-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaDifferential Function of Lip Residues in the Mechanism and Biology of an Anthrax Hemophore
To replicate in mammalian hosts, bacterial pathogens must acquire iron. The majority of iron is coordinated to the protoporphyrin ring of heme, which is further bound to hemoglobin. Pathogenic bacteria utilize secreted hemophores to acquire heme from heme sources such as hemoglobin. Bacillus anthracis, the causative agent of anthrax disease, secretes two hemophores, IsdX1 and IsdX2, to acquire heme from host hemoglobin and enhance bacterial replication in iron-starved environments. Both proteins contain NEAr-iron Transporter (NEAT) domains, a conserved protein module that functions in heme acquisition in Gram-positive pathogens. Here, we report the structure of IsdX1, the first of a Gram-positive hemophore, with and without bound heme. Overall, IsdX1 forms an immunoglobin-like fold that contains, similar to other NEAT proteins, a 310-helix near the heme-binding site. Because the mechanistic function of this helix in NEAT proteins is not yet defined, we focused on the contribution of this region to hemophore and NEAT protein activity, both biochemically and biologically in cultured cells. Site-directed mutagenesis of amino acids in and adjacent to the helix identified residues important for heme and hemoglobin association, with some mutations affecting both properties and other mutations affecting only heme stabilization. IsdX1 with mutations that reduced the ability to associate with hemoglobin and bind heme failed to restore the growth of a hemophore-deficient strain of B. anthracis on hemoglobin as the sole iron source. These data indicate that not only is the 310-helix important for NEAT protein biology, but also that the processes of hemoglobin and heme binding can be both separate as well as coupled, the latter function being necessary for maximal heme-scavenging activity. These studies enhance our understanding of NEAT domain and hemophore function and set the stage for structure-based inhibitor design to block NEAT domain interaction with upstream ligands.
Published in the journal: . PLoS Pathog 8(3): e32767. doi:10.1371/journal.ppat.1002559
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1002559Summary
To replicate in mammalian hosts, bacterial pathogens must acquire iron. The majority of iron is coordinated to the protoporphyrin ring of heme, which is further bound to hemoglobin. Pathogenic bacteria utilize secreted hemophores to acquire heme from heme sources such as hemoglobin. Bacillus anthracis, the causative agent of anthrax disease, secretes two hemophores, IsdX1 and IsdX2, to acquire heme from host hemoglobin and enhance bacterial replication in iron-starved environments. Both proteins contain NEAr-iron Transporter (NEAT) domains, a conserved protein module that functions in heme acquisition in Gram-positive pathogens. Here, we report the structure of IsdX1, the first of a Gram-positive hemophore, with and without bound heme. Overall, IsdX1 forms an immunoglobin-like fold that contains, similar to other NEAT proteins, a 310-helix near the heme-binding site. Because the mechanistic function of this helix in NEAT proteins is not yet defined, we focused on the contribution of this region to hemophore and NEAT protein activity, both biochemically and biologically in cultured cells. Site-directed mutagenesis of amino acids in and adjacent to the helix identified residues important for heme and hemoglobin association, with some mutations affecting both properties and other mutations affecting only heme stabilization. IsdX1 with mutations that reduced the ability to associate with hemoglobin and bind heme failed to restore the growth of a hemophore-deficient strain of B. anthracis on hemoglobin as the sole iron source. These data indicate that not only is the 310-helix important for NEAT protein biology, but also that the processes of hemoglobin and heme binding can be both separate as well as coupled, the latter function being necessary for maximal heme-scavenging activity. These studies enhance our understanding of NEAT domain and hemophore function and set the stage for structure-based inhibitor design to block NEAT domain interaction with upstream ligands.
Introduction
An important determinant in the outcome of a bacterial infection is how well the invading pathogen can acquire host iron. Hosts with high levels of free iron are more susceptible to infection, and deletion of iron acquisition systems in a wide range of bacterial species generally attenuates virulence [1]. The low free iron concentration in host tissues (10−18–24 M) likely acts as a barrier to efficient bacterial replication [2]. However, pathogenic bacteria have evolved at least two distinct uptake systems to attain iron. One such mechanism is to secrete siderophores, small molecules that chelate ferric iron with very high affinity [3]. The iron-bound siderophore binds to the bacterial surface and specific ferric iron receptors next deliver the iron or iron-siderophore complex into the cell [4]. The genetic deletion of biosynthetic systems that make siderophores, or the surface receptors that recognize siderophores, decreases the virulence of several pathogens, including the Gram-positive bacteria B. anthracis [5] and S. aureus [6].
The second system bacteria employ to attain host iron targets iron-protoporphyrin IX, or heme. Although heme constitutes up to 80% of the bodily iron reserves, free heme is rare. Most heme is tightly bound to hemoproteins such as hemoglobin. Hemoglobin's important role as an oxygen carrier protein means it is in high abundance and thus a target for bacterial iron uptake [7]–[9]. Bacterial proteins that acquire heme from hemoglobin are called hemophores [10]–[13]. Hemophores are generally secreted into the external milieu where they extract heme, via an undefined mechanism, from heme sources such as hemoglobin [14]. The heme-bound (holo) hemophore then delivers its bound iron-porphyrin to a cognate receptor on the bacteria surface, which leads to heme import into the bacterial cell [15], [16]. Heme from hemoglobin can also be attained at the bacterial surface through similar mechanisms involving receptors on the cell wall (Gram positive) or outer membrane (Gram negative). Heme import by bacterial pathogens is important for the establishment or maintenance of infections caused by Bordetella [17], Haemophilus [18], Brucella [19], Vibrio [20], Streptococcus [21], and Staphylococcus [22] species. Further, more recent studies suggest heme is a major determinant in the promotion and severity of bacterial sepsis [23]. Collectively, these studies highlight the important role of heme acquisition during infection of mammalian hosts and support the contention that the inhibition of iron uptake systems is a promising direction for the development of new therapeutics. However, despite this knowledge, no clinically-used antibiotics have been made that directly target bacterial iron import. An understanding of the molecular mechanism by which heme acquisition systems extract, bind, and transfer heme into bacterial cells would be an important first step in creating new drugs to treat deadly infections.
The first discovered hemophore was HasA from the Gram-negative pathogen S. marcescens [10]. HasA is a small ∼15 kDa protein that seems to passively acquire heme from hemoglobin by virtue of its high affinity for the iron-porphyrin [24]. HasA delivers its bound heme to HasR, an outer membrane surface receptor that delivers the heme into the cell [25]. Whereas the discovery of HasA set the precedence for the study of hemophores, recent studies indicate that Gram-positive bacteria also utilize hemophores. B. anthracis, a Gram-positive pathogen that is a potential weapon for bioterrorism, secretes two hemophores (IsdX1 and IsdX2), that acquire heme from hemoglobin and promote bacterial growth in low-iron environments [26]–[28]. IsdX1 transfers its bound heme to IsdC, a surface protein covalently attached to the peptidoglycan of the cell wall [29], [30]. The heme acquisition functions of IsdX1, IsdX2, and IsdC are dependent on the activity of their NEAT (near-iron transporter) domain [26], [29]. This approximately 125 amino acid domain is conserved in several Gram-positive bacteria, and the collective action of several NEATs on the bacterial surface is hypothesized to lead to transfer of heme through the cell wall [27], [31]–[33]. The deletion of genes encoding for NEAT proteins leads to reduction in virulence in both B. anthracis and S. aureus [34]–[36]. Further, vaccination of mice with recombinant proteins containing NEAT domains induces adaptive immunity to staphylococcal infection. [37]–[39]. However, the development of a safe vaccine or a small molecule inhibitor of NEATs will require a detailed understanding of how NEAT proteins perform their various heme transport functions, including a structural appreciation of the residues required for activity. At the heart of this activity is an understanding of how bacterial hemophores like IsdX1 mediate the specific extraction of heme from host hemoglobin, and whether the ability to take up heme is coupled to the process of heme removal from the globin donor. The NEAT domain of IsdX1 is the only known NEAT domain to bind both heme and hemoglobin, as well as acquire heme from hemoglobin, thus making IsdX1 a useful model protein to study the relationship of these processes to one another. To better understand the molecular basis of hemophore activity and provide mechanistic insights into NEAT protein function, we solved the structures of heme free and heme bound IsdX1 and used these structures to understand the functional roles of residues in a small 310-helix in the vicinity of the heme binding site.
Results
The apo and holo structure of IsdX1 highlights residues involved in hemophore function
To gain an appreciation of the residues in IsdX1 necessary for the hemophore activity of this protein, we solved the crystal structure of apo-IsdX1 to 1.8 Å resolution (Table 1). Overall, the backbone structure of IsdX1 is similar to the solved structures of NEAT domains from S. aureus, albeit with a unique surface charge distribution close to the heme binding pocket (Figure S1.) [40]–[44]. The structure consists of an immunoglobulin-like fold with eight β-strands arranged in two antiparallel β-sheets of a β-sandwich (Figure 1A).
