-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaFungal Morphology, Iron Homeostasis, and Lipid Metabolism Regulated by a GATA Transcription Factor in
Blastomyces dermatitidis belongs to a group of human pathogenic fungi that convert between two forms, mold and yeast, in response to temperature. Growth as yeast (37°C) in tissue facilitates immune evasion, whereas growth as mold (22°C) promotes environmental survival, sexual reproduction, and generation of transmissible spores. Despite the importance of dimorphism, how fungi regulate temperature adaptation is poorly understood. We identified SREB, a transcription factor that regulates disparate processes including dimorphism. SREB null mutants, which lack SREB, fail to fully complete the conversion to mold at 22°C. The goal of our research was to characterize how SREB regulates transcription during the switch to mold. Gene expression microarray along with chromatin binding and biochemical analyses indicated that SREB affected several processes including iron homeostasis, lipid biosynthesis, and lipid droplet formation. In vivo, SREB directly bound and regulated genes involved with iron uptake, lipid biosynthesis, and transcription. Functional analysis suggested that lipid metabolism may influence filamentous growth at 22°C. In addition, SREB interacted with another transcription factor, HAPX.
Published in the journal: . PLoS Pathog 11(6): e32767. doi:10.1371/journal.ppat.1004959
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1004959Summary
Blastomyces dermatitidis belongs to a group of human pathogenic fungi that convert between two forms, mold and yeast, in response to temperature. Growth as yeast (37°C) in tissue facilitates immune evasion, whereas growth as mold (22°C) promotes environmental survival, sexual reproduction, and generation of transmissible spores. Despite the importance of dimorphism, how fungi regulate temperature adaptation is poorly understood. We identified SREB, a transcription factor that regulates disparate processes including dimorphism. SREB null mutants, which lack SREB, fail to fully complete the conversion to mold at 22°C. The goal of our research was to characterize how SREB regulates transcription during the switch to mold. Gene expression microarray along with chromatin binding and biochemical analyses indicated that SREB affected several processes including iron homeostasis, lipid biosynthesis, and lipid droplet formation. In vivo, SREB directly bound and regulated genes involved with iron uptake, lipid biosynthesis, and transcription. Functional analysis suggested that lipid metabolism may influence filamentous growth at 22°C. In addition, SREB interacted with another transcription factor, HAPX.
Introduction
Blastomyces dermatitidis belongs to a group of medically important ascomycetes that adapt to shifts in temperature by undergoing a morphologic switch known as the phase transition [1]. In soil (22°C), these pathogens grow as filamentous mold, which produce infectious conidia. Following soil disruption, aerosolized conidia and mold fragments inhaled into the lungs of a mammalian host (37°C) convert into pathogenic yeast [1,2]. In the yeast form, B. dermatitidis is able to evade host immune defenses to cause pneumonia and disseminate to other organs such as the bone or brain [3–5]. This adaptive response, which is essential for virulence [2], is conserved in Histoplasma capsulatum, Coccidioides immitis, Coccidioides posadasii, Paracoccidioides brasiliensis, Penicillium marneffei, and Sporothrix schenckii [1]. The transition in the opposite direction, yeast to mold, is postulated to facilitate survival outside the mammalian host, sexual reproduction by mating, and geographic dispersion through production of conidia [6].
The phase transition is a complex process that involves alteration of transcription, metabolism, lipid content, and cell wall carbohydrate composition [7–13]. In B. dermatitidis and H. capsulatum, DRK1 (dimorphism-regulating kinase-1) and RYP1-4 (required for yeast phase) promote the temperature-dependent conversion from mold to yeast, respectively [2,7,14,15]. The transition to yeast at 37°C is essential for virulence; deletion of DRK1 renders B. dermatitidis and H. capsulatum avirulent during experimental murine pulmonary infection [2]. In Penicillium marneffei, HGRA (hyphal growth regulator) facilitates the conversion of yeast to hyphae at 25°C and TUPA promotes maintenance of mycelial phase morphology [16,17]. In B. dermatitidis, we discovered SREB (siderophore biosynthesis repressor in Blastomyces), which encodes a GATA transcription factor that affects the temperature-dependent morphologic switch and iron homeostasis [18]. SREB null (SREB∆) and insertional mutants fail to fully complete the conversion from yeast to sporulating mold after a drop in temperature from 37°C to 22°C, and under iron-replete conditions cannot repress the biosynthesis of iron-gathering siderophores [18]. The regulatory role of SREB on the phase transition appears conserved in the dimorphic fungi; knockdown of an SREB homolog in H. capsulatum led to failure of yeast cells to fully convert to mold at ambient temperature [19]. Deletion of VMA1, which encodes a catalytic subunit in a vacuolar ATPase involved with iron homeostasis and virulence, also impairs the conversion of H. capsulatum yeast to mold [20]. The kinetics of the temperature-dependent switch from yeast to mold is influenced by N-acetylglucosamine (GlcNAc), which is mediated by NGT1 and NGT2 transmembrane transporters [21]. Exogenous GlcNAc accelerated the conversion of B. dermatitidis and H. capsulatum yeast to mold at room temperature [21]. Although several genes have been identified to govern the morphologic switch, how these genes coordinate proper adaptation to temperature, which is reflected by growth as yeast at 37°C or mold at 22–25°C remains poorly understood.
GATA transcription factors bind DNA via zinc finger motifs to induce or repress transcription in response to environmental stimuli. In fungi, they regulate adaptive responses to light, temperature, nitrogen, and iron [22–28]. Moreover, GATA transcription factors are capable of governing disparate functions. ASH1 in Saccharomyces cerevisiae inhibits mating type switching and under nitrogen poor conditions induces pseudohyphal growth [22,23]. Cryptococcus neoformans CIR1, which is a homolog of SREB, regulates genes important for thermotolerance at 37°C, capsule formation, iron uptake, and mating [24,25]
The goals of this study were to characterize the SREB regulon on a genome-wide scale using gene expression microarrays, identify a subset of genes directly bound and regulated by SREB in vivo, and functionally test the impact of SREB-regulated genes on the phase transition. Because the morphologic defect in SREB∆ occurred within 24–48 hours of a drop in temperature, we focused our attention to the early part of the phase transition where B. dermatitidis yeast adapt to 22°C and begin the conversion to mold. The data generated from our approach indicated that the GATA transcription factor encoded by SREB affects transcription at 37°C and 22°C, and binds to genes involved with diverse (iron and non-iron) processes. In addition to functioning as a major regulator of iron assimilation, SREB affected the biosynthesis of lipids at 37°C and 22°C, and lipid droplet formation at 22°C. Similar to filamentous fungi such as Aspergillus fumigatus, SREB was in a regulatory circuit with the bZIP transcription factor HAPX. Deletion of SREB or overexpression of HAPX resulted in a defect in the morphologic switch at 22°C.
Results
The morphologic defect in SREB∆ occurs early in the phase transition
To investigate the kinetics of the phase transition defect in SREB∆, we compared it to an isogenic wild-type (WT) control at 37°C and at 6, 24, and 48-hrs following a drop in temperature to 22°C (Fig 1A–1C). At 37°C, WT and SREB∆ grew as budding yeast (Fig 1A and 1B). After 6-hrs at 22°C, WT and SREB∆ continued to have a yeast morphology with 22% and 4% cells exhibiting early germ tube development, respectively (Fig 1A and 1B). At 24 and 48-hrs at 22°C, the germ tubes of WT cells had elongated to become hyphae (Fig 1A and 1B). In contrast, sharp morphologic differences between SREB∆ and WT became apparent at 24 and 48-hrs at 22°C with SREB∆ demonstrating a delay in germ tube formation and stunted, misshapen germ tubes and hyphae (Fig 1A and 1B). Deletion of SREB did not affect cell viability (Fig 1C).
Fig. 1. Cell morphology during the early stages following a drop in temperature. 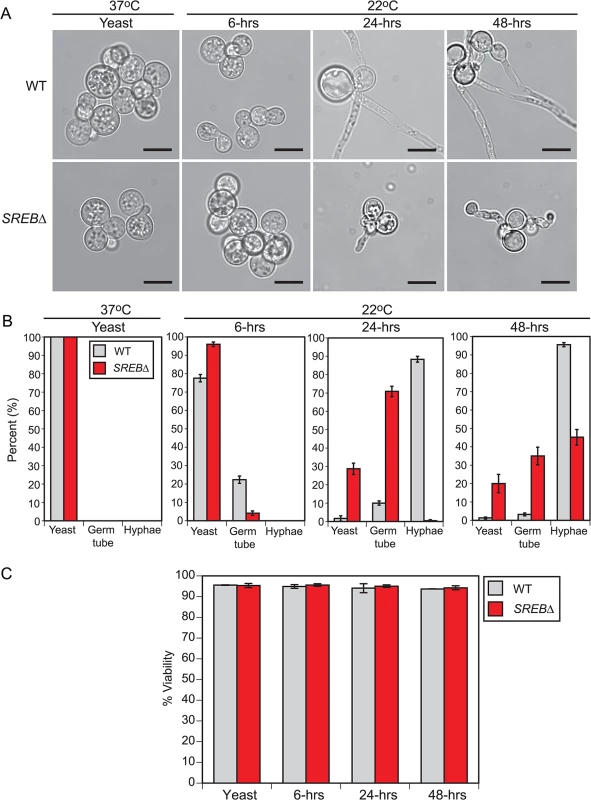
(A) Morphology of wild-type (WT) and SREB∆ at 37°C and at 6, 24, and 48-hrs after a drop in temperature at 22°C. When compared to WT, SREB∆ cells exhibited a delay in the filamentous growth (germ tube and hyphal formation). At 22°C, SREB∆ filaments were abnormal in morphology. Scale bar equals 10 μm. (B) Percentage of WT and SREB∆ cells with yeast morphology, germ tube development, and hyphal growth at 37°C and 22°C. Results were averaged from 2 independent experiments (> 200 cells counted in duplicate). (C) Percent of WT and SREB∆ cells that are viable at 37°C and 22°C. Results were averaged from 2 independent experiments (> 200 cells counted in duplicate) On the basis of the time-course data, we postulated that transcriptional alterations responsible for the morphologic defect in SREB∆ occurred early in the phase transition (conversion of wild-type B. dermatitidis to sporulating mold requires 10–14 days incubation at 22°C). To optimize detection of transcriptional events associated with the phase transition (e.g., germ tube formation and elongation), we focused on the first 48-hrs at 22°C, which was when the morphologic defect in SREB∆ first became apparent. Microarray analyses of later time points (e.g., 7 or 14 days at 22°C) were not performed because the transcriptional differences between SREB∆ and WT would reflect differences in filamentous growth rather than the morphologic switch. We harvested RNA from 3 biological replicates for WT and SREB∆ at 37°C, and 6, 24, and 48-hrs following a drop in temperature to 22°C. The 37°C time point served a baseline, whereas the 22°C time points provided insight into transcription at the beginning of germ tube formation (6-hrs) and during the transition from germ tubes to hyphae (24-hrs and 48-hrs) (Fig 1A and 1B). All cultures were grown under iron-replete conditions (10 μM FeSO4) to optimize transcription of SREB in the isogenic WT isolate. Iron induces transcription of SREB and is required for the binding of SREB homologs, such as H. capsulatum SRE1, to DNA [18,29].
LIMMA and EBarrays statistical packages were used to identify differentially expressed genes in SREB∆ versus WT isolates at each time point, [30,31]. Two statistical methods were used because each makes slightly different assumptions, and we wanted to ensure that our results were robust to the statistical method used. Both methods identified differentially expressed (DE) genes at all time points (Table 1 and S1 Table). All DE genes identified by EBarrays were also DE by LIMMA and, consequently, to be conservative, subsequent analyses herein include genes that were classified as DE by EBarrays (S1 Table). The majority of DE genes in SREB∆ exhibited a 1.5-fold or greater change in transcript abundance when controlling the false discovery rate at 5% (Table 1). In the yeast phase (37°C), the ratio of DE genes with increased to decreased transcription was 1.56 : 1. At 22°C, this ratio was nearly equal (1.16 : 1 at 6-hrs, 0.87 : 1 at 24-hrs; 1.02 : 1 at 48-hrs). At 22°C, the majority of DE genes (570 / 1109 for 6-hrs, 1196 / 1772 for 24-hrs, 1345 / 1910 for 48-hrs) did not overlap with DE genes found at baseline (37°C). Thus, differential expression of these genes was unique to the 22°C time points. Collectively, these data indicate that deletion of SREB affected transcription at 37°C and 22°C.
Tab. 1. Differentially expressed (DE) genes in SREB∆ versus wild type at 37°C and 22°C. 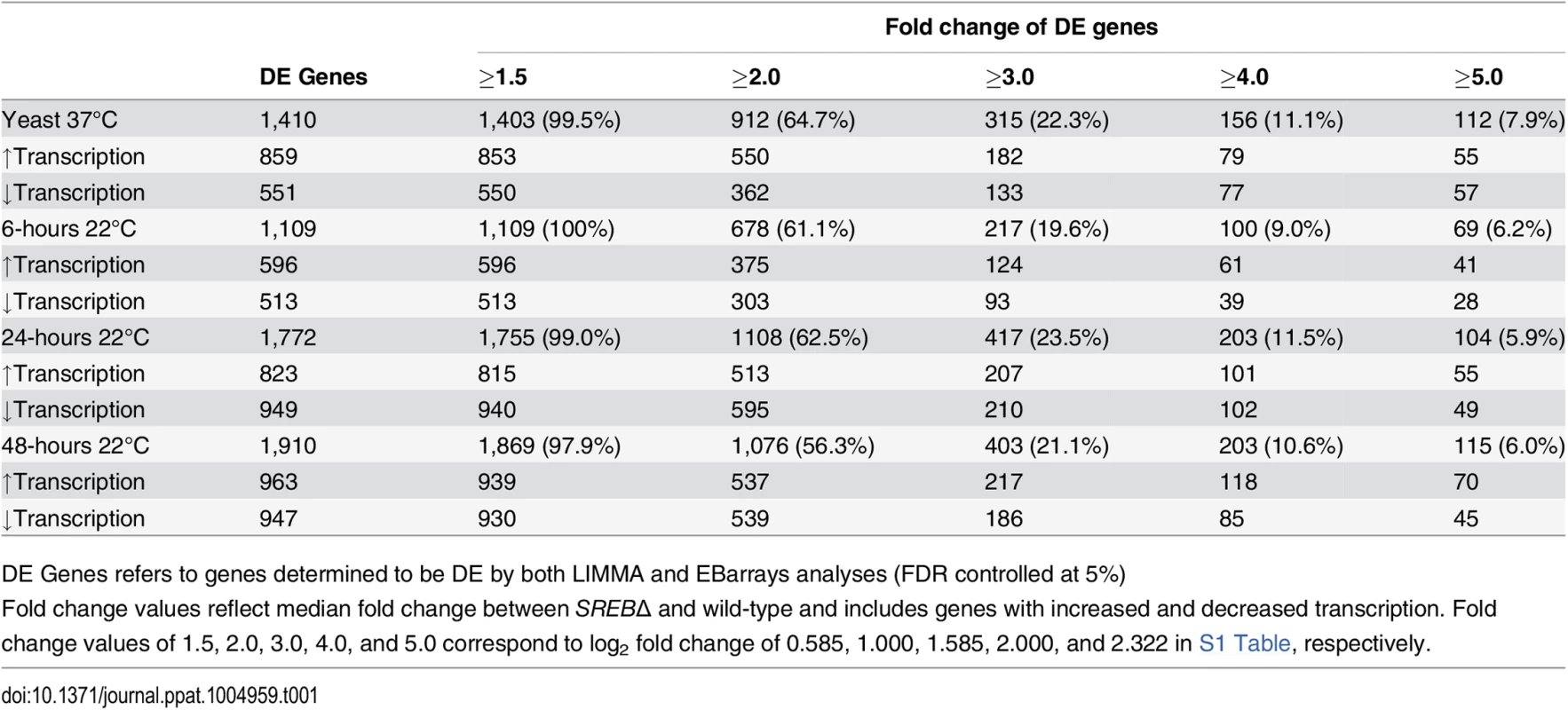
DE Genes refers to genes determined to be DE by both LIMMA and EBarrays analyses (FDR controlled at 5%) Deletion of SREB affected the transcription of genes involved with diverse processes
To facilitate functional classification of DE genes, gene ontology (GO) terms were assigned to proteins encoded by the B. dermatitidis genome using InterProScan. GO analysis revealed DE genes in SREB∆ were enriched for 17 GO terms and the number of enriched GO terms increased across the time course, which likely reflected the sharp morphologic differences between SREB∆ and WT at 22°C (Table 2; S2 Table, and S3 Table). Catalytic activity (GO:0003824), metabolic process (GO:0008152), oxidoreductase activity (GO:0016491), and oxidation-reduction process (GO:0055114) contained the largest number of DE genes that were enriched across all 22°C time points (Table 2, S3 Table, and S1 Fig). To assess potential function for genes within GO categories, we integrated GO term, TBLASTN (PubMed and Saccharomyces cerevisiae databases), and PFAM analyses (S2 Table). The predicted functions these genes were diverse (S2 Table) and many DE genes were shared among the enriched gene ontologies (S2 Table and S4 Table). Integrative analysis suggested several biological themes including iron ion binding, lipid biosynthesis, and amino acid metabolism.
