controlled by σG [38], it seemed plausible that transcription of csfB in pre-divisional cells could be at least in part, controlled by σG. As a first test to this idea, we investigated whether expression of csfB and the activity of σGN45E co-localized. We first replaced the wild type csfB allele by a csfB-yfp fusion. This csfB-yfp fusion was subsequently transferred to strains carrying either the wild type or the N45E alleles of sigG and in addition, the σG reporter PsspE-cfp [10]. In the wt strain grown in DSM, organisms began to display forespore-specific yfp fluorescence between hour 1 and 2 after the onset of stationary phase (Table 2), consistent with the timing of septation and the activation of σF. No fluorescing organisms were observed before hour 1, or in cells of a ΔsigF mutant, as expected for a σF-controlled gene (Table 2; data not shown). Conversely, cfp fluorescence was only observed in the forespore 2 hours after the onset of sporulation, as expected for a σG-controlled gene and demonstrating the functionality of the csfB-yfp fusion. In the N45E mutant, between 1% to 4% of the bacteria displayed both whole-cell YFP and CFP fluorescence during the first hours of stationary phase in DSM (Table 2). Around hour 2, the first cells showing forespore-specific expression of csfB-yfp were detected followed, around hour 3, by cells with engulfed forespores showing PsspE-cfp expression. From this analysis, it is clear that the whole-cell expression of csfB-yfp early in stationary phase is confined to cells that also show activity of σGN45E, suggesting that csfB-yfp was transcribed under the direction of σG. As a further test to the possibility that σG controlled transcription of csfB, we made use of the sigG inactive allele described above in which the N45E mutation was combined with the F91A and Y94A “promoter-melting” mutations (Figure S1). In cells of the triple sigG mutant, no whole-cell YFP or CFP fluorescence was detected. In addition, and as expected, cells of the triple mutant did not display forespore-specific CFP fluorescence (which is σG-dependent) but showed forespore-specific YFP fluorescence (which is σF-dependent) (Table 2). These results strongly suggest that the expression of csfB in pre-divisional cells is σG-dependent, a conclusion reinforced by the observation that deletion of sigF in a N45E background abolished both the forespore-specific YFP and CFP fluorescence (σF - and σG - dependent) while maintaining the early whole-cell expression of yfp and cfp (Table 2). Lastly, as a more direct test for the ability of σG to control the expression of csfB, we monitored the expression of a PcsfB-lacZ fusion upon artificial induction of σG production from PxylA in vegetatively growing cells. The results in Figure 5A show that addition of xylose resulted in the induction of csfB-lacZ expression, even in the presence of a sigF deletion mutation, consistent with the view that σG can also drive expression of csfB, and with the similarity of the −10 and −35 promoter elements recognized by σF and σG [39]–[41].
Fig. 5. σG drives expression of csfB under non-sporulation conditions. 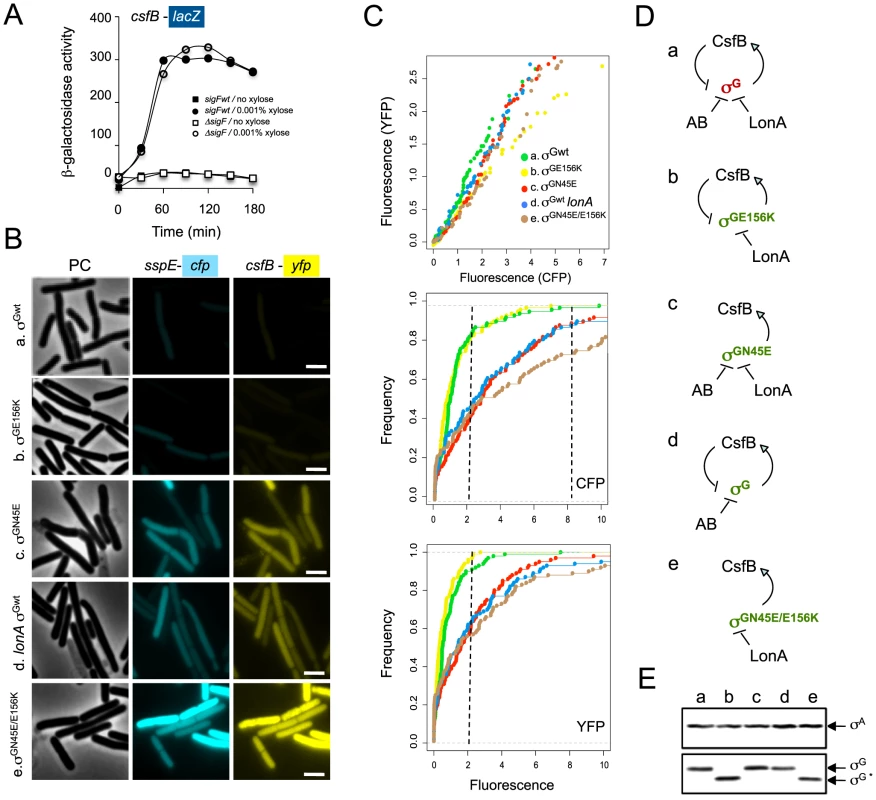
Panel A: Strains carrying a PcsfB-lacZ fusion, a xylose-inducible PxylA-sigG construct and the wild type sigF gene or a ΔsigF deletion, were grown to mid-log phase in LB medium and induced (circles) or not (squares) with 0.001% xylose. Samples were collected at the indicated time intervals and assayed for β-galactosidase activity (shown in Miller units). Panel B, σG activity and csfB expression was monitored in the same cells, by fluorescence microscopy, at the onset of stationary phase in LB. The strains used carry PsspE-cfp and PcsfB-yfp reporter fusions, a deletion of the sigG gene, and a copy of the wild type sigG gene (a), sigGN45E (b), sigGE156K (c), wild type sigG in a lonA deletion mutant (d), or sigGN45E/E156K (e) at the amyE locus under the control of PxylA. The strains were grown in the presence of 0.001% xylose. Scale bar, 2 µm. Panel C: quantitative analysis of CFP and YFP expression for the strains (a to e) examined in B. The top plot shows the correlation between the YFP (csfB-yfp) and CFP (PsspE-cfp) signals. The middle and bottom graphs show a cumulative frequency distribution of the YFP and CFP signals across the population. Fluorescence intensity is shown in arbitrary units; 100 cells were scored. Panel D shows the schematic representation of the regulatory interactions in the experimental situations shown in panels A and B (inactive and active σG in brown and green, respectively). Panel E: immunoblot analysis of σG accumulation in the strains (a-e) defined in panel B. Strains were grown in LB medium and samples taken at the onset of the stationary phase for microscopy or immunoblot analysis. “σG” represents the position of the wild type protein or σGN45E. The asterisk denotes σGE156K or σGN45E/E156K, both of which, because of the E156K substitution, migrate slightly faster than the wild type or σGN45E proteins [24]. Tab. 2. Localization of csfB-yfp and PsspE-cfp expression. 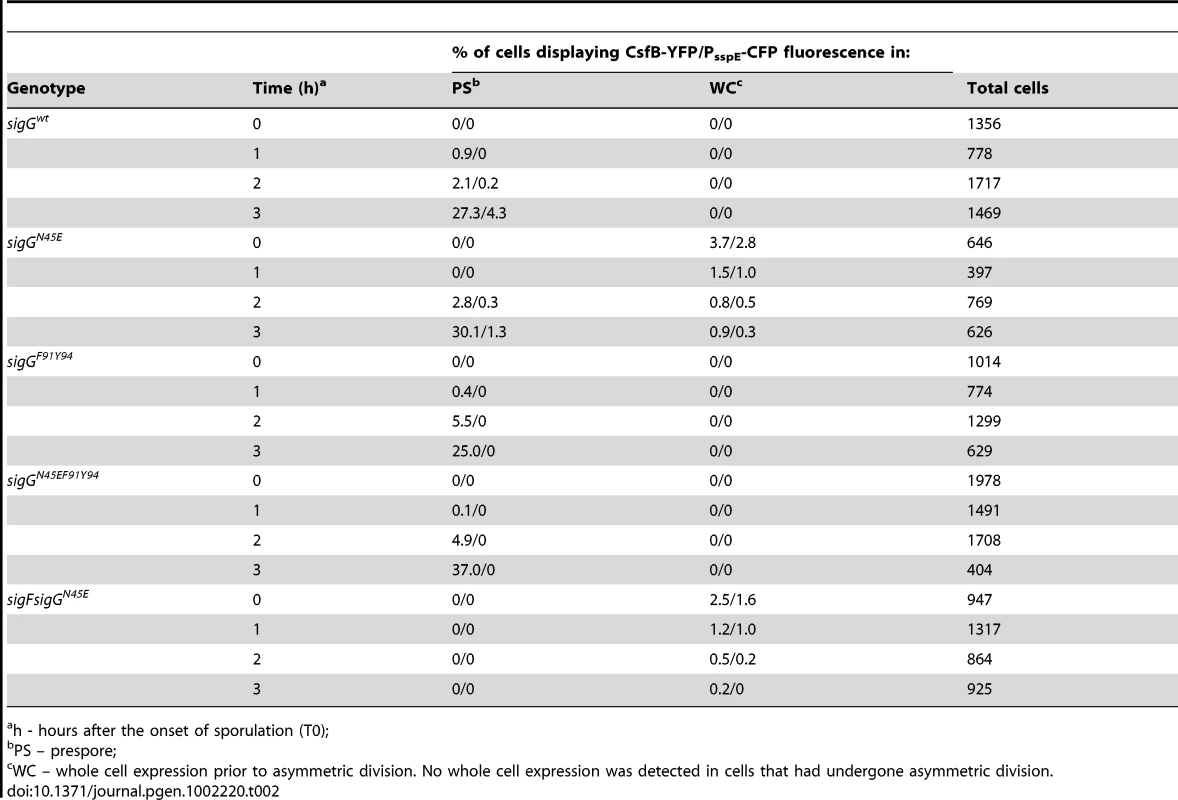
h - hours after the onset of sporulation (T0); Taken as a whole, the results suggest that the capacity of σG to drive production of CsfB in pre-divisional cells may be part of a mechanism to limit the ectopic activation of σG should any condition promote its activation.
Genetic lesions that relax the regulation of σG also result in CsfB production
If production of CsfB is part of a regulatory circuit that self-restrains the activity of σG, then mutations in other factors known to negatively regulate σG should also induce expression of csfB, and the extent of the effect across the population should reflect the contribution of the affected regulator to the regulation of σG. Two such factors are known, the LonA protease and the SpoIIAB anti-sigma factor, which act independently to negatively regulate the activity of σG, mainly under non-sporulation conditions [21], [22]. To determine the relative impact of mutations known to affect the regulation of σG on its activity across the population, and whether those mutations also increased the expression of csfB, we used fluorescence microscopy to simultaneously quantify the expression of PsspE-cfp and csfB-yfp at the onset of stationary phase in LB, in a panel of strains carrying PxylA fusions to wild type sigG, sigGN45E, sigGE156K (coding for a form of σG refractory to SpoIIAB; [24]), sigGN45E/E156K or a PxylA-sigGwt construct in combination with a lonA deletion mutation (Figure 5B). The growth medium was supplemented with 0.001% xylose, as in preliminary experiments (Text S1 and Figure S5) this was the highest concentration at which wild type σG could be induced without causing significant cell lysis. In control experiments, no fluorescence could be detected in strains lacking either of the PsspE-cfp and csfB-yfp fusions (not shown).
The results in Figure 5C (top graph) show a clear correlation between the YFP and CFP signals for all strains tested. Cells that produce CFP also produce YFP, and an increase in the expression of one reporter is accompanied by an increase in the expression of the other (Figure 5C, top), highlighting the link between the activity of σG and the production of its negative regulator, CsfB. The middle and lower graphs of Figure 5C are cumulative frequency distributions of the CFP and YFP signals for the various strains. For the N45E, lonA, and N45E/E156K strains, about 50% and 40% of the population shows CFP and YFP signals, respectively, above 2 arbitrary units. In contrast, only 10% of the wt or E156K populations show CFP or YFP signal intensities above this value (Figure 5C). Induction of σGN45E/E156K increased the number of cells with high CFP fluorescence (above 8 arbitrary units) to 20% of the population, as compared to 10% for the strains bearing the single N45E, E156K or lonA mutations. This observation is in agreement with the idea (see above) that σGN45E is still sensitive to SpoIIAB. Smaller differences in the YFP signal distribution were seen between the double N45E/E156K mutant and the single N45E, E156K and lonA mutants, possibly reflecting reduced YFP stability. While CsfB, mainly, and LonA emerge as the principal regulators of σG activity during entry into the stationary phase of growth, SpoIIAB per se seems to have only a minor role (Figure 5C). Importantly, we were unable to combine the sigGN45E/E156K allele with a lonA deletion, highlighting the convergent action of CsfB, SpoIIAB and LonA in the negative regulation of σG, and suggesting that these are likely to be the main, if not the only, negative regulators of σG at play. The results also unravel a negative σG autoregulatory loop (Figure 5D), in which by commanding the expression of csfB, the fraction of cells with ectopic activity of σG is curtailed.
Since production of σG in the strains above was driven from the PxylA promoter, we expected the various forms of σG to accumulate to similar levels, independently of the number of cells showing σG activity. This was verified by immunobloting analysis with an anti-σG antibody, for the N45E, E156K, N45E/E156K and wild type σG in the lonA background (Figure 5E).
CsfB discriminates between σF and σG via one single amino acid
Because changing N45 of σG for the residue found at the equivalent position in σF, E39, makes σG less efficiently bound by CsfB, and since CsfB does not bind to σF [20], we reasoned that perhaps this position was essential for the discrimination by CsfB between the two forespore-specific sigma factors in vivo. This inference was strengthened by the observation that N45 is invariant in σG orthologues, whereas E39 is highly conserved among σF proteins (Figure 1C).
Therefore, we decided to investigate whether E39 was important for the resistance of σF to CsfB, and for the regulation of its activity in vivo. We conducted GAL4-based yeast two-hybrid experiments to test the interaction between wild type σF and a mutant form of the protein with the E residue at position 39 replaced by an N (E39N; Figure 1B). In agreement with the results of an earlier study [20], CsfB did not interact with σF in our assay (Figure 6A and 6B). In contrast, CsfB interacted efficiently with σFE39N (Figure 6A and 6B). Thus, the E39N substitution is sufficient to allow binding of CsfB to σF.
Fig. 6. Interactions of CsfB with σF and σFE39N. 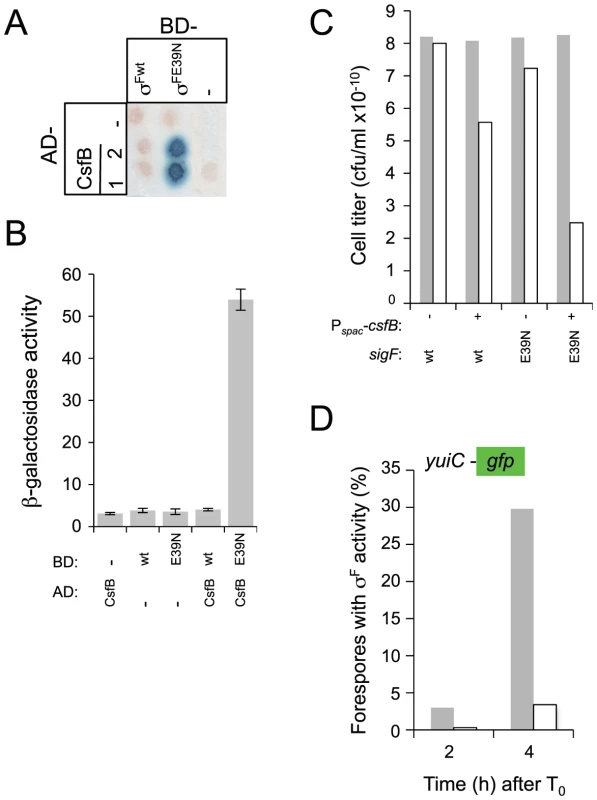
Panels A and B show the results of a colony lift (A) or a quantitative assay on liquid medium (B) for the detection of lacZ transcription in yeast strains expressing fusions of CsfB to the activation domain (AD; two colonies are shown) and fusions of σF or σFE39N to the binding domain (BD) of GAL4. Negative controls (“−”) included BD and AD expressed from empty vectors. β-galactosidase activity was detected as described in the legend for Figure 4. The results in B are the average of three independent experiments. Panel C shows the efficiency of sporulation of strains bearing a wild type copy of the sigF operon, or a copy with the sigFE39N allele, at amyE. In addition, the strains carry the csfB gene inserted at the thrC locus under the control of the IPTG inducible Pspac promoter (Table S1). The two strains (Pspac-csfB spoIIAABC wt and Pspac-csfB spoIIAABCE39N; NB: the third cistron of the spoIIA operon, also called sigF, codes for σF) were grown in DSM in the presence (+) or in the absence (−) of IPTG and the titer of heat resistant spores measured 18 hours after the onset of stationary phase (gray bars, viable cells; white bars, heat resistant cells). Panel D, shows the quantitative analysis of GFP expression in strains similar to those described for panel C, except that they additionally carry a fusion of the σF-controlled yuiC promoter to gfp. The two strains (Pspac-csfB sigFwt PyuiC-gfp, gray bars; Pspac-csfB sigFE39N PyuiC-gfp, white bars) were grown in sporulation medium (DSM) in the presence of IPTG and samples taken at the indicated times (in hours) after the onset of stationary phase for scoring the number of cells showing GFP expression by fluorescence microscopy. We next wanted to test whether the presence of an N at position 39 of σF, expected to make it susceptible to CsfB, would affect spore development. We found the E39N substitution to cause a 5-fold decrease in the efficiency of sporulation (data not shown). Chary et al. found that when csfB is expressed from the IPTG-inducible Pspac(Hy) promoter prior to the activation of σF, spore formation was severely reduced [27]. However, in this strain the activity of σF was not impaired and spore formation was blocked in the developmental pathway just after engulfment completion [27]. We used a similar assay to test for the effect of the E39N mutation on the activity of σF. We transferred the IPTG-inducible Pspac(Hy)-csfB fusion to strains mutant for sigF and with a second copy of the entire sigF operon (with either the wild type sigF cistron or sigFE39N) integrated at the amyE locus under the control of its native promoter. In the strain carrying the wild type allele of sigF grown in the presence of IPTG to induce csfB expression prior to the activation of σF, spore formation showed the reported 103-fold reduction relative to cultures without IPTG (Figure 6C) confirming the results of Chary et al. (2007). Strikingly, induction of csfB expression in the strain carrying the sigFE39N allele of sigF reduced spore formation 106-fold compared to the uninduced cultures (Figure 6C). To investigate whether the more drastic sporulation defect observed in the strain carrying the E39N allele was due to impaired σF activity, we used a fusion of the σF-dependent yuiC promoter to gfp [42]. GFP fluorescence was monitored by microscopy during sporulation in the strains with IPTG inducible expression of csfB and bearing either the wild type or sigGE39N alleles. We found that the induction of csfB reduced the activity of σFE39N to 10% of the levels observed when CsfB was produced prior to asymmetric division in the presence of wild type σF (Figure 6D). Thus, CsfB can interfere strongly with the activity of σFE39N in vivo.
Discussion
CsfB and the feedback inhibition of σG
The production of transcription factors often leads to the activation of gene expression during cell differentiation and development. In some instances, positive auto-regulation of the transcription factor drives gene expression in the differentiating cell down a specific developmental path. However, the power of these positive feedback loops raises the potential for inappropriate expression of the transcription factor in the wrong cell or at the wrong time. Therefore, the expression of autoregulatory transcriptional activators must be tightly controlled. We show here that CsfB has a function in preventing the activation of the forespore-specific, auto-regulatory σG factor, in stationary phase cells, in either a medium that does not support sporulation or in a sporulation medium, prior to the asymmetric division that initiates the program of compartment-specific gene expression that leads to differentiation of the spore.
This role of CsfB was uncovered because the N45E substitution in σG, which reduces binding by CsfB, also results in activation of the sigma factor in a fraction of stationary phase cells. We show that CsfB is also produced, under σG control, in the same stationary phase cells where σGN45E becomes active. Hence, a negative feedback loop is established which, with the help of SpoIIAB and LonA, dominates the positive feedback loop involving σG, and keeps its activity low. The role of LonA and SpoIIAB in the negative regulation of σG in stationary phase cells was shown before [9], [12], [17], [21], [22], but our analysis suggests that CsfB and LonA are the main regulators of σG. Nevertheless, the role of SpoIIAB is evidenced when the N45E and E156K substitutions are combined, and by our inability to construct a strain additionally carrying a lonA deletion. The lethality of this triple mutant further suggests that CsfB, SpoIIAB and LonA may be the only negative regulators of σG at play in stationary phase cells.
Although mutations that interfere with the function of CsfB, SpoIIAB or LonA may cause strong expression of σG-dependent genes in pre-divisional cells, this only occurs in a fraction of the population (Figure 5). We do not presently know whether the cells which show σG activity are somehow different from the rest of the population at some fundamental level, or whether σG activity arises because of random fluctuations in the levels of σG itself, and its negative regulators. In any case, high-level expression of even wild type σG in stationary phase cells causes cell lysis, emphasizing the importance of limiting the potential for σG activation (Figure S6). Lysis may be a consequence of high levels of σG activity [26], an indirect effect of the release of σF through titration of SpoIIAB by σG [20] or both, as induction of σG production in LB leads to lysis even in the absence of σF (data not show).
CsfB and the activation of σG
CsfB was initially proposed to be a key factor in keeping σG inactive in the forespore prior to engulfment completion [20]. However, a more consensual view of the role of CsfB is that the anti-sigma factor acts as a timing device, to help prevent σG activity prior to engulfment completion [8], [9], [27], [38]. The results of our investigation are in line with this view, as in our hands deletion of csfB or the N45E substitution in σG (which we show prevents binding of CsfB to σG) did not bypass the genetic and morphological controls that link the activity of σG to engulfment completion. We postulate that during sporulation the negative feedback loops contributes to counter the positive feedback loop involving σG until CsfB is inactivated, or its synthesis is reduced by an unknown second regulator, or σG accumulation overwhelms that of CsfB. It is not known if CsfB is inactivated in the forespore following engulfment completion, but the anti-sigma factor seems to rapidly disappear from the forespore once σG becomes active (our unpublished results). It is also likely that an additional factor prevents expression of csfB in the engulfed forespore. For example, σG drives production of SpoVT a forespore-specific transcription factor, which represses at least 27 σG-dependent transcriptional units [41], [43]. SpoVT has a C-terminal GAF (cGMP-specific and cGMP-stimulated phosphodiesterases, Anabaena adenylate cyclases, and Escherichia coli FhlA)-like domain, which is essential to modulate the DNA-binding activity of the N-terminal domain, and may respond to nucleotides or other small molecules [44]. The accumulation of nucleotides in the engulfed forespore in turn, may be essential for the activity of σG and may depend on the action of the SpoIIIA-Q channel [9].
Binding of CsfB to σG
Two observations are consistent with the interpretation that the N45E substitution interferes with binding of CsfB to σG. First, wild type σG could pull down CsfB from extracts of B. subtilis in sporulation medium, but σGN45E did so less efficiently, a difference that was amplified when purified CsfB was used (Figure 4; see also below). Second, CsfB interacted with wild type σG but not with σG N45E in a yeast two-hybrid system (Figure 4). CsfB was purified from E. coli cells as a C-terminal fusion to the Strep II tag because in vivo a CsfB-GFP fusion was fully functional (this work). The CsfB-Strep II protein had Zn2+ bound with a stoichiometry of 1∶1. The Zn2+ was released by oxidation of the protein with H2O2, suggesting the involvement of the conserved Cys residues in CsfB in its coordination (see Figure S4). Recently, a MalE-CsfB fusion protein was purified from sporulating cells of B. subtilis with Zn2+ bound with a stoichiometry of 0.5 mol/mol [28]. Together with genetic data, this suggested that CsfB could act as a dimer (or higher order multimer) and possibly alternate between an active and an inactive state [28]. Importantly, the activity of σG was efficiently inhibited in E. coli cells, when co-produced with CsfB [28], and the CsfB-Strep II protein purified from E. coli cells clearly discriminated σG and σGN45E in our pull-down assays (Figure 4E).
The N45 residue in σG may contribute to the interaction with CsfB. If so, however, this contact does not seem to be essential because the N45A substitution did not result in increased activity of σG in vivo, and caused only a small reduction in the ability of σG to interact with CsfB in yeast two-hybrid and pull-down assays (Figure 4). Additional mutagenesis studies may illuminate if and how the N45 residue contributes to the interaction with CsfB, and how the N45E substitution interferes with the interaction. CsfB is likely to contact σG at other positions, and these other contact sites are likely to be present in σF as well. First, because no other single mutation was found within the first 150 residues of σG that would affect its activity in vivo and second, because while incapable of binding to wild type σF ([20]; this work), CsfB bound efficiently to σFE39N (see also below). The location of the N45 residue within region 2.2 of σG and its role in permitting binding by CsfB, is also consistent with previous work in which a σF/G chimeric protein allowed the target for CsfB to be mapped within the first 77 residues of σG [20]. The N45 residue is invariant among σG orthologues of Bacillus species and related organisms but less conserved among the σG proteins of the Clostridia (Figure 1C). These observations highlight the importance of N45 (and homologous residues) in a sub-group of sporeformers including B. subtilis and related organisms, in which σG is regulated by the anti-sigma factor CsfB.
The N45E substitution may also affect a contact with the β′subunit of core RNA polymerase
The observation that the N45E substitution reduces binding of CsfB to σG provides a plausible explanation for the increased activity of σGN45E in vivo. However, we cannot at present discard the possibility that the N45E alteration, which affects a residue positioned within conserved region 2.2, also increases binding of σG to core RNA polymerase. The position homologous to N45 is often occupied by an acidic residue in proteins of the σ70 family of sigma factors (the σG orthologues of Bacillus species and related organisms being a conspicuous exception), and in the crystal structure of the σ70-containing RNA polymerase holoenzyme from Thermus aquaticus [45], E189 (homologous to N45 in the σG protein of B. subtilis) is involved, with other neighboring residues, in a direct contact with residue K159 in the β′subunit (Text S1 and Figure S6). An asparagine residue, as is found in σG, could also contribute to the interaction with β′ at this site. However, an acidic residue would most likely make a stronger, electrostatic, contribution to the interaction. This in turn suggests that the N45E substitution could also enhance the activity of σG by favoring its interaction with the β′subunit of core RNA polymerase. If so, then the regulation of σG activity in vivo could involve competition between CsfB and β′ for binding to σG. In any event, the possible contact involving N45 and β′ is in line with the view that one mechanism by which anti-sigma factors function is by occluding sigma-core binding interfaces [46], [47]. Two of the mutations known to impair binding of SpoIIAB to σF map within region 2.2, and mark residues that are conserved in σG ([24], [48]; see also Figure 1B). This suggests that SpoIIAB and CsfB may use partially overlapping interfaces in binding to σG and may explain the competition between the two anti-sigma factors for binding to σG under certain conditions [20], [28], [48]. However, σGN45E was still bound by SpoIIAB and was still susceptible to SpoIIAB in vivo. Therefore binding of SpoIIAB to σG does not seem to require the N45 residue.
Discrimination between σF and σG
The strict conservancy of N45 among σG proteins of other Bacillus species and related organisms, together with its nearly absolute exclusion from orthologues of σF, suggests an important, conserved role for this residue. CsfB does not seem to negatively modulate the activity of σF, consistent with its inability to bind to this σ factor [20], [27], [28], [38]; this work). Because the E39N substitution is sufficient to allow binding of CsfB to σF, the E39/N45 position in the σF/σG families of proteins seems critical for the discrimination by the CsfB anti-sigma factor. Perhaps strengthening this idea, the only exception to the rule that an N is excluded from the critical position in σF is B. clausii (Figure 1C), but in this organism no csfB orthologue could be identified (not shown).
Recently, a protein related to CsfB, and termed Fin, was shown to inhibit the activity of σF and to play an important role in promoting the switch from σF to σG in the forespore [38]. It is possible that the N45/E39 residues help enforcing the specific regulation of σF by Fin and of σG by CsfB. It is not known whether σFE39N is susceptible to Fin. However, the E39N substitution did not seem to affect the activity of σF and caused only a 5 fold reduction in the efficiency of sporulation (this work), whereas deletion of fin increased the window of expression of σF-dependent genes, and caused a 50-fold reduction in the efficiency of sporulation [38]. Perhaps then, σFE39N is still regulated by Fin. CsfB was also proposed recently, to antagonize low levels of σE resulting from inappropriate activation in the forespore, thus contributing to the confinement of its activity to the mother cell [49]. It is not yet known whether CsfB interacts with σE. However, if so, and because an acidic residue (E) is found at the position equivalent to N45 in σG, it follows that in the context of the σE protein binding by CsfB is likely to involve other residues.
Materials and Methods
Strains and general methods
The B. subtilis strains used in this work are congenic derivatives of the Spo+ strain MB24 (trpC2 metC3), and are listed in Table S1. The plasmids used in strain construction are described in the sections below and in Text S1. LB medium was used for growth or maintenance of E. coli and B. subtilis, and sporulation was induced by growth and exhaustion in Difco sporulation medium (DSM) [24]. The Quick Change site-directed mutagenesis system (Stratagene) was used for the generation of all site-specific mutations, which were always confirmed by sequencing.
Mutagenesis of the sigG gene
We used pMS45, containing the sigG gene [24] and sigG-specific primers (all primers are listed in Table S2) to convert the residues highlighted in Figure 1B (orange and red circles) into the aminoacid found in σF. The various mutations were then transferred to the sigG locus by congression as it has been observed that certain mutations cause a greater increase in the activity of σG when sigG is inserted at an heterologous locus such as amyE ([27]; our unpublished observations). For congression, pMS45 and its derivatives carrying the different sigG mutations, together with chromosomal DNA from AH6566 (ΔsigG ΔyycR:: PsspE-cfp ΔsspE::PsspE-lacZ), was used to co-transform strain AH2452 (ΔsigG ΔsspE::PsspE-lacZ) with selection to CmR. Spo+ congressants appeared at a frequency of about 3%. One congressant for each sigG allele for which the presence of the desired mutation was confirmed by PCR and sequencing was kept for further study. Mutants that showed increased b-galactosidase production from the PsspE-lacZ reporter fusion were identified on LB plates containing 5-bromo-4-chloro-3-indolyl-b-D-galactopyranoside (X-Gal).
Promoter and reporter gene fusions
The construction of fusions of the xylose-inducible xylA promoter to different sigG alleles, and of csfB-gfp, -yfp and lacZ fusions is described in detail in Text S1, accompanying this article.
Construction of the sigF E39N mutant
First, the entire spoIIA operon was PCR amplified with primers sigF219D and sigF2032R (Table S2 lists all primers used in this study), the 1813 bp product digested with BamHI and HindIII and inserted between the same sites of pDG364 [50], This created pMS393. Next, primers sigFE39ND and sigFE39NR were used to substitute the glutamate codon at position 39 of the sigF gene in pMS393 by an asparagine codon, which resulted in pMS394.
Yeast two-hybrid analysis
The coding regions of sigG, sigF and csfB were PCR amplified with primers sigG2016D and sigG2862R, sigF493D and sigF1318R, and primers csfB191D and csfB480R. The sigG and sigF PCR products were digested with NcoI and EcoRI and inserted between the same sites of pAS2-1 (Clontech) yielding plasmids pMS358 and pMS357, respectively. We used pMS358 and primers sigGN45ED and sigGN45ER to substitute the asparagine codon at position 45 of σG by a glutamate codon. This resulted in plasmid pMS360. We used pMS358 and primers sigGN45AD and sigGN45AR to substitute the asparagine codon at position 45 of σG by an alanine codon. This resulted in plasmid pMS429. We used pMS357 and primers sigFE39ND and sigFE39NR to substitute the glutamate codon at position 39 of σF by an asparagine codon. This resulted in plasmid pMS387. The csfB PCR product was digested with NcoI and SalI and inserted between the same sites of pACT2 (Clontech) yielding plasmid pMS356. Mating of Sacharomyces cerevisiae strains and detection of β-galactosidase activity were performed as described before [51].
Overproduction and purification of SpoIIAB
The spoIIAB coding region was PCR amplified with primers spoIIAB189D and spoIIAB698R. The PCR product was digested with BamHI and XhoI and inserted between the same sites of pET30a (+) (Novagen) creating pMS111, in which the sequence for the His6 tag was introduced between the first and second codons of spoIIAB. pMS111 was introduced into competent cells of BL21 (DE3) pLysS (Novagen). Growth, induction, and lysate preparation was essentially as described [52]. The His6-SpoIIAB fusion protein was partially purified on His-Trap chelating columns as described by the manufacturer (Amersham Pharmacia Biotech) and used to raise a polyclonal anti-SpoIIAB antibody in rabbits (Eurogentec, Belgium).
Overproduction and purification of CsfB
First, primers csfB191D and csfBstrepR, which include the sequence coding for the Strep II tag (IBA GmbH) were used to PCR amplify the coding region of csfB. The resulting PCR product was digested with NcoI and BamHI and cloned between the same sites of pET16b (Novagen) to create pMS350, which was then transformed into E. coli strain BL21(DE3). The E. coli expression strain was grown to mid-log phase in LB (0.6 optical density at 600 nm), induced with 1 mM isopropyl-D-thiogalactopyranoside (IPTG), and grown for 3 h before harvesting the cells. The cell pellets were resuspended in 3 ml portions of buffer A (100 mM NaCl, 10 mM Tris pH 8.0, 10% glycerol) per 50 ml of induced culture and lysed in a French pressure cell (18,000 lb/in2). The lysate was centrifuged to remove cell debris. CsfB-Strep II tag was purified on Strep-Tactin Sepharose columns following the manufacturer instructions (IBA GmbH). The metal content of the purified protein was analyzed by atomic absorption.