Fig. 1. Apo and holo IsdX1 crystal structures. 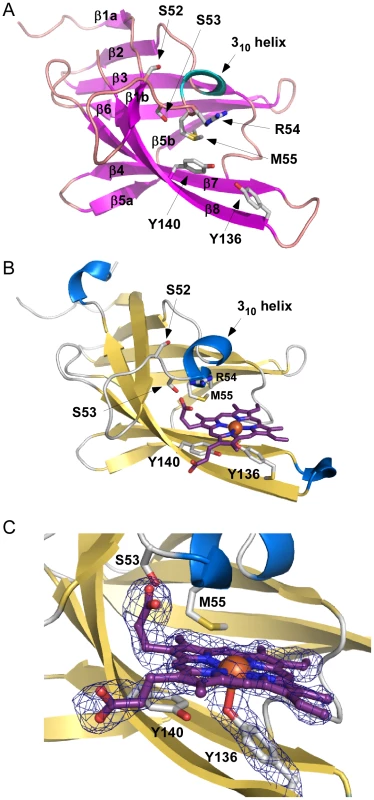
(A) Ribbon representation of apo-IsdX1 where β-strands and helices are colored pink and cyan, respectively. Important heme-binding residues are represented in stick model with carbon, sulfur, nitrogen, and oxygen atoms colored white, yellow, blue and red, respectively. (B) Cartoon representation of holo-IsdX1 where β-strands and helices are colored gold and blue. Heme (purple) is depicted in stick model with carbon (purple), nitrogen (blue), oxygen (red) and Fe (orange sphere). (C) Heme-binding pocket with Fo−Fc omit map contoured at 3.0 σ (dark blue mesh). Hydrogen bonds are indicated by black dashed lines. Tab. 1. Crystallography statistics. 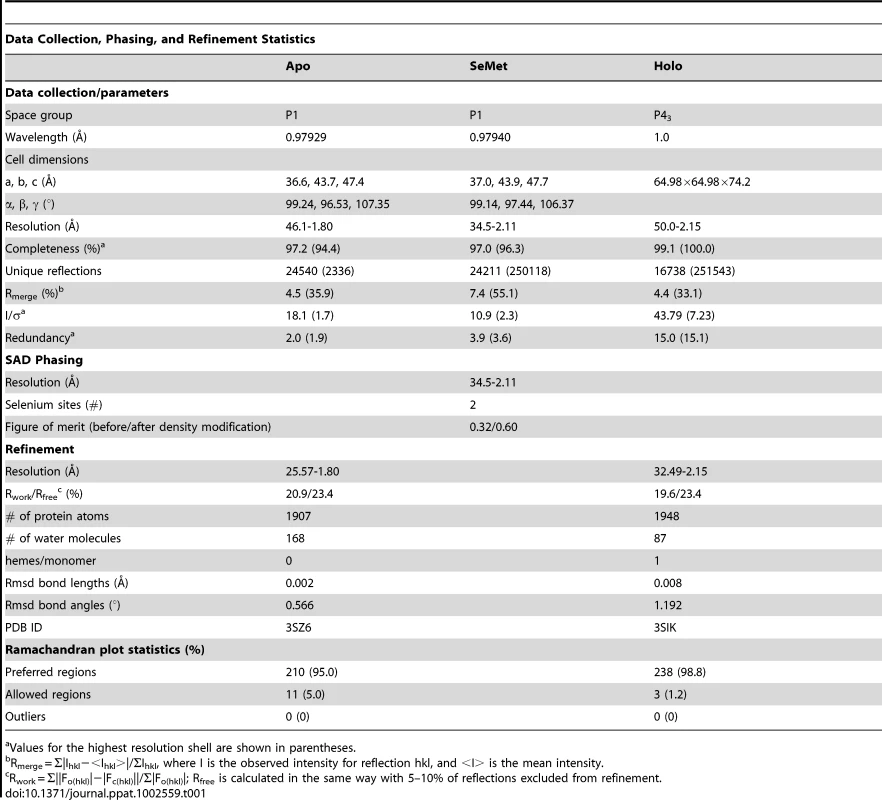
Values for the highest resolution shell are shown in parentheses. The heme-binding pocket is enclosed primarily by a 310-helix (residues Arg-54 to Tyr-58) on one side of the heme and a long β-hairpin (β7–β8) on the other. The 310-helix is sometimes referred to as the “lip” because it seems to protrude over the heme-binding pocket [42]. The backbone of the 310-helix in IsdX1 is fairly well-ordered even in the absence of heme. The integrity of the 310-helix without heme could be due to the hydrogen bonding network between Ser-52, Ser-53, Arg-54 and Asn-56 and Met-55 from the helix making van der Waals contacts with residues in β4, β7, and β8.
The heme bound form of IsdX1 was solved to 2.15 Å by molecular replacement using the structure of apo-IsdX1 as the search model. Heme-iron coordination is achieved through Tyr-136, which is conserved among all heme-binding NEAT domain proteins (Figure S2, upper panel). The distance between the iron atom and the tyrosine oxygen ligand is 2.3 Å, which is typical for an Fe-O bond of NEAT domains (Figures 1B and 1C) [40]–[45]. The coordination bond to iron is stabilized by the conserved residue Tyr-140, which forms a hydrogen bond with the phenolate oxygen of Tyr-136. The aromatic ring of Tyr-140 further stabilizes the pyrrole ring through π-stacking. Both of these residues lie on a β-hairpin region, a conserved region in bacterial NEAT proteins that provides a structural platform for the heme [40], [42]. The least solvent exposed heme propionate forms a hydrogen bond with the hydroxyl group of Ser-53, as well as the backbone nitrogen of Arg-54. Additionally, the side chain NH1 group of Arg-54 from chain A also hydrogen bonds to the least solvent exposed heme propionate, and also to the hydroxyl group of Ser-53 (Figure 2A, B). However, this hydrogen-bonding network is not observed for the side chain of Arg-54 from chain B. The heme molecule is further stabilized by Tyr-58, that weakly π-stacks with the buried heme pyrrole ring. The other residues lining the heme-binding pocket are Arg-54, Met-55, Phe-59, Ile-84, Val-127, Ile-129, Ile-131, Ile-142 and Phe-144, which mainly form an aliphatic environment to accommodate the hydrophobic regions of the heme and its side chains.
Fig. 2. Superimposition of apo-IsdX1 and holo-IsdX1. 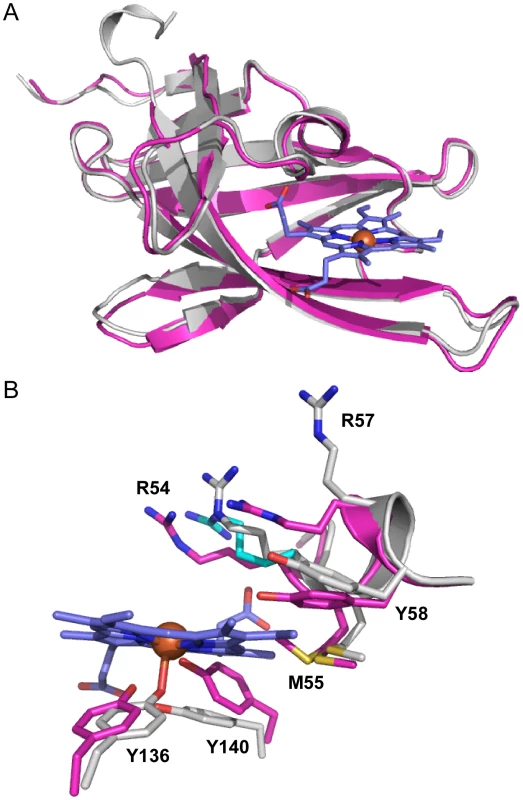
(A) Ribbon representation of superimposition of apo-IsdX1 (pink) and holo-IsdX (grey). (B) Heme-binding pocket with stick representation of heme (carbon, blue and Fe, orange sphere), apo-IsdX1 residues (carbon, pink) and holo-IsdX1 residues (carbon, grey), and sulfur (yellow), nitrogen (blue) and oxygen (red). Arg-54 in the holo structure had two conformations and the alternate conformation has cyan carbon atoms. The role of the 310-helix in heme binding
The location of the 310-helix in the structures of the NEAT domains suggests this region is important for NEAT protein function, including heme binding, hemoglobin association, heme extraction and NEAT-NEAT heme transfer [41]–[43], [45]. There is some conservation in this region, as noted by Pilpa et al, with aromatic residues common in the equivalent positions of amino acids 54 and 58 for IsdX1 [46]. Interestingly, a serine residue (Ser-53 in IsdX1), which is immediately adjacent to the first residue of the helix (Arg-54), is well conserved in these NEATs, including every NEAT domain from B. anthracis (Figure 3, bold) [47]. To determine the role of this and adjacent residues in the ability of IsdX1 to bind heme, each amino acid in 52-SSRM-55 was changed to alanine and mutant proteins purified from E. coli. Whereas wild-type IsdX1 co-purified with a significant amount of endogenous heme from E. coli, IsdX1(SSRM→AAAA) bound approximately 10-fold less heme after purification (Figure 4A, quantitated in 4C). Removal of the bound heme by organic extraction (Figure 4B) and quantitation of the Soret band intensity after titration of the apo protein with hemin confirmed the heme-binding defect of the mutant protein (Figure 4D).
Fig. 3. Alignment of the 310-helix and adjacent residues of NEAT proteins in Gram-positive bacteria. 
ClustalW was used to align the amino acid sequences of the annotated NEAT proteins from the Gram-positive bacterial pathogens Bacillus anthracis, Bacillus cereus, Staphylococcus aureus, Streptococcus pyogenes, Clostridium (botulinum and perfringens), and Listeria monocytogenes. A serine (Ser-53 in IsdX1) is conserved in all nine NEAT domains from B. anthracis and in several NEAT domains from other Gram-positive bacteria. Although an arginine occupies position 54 in IsdX1, a methionine is commonly observed in related NEATs and may serve as a sixth axial ligand to the heme-iron, as described by Gaudin et al [62]. Fig. 4. Functional role of the 310-helix and adjacent residues: heme binding. 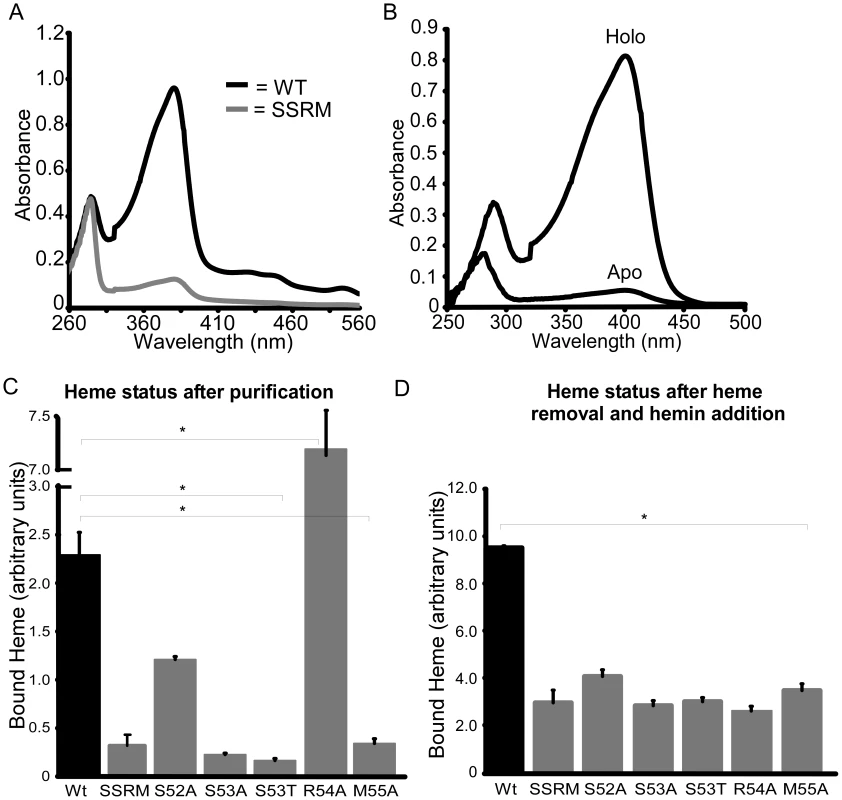
(A) Ser-52, Ser-53, Arg-54, and Met-55 of IsdX1, designated SSRM, were each substituted to alanine and recombinant protein purified from E. coli as described in the Experimental Procedures. The absorbance properties immediately after purification from E. coli of wild-type (black) and SSRM (grey) IsdX1 were analyzed from 260–560 nm. (B) Recombinant IsdX1 was treated with low pH to remove co-purifying heme and the absorbance (250–500 nm) compared to the same preparation that was not acid treated. (C, D) Wild-type IsdX1, IsdX1-SSRM, or IsdX1 harboring mutations in Ser-52, Ser-53, Arg-54, or Met-55 were purified from E. coli and the heme content assessed by determining the ratio of the heme (399 nm) to protein (280 nm) absorbance (referred to as “bound heme”). In (C), the relative amount of associated heme is recorded following the purification of each IsdX1 variant from E. coli. In (D), all endogenous heme was removed from the preparations as described in (B) and apo-proteins incubated with 5 µM heme for 10 minutes at 25°C, followed by absorbance measurements. The absorbance value of a heme-only control (5 µM) was subtracted from all IsdX1 plus heme reaction readings. The values in (C) and (D) represent the mean and standard deviation of three independent experiments. The asterisk (*) means the differences were significant (p<0.05). To determine the residues responsible for this defect, single substitution changes in 52-SSRM-55 were generated and each purified mutant assessed for heme binding activity. As demonstrated in Figure 4C and D, mutation of Ser-52, Ser-53, or Met-55 decreased the ability of each of these mutant proteins to either co-purify with heme (Figure 4C) or bind exogenously added hemin (Figure 4D). The raw spectra for these mutants are illustrated in Figure S3. These effects are not due to gross disruption of IsdX1 secondary structure since the mutant proteins retained a similar overall β-sheet content as the wild-type protein when assessed by far-UV circular dichroism (Figure S4). Further, the heme binding site seems to be somewhat intolerant of even small structural changes, since substitution of Ser-53 to a threonine, which differs only in the length of the side chain (extra methyl group), also abolished the interaction with heme (Figure 4 C,D). Interestingly, mutation of Arg-54 led to a purified IsdX1 preparation with a Soret band approximately three times greater than wild-type protein (Figure 4C). The high heme content in this sample was confirmed using the pyridine hemochrome method, which demonstrated the molar amounts of heme, on average, were 75–90% the molar concentration of protein, suggesting binding was stoichiometric for this preparation (data not shown). However, upon removal of the heme and incubation of R54A with hemin, very little heme bound the protein, despite a far-UV spectrum indistinguishable from wild-type protein (Figure 4D, Figure S4). In the structure of apo and holo-IsdX1, the side chain of Arg-54 shifts 2.5–2.8 Å to accommodate the heme (Figure 2B). This suggests placement of a less bulky residue (alanine) in place of Arg-54 may allow heme access to the heme-binding site, but potentially only during translation of the protein when partially unfolded. Regardless of the exact reason for this, these results demonstrate residues in and around the 310-helix are involved in the binding of heme to IsdX1.