Tab. 2. Gene ontology (GO) enrichment analysis. 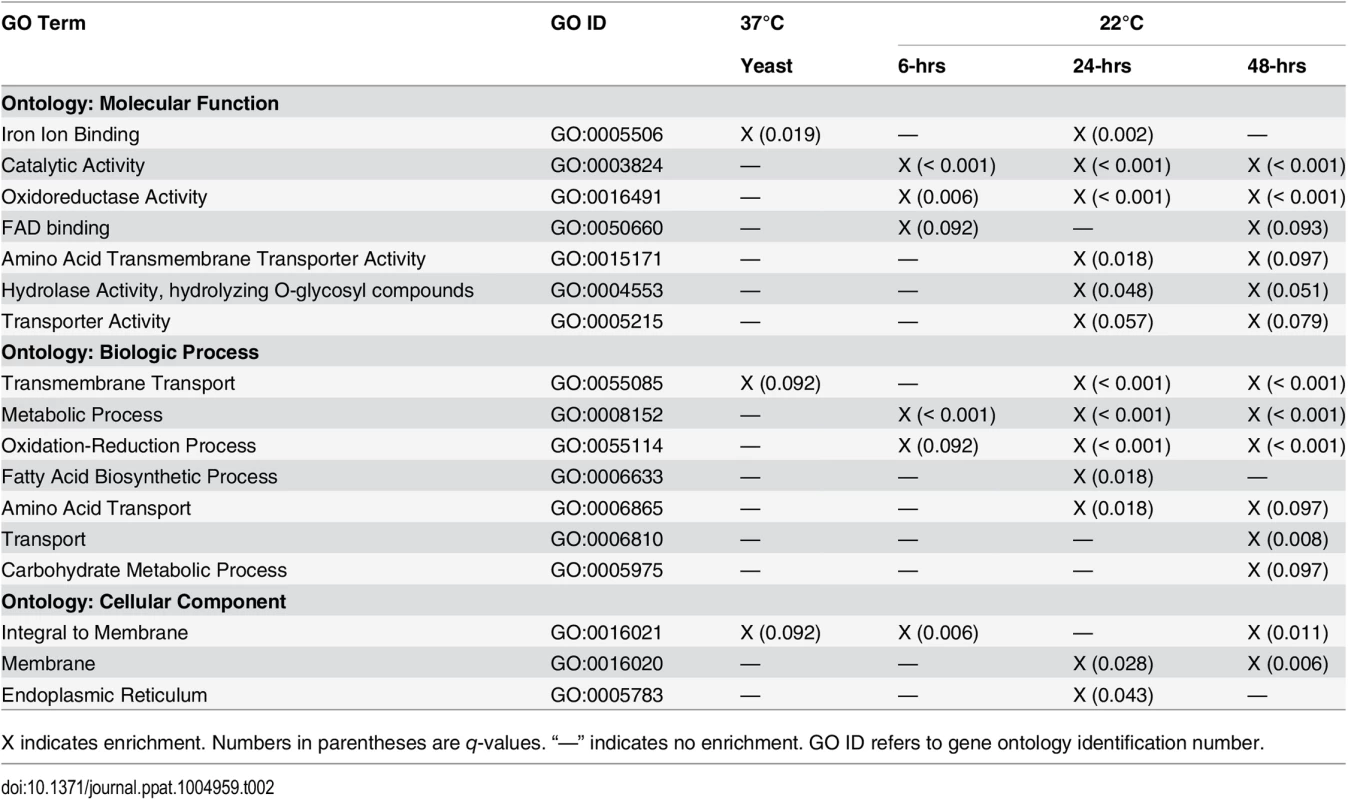
X indicates enrichment. Numbers in parentheses are q-values. “—” indicates no enrichment. GO ID refers to gene ontology identification number. Differentially expressed genes predicted to encode iron-binding proteins included those with electron-carrier activity and stress response. The majority of DE genes encoding proteins with electron carrier functions contained cytochrome (19 / 48 genes, 39.6%), acyl-CoA dehydrogenase/oxidase (11 / 48, 22.3%), and ferric reductase (4 / 48 genes, 8.3%) domains. Cytochrome and acyl-CoA dehydrogenase/oxidase encoding genes exhibited reduced transcription (22 / 30 genes, 73.3%) and known functions were limited to sterol biosynthesis (BDBG_01678, BDBG_03201, BDBG_04743, BDBG_06540) and beta-oxidation (BDBG_02355, BDBG_00463), respectively. Transcription of cytochromes involved with oxidative phosphorylation were unaffected by deletion of SREB∆ (BDBG_01291, BDBG_09317, BDBG_06453, BDBG_08893, BDBG_05852, BDBG_07205, BDBG_04875, 08630, BDBG_03418, BDBG_04645, BDBG_05869, BDBG_01464, BDBG_05537, BDBG_06649). Genes encoding proteins with ferric reductase domains had increased transcription in SREB∆ (3 / 4 genes, 75%) including an NADPH oxidase (BDBG_06745), a metalloreductase (BDBG_07462), and a FRE family reductase (BDBG_09322). Genes involved with stress response included those encoding peroxidases and catalases. Four peroxidases were DE including 3 with unknown function and 1 with a haem peroxidase domain. Similar to H. capsulatum, B. dermatitidis encodes 3 catalase isoforms (A, B, and P). In SREB∆, these isoenzymes (BDBG_05307, BDBG_08680, BDBG_07107) are DE at select time points and not throughout the time course (e.g., 0.75 and 1.38 log2 increase for catalase A at 24 and 48-hrs 22°C; 1.06 and 1.24 log2 increase for catalase B at 37°C and 48-hrs 22°C; log2 1.21 and 0.62 decrease for catalase P at 24 and 48-hrs 22°C). Similarly, superoxide dismutase was DE only at 48-hrs 22°C (BDBG_07234; log2 0.68 decrease at 48-hrs).
Deletion of SREB decreased transcription of genes in the triacylglycerol (TAG) and ergosterol biosynthetic pathways, but had limited impact on genes involved with phospholipid biosynthesis.
In SREB∆, 57.1% (8 / 14) of genes in the triacylglycerol (TAG) biosynthetic pathway and 66.7% (14 / 21) of genes in the ergosterol biosynthetic pathways were DE. The majority of these DE genes (7 of 8 for glycerolipid pathway, 12 of 14 for ergosterol pathway) had reduced transcript abundance in SREB∆ versus WT. In contrast, deletion of SREB had limited affect on phosopholipid biosynthesis in the Kennedy and cytidine diphosphate diacylglycerol (CDP-DAG) pathways (S2 Fig). In the Kennedy pathway, two choline transporters and two biosynthetic genes for phosphatidylcholine (PC) were DE in SREB∆, but the impact on transcription was divergent (S2 Fig). Transcription of genes for phosphadidylserine (PS), phosphatidylethanolamine (PE), and phosphatidylinositol (PI) biosynthesis were not altered. Moreover, homologs of S. cerevisiae OPI3 and PIS1, which are involved with conversion of PE to PC had similar transcript abundance in SREB∆ and WT (S2 Fig). The only genes with altered transcription in the CPD-DAG pathway were involved in the biosynthesis of cardiolipin. These two genes, which were homologs of S. cerevisiae CRD1 (BDBG_07720) and PGS1 (BDBG_03699) had 0.77 and 0.95 log2 decrease in transcription in SREB∆ (versus WT) at 24 and 48-hrs 22°C, respectively (S2 Fig). Homologs for S. cerevisiae INO2, INO4, and OPI1, which are transcription factors that regulate phospholipid biosynthesis [32], were not identified in the B. dermatitidis genome by tBLASTn analysis.
SREB∆ cells had reduced transcription for genes in 16 of 21 amino acid biosynthetic pathways at 24 and 48-hrs 22°C (S3 Fig). For example, nearly all genes (12 / 14, 85.7%) involved with branched chain amino acid biosynthesis (i.e., valine, leucine, isoleucine) had decreased transcript abundance (S3 Fig). Biosynthetic genes for methionine, glutamate, and serine were not altered by deletion of SREB∆ at 24 or 48-hrs 22°C, whereas cysteine and asparagine biosynthetic genes had increased transcription (S3 Fig). At 37°C, deletion of SREB had limited effect on biosynthetic genes; however, an amino acid acetyltransferase (BDBG_00643) involved with first step in ornithine biosynthesis had a 1.1 log2 increase (S3 Fig); ornithine serves as a substrate for siderophore biosynthesis.
Transcript abundance for DE genes encoding amino acid transporters was also altered at 24-hrs 22°C (14 / 20 genes) and 48-hrs 22°C (13 / 20 genes). At these time points, transcript abundance for transporters was decreased for proline (BDBG_04825), increased for cationic amino acids (BDBG_03808, BDBG_08628) and γ-aminobutyrate (BDBG_03404), and heterogenous for methionine/cysteine (BDBG_00563, BDBG_03572, BDBG_00074) as well as for transporters with unknown function (BDBG_00367, BDBG_02461, BDBG_03769, BDBG_06532, BDBG_07277, BDBG_09387). In addition, 2 putative mitochondrial transporters, BDBG_04548, BDBG_05901, had decreased transcription. Of the predicted amino acid transporters, BDBG_08000, which is a homolog of S. cerevisiae GAP1 (general amino acid permease), had the largest change in transcript abundance with log2 1.76–2.65 increase at 22°C in SREB∆ (versus WT).
In SREB∆, DE genes involved with carbohydrate metabolism, including hydrolysis of O-glycosyl linkages, were enriched at 22°C (Table 2). Approximately 73% (33 / 45) of DE carbohydrate metabolic genes had hydrolase activity and nearly 65% (29 / 45) of these were involved with hydrolysis of O-glycosyl compounds with 72.4% (21 / 29) predicted to hydrolyze O-glycosyl linkages in chitin or glucan (Table 2 and S4 Table). In SREB∆, the majority of DE chitinases (70%; 7/10 genes) demonstrated increased expression, whereas most DE glucanases (64%; 7/11 genes) were down-regulated (S1 Table and S2 Table). These genes were predicted to be involved with cell wall remodeling or maintenance. Deletion of SREB did not alter the transcription of genes involved with the biosynthesis of beta-(1,3)-glucan, beta-(1,6)-glucan, and alpha-(1,3)-glucan including FKS1 (BDBG_00352), RHO1 (BDBG_02597), KRE6 (BDBG_05692), AGS1 (BDBG_00595), and AMY1 (BDBG_07493). Similarly, deletion of SREB had limited impact on chitin biosynthesis, affecting 3 of 8 chitin synthases (1.03 log2 decrease at 37°C for BDBG_03232, a homolog of A. fumigatus csmB; 0.84 log2 decrease at 24-hrs 22°C for BDBG_00955, a homolog of A. fumigatus chsD; and 1.12 log2 increase at 48-hrs 22°C for BDBG_07172, a homolog of A. fumigatus chsB).
Although GO analysis illuminated some common themes, detailed functional information for many DE genes was limited (S2 Table). For example, the majority of short chain dehydrogenases/reductases (SDRs) had unknown function (36 / 47 for catalytic activity; 37 / 49 for metabolic process ontology; 29 / 35 for oxidation-reduction process; 31 / 36 for oxidoreductase activity) (S2 Table). Similarly, functions for most medium chain dehydrogenases (MDRs), lyases, hydrolases, acyl carrier proteins, and acetyltransferases were unknown (S2 Table). Notable exceptions included an acyl carrier protein (BDBG_00048) and a GNAT acetyltransferase (BDBG_00051) involved with siderophore biosynthesis, and isocitrate lyase (BDBG_04918). B. dermatitidis encodes 2 isoforms of isocitrate lyase (non-mitochondrial—BDBG_04918, mitochondrial—BDBG_05333) and 1 malate synthase (BDBG_05338). At 24 - and 48-hrs 22°C, non-mitrochondrial isocitrate lyase (BDBG_04918) and malate synthase (BDBG_05338) had > 1 log2 reduction in transcript compared to WT.
Weighted gene co-expression network analysis (WGCNA)
WGCNA was performed to identify groups of co-expressed genes. All genes across all time points were included in the analysis. We identified 30 color-coded modules including 11 modules that were enriched for GO terms (Table 3). In 4 of these 11 modules, the majority of genes were differentially expressed (53.5% – 85.0%; Table 3). Moreover, 3 modules were enriched for GO terms previously identified including metabolic process (midnight blue module), fatty acid biosynthesis / catalytic activity (green-yellow module), and transmembrane transport (blue module) (Tables 2 and 3 and Fig 2A–2C). Although these modules were enriched for specific GO terms, the predicted functions for DE genes within each module was diverse. Moreover, a substantial number of DE genes in each module had unknown function (Fig 2A–2D). In the midnight blue module, transcription of DE genes in SREB∆ was lower at baseline (37°C) and failed to properly increase across the 22°C time points when compared to WT (Fig 2A and S5 Table). In contrast, DE genes in SREB∆ exhibited a more precipitous decrease in transcription across the time course than WT in the green-yellow module (Fig 2B and S5 Table). For the blue module, the increase in transcription across the time points at 22°C was blunted for DE genes in SREB∆ versus WT (Fig 2C and S5 Table). Transcription of DE genes in SREB∆ in the turquoise module increased during the time course compared to WT (Fig 2D and S5 Table), which suggested these genes were derepressed. Collectively, this analysis indicated that deletion of SREB resulted in specific transcriptional changes at 37°C and 22°C.
Fig. 2. Weighted gene co-expression network analysis (WGCNA). 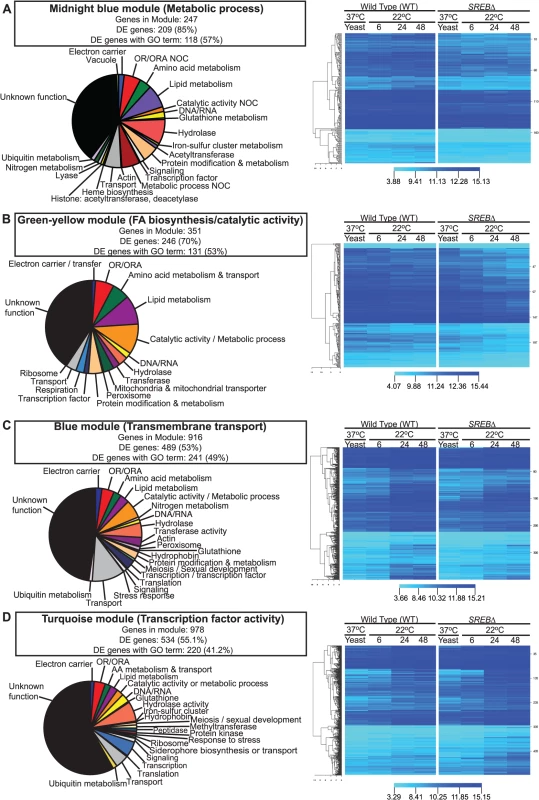
WGCNA identified 30 color-coded modules including 4 modules in which the majority of genes were differentially expressed and enriched for GO terms. (A) Midnight blue, (B) green-yellow, (C) blue, and (D) turquoise modules were enriched metabolic process, fatty acid biosynthesis / catalytic activity, transmembrane transport, and transcription factor activity GO terms, respectively. Putative gene function was assigned by integrating GO term, tBLASTn, and pFAM analyses. Fluorescent intensity values from microarray analysis for heat maps were clustered using Euclidean distance with average linkage. Heat map analysis demonstrated distinct transcriptional patterns between SREB∆ and WT for each module across the time course: (A) midnight blue–failure of transcript for genes in SREB∆ to increase; (B) green yellow–decrease in transcript for genes in SREB∆; (C) blue–increase in transcription was attenuated for genes in SREB∆; and (D) turquoise–increased transcription for genes in SREB∆. Genes for the heat maps are in Supplemental Table 5. Tab. 3. Weighted gene co-expression network analysis. 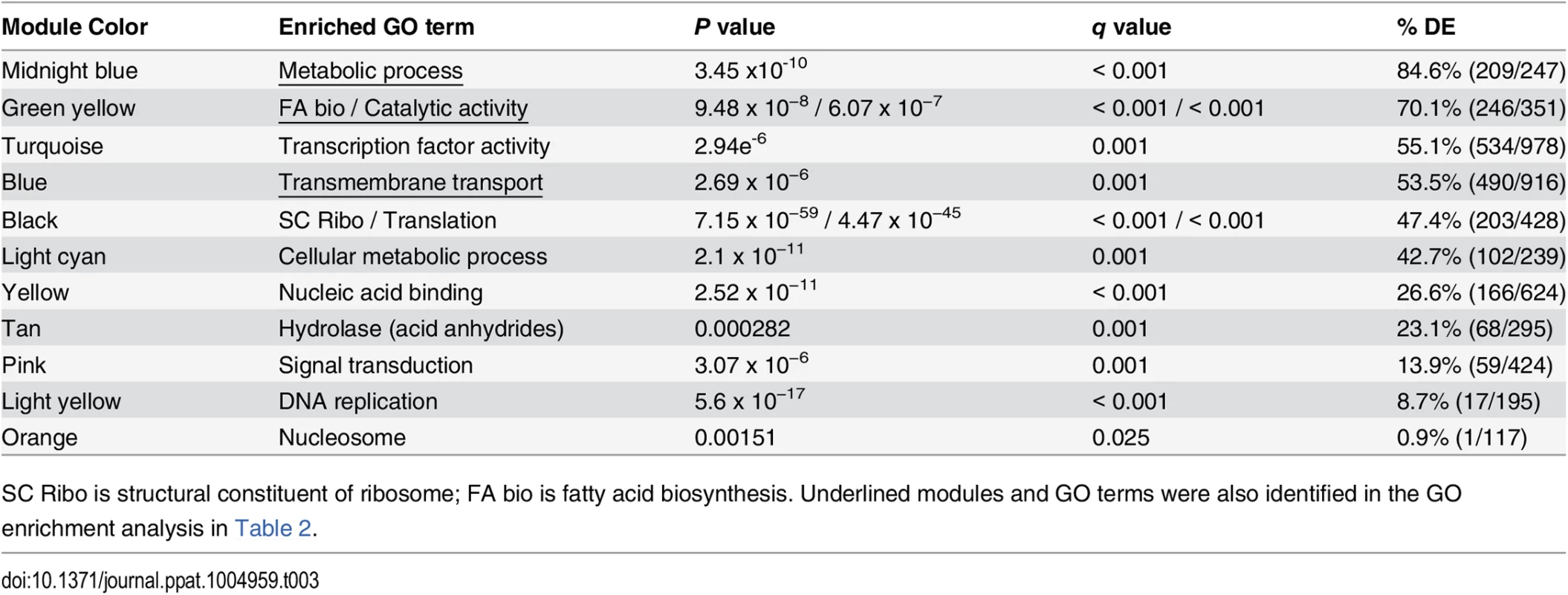
SC Ribo is structural constituent of ribosome; FA bio is fatty acid biosynthesis. Underlined modules and GO terms were also identified in the GO enrichment analysis in Table 2. The paucity of DE genes in modules enriched for nucleic acid binding, acid anhydride hydrolases, signal transduction, DNA replication, and nucleosome suggested that the deletion of SREB has limited affect on genes involved with these processes (Table 3).
SREB is a major regulator of genes involved with iron assimilation by siderophores, but not ferric reduction
The acquisition of iron from the host or the environment is important for growth, and fungi have developed several mechanisms for iron uptake [33]. The influence of iron on the growth of B. dermatitidis is unclear and one study concluded that iron failed to enhance growth of yeast during iron starvation; however, a high concentration of ferric iron was used [34]. To further characterize the effect of iron on B. dermatitidis, yeast were grown under iron-poor media or media containing 3, 5, and 10 μM FeCl3. B. dermatitidis yeast exhibited slow growth under iron-deplete conditions, whereas the addition of 3 μM FeCl3 improved growth (Fig 3A). Treatment of yeast with deferiprone (DFP), an iron chelator, impaired growth in a dose-dependent manner and the degree of growth inhibition was influenced by the concentration of exogenous iron (Fig 3B–3D). Collectively, these findings indicate that the acquisition of iron is important for the growth of B. dermatitidis.
Fig. 3. Growth of B. dermatitidis yeast is influenced by exogenous iron. 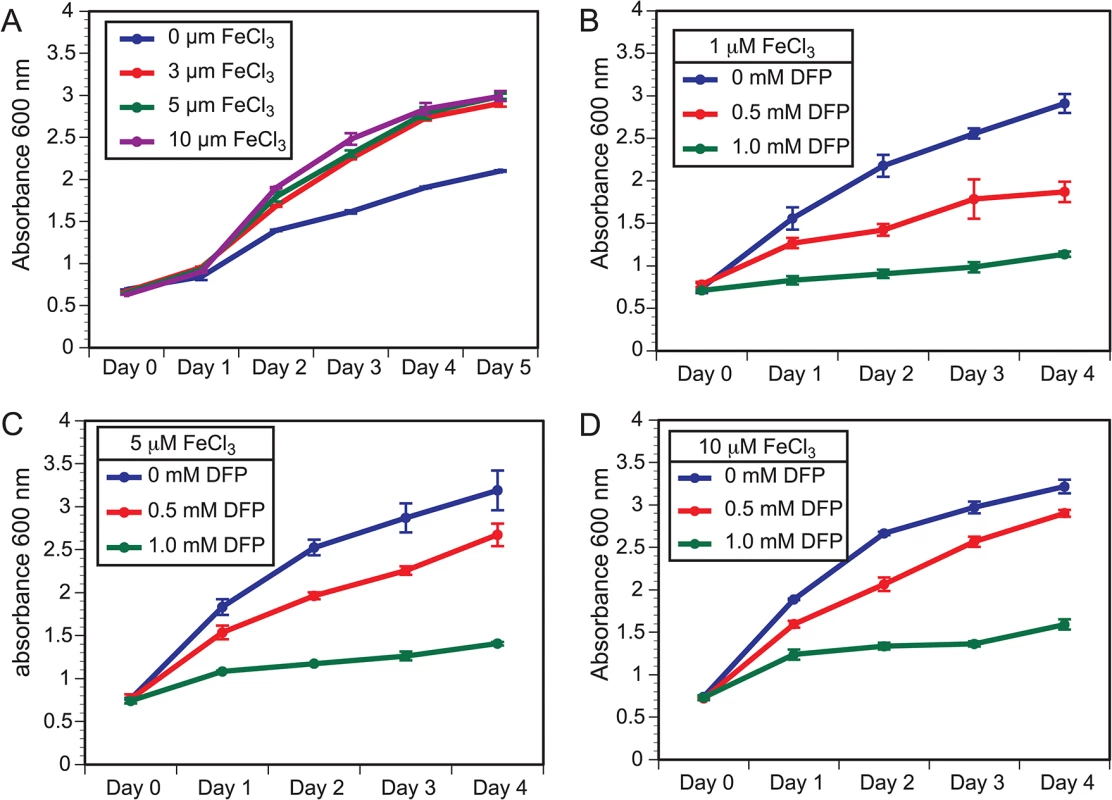
(A) B. dermatitidis yeast were grown in iron deplete and replete liquid HMM at 37°C. (B–D) B. dermatitidis yeast were grown in media supplemented with 1, 5, and 10 μM FeCl3 along with 0–1.0 mM deferiprone (DFP), an iron chelator. Gene expression microarray analysis indicated SREB functioned as a major regulator of iron assimilation. SREB∆ failed to repress genes involved with siderophore biosynthesis and transport under iron-replete conditions at 37°C and 22°C (Fig 4A–4C). Moreover, a siderophore biosynthetic gene cluster (SBGC) containing 10 genes across 31.295 kb was identified (Fig 4B). With the exception of a gene predicted to encode a conserved hypothetical protein with unknown function (BDBG_00049), all genes in the predicted SBGC were differentially expressed in SREB∆ when compared to a WT isolate (Fig 4A–4C). DELTA-BLAST analysis against the NCBI (National Center for Biotechnology Information) database predicted that genes in the SBGC were involved with the biosynthesis of siderophores (dimerum acid and coprogen) secreted into the extracellular environment. Quantitative real-time PCR confirmed derepression of DE genes in the SBGC under iron-replete conditions in SREB∆ (Fig 4C). Two genes located outside the SBGC, BDBG_09503, BDBG_08208, were predicted to biosynthesize ferricrocin, an intracellular storage siderophore. Transcript abundance for BDBG_09503, which encodes a siderophore biosynthetic protein, was similar in SREB∆ and WT (Fig 4A and 4C). In contrast, transcript for a non-ribosomal siderophore biosynthesis peptide synthase SIDC (BDBG_08208) was 1.4 and 2.5-fold higher in SREB∆ than WT by microarray and qRT-PCR analyses, respectively (Fig 4A and 4C). Reverse-phase HPLC demonstrated elevated concentration of intracellular ferricrocin in SREB∆ compared to WT under iron-replete conditions (Fig 4D).
Fig. 4. Genes in the SREB regulon involved with iron acquisition and homeostasis. 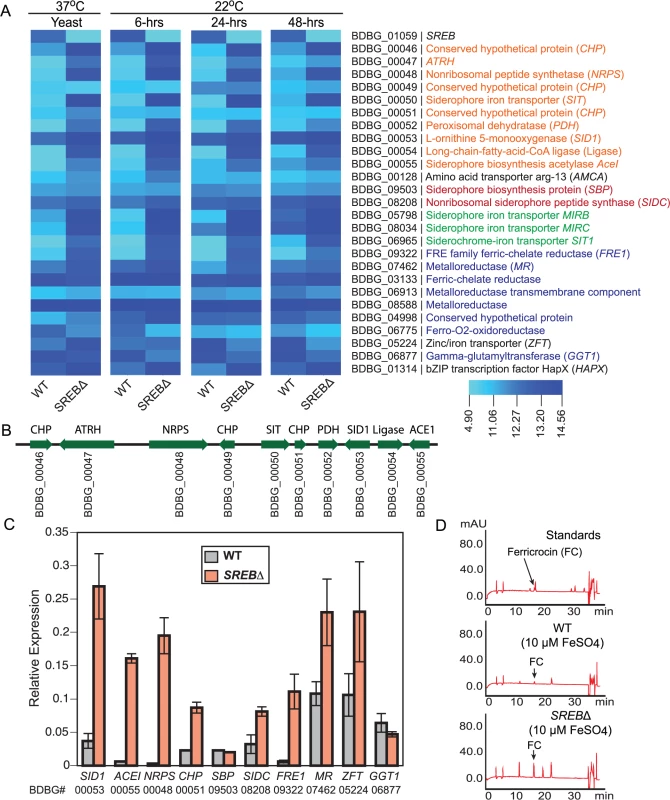
(A) Heat map of genes in the SREB regulon involved with siderophore biosynthesis (orange text for the siderophore biosynthetic gene cluster and red text for ferricrocin biosynthetic genes), siderophore transport (green text), and ferric reduction (blue text). (B) Schematic of putative siderophore biosynthetic gene cluster. Locus number (BDBG #) for genes in the Blastomyces genome database at the Broad Institute (www.broadinstitute.org) is listed below the coding region of each gene. (C) Quantitative real-time PCR analysis of yeast (37°C) for a subset of genes in the SREB regulon involved with iron acquisition and homeostasis. The results were averaged from 2 experiments. (D) Reverse-phase high-pressure liquid chromatography analysis for ferricrocin, an intracellular storage siderophore, in wild-type and SREB∆ yeast. Four genes in the B. dermatitidis genome were predicted to be involved with siderophore uptake (BDBG_00050, BDBG_05798, BDBG_08034, BDBG_06965) and were derepressed in SREB∆ under iron-replete conditions when compared to WT (Fig 4A–4C). One of the siderophore transporters (BDBG_00050) was located in the SBGC (Fig 4B).
The reduction of ferric to ferrous iron represents an important mechanism of iron assimilation for several ascomycete fungi including Saccharomyces cerevisiae, Cryptococcus neoformans, Candida albicans, and A. fumigatus [35–38]. The B. dermatitidis genome encodes 6 putative ferric reductases and a glutathione-dependent ferric reductase (GGT1) (Fig 4A and 4C). Ferric reductases are often involved with reductive iron assimilation (RIA) and function as part of a complex that includes a ferrous transporter/permease and a multicopper oxidase [39]. Deletion of SREB resulted in derepression of 2 of the 6 putative ferric reductases (BDBG_09322, BDBG_07462) under iron-replete conditions (Fig 4A and 4C). To assess the effect of SREB on cell surface ferric reductase activity, SREB∆ and WT yeast were grown in the presence of triphenyltetrazolium chloride (TTC). No enhancement of red color was observed for SREB∆ compared to WT under iron-replete conditions (S4 Fig). Translated BLAST analysis against the B. dermatitidis genome failed to yield homologs of S. cerevisiae FTR1 and A. fumigatus FTRA, which encode ferrous permeases involved with RIA. Similar analysis for Coccidioides immitis, Coccidioides posadasii, and Paracoccidioides brasiliensis also failed to identify FTR1/FTRA homologs. H. capsulatum strains G186AR, H88, and H143 contained FTR1/FTRA homologs, whereas NAM1 and G217B did not. Microarray analysis identified a putative B. dermatitidis Zn2+-Fe2+ transporter (BDBG_05224) that was DE in SREB∆ (derepressed at 37°C, repressed at 22°C) (Fig 4A and 4C). The transcription of B. dermatitidis ferro-O2-oxidoreductase (BDBG_06775), which is a homolog of S. cerevisiae FET5 multicopper oxidase, was repressed in SREB∆ (versus WT) at 37°C and 22°C under iron-replete conditions (Fig 4A). Collectively, these data indicate that SREB has a limited impact on the transcription of genes involved with ferric reduction.
Similar to H. capsulatum, the genome of B. dermatitidis encodes a γ-glutamyltransferase (GGT1; BDBG_06877), a glutathione-dependent ferric reductase, whose mechanism for iron acquisition is independent of RIA [40]. B. dermatitidis GGT1 transcript abundance in SREB∆ is similar to WT at 37°C, but is decreased at 22°C under iron-replete conditions (Fig 4A and 4C). These data suggest that unlike siderophore biosynthesis and transport, SREB does not repress GGT1 transcription under iron-replete conditions.
Deletion of SREB alters lipid metabolism
GO enrichment and WGCNA analyses suggested that deletion of SREB affected lipid metabolism at 37°C and 22°C. To investigate for defects in lipid biosynthesis, glycerolipids (triacylglycerol (TAG) and diacylglycerol (DAG)), phospholipids, free fatty acids, and ergosterol were extracted from WT and SREB∆ at 37°C and 6, 24, and 48-hrs at 22°C. Gene expression microarray analysis indicated 8 of 14 genes in the glycerolipid pathway involved with TAG biosynthesis were DE in SREB∆ versus WT (Fig 5A and 5B). Seven DE genes had decreased transcript abundance at 22°C, whereas a homolog of S. cerevisiae AYR1 (BDBG_09292) had increased transcription (Fig 5A and 5B). Quantitative RT-PCR analysis validated a subset of these DE genes; log2 fold changes were similar for qRT-PCR and microarray analyses at 48-hrs 22°C (Fig 5C). Gas chromatographic analysis of extracted lipids from SREB∆ suggested decreased TAG concentrations at 37°C and 22°C, and similar levels of DAG compared to WT (Fig 6A and 6B).
Fig. 5. Deletion of SREB affects the transcription of genes in the glycerolipid biosynthetic pathway. 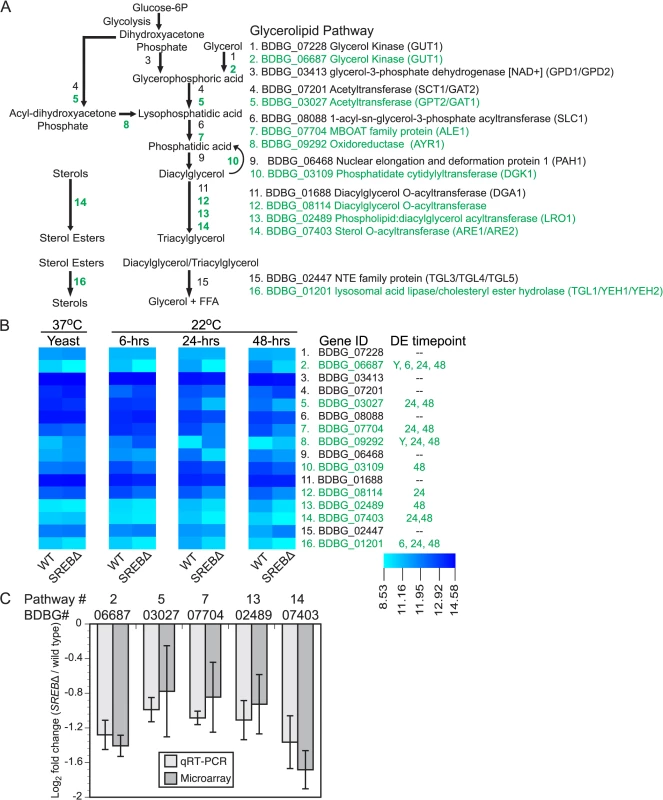
(A) Schematic of the glycerolipid biosynthetic pathway in B. dermatitidis. Putative lipases and esterases involved with sterol, diacylglycerol, and triacylglycerol breakdown are also shown. Differentially expressed (DE) genes are labeled in green. (B) Heat map of fluorescent intensity values (log2) for genes in glycerolipid biosynthetic pathway in wild-type (WT) and SREB∆ isolates. Genes are arranged in the same order as in (A). The time point at which gene transcription was DE in SREB∆ versus WT is adjacent to the heat map. DE genes are labeled in green. (C) Quantitative RT-PCR analysis of a subset of DE glycerolipid biosynthetic genes at 48-hrs 22°C. Log2 fold change values (SREB∆ versus wild-type) for qRT-PCR were compared to gene expression microarray data for BDBG_06687 (glycerol kinase), BDBG_03027 (acetyltransferase), BDBG_07704 (MBOAT family protein), BDBG_02489 (Phospholipid:diacylglycerol acetyltansferase), BDBG_07403 (Sterol-O-acyltransferase). The qRT-PCR results were averaged from 2 experiments. Fig. 6. Impact of SREB on lipid biosynthesis. 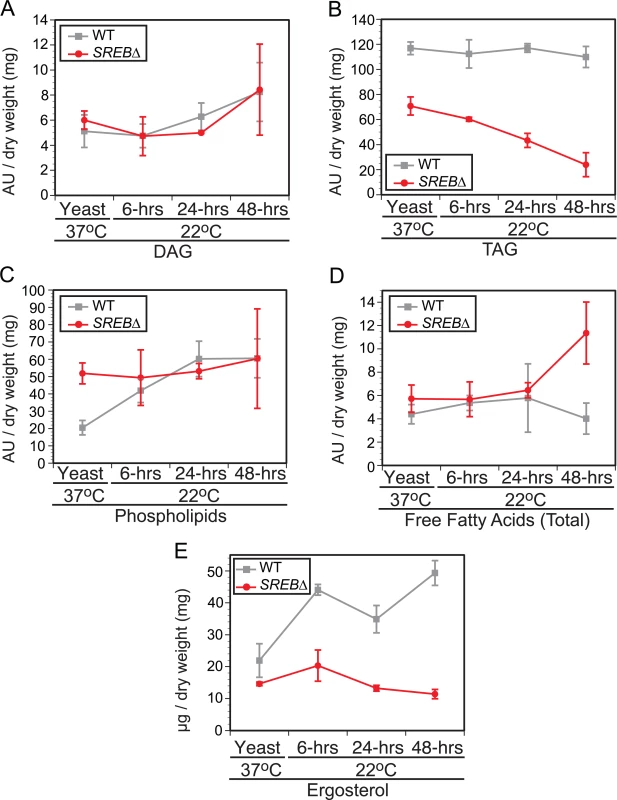
Gas chromatographic analysis of (A) diacylglycerol (DAG), (B) triacylglycerol (TAG), (C) phospholipids, (D) free fatty acids, and (E) ergosterol concentrations in wild-type (WT) and SREB∆ isolates. The results were averaged from 2 biological replicates for WT and SREB∆. Each biological replicate consisted of 2 technical replicates. The fatty acid abundance reported reflects the fatty acids present at end of protocol. Peak areas, which are denoted as arbitrary units (AU) were normalized by dry weight in milligrams. Deletion of SREB altered the transcription for 14 of 21 genes predicted to be involved with the biosynthesis of ergosterol (S5 Fig), a medically important sterol in the plasma membrane targeted by polyene and azole antifungals. Nearly all DE genes (12 / 14) in the ergosterol biosynthetic pathway had decreased transcription. In SREB∆, ergosterol concentrations were decreased at 37°C and 22°C compared to the WT isolate (Fig 6E). Moreover, ergosterol concentrations failed to increase in SREB∆ following a drop in temperature to 22°C (Fig 6E). To assess the effect of decreased ergosterol concentrations in SREB∆ to antifungal drugs, disc diffusion testing using amphotericin B deoxycholate (0, 0.5, 1, 2.5, 5, 10 μg) and voriconazole (0, 0.06, 0.125, 0.25, 0.5, 1 μg) was performed. No difference in antifungal drug susceptibility was observed in SREB∆ versus WT at 37°C (S6 Fig). Antifungal susceptibility testing at 22°C was precluded because SREB∆ exhibited a growth defect at this temperature; it failed to expand by radial growth or accumulate significant biomass [18].
In contrast to TAG and ergosterol, the concentration of phospholipids in SREB∆ and WT were similar following a drop in temperature from 37°C to 22°C (Fig 6C). Alterations in free fatty acid content were restricted to a single time point (48-hrs 22°C) in SREB∆ (Fig 6D), which was due to an increase in stearic acid (18 : 0) without alteration in oleic acid (18 : 1n9).
The reduction in TAG and ergosterol in SREB∆ prompted investigation for alteration in lipid droplets at 37°C and 22°C. Lipid droplets are organelles that consist of a neutral lipid core of TAG and sterol esters surrounded by a phospholipid monolayer intercalated with proteins [41]. Sterol esters are derived from ergosterol and intermediates in the ergosterol biosynthetic pathway [42]. SREB∆ was analyzed for defective lipid droplet formation using BODIPY, a fluorescent dye that is specific for lipid droplets (LDs) [43]. LDs were quantified in cells with yeast morphology at 37°C and 6-hrs 22°C. At 24 and 48-hrs 22°C, LDs in the yeast cell body as well as the emerging filament (germ tube or hyphae) were quantified. To adjust for the differences in filament length for SREB∆ compared to WT, LDs were quantified per 10 μm segments along the length of the filament. At 37°C and 6-hrs 22°C, there were no differences in LD abundance in yeast cells between SREB∆ and WT (Fig 7A and 7B). At 24-hrs 22°C, the median LD abundance was sharply reduced in SREB∆ filaments versus WT (0 versus 7); however, the median number of LD per yeast cell body for SREB∆ was similar to WT (16.5 versus 18) (Fig 7A and 7B). At 48-hrs 22°C, the median number of LDs per 10 μm segment (0 versus 6) and yeast cell body (10 versus 17) was decreased in SREB∆ compared to WT (Fig 7A and 7B). At 24 and 48-hrs 22°C, more than 80% of 10 μm segments analyzed for SREB∆ had ≤ 4 LD, whereas for WT, less than 33% of 10 μm segments had 4 or fewer LD (Fig 7C).
Fig. 7. Lipid droplets in wild type and SREB∆. 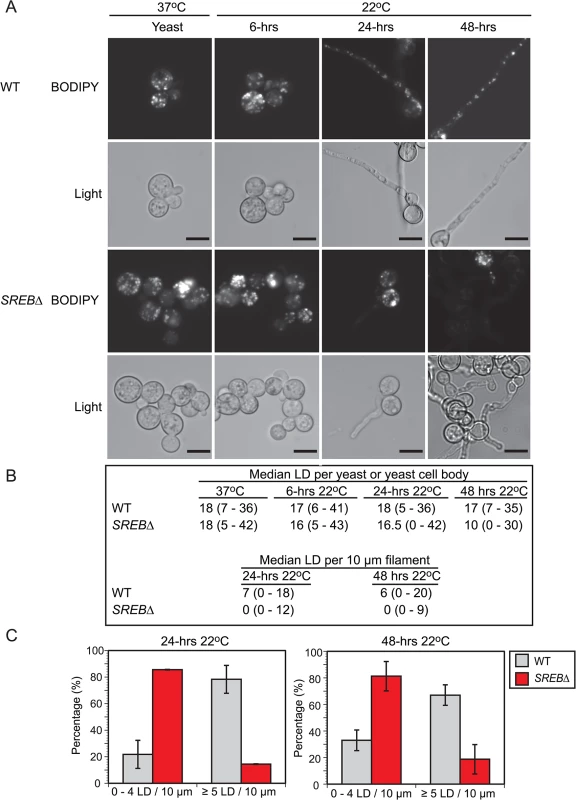
(A) BODIPY 493/503 was used to stain for lipid droplets (LD) in wild type (WT) and SREB∆ at 37°C and 6, 24, and 48-hrs following a drop in temperature to 22°C. Corresponding bright field microscopic images (light) are below the fluorescent images. Scale bar is 10 μm (B) Median number of lipid droplets per yeast, yeast cell body, and 10 μm filament were quantified at 37°C and 22°C (range is in parentheses). To adjust for differences in filamentous growth at 24 and 48-hrs 22°C, LDs were quantified per 10 μm segment along the length of the filament. LDs were quantified from 50 cells in duplicate. (C) Percentage of filaments that contained 0–4 or ≥ 5 LDs per 10 μm segment at 24 and 48-hrs 22°C. In eukaryotic cells, fatty acids (FAs) serve as an important substrate for TAG and sterol ester biosynthesis [44,45]. To investigate if exogenous fatty acids can influence the phase transition or LD formation, SREB∆ cells were treated with saturated (16 : 0, 18 : 0) and unsaturated (16 : 1n7, 18 : 1n9) fatty acids. Experiments with exogenous TAG were not performed because fungal cells are unable to uptake this glycerolipid. The poor solubility of saturated FAs in aqueous solutions necessitated dissolving 16 : 0 and 18 : 0 in DMSO prior to supplementation of iron-replete HMM. Treatment of SREB∆ with 0.5 mM palmitic (16 : 0) or 0.5 mM stearic (18 : 0) acid accelerated the morphologic switch at 22°C compared to control strains (SREB∆ untreated, SREB∆ DMSO) without affecting the phase transition of WT cells (untreated, DMSO, 16 : 0, 18 : 0) (Fig 8A). At 24-hrs 22°C, exogenous 16 : 0 and 18 : 0 increased germ tube formation in SREB∆ versus SREB∆ untreated and SREB∆ DMSO (Fig 8A). At 24 and 48-hrs 22°C, SREB∆ treated with 16 : 0 or 18 : 0 exhibited increased conversion to hyphae and decreased number of yeast cells compared to controls (SREB∆ untreated, SREB∆ DMSO) (Fig 8A). The morphology of yeast cells at 37°C was unaffected by 16 : 0 or 18 : 0 (S7 Fig). Although DMSO did not affect the phase transition (Fig 8A), higher concentrations of DMSO needed to solubilize ≥ 1 mM 16 : 0 or 18 : 0 hindered the morphologic switch to mold for WT and SREB∆. BODIPY staining demonstrated that exogenous 16 : 0 and 18 : 0 restored LDs in the growing filaments of SREB∆ at 24-hrs 22°C (Fig 8B and 8C). Median LD per 10 μm increased from 1 for SREB∆ controls (untreated, DMSO) to 5 for SREB∆ 16 : 0 and 2.5 for SREB∆ 18 : 0. Moreover, the percentage of 10 μm filament segments with 5 LDs increased 4-fold for SREB∆ 16 : 0 and 2.2-fold for SREB∆ 18 : 0 compared to controls (SREB∆ untreated, SREB∆ DMSO) (Fig 8B). The increase in LDs was transient and limited to the 24-hr 22°C time point (Fig 8A–8C). At 48-hrs 22°C, the median number of LDs per 10 μm for SREB∆ 16 : 0 and 18 : 0 declined to 2 and the percentage of 10 μm filament segments with ≤ 4 LDs increased (Fig 8A–8C). Exogenous 16 : 0 or 18 : 0 did not affect the number of lipid droplets for SREB∆ at 37°C and 6-hrs 22°C (Fig 8B and 8C, and S7 Fig). WT cells treated with 16 : 0 or 18 : 0 had similar number of LDs as controls (median 6–7 LD per 10 μm filament segment at 24 and 48-hrs 22°C) (Fig 8B and 8C). SREB∆ cells treated with 0.5 mM oleic acid (18 : 1n9) exhibited similar morphologic and LD defects as untreated SREB∆ cells at 48-hrs 22°C (S8 Fig). Treatment of cells with 0.5 mM palmitoleic acid (16 : 1n7) was lethal for WT and SREB∆; reducing 16 : 1n7 concentrations to 0.250 mM or 0.125 mM did not improve cell viability (S8 Fig).
Fig. 8. Fatty acid supplementation for wild type and SREB∆. 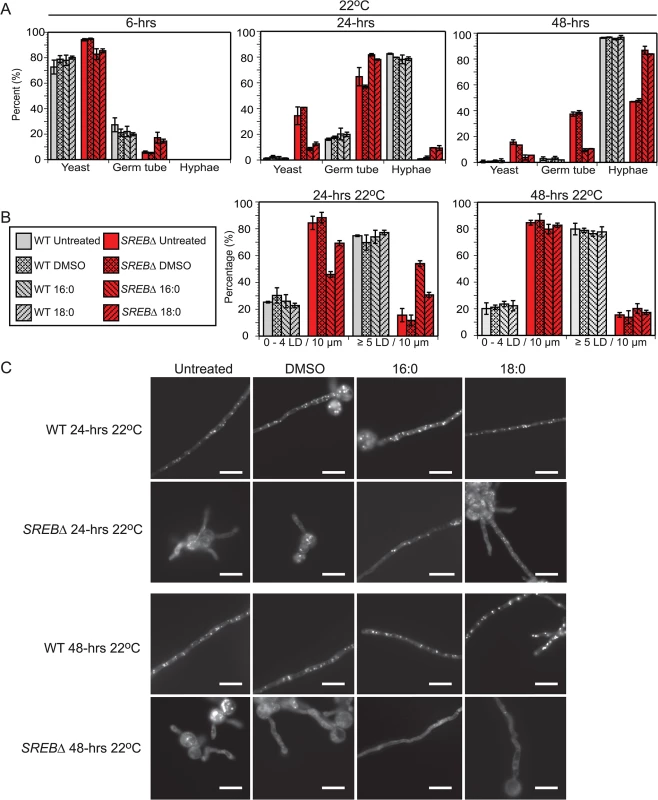
(A) Percentage of WT and SREB∆ with yeast morphology, germ tube development, and hyphae at 22°C for cells grown in media supplemented with 0.5 mM palmitic acid (16:0) or 0.5 mM stearic acid (18:0). Controls included DMSO only and cells grown in media without DMSO or saturated fatty acid (untreated). At least 200 cells were counted in duplicate. Results were averaged from 2 independent experiments. (B) Percentage of filaments that contained 0–4 or 5 LDs per 10 μm segment at 24 and 48-hrs 22°C for WT and SREB∆ cells (untreated, DMSO only, 16:0 and 18:0). LDs were quantified per 10 μm segment along the length of filaments from at least 30 cells. Results were averaged from 2 independent experiments. (C) BODIPY 493/503 staining of lipid droplets at 24 and 48-hrs 22°C for WT and SREB∆ cells (untreated, DMSO only, 16:0 and 18:0). Scale bar equals 10 μm. SREB directly binds and regulates the transcription of genes with disparate functions
ChIP-qPCR was used to identify genes bound and directly regulated by SREB in vivo. For ChIP, SREB was engineered to contain an in-frame 3x-hemagglutinin (HA) epitope tag at the C-terminus. To minimize mis-expression or abnormal cellular localization, SREB-3xHA was placed under the control of its native promoter and contained the 3’-untranslated region, respectively. Retransformation of SREB∆ with SREB-3xHA complemented the defects in phase transition (S9 Fig) and siderophore biosynthesis indicating the construct was functional. Western blot analysis using a ChIP-grade rabbit polyclonal HA antibody demonstrated specificity for 26199 strains transformed with SREB-3xHA and absence of antibody hybridization for wild type 26199 (S9 Fig). To identify potential sites of DNA binding by SREB, gene expression microarray data were integrated with genome-wide in silico analysis of extended GATA binding motifs (ATC-w-gAta-a) in promoters of differentially expressed genes; approximately 12% of DE genes possessed an extended GATA motif. Use of the classic GATA motif (A/T-GATA-A/G) was limited because this motif was found upstream of nearly all genes (DE and non-DE) in the B. dermatitidis genome [18].
Using this integrated approach, we identified a subset of genes bound and regulated by SREB-3xHA under iron-replete conditions (10 μM FeSO4) using ChIP-qPCR (Fig 9A and 9B). These included genes involved with extracellular (SID1, ACE1) and intracellular (SIDC) siderophore biosynthesis, siderophore transport (MIRB), ferric reduction (FRE1 and MR), ferrous iron/zinc transporter (ZFT), a WD-repeat protein of unknown function (WD), and a bZIP transcription factor (HAPX) (Fig 9A and 9B). For ChIP-qPCR, SREB-3xHA did not bind the GAPDH promoter, which lacked GATA motifs and served as a negative control. ChIP-seq analysis of yeast grown in iron-replete media (10 μM FeSO4) also demonstrated enrichment of SREB-3xHA binding for genes located in the siderophore biosynthetic gene cluster including BDBG_00046 (conserved hypothetical protein), BDBG_00047 (ATRH), BDBG_00048 (NRPS), BDBG_00053 (SID1), BDBG_00054 (long chain fatty acid CoA ligase), and BDBG_00055 (ACE1) (S10 Fig and S6 Table). In addition, SREB-3xHA bound to the promoters of SIDC (BDBG_08208) and siderophore transporters MIRB (BDBG_05798), MIRC (BDBG_08034), and SIT1 (BDBG_06965) (S10 Fig and S6 Table). ChIP-seq also showed enrichment of SREB-3xHA upstream of genes unrelated to iron homeostasis including a glycolipid surface protein predicted to have 1,3-beta-glucanosyltransferase activity (BDBG_000734), GNAT acyltransferase (BDBG_06912), a thioesterase-domain containing protein (BDBG_07971), and a cation diffusion facilitator (BDBG_07469) (S11 Fig and S6 Table). These genes were DE in the microarray analysis (S6 Table). In addition, SREB-3xHA was enriched upstream of an amino acid acetyltransferase, BDBG_00643, which is derepressed in SREB∆ and is predicted to be involved in the biosynthesis of ornithine. ChIP-qPCR confirmed enrichment of a subset of these genes including the glycolipid surface protein (BDBG_000734) and amino acid acetyltransferase (BDBG_00643) (Fig 9A and 9B, and S6 Table).
Fig. 9. Chromatin immunoprecipitation with quantitative real-time PCR (ChIP-qPCR). 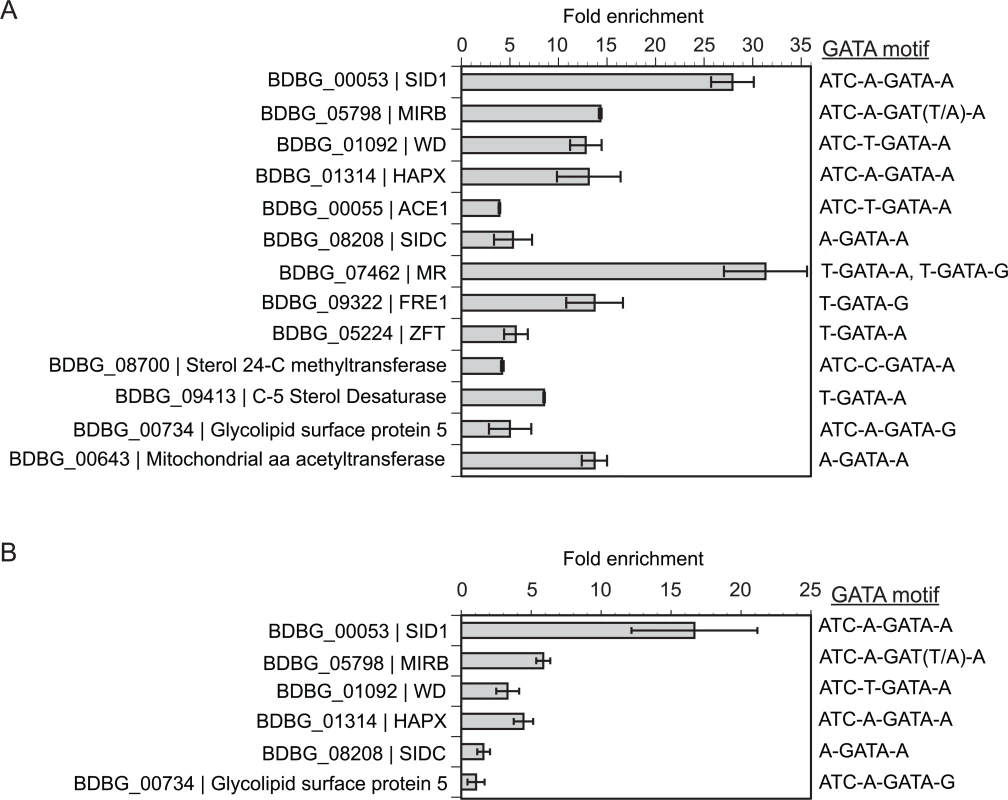
(A) Enrichment for binding of SREB-3xHA to promoter regions containing GATA binding motifs in B. dermatitidis yeast at 37°C. (B) Enrichment binding of SREB-3xHA to promoter regions containing GATA binding motifs in B. dermatitidis cells at 48-hrs following a drop in temperature from 37°C to 22°C. ChIP-qPCR was performed using SREB that was engineered to contain a C-terminal 3x-hemagglutinin tag. Integration of gene expression microarray data with genome-wide GATA motif analysis was used to identify putative GATA transcription factor binding sites. All genes analyzed were differentially expressed and contained a GATA motif (ATC-w-gAta-a or A/T-GATA-A/G) in the upstream promoter. For MIRB, the upstream motif in strain 26199 was ATC-A-GATT-A, whereas in other strains including SLH14081, ER-3, and ATCC 18188, the motif was ATC-A-GATA-A. BDBG is the locus number for genes in the B. dermatitidis genome database at the Broad Institute (www.broadinstitute.org). The reduction of ergosterol and TAG concentrations in SREB∆ prompted investigation of SREB-3xHA binding to a subset of differentially expressed genes in the ergosterol and glycerolipid biosynthetic pathways. Of the 14 DE genes in the ergosterol pathway, only ERG6, which encodes a sterol 24-C methyltransferase (BDBG_08700) contained an extended GATA motif in the promoter region. This gene was DE at 37°C (0.80 log2 decrease) and ChIP-qPCR demonstrated 4.2-fold enrichment in yeast (Fig 9A). Surprisingly, enrichment for this gene was not observed by ChIP-seq analysis at 37°C. This discrepancy may be related to differences in chromatin shearing. However, SREB-3xHA was enriched by ChIP-seq analysis upstream of a C-5 sterol desaturase (BDBG_09413, -1.0 Log2 fold change at 37°C) in the ergosterol biosynthetic pathway (S5 Fig) and binding was confirmed by ChIP-qPCR (Fig 9A and S6 Table). Differentially expressed genes involved with conversion of DAG to TAG in the glycerolipid biosynthetic pathway (BDBG_02489, BDBG_07403, BDBG_08114) were analyzed by ChIP-qPCR at 48-hrs 22°C. These genes possessed classic GATA motifs but lacked extended motifs. No enrichment was observed for BDBG_02489 and BDBG_07403 at 22°C. Analysis of BDBG_08114 was precluded due to difficulty in designing high-quality primers targeting the region containing the GATA motif.
Overexpression of HAPX alters the phase transition
Gene expression microarray data indicated that SREB altered the phase transition independent of VMA1 and HGRA; these genes are not DE in SREB∆. To test the importance of a subset of SREB-bound genes on the phase transition, we overexpressed WD and HAPX in wild-type strain 26199 under the control of an H2B promoter. WD was selected for overexpression because WD-repeat proteins can be involved with transcriptional regulation and affect fungal development [46,47]. HAPX is also involved with transcriptional regulation and in Aspergillus spp., it is in a negative regulatory circuit with SREA, a homolog of SREB [48,49]. These genes were overexpressed rather than silenced because they were derepressed in SREB∆. The H2B promoter was active at 37°C, 22°C, and during the phase transition when measured by qRT-PCR (Ct values 21 at 37°C and 22°C for H2B transcript). WD overexpression strains grew as yeast and converted normally to mold at 22°C. HAPX overexpression strains (OE 8, 18, 22) were morphologically similar to SREB∆ following a drop in temperature from 37°C to 22°C (Fig 10A). Quantitative RT-PCR analysis demonstrated 15.5–53.1-fold increase in HAPX transcript abundance in OE strains compared to WT (Fig 10B). SREB transcript abundance was decreased in OE 8 and OE 18, but was similar to WT and empty vector for OE 22 (Fig 10C). In HAPX OE 8 and 18, the reduction in SREB transcript was unstable and normalized to WT levels over serial passage; however, the morphologic defect and HAPX overexpression persisted. Unlike deletion of SREB, overexpression of HAPX did not adversely affect LD formation at 24 and 48-hrs 22°C.
Fig. 10. HAPX overexpression affects the phase transition in B. dermatitidis. 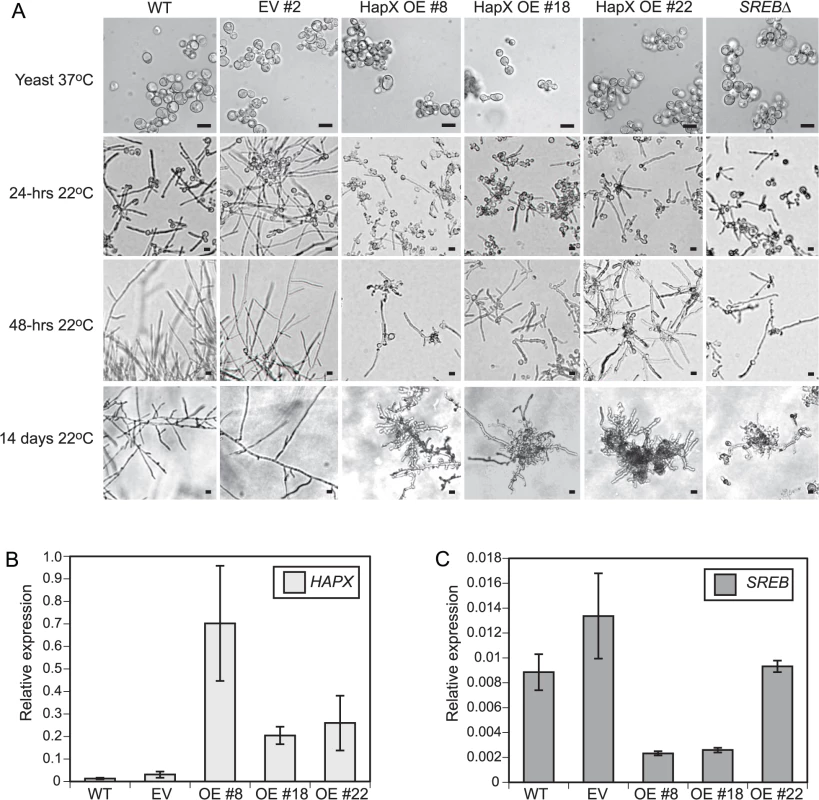
(A) Similar to SREB∆, HAPX overexpression (OE) strains #8, #18, and #22 grew as yeast at 37°C, but failed to complete conversion to mold following a downshift in temperature to 22°C. In contrast, wild-type (WT) and empty vector (EV) strains converted to mold at 22°C. Scale bar is 10 μm. (B) Quantitative RT-PCR analysis of HAPX transcript abundance in controls (WT, EV) and overexpression (OE# 8, OE#18, OE#22) strains. HAPX OE strains had 15.5–53.1-fold increased HAPX transcript abundance compared to controls. (C) Quantitative RT-PCR demonstrated a 3.4–5.8-fold decrease in SREB transcript abundance in OE strains #8 and #18 compared to WT and EV controls. SREB transcript abundance in OE #22 exhibited a 0.9–1.4-fold change compared to controls (WT, EV). All strains were grown in liquid HMM supplemented with 10 μM FeSO4. Quantitative RT-PCR results were averaged from 2 experiments. Integration of microarray, qRT-PCR, and ChIP data suggested that B. dermatitidis SREB and HAPX are in a negative regulatory circuit similar to Aspergillus nidulans and A. fumigatus [48,49]. B. dermatitidis does not encode a homolog to C. albicans SEF1, which forms a regulatory circuit with SFU1 and HAP43 [50]. Moreover, deletion of SREB did not alter the transcription of HAPB, HAPC, or HAPE homologs, which form a DNA binding complex that recruits HAPX to CCAAT promoter sites of target genes [48]. HAPX transcript abundance was substantially higher in the OE strains (15.5–53.1-fold) than SREB∆ (1.7–3.5-fold) compared to WT (Figs 4A and 10B). To investigate if less extreme changes in transcript abundance for either HAPX or SREB affected the phase transition, SREB was targeted for gene silencing using a GFP sentinel RNAi system [51]. SREB-GFP silenced strains had a 1.8–2.5-fold reduction in transcript abundance compared to controls (GFP reporter, empty vector, GFP-only silenced) (Fig 11A). SREB-GFP RNAi strains had increased SID1 transcript abundance under iron-replete (10 um FeSO4) conditions compared to controls (Fig 11B). HAPX transcript was increased 1.7–3.2-fold in SREB RNAi versus controls (Fig 11C). Following a drop in temperature, SREB-GFP silenced strains converted to mold at 22°C (Fig 11D). Moreover, there was no delay in the phase transition. This data suggested that the 1.7–3.5-fold increase in HAPX transcript in SREB∆ was not responsible for the defect in phase transition at 22°C; larger changes in HAPX transcript abundance are required to impact morphologic development at 22°C.
Fig. 11. Silencing of SREB using RNA interference. 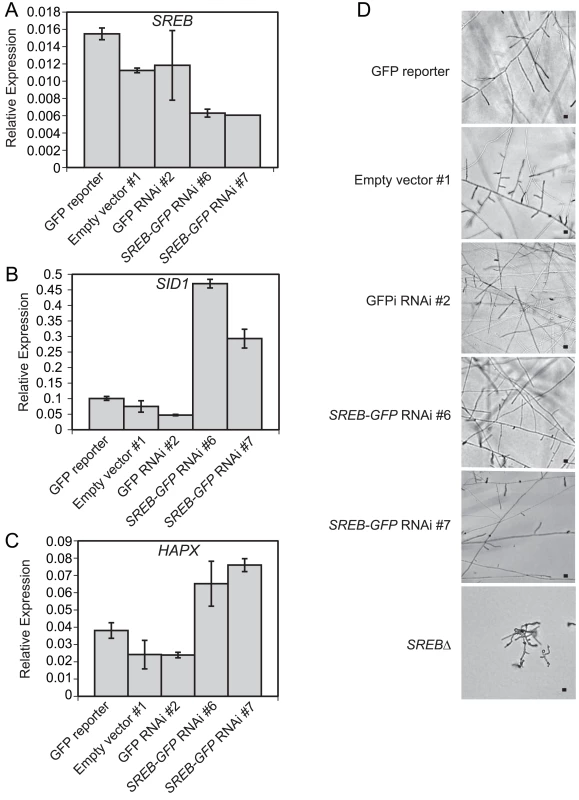
(A-C) Quantitative real-time PCR analysis demonstrated a 1.8–2.5-fold decrease in SREB transcript and derepression of SID1 (2.9 6.2-fold increase) and HAPX (1.7 3.2-fold increase) in SREB RNAi strains #6 and #7 compared to controls (GFP reporter, empty vector, GFP-RNAi). SID1 encodes L-ornithine-N5-oxygenase, which encodes the first enzyme in siderophore biosynthesis. Quantitative RT-PCR results were averaged from 2 experiments. (D) SREB-GFP knockdown strains along with empty vector and GFP RNAi only controls converted to mycelia at 22°C. Images were photographed at 9 days incubation 22°C. Scale bar equals 10 μm. Discussion
We used whole genome gene expression microarrays and bioinformatics analyses to identify and characterize the SREB regulon in B. dermatitidis following a drop in temperature from 37°C to 22°C. The microarray experiments captured the transcriptional response early in the phase transition (0 48-hrs) and under conditions (10 μM FeSO4) that promote binding of SREB to DNA. This facilitated analysis for how SREB affected disparate processes including the morphologic switch and iron assimilation; prior research demonstrated exogenous iron neither suppressed nor enhanced the defect in phase transition observed for SREB∆ [18]. Further delineation of the SREB regulon was accomplished using ChIP-qPCR integrated with microarray and MAST analyses (along with ChIP-seq) to identify a subset of genes directly bound and regulated by SREB in vivo. This included genes involved with the regulation of siderophore biosynthesis and uptake, iron homeostasis, and genes unrelated to iron assimilation. The results herein represent the first genome-wide transcriptional analysis integrated with in vivo transcription factor binding data during the transition from yeast to mold in B. dermatitidis. Functional analysis of SREB∆ suggested that alterations in lipid metabolism contribute to the defect in the phase transition. In addition, chromatin immunoprecipitation data along with RNAi and overexpression analyses indicated that SREB was in a regulatory circuit with HAPX. Alteration of this circuit by either deletion of SREB or strong overexpression of HAPX impaired the conversion to mold at 22°C.
Analysis of the microarray data demonstrated SREB had pleiotropic effects on transcription during growth as yeast at 37°C and during the phase transition following a drop in temperature to 22°C. Deletion of SREB affected the transcription of 11.6–19.9% of genes (1109–1910 / 9,583) across the time course. Similarly, deletion of CIR1, a homolog of SREB important for thermotolerance at 37°C, impacted transcription for 21.0% of genes (1,623 / 7,738) in the C. neoformans genome under iron-replete conditions [24]. RNA interference targeting SRE1 in H. capsulatum, which also affects the conversion from yeast to mycelia, altered transcription of 9.6% genes (364 / 3810) [19]. In contrast, deletion of SREA in A. fumigatus and SFU1 in Candida albicans altered transcription for 0.6% (49 / 8975) and 2.0% (139 / 7116) genes when iron was abundant, respectively [52,53]. Unlike CIR1∆ or SRE1 knockdowns, SREA∆ mutants are viable at 37°C and SFU1∆ mutants convert to hyphae under inducing conditions [52,53]. A shared feature among these GATA transcription factors is the regulation of genes involved with iron assimilation [19,24,52,53]. However, stark differences in transcriptional regulation exist between GATA transcription factors important for temperature adaptation and those that do not affect thermotolerance or morphology: SREB, SRE1, and CIR1 affect the transcription of genes involved with diverse processes, whereas SREA and SFU1 regulate a limited set of genes.
Similar to SREB, the transcription factors encoded by SRE1, CIR1, SREA, and SFU1 can either induce or repress gene transcription [19,24,52,53]. For CIR1∆ and SREB∆ yeast, the ratio of genes with increased to decreased transcription was slightly greater than 1 [24]; at 22°C, the ratio for DE genes in SREB∆ was close to 1. In contrast, the majority of DE genes in A. fumigatus SREA∆ were increased [52]. In C. albicans, conflicting results have been obtained from different SFU1∆ strains [50,53]. To further illuminate how deletion of SREB affected transcription during the phase transition, WGCNA was performed to identify groups of co-expressed genes. This demonstrated that deletion of SREB affected transcription differently among co-expressed gene sets. This included subsets of DE genes in SREB∆ that failed to properly increase or decrease following a drop in temperature as well as genes with reduced transcript abundance at 37°C and 22°C.
GO analysis offered additional support that deletion of SREB affected the transcription of genes with diverse functions. Analysis of DE genes identified 17 GO terms enriched in SREB∆ ranging from iron ion binding, catalytic activity, fatty acid biosynthetic process to carbohydrate metabolic process. Integrative analysis of genes within the enriched GO categories suggested several themes including iron ion binding, lipid biosynthesis, and amino acid metabolism. Genes that encoded iron ion binding proteins were involved with oxidative stress response and lipid metabolism. The absence of a coordinated or sustained transcriptional pattern for catalase isoforms and superoxide dismutase suggested that SREB∆ cells were not under substantial iron-related oxidative stress. Moreover, prior research demonstrated that exogenous iron concentrations did not affect the defect in the morphologic switch in SREB∆ [18]. GO enrichment performed by Jung and colleagues on CIR1∆ and our analysis of SREB∆ indicated involvement of GATA transcription factors with lipid metabolism; CIR1∆ was enriched for fatty acid metabolism (GO:0006631) and SREB∆ for fatty acid biosynthetic process (GO:0006633) [24]. In addition to altering lipid metabolism, deletion of SREB affected genes involved with the biosynthesis of 16 amino acids. Strikingly, the majority of DE genes had reduced transcript abundance at 24 and 48-hrs 22°C. This finding may reflect the distinct morphologic differences between SREB∆ and WT at these time points. B. dermatitidis and H. capsulatum amino acid concentrations are approximately 3-fold higher in mycelia than yeast [54,55].
Genes involved with carbohydrate metabolism, including hydrolysis of O-glycosyl linkages, were enriched at 24 and 48-hrs 22°C. Three patterns were observed: increased transcription of chitinases, reduced transcription of glucanases, and no impact on biosynthetic genes for alpha or beta-glucan. Homologs of these genes in S. cerevisiae are involved with cell wall remodeling or maintenance. The impact of altered cell wall remodeling on SREB∆ at 22°C was likely limited because the addition of exogenous saturated fatty acids was able to accelerate filamentous growth at 22°C. Similarly, the effect of reduced transcription of BDBG_00734, a homolog of S. cerevisiae GAS5, which is bound and regulated by SREB at 37°C had limited impact; prior research demonstrated similar growth rate of SREB∆ and WT as yeast [18].
One of the major functions of SREB was to regulate iron homeostasis by repressing the transcription of genes important for the biosynthesis and transport of siderophores when iron was abundant. Genes predicted to be involved with the biosynthesis of secreted siderophores were located in a gene cluster, whereas biosynthetic genes for ferricrocin, an intracellular storage siderophore, were not clustered. This arrangement is conserved in other fungi including H. capsulatum and A. fumigatus [29,56]. Using ChIP-qPCR and ChIP-seq, we demonstrated in vivo that SREB bound upstream of clustered (e.g., SID1, NRPS, ACE1) and non-clustered (e.g., SIDC) siderophore biosynthetic genes. For genes in the SBGC, SREB bound regions with extended GATA motifs (ATC-w-gAta-a) as predicted by gel electrophoretic-mobility shift [29] and GFP reporter [19] assays. We extended these findings by demonstrating in vivo that SREB binds the promoter SIDC, which has the classic GATA motif, A-GATA-A. Thus, SREB regulates extracellular and intracellular siderophore biosynthetic gene transcription by binding to regions with extended and classic GATA motifs.
Reductive iron assimilation (RIA) serves as a primary mechanism for iron uptake in C. neoformans, C. albicans, and S. cerevisiae, which do not produce siderophores [24,38]. Some fungi such as Schizosaccharomyces pombe can synthesize siderophores and use RIA to acquire iron [57]. The tripartite machinery for RIA consists of a ferric reductase (e.g. FRE1), ferrous permease (e.g. FTR1), and multicopper oxidase (e.g. FET3) [39]. Despite encoding for a putative ferric reductase and a multicopper oxidase, which are under the transcriptional control of SREB, homologs to FTR1/FTRA are absent in the sequenced B. dermatitidis genomes (SLH14081, ER-3, 18188, 18187, 26199). Similarly, the genomes of other closely related dimorphic fungi did not encode FTR1/FTRA homologs; however, some H. capsulatum genomes were predicted to contain FTR1/FTRA homologs. Collectively, this suggests that ferrous transport for RIA in B. dermatitidis occurs either through an unrecognized transmembrane transporter such as ZFT (BDBG_05224) or that RIA is dispensable. Moreover, B. dermatitidis has an RIA-independent mechanism for acquiring iron by secreting a glutathione-dependent ferric reductase (GGT1) [58]. In H. capsulatum, GGT1-mediated ferric reduction is important for virulence [40]. Unlike siderophore-mediated iron uptake, microarray analysis indicated that GGT1 transcription was not under the control of SREB at 37°C. This indicated SREB regulates some, but not all mechanisms for iron assimilation in B. dermatitidis.
The morphologic switch between yeast and mold in B. dermatitidis, H. capsulatum, and Paracoccidioides brasiliensis is associated with changes in lipid concentration. Lipid content is often ≥ 2-fold higher in mycelia than yeast [59,60]. Microarray and quantitative lipid analyses demonstrated that deletion of SREB reduced TAG and ergosterol concentrations, but had limited effect on phospholipids and free fatty acids (except for an increase in stearic acid at 48-hrs 22°C). As the time course progressed, the quantitative differences for ergosterol and TAG between SREB∆ and WT became more pronounced, which likely reflected the increased number of genes in these pathways that had reduced transcript abundance at 22°C versus 37°C. ChIP-qPCR and ChIP-seq analyses suggested that SREB had direct and indirect effects on genes involved with the biosynthesis of neutral lipids (TAG, ergosterol).
The decrease of neutral lipids was likely responsible for the reduction in LDs at 22°C in SREB∆; hydrolases involved with LD breakdown were not upregulated. Lipid droplet formation in eukaryotes including fungi is linked to TAG content as well as fatty acids, which serve as a substrate for TAG and sterol ester biosynthesis [44,45,61–63]. In S. cerevisiae, the biosynthesis of TAG contributes to lipid droplet formation to a greater degree than the biosynthesis of sterol esters [63]. The addition of exogenous saturated fatty acids (16 : 0 or 18 : 0) to SREB∆ transiently restored LD formation and accelerated the phase transition at 22°C. Importantly, exogenous 16 : 0 and 18 : 0 did not increase LD content or accelerate the conversion to hyphae in WT cells. The increase in LD content at 24-hrs followed by a decrease at 48-hrs 22°C in SREB∆ suggested that LDs are biosynthesized and degraded during the phase transition. The presence of LDs during filamentous growth in SREB∆ cells treated with 16 : 0 or 18 : 0 indicated that the loss of LDs in untreated cells was due to a reduction in biosynthesis and not from impaired intracellular LD transport. Thus, deletion of SREB disrupted LD homeostasis at 22°C in which reduced biosynthesis could not meet catabolic demands during filamentous growth. TAG and sterol esters in LDs can serve as a source of carbon for energy and lipids for membrane biosynthesis [64]. The increase in stearic acid (18 : 0) at 48-hrs 22°C in SREB∆ may represent a compensatory response to altered LD biosynthesis; however, it was not enough to overcome reduced concentrations of neutral lipids. Collectively, these data suggest that lipid metabolism including LD homeostasis may contribute to the morphologic switch from yeast to mold following a drop in temperature to 22°C.
Prior research on saturated and unsaturated fatty acids in the dimorphic fungi has focused on the heat shock response and thermotolerance of H. capsulatum during the transition to yeast following an increase in temperature to 37°C [65,66]. The addition of saturated fatty acids rescued the heat shock response by increasing HSP82 transcription in a temperature sensitive strain, whereas unsaturated fatty acids blunted the transcription of HSP82 in response to elevated temperature in a thermotolerant strain [66]. For SREB∆, the addition of saturated fatty acids enhanced the morphologic shift following a drop in temperature to 22°C, whereas unsaturated fatty acids did not impact the phase transition. Taken together, these data suggest that fatty acids and lipid metabolism influence the response to temperature at 37°C and 22°C.
B. dermatitidis SREB and HAPX likely form a negative regulatory circuit similar to A. fumigatus and A. nidulans. Functional analysis demonstrated a novel role for this circuit in B. dermatitidis. Deletion of SREB or overexpression of HAPX impaired the temperature-dependent morphologic switch. In A. fumigatus and A. nidulans, the SREA-HAPX circuit governs the cellular response to exogenous iron with SREA repressing HAPX transcription when iron is abundant, and HAPX inhibiting SREA transcription under iron poor conditions [48,49]. In C. albicans, the regulatory circuit involving HAP43, SFU1, and SEF1 mediates the ability of this pathogen to adapt to niches within the mammalian host that have radically different iron concentrations [46,67,68]. Similar to A. fumigatus, A. nidulans, and C. albicans, transcript abundance of B. dermatitidis HAPX and SREB is influenced by the presence or absence of exogenous iron [18]. In the broadest sense, the SREA-HAPX and SFU1-SEF1-HAP43 regulatory circuits mediate adaptation to changing environmental conditions. In addition to responding to exogenous iron, B. dermatitidis SREB and HAPX influence morphogenesis at 22°C. The defect in the phase transition in SREB∆ is unlikely to be due to HAPX derepression because SREB knockdown strains, which had similar levels of HAPX transcript as SREB∆, converted to mold. Moreover, the morphologic defect in HAPX OE strains was unlikely to be mediated through SREB because its repression was transient. These findings suggest that the amount of SREB and HAPX transcript influenced the morphologic response to ambient temperature.
In summary, we characterized the SREB regulon in B. dermatitidis during growth as yeast at 37°C and during the early part of the phase transition at 22°C. Deletion of SREB affected the transcription of genes involved with iron homeostasis, iron ion binding, amino acid biosynthesis, carbohydrate metabolism, lipid biosynthesis, and the transcriptional regulator HAPX. Moreover, deletion of SREB altered transcript differently among subsets of DE genes including those that failed to properly increase or decrease following a shift in temperature to 22°C. Integration of gene expression microarray and chromatin immunoprecipitation studies demonstrated that SREB binds and regulates genes involved with siderophore biosynthesis and iron uptake as well as genes unrelated to iron assimilation. SREB∆ cells had reduced TAG and ergosterol concentration along with decreased numbers of lipid droplets at 22°C. Moreover, experimental analysis suggested that LDs are biosynthesized and degraded during the phase transition and that lipid metabolism may contribute to filamentous growth at 22°C. Exogenous palmitic (16 : 0) and stearic (18 : 0) accelerated the morphologic shift and transiently restored LD formation in SREB∆ at 22°C. Deletion of SREB resulted in derepression of HAPX and functional analysis suggested that SREB and HAPX are in a negative regulatory circuit. Both SREB and HAPX affected the morphogenesis of B. dermatitidis at 22°C; however, large changes in transcript abundance by gene deletion for SREB or strong overexpression for HAPX were required.
Methods
Strains and media
B. dermatitidis strains used included American Type Culture Collection (ATCC) strain 26199, SREB∆, SREB-3xHA, and HAPX-OE. B. dermatitidis 26199 was originally isolated from a patient from South Carolina and is virulent in a murine model of pulmonary infection (ATCC 26199 has been used as a standard laboratory strain since the 1970’s and was a gift from the laboratory of Bruce S. Klein. This strain was not isolated from a patient for this specific study) [69–71]. SREB∆ was generated by targeted deletion of SREB in strain 26199 [18]. SREB-3xHA was engineered to contain an in-frame 3x-hemagglutinin (HA) epitope tag at the C-terminus in strain 26199. HAPX-OE strains were engineered to overexpress HAPX in strain 26199. B. dermatitidis cultures were maintained as yeast on Histoplasma macrophage medium (HMM) at 37°C containing 3 μM or 10 μM FeSO4 with or without hygromycin 25 μg ml-1 [18,72]. Cultures grown in liquid HMM at 37°C or 22°C were supplemented with 0, 1, 5, or 10 μM FeSO4. F-12 Ham’s nutrient mixture lacking FeSO4 was used for preparation of HMM. For iron chelation experiments, deferiprone (3-hydroxy-1,2-dimethyl-4(1H)-pyridone; Sigma-Aldrich, Saint Louis, MO) 0.5 mM or 1.0 mM was added to liquid HMM supplemented with 1, 5, or 10 μM FeSO4. Agrobacterium tumefaciens strain LBA1100 harboring the Ti helper plasmid pAL1100 (gift from C. van den Hondel; Leiden University, the Netherlands) was maintained on Luria-Bertani (LB) medium supplemented with 0.1% glucose, spectinomycin 100 ug/ml, and kanamycin 100 ug/ml after transformation with a binary vector [18,73].
Quantification of cell morphology and viability
To quantify the percentage of cells with yeast morphology, germ tube development, and hyphal growth, cells were stained with calcofluor white (Sigma Aldrich) [74]. Calcofluor white staining facilitated differentiation of germ tubes and hyphae by the absence or presence of septae, respectively. Eosin exclusion staining was performed to assess cell viability [18].
Whole genome gene expression microarrays
To compare transcription between SREB∆ and an isogenic WT isolate, 385K whole genome gene expression microarrays (Roche NimbleGen, Inc., Madison, WI) were custom designed for B. dermatitidis using the annotated sequence of strain SLH14081 (9,587 genes; www.broadinstitute.org). A total of 9,583 genes (nuclear and mitochondrial) were represented on each array in quadruplicate with 9,571 genes having a probe-set comprised of 10 unique oligonucleotide probes; 12 genes had less than 10 unique oligonucleotide probes. Oligonucleotide probes with sufficient specificity were unable to be designed for a mitochondrial laglidadg endonuclease (BDBG_17014), mitochondrial ATP synthase F0 subunit 8 (BDBG_17017), mitochondrial NADH dehydrogenase subunit 5 (BDBG_17032), and a predicted protein (BDBG_03106). Empty features on the array were filled with randomly generated probes with similar length and G+C content as the NimbleGen internal controls.
Experimental conditions and isolation of RNA for microarray analysis
Total RNA was isolated from 3 biological replicates of SREB∆ and the isogenic wild-type control at 37°C and 6 hours, 24 hours, and 48 hours following a drop in temperature to 22°C (24 total samples). All cultures were grown in liquid HMM under iron replete conditions (10 μM FeSO4). All glassware was pre-treated with 2N HCl and extensively washed with ddH2O. Prior investigation of SREB demonstrated that transcription was induced in the presence of exogenous iron [18]. In addition, optimal binding of SREB homologs to DNA requires coordination of iron by its cysteine-rich region [29,75]. The phenol-guanidinium thiocyanate-1-bromo-3-chloropropane method was used to extract total RNA [76]. In brief, B. dermatitidis cells were washed with PBS (pre-warmed to 37°C or 22°C), frozen in liquid nitrogen, lysed by grinding in a mortar and pestle, and treated with TRI Reagent followed by 1-bromo-3-chloropropane (Molecular Research Center Inc., Cincinnati, OH). Total RNA was precipitated using a 1 : 1 concentration of isopropanol and high salt solution (Molecular Research Center, Inc., Cincinnati, OH), and washed with 75% ethanol. Following resuspension in water pre-treated with diethyl pyrocarbonate (DEPC; Calbiochem, San Diego, CA), total RNA was treated with turboDNAase (Applied Biosystems/Ambion, Austin Texas), and purified using an RNeasy kit (Qiagen, Valencia, CA). To optimally remove guanidine salts, an extra wash of RPE buffer was performed during RNAeasy purification. RNA quality was assessed using an Agilent bioanalyzer (Agilent Technoligies, Santa Clara, CA). Double-stranded cDNA was synthesized and purified at the Gene Expression Center at the University of Wisconsin–Madison. Roche NimbleGen performed Cy3 labeling, hybridization, scanning, and RMA (Robust Multi-Array analysis) normalization for all 24 microarrays at their facility in Reykjavík, Iceland.
Statistical analysis of microarray data
To identify differentially expressed genes in SREB∆ versus WT at each time point, two different statistical programs were used, LIMMA (Linear Models for Microarray data) and EBarrays (Empirical Bayes Methods for Microarrays) [30,31]. Both programs were executed using R-based software. LIMMA used a Bayesian approach to fit fluorescent intensity data for all genes across the different arrays into a linear model, which was followed by analysis for differential gene expression between SREB∆ and WT using a moderated t-statistic [30]. We used q-values to adjust Limma results in order to remain at a 5% false discovery rate [77]. EBarrays used fluorescent intensity for all genes across all microarrays to model the posterior probability of a single gene to determine whether it had equivalent (µ1 = µ2) or different expression (µ1 µ2) in SREB∆ versus WT [31]. A lognormal-normal model with moderated variance was used in the EBarrays analysis because there were 3 biological replicates for SREB∆ and WT at each time point. An individual gene was considered differentially expressed if statistical significance was met for both LIMMA (q-value < 0.05) and EBarrays (posterior probability > 0.95) analyses. Heat maps were generated using CIMminer from the Genomics and Bioinformatics Group (Laboratory of Molecular Pharmacology, Center for Cancer Research, NCI, NIH, DHHS) (http://discover.nci.nih.gov). Gene expression microarray data have been deposited to the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) (accession number: GSE67724).
Gene ontology (GO) annotation and enrichment analysis
GO terms were assigned to the proteome encoded by the B. dermatitidis genome using InterProScan [78]. The annotation file contained the gene identification number with the associated GO term and GO identification number. Fisher’s exact test was used to test whether the number of DE genes as determined by EBarrays (posterior probability > 0.95) was higher for genes inside the GO category than outside the GO category. Q-values were calculated from the Fisher p-values in order to remain at a 5% false discovery rate [77]. The enrichment test suggests functionality of a group of genes that are differentially expressed under a particular condition.
Weighted gene co-expression network analysis (WGCNA)
We used WGCNA [79] to find clusters (or modules) of highly correlated genes, and for creating a module eigengene to relate modules to one another or to a clinical trait. In the first step of the analysis, we calculated a correlation matrix for the genes, and raised it to a power (β = 12) to create an adjacency matrix of connection strength. The sum of the connection strengths with the other network genes was defined as connectivity, and a topological overlap measure was calculated based on the number of shared neighbors. The dendrogram was created by hierarchical clustering 1 minus the topological overlap, and branches of the tree were cut using a dynamic tree cut algorithm to define modules [80]. Module eigengenes are the first principal component of a module, and can also be thought of as the average gene expression for all genes in a module for a sample. We used allez to determine GO and KEGG enrichment in each module, and calculated correlation between module eigengenes and time points.
Extraction of ferricrocin and reverse-phase high-pressure liquid chromatography (HPLC)
SREB∆ and WT yeast were grown in liquid HMM supplemented with 10 μM FeSO4 at 37°C. Once cells reached stationary growth when measured by spectrophotometric absorbance at 600 nm, they were washed 3 times in 1x phosphate buffered saline (PBS) to remove extracellular siderophores from the medium, flash frozen in liquid nitrogen, ground in a mortar and pestle, resuspended in PBS, and centrifuged to pellet the cellular debris. The supernatant was collected, treated with 2% ferric chloride, and applied to a column packed with Amberlite XAD-2 resin (Supelco, Bellefonte, PA). Resin and column preparation along with siderophore isolation was performed as previously described [18]. Following XAD-2 chromatographic isolation, siderophores were separated on a C18 column (Agilent Eclipse XDB-C18 column; 4.6x150 mm) using a water-acetonitrile gradient containing 0.1% trifluoroacetic acid (Sigma-Aldrich). The gradient of acetonitrile was increased from 5% to 15% over 10 minutes, then to 25% over 22 minutes. Flow rate was 0.75 ml/min and absorbance was measured at 465 nm. Retention times were compared to a ferricrocin siderophore standard (HPLC calibration kit–ferrichromes; EMC microcollections, Tubingen, Germany).
Triphenyltetrazolium chloride (TTC) overlay agar assay
Wild type and SREB∆ were incubated (5 x 105 cells) on iron poor HMM (IPM) and iron-replete (10 μM FeCl3) HMM media supplemented with 50 μM CuSO4 at 37°C for 4 days prior to exposure to 0.1% triphenyltetrazolium chloride (TTC; Sigma-Aldrich) [81,82]. TTC as added to overlay agar (1.5% bactoagar, 0.067 M phosphate buffer, pH 7) after autoclaving.
Isolation and characterization of lipids
SREB∆ and WT were grown at 37°C and 22°C in liquid HMM supplemented with 10 μM FeSO4. Cells were harvested, washed with H2O twice, resuspended in 0.97% KCl, and beaten with beads prewashed with methanol-chloroform (1 : 1). Lipids were extracted using chloroform-methanol (2 : 1) containing 5mg/L butylated hydroxytoluene (Sigma-Aldrich) in organic solvent-washed glassware (no plastics were used). Samples were mixed by vortexing for 3 minutes and incubated at 4°C for 2 hours. The samples were then treated with 0.97% KCl, vortexed for 30 seconds, and incubated at 4°C for 1 hour. Following centrifugation (300 x g for 5 minutes), the organic layer was removed and washed x 3 with 0.97% KCl. The collected lipid extracts were dried under nitrogen.
After drying, samples were reconstituted in 2 : 1 chloroform:methanol and lipid classes were separated by thin layer chromatography (TLC; 20 cm × 20 cm silica gel Si60 plates; Merck KGaA, Darmstadt, Germany) using heptane:isopropyl ether:acetic acid (60 : 40 : 3). After spraying TLC plates with 0.2% 2’,7’-dichlorofluorescein in ethanol, lipid classes (phospholipid, DAG, free fatty acids, TAG, sterol esters) were visualized by UV light and scraped off the TLC plate. Lipids were reextracted overnight at room temperature with chloroform: methanol (4 : 1) containing 100 mg/L butylated hydroxytoluene (Sigma-Aldrich), and centrifuged 850 x g to remove silica gel particles. The internal standard, pentadecanoic acid (100 μl of 0.05 mg/ml in chloroform) was added to each sample and the organic solvents were evaporated under nitrogen. Isolated lipids were then methylated using 0.5 ml 14% boron trifluoride-methanol solution (Sigma-Aldrich), boiled for 30 minutes at 100°C, cooled, mixed with 1 ml hexane, vortexed for 30 seconds, mixed with 0.5 ml H2O and centrifuged (850 x g for 5 minutes) at room temperature. The top hexane layer, which contained methyl ester lipid derivatives, was transferred to a new glass vial, dried under nitrogen, resuspended in 100 μl hexane, and transferred to gas chromatography vials.
Fatty acid methyl esters were identified by gas chromatography using a Hewlett-Packard 6890 (Hewlett Packard, Palo Alto, CA) equipped with a capillary column coated with DB-225 (30 m × 0.25 mm, internal diameter of 0.25 μm) (Agilent Technologies Inc., Wilmington, DE). Column temperature was kept at 70°C for 1 min, increased to 180°C at a rate of 20°C/min and then to 220°C at a rate of 3°C/min. The temperature was kept at 220°C for 15 min. The front inlet was set at 250ºC and the detector temperature was set at 300ºC. Peaks were identified by a comparison of retention times with a set of authentic FA standards (Supelco, Bellefonte, PA). The abundance of FAs was calculated from the relative peak areas [83]. The fatty acid abundance reported reflects the fatty acids present at the end of the protocol.
Sterols were extracted and analyzed according to the protocol described by Zarnowski et al. [84]. Briefly, sterols were reextracted by direct saponification with 1 ml of 6% (w/v) KOH in methanol for 1 h at 90°C. After addition of 1 vol of H2O, the unsaponifiable fraction was extracted three times with 3 volumes of C6H6. The sterols were identified by FAST GC-MS and quantified by the peak area ratio of sterol/internal standard (cholesterol) in the GC-FID chromatogram. The characterization of sterol was performed on a Shimadzu 2010 Plus system equipped with a Shimadzu QP 2010 Ultra mass spectrometer detector. The separation was performed on a Zebron ZB-5MS capillary column (10 m × 0.1 mm i.d., 0.1 μm film thickness, Phenomemex). The system was operated in the constant linear velocity (70 cm/s) using helium as a carrier gas and the sample was injected in the split mode (Split ratio 20 : 1). The GC oven temperature was started at 80 °C, increased to 220 °C at a rate of 60 °C min−1, then to 300 °C at a rate of 20 °C min−1 and finally held to 300 °C for 1.5 min. The injection port, transfer line and ion source temperature were maintained at 285, 300 and 250 °C, respectively. The MS was run in the mode of electron impact (EI) mode with an electron energy of 70 eV and the spectra were scanned in the range of m/z 50–500. The sterols were identified by comparing their mass spectra with the standard mass spectra in the NIST MS library. Additional FID detector was allowed the quantitative analysis of sterol. The FID temperature was 320 °C and the oven temperature program was the same as to the GC–MS operation.
Antifungal drug susceptibility testing
B. dermatitidis SREB∆ and WT yeast (1.2 x 106) were uniformly spread on 6-well plates (Corning Inc., Corning, NY) containing HMM supplemented with 10 μM FeSO4. Antibiotic assay paper discs (6 mm; Whatmann/GE Healthcare Life Sciences, Pittsburgh, PA) were impregnated with 0–10 μg Amphotericin B deoxycholate dissolved in dimethyl sulfoxide (DMSO; Sigma-Aldrich) or 0–1 μg voriconazole dissolved in water (Pfizer, New York, NY), placed in the center of each well, overlaid with 0.5% agarose top agar, and incubated at 37°C. Antifungal susceptibility was assessed by the diameter of the clear zone surrounding the discs impregnated with amphotericin B deoxycholate or voriconazole.
Lipid droplet staining and fatty acid supplementation
Lipid droplets in SREB∆ and WT cells were stained with BODIPY 493/503 (Life Technologies, Carlsbad, CA) at 37°C and 22°C [43]. Cultures were grown in liquid HMM supplemented with 10 μM FeSO4, washed in 1x PBS, and fixed in 2% paraformaldehyde to optimize intracellular entry of BODIPY 493/503. Following fixation, cells were washed and resuspended in 1x PBS. Yeast cells grown at 37°C (and 6-hrs 22°C) were stained with 1 : 50 dilution of 0.5 mM BODIPY 493/503 and cells grown at 22°C (24-hrs and 48-hrs) were stained with a 1 : 25 diluation of 0.5 mM BODIPY 493/503. Optimal image capture of LDs by the microscopy camera (EXi aqua, Qimaging, Surrey, British Columbia, Canada) required a higher concentration of BODIPY 493/503 for WT and SREB∆ cells harvested at 24 and 48-hrs 22°C versus 37°C. Intracellular lipid droplets were visualized using fluorescent microscopy (Olympus BX60; Olympus America, Center Valley, PA) using an excitation filter with a range of 460–490 nm. ImageJ was used to facilitate quantitative LD analysis.
SREB∆ and WT cells grown in iron-replete (10 μM FeSO4) liquid HMM at 37°C were supplemented with 0.5–1.0 mM palmitic acid (16 : 0), palmitoleic acid (16 : 1n7), stearic acid (18 : 0), and oleic acid (18 : 1n9) (Cayman Chemical, Ann Arbor, MI) prior to incubation at 22°C. Palmitic and stearic acid were nearly insoluble in aqueous solutions and required solubilization in DMSO (Sigma-Aldrich) prior to addition to iron-replete liquid HMM (final DMSO concentration was 1.43%). Palmitoleic and oleic acid were soluble in liquid HMM. SREB∆ and WT cells treated with fatty acids along with controls (untreated, DMSO only) were harvested at 37°C and 6, 24, and 48-hrs at 22°C. Harvested cells were washed twice with 1x PBS, fixed in 2% paraformaldehyde, and stained with either BODIPY 493/503 or calcofluor for analysis of LDs and morphology, respectively.
Chromatin immunoprecipitation with quantitative real-time PCR (ChIP-qPCR)
To perform ChIP, we engineered SREB to contain a 3x-HA tag located in-frame at the C-terminus. Following PCR amplification of the SREB coding region using primers SREB_F1 and 3xHA_RC (S7 Table), the PCR product was purified using QIAquick gel extraction kit (Qiagen, Valencia, CA), sequenced by the DNA Sequencing Laboratory at the University of Wisconsin Biotechnology Center, and ligated into a vector that contained the native SREB promoter and 3’ UTR to create pBTS47-SREB-3xHA. Agrobacterium tumefaciens-mediated transformation was used to transform B. dermatitidis strain 26199 [73]. Transformants were selected on HMM supplemented with 25 ug/ml nourseothricin (Werner Bioagents, Jena, Germany). Western blot analysis using a ChIP-grade rabbit polyclonal HA antibody (Abcam, Cambridge, MA) was performed to assess specificity of the antibody for SREB-3xHA (S8 Fig). In brief, wild-type and SREB-3xHA transformed cells were beaten with beads, suspended in 1x SDS-PAGE loading buffer, boiled for 10 minutes, and loaded onto an SDS-PAGE gel (6% stacking, 10% resolving) for separation by electrophoresis. Separated proteins were transferred to Immobilon-P PVDF membrane (Millipore), probed with primary (rabbit polyclonal HA antibody; 1 : 1000 dilution) and secondary (1 : 2000 dilution of peroxidase affinipure goat anti-rabbit IgG (H+L); Jackson ImmunoResearch Laboratories, West Grove, PA), developed using ECL western blotting substrate (Pierce, Rockford, IL), and imaged using VersaDoc imaging systems (Bio-Rad). To confirm the SREB-3xHA construct was functional, SREB∆ was transformed with pBTS47-SREB-3xHA using A. tumefaciens-mediated DNA transfer.
