-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaPositive Signature-Tagged Mutagenesis in : Tracking Patho-Adaptive Mutations Promoting Airways Chronic Infection
The opportunistic pathogen Pseudomonas aeruginosa can establish life-long chronic infections in the airways of cystic fibrosis (CF) patients. Persistent lifestyle is established with P. aeruginosa patho-adaptive variants, which are clonal with the initially-acquired strains. Several reports indicated that P. aeruginosa adapts by loss-of-function mutations which enhance fitness in CF airways and sustain its clonal expansion during chronic infection. To validate this model of P. aeruginosa adaptation to CF airways and to identify novel genes involved in this microevolution, we designed a novel approach of positive-selection screening by PCR-based signature-tagged mutagenesis (Pos-STM) in a murine model of chronic airways infection. A systematic positive-selection scheme using sequential rounds of in vivo screenings for bacterial maintenance, as opposed to elimination, generated a list of genes whose inactivation increased the colonization and persistence in chronic airways infection. The phenotypes associated to these Pos-STM mutations reflect alterations in diverse aspects of P. aeruginosa biology which include lack of swimming and twitching motility, lack of production of the virulence factors such as pyocyanin, biofilm formation, and metabolic functions. In addition, Pos-STM mutants showed altered invasion and stimulation of immune response when tested in human respiratory epithelial cells, indicating that P. aeruginosa is prone to revise the interaction with its host during persistent lifestyle. Finally, sequence analysis of Pos-STM genes in longitudinally P. aeruginosa isolates from CF patients identified signs of patho-adaptive mutations within the genome. This novel Pos-STM approach identified bacterial functions that can have important clinical implications for the persistent lifestyle and disease progression of the airway chronic infection.
Published in the journal: . PLoS Pathog 7(2): e32767. doi:10.1371/journal.ppat.1001270
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1001270Summary
The opportunistic pathogen Pseudomonas aeruginosa can establish life-long chronic infections in the airways of cystic fibrosis (CF) patients. Persistent lifestyle is established with P. aeruginosa patho-adaptive variants, which are clonal with the initially-acquired strains. Several reports indicated that P. aeruginosa adapts by loss-of-function mutations which enhance fitness in CF airways and sustain its clonal expansion during chronic infection. To validate this model of P. aeruginosa adaptation to CF airways and to identify novel genes involved in this microevolution, we designed a novel approach of positive-selection screening by PCR-based signature-tagged mutagenesis (Pos-STM) in a murine model of chronic airways infection. A systematic positive-selection scheme using sequential rounds of in vivo screenings for bacterial maintenance, as opposed to elimination, generated a list of genes whose inactivation increased the colonization and persistence in chronic airways infection. The phenotypes associated to these Pos-STM mutations reflect alterations in diverse aspects of P. aeruginosa biology which include lack of swimming and twitching motility, lack of production of the virulence factors such as pyocyanin, biofilm formation, and metabolic functions. In addition, Pos-STM mutants showed altered invasion and stimulation of immune response when tested in human respiratory epithelial cells, indicating that P. aeruginosa is prone to revise the interaction with its host during persistent lifestyle. Finally, sequence analysis of Pos-STM genes in longitudinally P. aeruginosa isolates from CF patients identified signs of patho-adaptive mutations within the genome. This novel Pos-STM approach identified bacterial functions that can have important clinical implications for the persistent lifestyle and disease progression of the airway chronic infection.
Introduction
Persistent bacterial infections involving the opportunistic pathogen Pseudomonas aeruginosa are responsible for much of the morbidity and mortality caused by cystic fibrosis (CF). After causing an initial acute disease state, which is restricted by an immune response, P. aeruginosa establishes persistent infection and colonizes the host by evading immune surveillance [1][2]. It has been shown that long-term colonization of the CF host is maintained by P. aeruginosa patho-adaptive lineages, which are clonal with the initially acquired strain and carried phenotypic variants [3]. A number of genetic mechanisms are responsible for generating clonal variants in P. aeruginosa, including acquisition or loss of genomic islands, genome rearrangements and recombinations [4]. Furthermore, point mutations lead to a wide spectrum of morphotypes with very minor genetic alteration [3]. Common mutations are consistently acquired by most CF strains as those in regulators of alginate biosynthesis (mucA and algU) [5] and virulence genes including motility (rpoN) [6], in the quorum-sensing regulator (lasR) [7][8], in the type-III secretion system [9], in the multidrug-efflux pump (mexA) and in mutator phenotypes (mutS) [10]. Most of these P. aeruginosa mutants have morphotypes which are easy-to-follow by established in vitro assays [11] while other patho-adaptive mutations have been discovered by whole-genome comparison [3]. The sequence analysis identified 68 mutations in the P. aeruginosa late isolate when compared with its clonal early strain [3]. Most were single-base pair changes and many were predicted to result in a change or loss of protein function. Interestingly, virulence factors required for the initiation of acute infections were selected against during chronic infection. This indicates reduced virulence of the late strains with regard to their ability to provoke acute infection [12][13]. This evolutionary scenario is similar to that of the genomes of other pathogens from other species. Genetic loss-of-function mutations confer enhanced fitness of the pathogen in a host-associated environment in Escherichia coli [14], Haemophilus influenzae [15] or Helicobacter pylori [16].
Despite the importance of P. aeruginosa clones with adapted virulence in the progression of CF airway disease, most patho-adaptative mutations and their role in the persistent lifestyle remain hidden in the genome due to the lack of genetic and functional tools for large-scale screens. The widening gap between the rapid progress in genome sequencing and the comparatively slow progress in the functional characterization of sequenced genomes represents a major concern to face the problem of P. aeruginosa chronic infection in patients with CF [17]. Furthermore, several different approaches, including signature tagged mutagenesis (STM), have been used to identify many bacterial genes required for virulence but they are restricted to certain stages of infection. STM is a genomics-based method for in vivo high-throughput screening based on transposon mutants tagged with a unique oligonucleotide. So far, STM has been based on a negative selection approach and applied to animal models of short-term acute infection. This approach selected and identified mutants with attenuated virulence in a variety of different pathogens [18], [19], [20]. According to the negative selection approach, mutants, present in the inoculum but not in bacterial pools recovered from short-term infected animals, are likely to be attenuated and therefore altered within virulence genes. On the contrary, a comprehensive screening of the bacterial genome for genes that identify the stage of chronic infection and can be the targets of phato-adaptative mutations has not been carried out. Progress in this field requires the development of a novel approach for the screening of STM libraries and sophisticated chronic infection models suited to directly identify functions whose inactivation promotes airways long-term chronic infection. Although several mouse models have been developed to mimic the chronic infection, the absence of CF-like lung disease, including long-term infection supported by a significant number of bacteria, has been disappointing [21]. We have recently reported that murine lung pathology associated with chronic infection induced by P. aeruginosa clinical strains isolated after years of colonization from CF patients, differs significantly to that of PAO1 reference strain [1]. In particular, exposure of murine airways to P. aeruginosa clinical strains embedded in the agar beads caused a long-term and higher rate of chronic infection than the challenge with the PAO1 prototype strain. The presence of patho-adaptive mutations in P. aeruginosa clinical strains and not in PAO1 may explain the different pathology in this model of murine chronic infection. Therefore, we designed a novel STM screening based on positive selection by using P. aeruginosa PAO1 STM library as source of mutants [22] and the validated agar beads mouse model of P. aeruginosa chronic infection mentioned above [23] [1]. This approach identified P. aeruginosa genes in PAO1 strain whose inactivation enhanced the establishment and maintenance of airways chronic infection similarly to clinical strains isolated from patients with CF. The features of the genes identified unmask novel strategies adopted by P. aeruginosa in the persistent lifestyle.
Results/Discussion
STM positive selection screening for P. aeruginosa mutations promoting long-term chronic airways infection
We wished to track patho-adaptive mutations promoting long-term P. aeruginosa infection in CF airways through the design of a STM screening based on positive selection (Pos-STM) in vivo. As tool to mimic the initial and progressive bronchopulmonary infection typical of CF patients, we used the validated agar beads-mouse model of chronic infection [23] [1]. The time course established previously with P. aeruginosa PAO1 [23] [1], and in this work with a PAO1 derivative PAO1293, demonstrated that both laboratory strains, given as doses ranging from 5×105 to 5×106 CFUs, induce low rates of acute mortality during the first three days (PAO1 : 24.1% and PAO1293 : 20%) and show capacity to establish chronic airways infection in some survivors (PAO1 : 24.7% and PAO1293 : 12.5%). Persistence vs clearance is taken as a simple read-out of an established chronic infection in the surviving mice at 14 days postinfection. To estimate bacterial levels in the chronic infection, surviving mice are sacrificed, lungs harvested, tissues homogenized and plated to evaluate bacterial load. The recovery of an output pool of more than 103 CFUs of P. aeruginosa is an indication of chronic infection as described previously [23] [1]. Therefore, our criterion for the selection of mutations in P. aeruginosa promoting chronic infection was the screening for persistence rates higher than that of PAO1293 (12.5%) as described above. Indeed, we established, as criteria, to consider positive in screening assay mutant output pools of bacteria which persisted after several passages and at 14 days time points (Fig. 1A).
Fig. 1. Implementation of the Pos-STM screening. 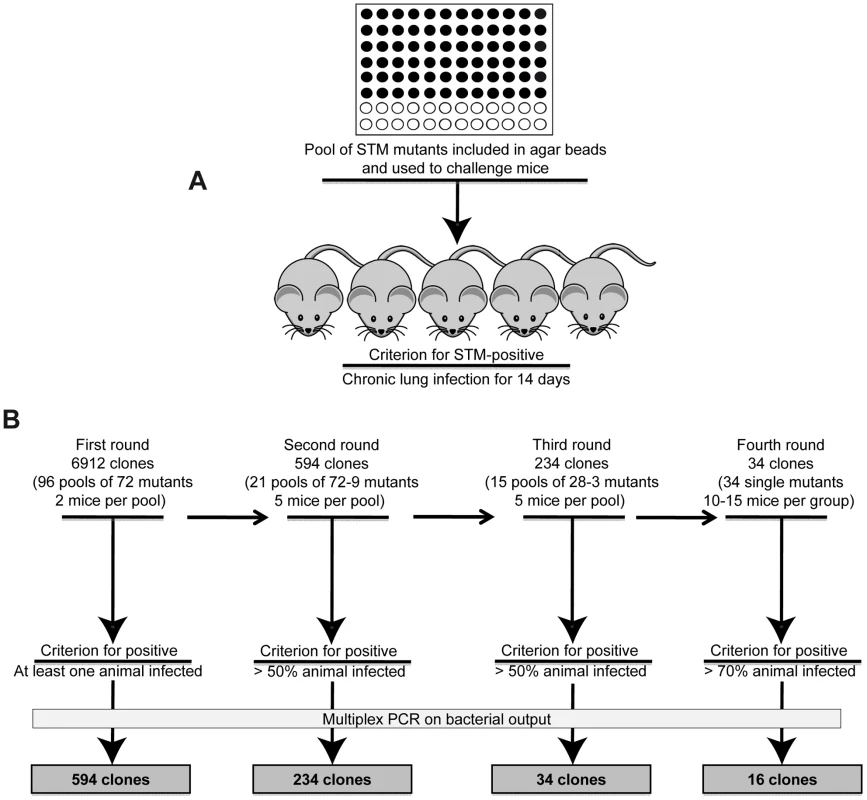
A) First round of screening of a P. aeruginosa PAO1293 STM mini-Tn5 library, consisting of 6912 mutants, arranged into 96 arrays of 72 mutants each [22]. The 72 P. aeruginosa mutants of each array were pooled, processed for agar beads preparation and injected into C57BL/6NCrlBR mice (input pool). After 14 days from challenge, mice were sacrificed, and bacteria recovered from lungs by plating homogenates. Bacterial cfu per lung and the percentage of infected mice were evaluated. B) In the first round of screening, clones that infected at least one of the two mice were identified as positive and selected for the following rounds of screening. For the second screening, we constructed 21 positive input pools with variable numbers of STM-mutants per pool, ranging from 72 to 9. Similarly, 15 positive input pools with variable numbers of STM-mutants per pool, ranging from 28 to 3 were constructed for the third screening. Mice were infected in second and third rounds of screening increasing to five the number of mice and selecting for more than 50% chronic infection. A total of 34 clones which passed the third round were tested singularly in the fourth round increasing the stringency for positive scoring and using 10–15 mice per group. STM mutants were identified by multiplex PCR. 16 mutants which increased significantly chronic infection (>70%) when compared to PAO1293 (12.5%) were considered positive Pos-STM mutants and analyzed further. The source of mutants was a P. aeruginosa PAO1293 STM mini-Tn5 library constructed previously [22], consisting of 6912 clones. In the first round of screening of the complete STM library (Fig. 1B), mice were challenged with 96 input pools of 72 mutants each. Since the large number of initial input pools, we decided to challenge only two mice per pool and to consider positive those challenges with at least one mouse with a chronic infection. Output pools of STM-mutants from positive challenges were further analyzed for clonal expansion of a given STM-mutant(s) by multiplex PCR (see Material and Methods for details) using as a template chromosomal DNA extracted from at least >103 pooled bacterial colonies grown on agar plates. Following this first round of screening, we identified from the initial 96 input pools, 21 positive output pools in which we detected variable numbers of STM-mutants per pool, ranging from 72 (i.e. all mutants of the pool) to 9, respectively, giving a total of 594 mutants. The number of STM-mutants within a pool recovered after this initial screening must take into consideration the potential of cooperative behavior of virulence in mixed infection [1] [24]. The 594 STM-mutants from the first screening were assembled into 21 novel input pools and subjected to further rounds of in vivo screening. Since the input pools were smaller, we decided to increase the number of animals to five (per input pool) and selected for pools of STM mutants which persisted in at least >50% of surviving mice. In the second screening, we obtained 234 STM mutants, which distributed over 15 positive output pools with variable numbers of STM-mutants per pool, ranging from 28 to 3. To proceed further, the 234 STM-mutants resulting from the second screening were assembled from the original STM library into 15 novel input pools and subjected to further step of screening. As shown in Fig. 1B, the third round of screening decreased the number of STM mutants to 34. Finally, in the last round of screening, the 34 STM-mutants were tested singly in larger groups of mice ranging from 10 to 15 animals and with stringency for positive scoring of persistence increasing to >70% of surviving mice. Sixteen of the 34 STM mutants were confirmed as Pos-STM mutants, i.e. they caused a significant increase in percentage of chronic infection in surviving mice when compared to the PAO1293 wt (Chi square analysis: PAO1293 wt vs Pos-STM mutants 12.5% vs 70–100%, p<0.05) (Fig. 2 and Table S2 in Text S1). During the first three days of infection, STM-mutants caused low mortality with no significant differences compared to PAO1293 wt (Chi square analysis: PAO1293 wt vs Pos-STM mutants 20% vs 10–53.3%, p>0.2).
Fig. 2. Virulence of P. aeruginosa Pos-STM mutants in a murine model of airways infection. 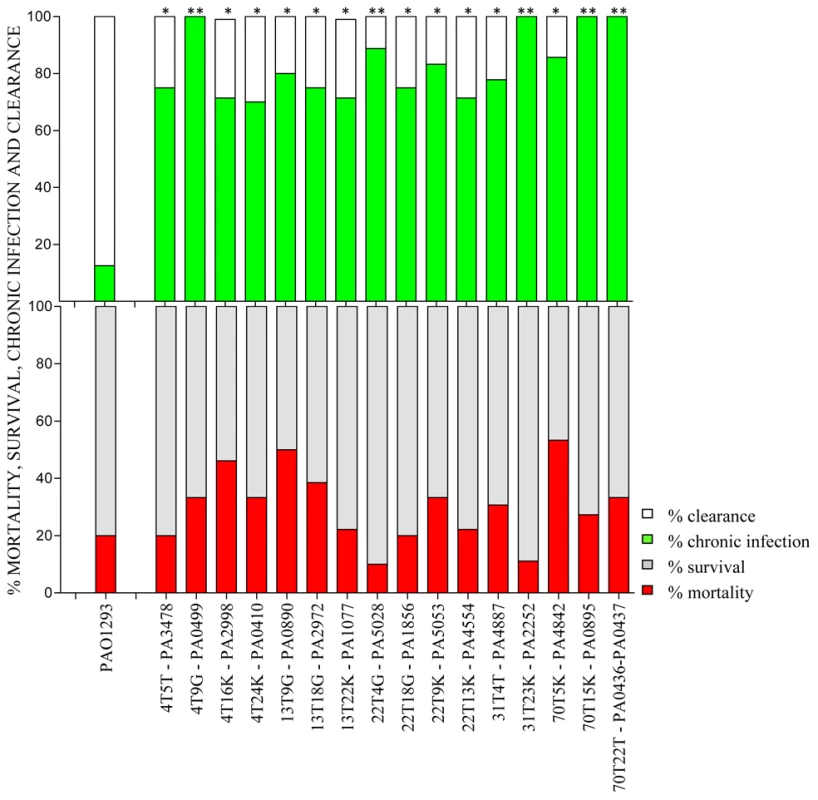
C57Bl/6NCrlBR mice were infected with Pos-STM P. aeruginosa mutants embedded in agar beads. Mortality induced by bacteremia (red) and survival (grey) were evaluated on challenged mice. Clearance (white) and capacity to establish chronic airways infection (green) after 14 days from challenge were determined on surviving mice. The data show the percentage of mice infected with P. aeruginosa pooling together two to three independent experiments (PAO1293 wt: n = 10; STM mutants n = 10–15). Statistical significance by Chi square is indicated: *p<0.05, **p<0.01, ***p<0.001. Histopathological lesions of P. aeruginosa Pos-STM mutants promoting long-term chronic airways infection
To assess clinical strain-traits of chronic infection, lung histopathology was performed on mice challenged singularly with the Pos-STM mutants for 14 days. The histopathological analysis of P. aeruginosa Pos-STM-induced chronic pneumonia indicates that in this model the lung was not totally compromised: the infection was pluri-focal and generally involved one or more lung lobes (Fig. 3A), whereas the others were unaffected or marginally involved (Fig. 3B). Bronchi were filled by agar beads surrounded by a massive neutrophilic infiltration (Fig. 3D) and the surrounding parenchyma was principally infiltrated by macrophages, lymphocytes and neutrophils (Fig. 3E). In other cases bronchial wall was disrupted with neutrophilic infiltration replaced by ‘foamy’ macrophages (Fig. 3F). The infected bronchi contained fewer and smaller agar beads at 14 days when compared to day 0, which were partially degraded by surrounding neutrophils and released bacterial cells. Immunofluorescence staining observed at confocal microscopy showed that the persisting bacterial cells were localized within the bronchial lumen inside the beads (Fig. 3H) and outside the beads as macrocolonies (Fig 3I). Therefore, some of the persisting bacterial cells of the Pos-STM mutants were protected from the murine respiratory defense system by the biofilm formation and not by the beads. Furthermore, Pos-STM mutant cells were presumably found invading inflammatory cells (Fig 3J). Severities of lesions in the lung were similar between all the Pos-STM mutants while most of the PAO1293 wt controls resolved the infection and inflammation (Fig 3C, G and K). Similar histopathological lesions were previously shown in murine lung infected for 14 days with late isolates from CF patients [1]. These results indicate that single gene inactivation in PAO1293 can favor the establishment of chronic infection reminiscent of the so-called “unique” strain lineages of P. aeruginosa pathogenic variants isolated from CF patients.
Fig. 3. Murine lung histology and localization of P. aeruginosa Pos-STM mutants after 14 days. 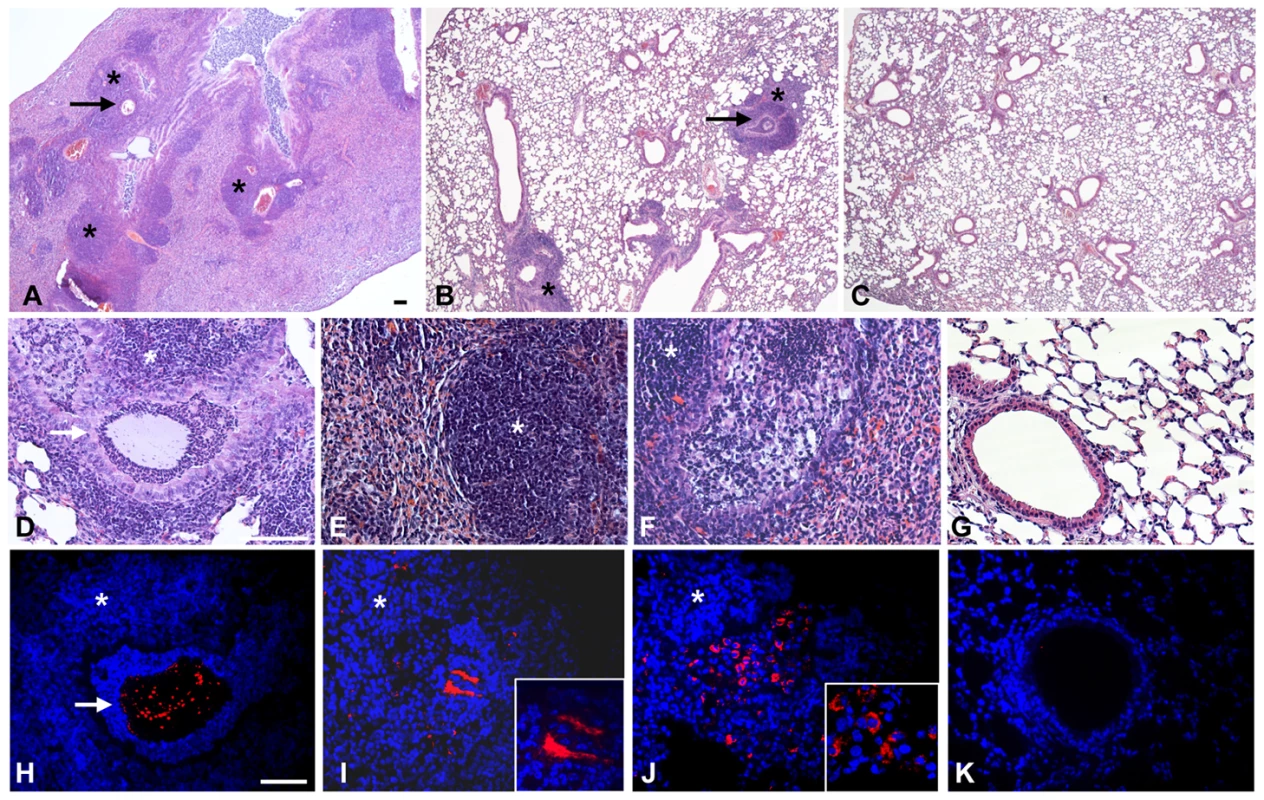
Mice were infected with 2×106 cfu/lung of P. aeruginosa Pos-STM strains embedded in agar beads. In this panel, PA5053- hslV (A and B), PA2998-nqrB (D and H), PA4554-pilY1 (E and I) and PA0499 (F and J) are taken as representative of the whole set of 16 Pos-STM mutants. Control mice were infected with PAO1293 wt (C, G and K). The lungs were stained with H&E (A–G) or with specific antibody against P. aeruginosa strains (red) (H–K). Counterstaining was performed with 4′,6-Diamidino-2-phenylindole dihydrochloride (DAPI) (blue) (H–K). Bronchi and pulmonary parenchyma are characterized by chronic lesion (*) infiltrated by macrophages, lymphocytes and neutrophils. Severity of lesions and lung involvement is heterogeneous in different lobes of the same mice (A and B). Bronchi contain massive granulocyte infiltration of bacteria and agar beads (arrow) (D and H) or bronchial wall is disrupted and the bronchial lumen is filled by foamy macrophages and bacteria presumably internalized by inflammatory cells (F and J). Bacterial macrocolonies are visible in the pulmonary parenchyma outside the beads (E and I). PAO1293 wt infected mice resolved the infection and inflammation (C, G and K). A–C: 2.5×; D–K: 20×; insert I–J: 63×. Confocal microscopy (H–K). Bars, 100 µm. Analysis of the Pos-STM mutants revealed transposon insertions within known virulence genes and other genes not previously associated to infection
Each Pos-STM mutant was subjected to mapping of the transposon insertion by sequencing of the mini-Tn5 flanking genomic regions (Table S1 in Text S1). This first analysis individuated single insertions within a specific gene in 15 Pos-STM mutants (Table 1) and a single insertion into the intergenic region between loci PA0436 and PA0437 in the remaining Pos-STM mutant. However, a further test by Southern blotting to ensure that the Pos-STM mutants did not carry more than one transposon insertion showed that mutants 70T5K and 70T15K carried one additional mini-Tn5 insertion that eluded the previous analysis of mapping (Table 1).