310-helix residues and heme stabilization
In an attempt to provide a more quantitative assessment of the importance of each residue in the 310-helix to the stabilization of bound heme, we measured the rates of heme dissociation from wild-type and mutant IsdX1 proteins. Each protein was purified from E. coli as described in the Materials and Methods, reconstituted with hemin, and holo-protein purified away from unbound hemin by gel filtration chromatography. The rate of heme dissociation was then assessed by mixing holo-IsdX1 preparations with excess H64Y/V68Y apo-myoglobin (Mb), a mutant globin with a high heme affinity (Kd∼10−12 M) and very low rate of heme dissociation [30], [48], [49]. The dissociation rate constant of heme loss from IsdX1 can be determined by measuring the spectral changes that occur with time as released heme is scavenged passively by the apo-Mb reagent.
As observed in Figure 5, IsdX1 containing mutations in Ser-53, Arg-54, and Met-55 all lose heme significantly faster than wild-type IsdX1, with S53A showing rates of heme loss that were greater than 400 times faster than the wild-type IsdX1 (see Table 2 for rates). Interestingly, S52A showed comparable rates of heme dissociation to that of wild-type, despite its apparent poor heme binding ability at equilibrium (Figure 4D). The best explanation for this is that while its rate of heme loss may be unaffected, its rate of heme association in the absence of hemoglobin may be poor. Taken together, the data are consistent with Ser-53 and Arg-54 of the 310-helix playing a substantial role in stabilizing the bound heme in IsdX1.
Fig. 5. Heme dissociation kinetics for wild-type and mutant IsdX1. 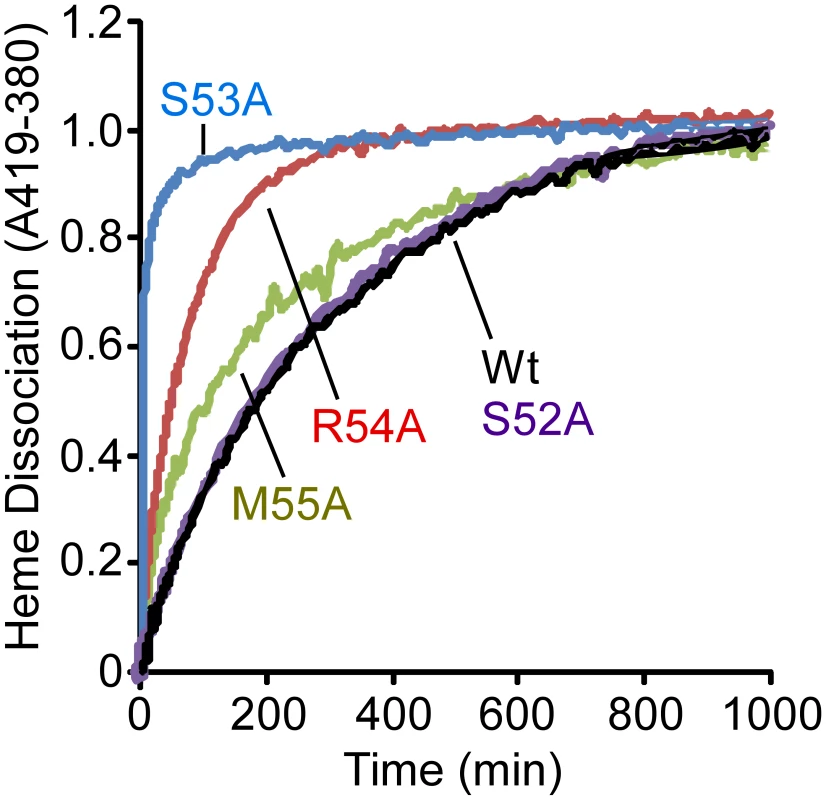
Wild-type or mutant (S52A, S53A, R54A, or M55A) IsdX1 were purified from E. coli and endogenous heme removed as described in the Materials and Methods. Proteins were re-constituted with heme, excess heme removed by gel filtration chromatography, and holo proteins (1 µM) mixed with H64Y/V68F apo-Mb (26 µM) at 25°C in PBS, pH 7.4. Time courses for hemin dissociation the IsdX1 variants were determined by measuring the difference between in the increase in absorbance at 419 nm (peak for holo H64Y/V68F holo-Mb) and the decrease in absorbance at 380 nm (strong absorbance by holo-IsdX1). Tab. 2. Rate constants for heme dissociation from IsdX1 variants. 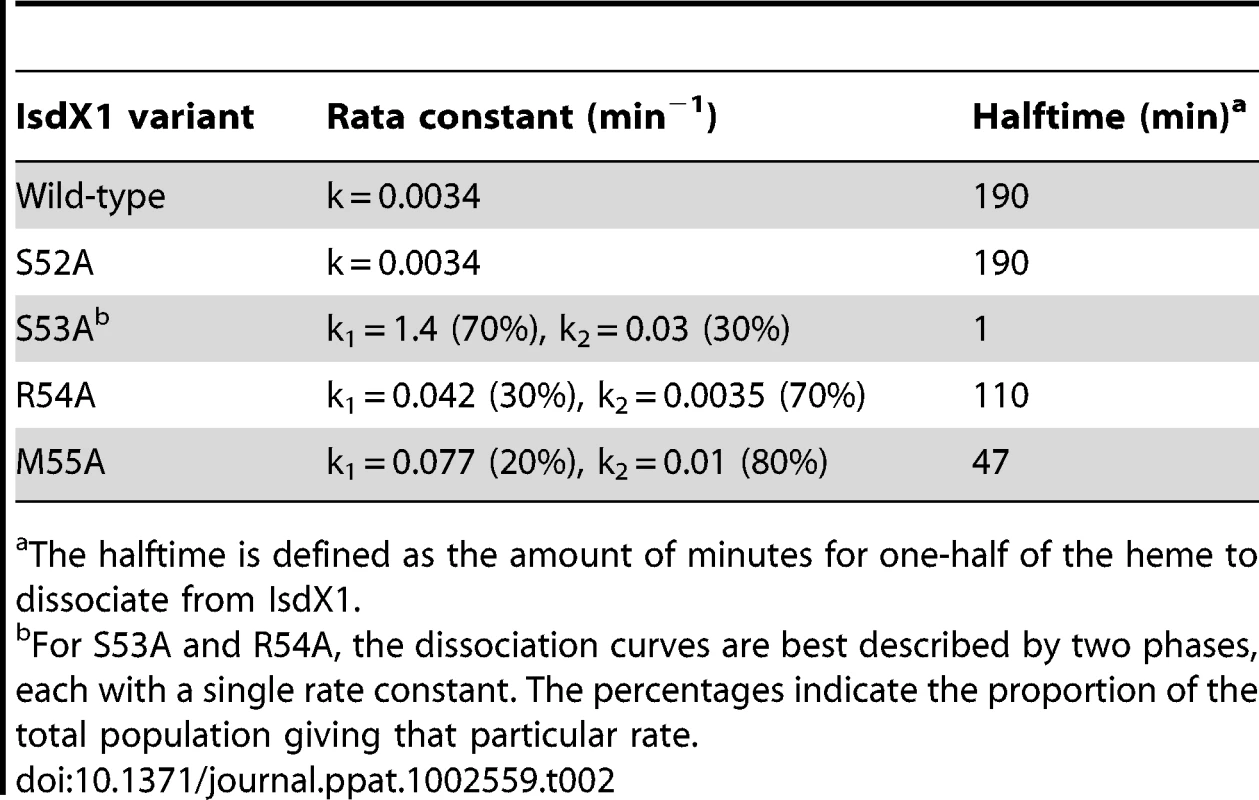
The halftime is defined as the amount of minutes for one-half of the heme to dissociate from IsdX1. The role of the 310-helix in hemoglobin association
The position of 52-SSRM-55 extending over the heme-binding site implies these residues may also be involved in the direct interaction with hemoglobin. To test this hypothesis, the association of each mutant with holo-hemoglobin was investigated by surface plasmon resonance spectroscopy. Apo forms of wild-type or mutant IsdX1 were infused over holo-hemoglobin covalently coupled to a carboxy-methyl chip and the kinetics of binding recorded by quantifying the change in response units (RUs) with time. Whereas similar responses were observed for wild-type, S52A, and M55A IsdX1, a significantly lower response was observed when S53A or R54A were infused over holo-hemoglobin (Figure 6, compare panels A, B, E to panels C, D). Indeed, estimations of the dissociation constants (KD – Figure 6 legend) from the association and dissociation phases of the response curves indicates that the S53A and R54A mutants, when compared to wild-type IsdX1, display an approximately 10 and 386-fold increase, respectively, in KD, signifying a lower affinity for hemoglobin. There was no interaction of the wild-type or mutant IsdX1 proteins with apo-hemoglobin, which infers binding is dependent on a heme-bound conformation of hemoglobin or is facilitated by interactions with the solvent exposed heme propionates in holo-hemoglobin (Figure 6F). These results indicate Ser-53 and Arg-54 are important in the interaction of the IsdX1 hemophore with holo-hemoglobin and collectively suggest the processes of heme binding and hemoglobin association in NEATs are coupled for some residues but not others.
Fig. 6. Functional role of the 310-helix and adjacent residues: hemoglobin association. 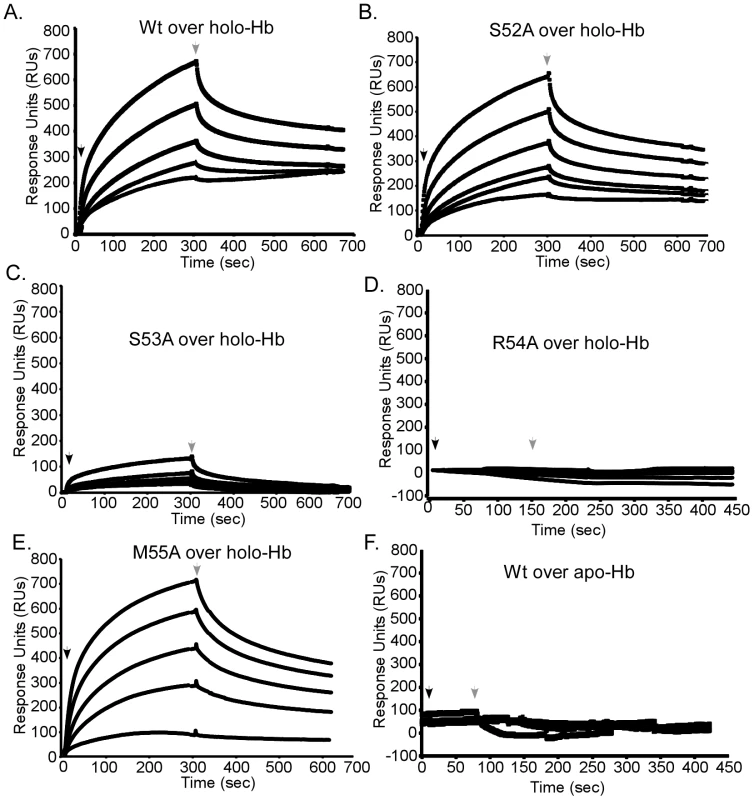
Wild-type (A), S52A (B), S53A (C), R54A (D), or M55A (E) IsdX1 were infused at 100, 200, 300, 350, or 500 nM over holo or apo-hemoglobin (wild-type only, panel F) coupled to a CM5 chip and response units recorded over 800 seconds. The dissociation constants (in nanomolar) were as follows: wild-type = 15.0±0.07, S52A = 14.0±0.17, S53A = 139.0±2.5, R54A = 5, 500±1, 800, and M55A = 18.0±0.4. Due to the weak response of R54A, the KD was calculated from response curves using concentrations approximately 100 times that injected for the wild-type protein. All other dissociation constants represent the mean and standard deviation of three independent measurements for the injection of 300 nM (final concentration) IsdX1 (χ2<than 1.0). Hb = hemoglobin. The role of the 310-helix in hemophore biology
IsdX1 and IsdX2 promote the growth of B. anthracis on hemoglobin as the sole source of iron [26]. To determine the functional contribution of the 52-SSRM-55 helix towards heme scavenging activity, we tested the ability of wild-type and mutant proteins to rescue a hemophore-dependent growth defect of a B. anthracis strain (ΔisdX1, ΔisdX2) lacking both hemophores grown in iron-deficient media with or without hemoglobin. Although little growth is observed in the absence of hemophore, the addition of wild-type IsdX1 to cultures with hemoglobin led to a 3 to 8-fold increase in growth (Figure 7, compare 7 to 8, from 4 to 8 hrs). This enhancement of growth is not due to heme or iron contamination of the protein preparation since only a marginal increase in growth was observed in the absence of hemoglobin (Figure 7, compare 8 to 1,2). Interestingly, whereas ΔisdX1, ΔisdX2 B. anthracis supplemented with S52A or M55A IsdX1 provided intermediate growth (Figure 7, compare 9,12 to 3,6 at 8 hrs), the level of replication in the S53A and R54A IsdX1 supplemented cultures was similar to the S53A/R54A-only controls, suggesting these proteins are unable to rescue a ΔisdX1, ΔisdX2-dependent growth defect on hemoglobin as the sole iron source (Figure 7, compare 10,11 to 4,5 at 8 hrs). Taken together, these results indicate Ser-53 and Arg-54-mediated association of IsdX1 with hemoglobin is important for heme scavenging in iron-limiting environments and provides the first experimental demonstration of the biologic function of this dynamic region in NEAT proteins in growing cells.
Fig. 7. Functional role of the 310-helix and adjacent residues: hemophore and NEAT-domain biology. 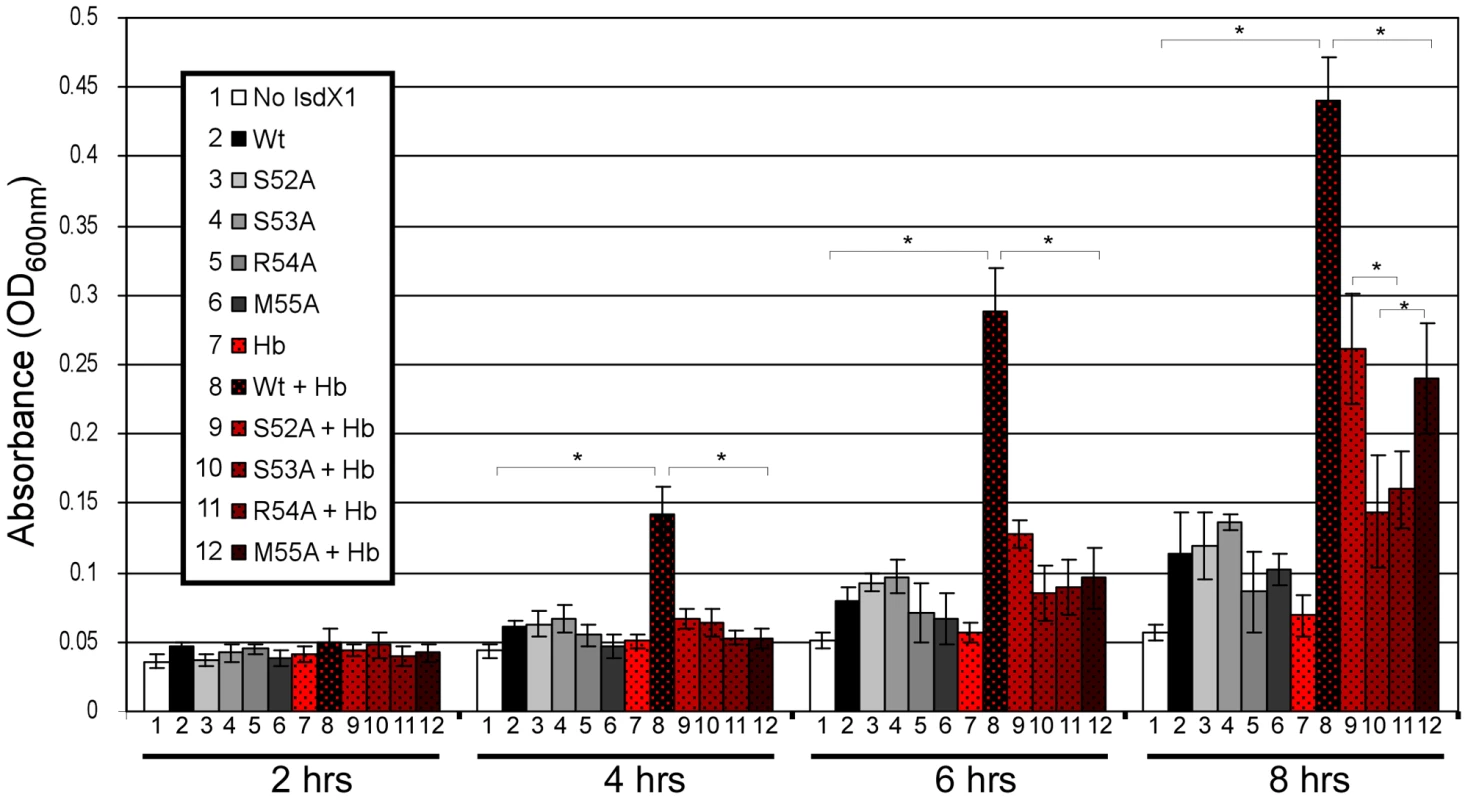
Purified wild-type or mutant IsdX1 were added to a final concentration of 1 µM to hemophore-deficient (ΔisdX1, ΔisdX2) B. anthracis Sterne 34F2 grown in iron-chelated RPMI with or without hemoglobin (10 µM) and the OD600 recorded at 2, 4, 6, and 8 hours. The results represent the mean and standard deviation of three independent experiments. The asterisk (*) means the differences were significant (p<0.05). Hb = hemoglobin. Discussion
Here, we report (i) the crystal structure of the apo and holo forms of a Gram-positive hemophore, (ii) the structure of a non-staphylococcal NEAT protein, (iii) that residues in and adjacent to the 310-helix contribute to the binding of the heme-iron in this hemophore, (iv) that the processes of heme binding and hemoglobin association can be delineated, and (v) that both optimal heme and hemoglobin binding are necessary for full hemophore activity for growing bacilli. Thus, these studies extend our knowledge of the molecular mechanism of NEAT protein function and provide evidence that abolishing a functional interface between the NEAT domain and hemoglobin can slow bacterial growth in iron-limiting environments.
Research into bacteria hemophores is a growing field, and several hemophores have been discovered [10], [16], [50], [51]. The most well-documented secreted hemophore is that of HasA from the Gram-negative pathogen S. marscescens [52]. Although functionally similar, it is likely that IsdX1 and HasA resulted from convergent evolution, as there is little structural similarity between the two proteins. Whereas HasA obtains heme from hemoglobin through a passive mechanism that does not seem to require a physical interaction, IsdX1 binds hemoglobin directly, an event that likely facilitates heme transfer [15], [24], [26]. Both proteins transfer the bound heme to their cognate cell surface receptors (IsdC for IsdX1 and HasR for HasA) through direct engagement. However, the IsdX1-IsdC interaction is dependent on the hemophore being heme loaded and is transient [30]. This contrasts with the HasA-HasR interaction, where both the apo and holo forms of HasA bind with similar affinities and to the same site on HasR [15], [53]. More recently, several secreted hemophores have been reported. HmuY, from P. gingivalis, binds heme and may deliver its heme to the surface receptor HmuR [54], [55]. Also in P. gingivalis, the recently described HusA is a heme-binding protein that is needed for growth under conditions of heme limitation [12]. A putative secreted hemophore from Mycobacterium tuberculosis has been characterized with a proposed heme-binding site consisting of one Tyr and two His, which has a similar heme-binding structural motif to that of HasA [56]. However, the overall fold of the Mtb hemophore is structurally diverse in comparison with both HasA and IsdX1 folds, and it was postulated that the Mtb hemophore may also be a product of convergent evolution [56].
Recent work has shed new insights into how Gram-positive bacteria acquire heme from mammalian hosts [57], [58]. The central structural unit is the NEAT domain, a protein module that mediates heme acquisition and import at the bacterial surface [26], [29], [31], [41], [57]–[61]. If NEAT proteins are to be targets for the development of anti-infectives, the molecular determinants of their mechanism of action need to be defined. In this context, the study of IsdX1 is particularly appropriate because this protein contains the only known NEAT domain that contains all the activities associated with hemophore activity (heme and hemoglobin association, heme extraction from hemoglobin, and heme transfer to receptors) [26]. Thus, the study of IsdX1 allows for insights into the relationship between heme coordination, hemoglobin binding, and heme extraction from mammalian globins.
To gain insights into these activities, we solved the 3-dimensional structure of apo and holo IsdX1. The overall structure is similar to the structures of the staphylococcal NEAT domains of IsdH (NEAT 1/3) [40], [44], IsdA [41], IsdC [42], [43], and IsdB (NEAT 2) [62], with three common features: (i) an immunoglobulin-like fold arranged into eight β-sheets, (ii) a small 4–5 residue 310-helix extending over the heme-binding pocket, and (iii) two anti-parallel β-sheets that house a conserved tyrosine (Tyr-136 in IsdX1) which coordinates the heme-iron (Figure S2A). However, surface charge calculations indicate a charge distribution on IsdX1 quite distinct from the other NEAT domains, including a net positively charged region near the 310-helix of the heme pocket (Figure S1). It is not known if this difference relates to the fact that IsdX1 acts extracellularly after secretion, as opposed to the staphylococcal NEAT proteins that are covalently anchored to the cell wall. Of note is the finding that the staphylococcal homolog of the putative receptor for IsdX1 (IsdC) contains overall anionic character in this region, a feature which leads one to postulate that opposing charges may partially dictate the association between these two proteins upon heme transfer.
The structures of apo-IsdX1 and holo-IsdX1 are highly similar with RMSD of 0.51 Å. However, superimposition of both apo and holo structures highlight subtle residue sidechain conformational differences surrounding the heme binding site. This finding suggests residues in the 310-helix may partially stabilize the heme-free form (discussed below), a requirement of this hemophore immediately after secretion into host tissues. To accommodate the heme molecule, there is a shift in the residue sidechains of Arg-54 (2.5–2.8 Å), Arg-57 (7.3 Å), Tyr-58 (2.1 Å), Tyr-136 (1.6 Å) and Tyr-140 (2.6 Å), away from the center of the heme binding pocket. Additionally, there is observable electron density for two side chain conformations for Met-55 within the apo structure, where one conformation is pointing toward the center of the heme binding pocket and the other away into the interior of IsdX1. Met-55 in the holo structure has its side chain pointing into the interior of the protein and makes hydrophobic contacts with the heme pyrrole ring. In contrast, there is no conformational change observed for either Ser-52 or Ser-53 between the holo and apo structures.
It is interesting to note that within the holo-IsdX1 structure, crystal lattice packing interactions occur through a heme-mediated protein interface between two holo-IsdX1 molecules from adjacent asymmetric units (Figure S5A, B). Two heme molecules from adjacent subunits planar stack upon one another (inter-iron distance of 5.7 Å) and the crystallographic interface is further stabilized by two hydrogen-bonds; one from the NH2 group of Arg-54 from Chain A to the most solvent exposed heme propionate from Chain B and the second from the NH1 group from Arg-57 from Chain B to the hydroxyl group of Asn-135 on the adjacent molecule. The apo-IsdX1 structure also has a similar protein interface near the vicinity of the heme-binding sites of adjacent monomers; however due to the lack of heme, there are more protein interactions between loop regions surrounding the active site (Figure S5A,B). We believe the heme stacking interaction, while caused by crystal packing, may reflect how IsdX1 transfers heme to downstream NEAT domain proteins such as B. anthracis IsdC. Indeed, Grigg et al (published during review of this paper) proposed that inter-NEAT domain interactions between S. aureus IsdB and IsdA may occur along the heme-mediated crystal symmetry interface based on in silico docking predictions [63]. Moreover, an NMR analysis by Villareal et al (also published during review) of the interaction between S. aureus IsdC and IsdA demonstrated that in the IsdC-IsdA complex the proteins are pseudosymmetrically arranged through a 180° rotation that is coplanar with the heme plane, which is identical to the symmetry relationship between IsdX1 monomers from adjacent asymmetric units [64]. Thus, our data also lend experimental support to these models.
With respect to the residues in and around the 310-helix of IsdX1, several interesting properties can be gleaned from this study. First, amino acids in this region contribute to the stabilization of heme, as demonstrated by the fact that the substitution of Ser-52, Ser-53, and Met-55 to alanine abrogated the ability of IsdX1 to attain heme from E. coli lysates as well as bind pure hemin when incubated with the apo protein. Mutation of Arg-54 led to significantly higher amounts of heme co-purifying with IsdX1. However, removal of this heme and assessment of hemin binding yielded a protein unable to subsequently coordinate heme. We cannot definitively explain this result, other than to propose that it is possible that while being synthesized in E. coli, the partially unfolded IsdX1 binds the heme and then folds thereby enclosing upon the iron-porphyrin. Upon heme removal, the resulting apo protein, potentially highly ordered, now becomes restricted and does not allow heme to access the binding pocket. Interestingly, this was not observed in an IsdA variant harboring an alanine substitution in the equivalent position (His-83) [41]. It was proposed this position in IsdA and IsdC (Ile-48) sterically hindered access to the sixth coordination position of the heme-iron. Indeed, cyanide and azide, two ligands often used to probe accessibility to the sixth position, did not bind IsdA or IsdC [42], [65]. The ability of the E. coli form of R54A to associate with more heme while expressed in E. coli may provide experimental support for this hypothesis, with Arg-54 sterically blocking access to Try-136 in the wild-type protein.
Second, mutation of this region decreases hemoglobin association; however, differential effects are observed. Substitution of Ser-53 and Arg-54 with alanine significantly reduced binding to hemoglobin, with R54A yielding the largest effect (greater than 300-fold). However, mutation of Ser-52 and Met-55, the two residues flanking Ser-53 and Met-54, produced hemoglobin-binding affinities similar to the wild-type protein. These findings support a model by which Arg-54, being rather forward in its location over the heme-binding site, provides initial contact with hemoglobin, perhaps stabilizing the initial interaction. The engagement of Arg-54 with hemoglobin may “peel” the side chain of this residue away from the heme-binding site, thus removing the steric block observed for residues in this position for other NEAT proteins. Further support for this hypothesis is that the hemoglobin-binding NEAT domain, IsdB-N2, has a Met in the same position as Arg-54, and the crystal structure of IsdB showed an alternate conformation of Met coordinated with heme-iron, whereby the authors suggest that this Met might be involved in heme transfer. If alternate conformations of Arg-54 are sampled, one conformation has the NH1 group of Arg-54 coordinated to heme-iron similar to IsdB (Figure S2, lower panel). Thus, Arg-54, by virtue of its unique orientation in the heme-binding pocket, may mediate several processes related to heme extraction from hemoglobin. In essence, this residue may function as a “molecular placeholder” by initially binding the heme during the first step of transfer. In support of this hypothesis, there is evidence that arginine can assist in heme binding in other systems [66]. Ser-53, whose side chain is directed towards the heme-binding site, would next be free to hydrogen bond with a heme propionate, thereby strengthening the IsdX1-hemoglobin interaction.
Once Ser-53 engages the heme, Tyr-136 of IsdX1, from the opposite side of the heme, can now displace hemoglobin's His-iron coordination to become the fifth axial ligand of IsdX1, with Tyr-140 providing additional strength for this coordination by H-bonding to the phenolate of Tyr-136. In addition to Tyr-136, the heme is also secured in the binding pocket by Ser-53 and Arg-54, since these residues show the highest rates of heme loss when mutated. Ser-52, whose mutation does not lead to greater heme dissociation, may also assist in drawing the heme into the heme-binding pocket but once the heme is in, does not contribute much to its stabilization. This hypothesis aligns well with the observation that the side chain of this residue sticks out into the solvent while the side chain of its neighbor, Ser-53, points into the heme-binding pocket.
During review of our manuscript, Kumar et al described the first ever co-crystal of a NEAT protein (the first NEAT domain of IsdH from S. aureus) in complex with the alpha-chain of hemoglobin [67]. The structure reveals several stabilizing contacts between the 310-helix of IsdH and hemoglobin, thereby confirming our predictions for the role of this helix in direct association with hemoglobin and heme scavenging. Perhaps most important is a hydrogen bond between a serine in IsdH NEAT 1 (Ser-130) with Lys-11 on hemoglobin. This serine is just 5 residues downstream of what would be the position of Ser-53 in IsdX1, leading us to speculate that the lack of functional interaction with hemoglobin observed upon mutation of this residue is because a key hydrogen bond is severed in the IsdX1-Hb interaction.
Whether or not the function of the 310-helix is confined only to heme acquisition from hemoglobin remains to be determined. For example, Grigg et al found the conserved coordinating tyrosine in the first NEAT domain of IsdA (the equivalent of Tyr-136 in IsdX1), and not residues in the 310-helix, played a role in NEAT to NEAT heme transfer [63]. In contrast, Villareal et al found that mutation of a single 310-helix residue in S. aureus IsdA did affect NEAT to NEAT heme transfer; however, this mutation was coupled to another mutation elsewhere in the protein that was also believed to mediate NEAT-NEAT binding [64]. Thus, more work is required to determine if the 310-helix also functions in downstream heme transfer processes.
Although this model of hemoglobin and heme binding is consistent with available data, we cannot rule out that there are additional residues outside of the 310-helix that participate in this process. Indeed, as demonstrated by Pilpa et al [46] for one of the staphylococcal hemoglobin receptors (NEAT 1 of IsdH) [68], [69], amino acids distal to this position (on the β3–β4 loop), also seem to be important for hemoglobin association. However, NEAT 1 of IsdH does not bind heme, and instead requires a third NEAT domain (NEAT 3), to acquire and stably bind the heme from the IsdH NEAT 1-hemoglobin complex. Thus, the data suggests residues in and around the 310-helix in IsdX1 evolved to bind both heme and hemoglobin, as well as those that are not interdependent, in order to provide several functionalities (hemoglobin binding coupled to heme extraction) within a single NEAT domain.
Finally, the exogenous addition of recombinant IsdX1 to culture restored the growth of a hemophore-deficient strain of B. anthracis on hemoglobin, suggesting all the information necessary for hemophore activity is contained within its amino acid sequence. The partial restoration of growth observed by S52A and M55A may be due to the ability of these mutants to still bind heme, albeit poorly, after association with hemoglobin. Although S53A and R54A bind heme at similar levels as S52A and M55A, it would seem their much lower affinity for hemoglobin precludes them from acquiring any heme in this assay (or alternatively, does not promote the release of enough heme from hemoglobin), meaning no or little free heme is available for transfer (or sequestration) at the bacterial surface. Because mutation of Ser-52, Ser-53, Arg-54, and Met-55 results in poor heme-binding activity, it is difficult to biochemically determine the exact role of these residues in the ability of IsdX1 to extract heme from hemoglobin. However, it is clear from the SPR and growth studies that mutations that effect the association with hemoglobin significantly compromises hemophore activity. Performing an identical experiment with a single mutant (ΔisdX1) strain did not yield a significant enough phenotype to evaluate these mutants for function (data not shown), likely because IsdX1 and IsdX2 are functionally redundant [26]. Taken as a whole, these experiments highlight the biological role of the 310-helix in NEAT protein function and hemophore biology and provide direct evidence that the mechanistic extraction and binding of heme from hemoglobin are important for B. anthracis replication in low-iron environments.
Although attempts to develop a universal inhibitor of heme uptake is likely to be challenging, the seemingly conserved functional properties shared by distinct NEAT domains does offer the prospect of targeting these processes in select Gram-positive bacteria. As we make advances in our understanding of NEAT mechanism of action, the elucidation of additional structures, an understanding of side chain chemistry in transfer reactions, and the identification of factors that drive ligand binding specificity, will all be required for the creation of novel anti-infectives that prevent iron-porphyrin uptake in these pathogenic bacteria.
Materials and Methods
Bacterial strains, reagents, and mutagenesis
DNA encoding for amino acids 26–146 of IsdX1 were amplified from the genome of B. anthracis strain Sterne 34F2 using PCR and primers containing EcoRI and BamHI restriction sites [70]. Amplified DNA was digested, ligated into pGEX2TK, and pGEX-IsdX1 transformed into E. coli BL21 for the expression of IsdX1 as a glutathione-S-transferase (GST) fusion protein as described [26], [29]. For the purification of IsdX1 used to generate the holo-protein crystals, DNA encoding residues 27–152 was cloned into pET28a (Novagen) encoding a fusion protein of IsdX1 with a His(6)-tag using NheI and NotI restriction enzyme sites. The 26–146 (apo) and 27–152 (holo) protein constructs were chosen because they yielded crystals that diffracted with the highest resolution. Site-directed mutagenesis of IsdX1 was performed on the 26–146 IsdX1 construct using QuikChange (Stratagene, Santa Clara, CA) according to the manufacturer's instructions. After DpnI digestion of reaction mixtures, DNA was transformed into E. coli BL21 and the resultant plasmid clones sequenced to confirm the presence of the mutation [71], [72]. All E. coli strains were grown in Luria-broth (LB) supplemented with 50 µg/mL ampicillin (Fisher Scientific, Waltham, MA) except for the pET28a construct that was supplemented with 30 µg/mL kanamycin (Fisher).
Purification of proteins for activity studies
Wild-type and mutant IsdX1 proteins were purified by GST-affinity chromatography as previously described [26], [29]. Briefly, 50-mL of overnight cultures of E. coli BL21 containing wild-type or mutant pgst-isdX1 were inoculated into 2-L of LB with ampicillin and rotated at 250 rpm at 37°C. After 2 hours, isopropyl - β-D-thiogalactopyranoside (IPTG - 1.5 mM) was added and cultures grown for an additional 2 hours. Bacteria were centrifuged at 6,000×g for 8 min, resuspended in phosphate buffered saline (PBS - 137 mM NaCl, 2.7 mM KCl, 10 mM sodium phosphate dibasic, 2 mM potassium phosphate monobasic, pH 7.4), and cells lysed by French press. The supernatant was obtained by centrifugation at 30,000×g for 15 min and filtered through a 0.45-µm pore-size cellulose filter. Lysates were next applied to 1-mL of glutathione-sepharose (GE Healthcare, Piscataway, NJ), washed with 40-mL of PBS, and bound protein incubated with thrombin (100 units, GE Healthcare) for 3 hours at 25°C. GST-free IsdX1 preparations were next incubated with 200-µL of aminobenzamidine sepharose (Sigma, St. Louis, MO) to remove thrombin. To remove endogenous heme, preparations were treated with HCl (final pH of 2.0) and methyl ethyl ketone was added to separate heme (organic layer) from IsdX1 (aqueous layer) as described [73]. Protein concentrations were either determined by UV/vis spectroscopy, the bicinchoninic acid method (Pierce, Rockford, IL), or by SDS-PAGE [74]. All protein preparations were stored at −20°C.
Purification of proteins for crystallography
apo-IsdX1 - To obtain an IsdX1 preparation for crystal seeding, IsdX1 was expressed and bound to glutathione-sepharose as described above [26], [29]. Bound protein was then cleaved from the column with Factor-XA (10 units, Amersham Biosciences) in 1 mM CaCl2, 100 mM NaCl, 50 mM Tris pH 7.9 for 16 hours. Heme was removed as described above and IsdX1 further purified by cation exchange chromatography using a Mono S column (GE Healthcare) and gel filtration chromatography using a Superdex 200 column (GE Healthcare). Selenomethionine-substituted protein was produced by inhibiting methionine biosynthesis and purified as above.
Holo-IsdX1 - IsdX1 was transformed into BL21-Gold (DE3) cells and grown at 37°C in LB medium containing 30 µg/mL kanamycin. Protein expression was induced when cells reached OD600 nm of 0.8 by the addition of 1 mM IPTG and cells harvested after 4 hours by centrifugation at 5, 100 rpm for 20 minutes, followed by resuspension in 50 mM Tris, pH 7.4, and 350 mM NaCl. Cells were next lysed by sonication after addition of egg hen lysozyme (5 mg, Sigma) with phenylmethylsulfonyl fluoride (40 µM, Sigma) and the cell lysate centrifuged at 14, 000 rpm for 20 minutes. After addition of 400 µL Proteoblock protease inhibitor cocktail (Fermentas), the supernatant was loaded onto a Ni2+-charged HisTrap column (GE Healthcare) and eluted with a linear imidazole gradient (between 100–250 mM imidazole). Fractions containing IsdX1 were identified by SDS-PAGE, pooled and concentrated using a Centricon centrifugal concentrator (Millipore). Further purification of IsdX1 was achieved by running the protein over an S75 gel filtration column (GE Healthcare) equilibrated with 50 mM Tris pH 7.4, 150 mM NaCl, which yielded nearly 100% homogeneous protein. Cleavage of the His(6)-tag was conducted in cleavage buffer (50 mM Tris pH 7.4, 150 mM NaCl, 10 mM CaCl2) by adding 1 mL of thrombin-agarose suspension (Sigma) to the protein, followed by removal of thrombin-agarose on a glass frit. IsdX1 was then run over an S75 gel filtration column equilibrated with 50 mM Tris pH 7.4, 150 mM NaCl to separate IsdX1 from the His(6)-tag.
Crystallization and structure determination
apo-IsdX1 - Purified IsdX1 in buffer A [10 mM MES pH 6.6, 200 mM NaCl] crystallized at room temperature using the hanging drop vapor diffusion method and a reservoir solution of 0.1 M citric acid pH 3.5 and 2 M NaCl. Crystals were frozen in N2 (l) following cryoprotection with the reservoir solution containing 16% glycerol. Data were collected to 1.8 Å at the Structural Biology Center beamline 19-BM at the Advanced Photon Source (APS), Argonne National Laboratory (ANL), and processed using HKL2000 [75]. SeMet-IsdX1 crystallized with a reservoir solution of 0.1 M citric acid pH 3.5 and 3.3 M NaCl and was frozen as above. Data were collected to 2.1 Å at the Life Sciences Collaborative Access Team (LS-CAT) beamline 21-ID-D at the APS, and processed with XDS and scaled using SCALA (Table 1) [76]. The structure was determined by the single-wavelength anomalous dispersion method using the anomalous scattering from two selenium atoms in the asymmetric unit cell with the program PHENIX [77]. The model was built automatically with PHENIX and manually with Coot[78] and refined with PHENIX.
Holo-IsdX1 - Approximately 4 mg of heme were dissolved in 0.4 mL of ice cold 0.1 M NaOH and vortexed periodically. After 30 minutes, 0.4 mL of 1 M Tris, pH 7.4 was added to the solution. The solution was subsequently centrifuged for 10 minutes at 4°C at 13,000 rpm. The heme solution was then diluted with 50 mM Tris, pH 7.4, 150 mM NaCl and centrifuged again at 5, 100 rpm to remove any heme aggregates. Final concentrations were determined using ε385 = 58.44 mM−1 cm−1. Heme solutions were used within 12 hours.
Holo-protein was reconstituted by slowly adding 1.5-fold excess heme to IsdX1 preparations in small increments. The UV/vis absorption spectrum was recorded after each addition of heme and saturation with heme was evident by the shift in the Soret maximum from 399 nm to a shorter wavelength. After 1-hour incubation at room temperature, excess heme was removed using an S200 gel filtration column (GE Healthcare) and the protein collected in 1-mL fractions. The UV/vis absorbance spectrum for each fraction was recorded and those with a Soret peak maximum at 399 nm and Abs399/Abs280 ratios greater than 3 were pooled. Protein concentrations were measured using the modified Lowry assay (Pierce, Rockford, IL). The extinction coefficient for the Soret peak at 399 nm was determined to be equal to 100 mM−1 cm−1 by the pyridine hemochrome assay and used for hemoprotein concentration determination [79]. Holo-IsdX1 was concentrated to 100 mg/mL in 50 mM Tris pH 7.4, 150 mM NaCl for crystallization trials.
Crystals of holo-IsdX1 were grown by the hanging drop, vapor diffusion method against a reservoir containing 0.1 M SPG buffer, pH 9.0 and 25% polyethylene glycol 1500 with crystallization drops containing 0.25 µL of protein to 0.25 µL of reservoir solution. Crystal growth was observed within 24 hours. The crystal was passed through a 1∶1 v/v solution containing the reservoir solution and glycerol for cryoprotection and the crystals harvested under cryoconditions. The diffraction data was collected at 77 K on beamline 9-2 at Stanford Synchrotron Radiation Lightsource. Two data sets were collected, a native set at 1.0 Å and a Fe-SAD set at the iron absorption edge (1.738 Å). Images were indexed, integrated and reduced using the HKL2000 suite resulting in a 99.1% complete dataset to 2.15 Å resolution for the native set and a 98.2% complete dataset to 2.3 Å resolution for the Fe-SAD set. The unit cell dimensions are equal to 65.1 Å×65.1 Å×74.4 Å in the space group P43 with two molecules of holo-IsdX1 per asymmetric unit (Table 1). The initial phases were calculated using Phaser [80] with apo-IsdX1 as the search model and the resulting electron density map was subjected to automated and manual model building procedures through a combination of PHENIX.autobuild [77], PHENIX.refine [77] and Coot [78]. Subsequently, two heme-iron sites per asymmetric unit were detected from the anomalous iron signal using SHELXC/D/E [81]. This additional phase information was combined with the molecular replacement model phases through SIGMAA and DM in CCP4i [82], which improved the resulting electron density map for a final round of refinement. Programs from the CCP4 package [82] as well as Phenix [77], Pymol [83], and Coot [84] were used to analyze the stereochemistry and geometry of the models and were found to be acceptable. Data collection and refinement statistics are presented in Table 1.
Heme-binding analysis
Apo forms of wild-type or mutant recombinant IsdX1 (5 µM) were incubated with hemin chloride (5 µM – Sigma) in PBS, pH 7.4 for 10 minutes at 25°C. Reactions were then subjected to a wavelength scan from 250–560 nm using a DU800 spectrophotometer (Beckman-Coulter, London, UK) [29]. The relative amount of bound heme was calculated as: (T399 nm−C399 nm)/T280 nm, where T399 nm = the absorbance maximum at 399 nm for samples containing IsdX1 with heme, C399 nm = the absorbance maximum at 399 nm for samples containing heme only, and T280 nm = the absorbance maximum at 280 nm for samples containing IsdX1 with heme. To measure the rates of heme dissociation, wild-type or mutant IsdX1 were re-constituted with heme, excess heme removed by gel filtration chromatography, and holo proteins (1 µM) mixed with apo-Mb (26 µM) at 25°C in PBS, pH 7.4. Dissociation rates are determined by measuring the increase in absorbance at 419 nm (apo-Mb) versus a reference, control wavelength (380 nm).
Hemoglobin-binding analysis
The interaction of wild-type and mutant IsdX1 proteins with hemoglobin was measured using a BIAcore 3000 biosensor (Amersham Biosciences, Piscataway, NJ) [30]. Briefly, 100 µL of holo - or apo-hemoglobin (Sigma-H2500, 10 µM in 50 mM Tris-HCl, pH 7.0) was covalently coupled to a CM5 sensor chip at 25°C to a density of 4000 response units (RUs) using amine chemistry as previously described [85], [86]. Wild-type or mutant IsdX1 proteins (100, 200, 300, 350, or 500 nM) in HBS-N (0.01 M HEPES, 0.15 M NaCl, pH 7.4) were injected at 20 µL/min for 300 s at 25°C and response curves followed for a total of 800 seconds. A parallel injection of IsdX1 over a blank CM5 surface (no hemoglobin) was used to control for non-specific binding. Injections at each concentration were performed in triplicate, and the data from the 300 nM injection used to calculate the dissociation constants, which were determined using BIAevaluation 4.1 software (Amersham Biosciences) after fitting the data to a 1∶1 Langmuir binding model with dR/dt = kaC(Rmax−R)−kdR, where R is the SPR signal (in response units), ka is the association rate constant (in M−1 s−1), kd is the dissociation rate constant (in s−1), C is the concentration of holo-IsdX1 (in M), Rmax is the maximum holo-IsdX1 binding capacity (in response units), and dR/dt is the rate of change of the SPR signal [87].
B. anthracis growth experiments
Purified IsdX1 (wild-type or mutant proteins) were treated with HCl and methyl ethyl ketone to remove any endogenous heme [88]. Preparations were then dialyzed against 2-L of PBS (pH 7.4) and treated with 100 mg/mL Chelex-100 (Sigma) to remove any contaminating free iron. B. anthracis Sterne strain (34F2) harboring deletions in both isdX1 and isdX2 [26] were subcultured into 3-mL of LB plus kanamycin at 30°C. After 12-hours, cells were washed 2× with PBS and 5-µL of washed, normalized cells inoculated into 500-µL of RPMI (iron chelated by treatment with 100 mg/mL Chelex-100 for 12 hours) with or without hemoglobin (10 µM) in a 48-well Costar tissue-culture plate. Wild-type or mutant proteins (1 µM) were next added to each well and OD600 recorded at 2, 4, 6, and 8 hours at 37°C using a Tecan 200 Pro microplater reader. The results represent the mean and standard deviation of three independent experiments.
Circular Dichroism Spectroscopy
Circular dicroism spectra of apo forms of wild-type and mutant IsdX1 were obtained using a JASCO-815 CD spectropolarimeter at 25°C [89], [90]. Protein samples were resuspended in PBS buffer, pH 7.4 at a concentration of approximately 50 µM. Far-UV spectra were recorded from 200–260 nm using a 1-mm path length at a scanning speed of 50 nm/min, with a bandwidth of 2 nm. Raw spectra are shown and represent the average accumulation of six scans.
Accession numbers
The NCBI accession number for isdX1 is NC_005945.1 (B. anthracis Sterne gene ID:2851614).
Supporting Information
Zdroje
1. SawatzkiGHoffmannFAKubanekB 1983 Acute iron overload in mice: pathogenesis of Salmonella typhimurium infection. Infect Immun 39 659 665
2. RaymondKNDertzEAKimSS 2003 Enterobactin: an archetype for microbial iron transport. Proc Natl Acad Sci U S A 100 3584 3588
3. WinkelmannG 2002 Microbial siderophore-mediated transport. Biochem Soc Trans 30 691 696
4. RaymondKN 2004 Biochemical and Physical Properties of Siderophores. JorgeHCrosaARMPayneShelleyM Iron Transport in Bacteria Washington, D.C. ASM Press 3 17
5. CendrowskiSMacArthurWHannaP 2004 Bacillus anthracis requires siderophore biosynthesis for growth in macrophages and mouse virulence. Mol Microbiol 51 407 417
6. DaleSEDoherty-KirbyALajoieGHeinrichsDE 2004 Role of siderophore biosynthesis in virulence of Staphylococcus aureus: identification and characterization of genes involved in production of a siderophore. Infect Immun 72 29 37
7. PonkaP 1999 Cell biology of heme. Am J Med Sci 318 241 256
8. AisenPEnnsCWessling-ResnickM 2001 Chemistry and biology of eukaryotic iron metabolism. Int J Biochem Cell Biol 33 940 959
9. WittenbergJBWittenbergBA 1990 Mechanisms of cytoplasmic hemoglobin and myoglobin function. Annu Rev Biophys Biophys Chem 19 217 241
10. LetoffeSGhigoJMWandersmanC 1994 Iron acquisition from heme and hemoglobin by a Serratia marcescens extracellular protein. Proc Natl Acad Sci U S A 91 9876 9880
11. WandersmanCDelepelaireP 2004 Bacterial iron sources: from siderophores to hemophores. Annu Rev Microbiol 58 611 647
12. GaoJLNguyenKAHunterN 2010 Characterization of a hemophore-like protein from Porphyromonas gingivalis. J Biol Chem 285 40028 40038
13. YuklETJepkorirGAlontagaAYPautschLRodriguezJC 2010 Kinetic and spectroscopic studies of hemin acquisition in the hemophore HasAp from Pseudomonas aeruginosa. Biochemistry 49 6646 6654
14. CescauSCwermanHLetoffeSDelepelairePWandersmanC 2007 Heme acquisition by hemophores. Biometals 20 603 613
15. LetoffeSNatoFGoldbergMEWandersmanC 1999 Interactions of HasA, a bacterial haemophore, with haemoglobin and with its outer membrane receptor HasR. Mol Microbiol 33 546 555
16. LetoffeSOmoriKWandersmanC 2000 Functional characterization of the HasA(PF) hemophore and its truncated and chimeric variants: determination of a region involved in binding to the hemophore receptor. J Bacteriol 182 4401 4405
17. BrickmanTJVanderpoolCKArmstrongSK 2006 Heme transport contributes to in vivo fitness of Bordetella pertussis during primary infection in mice. Infect Immun 74 1741 1744
18. MortonDJBakaletzLOJurcisekJAVanWagonerTMSealeTW 2004 Reduced severity of middle ear infection caused by nontypeable Haemophilus influenzae lacking the hemoglobin/hemoglobin-haptoglobin binding proteins (Hgp) in a chinchilla model of otitis media. Microb Pathog 36 25 33
19. PaulleyJTAndersonESRoopRM2nd 2007 Brucella abortus requires the heme transporter BhuA for maintenance of chronic infection in BALB/c mice. Infect Immun 75 5248 5254
20. HendersonDPPayneSM 1994 Vibrio cholerae iron transport systems: roles of heme and siderophore iron transport in virulence and identification of a gene associated with multiple iron transport systems. Infect Immun 62 5120 5125
21. TaiSSLeeCJWinterRE 1993 Hemin utilization is related to virulence of Streptococcus pneumoniae. Infect Immun 61 5401 5405
22. MazmanianSKTon-ThatHSuKSchneewindO 2002 An iron-regulated sortase enzyme anchors a class of surface protein during Staphylococcus aureus pathogenesis. Proc Natl Acad Sci USA 99 2293 2298
23. LarsenRGozzelinoRJeneyVTokajiLBozzaFA 2011 A central role for free heme in the pathogenesis of severe sepsis. Sci Transl Med 2 51ra71
24. IzadiNHenryYHaladjianJGoldbergMEWandersmanC 1997 Purification and characterization of an extracellular heme-binding protein, HasA, involved in heme iron acquisition. Biochemistry 36 7050 7057
25. Izadi-PruneyreNHucheFLukat-RodgersGSLecroiseyAGilliR 2006 The heme transfer from the soluble HasA hemophore to its membrane-bound receptor HasR is driven by protein-protein interaction from a high to a lower affinity binding site. J Biol Chem 281 25541 25550
26. MaressoAWGarufiGSchneewindO 2008 Bacillus anthracis secretes proteins that mediate heme acquisition from hemoglobin. PLoS Pathog 4 e1000132
27. HonsaESMaressoAW 2011 Mechanisms of iron import in anthrax. Biometals 24 533 545
28. GatOZaideGInbarIGrosfeldHChitlaruT 2008 Characterization of Bacillus anthracis iron-regulated surface determinant (Isd) proteins containing NEAT domains. Mol Microbiol 70 983 999
29. MaressoAWChapaTJSchneewindO 2006 Surface protein IsdC and sortase B are required for heme-iron scavenging of Bacillus anthracis. J Bacteriol 188 8145 8152
30. FabianMSolomahaEOlsonJSMaressoAW 2009 Heme transfer to the bacterial cell envelope occurs via a secreted hemophore in the gram-positive pathogen Bacillus anthracis. J Biol Chem 284 32138 46
31. AndradeMACiccarelliFDPerez-IratxetaCBorkP 2002 NEAT: a domain duplicated in genes near the components of a putative Fe3+ siderophore transporter from Gram-positive pathogenic bacteria. Genome Biol 3 research0047.1 research0047.5
32. GriggJCUkpabiGGaudinCFMurphyME 2010 Structural biology of heme binding in the Staphylococcus aureus Isd system. J Inorg Biochem 104 341 348
33. OuattaraMCunhaEBLiXHuangYSDixonD 2011 Shr of group A streptococcus is a new type of composite NEAT protein involved in sequestering haem from methaemoglobin. Mol Microbiol 78 739 756
34. TorresVJPishchanyGHumayunMSchneewindOSkaarEP 2006 Staphylococcus aureus IsdB is a hemoglobin receptor required for heme iron utilization. J Bacteriol 188 8421 8429
35. KimHKDeDentAChengAGMcAdowMBagnoliF 2010 IsdA and IsdB antibodies protect mice against Staphylococcus aureus abscess formation and lethal challenge. Vaccine 28 6382 6392
36. CarlsonPEJrCarrKAJanesBKAndersonECHannaPC 2009 Transcriptional profiling of Bacillus anthracis Sterne (34F2) during iron starvation. PLoS One 4 e6988
37. Stranger-JonesYKBaeTSchneewindO 2006 Vaccine assembly from surface proteins of Staphylococcus aureus. Proc Natl Acad Sci U S A 103 16942 16947
38. ChengAGKimHKBurtsMLKrauszTSchneewindO 2009 Genetic requirements for Staphylococcus aureus abscess formation and persistence in host tissues. Faseb J 23 3393 3404
39. KimHKDedentAChengAGMcAdowMBagnoliF 2010 IsdA and IsdB antibodies protect mice against Staphylococcus aureus abscess formation and lethal challenge. Vaccine 28 6382 92
40. PilpaRMFadeevEAVillarealVAWongMLPhillipsM 2006 Solution structure of the NEAT (NEAr Transporter) domain from IsdH/HarA: the human hemoglobin receptor in Staphylococcus aureus. J Mol Biol 360 435 447
41. GriggJCVermeirenCLHeinrichsDEMurphyME 2007 Haem recognition by a Staphylococcus aureus NEAT domain. Mol Microbiol 63 139 149
