-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaGC-Rich Sequence Elements Recruit PRC2 in Mammalian ES Cells
Polycomb proteins are epigenetic regulators that localize to developmental loci in the early embryo where they mediate lineage-specific gene repression. In Drosophila, these repressors are recruited to sequence elements by DNA binding proteins associated with Polycomb repressive complex 2 (PRC2). However, the sequences that recruit PRC2 in mammalian cells have remained obscure. To address this, we integrated a series of engineered bacterial artificial chromosomes into embryonic stem (ES) cells and examined their chromatin. We found that a 44 kb region corresponding to the Zfpm2 locus initiates de novo recruitment of PRC2. We then pinpointed a CpG island within this locus as both necessary and sufficient for PRC2 recruitment. Based on this causal demonstration and prior genomic analyses, we hypothesized that large GC-rich elements depleted of activating transcription factor motifs mediate PRC2 recruitment in mammals. We validated this model in two ways. First, we showed that a constitutively active CpG island is able to recruit PRC2 after excision of a cluster of activating motifs. Second, we showed that two 1 kb sequence intervals from the Escherichia coli genome with GC-contents comparable to a mammalian CpG island are both capable of recruiting PRC2 when integrated into the ES cell genome. Our findings demonstrate a causal role for GC-rich sequences in PRC2 recruitment and implicate a specific subset of CpG islands depleted of activating motifs as instrumental for the initial localization of this key regulator in mammalian genomes.
Published in the journal: . PLoS Genet 6(12): e32767. doi:10.1371/journal.pgen.1001244
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1001244Summary
Polycomb proteins are epigenetic regulators that localize to developmental loci in the early embryo where they mediate lineage-specific gene repression. In Drosophila, these repressors are recruited to sequence elements by DNA binding proteins associated with Polycomb repressive complex 2 (PRC2). However, the sequences that recruit PRC2 in mammalian cells have remained obscure. To address this, we integrated a series of engineered bacterial artificial chromosomes into embryonic stem (ES) cells and examined their chromatin. We found that a 44 kb region corresponding to the Zfpm2 locus initiates de novo recruitment of PRC2. We then pinpointed a CpG island within this locus as both necessary and sufficient for PRC2 recruitment. Based on this causal demonstration and prior genomic analyses, we hypothesized that large GC-rich elements depleted of activating transcription factor motifs mediate PRC2 recruitment in mammals. We validated this model in two ways. First, we showed that a constitutively active CpG island is able to recruit PRC2 after excision of a cluster of activating motifs. Second, we showed that two 1 kb sequence intervals from the Escherichia coli genome with GC-contents comparable to a mammalian CpG island are both capable of recruiting PRC2 when integrated into the ES cell genome. Our findings demonstrate a causal role for GC-rich sequences in PRC2 recruitment and implicate a specific subset of CpG islands depleted of activating motifs as instrumental for the initial localization of this key regulator in mammalian genomes.
Introduction
Polycomb proteins are epigenetic regulators required for proper gene expression patterning in metazoans. The proteins reside in two main complexes, termed Polycomb repressive complex 1 and 2 (PRC1 and PRC2). PRC2 catalyzes histone H3 lysine 27 tri-methylation (K27me3), while PRC1 catalyzes histone H2A ubiquitination and mediates chromatin compaction [1], [2]. PRC1 and PRC2 are initially recruited to target loci in the early embryo where they subsequently mediate lineage-specific gene repression. In embryonic stem (ES) cells, the complexes localize to thousands of genomic sites, including many developmental loci [3]–[5]. These target loci are not yet stably repressed, but instead maintain a “bivalent” chromatin state, with their chromatin enriched for the activating histone mark, H3 lysine 4 tri-methylation (K4me3), together with the repressive K27me3 [6], [7]. In the absence of transcriptional induction, PRC1 and PRC2 remain at target loci and mediate repression through differentiation. The mechanisms that underlie stable association of the complexes remain poorly understood, but likely involve interactions with the modified histones [8]–[12].
Proper localization of PRC1 and PRC2 in the pluripotent genome is central to the complex developmental regulation orchestrated by these factors. However, the sequence determinants that underlie this initial landscape remain obscure. Polycomb recruitment is best understood in Drosophila, where sequence elements termed Polycomb response elements (PREs) are able to direct these repressors to exogenous locations [13]. PREs contain clusters of motifs recognized by DNA binding proteins such as Pho, Zeste and GAGA, which in turn recruit PRC2 [14]–[17]. Despite extensive study, neither PRE sequence motifs nor binding profiles of PRC2-associated DNA binding proteins are sufficient to fully predict PRC2 localization in the Drosophila genome [1], [16], [18], [19].
While protein homologs of PRC1 and PRC2 are conserved in mammals, DNA sequence homologs of Drosophila PREs appear to be lacking in mammalian genomes [13]. Moreover, it remains controversial whether the DNA binding proteins associated with PRC2 in Drosophila have functional homologs in mammals. The most compelling candidate has been YY1, a Pho homolog that rescues gene silencing when introduced into Pho-deficient Drosophila embryos [20]. YY1 has been implicated in PRC2-dependent silencing of tumor suppressor genes in human cancer cells [21]. However, this transcription factor has also been linked to numerous other functions, including imprinting, DNA methylation, B-cell development and ribosomal protein gene transcription [22]–[26].
Recently, researchers identified two DNA sequence elements able to confer Polycomb repression in mammalian cells. Sing and colleagues identified a murine PRE-like element that regulates the MafB gene during neural development [27]. These investigators defined a critical 1.5 kb sequence element that is able to recruit PRC1, but not PRC2 in a transgenic cell assay. Woo and colleagues identified a 1.8 kb region of the human HoxD cluster that recruits both PRC1 and PRC2 and represses a reporter construct in mesenchymal tissues [28]. Both groups note that their respective PRE regions contain YY1 motifs. Mutation of the YY1 sites in the HoxD PRE resulted in loss of PRC1 binding and partial loss of repression, while comparatively, deletion of a separate highly conserved region from this element completely abrogated PRC1 and PRC2 binding as well as repression [28].
In addition to these locus-specific investigations, genomic studies have sought to define PRC2 targets and determinants in a systematic fashion. The Ezh2 and Suz12 subunits have been mapped in mouse and human ES cells by chromatin immunoprecipitation and microarrays (ChIP-chip) or high-throughput sequencing (ChIP-Seq) [3]–[5],[29]. Such studies have highlighted global correlations between PRC2 targets and CpG islands [5], [30] as well as highly-conserved genomic loci [4], [7], [31]. Recently, Jarid2 has been shown to associate with PRC2 and to be required for proper genome-wide localization of the complex [32]–[35]. Intriguingly, Jarid2 contains an ARID and a Zinc-finger DNA-binding domain. However, it is unclear how Jarid2 could account for PRC2 targeting given the lack of sequence specificity and the low affinity of its DNA binding domains [33], [36]. In summary, a variety of sequence elements including CpG islands, conserved elements and YY1 motifs have been implicated in Polycomb targeting in mammalian cells. Causality has only been demonstrated in two specific instances and a unifying view of the determinants of Polycomb recruitment remains elusive.
