-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaConserved Regulation of p53 Network Dosage by MicroRNA–125b Occurs through Evolving miRNA–Target Gene Pairs
MicroRNAs regulate networks of genes to orchestrate cellular functions. MiR-125b, the vertebrate homologue of the Caenorhabditis elegans microRNA lin-4, has been implicated in the regulation of neural and hematopoietic stem cell homeostasis, analogous to how lin-4 regulates stem cells in C. elegans. Depending on the cell context, miR-125b has been proposed to regulate both apoptosis and proliferation. Because the p53 network is a central regulator of both apoptosis and proliferation, the dual roles of miR-125b raise the question of what genes in the p53 network might be regulated by miR-125b. By using a gain - and loss-of-function screen for miR-125b targets in humans, mice, and zebrafish and by validating these targets with the luciferase assay and a novel miRNA pull-down assay, we demonstrate that miR-125b directly represses 20 novel targets in the p53 network. These targets include both apoptosis regulators like Bak1, Igfbp3, Itch, Puma, Prkra, Tp53inp1, Tp53, Zac1, and also cell-cycle regulators like cyclin C, Cdc25c, Cdkn2c, Edn1, Ppp1ca, Sel1l, in the p53 network. We found that, although each miRNA–target pair was seldom conserved, miR-125b regulation of the p53 pathway is conserved at the network level. Our results lead us to propose that miR-125b buffers and fine-tunes p53 network activity by regulating the dose of both proliferative and apoptotic regulators, with implications for tissue stem cell homeostasis and oncogenesis.
Published in the journal: . PLoS Genet 7(9): e32767. doi:10.1371/journal.pgen.1002242
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002242Summary
MicroRNAs regulate networks of genes to orchestrate cellular functions. MiR-125b, the vertebrate homologue of the Caenorhabditis elegans microRNA lin-4, has been implicated in the regulation of neural and hematopoietic stem cell homeostasis, analogous to how lin-4 regulates stem cells in C. elegans. Depending on the cell context, miR-125b has been proposed to regulate both apoptosis and proliferation. Because the p53 network is a central regulator of both apoptosis and proliferation, the dual roles of miR-125b raise the question of what genes in the p53 network might be regulated by miR-125b. By using a gain - and loss-of-function screen for miR-125b targets in humans, mice, and zebrafish and by validating these targets with the luciferase assay and a novel miRNA pull-down assay, we demonstrate that miR-125b directly represses 20 novel targets in the p53 network. These targets include both apoptosis regulators like Bak1, Igfbp3, Itch, Puma, Prkra, Tp53inp1, Tp53, Zac1, and also cell-cycle regulators like cyclin C, Cdc25c, Cdkn2c, Edn1, Ppp1ca, Sel1l, in the p53 network. We found that, although each miRNA–target pair was seldom conserved, miR-125b regulation of the p53 pathway is conserved at the network level. Our results lead us to propose that miR-125b buffers and fine-tunes p53 network activity by regulating the dose of both proliferative and apoptotic regulators, with implications for tissue stem cell homeostasis and oncogenesis.
Introduction
MicroRNAs (miRNAs) are short non-coding RNA molecules that were first discovered as regulators of developmental timing, and later found to regulate complex networks of genes to orchestrate cellular functions. Lin-4 was the first miRNA gene to be discovered, and shown to regulate developmental timing by repressing its target genes at the post-transcriptional level [1]. Subsequently, miRNAs were found to regulate processes ranging from proliferation and apoptosis, to cell differentiation and signal transduction [2]–[4]. Several miRNAs are conserved in metazoan evolution, one prominent example being lin-4 whose vertebrate homologues comprise the miR-125a/b family [5]. Much like lin-4′s role of regulating the homeostasis of reiterative or self-renewing stem cells in C. elegans [6], recent studies have shown that miR-125a/b regulates mammalian neural stem cell commitment, as well as the mammalian hematopoietic stem cell (HSC) pool size [7]–[10]. Although Lin28 and Bak1 have been proposed as the critical targets of miR-125a/b for regulating these stem cell compartments [8], [9], the hundreds of predicted targets for miR-125a/b suggest a more complex interplay between miR-125a/b and its targets in regulating proliferation and differentiation.
Depending on the cell context, miR-125b has been proposed to regulate both apoptosis and proliferation. miR-125b has been shown to downregulate apoptosis in many contexts, in some cases by repressing Tp53 and Bak1. Examples include mammalian hematopoietic stem cells, human leukemia cells, neuroblastoma cells, breast cancer and prostate cancer cells [9]–[18]. During zebrafish embryogenesis, loss of miR-125b leads to widespread apoptosis in a p53-dependent manner, causing severe defects in neurogenesis and somitogenesis [16]. On the other hand, miR-125b can also downregulate proliferation in a variety of human cancer cell-lines [19]–[23] and one of its bona fide targets Lin28, also promotes cancer cell proliferation [24]. Therefore in different contexts, miR-125b appears to be able to regulate both apoptosis and proliferation.
Another molecular pathway that regulates both apoptosis and proliferation is the highly conserved p53 network [25]–[28]. Due to the central role of the p53 network in these two processes, and because we found that miR-125b regulates both human and zebrafish Tp53 but not mouse Tp53 [16], we sought to examine if miR-125b regulates the p53 network in a conserved manner in vertebrates. To address this question, we used a gain - and loss-of-function screen for miR-125b targets in different vertebrates, and validated these targets with the luciferase assay and a novel miRNA-target pull-down assay. We demonstrate that miR-125b directly represses 20 novel targets in the p53 network, including both apoptosis regulators like Bak1, Igfbp3, Itch, Puma, Prkra, Tp53inp1, Tp53, Zac1, and also cell-cycle regulators like cyclin C, Cdc25c, Cdkn2c, Edn1, Ppp1ca, Sel1l. We found that although individual miRNA-target pairs were seldom conserved, regulation of the p53 network by miR-125b appears to be conserved at the network-level. This led us to propose that miR-125b buffers and fine-tunes p53 network dosage, with implications for the role of miR-125b in tissue stem cell homeostasis and oncogenesis.
Results
Identifying direct targets of miR-125b in the p53 network
To systematically identify direct targets of miR-125b in the p53 network of vertebrates, we first employed a bioinformatics approach by identifying all predicted miR-125b targets in the p53 network, followed by three complementary methods to screen and validate these targets for both direct binding and repression by miR-125b (Figure 1). Existing databases and prediction algorithms were used to shortlist a set of p53 network genes predicted to possess miR-125b-binding sites in their 3′ UTRs. We analyzed the Ingenuity Pathways Analysis™ (IPA) database and the p53 Knowledgebase [29], [30] for a list of genes and proteins that participate in the p53 network, either by regulating p53 upstream, by direct interaction with p53 protein, or by serving as effectors of p53 function downstream. We then analyzed the TargetScan and MicroCosm Target databases [31], [32] for genes that are predicted to possess miR-125b-binding sites in their 3′ UTRs, in three vertebrate genomes: human, mouse and zebrafish. The genes at the intersection of the predicted miR-125b target list and the list of p53 network genes constituted our list of predicted miR-125b targets in the p53 network (Table S1).