42. SharpKHSchneiderSCockayneAPaoliM 2007 Crystal structure of the heme-IsdC complex, the central conduit of the Isd iron/heme uptake system in Staphylococcus aureus. J Biol Chem 282 10625 10631
43. VillarealVAPilpaRMRobsonSAFadeevEAClubbRT 2008 The IsdC protein from Staphylococcus aureus uses a flexible binding pocket to capture heme. J Biol Chem 283 31591 31600
44. WatanabeMTanakaYSuenagaAKurodaMYaoM 2008 Structural basis for multimeric heme complexation through a specific protein-heme interaction: the case of the third neat domain of IsdH from Staphylococcus aureus. J Biol Chem 283 28649 28659
45. GaudinCFGriggJCArrietaALMurphyME 2011 Unique Heme-Iron Coordination by the Hemoglobin Receptor IsdB of Staphylococcus aureus. Biochemistry 50 5443 5452
46. PilpaRMRobsonSAVillarealVAWongMLPhillipsM 2009 Functionally distinct NEAT (NEAr Transporter) domains within the Staphylococcus aureus IsdH/HarA protein extract heme from methemoglobin. J Biol Chem 284 1166 1176
47. HonsaESFabianMCardenasAMOlsonJSMaressoAW 2011 The five near-iron transporter (NEAT) domain anthrax hemophore, IsdX2, scavenges heme from hemoglobin and transfers heme to the surface protein IsdC. J Biol Chem 286 33652 33660
48. HargroveMSSingletonEWQuillinMLOrtizLAPhillipsGNJr 1994 His64(E7)→Tyr apomyoglobin as a reagent for measuring rates of hemin dissociation. J Biol Chem 269 4207 4214
49. LiuMTanakaWNZhuHXieGDooleyDM 2008 Direct hemin transfer from IsdA to IsdC in the iron-regulated surface determinant (Isd) heme acquisition system of Staphylococcus aureus. J Biol Chem 283 6668 6676
50. RossiMSFetherstonJDLetoffeSCarnielEPerryRD 2001 Identification and characterization of the hemophore-dependent heme acquisition system of Yersinia pestis. Infect Immun 69 6707 6717
51. LetoffeSRedekerVWandersmanC 1998 Isolation and characterization of an extracellular haem-binding protein from Pseudomonas aeruginosa that shares function and sequence similarities with the Serratia marcescens HasA haemophore. Mol Microbiol 28 1223 1234
52. ArnouxPHaserRIzadiNLecroiseyADelepierreM 1999 The crystal structure of HasA, a hemophore secreted by Serratia marcescens. Nat Struct Biol 6 516 520
53. LetoffeSDeniauCWolffNDassaEDelepelaireP 2001 Haemophore-mediated bacterial haem transport: evidence for a common or overlapping site for haem-free and haem-loaded haemophore on its specific outer membrane receptor. Mol Microbiol 41 439 450
54. OlczakTSiudejaKOlczakM 2006 Purification and initial characterization of a novel Porphyromonas gingivalis HmuY protein expressed in Escherichia coli and insect cells. Protein Expr Purif 49 299 306
55. WojtowiczHGuevaraTTallantCOlczakMSrokaA 2009 Unique structure and stability of HmuY, a novel heme-binding protein of Porphyromonas gingivalis. PLoS Pathog 5 e1000419
56. TulliusMVHarmstonCAOwensCPChimNMorseRP 2011 Discovery and characterization of a unique mycobacterial heme acquisition system. Proc Natl Acad Sci U S A 108 5051 5056
57. MazmanianSKSkaarEPGasperAHHumayunMGornickiP 2003 Passage of heme-iron across the envelope of Staphylococcus aureus. Science 299 906 909
58. MaressoAWSchneewindO 2006 Iron acquisition and transport in Staphylococcus aureus. Biometals 19 193 203
59. MazmanianSKLiuGJensenERLenoyESchneewindO 2000 Staphylococcus aureus mutants defective in the display of surface proteins and in the pathogenesis of animal infections. Proc Natl Acad Sci U S A 97 5510 5515
60. MackJVermeirenCHeinrichsDEStillmanMJ 2004 In vivo heme scavenging by Staphylococcus aureus IsdC and IsdE proteins. Biochem Biophys Res Commun 320 781 788
61. DaouNBuissonCGoharMVidicJBierneH 2009 IlsA, a unique surface protein of Bacillus cereus required for iron acquisition from heme, hemoglobin and ferritin. PLoS Pathog 5 e1000675
62. GaudinCFGriggJCArrietaALMurphyME 2011 Unique Heme-Iron Coordination by the Hemoglobin Receptor IsdB of Staphylococcus aureus. Biochemistry 50 5443 5452
63. GriggJCMaoCXMurphyME 2011 Iron-coordinating tyrosine is a key determinant of NEAT domain heme transfer. J Mol Biol 413 684 698
64. VillarealVASpirigTRobsonSALiuMLeiB 2011 Transient weak protein-protein complexes transfer heme across the cell wall of Staphylococcus aureus. J Am Chem Soc 133 14176 14179
65. VermeirenCLPluymMMackJHeinrichsDEStillmanMJ 2006 Characterization of the heme binding properties of Staphylococcus aureus IsdA. Biochemistry 45 12867 12875
66. Gilles-GonzalezMACaceresAISousaEHTomchickDRBrautigamC 2006 A proximal arginine R206 participates in switching of the Bradyrhizobium japonicum FixL oxygen sensor. J Mol Biol 360 80 89
67. Krishna KumarKJacquesDAPishchanyGCaradoc-DaviesTSpirigT 2011 Structural basis for hemoglobin capture by Staphylococcus aureus cell-surface protein, IsdH. J Biol Chem 286 38439 38447
68. DrylaAGelbmannDvon GabainANagyE 2003 Identification of a novel iron regulated staphylococcal surface protein with haptoglobin-haemoglobin binding activity. Mol Microbiol 49 37 53
69. DrylaAHoffmannBGelbmannDGiefingCHannerM 2007 High-affinity binding of the staphylococcal HarA protein to haptoglobin and hemoglobin involves a domain with an antiparallel eight-stranded beta-barrel fold. J Bacteriol 189 254 264
70. SterneM 1937 Avirulent anthrax vaccine. Onderstepoort J Vet Sci Animal Ind 21 41 43
71. KunkelTA 1985 Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A 82 488 492
72. BramanJPapworthCGreenerA 1996 Site-directed mutagenesis using double-stranded plasmid DNA templates. Methods Mol Biol 57 31 44
73. AscoliFFanelliMRAntoniniE 1981 Preparation and properties of apohemoglobin and reconstituted hemoglobins. Methods Enzymol 76 72 87
74. LaemmliUK 1970 Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 680 685
75. MinorW 1997 Processing of X-ray Diffraction Data Collected in Oscillation Mode. Method Enzymol 276 307 326
76. EvansP 2006 Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr 62 72 82
77. AdamsPDAfoninePVBunkocziGChenVBDavisIW 2010 PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66 213 221
78. EmsleyPCowtanK 2004 Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60 2126 2132
79. BartschRG 1971 Cytochromes: Bacterial. Method Enzymol 23 344 363
80. McCoyAJ 2007 Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D Biol Crystallogr 63 32 41
81. SheldrickGM 2010 Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta Crystallogr D Biol Crystallogr 66 479 485
82. Collaborative Computational Project 1994 The CCP4Suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50 484 491
83. DeLanoWL 2010 The PyMOL Molecular Graphics System, 1.3 ed Schro€dinger, LLC
84. EmsleyPLohkampBScottWGCowtanK 2010 Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66 486 501
85. MurphyMJason-MollerLBrunoJ 2006 Using Biacore to measure the binding kinetics of an antibody-antigen interaction. Curr Protoc Protein Sci Chapter 19 Unit 19 14
86. HowellSKenmoreMKirklandMBadleyRA 1998 High-density immobilization of an antibody fragment to a carboxymethylated dextran-linked biosensor surface. J Mol Recognit 11 200 203
87. DementievaISTereshkoVMcCrossanZASolomahaEArakiD 2009 Pentameric assembly of potassium channel tetramerization domain-containing protein 5. J Mol Biol 387 175 191
88. BerryEATrumpowerBL 1987 Simultaneous determination of hemes a, b, and c from pyridine hemochrome spectra. Anal Biochem 161 1 15
89. JohnsonWCJr 1988 Secondary structure of proteins through circular dichroism spectroscopy. Annu Rev Biophys Biophys Chem 17 145 166
90. GreenfieldNJ 2006 Using circular dichroism spectra to estimate protein secondary structure. Nat Protoc 1 2876 2890
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2012 Číslo 3- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- An Entomopathogenic Nematode by Any Other Name
- Induced Release of a Plant-Defense Volatile ‘Deceptively’ Attracts Insect Vectors to Plants Infected with a Bacterial Pathogen
- A Foot in the Door for Dermatophyte Research
- New Insights into spp.: A Potential Link with Irritable Bowel Syndrome
- Indifferent, Affectionate, or Deceitful: Lifestyles and Secretomes of Fungi
- Mutation and Selection of Prions
- Sleeping with the Enemy: How Intracellular Pathogens Cope with a Macrophage Lifestyle
- Taste for Blood: Hemoglobin as a Nutrient Source for Pathogens
- Direct Recognition of by the NK Cell Natural Cytotoxicity Receptor NKp46 Aggravates Periodontal Disease
- A20 () Deficiency in Myeloid Cells Protects against Influenza A Virus Infection
- Anthrax Lethal Factor Cleavage of Nlrp1 Is Required for Activation of the Inflammasome
- Differential Function of Lip Residues in the Mechanism and Biology of an Anthrax Hemophore
- PK-sensitive PrP Is Infectious and Shares Basic Structural Features with PK-resistant PrP
- A Peptidoglycan Fragment Triggers β-lactam Resistance in
- Capsule Type of Determines Growth Phenotype
- Additive Function of MARTX and VvhA Cytolysins Promotes Rapid Growth and Epithelial Tissue Necrosis During Intestinal Infection
- A Novel Mouse Model of Egg-Induced Immunopathology
- Redundant Notch1 and Notch2 Signaling Is Necessary for IFNγ Secretion by T Helper 1 Cells During Infection with
- The Novel Transporter Dur31 Is a Multi-Stage Pathogenicity Factor
- Short ORF-Dependent Ribosome Shunting Operates in an RNA Picorna-Like Virus and a DNA Pararetrovirus that Cause Rice Tungro Disease
- Structural Insights into a Unique Effector LidA Recognizing Both GDP and GTP Bound Rab1 in Their Active State
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- A Foot in the Door for Dermatophyte Research
- Structural Insights into a Unique Effector LidA Recognizing Both GDP and GTP Bound Rab1 in Their Active State
- An Entomopathogenic Nematode by Any Other Name
- New Insights into spp.: A Potential Link with Irritable Bowel Syndrome
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