Here we present the identification of multiple sequence elements capable of recruiting PRC2 in mammalian ES cells. This was achieved through an experimental approach in which engineered bacterial artificial chromosomes (BACs) were stably integrated into the ES cell genome. Evaluation of a series of modified BACs specifically identified a 1.7 kb DNA fragment that is both necessary and sufficient for PRC2 recruitment. The fragment does not share sequence characteristics of Drosophila PREs and lacks YY1 binding sites, but rather corresponds to an annotated CpG island. Based on this result and a genome-wide analysis of PRC2 target sequences we hypothesized that large GC-rich sequence elements lacking transcriptional activation signals represent general PRC2 recruitment elements. We tested this model by assaying the following DNA sequences: (i) a ‘housekeeping’ CpG island which was re-engineered by removal of a cluster of activating motifs; and (ii) two large GC-rich intervals from the E. coli genome that satisfy the criteria of mammalian CpG islands. We found that all three GC-rich elements robustly recruit PRC2 in ES cells. We propose that a class of CpG islands distinguished by a lack of activating motifs play causal roles in the initial localization of PRC2 and the subsequent coordination of epigenetic controls during mammalian development.
Results
Recruitment of Polycomb repressors to a bacterial artificial chromosome integrated into ES cells
To identify DNA sequences capable of recruiting Polycomb repressors in mammalian cells, we engineered human BACs that correspond to genomic regions bound by these proteins in human ES cells.
We initially targeted a region of the human Zfpm2 (hZfpm2) locus, which encodes a developmental transcription factor involved in heart and gonad development [37]. In ES cells, the endogenous locus recruits PRC1 and PRC2, and is enriched for the bivalent histone modifications, K4me3 and K27me3 (Figure 1A). We used recombineering to engineer a 44 kb BAC containing this locus and a neomycin selection marker. The modified BAC was electroporated into mouse ES cells, and individual transgenic ES cell colonies containing the full length BAC were expanded (Figure S1). Fluorescent in situ hybridization (FISH) confirmed integration at a single genomic location (Figure S2).
Fig. 1. Recruitment of Polycomb repressors to a BAC integrated into ES cells. 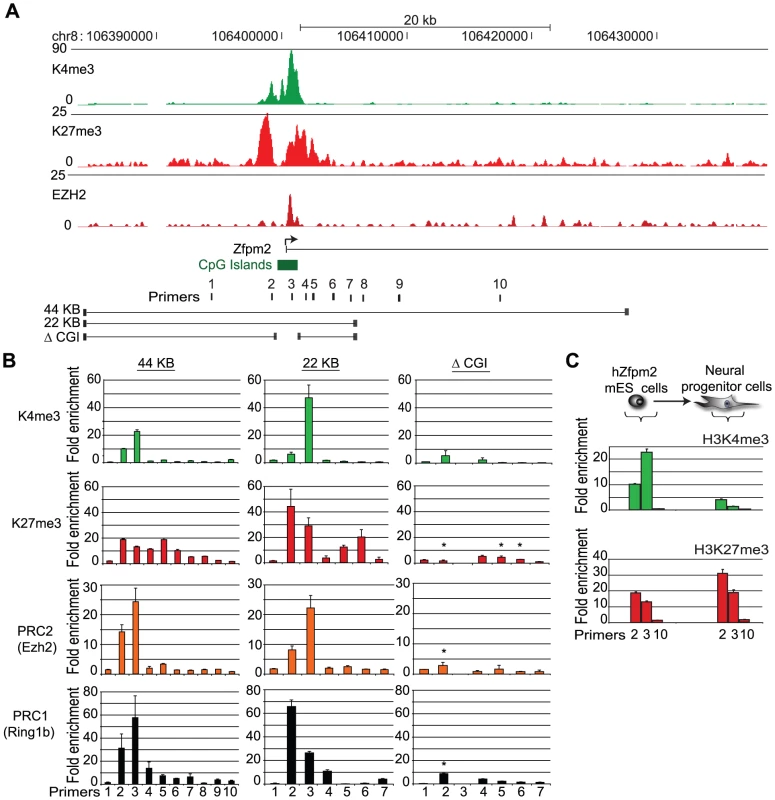
(A) ChIP-Seq tracks depict enrichment of K27me3 (the modification catalyzed by PRC2), Ezh2 (the enzymatic component of PRC2), and K4me3 across the endogenous hZfpm2 locus in human ES cells. Primers and constructs used in this study are indicated below the gene track. (B) BAC constructs from (A) containing the hZfpm2 locus were stably integrated into mouse ES cells. ChIP-qPCR enrichments are shown for K4me3, K27me3, Ezh2, and the PRC1 component Ring1b across the locus. The integrated locus adopts a ‘bivalent’ chromatin state with K27me3 and K4me3 in all constructs except the ΔCGI BAC. The locations of PCR amplicons are designated on the horizontal axis. (C) Transgenic ES cells differentiated along a neural lineage show enrichment for K27me3 but not K4me3 in NP cells. Error bars show standard error of the mean (SEM) for n = 3 (44 kb) or n = 2 (22 kb; ΔCGI) biological replicates. We used ChIP and quantitative PCR (ChIP-qPCR) with human specific primers to examine the chromatin state of the newly incorporated hZfpm2 locus. This analysis revealed strong enrichment for K27me3 and K4me3 (Figure 1B). In addition, we explicitly tested for direct binding of the Polycomb repressive complexes using antibody against the PRC1 subunit, Ring1B, or the PRC2 subunit, Ezh2. We detected robust enrichment for both complexes in the vicinity of the hZfpm2 gene promoter (Figure 1B). To confirm this result and eliminate the possibility of integration site effects, we tested two additional transgenic hZfpm2 ES cell clones with unique integration sites as well as a fourth transgenic ES cell line containing a distinct Polycomb target locus, Pax5. In each case, we observed a bivalent chromatin state analogous to the endogenous loci (Figure S3). Similar to endogenous bivalent CpG islands, we found the Zfpm2 CpG island was DNA hypomethylated (Figure S4). These results suggest that DNA sequence is sufficient to initiate de novo recruitment of Polycomb in ES cells.
The Zfpm2 BAC maintains K27me3 through ES cell differentiation
A key function of Polycomb repressors is to maintain a repressive chromatin state through cellular differentiation. To determine if the integrated BAC is capable of maintaining K27me3, the hZfpm2 transgenic ES cells were differentiated to neural progenitor (NP) cells in vitro [38]. ChIP-qPCR analysis revealed continued enrichment of K27me3 but loss of K4me3 (Figure 1C), a pattern frequently observed at endogenous loci that are not activated during differentiation [39].This indicates that DNA sequence at the hZfpm2 locus is sufficient to initiate K27me3 chromatin modifications in ES cells, and maintain the repressive chromatin state through neural differentiation.
Distinguishing Polycomb recruiting sequences in the Zfpm2 BAC
We next sought to define the sequences within the hZfpm2 BAC required for recruitment of Polycomb repressors. First, we re-engineered the 44 kb hZfpm2 BAC to remove 20 kb of flanking sequences that contained distal non-coding conserved sequence elements (Figure 1A). When we integrated the resulting 22 kb construct into ES cells we found that it robustly enriches for PRC1, PRC2, K4me3 and K27me3 (Figure 1B). Hence, these particular distal elements do not appear to be required for the recruitment of the complexes. Next, we considered the necessity of the CpG island which corresponds to the peak of Ezh2 enrichment in ChIP-Seq profiles (Figure 1A). We excised a 1.7 kb fragment containing the CpG island, and integrated the resulting BAC (ΔCGI) into ES cells. The ΔCGI BAC failed to recruit PRC1 or PRC2, and showed significantly reduced K27me3 levels relative to the other constructs (Figure 1B). This suggests that the CpG island is essential for recruitment of Polycomb proteins to the hZfpm2 locus.
A 1.7 kb CpG island is sufficient to recruit PRC2 to an exogenous locus
We next asked whether the hZfpm2 CpG island is sufficient to recruit Polycomb repressors to an exogenous locus. To test this, we selected an unremarkable gene desert region on human chromosome 1 that shows no enrichment for PRC1, PRC2 or K27me3 in ES cells (Figure 2A). We also verified that the gene desert BAC alone does not show any enrichment for K27me3 or Ezh2 when integrated into ES cells (Figure 2B). Using recombineering, we inserted the 1.7 kb sequence that corresponds to the hZfpm2 CpG island into the gene desert BAC. The resulting construct was integrated into mouse ES cells and three independent clones were evaluated. ChIP-qPCR analysis revealed strong enrichment for K27me3, K4me3 and PRC2 over the inserted CpG island (Figure 2C, Figure S5). In contrast, we observed relatively little enrichment for the PRC1 subunit Ring1B (Figure 2C). We confirmed the specificity of these enrichments with primers that span the boundary between the insertion and adjacent gene desert sequence. Notably, K27me3 enrichment was detected across the gene desert locus up to 2.5 kb from the inserted CpG island (Figure 2C). This indicates that the localized CpG island can initiate K27me3 that then spreads into adjacent sequence. Lastly we found no YY1 enrichment across the CpG island by ChIP-qPCR (Figure S5). Together, these data suggest that the hZfpm2 CpG island contains the necessary signals for PRC2 recruitment but is insufficient to confer robust PRC1 association.
Fig. 2. A 1.7 kb GC-rich sequence element is sufficient to recruit PRC2. 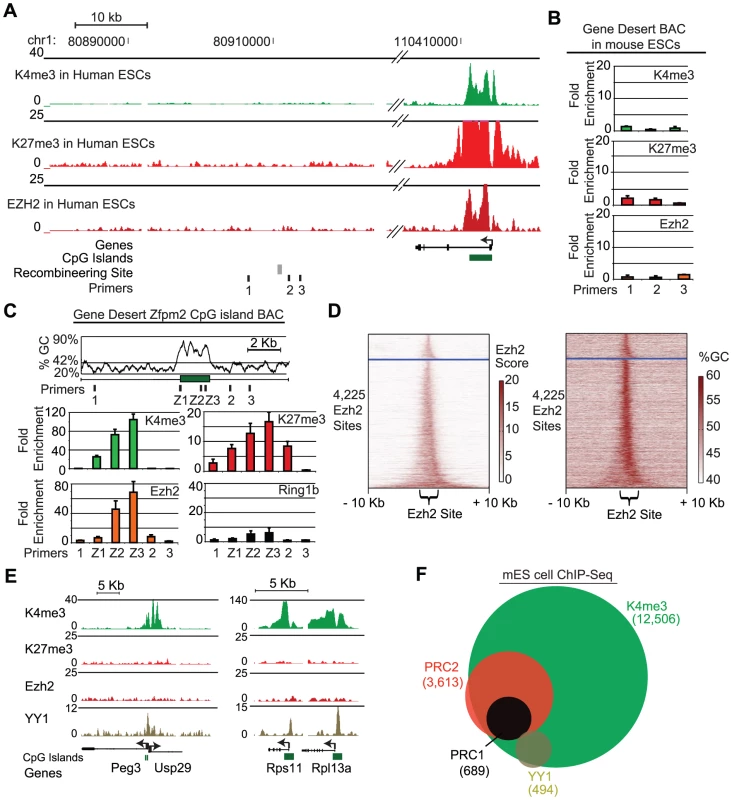
(A) ChIP-Seq tracks show no enrichment for K4me3, K27me3 or Ezh2 in human ES cells across the gene desert region. For comparison a nearby locus is shown. The recombineering site and primers used in this study are indicated below the tracks. (B) The gene desert BAC shows no enrichment of K4me3, K27me3 or PRC2 upon integration in mouse ES cells. (C) The hZfpm2 CpG island is depicted at the site of insertion into the gene desert BAC, along with the corresponding GC percentage (42% indicates genome average) and primers used for qPCR. Underlying plots represent ChIP-qPCR enrichment of K4me3, K27me3, PRC2 (Ezh2), and PRC1 (Ring1b) at the indicated sites (n = 2 biological replicates). (D) Heat maps show Ezh2 ChIP-Seq signal (left panel) or GC-percentage (right panel) for all Ezh2-bound regions in ES cells. Each row depicts a 20 kb region centered on the Ezh2 signal. Rows are separated into two groups based on whether the site overlaps a CpG island (below the blue line) and are then sorted based on the width of Ezh2 enrichment (see Methods). (E) ChIP-Seq was used to profile the mammalian Pho homolog YY1 in mouse ES cells. Genome browser views show ChIP-Seq enrichment signals for K4me3, K27me3, Ezh2 and YY1 for YY1 target loci. (F) Venn diagram shows overlap of K4me3, Ezh2, Ring1b, and YY1 at promoters in mES cells. Consideration of sequence determinants of PRC2 recruitment
The functionality of a CpG island in PRC2 recruitment is consistent with prior observations that a majority of PRC2 sites in ES cells correspond to CpG islands [4], [5] and with the striking correlation between intensity of PRC2 binding and the GC-richness of the underlying sequence (Figure 2D). We therefore considered whether specific signals within the Zfpm2 CpG island might underlie its capacity to recruit PRC2.
First, we searched for sequence motifs analogous to the PREs that recruit PRC2 in Drosophila. We focused on motifs recognized by YY1, the nearest mammalian homolog of the Drosophila recruitment proteins. Notably, both of the recently described mammalian PREs contain YY1 motifs [27], [28]. The 44 kb hZfpm2 BAC contains 11 instances of the consensus YY1 motif. However, none of these reside within the CpG island (Figure S6) (see Methods). We also examined YY1 binding directly in ES cells and NS cells using ChIP-Seq. Consistent with prior reports, YY1 binding is evident at the 5′ ends of many highly expressed genes, including those encoding ribosomal proteins, and is also seen at the imprinted Peg3 locus (Figure 2E, Table S1) [26]. However, no YY1 enrichment is evident at the Zfpm2 locus. Moreover, at a global level, YY1 shows almost no overlap with PRC2 or PRC1, but instead co-localizes with genomic sites marked exclusively by K4me3 (Figure 2F, Figure S6, and Table S1). Thus, although YY1 may contribute to Polycomb-mediated repression through distal interactions or in trans, it does not appear to be directly involved in PRC2 recruitment in ES cells.
We previously reported that CpG islands bound by PRC2 in ES cells could be predicted based on a relative absence of activating transcription factor motifs (AMs) in their DNA sequence [5]. We reasoned that transcriptional inactivity afforded by this absence of AMs is a requisite for PRC2 association [40], [41]. This could explain why PRC2 is absent from a majority of CpG islands, many of which are found at highly active promoters. Consistent with this model, when we examined a recently published RNA-Seq dataset for poly-adenylated transcripts in ES cells, we found that virtually all of the high-CpG promoters (HCPs) lacking Ezh2 are detectably transcribed (Figure S7). The small proportion of HCPs that are neither Ezh2-bound nor transcribed may reflect false-negatives in the ChIP-Seq or RNA-Seq data. Alternatively, these HCPs tend to correspond to CpG islands with relatively low GC-contents and lengths and may therefore have insufficient GC-richness to promote PRC2 binding (Figure S7). Thus, correlative analyses implicate large GC-rich elements that lack transcriptional activation signals as general PRC2 recruitment elements in mammals.
