-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaPathogenic Yeasts Deploy Cell Surface Receptors to Acquire Iron in Vertebrate Hosts
article has not abstract
Published in the journal: . PLoS Pathog 9(8): e32767. doi:10.1371/journal.ppat.1003498
Category: Pearls
doi: https://doi.org/10.1371/journal.ppat.1003498Summary
article has not abstract
Introduction
Brown et al. (2012) [1] recently highlighted the growing threat that fungal pathogens pose for humans, as well as the pressing need for additional antifungal drugs and efficacious vaccines. In this context, the process of iron acquisition presents compelling opportunities to prevent or treat fungal diseases because iron is an essential nutrient for pathogen proliferation in vertebrate hosts. Fungi and other pathogens must deploy competitive uptake mechanisms to steal iron from host sources and overcome the iron sequestration associated with nutritional immunity [2]. These extracellular and surface uptake functions may provide readily accessible targets for drugs to block iron uptake, and iron transporters might also be exploited to introduce antifungal agents into fungal cells [3], [4]. Additionally, extracellular or exposed transporters may be useful vaccine targets to block iron uptake and pathogen proliferation.
Mechanisms of iron acquisition have been well characterized in many microbial pathogens, and information is rapidly accumulating for fungal pathogens [5]–[9]. Fungi generally acquire iron by several mechanisms including: 1) the production and uptake of siderophores; 2) the use of a ferroxidase-iron permease complex for high-affinity uptake; 3) the transport of ferrous iron; and 4) the acquisition of iron from heme and hemoglobin [7]–[9]. Reductases in the plasma membrane and secreted reductants facilitate the reduction of ferric iron to ferrous iron for high - or low-affinity uptake [7]–[9]. The exploration of these mechanisms in fungal pathogens has revealed intriguing connections between cell surface molecules (cell wall and secreted proteins, capsular polysaccharide, and biofilms) and functions for iron acquisition from vertebrate sources (Figure 1A, B). Here we highlight these connections for the pathogenic yeasts Candida albicans and Cryptococcus neoformans. We also discuss the extent to which the newly identified functions add depth to our understanding of iron acquisition by fungi and illustrate the complex integration of iron sensing and virulence.
Fig. 1. Cell surface functions for iron acquisition in C. albicans and C. neoformans. 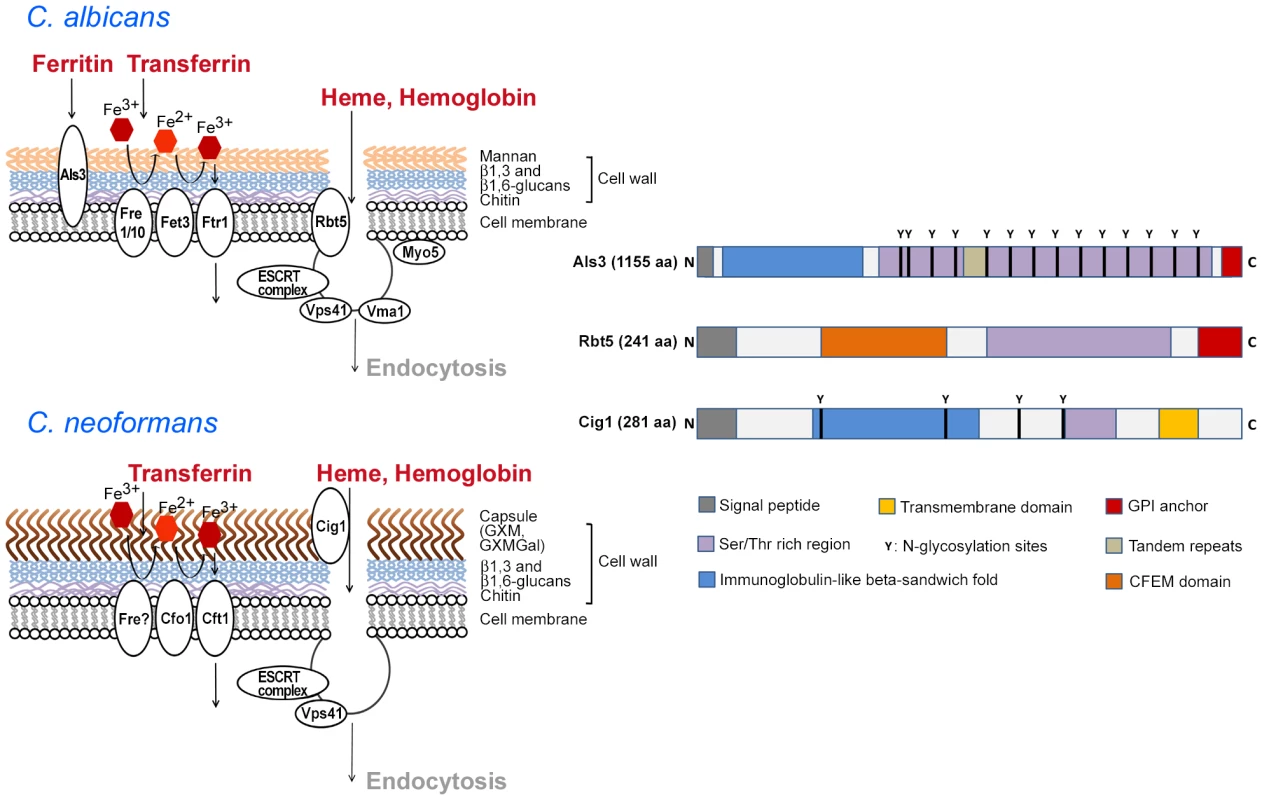