For ChIP-qPCR, we modified the Caenorhabditis elegans protocol published by Mukhopadhyay et al. [85]. B. dermatitidis SREB-3xHA and WT cells grown in liquid HMM supplemented with 10 μM FeSO4 at 37°C and 22°C were treated with 1% formaldehyde (Sigma-Aldrich) for 30 minutes to crosslink HA-tagged SREB to its DNA binding site. Formaldehyde crosslinking was quenched with 0.125 M glycine for 5 minutes. Cells were washed twice in ice-cold 1x PBS, washed once in ice-cold 1x PBS containing protease inhibitor cocktail (PIC; Sigma-Aldrich), flash frozen in liquid nitrogen, ground to a fine powder using a mortar and pestle, and resuspended in FA lysis buffer containing PIC plus phenylmethylsulfonyl fluoride (PMSF). The cell lysate was centrifuged 13,600 x g for 15 minutes at 4°C, supernatant discarded, pellet resuspended in FA lysis buffer with PIC plus PMSF, and stored at -80°C. Chromatin was sheared using probe sonication (Misonix S-4000; Qsonica, Newton, CT). Cells grown at 37°C were sonicated for a total of 25 minutes of “on time”; whereas cells at 22°C were sonicated for a total of 15 minutes of “on time”. Each round of sonication consisted of 1 minute of “on time”, which was divided into 15 seconds of sonication (amplitude 30) x 4 with intervening rest periods of 30 seconds. Cells were kept ice cold during sonication and between each round of sonication, the cells were chilled for 10 seconds in a dry ice-ethanol bath to prevent overheating. From each sample, 50 μl was saved as an “input” sample and 50 μl was analyzed for efficacy of chromatin shearing by 0.8% agarose gel electrophoresis.
For ChIP, protein A agarose/salmon sperm DNA beads were prepared according to the manufacturer’s instructions (Millipore, Billerica, MA) and resuspended in FA lysis buffer containing PIC plus PMSF. Sonicated chromatin (300 μg measured by nanodrop) was pre-cleared for 1 hour at 4°C prior to adding 10 μl anti-HA along with freshly prepared beads. Following an overnight incubation at 4°C with orbital rotation, the samples were washed twice with buffer #1 (FA lysis buffer), once with buffer #2 (FA lysis buffer containing 0.5 M NaCl), once with buffer #3 (10 mM Tris pH8, 0.25M LiCl, 1 mM EDTA, 0.5% Nonidet P-40, 0.5% sodium deoxycholate), and thrice with Tris-EDTA pH 8. All washes were 3 minutes at 4°C with orbital rotation. To reverse cross-linking, ChIP’d samples were treated with proteinase K for 2 hours at 45°C, and then 65°C overnight. Input samples were treated with proteinase K at 55°C for 4 hours then 65°C overnight. DNA was isolated and purified according to the protocol in Mukhopadhyay et al., and resuspended in Tris-HCl pH 8 [85]. For qPCR, primers were designed against the GATA motif and coding region of target genes (S7 Table). Potential GATA motifs were identified by MAST analysis [18]. Fold enrichment was determined by calculation of ∆Ct of SREB-3xHA and WT (∆Ct = ChIP Ct–input Ct) followed by 2^-∆∆CT (∆∆CT = ∆Ct SREB-3xHA - ∆Ct WT) [85].
ChIP with DNA sequencing (ChIP-seq)
For ChIP-seq, chromatin from yeast cells grown under iron-replete conditions (10 μM FeSO4) was prepared, sheared and immunoprecipitated similar to the protocol described for ChIP-qPCR; however, protein A agarose beads without salmon sperm DNA were used (otherwise the salmon sperm DNA would be sequenced in addition to B. dermatitidis DNA). Immunoprecipitated DNA (along with input DNA) from WT and SREB-3xHA strains were submitted to the University of Wisconsin Biotechnology Center for sequencing using Illumina HiSeq 2000 platform. Two biological replicates were analyzed for WT and SREB-3xHA. We aligned the ChIP and input reads using Bowtie (version 0.12.7 with parameters “-p 4-q-v 2-a—best—strata-m 2”) to the genome sequence of Blastomyces dermatitidis (blastomyces_dermatitidis_atcc_26199_1_supercontigs.fasta downloaded from www.broadinstitute.org/annotation/genome/blastomyces_dermatitidis/MultiDownloads.html [86–88]. We performed quality control analysis of the data using the quality metrics from Landt et al. [89]. We then used the R package mosaics in the two sample analysis with input data mode to call peaks [88]. The specific parameters of mosaics were set as follows: bin size = 150; fragment size = 150, fdr = 0.1; thres = 97.5th percentile of the bin-level ChIP read count distribution. This approach identified 37 26199-specific, 847 3xHA-specific, and 242 common peaks. We further filtered these peaks by taking into account the ChIP signal at the summit. Specifically, for each peak, we considered a window size of 1000 bps around the mid-point of the original peak and defined the highest read coverage point as the summit of the peak. We further filtered peaks with less than 20 reads per one million reads at the summit. We annotated the resulting peaks by overlapping them with the 500 bps upstream of the transcription start sites using the 9180 TSS groups defined in blastomyces_dermatitidis_atcc_26199_1_transcripts.gtf (www.broadinstitute.org). ChIP-seq data have been deposited to the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) (accession number: GSE68680).
B. dermatitidis overexpression strains
Plasmid pLBZ1, which was a gift from Dr. Jon Woods, was used to generate pBTS47-Woods to overexpress candidate genes in B. dermatitidis. pLBZ1 contains the H. capsulatum H2B promoter and a CatB terminator with intervening AscI and SbfI restriction sites. The H2B promoter through the CatB terminator in pLBZ1 was amplified using forward and reverse primers containing ClaI and BamHI restriction site sequences, respectively. The amplicon was separated by 1% agarose gel electrophoresis, purified using a Qiaquick PCR purification kit (Qiagen, Valencia, CA), digested with ClaI and BamHI-HF (New England Biolabs, Ipswich, MA), and ligated into pBTS47 using T4 DNA ligase (Promega, Madison, WI) resulting in pBTS47-Woods. pBTS47 contains a nousethricin resistance cassette and was a gift from Dr. Bruce Klein. Open reading frames of candidate genes were amplified using forward and reverse primers containing AscI and SbfI restriction site sequences, respectively (S7 Table). The amplicons were separated by gel electrophoresis, purified by QiaQuick PCR purification kit, digested with AscI and SbfI, and ligated into pBTS47-Woods using T4 DNA ligase. A. tumefaciens-mediated DNA transfer was used to transform B. dermatitidis strain 26199 with pBTS47-Woods containing the candidate gene of interest (S7 Table). Transformants were selected on HMM supplemented with 25 μg/ml nourseothricin and 200 mM cefotaxime.
Quantitative real-time PCR
Total RNA was extracted from B. dermatitidis cells grown in liquid HMM supplemented with 10 μM FeSO4 at 37°C and 22°C as described in the microarray section. One microgram of DNase-treated total RNA was converted to cDNA using iScript cDNA synthesis kit (Bio-Rad). Quantitative real-time PCR was performed with SsOFast EvaGreen Supermix (Bio-Rad) using a MyiQ real-time PCR detection system (Bio-Rad). Reactions were performed in triplicate using the following conditions: 1 cycle 95°C x 30 sec, followed by 40 cycles at 95°C for 5 sec, 60°C for 10 sec. Primers used for qRT-PCR are listed in S7 Table. Transcript abundance of genes involved with iron assimilation at 37°C and HAPX were normalized relative to the transcript abundance of GAPDH, whereas genes involved with glycerolipid biosynthesis analyzed at 37°C and 48-hrs 22°C were normalized to L34. For analyzing gene transcription at two different temperatures, 37°C and 22°C, L34 transcript abundance demonstrated the most stability (versus GAPDH and alpha-tubulin). Relative expression (RE) was calculated as RE = 2-∆Ct, ∆Ct = Cttarget gene−CtGAPDH or L34 [90].
Supporting Information
Zdroje
1. Gauthier GM & Klein BS. Insights into fungal morphogenesis and immune evasion. Microbe. 2008; 3 : 416–423. 20628478
2. Nemecek JC, Wüthrich M, Klein BS. Global control of dimorphism and virulence in fungi. Science. 2006;312 : 583–588. 16645097
3. Finkel-Jimenez B, Wüthrich M, Klein BS. BAD1, an essential virulence factor of Blastomyces dermatitidis, suppresses host TNF-alpha production through TGF-beta-dependent and–independent mechanisms. J Immunol. 2002;168 : 5746–5755. 12023375
4. Chapman SW, Dismukes WE, Proia LA, Bradsher RW, Pappas PG, et al. Clinical practice guidelines for the management of blastomycosis: 2008 update by the Infectious Diseases Society of America. Clin Infect Dis 2008;46 : 1801–1812. doi: 10.1086/588300 18462107
5. Gauthier GM, Safdar N, Klein BS, Andes DR. Blastomycosis in solid organ transplant recipients. Transpl Infect Dis. 2007;9 : 310–317. 17428278
6. Fisher MC, Koenig GL, White TL, San-Blas G, Negroni R, et al. Biogeographic range expansion into South America by Coccidioides immitis mirrors New World patterns of human migration. Proc Natl Acad Sci USA. 2001;98 : 4558–4562. 11287648
7. Nguyen VQ and Sil A. Temperature-induced switch to the pathogenic yeast form of Histoplasma capsulatum requires Ryp1, a conserved transcriptional regulator. Proc Natl Acad Sci USA 2008;105 : 4880–4885. doi: 10.1073/pnas.0710448105 18339808
8. Mahvi TA. A comparative study of the yeast and mycelial phases of Histoplasma capsulatum. I. pathways of carbohydrate dissimilation. J Infect Dis. 1965;115 : 226–232. 14331704
9. Kanetsuna F, and Carbonell LM. Enzymes in glycolysis and the citric acid cycle in the yeast and mycelial forms of Paracoccidioides brasiliensis. J Bacteriol. 1966;92 : 1315–1320. 5924267
10. Arraes EF, Benoliel B, Burtet RT, Costa PL, Galdino AS, et al. General metabolism of the dimorphic and pathogenic fungus Paracoccidioides brasiliensis. Gen Mol Res. 2005;4 : 290–308.
11. Domer JE, and Hamilton JG. The readily extracted lipids of Histoplasma capsulatum and Blastomyces dermatitidis. Biochim Biophys Acta. 1971; 231 : 465–478. 5089693
12. Toledo MS, Levery SB, Suzuki E, Straus AH, Takahashi HK. Characterization of cerebrosides from the thermally dimorphic mycopathogen Histoplasma capsulatum: expression of 2-hydroxy fatty N-acyl (E)-Δ3-unsaturation correlates with the yeast-mycelium phase transition. Glycobiology. 2001;11 : 113–124. 11287398
13. Kanetsuna F, and Carbonell LM. Cell wall composition of the yeast-like and mycelial forms of Blastomyces dermatitidis. J Bacteriol 1971;106 : 946–948. 5557599
14. Webster RH and Sil A. Conserved factors Ryp2 and Ryp3 control cell morphology and infectious spore formation in the fungal pathogen Histoplasma capsulatum. Proc Natl Acad Sci USA. 2008;105 : 14573–14578. doi: 10.1073/pnas.0806221105 18791067
15. Beyhan S, Gutierrez M, Voorhies M, Sil A. A temperature-responsive network links cell shape and virulence traits in a primary fungal pathogen. PLoS Biol. 2013;11 : 31001614.
16. Bugeja HE, Hynes MJ, Andrianopoulos A. HgrA is necessary and sufficient to drive hyphal growth in the dimorphic pathogen Penicillium marneffei. Mol Microbiol. 2013;88 : 998–1014. doi: 10.1111/mmi.12239 23656348
17. Todd RB, Greenhalgh JR, Hynes MJ, Andrianopoulos A. TupA, the Penicillium marneffei Tup1p homolog, represses both yeast and spore development. Mol Microbiol. 2003;48 : 85–94. 12657047
18. Gauthier GM, Sullivan TD, Gallardo SS, Brandhorst TT, Vanden Wymelenberg AJ, et al. SREB, a GATA transcription factor that directs disparate fates in Blastomyces dermatitidis including morphogenesis and siderophore biosynthesis. PLoS Pathog. 2010;6: e1000846. doi: 10.1371/journal.ppat.1000846 20368971
19. Hwang LH, Seth E, Gilmore SA, Sil A. SRE1 regulates iron-dependent and–independent pathways in the fungal pathogen Histoplasma capsulatum. Eukaryot Cell. 2012;11 : 16–25. doi: 10.1128/EC.05274-11 22117028
20. Hilty J, Smulian AG, Newman SL. The Histoplasma capsulatum vacuolar ATPase is required for iron homeostasis, intracellular replication in macrophages, and virulence in a murine model of histoplasmosis. Mol Microbiol. 2008;70 : 127–139. doi: 10.1111/j.1365-2958.2008.06395.x 18699866
21. Gilmore SA, Naseem S, Konopka JB, Sil A. N-acetylglucosamine (GlcNAc) triggers a rapid, temperature-responsive morphogenetic program in thermally dimorphic fungi. PLoS Genetics. 2013;9: e1003799. doi: 10.1371/journal.pgen.1003799 24068964
22. Chandarlapaty S and Errede B. Ash1, a daughter cell-specific protein, is required for pseudohyphal growth of Saccharomyces cerevisiae. Mol Cell Biol. 1998;18 : 2884–2891. 9566907
23. Long RM, Singer RH, Meng X, Gonzalez I, Nasmyth K, Jansen RP. Mating type switching in yeast controlled by asymmetric localization of ASH1 mRNA. Science. 1997;277 : 383–387. 9219698
24. Jung WH, Sham A, White R, Kronstad JW. Iron regulation of the major virulence factors in the AIDS-associated pathogen Cryptococcus neoformans. PLoS Biol. 2006; 4: e410. 17121456
25. Jung WH and Kronstad JW. The iron-responsive, GATA-type transcription factor cir1 influences mating in Cryptococcus neoformans. Mol Cells. 2011;31 : 73–7. doi: 10.1007/s10059-011-0011-0 21120626
26. He Q, Cheng P, Yang Y, Wang L, Garnder KH, Lui Y. (2002) White collar-1, a DNA binding transcription factor and a light sensor. Science 297 : 840–843. 12098705
27. Todd RB, Fraser JA, Wong KH, Davis MA, Hynes MJ. Nuclear accumulation of GATA factor AreA in response to complete nitrogen starvation by regulation of nuclear export. Eukaryot Cell. 2005;4 : 1646–53. 16215172
28. Haas H, Zadra I, Stoffler G, Angermayr K. The Aspergillus nidulans GATA factor SREA is involved in regulation of siderophore biosynthesis and control of iron uptake. J Biol Chem. 1999;274 : 4613–9. 9988696
29. Chao LY, Marletta MA, Rine J. Sre1, an iron-modulated GATA DNA-binding protein of iron-uptake genes in the fungal pathogen Histoplasma capsulatum. Biochemistry. 2008;47 : 7274–7283. doi: 10.1021/bi800066s 18549241
30. Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Statistical Applications in Genetics and Molecular Biology 2004;3: Article 3.
31. Kendziorski CM, Newton MA, Lan H, Gould MN. On parametric empirical Bayes methods for comparing multiple groups using replicated gene expression profiles. Stat Med. 2003;22 : 3899–3914. 14673946
32. Kumme J, Dietz M, Wagner C, Schuller HJ. Dimerization of yeast transcription factors Ino2 and Ino4 is regulated by precursors of phospholipid biosynthesis mediated by Opi1 repressor. Curr Genet. 2008;54 : 35–45. doi: 10.1007/s00294-008-0197-7 18542964
