-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaUtilize Host Actin for Efficient Maternal Transmission in
The world’s most common intracellular infection, Wolbachia pipientis, infects 40% of insect species and is currently used to prevent transmission of Dengue by mosquitoes. The bacterium targets the germline of insects, where it is faithfully transmitted to the developing oocyte and the next generation. Here we identify host cytoskeletal proteins required by Wolbachia in order to be efficiently transmitted between Drosophila melanogaster generations. We show that after only two generations in a phenotypically wild type, heterozygous mutant fly, Wolbachia infections are cleared or reduced in titer. Characterization of the mutants suggests that Wolbachia is sensitive to the regulation of actin in the ovary and that actin may be used by Wolbachia to both target and proliferate within host tissues and to be faithfully, maternally transmitted.
Published in the journal: . PLoS Pathog 11(4): e32767. doi:10.1371/journal.ppat.1004798
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1004798Summary
The world’s most common intracellular infection, Wolbachia pipientis, infects 40% of insect species and is currently used to prevent transmission of Dengue by mosquitoes. The bacterium targets the germline of insects, where it is faithfully transmitted to the developing oocyte and the next generation. Here we identify host cytoskeletal proteins required by Wolbachia in order to be efficiently transmitted between Drosophila melanogaster generations. We show that after only two generations in a phenotypically wild type, heterozygous mutant fly, Wolbachia infections are cleared or reduced in titer. Characterization of the mutants suggests that Wolbachia is sensitive to the regulation of actin in the ovary and that actin may be used by Wolbachia to both target and proliferate within host tissues and to be faithfully, maternally transmitted.
Introduction
Wolbachia pipientis is an intracellular α-proteobacterium that forms symbioses with an extremely broad array of hosts, including isopods, nematodes, and insects [1]. Wolbachia were first noted in the tissues of the mosquito, Culex pipiens, by Hertig and Wolbach in 1924, but subsequently, many more insects were found to harbor Wolbachia. Current estimates suggest that upwards of 40% of insect species may be infected by the parasite, making Wolbachia one of the most common intracellular bacteria on the planet [2]. Wolbachia are well known for the reproductive effects induced in the host, which range from the exotic (male killing) to the most common of reproductive effects, cytoplasmic incompatibility (CI) [1]. This recalcitrant, obligate symbiont has received much attention recently due to medical relevance. Wolbachia are heavily studied as potential drug targets for filarial nematode infection [3,4] and are currently being implemented to prevent transmission of Dengue fever from mosquitoes to humans [5,6]. Wolbachia may be one answer to controlling some vector borne human diseases—indeed mosquitoes harboring a virus-blocking strain of Wolbachia are presently being released in underdeveloped parts of the world with this hope in mind [6–8]. Given the ubiquity of Wolbachia in the insect world, and its relevance to human health, it is essential to understand the biological basis of transmission of the symbiont between host generations.
Wolbachia are maternally transmitted bacteria that infect the germline of their hosts such that their transmission fidelity in wild populations is extraordinarily high. Although physiologically stressful conditions are known to induce the loss of superinfections [9], perfect transmission has been measured in control laboratory Drosophila populations as well as in insects harboring transferred Wolbachia infections [10–12]. Localization in the germline, and in the developing oocyte, is critical to Wolbachia’s maternal transmission and in addition, densities in the embryo, and posterior localization, are correlated with reproductive phenotype (e.g. CI) [13,14].
Previous studies have provided some support for Wolbachia interactions with host cytoskeletal elements. Specifically, in Drosophila, Wolbachia require host microtubules and the motors Dynein and Dynactin for anterior localization early in development and Kinesin-1 for posterior localization in mid oogenesis, positioning them for inclusion in the germline [15,16]. This localization is thought to be crucial to the bacterium’s faithful transmission to subsequent generations at the appropriate densities. Additionally, Wolbachia use astral microtubules during asymmetric divisions in the developing embryo, leading to the widespread, but uneven, pattern of localization of the bacteria in adult tissues [17]. In both worms and flies, Wolbachia undergo somatic cell to germline transmission, suggesting an ability for the bacterium to alter the host actin cytoskeleton to facilitate uptake by germ cells [18,19]. More recently, work has suggested interactions between Wolbachia proteins from the Brugia malayi symbiont and host actin [20], although Wolbachia ultrastructure in Brugia does not reveal any obvious mechanism (such as actin comet tails produced during infection in other Rickettsiales) [21]. These previous studies have relied on microscopy and in vitro biochemistry and until now, no genetic evidence of interaction between Wolbachia and actin has been reported.
Here we present data showing that Wolbachia persistence and transmission within Drosophila melanogaster is sensitive to mutations affecting the actin cytoskeleton. The importance of actin during Wolbachia infection was investigated by acquiring Drosophila mutants in actin binding proteins, both involved in the regulation of F-actin filaments: the homologs of profilin (chickadee), which regulates the formation of filamentous actin, and villin (quail), which bundles actin filaments. We show that flies heterozygous for mutations in profilin (chic221/+ and chic1320/+) or villin (qua6-396/+) lose Wolbachia infection after only a few generations. Importantly, the effect is due to both an inability of Wolbachia to efficiently colonize germaria in heterozygous mutant hosts and by a reduction in titer when the host is infected. Importantly, both the less severe chic allele (chic1320), known to decrease an oocyte specific isoform of Drosophila profilin chickadee [22], as well as the null chic allele (chic221) produced a Wolbachia titer phenotype. We identified two different actin binding proteins (profilin and villin) that affect Wolbachia transmission and maintenance, supporting the conclusion that Wolbachia persistence within the host is sensitive to actin.
Materials and Methods
Drosophila stocks
Standard methods were used for all crosses and culturing. The following stocks were obtained from the Bloomington Drosophila Stock Center (BDSC) at Indiana University (http://flystocks.bio.indiana.edu/): stock number 145, which carries w1 was used as the Wolbachia infected control line. Two chickadee mutant fly stocks were used in this study. The chic221 cn1/CyO; ry506 flies carry a null recessive allele resulting from the deletion of 5’ non-coding and some chic-coding sequences [22]. The P{PZ}chic01320 cn1/CyO; ry506 flies carry a strong homozygous infertile loss-of-function allele in chickadee, generated by P-element insertion [23]. The quail mutant flies, qua6-396/SM1, carry a female sterile, recessive mutation induced by ethyl methanesulfonate [24]. We also utilized two chromosomal deficiency stocks: #9507, w1118; Df(2L)BSC148/CyO, is a chromosomal deletion of segments 36C8-36E3, covering the region containing the quail locus. The second of these stocks #24377, w1118; Df(2L)BSC353/CyO, covers segments 26A3-26B3, the region containing the chic locus. Both of these chromosomal deletions are part of the aberration stock collection and were created by FLP-mediated recombination between FRT-bearing transposon insertions [25]. Wolbachia were introduced into the heterozygous mutant backgrounds through crosses between w1 infected females (stock 145) and uninfected heterozygous males (mutant/CyO). In order to control for genetic background, we also created isogenized lines by backcrossing stock 145 and each mutant line to an uninfected w; Sco/Cyo stock for three generations (as per [26], S1 Fig). We used sibling controls to identify Wolbachia titer differences related to genotype.
In addition to these isogenized lines, and to examine the effect on Wolbachia titer of profilin knockdown during development, we utilized a fly stock carrying a UAS inducible profilin-specific short hairpin silencing trigger (RNAi; stock #34523, genotype y1 sc* v1; P{TRiP.HMS00550}attP2) [27]. In order to test the effect of induction on fly development (to recapitulate the developmental lethality of the profilin null) we crossed homozygous females from this line to w; P{w+, Act GAL4} /TM3 males. In order to knock down profilin, we then crossed homozygous females from this line to a homozygous Hsp70:Gal4 driver (a generous gift from Brian Calvi). An additional control for expression from the Hsp70:Gal4 driver included a UAS:GFP stock (also a gift from Brian Calvi). Flies were shocked at 37C for 10 minutes to induce the short hairpin. Wobachia infection status for stocks acquired from the BDSC was determined via PCR and Western blot targeting the gene wsp or its product (see methods below). All flies examined for Wolbachia infection in the experiments below were age matched in order to avoid confounding correlations between fly age and Wolbachia titer.
