-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaReduction of Protein Translation and Activation of Autophagy Protect against PINK1 Pathogenesis in
Mutations in PINK1 and Parkin cause familial, early onset Parkinson's disease. In Drosophila melanogaster, PINK1 and Parkin mutants show similar phenotypes, such as swollen and dysfunctional mitochondria, muscle degeneration, energy depletion, and dopaminergic (DA) neuron loss. We previously showed that PINK1 and Parkin genetically interact with the mitochondrial fusion/fission pathway, and PINK1 and Parkin were recently proposed to form a mitochondrial quality control system that involves mitophagy. However, the in vivo relationships among PINK1/Parkin function, mitochondrial fission/fusion, and autophagy remain unclear; and other cellular events critical for PINK1 pathogenesis remain to be identified. Here we show that PINK1 genetically interacted with the protein translation pathway. Enhanced translation through S6K activation significantly exacerbated PINK1 mutant phenotypes, whereas reduction of translation showed suppression. Induction of autophagy by Atg1 overexpression also rescued PINK1 mutant phenotypes, even in the presence of activated S6K. Downregulation of translation and activation of autophagy were already manifested in PINK1 mutant, suggesting that they represent compensatory cellular responses to mitochondrial dysfunction caused by PINK1 inactivation, presumably serving to conserve energy. Interestingly, the enhanced PINK1 mutant phenotype in the presence of activated S6K could be fully rescued by Parkin, apparently in an autophagy-independent manner. Our results reveal complex cellular responses to PINK1 inactivation and suggest novel therapeutic strategies through manipulation of the compensatory responses.
Published in the journal: . PLoS Genet 6(12): e32767. doi:10.1371/journal.pgen.1001237
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1001237Summary
Mutations in PINK1 and Parkin cause familial, early onset Parkinson's disease. In Drosophila melanogaster, PINK1 and Parkin mutants show similar phenotypes, such as swollen and dysfunctional mitochondria, muscle degeneration, energy depletion, and dopaminergic (DA) neuron loss. We previously showed that PINK1 and Parkin genetically interact with the mitochondrial fusion/fission pathway, and PINK1 and Parkin were recently proposed to form a mitochondrial quality control system that involves mitophagy. However, the in vivo relationships among PINK1/Parkin function, mitochondrial fission/fusion, and autophagy remain unclear; and other cellular events critical for PINK1 pathogenesis remain to be identified. Here we show that PINK1 genetically interacted with the protein translation pathway. Enhanced translation through S6K activation significantly exacerbated PINK1 mutant phenotypes, whereas reduction of translation showed suppression. Induction of autophagy by Atg1 overexpression also rescued PINK1 mutant phenotypes, even in the presence of activated S6K. Downregulation of translation and activation of autophagy were already manifested in PINK1 mutant, suggesting that they represent compensatory cellular responses to mitochondrial dysfunction caused by PINK1 inactivation, presumably serving to conserve energy. Interestingly, the enhanced PINK1 mutant phenotype in the presence of activated S6K could be fully rescued by Parkin, apparently in an autophagy-independent manner. Our results reveal complex cellular responses to PINK1 inactivation and suggest novel therapeutic strategies through manipulation of the compensatory responses.
Introduction
Parkinson's disease (PD) is the most common neurodegenerative disease affecting movement and currently there is no cure. A pathological hallmark of PD is the reduction of dopamine content in the brain, caused by the selective dysfunction and degeneration of DA neurons in the substantia nigra. The causes of DA neuron loss are complex and likely involve both environmental insults and genetic predisposition. Increasing evidences suggest that mitochondrial dysfunction may be linked to the pathogenesis of both sporadic and familial forms of PD.
Recent genetic studies of rare familial forms of PD identified multiple disease genes, including PINK1 and Parkin [1], [2]. PINK1 encodes a Ser/Thr kinase with a mitochondrial targeting sequence and is partially localized to the mitochondria [3], [4]. Parkin is an E3 ubiquitin ligase that is largely cytosolic under normal conditions. The inactivation of Drosophila orthologs of PINK1 or Parkin resulted in similar phenotypes, with the formation of enlarged, swollen mitochondria preceding muscle degeneration, DA neuron loss and spermatogenesis failure [5]–[7]. Further analysis showed that overexpression (OE) of Parkin could rescue PINK1 mutant phenotype, but not vice versa, suggesting that PINK1 and Parkin may function in the same pathway, with Parkin acting downstream of PINK1 [5]–[7]. Interestingly, promoting mitochondrial fission by either overexpression of mitochondrial fission protein Drp1 or downregulation of mitochondrial fusion proteins Marf (the D. melanogaster homolog of mammalian mitofusin) or Opa1 could completely rescue PINK1 or Parkin mutant phenotypes, suggesting that PINK1 and Parkin might regulate mitochondrial dynamics by interacting with the mitochondrial fusion/fission machinery [8]–[10].
PINK1 and Parkin have also been suggested to collaborate to form a mitochondrial quality control system [11], [12]. Despite being mainly cytosolic under normal conditions, Parkin can be mobilized to damaged mitochondria that have decreased membrane potential [11]. This translocation of Parkin requires the function of PINK1, which is stabilized and accumulates on damaged mitochondria [12]. Parkin recruited to damaged mitochondria can further ubiquitinate mitochondrial proteins to mark the damaged mitochondria for degradation by autophagy [11], [13], [14]. These studies offered an attractive molecular mechanism linking the inactivation of PINK1 or Parkin to the accumulation of dysfunctional mitochondria. However, most of these studies were carried out in cell culture. Their in vivo relevance remains to be determined.
The target of rapamycin (TOR) protein is an evolutionarily conserved serine/threonine protein kinase that functions as a master regulator of many crucial cellular processes, including protein translation, mRNA transcription, autophagy and cytoskeletal organization [15]. TOR exerts its regulatory function by integrating diverse cues ranging from extracellular growth factors to intracellular levels of ATP, amino acids and oxygen [15]. In response to ATP depletion, for example, TOR signaling is suppressed by the AMP-activated protein kinase (AMPK), leading to subsequent inhibition of S6 kinase (S6K)-mediated protein translation and activation of Atg1-mediated autophagy [16]–[18].
In an effort to further understand the mechanisms of PINK1 and Parkin pathogenesis, we performed genetic screens to find modifiers of PINK1 mutant phenotypes. We identified S6K and Atg1 as strong modifiers of PINK1. We found that activated S6K acts through protein translational regulation to significantly enhance muscle degeneration and DA neuron loss in PINK1 mutant. Together with our previous study of LRRK2 [19], this result supports that impaired translational control is an integral part of PD pathogenesis. We also found that Atg1 OE could suppress PINK1 mutant phenotypes even in the presence of constitutively active S6K, and the rescuing effect of Atg1 OE was dependent on its ability to directly promote autophagy, suggesting that enhancing autophagy represents another efficient way to combat PINK1-related Parkinson's disease. Since reduced S6K activation and enhanced autophagy were already observed in the PINK1 mutant background, the protective effects observed after further strengthening these processes suggest that they represent compensatory responses to PINK1 inactivation. Pharmacological augmentation of these responses thus holds significant therapeutic value.
Results
Gain - and loss-of-function analyses reveal strong genetic interactions between PINK1 and the TOR and autophagy pathways
To better understand PINK1 pathogenesis, we performed both gain-of-function and loss-of-function genetic screens to find genetic enhancers and suppressors of PINK1 mutant phenotypes. PINK1 mutant flies exhibit an easily observable abnormal wing posture (either held-up or drooped) caused by indirect flight muscle degeneration. Therefore, we used the Mhc-Gal4 driver to direct the expression of UAS-PINK1 RNAi transgenes specifically in the muscle and used the penetrance of the abnormal wing posture phenotype as an indicator of genetic interaction in our screens. PINK1 RNAi line, which exhibited weaker wing posture phenotype than the PINK1 null mutant, allowed us to screen for both enhancers and suppressors in the same screens by scoring the percentage of flies exhibiting abnormal wing posture at young and old ages.
In our genetic screens, we first took an unbiased approach by screening a collection of ∼300 EP lines, and based on the information obtained from this unbiased screen we implemented a more targeted approach by screening genes involved in specific pathways or processes. In our unbiased screen, we uncovered ∼30 lines that either enhanced or suppressed PINK1 RNAi phenotype (data not shown). These lines are associated with genes that have diverse functions, suggesting that PINK1 may functionally interact with many different cellular pathways. The strongest modifiers of PINK1 RNAi phenotype were identified in our targeted screen and one of the strongest enhancers of PINK1 RNAi-induced abnormal wing posture is S6 kinase (S6K). S6K is one of the downstream effectors of TOR, a master regulator of cell growth and proliferation. In response to cellular growth stimuli, TOR phosphorylates S6K to upregulate the synthesis of ribosomal proteins and translation initiation and elongation factors [20]. When wild type (WT) S6K was overexpressed in the muscle of PINK1 RNAi flies, the penetrance of the abnormal wing posture phenotype was greatly enhanced in an age-dependent manner (Figure 1A). Even more dramatic enhancement of the abnormal wing posture was observed when S6K-TE, S6K-STDE, or S6K-STDETE, the phosphomimetic, constitutively active forms of S6K [21], were co-expressed (Figure 1A). In these cases, more than 50% of the flies had abnormal wing posture at 1-day old, whereas virtually none of the PINK1 RNAi flies of the same age showed the phenotype. The phenotype became stronger in 14-day-old flies. Overexpression of WT or constitutively active S6Ks in wild type background did not affect wing posture, even after the flies were aged for weeks, suggesting that the effect on wing posture caused by S6K was specific to the PINK1 RNAi background (Figure 1B). On the other hand, when we reduced S6K function through S6K RNAi, it effectively attenuated PINK1 RNAi effects (Figure 1A). Similar genetic interaction between PINK1 and constitutively active S6K was observed in PINKB9 mutant background (Figure S1). This result, together with the observation that S6K OE did not affect PINK1 protein level (Figure S2), suggested that the genetic interaction between PINK1 and S6K was not due to a possible regulation of PINK1 expression by S6K.
Fig. 1. S6K and Atg1 act as genetic modifiers of PINK1 RNAi. 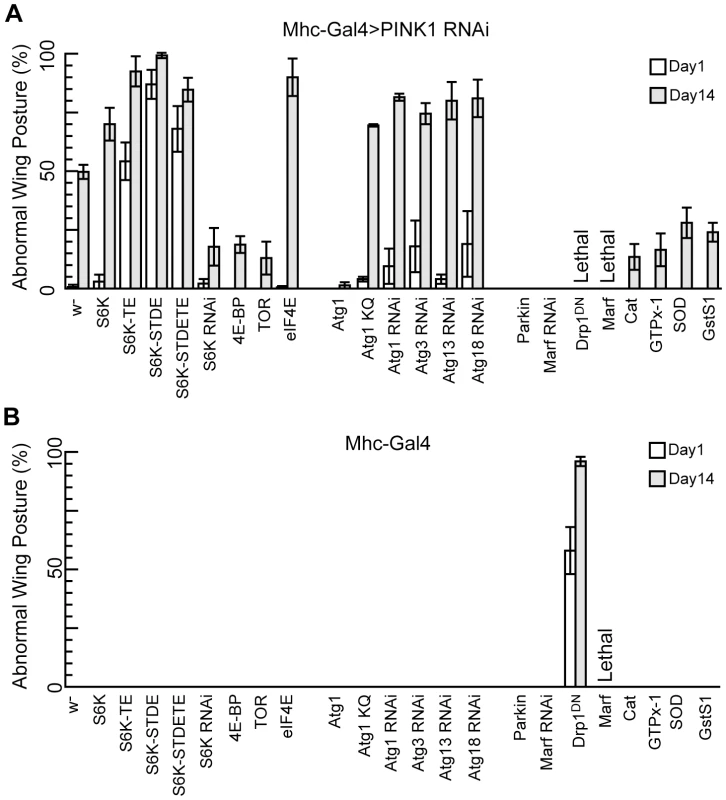
The flies of each indicated genotype were crossed to Mhc-Gal4>PINK1 RNAi flies (A) or Mhc-Gal4 flies (B), and the percentage of male offspring with abnormal wing posture phenotype was scored at 1-day and 14-day after eclosion. The flies were aged at 29°C. Data are presented as mean ± s.e.m. The genetic interactions between PINK1 and genes of the TOR pathway, autophagy pathway, mitochondrial fusion and fission machinery or antioxidant genes are demonstrated. The differences in abnormal wing posture phenotype between the genetic interaction flies and control flies shown in (A) are all statistically significant (P<0.005 in Student's t-test). The stronger effects of the phosphomimetic, constitutively active forms of S6K than WT S6K is consistent with the fact that S6K function is tightly controlled by TOR through ordered phosphorylation of multiple Ser/Thr residues [22], [23]. As S6K is one of the downstream effectors of TOR that regulate protein translation, we further tested whether other components of the TOR pathway that regulate translation also interact with PINK1 genetically. The eukaryotic translation initiation factor 4E (eIF4E)/eIF4E-binding protein (4E-BP) axis of translational control is also regulated by TOR. TOR signaling leads to 4E-BP phosphorylation, weakening its binding to eIF4E and releasing its inhibition on translational initiation [24]. Reducing translation by 4E-BP OE mildly suppressed the abnormal wing posture phenotype in PINK1 RNAi background. Conversely, eIF4E OE greatly enhanced the abnormal wing posture phenotype in aged PINK1 RNAi flies. However, these flies at 1-day old showed relatively normal wing posture, indicating that the effect of eIF4E is milder compared to constitutively active S6K. Consistent with the above genetic interactions, dTOR OE, which phenocopies dTOR loss-of-function effects in Drosophila [25], also suppressed PINK1 RNAi phenotypes. Together, these results support a strong functional interaction between PINK1 and TOR-mediated translational regulation.
In addition to enhancers, we also recovered strong suppressors of PINK1 RNAi phenotypes, with Atg1 being one of them. Atg1 is a kinase that has been suggested to play an essential role in the initiation of autophagy. Previous studies have shown that Atg1 OE in Drosophila fat body was sufficient to induce autophagy, and high level of Atg1 expression could cause growth arrest and apoptosis [26]. When we expressed high level of Atg1 in the muscle using the Mhc-Gal4 driver and strong UAS-Atg1 transgenes, the flies either failed to eclose or showed strong abnormal wing posture, possibly due to excess apoptosis in the flight muscle (data not shown). However, when we used a weaker Atg1 OE line (UAS-Atg1GS10797) [26], the flies enclosed with normal wing posture and flight ability (Figure 1B, data not shown). Interestingly, this mild overexpression of Atg1 completely suppressed the abnormal wing posture caused by PINK1 knockdown (Figure 1A), which was not due to a change in PINK1 expression level (Figure S2). Conversely, PINK1 RNAi phenotypes were exacerbated by the overexpression of Atg1 RNAi or Atg1K38Q, a kinase-dead form of Atg1 that acted in a dominant-negative fashion [26]. Similarly, RNAi-mediated knockdown of Atg3, Atg13 and Atg18, all of which are essential for autophagy in Drosophila [27], [28], also enhanced the abnormal wing posture phenotype in PINK1 RNAi background (Figure 1A). These data indicate that autophagy is physiologically relevant to PINK1 pathogenesis and that mild induction of autophagy protects against PINK1 pathogenesis.
To further validate the effectiveness of our screen, we tested genes that were known to genetically interact with PINK1. We found that Parkin OE and Marf RNAi could completely suppress the PINK1 RNAi phenotypes (Figure 1A), consistent with previous findings [5]–[10]. In contrast to the rescuing effect of Marf RNAi, overexpression of Marf alone in the muscle, which would lead to excessive mitochondrial fusion, caused lethality at third instar larval stage (Figure 1B). Flies overexpressing a dominant-negative form of Drp1 (Drp1DN), which inhibited mitochondrial fission, were viable but showed strong abnormal wing posture phenotype (Figure 1B). When Drp1DN was expressed in PINK1 RNAi background using the Mhc-Gal4 driver, a synthetic lethality phenotype was observed (Figure 1A). These results are consistent with previously reported genetic interactions between PINK1 and the mitochondrial fusion/fission machinery [8]-[10]. In addition, we tested the effects of antioxidant genes, some of which have been shown to rescue PINK1 and Parkin mutant phenotypes [29], [30]. All four antioxidant genes tested, catalase (Cat), glutathione peroxidase homolog with thioredoxin peroxidase activity (GTPx-1), Glutathione S transferase S1 (GstS1), and superoxide dismutase (SOD), could partially rescue the abnormal wing posture in PINK1 RNAi background when overexpressed. However, they were less effective compared to Atg1 and Parkin OE or Marf knockdown (Figure 1A).
Activated S6K enhances muscle degeneration and DA neuron loss in PINK1 deficiency flies
In addition to abnormal wing posture, PINK1 mutant flies typically exhibit enlarged mitochondria, energy depletion, muscle degeneration and DA neuron loss [5], [8]. To better understand the genetic interaction between S6K and PINK1, we tested whether S6K affected these phenotypes as well. In one-day-old flies, Mhc-Gal4-directed co-expression of constitutively active forms of S6Ks (S6K-TE, S6K-STDE and S6K-STDETE) in PINK1 RNAi background completely abolished their flight ability (Figure 2A), significantly decreased ATP level in the muscle (Figure 2B), and dramatically increased thoracic indentation (Figure 2C, 2D), which all indicate increased muscle degeneration. In contrast, overexpression of a S6K RNAi transgene in PINK1 RNAi background partially rescued these phenotypes (Figure 2A, 2B, 2C). Overexpression of constitutively active S6K or S6K RNAi transgenes in wild type flies had no obvious effect in these assays (Figure 2A, 2B), suggesting that their effects on muscle degeneration were specific to the PINK1 RNAi background. We further used transmission electron microscopy (TEM) to examine muscle degeneration in detail. At one-day after eclosion, the thoracic muscle of PINK1 RNAi flies showed only small lesions, while large areas devoid of muscle tissues were observed in the thoraces of PINK1 RNAi flies expressing constitutively active S6K, supporting the conclusion that S6K enhances the muscle degeneration in PINK1 RNAi background (Figure 2E). In comparison, the muscle morphology of flies expressing constitutively active S6K alone appeared indistinguishable from that of wild type flies, with healthy, electron-dense mitochondria laying in between muscle fibers in an organized fashion (Figure 2E).
