-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaGenomics Meets Glycomics—The First GWAS Study of Human N-Glycome Identifies HNF1α as a Master Regulator of Plasma Protein Fucosylation
Over half of all proteins are glycosylated, and alterations in glycosylation have been observed in numerous physiological and pathological processes. Attached glycans significantly affect protein function; but, contrary to polypeptides, they are not directly encoded by genes, and the complex processes that regulate their assembly are poorly understood. A novel approach combining genome-wide association and high-throughput glycomics analysis of 2,705 individuals in three population cohorts showed that common variants in the Hepatocyte Nuclear Factor 1α (HNF1α) and fucosyltransferase genes FUT6 and FUT8 influence N-glycan levels in human plasma. We show that HNF1α and its downstream target HNF4α regulate the expression of key fucosyltransferase and fucose biosynthesis genes. Moreover, we show that HNF1α is both necessary and sufficient to drive the expression of these genes in hepatic cells. These results reveal a new role for HNF1α as a master transcriptional regulator of multiple stages in the fucosylation process. This mechanism has implications for the regulation of immunity, embryonic development, and protein folding, as well as for our understanding of the molecular mechanisms underlying cancer, coronary heart disease, and metabolic and inflammatory disorders.
Published in the journal: . PLoS Genet 6(12): e32767. doi:10.1371/journal.pgen.1001256
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1001256Summary
Over half of all proteins are glycosylated, and alterations in glycosylation have been observed in numerous physiological and pathological processes. Attached glycans significantly affect protein function; but, contrary to polypeptides, they are not directly encoded by genes, and the complex processes that regulate their assembly are poorly understood. A novel approach combining genome-wide association and high-throughput glycomics analysis of 2,705 individuals in three population cohorts showed that common variants in the Hepatocyte Nuclear Factor 1α (HNF1α) and fucosyltransferase genes FUT6 and FUT8 influence N-glycan levels in human plasma. We show that HNF1α and its downstream target HNF4α regulate the expression of key fucosyltransferase and fucose biosynthesis genes. Moreover, we show that HNF1α is both necessary and sufficient to drive the expression of these genes in hepatic cells. These results reveal a new role for HNF1α as a master transcriptional regulator of multiple stages in the fucosylation process. This mechanism has implications for the regulation of immunity, embryonic development, and protein folding, as well as for our understanding of the molecular mechanisms underlying cancer, coronary heart disease, and metabolic and inflammatory disorders.
Introduction
Glycosylation is a post-translational modification that enriches protein complexity and function. Over half of all known proteins are modified by covalently bound glycans, which are important for normal physiological processes, including protein folding, degradation and secretion, cell signalling, immune function and transcription [1]–[4]. Configuration and composition of attached glycans significantly change the structure and activity of polypeptide portions of glycoproteins [5] and since this process is not template driven, complexity of the glycoproteome is estimated to be several orders of magnitude greater than for the proteome itself [6]. Disregulation of glycosylation is associated with a wide range of diseases, including cancer, diabetes, cardiovascular, congenital, immunological and infectious disorders [1], [3], [7]. Enzymes that are involved in glycosylation may therefore be promising targets for therapy [8]. The most prominent example of the importance of N-glycosylation is the group of rare diseases named congenital disorders of glycosylation [9] where different mutations in the biosynthesis pathway of N-glycans cause significant mortality and extensive motor, immunological, digestive and neurological symptoms [10], [11].
Due to experimental limitations in quantifying glycans in complex biological samples, our understanding of the genetic regulation of glycosylation is currently very limited [12]. However, recent technological advances have allowed reliable, high-throughput quantification of N-glycans [13], which now permits investigation of the genetic regulation and biological roles of glycan structures and brings glycomics into line with genomics, proteomics and metabolomics [14]. Recently we completed the first comprehensive population study of human plasma N-glycome which revealed variability that by far exceeds the variability of proteins and DNA [15]. However, within a single individual composition of plasma glycome is rather stable [16] and environmental factors have limited impact on the majority of glycans [17]. Specific altered glyco-phenotypes that can be associated with specific pathologies were also identified to exist in a population [18].
Variations in glycosylation are of great physiological significance as alterations in glycans significantly change the structure and function of polypeptide parts of glycoproteins [5]. A particularly interesting element of protein glycosylation is the addition of fucose to non-reducing ends of N-glycans. Fucose is a relatively novel sugar in evolutionary terms with two important structural features that distinguishes it from all other mammalian six-carbon monosaccharides; it lacks a hydroxyl group on the carbon at the 6-position and is the only monosaccharide that is in the L-configuration. The conversion of GDP-mannose to GDP-fucose is catalyzed by two enzymes (GMD and FX) that display remarkable evolutionary conservation [19], [20]. On the other hand, the large family of genes that add fucose to proteins and lipids (fucosyltranferases, FUTs) has a very complex evolutionary history, including several more recent events specific to primates [21]. In mammals, fucose-containing glycans have important roles in blood transfusion reactions, in the selectin-mediated leukocyte-endothelial adhesion that initiates an inflammatory response, in host-microbe interactions, and numerous ontogenic events [10], [19]. Acute phase proteins have altered fucosylation in many diseases [22] and changes in the levels of fucosylated glycans have been shown to be associated with several important pathological processes, including cancer [23].
Hepatocyte nuclear factor 1α (HNF1α) and its downstream target HNF4α are transcription factors that regulate gene expression in both the liver and pancreas in a tissue-specific manner and are key regulators of metabolic genes [24]. Mutations in the encoding genes HNF1α and HNF4α cause Maturity Onset Diabetes of the Young (MODY) types 3 and 1 respectively [25], [26]. Recently, HNF1α single nucleotide polymorphisms (SNPs) have been associated with plasma C-reactive protein (CRP) [27], LDL cholesterol and gamma glutamyltransferase (GGT) [28], and coronary heart disease [29]. HNF4α variants have been associated with ulcerative colitis [30] and with the plasma concentrations of CRP and apolipoprotein A1 (APOA1) [31]. Currently there is little evidence to link these transcription factors with fucose metabolism and the upstream mechanisms regulating fucosylation pathways are unknown.
Results
Common variants in fucosyltransferase genes affect the relative proportions of plasma N-glycans
We performed the first systematic analysis of the genetic regulation of individual N-glycans in plasma from 2,705 individuals in three population cohorts, from Croatia and Scotland, which have previously been characterized in great detail [32]. Desialylated 2AB-labelled human plasma N-glycans were separated into 13 structurally related groups of glycans, referred to as DG1–DG13 (see Table S1 for a list of specific glycans found within each DG group) [13]. The concentration of plasma N-glycans measured in each of these groups was then expressed as a proportion of the total plasma N-glycome to obtain 13 quantitative variables in each examinee. All N-glycans contain two core N-acetylglucosamine (GlcNAc) residues, to which a “core” fucose can be α1,6-linked to the inner GlcNAc, which is directly linked to an asparagine residue on the protein. Additional fucose residues can be transferred to different positions on antennas that have been added to the core glycan structure (Table S1). Two further traits were derived from the original variables to calculate the percentage of glycan structures containing core (FUC-C) or antennary (FUC-A) fucose, yielding a total of 15 glycan traits for analysis.
We conducted a meta-analysis of genome-wide association study (GWAS) data for the fifteen plasma N-glycan traits measured in three population-based cohorts, CROATIA-VIS (n = 924), CROATIA-KORCULA (n = 898) and ORCADES (n = 737). Additive SNP effects were tested in each cohort independently and then combined in an inverse-variance weighted meta-analysis. The genome-wide significance threshold for the meta-analysis was set at 5×10−08.
Genome-wide significant associations were found for DG1, DG6, DG7, DG9, DG11, as well as FUC-A (Table 1; Figure 1 and Figure 2). Association profiles for DG1, DG7, and DG9 are represented in their genomic context in Figure 1 for the associated region. Quantile–quantile plots for each association were consistent with an excess of true genetic associations, with modest genomic control inflation for each population (inflation factor <1.04 for all traits and each population as well as the meta-analysis), suggesting that the observed results were not due to population stratification (Figure 2A–2C).
Fig. 1. Significance plots. 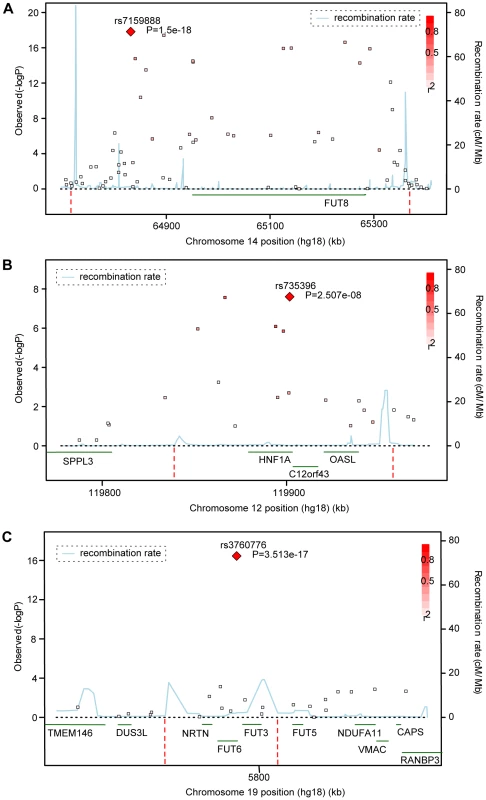
Significance plots for regions of interest from the meta-analysis of (a) DG1, (b) DG7 and (c) DG9. A region around the most significant genotyped SNP (represented by a red diamond) is displayed with the –log10 of the association p-values plotted against chromosome position. The degree of linkage disequilibrium between the most significant SNP and any SNP tested is indicated by a gradient of red shading. Recombination rate is displayed by a blue line with scale on the right-hand axis. Characterized genes in the region are represented with an arrow (showing the direction of transcription). The accompanying association interval as defined in the methods is marked by vertical red dotted lines. Fig. 2. Quantile–quantile plots and forest plots. 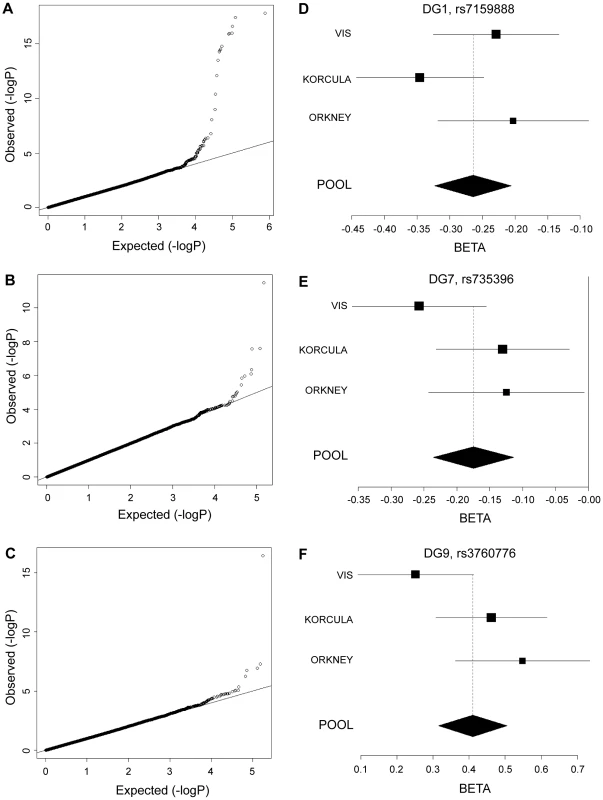
Quantile-quantile plots for test statistics (a–c) and forest plots of the most significant SNPs (d–f) from the meta-analysis of DG1, DG7, and DG9. −Log10 of the association p-values are plotted against −log10 of the expected p-value under the null hypothesis of no association for the meta-analysis. The effect size estimates for each individual population and the meta-analysis are shown along with their standard errors. The effect size presented is the β-coefficient, which represents a change in glycan levels measured in standard deviation units (adjusted for covariates) per copy of the allele modelled. The mean effect size estimates are represented by a square for individual cohorts where the size is proportional to its contribution to the pooled effect size. Tab. 1. Genetic markers associated with plasma N-glycan levels. 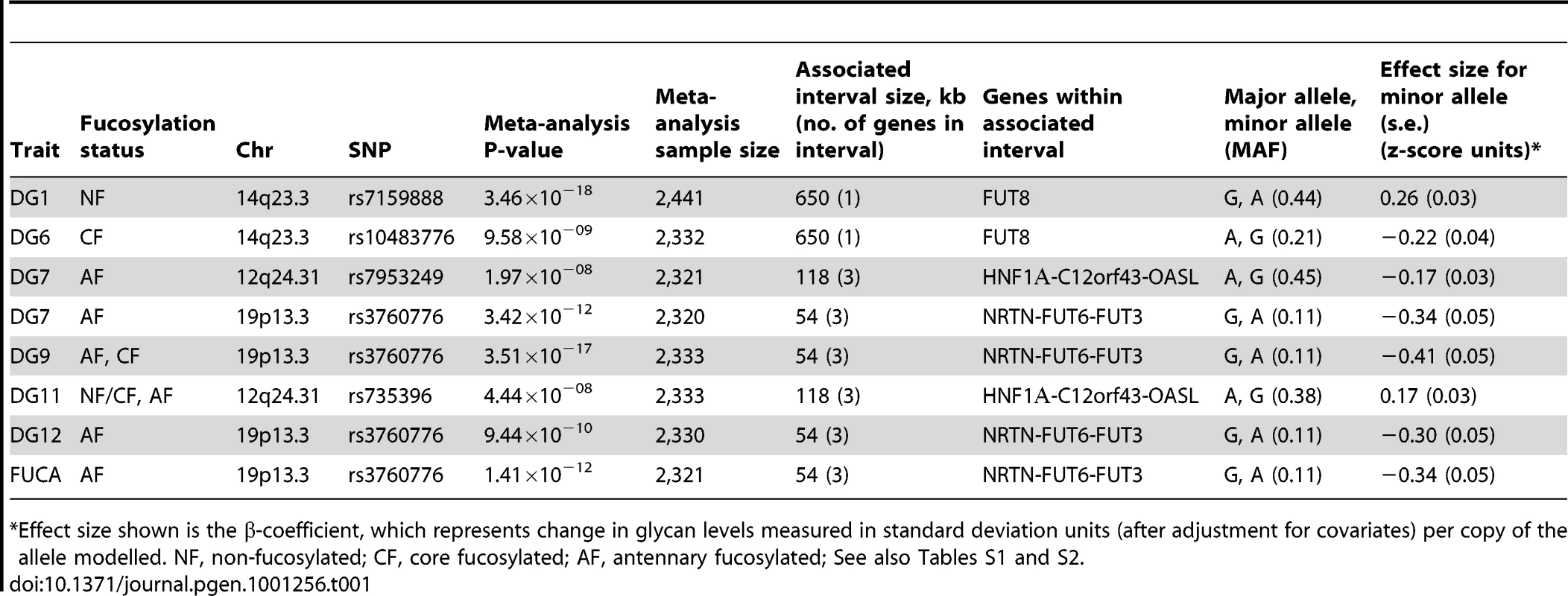
*Effect size shown is the β-coefficient, which represents change in glycan levels measured in standard deviation units (after adjustment for covariates) per copy of the allele modelled. NF, non-fucosylated; CF, core fucosylated; AF, antennary fucosylated; See also Tables S1 and S2. Fifteen SNPs located in the region encompassing the fucosyltransferase 8 gene (FUT8, Entrez GeneID: 2530) on chromosome 14 were significantly associated with plasma concentrations of desialylated glycan (DG) 1, the most significant being rs7159888 (p = 3.46×10−18) located 5′ of the gene. FUT8 was also associated with DG6, however for this trait only one SNP, rs10483776, reached genome-wide significance (p = 9.58×10−09). All SNPs significantly associated with DG1 levels were in high LD (r2>0.5) and located between two recombination hotspots, while no associations were found with SNPs located outside these boundaries nor with other genes located within this association interval (Figure 1A). The effect size of the G allele of rs7159888 was −0.2617 (s.e. 0.0301) for DG1 in the meta-analysis of the 3 populations studied (standard deviation units, after adjustment for sex and age; Figure 2D). All significant SNPs in this region had a similar effect size (absolute value of the range: 0.1828–0.3251), accounting for between 1 and 6 percent of the trait variance after adjustment for age and sex. The effect of rs7159888 on DG1 was consistent across populations with similar amplitude and direction of effect (Figure 2D) with the effect for each population plotted separately along with the pooled effect. Haplotype analysis found that a single SNP model performed better than the 3 - or 5-SNP haplotype model in every population.