33. Howard DH. Acquisition, transport, and storage of iron by pathogenic fungi. Clin Microbiol Rev. 1999;23 : 394–404.
34. Giles SS and Czuprynski CJ. Extracellular calcium and magnesium, but not iron, are needed for optimal growth of Blastomyces dermatitidis yeast form cells in vitro. Clin Diagn Lab Immunol. 2004;11 : 426–429. 15013999
35. Stearman R, Yuan DS, Yamaguchi-Iwai Y, Klausner RD, Dancis A. A permease-oxidase complex involved in high-affinity iron uptake in yeast. Science. 1996;271 : 1552–1557. 8599111
36. Jung WH, Sham A, Lian T, Singh A, Kosman DJ, Kronstad JW. Iron source preference and regulation of iron uptake in Cryptococcus neoformans. PLoS Pathog. 2008; 4: e45. doi: 10.1371/journal.ppat.0040045 18282105
37. Knight SA, Vilaire G, Lesulsse E, Dancis A. Iron acquisition from transferrin by Candida albicans depends on the reductive pathway. Infect Immun. 2005;73 : 5482–5492. 16113264
38. Schrettl M, Bignell E, Kragl C, Joechl C, Rogers T, et al. Siderophore biosynthesis but not reductive iron assimilation is essential for Aspergillus fumigatus virulence. J Exp Med. 2004;200 : 1213–1219. 15504822
39. Labbé S, Khan MG, Jacques JF. Iron uptake and regulation in Schizosaccharomyces pombe. Curr Opin Microbiol. 2013;16 : 669–676. doi: 10.1016/j.mib.2013.07.007 23916750
40. Zarnowski R, Cooper KG, Brunold LS, Calaycay J, Woods JP. Histoplasma capsulatum secreted gamma-glutamyltransferase reduces iron by generating an efficient ferric reductant. Mol Microbiol 2008;70 : 352–368. doi: 10.1111/j.1365-2958.2008.06410.x 18761625
41. Czabany T, Wagner A, Zweytick D, Lohner K, Leitner E, et al. Structural and biochemical properties of lipid particles from the yeast Saccharomyces cerevisiae. J Biol Chem. 2008;283 : 17065–17074. doi: 10.1074/jbc.M800401200 18430725
42. Kohlwein SD, Veenhuis M, van der Klei IJ. Lipid droplets and peroxisomes: key players in cellular lipid homeostasis or a matter of–fatstore ‘em up or burn ‘em down. Genetics. 2013;193 : 1–50. doi: 10.1534/genetics.112.143362 23275493
43. Radulovic M, Knittelfelder O, Cristobal-Sarramian A, Kolb D, Wolinski H et al. The emergence of lipid droplets in yeast: current status and experimental approaches. Curr Genet. 2013;59 : 231–242. doi: 10.1007/s00294-013-0407-9 24057105
44. Manickam E, Sinclair AJ, Cameron-Smith D. Suppressive actions of eicosapentaenoic acid on lipid droplet formation in 3T3-L1 adipocytes. Lipids Health Dis. 2010;9 : 57. doi: 10.1186/1476-511X-9-57 20525346
45. Natter K and Kohlwein SD. Yeast and cancer cells–common principles in lipid metabolism. Biochem Biophys Acta. 2013;1831 : 314–326. doi: 10.1016/j.bbalip.2012.09.003 22989772
46. Komachi K, Johnson AD. Residues in the WD repeats of Tup1 required for interaction with alpha2. Mol Cell Biol. 1997;17 : 6023–6028. 9315661
47. DeSilva H, Lee K, Osley MA. Functional dissection of yeast Hir1p, a WD repeat-containing transcriptional corepressor. Genetics. 1998;148 : 657–667. 9504914
48. Hortschansky P, Eisendle M, Al-Abadallah Q, Schmidt AD, Bergmann S et al. Interaction of HapX with the CCAAT-binding complex—a novel mechanism of gene regulation by iron. EMBO J. 2007;26 : 3157–3168. 17568774
49. Schrettl M, Beckmann N., Varga J., Heinekamp T, Jacobsen ID et al. HapX-mediated adaptation to iron starvation is crucial for virulence of Aspergillus fumigatus. PLoS Pathog. 2010;6: e1001124. doi: 10.1371/journal.ppat.1001124 20941352
50. Chen C, Kalyan P, French SD, Tuch BB, Noble SM. An iron homeostasis regulatory circuit in Candida albicans commensalism and pathogenesis. Cell Host Microbe 2011;10 : 118–135. doi: 10.1016/j.chom.2011.07.005 21843869
51. Krajaejun T, Gauthier GM, Rappleye CA, Sullivan TD, Klein BS. Development and application of a green fluorescent protein sentinel system for identification of RNA interference in Blastomyces dermatitidis illuminates the role of septin in morphogenesis and sporulation. Eukaryot Cell. 2007;6 : 1299–1309. 17496124
52. Schrettl M, Kim HS, Eisendle M, Kragl C, Nierman WC, et al. SreA-mediated iron regulation in Aspergillus fumigatus. Mol Microbiol. 2008;70 : 27–43. doi: 10.1111/j.1365-2958.2008.06376.x 18721228
53. Lan CY, Rodarte G, Murillo LA, Jones T, Davis RW, et al. Regulatory networks affected by iron availability in Candida albicans. Mol Microbiol. 2004;53 : 1451–1469. 15387822
54. Roy I and Landau JW. Composition of the alkali resistant cell wall material of dimorphic Blastomyces dermatitidis. Sabouraudia. 1972;10 : 107–112. 4557874
55. Kobayashi GS and Guiliacci PL. Cell wall studies of Histoplasma capsulatum. Sabouraudia. 1967;5 : 180–188. 6036225
56. Blatzer M, Schrettl M., Sarg B, Lindner HH, Pfaller K, et al. SidL, an Aspergillus fumigatus transacetylase involved in the biosynthesis of the siderophores ferricrocin and hydroxyferricrocin. Appl Environ Microbiol. 2011;77 : 4959–4966. doi: 10.1128/AEM.00182-11 21622789
57. Labbé S, Pelletier B, Mercier A. Iron homeostasis in the fission yeast Schizosaccharomyces pombe. Biometals. 2007;20 : 523–537. 17211681
58. Zarnowski R and Woods JP. Glutathione-dependent extracellular ferric reductase activities in dimorphic zoopathogenic fungi. Microbiology 2005;151 : 2244–2240.
59. Di Salvo AF and Denton JF. Lipid content of four strains of Blastomyces dermatitidis of different mouse virulence. J. Bacteriol. 1963;85 : 927–931. 14044964
60. Kanetsuna F, Carbonell LM, Moreno RE, Rodriguez J. Cell wall composition of the yeast and mycelial forms of Paracoccidioides brasiliensis. J. Bacteriol. 1969;97 : 1036–1041. 5776517
61. Wilfling F, Wang J, Haas JT, Krahmer N, Gould TJ et al. Triacylglycerol synthesis enzymes mediate lipid droplet growth by relocalizing from ER to lipid droplets. Dev Cell. 2013;24 : 384–399. doi: 10.1016/j.devcel.2013.01.013 23415954
62. Oelkers P, Cromley D, Padamsee M, Billheimer JT. The DGA1 gene determines a second triglyceride synthetic pathway in yeast. J Biol Chem 2002;277 : 8877–8881. 11751875
63. Sorger D, Athenstaedt K, Hrastnik H, Daum G. A yeast strain lacking lipid particles bears a defect in ergosterol formation. J Biol Chem. 2004;279 : 31190–31196. 15155725
64. Wolinski H, Kolb D, Hermann S, Koning RI, Kohlwein SD. A role for seipin in lipid droplet dynamics and inheritance in yeast. J Cell Sci. 2011;124 : 3894–3904. doi: 10.1242/jcs.091454 22100922
65. Maresca B, and Kobayashi G. Changes in membrane fluidity modulate heat shock gene expression and produced attenuated strains in the dimorphic fungus Histoplasma capsulatum. Arch Med Res. 1993;24 : 247–249. 8298273
66. Carratù L, Franceschelli S, Pardini CL, et al. Membrane lipid perturbation modified the set point of the temperature of heat shock response in yeast. Proc Natl Acad Sci USA. 1996;93 : 3870–3875. 8632982
67. Chen C and Noble SM. Post-transcriptional regulation of the Sef1 transcription factor controls the virulence of Candida albicans in its mammalian host. PLoS Pathog. 2012;8: e1002956. doi: 10.1371/journal.ppat.1002956 23133381
68. Singh RP, Prasad HK, Sinha I, Agarwal N, Ntarajan K. Cap2-HAP complex is a critical transcriptional regulator that has dual but contrasting roles in regulation of iron homeostasis in Candida albicans. J Biol Chem. 2011;286 : 25154–25170. doi: 10.1074/jbc.M111.233569 21592964
69. Brandhorst TT, Wüthrich M, Warner T, Klein B. Targeted gene disruption reveals an adhesin indispensable for pathogenicity of Blastomyces dermatitidis. J Exp Med 1999;189 : 1207–1216. 10209038
70. Wüthrich M, Filutowicz HI, Warner T, Klein BS. Requisite elements in vaccine immunity to Blastomyces dermatitidis: plasticity uncovers vaccine potential in immune-deficient hosts. J Immunol. 2002;169 : 6969–6976. 12471131
71. Harvey RP, Schmid ES, Carrington CC, Stevens DA. Mouse model of pulmonary blastomycosis: utility, simplicity, and quantitative parameters. Am Rev Respir Dis. 1978;117 : 695–703 646221
72. Worsham PL and Goldman WE. Quantitative plating of Histoplasma capsulatum without addition of conditioned medium or siderophores. J Med Vet Mycol. 1988;26 : 137–143. 3171821
73. Sullivan TD, Rooney PJ, Klein BS. Agrobacterium tumefaciens integrates transfer DNA into single chromosomal sites of dimorphic fungi and yields homokaryotic progeny from multinucleate yeast. Eukaryot Cell. 2002;1 : 895–905. 12477790
74. Marty AJ and Gauthier GM. Blastomyces dermatitidis septins CDC3, CDC10, and CDC12 impact the morphology of yeast and hyphae, but are not required for the phase transition. Med Mycol. 2013;51 : 93–102. doi: 10.3109/13693786.2012.699685 22783804
75. Jbel M., Mercier A, Pelletier B, Beaudoin J, Labbé S. Iron activates in vivo DNA binding of Schizosaccharomyces pombe transcription factor Fep1 through its amino-terminal region. Eukaryot Cell. 2009;8 : 649–664. doi: 10.1128/EC.00001-09 19252122
76. Sambrook J and Russell DW. Molecular cloning: a laboratory manual, 3rd Ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. pp. 7.4–7.8.
77. Storey JD and Tibshirani R. Statistical significance for genome-wide studies. Proc Natl Acad Sci USA. 2003;100 : 9440–9445. 12883005
78. Zdobnov EM and Apweiler R. InterProScan—an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17 : 847–848. 11590104
79. Zhang B. and Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4: Article 17.
80. Langfelder P., Zhang B., Horvath S. Defining clusters from a hierarchical cluster tree: the Dynamic Tree Cut package for R. Bioinformatics. 2008;24 : 719–720. 18024473
81. Hassett R and Kosman DJ. Evidence for Cu(II) reduction as a component of copper uptake by Saccharomyces cerevisiae. J Biol Chem. 1995;270 : 128–134. 7814363
82. Pelletier B, Beaudoin J, Mukai Y, Labbé S. FEP1, an iron sensor regulating iron transporter gene expression in Schizosaccharomyces pombe. J Biol Chem. 2002;277 : 22950–22958. 11956219
83. Zarnowski R, Miyazaki M, Dobrzyn A, Ntambi JM, Woods JP. Typing of Histoplasma capsulatum strains by fatty acid profile analysis. J Med Microbiol. 2007;56 : 788–797. 17510264
84. Zarnowski R., Jaromin A., Certik M., Czabany T., Fontaine J., Jakubik T., et al. The oil of Adenanthera pavonina L. seeds and its emulsions. Z Naturforsch C. 2004;59 : 321–326. 18998394
85. Mukhopadhyay A, Deplancke B, Walhout AJM, Tissenbaum HA. Chromatin immunoprecipitation by quantitative real-time PCR to study transcription factor binding to DNA in Caenorhabditis elegans. Nat Protocol. 2008;3 : 698–709.
86. Langmead B., Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10: R25. doi: 10.1186/gb-2009-10-3-r25 19261174
87. Kuan P, Chung D, Pan G, Thomson JA, Stewart R, Keles S. A statistical framework for the analysis of ChIP-seq data. J Am Stat Assoc 2011;106 : 891–903.
88. Sun G, Chung D, Liang K, Keles S. Statistical analysis of ChIP-seq data with MOSAiCS. Methods Mol Biol 2013;1038 : 193–212. doi: 10.1007/978-1-62703-514-9_12 23872977
89. Landt SG, Marinov GK, Kundaje A et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res 2012; 22 : 1813–1831. doi: 10.1101/gr.136184.111 22955991
90. Livak KF, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-∆∆Ct method. Methods. 2001;25 : 402–408. 11846609
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Clearance of Pneumococcal Colonization in Infants Is Delayed through Altered Macrophage TraffickingČlánek An Model of Latency and Reactivation of Varicella Zoster Virus in Human Stem Cell-Derived NeuronsČlánek Protective mAbs and Cross-Reactive mAbs Raised by Immunization with Engineered Marburg Virus GPsČlánek Specific Cell Targeting Therapy Bypasses Drug Resistance Mechanisms in African TrypanosomiasisČlánek Peptidoglycan Branched Stem Peptides Contribute to Virulence by Inhibiting Pneumolysin ReleaseČlánek HIV Latency Is Established Directly and Early in Both Resting and Activated Primary CD4 T CellsČlánek Sequence-Specific Fidelity Alterations Associated with West Nile Virus Attenuation in Mosquitoes
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2015 Číslo 6- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Introducing “Research Matters”
- Exploring Host–Pathogen Interactions through Biological Control
- Analysis of Bottlenecks in Experimental Models of Infection
- Expected and Unexpected Features of the Newly Discovered Bat Influenza A-like Viruses
- Clearance of Pneumococcal Colonization in Infants Is Delayed through Altered Macrophage Trafficking
- Recombinant Murine Gamma Herpesvirus 68 Carrying KSHV G Protein-Coupled Receptor Induces Angiogenic Lesions in Mice
- TRIM30α Is a Negative-Feedback Regulator of the Intracellular DNA and DNA Virus-Triggered Response by Targeting STING
- Targeting Human Transmission Biology for Malaria Elimination
- Two Cdc2 Kinase Genes with Distinct Functions in Vegetative and Infectious Hyphae in
- An Model of Latency and Reactivation of Varicella Zoster Virus in Human Stem Cell-Derived Neurons
- Protective mAbs and Cross-Reactive mAbs Raised by Immunization with Engineered Marburg Virus GPs
- Virulence Factors of Induce Both the Unfolded Protein and Integrated Stress Responses in Airway Epithelial Cells
- Peptide-MHC-I from Endogenous Antigen Outnumber Those from Exogenous Antigen, Irrespective of APC Phenotype or Activation
- Specific Cell Targeting Therapy Bypasses Drug Resistance Mechanisms in African Trypanosomiasis
- An Ultrasensitive Mechanism Regulates Influenza Virus-Induced Inflammation
- The Role of Human Transportation Networks in Mediating the Genetic Structure of Seasonal Influenza in the United States
- Host Delivery of Favorite Meals for Intracellular Pathogens
- Complement-Opsonized HIV-1 Overcomes Restriction in Dendritic Cells
- Inter-Seasonal Influenza is Characterized by Extended Virus Transmission and Persistence
- A Critical Role for CLSP2 in the Modulation of Antifungal Immune Response in Mosquitoes
- Twilight, a Novel Circadian-Regulated Gene, Integrates Phototropism with Nutrient and Redox Homeostasis during Fungal Development
- Surface-Associated Lipoproteins Link Virulence to Colitogenic Activity in IL-10-Deficient Mice Independent of Their Expression Levels
- Latent Membrane Protein LMP2A Impairs Recognition of EBV-Infected Cells by CD8+ T Cells
- Bank Vole Prion Protein As an Apparently Universal Substrate for RT-QuIC-Based Detection and Discrimination of Prion Strains
- Neuronal Subtype and Satellite Cell Tropism Are Determinants of Varicella-Zoster Virus Virulence in Human Dorsal Root Ganglia Xenografts
- Molecular Basis for the Selective Inhibition of Respiratory Syncytial Virus RNA Polymerase by 2'-Fluoro-4'-Chloromethyl-Cytidine Triphosphate
- Structure of the Virulence Factor, SidC Reveals a Unique PI(4)P-Specific Binding Domain Essential for Its Targeting to the Bacterial Phagosome
- Activated Brain Endothelial Cells Cross-Present Malaria Antigen
- Fungal Morphology, Iron Homeostasis, and Lipid Metabolism Regulated by a GATA Transcription Factor in
- Peptidoglycan Branched Stem Peptides Contribute to Virulence by Inhibiting Pneumolysin Release
- A Macrophage Subversion Factor Is Shared by Intracellular and Extracellular Pathogens
- A Novel AT-Rich DNA Recognition Mechanism for Bacterial Xenogeneic Silencer MvaT
- Reovirus FAST Proteins Drive Pore Formation and Syncytiogenesis Using a Novel Helix-Loop-Helix Fusion-Inducing Lipid Packing Sensor
- The Role of ExoS in Dissemination of during Pneumonia
- IRF-5-Mediated Inflammation Limits CD8 T Cell Expansion by Inducing HIF-1α and Impairing Dendritic Cell Functions during Infection
- Discordant Impact of HLA on Viral Replicative Capacity and Disease Progression in Pediatric and Adult HIV Infection
- Crystal Structure of USP7 Ubiquitin-like Domains with an ICP0 Peptide Reveals a Novel Mechanism Used by Viral and Cellular Proteins to Target USP7
- HIV Latency Is Established Directly and Early in Both Resting and Activated Primary CD4 T Cells
- HPV16 Down-Regulates the Insulin-Like Growth Factor Binding Protein 2 to Promote Epithelial Invasion in Organotypic Cultures
- The νSaα Specific Lipoprotein Like Cluster () of . USA300 Contributes to Immune Stimulation and Invasion in Human Cells
- RSV-Induced H3K4 Demethylase KDM5B Leads to Regulation of Dendritic Cell-Derived Innate Cytokines and Exacerbates Pathogenesis
- Leukocidin A/B (LukAB) Kills Human Monocytes via Host NLRP3 and ASC when Extracellular, but Not Intracellular
- Border Patrol Gone Awry: Lung NKT Cell Activation by Exacerbates Tularemia-Like Disease
- The Curious Road from Basic Pathogen Research to Clinical Translation
- From Cell and Organismal Biology to Drugs
- Adenovirus Tales: From the Cell Surface to the Nuclear Pore Complex
- A 21st Century Perspective of Poliovirus Replication
- Is Development of a Vaccine against Feasible?
- Waterborne Viruses: A Barrier to Safe Drinking Water
- Battling Phages: How Bacteria Defend against Viral Attack
- Archaea in and on the Human Body: Health Implications and Future Directions
- Degradation of Human PDZ-Proteins by Human Alphapapillomaviruses Represents an Evolutionary Adaptation to a Novel Cellular Niche
- Natural Variants of the KPC-2 Carbapenemase have Evolved Increased Catalytic Efficiency for Ceftazidime Hydrolysis at the Cost of Enzyme Stability
- Potent Cell-Intrinsic Immune Responses in Dendritic Cells Facilitate HIV-1-Specific T Cell Immunity in HIV-1 Elite Controllers
- The Mammalian Cell Cycle Regulates Parvovirus Nuclear Capsid Assembly
- Host Reticulocytes Provide Metabolic Reservoirs That Can Be Exploited by Malaria Parasites
- The Proteome of the Isolated Containing Vacuole Reveals a Complex Trafficking Platform Enriched for Retromer Components
- NK-, NKT- and CD8-Derived IFNγ Drives Myeloid Cell Activation and Erythrophagocytosis, Resulting in Trypanosomosis-Associated Acute Anemia
- Successes and Challenges on the Road to Cure Hepatitis C
- BRCA1 Regulates IFI16 Mediated Nuclear Innate Sensing of Herpes Viral DNA and Subsequent Induction of the Innate Inflammasome and Interferon-β Responses
- A Structural and Functional Comparison Between Infectious and Non-Infectious Autocatalytic Recombinant PrP Conformers
- Phosphorylation of the Peptidoglycan Synthase PonA1 Governs the Rate of Polar Elongation in Mycobacteria
- Human Immunodeficiency Virus Type 1 Nef Inhibits Autophagy through Transcription Factor EB Sequestration
- Sequence-Specific Fidelity Alterations Associated with West Nile Virus Attenuation in Mosquitoes
- EBV BART MicroRNAs Target Multiple Pro-apoptotic Cellular Genes to Promote Epithelial Cell Survival
- Single-Cell and Single-Cycle Analysis of HIV-1 Replication
- TRIM32 Senses and Restricts Influenza A Virus by Ubiquitination of PB1 Polymerase
- The Herpes Simplex Virus Protein pUL31 Escorts Nucleocapsids to Sites of Nuclear Egress, a Process Coordinated by Its N-Terminal Domain
- Host Transcriptional Response to Influenza and Other Acute Respiratory Viral Infections – A Prospective Cohort Study
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- HIV Latency Is Established Directly and Early in Both Resting and Activated Primary CD4 T Cells
- Battling Phages: How Bacteria Defend against Viral Attack
- A 21st Century Perspective of Poliovirus Replication
- Adenovirus Tales: From the Cell Surface to the Nuclear Pore Complex
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



