-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaUbiquitous Promoter-Localization of Essential Virulence Regulators in
Most transcription regulators are found only at those promoters they control. Here we show that the most prominent regulators of virulence gene expression in Francisella tularensis are found ubiquitously at promoters including those they do control and those they do not. Furthermore, we present evidence that these regulators—the RNA polymerase-associated SspA family members MglA and SspA, and the putative DNA-binding protein PigR—exert their coordinate regulatory effects only at promoters that contain a small DNA sequence element. Our findings reveal how transcription factors can associate with many promoters but only exert regulatory effects at a few. They also have implications for how SspA family members and other RNAP-associated transcription regulators might exert their effects in other pathogens.
Published in the journal: . PLoS Pathog 11(4): e32767. doi:10.1371/journal.ppat.1004793
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1004793Summary
Most transcription regulators are found only at those promoters they control. Here we show that the most prominent regulators of virulence gene expression in Francisella tularensis are found ubiquitously at promoters including those they do control and those they do not. Furthermore, we present evidence that these regulators—the RNA polymerase-associated SspA family members MglA and SspA, and the putative DNA-binding protein PigR—exert their coordinate regulatory effects only at promoters that contain a small DNA sequence element. Our findings reveal how transcription factors can associate with many promoters but only exert regulatory effects at a few. They also have implications for how SspA family members and other RNAP-associated transcription regulators might exert their effects in other pathogens.
Introduction
Francisella tularensis is a Gram-negative bacterium and the aetiological agent of tularemia, a disease that can be fatal in humans [1]. This pathogen is highly infectious, with as few as 10 organisms constituting an infectious dose, and is a potential bioweapon [2]. The ability of F. tularensis to cause disease is dependent principally upon its ability to grow within macrophages [1,3–5]. Prominent amongst those genes that are essential for the intramacrophage growth and survival of F. tularensis are those located on the Francisella pathogenicity island (FPI), which are thought to have been acquired through horizontal transfer [6–8]. Genes on the FPI encode a type VI secretion system that may secrete effector proteins into cells of the host [9,10], thereby enabling the organism to escape the so-called Francisella-containing vacuole and to replicate freely within the macrophage cytosol [4,11,12].
Expression of the genes on the FPI is dependent upon the coordinate activities of three regulators [13–18]. Two of these, MglA and SspA, belong to the stringent starvation protein A (SspA) family of proteins and form a heteromeric complex that associates with RNA polymerase (RNAP) [13,15,19]. The other is a putative DNA-binding protein called PigR (also known as FevR in F. novicida) that works in concert with MglA and SspA by contacting the RNAP-associated MglA-SspA complex directly [16–18]. MglA, SspA, and PigR also positively control the expression of virulence genes outside the FPI, and are thought to control the expression of ~100 genes in total, including many whose roles in virulence are not yet known [14–17]. The findings that MglA, SspA, and PigR are essential for intramacrophage growth and for virulence underscores the indispensible roles these regulators play in the coordinate control of virulence gene expression in F. tularensis [16,17,20].
According to the current view for how MglA, SspA, and PigR control the expression of virulence genes, PigR functions like a classical transcription activator, binding specifically to a DNA sequence present at the promoters of regulated genes; thus, contact between DNA-bound PigR and the RNAP-associated MglA-SspA complex would stabilize the binding of RNAP to those promoters that contain a PigR binding site [17,18]. However, it is not known whether the promoters of MglA/SspA/PigR-regulated genes contain a specific sequence element that confers responsiveness to PigR. If PigR were indeed to function like a classical transcription activator it would be expected to be located at only those promoters it regulates, and it would be predicted to bind to DNA recognition sites associated with target promoters regardless of whether or not the MglA-SspA complex were present in the cell. Indeed, most classical transcription activators are thought to bind specific sites on the DNA prior to interacting with RNAP [21,22]. Another prediction from the current model is that PigR interacts with the MglA-SspA complex that is associated with the RNAP holoenzyme (the form of the enzyme that contains the σ factor) during transcription initiation [17,18]. However, it is unknown whether the MglA-SspA complex is associated with the RNAP holoenzyme during transcription initiation or the RNAP core enzyme during transcription elongation, or both.
Using chromatin immunoprecipitation followed by high-throughput DNA sequencing (ChIP-Seq), we show that PigR, MglA, and SspA are present at virtually all detected promoters in F. tularensis. We also demonstrate that PigR requires MglA (and thus presumably the MglA-SspA complex) in order to specifically associate with promoters. Finally, we present evidence that the promoters of PigR-regulated genes contain a specific sequence motif that is both necessary and sufficient for PigR-mediated control. Our findings reveal that the most prominent regulators of virulence gene expression in F. tularensis are found at essentially all promoters but only positively control those that contain a specific sequence element.
Results
Defining promoter regions in F. tularensis using ChIP-Seq
In order to address the question of whether PigR specifically associates with the promoters of regulated genes, we first sought to define the locations of promoters on a genome-wide basis in F. tularensis. To do this, we determined the locations of the β′ subunit of RNA polymerase (RNAP) on the F. tularensis chromosome using ChIP-Seq. To immunoprecipitate the β′ subunit of RNAP, we constructed a strain of F. tularensis LVS in which the chromosomal copy of the rpoC gene was modified to encode β′ with a vesicular stomatitis virus-glycoprotein (VSV-G) epitope tag fused to its C-terminus (Fig 1A). This results in cells of LVS which synthesize the β′ subunit of RNAP with a VSV-G tag (β′-V) at native levels. Because β′ is a core subunit of RNAP, β′ is expected to be found at both promoter regions and within actively transcribed genes. Thus, in order to use β′-V to specifically identify the locations of promoters, we performed ChIP-Seq after treatment of the LVS β′-V cells with the RNAP inhibitor rifampicin (rif) to effectively trap RNAP at promoters [23,24] (Fig 1B). By determining the location of the β′ subunit of RNAP in cells treated with rif, we identified 526 promoter regions in F. tularensis LVS (S1 Table).
Fig. 1. Identification of promoters in F. tularensis using ChIP-Seq. 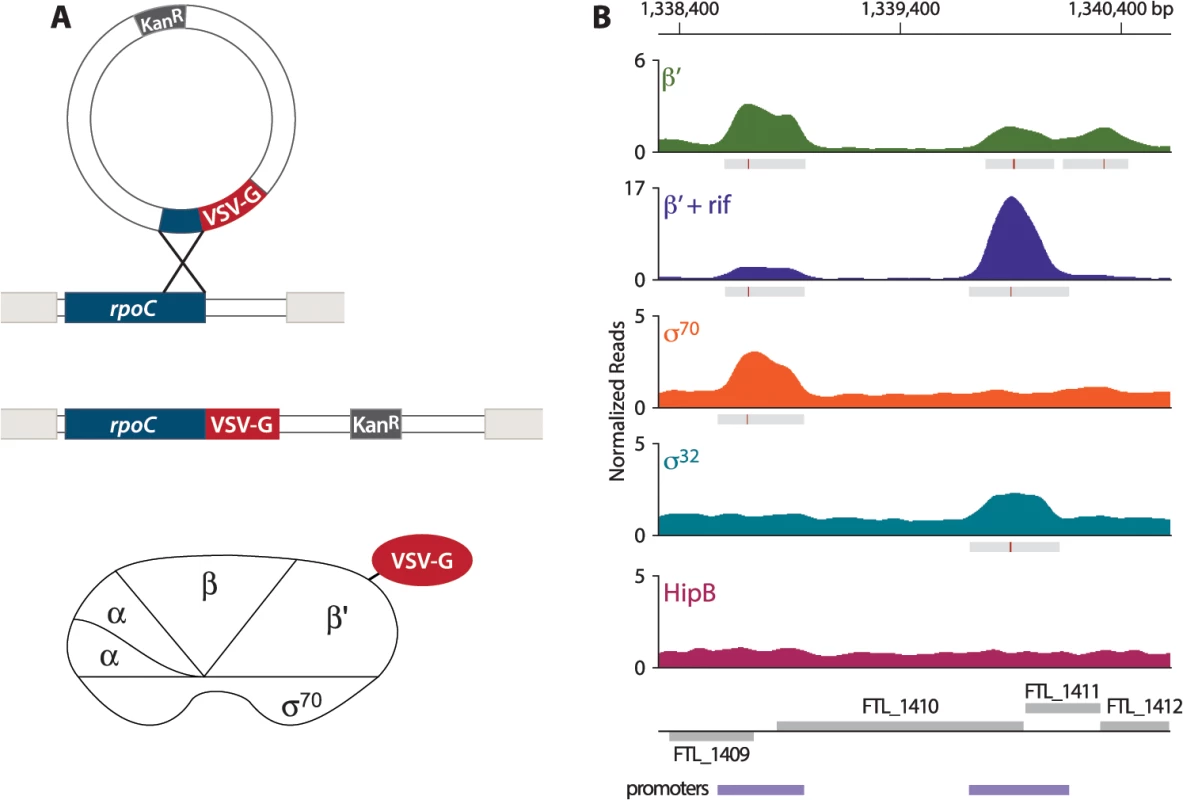
(A) Schematic representation of the VSV-G tag integration vector and its use to construct the LVS β′-V strain that synthesizes the β′ subunit of RNAP with a VSV-G tag (β′-V) at native levels. (B) A representative illustration of the density of normalized mapped sequencing reads (Y-axis) along a region of the chromosome (X-axis) after ChIP-Seq of each epitope-tagged factor: β′ (green), β′+ rif (purple), σ70 (orange), σ32 (cyan), and HipB (dark pink). Gray boxes below the read density plot indicate areas of significantly enriched reads; red lines indicate sites of maximum enrichment. Promoter regions, defined as areas with significant enrichment of β′ + rif, σ32, or σ70 with the chromosome, are indicated by the purple boxes below the gene annotations. F. tularensis encodes two σ factors: σ70, the so-called housekeeping σ factor, and σ32, the so-called heat-shock σ factor [25]. As a complementary approach to identify promoters in F. tularensis, and to determine which promoters are σ70-dependent and which are controlled by σ32, we performed ChIP-Seq with cells that synthesized epitope-tagged versions of each σ factor. To do this we constructed a strain of LVS that synthesized σ70 with a VSV-G epitope tag fused to its C-terminus (LVS σ70-V), and another strain of LVS that synthesized σ32 with a VSV-G tag fused to its C-terminus (LVS σ32-V). As a control we also constructed a strain that synthesized HipB, a predicted site-specific DNA-binding protein [26], with a VSV-G tag fused to its C-terminus (LVS HipB-V). ChIP-Seq with cells of the LVS σ70-V strain identified 333 promoter regions, of which 277 (83.2%) overlap with the locations of promoters defined by determining the location of the β′ subunit of RNAP in cells grown in the presence of rif (Fig 1B, S2 Table). ChIP-Seq with cells of the LVS σ32-V strain identified only 4 promoter regions (Fig 1B, S3 Table). ChIP-Seq with cells of the LVS HipB-V strain revealed that HipB associates with 26 regions of the chromosome (S4 Table). By defining a promoter as a region with significant enrichment of σ70, σ32, or the β′ subunit of RNAP in cells grown in the presence of rif, we identified 581 promoter regions in F. tularensis LVS, 495 (85%) of which were intergenic and 86 (15%) of which were intragenic (S5 Table).
MglA, SspA, and PigR are found at the majority of promoters in F. tularensis
Having determined the locations of promoters in F. tularensis on a genome-wide basis, we next sought to determine at which promoters PigR, MglA, and SspA were located. To do this we utilized a previously constructed strain in which the native chromosomal copy of mglA is altered such that it specifies MglA with a TAP (tandem affinity purification) tag fused to its C-terminus [15]. We also constructed two additional strains of LVS: one in which the native chromosomal copy of pigR had been altered such that it specified PigR with a VSV-G epitope tag fused to its C-terminus (LVS PigR-V); and another in which the native chromosomal copy of sspA had been altered such that it specified SspA with a VSV-G epitope tag fused to its C-terminus (LVS SspA-V).
ChIP-Seq with cells of the LVS PigR-V strain, cells of the LVS MglA-TAP strain, and cells of the LVS SspA-V strain revealed that PigR, MglA, and SspA are located at the majority of promoters in F. tularensis and not just at the promoters of regulated genes (Fig 2, S6 Table). The finding that PigR, MglA, and SspA are found at the promoters of both regulated and non-regulated genes is illustrated in Fig 2A and 2B which show the occupancies of the β′ subunit of RNAP (in the presence of rif), σ70, MglA, SspA, PigR, and HipB at the FTL_0491, FTL_0650, and FTL_0651 promoter regions as determined by ChIP-Seq. Specifically, Fig 2A shows that PigR, MglA, and SspA are found at the promoter of the FTL_0491 gene, which is an example of a gene that is positively regulated by MglA, SspA, and PigR [14,16] (see also S6 Table), whereas Fig 2B shows that PigR, MglA, and SspA are found at the promoters of the FTL_0650 and FTL_0651 genes, which are examples of genes that are known not to be positively regulated by MglA, SspA, and PigR [14–17]. HipB was not detected at any of these promoters by ChIP-Seq (Fig 2A and 2B) indicating the specificity of the observed associations of PigR, MglA, and SspA with these promoters. In contrast, HipB is specifically enriched upstream of the hipB gene (S1 Fig), suggesting that in F. tularensis HipB may control its own expression.
Fig. 2. MglA, SspA, and PigR are found ubiquitously at promoter regions. 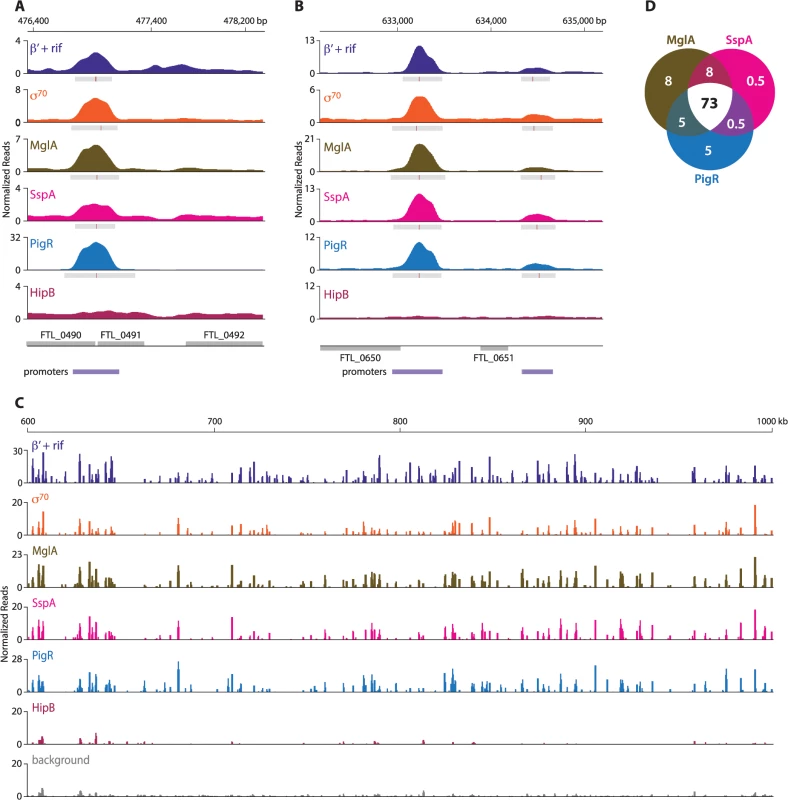
A representative illustration of the density of the normalized mapped sequencing reads after ChIP-Seq of β′+rif (purple), σ70 (orange), MglA (brown), SspA (light pink), PigR (blue), and HipB (dark pink) (A) at the FTL_0491 promoter, which is known to be regulated by MglA, SspA, and PigR; (normalized reads are displayed on a linear scale) (B) at the FTL_0650 and FTL_0651 promoters, which are not under the control of MglA, SspA, or PigR (normalized reads are displayed on a linear scale); and (C) across a 400 kb region of the F. tularensis chromosome (normalized reads are displayed on a log scale). There is significant concordance between the enrichment profiles of β′+rif, σ70, MglA, SspA, and PigR. HipB is not specifically enriched at these regions. (D) Venn diagram representing the overlap between MglA, SspA, and PigR peaks at σ70-associated promoters. Numbers indicate percent of promoters that are enriched for the indicated transcription factor. The finding that PigR, MglA, and SspA are found at the majority of promoters in F. tularensis is illustrated in Fig 2C which shows the locations and degrees of occupancy of the β′ subunit of RNAP (in cells grown in the presence of rif), σ70, MglA, SspA, PigR, and HipB over a representative 400 kb region of the F. tularensis chromosome. Comparison between the regions enriched for σ70, MglA, SspA, and PigR, together with the relative degree of enrichment, revealed a striking correspondence between the four (Fig 2C). Note that the degree of occupancy of the β′ subunit of RNAP at a particular promoter can differ in the presence and absence of rifampicin (Fig 1B) [27], which may explain why the ChIP-Seq enrichment profile for β′ in cells grown in the presence of rif differs slightly from that of σ70 in certain locations (Fig 2C). The concordance among the localization of σ70, MglA, SspA, and PigR across the entire F. tularensis chromosome is demonstrated in Fig 2D which represents the 98% of promoter regions identified by ChIP-Seq of σ70 at which at least one of the three factors, MglA, SspA, or PigR, is found; the Venn diagram shows that MglA, SspA, and PigR are found at the majority of promoters identified by detection of σ70 (Fig 2D). The identification of PigR at the majority of promoters suggests that PigR is not a regulator that is only found at the promoters of specific target genes.
ChIP-Seq with cells of the LVS PigR-V strain, cells of the LVS MglA-TAP strain, and cells of the LVS SspA-V strain also revealed that PigR, MglA, and SspA are present at promoters together with σ70 and are not detected in transcribed regions. This is in contrast to the situation with the β′ subunit of RNAP, which is found both at promoters and within transcribed regions in cells grown in the absence of rif (Fig 1B). These findings suggest that PigR, MglA, and SspA might not be components of the transcription elongation complex and that PigR, MglA, and SspA likely exert their regulatory effects at the level of transcription initiation.
PigR requires MglA in order to specifically associate with promoters
Interaction between PigR and the RNAP-associated MglA-SspA complex is required in order for PigR to function coordinately with MglA and SspA [18]. We therefore next asked whether PigR requires the MglA-SspA complex in order to associate with promoter regions in F. tularensis. Because the expression of pigR is dependent upon the presence of MglA [16,17], in order to address this question we performed ChIP-Seq with cells of a ∆pigR mutant strain and cells of a ∆pigR ∆mglA mutant strain that ectopically synthesized similar amounts of plasmid-encoded PigR-V. We found that when supplied from plasmid pF under the control of the strong heterologous groES promoter, PigR-V was significantly less abundant in cells of the ∆pigR ∆mglA mutant strain than in cells of the ∆pigR mutant strain (Fig 3A). The groES promoter is not positively controlled by MglA, so it is possible that the PigR-V protein is less abundant in cells of a ∆mglA mutant strain because it is less stable in the absence of the MglA-SspA complex. Therefore, to be able to compare cells containing similar amounts of PigR, we used the strong groES promoter on plasmid pF to drive the synthesis of PigR-V in cells of the ∆pigR ∆mglA mutant strain and a weakened groES promoter lacking an UP-element on plasmid pF2 to drive the synthesis of PigR-V in cells of the ∆pigR mutant strain [17] (Figs 3A and S2).
Fig. 3. PigR requires MglA to specifically associate with promoter regions. 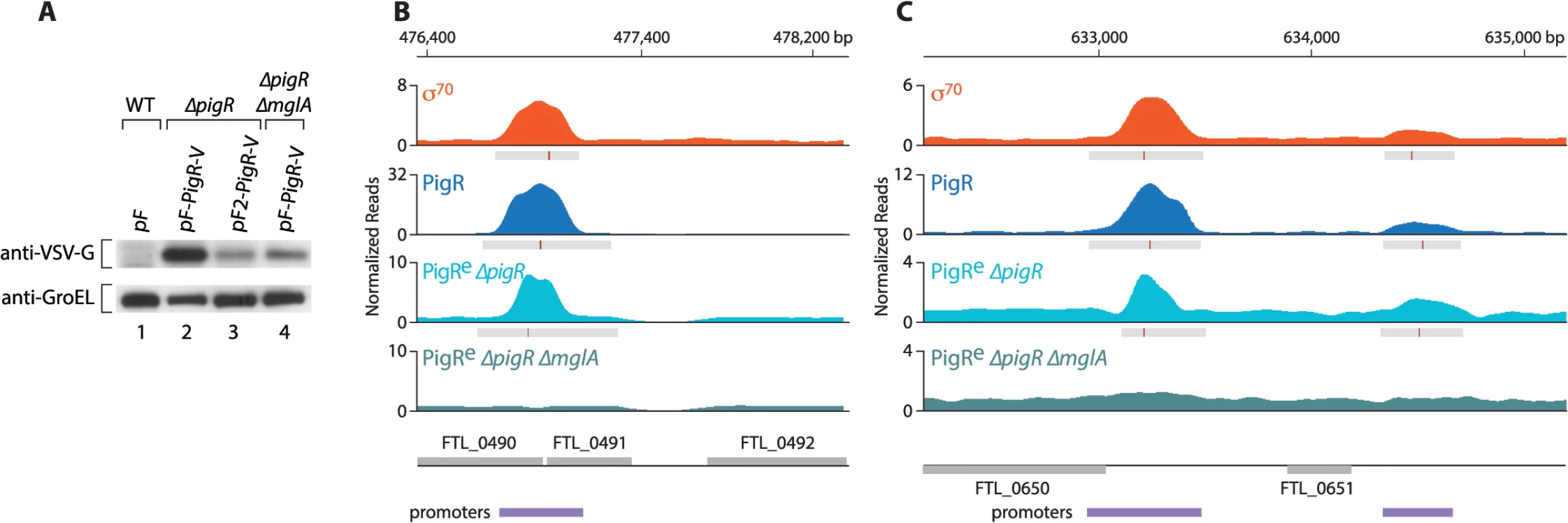
(A) Abundance of ectopically expressed PigR-V as analyzed by Western blot. (Upper) Western blot probed with antibody against the VSV-G tag. (Lower) Western blot probed with antibody against GroEL serves as a loading control. Wild-type LVS cells containing the empty control vector pF (lane 1); LVS ∆pigR mutant cells containing either pF-PigR-V (lane 2) or pF2-PigR-V (lane 3); LVS ∆pigR ∆mglA mutant cells containing pF-PigR-V (lane 4). (B) and (C) Representative datasets illustrating the density of the normalized mapped sequencing reads after ChIP-Seq with cells of the LVS σ70-V strain (orange), cells of the LVS PigR-V strain (blue), cells of the LVS ∆pigR mutant strain that ectopically synthesize PigR-V from plasmid pF2-PigR-V (PigRe ∆pigR, light blue), and cells of the LVS ∆pigR ∆mglA mutant strain that ectopically synthesize PigR-V from plasmid pF-PigR-V (PigRe ∆pigR ∆mglA, blue-green). (B) Ectopically produced PigR-V occupies the promoter of the PigR/MglA/SspA-regulated FTL_0491 gene in cells containing MglA, but not in cells lacking MglA. (C) Ectopically produced PigR-V occupies the promoters of the FTL_0650 and FTL_0651 genes, which are not under the control of PigR/MglA/SspA, only in cells that contain MglA. Comparison of the ChIP-Seq results obtained with ectopically produced PigR with those obtained with native PigR revealed that the ectopic synthesis of PigR does not significantly alter the genome-wide locations of this protein. This is illustrated at the promoter for the PigR/MglA/SspA-regulated FTL_0491 gene (Fig 3B), and illustrated at the promoters for the FTL_0650 and FTL_0651 genes, which are not PigR/MglA/SspA-regulated (Fig 3C). Comparison of the ChIP-Seq results obtained with ectopically produced PigR in the presence and absence of MglA revealed a striking difference; we found no specific enrichment of PigR at any promoter in the absence of MglA, or at any other region of the chromosome. This is illustrated at the FTL_0491, FTL_0650 and FTL_0651 promoters in Fig 3B and 3C. These findings indicate that MglA, and by inference the MglA-SspA complex, is required for PigR to specifically associate with promoter regions in F. tularensis.
A specific sequence motif is found at the promoters of PigR/MglA/SspA-regulated genes
Although PigR (together with MglA and SspA) is present at the majority of promoters, it appears to only positively regulate the expression of a fraction of the corresponding genes. We therefore reasoned that PigR might function as an activator at specific promoters through recognition of a specific sequence element.
To search for a conserved sequence motif in the promoters of genes that are regulated by PigR we first tested whether certain genes previously shown to be regulated by MglA and SspA were also regulated by PigR in F. tularensis. To do this we quantified specific candidate transcripts in both wild-type LVS cells and in cells of a LVS ∆pigR mutant strain using Nanostring (see Materials and Methods; S7 Table). Using MEME [28], we then searched for a specific motif in the promoter regions of genes that (i) were either previously shown to be positively regulated by PigR in LVS by DNA microarray [17], or shown to be positively regulated by PigR in our Nanostring assays (S7 Table), or both, and (ii) contained a region of PigR enrichment upstream from the translation start site, as determined by our ChIP-Seq studies with cells of our LVS PigR-V strain. Eleven genes fit these criteria and a 7 bp motif was found to be present in all 11 of the putative promoter regions analyzed. A logo representing this 7 bp motif, which we have named the PigR response element (PRE) is depicted in Fig 4A.
Fig. 4. The PigR response element (PRE) is necessary and sufficient for promoters to be controlled by PigR. 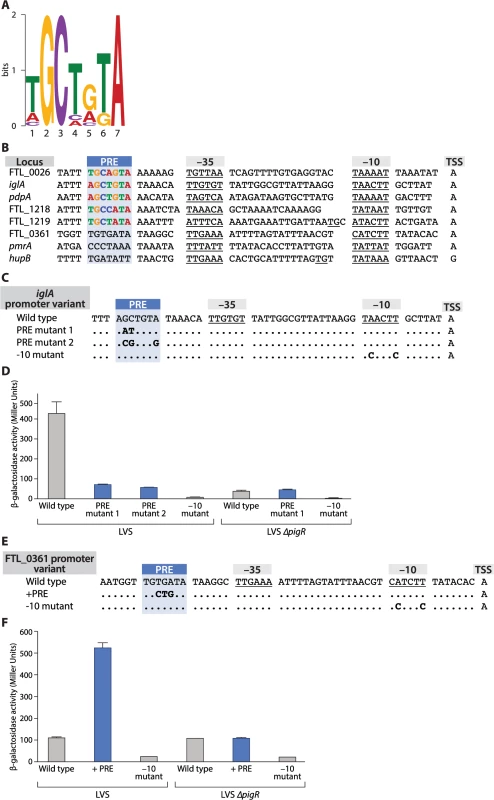
(A) A logo of the 7 bp consensus PRE sequence motif generated by MEME. (B) Alignment of promoters with mapped transcription start sites and predicted -10 and -35 elements (underlined), including the promoters of five PigR-regulated genes (FTL_0026, iglA pdpA, FTL_1218, FTL_1219, FTL_0361) and the promoters of three non-PigR regulated genes (FTL_0361, pmrA, hupB). The conserved PRE is found 6–7 bp upstream of the -35 element only in those promoters known to be controlled by PigR. (C) Alignment of iglA promoter variants. Nucleotide substitutions (in bold) were introduced in the iglA promoter fused to the lacZ reporter gene and integrated into the FTL_0111 locus. (D) Quantification of lacZ expression in strains LVS and LVS ∆pigR containing the indicated promoter variants (indicated along the X-axis) by β-galactosidase assay, as measured in Miller Units (Y-axis). (E) Alignment of FTL_0361 promoter variants. Nucleotide substitutions (in bold) were introduced into the FTL_0361 promoter fused to the lacZ reporter gene and integrated into the FTL_0361 locus. (F) Quantification of lacZ expression in strains LVS and LVS ∆pigR containing the indicated promoter variants (indicated along the X-axis) by β-galactosidase assay, as measured in Miller Units (Y-axis). Error bars for the -10 mutant in the LVS strain, the wild type FTL_0361 promoter and the -10 mutant in the LVS ∆pigR strain are too small to be illustrated. We next asked whether the PRE was found at a specific location relative to the transcription start site of a regulated promoter. To do this we first determined transcription start sites on a genome-wide basis using RNA-Seq [29,30]. This gave us 453 candidate transcription start sites (see Materials and Methods). This list was parsed further to include only those start sites found within 1 kb upstream of the translational start site of an annotated ORF, and remove from consideration those start sites associated with rRNAs and tRNAs, and those found within repeated sequences annotated as encoding transposases. To obtain a list of transcription start sites that could be independently verified as originating from a detectable promoter we further parsed this list of 197 start sites to include only those found within a region of enrichment for σ70, σ32, or the β′ subunit of RNAP (in cells grown in the presence of rif) as determined by ChIP-Seq. This gave us transcription start sites with high confidence for 110 promoters, including 3 of the 11 putative promoter regions used to initially identify the PRE through MEME (S8 Table). We then used primer extension to determine transcription start sites for 2 additional promoters of PigR-regulated genes used in the MEME analysis. Through determining the transcription start sites for the promoters driving the expression of 5 independent PigR-regulated genes we were able to infer the sequences and locations of putative -10 and -35 elements for each of these promoters and found that the PRE was either 6 or 7 bp upstream from the predicted -35 element in each case (Fig 4B). Analysis of the 110 promoters from our high quality data set revealed that only 3 of these contained a PRE (see Materials and Methods) and are known to be PigR-regulated, whereas 107 do not contain a PRE and are not known to be PigR-regulated [17] (Fig 4B, S7 and S8 Tables). This suggests that the presence and location (6 or 7 bp upstream of the putative -35 element) of the PRE is specific to PigR-regulated promoters, raising the possibility that PigR may bind directly to this site to activate transcription from those promoters that contain it.
The PRE is both necessary and sufficient to confer responsiveness to PigR
Having identified a specific conserved sequence element in the same location in the promoters of PigR-regulated genes we sought to determine whether this sequence rendered a particular promoter responsive to PigR. To do this we first constructed a reporter strain of LVS in which one of the two copies of the PigR/MglA/SspA-regulated iglA promoter is transcriptionally fused to lacZ using a chromosomal integration vector [31]. We also made three additional reporter strains of LVS. Two of these contained different mutations at conserved base pairs in the PRE of the iglA promoter-lacZ fusion (Fig 4C), whereas the third reporter strain contained mutations in the predicted -10 element of the iglA promoter-lacZ fusion that would be predicted to abolish promoter activity (Fig 4C) [32]. Finally, we made an additional three reporter strains in cells of the LVS ∆pigR mutant strain that contained the wild-type version, a PRE mutant version, or the -10 mutant version of the iglA promoter-lacZ fusion.
The results depicted in Fig 4D show that mutations in the PRE of the iglA promoter reduce expression of the linked lacZ reporter gene only when PigR is present (i.e. in cells of LVS but not in cells of the LVS ∆pigR mutant strain). Consistent with the idea that these differences are due to a decrease in the activity of the iglA promoter, cells of the reporter strains containing mutations in the -10 element that are predicted to decrease the activity of the promoter exhibit dramatically reduced lacZ expression (Fig 4D). Note that there are two identical copies of the iglA gene in LVS because there are two copies of the FPI in this organism. Only reporter strains carrying wild-type and mutant versions of the iglA promoter-lacZ fusion integrated at the FTL_0111 locus were used in these experiments, ruling out the possibility that any of the observed differences in lacZ expression were due to differences in the location of the reporter in the different strains. Taken together, these findings suggest that residues within the PRE of the iglA promoter are important in order for PigR to exert a positive effect on expression of the iglA gene.
Having established that conserved base pairs within the PRE are important for expression of a PigR-regulated gene we next asked whether the PRE was sufficient to confer control on a promoter that did not ordinarily contain a PRE. To do this we introduced 3 mutations into the FTL_0361 promoter that generated a consensus PRE 6 bp upstream of the putative -35 element (Fig 4E). We then made reporter strains of LVS and the LVS ∆pigR mutant strain that contained the wild-type version, a PRE-containing version, or a -10 mutant version of a FTL_0361 promoter-lacZ fusion. The results depicted in Fig 4F show that addition of a PRE to the FTL_0361 promoter results in an increase in expression of the FTL_0361 promoter-lacZ fusion only in the presence of PigR (i.e. in cells of the LVS wild-type strain but not in cells of the LVS ∆pigR mutant strain). These findings demonstrate that the PRE is sufficient to confer on a promoter the ability to respond to PigR, and by inference, the ability to respond to MglA and SspA.
Discussion
Using ChIP-Seq we have found that PigR, MglA, and SspA are found at the majority of promoters in F. tularensis. We have also found that PigR requires the MglA-SspA complex in order to specifically localize to promoter regions. We infer from this that interaction between PigR and the RNAP-associated MglA-SspA complex directs PigR specifically to promoter regions. Despite their ubiquitous presence at promoters, PigR, MglA, and SspA coordinately control the expression of approximately 5% of known genes and we have uncovered the molecular basis for this specificity. In particular, we have identified a 7 bp sequence element that we have called the PRE (the PigR response element), located approximately 6 bps upstream of the putative -35 element of promoters that are positively regulated by PigR/MglA/SspA. The PRE is both necessary and sufficient to confer control by PigR. Our findings indicate that although PigR, MglA, and SspA are present at essentially all promoters, they control the activities of only those promoters that contain a specific sequence element.
Finding PigR, MglA, and SspA at promoters but not within transcribed regions suggests that these proteins are associated with the RNAP holoenzyme and likely exert their regulatory effects at the level of transcription initiation. Consistent with the idea that the MglA-SspA complex interacts with the σ70-containing RNAP holoenzyme, σ70 together with the core subunits of RNAP and SspA were found to co-purify with MglA in LVS in stoichiometric amounts [15].
PigR contains a putative helix-turn-helix motif, suggesting it might exert its regulatory effects through interaction with the DNA [16,17]. Based on our findings that PigR, MglA, and SspA are present at the majority of σ70-dependent promoters in F. tularensis, together with our identification of the PRE, we propose a model for how PigR works in concert with the MglA-SspA complex to positively regulate the expression of a specific set of genes, including many that are required for virulence (Fig 5). According to this model, PigR is a transcription activator that associates with all promoters through its interaction with the RNAP-associated MglA-SspA complex. However, only at those promoters that contain a PRE does PigR make sufficiently strong contact with the DNA to further stabilize the binding of RNAP and activate transcription. In essence, the model specifies that PigR is an RNAP-associated transcription activator that functions by providing RNAP with an additional DNA-binding domain, conferring on RNAP the ability to form especially stable complexes at promoters that contain a PRE. Note that in this model, PigR/MglA/SspA-regulated promoters are depicted as being recognized by RNAP holoenzyme containing σ70 (i.e. are σ70-dependent promoters), since our ChIP-Seq studies reveal PigR, MglA, SspA, and σ70 are present at many of the same promoter regions. Note also that our model explains only how PigR, together with the MglA-SspA complex, exerts positive effects on gene expression; the small number of genes that are negatively regulated by PigR/MglA/SspA [15–17], may be controlled directly or indirectly by these factors. It is possible that some genes are regulated by PigR/MglA/SspA because they are subject to control by another regulator that is in turn regulated by PigR/MglA/SspA. However, to the best of our knowledge, pigR is the only gene encoding a putative DNA-binding protein that is positively regulated by PigR/MglA/SspA [15–17].
Fig. 5. Model for how PigR functions coordinately with the MglA-SspA complex to positively control the expression of genes. 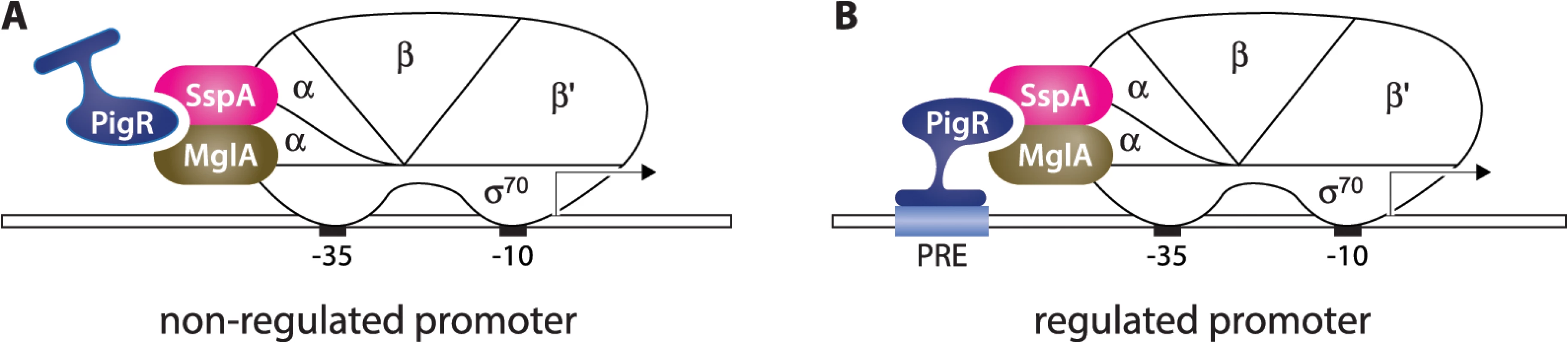
PigR associates with RNAP through interaction with the RNAP-associated MglA-SspA complex and is consequently found associated with RNAP at the promoters of both non-regulated (A) and regulated (B) genes. (B) PigR is a DNA-binding protein that binds the PRE present within the promoters of regulated genes; contact between RNAP-bound PigR and the DNA stabilizes the binding of RNAP to the promoter, thereby activating transcription specifically from promoters that contain a PRE. Although for convenience, MglA and SspA are depicted here as interacting with the α subunit of RNAP, it is not known which subunit(s) of RNAP are contacted by the MglA-SspA complex. Our model specifies that PigR is a DNA-binding protein that associates with RNAP via the MglA-SspA complex prior to promoter binding, and is therefore associated with all promoters, as is supported by our ChIP-Seq data. According to the classical view, transcription activators that bind the DNA and contact RNAP are found only at the promoters of regulated genes and function by first recognizing their respective target sites on the DNA and then, once tethered to the DNA, by contacting RNAP [21,22]. However, there is at least one precedent in the literature for a regulator that appears to manifest ubiquitous promoter localization, while exerting effects at only a subset of promoters. In particular, CarD is an essential RNAP-associated transcription regulator in Mycobacterium tuberculosis that has been found to associate with the majority of promoters [33,34]. Although the ability of CarD to bind the DNA is necessary in order for this regulator to stimulate transcription initiation, it is not yet known whether CarD exerts its regulatory effects at promoters through recognition of a specific sequence element [34]. In addition, in E. coli, members of the MarA family of transcription activators are thought to associate with RNAP prior to DNA-binding and to contact their DNA target sites as a pre-assembled activator-RNAP complex [35–37]. However, to the best of our knowledge, genome-wide location analyses have not been performed on members of the MarA family, and so it is not yet known whether these regulators are found at the majority of promoters in E. coli.
We note that although PigR is predicted to be a DNA-binding protein, PigR has yet to be shown to be capable of binding the DNA, and need not necessarily exert its regulatory effects through direct interaction with the PRE. It is formally possible that interaction between PigR and the RNAP-associated MglA-SspA complex may enable some other portion of RNAP, such as the C-terminal domain of one of the α subunits [15,38], or perhaps the MglA-SspA complex itself, to interact productively with the PRE, resulting in transcription activation. However, in relation to the latter possibility, SspA family members do not contain any obvious DNA-binding determinants and have not been shown to bind the DNA directly [39,40]. It is important to note that although we found PigR does not specifically associate with the DNA in the absence of MglA, this does not mean that PigR is not a site-specific DNA-binding protein. PigR may need to interact with the MglA-SspA complex in order to specifically interact with the PRE, either because the protein-protein interaction promotes a structural change within PigR that is essential for DNA-binding, or because interaction between PigR and the DNA is too weak to be able to occur in the absence of stabilizing interactions provided by promoter-bound RNAP. Indeed, most sequence-specific transcription regulators bind as dimers to their cognate recognition sites, which are typically 20 bp in length. If PigR does bind the 7 bp PRE directly, this would be more reminiscent of a monomer of a dimeric regulator binding a half-site. Regardless of whether or not the ability of PigR to interact directly with the PRE is essential in order for PigR to exert its regulatory effects, our findings clearly establish the PRE as the sequence element that renders a promoter subject to control by PigR, and thus presumably MglA and SspA as well.
The virulence genes present on the horizontally acquired FPI are the ones that are most strongly regulated by PigR, MglA, and SspA, and it is tempting to speculate that the limited size of the PRE (at 7 bp) may have facilitated the expansion of the PigR/MglA/SspA regulatory network to include these. Only three mutations were required in order to generate a consensus PRE within the FTL_0361 promoter (Fig 4E). More than three mutations would likely have been required had the PRE been closer to 20 as opposed to 7 bp. The relatively short length of the PRE means that relatively few changes would be required to place a particular promoter under the control of PigR/MglA/SspA, including any promoter that might have been acquired from a foreign source through horizontal transfer.
Our ChIP-Seq studies suggest that PigR interacts with the MglA-SspA complex at the majority of promoters. This raises the possibility that through interaction with the MglA-SspA complex, PigR may modulate the activity of any other regulator that functions through interaction with the RNAP-bound MglA-SspA complex. Indeed, it has been suggested that PmrA, another important regulator of virulence gene expression in Francisella, might function through interaction with MglA and SspA [41]. It is therefore conceivable that at some promoters PigR may modulate the activity of PmrA, or vice versa, through competition for a binding surface on the MglA-SspA complex.
The role of the MglA-SspA complex in positively controlling the expression of virulence genes appears to be to simply serve as a contact site on RNAP for PigR. Evidence for other SspA family members serving as contact sites for transcription activators comes from studies of bacteriophage P1 late gene expression; in E. coli, SspA evidently functions as a co-activator of P1 late gene expression by making simultaneous contact with RNAP and the phage-encoded sequence-specific DNA-binding protein Lpa [39]. However, in the case of Lpa, it is not known whether this regulator is associated with the majority of promoters in E. coli or just those driving expression of the P1 late genes. SspA family members have been shown to be important for the virulence of a variety of pathogens [42–47]. Serving as a contact site on RNAP for a transcription activator may represent a common mechanism by which SspA family members control the expression of virulence genes in numerous pathogens.
Materials and Methods
Growth conditions
F. tularensis subsp. holarctica LVS and its derivatives were grown at 37°C in either Mueller Hinton (MH) broth (Difco), supplemented with glucose (0.1%), ferric pyrophosphate (0.025%), and Isovitalex (2%), or on cysteine heart agar (Difco) medium supplemented with 1% hemoglobin solution (VWR); when appropriate, kanamycin was used for selection at either 5 μg/ml or 10 μg/ml. Escherichia coli strain XL1-blue (Stratagene) was used for plasmid construction and, when appropriate, kanamycin was used to select for resistance at 50 μg/ml. E. coli containing plasmid pBSK iglA-lacZ, or its derivatives, were grown at 30°C.
VSV-G tagging integration vectors
A modified version of pEX18Kan (provided by Shite Sebastian and Simon Dillon, Harvard Medical School, Boston, Massachusetts, United States) was used as the vector for VSV-G tagging integration constructs. We have used pEX18Kan for deletion constructs [15,17]; it utilizes a ColE1 origin of replication, which is nonfunctional in LVS, and contains the Tn903 kanamycin resistance gene (Epicentre) driven by the LVS groES promoter. The plasmid pKL01 was generated by first amplifying the last 400 base pairs (bp) of the FTL_1743 locus (rpoC), minus the stop codon, by PCR. The 5′ primer contained DNA specifying a KpnI site upstream of the gene fragment. The 3’ primer included DNA containing a NotI site and one extra base pair, encoding a 3 amino acid alanine linker. The linker is followed by DNA specifying the 11 amino acid vesicular stomatitis virus-glycoprotein (VSV-G) epitope tag, followed by a stop codon and DNA specifying an EcoRI site. The corresponding PCR product was digested with KpnI and EcoRI and cloned into pEX18Kan that had been digested with KpnI and EcoRI, generating pKL01. We largely removed the sacB gene by digesting with MscI and EcoRV and re-ligating the vector together, resulting in pKL02.
VSV-G tagging integration constructs for rpoD, rpoH, sspA, pigR, and hipB were generated by amplifying the last 250–400 bp (depending on gene size) of the gene using a 5′ primer containing a KpnI site and a 3′ primer containing a NotI site, which allows each fragment to be fused with DNA specifying the alanine linker and VSV-G epitope tag. Fragments were subcloned into pKL02 that had been digested with KpnI and NotI. Plasmid pKL05 contains the DNA specifying the 3’ end of rpoD and was used to generate strain LVS σ70-V. Plasmid pKL04 contains the DNA specifying the 3’ end of rpoH and was used to generate strain LVS σ32-V. Plasmid pKL08 contains the DNA specifying the 3’ end of pigR and was used to generate strain LVS PigR-V. Plasmid pCS05 contains the DNA specifying the 3’ end of hipB and was used to generate strain LVS HipB-V. Plasmid pKL07 contains the DNA specifying the 3’ end of sspA. Because expression of the putative sspA operon could potentially be interrupted by plasmid integration, pKL07 was modified to contain an outward facing promoter after plasmid integration. To do this, another groES promoter was amplified from LVS genomic DNA and cloned upstream of the sspA gene fragment, into the BamHI and PstI sites, resulting in plasmid pKL13, which was used to generate strain LVS SspA-V.
Plasmids for Francisella β-galactosidase reporter assays
The pBSK iglA-lacZ plasmid (provided by Thomas Kawula, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina, United States) utilizes a ColE1 origin of replication, which is nonfunctional in LVS, contains a kanamycin resistance determinate (aphA1), and contains a transcriptional fusion between the iglA promoter and the lacZ gene [31]. The pBSK iglA-lacZ plasmid contained two tandem PacI sites, so pMO1, which contains DNA specifying the wild-type iglA promoter with a single PacI site before the lacZ gene, was generated by digesting pBSK iglA-lacZ with PacI and NotI and recloning the wild-type iglA promoter fragment into the plasmid backbone. The pMO1 plasmid was used to generate LVS PiglA-lacZ and LVS ∆pigR PiglA-lacZ. Mutations in the PRE of the iglA promoter were generated using splicing by overlap extension PCR [48]. The corresponding PCR products were digested with PacI and NotI and cloned into pBSK iglA-lacZ that had been digested with PacI and NotI to replace the wild-type iglA promoter. The pMO2 plasmid contains DNA specifying the iglA promoter containing the PRE mutant 1 and was used to generate LVS PiglA-M1-lacZ and LVS ∆pigR PiglA-M1-lacZ. The pMO3 plasmid contains DNA specifying the iglA promoter containing the PRE mutant 2 and was used to generate LVS PiglA-M2-lacZ. The pMO4 plasmid contains DNA specifying the iglA promoter containing the -10 mutant and was used to generate LVS PiglA-10M-lacZ and LVS ∆pigR PiglA-10M-lacZ.
The promoter region of FTL_0361 was amplified from F. tularensis LVS genomic DNA using a 5′ primer containing DNA specifying a NotI site upstream of the promoter and the 3’ primer including DNA containing a PacI site downstream of the promoter. The resulting PCR product was digested with PacI and NotI and cloned into pBSK iglA-lacZ that had been digested with PacI and NotI to replace the iglA promoter, generating pMO5. The pMO5 plasmid was used to generate LVS PFTL_0361-lacZ and LVS ∆pigR PFTL_0361-lacZ. Plasmids containing mutations in the FTL_0361 promoter were generated in the same manner as plasmids containing mutations in the iglA promoter. The pMO6 plasmid contains DNA specifying the FTL_0361 promoter containing the PRE and was used to generate LVS PFTL_0361-PRE-lacZ and LVS ∆pigR PFTL_0361-PRE-lacZ. The pMO7 plasmid contains DNA specifying the FTL_0361 promoter containing the -10 mutant and was used to generate LVS PFTL_0361-10M-lacZ and LVS ∆pigR PFTL_0361-10M-lacZ.
Strain construction
Electroporation of integration plasmids into LVS was performed as described [49]. Cells in which a single homologous recombination event had occurred between the integration vector and the chromosome were selected on cysteine heart agar with 1% hemoglobin and either 5 μg/ml (for VSV-G tagging integration vectors) or 10 μg/ml kanamycin (for lacZ reporter integration vectors). Strains containing the correct integration were confirmed by colony PCR, by Western blotting and/or Southern blotting. Strain LVS βʹ-V, which synthesizes the βʹ subunit of RNAP with a C-terminal VSV-G tag, was generated by electroporation of plasmid pKL02 into LVS. Strain LVS σ70-V, which synthesizes the σ70 protein with a C-terminal VSV-G tag, was generated by electroporation of plasmid pKL05 into LVS. Strain LVS σ32-V, which synthesizes the σ32 protein with a C-terminal VSV-G tag, was generated by electroporation of plasmid pKL04 into LVS. Strain LVS SspA-V, which synthesizes the SspA protein with a C-terminal VSV-G tag, was generated by electroporation of plasmid pKL13 into LVS. Strain LVS PigR-V, which synthesizes the PigR protein with a C-terminal VSV-G tag, was generated by electroporation of plasmid pKL08 into LVS. Strain LVS HipB-V, which synthesizes the HipB protein with a C-terminal VSV-G tag, was generated by electroporation of plasmid pCS05 into LVS.
Strains containing the iglA-lacZ transcriptional fusion and derivatives were integrated at the FTL_0111 iglA locus, as determined by Southern blotting; the probe was synthesized using the PCR DIG Probe Synthesis Kit (Roche), hybridized to digested chromosomal DNA that had been transferred to a positively charged nylon membrane, and detected using CDP-Star (Roche). Strains PiglA-lacZ and LVS ∆pigR PiglA-lacZ, which contain lacZ under the control of the wild-type iglA promoter, were generated by electroporation of pMO1 into LVS and LVS ∆pigR, respectively. Strains LVS PiglA-M1-lacZ and LVS ∆pigR PiglA-M1-lacZ, which contain lacZ under the control of the iglA promoter containing the two mutations in the PRE (PRE mutant 1), were generated by electroporation of pMO2 into LVS and LVS ∆pigR, respectively. Strain LVS PiglA-M2-lacZ, which contain lacZ under the control of the iglA promoter containing three mutations in the PRE (PRE mutant 2), was generated by electroporation of pMO3 into LVS. Strains LVS PiglA-10M-lacZ and LVS ∆pigR PiglA-10M-lacZ, which contain lacZ under the control of the iglA promoter containing two mutations in the -10 element, were generated by electroporation of pMO4 into LVS and LVS ∆pigR, respectively. Strains LVS PFTL_0361-lacZ and LVS ∆pigR PFTL_0361-lacZ, which contain lacZ under the control of the FTL_0361 promoter, were generated by electroporation of pMO5 into LVS and LVS ∆pigR, respectively. Strains LVS PFTL_0361-PRE-lacZ and LVS ∆pigR PFTL_0361-PRE-lacZ, which contain lacZ under the control of the FTL_0361 promoter containing the PRE, were generated by electroporation of pMO6 into LVS and LVS ∆pigR, respectively. Strains LVS PFTL_0361-10M-lacZ and LVS ∆pigR PFTL_0361-10M-lacZ, which contain lacZ under the control of the FTL_0361 promoter containing mutations in the -10 element, were generated by electroporation of pMO7 into LVS and LVS ∆pigR, respectively.
Ectopic pigR expression
Plasmids pF and pF-PigR-V have been described previously [17] and were used as a negative control vector and to drive ectopic expression of PigR with a C-terminal VSV-G tag, respectively. The pF-PigR-V plasmid contains DNA encoding the PigR protein fused to the VSV-G epitope, which is driven from the groES promoter; the pF plasmid does not contain the pigR gene or DNA encoding the VSV-G epitope tag. Plasmid pF2-PigR-V synthesizes PigR-V under the control of a weakened groES promoter lacking its putative UP-element and was made by replacing sspA in the plasmid pF2-SspA [15] with DNA encoding PigR-V. These plasmids were electroporated into either cells of the previously described LVS ∆pigR mutant strain [17], or cells of a LVS ∆pigR ∆mglA mutant strain; LVS ∆pigR ∆mglA was created by using the pEX2-∆mglA vector [15] in the ∆pigR background, by allelic exchange and confirmed by Southern blotting.
ChIP-Seq
ChIP-Seq was performed with cells of the following strains: LVS βʹ-V; LVS σ70-V; LVS σ32-V; LVS PigR-V; LVS SspA-V; LVS HipB-V; LVS (as a mock control); LVS containing plasmid pF (as a mock control); LVS ∆pigR containing plasmid pF2-PigR-V; and LVS ∆pigR ∆mglA containing plasmid pF-PigR-V. In order to perform ChIP-Seq on MglA, we used cells of the LVS strain synthesizing MglA with a C-terminal TAP tag (LVS MglA-TAP) at native levels, which has been described previously [15]. Cells were grown at 37°C in 100 mL of supplemented MH to mid-log (OD600 0.3–0.4), and when indicated, rifampicin (Sigma) was added to a final concentration of 50 μg/mL for 30 minutes before crosslinking. Cells were incubated in a final concentration of 1% formaldeyhyde (Sigma) for 30 minutes, after which glycine (Sigma) was added to a final concentration of 250 mM. ChIP was performed in biological triplicate (excepting β′ + rifampicin, the LVS pF empty vector control, and LVS ∆pigR ∆mglA pF-PigR-V, which were performed in duplicate, and σ70, which was performed in quadruplicate) with either 40 mL or 80 mL of culture using anti-VSV-G agarose beads (Sigma) for cells synthesizing VSV-G tagged transcription factors or IgG Sepharose beads (GE Healthcare) for cells synthesizing TAP-tagged MglA essentially as described previously [50], except that a water bath sonicator (Biorupter, Diagenode) was used to lyse cells and shear chromosomal DNA to 200 to 500 bp. Immunoprecipitated DNA was quantified using the Quant-iT PicoGreen dsDNA Assay Kit (Invitrogen). The same protocol was performed with the untagged LVS strain as a mock immunoprecipitation (mock IP) control.
Illumina libraries were constructed with approximately 2 to 160 ng immunoprecipitated DNA using either the TruSeq DNA Sample Prep Kit (Illumina) or the NEBNext Ultra DNA Library Prep Kit for Illumina (NEB), generally following the supplied protocols. In using the TruSeq DNA Sample Prep Kit, adapters were diluted 1 : 10 before ligation and libraries were gel-purified after 11 cycles of amplification. When using the NEBNext Ultra DNA Library Prep Kit, adapters were diluted 1 : 10 before ligation and libraries were size-selected using Agencourt AMPure XP beads prior to 12 cycles of amplification. Libraries were quantified using the Quant-iT PicoGreen dsDNA Assay Kit (Life Technologies) and sequenced by Elim Biopharmaceuticals, Inc. (Hayward, CA), using an Illumina Genome Analyzer Ilx generating 36 bp single-end reads or an Illumina HiSeq 2500 generating 50 bp single-end reads. Sequencing reads have been submitted to the NCBI Sequence Read Archive (SRA, http://www.ncbi.nlm.nih.gov/Traces/sra) with the accession number SRP055716.
ChIP-Seq data analysis
For each strain, the reads were mapped to the F. tularensis subsp. holarctica LVS genome (NCBI locus AM233362) and the sequence of the integrated plasmid, if applicable, using bowtie2-2.0.6 [51]. Regions of enrichment were called using QuEST, version 2.42 [52]. The three mock IP biological replicates, consisting of approximately 30.6 million reads, were merged and used as a background control for each biological replicate. The two pF empty vector control IP biological replicates, consisting of approximately 42.7 million reads, were merged and used as a background control for the ectopic PigR-V experiments. Peaks in each biological replicate are regions that fit the following criteria: they are 1.5-fold enriched for reads over background, with a positive peak shift and strand correlation, and a q-value of less than 0.01. Peaks for each immunoprecipitated protein were defined as the maximal region identified in at least two biological replicates. Promoter regions are defined as the maximal regions of enrichment of βʹ plus rif, σ70 or σ32. Tracks were visualized using the Integrative Genomics Viewer (IGV), version 2.3 [53]. Peak analyses were carried out using Perl scripts, samtools, version 0.1.17 [54], and BEDtools, version 2.17.0 [55].
Immunoblots
Cell lysates were separated by SDS-PAGE on 4–12% or 12% Bis-Tris NuPAGE gels in MES or MOPS running buffer (Life Technologies). Either the iBlot dry blotting system or the XCell II Blot Module (Life Technologies) was used to transfer proteins to either PVDF or nitrocellulose. Membranes were blocked with SuperBlock Blocking Buffer (Pierce) with 0.25% Surfact-Amps 20 (Pierce) for 1 hour to overnight. Membranes were then probed with polyclonal anti-VSV-G (diluted 1 : 1,500; Sigma) or anti-GroEL (diluted 1 : 160,000; provided by Karsten Hazlett, Albany Medical College, Albany, New York, United States) for one hour, washed (10 minutes incubations in TBST plus 0.25% Surfact-Amps 20, 4 times) and re-blocked for 1 hour. After membranes were incubated with polyclonal goat anti-rabbit (diluted 1 : 10,000; Pierce) and washed, proteins were detected using SuperSignal West Pico Chemiluminescent Substrate (Life Technologies).
Transcriptomic analysis
Cells of the LVS wild-type strain and cells of the LVS ∆pigR mutant strain (described in 17) were grown to mid-log in biological triplicate. 1 mL of each sample was pelleted (20,000 rcf for 5 minutes), resuspended in 500 μL Qiagen buffer RLT, frozen on dry ice, and stored at -80°C. Equal amounts of lysate, normalized to OD600, in 4 μL Qiagen buffer RLT and 1 μL water were submitted to the Epithelial Cell Biology Core Facility (Boston Children’s Hospital) for processing using the Nanostring nCounter Prep Station and Digital Analyzer according to the manufacturer’s instructions. For each replicate, total transcript counts were normalized using internal controls with background subtraction, as per manufacturer’s instructions. Transcript abundance was determined by averaging biological triplicates. Criteria indicating a significant change in gene expression are a 2-fold change in transcript abundance and p-value < 0.05 in a two-tailed Student’s t-test.
Identifying the PRE
Genes with significant changes in expression in cells lacking PigR in comparison to wild-type cells (greater than 3-fold by microarray [17] or 2-fold by Nanostring) were examined for promoter regions with detectable PigR, identifying 11 genes (FTL_0026, iglA, pdpA, FTL_0131, FTL_0207, FTL_0449, FTL_0491, FTL_1218, FTL_1219, FTL_1678, FTL_1790). The 400 bp region surrounding the maximal PigR binding site, upstream from 11 genes, excluding coding regions, was searched for a common motif using MEME, version 4.9.1 [28]. We have named the second result, which was present in all 11 promoters and consisted of the consensus sequence TGCTGTA, the PigR response element (PRE).
Identifying transcription start sites using RNA-Seq
LVS cells were grown in aerated liquid culture at 37°C in supplemented MH to mid-log (OD600 0.3–0.4), and RNA was isolated from 10mL of cells in triplicate as described previously [18]. RNA-Seq was used to identify transcription start sites. In particular, from each sample we prepared three cDNA libraries derived from the 5’ ends of RNAs. The first library was generated from RNAs carrying a 5’ triphosphate (prepared as described in [56]), the second library was generated from RNAs carrying a 5’ monophosphate (prepared as described in [56]), while the third library was generated from RNAs carrying either a 5’ triphosphate or a 5’ monophosphate. The third library was prepared by omitting a single step (treatment with Terminator 5’ exonuclease) from the procedure used to generate RNAs carrying a 5’ triphosphate. To identify high quality transcription start sites we first identified 3,120 genomic loci where 50 or more sequencing reads aligned in one of three libraries generated from RNAs carrying a 5’ triphosphate. Of these 3,120 loci, we identified 452 sites that were significantly enriched in a comparison of libraries generated from RNAs carrying a 5’ triphosphate with libraries generated from RNAs carrying a 5’ monophosphate and significantly enriched in a comparison of libraries generated from RNAs carrying a 5’ triphosphate or a 5’ monophosphate with libraries generated from RNAs carrying a 5’ monophosphate. These 452 sites were further filtered to remove those associated with rRNAs, tRNAs, or repeated sequences annotated as transposases. Among the remaining sites, we identified 110 high quality start sites as those found within 1 kb of a translational start site for an annotated ORF and located within a promoter region defined by ChIP-Seq (S8 Table).
Primer extension analysis
Primer extension was used essentially as described previously [57] to determine the putative transcription start sites for pdpA and FTL_1219.
Identifying the location of the PRE
For each transcription start site identified by RNA-Seq, putative -10 and -35 elements were predicted based on homology to the E. coli consensus sequence. For each promoter, 11 bp of sequence, 5 bp upstream from the -35 element and extending upstream, was submitted to FIMO [58] to search for the PRE. Only three promoters were found to contain the PRE with a p-value <0.001, all of which are known to be PigR-regulated. None of the remaining promoters are known to be PigR-regulated and none of them contained the PRE (p<0.001).
β-galactosidase assays
Cells were grown to mid-log phase, and β-galactosidase activity was assessed essentially as described previously [17]. Assays were performed at least twice in triplicate on separate occasions. Representative data sets are shown. Values are averages based on one experiment.
Supporting Information
Zdroje
1. Sjöstedt A. Tularemia: history, epidemiology, pathogen physiology, and clinical manifestations. Ann N Y Acad Sci. 2007 Jun;1105 : 1–29. 17395726
2. Oyston PCF, Sjöstedt A, Titball RW. Tularaemia: bioterrorism defence renews interest in Francisella tularensis. Nat Rev Microbiol. 2004 Dec;2(12):967–78. 15550942
3. Gray CG, Cowley SC, Cheung KKM, Nano FE. The identification of five genetic loci of Francisella novicida associated with intracellular growth. FEMS Microbiology Letters. 2002 Sep 24;215(1):53–6. 12393200
4. Chong A, Celli J. The Francisella intracellular life cycle: toward molecular mechanisms of intracellular survival and proliferation. Front Microbiol. 2010;1 : 138. doi: 10.3389/fmicb.2010.00138 21687806
5. Barel M, Charbit A. Francisella tularensis intracellular survival: to eat or to die. Microbes and Infection. 2013 Oct 15;15(14–15):989–97. doi: 10.1016/j.micinf.2013.08.005 23999313
6. Nano FE, Zhang N, Cowley SC, Klose KE, Cheung KKM, Roberts MJ, et al. A Francisella tularensis pathogenicity island required for intramacrophage growth. J Bacteriol. 2004 Oct;186(19):6430–6. 15375123
7. Larsson P, Oyston PCF, Chain P, Chu MC, Duffield M, Fuxelius H-H, et al. The complete genome sequence of Francisella tularensis, the causative agent of tularemia. Nat Genet. 2005 Feb;37(2):153–9. 15640799
8. Nano FE, Schmerk C. The Francisella pathogenicity island. Ann N Y Acad Sci. 2007 Jun;1105 : 122–37. 17395722
9. Barker JR, Chong A, Wehrly TD, Yu J-J, Rodriguez SA, Liu J, et al. The Francisella tularensis pathogenicity island encodes a secretion system that is required for phagosome escape and virulence. Mol Microbiol. 2009 Dec;74(6):1459–70. 20054881
10. Russell AB, Wexler AG, Harding BN, Whitney JC, Bohn AJ, Goo YA, et al. A type VI secretion-related pathway in Bacteroidetes mediates interbacterial antagonism. Cell Host Microbe. 2014 Aug 13;16(2):227–36. doi: 10.1016/j.chom.2014.07.007 25070807
11. Golovliov I, Baranov V, Krocova Z, Kovarova H, Sjöstedt A. An attenuated strain of the facultative intracellular bacterium Francisella tularensis can escape the phagosome of monocytic cells. Infect Immun. 2003 Oct;71(10):5940–50. 14500514
12. Clemens DL, Lee B-Y, Horwitz MA. Virulent and avirulent strains of Francisella tularensis prevent acidification and maturation of their phagosomes and escape into the cytoplasm in human macrophages. Infect Immun. American Society for Microbiology; 2004 Jun;72(6):3204–17. 15155622
13. Lauriano CM, Barker JR, Yoon S-S, Nano FE, Arulanandam BP, Hassett DJ, et al. MglA regulates transcription of virulence factors necessary for Francisella tularensis intraamoebae and intramacrophage survival. Proc Natl Acad Sci USA. 2004 Mar 23;101(12):4246–9. 15010524
14. Brotcke A, Weiss DS, Kim CC, Chain P, Malfatti S, Garcia E, et al. Identification of MglA-regulated genes reveals novel virulence factors in Francisella tularensis. Infect Immun. 2006 Dec;74(12):6642–55. 17000729
15. Charity JC, Costante-Hamm MM, Balon EL, Boyd DH, Rubin EJ, Dove SL. Twin RNA polymerase-associated proteins control virulence gene expression in Francisella tularensis. PLoS Pathog. 2007 Jun;3(6):e84. 17571921
16. Brotcke A, Monack DM. Identification of fevR, a novel regulator of virulence gene expression in Francisella novicida. Infect Immun. 2008 Aug;76(8):3473–80. doi: 10.1128/IAI.00430-08 18559431
17. Charity JC, Blalock LT, Costante-Hamm MM, Kasper DL, Dove SL. Small molecule control of virulence gene expression in Francisella tularensis. PLoS Pathog. 2009 Oct;5(10):e1000641. doi: 10.1371/journal.ppat.1000641 19876386
18. Rohlfing AE, Dove SL. Coordinate Control of Virulence Gene Expression in Francisella tularensis Involves Direct Interaction between Key Regulators. J Bacteriol. 2014 Oct 1;196(19):3516–26. doi: 10.1128/JB.01700-14 25070738
19. Baron GS, Nano FE. MglA and MglB are required for the intramacrophage growth of Francisella novicida. Mol Microbiol. 1998 Jul;29(1):247–59. 9701818
20. Wehrly TD, Chong A, Virtaneva K, Sturdevant DE, Child R, Edwards JA, et al. Intracellular biology and virulence determinants of Francisella tularensis revealed by transcriptional profiling inside macrophages. Cell Microbiol. 2009 Jul;11(7):1128–50. doi: 10.1111/j.1462-5822.2009.01316.x 19388904
21. Hochschild A, Dove SL. Protein-protein contacts that activate and repress prokaryotic transcription. Cell. 1998 Mar 6;92(5):597–600. 9506513
22. Browning DF, Busby SJ. The regulation of bacterial transcription initiation. Nat Rev Microbiol. 2004 Jan;2(1):57–65. 15035009
23. Campbell EA, Korzheva N, Mustaev A, Murakami K, Nair S, Goldfarb A, et al. Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell. 2001 Mar 23;104(6):901–12. 11290327
24. Herring CD, Raffaelle M, Allen TE, Kanin EI, Landick R, Ansari AZ, et al. Immobilization of Escherichia coli RNA polymerase and location of binding sites by use of chromatin immunoprecipitation and microarrays. J Bacteriol. 2005 Sep;187(17):6166–74. 16109958
25. Grall N, Livny J, Waldor M, Barel M, Charbit A, Meibom KL. Pivotal role of the Francisella tularensis heat-shock sigma factor RpoH. Microbiology. 2009 Aug;155(Pt 8):2560–72. doi: 10.1099/mic.0.029058-0 19443547
26. Black DS, Irwin B, Moyed HS. Autoregulation of hip, an operon that affects lethality due to inhibition of peptidoglycan or DNA synthesis. J Bacteriol. 1994 Jul;176(13):4081–91. 8021189
27. Raffaelle M, Kanin EI, Vogt J, Burgess RR, Ansari AZ. Holoenzyme switching and stochastic release of sigma factors from RNA polymerase in vivo. Molecular Cell. 2005 Nov 11;20(3):357–66. 16285918
28. Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2 : 28–36. 7584402
29. Vvedenskaya IO, Sharp JS, Goldman SR, Kanabar PN, Livny J, Dove SL, et al. Growth phase-dependent control of transcription start site selection and gene expression by nanoRNAs. Genes Dev. 2012 Jul 1;26(13):1498–507. doi: 10.1101/gad.192732.112 22751503
30. Sharma CM, Vogel J. Differential RNA-seq: the approach behind and the biological insight gained. Curr Opin Microbiol. 2014 Jun;19 : 97–105. doi: 10.1016/j.mib.2014.06.010 25024085
31. Fuller JR, Kijek TM, Taft-Benz S, Kawula TH. Environmental and intracellular regulation of Francisella tularensis ripA. BMC Microbiol. 2009;9 : 216. doi: 10.1186/1471-2180-9-216 19821974
32. Roberts CW, Roberts JW. Base-specific recognition of the nontemplate strand of promoter DNA by E. coli RNA polymerase. Cell. 1996 Aug 9;86(3):495–501. 8756731
33. Stallings CL, Stephanou NC, Chu L, Hochschild A, Nickels BE, Glickman MS. CarD is an essential regulator of rRNA transcription required for Mycobacterium tuberculosis persistence. Cell. 2009 Jul 10;138(1):146–59. doi: 10.1016/j.cell.2009.04.041 19596241
34. Srivastava DB, Leon K, Osmundson J, Garner AL, Weiss LA, Westblade LF, et al. Structure and function of CarD, an essential mycobacterial transcription factor. Proc Natl Acad Sci USA. 2013 Jul 15;110(31):12619–24. doi: 10.1073/pnas.1308270110 23858468
35. Martin RG, Gillette WK, Martin NI, Rosner JL. Complex formation between activator and RNA polymerase as the basis for transcriptional activation by MarA and SoxS in Escherichia coli. Mol Microbiol. 2002 Jan;43(2):355–70. 11985714
36. Griffith KL, Shah IM, Myers TE, O'Neill MC, Wolf RE. Evidence for “pre-recruitment” as a new mechanism of transcription activation in Escherichia coli: the large excess of SoxS binding sites per cell relative to the number of SoxS molecules per cell. Biochem Biophys Res Commun. 2002 Mar 8;291(4):979–86. 11866462
37. Griffith KL, Wolf RE. Genetic evidence for pre-recruitment as the mechanism of transcription activation by SoxS of Escherichia coli: the dominance of DNA binding mutations of SoxS. J Mol Biol. 2004 Nov 12;344(1):1–10. 15504398
38. Gourse RL, Ross W, Gaal T. UPs and downs in bacterial transcription initiation: the role of the alpha subunit of RNA polymerase in promoter recognition. Mol Microbiol. 2000 Aug;37(4):687–95. 10972792
39. Hansen A-M, Lehnherr H, Wang X, Mobley V, Jin DJ. Escherichia coli SspA is a transcription activator for bacteriophage P1 late genes. Mol Microbiol. 2003 Jun;48(6):1621–31. 12791143
40. Hansen A-M, Gu Y, Li M, Andrykovitch M, Waugh DS, Jin DJ, et al. Structural Basis for the Function of Stringent Starvation Protein A as a Transcription Factor. J Biol Chem. 2005 Apr 29;280(17):17380–91. 15735307
41. Bell BL, Mohapatra NP, Gunn JS. Regulation of virulence gene transcripts by the Francisella novicida orphan response regulator PmrA: role of phosphorylation and evidence of MglA/SspA interaction. Infect Immun. 2010 May;78(5):2189–98. doi: 10.1128/IAI.00021-10 20231408
42. De Reuse H, Taha MK. RegF, an SspA homologue, regulates the expression of the Neisseria gonorrhoeae pilE gene. Res Microbiol. 1997 May;148(4):289–303. 9765808
43. Badger JL, Miller VL. Expression of invasin and motility are coordinately regulated in Yersinia enterocolitica. J Bacteriol. 1998 Feb;180(4):793–800. 9473031
44. Merrell DS, Hava DL, Camilli A. Identification of novel factors involved in colonization and acid tolerance of Vibrio cholerae. Mol Microbiol. 2002 Mar;43(6):1471–91. 11952899
45. Hansen A-M, Jin DJ. SspA up-regulates gene expression of the LEE pathogenicity island by decreasing H-NS levels in enterohemorrhagic Escherichia coli. BMC Microbiol. 2012;12 : 231. doi: 10.1186/1471-2180-12-231 23051860
46. Yin Y, Withers TR, Wang X, Yu HD. Evidence for sigma factor competition in the regulation of alginate production by Pseudomonas aeruginosa. PLoS ONE. 2013;8(8):e72329. doi: 10.1371/journal.pone.0072329 23991093
47. Skurnik D, Roux D, Aschard H, Cattoir V, Yoder-Himes D, Lory S, et al. A comprehensive analysis of in vitro and in vivo genetic fitness of Pseudomonas aeruginosa using high-throughput sequencing of transposon libraries. PLoS Pathog. 2013 Sep;9(9):e1003582. doi: 10.1371/journal.ppat.1003582 24039572
48. Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989 Apr 15;77(1):51–9. 2744487
49. Maier TM, Havig A, Casey M, Nano FE, Frank DW, Zahrt TC. Construction and characterization of a highly efficient Francisella shuttle plasmid. Appl Environ Microbiol. 2004 Dec;70(12):7511–9. 15574954
50. Castang S, McManus HR, Turner KH, Dove SL. H-NS family members function coordinately in an opportunistic pathogen. Proc Natl Acad Sci USA. 2008 Dec 2;105(48):18947–52. doi: 10.1073/pnas.0808215105 19028873
51. Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Meth. 2012 Apr;9(4):357–9.
52. Valouev A, Johnson DS, Sundquist A, Medina C, Anton E, Batzoglou S, et al. Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nat Meth. 2008 Sep;5(9):829–34.
53. Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinformatics. 2013 Mar;14(2):178–92. doi: 10.1093/bib/bbs017 22517427
54. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. Oxford University Press; 2009 Aug 15;25(16):2078–9. doi: 10.1093/bioinformatics/btp352 19505943
55. Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010 Mar 15;26(6):841–2. doi: 10.1093/bioinformatics/btq033 20110278
56. Goldman SR, Sharp JS, Vvedenskaya IO, Livny J, Dove SL, Nickels BE. NanoRNAs prime transcription initiation in vivo. Molecular Cell. 2011 Jun 24;42(6):817–25. doi: 10.1016/j.molcel.2011.06.005 21700226
57. Dove SL, Joung JK, Hochschild A. Activation of prokaryotic transcription through arbitrary protein-protein contacts. Nature 1997 Apr 10;386(6625):627–30. 9121589
58. Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011 Apr 1;27(7):1017–8. doi: 10.1093/bioinformatics/btr064 21330290
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Relay of Herpes Simplex Virus between Langerhans Cells and Dermal Dendritic Cells in Human SkinČlánek Does the Arthropod Microbiota Impact the Establishment of Vector-Borne Diseases in Mammalian Hosts?Článek The Ebola Epidemic Crystallizes the Potential of Passive Antibody Therapy for Infectious DiseasesČlánek Hepatitis D Virus Infection of Mice Expressing Human Sodium Taurocholate Co-transporting PolypeptideČlánek A Redox Regulatory System Critical for Mycobacterial Survival in Macrophages and Biofilm Development
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2015 Číslo 4- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Pathogens as Biological Weapons of Invasive Species
- Selection and Spread of Artemisinin-Resistant Alleles in Thailand Prior to the Global Artemisinin Resistance Containment Campaign
- Endopeptidase-Mediated Beta Lactam Tolerance
- Prospective Large-Scale Field Study Generates Predictive Model Identifying Major Contributors to Colony Losses
- Relay of Herpes Simplex Virus between Langerhans Cells and Dermal Dendritic Cells in Human Skin
- Structural Determinants of Phenotypic Diversity and Replication Rate of Human Prions
- Sigma Factor SigB Is Crucial to Mediate Adaptation during Chronic Infections
- EphrinA2 Receptor (EphA2) Is an Invasion and Intracellular Signaling Receptor for
- Toxin-Induced Necroptosis Is a Major Mechanism of Lung Damage
- Heterologous Expression in Remodeled . : A Platform for Monoaminergic Agonist Identification and Anthelmintic Screening
- Novel Disease Susceptibility Factors for Fungal Necrotrophic Pathogens in Arabidopsis
- Interleukin 21 Signaling in B Cells Is Required for Efficient Establishment of Murine Gammaherpesvirus Latency
- Phosphorylation at the Homotypic Interface Regulates Nucleoprotein Oligomerization and Assembly of the Influenza Virus Replication Machinery
- Human Papillomaviruses Activate and Recruit SMC1 Cohesin Proteins for the Differentiation-Dependent Life Cycle through Association with CTCF Insulators
- Ubiquitous Promoter-Localization of Essential Virulence Regulators in
- TGF-β Suppression of HBV RNA through AID-Dependent Recruitment of an RNA Exosome Complex
- The Immune Adaptor ADAP Regulates Reciprocal TGF-β1-Integrin Crosstalk to Protect from Influenza Virus Infection
- Antagonism of miR-328 Increases the Antimicrobial Function of Macrophages and Neutrophils and Rapid Clearance of Non-typeable (NTHi) from Infected Lung
- The Epigenetic Regulator G9a Mediates Tolerance to RNA Virus Infection in
- Does the Arthropod Microbiota Impact the Establishment of Vector-Borne Diseases in Mammalian Hosts?
- Hantaan Virus Infection Induces Both Th1 and ThGranzyme B+ Cell Immune Responses That Associated with Viral Control and Clinical Outcome in Humans
- Viral Inhibition of the Transporter Associated with Antigen Processing (TAP): A Striking Example of Functional Convergent Evolution
- Plasma Membrane Profiling Defines an Expanded Class of Cell Surface Proteins Selectively Targeted for Degradation by HCMV US2 in Cooperation with UL141
- Optineurin Regulates the Interferon Response in a Cell Cycle-Dependent Manner
- IFIT1 Differentially Interferes with Translation and Replication of Alphavirus Genomes and Promotes Induction of Type I Interferon
- The EBNA3 Family of Epstein-Barr Virus Nuclear Proteins Associates with the USP46/USP12 Deubiquitination Complexes to Regulate Lymphoblastoid Cell Line Growth
- Hepatitis C Virus RNA Replication Depends on Specific and -Acting Activities of Viral Nonstructural Proteins
- A Neuron-Specific Antiviral Mechanism Prevents Lethal Flaviviral Infection of Mosquitoes
- The Aspartate-Less Receiver (ALR) Domains: Distribution, Structure and Function
- Global Genome and Transcriptome Analyses of Epidemic Isolate 98-06 Uncover Novel Effectors and Pathogenicity-Related Genes, Revealing Gene Gain and Lose Dynamics in Genome Evolution
- The Ebola Epidemic Crystallizes the Potential of Passive Antibody Therapy for Infectious Diseases
- Ebola Virus Entry: A Curious and Complex Series of Events
- Conserved Spirosomes Suggest a Single Type of Transformation Pilus in Competence
- Spatial Structure, Transmission Modes and the Evolution of Viral Exploitation Strategies
- Bacterial Cooperation Causes Systematic Errors in Pathogen Risk Assessment due to the Failure of the Independent Action Hypothesis
- Transgenic Fatal Familial Insomnia Mice Indicate Prion Infectivity-Independent Mechanisms of Pathogenesis and Phenotypic Expression of Disease
- Cerebrospinal Fluid Cytokine Profiles Predict Risk of Early Mortality and Immune Reconstitution Inflammatory Syndrome in HIV-Associated Cryptococcal Meningitis
- Utilize Host Actin for Efficient Maternal Transmission in
- Borna Disease Virus Phosphoprotein Impairs the Developmental Program Controlling Neurogenesis and Reduces Human GABAergic Neurogenesis
- An Effector Peptide Family Required for Toll-Mediated Immunity
- Hepatitis D Virus Infection of Mice Expressing Human Sodium Taurocholate Co-transporting Polypeptide
- A Redox Regulatory System Critical for Mycobacterial Survival in Macrophages and Biofilm Development
- Quadruple Quorum-Sensing Inputs Control Virulence and Maintain System Robustness
- Leukocyte-Derived IFN-α/β and Epithelial IFN-λ Constitute a Compartmentalized Mucosal Defense System that Restricts Enteric Virus Infections
- A Strategy for O-Glycoproteomics of Enveloped Viruses—the O-Glycoproteome of Herpes Simplex Virus Type 1
- Macrocyclic Lactones Differ in Interaction with Recombinant P-Glycoprotein 9 of the Parasitic Nematode and Ketoconazole in a Yeast Growth Assay
- Neofunctionalization of the α1,2fucosyltransferase Paralogue in Leporids Contributes to Glycan Polymorphism and Resistance to Rabbit Hemorrhagic Disease Virus
- The Extracytoplasmic Linker Peptide of the Sensor Protein SaeS Tunes the Kinase Activity Required for Staphylococcal Virulence in Response to Host Signals
- Murine CMV-Induced Hearing Loss Is Associated with Inner Ear Inflammation and Loss of Spiral Ganglia Neurons
- Dual miRNA Targeting Restricts Host Range and Attenuates Neurovirulence of Flaviviruses
- GATA-Dependent Glutaminolysis Drives Appressorium Formation in by Suppressing TOR Inhibition of cAMP/PKA Signaling
- Role of Hypoxia Inducible Factor-1α (HIF-1α) in Innate Defense against Uropathogenic Infection
- Genetic Analysis Using an Isogenic Mating Pair of Identifies Azole Resistance Genes and Lack of Locus’s Role in Virulence
- A Temporal Gate for Viral Enhancers to Co-opt Toll-Like-Receptor Transcriptional Activation Pathways upon Acute Infection
- Neutrophil Recruitment to Lymph Nodes Limits Local Humoral Response to
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Toxin-Induced Necroptosis Is a Major Mechanism of Lung Damage
- Transgenic Fatal Familial Insomnia Mice Indicate Prion Infectivity-Independent Mechanisms of Pathogenesis and Phenotypic Expression of Disease
- Role of Hypoxia Inducible Factor-1α (HIF-1α) in Innate Defense against Uropathogenic Infection
- EphrinA2 Receptor (EphA2) Is an Invasion and Intracellular Signaling Receptor for
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



