-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaAvrBsT Acetylates ACIP1, a Protein that Associates with Microtubules and Is Required for Immunity
Bacterial pathogens of plant and animals share a homologous group of virulence factors, referred to as the YopJ effector family, which are translocated by the type III secretion (T3S) system into host cells during infection. Recent work indicates that some of these effectors encode acetyltransferases that suppress host immunity. The YopJ-like protein AvrBsT is known to activate effector-triggered immunity (ETI) in Arabidopsis thaliana Pi-0 plants; however, the nature of its enzymatic activity and host target(s) has remained elusive. Here we report that AvrBsT possesses acetyltransferase activity and acetylates ACIP1 (for ACETYLATED INTERACTING PROTEIN1), an unknown protein from Arabidopsis. Genetic studies revealed that Arabidopsis ACIP family members are required for both pathogen-associated molecular pattern (PAMP)-triggered immunity and AvrBsT-triggered ETI during Pseudomonas syringae pathovar tomato DC3000 (Pst DC3000) infection. Microscopy studies revealed that ACIP1 is associated with punctae on the cell cortex and some of these punctae co-localize with microtubules. These structures were dramatically altered during infection. Pst DC3000 or Pst DC3000 AvrRpt2 infection triggered the formation of numerous, small ACIP1 punctae and rods. By contrast, Pst DC3000 AvrBsT infection primarily triggered the formation of large GFP-ACIP1 aggregates, in an acetyltransferase-dependent manner. Our data reveal that members of the ACIP family are new components of the defense machinery required for anti-bacterial immunity. They also suggest that AvrBsT-dependent acetylation in planta alters ACIP1's defense function, which is linked to the activation of ETI.
Published in the journal: . PLoS Pathog 10(2): e32767. doi:10.1371/journal.ppat.1003952
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1003952Summary
Bacterial pathogens of plant and animals share a homologous group of virulence factors, referred to as the YopJ effector family, which are translocated by the type III secretion (T3S) system into host cells during infection. Recent work indicates that some of these effectors encode acetyltransferases that suppress host immunity. The YopJ-like protein AvrBsT is known to activate effector-triggered immunity (ETI) in Arabidopsis thaliana Pi-0 plants; however, the nature of its enzymatic activity and host target(s) has remained elusive. Here we report that AvrBsT possesses acetyltransferase activity and acetylates ACIP1 (for ACETYLATED INTERACTING PROTEIN1), an unknown protein from Arabidopsis. Genetic studies revealed that Arabidopsis ACIP family members are required for both pathogen-associated molecular pattern (PAMP)-triggered immunity and AvrBsT-triggered ETI during Pseudomonas syringae pathovar tomato DC3000 (Pst DC3000) infection. Microscopy studies revealed that ACIP1 is associated with punctae on the cell cortex and some of these punctae co-localize with microtubules. These structures were dramatically altered during infection. Pst DC3000 or Pst DC3000 AvrRpt2 infection triggered the formation of numerous, small ACIP1 punctae and rods. By contrast, Pst DC3000 AvrBsT infection primarily triggered the formation of large GFP-ACIP1 aggregates, in an acetyltransferase-dependent manner. Our data reveal that members of the ACIP family are new components of the defense machinery required for anti-bacterial immunity. They also suggest that AvrBsT-dependent acetylation in planta alters ACIP1's defense function, which is linked to the activation of ETI.
Introduction
It is well established that bacterial pathogens utilize type III secretion (T3S) systems to translocate virulence factors (referred to as T3S effectors) into eukaryotic hosts to modulate immune signaling during infection [1]. The T3S effector proteome reflects the coevolution of specific host-pathogen interactions as well as microbe-microbe interactions within a given environment. Few T3S effector homologs are conserved among bacterial pathogens that colonize plant or animals hosts. One exception is the YopJ effector family, which is shared by a number of bacterial species in different genera (e.g. Yersinia, Salmonella, Vibrio, Pseudomonas, Xanthomonas, and Sinorhizobium) [2].
The YopJ effector family is named after the archetypal protein YopJ, first identified in Yersinia pseudotuberculosis [3]. These effectors belong to the C55 peptidase family because they share putative structural folds characteristic of cysteine proteases and contain the conserved catalytic triad – His, Glu and Cys [4]. Mutation of this catalytic triad destroyed effector-triggered phenotypes in host cells [5], providing the first clue that enzyme activity is critical for the virulence of the YopJ effector family. Biochemical studies revealed however that YopJ has potent acetyltransferase activity [6]. In subsequent work, several effectors from this family were shown to have acetyltransferase activity important for host-pathogen interactions, including VopA from Vibrio parahemeolyticus [7], AvrA from Salmonella typhimurium [8], PopP2 from Ralstonia solanacearum [9], and HopZ1a from Pseudomonas syringae [10]. These data indicate that a predominant virulence activity for the YopJ effector family is the post-translational acetylation of host proteins.
Resistance to YopJ-like effectors (i.e. AvrBsT, AvrRxv, AvrXv4, HopZ1a, and PopP2) has been reported in several plant hosts [11]–[13]; however, only two disease resistance (R) proteins have been characterized to date [14], [15]. Arabidopsis RRS1-R (for RESISTANCE TO RALSTONIA SOLANACEARUM1) is a Toll-IL-1-receptor-nucleotide binding site-leucine rich repeat-WRKY motif (TIR-NBS-LRR-WRKY)-type R protein that recognizes the PopP2 effector from Ralstonia solanacearum [14]. RRS1-R directly interacts with PopP2 in the plant nucleus [16]. Arabidopsis ZAR1 (for HOPZ ACTIVATED RESISTANCE1) is a coiled-coil (CC)-NBS-LRR-type disease R protein that recognizes the HopZ1a effector from Pseudomonas syringae and activates immune signaling that is distinct from most R protein pathways and independent of salicylic acid [15]. Neither RRS1-R nor ZAR1 were reported to be acetylated by the corresponding acetyltransferase [9], [10] suggesting that acetylation of other plant targets is required for recognition and/or initiation of defense signaling by these R proteins. Interestingly, a recent study revealed that HopZ1a acetylates the Arabidopsis ZED1 (for HOPZ-ETI DEFICIENT1), a pseudokinase that is required for ZAR1-mediated immunity [17]. ZED1 is proposed to act as a decoy in a ZAR1 defense complex.
Notably in mammals, YopJ acetylation suppresses innate immune signaling by exclusively targeting kinases in mitogen-activated protein kinase (MAPK) and/or NF–κB pathways. For example, YopJ catalyzes the O-acetylation of Ser or Thr residues in the activation loop of MAPKK6 [6], MEK2 [18], inhibitor of kappa B kinase [18], and MAP3K transforming growth factor β-activated kinase 1 (TAK1) [19]. Similarly in flies, AvrA inhibits c-Jun N-terminal kinase signaling by O-acetylation of the Thr residue in the activation loop of the MAPKK JNK-K [8].
In plants, a direct link between YopJ-like effector acetylation and suppression of disease resistance has not been made. HopZ1a was reported to acetylate tubulin in vitro, suggesting that the plant cytoskeleton may be disrupted during infection [10]. Consistent with this hypothesis, P. syringae pathovar tomato strain DC3000 (Pst DC3000) infection reduced microtubule density in a HopZ1a catalytic-dependent manner [10]. Interestingly, the mammalian tubulin acetyltransferase TAT1 acetylates Lys40 in α-tubulin (Nε-acetylation) [20], [21] and this modification is commonly found in less dynamic microtubules. The type of tubulin acetylation mediated by HopZ1a in planta has not yet been reported.
In previous work, we exploited the use of the Pseudomonas-Arabidopsis pathosystem to elucidate the biochemical function of the AvrBsT effector from Xanthomonas euvesicatoria. AvrBsT was engineered to be delivered into plant cells by Pst DC3000's T3S system [22] because Arabidopsis is not a host for X. euvesicatoria. Two Arabidopsis ecotypes were identified that differentially respond to Pst DC3000 AvrBsT infection. The Col-0 ecotype is susceptible to Pst DC3000 AvrBsT infection whereas the Pi-0 ecotype is resistant. Pi-0 resistance is due to a recessive, loss of function mutation in SOBER1 (for SUPPRESSOR OF AVRBST-ELICITED RESISTANCE1). SOBER1 encodes a α/β-hydrolase that negatively regulates the accumulation of phosphatidic acid (PA) triggered by AvrBsT activity during bacterial infection [23]. High PA levels in Pst DC3000 AvrBsT-infected Pi-0 leaves correlate with ETI-like defense responses [22], [23]. These data suggest that AvrBsT interferes with lipid homeostasis during infection and that this interference induces strong immune responses in the absence of SOBER1 activity.
Given that PA is a multifunctional stress signal [24], we hypothesized that AvrBsT-triggered PA bursts may directly lead to the local activation of defense signaling. Moreover, we hypothesized that AvrBsT host targets may be linked to the generation or perception of lipid signals during AvrBsT-triggered immunity. To begin to test these hypotheses, we sought to identify AvrBsT interacting proteins from Arabidopsis and elucidate their function(s) in the Pi-0 sober1-1 background [22]. Importantly, the availability of putative host substrates also enabled us to determine if AvrBsT possesses acetyltransferase activity, as reported for other effectors in the YopJ family [6], [9], [10].
Here we report that AvrBsT has acetyltransferase activity. We provide evidence that AvrBsT-dependent trans-acetylation activity is required for the activation of ETI in Arabidopsis Pi-0 leaves and that AvrBsT trans-acetylates Arabidopsis ACIP1 (for ACETYLATED INTERACTING PROTEIN1). ACIP1 is an unknown protein that localizes to punctae on the cell cortex and some of these punctae co-localize with cortical microtubules. We provide evidence that ACIP1 is a new component of the defense machinery required for anti-bacterial immunity. These data support the model that AvrBsT-dependent acetylation in planta alters ACIP1's defense function, which is linked to the activation of ETI.
Results
AvrBsT interacts with Arabidopsis ACIP1
To identify potential AvrBsT-interacting proteins in Arabidopsis, we performed a yeast two-hybrid screen using the GAL4 DNA-binding domain (BD) fused to AvrBsT (i.e. BD-AvrBsT) and an Arabidopsis cDNA library fused to the GAL4 activation domain (AD). We screened ∼7 million primary yeast transformants and isolated 11 independent clones with a candidate cDNA encoded by At3g09980 (Figure 1A and S1A). Given that AvrBsT is predicted to encode an acetyltransferase, we named the At3g09980-encoded protein ACIP1, for putative acetylated-interacting protein 1 (Figure 1A).
