-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaLarge-Scale Gene-Centric Analysis Identifies Novel Variants for Coronary Artery Disease
Coronary artery disease (CAD) has a significant genetic contribution that is incompletely characterized. To complement genome-wide association (GWA) studies, we conducted a large and systematic candidate gene study of CAD susceptibility, including analysis of many uncommon and functional variants. We examined 49,094 genetic variants in ∼2,100 genes of cardiovascular relevance, using a customised gene array in 15,596 CAD cases and 34,992 controls (11,202 cases and 30,733 controls of European descent; 4,394 cases and 4,259 controls of South Asian origin). We attempted to replicate putative novel associations in an additional 17,121 CAD cases and 40,473 controls. Potential mechanisms through which the novel variants could affect CAD risk were explored through association tests with vascular risk factors and gene expression. We confirmed associations of several previously known CAD susceptibility loci (eg, 9p21.3:p<10−33; LPA:p<10−19; 1p13.3:p<10−17) as well as three recently discovered loci (COL4A1/COL4A2, ZC3HC1, CYP17A1:p<5×10−7). However, we found essentially null results for most previously suggested CAD candidate genes. In our replication study of 24 promising common variants, we identified novel associations of variants in or near LIPA, IL5, TRIB1, and ABCG5/ABCG8, with per-allele odds ratios for CAD risk with each of the novel variants ranging from 1.06–1.09. Associations with variants at LIPA, TRIB1, and ABCG5/ABCG8 were supported by gene expression data or effects on lipid levels. Apart from the previously reported variants in LPA, none of the other ∼4,500 low frequency and functional variants showed a strong effect. Associations in South Asians did not differ appreciably from those in Europeans, except for 9p21.3 (per-allele odds ratio: 1.14 versus 1.27 respectively; P for heterogeneity = 0.003). This large-scale gene-centric analysis has identified several novel genes for CAD that relate to diverse biochemical and cellular functions and clarified the literature with regard to many previously suggested genes.
Published in the journal: . PLoS Genet 7(9): e32767. doi:10.1371/journal.pgen.1002260
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002260Summary
Coronary artery disease (CAD) has a significant genetic contribution that is incompletely characterized. To complement genome-wide association (GWA) studies, we conducted a large and systematic candidate gene study of CAD susceptibility, including analysis of many uncommon and functional variants. We examined 49,094 genetic variants in ∼2,100 genes of cardiovascular relevance, using a customised gene array in 15,596 CAD cases and 34,992 controls (11,202 cases and 30,733 controls of European descent; 4,394 cases and 4,259 controls of South Asian origin). We attempted to replicate putative novel associations in an additional 17,121 CAD cases and 40,473 controls. Potential mechanisms through which the novel variants could affect CAD risk were explored through association tests with vascular risk factors and gene expression. We confirmed associations of several previously known CAD susceptibility loci (eg, 9p21.3:p<10−33; LPA:p<10−19; 1p13.3:p<10−17) as well as three recently discovered loci (COL4A1/COL4A2, ZC3HC1, CYP17A1:p<5×10−7). However, we found essentially null results for most previously suggested CAD candidate genes. In our replication study of 24 promising common variants, we identified novel associations of variants in or near LIPA, IL5, TRIB1, and ABCG5/ABCG8, with per-allele odds ratios for CAD risk with each of the novel variants ranging from 1.06–1.09. Associations with variants at LIPA, TRIB1, and ABCG5/ABCG8 were supported by gene expression data or effects on lipid levels. Apart from the previously reported variants in LPA, none of the other ∼4,500 low frequency and functional variants showed a strong effect. Associations in South Asians did not differ appreciably from those in Europeans, except for 9p21.3 (per-allele odds ratio: 1.14 versus 1.27 respectively; P for heterogeneity = 0.003). This large-scale gene-centric analysis has identified several novel genes for CAD that relate to diverse biochemical and cellular functions and clarified the literature with regard to many previously suggested genes.
Introduction
Coronary artery disease (CAD) has a substantial genetic component which is incompletely characterised. Genomewide association (GWA) studies have recently identified several novel susceptibility loci for CAD [1]–[9]. Because GWA studies involve assumption-free surveys of common genetic variation across the genome, they can identify genetic regions responsible for previously unsuspected or unknown disease mechanisms. However, despite the success of the GWA approach, it has potential limitations. Because CAD loci identified through GWA studies have predominantly been found in regions of uncertain biological relevance, further work is required to determine their precise contribution to disease aetiology. Furthermore, in contrast with their high coverage of common genetic variation, GWA studies tend to provide limited coverage of genes with well-characterised biological relevance (“candidate genes”) [2], particularly in relation to lower frequency genetic variants (such as those with minor allele frequencies of 1–5%). Such variants are also often difficult to impute from GWA data. Although candidate gene studies should provide more comprehensive coverage of lower frequency and functional variants than GWA studies, most have been inadequately powered.
To complement GWA studies, we undertook a large-scale gene-centric analysis of CAD using a customised gene array enriched with common and low frequency variants in ∼2,100 candidate cardiovascular genes reflecting a wide variety of biological pathways [10]. The array's potential to identify disease-associated lower frequency variants has been demonstrated by previous identification of strong independent associations with 2 variants in the LPA gene - rs3798220 (minor allele frequency 2%), and rs10455872 (7%) - and CAD risk [11]. We have now investigated this gene array in a further 13 studies comprising a total of 15,596 CAD cases and 34,992 controls. To enable interethnic comparisons, participants included 4,394 cases and 4,259 controls of South Asian descent, an ethnic group with high susceptibility to CAD. For further evaluation of putative novel associations, we attempted to replicate them in an additional 17,121 cases and 40,473 controls.
Results
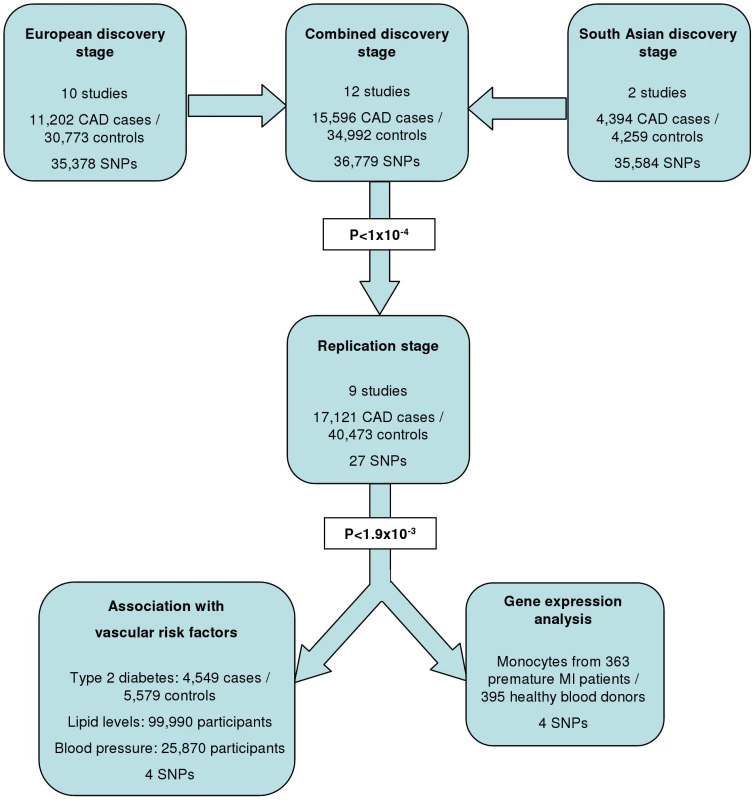
The experimental strategy used is shown in Figure 1. In the discovery phase we genotyped participants from 12 association studies of CAD/myocardial infarction (MI), including a total of 11,202 cases and 30,733 controls of European descent (10 studies), plus 4,394 South Asian cases and 4,259 South Asian controls (2 studies) (Table 1, Table S1, with further details of the studies given in Text S1).
Tab. 1. Summary details of discovery and replication stage studies. 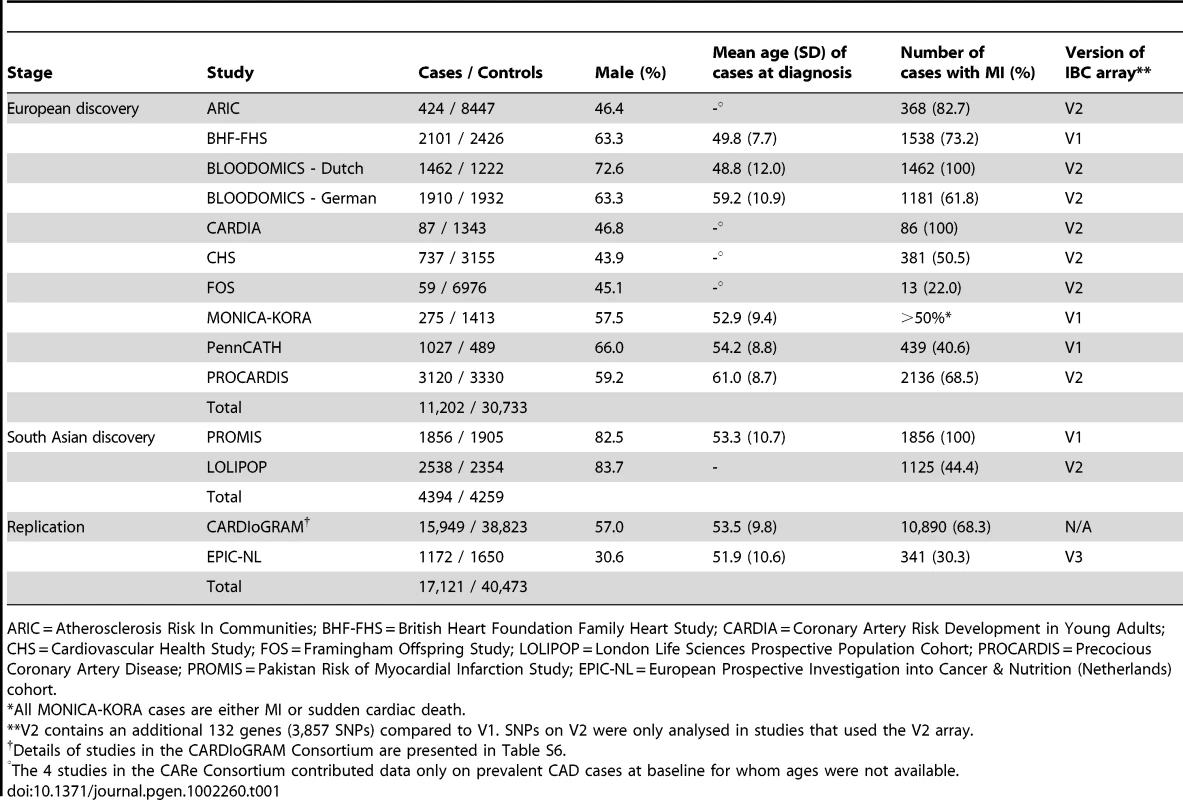
ARIC = Atherosclerosis Risk In Communities; BHF-FHS = British Heart Foundation Family Heart Study; CARDIA = Coronary Artery Risk Development in Young Adults; CHS = Cardiovascular Health Study; FOS = Framingham Offspring Study; LOLIPOP = London Life Sciences Prospective Population Cohort; PROCARDIS = Precocious Coronary Artery Disease; PROMIS = Pakistan Risk of Myocardial Infarction Study; EPIC-NL = European Prospective Investigation into Cancer & Nutrition (Netherlands) cohort. Associations with known CAD loci
36,799 SNPs passed QC and frequency checks and were included in the meta-analysis (reasons for exclusion of variants in each study are given in Table S2). The distribution of association P values in the discovery stage analyses are shown in Figure 2. We found significant associations with CAD for several previous GWA-identified loci contained on the array including 9p21.3 (rs1333042, combined European and South Asian P = 1.1×10−37) and 1p13.3 (rs646776, 3.1×10−17; Table S3). We also confirmed associations of other genes with strong prior evidence including the first association of a variant at the apolipoprotein E locus at genomewide significance (APOE/TOMM40, rs2075650, P = 3.2×10−8), as well as associations at apolipoprotein (a) (LPA, rs10455872, P = 1.2×10−20), and low density lipoprotein receptor (LDLR, rs6511720, P = 1.1×10−8; Table S3). However, we found no persuasive evidence of association of several prominently-studied genes and variants for which the previous epidemiological evidence has been inconclusive, even though the majority of these loci were well-tagged (Table S4) and the current study was well-powered to detect associations of modest effect (Figure S1). Notable variants that did not show significant association included the angiotensin converting enzyme (ACE) insertion/deletion polymorphism, the cholesteryl-ester transfer protein (CETP) Taq1B polymorphism and the paraoxonase 1 (PON1) Q192R polymorphism (Table S4). Perhaps contrary to expectation, apart from the LPA variant rs3798220, we did not observe any other strong association (odds ratio >1.5) among the ∼4,500 low frequency (1–5%) variants and/or variants with suspected or known functional impact on protein structure/function or gene expression specifically selected for the inclusion on the array (Table S3).
Fig. 2. Manhattan plots for discovery stage meta-analyses. 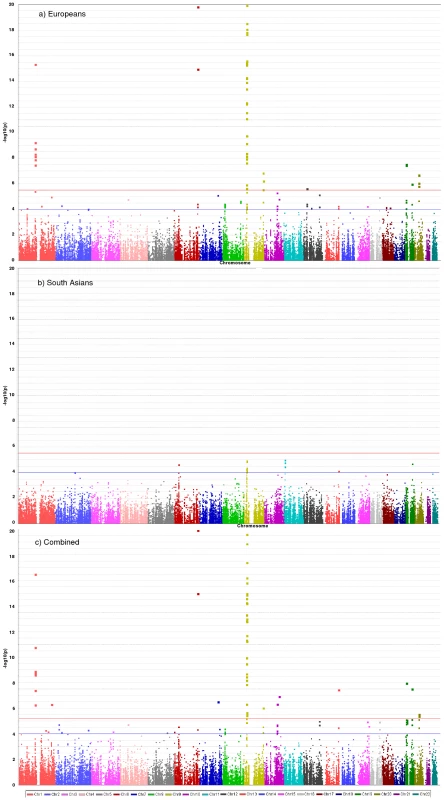
Y-axis shows unadjusted −log10(P values) from fixed-effect meta-analysis of discovery stage studies. NB: European and Combined plots are truncated at P = 10−20. Blue horizontal line at P = 10−4 indicates threshold for replication; Red horizontal line at P = 3×10−6 indicates array-wide significance level. Novel CAD loci
Based on simulations conducted prior to the analysis (Figure S2), loci were eligible for replication if unadjusted P-values for CAD were <1×10−4 in either the primary (each ethnic group analysed separately) or secondary (combined) analyses and the loci had not been previously established with CAD. This identified 27 loci in total: 15 in the European only analysis, 3 in the South Asian only analysis, and 9 in the combined analysis (Table S5). A recent GWA meta-analysis from the CARDIoGRAM Consortium with some overlapping cohorts to those in our study, reports discovery of three of these loci [12]: COL4A1/COL4A2, ZC3HC1, CYP17A1. The P values observed for the lead variants at these loci in the current study were: COL4A1/COL4A2: rs4773144, P = 3.5×10−8; ZC3HC1: rs11556924, P = 3.1×10−7; CYP17A1: rs3824755, P = 1.2×10−7, providing further strong evidence for the association of these loci with CAD. Hence, only the lead SNPs at the 24 remaining loci were taken forward for replication. This was done in silico in 17,121 CAD cases and 40,473 controls, all of whom were of white European ancestry and derived from non-overlapping cohorts from CARDIoGRAM and EPIC-NL (Text S1, Table S6). The power of our replication sample to confirm significant associations is shown in Figure S1. Of the 24 variants taken forward, four were independently replicated (1-tailed Bonferroni-corrected P<0.05 is P<1.9×10−3; Figure 3, Table S5), comprising variants in or adjacent to: LIPA, IL5, TRIB1 and ABCG5/ABCG8 (Figure 4). For the variant at the LIPA locus, the combined P-value was 4.3×10−9, exceeding conventional thresholds for GWA studies. For each of the IL5, TRIB1 and ABCG5/ABCG8 variants, the P-value was <3×10−6, exceeding array-wide levels of significance (Figure 3). CAD associations in the individual component studies are shown in Figure S3. The CAD associations for these loci did not vary materially by age, sex or when restricted to the MI subphenotype (Figure S4).
Fig. 3. Novel loci identified in the current study. 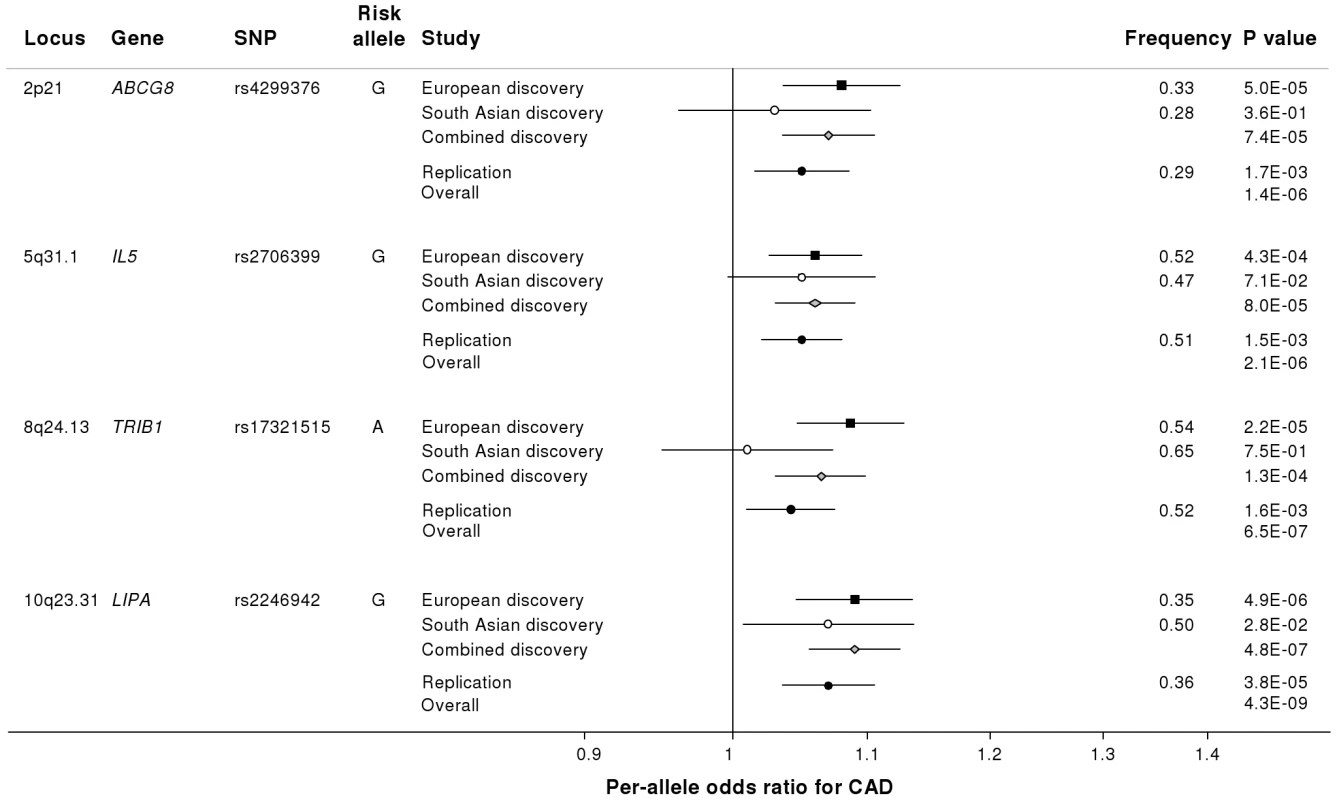
Loci ordered by chromosomal position. SNP = SNP showing strongest evidence of association in discovery stage studies; Frequency = pooled frequency of risk allele across controls; European discovery = per-allele odds ratio, confidence interval and 2-tailed P value from fixed-effect meta-analysis of European discovery stage studies; South Asian discovery = per-allele odds ratio, confidence interval and 2-tailed P value from fixed-effect meta-analysis of South Asian discovery stage studies; Combined discovery = per-allele odds ratio, confidence interval and 2-tailed P value from fixed-effect meta-analysis of all European and South Asian discovery stage studies combined; Replication = per-allele odds ratio, confidence interval and 1-tailed P value from fixed-effect meta-analysis of replication stage studies comprising non-overlapping participants from CARDIoGRAM plus all participants from EPIC-NL; Overall = P value from relevant discovery stage studies combined with the replication stage P value using Fisher's method. Fig. 4. Regional association plots for novel loci identified. 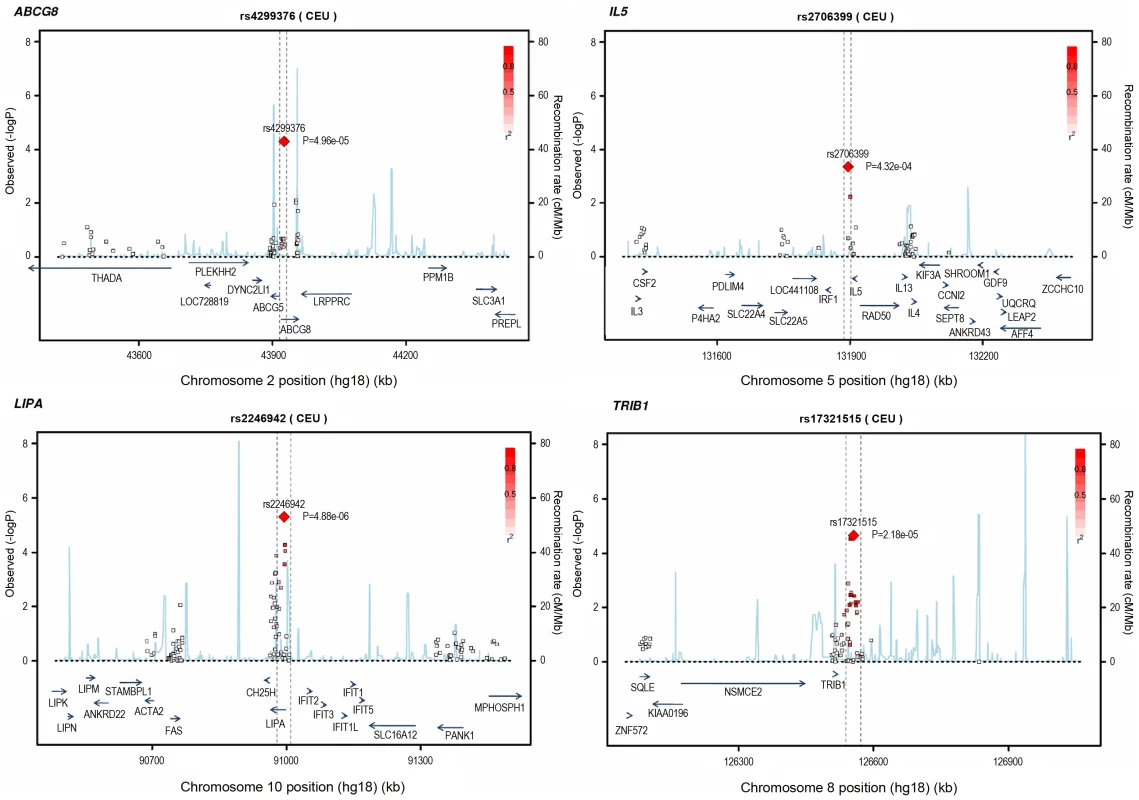
All SNPs included in meta-analysis of the European discovery stage studies are represented by diamonds, with the lead SNP (lowest P value) at each locus represented by a large red diamond. Genes are represented as horizontal arrows, with the direction of the arrow reflecting the direction of transcription. Recombination rates are represented as vertical blue peaks based on the Hapmap 2 CEU population. P values are from fixed-effect meta-analysis. LD, represented as r2, is estimated using the controls from the BHF-FHS study, or Hapmap 2 CEU population where data were not available in BHF-FHS. Vertical dashed lines represent the extent of LD with the lead SNP, based on an r2 threshold of 0.5 in the Hapmap 2 CEU population. The genes between these lines represent the most likely candidate genes for each association signal. Potential mechanisms
To investigate whether the 4 newly identified loci associate with cardiovascular risk traits, we interrogated available data from previous GWA meta-analyses of diabetes mellitus (n = 10,128 individuals) [13], systolic blood pressure (n = 25,870) [14], and low-density (LDL) and high-density (HDL) lipoprotein-cholesterol and triglycerides (n = 99,900) [15]. This showed that the risk allele at the TRIB1 locus was associated with higher triglyceride (P = 3.2×10−53), higher LDL-C (P = 6.7×10−29) and lower HDL-C (P = 9.9×10−17) and that the ABCG5/ABCG8 risk allele was associated with higher LDL-C (P = 1.7×10−47; Figure 5). We also examined the association of the novel risk variants with gene expression in full transcriptomic profiles of circulating monocytes derived from 363 patients with premature myocardial infarction and 395 healthy blood donors from the Cardiogenics study (Text S1). We found a highly significant association (P = 1.0×10−124) of the risk allele at the LIPA locus with LIPA mRNA levels in these cells explaining ∼50% of the variance in the expression of the gene (Figure 6).There were no other highly significant associations between CAD risk alleles and gene expression at the novel loci (Table S7a and S7b).
Fig. 5. Effects of novel CAD loci on known cardiovascular risk factors. 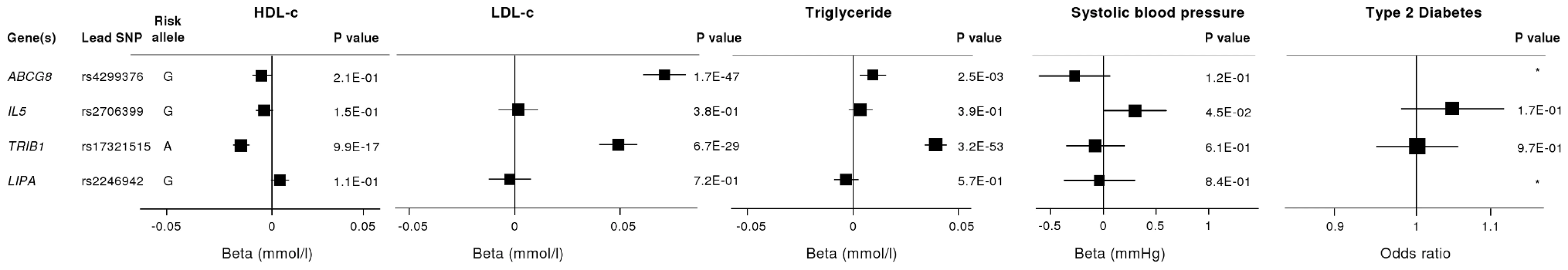
HDL-c = high-density lipoprotein cholesterol; LDL-c = low-density lipoprotein cholesterol; Beta/odds ratio = combined effect from meta-analysis of SNP versus blood pressure/lipids/T2D. Results for lipids from meta-analysis of 46 GWA studies containing up to 99,900 individuals [15]. Results for blood pressure from the Global BPGen Consortium: a meta-analysis of 17 GWA studies containing 25,870 individuals [14]. Results for diabetes from the DIAGRAM Consortium: a meta-analysis of 3 GWA studies containing 4,549 T2DM cases and 5,579 controls [13]. * No results available due to poor quality of SNP imputation. Fig. 6. Evidence for an eQTL association in the LIPA gene. 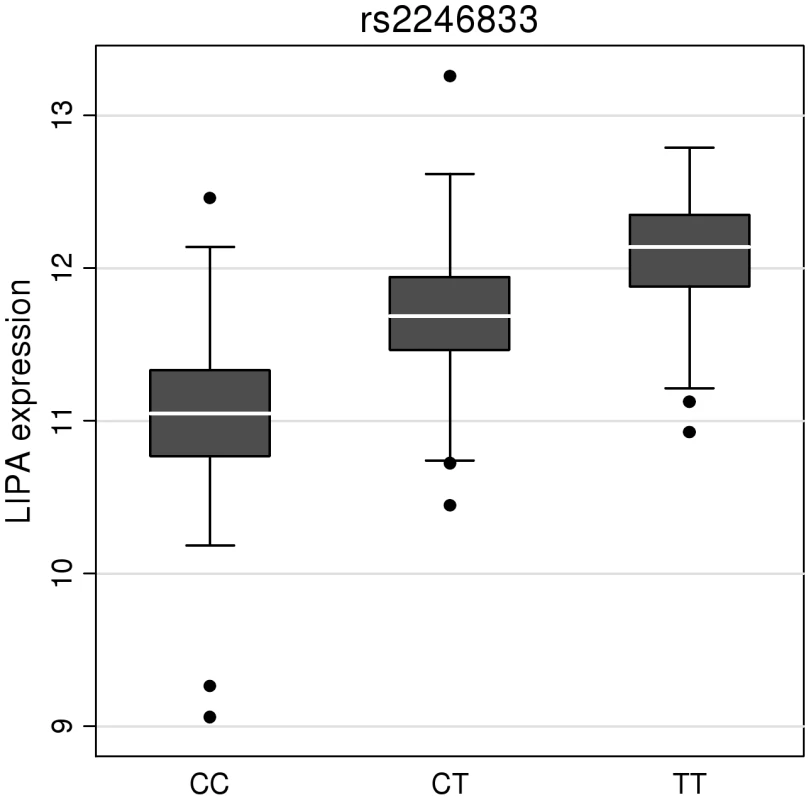
Expression levels of LIPA in monocytes taken from 758 individuals assembled by the Cardiogenics Consortium partitioned by genotype of SNP rs2246833. Boxes indicate interquartile ranges with a white horizontal line indicating the median. Error bars represent absolute minimum and maximum levels with dots showing those levels considered to be outliers. rs2246833 is in strong linkage disequilibrium (r2 = 0.93; D′ = 1) with the CAD-associated variant at the LIPA locus (rs2246942). The T allele, which is associated with increased LIPA expression, is inherited with the G allele of rs2246942, which is associated with increased risk of coronary disease. Ethnic-specific analyses
We explored whether associations of loci with CAD differed between individuals of white European ancestry and South Asian ancestry. For most loci, frequency of risk alleles and pattern of risk associations did not differ qualitatively by ethnicity, although the evidence of association was often weaker in South Asians, perhaps due to lower power (Figure 3, Tables S3 and S5). For the 9p21.3 locus, despite similar risk allele frequencies (Table S3), odds ratios were higher in Europeans than South Asians (rs1333042 : 1.27 v 1.14; P = 0.003 for difference), though common haplotype frequencies did not vary by ethnicity (Table S8). The three variants at the TUB, LCT and MICB loci selected for replication on the basis of South Asian-specific results did not show evidence of association in Europeans (Table S5).
Discussion
Our in-depth study of ∼2,100 candidate genes has yielded several novel and potentially important findings, adding to the emerging knowledge on the genetic determination of CAD. First, we have identified several novel genes for CAD. These genes relate to diverse biochemical and cellular functions: LIPA for the locus on 10q23.3; IL5 (5q31.1); ABCG5/ABCG8 (2p21); TRIB1 (8q24.13); COL4A1/COL4A2 (13q34); Z3HC1 (7q32.3); and CYP17A1 (10q24.3). We have furnished evidence directly implicating the candidacy of these genes, either because the locations of the signals discovered are within a narrow window of linkage disequilibrium or because there is evidence of a mechanistic effect, or both. Second, we have provided large-scale refutation of the relevance of many prominent candidate gene hypotheses in CAD, thereby clarifying the literature. Third, contrary to expectation, we did not observe highly significant novel associations between low frequency variants and CAD risk, despite study of >4,500 such variants. Fourth, we have confirmed the relevance of several previously established CAD genes to both Europeans and South Asians, without finding qualitative differences in results by ethnicity.
LIPA (lipase A) encodes a lysosomal acid lipase involved in the breakdown of cholesteryl esters and triglycerides. Mutations in LIPA cause Wolman's disease [16], a rare disorder characterized by accumulation of these lipids in multiple organs. However, despite evidence that the risk allele was associated with higher LIPA gene expression (suggesting that both under - and over-activity of LIPA increase CAD risk), it was not significantly associated with altered lipid levels. This finding suggests that the impact on CAD risk is either through an alternative pathway, or that the mechanism is more complex than reflected through conventionally measured plasma lipid levels. Two recent studies have also found associations of variants in the LIPA gene with CAD using a GWA approach, strengthening the evidence for this association [17], [18].
Our identification of the association of variants near interleukin 5 (IL5), an interleukin produced by T helper-2 cells, is interesting given the evidence that both acute and chronic inflammation may play important roles in the development and progression of CAD [19]. Most previous human association studies of inflammatory genes and CAD have focused on other cytokines and acute-phase reactants. Nevertheless, some experimental data predict that IL-5 has an atheroprotective effect and this has been supported by association between higher circulating IL-5 levels and lower carotid intimal-medial thickness [20]–[22]. Our findings now highlight the potential importance of IL-5 in CAD, especially as the IL-5 receptor is already a viable therapeutic target in allergic diseases, although we can not rule out the possibility that another gene at this locus may be mediating the association with CAD risk.
The ATP-binding cassette sub-family G proteins ABCG5 and ABCG8 are hemi-transporters that limit intestinal absorption and promote biliary excretion of sterols. Mutations in either gene are associated with sitosterolaemia, accumulation of dietary cholesterol and premature atherosclerosis [23]. Recently, common variants in ABCG8 have also been shown to be associated with circulating LDL-C and altered serum phytosterol levels with concordant changes in risk of CAD [15], [24]. Our findings confirm that this locus affects CAD risk either directly through its effect on plasma phytosterol levels or through primary/secondary changes in LDL-cholesterol.
The association signal on 8q24.13 maps near the TRIB1 gene which encodes the Tribbles homolog 1 protein. Tribbles are a family of phosphoproteins implicated in regulation of cell function, although their precise roles are unclear [25]. However, SNPs in or near TRIB1 - including the lead SNP in our study (rs17321515) - have recently been shown to have highly significant associations with levels of several major lipids [15], providing a possible mechanism for their association with CAD. Our findings confirm the previous suggestion that this variant is also associated with CAD risk [15], [26]. Hepatic over-expression of TRIB1 in mice has been shown to lower circulating triglycerides; conversely, targeted deletion of the TRIB1 gene in mice led to higher circulating triglycerides [27]. The location of the CAD-associated variant downstream of TRIB1 suggests that its effect may be mediated by regulation of TRIB1 expression leading to adverse lipid profiles, although we did not find an eQTL at this locus in monocytes.
Our study brings to 33 the number of confirmed loci with common variants affecting risk of CAD (Figure 7). We estimate that in aggregate these variants explain about 9% of the heritability of CAD which is consistent with the recent analysis by CARDIoGRAM [12]. Interestingly, the odds ratios that we observed for the novel loci were generally lower than those of previously identified loci. This suggests that most of the common variants with moderate effects have been identified and that increasingly larger sample sizes will be required to detect further common variants that affect risk of CAD. However, the modest odds ratios associated with such variants do not necessarily imply that they are not of potential clinical or therapeutic relevance. For example, there are only modest effects of common variants in the LDLR gene on CAD risk (Figure 7); yet this pathway has become a major target for the prevention and treatment of CAD with the development of statins.
Fig. 7. Novel loci identified in this study placed in the context of previously confirmed CAD loci. 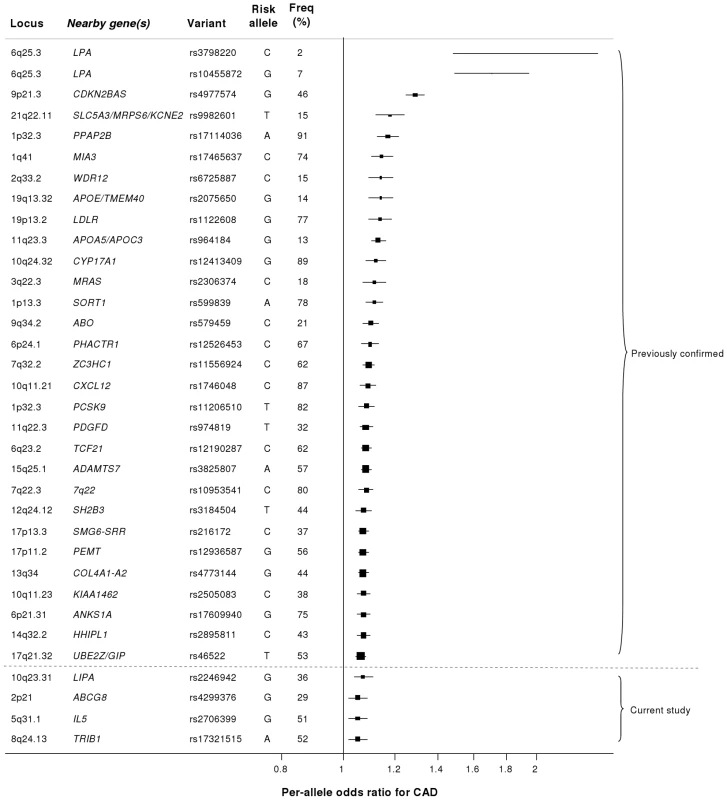
Previously reported variants listed are those from the NHGRI GWA studies catalogue [32] reported as having P<5×10−8 with CAD. Per-allele odds ratios and percentage risk allele frequencies (‘Freq’) are those listed in the catalogue. Frequencies and per-allele odds ratios for the novel variants reported in this study (appearing below the dashed line) are from the CARDIoGRAM replication stage. Despite the success of the GWA approach in identifying several common variants that affect risk of CAD, such loci explain only a small proportion of the heritability of CAD [5]. It has been hypothesized that some of the unexplained heritability resides in lower frequency (1–5%) variants which are not adequately represented on current genomewide arrays and/or are difficult to impute from GWA data. Because the gene array used in the current study included ∼4500 lower frequency variants as well as known functional variants for the majority of the genes on the array, we were able to examine this issue for CAD, at least in relation to candidate cardiovascular genes. Although we confirmed the previously reported associations of lower frequency variants in LPA and PCSK9 with CAD risk, we did not detect any other strongly associated variants in the 1–5% range or an enrichment of low frequency variants amongst SNPs that showed nominal association with CAD. However, it is important to note that rare variants in the genome (minor allele frequency <1%) were not addressed in this study.
CAD is more common in South Asians and tends to occur at an earlier age than in Europeans, perhaps partly due to genetic factors [28]. Our study provides the first systematic exploration of this issue. We observed a weaker effect size for the 9p21.3 locus in South Asians compared with Europeans, although this did not appear to be related to any obvious differences in haplotype structure at the locus, confirming recent findings in Pakistanis [29]. This difference in effect size between ethnic groups will require further evaluation and replication as other differences between the European and South Asian studies (eg, different sex distributions) could explain this finding. Most of the other disease-associated variants we found had slightly weaker effects in South Asians, although, because power to detect heterogeneity of effect between the ethnicities was low and there were only 2 South Asian studies, this finding will require further evaluation. We observed variants at 3 loci (TUB, LCT and MICB, Table S5) which showed modest (P<10−4) associations in South Asians but were convincingly null in Europeans and will therefore require replication in additional South Asian samples. Overall, we did not find clear evidence of major variation in genetic risk factors for CAD between Europeans and South Asians.
In summary, using a large-scale gene-centric approach we have identified novel associations of several genes for CAD that relate to diverse biochemical and cellular functions, including inflammation and novel lipid pathways, as well as genes of less certain function. Together, these findings indicate that previously unsuspected biological mechanisms operate in CAD, raising prospects for novel approaches to intervention.
Materials and Methods
Participants
Characteristics of the discovery phase studies are summarised in Table 1, Table S1 and the replication studies in Table S6. Further details of all the studies are given in Text S1. All individuals provided informed consent and all studies were approved by local ethics committees.
Genotyping in discovery cohorts
Using the HumanCVD BeadChip array (Illumina), which is also known as the “ITMAT-Broad-CARe” (IBC) 50K array, we genotyped 49,094 single nucleotide polymorphisms (SNPs) in ∼2,100 candidate genes identified in previous studies of cardiovascular disease, pathway-based approaches (including genes related to metabolism, lipids, thrombosis, circulation and inflammation), early access to GWA datasets for CAD, type 2 diabetes, lipids and hypertension, as well as human and mouse gene expression data [10]. Variants in genes suspected to be associated with sleep, lung and blood disease phenotypes were also included, along with SNPs that were related in GWA datasets to rheumatoid arthritis, Crohn's disease and type 1 diabetes. Human and mouse gene expression data was also used to select variants. Genes were then prioritised by investigators, with ‘high priority genes’ densely tagged (all SNPs with MAF>2% tagged at r2>0.8), ‘intermediate priority genes’ moderately covered (all SNPs with MAF>5% tagged at r2>0.5), and ‘low priority genes’ tagged using only non-synonymous SNPs and known functional variants with MAF>1%.
A “cosmopolitan tagging” approach was used to select SNPs providing high coverage of selected genes in 4 HapMap populations (CEPH Caucasians, Han Chinese, Japanese, Yorubans). For all genes, non-synonymous SNPs and known functional variants were prioritised on the array. Genotypes were called using standard algorithms (eg, GenCall Software and Illuminus) and standard quality control methods were applied to filter out poorly performing or rare (<1% minor allele frequency) SNPs (Text S1). After exclusion of low frequency variants (average 8,354 in each study), non-autosomal variants (average 1,224) and variants that failed quality control (average 842 – predominantly due to high missingness or failure of HWE), the number of SNPs taken forward for analysis in each study ranged from 30,550–39,027 (Table S2).
Statistical analysis
In each study, unadjusted logistic regression tests using a case-control design assuming an additive genetic model were conducted, with most studies using PLINK [30]. All studies made attempts to reduce over-dispersion. The genomic inflation factor for each study after adjustment was <1.10 with one exception (Table S2). The primary analysis was a fixed-effect inverse-variance-weighted meta-analysis performed separately for each ethnic group using STATA v11. A chi-squared test for between-ethnicity heterogeneity was performed. A secondary analysis combined European and South Asian studies to identify additional variants common to both ethnicities (Text S1).
Replication
Based on a simulation study conducted prior to the analysis (Figure S2), variants were selected for the replication stage if they had an unadjusted P<1×10−4 in either the primary analysis or the combined ethnicity analysis. Only the most significant (“lead”) SNP from each locus was taken forward for replication. SNPs at known coronary disease risk loci (eg, 9p21.3, LPA, APOE) were excluded from the replication stage, leaving 27 SNPs to take forward. In silico replication was conducted using non-overlapping participants from the CARDIoGRAM GWA meta-analysis [12] of CAD plus an additional study, EPIC-NL [31] (details in Table S6). In total, the replication stage comprised up to 17,121 coronary disease cases and 40,473 controls. The threshold for independent replication was a 1-tailed Bonferroni-corrected P<0.05 (P<1.9×10−3) from a Cochran-Armitage trend test. P values from the replication and discovery stages were combined using Fisher's method with a chip-wide value of P<3×10−6 considered to be statistically significant based on the simulation study (Figure S2). Adjusted P values accounting for both over-dispersion and heterogeneity in the discovery stage studies were also estimated through correction for study - and meta-analysis-specific inflation factors.
Additional analyses
To check for consistency of effect of variants that replicated, subgroup analyses were performed in the discovery stage studies for MI cases only, CAD cases aged less than 50, males only and females only. Replicating SNPs were tested for association with known cardiovascular risk factors such as blood pressure, lipids levels and type 2 diabetes mellitus using existing large-scale GWA meta-analyses data of these traits [13]–[15]. We also assessed the association of these variants with gene expression in circulating monocytes taken from 363 patients with premature myocardial infarction and 395 healthy blood donors (Text S1). To put novel findings from this study in the context of existing knowledge, we summarised associations of common variants established in CAD (P<5×10−8) using available information from the NHGRI's GWA studies catalogue [32].
Supporting Information
Zdroje
1. Wellcome Trust Case Control Consortium 2007 Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447 661 678
2. SamaniNJErdmannJHallASHengstenbergCManginoM 2007 Genomewide association analysis of coronary artery disease. N Engl J Med 357 443 453
3. HelgadottirAThorleifssonGManolescuAGretarsdottirSBlondalT 2007 A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science 316 1491 1493
4. McPhersonRPertsemlidisAKavaslarNStewartARobertsR 2007 A common allele on chromosome 9 associated with coronary heart disease. Science 316 1488 1491
5. KathiresanSVoightBFPurcellSMusunuruKArdissinoD 2009 Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat Genet 41 334 341
6. ErdmannJGrosshennigABraundPSKönigIRHengstenbergC 2009 New susceptibility locus for coronary artery disease on chromosome 3q22.3. Nat Genet 41 280 282
7. TregouetDAKönigIRErdmannJMunteanuABraundPS 2009 Genome-wide haplotype association study identifies the SLC22A3-LPAL2-LPA gene cluster as a risk locus for coronary artery disease. Nat Genet 41 283 285
8. ErdmannJWillenborgCNahrstaedtJPreussMKönigIR 2011 Genome-wide association study identifies a new locus for coronary artery disease on chromosome 10p11.23. Eur Heart J 32 158 168
9. ReillyMPLiMHeJFergusonJFStylianouIM 2011 Identification of ADAMTS7 as a novel locus for coronary atherosclerosis and association of ABO with myocardial infarction in the presence of coronary atherosclerosis: two genome-wide association studies. Lancet 377 383 392
10. KeatingBJTischfieldSMurraySSBhangaleTPriceTS 2008 Concept, design and implementation of a cardiovascular gene-centric 50 k SNP array for large-scale genomic association studies. PLoS ONE 3 e3583 doi:10.1371/journal.pone.0003583
11. ClarkeRPedenJFHopewellJCKyriakouTGoelA 2009 Genetic variants associated with Lp(a) lipoprotein level and coronary disease. N Engl J Med 361 2518 2528
12. SchunkertHKönigIRKathiresanSReillyMPAssimesTL 2011 Large-scale association analyses identifies 13 new susceptibility loci for coronary artery disease. Nat Genet 43 333 338
13. ZegginiEScottLJSaxenaRVoightBFMarchiniJL 2008 Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet 40 638 645
14. Newton-ChehCJohnsonTGatevaVTobinMDBochudM 2009 Genome-wide association study identifies eight loci associated with blood pressure. Nat Genet 41 666 676
15. TeslovichTMMusunuruKSmithAVEdmondsonACStylianouIM 2010 Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466 707 713
16. PisciottaLFresaRBellocchioAPinoEGuidoV 2009 Cholesteryl Ester Storage Disease (CESD) due to novel mutations in the LIPA gene. Mol Genet Metab 97 143 148
17. The Coronary Artery Disease (C4D) Genetics Consortium 2011 A genome-wide association study in Europeans and South Asians reveals five novel loci for coronary disease. Nat Genet 43 339 344
18. WildPSZellerTSchillertASzymczakSSinningCR 2011 A genome-wide association study identifies LIPA as a susceptibility gene for coronary artery disease. Circ Cardiovasc Genet Epub ahead of print
19. HanssonGK 2005 Inflammation, atherosclerosis, and coronary artery disease. N Engl J Med 352 1685 1695
20. BinderCJHartvigsenKChangMKMillerMBroideD 2004 IL-5 links adaptive and natural immunity specific for epitopes of oxidized LDL and protects from atherosclerosis. J Clin Invest 114 427 437
21. SampiMUkkolaOPaivansaloMKesaniemiYABinderCJHorkkoS 2008 Plasma interleukin-5 levels are related to antibodies binding to oxidized low-density lipoprotein and to decreased subclinical atherosclerosis. J Am Coll Cardiol 52 1370 1378
22. TalebSTedguiAMallatZ 2010 Adaptive T cell immune responses and atherogenesis. Curr Opin Pharmacol 10 197 202
23. BergeKETianHGrafGAYuLGrishinNV 2000 Accumulation of dietary cholesterol in sitosterolemia caused by mutations in adjacent ABC transporters. Science 290 1771 1775
24. TeupserDBaberRCeglarekUScholzMIlligT 2010 Genetic regulation of serum phytosterol levels and risk of coronary artery disease. Circ Cardiovasc Genet 3 331 339
25. HegedusZCzibulaAKiss-TothE 2007 Tribbles: a family of kinase-like proteins with potent signalling regulatory function. Cell Signal 19 238 250
26. WaterworthDMRickettsSLSongKChenLZhaoJH 2010 Genetic variants influencing circulating lipid levels and risk of coronary artery disease. Arterioscler Thromb Vasc Biol 30 2264 2276
27. BurkhardtRTohS-ALagorWRBirkelandALevinM 2010 Trib1 is a lipid - and myocardial infarction-associated gene that regulates hepatic lipogenesis and VLDL production in mice. J Clin Invest 120 4410 4414
28. BhopalR 2000 What is the risk of coronary heart disease in South Asians? A review of UK research. J Public Health Med 22 375 385
29. SaleheenDAlexanderMRasheedAWormserDSoranzoN 2010 Association of the 9p21.3 locus with risk of first-ever myocardial infarction in Pakistanis: case-control study in South Asia and updated meta-analysis of Europeans. Arterioscler Thromb Vasc Biol 30 1467 1473
30. PurcellSNealeBTodd-BrownKThomasLFerreiraMA 2007 PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81 559 575
31. BeulensJWMonninkhofEMVerschurenWMvan der SchouwYTSmitJ 2010 Cohort Profile: The EPIC-NL study. Int J Epidemiol 39 1170 1178
32. HindorffLASethupathyPJunkinsHARamosEMMehtaJP 2009 Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A 106 9362 9367
Štítky
Genetika Reprodukční medicína
Článek Produces a Long Noncoding RNA and Has General Functions in the Germline, Stem Cells, and BrainČlánek Inference of Relationships in Population Data Using Identity-by-Descent and Identity-by-StateČlánek A Genome-Wide Meta-Analysis of Six Type 1 Diabetes Cohorts Identifies Multiple Associated LociČlánek Retrotransposon-Induced Heterochromatin Spreading in the Mouse Revealed by Insertional PolymorphismsČlánek Essential Roles of BCCIP in Mouse Embryonic Development and Structural Stability of ChromosomesČlánek The Receptor Tyrosine Kinase Alk Controls Neurofibromin Functions in Drosophila Growth and LearningČlánek PCNA Ubiquitination Is Important, But Not Essential for Translesion DNA Synthesis in Mammalian CellsČlánek Genome-Wide Association Study Identifies Four Loci Associated with Eruption of Permanent Teeth
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2011 Číslo 9
-
Všechny články tohoto čísla
- Temporal Trends in Results Availability from Genome-Wide Association Studies
- Association of eGFR-Related Loci Identified by GWAS with Incident CKD and ESRD
- Genetic Variants at Chromosomes 2q35, 5p12, 6q25.1, 10q26.13, and 16q12.1 Influence the Risk of Breast Cancer in Men
- Large-Scale Gene-Centric Analysis Identifies Novel Variants for Coronary Artery Disease
- Genetic Association for Renal Traits among Participants of African Ancestry Reveals New Loci for Renal Function
- Transcriptome Kinetics Is Governed by a Genome-Wide Coupling of mRNA Production and Degradation: A Role for RNA Pol II
- Conserved Regulation of p53 Network Dosage by MicroRNA–125b Occurs through Evolving miRNA–Target Gene Pairs
- Heterozygous Mutations of Are Associated with an Increased Risk of Isolated Metopic Craniosynostosis in Humans and Mice
- Study of FoxA Pioneer Factor at Silent Genes Reveals Rfx-Repressed Enhancer at and a Potential Indicator of Esophageal Adenocarcinoma Development
- Cholesterol Metabolism Is Required for Intracellular Hedgehog Signal Transduction
- Produces a Long Noncoding RNA and Has General Functions in the Germline, Stem Cells, and Brain
- Age-Dependent Recombination Rates in Human Pedigrees
- Sequence Conservation and Functional Constraint on Intergenic Spacers in Reduced Genomes of the Obligate Symbiont
- Sex Chromosome Mosaicism and Hybrid Speciation among Tiger Swallowtail Butterflies
- A Negative Feedback Loop That Limits the Ectopic Activation of a Cell Type–Specific Sporulation Sigma Factor of
- Phased Whole-Genome Genetic Risk in a Family Quartet Using a Major Allele Reference Sequence
- Mutations in or near the Transmembrane Domain Alter PMEL Amyloid Formation from Functional to Pathogenic
- Inactivation of Alters Melanosome Shape But Has Only a Subtle Effect on Visible Pigmentation
- Novel Interactions between Actin and the Proteasome Revealed by Complex Haploinsufficiency
- Germline Genetic Variants Disturbing the /LIN28 Double-Negative Feedback Loop Alter Breast Cancer Susceptibility
- Separation of Recombination and SOS Response in RecA Suggests LexA Interaction Sites
- Inference of Relationships in Population Data Using Identity-by-Descent and Identity-by-State
- Misregulation of Scm3p/HJURP Causes Chromosome Instability in and Human Cells
- A Noncoding Point Mutation of Causes Multiple Developmental Malformations and Obesity in Twirler Mice
- The Evolutionarily Conserved Longevity Determinants HCF-1 and SIR-2.1/SIRT1 Collaborate to Regulate DAF-16/FOXO
- A Genome-Wide Metabolic QTL Analysis in Europeans Implicates Two Loci Shaped by Recent Positive Selection
- Bacterial Communities of Diverse Species: Ecological Context of a Host–Microbe Model System
- A Genome-Wide Meta-Analysis of Six Type 1 Diabetes Cohorts Identifies Multiple Associated Loci
- Elongator Complex Influences Telomeric Gene Silencing and DNA Damage Response by Its Role in Wobble Uridine tRNA Modification
- Elevated Proteasome Capacity Extends Replicative Lifespan in
- Genome-Wide Analysis of Heteroduplex DNA in Mismatch Repair–Deficient Yeast Cells Reveals Novel Properties of Meiotic Recombination Pathways
- MicroRNA Predictors of Longevity in
- An Iterative Genetic and Dynamical Modelling Approach Identifies Novel Features of the Gene Regulatory Network Underlying Melanocyte Development
- Atypical AT Skew in Firmicute Genomes Results from Selection and Not from Mutation
- Retrotransposon-Induced Heterochromatin Spreading in the Mouse Revealed by Insertional Polymorphisms
- Genomic Analysis of QTLs and Genes Altering Natural Variation in Stochastic Noise
- The Abnormal Phenotypes of Cartilage and Bone in Calcium-Sensing Receptor Deficient Mice Are Dependent on the Actions of Calcium, Phosphorus, and PTH
- Cell Type–Specific Transcriptome Analysis Reveals a Major Role for and miR-200b in Mouse Inner Ear Morphogenesis
- Essential Roles of BCCIP in Mouse Embryonic Development and Structural Stability of Chromosomes
- IAP1-Mediated Ubiquitylation Controls Activation of the Initiator Caspase DRONC Independent of Protein Degradation
- VANG-1 and PRKL-1 Cooperate to Negatively Regulate Neurite Formation in
- The Receptor Tyrosine Kinase Alk Controls Neurofibromin Functions in Drosophila Growth and Learning
- Comparative and Functional Genomics of PD630 for Biofuels Development
- Identification of Type 1 Diabetes–Associated DNA Methylation Variable Positions That Precede Disease Diagnosis
- PCNA Ubiquitination Is Important, But Not Essential for Translesion DNA Synthesis in Mammalian Cells
- Genetic Effects at Pleiotropic Loci Are Context-Dependent with Consequences for the Maintenance of Genetic Variation in Populations
- Genome-Wide Association Study Identifies Four Loci Associated with Eruption of Permanent Teeth
- Bmp and Nodal Independently Regulate Expression to Maintain Unilateral Nodal Activity during Left-Right Axis Specification in Zebrafish
- Inter-Allelic Prion Propagation Reveals Conformational Relationships among a Multitude of [] Strains
- Emergence and Modular Evolution of a Novel Motility Machinery in Bacteria
- Histone Methyltransferase MET-2 Shields the Male X Chromosome from Checkpoint Machinery and Mediates Meiotic Sex Chromosome Inactivation
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Retrotransposon-Induced Heterochromatin Spreading in the Mouse Revealed by Insertional Polymorphisms
- The Evolutionarily Conserved Longevity Determinants HCF-1 and SIR-2.1/SIRT1 Collaborate to Regulate DAF-16/FOXO
- Genome-Wide Analysis of Heteroduplex DNA in Mismatch Repair–Deficient Yeast Cells Reveals Novel Properties of Meiotic Recombination Pathways
- Association of eGFR-Related Loci Identified by GWAS with Incident CKD and ESRD
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání