-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaDoes the Arthropod Microbiota Impact the Establishment of Vector-Borne Diseases in Mammalian Hosts?
The impact of the microbiota on the immune status of its host is a source of intense research and publicity. In comparison, the effect of arthropod microbiota on vector-borne infectious diseases has received little attention. A better understanding of the vector microbiota in relation to mammalian host immune responses is vital, as it can lead to strategies that affect transmission and improve vaccine design in a field of research where few vaccines exist and effective treatment is rare. Recent demonstrations of how microbiota decrease pathogen development in arthropods, and thus alter vector permissiveness to vector-borne diseases (VBDs), have led to renewed interest. However, hypotheses on the interactions between the arthropod-derived microbiota and the mammalian hosts have yet to be addressed. Advances in DNA sequencing technology, increased yield and falling costs, mean that these studies are now feasible for many microbiologists and entomologists. Here, we distill current knowledge and put forward key questions and experimental designs to shed light on this burgeoning research topic.
Published in the journal: . PLoS Pathog 11(4): e32767. doi:10.1371/journal.ppat.1004646
Category: Opinion
doi: https://doi.org/10.1371/journal.ppat.1004646Summary
The impact of the microbiota on the immune status of its host is a source of intense research and publicity. In comparison, the effect of arthropod microbiota on vector-borne infectious diseases has received little attention. A better understanding of the vector microbiota in relation to mammalian host immune responses is vital, as it can lead to strategies that affect transmission and improve vaccine design in a field of research where few vaccines exist and effective treatment is rare. Recent demonstrations of how microbiota decrease pathogen development in arthropods, and thus alter vector permissiveness to vector-borne diseases (VBDs), have led to renewed interest. However, hypotheses on the interactions between the arthropod-derived microbiota and the mammalian hosts have yet to be addressed. Advances in DNA sequencing technology, increased yield and falling costs, mean that these studies are now feasible for many microbiologists and entomologists. Here, we distill current knowledge and put forward key questions and experimental designs to shed light on this burgeoning research topic.
Introduction
Recent advances in DNA sequencing technologies are leading to dramatic discoveries across a broad range of biological and medical themes. For example, it is now possible to survey the microbial communities present in a wide array of ecological and biological niches. This work has led to the terms “metagenomics” and “microbiome” (Box 1) appearing frequently in both the scientific and popular press. Understandably, the focus has been on the human microbiome, with sequencing of various tissues revealing a significant diversity in composition within and between individuals. Differences in the human microbiota (Box 1) influence the immune system and are associated with various diseases, including chronic conditions such as inflammatory bowel disease, as well as infectious diseases [1]. However, comparatively little is currently known about the microbiota of invertebrates. These microbe-invertebrate interactions may be of particular relevance in aiding our understanding of vector-borne diseases (VBDs, Box 1).
Box 1. Glossary
Metagenomics
This term has been defined as “the application of modern genomics techniques to the study of communities of microbial organisms directly in their natural environments, bypassing the need for isolation and lab cultivation of individual species” [33].
Microbiome
Many scientific articles distinguish “microbiome” and “microbiota” to describe either the collective genomes of the microorganisms that reside in an environmental niche or the microorganisms themselves, respectively.
Microbiota
This term refers to both the microflora and microfauna in an ecosystem.
Paratransgenesis
This technique aims to decrease transmission of VBDs by eliminating pathogens from arthropod vectors through transgenesis (introduction of exogenous genes) of a vector symbiont.
Symbiotic species
Symbionts are species that have a long-term interaction. Specifically for this review, we refer to symbionts as commensal, mutualistic, and parasitic relationships that exist between two or more groups of organisms. They may be obligate or not.
Vector-borne disease (VBD)
For the purposes of this review, this term refers to infections due to pathogens transmitted by arthropod vectors.
Arthropods act as vectors for a large array of pathogens (defined here as causative agents of VBDs), transmitting them between animal or plant hosts. The direct impact on human health is considerable; tick-borne Lyme disease, as well as malaria, chikungunya, and dengue, all transmitted by mosquitoes, represent newsworthy VBDs.
Many arthropods harbor large communities of diverse microorganisms [2] that, as with human hosts, almost certainly exceed the number of cells that make up the host itself [3]. These microorganisms live within the digestive tract and/or salivary glands of arthropods where they can interact with vector-borne pathogens and/or influence their lifecycle. The microbiota and/or microbiomes within certain arthropod vectors have been extensively catalogued and reveal a large diversity (reviewed in [4]).
In mammals, pathogen loads are greatly affected by the microbiome. Similarly, vector microbiomes have been shown to naturally diminish pathogen transmission from the vector to the host by decreasing pathogen loads in the vector [5–11]. However, this is dependent on the type of bacteria (e.g., Gram + versus Gram-) [12]. These findings are a first step towards creating tools that can alter the arthropod microbiota towards reducing vector-borne pathogen transmission. Indeed, paratransgenic studies (Box 1) are focusing on transforming specific arthropod microbiota to express gene products that interfere with pathogen transmission [13]. Although this work will help curb VBD transmission, the effect of arthropod microbiota on the establishment of VBDs—once they have been transmitted to their mammalian hosts—urgently requires further investigation to help researchers understand how these microorganisms affect the mammalian host’s immune response to the vector-borne pathogen.
Despite the research to date, and the increasing amount of data generated through new sequencing techniques, we are still not at a stage where we have comprehensively studied arthropod vector microbiomes. Moreover, reports of the vector microbiomes of the same arthropods have varied, depending on factors such as life-stage [14,15], sex [16,17] and geographical distribution/sampling time [6,18–21]. Whether the differences observed so far are due to differences in research methods (Box 2) remains under investigation. An additional shortcoming is that studies have typically focused on bacteria and ignored viruses, protozoa, and fungal species, and few studies have distinguished resident from transient microbial populations [14], though symbionts (Box 1) have been observed in the salivary glands of certain vectors such as Glossina flies, ticks, and mosquitoes [22–24]. Additionally, most research groups sequencing vector microbiomes have concentrated on characterizing whole arthropod or gut microbiomes [14,25]. In human medicine, there is a desire to move beyond cataloguing bacteria species to consideration of their functional interactions with the host [26]; the latter is likely to result in a more robust assessment of microbe-host interactions. Highlighting potentially overlapping functions may also help pinpoint the relevant variability in the microbiome. A similar approach is essential if we are to understand the impact of the microbiota on the transmission and establishment of VBDs.
Box 2. Methods used to study microbiomes
Traditionally, microbial studies of arthropod vectors have had to rely on culture-based techniques and have been more interested in identifying pathogens than commensals/mutualistic species (reviewed in [4]). However, only a proportion of bacterial species grow under culture conditions, which has severely limited our knowledge of the microbial communities present in a given ecological niche. The advances in DNA sequencing technologies, termed next-generation sequencing (NGS), are now allowing the study of mixed communities. NGS is now being used to understand the microbiota of arthropod vectors.
Microbiome amplicon sequencing targets the 16S ribosomal RNA gene, in a high throughput manner, with significantly reduced bias: redundant PCR primers are designed against constant regions of the 16S rRNA gene, which flank the more variable regions. Millions of sequence reads are generated, which can then be clustered using any of a growing number of algorithms and software. The result is a clearer idea of bacterial species richness and relative abundance in arthropod vectors. However, caution must be taken in interpreting data generated by NGS. Few studies have combined culture and PCR-based approaches to characterize arthropod microbiota [34–36], even though both methods yield different, yet complementary results.
So far, microbiome projects have concentrated on bacterial elements. Indeed, these are relatively easy to identify, originally grown in culture and more recently through 16S rRNA sequencing. However, in vivo, the microbiota of arthropods is composed of bacteria, viruses, fungi, and other eukaryotic parasites. These other microbes should not be dismissed when cataloguing the microbiota. Fungal species (symbiotic yeast species and the potentially pathogenic Aspergillus respectively) have already been identified in the guts of Aedes and Culex mosquitoes [37,38]. These could impact VBD transmission just as much as symbiotic bacteria, such as the Enterobacteriaceae family for malaria [6]. It will be crucial to discriminate between microbes that are resident, i.e., relatively static in a niche, and those that are transient, which may play the most important role in transmission and establishment of a VBD. Here, alternative technologies can provide additional and supporting information. For example, fluorescent in situ hybridization (FISH), reveals the location and abundance of a targeted microorganism [39]. Abundance data is considerably harder to determine with 16S sequencing because of dramatic gene copy number variation even between relatively closely related species; within the genus Bacillus the 16S copy number of different species varies between seven and 14 [40].
