-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaContinuous and Periodic Expansion of CAG Repeats in Huntington's Disease R6/1 Mice
Huntington's disease (HD) is one of several neurodegenerative disorders caused by expansion of CAG repeats in a coding gene. Somatic CAG expansion rates in HD vary between organs, and the greatest instability is observed in the brain, correlating with neuropathology. The fundamental mechanisms of somatic CAG repeat instability are poorly understood, but locally formed secondary DNA structures generated during replication and/or repair are believed to underlie triplet repeat expansion. Recent studies in HD mice have demonstrated that mismatch repair (MMR) and base excision repair (BER) proteins are expansion inducing components in brain tissues. This study was designed to simultaneously investigate the rates and modes of expansion in different tissues of HD R6/1 mice in order to further understand the expansion mechanisms in vivo. We demonstrate continuous small expansions in most somatic tissues (exemplified by tail), which bear the signature of many short, probably single-repeat expansions and contractions occurring over time. In contrast, striatum and cortex display a dramatic—and apparently irreversible—periodic expansion. Expansion profiles displaying this kind of periodicity in the expansion process have not previously been reported. These in vivo findings imply that mechanistically distinct expansion processes occur in different tissues.
Published in the journal: . PLoS Genet 6(12): e32767. doi:10.1371/journal.pgen.1001242
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1001242Summary
Huntington's disease (HD) is one of several neurodegenerative disorders caused by expansion of CAG repeats in a coding gene. Somatic CAG expansion rates in HD vary between organs, and the greatest instability is observed in the brain, correlating with neuropathology. The fundamental mechanisms of somatic CAG repeat instability are poorly understood, but locally formed secondary DNA structures generated during replication and/or repair are believed to underlie triplet repeat expansion. Recent studies in HD mice have demonstrated that mismatch repair (MMR) and base excision repair (BER) proteins are expansion inducing components in brain tissues. This study was designed to simultaneously investigate the rates and modes of expansion in different tissues of HD R6/1 mice in order to further understand the expansion mechanisms in vivo. We demonstrate continuous small expansions in most somatic tissues (exemplified by tail), which bear the signature of many short, probably single-repeat expansions and contractions occurring over time. In contrast, striatum and cortex display a dramatic—and apparently irreversible—periodic expansion. Expansion profiles displaying this kind of periodicity in the expansion process have not previously been reported. These in vivo findings imply that mechanistically distinct expansion processes occur in different tissues.
Introduction
Huntington's disease (HD) is a genetically determined neurodegenerative disorder, the onset of which is known to depend upon the length of glutamine-encoding CAG-repeat sequences lying within the Huntingtin (HTT) gene [1]. Humans may develop the disease if they have more than 36 repeats and disease onset usually starts during mid-life. An inverse relationship has been shown between CAG repeat length and age of onset in HD [2]–[5]. Additionally, somatic instability in human cortex has recently been shown to be a good predictor of disease onset [6]. Children with 108–256 CAG repeats are reported to show disease onset from one and a half years to six years of age [7].
Trinucleotide repeat (TNR) instability varies between organs in a variety of neurodegenerative disorders which are caused by expansion of CAG repeats in a coding gene, with the greatest instability observed in the brain [8]–[11]. In HD, striatum tissue shows the most severe neuropathology, followed by cortex. CAG length expansion is correlated with neuropathology and probably precedes the onset of symptoms [12]. The CAG repeat length is unstable in most cell types of the brain, but neurons tend to show the greatest mutation lengths in both humans and mice [13]–[15]. Meanwhile, minimal expansion is considered to occur in many other somatic tissues.
TNR sequences may form slipped strands during replication or repair, creating loops or hairpins, which protrude from the DNA duplex [16].
In the earliest model for repeat expansion the DNA polymerase forms slip-outs on the nascent strand leading to small-scale repeat expansion in repetitive sequences [17]. Loops of repeat-containing DNA are believed to cause either expansions or contractions during replication, when the slip-out occurs in the nascent or template strand, respectively [18], [19]. Several models have been suggested to explain TNR expansion during replication, such as folding of the lagging strand template into a hairpin, stalled replication forks and the orientation of the TNR in the genome, as well as the location of the origin of replication, as shown in several experiments in bacteria, yeast and human cells (Reviewed in [20]). More recently, a pertinent role of DNA repair proteins in CAG repeat expansion has been demonstrated in vivo. In particular, deletion of the mismatch repair (MMR) proteins, Msh2 and Msh3 [21]–[24] has been shown to abolish age-dependent somatic CAG repeat expansion in mouse models for HD. MMR has also been shown to be involved in TNR expansion in mouse models of myotonic dystrophy (DM1) [25]–[27]. Furthermore, the age-dependent expansion of TNR sequences in somatic cells was shown to be modified by the base excision repair (BER) 8-oxoguanine DNA glycosylase (Ogg1) in the R6/1 mouse model, demonstrating that there may be a link between oxidative DNA damage and TNR instability [13]. The flap endonuclease 1 (FEN1), which removes 5′-flaps during replication [28] and is involved in long-patch BER [29] is also implicated in expansion. Secondary TNR structures have been shown to inhibit FEN1 activity [30]. In addition to flap endonuclease activity, the EXO [31] and GAP activities of FEN1 have been shown to contribute to the resolution of TNR secondary structures in vitro [32]. Recently, it was shown that the stoichiometry of BER proteins, such as Ogg1, polymerase β and FEN1, may contribute to the tissue-selectivity of somatic HD CAG repeat expansion [33].
Nevertheless, the processes causing this expansion remain poorly understood, particularly in mammalian systems, although the formation of secondary DNA structures within the repeat sequence is thought to underlie the process [34]. Here we present evidence for two distinct modes of somatic expansion identified by the analysis of CAG repeat fragments from 103 HD R6/1 mice; a continuous slight expansion in tail, lung, heart and spleen, and a dramatic periodic expansion in striatum, and cortex, which we also compare to the expansions observed in liver. The continuous expansion process is shown here to conform to a bi-directional, forward-biased model that represents the occurrence of multiple short – tending towards unitary – CAG repeat insertions and deletions, at random moments as the mouse ages. In contrast, the dramatic expansion seen in brain tissues demonstrates a periodicity centred around seven repeats, which correlates with the stochastic insertion of stable TNR segments of consistent length. Meanwhile liver tissue shows a comparable average increase in CAG repeat length to striatum, but with a much weaker inclination to exhibit periodicity, tending more towards a continuum. This suggests either a much less controlled insertion length when compared to expansions in striatum, or that liver tissue undergoes both types of expansion simultaneously. We also present discursive models for these two expansion mechanisms. Identification of these two independent modes of expansion, in particular the tight mechanistic control implicit in the expansions within neuropathologically relevant tissues, increases our understanding of the tissue-dependent progress of HD. This brings us a step closer to inferring the in vivo mechanisms of the molecular components involved, by showing that only a limited selection of the existing models for expansion are able to explain the age-dependent CAG repeat expansions we observe.
Results
Fragment analysis shows tissue-specific modes of somatic CAG expansion
In order to understand the mechanisms underlying somatic CAG instability, 42 R6/1 HD exon 1 transgenic mice were sacrificed at either 10 or 21 weeks of age, whereupon tail, heart, lung, spleen, liver, cortex and striatum samples were taken for analysis of HD CAG repeat length. A tail biopsy at 3 weeks of age represents the reference level of CAG repeats present near birth in all tissues for each mouse [35] (Figure S3). Thus changes in the CAG composition of tissues in an individual mouse could be compared over a 7 - or 18-week period. A slight expansion was observed in tail (Figure 1A), whereas cortex and striatum demonstrated a dramatic and periodic expansion process, with no significant difference between genders (Figure 1B and 1C). Liver demonstrated an equally rapid, but apparently more continuous expansion. Heart, lung and spleen displayed a slight expansion that was identical to tail (Figure S11). A parallel dataset from 61 hHD+/−Fen1E160D/E160D mutant mice, in which flap endonuclease activity of FEN1 is reduced to ∼20% [36] was included in the study. Reduced FEN1 endonuclease activity did not affect the rate of CAG repeat expansions measured in any tissues, implying that this mutation did not affect any role FEN1 plays in expansion. The two datasets were qualitatively and quantitatively identical with regard to the following analysis in all organs tested and were therefore combined, such that our analysis covers observations across two HD genotypes, reinforcing the ubiquity of the results.
Fig. 1. Fragment analysis in tail, cortex, striatum, and liver. 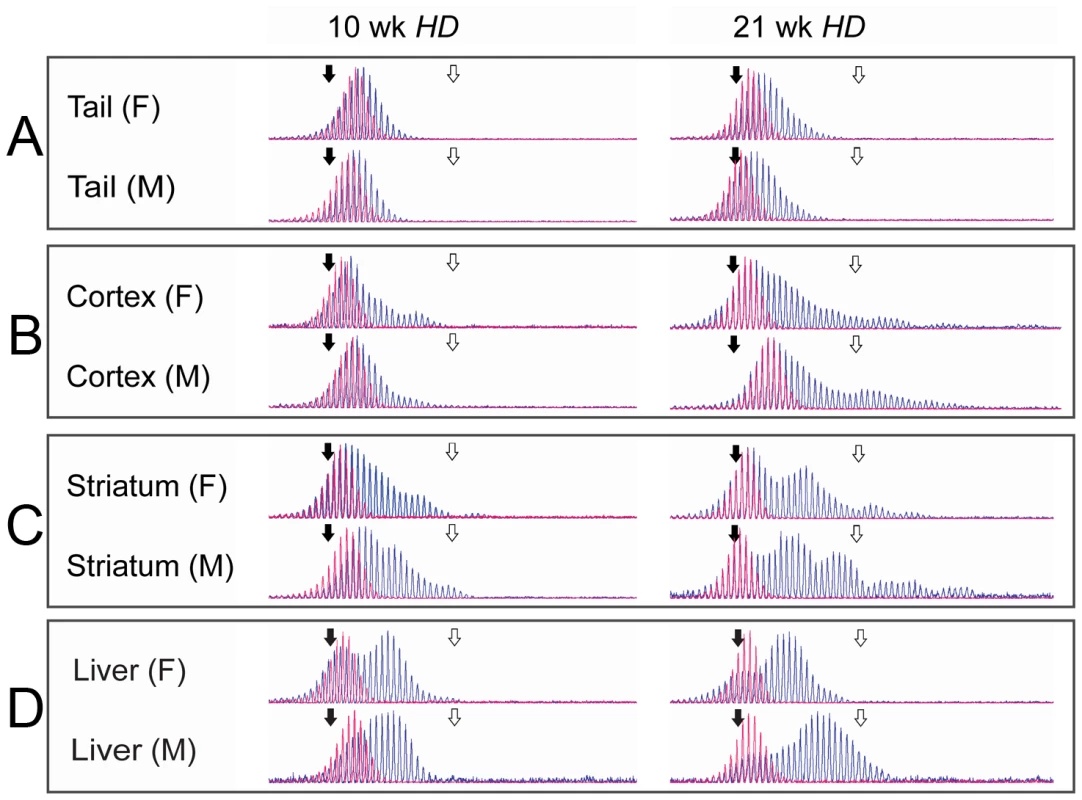
Representative examples of raw data from CAG-repeat sequences in tail (A), cortex (B) striatum (C) and liver (D) from individual male and female HD mice, aged 10 and 21 weeks are shown (blue) with the tail biopsy from the same 3-week old mouse overlaid (red). All traces demonstrate the increase in mean length of repeat sequences with time and the differing rates of expansion between tissue types. Of particular note is the strong periodicity shown in the older striatum samples. Size standard markers are shown for 118 (solid black arrow) and 138 (open white arrow) CAG repeats respectively. Continuous expansion of CAG repeats in tail
Individual fragments from tail fit well to a normal distribution and are thus described by the mean (μ) and standard deviation (σ) of the curves fitted to raw data (Figure 2A, 2B; see Methods). Expansion within the population of 59 mice (10 week old mice excluded) is clearly shown (Figure 2C) by the relative difference in μ of the 21-week and 3-week groups. The median expansion found in 59 tails of 21-week mice was 1.97 CAG triplets (Figure 2E). In addition, σ of individual tail data sets is shown to increase from 1.98 triplets at 3-weeks to 2.87 triplets at 21-weeks (Figure 2D). The increasing σ is not an artefact of PCR errors, as is demonstrated in Figure 3A.
Fig. 2. Slight continuous expansion measured in tail tissue. 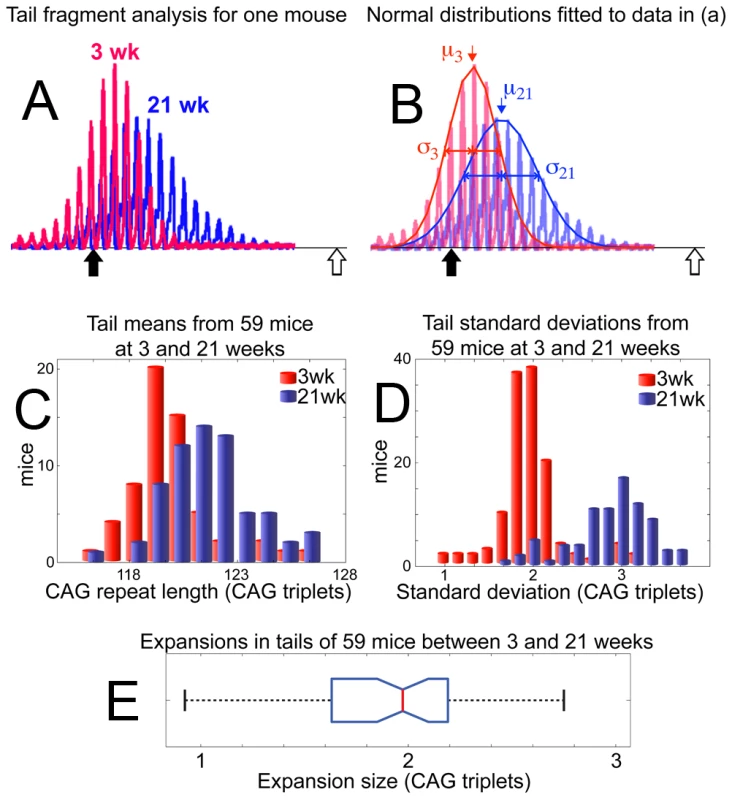
Size standard markers, are placed at 118 (solid black arrow) and 138 (open white arrow) CAG repeats. (A) A representative example of raw data from the 3-week biopsy (red) and 21-week sample (blue) are shown for one mouse. (B) Normal distributions are fitted to the data presented in (A), coloured as previously for 3-week (red) and 21-week (blue) samples. The resulting means (μ) and standard deviations (σ) are used to define the temporal change in repeat distribution within the sample, clearly demonstrating an increase in the mean number of repeats between 3 weeks (μ3) and 21 weeks of age (μ21). Likewise, broadening of the distribution is evident from the increase in standard deviation at 3 weeks (σ3) to that at 21 weeks (σ21). (C) Mean values (μ) for repeat lengths from the 3-week (red) and 21-week (blue) tails samples of 59 mice are compiled into a histogram, showing the systematic increase in repeat length with age. (D) Standard deviations of all 59 tail samples at 3-weeks (red) and 21-weeks (blue) are similarly compiled, with the histogram showing age dependent peak broadening. (E) A boxplot of expansions measured in the tails of 59 mice between 3 and 21 weeks of age shows a median expansion of 1.97 CAG repeats. Fig. 3. Age-dependent increase in mean and standard deviation for tail tissue. 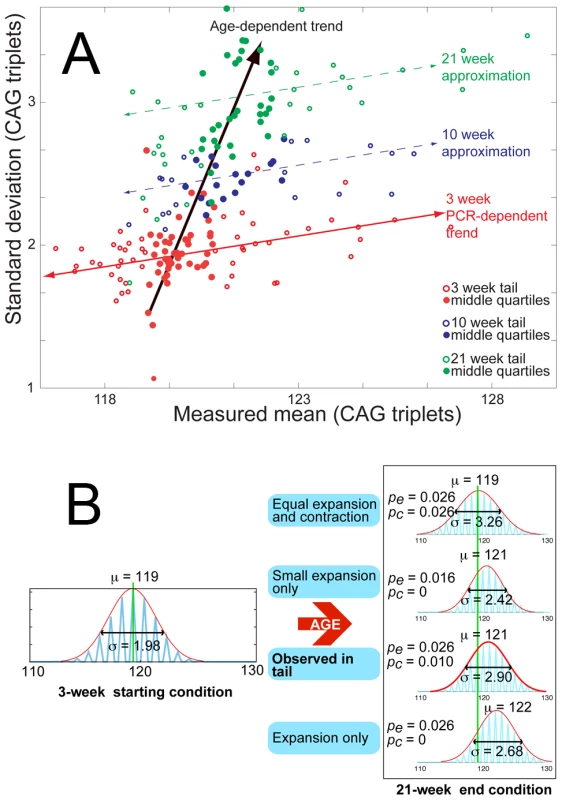
(A) Mean versus standard deviation points are plotted for all 103 mice in the data set, with the data divided into 3-week (red), 10-week (blue) and 21-week (green) age groups. To highlight the general trends in the data, the points representing the middle quartiles (by mean value) for each age group are shown with solid circles. The 3-week age group is shown with the weak positively-correlated trend line (red) implying a loose relation between standard deviation and mean value at a fixed age, which we assume to be caused by polymerase errors – during PCR of repeat sequences – which increase with sequence length. This trait is also present in 10-week and 21-week data, with parallel approximate trends shown (dashed) for emphasis. The standard deviations and means for each age group are, however, shown to increase systematically with age, along a trend-line (black arrow) that is completely separate from the PCR-dependent trend. This demonstrates the independence of the age-dependent increase in standard deviation and mean from the PCR induced variation. (B) Monte Carlo simulation of the proposed model mechanism for expansion is shown with a range of parameters for expansion and contraction probabilities pe and pc, to illustrate the change in mean and standard deviation of a distribution from a given starting point, dependent upon the relative expansion and contraction probabilities. It is clear from the results that only the combination of probabilities calculated from tail data can combine to generate the measured simultaneous change in mean and standard deviation. Having made these observations, it is necessary to consider them in the context of potential models for expansion, in order to fully investigate their implications and attempt to parameterize the processes involved. A continuous increase in both mean and standard deviation can be generally accounted for by multiple stochastic unitary (single CAG-repeats) extension and contraction events on the CAG tracts within the sample (Figure 3B). A full discussion of the potentially applicable models and considerations is presented as supporting information (Figure S8, Text S1, and Videos S1, S2, S3, S4, S5, S6, S7, S8) and we confine ourselves here to a simple application of the most probable hypothesis, yielding upper estimates for expansion and contraction rates. Assigning probabilities to non-simultaneous unitary expansions and contractions, and respectively, allows the measured temporal change in the mean () and variance () of tail samples to be defined by (1) and (2).(1)(2)Using a time interval () of one day, the measured expansion of 1.97 repeats and the concomitant increase in average standard deviation from 1.98 to 2.87, the values of and are calculated to be 0.026 and 0.010 respectively. This gives a maximum expectation of ∼0.036 (pe+pc) events per repeat tract per day.
Periodic expansions in striatum and cortex
In contrast to fragment data from tail, heart, lung and spleen, raw data from 10 - and 21-week cortex and striatum tissue show a peak retained at the 3-week repeat level alongside an age-dependent number of periodically spaced subsequent peaks (Figure 4A, Figures S1, S2, and S10), to which a series of normal distributions were fitted (see Methods). Knowing that the relative areas of overlapping distributions define the proportion of each mean CAG repeat length present (demonstrated with mixed samples and serial dilutions of a 21-week striatum sample in Figures S4 and S5), we infer – on account of the regularity of the intervals between neighbouring peaks at both 10 - and 21-weeks of age – that expansion involves a proportion of the brain tissue undergoing insertions of consistent-length CAG repeat fragments over time. If expansion events inserted CAG fragments of uncontrolled length, the clarity of subsequent peaks would be lost. Likewise if different cell-types within one sample expanded at different rates, one would expect a continuum of peak separations in the collected accumulated data from many mice, and would have little reason to expect a consistent periodicity between peaks at 10-weeks and 21-weeks. This argues for the stochastic step-wise insertion of CAG fragments with an average length μb−μa (Figure 4A (10wk and 21wk) and 4B) by a mechanism that may recur within the same cell. To measure the periodicity seen in brain tissues, we compiled histograms of all intervals between identifiable peaks, binned by size, from the individual cortex and striatum samples of mice aged 10 - and 21-weeks (Figure 4C). The measured intervals are clearly shown to be distributed around a peak at 7 CAG repeats, with a mean length of 7.14 (σ = 1.78 with a cut-off for doublet measurements set at intervals ≥12). The median interval of 7 also confirms that this expansion process is centred around the insertion of 7-repeat fragments into the CAG tract, although the width of the starting distribution indicates insertion of 5 to 9 repeat-fragments (see Figure S9 and Videos S1, S2, S3, S4, S5, S6, S7, S8 for further discussion). The relative sizes of peaks in 21-week striatum (see Methods) imply that on average, a total of ∼10,000 7-repeat insertions occurred in each dramatically expanding striatal sample (on our timescale probably mainly within neurons; however, glial cells also undergo expansion and not all neuronal cells are guaranteed to show expansion [14], [15], giving a periodic expansion probability estimate of 0.018 events per repeat tract per day. The fact that efficiency of PCR amplification of longer CAG tracts is reduced, may result in some measure of underestimation with this value. This is ∼70% of the estimated probability for unitary expansion events in tail, however the 7-repeat average insertion size renders the resulting expansion more dramatic in striatum. In contrast, liver data while showing comparable levels of average expansion to brain tissues, lacks a clear signature of periodicity (see Figure 1D) tending towards bimodality, with a more continuously located second peak. This would imply a much less controlled insertion length during the expansion process, or possibly a combination of expansion mechanisms.
Fig. 4. Periodic expansions in striatum and cortex. 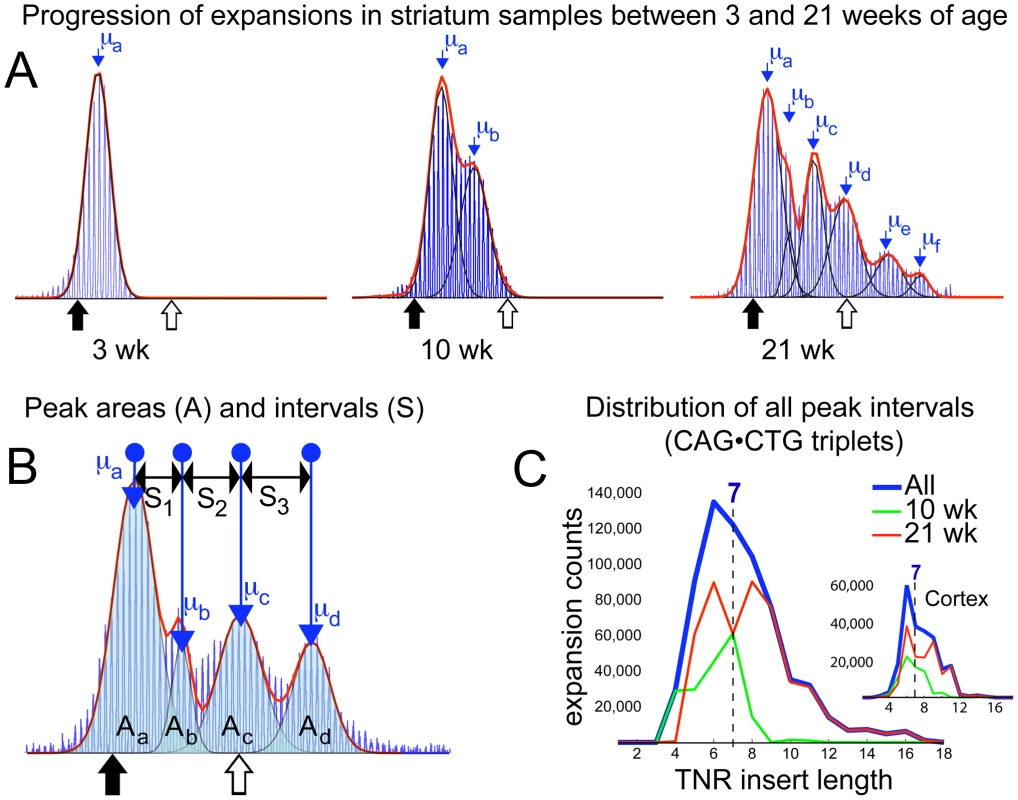
(A) Fragment analysis curves from 3-week tail, 10- and 21-week striatum are shown sequentially, with fitted normal distributions (black) overlaid. The sum of all fits (red) demonstrates the curve-fit accuracy. Mean values (μa etc) for each fitted peak are shown. Mean μa coincides with the mean of the corresponding 3-week tail at all ages. This 21-week striatum sample is best-fit by six consecutive normal distributions. Periodicity is clear from the regularity of the intervals between consecutive means. (B) Multiple peaks fitted to another 21-week striatum dataset are shown, with the areas under each fitted distribution (Aa .. Ad), the mean values and the separations between the four peaks (S1 .. S3). The area of a peak represents the proportion of tissue containing the measured mean number of repeats, thus separation values represent a step-wise expansion from the previous mean. The age-dependent propagation of peaks with higher means, as seen in (A), is due to the stochastic insertion of short repeat sequences, of consistent length, into the CAG tracts of individual cells, which, over time, generates the observed periodicity. For analysis purposes, the sum of all areas (Aa,b,c,d) was rescaled to 10,000 (the approximate number of cells forming a sample), allowing the number and length of insertion events to be estimated. (C) A histogram of insertion lengths for all expansion events measured in 69 separate striatum samples is shown (blue). Both mean and median values of the distribution point to a dominant insertion length of seven CAG repeats. The separate contributions from 10-week (green) and 21-week (red) data are also shown. Inset figure shows a similar result for cortex, with the insertion length distributed at 7 repeats, despite the smaller number of insertion events observed in total. Discussion
Having shown data and analysis to define these two distinct modes of expansion, we place our findings into the context of existing literature, in order to develop reasonable hypothetical models for these two types of expansion.
The continuous expansion we observe shows a progressive increase in CAG tract length, which is comparable with the expansions observed in fibroblasts derived from an adult HD mouse [37]. In the debate regarding the relative roles of replication and repair in TNR instability, these results present an interesting question, since the continuous expansion process occurs in organs containing dividing cells. A process occurring with the previously calculated expectation rate of ∼0.036 events per repeat tract per day on the ∼360 nucleotide CAG segment of the 2.5 gigabase mouse genome, would correlate to ∼250,000 genome-wide events per cell per day. This is well above the upper estimate for daily DNA damage events, making it unlikely that these are the sole initiator of expansions in tail tissue. Several replication-based models for TNR expansion in dividing cells [18], [19] have been proposed. The potential for replication to be entirely responsible for this expansion is considered in detail elsewhere (Figures S7, S8, and Text S1) and is considered to be unlikely, particularly in light of the fact that lymphoma tissues (with necessarily higher replication rates) isolated from several mice showed no increase in TNR instability (Figure S7). Therefore we infer that other mechanisms must also prevail, and propose that slipped strand structures [16], [38] generated by out of register rehybridization of CAG repeats [27] during transcription, or genome maintenance, may spontaneously form unstable loops or cruciforms which may subsequently stabilize by migrating apart. Similar small loops can also be formed by polymerase slippage during replication [17]. We propose that two separate pathways may repair these loops, leading to single repeat expansions or contractions (Figure S8). Further research is needed to resolve the specific origin of this mode of expansion.
A hypothesis for periodic expansion is also presented (Figure S9). The previously calculated periodic expansion probability of 0.018 events per repeat tract per day would correlate to ∼100,000 genome-wide events per cell per day. This lies in the vicinity of upper estimates for accumulated oxidative DNA damage [39]. It is therefore possible that DNA damage contributes as catalyst for periodic expansions in brain tissues. However, oxidative damage is not sufficient to trigger somatic instability [33]. Of particular interest here, are the potential molecular components that could repeatedly generate a regularly sized repeat insertion that is dominantly seven repeats in length. We have therefore chosen to briefly review the relevant literature in search of further insight.
Theory suggests that CAG flaps ranging from 4 to 16 triplet repeats in length can form thermodynamically unstable hairpin structures with an even number of repeats [40]. However, under physiological conditions, 6 triplet repeats have been shown to form hairpins irreversibly [41], [42]. This implies that a progressively generated triplet repeat flap can stabilise into a 6-repeat hairpin at the free end that could be cleaved by FEN1 causing no net expansion [43] (Figure S9). However, the presence of metastable intermediates during flap generation may allow the flap length to increase beyond 6 repeats before a stable structure is formed, thereby producing a hairpin with an overhanging CAG at the 5′-end. This free CAG repeat can hybridise back to the DNA duplex, and it has been shown by Liu et al. [31] that such hybridisation facilitates bubble formation followed by ligation and expansion. In a few instances, two or even three free CAG repeats in the 5′-end of the hairpin would produce a periodicity of eight or nine CAG repeat steps. In addition, the size of the hairpin could vary with a few repeats. However, this occurs more rarely, as observed in Figure 4C. After gap filling and ligation, a loop of excess CAG repeats in one strand would be produced (Figure S9) that can be recognized by the MMR complex Msh2-Msh3 with high affinity [44]. This binding might further stabilize the CAG loop and additionally explain why Msh2-Msh3 is necessary for large expansions to occur in striatum [21]–[24], although the role of MMR in causing TNR expansion is not understood. Moreover, Msh2-Msh3 function ceases due to impaired ATPase activity on loops exceeding 16 nucleotides in lengths [45] - probably due to the presence of A•A mispaired bases in the loop [22] - which may explain why the MMR system fails to repair longer extraneous CAG loops. Subsequently, a nick generated on the strand opposite to the loop could result in faulty repair of the CTG strand along the CAG slip-out, causing expansion by a repair process independent of MMR [46] (Figure S9). Thus, oxidative damage on the CTG strand could result in base excision repair (BER) and Ogg1 moderated expansion, which is also in concordance with the proposed ‘toxic oxidation cycle’ by Kovtun et al., 2007 [13]. We therefore propose that a coincidental cooperation can occur between MMR and BER in cases where a long CAG flap is able to stabilize itself, which can form the basis of the consistent periodic insertion we observe.
It should be pointed out that it is possible that other stabilized structures, such as loop-outs or a stabilizing interaction with one of the many proteins and complexes that are in contact with the DNA may also serve to cause the observed periodicity. The majority of the potentially applicable models are covered in the literature [13], [24], [27], [34]. Further work is necessary to resolve this completely.
Cell proliferation in neurons and glial cells has been observed in the subependymal layer adjacent to the caudate nucleus in human HD brains [47]. However, polymerase slippage usually forms small expansions [17], and the repetitive uniformity of the periodic expansion makes it unlikely that polymerase slippage is responsible for the dramatic expansions seen in cortex and striatum. Furthermore, the lack of periodically spaced peaks containing fewer repeats than the mice were born with – as would be expected from a bi-directional process - means that 7-repeat hairpin-based contraction events occur either at a negligible rate in comparison to expansions, or do not occur at all. During the 18 week period, tail-type expansion events are not evident in striatum since they would obfuscate latter periodic peaks (Figure S6). Therefore, the two expansion mechanisms seem to be either entirely independent – not occurring simultaneously in the same cell – or that if they do share common elements, they progress along mutually exclusive pathways. However, it is important to notice that both expansion mechanisms must be dependent on proteins from the MMR complex, since expansions are eliminated in all tissues in either Msh2 or Msh3 nulls in HD [21]–[24], and also in DM1 [25], [26]. To date, we have not managed to define the individual cell types that are specific to these expansion mechanisms. It will be of great interest to compare the expansions observed in animal models to those in cultured HD mouse fibroblasts, as a way of identifying the cell specificity of these modes of expansion.
The liver may be particularly interesting in this regard, since this tissue potentially exhibits a mix of both types of expansion mechanisms. This could be attributed to different modes of expansion occurring in different cell types. Indeed, instability of the DM1 CTG•CAG repeat is known to occur in liver hepatocytes with high DNA ploidy [48].
The question also arises as to why FEN1 did not influence CAG repeat expansion in the organs tested. One might expect a difference, since a recent in vitro study has shown that FEN1, together with long-patch BER of long repeat sequences by polymerase β, promotes expansion by facilitating the ligation of hairpins formed by strand slippage [49]. However, FEN1 flap activity has shown to be circumstantial, with much lower activity in the striatum than in the cerebellum of R6/1 mice [33]. In yeast the capture of flap structures by FEN1, rather than the endonuclease activity, is the most important function of FEN1 in preventing TNR expansion [50]. EXO - and GAP activity of FEN1 are also involved in in vitro triplet repeat expansion in yeast [32], and these activities are probably not influenced by the Fen1E160D/E160D mutation. Therefore, it seems that the 20% endonuclease activity [36] of the Fen1E160D/E160D mutation does not affect the rate of CAG repeat expansions. In concordance with our finding Fen1 did not control instability of (CTG)n*(CAG)n repeats in a knock-in mouse model for DM1 [51].
It is worth considering briefly why this periodicity has not been described before, since there are numerous potential reasons. One possibility could be that the mice used in the present study exhibit more instability due to environmental factors [52] or genetic background [53]. Perhaps the periodic signal becomes more disperse in older mice used elsewhere; degrading the quality of the data, and that the age-range, as well as the relatively long starting CAG lengths, of our samples is optimal for observing this periodicity. Another possibility is simply that later versions of the GeneMapper system are more sensitive, in comparison to the GeneScan method applied in some older studies, allowing us to see more detail. While periodicity has not featured in other studies of similar tissues and disease models, it is difficult to state unequivocally that it was unobservable in their data. The small volume of data presented in articles, uncertainty over the precise PCR conditions used and the simple fact that periodicity was not the focus of these investigations can be sufficient cause for this phenomenon to have been previously overlooked. There is some variability among replicate striatum samples as shown in Figure S10. This could be explained by sampling error or polymerase slippage in early PCR cycles. Sampling error is however unlikely to be the reason behind the periodicity as explained in Figure S12 and Text S2.
So far, we have only studied periodicity in the R6/1 mouse model and without specific studies of other HD CAG mice models the generality of our data is unknown. Yet, the R6/1 transgenic mouse is a widely accepted and commonly used model for human HD that exhibits a progressive neurological phenotype that exhibits many of the features of HD [54]. HD CAG repeat instability has shown to be similar in humans and mice, with the longest expansion lengths occurring in striatum, followed by cortex, and little expansion in cerebellum and most other tissues [8], [14], [35]. In addition, the HD CAG repeat length appears to be expand most in neuronal cells rather than glial cells in both species [14], [15]. Due to the long starting CAG repeat length in the transgenic mouse, the model may be most relevant as a model for juvenile HD. Given the stated similarities, there are grounds to suspect that the mechanisms of expansion are identical in mice and humans, only occurring at an accelerated pace in the mouse on account of the long repeat tract. In this case, the mouse model would function as a good model for human cases with mid-life age of onset as well. However, periodicity has not previously been reported in HD patients. A possible explanation could be that the repeat length in the R6/1 mouse is longer than the repeat length that has been analyzed in any HD human tissue with regard to somatic instability. The genomic localization of the randomly integrated HD gene fragment in R6/1 mice might modulate the CAG stability. Furthermore, CAG instability in HD patient brain cannot be analyzed at an early time-point that would allow for a direct comparison to the R6/1 data.
Importantly, CAG repeat expansion in human cortex is associated with an earlier age of disease onset, in addition to the role of the constitutional CAG repeat length [6]. This implies that there are disease modifiers that influence somatic instability, and conversely, factors that determine somatic instability which may modify disease pathogenesis. It is therefore critical to understand the mechanism of the different expansion processes and the factors involved.
To summarize, we present two different mechanisms of somatic CAG repeat expansion; a continuous bi-directional expansion in tail, lung, heart and spleen tissues, and a dramatic periodic expansion centred around 7-repeat insertions in striatum and cortex. Further experiments are needed to determine whether the 7-repeat step-size is independent of CAG tract length and species and it remains to be shown whether these two models can explain the expansions observed in other brain tissues and organs, as well as in humans. Nevertheless, these results provide significant new insights into in vivo expansion mechanisms, which may also be relevant to other triplet repeat disorders.
Methods
Ethics statement
All animal experiments have been approved by the local and national animal - and are carried out according to the regulation by FELASA (Federation of European Laboratory Animal Science Associations).
Animals and tissues
B6CBA-Tg(HDexon1)61pb/J mice of the R6/1 line [54] with ∼119 CAG repeats in exon 1 of the HTT gene, were purchased from The Jackson Laboratories and either interbred or crossed with C57BL/6J Fen1E160D/E160D mice [36]. The mice were fed Rat and Mouse No.3 breeding diet (Special Diet Services) and tap water ad libitum. At 3 weeks of age the mice were anesthetized by i.p. injection of a combination of Midazolam (Dormicum “Roche”) and Fentanyl/Fluanisone (Hypnorm) solutions, and tail biopsies were taken. DNA from tail biopsies of the first two generations were lysed as described [36] and DNA isolated using standard NaCl precipitation or phenol/chloroform procedure. At 10 and 21 weeks of age the mice were sacrificed by cervical dislocation. The organs were harvested, frozen on dry ice and stored at −70°C. During dissection of striatum we lost 9 samples. DNA from all tissues and tails from the F2 and F3 generation was isolated according to the DNeasy Blood & Tissue kit (Qiagen GmbH, Germany).
Genotyping
DNA from tail biopsies of 3-week old mice were used for genotyping. HD mice were genotyped with forward 5′-cggctgaggcagcagcggctgt-3′ and reverse 5′-gcagcagcagcagcaacagccgccaccgcc-3′ PCR primers [54] according to the Advantage GC 2 PCR Kit & Polymerase Mix (Clontech, CA). The Fen1+/+ and/or Fen1E160D/E160D knock-in allele was genotyped as described [36].
Sizing of CAG repeats
CAG repeats were sized by PCR with primers 5′-FAM-atgaaggccttcgagtccctcaagtccttc-3′ and 5′-ggcggctgaggaagctgagga-3′ according to [54] with slight modifications. Approximately 75ng of genomic DNA (this approximates to DNA extracts from ∼10,000 cells) was amplified with AmpliTaq Gold DNA polymerase with PCR Buffer II, 1.25 mM MgCl2 (Applied Biosystems, CA), and 2.5 mM dNTPs (GE Healthcare). The cycling conditions were 94°C for 10 min, 35 cycles of 94°C for 30 sec, 64°C for 30 sec, 72°C for 2 min, and a final extension at 72°C for 10 min. The FAM-labeled PCR products were mixed with GeneScan - 600 LIZ Size Standard and HiDi Formamide (Applied Biosystems) and run on an ABI 3730 Genetic Analyzer (Applied Biosystems). Sizing of the PCR fragments was performed by using the GeneMapper Software Version 3.7 (Applied Biosystems).
Data analysis
All raw data was processed through a masked Nelder-Mead simplex fitting method, optimising free parameters of standard deviation, mean and amplitude to fit consecutive normal distributions sufficient to account for ≥98% of the total area of the raw data set. In the case of tail data, only a single normal distribution was required. These optimised parameters were returned as the means (μ), and standard deviations (σ), which were used to define the TNR lengths present in each data set. CAG repeat tracts were flanked by sequences 86 bp in length as verified by sequencing. Thus, the mean number of CAG triplets (μt) present in a fragment analysis sample with a measured mean (μm) is defined by μt = (μm−86)/3. When analysing the periodicity present in striatum and cortex data, standard frequency analysis methods are not suitable, therefore our peak fitting method was used to fit consecutive normal distributions to the raw data (Figure 4A). We were unable to perform fitting analysis on 25 striatum samples due to the quality of the PCR product. The means (μa, μb etc) and relative areas (Aa, Ab etc) of each peak were calculated (Figure 4B). The intervals between neighbouring means (S1, S2 etc) were also recorded at both 10 and 21 weeks. The area (A) of each peak was used to estimate the number of cells containing triplet repeats that had expanded with a step size defined by the separation interval (S). The average area of the first peak in all 21-week data (Aa from Figure 4B) was used to estimate the proportion of non-expanding cells in the striatum as 54% (σ = 15.9), implying that approximately 45% of each striatum sample underwent periodic expansions within the first 21 weeks. By comparison, the proportion of dramatic expanding tissue observed in cortex samples was ∼20%. Previous work has shown that the dramatic expansion observed in the striatum of adult mouse brain tissue largely occur in the neuronal cells [15], [14], although slower expansion can be observed in glial cells and we used this to approximate the level of expansion in neuronal cells in combination with an estimate of the average number of expansion events that were measured in 21-week mice.
Supporting Information
Zdroje
1. The Huntington's Disease Collaborative Research Group 1993 A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell 72 971 983
2. AndrewSE
GoldbergYP
KremerB
TeleniusH
TheilmannJ
1993 The relationship between trinucleotide (CAG) repeat length and clinical features of Huntington's disease. Nat Genet 4 398 403
3. DuyaoM
AmbroseC
MyersR
NovellettoA
PersichettiF
1993 Trinucleotide repeat length instability and age of onset in Huntington's disease. Nat Genet 4 387 392
4. AmbroseC
MyersR
NovellettoA
PersichettiF
FrontaliM
1993 Relationship between trinucleotide repeat expansion and phenotypic variation in Huntington's disease. Nat Genet 4 393 397
5. StineOC
PleasantN
FranzML
AbbottMH
FolsteinSE
1993 Correlation between the onset age of Huntington's disease and length of the trinucleotide repeat in IT-15. Hum Mol Genet 2 1547 1549
6. SwamiM
HendricksAE
GillisT
MassoodT
MysoreJ
2009 Somatic expansion of the Huntington's disease CAG repeat in the brain is associated with an earlier age of disease onset. Hum Mol Genet 18 3039 3047
7. SakazumeS
YoshinariS
OgumaE
UtsunoE
IshiiT
2009 A patient with early onset Huntington disease and severe cerebellar atrophy. Am J Med Genet A 149A 598 601
8. TeleniusH
KremerB
GoldbergYP
TheilmannJ
AndrewSE
1994 Somatic and gonadal mosaicism of the Huntington disease gene CAG repeat in brain and sperm. Nat Genet 6 409 414
9. ChongSS
McCallAE
CotaJ
SubramonySH
OrrHT
1995 Gametic and somatic tissue-specific heterogeneity of the expanded SCA1 CAG repeat in spinocerebellar ataxia type 1. Nat Genet 10 344 350
10. UenoS
KondohK
KotaniY
KomureO
KunoS
1995 Somatic mosaicism of CAG repeat in dentatorubral-pallidoluysian atrophy (DRPLA). Hum Mol Genet 4 663 666
11. ItoY
TanakaF
YamamotoM
DoyuM
NagamatsuM
1998 Somatic mosaicism of the expanded CAG trinucleotide repeat in mRNAs for the responsible gene of Machado-Joseph disease (MJD), dentatorubral-pallidoluysian atrophy (DRPLA), and spinal and bulbar muscular atrophy (SBMA). Neurochem Res 23 25 32
12. KennedyL
EvansE
ChenCM
CravenL
DetloffPJ
2003 Dramatic tissue-specific mutation length increases are an early molecular event in Huntington disease pathogenesis. Hum Mol Genet 12 3359 3367
13. KovtunIV
LiuY
BjorasM
KlunglandA
WilsonSH
2007 OGG1 initiates age-dependent CAG trinucleotide expansion in somatic cells. Nature 447 447 452
14. ShelbournePF
Keller-McGandyC
BiWL
YoonSR
DubeauL
2007 Triplet repeat mutation length gains correlate with cell-type specific vulnerability in Huntington disease brain. Hum Mol Genet 16 1133 1142
15. GonitelR
MoffittH
SathasivamK
WoodmanB
DetloffPJ
2008 DNA instability in postmitotic neurons. Proc Natl Acad Sci U S A 105 3467 3472
16. SindenRR
Pytlos-SindenMJ
PotamanVN
2007 Slipped strand DNA structures. Front Biosci 12 4788 4799
17. KunkelTA
1993 Nucleotide repeats. Slippery DNA and diseases. Nature 365 207 208
18. MirkinSM
2005 Toward a unified theory for repeat expansions. Nat Struct Mol Biol 12 635 637
19. MirkinSM
2006 DNA structures, repeat expansions and human hereditary disorders. Curr Opin Struct Biol 16 351 358
20. KovtunIV
McMurrayCT
2008 Features of trinucleotide repeat instability in vivo. Cell Res 18 198 213
21. ManleyK
ShirleyTL
FlahertyL
MesserA
1999 Msh2 deficiency prevents in vivo somatic instability of the CAG repeat in Huntington disease transgenic mice. Nat Genet 23 471 473
22. OwenBA
YangZ
LaiM
GajekM
BadgerJD
2005 (CAG)(n)-hairpin DNA binds to Msh2-Msh3 and changes properties of mismatch recognition. Nat Struct Mol Biol 12 663 670
23. DragilevaE
HendricksA
TeedA
GillisT
LopezET
2009 Intergenerational and striatal CAG repeat instability in Huntington's disease knock-in mice involve different DNA repair genes. Neurobiol Dis 33 37 47
24. WheelerVC
LebelLA
VrbanacV
TeedA
teRH
2003 Mismatch repair gene Msh2 modifies the timing of early disease in Hdh(Q111) striatum. Hum Mol Genet 12 273 281
25. van den BroekWJ
NelenMR
WansinkDG
CoerwinkelMM
teRH
2002 Somatic expansion behaviour of the (CTG)n repeat in myotonic dystrophy knock-in mice is differentially affected by Msh3 and Msh6 mismatch-repair proteins. Hum Mol Genet 11 191 198
26. SavouretC
BrissonE
EssersJ
KanaarR
PastinkA
2003 CTG repeat instability and size variation timing in DNA repair-deficient mice. EMBO J 22 2264 2273
27. Gomes-PereiraM
FortuneMT
IngramL
McAbneyJP
MoncktonDG
2004 Pms2 is a genetic enhancer of trinucleotide CAG.CTG repeat somatic mosaicism: implications for the mechanism of triplet repeat expansion. Hum Mol Genet 13 1815 1825
28. RumbaughJA
HenricksenLA
DeMottMS
BambaraRA
1999 Cleavage of substrates with mismatched nucleotides by Flap endonuclease-1. Implications for mammalian Okazaki fragment processing. J Biol Chem 274 14602 14608
29. KlunglandA
LindahlT
1997 Second pathway for completion of human DNA base excision-repair: reconstitution with purified proteins and requirement for DNase IV (FEN1). EMBO J 16 3341 3348
30. SpiroC
PelletierR
RolfsmeierML
DixonMJ
LahueRS
1999 Inhibition of FEN-1 processing by DNA secondary structure at trinucleotide repeats. Mol Cell 4 1079 1085
31. LiuY
BambaraRA
2003 Analysis of human flap endonuclease 1 mutants reveals a mechanism to prevent triplet repeat expansion. J Biol Chem 278 13728 13739
32. SinghP
ZhengL
ChavezV
QiuJ
ShenB
2007 Concerted action of exonuclease and Gap-dependent endonuclease activities of FEN-1 contributes to the resolution of triplet repeat sequences (CTG)n - and (GAA)n-derived secondary structures formed during maturation of Okazaki fragments. J Biol Chem 282 3465 3477
33. GoulaAV
BerquistBR
WilsonDMIII
WheelerVC
TrottierY
2009 Stoichiometry of base excision repair proteins correlates with increased somatic CAG instability in striatum over cerebellum In Huntington's disease transgenic mice. PLoS Genet 5 e1000749 doi:10.1371/journal.pgen.1000749
34. MirkinSM
2007 Expandable DNA repeats and human disease. Nature 447 932 940
35. MangiariniL
SathasivamK
MahalA
MottR
SellerM
1997 Instability of highly expanded CAG repeats in mice transgenic for the Huntington's disease mutation. Nat Genet 15 197 200
36. LarsenE
KleppaL
MezaTJ
Meza-ZepedaLA
RadaC
2008 Early-onset lymphoma and extensive embryonic apoptosis in two domain-specific Fen1 mice mutants. Cancer Res 68 4571 4579
37. ManleyK
PughJ
MesserA
1999 Instability of the CAG repeat in immortalized fibroblast cell cultures from Huntington's disease transgenic mice. Brain Res 835 74 79
38. PearsonCE
TamM
WangYH
MontgomerySE
DarAC
2002 Slipped-strand DNAs formed by long (CAG)*(CTG) repeats: slipped-out repeats and slip-out junctions. Nucleic Acids Res 30 4534 4547
39. FragaCG
ShigenagaMK
ParkJW
DeganP
AmesBN
1990 Oxidative damage to DNA during aging: 8-hydroxy-2′-deoxyguanosine in rat organ DNA and urine. Proc Natl Acad Sci U S A 87 4533 4537
40. ZukerM
2003 Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31 3406 3415
41. VolkerJ
MakubeN
PlumGE
KlumpHH
BreslauerKJ
2002 Conformational energetics of stable and metastable states formed by DNA triplet repeat oligonucleotides: implications for triplet expansion diseases. Proc Natl Acad Sci U S A 99 14700 14705
42. AmraneS
SaccaB
MillsM
ChauhanM
KlumpHH
2005 Length-dependent energetics of (CTG)n and (CAG)n trinucleotide repeats. Nucleic Acids Res 33 4065 4077
43. HenricksenLA
TomS
LiuY
BambaraRA
2000 Inhibition of flap endonuclease 1 by flap secondary structure and relevance to repeat sequence expansion. J Biol Chem 275 16420 16427
44. PearsonCE
EwelA
AcharyaS
FishelRA
SindenRR
1997 Human MSH2 binds to trinucleotide repeat DNA structures associated with neurodegenerative diseases. Hum Mol Genet 6 1117 1123
45. WilsonT
GuerretteS
FishelR
1999 Dissociation of mismatch recognition and ATPase activity by hMSH2-hMSH3. J Biol Chem 274 21659 21664
46. PanigrahiGB
ClearyJD
PearsonCE
2002 In vitro (CTG)*(CAG) expansions and deletions by human cell extracts. J Biol Chem 277 13926 13934
47. CurtisMA
PenneyEB
PearsonAG
van Roon-MomWM
ButterworthNJ
2003 Increased cell proliferation and neurogenesis in the adult human Huntington's disease brain. Proc Natl Acad Sci U S A 100 9023 9027
48. van den BroekWJ
WansinkDG
WieringaB
2007 Somatic CTG*CAG repeat instability in a mouse model for myotonic dystrophy type 1 is associated with changes in cell nuclearity and DNA ploidy. BMC Mol Biol 8 61
49. LiuY
PrasadR
BeardWA
HouEW
HortonJK
2009 Coordination between polymerase beta and FEN1 can modulate CAG repeat expansion. J Biol Chem 284 28352 28366
50. LiuY
ZhangH
VeeraraghavanJ
BambaraRA
FreudenreichCH
2004 Saccharomyces cerevisiae flap endonuclease 1 uses flap equilibration to maintain triplet repeat stability. Mol Cell Biol 24 4049 4064
51. van den BroekWJ
NelenMR
van der HeijdenGW
WansinkDG
WieringaB
2006 Fen1 does not control somatic hypermutability of the (CTG)(n)*(CAG)(n) repeat in a knock-in mouse model for DM1. FEBS Lett 580 5208 5214
52. vanDA
BlakemoreC
DeaconR
YorkD
HannanAJ
2000 Delaying the onset of Huntington's in mice. Nature 404 721 722
53. LloretA
DragilevaE
TeedA
EspinolaJ
FossaleE
2006 Genetic background modifies nuclear mutant huntingtin accumulation and HD CAG repeat instability in Huntington's disease knock-in mice. Hum Mol Genet 15 2015 2024
54. MangiariniL
SathasivamK
SellerM
CozensB
HarperA
1996 Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell 87 493 506
Štítky
Genetika Reprodukční medicína
Článek Genome-Wide Interrogation of Mammalian Stem Cell Fate Determinants by Nested Chromosome DeletionsČlánek Season of Conception in Rural Gambia Affects DNA Methylation at Putative Human Metastable EpiallelesČlánek A Quantitative Systems Approach Reveals Dynamic Control of tRNA Modifications during Cellular StressČlánek Reduction of Protein Translation and Activation of Autophagy Protect against PINK1 Pathogenesis inČlánek The Loss of PGAM5 Suppresses the Mitochondrial Degeneration Caused by Inactivation of PINK1 inČlánek Cleavage of Phosphorothioated DNA and Methylated DNA by the Type IV Restriction Endonuclease ScoMcrAČlánek Competitive Repair by Naturally Dispersed Repetitive DNA during Non-Allelic Homologous Recombination
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2010 Číslo 12
-
Všechny články tohoto čísla
- Genome-Wide Interrogation of Mammalian Stem Cell Fate Determinants by Nested Chromosome Deletions
- Whole-Genome and Chromosome Evolution Associated with Host Adaptation and Speciation of the Wheat Pathogen
- Association of Variants at 1q32 and with Ankylosing Spondylitis Suggests Genetic Overlap with Crohn's Disease
- Initiator Elements Function to Determine the Activity State of BX-C Enhancers
- Identification of Genes Required for Neural-Specific Glycosylation Using Functional Genomics
- A Young Duplicate Gene Plays Essential Roles in Spermatogenesis by Regulating Several Y-Linked Male Fertility Genes
- The EpsE Flagellar Clutch Is Bifunctional and Synergizes with EPS Biosynthesis to Promote Biofilm Formation
- Histone H2A C-Terminus Regulates Chromatin Dynamics, Remodeling, and Histone H1 Binding
- Season of Conception in Rural Gambia Affects DNA Methylation at Putative Human Metastable Epialleles
- A Quantitative Systems Approach Reveals Dynamic Control of tRNA Modifications during Cellular Stress
- GC-Rich Sequence Elements Recruit PRC2 in Mammalian ES Cells
- A Single Enhancer Regulating the Differential Expression of Duplicated Red-Sensitive Opsin Genes in Zebrafish
- Investigation and Functional Characterization of Rare Genetic Variants in the Adipose Triglyceride Lipase in a Large Healthy Working Population
- Reduction of Protein Translation and Activation of Autophagy Protect against PINK1 Pathogenesis in
- Noisy Splicing Drives mRNA Isoform Diversity in Human Cells
- The Loss of PGAM5 Suppresses the Mitochondrial Degeneration Caused by Inactivation of PINK1 in
- Thymus-Associated Parathyroid Hormone Has Two Cellular Origins with Distinct Endocrine and Immunological Functions
- An ABC Transporter Mutation Is Correlated with Insect Resistance to Cry1Ac Toxin
- Role of Individual Subunits of the CSN Complex in Regulation of Deneddylation and Stability of Cullin Proteins
- The C-Terminal Domain of the Bacterial SSB Protein Acts as a DNA Maintenance Hub at Active Chromosome Replication Forks
- The DNA Damage Response Pathway Contributes to the Stability of Chromosome III Derivatives Lacking Efficient Replicators
- Cleavage of Phosphorothioated DNA and Methylated DNA by the Type IV Restriction Endonuclease ScoMcrA
- LaeA Control of Velvet Family Regulatory Proteins for Light-Dependent Development and Fungal Cell-Type Specificity
- Competitive Repair by Naturally Dispersed Repetitive DNA during Non-Allelic Homologous Recombination
- Distinct Functions for the piRNA Pathway in Genome Maintenance and Telomere Protection
- MOS11: A New Component in the mRNA Export Pathway
- Self-Mating in the Definitive Host Potentiates Clonal Outbreaks of the Apicomplexan Parasites and
- A Role for ATF2 in Regulating MITF and Melanoma Development
- Ancestral Regulatory Circuits Governing Ectoderm Patterning Downstream of Nodal and BMP2/4 Revealed by Gene Regulatory Network Analysis in an Echinoderm
- Cancer and Neurodegeneration: Between the Devil and the Deep Blue Sea
- Functional Comparison of Innate Immune Signaling Pathways in Primates
- Linking Crohn's Disease and Ankylosing Spondylitis: It's All about Genes!
- Genomics Meets Glycomics—The First GWAS Study of Human N-Glycome Identifies HNF1α as a Master Regulator of Plasma Protein Fucosylation
- Continuous and Periodic Expansion of CAG Repeats in Huntington's Disease R6/1 Mice
- Expression of Linear and Novel Circular Forms of an -Associated Non-Coding RNA Correlates with Atherosclerosis Risk
- Endocytic Sorting and Recycling Require Membrane Phosphatidylserine Asymmetry Maintained by TAT-1/CHAT-1
- Histone Deacetylases Suppress CGG Repeat–Induced Neurodegeneration Via Transcriptional Silencing in Models of Fragile X Tremor Ataxia Syndrome
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Functional Comparison of Innate Immune Signaling Pathways in Primates
- Expression of Linear and Novel Circular Forms of an -Associated Non-Coding RNA Correlates with Atherosclerosis Risk
- Genome-Wide Interrogation of Mammalian Stem Cell Fate Determinants by Nested Chromosome Deletions
- Histone H2A C-Terminus Regulates Chromatin Dynamics, Remodeling, and Histone H1 Binding
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



