-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaIdentification of a Mutant PfCRT-Mediated Chloroquine Tolerance Phenotype in
Mutant forms of the Plasmodium falciparum transporter PfCRT constitute the key determinant of parasite resistance to chloroquine (CQ), the former first-line antimalarial, and are ubiquitous to infections that fail CQ treatment. However, treatment can often be successful in individuals harboring mutant pfcrt alleles, raising questions about the role of host immunity or pharmacokinetics vs. the parasite genetic background in contributing to treatment outcomes. To examine whether the parasite genetic background dictates the degree of mutant pfcrt-mediated CQ resistance, we replaced the wild type pfcrt allele in three CQ-sensitive strains with mutant pfcrt of the 7G8 allelic type prevalent in South America, the Oceanic region and India. Recombinant clones exhibited strain-dependent CQ responses that ranged from high-level resistance to an incremental shift that did not meet CQ resistance criteria. Nonetheless, even in the most susceptible clones, 7G8 mutant pfcrt enabled parasites to tolerate CQ pressure and recrudesce in vitro after treatment with high concentrations of CQ. 7G8 mutant pfcrt was found to significantly impact parasite responses to other antimalarials used in artemisinin-based combination therapies, in a strain-dependent manner. We also report clinical isolates from French Guiana that harbor mutant pfcrt, identical or related to the 7G8 haplotype, and manifest a CQ tolerance phenotype. One isolate, H209, harbored a novel PfCRT C350R mutation and demonstrated reduced quinine and artemisinin susceptibility. Our data: 1) suggest that high-level CQR is a complex biological process dependent on the presence of mutant pfcrt; 2) implicate a role for variant pfcrt alleles in modulating parasite susceptibility to other clinically important antimalarials; and 3) uncover the existence of a phenotype of CQ tolerance in some strains harboring mutant pfcrt.
Published in the journal: . PLoS Pathog 6(5): e32767. doi:10.1371/journal.ppat.1000887
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1000887Summary
Mutant forms of the Plasmodium falciparum transporter PfCRT constitute the key determinant of parasite resistance to chloroquine (CQ), the former first-line antimalarial, and are ubiquitous to infections that fail CQ treatment. However, treatment can often be successful in individuals harboring mutant pfcrt alleles, raising questions about the role of host immunity or pharmacokinetics vs. the parasite genetic background in contributing to treatment outcomes. To examine whether the parasite genetic background dictates the degree of mutant pfcrt-mediated CQ resistance, we replaced the wild type pfcrt allele in three CQ-sensitive strains with mutant pfcrt of the 7G8 allelic type prevalent in South America, the Oceanic region and India. Recombinant clones exhibited strain-dependent CQ responses that ranged from high-level resistance to an incremental shift that did not meet CQ resistance criteria. Nonetheless, even in the most susceptible clones, 7G8 mutant pfcrt enabled parasites to tolerate CQ pressure and recrudesce in vitro after treatment with high concentrations of CQ. 7G8 mutant pfcrt was found to significantly impact parasite responses to other antimalarials used in artemisinin-based combination therapies, in a strain-dependent manner. We also report clinical isolates from French Guiana that harbor mutant pfcrt, identical or related to the 7G8 haplotype, and manifest a CQ tolerance phenotype. One isolate, H209, harbored a novel PfCRT C350R mutation and demonstrated reduced quinine and artemisinin susceptibility. Our data: 1) suggest that high-level CQR is a complex biological process dependent on the presence of mutant pfcrt; 2) implicate a role for variant pfcrt alleles in modulating parasite susceptibility to other clinically important antimalarials; and 3) uncover the existence of a phenotype of CQ tolerance in some strains harboring mutant pfcrt.
Introduction
The massive use of chloroquine (CQ) in the 20th century heralded substantial gains in the global fight against malaria. These advances were later lost as CQ resistance (CQR) arose and spread throughout malaria-endemic areas [1], [2]. Today, CQ and the alternative first-line antimalarial sulfadoxine-pyrimethamine have officially been mostly replaced by artemisinin-based combination therapies (ACTs) [3]. Nevertheless, CQ continues to be widely used in parts of sub-Saharan Africa at the household level, presumably because of its ability to provide temporary relief from symptoms for patients unable to afford ACTs or other expensive drugs [4], [5]. Recent findings also suggest the possibility of reintroducing CQ-based combination therapies into African regions where an extended hiatus from CQ use has led to the dominance of CQ-sensitive Plasmodium falciparum parasites that have outcompeted the less-fit CQ-resistant strains [6]. At the cellular level, CQ is thought to act by accumulating to low millimolar concentrations in the acidic digestive vacuole of asexual intra-erythrocytic Plasmodium parasites, wherein it interferes with the detoxification of iron-bound heme moieties produced as a result of hemoglobin degradation [7].
Clinical and epidemiological studies reveal that CQR emerged on very few occasions despite its abundant use, leading researchers to initially posit a multigenic basis of resistance [8]. This theory was challenged by the finding that CQR was inherited as a single locus in a genetic cross between the CQ-resistant Dd2 (Indochina) and the CQ-sensitive HB3 (Honduras) clones [9], [10]. The causal determinant in this locus was ultimately identified as the P. falciparum chloroquine resistance transporter (pfcrt), whose 49 kDa protein product PfCRT resides on the DV membrane [11], [12]. Comparison of the Dd2 and HB3 sequence revealed eight point mutations that all mapped to sites within or near several of the 10 putative transmembrane domains [11].
Quantitative trait loci analysis of the HB3×Dd2 cross has revealed that mutant pfcrt from the Dd2 parent accounts for >95% of the CQ response variation among the progeny [13]. Further evidence supporting pfcrt as the primary determinant of CQR has come from studies of culture-adapted field isolates, which show extensive linkage disequilibrium surrounding the pfcrt locus in CQ-resistant isolates [14]. These data suggest that strong selective sweeps drove mutant pfcrt through P. falciparum populations across the globe, a notion also supported by more recent studies of nucleotide diversity in geographically distinct strains [15], [16]. The PfCRT K76T mutation, ubiquitous to CQ-resistant strains, has proven to be a highly sensitive marker of CQR in vitro and is associated with a significantly increased risk of CQ treatment failure in vivo [17]–[19].
While these studies have demonstrated the primary importance of pfcrt in CQR, other evidence suggests that additional genes might contribute to the CQR phenotype. Most notably, a strain-dependent association has been demonstrated between mutant pfcrt and point mutations in pfmdr1. This may reflect parasite physiologic adaptations to counteract the fitness cost of mutant PfCRT, or an independent role for pfmdr1 in CQR [1], [8], [19]–[22]. Nevertheless, even with identical pfcrt and pfmdr1 alleles, large variations in response to CQ can exist, suggestive of a secondary effect of additional parasite factors [13], [23]–[25].
Clinically, resistance to CQ is graded by the World Health Organization ETF-LTF-ACPR system (corresponding to early treatment failure, late treatment failure, or adequate clinical and parasitological response), based on the time to manifest clinical or parasitological evidence of treatment failure [26]. Studies aimed at dissecting the roles of pfcrt and pfmdr1 mutations in modulating the different grades of in vivo resistance have shown an increased risk of early treatment failure with PfCRT K76T, which in some reports is augmented with PfMDR1 N86Y [27], [28]. However, the PfCRT K76T molecular marker cannot reliably predict CQ treatment failure, revealing moderate specificity of this marker. Discordance between in vitro parasite responses and in vivo patient outcomes following CQ treatment can be as high as 20% [17], [29]. This discordance can be partially attributed to host and environmental factors, including patient immunity, individual pharmacokinetic differences, polyclonal infections, and limitations in obtaining repeated measurements of drug susceptibilities with patient isolates [30]. An additional explanation could be the variable presence of additional parasite determinants.
We have previously adopted allelic exchange strategies to show that different mutant pfcrt alleles could confer verapamil (VP)-reversible CQR in a single, defined genetic background, the CQ-sensitive strain GC03 [31]. A separate transfection-based study found that pfcrt-mediated CQR in two geographically distinct strains, Dd2 (from Indochina) and 7G8 (from Brazil), was entirely dependent on the presence of the K76T mutation [32]. These strains were chosen as they encode a PfCRT haplotype frequently observed in Africa and Asia (Dd2) or in Papua New Guinea, South America and India (7G8). Both alleles have been documented in multiple clinical trials to be highly specific for CQ treatment failures, with repeated evidence of significant selection for mutant pfcrt of either allelic type in early or late treatment failures. Frequencies of mutant alleles in those cases often attained 100% [17], [33]–[37]. Trials were conducted in Africa, Southeast Asia, South America or the Oceanic region.
Here, we have assessed the effect of mutant pfcrt on the CQ response of three CQ-sensitive strains. We also describe two isolates from French Guiana that provide clinical validation of our genetic investigations. Our data reveal the existence of a mutant PfCRT-mediated CQ tolerance phenotype in some strains of P. falciparum.
Results
Generation of recombinant lines expressing mutant pfcrt
To define the impact of mutant pfcrt on CQ response in diverse genetic backgrounds, we developed an allelic exchange strategy based on a single round of homologous recombination and single-site crossover integration (Figure 1A), and applied this to the CQ-sensitive P. falciparum strains 3D7 (isolated in the Netherlands), D10 (Papua New Guinea), and GC03 (a progeny of the HB3×Dd2 genetic cross). This strategy differed from an earlier approach that required two rounds of allelic exchange to generate the desired recombinants [31]. Briefly, we constructed selectable transfection plasmids that contained a 2.9 kb pfcrt insert consisting of 0.5 kb of the endogenous 5′ untranslated region (UTR), exon 1, intron 1, and the remaining exons 2–13 (Figure 1A). This truncated 5′ UTR fragment (termed Δ5′) was previously observed by luciferase assays to give insignificant levels of activity (A. Sidhu, unpublished data). Single-site crossover between the pfcrt insert and the homologous pfcrt sequence upstream of codons 72–76 was predicted to replace the endogenous pfcrt gene with a recombinant allele harboring all the single nucleotide polymorphisms from the 7G8 or Dd2 pfcrt allele. Expression of this recombinant allele was driven by the endogenous full-length (3.0 kb) 5′ UTR and a previously characterized, functional 0.7 kb 3′ UTR (termed Py3′) from the pfcrt ortholog in Plasmodium yoelii [31]. In addition to these pBSD-7G8 and pBSD-Dd2 constructs, we also generated the control pBSD-GC03 plasmid that encoded the wild type (WT) pfcrt sequence in order to obtain recombinant control parasites.