Fig. 2. Overexpression or knockdown of S6K strongly modifies PINK1 RNAi phenotypes in the muscle. 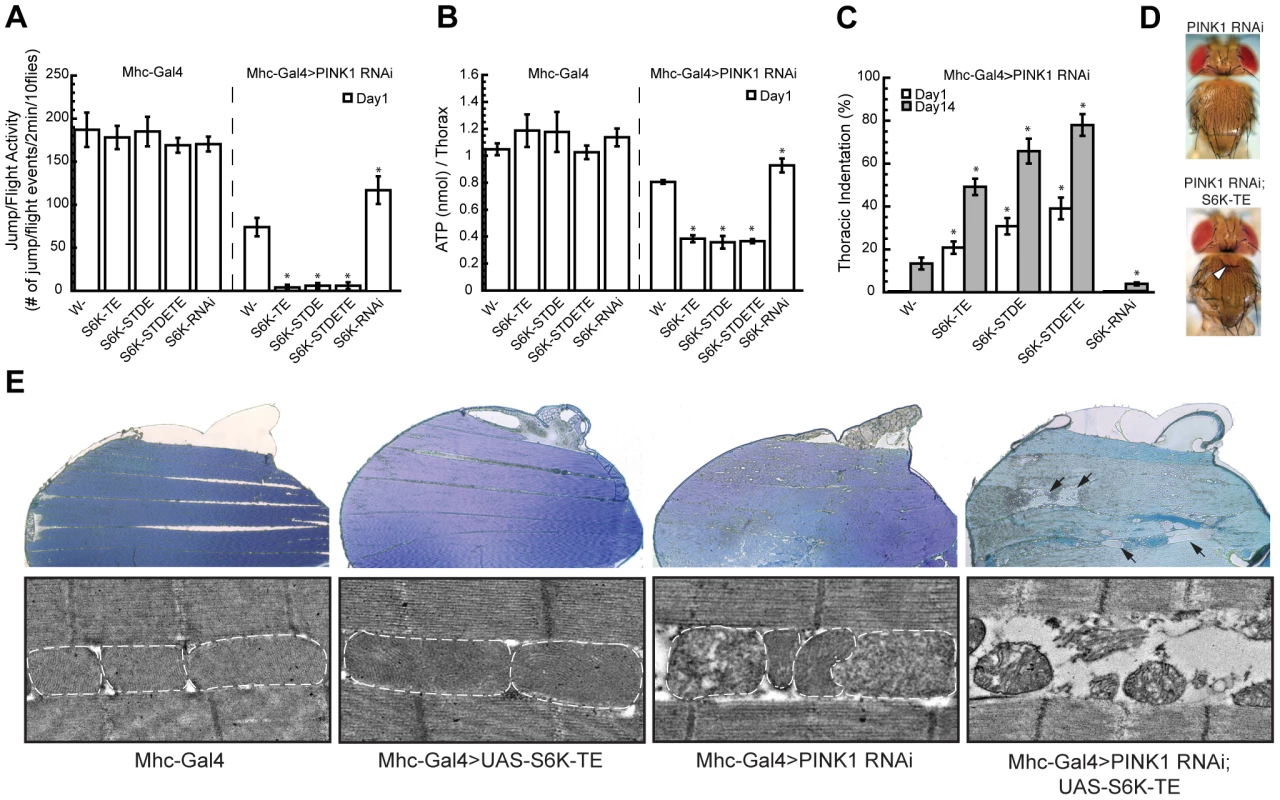
Overexpression of constitutively active S6Ks (S6K-TE, S6K-STDE and S6K-STDETE) in the muscle of PINK1 RNAi flies completely abolished their jump/flight ability (A), significantly decreased their muscle ATP level (B) and dramatically increased their thoracic indentation (C). In contrast, the overexpression of S6K RNAi transgene partially rescued these phenotypes in PINK1 RNAi flies. Open and closed bars represented data scored on Day 1 and Day 14 after eclosion, respectively. Data are presented as mean ± s.e.m. Significance was determined by Student's t test (*P<0.005). (D) Representative image of thoracic indentation (indicated by the white arrowhead) of Mhc-Gal4; PINK1 RNAi; S6K-TE fly at 1-day old (bottom) compared to the normal thoracic phenotype of Mhc-Gal4, PINK1 RNAi fly of the same age (top). (E) Overexpression of constitutively active S6K in PINK1 RNAi flies dramatically increased muscle degeneration. Sections from resin-embedded thoraces of 1-day-old adult flies were either stained with toluidine blue to visualize overall muscle structure (top panel) or directly visualized using TEM for mitochondrial morphology (bottom panel). WT flies or flies expressing constitutively active S6K show normal muscle structure with healthy, electron-dense mitochondria. Flies expressing PINK1 RNAi transgene had small lesions in the muscle with dysfunctional mitochondria showing broken cristae. Co-expression of S6K-TE in PINK1 RNAi flies caused more severe degeneration of mitochondria and muscle fibers, generating large lesions in the muscle that were filled with resin during embedding and are readily recognizable (indicated by black arrows). We further examined mitochondrial morphology in DA neurons using a mitochondrially targeted GFP (mitoGFP) as a marker. When we used the tyrosine hydroxylase (TH)-Gal4 driver to induce the expression of mitoGFP in wild type DA neurons, most mitochondria exhibited tubular-shaped mitochondrial network (Figure 3A). Overexpression of constitutively active S6K-TE did not change the overall mitochondrial shape, although mitochondrial content appeared increased. PINK1 mutant (PINK1B9) showed enlarged mitochondria in DA neurons (Figure 3A), consistent with previous reports [6], [8]. When S6K-TE was expressed in PINK1B9 background, mitochondrial sizes were further increased, frequently doubling those in the PINK1 mutant in diameter (Figure 3A, 3B). As enlarged mitochondria or mitochondrial clusters are hallmarks of PINK1-related parkinsonism, this further increase of mitochondrial size after S6K-TE overexpression, which is possibly the consequence of inefficient mitophagy (see Discussion and Figure S3), suggests exacerbation of the disease process. Consistent with this idea, the expression of S6K-TE in PINK1B9 mutant background also promoted DA neuron death, as the number of DA neurons in the protocerebral posterior lateral 1 (PPL1) cluster was further decreased compared to PINK1B9 mutants in aged flies (Figure 3C). In summary, the overexpression of constitutively active S6K in PINK1 mutants significantly enhanced muscle degeneration and DA neuron loss.
Fig. 3. Constitutively active S6K increases mitochondrial aggregation and DA neuron loss in PINK1 mutants. 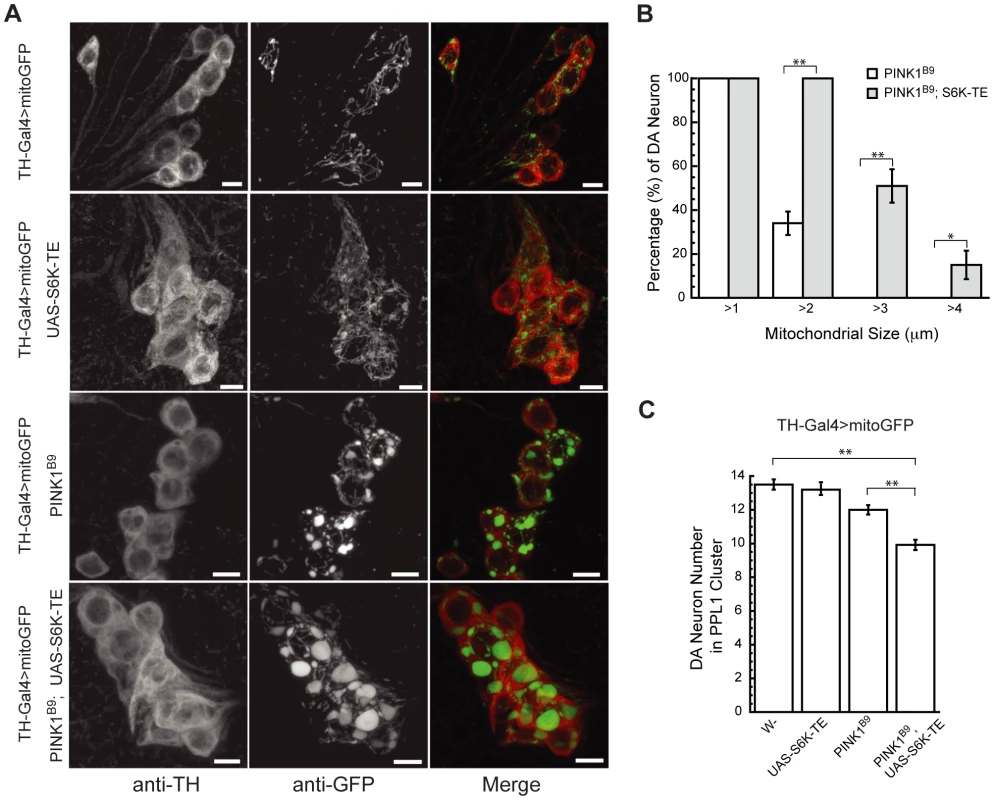
(A) Overexpression of constitutively active S6K increased the size of swollen or aggregated mitochondria in the DA neurons of PINK1 mutants. Mitochondrially targeted GFP (mitoGFP) was expressed in the DA neurons using TH-Gal4 driver [44] to help visualize mitochondrial morphology. Brains of 7-day-old adult flies of the indicated genotypes were immunostained with anti-TH antibody (red) to label DA neuron and anti-GFP antibody (green) to label mitochondria. Images of DA neurons in the PPL1 cluster were shown. Overexpression of S6K-TE in PINK1 mutant significantly increased the size of mitochondrial aggregates in DA neurons. The scale bar represents 5 µm. (B) Comparison of mitochondrial size distribution in PINK1 mutants with or without S6K-TE overexpression. Statistical significance was determined by Student's t test (**P<0.001, *P<0.05). (C) Overexpression of constitutively active S6K increased DA neuron loss in the PPL1 cluster of PINK1 mutant. DA neuron number was scored in flies aged for 14 days at 25°C. At least 7 flies were used for each genotype. Statistical significance was determined by Student's t test (**P<0.001). Data are presented as mean ± s.e.m. S6K modifies PINK1 mutant phenotypes through its regulation of translation
Although S6 kinase has been shown to have multiple substrates [31], its best-known function is to phosphorylate the 40S ribosomal subunit S6 (RpS6) and upregulate the translation of proteins involved in ribosomal biogenesis and protein synthesis [20]. To directly examine whether S6K enhances PINK1 mutant phenotype through increasing translation, we expressed an RpS6 RNAi transgene together with S6K-TE in the PINK1 RNAi background. Significantly, RpS6 RNAi efficiently blocked S6K-TE's enhancing effect on PINK1 RNAi-induced abnormal wing posture (Figure 4A), thoracic indentation (Figure 4B), increased mitochondrial aggregation (Figure 4C) and ATP depletion in the muscle (Figure 4D). RpS6 RNAi also partially suppressed such phenotypes in PINK1 RNAi background without S6K-TE co-expression (Figure 4A, data not shown). The effects of RpS6 RNAi were more obvious in young flies. In aged flies, the rescuing effect of RpS6 RNAi was mild in both the PINK1 RNAi and PINK1 RNAi/S6K-TE OE backgrounds (Figure 4A, 4D, data not shown). These results suggested that S6K did act through RpS6 to genetically interact with PINK1; however, reduction of translation through RpS6 RNAi only provided partial suppression of PINK1 RNAi phenotypes. This could be due to either inefficient knockdown of RpS6 function by RNAi or the involvement of other pathogenic pathway(s).
Fig. 4. RpS6 or RpS9 RNAi blocks the enhancing effects of S6K-TE in PINK1 RNAi background. 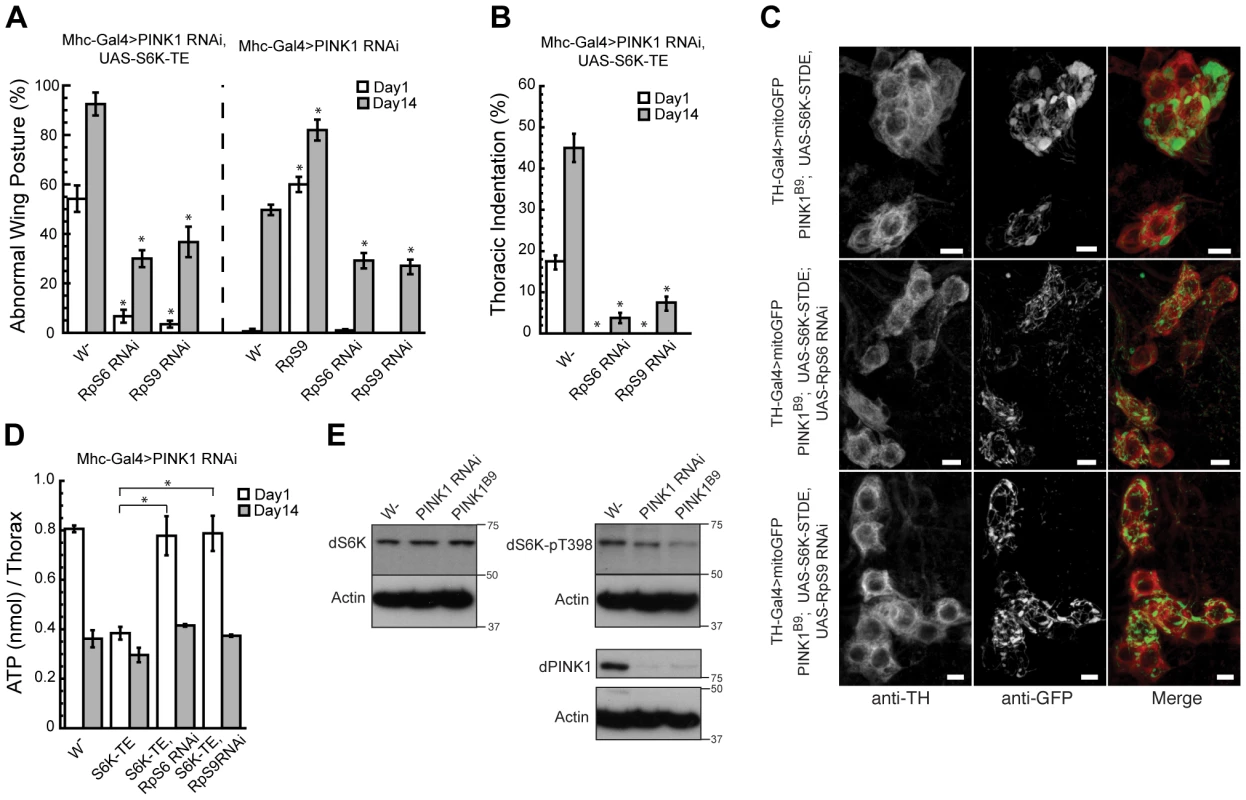
Overexpression of RpS6 or RpS9 RNAi transgenes in PINK1 RNAi or PINK1 RNAi/UAS-S6K-TE flies efficiently rescued the abnormal wing posture (A), thoracic indentation (B), and energy depletion (D) phenotypes in 1-day-old flies and partially suppressed these phenotypes in 14-day-old flies. Data are presented as mean ± s.e.m. Statistical significance was determined by Student's t test (*P<0.001). (C) RpS6 or RpS9 RNAi blocked increased mitochondrial aggregation in PINK1 RNAi, UAS-S6K-TE flies. The scale bar represents 5 µm. (E) Western blot analysis comparing the levels of dS6K and phosphorylated S6K (T398) in wild type, Mhc-Gal4>PINK1 RNAi and PINK1B9 mutant flies. The phosphorylation of S6K was significantly decreased in PINK1 RNAi or mutant flies. To further confirm the genetic interaction between PINK1 and the protein translational control pathway, we screened more than 20 EP lines expressing cytosolic or mitochondrial ribosomal subunits to see if any of these lines could also modify PINK1 RNAi phenotypes. Interestingly, one line that overexpresses ribosomal protein S9 (RpS9) greatly enhanced the PINK1 RNAi phenotypes (Figure 4A). Further, RpS9 RNAi was as effective as RpS6 RNAi in blocking S6K-TE's enhancing effects on PINK1 RNAi phenotypes (Figure 4A–4D). Previously, RpS9 knockdown was shown to significantly reduce the rate of protein synthesis and cell proliferation in primary human fibroblasts and tumor cell lines [32]. It is therefore likely that RpS9 RNAi mitigates the effects of constitutively active S6Ks through downregulating translation.
To test whether S6K and the related translational control pathway is normally involved in PINK1 pathogenesis, we examined the levels of S6K and phosphorylated S6K in PINK1 mutants. We found that the level of total S6K in PINK1 RNAi or PINK1B9 mutant flies was largely unchanged. However, the level of phosphorylated, active form of S6K was significantly decreased (Figure 4E), suggesting that there is decreased TOR signaling and protein translation in PINK1 mutant background. Since protein translation is a very energy-consuming process, reduction of translation could serve as a compensatory response to the mitochondrial dysfunction caused by PINK1 inactivation.
Atg1 activation partially rescues muscle degeneration in PINK1 mutant
As mentioned earlier, Atg1 emerged as a strong suppressor of PINK1 RNAi phenotype in our screen. To confirm this result, we tested the effect of Atg1 OE in PINK1B9 mutant, which exhibited stronger phenotypes than PINK1 RNAi flies. Similar to the results obtained using PINK1 RNAi flies, Atg1 OE could efficiently suppress the abnormal wing posture and thoracic indentation phenotypes of PINK1B9 flies (Figure 5A, 5B). Of note, our behavior test and muscle ATP level analysis indicated that the rescuing effect of Atg1 OE was not as strong as Parkin OE or Marf RNAi (Figure 5C, 5D). When we used mitoGFP to examine mitochondrial morphology in the muscle, Parkin OE and Marf RNAi could almost completely suppress the mitochondrial aggregation phenotype, but we could still see enlarged mitochondria in PINK1 mutant overexpressing Atg1 (Figure 5E).
Fig. 5. Overexpression of Atg1 rescues PINK1 mutant phenotypes. 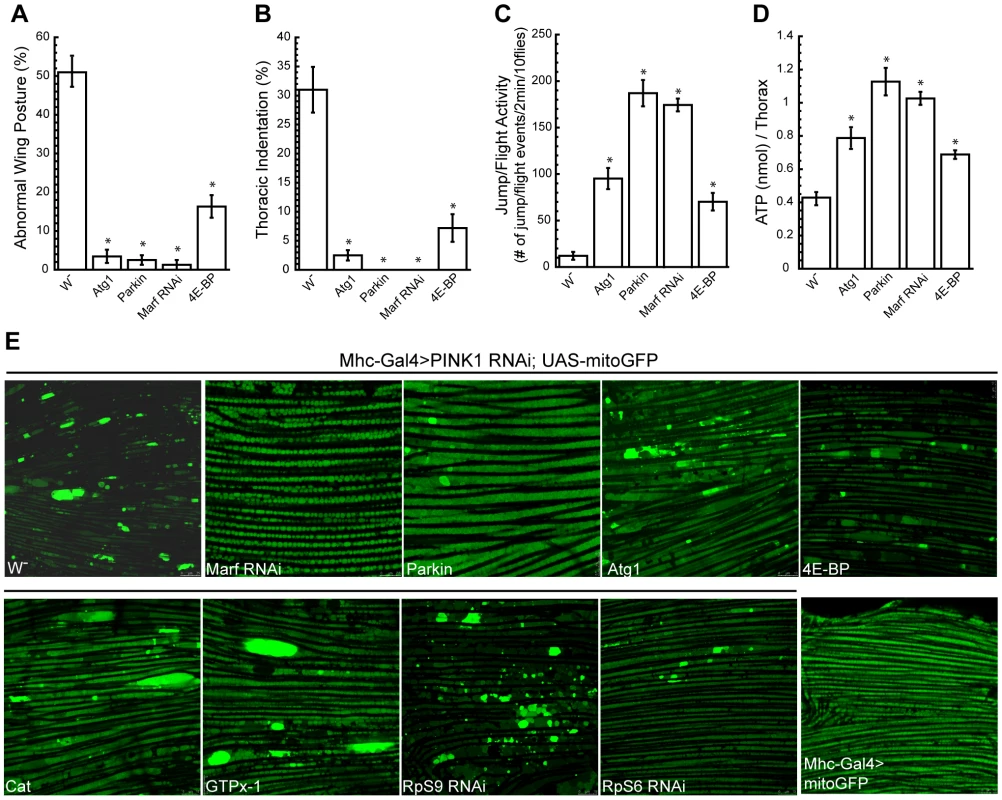
Overexpression of Atg1 in the muscle of PINK1B9 mutants rescued their abnormal wing posture (A), thoracic indentation (B), jump/flight activity (C) and muscle ATP level (D). Data are presented as mean ± s.e.m. Statistical significance was determined by Student's t test (*P<0.001). (E) Atg1 overexpression did not completely rescue the mitochondrial aggregation phenotype in the muscle of PINK1 RNAi flies. mitoGFP was expressed in the muscle using Mhc-Gal4 driver to visualize mitochondrial morphology by live imaging. Wild type flies showed mitochondria of relatively uniform sizes (bottom right), while PINK1 RNAi flies had bright mitochondrial aggregates. Only the co-expression of Marf RNAi or Parkin OE was able to efficiently rescue the mitochondrial aggregation phenotype in the PINK1 RNAi background. Similar to Atg1 OE, 4E-BP OE only partially rescued PINK1B9 mutant phenotypes (Figure 5A–5D). The overexpression of Catalase, GTPx-1, RpS6 RNAi or RpS9 RNAi also could not fully rescue the abnormal mitochondrial morphology phenotype in the PINK1 mutant (Figure 5E). Therefore, these genes modified PINK1 mutant phenotypes without effectively rescuing mitochondrial morphology, suggesting that they might act downstream or in parallel to the mitochondrial dynamics pathway.
Atg1 OE rescues PINK1 mutant phenotype by inducing autophagy
Atg1 is a Ser/Thr protein kinase involved in the initiation of autophagosome formation, which is under the control of TOR signaling. The loss of TOR signaling promotes the association of Atg1 with Atg13 and Atg17, which further recruit other Atg proteins to the pre-autophagosomal structure to mediate the formation of autophagosome [33]. In Drosophila, Atg1 OE alone is sufficient to induce autophagy in the fat body [26]. In addition to being a downstream effector of TOR, Atg1 can also exert feedback inhibitory effect on TOR. Atg1 OE has been shown to cause reduced phosphorylation of Drosophila S6K (dS6K) at T398, indicating downregulation of TOR signaling by Atg1 [26], [34]. Since S6K OE and Atg1 OE exert opposite effects in PINK1 RNAi background, we next tried to distinguish whether the rescuing effect of Atg1 OE was due to the inhibition of S6K function or induction of autophagy. To test whether Atg1 suppressed PINK1 RNAi-induced abnormal wing posture through inhibition of S6K, we expressed Atg1 together with S6K-TE in the PINK1 RNAi background. S6K-TE harbors the T398E mutation that mimics the phosphorylated form of S6K, which is constitutively active and cannot be suppressed by Atg1 [21]. Strikingly, Atg1 OE could strongly rescue the abnormal wing posture and ATP depletion phenotypes in the PINK1 RNAi/S6K-TE OE background (Figure 6A, 6B), suggesting that the rescuing effect of Atg1 did not rely on its known effect on S6K phosphorylation. Similarly, Parkin OE or Marf RNAi also significantly suppressed the abnormal wing posture and energy depletion phenotypes in PINK1 RNAi/S6K-TE OE background (Figure 6A, 6B). In contrast, overexpression of 4E-BP or the anti-oxidant genes, such as GTPx-1, Cat and SOD, were not as effective (Figure 6A, 6B).
Fig. 6. Atg1 OE rescues PINK1 RNAi phenotype by inducing autophagy. 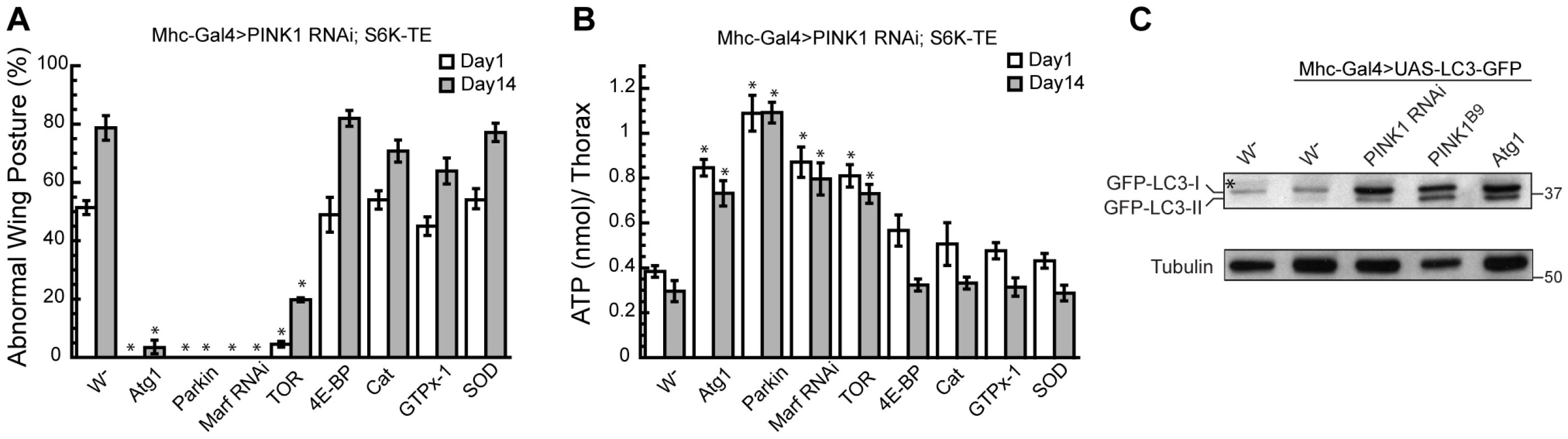
(A, B) The rescuing effect of Atg1 OE in PINK1 RNAi flies was not dependent on S6K inhibition. Atg1 OE, as well as Parkin OE or Marf RNAi, efficiently rescued the abnormal wing posture (A) and muscle energy depletion (B) in PINK1 RNAi/S6K-TE flies. In contrast, overexpression of 4E-BP and the antioxidant genes were not as effective. Data are presented as mean ± s.e.m. Statistical significance was determined by Student's t test (*P<0.001). (C) Overexpression of Atg1 was sufficient to induce autophagy in fly muscle. UAS-LC3-GFP was expressed in the muscle of flies with the indicated genetic background, and the level of autophagy was determined by Western Blot using anti-GFP antibody. Overexpression of Atg1 significantly increased the level of LC3-II in the muscle, indicating increased autophagy. Increased autophagy was also observed in PINK1 RNAi and PINK1 mutant flies. (* indicates a cross-reaction band). To test whether Atg1 OE rescued PINK1 RNAi phenotype by inducing autophagy, we first tested whether Atg1 OE could directly induce autophagy in the muscle, as observed in the fat body [26]. We used LC3-GFP as a marker to examine the lipidation of LC3 in different genetic backgrounds and used GFP antibody to detect mobility shift of LC3-GFP. Compared to the control, Atg1 OE led to an increased level of LC3-II, indicating induction of autophagy (Figure 6C). To test whether PINK1 deficiency affects autophagy in vivo, we introduced LC3-GFP into PINK1 RNAi and PINK1B9 backgrounds. Elevated autophagy was observed in both cases (Figure 6C). Thus, autophagy is basally induced in PINK1 mutant and further enhancement of autophagy is protective, suggesting that similar to decreased protein translation, increased autophagy also represents a compensatory response in PINK1 loss-of-function background.
To further prove that Atg1 OE-induced autophagy was critical for the suppression of PINK1 RNAi phenotype, we attempted to block the Atg1 OE effects with Atg18 RNAi. The co-expression of Atg18 RNAi could largely abolish the rescuing effects of Atg1 OE in PINK1B9 mutant or PINK1 RNAi/S6K-TE OE backgrounds (Figure 7A, 7B), suggesting that Atg1 OE rescued PINK1 mutant phenotypes mainly through inducing autophagy.
Fig. 7. Atg18 RNAi blocks the rescuing effect of Atg1 OE but not that of Parkin OE or Marf RNAi. 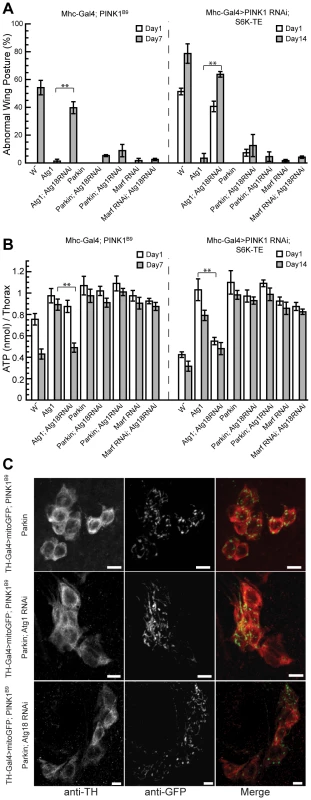
The rescuing effect of Atg1 OE on the abnormal wing posture (A) and muscle energy depletion (B) phenotypes could be blocked by the co-expression of Atg18 RNAi, suggesting that Atg1 functions through inducing autophagy to rescue PINK1 RNAi phenotype. In contrast, the rescue of PINK1 RNAi phenotypes by Parkin OE or Marf RNAi were largely unaffected by the disruption of Atg1 or Atg18 through RNAi. The tests were carried out in both PINK1B9 mutant and Mhc-Gal4>PINK1 RNAi/S6K-TE backgrounds. Data are presented as mean ± s.e.m. Statistical significance was determined by Student's t test (**P<0.001). (C) Atg1 or Atg18 RNAi did not abolish the rescuing effect of Parkin OE in DA neurons. mitoGFP was expressed in DA neurons using the TH-Gal4 driver to visualize mitochondrial morphology. Overexpression of Parkin efficiently rescued the mitochondrial aggregation phenotype in PINK1 mutant (top panel). Similar rescuing effect was observed when Atg1 RNAi or Atg18 RNAi was co-expressed with Parkin (middle and bottom panels). The scale bar represents 5 µm. Blocking autophagy does not abolish the rescuing effects of Parkin OE or Marf RNAi in PINK1 mutant background
Recent cell culture studies showed that Parkin is specifically recruited to dysfunctional mitochondria to mediate their elimination by the autophagy pathway, and the lack of PINK1 prevented this process, suggesting that Parkin plays an essential role in the selective elimination of damaged mitochondria [11]. We sought to directly test the role of autophagy in Parkin's rescue of PINK1 mutant phenotypes in vivo. In stark contrast to the efficient blockage of Atg1's rescuing effect by Atg18 RNAi, Atg18 RNAi failed to block Parkin's rescue of the abnormal wing posture (Figure 7A) and energy depletion (Figure 7B) phenotypes in PINK1B9 and PINK1 RNAi/S6K-TE OE flies. Similarly, Atg1 RNAi also could not block the rescuing effect of Parkin (Figure 7A, 7B). Consistent with these results obtained in the muscle, Atg1 RNAi or Atg18 RNAi failed to block the ability of Parkin to rescue the mitochondrial morphology phenotype in PINK1 mutant DA neurons (Figure 7C). These results suggest that Parkin might act through other mechanisms to rescue PINK1 mutant phenotype than solely promoting selective autophagy. Atg18 RNAi also failed to block the rescuing effects of Marf RNAi in PINK1B9 and PINK1 RNAi/S6K-TE OE backgrounds (Figure 7A, 7B), suggesting that decreased mitochondrial fusion can rescue PINK1 mutant phenotype independent of the autophagy pathway.
Discussion
The occurrence and the progression of Parkinson's disease can be determined by both genetic predisposition and environmental insults. Recent human genetic studies have identified many genes responsible for the heritable forms of the disease, greatly enhancing our understanding of disease pathogenesis [35]. By analyzing the cellular pathways that interact with these genes, hopefully we will ultimately find ways to better understand and treat this devastating disease.
Previously, PINK1 and Parkin have been suggested to interact with mitochondrial fusion/fission machinery and the autophagy pathway [8]–[11]. In this study, we found that PINK1 also genetically interacted with the protein translation pathway. Increased global protein translation with S6K or eIF4E OE exacerbated PINK1 mutant phenotypes, while decreased translation had the opposite effects. Overexpression of constitutively active S6Ks dramatically enhanced muscle and DA neuron degeneration in PINK1 mutant flies, which could be mitigated by the co-expression of RpS6 RNAi or RpS9 RNAi, supporting that the TOR/S6K pathway modifies PINK1 mutant phenotypes through regulating global translation. Recently, we have reported that pathogenic leucine-rich repeat kinase 2 (LRRK2), which represents the most frequent molecular lesions found in Parkinson's disease, promotes 4E-BP phosphorylation, resulting in increased eIF4E-mediated translation, enhanced sensitivity to oxidative stress, and DA neuron loss [19]. Taken together, our results support the idea that deregulated protein translation is generally involved in the pathogenesis of Parkinson's disease.
Deregulated translation affects Parkinson's disease pathogenesis most likely at the level of energy metabolism, since protein translation is a very energy-consuming process, of which ribosomal biogenesis is the most costly, consuming approximately 80% of the energy in proliferating cells [36]. Here we show that forced upregulation of ribosomal biogenesis in the fly muscle by the overexpression of constitutively active S6K was well tolerated in WT flies; however, such manipulation in PINK1 RNAi flies completely abolished their flight ability, depleted ATP in the muscle and enhanced muscle and DA neuron degeneration. The tolerance of increased protein translation by wild type flies is probably due to the existence of an intact mitochondrial quality control system containing PINK1 and Parkin, which can either eliminate damaged mitochondria generated during elevated energy production or minimize damages caused by increased ROS generated during energy production. However, in PINK1 or Parkin mutants that lack a functional mitochondrial quality control system, increased protein translation and the corresponding energy demand will translate into increased ROS generation, accumulation of dysfunctional mitochondria, and eventual energy depletion and tissue degeneration. Since downregulation of translation through knockdown of S6K, RpS6, or RpS9 is beneficial to PINK1 mutant flies, and S6K activity is already tuned down in PINK1 mutant flies, reduction of translation likely represents one of the cellular compensatory responses to the energy deficit caused by mitochondrial dysfunction in PINK1 mutants. Interestingly, partial reduction of S6K activity prolonged fly lifespan, whereas increased S6K activity had the opposite effects on longevity [37]. The effects of S6K on animal lifespan and PINK1 mutant phenotypes can both be explained by the energy metabolism hypothesis and they offer a tantalizing link between aging and the pathogenesis of Parkinson's disease.
Supporting the energy metabolism model, we show that downregulation of protein translation by knocking down positive regulators of translation (S6K, RpS6, RpS9) or overexpressing a negative regulator (4E-BP) could rescue PINK1 mutant phenotypes. These manipulations presumably act by preserving cellular energy and reducing the workload and ROS production of mitochondria. Previously, 4E-BP OE was suggested to rescue PINK1 mutant phenotype by upregulating Cap-independent translation of stress related genes, including antioxidant genes [30], and boosting antioxidant gene activity has been suggested as a therapeutic strategy in the PINK1 and Parkin models of Parkinson's disease [38]. We found that although overexpression of antioxidant genes, such as Catalase, GTPx-1, SOD and GstS1, all showed some degree of rescue of PINK1 mutant phenotypes, their effects were in general weaker than that of Atg1 OE, Parkin OE, or Marf RNAi, particularly in the PINK1 RNAi/S6K-TE OE background. These data suggest that increasing autophagy and mitochondrial fission might be better choices to combat PINK1-related Parkinson's disease.
Autophagy is a conserved cellular process through which cytoplasmic content or defective intracellular organelles can be eliminated or recycled. Although autophagy is usually induced under adverse conditions to provide means for survival, basal level of autophagy in the cell is just as critical to the physiological health of the organism, since defects in autophagy are frequently associated with cancer, neurodegeneration, and aging [39]. The induction of autophagy leads to the de novo formation of double membrane structure called isolation membrane, which expands to form a sealed compartment named autophagosome that will engulf materials destined for degradation. The large size of mitochondria likely poses a challenge for the autophagy machinery, as engulfment of an entire mitochondrion requires a significant amount of building materials for autophagosome formation. This is especially the case in PINK1 mutant where dysfunctional mitochondria becomes grossly swollen or aggregated. Previously, we and others showed that increased mitochondrial fission or Parkin OE could efficiently rescue the enlarged mitochondria phenotype in PINK1 mutants [5]–[10]. The rescuing effect by increased mitochondrial fission could be due to the fact that it decreases mitochondrial size and makes it easier for the autophagosome to engulf the entire mitochondrion during mitophagy (Figure S3). In addition, increased mitochondrial fission could facilitate the segregation of the healthy part of a mitochondrion from the unhealthy part, thus enhancing the selective elimination of dysfunctional mitochondria through mitophagy [40]. Supporting the mitophagy model, Parkin has been proposed to promote the efficient removal of damaged mitochondria by selectively ubiquitinating proteins on damaged mitochondria [11]. A key prediction of the mitophagy model is that the protective effects of Parkin OE and increased mitochondrial fission as in the case of Marf RNAi will depend on the autophagy pathway. Surprisingly, we found that blocking autophagy through Atg1 RNAi or Atg18 RNAi failed to block Parkin OE or Marf RNAi's rescuing abilities in PINK1 mutant, although Atg18 RNAi was effective in blocking the rescuing ability of Atg1 OE. This result suggests that the rescuing effect of Parkin OE or Marf RNAi is not entirely dependent on autophagy, and that other processes are likely involved. For example, Parkin has been suggested to promote mitochondrial biogenesis [41] and regulate protein translation [42]. Further studies are needed to elucidate the exact molecular functions of Parkin that are critically involved in mitochondrial function and tissue maintenance in vivo.
Given the well-established catabolic role of autophagy in degrading cytoplasmic contents, it helps recycle nutrients and provide energy source needed for survival under harsh conditions. In PINK1 mutants that suffer energy deficit due to mitochondrial dysfunction, induction of autophagy would present as a compensatory response to cope with the limited energy supply. Indeed, we found that basal autophagy is induced in PINK1 mutant, and further increase of autophagy through Atg1 OE protects against PINK1 pathogenesis. Thus, decreased translation and increased autophagy both represent compensatory responses in PINK1 mutant flies, and further augmentation of these responses can effectively protect against the toxic effects of PINK1 inactivation. A previous study in cultured mammalian cells also indicated that autophagy is induced in response to PINK1 inactivation [43]. Thus, the in vivo compensatory responses revealed in this study are likely relevant to PINK1 pathogenesis in mammals. Pharmacological interventions that promote these responses offer potential new treatment strategies for Parkinson's disease.
Materials and Methods
Fly strains
Flies were raised according to standard procedures at indicated temperatures. dPINK1 null mutant line dPINK1B9 was a gift from Dr. Jongkeong Chung [6]. The TH-GAL4 line was a gift from Dr. Serge Birman [44]. UAS-mitoGFP line was a gift from Dr. William Saxton. UAS-Atg1[6A], UAS-Atg1[6B], UAS-Atg1KQ, and UAS-Atg13 RNAi lines were gifts from Dr. Thomas Neufeld [26]. UAS-Atg1GS10797 line was a gift from Dr. Eric Baehrecke. UAS-Marf line was a gift from Dr. Alex Whitworth. UAS-PINK1 RNAi and UAS-Parkin were generated as described [5], [45]. UAS-Atg1 RNAi, UAS-Atg3 RNAi, UAS-Atg18 RNAi, UAS-RpS6 RNAi, UAS-RpS9 RNAi, and UAS-S6K RNAi lines were from Vienna Drosophila RNAi Center. All the other lines were from Bloomington Stock Center.
Muscle histology and transmission electron microscopy analysis
Muscle histology with toluidine blue staining and transmission electron microscopy analysis was performed essentially as described [45], except that Epon resin was used for embedding. For mitochondrial morphology analysis using mitoGFP, indirect fly muscle was dissected out in PBS and examined by live imaging.
Abnormal wing posture and behavior analyses
For abnormal wing posture analysis, male flies were aged at indicated temperature with 20 flies per vial. The abnormal wing posture penetrance was calculated as the percentage of flies with either held-up or drooped wing posture [5]. For each experiment, at least 60 flies were scored for their wing posture phenotype for each genotype. Each experiment was repeated at least three times. For jump/flight ability analyses, 5 to 10 flies were put into each vial. The jump/flight events were counted for two consecutive minutes, during which vials were gently tapped to initiate those events. Data were normalized and represented to reflect the jump/fly activity of 10 animals. Each of these analyses had been repeated at least three times.
ATP measurement
The thoracic ATP level was measured using a luciferase based bioluminescence assay (ATP Bioluminescence Assay Kit HS II, Roche Applied Science). For each measurement, two thoraces were dissected out (with wings and legs removed) and immediately homogenized in 100 µl lysis buffer. The lysate was boiled for 5 min and cleared by centrifugation at 20,000 g for 1 min. 2.5 µl of cleared lysate was added to 187.5 µl dilution buffer and 10 µl luciferase, and the luminescence was immediately measured using a Lumat LB 9507 tube luminometer (Berthold Technologies). Each reading was converted to the amount of ATP per thorax based on the standard curve generated with ATP standards. At least 3 measurements were made for each genotype.
Immunohistochemistry and western blot
Whole-mount immunohistochemistry for TH and mitoGFP was performed as described [46]. Rabbit anti-TH antibody (1∶500)[5] and chicken anti-GFP antibody (1∶5000) (Chemicon International) were used. The images of DA neurons of the protocerebral posterior lateral 1 (PPL1) cluster were collected at 0.5 mm steps along Z-axis on a Leica confocal microscope, and all the images shown were generated by Z-stack deconvolution. Western Blot was performed following standard protocol. Rabbit anti-dS6K antibody was a gift from Dr. George Thomas. Rabbit anti-Phospho-S6K (Thr398) antibody was from Cell Signaling Technology, and chicken anti-GFP antibody was purchased from Chemicon International. Rabbit anti-dPINK1 antibody was generated as described [5].
Supporting Information
Zdroje
1. ValenteEM
Abou-SleimanPM
CaputoV
MuqitMM
HarveyK
2004 Hereditary early-onset Parkinson's disease caused by mutations in PINK1. Science 304 1158 1160
2. KitadaT
AsakawaS
HattoriN
MatsumineH
YamamuraY
1998 Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature 392 605 608
3. GandhiS
MuqitMM
StanyerL
HealyDG
Abou-SleimanPM
2006 PINK1 protein in normal human brain and Parkinson's disease. Brain 129 1720 1731
4. ZhouC
HuangY
ShaoY
MayJ
ProuD
2008 The kinase domain of mitochondrial PINK1 faces the cytoplasm. Proc Natl Acad Sci U S A 105 12022 12027
5. YangY
GehrkeS
ImaiY
HuangZ
OuyangY
2006 Mitochondrial pathology and muscle and dopaminergic neuron degeneration caused by inactivation of Drosophila Pink1 is rescued by Parkin. Proc Natl Acad Sci U S A 103 10793 10798
6. ParkJ
LeeSB
LeeS
KimY
SongS
2006 Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature 441 1157 1161
7. ClarkIE
DodsonMW
JiangC
CaoJH
HuhJR
2006 Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature 441 1162 1166
8. YangY
OuyangY
YangL
BealMF
McQuibbanA
2008 Pink1 regulates mitochondrial dynamics through interaction with the fission/fusion machinery. Proc Natl Acad Sci U S A 105 7070 7075
9. PooleAC
ThomasRE
AndrewsLA
McBrideHM
WhitworthAJ
2008 The PINK1/Parkin pathway regulates mitochondrial morphology. Proc Natl Acad Sci U S A 105 1638 1643
10. DengH
DodsonMW
HuangH
GuoM
2008 The Parkinson's disease genes pink1 and parkin promote mitochondrial fission and/or inhibit fusion in Drosophila. Proc Natl Acad Sci U S A 105 14503 14508
11. NarendraD
TanakaA
SuenDF
YouleRJ
2008 Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol 183 795 803
12. NarendraDP
JinSM
TanakaA
SuenDF
GautierCA
2010 PINK1 Is Selectively Stabilized on Impaired Mitochondria to Activate Parkin. PLoS Biol 8 e1000298 doi:10.1371/journal.pbio.1000298
13. ZivianiE
TaoRN
WhitworthAJ
2010 Drosophila Parkin requires PINK1 for mitochondrial translocation and ubiquitinates Mitofusin. Proc Natl Acad Sci U S A 107 5018 5023
14. GeislerS
HolmstromKM
SkujatD
FieselFC
RothfussOC
2010 PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol 12 119 131
15. WullschlegerS
LoewithR
HallMN
2006 TOR signaling in growth and metabolism. Cell 124 471 484
16. CardenasC
MillerRA
SmithI
BuiT
MolgoJ
2010 Essential regulation of cell bioenergetics by constitutive InsP3 receptor Ca2+ transfer to mitochondria. Cell 142 270 283
17. HardieDG
2007 AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat Rev Mol Cell Biol 8 774 785
18. DingWX
NiHM
LiM
LiaoY
ChenX
2010 Nix is critical to two distinct phases of mitophagy, reactive oxygen species-mediated autophagy induction and Parkin-ubiquitin-p62-mediated mitochondrial priming. J Biol Chem 285 27879 27890
19. ImaiY
GehrkeS
WangHQ
TakahashiR
HasegawaK
2008 Phosphorylation of 4E-BP by LRRK2 affects the maintenance of dopaminergic neurons in Drosophila. Embo J 27 2432 2443
20. JefferiesHB
FumagalliS
DennisPB
ReinhardC
PearsonRB
1997 Rapamycin suppresses 5'TOP mRNA translation through inhibition of p70s6k. Embo J 16 3693 3704
21. BarceloH
StewartMJ
2002 Altering Drosophila S6 kinase activity is consistent with a role for S6 kinase in growth. Genesis 34 83 85
22. PearsonRB
DennisPB
HanJW
WilliamsonNA
KozmaSC
1995 The principal target of rapamycin-induced p70s6k inactivation is a novel phosphorylation site within a conserved hydrophobic domain. Embo J 14 5279 5287
23. HanJW
PearsonRB
DennisPB
ThomasG
1995 Rapamycin, wortmannin, and the methylxanthine SQ20006 inactivate p70s6k by inducing dephosphorylation of the same subset of sites. J Biol Chem 270 21396 21403
24. RaughtB
GingrasAC
SonenbergN
2000 Regulation of Ribosomal Recruitment in Eukaryotes.
SonenbergN
HersheyJWB
MathewsMB
Translational Control of Gene Expression Cold Spring Harbor Cold Spring Harbor Laboratory Press 245 293
25. HennigKM
NeufeldTP
2002 Inhibition of cellular growth and proliferation by dTOR overexpression in Drosophila. Genesis 34 107 110
26. ScottRC
JuhaszG
NeufeldTP
2007 Direct induction of autophagy by Atg1 inhibits cell growth and induces apoptotic cell death. Curr Biol 17 1 11
27. BerryDL
BaehreckeEH
2007 Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell 131 1137 1148
28. ChangYY
NeufeldTP
2009 An Atg1/Atg13 complex with multiple roles in TOR-mediated autophagy regulation. Mol Biol Cell 20 2004 2014
29. WhitworthAJ
TheodoreDA
GreeneJC
BenesH
WesPD
2005 Increased glutathione S-transferase activity rescues dopaminergic neuron loss in a Drosophila model of Parkinson's disease. Proc Natl Acad Sci U S A 102 8024 8029
30. TainLS
MortiboysH
TaoRN
ZivianiE
BandmannO
2009 Rapamycin activation of 4E-BP prevents parkinsonian dopaminergic neuron loss. Nat Neurosci 12 129 135
31. RuvinskyI
MeyuhasO
2006 Ribosomal protein S6 phosphorylation: from protein synthesis to cell size. Trends Biochem Sci 31 342 348
32. LindstromMS
ZhangY
2008 Ribosomal protein S9 is a novel B23/NPM-binding protein required for normal cell proliferation. J Biol Chem 283 15568 15576
33. NakatogawaH
SuzukiK
KamadaY
OhsumiY
2009 Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol 10 458 467
34. LeeSB
KimS
LeeJ
ParkJ
LeeG
2007 ATG1, an autophagy regulator, inhibits cell growth by negatively regulating S6 kinase. EMBO Rep 8 360 365
35. LinMT
BealMF
2006 Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature 443 787 795
36. SchmidtEV
1999 The role of c-myc in cellular growth control. Oncogene 18 2988 2996
37. KapahiP
ZidBM
HarperT
KosloverD
SapinV
2004 Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr Biol 14 885 890
38. ChaturvediRK
BealMF
2008 Mitochondrial approaches for neuroprotection. Ann N Y Acad Sci 1147 395 412
39. MizushimaN
LevineB
CuervoAM
KlionskyDJ
2008 Autophagy fights disease through cellular self-digestion. Nature 451 1069 1075
40. TwigG
ElorzaA
MolinaAJ
MohamedH
WikstromJD
2008 Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. Embo J 27 433 446
41. KurodaY
MitsuiT
KunishigeM
ShonoM
AkaikeM
2006 Parkin enhances mitochondrial biogenesis in proliferating cells. Hum Mol Genet 15 883 895
42. CortiO
HampeC
KoutnikovaH
DariosF
JacquierS
2003 The p38 subunit of the aminoacyl-tRNA synthetase complex is a Parkin substrate: linking protein biosynthesis and neurodegeneration. Hum Mol Genet 12 1427 1437
43. DagdaRK
CherraSJ3rd
KulichSM
TandonA
ParkD
2009 Loss of PINK1 function promotes mitophagy through effects on oxidative stress and mitochondrial fission. J Biol Chem 284 13843 13855
44. Friggi-GrelinF
CoulomH
MellerM
GomezD
HirshJ
2003 Targeted gene expression in Drosophila dopaminergic cells using regulatory sequences from tyrosine hydroxylase. J Neurobiol 54 618 627
45. PesahY
PhamT
BurgessH
MiddlebrooksB
VerstrekenP
2004 Drosophila parkin mutants have decreased mass and cell size and increased sensitivity to oxygen radical stress. Development 131 2183 2194
46. DavisRJ
TavsanliBC
DittrichC
WalldorfU
MardonG
2003 Drosophila retinal homeobox (drx) is not required for establishment of the visual system, but is required for brain and clypeus development. Dev Biol 259 272 287
Štítky
Genetika Reprodukční medicína
Článek Genome-Wide Interrogation of Mammalian Stem Cell Fate Determinants by Nested Chromosome DeletionsČlánek Season of Conception in Rural Gambia Affects DNA Methylation at Putative Human Metastable EpiallelesČlánek A Quantitative Systems Approach Reveals Dynamic Control of tRNA Modifications during Cellular StressČlánek The Loss of PGAM5 Suppresses the Mitochondrial Degeneration Caused by Inactivation of PINK1 inČlánek Cleavage of Phosphorothioated DNA and Methylated DNA by the Type IV Restriction Endonuclease ScoMcrAČlánek Competitive Repair by Naturally Dispersed Repetitive DNA during Non-Allelic Homologous Recombination
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2010 Číslo 12
-
Všechny články tohoto čísla
- Genome-Wide Interrogation of Mammalian Stem Cell Fate Determinants by Nested Chromosome Deletions
- Whole-Genome and Chromosome Evolution Associated with Host Adaptation and Speciation of the Wheat Pathogen
- Association of Variants at 1q32 and with Ankylosing Spondylitis Suggests Genetic Overlap with Crohn's Disease
- Initiator Elements Function to Determine the Activity State of BX-C Enhancers
- Identification of Genes Required for Neural-Specific Glycosylation Using Functional Genomics
- A Young Duplicate Gene Plays Essential Roles in Spermatogenesis by Regulating Several Y-Linked Male Fertility Genes
- The EpsE Flagellar Clutch Is Bifunctional and Synergizes with EPS Biosynthesis to Promote Biofilm Formation
- Histone H2A C-Terminus Regulates Chromatin Dynamics, Remodeling, and Histone H1 Binding
- Season of Conception in Rural Gambia Affects DNA Methylation at Putative Human Metastable Epialleles
- A Quantitative Systems Approach Reveals Dynamic Control of tRNA Modifications during Cellular Stress
- GC-Rich Sequence Elements Recruit PRC2 in Mammalian ES Cells
- A Single Enhancer Regulating the Differential Expression of Duplicated Red-Sensitive Opsin Genes in Zebrafish
- Investigation and Functional Characterization of Rare Genetic Variants in the Adipose Triglyceride Lipase in a Large Healthy Working Population
- Reduction of Protein Translation and Activation of Autophagy Protect against PINK1 Pathogenesis in
- Noisy Splicing Drives mRNA Isoform Diversity in Human Cells
- The Loss of PGAM5 Suppresses the Mitochondrial Degeneration Caused by Inactivation of PINK1 in
- Thymus-Associated Parathyroid Hormone Has Two Cellular Origins with Distinct Endocrine and Immunological Functions
- An ABC Transporter Mutation Is Correlated with Insect Resistance to Cry1Ac Toxin
- Role of Individual Subunits of the CSN Complex in Regulation of Deneddylation and Stability of Cullin Proteins
- The C-Terminal Domain of the Bacterial SSB Protein Acts as a DNA Maintenance Hub at Active Chromosome Replication Forks
- The DNA Damage Response Pathway Contributes to the Stability of Chromosome III Derivatives Lacking Efficient Replicators
- Cleavage of Phosphorothioated DNA and Methylated DNA by the Type IV Restriction Endonuclease ScoMcrA
- LaeA Control of Velvet Family Regulatory Proteins for Light-Dependent Development and Fungal Cell-Type Specificity
- Competitive Repair by Naturally Dispersed Repetitive DNA during Non-Allelic Homologous Recombination
- Distinct Functions for the piRNA Pathway in Genome Maintenance and Telomere Protection
- MOS11: A New Component in the mRNA Export Pathway
- Self-Mating in the Definitive Host Potentiates Clonal Outbreaks of the Apicomplexan Parasites and
- A Role for ATF2 in Regulating MITF and Melanoma Development
- Ancestral Regulatory Circuits Governing Ectoderm Patterning Downstream of Nodal and BMP2/4 Revealed by Gene Regulatory Network Analysis in an Echinoderm
- Cancer and Neurodegeneration: Between the Devil and the Deep Blue Sea
- Functional Comparison of Innate Immune Signaling Pathways in Primates
- Linking Crohn's Disease and Ankylosing Spondylitis: It's All about Genes!
- Genomics Meets Glycomics—The First GWAS Study of Human N-Glycome Identifies HNF1α as a Master Regulator of Plasma Protein Fucosylation
- Continuous and Periodic Expansion of CAG Repeats in Huntington's Disease R6/1 Mice
- Expression of Linear and Novel Circular Forms of an -Associated Non-Coding RNA Correlates with Atherosclerosis Risk
- Endocytic Sorting and Recycling Require Membrane Phosphatidylserine Asymmetry Maintained by TAT-1/CHAT-1
- Histone Deacetylases Suppress CGG Repeat–Induced Neurodegeneration Via Transcriptional Silencing in Models of Fragile X Tremor Ataxia Syndrome
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Functional Comparison of Innate Immune Signaling Pathways in Primates
- Expression of Linear and Novel Circular Forms of an -Associated Non-Coding RNA Correlates with Atherosclerosis Risk
- Genome-Wide Interrogation of Mammalian Stem Cell Fate Determinants by Nested Chromosome Deletions
- Histone H2A C-Terminus Regulates Chromatin Dynamics, Remodeling, and Histone H1 Binding
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



