-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaCombining ChIP-chip and Expression Profiling to Model the MoCRZ1 Mediated Circuit for Ca/Calcineurin Signaling in the Rice Blast Fungus
Significant progress has been made in defining the central signaling networks in many organisms, but collectively we know little about the downstream targets of these networks and the genes they regulate. To reconstruct the regulatory circuit of calcineurin signal transduction via MoCRZ1, a Magnaporthe oryzae C2H2 transcription factor activated by calcineurin dephosphorylation, we used a combined approach of chromatin immunoprecipitation - chip (ChIP-chip), coupled with microarray expression studies. One hundred forty genes were identified as being both a direct target of MoCRZ1 and having expression concurrently differentially regulated in a calcium/calcineurin/MoCRZ1 dependent manner. Highly represented were genes involved in calcium signaling, small molecule transport, ion homeostasis, cell wall synthesis/maintenance, and fungal virulence. Of particular note, genes involved in vesicle mediated secretion necessary for establishing host associations, were also found. MoCRZ1 itself was a target, suggesting a previously unreported autoregulation control point. The data also implicated a previously unreported feedback regulation mechanism of calcineurin activity. We propose that calcium/calcineurin regulated signal transduction circuits controlling development and pathogenicity manifest through multiple layers of regulation. We present results from the ChIP-chip and expression analysis along with a refined model of calcium/calcineurin signaling in this important plant pathogen.
Published in the journal: . PLoS Pathog 6(5): e32767. doi:10.1371/journal.ppat.1000909
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1000909Summary
Significant progress has been made in defining the central signaling networks in many organisms, but collectively we know little about the downstream targets of these networks and the genes they regulate. To reconstruct the regulatory circuit of calcineurin signal transduction via MoCRZ1, a Magnaporthe oryzae C2H2 transcription factor activated by calcineurin dephosphorylation, we used a combined approach of chromatin immunoprecipitation - chip (ChIP-chip), coupled with microarray expression studies. One hundred forty genes were identified as being both a direct target of MoCRZ1 and having expression concurrently differentially regulated in a calcium/calcineurin/MoCRZ1 dependent manner. Highly represented were genes involved in calcium signaling, small molecule transport, ion homeostasis, cell wall synthesis/maintenance, and fungal virulence. Of particular note, genes involved in vesicle mediated secretion necessary for establishing host associations, were also found. MoCRZ1 itself was a target, suggesting a previously unreported autoregulation control point. The data also implicated a previously unreported feedback regulation mechanism of calcineurin activity. We propose that calcium/calcineurin regulated signal transduction circuits controlling development and pathogenicity manifest through multiple layers of regulation. We present results from the ChIP-chip and expression analysis along with a refined model of calcium/calcineurin signaling in this important plant pathogen.
Introduction
Rice blast, caused by the fungal pathogen Magnaporthe oryzae, is a recurrent and devastating problem worldwide [1]. Severe disease outbreaks can destroy upwards of 90% of rice yields for an entire field, region, or country resulting in a dramatic impact on human welfare and regional economies. The process starts when an asexual spore lands on a rice leaf. Given suitable moisture and temperature, the spore germinates with a short germination tube at the tip of which a specialized infection structure, an appressorium, emerges. The formation of an appressorium is essential for successful disease as it facilitates breaching the plant cuticle and cell wall, allowing access to the underlying tissues. Following penetration, the entry peg forms an un-branched hyphal strand that subsequently matures into branched bulbous infection hyphae. The fungus fills the infected cell in what is considered a biotrophic state before invading adjacent cells and switching to a necrotrophic state where it ramifies through the host tissues killing cells. In the final stage, the fungus produces new asexual spores that are spread to neighboring plants [2], [3]. While knowledge of the core signal pathways regulating each phase of this process continues to resolve, the key determinants controlling environmental perception and cellular response are as yet not fully understood [4]. Specifically, we have little knowledge of the upstream receptors used by the fungus to detect stimuli, nor do we know how downstream factors specifically interact to affect expression of genes deployed during infection related development, establishment of host associations, and invasive growth.
Calcium signaling has been implicated in regulating growth and development in M. oryzae including the infection process [5]–[9]. The components of Ca2+ signaling have been studied in many organisms and are relatively well understood. Ca2+ signaling starts when G-protein coupled receptors are activated by an external stimulus. Phospholipase C (PLC) is activated to hydrolyze phosphatidyl inositol-1,4-bisphosphate (PIP2) into inositol 1,4,5-triphosphate (IP3) and diacylglycerol. IP3 activates Ca2+ release from intracellular stores into the cytosol. Ca2+ ions bind to and activate calmodulin, which in turn, activates the Ca2+/calmodulin-dependent serine/threonine protein phosphatase calcineurin. Calcineurin is a heterodimer consisting of catalytic (CNA) and regulatory (CNB) subunits. In fungi, calcineurin mediated Ca2+ signaling has been shown to be required for growth, development, response to stress, and pathogenesis [10]. It was necessary for survival during environmental stresses such as ions (Mn2+, Li+, Na+), high pH, high temperature, ER stress, and prolonged incubation with mating pheromone α-factor in Saccharomyces cerevisiae [11], [12]. It is essential for growth and virulence of Candida albicans and Cryptococcus neoformans [13]–[16], and controls the dimorphic transition from mycelia to yeast in Paracoccidioides basiliensis [17]. Effects of gene deletion or chemical inhibition in filamentous fungi typically have pleiotropic effects. For example, a cnaA deletion mutant in Aspergillus fumigatus was viable but severely affected in hyphal morphology, sporulation, conidial architecture, pathogenicity, and invasive growth [18], [19]. Reduction of calcineurin activity by the immunosuppressant drug cyclosporine A, resulted in reduction of mycelial growth and alteration in hyphal morphology as shown in Neurospora crassa [20], [21], A. nidulans [22], A. oryzae [23], and M. oryzae [24]. RNA silencing in M. oryzae showed similar effects, specifically a reduction in mycelial growth, sporulation, and appressorium formation in MCNA knock down mutants [8].
Calcineurin functions mainly through the activation of the transcription factor CRZ1 (Calcineurin Responsive Zinc Finger 1). Upon activation by increased intracellular Ca2+ and calmodulin, calcineurin dephosphorylates CRZ1 leading to its nuclear localization. As a major mediator of calcineurin signaling, crz1 deletion mutants in a variety of fungi showed similar phenotypes as calcineurin mutants [5], [25]–[28]. However, differences have been noted. CRZ1 in C. albicans was not involved in tolerance to antifungal agents (fluconazole, terbinafine) and only slightly affected in virulence, which is in contrast to the calcineurin mutants [28]. On the other hand, CRZ1 is strongly associated with virulence both in human and plant pathogenic fungi [5], [25], [26], [28], [29]. The B. cinerea CRZ1 ortholog BcCRZ1 was required for growth, conidial and sclerotial development, and full virulence while being dispensable for conidia-derived infection of bean plants [25]. In M. oryzae, the Δmocrz1 deletion mutant showed decreased conidiation and was not able to cause disease when spray inoculated. Mutant conidia were not distinguishable from that of wild type and formed appressoria at a similar level as wild type. Importantly, a significant portion of appressoria failed to penetrate rice sheath tissue. The mutant could colonize and cause disease when the conidia were infiltrated directly into the host tissue, thus bypassing the penetration process and suggesting that MoCRZ1 plays a role in appressorium mediated penetration and establishing a biotrophic association with its host [5].
Comprehensive genome-wide expression analysis in S. cerevisiae identified 163 genes regulated in a calcineurin/CRZ1 dependent manner by the stimulation of Ca2+ or Na+. These genes were associated with a diverse range of cellular processes including signaling pathways, ion/small molecule transport, cell wall synthesis/maintenance, and vesicular trafficking [30]. In C. albicans, microarray analysis revealed 60 genes to be transcriptionally activated by exogenous Ca2+ treatment through calcineurin/CRZ1 regulation using cnaΔ/Δ and crz1Δ/Δ mutants. Analysis of putative functions revealed that about 60% of these genes were involved in cell wall organization, cellular organization, cellular transport and homeostasis, cell metabolism, and protein fate [28]. Many of the genes regulated through calcineurin signaling in these two species belonged to similar functional groups, although only 9 genes were found to be commonly regulated [28]. To date, no genome-wide study has been conducted to identify regulated genes by direct binding of CRZ1 to promoter regions.
Here we report the use of chromatin immunoprecipitation coupled with non-coding region tiling arrays (ChIP-chip) analysis and whole-genome expression studies to identify target genes directly regulated by MoCRZ1. To our knowledge, this is the first report of ChIP-chip technology being applied to filamentous fungi. From this analysis, we can model the Ca2+/calcineurin signaling and control pathways that in-part influence infection related development and establishment of a compatible host association for this devastating plant pathogen. Further, this work reveals divergence within the fungal kingdom of the suites of genes and processes directly regulated by this ubiquitous signaling pathway.
Results
GFP tagging of MoCRZ1 and localization
A MoCRZ1::eGFP construct was co-transformed into fungal protoplasts along with a hygromycin resistance conferring vector. Transformants were single spore isolated and screened under the epi-fluorescent microscope. MoCRZ1-eGFP fluorescence was faint and evenly dispersed through the cytosol in mycelia with nuclear localization detected in hyphal tips (data not shown). Nuclear translocation of CRZ1 in response to Ca2+ is a well conserved phenomenon and was shown previously to occur in M. oryzae [5]. Following mycelia treatment with CaCl2, eGFP fluorescence was localized to the nucleus (Figure 1A), as expected. Addition of the calcineurin inhibitor FK506 completely blocked nuclear accumulation of the fluorescent protein. The MoCRZ1::eGFP over-expression line showed normal growth, appressoria development, and virulence to susceptible rice cultivar Nipponbare (data not shown), and was selected for subsequent ChIP-chip analysis.