Noting the type of symbiont identified is also paramount. Commensal, mutualistic, and parasitic species may have very different impacts on VBD transmission. Indeed the impact of commensals of humans on the human host has been shown to depend on the density of the species: beneficial at some levels, harmful at others [41]. Also, the transmission of the bacterial species between vectors (horizontal or vertical) will affect how useful they are as a tool in curbing VBD transmission. Indeed, obligate species like Wolbachia, which are parasitic in many insects, and which are transmitted vertically have already been introduced into vector populations and shown to reduce VBD transmission and remain stable in the vector population [42]. Even genotypes of the same strain can have different impacts on VBD transmission. Differences in genotypes of Sodalis, a symbiont of the tsetse fly, result in differences in transmission of trypanosomes [43].
At this point, our knowledge of vector microbiomes is still limited and consists mostly of an emerging appreciation of their impact on pathogens carried by the vectors. Research has so far only focused on the role of the vector microbiota within their arthropod host, where they can strongly influence the transmission of vector-borne pathogens [5–11].
What is entirely missing is an understanding of how vector microbiomes may affect the immune response of the mammalian host to the pathogen following the bite of an infected arthropod. The impact of vector microbiota on skin-mediated host immunity and on the establishment of VBDs within the mammalian host have yet to be investigated in any depth, despite direct relevance to disease control (Fig 1). Infection transmission routes make the transmission of vector-borne microbiota along with vector-borne pathogens likely. Injection (e.g., ticks, fleas, mosquitoes, sand flies) could lead to the transmission of salivary gland and gut microbiomes through feeding and regurgitation cycles. A number of vector salivary components delivered to the host during arthropod feeding strongly impact VBD dynamics and outcome [27]. If transmitted through saliva or co-deposited with the pathogen and saliva, could the vector microbiome impact downstream host immune responses to the arthropod bite and any vector-borne pathogens? Since the mammalian and vector microbiomes can stimulate the immune system of their hosts [8,7,28], this seems likely. The composition of the biofilm growth of Yersinia pestis transmitted to the mammalian host is thought to contain flea gut and bacterially derived components [29]. Defecation (e.g., for reduviids, which transmit Chagas disease–causing trypanosome parasites) could also lead to the transmission of gut microbiome components to the vertebrate host.
Fig. 1. How arthropod microbiota could enhance/interfere with the transmission/establishment of VBDs. 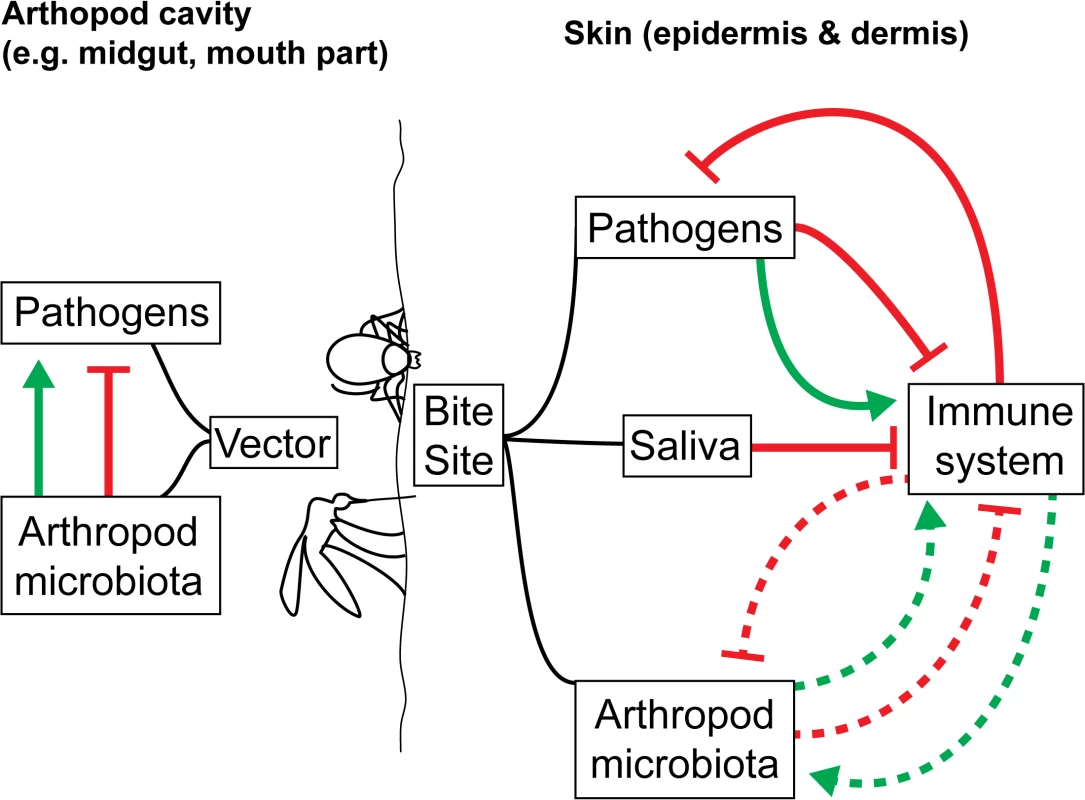
In the arthropod cavity, the arthropod microbiota can alter pathogen development, resulting in decreased or increased loads in the vectors and reduced or increased transmission. However, the impact of the pathogens on the microbiota has yet to be assessed. Once transmission has occurred, the host immune system generates a response to destroy the pathogens in the skin. Components from the pathogens themselves and the arthropod saliva are known to actively inhibit this process. The role of the arthropod microbiota, likely transmitted along with the