Western blots
Flies were ground in 1.5ml centrifuge tubes using an electric hand drill and disposable pestle in lysis buffer: 150mM NaCl, 1% Triton X-100, 50mM TrisHCl (pH8) containing HALT protease inhibitor cocktail (Thermo Scientific) and 5 mM EDTA. The lysates were centrifuged for 1 minute at 8000 X g to pellet debris. Samples were heated for 5 minutes at 95°C in Laemmli sample buffer containing 5% β-mercaptoethanol (Bio-Rad) prior to SDS-PAGE electrophoresis. Proteins were separated on 4–20% Tris-Glycine NB precast gels (NuSep) in 1X Tris/Glycine/SDS running buffer (Bio-Rad) and transferred to PVDF membrane in Tris-Glycine transfer buffer with 15% methanol at 40v on ice for 3–4 hours. The membrane was blocked for 5 minutes in Starting Block T20 (TBS) Blocking Buffer (Thermo Scientific), followed by incubation in primary antibody (for 1 hour at RT or O/N at 4°C) according to standard protocols. SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific) was used according to the manufacturer’s instructions to detect HRP (after incubation with secondary antibodies) on the immunoblots. Blots were re-probed after stripping in 100mM Glycine, 0.15 ND-40, 1% SDS, pH 2 for 1 hour at RT, then overnight at 4°C. PageRuler prestained protein ladder (Thermo Scientific) was used as a molecular mass marker. The following antibody was obtained through BEI Resources, NIAID, NIH: Monoclonal Anti-Wolbachia Surface Protein (WSP), NR-31029, and used at a dilution of 1 : 1000. Additionally, we used anti-actin monoclonal at 1 : 10,000 (Seven Hills Bioreagents) as a loading control as well as secondary antibodies: HRP enzyme conjugates (Invitrogen) at 1 : 5000. Densitometry measures were made in ImageJ using scanned film with same exposure times across multiple experiments. Control and experimental flies were included on the same blot in order to ensure consistencies in measured ratios.
Immunohistochemistry, fluorescence in situ hybridization, and microscopy
Immunohistochemistry was performed as follows: ovaries for immunolocalization were dissected in Ringer’s solution 3–5 days after fly eclosion, then fixed as previously described [28] with following modification: 6% formaldehyde devitellinizing buffer was replaced with 5.3% paraformaldehyde in same (Electron Microscopy Sciences). After a series of washes in PBS buffer, ovaries were blocked with 0.5% BSA in PBST for 10 min. The monoclonal anti-Heat Shock Protein 60 (HSP60), clone LK2, H 3524 (Sigma) was diluted 1 : 150 in PBST with 1% BSA or a custom antibody created against full length Wolbachia FtsZ was diluted 1 : 150 in PBST with 1% BSA. Cy3 conjugated to goat anti-mouse secondary antibody (Jackson Immunoresearch) or rabbit secondary antibody (Jackson Immunoresearch) diluted 1 : 250 in PBST + BSA was used to detect the primary antibody. For F-actin detection we used Acti-stain 488 Fluorescent Phalloidin (Cytoskeleton, Inc). Tissues were mounted in Slow Fade “Gold” antifade reagent (Invitrogen) and stored at 4°C.
To confirm staining by immunohistochemistry, we also used fluorescent in situ hybridization, following published protocols [18] with the following modifications: post-fixation in 4% paraformaldehyde in DEPC treated PBS, ovaries were dehydrated in methanol and stored overnight at -20°C. In the morning, washes in DEPC-PBST preceded a 5 minute proteinase K treatment (0.05 mg/mL) at 37C before prehybridization in hyb buffer (50% formamide, 5X SSC, 250 mg/L SS DNA, 0.5x Denhardts, 20 mM Tris-HCl and 0.1% SDS). Universal bacterial probe EUB338 conjugated to Alexa488 (Molecular Probes) was used to detect Wolbachia in the ovarioles. Hybridized ovaries were mounted in Slow Fade “Gold” antifade reagent (Invitrogen).
Images were taken as Z-series stacks at 1.5 um intervals using a Nikon E800 fluorescent microscope with 40x oil objective and processed using Metamorph imaging software (Molecular Devices). Care was taken such that exposure times were normalized across all experiments. For quantification of Wolbachia and F-actin within the germarium z-sections maximum projections were used and regions of the germarium demarcated using masks (S2 Fig). We were careful to exclude the peritoneal sheath for F-actin quantification and for Z-stacks where the sheath was difficult to exclude (due to placement of the sections), the images were not included in the F-actin quantification. Germaria showing aggregates of Wolbachia were scored based on a striking pixel intensity in the presumed somatic stem cell niche.
DNA and RNA extractions and polymerase chain reactions
DNA was extracted from flies utilizing the Qiagen DNeasy Blood and Tissue Kit (Qiagen) according to directions with the following modification. Flies were ground in a 1.5ml centrifuge tube using a disposable pestle and an electric hand drill in 180 ul PBS, 200 ul ALT buffer, and 20 ul Proteinase K solution. The samples were incubated at 56°C for 10 minutes with vigorous shaking and then centrifuged briefly to pellet debris before continuing with the ethanol precipitation in the kit protocol. DNAs were quantified by measuring absorbance at 260nm using an Epoch spectrophotometer (Biotek). Semi-quantitative PCR was performed by standardizing the amount of DNA in each reaction. We utilized Phusion High Fidelity PCR Master Mix with HF buffer (New England Biolabs). The protocol for amplification was: 98°C for 3 minutes, followed by 25 cycles of 98°C for 10 seconds, 56°C for 45 seconds, 72°C for 1 minute 30 seconds with a final 10 minute extension at 72°C. Primers were as follows: wsp F1 5’-GTC CAA TAR STG ATG ARG AAA C—3’ and wsp R1 5’ - CYG CAC CAA YAG YRC TRT AAA -3’ [29]. RNA and DNA were extracted from individual flies or pupae using a modified Trizol extraction protocol. Briefly, 500 uL of Trizol was added to flies and samples homogenized using a pestle. After a 5 minute incubation at room temperature, a 12,000 rcf centrifugation (at 4C for 10 min) was followed by a chloroform extraction. Aqueous phase containing RNA was extracted a second time with phenol:chloroform before isopropanol precipitation of RNA. This RNA pellet was washed and resuspended in The RNA Storage Solution (Ambion). DNA extraction from the same flies or pupae was performed using ethanol precipitation of the organic phase during the first chloroform extraction. Quantitative PCR was performed on the DNA to detect the Wolbachia titer (with reference to the host) using an Applied Biosystems StepOne Real-time PCR system and SybrGreen chemistry (Applied Biosystems). We used wsp primers for Wolbachia (Forward: CATTGGTGTTGGTGTTGGTG; Reverse: ACCGAAATAACGAGCTCCAG) and Rpl32 primers for the host (Forward: CCGCTTCAAGGGACAGTATC; Reverse: CAATCTCCTTGCGCTTCTTG) at the following temperatures: 95°C for 10 min, then 40 cycles of 95°C for 15 seconds and 60°C for 1 minute. To detect number of profilin transcripts we utilized the RNA extracted from these flies and the SensiFAST SYBER Hi-ROX One-step RT mix (Bioline) and the following primer set: chicF: TGCACTGCATGAAGACAACA, chicR: GTTTCTCTACCACGGAAGCG (FlyPrimerBank, DRSC). Reactions were performed in a 96-well plate and calibration standards were used in every run to calculate primer efficiencies. These efficiencies, along with the CT values generated by the machine, were used to calculate the relative amounts of Wolbachia using the ΔΔ Ct (Livak) and Pfaffl methods [30].
F and G-actin quantification
In order to identify the ratio of filamentous to globular actin in ovaries from age matched flies, we used ultracentrifugation coupled to SDS-PAGE and Western blots using an in vivo F/G actin assay kit (Cytoskeleton, Inc). Age-matched, virgin female flies from chic221/Cyo or control (stock #145) were dissected in LAS2 buffer at 37°C and incubated for 10 minutes at 37°C. A brief 300 g centrifugation step (5 minutes) was followed by a 1 hour ultracentrifugation at 100,000 g at 37°C. Supernatants containing globular actin were removed and pellets resuspended in actin depolymerization buffer on ice, by pipetting up and down every 15 minutes for 1 hour. Pellets containing F-actin fractions and supernatants containing G-actin fractions were run on an SDS-PAGE gel and Western blots performed (as above) using a primary mouse monoclonal anti-actin antibody. Bands were quantified using densitometric analysis in ImageJ (as above).
Results
Wolbachia infection is lost or reduced in fly mutants heterozygous for actin binding proteins chickadee and quail
All three actin binding protein mutant fly stocks used in this study were uninfected with Wolbachia upon receipt from the Bloomington Drosophila Stock Center. In order to establish an infection in the flies, infected control females were crossed with mutant uninfected males to generate F1 progeny, half of which carried the mutation, and half of which carried the Cyo balancer (a second chromosome containing inversion breakpoints and a dominant visible mutation of curly wings). F1 heterozygous mutants for the actin binding protein alleles were then back-crossed to the paternal mutant line (mutant/Cyo) and F2 progeny from that cross, carrying the mutation and harboring straight wings, were collected. We screened both the F1 and F2 progeny for Wolbachia infection using PCR against the Wolbachia surface protein gene (wsp) (Fig 1A). We observed a trend where Wolbachia transmission was not complete in these crosses. For example, the bacterium could be introduced into some heterozygous mutant backgrounds; F1 progeny were infected if they resulted from crosses between control females and chic1320/CyO as well as qua6-396/CyO fathers, but the bacterium failed to colonize chic221/+ F1 progeny efficiently. We were unable to detect Wolbachia in many of the F2 progeny (Fig 1A). In order to quantify this reduction in titer, we performed qPCR on DNA extracts from F1 progeny from each of five individuals from the heterozygous mutants and compared these results to the quantified Wolbachia loads found in control flies (Fig 1B). Progeny from each F1 cross have a statistically significant reduction in Wolbachia titer (as quantified through qPCR) compared to the control lines (p < 0.01 for all pairwise comparisons, using a Bonferroni correction for df = 8). As additional support for the importance of the chic and qua loci in the Wolbachia titer defects we observed, we also quantified the amount of Wolbachia within two chromosomal deficiency stocks (deletions in the same region as either chic or qua in isogenic backgrounds)[25]. These deficiencies showed the same phenotype as our chic and qua mutants, supporting our observation that these genomic loci are responsible for the Wolbachia titer defect (Fig 1B). In addition to reductions in the F1 progeny, we also quantified a reduction in F2 progeny for the three actin mutant lines. For flies in which we can detect Wolbachia, the F2 progeny are further reduced in titer compared to the F1 lines (ratio of expression F1 versus F2: min = 0.56, max = 0.78).
Fig. 1. Presence of Wolbachia within various Drosophila melanogaster genotypes and their offspring assessed using polymerase chain reaction (A) and qPCR targeting the wsp gene on individual flies (B) or Western blot using antibodies against Wsp on both pooled fly lysates (C) and individual flies (D). 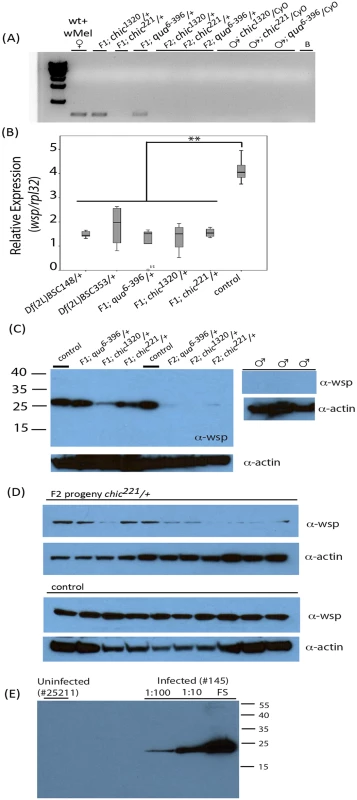
Anti-actin loading controls also shown. Consistency of Wolbachia Wsp production in wild-type control flies shown in panel (D) using 10 age-matched female flies (stock 145). Variability in the maintenance of the Wolbachia infection is shown in panel (D) where F2 chic221/+ progeny are probed with anti-wsp antibody. Specificity of the anti-Wsp antibody shown in panel (E) where uninfected flies are unreactive and intensity of reactivity is directly associated with amount of fly lysate loaded (FS = full strength). In order to control for effects of host genetic background on Wolbachia titer, we created isogenized lines from the control stock (145) and each of the mutant stocks by backcrossing to an uninfected w; Sco/Cyo line for three generations. We then crossed these Wolbachia infected F3 females (w; Sco/Cyo) to Wolbachia uninfected w; mutant/Cyo males (S1 Fig). In the F5 generation, we observed a significant effect of genotype on Wolbachia titer. Specifically, and regardless of mutant allele, mutant/Cyo progeny were reduced in Wolbachia titer by 1/3 compared to their w; Sco/Cyo siblings (mean relative ratio wsp/rpl32; t = -4.514; df = 9; p = 0.001). This result suggested to us that the reduction in titer was at least partially due to a result of a developmental defect in Wolbachia maintenance and persistence within the heterozygous mutant hosts.
As an additional control for host genetic background and to explore direct effects on profilin knockdown during development, we took advantage of an infected fly stock carrying a UAS inducible profilin-specific short hairpin silencing trigger (RNAi; stock #34523, genotype y1 sc* v1; P{TRiP.HMS00550}attP2) [27]. In order to test the effect of induction on fly development (to recapitulate the developmental lethality of the profilin null) we crossed homozygous females from this line to w; P{w+, Act GAL4} /TM3 males. From this cross we only recovered stubble progeny, suggesting that this particular RNAi line, which hadn’t previously been utilized in a publication to knock down profilin expression, is effective. In order to test the effect of induction on fly development we crossed homozygous females (y1 sc* v1; P{TRiP.HMS00550}attP2) to a homozygous Hsp70:Gal4 driver [2–5]. Third instar larvae were shocked at 37C for 10 minutes to induce the short hairpin and late pupae collected for RNA and DNA extraction (N = 8 for each treatment and genotype; y1 sc* v1; P{TRiP.HMS00550}attP2 with or without Hsp70:Gal4 and with or without heat shock). In the maternal y1 sc* v1; P{TRiP.HMS00550}attP2 background, heat shock did not affect either Wolbachia titers (t = 1.207, df = 2, p = 0.351) nor profilin expression (t = -1.144, df = 2, p = 0.371). In contrast, profilin expression was statistically significantly reduced in flies expressing the RNAi construct compared to non-heat shocked siblings (the mean expression ratio chic/rpl32 = 0.57; t = -6.240; df = 2; p = 0.025). In addition, knockdown of profilin did have a significant and measurable effect on Wolbachia titers in these same flies; the fly Wolbachia titers were reduced by 1/3 compared to their non-heat shocked siblings (mean relative ratio wsp/rpl32 = 0.66, t = -8.593; df = 2; p = 0.013).
To provide additional support for the reduction in titer observed via PCR, we probed Western blots of pooled or individual fly lysates produced from the F1 and F2 progeny and their parents for Wsp (Fig 1C, 1D and 1E). Results corroborated our previous finding that Wolbachia transmission was imperfect in the mutant flies (Fig 1A and 1B). Specifically, infected F1 progeny, especially in the chic mutant backgrounds, appeared to carry a reduced titer of Wolbachia when compared to the maternal, infected line (Fig 1). Indeed, flies from control crosses are consistently higher titer in Wolbachia, as based on densitometric quantitation of Western blot bands (Average +/ - STERR over 5 experiments for Control = 13,106 +/ - 3,294; chic1320/+ = 6,418 +/ - 4,890; chic221/+ = 6,545 +/ - 1,576; qua6-396/+ = 6,179 +/ - 645; t-test; p = 0.036, 0.001, 0.002 for each heterozygous mutant compared to control). Additionally, we could detect a statistically significant reduction between the F1 and F2 heterozygous mutant flies (p = 0.012). As observed in our results based on PCR, Wolbachia titer (based on quantity of protein on a Western blot) is also reduced, with some variability, in the F2 progeny (Fig 1D). We hypothesized that the loss of Wolbachia in some F2 progeny was a result of a reduction in Wolbachia titer in F1 females during oogenesis. We therefore visualized the Wolbachia infection in the germarium in F1 females (mutant/+; below).
Wolbachia is reduced in the germarium and early egg chamber when hosts are heterozygous mutants in chickadee or quail
To colonize the oocyte, and therefore complete maternal transmission, Wolbachia occupy the germline and somatic stem cell niches (SSCN) in their hosts [18,26,31]. Wolbachia can achieve this localization after injection into the fly abdomen, suggesting that the stem cell niche targets are essential for Wolbachia infection [31]. The Drosophila ovariole provides an opportunity to view oocyte development and Wolbachia localization within each progressive stage. Wolbachia concentrate preferentially in the somatic stem cell niche, which is thought to serve as a source of infection for the germline. As germline development progresses from regions 2a to 2b, Wolbachia are thought to infect via the somatic stem cell niche, increasing the numbers of bacteria found within the germline after association with the SSCN [31]. We utilized immunohistochemistry to detect Wolbachia in the germarium of our flies, producing localizations expected based on previous publications [15,18,26,31](S1 Table and S2 and S3 Figs). Wolbachia infection within the entire germarium is significantly reduced in heterozygous mutant flies (when comparing the amount of fluorescence observed in control flies to that found in either chic221/+, chic1320/+ or qua6-396/+, respectively; Mann-Whitney U = 171.5, Z = -3.995, p < 0.001; Mann-Whitney U = 98, Z = -5.295, p < 0.001; Mann-Whitney U = 55.5, Z = -5.496, p < 0.001; Figs 2 and 3). When either chic1320/+ or qua6-396/+ heterozygous mutant flies are infected, the Wolbachia titer in region 2 (as quantified by anti-Hsp60 staining) is also significantly reduced, compared to the control maternal line (Mann-Whitney U = 194, Z = -3.78, p < 0.001; Mann-Whitney U = 134; Z = -4.097, p < 0.001, pairwise comparison between control and chic1320/+ or qua6-396/+ heterozygous mutants, respectively; Fig 3). Additionally, Wolbachia infection within early egg chambers (stage 1) is significantly reduced in all heterozygous mutant flies (when comparing the amount of fluorescence observed in control flies to that found in either chic221/+, chic1320/+ or qua6-396/+, respectively; Mann-Whitney U = 74, Z = -5.74, p < 0.001; Mann-Whitney U = 58, Z = -5.872, p < 0.001; Mann-Whitney U = 39, Z = -5.767, p < 0.001; Figs 2 and 3). In order to quantify this reduction, for each germarium, we calculated the ratio of fluorescence intensity in the earliest egg chamber over that found in region 2 (as quantified by anti-Hsp60 staining). Each of the three mutant lines showed a statistically significant reduction in this ratio when compared to control germaria (average ratios for control flies: 1.72; chic221/+: 0.52; chic1320/+: 0.64; qua6-396/+: 0.80, t-test; p < 0.0001). The reductions in infection in the germaria suggest two things: (1) that Wolbachia has difficulties in transiting or maintenance in a population within the germarium during development in the heterozygous mutant flies and (2) even when region two, the location of the SSCN, is occupied by Wolbachia, the bacteria are deficient in colonization of the early egg chamber in the heterozygous mutant flies (Fig 3). We did not quantify differences in staining of the presumed germline stem cell niche due to variability in staining in this region within the control flies.
Fig. 2. Mutations in actin binding proteins reduce the titer of Wolbachia within the region 2 and early egg chambers in heterozygous mutant flies. 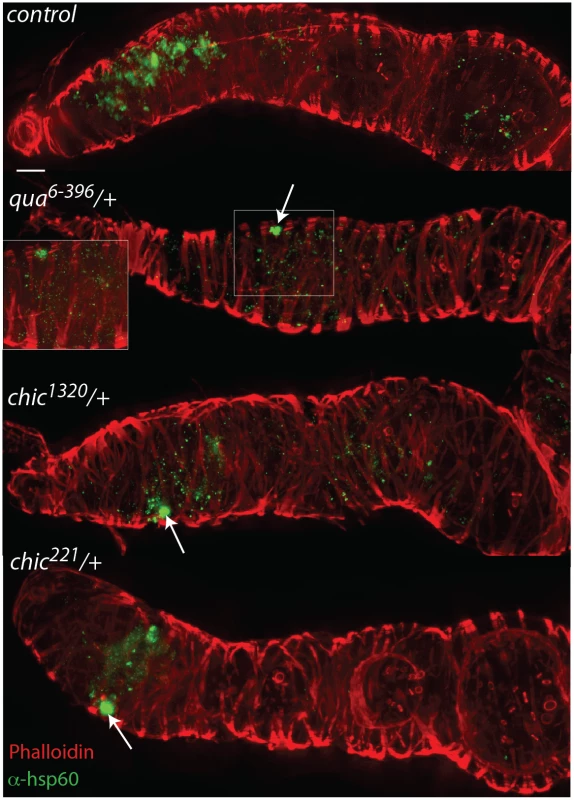
Drosophila melanogaster germaria from control, qua6-396/+, chic1320/+, and chic221/+ backgrounds, stained with α-Hsp60 for Wolbachia (green) and Acti-stain phalloidin conjugate for actin (red). Arrows point to Wolbachia aggregates within heterozygous mutants. Scale bar = 10 μm. Inlays are 100x magnification of aggregates within the tissue. Fig. 3. Quantification of reduction in Wolbachia titer during oogenesis, within the entire germarium (in blue), region 2, including the somatic stem cell niche (SSCN, green) and early egg chambers (stage 1, red). 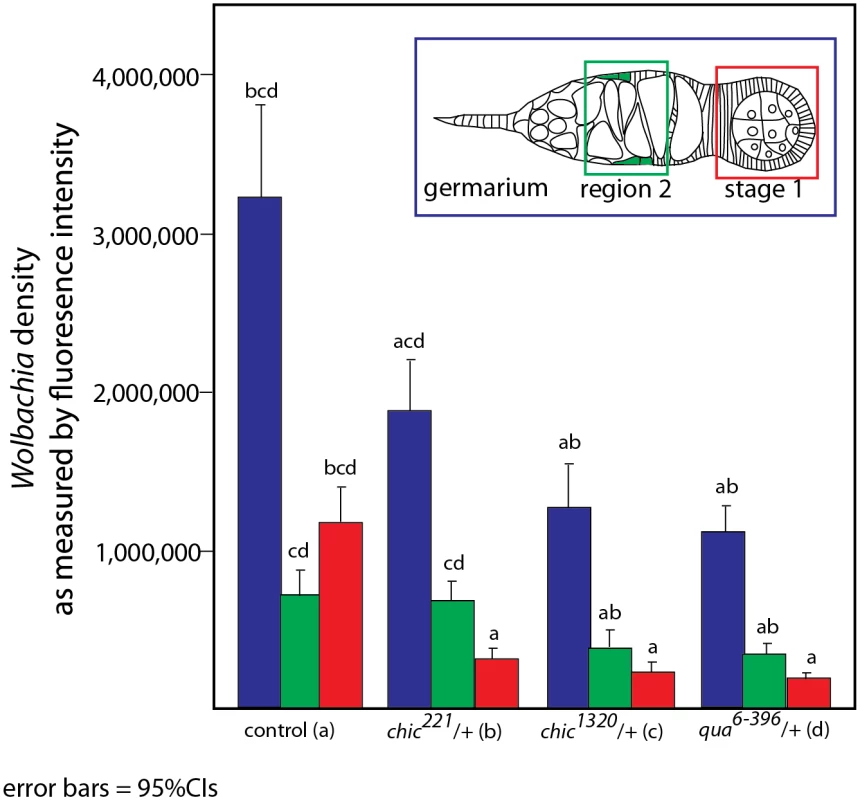
Flattened, maximum projections from z-sections were utilized to compare between control flies and chic221/+, chic1320/+ and qua6-396/+ F1 progeny stained with Hsp-60 for Wolbachia. Error bars = 95% confidence intervals and statistical significance in pairwise comparisons is shown above each bar (Kruskal Wallis tests followed by pairwise Mann-Whitney U tests between genotypes; p < 0.05). Within heterozygous mutant flies, we observed that the Wolbachia that successfully manage to colonize the germarium do so with a distinctive localization; these Wolbachia appear as aggregates, in sharp contrast to the more even distribution of Wolbachia within control germaria (Table 1 and Fig 2). Under high magnification (100x), the Wolbachia aggregates within the heterozygous mutant flies appear to be multiple Wolbachia forming micro-colonies within the tissue, based on shape and size and consistent localization within the genotypes.
Tab. 1. Wolbachia aggregates within the germarium of Drosophila melanogaster. 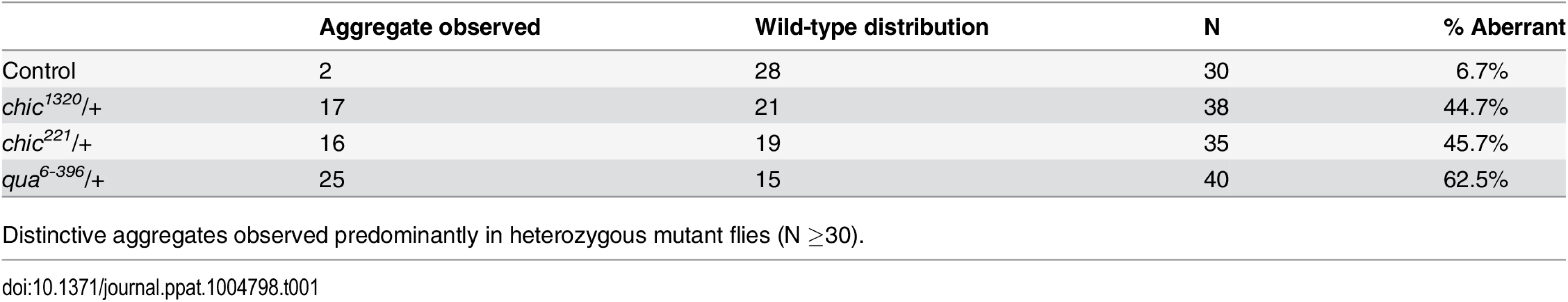
Distinctive aggregates observed predominantly in heterozygous mutant flies (N ≥30). Characterization of F/G-actin and profilin expression within heterozygous mutant flies
Both Drosophila profilin (chickadee) and villin (quail) are important in the regulation of F-actin during oogenesis. Because profilin promotes the polymerization of F-actin filaments and villin stabilizes these filaments through bundling, we were curious to know whether or not the heterozygous mutant flies differed in the quantity of F-actin found in the germarium, when compared to control flies. In addition, visualization of the F-actin cytoskeleton allowed us to examine the actin ring canals in the heterozygous mutant flies at all stages of oocyte development. At no point were ring canals occluded by nuclei, supporting our finding that cytoplasmic streaming and maternal dumping are unaffected in heterozygous mutant flies (N = 300, scored by eye). Using quantified fluorescence of F-actin in the images we were unable to detect a statistically significant difference between median levels of phalloidin staining in control flies compared to the heterozygous mutants (Kruskal Wallis test: χ2 = 4.005, df = 3, p = 0.261; S4A Fig). Because F-actin levels in the germarium (observed through phalloidin staining) did not correlate with Wolbachia intensity (as quantified by anti-Hsp60 staining), the Wolbachia titer phenotype observed in these flies may not be directly related to the F-actin network in the germarium. We therefore examined the in vivo amounts of filamentous and globular actin in ovaries from control flies and compared this to that seen in heterozygous mutant chic221 flies. Using ultracentrifugation coupled to Western blot, we found that we could consistently detect globular actin in the ovaries of control flies. In contrast, we found a statistically significant decrease in total amount of globular actin detected in the heterozygous mutant lines (Kruskal Wallis: χ2 = 4.192, df = 1, p = 0.041; S4B Fig).
The difference in G actin between control and heterozygous chic221 flies prompted us to investigate expression of profilin in the chic221/+ F1 mutants and control flies. The rationale was that although these flies are phenotypically wild type, the dosage effect of a single, wild type chromosome in the chic221/+ F1 mutants might be significant and correlate with Wolbachia absence. We extracted both RNA and DNA from individual F1 chic221/+ female flies as well as age-matched control flies and used quantitative RT-PCR to detect profilin transcript levels (in total RNA) and Wolbachia surface protein (in total DNA) relative to Rpl32. Control flies express, on average, 2x as much profilin as heterozygous mutant F1 progeny (means control μ = 4.03; mutant μ = 2.28; t = 2.590, df = 11.31, p = 0.025). Additionally, although we could detect Wolbachia in each of the wild type flies included (N = 10), we were only able to detect a Wolbachia infection in three of the heterozygous mutant F1 flies (S4C Fig). Wolbachia may have been present in these flies, but at titers below the limit of detection for this method.
Heterozygous mutants in chickadee and quail produce the same number of progeny as control flies and lay eggs of normal size and shape
Because Wolbachia target the germline, and within the germarium, the stem cell niche [18,31], the number of egg chambers produced by the host may affect Wolbachia’s ability to be transmitted between generations. Flies that are homozygous mutants in chickadee show defects in germline stem cell proliferation as well as enclosure by somatic cyst cells [32,33] so it was therefore important to confirm that the heterozygous mutant flies do not display similar defects. We counted the number of viable progeny resulting from individual crosses within mutant fly lines and compared the number of resulting offspring to those from control crosses. Heterozygous villin or profilin mutant flies do not show a defect in fertility when compared to control flies (S5 Fig). Additionally, we observed over 300 eggs for each of the fly mutant stocks and did not see any morphological abnormalities when compared to the control stock (N = 300, scored by eye).
Discussion
Wolbachia maternal transmission in Drosophila melanogaster is normally extremely effective, with perfect transmission observed in laboratory populations and near perfect transmission in the wild [10–12]. Wolbachia are thought to localize in the germarium, and ultimately in the oocyte, in order to accomplish this maternal transmission. Previous work has shown that Wolbachia use host microtubules to localize preferentially to the oocyte during development [15–17]. The striking anterior localization of Wolbachia during oogenesis can be perturbed by feeding Drosophila microtubule inhibitors such as colchicine, or by mutations that perturb the microtubule cytoskeleton [15]. In contrast, direct treatment of dissected ovaries with actin disrupting drugs (such as cytochalasin-D) does not alter this localization [15]. However, other pieces of evidence suggest that Wolbachia manipulate the host actin cytoskeleton. For example, Wolbachia injected into the abdominal cavity of Drosophila migrate to the germline stem cell niche, a feat that requires traversing several host tissues and cell types [31]. Also, Wolbachia in the terrestrial isopod Armadillidium vulgare are not found in all primary oocytes and instead, enrichment of Wolbachia is seen during the course of development [34]. Finally, Wolbachia are associated with areas of weak cortical actin staining in filarial nematodes, suggestive of a mechanism for entry into the germline from somatic cells [17,19]. Therefore, it is likely that Wolbachia use both microtubules and actin for persistence in the host and maintenance across host generations.
Oogenesis in Drosophila relies on rearrangements of both the actin and microtubule networks [35]. We were therefore careful in our analysis to separate direct effects of actin modulation from indirect effects resulting from perturbations of the reproductive biology of the fly. Products of both the quail and chickadee loci are necessary for fly reproduction [22,36–38]; homozygous or hemizygous mutants in either gene result in fertility defects or are lethal. Importantly, in this study we followed Wolbachia infections in phenotypically wild type flies harboring a functional copy of the actin binding protein in question. These heterozygous mutant flies produce the same number of offspring as the control flies and produce eggs with the same morphology as controls, however the flies do not faithfully maintain a Wolbachia infection. Several hypotheses partially explain our data, and below we delineate our hypothesis and alternative hypotheses and summarize our evidence to support or refute them.
Heterozygous mutant flies are phenotypically wild type with respect to oocyte polarization and number of progeny
The developing oocyte is loaded with maternal determinants (e.g. mRNA and protein), a process which begins early (stage 1), and continues until about stage 10 when maternal nurse cells dump their remaining cytoplasmic contents into the oocyte [35]. The actin cytoskeleton is critical to this process, as mutations in actin binding proteins have been known to cause severe defects. Specifically, cytoplasmic actin bundles are required to restrain the nurse cell nuclei during transport; mutations in quail, which regulates bundling of cytoplasmic actin, cause a dumpless phenotype [39,40]. In quail mutant flies, nurse cell nuclei can be observed extending through the actin ring canals [39]. We reasoned that although heterozygous mutant flies (chic221/+, chic1320/+ and qua6-396/+) produce viable progeny, and we found no occluded ring canals in any of these backgrounds, a subtle defect in maternal cytoplasmic dumping could alter the ability of Wolbachia to be transmitted faithfully to the oocyte. Wolbachia has been suggested to utilize cytoplasmic dumping to increase titer in the oocyte (as compared to the nurse cells) [15]. In addition to regulating the bundling of microtubules and therefore cytoplasmic streaming, profilin is also required for posterior patterning in the oocyte as chic mutants fail to localize STAUFEN and oskar mRNA [41]. Wolbachia utilizes these posterior determinants to localize in the oocyte, as disruption of osk and stau results in mislocalization of Wolbachia in D. melanogaster [16]. If heterozygous mutant flies are defective in cytoplasmic dumping or polarization, we should observe both egg size and morphology defects. Over 300 eggs were scored for each of the mutant lines, as well as control flies, without any phenotypic differences detected. Importantly, however, the primary loss of Wolbachia in these heterozygous mutants occurs in the germarium, before defects would begin to affect Wolbachia titers. Therefore, although our fly mutants could conceivably exhibit subtle polarization defects, these defects alone would not entirely explain the observed phenotype.
In addition to serving important roles during maternal loading in the late stage oocyte, profilin functions in germline stem cell (GSC) maintenance and germ cell enclosure by somatic cyst cells [32,33]—homozygous chickadee mutants fail to maintain germline stem cell number. However, chic221/+ flies are equivalent to wild type [32]; that is to say, heterozygous mutant flies do not have a GSC deficiency. Importantly, although Wolbachia are known to alter germline stem cell proliferation [26] and some Wolbachia colonize the germline stem cell niche [18], wMel colonizes the somatic stem cell niche in Drosophila melanogaster (Fig 2). Regardless, a defect in fertility, resulting from defects in GSC maintenance might affect Wolbachia proliferation in these mutant flies. We therefore counted the number of viable progeny (a measure of fertility) for each of the mutant lines. No statistically significant difference was observed for any of the heterozygous, mutant flies, when compared to the control (S5 Fig). We therefore did not find support for this hypothesis to explain the Wolbachia clearing phenotype of profilin and villin heterozygous mutants.
Wolbachia localization during development impacts maternal transmission
There is significant evidence that Wolbachia colonize the primordial germ cells and the posterior pole of developing embryos in numerous insect hosts. In D. melanogaster, for example, strain wMel concentrates at the posterior pole in a poleplasm dependent fashion [16,42,43]. However, this posterior concentration of Wolbachia is not universal in insects nor in Drosophila. Wolbachia strain wRi infects the entire embryo uniformly while B group Wolbachia actually show exhibit anterior localization [14]. Similarly, in other Drosophila species there are different patterns of Wolbachia colonization: although wWil infects primordial germ cells, wAu infects the entire embryo [44]. This posterior localization is clearly important—the extent of CI is correlated with the number of Wolbachia in the posterior of the embryo [14]. However, this posterior localization is not necessarily correlated with maternal transmission, which is near 100% for some Drosophila species and quite low for others [45–47]. This result suggests that high titer localization to primordial germ cells and the posterior pole does not guarantee maternal transmission. However, if our heterozygous mutant flies induce defects in these early localization patterns (to the posterior pole or to the developing germ line), we might expect the inefficient transmission phenotype observed.
What other ways might Wolbachia use to eventually colonize the germline? Wolbachia colonization of somatic tissues has been known for some time [48] but recently, it has been suggested that Wolbachia infection of the soma may serve as a reservoir for germline infection. In the terrestrial isopod, Armadillidium vulgare, Wolbachia is absent from many early oocytes and infects the older oocytes late in development, an enrichment that is thought to come from a somatic reservoir (the follicle cells) [34]. In nematodes, Wolbachia initially are concentrated in the posterior of the P2 blastomere, the precursor of the adult germ line. However, Wolbachia are subsequently excluded from the germ line in the next cell division and instead, invade the germ cells later, from the surrounding somatic gonadal cells [19]. This soma to germ cell invasion in Brugia is correlated with a disruption in polymerized actin at those foci [19]. Because we observed a reduction in anti-Hsp60 staining in stage 1 egg chambers of heterozygous mutant flies as well as transmission defects, one interpretation of our data is that Wolbachia require actin for soma to germline transmission. Importantly, however, we did not observe actin disruptions (similar to those seen in Brugia) within Drosophila germaria.
Actin regulation impacts Wolbachia titers during development, affecting transmission efficiency
Our data suggest that Wolbachia rely on the actin cytoskeleton to achieve adequate titer in the Drosophila host during development. First, we observe reductions in titer of Wolbachia in heterozygous mutants compared to both their non-mutant sibling controls as well as parental controls (Fig 1). Second, knockdown of profilin in third instar larvae reduces Wolbachia titer in pupae, suggesting that the regulation of actin is important to the maintenance of a Wolbachia infection during development. Additionally, passage of Wolbachia through heterozygous mutant lines for multiple generations results in the enrichment for mutant Wolbachia; the heterozygous mutant flies bottleneck the Wolbachia infection, increasing the stochastic segregation of variants [49]. This decrease in titer may explain the inefficient transmission of Wolbachia observed in the mutant flies. Actin may be used by Wolbachia to properly localize during development, or may support the infection via other unknown mechanisms.
Potential mechanisms to explain the Wolbachia phenotype in mutant flies
Both of the proteins investigated here (profilin and villin), are known to increase the amount and stability of F-actin in the Drosophila egg chamber. Profilin promotes F - actin in the follicular epithelium while villin bundles and binds to filamentous actin [37,50]. One potential cause of the Wolbachia phenotype in these backgrounds is a mis-regulation in F-actin content. Interestingly, chic mutants have been previously observed to exhibit decreased F-actin levels in the follicle cells [50]. Both the somatic stem cell niche and the follicular epithelium have been suggested to be a source of Wolbachia during oogenesis [18,34]. Because Wolbachia densely colonize the follicular epithelium tissue, and because it surrounds the oocyte throughout development, this tissue may be a candidate for the source of the infection. We detected a significant reduction in the amount of actin in heterozygous mutant chic221 flies compared to controls, which corresponded to a decrease in profilin transcripts and a decrease in detected Wolbachia (S4 Fig). These data are suggestive of a role for actin in Wolbachia maintenance and transmission but do not elucidate an exact mechanism.
We have shown that the host actin cytoskeleton is clearly important for the maintenance of a Wolbachia infection. Perhaps this reproductive parasite secretes proteins that interact directly with eukaryotic actin or host actin binding proteins. Indeed, other members of the Rickettsiales are known for their striking coopting of host actin in the production of comet tails [51]. However, when intracellular, Wolbachia persist within membrane-bound compartments and no such comet-like structures have been observed to be associated with the vacuole [21]. That said, our results here and the work of others strongly suggest that Wolbachia is able to enter and exit eukaryotic cells; Wolbachia transit to the germline from the fly abdomen and are loaded into the germ cells from surrounding somatic cells [18,26,31]. Wolbachia’s success likely depends upon an ability to secrete proteins that modify host actin to promote internalization by non-phagocytic cells. Recently, in vitro biochemical associations between the filarial nematode Wolbachia (wBm) PAL-like protein wBm0152 and actin have been observed, although results do not conclusively implicate this particular protein in interactions with host actin during infection [20]. Regardless, as is clear from our work, a Wolbachia infection depends on the actin cytoskeleton. Therefore, future work to identify and characterize Wolbachia proteins that bind to or alter host actin dynamics will be important for understanding the molecular basis of the interaction between the host and the symbiont.
Summary
In order for intracellular, maternally transmitted symbionts to successfully infect the next generation, the bacteria must target the oocyte. Wolbachia achieves this through a specific infection of the somatic stem cell niche in the germarium of Drosophila melanogaster [18]. Here we show that Wolbachia is extraordinarily sensitive to the regulation of actin, such that phenotypically wild type heterozygous mutant flies cannot faithfully transmit the bacterium to their progeny. Our results, particularly that titer is significantly reduced in the germaria of chic221/+, chic1320/+, and qua6-396/+ flies, suggest that Wolbachia utilize host actin to enter and persist within host tissues during Drosophila development. Additionally, our finding that these heterozygous mutant flies cannot transmit the infection suggests that Wolbachia titers within a host are reduced when actin regulation is disrupted, impacting transmission efficiency.
Supporting Information
Zdroje
1. Werren JH, Baldo L, Clark ME (2008) Wolbachia: master manipulators of invertebrate biology. Nat Rev Microbiol 6 : 741–751. doi: 10.1038/nrmicro1969 18794912
2. Zug R, Hammerstein P (2012) Still a host of hosts for wolbachia: analysis of recent data suggests that 40% of terrestrial arthropod species are infected. PLoS One 7: e38544. doi: 10.1371/journal.pone.0038544 22685581
3. Hoerauf A, Specht S, Buttner M, Pfarr K, Mand S, et al. (2008) Wolbachia endobacteria depletion by doxycycline as antifilarial therapy has macrofilaricidal activity in onchocerciasis: a randomized placebo-controlled study. Medical Microbiology and Immunology 197 : 295–311. 17999080
4. Taylor MJ, Bandi C, Hoerauf AM, Lazdins J (2000) Wolbachia bacteria of filarial nematodes: A target for control? Parasitology Today 16 : 179–180. 10782070
5. Moreira LA, Iturbe-Ormaetxe I, Jeffery JA, Lu GJ, Pyke AT, et al. (2009) A Wolbachia Symbiont in Aedes aegypti Limits Infection with Dengue, Chikungunya, and Plasmodium. Cell 139 : 1268–1278. doi: 10.1016/j.cell.2009.11.042 20064373
6. Turley AP, Moreira LA, O'Neill SL, McGraw EA (2009) Wolbachia Infection Reduces Blood-Feeding Success in the Dengue Fever Mosquito, Aedes aegypti. Plos Neglected Tropical Diseases 3(9):e516. doi: 10.1371/journal.pntd.0000516 19753103
7. LePage D, Bordenstein SR (2013) Wolbachia: Can we save lives with a great pandemic? Trends Parasitol 29 : 385–393. doi: 10.1016/j.pt.2013.06.003 23845310
8. Moreira LA, Iturbe-Ormaetxe I, Jeffery JA, Lu G, Pyke AT, et al. (2009) A Wolbachia symbiont in Aedes aegypti limits infection with dengue, Chikungunya, and Plasmodium. Cell 139 : 1268–1278. doi: 10.1016/j.cell.2009.11.042 20064373
9. Kittayapong P, Baisley KJ, Sharpe RG, Baimai V, O'Neill SL (2002) Maternal transmission efficiency of Wolbachia superinfections in Aedes albopictus populations in Thailand. Am J Trop Med Hyg 66 : 103–107. 12135258
10. Turelli M, Hoffmann AA (1995) Cytoplasmic incompatibility in Drosophila simulans: dynamics and parameter estimates from natural populations. Genetics 140 : 1319–1338. 7498773
11. McGraw EA, Merritt DJ, Droller JN, O'Neill SL (2002) Wolbachia density and virulence attenuation after transfer into a novel host. Proc Natl Acad Sci U S A 99 : 2918–2923. 11880639
12. Weeks AR, Turelli M, Harcombe WR, Reynolds KT, Hoffmann AA (2007) From parasite to mutualist: rapid evolution of Wolbachia in natural populations of Drosophila. PLoS Biol 5: e114. 17439303
13. Perrot-Minnot MJ, Werren JH (1999) Wolbachia infection and incompatibility dynamics in experimental selection lines. Journal of Evolutionary Biology 12 : 272–282.
14. Veneti Z, Clark ME, Karr TL, Savakis C, Bourtzis K (2004) Heads or tails: host-parasite interactions in the Drosophila-Wolbachia system. Appl Environ Microbiol 70 : 5366–5372. 15345422
15. Ferree PM, Frydman HM, Li JM, Cao J, Wieschaus E, et al. (2005) Wolbachia utilizes host microtubules and Dynein for anterior localization in the Drosophila oocyte. PLoS Pathog 1: e14. 16228015
16. Serbus LR, Sullivan W (2007) A cellular basis for Wolbachia recruitment to the host germline. PLoS Pathog 3: e190. 18085821
17. Albertson R, Casper-Lindley C, Cao J, Tram U, Sullivan W (2009) Symmetric and asymmetric mitotic segregation patterns influence Wolbachia distribution in host somatic tissue. J Cell Sci 122 : 4570–4583. doi: 10.1242/jcs.054981 19934219
18. Toomey ME, Panaram K, Fast EM, Beatty C, Frydman HM (2013) Evolutionarily conserved Wolbachia-encoded factors control pattern of stem-cell niche tropism in Drosophila ovaries and favor infection. Proc Natl Acad Sci U S A 110 : 10788–10793. doi: 10.1073/pnas.1301524110 23744038
19. Landmann F, Bain O, Martin C, Uni S, Taylor MJ, et al. (2012) Both asymmetric mitotic segregation and cell-to-cell invasion are required for stable germline transmission of Wolbachia in filarial nematodes. Biol Open 1 : 536–547. doi: 10.1242/bio.2012737 23213446
20. Melnikow E, Xu S, Liu J, Bell AJ, Ghedin E, et al. (2013) A potential role for the interaction of Wolbachia surface proteins with the Brugia malayi glycolytic enzymes and cytoskeleton in maintenance of endosymbiosis. PLoS Negl Trop Dis 7: e2151. doi: 10.1371/journal.pntd.0002151 23593519
21. Fischer K, Beatty WL, Weil GJ, Fischer PU (2014) High pressure freezing/freeze substitution fixation improves the ultrastructural assessment of Wolbachia endosymbiont—filarial nematode host interaction. PLoS One 9:e86383. doi: 10.1371/journal.pone.0086383 24466066
22. Verheyen EM, Cooley L (1994) Profilin Mutations Disrupt Multiple Actin-Dependent Processes during Drosophila Development. Development 120 : 717–728. 7600952
23. Cooley L, Verheyen E, Ayers K (1992) chickadee encodes a profilin required for intercellular cytoplasm transport during Drosophila oogenesis. Cell 69 : 173–184. 1339308
24. Bakken AH (1973) A cytological and genetic study of oogenesis in Drosophila melanogaster. Dev Biol 33 : 100–122. 4363796
25. Cook RK, Christensen SJ, Deal JA, Coburn RA, Deal ME, et al. (2012) The generation of chromosomal deletions to provide extensive coverage and subdivision of the Drosophila melanogaster genome. Genome Biol 13: R21. doi: 10.1186/gb-2012-13-3-r21 22445104
26. Fast EM, Toomey ME, Panaram K, Desjardins D, Kolaczyk ED, et al. (2011) Wolbachia enhance Drosophila stem cell proliferation and target the germline stem cell niche. Science 334 : 990–992. doi: 10.1126/science.1209609 22021671
27. Ni JQ, Zhou R, Czech B, Liu LP, Holderbaum L, et al. (2011) A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat Methods 8 : 405–407. doi: 10.1038/nmeth.1592 21460824
28. Verheyen E, Cooley L (1994) Looking at Oogenesis. Methods in Cell Biology, Vol 44 44 : 545–561. 7707970
29. Baldo L, Hotopp JCD, Jolley KA, Bordenstein SR, Biber SA, et al. (2006) Multilocus sequence typing system for the endosymbiont Wolbachia pipientis. Applied and Environmental Microbiology 72 : 7098–7110. 16936055
30. Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research 29: e(45).
31. Frydman HM, Li JM, Robson DN, Wieschaus E (2006) Somatic stem cell niche tropism in Wolbachia. Nature 441 : 509–512. 16724067
32. Shields AR, Spence AC, Yamashita YM, Davies EL, Fuller MT (2014) The actin-binding protein profilin is required for germline stem cell maintenance and germ cell enclosure by somatic cyst cells. Development 141 : 73–82. doi: 10.1242/dev.101931 24346697
33. Gonczy P, DiNardo S (1996) The germ line regulates somatic cyst cell proliferation and fate during Drosophila spermatogenesis. Development 122 : 2437–2447. 8756289
34. Genty LM, Bouchon D, Raimond M, Bertaux J (2014) Wolbachia infect ovaries in the course of their maturation: last minute passengers and priority travellers? PLoS One 9: e94577. doi: 10.1371/journal.pone.0094577 24722673
35. Robinson DN, Cooley L (1997) Genetic analysis of the actin cytoskeleton in the Drosophila ovary. Annual Review of Cell and Developmental Biology 13 : 147–170. 9442871
36. Castrillon DH, Gonczy P, Alexander S, Rawson R, Eberhart CG, et al. (1993) Toward a molecular genetic analysis of spermatogenesis in Drosophila melanogaster: characterization of male-sterile mutants generated by single P element mutagenesis. Genetics 135 : 489–505. 8244010
37. Matova N, Cooley L (1998) Quail, a Drosophila villin-like protein, bundles actin filaments in apoptotic nurse cells. Molecular Biology of the Cell 9 : 18A–18A.
38. Matova N, Foley K, Cooley L (1998) Death and actin dynamics during Drosophila egg chamber development. Molecular Biology of the Cell 9 : 382A–382A.
39. Mahajanmiklos S, Cooley L (1994) The Villin-Like Protein Encoded by the Drosophila Quail Gene Is Required for Actin Bundle Assembly during Oogenesis. Cell 78 : 291–301. 8044841
40. Matova N, Mahajan-Miklos S, Mooseker MS, Cooley L (1999) Drosophila Quail, a villin-related protein, bundles actin filaments in apoptotic nurse cells. Development 126 : 5645–5657. 10572041
41. Manseau L, Calley J, Phan H (1996) Profilin is required for posterior patterning of the Drosophila oocyte. Development 122 : 2109–2116. 8681792
42. Hadfield SJ, Axton JM (1999) Reproduction—Germ cells colonized by endosymbiotic bacteria. Nature 402 : 482–482. 10591206
43. Kose H, Karr TL (1995) Organization of Wolbachia-Pipientis in the Drosophila Fertilized Egg and Embryo Revealed by an Anti-Wolbachia Monoclonal-Antibody. Mechanisms of Development 51 : 275–288. 7547474
44. Miller WJ, Riegler M (2006) Evolutionary dynamics of wAu-Like Wolbachia variants in neotropical Drosophila spp. Applied and Environmental Microbiology 72 : 826–835. 16391124
45. Turelli M, Hoffmann AA (1991) Rapid spread of an inherited incompatibility factor in California Drosophila. Nature 353 : 440–442. 1896086
46. Hamm CA, Begun DJ, Vo A, Smith CC, Saelao P, et al. (2014) Wolbachia do not live by reproductive manipulation alone: infection polymorphism in Drosophila suzukii and D. subpulchrella. Mol Ecol.
