-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaMassive APOBEC3 Editing of Hepatitis B Viral DNA in Cirrhosis
DNA viruses, retroviruses and hepadnaviruses, such as hepatitis B virus (HBV), are vulnerable to genetic editing of single stranded DNA by host cell APOBEC3 (A3) cytidine deaminases. At least three A3 genes are up regulated by interferon-α in human hepatocytes while ectopic expression of activation induced deaminase (AICDA), an A3 paralog, has been noted in a variety of chronic inflammatory syndromes including hepatitis C virus infection. Yet virtually all studies of HBV editing have confined themselves to analyses of virions from culture supernatants or serum where the frequency of edited genomes is generally low (≤10−2). We decided to look at the nature and frequency of HBV editing in cirrhotic samples taken during removal of a primary hepatocellular carcinoma. Forty-one cirrhotic tissue samples (10 alcoholic, 10 HBV+, 11 HBV+HCV+ and 10 HCV+) as well as 4 normal livers were studied. Compared to normal liver, 5/7 APOBEC3 genes were significantly up regulated in the order: HCV±HBV>HBV>alcoholic cirrhosis. A3C and A3D were up regulated for all groups while the interferon inducible A3G was over expressed in virus associated cirrhosis, as was AICDA in ∼50% of these HBV/HCV samples. While AICDA can indeed edit HBV DNA ex vivo, A3G is the dominant deaminase in vivo with up to 35% of HBV genomes being edited. Despite these highly deleterious mutant spectra, a small fraction of genomes survive and contribute to loss of HBeAg antigenemia and possibly HBsAg immune escape. In conclusion, the cytokine storm associated with chronic inflammatory responses to HBV and HCV clearly up regulates a number of A3 genes with A3G clearly being a major restriction factor for HBV. Although the mutant spectrum resulting from A3 editing is highly deleterious, a very small part, notably the lightly edited genomes, might help the virus evolve and even escape immune responses.
Published in the journal: . PLoS Pathog 6(5): e32767. doi:10.1371/journal.ppat.1000928
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1000928Summary
DNA viruses, retroviruses and hepadnaviruses, such as hepatitis B virus (HBV), are vulnerable to genetic editing of single stranded DNA by host cell APOBEC3 (A3) cytidine deaminases. At least three A3 genes are up regulated by interferon-α in human hepatocytes while ectopic expression of activation induced deaminase (AICDA), an A3 paralog, has been noted in a variety of chronic inflammatory syndromes including hepatitis C virus infection. Yet virtually all studies of HBV editing have confined themselves to analyses of virions from culture supernatants or serum where the frequency of edited genomes is generally low (≤10−2). We decided to look at the nature and frequency of HBV editing in cirrhotic samples taken during removal of a primary hepatocellular carcinoma. Forty-one cirrhotic tissue samples (10 alcoholic, 10 HBV+, 11 HBV+HCV+ and 10 HCV+) as well as 4 normal livers were studied. Compared to normal liver, 5/7 APOBEC3 genes were significantly up regulated in the order: HCV±HBV>HBV>alcoholic cirrhosis. A3C and A3D were up regulated for all groups while the interferon inducible A3G was over expressed in virus associated cirrhosis, as was AICDA in ∼50% of these HBV/HCV samples. While AICDA can indeed edit HBV DNA ex vivo, A3G is the dominant deaminase in vivo with up to 35% of HBV genomes being edited. Despite these highly deleterious mutant spectra, a small fraction of genomes survive and contribute to loss of HBeAg antigenemia and possibly HBsAg immune escape. In conclusion, the cytokine storm associated with chronic inflammatory responses to HBV and HCV clearly up regulates a number of A3 genes with A3G clearly being a major restriction factor for HBV. Although the mutant spectrum resulting from A3 editing is highly deleterious, a very small part, notably the lightly edited genomes, might help the virus evolve and even escape immune responses.
Introduction
The human genome harbours a group of 11 genes encoding cytidine deaminases, the majority having substrate specificity for single stranded DNA (ssDNA) [1], [2], [3], [4], [5], [6], [7], [8], [9], [10]. These include the prototypical enzyme APOBEC1 (A1) and activation induced deaminase (AICDA). The large seven gene APOBEC3 cluster spans ∼150kb at ch22q13.1 [11]. Two additional genes, APOBEC2 and APOBEC4, show homology to the above, although no editing activity has been described so far for either. Many of the human APOBEC3 (A3) enzymes can edit the cDNA of numerous retroviruses, retrovirus elements and hepadnaviruses in tissue culture experiments [3], [4], [6], [12], [13], [14], [15], [16], [17], [18], [19], [20], [21]. Yet in vivo only the lentiviruses, of which human immunodeficiency virus (HIV) is the most notorious, hepatitis B virus (HBV), human papillomaviruses (HPV) and TTV genomes have proven to be edited [21], [22], [23], [24], [25], [26], [27].
The outcome of cytidine deamination is oxidation of the C4 amino group yielding uridine - in short cytidine deamination is mutagenic. The degree of editing can be as little as a few bases per kilobase or up to 90% of all cytidine residues, APOBEC3A deamination of hepatitis B virus DNA in tissue culture being a case in point [16]. In virology, mutations are generally related to the plus strand. Hence, so called G→A hypermutants reflect cytidine deamination of (−) stand DNA while C→T hypermutants reflect editing of the viral (+) strand. Due to the extent of editing, hypermutation can be seen as part of an innate anti-viral response. This theme is echoed by the fact that some APOBEC3 genes are up regulated by interferon-α and –γ in a wide variety of cells including primary human hepatocytes [28], [29], [30], [31], [32], [33], [34], [35].
While AICDA expression is chiefly expressed in germinal centre centroblasts [36], ectopic expression of human AICDA has been shown in at least four settings, all involving chronic inflammation – HCV associated chronic hepatitis, Helicobacter pylori associated stomach cancer, human colitis and chronic inflammatory bile duct disease [37], [38], [39], [40], [41]. Transgenic mice bearing the human AICDA or A1 genes invariably induce cancers, the organ specificity being dependent on the promoter [42], [43], [44]. As there is a long historical link between chronic inflammation and cancer [45], [46], the ensemble suggested a link between aberrant AICDA expression and, by inference, expression of other human APOBEC genes and editing of the nuclear genome [41], [44], [47]. Cancer frequently emerges from a background of cellular dysplasia. For the liver, cirrhosis is seen as a polyclonal proliferative disease, a prodrome that generally precedes the HCC. Yet virtually all APOBEC editing studies of the HBV genome have confined themselves to analyses of virions from culture supernatants or serum where the frequency of edited genomes is generally low (≤10−2, [21], [25]). In view of the above observations, we decided to look at the nature and frequency of HBV editing in cirrhotic liver samples taken during removal of a primary HCC. Given the finding of ectopic AICDA expression in HCV associated chronic hepatitis [39], [48], we chose to work with cirrhotic tissue from HBV mono - and HBV plus HCV double infections.
It is shown here that while human AICDA can indeed hyperedit HBV genomes in an in vitro setting, editing by AICDA is a rare event in vivo. By contrast, up to 35% of HBV genomes are edited in vivo by several A3 enzymes including A3G indicating that human A3 deaminases represent major restriction factors for HBV replication. Yet through HBV editing, A3 deaminases generate a mutant spectrum upon which selection can occur. It is suggested that APOBEC3 editing may contribute towards loss of HBeAg antigenemia and immune escape.
Results
Up regulation of APOBEC deaminases in cirrhosis
A succinct description of the 41 cirrhotic samples is given in Table S1. Ten DNA samples from patients with alcoholic cirrhosis, 10 HBV+, 11 HBV+HCV+ and 10 from HCV+ patients were analysed. Complementary DNA was made to total RNA extracted in parallel with the DNA samples and hybridized to a custom made TaqMan PCR chip comprising all 11 human cytidine deaminases related genes (A1, A2, A3A–H, A4 and AICDA), a number of pro-inflammatory, apoptotic and mismatch repair genes as well as 3 reference genes (TRIM44, HMBS and LMF2) that have been validated for analyses of liver tissue.
Compared to the 4 normal livers, there was significant up regulation of 2–5 A3 genes in the order: HCV±HBV>HBV>alcoholic cirrhosis. A3C and A3D were up regulated for all groups, while the interferon inducible A3G was over expressed in virus associated, as opposed to alcoholic cirrhosis (Figure 1, Figure S1 & Table S2). While there was a trend towards increased A3A and A3F expression in all four groups, it never reached statistical significance. AICDA was up regulated in 15/31 HBV and/or HCV samples, indicating that ectopic expression of AICDA is also a feature of HBV liver disease (Figure 1 inset). By contrast expression was undetectable in the normal liver, which is why the data are not normalized as per A3 data (Figure S1). APOBEC1 was phenomenally expressed in one sample (#146) yet again normalization wasn't possible as it was weakly expressed in just one normal liver sample (Figure 1 inset). APOBEC2 transcripts were relatively absent while APOBEC4 was undetectable throughout. A number of cellular genes associated with inflammation were significantly up regulated, notably FAS, FASLG, BCL2, IFNγ and LTA (Table S2).
Fig. 1. Transcription profiling of all 11 human cytidine deaminases in cirrhosis. 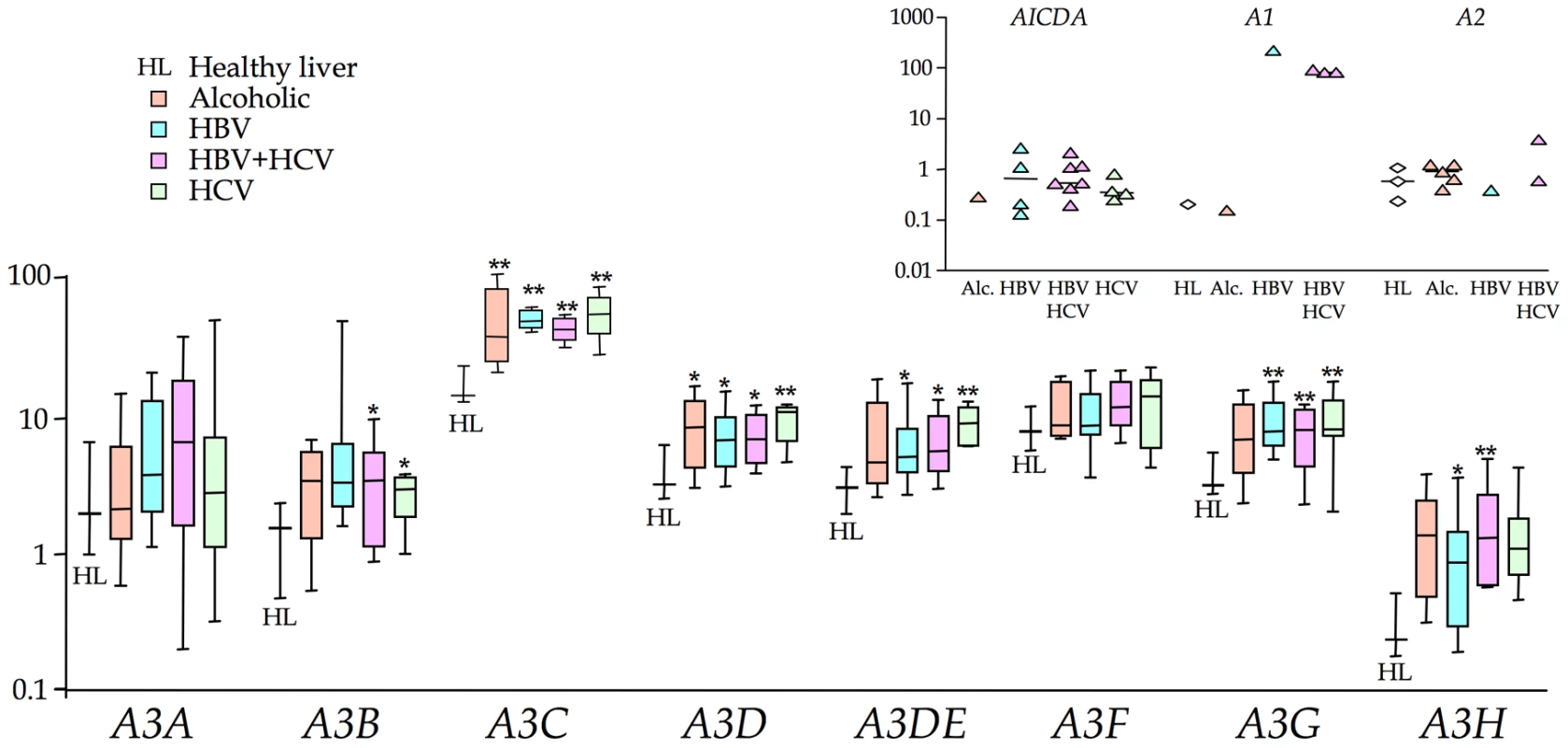
The A3 transcription data are in the form of box-whisker plots with the mean, quartiles, maxima and minima. Data are normalized to the expression levels of three invariable reference genes (TRIM44, HMBS and LMF2). Asterisks indicate statistically significant up regulation: ** 0.01<p<0.001; * 0.05<p<0.01. Inset) AICDA, APOBEC1 and APOBEC2 transcripts were detected in 0, 1 and 1 HL samples respectively but present in several HBV±HCV samples. APOBEC4 transcripts were undetectable in all samples tested. AICDA can hyperedit HBV genomes ex vivo
An infectious molecular clone of HBV was transfected into the hepatocyte derived Huh7 cell line along with human AICDA construct and A3G as positive control. HBsAg secretion in the supernatant was used as readout for editing at a macroscopic level. Figure 2A shows that both AICDA and A3G expression reduced HBsAg to levels ≤50% of control. Given that transfection frequencies were ∼30–40%, this suggests that the majority of genomes were edited. At 72 hours, total DNA was recovered from the supernatants and 3DPCR performed on a segment of the X gene [16], [21], [49], [50]. 3DPCR products were recovered as low as 85.2°C for the AICDA cotransfection compared to 86.6°C for the A3G control (Figure 2B). The 88.7°C 3DPCR products were cloned and sequenced. As can be seen from the mutation matrices (Figure 2C), AICDA (mean cytidine deamination frequency, fc = 42%, range 16–61%) was just as good as A3G at hyperediting HBV DNA (fc = 33%, range 7–98%).
Fig. 2. AICDA impacts HBV replication by hyperediting the genome. 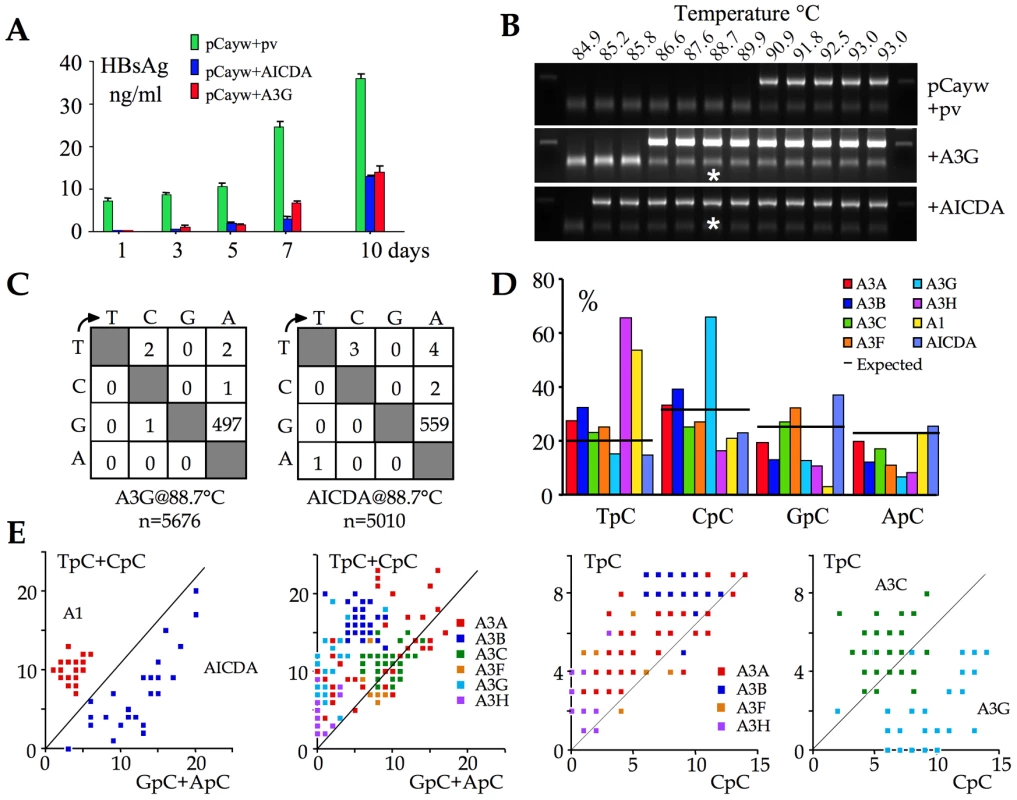
A) Macroscopic impact of AICDA and A3G on HBsAg secretion into the culture supernatant following transfection of the infectious molecular clone, pCayw. B) 3DPCR recovered A3G- and AICDA-edited HBV genomes down to 86.6°C and 85.2°C respectively. pv: empty plasmid vector and HBV alone. Asterisks refer to the samples cloned and sequenced. C) Mutation matrices for hyperedited X gene sequences derived from cloned 88.7°C 3DPCR products. n indicates the number of bases sequenced. D) Bulk dinucleotide context of HBV X region minus strand DNA by eight human cytidine deaminases. E) Clonal analysis of editing for individual edited sequences. The number of TpC+CpC vs. GpC+ApC targets edited per sequence are computed and represented on the y and x axes respectively. As the A3C and A3G genes were strongly up regulated (Figure 1) they have been separated from the others. Clonal analysis using TpC vs. CpC allows clear isolation of A3G from other A3 enzymes. As expected from extant data, AICDA editing of the HBV target was concentrated in GpC and ApC sites [1], [51] unlike its A3 counterparts (Figure 2D). Combined with previous data we now have a reference set for the HBV X region edited by 8/11 human cytidine deaminases [16]. Four deaminases (A3G, A3H, A1 and AICDA) showed polarized editing biases (Figure 2D) that can be used as hallmarks for specific deaminase activity in vivo. The singularity of AICDA and A3G editing with respect to other A3 enzymes becomes apparent when plotting the number of edited cytidine residues in selected dinucleotide contexts for single sequences (Figures 2E).
A3G and not AICDA is the major restriction factor in vivo
With these metrics in hand, 3DPCR was used to identify hyperedited HBV genomes from the cirrhotic samples. First round PCR DNA was performed on ∼0.5µg of total DNA using the X gene specific primers. In a second round, 3DPCR was performed at the restricting temperature of 88.7°C (Figure 3A). Fifteen of 17 DNA samples (88%) yielded robust amplification at the restrictive temperature. Five DNA samples – 2 HBV and 3 HBV+HCV - indicated by asterisks were cloned and sequenced. Hyperediting of the HBV genome was essentially confined to the minus DNA strand with only a handful of plus strand hypermutants, a finding in keeping with previous reports [21]. The mean cytidine editing frequencies were somewhat higher in the case of HCV co-infections (fc = 38.8%, 41.7% and 46.9%) compared to HBV monoinfection (fc = 23.3% and 31.6%, Figure 3B).
Fig. 3. A3 deaminases are the major editors of HBV DNA in vivo. 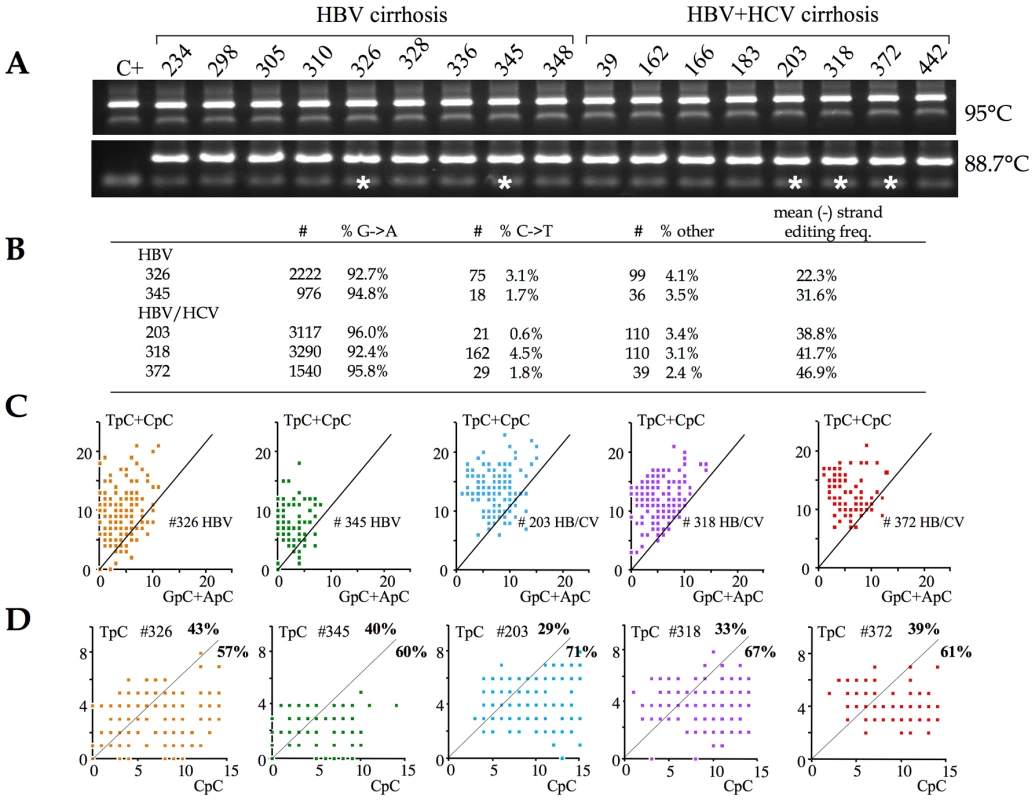
A) PCR and 3DPCR amplification at the normal and restrictive temperatures of 95°C and 88.7°C respectively. Sample codes are given above. Five 3DPCR samples identified by asterisks were chosen for cloning and sequencing. B) Summary of the unique hyperedited sequences in the form of the number and fraction of G→A (minus stand) edits, C→T (plus strand) and all other point mutations. The excess of GC→AT transitions over all other mutations varied from 23–40 fold. C) Clonal analysis using the number of TpC+CpC vs. GpC+ApC targets edited. The vast majority of patient sequences map to the area typical of APOBEC deaminases. D) Clonal analysis using the number of TpC vs. CpC targets edited. The majority of sequences map to the area typical of APOBEC3G (between 57–71% marked in bold face). The dinucleotide editing context for these hyperedited genomes was remarkably uniform with a strong preference for CpC and an aversion for ApC, which fits rather well with the profile for A3G (not shown). However, it is possible that such averaging could mask the occasional AICDA edited genome. Accordingly, a clonal analysis was used to highlight editing by distinct enzymes (Figure 3C). Very few patient sequences mapped to the area characteristic of AICDA (Figure 3C. vs. Figure 2E), indicating that it is not a major editor in vivo. By contrast, between 57–71% of patient sequences fell within the area covered by A3G (Figure 3D vs. Figure 2E). The remaining sequences mapped to regions where there was considerable overlap between A3 deaminases (Figure 2E). As A3C was significantly up regulated in these liver samples and expressed at greater levels than any other A3 gene (Figure 1), editing by A3C alone could explain the remainder.
In order to determine accurately the overall hyperediting frequencies in vivo, we performed deep sequencing on cloned first round PCR DNA (95°C). To our surprise for 4/5 samples a sizeable proportion (10–35%) of X gene segments showed unmistakable signs of hyperediting. The frequency distribution of editing is shown in Figure 4A. Some sequences harboured up to 50/58 (86%) edited cytidines (Figure 4A inset), comparable to those recovered from a transfection experiment. Figure 4B shows the frequency distribution of hyperedited genomes from patients recovered by 3DPCR. That the two frequency distributions are distinct reflects selection against lightly edited genomes during 3DPCR, as previously noted [21]. For the 95°C derived sequences, dinucleotide and clonal analysis indicated that the edited bases showed the same hallmarks as those recovered by 3DPCR, that is a penchant for CpC typical of A3G editing with at least one other A3 enzyme explaining the remainder (Figure 4C–E). Hence, Figure 4B represents a subset of a more general distribution typified by Figure 4A.