Fig. 1. AvrBsT interacts with Arabidopsis ACIP1. 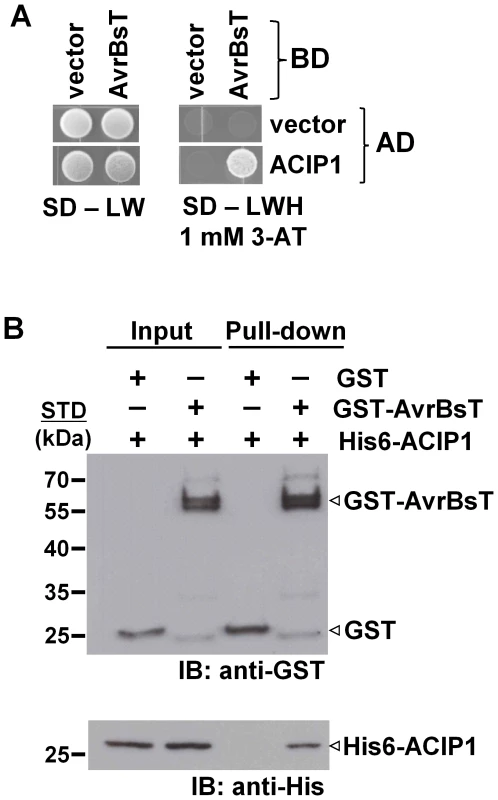
(A) Yeast two-hybrid assay showing AvrBsT binding to Arabidopsis ACIP1. Yeast strain AH109 carrying pXDGATcy86 (vector) or pXDGATcy86(AvrBsT) were independently transformed with the pGADT7 (vector) or pGADT7 containing ACIP1. pXDGATcy86 contains the GAL4 DNA-binding domain (BD) and pGADT7 contains the GAL4 activation domain (AD). Strains were spotted on nonselective (SD – LW) and selective (SD –LWH +1 mM 3-AT) media and then incubated at 30°C for 3 d. (B) GST-AvrBsT affinity purification of His6-ACIP1 in vitro. GST or GST-AvrBsT was incubated with E. coli extracts containing His6-ACIP1. Proteins were purified by using glutathione sepharose and analyzed by immunoblot (IB) analysis using anti-GST and anti-His sera. Protein input is shown on left and pull-down on right. Expected protein MW: GST = 28 kDa; GST-AvrBsT = 65 kDa; and His6-ACIP1 = 28 kDa. +, protein expressed; −, vector control. STD, molecular weight standard. Similar phenotypes were observed in at least 3 independent experiments. ACIP1 is predicted to encode a protein with 178 amino acids and molecular weight of ∼20.6 kDa. ACIP1's only distinguishing feature is that it is predicted to be a small, α-helical protein [25] that contains the widely conserved domain of unknown function, DUF662 [26]. It was first identified as a tubulin-binding protein [27]. ACIP1 belongs to a small Arabidopsis protein family containing six ACIP-like isoforms (ACIP-L1 to ACIP-L6, Figure S1A). ACIP-L4 and its wheat ortholog TaSRG are required for salt tolerance [28], although their biochemical function(s) are not known. ACIP1 shares 79% identity and 87% similarity with ACIP-L1, the closest isoform. A tree for the Arabidopsis ACIP protein family is shown in Figure S1B. None of the ACIP-like isoforms were isolated in the primary AvrBsT interaction screen.
A candidate yeast interaction screen comparing AvrBsT binding to ACIP1 or the six ACIP-like isoforms revealed that AvrBsT strongly interacts with ACIP1 but only weakly interacts with ACIP-L1 on selection media containing 1 mM 3-AT (Figure S1C,D). In the presence of 5 mM 3-AT, AvrBsT only interacted with ACIP1 (data not shown). Taken together, these data suggest that AvrBsT preferentially binds to ACIP1 in yeast.
Next, we used GST pull-down assays to independently monitor the physical association of AvrBsT and ACIP1 in vitro. Recombinant GST and GST-AvrBsT were expressed in E. coli and then purified using glutathione sepharose. Purified GST-AvrBsT migrated as a doublet in protein gels, suggesting that proteolysis of the full-length polypeptide likely occurred during extraction and/or affinity purification. His-tagged ACIP1 was expressed in E. coli and soluble protein extracts were incubated with the GST or GST-AvrBsT in a standard GST pull-down assay. His6-ACIP1 was affinity purified by GST-AvrBsT but not GST (Figure 1B). These findings are in agreement with the yeast two-hybrid data and provide additional evidence that AvrBsT interacts with Arabidopsis ACIP1.
We attempted to verify AvrBsT-ACIP1 physical interaction in planta; however, the assays were not successful. Transient or inducible expression of AvrBsT in Arabidopsis Pi-0 leaves or Nicotiana benthamiana leaves results in localized cell death. It was difficult to obtain reproducible, conclusive binding data under these cellular conditions.
AvrBsT has acetyltransferase activity
AvrBsT belongs to the YopJ family of T3S effector proteins, some of which have been shown to exhibit acetyltransferase activity [6], [9], [10]. To ascertain if AvrBsT acetylates ACIP1, we first sought to determine if AvrBsT possesses auto-acetylation activity in vitro. Recombinant wild-type GST-AvrBsT, GST (negative control) and GST-HopZ1a (positive control) [10] were over-expressed in E. coli and then purified using glutathione sepharose. Purified proteins were incubated with 14C-acetyl-coenzyme A (acetyl-CoA) ±100 nM inositol hexakisphosphate (IP6) for 30 minutes at room temperature and then separated by SDS-PAGE analysis followed by autoradiography. IP6 is a eukaryotic cofactor that stimulates the acetyltransferase activity of effectors in the YopJ family [9], [10], [29].
Auto-acetylation of GST-AvrBsT was detected in the presence of IP6 but not its absence (Figure 2A and S2). As expected, similar IP6-dependent activation and auto-acetylation of GST-HopZ1a was observed, and GST was not modified (Figure 2A and S2). Mutation of the conserved catalytic Cys residue (C222) or His residue (H154) to Ala inactivated AvrBsT-dependent acetyltransferase activity but did not affect protein expression levels (Figure 2A). By contrast, mutation of the conserved Lys residue (K282) (Figure S3A) to Arg, which has been shown to be an auto-acetylation site for some effectors in the YopJ family [9], [10], did not affect AvrBsT's acetylation state or protein accumulation (Figure 2A). The auto-acetylation activity of GST-AvrBsT(K282R) was comparable to that of wild-type GST-AvrBsT in reactions with varying concentrations of enzyme (Figure S3B). All GST-AvrBsT protein (wild type and mutant) analyzed migrated as a doublet and both of these species were auto-acetylated (Figure 2). Taken together, these data indicate that AvrBsT possesses auto-acetylation activity in vitro that is dependent on the conserved catalytic residues H154 and C222, but this activity is independent of K282.
Fig. 2. AvrBsT is an acetyltransferase that specifically acetylates ACIP1. 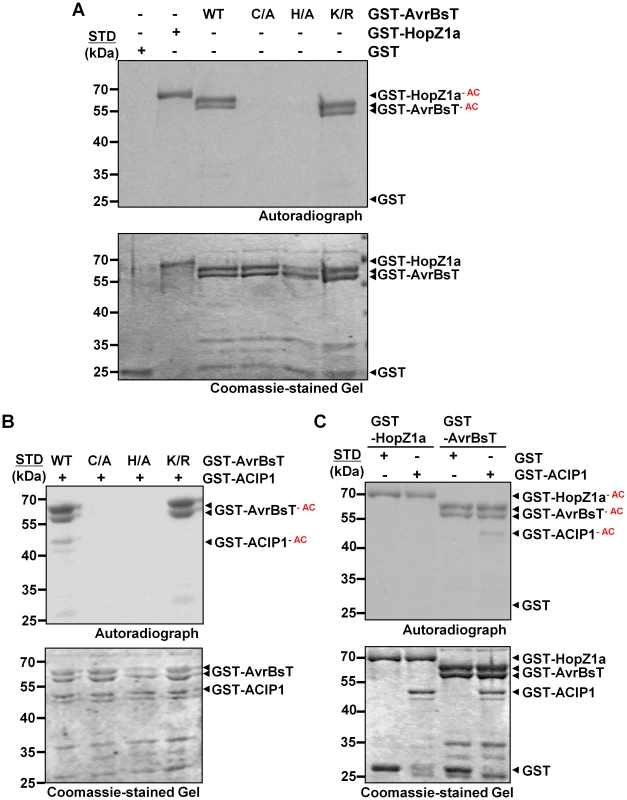
(A) AvrBsT auto-acetylation activity in vitro. Acetylation reactions using GST and GST-AvrBsT (wild-type, C222A, H154A, and K282R) proteins. (B) AvrBsT trans-acetylates ACIP1 in vitro. Acetylation reactions using GST-ACIP1 or GST with GST-AvrBsT, GST-AvrBsT(C222A), GST-AvrBT(H154A) or GST-AvrBsT(K282R). (C) Substrate specificity of AvrBsT and HopZ1a. Acetylation reactions using GST-ACIP1 or GST with GST-HopZ1a or GST-AvrBsT. For acetylation reactions, proteins were incubated with 0.4 µCi 14C-acetyl CoA and 100 nM inositol hexakisphosphate (IP6) for 30 min at RT. Proteins were then separated by SDS-PAGE. Gels were stained with Coomassie and then analyzed by autoradiography. GST and GST-HopZ1a were used as negative and positive acetyltransferase enzyme controls, respectively. Acetylated proteins (GST-HopZ1a-AC, GST-AvrBsT-AC, and GST-ACIP1-AC) are labeled in the autoradiograph. STD, molecular weight standard in kDa. GST = 28 kDa; GST-HopZ1a = 70 kDa; GST-AvrBsT = 65 kDa; GST-ACIP1 = 50 kDa. Similar results were obtained in three independent experiments. AvrBsT acetylates ACIP1 in vitro
Next, we tested if AvrBsT directly acetylates ACIP1 using similar reaction conditions to those described above. Wild-type GST-AvrBsT activity resulted in auto-acetylation of the enzyme and trans-acetylation of GST-ACIP1 (Figure 2B), whereas the catalytic core mutants GST-AvrBsT(C222A) or GST-AvrBsT(H154A) exhibited neither activities (Figure 2B). Although the GST-AvrBsT(K282R) mutant possessed auto-acetylation activity, trans-acetylation of ACIP1 was not detected under the same reaction conditions (Figure 2B). Importantly, mutation of C222 or K282 did not disrupt AvrBsT binding to ACIP1 in vitro (Figure S4).
To gain insight to the specificity of acetyltransferases in the YopJ effector family, we determined if HopZ1a could acetylate AvrBsT's substrate ACIP1. Conversely, we determined if AvrBsT could acetylate HopZ1a's substrate tubulin [10]. Incubation of GST-HopZ1a with GST-ACIP1 did not result in detectable acetylation of ACIP1 (Figure 2C). Moreover, neither HopZ1a nor other members of the HopZ family could physically associate with ACIP1 in targeted yeast two-hybrid screens (Figure S5A, B). Similarly, we could not detect AvrBsT-dependent acetylation of tubulin in vitro (Figure S5C) or direct physical interaction between AvrBsT and tubulin in yeast (Figure S5A,B). These data suggest that AvrBsT and HopZ1a possess distinct substrate specificity.
AvrBsT(K282R) does not elicit resistance in Pi-0 leaves
We assessed the biological activity of the AvrBsT(K282R) mutant in Arabidopsis Pi-0 leaves, given that mutation of the analogous Lys residue in PopP2 and HopZ1a inhibits effector auto-acetylation activity and effector-dependent phenotypes in planta [9], [10]. Bacterial growth curve analyses showed that the K282R mutation attenuated the ability of AvrBsT to activate defense in Pi-0, similar to that observed for the H154A mutation in the catalytic core (Figure 3A). Furthermore, Pst DC3000 expressing AvrBsT(K282R) did not elicit HR in Pi-0 leaves (Figure 3B) despite stable protein expression (Figure S3C). These data indicate that the K282R mutation affects AvrBsT's trans-acetylation activity in vitro (Figure 2B) and its defense eliciting activity in planta (Figure 3). Moreover, these data indicate that the auto-acetylation activity of AvrBsT(K282R) is not sufficient to activate ETI in Arabidopsis.
