-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaRegulation of Mutagenic DNA Polymerase V Activation in Space and Time
Escherichia coli, and many other bacteria, respond to high levels of DNA damage with an inducible system called the SOS response. In this response, bacteria first try to restart replication using non-mutagenic DNA repair strategies. If that fails, replication can be restored using DNA polymerases that simply replicate over DNA lesions, a desperation strategy that results in mutations. DNA polymerase V (pol V) is responsible for most mutagenesis that accompanies the SOS response. Because of the risk inherent to elevated mutation levels, pol V activation is tightly constrained. This report introduces a new layer of regulation on pol V activation, with a novel spatial component. After synthesis, the UmuC subunit of pol V is sequestered transiently at the membrane. Release into the cytosol and final activation depends on the activity of RecA protein and the autocatalytic cleavage of UmuD to generate the UmuD' subunit of pol V. The resulting delay in activation represents an additional molecular mechanism that limits the amount of time that this sometimes necessary but potentially detrimental enzyme spends on the DNA.
Published in the journal: . PLoS Genet 11(8): e32767. doi:10.1371/journal.pgen.1005482
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005482Summary
Escherichia coli, and many other bacteria, respond to high levels of DNA damage with an inducible system called the SOS response. In this response, bacteria first try to restart replication using non-mutagenic DNA repair strategies. If that fails, replication can be restored using DNA polymerases that simply replicate over DNA lesions, a desperation strategy that results in mutations. DNA polymerase V (pol V) is responsible for most mutagenesis that accompanies the SOS response. Because of the risk inherent to elevated mutation levels, pol V activation is tightly constrained. This report introduces a new layer of regulation on pol V activation, with a novel spatial component. After synthesis, the UmuC subunit of pol V is sequestered transiently at the membrane. Release into the cytosol and final activation depends on the activity of RecA protein and the autocatalytic cleavage of UmuD to generate the UmuD' subunit of pol V. The resulting delay in activation represents an additional molecular mechanism that limits the amount of time that this sometimes necessary but potentially detrimental enzyme spends on the DNA.
Introduction
Replication of the Escherichia coli chromosome is carried out by replisomes: dynamic multi-protein complexes that coordinate genome duplication by DNA polymerase (pol) III [1]. Pol III replisomes are both exceptionally fast [1] and accurate [2], but are inefficient at synthesising DNA on damaged templates [3]. High levels of DNA damage lead to replication-fork collapse, which can be lethal if not resolved. This situation is usually addressed by the approximately 40 genes of the bacterial SOS response, induced in two stages that reflect two different strategies for restoring replication. The earliest stage features induction of proteins involved in several pathways of error-free DNA repair. If this initial repair does not suffice to restart DNA replication, a later mutagenic process ensues with the induction of the umuDC operon [4]. This operon encodes the translesion synthesis polymerase, pol V [5]. Pol V is responsible for UV - and most chemical-induced chromosomal mutagenesis [6]. SOS mutagenesis also results in more rapid adaptation to stress and development of resistance to antibiotics in the absence of exogenous DNA damage [7,8]. Translesion DNA synthesis by pol V allows for resumption of replication on heavily damaged chromosomes, but dramatically increases mutation rates. Pol V is activated only after the cell’s capacity for non-mutagenic DNA repair has been exceeded. An elaborate regulatory regime should not be surprising, but the cellular constraints on pol V activity remain imperfectly understood.
The active form of the enzyme, pol V Mut, is produced through a series of steps dependent on nucleoprotein filaments of RecA, denoted as RecA*, that are formed on single-stranded DNA after damage (Fig 1A). The umuDC operon, which encodes the pol V precursors UmuD2 and UmuC, contains a particularly high-affinity binding site for the LexA repressor that limits transcription of the umuDC genes [9]. Intracellular levels of UmuD and UmuC are further kept to a minimum through Lon-mediated proteolytic degradation [10]. As a result UmuD and UmuC only accumulate approximately 30 min after DNA damage [4]. However, UmuD2 and UmuC are not active for DNA synthesis. UmuD2 must first undergo a RecA*-mediated autocatalytic cleavage reaction that removes the N-terminal 24 amino acid residues of each subunit to generate UmuD′2. However, this reaction is inefficient and leads to the formation of UmuD/D' heterodimers. UmuD' in the UmuD/D' heterodimer is then rapidly targeted for degradation by the ClpXP protease [10]. As a consequence, UmuD' homodimers only accumulate in response to a persistent damage-inducing signal. UmuD'2 then associates with UmuC to form pol V (UmuD′2C) [5]. Pol V will only accumulate if the damage, and thus RecA*, persists until after the initial 45 minutes of error-free repair [4]. Pol V has weak catalytic activity in vitro [5,11] and is unable to promote translesion DNA synthesis in the absence of RecA in vivo [12]. To facilitate translesion synthesis, pol V must physically interact with RecA* and remove a single RecA-ATP molecule from the 3'-proximal end of the filament to form the mutagenic and highly active pol V Mut (UmuD'2-UmuC-RecA-ATP) [13,14].
Fig. 1. Construction of a fluorescent UmuC reporter strain to visualize pol V regulation. 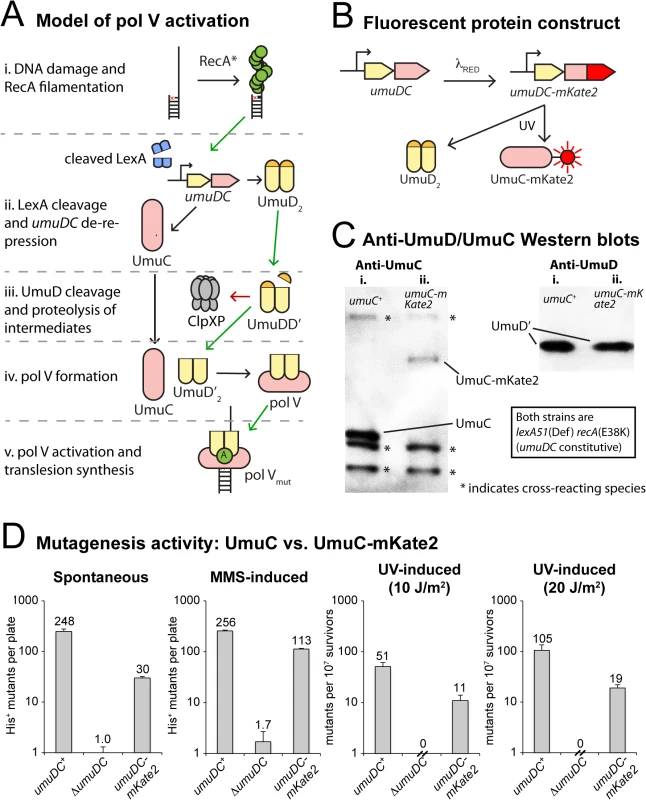
(A) Sketch depicting the regulation of pol V activity in E. coli. Transcriptional repression, targeted proteolysis, posttranslational modification and protein-protein interactions regulate levels of pol V Mut (UmuD’2-UmuC-RecA-ATP), which is capable of mutagenic translesion DNA synthesis. The symbol “A” represents a molecule of ATP bound to RecA in the activated pol V Mut complex. (B) Replacement of the wild-type chromosomal umuC locus with umuC-mKate2 using λRED recombineering. (C) Western blots with anti-UmuC and anti-UmuD antibodies confirming the expression of full length UmuC-mKate2. Note that steady-state levels of the chimeric UmuC-mKate2 protein (lane ii, umuC-mKate2 background, strain RW1314) are approximately 20% of the wild-type UmuC protein (lane i, wild-type umuC background, RW574). Bands resulting from cross-reaction of the anti-UmuC antibody (see S1A Fig) with other proteins are also observed (marked with *) and indicate that similar amounts of lysate were loaded in each lane. Both strains are lexA(Def) recA(E38K) and thus constitutively express UmuD and UmuC and convert UmuD to UmuD'. Steady-state levels of UmuD' are similar in the wild-type umuC+ (lane i) and umuC-mKate2 cells (lane ii). (D) In vivo mutagenesis assays performed on RW1314 show that UmuC-mKate2 is active for both spontaneous and damage-induced mutagenesis. Levels of mutagenesis are lower than that of the wild-type UmuC strain (RW574), but are consistent with overall lower steady state levels of the chimera. Direct observation of the various activation steps of pol V inside living cells with fluorescence microscopy would allow for the testing of various hypotheses and potentially the construction of a global model for mutasome activity. However, analysis of pol V activity in vivo has long proven difficult due its very low expression levels [15]. Here, we report the use of single-molecule fluorescence microscopy to directly visualize the dynamics of pol V inside living cells. Surprisingly, we observe that pol V precursors are temporarily sequestered at the cellular membrane, adding another prominent factor to those limiting pol V activity during early stages of the DNA damage response. Spatial regulation of a DNA processing enzyme is unprecedented in bacteria and suggests the presence of several layers of spatiotemporal partitioning to regulate the molecular processes underlying genomic maintenance.
Results
Single-molecule observation of pol V in live E. coli cells
In order to visualize the regulation of pol V, we altered the umuDC operon so that the bright red fluorescent protein mKate2 is fused to the C-terminus of UmuC (Fig 1B). Western blotting of the chromosomally expressed UmuC-mKate2 protein revealed expression of the chimeric protein, albeit at steady-state levels somewhat lower than untagged UmuC (Fig 1C, S1A Fig). Nevertheless, the chimeric umuC-mKate2 allele promoted both spontaneous and damage-induced reversion of the hisG4 allele, confirming it is functionally active (Fig 1D, S1B Fig). The level of mutagenesis promoted by UmuC-mKate2 was lower than that of wild-type UmuC, which is consistent with the overall lower steady-state levels of the chimera in the cell.
We imaged E. coli K12 MG1655 UmuC-mKate2 cells on a home-built wide-field single-molecule fluorescence microscope. To monitor changes in the cellular levels and location of UmuC as a function of time after UV-induced damage, E. coli was immobilized inside flow cells and a series of time-lapse images were recorded. We developed a novel flow-cell design that allowed for imaging of individual cells with single-molecule sensitivity, while also providing space for cells to grow into filaments (a hallmark of the SOS response) and allowing for in situ UV irradiation. Cells were irradiated with UV light from a mercury lamp (fluence = 1–100 J/m2, λ = 254 nm) and UmuC-mKate2 fluorescence was recorded over a course of 3h, imaging once every 5 min.
We first measured changes in fluorescence intensity using time-lapse measurements, allowing us to monitor the production of UmuC-mKate2 (Fig 2A and 2B). At all UV doses, very little expression of UmuC-mKate2 was observed during the first 30 min following UV irradiation (Fig 2A and 2B), in good agreement with the previously measured value of 30 min required for expression of UmuC and UmuD [4]. Furthermore, UmuC-mKate2 levels increased at all doses after this initial delay, albeit with different kinetics. While at doses ≤ 3 J/m2, UmuC-mKate2 levels gradually increased throughout the duration of the experiment; at higher doses the protein levels decreased after reaching a maximum at 90–120 min. At the highest dose, 100 J/m2, UmuC-mKate2 was produced more slowly than at 30 J/m2 and ultimately reached lower levels, presumably due to inhibition of protein synthesis as the result of extensive DNA damage.
Fig. 2. Monitoring DNA damage induced expression of UmuC-mKate2 using time-lapse and time-sampling analysis. 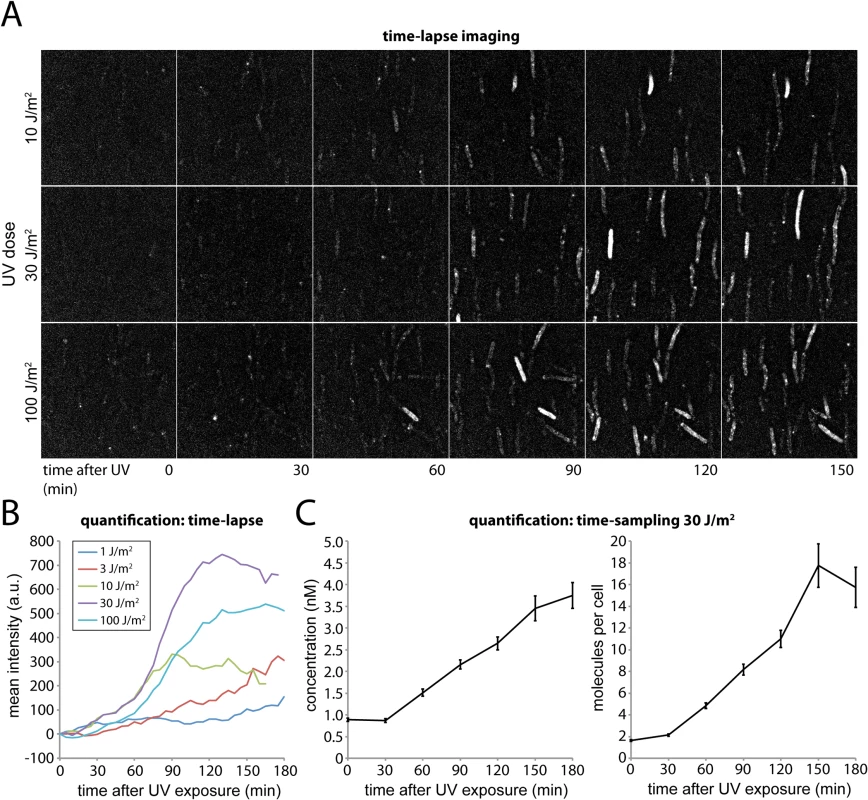
(A) Time-lapse imaging of UmuC-mKate2 expressing cells (EAW282). Cells were grown at 37°C in flow cells and irradiated in situ with 10, 30 or 100 J.m-2 of UV light (λ = 254 nm). Following irradiation fluorescence images were recorded every 5 min for 180 min. These images reveal that UmuC induction begins around 60 mins after DNA damage. (B) Quantification of the mean fluorescence signal of cell-containing regions within time-lapse image series after various does of UV-light. (C) Quantification of the mean number of molecules per cell and mean concentration of UmuC-mKate2 in time-sampling measurements after treatment with 30 J/m2 UV light. The amount of UmuC-mKate2 inside cells was quantified using time-sampling measurements (Fig 2C). Similar to the time-lapse experiments, cells were grown, irradiated and imaged within flow cells. Rather than periodically recording single fluorescence images to monitor the response of the same set of individual cells in time, we recorded video-rate movies of UmuC-mKate2 fluorescence using a new population of cells for every time point after UV irradiation. This procedure provided higher-quality intensity information but led to significant photobleaching. We therefore chose a new field of view with unbleached cells for each new measurement. Movies (296 × 34 ms frames) were recorded over 10 fields of view every 30 min for 3 h, capturing between 70 and 236 cells. For each field of view we also recorded a bright-field image, allowing us to define the outline of each cell. The fluorescence movies were used to determine the concentration of UmuC-mKate2 inside each cell (S2 Fig).