pathogens, on the host immune system is currently unknown (dotted lines). Establishing the transfer of vector microbiota to the host represents an important step towards understanding the initial host immune response to arthropod bites and vector-borne pathogens. As a first step, available genomic and transcriptomic data from vector saliva or midguts (e.g., ticks [30] and sand flies [31]) can be used to identify the presence of vector microbiota in arthropod saliva or midgut tissue. Once key species are identified in the vector, their presence can be confirmed at the bite site. This is a crude, indirect method, in which the host skin microbiome may act as a confounder, but it serves as a preliminary indication of the presence of vector microbiota in arthropod bites. For a more in-depth investigation, fluorescently tagged species can be introduced into the vector salivary glands and/or midguts. After a feed, fluorescence can be tracked in the host. If the microbiota are found to be transferred from vector to host, using germ-free arthropods (through temporary [9,8,7,10] or permanent removal of microbes [32]) may provide insights into whether the immune response to VBDs is modulated by the vector microbiota.
We are only just beginning to uncover the full impact of the vector microbiota on VBD disease dynamics in arthropod vectors. Their impact on the disease transmission process and mammalian immune responses to vector-borne pathogens (Fig 1) are topics that have been under-studied and largely ignored until now. However, with DNA sequencing approaches now generating huge volumes of data with which to study vector microbiomes, a new wealth of data is becoming available. It will need to be carefully collected, compiled, and analyzed in order to generate meaningful interpretations that can be applied to understanding and perhaps controlling VBDs.
Zdroje
1. Molloy MJ, Bouladoux N, Belkaid Y (2012) Intestinal microbiota: shaping local and systemic immune responses. Semin Immunol 24 : 58–66. doi: 10.1016/j.smim.2011.11.008 22178452
2. Engel P, Moran NA (2013) The gut microbiota of insects—diversity in structure and function. FEMS Microbiol Rev 37 : 699–735. doi: 10.1111/1574-6976.12025 23692388
3. Dillon RJ, Dillon VM (2004) The gut bacteria of insects: nonpathogenic interactions. Annu Rev Entomol 49 : 71–92. 14651457
4. Azambuja P, Garcia ES, Ratcliffe NA (2005) Gut microbiota and parasite transmission by insect vectors. Trends Parasitol 21 : 568–572. 16226491
5. Pumpuni CB, Demaio J, Kent M, Davis JR, Beier JC (1996) Bacterial population dynamics in three anopheline species: the impact on Plasmodium sporogonic development. Am J Trop Med Hyg 54 : 214–218. 8619451
6. Gonzalez-Ceron L, Santillan F, Rodriguez MH, Mendez D, Hernandez-Avila JE (2003) Bacteria in midguts of field-collected Anopheles albimanus block Plasmodium vivax sporogonic development. J Med Entomol 40 : 371–374. 12943119
7. Xi Z, Ramirez JL, Dimopoulos G (2008) The Aedes aegypti toll pathway controls dengue virus infection. PLoS Pathog 4: e1000098. doi: 10.1371/journal.ppat.1000098 18604274
8. Dong Y, Manfredini F, Dimopoulos G (2009) Implication of the mosquito midgut microbiota in the defense against malaria parasites. PLoS Pathog 5: e1000423. doi: 10.1371/journal.ppat.1000423 19424427
9. Mourya DT, Soman RS (1985) Effect of gregarine parasite, Ascogregarina culicis & tetracycline on the susceptibility of Culex bitaeniorhynchus to JE virus. Indian J Med Res 81 : 247–250. 3926641
10. Castro DP, Moraes CS, Gonzalez MS, Ratcliffe NA, Azambuja P, et al. (2012) Trypanosoma cruzi Immune Response Modulation Decreases Microbiota in Rhodnius prolixus Gut and Is Crucial for Parasite Survival and Development. PLoS One 7: e36591. doi: 10.1371/journal.pone.0036591 22574189
11. Sant’Anna MR V, Diaz-Albiter H, Aguiar-Martins K, Al Salem WS, Cavalcante RR, et al. (2014) Colonisation resistance in the sand fly gut: Leishmania protects Lutzomyia longipalpis from bacterial infection. Parasit Vectors 7 : 329. doi: 10.1186/1756-3305-7-329 25051919
12. Straif SC, Mbogo CN, Toure AM, Walker ED, Kaufman M, et al. (1998) Midgut bacteria in Anopheles gambiae and An. funestus (Diptera: Culicidae) from Kenya and Mali. J Med Entomol 35 : 222–226. 9615538
13. Hurwitz I, Fieck A, Read A, Hillesland H, Klein N, et al. (2011) Paratransgenic control of vector borne diseases. Int J Biol Sci 7 : 1334–1344. 22110385
14. Wang Y, Gilbreath TM 3rd, Kukutla P, Yan G, Xu J (2011) Dynamic gut microbiome across life history of the malaria mosquito Anopheles gambiae in Kenya. PLoS One 6: e24767. doi: 10.1371/journal.pone.0024767 21957459
15. Akhoundi M, Bakhtiari R, Guillard T, Baghaei A, Tolouei R, et al. (2012) Diversity of the bacterial and fungal microflora from the midgut and cuticle of phlebotomine sand flies collected in North-Western Iran. PLoS One 7: e50259. doi: 10.1371/journal.pone.0050259 23226255
16. Rani A, Sharma A, Rajagopal R, Adak T, Bhatnagar RK (2009) Bacterial diversity analysis of larvae and adult midgut microflora using culture-dependent and culture-independent methods in lab-reared and field-collected Anopheles stephensi-an Asian malarial vector. BMC Microbiol 9 : 96. doi: 10.1186/1471-2180-9-96 19450290
17. Andreotti R, Perez de Leon AA, Dowd SE, Guerrero FD, Bendele KG, et al. (2011) Assessment of bacterial diversity in the cattle tick Rhipicephalus (Boophilus) microplus through tag-encoded pyrosequencing. BMC Microbiol 11 : 6. doi: 10.1186/1471-2180-11-6 21211038
18. Lalzar I, Harrus S, Mumcuoglu KY, Gottlieb Y (2012) Composition and Seasonal Variation of Rhipicephalus turanicus and Rhipicephalus sanguineus Bacterial Communities. Appl Env Microbiol 78 : 4110–4116. doi: 10.1128/AEM.00323-12 22467507
19. Jones RT, Knight R, Martin AP (2010) Bacterial communities of disease vectors sampled across time, space, and species. ISME J 4 : 223–231. doi: 10.1038/ismej.2009.111 19865184
20. Boissière A, Tchioffo MT, Bachar D, Abate L, Marie A, et al. (2012) Midgut microbiota of the malaria mosquito vector Anopheles gambiae and interactions with Plasmodium falciparum infection. PLoS Pathog 8: e1002742. doi: 10.1371/journal.ppat.1002742 22693451
21. Sant’Anna MR V, Darby AC, Brazil RP, Montoya-Lerma J, Dillon VM, et al. (2012) Investigation of the bacterial communities associated with females of Lutzomyia sand fly species from South America. PLoS One 7: e42531. doi: 10.1371/journal.pone.0042531 22880020
22. Cheng Q, Aksoy S (1999) Tissue tropism, transmission and expression of foreign genes in vivo in midgut symbionts of tsetse flies. Insect Mol Biol 8 : 125–132. 9927181
23. Klyachko O, Stein BD, Grindle N, Clay K, Fuqua C (2007) Localization and visualization of a coxiella-type symbiont within the lone star tick, Amblyomma americanum. Appl Environ Microbiol 73 : 6584–6594. 17720830
24. Crotti E, Damiani C, Pajoro M, Gonella E, Rizzi A, et al. (2009) Asaia, a versatile acetic acid bacterial symbiont, capable of cross-colonizing insects of phylogenetically distant genera and orders. Environ Microbiol 11 : 3252–3264. doi: 10.1111/j.1462-2920.2009.02048.x 19735280
25. Carpi G, Cagnacci F, Wittekindt NE, Zhao F, Qi J, et al. (2011) Metagenomic profile of the bacterial communities associated with Ixodes ricinus ticks. PLoS One 6: e25604. doi: 10.1371/journal.pone.0025604 22022422
26. Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, et al. (2009) A core gut microbiome in obese and lean twins. Nature 457 : 480–484. doi: 10.1038/nature07540 19043404
27. Leitner WW, Costero-Saint Denis A, Wali T (2011) Immunological consequences of arthropod vector-derived salivary factors. Eur J Immunol 41 : 3396–3400. doi: 10.1002/eji.201190075 22125007
28. Proal AD, Albert PJ, Marshall TG (2013) The human microbiome and autoimmunity. Curr Opin Rheumatol 25 : 234–240. doi: 10.1097/BOR.0b013e32835cedbf 23370376
29. Jarrett CO, Sebbane F, Adamovicz JJ, Andrews GP, Hinnebusch BJ (2004) Flea-borne transmission model to evaluate vaccine efficacy against naturally acquired bubonic plague. Infect Immun 72 : 2052–2056. 15039326
30. VALENZUELA JG (2004) Exploring tick saliva: from biochemistry to “sialomes” and functional genomics. Parasitology 129: S83–S94. 15938506
31. Oliveira F, Jochim RC, Valenzuela JG, Kamhawi S (2009) Sand flies, Leishmania, and transcriptome-borne solutions. Parasitol Int 58 : 1–5. doi: 10.1016/j.parint.2008.07.004 18768167
32. Epstein JE, Tewari K, Lyke KE, Sim BK, Billingsley PF, et al. (2011) Live attenuated malaria vaccine designed to protect through hepatic CD8(+) T cell immunity. Science (80-) 334 : 475–480.
33. Chen K, Pachter L (2005) Bioinformatics for whole-genome shotgun sequencing of microbial communities. PLoS Comput Biol 1 : 106–112. 16110337
34. Lindh JM, Terenius O, Faye I (2005) 16S rRNA gene-based identification of midgut bacteria from field-caught Anopheles gambiae sensu lato and A. funestus mosquitoes reveals new species related to known insect symbionts. Appl Env Microbiol 71 : 7217–7223. 16269761
35. Zahner V, Lucarotti CJ, McIntosh D (2008) Application of 16S rDNA-DGGE and plate culture to characterization of bacterial communities associated with the sawfly, Acantholyda erythrocephala (Hymenoptera, Pamphiliidae). Curr Microbiol 57 : 564–569. doi: 10.1007/s00284-008-9243-4 18769850
36. Lindh JM, Lehane MJ (2011) The tsetse fly Glossina fuscipes fuscipes (Diptera: Glossina) harbours a surprising diversity of bacteria other than symbionts. Antonie Van Leeuwenhoek 99 : 711–720. doi: 10.1007/s10482-010-9546-x 21203841
37. Gusmao DS, Santos A V, Marini DC, Bacci M Jr., Berbert-Molina MA, et al. (2010) Culture-dependent and culture-independent characterization of microorganisms associated with Aedes aegypti (Diptera: Culicidae) (L.) and dynamics of bacterial colonization in the midgut. Acta Trop 115 : 275–281. doi: 10.1016/j.actatropica.2010.04.011 20434424
38. Vasanthi V, Hoti SL (1992) Microbial flora in gut of Culex quinquefasciatus breeding in cess pits. Southeast Asian J Trop Med Public Heal 23 : 312–317. 1439987
39. Collins AJ, LaBarre BA, Won BSW, Shah M V, Heng S, et al. (2012) Diversity and partitioning of bacterial populations within the accessory nidamental gland of the squid Euprymna scolopes. Appl Environ Microbiol 78 : 4200–4208. doi: 10.1128/AEM.07437-11 22504817
40. Lee ZM-P, Bussema C, Schmidt TM (2009) rrnDB: documenting the number of rRNA and tRNA genes in bacteria and archaea. Nucleic Acids Res 37: D489–D493. doi: 10.1093/nar/gkn689 18948294
41. Littman DR, Pamer EG (2011) Role of the commensal microbiota in normal and pathogenic host immune responses. Cell Host Microbe 10 : 311–323. doi: 10.1016/j.chom.2011.10.004 22018232
42. Moreira LA, Iturbe-Ormaetxe I, Jeffery JA, Lu G, Pyke AT, et al. (2009) A Wolbachia symbiont in Aedes aegypti limits infection with dengue, Chikungunya, and Plasmodium. Cell 139 : 1268–1278. doi: 10.1016/j.cell.2009.11.042 20064373
43. Geiger A, Ravel S, Mateille T, Janelle J, Patrel D, et al. (2007) Vector competence of Glossina palpalis gambiensis for Trypanosoma brucei s.l. and genetic diversity of the symbiont Sodalis glossinidius. Mol Biol Evol 24 : 102–109. 17012373
44. Leitner WW, Costero-Saint Denis A, Wali T (2012) Role of immune cell subsets in the establishment of vector-borne infections. Eur J Immunol 42 : 3110–3115. doi: 10.1002/eji.201270102 23255007
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Relay of Herpes Simplex Virus between Langerhans Cells and Dermal Dendritic Cells in Human SkinČlánek The Ebola Epidemic Crystallizes the Potential of Passive Antibody Therapy for Infectious DiseasesČlánek Hepatitis D Virus Infection of Mice Expressing Human Sodium Taurocholate Co-transporting PolypeptideČlánek A Redox Regulatory System Critical for Mycobacterial Survival in Macrophages and Biofilm Development
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2015 Číslo 4- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Pathogens as Biological Weapons of Invasive Species
- Selection and Spread of Artemisinin-Resistant Alleles in Thailand Prior to the Global Artemisinin Resistance Containment Campaign
- Endopeptidase-Mediated Beta Lactam Tolerance
- Prospective Large-Scale Field Study Generates Predictive Model Identifying Major Contributors to Colony Losses
- Relay of Herpes Simplex Virus between Langerhans Cells and Dermal Dendritic Cells in Human Skin
- Structural Determinants of Phenotypic Diversity and Replication Rate of Human Prions
- Sigma Factor SigB Is Crucial to Mediate Adaptation during Chronic Infections
- EphrinA2 Receptor (EphA2) Is an Invasion and Intracellular Signaling Receptor for
- Toxin-Induced Necroptosis Is a Major Mechanism of Lung Damage
- Heterologous Expression in Remodeled . : A Platform for Monoaminergic Agonist Identification and Anthelmintic Screening
- Novel Disease Susceptibility Factors for Fungal Necrotrophic Pathogens in Arabidopsis
- Interleukin 21 Signaling in B Cells Is Required for Efficient Establishment of Murine Gammaherpesvirus Latency
- Phosphorylation at the Homotypic Interface Regulates Nucleoprotein Oligomerization and Assembly of the Influenza Virus Replication Machinery
- Human Papillomaviruses Activate and Recruit SMC1 Cohesin Proteins for the Differentiation-Dependent Life Cycle through Association with CTCF Insulators
- Ubiquitous Promoter-Localization of Essential Virulence Regulators in
- TGF-β Suppression of HBV RNA through AID-Dependent Recruitment of an RNA Exosome Complex
- The Immune Adaptor ADAP Regulates Reciprocal TGF-β1-Integrin Crosstalk to Protect from Influenza Virus Infection
- Antagonism of miR-328 Increases the Antimicrobial Function of Macrophages and Neutrophils and Rapid Clearance of Non-typeable (NTHi) from Infected Lung
- The Epigenetic Regulator G9a Mediates Tolerance to RNA Virus Infection in
- Does the Arthropod Microbiota Impact the Establishment of Vector-Borne Diseases in Mammalian Hosts?
- Hantaan Virus Infection Induces Both Th1 and ThGranzyme B+ Cell Immune Responses That Associated with Viral Control and Clinical Outcome in Humans
- Viral Inhibition of the Transporter Associated with Antigen Processing (TAP): A Striking Example of Functional Convergent Evolution
- Plasma Membrane Profiling Defines an Expanded Class of Cell Surface Proteins Selectively Targeted for Degradation by HCMV US2 in Cooperation with UL141
- Optineurin Regulates the Interferon Response in a Cell Cycle-Dependent Manner
- IFIT1 Differentially Interferes with Translation and Replication of Alphavirus Genomes and Promotes Induction of Type I Interferon
- The EBNA3 Family of Epstein-Barr Virus Nuclear Proteins Associates with the USP46/USP12 Deubiquitination Complexes to Regulate Lymphoblastoid Cell Line Growth
- Hepatitis C Virus RNA Replication Depends on Specific and -Acting Activities of Viral Nonstructural Proteins
- A Neuron-Specific Antiviral Mechanism Prevents Lethal Flaviviral Infection of Mosquitoes
- The Aspartate-Less Receiver (ALR) Domains: Distribution, Structure and Function
- Global Genome and Transcriptome Analyses of Epidemic Isolate 98-06 Uncover Novel Effectors and Pathogenicity-Related Genes, Revealing Gene Gain and Lose Dynamics in Genome Evolution
- The Ebola Epidemic Crystallizes the Potential of Passive Antibody Therapy for Infectious Diseases
- Ebola Virus Entry: A Curious and Complex Series of Events
- Conserved Spirosomes Suggest a Single Type of Transformation Pilus in Competence
- Spatial Structure, Transmission Modes and the Evolution of Viral Exploitation Strategies
- Bacterial Cooperation Causes Systematic Errors in Pathogen Risk Assessment due to the Failure of the Independent Action Hypothesis
- Transgenic Fatal Familial Insomnia Mice Indicate Prion Infectivity-Independent Mechanisms of Pathogenesis and Phenotypic Expression of Disease
- Cerebrospinal Fluid Cytokine Profiles Predict Risk of Early Mortality and Immune Reconstitution Inflammatory Syndrome in HIV-Associated Cryptococcal Meningitis
- Utilize Host Actin for Efficient Maternal Transmission in
- Borna Disease Virus Phosphoprotein Impairs the Developmental Program Controlling Neurogenesis and Reduces Human GABAergic Neurogenesis
- An Effector Peptide Family Required for Toll-Mediated Immunity
- Hepatitis D Virus Infection of Mice Expressing Human Sodium Taurocholate Co-transporting Polypeptide
- A Redox Regulatory System Critical for Mycobacterial Survival in Macrophages and Biofilm Development
- Quadruple Quorum-Sensing Inputs Control Virulence and Maintain System Robustness
- Leukocyte-Derived IFN-α/β and Epithelial IFN-λ Constitute a Compartmentalized Mucosal Defense System that Restricts Enteric Virus Infections
- A Strategy for O-Glycoproteomics of Enveloped Viruses—the O-Glycoproteome of Herpes Simplex Virus Type 1
- Macrocyclic Lactones Differ in Interaction with Recombinant P-Glycoprotein 9 of the Parasitic Nematode and Ketoconazole in a Yeast Growth Assay
- Neofunctionalization of the α1,2fucosyltransferase Paralogue in Leporids Contributes to Glycan Polymorphism and Resistance to Rabbit Hemorrhagic Disease Virus
- The Extracytoplasmic Linker Peptide of the Sensor Protein SaeS Tunes the Kinase Activity Required for Staphylococcal Virulence in Response to Host Signals
- Murine CMV-Induced Hearing Loss Is Associated with Inner Ear Inflammation and Loss of Spiral Ganglia Neurons
- Dual miRNA Targeting Restricts Host Range and Attenuates Neurovirulence of Flaviviruses
- GATA-Dependent Glutaminolysis Drives Appressorium Formation in by Suppressing TOR Inhibition of cAMP/PKA Signaling
- Role of Hypoxia Inducible Factor-1α (HIF-1α) in Innate Defense against Uropathogenic Infection
- Genetic Analysis Using an Isogenic Mating Pair of Identifies Azole Resistance Genes and Lack of Locus’s Role in Virulence
- A Temporal Gate for Viral Enhancers to Co-opt Toll-Like-Receptor Transcriptional Activation Pathways upon Acute Infection
- Neutrophil Recruitment to Lymph Nodes Limits Local Humoral Response to
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Toxin-Induced Necroptosis Is a Major Mechanism of Lung Damage
- Transgenic Fatal Familial Insomnia Mice Indicate Prion Infectivity-Independent Mechanisms of Pathogenesis and Phenotypic Expression of Disease
- Role of Hypoxia Inducible Factor-1α (HIF-1α) in Innate Defense against Uropathogenic Infection
- EphrinA2 Receptor (EphA2) Is an Invasion and Intracellular Signaling Receptor for
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