Fig. 3. Mutation of K282 attenuates AvrBsT-triggered resistance. 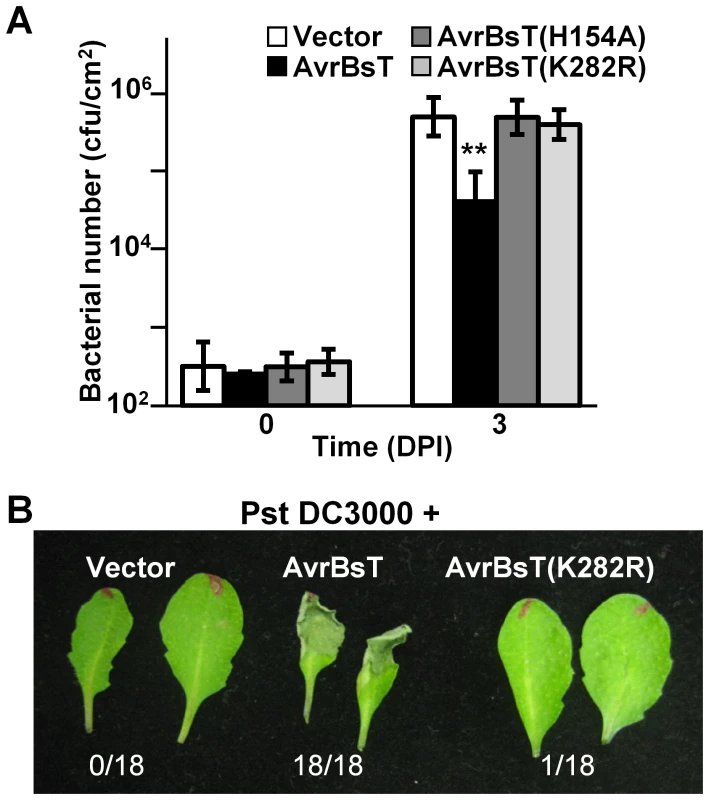
(A) Growth of Pst DC3000 in Arabidopsis Pi-0 leaves. Leaves were syringe infiltrated with a 1×105 cells/mL suspension of bacteria: Pst DC3000 carrying vector (black bars), AvrBsT (white bars), AvrBsT(H154A) (dark grey bars) or AvrBsT(K282R) (light grey bars). Titers were assessed at 0 and 3 days post-inoculation. Data are mean cfu/cm2 ± SD (n = 6). Asterisks indicate statistically significant differences from Pi-0 (student's t-test, **p<0.01). Similar results were obtained in three independent experiments. (B) HR phenotypes in Pi-0 leaves. Leaves were infiltrated with a 3×108 cells/mL suspension of Pst DC3000 carrying vector, AvrBsT or AvrBsT(K282R). Photograph was taken at 12 hours post-inoculation (HPI). Number of leaves exhibiting confluent HR at 10 HPI out of 18 inoculated leaves is shown at bottom. ACIP1 is a positive regulator of immunity
Given that nothing was known about ACIP1 function, we first sought to elucidate its potential role in immunity. Previously we showed that the Pi-0 ecotype of Arabidopsis is resistant to Pst DC3000 expressing AvrBsT, whereas the Col-0 ecotype is susceptible [22]. Interestingly, ACIP1 mRNA abundance was significantly reduced at 3 and 6 HPI in Pi-0 (Figure S6A) and Col-0 (data not shown) leaves inoculated with a 2×108 cells/mL suspension of Pst DC3000 or Pst DC3000 AvrBsT compared to leaves inoculated with mock solution of 1 mM MgCl2. By contrast, endogenous ACIP1 protein levels appeared to remain constant (Figure S6B). These data suggest that ACIP1 may be transcriptionally or post-transcriptionally regulated during pathogen attack and potentially linked to PTI and/or ETI.
To explore this further, we first analyzed the growth of virulent Pst DC3000 in a homozygous Col-0 acip1 null mutant (SALK_028810) to determine if ACIP1 is required to limit pathogen growth. Pst DC3000 grew equally well in wild-type Col-0 and acip1 mutant leaves (data not shown). Similar results were observed when the acip1 null allele was crossed into the Pi-0 background (data not shown). We speculated that the lack of a bacterial growth phenotype in the Col-0 acip1 and Pi-0 acip1 mutants may be due to genetic redundancy since ACIP1 belongs to a small gene family in Arabidopsis (Figure S1A,B). Since the nucleotide sequences between ACIP1 and ACIP-like genes are highly conserved (Figure S7A), we engineered RNAi lines to target multiple ACIP family members in attempt to uncover an immune phenotype linked to this gene family. Notably, we silenced the ACIP gene family in the Pi-0 background to be able to monitor both PTI and ETI, considering that AvrBsT induces ETI in the Pi-0 ecotype but not the Col-0 ecotype [22]. A 365-bp hairpin ACIP binary construct (hp-ACIP) was designed using the ACIP1 gene, which included the most conserved region shared by the entire gene family (Figure S7A,B), and then it was transformed into Pi-0 plants. Five independent transgenic RNAi lines were characterized. The hp-ACIP construct significantly reduced the mRNA levels for 4 of the 7 family members (i.e. ACIP1, ACIP-L1, ACIP-L2, and ACIP-L3) in two T2 ACIP RNAi lines (i.e. lines 1 and 29; Figure S7C). Of these 4 genes, ACIP1 mRNA was the most abundant transcript in 4-week old Pi-0 leaves (Figure S7D), suggesting that it may be the major isoform expressed in leaves.
To monitor ACIP1 protein expression in leaves, we generated rabbit polyclonal antisera using recombinant ACIP1-His6 protein purified from E. coli. The resulting antisera recognized multiple, recombinant purified ACIP isoforms with distinct molecular weights by immunoblot analysis (data not shown). However in wild-type Pi-0 leaf extracts, the antisera only detected a single 20 kDa protein band (Figure 4A, inset). Three of the isoforms have predicted molecular weights in this range: ACIP1 = 20.5 kDa, ACIP-L1 = 20.2 kDa, and ACIP-L3 = 20.9 kDa. The 20 kDa protein band was not detected in the two ACIP RNAi lines (Figure 4A, inset) suggesting that ACIP1, ACIP-L1 and/or ACIP-L2 protein accumulation was significantly reduced.
Fig. 4. Members of Arabidopsis ACIP family are required for AvrBsT-triggered ETI. 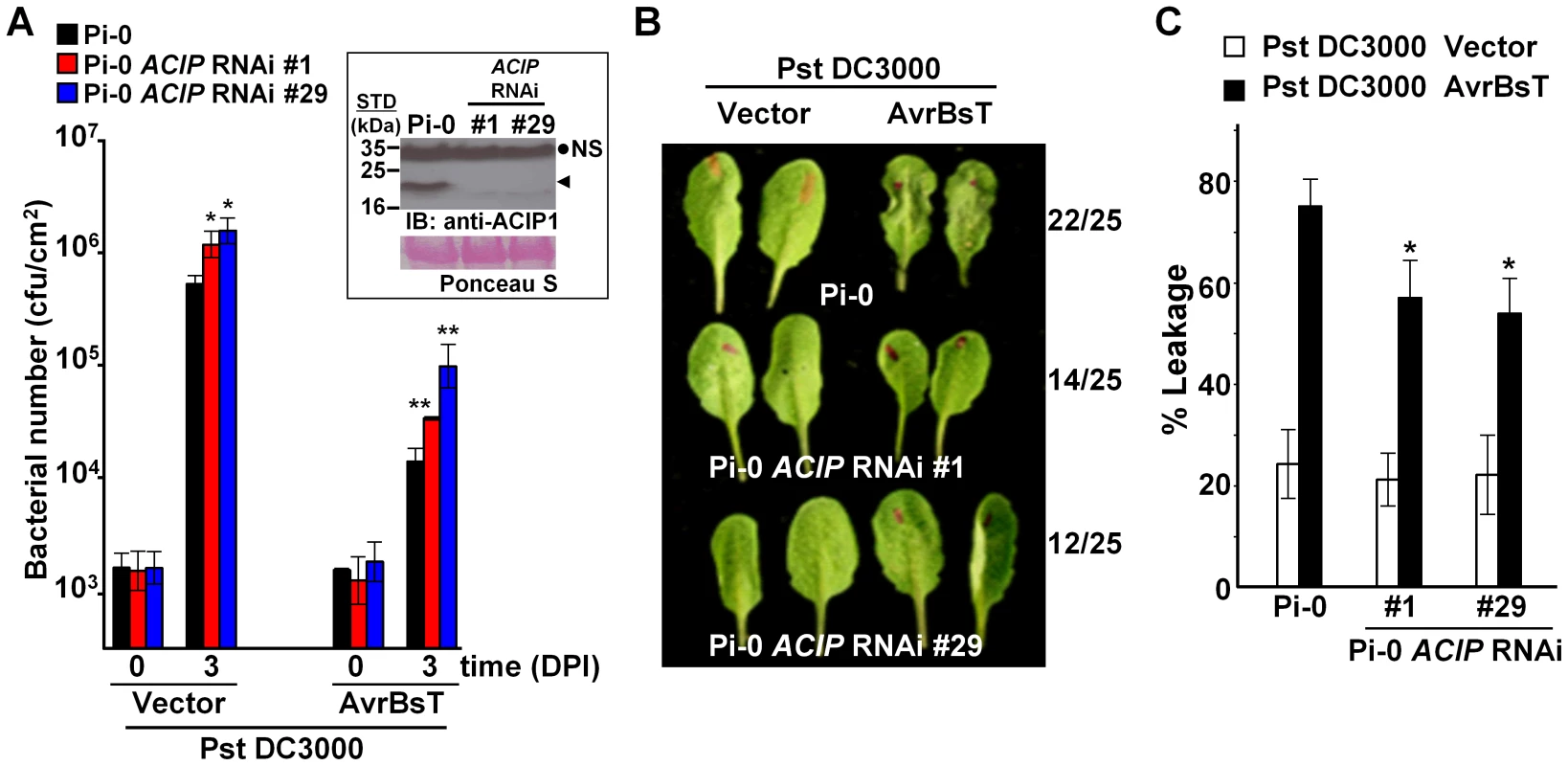
(A) Increased growth of Pst DC3000 and Pst DC3000 AvrBsT in Pi-0 ACIP RNAi line #1 (red bars) and line #29 (blue bars) compared to wild-type Pi-0 (black bars). Leaves were syringe-infiltrated with a 1×105 cells/mL suspension of bacteria. Titers were assessed at 0 and 3 days post-inoculation (DPI). Data are mean cfu/cm2 ± SD (n = 4). Asterisks indicate statistically significant differences from Pi-0 (student t-test, *p<0.05, **p<0.01). Experiment was repeated three times with similar results. Inset: Immunoblot analysis of protein extracted from Pi-0 and Pi-0 ACIP RNAi leaves using anti-ACIP1 sera. Black dot, non-specific band (NS); arrowhead, detected ∼20 kDa protein band expected to correspond to ACIP1, ACIP-L1, and/or ACIP-L3. STD, molecular weight standard in kDa. Ponceau S-stained Rubisco large subunit was used as loading control. (B) AvrBsT-elicited HR phenotype in Pi-0 and Pi-0 ACIP RNAi lines. Leaves were infiltrated with a 3×108 cells/mL suspension of Pst DC3000 alone (vector) or Pst DC3000 AvrBsT (AvrBsT). Photograph was taken at 9 hours post-inoculation (HPI). Number of leaves exhibiting confluent HR at 10 HPI out of 25 inoculated leaves is shown at right. (C) Quantification of electrolyte leakage in the leaves described in (B) at 10 HPI. Error bars represent SD (n = 10). Asterisks indicate statistically significant differences from Pi-0 (student's t-test, *p<0.05). Experiment was repeated three times with similar results. Bacterial growth curves were then performed using a 1×105 cells/mL suspension of Pst DC3000 expressing AvrBsT and the two Pi-0 ACIP RNAi transgenic lines to determine if ACIP expression is required for AvrBsT-triggered ETI. The phenotypes of the ACIP-silenced lines were compared with an unsilenced Pi-0 control plant (Figure 4).
We found that the titer of Pst DC3000 AvrBsT was significantly higher in infected Pi-0 ACIP RNAi leaves compared to that in wild-type Pi-0 leaves (Figure 4A). Notably, the Pi-0 ACIP RNAi leaves were also more susceptible to Pst DC3000. These data suggested that the silenced ACIP isoforms might function in PTI as well as ETI.