Plotting the average cellular concentration of UmuC-mKate2 over time, we observed a sharp increase (Fig 2C), with dynamics consistent with those measured by time-lapse measurements (Fig 2B). At 30 J/m2, the concentration of UmuC-mKate2 increased from 0.9 ± 0.04 nM (S.E.M, n = 236 cells) to 3.7 ± 0.3 nM (n = 70), 180 min after irradiation with UV light. These concentrations correspond to an increase from an average of 1.6 ± 0.08 (n = 236) molecules per cell to 15.7 ± 1.8 (n = 70) molecules per cell at 180 min. The numbers of UmuC molecules measured by fluorescence microscopy (16 per cell) are lower than measurements obtained by Western blotting of wild-type cells with anti-UmuC antibodies (~ 60 per cell) [4, 15], but again, this observation is consistent with overall lower intracellular steady-state levels of the UmuC-mKate2 chimera compared to the wild-type UmuC protein (Fig 1C). Quantification of UmuC-mKate2 levels in cells periodically sampled from shaking culture showed similar expression dynamics and concentration levels as cells grown in flow cells: no UmuC-mKate2 is produced during the first 30 min, then levels rise to 3 nM in the period 40–120 min after irradiation (S3 Fig).
UmuC appears at the cell membrane and is then released into the cytosol
The fast time-sampling movies showed foci of UmuC-mKate2 that move within a thin band along the cell periphery (Fig 3A, S1 movie). This observation suggested that many molecules of UmuC associate with the cell membrane, diffusing slowly enough to result in sharply defined foci on a single image (34 ms frame duration), but sufficiently mobile to show movement on longer timescales. To examine this apparent membrane association further, we used a plasmid encoding a fluorescently labelled inner membrane protein (LacY-eYFP) as a reference for the position of the membrane. Super-resolution images were produced for both UmuC-mKate2 and LacY-eYFP, which revealed that many of the UmuC-mKate2 foci were indeed located with LacY on the cell membrane (Fig 3B). Control experiments with cells expressing free mKate2 (including the same linker as the UmuC-mKate2 construct) confirmed that membrane association is a property of UmuC, as opposed to mKate2 or the linker region (S4 Fig).
Fig. 3. Changes in the cellular location of UmuC in response to UV irradiation. 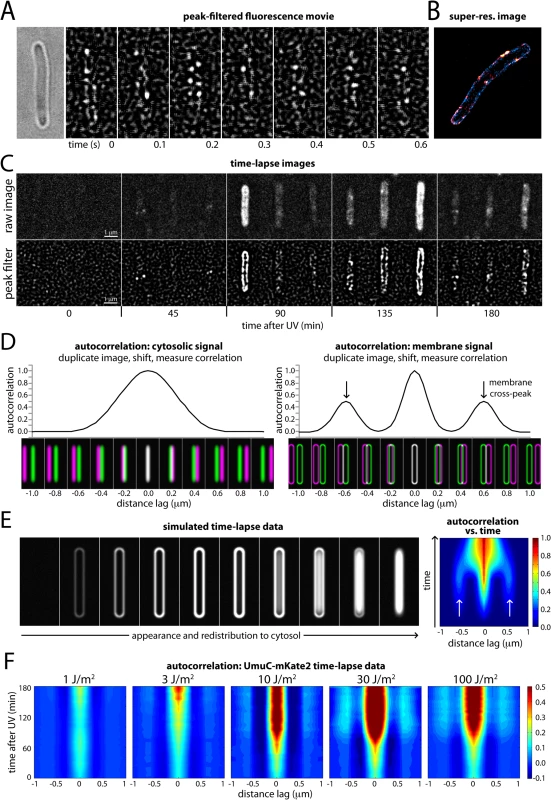
(A) Montage of frames from a time-sampling movie showing UmuC-mKate2 diffusing along the cell periphery (EAW282). A peak-enhancing filter was applied to fluorescence images [57]. (B) Two-color super-resolution reconstruction showing co-localization of UmuC-mKate2 (red) and LacY-eYFP (blue) on the cell membrane (EAW191 containing pBAD-LacY-eYFP). (C) Individual cells cropped from time-lapse series showing UmuC transitioning from a membrane-associated to a cytosolic localization (EAW282). (D) Autocorrelation analysis of simulated images of E. coli cells displaying cytosolic and membrane-associated signals. Autocorrelation measurement can be conceptualised as follows: correlations are measured between an image and its distance-shifted duplicate. Correlations are measured as a function of shift distance (or distance lag). Autocorrelation analysis of cells displaying cytosolic signals produces a broad origin peak, whereas cells with membrane-associated signal produce a distinctive cross-peak (indicated with arrows). (E) Autocorrelation analysis of a simulated time-lapse series for an E. coli cell expressing a redistributing fluorescent protein. The simulation begins with no fluorescent protein signal. The protein is then induced, producing a membrane-associated signal before redistributing to the cytosol. Here the autocorrelation analysis is presented as a 2D contour plot. Blue areas indicate low correlation, whereas red areas indicate high correlation. Arrows indicate the membrane cross-peak that is visible when the cell has membrane-associated signal. (F) Similar autocorrelation analysis of experimentally acquired time-lapse series showing dose-dependent changes in UmuC localization (EAW282). At higher doses of UV the membrane cross-peaks begin to decline at ~90 min post-irradiation, while the origin peak becomes broader, indicating redistribution of UmuC-mKate2 into the cytosol. A peak-enhancing filter was applied to fluorescence images prior to autocorrelation analysis [57]. In both the time-lapse and time-sampling datasets, visual inspection of the movies indicated a change in the localization behaviour of UmuC during the course of the DNA-damage response. For the first 90–120 min after UV exposure, UmuC-mKate2 was predominantly located at the cell periphery, while in later images UmuC-mKate2 was distributed throughout the cytosol (Fig 3C, S5 Fig).
We developed a novel autocorrelation-based approach to quantify the localization behaviour of UmuC across all cells in the time-lapse measurements. Autocorrelation of images can be conceptualized as follows. First, the cell image to be analyzed is duplicated and superimposed with the original (schematically depicted in Fig 3D) so that the intensities of each pixel in the two images are perfectly correlated. One of the images is then shifted sideways in increments (presented in the figures as the distance lag) relative to the other image and the correlation between pixel values is re-calculated. This procedure is repeated over a range of different shift sizes (or distance lags) to produce an autocorrelation function.
The highest correlation of pixels is seen when the duplicate images are perfectly superimposed (zero lag). If significant amounts of UmuC are concentrated in the membranes, additional signal peaks will be seen at two non-zero shift increments where the right membrane of one image passes over the left membrane of the other, and vice versa (schematically shown in Fig 3D). Cytosolic UmuC with its distribution centered along the central cellular axis should not generate such secondary peaks.
To illustrate this approach, we simulated images of E. coli cells with a width of 0.6 μm, expressing cytosolic or membrane-localized fluorescent proteins. Cells containing cytosolic signal gave rise to an autocorrelation function with a single, broad peak at zero lag (Fig 3D). Cells with membrane-associated fluorescence instead produced a narrow peak at zero lag (perfect superimposition), as well as two distinctive cross peaks when the duplicate images were shifted 0.6 μm, arising from correlations between signals on the two sides of the cell.
To test if this autocorrelation approach could be used to monitor changes over time in cellular distribution of UmuC-mKate2, we simulated a time-lapse series with an E. coli cell transitioning from a state with no signal, to a membrane-associated signal, to a cytosolic signal (Fig 3E; left). We graphed autocorrelation functions measured at each time point of the simulation as a 2D contour plot, with blue-to-red coloring indicating low-to-high levels of correlation (Fig 3E; right). At each time point, autocorrelation functions were normalized to the fluorescence intensity of the cell. The amplitude of the central peak (zero lag) thus reflects relative changes in fluorescent protein concentration: the amplitude is low (blue) at lower concentrations and high (red) at high cellular concentrations. When present, membrane-bound fluorescent protein gives rise to weaker secondary signals (light blue) to either side of the central peak. Analysis of experimentally acquired data produced similar results: time-lapse images of LacY-eYFP (an inner membrane protein) produced an autocorrelation plot with a strong central peak and weaker secondary peaks, while DnaX-YPet (which is cytosolic/nucleoid associated) produced only a strong central peak (S6 Fig). Our autocorrelation approach thus provides a means to monitor the distribution of fluorescent proteins across the short-axis of the cell during time-lapse measurements: membrane-associated protein produces distinctive cross-peaks, whereas cytosolic/nucleoid associated protein does not.
We then applied autocorrelation analysis to our experimentally acquired UmuC-mKate2 time-lapse images (Fig 3F), beginning at the time in which cells were irradiated at the UV dose indicated. Within the time-lapse images, the orientation of E. coli cells was biased by the flow of medium through the flow cell: the majority of cells aligned with the vertical axis of the images. This orientation bias allowed us to look for the repeating pattern of membrane-localized UmuC-mKate2 signals by measuring image-wide autocorrelation functions across the horizontal axis of the images. The autocorrelation functions were normalized by cellular intensity across the entire dataset of UV doses. At 1 J/m2, there was little change in the autocorrelation function during the time-course and little detectable membrane localization. At 3 J/m2, a modest signal from membrane-localized UmuC appears late in the time-lapse. At 10 J/m2, two distinctive membrane-bound cross peaks appear at about 90 min and persist until 180 min. At 30 J/m2 and 100 J/m2, membrane localization is prominent from 90 min. From 120–180 min, the autocorrelation is significantly broader, with the valley between the central peak and the membrane peak becoming filled in. This broadening is indicative of UmuC entering the cytosol.
Evidence of redistribution is also visible in cross-correlation analysis between UmuC-mKate2 and the inner membrane protein LacY-eYFP (Fig 4A). We found that expression of the LacY-eYFP protein partially induced the SOS response: cells were longer than usual and produced an elevated level of UmuC-mKate2 prior to UV irradiation. At early time-points after UV irradiation, clear cross-peaks are visible due to co-localization of UmuC-mKate2 and LacY-eYFP on the membrane. Each UmuC-mKate2 focus correlates with LacY-eYFP signal on both the same side of the cell (producing a central peak at lag = 0) and the opposite side of the cell (producing cross-peaks at lag = ± 0.8 μm). From 90–180 min after irradiation, these cross-peaks angle in towards the central peak and become broader. This is consistent with UmuC-mKate2 foci forming in the cytosol, where the distance from each focus to either side of the membrane falls between 0 and 0.8 μm. As UmuC-mKate2 is no longer confined to the membrane, the distance to each side of the membrane becomes more variable, causing the cross-peaks to broaden.
Fig. 4. Visualization of UmuC redistribution by cross-correlation analysis and Western blotting. 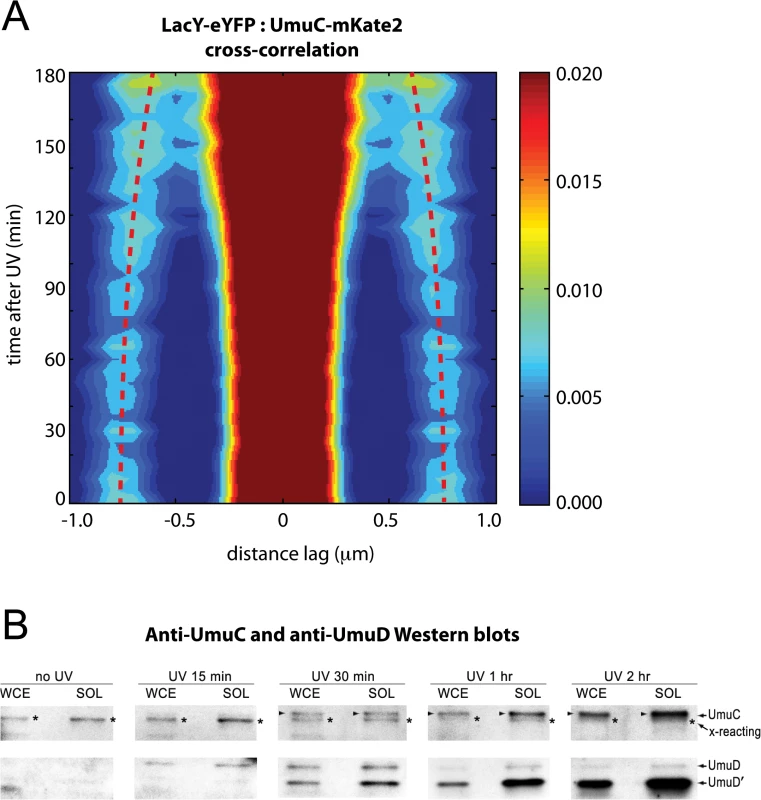
(A) Cross-correlation analysis of LacY-eYFP and UmuC-mKate2 signals from time-lapse measurements (EAW191 containing pBAD-LacY-eYFP). Distance-dependent correlation values between LacY-eYFP and UmuC-mKate2 signals are presented as 2D contour plots. Blue areas indicate low correlation, whereas red areas indicate high correlation. Image pairs containing membrane-associated UmuC-mKate2 foci, thus co-localizing with membrane-defining LacY-eYFP signals, produce sharp cross-correlation peaks at lag = 0 and ± 0.8 μm. Image pairs containing cytosolic UmuC-mKate2 foci produce broader cross-correlation peaks at values between 0 and 0.8 μm. Red dotted lines mark the position of the secondary peaks in time and highlight a time-dependent trend towards lag values < 0.8 μm. A peak-enhancing filter was applied to fluorescence images prior to autocorrelation analysis. (B) UmuD, UmuD' and UmuC expressed from the recA+ lexA+ strain, RW118, after exposure to 30 Jm-2 was monitored in whole cell extracts (WCE) and soluble fraction (SOL) at various time points after irradiation. The protein that is slightly smaller than UmuC and cross-reacts with the UmuC antibodies is indicated with an asterisk to the right of each lane. When detectible, UmuC is indicated by an arrow on the left of each lane. The UmuD and UmuD' proteins are indicated on the right hand side of the image The redistribution of native chromosomally expressed UmuC into the cytosol was also followed by Western blotting of whole-cell extracts (WCE) and soluble fractions at various time points after UV irradiation. (Fig 4B). As previously reported, basal levels of UmuD and UmuC in a recA+ lexA+ strain are undetectable [15]. The affinity purified UmuC antibodies do, however, cross-react with a protein that is slightly smaller than UmuC (S1A Fig) that can be detected in both the WCE and soluble fractions (Fig 4B). Importantly, the solubility of this cross-reacting protein does not change after UV. We can therefore use this cross-reacting protein as an internal control for the amount of cellular material loaded in each lane. We begin to observe the appearance of soluble UmuC 30 minutes after UV irradiation. The amount of UmuC in the soluble fraction increases further 1 hour and 2 hours post-UV, consistent with the release of UmuC into the cytosol. The extent of soluble UmuC shows an excellent correlation to the amount of soluble UmuD' in the cell. Fifteen minutes post-UV, we observe the induction of UmuD and very little conversion to UmuD'. By 30 minutes post-UV, roughly 50% of the UmuD is converted to UmuD' in the WCE. Much more UmuD' is found in the soluble fraction, indicating that UmuD' is inherently more soluble than UmuD. The amount of UmuD' in the soluble fraction continues to accumulate 1 hour and 2 hours post-UV.
Finally, mapping the cellular location of individual UmuC molecules visualized in cells grown in shaking culture also indicates redistribution of UmuC-mKate2 from the membrane to the cytosol beginning 90 min after UV irradiation (S7 Fig). These results confirm that the membrane localization is observed independent of whether the cells grow in culture or on cover slips, and independent on whether autocorrelation analysis is used or direct mapping.
We observed that at large doses of UV light (> 30 J/m2), many cells produced a single large burst of UmuC-mKate2 and that the change from membrane-associated to cytosolic distribution seemed to initiate when the level of UmuC reached a maximum. This maximum always occurred more than 90 min after UV. To examine this phenomenon further, we cropped movies of single cells from our time-lapse series (100 J/m2) and post-synchronized them each individually to the time point with the maximum fluorescence intensity (Fig 5A and 5B). Where appropriate, the cells were also rotated to align with the y-axis of the image to further enhance the sensitivity of autocorrelation analysis. These post-synchronized movies reveal that during bursts of production (lasting typically 30–60 min) UmuC is membrane-associated and gradually becomes cytosolic after reaching peak intensity (Fig 5A).
Fig. 5. Post-synchronization of individual cells within time-lapse series (EAW282). 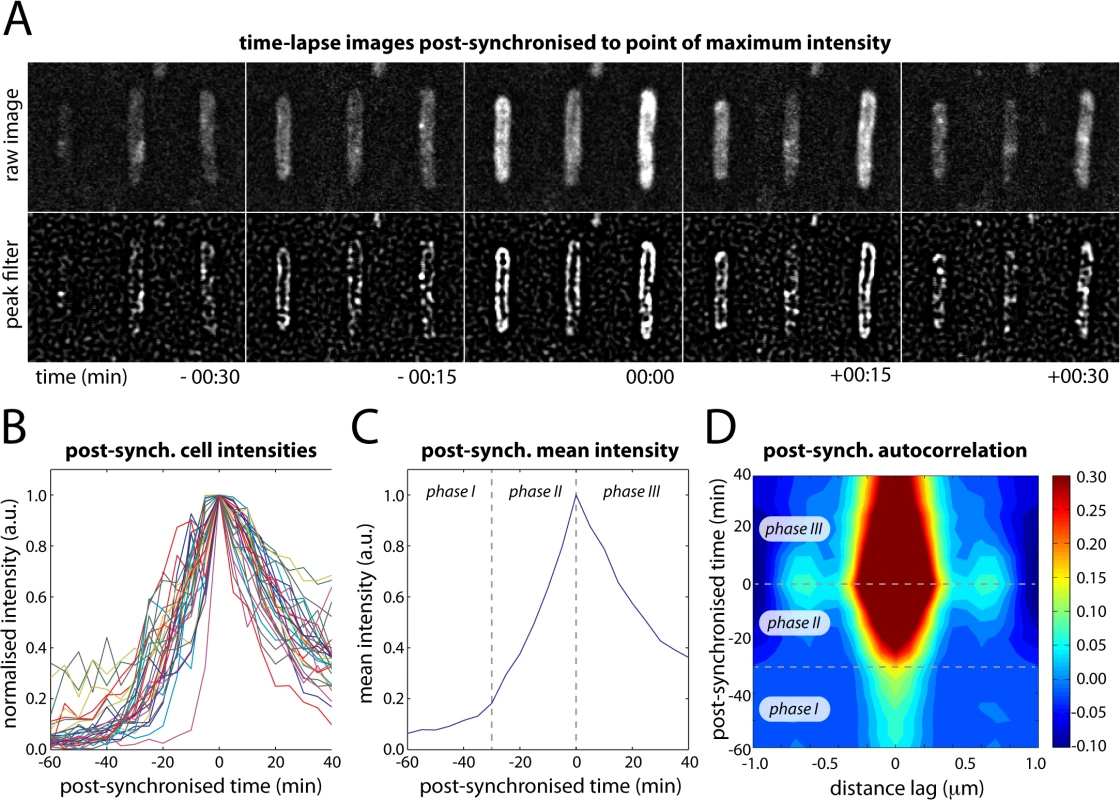
(A) Time-lapse images of individual cells post-synchronized to the point at which the intensity of UmuC-mKate2 signal reached a maximum (typically 90–150 min after treatment with 100 J/m2 UV light). (B) Post-synchronized intensity vs time trajectories for 31 individual cells (of 100 total) that produced a single well-defined burst of UmuC-mKate2 synthesis. The remaining 69 cells either produced no detectable bursts of UmuC-mKate2 synthesis or produced multiple bursts of synthesis, preventing post-synchronization. (C) Mean intensity vs time trajectory over all cells showing three phases in the production of UmuC: (phase I) little UmuC-mKate2 produced; (phase II) increased production of UmuC-mKate2; (phase III) production of UmuC-mKate2 ceases and cellular levels decline. (D) Autocorrelation analysis of post-synchronized time-lapse series showing that the three phases in the production of UmuC correspond with changes in its cellular location: (phase I) weak membrane cross-peaks indicate low levels of membrane-associated UmuC-mKate2; (phase II) stronger membrane cross-peaks indicate production of membrane-associated UmuC-mKate2; (phase III) decreasing membrane cross-peaks and broadening of origin peak indicating redistribution of UmuC-mKate2 to the cytosol. A peak-enhancing filter was applied to fluorescence images prior to autocorrelation analysis [57]. Autocorrelation analysis on the post-synchronized movies revealed three distinct phases in the behaviour of UmuC (Fig 5C and 5D). In phase I, very little UmuC is produced and those molecules that are present associate with the cell membrane. In phase II, substantially more UmuC is produced and these molecules accumulate at the membrane. In phase III, production ceases and UmuC redistributes into the cytosol. The redistribution of UmuC into the cytosol is essentially complete 30 min after the cell reaches peak intensity. The fact that UmuC only redistributes to the cytosol during phase III and after its production has ceased indicates that UmuC is being released from the membrane, as opposed to newly synthesized UmuC becoming sequestered in the cytosol.
UmuC membrane release requires UmuD cleavage
The change in abundance of soluble UmuC exhibits a strong correlation to the appearance of UmuD' (Fig 4B). To test the hypothesis that the interaction between UmuD' and UmuC allows UmuC to relocate to the cytosol, we employed a umuD(K97A) strain that cannot convert UmuD to UmuD′ [12]. Compared with the wild-type background (Fig 6A), UmuC-mKate2 in the umuD(K97A) strain appeared at about the same time after UV irradiation (Fig 6B). However, in this strain the UmuC was more strongly membrane-associated in images and produced stronger and more persistent membrane cross-peaks upon autocorrelation analysis (Fig 6B). Note that UmuC is stabilized in the cell when in a complex with UmuD′ [10,16], such that degradation of UmuC by the Lon protease limits the formation of UmuC signals when UmuD′ cannot be formed. This indicated a strong tendency for UmuC-mKate2 to accumulate at the membrane, both before and after UV irradiation. Analysis of umuD(K97A) cells grown in shaking culture similarly show a strong bias for UmuC-mKate2 to be localized on the cell membrane (Fig 6B). Together, these observations support the hypothesis that conversion of UmuD to UmuD′ is critical for re-localisation of UmuC.
Fig. 6. Identifying the cellular location of UmuC-containing species using mutants with defects in the pol V activation pathway. 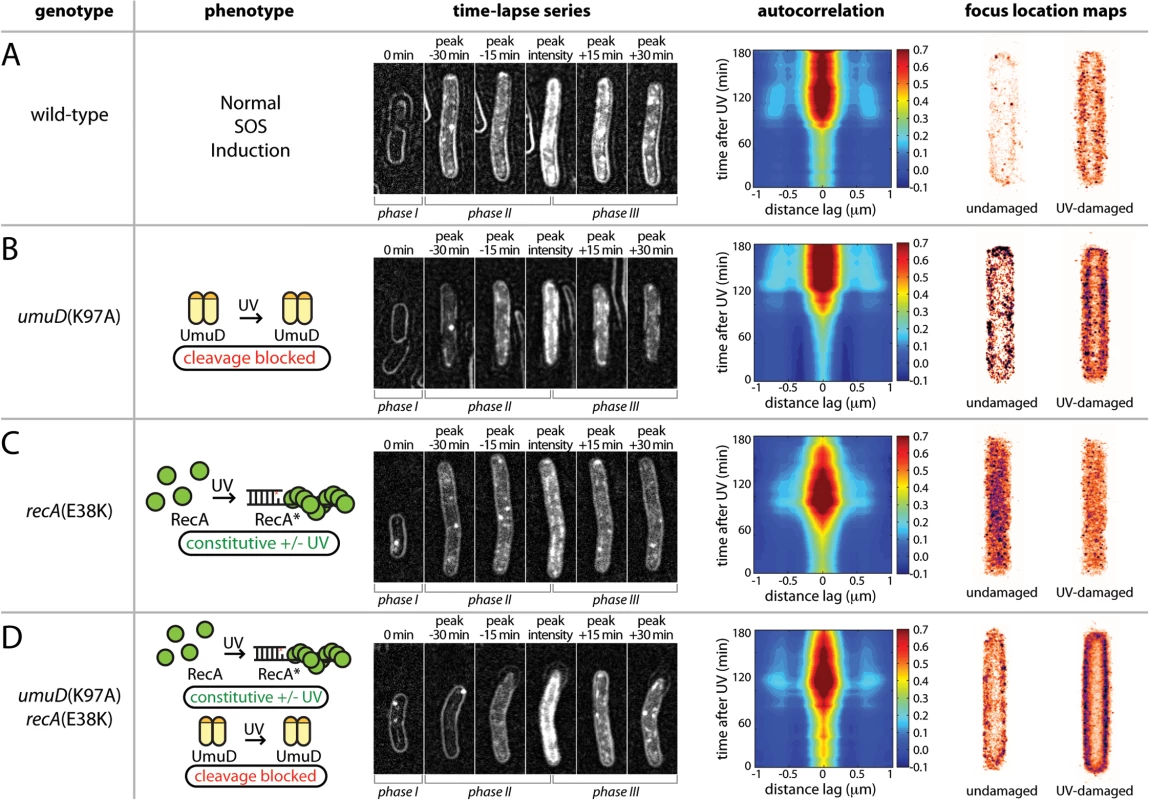
(A-D) Analysis of UmuC-mKate2 localization in strains with different genetic backgrounds; EAW282 (recA+ lexA+); EAW329 [recA+ lexA+ umuD(K97A)]; EAW307 (recA(E38K) lexA+); and EAW455 [recA(E38K) lexA+ umuD(K97A)]. For time-lapse/autocorrelation analysis, cells were treated at t = 0 min with UV light at 30 J/m2. The images shown in time-lapse series are composites of bright-field images (cell outlines), raw fluorescence images and peak-filtered fluorescence images. The intensity range used for each channel was the same for all composites. The images represent stages in the UV-damage response (left to right: immediately after irradiation; peak signal—30 min; peak signal—15 min; peak signal; peak signal + 15 min; peak signal + 30 min). Autocorrelation analyses are again presented as a 2D contour plots, with the time course beginning at 0 min at the bottom. A peak-enhancing filter was applied to fluorescence images prior to autocorrelation analysis [57]. Side peaks in the autocorrelation function at -0.6 μm and 0.6 μm indicate membrane-associated UmuC-mKate2. More intense cross-peaks indicate a higher proportion of cells with membrane-localised distribution. A broad peak at the origin with no discernable membrane peaks is indicative of cytosolic UmuC-mKate2. Focus location maps (far right panels) were produced from analysis of shaking-culture cells. Cells were grown in EZ medium with glucose at 37°C. The entire 0.5 ml culture was irradiated with 10 J/m2 UV light while sandwiched between two quartz plates then returned to shaking culture. Aliquots were taken every 10 min, placed on an APTES-treated coverslip, closed in with a second plain glass coverslip and imaged. 96 × 34 ms frames were recorded using 568 nm excitation light at a power of 1800 W/cm2. All UmuC-mKate2 foci from all cells were mapped as Gaussians to a cell of standard size and shape. Maps for UV-irradiated cells include images recorded 10–120 min after irradiation. To further test this hypothesis, we determined the location of UmuC-mKate2 in a recA(E38K) (also known as recA730) background, in which active RecA nucleoprotein filaments (RecA E38K*) are formed constitutively in the absence of DNA damage [17]. Under these conditions, the SOS regulon is partially derepressed and much of the temporal regulation normally imposed on pol V is circumvented: pol V is constitutively formed through UmuD cleavage and activated to pol V Mut. However, the ability of RecA(E38K) to spontaneously form nucleoprotein filaments depends upon the cellular concentration of the RecA mutant [15] and RecA(E38K) can be further activated under various conditions [18]. Indeed, UV treatment results in the apparent induction of UmuC in the recA(E38K) lexA+ background, where all LexA-regulated proteins are expected to already be expressed in the absence of DNA damage. As expected, and in contrast to the recA+ lexA+, background (Figs 3–5 and 6A), in the recA(E38K) lexA+ strain UmuC was present at all times after irradiation (Fig 6C). The UmuC signal increased over the first 90 min after irradiation, with the brief appearance of a signature for membrane-associated UmuC-mKate2 at the peak of UmuC production (~90 min). This observation suggests that higher levels of UmuC after UV treatment went to the membrane first and then became cytosolic. While recA(E38K) cells efficiently cleave UmuD to UmuD' in the absence of DNA damage, UmuD' can only be generated from full-length UmuD and it is possible that there is sufficient uncleaved UmuD in these cells to transiently allow binding of UmuC to the membrane. The same cells grown in shaking culture show a general cytosolic distribution of UmuC-mKate2 before and 10–120 min after UV irradiation. These observations again indicate that fully activated pol V Mut is largely cytosolic and that the membrane-localized state corresponds to an intermediate species preceding pol V Mut formation.
