-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaThe Post-transcriptional Regulator / Activates T3SS by Stabilizing the 5′ UTR of , the Master Regulator of Genes, in
The RsmA/CsrA family of the post-transcriptional regulators of bacteria is involved in the regulation of many cellular processes, including pathogenesis. In this study, we demonstrated that rsmA not only is required for the full virulence of the phytopathogenic bacterium Xanthomonas citri subsp. citri (XCC) but also contributes to triggering the hypersensitive response (HR) in non-host plants. Deletion of rsmA resulted in significantly reduced virulence in the host plant sweet orange and a delayed and weakened HR in the non-host plant Nicotiana benthamiana. Microarray, quantitative reverse-transcription PCR, western-blotting, and GUS assays indicated that RsmA regulates the expression of the type 3 secretion system (T3SS) at both transcriptional and post-transcriptional levels. The regulation of T3SS by RsmA is a universal phenomenon in T3SS-containing bacteria, but the specific mechanism seems to depend on the interaction between a particular bacterium and its hosts. For Xanthomonads, the mechanism by which RsmA activates T3SS remains unknown. Here, we show that RsmA activates the expression of T3SS-encoding hrp/hrc genes by directly binding to the 5′ untranslated region (UTR) of hrpG, the master regulator of the hrp/hrc genes in XCC. RsmA stabilizes hrpG mRNA, leading to increased accumulation of HrpG proteins and subsequently, the activation of hrp/hrc genes. The activation of the hrp/hrc genes by RsmA via HrpG was further supported by the observation that ectopic overexpression of hrpG in an rsmA mutant restored its ability to cause disease in host plants and trigger HR in non-host plants. RsmA also stabilizes the transcripts of another T3SS-associated hrpD operon by directly binding to the 5′ UTR region. Taken together, these data revealed that RsmA primarily activates T3SS by acting as a positive regulator of hrpG and that this regulation is critical to the pathogenicity of XCC.
Published in the journal: . PLoS Pathog 10(2): e32767. doi:10.1371/journal.ppat.1003945
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1003945Summary
The RsmA/CsrA family of the post-transcriptional regulators of bacteria is involved in the regulation of many cellular processes, including pathogenesis. In this study, we demonstrated that rsmA not only is required for the full virulence of the phytopathogenic bacterium Xanthomonas citri subsp. citri (XCC) but also contributes to triggering the hypersensitive response (HR) in non-host plants. Deletion of rsmA resulted in significantly reduced virulence in the host plant sweet orange and a delayed and weakened HR in the non-host plant Nicotiana benthamiana. Microarray, quantitative reverse-transcription PCR, western-blotting, and GUS assays indicated that RsmA regulates the expression of the type 3 secretion system (T3SS) at both transcriptional and post-transcriptional levels. The regulation of T3SS by RsmA is a universal phenomenon in T3SS-containing bacteria, but the specific mechanism seems to depend on the interaction between a particular bacterium and its hosts. For Xanthomonads, the mechanism by which RsmA activates T3SS remains unknown. Here, we show that RsmA activates the expression of T3SS-encoding hrp/hrc genes by directly binding to the 5′ untranslated region (UTR) of hrpG, the master regulator of the hrp/hrc genes in XCC. RsmA stabilizes hrpG mRNA, leading to increased accumulation of HrpG proteins and subsequently, the activation of hrp/hrc genes. The activation of the hrp/hrc genes by RsmA via HrpG was further supported by the observation that ectopic overexpression of hrpG in an rsmA mutant restored its ability to cause disease in host plants and trigger HR in non-host plants. RsmA also stabilizes the transcripts of another T3SS-associated hrpD operon by directly binding to the 5′ UTR region. Taken together, these data revealed that RsmA primarily activates T3SS by acting as a positive regulator of hrpG and that this regulation is critical to the pathogenicity of XCC.
Introduction
Pathogenic bacteria belonging to the genus Xanthomonas cause diseases in many economically important plants throughout the world. The virulence of these bacteria depends on a type 3 protein secretion system (T3SS) [1], [2], [3]. T3SS mediates the translocation of bacterial effector proteins into the host cell, where they may interfere with host metabolic pathways and/or suppress plant defense reactions [4], [5], [6], [7].Genes encoding the T3SS are referred to as Hypersensitive Response and Pathogenicity (hrp) genes because hrp mutants lost the abilities to trigger the HR in non-host plants and pathogenesis in host plants [8]. In Xanthomonads, the hrp gene cluster contains at least 22 genes, nine of which are highly conserved and therefore have been termed hrc (hrp conserved) genes [4]. The expression of the hrp cluster of genes in Xanthomonas has been shown to be activated in planta and in the minimal medium XVM2 by the transcriptional regulators HrpG and HrpX [9], [10]. HrpG is a response regulator belonging to the OmpR family of two-component system response regulators that contains an N-terminal response receiver (RR) domain and a C-terminal DNA-binding motif [10], [11]. Recently, a putative cognate sensor histidine kinase was reported for HrpG in Xanthomonas campestris, but further studies are needed to identify the activation signal and whether homologs of this sensor kinase are functional in other Xanthomonas species [1], [7], [12]. Phosphorylated HrpG is predicted to activate the expression of hrpX, which encodes an AraC-type regulator [11]. HrpX binds to a cis-regulatory element, the plant inducible promoter (PIP) conserved motif (TTCGC-N15-TTCGC) or a less-conserved PIP-like motif (TTCGC-N8-TTCGT), which is present in the promoter regions of four operons (hrpB, hrpC, hrpD and hrpE), hrpF within the hrp cluster (Fig. S1), and a set of genes involved in the virulence of Xanthomonads [4], [9], [13], [14]. Likewise, HrpG and HrpX homologs have been reported to mediate the expression of genes in the hrp cluster of the plant pathogenic bacteria Ralstonia solanacearum and the animal pathogen Burkholderia pseudomallei [15], [16]. The trigger signal and post-transcriptional regulation of HrpG/HrpX-dependent signaling pathway, which is different from that in Erwinia carotovora and Pseudomonas spp., denominated type I regulatory system, are poorly understood [7], [17], [18].
Further studies in Xanthomonads indicate that the post-transcriptional regulator RsmA (repressor of secondary metabolism) positively regulates pathogenicity [19], [20]. RsmA belongs to a conserved family of RNA-binding proteins that were initially identified as repressors of carbon metabolism (carbon storage regulator [CsrA]) [21], [22]. Members of the RsmA/CsrA family are widely distributed among eubacteria and control various traits including biofilm formation, motility, carbon flux, secondary metabolism, quorum-sensing, and virulence to animal and plant hosts [22], [23], [24], [25], [26], [27], [28]. RsmA/CsrA has been shown to regulate the T3SS genes of many animal and plant pathogenic bacteria including Pseudomonas aeruginosa [29], Salmonella enterica Serovar Typhimurium [28], Erwinia carotovora [30], [31], Dickeya dadantii [32], and Xanthomonas spp. [19], [20]. In X. campestris pv. campestris and X. oryzae pv. oryzae, deletion of an rsmA homolog resulted in a complete loss of virulence in host and a loss of HR induction in non-host plants, impairment of endoglucanase production, and enhanced biofilm formation [19], [20]. While T3SS genes have been shown to be regulated by RsmA homologs in many animal and plant pathogenic bacteria, the mechanistic understanding of RsmA-mediated regulation of T3SS genes in Xanthomonads remained unknown.
RsmA can bind to the 5′ untranslated regions (UTRs) of specific mRNAs by recognizing a wide variety of sequences containing the conserved trinucleotide motif GGA in the loop structures [22], [33], [34], [35]. By directly binding to the target mRNAs, RsmA can either down - or up-regulate the expression of target genes or inhibit translation by blocking the access of the ribosome to the Shine-Dalgarno sequence of the specific mRNA [25], [36], [37]. In animal and plant pathogenic bacteria including Escherichia coli, S. enterica Serovar Typhimurium, P. aeruginosa, and E. carotovora, RsmA/CsrA proteins are inhibited by small noncoding RNAs (rsmB family) with various GGA motifs present in the loops of the secondary structure [25], [38], [39], [40], [41]. The rsmB RNA can bind and sequester multiple copies of RsmA/CsrA [25], [42]. However, rsmB homologs have not been identified in Xanthomonas spp. genomes, suggesting that RsmA regulation involves a novel, as-yet-undiscovered mechanism.
Despite that RsmA/CsrA represses the translation initiation of a variety of genes, a molecular mechanism by which RsmA/CsrA mediates the activation of the flhDC operon, which encodes the master regulator of flagellum synthesis, has recently been identified in E. coli [43], [44]. RsmA/CsrA binds to two sites of the flhDC leader sequence, and by interfering with the 5′ end-dependent RNAse E cleavage pathway, RsmA/CsrA can stabilize the flhDC transcript [44].
To advance the mechanistic understanding of the RsmA regulation of T3SS genes in Xanthomonads, we investigated how RsmA activates the hrp/hrc genes of Xanthomonas citri subsp. citri (XCC), which causes the important citrus canker disease throughout the world [45]. In this paper, we present a model in which RsmA positively regulates the hrp/hrc genes in Xanthomonas, and discuss the implications of these findings in plant host-pathogen interactions.
Results
RsmA plays a central role in the activation of the pathogenicity and HR of XCC
The rsmA gene of XCC (XAC1743, GenBank accession number NP_642074) consists of 213 bp nucleotides and encodes a protein comprising 70 amino acids [46]. RsmA of XCC displays 74% identity with E. coli K12 CsrA (NP_417176.1), 93% with X. campestris pv. campestris strain 8004 RsmA (AAY49556), and 100% with X. oryzae pv. oryzae strain 13751 XOO_2760 (NC_007705.1).
The sequence alignment of Xanthomonas RsmA with homologous proteins exhibited many highly conserved residues in its primary structure [19]. One notable exception is the presence of nine additional residues in the carboxy-terminal region of RsmA of Xanthomonas spp. that do not exist in RsmA homologs in Enterobacteria, Pseudomonas spp. and Erwinia spp. (Fig. S2A) [30], [47], [48]. An unusual C-terminal extension is also present in an RsmA homolog in Sinorhizobium meliloti (RsmAsm) which is 81% identical to XCC RsmA [49]. Deletion of the extended C-terminal of RsmAsm seems to affect its cellular concentration, but enhances its relative RNA binding activity. Interestingly, computational analysis of the predicted secondary structure of XCC RsmA by Phyre [50] revealed that a unique α-helix motif is formed between residues 44–60 (Fig. S2A), in contrast to RsmA homologs from other species with α-helix motifs restricted to residues 44–53 [33], [34]. This difference could be caused by the extended C-terminal sequence in the Xanthomonas RsmA. Furthermore, comprehensive alanine-scanning mutagenesis and structural studies of RsmA homologs identified the residues Leu2, Leu4, Arg6, Arg7, Val40, Val42, Arg44 and Ile47, but not the motif GxxG as hypothesized in previous reports [33], [34], [48], [51], as required for interactions with mRNA targets [33], [34]. We analyzed the three-dimensional structure model of XCC RsmA using the Swiss-Model Repository program [52], [53], [54] (http://swissmodel.expasy.org) with E. coli CsrA structure as a model (PDB: 1Y00, [48]), and visualized it in Pymol [55] (Fig. S2B). As the determinate structures of RsmA homologs in E. coli, P. aeruginosa and Yersinia enterocolitica (PDBs: 1Y00, 1VPZ and 2BTI, respectively), the structural model of XCC RsmA is a homodimer with identical RNA-binding surfaces that can simultaneously bind to two sites within a transcript. The critical RsmA/CsrA residues involved in RNA-binding are conserved in the Xanthomonas RsmA and highlighted in the XCC RsmA structure model (Fig. S2).
To study the contribution of rsmA to the pathogenicity of XCC, we employed an allelic exchange protocol to construct the ΔrsmA mutant of XCC strain 306 containing an in-frame deletion of rsmA codons 5–66 (Table S1). The deletion of rsmA was confirmed by PCR, and the complementation with wild-type rsmA restored the virulence in the host plants (Fig. 1).
Fig. 1. rsmA is required for the pathogenicity of Xanthomonas citri subsp. citri in the host plant sweet orange and contributes to the hypersensitive response (HR) in tobacco leaves (Nicotiana benthamiana). 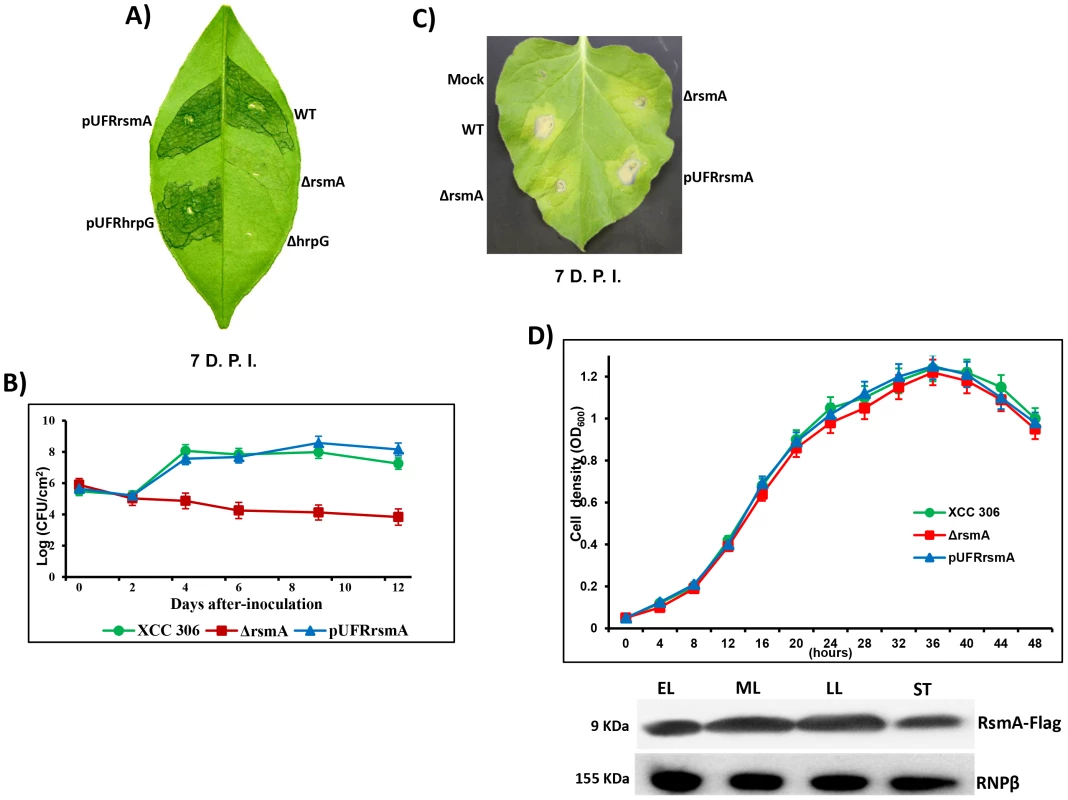
A) Disease symptoms on host sweet orange (Citrus sinensis) leaves 7 days post inoculation (D.P.I.) of bacterial cells at a concentration of 106 CFU/ml. B) Growth assay in planta. Bacterial cells were inoculated into sweet orange leaves at a concentration of 106 CFU/ml and recovered at different time points. The values represent the means of three replicates. The experiment was repeated three times with similar results. Means ± standard deviations are plotted. C) Macroscopic symptoms induced 7 D.A.I of tobacco leaves by infiltrating bacterial cells at a concentration of 106 CFU/ml. D) Growth curve in the minimal medium XVM2 and Western-blotting assay using protein extracts of rsmA mutant cells harboring the pUFR047-rsmA-Flag construct. Cells were collected in different growth stages: EL, early log; ML, medium log); LL, late log; and ST, stationary phase. Wt = X. citri subsp. citri strain 306, ΔrsmA = mutant with a deletion of XAC1743 (rsmA) harboring the empty plasmid pUFR047, pUFRrsmA = complementation of ΔrsmA with rsmA cloned in pUFR047, ΔhrpG = hprG mutant [9], pUFRhrpG = complementation of ΔhrpG with hrpG cloned into pUFR047, and Mock, 10 mM MgCl2. RNPβ: antibody to the β-subunit of RNA polymerase. While infection of the host plant sweet orange (Citrus sinensis) with wild-type XCC strain resulted in clear canker disease symptoms, the ΔrsmA strain caused less hypertrophy and hyperplasia and failed to produce water-soaking and necrosis symptoms in the susceptible citrus host (Fig. 1A). Consistent with the reduced symptom development caused by the rsmA mutant, our results also indicated that rsmA is essential for the proliferation of XCC in host plants (Fig. 1B). The deletion of rsmA impaired the growth of XCC inside the host, whereas the complemented strain multiplied to the same level as wild-type (Fig. 1B). These results are consistent with previous studies showing the requirement of rsmA homologs for the virulence of X. campestris pv. campestris and X. oryzae pv. oryzae in their specific hosts [19], [20]. In addition, it was observed that rsmA contributes to HR induction in the non-host tobacco plant (Nicotiana benthamiana) (Fig. 1C).