GST pull-down experiments
Primers sigG2016D and sigG2964R were used to PCR amplify the coding regions of sigG, sigGN45A and sigGN45E from pMS45, pJS4, and pJS2 (see above). The PCR products were digested with BglII and XhoI and cloned between the BamHI and XhoI sites of pGex4T-3 (GE Healthcare) to create pMS375, pMS376, and pMS377, respectively, which bear in-frame N-terminal GST fusions to the different forms of σG. Derivatives of BL21(DE3) bearing each of these plasmids or pGex4T-3 (GST-alone) were grown to mid-log phase (O.D.600≈0.6) in LB, and induced with 1 mM IPTG for 3 h before the cells were harvested. The cell pellets were resuspended in 1 ml portions of buffer A [100 mM NaCl, 10 mM Tris-HCl (pH 8.0), 10% glycerol] per 50 ml of induced culture and lysed in a French pressure cell (18,000 lb/in2). The lysate was cleared by centrifugation. One milliliter of cleared lysate was bound to 50 µl of a 50% slurry of glutathione Sepharose beads (GE Healthcare) at room temperature for 30 min. The beads were washed three times in buffer B (same as A but with 200 mM NaCl).
For the SpoIIAB and CsfB interaction assays, 1 ml portions of soluble extracts prepared from cultures of B. subtilis AH6608 (csfB::km ΔsigG ΔamyE::csfB-gfp) 2 h after the onset of sporulation were incubated for 30 min at room temperature with GST or the various GST fusions proteins bound to glutathione Sepharose beads or with the beads alone. The mixtures were washed three times with buffer B (above), resuspended in a final volume of 30 µl, and subjected to SDS-PAGE and immunoblotting. Rabbit anti-GFP (A.L. Isidro and A.O. Henriques, unpublished) and anti-SpoIIAB (above) antibodies were used at dilutions of 1∶1000 and 1∶500, respectively. An anti-σG antibody, at a 1∶1000 dilution, was used to control for the level of the GST-σG fusions immobilized [24].
For the CsfB interaction assay, 100 nM of purified CsfB-Strep II tag was incubated for 30 min at room temperature with the glutathione Sepharose beads complexed with the GST fusion proteins or with glutathione Sepharose beads alone. The mixtures were washed three times with buffer B (above) and resuspended in a final volume of 30 µl. The samples were subjected to SDS-PAGE and immunoblotting. An anti-Strep II tag polyclonal antibody was used at a 1∶1000 dilution (IBA GmbH). For graphical representation of the data, the immunoblots were scanned and analyzed using the ImageJ software (http://rsbweb.nih.gov/ij).
Accumulation of σG
Immunoblot analysis was used to monitor the accumulation of σG during growth or sporulation as previously described [17]. An anti-σA antibody was used as described before [36].
β-Galactosidadse assays
β-Galactosidadse activity was assayed with the substrate o-nitro-β-D-galactopyranoside (ONPG), with enzyme activity expressed in Miller units [52].
Fluorescence microscopy
Samples (0.6 ml) of LB or DSM cultures were collected, resuspended in 0.2 ml of phosphate-buffered saline (PBS) and the membrane dye FM4-64 (Molecular Probes) added to a final concentration of 10 µg ml−1. Microscopy was carried out as described previously [53]. Quantitative analysis of fluorescence intensity was done using the MetaMorph software package (MDS Analytical Technologies). Data was analyzed and plotted using the “R” statistical computing and graphics software package (www.r-project.org).
Supporting Information
Zdroje
1. HilbertDWPiggotPJ 2004 Compartmentalization of gene expression during Bacillus subtilis spore formation. Microbiol Mol Biol Rev 68 234 262
2. RudnerDZLosickR 2001 Morphological coupling in development: lessons from prokaryotes. Dev Cell 1 733 742
3. StragierPLosickR 1996 Molecular genetics of sporulation in Bacillus subtilis. Annu Rev Genet 30 297 241
4. Karmazyn-CampelliCBonamyCSavelliBStragierP 1989 Tandem genes encoding sigma-factors for consecutive steps of development in Bacillus subtilis. Genes Dev 3 150 157
5. SunDXCabrera-MartinezRMSetlowP 1991 Control of transcription of the Bacillus subtilis spoIIIG gene, which codes for the forespore-specific transcription factor sigma G. J Bacteriol 173 2977 2984
6. EvansLFeuchtAErringtonJ 2004 Genetic analysis of the Bacillus subtilis sigG promoter, which controls the sporulation-specific transcription factor sigma G. Microbiology 150 2277 2287
7. PartridgeSRErringtonJ 1993 The importance of morphological events and intercellular interactions in the regulation of prespore-specific gene expression during sporulation in Bacillus subtilis. Mol Microbiol 8 945 955
8. CampAHLosickR 2008 A novel pathway of intercellular signalling in Bacillus subtilis involves a protein with similarity to a component of type III secretion channels. Mol Microbiol 69 402 417
9. CampAHLosickR 2009 A feeding tube model for activation of a cell-specific transcription factor during sporulation in Bacillus subtilis. Genes Dev 23 1014 1024
10. DoanTMorlotCMeisnerJSerranoMHenriquesAO 2009 Novel secretion apparatus maintains spore integrity and developmental gene expression in Bacillus subtilis. PLoS Genet 5 e1000566 doi:10.1371/journal.pgen.1000566
11. IllingNErringtonJ 1991 The spoIIIA operon of Bacillus subtilis defines a new temporal class of mother-cell-specific sporulation genes under the control of the sigma E form of RNA polymerase. Mol Microbiol 5 1927 1940
12. KellnerEMDecaturAMoranCPJr 1996 Two-stage regulation of an anti-sigma factor determines developmental fate during bacterial endospore formation. Mol Microbiol 21 913 924
13. Londono-VallejoJAFrehelCStragierP 1997 SpoIIQ, a forespore-expressed gene required for engulfment in Bacillus subtilis. Mol Microbiol 24 29 39
14. MeisnerJWangXSerranoMHenriquesAOMoranCPJr 2008 A channel connecting the mother cell and forespore during bacterial endospore formation. Proc Natl Acad Sci U S A 105 15100 15105
15. ErringtonJApplebyLDanielRAGoodfellowHPartridgeSR 1992 Structure and function of the spoIIIJ gene of Bacillus subtilis: a vegetatively expressed gene that is essential for sigma G activity at an intermediate stage of sporulation. J Gen Microbiol 138 2609 2618
16. MurakamiTHagaKTakeuchiMSatoT 2002 Analysis of the Bacillus subtilis spoIIIJ gene and its Paralogue gene, yqjG. J Bacteriol 184 1998 2004
17. SerranoMCorteLOpdykeJMoranCPJrHenriquesAO 2003 Expression of spoIIIJ in the prespore is sufficient for activation of sigma G and for sporulation in Bacillus subtilis. J Bacteriol 185 3905 3917
18. SerranoMVieiraFMoranCPJrHenriquesAO 2008 Processing of a membrane protein required for cell-to-cell signaling during endospore formation in Bacillus subtilis. J Bacteriol 190 7786 7796
19. BlaylockBJiangXRubioAMoranCPJrPoglianoK 2004 Zipper-like interaction between proteins in adjacent daughter cells mediates protein localization. Genes Dev 18 2916 2928
20. Karmazyn-CampelliCRhayatLCarballido-LopezRDuperrierSFrandsenN 2008 How the early sporulation sigma factor sigmaF delays the switch to late development in Bacillus subtilis. Mol Microbiol 67 1169 1180
21. RatherPNCoppolecchiaRDeGraziaHMoranCPJr 1990 Negative regulator of sigma G-controlled gene expression in stationary-phase Bacillus subtilis. J Bacteriol 172 709 715
22. SchmidtRDecaturALRatherPNMoranCPJrLosickR 1994 Bacillus subtilis lon protease prevents inappropriate transcription of genes under the control of the sporulation transcription factor sigma G. J Bacteriol 176 6528 6537
23. SerranoMHovelSMoranCPJrHenriquesAOVolkerU 2001 Forespore-specific transcription of the lonB gene during sporulation in Bacillus subtilis. J Bacteriol 183 2995 3003
24. SerranoMNevesASoaresCMMoranCPJrHenriquesAO 2004 Role of the anti-sigma factor SpoIIAB in regulation of sigmaG during Bacillus subtilis sporulation. J Bacteriol 186 4000 4013
25. EvansLClarksonJYudkinMDErringtonJFeuchtA 2003 Analysis of the interaction between the transcription factor sigmaG and the anti-sigma factor SpoIIAB of Bacillus subtilis. J Bacteriol 185 4615 4619
26. KirchmanPADeGraziaHKellnerEMMoranCPJr 1993 Forespore-specific disappearance of the sigma-factor antagonist spoIIAB: implications for its role in determination of cell fate in Bacillus subtilis. Mol Microbiol 8 663 671
27. CharyVKXenopoulosPPiggotPJ 2007 Expression of the sigmaF-directed csfB locus prevents premature appearance of sigmaG activity during sporulation of Bacillus subtilis. J Bacteriol 189 8754 8757
28. RhayatLDuperrierSCarballido-LopezRPellegriniOStragierP 2009 Genetic dissection of an inhibitor of the sporulation sigma factor sigma(G). J Mol Biol 390 835 844
29. DecaturALosickR 1996 Identification of additional genes under the control of the transcription factor sigma F of Bacillus subtilis. J Bacteriol 178 5039 5041
30. HackettRHSetlowP 1988 Properties of spores of Bacillus subtilis strains which lack the major small, acid-soluble protein. J Bacteriol 170 1403 1404
31. MasonJMHackettRHSetlowP 1988 Regulation of expression of genes coding for small, acid-soluble proteins of Bacillus subtilis spores: studies using lacZ gene fusions. J Bacteriol 170 239 244
32. SunDXStragierPSetlowP 1989 Identification of a new sigma-factor involved in compartmentalized gene expression during sporulation of Bacillus subtilis. Genes Dev 3 141 149
33. FeklistovADarstSA 2009 Promoter recognition by bacterial alternative sigma factors: the price of high selectivity? Genes Dev 23 2371 2375
34. MoreiraISFernandesPARamosMJ 2007 Hot spot occlusion from bulk water: a comprehensive study of the complex between the lysozyme HEL and the antibody FVD1.3. J Phys Chem B 111 2697 2706
35. ChastanetAVitkupDYuanGCNormanTMLiuJS 2010 Broadly heterogeneous activation of the master regulator for sporulation in Bacillus subtilis. Proc Natl Acad Sci U S A 107 8486 8491
36. FujitaMLosickR 2005 Evidence that entry into sporulation in Bacillus subtilis is governed by a gradual increase in the level and activity of the master regulator Spo0A. Genes Dev 19 2236 2244
37. LevinPALosickR 1996 Transcription factor Spo0A switches the localization of the cell division protein FtsZ from a medial to a bipolar pattern in Bacillus subtilis. Genes Dev 10 478 488
38. CampAHWangAFLosickR 2011 A small protein required for the switch from {sigma}F to {sigma}G during sporulation in Bacillus subtilis. J Bacteriol 193 116 124
39. AmayaEKhvorovaAPiggotPJ 2001 Analysis of promoter recognition in vivo directed by sigma(F) of Bacillus subtilis by using random-sequence oligonucleotides. J Bacteriol 183 3623 3630
40. SunDFajardo-CavazosPSussmanMDTovar-RojoFCabrera-MartinezRM 1991 Effect of chromosome location of Bacillus subtilis forespore genes on their spo gene dependence and transcription by E sigma F: identification of features of good E sigma F-dependent promoters. J Bacteriol 173 7867 7874
41. WangSTSetlowBConlonEMLyonJLImamuraD 2006 The forespore line of gene expression in Bacillus subtilis. J Mol Biol 358 16 37
42. SteilLSerranoMHenriquesAOVolkerU 2005 Genome-wide analysis of temporally regulated and compartment-specific gene expression in sporulating cells of Bacillus subtilis. Microbiology 151 399 420
43. BagyanIHobotJCuttingS 1996 A compartmentalized regulator of developmental gene expression in Bacillus subtilis. J Bacteriol 178 4500 4507
44. AsenIDjuranovicSLupasANZethK 2009 Crystal structure of SpoVT, the final modulator of gene expression during spore development in Bacillus subtilis. J Mol Biol 386 962 975
45. MurakamiKSMasudaSDarstSA 2002 Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 A resolution. Science 296 1280 1284
46. CampbellEAWestbladeLFDarstSA 2008 Regulation of bacterial RNA polymerase sigma factor activity: a structural perspective. Curr Opin Microbiol 11 121 127
47. HelmannJD 1999 Anti-sigma factors. Curr Opin Microbiol 2 135 141
48. DecaturALLosickR 1996 Three sites of contact between the Bacillus subtilis transcription factor sigmaF and its antisigma factor SpoIIAB. Genes Dev 10 2348 2358
49. CharyVKXenopoulosPEldarAPiggotPJ 2010 Loss of compartmentalization of sigma(E) activity need not prevent formation of spores by Bacillus subtilis. J Bacteriol 192 5616 5624
50. CuttingSaHPB 1990 Genetic Analysis; HarwoodCRaCSM John Wiley and sons, Lta
51. ZilhaoRSerranoMIsticatoRRiccaEMoranCPJr 2004 Interactions among CotB, CotG, and CotH during assembly of the Bacillus subtilis spore coat. J Bacteriol 186 1110 1119
52. SerranoMZilhaoRRiccaEOzinAJMoranCPJr 1999 A Bacillus subtilis secreted protein with a role in endospore coat assembly and function. J Bacteriol 181 3632 3643
53. RealGAutretSHarryEJErringtonJHenriquesAO 2005 Cell division protein DivIB influences the Spo0J/Soj system of chromosome segregation in Bacillus subtilis. Mol Microbiol 55 349 367
Štítky
Genetika Reprodukční medicína