To rule out the possibility that the cytosolic localization of UmuC-mKate2 in the recA(E38K) strain is due to constitutive production of RecA E38K* as opposed to UmuD cleavage, we then determined the location of UmuC-mKate2 in a recA(E38K) umuD(K97A) background, in which UmuC and UmuD are produced constitutively but UmuD cannot be cleaved to form UmuD'. As already indicated, overall UmuC levels decline because of Lon-dependent degradation of UmuC in the absence of UmuD′. In this strain, UmuC-mKate2 is seen at the membrane at all time points, increasing after UV induction to a higher and persistent level 90 min after UV induction (Fig 6D). Autocorrelation analysis revealed clear membrane cross-peaks throughout the measurement, indicating that in this background UmuC-mKate2 remains associated with the membrane in both the presence and absence of damage. The membrane association in this strain is corroborated by a clean membrane-associated distribution of UmuC-mKate2 both before and 10–120 after UV irradiation seen in the same cells in shaking culture (Fig 6D).
The membrane localization of wild-type UmuC when co-expressed with a non-cleavable UmuD protein was also confirmed directly by immuno-electron microscopy (Fig 7A and 7B). Furthermore, in experiments measuring the solubility of wild-type UmuC in E. coli cell extracts, there appeared to be significantly less soluble UmuC when it was co-expressed with UmuD(K97A) than when expressed alone, or with UmuD' (Fig 7C). This is consistent with an active role for UmuD in sequestering UmuC in an insoluble membrane-bound fraction, and/or a requirement for UmuC association with UmuD' to effect release to the cytosol. Together, these results provide clear evidence that conversion of UmuD to UmuD' is a requirement for the spatial relocalization of UmuC to the cytosol.
Fig. 7. Detection of UmuC location by immuno-electron microscopy and solubility in cell lysates. 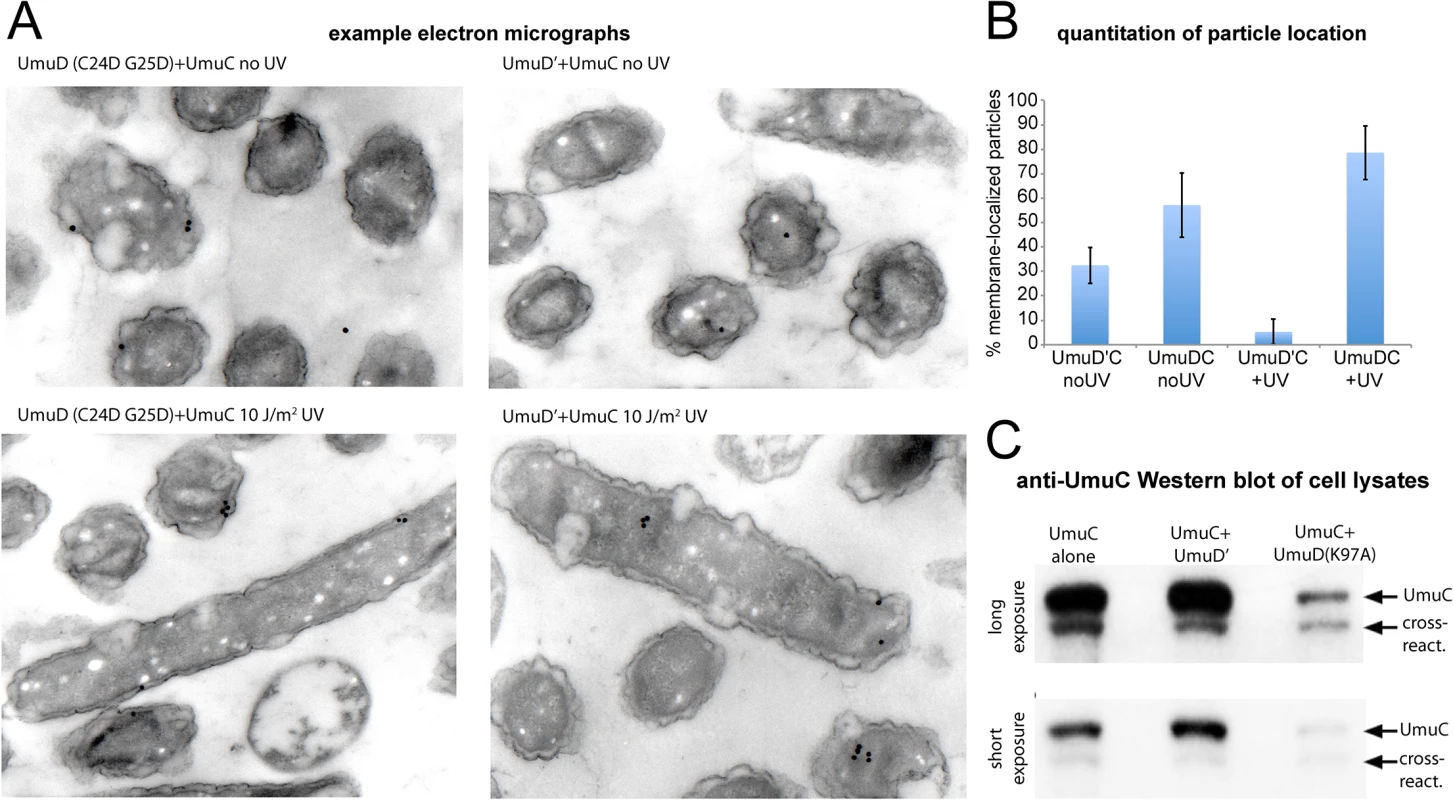
(A–B) Immuno-electron microscopy of plasmid-expressed UmuC in the presence of umuD(C24D G25D) and umuD' in the E.coli strain RW1394. Sectioned electron-microscopy grids were incubated with anti-UmuC antibody and subsequently labeled with 30 nm gold beads conjugated to a secondary goat-anti-rabbit antibody. (A) Representative micrographs. Although the labeling density was low, gold beads were clearly visible for cells expressing UmuC, whereas control cells lacking UmuC showed virtually no beads. (B) Quantification of the proportion of beads associated with the membrane each sample. Beads found to be within 100 nm of the cell membrane were classed as membrane-associated. (C) Levels of soluble UmuC detected by western blotting (E. coli strain, TCH03). The two images are identical, except that upper panel is a darker exposure of the lower panel. UmuC was expressed alone, or co-expressed with UmuD' or UmuD(K97A). The images clearly show that UmuC is most soluble when co-expressed with UmuD' and least soluble when co-expressed with UmuD(K97A). Co-localization of pol V mutasomes with replication forks depends on RecA
Having revealed a novel membrane binding and release mechanism underlying the regulation of pol V, we next studied the positioning of pol V molecules after they were released from the membrane, in the context of translesion synthesis. For many years, the prevailing model for pol V-catalysed translesion synthesis was one that described a highly concerted series of events: (i) replisomes stall at lesion sites, exposing ssDNA; (ii) RecA* filaments form on the ssDNA, triggering the SOS response and activating pol V; (iii) pol V displaces pol III through mass action and synthesises over the lesion; (iv) replisomes restart and the cells return to normal. However, a series of recent in vitro observations has brought this model into question. First, RecA*-dependent activation of pol V can occur in trans, i.e., remote from the site of DNA synthesis [13,19]. Second, replisomes do not necessarily stall at lesions, but can instead skip over them leaving a ssDNA gap in their wake [20]. Finally, the presence of RecA* on the template increases the displacement of pol III by translesion synthesis polymerases, indicating additional complexity above a simple mass action-driven exchange mechanism [21].
Our single-molecule observations allow us to address whether translesion synthesis takes place at replication forks by simultaneously imaging UmuC fused to the red mKate2 and the τ subunit of the DNA pol III holoenzyme fused to the yellow YPet; DnaX-YPet) [22]. We recorded two-color time-lapse series of wild-type recA+ cells and constitutively active recA(E38K) cells following irradiation with UV light at 30 J/m2. We then measured cross-correlation functions between the two image channels as a function of time. This procedure was similar to the autocorrelation-based one described above, except that image color pairs were correlated with each other rather than each image being correlated against itself. Image pairs containing co-localized fluorescent spots should return a strong, sharp cross-correlation signature, whereas spatially unrelated spots should return weak or no cross-correlation.
In the wild-type background, a broad and relatively weak cross-correlation peak was observed, suggesting little co-localization between DnaX-YPet and UmuC-mKate2 molecules beyond their presence within the same cell (Fig 8A). In the recA(E38K) background, stronger cross-correlation was observed across a relatively narrow distance regime, consistent with co-localizing spots of diffraction-limited size (Fig 8A). However, the cross-correlation declines with time after UV treatment. This observation suggests that in contrast with the wild-type background, a significant proportion of UmuC-mKate2 molecules (primarily in the form of pol V Mut in this background) co-localize with replisomes in the recA(E38K) background under normal growth, but that the co-localization declines at later times after UV irradiation.
Fig. 8. Colocalization of replisomes and mutasomes in wild-type (EAW282) and recAE38K cells (EAW307). 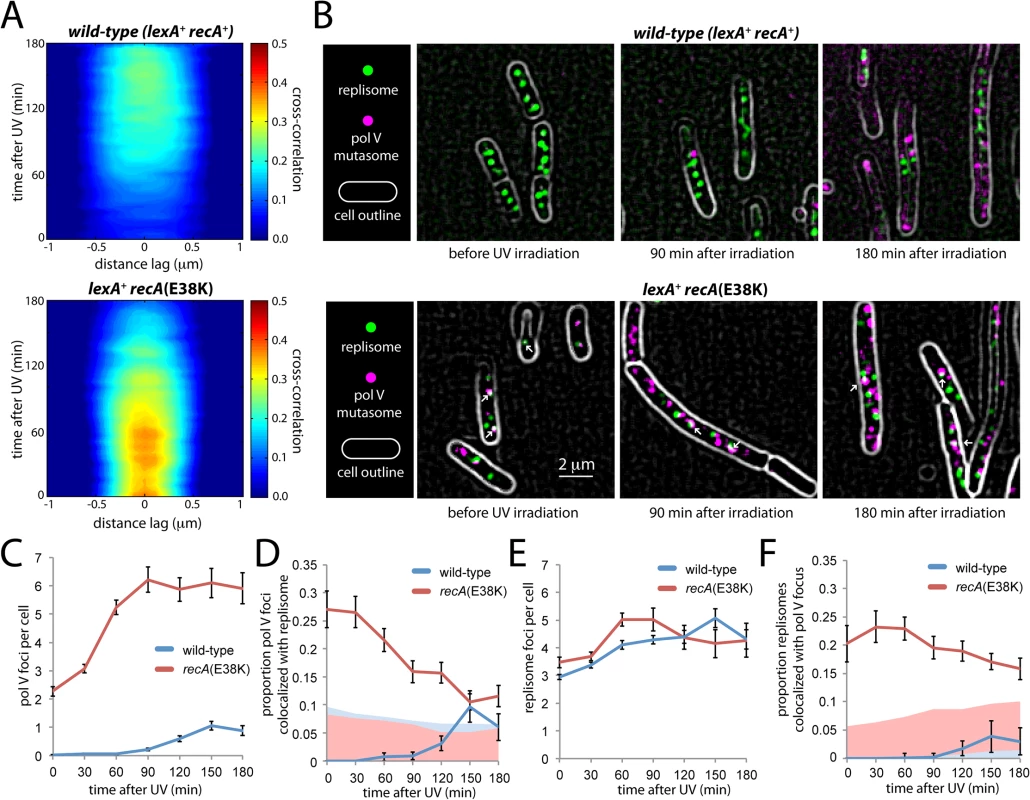
(A) Cross-correlation analysis of DnaX-YPet and UmuC-mKate2 signals from time-lapse measurements. Distance-dependent correlation between DnaX-YPet and UmuC-mKate2 signals are presented as 2D contour plots. Blue areas indicate low correlation, whereas red areas indicate high correlation. Image pairs containing co-localized replisome and mutasome foci produce high cross-correlation values over a short distance range. A peak-enhancing filter was applied to fluorescence images prior to autocorrelation analysis [57]. (B) Average projections of time-sampling movies showing replisome and mutasome foci. Arrows indicate co-localized replisome-mutasome pairs. A peak-enhancing filter was applied to fluorescence images prior to autocorrelation analysis [57]. (C-F) Changes in replisome and mutasome foci following 30 J/m2 UV irradiation. (C) Number of mutasome foci per cell. (D) Proportion of mutasome foci that colocalize with a replisome focus. (E) Number of replisome foci per cell. (F) Proportion of replisome foci that colocalize with a mutasome focus. To probe this co-localization further, we analysed individual cells within the time-sampled movies. Pol V Mut synthesizes DNA at a very low rate (0.3–1 nt·s-1) [11]. It can therefore be assumed that pol V Mut molecules forming active mutasomes must remain immobilized on the DNA for at least a few seconds. These molecules should present as static foci in our fluorescence movies (at 34 ms per frame). Making average projections of these movies allowed us to highlight static foci while blurring both transient DNA binders and mobile membrane-associated molecules into the diffuse cellular background (Fig 8B). Foci that persisted for > 300 ms were identified as static foci.
Static foci were observed in UV-irradiated, wild-type recA+ cells as well as in both untreated and UV-irradiated recA(E38K) cells (Fig 8B). In both backgrounds, the number of static foci increases in response to UV-irradiation in a dose-dependent fashion (Fig 8C). In the recA(E38K) background, a large proportion of static foci co-localize with replisomes in the absence of UV (Fig 8D). These observations strongly support the notion that the static foci we observe represent DNA-bound pol V Mut molecules. From this point forward we will refer to these static foci as mutasome foci.
In the wild-type background, mutasome foci appear in the cytosol 90 min after damage was induced (Fig 8B and 8C), 30 min after the point where UmuC levels begin to increase (Fig 2). On average, cells contained 1.1 ± 0.2 mutasome foci at the peak time of 150 min. Very few of these co-localized with replisomes: the percentage detected as being co-localized fell within the range expected by chance (based on the area of the cell occupied by replisome foci, ~5–10%, Fig 8D). These observations are consistent with the weak cross-correlation signature we observed in the time-lapse analysis (Fig 8A).
In the recA(E38K) strain, pol V Mut is produced constitutively and is evenly distributed throughout the cytosol both before and after inducing damage (Fig 6B, S8 Fig). Prior to UV irradiation, static foci are observed in the cytosol, averaging 2.3 ± 0.2 per cell (Fig 8B and 8C). In contrast to the behavior observed in wild-type cells, a significant proportion of these foci (27 ± 3%, n = 111 cells) co-localize with replisomes (Fig 8D). This observation is consistent with the strong cross-correlation observed between DnaX-YPet and UmuC-mKate2 in the corresponding time-lapse measurements (Fig 8A) and indicates that pol V Mut gains access to replisomes under these conditions. This observation is consistent with the elevated rate of mutagenesis (~ 100-fold) observed in the recA(E38K) (recA730) background in the absence of damage [17]. As seen in the cross-correlation (Fig 8A), we observe that after UV irradiation, the co-localization with replisomes gradually decreases (Fig 8D). By 90 min after UV, relatively few pol V Mut foci colocalize with replisomes (16 ± 2%, n = 84) and by 150 min co-localization (11 ± 1%, n = 90) approaches levels expected to occur by chance (~5–10%, Fig 8D). The proportion of replisomes that co-localized with a static pol V focus remained approximately the same throughout the measurement, indicating that pol V Mut gains similar levels of access to replisomes in both the presence and absence of damage in the recA(E38K) background (Fig 8E and 8F). Analysis of cells grown in shaking culture returned similar results: there is no significant co-localization of mutasomes and replisomes in the wild-type background; a significant proportion of mutasomes do co-localize with replisomes in undamaged recA(E38K) cells; this co-localization gradually diminishes after UV irradiation (S9 Fig).
Overall, our results show that mutasomes form at replisomes in undamaged recA(E38K) cells in which the SOS response is constitutive, but few sites for DNA synthesis exist outside of replisomes. After damage, additional mutasomes are assembled at sites that are spatially distinct from replication forks. In the wild-type background, static foci rarely form at replication forks, clearly questioning the canonical view of mass-action driven binding of pol V at stalled replication forks.
Discussion
This work reveals a new level of regulation in the activation of DNA polymerase V. Previous work has defined two molecular mechanisms that limit the amount of time that pol V has to access DNA templates. The first is the transcriptional, posttranslational and proteolytic regulation of pol V, ensuring that the polymerase accumulates relatively late in the SOS response [4,9]. The second consists of an intrinsic ATPase activity that limits the number of nucleotides incorporated by pol V Mut once TLS commences [14]. The work described here reveals that the UmuC and UmuD proteins are not activated as soon as they appear. Instead, activation is further delayed by sequestration of UmuC (and probably UmuD as well) at the inner cell membrane. The resulting delay constitutes another time-limiting mechanism that functions between the other two (Fig 9). This new layer of regulation is not merely temporal, but also introduces a novel spatial dimension to the system. Release from the membrane requires the conversion of UmuD to UmuD'. These many layers of control help ensure that pol V activation is limited to circumstances where the extent of damage exceeds the cell’s capacity for error-free repair. Once activated, many pol V Mut foci do not co-localize with DNA pol III replisomes, suggesting that pol V Mut may often function at lesion-containing gaps left behind by the replicative polymerase.
Fig. 9. Multiple mechanisms limit pol V activity on DNA. 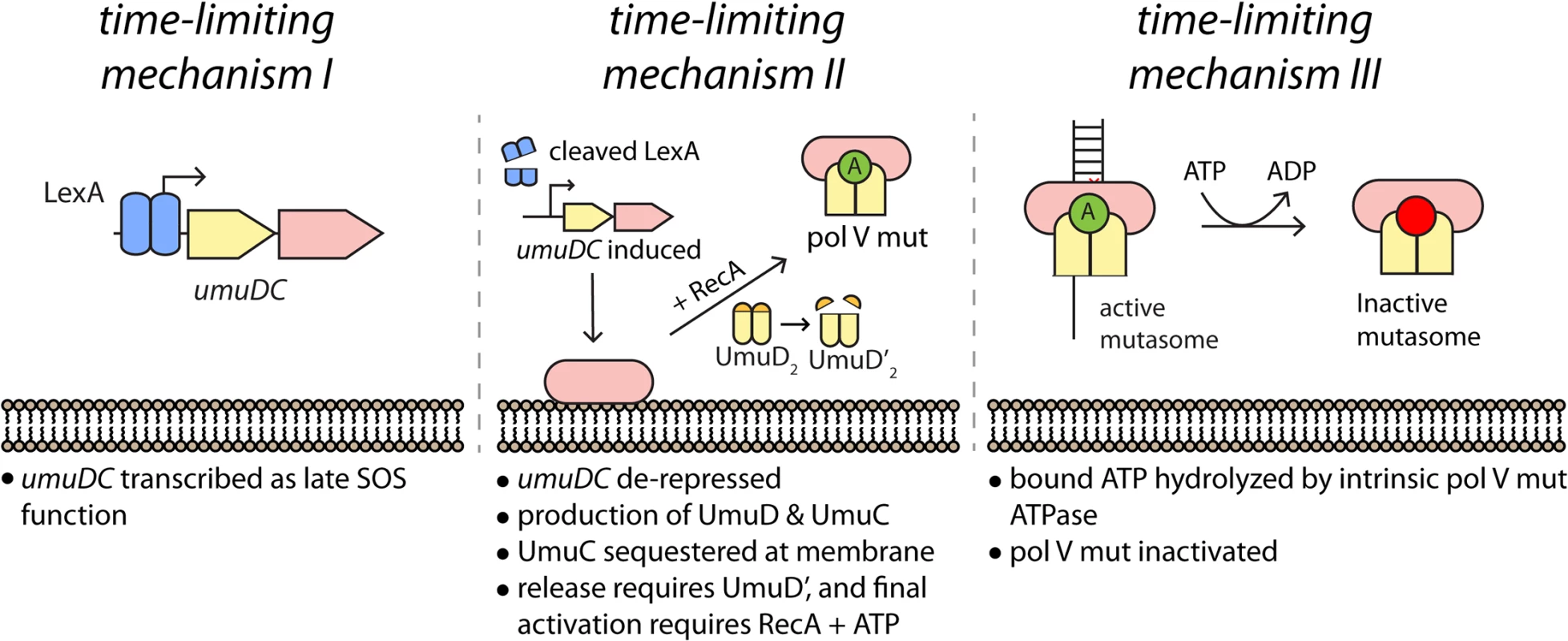
(I) The activity of low fidelity pol V is regulated at the transcriptional level, with gene transcription and translation delayed by tight binding to the umuDC operator by LexA protein. (II) As documented here, activation of pol V is further delayed by sequestration of UmuC (and possibly UmuD) in the membrane. Release and final activation requires the conversion of UmuD to UmuD'. (III) Once activated, the length of time that pol V Mut is operative in TLS DNA replication is dependent on an ATPase activity intrinsic to the enzyme [14]. The symbol “A” represents a molecule of ATP bound to RecA in the activated pol V Mut complex. Time-lapse analyses revealed three distinct phases in the regulation of pol V activity within UV-irradiated cells. In phase I, little UmuC is produced. The few molecules that are present at this stage are primarily associated with the cell membrane. In phase II, the number of UmuC molecules in the cell increases sharply as the umuDC operon is derepressed by cleavage of LexA. Presumably, the majority of other SOS-regulated proteins are also induced during this phase. During phase II, UmuC associates with the cell membrane. In phase III, UmuC is no longer expressed and its cellular levels decrease. During this phase, UmuC gradually enters the cytosol as pol V Mut, forming mutasomes on the DNA at sites spatially distinct from replisomes. The transition from phase II to III requires UmuD cleavage to UmuD'. This represents not only a biochemical switch that allows formation of active pol V Mut, but also a spatial switch that releases UmuC from the membrane.
Spatial organization is a common feature of regulation in eukaryotes, where many processes are tied together by protein scaffolds or segregated into compartments [23–32]. Well-characterized examples are much less common in bacteria. The transient sequestration of UmuC in the bacterial inner membrane provides the first example of spatial regulation for a bacterial DNA processing enzyme. It is interesting to speculate that the inner membrane may be a more general repository of DNA metabolism enzymes that are temporarily not needed but still readily available if required.
Our unexpected observation of prescribed changes in the spatial distribution of UmuC with time after exposure to UV raises new questions about the mechanisms of translesion synthesis in E. coli. The results show that in vivo, UmuC interacts with the cell membrane and is released as soluble pol V following RecA*-mediated conversion of UmuD2 to UmuD′2. How does UmuC become associated with the membrane? When expressed alone, the vast majority of UmuC is naturally insoluble, yet a small fraction can nevertheless be recovered in soluble cell extracts (Fig 7C). The amount of soluble UmuC increases when co-expressed with UmuD′ (Fig 7C) [11,33], suggesting that the insoluble UmuC is not mis-folded, but rather is simply too hydrophobic to enter aqueous solution by itself. It would therefore be reasonable to hypothesize that such hydrophobicity might promote a direct interaction of UmuC with the cell membrane. Our measurements, however, indicate that other factors are involved. For example, the amount of soluble UmuC decreases significantly when co-expressed with non-cleavable UmuD(K97A) (Fig 7C). This observation suggests that full-length UmuD also plays a role in helping to sequester UmuC in an insoluble form, (most likely on the inner membrane), an issue that is under further investigation.
It is possible that other DNA damage-induced proteins might also be involved in sequestering UmuC at the membrane. Thirty years ago, evidence was published for DNA damage-dependent association of RecA with the cell membrane [34]. Both UmuC and UmuD are known to interact with RecA [35–37], thus it would be interesting to determine if membrane-localized RecA also plays a role in sequestering UmuC. Because of the many roles that RecA plays in the regulation of pol V, however, examining this possibility would be extremely challenging experimentally and is beyond the scope of the current study.
How might membrane association provide control over mutagenic pol V activity? At the simplest level, sequestration of the bulk of UmuC at the membrane would leave few pol V Mut molecules free in the cytosol with access to DNA. A delay between production of UmuC and formation of cytosolic pol V Mut provides an expanded window of time in which error-free DNA repair proteins (which have weaker affinity LexA-binding sites and thus are highly unlikely to be co-expressed with UmuC) can repair the damaged DNA before mutagenic pol V lesion bypass takes place. Ideally, one would directly examine the links between membrane-sequestration of UmuC and pol V-dependent mutagenesis rates, independently of biochemical switches that regulate pol V activity. Our results show, however, that at least one of these biochemical switches, the conversion of UmuD to UmuD', is intimately tied to changes in UmuC localization. Currently, no mutants exist that would allow for separation of these functions. What is clear, however, is that the umuD(K97A) mutation, which prevents release of pol V from the membrane, strongly inhibits pol V-induced mutagenesis [12].
In UV-irradiated wild-type recA+ cells, pol V mutasomes rarely co-localize with DnaX-YPet at replisomes. Co-localization is frequently observed for recA(E38K) cells exhibiting constitutive RecA* activity, but decreases after UV treatment. Previous models for TLS have posited that at stalled replisomes, TLS polymerases displace pol III core (αεθ) from the β sliding clamp, while the rest of replisome, including the clamp loader complex, remains associated [38]. Our observations suggest that for pol V Mut, such an ‘alternative replisome’ mechanism is the exception rather than the rule.
Recent elucidation of pol V Mut as a mobile mutasome complex when first activated [13, 39] provides a facile mechanism for TLS-mediated repair of gaps that are not near the replisome. The fact that UmuC co-localizes with replisomes in an undamaged recA(E38K) strain is in excellent agreement with the suggestion that the pol V-dependent spontaneous mutator phenotype of recA(E38K) strains is due to a competition between pol III and pol V at a nascent replication fork [40], with most pol V mutator activity occurring on the lagging strand [41]. In an irradiated cell, pol III motion is blocked by the presence of a UV-induced photoproduct, resulting in the replisome dissociating and re-initiating synthesis downstream of the lesion [20,42]. Such a mechanism would leave a lesion-containing single-stranded gap in the chromosome, complete with a β2 sliding clamp left behind by the departing replisome. Translesion DNA synthesis in such a gap may represent a major function of pol V Mut.
Experimental procedures
A complete list of the Escherichia coli strains used in this study appears in Table 1.
Tab. 1. Strains used in this study. 
Strain and plasmids for mutagenesis assays
The E. coli K-12 strain RW118 [full genotype:thr-1 araD139 Δ(gpt-proA)62 lacY1 tsx-33 supE44 galK2 hisG4 rpsL31 xyl-5 mtl-1 argE3 thi-1 sulA211] and the isogenic ΔumuDC595::cat strain, RW120, have been described previously [43]. An isogenic strain, RW1480, expressing umuC-mKate2:: Kan was made by P1 transduction of the allele from EAW191 into RW120 by selecting for kanamycin resistance and screening for chloramphenicol sensitivity. RW574 and RW578 are isogenic recA(E38K) lexA51(Def) derivatives of RW118 and RW120 respectively [44]. An isogenic recA(E38K) lexA51(Def) strain, RW1314, expressing umuC-mKate2:: Kan was made by P1 transduction of the allele from EAW191 into RW578 by selecting for kanamycin resistance and screening for chloramphenicol sensitivity.
Spontaneous and chemically induced mutagenesis assays
Bacterial cultures were grown overnight in LB medium containing the appropriate antibiotics. Aliquots (1 ml) were centrifuged and resuspended in an equal volume of SM buffer [45]. The ability of each strain to promote Umu-dependent chemically induced mutagenesis was judged by plating 100 μl aliquots on minimal agar plates supplemented with a trace amount of histidine (1 μg/ml) [46]. Immediately after plating, 5 μl of a 1 : 5 dilution of methyl methane sulfonate (MMS, Sigma) in dimethyl sulfoxide (DMSO, Sigma) was applied to a small sterile disk in the center of the plate. His+ revertants were scored after 4 days of incubation at 37°C. The number of His+ revertants is expressed as the mean number of MMS-induced mutants per plate after subtracting the mean number of spontaneously arising His+ revertants on plates that were not exposed to MMS and were obtained from 5 individual cultures plated in triplicate for each assay.
Quantitative UV-induced mutagenesis assays
Spontaneous and damage induced reversion of the hisG4 locus was assayed as previously described [44]. Cells were grown overnight at 37°C in LB media. The overnight culture was diluted 1 : 100 in fresh LB media and grown until an OD600 ~0.4. Cells were harvested by centrifugation and resuspended in an equal volume of SM buffer [45]. Cells were irradiated with UV-light (254nm) in a petri dish at a UV-fluence of 0.5 J/m2 per second. Aliquots were removed at each UV dose and serial dilutions of the culture spread in triplicate on minimal agar plates supplemented with a trace amount of histidine (1 μg/ml). On these plates, less than 1000 viable bacteria grew on the low level of histidine to form small detectable colonies after 2 days. When ~ 4x107 bacteria were seeded, they grew to form a lawn, concomitantly exhausting the low level of histidine. His+ mutants grew up through the lawn and were counted after 4 days. The number of pre-existing spontaneous mutants was determined by seeding on minimal plates lacking histidine. The UV-induced mutation frequencies were calculated as previously described [47] and takes into account spontaneously arising mutants, as well as any cell killing that reduces the number of pre-existing His+ mutants in the culture. Experiments were carried out at least three times and were performed under yellow light to avoid unwanted photoreactivation.
Strains for expression and solubility assays
The expression strain, RW644 (BL21(λDE3) ΔpolB::Spec ΔdinB::Zeo ΔumuDC::Erm) has been described previously [11]. TCH03 is isogenic with RW644 and was made by transducing ΔpolB770::Kan from JW0059 (E. coli Genetic Stock Center) into RW644 selecting for kanamycin resistance and screening for sensitivity to spectinomycin. The wild-type umuC gene was chemically synthesized by Genscript (Piscatawy, NJ) and cloned into pET22b+ as an ~1.3kb NdeI-XhoI fragment, to generate pJM1113. A plasmid expressing mKate2-UmuC was generated by the chemical synthesis of a DNA fragment containing the 3' end of umuC in frame with the mKate2 gene (Genscript), and was cloned into the BamHI-XhoI sites of pJM1113 to generate pJM1115. pJM1116 and pJM1117 are low-copy spectinomycin resistant plasmids that constitutively express UmuD' or UmuD(K97A) from the RecA(Oc) promoter. They were constructed by subcloning the umuD' gene from pJM105 [48] as an NdeI-PstI fragment into pJM1071 and the umuD(K97A) gene as an NdeI-BglII fragment from pEC69 [49] into NdeI-BamHI digested pJM1071, respectively.
Western blotting for UmuC expression and solubility
Intracellular levels of UmuD' and UmuC proteins was determined by western blotting using affinity purified polyclonal UmuD' and UmuC antisera using the protocols previously described [15]. Overnight cell cultures were diluted 1 : 100 in fresh media and grown at 37°C until they reached an OD600 ~0.5. Cultures were centrifuged and resuspended in 8 ml of Buffer A (50mM Tris pH 7.5, 300 mM NaCl and 10% glycerol). Cells were lysed by sonication on ice for a total processing time of 3 min. The suspension was then centrifuged at 35,000 RPM for 30 minutes and the supernatant saved as the soluble cell extract. Protein extracts were separated on a NuPAGE 4–12% Bis-Tris gel and transferred to a Novex PVDF membrane. The membrane was incubated overnight with a 1 : 1000 dilution of affinity purified polyclonal rabbit antisera raised against UmuC. The next day, it was washed twice and subsequently incubated for 90 minutes with a 1 : 10,000 dilution of goat anti-rabbit IgG conjugated to alkaline phosphatase (Applied Biosystems; cat # T2191). Finally, the membrane was treated with a 1 : 50 dilution of Tropix CDP-Star substrate (Applied Biosystems) in 1X assay buffer for 10 min. Proteins were visualized by exposing the membrane to X-ray film for various time periods.
Strain and plasmids for electron microscopy assays
Strain RW1394 is isogenic with RW578 but also carries the ΔrcsA::Kan and Δlon::Tet alleles that were sequentially transduced from SG22094 and SG21047 respectively into RW578. pRW134 expressing UmuD'C and pJM155 expressing a non-cleavable cleavage site mutant of UmuD together with UmuC, are derivatives of the low-copy number spectinomycin resistant vector, pGB2 [50] and have been described previously [49,51].
Strain and plasmids for fluorescence assays
EAW282 is E. coli MG1655 [52] dnaX-YPet, umuC-mKate2. It was made by P1 transduction of JJC5945 to umuC-mKate2 with P1 grown on EAW191. JJC5945 is MG1655 dnaX-YPet and was obtained from Benedicte Michel. EAW191 was made by replacing the MG1655 umuC gene in its native chromosome location with a umuC gene with a C-terminal 11 amino acid spacer followed by mKate2 and a mutant FRT-Kanamycin resistance-wt FRT cassette, via λ RED recombination [53]. The sequence encoding the 11 amino acid spacer (TCCGCTGGCTCCGCTGCTGGTTCTGGCGAGTTC) is that used by Rodrigo Reyes-Lamothe [22] with changes for better codon use and the EcoRI site removed. EAW307 is E. coli MG1655 sulA–, umuC-mKate2, recA(E38K), dnaX-YPet. EAW191 was transduced to sulA− with P1 grown on EAW13. EAW13 is MG1655 with a FRT-Kanamycin resistance-FRT cassette replacing the sulA gene between the SphI and HpaI sites.
The sulA−umuC-mKate2 strain, designated EAW297, was P1 transduced to recA(E38K) with P1 grown on EAW287. EAW287 is MG1655 sulA–, recA(E38K) with a mutant FRT-Kanamycin resistance-wt FRT located after the C-terminus of the downstream recX gene. MG1655 sulA–, umuC-mKate2, recA(E38K) was transduced to dnaX-YPet with P1 grown on JJC5945. EAW309 is E. coli MG1655 umuC-mKate2 sulA - lexA(Def). It was made by transducing EAW297 with P1 grown on EAW26. EAW26 was made by lambda RED recombination of EAW13 to replace the wild-type lexA gene with a deficient lexA that had the gene segment between the PshAI and AflIII sites replaced by the KanR gene. EAW329 is MG1655 umuD(K97A) umuC-mKate2 and was made by replacing the umuDC genes with umuD(K97A) umuC-mKate2 by lambda RED recombination. Both have the same arrangement of umuC-spacer-mKate2-mutant FRT-KanR-wt FRT as EAW191.
The plasmid pBAD-LacY-eYFP was a generous gift from J. T. Mika (University of Groningen). It was constructed by inserting lacY-eYFP (encoding the lactose transporter LacY fused to the N-terminus of eYFP) into the plasmid pBAD/His (Invitrogen) and allows for L-arabinose-inducible expression of LacY-eYFP. The plasmid pBAD-mKate2 was constructed by inserting mKate2 (including an 11 amino acid linker at its N-terminus, sequence TCCGCTGGCTCCGCTGCTGGTTCT-GGCGAGTTC) into pBAD/His (Invitrogen). The linker-mKate2 fragment was created by PCR amplifying (KOD polymerase, Novagen) the mKate2 gene (Evrogen) using forward primer containing the linker sequence (SAGSAAGEF; ATCCGAGCTCGAG ATG TCG GCT GGC TCC GCT GCT GGT TCT GGC GAA TTC ATG GTG AGC GAG CTG ATT AAG GAG) and reverse primer (GTT CCT ATT CTC TAG AAA CTA TAG GAA CTT CTC ATC TGT GC). The amplified fragment and pBAD-myc-hisB were digested with XhoI and XbaI (New England Biolabs) and ligated together with T4 DNA ligase (New England Biolabs, 16°C, overnight) followed by transformation into E coli DH5α. Plasmid DNA was isolated from bacterial colonies containing the pBAD-myc-hisB with the ligated linker-mKate2 construct and sequenced to verify the sequence of the linker (GATC biotech).
Electron microscopy
E. coli strain RW1394 (relevant genotype; recA(E38K) lexA51(Def) ΔumuDC ΔrcsA Δlon) harboring low-copy-number spectinomycin resistant plasmids pGB2 (vector) [50], pRW134 (recombinant UmuD'C) [51], or pJM155 (non-cleavable cleavage site UmuD mutant, UmuD(C24D-G25D)/UmuC) [49] were grown overnight at 37°C in Davis and Mingioli (DM) minimal media. The overnight culture was diluted 1 : 100 into 25ml fresh DM media and grown approximately 4 h at 37°C until an OD600 ~0.4–0.6. 10 ml of the culture was transferred to a sterile petri-dish and irradiated with 10 J/m2 UV-light (254nm) at a fluence of 0.5 Jm-2/sec and then returned to a new culture flask. Both unirradiated and irradiated cultures were incubated at 37°C for a further 45 minutes before 1 ml of the culture was transferred to a microfuge tube and centrifuged at ~14,000 x g for 5 mins. The supernatant was decanted and the pellet resuspended in an equal volume of PBS containing 0.5% glutaraldehyde. The suspension was kept on ice for 25 mins before centrifugation and washed 2x with an equal volume of PBS. Cells were embedded in resin and sectioned by Jan Endlich (JFE Enterprises, Beltsville, MD) under a custom service contract. This process entailed lightly centrifuging the cells into a pellet, which were dehydrated in a series of ethanol solutions, (30% EtOH, 50% EtOH and 70% EtOH). Cells were then infiltrated 1 : 1 with LR White to 70% EtOH, 1 : 2 LR White to 70% EtOH, 1 : 3 LR White to 70% EtOH and 3 changes of pure resin. The infiltrated samples were then transferred in gelatin capsules with fresh LR White and then cured for 48 hours at 50°C. Blocks were removed and gelatin capsule sections were cut approximately 60–80nm thin. Sectioned EM grids were returned to us and were placed on 50 μl droplets of sterile PBS containing 0.5% BSA and 0.1% gelatin (PBSBG) for 3 hours at room temperature to reduce any non-specific binding of the UmuC antibodies. After which time, the grids were placed on a 25 μl droplet of a 1 : 20 dilution of affinity purified polyclonal rabbit antisera diluted in PBSBG and incubated overnight at 4°C. Grids were washed 3x by placing on sequential drops of 50 μl PBSG for 10 mins each. Grids were then placed on a 25 μl droplet of a 1 : 40 dilution of goat-anti-rabbit antibody (in PBSBG) conjugated to 30nm gold particles (Abcam, cat # ab119178) and incubated overnight at 4°C. The grids were washed 3x with PBSG for 10 mins each and dried by blotting on filter paper. Grids were then returned to Jan Endlich (JFE enterprises, Beltsville, MD), who stained them for 5 minutes with 2% aqueous uranyl acetate and then examined and imaged the cells with a Ziess EM 10C transmission electron microscope.
Fluorescence microscopy
We constructed a wide-field single-molecule fluorescence microscope by coupling high power laser excitation into a commercially available inverted fluorescence microscope body (IX-81, Olympus) equipped with a 1.49 NA 100x objective and a 512 × 512 pixel EM-CCD camera (C9100-13, Hamamatsu). Excitation light was provided continuous-wave optically pumped semidiode lasers (Sapphire LP, Coherent) of wavelength 514 nm (150 mW max. output) and 568 nm (200 mW max. output). For imaging UmuC-mKate2 fusions we used 568 nm excitation light at high intensity (180–1800 W.cm-2) and collected light emitted between 610–680 nm (ET 645/75m filter, Chroma). For imaging DnaX-YPet we used 514 nm laser excitation at lower power (10 W.cm-2) and collected between 525–555 nm (ET540/30m filter, Chroma). For LacY-eYFP imaging we used 514 nm excitation at high power (1800 W.cm-2). Image analysis was performed with ImageJ [54].
For imaging we used glass coverslips derivatized with 3-aminopropyl triethoxy silane (APTES, Sigma). Coverslips were first cleaned by sonicating for 60 min in 5M KOH and then thoroughly washing with water. The coverslips were then treated for 2 h with 2% APTES in water before being washed thoroughly with water, then acetone and dried before assembling flow cells. Our flow cell design was that used in reference [55]. Typical channel dimensions were 3 × 30 × 0.03 mm (length × width × height).
UV irradiation and imaging
For most imaging experiments, cells were grown at 37°C in EZ rich defined medium (Teknova) that included 0.2% (w/v) glucose. Cells containing pBAD-LacY-eYFP were grown in EZ that contained 0.2%(v/v) glycerol and 0.001% (w/v) L-arabinose. For time-lapse and time-sampling imaging we mounted APTES flow cells to our microscope, maintaining the temperature at 37°C by a combination of stage heating and objective lens heating. Cells were grown in shaking culture until reaching mid-log phase (OD600 ~ 0.5) before being loaded into flow cells by pulling with a syringe. The inlet was then placed into fresh medium that was constantly aerated using an aquarium pump. Medium was then pulled through the flow cell using a syringe pump, at a rate of 30 μl/min. Cells were irradiated in situ with 254 nm UV light from a mercury lamp (UVP) at a fluence of 1–100 J.m-2.
Super-resolution images of UmuC-mKate2 and LacY-eYFP
Super-resolution reconstructions (e.g. PALM images) require that fluorescent protein molecules are observed one at a time as spatially separate foci. For typical, densely labeled samples this requires the use of a photoswitchable fluorescent protein. Because of the low cellular levels of UmuC-mKate2 in the wild-type (recA+ lexA+) cells, however, it was possible to reconstruct super-resolution images without photoswitching. For LacY-eYFP we used a strategy described previously [56], driving molecules into a non-bleached dark state before imaging spontaneous single-molecule returns to the fluorescent state (4000 × 34 ms frames).
To reconstruct images we identified single-molecule foci by applying a discoidal averaging filter [57] to the movies and selecting those peaks above a defined threshold value. Returning to the untreated movies, we then fit each focus with a 2D Gaussian function in order to accurately determine their center positions (non-linear least squares fitting with Levenberg-Marquadt algorithm). We reconstructed images by drawing each position as 2D Gaussian-shaped spots as described by the formula:
In this way the volume under each spot remained constant, whereas the width was determined by the fitting error in x and y (σx and σy).
Determining cellular concentrations of UmuC-mKate2
It is possible to determine the number of molecules of a fluorescent protein in a cell by dividing its total fluorescence by the average signal arising from individually measured single molecules. When measuring cells that only contain a small number of fluorescent proteins, however, individual fluorescence images tend to contain a lot of noise. We therefore decided on a different strategy for analysis of our time-sampling data. Having removed contributions from background fluorescence, we fit the loss of fluorescence signal due to photobleaching of mKate2 with an exponential decay function and used the fit amplitudes as a measure for the initial fluorescence. In this way, the total fluorescence of the cell is derived from measurements over all 296 frames of the movie, reducing the contribution of noise dramatically. The concentration of UmuC-mKate2 could then be determined for each cell using its volume as calculated from its corresponding bright field image.
For each all fluorescence movies we first flattened the images to correct for uneven illumination profile of the excitation light. To produce and of the illumination beam we averaged together each slice from all 70 movies recorded during a 3h UV-damage experiment, smoothed the resulting image by Gaussian blurring (10 pixel radius) and normalised the intensity as a fraction of the brightest pixel. This showed that illumination was already relatively flat (at the corners of the image was 70% as intense as that at the centre of the image). We then divided each movie by this “beam” movie.
For each cell we measured the change in mean fluorescence per pixel due to photobleaching. The raw data included contributions not only from UmuC-mKate2, but also cellular autofluorescence (S1A Fig) and background fluorescence from the glass coverslip (S1B Fig), which were removed prior to quantitating mKate2 fluorescence. Under our conditions, mKate2 follows a single-exponential photobleaching decay with time constant t ≈ 5 s (S1C Fig). We quantified the total mKate2 fluorescence of individual cells by fitting the amplitude of each decay curve. We note that by focussing on the middle portion of the cell, signal arising from the top and bottom of the cell may not be completely captured and thus measured signals may be slightly underestimated. Cell outlines were assigned using MicrobeTracker [58].
To calibrate our measurements for number of UmuC-mKate2 molecules per cell, we measured the fluorescence signals of individual molecules. To do this, we examined the final 50 frames of our movies. Here most of the cellular fluorescence had been bleached, however individual molecules could occasionally be observed as foci as they spontaneously returned to a bright state. We fit these foci with 2D Gaussian functions and made a histogram of the integrated volumes under each curve (S1D Fig). We could then calibrate our cellular measurements by dividing the total fluorescence of each cell by the mean value for single molecules.
Imaging and analysis of cells grown in shaking culture
Cells were grown in shaking culture at 37°C in EZ rich defined medium (Teknova) that included 0.2% (w/v) glucose. To induce DNA damage cells were grown until reaching mid-log phase (OD600 ~ 0.5), placed between two quartz sheets separated by 170 μm spacers and irradiated with 254 nm UV light from a mercury lamp (UVP) at a fluence of 10 J.m-2. Cells were then put back into shaking culture in preparation for imaging.
Microscopy samples were prepared by transferring 30 μl of cell suspension onto an APTES-treated coverslip and placing a clean coverslip on top. The sample was allowed to stand for 1–2 min, before excess medium was removed by gently pressing down on the top coverslip. Imaging was started within 3 min. For each sample, 10 fields were imaged taking 5–10 min in total. Cells grew and divided at similar rates before and after imaging.
In order to characterize the spatial distributions of UmuC-mKate2 foci we produced maps of their cellular locations. We achieved this by identifying every focus that presented during a movie and pinpointing its centre position using 2D Gaussian fitting. The position of each focus was then mapped to its location along the long and short axes of the cell using MicrobeTracker [58]. For each time point of our experiment, we produced a “location map” by displaying the cellular locations of all foci from all cells as Gaussian-shaped spots on a “cell” of average width and length for that time point. To correct for variation in sample size, the intensity of each resulting image was then divided by the total number of cells analyzed.
Analysis of fluorescent protein distribution by autocorrelation and cross-correlation
To analyze the distribution of UmuC-mKate2 in time-lapse experiments we measured autocorrelation functions across the horizontal axis of the images, which was perpendicular to the direction of flow. Images were subjected to peak filtering prior to analysis [57]. As cellular UmuC-mKate2 levels were low, foci were sparsely distributed in images. We therefore averaged along the vertical axis of each image in 20 μm wide bins. Autocorrelation was measured over a 1 μm distance lag range for each bin in each image and the resulting autocorrelation functions averaged. These autocorrelation functions were plotted as a function of time after UV exposure as a 2D contour plot. Each autocorrelation function was normalized against the mean cellular fluorescence intensity (I), such that the autocorrelation value at zero lag and Imax = 1. Cross-correlation functions were determined similarly, except that images of both DnaX-YPet and UmuC-mKate2 were used as input and no intensity-based normalization was used. All correlations were measured using the xcorr function in Matlab.
Supporting Information
Zdroje
1. Kornberg A, Baker TA (1992) DNA Replication. New York: W. H. Freeman & Co.
2. Bloom LB, Chen X, Fygenson DK, Turner J, O'Donnell M, et al. (1997) Fidelity of Escherichia coli DNA polymerase III holoenzyme. The effects of beta, gamma complex processivity proteins and epsilon proofreading exonuclease on nucleotide misincorporation efficiencies. J Biol Chem 272 : 27919–27930. 9346941
3. Borden A, O'Grady PI, Vandewiele D, Fernandez de Henestrosa AR, Lawrence CW, et al. (2002) Escherichia coli DNA Polymerase III Can Replicate Efficiently past a T-T cis-syn Cyclobutane Dimer if DNA Polymerase V and the 3' to 5' Exonuclease Proofreading Function Encoded by dnaQ Are Inactivated. J Bacteriol 184 : 2674–2681. 11976296
4. Sommer S, Boudsocq F, Devoret R, Bailone A (1998) Specific RecA amino acid changes affect RecA-UmuD'C interaction. Mol Microbiol 28 : 281–291. 9622353
5. Tang M, Shen X, Frank EG, O'Donnell M, Woodgate R, et al. (1999) UmuD'(2)C is an error-prone DNA polymerase, Escherichia coli pol V. Proc Natl Acad Sci U S A 96 : 8919–8924. 10430871
6. Kato T, Shinoura Y (1977) Isolation and characterization of mutants of Escherichia coli deficient in induction of mutations by ultraviolet light. Mol Gen Genet 156 : 121–131. 340898
7. Corzett CH, Goodman MF, Finkel SE (2013) Competitive Fitness During Feast and Famine: How SOS DNA Polymerases Influence Physiology and Evolution in Escherichia coli. Genetics 194 : 409–420. doi: 10.1534/genetics.113.151837 23589461
8. MacLean RC, Torres-Barcelo C, Moxon R (2013) Evaluating evolutionary models of stress-induced mutagenesis in bacteria. Nat Rev Genet 14 : 221–227. doi: 10.1038/nrg3415 23400102
9. Fernandez De Henestrosa AR, Ogi T, Aoyagi S, Chafin D, Hayes JJ, et al. (2000) Identification of additional genes belonging to the LexA regulon in Escherichia coli. Mol Microbiol 35 : 1560–1572. 10760155
10. Frank EG, Ennis DG, Gonzalez M, Levine AS, Woodgate R (1996) Regulation of SOS mutagenesis by proteolysis. Proc Natl Acad Sci U S A 93 : 10291–10296. 8816793
11. Karata K, Vaisman A, Goodman MF, Woodgate R (2012) Simple and efficient purification of Escherichia coli DNA polymerase V: cofactor requirements for optimal activity and processivity in vitro. DNA Repair (Amst) 11 : 431–440.
12. Nohmi T, Battista JR, Dodson LA, Walker GC (1988) RecA-mediated cleavage activates UmuD for mutagenesis: mechanistic relationship between transcriptional derepression and posttranslational activation. Proc Natl Acad Sci U S A 85 : 1816–1820. 3279418
13. Jiang Q, Karata K, Woodgate R, Cox MM, Goodman MF (2009) The active form of DNA polymerase V is UmuD'(2)C-RecA-ATP. Nature 460 : 359–363. doi: 10.1038/nature08178 19606142
14. Erdem AL, Jaszczur M, Bertram JG, Woodgate R, Cox MM, et al. (2014) DNA polymerase V activity is autoregulated by a novel intrinsic DNA-dependent ATPase. Elife 3: e02384. doi: 10.7554/eLife.02384 24843026
15. Woodgate R, Ennis DG (1991) Levels of chromosomally encoded Umu proteins and requirements for in vivo UmuD cleavage. Mol Gen Genet 229 : 10–16. 1654503
16. Frank EG, Gonzalez M, Ennis DG, Levine AS, Woodgate R (1996) In vivo stability of the Umu mutagenesis proteins: a major role for RecA. J Bacteriol 178 : 3550–3556. 8655553
17. Witkin EM, McCall JO, Volkert MR, Wermundsen IE (1982) Constitutive expression of SOS functions and modulation of mutagenesis resulting from resolution of genetic instability at or near the recA locus of Escherichia coli. Mol Gen Genet 185 : 43–50. 6211591
18. Vlasic I, Simatovic A, Brcic-Kostic K (2011) Genetic requirements for high constitutive SOS expression in recA730 mutants of Escherichia coli. J Bacteriol 193 : 4643–4651. doi: 10.1128/JB.00368-11 21764927
19. Schlacher K, Cox MM, Woodgate R, Goodman MF (2006) RecA acts in trans to allow replication of damaged DNA by DNA polymerase V. Nature 442 : 883–887. 16929290
20. Yeeles JT, Marians KJ (2011) The Escherichia coli replisome is inherently DNA damage tolerant. Science 334 : 235–238. doi: 10.1126/science.1209111 21998391
21. Indiani C, Patel M, Goodman MF, O'Donnell ME (2013) RecA acts as a switch to regulate polymerase occupancy in a moving replication fork. Proc Natl Acad Sci U S A 110 : 5410–5415. doi: 10.1073/pnas.1303301110 23509251
22. Reyes-Lamothe R, Sherratt DJ, Leake MC (2010) Stoichiometry and architecture of active DNA replication machinery in Escherichia coli. Science 328 : 498–501. doi: 10.1126/science.1185757 20413500
23. Elbaz Y, Schuldiner M (2011). Staying in touch: the molecular era of organelle contact sites. Trends Biochem Sci 36 : 616–623. doi: 10.1016/j.tibs.2011.08.004 21958688
24. Fu MM, Holzbaur ELF (2014). Integrated regulation of motor-driven organelle transport by scaffolding proteins. Trends Cell Biol 24 : 564–574. doi: 10.1016/j.tcb.2014.05.002 24953741
25. Kim YJ, Hernandez MLG, Balla T (2013). Inositol lipid regulation of lipid transfer in specialized membrane domains. Trends Cell Biol 23 : 270–278. doi: 10.1016/j.tcb.2013.01.009 23489878
26. Langeberg LK, Scott JD (2015). Signalling scaffolds and local organization of cellular behaviour. Nature Rev Mol Cell Biol 16 : 232–244.
27. Schuh AL, Audhya A (2014). The ESCRT machinery: From the plasma membrane to endosomes and back again. Crit Rev Biochem Mol Biol 49 : 242–261. doi: 10.3109/10409238.2014.881777 24456136
28. Sweetlove LJ, Fernie AR (2013). The Spatial Organization of Metabolism Within the Plant Cell. Annu Rev Plant Biol, 64 : 723–746. doi: 10.1146/annurev-arplant-050312-120233 23330793
29. Tutucci E, Stutz F (2011). Keeping mRNPs in check during assembly and nuclear export. Nature Rev Mol Cell Biol 12 : 376–383.
30. Xu HX, Ren DJ (2015). Lysosomal Physiology. Annu Rev Physiol 77 : 57–80. doi: 10.1146/annurev-physiol-021014-071649 25668017
31. Zattas D, Hochstrasser M (2015). Ubiquitin-dependent protein degradation at the yeast endoplasmic reticulum and nuclear envelope. Crit Rev Biochem Mol Biol 50 : 1–17. doi: 10.3109/10409238.2014.959889 25231236
32. Chan YW, West SC (2014) Nat Commun 11 : 4844.
33. Bruck I, Woodgate R, McEntee K, Goodman MF (1996) Purification of a soluble UmuD'C complex from Escherichia coli: Cooperative binding of UmuD'C to single-stranded DNA. Journal of Biological Chemistry 271 : 10767–10774. 8631887
34. Garvey N, St John AC, Witkin EM (1985). Evidence for RecA protein association with the cell membrane and for changes in the levels of major outer membrane proteins in SOS-induced Escherichia coli cells. J Bacteriol 163 : 870–876. 3897198
35. Frank EG, Hauser J, Levine AS, Woodgate R (1993). Targeting of the UmuD, UmuD', and MucA' mutagenesis proteins to DNA by RecA protein. Proc Natl Acad Sci U S A 90 : 8169–8173. 8367479
36. Frank EG, Cheng N, Do CC, Cerritelli ME, Bruck I, Goodman MF, Egelman EH, Woodgate R, Steven AC (2000). Visualization of two binding sites for the Escherichia coli UmuD'(2)C complex (DNA pol V) on RecA-ssDNA filaments. J Mol Biol 297 : 585–597. 10731413
37. Gruber AJ, Erdem AL, Sabat G, Karata K, Jaszczur MM, Vo DD, Olsen TM, Woodgate R, Goodman MF, Cox MM (2015). A RecA protein surface required for activation of DNA polymerase V. PLoS Genet 11: e1005066. doi: 10.1371/journal.pgen.1005066 25811184
38. Langston LD, Indiani C, O'Donnell M (2009). Whither the replisome: emerging perspectives on the dynamic nature of the DNA replication machinery. Cell Cycle 8 : 2686–2691. 19652539
39. Patel M, Jiang Q, Woodgate R, Cox MM, Goodman MF (2010). A new model for SOS-induced mutagenesis: how RecA protein activates DNA polymerase V. Crit Rev Biochem Mol Biol 45 : 171–184. doi: 10.3109/10409238.2010.480968 20441441
40. Fijalkowska IJ, Dunn RL, Schaaper RM (1997) Genetic requirements and mutational specificity of the Escherichia coli SOS mutator activity. J Bacteriol 179 : 7435–7445. 9393709
41. Fijalkowska IJ, Schaaper RM, Jonczyk P (2012) DNA replication fidelity in Escherichia coli: a multi-DNA polymerase affair. FEMS Microbiol Rev 36 : 1105–1121. doi: 10.1111/j.1574-6976.2012.00338.x 22404288
42. Yeeles JT, Marians KJ (2013) Dynamics of leading-strand lesion skipping by the replisome. Mol Cell 52 : 855–865. doi: 10.1016/j.molcel.2013.10.020 24268579
43. Ho C, Kulaeva OI, Levine AS, Woodgate R (1993). A rapid method for cloning mutagenic DNA repair genes: isolation of umu-complementing genes from multidrug resistance plasmids R391, R446b, and R471a. J Bacteriol 175 : 5411–5419. 8366028
44. Mead S, Vaisman A, Valjavec-Gratian M, Karata K, Vandewiele D, et al. (2007) Characterization of polVR391: a Y-family polymerase encoded by rumA'B from the IncJ conjugative transposon, R391. Mol Microbiol 63 : 797–810. 17302804
45. Miller JH (1992) A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press.
46. Davis BD, Mingioli ES (1950) Mutants of Escherichia coli requiring methionine or vitamin B12. JBacteriol 60 : 17–28.
47. Sedgwick SG, Bridges BA (1972) Evidence for indirect production of DNA strand scissions during mild heating of Escherichia coli. J Gen Microbiol 71 : 191–193. 4556972
48. Gonzalez M, Frank EG, McDonald JP, Levine AS, Woodgate R (1998) Structural insights into the regulation of SOS mutagenesis. Acta Biochim Pol 45 : 163–172. 9701508
49. McDonald JP, Frank EG, Levine AS, Woodgate R (1998) Intermolecular cleavage by UmuD-like mutagenesis proteins. Proc Natl Acad Sci U S A 95 : 1478–1483. 9465040
50. Churchward G, Belin D, Nagamine Y (1984) A pSC101-derived plasmid which shows no sequence homology to other commonly used cloning vectors. Gene 31 : 165–171. 6098521
51. Szekeres ES Jr., Woodgate R, Lawrence CW (1996) Substitution of mucAB or rumAB for umuDC alters the relative frequencies of the two classes of mutations induced by a site-specific T-T cyclobutane dimer and the efficiency of translesion DNA synthesis. J Bacteriol 178 : 2559–2563. 8626322
52. Blattner FR, Plunkett G 3rd, Bloch CA, Perna NT, Burland V, et al. (1997) The complete genome sequence of Escherichia coli K-12. Science 277 : 1453–1462. 9278503
53. Huang LC, Wood EA, Cox MM (1997) Convenient and reversible site-specific targeting of exogenous DNA into a bacterial chromosome by use of the FLP recombinase: the FLIRT system. J Bacteriol 179 : 6076–6083. 9324255
54. Schneider CA, Rasband WS, Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9 : 671–675. 22930834
55. Tanner NA, van Oijen AM (2010) Visualizing DNA replication at the single-molecule level. Methods Enzymol 475 : 259–278. doi: 10.1016/S0076-6879(10)75011-4 20627161
56. Biteen J.S., Thompson M.A., Tselentis N.K., Bowman G.R., Shapiro L., Moerner W.E. (2008). Super-resolution imaging in live Caulobacter crescentus cells using photoswitchable EYFP. Nat. Methods 5, 947–949. doi: 10.1038/nmeth.1258 18794860
57. Hedde PN, Fuchs J, Oswald F, Wiedenmann J, Nienhaus GU (2009) Online image analysis software for photoactivation localization microscopy. Nat Methods 6 : 689–690. doi: 10.1038/nmeth1009-689 19789527
58. Sliusarenko O, Heinritz J, Emonet T, Jacobs-Wagner C (2011) High-throughput, subpixel precision analysis of bacterial morphogenesis and intracellular spatio-temporal dynamics. Mol Microbiol 80 : 612–627. doi: 10.1111/j.1365-2958.2011.07579.x 21414037
59. Watkins LP, Yang H (2005) Detection of intensity change points in time-resolved single-molecule measurements. J Phys Chem B 109 : 617–628. 16851054
60. Yang H (2011) Change-Point Localization and Wavelet Spectral Analysis of Single-Molecule Time Series. Hoboken, NJ, USA: John Wiley & Sons, Inc.
Štítky
Genetika Reprodukční medicína
Článek Loss and Gain of Natural Killer Cell Receptor Function in an African Hunter-Gatherer PopulationČlánek Let-7 Represses Carcinogenesis and a Stem Cell Phenotype in the Intestine via Regulation of Hmga2Článek Binding of Multiple Rap1 Proteins Stimulates Chromosome Breakage Induction during DNA ReplicationČlánek SLIRP Regulates the Rate of Mitochondrial Protein Synthesis and Protects LRPPRC from DegradationČlánek Protein Composition of Infectious Spores Reveals Novel Sexual Development and Germination Factors inČlánek The Formin Diaphanous Regulates Myoblast Fusion through Actin Polymerization and Arp2/3 RegulationČlánek Runx1 Transcription Factor Is Required for Myoblasts Proliferation during Muscle Regeneration
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 8
-
Všechny články tohoto čísla
- Putting the Brakes on Huntington Disease in a Mouse Experimental Model
- Identification of Driving Fusion Genes and Genomic Landscape of Medullary Thyroid Cancer
- Evidence for Retromutagenesis as a Mechanism for Adaptive Mutation in
- TSPO, a Mitochondrial Outer Membrane Protein, Controls Ethanol-Related Behaviors in
- Evidence for Lysosome Depletion and Impaired Autophagic Clearance in Hereditary Spastic Paraplegia Type SPG11
- Loss and Gain of Natural Killer Cell Receptor Function in an African Hunter-Gatherer Population
- Trans-Reactivation: A New Epigenetic Phenomenon Underlying Transcriptional Reactivation of Silenced Genes
- Early Developmental and Evolutionary Origins of Gene Body DNA Methylation Patterns in Mammalian Placentas
- Strong Selective Sweeps on the X Chromosome in the Human-Chimpanzee Ancestor Explain Its Low Divergence
- Dominance of Deleterious Alleles Controls the Response to a Population Bottleneck
- Transient 1a Induction Defines the Wound Epidermis during Zebrafish Fin Regeneration
- Systems Genetics Reveals the Functional Context of PCOS Loci and Identifies Genetic and Molecular Mechanisms of Disease Heterogeneity
- A Genome Scale Screen for Mutants with Delayed Exit from Mitosis: Ire1-Independent Induction of Autophagy Integrates ER Homeostasis into Mitotic Lifespan
- Non-synonymous FGD3 Variant as Positional Candidate for Disproportional Tall Stature Accounting for a Carcass Weight QTL () and Skeletal Dysplasia in Japanese Black Cattle
- The Relationship between Gene Network Structure and Expression Variation among Individuals and Species
- Calmodulin Methyltransferase Is Required for Growth, Muscle Strength, Somatosensory Development and Brain Function
- The Wnt Frizzled Receptor MOM-5 Regulates the UNC-5 Netrin Receptor through Small GTPase-Dependent Signaling to Determine the Polarity of Migrating Cells
- Nbs1 ChIP-Seq Identifies Off-Target DNA Double-Strand Breaks Induced by AID in Activated Splenic B Cells
- CCNYL1, but Not CCNY, Cooperates with CDK16 to Regulate Spermatogenesis in Mouse
- Evidence for a Common Origin of Blacksmiths and Cultivators in the Ethiopian Ari within the Last 4500 Years: Lessons for Clustering-Based Inference
- Of Fighting Flies, Mice, and Men: Are Some of the Molecular and Neuronal Mechanisms of Aggression Universal in the Animal Kingdom?
- Hypoxia and Temperature Regulated Morphogenesis in
- The Homeodomain Iroquois Proteins Control Cell Cycle Progression and Regulate the Size of Developmental Fields
- Evolution and Design Governing Signal Precision and Amplification in a Bacterial Chemosensory Pathway
- Rac1 Regulates Endometrial Secretory Function to Control Placental Development
- Let-7 Represses Carcinogenesis and a Stem Cell Phenotype in the Intestine via Regulation of Hmga2
- Functions as a Positive Regulator of Growth and Metabolism in
- The Nucleosome Acidic Patch Regulates the H2B K123 Monoubiquitylation Cascade and Transcription Elongation in
- Rhoptry Proteins ROP5 and ROP18 Are Major Murine Virulence Factors in Genetically Divergent South American Strains of
- Exon 7 Contributes to the Stable Localization of Xist RNA on the Inactive X-Chromosome
- Regulates Refractive Error and Myopia Development in Mice and Humans
- mTORC1 Prevents Preosteoblast Differentiation through the Notch Signaling Pathway
- Regulation of Gene Expression Patterns in Mosquito Reproduction
- Molecular Basis of Gene-Gene Interaction: Cyclic Cross-Regulation of Gene Expression and Post-GWAS Gene-Gene Interaction Involved in Atrial Fibrillation
- The Spalt Transcription Factors Generate the Transcriptional Landscape of the Wing Pouch Central Region
- Binding of Multiple Rap1 Proteins Stimulates Chromosome Breakage Induction during DNA Replication
- Functional Divergence in the Role of N-Linked Glycosylation in Smoothened Signaling
- YAP1 Exerts Its Transcriptional Control via TEAD-Mediated Activation of Enhancers
- Coordinated Evolution of Influenza A Surface Proteins
- The Evolutionary Potential of Phenotypic Mutations
- Genome-Wide Association and Trans-ethnic Meta-Analysis for Advanced Diabetic Kidney Disease: Family Investigation of Nephropathy and Diabetes (FIND)
- New Routes to Phylogeography: A Bayesian Structured Coalescent Approximation
- SLIRP Regulates the Rate of Mitochondrial Protein Synthesis and Protects LRPPRC from Degradation
- Satellite DNA Modulates Gene Expression in the Beetle after Heat Stress
- SHOEBOX Modulates Root Meristem Size in Rice through Dose-Dependent Effects of Gibberellins on Cell Elongation and Proliferation
- Reduced Crossover Interference and Increased ZMM-Independent Recombination in the Absence of Tel1/ATM
- Suppression of Somatic Expansion Delays the Onset of Pathophysiology in a Mouse Model of Huntington’s Disease
- Protein Composition of Infectious Spores Reveals Novel Sexual Development and Germination Factors in
- The Evolutionarily Conserved LIM Homeodomain Protein LIM-4/LHX6 Specifies the Terminal Identity of a Cholinergic and Peptidergic . Sensory/Inter/Motor Neuron-Type
- SmD1 Modulates the miRNA Pathway Independently of Its Pre-mRNA Splicing Function
- piRNAs Are Associated with Diverse Transgenerational Effects on Gene and Transposon Expression in a Hybrid Dysgenic Syndrome of .
- Retinoic Acid Signaling Regulates Differential Expression of the Tandemly-Duplicated Long Wavelength-Sensitive Cone Opsin Genes in Zebrafish
- The Formin Diaphanous Regulates Myoblast Fusion through Actin Polymerization and Arp2/3 Regulation
- Genome-Wide Analysis of PAPS1-Dependent Polyadenylation Identifies Novel Roles for Functionally Specialized Poly(A) Polymerases in
- Runx1 Transcription Factor Is Required for Myoblasts Proliferation during Muscle Regeneration
- Regulation of Mutagenic DNA Polymerase V Activation in Space and Time
- Variability of Gene Expression Identifies Transcriptional Regulators of Early Human Embryonic Development
- The Drosophila Gene Interacts Genetically with and Shows Female-Specific Effects of Divergence
- Functional Activation of the Flagellar Type III Secretion Export Apparatus
- Retrohoming of a Mobile Group II Intron in Human Cells Suggests How Eukaryotes Limit Group II Intron Proliferation
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Exon 7 Contributes to the Stable Localization of Xist RNA on the Inactive X-Chromosome
- YAP1 Exerts Its Transcriptional Control via TEAD-Mediated Activation of Enhancers
- SmD1 Modulates the miRNA Pathway Independently of Its Pre-mRNA Splicing Function
- Molecular Basis of Gene-Gene Interaction: Cyclic Cross-Regulation of Gene Expression and Post-GWAS Gene-Gene Interaction Involved in Atrial Fibrillation
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání




