-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaPhosphorylation of the Peptidoglycan Synthase PonA1 Governs the Rate of Polar Elongation in Mycobacteria
Bacterial infections rely on continued bacterial growth. Studying cell growth is particularly important for pathogens such as Mycobacterium tuberculosis that grow differently than model organisms. Unlike Escherichia coli or Bacillus subtilis, which grow by incorporating cell wall material along their body, mycobacteria grow from the pole. It remains unclear how mycobacteria construct and extend their poles. Our work identifies a cell wall synthase, PonA1, as a key determinant of mycobacterial polar growth. PonA1 governs polar growth through two enzymatic activities that build the cell wall’s peptidoglycan (PG); both of these activities are required for normal cell growth. Changes in the amount or activity of PonA1 leads to misplaced cell poles and inhibition of cell proliferation. PonA1 is phosphorylated, an unusual modification for PG synthases. This phosphorylation tunes the rate of cell elongation. Changing PonA1’s regulatory or enzymatic activity impacts the survival of cells in the host or when treated with antibiotics. Our work shows how mycobacterial cell pole construction and cell fitness is governed by a major cell wall synthase; these findings may have implications for other bacteria that elongate from their poles.
Published in the journal: . PLoS Pathog 11(6): e32767. doi:10.1371/journal.ppat.1005010
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1005010Summary
Bacterial infections rely on continued bacterial growth. Studying cell growth is particularly important for pathogens such as Mycobacterium tuberculosis that grow differently than model organisms. Unlike Escherichia coli or Bacillus subtilis, which grow by incorporating cell wall material along their body, mycobacteria grow from the pole. It remains unclear how mycobacteria construct and extend their poles. Our work identifies a cell wall synthase, PonA1, as a key determinant of mycobacterial polar growth. PonA1 governs polar growth through two enzymatic activities that build the cell wall’s peptidoglycan (PG); both of these activities are required for normal cell growth. Changes in the amount or activity of PonA1 leads to misplaced cell poles and inhibition of cell proliferation. PonA1 is phosphorylated, an unusual modification for PG synthases. This phosphorylation tunes the rate of cell elongation. Changing PonA1’s regulatory or enzymatic activity impacts the survival of cells in the host or when treated with antibiotics. Our work shows how mycobacterial cell pole construction and cell fitness is governed by a major cell wall synthase; these findings may have implications for other bacteria that elongate from their poles.
Introduction
Mycobacterium tuberculosis, the causative agent of tuberculosis (TB), remains a significant threat to human health. The World Health Organization estimates that 1.5 million individuals die of TB every year, and that roughly 2 billion people are latently infected with M. tuberculosis (Mtb)[1]. Despite the substantial global burden of TB, little is known about the bacterium’s basic physiological pathways, including cell growth and division.
Two processes govern growth of rod-shaped bacterial cells: elongation of the cell and then division of that cell into two daughter cells. This growth and division requires extensive remodeling of the cell wall, a layer or layers of complex saccharides that surrounds the cell’s plasma membrane. Cell division involves the synthesis and then splitting at mid-cell of a peptidoglycan layer (a macromolecule composed of disaccharides crosslinked by short peptides), called the septum. Bacteria elongate by incorporating new cell wall material into their existing cell wall. Many rod-shaped bacteria grow by elongating their mid-section; however, mycobacteria have a different body plan and elongate by incorporating new cell wall precursors at the cell poles[2,3].
Cell growth and division rely on the synthesis of new cell wall. The bacterial cell wall is required for cell survival and determines bacterial shape. Peptidoglycan (PG) is a major component of bacterial envelopes and is central to the integrity of the cell wall. PG is remodeled in response to stress and undergoes rounds of synthesis and hydrolysis every cell cycle to promote cell elongation and division[2,3]. Although coordination of PG synthesis has been extensively studied in other bacterial species, this process, particularly during elongation, remains relatively unexplored in mycobacteria.
One key enzyme that plays a central role in elongation and septation is PonA1, which localizes to the cell pole and septum, where it interacts with the essential PG hydrolase RipA[4,5]. Our previous work suggested that PonA1 was essential in Mycobacterium smegmatis (Msm)[4], indicating that we could use Msm as a convenient tool to define cellular activity specific to PonA1. Studying PonA1 enables us to determine how the cell governs one of the most critical steps in PG synthesis—elongation of the cell pole.
PonA1 is a penicillin binding protein (PBP), a member of a family of proteins that promotes cell growth and division through the synthesis of PG[2,3]. PonA1 has two extracellular catalytic domains that carry out the two necessary reactions for peptidoglycan synthesis: transglycosylation (TG) and transpeptidation (TP). TG reactions link the repeating disaccharide units that form the glycan backbone of peptidoglycan. Pentapeptide tails descend from the glycan chains and are crosslinked by TP reactions[2,3]. In addition to its two catalytic domains, PonA1 contains a long unconserved cytoplasmic tail. The cytoplasmic tail is phosphorylated[6], an unusual modification for PBPs that may play a role in PonA1’s cellular function.
Here we provide evidence that PonA1 is required for cell proliferation in Msm and is necessary for normal survival of Mtb during infection. It plays multiple roles in determining cell length and defining new growth. PonA1 is an early polar localizing factor that can nucleate elongation complexes to construct new cell poles, whose elongation rates are modulated by PonA1’s phosphorylation. Changes in PonA1 activity impact cell shape and growth, likely through insults to the cell wall peptidoglycan, which ultimately results in reduced cell fitness during infection and stress. Collectively, our data suggest that cell elongation in mycobacteria requires PonA1, whose catalytic and regulatory activities modulate the function of cell growth complexes.
Results
Normal growth of M. smegmatis and M. tuberculosis requires PonA1
Transcriptional depletion of ponA1 (MSMEG_6900, Entrez Gene ID 4536904) was previously shown to severely impact proliferation of Msm and induces the formation of lemon-shaped or ballooning cells, suggesting that PonA1 plays a critical role in cell growth in mycobacteria[4]. To determine whether PonA1 is essential for growth of Msm, we used an allelic replacement system[7] that allows exchange of PonA1 alleles on the bacterial chromosome (Fig 1A). We generated two exchange vectors, one encoding wildtype ponA1 and a negative control vector lacking ponA1. Exchanging wildtype ponA1 with a similar wildtype ponA1 fully restored bacterial growth; however, exchange of ponA1 with the negative control vector abolished bacterial growth (Fig 1A). The few remaining colonies on the negative control plate were confirmed ponA1+ by PCR (seven colonies from approximately 4x108 transformed cells). These data show that growth of Msm requires ponA1. However, transposon mutagenesis data from M. tuberculosis H37Rv (Mtb) suggested that ponA1 (Rv0050, Entrez Gene ID 887065) could be disrupted without compromising proliferation of Mtb in culture[8], but that such a disruption would impact survival in a mouse model of TB[9]. We therefore tested whether ponA1 could be deleted from the Mtb genome by recombineering (see Materials and Methods for details). We successfully isolated a ΔponA1 Mtb mutant and confirmed that ΔponA1 Mtb cells grew similarly to wildtype Mtb under standard laboratory culture conditions (S1A and S1B Fig). However, ΔponA1 Mtb cells replicated less robustly than wildtype Mtb in the lungs of C57Bl6 mice at 15 and 42 days post infection (dpi) and were moderately impaired in dissemination to the spleen at 42 dpi (Fig 1B), indicating that loss of PonA1 impacted Mtb fitness during infection. Together, these data show that PonA1 is necessary for normal growth of Msm in culture and promotes normal fitness of Mtb during infection.
Fig. 1. PonA1 is essential in M. smegmatis and required for normal growth of M. tuberculosis. 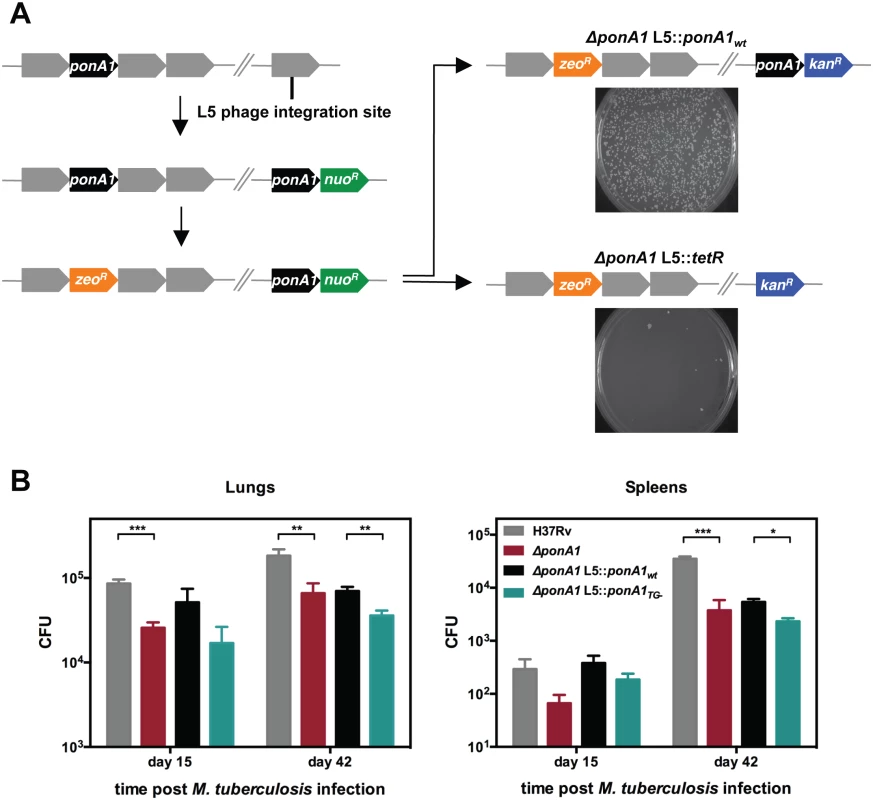
(A) An allelic exchange system in M. smegmatis provides an efficient method to test the importance of PonA1 for bacterial survival. PonA1 is essential in M. smegmatis, as allelic exchange with a vector encoding ponA1 complements bacterial growth, while allelic exchange with a negative control vector fails to rescue growth. (B) C57Bl6 mice were aerosol infected with H37Rv wildtype, ΔponA1, ΔponA1::ponA1wt, and ΔponA1::ponA1TG- (an allele of PonA1 with a catalytic active site mutation in the transglycosylase (TG) domain), and CFU were enumerated were from lung and spleen homogenates at 15 and 42 days post infection (dpi). ΔponA1 and ΔponA1::ponA1TG- cells are less fit than H37Rv wildtype or isogenic wildtype, respectively, at 15 and 42 dpi. Statistical significance was calculated by a one-tailed t-test (lungs at 15 dpi, H37Rv compared to ΔponA1 *** indicates p-value = 0.0005; lungs 42 dpi, H37Rv compared to ΔponA1 ** indicates p-value = 0.0089, and ΔponA1::ponA1wt compared to ΔponA1::ponA1TG- ** indicates p-value = 0.0042. Spleens at 42 dpi, H37Rv compared to ΔponA1 *** indicates p-value = 0.0001, and ΔponA1::ponA1wt compared to ΔponA1::ponA1TG- * indicates p-value = 0.0122). PonA1, while not required for growth of M. tuberculosis in culture, is required for normal bacterial multiplication and dissemination during infection in a mouse model of tuberculosis. PonA1’s synthesis of glycan chains is essential for M. smegmatis viability and for normal cell length of M. tuberculosis
PonA1’s essentiality in Msm is unusual, since bifunctional PBPs in many bacterial species, including Escherichia coli[10], Bacillus subtilis[11], Vibrio cholerae[12], and even the closely related, polar growing actinomycete Corynebacterium glutamicum[13] have largely redundant roles. The Msm genome encodes two other bifunctional PBPs, PonA2 and PonA3, with presumably similar catalytic activities[14], whereas Mtb encodes just PonA1 and PonA2[15]. To identify which function of PonA1 was essential in Msm, we produced a panel of PonA1 mutants with varying catalytic functionality (S2A Fig) for use in the allelic exchange system shown in Fig 1A. Mutations in homologous active site residues eliminated catalytic activity in in vitro assays for E. coli PBP1a[16] and PBP1b[17]. In E. coli, genetic ablation of TP activity did not alter TG activity[16,17], whereas elimination of TG activity also abolished the enzyme’s TP activity in vitro, presumably because the substrate for the TP reaction was not produced[16,17]. However, whether loss of TP activity also abolishes TG activity in the cell remains unclear.
To test whether TG, TP or both activities were required to sustain growth, we transformed Msm with the exchange vectors and tested for appropriate chromosomal integration by antibiotic selection (see Materials and Methods). We found that only a PonA1TP- allele (predicted to lack TP activity) was capable of rescuing bacterial growth while PonA1TG- and PonA1TG-TP- alleles (which lack either TG or both activities) failed to rescue bacterial survival (Fig 2A). These data suggest that PonA1’s polymerization of glycan strands is required in Msm, but that its crosslinking of PG is dispensable. This is consistent with a previous report of an Msm mutant with a transposon insertion in ponA1 that generates a truncated protein that includes the TG domain but lacks the TP domain[18]. This mutant is likely still capable of transglycosylation but lost the ability to catalyze transpeptidation [18].
Fig. 2. PonA1’s glycan synthesis is required in M. smegmatis and catalytic activity promotes normal cell elongation. 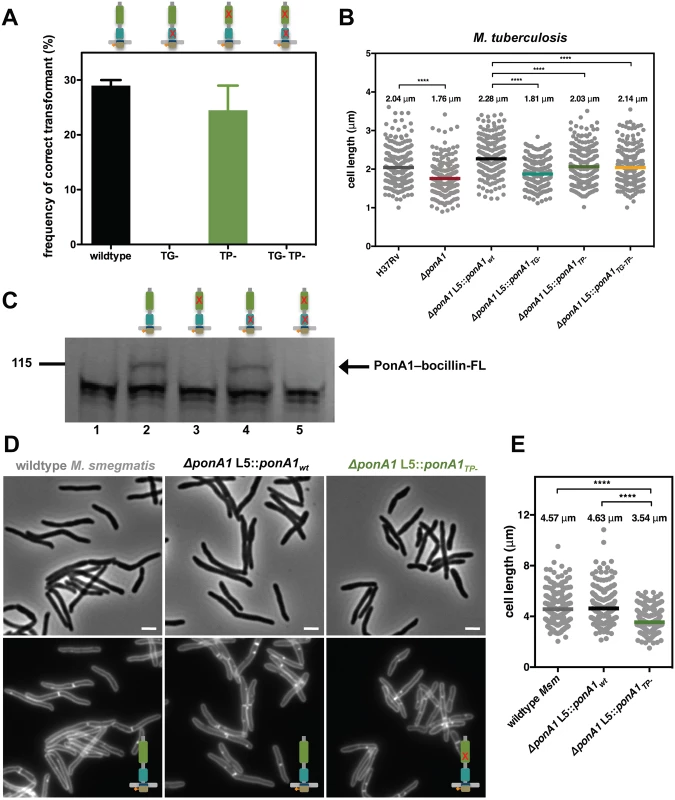
(A) Allelic exchange (see Fig 1A) with vectors encoding alleles of PonA1 with varying catalytic activities (TG-, transglycosylase mutant; TP-, transpeptidase mutant; TG-TP-, transglycosylase and transpeptidase double mutant) demonstrates that a TG- allele fails to rescue bacterial survival. In contrast, a TP- allele complements bacterial growth. It is likely that PonA1’s synthesis of the glycan backbone of peptidoglycan is required for cell survival while other enzymes coordinate with PonA1 to crosslink those glycan strands into the existing sacculus. (B) M. tuberculosis strains were grown in culture and total cell length was measured: H37Rv wildtype (234 cells), ΔponA1 (241 cells; approximate p-value < 0.0001 by the Kolmogorov-Smirnov test), ΔponA1 L5::ponA1wt (212 cells), ΔponA1 L5::ponA1TG- (220 cells; approximate p-value < 0.0001 by the Kolmogorov-Smirnov test), ΔponA1 L5::ponA1TP- (227 cells; approximate p-value < 0.0001 by the Kolmogorov-Smirnov test), and ΔponA1 L5::ponA1TG-TP- (209 cells; approximate p-value < 0.0001 by the Kolmogorov-Smirnov test). Loss of PonA1 or expression of a TG- allele of PonA1 reduces cell length compared to their isogenic wildtypes, supporting the role of PonA1’s TG activity in regulating cell growth in M. tuberculosis. Loss of TP activity had a more modest effect on cell length. (C) Msm PonA1 isoforms were produced in E. coli and assessed for their ability to bind bocillin-FL. Lane 1, negative control (wildtype E. coli); lane 2, PonA1wt; lane 3, PonA1TP-; lane 4, PonA1TG-; lane 5, PonA1TG-TP-. Mutants that lack TP activity cannot bind bocillin-FL. However, PonA1TG- binds bocillin-FL at a level similar to wildtype PonA1, showing that PonA1’s catalytic activities can be uncoupled. (D) Msm cells that express PonA1TP- do not exhibit gross cell shape changes, suggesting another enzyme coordinates peptidoglycan crosslinking with PonA1 to maintain cell integrity. Scale bar, 2 μm. (E) ΔponA1 L5::ponA1TP- cells (229 cells) are shorter than wildtype M. smegmatis (wt Msm, 251 cells; approximate p-value < 0.0001 by the Kolmogorov-Smirnov test) or isogenic wildtype (ΔponA1 L5::ponA1wt, 247 cells; approximate p-value < 0.0001 by the Kolmogorov-Smirnov test), indicating that PonA1’s crosslinking influences cell length. In Mtb, PonA1’s synthesis of glycan chains is required for normal bacterial growth. Mtb cells that expressed a PonA1TG- allele exhibited moderately impaired viability and dissemination during infection, similar to ΔponA1 cells, when compared to isogenic wildtype (Fig 1B). This fitness defect could be associated with abnormal cell length, as ΔponA1 and ΔponA1 L5::ponA1TG- cells are shorter than their isogenic wildtypes by 14% and 21%, respectively (Fig 2B) but showed no gross cell shape changes or substantial population doubling defects (S1A, S1B and S1C Fig). The ΔponA1 L5::ponA1TG- cells also produced somewhat less phthiocerol dimycocerosate (PDIM) (S3 Fig), a cell wall lipid that is important for robust growth during infection. Lower PDIM production by this strain may contribute to the decreased fitness of these cells in the host, although we do not suspect that low PDIM is related to TG - PonA1 as low PDIM production has been described in unrelated genetic mutants[19]. Furthermore, ΔponA1 Mtb cells exhibit a cell length defect but produce PDIM at levels similar to or slightly above wildtype H37Rv, suggesting that PDIM loss is not correlated with decreased cell length. The change in cell length observed with ΔponA1 and ΔponA1 L5::ponA1TG- cells suggests that PonA1’s polymerization of glycan chains is required for normal cell length in Mtb. Altered peptidoglycan structure in these shorter cells likely underlies their decreased ability to resist host-induced stresses.
Correct cell length depends on PonA1’s crosslinking of peptidoglycan
The differential essentiality of PonA1’s TG and TP activities in the cell suggested that they could be uncoupled. Uncoupling of the TG and TP activities for PonA1 may be unusual. in vitro studies have shown that in other bifunctional PBPs, the TP domain crosslinks the nascent glycan strands produced by the cis-TG domain, and it cannot function in the absence of TG activity[16,17,20]. To determine whether PonA1’s crosslinking activity depended on its ability to polymerize glycan strands, we tested whether the PonA1TG- allele could bind to a fluorescent penicillin analog, bocillin-FL, which covalently interacts with the transpeptidase domain active site serine[21]. Using FLAG-tagged PonA1 isoforms, we found that PonA1TG- binds to bocillin at a level similar to wildtype PonA1, whereas PonA1TP- and PonA1TG-TP- did not bind to bocillin, as expected (Fig 2C). FLAG immunoblotting confirmed the bocillin-labeled band was PonA1-FLAG (S2B Fig). These data suggest that PonA1’s catalytic activities can be uncoupled, and specifically that its transpeptidase activity does not require active cis-transglycosylation.
Accordingly, we hypothesized that the uncoupling of PonA1’s catalytic activities could impact cell elongation and/or division, resulting in altered cell morphology. We imaged Msm cells that expressed PonA1wt or PonA1TP- and quantified cell length. We found that PonA1TP- Msm were 24% shorter on average than isogenic wildtype (Fig 2D and 2E) and had impaired population doubling (S2C and S2D Fig) although similar levels of protein were expressed (S2E Fig). These data suggest that PonA1’s crosslinking of PG is required for normal cell length, and that the balance of PonA1 catalytic activities could have a role in controlling cell length and withstanding stress. Impaired PonA1 crosslinking in Mtb decreased cell length as well. Cells that expressed a PonA1TP- or PonA1TG-TP- allele were 11% or 6% shorter on average, respectively, than isogenic wildtype (Fig 2B). These data indicate that while PonA1’s TG activity is critical for cell viability and normal cell length (Figs 1B and 2), its TP activity also has a role in maintaining correct cell length in Mtb, although these mutations do not significantly alter population doubling rates (S1B, S1C and S1D Fig).
PonA1’s transpeptidase domain is dispensable for cell survival
As PonA1’s TG activity is necessary for the viability of Msm, we hypothesized that the TG domain as the sole periplasmic domain would be sufficient to rescue bacterial growth. Furthermore, our previous work indicated that PonA1 dampened the hydrolytic activity of the RipA-RpfB complex in vitro[4], suggesting that PonA1’s modulation of RipA’s lytic activity could also contribute to the requirement of PonA1 in Msm. PonA1’s TP domain mediates the interaction with RipA[4]. To test whether Msm could survive with only PonA1’s TG domain in the periplasm, we generated a truncation mutant of PonA1 that eliminated the TP domain (PonA11-360) (S2A Fig). We tested whether this truncation could complement loss of PonA1 via allelic exchange (Fig 1A). We found that PonA11-360 could support bacterial survival and growth. However, cells that expressed PonA11-360 exhibited cell shape defects (Fig 3A) and were 38% shorter than isogenic wildtype cells (Fig 3B). These morphological changes translated to diminished population growth rates (S2F and S2G Fig); however, these phenotypic differences were not due to difference in protein expression of the truncated PonA1 allele (S2H Fig). In sum, these data suggest that PonA1’s TG activity is necessary and that the TG domain is the critical periplasmic domain required for the survival of Msm. PonA1 modulation of RipA activity may be an important cellular role for PonA1, but it is not absolutely required for viability of Msm, which has other means of controlling RipA lytic activity[5].
Fig. 3. PonA1’s periplasmic domains modulate cell shape in mycobacteria. 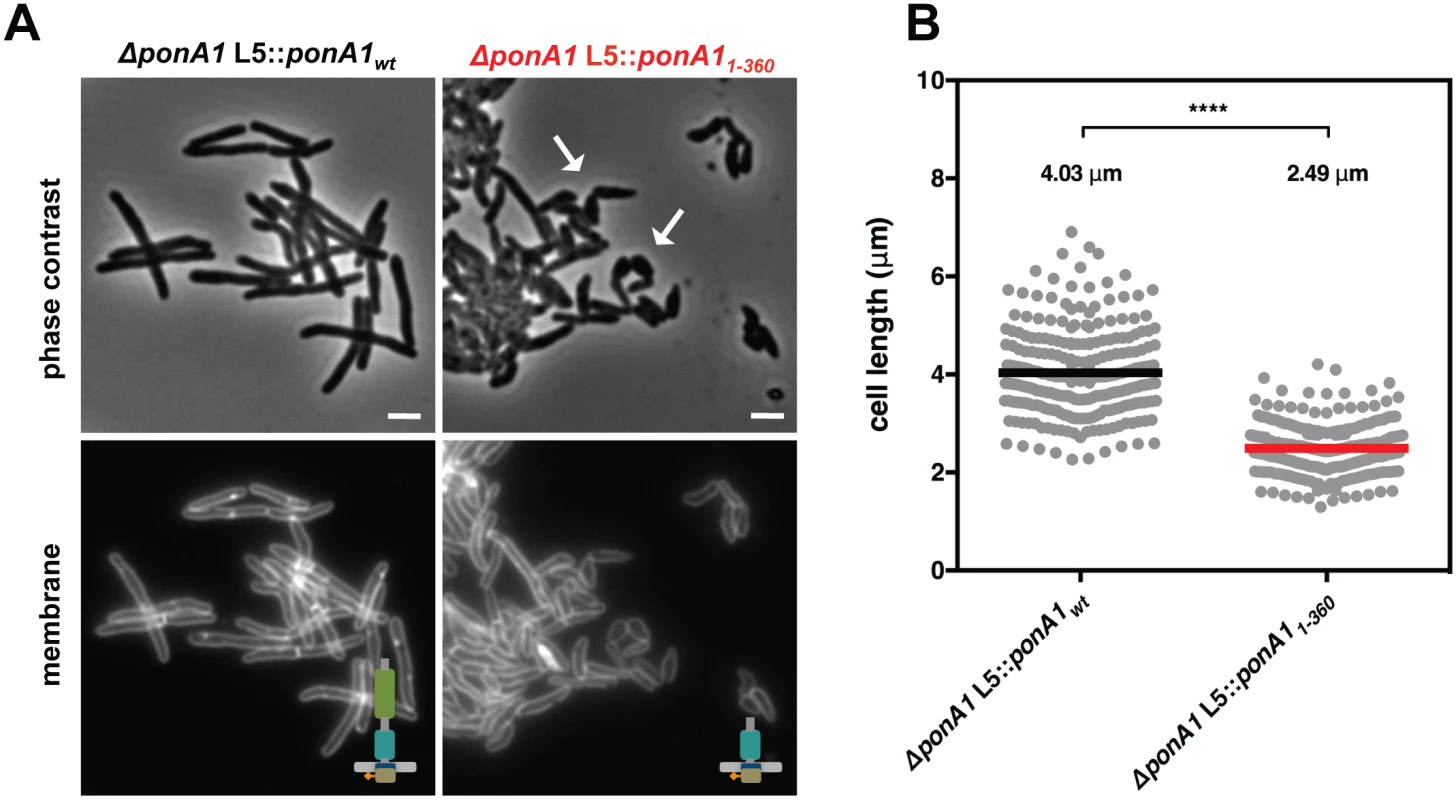
(A) Replacement of wildtype PonA1 with a PonA1 truncation containing the cytoplasmic tail and TG domain (PonA11-360) rescues bacterial survival. Cells exhibit shape changes, but survive, suggesting that PonA1’s TG domain is the sufficient periplasmic domain required for bacterial growth. Cell shape may change due to missing protein-protein interactions that occur along the TP domain. Scale bar, 2 μm. (B) PonA11-360 cells (266 cells) are shorter than isogenic wildtype (251 cells; approximate p-value < 0.0001 by the Kolmogorov-Smirnov test), which may indicate that the TP domain plays a role in complex formation that influences cell length. These measurements were taken on a different day than Fig 2C and 2D, hence the slight difference in isogenic wildtype cell length. Linear polar elongation requires balanced PonA1 activity
Our data together suggest that PonA1 is critical for robust growth and that its catalytic activity is required for normal cell length. Do these critical catalytic activities lead to a dominant negative phenotype such that replacing wildtype PonA1 in cell growth complexes induces cellular toxicity? We tested this hypothesis by overproducing PonA1 in Msm. The overproduction of the PonA1TG-TP- double mutant induced the formation of ectopic cell poles (Fig 4A) and severely inhibited population growth rates (Fig 4B), as did the single catalytic mutants. However, we observed the same phenotypes with overproduction of wildtype PonA1 (Fig 4A and 4B). This is in contrast to overproduction of the PonA1 homologue PBP1b in E. coli, which exhibits a dominant negative phenotype when overexpressed; namely, catalytically inactive PBP1b, but not wildtype PBP1b, inhibited growth and induced cell lysis[22]. These data suggest the cellular activity of PonA1 is different than its PBP1b homologue in E. coli. The growth inhibition with PonA1 overproduction in Msm correlated with the observation that the wildtype ponA1 complement in Mtb (ΔponA1 L5::PonA1wt), which is driven by a strong promoter, did not fully complement wildtype growth at later time points in both lungs and spleens (Fig 1B). These data suggest that overproduction of PonA1 in Mtb also impacted cell fitness. Collectively, these results indicate that the cell must tightly regulate PonA1 levels and activity to maintain robust cell growth.
Fig. 4. Excess PonA1 induces ectopic polar growth in M. smegmatis. 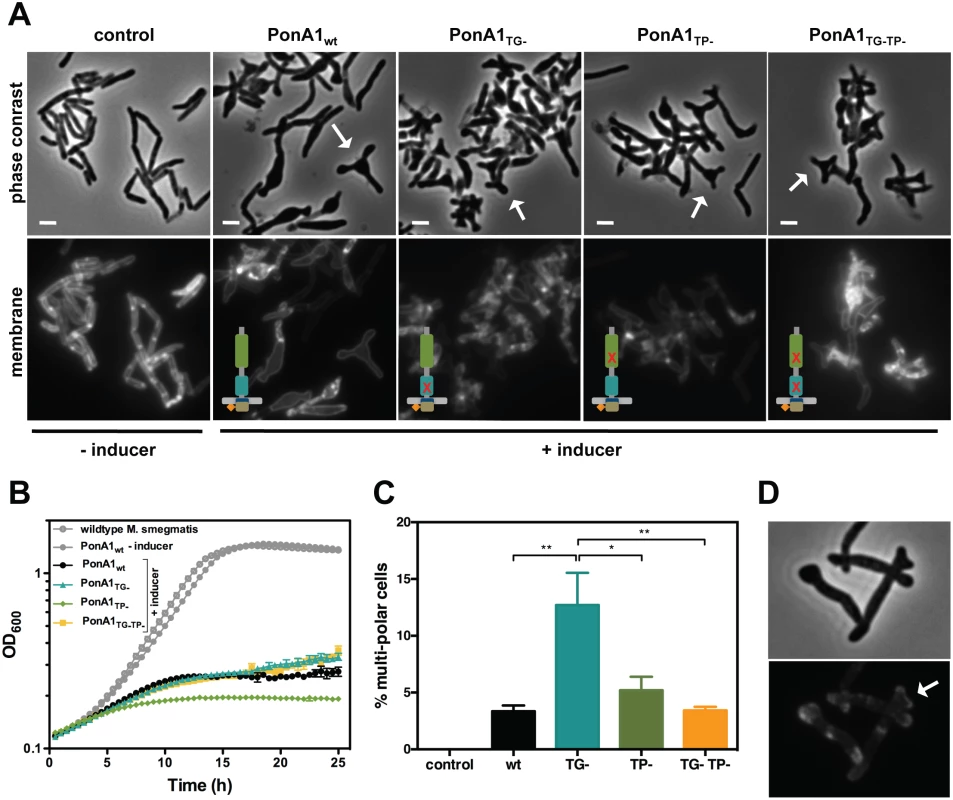
(A) Overexpression of PonA1 causes the formation of ectopic growth poles, regardless of PonA1’s catalytic activity, suggesting that PonA1’s complex formation is sufficient to induce ectopic polar growth. Scale bar, 2 μm. (B) Excess levels of PonA1, including wildtype PonA1, inhibit bacterial multiplication, likely because of ectopic polar growth. (C) Although PonA1’s catalytic activity is not required for ectopic pole formation, it does influence the frequency at which ectopic poles form. Cells that express a TG- allele form ectopic poles at more than 2 times the frequency of other PonA1 alleles, suggesting that local peptidoglycan architecture could influence ectopic pole formation (statistical significance was assessed by one-way analysis of variance with Bonferroni’s multiple comparison test, and multiplicity adjusted p-values are reported. PonA1wt compared to PonA1TG-, p-value = 0.0039; PonA1TG- compared to PonA1TP-, p-value = 0.0225; PonA1TG- compared to PonA1TG-TP-, p-value = 0.0042). (D) PonA1-RFP localizes to both growing tips of the ectopic pole, indicating that PonA1 is involved in nucleating elongation complexes at these growth tips. Although PonA1’s enzymatic activity is not required for ectopic pole formation in Msm, it influenced the frequency of ectopic poles. Cells that overproduced PonA1TG- induced the formation of multi-poled cells at a higher frequency than other alleles of PonA1 (Fig 4C). Taken together, these results suggest that some balance between TG and TP activities is required to induce pole formation. However, no enzymatic activity is required for this phenomenon, suggesting another role for PonA1, perhaps as a scaffold or recruitment factor within protein complexes that promote cell pole growth.
Previous work showed that ectopically-produced PonA1-RFP localized to the pole and septum[4]. A chromosomally RFP-tagged ponA1 also exhibits polar and mid-cell localization (S3D Fig). To determine whether PonA1 localized at the ectopic growth pole, we imaged Msm cells that overproduced PonA1-RFP. We observed that PonA1-RFP localized to each ectopic tip of the growing pole (Fig 4D). Furthermore, PonA1 localized to the pole prior to its degeneration into ectopic poles (S5A Fig, follow pole with white arrow, and S1 Video), suggesting that PonA1 is an early localizing factor that promotes elongation of the cell pole. Ectopic polar growth occurred predominantly from one pole and not both poles (Figs 4A, S4 and S5), which correlates with the expected pattern of asymmetric polar elongation[23,24], and cells that did not overexpress PonA1 did not exhibit ectopic polar growth (Figs 4A, S5B and S2 Video). Occasional cells with bulges or ectopic poles exhibited no RFP signal at those foci. PonA1’s localization to the pole prior to the generation of ectopic poles indicates that PonA1 is intimately involved in not just elongating the pole, but in formation of the cell pole itself, potentially through interaction with DivIVA, which is required for pole formation in mycobacteria[24]. Furthermore, overexpression of PonA1 did not induce misplaced septa, suggesting that the majority of PonA1 is targeted to the cell pole during normal growth.
Ectopic polar growth upon overproduction of PonA1 in Msm is distinct from that observed in E. coli, where ectopic branches appeared when combinations of PBP1a and several low molecular weight PBPs were jointly deleted[25,26]. These data suggest that PonA1’s role in mycobacterial growth is distinct from the homologous bifunctional PBPs in E. coli. This phenotype may be due to the fundamentally different way that mycobacteria elongate—from the cell pole and not along the lateral body as in E. coli.
Mycobacteria control cell elongation through post-translational modification of PonA1
One method the cell may use to regulate PonA1 activity is through post-translational modification. The cytoplasmic tail of PonA1 from Mtb was recently identified as a substrate for the serine-threonine protein kinase PknB[6]. Prisic et al suggested the H37Rv genome had a misannotated start site for rv0050, which was corroborated by another report that identified an alternative protein translation start site in the 5’ UTR of ponA1[27]. We have used a construct beginning 426 nucleotides upstream of the annotated start site, which captures the phosphorylation site, reported translational start site, and yields a predicted transmembrane domain[28] (S6A Fig), a necessary feature of all PBPs[29]. We accordingly aligned the mc2155 genome for Msm PonA1; the start site was shifted 126 nucleotides upstream, which aligned well with -426 Mtb PonA1 (S6A and S6B Fig). Furthermore, the -426 Mtb PonA1 complemented growth of Msm lacking endogenous PonA1, which demonstrated that the -426 Mtb PonA1 allele was functional (S6A and S6B Fig). The -426 Mtb PonA1 protein was also produced at equivalent levels to Msm PonA1 (S6C Fig). To test whether -426 Mtb PonA1 was phosphorylated, we purified from E. coli the Mtb PonA1 cytoplasmic domain tagged to MBP along with his-MBP-tagged kinase domains of all the functional serine-threonine kinases of Mtb. Phospho-transfer profiling of MBP-Mtb PonA1cyto with the kinases revealed that PknB efficiently phosphorylated -426 Mtb PonA1 in vitro (Fig 5A). The specificity of the phosphorylation on MBP-Mtb PonA1cyto’s T34 residue (S6B Fig) was verified by mass spectrometric analysis (S7D Fig).
Fig. 5. Phosphorylation regulates the rate of cell elongation. 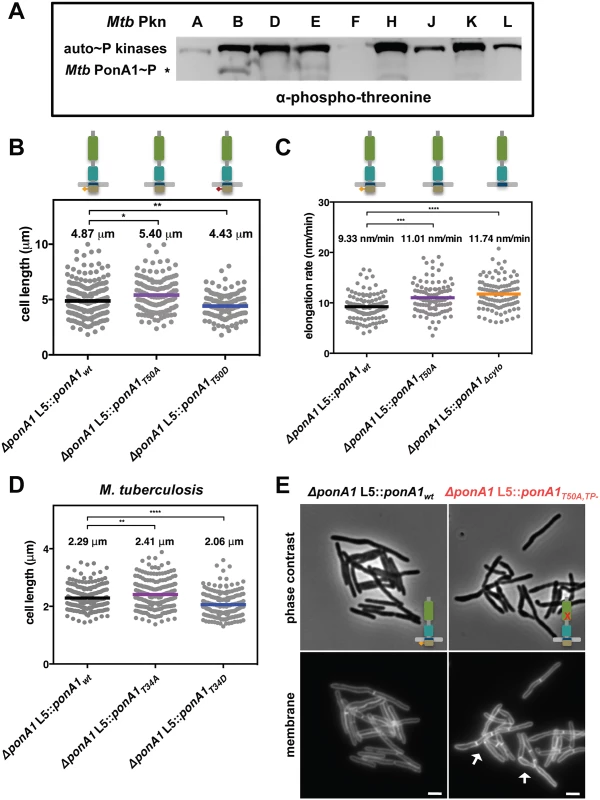
(A) Phospho-transfer profiling with the kinase domains of the major serine-threonine protein kinases of M. tuberculosis (Mtb Pkn) reveals that PknB efficiently phosphorylates Mtb MBP-PonA1cyto. (B) M. smegmatis cells that express a T50A allele of ponA1 (134 cells; approximate p-value = 0.0145 by the Kolmogorov-Smirnov test) are longer than isogenic wildtype cells (219 cells), while cells that express a T50D allele (139 cells; approximate p-value = 0.0082 by the Kolmogorov-Smirnov test) are shorter than isogenic wildtype, suggesting PonA1’s phosphorylation regulates cell elongation or division. (C) Timelapse microscopy revealed that cells that expressed a T50A (127 cells; approximate p-value = 0.0002 by the Kolmogorov-Smirnov test) allele elongated faster than isogenic wildtype cells (174 cells), which was phenocopied by a truncation of the cytoplasmic tail (Δcyto; 202 cells; approximate p-value < 0.0001 by the Kolmogorov-Smirnov test). These data suggest that PonA1’s phosphorylation negatively regulates cell elongation. (D) Similarly, phosphorylation status of PonA1 affects total cell length in M. tuberculosis. Cells that expressed a T34A allele (211 cells; approximate p-value = 0.0066 by the Kolmogorov-Smirnov test) exhibited an average cell length 5% longer than isogenic wildtype (202 cells), and cells that expressed a T34D allele (207 cells; approximate p-value < 0.0001 by the Kolmogorov-Smirnov test) were 11% shorter than isogenic wildtype. This suggests that PonA1’s unusual phosphorylation negatively regulates cell elongation in M. tuberculosis as in M. smegmatis. (E) Msm cells that encode a T50A,TP- allele of PonA1 are defective for normal cell separation. These cells form short chains of cells with multiple septa (white arrows). These data suggest that PonA1’s phosphorylation may regulate PonA1 TG activity, the remaining functional catalytic activity for this allele, and that alterations to PonA1’s TG activity impact the cell’s peptidoglycan and consequent cleavage of that peptidoglycan. Because PknB’s phosphorylation of other targets modulates key steps in cell growth and division[2], we hypothesized that PonA1’s phosphorylation might play a role in regulating cell elongation or division. We tested this hypothesis by constructing two different alleles of Msm PonA1, one in which phosphorylation is blocked by substitution of an alanine residue for the targeted threonine (T50A) and another where an aspartate substitution might mimic the effects of phosphorylation (T50D) and confirmed these isoforms were stable (S6C Fig). We swapped these for the native allele and measured Msm cell length. Cells that expressed the T50A allele were longer than isogenic wildtype, whereas cells that expressed the T50D allele were shorter than isogenic wildtype (Fig 5B). Because the observed changes in cell length did not alter gross optical density measurements of these cell populations (S8A Fig), we used timelapse microscopy to measure single cell elongation and division events. Cells were labeled with a green fluorescent amine-reactive dye that stains the cell surface and does not diffuse over time. This allows the visualization of new cell growth (S8B Fig)[23,30]. After labeling, cells were imaged in custom microfluidic devices, and cell elongation and cell division events were quantified over time (S8B Fig; see Materials and Methods for details). Msm cells that expressed PonA1T50A allele elongated 15% faster than isogenic wildtype (Fig 5C). This faster elongation rate correlated with increased cell length at division (S8C Fig), and did not alter generation time at the single cell level (S8D Fig). Timelapse microscopy analysis did not show substantial growth differences between the PonA1T50D mutant and isogenic wildtype cells. Truncation of the cytoplasmic tail of PonA1 (PonA1Δcyto), which generated a stable protein (S6C Fig), phenocopied the increased elongation rate (Fig 5C), supporting the role of PonA1’s cytoplasmic domain, and likely its phosphorylation, in downregulating cell elongation. Cells that expressed PonA1T50A or PonA1Δcyto elongated predominantly from the old pole (S8E Fig) instead of the new pole (S8F Fig), in agreement with the expected growth pattern for mycobacteria[23,24]. These data indicate that loss of PonA1’s phosphorylation does not alter the subcellular localization of elongation complexes or PonA1’s localization in the elongation complex itself. Thus, it appears that PonA1’s cytoplasmic domain is not required for localization of PonA1 in the elongation complex or for PonA1’s function in that complex. However, PonA1’s cytoplasmic tail could interact with cytoplasmic factors that are involved in modulating or are required for the activity of the elongation complex. For example, it could mediate interaction with DivIVA, an essential determinant of polar growth in mycobacteria [24].
PonA1’s phosphorylation plays a similar role in Mtb. We measured the length of cells that expressed a PonA1 T34A or T34D isoform. We found that the T34A mutant increased cell length by 5% compared to isogenic wildtype, while Mtb that encoded the T34D allele were 11% shorter than isogenic wildtype (Fig 5D). These data correlate with the observation in Msm. Furthermore, expression of T34A Mtb PonA1 in Msm increased cell length of Msm while expression of T34D Mtb PonA1 decreased Msm cell length. Collectively, these data suggest that PonA1’s phosphorylation reduces elongation in both Mtb and Msm and provides the cell with a facile method to modulate cell length, an important response to certain stress conditions, including in-host survival[31].
Because PonA1’s phosphorylation was not essential for protein function, but did modulate cell elongation rate, we hypothesized that PonA1’s phosphorylation regulated its enzymatic activity. To test this, we generated a double mutant in which the phosphorylation and transpeptidase active sites were ablated. Msm cells that expressed this double PonA1 mutant (PonA1T50A,TP-) were defective for normal cell separation. Cells formed short chains with cells of mixed length containing several septa (Fig 5E). This is distinct from the short cell phenotype of TP - cells (Fig 2C and 2D) and the elongated cell phenotype of T50A cells (Fig 5B), neither of which form short chains. Additionally, the Msm PonA1T50A,TP- cells had diminished growth rates compared to either of the single mutants or isogenic wildtype (S9A and S9B Fig), although similar protein levels were produced (S9C Fig). These data suggest that the peptidoglycan synthesized by this PonA1 mutant diminishes efficient cleavage. This could indicate that PonA1’s phosphorylation regulates its TG activity—the remaining enzymatic activity in this mutant—and that the observed division defects result from imbalanced synthesis of peptidoglycan.
Altering PonA1 activity changes antibiotic susceptibility
Because changes to PonA1’s phosphorylation status or synthetic ability altered cell shape and viability, we investigated whether these mutations would impact cell physiology under stress conditions, particularly antibiotic treatment. We previously demonstrated that single nucleotide polymorphisms in ponA1 identified in clinical isolates alter Mtb’s fitness during rifampicin treatment[32]. We tested if altered Mtb PonA1 enzymatic or regulatory activity would also impact susceptibility to rifampicin. Indeed, Mtb cells that expressed PonA1TG- or PonA1T34D were 5 and 4 fold more tolerant to rifampicin treatment than isogenic wildtype, respectively (S10A Fig). In contrast, the inactivation of PonA1’s TP activity or blockade of phosphorylation did not alter cell survival in the presence of rifampicin (S10A Fig). In sum, these data suggest that altering PonA1 catalytic and regulatory activity changes cell fitness in the presence of the clinically relevant antibiotic rifampicin, although the mechanism remains unclear.
We also hypothesized that changes in PonA1 activity would alter susceptibility to cell wall stress, including antibiotics that target PG synthesis. Genetically ablating Mtb PonA1’s TP activity or its phosphorylation site decreased by 8 - or 4-fold, respectively, the minimum inhibitory concentration (MIC) of teicoplanin, a glycopeptide that inhibits PG crosslinking[33] (Fig 6). The increased sensitivity to teicoplanin suggests that imbalanced peptidoglycan synthesis occurred in TP - as well as T34A cells, supporting a model in which phosphorylation may regulate PonA1’s TG activity (the remaining catalytic activity of the TP - mutant). A shifted teicoplanin MIC was also observed in Msm where the TP - allele exhibits a 16-fold change in MIC (Fig 6), although the Msm T50A allele did not phenocopy the shifted teicoplanin MIC observed in Mtb. However, Msm cells that expressed PonA1T50A alleles were more susceptible to a range of meropenem concentrations (S10B Fig), supporting the idea that balanced PonA1 activity is required for architecturally sound peptidoglycan to resist stress. We also tested whether inhibiting glycan synthesis would impact cellular fitness of PonA1 mutants, but observed minimal efficacy of the small molecule inhibitor moenomycin A (S10C Fig).
Fig. 6. PonA1’s transpeptidase activity is required for normal teicoplanin tolerance in mycobacteria. 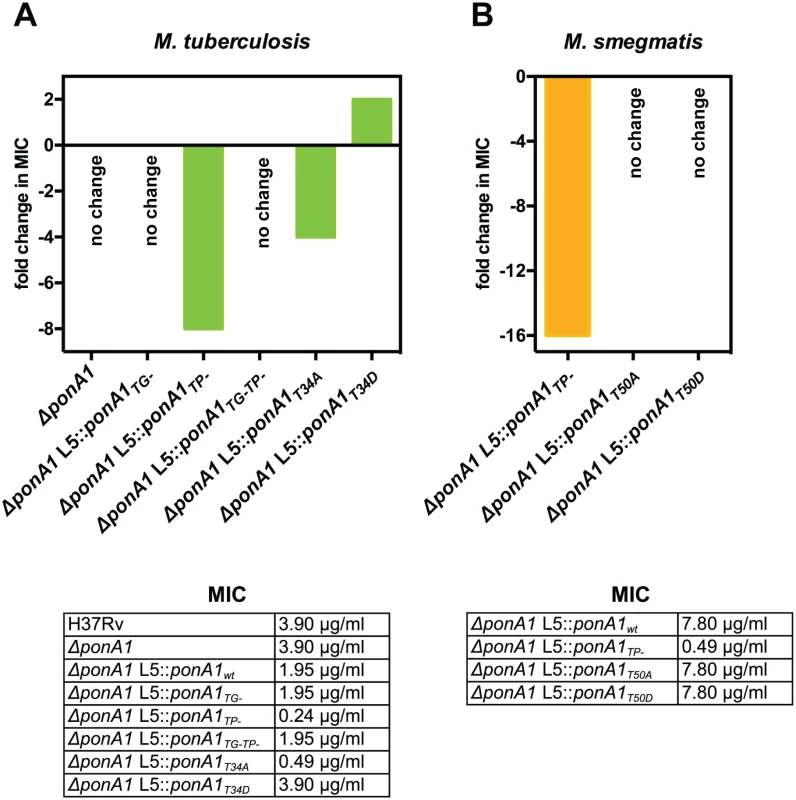
(A) Minimum inhibitory concentration (MIC) of teicoplanin for wildtype and different PonA1 mutant M. tuberculosis strains is tabulated. Fold change in MIC is calculated from wildtype Mtb (for ΔponA1) or isogenic wildtype (for the TG-, TP-, TG-TP-, T34A, and T34D strains). Cells that express a PonA1 TP mutant or the T34A allele are more sensitive to teicoplanin than isogenic wildtype. No change, no fold change in MIC from appropriate wildtype strain. (B) Teicoplanin MICs for M. smegmatis mutant strains are tabulated. Fold change is calculated from isogenic wildtype. Cells that express a PonA1 TP mutant are more sensitive to teicoplanin than isogenic wildtype. Our data suggested that PonA1 activity modulated cell fitness in the presence of drugs. We tested whether changes to PonA1 activity would alter cell fitness under other chemical stresses that target the cell wall. Upon treatment with SDS, Msm cells that expressed PonA1TP- and PonA1T50A exhibited a modest decrease in survival (S11A and S11B Fig). D-amino acids have also been shown to modulate cell wall homeostasis[34], and we tested the fitness of PonA1TP- cells in the presence of D-Met. Msm PonA1TP- cells grew less robustly and did not reach the same optical density at stationary phase as compared to isogenic wildtype cells (S11C and S11D Fig). Furthermore, PonA1TP- cells exhibited aberrant cell shape in deep stationary phase. After four days of growth in culture, Msm PonA1TP- cells became wider and shorter compared to isogenic wildtype (S11E Fig, white arrows). These data together indicate that normal PonA1 catalytic and regulatory activity is required for the synthesis of structurally robust peptidoglycan to ensure cellular survival in the face of multiple stresses.
Discussion
The paradigms of cell growth and division in rod-shaped bacteria are based on organisms that grow fundamentally differently than mycobacteria. Consequently, our understanding of mycobacterial growth—and the cell’s ability to repress growth—remains elementary. We have addressed this knowledge gap by investigating how a key cell wall synthase, PonA1, promotes and regulates cell growth in mycobacteria.
PonA1 is non-redundant during cell growth
We found that PonA1 is essential in Msm and required for normal proliferation in the lung as well as dissemination outside of the lung during an Mtb infection (Fig 1). These data suggest that the major PBPs in mycobacteria do not have truly redundant functions. This is similar to E. coli where distinct complexes employ PBP1a or PBP1b in E. coli[3], as well as recent work in V. cholerae[12] and Listeria monocytogenes[35] that show PBP1a and PBP1b have specialized biological roles during infection. Furthermore, this implies a revision of the classic understanding of the PBPs as redundant factors. Imperfect redundancy during stress could be exploited to identify unique physiological roles for the PBPs in a variety of bacterial species.
The differential essentiality of PonA1 in Msm and Mtb for growth in culture is somewhat surprising; it suggests a difference in expression or functionality of other PBPs, such as the highly homologous protein PonA2, in these species. The role for bifunctional PBPs in Mtb cell elongation is probably jointly filled by PonA1 and PonA2, whereas Msm has evolved to solely depend on PonA1. Even though PonA1 exhibits different essentiality in Mtb and Msm, the role of PonA1 is likely highly similar in the two species. We found that Mtb PonA1 complements survival of Msm depleted of endogenous PonA1 (S7 Fig), and the two proteins are 70% identical at the sequence level (S6 Fig). Furthermore, our data suggest that the physiological role of PonA1 in both Msm and Mtb is very similar.
Governance of cell length and shape
Our data suggest that PonA1’s polymerization of glycan strands plays a key role in determining cell length (Figs 2 and 7A). Indeed, overproduction of TG-inactive PonA1 in Msm yields cells 38% shorter when compared to isogenic wildtype (Figs 4A, S4A and S4B). These cells’ length defect is not solely due to their higher frequency of ectopic pole formation, as even cells without ectopic poles are noticeably shorter than isogenic wildtype (S4A Fig). Balanced PonA1 catalytic activity, however, is required for normal cell length. Whereas TG seems to play a large role in modulating cell length, PonA1’s TP activity is also required for normal cell length (Fig 2). This supports the model wherein efficient PG synthesis occurs when one enzyme is catalytically capable of both synthesizing and crosslinking nascent glycan strands[3]. The linking of PonA1’s activities provides tight control over cell length and the protection of cell integrity and growth.
Fig. 7. A model for how PonA1 promotes cell elongation in mycobacteria. 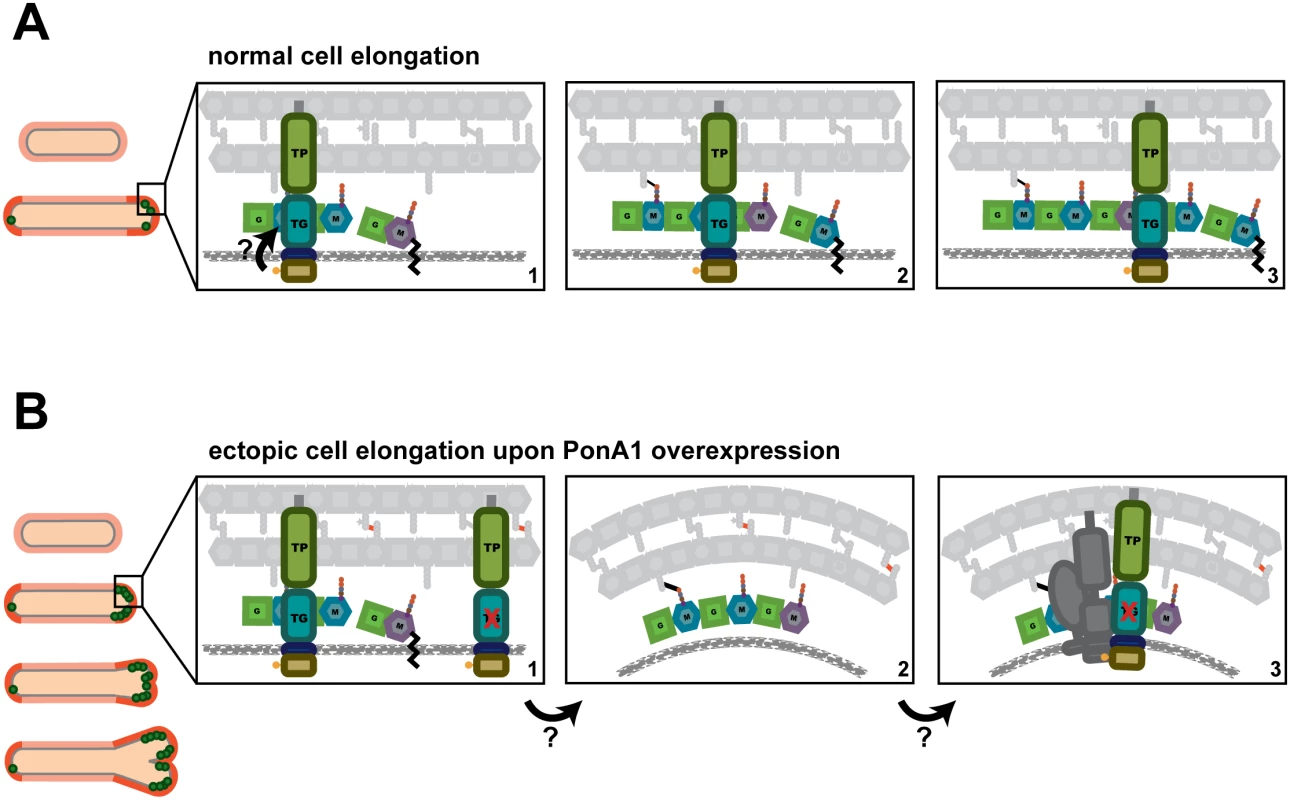
(A) We propose a model wherein PonA1 localizes early to the growth tip and promotes pole elongation through PG synthesis. PonA1 may recruit other factors to form the elongation complex or they are recruited independently of PonA1 (factors not shown). Together with these factors, polar elongation proceeds (orange cells). PonA1 furthers cell elongation through its synthesis of PG. PonA1’s synthesis of glycan strands (colored subunits, panels 1–3) and its crosslinking of those glycan strands (black lines, panels 2–3) promote normal cell length. These catalytic activities are potentially modulated through PonA1’s phosphorylation (arrow), which acts to regulate the rate of cell elongation. (B) Excess PonA1 stimulates ectopic cell elongation (orange cells). The frequency of ectopic pole formation increases with imbalanced PG crosslinking and may result from changes in local peptidoglycan architecture (panels 1–2, factors removed from panel 2 for clarity). These architectural changes may act like a sink that ‘recruits’ additional protein-interactors of PonA1 or additional elongation complex components to spur cell elongation at ectopic sites (panel 3, dark gray factors). We found that PonA1-specific PG crosslinking is not required in mycobacteria, although it does promote normal cell length. This suggests that other factors either can compensate for lack of PonA1 crosslinking when absent or that PonA1 normally functions in the elongation complex with another crosslinking enzyme[2], as is true for bifunctional PBPs in other bacterial species[3,20]. Because mycobacterial peptidoglycan is estimated to contain predominantly 3–3 crosslinks[36,37] formed by l,d-transpeptidases, these ‘non-classical’ 3–3 crosslinking enzymes may be active in the elongation complex under wildtype conditions. Indeed, deletion of LdtB in Mtb results in shorter cells[38].
Linear cell growth
We found that PonA1 catalytic activity is required for normal cell length and that PonA1 is an early acting factor that promotes extension of the cell pole. Overproduced PonA1-RFP localizes to the cell pole when the cell is still rod-shaped (S5 Fig). These cells then generate an ectopic pole, which is also marked by PonA1-RFP. We also see the formation of ectopic poles when PonA1-FLAG is overproduced (Figs 4 and S4), suggesting that this observation is not an artifact of the fusion partner. Combined with the fact that enzymatically inactive PonA1 induces ectopic pole formation, these data suggest that PonA1 is important for nucleating elongation complexes, and that PonA1 is a major factor that promotes pole elongation in mycobacteria. Because overproduction of enzymatically inactive PonA1 still results in ectopic pole formation, it is clear that other factors are involved in physically synthesizing new cell wall at these foci. This could be endogenous PonA1, but other factors are also likely involved in elongation. Identification of PonA1’s interacting partners would illuminate the composition of the elongation complex in mycobacteria and provide further understanding of how cell length and cell shape maintenance are controlled. For example, PonA1 and DivIVA might interact, even transiently, in the elongation complex. DivIVA is required for the rod-shape of mycobacteria [24] but has no enzymatic activity. It must coordinate cell growth with the enzymes that synthesize new cell wall [24]. PonA1, as a key factor that promotes cell elongation, may be one such factor that DivIVA helps position or facilitate its activity at the pole.
Even at hyper-physiological levels, PonA1 is properly recruited to the cell pole, as PonA1-RFP localizes at the pole before it degenerates into ectopic poles (S5 Fig). Furthermore, ectopic poles are not produced along the cell body. This suggests that the machinery that recruits PonA1 to the cell pole is not saturated even at increased levels of PonA1 nor is PonA1’s recruitment mechanism rate-limiting for elongation under physiological levels of PonA1. Additionally, the ectopic growth pole normally forms at one cell pole, although occasional cells with both cell ends sporting ectopic growth poles have been observed; however, these cells tend to have multiple septa (S4C Fig, white arrows). These data support a model of mycobacterial elongation where the cell poles are asymmetrically active, at least for a time[2,23,30].
Interestingly, the frequency of ectopic poles correlates with specific imbalanced PonA1 activity. The development of ectopic poles was highest with a PonA1 isoform that can crosslink PG but cannot polymerize glycan strands. The retention of transpeptidase activity in these mutants might lead to abnormal numbers of PG crosslinks. These data suggest that imbalanced crosslinking might contort local PG architecture, which may change the curvature of the membrane enough to misdirect elongation complexes to those foci (Fig 7B). Alternatively, PonA1 may nucleate elongation complexes at these contorted foci to then extend a functional growth pole. Although these ectopic poles elongate over time, they eventually block cell growth. These data suggest that PonA1’s coordination of polar growth must be tightly regulated to maintain linearity of the bacterial cell. This may be accomplished through proper protein-protein interactions and regulation of PonA1 activity.
Phosphorylation regulates elongation rate
We found that cell growth is regulated through PonA1’s phosphorylation. Phosphorylation of PBPs is unusual, and only mycobacteria have so far been reported to phosphorylate their PBPs[6], suggesting that phosphorylation could regulate the activity of these proteins in mycobacteria. We found that the absence of PonA1’s phosphorylation increased the rate of single cell elongation (Fig 5), suggesting that PonA1’s phosphorylation normally dampens cell elongation in both Msm and Mtb. Cells of both species that harbor PonA1 alleles that cannot be phosphorylated (Msm T50A and Mtb T34A) are longer, suggesting that phosphorylation negatively regulates cell elongation (Fig 5). PonA1’s phosphorylation by PknB is consistent with PknB’s phosphorylation of other members of the PG biosynthetic pathway[39,40]. In fact, PknB regulates PG synthesis at each key point of the pathway: precursor synthesis (GlmU)[40], export (MviN)[39] and polymerization (PonA1). Interestingly, PknB’s phosphorylation of these three distinct factors is inhibitory, providing consistent regulation of PG biogenesis at multiple steps. It would be of interest to determine if PonA1 phosphorylation correlates with stages of the cell cycle; PonA1 may be phosphorylated as the cells begin the switch from elongation to septation, where cells need to slow down elongation. Alternatively, PonA1’s phosphorylation status may cycle with rounds of peptidoglycan synthesis during growth. Further work is required to test these models in addition to determining if PonA1’s phosphorylation impacts Mtb fitness during growth during infection.
How PonA1’s phosphorylation specifically regulates cell elongation is unknown, although our data suggest that it could act to regulate TG activity. In the absence of PonA1 phosphorylation, TG functionality could be hyperactive. This could lead to increased cell length observed with T50A cells (Fig 5) and correlates with the shortness of Mtb cells expressing TG - (Fig 1) or overexpressing TG - Msm cells (Figs 4 and S4). Additionally, when the T50A mutation is combined with TP-, Msm cells exhibit substantial defects in division (Fig 5), which could be a result of hyper-active glycan chain synthesis without corresponding crosslinking by PonA1’s cis-TP domain, altering the normal cellular response to PG synthesis. Differences in antibiotic susceptibility correlate between PonA1’s phosphorylation and enzymatic activity as well. In Mtb, PonA1T34D and PonA1TG- cells exhibit the same rifampicin tolerance (S10A Fig), whereas T34A and TP - have the same teicoplanin profile (Fig 6). In sum, these data suggest a model wherein PonA1’s phosphorylation regulates its TG activity (Fig 7A). How this could be achieved remains unclear, although it is possible that conformational changes occur upon phosphorylation. Alternatively, PonA1 phosphorylation may impact the stability of the elongation complex or of PonA1’s retention in the elongation complex, which could underlie the changes in cell length observed when cells express phosphorylation mutants. In any case, this is an unusual example of cytoplasmic regulation of periplasmic protein activity, though, given the number of phosphorylated proteins found in mycobacteria, this might be a common theme in these organisms.
Altering PonA1 activity changes antibiotic susceptibility
Changes to PonA1 regulatory or enzymatic activity are required for robust growth during antibiotic treatment, suggesting that PonA1 mediates tolerance to drugs that target penicillin binding proteins or PG synthesis. We found markedly enhanced susceptibility of Mtb and Msm TP - cells to teicoplanin. Enhanced sensitivity of PonA1 TP - cells may reflect an increased cellular toxicity from imbalanced peptidoglycan synthesis. Recent work suggested that beta-lactams are particularly effective because they not only target TP domains of penicillin binding proteins but they also block crosslinking of nascent glycan strands, leading to an accumulation of non-crosslinked or improperly crosslinked strands that are then degraded, causing a futile cycle of synthesis and degradation that consumes resources[41]. Teicoplanin treated PonA1 TP - cells may experience this cellular toxicity. Because the TP - allele is likely still capable of transglycosylation, it can synthesize nascent glycan strands that will then be blocked from crosslinking when cells are treated with a crosslinking inhibitor. This could lead to an accumulation of uncrosslinked glycan strands and induce cellular toxicity, resulting in the increased teicoplanin sensitivity these cells exhibit. Because the T34A allele demonstrates a similar shifted MIC, it suggests that glycan strand synthesis is also altered in this mutant. Loss of phosphorylation may lead to enhanced TG activity, with a consequent imbalance of crosslinking of the new glycan chains when cells are treated with teicoplanin. Furthermore, the endogenous mycobacterial beta-lactamase is ineffective with drugs like teicoplanin, increasing their potential usefulness. These data suggest that transpeptidation inhibitors, such as teicoplanin, could be a fruitful avenue for TB drug development.
Materials and Methods
Bacterial strains and culture conditions
M. smegmatis mc2155 was cultured in Middlebrook 7H9 salts (Becto-Dickinson) supplemented with ADC (5 g/L albumin, 2 g/L dextrose, 3 g/L catalase), 0.25% glycerol, and 0.05% Tween-80 or plated on LB agar. M. tuberculosis H37Rv was cultured in Middlebrook 7H9 salts supplemented with OADC (oleic acid, albumin, dextrose, catalase [BD Biosciences, Franklin Lakes, NJ]), 0.25% glycerol, and 0.05% Tween-80 or plated on 7H10 agar. Selection was performed at 50 μg/ml hygromycin, 25 μg/ml kanamycin, 20 μg/ml zeocin or 20 μg/ml nourseothricin for both liquid and solid media. E. coli XL-1 Blue (Stratagene, Santa Clara, CA) or TOP10 (Invitrogen, Carlsbad, CA) were used for cloning and were cultured in LB broth or agar. Selection for E. coli occurred at 100 μg/ml hygromycin, 50 μg/ml kanamycin, 20 μg/ml zeocin or 50 μg/ml nourseothricin for both liquid and solid media. All strains were grown at 37°C. The generation of single copy PonA1 mutant strains is described further below. To generate multicopy PonA1 strains, alleles of ponA1 were subcloned into an anhydrotetracycline inducible pMS2 derivative plasmid[42]. These plasmids were transformed into an M. smegmatis mc2155 strain with the pMC1s plasmid[42], encoding the tetR repressor, integrated at the L5 phage integration site.
Recombinant DNA constructs
M. smegmatis PonA1 (MSMEG_6900) and M. tuberculosis PonA1 (Rv0050) catalytic mutants were generated using site directed mutagenesis and PCR stitching with the primers in S1 Table. M. smegmatis PonA1 truncation mutants were generated with primers listed in S1 Table. The double T50A,TP - Msm PonA1 construct was generated via PCR stitching using the appropriate primers in S1 Table. For all Mtb PonA1 constructs, the -426 start site was used. All constructs used in this study were cloned under the strong promoter pUV15 modified to contain TetR operator sites[42] (although no TetR is encoded in the cells’ genomes so expression of ponA1 is constitutive). All PCR reactions were performed with KOD Xtreme Hot Start DNA polymerase (EMD Millipore, Billerica, MA). All M. smegmatis and M. tuberculosis PonA1 constructs listed above were cloned as translational fusions with either the FLAG epitope or monomeric red fluorescent protein (RFP). The FLAG epitope and RFP do not obstruct PonA1 function, as both tags recombineered onto the C-terminus of ponA1 on the chromosome complement Msm survival.
Deletion of ponA1 in M. tuberculosis
A ponA1 (Rv0050) knockout construct was generated by PCR and subcloning. Because ponA1 exists in an operon, the 5’ and 3’ 25 nucleotides of ponA1 were preserved in the knockout construct. The 500 nucleotides upstream of ponA1 were PCR amplified with a 3’ NdeI restriction endonuclease site. The 500 nucleotides downstream of ponA1 were PCR amplified with a 5’ PvuI restriction endonuclease site. A hygromycin resistance cassette flanked by loxP sites was amplified with a 5’ NdeI site and a 3’ PvuI site. These three fragments were subcloned by digesting with NdeI and/or PvuI. The final ligated product was PCR amplified and used to replace ponA1 in the Mtb H37Rv genome via recombineering[43]. Cells were plated on 7H10 plates with 50 μg/ml hygromycin. After incubation at 37°C for three weeks, colonies were inoculated into 2 ml of selective 7H9. Cell lysates were generated and screened by PCR for correct recombinants. One out of 11 colonies was positive for recombination; loss of ponA1 from the genome was verified by PCR and whole genome sequencing.
Ethics statement
The Institutional Animal Care and Use Committee of Harvard University approved and monitored all protocols, personnel, and animal use. The animal facilities are AAALAC accredited, and work was performed under the NIH Office of Laboratory Animal Welfare (OLAW) permit number A-3431-01.
Mouse infections
Six week old wildtype female C57Bl6 mice (Jackson Laboratories, Bar Harbor, ME) were aerosol infected with low doses of M. tuberculosis H37Rv wildtype, H37Rv ΔponA1::Hyg, H37Rv ΔponA1::Hyg L5::ponA1wt, and H37Rv ΔponA1::Hyg L5::ponA1TG-. Strains were confirmed as PDIM positive by mass spectrometry prior to infection using established protocols[44]. Fifteen mice were infected per strain, and five mice were sacrificed at 1 day, 15 days, and 42 days post infection, respectively. To minimize suffering, animals were first put to sleep with isoflurane before sacrifice. Lungs and spleens were homogenized and serially diluted on selective 7H10 plates (for the genetically modified strains) or 7H10 plates lacking antibiotics (for H37Rv wildtype) for CFU enumeration.
Allelic exchange in M. smegmatis
A marked copy of PonA1 was first integrated in the chromosome at the L5 phage integration site. The endogenous locus of PonA1 was then replaced with a different antibiotic cassette via recombineering. This dual marked strain was then transformed with an L5-integrating vector that is marked with a third antibiotic cassette. This second integration event replaces the original integrated allele, and selection for the third antibiotic recovers cells with the desired allele (Fig 1A). Exchange at the L5 site is imperfect and to confirm desired transformants, cells are patched onto plates containing the first L5 integrant marker or the second L5 integrant marker. Cells that are mono-resistant to the second L5 integrant antibiotic are counted as correct transformants; frequencies of correct transformation are calculated by comparison to wildtype ponA1 control transformations.
Bocillin labeling
FLAG-tagged isoforms of M. smegmatis PonA1 were produced in TOP10 E. coli. Untransformed TOP10 cells were used as a negative control. Cells were grown overnight in LB and back-diluted 1 : 100 into 10ml of LB. Once cells reached 0.5 OD600, they were pelleted. Cell pellets were washed once in 1ml of 1x PBS, resuspended in 1ml of 1x PBS, and sonicated twice for 30 seconds each. Bocillin-FL (Life Technologies) was added to the cell solutions at a final concentration of 15 μg/ml and allowed to label protein for 30 minutes at room temperature in the dark. The labeled cell solutions were then pelleted at 100,000 g for 20 minutes to concentrate membrane proteins. The pellets were resuspended in 100 μl of sample buffer containing βME. Proteins were resolved by SDS-PAGE using a 4–15% Tris-Glycine gel. Bocillin-labeled proteins were visualized by imaging with a Typhoon 9400 Variable Mode Imager (GE Healthcare).
Timelapse microscopy and data analysis
For the data in Figs 5 and S8, vegetatively growing M. smegmatis cells were washed twice in 1X PBS with 0.1% Tween 20, stained with 50 μg/ml Alexa488 (Invitrogen), and filtered twice through a 10 μm filter to obtain single cells. Cells were inserted into custom microfluidic devices (see Reference [23]) and imaged at 37°C with constant flow of selective 7H9. Cells were imaged with a DeltaVision PersonalDV microscope using the 60x objective (Plan APO NA1.42) every 10 minutes for a minimum of 18 hours. Images were captured with a CoolSnap HQ2 camera (Photometric). Cell elongation and division events were annotated using ImageJ (National Institutes of Health) with the ObjectJ plugin (Norbert Vischer and Stelian Nastase, University of Amsterdam, http://simon.bio.uva.nl/objectj/index.html). Cell elongation was measured as new unstained cell wall material; cell division was defined as the first frame when physical invagination of the cell wall was visible. A custom Python script was used to analyze the annotations.
For the data in S4 Fig, M. smegmatis cells were imaged using a CellASIC microfluidic system (EMD Millipore, Billerica, MA). Vegetatively growing cells were cultured for four hours ± 100 ng/mL anhydrotetracycline (aTc) inducer to overexpress PonA1TG—RFP prior to growth in the CellASIC chambers. Cells were imaged every 15 minutes for 17 hours with constant flow of selective 7H9 ± 100 ng/mL aTc. Images were captured by a Nikon Eclipse TI microscope using the 60x objective, which is maintained at 37°C with an objective heater. Images were captured with a CoolSNAP SQ2 camera. The image montage in S4 Fig was compiled with FIJI[44].
Light microscopy and image analysis
For imaging, cells were washed once in 1x PBS and resuspended in 1X PBS ± 2.5 μg/ml FM4-64Fx (Invitrogen) to visualize the plasma membrane. After resuspension, cells were incubated in the dark for 10 minutes prior to imaging. For M. tuberculosis, cells were first fixed overnight in 1% formalin in the biosafety level 3 facility before removal and consequent imaging. Cells were imaged on a Nikon TE-200E microscope using the 100x (NA1.40) objective. Images were captured with an Orca-II ER cooled CCD camera (Hamamatsu, Japan). Exposure and image acquisition were controlled with Metamorph Software (Molecular Devices). Final images were prepared in Adobe Photoshop CS3. To quantitate cell length, the fluorescent images of cells stained with FM4-64Fx were used. Cell length was calculated using ImageJ software (National Institutes of Health) and converted to microns using the appropriate pixel to micron conversion. Length was measured from cell pole to the opposite cell pole or from cell pole to septum, if present. Cell synchronization is not yet possible for mycobacteria; hence, cell lengths were measured for unsynchronized populations. To quantitate the frequency of multi-poled cells, cells were both imaged and scored blind to eliminate potential bias. The figure shows data from five combined experiments. 2652 cells were imaged and counted for PonA1wt, 2910 cells for PonA1TG- cells, 2054 for PonA1TP- cells, and 2379 for PonA1TG-TP- cells. The control represents data collected from uninduced cells. Overexpression of PonA1 was induced with 100 ng/mL aTc.
In vitro kinase assays and detection of PonA1’s phosphorylation site
N-terminally his-MBP-tagged kinase domains of the nine canonical serine-threonine protein kinases from Mtb were expressed and purified from E. coli using a nickel column and then an S75 size exclusion column, in kinase buffer (50 mM Tris pH7.5, 150 mM NaCl, 20% glycerol). His-MBP-PonA1 (cytoplasmic domain) was expressed and purified from E. coli using Ni-NTA beads (Novagen) according to the protocol recommended by the manufacturer. Protein levels were measured by A280, and kinase reactions were started by mixing 1 μg of kinase, 10 μg of his-MBP-PonA1(cyto), 1mM MnCl2 and 1mM ATP. Reactions were incubated at room temperature for 1 hour and stopped by addition of Laemmli buffer. Samples were heated, separated by SDS-PAGE, and detected by western blot using α-phospho-threonine antibody (Cell Signaling Technology). Kinase reactions for mass spectrometry were performed the same way, using only his-MBP-PknB and his-MBP-PonA1(cyto). The his-MBP-PonA1 protein band was cut out of the polyacrylamide gel and in-gel trypsin digested. Samples were analyzed by liquid chromatography (LC)/MS/MS with an Agilent 6520 Accurate-Mass Quadrupole Time-of-Flight instrument. Peptides were separated on a POROSHELL 300SB-C18 (2.1 × 75 mm, 5 μm) at a 0.5 mL/min flow rate, by using a linear gradient of increasing acetonitrile in water. Spectrum Mill software (Agilent) was used to identify peptides.
Antibiotic treatment and determination of bacterial cell fitness
The antibacterial effects of teicoplanin (Sigma Aldrich, St. Louis, MO) and moenomycin A (Sigma Aldrich, St. Louis, MO) were determined by culturing exponentially growing M. tuberculosis or M. smegmatis cells in complete 7H9 in non-treated 96 well plates (Genesee Scientific, San Diego, CA) in the presence of serially diluted drug, in duplicate. Plates also contained duplicate untreated wells as controls. M. tuberculosis was grown to OD600 of 0.5 before culturing ± drug at a calculated starting OD600 of 0.006 for six days at 37°C with shaking. M. smegmatis was grown to OD600 of 0.5 before culturing ± drug at a calculated starting OD600 of 0.05 for 16 hours at 37°C with shaking. Bacterial growth was evaluated by adding 0.002% resazurin (Alamar Blue, Sigma Aldrich, St. Louis, MO) to each well. M. tuberculosis plates were incubated with resazurin for 18–24 hours. M. smegmatis plates were incubated with resazurin for 3–5 hours. Wells were scored as blue or purple indicating metabolically inactive cells and pink as metabolically active cells. The inhibitory effects of meropenem (Sigma Aldrich, St. Louis, MO) were determined by growing M. smegmatis to an OD600 of 0.5 before culturing ± drug at a calculated starting OD600 of 0.1. Cells were grown in complete 7H9 in 96 well honeycomb plates (Growth Curves USA, Piscataway, NJ) ± drug, in triplicate, at 37°C with shaking in a Bioscreen growth curve machine (Growth Curves USA, Piscataway, NJ). Optical density was measured by absorbance at 600nm every 30 minutes for 48 hours. The inhibitory effects of rifampicin (Sigma Aldrich, St. Louis, MO) were determined by growing M. tuberculosis to an OD600 of 0.5 before culturing ± drug at a calculated starting OD600 of 0.1. Cells were grown in complete 7H9 in 24 well untreated plates ± drug, in triplicate, at 37°C with shaking. Optical density was measured after six days, and growth was normalized to untreated control wells.
Optical density measurements
Population growth curves for M. smegmatis strains were performed in 96 well honeycomb plates (Growth Curves USA, Piscataway, NJ), in triplicate, at 37°C with shaking in a Bioscreen growth curve machine (Growth Curves USA, Piscataway, NJ). Cells were grown to an OD600 of 0.5 before beginning the growth curve at a calculated starting OD600 of 0.1. Cells were grown in complete 7H9 with the appropriate antibiotic selection. Optical density was measured by absorbance at 600nm every 30 minutes for 48 hours. Error bars are often too small to see. Population growth curves for M. tuberculosis strains were performed in 30 ml inkwells (Corning Life Sciences, Corning, NY), in triplicate, at 37°C with shaking. Cells were grown to an OD600 of 0.5 before beginning the growth curve at a calculated starting OD600 of 0.1. Cells were grown in complete 7H9 with the appropriate antibiotic selection. Optical density was measured daily by absorbance at 600nm.
Immunoblotting
Cells were pelleted, washed once in 1X PBS, and lysed by bead-beating. 6X Laemmli buffer was added to the whole cell lysate and then boiled for 10 minutes. Proteins were separated on a 12–14% Tris-Glycine SDS PAGE gels (BioRad, Hercules, CA), transferred to PVDF membrane (Pall Corp, Pensacola, FL), incubated with anti-FLAG antibodies (Sigma Aldrich, St. Louis, MO) at 1 : 15,000 dilution, washed thrice in 1X PBST, and incubated with a 1 : 1000 dilution of a secondary antibody conjugated to HRP (Pierce, Rockford, IL). Membranes were incubated with SuperSignal chemiluminescent reagent (Thermo Fisher Scientific, Rockford, IL) for 5 minutes in the dark and imaged on a FluorChem8900 (AlphaInnotech, Santa Clara, CA).
Data representation and statistical analysis
Prism 6.0 software (GraphPad Software, La Jolla, CA) was used to graph and analyze numerical data. Statistical tests in the Prism software were used to calculate significance of measurements as described in the figure legends. The doubling times in S2 Fig and S9 Fig were generated by fitting an exponential growth function (in Prism) to all datapoints between 5 and 13 hours of OD600 measurements.
Supporting Information
Zdroje
1. World Health Organization. Global tuberculosis report 2014. World Health Organization; 2014.
2. Kieser KJ, Rubin EJ. How sisters grow apart: mycobacterial growth and division. Nat Rev Micro. 2014;12 : 550–62.
3. Typas A, Banzhaf M, Gross CA, Vollmer W. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Micro.; 2012;10 : 123–36.
4. Hett EC, Chao MC, Rubin EJ. Interaction and Modulation of Two Antagonistic Cell Wall Enzymes of Mycobacteria. PLoS Pathog. 2010;6:e1001020. doi: 10.1371/journal.ppat.1001020 20686708
5. Chao MC, Kieser KJ, Minami S, Mavrici D, Aldridge BB, Fortune SM, Alber T, Rubin EJ. Protein Complexes and Proteolytic Activation of the Cell Wall Hydrolase RipA Regulate Septal Resolution in Mycobacteria. PLoS Pathog. 2013;9:e1003197. doi: 10.1371/journal.ppat.1003197 23468634
6. Prisic S, Dankwa S, Schwartz D, Chou MF, Locasale JW, Kang C-M, et al. Extensive phosphorylation with overlapping specificity by Mycobacterium tuberculosis serine/threonine protein kinases. Proc Natl Acad Sci USA. 2010;107 : 7521–6. doi: 10.1073/pnas.0913482107 20368441
7. Pashley CA, Parish T. Efficient switching of mycobacteriophage L5-based integrating plasmids in Mycobacterium tuberculosis. FEMS Microbiol Lett. 2003;229 : 211–5. 14680701
8. Zhang YJ, Ioerger TR, Huttenhower C, Long JE, Sassetti CM, Sacchettini JC, Rubin EJ. Global Assessment of Genomic Regions Required for Growth in Mycobacterium tuberculosis. PLoS Pathog. 2012;8:e1002946. doi: 10.1371/journal.ppat.1002946 23028335
9. Zhang YJ, Reddy MC, Ioerger TR, Rothchild AC, Dartois V, Schuster BM, et al. Tryptophan Biosynthesis Protects Mycobacteria from CD4 T-Cell-Mediated Killing. Cell.; 2013;155 : 1296–308. doi: 10.1016/j.cell.2013.10.045 24315099
10. Paradis-Bleau C, Markovski M, Uehara T, Lupoli TJ, Walker S, Kahne DE, et al. Lipoprotein Cofactors Located in the Outer Membrane Activate Bacterial Cell Wall Polymerases. Cell. 2013;143 : 1110–20.
11. McPherson DC, Popham DL. Peptidoglycan synthesis in the absence of class A penicillin-binding proteins in Bacillus subtilis. J Bacteriol. 2003;185 : 1423–31. 12562814
12. Dörr T, Möll A, Chao MC, Cava F, Lam H, Davis BM, et al. Differential requirement for PBP1a and PBP1b in in vivo and in vitro fitness of Vibrio cholerae. Infection and Immunity. 2014;82 : 2115–24. doi: 10.1128/IAI.00012-14 24614657
13. Valbuena N, Letek M, Ordóñez E, Ayala JA, Daniel RA, Gil JA, et al. Characterization of HMW-PBPs from the rod-shaped actinomycete Corynebacterium glutamicum: peptidoglycan synthesis in cells lacking actin-like cytoskeletal structures. Mol Microbiol. 2007;66 : 643–57. 17877698
14. Patru M-M, Pavelka MS. A Role for the Class A Penicillin-Binding Protein PonA2 in the Survival of Mycobacterium smegmatis under Conditions of Nonreplication. J Bacteriol. 2010;192 : 3043–54. doi: 10.1128/JB.00025-10 20400545
15. Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393 : 537–44. 9634230
16. Born P, Breukink E, Vollmer W. In vitro synthesis of cross-linked murein and its attachment to sacculi by PBP1A from Escherichia coli. J Biol Chem. 2006;281 : 26985–93. 16840781
17. Terrak M, Ghosh TK, van Heijenoort J, Van Beeumen J, Lampilas M, Aszodi J, et al. The catalytic, glycosyl transferase and acyl transferase modules of the cell wall peptidoglycan-polymerizing penicillin-binding protein 1b of Escherichia coli. Mol Microbiol. 1999;34 : 350–64. 10564478
18. Billman-Jacobe H, Haites RE, Coppel RL. Characterization of a Mycobacterium smegmatis mutant lacking penicillin binding protein 1. Antimicrob Agents Ch. 1999;43 : 3011–3.
19. Domenech P, Reed MB. Rapid and spontaneous loss of phthiocerol dimycocerosate (PDIM) from Mycobacterium tuberculosis grown in vitro: implications for virulence studies. Microbiology. 2009;155 : 3532–43. doi: 10.1099/mic.0.029199-0 19661177
20. Banzhaf M, van den Berg van Saparoea B, Terrak M, Fraipont C, Egan A, Philippe J, et al. Cooperativity of peptidoglycan synthases active in bacterial cell elongation. Mol Microbiol. 2012;85 : 179–94. doi: 10.1111/j.1365-2958.2012.08103.x 22606933
21. Zhao G, Meier TI, Kahl SD, Gee KR, Blaszczak LC (1999) BOCILLIN FL, a sensitive and commercially available reagent for detection of penicillin-binding proteins. Antimicrob Agents Chemother 43 : 1124–8. 10223924
22. Meisel U, Holtje J-V, Vollmer W. Overproduction of Inactive Variants of the Murein Synthase PBP1B Causes Lysis in Escherichia coli. J Bacteriol. 2003;185 : 5342–8. 12949085
23. Aldridge BB, Fernandez-Suarez M, Heller D, Ambravaneswaran V, Irimia D, Toner M, Fortune SM. Asymmetry and Aging of Mycobacterial Cells Lead to Variable Growth and Antibiotic Susceptibility. Science. 2012;335 : 100–4. doi: 10.1126/science.1216166 22174129
24. Meniche X, Otten R, Siegrist MS, Baer CE, Murphy KC, Bertozzi CR, Sassetti CM. Subpolar addition of new cell wall is directed by DivIVA in mycobacteria. Proc Natl Acad Sci USA. 2014;111:E3243–51. doi: 10.1073/pnas.1402158111 25049412
25. Nelson DE, Young KD. Penicillin binding protein 5 affects cell diameter, contour, and morphology of Escherichia coli. J Bacteriol. 2000;182 : 1714–21. 10692378
26. De Pedro MA, Young KD, Höltje J-V, Schwarz H. Branching of Escherichia coli cells arises from multiple sites of inert peptidoglycan. J Bacteriol. 2003;185 : 1147–52. 12562782
27. Schubert OT, Mouritsen J, Ludwig C, Röst HL, Rosenberger G, Arthur PK, et al. The Mtb Proteome Library: A Resource of Assays to Quantify the Complete Proteome of Mycobacterium tuberculosis. Cell Host Microbe.; 2013;13 : 602–12. doi: 10.1016/j.chom.2013.04.008 23684311
28. Krogh A, Larsson B, Heijne von G, Sonnhammer ELL. Predicting transmembrane protein topology with a hidden markov model: application to complete genomes. J Mol Biol. 2001;305 : 567–80. 11152613
29. Sauvage E, Kerff F, Terrak M, Ayala JA, Charlier P. The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol Rev. 2008;32 : 234–58. doi: 10.1111/j.1574-6976.2008.00105.x 18266856
30. Santi I, Dhar N, Bousbaine D, Wakamoto Y, McKinney JD. Single-cell dynamics of the chromosome replication and cell division cycles in mycobacteria. Nat Commun.; 2013;4 : 1–10.
31. Chauhan A, Madiraju MVVS, Fol M, Lofton H, Maloney E, Reynolds R, Rajagopalan M. Mycobacterium tuberculosis Cells Growing in Macrophages Are Filamentous and Deficient in FtsZ Rings. J Bacteriol. 2006;188 : 1856–65. 16484196
32. Farhat MR, Shapiro BJ, Kieser KJ, Sultana R, Jacobson KR, Victor TC, et al. Genomic analysis identifies targets of convergent positive selection in drug-resistant Mycobacterium tuberculosis. Nat Genet.; 2013;45 : 1183–9. doi: 10.1038/ng.2747 23995135
33. Binda E, Marinelli F, Marcone G. Old and New Glycopeptide Antibiotics: Action and Resistance. Antibiotics. 2014;3 : 572–94.
34. Lam H, Oh D-C, Cava F, Takacs CN, Clardy J, De Pedro MA, et al. D-amino acids govern stationary phase cell wall remodeling in bacteria. Science. 2009;325 : 1552–5. doi: 10.1126/science.1178123 19762646
35. Rismondo J, Möller L, Aldridge C, Gray J, Vollmer W, Halbedel S. Discrete and overlapping functions of peptidoglycan synthases in growth, cell division and virulence of Listeria monocytogenes. Mol Microbiol. 2015;95 : 332–51. doi: 10.1111/mmi.12873 25424554
36. Cho H, Uehara T, Bernhardt TG. Beta-Lactam Antibiotics Inducea Lethal Malfunctioning of the Bacterial Cell Wall Synthesis Machinery. Cell.; 2014;159 : 1300–11. doi: 10.1016/j.cell.2014.11.017 25480295
37. Lavollay M, Arthur M, Fourgeaud M, Dubost L, Marie A, Veziris N, et al. The Peptidoglycan of Stationary-Phase Mycobacterium tuberculosis Predominantly Contains Cross-Links Generated by L,D-Transpeptidation. J Bacteriol. 2008;190 : 4360–6. doi: 10.1128/JB.00239-08 18408028
38. Kumar P, Arora K, Lloyd JR, Lee IY, Nair V, Fischer E, et al. Meropenem inhibits D,D-carboxypeptidase activity in Mycobacterium tuberculosis. Mol Microbiol. 2012;86 : 367–81. doi: 10.1111/j.1365-2958.2012.08199.x 22906310
39. Schoonmaker MK, Bishai WR, Lamichhane G. Nonclassical transpeptidases of Mycobacterium tuberculosis alter cell size, morphology, cytosolic matrix, protein localization, virulence and resistance to β-lactams. J Bacteriol. 2014;196 : 1394–402. doi: 10.1128/JB.01396-13 24464457
40. Gee CL, Papavinasasundaram KG, Blair SR, Baer CE, Falick AM, King DS, et al. A Phosphorylated Pseudokinase Complex Controls Cell Wall Synthesis in Mycobacteria. Sci Signal. 2012;5:ra7–ra7. doi: 10.1126/scisignal.2002525 22275220
41. Parikh A, Verma SK, Khan S, Prakash B, Nandicoori VK. PknB-Mediated Phosphorylation of a Novel Substrate, N-Acetylglucosamine-1-Phosphate Uridyltransferase, Modulates Its Acetyltransferase Activity. J Mol Biol.; 2009;386 : 451–64. doi: 10.1016/j.jmb.2008.12.031 19121323
42. Ehrt S, Guo XV, Hickey CM, Ryou M, Monteleone M, Riley LW, et al. Controlling gene expression in mycobacteria with anhydrotetracycline and Tet repressor. Nucleic Acids Res. 2005;33:e21. 15687379
43. Van Kessel JC, Hatfull GF. Recombineering in Mycobacterium tuberculosis. Nat Meth. 2007;4 : 147–52.
44. Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, et al. Fiji: an open-source platform for biological-image analysis. Nat Meth.; 2012;9 : 676–82.
45. Layre E, Sweet L, Hong S, Madigan CA, Desjardins D, Young DC, et al. A comparative lipidomics platform for chemotaxonomic analysis of Mycobacterium tuberculosis. Chem Biol. 2011; 18 : 1537–49. doi: 10.1016/j.chembiol.2011.10.013 22195556
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Clearance of Pneumococcal Colonization in Infants Is Delayed through Altered Macrophage TraffickingČlánek An Model of Latency and Reactivation of Varicella Zoster Virus in Human Stem Cell-Derived NeuronsČlánek Protective mAbs and Cross-Reactive mAbs Raised by Immunization with Engineered Marburg Virus GPsČlánek Specific Cell Targeting Therapy Bypasses Drug Resistance Mechanisms in African TrypanosomiasisČlánek Peptidoglycan Branched Stem Peptides Contribute to Virulence by Inhibiting Pneumolysin ReleaseČlánek HIV Latency Is Established Directly and Early in Both Resting and Activated Primary CD4 T CellsČlánek Sequence-Specific Fidelity Alterations Associated with West Nile Virus Attenuation in Mosquitoes
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2015 Číslo 6- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Introducing “Research Matters”
- Exploring Host–Pathogen Interactions through Biological Control
- Analysis of Bottlenecks in Experimental Models of Infection
- Expected and Unexpected Features of the Newly Discovered Bat Influenza A-like Viruses
- Clearance of Pneumococcal Colonization in Infants Is Delayed through Altered Macrophage Trafficking
- Recombinant Murine Gamma Herpesvirus 68 Carrying KSHV G Protein-Coupled Receptor Induces Angiogenic Lesions in Mice
- TRIM30α Is a Negative-Feedback Regulator of the Intracellular DNA and DNA Virus-Triggered Response by Targeting STING
- Targeting Human Transmission Biology for Malaria Elimination
- Two Cdc2 Kinase Genes with Distinct Functions in Vegetative and Infectious Hyphae in
- An Model of Latency and Reactivation of Varicella Zoster Virus in Human Stem Cell-Derived Neurons
- Protective mAbs and Cross-Reactive mAbs Raised by Immunization with Engineered Marburg Virus GPs
- Virulence Factors of Induce Both the Unfolded Protein and Integrated Stress Responses in Airway Epithelial Cells
- Peptide-MHC-I from Endogenous Antigen Outnumber Those from Exogenous Antigen, Irrespective of APC Phenotype or Activation
- Specific Cell Targeting Therapy Bypasses Drug Resistance Mechanisms in African Trypanosomiasis
- An Ultrasensitive Mechanism Regulates Influenza Virus-Induced Inflammation
- The Role of Human Transportation Networks in Mediating the Genetic Structure of Seasonal Influenza in the United States
- Host Delivery of Favorite Meals for Intracellular Pathogens
- Complement-Opsonized HIV-1 Overcomes Restriction in Dendritic Cells
- Inter-Seasonal Influenza is Characterized by Extended Virus Transmission and Persistence
- A Critical Role for CLSP2 in the Modulation of Antifungal Immune Response in Mosquitoes
- Twilight, a Novel Circadian-Regulated Gene, Integrates Phototropism with Nutrient and Redox Homeostasis during Fungal Development
- Surface-Associated Lipoproteins Link Virulence to Colitogenic Activity in IL-10-Deficient Mice Independent of Their Expression Levels
- Latent Membrane Protein LMP2A Impairs Recognition of EBV-Infected Cells by CD8+ T Cells
- Bank Vole Prion Protein As an Apparently Universal Substrate for RT-QuIC-Based Detection and Discrimination of Prion Strains
- Neuronal Subtype and Satellite Cell Tropism Are Determinants of Varicella-Zoster Virus Virulence in Human Dorsal Root Ganglia Xenografts
- Molecular Basis for the Selective Inhibition of Respiratory Syncytial Virus RNA Polymerase by 2'-Fluoro-4'-Chloromethyl-Cytidine Triphosphate
- Structure of the Virulence Factor, SidC Reveals a Unique PI(4)P-Specific Binding Domain Essential for Its Targeting to the Bacterial Phagosome
- Activated Brain Endothelial Cells Cross-Present Malaria Antigen
- Fungal Morphology, Iron Homeostasis, and Lipid Metabolism Regulated by a GATA Transcription Factor in
- Peptidoglycan Branched Stem Peptides Contribute to Virulence by Inhibiting Pneumolysin Release
- A Macrophage Subversion Factor Is Shared by Intracellular and Extracellular Pathogens
- A Novel AT-Rich DNA Recognition Mechanism for Bacterial Xenogeneic Silencer MvaT
- Reovirus FAST Proteins Drive Pore Formation and Syncytiogenesis Using a Novel Helix-Loop-Helix Fusion-Inducing Lipid Packing Sensor
- The Role of ExoS in Dissemination of during Pneumonia
- IRF-5-Mediated Inflammation Limits CD8 T Cell Expansion by Inducing HIF-1α and Impairing Dendritic Cell Functions during Infection
- Discordant Impact of HLA on Viral Replicative Capacity and Disease Progression in Pediatric and Adult HIV Infection
- Crystal Structure of USP7 Ubiquitin-like Domains with an ICP0 Peptide Reveals a Novel Mechanism Used by Viral and Cellular Proteins to Target USP7
- HIV Latency Is Established Directly and Early in Both Resting and Activated Primary CD4 T Cells
- HPV16 Down-Regulates the Insulin-Like Growth Factor Binding Protein 2 to Promote Epithelial Invasion in Organotypic Cultures
- The νSaα Specific Lipoprotein Like Cluster () of . USA300 Contributes to Immune Stimulation and Invasion in Human Cells
- RSV-Induced H3K4 Demethylase KDM5B Leads to Regulation of Dendritic Cell-Derived Innate Cytokines and Exacerbates Pathogenesis
- Leukocidin A/B (LukAB) Kills Human Monocytes via Host NLRP3 and ASC when Extracellular, but Not Intracellular
- Border Patrol Gone Awry: Lung NKT Cell Activation by Exacerbates Tularemia-Like Disease
- The Curious Road from Basic Pathogen Research to Clinical Translation
- From Cell and Organismal Biology to Drugs
- Adenovirus Tales: From the Cell Surface to the Nuclear Pore Complex
- A 21st Century Perspective of Poliovirus Replication
- Is Development of a Vaccine against Feasible?
- Waterborne Viruses: A Barrier to Safe Drinking Water
- Battling Phages: How Bacteria Defend against Viral Attack
- Archaea in and on the Human Body: Health Implications and Future Directions
- Degradation of Human PDZ-Proteins by Human Alphapapillomaviruses Represents an Evolutionary Adaptation to a Novel Cellular Niche
- Natural Variants of the KPC-2 Carbapenemase have Evolved Increased Catalytic Efficiency for Ceftazidime Hydrolysis at the Cost of Enzyme Stability
- Potent Cell-Intrinsic Immune Responses in Dendritic Cells Facilitate HIV-1-Specific T Cell Immunity in HIV-1 Elite Controllers
- The Mammalian Cell Cycle Regulates Parvovirus Nuclear Capsid Assembly
- Host Reticulocytes Provide Metabolic Reservoirs That Can Be Exploited by Malaria Parasites
- The Proteome of the Isolated Containing Vacuole Reveals a Complex Trafficking Platform Enriched for Retromer Components
- NK-, NKT- and CD8-Derived IFNγ Drives Myeloid Cell Activation and Erythrophagocytosis, Resulting in Trypanosomosis-Associated Acute Anemia
- Successes and Challenges on the Road to Cure Hepatitis C
- BRCA1 Regulates IFI16 Mediated Nuclear Innate Sensing of Herpes Viral DNA and Subsequent Induction of the Innate Inflammasome and Interferon-β Responses
- A Structural and Functional Comparison Between Infectious and Non-Infectious Autocatalytic Recombinant PrP Conformers
- Phosphorylation of the Peptidoglycan Synthase PonA1 Governs the Rate of Polar Elongation in Mycobacteria
- Human Immunodeficiency Virus Type 1 Nef Inhibits Autophagy through Transcription Factor EB Sequestration
- Sequence-Specific Fidelity Alterations Associated with West Nile Virus Attenuation in Mosquitoes
- EBV BART MicroRNAs Target Multiple Pro-apoptotic Cellular Genes to Promote Epithelial Cell Survival
- Single-Cell and Single-Cycle Analysis of HIV-1 Replication
- TRIM32 Senses and Restricts Influenza A Virus by Ubiquitination of PB1 Polymerase
- The Herpes Simplex Virus Protein pUL31 Escorts Nucleocapsids to Sites of Nuclear Egress, a Process Coordinated by Its N-Terminal Domain
- Host Transcriptional Response to Influenza and Other Acute Respiratory Viral Infections – A Prospective Cohort Study
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- HIV Latency Is Established Directly and Early in Both Resting and Activated Primary CD4 T Cells
- Battling Phages: How Bacteria Defend against Viral Attack
- A 21st Century Perspective of Poliovirus Replication
- Adenovirus Tales: From the Cell Surface to the Nuclear Pore Complex
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