Fig. 4. High frequency of A3 editing recovered by standard PCR. 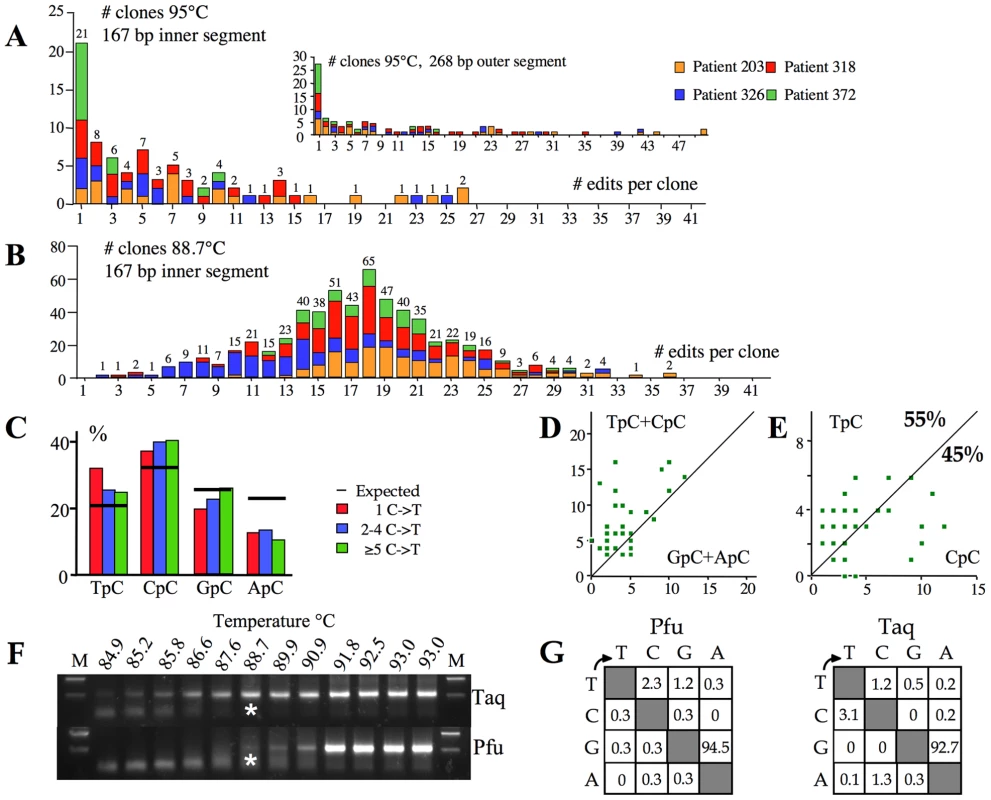
A) Frequency analysis of edited genomes as a function of the number of edits per sequence for 95°C derived PCR products. The 268 bp sequences were derived from the first round product. The sequences in insert to Figure 4A were stripped down to the size of the inner 167 bp locus and reanalyzed to allow comparison with the 3DPCR products. The numbers above the columns indicate the combined numbers of sequences across the four samples. B) Frequency distribution of edited sequences for the 3DPCR products obtained at 88.7°C. In order to calculate the bias resulting from PCR close to the denaturation temperature, let's assume that the frequency of clones with 1–17 edited cytidines reflected sub optimal amplification, while the profile in Figure 4A is close to the true distribution. By summing the number of clones with 1–17 and ≥18 edits for Figures 4A and 4B the estimated number with between 1–17 edits in Figure 4B is n, where 79/n = 7/300; n = 3386. As the number of clones in Figure 4B with 1–17 edits is 284, 3DPCR underestimates the true frequency by a factor of 3386/284, or ∼12. C) Bulk dinucleotide analysis of the 95°C sequences harbouring 1, 2–4 and ≥5 G→A transitions reveals the CpC hallmark of A3G editing. D & E) Clonal analysis reveals that editing was due to an APOBEC deaminase, approximately half being due to A3G. The smaller number of sequences used (n = 40) in these figures means that the values of 55% and 45% are less robust than for Figure 3C & 3D. F) 3DPCR amplification across a 85–93°C gradient using either Taq or Pfu DNA polymerase, the latter fails to amplify DNA containing dU, the product of A3 deamination. Asterisks indicate the PCR products cloned and sequenced. G) Mutation matrices of Pfu and Taq amplified HBV hypermutants given as percentages. Assuming that the highly edited part of the distribution (n≥18, Figure 4B) is unaffected by suboptimal amplification, then the expected number of edited genomes can be calculated to be 300×79/7 = 3386 (see legend to Figure 4). In other words ∼12 fold more genomes are edited than suggested by 3DPCR. Yet the actual number is probably higher as the X gene segment analyzed represents only ∼5% of the HBV genome. A genome with a wild type X gene sequence could be edited elsewhere. Hence, the real frequencies of slightly edited genomes are almost certainly higher.
A subset of A3 edited genomes can be repaired
A sizeable proportion of neo-synthesized HBV DNA returns to the nucleus to augment the pool of cccDNA, the viral replication template. DNA bearing multiple dU residues might be degraded by the UNG-APE1 pathway or, if copied on the opposite strand, the resulting dU(−)∶dA(+) base pair might be corrected to dT∶dA. To explore this issue we performed first round and 3DPCR on sample #326 using Pfu polymerase. Like all archaeal DNA polymerases, Pfu is unable to amplify DNA templates bearing dU [52]. As can be seen, Pfu amplification recovered much less hyperedited DNA than Taq polymerase (Figure 4F). There were enough 3DPCR products from the 88.7°C amplification to allow cloning and sequencing. The genomes were exclusively edited on the minus strand (mean fc = 25%, Figure 4G) indicating that multiple dU∶dA pairs can be repaired to dT∶dA. The corresponding amplification using Taq (Figure 4G) yielded a comparable frequency (fc = 32%). These observations indicate that substantial repair does occur for a small number of hyperedited genomes.
Precore/core mutations
The HBV core orf allows translation from the first, suboptimal AUG giving rise to the HBeAg precursor. Serum HBeAg is a marker for high viremia. Initiation from the second, optimal AUG, leads to synthesis of the capsid monomer, HBcAg. Over the course of a chronic infection, G→A mutations arise in the precore region, particularly at residue G1896 and to a lesser extent G1897 resulting in the loss of HBeAg synthesis and HBeAg seroconversion [53], [54]. These mutations result in a W28Stop change. There is no consensus as to whether this is due to A3 editing [20], [25], [55] despite the fact that G→A transitions in a run of four Gs (4Cs on the edited minus strand) are reminiscent of a hot spot for A3 deamination [21].
To explore this issue, a region spanning the precore region was analyzed by PCR and 3DPCR from a HBV/A3G transfection and patient #326. As can be seen from Figure 5A, G1896 was indeed a hot spot for A3 editing (70–80% editing on the minus strand) both in vivo and for the A3G transfection experiment. Editing of the plus strand was also evident. This led us to analyze the core region for samples #203, #318 and #372 at 85.5°C (3DPCR) and 95°C (PCR). The data are summarized in Figure 5B. A gradient of editing is apparent with G1896 and G1897 apparently hot spots for A3 editing. Focussing on mutations identified at 95°C, which can be considered as relatively high frequency events compared to those recovered by 3DPCR, they overlap nicely with those reported in a number of previous studies (Figure 5C) suggesting that A3 editing may well explain their origin [54], [55].
Fig. 5. A3 editing of the precore and HBsAg coding regions. 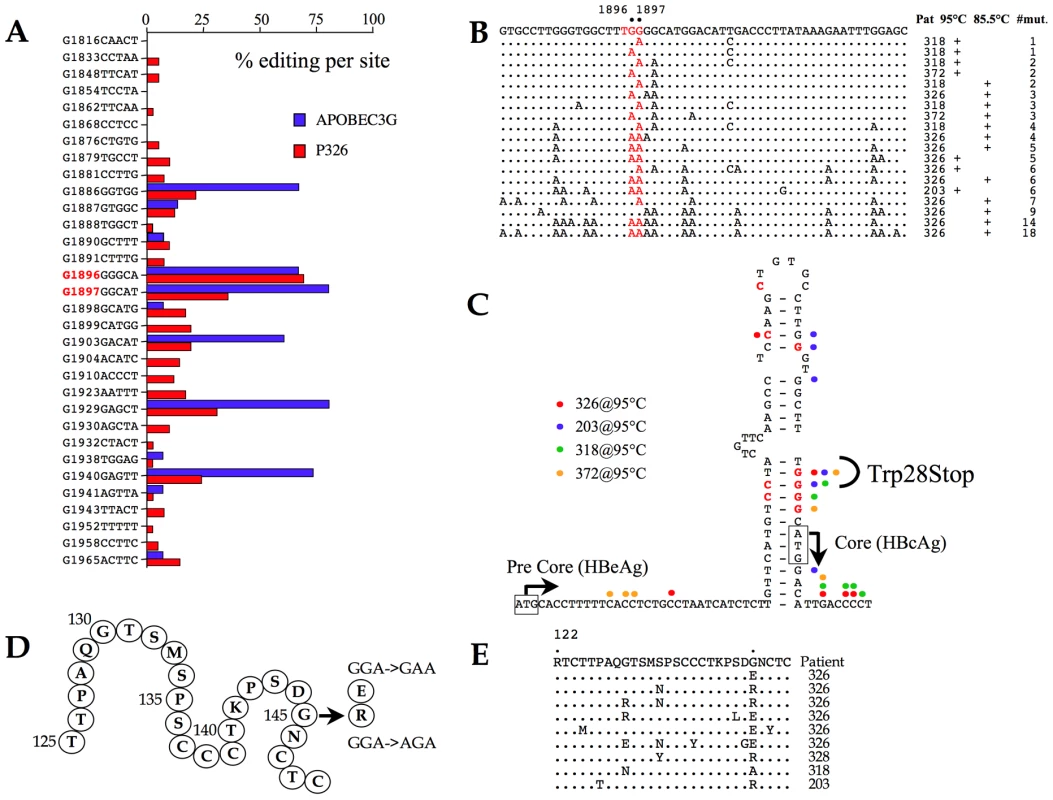
A) Site specific frequency analysis of A3G editing in the precore region from a tissue culture transfection experiment and from patient #326 recovered by 3DPCR at the restrictive temperature of 85.5°C. B) A selection of edited precore sequences in vivo. C) Correlation of G→A and C→T edited sites observed from the 95°C amplifications of samples #203, #318, #326 and #372 with those described from other studies with respect to the RNA stem loop structure implicit to DNA replication. D) Secondary structure of the common “a” determinant of HBsAg. The frequent G145R substitution and the G145R variant are noted along with the corresponding codon changes. E) A selection of A3 edited S region sequences bearing mutations in the G145 codon (standard PCR (95°C)). Such sequences represent <10% of the total. HBsAg mutants
The major viral surface antigen, HBsAg, is the basis of the highly successful subunit vaccine. Vaccine escape mutants are known, with a glycine to arginine (G145R) being the most frequent substitution mapping to the common double loop “a” determinant (residues 110–149, Figure 5D) [56], [57]. A G145E substitution is also known, as are a few other loop changes. The G145R mutant also emerges among HBV-immunoglobulin (HBIg) treated chronic carriers [58], [59] and goes undetected by standard diagnostic kits [60]. The local dinucleotide context associated with the G145 substitutions (Figure 5D) are again reminiscent of A3 editing. DNA spanning the “a” determinant was analyzed using the two-step PCR/3DPCR approach. Not surprisingly 3DPCR recovered hypermutants from all 4 samples tested at a restricting temperature of 85.5°C. The G145 codon was not a hotspot in vivo. As expected, the degree of editing was lower for genomes recovered at 95°C. Among sequences harbouring substitutions in codon 145 most were hypermutants with the G145E substitution while the G145R substitution was accompanied by other mutations (Figure 5E).
Discussion
The transcriptome data shows that numerous A3 genes are up regulated in virus associated cirrhosis, notably A3B, A3C, A3G, A3H and AICDA accompanied by a number of genes frequently up regulated in inflammatory tissues. By contrast, for alcoholic cirrhosis only two A3 genes (A3C and A3D) were significantly up regulated. Using the dinucleotide context as a hallmark to identify a posteriori the deaminase in vivo, A3G clearly emerges as a major restriction factor for HBV replication in cirrhosis along with at least one other A3 deaminase, which could be A3C (Figure 3C, D). The degree of editing is every bit as extensive as in co-transfection experiments using the powerful CMV-IE promoter in tissue culture [16], [21]. The up regulation of AICDA doesn't impact HBV editing in vivo despite the fact that AICDA shows itself to be a potent editor of replicating HBV genomes (Figure 2). This apparent dichotomy could be explained by the neogenesis of lymphoid follicles harbouring AICDA+ centroblasts, a feature associated with chronic inflammation [61], [62], [63], or circulating AICDA+ B cells as has been described for chronic HCV infection [64], [65], [66].
That A3G is the dominant A3 enzyme fits nicely with the fact that of all the A3 genes, A3G is the most strongly up regulated by interferon-α in primary hepatocytes [29]. Other reports show that the gene is also sensitive to induction by interferon–γ [28], [30], [34]. Pegylated interferon–α is used to treat a proportion of HBV infected individuals and the present data may explain part of that effect [67]. A recent report made a link between IFN-α treatment and HBeAg seroconversion, although they concluded that the link was tenuous given the low levels of editing in sera [25]. Although they didn't sequence precore DNA they used a 3DPCR approach. As shown here, the technique tends to underestimate levels by ∼10 fold (Figure 4). Accordingly hyperedited mutant frequencies reported in that study [25] can be revised upwards to ∼2–33%, in excellent agreement with the present findings (<2–35%). Given the strong impact of A3 deaminases on HBV replication in late stage disease, where does the virus replicate, especially as it is not known to encode an IFN or A3 antagonist? Some simple possibilities could be interferon resistant or A3low hepatocytes.
On a background of HBV+HCV double infection, HBV titres are generally lower [68], [69], as though HCV infection rendered the liver an even more hostile environment for HBV. It might be that the strong pro-inflammatory responses associated with HCV immune responses induce A3 genes with their detrimental effect on HBV. That the mean HBV cytidine deamination frequency was higher from the double infection compared to the monoinfection is the result expected if this hypothesis is correct (Figure 3B). As there has been debate as to the importance of A3G alleles in HIV disease (H186R [70]), it is possible that polymorphisms in the A3G gene impact the outcome of HBV infection. Certainly the most striking of all A3 polymorphisms, ΔA3B−/−, doesn't impact HBV disease [22], [71].
Given their ability to hypermutate DNA, A3 enzymes generate complex mutant spectra, the vast majority probably being deleterious. Even so, lightly A3 edited genomes predominated (Figure 4A) while a small fraction resists degradation and are repaired to standard DNA as the Pfu/Taq comparison shows (Figure 4F). Thereafter selection will operate on the remaining genomes. It would seem that IFN induced A3 editing may indeed lead to the occasional emergence of variants, of which the precore C1896T and/or C1897T and HBsAg G145R,E mutants are tangible examples. In this respect, there are remarkable parallels between some RNA viruses and IFN-α induction of the dsRNA adenosine deaminase, ADAR-1L. Editing by this enzyme of A1012 in the hepatitis D virus genome is essential to complete replication [72], [73]. Similarly, a handful of edited adenosine residues allows escape of respiratory syncytial viruses from monoclonal antibodies [74], [75].
In conclusion, the cytokine storm associated with chronic inflammatory responses to HBV and HCV clearly up regulates a number of A3 genes with A3G clearly being a major restriction factor for HBV. Could this also be a feature of other chronic inflammatory syndromes or even autoimmune diseases? Although the mutant spectrum resulting from A3 editing is highly deleterious, a very small part, notably the lightly edited genomes might help the virus evolve and even escape immune responses.
Materials and Methods
Patients, samples, RNA extraction, integrity and amplification
Patients were predominantly males, the mean ages being 60 years (HBV), 63 years (HBV+HCV), 64 years (HCV) and 67 years (alcoholic). All were negative for HIV. No patient was on interferon therapy in the months prior surgery. The study was approved by an Institutional Human Research Review Board (RBM 2005–019) and by Institut Pasteur. Written informed consent was obtained for each patient.
Forty-one cirrhotic samples as well as 4 normal livers were dissected and directly frozen in liquid nitrogen after surgical removal. Healthy samples represent tissue surrounding benign tumours such as angioma or focal nodular hyperplasia. Total RNA extraction was performed by a phenol-based method (Euromedex, Souffelweyersheim, France). DNase treatment was performed on 10 µg of total RNA using a DNA-free kit (Ambion). The RNA concentration and integrity were assessed using 100 ng of each RNA isolate to perform a capillary gel electrophoresis analysis (RNA 6000 Nano chip kit, Agilent Technologies, Palo Alto, CA) to establish an RNA integrity index (RIN). All the samples had acceptable quality, 86% with 7.0<RIN<10.0 and 14%, 4.1<RIN<6.9. Reverse transcription of 1 µg RNA was performed in a final volume of 20 µl (High-Capacity cDNA Archive kit, Applied Biosystems).
Real time quantitative PCR based gene expression
Messenger RNA were quantified by the TaqMan Low Density Array (TLDA) technology (Applied Biosystems, Courtaboeuf, France). Pre-designed hydrolysis probe and primer sets for target genes were factory loaded into the 384 wells of TLDA configured to contain duplicates per target gene (Table S2). Quantitative PCR was performed using cDNA samples corresponding to 400 ng of starting RNA and TaqMan Universal PCR Master Mix (Applied Biosystems) for 48 target genes in duplicate. QPCR conditions were one step of 94.5°C for 10 min. followed by 40 cycles at 97°C for 30 sec. and 59.7°C for 1 min. on a 7900HT Micro Fluidic Card instrument (Applied Biosystems).
For data analysis, gene expression values were determined using the calculation of the relative quantitation (RQ) of target genes normalized to a calibrator corresponding to 4 normal livers. RQ calculation was performed using the {Delta}{Delta}CT method using the geometric mean of three reference genes (TRIM44, HMBS and LMF2). The three references genes were selected among 12 constant genes arising from a previous array analysis of 70 HCC samples and 9 normal livers for which we applied the algorithms described [76] in the geNorm manual available on the web site http://medgen.ugent.be/~jvdesomp/genorm The study was performed according to the Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) and redaction of the manuscript according to the RDML (Real-Time PCR Data Markup Language) data standard (http://www.rdml.org).
Molecular biology
The pCayw plasmid and all APOBEC and AICDA expression plasmids have been previously described as have the cell lines and transfection protocols [16], [21]. Transfections were performed independently in triplicate on Huh7 cells and supernatant HBsAg was measured every day with the Monalisa HBsAg PLUS Kit (Bio-Rad). 3DPCR [50] was performed on a Eppendorf gradient Mastercycler S programmed to generate a 4–12°C gradient in the denaturation temperature. A fragment of the X region was amplified by employing a nested procedure. The first-round primers were: 5′Xout: 5′CGCAAATATACATCGTATCCAT and 3′Xout: AAGAGTYYTYTTATGTAAGACYTT, where Y is T,C and R is A,G. First PCR, conditions were: 5 min 95°C then (30 sec, 95°C; 30 sec, 60°C; 1 min, 72°C) ×35. Nested PCR was performed with 1/50 of the first round, primers were: 5′Xin: ATGGCTGCTARGCTGTGCTGCCAA and 3′Xin: AAGTGCACACGGTYYGGCAGAT, amplification conditions were: 5 min (82–93°C), then (1 min, 82–93°C; 30 sec, 60°C; 1 min, 72°C) ×35 then at 10 min 72°C.
The precore amplification was performed as following, first PCR, conditions were: 5 min 95°C then (30 sec, 95°C; 30 sec, 55°C; 1 min, 72°C) ×35 with primers 5′PreCout: GTACTAGGAGGCTGTAGGCATA and 3′PreCout: 5′ AGAGCTGAGGCGGTATCTAGAA. Nested PCR was performed with 1/50 of the first round, conditions were: 5 min (82–93°C), then (1 min, 82–93°C; 30 sec, 55°C; 1 min, 72°C) ×35, then 10 min at 72°C and primers were: 5′PreCin: TAAATTGGTCTGCGCACCAGCA and 3′PreCin: GATCTCGTACTGAAGGAAAGAA.
Amplification of the HBsAg was performed with a nested procedure, first PCR conditions were: 5 min 95°C then (30 sec, 95°C; 30 sec, 60°C; 1 min, 72°C) ×35, primers were: 5′HBsout: CGGCGTTTTATCATCTTCCTCTTCAT and 3′HBsout: CATCCATATAACTGAAAGCCAAACAGT. Nested PCR was performed with 1/50 of the first round, conditions were 5 min (82–93°C), then (1 min, 82–93°C; 30 sec, 60°C; 1 min, 72°C) ×35 then 10 min at 72°C with primers, 5′HBsin: TCTTCATCCTGCTGCTATGCCTCAT and 3′HBsin: AAAGCCCTACGAACCACTGAACAAAT.
PCR and 3DPCR products were purified from agarose gels (Qiaex II kit, Qiagen, France) and ligated into the TOPO TA cloning vector (Invitrogen, France). The 88.7°C 3DPCR products obtained from X gene amplification were chosen as experience shows they provide a wide range of edited HBV genomes. While products analyzed at lower temperatures are more edited, they proved to be more homogeneous. Sequencing was outsourced to Cogenics. All mutations were verified on the chromatogram.
Supporting Information
Zdroje
1. BealeRC
Petersen-MahrtSK
WattIN
HarrisRS
RadaC
2004 Comparison of the differential context-dependence of DNA deamination by APOBEC enzymes: correlation with mutation spectra in vivo. J Mol Biol 337 585 596
2. BishopKN
HolmesRK
SheehyAM
DavidsonNO
ChoSJ
2004 Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr Biol 14 1392 1396
3. ConticelloSG
HarrisRS
NeubergerMS
2003 The Vif protein of HIV triggers degradation of the human antiretroviral DNA deaminase APOBEC3G. Curr Biol 13 2009 2013
4. LecossierD
BouchonnetF
ClavelF
HanceAJ
2003 Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science 300 1112
5. HarrisRS
LiddamentMT
2004 Retroviral restriction by APOBEC proteins. Nat Rev Immunol 4 868 877
6. MangeatB
TurelliP
CaronG
FriedliM
PerrinL
2003 Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424 99 103
7. MarianiR
ChenD
SchrofelbauerB
NavarroF
KonigR
2003 Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 114 21 31
8. SuspèneR
SommerP
HenryM
FerrisS
GuétardD
2004 APOBEC3G is a single-stranded DNA cytidine deaminase and functions independently of HIV reverse transcriptase. Nucleic Acids Res 32 2421 2429
9. BogerdHP
DoehleBP
WiegandHL
CullenBR
2004 A single amino acid difference in the host APOBEC3G protein controls the primate species specificity of HIV type 1 virion infectivity factor. Proc Natl Acad Sci USA 101 3770 3774
10. ZhangH
YangB
PomerantzRJ
ZhangC
ArunachalamSC
2003 The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature 424 94 98
11. JarmuzA
ChesterA
BaylissJ
GisbourneJ
DunhamI
2002 An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics 79 285 296
12. BogerdHP
WiegandHL
HulmeAE
Garcia-PerezJL
O'SheaKS
2006 Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc Natl Acad Sci USA 103 8780 8785
13. ChiuYL
WitkowskaHE
HallSC
SantiagoM
SorosVB
2006 High-molecular-mass APOBEC3G complexes restrict Alu retrotransposition. Proc Natl Acad Sci USA 103 15588 15593
14. DelebecqueF
SuspeneR
CalattiniS
CasartelliN
SaibA
2006 Restriction of foamy viruses by APOBEC cytidine deaminases. J Virol 80 605 614
15. EsnaultC
HeidmannO
DelebecqueF
DewannieuxM
RibetD
2005 APOBEC3G cytidine deaminase inhibits retrotransposition of endogenous retroviruses. Nature 433 430 433
16. HenryM
GuétardD
SuspèneR
RusniokC
Wain-HobsonS
2009 Genetic editing of HBV DNA by monodomain human APOBEC3 cytidine deaminases and the recombinant nature of APOBEC3G. PLoS One 4 e4277
17. PetitV
GuétardD
RenardM
KerielA
SitbonM
2009 Murine APOBEC1 is a powerful mutator of retroviral and cellular RNA in vitro and in vivo. J Mol Biol 385 65 78
18. RussellRA
WiegandHL
MooreMD
SchaferA
McClureMO
2005 Foamy virus Bet proteins function as novel inhibitors of the APOBEC3 family of innate antiretroviral defense factors. J Virol 79 8724 8731
19. MahieuxR
SuspèneR
DelebecqueF
HenryM
SchwartzO
2005 Extensive editing of a small fraction of human T-cell leukemia virus type 1 genomes by four APOBEC3 cytidine deaminases. J Gen Virol 86 2489 2494
20. TurelliP
MangeatB
JostS
VianinS
TronoD
2004 Inhibition of Hepatitis B Virus Replication by APOBEC3G. Science 303 1829
21. SuspèneR
GuétardD
HenryM
SommerP
Wain-HobsonS
2005 Extensive editing of both hepatitis B virus DNA strands by APOBEC3 cytidine deaminases in vitro and in vivo. Proc Natl Acad Sci USA 102 8321 8326
22. AbeH
OchiH
MaekawaT
HatakeyamaT
TsugeM
2009 Effects of structural variations of APOBEC3A and APOBEC3B genes in chronic hepatitis B virus infection. Hepatol Res 39 1159 1168
23. BaumertTF
RoslerC
MalimMH
von WeizsackerF
2007 Hepatitis B virus DNA is subject to extensive editing by the human deaminase APOBEC3C. Hepatology 46 682 689
24. JaniniM
RogersM
BirxDR
McCutchanFE
2001 Human immunodeficiency virus type 1 DNA sequences genetically damaged by hypermutation are often abundant in patient peripheral blood mononuclear cells and may be generated during near-simultaneous infection and activation of CD4(+) T cells. J Virol 75 7973 7986
25. NoguchiC
ImamuraM
TsugeM
HiragaN
MoriN
2009 G-to-A hypermutation in hepatitis B virus (HBV) and clinical course of patients with chronic HBV infection. J Infect Dis 199 1599 1607
26. TsugeM
NoguchiC
AkiyamaR
MatsushitaM
KunihiroK
2010 G to A hypermutation of TT virus. Virus Res 149 211 216
27. VartanianJP
GuétardD
HenryM
Wain-HobsonS
2008 Evidence for editing of human papillomavirus DNA by APOBEC3 in benign and precancerous lesions. Science 320 230 233
28. ArgyrisEG
AcheampongE
WangF
HuangJ
ChenK
2007 The interferon-induced expression of APOBEC3G in human blood-brain barrier exerts a potent intrinsic immunity to block HIV-1 entry to central nervous system. Virology 367 440 451
29. BonvinM
AchermannF
GreeveI
StrokaD
KeoghA
2006 Interferon-inducible expression of APOBEC3 editing enzymes in human hepatocytes and inhibition of hepatitis B virus replication. Hepatology 43 1364 1374
30. KoningFA
NewmanEN
KimEY
KunstmanKJ
WolinskySM
2009 Defining APOBEC3 expression patterns in human tissues and hematopoietic cell subsets. J Virol 83 9474 9485
31. PengG
LeiKJ
JinW
Greenwell-WildT
WahlSM
2006 Induction of APOBEC3 family proteins, a defensive maneuver underlying interferon-induced anti-HIV-1 activity. J Exp Med 203 41 46
32. StengleinMD
BurnsMB
LiM
LengyelJ
HarrisRS
2010 APOBEC3 proteins mediate the clearance of foreign DNA from human cells. Nat Struct Mol Biol 17 222 229
33. TanakaY
MarusawaH
SenoH
MatsumotoY
UedaY
2006 Anti-viral protein APOBEC3G is induced by interferon-alpha stimulation in human hepatocytes. Biochem Biophys Res Commun 341 314 319
34. VetterML
JohnsonME
AntonsAK
UnutmazD
D'AquilaRT
2009 Differences in APOBEC3G expression in CD4+ T helper lymphocyte subtypes modulate HIV-1 infectivity. PLoS Pathog 5 e1000292
35. WangFX
HuangJ
ZhangH
MaX
2008 APOBEC3G upregulation by alpha interferon restricts human immunodeficiency virus type 1 infection in human peripheral plasmacytoid dendritic cells. J Gen Virol 89 722 730
36. MuramatsuM
SankaranandVS
AnantS
SugaiM
KinoshitaK
1999 Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J Biol Chem 274 18470 18476
37. EndoY
MarusawaH
KinoshitaK
MorisawaT
SakuraiT
2007 Expression of activation-induced cytidine deaminase in human hepatocytes via NF-kappaB signaling. Oncogene 26 5587 5595
38. EndoY
MarusawaH
KouT
NakaseH
FujiiS
2008 Activation-induced cytidine deaminase links between inflammation and the development of colitis-associated colorectal cancers. Gastroenterology 135 889 898
39. MachidaK
KondoY
HuangJY
ChenYC
ChengKT
2008 Hepatitis C virus (HCV)-induced immunoglobulin hypermutation reduces the affinity and neutralizing activities of antibodies against HCV envelope protein. J Virol 82 6711 6720
40. MatsumotoY
MarusawaH
KinoshitaK
EndoY
KouT
2007 Helicobacter pylori infection triggers aberrant expression of activation-induced cytidine deaminase in gastric epithelium. Nat Med 13 470 476
41. MorisawaT
MarusawaH
UedaY
IwaiA
OkazakiIM
2008 Organ-specific profiles of genetic changes in cancers caused by activation-induced cytidine deaminase expression. Int J Cancer 123 2735 2740
42. OkazakiIM
HiaiH
KakazuN
YamadaS
MuramatsuM
2003 Constitutive expression of AID leads to tumorigenesis. J Exp Med 197 1173 1181
43. YamanakaS
BalestraME
FerrellLD
FanJ
ArnoldKS
1995 Apolipoprotein B mRNA-editing protein induces hepatocellular carcinoma and dysplasia in transgenic animals. Proc Natl Acad Sci USA 92 8483 8487
44. TakaiA
ToyoshimaT
UemuraM
KitawakiY
MarusawaH
2009 A novel mouse model of hepatocarcinogenesis triggered by AID causing deleterious p53 mutations. Oncogene 28 469 478
45. AggarwalBB
GehlotP
2009 Inflammation and cancer: how friendly is the relationship for cancer patients? Curr Opin Pharmacol 9 351 369
46. MantovaniA
AllavenaP
SicaA
BalkwillF
2008 Cancer-related inflammation. Nature 454 436 444
47. FeldhahnN
HenkeN
MelchiorK
DuyC
SohBN
2007 Activation-induced cytidine deaminase acts as a mutator in BCR-ABL1-transformed acute lymphoblastic leukemia cells. J Exp Med 204 1157 1166
48. MachidaK
ChengKT
SungVM
ShimodairaS
LindsayKL
2004 Hepatitis C virus induces a mutator phenotype: enhanced mutations of immunoglobulin and protooncogenes. Proc Natl Acad Sci USA 101 4262 4267
49. Cervantes-GonzalezMC
SuspèneR
HenryM
GuétardD
Wain-HobsonS
2009 Human APOBEC1 cytidine deaminase edits HBV DNA. Retrovirology 6 96
50. SuspèneR
HenryM
GuillotS
Wain-HobsonS
VartanianJP
2005 Recovery of APOBEC3-edited human immunodeficiency virus G→A hypermutants by differential DNA denaturation PCR. J Gen Virol 86 125 129
51. PhamP
BransteitterR
PetruskaJ
GoodmanMF
2003 Processive AID-catalysed cytosine deamination on single-stranded DNA simulates somatic hypermutation. Nature 424 103 107
52. GreaggMA
FoggMJ
PanayotouG
EvansSJ
ConnollyBA
1999 A read-ahead function in archaeal DNA polymerases detects promutagenic template-strand uracil. Proc Natl Acad Sci USA 96 9045 9050
53. EvansA
RivaA
CooksleyH
PhillipsS
PuranikS
2008 Programmed death 1 expression during antiviral treatment of chronic hepatitis B: Impact of hepatitis B e-antigen seroconversion. Hepatology 48 759 769
54. GuarnieriM
KimKH
BangG
LiJ
ZhouY
2006 Point mutations upstream of hepatitis B virus core gene affect DNA replication at the step of core protein expression. J Virol 80 587 595
55. ZhongS
ChanJY
YeoW
TamJS
JohnsonPJ
2000 Frequent integration of precore/core mutants of hepatitis B virus in human hepatocellular carcinoma tissues. J Viral Hepat 7 115 123
56. CarmanWF
ZanettiAR
KarayiannisP
WatersJ
ManzilloG
1990 Vaccine-induced escape mutant of hepatitis B virus. Lancet 336 325 329
57. ZuckermanAJ
2000 Effect of hepatitis B virus mutants on efficacy of vaccination. Lancet 355 1382 1384
58. ChiouHL
LeeTS
KuoJ
MauYC
HoMS
1997 Altered antigenicity of ‘a’ determinant variants of hepatitis B virus. J Gen Virol 78 2639 2645
59. CooremanMP
Leroux-RoelsG
PaulijWP
2001 Vaccine - and hepatitis B immune globulin-induced escape mutations of hepatitis B virus surface antigen. J Biomed Sci 8 237 247
60. LouisirirotchanakulS
KanoksinsombatC
TheamboonlertA
PuthavatanaP
WasiC
2004 Mutation of the “a” determinant of HBsAg with discordant HBsAg diagnostic kits. Viral Immunol 17 440 444
61. AloisiF
Pujol-BorrellR
2006 Lymphoid neogenesis in chronic inflammatory diseases. Nat Rev Immunol 6 205 217
62. Moyron-QuirozJE
Rangel-MorenoJ
KusserK
HartsonL
SpragueF
2004 Role of inducible bronchus associated lymphoid tissue (iBALT) in respiratory immunity. Nat Med 10 927 934
63. LeeY
ChinRK
ChristiansenP
SunY
TumanovAV
2006 Recruitment and activation of naive T cells in the islets by lymphotoxin beta receptor-dependent tertiary lymphoid structure. Immunity 25 499 509
64. ItoM
MurakamiK
SuzukiT
MochidaK
SuzukiM
2010 Enhanced expression of lymphomagenesis-related genes in peripheral blood B cells of chronic hepatitis C patients. Clin Immunol in press
65. SagnelliE
PasqualeG
CoppolaN
MarroccoC
ScaranoF
2005 Liver histology in patients with HBsAg negative anti-HBc and anti-HCV positive chronic hepatitis. J Med Virol 75 222 226
66. VillariD
PerniceM
SpinellaS
SquadritoG
RodinoG
1995 Chronic hepatitis in patients with active hepatitis B virus and hepatitis C virus combined infections: a histological study. Am J Gastroenterol 90 955 958
67. TaylorMW
GrosseWM
SchaleyJE
SandaC
WuX
2004 Global effect of PEG-IFN-alpha and ribavirin on gene expression in PBMC in vitro. J Interferon Cytokine Res 24 107 118
68. SchuttlerCG
FiedlerN
SchmidtK
ReppR
GerlichWH
2002 Suppression of hepatitis B virus enhancer 1 and 2 by hepatitis C virus core protein. J Hepatol 37 855 862
69. ShihCM
LoSJ
MiyamuraT
ChenSY
LeeYH
1993 Suppression of hepatitis B virus expression and replication by hepatitis C virus core protein in HuH-7 cells. J Virol 67 5823 5832
70. AnP
BleiberG
DuggalP
NelsonG
MayM
2004 APOBEC3G genetic variants and their influence on the progression to AIDS. J Virol 78 11070 11076
71. KiddJM
NewmanTL
TuzunE
KaulR
EichlerEE
2007 Population stratification of a common APOBEC gene deletion polymorphism. PLoS Genet 3 e63
72. PolsonAG
BassBL
CaseyJL
1996 RNA editing of hepatitis delta virus antigenome by dsRNA-adenosine deaminase. Nature 380 454 456
73. WongSK
LazinskiDW
2002 Replicating hepatitis delta virus RNA is edited in the nucleus by the small form of ADAR1. Proc Natl Acad Sci USA 99 15118 15123
74. RuedaP
Garcia-BarrenoB
MeleroJA
1994 Loss of conserved cysteine residues in the attachment (G) glycoprotein of two human respiratory syncytial virus escape mutants that contain multiple A–G substitutions (hypermutations). Virology 198 653 662
75. MartinezI
DopazoJ
MeleroJA
1997 Antigenic structure of the human respiratory syncytial virus G glycoprotein and relevance of hypermutation events for the generation of antigenic variants. J Gen Virol 78 2419 2429
76. VandesompeleJ
De PreterK
PattynF
PoppeB
Van RoyN
2002 Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3 RESEARCH0034
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Mouse Senile Amyloid Fibrils Deposited in Skeletal Muscle Exhibit Amyloidosis-Enhancing ActivityČlánek The Role of Intestinal Microbiota in the Development and Severity of Chemotherapy-Induced MucositisČlánek Crystal Structure of HIV-1 gp41 Including Both Fusion Peptide and Membrane Proximal External RegionsČlánek Demonstration of Cross-Protective Vaccine Immunity against an Emerging Pathogenic Ebolavirus Species
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2010 Číslo 5- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Quorum Sensing Inhibition Selects for Virulence and Cooperation in
- The HMW1C Protein Is a Glycosyltransferase That Transfers Hexose Residues to Asparagine Sites in the HMW1 Adhesin
- Analysis of Virion Structural Components Reveals Vestiges of the Ancestral Ichnovirus Genome
- Mouse Senile Amyloid Fibrils Deposited in Skeletal Muscle Exhibit Amyloidosis-Enhancing Activity
- Global Migration Dynamics Underlie Evolution and Persistence of Human Influenza A (H3N2)
- The Type III Effectors NleE and NleB from Enteropathogenic and OspZ from Block Nuclear Translocation of NF-κB p65
- VEGF Promotes Malaria-Associated Acute Lung Injury in Mice
- Identification of a Mutant PfCRT-Mediated Chloroquine Tolerance Phenotype in
- The Early Stage of Bacterial Genome-Reductive Evolution in the Host
- Host-Detrimental Role of Esx-1-Mediated Inflammasome Activation in Mycobacterial Infection
- Elevation of Intact and Proteolytic Fragments of Acute Phase Proteins Constitutes the Earliest Systemic Antiviral Response in HIV-1 Infection
- The Pleiotropic CymR Regulator of Plays an Important Role in Virulence and Stress Response
- Alternative Sigma Factor σ Modulates Prophage Integration and Excision in
- Effect of Neuraminidase Inhibitor–Resistant Mutations on Pathogenicity of Clade 2.2 A/Turkey/15/06 (H5N1) Influenza Virus in Ferrets
- Massive APOBEC3 Editing of Hepatitis B Viral DNA in Cirrhosis
- NK Cells and γδ T Cells Mediate Resistance to Polyomavirus–Induced Tumors
- Is Genetically Diverse in Animals and Appears to Have Crossed the Host Barrier to Humans on (At Least) Two Occasions
- Adenylate Cyclase Toxin Mobilizes Its β Integrin Receptor into Lipid Rafts to Accomplish Translocation across Target Cell Membrane in Two Steps
- The Role of Intestinal Microbiota in the Development and Severity of Chemotherapy-Induced Mucositis
- HIV-1 Transmitting Couples Have Similar Viral Load Set-Points in Rakai, Uganda
- Few and Far Between: How HIV May Be Evading Antibody Avidity
- Galectin-9/TIM-3 Interaction Regulates Virus-Specific Primary and Memory CD8 T Cell Response
- Perforin Expression Directly by HIV-Specific CD8 T-Cells Is a Correlate of HIV Elite Control
- The Set3/Hos2 Histone Deacetylase Complex Attenuates cAMP/PKA Signaling to Regulate Morphogenesis and Virulence of
- Infidelity of SARS-CoV Nsp14-Exonuclease Mutant Virus Replication Is Revealed by Complete Genome Sequencing
- Combining ChIP-chip and Expression Profiling to Model the MoCRZ1 Mediated Circuit for Ca/Calcineurin Signaling in the Rice Blast Fungus
- Internalin B Activates Junctional Endocytosis to Accelerate Intestinal Invasion
- A Complex Small RNA Repertoire Is Generated by a Plant/Fungal-Like Machinery and Effected by a Metazoan-Like Argonaute in the Single-Cell Human Parasite
- Opc Invasin Binds to the Sulphated Tyrosines of Activated Vitronectin to Attach to and Invade Human Brain Endothelial Cells
- Muc2 Protects against Lethal Infectious Colitis by Disassociating Pathogenic and Commensal Bacteria from the Colonic Mucosa
- PdeH, a High-Affinity cAMP Phosphodiesterase, Is a Key Regulator of Asexual and Pathogenic Differentiation in
- Isolates with Antimony-Resistant but Not -Sensitive Phenotype Inhibit Sodium Antimony Gluconate-Induced Dendritic Cell Activation
- The Microbiota and Allergies/Asthma
- Environmental Factors Determining the Epidemiology and Population Genetic Structure of the Group in the Field
- Prolonged Antigen Presentation Is Required for Optimal CD8+ T Cell Responses against Malaria Liver Stage Parasites
- Crystal Structure of HIV-1 gp41 Including Both Fusion Peptide and Membrane Proximal External Regions
- Susceptibility to Anthrax Lethal Toxin-Induced Rat Death Is Controlled by a Single Chromosome 10 Locus That Includes
- Demonstration of Cross-Protective Vaccine Immunity against an Emerging Pathogenic Ebolavirus Species
- Effective, Broad Spectrum Control of Virulent Bacterial Infections Using Cationic DNA Liposome Complexes Combined with Bacterial Antigens
- High Multiplicity Infection by HIV-1 in Men Who Have Sex with Men
- The -Specific Human Memory B Cell Compartment Expands Gradually with Repeated Malaria Infections
- EBV Promotes Human CD8 NKT Cell Development
- Persistent Growth of a Human Plasma-Derived Hepatitis C Virus Genotype 1b Isolate in Cell Culture
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Quorum Sensing Inhibition Selects for Virulence and Cooperation in
- The Role of Intestinal Microbiota in the Development and Severity of Chemotherapy-Induced Mucositis
- Crystal Structure of HIV-1 gp41 Including Both Fusion Peptide and Membrane Proximal External Regions
- Susceptibility to Anthrax Lethal Toxin-Induced Rat Death Is Controlled by a Single Chromosome 10 Locus That Includes
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