To determine whether mutation in rsmA has any effect on the growth of XCC in media, we examined the growth of the rsmA mutant strain (ΔrsmA), the wild-type strain XCC and the complemented strain pUFRrsmA in the minimal medium XVM2 [56], [57]. The results showed that the ΔrsmA and wild-type XCC strains grew to similar levels in the XVM2 medium (Fig. 1D), indicating that rsmA is not required for the growth of XCC in the XVM2 minimal medium. We also observed an increase in RsmA protein levels in the mid-log and late-log growth stages of wild-type XCC carrying the pUFRrsmA-flag construct (Fig. 1D).
RsmA positively regulates the expression of hrp/hrc genes
The significant contribution of RsmA to the pathogenicity and HR-triggering activity of XCC prompted us to investigate its regulatory effect on the hrp/hrc genes by confirming whether RsmA controls the expression of T3SS genes as has been observed in other pathogenic bacteria. The expression of hrp/hrc genes in Xanthomonads has been reported to be repressed in nutrient-rich media but strongly induced in planta and in the XVM2 medium [10], [56]. A previous microarray analysis showed that the whole XCC hrp gene cluster, which contains 24 genes (XAC0393 to XAC0417) (Fig. S1), was down-regulated in hrpG and hrpX mutants of XCC grown in XVM2 medium [9].
To test whether RsmA regulates the expression of hrp/hrc genes in XCC, microarray analyses were conducted to compare the gene expression of the ΔrsmA mutant with that of wild-type XCC strain 306 grown in XVM2 medium to OD600 nm = 0.5. In this study, false discovery rate (FDR) = 0.05 and absolute value of log2-fold change = 1 (equivalent to a fold change of 2) were used as the cutoff values. Compared to wild-type XCC strain 306, a total of 82 genes were down-regulated and 88 genes were up-regulated in the ΔrsmA mutant (Table S2). Overall, 23 genes in the hrp cluster and 16 genes encoding putative T3SS effectors were down-regulated in the ΔrsmA mutant (Table S2). The microarray data were validated by quantitative reverse-transcription PCR (qRT-PCR) of 11 hrp genes. Although we observed differences in the amplitude of fold changes determined using the two techniques, the general trends in gene expression were consistent for all 11 genes that were tested (Table 1).
Tab. 1. rsmA regulation of the expression of the genes in the hrp cluster of Xanthomonas citri subsp. citri. 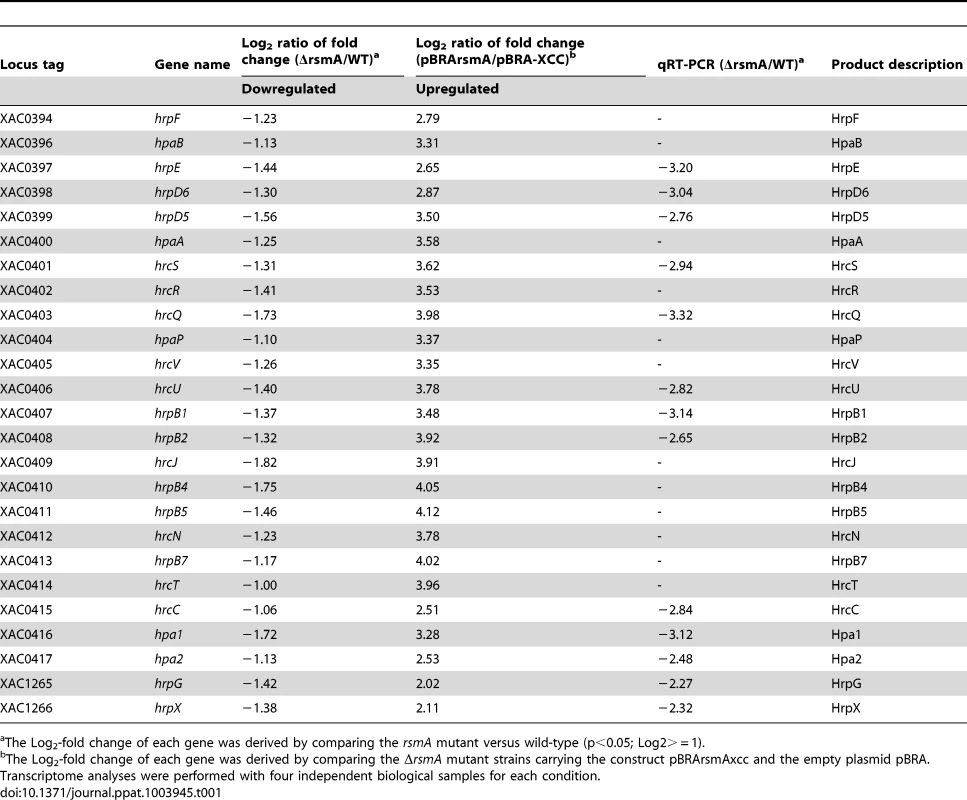
The Log2-fold change of each gene was derived by comparing the rsmA mutant versus wild-type (p<0.05; Log2> = 1). Induced overexpression of rsmA positively regulates hrp/hrc genes
To further investigate the regulation of hrp/hrc genes by RsmA, a recombinant construct named pBRA-rsmA, which contains rsmA fused to an arabinose-inducible promoter, was used to induce the overexpression of 6HisRsmA in the ΔrsmA mutant strain (Table 1). After induction with 0.3% L-arabinose in the XVM2 medium, the expression and functionality of 6HisRsmA were confirmed by Western-blotting and by measuring the extracellular endoglucanase activity (Fig. S3). The endoglucanase activity in the ΔrsmA mutant was restored to 67% of the wild-type level after the induction of 6HisRsmA (Fig. S3). Microarray analysis was also conducted to evaluate gene expression profiles in the ΔrsmA mutant bacterial cells transformed with pBRA-rsmA (strain pBRA-rsmA) or with the empty plasmid pBRA (strain pBRA-XCC). Again, the bacterial cells were cultured in the XVM2 medium supplemented with 0.3% L-arabinose at an OD600 nm of 0.5. Using the same cutoff values, our microarray data showed that the overexpression of rsmA led to the activation of 72 genes and the down-regulation of 81 genes, corresponding to a 90% overlap with the differentially expressed genes identified in the ΔrsmA/WT microarray analysis (Table S2). The patterns of differential gene expression were inversely related between the the ΔrsmA mutant and pBRA-rsmA strains (Tables 1 and S2). The hrp/hrc genes exhibited the strongest activation after 6HisRsmA induction in the pBRA-rsmA strain (Table 1). While the deletion of rsmA resulted in the down-regulation of the hrp/hrc genes and significantly reduced virulence, the expression of T3SS genes was significantly activated by the overexpression of RsmA in the ΔrsmA mutant (Table 1). Taken together, these data confirmed that rsmA contributes to virulence by activating the expression of hrp/hrc genes.
Deletion of rsmA reduces protein levels of T3SS components
To investigate whether the deletion of rsmA also caused a reduction in the protein levels of T3SS, we performed Western-blotting experiments to evaluate the abundances of HrcU, HrpB1, HrpB2 and HrpD6 in total cell extracts of wild-type XCC strain 306, the ΔrsmA mutant, and the complementation strain grown in the XVM2 medium. Based on the immunobloting results, the relative protein levels in the ΔrsmA mutant was estimated in reference to the wild-type strain and normalized according to unspecific protein bands also recognized by the polyclonal antibodies (Figs. 2A, 2B and 2C). The protein levels of HrcU, HrpB1, HrpB2, and HrpD6 were significantly reduced to 14%, 40%, 3% and 10%, respectively, in the ΔrsmA mutant (Figs. 2A, 2B and 2C). Interestingly, the antibodies generated against HrcU recognized two peptides of approximately 28 and 10 KDa, respectively (Fig. 2C). Indeed, HrcU can undergo autocatalytic cleavage between the asparagine and the proline residues of a conserved NPTH motif (amino acids 264–267), resulting in a reorientation of the PTH loop, which is required for the secretion of late T3SS substrates [58], [59]. The relative protein levels of HrcU, HrpB1, HrpB2 and HrpD6 were restored by the expression of rsmA in the complemented strain (Figs. 2A, 2B and 2C).
Fig. 2. RsmA regulates protein levels of T3SS in X. citri subsp. citri. 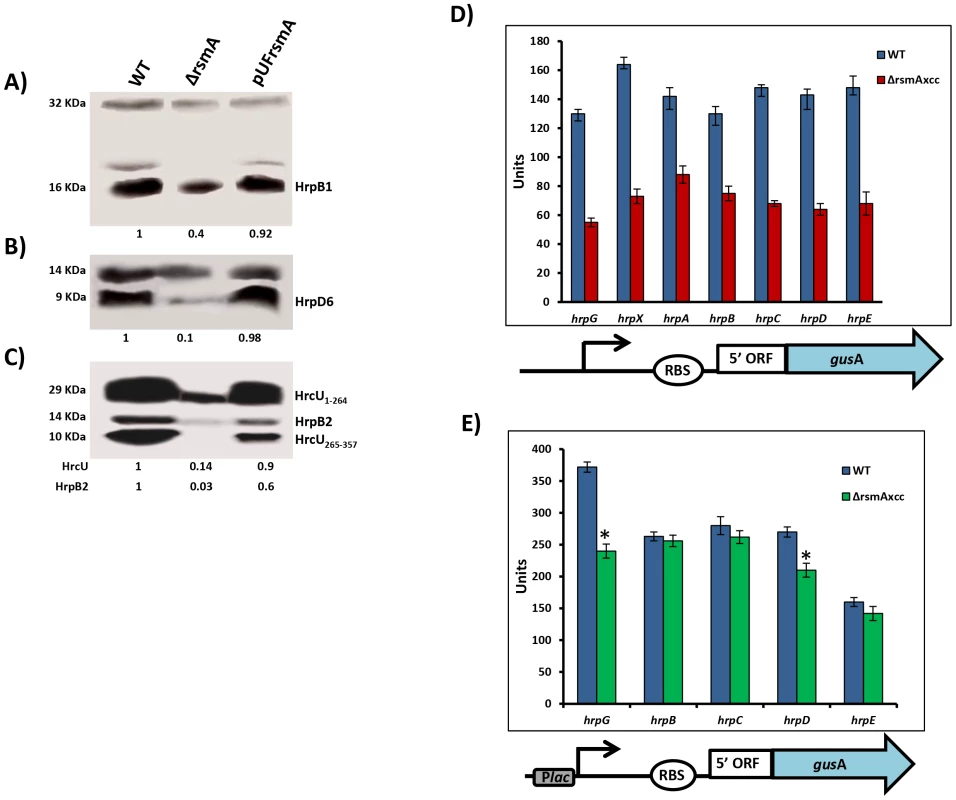
Immunoblotting analyses of the total protein extracts of the wild-type (Wt), the rsmA mutant (ΔrsmA) harboring the empty plasmid pUFR047 and the complemented strain (pUFRrsmA) are shown. Bacterial cells were grown in the XVM2 medium and collected at OD600 nm = 0.5. Proteins were separated by SDS-PAGE and transferred to a nitrocellulose membrane. The blots were probed with A) HrpB1, B) HrpD6 or C) HrcU and HrpB2 polyclonal antibodies, respectively. Protein-A conjugated with horseradish peroxidase was used to detect the blots. Beneath the panels are presented the values of the relative levels of detected proteins in rsmA mutant and complemented strains which were estimated according to wild-type results. The estimated values were normalized with the values obtained to unspecific protein bands also recognized by the antibodies. D) and E) GUS assays using translational fusion constructs. The different constructs used in this assay are represented by diagrams bellow of the graphics. D) Wild-type and rsmA mutant cells harboring plasmid-borne promoterless gusA in-frame fused to the native promoters and the first codons of the hrp genes. E) Wild-type and rsmA mutant cells transformed with translational fusions driven by the constitutive Plac promoter. Values presented are means ± standard deviations of three independent experiments. * represents the significant difference between the wild-type and ΔrsmA values by using ANOVA. The GUS assay was repeated twice with similar results. The positive regulation of hrp/hrc genes by RsmA in XCC was further investigated using translational fusion constructs with a gusA reporter gene [60], [61]. The translational fusion constructs harbor the native promoter, the 5′ leader region and at least the first three codons of the coding DNA sequences of hrpG, hrpX, hrpA genes, as well as hrpC, hrpD and hrpE operons fused in-frame to the ORF (open reading frame) of gusA (Fig. 2D). To differentiate transcriptional and post-transcriptional regulation, we investigated whether replacing the original promoters of the hrp/hrc genes by constitutively expressed Plac promoter affects the GUS activities in wide type and the rsmA mutant strains (Fig. 2E). Results of the GUS activities demonstrated that the rsmA mutation significantly affected the expression of all hrp genes from their native promoters, but it specially affected the expression of hrpG and hrpD under the control of Plac (Figs. 2D, 2E). Taken together, these results indicated that RsmA regulates the expression of hrp/hrc genes at the transcriptional level, and may also control hrpG and hrpD expression at the post-transcriptional level.
Mapping of the 5′-UTR sequences of hrp/hrc transcripts
Previous studies have suggested that RsmA binds to the consensus sequence, ACARGGAUG, with the GGA motif representing the most conserved nucleotides [35], [62]. RsmA can bind to the 5′-UTR of a specific mRNA and negatively or positively regulate the transcript stability and thus the translation [22]. Because RsmA activates all hrp/hrc genes including hrpG and hrpX (Table 1), the two master regulators of hrp/hrc genes, we hypothesized that RsmA exerts its regulation of the hrp/hrc genes via hrpG, the upmost in the regulatory hierarchy. Alternatively, RsmA could bind to the 5′ UTR of each individual transcription unit of the hrp cluster.
The transcriptional initiation sites of the genes hrpG, hrpX, hrpF, and also of the operons hrpB, hrpD, hrpD and hrpE in XCC were determined by rapid amplification of 5′-cDNA Ends (5′ RACE) (Fig. 3A). The RACE PCR products were then sequenced to identify specific nucleotides, indicated by arrows in the Figure 3B, as the transcription start sites of individual transcription units. The analysis of the 5′ RACE results allowed us to determine the PIP-box promoters, -35 and -10 regions, leader sequences and the first codon for each hrp/hrc transcript in XCC (Fig. 3B).
Fig. 3. Determination of hrp/hrc transcriptional start sites of X. citri subsp. citri. 
The transcriptional start sites for the genes hrpG, hrpX and hrpF, and the operons hrpB, hrpC, hrpD and hrpE of XCC were determined by 5′RACE. A) Specific PCR products were detected after the amplification of reverse-transcribed cDNA with the gene-specific primers for hrp/hrc genes together with an adapter primer (Roche), respectively. B) Sequencing of the PCR products identified the nucleotides indicated by arrow as the transcription start sites of hrpG, hrpX, hrpB, hrpC, hrpD, hrpE and hrpF of XCC. Analysis of the 5′ leader sequences of the hrp/hrc transcripts suggests potential RsmA binding sites (highlighted) in hrpG, hrpC, hrpD and hrpE. However, the 5′ leader regions of the hrpB, hrpF and hrpX do not contain the GGA motifs. The +1 nucleotide is indicated with an arrow, putative RsmA binding sites (GGA) in the leader sequences are highlighted in red color, and the PIP-box motif within each promoter are in bold. The −35 and −10 sequences are shown in bold and italics. ATG or GTG are indicated in bold and underlined. The specific primers used to amplify the fragments are underlined. The PIP-box promoters and leader sequences for the hrp/hrc transcripts were previously experimentally identified in X. campestris pv. vesicatoria and X. oryzae pv. oryzae [4], [13], [46], [63], [64]. Our results confirmed that the hrp/hrc transcriptional start sites identified in XCC are similar to those reported in X. campestris pv. vesicatoria [4], [57], [63], [65]. To identify the putative RsmA binding sites in the hrp/hrc transcripts, we analyzed the presence of the GGA motifs in the 5′ leader sequences of the hrpB, hrpC, hrpD, hrpE, hrpF, hrpG, and hrpX transcripts of XCC (Fig. 3B). We were able to find putative RsmA binding sites in the 5′ leader sequences of the hrpC, hrpD, hrpE and hrpG transcripts. No GGA motifs were found in the 5′ UTRs of hrpB, hrpF or hrpX (Fig. 3B).
RsmA directly interacts with the 5′ UTRs of hrpG and hrpD
To experimentally determine whether RsmA of XCC directly binds to the GGA motifs identified in the leader sequences of hrpC, hrpD, hrpE and hrpG transcripts, RNA gel mobility shift assays were performed. RNA gel mobility shift assays were also conducted to test whether RsmA binds to the leader sequences of hrpB, hrpF and hrpX, which do not contain putative RsmA binding sites using RNA oligonucleotides designed based on the predicted secondary structures of the hrpB, hrpF and hrpX 5′ leader sequences by Mfold analysis (Table S4) [66].
The recombinant protein 6HisRsmA was purified by nickel affinity chromatography and tested for binding to a 3′-end-biotin-labeled R9-43 probe, which was previously identified as an E. coli RsmA high-affinity ligand (Table S4) [35]. Our results demonstrated that the XCC 6HisRsmA was able to promote a mobility shift of the 3′-biotin-labeled R9-43 RNA probe (Fig. 4A). We then performed the competition assay by adding unlabeled R9-43 RNA in the reaction, which led to a reduction in the intensity of the shifted band, confirming the specificity of the 6HisRsmA/R9-43 interaction (Fig. 4A).
Fig. 4. RNA mobility shift assays with purified 6HisRsmA of X. citri subsp. citri 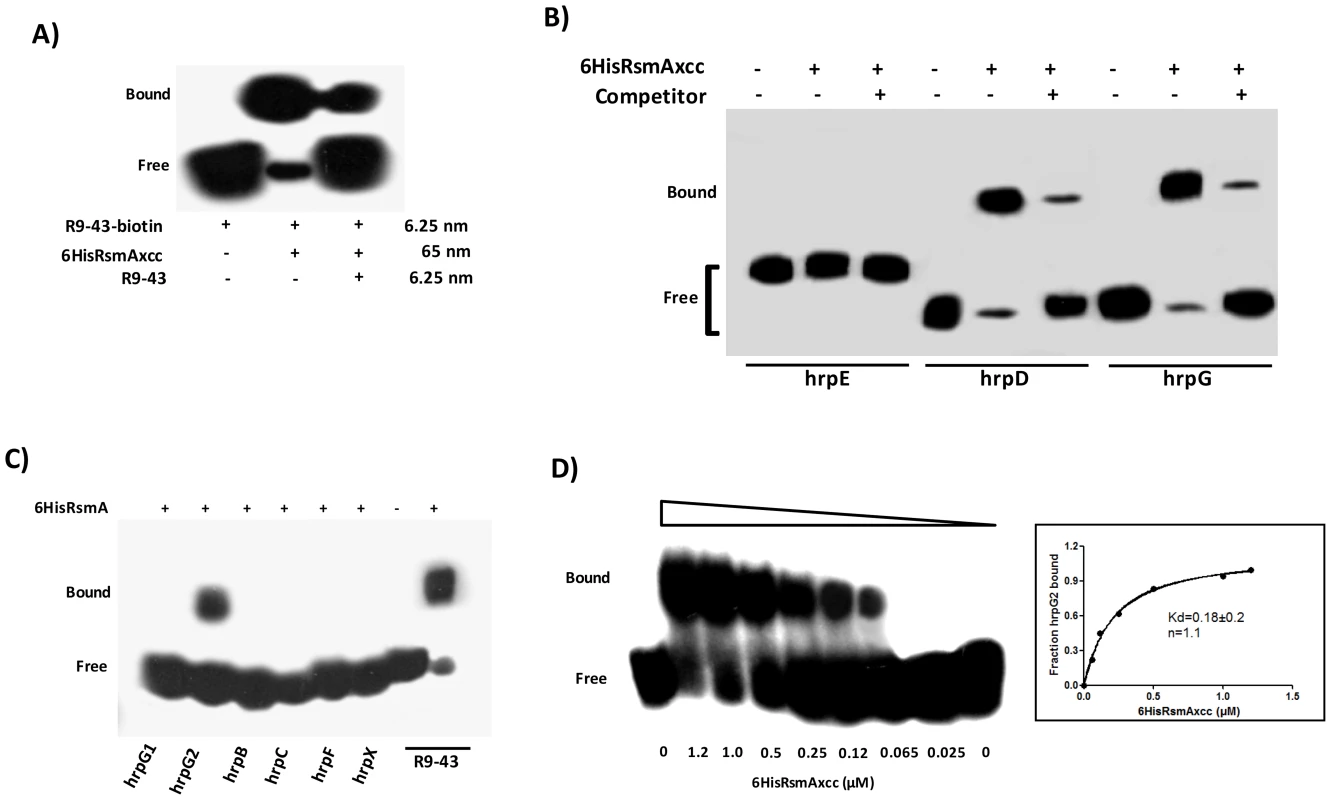
A) 6HisRsmAxcc (65 nM) binds to the high affinity RNA target R9-43. Biotin 3′-end-labeled R9-43 (6.25 nM) was incubated with 6HisRsmAxcc (65 nM) for 30 minutes at room temperature, followed by analysis on a 5% native polyacrylamide gel. A competitive assay in which unlabeled R9-43 RNA (6.25 nM) was added to the reaction reduced the signal resulting from the biotinylated nucleotide. B) 6HisRsmAxcc directly interacts with the 5′ UTRs of hrpG and hrpD. The leader sequences of hrpD, hrpE and hrpG cloned were transcribed in vitro and biotinylated with RNA Labeling kit (Roche). Biotinylated RNA probes were incubated with 6HisRsmAxcc and resolved in a 5% native polyacrylamide gel. The addition of unlabeled competitor R9-43 to the reactions reduced the intensity of the shifted band, which confirmed the specificity of the RsmAxcc-hrpG and RsmAxcc-hrpD interactions. C) 3′-end-biotin-labeled RNA probes encoding the leader sequences of hrpB, hrpC, hrpF and hrpX were tested for interactions with 6HisRsmAxcc (Table S4). In addition, 3′-end-biotin–labeled RNA probes hrpG1 and hrpG2, which bear the GGA motifs encoded by the 5′ leader sequence of hrpG, were used to map the interaction RsmAxcc-hrpG. Only the GGA motif between nucleotides 80 and 120 in the hrpG leader sequence (hrpG2 probe) interacted with 6HisRsmAxcc. D) To determinate the apparent equilibrium binding constant (Kd), 3′ end-labeled hrpG2 RNA (6.25 nM) was incubated with increasing concentrations of 6HisRsmAxcc as noted at the bottom of each lane. The binding curve for the 6HisRsmAxcc-hrpG2 interaction was determined as a function of 6HisRsmAxcc concentration and shifted band intensity. The average pixel value of each shifted band was calculated with ImageJ software [68], [69], [112]. The apparent equilibrium binding constant (Kd) for this reaction was 0.18±0.2 µM 6HisRsmAxcc. Samples were loaded and resolved onto a 5% native polyacrylamide gel. All probes were transferred and cross-linked to a nylon membrane, incubated with streptavidin conjugated with horseradish peroxidase, and detected according to manufacturer's instructions (LightShiftChemiluminescent RNA EMSA Kit, Thermo Scientific). Signals + and − correspond to the presence and absence in the reaction, respectively. Positions of bound and free probes are shown. Next, gel mobility shift analyses of the interactions between 6HisRsmA and biotinylated hrp/hrc leader RNA probes were performed at a concentration at least ten-fold lower than the lowest protein concentration used in the binding reactions as in Yakhnin et al. [67]. This enables us to assume that the concentration of free 6HisRsmA remained constant during the binding reaction [34], [35]. Interestingly, the recombinant protein 6HisRsmA caused mobility shift in the hrpG and hrpD biotinylated transcripts (Fig. 4B). 6HisRsmA did not interact with the hrpC or hrpE transcripts, which also contain potential GGA motifs in the 5′ untranslated mRNA sequences, or with hrpB, hrpF or hrpX, which do not contain GGA motifs in their 5′ UTRs (Figs. 4B and 4C). The biotinylated RNA probe R9-43 was used as a positive control in these assays (Fig. 4C).
Furthermore, the specificities of RsmA-hrpG and RsmA-hrpD 5′ UTRs interactions were investigated by performing competition experiments using the unlabeled specific competitor R9-43 (Fig. 4B). Also, the RsmA-hrpG 5′ UTR interaction was examined by gel mobility shift using two different biotinylated RNA oligonucleotides hrpG1 and hrpG2 (Table S4), which bear two potential RsmA binding sites found in the hrpG leader sequence. The recombinant protein 6HisRsmA was able to retard the hprG2 RNA probe (carrying the second GGA motif of the hrpG leader sequence), but not the hrpG1 RNA probe which carries the first GGA motif (Fig. 4C).
The fraction of bound hrpG2 increased when we raised the 6HisRsmA concentration in the binding reactions (Fig. 4D). Using the biotin-labeled hrpG2 probe, only one complex was observed with 0.065 µM of 6HisRsmA, and essentially most of the starting RNA was shifted with 1.2 µM of 6HisRsmA. The binding curves, Hill coefficient (n), and apparent equilibrium binding constant (Kd) for the 6HisRsmA-hrpG2 RNA interaction were estimated by a nonlinear least-squares analysis using the average pixel values of the shifted bands determined with the ImageJ software [68], [69], [70] in three independent experiments, as described previously [70], [71]. The apparent Kd value was calculated as 0.18±0.2 µM 6HisRsmA (Fig. 4D). This value is around 3.5-fold and 7.5-fold higher than the Kd values of E. coli RsmA/CsrA when interacting with the 5′ leader regions of cstA (Kd 40 nM) and flhDC or pgaA (Kd ∼21 nM), respectively [44], [70], [72].
The RsmA binding site predicted within hrpG2 -AGAUGGAUUU - has an 80% nucleotide identity with the RsmA target site in the high-affinity ligand sequence R9-43 -ACARGGAUGU-. As the interaction of RsmA-RNA depends on RNA sequence and secondary structure and because hrpG1 and hrpG2 RNA probes bear only partial sequences of the hrpG leader region, our data could not rule out the possibility that RsmA also can interact with the potential binding site 1 in the hrpG 5′-UTR.
Determination of the potential RsmA binding sites in the hrpG transcript
The specificity of RsmA interaction with full length hrpG1-189 leader sequence was investigated by performing a competition experiment with unlabeled in vitro transcribed 5′-UTR of the hrpG transcript. The concentrations of 6HisRsmA and the biotinylated hrpG transcript were maintained at 0.5 µM and 6.25 nM, respectively, but the concentrations of unlabeled hrpG transcript were ranged from 0 to 1 µM. The unlabeled in vitro transcribed 5′-UTR of the hrpG transcript was able to compete for 6HisRsmA binding as indicated by the disappearance of the mobility shift at higher concentrations of the competitor (Fig. 5A). Also, the nonspecific yeast tRNA was added to all reactions to verify the specificity of the RsmA-hrpG transcript interaction. These results strengthen our hypothesis that RsmA binds specifically to the leader sequence of hrpG mRNA in XCC.
Fig. 5. Gel mobility shift analysis for mapping the RsmA binding sites within the hrpG transcript. 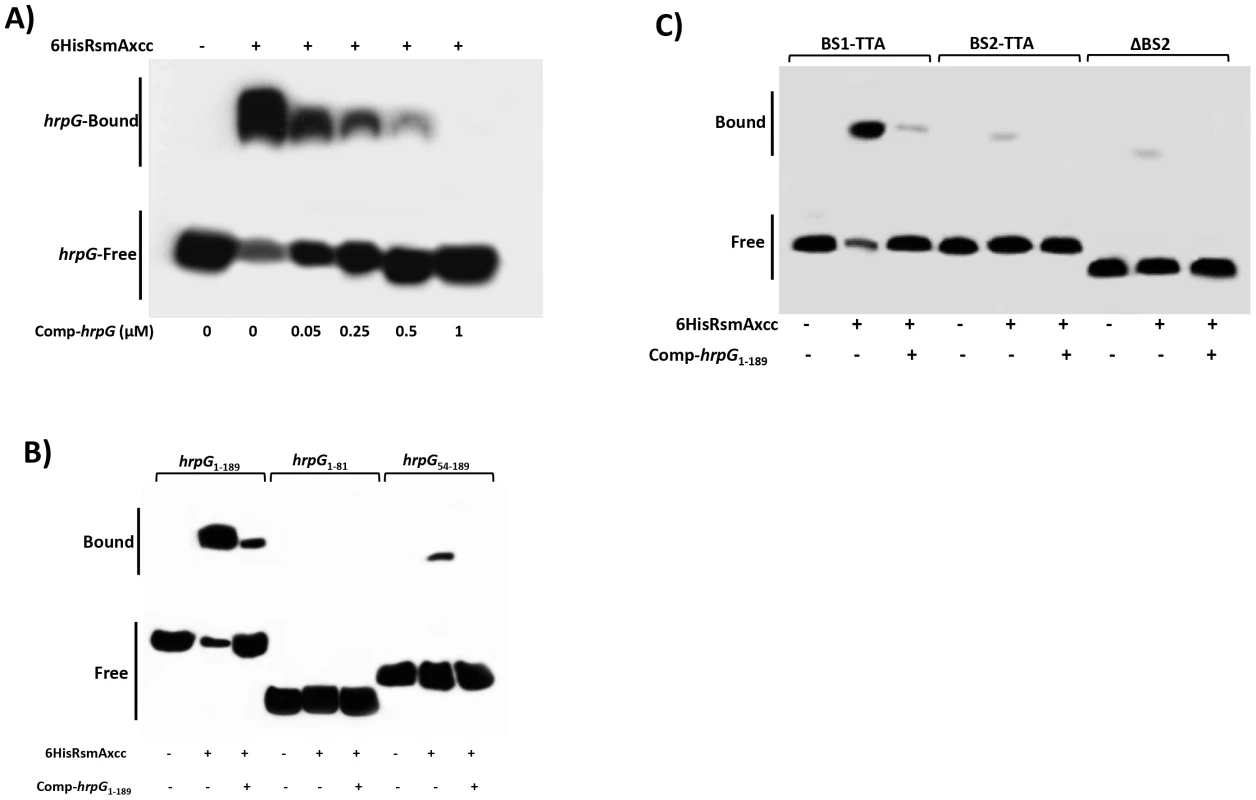
A) For a competition assay, interactions between 6HisRsmAxcc (0.5 µM) and biotinylated hprG1-189 transcript were tested in the presence of the competitor hprG1-189 non-biotinylated (Comp-hrpG). The specific concentrations of the unlabeled Comp-hprG1-189 are indicated at below of each lane. B) Biotinylated hrpG1-189 transcripts carrying mutations in the potential RsmA-binding sites 1 and 2 (BS1-TTA and BS2-TTA) or a deletion of the GGA motif plus 20 nucleotides in the BS2 (ΔBS2) were incubated with 6HisRsmA (0.5 µM) and competitor hprG1-189 non-biotinylated (Comp-hprG1-189). C) The interaction with 6HisRsmA (0.5 µM) was tested using the full length hrpG leader sequence (hrpG1-189) and two fragments hrpG1-81 and hrpG54-189 in vitro transcribed and biotinylated. Signals + and − correspond to the presence and absence in the reaction, respectively. Positions of bound and free hrpG transcripts are shown. To determine whether the potential RsmA-binding sites were responsible for the observed mobility shift, we used mutated hrpG1-189 transcripts in which the GGA binding sites (BS1 and BS2) were mutagenized to TTA. Interestingly, the hrpG transcript mutated in the potential binding site 1 (BS1) did not prevent the mobility shift by 6HisRsmA (Fig. 5B). However, a significant reduction in the mobility shift was observed when the BS2 of the hrpG transcript was changed to TTA (Fig. 5B). In addition, deletion of a 23 nt fragment containing the GGA motif in the BS2 (ten nt were upstream and the other ten nt were downstream of the GGA motif) led to impairment of the mobility shift of the biotinylated hrpG transcript (Fig. 5B).
Furthermore, different fragments of the hrpG leader sequence were synthesized by in vitro transcription, and after biotinylation they were incubated with 6HisRsmA (0.5 µM) to verify binding activity. We did not observe mobility shift of the hrpG1-81 transcript carrying BS1 of the hrpG. In contrast, we observed a shift for the biotinylated hrpG54-189 transcript, although it is weaker (about 8-fold less) than that of the full length hrpG1-189 transcript (Fig. 5C). Consistently, the addition of the unlabeled hrpG1-189 transcript at high concentrations in the reactions impaired the binding of both hrpG1-189 and hrpG54-189 with 6HisRsmA (Fig. 5C). These results suggest that not only the sequence, but also the secondary structure of the hrpG transcript are critical for its interaction with RsmA.
Effects of RsmA on hrpG and hrpD mRNA stability
As was demonstrated for flhDC in E. coli, the interaction of RsmA with the leader region of hrpG suggests that RsmA may stabilize and protect hrpG transcripts against 5′-end - dependent RNase E cleavage in XCC [44], [73], [74]. Previous studies revealed that differences in the steady-state levels of flhDC and glgC transcripts between E. coli wild-type and rsmA mutant strains are resulted from the effect of RsmA on the rates of mRNA decay [43], [44], [75]. To determine whether the stability of the hrpG and hrpD transcripts are affected by rsmA in vivo, the abundances of the hrpG and hrpD transcripts and the 16S transcript as a control were analyzed by RT-PCR after the addition of 10 µg/µL ciprofloxacin (Sigma, USA) in XCC cell cultures (Fig. 6A). Ciprofloxacin targets DNA gyrase and topoisomerase IV on DNA, forming ternary complexes that block the movement of replication forks and transcription complexes [76], [77]. We used ciprofloxacin in this assay because Xanthomonas spp. exhibit resistance to rifampicin [78], [79]. The relative abundance of the hrpG and hrpD transcripts was estimated from the specific PCR products on the agarose gel by calculating the average pixel values of the bands, subtracting the background, and determining the area of the bands by integration in the ImageJ software [68], [69], [71]. The calculated average pixel values of hrpG and hrpD were normalized based on the pixel value of the corresponding 16S amplified band, the transcript levels of which did not vary in response to the treatment. The hrpG and hrpD transcripts were at least two-fold more stable in wild-type XCC (hrpG and hrpD half-life ∼7.8 min and 6 min, respectively) than in the ΔrsmA mutant cells (hrpG and hrpD half-life ∼3.2 min and ∼2.8 min, respectively) (Fig. 6B), which likely contributes to the differences in the transcript levels between the ΔrsmA mutant and the wild-type XCC strain observed in the microarray and qRT-PCR experiments (Table 1).
Fig. 6. Analysis of hrpG and hrpD mRNA stability in the wild-type and rsmA mutant strains by RT-PCR. 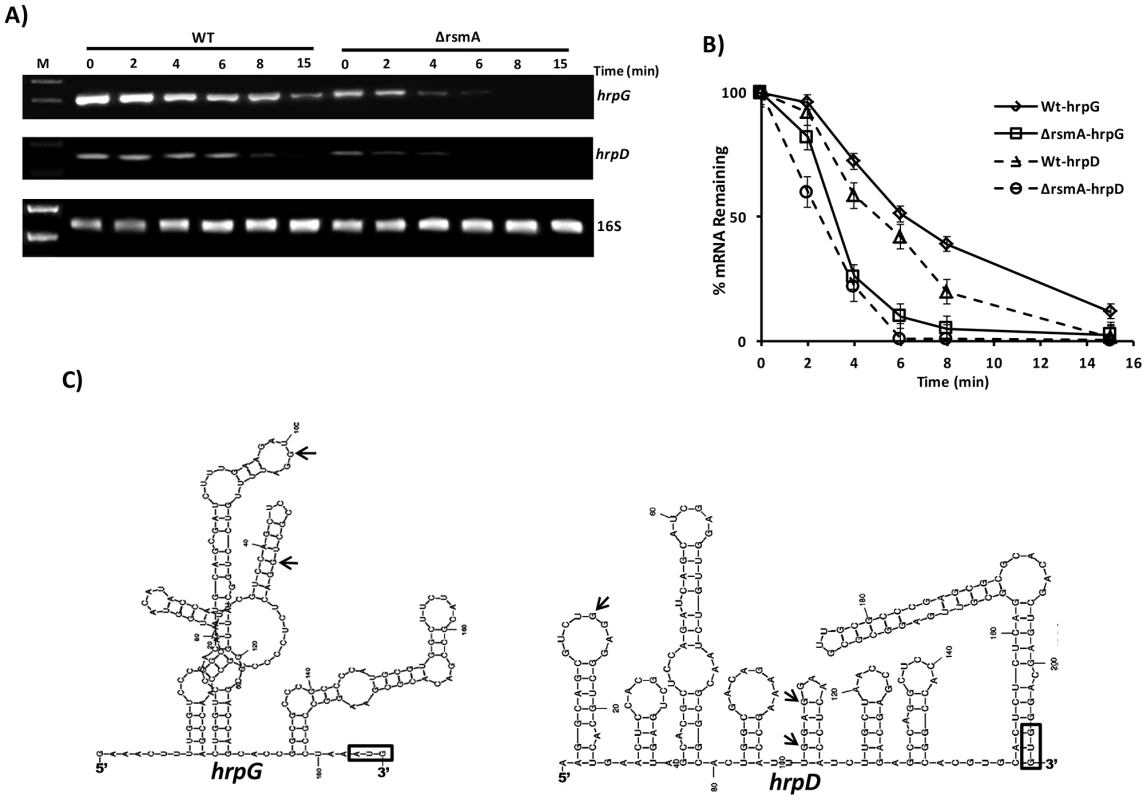
A) Cells of the XCC wild-type and ΔrsmA strains were grown in XVM2 medium to OD600 nm = 0.6, treated with 10 µg/µL ciprofloxacin and harvested at several time points after treatment. Total RNA was isolated, and 2 µg of RNA was used for One Step RT-PCR (Qiagen) in 25 µL reactions. Reactions were subjected to PCR amplification for 26 cycles. Ten microliters of each reaction were resolved on a 1.5% agarose gel. The stability of the hrpD transcript was evaluated using primers annealing within the first orf hrpQ. The 16S RNA was analyzed as a control for normalizing the hrpG and hrpD amplification products. B) The relative values of hrpG and hrpD mRNA half-lives were estimated by determinating the average pixel value of each amplified product and subtracting the background using ImageJ software [68], [112]. The mean values were normalized to the corresponding 16S amplification product. Mean values derived from two independent experiments are shown. C) Model for the predicted secondary structure of the hrpG and hrpD leader sequences obtained with MFold software [66]. The positions of GGA motifs in the structures are indicated with arrows. AUG is shown in an open box. The 5′ UTRs of hrpG and hrpD are 182 nt and 204 nt in length, respectively, which are considered unusually long for bacterial transcripts. RNA structure predictions using Mfold [66] suggested that the hrpG and hrpD 5′ UTRs were highly structured, containing loops (Fig. 6C). Additionally, these results suggest that hrpG and hrpD might be targeted by small RNAs in Xanthomonads. The RsmA binding site identified in the 5′ UTR of hrpG was found in a loop between nucleotides 80 and 120 (Fig. 6C). The predicted structure of hrpD 5′ UTR exhibited three GGA motifs between the nucleotides 1 and 120 and within two different loops (Fig. 6D).
RsmA positively regulates HrpG and HrcQ protein levels
The experiments above showed that RsmA can bind and stabilize the hrpG and hrpD transcripts (Figs. 4B and 6A). The first ORF encoded by the hrpD operon (hrcQRShpaA) is hrcQ (Fig. S1), which encodes for an essential C-ring component of the T3SS [80]. Recent findings suggest that HrcQ might act as a general substrate acceptor for T3SS effector proteins due to its interactions with the T3S-ATPase HrcN, the T3S-membrane proteins HrcV and HrcU, and the T3SS effectors [80].
To verify whether the deletion of rsmA in XCC leads to reductions in HrpG and HrcQ protein levels, recombinant constructs encoding HrpG-6His and HrcQ-Flag with native promoters and full-length 5′ UTRs were inserted into the chromosomes of wild-type and the ΔrsmA mutant XCC strains using the integrative plasmid pPm7g [81]. The plasmid pPm7g harbors an 800 bp fragment of the α-amylase gene (106–912) of XCC (XAC0798), and its insertion into the XCC genome did not affect pathogenicity [81]. Additionally, ΔrsmA carrying the HrpG-6His construct was transformed with a plasmid-borne pUFRrsmA-flag to generate a complementation strain. All strains were grown in the XVM2 medium for 20 h, and total proteins were extracted for Western-blotting analysis. The deletion of rsmA resulted in reduced levels of HrpG-6His and HrpQ-Flag to 38% and 32%, respectively, in relation to the wild-type protein levels (Figs. 7A and 7B, respectively). The complemented strain restored the HrpG-6His and HrpQ-Flag levels to that of the wild-type strain (Fig. 7A and 7B). The relative protein values estimated for HrpG-6His and HrpQ-Flag were normalized with the bands detected in the total protein extracts probed with antibodies against the β-subunit of E. coli RNA polymerase (Abcam, ab12087), which was used as a loading control (Figs. 7A and 7B).
Fig. 7. Immunoblotting experiments showed reduction of HrpG-6His and HrcQ-Flag protein levels in the rsmA mutant cells of Xanthomonas citri subsp. citri. 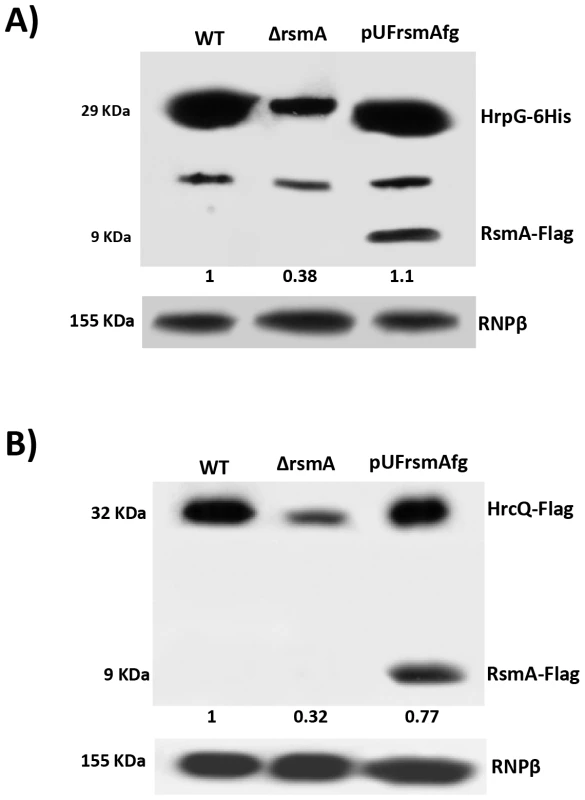
The wild-type, ΔrsmA and complemented strains of XCC carrying the chromosomally inserted recombinant constructs pPM7-hrpG-6His and pPM7-hrcQ-Flag were grown in XVM2 medium to OD600 nm = 0.6. Total cell extracts were analyzed by SDS-PAGE and immunoblotting using specific antibodies. A) For detection of HrpG-6His, after transferred to membrane protein extracts were incubated with anti-6HisTag antibodies (Medical and Biology Lab, MBL). Also, expression of RsmA-Flag was verified only in protein extracts of complemented strain (pUFrsmAfg) probed with anti-FlagTag antibodies (Sigma). B) For detection of HrcQ-Flag, protein extracts were incubated with anti-FlagTag antibodies (Sigma). Ectopic expression of hrpG in the ΔrsmA mutant restores its virulence and HR-triggering activity
The results presented above suggest that RsmA contributes to the virulence of XCC by stabilizing the transcripts of hrpG, the master regulator of T3SS, and it also has a direct effect on the expression of hrpD operon. To further test our hypothesis that RsmA mostly exerts its regulation of the hrp/hrc genes via hrpG, we ectopically expressed hrpG-6His in the ΔrsmA mutant of XCC. The ectopic expression of hrpG-6His in the XCC ΔrsmA mutant not only restored its virulence and bacterial population levels in the host plant sweet orange, but also contributed to HR elicitation ability in the non-host plant N. benthamiana (Figs. 8A, 8B and 8C). Previously, the XCC wild-type strain 306 was shown to be able to elicit a visible HR in tobacco seven days after inoculation (Fig. 1C). However, the constitutive expression of hrpG-6His in the wild-type strain carrying pBBR5Lac-hrpG-6His can trigger a strong HR in tobacco within two days after inoculation (Fig. 8B).
Fig. 8. Ectopic expression of hrpG under control of a constitutive promoter restores full pathogenicity and HR of the rsmA mutant of Xanthomonas citri subsp. citri. 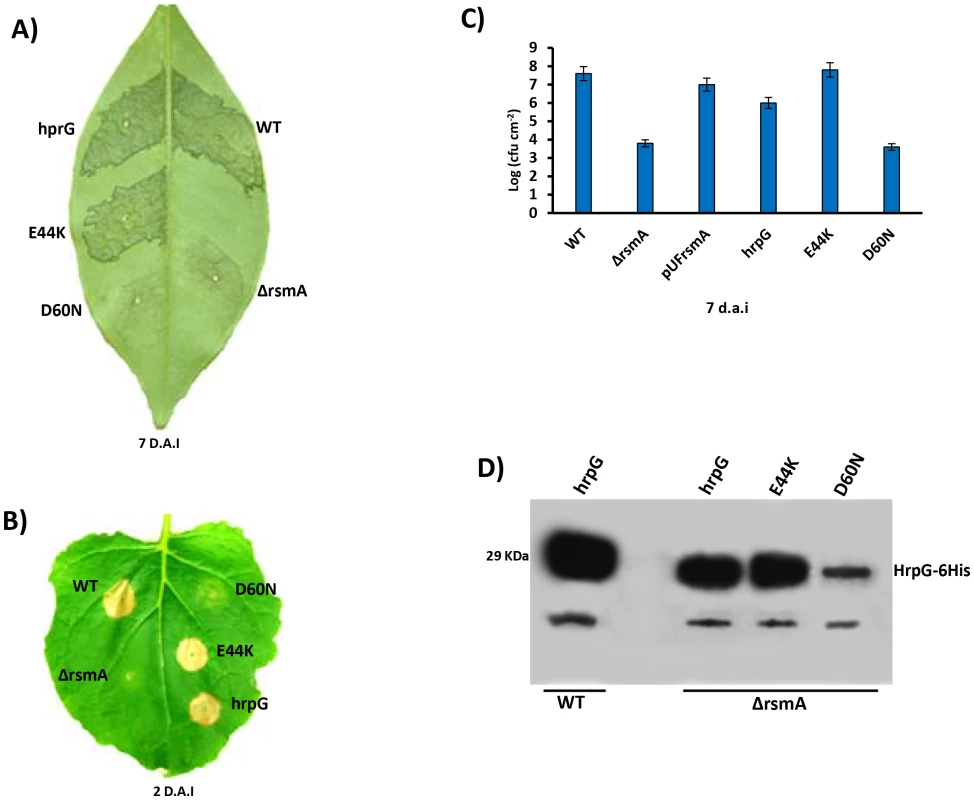
A) XCC strains 306 (WT) and the rsmA mutant carrying the empty plasmid pBBR5 (ΔrsmA) or pBBR5 with hrpG wild-type (hrpG) or hrpG alleles with E44K and D60N mutations were inoculated into A) sweet orange (Citrus sinensis) leaves or B) tobacco leaves by infiltrating bacterial cells at a concentration of 106 CFU/ml. Sweet orange leaves inoculated with WT and rsmA mutant strains harboring both hrpG and hrpG-E44K alleles showed canker symptoms 7 days after inoculation, while a strong HR was observed in tobacco leaves only 2 days after infiltration. However, constitutive expression of the hrpGD60N allele was not able to recover the pathogenicity and HR in the rsmA mutant. C) In plant growth curve experiments confirmed that the rsmA mutant transformed with both empty pBBR5 (ΔrsmA) and pBBR5Lac-hrpGD60N-6His (D60N) have impaired growth in host plants. Error bars represent standard deviations. D) Western-blotting analysis of HrpG-His protein levels in the wild-type and rsmA mutant strains carrying the hrpG (hrpG) or mutated hrpG alleles (E44K and D60N) under control of a constitutive promoter. Equal amounts of total cell extracts were analyzed by immunoblotting with anti-6HisTag antibodies (MBL, USA). To determine whether the post-translational modification of HrpG is affected by the deletion of rsmA in XCC, we generated mutations in the critical residues E44 and D60 within the response regulator domain of the HrpG. The hrpG-6His constructs bearing the E44K and D60N mutations were fused to the Lac promoter of the pBBR5 plasmid to obtain recombinant constructs pBBR5Lac-hrpGE44K-6His and pBBR5Lac-hrpGD60N-6His. It was observed that pBBR5Lac-hrpG-6His and pBBR5Lac-hrpGE44K-6His recovered the virulence and the ability to elicit HR of the ΔrsmA mutant (Figs. 8A, 8B and 8C). The mutant HrpGE44K was previously demonstrated to be constitutively active in Xanthomonas by an unknown mechanism that might involve an increase in the affinity between the HrpG response regulator domain and the α-subunit of RNA polymerase [11], [82]. On the other hand, Lac-hrpGD60N-6His did not restore the virulence or the HR-triggering activity of the ΔrsmA mutant (Figs. 8A, 8B and 8C). These findings support previous studies which had suggested that the residue D60 might be essential for phosphorylation and activation of HrpG [11]. Western-blotting experiments confirmed that HrpG-6His, HrpGE44K-6His and HrpGD60N-6His were expressed in the ΔrsmA mutant harboring the corresponding pBBR5 constructs, although the D60N mutant contained a lower amount of protein (Fig. 8D).
In vivo phosphorylation of HrpG Asp60 residue is critical to restore the virulence of the ΔrsmA mutant, and the deletion of rsmA did not affect HrpG phosphorylation
As shown previously, the constitutive expression of hrpG-6His completely restored the virulence and HR in the ΔrsmA mutant, although the mutated hrpGD60N-6His failed to elicit canker symptoms and restore impaired bacterial growth in host plants (Figs. 8A and 8C). We next tested whether the loss in the function of hrpGD60N-6His was caused by impaired phosphorylation in the ΔrsmA mutant. We utilized the Manganese(II)-Phos-tag (Wako, USA) acrylamide gel system to analyze the in vivo levels of HrpG6His∼P. Both HrpG-6His and HrpG-6His∼P were detected in the cytoplasm of the XCC wild-type and ΔrsmA mutant strains (Fig. 9A). These two HrpG6His isoforms were also present in the mutant D41N and E44K backgrounds, but only the non-phosphorylated form was observed for the D60N mutant (Fig. 9A). A small amount of HrpG6His∼P was observed in the protein extract of wild-type cells grown in the nutrient broth medium, which represses T3SS gene expression (Fig. 9A), but the major fraction of HrpG-6His was not phosphorylated under this condition. These findings suggest that T3SS activation depends on switching the phosphorylated/non-phosphorylated ratio of HrpG isoforms in Xanthomonas cells. Interestingly, phosphorylated isoforms of HrpG6His were observed in the XCC wild-type and ΔrsmA strains transformed with the hrpG wild-type allele, as well as in the ΔrsmA strain carrying the hrpG mutations D41N and E44K. The hrpG mutation D60N impaired HrpG phosphorylation under T3SS-inducible conditions. Taken together, these results indicate that the in vivo phosphorylation of HrpG6His occurred at the Asp60 residue and that the deletion of rsmA did not affect HrpG phosphorylation under T3SS-permissive conditions (Fig. 9A). The same protein samples were separated by SDS-PAGE without Manganese(II)-Phos-tag, and then analyzed by Western-blotting to detect only the HrpG-6His isoform as a control (Fig. 9B).
Fig. 9. In vivo phosphorylation of HrpG Asp60 residue is critical to restore the virulence in the ΔrsmA mutant of XCC. 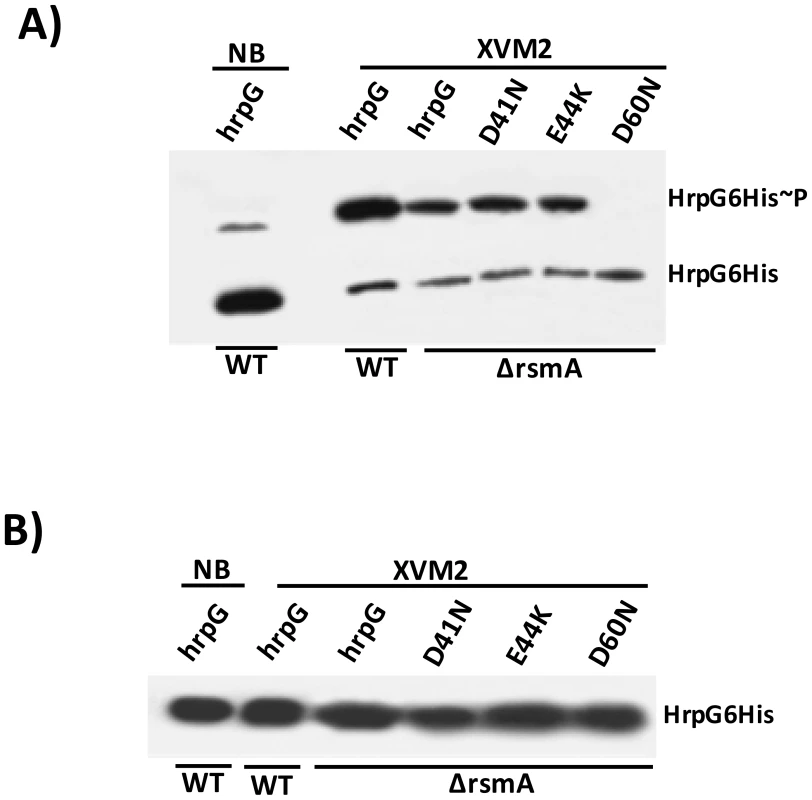
A) Western-blotting analysis of XCC lysates on a 25 µM Phos-tag acrylamide gel (Wako, USA). Bacteria cells were grown in nutrient broth or XVM2 medium (T3SS inducible medium) to OD600 nm = 0.6, and equal amounts of total cell extracts were analyzed by immunoblotting with anti-6HisTag antibodies (MBL, USA). B) Western-blotting analysis to determine HrpG-His protein levels in XCC cell extracts resolved in a 12% acrylamide gel system without Manganese(II)-Phos-tag. Strains analyzed: WT harboring the wild-type hrpG-6His allele; ΔrsmA carrying the wild-type hrpG-6His allele and the mutated hrpG-6His alleles D41N, E44K and D60N. All hrpG-6His constructs were cloned into the pBBR5 plasmid and placed under the control of a constitutive promoter. Discussion
In this study, we demonstrated that rsmA of XCC is critical for activating full virulence in host plants and contributing to the elicitation of HR in non-host plants by positively regulating the expression of the hrp/hrc genes and T3SS effector genes. Microarray and qRT-PCR analyses indicated that the hrp/hrc genes and T3SS effector genes are down-regulated in the rsmA mutant (Tables 1 and S2). Western-blotting assays and GUS assays also showed that RsmA activates the hrp/hrc genes at the transcriptional level (Table 1), and the hrpG and hrpD genes at the post-transcriptional level (Fig. 2). These findings are consistent with the previous transcriptomic analyses of rsmA mutants in X. campestris pv. campestris and X. oryzae pv. oryzae [19], [20], indicating that RsmA-mediated activation of hrp/hrc genes and T3SS effector genes is conserved in different species of Xanthomonads.
We further showed that RsmA binds specifically to the 5′ UTRs of the hrpG and hrpD mRNAs. Using gel shift mobility assays, we revealed that RsmA binds to the second GGA motif within a loop between nucleotide 80 and 120. Consistently, mutation or deletion of this GGA motif led to diminution of the binding of the hrpG transcript with RsmA (Fig. 5). Interaction of the hrpG and hrpD 5′ UTRs with RsmA seems to stabilize these transcripts by protecting them from RNA decay in XCC cells (Fig. 6). Likewise, the deletion of the rsmA homolog in E. coli reduced flhDC mRNA stability and flagellum gene expression, resulting in impaired swimming motility [43]. A recent report demonstrated that E. coli CsrA binding activates flhDC expression by inhibiting the 5′ end-dependent RNase E cleavage pathway [44]. Although lower abundance was observed for hrpG and hrpD mRNAs in the XCC ΔrsmA mutant, it remains unclear whether these transcripts are targeted by the 5′ end-dependent RNase E pathway in the absence of RsmA binding, which may lead to down-regulation of hrp genes expression in Xanthomonads.
Studies on the regulation of protein synthesis have shown that mRNA secondary structures present in the 5′ UTR can dramatically influence both mRNA stability and translation initiation [83], [84], [85]. Therefore, protecting the 5′ UTR of mRNA might avoid endonucleolytic cleavage, a rate-limiting step for mRNA decay [85]. Furthermore, similar to flhDC mRNAs that lack a strong Shine-Dalgarno sequence (GGGUGG) [44], hrpG and hrpD also contain weak Shine-Dalgarno sequences (AGGCCG and AGCUGA, respectively). Thus, the interactions between RsmA and the hrpG and hrpD leader sequences can not only promote mRNA stability, but also increase the translation efficiency of these mRNAs in XCC cells. In fact, the translational fusion of hrpG and hrcQ (the first ORF of the hrpD operon) to the gusA gene driven by a constitutive promoter resulted in reduced β-glucuronidase activity in the ΔrsmA mutant compared to wild-type XCC cells under T3SS-permissive conditions (Fig. 2). In addition, the proteins levels of HrpG and HrcQ were reduced in the ΔrsmA mutant, suggesting that RsmA may also regulate translation efficiency (Fig. 7).
The results presented here support our hypothesis that RsmA exerts its positive regulation on the hrp/hrc genes via hrpG, the upmost regulator in the regulatory hierarchy of hrp/hrc genes. HrpG is the master transcriptional regulator of hrp/hrc and T3SS effector genes by controlling the expression of the second transcriptional factor HrpX, which can recognize and directly bind to the PIP-box within the hrp cluster and most T3SS effector genes [9], [57]. Of all the hrp/hrc transcripts tested here, RsmA directly interacts with the 5′UTRs of hrpG and hrpD but not with that of hrpB, hrpC and hrpE, all of which carry potential GGA motifs in their leader sequences, or hrpF and hrpX, which do not carry potential GGA motifs in their 5′ UTRs. Although the significance of the direct binding of RsmA to the hrpD operon, which contains the T3SS structural genes hrcQ, hrcR, and hrcS, remains to be determined, this function was not able to explain the overall regulation of RsmA on hrp/hrc and T3SS effector genes. Instead, hrpG as the master regulator of T3SS genes, could link the overall positive regulation of T3SS genes by RsmA. This hypothesis is strengthened by the observation that the constitutive expression of hrpG-6His in the ΔrsmA mutant restored its full virulence in host plants and HR-triggering activity in the non-host plant N. benthamiana.
Our results further indicate that RsmA controls hrpG mRNA stability but not the signaling pathway involving the phosphorylation of HrpG protein. The phosphorylation of HrpG is required to activate the expression of hrpX and regulate downstream hrp/hrc and T3SS effector genes [9]. Wild-type HrpG, and the mutants HrpGE44K, and HrpGD41N could all be phosphorylated, while HrpGD60N could not be phosphorylated in the XCC cells grown in the XVM2 medium. Consequently, unlike the phosphorylated wild-type, E44K, and D41N mutants, non-phosphorylated HrpGD60N lost the ability to recover pathogenicity or HR-triggering activity of the ΔrsmA mutant. However, we cannot rule out the possibility that the mutation D60N might reduce HrpG protein stability, resulting in a reduction in virulence and HR-triggering activity. The deletion of rsmA in XCC did not affect the phosphorylation of HrpG under the conditions tested here, suggesting that RsmA can control hrpG expression and stability but not the signaling pathway that triggers HrpG phosphorylation.
Interestingly, in Salmonella, the expression of the transcriptional activator of T3SS genes, hilA, is directly controlled by three AraC-like regulators: HilD, HilC, and RtsA [86]. The CsrA protein binds to and prevents the translation of the hilD mRNA [87]. However, during the infection process, the two-component system BarA/SirA activates the expression of the csrB and csrC small RNAs, which inhibit the action of RsmA/CsrA [86], [88]. Likewise, in the phytopathogenic bacteria E. carotovora, RsmA targets and destabilizes the transcripts that encode plant-cell-wall degrading enzymes (PCWDEs), which are crucial virulence factors, and HrpL, an alternative sigma factor that regulates the expression of T3SS genes. The small noncoding RNA rsmB, which is a homolog of E. coli and Salmonella csrB, inhibits RsmA and enables the translation of the PCWDE encoding genes and hrpL [89], [90]. Moreover, RsmA plays an important role in the interaction between P. aeruginosa and human airway epithelial cells, as shown by the fact that an rsmA mutant failed to induce actin depolymerization and cytotoxicity in epithelial cells due to the decreased transcription of several regulators of T3SS [91].
In contrast to Enterobacteria, E. carotovora, and Pseudomonas spp., the signaling pathway involving RsmA in Xanthomonads is poorly understood as Xanthomonads lacks the small inhibitory RNA rsmB. Recently, a differential RNA sequencing (dRNA-seq) approach was used to identify eight Xanthomonas small RNAs whose expression levels were controlled by HrpG [18]. However, it remains to be determined whether these small RNAs target and activate hrp genes. Furthermore, Schmidtke et al. identified transcripts with unusually long 5′-UTRs, which could indicate extensive post-transcriptional regulation in Xanthomonads [18]. The RsmA target mRNAs hrpG and hrpD identified here also contain long leader regions with loops, suggesting that the expression of T3SS genes may be under several post-transcriptional controls in Xanthomonads. Recent findings demonstrated that in E. coli, the 5′ UTR of the flagellum master regulator flhDC mRNA is targeted for repression by four small RNAs (ArcZ, OmrA, OmrB and OxyS), and for activation by one small RNA (McaS) [92], [93].
RsmA seems to positively regulate critical genes involved in pathogen-host interactions such as hrp/hrc and T3SS effector genes in plant pathogenic bacteria. RsmA also regulates features that are controlled by the quorum-sensing signaling pathway such as extracellular enzymes production, cell motility and biofilm formation in bacteria. Fine-tuning of RsmA activity allows bacteria to overcome a variety of environmental challenges such as host defenses by promptly modifying the mRNA stability and regulating the translation initiation of specific transcripts. Future studies designed to identify new components of RsmA regulatory circuits in Xanthomonads, such as small RNAs, two-component system proteins, and 5′ end-dependent RNAse E cleavage pathways, will shed light on the signaling network of the HrpG-HrpX-mediated expression of the T3S machinery, and also the overlapping of regulation of the virulence traits driven by the quorum-sensing system in plant pathogenic bacteria.
Materials and Methods
Bacterial strains, plasmids, and growth conditions
The plasmids, oligonucleotides and bacterial strains used in this study are listed in Tables S1 and S4. E. coli cells were grown at 37°C in lysogeny broth (LB) medium (1% (w/v) tryptone, 0.5% (w/v) yeast extract, 1% (w/v) sodium chloride, pH 7.5). The XCC wild-type strain 306 (rifampicin resistant) [46] and the mutant strains were routinely grown in nutrient broth (NB), on nutrient agar (NA) at 28°C. For induction of hrp genes, XCC cells were grown in the minimal medium XVM2 (pH 6.7, 20 mM NaCl, 10 mM (NH4)2SO4, 5 mM MgSO4, 1 mM CaCl2, 0.16 mM KH2PO4, 0.32 mM K2HPO4, 0.01 mM FeSO4, 10 mM fructose, 10 mM sucrose, 0.03% casamino acid) as previously described [57]. Endoglucanase assays were performed as previously described in [94], [95], but with the addition of 0.25% L-arabinose. Plasmids were introduced into E. coli by heat-shock at 42°C and into XCC by electroporation [96]. XCC deletion mutants were generated using the suicide vectors pNPTS138 as described elsewhere [97]. Antibiotics were used at the following concentrations: rifampicin, 50 mg/ml; kanamycin, 50 mg/mL; ampicillin, 100 mg/mL; spectinomycin, 50 mg/mL; streptomycin, 50 mg/mL; gentamicin, 10 mg/mL; chloramphenicol, 35 mg/mL; tetracycline, 10 mg/mL and ciprofloxacin 10 mg/mL.
Construction of the rsmA deletion mutant and complemented strains of X. citri subsp. citri
DNA manipulations and PCR were performed according to standard procedures [98]. Restriction digestion and DNA ligation were performed in accordance with the manufacturer's instructions (New England Biolabs, USA). To construct the rsmA deletion mutant, approximately 1 kb of the upstream and downstream regions of the rsmA gene (XAC1743) were amplified by PCR from XCC genomic DNA, and the two fragments were ligated to produce an in frame deletion, leaving only the region coding for the first five and last four codons. This sequence was then cloned into the EcoRI restriction site of the pNPTS138 suicide vector (M. R. Alley, unpublished), thus generating the plasmid pNPTS-rsmA. This vector was introduced into XCC by electroporation, and the wild-type copy was replaced by the deleted version after two recombination events as described previously [97]. To complement the rsmA knockout mutant, a fragment including the rsmA gene plus 1000 bp of upstream sequence was amplified by PCR from XCC genomic DNA and inserted into the pUFR047 vector [99] at the EcoRI restriction site, creating the plasmid pUFR-rsmA. This plasmid was then transferred to the ΔrsmA strain by electroporation and selection for gentamicin resistance. To generate the complementation plasmid for the hrpG mutant, the fragment containing the entire hrpG gene and its promoter was amplified using XCC genomic DNA as a template and the primers F-hrpG and R-hrpG. The fragment was purified from a 1% agarose gel, digested with BamHI and HindIII restriction enzymes and cloned into the BamHI/HindIII enzyme sites of pUFR047. The derivative construct, pUFR-hrpG, was transferred into the hrpG mutant by electroporation (Table S1). The transformed XCC cells were selected on NA with gentamicin. All constructs were sequenced for confirmation.
Generation of expression plasmids in XCC
To produce the fusion of rsmA to an L-arabinose inducible promoter (BAD) in the plasmid pBRA-6HisrsmA, pET-6HisrsmA (Table S1) was digested with the NcoI/XhoI restriction enzymes, and the DNA fragment encoding 6HisRsmA was inserted into the NcoI/SalI enzyme sites of pBRA (Table S1). To generate a Flag epitope-tagged RsmA containing its native promoter, a fragment encoding the rsmA gene plus 1 kb upstream sequence was amplified by PCR using the primers F1-rsmAflag and R1-rsmAflag (Table S3), and then a second PCR using F1-rsmAflag and R2-rsmAflag created the fragment rsmA-flag, which was digested with the BamHI and XhoI restriction enzymes and cloned into the BamHI/SalI enzymatic sites of pUFR047. The resulting construct pUFR-rsmAflag was used to transform the XCC ΔrsmA mutant strain by electroporation. To fuse hrpG to a constitutive promoter, the construct pBBRLac-hrpG6his was produced by PCR amplification of the hrpG orf plus 50 nt upstream using the Fwconst-hrpG and R2-hrpG6His primers (Table S3). The generated fragment was digested with the KpnI/XhoI restriction enzymes and cloned into the same enzymatic sites of pBBR1-MCS-5. The plasmid pBBR1-MCS-5 contains a lac promoter that promotes constitutive gene expression in Xanthomonas [100]. The construct pBBRLac-hrpG6his was then used as a template to produce the HrpG point mutants hrcGD41N-6His, hrcGE44K-6His and hrcGD60N-6His using the QuikChange Site-Directed Mutagenesis Kit and the specific primers as described in Table S3 (Stratagene). The constructs hrpG-6his and hrcQ-flag with their native promoters were produced by two parallel PCR reactions using XCC genomic DNA as the template and the primers F-hrpG/R1-hrpGflag and F-hrcQ/R1-hrcQflag, respectively. In a second PCR, the hrpG and hrcQ fragments amplified in the previous step were used as templates with the primers F-hrpG/R2/hrpGflag and F-hrcQ/R2-hrcQflag generating the restriction enzyme sites within the amplified fragments (Table S3). The recombinant constructs hrpG-his and hrcQ-Flag were digested and cloned into the BamHI/XhoI enzymatic sites of the pPm7g plasmid producing hrpG6his-pPm7g and hrcQflag-pP7mg (Table S1). These pP7mg constructs were used to insert hrpG6his and hrcQflag into the XCC chromosome. The pP7mg plasmid harbors an 800 bp fragment extracted from the α-amylase gene (106–912) of XCC (XAC0798), and its insertion into XCC chromosome did not affect pathogenicity [81]. XCC cells transformed with pPM7-hrpG-6His and pPM7-hrcQ-Flag constructs were selected in NA with kanamycin and confirmed by PCR. All constructs were confirmed by sequencing.
Pathogenicity and HR assays
Pathogenicity assays were conducted in a quarantine greenhouse facility at the Citrus Research and Education Center, Lake Alfred, FL, U.S.A. Assays were performed using fully expanded, immature leaves of sweet orange (Citrus sinensis). XCC wild-type and mutant strains used in this assay were grown with shaking overnight at 28°C in NB, centrifuged, and suspended in sterile tap water, and the concentrations were adjusted to 108 CFU/ml. For the pathogenicity assays, bacterial solutions of both 108 and 105 CFU/ml were infiltrated into the leaves with needleless syringes [9], [101]. Disease symptoms were photographed at 7 days post-inoculation. The strains were also tested for their ability to elicit an HR on Nicotiana benthamiana by infiltration of plant tissue with strains adjusted to 108 CFU/ml with a needleless syringe. Plant responses were scored for HR in tobacco 2 days and 7 days post-inoculation. Tobacco plants were grown in growth chambers at 25°C with a 12 h photoperiod. Experiments were repeated three times with similar results.
RNA extraction and qRT-PCR assays
Single bacterial colonies were picked and grown in 5 ml of NB at 28°C for 24 h with shaking, and then transferred into 50 ml of NB for overnight incubation. The bacterial cultures in the middle exponential stage were centrifuged, washed twice with XVM2 medium, and then inoculated in XVM2 medium at an initial concentration of OD600 nm 0.03. Bacteria were grown in XVM2 medium or XMV2 plus 0.25% L-arabinose with shaking at 200 rpm at 28°C, and samples of the cultures were collected at OD600 nm 0.5. Four biological replicates were used for each strain for qRT-PCR and microarray analyses. RNA was stabilized immediately by mixing the bacterial culture with two volumes of RNA protect bacterial reagent (Qiagen, CA, U.S.A.) and incubated at room temperature for 5 min. Bacterial cells were centrifuged at 5000× g for 10 min and cell pellets were used for RNA extraction. Cell pellets were treated with lysozyme, and RNA extractions were performed using an RNeasy Mini kit (Qiagen). Purified RNA was treated with DNAse I-Free (Qiagen), and successful removal of genomic DNA was confirmed by PCR. RNA quantity was initially determined on a ND-8000 Nanodrop spectrophotometer (NanoDrop Technologies, U.S.A.) and RNA quality was assessed using the Agilent 2100 bioanalyzer (Agilent Technologies, CA, U.S.A.). For qRT-PCR assays, reverse transcription was performed using 1 µg of DNaseI-treated RNA and the RevertAid H-Minus First Strand cDNA Synthesis Kit, following the manufacturer's protocol (Fermentas). Quantitative amplification of the resulting cDNA (40 ng) was performed using 0.3 µM of each primer (Table S4) and SYBR Green/ROX qPCR Master Mix in the ABI7300 Real-Time System (Applied Biosystems). Relative quantification of gene expression was performed using 16S and XAC1631 (which codes for subunit A of DNA gyrase) as endogenous controls and the 2−ΔΔCT method [102]. Primers were designed using the Primer Express Software (Applied Biosystems).
Microarray analyses
For transcriptome studies of the ΔrsmA mutant and ΔrsmA mutant carrying pBRA-rsmA, an Agilent 8-by-15K microarray containing 60-mer oligonucleotides for all predicted orfs in the XCC strain 306 genome were used as described previously [9]. Labeled cDNA was generated using a Fairplay III microarray labeling kit (Agilent Technologies). Total RNA input (5 µg) was used to generate labeled cDNA according to the manufacturer's protocol. Briefly, cDNA was synthesized from 5 µg of the total RNA with Affinity Script HC and random primer, and then modified cDNA was labeled with either cy3 or cy5. The labeled cDNA was purified following the manufacturer's instructions [9], [103]. A total of 300 ng of labeled cDNA per sample was used for the hybridization as previously described [9]. A dye swap was performed to remove any bias resulting from the labeling dyes. Hybridization was performed using a Gene Expression Hybridization Kit (Agilent Technologies) according to microarray protocols recommended by Agilent. The arrays were scanned using a dual-laser DNA microarray scanner (Model G2505C; Agilent Technologies). The microarray analyses, normalization of the mean signal intensities and statistics of the data were carried out as described previously [9]. Log2-transformed values were used for statistical analysis. A linear modeling approach and the empirical Bayes statistics for differential expression analysis were employed as in the report by [104]. The P values were adjusted using the Benjamini and Hochberg method, designated as FDR [105]. Differentially expressed genes were ranked based on FDR, and genes with FDR<0.05 and a minimum absolute value of log2-fold-change > = 1 (equivalent to 2-fold) were considered to be significantly differentially expressed.
Western-blotting and in vivo phosphorylation analyses
To analyze the protein levels in XCC, cells were grown in XVM2 medium to an OD600 nm of 0.5. Bacterial cells were harvested and pellets were resuspended in 50 mM Tris-HCl (pH 8.0), 100 mM NaCl, 0.1% Tween-20, and 1 mg ml−1 lysozyme. Cells were ruptured by sonication. Cell-free extract was prepared by centrifugation at 15, 000 g at 4°C for 45 min. Protein extracts were quantified by the BCA protein assay (Pierce). Equal amount of proteins were loaded and separated by 15% SDS-PAGE, transferred to a nitrocellulose membrane (GE Healthcare) followed by immunoblotting analysis. Rabbit polyclonal antibodies raised against XCC HrcU, HrpB1, HrpB2 and HrpD6 were previously reported in Cappelletti et al. [57]. The rabbit sera against Hrp proteins were diluted to 1∶1000, while anti-Flag (Sigma) and anti-6His (BML, USA) were diluted to 1∶5000. Rabbit antibodies were detected with staphylococcal protein-A conjugated to horseradish peroxidase (Sigma). The in vivo levels of phosphorylated HrpG were detected using the Manganese(II)-Phos-tag (Wako, USA) acrylamide gel system. Formic acid-lysed XCC cells grown in XVM2 media to mid-log phase (OD600 nm = 0.6) at 30°C were fractionated on a 12.5% acrylamide gel as described previously [106], [107]. After electrophoresis, the gel was washed twice with transfer buffer plus 5 mM EDTA pH 8, and the resolved proteins were then transferred to a nitrocellulose membrane. The membrane was then incubated with anti-6HisTag antibody (BML, USA) followed by protein A-peroxidase. As a control to these experiments, immunoblotting was carried out for the same protein extracts resolved on a 12.5% acrylamide gel without Manganese(II)-Phos-tag. Immunoblots were developed with SuperSignal West Femto Chemiluminescent Substrate (Pierce) and exposed to CL-XPosure Film (Pierce).
GUS activity assay
To generate the translational constructs with hrp gene-gusA fusion under control of the hrp genes native promoters, the promoter and 5′ UTRs plus three first codons of the hrp genes were fused in frame to the promoterless gusA gene without its ribosome binding site [108]. The upstream sequence and first codons of the hrp genes were amplified by PCR using the total DNA of the XCC wild-type strain 306 as the template. The gusA orf was amplified from the vector pBI121 (Table S1). The primer pairs used in these PCR reactions are listed in Table S3. Fragments containing the translational hrp gene-gusA fusions were first cloned into the pGEM T-easy vector, and then digested with EcoRI/HindIII and inserted into the same enzymatic sites of pLAFR3 [109] (Table S1). Further, the translational constructs with hrp gene-gusA fusion driven by a constitutive promoter were produced after amplifying the leader sequence of hrp genes plus gusA from the constructs described above (Table S3). The PCR products were purified, digested with BamHI/HindIII and inserted downstream of the lac promoter in the vector pUC18-mini-Tn7T-LAC [110]. Those constructs were used to transform XCC wild-type and rsmA mutant strains. Transformed colonies were selected on NA medium plus tetracycline or gentamicin. Wild-type and rsmA mutant cells harboring plasmid-borne promoterless gusA fused in frame to the hrp genes were cultured in XVM2 for 20 h and assayed for β-glucuronidase (GUS) activity. Bacteria cells were diluted and disrupted in a GUS assay buffer (20 mM Tris-HCl, pH 7.0, 10 mM 2-mercaptoethanol, 5 mM EDTA, and 1% TritonX-100). GUS activities were determined at intervals of 15 min for 4 h by measuring the OD420 nm using PNPG (p-Nitrophenyl-β-D-glucuronide) as the substrate [60]. The relative GUS activity was expressed as (1,000×A420)/(time in min×OD600) in Miller units (U) [27], [111]. Values presented are means ± standard deviations of three independent experiments. The GUS assay was repeated twice with similar results.
5′-RACE
The transcriptional start sites of hrpG and hrpX were determined using the 3′/5′ RACE Kit (Roche), according to manufacturer's instructions. Briefly, total RNA was obtained from XCC wild-type cell cultures grown in XVM2 medium to an OD600 nm of 0.5. After treatment with DNase I-free (Qiagen), the RNA was reverse transcribed using a gene-specific primer (SP1, Table S4), purified and poly(dA) tailed at the 3′ end by reaction with terminal transferase enzyme. The resulting cDNA was amplified by PCR using the poly-dT primer provided by the kit to anneal at the poly(dA) tail and a gene-specific primer (SP2, Table S3), complementary to a region upstream of the original cDNA primer. The amplicons from the first PCR were submitted to a second PCR reaction using the poly dT primer and a distinct gene-specific nested primer (SP3, Table S3) that was internal to the first. PCR products were ligated into the pGEM-T vector (Promega) and several distinct clones were sequenced.
Expression and purification of 6HisRsmA in E. coli
For expression of the recombinant protein 6HisRsmA in E. coli, a fragment containing the rsmA gene sequence was amplified by PCR from XCC genome using primers and F-peTrsmA and R-peTrsmA (Table S4), digested with NdeI/EcoRI and inserted into pET-28a (Novagen) resulting in pET-6HisrsmA. Recombinant 6HisRsmA protein was over-expressed in E. coli BL21 Star (DE3) pLysS (Novagen) by addition of 1 mM isopropyl-b-D-thiogalactopyranoside to cells grown to mid-exponential phase. Bacterial cells were harvested 4 h later. Cell pellets were resuspended in 50 mM NaH2PO4 (pH 8.0), 300 mM NaCl, 10 mM imidazole and 1 mg ml−1 lysozyme. Cells were ruptured by sonication. Cell-free extract was prepared by centrifugation at 14,000 g at 4°C for 45 min. 6HisRsmA was purified by Ni-NTA affinity chromatography (Pierce) according to the manufacturer's recommendations. The protein-containing fractions were concentrated and the buffer was exchanged with 10 mM HEPES pH 7.3, 20 mM KCl, 1 mM MgCl2, 1 mM DTT, and 10% Glycerol. The protein concentration was estimated using a BCA method (Pierce) with bovine serum albumin as the standard. The purified protein was stored at −20°C.
RNA gel mobility shift assay
RNA gel mobility shift assays were carried out as previously reported by Yakhnin et al. [67] with some modifications. The leader sequences of the hrpD, hrpE and hrpG were transcribed in vitro with the T7 RNA polymerase (Roche) and biotinylated using the Biotin RNA Labeling Mix (Roche). Further, 3′-end-biotin-labeled RNA oligonucleotides encoding the leader sequences of hrpC, which contain a GGA motif, and hrpB, hrpF and hrpX, which do not contain putative RsmA binding sites, were synthesized by IDT, Inc. (Integrated DNA Technology, Coralville, Iowa, USA) to test also for interactions with 6HisRsmA (Table S4). The RNA oligonucleotides were designed based on the predicted secondary structures of the hrpB, hrpC, hrpF and hrpX 5′ leader sequences by analysis in MFold [66]. Briefly, DNA templates for the generation of the targeted RNA transcripts were produced using XCC wild-type strain 306 genomic DNA as template and the specific primer pairs shown in Table S3. These DNA fragments were cloned into pGEM T-vector producing pGEMT-5UTRhrpG, pGEMT-5UTRhrpD and pGEMT-5UTRhrpE (Table S1). These plasmids were linearized with PstI restriction enzyme and purified using DNA gel extraction kit (Qiagen). The hrp transcripts were produced in vitro by using the T7 Transcription kit (Roche) and Biotin RNA labeling Kit (Roche) following the manufacturer's instructions. Transcripts were purified by using the Zymoclean Gel RNA Recovery kit (Zymo Research, USA), re-suspended in DEPC water and quantified using Nanodrop. Biotin-labeled RNA probes, in vitro transcribed (Table S3) or synthesized by IDT corporation (Table S4), were heated to 95°C for 3 minutes and reconstituted by incubating 15 minutes at room temperature. The binding assays were performed using the LightShift Chemiluminescent RNA EMSA Kit (Thermo Scientific, USA) according to the manufacturer's instructions. Then approximately 65 nM 6HisRsmA protein and 6.25 nM Biotin-labeled RNA were mixed into a tube with binding buffer [(10 mM HEPES pH 7.3, 20 mM KCl, 1 mM MgCl2, 1 mM DTT, 5% Glycerol, 0.1 µg/µL yeast tRNA, 20 U RNasin (Promega)] to a total volume of 20 µL reaction. The binding reactions were incubated at 25°C for 20 min to allow RsmA–RNA complex formation. A total of 5 µL loading buffer (97% glycerol, 0.01% bromophenol blue, 0.01% xylene cyanol) was added to the binding reaction and immediately loaded and separated using 5% native polyacrylamide gels. Signal bands were visualized using the LightShift Chemiluminescent RNA EMSA Kit (Thermo Scientific) according to the manufacturer's instructions. In the competition assays and for calculating apparent Kd of RsmA-hrpG2 complex the concentrations of the 6HisRsmA and RNA probes are indicated in the respective figures and legends. The binding curve for 6HisRsmAxcc-hrpG2 interaction was defined as a function of 6HisRsmA concentrations and relative values of shifted band intensity. The apparent equilibrium binding constant (Kd) was estimated based on the average pixel value of each shifted band calculated with ImageJ software [68], [69], [112]. The calculated mean pixel values (P-value <0.05) were obtained as results of three independent assays.
mRNA stability assay
Bacterial cultures of XCC wild-type and rsmA mutant were grown at 28°C in XVM2 medium to OD600 nm of 0.6 and treated with ciprofloxacin at a final concentration of 10 mg/ml to inhibit transcription. Samples were collected at 0, 2, 4, 6, 8 and 15 min after ciprofloxacin treatment. The cells were harvested by centrifugation at 10,000 r.p.m. and used immediately for RNA extraction by addition of Trizol (Life, USA) in the pellets, and heating at 65°C for 10 min. After the separation of organic and aqueous phase, the supernatants containing total RNA were transferred to new eppendorf tubes and RNA extraction was continued by using RNeasy Mini kit (Qiagen). Total RNA samples were treated with DNase I-Free (Qiagen) and quantified by using Nanodrop. A total of 2 µg of treated RNA was used for One-Step RT-PCR (Qiagen) in 25 µL reactions following the manufacturer's protocol. Reactions were subjected to PCR amplification for 26 cycles. Ten microliters of each reaction was resolved in 1.5% agarose gel. Stability of hrpD transcript was evaluated with primers annealing within the first orf hrpQ (Table S4). Analysis of 16S RNA was performed as control for normalizing the hrpG and hrpD amplified products. The relative value of hrpG and hrpD mRNA half-life was estimated with determination of the average pixel value of each amplified products and subtracting the background by ImageJ software [68], [112]. The obtained mean values were normalized with 16S correspondent product of amplification. Mean values derived from two independent experiments are shown.
Supporting Information
Zdroje
1. ButtnerD, BonasU (2010) Regulation and secretion of Xanthomonas virulence factors. FEMS microbiology reviews 34 : 107–133.
2. KayS, BonasU (2009) How Xanthomonas type III effectors manipulate the host plant. Current opinion in microbiology 12 : 37–43.
3. LaiaML, MoreiraLM, DezajacomoJ, BrigatiJB, FerreiraCB, et al. (2009) New genes of Xanthomonas citri subsp. citri involved in pathogenesis and adaptation revealed by a transposon-based mutant library. BMC microbiology 9 : 12.
4. KoebnikR, KrugerA, ThiemeF, UrbanA, BonasU (2006) Specific binding of the Xanthomonas campestris pv. vesicatoria AraC-type transcriptional activator HrpX to plant-inducible promoter boxes. Journal of bacteriology 188 : 7652–7660.
5. BochJ, BonasU (2010) Xanthomonas AvrBs3 family-type III effectors: discovery and function. Annual review of phytopathology 48 : 419–436.
6. Akimoto-TomiyamaC, FurutaniA, TsugeS, WashingtonEJ, NishizawaY, et al. (2012) XopR, a type III effector secreted by Xanthomonas oryzae pv. oryzae, suppresses Microbe-Associated Molecular Pattern-triggered immunity in Arabidopsis thaliana. Molecular plant-microbe interactions 25 : 505–514.
7. MoleBM, BaltrusDA, DanglJL, GrantSR (2007) Global virulence regulation networks in phytopathogenic bacteria. Trends in microbiology 15 : 363–371.
8. BonasU (1994) hrp genes of phytopathogenic bacteria. Current topics in microbiology and immunology 192 : 79–98.
9. GuoY, FigueiredoF, JonesJ, WangN (2011) HrpG and HrpX play global roles in coordinating different virulence traits of Xanthomonas axonopodis pv. citri. Molecular plant-microbe interactions 24 : 649–661.
10. WengelnikK, Van den AckervekenG, BonasU (1996) HrpG, a key hrp regulatory protein of Xanthomonas campestris pv. vesicatoria is homologous to two-component response regulators. Molecular plant-microbe interactions 9 : 704–712.
11. WengelnikK, RossierO, BonasU (1999) Mutations in the regulatory gene hrpG of Xanthomonas campestris pv. vesicatoria result in constitutive expression of all hrp genes. Journal of bacteriology 181 : 6828–6831.
12. LiRF, LuGT, LiL, SuHZ, FengGF, et al. (2013) Identification of a putative cognate sensor kinase for the two-component response regulator HrpG, a key regulator controlling the expression of the hrp genes in Xanthomonas campestris pv. campestris. Environ Microbiol [epub ahead of print].
13. FurutaniA, NakayamaT, OchiaiH, KakuH, KuboY, et al. (2006) Identification of novel HrpXo regulons preceded by two cis-acting elements, a plant-inducible promoter box and a −10 box-like sequence, from the genome database of Xanthomonas oryzae pv. oryzae. FEMS microbiology letters 259 : 133–141.
14. JiangW, JiangBL, XuRQ, HuangJD, WeiHY, et al. (2009) Identification of six type III effector genes with the PIP box in Xanthomonas campestris pv. campestris and five of them contribute individually to full pathogenicity. Molecular plant-microbe interactions 22 : 1401–1411.
15. LipscombL, SchellMA (2011) Elucidation of the regulon and cis-acting regulatory element of HrpB, the AraC-type regulator of a plant pathogen-like type III secretion system in Burkholderia pseudomallei. J Bacteriol 193 : 1991–2001.
16. CunnacS, BoucherC, GeninS (2004) Characterization of the cis-acting regulatory element controlling HrpB-mediated activation of the type III secretion system and effector genes in Ralstonia solanacearum. Journal of bacteriology 186 : 2309–2318.
17. TangX, XiaoY, ZhouJM (2006) Regulation of the type III secretion system in phytopathogenic bacteria. Mol Plant Microbe Interact 19 : 1159–1166.
18. SchmidtkeC, FindeissS, SharmaCM, KuhfussJ, HoffmannS, et al. (2012) Genome-wide transcriptome analysis of the plant pathogen Xanthomonas iden Nucleic acids research40 : 2020–2031.
19. ChaoNX, WeiK, ChenQ, MengQL, TangDJ, et al. (2008) The rsmA-like gene rsmA(Xcc) of Xanthomonas campestris pv. campestris is involved in the control of various cellular processes, including pathogenesis. Molecular plant-microbe interactions 21 : 411–423.
20. ZhuPL, ZhaoS, TangJL, FengJX (2011) The rsmA-like gene rsmA(Xoo) of Xanthomonas oryzae pv. oryzae regulates bacterial virulence and production of diffusible signal factor. Molecular plant pathology 12 : 227–237.
21. LapougeK, SchubertM, AllainFH, HaasD (2008) Gac/Rsm signal transduction pathway of gamma-proteobacteria: from RNA recognition to regulation of social behaviour. Molecular microbiology 67 : 241–253.
22. RomeoT, VakulskasCA, BabitzkeP (2012) Post-transcriptional regulation on a global scale: form and function of Csr/Rsm systems. Environmental microbiology 15 (2) 313–24.
23. WhiteD, HartME, RomeoT (1996) Phylogenetic distribution of the global regulatory gene csrA among eubacteria. Gene 182 : 221–223.
24. RomeoT (1998) Global regulation by the small RNA-binding protein CsrA and the non-coding RNA molecule CsrB. Molecular microbiology 29 : 1321–1330.
25. BabitzkeP, RomeoT (2007) CsrB sRNA family: sequestration of RNA-binding regulatory proteins. Current opinion in microbiology 10 : 156–163.
26. JonasK, EdwardsAN, SimmR, RomeoT, RomlingU, et al. (2008) The RNA binding protein CsrA controls cyclic di-GMP metabolism by directly regulating the expression of GGDEF proteins. Molecular microbiology 70 : 236–257.
27. LuXH, AnSQ, TangDJ, McCarthyY, TangJL, et al. (2012) RsmA regulates biofilm formation in Xanthomonas campestris through a regulatory network involving cyclic di-GMP and the Clp transcription factor. PloS one 7: e52646.
28. AltierC, SuyemotoM, LawhonSD (2000) Regulation of Salmonella enterica serovar typhimurium invasion genes by csrA. Infection and immunity 68 : 6790–6797.
29. MulcahyH, O'CallaghanJ, O'GradyEP, AdamsC, O'GaraF (2006) The posttranscriptional regulator RsmA plays a role in the interaction between Pseudomonas aeruginosa and human airway epithelial cells by positively regulating the type III secretion system. Infect Immun 74 : 3012–3015.
30. CuiY, ChatterjeeA, LiuY, DumenyoCK, ChatterjeeAK (1995) Identification of a global repressor gene, rsmA, of Erwinia carotovora subsp. carotovora that controls extracellular enzymes, N-(3-oxohexanoyl)-L-homoserine lactone, and pathogenicity in soft-rotting Erwinia spp. Journal of bacteriology 177 : 5108–5115.
31. LiuY, CuiY, MukherjeeA, ChatterjeeAK (1998) Characterization of a novel RNA regulator of Erwinia carotovora ssp. carotovora that controls production of extracellular enzymes and secondary metabolites. Molecular microbiology 29 : 219–234.
32. ZouL, ZengQ, LinH, GyaneshwarP, ChenG, et al. (2012) SlyA regulates type III secretion system (T3SS) genes in parallel with the T3SS master regulator HrpL in Dickeya dadantii 3937. Appl Environ Microbiol 78 : 2888–2895.
33. SchubertM, LapougeK, DussO, OberstrassFC, JelesarovI, et al. (2007) Molecular basis of messenger RNA recognition by the specific bacterial repressing clamp RsmA/CsrA. Nature structural & molecular biology 14 : 807–813.
34. MercanteJ, SuzukiK, ChengX, BabitzkeP, RomeoT (2006) Comprehensive alanine-scanning mutagenesis of Escherichia coli CsrA defines two subdomains of critical functional importance. The Journal of biological chemistry 281 : 31832–31842.
35. DubeyAK, BakerCS, RomeoT, BabitzkeP (2005) RNA sequence and secondary structure participate in high-affinity CsrA-RNA interaction. RNA 11 : 1579–1587.
36. MercanteJ, EdwardsAN, DubeyAK, BabitzkeP, RomeoT (2009) Molecular geometry of CsrA (RsmA) binding to RNA and its implications for regulated expression. Journal of molecular biology 392 : 511–528.
37. WangX, DubeyAK, SuzukiK, BakerCS, BabitzkeP, et al. (2005) CsrA post-transcriptionally represses pgaABCD, responsible for synthesis of a biofilm polysaccharide adhesin of Escherichia coli. Molecular microbiology 56 : 1648–1663.
38. KerrinnesT, ZelasZB, StreckelW, FaberF, TietzeE, et al. (2009) CsrA and CsrB are required for the post-transcriptional control of the virulence-associated effector protein AvrA of Salmonella enterica. International journal of medical microbiology : IJMM 299 : 333–341.
39. LiuMY, GuiG, WeiB, PrestonJF3rd, OakfordL, et al. (1997) The RNA molecule CsrB binds to the global regulatory protein CsrA and antagonizes its activity in Escherichia coli. The Journal of biological chemistry 272 : 17502–17510.
40. BurrowesE, BaysseC, AdamsC, O'GaraF (2006) Influence of the regulatory protein RsmA on cellular functions in Pseudomonas aeruginosa PAO1, as revealed by transcriptome analysis. Microbiology 152 : 405–418.
41. ChatterjeeA, CuiY, ChatterjeeAK (2002) RsmA and the quorum-sensing signal, N-[3-oxohexanoyl]-L-homoserine lactone, control the levels of rsmB RNA in Erwinia carotovora subsp. carotovora by affecting its stability. Journal of bacteriology 184 : 4089–4095.
42. LapougeK, SinevaE, LindellM, StarkeK, BakerCS, et al. (2007) Mechanism of hcnA mRNA recognition in the Gac/Rsm signal transduction pathway of Pseudomonas fluorescens. Molecular microbiology 66 : 341–356.
43. WeiBL, Brun-ZinkernagelAM, SimeckaJW, PrussBM, BabitzkeP, et al. (2001) Positive regulation of motility and flhDC expression by the RNA-binding protein CsrA of Escherichia coli. Molecular microbiology 40 : 245–256.
44. YakhninAV, BakerCS, VakulskasCA, YakhninH, BerezinI, et al. (2013) CsrA activates flhDC expression by protecting flhDC mRNA from RNase E-mediated cleavage. Mol Microbiol 87 : 851–866.
45. GottwaldTR, SunX, RileyT, GrahamJH, FerrandinoF, et al. (2002) Geo-referenced spatiotemporal analysis of the urban citrus canker epidemic in Florida. Phytopathology 92 : 361–377.
46. da SilvaAC, FerroJA, ReinachFC, FarahCS, FurlanLR, et al. (2002) Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 417 : 459–463.
47. RifeC, SchwarzenbacherR, McMullanD, AbdubekP, AmbingE, et al. (2005) Crystal structure of the global regulatory protein CsrA from Pseudomonas putida at 2.05 A resolution reveals a new fold. Proteins 61 : 449–453.
48. GutierrezP, LiY, OsborneMJ, PomerantsevaE, LiuQ, et al. (2005) Solution structure of the carbon storage regulator protein CsrA from Escherichia coli. Journal of bacteriology 187 : 3496–3501.
49. AgarasB, SobreroP, ValverdeC (2013) A CsrA/RsmA translational regulator gene encoded in the replication region of a Sinorhizobium meliloti cryptic plasmid complements Pseudomonas fluorescens rsmA/E mutants. Microbiology 159 : 230–242.
50. KelleyLA, SternbergMJ (2009) Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc 4 : 363–371.
51. MardenJN, DiazMR, WaltonWG, GodeCJ, BettsL, et al. (2013) An unusual CsrA family member operates in series with RsmA to amplify posttranscriptional responses in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 110 : 15055–15060.
52. BordoliL, KieferF, ArnoldK, BenkertP, BatteyJ, et al. (2009) Protein structure homology modeling using SWISS-MODEL workspace. Nat Protoc 4 : 1–13.
53. ArnoldK, BordoliL, KoppJ, SchwedeT (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22 : 195–201.
54. SchwedeT, KoppJ, GuexN, PeitschMC (2003) SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res 31 : 3381–3385.
55. OrdogR (2008) PyDeT, a PyMOL plug-in for visualizing geometric concepts around proteins. Bioinformation 2 : 346–347.
56. Astua-MongeG, Freitas-AstuaJ, BacocinaG, RoncolettaJ, CarvalhoSA, et al. (2005) Expression profiling of virulence and pathogenicity genes of Xanthomonas axonopodis pv. citri. Journal of bacteriology 187 : 1201–1205.
57. WengelnikK, BonasU (1996) HrpXv, an AraC-type regulator, activates expression of five of the six loci in the hrp cluster of Xanthomonas campestris pv. vesicatoria. Journal of bacteriology 178 : 3462–3469.
58. LorenzC, ButtnerD (2011) Secretion of early and late substrates of the type III secretion system from Xanthomonas is controlled by HpaC and the C-terminal domain of HrcU. Molecular microbiology 79 : 447–467.
59. CappellettiPA, dos SantosRF, do AmaralAM, HomemRA, Souza TdosS, et al. (2011) Structure-function analysis of the HrpB2-HrcU interaction in the Xanthomonas citri type III secretion system. PloS one 6: e17614.
60. JeffersonRA (1989) The GUS reporter gene system. Nature 342 : 837–838.
61. AtanassovII, EtchellsJP, TurnerSR (2009) A simple, flexible and efficient PCR-fusion/Gateway cloning procedure for gene fusion, site-directed mutagenesis, short sequence insertion and domain deletions and swaps. Plant Methods 5 : 14.
62. HerovenAK, BohmeK, DerschP (2012) The Csr/Rsm system of Yersinia and related pathogens: A post-transcriptional strategy for managing virulence. RNA biology 9 : 379–391.
63. WeberE, BergerC, BonasU, KoebnikR (2007) Refinement of the Xanthomonas campestris pv. vesicatoria hrpD and hrpE operon structure. Molecular plant-microbe interactions 20 : 559–567.
64. KogenaruS, QingY, GuoY, WangN (2012) RNA-seq and microarray complement each other in transcriptome profiling. BMC genomics 13 : 629.
65. FurutaniA, NakayamaT, OchiaiH, KakuH, KuboY, et al. (2006) Identification of novel HrpXo regulons preceded by two cis-acting elements, a plant-inducible promoter box and a −10 box-like sequence, from the genome database of Xanthomonas oryzae pv. oryzae. FEMS Microbiol Lett 259 : 133–141.
66. ZukerM (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31 : 3406–3415.
67. YakhninAV, YakhninH, BabitzkeP (2012) Gel mobility shift assays to detect protein-RNA interactions. Methods Mol Biol 905 : 201–211.
68. GirishV, VijayalakshmiA (2004) Affordable image analysis using NIH Image/ImageJ. Indian J Cancer 41 : 47.
69. HartigSM (2013) Basic image analysis and manipulation in ImageJ. Curr Protoc Mol Biol Chapter 14: Unit14 15.
70. DubeyAK, BakerCS, SuzukiK, JonesAD, PanditP, et al. (2003) CsrA regulates translation of the Escherichia coli carbon starvation gene, cstA, by blocking ribosome access to the cstA transcript. Journal of bacteriology 185 : 4450–4460.
71. GoldsboroughAS, KornbergTB (1994) Allele-specific quantification of Drosophila engrailed and invected transcripts. Proc Natl Acad Sci U S A 91 : 12696–12700.
72. BakerCS, EoryLA, YakhninH, MercanteJ, RomeoT, et al. (2007) CsrA inhibits translation initiation of Escherichia coli hfq by binding to a single site overlapping the Shine-Dalgarno sequence. Journal of bacteriology 189 : 5472–5481.
73. WeiBL, Brun-ZinkernagelAM, SimeckaJW, PrussBM, BabitzkeP, et al. (2001) Positive regulation of motility and flhDC expression by the RNA-binding protein CsrA of Escherichia coli. Mol Microbiol 40 : 245–256.
74. MackieGA (1998) Ribonuclease E is a 5′-end-dependent endonuclease. Nature 395 : 720–723.
75. LiuMY, YangH, RomeoT (1995) The product of the pleiotropic Escherichia coli gene csrA modulates glycogen biosynthesis via effects on mRNA stability. Journal of bacteriology 177 : 2663–2672.
76. DrlicaK, ZhaoX (1997) DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol Mol Biol Rev 61 : 377–392.
77. DrlicaK, HiasaH, KernsR, MalikM, MustaevA, et al. (2009) Quinolones: action and resistance updated. Curr Top Med Chem 9 : 981–998.
78. XuY, LuoQQ, ZhouMG (2013) Identification and characterization of integron-mediated antibiotic resistance in the phytopathogen Xanthomonas oryzae pv. oryzae. PLoS One 8: e55962.
79. ChengCM, TuJ, YangCC, KuoTT (1996) Rifampicin: an inhibitor of Xp12-specific protein phosphorylation in Xanthomonas oryzae pv. oryzae. FEMS Microbiol Lett 143 : 141–149.
80. LorenzC, HausnerJ, ButtnerD (2012) HrcQ provides a docking site for early and late type III secretion substrates from Xanthomonas. PLoS One 7: e51063.
81. MartinsPM, LauIF, BacciM, BelasqueJ, do AmaralAM, et al. (2010) Subcellular localization of proteins labeled with GFP in Xanthomonas citri ssp. citri: targeting the division septum. FEMS Microbiol Lett 310 : 76–83.
82. KondoH, NakagawaA, NishihiraJ, NishimuraY, MizunoT, et al. (1997) Escherichia coli positive regulator OmpR has a large loop structure at the putative RNA polymerase interaction site. Nat Struct Biol 4 : 28–31.
83. IngleCA, KushnerSR (1996) Development of an in vitro mRNA decay system for Escherichia coli: poly(A) polymerase I is necessary to trigger degradation. Proc Natl Acad Sci U S A 93 : 12926–12931.
84. MohantyBK, GiladiH, MaplesVF, KushnerSR (2008) Analysis of RNA decay, processing, and polyadenylation in Escherichia coli and other prokaryotes. Methods Enzymol 447 : 3–29.
85. KushnerSR (2002) mRNA decay in Escherichia coli comes of age. J Bacteriol 184 : 4658–4665 discussion 4657.
86. MartinezLC, YakhninH, CamachoMI, GeorgellisD, BabitzkeP, et al. (2011) Integration of a complex regulatory cascade involving the SirA/BarA and Csr global regulatory systems that controls expression of the Salmonella SPI-1 and SPI-2 virulence regulons through HilD. Mol Microbiol 80 : 1637–1656.
87. KerrinnesT, ZelasZB, StreckelW, FaberF, TietzeE, et al. (2009) CsrA and CsrB are required for the post-transcriptional control of the virulence-associated effector protein AvrA of Salmonella enterica. Int J Med Microbiol 299 : 333–341.
88. FortuneDR, SuyemotoM, AltierC (2006) Identification of CsrC and characterization of its role in epithelial cell invasion in Salmonella enterica serovar Typhimurium. Infect Immun 74 : 331–339.
89. ChatterjeeA, CuiY, ChakrabartyP, ChatterjeeAK (2010) Regulation of motility in Erwinia carotovora subsp. carotovora: quorum-sensing signal controls FlhDC, the global regulator of flagellar and exoprotein genes, by modulating the production of RsmA, an RNA-binding protein. Molecular plant-microbe interactions 23 : 1316–1323.
90. LiuY, CuiY, MukherjeeA, ChatterjeeAK (1998) Characterization of a novel RNA regulator of Erwinia carotovora ssp. carotovora that controls production of extracellular enzymes and secondary metabolites. Mol Microbiol 29 : 219–234.
91. O'GradyEP, MulcahyH, O'CallaghanJ, AdamsC, O'GaraF (2006) Pseudomonas aeruginosa infection of airway epithelial cells modulates expression of Kruppel-like factors 2 and 6 via RsmA-mediated regulation of type III exoenzymes S and Y. Infection and immunity 74 : 5893–5902.
92. JorgensenMG, ThomasonMK, HavelundJ, Valentin-HansenP, StorzG (2013) Dual function of the McaS small RNA in controlling biofilm formation. Genes Dev 27 : 1132–1145.
93. De LayN, GottesmanS (2012) A complex network of small non-coding RNAs regulate motility in Escherichia coli. Mol Microbiol 86 : 524–538.
94. SlaterH, Alvarez-MoralesA, BarberCE, DanielsMJ, DowJM (2000) A two-component system involving an HD-GYP domain protein links cell-cell signalling to pathogenicity gene expression in Xanthomonas campestris. Mol Microbiol 38 : 986–1003.
95. RyanRP, McCarthyY, AndradeM, FarahCS, ArmitageJP, et al. (2010) Cell-cell signal-dependent dynamic interactions between HD-GYP and GGDEF domain proteins mediate virulence in Xanthomonas campestris. Proc Natl Acad Sci U S A 107 : 5989–5994.
96. WhiteTJ, GonzalezCF (1995) Electroporation of Xanthomonas. Methods Mol Biol 47 : 135–141.
97. SouzaDP, AndradeMO, Alvarez-MartinezCE, ArantesGM, FarahCS, et al. (2011) A component of the Xanthomonadaceae type IV secretion system combines a VirB7 motif with a N0 domain found in outer membrane transport proteins. PLoS Pathog 7: e1002031.
98. Sambrook J, Fritsch EF (1989) Molecular cloning : a laboratory manual. Cold Spring Harbor, N.Y. Cold Spring Harbor Laboratory Press.
99. De FeyterR, YangY, GabrielDW (1993) Gene-for-genes interactions between cotton R genes and Xanthomonas campestris pv. malvacearum avr genes. Mol Plant Microbe Interact 6 : 225–237.
100. SzczesnyR, JordanM, SchrammC, SchulzS, CogezV, et al. (2010) Functional characterization of the Xcs and Xps type II secretion systems from the plant pathogenic bacterium Xanthomonas campestris pv vesicatoria. The New phytologist 187 : 983–1002.
101. RybakM, MinsavageGV, StallRE, JonesJB (2009) Identification of Xanthomonas citri ssp. citri host specificity genes in a heterologous expression host. Mol Plant Pathol 10 : 249–262.
102. LivakKJ, SchmittgenTD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods 25 : 402–408.
103. GuoY, ZhangY, LiJL, WangN (2012) Diffusible signal factor-mediated quorum sensing plays a central role in coordinating gene expression of Xanthomonas citri subsp. citri. Molecular plant-microbe interactions 25 : 165–179.
104. SmythGK, MichaudJ, ScottHS (2005) Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics 21 : 2067–2075.
105. HochbergY, BenjaminiY (1990) More powerful procedures for multiple significance testing. Stat Med 9 : 811–818.
106. BoulangerA, ChenQ, HintonDM, StibitzS (2013) In vivo phosphorylation dynamics of the Bordetella pertussis virulence-controlling response regulator BvgA. Mol Microbiol 88 : 156–172.
107. BarbieriCM, StockAM (2008) Universally applicable methods for monitoring response regulator aspartate phosphorylation both in vitro and in vivo using Phos-tag-based reagents. Anal Biochem 376 : 73–82.
108. MitsuharaI, UgakiM, HirochikaH, OhshimaM, MurakamiT, et al. (1996) Efficient promoter cassettes for enhanced expression of foreign genes in dicotyledonous and monocotyledonous plants. Plant Cell Physiol 37 : 49–59.
109. StaskawiczB, DahlbeckD, KeenN, NapoliC (1987) Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J Bacteriol 169 : 5789–5794.
110. ChoiKH, GaynorJB, WhiteKG, LopezC, BosioCM, et al. (2005) A Tn7-based broad-range bacterial cloning and expression system. Nat Methods 2 : 443–448.
111. TsugeS, FurutaniA, FukunakaR, OkuT, TsunoK, et al. (2002) Expression of Xanthomonas oryzae pv. oryzae hrp Genes in XOM2, a Novel Synthetic Medium J Gen Plant Pathol 68 : 363–371.
112. JensenEC (2013) Quantitative analysis of histological staining and fluorescence using ImageJ. Anat Rec (Hoboken) 296 : 378–381.
113. CrooksGE, HonG, ChandoniaJM, BrennerSE (2004) WebLogo: a sequence logo generator. Genome Res 14 : 1188–1190.
114. BramucciE, PaiardiniA, BossaF, PascarellaS (2012) PyMod: sequence similarity searches, multiple sequence-structure alignments, and homology modeling within PyMOL. BMC Bioinformatics 13 Suppl 4: S2.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Structure of the Membrane Anchor of Pestivirus Glycoprotein E, a Long Tilted Amphipathic HelixČlánek Iron Acquisition in : The Roles of IlsA and Bacillibactin in Exogenous Ferritin Iron MobilizationČlánek AvrBsT Acetylates ACIP1, a Protein that Associates with Microtubules and Is Required for ImmunityČlánek Viral MicroRNA Effects on Pathogenesis of Polyomavirus SV40 Infections in Syrian Golden HamstersČlánek Genome-Wide RNAi Screen Identifies Broadly-Acting Host Factors That Inhibit Arbovirus Infection
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2014 Číslo 2- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Viral Enhancer Mimicry of Host Innate-Immune Promoters
- The Epstein-Barr Virus-Encoded MicroRNA MiR-BART9 Promotes Tumor Metastasis by Targeting E-Cadherin in Nasopharyngeal Carcinoma
- Implication of PMLIV in Both Intrinsic and Innate Immunity
- The Consequences of Reconfiguring the Ambisense S Genome Segment of Rift Valley Fever Virus on Viral Replication in Mammalian and Mosquito Cells and for Genome Packaging
- Substrate-Induced Unfolding of Protein Disulfide Isomerase Displaces the Cholera Toxin A1 Subunit from Its Holotoxin
- Male-Killing Induces Sex-Specific Cell Death via Host Apoptotic Pathway
- Highly Active Antiretroviral Therapies Are Effective against HIV-1 Cell-to-Cell Transmission
- The microRNAs in an Ancient Protist Repress the Variant-Specific Surface Protein Expression by Targeting the Entire Coding Sequence
- Transmission-Blocking Antibodies against Mosquito C-Type Lectins for Dengue Prevention
- Type III Secretion Protein MxiI Is Recognized by Naip2 to Induce Nlrc4 Inflammasome Activation Independently of Pkcδ
- Lundep, a Sand Fly Salivary Endonuclease Increases Parasite Survival in Neutrophils and Inhibits XIIa Contact Activation in Human Plasma
- Induction of Type I Interferon Signaling Determines the Relative Pathogenicity of Strains
- Structure of the Membrane Anchor of Pestivirus Glycoprotein E, a Long Tilted Amphipathic Helix
- Foxp3 Regulatory T Cells Delay Expulsion of Intestinal Nematodes by Suppression of IL-9-Driven Mast Cell Activation in BALB/c but Not in C57BL/6 Mice
- Iron Acquisition in : The Roles of IlsA and Bacillibactin in Exogenous Ferritin Iron Mobilization
- MicroRNA Editing Facilitates Immune Elimination of HCMV Infected Cells
- Reversible Silencing of Cytomegalovirus Genomes by Type I Interferon Governs Virus Latency
- Identification of Host-Targeted Small Molecules That Restrict Intracellular Growth
- A Cyclophilin Homology Domain-Independent Role for Nup358 in HIV-1 Infection
- Engagement of NKG2D on Bystander Memory CD8 T Cells Promotes Increased Immunopathology following Infection
- Suppression of RNA Silencing by a Plant DNA Virus Satellite Requires a Host Calmodulin-Like Protein to Repress Expression
- CIB1 Synergizes with EphrinA2 to Regulate Kaposi's Sarcoma-Associated Herpesvirus Macropinocytic Entry in Human Microvascular Dermal Endothelial Cells
- A Gammaherpesvirus Bcl-2 Ortholog Blocks B Cell Receptor-Mediated Apoptosis and Promotes the Survival of Developing B Cells
- Metabolic Reprogramming during Purine Stress in the Protozoan Pathogen
- The Post-transcriptional Regulator / Activates T3SS by Stabilizing the 5′ UTR of , the Master Regulator of Genes, in
- Tailored Immune Responses: Novel Effector Helper T Cell Subsets in Protective Immunity
- AvrBsT Acetylates ACIP1, a Protein that Associates with Microtubules and Is Required for Immunity
- Epstein-Barr Virus Large Tegument Protein BPLF1 Contributes to Innate Immune Evasion through Interference with Toll-Like Receptor Signaling
- The Major Cellular Sterol Regulatory Pathway Is Required for Andes Virus Infection
- Insights into the Initiation of JC Virus DNA Replication Derived from the Crystal Structure of the T-Antigen Origin Binding Domain
- Domain Shuffling in a Sensor Protein Contributed to the Evolution of Insect Pathogenicity in Plant-Beneficial
- Lectin-Like Bacteriocins from spp. Utilise D-Rhamnose Containing Lipopolysaccharide as a Cellular Receptor
- A Compositional Look at the Human Gastrointestinal Microbiome and Immune Activation Parameters in HIV Infected Subjects
- Exploits Asparagine to Assimilate Nitrogen and Resist Acid Stress during Infection
- Interleukin-33 Increases Antibacterial Defense by Activation of Inducible Nitric Oxide Synthase in Skin
- Protective Vaccination against Papillomavirus-Induced Skin Tumors under Immunocompetent and Immunosuppressive Conditions: A Preclinical Study Using a Natural Outbred Animal Model
- Gem-Induced Cytoskeleton Remodeling Increases Cellular Migration of HTLV-1-Infected Cells, Formation of Infected-to-Target T-Cell Conjugates and Viral Transmission
- Viral MicroRNA Effects on Pathogenesis of Polyomavirus SV40 Infections in Syrian Golden Hamsters
- Genome-Wide RNAi Screen Identifies Broadly-Acting Host Factors That Inhibit Arbovirus Infection
- Inflammatory Monocytes Orchestrate Innate Antifungal Immunity in the Lung
- Quantitative and Qualitative Deficits in Neonatal Lung-Migratory Dendritic Cells Impact the Generation of the CD8+ T Cell Response
- Human Genome-Wide RNAi Screen Identifies an Essential Role for Inositol Pyrophosphates in Type-I Interferon Response
- The Master Regulator of the Cellular Stress Response (HSF1) Is Critical for Orthopoxvirus Infection
- Code-Assisted Discovery of TAL Effector Targets in Bacterial Leaf Streak of Rice Reveals Contrast with Bacterial Blight and a Novel Susceptibility Gene
- Competitive and Cooperative Interactions Mediate RNA Transfer from Herpesvirus Saimiri ORF57 to the Mammalian Export Adaptor ALYREF
- The Type III Secretion Chaperone Slc1 Engages Multiple Early Effectors, Including TepP, a Tyrosine-phosphorylated Protein Required for the Recruitment of CrkI-II to Nascent Inclusions and Innate Immune Signaling
- Yeasts: How Many Species Infect Humans and Animals?
- Clustering of Pattern Recognition Receptors for Fungal Detection
- Distinct Antiviral Responses in Pluripotent versus Differentiated Cells
- Igniting the Fire: Virulence Factors in the Pathogenesis of Sepsis
- Inactivation of the Host Lipin Gene Accelerates RNA Virus Replication through Viral Exploitation of the Expanded Endoplasmic Reticulum Membrane
- Inducible Deletion of CD28 Prior to Secondary Infection Impairs Worm Expulsion and Recall of Protective Memory CD4 T Cell Responses
- Clonal Expansion during Infection Dynamics Reveals the Effect of Antibiotic Intervention
- The Secreted Triose Phosphate Isomerase of Is Required to Sustain Microfilaria Production
- Unifying Viral Genetics and Human Transportation Data to Predict the Global Transmission Dynamics of Human Influenza H3N2
- ‘Death and Axes’: Unexpected Ca Entry Phenologs Predict New Anti-schistosomal Agents
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Reversible Silencing of Cytomegalovirus Genomes by Type I Interferon Governs Virus Latency
- Implication of PMLIV in Both Intrinsic and Innate Immunity
- Transmission-Blocking Antibodies against Mosquito C-Type Lectins for Dengue Prevention
- Lundep, a Sand Fly Salivary Endonuclease Increases Parasite Survival in Neutrophils and Inhibits XIIa Contact Activation in Human Plasma
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