The diagrams on the left show the polysaccharides and iron-related uptake functions at the fungal cell surface. For C. albicans, the iron-uptake proteins include the ferritin-binding protein Als3, ferric reductases (Fre1/10), a multicopper oxidase (Fet3), and an iron permease (Ftr1) [8]–[10]. The components of a heme uptake pathway include the receptor Rbt5 and some of the proteins involved in internalization [18], [19]. These proteins are depicted schematically as contributing to endocytosis of heme and/or hemoglobin. For C. neoformans, surface polysaccharides are shown for the capsule (GXM, GXMGal) and the cell wall, and iron-uptake functions include putative ferric reductase activity (Fre), a multicopper oxidase (Cfo1), and an iron permease (Cft1) [7], [37]. The role of Cig1 and known or hypothesized functions for endocytosis are also shown in a schematic depiction [27], [30]. The diagrams on the right present the structures of Als3 [10], Rbt5 [17]–[19], and Cig1 ([27]; B. Cadieux, unpublished) to depict shared and distinct domains. Note that the polypeptides are not draw to scale and the actual amino acid length of each is indicated. Molecular modeling suggests that the immunoglobulin-like fold of Als3 may interact with cadherins [10]. However, the structural features that contribute to iron acquisition have not been defined for any of the proteins. Cell Surface Proteins Link the Use of Ferritin, Heme, and Hemoglobin with Biofilm Formation in C. albicans
The adhesin Als3 and the CFEM domain protein Rbt5 are two notable examples of proteins that link cell surface activities related to morphogenesis and/or biofilm formation with iron acquisition in C. albicans (Figure 1C). Als3 is a hypha-specific surface protein that functions as an adhesin for epithelial and endothelial cells, and that also mediates adherence to extracellular matrix proteins (reviewed in [10]). The ability of C. albicans to switch its morphology between yeast and hyphal forms is important for virulence, as is the ability of the fungus to form biofilms on implanted medical devices. Als3 contributes to virulence in mouse models of candidiasis, although the impact of the protein depends on the method of inoculation and the immune status of the host [10]. Expression of ALS3 is transcriptionally controlled in a complex manner by regulators of the yeast-hyphal transition, by the major regulator of biofilm formation Bcr1, and by the alkaline response transcription factor Rim101 [10]–[14].
Als3 is an interesting multifunctional protein because, in addition to binding to and provoking endocytosis of the fungus by host cells, it also plays a role in biofilm formation and it binds the host iron-storage protein ferritin. Specifically, Almeida et al. (2008) [15] found that C. albicans can use ferritin as an iron source at physiological pH, and that Als3 is required for this process. Ferritin is bound by hyphal cells that express Als3 (but not by yeast cells), and deletion of the ALS3 alleles eliminates both binding and growth on ferritin. The ferritin binding by hyphae is also observed when these cells invade epithelial cells in vitro, and Als3 is required for C. albicans to damage these cells [15]. Overall, Als3 functionally links biofilm formation, the yeast-hyphal transition, and the use of a specific host iron source.
The second example of the connection between biofilm formation, morphology, and C. albicans iron acquisition involves Rbt5, an O-mannosylated, glycosylphosphatidylinositol (GPI)-anchored protein (Figure 1C) [16]–[18]. Rbt5 is located in the plasma membrane and the cell wall, the protein binds heme and hemoglobin, and deletion of RBT5 reduces the use of these host iron sources by C. albicans [16]–[18]. RBT5 expression is induced by iron limitation and negatively regulated by the Tup1 regulator of morphology [17], [19]. Related genes encoding CFEM domain proteins are present in C. albicans, and these include RBT51/PGA10, CSA1, CSA2, and PGA7. Rbt51 also participates in hemoglobin binding and, in fact, the protein was originally identified in a screen for C. albicans genes that allowed S. cerevisiae to use iron from hemoglobin [17]. Weissman et al. (2008) [18] exploited this property of Rbt51 to identify mutations in S. cerevisiae that blocked hemoglobin use. The identified functions included subunits of the vacuolar ATPase, components of the ESCRT system, HOPS complex proteins and t-SNAREs, and several other functions. Notably, the ESCRT and HOPS complexes, and the t-SNAREs, contribute to endocytosis. An examination of C. albicans mutants with defects in the corresponding genes confirmed that many of these functions played roles in iron use from hemoglobin (e.g., Vma11, Vps41, ESCRT complex components, Myo5). Overall, these studies revealed an endocytic pathway for heme/hemoglobin internalization and trafficking to the vacuole for processing.
Similar to Als3, mutants lacking Rbt5, Rbt51, or the related protein Csa1 form thinner and more fragile biofilms with less extracellular matrix compared with the wild-type strain [20], [21]. These mutants also displayed changes in their cell surface as examined microscopically and by measurements of cell surface hydrophobicity. Interestingly, conserved transcriptional regulation by the biofilm regulator Bcr1 is observed for three CFEM genes in C. albicans (RBT5, PGA7, and CSA1) and for three genes (CFEM2, CFEM3, and CFEM6) in the related pathogen Candida parapsilosis [22]. However, these proteins have divergent roles in that the three CFEM proteins are not involved in biofilm formation in C. parapsilosis [22].
Links between the Cell Surface and Heme Uptake in C. neoformans
The role of extracellular proteins in iron acquisition and connections with the cell surface extends to C. neoformans. In this fungus, the CIG1 gene was identified by transcriptional profiling as the most abundant transcript in iron-starved cells, and the protein had previously been identified as an extracellular cytokine-inducing glycoprotein (Figure 1C) [23], [24]. Glycoproteins (mannoproteins) associated with the polysaccharide capsule have been proposed as vaccine candidates for C. neoformans [24], [25]. The polysaccharide capsule is a major virulence factor for C. neoformans, and it has long been known that the fungus produces a large capsule upon iron deprivation and a small capsule in iron-replete conditions [26]. Interestingly, disruption of the CIG1 gene in a strain with the D capsular serotype resulted in altered capsule regulation by iron [23]. More recently, it was found that deletion of CIG1 in both serotype A or D strains resulted in delayed growth on heme as the sole iron source ([27]; B. Cadieux, unpublished). This growth delay was observed at physiological pH but not at acidic pH, and this result was consistent with the observation of O'Meara et al. (2010) [28] that the pH-responsive transcription factor Rim101 activates expression of CIG1. Furthermore, a rim101 mutant also had delayed growth on heme [27].
Cig1 may participate in an uptake mechanism that involves heme accumulation at the cell surface because recombinant Cig1 binds heme, suggesting that it may be a hemophore [27]. A hemophore would potentially allow access to heme, which contains the majority of the iron present in vertebrate hosts. A role in heme uptake is supported by the finding that a cig1 mutant displays reduced susceptibility to non-iron metalloporphyrins such as gallium protoporphyrin IX (Ga-PPIX). In bacterial pathogens, these compounds rely on heme uptake to cause toxicity, and they serve as reagents to distinguish heme uptake from processing at the cell surface [29]. Functions downstream of Cig1 that appear to participate in heme uptake may include components of the ESCRT pathway because the ESCRT-I protein Vps23 is required for heme uptake, susceptibility to Ga-PPIX, capsule attachment, and virulence [30]. In this case, ESCRT complexes may contribute to endocytosis and intracellular trafficking of heme in C. neoformans, as was found in C. albicans.
A mutant lacking Cig1 was equivalent in virulence to the wild-type parental strain in a mouse inhalation model of cryptococcosis, and a contribution to virulence was only revealed when the cig1 mutation was combined with a mutation in the CFO1 gene encoding a ferroxidase for high-affinity uptake. This latter finding reinforces a key conclusion about iron acquisition and virulence: pathogens appear to have multiple redundant mechanisms to acquire iron because of the critical requirement for the metal and the need to overcome host defense by sequestration. However, it is curious that Cig1 makes a positive contribution to virulence while loss of its regulator, Rim101, results in hypervirulence. O'Meara et al. (2013) [31] recently investigated the virulence contribution of Rim101 in more detail and found that the protein regulates the composition of the cell wall, leading to increased stimulation of the host immune response. Part of the surface change in a rim101 mutant includes release of approximately four times more capsule polysaccharide into the culture medium than the wild-type strain, and this may result from changes in the cell wall that reduce attachment. The regulation of cell wall functions by Rim101 has been observed previously in other fungi [14].
Perspective
The integration of iron sensing, virulence factor expression, and biofilm formation has been previously documented in bacterial pathogens such as Pseudomonas aeruginosa [32]. The studies outlined above reveal that the pathogenic yeasts C. albicans and C. neoformans also have intriguing connections between mechanisms to sense and regulate the response to host conditions, acquire iron from host sources, and remodel cell surface features. In particular, a growing list of proteins have been found to contribute to iron acquisition and to also influence non-protein surface components such as cell wall glucans, capsular polysaccharides, and perhaps the extracellular matrix in biofilms. These contributions may simply indicate that the proteins have more than one function or there may be underlying functional implications. In the latter case, it is interesting to speculate that one role of the capsule polysaccharide in C. neoformans and the biofilm-associated extracellular matrix in C. albicans may be to sequester iron and other nutrients close to the cell surface to facilitate uptake. This idea is supported by a growing body of evidence that extracellular polysaccharides bind small molecules. For example, β-1,3 glucans in C. albicans biofilms bind antifungal drugs, and glucuronoxylomannan in the C. neoformans capsule binds divalent cations [33]–[35]. Of course, it is also possible that pathogens must coincidently express iron-uptake functions to counteract possible interference of polysaccharides with iron uptake. The challenge now is to find approaches to test the positive and (potentially) negative contributions of the extracellular matrix in C. albicans and the polysaccharide capsule in C. neoformans for acquisition of iron and other nutrients. The ongoing identification of mutants with defects in polysaccharide synthesis, modification, and delivery will provide opportunities to meet this challenge [36], [37]. Furthermore, more detailed studies are needed to test the relationship between iron and biofilm formation by C. albicans. Hameed et al. (2008) [38] suggested that iron deprivation does not influence biofilm formation because the wild-type strain and an ftr1 (iron permease) mutant were still able to form biofilms in the presence of an iron chelator. However, more work is needed because the mutant may not have been sufficiently starved for iron given that it still grew under the conditions tested.
Finally, we note that the iron-acquisition proteins are particularly good candidate vaccine targets because of their contributions to more than one virulence-related function. In this context, the CFEM proteins may be useful vaccine targets for C. albicans because Mochon et al. (2010) [39] found Rbt5 and Csa1 (and a ferric reductase) among 33 antigens recognized by sera from convalescent candidemia patients. Als3 is also a promising vaccine candidate for C. albicans because recombinant Als3 protects immunocompetent mice from candidiasis [10], [16].
Zdroje
1. BrownGD, DenningDW, GowNA, LevitzSM, NeteaMG, et al. (2012) Hidden killers: human fungal infections. Sci Transl Med 4 : 165rv13 doi:10.1126/scitranslmed.3004404
2. JohnsonEE, Wessling-ResnickM (2012) Iron metabolism and the innate immune response to infection. Microbes Infect 14 : 207–216.
3. FoleyTL, SimeonovA (2012) Targeting iron assimilation to develop new antibacterials. Expert Opin Drug Discov 7 : 831–847.
4. MöllmannU, HeinischL, BauernfeindA, KöhlerT, Ankel-FuchsD (2009) Siderophores as drug delivery agents: application of the “Trojan Horse” strategy. Biometals 22 : 615–624.
5. SchaibleUE, KaufmannSH (2004) Iron and microbial infection. Nat Rev Microbiol 2 : 946–953.
6. SutakR, LesuisseE, TachezyJ, RichardsonDR (2008) Crusade for iron: iron uptake in unicellular eukaryotes and its significance for virulence. Trends Microbiol 16 : 261–268.
7. JungWH, KronstadJW (2008) Iron and fungal pathogenesis: a case study with Cryptococcus neoformans. Cell Microbiol 10 : 277–284.
8. AlmeidaRS, WilsonD, HubeB (2009) Candida albicans iron acquisition within the host. FEMS Yeast Res 9 : 1000–1012.
9. KornitzerD (2009) Fungal mechanisms for host iron acquisition. Curr Opin Microbiol 12 : 377–83.
10. LiuY, FillerSG (2011) Candida albicans Als3, a multifunctional adhesin and invasin. Eukaryot Cell 10 : 168–173.
11. NobileCJ, AndesDR, NettJE, SmithFJJr, YueF, et al. (2006) Critical role of Bcr1-dependent adhesins in C. albicans biofilm formation in vitro and in vivo. PLoS Pathog 2: e63 doi:10.1371/journal.ppat.0020063
12. FanningS, XuW, SolisN, WoolfordCA, FillerSG, et al. (2012) Divergent targets of Candida albicans biofilm regulator Bcr1 in vitro and in vivo. Eukaryot Cell 11 : 896–904.
13. FinkelJS, MitchellAP (2011) Genetic control of Candida albicans biofilm development. Nat Rev Microbiol 9 : 109–118.
14. NobileCJ, SolisN, MyersCL, FayAJ, DeneaultJS, et al. (2008) Candida albicans transcription factor Rim101 mediates pathogenic interactions through cell wall functions. Cell Microbiol 10 : 2180–2196.
15. AlmeidaRS, BrunkeS, AlbrechtA, ThewesS, LaueM, et al. (2008) The hyphal-associated adhesin and invasin Als3 of Candida albicans mediates iron acquisition from host ferritin. PLoS Pathog 4: e1000217 doi:10.1371/journal.ppat.1000217
16. HeilmannCJ, SorgoAG, KlisFM (2012) News from the fungal front: wall proteome dynamics and host–pathogen interplay. PLoS Pathog 8: e1003050 doi:10.1371/journal.ppat.1003050
17. WeissmanZ, KornitzerD (2004) A family of Candida cell surface haem-binding proteins involved in haemin and haemoglobin-iron utilization. Mol Microbiol 53 : 1209–1220.
18. WeissmanZ, ShemerR, ConibearE, KornitzerD (2008) An endocytic mechanism for haemoglobin-iron acquisition in Candida albicans. Mol Microbiol 69 : 201–217.
19. BraunBR, HeadWS, WangMX, JohnsonAD (2000) Identification and characterization of TUP1-regulated genes in Candida albicans. Genetics 156 : 31–44.
20. PérezA, PedrósB, MurguiA, CasanovaM, López-RibotJL, et al. (2006) Biofilm formation by Candida albicans mutants for genes coding fungal proteins exhibiting the eight-cysteine-containing CFEM domain. FEMS Yeast Res 6 : 1074–1084.
21. PérezA, RamageG, BlanesR, MurguiA, CasanovaM, et al. (2011) Some biological features of Candida albicans mutants for genes coding fungal proteins containing the CFEM domain. FEMS Yeast Res 11 : 273–284.
22. DingC, VidanesGM, MaguireSL, GuidaA, SynnottJM, et al. (2011) Conserved and divergent roles of Bcr1 and CFEM proteins in Candida parapsilosis and Candida albicans. PLoS ONE 6: e28151 doi:10.1371/journal.pone.0028151
23. LianTS, SimmerMI, D'SouzaCA, SteenBR, ZuyderduynSD, et al. (2005) Iron-regulated transcription and capsule formation in the fungal pathogen Cryptococcus neoformans. Mol Microbiol 55 : 1452–1472.
24. BiondoC, MancusoG, MidiriA, BombaciM, MessinaL, et al. (2006) Identification of major proteins secreted by Cryptococcus neoformans. FEMS Yeast Res 6 : 645–651.
25. VecchiarelliA (2000) Immunoregulation by capsular components of Cryptococcus neoformans. Med Mycol 38 : 407–417.
26. VartivarianSE, AnaissieEJ, CowartRE, SpriggHA, TinglerMJ, et al. (1993) Regulation of cryptococcal capsular polysaccharide by iron. J Infect Dis 167 : 186–190.
27. CadieuxB, LianT, HuG, WangJ, BiondoC, et al. (2013) The mannoprotein Cig1 supports iron acquisition from heme and virulence in the pathogenic fungus Cryptococcus neoformans. J Infect Dis 207 : 1339–1347.
28. O'MearaTR, NortonD, PriceMS, HayC, ClementsMF, et al. (2010) Interaction of Cryptococcus neoformans Rim101 and protein kinase A regulates capsule. PLoS Pathog 6: e1000776 doi:10.1371/journal.ppat.1000776
29. StojiljkovicI, KumarV, SrinivasanN (1999) Non-iron metalloporphyrins: potent antibacterial compounds that exploit haem/Hb uptake systems of pathogenic bacteria. Mol Microbiol 31 : 429–442.
30. HuG, CazaM, CadieuxB, ChanV, LiuV, et al. (2013) Cryptococcus neoformans requires the ESCRT protein Vps23 for iron acquisition from heme, for capsule formation, and for virulence. Infect Immun 81 : 292–302.
31. O'MearaTR, HolmerSM, SelvigK, DietrichF, AlspaughJA (2013) Cryptococcus neoformans Rim101 is associated with cell wall remodeling and evasion of the host immune responses. mBio 4: e00522–12 doi:10.1128/mBio.00522-12
32. BaninE, VasilML, GreenbergEP (2005) Iron and Pseudomonas aeruginosa biofilm formation. Proc Natl Acad Sci U S A 102 : 11076–11081.
33. VediyappanG, RossignolT, d'EnfertC (2010) Interaction of Candida albicans biofilms with antifungals: transcriptional response and binding of antifungals to beta-glucans. Antimicrob Agents Chemother 54 : 2096–2111.
34. NettJE, SanchezH, CainMT, AndesDR (2010) Genetic basis of Candida biofilm resistance due to drug-sequestering matrix glucan. J Infect Dis 202 : 171–175.
35. RobertsonEJ, WolfJM, CasadevallA (2012) EDTA inhibits biofilm formation, extracellular vesicular secretion, and shedding of the capsular polysaccharide glucuronoxylomannan by Cryptococcus neoformans. Appl Environ Microbiol 78 : 7977–7984.
36. TaffHT, NettJE, ZarnowskiR, RossKM, SanchezH, et al. (2012) A Candida biofilm-induced pathway for matrix glucan delivery: implications for drug resistance. PLoS Pathog 8: e1002848 doi:10.1371/journal.ppat.1002848
37. KumarP, YangM, HaynesBC, SkowyraML, DoeringTL (2011) Emerging themes in cryptococcal capsule synthesis. Curr Opin Struct Biol 21 : 597–602.
38. HameedS, PrasadT, BanerjeeD, ChandraA, MukhopadhyayCK, et al. (2008) Iron deprivation induces EFG1-mediated hyphal development in Candida albicans without affecting biofilm formation. FEMS Yeast Res 8 : 744–755.
39. MochonAB, JinY, KayalaMA, WingardJR, ClancyCJ, et al. (2010) Serological profiling of a Candida albicans protein microarray reveals permanent host-pathogen interplay and stage-specific responses during candidemia. PLoS Pathog 6: e1000827 doi:10.1371/journal.ppat.1000827
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Molecular Cloning and Characterization of Novel Glutamate-Gated Chloride Channel Subunits fromČlánek X-Box Binding Protein 1 (XBP1s) Is a Critical Determinant of Homoserine Lactone-Mediated Apoptosis
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2013 Číslo 8- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Biosecurity Implications of New Technology and Discovery in Plant Virus Research
- Pathogenic Yeasts Deploy Cell Surface Receptors to Acquire Iron in Vertebrate Hosts
- Asparagine Repeats in Proteins: Good for Nothing?
- Role of the Nervous System in the Control of Proteostasis during Innate Immune Activation: Insights from
- Intact Type I Interferon Production and IRF7 Function in Sooty Mangabeys
- Prion Replication Occurs in Endogenous Adult Neural Stem Cells and Alters Their Neuronal Fate: Involvement of Endogenous Neural Stem Cells in Prion Diseases
- Relevance of Trehalose in Pathogenicity: Some General Rules, Yet Many Exceptions
- HIV-1 Superinfection Occurs Less Frequently Than Initial Infection in a Cohort of High-Risk Kenyan Women
- Crystal Structure of the Full-Length Japanese Encephalitis Virus NS5 Reveals a Conserved Methyltransferase-Polymerase Interface
- Quantitative Models of the Dose-Response and Time Course of Inhalational Anthrax in Humans
- Stabilization of Myc through Heterotypic Poly-Ubiquitination by mLANA Is Critical for γ-Herpesvirus Lymphoproliferation
- The Extracellular Matrix Component Psl Provides Fast-Acting Antibiotic Defense in Biofilms
- A Novel Role for Pro-Coagulant Microvesicles in the Early Host Defense against
- Molecular Cloning and Characterization of Novel Glutamate-Gated Chloride Channel Subunits from
- CD36 Recruits αβ Integrin to Promote Cytoadherence of -Infected Erythrocytes
- X-Box Binding Protein 1 (XBP1s) Is a Critical Determinant of Homoserine Lactone-Mediated Apoptosis
- Discovery of Anthelmintic Drug Targets and Drugs Using Chokepoints in Nematode Metabolic Pathways
- Chikungunya Virus 3′ Untranslated Region: Adaptation to Mosquitoes and a Population Bottleneck as Major Evolutionary Forces
- Mucin Gene (PoMuc) Expression: Epigenetic Control to Shape Adaptation to a New Host
- Determinants of GPI-PLC Localisation to the Flagellum and Access to GPI-Anchored Substrates in Trypanosomes
- The Smallest Capsid Protein Mediates Binding of the Essential Tegument Protein pp150 to Stabilize DNA-Containing Capsids in Human Cytomegalovirus
- Understanding Human Variation in Infectious Disease Susceptibility through Clinical and Cellular GWAS
- Human Genetic Susceptibility to Invasive Aspergillosis
- Host Immune Response to Intestinal Amebiasis
- Fungi Infecting Plants and Animals: Killers, Non-Killers, and Cell Death
- Bed Bugs and Infectious Disease: A Case for the Arboviruses
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Discovery of Anthelmintic Drug Targets and Drugs Using Chokepoints in Nematode Metabolic Pathways
- Host Immune Response to Intestinal Amebiasis
- Bed Bugs and Infectious Disease: A Case for the Arboviruses
- Relevance of Trehalose in Pathogenicity: Some General Rules, Yet Many Exceptions
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