47. Carrington LB, Lipkowitz JR, Hoffmann AA, Turelli M (2011) A re-examination of Wolbachia-induced cytoplasmic incompatibility in California Drosophila simulans. PLoS One 6: e22565. doi: 10.1371/journal.pone.0022565 21799900
48. Dobson SL, Bourtzis K, Braig HR, Jones BF, Zhou WG, et al. (1999) Wolbachia infections are distributed throughout insect somatic and germ line tissues. Insect Biochemistry and Molecular Biology 29 : 153–160. 10196738
49. Newton IL, Sheehan KB (2014) Passage of Wolbachia through mutant Drosophila melanogaster induces phenotypic and genomic changes. Appl Environ Microbiol.
50. Baum B, Perrimon N (2001) Spatial control of the actin cytoskeleton in Drosophila epithelial cells. Nat Cell Biol 3 : 883–890. 11584269
51. Gouin E, Gantelet H, Egile C, Lasa I, Ohayon H, et al. (1999) A comparative study of the actin-based motilities of the pathogenic bacteria Listeria monocytogenes, Shigella flexneri and Rickettsia conorii. J Cell Sci 112 (Pt 11): 1697–1708. 10318762
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Relay of Herpes Simplex Virus between Langerhans Cells and Dermal Dendritic Cells in Human SkinČlánek Does the Arthropod Microbiota Impact the Establishment of Vector-Borne Diseases in Mammalian Hosts?Článek The Ebola Epidemic Crystallizes the Potential of Passive Antibody Therapy for Infectious DiseasesČlánek Hepatitis D Virus Infection of Mice Expressing Human Sodium Taurocholate Co-transporting PolypeptideČlánek A Redox Regulatory System Critical for Mycobacterial Survival in Macrophages and Biofilm Development
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2015 Číslo 4- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Pathogens as Biological Weapons of Invasive Species
- Selection and Spread of Artemisinin-Resistant Alleles in Thailand Prior to the Global Artemisinin Resistance Containment Campaign
- Endopeptidase-Mediated Beta Lactam Tolerance
- Prospective Large-Scale Field Study Generates Predictive Model Identifying Major Contributors to Colony Losses
- Relay of Herpes Simplex Virus between Langerhans Cells and Dermal Dendritic Cells in Human Skin
- Structural Determinants of Phenotypic Diversity and Replication Rate of Human Prions
- Sigma Factor SigB Is Crucial to Mediate Adaptation during Chronic Infections
- EphrinA2 Receptor (EphA2) Is an Invasion and Intracellular Signaling Receptor for
- Toxin-Induced Necroptosis Is a Major Mechanism of Lung Damage
- Heterologous Expression in Remodeled . : A Platform for Monoaminergic Agonist Identification and Anthelmintic Screening
- Novel Disease Susceptibility Factors for Fungal Necrotrophic Pathogens in Arabidopsis
- Interleukin 21 Signaling in B Cells Is Required for Efficient Establishment of Murine Gammaherpesvirus Latency
- Phosphorylation at the Homotypic Interface Regulates Nucleoprotein Oligomerization and Assembly of the Influenza Virus Replication Machinery
- Human Papillomaviruses Activate and Recruit SMC1 Cohesin Proteins for the Differentiation-Dependent Life Cycle through Association with CTCF Insulators
- Ubiquitous Promoter-Localization of Essential Virulence Regulators in
- TGF-β Suppression of HBV RNA through AID-Dependent Recruitment of an RNA Exosome Complex
- The Immune Adaptor ADAP Regulates Reciprocal TGF-β1-Integrin Crosstalk to Protect from Influenza Virus Infection
- Antagonism of miR-328 Increases the Antimicrobial Function of Macrophages and Neutrophils and Rapid Clearance of Non-typeable (NTHi) from Infected Lung
- The Epigenetic Regulator G9a Mediates Tolerance to RNA Virus Infection in
- Does the Arthropod Microbiota Impact the Establishment of Vector-Borne Diseases in Mammalian Hosts?
- Hantaan Virus Infection Induces Both Th1 and ThGranzyme B+ Cell Immune Responses That Associated with Viral Control and Clinical Outcome in Humans
- Viral Inhibition of the Transporter Associated with Antigen Processing (TAP): A Striking Example of Functional Convergent Evolution
- Plasma Membrane Profiling Defines an Expanded Class of Cell Surface Proteins Selectively Targeted for Degradation by HCMV US2 in Cooperation with UL141
- Optineurin Regulates the Interferon Response in a Cell Cycle-Dependent Manner
- IFIT1 Differentially Interferes with Translation and Replication of Alphavirus Genomes and Promotes Induction of Type I Interferon
- The EBNA3 Family of Epstein-Barr Virus Nuclear Proteins Associates with the USP46/USP12 Deubiquitination Complexes to Regulate Lymphoblastoid Cell Line Growth
- Hepatitis C Virus RNA Replication Depends on Specific and -Acting Activities of Viral Nonstructural Proteins
- A Neuron-Specific Antiviral Mechanism Prevents Lethal Flaviviral Infection of Mosquitoes
- The Aspartate-Less Receiver (ALR) Domains: Distribution, Structure and Function
- Global Genome and Transcriptome Analyses of Epidemic Isolate 98-06 Uncover Novel Effectors and Pathogenicity-Related Genes, Revealing Gene Gain and Lose Dynamics in Genome Evolution
- The Ebola Epidemic Crystallizes the Potential of Passive Antibody Therapy for Infectious Diseases
- Ebola Virus Entry: A Curious and Complex Series of Events
- Conserved Spirosomes Suggest a Single Type of Transformation Pilus in Competence
- Spatial Structure, Transmission Modes and the Evolution of Viral Exploitation Strategies
- Bacterial Cooperation Causes Systematic Errors in Pathogen Risk Assessment due to the Failure of the Independent Action Hypothesis
- Transgenic Fatal Familial Insomnia Mice Indicate Prion Infectivity-Independent Mechanisms of Pathogenesis and Phenotypic Expression of Disease
- Cerebrospinal Fluid Cytokine Profiles Predict Risk of Early Mortality and Immune Reconstitution Inflammatory Syndrome in HIV-Associated Cryptococcal Meningitis
- Utilize Host Actin for Efficient Maternal Transmission in
- Borna Disease Virus Phosphoprotein Impairs the Developmental Program Controlling Neurogenesis and Reduces Human GABAergic Neurogenesis
- An Effector Peptide Family Required for Toll-Mediated Immunity
- Hepatitis D Virus Infection of Mice Expressing Human Sodium Taurocholate Co-transporting Polypeptide
- A Redox Regulatory System Critical for Mycobacterial Survival in Macrophages and Biofilm Development
- Quadruple Quorum-Sensing Inputs Control Virulence and Maintain System Robustness
- Leukocyte-Derived IFN-α/β and Epithelial IFN-λ Constitute a Compartmentalized Mucosal Defense System that Restricts Enteric Virus Infections
- A Strategy for O-Glycoproteomics of Enveloped Viruses—the O-Glycoproteome of Herpes Simplex Virus Type 1
- Macrocyclic Lactones Differ in Interaction with Recombinant P-Glycoprotein 9 of the Parasitic Nematode and Ketoconazole in a Yeast Growth Assay
- Neofunctionalization of the α1,2fucosyltransferase Paralogue in Leporids Contributes to Glycan Polymorphism and Resistance to Rabbit Hemorrhagic Disease Virus
- The Extracytoplasmic Linker Peptide of the Sensor Protein SaeS Tunes the Kinase Activity Required for Staphylococcal Virulence in Response to Host Signals
- Murine CMV-Induced Hearing Loss Is Associated with Inner Ear Inflammation and Loss of Spiral Ganglia Neurons
- Dual miRNA Targeting Restricts Host Range and Attenuates Neurovirulence of Flaviviruses
- GATA-Dependent Glutaminolysis Drives Appressorium Formation in by Suppressing TOR Inhibition of cAMP/PKA Signaling
- Role of Hypoxia Inducible Factor-1α (HIF-1α) in Innate Defense against Uropathogenic Infection
- Genetic Analysis Using an Isogenic Mating Pair of Identifies Azole Resistance Genes and Lack of Locus’s Role in Virulence
- A Temporal Gate for Viral Enhancers to Co-opt Toll-Like-Receptor Transcriptional Activation Pathways upon Acute Infection
- Neutrophil Recruitment to Lymph Nodes Limits Local Humoral Response to
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Toxin-Induced Necroptosis Is a Major Mechanism of Lung Damage
- Transgenic Fatal Familial Insomnia Mice Indicate Prion Infectivity-Independent Mechanisms of Pathogenesis and Phenotypic Expression of Disease
- Role of Hypoxia Inducible Factor-1α (HIF-1α) in Innate Defense against Uropathogenic Infection
- EphrinA2 Receptor (EphA2) Is an Invasion and Intracellular Signaling Receptor for
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