Fig. 1. Establishment of Chromatin immunoprecipitation. 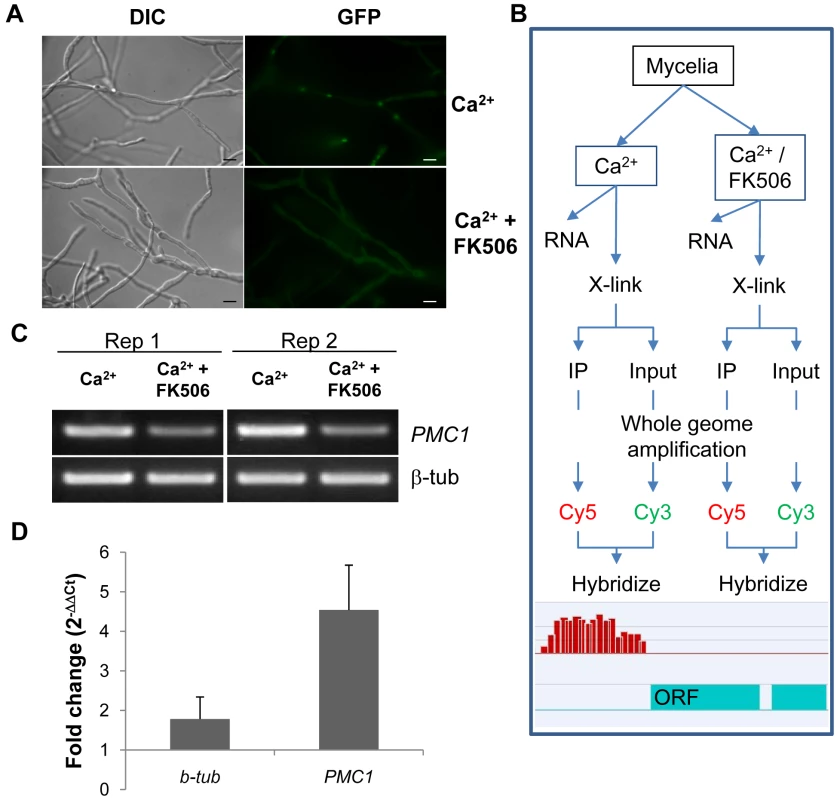
(A) Nuclear localization of MoCRZ1 was visualized in the eGFP tagged strain under the native promoter. FK506 blocks nuclear localization of MoCRZ1::eGFP. Bar indicates 10 µm. (B) ChIP-chip experimental design to identify MoCRZ1 targets activated by calcium treatment. The Ca2+/FK506 treated sample served as the negative control treatment. (C) RT-PCR to verify that PMC1 was up-regulated in the Ca2+ treated but not in Ca2+/FK506 treated sample. (D) Quantitative PCR was conducted with DNA after ChIP with antiGFP antibody. 30% input DNA collected prior to pull down was used as control. 1 µl each of ChIPed and input DNA was used for real-time PCR. Fold changes were calculated by 2ΔΔCt, where ΔΔCt = (Ctinput DNA-CtChIPed DNA) Ca2+ treated sample - (Ctinput DNA-CtChIPed DNA) Ca2+/FK506 treated sample. Chromatin immunoprecipitation of MoCRZ1 bound sequences
Experimental design for ChIP-chip analysis is depicted in Figure 1B. CaCl2 treated mycelia were used to enrich MoCRZ1 occupied genomic fragments, while mycelia treated with CaCl2 and FK506 were used as the negative control. Expression of PMC1 (P-type ATPase) was analyzed for each sample by RT-PCR as shown in Figure 1C to confirm the effect of each treatment, i.e., up-regulated in the calcium treated mycelium and blocked by FK506. PMC1 is a previously described target of MoCRZ1 [5] and was used throughout this study as positive control marker. Enrichment of MoCRZ1 bound DNA fragments in ChIPed fractions when compared to input DNA (non-ChIPed) was confirmed by real-time PCR (Figure 1D). As expected, the fold change of PMC1 was 4.54±1.13 in ChIPed DNA from Ca2+ treated mycelia over input DNA from Ca2+ + FK506 treated mycelia, while that of β-tubulin was 1.78±0.56 (Figure 1D). Input and IPed DNA was amplified and subsequently re-amplified to amass sufficient DNA (∼8.5 ug) for labeling and hybridization to the array. Enrichment of MoCRZ1 bound DNA in the ChIPed fraction was validated by PCR amplification of PMC1 at each step (data not shown). ChIPed DNA labeled with Cy5 was co-hybridized with Cy3 labeled input DNA to the NimbleGen M. oryzae intergenic region specific tiling array (see Materials and Methods for array description).
Genome-wide identification of MoCRZ1 binding sites and downstream genes
Two complementary approaches were applied to analyze ChIP-chip hybridization data to identify putative MoCRZ1 binding sites and the genes it regulates. Initially, 42 peaks were identified as having a false discovery rate less than 0.2 and being common in both biological replications of the Ca2+ treatment but not in the Ca2++FK506 treatment. The 42 peaks were within 1 kb upstream of 37 predicted genes. Following this initial analysis, we used NimbleGen's SignalMap software to manually interrogate the ratio signal tracks across the genome to identify short sequence stretches showing a normal distribution profile of signal intensities upstream of annotated ORFs (Figure 1B bottom). Sequence tracks showing this profile were accepted only if they appeared in both biological replications of the Ca2+ treatment with no or lower signal intensities in the Ca2++FK506 treatment. If the binding signals were located between two divergently transcribed ORFs, both ORFs were regarded as possible candidates. This manual analysis resulted in the identification of 346 genes evenly distributed through the genome with no apparent bias (Figure 2, Table S1). Importantly, the 37 genes resulting from the first automated analysis were captured in the set of 346. Manual analysis produced more putative targets than the automated because SignalMap reports the probe ID with the highest signal intensity. In most cases, a single binding site did not share the identical probe as having the highest signal between biological replicates, thereby disqualifying them from the automated analysis. Data was deposited in the Gene Expression Omnibus (GEO) at NCBI under the accession number of GSE18180 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=dlarvwwmsoquoxs&acc=GSE18080).
Fig. 2. Genome-wide distribution of putative MoCRZ1 targets. 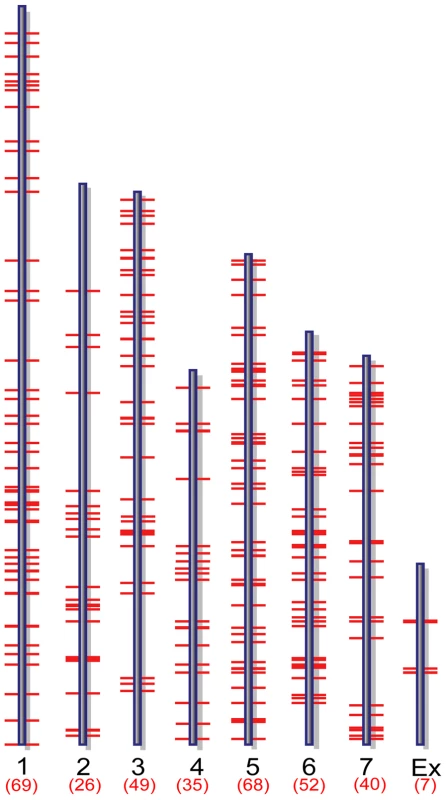
Target genes mapped to M. oryzae supercontigs version 5. Microarray validation of ChIP-chip results
Expression microarray analysis was conducted to corroborate genes predicted by ChIP-chip to be regulated by MoCRZ1. The experimental design is described in Figure 3A. Wild type strain KJ201 was treated with CaCl2 without or with FK506 to identify calcium and calcineurin dependent genes, respectively. MoCRZ1 deletion mutant (Δmocrz1) was also treated with CaCl2 to identify MoCRZ1 regulated genes. Four biological replicates for each of the 4 treatments were selected for hybridization to the Agilent M. oryzae whole genome microarray chip version 2. Signal intensities from the single channel hybridization were normalized to the average expression level of all probes among the 16 data sets. Pair wise comparison between treatments was conducted as depicted in Figure 3A, in which (a), compared Ca2+ treated/no treatment in wild type strain KJ201 (CA/CK); (b), Ca2+ treated/Ca2++FK506 treated in KJ201 (CA/CAFK); (c), Ca2+ treatments in KJ201/Ca2+ treatments in Δmocrz1 (CA/CRZ). Genes were regarded as differentially expressed if their average signal intensity among 4 replicates was above 20 in a minimum of one condition and the expression ratio is greater than 2 fold with P<0.05 (Student's t-test). Changes in gene expression of the 346 genes identified from ChIP-chip were analyzed. Of the 346, we found 309 with expression in each condition, with 121 and 19 genes up - or down - regulated, respectively, in the Ca2+ treated KJ201 condition in at least one comparison (Table S1, Figure 3B). It was noteworthy that the expression level of some MoCRZ1 target genes was lower in the Ca2+ activated wild type condition than in calcineurin and/or MoCRZ1 defective conditions suggesting that MoCRZ1 can act as repressor. These 140 (121+19) genes represent those directly bound to and regulated by MoCRZ1, and form the set used for analyses described below. The full dataset was deposited in NCBI GEO with the accession number of GSE18185 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=ttmdlgumsmsoury&acc=GSE18185). SuperSeries GSE18193 combining the ChIP-chip and microarray data were also generated (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=tnutnsemyikamlm&acc=GSE18193).
Fig. 3. Expression dynamics of MoCRZ1 targets. 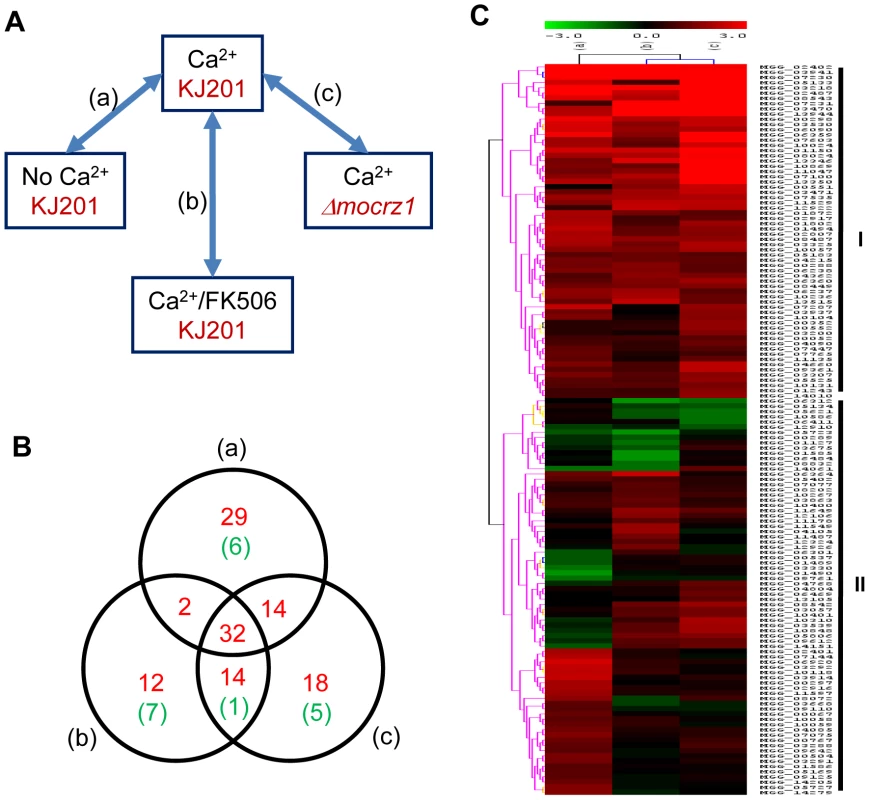
(A) Expression microarray design. Wild type strain KJ201 and MoCRZ1 deletion mutant (Δmocrz1) were treated with Ca2+ and/or FK506 as depicted. Agilent M. oryzae whole genome microarray chip ver. 2 was hybridized in a single channel design with four biological replications per treatment. After global normalization of signal intensities to the average expression level of all probes among the 16 data sets, pairwise comparison between treatments was conducted. (B) Venn diagram showing number of genes identified from ChIP-chip and up-regulated in transcriptome profiling described in panel A. Number of genes with more than 2 fold differential expression with P<0.05 were noted as red for up-regulation and green for down-regulation in Ca2+ treated wild type samples in each comparison. (C) Hierachical clustering resulted in two large groups with differential expression. Validation of gene expression by real-time RT-PCR
The 15 most up-regulated genes in calcium treated wild type strain KJ201compared to that of the Δmocrz1 mutant were selected for real-time RT-PCR to validate expression data. The results in Figure 4 show that each gene is transcriptionally more activated in all three comparison, i.e., Ca2+ treated vs. untreated control (a), Ca2+ treated vs. Ca2+ + FK506 treatment (b), and Ca2+ treated wild type vs. Δmocrz1 (c). Although the magnitude of fold changes was much higher than that from microarray in most cases, real-time RT-PCR data supported the microarray results.
Fig. 4. Real-time RT-PCR to validate differential expression. 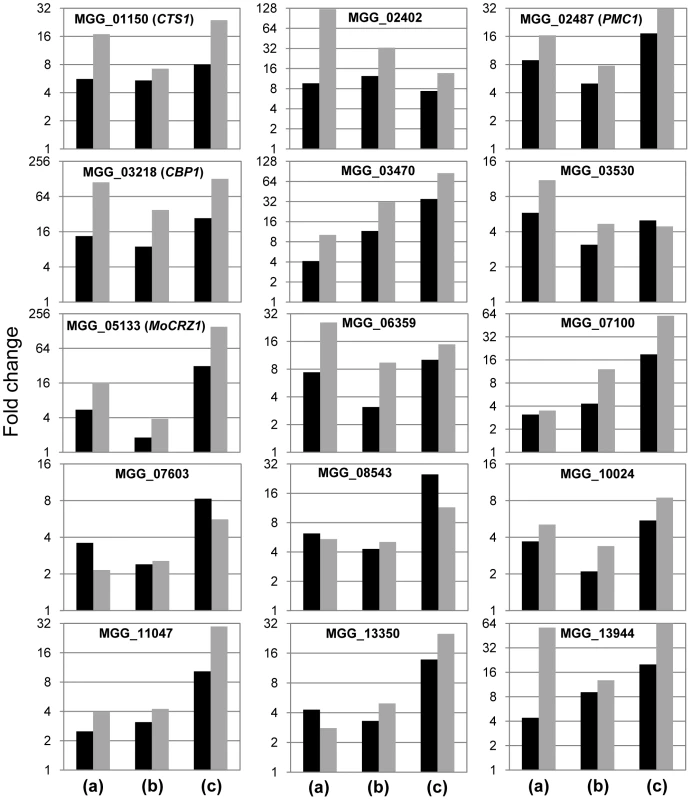
cDNA relevant to 25 ng total RNA was used to run real-time RT-PCR. Log2(Fold changes) calculated by 2−ΔΔCt were displayed. ΔΔCt = (Ctgene of interest-Ctcontrol gene)test condition-(Ctgene of interest-Ctcontrol gene)control condition. Black bar represents signal ratio from microarray data, while grey bar shows fold changes from real-time RT-PCR. Fold changes were calculated by normalizing Ct values as in Figure 3A, where (a) compares expression level between Ca2+ treated vs. no treatment in wild type strain KJ201; (b), Ca2+ vs. Ca2++FK506 in KJ201; (c), Ca2+ treatments in KJ201 vs. in Δmocrz1. Identification of novel and known MoCRZ1 regulated genes
Functional categorization was conducted in two ways. At first, hierarchical classification according to the expression pattern resulted in two groups. This analysis was followed by GO annotation using an InterPro to GO module incorporated in the Comparative Functional Genomics Platform (Figure 3C and Table S2, S3) [31]. Group I contains 64 genes of which expression was tightly regulated in a Ca2+/calcineurin/MoCRZ1 dependent fasion, i.e., up-regulated in all three comparisons. Group II comprises 76 genes whose expression was differentially regulated in three comparisons. Twenty four genes of group I and 36 of group II were assigned with GO terms, 14 and 22 to biological process, 20 and 33 to molecular function, 10 and 19 to cellular component, respectively (Table S2). Second, genes were annotated through literature with their specific functions assigned according to the functional classes of Cyert [32] (Table 1). Sixty-two of 140 genes identified by both ChIP-chip and microarray analyses could be assigned to one of 7 functional groups (Table 1). Consistent with the role of MoCRZ1 in providing tolerance to ionic and cell wall stress, genes involved in small molecule transport or ion homeostasis and cell wall synthesis/maintenance were highly represented. Among them was PMC1, which provides support for the validity of these results. Furthermore, the AAA family of ATPase as a whole was highly represented as direct targets of MoCRZ1, as well as members of major facilitator superfamily of multidrug-resistance proteins. Considering cell wall synthesis/maintenance genes, a number of GPI-anchored cell surface glycoproteins were captured in addition to the previously known downstream genes like chitin synthase activator (Chs3) and chitin syntase 1. Small secretory proteins, including effectors and cell wall degrading proteins, are regarded as key molecules acting at the interface between the plant and microbe. Efficient secretion of these proteins is assumed to be essential during the interaction between host and pathogen. Among the targets identified were genes comprising the vesicle mediated secretory pathway, including rhomboid family membrane protein (MGG_07535), Sso1/2 type SNARE protein (MGG_04090) known to be localized at secretory vesicles from Golgi to plasmamembrane, homocysteine S-methyltransferase (MGG_04215), golgi apyrase (MGG_07077), and a protein required for assembly of ER-to-Golgi SNARE complex (MGG_01489). Proper protein folding in the ER mediated by co-chaperone LHS1 [33], and efficient Golgi performance involving exocytosis entailing functions of the integral membrane P-type ATPase encoded by MgAPT2 [34], have been reported to be necessary for protein secretion and biotrophic phase infection in this fungus.
Tab. 1. Functional classification of MoCRZ1 direct targets. 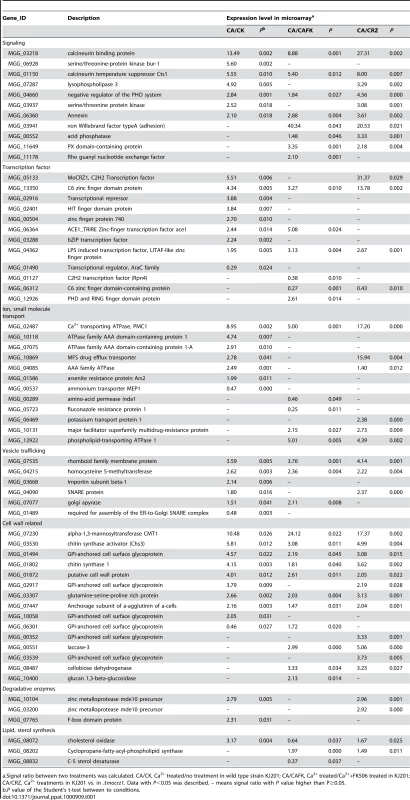
a.Signal ratio between two treatments was calculated. CA/CK, Ca2+ treated/no treatment in wild type strain KJ201; CA/CAFK, Ca2+ treated/Ca2++FK506 treated in KJ201; CA/CRZ, Ca2+ treatments in KJ201 vs. in Δmocrz1. Data with P<0.05 was described. – means signal ratio with P value higher than P≥0.05. A major group of genes found in this study to be regulated by MoCRZ1 are those involved in cellular signaling and transcription. Among them were genes encoding serine/threonine protein kinases (MGG_04660, MGG_07287, MGG_06928), a phosphatase (MGG_00552), and a Rho guanyl nucleotide exchange factor (MGG_11178). In addition, genes comprising calcium signaling machinery were also common. Genes encoding annexin (MGG_06360), lysophospholipase 3 (MGG_07287), PX domain-containing protein (MGG_11649), calcineurin binding protein (CBP1; MGG_03218) and the calcineurin temperature suppressor CTS1 (MGG_01150) were identified. Of note, the expression of CBP1 and CTS1 was highly increased in all three comparisons. Binding in the promoter of these two genes and regulation of their transcription strongly suggests a previously uncharacterized level of negative feedback regulating calcineurin activity. MoCRZ1 functions to activate 12 genes from diverse families of transcription factors. Significantly, MoCRZ1 itself was identified, suggesting an autoregulatory role not previously reported. The expression of MoCRZ1 was induced by exogenous calcium treatment, but not altered by the inhibition of calcineurin activity with FK506. Regarding the fact that calcineurin dephosphorylates MoCRZ1 upon activation by calcium and that FK506 blocks nuclear localization of MoCRZ1, inactivation of calcineurin regulates function only at post-translational level. This data suggests an additional level of regulation at transcription.
Classification of pathogenicity related genes regulated by MoCRZ1
Δmocrz1 mutants are defective in post appressoria formation penetration and establishment of biotrophic host association. Appressoria from this mutant background have defects in penetration, however those that successfully penetrated fail to incite disease. We examined MoCRZ1 target genes for previously defined roles in fungal virulence. Of the 140 target genes queried to pathogen-host interactions database (PHI-base) version 3.1 [35], [36], 16 had matches to more than one entry from plant or human pathogens using a stringent cut-off value e<−20 (Table 2). Three proteins, MoCRZ1 itself [5], MGG_03530 encoding chitin syntase activator Chs3 [37] and PMC1 [8] were previously characterized to regulate virulence in M. oryzae. Similarly, members of ATPases family, PDE1 [38] and MgAPT2 [34], are known to be involved in M. oryzae pathogenicity. Two genes encoding serine/threonine protein kinases (MGG_04660 and MGG_06928) had 45 and 27 hits, respectively. Genes involved in drug resistance (MGG_05723 and MGG_10869), MGG_07230 alpha-1,3-mannosyltransferase CMT1, MGG_07287 lysophospholipase 3, MGG_05727 ankyrin repeat protein, MGG_03288 bZIP transcription factor, and MGG_09361 homolog of CgDN3 were similarly listed as being involved in fungal virulence.
Tab. 2. Genes implicated in fungal virulence. 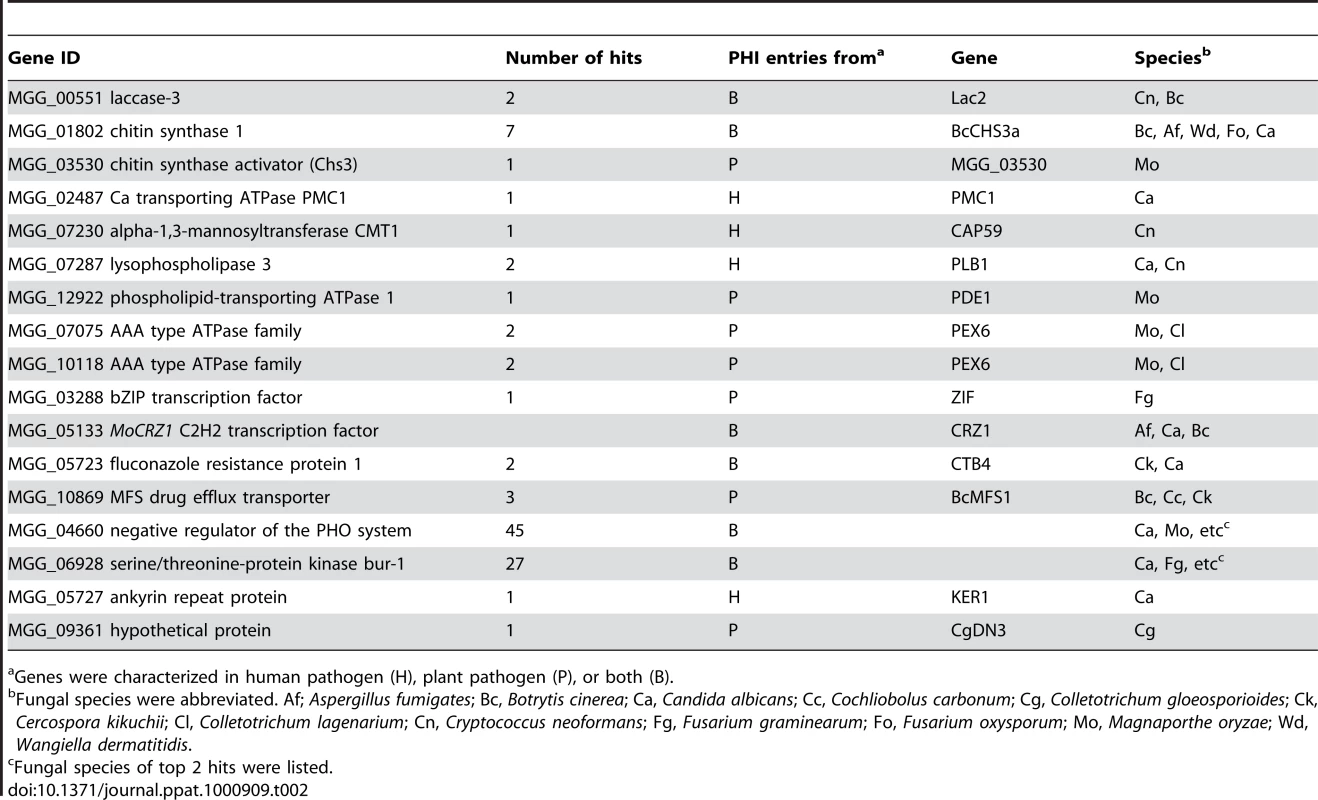
Genes were characterized in human pathogen (H), plant pathogen (P), or both (B). Analysis of the MoCRZ1 binding motif
To identify the MoCRZ1 binding motif, the exact binding sequences of MoCRZ1 (∼50–1247 bp) revealed from ChIP-chip studies were retrieved from the promoters of genes in common between ChIP-chip and microarray expression studies and subjected to motif signature analyses (Figure 5A). Initially, 106 sequences from each of the 83 genes differentially regulated in the WT/Δmocrz1 comparison (Figure 3B) were analyzed with MEME [39] and MDScan [40] (Figure 5A). There were more sequences than genes as 21 genes had 2 peaks in their promoter region and 2 genes had 3. Candidate motifs from both algorithms were manually interrogated and enumerated to identify the top 3 candidates, which were subsequently screened against randomly retrieved 106 intergenic sequences with an average length of 509 bp (Figure 5A). The most enriched motif of CAC[AT]GCC was identified in 33 sequences in front of 24 genes, a 16X enrichment in MoCRZ1 bound sequences. The most common motif of TTGNTTG was found in 68 promoter sequences in front of 42 genes with 4X enrichment. Motif TAC[AC]GTA occurred in 22 promoter sequences of 18 genes with 4X enrichment. Fifty-six genes had at least one motif, while all three of these motifs occurred in front of 5 genes including PMC1, CTS1, MGG_01494 encoding a cell wall protein, and two genes encoding conserved hypothetical proteins MGG_03539 and MGG_06359 (Table S4). These motifs were searched against yeast motif database using TOMTOM [41]. The top match for CAC[AT]GCC was MET28 (p-value = 0.0013), while the second match was CRZ1 with significant p-value (0.0022), showing Crz1p of S. cerevisiae has this motif in its promoters although it was not previously identified as a calcineurin dependent response element (CDRE) (Figure 5B). Pbx1b (p-value = 0.00039) and Zec (p-value = 0.00045) were best two matches for TTGNTTG, while no significant match was returned for TAC[AC]GTA. Binding of MoCRZ1 to the promoter region was confirmed by Electrophoretic Mobility Shift Assay (EMSA). A 209 bp PCR fragment having 1 TTGNTTG and 2 CAC[AT]GCC motifs from the MoCRZ1 promoter region was bound to purified MoCRZ1 protein (Figure 5C, left panel). A 325 bp fragment of CBP1 (MGG_03218) having 1 TTGNTTG and 1 CAC[AT]GCC motifs was also shown to bind to purified MoCRZ1 (Figure 5C, right panel).
Fig. 5. Putative MoCRZ1 binding motifs identified. 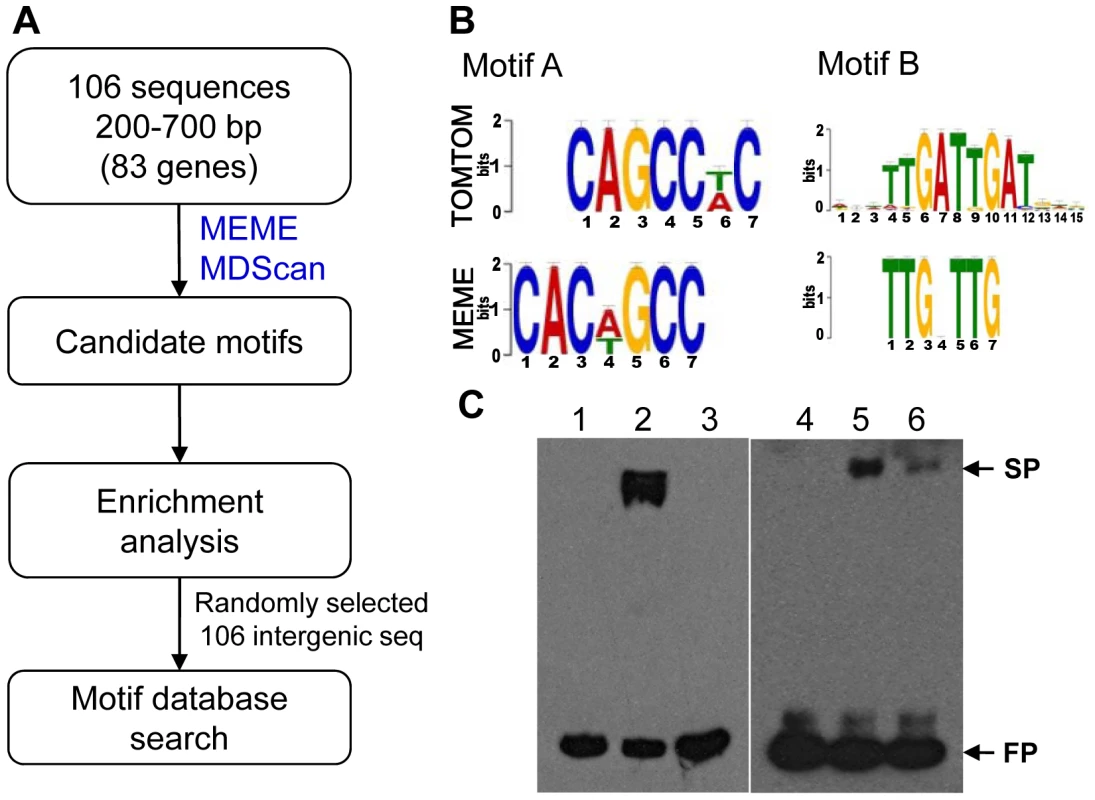
(A) Analysis schema to identify putative MoCRZ1 binding motifs. (B) WebLogo of Top 2 motifs and the best hits in the yeast motif database. The consensus sequences of the putative motif sequences (MEME) calculated using WebLogo server were displayed in order of predominance from top to bottom at each position with large letters being higher frequency. Best hit from yeast motif database was displayed for comparison (TOMTOM). (C) Electrophoretic mobility shift assay. Probe DNA was amplified by PCR from the promoter regions of MoCRZ1 (left panel, lanes 1 to 3) and CBP1 (right panel, lanes 4 to 6) with 5′-Biotin-labeled primer pairs, purified by gel isolation, and allowed to bind to purified MoCRZ1 protein. Lanes 1 and 4, Biotin-labeled probe DNA; lanes 2 and 5, probe DNA with purified MoCRZ1 protein; lanes 3 and 6, competition reaction with 200 fold molar excess of unlabeded DNA. FP, free probe; SP, shifted probe. Reaction mixture was run on 5% polyacrylamide gel in 0.5X TBE and transferred onto Hybond N+ (GE Healthcare) membrane. Signals were detected using chemiluminescence. Discussion
The most common signal transduction pathways in nature, MAPK, cAMP, and calcium mediated, have been shown to be involved in all aspects of growth, development, and pathogenicity of M. oryzae and several other fungal pathogens of plants. Recently, a transcription factor, MoCRZ1, relaying calcium signals from calcineurin has been characterized [5], [42]. As a major mediator of calcineurin signaling, crz1 deletion mutants in a variety of fungi showed similar phenotypes as calcineurin mutants such as sensitivity to Ca2+ and other ionic stresses as well as cell wall stresses. In addition, Δmocrz1 showed defects in development and pathogenicity, including reduced conidiation and defective appressorium mediated penetration [5], [42]. MoCRZ1 is required for the calcineurin-dependent transcriptional induction of downstream genes such as PMC1, encoding P-type ATPase, CHS2 and CHS4, encoding chitin synthase. In this study, we gained a genome-wide perspective of the direct downstream targets of MoCRZ1 signaling using a combined approach of ChIP-chip and expression microarray analysis. CRZ1 in filamentous fungi have common as well as unique roles compared to those of their yeast ortholog. As such, we find that the suite of genes regulated by MoCRZ1 contains a high percentage specific to M. oryzae.
Novel and expanded findings in the CRZ1 regulatory circuitry
Our combined approach identified 140 direct targets of MoCRZ1 whose expression was concurrently regulated in a calcineurin/CRZ1 dependent manner. Sixty-two of these genes could be grouped in the same categories that were used to functionally assign yeast genes [32]. These groups include cell wall synthesis, ion or small molecule transport, vesicle transport, lipid or sterol synthesis, degradative enzymes, and signaling and transcription. This is expected as the Δmocrz1 mutant also exhibited sensitivity to calcium ions and chemicals perturbing cell wall integrity as did the yeast crz1 mutant [5]. However, the large diversity in the suite of individual target genes is compelling. In S. cerevisiae, genome wide expression profiling identified 153 calcium/calcineurin/Crz1p dependent genes [30]. Comparison between these two sets showed that only 15 out of 140 MoCRZ1 targets had 12 yeast orthologs whose expression was regulated by calcium/calcineurin/Crz1p (Table 3). When we compared our gene list to the 120 A. fumigatus genes whose expression was changed by exposure to calcium for 10 min [26], only 21 matched to 14 A. fumigatus genes, with only 6 having reciprocal best blast hits (Table 3). When the same analysis was applied to the 141 A. fumigatus genes whose expression was modulated by AfcrzA, as recently reported by Soriani et al. [43], 28 MoCRZ1 targets having 15 A. fumigatus orthologs were found with only 3 matching the 14 genes identified previously (data not shown). The observed diversity may reflect divergently evolved molecular features modulated by CRZ1 in each species to cope with its unique environment. This diversity was also reflected in the crz1 mutant phenotypes across the species. For example, crz1 mutants in different species showed a spectrum of ion sensitivity; ΔcrzA of A. fumigatus was more sensitive to Mn2+, but Δmocrz1 of M. oryzae was not. BcCRZ1 of B. cinerea was dispensable for conidia mediated infection, but MoCRZ1 was necessary. These data suggest that although the core calcium signaling machinery including calmodulin and calcineurin is highly conserved across the species, their mechanism of action has diverged. Similar suggestions have been proposed by Karababa et al. [28]. Kraus and Heitman [44] also made note of species-specific action mechanisms through which calcium signaling involving calmodulin and calcineurin was manifested in three different species, S. cerevisiae, C. albicans, and C. neoformans. Additional evidence supporting this hypothesis is found in the differences in CDRE (calcineurin-dependent response elements) sequences [27], [30], [45], [46]. In S. cerevisiae, nucleotide stretches of 5′-GNGGC(G/T)CA-3′ was reported as the Crz1p-binding site by in vitro site selection. A similar sequence, 5′-GAGGCTG-3′, was also identified as a common motif in the upstream 500 bp regions of 40 genes with ≥4.0 fold Crz1-dependent expression profile [30]. Two similar sequences, 5′-GTGGCTC-3′ and 5′-GAGGCTC-3′, were reported as CDREs from the genus Aspergillus in the A. nidulans chsB and A. giganteus afp promoters [47]. A slightly divergent motif, 5′-G(C/T)GGT-3′, was identified as a common regulatory sequence from the 60 upstream regions of calcineurin/Crz1p-dependent genes of C. albicans [28]. In contrast, the motif sequences obtained in this study shows further divergence from that of S. cerevisiae or C. albicans. Although, core hepta nucleotide, 5′-GGCTC-3′, was found in the probe sequence of 10 genes including PMC1 (MGG_02487) and MGG_03530 (chitin synthase activator), the occurrence of the full S. cerevisiae or C. albicans motif sequences is not enriched/overrepresented among the 106 sequences we analyzed (data not shown).
Tab. 3. List of common genes regulated by calcium/calcineurin/CRZ1 across species. 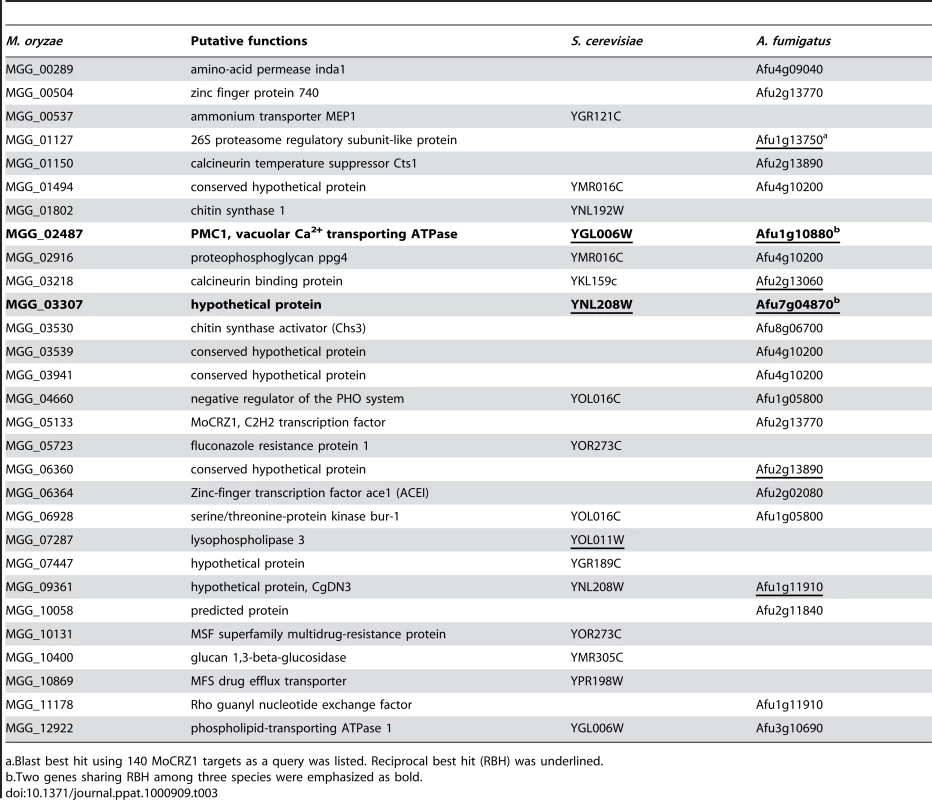
a.Blast best hit using 140 MoCRZ1 targets as a query was listed. Reciprocal best hit (RBH) was underlined. Feedback regulation of the calcium signaling network by CRZ1
In addition to PMC1, several other genes implicated in the calcium signaling pathway regulating calcineurin activity were identified. Among them were calcineurin binding protein CBP1 (MGG_03218) and the calcineurin temperature suppressor CTS1 (MGG_01150), suggesting feedback regulation of calcineurin activity mediated by MoCRZ1. In addition, MoCRZ1 bound its own promoter to activate expression. CBP1 shows homology to CbpA in A. fumigatus and Cbp1 of C. neoformans, which in turn, are orthologous to RCN1 of S. cerevisiae, an inhibitor of calcineurin called calcipressin. Over-expression of RCN1 in a pmc1 mutant background conferred Ca2+ tolerance by activation of vacuolar Ca2+/H+ exchanger Vcx1p, expression of which was negatively regulated by calcineurin. Expression of RCN1 (YKL159c) was regulated by calcineurin and Crz1p, suggesting negative feedback regulation [30], [48]. However, both stimulatory and inhibitory regulation of calcineurin by RCN1 was reported [49]. RCN1 expression at endogenous level and phosphorylation by GSK-3 kinase, positively regulated calcineurin activity [49]. Degradation of phosphorylated RCN1 is required for precise calcineurin activity in response to changes in Ca2+ concentration [50]. Negative feedback regulation of calcineurin by CbpA was also reported in A. fumigatus, where it down regulates cnaA expression as well as that of downstream genes vcxA and chsA encoding vacuolar Ca2+/H+ exchanger and chitin synthase A, respectively [51]. Expression of CbpA (Afu2g13060) was known to be up-regulated in response to Ca2+, which was CrzA dependent [26]. This feedback loop seems to extend at least to the level of calcineurin. Roles and action mechanisms of Cbp1 in the calcium/calcineurin signaling pathway also seems to be diverged in a species specific manner. In C. neoformans, Cbp1 does not stimulate or inhibit calcineurin expression, and does not seem to participate in a feedback loop. Taken together, these data lead us to propose that a negative or positive feedback loop, which includes MGG_03218, regulates the calcium/calcineurin signal transduction pathway in M. oryzae.
Phospholipid-binding protein Cts1 (calcineurin temperature suppressor) was identified and characterized in C. neoformans as able to restore growth defect at 37°C in calcineurin-deficient strains and to confer resistance to the calcineurin inhibitor FK506 [52]. Δcts1 mutants were synthetically lethal in combination with a calcineurin mutation. However, no direct interaction between Cts1 with either the catalytic or regulatory subunit of calcineurin was reported. With these data, they suggested that Cts1 acted in either parallel pathways or a branched pathway to compensate, at least in part, for the loss of calcineurin function [52]. MGG_01150, an ortholog of Cts1, was found to be a direct target of MoCRZ1. Its calcineurin dependent expression pattern is opposite to that of Cts1. Unlike the elevated expression of Cts1in calcineurin deficient strains, MGG_01150 expression was activated by Ca2+ treatment, which was blocked by calcineurin inhibitor FK506 and abolished in the Δmocrz1 mutant. Therefore, it is evident that MGG_01150 is a component of the calcineurin signaling pathway in M. oryzae unlike its counterpart in C. neoformans.
Our data revealed that MoCRZ1 binds to its own promoter to activate expression in a Ca2+ /calcineurin dependent manner. Therefore, MoCRZ1 regulation appears to occur at the posttranslational and transcriptional levels via activation by calcineurin and positive autoregulation respectively. Calcineurin/CRZ1 dependent expression of CRZ1 was also reported in C. albicans suggesting a common mechanism of regulation across the fungal species [28]. However, expression dynamics of the catalytic (MCNA: MGG_07456) and regulatory (MCNB: MGG_06933) subunit were not altered in the Δmocrz1 mutant compared to wild type in response to Ca2+ treatment (data not shown). This suggests that the proposed feedback regulation does not include direct regulation of calcineurin expression by CRZ1.
Fungal pathogenicity
Involvement of CRZ1 in fungal virulence has been recently demonstrated in both human and plant pathogenic fungi [5], [10], [25], [26], [28], [29]. Signals related to these virulence traits seemed to be transmitted to a diverse range of downstream genes, as 18 genes out of 140 direct targets including MoCRZ1 have been found to be related to fungal virulence in both human and plant pathogens. Gene repertoire ranges from cell wall synthetic enzymes, proteins conferring resistance to antifungal agents encoded by major facilitator type transporter, calcium homeostasis to transcription factors. Three genes (MoCRZ1, MGG_03530 encoding chitin synthase activator 3, and MGG_02487) have been functionally characterized in M. oryzae [5], [8], [37]. Association of MGG_03530 encoding chitin synthase activator 3 (Chs3) with virulence was found in a T-DNA insertion strain with reduced growth rate on nutrient rich media, reduced conidiation rate with aberrant conidia morphology, and reduced appressorium formation and virulence [37]. Filamentous fungal genomes contain up to 10 chitin synthase genes of 7 classes [53]. Different CHS were regarded as to have functional redundancy in a variety of developmental processes because single mutation of a class I or II gene did not result in a marked phenotype. Therefore, specific roles for individual genes have not yet been clearly assigned and the association with fungal virulence has been controversial. However, several lines of evidence implicate class V myosin-like CHS in virulence in the maize anthracnose pathogen Colletotrichum graminearum [54] and in Fusarium oxysporum, the tomato wilt fungus [55]. One Class III chitinases, BcChs3a of Botrytis cinerea had important roles in virulence, especially on leaf tissue colonization, grape vines and Arabidopsis thaliana [56]. The M. oryzae genome contains 7 predicted chitin synthases, of which the expression of 5 were induced and 1 repressed in response to exogenous calcium treatment (data not shown). Only one (MGG_01802 encoding class II chitinase) was directly regulated by MoCRZ1. Therefore, calcium seems to regulate expression of most chitin synthases in diverse pathways.
Two genes encoding small molecule transporting P-type ATPases (MGG_02487 Ca transporting ATPase and MGG_12922 phospholipid-trasnporting ATPase) were found to have homologs implicated in fungal pathogenicity. Knock-down of MGG_02487 encoding PMC1 by RNAi technology resulted in no conidiation, growth retardation, and reduced melanization [8]. The association of PMC1 and fungal virulence was not investigated in other fungi. Other evidence on the involvement of Ca2+ transporting ATPase in fungal virulence arises from the study of PMR1 of C. albicans [57]. PMR1 is a Golgi membrane located Ca2+ transporting P-type ATPase, and is known to work cooperatively with PMC1 in the maintenance of cytosolic Ca2+ homeostasis. Capmr1Δ mutant of C. albicans had a weakened cell wall, probably due to the glycosylation defect and showed severely attenuated virulence in a murine model of systemic infection [57]. Two genes encoding Drs2 family of P-type ATPases, PDE1 and MgAPT2, were functionally characterized to act in appressorium formation and invasive growth [34], [38]. MgAPT2 was necessary for the normal development of Golgi apparatus that is required for secretion of a subset of extracellular enzymes via exocytosis [34], [58].
This study is the first of its kind where ChIP-chip technology has been applied to filamentous fungi. The correlation of comprehensive whole genome expression data with results from ChIP-chip have allowed for significant refinement of the predicted targets of MoCRZ1. This refinement alone allowed for the identification of a predicted signature binding motif for this transcription factor. This study reveals conserved elements of the calcium/calcineurin signaling pathway, as well as elucidates species specific differences that we propose function to regulate the system and allow for responses tailored to biology of the organism. Calcium signaling is a ubiquitous and complicated aspect of cell physiology. This study represents a major advance in our understanding of this pathway in M. oryzae and provides the launching point for the functional characterization of the genes and interactions it implicates. Figure 6 depicts our proposed model resulting from this work and includes our new findings of predictive roles for MoCRZ1 in autoregulation, feedback inhibition, and secretion.
Fig. 6. Proposed model of MoCRZ1 regulation of downstream genes. 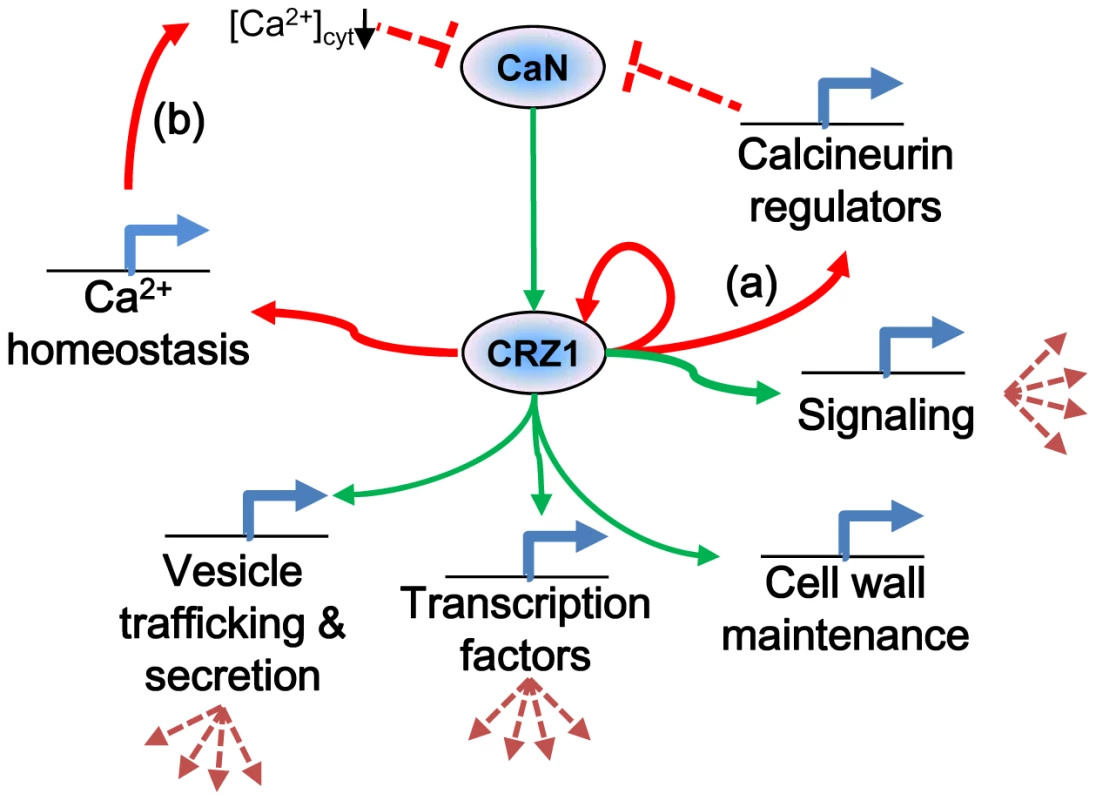
Consensus model of calcineurin/CRZ-1 signaling with green arrows indicating feedforward regulation and red arrows feedback regulation. Regulation of calcineurin occurs in multiple layers. (a) direct feedback by induction of expression of calcineurin regulators CBP1 or CTS1. (b) indirect feedback by induction of PMC1 leading to sequestration of cytosolic Ca2+ ions to inactivate calcineurin. Materials and Methods
Fungal cultures and chemical treatment
Strains of M. oryzae were maintained on oatmeal agar (50 g of oatmeal per liter) and grown at 22 °C under constant fluorescent light to promote conidiation. Mycelial blocks from actively growing margins of colony were inoculated into complete media (CM) liquid media at 25 °C by shaking for 3 days. After thorough washing with sterile distilled water, the mycelia were treated with 200 mM CaCl2 with or without 10 µg/ml FK506 for the indicated time. Mycelia were harvested and immediately frozen with liquid nitrogen and stored at −80°C before use. FK506 (Sigma, St. Louis, MO) stock solution in 5 mg/ml was prepared with DMSO, and stored at −20°C until used.
Fungal transformation and microscopy
Protoplasts generation and transformation were performed following established protocols [59]. Protoplasts were generated from young mycelia grown in complete media with 10 mg/ml Lysing Enzyme (Sigma, St. Louis, MO) in 20% sucrose. Protoplasts were harvested by filtration through 4 layers of miracloth (Calbiochem, Darmstadt, Germany), washed twice with STC (20% sucrose, 50 mM Tris-HCl, 50 mM CaCl2, pH 8.0) followed by centrifugation at 5,000 rpm for 15 min at 4°C, and resuspended to 5×107 protoplasts/ml. GFP tagging construct in TOPO cloning vector (PMoCRZ1::MoCRZ1::GFP) was co-transformed with pCX63 containing hygromycin resistance cassette by the mediation of 40% polyethyleneglycol. After incubation for 7–10 days at 25°C, hygromycin resistant colonies were transferred to V8 juice agar media. The fluorescing transformants observed under the microscope with epifluorescent optics (Nikon eclipse 80i, Melville, NY) were purified through single spore isolation. Nuclear translocalization of MoCRZ1 was observed after treatment with 200 mM CaCl2 with or without 10 µg/ml FK506 for 1 hour at room temperature.
RNA isolation and real-time RT-PCR
Total RNA was isolated from frozen mycelial powder using an Easy-Spin RNA extraction kit (iNtRON Biotechnology, Seoul, Korea). Five micrograms of total RNA was reverse-transcribed into first-strand cDNA by oligo dT priming using the SuperScript first-strand cDNA synthesis kit according to the manufacturer's instructions (Invitrogen Life Technologies, Carlsbad, CA). Resulting cDNA was diluted to 1∶20 with sterile water. Real-time RT-PCR was performed according to the established protocol [59] using iQ SYBR Green Supermix (Bio-rad, Hercules, CA) on an iCycler iQ5 Real-Time PCR Detection System (Bio-rad). Fold changes were calculated by 2−ΔΔCt, where ΔΔCt = (Ctgene of interest-Ctcontrol gene)test condition-(Ctgene of interest-Ctcontrol gene)control condition. Test and control conditions are as same as in Figure 3A, where (a) compares expression level between Ca2+ treated vs. no treatment in wild type strain KJ201, (b) Ca2+ vs. Ca2++FK506 in KJ201, (c) Ca2+ treatments in KJ201 vs. in Δmocrz1. Primer sequences were listed in Table S5.
Chromatin immunoprecipitation-chip and analysis
Young mycelia grown in liquid media were treated with 200 mM CaCl2 with or without 10 µg/ml FK506 for 1 hour with shaking. The harvested mycelia were divided with one part being immediately frozen in liquid nitrogen for future RNA isolation and the other treated with 1% formaldehyde in buffer A (0.4 M sucrose, 10 mM Tris-HCl, pH 8.0, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, and 1% formaldehyde) for cross-linking for 20 min. Mycelia were harvested with excess amount of distilled water after stopping cross-link with 0.1 M glycine for 10 min, frozen in liquid nitrogen, ground into a fine powder in a chilled mortar and pestle, and stored at −80°C until used. Chromatin immunoprecipitation was conducted according to published procedures with modification [60], [61]. Nuclear DNA was then isolated from cross-linked mycelia with Plant Nuclear Isolation Kit (Sigma, St. Louis, MO) and sheared into fragments by sonication to 200 - to 1,000-bp with an average size of 500 bp. Sonication was conducted on ice with an amplitude of 10% using 30×30 s pulses (30 s between bursts) using Biorupter (Cosmo Bio, Tokyo, Japan). After pre-clearing nuclear lysates with Salmon sperm/protein A agarose (Upstate, Temecula, CA) for 4 hours at 4°C, immunoprecipitations were performed with either 1 µg of rabbit control IgG (ab46540-1, Abcam, Cambridge, MA) or 0.5 µl of antiGFP antibody (ab290, Abcam) for overnight at 4°C. A small aliquot of DNA (30%) was saved for input DNA (input). Immunoprecipitated DNA was captured with proteinA agarose beads (Upstate, Temecula, CA) for 4 hours, and then washed twice with LNDET buffer (0.25 M LiCl, 1% NP40, 1% deoxycholate, 1 mM EDTA) and twice with TE buffer. The DNAs were reverse-cross linked at 65 °C overnight in elution buffer (1% SDS and 0.1 M NaHCO3) containing 1 mg/ml proteinase K, and purified using PCR purification kit (Qiagen). Real-time PCR was performed with 1 µl each of pulled-down DNA and input DNA as template following the procedures described above. Fold changes for control gene (β-tubulin) and putative target gene (PMC1) were calculated by 2−ΔΔCt, where ΔΔCt = (Ctinput DNA-CtChIPed DNA) Ca2+ treated sample - (Ctinput DNA-CtChIPed DNA) Ca2+/FK506 treated sample. Primer sequences for the promoter region of PMC1 and β-tubulin were listed in Table S5. For ChIP-chip experiments, 10 µl ChIPed DNA and 10 ng input DNA were amplified using GenomePlex Whole Genome Amplification Kit (Sigma). Amplified DNA was then labeled with Cy3 or Cy5 fluorescent dyes for input or immunoprecipitated DNA, respectively, and hybridized to NimbleGen Magnaporthe grisea promoter tiling arrays according to the manufacturer's instruction (NimbleGen Systems of Iceland). Probes for tiling array were designed to have about 70 nucleotides per 100 bp of promoter and intergenic region based on annotation of M. grisea genome version 5. After getting peak intensity, peak data files (.gff) were generated from the scaled log2-ratio data using NimbleScan. It detects peaks by searching for 4 or more probes whose signals are above the cutoff values using a 500 bp sliding window. The ratio data was then randomized 20 times to evaluate the probability of “false positives”. Each peak was then assigned a false discovery rate (FDR) score based on the randomization. Peaks with FDR score ≤0.2 were regarded as positive.
Expression microarray and analysis
Mycelia of wild type KJ201 and Δmocrz1 strain were treated with 200 mM CaCl2 with or without 10 µg/ml FK506, with water as control. Initially, samples were harvested at 0, 15, 30, and 60 min. after treatment. PMC1 expression level was checked by RT-PCR with the highest expression at the 30 min. time point. Four biological replicates of wild type and mutant mycelia were harvested after 30 min. treatment with chemicals, frozen immediately with liquid nitrogen. Total RNA was isolated described as above. After validation of sample quality by RT-PCR, total RNA was sent to Cogenics (Morrisville, NC) for hybridization to the Agilent M. grisea whole genome microarray chip version 2.0 using the single channel hybridization design. Quality of RNA was determined with Agilent Bioanalyzer. Five hundred nanograms of total RNA was converted into labeled cRNA with nucleotides coupled to fluorescent dye Cy3 using the Quick Amp Kit following the manufacturer's instructions (Agilent Technologies, Palo Alto, CA). After analyzing the quality with Agilent Bioanalyzer, Cy3-labeled cRNA (1.65 µg) was hybridized to M. grisea 2.0 4×44 k microarrays. The hybridized array was washed and scanned, and the data were extracted from the scanned image using Feature Extraction version 10.2 (Agilent Technologies). An error-weighted average signal intensity of two probes within a chip was used for normalization with Lowess normalization module implanted in JMP Genomics software. An average expression of all probes among 16 data sets was used as the baseline. Pairwise comparison between treatments was conducted to get the expression profiles of each probe. Genes were regarded as differentially expressed if their average signal intensity among 4 replicates was above 20 in a minimum of one condition and expression ratio is greater than 2 fold with P<0.05 (Student's t-test).
Motif analysis
The two commonly used motif discovery programs, MEME [39] and MDScan [40], were used to identify the MoCRZ1 binding motif. Input data consisted of the exact binding sequences retrieved from the promoters of 83 genes with differential expression in the WT/Δmocrz1 comparison (Figure 3A). Candidate motifs from both algorithms were manually interrogated and enumerated to identify the 3 top candidates, and queried against the yeast motif database using TOMTOM [41]. Enrichment was calculated over the 106 background sequence set which was randomly retrieved from intergenic region of the whole genome. Consensus motif sequences were calculated using WebLogo server at http://weblogo.berkeley.edu [62].
Protein expression and electrophoretic mobility shift assay
Protein expression vector was constructed by ligation of MoCRZ1 cDNA encompassing full ORF into pGEX-6P-1 (Invitrogen, Carlsbad, CA) having GST tag at the N terminus. The resulting construct was transformed into the E. coli strain BL21 (DE3) pLysS (Novagen) after verifying the cDNA sequences. Induced protein was purified with GST agarose beads (Sigma) based on the procedures of Frangioni et al.[63].
Probe DNA was prepared by PCR with Biotin labeled primer at the 5′ end, followed by gel purification. Cold probe was amplified with the same primer sequence without Biotin labeling. Primer sequences were listed in Table S5. Binding of putative motif sequences to MoCRZ1 protein was performed using LightShift Chemiluminescent EMSA kit following the manufacturer's manual (PIERCE, Rockford, IL). Reaction mixtures containing 10 ng of purified MoCRZ1 and biotin labeled probe were incubated for 20 min. at room temperature. The reactions were electrophoresed on 5% polyacrylamide gel in 0.5×TBE, and transferred to a positively charged nylon membrane (Hybond N+, GE Healthcare). Signals were detected using Chemiluminescent Nucleic Acid Detection Module (PIERCE) according to the manufacturer's instruction.
Supporting Information
Zdroje
1. EbboleDJ
2007 Magnaporthe as a model for understanding host-pathogen interactions. Annu Rev Phytopathol 45 437 456
2. KankanalaP
CzymmekK
ValentB
2007 Roles for rice membrane dynamics and plasmodesmata during biotrophic invasion by the blast fungus. Plant Cell 19 706 724
3. WilsonRA
TalbotNJ
2009 Under pressure: investigating the biology of plant infection by Magnaporthe oryzae. Nat Rev Microbiol 7 185 195
4. XuJR
ZhaoX
DeanRA
2007 From genes to genomes: a new paradigm for studying fungal pathogenesis in Magnaporthe oryzae. Adv Genet 57 175 218
5. ChoiJ
KimY
KimS
ParkJ
LeeYH
2009 MoCRZ1, a gene encoding a calcineurin-responsive transcription factor, regulates fungal growth and pathogenicity of Magnaporthe oryzae. Fungal Genet Biol 46 243 254
6. ChoiJH
KimY
LeeYH
2009 Functional analysis of MCNA, a gene encoding a catalytic subunit of calcineurin, in the rice blast fungus Magnaporthe oryzae. J Microbiol Biotechnol 19 11 16
7. LeeSC
LeeYH
1998 Calcium/calmodulin-dependent signaling for appressorium formation in the plant pathogenic fungus Magnaporthe grisea. Mol Cells 8 698 704
8. NguyenQB
KadotaniN
KasaharaS
TosaY
MayamaS
2008 Systematic functional analysis of calcium-signalling proteins in the genome of the rice-blast fungus, Magnaporthe oryzae, using a high-throughput RNA-silencing system. Mol Microbiol 68 1348 1365
9. RhoHS
JeonJ
LeeYH
2009 Phospholipase C-mediated calcium signalling is required for fungal development and pathogenicity in Magnaporthe oryzae. Mol Plant Pathol 10 337 346
10. SteinbachWJ
ReedyJL
CramerRAJr
PerfectJR
HeitmanJ
2007 Harnessing calcineurin as a novel anti-infective agent against invasive fungal infections. Nat Rev Microbiol 5 418 430
11. CarafoliE
2002 Calcium signaling: a tale for all seasons. Proc Natl Acad Sci U S A 99 1115 1122
12. MiyakawaT
MizunumaM
2007 Physiological roles of calcineurin in Saccharomyces cerevisiae with special emphasis on its roles in G2/M cell-cycle regulation. Biosci Biotechnol Biochem 71 633 645
13. BaderT
BodendorferB
SchroppelK
MorschhauserJ
2003 Calcineurin is essential for virulence in Candida albicans. Infect Immun 71 5344 5354
14. BlankenshipJR
WormleyFL
BoyceMK
SchellWA
FillerSG
2003 Calcineurin is essential for Candida albicans survival in serum and virulence. Eukaryot Cell 2 422 430
15. OdomA
MuirS
LimE
ToffalettiDL
PerfectJ
1997 Calcineurin is required for virulence of Cryptococcus neoformans. EMBO J 16 2576 2589
16. SanglardD
IscherF
MarchettiO
EntenzaJ
BilleJ
2003 Calcineurin A of Candida albicans: involvement in antifungal tolerance, cell morphogenesis and virulence. Mol Microbiol 48 959 976
17. CamposCB
Di BenedetteJP
MoraisFV
OvalleR
NobregaMP
2008 Evidence for the role of calcineurin in morphogenesis and calcium homeostasis during mycelium-to-yeast dimorphism of Paracoccidioides brasiliensis. Eukaryot Cell 7 1856 1864
18. da Silva FerreiraME
HeinekampT
HartlA
BrakhageAA
SemighiniCP
2007 Functional characterization of the Aspergillus fumigatus calcineurin. Fungal Genet Biol 44 219 230
19. SteinbachWJ
CramerRAJr
PerfectBZ
AsfawYG
SauerTC
2006 Calcineurin controls growth, morphology, and pathogenicity in Aspergillus fumigatus. Eukaryot Cell 5 1091 1103
20. KotheGO
FreeSJ
1998 Calcineurin subunit B is required for normal vegetative growth in Neurospora crassa. Fungal Genet Biol 23 248 258
21. ProkischH
YardenO
DiemingerM
TropschugM
BarthelmessIB
1997 Impairment of calcineurin function in Neurospora crassa reveals its essential role in hyphal growth, morphology and maintenance of the apical Ca2+ gradient. Mol Gen Genet 256 104 114
22. RasmussenC
GarenC
BriningS
KincaidRL
MeansRL
1994 The calmodulin-dependent protein phosphatase catalytic subunit (calcineurin A) is an essential gene in Aspergillus nidulans. EMBO J 13 2545 2552
23. JuvvadiPR
KurokiY
AriokaM
NakajimaH
KitamotoK
2003 Functional analysis of the calcineurin-encoding gene cnaA from Aspergillus oryzae: evidence for its putative role in stress adaptation. Arch Microbiol 179 416 422
24. ViaudMC
BalhaderePV
TalbotNJ
2002 A Magnaporthe grisea cyclophilin acts as a virulence determinant during plant infection. Plant Cell 14 917 930
25. SchumacherJ
de LarrinoaIF
TudzynskiB
2008 Calcineurin-responsive zinc finger transcription factor CRZ1 of Botrytis cinerea is required for growth, development, and full virulence on bean plants. Eukaryot Cell 7 584 601
26. SorianiFM
MalavaziI
da Silva FerreiraME
SavoldiM
Von Zeska KressMR
2008 Functional characterization of the Aspergillus fumigatus CRZ1 homologue, CrzA. Mol Microbiol 67 1274 1291
27. StathopoulosAM
CyertMS
1997 Calcineurin acts through the CRZ1/TCN1-encoded transcription factor to regulate gene expression in yeast. Genes Dev 11 3432 3444
28. KarababaM
ValentinoE
PardiniG
CosteAT
BilleJ
2006 CRZ1, a target of the calcineurin pathway in Candida albicans. Mol Microbiol 59 1429 1451
29. CramerRAJr
PerfectBZ
PinchaiN
ParkS
PerlinDS
2008 Calcineurin target CrzA regulates conidial germination, hyphal growth, and pathogenesis of Aspergillus fumigatus. Eukaryot Cell 7 1085 1097
30. YoshimotoH
SaltsmanK
GaschAP
LiHX
OgawaN
2002 Genome-wide analysis of gene expression regulated by the calcineurin/Crz1p signaling pathway in Saccharomyces cerevisiae. J Biol Chem 277 31079 31088
31. ParkJ
ParkB
JungK
JangS
YuK
2008 CFGP: a web-based, comparative fungal genomics platform. Nucleic Acids Res 36 D562 571
32. CyertMS
2003 Calcineurin signaling in Saccharomyces cerevisiae: how yeast go crazy in response to stress. Biochem Biophys Res Commun 311 1143 1150
33. YiM
ChiMH
KhangCH
ParkSY
KangS
2009 The ER chaperone LHS1 is involved in asexual development and rice infection by the blast fungus Magnaporthe oryzae. Plant Cell 21 681 695
34. GilbertMJ
ThorntonCR
WakleyGE
TalbotNJ
2006 A P-type ATPase required for rice blast disease and induction of host resistance. Nature 440 535 539
35. WinnenburgR
BaldwinTK
UrbanM
RawlingsC
KohlerJ
2006 PHI-base: a new database for pathogen host interactions. Nucleic Acids Res 34 D459 464
36. WinnenburgR
UrbanM
BeachamA
BaldwinTK
HollandS
2008 PHI-base update: additions to the pathogen host interaction database. Nucleic Acids Res 36 D572 576
37. JeonJ
ParkSY
ChiMH
ChoiJ
ParkJ
2007 Genome-wide functional analysis of pathogenicity genes in the rice blast fungus. Nat Genet 39 561 565
38. BalhaderePV
TalbotNJ
2001 PDE1 encodes a P-type ATPase involved in appressorium-mediated plant infection by the rice blast fungus Magnaporthe grisea. Plant Cell 13 1987 2004
39. BaileyTL
ElkanC
1994 Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2 28 36
40. LiuXS
BrutlagDL
LiuJS
2002 An algorithm for finding protein-DNA binding sites with applications to chromatin-immunoprecipitation microarray experiments. Nat Biotechnol 20 835 839
41. GuptaS
StamatoyannopoulosJA
BaileyTL
NobleWS
2007 Quantifying similarity between motifs. Genome Biol 8 R24
42. ZhangH
ZhaoQ
LiuK
ZhangZ
WangY
2009 MgCRZ1, a transcription factor of Magnaporthe grisea, controls growth, development and is involved in full virulence. FEMS Microbiol Lett 293 160 169
43. SorianiFM
MalavaziI
SavoldiM
EspesoE
DinamarcoTM
2010 Identification of possible targets of the Aspergillus fumigatus CRZ1 homologue, CrzA. BMC Microbiol 10 12
44. KrausPR
HeitmanJ
2003 Coping with stress: calmodulin and calcineurin in model and pathogenic fungi. Biochem Biophys Res Commun 311 1151 1157
45. KumarKS
Ravi KumarB
SiddavattamD
SubramanyamC
2006 Characterization of calcineurin-dependent response element binding protein and its involvement in copper-metallothionein gene expression in Neurospora. Biochem Biophys Res Commun 345 1010 1013
46. DengL
SugiuraR
TakeuchiM
SuzukiM
EbinaH
2006 Real-time monitoring of calcineurin activity in living cells: evidence for two distinct Ca2+-dependent pathways in fission yeast. Mol Biol Cell 17 4790 4800
47. SpielvogelA
FindonH
ArstHN
Araujo-BazanL
Hernandez-OrtizP
2008 Two zinc finger transcription factors, CrzA and SltA, are involved in cation homoeostasis and detoxification in Aspergillus nidulans. Biochem J 414 419 429
48. KingsburyTJ
CunninghamKW
2000 A conserved family of calcineurin regulators. Genes Dev 14 1595 1604
49. HiliotiZ
GallagherDA
Low-NamST
RamaswamyP
GajerP
2004 GSK-3 kinases enhance calcineurin signaling by phosphorylation of RCNs. Genes Dev 18 35 47
50. KishiT
IkedaA
NagaoR
KoyamaN
2007 The SCFCdc4 ubiquitin ligase regulates calcineurin signaling through degradation of phosphorylated Rcn1, an inhibitor of calcineurin. Proc Natl Acad Sci U S A 104 17418 17423
51. PinchaiN
PerfectBZ
JuvvadiPR
FortwendelJR
CramerRAJr
2009 Aspergillus fumigatus calcipressin CbpA is involved in hyphal growth and calcium homeostasis. Eukaryot Cell 8 511 519
52. FoxDS
CoxGM
HeitmanJ
2003 Phospholipid-binding protein Cts1 controls septation and functions coordinately with calcineurin in Cryptococcus neoformans. Eukaryot Cell 2 1025 1035
53. ChoquerM
BoccaraM
GoncalvesIR
SoulieMC
Vidal-CrosA
2004 Survey of the Botrytis cinerea chitin synthase multigenic family through the analysis of six euascomycetes genomes. Eur J Biochem 271 2153 2164
54. WernerS
SuguiJA
SteinbergG
DeisingHB
2007 A chitin synthase with a myosin-like motor domain is essential for hyphal growth, appressorium differentiation, and pathogenicity of the maize anthracnose fungus Colletotrichum graminicola. Mol Plant Microbe Interact 20 1555 1567
55. Martin-UrdirozM
RonceroMI
Gonzalez-ReyesJA
Ruiz-RoldanC
2008 ChsVb, a class VII chitin synthase involved in septation, is critical for pathogenicity in Fusarium oxysporum. Eukaryot Cell 7 112 121
56. SoulieMC
PerinoC
PiffeteauA
ChoquerM
MalfattiP
2006 Botrytis cinerea virulence is drastically reduced after disruption of chitin synthase class III gene (Bcchs3a). Cell Microbiol 8 1310 1321
57. BatesS
MacCallumDM
BertramG
MunroCA
HughesHB
2005 Candida albicans Pmr1p, a secretory pathway P-type Ca2+/Mn2+-ATPase, is required for glycosylation and virulence. J Biol Chem 280 23408 23415
58. Ramos-PamplonaM
NaqviNI
2006 Host invasion during rice-blast disease requires carnitine-dependent transport of peroxisomal acetyl-CoA. Mol Microbiol 61 61 75
59. KimS
AhnIP
RhoHS
LeeYH
2005 MHP1, a Magnaporthe grisea hydrophobin gene, is required for fungal development and plant colonization. Mol Microbiol 57 1224 1237
60. MorohashiK
ZhaoM
YangM
ReadB
LloydA
2007 Participation of the Arabidopsis bHLH factor GL3 in trichome initiation regulatory events. Plant Physiol 145 736 746
61. MorohashiK
GrotewoldE
2009 A systems approach reveals regulatory circuitry for Arabidopsis trichome initiation by the GL3 and GL1 selectors. PLoS Genet 5 e1000396
62. CrooksGE
HonG
ChandoniaJM
BrennerSE
2004 WebLogo: a sequence logo generator. Genome Res 14 1188 1190
63. FrangioniJV
NeelBG
1993 Solubilization and purification of enzymatically active glutathione S-transferase (pGEX) fusion proteins. Anal Biochem 210 179 187
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Mouse Senile Amyloid Fibrils Deposited in Skeletal Muscle Exhibit Amyloidosis-Enhancing ActivityČlánek The Role of Intestinal Microbiota in the Development and Severity of Chemotherapy-Induced MucositisČlánek Crystal Structure of HIV-1 gp41 Including Both Fusion Peptide and Membrane Proximal External RegionsČlánek Demonstration of Cross-Protective Vaccine Immunity against an Emerging Pathogenic Ebolavirus Species
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2010 Číslo 5- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Quorum Sensing Inhibition Selects for Virulence and Cooperation in
- The HMW1C Protein Is a Glycosyltransferase That Transfers Hexose Residues to Asparagine Sites in the HMW1 Adhesin
- Analysis of Virion Structural Components Reveals Vestiges of the Ancestral Ichnovirus Genome
- Mouse Senile Amyloid Fibrils Deposited in Skeletal Muscle Exhibit Amyloidosis-Enhancing Activity
- Global Migration Dynamics Underlie Evolution and Persistence of Human Influenza A (H3N2)
- The Type III Effectors NleE and NleB from Enteropathogenic and OspZ from Block Nuclear Translocation of NF-κB p65
- VEGF Promotes Malaria-Associated Acute Lung Injury in Mice
- Identification of a Mutant PfCRT-Mediated Chloroquine Tolerance Phenotype in
- The Early Stage of Bacterial Genome-Reductive Evolution in the Host
- Host-Detrimental Role of Esx-1-Mediated Inflammasome Activation in Mycobacterial Infection
- Elevation of Intact and Proteolytic Fragments of Acute Phase Proteins Constitutes the Earliest Systemic Antiviral Response in HIV-1 Infection
- The Pleiotropic CymR Regulator of Plays an Important Role in Virulence and Stress Response
- Alternative Sigma Factor σ Modulates Prophage Integration and Excision in
- Effect of Neuraminidase Inhibitor–Resistant Mutations on Pathogenicity of Clade 2.2 A/Turkey/15/06 (H5N1) Influenza Virus in Ferrets
- Massive APOBEC3 Editing of Hepatitis B Viral DNA in Cirrhosis
- NK Cells and γδ T Cells Mediate Resistance to Polyomavirus–Induced Tumors
- Is Genetically Diverse in Animals and Appears to Have Crossed the Host Barrier to Humans on (At Least) Two Occasions
- Adenylate Cyclase Toxin Mobilizes Its β Integrin Receptor into Lipid Rafts to Accomplish Translocation across Target Cell Membrane in Two Steps
- The Role of Intestinal Microbiota in the Development and Severity of Chemotherapy-Induced Mucositis
- HIV-1 Transmitting Couples Have Similar Viral Load Set-Points in Rakai, Uganda
- Few and Far Between: How HIV May Be Evading Antibody Avidity
- Galectin-9/TIM-3 Interaction Regulates Virus-Specific Primary and Memory CD8 T Cell Response
- Perforin Expression Directly by HIV-Specific CD8 T-Cells Is a Correlate of HIV Elite Control
- The Set3/Hos2 Histone Deacetylase Complex Attenuates cAMP/PKA Signaling to Regulate Morphogenesis and Virulence of
- Infidelity of SARS-CoV Nsp14-Exonuclease Mutant Virus Replication Is Revealed by Complete Genome Sequencing
- Combining ChIP-chip and Expression Profiling to Model the MoCRZ1 Mediated Circuit for Ca/Calcineurin Signaling in the Rice Blast Fungus
- Internalin B Activates Junctional Endocytosis to Accelerate Intestinal Invasion
- A Complex Small RNA Repertoire Is Generated by a Plant/Fungal-Like Machinery and Effected by a Metazoan-Like Argonaute in the Single-Cell Human Parasite
- Opc Invasin Binds to the Sulphated Tyrosines of Activated Vitronectin to Attach to and Invade Human Brain Endothelial Cells
- Muc2 Protects against Lethal Infectious Colitis by Disassociating Pathogenic and Commensal Bacteria from the Colonic Mucosa
- PdeH, a High-Affinity cAMP Phosphodiesterase, Is a Key Regulator of Asexual and Pathogenic Differentiation in
- Isolates with Antimony-Resistant but Not -Sensitive Phenotype Inhibit Sodium Antimony Gluconate-Induced Dendritic Cell Activation
- The Microbiota and Allergies/Asthma
- Environmental Factors Determining the Epidemiology and Population Genetic Structure of the Group in the Field
- Prolonged Antigen Presentation Is Required for Optimal CD8+ T Cell Responses against Malaria Liver Stage Parasites
- Crystal Structure of HIV-1 gp41 Including Both Fusion Peptide and Membrane Proximal External Regions
- Susceptibility to Anthrax Lethal Toxin-Induced Rat Death Is Controlled by a Single Chromosome 10 Locus That Includes
- Demonstration of Cross-Protective Vaccine Immunity against an Emerging Pathogenic Ebolavirus Species
- Effective, Broad Spectrum Control of Virulent Bacterial Infections Using Cationic DNA Liposome Complexes Combined with Bacterial Antigens
- High Multiplicity Infection by HIV-1 in Men Who Have Sex with Men
- The -Specific Human Memory B Cell Compartment Expands Gradually with Repeated Malaria Infections
- EBV Promotes Human CD8 NKT Cell Development
- Persistent Growth of a Human Plasma-Derived Hepatitis C Virus Genotype 1b Isolate in Cell Culture
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Quorum Sensing Inhibition Selects for Virulence and Cooperation in
- The Role of Intestinal Microbiota in the Development and Severity of Chemotherapy-Induced Mucositis
- Crystal Structure of HIV-1 gp41 Including Both Fusion Peptide and Membrane Proximal External Regions
- Susceptibility to Anthrax Lethal Toxin-Induced Rat Death Is Controlled by a Single Chromosome 10 Locus That Includes
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