Článek Produces a Long Noncoding RNA and Has General Functions in the Germline, Stem Cells, and BrainČlánek Inference of Relationships in Population Data Using Identity-by-Descent and Identity-by-StateČlánek A Genome-Wide Meta-Analysis of Six Type 1 Diabetes Cohorts Identifies Multiple Associated LociČlánek Retrotransposon-Induced Heterochromatin Spreading in the Mouse Revealed by Insertional PolymorphismsČlánek Essential Roles of BCCIP in Mouse Embryonic Development and Structural Stability of ChromosomesČlánek The Receptor Tyrosine Kinase Alk Controls Neurofibromin Functions in Drosophila Growth and LearningČlánek PCNA Ubiquitination Is Important, But Not Essential for Translesion DNA Synthesis in Mammalian CellsČlánek Genome-Wide Association Study Identifies Four Loci Associated with Eruption of Permanent Teeth
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2011 Číslo 9
-
Všechny články tohoto čísla
- Temporal Trends in Results Availability from Genome-Wide Association Studies
- Association of eGFR-Related Loci Identified by GWAS with Incident CKD and ESRD
- Genetic Variants at Chromosomes 2q35, 5p12, 6q25.1, 10q26.13, and 16q12.1 Influence the Risk of Breast Cancer in Men
- Large-Scale Gene-Centric Analysis Identifies Novel Variants for Coronary Artery Disease
- Genetic Association for Renal Traits among Participants of African Ancestry Reveals New Loci for Renal Function
- Transcriptome Kinetics Is Governed by a Genome-Wide Coupling of mRNA Production and Degradation: A Role for RNA Pol II
- Conserved Regulation of p53 Network Dosage by MicroRNA–125b Occurs through Evolving miRNA–Target Gene Pairs
- Heterozygous Mutations of Are Associated with an Increased Risk of Isolated Metopic Craniosynostosis in Humans and Mice
- Study of FoxA Pioneer Factor at Silent Genes Reveals Rfx-Repressed Enhancer at and a Potential Indicator of Esophageal Adenocarcinoma Development
- Cholesterol Metabolism Is Required for Intracellular Hedgehog Signal Transduction
- Produces a Long Noncoding RNA and Has General Functions in the Germline, Stem Cells, and Brain
- Age-Dependent Recombination Rates in Human Pedigrees
- Sequence Conservation and Functional Constraint on Intergenic Spacers in Reduced Genomes of the Obligate Symbiont
- Sex Chromosome Mosaicism and Hybrid Speciation among Tiger Swallowtail Butterflies
- A Negative Feedback Loop That Limits the Ectopic Activation of a Cell Type–Specific Sporulation Sigma Factor of
- Phased Whole-Genome Genetic Risk in a Family Quartet Using a Major Allele Reference Sequence
- Mutations in or near the Transmembrane Domain Alter PMEL Amyloid Formation from Functional to Pathogenic
- Inactivation of Alters Melanosome Shape But Has Only a Subtle Effect on Visible Pigmentation
- Novel Interactions between Actin and the Proteasome Revealed by Complex Haploinsufficiency
- Germline Genetic Variants Disturbing the /LIN28 Double-Negative Feedback Loop Alter Breast Cancer Susceptibility
- Separation of Recombination and SOS Response in RecA Suggests LexA Interaction Sites
- Inference of Relationships in Population Data Using Identity-by-Descent and Identity-by-State
- Misregulation of Scm3p/HJURP Causes Chromosome Instability in and Human Cells
- A Noncoding Point Mutation of Causes Multiple Developmental Malformations and Obesity in Twirler Mice
- The Evolutionarily Conserved Longevity Determinants HCF-1 and SIR-2.1/SIRT1 Collaborate to Regulate DAF-16/FOXO
- A Genome-Wide Metabolic QTL Analysis in Europeans Implicates Two Loci Shaped by Recent Positive Selection
- Bacterial Communities of Diverse Species: Ecological Context of a Host–Microbe Model System
- A Genome-Wide Meta-Analysis of Six Type 1 Diabetes Cohorts Identifies Multiple Associated Loci
- Elongator Complex Influences Telomeric Gene Silencing and DNA Damage Response by Its Role in Wobble Uridine tRNA Modification
- Elevated Proteasome Capacity Extends Replicative Lifespan in
- Genome-Wide Analysis of Heteroduplex DNA in Mismatch Repair–Deficient Yeast Cells Reveals Novel Properties of Meiotic Recombination Pathways
- MicroRNA Predictors of Longevity in
- An Iterative Genetic and Dynamical Modelling Approach Identifies Novel Features of the Gene Regulatory Network Underlying Melanocyte Development
- Atypical AT Skew in Firmicute Genomes Results from Selection and Not from Mutation
- Retrotransposon-Induced Heterochromatin Spreading in the Mouse Revealed by Insertional Polymorphisms
- Genomic Analysis of QTLs and Genes Altering Natural Variation in Stochastic Noise
- The Abnormal Phenotypes of Cartilage and Bone in Calcium-Sensing Receptor Deficient Mice Are Dependent on the Actions of Calcium, Phosphorus, and PTH
- Cell Type–Specific Transcriptome Analysis Reveals a Major Role for and miR-200b in Mouse Inner Ear Morphogenesis
- Essential Roles of BCCIP in Mouse Embryonic Development and Structural Stability of Chromosomes
- IAP1-Mediated Ubiquitylation Controls Activation of the Initiator Caspase DRONC Independent of Protein Degradation
- VANG-1 and PRKL-1 Cooperate to Negatively Regulate Neurite Formation in
- The Receptor Tyrosine Kinase Alk Controls Neurofibromin Functions in Drosophila Growth and Learning
- Comparative and Functional Genomics of PD630 for Biofuels Development
- Identification of Type 1 Diabetes–Associated DNA Methylation Variable Positions That Precede Disease Diagnosis
- PCNA Ubiquitination Is Important, But Not Essential for Translesion DNA Synthesis in Mammalian Cells
- Genetic Effects at Pleiotropic Loci Are Context-Dependent with Consequences for the Maintenance of Genetic Variation in Populations
- Genome-Wide Association Study Identifies Four Loci Associated with Eruption of Permanent Teeth
- Bmp and Nodal Independently Regulate Expression to Maintain Unilateral Nodal Activity during Left-Right Axis Specification in Zebrafish
- Inter-Allelic Prion Propagation Reveals Conformational Relationships among a Multitude of [] Strains
- Emergence and Modular Evolution of a Novel Motility Machinery in Bacteria
- Histone Methyltransferase MET-2 Shields the Male X Chromosome from Checkpoint Machinery and Mediates Meiotic Sex Chromosome Inactivation
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Retrotransposon-Induced Heterochromatin Spreading in the Mouse Revealed by Insertional Polymorphisms
- The Evolutionarily Conserved Longevity Determinants HCF-1 and SIR-2.1/SIRT1 Collaborate to Regulate DAF-16/FOXO
- Genome-Wide Analysis of Heteroduplex DNA in Mismatch Repair–Deficient Yeast Cells Reveals Novel Properties of Meiotic Recombination Pathways
- Association of eGFR-Related Loci Identified by GWAS with Incident CKD and ESRD
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