To confirm that AvrBsT-triggered ETI is impaired in the RNAi lines, we performed HR and electrolyte leakage assays in leaves challenged with a high titer (3×108 cells/mL) of Pst DC3000 AvrBsT. ETI in the Pi-0 ACIP RNAi lines was delayed but not fully inhibited (Figure 4B). In control Pi-0 leaves, AvrBsT-dependent HR was visible at 9 HPI in many leaves and by 10 HPI, 22/25 leaves exhibited HR. By contrast, HR was not observed in similarly inoculated RNAi leaves at 9 HPI; however, HR started to develop at 10 HPI in 14/25 leaves for line 1 and 12/25 leaves for line 29. Consistent with these findings, electrolyte leakage was significantly reduced in the Pst DC3000 AvrBsT-inoculated Pi-0 ACIP RNAi leaves relative to the inoculated Pi-0 leaves at 10 HPI (Figure 4C). These data suggest that multiple ACIP isoforms are required for AvrBsT-triggered ETI symptoms in Pi-0.
The Pi-0 ACIP RNAi lines were also examined for their ability to mount ETI in response to two other Pseudomonas effectors – AvrB and AvrRpt2 [30], [31]. As observed for AvrBsT, HR symptom development was slower in the RNAi lines infected with a high titer of Pst DC3000 AvrB or Pst DC3000 AvrRpt2 (data not shown). Subsequent bacterial growth curve analyses revealed that the Pi-0 ACIP RNAi lines were more susceptible to both Pst DC3000 AvrB and Pst DC3000 AvrRpt2 (Figure S8). These data suggest that the ACIP isoforms play a general role in ETI and are not specific to defense responses elicited by AvrBsT.
Given that the RNAi lines were also more susceptible to Pst DC3000, we next examined the potential role of the ACIP family in PTI. First, we analyzed the responsiveness of the Pi-0 ACIP RNAi lines to the PAMP elicitor flg22 (Figure 5). Perception of flg22 by the PRR FLS2 results in the production of reactive oxygen species (ROS) [32], one the first measurable PTI responses, followed by changes in PTI gene induction [33]. Flg22-induced ROS production was significantly reduced in both Pi-0 ACIP RNAi lines (Figure 5A). Similarly, flg22-induced mRNA accumulation for WRKY22 and WRKY29, two genes encoding transcription factors that positively regulate PTI, was significantly reduced at 3 hr post-treatment in both Pi-0 ACIP RNAi lines (Figure 5B). Consistently, the RNAi line 29 exhibited the least responsiveness to flg22 elicitation (Figure 5A,B).
Fig. 5. Members of Arabidopsis ACIP family are required for PTI. 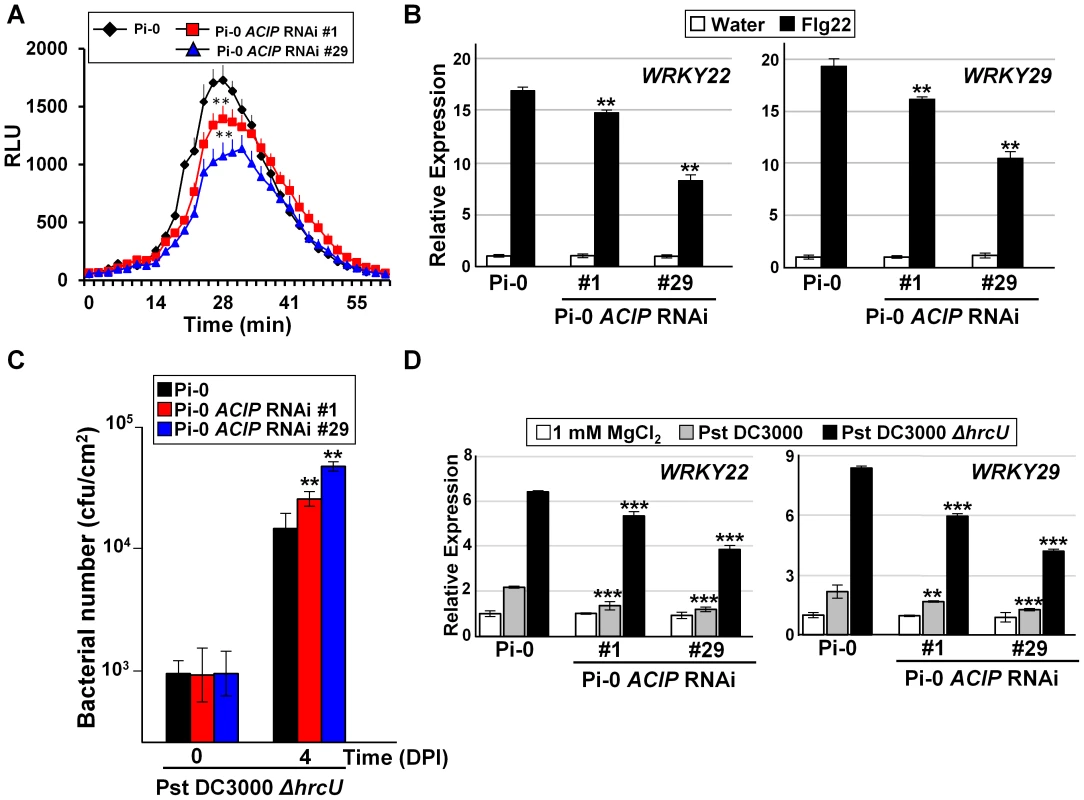
(A) Flg22-stimulated oxidative burst response in Pi-0 and Pi-0 ACIP RNAi leaves. RLU = relative luminescence unit. Error bars represent SD (n = 9). Response in both RNAi lines (#1 and #29) was significantly different from that in Pi-0 between time interval 28–32 minutes (student's t-test, **p<0.01). (B) Flg22-stimulated PTI marker gene induction in Pi-0 and Pi-0 ACIP RNAi leaves. Leaves of three plants were infiltrated with water (control) or 100 nM flg22 and then pooled for RNA extraction. WRKY22 and WRKY29 mRNA levels were quantified by qPCR. UBQ5 was used to normalize the expression value for each sample. Relative expression (mean ± SD; n = 4) is shown. (C) Growth of Pst DC3000 ΔhrcU in Pi-0 (black bars) and Pi-0 ACIP RNAi leaves (red and blue bars). Leaves were inoculated with a 1×105 cells/mL suspension of bacteria. Titers were assessed at 0 and 4 DPI. Data are mean cfu/cm2 ± SD (n = 4). (D) Pst DC3000 ΔhrcU-stimulated PTI marker gene induction in Pi-0 and Pi-0 ACIP RNAi lines. Leaves were infiltrated with a 2×108 cells/mL suspension of Pst DC3000 (grey bar), Pst DC3000 (ΔhrcU) (black bar) or 1 mM MgCl2 (white bar). Samples were collected at 6 HPI and then analyzed as described in (B). Asterisks indicate statistically significant differences from Pi-0 (student's t-test,*p<0.05,**p<0.01, ***p<0.001). Similar results were obtained in three independent experiments for (A–C), and two independent experiments for (D). We also examined the responsiveness of the Pi-0 ACIP RNAi lines to Pst DC3000 ΔhrcU, a Pseudomonas strain known to elicit PTI. Pst DC3000 ΔhrcU lacks a functional T3S apparatus [34] and does not suppress PTI because T3S effectors are not secreted. Leaves were infected with a 1×105 cells/mL suspension of bacteria and titers were determined at 4 DPI. Pi-0 ACIP RNAi leaves were significantly more susceptible to Pst DC3000 ΔhrcU (Figure 5C). Consistent with these findings, accumulation of WRKY22 and WRKY29 mRNA was significantly reduced at 6 HPI in Pi-0 ACIP RNAi leaves compared to wild-type Pi-0 leaves inoculated with a high titer (2×108 cells/mL suspension) of Pst DC3000 ΔhrcU (Figure 5D). Taken together, these data suggest that a subset of the ACIP family (i.e. ACIP1, ACIP-L1, ACIP-L2, and ACIP-L3) collectively contribute to anti-bacterial immunity in Arabidopsis.
ACIP1 co-localizes with microtubules
To begin to address ACIP1's function, we examined ACIP1 protein localization in Arabidopsis seedlings and mature plants. We generated homozygous transgenic Pi-0 plants expressing a GFP-ACIP1 protein fusion under the control of the native ACIP1 promoter (i.e. PACIP1::GFP-ACIP1). Using confocal microscopy, we observed a low level of GFP-ACIP1 fluorescence in 4-day old etiolated seedlings and juvenile leaves. Little or no detectable fluorescence was observed in mature, senescing leaves. In hypocotyl epidermal cells, GFP-ACIP1 was predominantly found as punctae at the cell cortex. A portion of these punctae was aligned, forming transverse cable-like structures (Figure 6A). ACIP1's subcellular localization pattern partially resembled that of cytoskeletal structures. Unlike TaSRG, the ACIP-L4 ortholog, GFP-ACIP1 was not observed in the plant nucleus, indicating that ACIP1 localization is distinct from this predicted transcription factor [28].
Fig. 6. GFP-ACIP1 co-localizes with microtubules. 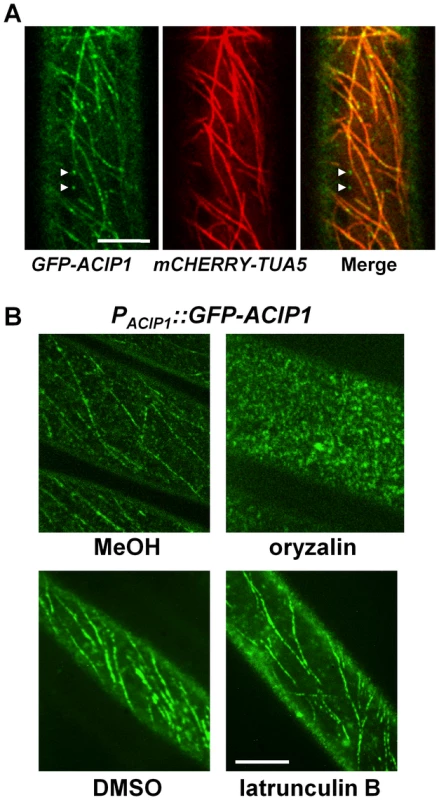
(A) Single plane images of periclinal surface of epidermal cells of 4-day old etiolated Pi-0 PACIP1::GFP-ACIP1/P35S::mCHERRY-TUA5 hypocotyl cells. Arrowheads show GFP-ACIP1 punctae that are not associated with microtubules. (B) Localization of GFP-ACIP1 in 4-day old etiolated Pi-0 PACIP1::GFP-ACIP1 hypocotyls treated with MeOH or MeOH +10 µM oryzalin (top panels), or DMSO or DMSO +1 µM latrunculin B (bottom panels). Cells were imaged using confocal microscopy. Bars = 10 µm. Next, we applied drugs to disrupt the cytoskeleton to determine if ACIP1 co-localizes with actin and/or microtubules. Treatment of the Pi-0 PACIP1::GFP-ACIP1 seedlings with oryzalin, a microtubule depolymerizing agent, disrupted the GFP-ACIP1 cable-like structures and caused the formation of numerous GFP-ACIP1 punctae throughout the cell (Figure 6B). By contrast, the actin depolymerizing agent latrunculin B did not appear to significantly disrupt these cables (Figure 6B).
To show ACIP co-localization with microtubules, we transformed the Pi-0 PACIP1::GFP-ACIP1 lines with P35S::mCHERRY-TUA5, a fluorescently tagged isoform of α-tubulin. A large portion of the GFP-ACIP1 punctae co-localized with mCHERRY-TUA5 microtubules (Figure 6A). Some of the cortical GFP-ACIP1 punctae were not associated with microtubules. Inspection of the literature revealed that ACIP1 was identified in the Arabidopsis proteome that co-purified with tubulin by affinity chromatography [27]. We did not detect a direct interaction between ACIP1 and TUA5 in a targeted yeast two-hybrid assay (Figure S5A,B). It is possible that ACIP1 association with tubulin might be indirect or via a weak electrostatic interaction. Or, ACIP1 might interact with another isoform of tubulin. Collectively, our findings indicate that GFP-ACIP1 signal forms punctae on the cell cortex and some of these punctae co-localize with the cortical microtubule network.
AvrBsT alters ACIP1 localization
We speculated that AvrBsT-binding to and acetylation of ACIP1 might interfere with ACIP1's stability and/or localization within plant leaves during pathogen infection. We did not detect by protein gel blot analysis any differences in endogenous ACIP1 protein abundance or mobility using extracts isolated from Pi-0 leaves infected with Pst DC3000 or Pst DC3000 AvrBsT (Figure S6B). However, we did notice that GFP-ACIP1 localization in 4-week old Pi-0 PACIP1::GFP-ACIP1 leaves was dramatically altered in response to both Pst DC3000 and Pst DC3000 AvrBsT (Figure 7). Unlike the signal in young hypocotyls, GFP-ACIP1 fluorescence at the cortex of epidermal cells in 4-week old leaves inoculated with the 1 mM MgCl2 mock control was diffuse and faint. This signal was difficult to capture in the image projection and varied among plants. By contrast, GFP-ACIP1 punctae were observed at or near the cell periphery of these cells (Figure 7A). Infection with Pst DC3000 for 6 h led to the formation of rod-shaped GFP-ACIP1 structures of various lengths (Figure 7B), which were difficult to detect in the 1 mM MgCl2 mock control (Figure 7A). The GFP-ACIP1 rods were also detected in response to Pst DC3000 ΔhrcU (Figure 7C), indicating that these structures are associated with PTI in a T3S effector-independent manner. Strikingly, Pst DC3000 AvrBsT infection for 6 h led to the formation of large, bright GFP-ACIP1 aggregates and fewer rod-like structures (Figure 7D). This localization pattern was dependent on AvrBsT's catalytic activity. Pst DC3000 AvrBsT (H154A) infection resulted in a GFP-ACIP1 signal (Figure 7E) similar to that induced by Pst DC3000 alone (Figure 7B). Similarly, Pst DC3000 AvrBsT(K282R) infection led to the formation of GFP-ACIP1 rods (Figure 7F), but not large aggregates.
Fig. 7. AvrBsT alters GFP-ACIP1's localization in a catalytic-dependent manner. 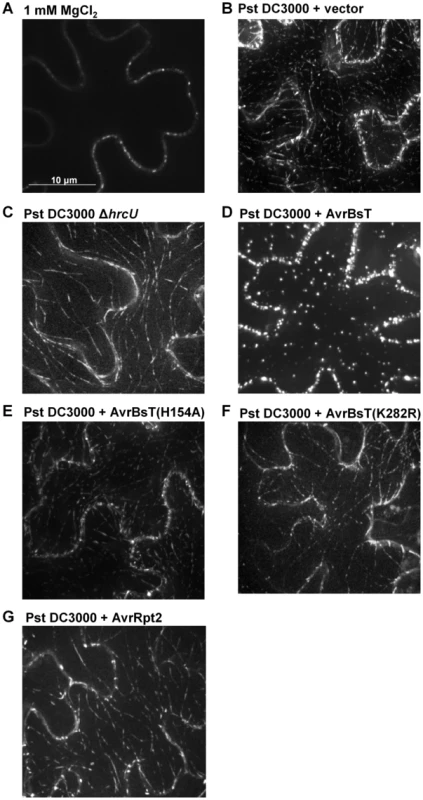
Pi-0 PACIP1::GFP-ACIP1 leaves were inoculated with (A) 1 mM MgCl2 or a 3×108 cells/mL suspension of (B) Pst DC3000 vector, (C) Pst DC3000 ΔhrcU, (D) Pst DC3000 AvrBsT, (E) Pst DC3000 AvrBsT(H154A), (F) Pst DC3000 AvrBsT(K282R), or (G) Pst DC3000 AvrRpt2. Spinning disk confocal images were recorded at 6–7 HPI. Bar = 10 µm. Similar results were obtained in more than 3 independent experiments. To determine if GFP-ACIP1 aggregates are generated specifically by AvrBsT and/or in response to PA production, we tested the phenotype of Pst DC3000 AvrRpt2. AvrRpt2 elicits ETI in Pi-0 leaves [23], which is dependent on PA production [23], [35]. Infection with Pst DC3000 AvrRpt2 induced the formation of GFP-ACIP1 rods (Figure 7G), similar to those formed in response to Pst DC3000 alone, Pst DC3000 AvrBsT(H154A), and Pst DC3000 AvrBsT(K282R) (Figure 7B,E,F). However, ACIP1 aggregates were not observed in response to Pst DC3000 AvrRpt2. Pi-0 PACIP1::GFP-ACIP1 leaves were inoculated with 50 µM PA and then imaged the leaves 1.5 hr later. Exogenous PA triggered the formation of several GFP-ACIP1 rods but only a few punctae (Figure S8C), whereas the mock control containing 0.2% DMSO did not (Figure S8D). These data suggest that PA exposure is sufficient to promote the formation of ACIP1 punctae and rods, but not the formation of ACIP1 aggregates. Moreover, they indicate that ACIP1 aggregate formation is a specific phenotype linked to AvrBsT acetyltransferase activity in planta.
Discussion
Pathogen-dependent acetylation of host targets has emerged as a key virulence strategy to alter eukaryotic defense responses. The study of several YopJ and YopJ-like effectors in animals and flies indicates that O-acetylation of Ser/Thr residues or Nε-acetylation of Lys residues in the activation loop of kinases in innate immune pathways directly interferes with residue phosphorylation or ATP binding, respectively [6]–[8], [18]. Both scenarios prevent the activation of kinases that are required to mediate innate immune signal transduction. In plants, the mechanism(s) by which YopJ-like effector acetylation of host substrates modulates immune signaling is less clear.
Based on this study, we propose that the YopJ-like effector AvrBsT acetylates Arabidopsis ACIP1. The role of ACIP1 in planta is not known; however, it is predicted to be a small α-helical protein [25]. ACIP1 emerged from an Arabidopsis screen looking for tubulin-binding proteins [27], suggesting that it might be a microtubule-associated protein. Our microscopy studies of Arabidopsis Pi-0 lines expressing GFP-ACIP1 revealed that ACIP1 is localized to punctae on the cell cortex and some of these punctae co-localize with the cortical microtubule network (Figure 6). These data are consistent with ACIP1 being a part of the tubulin proteome [27].
Importantly, we discovered that GFP-ACIP1 organization and accumulation changed significantly during bacterial infection (Figure 7). Numerous small GFP-ACIP1 punctae and rod-like structures formed throughout the cell in response to Pst DC3000 infection. These structures also formed during Pst DC3000 ΔhrcU infection, indicating that changes in ACIP1 localization are coincident with PTI. Strikingly, Pst DC3000 AvrBsT infection, but not Pst DC3000 AvrRpt2 infection, dramatically altered GFP-ACIP1 localization. AvrBsT activity triggered the accumulation of large GFP-ACIP1 aggregates throughout the plant cell. The aggregates did not appear to be aligned like those observed in leaves infected with Pst DC3000 or uninfected hypocotyl epidermal cells (Figure 6). Interestingly, a mutation (K282R) that disrupted AvrBsT's ability to acetylate ACIP1 in vitro (Figure 2B) also prevented the formation of GFP-ACIP1 aggregates (Figure 7) and activation of ETI during Pst DC3000 AvrBsT infection (Figure 3). Taken together, these data suggest the model that AvrBsT acetyltransferase activity in planta uniquely alters ACIP1's localization, which is linked to AvrBsT-dependent activation of ETI.
The nature of the large GFP-ACIP1 aggregates and their function during pathogen infection remains to be determined. Given the requirement for ACIP1 for both PTI (Figure 5) and the formation of ACIP1 punctae and rods during Pst DC3000 infection (Figure 7), we speculate that ACIP1 association with microtubules and/or the cell cortex is important for plant immunity. We also speculate that AvrBsT acetyltransferase activity either directly or indirectly alters ACIP1 association with microtubules. Association of ACIP1 with microtubules may play a direct role in microtubule organization, or it may be involved in microtubule-dependent processes such as vesicle and protein trafficking. Alternatively, ACIP1 may simply use microtubules to position itself and its interacting proteins at the cell cortex, where plant cells first encounter injected bacterial effectors. Our ACIP RNAi plants (silenced for ACIP1, ACIP-L1, ACIP-L2, and ACIP-L3, Figure S7) did not show cell shape or cell growth phenotypes, which are caused by microtubule cytoskeleton defects, suggesting that four ACIP family members do not regulate microtubule cytoskeleton structure. Future functional studies will test if ACIP1 and/or other isoforms expressed in leaves play a role in suppressing bacterial growth by regulating microtubule-dependent trafficking or by regulating other processes at the plasma membrane or cell cortex.
Notably, AvrBsT catalysis in Arabidopsis Pi-0 leaves leads to the accumulation of PA, a lipid signal associated with plant adaptation to biotic and abiotic stress [24]. Elevated PA levels in Pi-0 leaves inoculated with Pst DC3000 AvrBsT correlate with ETI [22], [23]. ACIP1 is required for AvrBsT-triggered ETI (Figure 4); however, the causal relationship between changes in PA production and ACIP1 localization in response to AvrBsT acetyltransferase activity is not clear. Furthermore, it is not known if PA is required to alter ACIP1 localization and/or function. Exogenous PA treatment (Figure S8C) or infection with Pst DC3000 AvrRpt2 (Figure 7G), which triggers a PA burst during ETI [35], induced the formation of small ACIP1 punctae and rods of various sizes, but large ACIP1 aggregates similar to those formed in response to AvrBsT-dependent catalysis (Figure 7D) were not observed. These data further highlight the specificity for AvrBsT in inducing ACIP1 aggregation during infection. They also suggest that a threshold concentration of PA or local production of PA relative to ACIP1 might be required to trigger the formation of large ACIP1 aggregates in planta.
PA is known to play a critical role in the regulation of cytoskeletal dynamics [36]. Recent data suggests that PA alters the microtubule network by directly binding to cytoskeletal components, including tubulin and the microtubule bundling protein MAP65-1 [37], [38]. Interestingly, elevated PA resulting from salt stress recruits Arabidopsis MAP65-1 to the membrane and enhances its ability to stabilize microtubules, which promotes cell survival [38]. How PA directly alters the microtubule network during ETI is not known. The link between PA, ACIP1, and the microtubule network during pathogen infection established in this study suggests that PA might regulate ACIP1 complex formation and/or association with microtubules.
Interestingly, the HopZ1a acetyltransferase was recently shown to disrupt plant cortical microtubule arrays and secretion during bacterial infection [10]. In this case, P. syringae HopZ1a infection led to reduced microtubule density, suggesting that HopZ1a acetylation in planta affects the stability or nucleation of microtubules. Acetylation of mammalian EB1, a microtubule-associated protein, which promotes microtubule assembly, was recently shown to compromise EB1 binding to other microtubule plus-end tracking proteins [39]. HopZ1a binds and acetylates tubulin in vitro. Whether or not HopZ1a modifies tubulin and/or affects microtubule properties (i.e. assembly, disassembly, and/or stability) during infection remains to be determined.
In terms of acetylation, our data suggests that AvrBsT trans-acetylation activity, not auto-acetylation activity, triggers ETI in Pi-0 leaves (Figure 3). Mutation of Lys 282 to Arg in AvrBsT, a conserved residue found in YopJ and YopJ-like effectors [9], did not affect AvrBsT's auto-acetyltransferase activity in vitro (Figure 2A), although it inhibited its ability to trans-acetylate ACIP1 (Figure 2B). We speculate that K282 is required for enzyme-substrate interactions, although acetyl-CoA docking or direct acetylation [9] is also possible. Importantly, Pst DC3000 expressing the AvrBsT(K282R) mutant failed to trigger ACIP1 aggregates (Figure 7) and elicit host resistance (Figure 3). These data suggest that acetylation is linked to changes in ACIP1 function and immunity. Whether or not acetylation of ACIP1 is directly linked to punctae formation, localization with microtubules, PA production and/or the activation of AvrBsT-triggered ETI awaits characterization of ACIP1's acetylation status in planta. Since acetylation can increase the electronegativity of proteins, it has the potential to disrupt ACIP1 interactions with the negatively charged microtubule lattice. Electrostatic interactions have been shown to play a significant role in microtubule binding of motor proteins and microtubule-associated proteins that often possess domains enriched in positively charged residues [40]–[42]. Future mapping of the ACIP1 microtubule-interaction domain in relation to residues acetylated by AvrBsT will allow us to test the functional significance of ACIP1 acetylation in planta.
A growing number of plant targets have been identified for YopJ-like effectors, questioning the specificity of these enzymes as acetyltransferases versus binding partners in immune complexes. Our work indicates that there is selectivity between AvrBsT and HopZ1a in vitro. AvrBsT acetylates ACIP1 whereas HopZ1a acetylates tubulin. In addition to ACIP1, AvrBsT has been recently shown to bind to arginine decarboxylase (ADC1), an enzyme proposed to mediate polyamine and γ-aminobutyric acid metabolism and impact cell death responses [43], and SNF-1 related kinase (SnRK1), a putative regulator of sugar metabolism [44]. Post-translational acetylation of these plant proteins has not yet been reported. The fact that a number of metabolic enzymes are normally regulated by acetylation warrants further investigation [45].
Similarly, HopZ1a appears to have multiple plant targets. In addition to tubulin, HopZ1a was shown to acetylate Arabidopsis ZED1, a pseudokinase required for HopZ1a-dependent ETI [17], and Arabidopsis jasmonate (JA) ZIM-domain proteins required to repress JA signaling during PTI [46]. HopZ1a was also shown to bind and destabilize an enzyme involved in isoflavonoid biosynthesis, 2-hydroxyisoflavanone dehydratase (HID1), by an unknown mechanism [47]. The diverse nature of these targets suggests that HopZ1a is a promiscuous enzyme capable of altering defense signaling at multiple nodes.
It is intriguing that HopZ2, the closest YopJ-like homolog to AvrBsT [48], was found to directly interact with Arabidopsis MLO2 in planta [49]. Arabidopsis mlo2-7 mutants are compromised for HopZ2-dependent virulence, further supporting the role of MLO2 as a negative regulator of immunity [49]. MLO2 is a plasma membrane protein of unknown function that interferes with vesicular trafficking mediated by the syntaxin PEN1 [50], [51]. It is too early to tell if there is a common theme for YopJ-like targets in plant cells. However, the identification of ACIP1, tubulin, and MLO2 as host targets suggests that some YopJ-like effectors might have undergone specialization to interfere with the trafficking function of the microtubule cytoskeleton in infected cells.
Does AvrBsT target ACIP1 or an ACIP1 complex to suppress immunity? This question has been difficult to answer because we have yet to detect an AvrBsT virulence phenotype in Arabidopsis during bacterial infection. This is not so surprising given the potential functional redundancy between AvrBsT and the suite of T3S effectors in Pst DC3000. Overexpression of AvrBsT in transgenic Arabidopsis lines however was recently shown to enhance susceptibility to Pst DC3000 [52]. In solanaceous plants, AvrBsT is known to suppress PTI in tomato [53] and ETI in pepper [44] during Xanthomonas infection. The study of the ACIP1 ortholog in tomato may provide insight to AvrBsT virulence, by specifically addressing how AvrBsT acetyltransferase activity interferes with ACIP1's function during PTI and/or ETI.
In summary, the study of AvrBsT-triggered defense responses in Arabidopsis Pi-0 plants has led to the identification of ACIP1, a member of a new protein family required for PTI and ETI. We demonstrate that the expression of four Arabidopsis ACIP isoforms (ACIP1, ACIP-L1, ACIP-L2, and ACIP-L3) is required for proper execution of PTI in response to Pst DC3000 (Figure 5) and ETI in response to Pst DC3000 expressing AvrBsT, AvrB or AvrRpt2 (Figure 4, S8). In addition, we show that AvrBsT is an acetyltransferase and provide evidence that acetyltransferase activity plays an important role in altering ACIP1 localization within the plant cell during infection and the activation of ETI. This study highlights an important link between ACIP1 and the microtubule network during plant defense.
Materials and Methods
Bacterial strains and growth
Escherichia coli DH5 alpha and Agrobacterium tumefaciens strain GV3101 were grown on Luria agar medium at 37 and 28°C, respectively. Pseudomonas syringae pathovar tomato (Pst) DC3000 strains were grown on nutrient yeast glycerol agar (NYGA) [54] at 28°C. E. coli antibiotic selection was 100 µg/mL carbenicillin and/or 50 µg/mL kanamycin. A. tumefaciens antibiotic selection was 100 µg/mL rifampicin, 50 µg/mL kanamycin, and/or 30 µg/mL gentamicin. Pst antibiotic selection was 100 µg/mL rifampicin, and/or 50 µg/mL kanamycin.
Plant lines and growth
Arabidopsis thaliana Col-0 and Pi-0 ecotypes were grown in growth chambers (22°C, 60% RH, 125 µE/m2/s fluorescent illumination) on an 8-h light/16-h dark cycle. Plants were transformed using the floral dip method [55].
Molecular constructions
Standard DNA cloning methods [56], PCR, and Gateway technology (Invitrogen) were used for plasmid construction. All primer sequences are listed in Table S1. For GST-AvrBsT, avrBsT (wild type, H154A, C222A, and K282R) was amplified by PCR, cloned into pJET1.2, and then sub-cloned into pGEX-5X-3 using BamHI and XhoI. For Gateway constructions, amplified PCR products (i.e. avrBsT, hopZ1a, hopZ1b, hopZ2, hopZ3, ACIP1, ACIP-like genes (ACIP-L1 to ACIP-L6), and TUA5) were cloned into pCR8 to create donor plasmids. The respective donor plasmids were recombined into: 1) pGADT7 to create AD-gene fusions and pGBKT7 or pXDGATcy86 to create BD-gene fusions for two-hybrid analysis; 2) pDEST15 for GST-fusions; and/or 3) pDEST17 for His6-fusions. For avrBsT mutagenesis, QuikChange Site –Directed Mutagenesis kit (Stratagene) was performed with pCR8(avrBsT) and PfuUltra II Fusion HS DNA polymerase (Agilent).
Yeast two-hybrid screen
Yeast strain AH109 carrying pXDGATcy86(avrBsT) was transformed with pAD-GAL4-2.1 containing the Horwitz and MA cDNA library isolated from A. thaliana inflorescence meristem, floral meristem, and floral buds (obtained from TAIR). Approximately 7 million transformants were screened and interaction with At3g09980 cDNA was confirmed.
Yeast protein extraction
Yeast cells were resuspended in lysis buffer (1.85 M NaOH and 7% 2-mercaptoethanol) and then proteins were precipitated in 10% trichloroacetic acid. Protein pellets were washed in 1 M Tris and then resuspended in 8 M urea sample buffer.
Protein gel blot analysis
Protein was extracted from plant cells as described [34], separated by SDS-PAGE, transferred to nitrocellulose, and then detected by ECL or ECL plus (GE Healthcare) using anti-ACIP1, anti-HA (Covance), anti-Myc (Covance), anti-His (Qiagen), or anti-GST (Santa Cruz) sera and horseradish peroxidase conjugated secondary antibodies (Bio-Rad). Membranes were stained with Ponceau S to control for loading.
ACIP1 antibody production
Recombinant His6-ACIP1 was expressed in E. coli BL21 tRNA cells and purified using Ni-NTA agarose under denaturing conditions following manufacturer's protocol (Qiagen). Polyclonal antisera were raised in rabbits using the purified His-ACIP1 protein (Covance).
In vitro GST pull-down assay
GST or GST-AvrBsT were expressed in E. coli BL21-CodonPlus(DE3) cells (Stratagene). Cells were lysed in buffer (1X PBS, pH 8, 1% Triton X-100, 0.1% 2-mercaptoethanol, and 1 mM phenylmethylsulfonyl fluoride (Sigma Aldrich)) with a sonicator (Branson). GST and GST-AvrBsT supernatants were incubated with 30 µL of pre-equilibrated Glutathione Sepharose 4B (GE Healthcare) for 1 h at 4°C with rotation. Sepharose beads were recovered by centrifugation and then washed with buffer for 5 min at 4°C with rotation. GST or GST-AvrBsT (WT, C222A, or K282R) bound beads were incubated with soluble E. coli lysates containing His6-ACIP1 for 2 h at 4°C with rotation. The beads were washed with buffer (50 mM TrisHCl, pH 7.5, 150 mM NaCl, 10 mM MgCl2, 0.1% Triton X-100, and 0.1% 2-mercaptoethanol) three times. Protein bound to the beads was separated by SDS-PAGE and analyzed by immunoblot analysis. Anti-GST and anti-His sera were used to detect GST-AvrBsT and His6-ACIP1.
Protein acetylation assay
Purified recombinant GST-tagged proteins (1 µg each) were incubated with 100 nM inositol hexakisphosphate (IP6) (Santa Cruz), 0.4 µCi 14C-acetyl-CoA (Perkin Elmer) in 50 mM TrisHCl pH 8 and 1 mM DTT for 30 min at RT. Urea sample buffer was added to stop the reactions. Proteins assayed included: GST, GST-AvrBsT, GST-AvrBsT(C222A), GST-AvrBsT(H154A), GST-AvrBsT(K282R), GST-HopZ1a, and GST-ACIP1. Proteins were separated in a 10% SDS-PAGE gel, stained with Coomassie blue, transferred to blotting paper, dried, treated with EN3HANCE (Perkin Elmer), and then exposed to film for 2–3 weeks at 80°C.
ACIP silencing construct and transgenic Arabidopsis plants
A 365 bp region of ACIP1 was PCR amplified using primer set JG616/JG617, and the product was cloned into pKANNIBAL to create pKANNIBAL(hp-ACIP). The NotI fragment was then subcloned into pART27 [57], creating pAR27(hp-ACIP). The resulting plasmid was then transformed into A. thaliana ecotypes Col-0 and Pi-0. Transformants were analyzed by quantitative RT-PCR to measure ACIP isoform mRNA levels using primer sets listed in Table S1.
Bacterial HR and growth assays
Fully expanded leaves of 4 - to 5-week-old plants were used for bacterial inoculations. A suspension of bacterial cells (Pst DC3000, Pst DC3000 AvrBsT, Pst DC3000 AvrB, Pst DC3000 AvrRpt2, or Pst DC3000 ΔhrcU; 3×108 cells/mL for HR and 1×105 cells/mL for growth curves) was infiltrated into the extracellular space of fully expanded leaves using a 1-mL syringe. For HR, plants were incubated at RT under lights and phenotypes were recorded 9–12 HPI. For growth curves, plants were incubated at high humidity in a growth chamber for 4 d. Leaf tissue was collected at 0–4 DPI, ground in 1 mM MgCl2, diluted and then plated on NYGA plates containing appropriate antibiotics and cycloheximide (50 µg/mL) in triplicate to determine bacterial load. Four plants were used and the experiment was repeated at least three times. The average bacterial titer ± SD is reported.
Electrolyte leakage assay
Three fully expanded leaves of 4 - to 5-week-old plants (n = 4) were inoculated with a 3×108 cells/mL suspension of Pst DC3000 (vector) or Pst DC3000 (AvrBsT). Ten HPI, three leaf discs (10 mm diameter) per plant were floated in 20 mL of water in petri dishes for 5 min and then transferred to a test tube containing 3 mL of water. Tubes were incubated for 1 h at RT with shaking. Conductivity of the solution was measured with an EC meter (Spectrum Technologies) before and after boiling for 30 min [58]. Percent electrolyte leakage was calculated as conductivity before boiling/conductivity after boiling ×100. Assay was repeated at least three times.
Oxidative burst assay
Three leaf discs (5 mm diameter) from the youngest fully expanded leaves of a 4-week-old plant (n = 9–18) were incubated in water in a 96-well plate (one leaf disc per well) for 24 h. To measure ROS, leaf discs were treated with ± flg22 (100 nM) in 10 µg/mL horseradish peroxidase and 100 µM Luminol (Sigma), and then luminescence was immediately measured with a 1420 Multilabel Counter (PerkinElmer) [32]. Relative luminescence units (RLU) are reported. Assay was repeated at least three times.
RNA isolation and quantitative RT-PCR
Total RNA was isolated from uninfected or infected leaves using Trizol reagent (Invitrogen) according to manufacturer's instructions. For infection, leaves were inoculated with 1 mM MgCl2 or bacterial strains (2×108 cells/mL in 1 mM MgCl2) and then one leaf from three plants was harvested, pooled, and total RNA was extracted. 2.5 µg of RNA were used for cDNA synthesis. Quantitative RT-PCR was performed using the cDNA and gene-specific primers (Table S1). Each cDNA was amplified by real-time PCR using SensiFAST SYBR Kit (Bioline) and the MJ Opticon 2 instrument (Bio-Rad). UBQ5 or ACTIN8 expression was used to normalize the expression value in each sample and relative expression values were determined against the average value of buffer or bacterially infected sample using the comparative Ct method (2−ΔΔCt).
Microscopy
To monitor ACIP1 protein expression and localization, the promoter region (1.5-kb upstream of start) was fused with GFP-ACIP1 in the backbone of pMDC43 to create pMDC43(PACIP1::GFP-ACIP1). The resulting plasmid was transformed into A. thaliana Pi-0 plants. Transgenic PACIP1::GFP-ACIP1 lines were then transformed with P35S::mCHERRY-HA-TUA5. This plasmid was construct by modifying pEarleygate 104 [59]. YFP was substituted with mCHERRY and Arabidopsis TUA5 genomic coding region was inserted after mCHERRY. Localization of GFP-ACIP1 and mCHERRY-TUA5 in 5-day old dark grown hypocotyls was determined using a Leica TCS SP5 confocal microscope (Leica Microsystems) with Leica LAS AF software and a Leica spinning disc confocal microscope with the Yokogawa CSUX-M1 confocal scanner. Seedlings were treated with 10 µM oryzalin in MeOH for 8 hr at RT or 1 µM latrunculin B in DMSO for 4 hr at RT and then imaged. For infection, Pi-0 PACIP1::GFP-ACIP1 leaves were inoculated with 1 mM MgCl2 or bacterial strains (3×108 cells/mL in 1 mM MgCl2) for 6 h. For exogenous PA treatment, Pi-0 PACIP1::GFP-ACIP1 leaves were inoculated with 50 µM PA in 0.2% DMSO or 0.2% DMSO. Images were analyzed using ImageJ [60].
Accession numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: At3G09980 (ACIP1), At5G03660 (ACIP-L1), At2G36410 (ACIP-L2), At3G52920 (ACIP-L3), At2G27740 (ACIP-L4), At3G52900 (ACIP-L5) and At2G36355 (ACIP-L6).
Supporting Information
Zdroje
1. ButtnerD (2012) Protein export according to schedule: architecture, assembly, and regulation of type III secretion systems from plant - and animal-pathogenic bacteria. Microbiol Mol Biol Rev 76 : 262–310.
2. LewisJD, LeeA, MaW, ZhouH, GuttmanDS, et al. (2011) The YopJ superfamily in plant-associated bacteria. Mol Plant Pathol 12 : 928–937.
3. MonackDM, MecsasJ, GhoriN, FalkowS (1997) Yersinia signals macrophages to undergo apoptosis and YopJ is necessary for this cell death. Proc Natl Acad Sci U S A 94 : 10385–10390.
4. Rawlings ND, Salvesen G (2013) Handbook of proteolytic enzymes. 3rd ed. London ; Boston: Academic Press.
5. OrthK, XuZ, MudgettMB, BaoZQ, PalmerLE, et al. (2000) Disruption of signaling by Yersinia effector YopJ, a ubiquitin-like protein protease. Science 290 : 1594–1597.
6. MukherjeeS, KeitanyG, LiY, WangY, BallHL, et al. (2006) Yersinia YopJ acetylates and inhibits kinase activation by blocking phosphorylation. Science 312 : 1211–1214.
7. TroskyJE, LiY, MukherjeeS, KeitanyG, BallH, et al. (2007) VopA inhibits ATP binding by acetylating the catalytic loop of MAPK kinases. J Biol Chem 282 : 34299–34305.
8. JonesRM, WuH, WentworthC, LuoL, Collier-HyamsL, et al. (2008) Salmonella AvrA Coordinates Suppression of Host Immune and Apoptotic Defenses via JNK Pathway Blockade. Cell host & microbe 3 : 233–244.
9. TassetC, BernouxM, JauneauA, PouzetC, BriereC, et al. (2010) Autoacetylation of the Ralstonia solanacearum effector PopP2 targets a lysine residue essential for RRS1-R-mediated immunity in Arabidopsis. PLoS pathogens 6: e1001202.
10. LeeAH, HurleyB, FelsensteinerC, YeaC, CkurshumovaW, et al. (2012) A bacterial acetyltransferase destroys plant microtubule networks and blocks secretion. PLoS pathogens 8: e1002523.
11. SharlachM, DahlbeckD, LiuL, ChiuJ, Jimenez-GomezJM, et al. (2013) Fine genetic mapping of RXopJ4, a bacterial spot disease resistance locus from Solanum pennellii LA716. Theor Appl Genet 126 : 601–609.
12. WhalenMC, WangJF, CarlandFM, HeiskellME, DahlbeckD, et al. (1993) Avirulence gene avrRxv from Xanthomonas campestris pv. vesicatoria specifies resistance on tomato line Hawaii 7998. Mol Plant Microbe Interact 6 : 616–627.
13. ZhouH, MorganRL, GuttmanDS, MaW (2009) Allelic variants of the Pseudomonas syringae type III effector HopZ1 are differentially recognized by plant resistance systems. Mol Plant Microbe Interact 22 : 176–189.
14. DeslandesL, OlivierJ, TheulieresF, HirschJ, FengDX, et al. (2002) Resistance to Ralstonia solanacearum in Arabidopsis thaliana is conferred by the recessive RRS1-R gene, a member of a novel family of resistance genes. Proc Natl Acad Sci U S A 99 : 2404–2409.
15. LewisJD, WuR, GuttmanDS, DesveauxD (2010) Allele-specific virulence attenuation of the Pseudomonas syringae HopZ1a type III effector via the Arabidopsis ZAR1 resistance protein. PLoS genetics 6: e1000894.
16. DeslandesL, OlivierJ, PeetersN, FengDX, KhounlothamM, et al. (2003) Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc Natl Acad Sci U S A 100 : 8024–8029.
17. LewisJD, LeeAH, HassanJA, WanJ, HurleyB, et al. (2013) The Arabidopsis ZED1 pseudokinase is required for ZAR1-mediated immunity induced by the Pseudomonas syringae type III effector HopZ1a. Proc Natl Acad Sci U S A 110 : 18722–18727.
18. MittalR, Peak-ChewSY, McMahonHT (2006) Acetylation of MEK2 and I kappa B kinase (IKK) activation loop residues by YopJ inhibits signaling. Proc Natl Acad Sci U S A 103 : 18574–18579.
19. MeinzerU, BarreauF, Esmiol-WelterlinS, JungC, VillardC, et al. (2012) Yersinia pseudotuberculosis effector YopJ subverts the Nod2/RICK/TAK1 pathway and activates caspase-1 to induce intestinal barrier dysfunction. Cell host & microbe 11 : 337–351.
20. AkellaJS, WlogaD, KimJ, StarostinaNG, Lyons-AbbottS, et al. (2010) MEC-17 is an alpha-tubulin acetyltransferase. Nature 467 : 218–222.
21. ShidaT, CuevaJG, XuZ, GoodmanMB, NachuryMV (2010) The major alpha-tubulin K40 acetyltransferase alphaTAT1 promotes rapid ciliogenesis and efficient mechanosensation. Proc Natl Acad Sci U S A 107 : 21517–21522.
22. CunnacS, WilsonA, NuwerJ, KirikA, BaranageG, et al. (2007) A Conserved Carboxylesterase Is a SUPPRESSOR OF AVRBST-ELICITED RESISTANCE in Arabidopsis. Plant Cell 19 : 688–705.
23. KirikA, MudgettMB (2009) SOBER1 phospholipase activity suppresses phosphatidic acid accumulation and plant immunity in response to bacterial effector AvrBsT. Proc Natl Acad Sci U S A 106 : 20532–20537.
24. TesterinkC, MunnikT (2011) Molecular, cellular, and physiological responses to phosphatidic acid formation in plants. J Exp Bot 62 : 2349–2361.
25. KelleyLA, SternbergMJ (2009) Protein structure prediction on the Web: a case study using the Phyre server. Nature protocols 4 : 363–371.
26. PuntaM, CoggillPC, EberhardtRY, MistryJ, TateJ, et al. (2012) The Pfam protein families database. Nucleic Acids Res 40: D290–301.
27. ChuongSD, GoodAG, TaylorGJ, FreemanMC, MoorheadGB, et al. (2004) Large-scale identification of tubulin-binding proteins provides insight on subcellular trafficking, metabolic channeling, and signaling in plant cells. Mol Cell Proteomics 3 : 970–983.
28. HeX, HouX, ShenY, HuangZ (2011) TaSRG, a wheat transcription factor, significantly affects salt tolerance in transgenic rice and Arabidopsis. FEBS Lett 585 : 1231–1237.
29. MittalR, Peak-ChewSY, SadeRS, VallisY, McMahonHT (2010) The acetyltransferase activity of the bacterial toxin YopJ of Yersinia is activated by eukaryotic host cell inositol hexakisphosphate. J Biol Chem 285 : 19927–19934.
30. MackeyD, BelkhadirY, AlonsoJM, EckerJR, DanglJL (2003) Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 112 : 379–389.
31. AxtellMJ, StaskawiczBJ (2003) Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell 112 : 369–377.
32. Gomez-GomezL, FelixG, BollerT (1999) A single locus determines sensitivity to bacterial flagellin in Arabidopsis thaliana. Plant J 18 : 277–284.
33. AsaiT, TenaG, PlotnikovaJ, WillmannMR, ChiuWL, et al. (2002) MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415 : 977–983.
34. MudgettMB, StaskawiczBJ (1999) Characterization of the Pseudomonas syringae pv. tomato AvrRpt2 protein: demonstration of secretion and processing during bacterial pathogenesis. Mol Microbiol 32 : 927–941.
35. AnderssonMX, KourtchenkoO, DanglJL, MackeyD, EllerstromM (2006) Phospholipase-dependent signalling during the AvrRpm1 - and AvrRpt2-induced disease resistance responses in Arabidopsis thaliana. Plant J 47 : 947–959.
36. PleskotR, LiJ, ZarskyV, PotockyM, StaigerCJ (2013) Regulation of cytoskeletal dynamics by phospholipase D and phosphatidic acid. Trends Plant Sci 18 : 496–504.
37. TesterinkC, DekkerHL, LimZY, JohnsMK, HolmesAB, et al. (2004) Isolation and identification of phosphatidic acid targets from plants. Plant J 39 : 527–536.
38. ZhangQ, LinF, MaoT, NieJ, YanM, et al. (2012) Phosphatidic acid regulates microtubule organization by interacting with MAP65-1 in response to salt stress in Arabidopsis. Plant Cell 24 : 4555–4576.
39. XiaP, WangZ, LiuX, WuB, WangJ, et al. (2012) EB1 acetylation by P300/CBP-associated factor (PCAF) ensures accurate kinetochore-microtubule interactions in mitosis. Proc Natl Acad Sci U S A 109 : 16564–16569.
40. GrantBJ, GheorgheDM, ZhengW, AlonsoM, HuberG, et al. (2011) Electrostatically biased binding of kinesin to microtubules. PLoS biology 9: e1001207.