Fig. 1. pfcrt allelic exchange strategy and molecular characterization of clones. 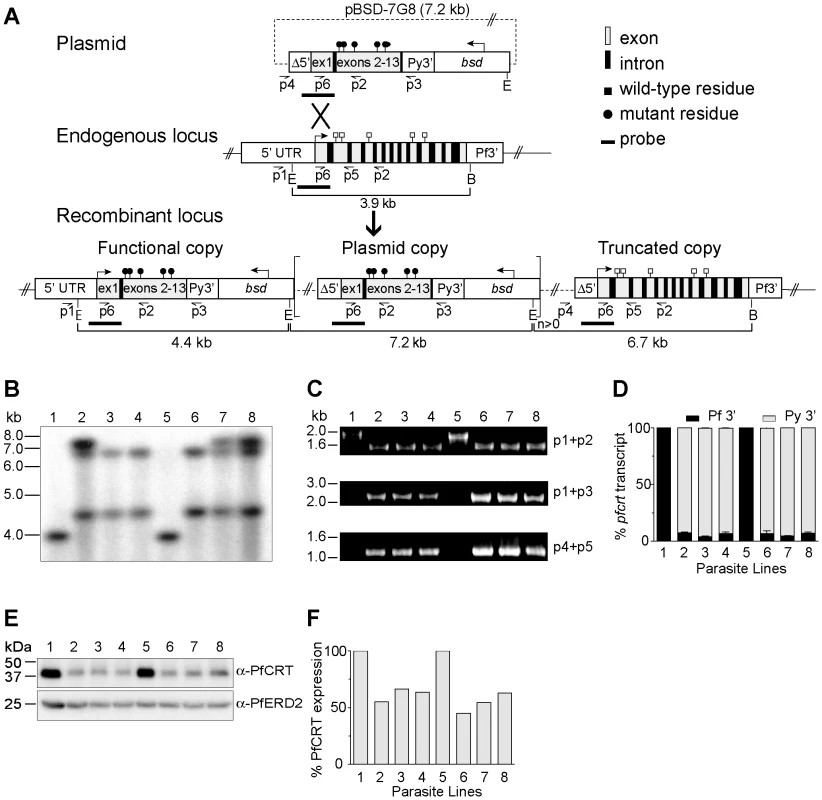
(A) Schematic representation of single-site crossover between the transfection plasmid pBSD-7G8 and the endogenous pfcrt allele, leading to expression of a recombinant allele containing the 7G8 polymorphisms (black circles), transcribed from the endogenous 3.0 kb full-length promoter. In some parasites the downstream plasmid sequence integrated as tandem linear copies (delineated as square brackets with a copy number n≥0). The distal truncated locus harbored the Δ5′ UTR, which was previously found in luciferase assays to have minimal activity. E, EcoRI; B, BglII. (B) Southern blot hybridization of EcoRI/BglII–digested genomic DNA samples hybridized with a pfcrt probe from the 5′ UTR and exon 1 region (depicted in panel A). (C) PCR analyses of the recombinant clones and parental lines. (D) Transcript levels from the functional and truncated pfcrt loci (terminated by Py3′ and Pf3′ UTRs respectively in the case of the recombinant clones). Data are presented as a percentage of total pfcrt transcript levels normalized against the respective WT control (3D7 or D10). (E) Western blot analysis of recombinant and parental lines, probed with antibodies to PfCRT or the ER-Golgi marker PfERD2 [80]. (F) Signals were quantified, normalized against PfERD2, and expressed as a proportion of the signals obtained in the appropriate parental line. (B–F) Lanes: 1-3D7, 2-3D7C, 3-3D77G8-1, 4-3D77G8-2, 5-D10, 6-D10C, 7-D107G8-1, and 8-D107G8-2. GC03 clones were also confirmed by PCR, sequencing, and Southern hybridization, and were found to have similar levels of pfcrt RNA and protein expression as compared with the 3D7 and D10 clones (data not shown). 3D7, D10 and GC03 parasites were transfected with the pBSD-7G8, pBSD-Dd2, or pBSD-GC03 plasmids and screened monthly by PCR for homologous recombination at the pfcrt locus. With the 7G8 and GC03 alleles, integration into the pfcrt locus was first detected within 60 days of electroporation, and subsequently cloned by limiting dilution. In contrast, the Dd2 allele failed to show PCR evidence of homologous recombination even after 200 days of continuous culture in 3 separate transfection experiments, suggesting that this allele was detrimental to the growth of 3D7 and D10 parasites (data not shown). Repeated efforts failed to transfect 7G8 and Dd2 pfcrt alleles into the CQ-sensitive strains MAD1 and Santa Lucia (from Madagascar and Santa Lucia, a kind gift of Drs Milijaona Randrianarivelojosia and Dennis Kyle respectively), as well as HB3. Recombinant parasites either never appeared following plasmid electroporation and drug selection, or the plasmids never integrated into the pfcrt locus.
Successful transfection of the 3D7, D10 and GC03 strains produced the recombinant mutant clones 3D77G8-1, 3D77G8-2, D107G8-1, D107G8-2, GC037G8-1 and GC037G8-2 (all generated from the plasmid containing the 7G8 pfcrt sequence) or the recombinant control clones 3D7c, D10c and GC03c clones (generated with the control plasmid harboring the WT pfcrt sequence; Table 1). Southern hybridization of EcoRI/BglII-digested genomic DNA samples with a pfcrt probe confirmed the expected recombinant locus, as evidenced by the loss of a 3.9 kb band present in the WT lines and the acquisition of 4.4 kb and 6.7 kb bands consistent with recombination in pfcrt (results shown for the 3D7 and D10 clones in Figure 1B). The 7.2 kb bands present in 3D7C, D107G8-1, and D107G8-2 were indicative of integration of tandem plasmid copies into the pfcrt locus.
Tab. 1. Summary of pfcrt-modified lines and reference strains. 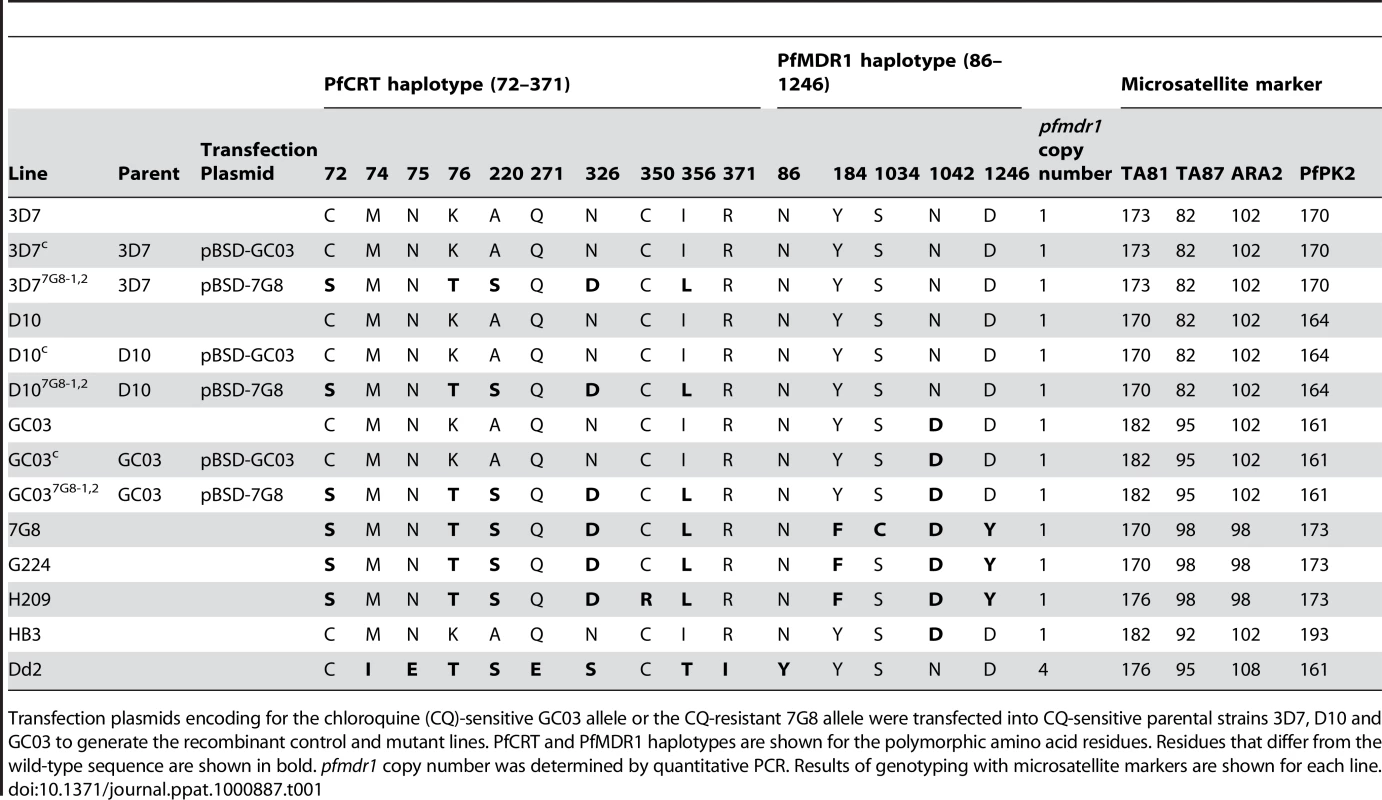
Transfection plasmids encoding for the chloroquine (CQ)-sensitive GC03 allele or the CQ-resistant 7G8 allele were transfected into CQ-sensitive parental strains 3D7, D10 and GC03 to generate the recombinant control and mutant lines. PfCRT and PfMDR1 haplotypes are shown for the polymorphic amino acid residues. Residues that differ from the wild-type sequence are shown in bold. pfmdr1 copy number was determined by quantitative PCR. Results of genotyping with microsatellite markers are shown for each line. We confirmed these recombination events using PCR analyses with a 5′ UTR-specific primer (p1) and an exon 5-specific primer (p2), which revealed a change in size from the 1.8 kb WT-specific band to a shorter 1.5 kb band in the recombinant controls and mutants reflecting the loss of introns 2–4 (Figure 1C). The recombinant controls and mutants also showed the acquisition of PCR bands specific for the full-length functional copy of the pfcrt locus (2.2 kb, p1+p3) and the downstream truncated copy (1.1 kb, p4+p5) (Figure 1C). Sequencing of these PCR products (data not shown) confirmed that the integration event placed the K76T mutation in the functional locus, and that the WT allele was displaced to the downstream non-functional locus. Reverse-transcriptase (RT)-PCR assays on synchronized ring stage RNA with primers specific to exons 2 and 5 (p6+p2) produced a single band corresponding to cDNA, with no evidence of genomic DNA contamination (data not shown). Sequence analysis of those products detected transcripts only from the functional recombinant locus (under the control of the 3.0 kb full-length 5′ UTR) and not from the downstream truncated locus (data not shown). No endogenous WT locus was detected in any recombinant clone.
To precisely assess the transcriptional status of the functional vs. the truncated pfcrt copies, we performed quantitative real-time RT-PCR utilizing primers specific for transcripts containing the Py3′ vs. Pf3′ UTRs respectively. Quantification of pfcrt steady state transcript levels was made by extrapolation from a standard curve generated from genomic DNA of D10C, which has a single copy of the pBSD-GC03 plasmid integrated into the pfcrt locus (Figure 1B). Results showed that transcription from the functional pfcrt allele with Py3′ accounted for 93–95% of the total pfcrt transcript within each line (Figure 1D). Western blot analysis showed that PfCRT protein levels in the recombinant 3D7 and D10 lines were 55–66% and 45–63% those observed in the parental controls respectively (Figures 1E and 1F). This finding of reduced pfcrt transcript and protein expression levels following allelic exchange is consistent with earlier pfcrt transfection studies [31], [32], [38]. Importantly, those studies have shown that reduced pfcrt expression in recombinant lines causes a concomitant reduction in CQ IC50 values, which thus become lower than the IC50 values observed in parasites harboring non-recombinant pfcrt. In our drug assays, the IC50 value refers to the drug concentration that inhibits incorporation of [3H]-hypoxanthine, a marker of in vitro parasite growth, by 50%.
Mutant pfcrt is insufficient to confer high-level chloroquine resistance in the 3D7 and D10 genetic backgrounds
Once the desired integration events were confirmed, we assessed the effect of mutant pfcrt on the CQ response in the recombinant lines. In the 3D7 background, mutant pfcrt was found to confer a 2.7-fold increase in CQ IC50 values (mean±SEM CQ IC50 values of 84±14 nM and 79±11 nM for 3D77G8-1 and 3D77G8-2 respectively) compared to the 3D7 recombinant control (29±2 nM, P<0.001; Figure 2A, Table S1). These values were 2.4-fold lower than the IC50 values for WT 7G8 (190±14 nM). For the D10 mutants, there was no significant increase in CQ IC50 values for D107G8-1 and D107G8-2 compared to D10C (63±11 nM, 71±16 nM, and 45±3 nM, respectively, P>0.05).
Fig. 2. In vitro response of pfcrt-modified clones to chloroquine and its primary metabolite monodesethylchloroquine. 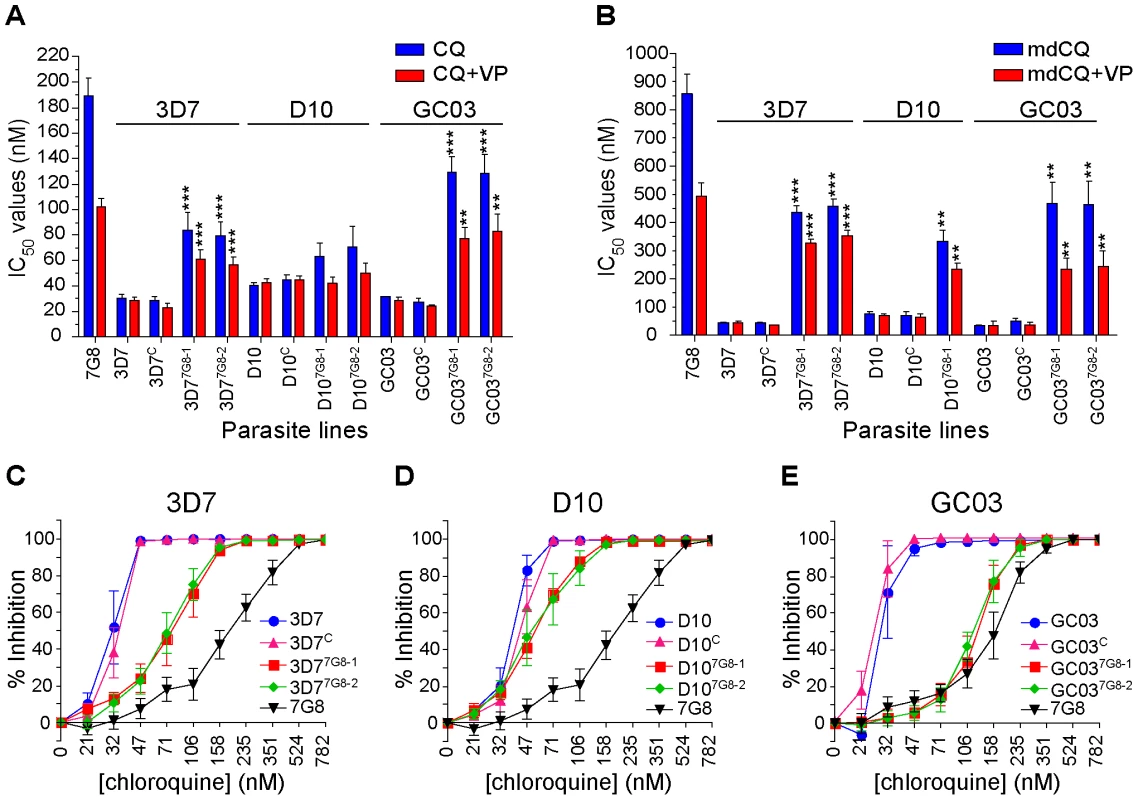
In vitro [3H]-hypoxanthine incorporation assays were performed with the WT, control, and mutant pfcrt clones. All lines were tested in duplicate against CQ±VP and mdCQ±VP an average of 7 times (range 4–11; see summary in Table S1). Mean±SEM IC50 values are presented for (A) CQ and (B) its metabolite mdCQ. Statistical comparisons comparing mutant pfcrt-modified lines against recombinant control lines of the same genetic backgrounds were performed using one-way ANOVA with a Bonferroni post-hoc test. **P<0.01; ***P<0.001. (C–E) Percent inhibition of growth (shown as means±SEMs derived from all assays) across a range of CQ concentrations for (C) 3D7, (D) D10, and (E) GC03 lines. When tested against the primary in vivo metabolite monodesethyl-chloroquine (mdCQ), a significant decrease in susceptibility was found in both genetic backgrounds. The 3D7 mutant clones demonstrated a 10-fold increase in mdCQ IC50 values compared to 3D7C (P<0.001, Figure 2B). In comparison, the IC50 values for the D107G8-1 mutant were 5-fold higher than D10C (P<0.01, Figure 2B, D107G8-2 was not tested). Nevertheless, the mdCQ IC50 values in both backgrounds were approximately 2–fold lower than those observed in WT 7G8, suggesting that mutant pfcrt was insufficient to confer high-level mdCQ resistance to 3D7 and D10 parasites.
These findings of a relatively moderate, strain-dependent decrease in CQ susceptibility in the 3D7 and D10 pfcrt mutants contrasted with our earlier observation that the introduction of 7G8 mutant pfcrt in the GC03 background resulted in CQ IC50 values >100 nM [31]. To directly compare the effects of mutant pfcrt between strains, and to assess for any potentially confounding differences in our transfection strategies, we generated recombinant control (GC03C) and mutant clones expressing the 7G8 allele (GC037G8-1 and GC037G8-2) using our single-round transfection strategy. These clones were confirmed by PCR, sequencing, and Southern hybridization, and were found to have similar levels of pfcrt RNA and protein expression compared to the 3D7 and D10 clones (data not shown). In the GC03 background, introduction of the 7G8 mutant pfcrt allele increased the CQ IC50 values 4.7-fold (P<0.001), from 27±3 nM for GC03C to ∼130±8 nM for both recombinant clones (Figure 2A, Table S1), and increased the mdCQ IC50 values by 9-fold (P<0.01; Figure 2B). These determinations included four independent assays that directly compared GC037G8-1 and GC037G8-2 with the C67G8 line. The latter was produced using our earlier pfcrt modification strategy involving consecutive rounds of allelic exchange [31]. C67G8 also expresses the 7G8 pfcrt allele in the GC03 background, yet differs from the clones produced in the current study in that C67G8 contains both the human dihydrofolate reductase and the bsd selectable markers, and lacks the 0.5 kb 5′UTR present in the downstream pfcrt loci in the GC037G8-1 and GC037G8-2 clones (see Figure 1A). Drug assays with these lines produced CQ IC50 values of 131±7, 129±8 and 130±7 nM for GC037G8-1, GC037G8 and C67G8 respectively (Table S1). These results are comparable to our published data with C67G8 (127±17 nM; [31]) and are consistent with both allelic exchange strategies producing the same CQ responses. Our data from all three strains also provide clear evidence that the degree of CQR conferred by mutant pfcrt is strain-dependent.
We also found that the genetic background influenced the degree of VP chemosensitization, a hallmark of P. falciparum CQR [39]. In 3D7 and D10, expression of mutant pfcrt conferred a VP reversibility of 24±1% and 28±1% (calculated as the mean±SEM of percent reversibility for all CQ and mdCQ values), compared to 44±2% for GC03 (Figure S1). Notably, significant VP reversibility occurred in the D10 mutants despite the lack of a significant increase in CQ IC50 values (Figure 2D, Table S1). By comparison, VP reversibility for 7G8 CQ and mdCQ responses was 46±3% (Figure 2B). This is lower than the degree of VP reversibility that results from expression of the Dd2 pfcrt allele [31], [40].
Analysis of the dose response curves generated during these studies revealed a more complex picture than was evident from the IC50 values alone. For all three genetic backgrounds, introduction of the 7G8 mutant allele into the CQ-sensitive strains caused a pronounced change in the slope of the dose-response profiles, with evidence of continued growth at high CQ concentrations (Figures 2C–E). This was particularly pronounced for the recombinant D107G8-1 and D107G8-2 lines, whose CQ IC50 values were similar to those of D10 and D10C, yet whose IC90 values (i.e. the drug concentrations that inhibited [3H]-hypoxanthine uptake into cultured parasites by 90%) were greatly elevated. Indeed, analysis of the CQ IC90/IC50 ratios for the lines in each genetic background revealed significant increases in the mean ratios of the mutant lines (Figure S2). For the 3D7 and D10 backgrounds in particular, the relatively modest increase in CQ IC50 values appeared to be compensated by an increased ability of these parasites to withstand high CQ concentrations.
The genetic background dictates whether mutant pfcrt confers chloroquine resistance or tolerance
We posited that these elevated IC90 values imparted by mutant pfcrt subtly reflected a CQ tolerance phenotype. To test this, we assayed our lines for the ability to survive treatment with 50 nM CQ, a concentration that was lethal after three generations of exposure for all three WT strains, and 80 nM CQ, which substantially exceeded each of their CQ IC90 values (Table S1).
Parental, control, and mutant lines were assayed for in vitro recrudescence (defined as 50% of cultures testing positive for growth) after a six-day exposure to CQ. The parental and recombinant control lines from the 3D7, D10, and GC03 backgrounds showed no signs of growth at 30 days post-exposure to 50 nM CQ (Figures 3A and 3B). In contrast, 3D77G8-1 recrudesced at 9 and 13 days post-treatment with 50 nM and 80 nM CQ respectively (Figure 2A). We also tested 3D77G8-1 that had been pretreated with 50 nM CQ for 3 generations approximately 30 days earlier (3D77G8-1/preCQ), and observed similar rates of recrudescence. All untreated lines were positive at day 7, as was WT 7G8 that showed no inhibition of growth with 80 nM CQ treatment.
Fig. 3. CQ recrudescence data for pfcrt -modified and parental clones. 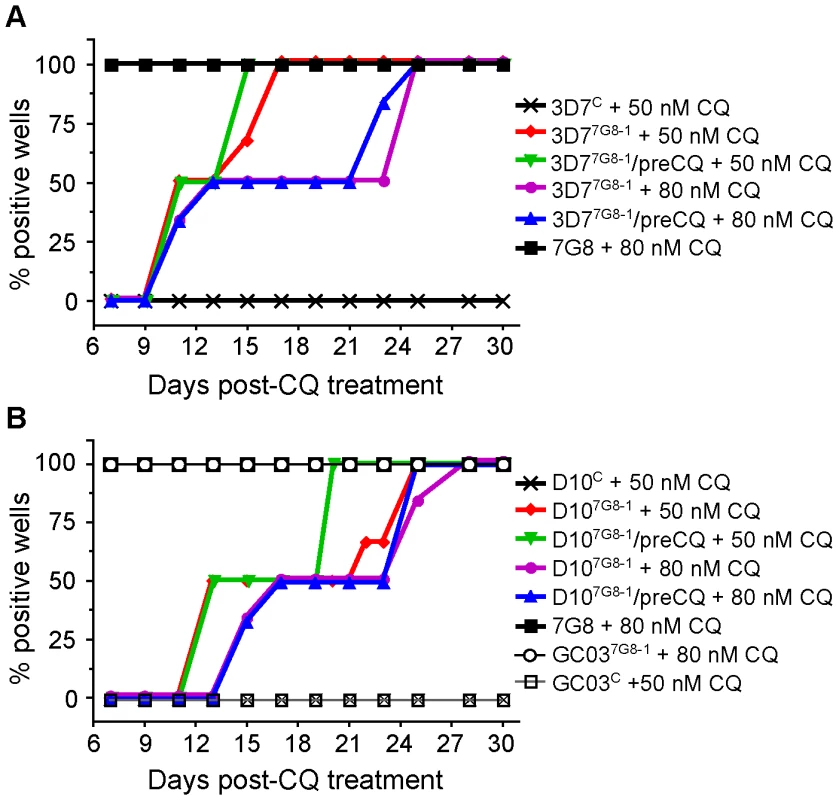
Lines were subjected to 50 nM or 80 nM CQ for 6 days and assayed for recrudescence every 2–3 days from days 7–30. Pooled data from two independent experiments were plotted as the percent of positive wells as a function of time post-CQ exposure. The panels show (A) 3D7 and (B) D10 clones and controls, including recombinant clones pretreated with 50 or 80 nM CQ. All no-treatment controls were positive on day 7 (as shown for 7G8+80 nM CQ), as was GC037G8-1 treated with both 50 nM and 80 nM CQ. Although the introduction of mutant pfcrt resulted in no significant increase in CQ IC50 values in the D10 background, both D107G8-1 and pretreated D107G8-1/preCQ recrudesced at days 13 and 17 with treatment with 50 nM and 80 nM CQ, respectively (Figure 3B). In the GC03 background, GC037G8-1 showed no inhibition of growth at 7 days with both 50 nM and 80 nM CQ treatments, reflecting the high-level CQR phenotype imparted by mutant pfcrt in this strain.
Characterization of chloroquine-sensitive P. falciparum clinical isolates from French Guiana that possess mutant pfcrt
Given the evidence that mutant pfcrt was insufficient to confer CQR in all genetic backgrounds, we asked whether there were CQ-sensitive parasites harboring mutant pfcrt in the field. After an extensive search, this led to the identification of two clinical isolates from French Guiana that express the PfCRT K76T marker for CQR but are sensitive to CQ. These isolates, G224 and H209, were harvested in 2003 and 2004, respectively, and were genotyped at the pfcrt and pfmdr1 loci. The PfCRT haplotype of G224 was found to be identical to that of 7G8, whereas H209 possessed a C350R mutation that has not been previously described (Table 1). Both G224 and H209 possessed a single copy of pfmdr1 with the same haplotype that differed from 7G8 only at position 1034. Western blot analyses revealed equivalent levels of PfCRT expression compared to 7G8 (data not shown).
Drug susceptibility assays using CQ and mdCQ showed that these strains had low IC50 values for CQ (mean IC50 values of 52±8 nM and 35±7 nM for G224 and H209, respectively) and mdCQ (mean IC50 values of 349±46 nM and 70±9 nM) (Figures 4A and 4B, Table S1). Further, both G224 and H209 demonstrated VP reversibility of their CQ and mdCQ response (averaging 37% and 35%, respectively; Table S1, Figure S1). Analysis of the CQ inhibition curves revealed that the IC90 values were skewed towards the IC90 of 7G8 (Figure 4C), reminiscent of the effect seen in our 3D7 and D10 mutant pfcrt lines (Figures 2C and 2D). This was particularly pronounced for G224, whose IC90 for CQ was 123±27 nM. When tested for in vitro recrudescence after a 6-day exposure to CQ, G224 recrudesced at days 11 and 17 when treated with 50 nM and 80 nM CQ respectively (Figure 3D). Interestingly, H209 showed recrudescence at days 21 and 25 for 50 nM and 80 nM CQ respectively, despite having a very low CQ IC90 value of 44±7 nM.
Fig. 4. Characterization of the French Guiana isolates G224 and H209. 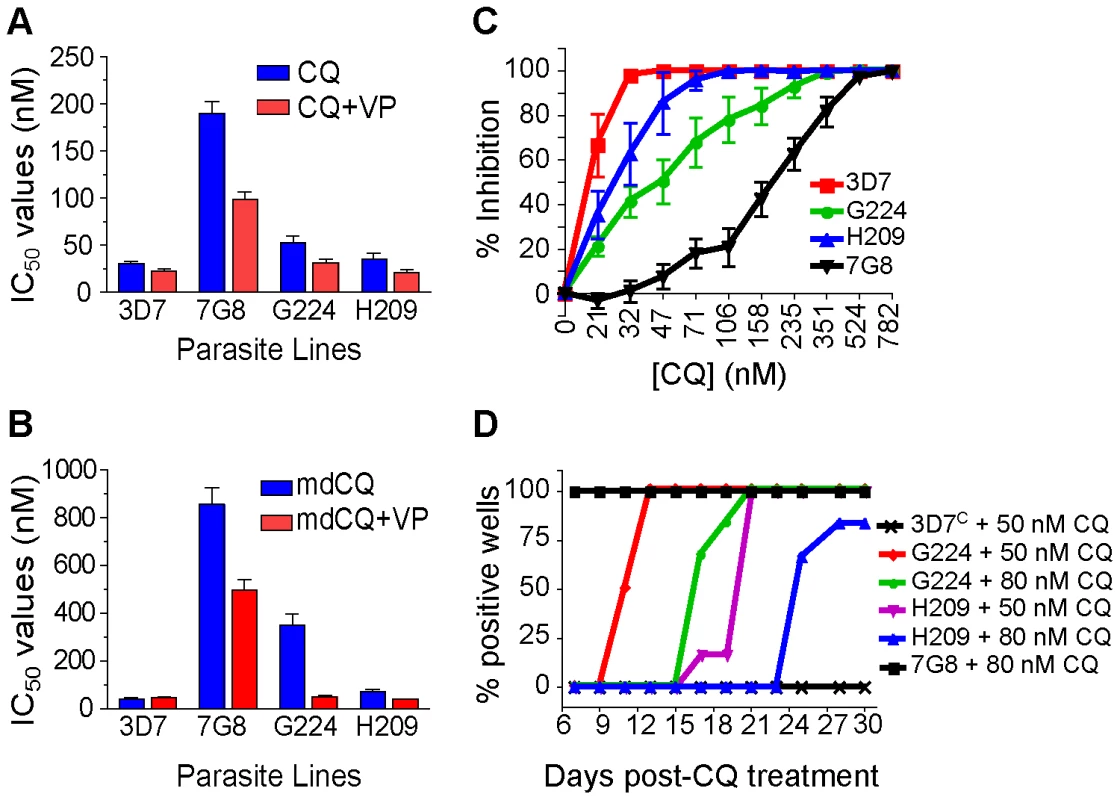
(A–B) In vitro [3H]-hypoxanthine incorporation assays against CQ±VP (A) and mdCQ±VP (B) were performed with a CQ-sensitive (3D7) and a CQ-resistant (Dd2) control line. All lines were tested in duplicate on average 6 times (range 3–8). Mean±SEM IC50 values were derived by linear extrapolation. (C) Percent inhibition of growth (means±SEM derived from all assays) determined across a range of CQ concentrations. (D) Lines were exposed to 50 nM or 80 nM CQ for 6 days and assayed for recrudescence every 2–3 days from days 7–30. All no-treatment controls were positive on day 7 (as shown for 7G8+80 nM CQ). The genetic background also determines the effect of mutant pfcrt on response to other antimalarials
To test whether the host strain also influenced the effect of mutant pfcrt on parasite response to other drugs, particularly those currently used in ACTs, we tested our lines against quinine (QN), artemisinin (ART), monodesethyl-amodiaquine (mdADQ, the potent in vivo metabolite of amodiaquine), lumefantrine (LMF), and piperaquine (PIP). The responses of the French Guiana isolates G224 and H209 were also assessed.
In the 3D7, D10 and GC03 backgrounds, we observed no effect of mutant pfcrt on QN response (Figure 5A, Table S1). Interestingly, the highest QN IC50 values were observed with H209, which showed a moderately high level of resistance (405±40 nM). When tested against ART, introduction of mutant pfcrt showed a significant 2–fold decrease in IC50 values in the D10 and GC03 backgrounds, when compared to recombinant clones expressing WT pfcrt (P<0.05 and P<0.01 respectively; Figure 5B). 3D77G8-1 also yielded a 33% lower ART IC50 compared to the 3D7C control, however this did not attain statistical significance (P = 0.06). Again, the highest ART IC50 values were observed with H209 (Table S1). For mdADQ, 3D77G8-1 had a 1.5-fold increase in IC50 value compared to 3D7C (P<0.05), and GC037G8-1 showed an even more pronounced (2.6-fold) increase compared to GC03C (P<0.01; Figure 5C). There was no effect of mutant pfcrt on mdADQ response in the D10 background. With this drug, G224 and H209 were both moderately resistant, as was 7G8.
Fig. 5. In vitro response of pfcrt-modified clones and French Guiana isolates to clinically important antimalarials. 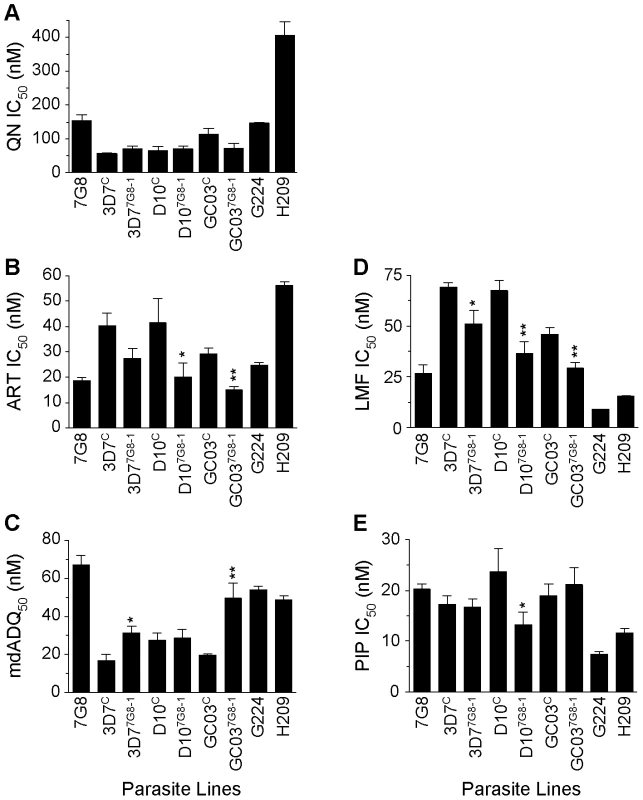
In vitro [3H]-hypoxanthine incorporation assays were performed in duplicate an average of 6 separate times (range 3–12 independent assays). Mean±SEM IC50 values are presented for (A) quinine (QN), (B) artemisinin (ART), (C) monodesethyl-amodiaquine (mdADQ), (D) lumefantrine (LMF), and (E) piperaquine (PIP). Statistical comparisons comparing mutant pfcrt-modified lines against recombinant control lines of the same genetic backgrounds were performed using unpaired student t tests. *P<0.05; **P<0.01. For brevity, a single recombinant mutant clone is presented for each host strain. Results from additional recombinant mutant lines are included in Table S1. Introduction of mutant pfcrt was also found to confer significantly increased sensitivity to LMF in all three strains, equating to a 23%, 44%, and 35% decrease in IC50 values for the 3D7, D10, and GC03 backgrounds respectively (Figure 5D). H209 was also found to be less susceptible to LMF than was G224, mirroring their responses to QN and ART. Finally, we found that mutant pfcrt had a significant effect on PIP response only in the D10 background, in which D107G8-1 was 1.7-fold less sensitive than D10C (P<0.05). G224 and H209 were found to be 2.7 - and 1.7-fold more sensitive to PIP when compared to 7G8.
Discussion
Here, we provide evidence that the genetic background of P. falciparum determines whether expression of mutant pfcrt allele confers a full CQR phenotype, as defined by CQ IC50 values that exceed the in vitro CQR threshold [30], or instead mediates increased tolerance to CQ, as evidenced by dose-response shifts manifesting primarily at the IC90 level. All our recombinant clones expressing mutant pfcrt recrudesced in vitro after being exposed for three generations to concentrations of CQ that were uniformly lethal to CQ-sensitive parasites; however the rate of recrudescence varied with the genetic background. The GC03 mutant lines, which had the highest CQ IC50 values, showed no growth inhibition. In contrast, the pfcrt-mutant lines generated in the 3D7 and D10 backgrounds, as well as the clinical isolates G224 and H209, required 1–3 weeks for the detection of recrudescent parasites.
Based on our findings, we propose that IC50 values, which typically constitute the sole measurement of CQ response in vitro, adequately identify high-level CQR but are insufficient to detect strains that have low-level resistance or manifest tolerance to CQ. Instead, our data suggest that accurate determinations of IC90 values provide a more predictive measure of whether parasites can recrudesce in the presence of CQ concentrations that are lethal to drug-sensitive parasites, a trait that we here refer to as CQ tolerance. Tolerance is also apparent in decreased parasite susceptibility to the primary drug metabolite mdCQ. We posit that pfcrt-mediated CQ tolerance might be an important component of late treatment failures in patients. These are classified as cases where symptoms occur during a follow-up period of 4–28 days post CQ treatment, or asymptomatic infection appearing 7–28 days post-treatment (see the WHO 2006 publication on malaria treatment: http://whqlibdoc.who.int/publications/2006/9241546948_eng_full.pdf). In contrast, early treatment failures might result more often from infections with parasites in which mutant pfcrt exerts a higher degree of CQR. Early treatment failures are classified as the development of clinical or parasitological symptoms during the first three days following CQ treatment. We note that clinically, care must be taken when evaluating early failures, as these can also include patients that respond relatively slowly to treatment yet progress to full cure. Moreover, the joint effects of low-level CQ resistance reported here and acquired protective immunity might help explain why CQ treatment can successfully cure some infections harboring mutant pfcrt parasites in semi-immune individuals [1], [18]. The importance of immunity in shaping the host's ability to resolve drug-resistant infections harboring mutant pfcrt was first demonstrated in work from Mali that found that successful CQ treatment of pfcrt mutant parasites was strongly dependent on age, a known surrogate for protective immunity in endemic areas [18]. These data complement other observations in the malaria literature indicating that the immune response can allow a relatively ineffective drug to clear an infection, and even at times clear infections without therapy [41], [42]. Our data extend these reports by suggesting that successful CQ treatment of drug-resistant parasites is dependent both on the level of host immunity and the strain-dependent extent to which mutant pfcrt imparts CQR.
A review of our CQ IC50 data reveals a relatively weak effect of mutant pfcrt, which attained the widely used in vitro CQR threshold of 80–100 nM only for GC03 (Figure 2, Table S1). This threshold, however, was based on studies with field isolates [30] and does not readily extrapolate to our pfcrt-modified parasite lines. Our data (Figure 1) show that these lines underexpress pfcrt, a consequence of allelic exchange into this locus that was earlier shown to cause artificially low CQ IC50 values whose level of reduction was concordant with the degree of reduced expression [31], [32], [38]. In our current study, the importance is not the absolute levels of CQR that we measured, but rather the finding that the genetic background of CQ-sensitive strains dictates a spectrum of mutant pfcrt-mediated changes in CQ response that ranges from tolerance to high-level resistance.
We note that our data were obtained with the 7G8 pfcrt allele, which is known to have appeared independently in South America and the Oceanic region in or near Papua New Guinea and has recently spread throughout India [43]. The 7G8 haplotype (C72S/K76T/A220S/N326D/I356L) shares only two mutations (K76T/A220S) with the Dd2 haplotype (I74E/N75E/K76T/A220S/Q271E/N326S/I356T/R371I) that is common to Africa and SE Asia [11], [14], [44]. Our earlier allelic exchange studies on recombinant lines generated in the GC03 strain found that the 7G8 pfcrt haplotype confers a lower degree of resistance than that imparted by the Dd2 allele (averaging 15% and 45% less or CQ and mdCQ respectively). This was consistent with the intrinsic differences observed between the parental 7G8 and Dd2 strains [31]. It is possible that in the D10 and 3D7 strains, higher degrees of resistance might have been observed with the Dd2 allele, however we were unable to test this. We note that D10 originates from Papua New Guinea where the 7G8 allele is highly prevalent, and our lack of success with introducing the Dd2 pfcrt allele into either this strain or 3D7 suggests a physiologic context that precludes expression and normal viability. Other evidence of a fitness cost imparted by the Dd2 allele comes from studies in Malawi showing that this allele is progressively lost from the parasite population in the absence of sustained CQ pressure [45], [46].
Field studies have sometimes reported discordance in the association of K76T and in vitro CQR, suggesting the contribution of other genetic loci [24], [47]–[49]. However, the interpretation of these results has been confounded by potential inaccuracies stemming from measuring one-time drug responses from frequently polyclonal fresh patient isolates. Our study provides, to the best of our knowledge, the first report of culture-adapted, monoclonal isolates that harbor mutant pfcrt and that, based on multiple drug susceptibility assays, show low CQ IC50 values that fail to meet the standard criteria for CQR. These findings, obtained with the G224 and H209 isolates from French Guiana, therefore provide indisputable evidence that mutant pfcrt is insufficient to confer CQR to all genetic backgrounds. Nevertheless, both isolates exhibited tolerance to high CQ concentrations and recrudesced under CQ pressure. Microsatellite typing revealed a close genetic similarity between G224 and 7G8 (Table 1), with the exception of the residue at PfMDR1 position 1034 that could potentially affect CQ response [22], [50].
Of particular interest, H209 was highly sensitive to CQ and yet demonstrated delayed recrudescence (Figure 4). This might in part be attributable to the PfCRT C350R charge substitution in transmembrane domain 9, a region postulated to function in substrate binding and translocation [51]. Studies are underway to introduce the H209 pfcrt allele, encoding the C350R mutation, into GC03 parasites to compare these to the GC037G8 parasites whose expressed pfcrt allele differs only at codon 350 (Table 1). We note that an adjacent charge substitution at residue 352 (Q352K/R) was previously selected by QN pressure in a CQ-resistant line, with a concomitant reversion to CQ-sensitivity [52]. The H209 line also showed elevated IC50 values for QN, as well as ART, when compared to G224 and 7G8 (Figure 4). Of note, QN-doxycycline, and more recently artemether-LMF, have been implemented as first line antimalarials in French Guiana since the cessation of CQ use for the treatment of P. falciparum malaria in the mid 1990s [53]. Indeed, a recent report from French Guiana documented the existence of several field isolates with elevated artemether IC50 values (>30 nM in 7 of 289 isolates), suggesting decreased susceptibility to this agent [54]. G224 was tested at that time and found to have an artemether IC50 value of ∼1 nM. H209, which yielded artemisinin IC50 values two-fold higher than G224 (Table S1), was isolated one year later. Our subsequent studies reveal comparable IC50 values between these two lines with the more potent clinical derivatives artemether, artesunate and artemether (values provided in Table S1).
ACTs are rapidly assuming the role of first line antimalarials around the world [55]. Our studies with isogenic pfcrt-modified lines confirm previous reports that mutations in PfCRT can significantly affect parasite susceptibility to many of the antimalarials that constitute these ACTs [19], [56], and provide evidence that for certain drugs this effect is strain-dependent (Figure 5). In the case of the fast-acting ART, all three strains displayed enhanced susceptibility upon introduction of mutant pfcrt. With the amodiaquine metabolite mdADQ, elevated IC50 values were noted in two of the three recipient strains, supporting earlier epidemiological evidence that mutant PfCRT might contribute to a multigenic basis of amodiaquine resistance ([57]–[59]; see below). The opposite effect was observed with the bisquinoline PIP, which is highly effective against CQ-resistant strains of P. falciparum [60], and for which we observed a strain-dependent increase in susceptibility. For LMF, significantly enhanced susceptibility was observed in all three genetic backgrounds, supporting recent field studies [59], [61]. The generally enhanced potency of LMF and artemisinin derivatives against mutant pfcrt parasites bodes well for the widely used LMF-artemether co-formulation. The enhanced susceptibility conferred by the mutant pfcrt 7G8 allele to the ACT partner drugs LMF and PIP, but not amodiaquine, has potentially important implications in regional antimalarial drug policy.
Our pfcrt and CQ data speak to a requirement for additional parasite factors that, at least in some strains, either augment the level of PfCRT-mediated CQR or on the contrary, create an intracellular physiologic environment in which PfCRT is unable to exert its full capacity to dictate CQR [62], [63]. pfmdr1 would appear to be one gene that contributes to this strain-dependent effect. Transfection-based studies have shown that in CQ-resistant strains that harbor mutant pfcrt, mutations in pfmdr1 can contribute to elevated CQ IC50 values, but only in a subset of strains. Mutant pfdmr1 alone shows no effect on CQ response in sensitive parasites harboring wild-type pfcrt [19], [20]. Evidence from CQ treatment trials in African, Southeast Asia and the Oceanic region show that mutant pfmdr1 is associated with an increased risk of CQ treatment failure, however this risk is usually substantially higher in the presence of mutant pfcrt [17], [28], [36], [64], [65]. Of note, while mutant pfcrt is virtually ubiquitous to CQ treatment failures, mutant pfmdr1 is often absent ([65] and references therein). Functional assays have yet to be developed to test whether pfmdr1 can directly reduce drug toxicity, or instead is associated with CQR because of its non-random association with mutant pfcrt, which potentially could relate to improved parasite fitness [66].
We note that in our study, pfmdr1 cannot account for differences in the extent to which mutant pfcrt affects CQ response, as both the resistant 3D7 and the tolerant D10 mutants (3D77G8 and D107G8 respectively) share the same wild-type pmfdr1 haplotype (Table 1). The highly resistant GC03 mutants (GC037G8) differ in having the pfmdr1 N1042D mutation that in allelic exchange studies had no impact on CQ response (although it did affect a number of other antimalarials including QN, mefloquine and ART; [21]). Clear evidence that mutant forms of PfCRT and PfMDR1 can combine in a region-specific manner to create higher levels of drug resistance comes from the recent study by Sa et al. [67], showing that the 7G8 South American haplotypes of these two determinants produce high-level resistance to mdADQ. This study also found that the Asian/African Dd2 haplotype of PfCRT was associated with high level CQR with minimal apparent contribution from variant PfMDR1 haplotypes.
Why has no gene other than pfmdr1 been found associated with CQR? In the case of the HB3×Dd2 genetic cross where mutant pfcrt was clearly the primary determinant, evidence that modulatory factors must exist was provided by the 2.7-fold spread in CQ IC50 values observed among the CQ-resistant progeny [13]. Such factors may be present within the 36 kb CQR-associated linkage group harboring pfcrt [10], [68], or potentially might already be present in the HB3 parent, thereby rendering this competent for CQR and masking the inheritance of a secondary determinant [8]. To test the latter hypothesis, we attempted to introduce mutant pfcrt into the HB3 strain, but were unable to obtain integrants in three independent transfection experiments (data not shown). Independent genomic approaches analyzing linkage disequilibrium in CQ-resistant isolates have also failed to identify any gene besides pfcrt [14]–[16], [69], as elaborated upon below.
The genetic identity of these secondary determinants associated with CQR may reflect the geographic distribution of distinct PfCRT haplotypes around the globe [19]. Indeed, the PfCRT 7G8 haplotype found in South America and the Pacific is typically associated with PfMDR1 N1042D/D1246Y (±S1034C), whereas the PfCRT Dd2 haplotype common to Asia and Africa is often associated in CQ-resistant isolates with PfMDR1 N86Y [43], [50]. Identifying additional genetic determinants has been complicated by the complexity of performing genome-wide association studies with large numbers of culture-adapted parasite lines from different geographic regions and comparing these to parasite drug responses [50], [70]. Major advances have recently been achieved in a seminal study by Mu et al. [69], who performed genome-wide association studies with a 3,000 single nucleotide diversity array probed with DNA from189 culture-adapted P. falciparum lines from Africa, Asia, Papua New Guinea and South America, and compared their genetic diversity with CQ response. When accounting for local population structures, the authors found associations between CQ response and changes in pfcrt, pfmdr1, and surprisingly a putative tyrosine kinase (PF11_0079). These associations could only readily be discerned in African populations where a sufficient number of CQ-sensitive strains could be identified; as opposed to South American, Asian and Papua New Guinean strains where mutant pfcrt remained at a high prevalence. Of the genes listed above, pfcrt stood out as being one of handful of genes in the parasite genome that were apparently under very substantial selection pressure in all three populations studied - Asia, Africa and South America. No other genes were convincingly associated with CQR, even though a number of genes potentially involved in drug transport (including the putative drug/metabolite transporter PF14_0260, and the ABC transporters PF13_0271 and PFA0590w) were found to be under lesser selection pressure in local populations. We note that evidence of selection was also observed in genes adjacent to pfcrt, although these may simply represent genetic hitchhiking and insufficient time for genetic recombination to have disrupted these associations.
Our conclusion from these studies is that mutant pfcrt has been the dominant genetic force that has driven CQR across the globe, with some degree of participation from mutant pfmdr1, and that even the phenotype of CQ tolerance observed herein in D10 parasites expressing mutant pfcrt would appear sufficient to confer substantial levels of viability during a course of CQ treatment. This level of protection against drug onslaught, while appearing modest in vitro, appears to have sufficed for selection and rapid mobility through parasite populations subjected to CQ treatment. Experiments to define secondary determinants that can augment CQR would require, as an example, deeper sequence coverage of the set of 189 genotypically and phenotypically characterized isolates mentioned above [69], followed by quantitative trait loci analysis that computationally subtracted the dominant effect of pfcrt to identify potential residual associations in local parasite population structures.
Other hypothesis-driven approaches to identify secondary parasite factors could involve investigations into the function of mutant PfCRT and the cellular basis of CQ mode of action. Recent studies based on heterologous expression of codon-harmonized, surface-expressed PfCRT in Xenopus laevis oocytes have recently provided compelling evidence that mutant PfCRT can transport CQ [71], a finding consistent with earlier evidence from Pichia pastoris and Dictyostelium discoideum [72], [73]. The Xenopus study also identified peptides that could interfere with transport of radiolabeled CQ through mutant PfCRT, raising the possibility that PfCRT is involved in transport of certain peptide sequences out of the DV and into the cytoplasm ([74] and references therein). Secondary factors could potentially alter the kinetics of peptide production (resulting from hemoglobin proteolysis in the DV) or their translocation into the parasite cytosol and subsequent conversion into amino acids that can be incorporated into newly synthesized proteins.
Other potential factors could relate to the tri-peptide glutathione (GSH) and redox regulation. Interestingly, an earlier study by Ginsburg and colleagues reported that altering the intracellular levels of GSH caused a corresponding shift in CQ susceptibility in P. falciparum [75]. Work from these authors led to the hypothesis that GSH could degrade iron-bound heme (a toxic byproduct of hemoglobin degradation) that might be released into the parasite cytosol as a result of CQ action [76]. Further support for a relationship between GSH and levels of CQR was recently obtained following the genetic disruption of the P. falciparum gene PfMRP (PFA0590w), whose ABC transporter product has been localized to the parasite surface. These knockout parasites, generated in the CQ-resistant W2 strain, accumulated more radioactive GSH and CQ and became less resistant to CQ as well as several other antimalarials [77]. Indirect additional evidence of a potential link between CQR and GSH comes from the recent report that PfCRT homologs in Arabidopsis thaliana can mediate GSH transport when assayed in Xenopus oocytes [78]. Collectively, these data suggest that GSH homeostasis is related to CQR, and possibly to PfCRT, in a strain-dependent manner. A multifactorial, and potentially region-specific basis for these differences would have precluded their identification to date. Further investigations into parasite cell biology, employing genomic, proteomic and metabolomic studies to compare CQ response phenotypes within regional populations, are warranted to identify these molecules and their determinants. French Guinea may well provide an ideal set of geographically restricted isolates in which to define these factors, because of its complex history of antimalarial drug usage and the existence of mutant pfcrt strains with both resistance and tolerance phenotypes.
Materials and Methods
Ethics statement
Informed consent was not required for this study as the collection of samples from malaria patients for drug susceptibility testing are part of the French national recommendations for the care and surveillance of malaria. As the Pasteur Institute French Guiana laboratory is the regional malaria reference center, blood samples are sent to the laboratory by practitioners (from health centers, private medical offices and hospitals) for drug susceptibility testing, as part of the national regular medical surveillance. This included in vitro drug susceptibility testing and assessments of molecular markers. This research is mandated by the French Ministry of Health, and has been approved by the Institutional Review Boards of the Pasteur Institute in Paris and in French Guiana.
Plasmid constructs
pfcrt plasmid inserts were assembled from two contiguous sequences. The first 800 bp sequence, spanning 0.5 kb of the pfcrt 5′ UTR (denoted Δ5′) through to the intron 1/exon 2 junction (nucleotides 22960–23747 of the GenBank accession number AF030694), was amplified from Dd2 genomic DNA with the primers p251 and 10AE1-3′A (a list of these and all other primers used in this study is provided in Table S2). A 2.1 kb fragment corresponding to pfcrt exons 2–13 and the 3′ UTR of the P. yoelii ortholog pycrt (termed Py3′) was released following AvrII/BamHI digestion of the plasmids pBSD/AE123 -7G8, -GC03, and -SC01 (the latter has the Dd2 sequence) [31]). These two sequences were assembled in pCR2.1 (Invitrogen) to generate a 2.9 kb pfcrt fragment containing Δ5′, exon 1, intron 1, exons 2–13, and Py3′. This insert was subcloned as a SacII/BamHI fragment into the pCAM-BSD transfection plasmid. This plasmid expresses the bsd selectable marker, which is under control of a 0.6 kb P. falciparum calmodulin (cam) 5′ UTR and a 0.6 kb P. falciparum hrp2 3′ UTR. The resulting 7.2 kb plasmids were designated pBSD-7G8, pBSD-GC03, and pBSD-Dd2.
Parasite transfections and DNA analysis
The P. falciparum 3D7, D10, and GC03 strains were cultured in human erythrocytes, transfected as described [21], and selected with 2.0 mg/ml blasticidin HCl (Invitrogen). Upon integration, recombinant parasites were cloned by limiting dilution and identified using Malstat assays [31]. The isolates from French Guyana were collected from malaria patients referred to the reference malaria laboratory of the Pasteur Institute of Guyana, in Cayenne, France. Each year this work was reviewed and approved by the Pasteur Institute Surveillance Committees of Guyana and Paris. The institutional review board of the Columbia University Medical Center also reviewed and approved the P. falciparum culture work.
PCR-based detection of plasmid integration into transfected parasites (Figure 1) used the pfcrt 5′ UTR-specific primer p1, the pfcrt exon 5-specific primer p2, the Py3′-specifc primer p3, the pfcrt intron 2-specific primer p4, and the plasmid-specific primer p5. For Southern blot analysis, 1 µg of DNA was digested with EcoRI/BglII, electrophoresed, and transferred onto nylon membranes. Hybridizations were performed with a hexamer-primed [32P]-labeled probe prepared from the 0.8 kb fragment spanning Δ5′, exon1 and intron 1, and released following SacII/AvrII digestion of the transfection plasmid pBSD-Dd2. The full-length sequence of pfcrt was determined from the complete coding sequence amplified from cDNA using the primers p251+BB116C and sequenced internally with the primers CF5C, BB84, AF12, AB22, AB25, and BB116B. For sequencing of the upstream pfmdr1 polymorphic residues at positions 86 and 184, genomic DNA was amplified with the primers p423+p231, and the resulting 0.7 kb products were sequenced with p231. For the downstream polymorphic residues at positions 1034, 1042, and 1246, the 0.8 kb amplification product of p426+p215 was sequenced with p238. pfmdr1 copy number was measured by Taqman quantitative real-time PCR and quantified with the ΔΔCt method as described elsewhere [79]. Genomic DNA samples were run twice in triplicate.
Quantitative real-time RT-PCR assays
The expression of pfcrt in the recombinant clones was assessed by quantitative real-time PCR assays performed with the QuantiTect SYBR Green PCR Kit (Qiagen) on an Opticon2 (BioRad). Expression from the different alleles (endogenous and genetically introduced) was analyzed utilizing primers specific for the two different 3′ UTRs, designated Py3′ and Pf3′. For the loci containing Py3′, the primers p1752 and p1753 were used to generate a 182 bp amplicon. For the locus containing Pf3′, a 191 bp amplicon was generated using the primers p1754 and p1756. PCR conditions were optimized so that the relative efficiencies of the Pf3′ and Py3′ amplifications were equal. Reactions were performed in 25 mL volumes with 300 nM of each primer, 3 mM Mg2+, and 1/80th of the oligo(dT) primed cDNA generated from 1.5 µg of total RNA. As a control for each sample, a 150 bp amplicon of β-actin was amplified using the primers A129 and A130, using the same conditions as for Py3′ and Pf3′ except that the Mg2+ concentration was 3.5 mM. All amplifications were performed with 15 minutes of hot start at 95°C, followed by 40 cycles of denaturing for 30 seconds at 95°C, annealing for 30 seconds at 49°C, and extension for 30 seconds at 62°C. Melting curve analysis was performed for each assay to verify that a single melting peak was produced, indicating a single specific PCR product for each reaction. A standard curve for each reaction was generated with 10-fold serial dilutions of genomic DNA, spanning the range of 5 to 5×105 genome copies). This genomic DNA was prepared from D10C, a recombinant clone shown by Southern hybridization to have a single copy of each locus (Py3′ and Pf3′, Figure 1B). Each sample was run in triplicate on three separate occasions.
Protein analysis
Protein extracts were prepared from sorbitol-synchronized trophozoite-stage parasites. For each sample, protein from ∼1×106 parasites was loaded per well, electrophoresed on 12% SDS-PAGE gels, and transferred onto polyvinylidene difluoride membranes. Membranes were probed with rabbit anti-PfCRT antibodies (diluted 1∶2,500) [11], followed by incubation with horseradish peroxidase-conjugated donkey anti-rabbit IgG (1∶10,000; Amersham Biosciences). Rabbit anti-PfERD2 antibodies (diluted 1∶1,000) [80] were used as an independent loading control. Bands were visualized by enhanced chemiluminescence (Amersham Biosciences) and quantified by densitometric analysis of autoradiograph data using NIH ImageJ 1.38× (http://rsb.info.nih.gov/ij). PfCRT band intensities were normalized against the PfERD2 bands to correct for minor differences in protein loading.
In vitro antimalarial drug assays
Parasite susceptibilities to antimalarial drugs were measured in vitro by [3H]-hypoxanthine incorporation assays, as described [81]. Briefly, predominately ring-stage cultures were seeded in duplicate in 96-well plates at 0.4% parasitemia and 1.6% hematocrit. Parasites were exposed to a range of drug concentrations, or no drug controls, for 72 hr, with 0.5 µCi per well of [3H]-hypoxanthine added at the 48 hr time point. IC50 and IC90 values were extrapolated by linear regression, as described [81]. Compounds were tested in duplicate on 4–11 separate occasions for CQ and mdCQ and 3–12 separate occasions for the other drugs. In some assays, VP was included at 0.8 µM final concentration. Statistical analyses comparing mutant pfcrt-modified lines against recombinant control lines of the same genetic backgrounds were performed using one-way ANOVA with a Bonferroni post-hoc test for CQ and mdCQ, or unpaired student t tests for quinine (QN), artemisinin (ART), monodesethyl-amodiaquine (mdADQ), lumefantrine (LMF), and piperaquine (PIP).
In vitro recrudescence assays
Parasites were assayed for their ability to grow under short-term exposure to high CQ concentrations. Predominately ring-stage cultures were seeded in 96-well plates at 0.2% parasitemia and 1.6% hematocrit. Parasites were exposed for 6 days to no drug, 50 nM CQ, or 80 nM CQ, with daily media changes. Drug pressure was then removed on day 7 and parasite growth was measured using Malstat assays ([31]). From days 7 through 30, media changes and Malstat assays were performed every two days, and the cultures cut 1∶2 into fresh erythrocytes weekly until the detection of positive wells. As part of this experiment, cultures of 3D77G8-1 and D107G8-1 were exposed to 50 nM CQ for 6 days and maintained until parasites became microscopically detectable, at days 15 and 20 respectively. These CQ-pretreated cultures were assayed for recrudescence alongside 7G8, 3D7C, 3D77G8-1, D10C, D107G8-1, GC03C, and GC037G8-1. Data were pooled from two independent experiments in which each line was assayed in duplicate for the no drug controls and in triplicate for the 50 nM and 80 nM CQ treatments.
Malstat assays
These were performed as described [82], with minor modifications. Briefly, 100 µL of Malstat reagent was added to 50 µL of culture supernatant and incubated for 1 hr. Absorbance at 595 nM was measured on a VICTOR3 Multilabel Plate Reader (Perkin-Elmer). Wells positive for parasite growth were identified based on absorbance values greater than twice those obtained from control wells with uninfected erythrocytes. Positive wells were verified by microscopic evaluation of Giemsa-stained thin smears.
Gene identification numbers
pfcrt: MAL7P1.27; pfmdr1: PFE1150w; pycrt: PY05061; b-actin: PFL2215w. Pfmrp: PFA0590w. All numbers are from www.plasmodb.org.
Supporting Information
Zdroje
1. WellemsTE
PloweCV
2001 Chloroquine-resistant malaria. J Infect Dis 184 770 776
2. WongsrichanalaiC
PickardAL
WernsdorferWH
MeshnickSR
2002 Epidemiology of drug-resistant malaria. Lancet Infect Dis 2 209 218
3. EastmanRT
FidockDA
2009 Artemisinin-based combination therapies: a vital tool in efforts to eliminate malaria. Nat Rev Microbiol 7 864 874
4. RieckmannKH
2006 The chequered history of malaria control: are new and better tools the ultimate answer? Ann Trop Med Parasitol 100 647 662
5. GardellaF
AssiS
SimonF
BogreauH
EggelteT
2008 Antimalarial drug use in general populations of tropical Africa. Malar J 7 124
6. LauferMK
ThesingPC
EddingtonND
MasongaR
DzinjalamalaFK
2006 Return of chloroquine antimalarial efficacy in Malawi. N Engl J Med 355 1959 1966
7. BanerjeeR
GoldbergDE
2001 The Plasmodium food vacuole.
RosenthalPJ
Antimalarial chemotherapy Totowa, NJ Humana Press 43 63
8. FooteSJ
KyleDE
MartinRK
OduolaAM
ForsythK
1990 Several alleles of the multidrug-resistance gene are closely linked to chloroquine resistance in Plasmodium falciparum. Nature 345 255 258
9. WellemsTE
PantonLJ
GluzmanIY
do RosarioVE
GwadzRW
1990 Chloroquine resistance not linked to mdr-like genes in a Plasmodium falciparum cross. Nature 345 253 255
10. SuX-Z
KirkmanLS
WellemsTE
1997 Complex polymorphisms in a ∼330 kDa protein are linked to chloroquine-resistant P. falciparum in Southeast Asia and Africa. Cell 91 593 603
11. FidockDA
NomuraT
TalleyAK
CooperRA
DzekunovSM
2000 Mutations in the digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol Cell 6 861 871
12. CooperRA
FerdigMT
SuXZ
UrsosLM
MuJ
2002 Alternative mutations at position 76 of the vacuolar transmembrane protein PfCRT are associated with chloroquine resistance and unique stereospecific quinine and quinidine responses in Plasmodium falciparum. Mol Pharmacol 61 35 42
13. FerdigMT
CooperRA
MuJ
DengB
JoyDA
2004 Dissecting the loci of low-level quinine resistance in malaria parasites. Mol Microbiol 52 985 997
14. WoottonJC
FengX
FerdigMT
CooperRA
MuJ
2002 Genetic diversity and chloroquine selective sweeps in Plasmodium falciparum. Nature 418 320 323
15. KidgellC
VolkmanSK
DailyJ
BorevitzJO
PlouffeD
2006 A systematic map of genetic variation in Plasmodium falciparum. PLoS Pathog 2 e57
16. VolkmanSK
SabetiPC
DeCaprioD
NeafseyDE
SchaffnerSF
2007 A genome-wide map of diversity in Plasmodium falciparum. Nat Genet 39 113 119
17. DjimdéA
DoumboMD
CorteseJF
KayentaoK
DoumboS
2001 A molecular marker for chloroquine resistant falciparum malaria. New Engl J Med 344 257 263
18. DjimdéAA
DoumboOK
TraoreO
GuindoAB
KayentaoK
2003 Clearance of drug-resistant parasites as a model for protective immunity in Plasmodium falciparum malaria. Am J Trop Med Hyg 69 558 563
19. ValderramosSG
FidockDA
2006 Transporters involved in resistance to antimalarial drugs. Trends Pharmacol Sci 27 594 601
20. DuraisinghMT
CowmanAF
2005 Contribution of the pfmdr1 gene to antimalarial drug-resistance. Acta Trop 94 181 190
21. SidhuAB
ValderramosSG
FidockDA
2005 pfmdr1 mutations contribute to quinine resistance and enhance mefloquine and artemisinin sensitivity in Plasmodium falciparum. Mol Microbiol 57 913 926
22. ReedMB
SalibaKJ
CaruanaSR
KirkK
CowmanAF
2000 Pgh1 modulates sensitivity and resistance to multiple antimalarials in Plasmodium falciparum. Nature 403 906 909
23. ChenN
RussellB
StaleyJ
KoteckaB
NasveldP
2001 Sequence polymorphisms in pfcrt are strongly associated with chloroquine resistance in Plasmodium falciparum. J Infect Dis 183 1543 1545
24. ChenN
RussellB
FowlerE
PetersJ
ChengQ
2002 Levels of chloroquine resistance in Plasmodium falciparum are determined by loci other than pfcrt and pfmdr1. J Infect Dis 185 405 407
25. KaddouriH
DjimdeA
DamaS
KodioA
TeketeM
2008 Baseline in vitro efficacy of ACT component drugs on Plasmodium falciparum clinical isolates from Mali. Int J Parasitol 38 791 798
26. PriceRN
DorseyG
AshleyEA
BarnesKI
BairdJK
2007 World Antimalarial Resistance Network I: clinical efficacy of antimalarial drugs. Malar J 6 119
27. HappiTC
ThomasSM
GbotoshoGO
FaladeCO
AkinboyeDO
2003 Point mutations in the pfcrt and pfmdr-1 genes of Plasmodium falciparum and clinical response to chloroquine, among malaria patients from Nigeria. Ann Trop Med Parasitol 97 439 451
28. MockenhauptFP
EhrhardtS
EggelteTA
Agana-NsiireP
StollbergK
2005 Chloroquine-treatment failure in northern Ghana: roles of pfcrt T76 and pfmdr1 Y86. Ann Trop Med Parasitol 99 723 732
29. RingwaldP
BascoLK
1999 Comparison of in vivo and in vitro tests of resistance in patients treated with chloroquine in Yaounde, Cameroon. Bull World Health Organ 77 34 43
30. EklandEH
FidockDA
2008 In vitro evaluations of antimalarial drugs and their relevance to clinical outcomes. Int J Parasitol 38 743 747
31. SidhuAB
Verdier-PinardD
FidockDA
2002 Chloroquine resistance in Plasmodium falciparum malaria parasites conferred by pfcrt mutations. Science 298 210 213
32. LakshmananV
BrayPG
Verdier-PinardD
JohnsonDJ
HorrocksP
2005 A critical role for PfCRT K76T in Plasmodium falciparum verapamil-reversible chloroquine resistance. Embo J 24 2294 2305
33. BascoLK
NdoungaM
NganeVF
SoulaG
2002 Molecular epidemiology of malaria in Cameroon. XIV. Plasmodium falciparum chloroquine resistance transporter (PfCRT) gene sequences of isolates before and after chloroquine treatment. Am J Trop Med Hyg 67 392 395
34. NageshaHS
CaseyGJ
RieckmannKH
FryauffDJ
LaksanaBS
2003 New haplotypes of the Plasmodium falciparum chloroquine resistance transporter (pfcrt) gene among chloroquine-resistant parasite isolates. Am J Trop Med Hyg 68 398 402
35. PatiSS
MishraS
MohantyS
MohapatraDN
SahuPK
2007 Pfcrt haplotypes and in-vivo chloroquine response in Sundergarh district, Orissa, India. Trans R Soc Trop Med Hyg 101 650 654
36. PillaiDR
LabbeAC
VanisavethV
HongvangthongB
PomphidaS
2001 Plasmodium falciparum malaria in Laos: chloroquine treatment outcome and predictive value of molecular markers. J Infect Dis 183 789 795
37. ValechaN
JoshiH
MallickPK
SharmaSK
KumarA
2009 Low efficacy of chloroquine: time to switchover to artemisinin-based combination therapy for falciparum malaria in India. Acta Trop 111 21 28
38. WallerKL
MuhleRA
UrsosLM
HorrocksP
Verdier-PinardD
2003 Chloroquine resistance modulated in vitro by expression levels of the Plasmodium falciparum chloroquine resistance transporter. J Biol Chem 278 33593 33601
39. MartinSK
OduolaAM
MilhousWK
1987 Reversal of chloroquine resistance in Plasmodium falciparum by verapamil. Science 235 899 901
40. MehlotraRK
FujiokaH
RoepePD
JannehO
UrsosLM
2001 Evolution of a unique Plasmodium falciparum chloroquine-resistance phenotype in association with pfcrt polymorphism in Papua New Guinea and South America. Proc Natl Acad Sci USA 98 12689 12694
41. GreenhouseB
SlaterM
Njama-MeyaD
NzarubaraB
Maiteki-SebuguziC
2009 Decreasing efficacy of antimalarial combination therapy in Uganda is explained by decreasing host immunity rather than increasing drug resistance. J Infect Dis 199 758 765
42. SchofieldL
MuellerI
2006 Clinical immunity to malaria. Curr Mol Med 6 205 221
43. MehlotraRK
MatteraG
BockarieMJ
MaguireJD
BairdJK
2008 Discordant patterns of genetic variation at two chloroquine resistance loci in worldwide populations of the malaria parasite Plasmodium falciparum. Antimicrob Agents Chemother 52 2212 2222
44. Mixson-HaydenT
JainV
McCollumAM
PoeA
NagpalAC
2010 Evidence of selective sweeps in genes conferring resistance to chloroquine and pyrimethamine in Plasmodium falciparum within India. Antimicrob Agents Chemother in press
45. KublinJG
CorteseJF
NjunjuEM
MukadamRA
WirimaJJ
2003 Reemergence of chloroquine-sensitive Plasmodium falciparum malaria after cessation of chloroquine use in Malawi. J Infect Dis 187 1870 1875
46. MitaT
KanekoA
LumJK
BwijoB
TakechiM
2003 Recovery of chloroquine sensitivity and low prevalence of the Plasmodium falciparum chloroquine resistance transporter gene mutation K76T following the discontinuance of chloroquine use in Malawi. Am J Trop Med Hyg 68 413 415
47. ThomasSM
NdirO
DiengT
MboupS
WypijD
2002 In vitro chloroquine susceptibility and PCR analysis of pfcrt and pfmdr1 polymorphisms in Plasmodium falciparum isolates from Senegal. Am J Trop Med Hyg 66 474 480
48. LimP
ChyS
ArieyF
IncardonaS
ChimP
2003 pfcrt polymorphism and chloroquine resistance in Plasmodium falciparum strains isolated in Cambodia. Antimicrob Agents Chemother 47 87 94
49. DurrandV
BerryA
SemR
GlaziouP
BeaudouJ
2004 Variations in the sequence and expression of the Plasmodium falciparum chloroquine resistance transporter (Pfcrt) and their relationship to chloroquine resistance in vitro. Mol Biochem Parasitol 136 273 285
50. MuJ
FerdigMT
FengX
JoyDA
DuanJ
2003 Multiple transporters associated with malaria parasite responses to chloroquine and quinine. Mol Microbiol 49 977 989
51. MartinRE
KirkK
2004 The malaria parasite's chloroquine resistance transporter is a member of the drug/metabolite transporter superfamily. Mol Biol Evol 21 1938 1949
52. CooperRA
LaneKD
DengB
MuJ
PatelJJ
2007 Mutations in transmembrane domains 1, 4 and 9 of the Plasmodium falciparum chloroquine resistance transporter alter susceptibility to chloroquine, quinine and quinidine. Mol Microbiol 64 1139
53. LegrandE
VolneyB
MeynardJB
Mercereau-PuijalonO
EsterreP
2008 In vitro monitoring of Plasmodium falciparum drug resistance in French Guiana: a synopsis of continuous assessment from 1994 to 2005. Antimicrob Agents Chemother 52 288 298
54. JambouR
LegrandE
NiangM
KhimN
LimP
2005 Resistance of Plasmodium falciparum field isolates to in-vitro artemether and point mutations of the SERCA-type PfATPase6. Lancet 366 1960 1963
55. WhiteNJ
2008 Qinghaosu (artemisinin): the price of success. Science 320 330 334
56. CooperRA
HartwigCL
FerdigMT
2005 pfcrt is more than the Plasmodium falciparum chloroquine resistance gene: a functional and evolutionary perspective. Acta Trop 94 170 180
57. DjimdéAA
FofanaB
SagaraI
SidibeB
ToureS
2008 Efficacy, safety, and selection of molecular markers of drug resistance by two ACTs in Mali. Am J Trop Med Hyg 78 455 461
58. EcheverryDF
HolmgrenG
MurilloC
HiguitaJC
BjorkmanA
2007 Short report: polymorphisms in the pfcrt and pfmdr1 genes of Plasmodium falciparum and in vitro susceptibility to amodiaquine and desethylamodiaquine. Am J Trop Med Hyg 77 1034 1038
59. NsobyaSL
DokomajilarC
JolobaM
DorseyG
RosenthalPJ
2007 Resistance-mediating Plasmodium falciparum pfcrt and pfmdr1 alleles after treatment with artesunate-amodiaquine in Uganda. Antimicrob Agents Chemother 51 3023 3025
60. NostenF
WhiteNJ
2007 Artemisinin-based combination treatment of falciparum malaria. Am J Trop Med Hyg 77 181 192
61. SisowathC
PetersenI
VeigaMI
MartenssonA
PremjiZ
2009 In vivo selection of Plasmodium falciparum parasites carrying the chloroquine-susceptible pfcrt K76 allele after treatment with artemether-lumefantrine in Africa. J Infect Dis 199 750 757
62. SanchezCP
SteinWD
LanzerM
2007 Is PfCRT a channel or a carrier? Two competing models explaining chloroquine resistance in Plasmodium falciparum. Trends Parasitol 23 332 339
63. LehaneAM
KirkK
2008 Chloroquine resistance-conferring mutations in pfcrt give rise to a chloroquine-associated H+ leak from the malaria parasite's digestive vacuole. Antimicrob Agents Chemother 52 4374 4380
64. HuamanMC
YoshinagaK
SuryanathaA
SuarsanaN
KanbaraH
2004 Short report: polymorphisms in the chloroquine resistance transporter gene in Plasmodium falciparum isolates from Lombok, Indonesia. Am J Trop Med Hyg 71 40 42
65. PicotS
OlliaroP
de MonbrisonF
BienvenuAL
PriceRN
2009 A systematic review and meta-analysis of evidence for correlation between molecular markers of parasite resistance and treatment outcome in falciparum malaria. Malar J 8 89
66. EklandEH
FidockDA
2007 Advances in understanding the genetic basis of antimalarial drug resistance. Curr Opin Microbiol 10 363 370
67. SaJM
TwuO
HaytonK
ReyesS
FayMP
2009 Geographic patterns of Plasmodium falciparum drug resistance distinguished by differential responses to amodiaquine and chloroquine. Proc Natl Acad Sci U S A 106 18883 18889
68. FidockDA
NomuraT
CooperRA
SuX
TalleyAK
2000 Allelic modifications of the cg2 and cg1 genes do not alter the chloroquine response of drug-resistant Plasmodium falciparum. Mol Biochem Parasitol 110 1 10
69. MuJ
MyersRA
JiangH
LiuS
RicklefsS
2010 Plasmodium falciparum genome-wide scans for positive selection, recombination hot spots and resistance to antimalarial drugs. Nat Genet in press
70. AndersonTJ
NairS
QinH
SinglamS
BrockmanA
2005 Are transporter genes other than the chloroquine resistance locus (pfcrt) and multidrug resistance gene (pfmdr) associated with antimalarial drug resistance? Antimicrob Agents Chemother 49 2180 2188
71. MartinRE
MarchettiRV
CowanAI
HowittSM
BroerS
2009 Chloroquine transport via the malaria parasite's chloroquine resistance transporter. Science 325 1680 1682
72. ZhangH
PaguioM
RoepePD
2004 The antimalarial drug resistance protein Plasmodium falciparum chloroquine resistance transporter binds chloroquine. Biochemistry 43 8290 8296
73. NaudeB
BrzostowskiJA
KimmelAR
WellemsTE
2005 Dictyostelium discoideum expresses a malaria chloroquine resistance mechanism upon transfection with mutant, but not wild-type, Plasmodium falciparum transporter PfCRT. J Biol Chem 280 22596 22603
74. RaghebD
BompianiK
DalalS
KlembaM
2009 Evidence for catalytic roles for Plasmodium falciparum aminopeptidase P in the food vacuole and cytosol. J Biol Chem 284 24806 24815
75. GinsburgH
FaminO
ZhangJ
KrugliakM
1998 Inhibition of glutathione-dependent degradation of heme by chloroquine and amodiaquine as a possible basis for their antimalarial mode of action. Biochem Pharmacol 56 1305 1313
76. GinsburgH
GolenserJ
2003 Glutathione is involved in the antimalarial action of chloroquine and its modulation affects drug sensitivity of human and murine species of Plasmodium. Redox Rep 8 276 279
77. RajDK
MuJ
JiangH
KabatJ
SinghS
2009 Disruption of a Plasmodium falciparum multidrug resistance-associated protein (PfMRP) alters its fitness and transport of antimalarial drugs and glutathione. J Biol Chem 284 7687 7696
78. MaughanSC
PasternakM
CairnsN
KiddleG
BrachT
2010 Plant homologs of the Plasmodium falciparum chloroquine-resistance transporter, PfCRT, are required for glutathione homeostasis and stress responses. Proc Natl Acad Sci U S A 107 2331 2336
79. PriceRN
UhlemannAC
BrockmanA
McGreadyR
AshleyE
2004 Mefloquine resistance in Plasmodium falciparum and increased pfmdr1 gene copy number. Lancet 364 438 447
80. ElmendorfHG
HaldarK
1993 Identification and localization of ERD2 in the malaria parasite Plasmodium falciparum: separation from sites of sphingomyelin synthesis and implications for organization of the Golgi. Embo J 12 4763 4773
81. FidockDA
NomuraT
WellemsTE
1998 Cycloguanil and its parent compound proguanil demonstrate distinct activities against Plasmodium falciparum malaria parasites transformed with human dihydrofolate reductase. Mol Pharmacol 54 1140 1147
82. GoodyerID
TaraschiTF
1997 Plasmodium falciparum: a simple, rapid method for detecting parasite clones in microtiter plates. Exp Parasitol 86 158 160
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Mouse Senile Amyloid Fibrils Deposited in Skeletal Muscle Exhibit Amyloidosis-Enhancing ActivityČlánek The Role of Intestinal Microbiota in the Development and Severity of Chemotherapy-Induced MucositisČlánek Crystal Structure of HIV-1 gp41 Including Both Fusion Peptide and Membrane Proximal External RegionsČlánek Demonstration of Cross-Protective Vaccine Immunity against an Emerging Pathogenic Ebolavirus Species
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2010 Číslo 5- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Quorum Sensing Inhibition Selects for Virulence and Cooperation in
- The HMW1C Protein Is a Glycosyltransferase That Transfers Hexose Residues to Asparagine Sites in the HMW1 Adhesin
- Analysis of Virion Structural Components Reveals Vestiges of the Ancestral Ichnovirus Genome
- Mouse Senile Amyloid Fibrils Deposited in Skeletal Muscle Exhibit Amyloidosis-Enhancing Activity
- Global Migration Dynamics Underlie Evolution and Persistence of Human Influenza A (H3N2)
- The Type III Effectors NleE and NleB from Enteropathogenic and OspZ from Block Nuclear Translocation of NF-κB p65
- VEGF Promotes Malaria-Associated Acute Lung Injury in Mice
- Identification of a Mutant PfCRT-Mediated Chloroquine Tolerance Phenotype in
- The Early Stage of Bacterial Genome-Reductive Evolution in the Host
- Host-Detrimental Role of Esx-1-Mediated Inflammasome Activation in Mycobacterial Infection
- Elevation of Intact and Proteolytic Fragments of Acute Phase Proteins Constitutes the Earliest Systemic Antiviral Response in HIV-1 Infection
- The Pleiotropic CymR Regulator of Plays an Important Role in Virulence and Stress Response
- Alternative Sigma Factor σ Modulates Prophage Integration and Excision in
- Effect of Neuraminidase Inhibitor–Resistant Mutations on Pathogenicity of Clade 2.2 A/Turkey/15/06 (H5N1) Influenza Virus in Ferrets
- Massive APOBEC3 Editing of Hepatitis B Viral DNA in Cirrhosis
- NK Cells and γδ T Cells Mediate Resistance to Polyomavirus–Induced Tumors
- Is Genetically Diverse in Animals and Appears to Have Crossed the Host Barrier to Humans on (At Least) Two Occasions
- Adenylate Cyclase Toxin Mobilizes Its β Integrin Receptor into Lipid Rafts to Accomplish Translocation across Target Cell Membrane in Two Steps
- The Role of Intestinal Microbiota in the Development and Severity of Chemotherapy-Induced Mucositis
- HIV-1 Transmitting Couples Have Similar Viral Load Set-Points in Rakai, Uganda
- Few and Far Between: How HIV May Be Evading Antibody Avidity
- Galectin-9/TIM-3 Interaction Regulates Virus-Specific Primary and Memory CD8 T Cell Response
- Perforin Expression Directly by HIV-Specific CD8 T-Cells Is a Correlate of HIV Elite Control
- The Set3/Hos2 Histone Deacetylase Complex Attenuates cAMP/PKA Signaling to Regulate Morphogenesis and Virulence of
- Infidelity of SARS-CoV Nsp14-Exonuclease Mutant Virus Replication Is Revealed by Complete Genome Sequencing
- Combining ChIP-chip and Expression Profiling to Model the MoCRZ1 Mediated Circuit for Ca/Calcineurin Signaling in the Rice Blast Fungus
- Internalin B Activates Junctional Endocytosis to Accelerate Intestinal Invasion
- A Complex Small RNA Repertoire Is Generated by a Plant/Fungal-Like Machinery and Effected by a Metazoan-Like Argonaute in the Single-Cell Human Parasite
- Opc Invasin Binds to the Sulphated Tyrosines of Activated Vitronectin to Attach to and Invade Human Brain Endothelial Cells
- Muc2 Protects against Lethal Infectious Colitis by Disassociating Pathogenic and Commensal Bacteria from the Colonic Mucosa
- PdeH, a High-Affinity cAMP Phosphodiesterase, Is a Key Regulator of Asexual and Pathogenic Differentiation in
- Isolates with Antimony-Resistant but Not -Sensitive Phenotype Inhibit Sodium Antimony Gluconate-Induced Dendritic Cell Activation
- The Microbiota and Allergies/Asthma
- Environmental Factors Determining the Epidemiology and Population Genetic Structure of the Group in the Field
- Prolonged Antigen Presentation Is Required for Optimal CD8+ T Cell Responses against Malaria Liver Stage Parasites
- Crystal Structure of HIV-1 gp41 Including Both Fusion Peptide and Membrane Proximal External Regions
- Susceptibility to Anthrax Lethal Toxin-Induced Rat Death Is Controlled by a Single Chromosome 10 Locus That Includes
- Demonstration of Cross-Protective Vaccine Immunity against an Emerging Pathogenic Ebolavirus Species
- Effective, Broad Spectrum Control of Virulent Bacterial Infections Using Cationic DNA Liposome Complexes Combined with Bacterial Antigens
- High Multiplicity Infection by HIV-1 in Men Who Have Sex with Men
- The -Specific Human Memory B Cell Compartment Expands Gradually with Repeated Malaria Infections
- EBV Promotes Human CD8 NKT Cell Development
- Persistent Growth of a Human Plasma-Derived Hepatitis C Virus Genotype 1b Isolate in Cell Culture
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Quorum Sensing Inhibition Selects for Virulence and Cooperation in
- The Role of Intestinal Microbiota in the Development and Severity of Chemotherapy-Induced Mucositis
- Crystal Structure of HIV-1 gp41 Including Both Fusion Peptide and Membrane Proximal External Regions
- Susceptibility to Anthrax Lethal Toxin-Induced Rat Death Is Controlled by a Single Chromosome 10 Locus That Includes
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