Tab. 1. Genotypic and phenotypic characterization of Pos-STM mutants. 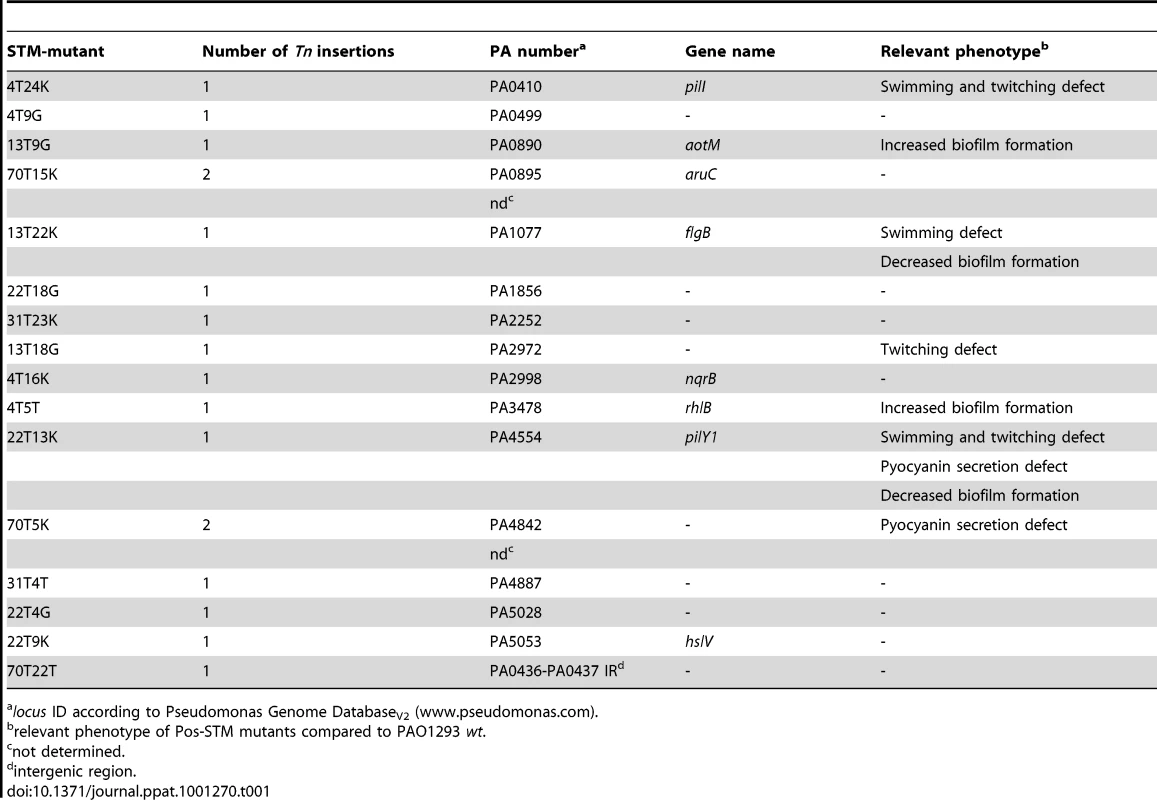
locus ID according to Pseudomonas Genome DatabaseV2 (www.pseudomonas.com). The 15 mini-Tn5 inserted genes encoded proteins from almost all functional classes (Table S3 in Text S1): hypothetical, unknown, unclassified proteins (PA2972, PA4842, PA5028), motility and attachment (PA0410-pilI, PA0499, PA1077-flgB, PA4554-pilY1), putative enzymes (PA1856), transport of small molecules (PA0890-aotM, PA2252, PA4887), amino acid biosynthesis and metabolism (PA0895-aruC), energy metabolism (PA2998-nqrB), secreted factors (PA3478-rhlB), chaperones and heat shock proteins (PA5053-hslV). However, we noted that some insertions map into either the first or internal genes of certain operons, as listed in Table S3 in Text S1, and polar effects on the expression of downstream genes could account for the observed phenotype. In the case of mutants 70T5K and 70T15K, the Pos-STM phenotype can be due to a combination effect caused by the double insertion and or recombination events. Finally, since the intergenic transposon insertion in the 70T22T STM mutant is between convergent PA loci, we speculate a negative effect of the insertion on transcription termination. Furthermore, inactivation of small non coding RNA gene(s) cannot be ruled out. Interestingly, previous report showed that eight different intergenic regions were mutated in the 96-month isolate from CF patients [3].
In a second step of analysis, the 16 Pos-STM mutants were tested for the following traits, known to play an important role in the P. aeruginosa pathogenesis of CF infections: motility, mucoidy, hypermutability, protease secretion, siderophore, hemolysis, autolysis, pyocyanin production, biofilm formation and LasR phenotype, consisting of visible accumulation of the iridescent intercellular signal 4-hydroxy-2-heptylquinoline due to loss-of-function inactivation of lasR. The results of these analyses are summarized in Table 1 and Table S4 in Text S1. No Pos-STM mutant showed mucoid and LasR phenotypes, or defects in protease secretion and siderophore production. On the contrary, a significant proportion of mutants in functions associated with motility emerged in this screening. In fact, three Pos-STM mutants carried insertions in PA0410-pilI and PA4554-pilY1, involved in type IV pilus biogenesis, and in PA1077-flgB, coding for the flagellar basal-body rod protein. Moreover, the Pos-STM mutant in PA2972 coding for an hypothetical protein was found to be defective in twitching motility. Our results, which indicated that a loss of motility favors the establishment and maintenance of chronic infection, are consistent with other studies. On the one hand, lack of motility is one of the phenotypes acquired by P. aeruginosa isolates over the course of CF airway infection and increases the risk of chronic infection in animal models [3] [1]. On the other hand, it was shown that the risk of bacteremia is increased by the presence of pili and flagella since they confer a selective advantage for spreading from the lung to other organs [25] [1] [26]. In addition to motility, flagella and pili also provide a ligand for phagocytic clearance [25]. Moreover, there is another striking consistency of our data with a previous P. aeruginosa STM screening in a polymorphonuclear neutrophil (PMN) phagocytosis assay [27]. In that case, two independent STM-mutants in pilY1 arose from the screening since they showed higher survival rates (i.e. more resistant to killing by PMNs) than the wild type and any of the 3500 tested STM-mutants. In murine model, inactivation of pilY1 impacted virulence promoting P. aeruginosa persistence in the airways [27]. Taken together, these consistencies validate our Pos-STM approach and strongly indicate its robustness for the identification of novel genes which impact the genetic adaptation of P. aeruginosa in CF airways.
Pyocyanin secretion defects were found in the PA4554-pilY1 mutant, as expected from a previous report [27], and in the PA4842 mutant. Finally, regarding biofilm formation, PA4554-pilY1 and PA1077-flgB mutants showed low levels of biofilm, respectively. This was not surprising given the involvement of flagella and pili in biofilm development by P. aeruginosa [28]. On the contrary, PA3478-rhlB and PA0890-aotM showed an improved ability to form biofilm. The remaining nine Pos-STM mutants showed no alteration in motility, pyocyanin secretion, and biofilm formation. As in the case of pilY1 mutants, higher fitness to colonize and persist in airways may be multifactorial. This may also be true for PA3478-rhlB and PA0890-aotM mutants. One key factor for chronic infection could be the improved capacity to form biofilm. Other factors may be directly related to the inactivation of rhlB and aotM genes, respectively, as described below. The rhlB gene encodes RhlB rhamnosyltransferase [29], an enzyme involved in the synthesis of the surfactant mono-rhamnolipids (RLs) [30]. Among their roles, RLs have been shown to act as immune modulators and virulence factors. It has been demonstrated that purified RLs act directly on immune cells. For instance, they have been shown to induce direct neutrophil chemotactic activity, to stimulate the copious release of interleukin IL-8, granulocyte-macrophage colony stimulating factor and IL-6, and to induce the lysis of PMNs. Therefore, the inactivation of rhlB gene could attenuate the bacterium and allow it to evade the immune system. The reduced ability to induce IL-8 secretion was assessed for the PA3478-rhlB mutant (see below). Whereas, the improved fitness for chronic infection of PA0890-aotM mutant may be more closely-related to bacterial metabolism. In fact, the aotM gene codes for a component of the arginine/histidine ABC transporter, involved in the catabolism of arginine and controlled by ArgR and L-arginine under aerobic conditions [31]. Inactivation of aotM may stimulate the anaerobic catabolism of arginine by the arginine deiminase (ADI) pathway [32] and thus enhance the fitness for the colonization of and the persistence in the airways. The same effect may be possible for PA0895-aruC mutant. In fact, aruC codes for N-succinylglutamate 5-semialdehyde dehydrogenase involved in the aerobic arginine utilization via the arginine succinyltransferase (AST) pathway [33]. Remarkably, P. aeruginosa anaerobic catabolism of arginine can be relevant for colonization of CF lung which is typically an anaerobic niche [34]. Mutation-induced metabolic rearrangements could also involve other Pos-STM mutants. For instance, inactivation of PA1856, coding for a probable cytochrome c oxidase cbb3-type (subunit I), or PA2998-nqrB, coding for Na(+)-translocating NADH-quinone reductase subunit B, may rearrange the P. aeruginosa branched (micro)aerobic respiratory chain [35]. Furthermore, inactivation of PA2252, coding for a probable AGCS sodium/alanine/glycine symporter, or PA4887, coding for a probable major facilitator superfamily (MFS) transporter, may induce a metabolic reset better suited to colonize and persist in airways. Interestingly, another probable MFS transporter (PA2092) was mutated in the 96-month isolates from CF patients [3].
The significantly high number of Pos-STM mutants in genes related to metabolic functions strongly supports a key role of bacterial metabolic adaptation during chronic infections. This notion was raised previously [7] for mutations inactivating lasR, one of the most common targets of mutation in P. aeruginosa CF isolates [3]. The lasR loss-of-function mutations in these strains confer a growth advantage with particular carbon and nitrogen sources, including amino acids [7]. This growth phenotype was supposed to contribute to positive selection of lasR mutants within CF airways.
On the whole, as suggested previously by the analysis of P. aeruginosa isolates over the course of CF airways infection, the characterization of the panel of the Pos-STM mutants strongly indicates that chronic-adapted phenotypes reflect alterations in different aspects of the bacterium's biology. Remarkably, our Pos-STM screening did not detect mutants in functions associated with the alginate biosynthesis resulting in a mucoid phenotype, or with mismatch-repair systems giving rise to hypermutability. These two phenotypes usually appear relatively late in the chronic infection [10] [5] and it is possible they might not be critical for the establishment and the early stages of chronic infection. Furthermore, the Pos-STM screening may be affected by other factors limiting the technology as discussed in the conclusion below.
Cellular invasion and stimulation of the host immune response by Pos-STM mutants
CF airway chronic infections occur with P. aeruginosa genetic variants and depend on the ability of bacteria to resist the host defense [1][2]. For these reasons, in addition to the bacterial phenotypes analyzed above, we tested the ability of the Pos-STM mutants to invade human respiratory epithelial cells and to stimulate an immune response. As shown in Figure 4A, 11 of 16 Pos-STM mutants were found to be significantly more invasive than the reference PAO1293 wt strain. On the contrary, PA3478-rhlB and PA0890-aotM mutants were found to have a non-invasive phenotype. This feature may be related to the production of higher levels of biofilm by these strains. Other Pos-STM mutants showing limited invasiveness as PAO1293 wt were found in PA2972 and PA5028. Finally, PA4554-pilY1 was found to be significantly less invasive in A549 cells than PAO1293 wt.
Fig. 4. Stimulation of A549 with P. aeruginosa Pos-STM mutants. 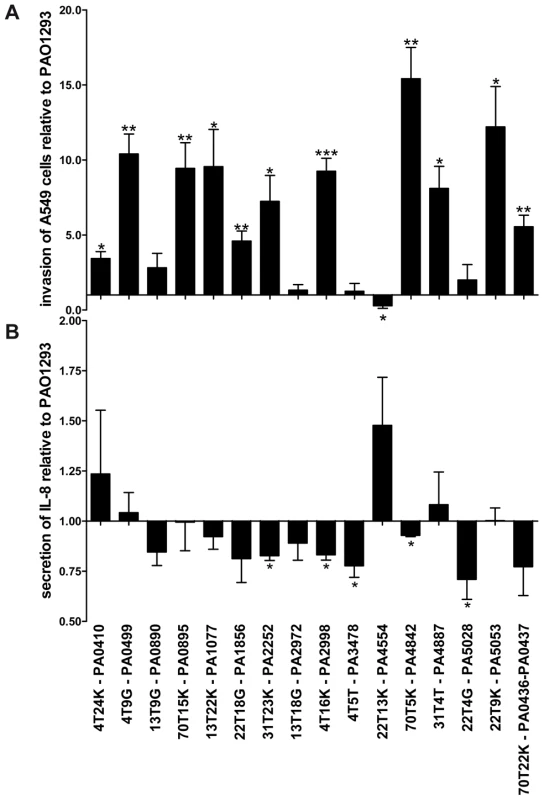
A) Fold of invasion relative to PAO1293 after 2 h of stimulation with Pos-STM mutants. B) IL-8 secretion was quantified by ELISA after stimulation with Pos-STM mutants for 2 h. Measurements were performed in triplicate. *p<0.05, **p<0.01, ***p<0.001 in the Student's t-test. When the bacteria-induced IL-8 secretion was tested in A549 cells, most of the Pos-STM mutants were found to induce lower stimulation of IL-8 secretion. Significant differences were obtained for PA2252, PA2998-nqrB, PA3478-rhlB, PA4842, and PA5028 when compared with PAO1293 wt (Figure 4B). Therefore, the majority of mutants showed increased invasiveness and/or lower stimulation of IL-8 secretion. Together, these results indicate that invasion of host cells and/or lower stimulation of IL-8 secretion can be instrumental to the higher fitness to colonize and persist in CF airways.
Consistently with data obtained in P. aeruginosa clinical strains [2] and strategies used by a number of other bacterial pathogens [36], [37], [38], it is feasible that Pos-STM mutants acquired mechanisms for cellular invasion. Internalization in a host could benefit bacterial survival through evasion of host phagocytic - and antibody/complement-mediated killing mechanisms. Intracellular survival is a key component in the pathogenic cycle of a number of bacterial pathogens, including Mycobacteria spp., Shigella flexneri, Salmonella spp., Legionella pneumophila and Burkholderia cenocepacia [39][40][41]. Furthermore, in CF disease, decreased internalization of P. aeruginosa in CFTR deficient cells may increase bacterial lung burdens and contribute to establish biofilm-mediated chronic infection [42][43].
Several lines of evidence indicate that, to permit long-term colonization, there has been selective pressure on pathogens to avoid triggering an intensive inflammatory reaction. Comparing lipid A and peptidoglycan of sequential strains isolated from CF patients, we recently showed that P. aeruginosa has evolved the capacity to evade immune system detection [2]. In this study, we extended this notion showing that the majority of Pos-STM mutants reduced the immune stimulatory potential. These findings emphasize further studies to establish whether P. aeruginosa adapted mutants evade immune system thus promoting survival and establishing favourable conditions for chronic persistence in CF patients.
Pathoadaptive mutations in P. aeruginosa clinical strains from CF patients
Next, we asked whether the mutations observed in our Pos-STM approach are, to the some extent, representative of patho-adaptive mutations associated with P. aeruginosa infections in CF patients. To address this issue, we set out to sequence the 15 genes which were inserted in the Pos-STM mutants in a collection of longitudinal isolates from seven different CF patients; they include early strains isolated at the onset of chronic colonization and late strains collected over a period of up to 16.3 years [5]. Out of 15, we could obtain the full-length sequence of 14 genes. In fact, we were unable to amplify by PCR the full-length pilY1 gene sequence throughout the collection of the P. aeruginosa longitudinally isolates. We attempted to amplify internal gene regions. The sequence analysis of the few PCR fragments that we obtained indicated several changes involving the pilY1 gene. Thus, this strongly indicated that pilY1 gene underwent extensive changes in the clinical isolates. Sequences analysis of the remaining genes by comparison between early and late strains revealed single base-pair synonymous and non-synonymous mutations in seven of 14 genes in six different clonal lineages (See Table 2 and Table S5 in Text S1 for the complete panel of mutations). Non-synonymous mutations were found in five genes (PA0895-aruC, PA1856, PA2252, PA3478-rhlB and PA4887) from five different clonal lineages of CF patients (Table 2). A computational method for predicting the effect of each non-synonymous mutations on protein function [44] suggested that the non-tolerated changes in PA0895-aruC, PA3478-rhlB and PA4887 are likely to affect protein function (Table S5 in Text S1). P. aeruginosa clonal isolates from a single time point after long-term chronic infection were heterogeneous, indicating a genetic diversification of a single strain during growth in CF airways as previously reported [3]. Overall, these data hint that some Pos-STM mutations, might recapitulate patho-adaptative mutations occurring in P. aeruginosa clinical strains. Previous studies have shown that well-known pathoadaptative mutations in loci including for instance mucA [5], LasR [7] or mutS [10] are common but not acquired by all P. aeruginosa isolates from CF patients. These observations have supported the notion that a number of genes in the genome are targets for patho-adaptative mutations promoting CF airways chronic infection, although most of these genes are mutated in only a fraction of bacterial strains isolated from infections. Therefore, the analysis of significant rates of the patho-adaptative mutations in Pos-STM genes identified in this study could require the study of broader collections of longitudinal P. aeruginosa isolates from CF patients.
Tab. 2. Gene sequence comparison between early and late P. aeruginosa isolates from CF patients. 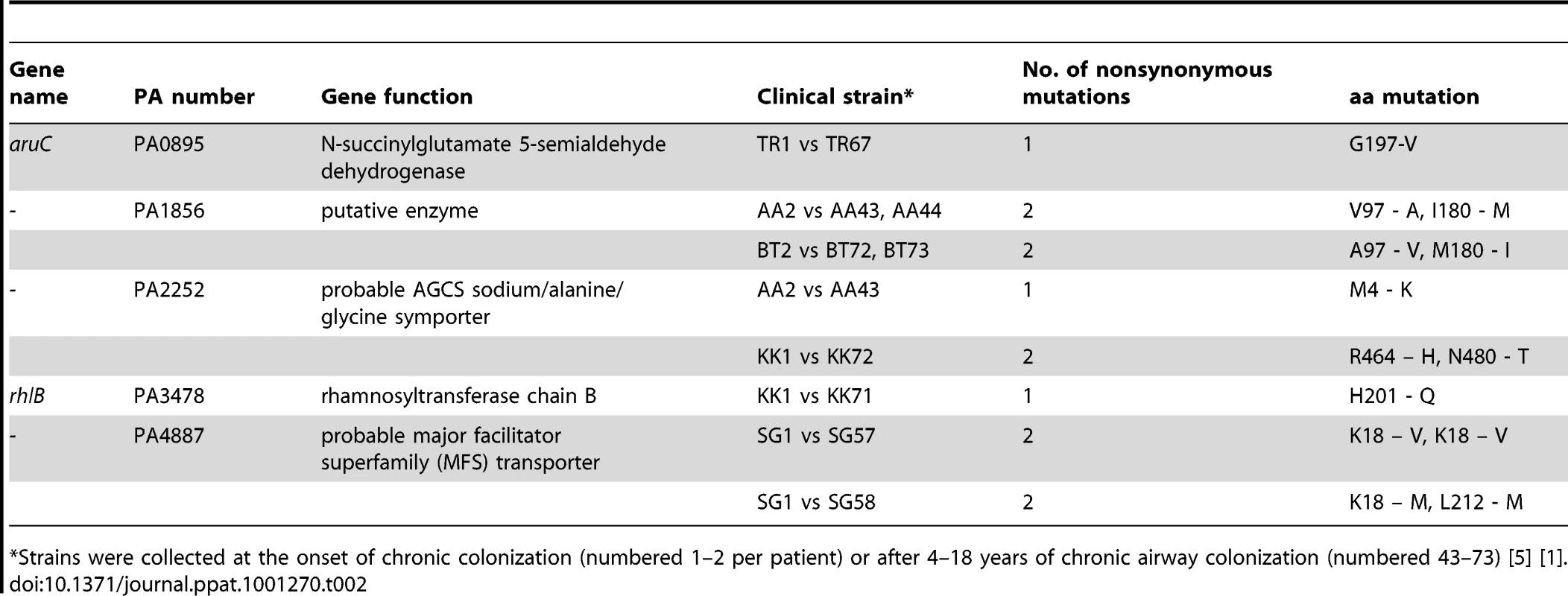
*Strains were collected at the onset of chronic colonization (numbered 1–2 per patient) or after 4–18 years of chronic airway colonization (numbered 43–73) [5] [1]. Conclusions
For the first time in an animal model of chronic lung infection by P. aeruginosa, an STM approach based on positive selection is presented. The main objective of this work was to address the genetic adaptation within a defined genetic system of P. aeruginosa in CF airways through loss-of function mutations in virulence factors and regulators. As with typical STM, the Pos-STM is a screening assay and has limitations. For instance, Pos-STM can be biased by factors such as pooling of mutants giving intra-complementation between cells growing in vivo. Bacteria cells that are not necessarily enhanced in establishing chronic infection can be maintained by those that are. This can be overcome by using several rounds of screening and diminishing the number of mutants per pool as done here. Other limitations include the concentration of bacteria in the inoculum, the limited number of initial STM mutants (6912 mutants in this case). Obviously, the collection of STM mutants used is not representative of the complete genome but covers a small fraction of genes. Genes not in the STM collection will not be selected by Pos-STM. Other factors limiting this technology include the time points selected after infection to recuperate bacteria and the selective pressure for the bacterial growth in the agar beads mouse model. As a result, we identified 16 insertion mutations which lead to an increase in the fitness to promote chronic infection. This panel of mutants shows directly that P. aeruginosa establishes and maintains long-term chronic infection with patho-adapted variants. The reliability and robustness of our Pos-STM approach is supported by the high proportion of mutations in motility-related functions which are known key players in the pathogenesis of P. aeruginosa. Remarkably, our findings rigorously support the previous hypothesis that virulence factors essential for acute infection are lost when P. aeruginosa establishes long-term chronic infection. In addition to this, our screenings resulted in the identification of novel genes which were not previously associated to infection. Thus, our novel approach provides the basis to investigate the P. aeruginosa pathogenesis further and to extrapolate the results to other bacterial pathogens. The significantly high number of these genes linked with metabolic functions reinforces the notion of a key role played by P. aeruginosa metabolic adaptation during chronic infection. Furthermore, the behaviour of the majority of our Pos-STM mutants on host cells suggests that invasiveness and/or altered stimulation of the immune response influence the fitness to colonize and persist in CF airways. The fact that many of these bacterial functions have been missed in the other screenings, including large-scale DNA sequencing [3] and morphological phenotypes assays [3], emphasizes the gap of tools for profiling chronic infection and points to the need to perform this novel Pos-STM approach.
Finally, some of the genes of this STM screening were found to be mutated throughout a collection of longitudinal isolates from CF patients. This hints that they may be targets of patho-adaptive mutations during the P. aeruginosa microevolution during CF lung infection with major implication in the progression of the lung disease. The novel approach of positive-selection screening, described for the first time in this study, should find general applicability to other pathogens that enhance fitness in their host through patho-adaptive mutations and so provide a basis for more comprehensive understanding of chronic infection diseases.
Materials and Methods
Ethics statement
Animal studies were conducted according to protocols approved by the San Raffaele Scientific Institute (Milan, Italy) Institutional Animal Care and Use Committee (IACUC) and adhered strictly to the Italian Ministry of Health guidelines for the use and care of experimental animals.
Research on the bacterial isolates from the individual with CF has been approved by the responsible physician at the CF center at Hannover Medical School, Germany. All patients gave informed consent before the sample collection. Approval for storing of biological materials was obtained by the Hannover Medical School, Germany.
Bacterial strains
The P. aeruginosa PAO1/PAO1293 STM library was described previously [22]. PAO1293 is a derivative of PAO1 from the Holloway laboratory [45] and carries the E79tv-2 giving chloramphenicol susceptibility. STM mutants were cultured at 37°C in brain-heart infusion (BHI) or on TSB agar plates containing 300 µg/ml kanamycin or 25 µg/ml tetracycline. For agar beads preparation, STM mutants were grown in BHI broth in a 96-well plate at 37°C for 18h, pooled, centrifuged at 4000 rpm for 10 min and re-suspended at a concentration of approximately 5×109 cfu/ml. P. aeruginosa clinical strains were isolated from CF patients as described previously [5] [1].
Pos-STM screening in a mouse model of chronic infection
C57Bl/6NCrlBR (Charles River) (6–8 weeks) male mice were infected with STM mutant loads ranging from 5×105 and 2.5×106 CFUs of P. aeruginosa embedded in agar beads and infected as previously described [23]. Fourteen days after infection, murine lungs were excised, homogenized and plated onto TSB-agar plates for cfu counting and identification. For STM mutant identification, at least >104 bacterial colonies grown on agar plates were pooled from each positive output pools and subjected to chromosomal DNA extraction and multiplex PCR analysis according to established protocol [22]. For confirmation, the PCR was repeated for each putative STM mutant.
Histological examination and immunofluorescence
Lungs were removed, fixed in 10% buffered formalin for at least 24 h and embedded in paraffin. Consecutive 2-µm sections from the middle of the five lung lobes were used for histological and immunofluorescence examination in each mouse. Sections for histological analysis were stained by Haematoxylin-Eosin and examined blindly. Localization of P. aeruginosa was performed in de-paraffinized lung sections by employing a rabbit antiserum specific for P.aeruginosa and Texas Red-labelled goat anti-rabbit IgG as described [1]. Immunofluorescence images were recorded with an EM-CCD Hamamatsu C9100 CCD camera (Hamamatsu Photonics, Hamamatsu City, Japan) mounted on an UltraVIEW Spinning Disk Confocal Microscope (Perkin Elmer, Waltham, MA, USA). Slides stained with haematoxylin and eosin were visualized with Axioplan2 (Zeiss, Jena, Germany) with AxioCam provided with the CCD MRc5 (Zeiss).
Sequencing transposon flanking regions
Transposon flanking regions were amplified by the Y-linker method [46]. Approximately 40 ng of genomic DNA, digested with NlaIII, was ligated to 1 µg of Y linker with T4 DNA ligase in a final volume of 20 µl. After overnight incubation at 16°C, 1 µl of the reaction mixture was used as template for hot-start PCR amplification, using a transposon specific oligos (pUTKana2; pUTgfpR2) [22] and the Y-linker specific Y-primer oligo (Table S1 in Text S1). The PCR product was purified from agarose gel with QIAquick Gel Extraction Kit (Qiagen) and then sequenced by Eurofins MWG-Operon (Germany).
Phenotypic characterization
Motility and analysis for LasR mutants were evaluated as described [28] [7]. Mutation frequency measurement were determined by rifampicin assay [10] [47]. Protease and siderophore secretion, pyocyanin, mucoidy, and biofilm formation assay haemolytic/autolytic activity were assayed as described in the online data supplement.
Gene sequencing and computational tools
P. aeruginosa clinical strains described previously [5] [1] were used to sequence the genes inserted in the Pos-STM mutants. PCR genes amplification was carried out using the primers listed in Table S1 in Text S1. The amplified DNA samples were sequenced by standard automated DNA sequence technology. The sequence results were compared within P. aeruginosa clonal lineages with BioEdit v7.0.5 to determine the occurrence of sequence variants within the selected genes. The program SIFT was used to predict the effect of non-synonymous mutations on protein function [44].
Cell cultures and invasion assay
The A549 (human type II pneumocytes) cell line was purchased from ATCC CCL-185 and cultured as described [48]. Bacteria invasion assay was performed using ceftazidime-amikacin protection assay with minor modifications [48]. P. aeruginosa strains, grown to the mid-exponential phase, were used to infect cell monolayers at a 100∶1 multiplicity of infection for 2h. The monolayers were washed with PBS, treated with antibiotic for 2 h, washed, lysed with H2O and plated on TSB-agar plates (Difco).
IL-8 secretion
IL-8 was determined in supernatants collected from the cell cultures described above using an ELISA kit (Biosource Europe). According to the manufacturer the sensitivity of the assay is less than 0.7 pg/ml. Values were normalized to 106 cells; results were expressed as mean ± SD.
Statistical analysis
Statistical calculations and tests were performed using Student's t-test and the Chi-square test considering p<0.05 as the limit of statistical significance. All data were expressed as mean +/ − standard deviation (SD).
Supporting Information
Zdroje
1. BragonziA
ParoniM
NonisA
CramerN
MontanariS
2009 Pseudomonas aeruginosa microevolution during cystic fibrosis lung infection establishes clones with adapted virulence. AJRCCM 180 138 145
2. CiganaC
CurcurùL
LeoneMR
IeranòT
LorèNI
2009 Pseudomonas aeruginosa exploits lipid A and muropeptides modification as a strategy to lower innate immunity during cystic fibrosis lung infection. PLoS One 4 e8439
3. SmithEE
BuckleyDG
WuZ
SaenphimmachakC
HoffmanLR
2006 Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc Natl Acad Sci U S A 30 8487 8492
4. TümmlerB
2006 Clonal variations in Pseudomonas aeruginosa.
RamosJ-L
LevesqueRC
Pseudomonas: molecular biology of emerging issues, vol. 4 New York Springer 35 68
5. BragonziA
WiehlmannL
KlockgetherJ
CramerN
WorlitzschD
2006 Sequence diversity of the mucABD locus in Pseudomonas aeruginosa isolates from patients with cystic fibrosis. Microbiology 152 3261 3269
6. MahenthiralingamE
CampbellME
SpeertDP
1994 Nonmotility and phagocytic resistance of Pseudomonas aeruginosa isolates from chronically colonized patients with cystic fibrosis. Infect Immun 62 596 605
7. D'ArgenioDA
WuM
HoffmanLR
KulasekaraHD
DézielE
2007 Growth phenotypes of Pseudomonas aeruginosa lasR mutants adapted to the airways of cystic fibrosis patients. Mol Microbiol 64 512 533
8. HoffmanL
KulasekaraHD
EmersonJ
HoustonLS
BurnsJL
2009 Pseudomonas aeruginosa lasR mutants are associated with cystic fibrosis lung disease progression. J Cyst Fibros 8 66 70
9. JainM
RamirezD
SeshadriR
CullinaJF
PowersCA
2004 Type III secretion phenotypes of Pseudomonas aeruginosa strains change during infection of individuals with cystic fibrosis. J Clin Microbiol 42 5229 5237
10. OliverA
CantónR
CampoP
BaqueroF
BlázquezJ
2000 High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 288 1251 1254
11. RakhimovaE
MunderA
WiehlmannL
BredenbruchF
TümmlerB
2008 Fitness of isogenic colony morphology variants of Pseudomonas aeruginosa in murine airway infection. PLoS One 3 e1685
12. YoungD
HussellT
DouganG
2002 Chronic bacterial infections: living with unwanted guests. Nat Immunol 3 1026 1032
13. NguyenD
SinghPK
2006 Evolving stealth: genetic adaptation of Pseudomonas aeruginosa during cystic fibrosis infections. Proc Natl Acad Sci U S A 30 8305 8306
14. SokurenkoE
ChesnokovaV
DykhuizenDE
OfekI
WuXR
1998 Pathogenic adaptation of Escherichia coli by natural variation of the FimH adhesin. Proc Natl Acad Sci U S A 95 8922 8926
15. MoxonE
MurphyPA
1978 Haemophilus influenzae bacteremia and meningitis resulting from survival of a single organism. Proc Natl Acad Sci U S A 75 1534 1536
16. AkopyantsN
EatonKA
BergDE
1995 Adaptive mutation and cocolonization during Helicobacter pylori infection of gnotobiotic piglets. Infect Immun 63 116 121
17. GalperinM
KooninEV
2010 From complete genome sequence to ‘complete’ understanding? Trends Biotechnol 28 398 406
18. HenselM
SheaJE
GleesonC
JonesMD
DaltonE
1995 Simultaneous identification of bacterial virulence genes by negative selection. Science 269 400 403
19. CoulterSN
SchwanWR
NgEY
LanghorneMH
RitchieHD
1998 Staphylococcus aureus genetic loci impacting growth and survival in multiple infection environments. Mol Microbiol 30 393 404
20. HerbertMA
HayesS
DeadmanME
TangCM
HoodDW
2002 Signature Tagged Mutagenesis of Haemophilus influenzae identifies genes required for in vivo survival. Microb Pathog 33 211 23
21. BragonziA
2010 Murine models of acute and chronic lung infection with cystic fibrosis pathogens. IJMM 300 584 93
22. PotvinE
LehouxDE
Kukavica-IbruljI
RichardKL
SanschagrinF
2003 In vivo functional genomics of Pseudomonas aeruginosa for high-throughput screening of new virulence factors and antibacterial targets. Environ Microbiol 5 1294 1308
23. BragonziA
WorlitzschD
PierGB
TimpertP
UlrichM
2005 Nonmucoid Pseudomonas aeruginosa expresses alginate in the lungs of patients with cystic fibrosis and in a mouse model. J Infect Dis 192 410 419
24. ReadAF
TaylorLH
2001 The ecology of genetically diverse infections. Science 292 1099 1102
25. FeldmanM
BryanR
RajanS
SchefflerL
BrunnertS
1998 Role of flagella in pathogenesis of Pseudomonas aeruginosa pulmonary infection. Infect Immun 66 43 51
26. TangHB
DiMangoE
BryanR
GambelloM
IglewskiBH
1996 Contribution of specific Pseudomonas aeruginosa virulence factors to pathogenesis of pneumonia in a neonatal mouse model of infection. Infect Immun 64 37 43
27. BohnYS
BrandesG
RakhimovaE
HoratzekS
SalunkheP
2009 Multiple roles of Pseudomonas aeruginosa TBCF10839 PilY1 in motility, transport and infection. Mol Microbiol 71 730 747
28. O'TooleGA
KolterR
1998 Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol 30 295 304
29. OchsnerUA
FiechterA
ReiserJ
1994 Isolation, characterization, and expression in Escherichia coli of the Pseudomonas aeruginosa rhlAB genes encoding a rhamnosyltransferase involved in rhamnolipid biosurfactant synthesis. J Biol Chem 269 19787 19795
30. Abdel-MawgoudAM
LépineF
DézielE
2010 Rhamnolipids: diversity of structures, microbial origins and roles. Appl Microbiol Biotechnol 86 1323 1336
31. LuCD
YangZ
LiW
2004 Transcriptome analysis of the ArgR regulon in Pseudomonas aeruginosa. J Bacteriol 186 3855 3861
32. HaasD
GalimandsM
GamperM
ZimmermannA
1990 Arginine network of Pseudomonas aeruginosa: specific and global controls.
SilverS
ChakrabartyA-M
IglewskiB
KaplanS
Pseudomonas: biotransformations, pathogenesis, and evolving biotechnology Washington, DC American Society for Microbiology 303 316
33. ItohY
1997 Cloning and characterization of the aru genes encoding enzymes of the catabolic arginine succinyltransferase pathway in Pseudomonas aeruginosa. J Bacteriol 179 7280 7729
34. WorlitzschD
TarranR
UlrichM
SchwabU
CekiciA
2002 Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J Clin Invest 109 317 325
35. Alvarez-OrtegaC
HarwoodCS
2007 Responses of Pseudomonas aeruginosa to low oxygen indicate that growth in the cystic fibrosis lung is by aerobic respiration. Mol Microbiol 65 153 165
36. MartinDW
MohrCD
2000 Invasion and intracellular survival of Burkholderia cepacia. Infection and Immunity 68 24 29
37. PhilpottDJ
BelaidD
TroubadourP
ThibergeJM
TankovicJ
2002 Reduced activation of inflammatory responses in host cells by mouse-adapted Helicobacter pylory isolates. Cell Microbiol 4 285 296
38. MonackD
MuellerA
FalkowS
2004 Persistent bacterial infections: the interface of the pathogen and the host immune system. Nat Rev Microbiol 2 747 765
39. FinlayB
FalkowS
1997 Common themes in microbial pathogenicity revisited. Microbiol Mol Biol Rev 61 136 169
40. KaufmannSHE
1993 Immunity to intracellular bacteria. Annu Rev Immunol 11 129 163
41. LamotheJ
HuynhKK
GrinsteinS
ValvanoMA
2007 Intracellular survival of Burkholderia cenocepacia in macrophages is associated with a delay in the maturation of bacter. Cell Microbiol 9 40 53
42. PierG
GroutM
ZaidiTS
OlsenJC
JohnsonLG
1996 Role of mutant CFTR in hypersusceptibility of cystic fibrosis patients to lung infections. Science 271 64 67
43. PierG
GroutM
ZaidiTS
1997 Cystic fibrosis transmembrane conductance regulator is an epithelial cell receptor for clearance of Pseudomonas aeruginosa from the lung. Proc Natl Acad Sci U S A 94 12088 12093
44. KumarP
HenikoffS
NgPC
2009 Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 4 1073 1081
45. HollowayB
KrishnapillaiV
MorganAF
1979 Chromosomal genetics of Pseudomonas. Microbiol Rev 43 73 102
46. KwonY
RickeSC
2000 Efficient amplification of multiple transposon-flanking sequences. J Microbiol Methods 41 195 199
47. MontanariS
OliverA
SalernoP
MenaA
BertoniG
2007 Biological cost of hypermutation in Pseudomonas aeruginosa strains from patients with cystic fibrosis. Microbiology 153 1445 1454
48. PironeL
BragonziA
FarcomeniA
ParoniM
AuricheC
2008 Burkholderia cenocepacia strains isolated from cystic fibrosis patients are apparently more invasive and more virulent than rhizosphere strains. Environ Microbiol 10 2773 2784
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Compensatory Evolution of Mutations Restores the Fitness Cost Imposed by β-Lactam Resistance inČlánek The C-Terminal Domain of the Arabinosyltransferase EmbC Is a Lectin-Like Carbohydrate Binding ModuleČlánek A Viral microRNA Cluster Strongly Potentiates the Transforming Properties of a Human Herpesvirus
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2011 Číslo 2- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- A Fresh Look at the Origin of , the Most Malignant Malaria Agent
- In Situ Photodegradation of Incorporated Polyanion Does Not Alter Prion Infectivity
- Highly Efficient Protein Misfolding Cyclic Amplification
- Positive Signature-Tagged Mutagenesis in : Tracking Patho-Adaptive Mutations Promoting Airways Chronic Infection
- Charge-Surrounded Pockets and Electrostatic Interactions with Small Ions Modulate the Activity of Retroviral Fusion Proteins
- Whole-Body Analysis of a Viral Infection: Vascular Endothelium is a Primary Target of Infectious Hematopoietic Necrosis Virus in Zebrafish Larvae
- Inhibition of Nox2 Oxidase Activity Ameliorates Influenza A Virus-Induced Lung Inflammation
- STAT2 Mediates Innate Immunity to Dengue Virus in the Absence of STAT1 via the Type I Interferon Receptor
- Uropathogenic P and Type 1 Fimbriae Act in Synergy in a Living Host to Facilitate Renal Colonization Leading to Nephron Obstruction
- Elite Suppressors Harbor Low Levels of Integrated HIV DNA and High Levels of 2-LTR Circular HIV DNA Compared to HIV+ Patients On and Off HAART
- DC-SIGN Mediated Sphingomyelinase-Activation and Ceramide Generation Is Essential for Enhancement of Viral Uptake in Dendritic Cells
- Short-Lived IFN-γ Effector Responses, but Long-Lived IL-10 Memory Responses, to Malaria in an Area of Low Malaria Endemicity
- Induces T-Cell Lymphoma and Systemic Inflammation
- The C-Terminus of RON2 Provides the Crucial Link between AMA1 and the Host-Associated Invasion Complex
- Critical Role of the Virus-Encoded MicroRNA-155 Ortholog in the Induction of Marek's Disease Lymphomas
- Type I Interferon Signaling Regulates Ly6C Monocytes and Neutrophils during Acute Viral Pneumonia in Mice
- Atypical/Nor98 Scrapie Infectivity in Sheep Peripheral Tissues
- Innate Sensing of HIV-Infected Cells
- BosR (BB0647) Controls the RpoN-RpoS Regulatory Pathway and Virulence Expression in by a Novel DNA-Binding Mechanism
- Compensatory Evolution of Mutations Restores the Fitness Cost Imposed by β-Lactam Resistance in
- Expression of Genes Involves Exchange of the Histone Variant H2A.Z at the Promoter
- The RON2-AMA1 Interaction is a Critical Step in Moving Junction-Dependent Invasion by Apicomplexan Parasites
- Epstein-Barr Virus Nuclear Antigen 3C Facilitates G1-S Transition by Stabilizing and Enhancing the Function of Cyclin D1
- Transcription and Translation Products of the Cytolysin Gene on the Mobile Genetic Element SCC Regulate Virulence
- Phosphatidylinositol 3-Monophosphate Is Involved in Apicoplast Biogenesis
- The Rubella Virus Capsid Is an Anti-Apoptotic Protein that Attenuates the Pore-Forming Ability of Bax
- Episomal Viral cDNAs Identify a Reservoir That Fuels Viral Rebound after Treatment Interruption and That Contributes to Treatment Failure
- Genetic Mapping Identifies Novel Highly Protective Antigens for an Apicomplexan Parasite
- Relationship between Functional Profile of HIV-1 Specific CD8 T Cells and Epitope Variability with the Selection of Escape Mutants in Acute HIV-1 Infection
- The Genotype of Early-Transmitting HIV gp120s Promotes αβ –Reactivity, Revealing αβ/CD4 T cells As Key Targets in Mucosal Transmission
- Small Molecule Inhibitors of RnpA Alter Cellular mRNA Turnover, Exhibit Antimicrobial Activity, and Attenuate Pathogenesis
- The bZIP Transcription Factor MoAP1 Mediates the Oxidative Stress Response and Is Critical for Pathogenicity of the Rice Blast Fungus
- Entrapment of Viral Capsids in Nuclear PML Cages Is an Intrinsic Antiviral Host Defense against Varicella-Zoster Virus
- NS2 Protein of Hepatitis C Virus Interacts with Structural and Non-Structural Proteins towards Virus Assembly
- Measles Outbreak in Africa—Is There a Link to the HIV-1 Epidemic?
- New Models of Microsporidiosis: Infections in Zebrafish, , and Honey Bee
- The C-Terminal Domain of the Arabinosyltransferase EmbC Is a Lectin-Like Carbohydrate Binding Module
- A Viral microRNA Cluster Strongly Potentiates the Transforming Properties of a Human Herpesvirus
- Infections in Cells: Transcriptomic Characterization of a Novel Host-Symbiont Interaction
- Secreted Bacterial Effectors That Inhibit Host Protein Synthesis Are Critical for Induction of the Innate Immune Response to Virulent
- Genital Tract Sequestration of SIV following Acute Infection
- Functional Coupling between HIV-1 Integrase and the SWI/SNF Chromatin Remodeling Complex for Efficient Integration into Stable Nucleosomes
- DNA Damage and Reactive Nitrogen Species are Barriers to Colonization of the Infant Mouse Intestine
- The ESCRT-0 Component HRS is Required for HIV-1 Vpu-Mediated BST-2/Tetherin Down-Regulation
- Targeted Disruption of : Invasion of Erythrocytes by Using an Alternative Py235 Erythrocyte Binding Protein
- Trivalent Adenovirus Type 5 HIV Recombinant Vaccine Primes for Modest Cytotoxic Capacity That Is Greatest in Humans with Protective HLA Class I Alleles
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Genetic Mapping Identifies Novel Highly Protective Antigens for an Apicomplexan Parasite
- Type I Interferon Signaling Regulates Ly6C Monocytes and Neutrophils during Acute Viral Pneumonia in Mice
- Infections in Cells: Transcriptomic Characterization of a Novel Host-Symbiont Interaction
- The ESCRT-0 Component HRS is Required for HIV-1 Vpu-Mediated BST-2/Tetherin Down-Regulation
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