41. ZimmermannD, Abdel MotaalB, Voith von VoithenbergL, SchliwaM, OktenZ (2011) Diffusion of myosin V on microtubules: a fine-tuned interaction for which E-hooks are dispensable. PLoS One 6: e25473.
42. CurrieJD, StewmanS, SchimizziG, SlepKC, MaA, et al. (2011) The microtubule lattice and plus-end association of Drosophila Mini spindles is spatially regulated to fine-tune microtubule dynamics. Mol Biol Cell 22 : 4343–4361.
43. KimNH, KimBS, HwangBK (2013) Pepper Arginine Decarboxylase Is Required for Polyamine and gamma-Aminobutyric Acid Signaling in Cell Death and Defense Response. Plant Physiol 162 : 2067–2083.
44. SzczesnyR, ButtnerD, EscolarL, SchulzeS, SeiferthA, et al. (2010) Suppression of the AvrBs1-specific hypersensitive response by the YopJ effector homolog AvrBsT from Xanthomonas depends on a SNF1-related kinase. New Phytol 187 : 1058–1074.
45. XingS, PoirierY (2012) The protein acetylome and the regulation of metabolism. Trends Plant Sci 17 : 423–430.
46. JiangS, YaoJ, MaKW, ZhouH, SongJ, et al. (2013) Bacterial effector activates jasmonate signaling by directly targeting JAZ transcriptional repressors. PLoS pathogens 9: e1003715.
47. ZhouH, LinJ, JohnsonA, MorganRL, ZhongW, et al. (2011) Pseudomonas syringae type III effector HopZ1 targets a host enzyme to suppress isoflavone biosynthesis and promote infection in soybean. Cell host & microbe 9 : 177–186.
48. MaW, DongFF, StavrinidesJ, GuttmanDS (2006) Type III effector diversification via both pathoadaptation and horizontal transfer in response to a coevolutionary arms race. PLoS genetics 2: e209.
49. LewisJD, WanJ, FordR, GongY, FungP, et al. (2012) Quantitative Interactor Screening with next-generation Sequencing (QIS-Seq) identifies Arabidopsis thaliana MLO2 as a target of the Pseudomonas syringae type III effector HopZ2. BMC Genomics 13 : 8.
50. AssaadFF, QiuJL, YoungsH, EhrhardtD, ZimmerliL, et al. (2004) The PEN1 syntaxin defines a novel cellular compartment upon fungal attack and is required for the timely assembly of papillae. Mol Biol Cell 15 : 5118–5129.
51. CollinsNC, Thordal-ChristensenH, LipkaV, BauS, KombrinkE, et al. (2003) SNARE-protein-mediated disease resistance at the plant cell wall. Nature 425 : 973–977.
52. HwangIS, KimNH, ChoiDS, HwangBK (2012) Overexpression of Xanthomonas campestris pv. vesicatoria effector AvrBsT in Arabidopsis triggers plant cell death, disease and defense responses. Planta 236 : 1191–1204.
53. KimNH, ChoiHW, HwangBK (2010) Xanthomonas campestris pv. vesicatoria effector AvrBsT induces cell death in pepper, but suppresses defense responses in tomato. Mol Plant Microbe Interact 23 : 1069–1082.
54. TurnerP, BarberC, DanielsM (1984) Behaviour of the transposons Tn5 and Tn7 in Xanthomonas campestris pv. campestris. Mol Gen Genet 195 : 101–107.
55. BentAF (2000) Arabidopsis in Planta Transformation. Uses, Mechanisms, and Prospects for Transformation of Other Species. Plant Physiology (Rockville) 124 : 1540–1547.
56. Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: A laboratory manual. Cold Spring Harbor: Cold Spring Harbor Laboratory Press.
57. GleaveAP (1992) A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol Biol 20 : 1203–1207.
58. RizhskyL, ShulaevV, MittlerR (2004) Measuring programmed cell death in plants. Methods Mol Biol 282 : 179–189.
59. EarleyKW, HaagJR, PontesO, OpperK, JuehneT, et al. (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45 : 616–629.
60. SchneiderCA, RasbandWS, EliceiriKW (2012) NIH Image to ImageJ: 25 years of image analysis. Nature methods 9 : 671–675.
61. TamuraK, DudleyJ, NeiM, KumarS (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) Software Version 4.0. Mol Biol Evol 24 : 1596–1599.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Structure of the Membrane Anchor of Pestivirus Glycoprotein E, a Long Tilted Amphipathic HelixČlánek Iron Acquisition in : The Roles of IlsA and Bacillibactin in Exogenous Ferritin Iron MobilizationČlánek Viral MicroRNA Effects on Pathogenesis of Polyomavirus SV40 Infections in Syrian Golden HamstersČlánek Genome-Wide RNAi Screen Identifies Broadly-Acting Host Factors That Inhibit Arbovirus Infection
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2014 Číslo 2- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Viral Enhancer Mimicry of Host Innate-Immune Promoters
- The Epstein-Barr Virus-Encoded MicroRNA MiR-BART9 Promotes Tumor Metastasis by Targeting E-Cadherin in Nasopharyngeal Carcinoma
- Implication of PMLIV in Both Intrinsic and Innate Immunity
- The Consequences of Reconfiguring the Ambisense S Genome Segment of Rift Valley Fever Virus on Viral Replication in Mammalian and Mosquito Cells and for Genome Packaging
- Substrate-Induced Unfolding of Protein Disulfide Isomerase Displaces the Cholera Toxin A1 Subunit from Its Holotoxin
- Male-Killing Induces Sex-Specific Cell Death via Host Apoptotic Pathway
- Highly Active Antiretroviral Therapies Are Effective against HIV-1 Cell-to-Cell Transmission
- The microRNAs in an Ancient Protist Repress the Variant-Specific Surface Protein Expression by Targeting the Entire Coding Sequence
- Transmission-Blocking Antibodies against Mosquito C-Type Lectins for Dengue Prevention
- Type III Secretion Protein MxiI Is Recognized by Naip2 to Induce Nlrc4 Inflammasome Activation Independently of Pkcδ
- Lundep, a Sand Fly Salivary Endonuclease Increases Parasite Survival in Neutrophils and Inhibits XIIa Contact Activation in Human Plasma
- Induction of Type I Interferon Signaling Determines the Relative Pathogenicity of Strains
- Structure of the Membrane Anchor of Pestivirus Glycoprotein E, a Long Tilted Amphipathic Helix
- Foxp3 Regulatory T Cells Delay Expulsion of Intestinal Nematodes by Suppression of IL-9-Driven Mast Cell Activation in BALB/c but Not in C57BL/6 Mice
- Iron Acquisition in : The Roles of IlsA and Bacillibactin in Exogenous Ferritin Iron Mobilization
- MicroRNA Editing Facilitates Immune Elimination of HCMV Infected Cells
- Reversible Silencing of Cytomegalovirus Genomes by Type I Interferon Governs Virus Latency
- Identification of Host-Targeted Small Molecules That Restrict Intracellular Growth
- A Cyclophilin Homology Domain-Independent Role for Nup358 in HIV-1 Infection
- Engagement of NKG2D on Bystander Memory CD8 T Cells Promotes Increased Immunopathology following Infection
- Suppression of RNA Silencing by a Plant DNA Virus Satellite Requires a Host Calmodulin-Like Protein to Repress Expression
- CIB1 Synergizes with EphrinA2 to Regulate Kaposi's Sarcoma-Associated Herpesvirus Macropinocytic Entry in Human Microvascular Dermal Endothelial Cells
- A Gammaherpesvirus Bcl-2 Ortholog Blocks B Cell Receptor-Mediated Apoptosis and Promotes the Survival of Developing B Cells
- Metabolic Reprogramming during Purine Stress in the Protozoan Pathogen
- The Post-transcriptional Regulator / Activates T3SS by Stabilizing the 5′ UTR of , the Master Regulator of Genes, in
- Tailored Immune Responses: Novel Effector Helper T Cell Subsets in Protective Immunity
- AvrBsT Acetylates ACIP1, a Protein that Associates with Microtubules and Is Required for Immunity
- Epstein-Barr Virus Large Tegument Protein BPLF1 Contributes to Innate Immune Evasion through Interference with Toll-Like Receptor Signaling
- The Major Cellular Sterol Regulatory Pathway Is Required for Andes Virus Infection
- Insights into the Initiation of JC Virus DNA Replication Derived from the Crystal Structure of the T-Antigen Origin Binding Domain
- Domain Shuffling in a Sensor Protein Contributed to the Evolution of Insect Pathogenicity in Plant-Beneficial
- Lectin-Like Bacteriocins from spp. Utilise D-Rhamnose Containing Lipopolysaccharide as a Cellular Receptor
- A Compositional Look at the Human Gastrointestinal Microbiome and Immune Activation Parameters in HIV Infected Subjects
- Exploits Asparagine to Assimilate Nitrogen and Resist Acid Stress during Infection
- Interleukin-33 Increases Antibacterial Defense by Activation of Inducible Nitric Oxide Synthase in Skin
- Protective Vaccination against Papillomavirus-Induced Skin Tumors under Immunocompetent and Immunosuppressive Conditions: A Preclinical Study Using a Natural Outbred Animal Model
- Gem-Induced Cytoskeleton Remodeling Increases Cellular Migration of HTLV-1-Infected Cells, Formation of Infected-to-Target T-Cell Conjugates and Viral Transmission
- Viral MicroRNA Effects on Pathogenesis of Polyomavirus SV40 Infections in Syrian Golden Hamsters
- Genome-Wide RNAi Screen Identifies Broadly-Acting Host Factors That Inhibit Arbovirus Infection
- Inflammatory Monocytes Orchestrate Innate Antifungal Immunity in the Lung
- Quantitative and Qualitative Deficits in Neonatal Lung-Migratory Dendritic Cells Impact the Generation of the CD8+ T Cell Response
- Human Genome-Wide RNAi Screen Identifies an Essential Role for Inositol Pyrophosphates in Type-I Interferon Response
- The Master Regulator of the Cellular Stress Response (HSF1) Is Critical for Orthopoxvirus Infection
- Code-Assisted Discovery of TAL Effector Targets in Bacterial Leaf Streak of Rice Reveals Contrast with Bacterial Blight and a Novel Susceptibility Gene
- Competitive and Cooperative Interactions Mediate RNA Transfer from Herpesvirus Saimiri ORF57 to the Mammalian Export Adaptor ALYREF
- The Type III Secretion Chaperone Slc1 Engages Multiple Early Effectors, Including TepP, a Tyrosine-phosphorylated Protein Required for the Recruitment of CrkI-II to Nascent Inclusions and Innate Immune Signaling
- Yeasts: How Many Species Infect Humans and Animals?
- Clustering of Pattern Recognition Receptors for Fungal Detection
- Distinct Antiviral Responses in Pluripotent versus Differentiated Cells
- Igniting the Fire: Virulence Factors in the Pathogenesis of Sepsis
- Inactivation of the Host Lipin Gene Accelerates RNA Virus Replication through Viral Exploitation of the Expanded Endoplasmic Reticulum Membrane
- Inducible Deletion of CD28 Prior to Secondary Infection Impairs Worm Expulsion and Recall of Protective Memory CD4 T Cell Responses
- Clonal Expansion during Infection Dynamics Reveals the Effect of Antibiotic Intervention
- The Secreted Triose Phosphate Isomerase of Is Required to Sustain Microfilaria Production
- Unifying Viral Genetics and Human Transportation Data to Predict the Global Transmission Dynamics of Human Influenza H3N2
- ‘Death and Axes’: Unexpected Ca Entry Phenologs Predict New Anti-schistosomal Agents
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Reversible Silencing of Cytomegalovirus Genomes by Type I Interferon Governs Virus Latency
- Implication of PMLIV in Both Intrinsic and Innate Immunity
- Transmission-Blocking Antibodies against Mosquito C-Type Lectins for Dengue Prevention
- Lundep, a Sand Fly Salivary Endonuclease Increases Parasite Survival in Neutrophils and Inhibits XIIa Contact Activation in Human Plasma
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