A single SNP located on chromosome 19, rs3760776, was associated with DG7, DG9, DG12 and FUC-A (p = 3.42×10−12, p = 3.51×10−17, p = 9.44×10−10, p = 1.41×10−12). This SNP is located at the 5′ end of the fucosyltranferase 6 gene (FUT6, Entrez GeneID: 2528). The association interval for this SNP contains the NRTN, FUT6 and FUT3 genes (see Figure 1C), of which FUT6 and FUT3 are both biologically plausible candidates to explain the observed associations. The effect size of the G allele of rs3760776 is 0.3387 (s.e. 0.0487) for DG7 (standard deviation units, after adjustment for significant covariates: sex, age and fibrinogen); and 0.4104 (s.e. 0.0487), 0.2974 (s.e. 0.0486), and 0.3446 (s.e. 0.0486) for DG9, DG12 and FUC-A respectively (standard deviation units, after adjustment for age and fibrinogen). These effects account for 2% (DG7), 3% (DG9), 2% (DG12) and 2% (FUC-A) of the trait variance. A forest plot of the effect size of rs3760776 in each population and the meta for DG7 is presented in Figure 2F. Haplotype analysis suggested that a 5-SNP haplotype across this region has a stronger effect on these glycan levels than a single SNP model. Another fucosyltransferase gene (FUT3) is also within the region, so the causal variant(s) may affect one or both of these genes. The best 5-SNP haplotype contained rs3760776 and encompassed FUT6 but not FUT3 in every population and for every glycan group tested which suggests that the association is with FUT6, not FUT3.
The glycan structures which were significantly associated with genetic variants in the FUT6 and FUT8 genes are summarised in Table 1. Glycan group DG1 consists of a single structure GlcNAc2Man3GlcNAc2 that is known to be a substrate for the α1-6-fucosyltransferase (FUT8) (Table S1) [15], [33]. Group DG6 contains three glycan structures, two of which are core fucosylated so the results are consistent with the known biological role of FUT8. In contrast, groups DG7, DG9 and DG12 include glycans containing antennary fucose while FUC-A was derived as an overall measure of antennary fucosylation. FUT6 encodes the enzyme fucosyltransferase VI which was reported to be the key enzyme responsible for the α3-fucosylation of plasma proteins [34]. The association of FUT8 and FUT6 genes with N-glycan structures containing core and antennary fucosylation is supported by their known biological functions [35] and the fact that they were identified in this study is an effective proof of principle that HPLC measured glycan levels can be used to identify genes that regulate protein glycosylation.
Novel association of HNF1α with N-glycans
Two SNPs on chromosome 12, rs7953249 and rs735396, showed genome-wide significant associations with DG7 (p = 1.97×10−08, p = 1.75×10−08). The latter SNP was also associated with DG11 (p = 4.44×10−08), with an effect in the opposite direction, and was close to genome-wide significance with DG9 (Table S2). Both SNPs are located in the HNF1α (Entrez GeneID: 6927) gene region: rs7953249 is found 13 kb 5′ to the gene and rs735396 is in intron 9. Two other genes are found between the recombination hotspots that comprise the boundaries of the association interval, C12orf43 and OASL (Figure 1B). However, none of the most significantly associated SNPs are located in these genes and all SNPs with suggestive p-values (p<1×10−05) are located within HNF1α (Table S2). The effect size of the G allele of rs735396 is −0.1767 (standard deviation units, after adjustment for sex, age and fibrinogen; s.e. 0.0314) for DG7, which only contains glycans with antennary fucose, and in the opposite direction (0.1699 standard deviation units, after adjustment for age and fibrinogen; s.e. 0.0310) for DG11, which has no antennary fucose (Table 1). All significant SNPs in this region had a similar effect size (absolute value of the range: 0.1396–0.1767), representing 1–3% of the trait variance. Figure 2E shows the effect size for rs735396 with DG7 for each population separately and the pooled meta-analysis. Comparison of models including rs7953249 and rs735396 separately and combined suggests that the causal variant is located between these two SNPs. This was confirmed by analysis of imputed data based on HapMap release 2 with the most significant SNPs located across intron 1 of HNF1α.
HNF1α regulates multiple stages in protein fucosylation: (1) regulation of GDP-fucose biosynthesis
The shared characteristic of all glycan groups that showed association with HNF1α SNPs was the presence or absence of antennary fucose (Table S2). We hypothesised that HNF1α transcriptionally regulates the expression of genes involved in the separates steps of fucosylation. This is supported by the fact that a functionally related transcription factor, HNF4α was previously shown to bind the regulatory elements of the GDP-mannose-4,6-dehydratase (GMDS) gene in a genome-wide ChIP-ChIP. GMDS is involved in the de novo pathway of L-fucose synthesis to produce GDP-fucose, the substrate used by both core and antennary fucosyltransferases to N-glycosylated proteins [24]. Moreover, HNF4α directly regulates the expression of the hepatic fucosyltransferase VI gene (FUT6) [36]. Therefore, we tested whether HNF1α and/or HNF4α might regulate other genes involved in GDP-fucose biosynthesis. To this end, HNF1α and HNF4α were transiently knocked-down in liver and pancreatic cell lines using RNA interference. Both HNF1α and HNF4α expression levels decreased upon knockdown of either of them in hepatocytes (Figure 3A). In pancreatic cells, HNF1α knockdown up-regulates HNF4α expression but the reverse is not true (Figure S1). This confirms the differential regulation of gene expression downstream of HNFs in liver vis-a-vis pancreas [28], [37]. It also corroborated recent findings in murine Hnf1α hetrozygote pancreas, where the levels of Hnf4α mRNA increase [38].
Fig. 3. HNF1α is major regulator of fucose synthesis pathway genes. 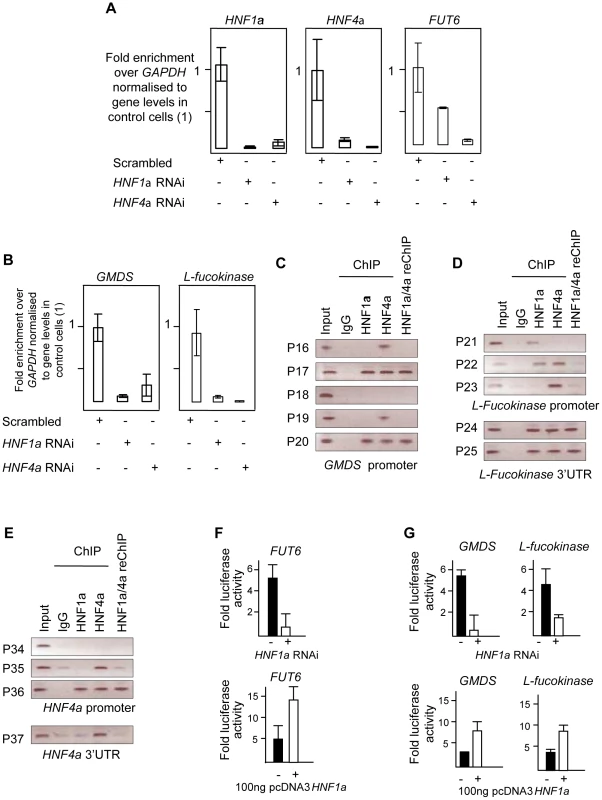
A). Real time PCR for HNF1α, HNF4α and FUT6 RNAs after HNF1α and HNF4α RNA interference (RNAi) in HepG2 cells. B). Real time PCR for GMDS and L-Fucokinase transcripts after HNF1α and HNF4α RNA interference (RNAi) in HepG2 cells. Expression levels were compared to GAPDH transcripts. All experiments were performed in triplicate. C). Chromatin immunoprecipitation (ChIP) analysis of regions as determined by the indicated primer sets for the GMDS promoter. D). ChIP analysis of regions as determined by the indicated primer sets for the L-Fucokinase promoter and 3′ UTRs. E). ChIP analysis of regions as determined by the indicated primer sets for the HNF4α promoter and 3′ UTRs. F). Reporter activity of the control FUT6 promoter upon HNF1α RNAi in HepG2 cells or pcDNA3-HNF1α expression in HEK293 cells. G). Reporter activity of the GMDS and L-Fucokinase promoters upon HNF1α RNAi in HepG2 cells or pcDNA3-HNF1α expression in HEK293 cells. Abbreviations: HNF: Hepatocyte nuclear factor; RNAi: RNA interference, GMDS: GDP-mannose 4,6-dehydratase; L-Fuc: L-Fucokinase; FUT: Fucosyltranferase. 3′UTR: 3′ un-translated region. reChIP: re-precipitation (or sequential) ChIP. See also Figure S2 for gene expression results in PANC1 cell line. As a positive control, the expression of FUT6, a known target of HNF4α in hepatocytes, was first analysed. The ablation of the HNF4α transcript abolished the expression of FUT6 in HepG2 cells confirming that the knockdown was effective. Surprisingly, knockdown of HNF1α resulted in 50% reduction in FUT6 transcript levels suggesting that HNF1α also regulate FUT6 expression in HepG2. This experiment suggested that our hypothesis may potentially explain and provide a direct link between HNF1α and the fucosylation genes. Therefore, we focused on the genes responsible for fucose biosynthesis, a rate limiting step in protein fucosylation. To this end, we analysed the expression of GMDS and L-Fucokinase which regulate de novo and salvage pathways of fucose synthesis, respectively. In HepG2 liver cells, HNF1α and HNF4α knockdown resulted in dramatic down-regulation in the expression of GMDS (91 and 77%, respectively) and L-Fucokinase (92 and 98%, respectively) (Figure 3B). In the pancreatic Panc1 cell lines, HNF4α RNAi resulted in a 70% decrease in GMDS and L-Fucokinase transcript levels (Figure 1). However, HNF1α RNAi led to a 90% reduction in GMDS transcript levels but did not affect L-Fucokinase mRNA abundance (Figure 1). This suggests that HNF1α regulates de novo synthesis of d-fucose in both cell lines tested (liver and pancreas), but only the salvage pathway in the liver cell line tested. HNF4α, on the other hand, regulates both pathways in both cell types tested.
We therefore focused on HNF1α direct transcriptional regulation of HNF4α, GMDS and L-Fucokinase in HepG2 cells. In order to investigate the latter, we performed a bioinformatics analysis to delineate in silico HNF1α and HNF4α binding sites. First, we assessed the conservation of regulatory elements (at the 5′ and 3′ end) between human and other primates as described previously [39]. It was recently shown the sites are not conserved between primates and rodents [40]. Second, the conserved regions were then mined for potential sites using ECR browser and the TRANSFAC database [41]. Finally, the potential sites were analysed manually to ascertain the likely binding sites based on homology to HNF1α and HNF4α consensus binding sites mined using genome-wide ChIP analyses [40], [42]. This limited our analysis to 5 sites (primer pairs P16 to P20, Figure 3C) in the GMDS promoter, 3 sites in the promoter (primer pairs P21 to P23, Figure 3D) as well as 2 sites at the 3′end (primer pairs P24 and P25, Figure 3D) of the L-Fucokinase gene and 3 sites in the promoter (primer pairs P34 to P36, Figure 3D) as well as a single site at the 3′ end (primer pair P37) of the HNF4α gene. The primer pairs are less than 1Kbps away from each other and some contained both HNF1α and HNF4α binding sites (or half sites) within the 200bps amplifiable regions.
Using these primer pairs, we performed chromatin immunoprecipitation (ChIP) assays to delineate the occupancy of these sites by HNF1α, HNF4α or both proteins as described earlier [43], [44].
In HepG2, both HNF1α and HNF4α bind the promoters of GMDS (P17, Figure 3C), L-Fucokinase (P22 although the two factors cannot be re-precipitated, Figure 3D) and HNF4α (P36, Figure 3E). Also, we show binding of HNF1α and HNF4α at the 3′UTR of L-Fucokinase as well as HNF4α binds the 3′UTR of HNF4α (Figure 3D and 3E, respectively). The interactions of these proteins is not affected by shearing as the primers acts as genomic controls for each other and no signal above background was apparent in the IgG isotype control antibody. Together, the data suggests a complex network of interactions between HNF4α and HNF1α to regulate fucose biosynthesis gene expression and point to a novel and an unappreciated role for HNF1α in regulating the two genes studied (GMDS and L-fucokinase). We further investigated the role of HNF1α in regulating the activity of the promoter regions bound by HNF factors (i.e. regions amplified by primer pairs P17, P22 and P37). We cloned these fragments into luciferase expressing vector (Promega's pGL4-basic) and assayed for reporter activity in two systems to delineate whether HNF1α is necessary to drive reporter expression (RNAi in HepG2 cells) and sufficient (expression of HNF1α in HEK293 cells that do not express endogenous HNF1α). Knockdown of HNF1α leads to a downregulation in the activity of both GMDS (5 fold reduction) and L-Fucokinase (2 fold reduction) promoter regions. Conversely, HNF1α overexpression leads to the induction of the luciferase activity in reporters driven by the two promoter regions. Put together, the expression data combined with the ChIP analysis and the reporter activity results strongly support a direct role for HNF1α in regulating the two key genes GMDS and L-Fucokinase that are responsible for de novo and salvage pathway of fucose synthesis.
HNF1α regulates multiple stages in protein fucosylation: (2) transcriptional regulation of core and antennary fucosyltransferases
After confirming the role of HNF1α in the biosynthesis of GDP-fucose, we analysed the role of HNF1α and HNF4α in the regulation of the expression of fucosyltransferase (FUT) genes (FUT3-11) in HepG2 and Panc1 cell lines to assess whether these hepatic factors regulate other stages of protein fucosylation. In HepG2 cells, HNF1α knockdown down-regulated the expression of all FUT genes (Figure 4A and 4B), except FUT8 whish was induced upon the loss of HNF1α (Figure 4C). HNF4α knockdown led to a statistically significant downregulation of FUT3, FUT5, FUT6, FUT10, FUT11 but not FUT7 or FUT9 (Figure 4A and 4B). Conversely, FUT8 expression levels increased 10 fold upon the loss of HNF4α ((Figure 4C). FUT4 was not expressed in HepG2 cells confirming earlier studies [45]. In the pancreas, all FUT genes were down-regulated (Figure 2) pointing to a key role for HNF1α in the regulation of fucosylation in the pancreas. Knockdown of HNF4α in liver cells reduced the expression of all FUT genes analysed except FUT7 or FUT9, but to a lesser extent than HNF1α knockdown (Figure 4A and 4B), however, FUT8 was again up-regulated (Figure 4A and 4B). The data supports a wider effect of HNF1α on the expression of the 8 fucosyltransferase genes compared to HNF4α. The data also suggests that HNF1α and HNF4α downregulate FUT8, which adds fucose to the core glycan, in contrast to all other FUTs that add fucose to the antennary arms of glycans [33]. We observed a rather high correlation between concentrations of antennary and core fucose in our population samples (r = 0.574, p = 4.01×10−85), indicating that the availability of the common substrate of both core and antennary FUTs, GDP-fucose, is a rate-limiting factor in protein fucosylation. It therefore appears that HNF1α not only enhances the activity of antennary FUTs but also, by down-regulating FUT8, increases the amount of GDP-fucose available for antennary fucosylation.
Fig. 4. HNF1α is major regulator of the fucosyltransferase genes. 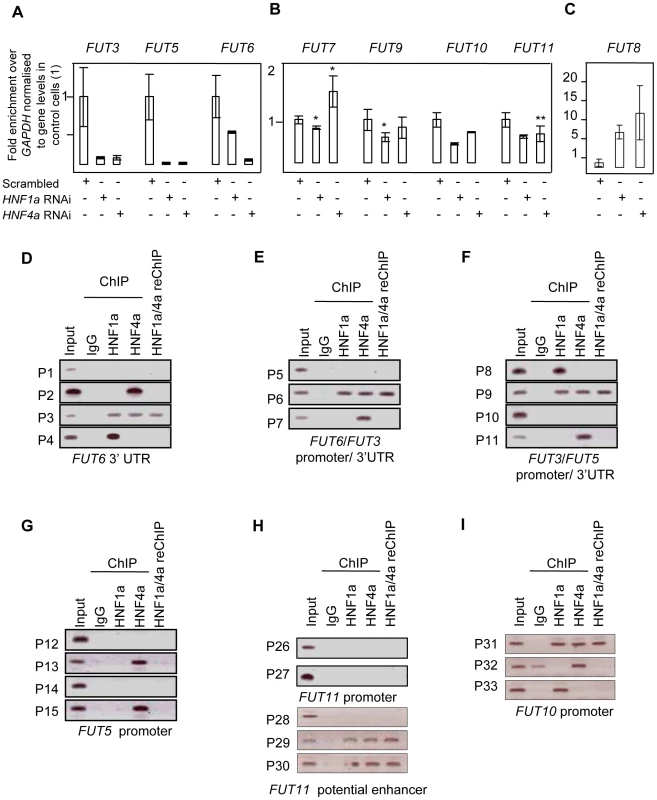
Real time PCR for FUT3, FUT5 and FUT6 RNAs (A); FUT7, FUT9, FUT10 and FUT11 (B); FUT8 (C) after HNF1α and HNF4α RNA interference (RNAi) in HepG2 cells. H) analyses show that Hnf1α (HNF1) and Hnf4α (HNF4) bind predicted regulatory elements for key fucosylation genes in liver cells. D), E), F), G), H) and I) Binding of HNF1α and HNF4α to predicted regulatory elements of the FUT3, FUT5, FUT6, FUT10 and FUT11. Abbreviations: the gene names are detailed in Figure 1; P1–P37: Primer sets 1 to 37; 3′UTR: 3′ un-translated region. reChIP: re-precipitation (or sequential) ChIP. FUT3, FUT6 and FUT5 were the only FUTs to be highly repressed (more than 3-fold) upon the loss of both HNF1α and HNF4α in liver cells (Figure 4A), suggesting a co-regulation of the three genes. In pancreatic cells, FUT3 and FUT6, but not FUT5 followed the same dynamics (Figure 2). FUT3 and FUT6 expression was not repressed upon HNF4α loss (Figure 2). This could be explained by a differential role for HNF4α in regulating FUT5 but not FUT3 or FUT6. These data suggest that HNF1α is the major regulator of the fucosylation pathway in both liver and pancreatic cell lines. While HNF4α also regulates the expression of these genes, its role is probably secondary to HNF1α. However, none of the genes studied here have previously been shown to be regulated in vivo by HNFs. Only the GMDS promoter has previously been shown to be chromatin immunoprecipitated with HNF4α antibody [28].
Bioinformatic analysis showed that FUT3, FUT5 and FUT6 are clustered in one locus in the human genome (see and Figure S3) [39]. This also corroborated our findings that FUT3, FUT5 and FUT6 are co-regulated downstream of HNF4α and HNF1α (Figure 4A). However, the FUT3/5/6 cluster was neither syntenic nor conserved in the mouse genome. We therefore focused on primate conservation only.
The promoter, intergenic and 3′ regulatory element conserved regions were analysed for HNF binding sites as detailed above for GMDS and L-Fucokinase. This analysis identified a limited number of sites in regulatory regions of FUT3, FUT5, FUT6, and FUT10. It did not identify any binding sites in silico in the FUT11 promoter, but a highly conserved long range enhancer was found within the ADK gene, that is 650 kb upstream and rich in HNF binding sites. We were unable to detect any HNF binding sequences within the FUT8 regulatory elements analysed.
Using ChIP, the binding of HNF1α and HNF4α to the putative response elements identified in silico was analysed. ChIP analysis showed that HNF1α and HNF4α bound multiple sequences within the predicted regulatory regions of multiple FUT genes, including FUT3, FUT5, FUT6, FUT10, FUT11 (Figure 4D–4I). HNF4α, and not HNF1α, bound the promoter of FUT5 (P13 and P15, Figure 4G). The unique binding of HNF4α to the promoter of FUT5 corroborated our findings that knockdown of HNF4α in pancreatic PANC1cells abolished the expression of FUT5 but not FUT3 or FUT6 (Figure 2).
Using re-precipitation (reChIP), we confirmed that both HNF transcription factors bound (i) the promoters of FUT3, FUT6 and FUT10 (Figure 4E, 4F, 4D, and 4I respectively); (ii) 3′UTRs of FUT6 (Figure 4D); and (iii) the long range enhancer 650 kb upstream of FUT11 (Figure 4H). This shows that HNF1α and HNF4α are potential regulators of the expression of these genes in vivo.
Discussion
By performing the first genome-wide association analysis (GWAS) of protein glycosylation we have taken the first steps towards the mapping of the complex network of genes that regulate protein N-glycosylation. We also identified common variants in three genes which exert a relatively strong influence on N-glycans in plasma (1–6% of variance explained). Importantly, all of the identified genes (FUT6, FUT8 and HNF1α) are involved in fucosylation, indicating that the addition of this unusual sugar may be a rate-limiting step in N-glycan synthesis. A gene encoding the transcription factor HNF1α, with previously unknown biological links to glycosylation, is shown to be strongly associated with the relative proportions of plasma N-glycans. The possible function(s) of HNF1α are a focus of intense current interest following its recently reported associations in GWAS with plasma C-reactive protein (CRP) [27], gamma-glutamyl transferase (GGT) [28], LDL cholesterol and apolipoprotein [29], [31] and coronary artery disease [29], [46]. Our analysis of gene knockdowns (RNAi) showed that HNF1α is an upstream regulator of several key genes involved in different stages of the fucosylation pathway. We have demonstrated that HNF1α binds the promoters in vivo, and is necessary and sufficient for the in vitro expression, of two genes, fucokinase and GMDS, required for de novo and salvage pathways of fucose synthesis, respectively (Figure 5C). Fucose synthesis is the rate limiting step for fucosylation in eukaryotes and prokaryotes [35] and, by up-regulating its synthesis, HNF1α increases the availability of fucose to the glycosylation machinery. In addition, HNF1α directly regulates the expression of several fucosyltransferase (FUT) genes (Figure 5D). Our results also demonstrate that HNF1α reciprocally regulates core versus antennary fucosylation; while activating FUTs involved in antennary fucosylation, HNF1α represses FUT8, which adds fucose to the core-GlcNAc. In this way, HNF1α decreases the consumption of GDP-fucose for core-fucosylation, and further increases the pool of fucose available for antennary fucosylation.
Fig. 5. HNF1α is at the heart of fucosylation regulation. 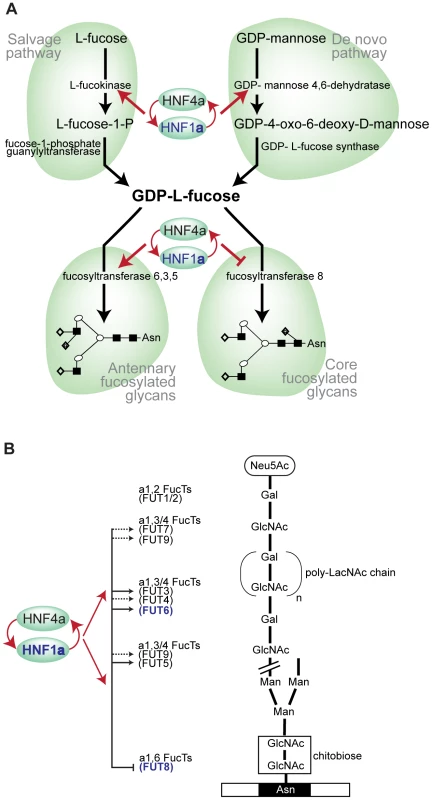
A). Diagram showing HNF1α and HNF4α regulation of pathways involved in fucose synthesis. B). Pathways of core (tri-mannosyl-chitobiose) and antennary fucosylation and their proposed regulation by the transcription factors Hnf1α and Hnf4α, encoded by the HNF1α and HNF4α genes respectively. Red arrows indicate genes that are targets of Hnf1α/Hnf4α. Solid lines show direct regulation and dashed lines show indirect regulation of the FUT genes shown. Genes shown in blue depict GWAS identified genes (HNF1α, FUT6 and FUT8). Abbreviations: Neu5Ac, N-Acetylneuraminic (sialic) acid; GlcNAc, N-acetylglucosamine; Man, mannose; Gal, galactose; poly-LacNAc, Galβ1,4GlcNAc (LacNAc) antennary chains; Asn, asparagine residue on glycosylated protein. Having shown this novel regulation of fucosylation genes, we scanned earlier genome wide studies for HNF factors to identify whether these genes were picked up. In fact, other genome wide studies support our findings. Boyd et al (2009) mapped HNF4α binding to both FUT2 and FUT5 in intestinal epithelial cells [42]. A genome wide prediction study for HNF4α functional binding sites identified FUT6, FUT5, FUT9, GMDS and FUT12 as functional targets [47].
We hypothesize that the role of HNF1α and its transcriptional co-factor HNF4α in the regulation of fucosylation is an essential part of mounting an acute phase response to infection in humans. Antennary fucosylation of their glycoprotein ligands is needed for binding of E-, L - and P-selectins to their target cells and the initiation of inflammation [48]. The decrease in fucosylation in the rare Leukocyte Adhesion Deficiency II (LAD II) impairs neutrophil function, which can be restored by oral administration of fucose [49]. Recently, we have reported moderate correlations between fucosylated plasma N-glycans and components of the acute phase response [15], which are also highly glycosylated and have high content of antennary-fucose [50]. Mounting a successful acute-phase response requires a rapid increase in the concentration of acute-phase proteins and this in turn is dependent on their efficient fucosylation. Our results indicate that fucosylation is a rate-limiting step in plasma protein glycosylation, and by both increasing de novo and salvage synthesis of GDP-fucose, up-regulation of antennary fucosyltransferases and down-regulation of core-fucosyltransferase, HNF1α appears to be a master regulator of this process. Variants in HNF1α and HNF4α genes were previously reported to be associated with concentrations of acute phase proteins in human plasma [24], [27]. Plasma protein fucosylation plays an important role in inflammation [22] and the central role of HNF1α in the regulation of multiple genes involved in fucosylation may be the molecular mechanism behind the reported association between common variants in HNF1α and inflammatory markers (such as CRP) as well as several diseases in which inflammation plays a key pathogenic role (such as coronary artery disease, inflammatory bowel disease and cancer).
Materials and Methods
Study populations and genotyping
All three populations recruited adult individuals within a community irrespective of any specific phenotype. The CROATIA-VIS and CROATIA-KORCULA studies are both cohorts from the Croatian Dalmatian islands recruited in 2003–2004 and 2007 respectively. The ORCADES study is ongoing with participants recruited from the Orkney islands in Scotland. Fasting blood samples were collected, biochemical and physiological measurements taken and questionnaires of medical history as well as lifestyle and environmental exposures collected following similar protocols.
The CROATIA-VIS study includes 1008 Croatians, aged 18–93 years, who were recruited from the villages of Vis and Komiza on the Dalmatian island of Vis during 2003 and 2004 within a larger genetic epidemiology program [32].
The CROATIA-KORCULA study includes 969 Croatians between the ages of 18 and 98 [32]. The field work was performed in 2007 in the eastern part of the island, targeting healthy volunteers from the town of Korčula and the villages of Lumbarda, Žrnovo and Račišće.
The Orkney Complex Disease Study (ORCADES) is an ongoing study in the isolated Scottish archipelago of Orkney [32]. Data for participants aged 18 to 100 years, from a subgroup of ten islands, were used for this analysis.
DNA samples were genotyped according to the manufacturer's instructions on Illumina Infinium SNP bead microarrays (HumanHap300v1 for the CROATIA-VIS cohort, HumanHap300v2 for the ORCADES cohort and HumanCNV370v1 for the CROATIA-KORCULA cohort). Genotypes were determined using Illumina BeadStudio software. Genotyping was successfully completed on 991 individuals from CROATIA-VIS, 953 from CROATIA-KORCULA and 761 from ORCADES.
Ethics statement
All studies conformed to the ethical guidelines of the 1975 Declaration of Helsinki and were approved by appropriate ethics boards with all respondents signing informed consent prior to participation.
Glycan release and labelling
The N-glycans from plasma sample (5 µl) proteins were released and labelled with 2-aminobenzamide (LudgerTag 2-AB labelling kit Ludger Ltd., Abingdon, UK) as described previously [13]. Labelled glycans were dried in a vacuum centrifuge and redissolved in known volume of water for further analysis.
Sialidase digestion
After initial HPLC quantification sialidase digestion was performed to improve measurement precision. Aliquots of the 2-AB-labeled glycan pool were dried down in 200-µl microcentrifuge tubes. To these, the following was added: 1 µl of 500 mM sodium acetate incubation buffer (pH 5.5), 1 µl (0.005 units) of ABS, Arthrobacter ureafaciens sialidase (releases α2–3, 6, 8 sialic acid, Prozyme) and H2O to make up to 10 µl. This was incubated overnight (16–18 h) at 37°C and then passed through a Micropure-EZ enzyme remover (Millipore, Billerica, MA, USA) before applying to the HPLC.
Hydrophilic interaction high-performance liquid chromatography (HILIC)
Released glycans were subjected to hydrophilic interaction high performance liquid chromatography (HILIC) on a 250×4.6 mm i.d. 5 µm particle packed TSKgel Amide 80 column (Tosoh Bioscience, Stuttgart, Germany) at 30°C with 50 mM formic acid adjusted to pH 4.4 with ammonia solution as solvent A and acetonitrile as solvent B. 60 min runs were on a 2795 Alliance separations module (Waters, Milford, MA). HPLCs were equipped with a Waters temperature control module and a Waters 2475 fluorescence detector set with excitation and emission wavelengths of 330 and 420 nm, respectively. The system was calibrated using an external standard of hydrolyzed and 2-AB-labeled glucose oligomerase from which the retention times for the individual glycans were converted to glucose units (GU) [51]. Glycans were analyzed on the basis of their elution positions and measured in glucose units then compared to reference values in NIBRT's “GlycoBase v3.0 ” database available at http://glycobase.nibrt.ie) for structure assignment [52].
HPLC analysis was performed partly in the National Institute for Biotechnology and Training (NIBRT) in Dublin, Ireland, and partly in the Glycobiology laboratory of Genos Ltd in Zagreb, Croatia. Both laboratories used the same columns and separation conditions. Duplicate analysis of a number of samples was performed and confirmed full reproducibility of the analytical results both within and between laboratories.
Glycan structural features
Levels of glycans sharing the same structural features were approximated by adding the structures having same characteristics: Core fucosylated glycans (FUC-C) = DG6/(DG5+DG6)*100; Antennary fucosylated glycans (FUC-A) = DG7/(DG5+DG7)*100.
Genotype and phenotype quality control
Genotyping quality control was performed using the same procedures for all cohorts. Individuals with a call rate less than 97% were removed as well as SNPs with a call rate less than 98% (95% for CROATIA-VIS), minor allele frequency less than 0.02% or Hardy-Weinburg equilibrium p-value less than 1×10−10. Differences in SNP call rate threshold were used to account for observed differences between genotyping arrays. 924 individuals passed all quality control thresholds from CROATIA-VIS, 898 from CROATIA-KORCULA and 737 from ORCADES.
Extreme outliers were removed for each glycan measure to account for errors in quantification and to remove individuals not representative of normal variation within the population. An individual was classified to be an extreme outlier if their measure for the trait was more than 3 interquartile distances away from the mean.
Genome-wide association analysis
Each trait was tested for normality within each cohort then the transformation that performed best for all cohorts was used. Models including sex, age and fibrinogen as covariates were tested for each cohort separately. Any covariate that was significant within any cohort was included as a covariate in the final model.
Genome-wide associations were performed for all glycan measures using the same transformation to normality and covariates for each cohort separately then combined in a meta-analysis. The “mmscore” function of the GenABEL package for R statistical software [53] was used for the association test under an additive model. This score test for family based association takes into account pedigree structure and allowed unbiased estimations of SNP allelic effect when relatedness is present between examinees [54]. The relationship matrix used in this analysis was generated by the “ibs” function of GenABEL which used IBS genotype sharing to determine the realised pairwise kinship coefficient. Meta-analysis was performed using the MetABEL package for R [53]. An association was considered statistically significant at the genome-wide level if the p-value for an individual SNP was less that 5×10−8 (based on Bonferroni correction to account for multiple testing). All identified SNPs that reached significance or seemed to be suggestive of significance were visualised using Haploview software [55].
Association interval
An associated interval for a region of interest was defined by determining the HapMap SNPs in linkage disequilibrium of r2 >0.5 with the most significantly associated SNP in the region using the web-based program SNAP [56]. The bounds of the associated interval were determined by the flanking HapMap recombination hotspots.
Haplotype analysis
Haplotype analysis was performed on “unrelated” individuals in each population separately to account for possible allele frequency and haplotype differences between populations. Individuals were considered to be unrelated with a kinship coefficient of less than 0.05 (first cousins once removed). This left 525 individuals in the CROATIA-VIS cohort, 568 in CROATIA-KORCULA and 263 in ORCADES. An EM based algorithm was used to infer haplotypes from genotypic data. The “scan.haplo” function of the GenABEL package for R [53], which calls the “haplo.score.slide” function of the haplo.stats package for R [57], was used to test a sliding window of 3 - and 5-SNP haplotypes across the associated interval. These results were compared to a single SNP model across the same region obtained using the “qtscore” function of the GenABEL package for R. A significant difference between haplotype and single-SNP analysis was determined using the Akaike information criterion [58].
ChIP identification of HNF binding sites in FUT genes
To establish whether HNF1α and HNF4α bind the regulatory elements of the fucosylation genes, their genomic loci were analysed using bioinformatics to identify HNF response elements. Conserved elements between human and mouse genomes [39] were analysed initially to delineate the binding sites of HNF1α and HNF4α using the TRANSFAC database and the ECR browser (http://ecrbrowser.dcode.org/). Primers for ChIP, reChIP and real-time PCR are listed in Text S1.
RNA interference
Production of the RNA duplexes for RNA interference was described in details earlier (Kittler et al., 2005). The target sequences (see Text S1) against HNF1α and HNF4α were designed using the siDESIGN Center (Dharmacon). The Trasnfection of HepG2 and PANC1 cells was carried out as described by (Yu et al., 2002).
Luciferase assays
The PCR products of ChIP primers (Sequences are detailed in the Text S1) were cloned into pGEM-T easy vector (Promega) and subcloned into pGL4 vectors (Promega) as described earlier (Essafi et al., 2005). pGL4-luc constructs (100 ng) and internal control of pRLTK (20 ng) renilla plasmid were transiently co-transfected into HepG2 and PANC1 cells (105) using the calcium phosphate co-precipitation. Cells were harvested 48 hr post-transfection for luciferase reporter assay using the Dual-Luciferase reporter assay system (Promega). The luciferase activity was normalized by Renilla luciferase activity. All assays were performed in three separate experiments done in triplicate.
ChIP
ChIP was carried out on HepG2 cells essentially as detailed earlier (Essafi et al., 2005). The antibodies used were HNF1α (sc-6547) and HNF4α (sc-6556) from Santa Cruz Biotechnology. The corresponding control IgG antibodies were from Sigma-Aldrich.
Real-time PCR
RNA isolation, cDNA synthesis and Real time PCR were performed as described earlier (Birkenkamp et al., 2007). PCR primer sequences are listed in Text S1.
Supporting Information
Zdroje
1. MarthJD
GrewalPK
2008 Mammalian glycosylation in immunity. Nature Reviews Immunology 8 874 887
2. DennisJW
LauKS
DemetriouM
NabiIR
2009 Adaptive Regulation at the Cell Surface by N-Glycosylation. Traffic 10 1569 1578
3. OhtsuboK
MarthJD
2006 Glycosylation in cellular mechanisms of health and disease. Cell 126 855 867
4. ApweilerR
HermjakobH
SharonN
1999 On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochimica et Biophysica Acta 1473 4 8
5. SkropetaD
2009 The effect of individual N-glycans on enzyme activity. Bioorganic & Medicinal Chemistry 17 2645 2653
6. HartGW
HousleyMP
SlawsonC
2007 Cycling of O-linked beta-N-acetylglucosamine on nucleocytoplasmic proteins. Nature 446 1017 1022
7. CrockerPR
PaulsonJC
VarkiA
2007 Siglecs and their roles in the immune system. Nature Reviews Immunology 7 255 266
8. BrownJR
CrawfordBE
EskoJD
2007 Glycan antagonists and inhibitors: a fount for drug discovery. Critical reviews in biochemistry and molecular biology 42 481 515
9. JaekenJ
2003 Komrower Lecture. Congenital disorders of glycosylation (CDG): it's all in it! J Inherit Metab Dis 26 99 118
10. FreezeHH
2006 Genetic defects in the human glycome. Nature Reviews Genetics 7 537 551
11. FreezeHH
2002 Human disorders in N-glycosylation and animal models. Biochimica et Biophysica Acta 1573 388 393
12. LaucG
RudanI
CampbellH
RuddPM
2010 Complex Genetic Regulation of Protein Glycosylation. Molecular Biosystems 6 329 335
13. RoyleL
CampbellMP
RadcliffeCM
WhiteDM
HarveyDJ
2008 HPLC-based analysis of serum N-glycans on a 96-well plate platform with dedicated database software. Analytical Biochemistry 376 1 12
14. RuddPM
RudanI
WrightAF
2009 High-Throughput Glycome Analysis Is Set To Join High-Throughput Genomics. Journal of Proteome Research 8 1105 1105
15. KneževićA
PolašekO
GornikO
RudanI
CampbellH
2009 Variability, Heritability and Environmental Determinants of Human Plasma N-Glycome. Journal of Proteome Research 8 694 701
16. GornikO
WagnerJ
PučićM
KneževićA
RedžićI
2009 Stability of N-glycan profiles in human plasma. Glycobiology 19 1547 1553
17. KneževićA
GornikO
PolašekO
PučićM
NovokmetM
2010 Effects of aging, body mass index, plasma lipid profiles, and smoking on human plasma N-glycans. Glycobiology 20 959 969
18. PučićM
PintoS
NovokmetM
KneževićA
GornikO
2010 Common aberrations from normal human N-glycan plasma profile. Glycobiology 20 970 975
19. BeckerDJ
LoweJB
2003 Fucose: biosynthesis and biological function in mammals. Glycobiology 13 41R 53R
20. TonettiM
SturlaL
BissoA
BenattiU
De FloraA
1996 Synthesis of GDP-L-fucose by the human FX protein. Journal of Biological Chemistry 271 27274 27279
21. JavaudC
DupuyF
MaftahA
JulienR
PetitJM
2003 The fucosyltransferase gene family: an amazing summary of the underlying mechanisms of gene evolution. Genetica 118 157 170
22. GornikO
LaucG
2008 Glycosylation of serum proteins in inflammatory diseases. Disease Markers 25 267 278
23. MiyoshiE
MoriwakiK
NakagawaT
2008 Biological function of fucosylation in cancer biology. Journal of Biochemistry 143 725 729
24. OdomDT
ZizlspergerN
GordonDB
BellGW
RinaldiNJ
2004 Control of pancreas and liver gene expression by HNF transcription factors. Science 303 1378 1381
25. YamagataK
FurutaH
OdaN
KaisakiPJ
MenzelS
1996 Mutations in the hepatocyte nuclear factor-4alpha gene in maturity-onset diabetes of the young (MODY1). Nature 384 458 460
26. YamagataK
OdaN
KaisakiPJ
MenzelS
FurutaH
1996 Mutations in the hepatocyte nuclear factor-1alpha gene in maturity-onset diabetes of the young (MODY3). Nature 384 455 458
27. ElliottP
ChambersJC
ZhangW
ClarkeR
HopewellJC
2009 Genetic Loci associated with C-reactive protein levels and risk of coronary heart disease. JAMA 302 37 48
28. YuanX
WaterworthD
PerryJR
LimN
SongK
2008 Population-based genome-wide association studies reveal six loci influencing plasma levels of liver enzymes. American Journal of Human Genetics 83 520 528
29. ReinerAP
GrossMD
CarlsonCS
BielinskiSJ
LangeLA
2009 Common coding variants of the HNF1A gene are associated with multiple cardiovascular risk phenotypes in community-based samples of younger and older European-American adults: the Coronary Artery Risk Development in Young Adults Study and The Cardiovascular Health Study. Circ Cardiovasc Genet 2 244 254
30. BarrettJC
LeeJC
LeesCW
PrescottNJ
AndersonCA
2009 Genome-wide association study of ulcerative colitis identifies three new susceptibility loci, including the HNF4A region. Nature Genetics 41 1330 1334
31. KathiresanS
WillerCJ
PelosoGM
DemissieS
MusunuruK
2009 Common variants at 30 loci contribute to polygenic dyslipidemia. Nature Genetics 41 56 65
32. VitartV
RudanI
HaywardC
GrayNK
FloydJ
2008 SLC2A9 is a newly identified urate transporter influencing serum urate concentration, urate excretion and gout. Nature Genetics 40 437 442
33. TaniguchiN
HonkeK
FukudaM
2002 Handbook of glycosyltransferases and related genes. Tokyo Springer Verlag
34. Brinkman-Van der LindenEC
MolliconeR
OriolR
LarsonG
Van den EijndenDH
1996 A missense mutation in the FUT6 gene results in total absence of alpha3-fucosylation of human alpha1-acid glycoprotein. J Biol Chem 271 14492 14495
35. MaB
Simala-GrantJL
TaylorDE
2006 Fucosylation in prokaryotes and eukaryotes. Glycobiology 16 158R 184R
36. HigaiK
MiyazakiN
AzumaY
MatsumotoK
2008 Transcriptional regulation of the fucosyltransferase VI gene in hepatocellular carcinoma cells. Glycoconjugate Journal 25 225 235
37. ServitjaJM
PignatelliM
MaestroMA
CardaldaC
BojSF
2009 Hnf1alpha (MODY3) controls tissue-specific transcriptional programs and exerts opposed effects on cell growth in pancreatic islets and liver. Molecular & Cellular Biology 29 2945 2959
38. BojSF
PetrovD
FerrerJ
2010 Epistasis of transcriptomes reveals synergism between transcriptional activators Hnf1alpha and Hnf4alpha. PLoS Genet 6 e1000970 doi:10.1371/journal.pgen.1000970
39. BejeranoG
SiepelAC
KentWJ
HausslerD
2005 Computational screening of conserved genomic DNA in search of functional noncoding elements. Nature Methods 2 535 545
40. OdomDT
DowellRD
JacobsenES
GordonW
DanfordTW
2007 Tissue-specific transcriptional regulation has diverged significantly between human and mouse. Nat Genet 39 730 732
41. LootsG
OvcharenkoI
2007 ECRbase: database of evolutionary conserved regions, promoters, and transcription factor binding sites in vertebrate genomes. Bioinformatics 23 122 124
42. BoydM
BressendorffS
MollerJ
OlsenJ
TroelsenJT
2009 Mapping of HNF4alpha target genes in intestinal epithelial cells. BMC Gastroenterol 9 68
43. BenkoS
FantesJA
AmielJ
KleinjanDJ
ThomasS
2009 Highly conserved non-coding elements on either side of SOX9 associated with Pierre Robin sequence. Nat Genet 41 359 364
44. EssafiA
Fernandez de MattosS
HassenYA
SoeiroI
MuftiGJ
2005 Direct transcriptional regulation of Bim by FoxO3a mediates STI571-induced apoptosis in Bcr-Abl-expressing cells. Oncogene 24 2317 2329
45. WithersDA
HakomoriSI
2000 Human alpha (1,3)-fucosyltransferase IV (FUTIV) gene expression is regulated by elk-1 in the U937 cell line. Journal of Biological Chemistry 275 40588 40593
46. ErdmannJ
GrosshennigA
BraundPS
KonigIR
HengstenbergC
2009 New susceptibility locus for coronary artery disease on chromosome 3q22.3. Nature Genetics 41 280 282
47. KelAE
NiehofM
MatysV
ZemlinR
BorlakJ
2008 Genome wide prediction of HNF4alpha functional binding sites by the use of local and global sequence context. Genome Biol 9 R36
48. BrandleyBK
KisoM
AbbasS
NikradP
SrivasatavaO
1993 Structure-function studies on selectin carbohydrate ligands. Modifications to fucose, sialic acid and sulphate as a sialic acid replacement. Glycobiology 3 633 641
49. LuhnK
MarquardtT
HarmsE
VestweberD
2001 Discontinuation of fucose therapy in LADII causes rapid loss of selectin ligands and rise of leukocyte counts. Blood 97 330 332
50. KleinA
2008 Human total serum N-glycome. Advances in Clinical Chemistry 46 51 85
51. RoyleL
RadcliffeCM
DwekRA
RuddPM
2006 Detailed structural analysis of N-glycans released from glycoproteins in SDS-PAGE gel bands using HPLC combined with exoglycosidase array digestions. Methods Mol Biol 347 125 143
52. CampbellMP
RoyleL
RadcliffeCM
DwekRA
RuddPM
2008 GlycoBase and autoGU: tools for HPLC-based glycan analysis. Bioinformatics 24 1214 1216
53. AulchenkoYS
RipkeS
IsaacsA
van DuijnCM
2007 GenABEL: an R library for genome-wide association analysis. Bioinformatics 23 1294 1296
54. ChenWM
AbecasisGR
2007 Family-based association tests for genomewide association scans. Am J Hum Genet 81 913 926
55. BarrettJC
FryB
MallerJ
DalyMJ
2005 Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21 263 265
56. JohnsonAD
HandsakerRE
PulitSL
NizzariMM
O'DonnellCJ
2008 SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics 24 2938 2939
57. SchaidDJ
RowlandCM
TinesDE
JacobsonRM
PolandGA
2002 Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet 70 425 434
58. AkaikeH
1974 New Look at Statistical-Model Identification. Ieee Transactions on Automatic Control AC19 716 723
Štítky
Genetika Reprodukční medicína
Článek Genome-Wide Interrogation of Mammalian Stem Cell Fate Determinants by Nested Chromosome DeletionsČlánek Season of Conception in Rural Gambia Affects DNA Methylation at Putative Human Metastable EpiallelesČlánek A Quantitative Systems Approach Reveals Dynamic Control of tRNA Modifications during Cellular StressČlánek Reduction of Protein Translation and Activation of Autophagy Protect against PINK1 Pathogenesis inČlánek The Loss of PGAM5 Suppresses the Mitochondrial Degeneration Caused by Inactivation of PINK1 inČlánek Cleavage of Phosphorothioated DNA and Methylated DNA by the Type IV Restriction Endonuclease ScoMcrAČlánek Competitive Repair by Naturally Dispersed Repetitive DNA during Non-Allelic Homologous Recombination
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2010 Číslo 12
-
Všechny články tohoto čísla
- Genome-Wide Interrogation of Mammalian Stem Cell Fate Determinants by Nested Chromosome Deletions
- Whole-Genome and Chromosome Evolution Associated with Host Adaptation and Speciation of the Wheat Pathogen
- Association of Variants at 1q32 and with Ankylosing Spondylitis Suggests Genetic Overlap with Crohn's Disease
- Initiator Elements Function to Determine the Activity State of BX-C Enhancers
- Identification of Genes Required for Neural-Specific Glycosylation Using Functional Genomics
- A Young Duplicate Gene Plays Essential Roles in Spermatogenesis by Regulating Several Y-Linked Male Fertility Genes
- The EpsE Flagellar Clutch Is Bifunctional and Synergizes with EPS Biosynthesis to Promote Biofilm Formation
- Histone H2A C-Terminus Regulates Chromatin Dynamics, Remodeling, and Histone H1 Binding
- Season of Conception in Rural Gambia Affects DNA Methylation at Putative Human Metastable Epialleles
- A Quantitative Systems Approach Reveals Dynamic Control of tRNA Modifications during Cellular Stress
- GC-Rich Sequence Elements Recruit PRC2 in Mammalian ES Cells
- A Single Enhancer Regulating the Differential Expression of Duplicated Red-Sensitive Opsin Genes in Zebrafish
- Investigation and Functional Characterization of Rare Genetic Variants in the Adipose Triglyceride Lipase in a Large Healthy Working Population
- Reduction of Protein Translation and Activation of Autophagy Protect against PINK1 Pathogenesis in
- Noisy Splicing Drives mRNA Isoform Diversity in Human Cells
- The Loss of PGAM5 Suppresses the Mitochondrial Degeneration Caused by Inactivation of PINK1 in
- Thymus-Associated Parathyroid Hormone Has Two Cellular Origins with Distinct Endocrine and Immunological Functions
- An ABC Transporter Mutation Is Correlated with Insect Resistance to Cry1Ac Toxin
- Role of Individual Subunits of the CSN Complex in Regulation of Deneddylation and Stability of Cullin Proteins
- The C-Terminal Domain of the Bacterial SSB Protein Acts as a DNA Maintenance Hub at Active Chromosome Replication Forks
- The DNA Damage Response Pathway Contributes to the Stability of Chromosome III Derivatives Lacking Efficient Replicators
- Cleavage of Phosphorothioated DNA and Methylated DNA by the Type IV Restriction Endonuclease ScoMcrA
- LaeA Control of Velvet Family Regulatory Proteins for Light-Dependent Development and Fungal Cell-Type Specificity
- Competitive Repair by Naturally Dispersed Repetitive DNA during Non-Allelic Homologous Recombination
- Distinct Functions for the piRNA Pathway in Genome Maintenance and Telomere Protection
- MOS11: A New Component in the mRNA Export Pathway
- Self-Mating in the Definitive Host Potentiates Clonal Outbreaks of the Apicomplexan Parasites and
- A Role for ATF2 in Regulating MITF and Melanoma Development
- Ancestral Regulatory Circuits Governing Ectoderm Patterning Downstream of Nodal and BMP2/4 Revealed by Gene Regulatory Network Analysis in an Echinoderm
- Cancer and Neurodegeneration: Between the Devil and the Deep Blue Sea
- Functional Comparison of Innate Immune Signaling Pathways in Primates
- Linking Crohn's Disease and Ankylosing Spondylitis: It's All about Genes!
- Genomics Meets Glycomics—The First GWAS Study of Human N-Glycome Identifies HNF1α as a Master Regulator of Plasma Protein Fucosylation
- Continuous and Periodic Expansion of CAG Repeats in Huntington's Disease R6/1 Mice
- Expression of Linear and Novel Circular Forms of an -Associated Non-Coding RNA Correlates with Atherosclerosis Risk
- Endocytic Sorting and Recycling Require Membrane Phosphatidylserine Asymmetry Maintained by TAT-1/CHAT-1
- Histone Deacetylases Suppress CGG Repeat–Induced Neurodegeneration Via Transcriptional Silencing in Models of Fragile X Tremor Ataxia Syndrome
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Functional Comparison of Innate Immune Signaling Pathways in Primates
- Expression of Linear and Novel Circular Forms of an -Associated Non-Coding RNA Correlates with Atherosclerosis Risk
- Genome-Wide Interrogation of Mammalian Stem Cell Fate Determinants by Nested Chromosome Deletions
- Histone H2A C-Terminus Regulates Chromatin Dynamics, Remodeling, and Histone H1 Binding
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



