-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaTRIM30α Is a Negative-Feedback Regulator of the Intracellular DNA and DNA Virus-Triggered Response by Targeting STING
Negative-feedback regulation is a broad and pivotal biological event to maintain the homeostasis of the host. Viral DNA species derived from DNA viruses or retroviruses can activate STING signaling to produce pro-inflammatory cytokines and type I interferon, which further recruit immune cells or induce interferon stimulated genes (ISGs) to clear viral infection respectively. However, excessive STING-signaling activation has been shown to induce autoimmune disorders. Thus, it is important to finely turn off STING signaling. Here we demonstrate that TRIM30α is rapidly induced followed by STING activation. Trim30α-deficient mice show more resistance to infection by DNA viruses. Meanwhile, knockdown or genetic ablation of TRIM30α augments the type I IFNs and IL-6 responses to intracellular DNA and DNA viruses. Biochemical analyses show that TRIM30α interacts with STING and promotes the degradation of STING via K48-linked ubiquitination at Lys275. These findings demonstrate that induced TRIM30α is a negative-feedback regulator of STING pathway activation triggered by DNA and DNA viruses, which helps the host to avoid excessive response and maintain homeostasis.
Published in the journal: . PLoS Pathog 11(6): e32767. doi:10.1371/journal.ppat.1005012
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1005012Summary
Negative-feedback regulation is a broad and pivotal biological event to maintain the homeostasis of the host. Viral DNA species derived from DNA viruses or retroviruses can activate STING signaling to produce pro-inflammatory cytokines and type I interferon, which further recruit immune cells or induce interferon stimulated genes (ISGs) to clear viral infection respectively. However, excessive STING-signaling activation has been shown to induce autoimmune disorders. Thus, it is important to finely turn off STING signaling. Here we demonstrate that TRIM30α is rapidly induced followed by STING activation. Trim30α-deficient mice show more resistance to infection by DNA viruses. Meanwhile, knockdown or genetic ablation of TRIM30α augments the type I IFNs and IL-6 responses to intracellular DNA and DNA viruses. Biochemical analyses show that TRIM30α interacts with STING and promotes the degradation of STING via K48-linked ubiquitination at Lys275. These findings demonstrate that induced TRIM30α is a negative-feedback regulator of STING pathway activation triggered by DNA and DNA viruses, which helps the host to avoid excessive response and maintain homeostasis.
Introduction
The innate immune system is the first barrier of defense against pathogens, and a variety of germline-encoded pattern recognition receptors (PRRs), including Toll-like receptors (TLRs), C-type lectin receptors (CLRs), RIG-I-like receptors (RLRs) and NOD-like receptors (NLRs), have evolved in this system [1]. PRRs recognize the pathogen-associated molecular patterns (PAMPs) of microbes, which activate the innate immune response. The major PAMPs include nucleic acids derived from diverse bacterial, viral and eukaryotic pathogens. Thus far, many RNA receptors have been verified, among which TLR3 and TLR7/8, which are located in endosomes, sense double-stranded (ds)RNA and single-stranded (ss)RNA [2,3]. Retinoic-acid-inducible gene I (RIG-I) and melanoma differentiation-associated gene 5 (MDA5), which are located in the cytoplasm, detect cytosolic RNA [4,5].
In the past 5 years, the molecular basis of DNA sensing by the innate immune system has begun to be revealed, and a variety of DNA receptors have been proposed [6]. For example, TLR9 senses the CpG motifs-contained DNA [7]. In addition, AIM2 (absent in melanoma 2) binds to DNA and activates the inflammasome complex via the adaptor protein ASC (apoptosis-associated speck-like protein containing a CARD) [8–10]. Furthermore, RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway [11,12]. Moreover, many other DNA receptors, including DAI, IFI16, DDX41 and cGAS, trigger type I interferon response via adaptor STING [13–16]. Although aspects of the molecular basis of STING-mediated DNA sensing by the innate immune system have been determined, the precise mechanism by which this pathway is fine-tuned remains unclear. Normal activation of the DNA signaling pathway contributes to appropriate host defense. However, uncontrolled stimulation by DNA results in excessive production of inflammatory cytokines and type I IFNs, which may cause severe autoimmune diseases, such as systemic lupus erythematosus (SLE) [17]. Several negative regulators of the STING-meditated DNA signaling pathway have been described. For example, the E3 ubiquitin ligase TRIM21 induces the Lys48 (K48)-linked ubiquitination and degradation of DDX41 and negatively regulates the innate immune response to intracellular dsDNA [18]. Another E3 ubiquitin ligase, RNF5, which is constitutive expressed, inhibits virus-triggered signaling by targeting STING for ubiquitination and degradation [19]. Hiroyasu Konno et al demonstrated that cyclic dinucleotides (CDN) trigger the phosphorylation of STING by UNC-51-like kinase (ULK1), which inhibits STING activity and thus prevents the persistent transcription of innate immune genes [20]. A recent study has identified NLRC3 as another negative regulator of STING-induced innate immune response [21].
Previous work in our laboratory has demonstrated that TRIM30α negatively regulates TLR4 signaling by targeting TAB2 and TAB3 for degradation [22]. TRIM30α is also found to negatively regulate NLRP3 inflammasome activation by modulating reactive oxygen species production [23]. In this study, we report that TRIM30α is a negative-feedback regulator of innate immune responses to intracellular DNA and DNA viruses by promoting degradation of STING via K48-linked ubiquitination at Lys275.
Results
TRIM30α knockdown increases the host response to cytoplasmic DNA and DNA virus infection in D2SC cells
TRIM30α has been reported to be induced by TLR ligands in an NF-κB-dependent way and then negatively regulates TLR signaling by a ‘feedback’ mechanism. We found TRIM30α can also be induced upon intracellular poly(dA:dT) stimulation in bone marrow-derived dendritic cells (BMDCs) (S1A Fig). To address whether TRIM30α regulates the immune response to DNA-mediated signaling, we used TRIM30α siRNA (T3) to silence endogenous TRIM30α expression in D2SC cells, a mouse mDC cell line. The results showed that the T3 siRNA efficiently reduced the TRIM30α protein expression compared with control siRNA (SC) (Fig 1A). Various DNA ligands were used to stimulate D2SC cells, including poly(dA:dT), interferon stimulatory DNA (ISD), the genomic DNA of C57BL/6 mice and cyclic di-GMP (c-di-GMP). Knockdown of TRIM30α enhanced the production of type I IFN and IL-6 compared with controls (Fig 1B). In contrast, the induction of type I IFN by the synthetic analog of viral dsRNA poly (I:C) (the signaling of which is largely governed by MDA5) was reduced when TRIM30α was knocked down. To further confirm the function of TRIM30α in the anti-DNA viral response, we infected D2SC cells with the DNA viruses HSV-1 or vaccinia virus (VACV). The data showed that IFN-β and IL-6 production was dramatically increased in D2SC cells transfected with TRIM30α-specific siRNA (Fig 1C). TRIM30α knockdown also promoted the type I IFN and IL-6 expression at the mRNA level after stimulation with poly(dA:dT), ISD or HSV-1 (S1B and S1C Fig). We next examined the IFN-stimulated genes, such as the chemokine IP-10. Notably, TRIM30α knockdown enhanced the production of IP-10 mRNA upon ISD stimulation, but had no effect on the poly(I:C)-mediated signaling (Fig 1D).
Fig. 1. TRIM30α knockdown promotes immune signaling by cytoplasmic DNA and DNA virus infection in D2SC cells. 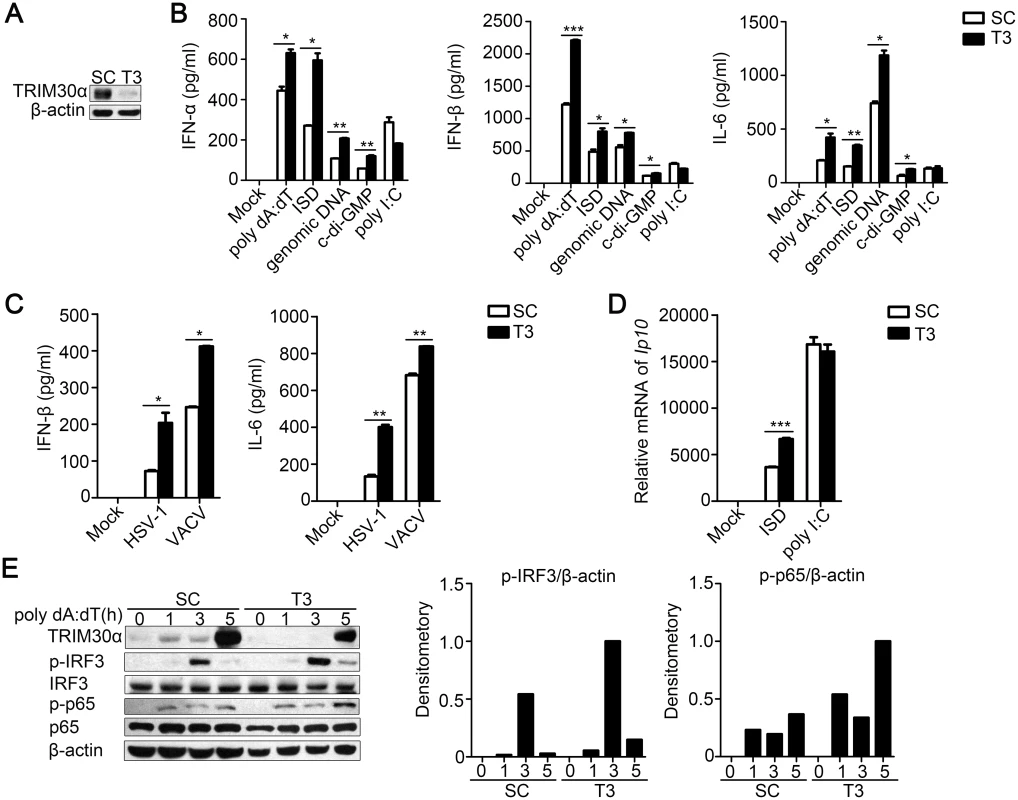
(A) Immunoblot analysis of the knockdown of exogenous TRIM30α in D2SC cells treated with control siRNA (SC) or TRIM30α siRNA (T3) for 24 h and then stimulated for 16 h with poly(dA:dT) (1 μg/ml). β-actin served as a loading control throughout the experiment. (B, C) ELISA of type I IFN and IL-6 in D2SC cells treated with siRNA SC or T3 and then stimulated for 16 h with poly(dA:dT) (1 μg/ml), ISD (1 μg/ml), genomic DNA (2 μg/ml), c-di-GMP (8 μg/ml) or poly(I:C) (5 μg/ml) (C) or with HSV-1 (MOI 10) or vaccinia virus (VACV) (MOI 10). (D) Real-time PCR of IP-10 mRNA in D2SC cells treated with siRNA SC or T3 and then stimulated for 8 h with ISD (1 μg/ml) or poly(I:C) (5 μg/ml). (E) Immunoblot analysis of p-IRF3, p-p65 and TRIM30α in lysates of D2SC cells treated with siRNA SC or T3 and then stimulated for 1–5 h with poly(dA:dT) (1 μg/ml). Densitometry analysis to quantify ratio of p-IRF3 or p-p65 to β-actin is shown on the right. The data are representative of three independent experiments and are presented as mean ± SEM. *p < 0.05, **p < 0.01 and ***p < 0.001. Intracellular nucleic acids and most viruses activate NF-κB, which is indispensable for TRIM30α expression. We found that poly(I:C), poly(dA:dT), ISD, c-di-GMP, genomic DNA and DNA viruses, including HSV-1 and VACV, facilitated the expression of TRIM30α (S1D Fig). Moreover, TRIM30α expression was considerably decreased by treatment with two NF-κB inhibitors, the IκB kinase-2 (IKK-2) inhibitor TPCA-1 (2-[(aminocarbonyl) amino]-5-(4-fluorophenyl)-3-thio-phenecarboxamide) and PDTC (pyrrolidine dithiocarbamate), in BMDCs stimulated for 6 h with poly(I:C), ISD or HSV-1 (S1E Fig).
Type I IFN and IL-6 expression is dependent upon the transcription factors IRF3 and NF-κB. Therefore, the amount of phosphorylated-IRF3 and-p65 was determined in D2SC cells after exposure to poly(dA:dT) at different time points. We found that TRIM30α knockdown resulted in high level of phosphorylation of IRF3 and p65 after poly(dA:dT) treatment (Fig 1E). Collectively, these results indicate that specifically knocking down TRIM30α considerably enhances type I IFN and IL-6 activation and the expression of IP-10 in response to cytoplasmic DNA and DNA viruses.
TRIM30α deficiency increases the host response to cytoplasmic DNA and DNA virus infection in dendritic cells
To further demonstrate the function of TRIM30α in immune response to DNA-mediated signaling, we generated Trim30α-deficient mice, in which the second exon was knocked out by homologous recombination (S2 Fig). Immunoblot analysis confirmed that TRIM30α expression was completely absent in Trim30α-deficient mice (Fig 2A). As shown in Fig 2B, TRIM30α deficiency resulted in much higher expression of type I IFN and IL-6 in response to multiple DNA ligands compared with wild-type DCs, similar to the results observed in D2SC cells. In contrast, TRIM30α deficiency did not affect poly(I:C) signaling (Fig 2B). Consistently with previous results, TRIM30α deficiency dramatically increased IFN-β and IL-6 expression induced by HSV-1 and VACV infection at protein level (Fig 2C). Beside the role in DNA sensing pathway, we also determine the function of TRIM30α against RNA viruses. As shown in S3 Fig, TRIM30α knockdown or deficiency dramatically potentiated IFN-β and IL-6 production by infection with VSV in D2SC cells and BMDCs (S3 Fig). The results suggest that TRIM30α negatively regulate immune response both to DNA and RNA viruses.
Fig. 2. TRIM30α deficiency promotes immune signaling by cytoplasmic DNA and DNA virus infection in BMDCs. 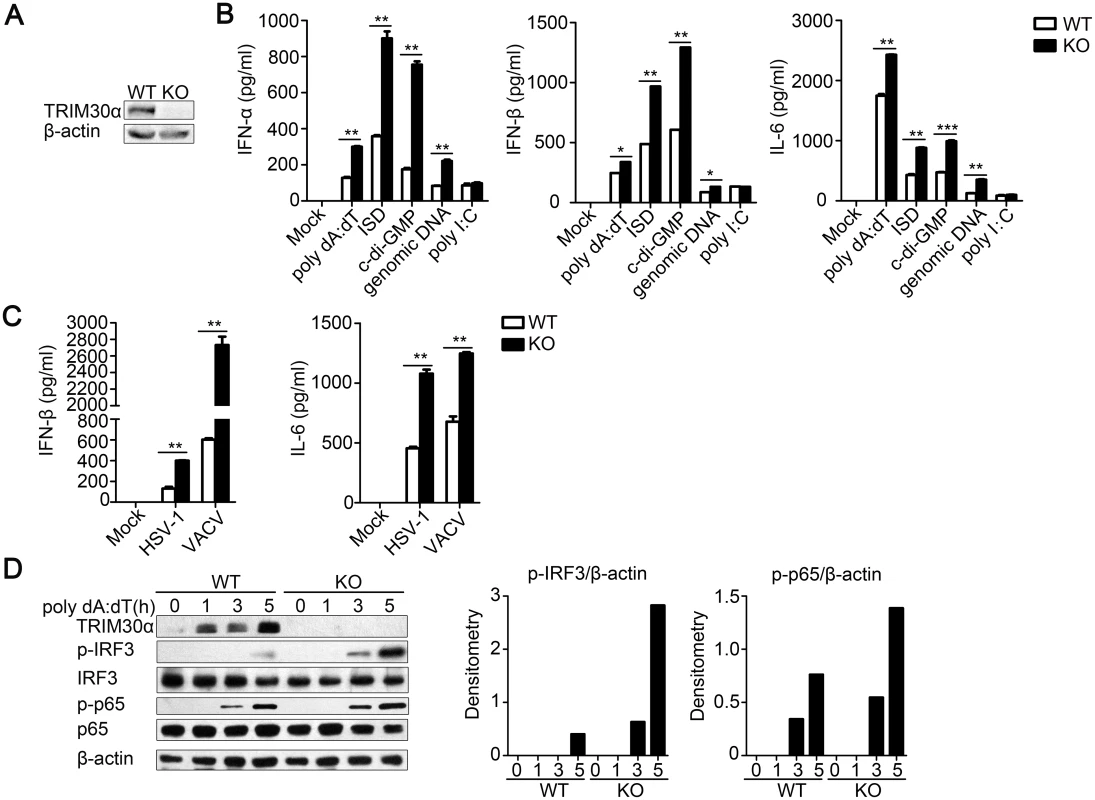
(A) Immunoblot analysis of TRIM30α expression in the splenocytes of wild type (WT) and Trim30α-/- (KO) mice. (B, C) ELISA of type I IFN and IL-6 in wild-type and Trim30α-/- BMDCs mock treated or stimulated for 16 h by transfection with poly(dA:dT) (1 μg/ml), ISD (1 μg/ml), c-di-GMP (8 μg/ml), genomic DNA (2 μg/ml) or poly(I:C) (5 μg/ml) (B); or with HSV-1 (MOI 10) or VACV (MOI 10) (C). (D) Immunoblot analysis of phosphorylated IRF3 (p-IRF3), p-p65 and TRIM30α in lysates of wild type and Trim30α-/- BMDCs stimulated for 1–5 h with poly(dA:dT) (1 μg/ml). Densitometry analysis to quantify ratio of p-IRF3 or p-p65 to β-actin is shown on the right. The data are representative of three independent experiments and are presented as mean ± SEM. *p < 0.05, **p < 0.01 and ***p < 0.001. Finally, we assessed IRF3 and p65 phosphorylation in BMDCs after poly(dA:dT) stimulation and found that TRIM30α deficiency promoted much higher expression of the phosphorylation of endogenous IRF3 and p65 (Fig 2D). These results suggest that TRIM30α plays a negative role in regulating type I IFN and IL-6 production in response to cytoplasmic DNA and DNA viruses.
TRIM30α deficiency inhibits HSV-1 infection
To evaluate the function of TRIM30α in host antiviral responses in vitro, we knocked down the expression of TRIM30α in mouse fibroblast L929 cells and then infected these cells with the DNA virus HSV-1. As expected, TRIM30α knockdown suppressed HSV-1 infection (Fig 3A). In contrast, overexpression of TRIM30α markedly facilitated HSV-1 replication (Fig 3B). These data suggest that TRIM30α is an important negative regulator in anti-viral defense. To further identify the role of TRIM30α in vivo host defense, wild type and Trim30α-deficient mice were challenged by intraperitoneal (i.p.) injection with HSV-1, and virus titers were examined 20 h later. Significantly more HSV-1 replication was detected in wild type mice (Fig 3C).
Fig. 3. TRIM30α deficiency inhibits DNA virus infection. 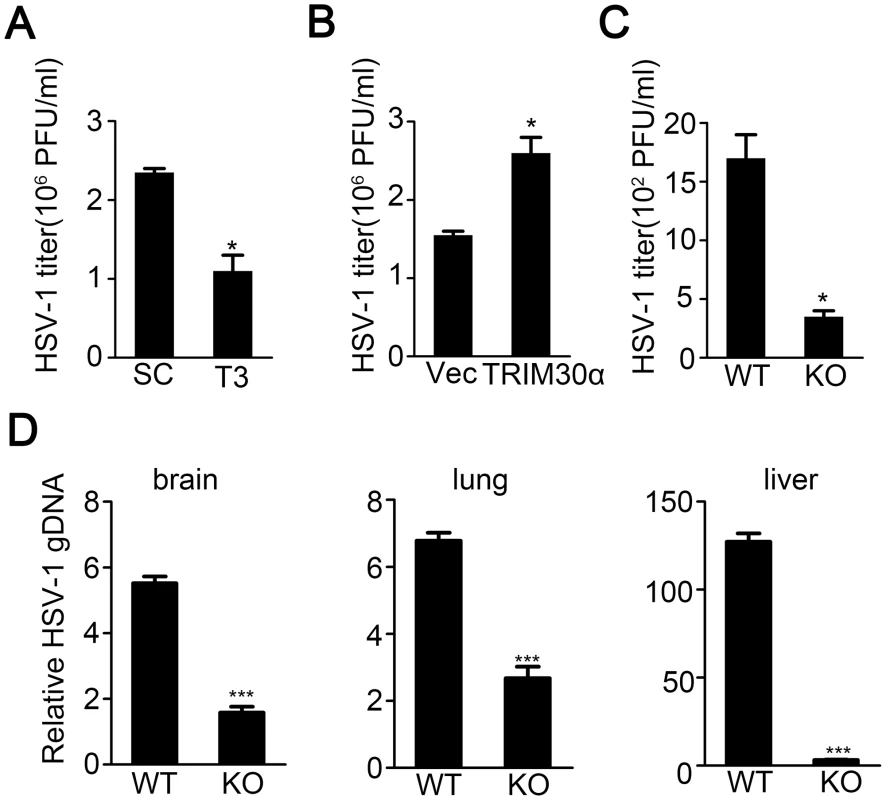
(A, B) Viral titers in L929 cells transfected with control siRNA SC and T3 (A), empty vector (Vec) or TRIM30α plasmid (B) for 24 h and then infected with HSV-1 (MOI 10) for 20 h. The titers of HSV-1 were determined by standard plaque assay. (C) Viral titers in wild type and Trim30α-/- mice intraperitoneally injected with HSV-1 (1×107 plaque-forming units (PFU)). HSV-1 titers were measured 20 h later by plaque assay of peritoneal wash fluid. (D) Real-time PCR of HSV-1 genomic DNA in the brain, lung and liver from wild type and Trim30α-/- mice infected with HSV-1 (2×107 PFU) (i.v.) for 2 days. The data are representative of three independent experiments and are presented as mean ± SEM. *p < 0.05, ***p < 0.001. Moreover, genomic DNA copies of HSV-1 were dramatically reduced in the brain, lung and liver of Trim30α-deficient mice in comparison to wild type mice upon HSV-1 infection for 2 days (Fig 3D). Interestingly, the liver showed much more difference between WT and KO mice compared to brain and lung, which was worthy to be explored. Together these data suggest that TRIM30α knockdown or deficiency inhibited HSV-1 replication both in vitro and in vivo.
TRIM30α deficiency protects mice from infection with HSV-1
To investigate the role of TRIM30α in host antiviral innate response, we isolated CD11c+ splenocytes from wild-type and Trim30α-deficient mice and treated these cells for 16 h with ISD, HSV-1 or poly(I:C). The Trim30α-deficient CD11c+ splenocytes produced high levels of IFN-β and IL-6 upon stimulation with ISD and HSV-1. However, TRIM30α deficiency had little effect on poly(I:C)-triggered response (Fig 4A and 4B). Besides DCs, macrophages are also involved in immune response triggered by intracellular DNA or DNA virus. We then obtained peritoneal macrophages (PM) from wild type and Trim30α-/- mice and then the above PM were stimulated with ISD or infected with HSV-1. The real-time PCR analysis demonstrated that Trim30α-/- PM produced more type I IFN and ISGs, suggesting that the negative role of TRIM30α in regulating DNA-sensing signaling pathway was not restricted in DCs (S4A and S4B Fig).
Fig. 4. TRIM30α deficiency protects the mice from DNA virus infection. 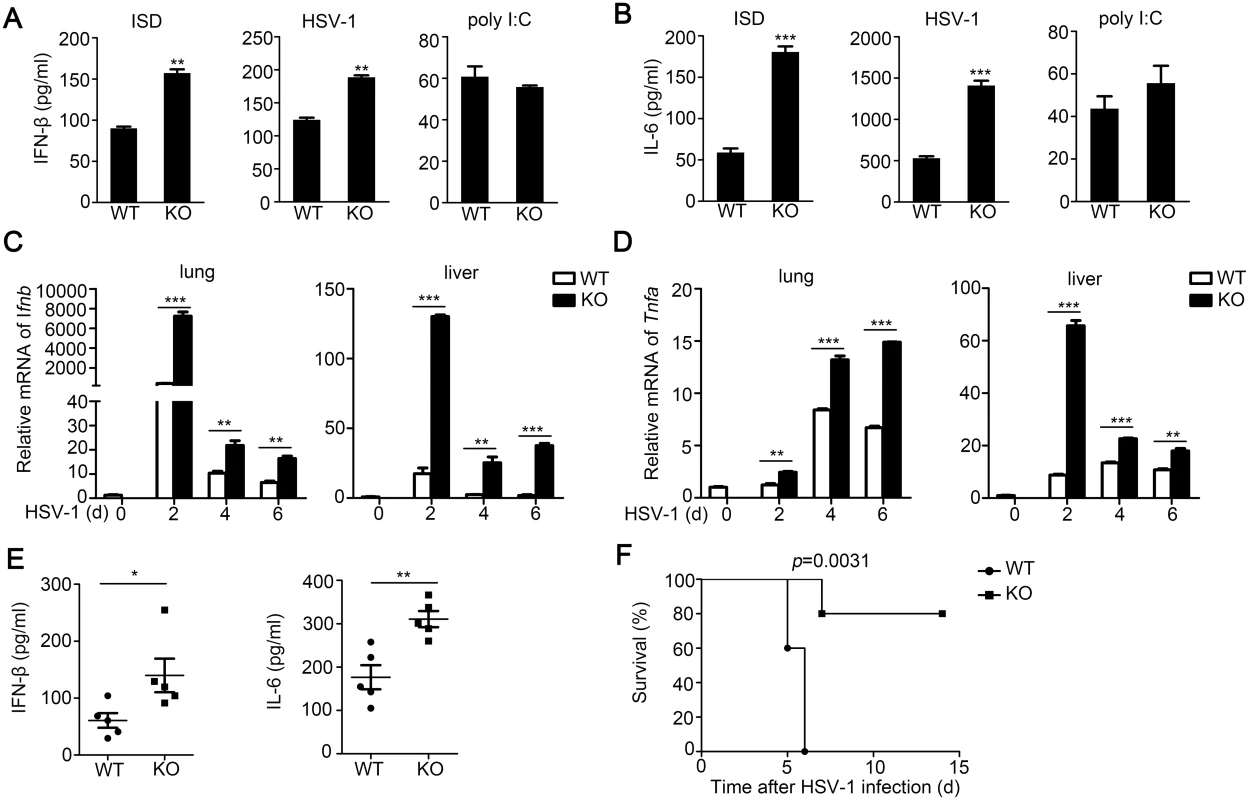
(A, B) ELISA of IFN-β and IL-6 in wild-type and Trim30α-/- CD11c+ splenocytes stimulated for 24 h with ISD (1 μg/ml), HSV-1 (MOI 10) or poly(I:C) (5 μg/ml). (C, D) Real-time PCR of IFN-β and TNF-α in the lung and liver from wild type and Trim30α-/- mice infected with HSV-1 (2×107 PFU) (i.v.) for the indicated times. (E) ELISA of IFN-β and IL-6 in serum of wild type and Trim30α-/- mice 6 h after intravenous infection with HSV-1 (1.2×107 PFU) (n = 5). (F) Survival of age- and sex-matched wild-type and Trim30α-/- mice (n = 5 per group) infected with HSV-1 (2×107 PFU) (i.v.) and monitored daily for 15 d. The data are representative of three independent experiments and are presented as mean ± SEM. *p < 0.05, **p < 0.01 and ***p < 0.001. To further elucidate the function of TRIM30α in vivo, wild-type and Trim30α-deficient mice were infected intravenously (i.v.) with HSV-1. Then we isolated various organs to evaluate immune response induced by DNA viruses. The levels of IFN-β and TNF-α mRNAs in liver and lung were markedly potentiated in Trim30α-deficient mice (Fig 4C and 4D). In addition, we observed higher IFN-β and IL-6 levels in the serum of Trim30α-deficient mice than in wild-type mice followed by HSV-1 infection (Fig 4E). Survival experiments demonstrated that the survival rate was significantly prolonged in the Trim30α-deficient mice infected with HSV-1 (Fig 4F). Thus, TRIM30α negatively controls DNA virus-triggered signaling, and TRIM30α deficiency protects mice from DNA virus infection.
TRIM30α interacts with STING
To identify the molecular mechanisms by which TRIM30α inhibits the DNA-triggered response, luciferase assays were performed to detect the target of TRIM30α. We found that TRIM30α inhibited the IFN-β reporter activation mediated by STING, a key adaptor protein for most DNA-sensing pathways [24,25]. However, TRIM30α did not affect the downstream kinase of STING, TBK1, and had no influence on MDA5 - and virus-induced signaling adaptor (VISA)-induced signaling (Fig 5A). In addition, TRIM30α overexpression significantly inhibited STING-induced NF-κB reporter activation but did not affect VISA (Fig 5B). It is hypothesized that TRIM30α targets STING to inhibit DNA-mediated response. As shown in Fig 5C, exogenous expression of TRIM30α inhibited STING-induced IFN-β and NF-κB reporter activation in a dose dependent manner (Fig 5C). In contrast, TRIM30α (C35A), which contains an enzymatically inactive mutant of the RING domain had no effect on STING-induced IFN-β and NF-κB reporter activation (Fig 5D). All these suggest that TRIM30α suppresses STING signaling dependent on its RING domain. Furthermore, co-immunoprecipitation assays were performed and the results showed that TRIM30α interacted with STING when co-transfected into HEK293T cells (Fig 5E). TGF beta-activated kinase (TAK1) interacts with TRIM30α, which has been previously reported, was used as a positive control [22]. We next performed endogenous co-immunoprecipitation experiments and verified that endogenous TRIM30α could interact with STING in D2SC cells after poly(dA:dT) or HSV-1 stimulation (Fig 5F and 5G). To further demonstrate the interaction, TRIM30α and STING were quickly translated in vitro, and immunoprecipitation analysis indicated that TRIM30α could interact with STING directly (S5 Fig).
Fig. 5. TRIM30α interacts with STING. 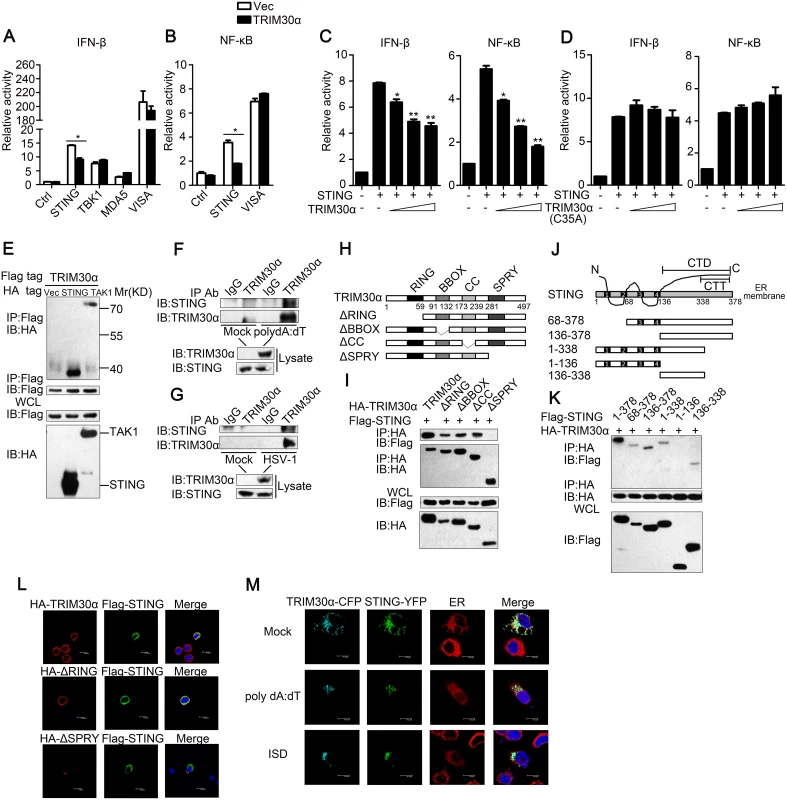
(A) Luciferase activity of MEF cells transfected with the IFN-β luciferase reporter, together with STING, TBK1, MDA5 or VISA, and the empty vector (Vec) or the TRIM30α plasmid. (B) Luciferase activity of MEF cells transfected with the NF-κB luciferase reporter, together with STING or VISA, and the empty vector (Vec) or the TRIM30α plasmid. (C and D) Luciferase activity of MEF cells cotransfected with the IFN-β promoter or the NF-κB promoter and empty vector or increasing amounts of TRIM30α (C) or C35A (D) (0.2, 0.4 and 0.6 μg) together with STING. (E) Immunoblot analysis of lysates from HEK293T cells transfected with Flag-TRIM30α together with HA-tagged vector, STING or TAK1 plasmids, followed by immunoprecipitation (IP) with anti-Flag Ab and immunoblot analysis with anti-HA Ab. (F, G) Immunoblot analysis in lysates of D2SC cells mock treated or stimulated with poly(dA:dT) (1 μg/ml) (F); or with HSV-1 (MOI 10) (G) for 8 h, followed by immunoprecipitation with anti-TRIM30α Ab and immunoblot analysis with anti-STING Ab. (H) A schematic presentation of full-length TRIM30α and its mutants. RING, ring-finger domain; BBOX, B-box domain; CC, coiled-coil domain; SPRY, SPRY domain. (I) Immunoblot analysis of lysates from HEK293T cells transfected with the indicated plasmids, followed by immunoprecipitation (IP) with anti-HA Ab and immunoblot analysis with anti-Flag Ab. (J) A schematic presentation of full-length STING and its mutants. (Transmembrane) TM1, 21-41aa; TM2, 47-67aa; TM3, 87-106aa; TM4, 115-135aa; CTD, carboxy-terminal domain; CTT, carboxy-terminal tail. (K) Immunoblot analysis of lysates from HEK293T cells transfected with the indicated plasmids and then performed as in I. (L) Confocal microscopy of L929 cells transfected for 24 h with Flag-tagged STING and HA-tagged full-length TRIM30α, ΔRING or ΔSPRY mutant of TRIM30α. Immunofluorescence was performed using anti-HA (red) and anti-Flag (green). (M) Confocal microscopy of L929 cells transfected for 24 h with cyan fluorescent protein-labeled TRIM30a (CFP-TRIM30α) and yellow fluorescent protein-labeled STING (YFP-STING) and then mock treated or stimulated for 4 h with poly(dA:dT) (1 μg/ml) or ISD (1 μg/ml). Nuclei were stained with the DNA-intercalating dye DAPI. Staining of calnexin served as a marker of the endoplasmic reticulum (ER). The data are representative of three independent experiments and are presented as mean ± SEM. *p < 0.05 and **p < 0.01. To explore the domains that govern the association between TRIM30α and STING, HA-tagged full-length TRIM30α and various TRIM30α truncations were expressed in HEK293T cells. Co-immunoprecipitation assays showed that only the SPRY domain of TRIM30α bound to STING (Fig 5H and 5I). Previous studies have reported that STING is a multiple transmembrane domain-containing protein and that the C-terminal amino acids of STING include a globular carboxy-terminal domain (CTD, amino acids 136–378) and carboxy-terminal tail (CTT, amino acids 338–378) [26,27]. We found that only the C-terminal amino acids 136–338 of STING were required for interaction with TRIM30α (Fig 5J and 5K). Flag-tagged STING, HA-tagged TRIM30α and TRIM30α truncations were expressed in L929 cells. Confocal microscopy experiments showed that STING was co-localized with full-length TRIM30α and ΔR, but not ΔSPRY, confirming that the SPRY domain of TRIM30α is required for interaction with STING (Fig 5L). We then expressed CFP-TRIM30α and YFP-STING in L929 cells and detected the co-localization of these two proteins (Fig 5M). Interestingly, it was also found that STING translocated from the endoplasmic reticulum (ER) to perinuclear sites in response to poly(dA:dT) and ISD, in agreement with previous studies [25]. These results suggest that TRIM30α interacts with STING to inhibit DNA-mediated immune response.
TRIM30α enhances the degradation of STING
It is very important to determine the mechanism by which interaction of TRIM30α with STING suppress the intracellular DNA-sensing pathway. As shown in Fig 6A, Trim30α-deficient BMDCs maintained a higher expression of STING than wild type BMDCs after poly(dA:dT) stimulations of different durations (Fig 6A). In contrast, the expression of TBK1 and IRF3, two adaptors downstream of STING, maintained unchanged in Trim30α-deficient BMDCs, suggesting that TRIM30α may suppress the stability of STING. We next treated wild type BMDCs with poly(dA:dT) in the presence or absence of MG132, the inhibitor of proteasome. We observed a high level expression of STING in BMDCs upon MG132 treatment than control, indicating that STING undergoes degradation after stimulation with intracellular DNA in a proteasome way (Fig 6B). Moreover, MG132 treatment significantly promoted much higher expression of phosphorylation of TBK1 and IRF3 than control cells, which mimics TRIM30α deficiency upon intracellular DNA stimulation. Confocal microscopy further demonstrated that STING was co-localized with proteasome during ISD stimulation, suggesting that STING may undergo degradation via proteasome in response to DNA stimulus (Fig 6C).
Fig. 6. TRIM30α enhances the degradation of STING. 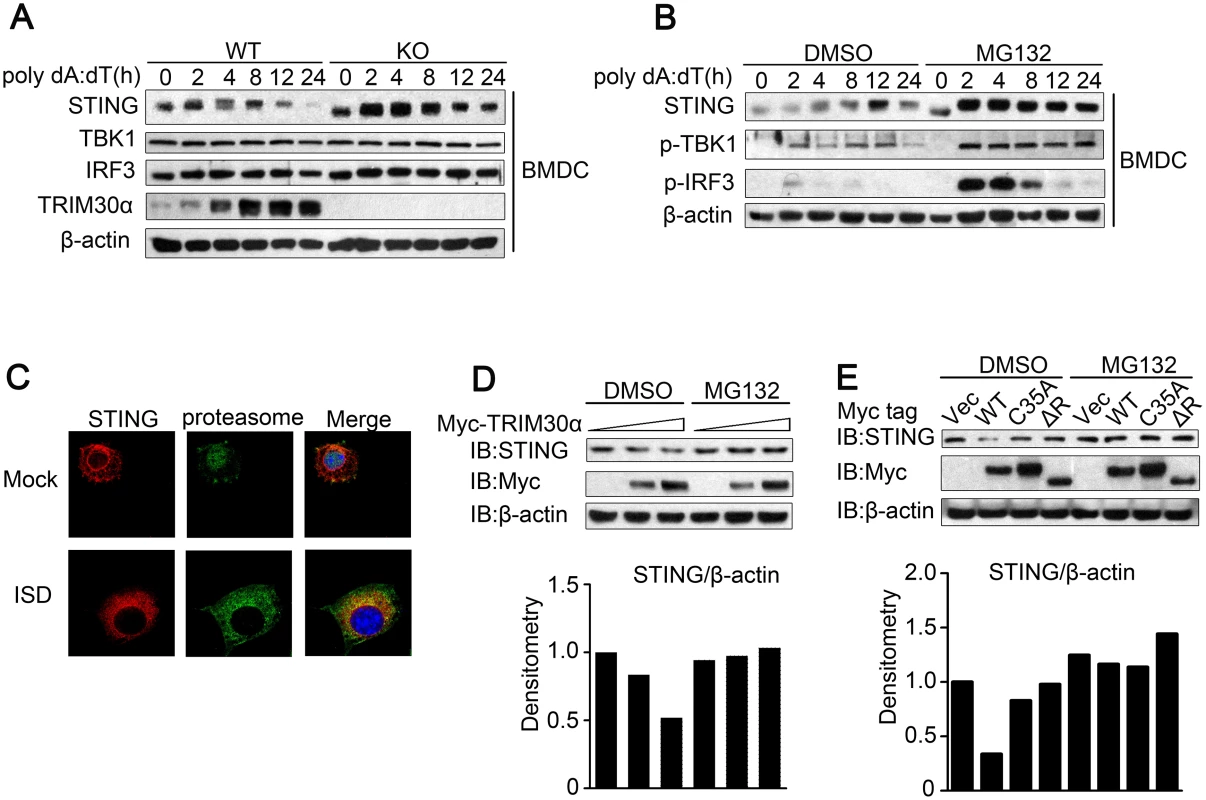
(A) Immunoblot analysis of STING, TBK1, IRF3 and TRIM30α in lysates from wild type and Trim30α-/- BMDCs stimulated for 2–24 h with poly(dA:dT) (1 μg/ml). (B) Immunoblot analysis of STING in lysates from wild type BMDCs stimulated for 2–24 h with poly(dA:dT) (1 μg/ml) in the presence or absence of 20 mM MG132. (C) Confocal microscopy of L929 cells transfected for 24 h with Flag-tagged STING and then mock treated or stimulated for 12 h with ISD (1 μg/ml). Immunofluorescence was performed using anti-Flag (red) and anti-20S proteasome β1 (green). Nuclei were stained with the DNA-intercalating dye DAPI. (D) Immunoblot analysis of STING in lysates of L929 cells transfected with increasing doses of Myc-tagged TRIM30α (0, 0.8 and 1.2 μg) and then treated for 6 h with DMSO (negative control) or 20 mM MG132. Densitometry analysis to quantify ratio of STING to β-actin is shown on the below. (E) Immunoblot analysis of STING in lysates of L929 cells transfected with full-length TRIM30α, C35A or ΔR (1μg) and then treated as in D. Because TRIM30α promotes the degradation of TAB2 and TAB3 [22], we hypothesized that TRIM30α also promotes the degradation of STING. To address this, TRIM30α was overexpressed in L929 cells. The expression of endogenous STING in L929 cells was diminished in a dose-dependent manner and rescued by MG132 (Fig 6D). Furthermore, we determined whether STING degradation is dependent upon the RING domain of TRIM30α, which confers it E3 ubiquitin ligase activity. TRIM30α (C35A) and TRIM30α (ΔR) attenuated STING degradation when expressed in L929 cells compared with full-length TRIM30α (Fig 6E). Collectively, these results indicated that the interaction of TRIM30α with STING enhances STING degradation in a proteasome pathway, which is dependent on its RING domain.
TRIM30α targets STING for K48-linked ubiquitination at Lys275
Most TRIM proteins contain RING domains, which allow them to mediate ubiquitylation events [28]. Therefore, we hypothesized that TRIM30α is an E3 ubiquitin ligase for STING. To address this, TRIM30α and STING were co-transfected in HEK293T cells. Immunoprecipitation and immunoblot analysis showed that TRIM30α could ubiquitinate STING in a dose-dependent manner (Fig 7A). Moreover, we observed that TRIM30α overexpression enhanced K48-linked ubiquitination of STING but not K63-linked ubiquitination (Fig 7B). K48-linked ubiquitination is normally linked to proteasomes-mediated degradation of proteins, which is in line with our early data (Fig 6D and 6E). As shown in Fig 7C, TRIM30α (C35A) and ΔR dramatically attenuated the ubiquitination of STING compared with full-length TRIM30α, suggesting that TRIM30α ubiquitinates STING dependent upon its RING domain activity (Fig 7C). Next, we examined the ubiquitination of STING in primary cells and observed that wild type and K48-linked ubiquitination of STING in wild type BMDCs was increased in comparison to Trim30α-deficient BMDCs stimulated with ISD for 8 h (Fig 7D). Collectively, these findings indicate that TRIM30α is an E3 ubiquitin ligase for STING and mediates STING degradation via the proteasome pathway. To further investigate whether TRIM30α directly ubiquitinates STING, TRIM30α and STING were quickly translated in vitro. In vitro ubiquitination showed that the TRIM30α protein directly mediated ubiquitination of STING (Fig 7E).
Fig. 7. TRIM30α is an E3 ubiquitin ligase and targets STING for K48-mediated ubiquitination at Lys275. 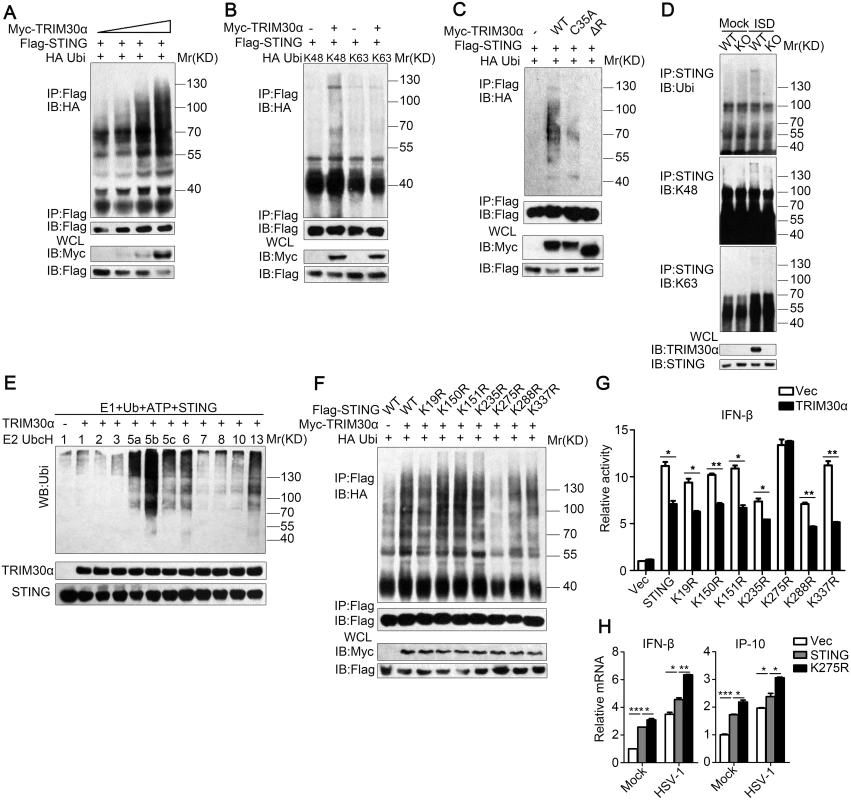
(A) Immunoblot analysis of lysates from HEK293T cells transfected with plasmids for Flag-STING, HA-ubiquitin and increasing concentrations of Myc-TRIM30α (0, 1, 1.5 and 2 μg), followed by immunoprecipitation with anti-Flag, and analyzed via immunoblot with anti-HA Ab. (B) Immunoblot analysis of lysates from HEK293T cells transfected with various combinations of plasmids for Myc-TRIM30α, Flag-STING, HA-K48-ubi and HA-K63-ubi and then performed as in A. (C) Immunoblot analysis of lysates from HEK293T cells transfected with the indicated plasmids and Myc-tagged TRIM30α (C35A), ΔR and then performed as in B. (D) Immunoblot analysis of lysates in wild-type and Trim30α-/- BMDCs stimulated for 8 h with ISD (1 μg/ml), followed by immunoprecipitation with anti-STING Ab and analyzed with anti-Ub, anti-K48- and anti-K63-linked ubiquitin. (E) Immunoblot analysis of STING ubiquitination in vitro. TRIM30α and STING were quickly translated in vitro, and then, the biotin-ubiquitin E1 and indicated E2s were added for the ubiquitination assays. Ubiquitination was assessed by anti-ubi. (F) Immunoblot analysis of lysates from HEK293T cells transfected with Myc-tagged TRIM30α, Flag-tagged wild-type and mutant STING together with HA-tagged ubiquitin, then performed as in A. (G) Luciferase assays of MEF cells cotransfected with the IFN-β promoter and empty vector or TRIM30α together with wild-type and mutant STING plasmids. (H) Real-time PCR of IFN-β and IP-10 mRNA in L929 cells transfected with Flag-tagged vector, wild type STING and K275R mutant, 24 h after transfection, then infected the above cells with HSV-1 (MOI 10) for 10 h. The data are representative of three independent experiments and are presented as mean ± SEM. *p < 0.05, **p < 0.01 and ***p < 0.001. In order to map the ubiquitination sites on STING that are targeted by TRIM30α, we replaced each of the seven STING lysine (K) residues that not in transmembrane region with arginine (R). We then expressed TRIM30α, wild-type and mutant STING in HEK293T cells. As shown in Fig 7F, the ubiquitination of mutation of K275 to arginine was obviously attenuated (Fig 7F). In addition, luciferase assays showed that TRIM30α significantly suppressed wild-type and STING mutants-induced IFN-β reporter activity except for STING K275R (Fig 7G). Moreover, STING K275R maintained high level in HSV-1 triggered IFN-β and IP-10 production in comparison to wild type STING (Fig 7H). Taken together, the results suggest that Lys275 was the ubiquitination site of STING targeted by TRIM30α.
Discussion
STING has been identified as an essential adaptor protein for controlling TLR-independent cytosolic DNA signaling [29]. Cytosolic DNA derived from DNA viruses, bacteria or parasites is sensed by DNA receptors that subsequently activate STING. Activated STING recruits and activates the cytosolic kinases IKK and TBK1, which in turn activate NF-κB and IRF3, respectively [30]. In addition, cyclic dinucleotides (CDNs) can directly bind and activate STING. However, the regulation of STING signaling remains to be fully elucidated. In this context, we discovered the novel function of TRIM30α in negatively regulating the STING pathway. TRIM30α deficiency or knockdown enhanced the production of type I IFNs and the inflammatory cytokine IL-6 upon stimulation with intracellular DNA or infection with the DNA viruses HSV-1 or VACV in BMDCs and D2SC cells. Moreover, Trim30α-deficient mice were resistant to HSV-1 compared with wild type mice. In vitro and in vivo studies demonstrated the negative role of TRIM30α in cytosolic DNA-mediated responses. Co-immunoprecipitation and immunoblot analyses indicated that TRIM30α interacted with STING through binding of the SPRY domain of TRIM30α with C-terminal amino acids 136–338 of STING. We further investigated that TRIM30α mediated K48-linked ubiquitination of STING at Lys275, which promoted STING degradation via the proteasome-dependent pathway. In our experiments, we found that TRIM30α expression was induced by DNA virus-triggered NF-κB activation, which then induced STING degradation, and that these events negatively regulated the STING-mediated signaling pathway.
Our laboratory has previously shown that TRIM30α expression is dependent on NF-κB activation and then targeting of TAB2 and TAB3 for degradation, thus attenuating the TLR signaling pathway [22]. Based on our previous work, we speculate that TRIM30α may act as a brake to prevent excessive immune response activation. In TLR and STING-mediated signaling, TRIM30α serves as an important feedback regulator and appropriately controls excessive inflammatory responses or type I IFNs production. Our results suggest that TRIM30α may be a negative regulator involved in other immune pathways in general. This hypothesis remains to be explored.
In the present study, we determined that TRIM30α is an E3 ubiquitin ligase for STING, the activity of which is dependent upon its RING domain. However, TRIM30α induces TAB2 degradation via lysosomes but not the ubiquitin-proteasome pathway. This observation suggests that TRIM30α is a multifunctional protein, degrading different substrates via distinct pathways. However, the characteristics of the target elements and the manner by which TRIM30α chooses the degradation pattern remain unclear.
Once activated, STING leads to increased expression of inflammatory cytokines and/or type I IFNs. Therefore, prevention of STING activity is of utmost importance for avoiding severe autoimmune disorders. Glen N. Barber demonstrated that when STING undergoes autophagy-dependent delivery, it is phosphorylated by serine/threonine UNC-51-like kinase [20]. Phosphorylation of Ser366 in STING was found to inhibit STING-dependent IRF3 activity but not NF-κB activity. However, TRIM30α can efficiently attenuate both the IRF3 and NF-κB pathways, dampening persistent immune activation. TRIM30α is a fine-tuned regulator in the down-regulation of STING-mediated signaling.
Materials and Methods
Ethics statement
C57BL/6 mice were purchased from the Shanghai Laboratory Animal Center (SLAC). All mice were bred and kept in specific pathogen-free (SPF) conditions in the Shanghai Institute of Biochemistry and Cell Biology. All animal care and use protocols were performed in accordance with the Regulations for the Administration of Affairs Concerning Experimental Animals approved by the State Council of People's Republic of China. The animal experiments were approved by the Institutional Animal Care and Use Committee of the Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences (Approval Number: IBCBSPF0028).
Mice
TRIM30α-deficient mice were generated on a 129 background and backcrossed to C57BL/6 for at least 7 generations by the Shanghai Research Center for Model Organisms. Mice 6–8 weeks of age that were matched by body weight and sex were used in the experiments.
Reagents and cDNA constructs
The poly(I:C), ISD and c-di-GMP were from Invitrogen. Poly(dA:dT) and lipopolysaccharide (LPS) were from Sigma. Genomic DNA from C57BL/6 mice was made in-house. The following antibodies were used for immunoblot analysis or immunoprecipitation: anti-HA (CO-MMS-101R; Covance), anti-Flag (F3165; Sigma), anti-STING (3337; Cell Signaling), anti-TRIM30α (previously described) [22], K63-specific anti-ubiquitin (05–1313; Millipore), K48-specific anti-ubiquitin (05–1307; Millipore), anti-IRF3 (sc-9082; Santa Cruz), anti-phosphorylated IRF3 (4947s; Cell Signaling), anti-p65 (4764s; Cell Signaling) and anti-phosphorylated p65 (3033s; Cell Signaling). The following antibodies were used for confocal microscopy: anti-20S proteasome β1 (SC-67345; Santa Cruz); anti-calnexin (C4731; Sigma) and Alexa Fluor 647 donkey anti-rabbit IgG (711-175-152; Molecular Probes). Lipofectamine 2000 was obtained from Invitrogen. The X-tremeGENE DNA transfection reagent was acquired from Roche. The Turbofect transfection reagent was acquired from ThermoFisher. The TNT Quick-coupled Transcription/Translation Systems kit was obtained from Promega. The ubiquitination kit (BML-uw9920-0001) was purchased from Enzo Life Sciences. The Protein A/G Plus-Agarose immunoprecipitation reagent was purchased from Santa Cruz. Anti-HA beads were purchased from Sigma. The ELISA kits were acquired from the following sources: murine IFN-β (PBL), murine IFN-α (PBL) and murine IL-6 (R&D). The mouse STING sequence was amplified by PCR using cDNA from BMDCs and subsequently cloned into a pcDNA3 vector (Invitrogen). All TRIM30α and STING deletion mutants/point mutants were constructed by PCR and subcloned into a pcDNA3 vector. The other plasmids were either generated or obtained as described previously [22,31].
Cell culture, transfection and stimulation
D2SC cells were provided by Yong-Jun Liu (UT MD Anderson Cancer Center, Texas, USA) and maintained in Iscove’s modified Dulbecco’s medium containing 10% (vol/vol) heat-inactivated FCS and 1% (vol/vol) penicillin-streptomycin (Invitrogen-Gibco). L929, Vero, MEF, HEK293T and HEK293 cells were cultured in DMEM supplemented with 10% (vol/vol) FBS, penicillin (100 U/ml) and streptomycin (100 U/ml). Single-cell suspensions of CD11c+ splenocytes were isolated from spleens with CD11c MicroBeads (130-052-001; Miltenyi Biotec) following the manufacturer’s instructions. The procedure for generating BMDCs has been described previously [32]. X-tremeGENE was used for transient transfection of plasmid DNA into L929 and MEF. Transfection of 293T was performed with Turbofect. For stimulation, poly(dA:dT) (1 μg/ml), ISD (1 μg/ml), c-di-GMP (8 μg/ml), genomic DNA (2 μg/ml) or poly(I:C) (5 μg/ml) were delivered into cells using Lipofectamine 2000.
Viruses and infection
HSV-1 and VACV were kindly provided by Xuetao Cao (Second Military Medical University, Shanghai, China) and Zhengfan Jiang (Peking University, Shanghai, China), respectively. Cells were infected with HSV-1 (MOI 10) or VACV (MOI 10) for 1.5 h and subsequently washed with PBS and cultured in fresh media. Cytokine production was analyzed 16 h or 24 h later. For the in vivo cytokine production study, age - and sex-matched groups of mice were intravenously infected with HSV-1 (1.2×107 PFU per mouse). HSV-1 viral titer was determined by the plaque-forming assay on Vero cells.
RNA-mediated interference
The TRIM30α siRNA T3 and negative control siRNA have been described previously [22]. D2SC cells were transfected with siRNA delivered by Lipofectamine 2000. At 24 h after transfection, the cells were used for further experiments.
Real-time PCR
Total RNA was extracted from cells or tissues using TRIzol reagent (Invitrogen). The RNA was then used in a RT reaction using the Prime Script RT Master Mix kit (TaKaRa). All gene transcripts were analyzed by quantitative PCR with SYBR Green
Master Mix (ABI) using an ABI PRISM 7900HT Sequence Detection System (PE Applied Biosystems).
Primers for PCR are listed as follows:
IP-10:
sp 5’-GGGCCAGTGAGAATGAGGG-3’,
as 5’-GCTCGCAGGGATGATTTCAA-3’;
HSV-1 genomic DNA:
sp 5’-TGGGACACATGCCTTCTTGG-3’,
as 5’-ACCCTTAGTCAGACTCTGTTACTTACCC-3’;
IFN-α1
sp 5’-CCTGAACATCTTCACATCAAAGGA-3’,
as 5’-AGCTGCTGGTGGAGGTCATT-3’;
IFN-β:
sp 5’-CCTGGAGCAGCTGAATGGAA-3’,
as 5’-TTGAAGTCCGCCCTGTAGGT-3’;
TNF-α:
sp 5’-AAGCCTGTAGCCCACGTCGTA-3’,
as 5’-GGCACCACTAGTTGGTTGTCTTTG-3’;
ISG12
sp 5’-TTGCCAATGGAGGTGGAGTT-3’,
as 5’-AGGACCCCTGCTGATTGGA-3’;
ISG20
sp 5’-CGCTGCAGCATTGTGAACA-3’,
as 5’-CGGGTCGGATGTACTTGTCA-3’;
ISG56
sp 5’-CTCAGAGCAGGTCCAGTTCCTT-3’,
as 5’-GGCCAGGAGGTTGTGCAT-3’;
HPRT:
sp 5’-TGCTCGAGATGTCATGAAGGAG-3’,
as 5’-CAGAGGGCCACAATGTGATG-3’;
Acta2:
sp 5’-ATGACCCAGATTATGTTTGAGACC-3’;
as 5’-CCAGAGTCCAGCACAATACC-3’.
In vitro ubiquitination assay
TRIM30α and STING proteins were expressed with a TNT Quick-coupled Transcription/Translation Systems kit (Promega). In vitro ubiquitination assay was performed with a ubiquitination kit (Enzo Life Science) following the manufacturer’s instructions.
Immunoprecipitation and immunoblot analysis
These experiments were performed as described previously [22]. In brief, HEK293T cells were transfected with various combinations of plasmids. At 24 h after transfection, lysates of the cells were prepared in lysis buffer and incubated with the Protein A/G Plus-Agarose immunoprecipitation reagent together with the indicated Ab overnight at 4°C. Complexes were washed three times with lysis buffer and analyzed by immunoblot. For endogenous co-immunoprecipitation experiments, lysates of D2SC cells stimulated with poly(dA:dT) for 12 h were incubated with anti-TRIM30α or rabbit IgG Ab and analyzed by immunoblot. For ubiquitination, BMDCs were stimulated with 1 μg/ml ISD for 8 h delivered by Lipofectamine 2000 and then collected for immunoblot analysis with anti-STING.
Confocal microscopy
L929 cells were transfected with expressing plasmids for cyan fluorescent protein-labeled TRIM30α and yellow fluorescent protein-labeled STING. After 24 h, cells were stimulated for 4 h with 1 μg/ml poly(dA:dT), 1 μg/ml ISD or left unstimulated. After stimulation, cells were fixed with 4% PFA in PBS and permeabilized with Triton X-100 and then blocked with 10% FBS in PBS, stained with anti-calnexin, followed by Alexa Fluor 647 donkey anti-rabbit IgG. Nuclei were stained with 4, 6-diamidino-2-phenylindole, and fluorescent images were captured with a Leica TCS SP2 laser confocal microscope.
Luciferase reporter gene assay
MEF cells were transfected with an IFN-β luciferase reporter plasmid and a Renilla luciferase plasmid as an internal control plus the indicated expression plasmids. Empty control vector was added so that a total of 1 μg of DNA was transfected into each well of cells. Then, 24 h after transfection, cells were lysed, and reporter activity was analyzed with the Dual-Luciferase Reporter Assay system (Promega).
Statistics
The data are presented as the mean ± SEM from at least three independent experiments. Student’s t-test was used to compare two independent groups. For all tests, values of p < 0.05 were considered statistically significant.
Proteins accession numbers
The accession numbers in the UniProtKB/SwissProt database for the proteins in the manuscript are followed: TRIM30α, P15533; IFN-α, P01572; IFN-β, P01575; IL-6, P08505; IRF3, P70671; p65, Q04207; STING, Q3TBT3; TBK1, Q9WUN2; MDA5,Q9BYX4; VISA, Q7Z434.
Supporting Information
Zdroje
1. Takeuchi O, Akira S (2010) Pattern Recognition Receptors and Inflammation. Cell 140 : 805–820. doi: 10.1016/j.cell.2010.01.022 20303872
2. Alexopoulou L, Holt AC, Medzhitov R, Flavell RA (2001) Recognition of double-stranded RNA and activation of NF-kappa B by Toll-like receptor 3. Nature 413 : 732–738. 11607032
3. Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, et al. (2004) Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science 303 : 1526–1529. 14976262
4. Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, et al. (2004) The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nature Immunology 5 : 730–737. 15208624
5. Yoneyama M, Kikuchi M, Matsumoto K, Imaizumi T, Miyagishi M, et al. (2005) Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. Journal of Immunology 175 : 2851–2858.
6. Paludan Søren R, Bowie Andrew G (2013) Immune Sensing of DNA. Immunity 38 : 870–880. doi: 10.1016/j.immuni.2013.05.004 23706668
7. Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, et al. (2000) A Toll-like receptor recognizes bacterial DNA. Nature 408 : 740–745. 11130078
8. Burckstummer T, Baumann C, Bluml S, Dixit E, Durnberger G, et al. (2009) An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nature Immunology 10 : 266–272. doi: 10.1038/ni.1702 19158679
9. Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, et al. (2009) AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature 458 : 514–U516. doi: 10.1038/nature07725 19158675
10. Rathinam VAK, Jiang ZZ, Waggoner SN, Sharma S, Cole LE, et al. (2010) The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nature Immunology 11 : 395–403. doi: 10.1038/ni.1864 20351692
11. Ablasser A, Bauernfeind F, Hartmann G, Latz E, Fitzgerald KA, et al. (2009) RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III—transcribed RNA intermediate. Nature Immunology 10 : 1065–1072. doi: 10.1038/ni.1779 19609254
12. Chiu Y-H, MacMillan JB, Chen ZJ (2009) RNA Polymerase III Detects Cytosolic DNA and Induces Type I Interferons through the RIG-I Pathway. Cell 138 : 576–591. doi: 10.1016/j.cell.2009.06.015 19631370
13. Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, et al. (2007) DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature 448 : 501–U514. 17618271
14. Zhang Z, Yuan B, Bao M, Lu N, Kim T, et al. (2011) The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat Immunol 12 : 959–965. doi: 10.1038/ni.2091 21892174
15. Sun L, Wu J, Du F, Chen X, Chen ZJ (2013) Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339 : 786–791. doi: 10.1126/science.1232458 23258413
16. Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, et al. (2010) IFI16 is an innate immune sensor for intracellular DNA. Nature Immunology 11 : 997–1004. doi: 10.1038/ni.1932 20890285
17. Stetson DB, Ko JS, Heidmann T, Medzhitov R (2008) Trex1 Prevents Cell-Intrinsic Initiation of Autoimmunity. Cell 134 : 587–598. doi: 10.1016/j.cell.2008.06.032 18724932
18. Zhang Z, Bao M, Lu N, Weng L, Yuan B, et al. (2013) The E3 ubiquitin ligase TRIM21 negatively regulates the innate immune response to intracellular double-stranded DNA. Nat Immunol 14 : 172–178. doi: 10.1038/ni.2492 23222971
19. Zhong B, Zhang L, Lei C, Li Y, Mao A-P, et al. (2009) The Ubiquitin Ligase RNF5 Regulates Antiviral Responses by Mediating Degradation of the Adaptor Protein MITA. Immunity 30 : 397–407. doi: 10.1016/j.immuni.2009.01.008 19285439
20. Konno H, Konno K, Barber GN (2013) Cyclic dinucleotides trigger ULK1 (ATG1) phosphorylation of STING to prevent sustained innate immune signaling. Cell 155 : 688–698. doi: 10.1016/j.cell.2013.09.049 24119841
21. Zhang L, Mo J, Swanson Karen V, Wen H, Petrucelli A, et al. (2014) NLRC3, a Member of the NLR Family of Proteins, Is a Negative Regulator of Innate Immune Signaling Induced by the DNA Sensor STING. Immunity.
22. Shi M, Deng W, Bi E, Mao K, Ji Y, et al. (2008) TRIM30 alpha negatively regulates TLR-mediated NF-kappa B activation by targeting TAB2 and TAB3 for degradation. Nat Immunol 9 : 369–377. doi: 10.1038/ni1577 18345001
23. Hu Y, Mao K, Zeng Y, Chen S, Tao Z, et al. (2010) Tripartite-motif protein 30 negatively regulates NLRP3 inflammasome activation by modulating reactive oxygen species production. J Immunol 185 : 7699–7705. doi: 10.4049/jimmunol.1001099 21048113
24. Ishikawa H, Barber GN (2008) STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455 : 674–678. doi: 10.1038/nature07317 18724357
25. Ishikawa H, Ma Z, Barber GN (2009) STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 461 : 788–792. doi: 10.1038/nature08476 19776740
26. Sun W, Li Y, Chen L, Chen H, You F, et al. (2009) ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc Natl Acad Sci U S A 106 : 8653–8658. doi: 10.1073/pnas.0900850106 19433799
27. Burdette DL, Vance RE (2012) STING and the innate immune response to nucleic acids in the cytosol. Nature Immunology 14 : 19–26.
28. Ozato K, Shin D-M, Chang T-H, Morse HC (2008) TRIM family proteins and their emerging roles in innate immunity. Nature Reviews Immunology 8 : 849–860. doi: 10.1038/nri2413 18836477
29. Barber GN (2014) STING-dependent cytosolic DNA sensing pathways. Trends Immunol 35 : 88–93. doi: 10.1016/j.it.2013.10.010 24309426
30. Wu J, Sun L, Chen X, Du F, Shi H, et al. (2013) Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 339 : 826–830. doi: 10.1126/science.1229963 23258412
31. Yang B, Wang J, Wang Y, Zhou H, Wu X, et al. (2013) Novel Function of Trim44 Promotes an Antiviral Response by Stabilizing VISA. The Journal of Immunology 190 : 3613–3619. doi: 10.4049/jimmunol.1202507 23460740
32. Hou W, Wu Y, Sun S, Shi M, Sun Y, et al. (2003) Pertussis toxin enhances Th1 responses by stimulation of dendritic cells. J Immunol 170 : 1728–1736. 12574336
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Clearance of Pneumococcal Colonization in Infants Is Delayed through Altered Macrophage TraffickingČlánek An Model of Latency and Reactivation of Varicella Zoster Virus in Human Stem Cell-Derived NeuronsČlánek Protective mAbs and Cross-Reactive mAbs Raised by Immunization with Engineered Marburg Virus GPsČlánek Specific Cell Targeting Therapy Bypasses Drug Resistance Mechanisms in African TrypanosomiasisČlánek Peptidoglycan Branched Stem Peptides Contribute to Virulence by Inhibiting Pneumolysin ReleaseČlánek HIV Latency Is Established Directly and Early in Both Resting and Activated Primary CD4 T CellsČlánek Sequence-Specific Fidelity Alterations Associated with West Nile Virus Attenuation in Mosquitoes
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2015 Číslo 6- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Introducing “Research Matters”
- Exploring Host–Pathogen Interactions through Biological Control
- Analysis of Bottlenecks in Experimental Models of Infection
- Expected and Unexpected Features of the Newly Discovered Bat Influenza A-like Viruses
- Clearance of Pneumococcal Colonization in Infants Is Delayed through Altered Macrophage Trafficking
- Recombinant Murine Gamma Herpesvirus 68 Carrying KSHV G Protein-Coupled Receptor Induces Angiogenic Lesions in Mice
- TRIM30α Is a Negative-Feedback Regulator of the Intracellular DNA and DNA Virus-Triggered Response by Targeting STING
- Targeting Human Transmission Biology for Malaria Elimination
- Two Cdc2 Kinase Genes with Distinct Functions in Vegetative and Infectious Hyphae in
- An Model of Latency and Reactivation of Varicella Zoster Virus in Human Stem Cell-Derived Neurons
- Protective mAbs and Cross-Reactive mAbs Raised by Immunization with Engineered Marburg Virus GPs
- Virulence Factors of Induce Both the Unfolded Protein and Integrated Stress Responses in Airway Epithelial Cells
- Peptide-MHC-I from Endogenous Antigen Outnumber Those from Exogenous Antigen, Irrespective of APC Phenotype or Activation
- Specific Cell Targeting Therapy Bypasses Drug Resistance Mechanisms in African Trypanosomiasis
- An Ultrasensitive Mechanism Regulates Influenza Virus-Induced Inflammation
- The Role of Human Transportation Networks in Mediating the Genetic Structure of Seasonal Influenza in the United States
- Host Delivery of Favorite Meals for Intracellular Pathogens
- Complement-Opsonized HIV-1 Overcomes Restriction in Dendritic Cells
- Inter-Seasonal Influenza is Characterized by Extended Virus Transmission and Persistence
- A Critical Role for CLSP2 in the Modulation of Antifungal Immune Response in Mosquitoes
- Twilight, a Novel Circadian-Regulated Gene, Integrates Phototropism with Nutrient and Redox Homeostasis during Fungal Development
- Surface-Associated Lipoproteins Link Virulence to Colitogenic Activity in IL-10-Deficient Mice Independent of Their Expression Levels
- Latent Membrane Protein LMP2A Impairs Recognition of EBV-Infected Cells by CD8+ T Cells
- Bank Vole Prion Protein As an Apparently Universal Substrate for RT-QuIC-Based Detection and Discrimination of Prion Strains
- Neuronal Subtype and Satellite Cell Tropism Are Determinants of Varicella-Zoster Virus Virulence in Human Dorsal Root Ganglia Xenografts
- Molecular Basis for the Selective Inhibition of Respiratory Syncytial Virus RNA Polymerase by 2'-Fluoro-4'-Chloromethyl-Cytidine Triphosphate
- Structure of the Virulence Factor, SidC Reveals a Unique PI(4)P-Specific Binding Domain Essential for Its Targeting to the Bacterial Phagosome
- Activated Brain Endothelial Cells Cross-Present Malaria Antigen
- Fungal Morphology, Iron Homeostasis, and Lipid Metabolism Regulated by a GATA Transcription Factor in
- Peptidoglycan Branched Stem Peptides Contribute to Virulence by Inhibiting Pneumolysin Release
- A Macrophage Subversion Factor Is Shared by Intracellular and Extracellular Pathogens
- A Novel AT-Rich DNA Recognition Mechanism for Bacterial Xenogeneic Silencer MvaT
- Reovirus FAST Proteins Drive Pore Formation and Syncytiogenesis Using a Novel Helix-Loop-Helix Fusion-Inducing Lipid Packing Sensor
- The Role of ExoS in Dissemination of during Pneumonia
- IRF-5-Mediated Inflammation Limits CD8 T Cell Expansion by Inducing HIF-1α and Impairing Dendritic Cell Functions during Infection
- Discordant Impact of HLA on Viral Replicative Capacity and Disease Progression in Pediatric and Adult HIV Infection
- Crystal Structure of USP7 Ubiquitin-like Domains with an ICP0 Peptide Reveals a Novel Mechanism Used by Viral and Cellular Proteins to Target USP7
- HIV Latency Is Established Directly and Early in Both Resting and Activated Primary CD4 T Cells
- HPV16 Down-Regulates the Insulin-Like Growth Factor Binding Protein 2 to Promote Epithelial Invasion in Organotypic Cultures
- The νSaα Specific Lipoprotein Like Cluster () of . USA300 Contributes to Immune Stimulation and Invasion in Human Cells
- RSV-Induced H3K4 Demethylase KDM5B Leads to Regulation of Dendritic Cell-Derived Innate Cytokines and Exacerbates Pathogenesis
- Leukocidin A/B (LukAB) Kills Human Monocytes via Host NLRP3 and ASC when Extracellular, but Not Intracellular
- Border Patrol Gone Awry: Lung NKT Cell Activation by Exacerbates Tularemia-Like Disease
- The Curious Road from Basic Pathogen Research to Clinical Translation
- From Cell and Organismal Biology to Drugs
- Adenovirus Tales: From the Cell Surface to the Nuclear Pore Complex
- A 21st Century Perspective of Poliovirus Replication
- Is Development of a Vaccine against Feasible?
- Waterborne Viruses: A Barrier to Safe Drinking Water
- Battling Phages: How Bacteria Defend against Viral Attack
- Archaea in and on the Human Body: Health Implications and Future Directions
- Degradation of Human PDZ-Proteins by Human Alphapapillomaviruses Represents an Evolutionary Adaptation to a Novel Cellular Niche
- Natural Variants of the KPC-2 Carbapenemase have Evolved Increased Catalytic Efficiency for Ceftazidime Hydrolysis at the Cost of Enzyme Stability
- Potent Cell-Intrinsic Immune Responses in Dendritic Cells Facilitate HIV-1-Specific T Cell Immunity in HIV-1 Elite Controllers
- The Mammalian Cell Cycle Regulates Parvovirus Nuclear Capsid Assembly
- Host Reticulocytes Provide Metabolic Reservoirs That Can Be Exploited by Malaria Parasites
- The Proteome of the Isolated Containing Vacuole Reveals a Complex Trafficking Platform Enriched for Retromer Components
- NK-, NKT- and CD8-Derived IFNγ Drives Myeloid Cell Activation and Erythrophagocytosis, Resulting in Trypanosomosis-Associated Acute Anemia
- Successes and Challenges on the Road to Cure Hepatitis C
- BRCA1 Regulates IFI16 Mediated Nuclear Innate Sensing of Herpes Viral DNA and Subsequent Induction of the Innate Inflammasome and Interferon-β Responses
- A Structural and Functional Comparison Between Infectious and Non-Infectious Autocatalytic Recombinant PrP Conformers
- Phosphorylation of the Peptidoglycan Synthase PonA1 Governs the Rate of Polar Elongation in Mycobacteria
- Human Immunodeficiency Virus Type 1 Nef Inhibits Autophagy through Transcription Factor EB Sequestration
- Sequence-Specific Fidelity Alterations Associated with West Nile Virus Attenuation in Mosquitoes
- EBV BART MicroRNAs Target Multiple Pro-apoptotic Cellular Genes to Promote Epithelial Cell Survival
- Single-Cell and Single-Cycle Analysis of HIV-1 Replication
- TRIM32 Senses and Restricts Influenza A Virus by Ubiquitination of PB1 Polymerase
- The Herpes Simplex Virus Protein pUL31 Escorts Nucleocapsids to Sites of Nuclear Egress, a Process Coordinated by Its N-Terminal Domain
- Host Transcriptional Response to Influenza and Other Acute Respiratory Viral Infections – A Prospective Cohort Study
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- HIV Latency Is Established Directly and Early in Both Resting and Activated Primary CD4 T Cells
- Battling Phages: How Bacteria Defend against Viral Attack
- A 21st Century Perspective of Poliovirus Replication
- Adenovirus Tales: From the Cell Surface to the Nuclear Pore Complex
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