Sufficiency of GC-rich sequences for PRC2 recruitment
To obtain direct experimental support for the general sufficiency of large GC-rich elements lacking AMs in PRC2 recruitment, we carried out the following experiments. First, we tested whether a K4me3-only CpG island could be turned into a PRC2 recruitment element by removing activating motifs. We targeted a 1.3 kb CpG island that overlaps the promoters of two ubiquitously expressed genes – Arl3 and Sfxn2. Neither gene carries K27me3 in ES cells, or in any other cell type tested (Figure S8, and data not shown). This CpG island was selected as it has many conserved AMs clustered in one half of the island (Figure 3A). We hypothesized that the portion of the Arl3/Sfxn2 CpG island lacking AMs would, in isolation, lack active transcription and recruit PRC2. In contrast, we predicted that the half containing multiple AMs would lack Polycomb. To test this, we generated two additional BAC constructs containing the respective portions of the Arl3/Sfxn2 CpG island positioned within the gene desert, and integrated these constructs into ES cells (Figure 3A). ChIP-qPCR shows that the portion of the CpG island lacking AMs is able to recruit PRC2 and becomes enriched for K27me3 (Figure 3B). In contrast, the AM-containing portion shows no enrichment for K27me3 or Ezh2, but is instead marked exclusively by K4me3, similar to the endogenous human locus (Figure 3C, Figure S8). Thus, a GC-rich sequence element with no known requirement for Polycomb regulation can recruit PRC2 when isolated from activating sequence features.
Fig. 3. Removal of activating transcription factor motifs initiates PRC2 recruitment. 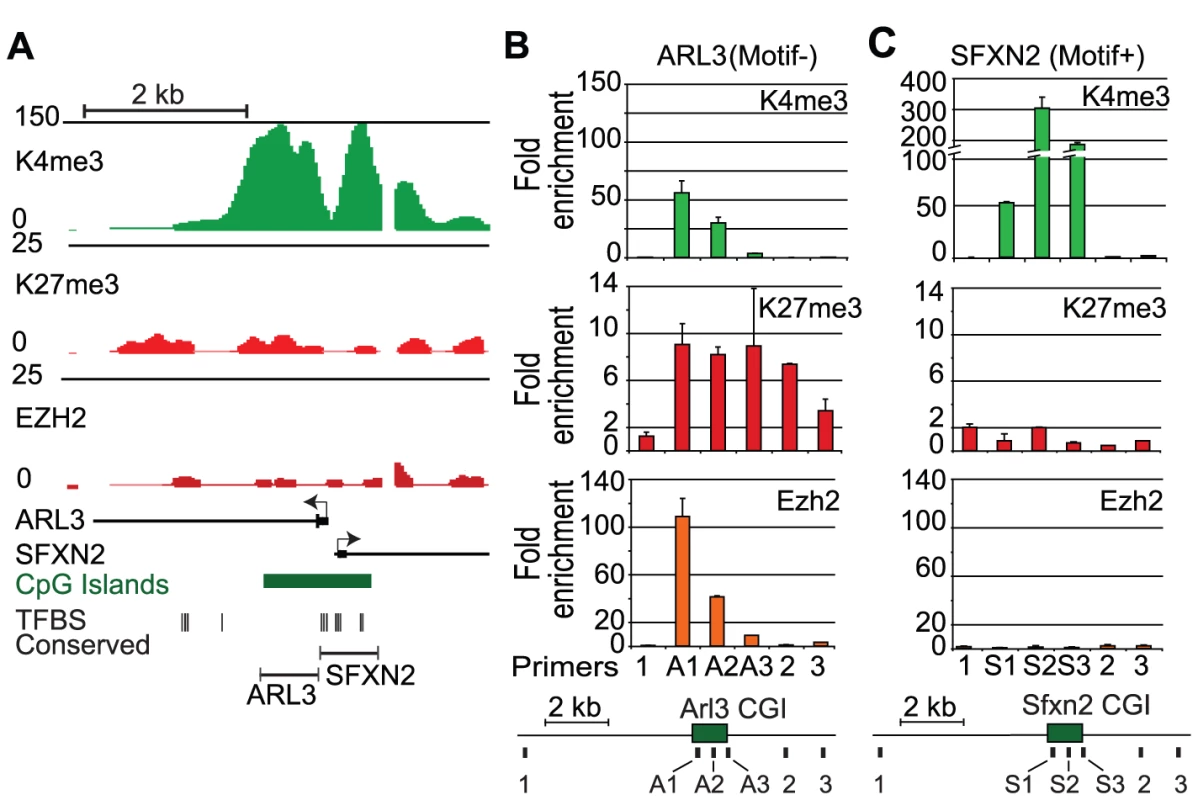
(A) Genome browser views shows a locus containing the promoters for the housekeeping genes Arl3 and Sfxn2 with ChIP-Seq enrichment signals for K4me3, K27me3, and Ezh2 in mouse ES cells. This region contains a 1.8 kb CpG island that has the transcription factor motifs clustered on one side. Below shows the regions used for integration into the gene desert BAC. (B) After integration into mouse ES cells, ChIP-qPCR was conducted using three primers from the CpG island inserts and 3 primers in the flanking gene desert sequence. The motif devoid Arl3 section shows de novo PRC2 (Ezh2) recruitment and K4me3 and K27me3 enrichment. (C) The motif containing Sfxn2 half shows no enrichment for K27me3 but significant enrichment for K4me3, similar to the endogenous locus shown in (A) (n = 2 biological replicates). Next, we tested whether even more generic GC-rich elements might also be capable of recruiting PRC2 in ES cells. Here, we focused on sequences derived from the genome of E. coli, reasoning that there would be no selection for PRC2 recruiting elements in this prokaryote given the complete lack of chromatin regulators. We arbitrarily selected three 1 kb segments of the E. coli genome. Two with GC contents above the threshold for a mammalian CpG island but that each contained few AMs, and one AT rich segment as a control (Table S3). We recombined each segment into the gene desert BAC and integrated the resulting constructs into ES cells. ChIP-qPCR confirmed that both GC-rich E. Coli segments recruit Ezh2 and form a bivalent chromatin state (Figure 4A, 4B, Figure S9). Notably, the GC-rich segment also enriches for Jarid2, a PRC2 component with DNA binding activity (Figure S10). In contrast, the AT-rich segment did not recruit Ezh2 or enrich for either K4me3 or K27me3 (Figure 4C, Figure S9). Together, our findings suggest that GC-rich sequence elements that lack signals for transcriptional activation have an innate capacity to recruit PRC2 in mammalian ES cells.
Fig. 4. PRC2 is recruited to E.coli GC-rich sequences in mouse ES cells. 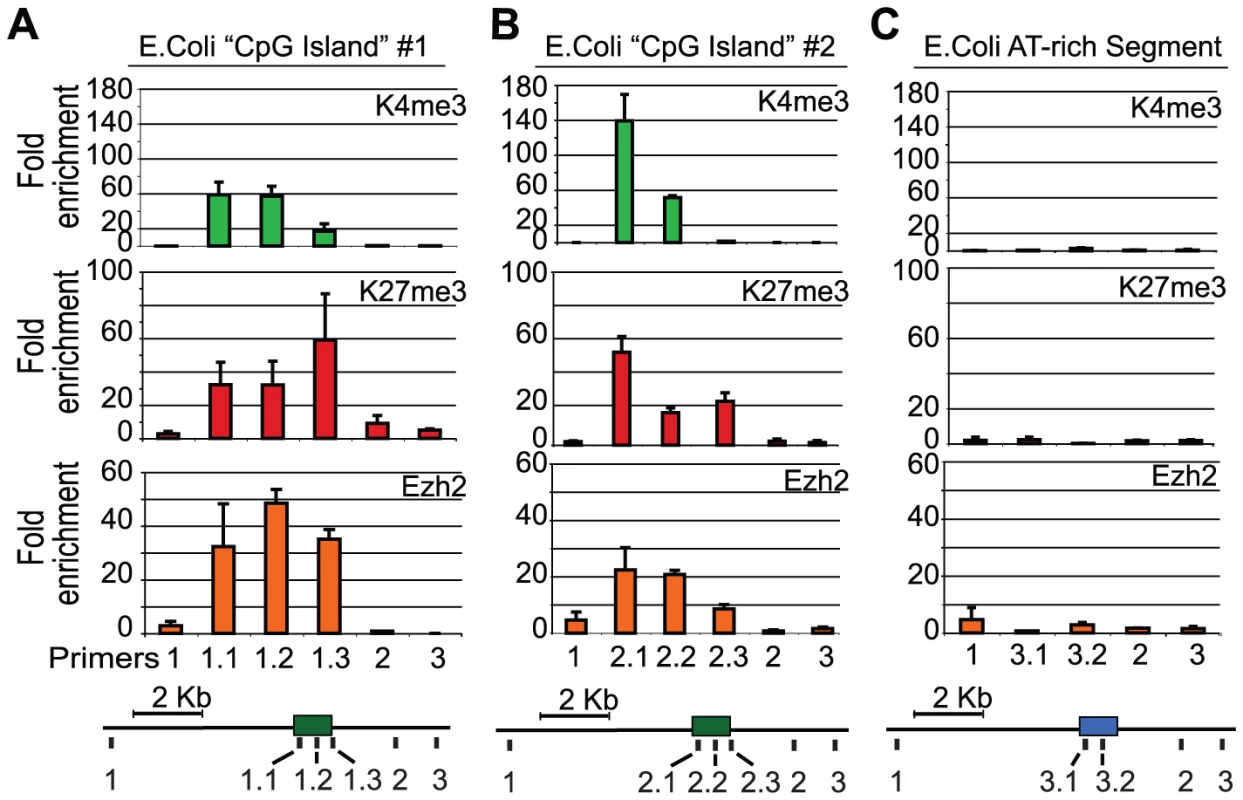
The E. coli genome was scanned for 1 kb regions that met the criteria for a mammalian CpG island and had few motifs for mammalian transcription factors (see Methods). (A,B) Both GC-rich segments adopt a ‘bivalent’ chromatin state with K27me3 and K4me3 and recruit PRC2 (Ezh2) upon integration in mouse ES cells. (C) A non-CG rich region of the E. coli genome failed to recruit Ezh2 and lacked K4me3 and K27me3 (n = 2 biological replicates). Discussion
Several lines of evidence suggest that the initial landscape of Polycomb complex binding is critical for proper patterning of gene expression in metazoan development [1], [2], [13]. Failure of these factors to engage their target loci in embryogenesis has been linked to a loss of epigenetic repression at later stages. Accordingly, the determinants that localize Polycomb complexes at the pluripotent stage are almost certainly essential to the global functions of these repressors through development.
We find that DNA sequence is sufficient for proper localization of Polycomb repressive complexes in ES cells, and specifically identify a CpG island within the Zfpm2 locus as being critical for recruitment. We provide evidence that GC-rich elements lacking activating signals suffice in general to recruit PRC2. This includes demonstrations (i) that a motif devoid segment of an active ‘housekeeping’ CpG island can recruit PRC2; and (ii) that arbitrarily selected GC-rich elements from the E. coli genome can themselves mediate PRC2 recruitment when integrated into the ES cell genome.
Several possible mechanistic models could explain the causality of GC-rich DNA elements in PRC2 recruitment (Figure 5). First, we note that CpG islands have been shown to destabilize nucleosomes in mammalian cells [42]. At transcriptionally inactive loci, this property could increase their accessibility to PRC2-associated proteins with DNA affinity but low sequence specificity, such as Jarid2 or AEBP2 [32]–[35], [43] (Figure S10). Although this association would be abrogated by transcriptional activity at most CpG islands, those lacking activation signals would remain permissive to PRC2 association (Figure 5). In support of this model, PRC2 targets in ES cells are also enriched for H2A.Z and H3.3, histone variants linked to nucleosome exchange dynamics [44], [45]. Alternatively or in addition, targeting could be supported by DNA binding proteins with affinity for low complexity GC-rich motifs or CpG dinucleotides, such as CXXC domain proteins [46]. Localization may also be promoted or stabilized by long and short non-coding RNAs [47]–[50] as well as by the demonstrated affinity of PRC2 for its product, H3K27me3 [11], [12]. Notably, PRC2 recruitment in ES cells appears distinct from that in Drosophila, as we do not find evidence for involvement of PRE-like sequence motifs or mammalian homologues such as YY1.
Fig. 5. A model showing CpG islands as a chromatin switch. 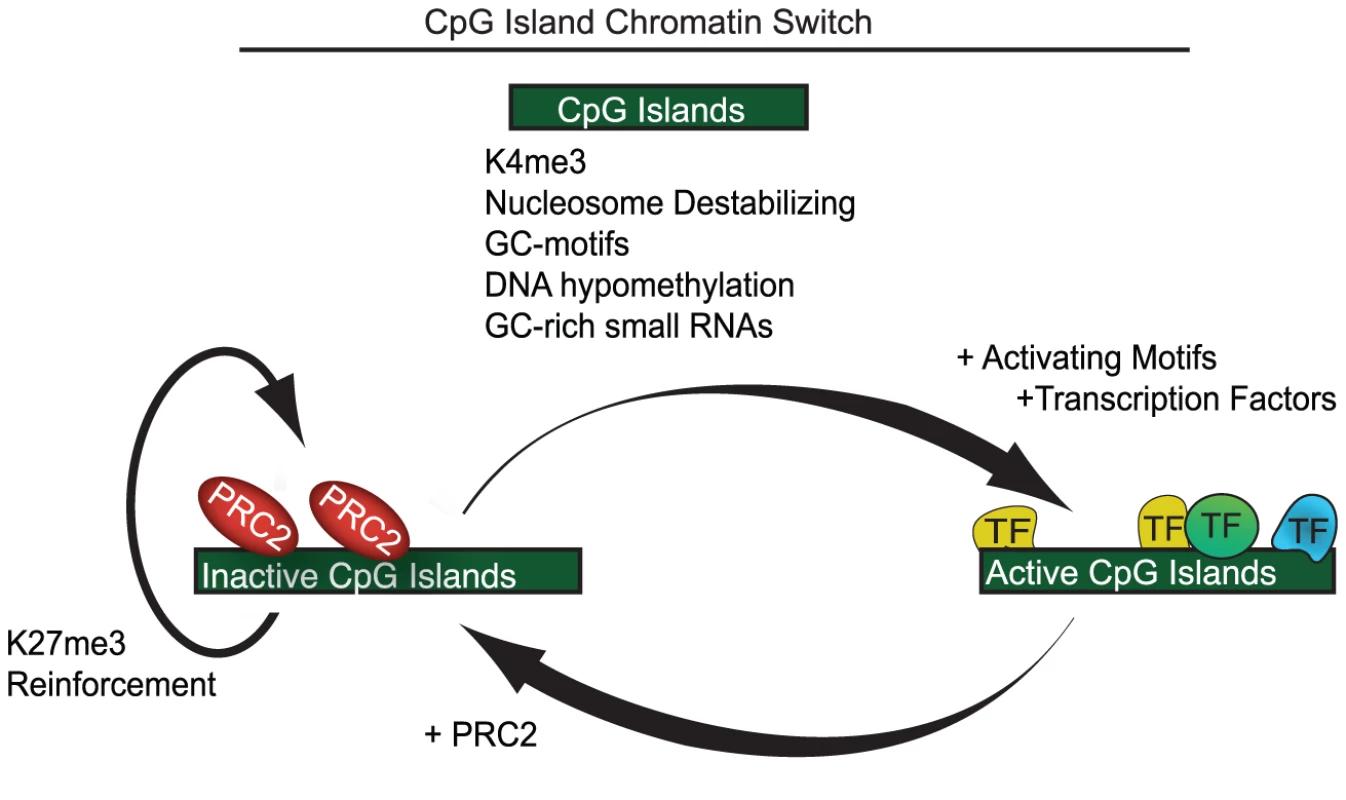
(A) Features common to both active and inactive CpG islands include destabalization of nucleosomes, simple GC-motifs, K4me3 and lack of DNA methylation. Additionally, many CpG island transcribe small non-coding GC-rich RNAs. Active CpG islands contain motifs associated with numerous activating transcription factors and transcriptional machinery, which likely prevent PRC2 from binding. In contrast, CpG islands lacking activating motifs are bound by PRC2 which, through a positive feedback loop with K27me3, maintains an inactive state. It should be emphasized that PRC2 localization does not necessarily equate with epigenetic repression. Indeed virtually all PRC2 bound sites in ES cells, and all CpG islands tested here, are also enriched for K4me3, and presumably poised for activation upon differentiation. Epigenetic repression during differentiation may require PRC1 and thus depend on additional binding determinants. YY1 remains an intriguing candidate in this regard, given prior evidence for physical and genetic interactions with PRC1 [51], [52]. YY1 consensus motifs are present in the Polycomb-dependent silencing elements recently identified in the MafB and HoxD loci. Interestingly, the HoxD element combines a CpG island with a cluster of conserved YY1 motifs. Mutation of the motifs abrogated PRC1 binding but left PRC2 binding intact. Still, the fact that only a small fraction of documented PRC2 and PRC1 sites have YY1 motifs or binding suggests that this transcription factor may act indirectly and/or explain only a subset of cases. Nonetheless, it is likely that a fully functional epigenetic silencer would require a combination of features, including a GC-rich PRC2 element as well as appropriate elements to recruit PRC1. Further study is needed to expand the rules for PRC2 binding to include a global definition of PRC1 determinants and ultimately, to understand how the initial landscape facilitates the maintenance of gene expression programs in the developing organism.
Methods
BAC construct design
BAC constructs CTD331719L (‘Zfpm2 44’), CTD-2535J16 (‘Pax5’) and CTD-3219L19 (‘Gene Desert’) were obtained from Open Biosystems. Recombineering was done using the RedET system (Open Biosystems) in DH10B cells. Homology arms 200–500 bp in length were PCR amplified and cloned into a PGK; Neomycin cassette (Gene Bridges). This cassette was used to recombineer all BACs to enable selection in mammalian cells. The 22 kb hZfpm2 BAC was created by restricting the hZfpm2 BAC at two sites using ClaI, and re-ligating the BAC lacking the intervening sequence. The CpG island was excised from the 22 kb hZfpm2 BAC by amplification of flanking homology arms, and cloned into a construct containing an adjacent ampicillin cassette (Frt-amp-Frt; Gene Bridges). After recombination, the ampicillin cassette was removed using Flp-recombinase and selection for clones that lost ampicillin resistance (Flp-706; Gene Bridges). PCR across the region confirmed excision of the CpG island. For the Gene Desert BACs, the Zfpm2, Arl3, Sfxn2 and E. coli CpG islands were amplified with primers containing XhoI sites and cloned into the Frt-amp-Frt vector that contains homology arms from the Gene Desert region. The final constructs were confirmed by sequencing across recombination junctions. All primers used for CpG islands and recombineering homology arms are listed in Table S2.
Transgenic ES cell and ChIP experiments
ES cells (V6.5) were maintained in ES cell medium (DMEM; Dulbecco's modified Eagle's medium) supplemented with 15% fetal calf serum (Hyclone), 0.1 mM ß-mercaptoethanol (Sigma), 2 mM Glutamax, 0.1 mM non-essential amino acid (NEAA; Gibco) and 1000U/ml recombinant leukemia inhibitory factor (ESGRO; Chemicon). Roughly 50 µg of linearized BAC was nucleofected using the mouse ES cell nucleofector kit (Lonza) into 106 mouse ES cells, and selected 7–10 days with 150 µg/ml Geneticin (Invitrogen) on Neomycin resistant MEFs (Millipore). Individual resistant colonies were picked, expanded and tested for integration of the full length BAC by PCR. Differentiation of hZfpm2 ES cell clone 1 into a population of neural progenitor (NP) cells was done as previously described [53]. FISH analysis was done as described previously [54]. DNA methylation analysis was done as previously described [55] and primers used to amplify bisulfite treated DNA are listed in Table S2.
For each construct, between one and three ES cell clones were expanded and subjected to ChIP using antibody against K4me3 (Abcam ab8580 or Upstate/Millipore 07-473), K27me3 (Upstate/Millipore 07-449), Ezh2 (Active Motif 39103 or 39639), or Ring1B (MBL International d139-3) as described previously [5], [7], [39]. ChIP DNA was quantified by Quant-iT Picogreen dsDNA Assay Kit (Invitrogen). ChIP enrichments were assessed by quantitative PCR analysis on an ABI 7500 with 0.25 ng ChIP DNA and an equal mass of un-enriched input DNA. Enrichments were calculated from 2 or 3 biologically independent ChIP experiments. For K27me3, and Ezh2 enrichment, background was subtracted by normalizing over a negative genomic control. Error bars represent standard error of the mean (SEM). We confirmed that the human specific primers do not non-specifically amplify mouse genomic DNA. Primers used for qPCR are listed in Table S2.
Genomic and computational analysis
Genomewide maps of YY1 binding sites were determined by ChIP-Seq as described previously [39]. Briefly, ChIP was carried out on 6×107 cells using antibody against YY1 (Santa Cruz Biotechnology sc-1703). ChIP DNA was used to prepare libraries which were sequenced on the Illumina Genome Analyzer. Density profiles were generated as described [39]. Promoters (RefSeq; http://genome.ucsc.edu) were classified as positive for YY1, H3K4me3 or H3K27me3 if the read density was significantly enriched (p<10−3) over a background distribution based on randomized reads generated separately for each dataset to account for the varying degrees of sequencing depth. ChIP-Seq data for YY1 are deposited to the NCBI GEO database under the following accession number GSE25197 (http://www.ncbi.nlm.nih.gov/projects/geo/query/acc.cgi?acc=GSE25197). Sites of Ezh2 enrichment (p<10−3) were calculated genomewide using sliding 1 kb windows, and enriched windows within 1 kb were merged. DNA methylation levels were calculated using previously published Reduced Representation Bisulphite Sequenced (RRBS) libraries [55]. Composite plots represent the mean methylation level in sliding 200 bp windows in the the 10 kb surrounding the TSSs of the indicated gene sets.
YY1 motifs were identified using the MAST algorithm [56] where a match to the consensus motif was defined at significance level 5×10−5. Candidate CpG islands for TF motif analysis were identified by scanning annotated CpG islands (http://genome.ucsc.edu) for asymmetric clustering of motifs related to transcriptional activation in ES cells [5]. Motifs shown in Figure 3A and Figure S6 are from UCSCs TFBS conserved track. GC-rich elements from the E. coli K12 genome were selected by calculating %GC and CpG O/E in sliding 1 kb windows. Sequences matching the criteria for mammalian CpG islands while simultaneously being depleted of motifs related to transcriptional activation [5] were chosen for insertion into mouse ES cells. Transcriptionally inactive HCPs were selected based on a lack of transcript enrichment by both expression arrays [39] and RNA-Seq data [57]. In the case of RNA-Seq, each gene was assigned the maximum read density within any 1 kb window of exonic sequence. To ease analysis of promoter CpG island statistics, only HCPs containing a single CpG island were considered.
Supporting Information
Zdroje
1. SchuettengruberB
GanapathiM
LeblancB
PortosoM
JaschekR
2009 Functional anatomy of polycomb and trithorax chromatin landscapes in Drosophila embryos. PLoS Biol 7 e1000013 doi:10.1371/journal.pbio.1000013
2. SchwartzYB
PirrottaV
2007 Polycomb silencing mechanisms and the management of genomic programmes. Nat Rev Genet 8 9 22
3. BoyerLA
PlathK
ZeitlingerJ
BrambrinkT
MedeirosLA
2006 Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441 349 353
4. LeeTI
JennerRG
BoyerLA
GuentherMG
LevineSS
2006 Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125 301 313
5. KuM
KocheRP
RheinbayE
MendenhallEM
EndohM
2008 Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet 4 e1000242 doi:10.1371/journal.pgen.1000242
6. AzuaraV
PerryP
SauerS
SpivakovM
JorgensenHF
2006 Chromatin signatures of pluripotent cell lines. Nat Cell Biol 8 532 538
7. BernsteinB
MikkelsenT
XieX
KamalK
HuebertD
2006 A Bivalent Chromatin Structure Marks Key Developmental Genes in Embryonic Stem Cells. Cell 125 315 326
8. CaoR
WangL
WangH
XiaL
Erdjument-BromageH
2002 Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298 1039 1043
9. CzerminB
MelfiR
McCabeD
SeitzV
ImhofA
2002 Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111 185 196
10. KuzmichevA
NishiokaK
Erdjument-BromageH
TempstP
ReinbergD
2002 Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev 16 2893 2905
11. HansenKH
BrackenAP
PasiniD
DietrichN
GehaniSS
2008 A model for transmission of the H3K27me3 epigenetic mark. Nat Cell Biol 10 1291 1300
12. MargueronR
JustinN
OhnoK
SharpeML
SonJ
2009 Role of the polycomb protein EED in the propagation of repressive histone marks. Nature 461 762 767
13. RingroseL
ParoR
2007 Polycomb/Trithorax response elements and epigenetic memory of cell identity. Development 134 223 232
14. SimonJ
ChiangA
BenderW
ShimellMJ
O'ConnorM
1993 Elements of the Drosophila bithorax complex that mediate repression by Polycomb group products. Dev Biol 158 131 144
15. DejardinJ
RappaillesA
CuvierO
GrimaudC
DecovilleM
2005 Recruitment of Drosophila Polycomb group proteins to chromatin by DSP1. Nature 434 533 538
16. TolhuisB
de WitE
MuijrersI
TeunissenH
TalhoutW
2006 Genome-wide profiling of PRC1 and PRC2 Polycomb chromatin binding in Drosophila melanogaster. Nat Genet 38 694 699
17. WangL
BrownJL
CaoR
ZhangY
KassisJA
2004 Hierarchical recruitment of polycomb group silencing complexes. Mol Cell 14 637 646
18. SchwartzYB
KahnTG
NixDA
LiXY
BourgonR
2006 Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat Genet 38 700 705
19. NegreN
HennetinJ
SunLV
LavrovS
BellisM
2006 Chromosomal distribution of PcG proteins during Drosophila development. PLoS Biol 4 e170 doi:10.1371/journal.pbio.0040170
20. AtchisonL
GhiasA
WilkinsonF
BoniniN
AtchisonML
2003 Transcription factor YY1 functions as a PcG protein in vivo. Embo J 22 1347 1358
21. KoCY
HsuHC
ShenMR
ChangWC
WangJM
2008 Epigenetic silencing of CCAAT/enhancer-binding protein delta activity by YY1/polycomb group/DNA methyltransferase complex. J Biol Chem 283 30919 30932
22. SuiG
Affar elB
ShiY
BrignoneC
WallNR
2004 Yin Yang 1 is a negative regulator of p53. Cell 117 859 872
23. YueR
KangJ
ZhaoC
HuW
TangY
2009 Beta-arrestin1 regulates zebrafish hematopoiesis through binding to YY1 and relieving polycomb group repression. Cell 139 535 546
24. LiuH
Schmidt-SupprianM
ShiY
HobeikaE
BartenevaN
2007 Yin Yang 1 is a critical regulator of B-cell development. Genes Dev 21 1179 1189
25. XiH
YuY
FuY
FoleyJ
HaleesA
2007 Analysis of overrepresented motifs in human core promoters reveals dual regulatory roles of YY1. Genome Res 17 798 806
26. KimJD
KangK
KimJ
2009 YY1's role in DNA methylation of Peg3 and Xist. Nucleic Acids Res 37 5656 5664
27. SingA
PannellD
KaraiskakisA
SturgeonK
DjabaliM
2009 A vertebrate Polycomb response element governs segmentation of the posterior hindbrain. Cell 138 885 897
28. WooCJ
KharchenkoPV
DaheronL
ParkPJ
KingstonRE
A region of the human HOXD cluster that confers polycomb-group responsiveness. Cell 140 99 110
29. BrackenAP
DietrichN
PasiniD
HansenKH
HelinK
2006 Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev 20 1123 1136
30. MohnF
WeberM
RebhanM
RoloffTC
RichterJ
2008 Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell 30 755 766
31. TanayA
O'DonnellAH
DamelinM
BestorTH
2007 Hyperconserved CpG domains underlie Polycomb-binding sites. Proc Natl Acad Sci U S A 104 5521 5526
32. PasiniD
CloosPA
WalfridssonJ
OlssonL
BukowskiJP
JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature 464 306 310
33. LiG
MargueronR
KuM
ChambonP
BernsteinBE
Jarid2 and PRC2, partners in regulating gene expression. Genes Dev 24 368 380
34. PengJC
ValouevA
SwigutT
ZhangJ
ZhaoY
2009 Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell 139 1290 1302
35. ShenX
KimW
FujiwaraY
SimonMD
LiuY
2009 Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells. Cell 139 1303 1314
36. KimTG
KrausJC
ChenJ
LeeY
2003 JUMONJI, a critical factor for cardiac development, functions as a transcriptional repressor. J Biol Chem 278 42247 42255
37. TevosianSG
AlbrechtKH
CrispinoJD
FujiwaraY
EicherEM
2002 Gonadal differentiation, sex determination and normal Sry expression in mice require direct interaction between transcription partners GATA4 and FOG2. Development 129 4627 4634
38. ContiL
PollardSM
GorbaT
ReitanoE
ToselliM
2005 Niche-independent symmetrical self-renewal of a mammalian tissue stem cell. PLoS Biol 3 e283 doi:10.1371/journal.pbio.0030283
39. MikkelsenTS
KuM
JaffeDB
IssacB
LiebermanE
2007 Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448 553 560
40. PouxS
McCabeD
PirrottaV
2001 Recruitment of components of Polycomb Group chromatin complexes in Drosophila. Development 128 75 85
41. SchmittS
PrestelM
ParoR
2005 Intergenic transcription through a polycomb group response element counteracts silencing. Genes Dev 19 697 708
42. Ramirez-CarrozziVR
BraasD
BhattDM
ChengCS
HongC
2009 A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell 138 114 128
43. KimH
KangK
KimJ
2009 AEBP2 as a potential targeting protein for Polycomb Repression Complex PRC2. Nucleic Acids Res 37 2940 2950
44. CreyghtonMP
MarkoulakiS
LevineSS
HannaJ
LodatoMA
2008 H2AZ is enriched at polycomb complex target genes in ES cells and is necessary for lineage commitment. Cell 135 649 661
45. GoldbergAD
BanaszynskiLA
NohKM
LewisPW
ElsaesserSJ
Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 140 678 691
46. TateCM
LeeJH
SkalnikDG
2009 CXXC Finger Protein 1 Contains Redundant Functional Domains That Support Embryonic Stem Cell Cytosine Methylation, Histone Methylation, and Differentiation. Mol Cell Biol
47. ZhaoJ
SunBK
ErwinJA
SongJJ
LeeJT
2008 Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 322 750 756
48. KanhereA
ViiriK
AraujoCC
RasaiyaahJ
BouwmanRD
Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2. Mol Cell 38 675 688
49. RinnJL
KerteszM
WangJK
SquazzoSL
XuX
2007 Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129 1311 1323
50. TsaiMC
ManorO
WanY
MosammaparastN
WangJK
Long noncoding RNA as modular scaffold of histone modification complexes. Science 329 689 693
51. LorenteM
PerezC
SanchezC
DonohoeM
ShiY
2006 Homeotic transformations of the axial skeleton of YY1 mutant mice and genetic interaction with the Polycomb group gene Ring1/Ring1A. Mech Dev 123 312 320
52. GarciaE
Marcos-GutierrezC
del Mar LorenteM
MorenoJC
VidalM
1999 RYBP, a new repressor protein that interacts with components of the mammalian Polycomb complex, and with the transcription factor YY1. Embo J 18 3404 3418
53. PollardSM
BenchouaA
LowellS
2006 Neural stem cells, neurons, and glia. Methods Enzymol 418 151 169
54. MrakRE
YasargilMG
MohapatraG
EarelJJr
LouisDN
2004 Atypical extraventricular neurocytoma with oligodendroglioma-like spread and an unusual pattern of chromosome 1p and 19q loss. Hum Pathol 35 1156 1159
55. MeissnerA
MikkelsenTS
GuH
WernigM
HannaJ
2008 Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 454 766 770
56. BaileyTL
GribskovM
1998 Combining evidence using p-values: application to sequence homology searches. Bioinformatics 14 48 54
57. CloonanN
ForrestAR
KolleG
GardinerBB
FaulknerGJ
2008 Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nat Methods 5 613 619
Štítky
Genetika Reprodukční medicína
Článek Genome-Wide Interrogation of Mammalian Stem Cell Fate Determinants by Nested Chromosome DeletionsČlánek Season of Conception in Rural Gambia Affects DNA Methylation at Putative Human Metastable EpiallelesČlánek A Quantitative Systems Approach Reveals Dynamic Control of tRNA Modifications during Cellular StressČlánek Reduction of Protein Translation and Activation of Autophagy Protect against PINK1 Pathogenesis inČlánek The Loss of PGAM5 Suppresses the Mitochondrial Degeneration Caused by Inactivation of PINK1 inČlánek Cleavage of Phosphorothioated DNA and Methylated DNA by the Type IV Restriction Endonuclease ScoMcrAČlánek Competitive Repair by Naturally Dispersed Repetitive DNA during Non-Allelic Homologous Recombination
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2010 Číslo 12
-
Všechny články tohoto čísla
- Genome-Wide Interrogation of Mammalian Stem Cell Fate Determinants by Nested Chromosome Deletions
- Whole-Genome and Chromosome Evolution Associated with Host Adaptation and Speciation of the Wheat Pathogen
- Association of Variants at 1q32 and with Ankylosing Spondylitis Suggests Genetic Overlap with Crohn's Disease
- Initiator Elements Function to Determine the Activity State of BX-C Enhancers
- Identification of Genes Required for Neural-Specific Glycosylation Using Functional Genomics
- A Young Duplicate Gene Plays Essential Roles in Spermatogenesis by Regulating Several Y-Linked Male Fertility Genes
- The EpsE Flagellar Clutch Is Bifunctional and Synergizes with EPS Biosynthesis to Promote Biofilm Formation
- Histone H2A C-Terminus Regulates Chromatin Dynamics, Remodeling, and Histone H1 Binding
- Season of Conception in Rural Gambia Affects DNA Methylation at Putative Human Metastable Epialleles
- A Quantitative Systems Approach Reveals Dynamic Control of tRNA Modifications during Cellular Stress
- GC-Rich Sequence Elements Recruit PRC2 in Mammalian ES Cells
- A Single Enhancer Regulating the Differential Expression of Duplicated Red-Sensitive Opsin Genes in Zebrafish
- Investigation and Functional Characterization of Rare Genetic Variants in the Adipose Triglyceride Lipase in a Large Healthy Working Population
- Reduction of Protein Translation and Activation of Autophagy Protect against PINK1 Pathogenesis in
- Noisy Splicing Drives mRNA Isoform Diversity in Human Cells
- The Loss of PGAM5 Suppresses the Mitochondrial Degeneration Caused by Inactivation of PINK1 in
- Thymus-Associated Parathyroid Hormone Has Two Cellular Origins with Distinct Endocrine and Immunological Functions
- An ABC Transporter Mutation Is Correlated with Insect Resistance to Cry1Ac Toxin
- Role of Individual Subunits of the CSN Complex in Regulation of Deneddylation and Stability of Cullin Proteins
- The C-Terminal Domain of the Bacterial SSB Protein Acts as a DNA Maintenance Hub at Active Chromosome Replication Forks
- The DNA Damage Response Pathway Contributes to the Stability of Chromosome III Derivatives Lacking Efficient Replicators
- Cleavage of Phosphorothioated DNA and Methylated DNA by the Type IV Restriction Endonuclease ScoMcrA
- LaeA Control of Velvet Family Regulatory Proteins for Light-Dependent Development and Fungal Cell-Type Specificity
- Competitive Repair by Naturally Dispersed Repetitive DNA during Non-Allelic Homologous Recombination
- Distinct Functions for the piRNA Pathway in Genome Maintenance and Telomere Protection
- MOS11: A New Component in the mRNA Export Pathway
- Self-Mating in the Definitive Host Potentiates Clonal Outbreaks of the Apicomplexan Parasites and
- A Role for ATF2 in Regulating MITF and Melanoma Development
- Ancestral Regulatory Circuits Governing Ectoderm Patterning Downstream of Nodal and BMP2/4 Revealed by Gene Regulatory Network Analysis in an Echinoderm
- Cancer and Neurodegeneration: Between the Devil and the Deep Blue Sea
- Functional Comparison of Innate Immune Signaling Pathways in Primates
- Linking Crohn's Disease and Ankylosing Spondylitis: It's All about Genes!
- Genomics Meets Glycomics—The First GWAS Study of Human N-Glycome Identifies HNF1α as a Master Regulator of Plasma Protein Fucosylation
- Continuous and Periodic Expansion of CAG Repeats in Huntington's Disease R6/1 Mice
- Expression of Linear and Novel Circular Forms of an -Associated Non-Coding RNA Correlates with Atherosclerosis Risk
- Endocytic Sorting and Recycling Require Membrane Phosphatidylserine Asymmetry Maintained by TAT-1/CHAT-1
- Histone Deacetylases Suppress CGG Repeat–Induced Neurodegeneration Via Transcriptional Silencing in Models of Fragile X Tremor Ataxia Syndrome
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Functional Comparison of Innate Immune Signaling Pathways in Primates
- Expression of Linear and Novel Circular Forms of an -Associated Non-Coding RNA Correlates with Atherosclerosis Risk
- Genome-Wide Interrogation of Mammalian Stem Cell Fate Determinants by Nested Chromosome Deletions
- Histone H2A C-Terminus Regulates Chromatin Dynamics, Remodeling, and Histone H1 Binding
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



