-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaDomain Shuffling in a Sensor Protein Contributed to the Evolution of Insect Pathogenicity in Plant-Beneficial
Pseudomonas protegens is a biocontrol rhizobacterium with a plant-beneficial and an insect pathogenic lifestyle, but it is not understood how the organism switches between the two states. Here, we focus on understanding the function and possible evolution of a molecular sensor that enables P. protegens to detect the insect environment and produce a potent insecticidal toxin specifically during insect infection but not on roots. By using quantitative single cell microscopy and mutant analysis, we provide evidence that the sensor histidine kinase FitF is a key regulator of insecticidal toxin production. Our experimental data and bioinformatic analyses indicate that FitF shares a sensing domain with DctB, a histidine kinase regulating carbon uptake in Proteobacteria. This suggested that FitF has acquired its specificity through domain shuffling from a common ancestor. We constructed a chimeric DctB-FitF protein and showed that it is indeed functional in regulating toxin expression in P. protegens. The shuffling event and subsequent adaptive modifications of the recruited sensor domain were critical for the microorganism to express its potent insect toxin in the observed host-specific manner. Inhibition of the FitF sensor during root colonization could explain the mechanism by which P. protegens differentiates between the plant and insect host. Our study establishes FitF of P. protegens as a prime model for molecular evolution of sensor proteins and bacterial pathogenicity.
Published in the journal: . PLoS Pathog 10(2): e32767. doi:10.1371/journal.ppat.1003964
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1003964Summary
Pseudomonas protegens is a biocontrol rhizobacterium with a plant-beneficial and an insect pathogenic lifestyle, but it is not understood how the organism switches between the two states. Here, we focus on understanding the function and possible evolution of a molecular sensor that enables P. protegens to detect the insect environment and produce a potent insecticidal toxin specifically during insect infection but not on roots. By using quantitative single cell microscopy and mutant analysis, we provide evidence that the sensor histidine kinase FitF is a key regulator of insecticidal toxin production. Our experimental data and bioinformatic analyses indicate that FitF shares a sensing domain with DctB, a histidine kinase regulating carbon uptake in Proteobacteria. This suggested that FitF has acquired its specificity through domain shuffling from a common ancestor. We constructed a chimeric DctB-FitF protein and showed that it is indeed functional in regulating toxin expression in P. protegens. The shuffling event and subsequent adaptive modifications of the recruited sensor domain were critical for the microorganism to express its potent insect toxin in the observed host-specific manner. Inhibition of the FitF sensor during root colonization could explain the mechanism by which P. protegens differentiates between the plant and insect host. Our study establishes FitF of P. protegens as a prime model for molecular evolution of sensor proteins and bacterial pathogenicity.
Introduction
Pseudomonas protegens is a beneficial root-associated bacterium of the Pseudomonas fluorescens group that is able to promote the growth of crop plants and to efficiently protect their roots against fungal and oomycete phytopathogens [1], [2]. P. protegens can also turn into an insect pathogen [3]–[5]. The bacterium produces a potent insecticidal toxin termed Fit (for P. fluorescens insecticidal toxin) which is required for its capacity to efficiently kill larvae of important agricultural pest insects upon oral or systemic infection [5], [6]. The gene encoding the Fit protein toxin is part of an eight-gene cluster which comprises also genes coding for a type I secretion system and three regulatory proteins (Figure S1 and [6], [7]). Expression of the insecticidal toxin is activated during infection of the insect host, but not on plant roots or in standard laboratory media [7]. We recently demonstrated that toxin expression is tightly controlled by two regulators, named FitG (an activator) and FitH (a repressor) [7]. The third regulatory protein encoded in the Fit cluster is named FitF and codes for a putative sensor histidine kinase-response regulator hybrid protein. We hypothesize that FitF is responsible for the detection of the host environment and for activating insecticidal toxin production via FitH and FitG specifically upon infection of the insect host (Figure S1).
Sensor proteins enable bacteria to sense the environment they live in and to adapt their behavior accordingly, which is particularly relevant for pathogen-host interactions [8]–[10]. The number of sensor protein types is particularly high in bacteria such as pseudomonads that inhabit diverse and changing environments [11], [12]. An important category of sensor proteins is that of the two-component regulatory systems, which couple extracellular stimuli to adaptive responses. A typical two-component system consists of a membrane-bound sensor histidine kinase, which perceives a stimulus, and a cytosolic response regulator, which transduces the signal into an output, such as altering specific gene expression. Signal transduction is achieved by phosphotransfer reactions between the sensor kinase and the response regulator. In some cases, like in the so-called phosphorelay system, the sensor histidine kinase is a hybrid response regulator protein undergoing multiple intramolecular phosphotransfer reactions, before finally activating a separate response regulator protein [13], [14].
Sensor and signal transduction proteins usually show a modular organization of conserved domains [14], which can be highly variable in their order and topological organization [8]. Not surprisingly, therefore, it has been proposed that the modularity of two-component systems enables rapid evolution and generation of new functional properties. Gene duplication and domain shuffling are considered to be driving mechanisms for the formation of new two-component systems in bacteria [10], [12]. More than 70% of estimated recently duplicated histidine kinases have input domains different from those of their closest paralogs, suggesting frequent domain shuffling events [10]. It was proposed that by shuffling of the sensor domain recently duplicated histidine kinases gained new sensing specificity and thus might have enabled the bacteria to respond to a broader range of environmental changes [12].
The major goal of our work is to understand the molecular mechanisms that allow P. protegens and related bacteria to survive within and to kill the insect host. Of particular interest for the underlying work was the question as to how insect pathogenicity may have evolved and has been selected for. Because sensory systems are essential for niche adaptation, we felt that an evolutionary analysis of the chemosensory systems enabling insect recognition in P. protegens and in particular of the Fit system would be fundamental to the understanding of host adaptation.
Here we thus report the detailed regulation of Fit toxin expression and in particular describe the role of the hybrid sensor kinase protein FitF. We noticed that the periplasmic region of FitF is strikingly similar to the sensor domain of the histidine kinase DctB, which regulates the uptake of C4-dicarboxylates in Proteobacteria [15]. The crystal structures of DctB of Vibrio cholerae and Sinorhizobium meliloti have been solved [16], [17] and show an inserted repeat of a Per-Arnt-Sim (PAS)-like fold (PASp) in the periplasmic sensory domain, which was later termed the PhoQ/DcuS/CitA (PDC) domain [18]. PAS domains are universally distributed among all kingdoms of life, are the most frequent type of signal sensors in bacteria, can fulfill several functions and can bind chemically diverse small-molecule ligands [9], [19]–[21]. The membrane distal PASp domain of DctB binds C4-dicarboxylates such as malate, fumarate and succinate [15].
We present several lines of evidence illustrating that the periplasmic sensory domain of FitF evolved from a common ancestor with DctB, enabling P. protegens to survive and switch on toxin expression only in the insect host. By expressing a chimeric DctB-FitF protein in P. protegens and thereby testing the proposed domain shuffling event, we show that the DctB sensor domain is effectively suitable to drive the expression of the insecticidal toxin in a similar way as wild-type FitF. We found that the periplasmic sensor region of FitF possesses an important and conserved peptide motif and demonstrate by site-directed mutagenesis that, as for DctB, it is essential for the function of the histidine kinase. Bioinformatic analyses further support that the specific tandem PASp domain probably served as a sensory module for numerous proteins in P. protegens and other bacterial species, highlighting its importance, mobility and evolutionary plasticity. Our work reveals how the FitF sensor kinase could have evolved into a crucial virulence gene expression regulator, and has contributed to the ability of P. protegens to exploit a new ecological niche by recruiting a functional domain from an ancestor of sensor proteins involved in the regulation of the primary metabolism. In addition, our evolutionary analysis of the Fit regulatory system could provide a unique model system to study the hypothesis of domain shuffling in sensor protein evolution, which so far had been postulated mainly on the basis of bioinformatic analysis of proteins [10], [22] and construction of artificial chimeric proteins [23]–[25].
Results
FitF is essential for Fit toxin expression in the insect host
The fit locus (EU400157) of P. protegens comprises three genes (fitF, fitG, and fitH) that code for regulatory proteins (Figure S1). We previously demonstrated that expression of the insecticidal Fit toxin can be activated in strain CHA0 in Lysogeny Broth (LB) by overexpression of fitG or deletion of fitH, thus identifying the encoded proteins as an activator and repressor of insect toxin expression, respectively [7]. The third gene fitF, which was predicted to code for a sensor histidine kinase-response regulator hybrid protein (Figure 1A), was hypothesized to function as a detector of the insect environment and a regulator of Fit toxin production [7].
Fig. 1. The hybrid sensor kinase FitF is essential for Fit toxin expression. 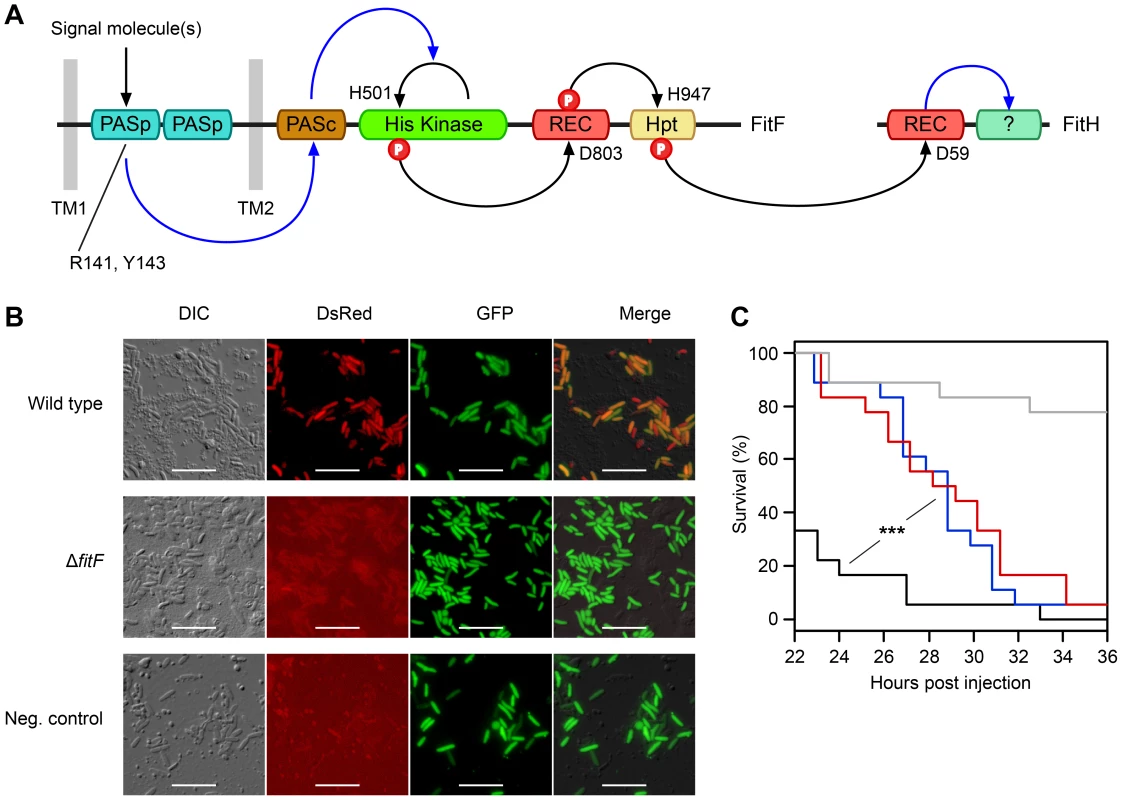
(A) Domain topology of FitF and FitH and putative signal transduction pathways (blue arrays) and phosphotransfer reactions (black arrows) between domains and proteins predicted by NCBI Conserved Domain Search [42] and SMART [43]. The conserved amino acid residues predicted by NCBI Conserved Domain Search to be phosphorylated or to be important for signal recognition are indicated with their respective amino acid positions. Hpt, phosphotransfer domain; PASc, cytoplasmic Per-Arnt-Sim (PAS) domain; PASp, periplasmic PAS domain; REC, receiver domain; TM, transmembrane region. (B) Epifluorescence microscopy of hemolymph extracts from larvae of G. mellonella infected with FitD-mCherry reporter strains with the wild-type (CHA1176) and ΔfitF mutant (CHA1174-gfp2) background for 24 h. The injected strains harbor a constitutive GFP cell tag for identification, expression of FitD-mCherry can be seen in the DsRed channel. Strain CHA0-gfp2 was used as a negative control. Bars represent 10 µm, micrographs are false-colored. The experiment was repeated twice with similar results. (C) Systemic virulence assay with injection of wild-type (in black, CHA0) and isogenic mutants (ΔfitF in red, CHA1154; ΔfitD in blue, CHA1151) of P. protegens CHA0 into last instar larvae of G. mellonella. Saline solution served as a negative control (in gray). Significant differences between the different treatments are indicated with *** (p-value<0.0001; Log-rank test). The experiment was repeated twice with similar results. To demonstrate that FitF is necessary for Fit toxin production, we used reporter strains of P. protegens CHA0 in which the full-length fitD gene was translationally fused at its native locus to mcherry by markerless gene replacement [7]. Epifluorescence microscopy confirmed that FitD-mCherry was visibly expressed in P. protegens CHA0 cells during infection of larvae of the greater wax moth Galleria mellonella, but was absent when fitF was inactivated by an in-frame deletion (Figure 1B). Also, the virulence of the CHA0 fitF deletion mutant in a Galleria injection assay was statistically significantly decreased compared to the wild type and was similar to a fitD deletion mutant (Fit toxin-deficient) (Figure 1C). These results demonstrate that FitF is essential for the activation of Fit toxin expression by P. protegens CHA0 in the insect host.
Activation of Fit toxin expression in an insect-mimicking medium
Although FitD-mCherry was readily expressed during infection of larvae, it was hardly detectable when P. protegens CHA0 was growing in standard bacterial culture media such as LB or Brain Heart Infusion (BHI) (Figure 2A). Fit toxin production was strongly induced when the bacteria were grown in Grace's Insect Medium (GIM), with on average 60-fold higher red fluorescence levels of individual cells than in LB. GIM, which is a defined medium rich in amino acids and C4-dicarboxylates, is widely used for insect cell cultures and reflects closely the composition of Lepidopteran hemolymph [26]. In GIM, wild-type bacteria expressed the Fit toxin mostly at the end of exponential growth but no longer produced it in stationary phase (Figure S2A). Compared to LB, FitD-mCherry expression was also significantly higher in M9 minimal medium supplemented with L-malate as sole carbon source, but not in fetal bovine serum or in marine broth, although both media provide conditions similar to insect hemolymph (Figure 2A). Interestingly, FitD-mCherry production was significantly lower in M9 or GIM supplemented with plant root extracts (Figure 2B). Also more than 20% (v/v) of LB mixed in with GIM abolished FitD-mCherry expression (data not shown). Altering pH in M9 medium did not impede FitD-mCherry expression (data not shown).
Fig. 2. Expression of the Fit insect toxin can be induced in an insect hemolymph-mimicking medium (GIM). 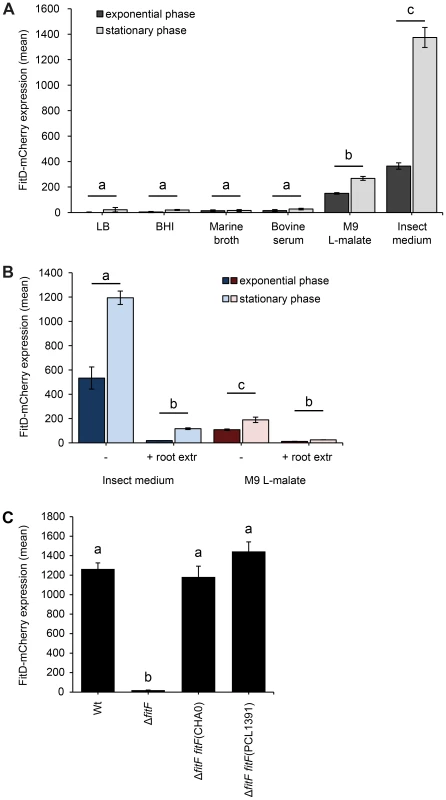
(A) The FitD-mCherry reporter strain of P. protegens CHA0 (CHA1163) was grown in different media and red fluorescence intensities of single cells were quantified by epifluorescence microscopy in the exponential (8 h post inoculation) and stationary (24 h post inoculation) growth phase. Results are the mean and standard deviation of population averages of single cell fluorescence intensities from three independent cultures (n = on average approx. 3200 cells per treatment and time point). Treatments labeled with a different letter are significantly different (p-values<0.0001; two-way ANOVA with Tukey's HSD test for post-hoc comparisons). The experiment was performed three times with similar results. (B) Quantification of the expression of FitD-mCherry in the wild-type background of CHA0 (CHA1163) in GIM and M9 L-malate with or without root extracts from field-grown wheat (n = on average approx. 2600 cells per treatment and time point). Characters indicate significant differences between the treatments (p-values<0.05; two-way ANOVA with Tukey's HSD test for post-hoc comparisons). The experiment was repeated twice with similar results. (C) Quantification of the expression of FitD-mCherry in the wild-type (CHA1163) and ΔfitF deletion mutant (CHA1174) background of strain CHA0 grown in GIM for 24 h at 25°C (n = 2768–3239 cells per strain). Re-introducing a single copy of fitF from CHA0 (CHA5066) or PCL1391 (CHA5073) in the bacterial chromosome rescued the expression of FitD-mCherry. Means labeled with a different letter are significantly different (p-value<0.05; one-way ANOVA with Tukey's HSD test for post-hoc comparisons). The experiment was performed three times with similar results. Expression levels of the FitD-mCherry fusion protein in GIM were similar in the P. protegens wild type and in a fitH deletion mutant, which constitutively expresses the toxin (Figure S3). Furthermore, deletion of fitF abolished the expression of FitD-mCherry in GIM (Figure 2C), but could be fully rescued by complementation of the mutant strain by insertion of a single copy of the fitF gene into the chromosome (Figure 2C). Interestingly, the fitF deletion mutant of strain CHA0 could also be fully complemented with the homologue fitF from P. chlororaphis strain PCL1391 (Figure 2C), even though P. chlororaphis FitF is predicted to harbor two cytoplasmic PAS domains instead of one for FitF from P. protegens [4], [5], [27]. Results of FitD-mCherry expression were confirmed by assaying the activity of the PfitA promoter, which drives the expression of toxin and type I transporter genes [7], using a GFP-based transcriptional reporter fusion (Figure S2B).
Using a hemolymph-mimicking medium, we were thus able to confirm the essential role of FitF in regulation of insect toxin production in a controlled and reproducible manner in an ex vivo environment.
FitF has a periplasmic region homologous to the C4-dicarboxylate-sensing PASp domains of DctB
FitF is predicted to possess two transmembrane domains, a periplasmic sensor domain, a cytoplasmic PAS domain, a histidine kinase domain (comprising a conserved phosphoacceptor domain and an ATPase domain), a CheY-homologous receiver domain, and a phosphotransfer domain (Figure 1A). BLAST comparisons with the amino acid sequence of the periplasmic region of FitF (FitFp) of P. protegens CHA0 indicated 54% amino acid sequence similarity (27% sequence identity) across the whole length to the double PASp domain of the C4-dicarboxylate sensor DctB (DctBp) of V. cholerae (Figure 3A). Phylogenetic analysis further indicated that FitFp homologues from various strains of P. protegens and P. chlororaphis group with DctBp homologues of different proteobacterial species, while the periplasmic regions of DctB-related CitA and DcuS proteins appear to be phylogenetically more distant (Figure 3B and Table S1). CLANS cluster analysis revealed similar results with FitFp clustering in close proximity to homologs of DctBp and CitA and DcuS clustering further away (Figure S4 and Table S1). We found a conserved “FRPYF” motif among the FitFp homologues (Figure 3A), which is similar to the previously reported signal molecule-binding “RXYF” motif in DctB homologues and other proteins with double-PASp domains [28], [29]. Protein threading and modeling approaches predicted a similar secondary and tertiary structure for FitFp as DctBp (Figure 3C). This suggests that the FitFp and DctBp domains share a common ancestor. Concurrently, FitF and DctB display different domain topologies in their cytoplasmic portions, which is in contrast to the similarity in the periplasmic region of the proteins.
Fig. 3. FitFp is homologous to the periplasmic DctB-like sensor domain. 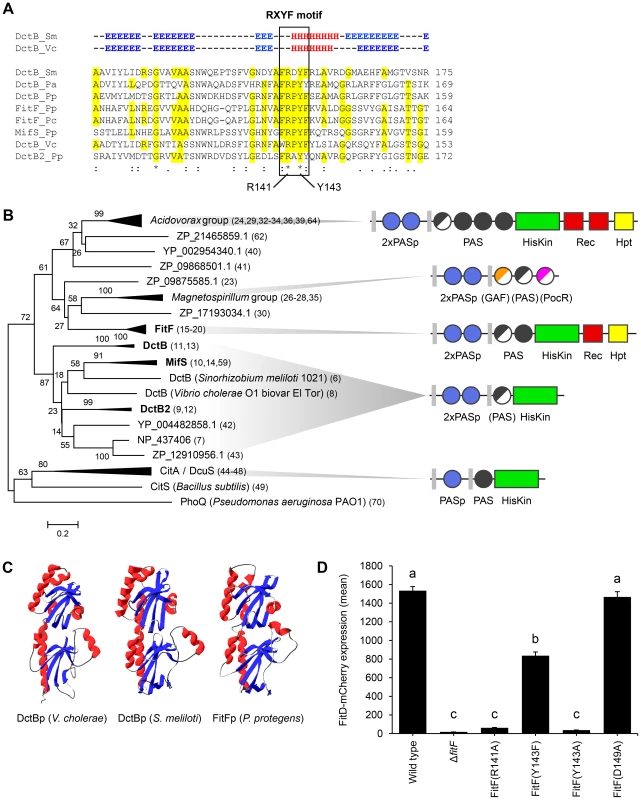
(A) Multiple sequence alignment of the periplasmic region of FitF and DctB homologs (selection). Amino acid residues that are identical to FitF are highlighted in yellow. Secondary structures of DctB were deduced from the corresponding crystal structures and are displayed on top (H, alpha helix; E, beta sheet; -, coil). Pa, P. aeruginosa PAO1; Pp, P. protegens CHA0; Pc, P. chlororaphis PCL1391; Sm, S. meliloti; Vc, V. cholerae. (B) Phylogenetic tree with sequences obtained from BLASTp searches using the periplasmic sequence of FitF of P. protegens CHA0 and of homologs of DctBp. MAFFT was used for sequence alignment and the Minimum Evolution method in MEGA [44] for inferring the evolutionary history of the proteins. The percentage of replicate trees in which the associated proteins clustered together in the bootstrap test (500 replicates) is shown next to the branches. Evolutionary distances, which were computed using the Poisson correction method, are drawn to scale and are in the units of the number of amino acid substitutions per site. The corresponding protein sequences can be found in File S1. The predicted domain topology of the entire proteins is depicted for groups of interest. Domains that are displayed in half do not exist in all proteins of the respective group. PhoQ was used as out group. (C) Tertiary structure prediction for P. protegens FitFp by Phyre2 in comparison with crystal structures of DctBp of V. cholerae (PDB code 3BY9) and S. meliloti (PDB code 3E4O). Other modeling programs predicted highly similar structures (data not shown). (D) Site-directed mutagenesis of the native fitF gene in the FitD-mCherry reporter strain CHA1163. The sites of the mutated residues are depicted in panel A and Figure 1C. Microscopic quantification of the expression of FitD-mCherry in the wild-type and individual mutant backgrounds of CHA0 grown for 24 h in GIM. Results are the mean and standard deviation of population averages of single cell fluorescence intensities from three independent cultures (n = on average approx. 2900 cells per strain). Characters indicate significant differences between the means (p-values<0.01; one-way ANOVA with Tukey's HSD test for post-hoc comparisons). The experiment was performed three times with similar results. By using in vivo site-directed mutagenesis, we replaced a number of residues in fitF and fitH and studied the effect on FitD-mCherry expression in P. protegens. Change of Arg141 and of Tyr143 in the RXYF motif of FitF to Ala following the mutagenesis of dctB described by Nan et al. [28], resulted in almost completely abolished FitD-mCherry production (Figure 3D). In contrast, change of Asp149 to Ala (used as an internal negative control) did not alter the expression of the insecticidal toxin. Changing Tyr143 to Phe reduced expression of FitD-mCherry by approximately 45%. Replacement of predicted conserved phosphorylation residues of the histidine kinase and receiver domains in FitF (H501 and D803) and FitH (D59) (Figure 1A) by alanine diminished the expression of FitD-mCherry (Figure 4). Together, these data demonstrate conspicuous structural and functional relatedness between the periplasmic domain of FitF and the sensor domain of DctB, with a conserved peptide motif being crucial for activation of Fit toxin expression.
Fig. 4. Site-directed mutagenesis of fitF and fitH. 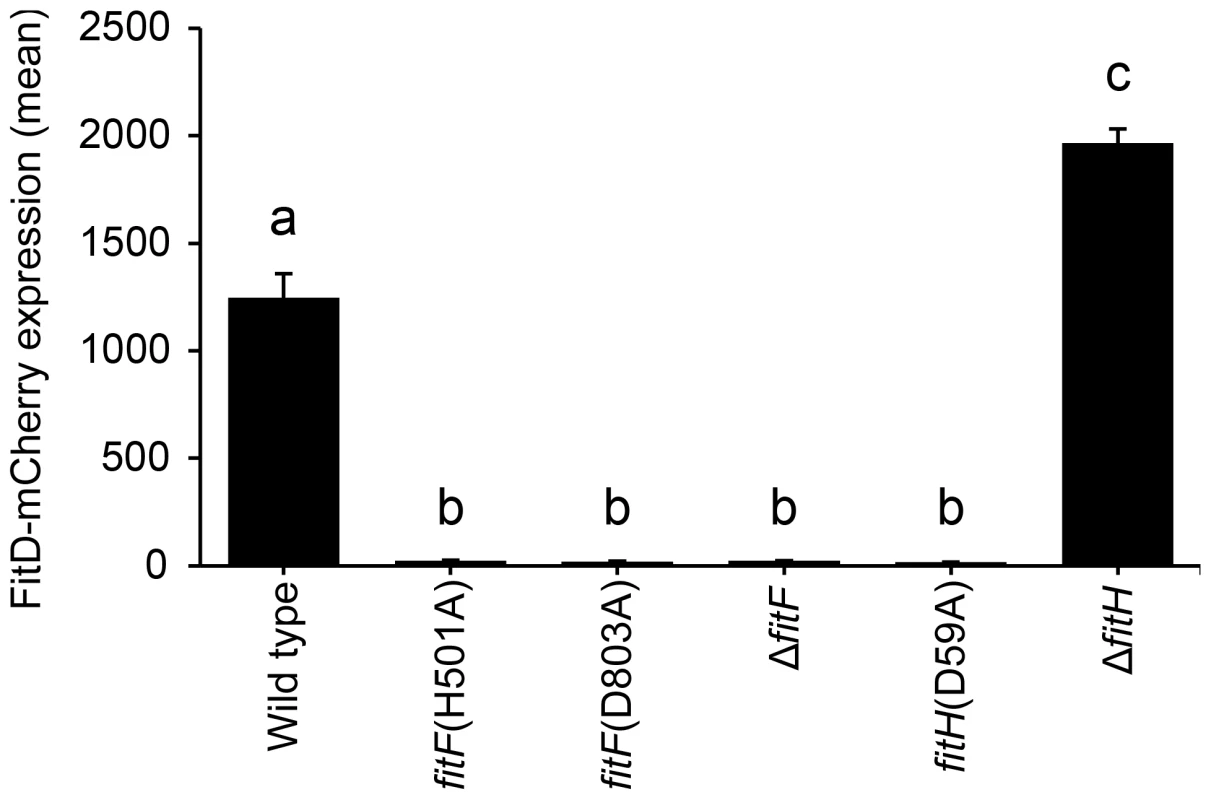
Site-directed mutagenesis of the native fitF and fitH genes in the FitD-mCherry reporter strain CHA1163. Quantification of the expression of FitD-mCherry in the wild-type (CHA1163) and individual mutant backgrounds of CHA0 (CHA5056, CHA5075, CHA1174, CHA5084, and CHA1175) grown for 24 h in GIM. Results are the mean and standard deviation of population averages of single cell fluorescence intensities from three independent cultures (n = on average approx. 2900 cells per strain). Characters indicate significant differences between the means (p-values<0.001; one-way ANOVA with Tukey's HSD test for post-hoc comparisons). The experiment was repeated twice with similar results. An artificial chimera of DctB and FitF is functional
Because of the conspicuous similarity between the FitFp and DctBp domains, we hypothesized that perhaps the actual FitF protein might have been the result of a fusion of an ancestor DctBp domain into a FitF precursor. To simulate the proposed domain shuffling event and to test experimentally whether the sensor module of DctB is effectively suitable to regulate the expression of the Fit toxin, we created an artificial DctBp-FitFc chimera in which the periplasmic domain of DctB of P. protegens CHA0 was fused to the cytoplasmic portion of FitF (FitFc) (Figure 5A).
Fig. 5. A DctBp-FitFc chimera regulates toxin expression similarly to wild-type FitF. 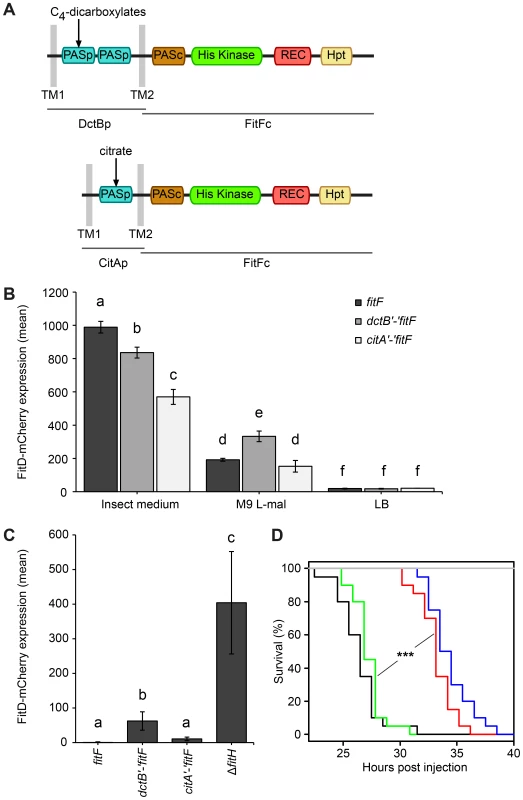
(A) A chimeric protein of the cytoplasmic portion of FitF and the N-terminal part of DctB including its double-PASp sensor domain and the transmembrane regions was constructed by fusing the respective P. protegens CHA0 genes using the conserved DNA sequence coding for the second transmembrane region as a linker. A CitAp-FitFc chimera was constructed analogously using E. coli citA. (B) Expression of FitD-mCherry in the ΔfitF reporter strain CHA1174 complemented with either wild-type fitF (CHA5066), the dctB‘-’fitF chimeric gene (CHA5093) or the citA‘-’fitF chimeric gene (CHA5151) in different media for 24 h. Results are the mean and standard deviation of population averages of single cell fluorescence intensities from three independent cultures (n = on average approx. 3590 cells per treatment). Characters indicate significant differences between the means (p-values<0.05; one-way ANOVA with Tukey's HSD test for post-hoc comparisons). The experiment was performed three times with similar results. (C) Quantification by epifluorescence microscopy of FitD-mCherry expression in reporter strains CHA5066, CHA5093, CHA5151, and CHA1175 (ΔfitH, positive control), all harboring the plasmid pPROBE-TT for GFP-tagging of the cells, grown for five days on roots of cucumber. Shown are means and standard deviations of population averages of single cell fluorescence intensities of bacteria isolated from six independent plants (n = on average approx. 1170 cells per strain). Characters indicate significant differences between the means (p-values<0.05; one-way ANOVA with Tukey's HSD test for post-hoc comparisons). The experiment was repeated twice with similar results. (D) Galleria injection assay with wild-type (in black, CHA0) and isogenic mutants (ΔfitF in red, CHA1154; ΔfitD in blue, CHA1151; ΔfitF dctB‘-’fitF in green, CHA5150) of P. protegens CHA0 into last instar larvae of G. mellonella. Saline solution served as a negative control (in gray). Significant differences between the different treatments are indicated with *** (p-value<0.0001; Log-rank test). The experiment was repeated twice with similar results. Indeed, expression of the DctBp-FitFc chimeric protein in a ΔfitF mutant background of strain CHA0 led to FitD-mCherry production in GIM, but not in LB (Figure 5B). Still, FitD-mCherry expression was significantly higher in GIM in the ΔfitF mutant complemented with wild-type fitF than with the dctB‘-’fitF chimeric gene. Remarkably, however, FitD-mCherry production was activated in CHA0 expressing the DctBp-FitFc chimeric protein when the bacteria were growing on plant roots, while toxin production was completely off in bacteria expressing wild-type FitF (Figure 5C). Furthermore, bacteria with the DctBp-FitFc background produced FitD-mCherry at significantly higher levels in minimal medium with L-malate as sole carbon source than bacteria expressing wild-type FitF (Figure 5B). In a Galleria injection assay the DctBp-FitFc chimera fully complemented the fitF mutant (Figure 5D). These results thus indicate that the DctB sensor domain can replace the FitFp domain of FitF. Yet, this causes a shift in sensor protein sensitivity resulting in a loss of responsiveness in an insect environment and a gain of responsiveness in a root environment.
A chimera of the more distantly related PASp sensor domain of CitA and FitFc was functional and even less responsive to the insect mimicking medium than the DctBp-FitFc chimera (Figure 5).
Activation of Fit toxin production is host-specific
In order to investigate whether toxin production is not only host-dependent but also specific toward certain insect orders, the expression of FitD-mCherry by P. protegens CHA0 was studied in additional insect species. The expression of the Fit toxin was activated in the hemocoel of the African cotton leafworm Spodoptera littoralis (Lepidoptera) and the mealworm Tenebrio molitor (Coleoptera) (Figure 6A). In contrast to the ΔfitH mutant of strain CHA0, however, the insecticidal toxin was hardly produced in the phylogenetically distant pea aphid Acyrthosiphon pisum (Hemiptera) (Figure 6A). In addition, as already shown for cucumber [7], no toxin expression was detectable on roots of wheat and tomato (Figure 6B). Moreover, the presence of a phytopathogenic fungus (Fusarium oxysporum) on tomato roots did not activate Fit toxin production in the bacteria (Figure 6B). These results suggest that P. protegens CHA0 is capable of expressing its insecticidal toxin in a host-specific manner.
Fig. 6. Fit toxin expression is controlled in a host-specific manner. 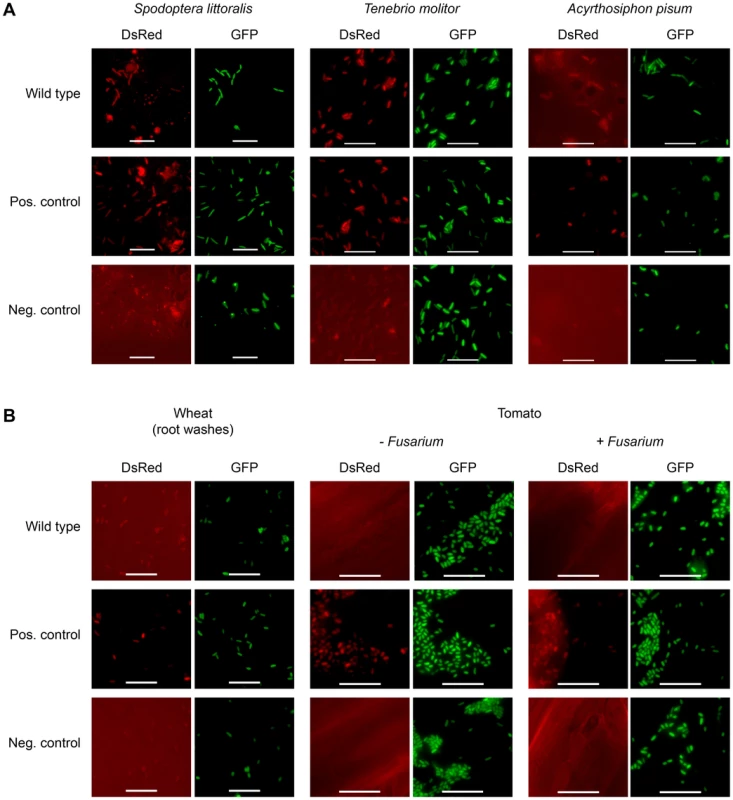
The insectidical toxin is expressed by P. protegens CHA0 only in certain insect species and not on plant roots. (A) Epifluorescence microscopy of hemolymph isolated from S. littoralis, T. molitor and A. pisum infected with FitD-mCherry reporter strains with the wild-type (CHA1176) and ΔfitH mutant (CHA1178, positive control) background. The bacteria harbor a constitutive GFP cell tag for identification, expression of FitD-mCherry can be seen in the DsRed channel. Strain CHA0-gfp2 was used as a negative control. Bars represent 10 µm, micrographs are false-colored. The experiments were performed at least twice with similar results. (B) Epifluorescence microscopy of plant roots (or root washes) three to five days after the inoculation with the same reporter strains as in panel A, with or without co-inoculation with the phytopathogen Fusarium oxysporum f. sp. radicis-lycopersici. The experiments were performed twice with similar results. Discussion
Fit toxin production is dependent on the sensor kinase FitF
Here we show that the histidine kinase FitF is responsible for activation of Fit toxin expression in P. protegens CHA0. We deleted fitF in the CHA0 genome and our results show unambiguously that FitF is essential for the induction of Fit toxin expression and for full virulence of the bacterial strain in the insect host (Figure 1 and Figure 2C). We assume that FitF is the primary sensor to signal P. protegens the appropriate conditions to start toxin expression, activating a phosphorelay from the histidine kinase to the receiver and phosphotransfer domain of FitF (Figure 1A). FitF then most likely inactivates FitH via phosphorylation of a conserved aspartate residue, since the substitution of this residue by alanine locked the protein in its repressing state (Figure 4). Inactivation of FitH might derepress FitG, which subsequently activates transcription of the fitABCDE operon (Figure S1).
FitF acquired the mobile DctB-like sensor domain by domain shuffling
The periplasmic region of FitF showed remarkable structural and functional similarity to the sensor domain of DctB (Figure 3 and Figure 5). In particular, a RXYF motif was found in FitFp and we could show by site-directed mutation analysis that this conserved and known peptide motif is crucial for the activation of Fit toxin expression in P. protegens (Figure 3D). However, these two proteins differ substantially in their domain topologies in the cytoplasmic portion (Figure 3D). This suggested that an ancestor DctBp domain was acquired through shuffling in a precursor FitF. We present experimental and bioinformatic evidence that FitF most likely evolved via a fusion of two genes coding for a histidine kinase-response regulator hybrid protein and a duplicated DctB homolog (Figure 7). We noticed that DctB and FitF share a high degree of primary sequence identity in the second transmembrane region. It may therefore be possible that the fusion occurred via homologous recombination within the DNA sequence coding for the second transmembrane alpha helix.
Fig. 7. Model for evolution of FitF via a domain shuffling event involving a DctB ancestor. 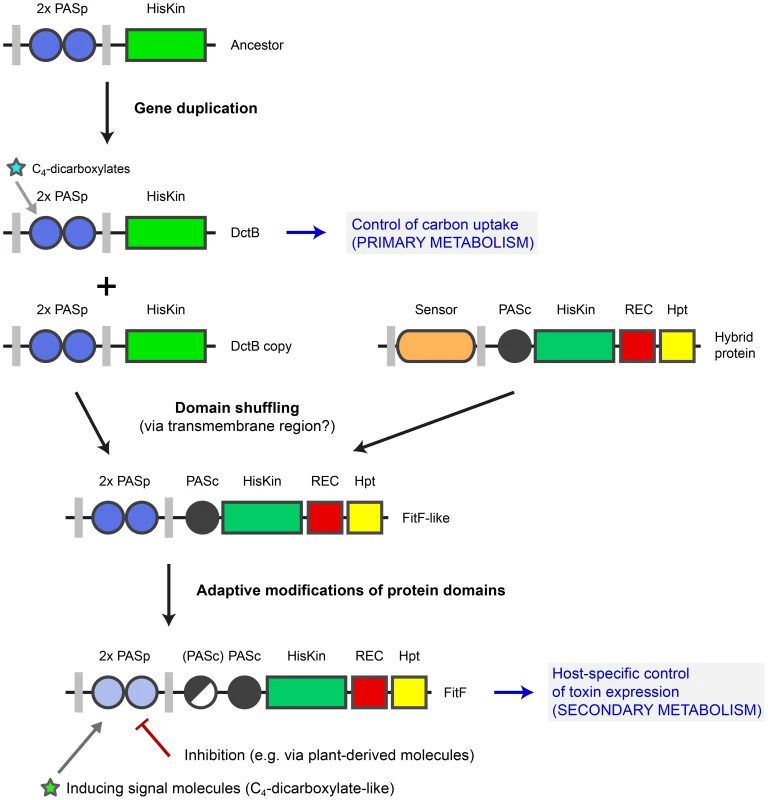
The ancestor of the gene coding for the sensor kinase DctB was duplicated several times in various proteobacterial species. One dctB gene copy underwent a fusion with a gene encoding a histidine kinase-response regulator hybrid protein, possibly by homologous recombination via a conserved region coding for the second transmembrane region of the sensor proteins. This domain shuffling event resulted in the expression of a hybrid histidine kinase with a dual PASp domain architecture in the periplasmic portion. Selective pressure then led to adaptive modifications in the protein sequence and domain topology (i.e. insertion of a second PASc domain in P. chlororaphis). Domain shuffling and subsequent modifications during the evolution of FitF significantly contributed to the ability of P. protegens CHA0 to produce its insecticidal toxin in a host-specific manner and as a result to the evolution of insect pathogenicity in this biocontrol bacterium. Inhibition of FitF by plant-derived molecules may be a mechanism helping the bacterium to distinguish between the plant and insect host. The evolution of FitF may have taken place in bacterial species other than P. protegens, implying horizontal gene transfer. Despite limited primary sequence conservation between DctBp and FitFp, a constructed DctBp-FitFc chimera was functional and, most interestingly, induced Fit toxin production in P. protegens in the insect medium, although to significantly lower expression levels than wild-type FitF. This strongly suggests that the tandem PASp sensor of DctB is functionally analogous to that of FitF and may have been at the basis of sensor specificity acquisition by FitF. This experiment is limited by the fact that the chimeric protein was constructed using sequences of extant proteins as it is not possible to reconstruct the sensor protein as it was shortly after the proposed domain shuffling event.
Protein comparisons further suggested that similar double-PASp domains occur widely among prokaryotes and in a variety of modular proteins (Figure 3B and Figure S4). Domains homologous to DctBp cannot only be found in histidine kinases but also in cyclic di-GMP modulating proteins (Figure 3B and Figure S4). PAS domains are known to be the most frequent type of sensor domains in bacteria [9], [20]. It is thus imaginable that such domains have been frequently interchanged and that such shuffling has been fundamental to evolution of FitF specificity.
In contrast to DctB, FitF possesses a cytoplasmic PAS domain as a linker between the sensor and kinase domain (Figure 1). We noticed that DctB proteins with an inserted PASc domain also occur in certain Acidovorax species. Furthermore, the C4-dicarboxylate sensing DcuS and CitA proteins of Escherichia coli possess a DctB-like PASp sensor domain in the periplasmic portion and a PASc domain as a linker between the sensor and the histidine kinase domain [15], [30]. These observations further support the notion that an ancestral DctB-like sensor domain served as an adaptable and mobile module for the evolution of diverse proteins, since it can be fused to a variety of other protein domains. This is further supported by our observation that a fusion of the periplasmic sensor domain of CitA to FitFc was functional (Figure 5).
Domain shuffling may require gene duplication and recombination [12]. In this respect, it is interesting to note that like many Pseudomonas species, P. protegens encodes three paralogs of the dctB gene (Figure 3). The dctB paralogs are functionally different. One of them (DctB) is involved in regulation of the uptake of C4-dicarboxylates (Figure S5 and [31]), whereas another (named MifS) was reported to be a regulator of biofilm formation in P. aeruginosa [32]. Pseudomonas fulva strain 12-X encodes four dctB paralogs (GeneBank CP002727), suggesting that duplications of dctB must have occurred frequently and could have been the basis for domain shuffling events in these bacteria.
The molecular mechanism of domain shuffling in the bacterial kingdom is still unknown. However, it has been reported that hybrid sensor kinases as is FitF show particularly high levels of DNA polymorphism and fast evolutionary rates [33]. Moreover, they are thought to have mostly evolved by lateral recruitment of individual protein domains [19]. Therefore, not only lineage-specific expansion but also recombination with horizontally acquired sequences could have played a role in the evolution of FitF. The sensor protein could have evolved by shuffling of functional domains that originated from different bacterial species.
Adaptive modifications to ensure host-specific expression of the insecticidal toxin
We discovered that Fit toxin expression in P. protegens CHA0 can be highly induced independently of the host organism in an insect hemolymph-mimicking medium (Figure 2A). The physicochemical conditions given by the insect medium are thus sufficient for the observed activation of toxin production during infection of the insect host. Despite extensive testing (not shown), however, we currently do not know the precise chemical structure of the signaling compound(s) that trigger FitF activation. The fact that the DctBp-FitFc chimera controlled Fit toxin production similarly to wild-type FitF, suggests that the signal molecule may be similar to C4-dicarboxylates. However, the chimera seemed to respond differentially to changing environmental conditions (Figure 5B and C). In addition and in contrast to DctB [28], the conservative replacement of the important tyrosine residue Y143 by phenylalanine did not diminish Fit toxin expression in the insect medium (Figure 3D). Moreover, certain cells within the population of bacteria with the DctBp-FitFc chimera expressed the insect toxin on plant roots, which was not the case with bacteria expressing wild-type FitF (Figure 5C). These results indicate that the signal molecules recognized by FitFp are no longer (only) C4-dicarboxylates. Molecules that bind to the sensor domain of FitF could be detected when solving the crystal structure of its periplasmic sensor domain in future studies, as it was demonstrated for several proteins with double-PASp sensor domains in the work of Zhang and Hendrickson [29].
Our findings suggest that even though a DctBp domain may have been at the basis of acquisition of FitF sensory capacity, further adaptive mutations occurred after the domain shuffling event, shifting the spectrum of recognized signals to ensure specificity of toxin production toward the insect environment. Indeed, we found indications that the Fit toxin is produced by wild type P. protegens CHA0 in a host-specific manner (Figure 6).
Competitive inhibition by plant molecules as a mechanism for host recognition?
Interestingly, FitD-mCherry expression by P. protegens diminished when induction media were supplemented with plant root extract (Figure 2B). We speculate that this may be the result of a competitive inhibition rather than of absence of inducer compounds, because the rest of the induction medium was kept the same. If FitF could be directly or indirectly inhibited by plant molecules, this would explain the observed loss in toxin expression on roots, and could form a mechanism for host (plant or insect) differentiation. Activation of toxin expression in the insect host via FitF would then be the result of absence of inhibiting plant-derived molecules and the simultaneous presence of specific activating signal molecules in insect hemolymph (Figure 7). Competitive interactions are known from studies on DctB, where it was reported that molecules structurally resembling C4-dicarboxylates (e.g. malonate) can bind to the membrane distal PASp domain of DctB but do not lead to an activation of the kinase by conformational change [17]. The possibility of competition between activating and inhibitory molecules for the signal binding pocket of DctB was not discussed so far, but would be an interesting aspect for future research on PAS sensor domains. Alternatively, the observed inhibition of toxin production on roots could be due to repression of FitF by another protein. In the case of DctB it was suggested that the activity of the sensor kinase can be controlled by the transporter DctA directly by protein-protein interaction [15]. The proposed inhibition of FitF could also be mediated indirectly through changes in the metabolism of the bacterium when growing on roots.
In summary, the present study provides evidence that a virulence-associated sensor histidine kinase, contributing to control the switch of the pseudomonad between a plant-beneficial and an insect pathogenic lifestyle, evolved by acquisition of a prominent sensory domain from a common ancestor of a protein, which regulates carbon uptake and primary carbon metabolism. This event was crucial for the ability of the microorganism to activate toxin expression in insects in a host-specific manner and thus to the adaptation of this bacterium to the insect environment.
To our best knowledge, P. protegens at first is well adapted to the life on plant roots. The microorganism acquired and evolved virulence determinants, such as the fit cluster, and adapted to the insect environment, allowing it to survive within and to kill larvae of certain insect species. Since two-component signal transduction pathways are often involved in sensing and responding to changing environments, they have played a fundamental role in the adaptation of bacteria to a range of ecological niches [12]. P. protegens has the ability to tightly control Fit toxin production in a way that the toxin is only expressed during infection of certain insects but not on plant roots (Figure 6 and [7]). As we show here, FitF thereby plays an important role as a regulatory protein. We recently demonstrated that the Fit toxin is required for full virulence upon oral or systemic infection of insect larvae [5]–[7]. Therefore, the proposed domain shuffling event during the evolution of FitF has significantly contributed to the adaptation of this bacterium to a new niche and thus to the evolution of insect pathogenicity.
With the existing molecular techniques, the provided reporter constructs, the possibility to induce the expression of the Fit toxin in vitro in an insect medium, and the current knowledge about the regulation of Fit toxin expression, the Fit regulatory system could serve as a prime example for future studies on domain shuffling and related molecular mechanisms driving the evolution of sensory systems involved in the regulation of bacterial virulence and on the evolution of pathogenesis in general.
Materials and Methods
Bacterial strains, plasmids, media, and culture conditions
All strains and plasmids used in this study are listed in Table S2. Bacteria were routinely cultured in LB (LB Broth Miller, BD Difco), or in nutrient yeast broth (NYB) or on nutrient agar (NA) [34]. E. coli cells were grown at 37°C while P. protegens was cultured at 25°C. When appropriate, growth media were supplemented with ampicillin (100 µg/ml), chloramphenicol (10 µg/ml), kanamycin (25 µg/ml), gentamicin (10 µg/ml), tetracycline (25 µg/ml or 125 µg/ml for E. coli and P. protegens, respectively), or isopropyl β-D-1-thiogalactopyranoside (IPTG) (0.1 mM).
For Fit toxin expression studies, the following media were used. LB; Brain Heart Infusion (BHI) (BD Bacto); sterile-filtered Grace's Insect Medium (GIM) (G9771, with L-glutamine, without sodium bicarbonate, adjusted to pH 5.5 with sodium bicarbonate) (Sigma-Aldrich); M9 minimal medium (50 mM Na2HPO4×2 H2O, 22 mM KH2PO4, 9 mM NaCl, 19 mM NH4Cl, 2 mM MgSO4, 0.1 mM CaCl2, 134 µM EDTA, 31 µM FeCl3×6 H2O, 6.2 µM ZnCl2, 760 nM CuCl2×2 H2O, 420 nM CoCl2×2 H2O, 1.62 µM H3BO3, 81 nM MnCl2×4 H2O, pH 7) with 10 mM L-malate, except for growth curve assays which were performed with 20 mM L-malate; sterile-filtered Fetal Bovine Serum (Invitrogen Gibco); and Marine Broth 2216 (BD Difco). Cold root extracts were prepared by adding 4 g/L of washed and cut roots of field-grown wheat to M9 L-malate or GIM. The mixture was aggitated for 30 min at 300 rpm and room temperature and sterilized by using 5 µm and 0.45 µm filters. Dose-response assays were performed with LB, GIM and different ratios of LB and GIM.
Recombinant DNA techniques
DNA manipulations and PCRs were conducted according to standard protocols [34]. Genomic DNA was extracted using the Promega Wizard Genomic DNA Purification Kit. Plasmid DNA was routinely extracted and purified using the QIAprep Spin Miniprep Kit (Qiagen). Larger scale plasmid preparations were performed with the Genomed JETStar Plasmid Purification Midi Kit. DNA gel extractions were conducted using the MinElute Gel Extraction Kit and the QIAquick Gel Extraction Kit (Qiagen). DNA restriction and modification enzymes were from Promega and were used according to the manufacturer's recommendations. DNA enzyme reaction cleanups were performed using the QIAquick PCR Purification Kit (Qiagen). PCR was routinely conducted using the PrimeSTAR HS high-fidelity DNA polymerase kit (Takara Bio Inc.) for molecular cloning and the GoTaq DNA Polymerase kit (Promega) for analytic purposes according to the recommendations of the manufacturer. Primers used for this study were obtained from Microsynth AG (Balgach, Switzerland) and are listed in Table S3. DNA sequencing was conducted at GATC Biotech (Konstanz, Germany). Sequences were analyzed using the DNASTAR Lasergene software suite.
In-frame deletion of fitF and integration of reporter constructs
For the construction of the ΔfitF mutant CHA1154, a 2982-bp fragment was deleted in-frame in the fitF gene as follows. Using CHA0 DNA as a template, a 722-bp KpnI-EcoRI fragment encompassing the first 42 codons of fitF and the adjacent upstream region was amplified by PCR with primers PfitF1 and PfitF2 (Table S3). An 884-bp EcoRI-XbaI fragment comprising the last 41 codons of fitF plus downstream region was amplified by PCR using primers PfitF3 and PfitF4. The fragments obtained were digested with KpnI and EcoRI and with EcoRI and XbaI, respectively, and cloned by triple ligation into pUK21 opened with KpnI and XbaI. The 1.6-kb KpnI-XbaI insert in the resulting plasmid was checked by sequencing, excised and cloned into the suicide plasmid pME3087 digested with the same enzymes, giving pME8256 (Table S2). The constructed replacement vector was then used to delete fitF in P. protegens CHA0 by D-cycloserine counterselection as described before [35], [36], resulting in strain CHA1154 (Table S2). The suicide plasmid pME8217 was used to replace the native fitD with the fitD-mcherry fusion in strain CHA1154 by homologous recombination, generating strain CHA1174 (Table S2). For insect assays, the strain CHA1174 additionally was marked with a constitutively expressed GFP tag using the Tn7 delivery vector pBKminiTn7-gfp2, producing CHA1174-gfp2 (Table S2).
In vivo site-directed mutagenesis of fitF and fitH
For the mutagenesis of the periplasmic region of FitF, a region of fitF of 979 bp length encompassing the site of interest in the centre was amplified by PCR with CHA0 DNA using the primers fitF-mut1-hr-F and fitF-mut1-hr-R (Table S3). The resulting fragment was digested with EcoRI and BamHI and ligated into the suicide vector pEMG [37] opened with the same enzymes. The insert of the resulting plasmid pME8271 was checked by DNA sequencing. To introduce mutations into the insert sequence of pME8271 to subsequently replace the single amino acid residues R141, Y143, and D149 of FitF, primer pairs fitF-R141A-F/fitF-R141A-R, fitF-Y143A-F/fitF-Y143A-R, fitF-Y143F-F/fitF-Y143F-R, and fitF-D149-F/fitF-D149-R (Table S3), respectively, were used to amplify the vector pME8271 by PCR. The template plasmids used for the PCR were degraded by DpnI for 1 h at 37°C and PCR-amplified vectors were obtained by electroporation of E. coli DH5α λpir cells with purified PCR reaction and selection for kanamycin resistance. The insert sequences of the resulting plasmids were controlled by DNA sequencing.
For the replacement of H501 of FitF by alanine, a 489-bp fragment of the upstream region was amplified by PCR with primers fitF-mut2-hr-F and fitF-mut2-R using CHA0 DNA (Table S3). A 524-bp fragment of the downstream region was amplified by PCR using primers fitF-mut2-F and fitF-mut2-hr-R using CHA0 DNA as template. The two fragments were combined by overlap extension PCR using the primers fitF-mut2-hr-F and fitF-mut2-hr-R, creating a 984-bp KpnI-HindIII fragment. The PCR product was digested by KpnI and HindIII and ligated into the plasmid pUK21. The insert was checked by sequencing, excised by digestion with KpnI and BamHI and cloned into the suicide plasmid pEMG by ligation. The resulting plasmid pME8265 was then used to create strain CHA5056 (Table S2).
An analogous approach (leaving out the cloning of the PCR fragment into the plasmid pUK21) was used to create the suicide vector for the replacement of D803 of FitF and D59 of FitH by alanine. For FitF(D803A) the primers fitF-REC-hr-F, fitF-REC-hr-R, fitF-D803A-F, and fitF-D803A-R were used to construct the suicide plasmid pME8302 and create strain CHA5075. For FitH(D59A) the primers fitH-REC-hr-F, fitH-REC-hr-R, fitH-D59A-F, and fitH-D59A-R were used to construct the suicide plasmid pME8303 and generate strain CHA5084.
Isogenic mutants of P. protegens strain CHA0 were constructed by allelic replacement using the I-SceI system with pEMG. The I-SceI system protocol described by Martinez-Garcia and de Lorenzo [37] was modified for P. protegens for this study. Briefly, the pEMG suicide vector bearing sequences homologous to genomic counterparts was integrated into the chromosome of P. protegens via homologous recombination after delivery by electroporation of competent cells. Bacteria were selected for kanamycin resistance on agar plates and competent cells were transformed with the expression plasmid pSW-2 by electroporation. Bacterial cells were selected for gentamicin resistance on agar plates and grown overnight at 30°C in LB supplemented with 10 µg/ml gentamicin. Ten milliliter of fresh LB was inoculated with 2 ml of overnight culture, supplemented with 2 mM m-toluate and 10 µg/ml gentamicin and incubated for 7 h at 30°C to allow second homologous recombinations to occur. Bacterial cultures were diluted and plated on nutrient agar plates without antibiotics. Isolated colonies were screened for kanamycin sensitivity and mutants were identified by specific PCR and sequencing of the respective genomic region.
In-frame deletion of dctB homologs
Deletions of the three dctB homologs in P. protegens CHA0 were performed based on homologous recombinations using the suicide vector pEMG and the I-SceI system.
For the construction of suicide vectors for in-frame gene deletions of CHA0 dctB (PFLCHA0_c03070), dctB2 (PFLCHA0_c48560) and mifS (PFLCHA0_c47820), upstream and downstream regions of 500–600 bp length flanking the region to be deleted, encompassing the first five codons and the last 7–18 codons of the open reading frames, were amplified by PCR using the primers listed in Table S3. The resulting BamHI-HindIII fragments were digested with BamHI and HindIII and cloned by triple ligation into pEMG opened with BamHI. Correct insert sequences of the obtained plasmids pME8307, pME8308 and pME8309 for ΔdctB1, ΔdctB2 and ΔmifS, respectively, were confirmed by DNA sequencing (Table S2). The constructed suicide plasmids then served to construct strains CHA5085, CHA5090 and CHA5089, respectively, using the I-SceI system (Table S2).
Inducible expression of fitF
For complementation of the ΔfitF mutant of CHA0, the fitF genes of strains P. protegens CHA0 and P. chlororaphis PCL1391 were cloned under the control of the Ptac/lacIq promoter and introduced into the unique chromosomal Tn7 attachment site of strain CHA1174 using the mini-Tn7 delivery vector pME9411 as follows. Primers fitF-F-SD-new and fitF-R-HindIII were used to amplify the fitF gene of strain CHA0 by PCR. The 3.2-kb EcoRI-HindIII fragment was digested with EcoRI and HindIII and ligated into plasmid pME4510 opened with the same restriction enzymes. After blunt-ending the EcoRI restriction site, the fragment was ligated into pME9411 opened with SmaI and HindIII, to obtain pME8288, and the correct insertion was confirmed by sequencing. The pME9411 derivative and the Tn7 transposition helper plasmid pUX-BF13 were co-electroporated into competent cells of the recipient strain CHA1174 to create strain CHA5066 (Table S2).
An analogous approach was taken to complement the ΔfitF mutant of CHA0 in trans with fitF of strain PCL1391 [5]. A 1188-bp EcoRI–BamHI fragment (primers PCL-fitF-F-SD and PCL-fitF-br-R), a 1704-bp BamHI–StuI fragment (primers PCL-fitF-br-F and PCL-fitF-StuI-R), and a 957-bp StuI–HindIII fragment (primers PCL-fitF-StuI-F and PCL-fitF-R) were amplified by PCR with the indicated primer pairs using chromosomal DNA from strain PCL1391. The individual fragments were digested with the respective restriction enzymes and ligated individually into plasmid pUK21 opened with the same enzymes. The inserts in the resulting plasmids were checked by sequencing. The insert fragments were excised from the plasmids with the respective enzymes and cloned by quadruple ligation into plasmid pME4510 opened with EcoRI and HindIII. After blunt-ending the EcoRI restriction site, the fragment was ligated into pME9411 opened with SmaI and HindIII, and the correct insertion was confirmed by sequencing. The resulting mini-Tn7-Ptac/lacIq-fitF(PCL1391) delivery plasmid pME8295 then served to generate strain CHA5073 (Table S2).
Construction of the dctB‘-’fitF and citA‘-’fitF chimeras
Primers ME8300-F and ME8300-SpeI-R were used to amplify the lacIq gene and the IPTG-inducible promoter region of the plasmid pME6032 by PCR. The PCR product was purified, digested with NcoI and HindIII, and ligated into the vector pME6182 opened with the same enzymes. The insert in the resulting plasmid pME8300 was checked by DNA sequencing.
Primers dctB-F-SpeI and dctB-R-overlap were used to amplify an 879-bp fragment of dctB using genomic DNA from strain CHA0. Primers fitFc-F and fitF-R-HindIII were used to amplify a 2271-bp fragment of fitF by PCR with CHA0 DNA. The two fragments were combined by overlap extension PCR using the primers dctB-F-SpeI and fitF-R-HindIII, creating a 3.3-kb SpeI-HindIII fragment. The PCR product was digested by SpeI and HindIII and ligated into the plasmid pME8300. The insert of the resulting plasmid pME8317 was checked by DNA sequencing. The Ptac/lacIq-dctB‘-’fitF construct was then integrated into the chromosome of the ΔfitF mutant of CHA1163 (CHA1174) using the mini-Tn7 delivery system, yielding strain CHA5093 (Table S2).
Analogously, the citA‘-’fitF chimera was constructed with primer pairs citA-F-SpeI/citA-R-overlap and fitFc-F2/fitF-R-HindIII using genomic DNA from E. coli K-12 and P. protegens CHA0, respectively, as a template. The resulting plasmid pME8354 was used to create strain CHA5151 (Table S2).
Quantification of Fit toxin expression in batch cultures using GFP reporters
For assays with transcriptional reporter strains, GFP fluorescence was measured with a BMG FLUOstar Galaxy multidetection microplate reader as detailed previously [7], [38].
Quantification of Fit toxin expression in batch cultures by epifluorescence microscopy
Bacterial strains were grown overnight in 10 ml of LB at 25°C and 180 rpm. Bacterial cells were washed once in 0.9% NaCl solution and the optical density at 600 nm was adjusted to 1, if not otherwise specified. Ten milliliters of the respective medium (LB, BHI, marine broth, FBS, M9 L-malate, or GIM) in 50-ml Erlenmeyer flasks was inoculated 1∶100 with the bacterial suspension and incubated for 8 h (exponential growth phase) and 24 h (stationary growth phase) at 25°C and 180 rpm. Quantification of red fluorescence intensities of single cells by epifluorescence microscopy was performed as described previously [7]. Exposure times were 2 sec for the DsRed channel and 80 msec for the Ph3 channel. The CHA0 wild-type strain was used to correct for autofluorescence of the bacterial cells.
Bacterial infection of insects and monitoring of Fit toxin expression by epifluorescence microscopy
Injection assays for virulence determination using last-instar larvae of G. mellonella (Reptile-food.ch GmbH, Dübendorf, Switzerland) were performed as described before [7]. For complementation assays, IPTG was added to the inoculi to a final concentration of 1 mM. Reporter strains of P. protegens CHA0 were in injected in and extracted from forth instar larvae of S. littoralis (Syngenta Crop Protection, Stein, Switzerland) and last instar larvae of T. molitor (The Animal House, Zuzwil, Switzerland) as described before for G. mellonella [7]. A. pisum (The Animal House) was infected with reporter strains of P. protegens CHA0 by placing 20 adult individuals in a small Petri dish on leaves of white beans (Phaseolus vulgaris) that contained drops of bacterial suspensions (at a concentration of 108 cfu per ml, 100 µl per dish). After three days of incubation at room temperature, adult aphids were shock frozen in liquid nitrogen, surface-sterilized with 70% ethanol for 2 min and hemolymph was extracted by crushing them on microscope slides. Extracted hemolymph was fixed on 1% agarose pads placed on microscope slides and observed by epifluorescence microscopy as described previously [7].
Monitoring of Fit toxin expression on roots by epifluorescence microscopy
Visualization of Fit toxin expression on tomato (Solanum lycopersicum cv. Marmande) and wheat (Triticum aestivum cv. Arina) roots was performed as described previously for cucumber [7]. Infection of tomato roots with the crown and root rot pathogen Fusarium oxysporum f. sp. radicis-lycopersici isolate Forl22 was done as detailed elsewhere [39]. Fit toxin expression on cucumber (Cucumis sativus cv. Chinese Snake) roots with the DctBp-FitFc chimera was studied as follows. Cucumber seedlings were grown axenically for three days at room temperature in the dark and inoculated with different reporter strains of P. protegens CHA0 by placing them for 30 min in bacterial suspension, which was prepared from an overnight culture in LB by washing them once in saline solution and adjusting the optical density at 600 nm to 1. The seedlings were then placed into 50-ml tubes (three plants per tube) containing 35-ml of 0.35% (w/v) water agar supplemented with 0.1 mM IPTG, 125 µg/ml tetracycline and 10 µg/ml gentamicin if necessary. The tubes were wrapped in aluminum foil for the lower part to protect roots from light and incubated in a growth chamber set to 80% relative humidity for 16 h with light (160 µE/m2/s) at 22°C, followed by an 8-h dark period at 18°C. After incubation for five days, roots were individually removed, cut into smaller pieces and placed into Eppendorf tubes containing 100 µl of saline solution supplemented with 0.1% Silwet L-77 for the isolation of the bacteria (GE Bayer Silicones Sàrl, Switzerland). The mixture was vigorously agitated for 2 min and 5 µl were used for epifluorescence microscopy as described above. Quantification of single cell fluorescence was performed by using the GFP (2 sec exposure time) and DsRed (2 sec exposure time) channels.
Bioinformatics
Homologs of the periplasmic domains of P. protegens FitF were identified from the NCBI nonredundant protein sequence database using PSI-BLAST and an E-value cutoff of 1e-12 [40]. Periplasmic regions of membrane-bound proteins were determined by predicting transmembrane regions using DAS [41] and PRED-TMR (http://athina.biol.uoa.gr/PRED-TMR/input.html). Functional domains of proteins were predicted using the NCBI Conserved Domain Search [42] and SMART [43] with default parameters. Multiple sequence alignments including sequences from reference proteins with known functions were performed with MAFFT version 7 (http://mafft.cbrc.jp/alignment/server) and phylogenetic analyses were conducted in MEGA5 using the Minimum Evolution method for inferring the evolutionary history [44]. Cluster analyses were performed with CLANS [45] as described earlier [46] using 2D clustering with default parameters.
Secondary and tertiary structure predictions of the periplasmic region of FitF were performed using ESyPred3D [47], I-TASSER [48], LOMETS [49], Phyre2 [50], SABLE (http://sable.cchmc.org), and SWISS-MODEL [51] using default parameters and the crystal structure of the V. cholerae DctB sensor domain (3BY9) as template if required. Structure models were visualized using the Swiss-PdbViewer version 4.0.3 (http://spdbv.vital-it.ch).
Statistical analysis
Significant differences between treatments or strains were calculated in R version 2.13.1 (http://www.r-project.org) by one-way or two-way analysis of variance (ANOVA) with Tukey's HSD test for post-hoc comparisons. The Log-Rank test of the Survival package of R was used to calculate significant differences in insect toxicity between P. protegens CHA0 and isogenic mutant strains in the Galleria injection assay.
Supporting Information
Zdroje
1. HaasD, DéfagoG (2005) Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat Rev Microbiol 3 : 307–319.
2. RametteA, FrapolliM, Fischer-Le SauxM, GruffazC, MeyerJM, et al. (2011) Pseudomonas protegens sp nov., widespread plant-protecting bacteria producing the biocontrol compounds 2,4-diacetylphloroglucinol and pyoluteorin. Syst Appl Microbiol 34 : 180–188.
3. KupferschmiedP, MaurhoferM, KeelC (2013) Promise for plant pest control: root-associated pseudomonads with insecticidal activities. Front Plant Sci 4 : 287.
4. LoperJE, HassanKA, MavrodiDV, DavisEW, LimCK, et al. (2012) Comparative genomics of plant-associated Pseudomonas spp.: insights into diversity and inheritance of traits involved in multitrophic nteractions. PLoS Genet 8: e1002784.
5. RuffnerB, Péchy-TarrM, RyffelF, HoeggerP, ObristC, et al. (2013) Oral insecticidal activity of plant-associated pseudomonads. Environ Microbiol 15 : 751–763.
6. Péchy-TarrM, BruckDJ, MaurhoferM, FischerE, KeelC (2008) Molecular analysis of a novel gene cluster encoding an insect toxin in plant-associated strains of Pseudomonas fluorescens. Environ Microbiol 10 : 2368–2386.
7. Péchy-TarrM, BorelN, KupferschmiedP, TurnerV, BinggeliO, et al. (2013) Control and host-dependent activation of insect toxin expression in a root-associated biocontrol pseudomonad. Environ Microbiol 15 : 736–750.
8. GaoR, StockAM (2009) Biological insights from structures of two-component proteins. Annu Rev Microbiol 63 : 133–154.
9. KrellT, LacalJ, BuschA, Silva-JimenezH, GuazzaroniME, et al. (2010) Bacterial sensor kinases: diversity in the recognition of environmental signals. Annu Rev Microbiol 64 : 539–559.
10. AlmE, HuangK, ArkinA (2006) The evolution of two-component systems in bacteria reveals different strategies for niche adaptation. PLoS Comput Biol 2: e143.
11. GalperinMY (2005) A census of membrane-bound and intracellular signal transduction proteins in bacteria: bacterial IQ, extroverts and introverts. BMC Microbiol 5 : 35.
12. CapraEJ, LaubMT (2012) Evolution of two-component signal transduction systems. Annu Rev Microbiol 66 : 325–347.
13. RaghavanV, GroismanEA (2010) Orphan and hybrid two-component system proteins in health and disease. Curr Opin Microbiol 13 : 226–231.
14. JungK, FriedL, BehrS, HeermannR (2012) Histidine kinases and response regulators in networks. Curr Opin Microbiol 15 : 118–124.
15. ScheuPD, KimOB, GriesingerC, UndenG (2010) Sensing by the membrane-bound sensor kinase DcuS: exogenous versus endogenous sensing of C(4)-dicarboxylates in bacteria. Future Microbiol 5 : 1383–1402.
16. CheungJ, HendricksonWA (2008) Crystal structures of C-4-dicarboxylate ligand complexes with sensor domains of histidine kinases DcuS and DctB. J Biol Chem 283 : 30256–30265.
17. ZhouYF, NanBY, NanJ, MaQJ, PanjikarS, et al. (2008) C(4)-dicarboxylates sensing mechanism revealed by the crystal structures of DctB sensor domain. J Mol Biol 383 : 49–61.
18. CheungJ, HendricksonWA (2010) Sensor domains of two-component regulatory systems. Curr Opin Microbiol 13 : 116–123.
19. ZhangW, ShiL (2005) Distribution and evolution of multiple-step phosphorelay in prokaryotes: lateral domain recruitment involved in the formation of hybrid-type histidine kinases. Microbiology 151 : 2159–2173.
20. ChangC, TesarC, GuM, BabniggG, JoachimiakA, et al. (2010) Extracytoplasmic PAS-like domains are common in signal transduction proteins. J Bacteriol 192 : 1156–1159.
21. HenryJT, CrossonS (2011) Ligand-binding PAS domains in a genomic, cellular, and structural context. Annu Rev Microbiol 65 : 261–286.
22. StephensonK, HochJA (2002) Evolution of signalling in the sporulation phosphorelay. Mol Microbiol 46 : 297–304.
23. RiechmannL, WinterG (2000) Novel folded protein domains generated by combinatorial shuffling of polypeptide segments. Proc Natl Acad Sci U S A 97 : 10068–10073.
24. MöglichA, AyersRA, MoffatK (2010) Addition at the molecular level: signal integration in designed Per-ARNT-Sim receptor proteins. J Mol Biol 400 : 477–486.
25. ChecaSK, ZurbriggenMD, SonciniFC (2012) Bacterial signaling systems as platforms for rational design of new generations of biosensors. Curr Opin Biotechnol 23 : 766–772.
26. GraceTD (1962) Establishment of four strains of cells from insect tissues grown in vitro. Nature 195 : 788–789.
27. ShenX, HuH, PengH, WangW, ZhangX (2013) Comparative genomic analysis of four representative plant growth-promoting rhizobacteria in Pseudomonas. BMC Genomics 14 : 271.
28. NanBY, LiuX, ZhouYF, LiuJW, ZhangL, et al. (2010) From signal perception to signal transduction: ligand-induced dimeric switch of DctB sensory domain in solution. Mol Microbiol 75 : 1484–1494.
29. ZhangZ, HendricksonWA (2010) Structural characterization of the predominant family of histidine kinase sensor domains. J Mol Biol 400 : 335–353.
30. EtzkornM, KneuperH, DunnwaldP, VijayanV, KramerJ, et al. (2008) Plasticity of the PAS domain and a potential role for signal transduction in the histidine kinase DcuS. Nat Struct Mol Biol 15 : 1031–1039.
31. ValentiniM, StorelliN, LapougeK (2011) Identification of C(4)-dicarboxylate transport systems in Pseudomonas aeruginosa PAO1. J Bacteriol 193 : 4307–4316.
32. PetrovaOE, SauerK (2009) A novel signaling network essential for regulating Pseudomonas aeruginosa biofilm development. PLoS Pathog 5: e1000668.
33. QianW, HanZJ, HeC (2008) Two-component signal transduction systems of Xanthomonas spp.: a lesson from genomics. Mol Plant Microbe Interact 21 : 151–161.
34. Sambrook J, Russel DW (2001) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor (New York): Cold Spring Harbor Laboratory Press.
35. Schnider-KeelU, SeematterA, MaurhoferM, BlumerC, DuffyB, et al. (2000) Autoinduction of 2,4-diacetylphloroglucinol biosynthesis in the biocontrol agent Pseudomonas fluorescens CHA0 and repression by the bacterial metabolites salicylate and pyoluteorin. J Bacteriol 182 : 1215–1225.
36. Défago G, Haas D (1990) Pseudomonads as antagonists of soilborne plant pathogens: modes of action and genetic analysis. In: Bollag JM, Stotzky G, editors. Soil Biochemistry. New York, USA: Marcel Dekker. pp. 249–291.
37. Martinez-GarciaE, de LorenzoV (2011) Engineering multiple genomic deletions in Gram-negative bacteria: analysis of the multi-resistant antibiotic profile of Pseudomonas putida KT2440. Environ Microbiol 13 : 2702–2716.
38. BaehlerE, BottiglieriM, Pechy-TarrM, MaurhoferM, KeelC (2005) Use of green fluorescent protein-based reporters to monitor balanced production of antifungal compounds in the biocontrol agent Pseudomonas fluorescens CHA0. J Appl Microbiol 99 : 24–38.
39. Sharifi-TehraniA, ZalaM, NatschA, Moenne-LoccozY, DefagoG (1998) Biocontrol of soil-borne fungal plant diseases by 2,4-diacetylphloroglucinol-producing fluorescent pseudomonads with different restriction profiles of amplified 16S rDNA. Eur J Plant Pathol 104 : 631–643.
40. AltschulSF, MaddenTL, SchafferAA, ZhangJ, ZhangZ, et al. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25 : 3389–3402.
41. CserzoM, WallinE, SimonI, von HeijneG, ElofssonA (1997) Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng 10 : 673–676.
42. Marchler-BauerA, LuS, AndersonJB, ChitsazF, DerbyshireMK, et al. (2011) CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res 39: D225–229.
43. SchultzJ, MilpetzF, BorkP, PontingCP (1998) SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci U S A 95 : 5857–5864.
44. TamuraK, PetersonD, PetersonN, StecherG, NeiM, et al. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28 : 2731–2739.
45. FrickeyT, LupasA (2004) CLANS: a Java application for visualizing protein families based on pairwise similarity. Bioinformatics 20 : 3702–3704.
46. KrämerJ, FischerJD, ZientzE, VijayanV, GriesingerC, et al. (2007) Citrate sensing by the C-4-dicarboxylate/citrate sensor kinase DcuS of Escherichia coli: Binding site and conversion of DcuS to a C-4-dicarboxylate - or citrate-specific sensor. J Bacteriol 189 : 4290–4298.
47. LambertC, LeonardN, De BolleX, DepiereuxE (2002) ESyPred3D: Prediction of proteins 3D structures. Bioinformatics 18 : 1250–1256.
48. RoyA, KucukuralA, ZhangY (2010) I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc 5 : 725–738.
49. WuS, ZhangY (2007) LOMETS: a local meta-threading-server for protein structure prediction. Nucleic Acids Res 35 : 3375–3382.
50. KelleyLA, SternbergMJ (2009) Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc 4 : 363–371.
51. ArnoldK, BordoliL, KoppJ, SchwedeT (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22 : 195–201.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Structure of the Membrane Anchor of Pestivirus Glycoprotein E, a Long Tilted Amphipathic HelixČlánek Iron Acquisition in : The Roles of IlsA and Bacillibactin in Exogenous Ferritin Iron MobilizationČlánek AvrBsT Acetylates ACIP1, a Protein that Associates with Microtubules and Is Required for ImmunityČlánek Viral MicroRNA Effects on Pathogenesis of Polyomavirus SV40 Infections in Syrian Golden HamstersČlánek Genome-Wide RNAi Screen Identifies Broadly-Acting Host Factors That Inhibit Arbovirus Infection
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2014 Číslo 2- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Viral Enhancer Mimicry of Host Innate-Immune Promoters
- The Epstein-Barr Virus-Encoded MicroRNA MiR-BART9 Promotes Tumor Metastasis by Targeting E-Cadherin in Nasopharyngeal Carcinoma
- Implication of PMLIV in Both Intrinsic and Innate Immunity
- The Consequences of Reconfiguring the Ambisense S Genome Segment of Rift Valley Fever Virus on Viral Replication in Mammalian and Mosquito Cells and for Genome Packaging
- Substrate-Induced Unfolding of Protein Disulfide Isomerase Displaces the Cholera Toxin A1 Subunit from Its Holotoxin
- Male-Killing Induces Sex-Specific Cell Death via Host Apoptotic Pathway
- Highly Active Antiretroviral Therapies Are Effective against HIV-1 Cell-to-Cell Transmission
- The microRNAs in an Ancient Protist Repress the Variant-Specific Surface Protein Expression by Targeting the Entire Coding Sequence
- Transmission-Blocking Antibodies against Mosquito C-Type Lectins for Dengue Prevention
- Type III Secretion Protein MxiI Is Recognized by Naip2 to Induce Nlrc4 Inflammasome Activation Independently of Pkcδ
- Lundep, a Sand Fly Salivary Endonuclease Increases Parasite Survival in Neutrophils and Inhibits XIIa Contact Activation in Human Plasma
- Induction of Type I Interferon Signaling Determines the Relative Pathogenicity of Strains
- Structure of the Membrane Anchor of Pestivirus Glycoprotein E, a Long Tilted Amphipathic Helix
- Foxp3 Regulatory T Cells Delay Expulsion of Intestinal Nematodes by Suppression of IL-9-Driven Mast Cell Activation in BALB/c but Not in C57BL/6 Mice
- Iron Acquisition in : The Roles of IlsA and Bacillibactin in Exogenous Ferritin Iron Mobilization
- MicroRNA Editing Facilitates Immune Elimination of HCMV Infected Cells
- Reversible Silencing of Cytomegalovirus Genomes by Type I Interferon Governs Virus Latency
- Identification of Host-Targeted Small Molecules That Restrict Intracellular Growth
- A Cyclophilin Homology Domain-Independent Role for Nup358 in HIV-1 Infection
- Engagement of NKG2D on Bystander Memory CD8 T Cells Promotes Increased Immunopathology following Infection
- Suppression of RNA Silencing by a Plant DNA Virus Satellite Requires a Host Calmodulin-Like Protein to Repress Expression
- CIB1 Synergizes with EphrinA2 to Regulate Kaposi's Sarcoma-Associated Herpesvirus Macropinocytic Entry in Human Microvascular Dermal Endothelial Cells
- A Gammaherpesvirus Bcl-2 Ortholog Blocks B Cell Receptor-Mediated Apoptosis and Promotes the Survival of Developing B Cells
- Metabolic Reprogramming during Purine Stress in the Protozoan Pathogen
- The Post-transcriptional Regulator / Activates T3SS by Stabilizing the 5′ UTR of , the Master Regulator of Genes, in
- Tailored Immune Responses: Novel Effector Helper T Cell Subsets in Protective Immunity
- AvrBsT Acetylates ACIP1, a Protein that Associates with Microtubules and Is Required for Immunity
- Epstein-Barr Virus Large Tegument Protein BPLF1 Contributes to Innate Immune Evasion through Interference with Toll-Like Receptor Signaling
- The Major Cellular Sterol Regulatory Pathway Is Required for Andes Virus Infection
- Insights into the Initiation of JC Virus DNA Replication Derived from the Crystal Structure of the T-Antigen Origin Binding Domain
- Domain Shuffling in a Sensor Protein Contributed to the Evolution of Insect Pathogenicity in Plant-Beneficial
- Lectin-Like Bacteriocins from spp. Utilise D-Rhamnose Containing Lipopolysaccharide as a Cellular Receptor
- A Compositional Look at the Human Gastrointestinal Microbiome and Immune Activation Parameters in HIV Infected Subjects
- Exploits Asparagine to Assimilate Nitrogen and Resist Acid Stress during Infection
- Interleukin-33 Increases Antibacterial Defense by Activation of Inducible Nitric Oxide Synthase in Skin
- Protective Vaccination against Papillomavirus-Induced Skin Tumors under Immunocompetent and Immunosuppressive Conditions: A Preclinical Study Using a Natural Outbred Animal Model
- Gem-Induced Cytoskeleton Remodeling Increases Cellular Migration of HTLV-1-Infected Cells, Formation of Infected-to-Target T-Cell Conjugates and Viral Transmission
- Viral MicroRNA Effects on Pathogenesis of Polyomavirus SV40 Infections in Syrian Golden Hamsters
- Genome-Wide RNAi Screen Identifies Broadly-Acting Host Factors That Inhibit Arbovirus Infection
- Inflammatory Monocytes Orchestrate Innate Antifungal Immunity in the Lung
- Quantitative and Qualitative Deficits in Neonatal Lung-Migratory Dendritic Cells Impact the Generation of the CD8+ T Cell Response
- Human Genome-Wide RNAi Screen Identifies an Essential Role for Inositol Pyrophosphates in Type-I Interferon Response
- The Master Regulator of the Cellular Stress Response (HSF1) Is Critical for Orthopoxvirus Infection
- Code-Assisted Discovery of TAL Effector Targets in Bacterial Leaf Streak of Rice Reveals Contrast with Bacterial Blight and a Novel Susceptibility Gene
- Competitive and Cooperative Interactions Mediate RNA Transfer from Herpesvirus Saimiri ORF57 to the Mammalian Export Adaptor ALYREF
- The Type III Secretion Chaperone Slc1 Engages Multiple Early Effectors, Including TepP, a Tyrosine-phosphorylated Protein Required for the Recruitment of CrkI-II to Nascent Inclusions and Innate Immune Signaling
- Yeasts: How Many Species Infect Humans and Animals?
- Clustering of Pattern Recognition Receptors for Fungal Detection
- Distinct Antiviral Responses in Pluripotent versus Differentiated Cells
- Igniting the Fire: Virulence Factors in the Pathogenesis of Sepsis
- Inactivation of the Host Lipin Gene Accelerates RNA Virus Replication through Viral Exploitation of the Expanded Endoplasmic Reticulum Membrane
- Inducible Deletion of CD28 Prior to Secondary Infection Impairs Worm Expulsion and Recall of Protective Memory CD4 T Cell Responses
- Clonal Expansion during Infection Dynamics Reveals the Effect of Antibiotic Intervention
- The Secreted Triose Phosphate Isomerase of Is Required to Sustain Microfilaria Production
- Unifying Viral Genetics and Human Transportation Data to Predict the Global Transmission Dynamics of Human Influenza H3N2
- ‘Death and Axes’: Unexpected Ca Entry Phenologs Predict New Anti-schistosomal Agents
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Reversible Silencing of Cytomegalovirus Genomes by Type I Interferon Governs Virus Latency
- Implication of PMLIV in Both Intrinsic and Innate Immunity
- Transmission-Blocking Antibodies against Mosquito C-Type Lectins for Dengue Prevention
- Lundep, a Sand Fly Salivary Endonuclease Increases Parasite Survival in Neutrophils and Inhibits XIIa Contact Activation in Human Plasma
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