Fig. 1. Identifying miR-125b targets in the p53 network of vertebrates. 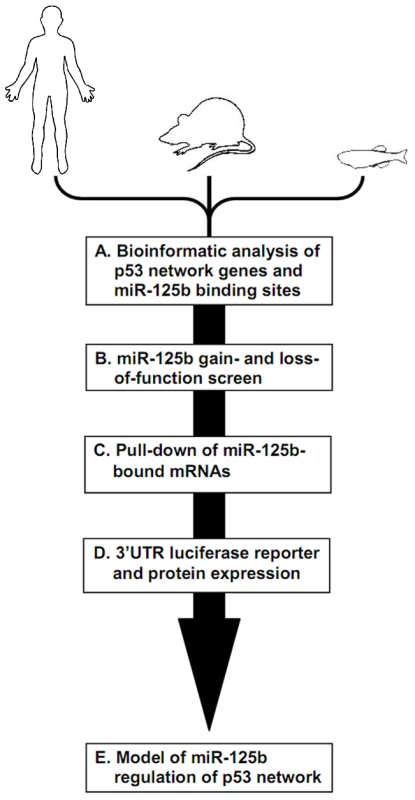
Schematic of experimental design and workflow. (A) Bioinformatic analysis was performed on p53 network genes listed in the Ingenuity Pathways Analysis database and p53 Knowledgebase, and miR-125b binding sites predicted by the TargetScan and MicroCosm databases. (B) p53 network genes were screened for miR-125b targets by using gain- (GOF) and loss-of-function (LOF) of miR-125b in human cells, mouse cells and zebrafish embryos, as indicated by effects on gene expression using qRT-PCR. (C) p53 network genes that were positive in either the GOF or LOF screen were assayed for direct binding to miR-125b using a biotinylated microRNA pull-down method. (D) p53 network genes that were also positive in the miR-125b pull-down were finally validated as miR-125b targets by 3′ UTR luciferase reporter assays and Western blots for protein expression. (E) A model of how miR-125b regulates the p53 network across vertebrates was constructed using our combined datasets for human, mouse and zebrafish cells. miR-125b gain - and loss-of-function screen in 3 vertebrates
Next we sought to screen our list of predicted targets for significant repression by miR-125b in cells, by performing a miR-125b gain - and loss-of-function screen. Gain-of-function (GOF) in miR-125b was achieved by transfection of miR-125b duplex into human SH-SY5Y or mouse N2A neuroblastoma cells, whereas loss-of-function (LOF) in miR-125b was achieved in human primary lung fibroblasts or mouse 3T3 fibroblasts by knocking down miR-125b with an antisense (AS) RNA (Figure 2A). We chose to perform a gain-of-function screen in human (SH-SY5Y) or mouse (N2A) neuroblastoma cells, because these cells possess low levels of endogenous miR-125b (Figure S1A, S1B). For the loss-of-function screen, we chose human fetal lung (hLF) or mouse (3T3) fibroblasts because they possess high levels of miR-125b (Figure S1C, S1D). miR-125a-AS was co-transfected with miR-125b-AS to achieve a complete silencing of the miR-125a/b family, because miR-125a, which shares the same seed sequence and the same predicted targets as miR-125b, is also highly expressed in human and mouse fibroblasts (Figure S1C, S1D). Genes that were either significantly repressed by miR-125b or significantly derepressed by miR-125a/b-AS with fold-changes within the range of microRNA regulation (P<0.05, fold change > 1.3), were selected as candidate miR-125b targets (Figure 2B–2D). For zebrafish embryos, which possess high levels of miR-125b, the loss-of-function (LOF) screen was performed using an antisense morpholino cocktail that blocks the loop regions of all 3 pre-miR-125b hairpin precursors [16]. The gain-of-function (GOF) screen was performed by co-injecting miR-125b duplex with the morpholino (Figure 2A). All gene expression changes were measured with at least three biological replicates using qRT-PCR.
Fig. 2. GOF/LOF screen for p53 network genes regulated by miR-125b. 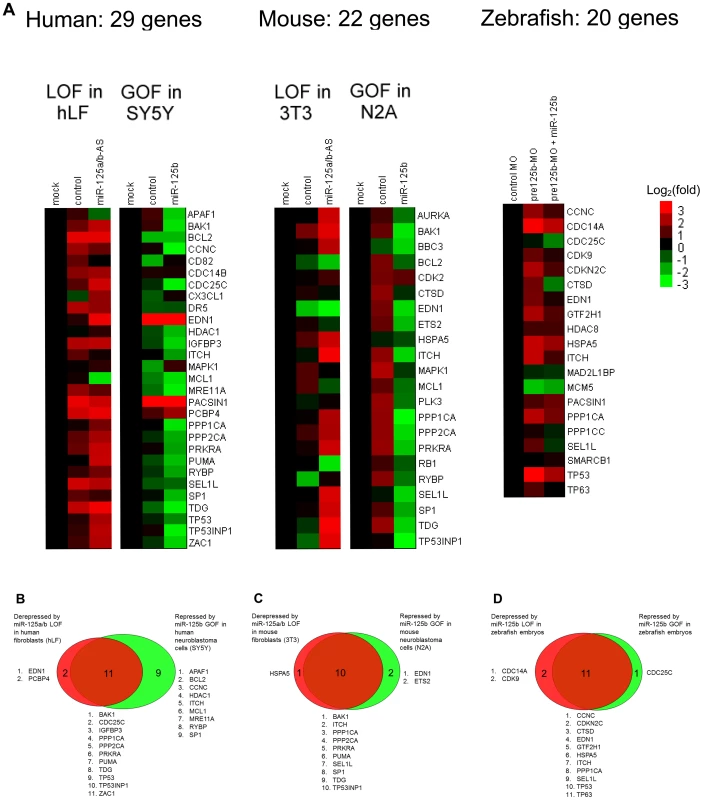
(A) Loss-of-function (LOF) screens were performed in human primary lung fibroblasts (hLF) or mouse 3T3 fibroblasts by transfecting an antisense RNA against both miR-125a and miR-125b (miR-125a/b-AS), or by microinjecting morpholinos (MO) against pre-mir-125b hairpin precursors (all 3 isoforms) into zebrafish embryos. Gain-of-function (GOF) screens were performed in human SH-SY5Y and mouse N2A neuroblastoma by transfecting the miR-125b duplex into cells in culture, or by coinjecting the miR-125b duplex with the morpholinos against pre-mir-125b into zebrafish embryos. Fold changes in gene expression were measured by qRT-PCR twenty-four hours after transfection or injection, relative to the mock and negative control miRNA or morpholino, and shown as log2(fold change) using a heat-map. (B) Human: 13 genes were significantly derepressed by a loss of miR-125b, while 20 genes were significantly repressed by a gain of miR-125b, making a total of 22 genes that passed the screen (P<0.05, fold change > 1.3, relative to mock control). (C) Mouse: 11 genes were significantly derepressed by a loss of miR-125b, while 12 genes were significantly repressed by a loss of miR-125b, making a total of 13 genes that passed the screen (P<0.05, fold change > 1.3, relative to mock control). (D) Zebrafish: 13 genes were significantly derepressed by a loss of pre-mir-125b (P<0.05, fold change > 1.3, relative to control MO), while 12 genes were significantly repressed/rescued by a gain of miR-125b (P<0.05, fold change > 1.3, relative to pre-mir-125b MO), making a total of 14 genes that passed the screen. All experiments were performed with at least three biological replicates. Our GOF/LOF screen revealed that in humans, out of 29 predicted targets in the p53 network, 13 genes were derepressed by miR-125a/b-AS in hLF cells and 20 genes were repressed by miR-125b in SH-SY5Y cells (Figure 2B). In mice, out of 22 predicted targets in the p53 network, 11 genes were derepressed by miR-125a/b-AS in 3T3 cells and 12 genes were repressed by miR-125b in N2A cells (Figure 2C). In zebrafish embryos, out of 20 predicted targets in the p53 network, 13 genes were derepressed by pre-miR-125b morpholino and 12 genes were repressed by the injection of miR-125b duplex (Figure 2D). In total, 22 human genes, 13 mouse genes and 14 zebrafish genes passed the gain - and loss-of-function qRT-PCR screen.
Direct binding interactions between miR-125b and mRNA targets from the p53 network
To assess which candidate miR-125b targets identified in the gain - and loss-of-function qRT-PCR screen are directly bound by miR-125b in cells, we employed a novel miRNA pull-down method developed by Lal et al. (manuscript in preparation). RNA transcripts bound to biotinylated-miR-125b were pulled down with streptavidin beads and quantified by qRT-PCR relative to mRNAs bound to biotinylated-control miRNA (log2 fold change > 0.5, P<0.05). In this assay, biotinylated miRNAs were shown to be loaded into the RNA-induced silencing complex (RISC) and fully functional in repressing their target mRNAs (Lal et al., manuscript in preparation). This method provides a robust and complementary method for detecting miRNAs bound to endogenous target mRNAs, and serves as a useful approach for distinguishing direct and indirect targets in the same pathway (Lal et al., manuscript in preparation). Quantification of the pulled down mRNA targets in hLF cells revealed that 13 out of 22 gene transcripts, Bak1, Cdc25c, Edn1, Igfbp3, Mre11a, Ppp1ca, Ppp2ca, Prkra, Puma, Tdg, Tp53, Tp53inp1 and Zac1, were direct binding targets of miR-125b in human cells (Figure 3A). In mouse 3T3 cells, 11 out of 13 gene transcripts, Bak1, Hspa5, Itch, Ppp1ca, Ppp2ca, Prkra, Puma, Sel1l, Sp1, Tdg and Tp53inp1, were found to be direct binding targets of miR-125b (Figure 3B). In zebrafish embryos, 8 out of 14 gene transcripts, Cdc25c, Cdkn2c, Gtf2h1, Hspa5, Itch, Ppp1ca, Sel1l, and Tp53, were pulled down by miR-125b (Figure 3C). Tp53 mRNA was pulled down by miR-125b only in human lung fibroblasts and zebrafish embryos but not in mouse fibroblasts, consistent with previously published results [16] and the Targetscan algorithmic prediction that miR-125b targets Tp53 in humans and zebrafish but not in mice.
Fig. 3. Direct binding of miR-125b to p53 network targets. 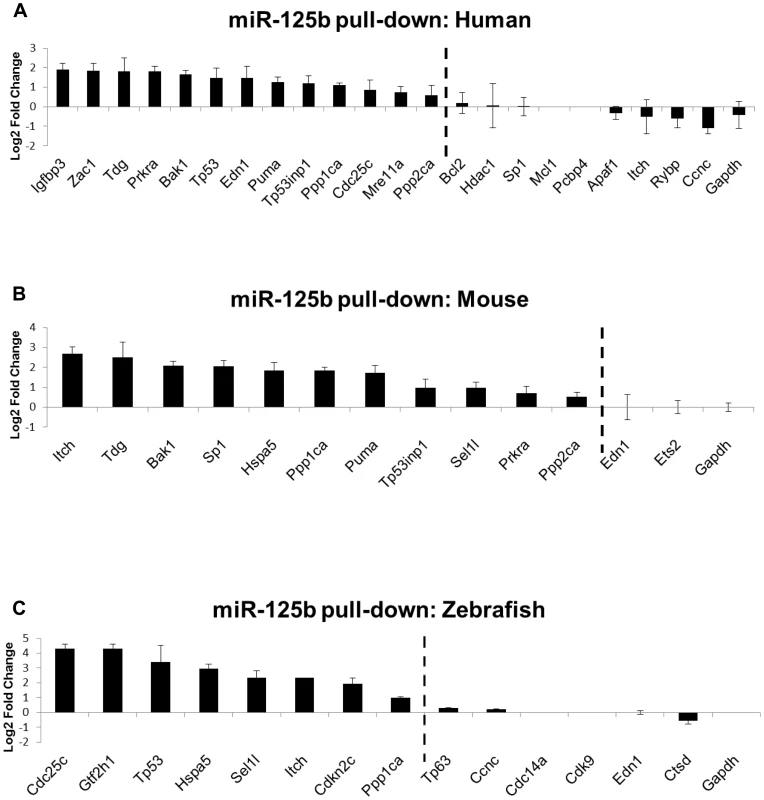
Biotinylated miR-125b was used as bait to pull-down mRNAs bound to miR-125b, using streptavidin-conjugated magnetic beads. The mRNAs were quantified by qRT-PCR, normalized to Gapdh, and then compared relative to the same mRNA species pulled down by a biotinylated C. elegans negative control microRNA. The enrichment of mRNAs bound to miR-125b is presented as mean log2 fold change ± s.e.m. (n≥3 biological replicates). (A) Human: 13 out of 22 candidate targets were significantly enriched by miR-125b pull-down in human primary lung fibroblasts (hLF) 24 hours after transfection. (B) Mouse: 11 out of 13 candidate targets were significantly enriched by miR-125b pull-down in mouse 3T3 fibroblasts 24 hours after transfection. (C) Zebrafish: 8 of 14 candidate targets were significantly enriched by miR-125b pull-down in zebrafish embryos 24 hours after injection. Dashed line: cutoff for genes that were significantly enriched (Log2 Fold change > 0.5, P<0.05). Validation of miR-125b targets in the p53 network
As a final validation of the candidate miR-125b targets we have identified in the p53 network, we tested our candidate target genes with the luciferase reporter assay. Where cloning was successful, we cloned the entire 3′ UTR of selected candidate target genes into a Renilla luciferase reporter, and assayed luciferase expression following co-transfection of miR-125b duplex into HEK-293T cells. Transfection of miR-125b significantly suppressed 40-60% (P<0.01) of the luciferase activity of many 3′ UTR reporters of the miR-125b targets we analyzed, relative to transfection of the negative control miRNA (Figure 4A). For humans, the 3′ UTR reporters of Bak1, Cdc25c, Ppp1ca, Ppp2ca, Prkra, Puma, Tdg, Tp53, Tp53inp1, and Zac1 were significantly suppressed by miR-125b. In mice, the 3′ UTR reporters of Bak1, Itch, Ppp1ca, Ppp2ca, Prkra, Puma, Sel1l, Tdg, and Tp53inp1 were significantly suppressed by miR-125b (Figure 4B). In zebrafish, the 3′ UTR reporters of Ccnc, Cdc25c, Cdkn2c, Gtf2h1, Hspa5, Ppp1ca, and Tp53 were significantly suppressed by miR-125b (Figure 4C). With the exception of zebrafish Ccnc, all genes tested were positive in the miR-125b-pull-down as well as the miR-125b gain - and loss-of-function screen. Amongst these targets, we found Ppp1ca, Prkra and Tp53 to be especially interesting from the evolutionary viewpoint, since all 3 vertebrate species possess these 3 genes, but each gene shows a different pattern of evolutionary conservation with respect to miR-125b-repression. Ppp1ca is repressed by miR-125b in all 3 species, Prkra is repressed by miR-125b in humans and mice, while Tp53 is repressed in humans and zebrafish. To examine the sequence evolution of these miRNA-mRNA pairs in greater detail, we compared the Targetscan-predicted miR-125b binding sites of these genes in humans, mice and zebrafish. In Ppp1ca, the predicted binding site is 95% identical between humans and mice and 55% identical between humans and zebrafish, while the seed binding sequence is 100% conserved in all 3 species (Figure 4D). In Prkra, the predicted binding site is 94% identical between humans and mice, but only 26% identical between humans and zebrafish, while the seed binding sequence is completely absent in zebrafish (Figure 4D). In contrast, the predicted binding site in Tp53 is 64% identical between humans and zebrafish, and the seed binding sequence is 100% conserved between humans and zebrafish, but only 36% identical between humans and mice, while the mouse seed binding sequence has acquired 2 point mutations (Figure 4D). The miR-125b-repression patterns we observed for each of these genes in the qPCR, pull-down and luciferase assays are consistent with these DNA sequence analyses, suggesting that evolution in the miRNA-mRNA binding is driving the evolution in miR-125b-repression patterns. Introduction of point mutations into the predicted seed binding sequences abrogated miR-125b-repression of each target 3′UTR luciferase reporter (P<0.05), validating the predicted miR-125b binding sites and confirming the miRNA-mRNA sequence evolution patterns we observed (Figure 4E).
Fig. 4. Validation of miR-125b targets. 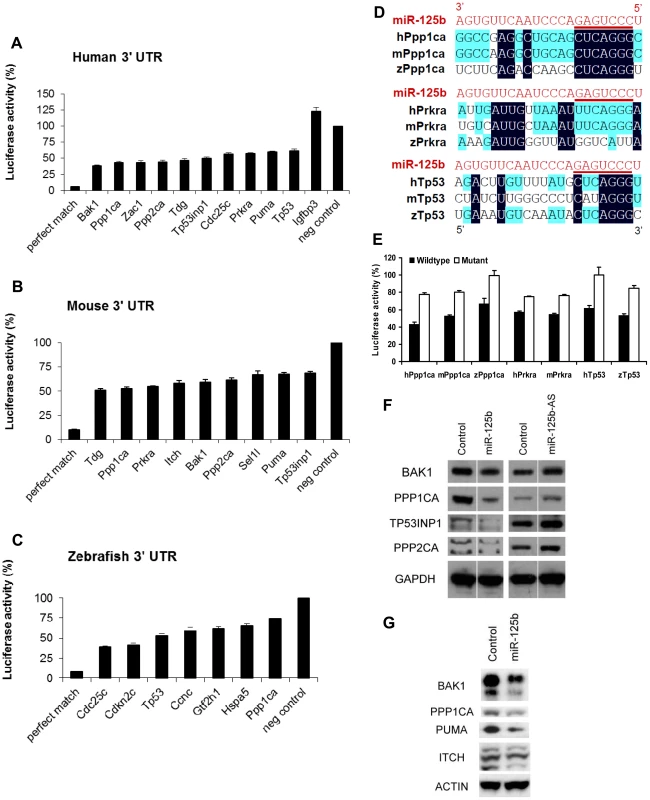
Candidate p53 network genes that were positive in both the GOF/LOF screen and miR-125b pull-down were validated for targeting by miR-125b using the 3′ UTR luciferase reporter assay and Western blots for protein expression. (A-C), Reporter genes containing the full-length 3′ UTRs of each selected target gene were co-transfected with miR-125b duplex into 293T cells. Luciferase readings were obtained 48 hours after transfection and presented here as the average percentage of luciferase activity ± s.e.m. (n≥3) relative to a scrambled duplex co-transfected control (100%). A reporter containing a 23-nucleotide-binding-site with perfect complementarity to miR-125b was used as the perfect match positive control, while the unmodified luciferase reporter was used as the empty negative control. (A) Human: 10 out of 13 candidate genes' 3′ UTRs showed significant repression by miR-125b relative to the control (p<0.01). (B) Mouse: 9 out of 11 candidate genes' 3′ UTRs showed significant repression by miR-125b relative to the control (p<0.01). (C) Zebrafish: 7 out of 8 candidate genes' 3′ UTRs showed significant repression by miR-125b relative to the control (p<0.01). (D) Alignment of predicted miR-125b binding sites in the 3′UTRs of Ppp1ca, Prkra and Tp53 across three species. Seed-binding sequences are underlined. Bases conserved in two (blue) or three (black) species are highlighted. (E) The 3′UTR seed-binding sequences of 7 target mRNAs were mutated and assayed for direct binding to miR-125b using the luciferase reporter assay, relative to wild-type 3′UTR sequences. (E) The seed-binding sequences in the 3′UTR of 7 predicted target mRNAs were mutated and compared to wild-type sequences for binding to miR-125b using luciferase reporter assay. (F-G) Western blot analysis of protein expression of selected target genes two days after a transfection of miR-125b duplex, miR-125b antisense (AS) or negative control duplex or negative control antisense. (F) Western blots showed that miR-125b repressed BAK1, PPP1CA, TP53, and PPP2CA levels in human SH-SY5Y neuroblastoma cells, while the antisense RNA miR-125b-AS derepressed expression of these proteins in human ReNcell VM neural progenitor cells. (G) Western blots showed that miR-125b repressed BAK1, PPP1CA, PUMA, and ITCH levels in mouse N2A neuroblastoma cells. Abbreviations: h, human; m, mouse; z, zebrafish. Finally, we checked miR-125b regulation of protein expression in a subset of p53 network targets for which reliable Western blotting was possible. miR-125b significantly downregulated the protein levels of human BAK1, PPP1CA, TP53INP1, PPP2CA, CDC25C, and TP53 in SH-SY5Y neuroblastoma cells (Figure 4F). In mouse N2A neuroblastoma cells, miR-125b significantly downregulated mouse BAK1, PPP1CA, PUMA, and ITCH protein (Figure 4G).
miR-125b regulation of the p53 network, but not individual miRNA-target pairs, is conserved
Our results reveal that miR-125b regulation of the p53 network is conserved at the network-level over the course of vertebrate evolution, but individual miRNA-target pairs are evolving rapidly. To summarize our results, our list of predicted miR-125b targets in the p53 network (Table S1) was filtered and reclassified according to the results of the screen and validation assays (Figure 5). From the GOF/LOF screen we were able to identify mRNAs perturbed by miR-125b. However these results did not discriminate between direct or indirect targets. To supplement these experiments the pull-down assay was used to uncover mRNAs physically associated with miR-125b. Of note, the pull-down might not identify mRNA targets that are rapidly degraded, and as such the luciferase reporter assay can complement its shortcomings. Taken together the three assays provide a powerful means to identify direct miR-125b targets. In order to minimize false positives, we counted the number of assays for which each gene target was positive, and gene targets that failed to pass at least 2 assays in at least one vertebrate species were filtered out. Predicted targets that passed 3 assays (red), 2 assays (orange), 1 assay (yellow), or predicted targets that failed all assays but whose orthologues in other species passed 3 assays of direct regulation by miR-125b (pink), were colored as indicated (Figure 5). Using our conservative estimate of miR-125b targets in the p53 network, we found that in all three vertebrates we examined – humans, mice and zebrafish – miR-125b regulates multiple p53 network genes. This shows that miR-125b regulation of the p53 network is conserved at least at the network level. However very few individual gene targets of miR-125b in the p53 network were conserved across all three vertebrates (Figure 5; Figure 6A-6C). Instead, conserved miR-125b regulation of the p53 network appears to occur through evolving miRNA-target pairs in the three vertebrates – zebrafish (Figure 6A), mouse (Figure 6B), and humans (Figure 6C). In general, we observe miR-125b regulating 2 general classes of genes in the p53 network: (i) apoptosis regulators like Bak1, Igfbp3, Itch, Puma, Prkra, Tp53inp1, Tp53, and Zac1, and (ii) cell-cycle regulators like cyclin C, Cdc25c, Cdkn2c, Edn1, Ppp1ca, and Sel1l.
Fig. 5. Summary of genes in p53 network that are directly targeted by miR-125b. 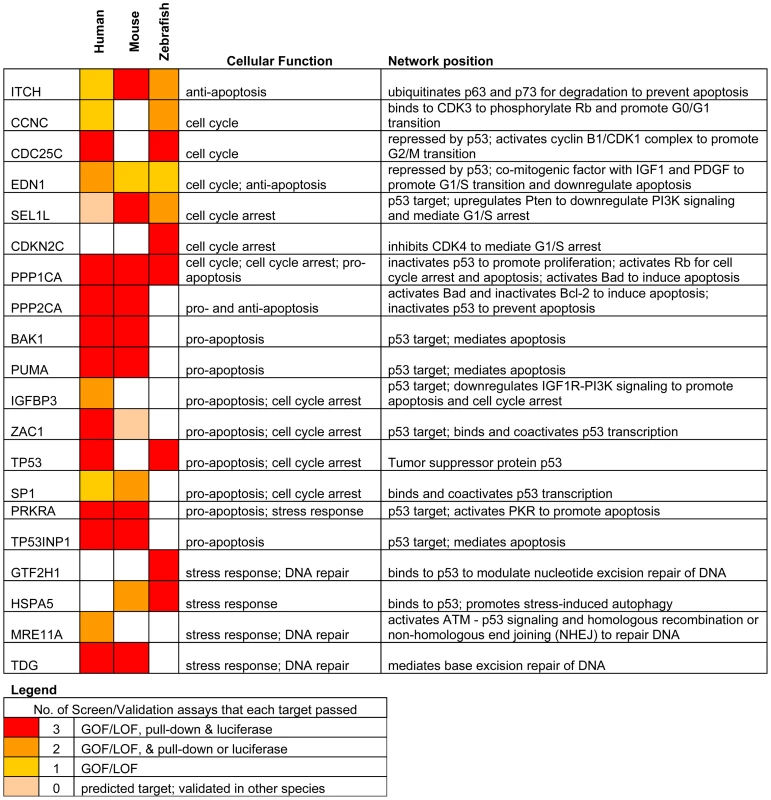
Only targets that passed ≥ 2 validation assays, in at least one species, are shown. Red: predicted targets validated by 3 assays; Orange: predicted targets validated by 2 assays; Yellow: predicted targets validated by 1 assay; Pink: predicted targets not validated by any assay, but validated by 3 assays in another species. Fig. 6. Models of miR-125b regulation of p53 networks in humans, mice, and zebrafish. 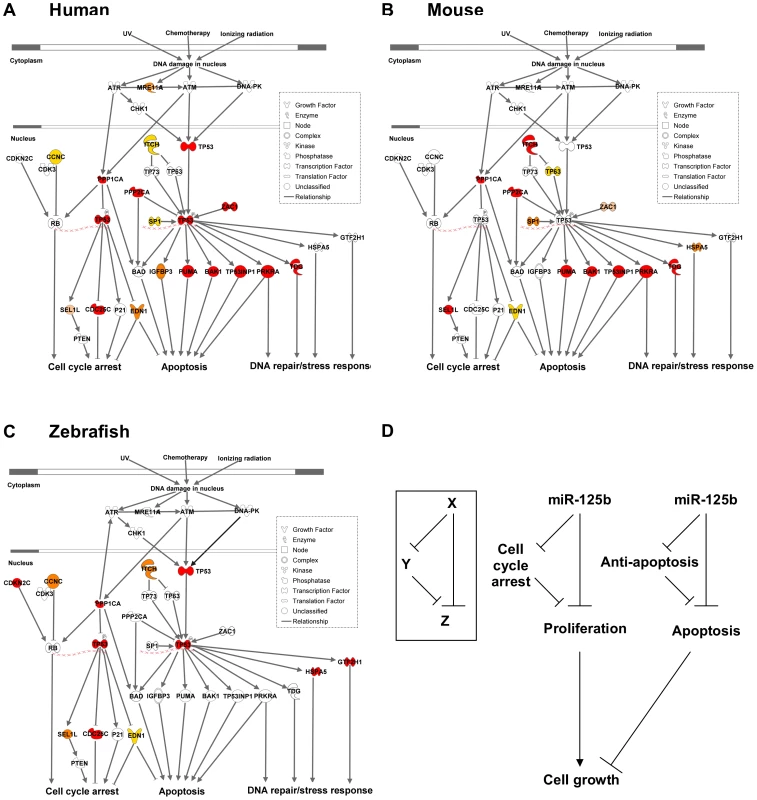
(A) Human p53 network. (B) Mouse p53 network. (C) Zebrafish p53 network. Models were constructed by Ingenuity Pathway Analysis. Red: predicted targets validated by 3 assays; Orange: predicted targets validated by 2 assays; Yellow: predicted targets validated by 1 assay; Pink: predicted targets not validated by any assay, but validated by 3 assays in another species. (D) Incoherent feedforward loop (FFL) motifs characterize miR-125b regulation of p53 network genes that mediate apoptosis or cell cycle arrest. Because miR-125b represses both pro-apoptosis and anti-apoptosis genes, as well as both proliferation and cell-cycle arrest genes in all three vertebrates (Figure 5), miR-125b appears to modulate the p53 network on the whole through an incoherent feedforward loop (FFL) [33], [34] acting on the cellular processes of apoptosis and cell proliferation (Figure 6D). An incoherent type-2 FFL is a regulatory pattern in which X represses a target Z and also represses Y, another repressor of Z (Figure 6D). Incoherent FFLs have been found in the transcription factor networks of human embryonic stem cells and hematopoietic stem cells, and have been shown to modulate E2F1 dosage in the Myc-E2F1 pathway [35]-[37]. Besides accelerating responses and acting as amplitude filters [38]-[40], the incoherent FFL motif is also a noise buffering motif that reduces the variance of network dosage [41]–[43]. Thus our finding that incoherent FFLs fit the overall structure of network relationships between miR-125b and the p53-mediated processes, suggests that miR-125b is fine-tuning and buffering p53 network dosage.
Discussion
In this study, we sought to identify direct targets of miR-125b in the p53 network of humans, mice and zebrafish, to better understand how miR-125b regulates the p53 network throughout evolution and how that might relate to its conserved role in regulating tissue stem cells.
We identified 20 direct targets of miR-125b in the p53 network, including 15 novel targets like Zac1, Puma, Itch and Cdc25c, and also targets like Bak1 and Tp53 that were identified in previous studies [9], [16]–[18]. In general, we found that miR-125b directly represses 2 classes of genes: apoptosis regulators and cell-cycle regulators. With the exception of Ppp1ca, Itch and Edn1, very few individual targets were strictly conserved throughout vertebrate evolution. Instead, we found that only the network-level of regulation was conserved, and miR-125b-regulation of individual apoptosis and proliferation regulators appears to be evolving rapidly from species to species. This observation suggests that, at least within the vertebrates, the 3′ UTR sequences of each gene target is evolving rapidly via neutral genetic drift. In other words, the loss or gain of a single miR-125b-binding site in the 3′ UTR of most genes appears to have a relatively insignificant effect on the fitness of an organism. On the other hand, the strict conservation of miR-125b-regulation at the network-level in humans, mice and zebrafish, suggests that natural selection acts on the network-level rather than the gene-level with regard to miRNA-target evolution. It will be interesting to see if this novel paradigm applies to other microRNAs or gene networks as well.
Previous studies on miRNA evolution have suggested that a relatively poor conservation of individual miRNA-target pairs but strong conservation of a miRNA-gene network relationship is consistent with miRNAs' role as buffers of gene expression [42], [44], [45]. Our observation that an incoherent FFL-like network motif fits the overall structure of the miR-125b - p53 network models with respect to apoptosis and cell proliferation, lends further support to this idea since incoherent FFL network motifs are well-adapted for noise filtering [41], [43], [46]. It is thought that miRNAs are at least partially responsible for the phenomenon of developmental or phenotypic stability within each species [41], [42], termed “canalization” by C. H. Waddington [47]. These studies suggest that miRNAs have a conserved role in regulating the overall stability of pathways/networks, a role which is relatively unaffected by the loss or gain of individual miRNA-targets over the course of evolution. A network buffering function has also been suggested for the regulation of muscle development by miR-1 throughout evolution, regulation of the Wnt pathway by miR-8, and fine-regulation of Pten dosage by a variety of miRNAs [48]–[50]. Our findings suggest that the fine-tuning of p53 network dosage by miR-125b is another example of this paradigm.
Fine-regulation of p53 network dosage by miR-125b may also explain miR-125b's conserved role in regulating tissue stem cell homeostasis. In C. elegans, loss-of-function mutations in lin-4 lead to a delay in differentiation and thus expansion of vulval precursor cells, seam stem cells in the lateral hypodermis and mesoblasts, causing multiple defects in larval development [6]. In zebrafish, loss of miR-125b leads to widespread p53-dependent apoptosis with consequent defects in early embryogenesis, especially in neurogenesis and somitogenesis [16]. Overexpression of miR-125a/b causes an expansion of mammalian hematopoietic stem cells (HSCs) and aberrant differentiation, leading to myeloid leukemia [9], [10] and also lymphoid leukemia if miR-125b is overexpressed in fetal liver HSC-enriched cells [12]. However, the molecular underpinnings of miR-125a/b's regulation of tissue stem cell homeostasis had remained unclear largely due to the complex nature of microRNA regulation of gene networks. The 2 classes of miR-125b targets in the p53 network, and the incoherent FFL network motifs that we found, may at least partially explain how miR-125b regulates tissue stem cells in vertebrates. By fine-tuning both apoptosis regulators and cell-cycle regulators, miR-125b may fine-tune the p53 network dosage to drive the self-renewal of tissue stem cells. It could explain how overexpression of miR-125b leads to an expansion of self-renewing hematopoietic stem cells while loss of miR-125b leads to aberrant apoptosis and proliferation, with consequent defects in tissue differentiation.
Several studies have implicated miR-125b as an oncogene in a variety of mammalian tissue compartments, e.g. leukemia, neuroblastoma, prostate cancer and breast cancer [9]–[18]. These studies have ascribed miR-125b's anti-apoptotic effect as an oncogene to its direct suppression of Bak1 or Tp53 [9], [16]-[18]. On the other hand, several research groups have also reported miR-125b's role as a potential tumor suppressor by suppressing proliferation in cell-culture models [19]–[23]. Our identification of 20 direct targets of miR-125b in the p53 network reconciles these findings because miR-125b modulates the expression of both apoptosis regulators and cell-cycle regulators. Although miR-125b's suppression of p53 itself is not conserved in mice, miR-125b's anti-apoptotic role – through suppression of multiple pro-apoptosis regulators in the p53 network – appears to be conserved in vertebrates. miR-125b's ability to fine-tune the subtle balance of apoptosis vs. cell-cycle regulators and thus buffer the p53 network dosage in different contexts, could explain why miR-125b dysregulation can lead to either tumor suppression or oncogenesis depending on the context. It is possible that this buffering feature of miR-125b represents a general principle of miRNA regulation of gene networks.
Materials and Methods
Prediction of miR-125b targets in the p53 network
A list of p53-associated genes was compiled from the p53 Knowledgebase website [30] and from the Ingenuity Pathway Analysis™ database [29]. The targets of miR-125b in human and mouse were predicted by TargetScan [31]. The targets of miR-125b in zebrafish were predicted by MicroCosm [32]. The human homologues of mouse and zebrafish targets were identified by the DAVID gene ID conversion tool.
Cell culture and transfection
Human lung fibroblast cells, human neuroblastoma SH-SY5Y cells, mouse neuroblastoma Neuro-2A cells, mouse fibroblast Swiss-3T3 cells and human HEK-293T cells were maintained in DMEM media, supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin (Invitrogen). Neuro-2A cells, 3T3 cells, SH-SY5Y cells and human lung fibroblast cells were transfected in suspension with 5×105 cells per well in 6-well plates using lipofectamin-2000 (Invitrogen). miRNA duplexes and antisense oligonucleotides (Ambion) were transfected at a final concentration of 80 nM.
Microinjection in zebrafish embryos
Wild-type zebrafish were maintained by standard protocols [51]. All injections were carried out at 1–4 cell stage with 2 nl of solution into each embryo. In the knockdown experiments, miR-125b morpholinos were injected at 0.75 pmole/embryo (lp125bMO1/2/3 indicates the co-injection of three lp125bMOs, 0.25 pmole each); miR-125b duplex was injected as 37.5 fmole/embryo.
Quantitative RT-PCR
RNA was extracted from cells or zebrafish embryos using Trizol reagent (Invitrogen) and subsequently column-purified with RNeasy kits (Qiagen). For qRT-PCR of miR-125a, miR-125b and RNU6B, 100 ng of total RNA was reverse-transcribed and subjected to Taqman microRNA assay (Applied Biosystems). For qRT-PCR of mRNAs, cDNA synthesis was performed with 1 µg of total RNA using the High Capacity cDNA Archive Kit (Applied Biosystems). The expression of all genes was analyzed by SYBR assay using the Applied Biosystems real-time PCR system or the Fluidigm 48x48 dynamic array system (Fluidigm) following the manufacturer's protocol.
miRNA–target pull-down assay
50 ul of streptavidin coated magnetic beads (Invitrogen) were blocked with 1 mg/ml yeast tRNA and 1 mg/ml BSA in 1 ml lysis buffer (20 mM Tris pH 7.5, 100 mM KCl, 5 mM MgCl2, 0.3% NP-40) for 2 hours at 4°C and wash twice with lysis buffer. hsa-miR-125b or cel-miR-67 (negative control) duplex was synthesized with a biotin conjugated at the 3′ end of the active strand by Dharmacon Research Inc. The miRNAs were transfected into human lung fibroblasts or mouse 3T3 fibroblasts at a final concentration of 80 nM as described above. The miRNAs were also injected into zebrafish embryos at 1 to 4-cell stage at a final concentration of 37.5 fmole/embryo. After 24 hours, cells from 3 wells of fibroblasts or 50 zebrafish embryos were incubated with 500 ul cold lysis buffer containing freshly added 100 units/ml RNase inhibitor (Invitrogen) and protease inhibitor cocktail (Roche) for 20 minutes on ice. After the cell debris is removed by centrifugation, the lysate was incubated with pre-blocked streptavidin coated beads for 2 hours at 4°C. Subsequently, the beads were washed 5 times with cold lysis buffer and incubated with Trizol for RNA extraction.
Luciferase reporter assay
The whole 3′ UTR of target genes were cloned into the psiCHECK-2 vector (Promega), between the XhoI and NotI site, immediately 3’ downstream of the Renilla luciferase gene. For selected targets, we introduced 3 point mutations into the 7-nt seed-binding sequence using inverse PCR with non-overlapping primers carrying the mutated sequences. 10 ng of each psiCHECK-2 construct was co-transfected with 10 nM miRNA duplexes or into HEK-293T cells in a 96-well plate using lipofectamin-2000 (Invitrogen). After 48 hours, the cell extract was obtained; Firefly and Renilla luciferase activities were measured with the Dual-Luciferase reporter system (Promega) according to the manufacturer's instructions.
Western blot assay
Cells were lysed in RIPA buffer (Pierce). Proteins were separated by a 10% polyacrylamide gel and transferred to a methanol-activated PVDF membrane (GE Healthcare). The membrane was blocked for one hour in PBST containing 7.5% milk and subsequently probed with primary antibodies (Santa Cruz) overnight at 4°C. After 1-hour incubation with goat-anti-mouse HRP-conjugated secondary antibody (Santa Cruz), the protein level was detected with luminol reagent (Santa Cruz).
Statistical analysis
Two-tail T-tests were used to determine the significance of differences between the treated samples and the controls where values were obtained from luciferase reporter assay or qRT-PCR. The tests were performed using Microsoft Excel where the test type is always set to two-sample equal variance.
Supporting Information
Zdroje
1. LeeRCFeinbaumRLAmbrosV 1993 The C-elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75 843 854
2. BartelDPChenCZ 2004 Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nature Reviews Genetics 5 396 400
3. HeLHannonGJ 2004 MicroRNAs: Small RNAs with a big role in gene regulation (vol 5, pg 522 2004). Nature Reviews Genetics 5 522-+
4. InuiMMartelloGPiccoloS 2010 MicroRNA control of signal transduction. Nature Reviews Molecular Cell Biology 11 252 263
5. Lagos-QuintanaMRauhutRYalcinAMeyerJLendeckelW 2002 Identification of tissue-specific microRNAs from mouse. Current Biology 12 735 739
6. ChalfieMHorvitzHRSulstonJE 1981 Mutations that lead to reiterations in the cell lineages of C-elegans. Cell 24 59 69
7. LeMTNXieHMZhouBYChiaPHRizkP 2009 MicroRNA-125b Promotes Neuronal Differentiation in Human Cells by Repressing Multiple Targets. Molecular and Cellular Biology 29 5290 5305
8. RybakAFuchsHSmirnovaLBrandtCPohlEE 2008 A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nature Cell Biology 10 987 993
9. GuoSQLuJSchlangerRZhangHWangJY 2010 MicroRNA miR-125a controls hematopoietic stem cell number. Proceedings of the National Academy of Sciences of the United States of America 107 14229 14234
10. O'ConnellRMChaudhuriAARaoDSGibsonWSJBalazsAB 2010 MicroRNAs enriched in hematopoietic stem cells differentially regulate long-term hematopoietic output. Proceedings of the National Academy of Sciences of the United States of America 107 14235 14240
11. BousquetMQuelenCRosatiRMansat-De MasRLa StarzaR 2008 Myeloid cell differentiation arrest by miR-125b-1 in myelodysplasic syndrome and acute myeloid leukemia with the t(2;11)(p21;q23) translocation. Journal of Experimental Medicine 205 2499 2506
12. BousquetMHarrisMHZhouBLodishHF 2010 MicroRNA miR-125b causes leukemia. Proceedings of the National Academy of Sciences of the United States of America 107 21558 21563
13. ChapiroERussellLJStruskiSCaveHRadford-WeissI 2010 A new recurrent translocation t(11;14)(q24;q32) involving IGH@ and miR-125b-1 in B-cell progenitor acute lymphoblastic leukemia. Leukemia 24 1362 1364
14. KlusmannJHLiZBohmerKMarozAKochML 2010 miR-125b-2 is a potential oncomiR on human chromosome 21 in megakaryoblastic leukemia. Genes & Development 24 478 490
15. SonokiTIwanagaEMitsuyaHAsouN 2005 Insertion of microRNA-125b-1, a human homologue of lin-4, into a rearranged immunoglobulin heavy chain gene locus in a patient with precursor B-cell acute lymphoblastic leukemia. Leukemia 19 2009 2010
16. LeMTNTehCShyh-ChangNXieHMZhouBY 2009 MicroRNA-125b is a novel negative regulator of p53. Genes & Development 23 862 876
17. ZhouMLiuZXZhaoYHDingYLiuH 2010 MicroRNA-125b Confers the Resistance of Breast Cancer Cells to Paclitaxel through Suppression of Pro-apoptotic Bcl-2 Antagonist Killer 1 (Bak1) Expression. Journal of Biological Chemistry 285 21496 21507
18. ShiXBXueLYangJMaAHZhaoJ 2007 An androgen-regulated miRNA suppresses Bak1 expression and induces androgen-independent growth of prostate cancer cells. Proceedings of the National Academy of Sciences of the United States of America 104 19983 19988
19. ScottGKGogaABhaumikDBergerCESullivanCS 2007 Coordinate suppression of ERBB2 and ERBB3 by enforced expression of micro-RNA miR-125a or miR-125b. Journal of Biological Chemistry 282 1479 1486
20. ShiLZhangJXPanTHZhouJFGongWY 2010 MiR-125b is critical for the suppression of human U251 glioma stem cell proliferation. Brain Research 1312 120 126
21. XiaHFHeTZLiuCMCuiYSongPP 2009 MiR-125b Expression Affects the Proliferation and Apoptosis of Human Glioma Cells by Targeting Bmf. Cellular Physiology and Biochemistry 23 347 358
22. HofmannMHHeinrichJRadziwilGMoellingK 2009 A Short Hairpin DNA Analogous to miR-125b Inhibits C-Raf Expression, Proliferation, and Survival of Breast Cancer Cells. Molecular Cancer Research 7 1635 1644
23. MizunoYYagiKTokuzawaYKanesaki-YatsukaYSudaT 2008 miR-125b inhibits osteoblastic differentiation by down-regulation of cell proliferation. Biochemical and Biophysical Research Communications 368 267 272
24. ViswanathanSPowersJEinhornWHoshidaYNgT 2009 Lin28 promotes transformation and is associated with advanced human malignancies. Nature Genetics 41 843 848
25. LevineAJ 1997 p53, the cellular gatekeeper for growth and division. Cell 88 323 331
26. AgarwalMLTaylorWRChernovMVChernovaOBStarkGR 1998 The p53 network. Journal of Biological Chemistry 273 1 4
27. MayoLDDonnerDB 2002 The PTEN, Mdm2, p53 tumor suppressor-oncoprotein network. Trends in Biochemical Sciences 27 462 467
28. HeXYHeLHannonGJ 2007 The guardian's little helper: MicroRNAs in the p53 tumor suppressor network. Cancer Research 67 11099 11101
29. SiuDCLauranceM 2004 Rapid generations of de novo biological pathways from large-scale gene expression data using the Ingenuity Pathways Analysis application. Proceedings of the American Association for Cancer Research Annual Meeting 45 756
30. LimYPLimTTChanYLSongACMYeoBH 2007 The p53 knowledgebase: an integrated information resource for p53 research. Oncogene 26 1517 1521
31. LewisBPBurgeCBBartelDP 2005 Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120 15 20
32. Griffiths-JonesSSainiHKvan DongenSEnrightAJ 2008 miRBase: tools for microRNA genomics. Nucleic Acids Research 36 D154 D158
33. Shen-OrrSSMiloRManganSAlonU 2002 Network motifs in the transcriptional regulation network of Escherichia coli. Nature Genetics 31 64 68
34. MiloRShen-OrrSItzkovitzSKashtanNChklovskiiD 2002 Network motifs: Simple building blocks of complex networks. Science 298 824 827
35. BoyerLALeeTIColeMFJohnstoneSELevineSS 2005 Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122 947 956
36. SwiersGPatientRLooseM 2006 Genetic regulatory networks programming hematopoietic stem cells and erythroid lineage specification. Developmental Biology 294 525 540
37. O'DonnellKAWentzelEAZellerKIDangCVMendellJT 2005 c-Myc-regulated microRNAs modulate E2F1 expression. Nature 435 839 843
38. ManganSAlonU 2003 Structure and function of the feed-forward loop network motif. Proceedings of the National Academy of Sciences of the United States of America 100 11980 11985
39. ManganSItzkovitzSZaslaverAAlonU 2006 The incoherent feed-forward loop accelerates the response-time of the gal system of Escherichia coli. Journal of Molecular Biology 356 1073 1081
40. KaplanSBrenADekelEAlonU 2008 The incoherent feed-forward loop can generate non-monotonic input functions for genes. Molecular Systems Biology 4
41. HornsteinEShomronN 2006 Canalization of development by microRNAs. Nature Genetics 38 S20 S24
42. WuCIShenYTangT 2009 Evolution under canalization and the dual roles of microRNAs-A hypothesis. Genome Research 19 734 743
43. OsellaMBosiaCCoráDCaselleM 2011 The Role of Incoherent MicroRNA-Mediated Feedforward Loops in Noise Buffering. PLoS Comput Biol 7 e1001101 doi:10.1371/journal.pcbi.1001101
44. CilibertiSMartinOCWagnerA 2007 Robustness can evolve gradually in complex regulatory gene networks with varying topology. PLoS Comput Biol 3 e15 doi:10.1371/journal.pcbi.0030015
45. WagnerA 2005 Circuit topology and the evolution of robustness in two-gene circadian oscillators. Proceedings of the National Academy of Sciences of the United States of America 102 11775 11780
46. TsangJZhuJvan OudenaardenA 2007 MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Molecular Cell 26 753 767
47. WaddingtonCH 1959 Canalization of development and genetic assimilation of acquired characters. Nature 183 1654 1655
48. StefaniGSlackFJ 2008 Small non-coding RNAs in animal development. Nature Reviews Molecular Cell Biology 9 219 230
49. KennellJAGerinIMacDougaldOACadiganKM 2008 The microRNA miR-8 is a conserved negative regulator of Wnt signaling. Proceedings of the National Academy of Sciences of the United States of America 105 15417 15422
50. PolisenoLSalmenaLZhangJWCarverBHavemanWJ 2010 A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 465 1033-U1090
51. Nuesslein-VolhardCDahmR 2002 Zebrafish: A practical approach. Zebrafish: A practical approach: i-xviii, 1-303
Štítky
Genetika Reprodukční medicína
Článek Produces a Long Noncoding RNA and Has General Functions in the Germline, Stem Cells, and BrainČlánek Inference of Relationships in Population Data Using Identity-by-Descent and Identity-by-StateČlánek A Genome-Wide Meta-Analysis of Six Type 1 Diabetes Cohorts Identifies Multiple Associated LociČlánek Retrotransposon-Induced Heterochromatin Spreading in the Mouse Revealed by Insertional PolymorphismsČlánek Essential Roles of BCCIP in Mouse Embryonic Development and Structural Stability of ChromosomesČlánek The Receptor Tyrosine Kinase Alk Controls Neurofibromin Functions in Drosophila Growth and LearningČlánek PCNA Ubiquitination Is Important, But Not Essential for Translesion DNA Synthesis in Mammalian CellsČlánek Genome-Wide Association Study Identifies Four Loci Associated with Eruption of Permanent Teeth
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2011 Číslo 9
-
Všechny články tohoto čísla
- Temporal Trends in Results Availability from Genome-Wide Association Studies
- Association of eGFR-Related Loci Identified by GWAS with Incident CKD and ESRD
- Genetic Variants at Chromosomes 2q35, 5p12, 6q25.1, 10q26.13, and 16q12.1 Influence the Risk of Breast Cancer in Men
- Large-Scale Gene-Centric Analysis Identifies Novel Variants for Coronary Artery Disease
- Genetic Association for Renal Traits among Participants of African Ancestry Reveals New Loci for Renal Function
- Transcriptome Kinetics Is Governed by a Genome-Wide Coupling of mRNA Production and Degradation: A Role for RNA Pol II
- Conserved Regulation of p53 Network Dosage by MicroRNA–125b Occurs through Evolving miRNA–Target Gene Pairs
- Heterozygous Mutations of Are Associated with an Increased Risk of Isolated Metopic Craniosynostosis in Humans and Mice
- Study of FoxA Pioneer Factor at Silent Genes Reveals Rfx-Repressed Enhancer at and a Potential Indicator of Esophageal Adenocarcinoma Development
- Cholesterol Metabolism Is Required for Intracellular Hedgehog Signal Transduction
- Produces a Long Noncoding RNA and Has General Functions in the Germline, Stem Cells, and Brain
- Age-Dependent Recombination Rates in Human Pedigrees
- Sequence Conservation and Functional Constraint on Intergenic Spacers in Reduced Genomes of the Obligate Symbiont
- Sex Chromosome Mosaicism and Hybrid Speciation among Tiger Swallowtail Butterflies
- A Negative Feedback Loop That Limits the Ectopic Activation of a Cell Type–Specific Sporulation Sigma Factor of
- Phased Whole-Genome Genetic Risk in a Family Quartet Using a Major Allele Reference Sequence
- Mutations in or near the Transmembrane Domain Alter PMEL Amyloid Formation from Functional to Pathogenic
- Inactivation of Alters Melanosome Shape But Has Only a Subtle Effect on Visible Pigmentation
- Novel Interactions between Actin and the Proteasome Revealed by Complex Haploinsufficiency
- Germline Genetic Variants Disturbing the /LIN28 Double-Negative Feedback Loop Alter Breast Cancer Susceptibility
- Separation of Recombination and SOS Response in RecA Suggests LexA Interaction Sites
- Inference of Relationships in Population Data Using Identity-by-Descent and Identity-by-State
- Misregulation of Scm3p/HJURP Causes Chromosome Instability in and Human Cells
- A Noncoding Point Mutation of Causes Multiple Developmental Malformations and Obesity in Twirler Mice
- The Evolutionarily Conserved Longevity Determinants HCF-1 and SIR-2.1/SIRT1 Collaborate to Regulate DAF-16/FOXO
- A Genome-Wide Metabolic QTL Analysis in Europeans Implicates Two Loci Shaped by Recent Positive Selection
- Bacterial Communities of Diverse Species: Ecological Context of a Host–Microbe Model System
- A Genome-Wide Meta-Analysis of Six Type 1 Diabetes Cohorts Identifies Multiple Associated Loci
- Elongator Complex Influences Telomeric Gene Silencing and DNA Damage Response by Its Role in Wobble Uridine tRNA Modification
- Elevated Proteasome Capacity Extends Replicative Lifespan in
- Genome-Wide Analysis of Heteroduplex DNA in Mismatch Repair–Deficient Yeast Cells Reveals Novel Properties of Meiotic Recombination Pathways
- MicroRNA Predictors of Longevity in
- An Iterative Genetic and Dynamical Modelling Approach Identifies Novel Features of the Gene Regulatory Network Underlying Melanocyte Development
- Atypical AT Skew in Firmicute Genomes Results from Selection and Not from Mutation
- Retrotransposon-Induced Heterochromatin Spreading in the Mouse Revealed by Insertional Polymorphisms
- Genomic Analysis of QTLs and Genes Altering Natural Variation in Stochastic Noise
- The Abnormal Phenotypes of Cartilage and Bone in Calcium-Sensing Receptor Deficient Mice Are Dependent on the Actions of Calcium, Phosphorus, and PTH
- Cell Type–Specific Transcriptome Analysis Reveals a Major Role for and miR-200b in Mouse Inner Ear Morphogenesis
- Essential Roles of BCCIP in Mouse Embryonic Development and Structural Stability of Chromosomes
- IAP1-Mediated Ubiquitylation Controls Activation of the Initiator Caspase DRONC Independent of Protein Degradation
- VANG-1 and PRKL-1 Cooperate to Negatively Regulate Neurite Formation in
- The Receptor Tyrosine Kinase Alk Controls Neurofibromin Functions in Drosophila Growth and Learning
- Comparative and Functional Genomics of PD630 for Biofuels Development
- Identification of Type 1 Diabetes–Associated DNA Methylation Variable Positions That Precede Disease Diagnosis
- PCNA Ubiquitination Is Important, But Not Essential for Translesion DNA Synthesis in Mammalian Cells
- Genetic Effects at Pleiotropic Loci Are Context-Dependent with Consequences for the Maintenance of Genetic Variation in Populations
- Genome-Wide Association Study Identifies Four Loci Associated with Eruption of Permanent Teeth
- Bmp and Nodal Independently Regulate Expression to Maintain Unilateral Nodal Activity during Left-Right Axis Specification in Zebrafish
- Inter-Allelic Prion Propagation Reveals Conformational Relationships among a Multitude of [] Strains
- Emergence and Modular Evolution of a Novel Motility Machinery in Bacteria
- Histone Methyltransferase MET-2 Shields the Male X Chromosome from Checkpoint Machinery and Mediates Meiotic Sex Chromosome Inactivation
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Retrotransposon-Induced Heterochromatin Spreading in the Mouse Revealed by Insertional Polymorphisms
- The Evolutionarily Conserved Longevity Determinants HCF-1 and SIR-2.1/SIRT1 Collaborate to Regulate DAF-16/FOXO
- Genome-Wide Analysis of Heteroduplex DNA in Mismatch Repair–Deficient Yeast Cells Reveals Novel Properties of Meiotic Recombination Pathways
- Association of eGFR-Related Loci Identified by GWAS with Incident CKD and ESRD
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



