-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamamTORC1 Prevents Preosteoblast Differentiation through the Notch Signaling Pathway
The coordinated activities of osteoblasts and osteoclasts in bone deposition and resorption form the internal structure of bone. Disruption of the balance between bone formation and resorption results in loss of bone mass and causes bone diseases such as osteoporosis. Current therapies for osteoporosis are limited to anti-resorptive agents, while bone diseases due to reduced osteoblast activity, such as senile osteoporosis, urgently require targeted treatment and novel strategies to promote bone formation. mTORC1 has emerged as a critical regulator of bone formation and is therefore a potential target in the development of novel bone-promoting therapeutics. Identifying the detailed function of mTORC1 in bone formation and clarifying the underlying mechanisms may uncover useful therapeutic targets. In this study, we reveal the role of mTORC1 in osteoblast formation. mTORC1 stimulated preosteoblast proliferation but prevented their differentiation and attenuated bone formation via activation of the Notch pathway. Pharmaceutical coordination of the pathways and agents in preosteoblasts may be beneficial in bone formation.
Published in the journal: . PLoS Genet 11(8): e32767. doi:10.1371/journal.pgen.1005426
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005426Summary
The coordinated activities of osteoblasts and osteoclasts in bone deposition and resorption form the internal structure of bone. Disruption of the balance between bone formation and resorption results in loss of bone mass and causes bone diseases such as osteoporosis. Current therapies for osteoporosis are limited to anti-resorptive agents, while bone diseases due to reduced osteoblast activity, such as senile osteoporosis, urgently require targeted treatment and novel strategies to promote bone formation. mTORC1 has emerged as a critical regulator of bone formation and is therefore a potential target in the development of novel bone-promoting therapeutics. Identifying the detailed function of mTORC1 in bone formation and clarifying the underlying mechanisms may uncover useful therapeutic targets. In this study, we reveal the role of mTORC1 in osteoblast formation. mTORC1 stimulated preosteoblast proliferation but prevented their differentiation and attenuated bone formation via activation of the Notch pathway. Pharmaceutical coordination of the pathways and agents in preosteoblasts may be beneficial in bone formation.
Introduction
The skeleton is a highly specialized and dynamic structure undergoing constant remodeling [1]. The remodeling process is executed by temporary cellular structures that comprise teams of coupled osteoblasts and osteoclasts. The rate of genesis as well as death of these two cell types is vital for the maintenance of bone homeostasis [2], and common metabolic bone disorders such as osteoporosis are largely caused by a derangement in the proliferation, differentiation or apoptosis of these cells [3].
Osteoblasts, which are the chief bone-making cells, differentiate and produce bone matrix during skeletal development [4]. The differentiation process of osteoblasts is often divided into stages of mesenchymal progenitors, preosteoblasts and osteoblasts (often called mature osteoblasts) [5]. Osteoblasts are often characterized by the expression of osteocalcin, while preosteoblasts are usually considered to express the transcription factor Runx2 or both Runx2 and osterix (Osx). Preosteoblasts have been shown to actively divide in vitro and are multipotent in differentiating, thus proliferative expansion and osteoblastic differentiation of preosteoblasts are essential for bone formation. Understanding the intracellular signaling pathways that control preosteoblast proliferation and differentiation is critical for preventing bone loss-related disease caused by impaired bone formation such as senile osteoporosis.
The mechanistic target of rapamycin (mTOR) is a conserved Ser/Thr kinase nucleating at least two distinct multi-protein complexes, mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2) [6]. mTORC1 uniquely contains raptor and is the sensitive target of rapamycin, it integrates both intracellular and extracellular signals, including growth factors, nutrients, energy levels, and cellular stress [7]. The tuberous sclerosis 1 (TSC1)-TSC2-TBC1D7 (TSC-TBC) complex is the major upstream inhibitory regulator of mTORC1 [8], and loss of this complex causes cells and tissues to display constitutive mTORC1 activation. TSC-TBC accelerates the intrinsic rate of GTP hydrolysis of Rheb, converting Rheb from the GTP-bound (active) to the GDP-bound (inactive) form. The active GTP-bound form of Rheb directly interacts with mTORC1 to stimulate its activity. mTORC1 phosphorylates the translational regulators, eukaryotic initiation factor 4E-binding protein-1 (4E-BP1) and S6 kinase 1 (S6K1), to regulate biosynthesis of proteins and modulates autophagy, biosynthesis of lipids and organelles and mitochondrial metabolism as well, through which mTORC1 exerts an essential role in regulating cell metabolism, survival, growth and proliferation [6,9].
mTORC1 signaling has emerged as a critical regulator of bone formation. Patients with tuberous sclerosis due to mutation in TSC1/2 present with sclerotic and lytic bone changes [10,11,12,13,14]. Moreover, mTOR has recently been identified among genes and pathways that are connected with human skeletal growth [15]. However, results from in vitro and in vivo studies on the function of mTORC1 in osteoblast lineage are inconsistent. The mTORC1 inhibitor, rapamycin, showed controversial capacity to influence the differentiation of various osteoblastic lineage cell lines in vitro [16,17,18,19,20,21,22] and demonstrated inconsistent potential for bone formation in vivo [23,24,25]. In addition to an undefined function in the osteoblast lineage, the mechanisms through which mTORC1 modulates osteoblast differentiation and bone formation are unknown. Using conditional Tsc1 knockout cell and mouse models, we demonstrated that mTORC1 activation is crucial for preosteoblast proliferation, but prevents their differentiation and maturation.
Notch signaling mediates communication between neighboring cells to control cell fate decision [26, 27]. Notch has been reported to be positively regulated by mTORC1 in various cell lines and be responsible for the impaired cell differentiation by mTORC1 [28, 29, 30]. We determined here that it is true in osteoblast lineage cells as well. Our mechanistic studies revealed that Notch signaling is strongly activated by mTORC1 to inhibit osteoblastic transcription factor Runx2 and prevent preosteoblast maturation and bone formation.
Results
mTORC1 is activated during preosteoblast proliferation but is suppressed during their differentiation
mTORC1 is known to promote cell proliferation in many types of cells. To examine the relationship between preosteoblast proliferation and mTORC1 activity, we analyzed the level of mTORC1 during the proliferative expansion of MC3T3-E1 cells, a murine preosteoblast cell line [31], and subsequent cessation of growth. We counted cells each day to monitor their growth and observed that the cells reached confluence after 6–7 days, when proliferation ceased due to contact inhibition (Fig 1A). Western blot analysis reflected this growth inhibition as a decrease in the levels of the cell cycle markers, cyclin D1 and proliferative cell nuclear antigen (PCNA). As expected, the high level of mTORC1 activity (indicated by P-S6K (T389) and P-S6 (S235/236)) decreased when proliferation of MC3T3-E1 cells slowed down and eventually ceased (Fig 1B). Cells did not differentiate during this growth period, as the level of osterix (Osx, a marker of early osteoblasts) was unchanged throughout. These data indicate that the level of mTORC1 activity was positively correlated with the rate of preosteoblast proliferation.
Fig. 1. mTORC1 is activated during preosteoblast proliferation but is suppressed during their differentiation. 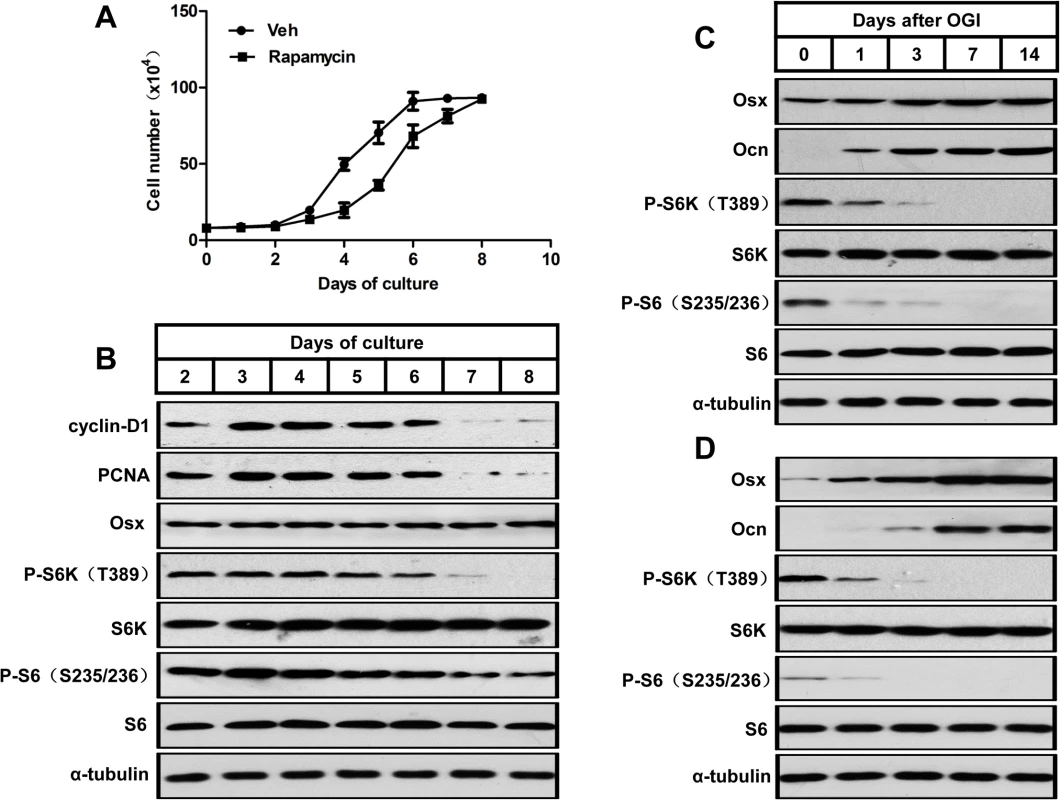
(A) Growth curve of proliferating MC3T3-E1 cells treated with vehicle (Veh) or rapamycin (0.1nM). (B) Western blot analysis of proliferative markers (cyclin D1 and PCNA), Osx and mTORC1 activity (indicated by P-S6K (T389) and P-S6 (S235/236)) during MC3T3-E1 proliferation. (C) Western blot analysis of osteoblastic markers (Osx and Ocn) and mTORC1 activity during differentiation of MC3T3-E1 cells. (D) Western blot analysis of osteoblastic markers (Osx and Ocn) and mTORC1 activity during differentiation of FRC. Osx: osterix; Ocn: osteocalcin; FRC: fetal rat calvarial cells, OGI: osteogenic induction. We then induced osteoblastic differentiation in confluent MC3T3-E1 cells. The induced cells showed a decreased mTORC1 activity during osteoblast differentiation in parallel with increases in markers of osteoblast differentiation (i.e. osterix (Osx) and osteocalcin (Ocn)) (Fig 1C). A similar pattern of mTORC1 activity was observed in osteoblastic induced fetal rat calvarial cells (Fig 1D), which implicated that mTORC1 activity is not required for osteoblastic differentiation of preosteoblasts.
Inactivation of mTORC1 prevents preosteoblast proliferation but enhances their differentiation in vitro and in vivo
We next investigated the role of reduced mTORC1 activity caused by rapamycin in the proliferation and differentiation of preosteoblasts. As shown in Fig 1A, the growth of cells treated with 0.1 nM rapamycin significantly lagged behind control cells, and the decreased level of proliferative markers (cyclin D1 and PCNA) in rapamycin-treated cells revealed the underlying mechanism (Fig 2A). We next determined the role of reduced mTORC1 activity in the differentiation of preosteoblasts. As seen in Fig 2B, a low concentration of rapamycin (0.1nM) increased the expression of osterix and osteocalcin. Separate sets of cells were tested for mineralization capacity, a terminal differentiation parameter for osteoblasts, by staining with alizarin red, and the results confirmed enhanced mineralization of the extracellular matrix (ECM) in MC3T3-E1 cells with impaired mTORC1 activity (Fig 2C). Increased mineralization of ECM was also observed in fetal rat calvarial cells treated with 0.1nM rapamycin (S1 Fig).
Fig. 2. Inactivation of mTORC1 prevents preosteoblast proliferation but enhances their differentiation in vitro and in vivo. 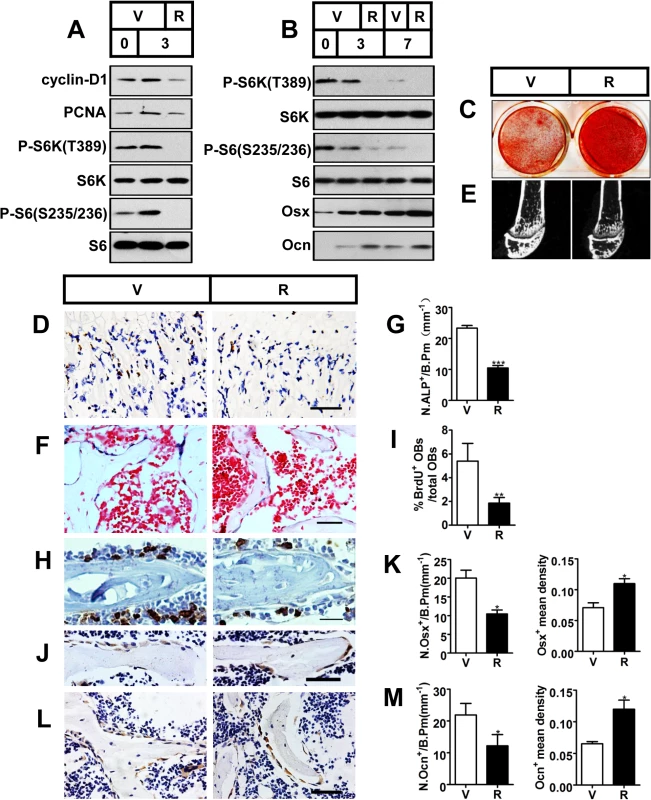
(A) Proliferating MC3T3-E1 cells were treated with vehicle (V) or 0.1 nM rapamycin (R) and underwent immunoblotting to detect proliferative markers (cyclin-D1 and PCNA) on the day of cell plating (day 0) and on the 3rd day. (B) Differentiating MC3T3-E1 cells were treated with vehicle or 0.1 nM rapamycin for the indicated time (3d, 7d) and underwent immunoblotting to detect osteoblastic markers (Osx and Ocn). (C) AR-S staining of differentiated MC3T3-E1 cells on the 14th day of osteogenic induction. (D) Immunohistochemistry staining for S6 phosphorylation (Ser235/236) in sections of distal femur of 10-week-old female C57BL/6 mice treated with vehicle (V) or rapamycin (R). (E) Micro-CT images of metaphyseal trabecular bone of the distal femur. (F) Alkaline phosphatase (ALP) staining of trabecular bone in sections from the distal femora (G) Quantification of osteoblast lineage cells stained with ALP. N.ALP+/B.Pm, number of ALP positive cells per bone perimeter (mm-1). Representative immunohistochemistry staining for (H) BrdU, (J) osterix, and (L) osteocalcin in femur sections. (I) Percentage of BrdU+ osteoblasts out of total osteoblasts on bone surface. (K, M) Number of Osx-positive cells (N.Osx+) and Ocn-positive cells (N.Ocn+) on the bone surface was measured as cells per millimeter of perimeter in sections (/B.Pm) and mean density of the corresponding positive cells was calculated as integrated optical density (IOD) per area of positive cells. All data are mean ± SD (n = 3 mice).Scale bar, 100 μm for (D) and 50 μm for (F), (H), (J), (L). *P < 0.05, **P < 0.01, ***P<0.001 by t test. To test the results in vivo, we subcutaneously injected newborn wild-type C57BL/6 mice with rapamycin daily (0.1 mg/kg body weight/day) for 10 weeks. mTORC1 activity was efficiently down-regulated in the primary spongiosa as observed by the reduction in immunohistochemistry staining for S6 phosphorylation (Ser235/236) in rapamycin-treated mice (Fig 2D). Micro-CT analysis of the distal femur showed a marked decrease in bone mass in the rapamycin-treated mice (Fig 2E), as demonstrated by a significant decrease in BV/TV, trabecular number or trabecular thickness, coupled with an increase in trabecular separation (Table 1). In line with the reduced trabecular bone mass, mice treated with rapamycin showed decreased cortical thickness as well as smaller outer and inner femoral mid-shaft bone perimeters when compared with controls (Table 1). The decrease in cortical thickness was due to deficient periosteal apposition, as evidenced by a decrease in the outer perimeter of the mid-shaft, while the inner perimeter was also decreased.
Tab. 1. Micro CT analysis of rapamycin-treated C57BL/6 mice at 10 weeks of age. 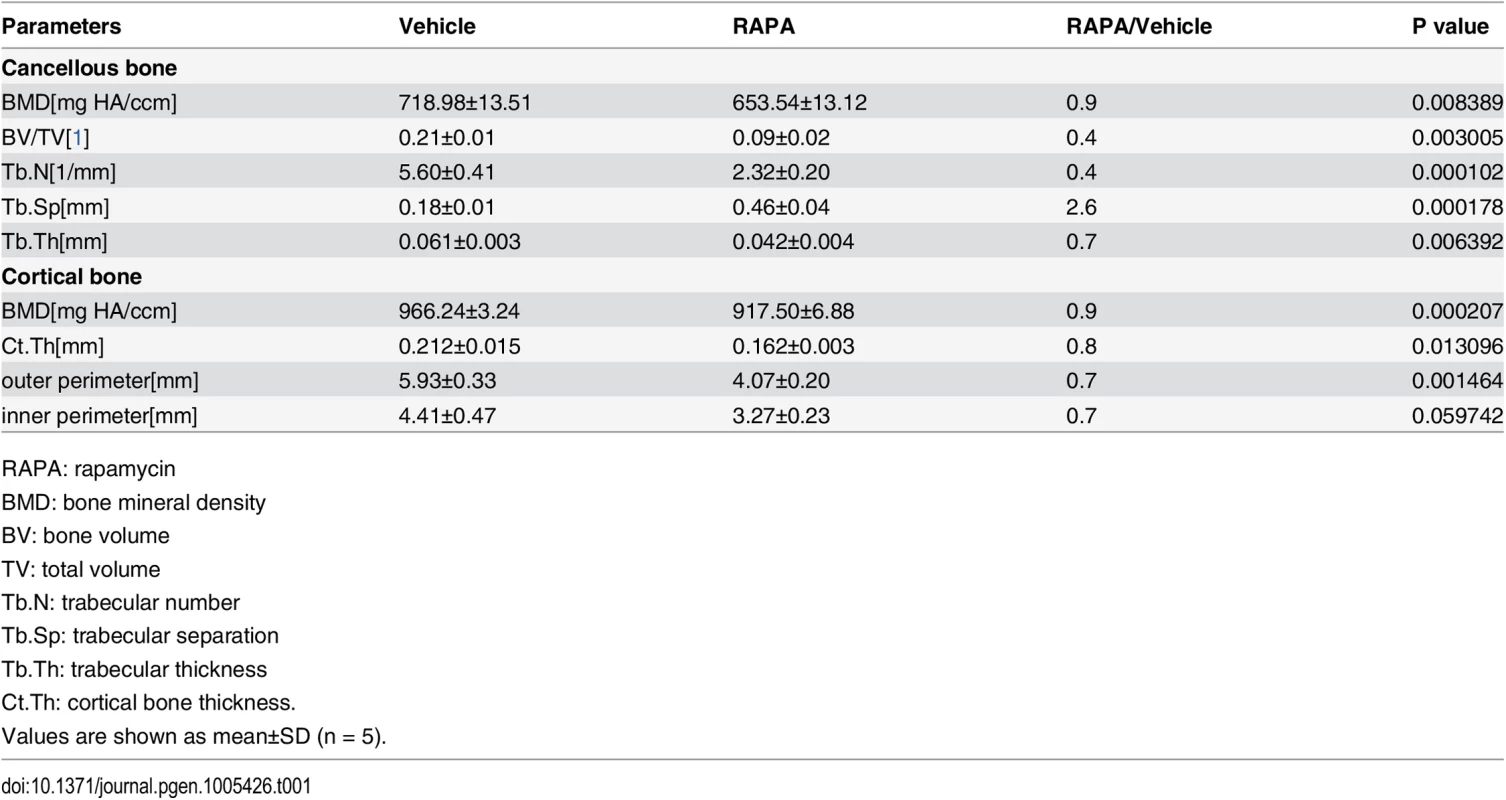
RAPA: rapamycin We then analyzed the cellular basis of decreased bone mass in rapamycin-treated mice. Tartrate-resistant acid phosphatase (TRAP) staining on femoral sections revealed a reduction in osteoclasts within the trabecular bone region in rapamycin-treated mice (S2 Fig). Thus, decreased bone mass in the rapamycin-treated mice was not caused by an overall decrease in bone resorption. We next investigated bone formation parameters by first examining the total number of osteoblasts. ALP staining of the distal femoral section from rapamycin-treated mice showed a marked reduction in osteoblastic lineage cells when normalized to the bone perimeter (Fig 2F and 2G). Immunohistochemistry staining for BrdU revealed that proliferation of osteoblastic lineage cells was attenuated in rapamycin-treated mice (Fig 2H and 2I). To determine if differentiation of osteoblasts was affected, we next measured their numbers at different stages of differentiation by immunostaining femur sections from rapamycin-treated mice and controls. The number of osterix-positive preosteoblasts (Fig 2J and 2K) and osteocalcin-positive mature osteoblasts (Fig 2L and 2M) on the bone surfaces of rapamycin-treated mice were less than those in the controls. However, the mean density of osterix and osteocalcin-positive cells was increased in mice treated with rapamycin (Fig 2K and 2M), indicating that expression of osterix and osteocalcin in single osteoblasts was increased in rapamycin-treated mice. Thus, in agreement with the in vitro results, rapamycin attenuated proliferation of preosteoblasts but promoted their differentiation in vivo.
mTORC1 activation in preosteoblasts produces immature woven bone
To characterize the role of mTORC1 activation in the regulation of proliferation and differentiation of osteogenic progenitors, we generated mice in which mTORC1 were selectively activated in osteoprogenitor cells committed to the osteoblast lineage. To achieve such cell type-specific knockout, we crossed floxed Tsc1 mice with Osx-GFP::Cre mice (which express a GFP-Cre fusion protein under the direction of the Osx1 promoter) to generate conditional Tsc1 knockout mice. We mated Osx-GFP::CreTG/+;Tsc1flox/+ mice and selected female mice with the genotype Osx-GFP::CreTG/+;Tsc1flox/flox (hereafter, referred to as ΔTsc1) for detailed analysis. Female Osx-GFP::CreTG/+;Tsc1+/+ littermates served as controls. ΔTsc1 mice were born at the expected Mendelian frequency, and recombination of Tsc1 alleles only occurred in skeletal tissues (i.e., legs and skull) as demonstrated by allele specific PCR (Fig 3A). Immunohistochemical staining of distal femur sections showed a dramatic increase in S6 phosphorylation (Ser235/236) in ΔTsc1 mice (Fig 3B), indicating that mTORC1 was activated by Tsc1 disruption.
Fig. 3. mTORC1 activation in preosteoblasts produces immature woven bone. 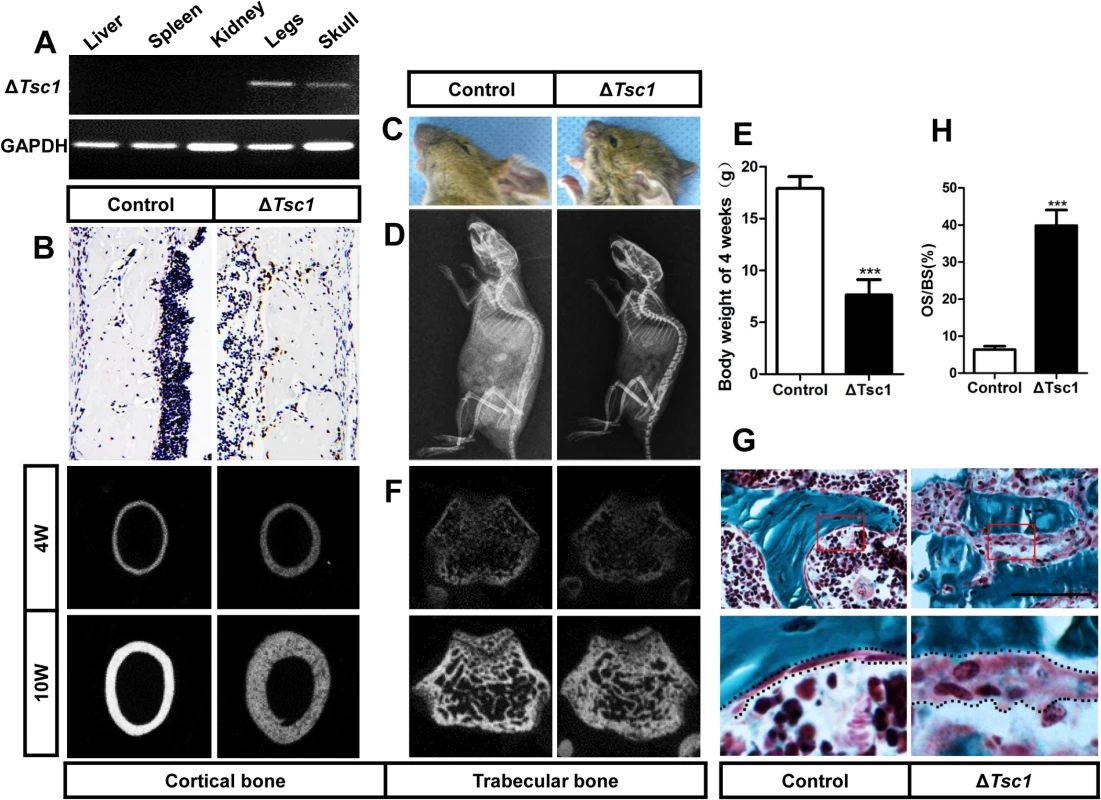
(A) PCR analysis of Tsc1 allele recombination in tissues from Osx-GFP::CreTG/+;Tsc1flox/flox mice (ΔTsc1 mice). Primers for GAPDH were used as a loading control. (B) Immunohistochemical staining of S6 phosphorylation (Ser235/236) in sections of distal femur from 10-week-old control and ΔTsc1 mice. (C) The 10-week-old ΔTsc1 mouse had a square skull (the result of hyperplasia of the osteoid, a typical manifestation of rickets) compared with the control littermate. (D) Plain X-ray examination of a 4-week-old ΔTsc1 mouse revealed a smaller skeleton and higher bone mass compared with the control littermate. (E) Body weight of 4-week-old control and ΔTsc1 mice. (F) Representative images of micro-CT analyses of the structure of metaphyseal trabecular bone and cortical bone in the distal femur showed increased bone mass, but more porous areas (indicating more hypomineralized areas) in ΔTsc1 mice. (G, H) Goldner’s Masson trichrome staining of distal femur showed more osteoid/hypomineralized areas (stained red) in bone of ΔTsc1 mice. Boxed area is enlarged in the panel below. The dotted line marked boundary of the red osteoid. OS/BS, osteoid per bone surface. Scale bar, 100μm. At the gross level, 4-week-old ΔTsc1 mice demonstrated square skulls and prominent dwarfism (Fig 3C and 3D). The body weight of the Tsc1-deficient mice was significantly lower than that of their control littermates, indicating retardation of growth in the mutant mice (Fig 3E). Whole body X-ray analysis showed increased bone mass in the skull, vertebrae and long bones (Fig 3D). Because of thickened cortical bone, pale bones were observed in ΔTsc1 mice (S3 Fig). Micro-CT analysis of the distal regions of the femur confirmed a marked increase in cancellous bone mass in ΔTsc1 mice (Fig 3F), as reflected in a 240%, 140% and 170% increase in BV/TV, trabecular number and trabecular thickness, respectively, coupled with a 50% decrease in trabecular separation. Analysis of femoral mid-shaft revealed a similar increase in cortical bone mass in ΔTsc1 mice, as reflected in a 230% and 130% increase in cortical thickness and outer perimeter, respectively, and a 90% decrease in inner perimeter (Table 2). Although bone mass was increased in ΔTsc1 mice, micro-CT radiograms showed more porous areas in the bones of mutant mice (Fig 3F), which resulted in a uniform 90% decrease in bone mineral density (BMD) in cancellous and cortical bone (Table 2). The appearance of more porous areas in mutant mice was the result of increased areas of hypomineralization. As shown by Goldner’s Masson trichrome staining, ΔTsc1 mice had 35% more osteoid/hypomineralized areas (stained red) in bone (Fig 3G and 3H). Together, these data suggest that mTORC1 activation in osteogenic progenitors stimulates these progenitors to produce increased amounts of immature woven bone.
Tab. 2. Micro CT analysis of ΔTsc1 mice at 10 weeks of age. 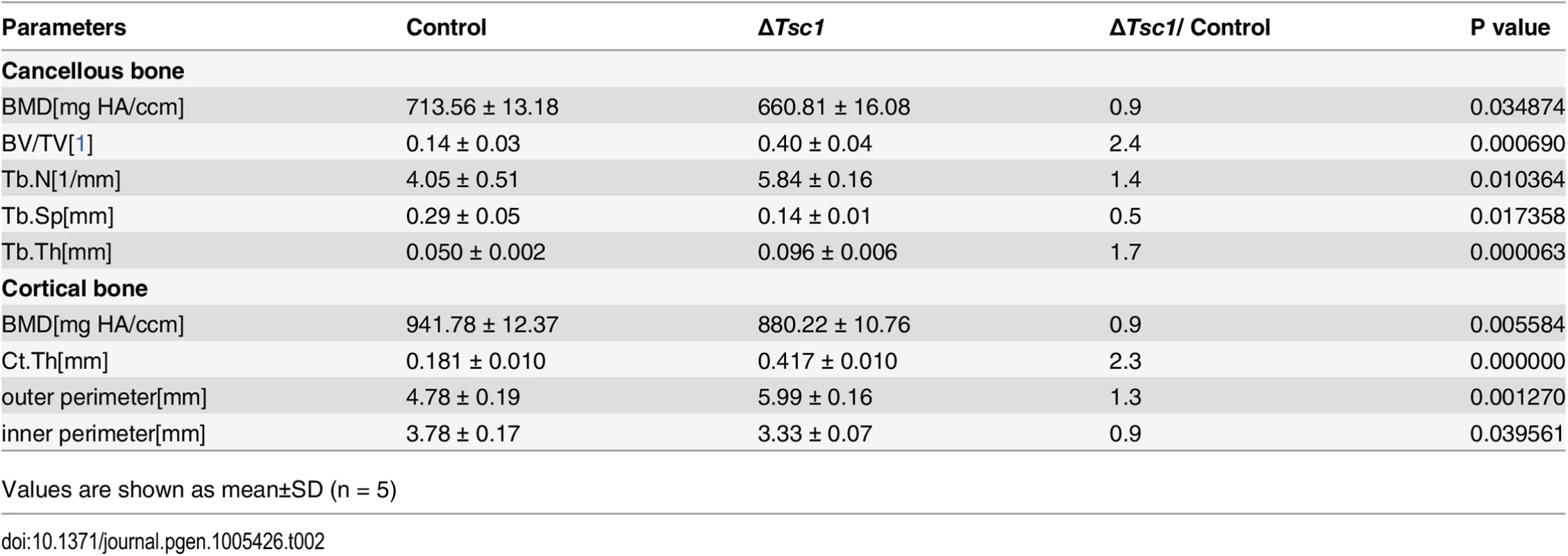
Values are shown as mean±SD (n = 5) mTORC1 activation promotes preosteoblast proliferation but prevents their differentiation
We then analyzed the cellular basis for the increased amounts of immature woven bone in ΔTsc1 mice. ALP staining of 10-week-old ΔTsc1 mouse femurs indicated an increased number of osteoblast lineage cells (Fig 4A and 4B). To determine the underlying molecular mechanism of the increase in osteoblasts in transgenic bone, we cultured postnatal day 3 calvarial osteoblasts and found a significant increase in BrdU-positive cells, indicating increased cellular proliferation in ΔTsc1 calvarial osteoblasts (S4 Fig). In addition, the percentage of proliferative osteoblasts on bone surface was increased in ΔTsc1 mice (Fig 4C and 4D).
Fig. 4. mTORC1 promotes proliferation of preosteoblasts but prevents their maturation. 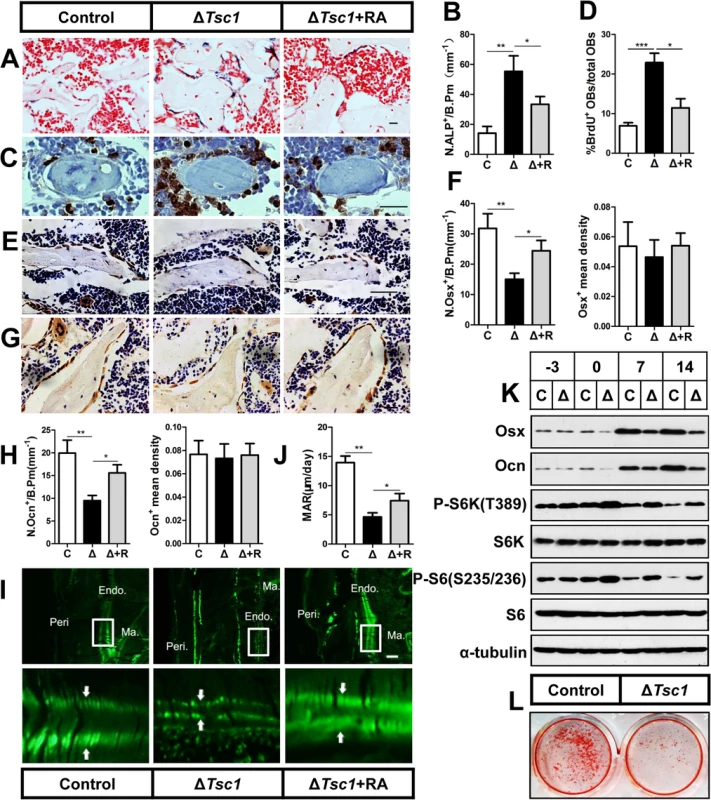
(A) ALP staining in sections of distal femur from 10-week-old control (C), ΔTsc1 (Δ) and rapamycin treated ΔTsc1 (Δ+R) mice. (B) Number of ALP positive cells per bone perimeter (mm-1) (N.ALP+/B.Pm). Immunohistochemistry staining for (C) BrdU, (E) osterix (Osx) and (G) osteocalcin (Ocn) in distal femur. (D) Percentage of BrdU+ osteoblasts out of total osteoblasts on bone surface. (F, H) Number of osterix positive cells (N.Osx+) and osteocalcin positive (N.Ocn+) on the bone surface was measured as cells per millimeter of perimeter in sections (/B.Pm) and the mean density of the corresponding positive cells was calculated as integrated optical density (IOD) per area of positive cells. (I) Calcin double labeling of cortical bone in distal femora. Boxed area is enlarged in the panel below. Ma.: marrow, Endo.: endosteum, Peri.: periosteum. (J) Mineral apposition rate (MAR). (K) Western blot analysis of differentiating primary calvarial preosteoblasts showed decreased expression of osterix and osteocalcin following doxycycline discontinuation in ΔTsc1 cells (0, 7th, 14th day). (L) Alizarin red staining of differentiated primary calvarial preosteoblasts on the 14th day showed decreased mineralized nodules in ΔTsc1 cells. All data are mean ± SD (n = 5 mice), scale bars represent 50 μm for (A), (C), (E), (G) and 100 μm for (I). *P < 0.05, ** P < 0.01, ***P<0.001 by t test. We next determined whether the differentiation of preosteoblasts was influenced in ΔTsc1 mice. Osteoblasts in ΔTsc1 mice lost normal morphology and appeared immature as shown by scanning electron microscopy (SEM) analyses (S5 Fig), indicating a faulty maturation process from precursors to osteoblasts. Immunohistochemical staining of distal femur sections showed decreased expression of osterix (Fig 4E and 4F) and osteocalcin (Fig 4G and 4H), indicating impaired differentiation of preosteoblasts in mutant mice. To detect osteoblast activity, we performed double fluorochrome labeling analyses. Incorporation of the two fluorochromes was evident in the control mice bone, while mutants’ bone displayed diffuse fluorochrome labeling, a characteristic feature of immature woven bone. Although mineralizing surface was dramatically increased abnormally in cortical bone, distance between calcin-labeled mineralization fronts at endosteum of the midshaft of femur was smaller in ΔTsc1 mice than that in the controls (Fig 4I). Histomorphometric measurements showed that the endosteum mineral apposition rate (MAR) of ΔTsc1 mice was lower than that of controls (Fig 4J), suggesting impaired performance of individual osteoblasts in ΔTsc1 mice.
To further examine the impact of mTORC1 activation on osteoblastic precursor differentiation, we used primary calvarial preosteoblast cultures in which Tsc1 was eliminated prior to the induction of osteoblast differentiation due to suppression of Osx-Cre by doxycycline to specifically delete Tsc1. On the day the cells reached confluence (day -3), mTORC1 was not activated (as indicated by unchanged phosphorylation of S6K and S6) and markers of differentiated osteoblasts (Osx and Ocn) remained normal in ΔTsc1 cells compared with the controls. Doxycycline was then discontinued to activate Osx-Cre and delete Tsc1 and osteoblastic differentiation was induced for another 14 days. Total proteins were extracted for western blot on different days after induction (day 0, 7, 14). Osteoblasts lacking Tsc1 exhibited the expected activation of mTORC1 and reduced expression of osterix and osteocalcin (Fig 4K). Alizarin red staining confirmed that ΔTsc1 mice exhibited a marked decreased capacity to form mineralized nodules (Fig 4L).
Notably, osteoclast numbers on the bone surface (OC.N/B.Pm) were significantly decreased in mutant mice (S6 Fig).These data suggest that mTORC1 activation in preosteoblasts promotes their proliferation but prevents their differentiation and decreases osteoclast number by an undefined mechanism.
Rapamycin reverses phenotypes in ΔTsc1 mice
To identify whether the increased amount of immature woven bone was mTORC1-dependent, ΔTsc1 mice were administered with rapamycin prenatally from E13.5 (the approximate day of Osx-Cre expression) and then after birth until 10 weeks old. After treatment with 0.1 mg/kg/day rapamycin, S6 phosphorylation (Ser235/236) was significantly decreased in the primary spongiosa (Fig 5A), accelerated proliferation was reduced to normal level (Fig 4C and 4D), increased osteoblastic lineage cells with ALP (Fig 4A and 4B) were reduced to normal, expression of osterix (Fig 4E and 4F) and osteocalcin (Fig 4G and 4H) was increased, diffuse and narrowed distance between fluorochrome labeling was distinct and broadened (Fig 4I and 4J), high bone mass was notably reduced (Fig 5B), narrowed bone marrow cavity was expanded (Fig 5C), areas of hypomineralization were decreased to normal (Fig 5D and 5E) and bone architecture was partially normalized (Table 3). These results indicate that preosteoblasts in ΔTsc1 mice re-acquired the ability to differentiate, and form normal bone. The results of rapamycin treatment in differentiating calvarial cells confirmed these findings. Following treatment with a low concentration (0.1 nM) of rapamycin for 14 days, decreased expression of osterix and osteocalcin was elevated (Fig 5F) and mineralized nodules were formed increasingly (Fig 5G) in ΔTsc1 cells.
Fig. 5. Rapamycin reverses phenotypes in ΔTsc1 mice. 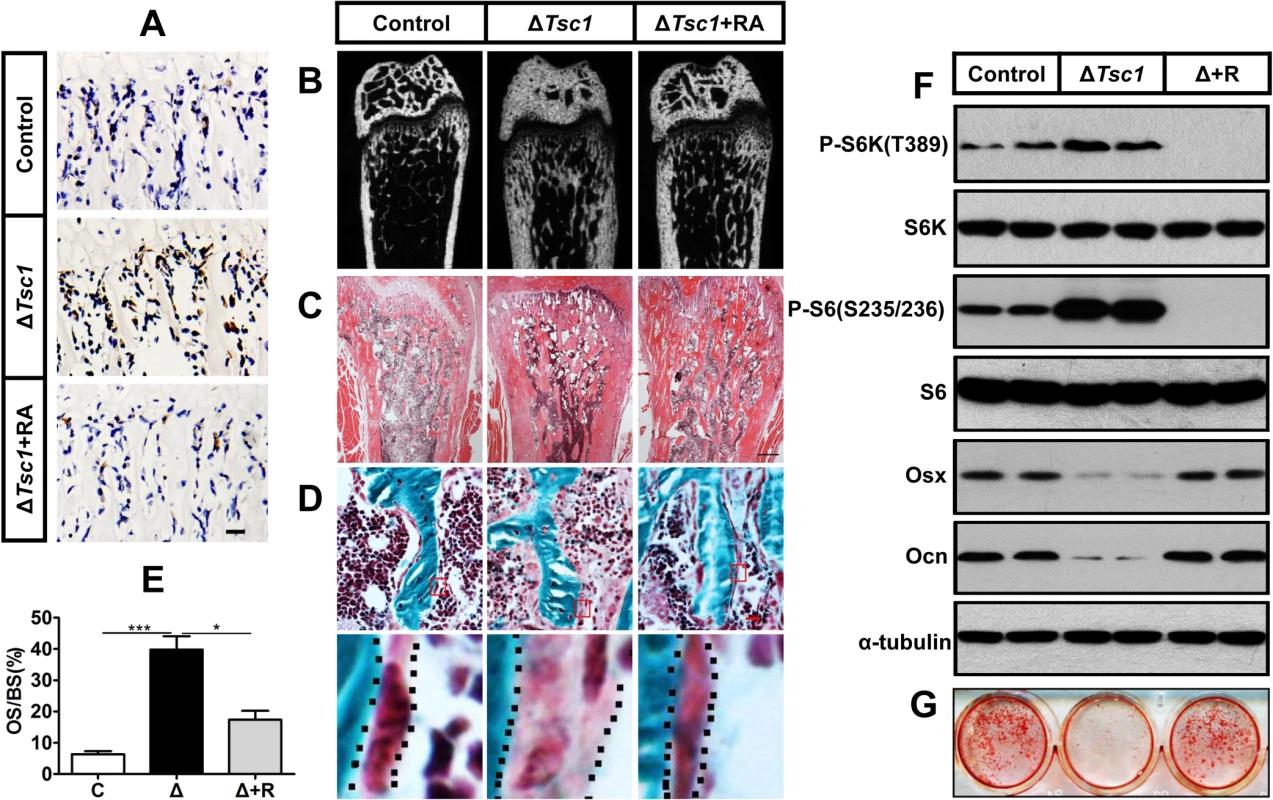
(A) Immunohistochemical staining of phosphorylation of S6 (Ser235/236) in sections of distal femur from 10-week-old control, ΔTsc1 and rapamycin treated ΔTsc1 mice. (B) Representative images of micro-CT analyses in the distal femur showed reduced bone mass in rapamycin treated ΔTsc1 mice. (C) HE staining of distal femur showed expanded bone marrow cavity in rapamycin treated ΔTsc1 mice. (D, E) Goldner’s Masson trichrome staining showed reduced osteoid in rapamycin treated ΔTsc1 mice. OS/BS, osteoid per bone surface. Scale bar, 100 μm. ***P < 0.001, * P < 0.05 by t test. (F) Western blot analysis showed elevated osterix and osteocalcin expression in ΔTsc1 calvarial cells treated with rapamycin (0.1nM). (G) Alizarin red staining of differentiated primary calvarial preosteoblasts on the 14th day showed increased mineralized nodules in rapamycin treated ΔTsc1 cells. Tab. 3. Micro CT analysis of rapamycin-treated ΔTsc1 mice at 10 weeks of age. 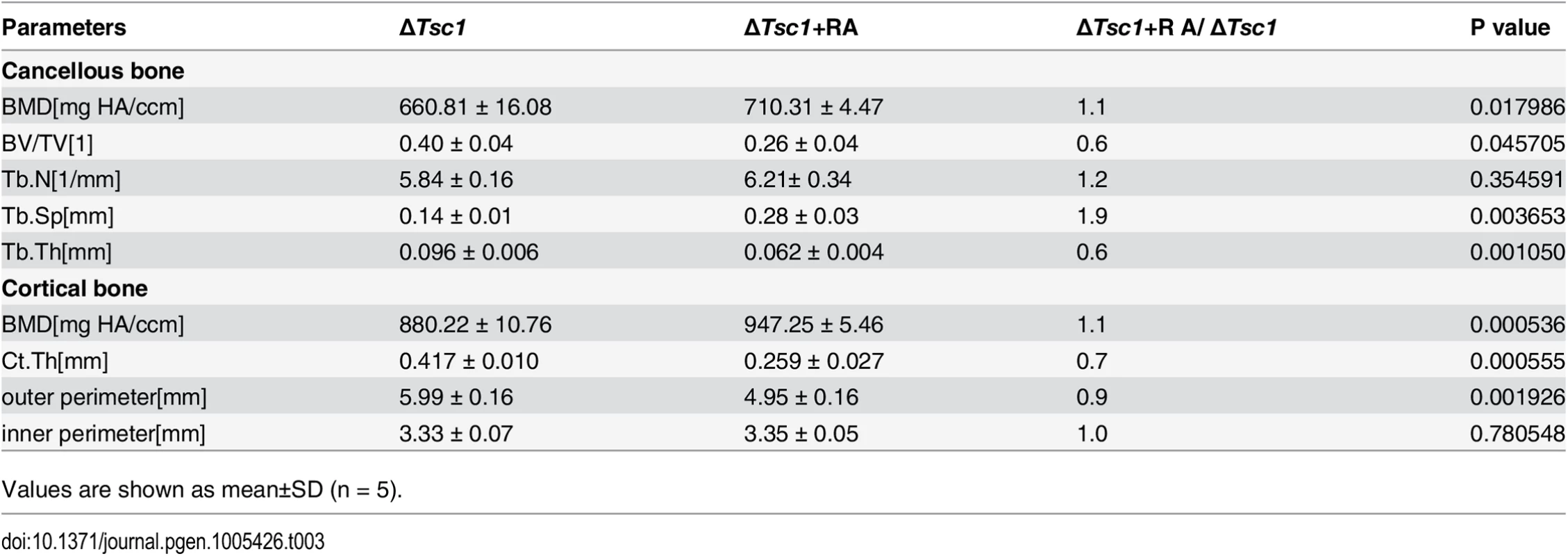
Values are shown as mean±SD (n = 5). mTORC1 inhibits Runx2 expression by activating Notch signaling in preosteoblasts
We next investigated the mechanism by which mTORC1 regulates osteoblast differentiation. Runx2, a Runt domain-containing transcription factor, is required for commitment of mesenchymal osteochondroprogenitors to the osteoblastic lineage, differentiation into mature osteoblasts and terminal differentiation into osteocytes [32]. ΔTsc1 calvarial cells exhibited a reduced Runx2 expression level, while rapamycin treatment led to an increase in Runx2 above baseline (Fig 6A). The regulation of Runx2 by mTORC1 was further confirmed in ΔTsc1 mice (Fig 6B and 6C). Thus, mTORC1 attenuated the expression of Runx2 in vitro and in vivo, and the delay in osteoblast differentiation was probably due to repression of Runx2 in ΔTsc1 mice.
Fig. 6. mTORC1 inhibits Runx2 expression by activating Notch signaling in preosteoblasts. 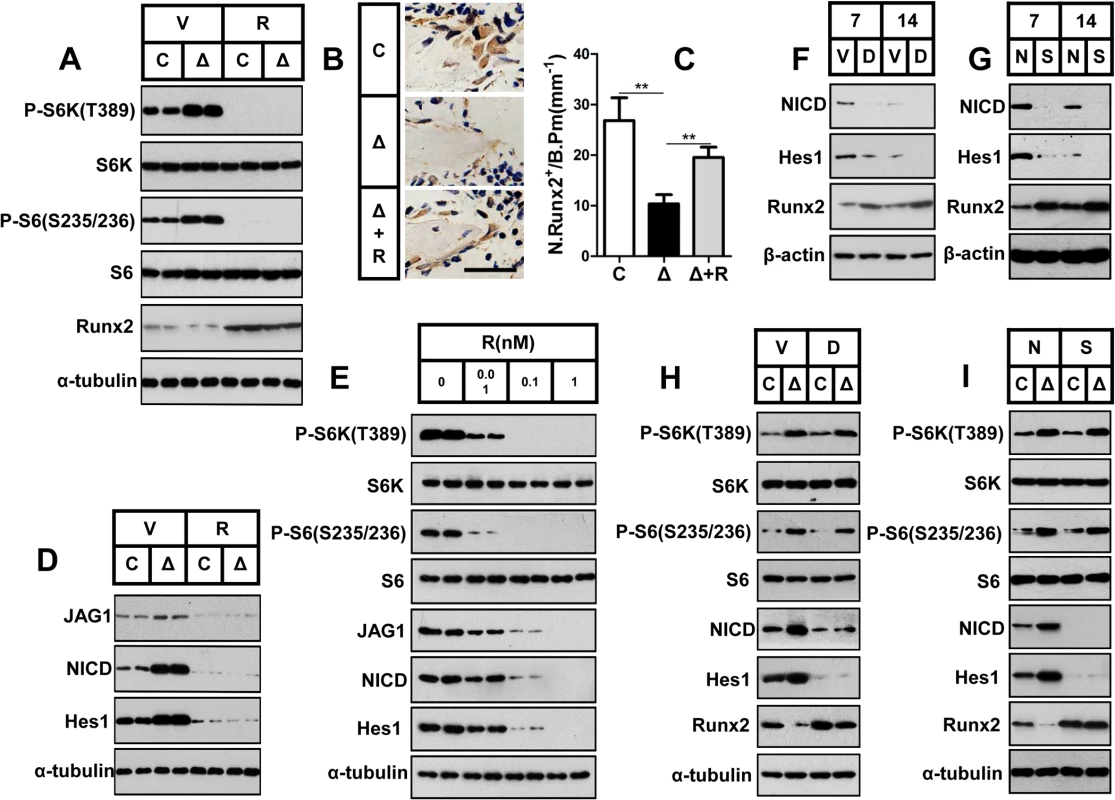
(A) Control (C) and ΔTsc1 (Δ) primary calvarial cells were treated with vehicle (V) or 0.1 nM rapamycin (R) for 24 hours and then underwent immunoblotting to detect Runx2. (B) Immunohistochemical staining of Runx2 in sections of distal femur from 10-week-old control (C), ΔTsc1 (Δ) and rapamycin treated ΔTsc1 (Δ+R) mice. Scale bar, 50μm. (C) Number of Runx2 positive cells per bone perimeter (mm-1) (N.Runx2+/B.Pm).** P < 0.01 by t test. (D) Deletion of Tsc1 caused elevated expression of Jagged1, NICD and Hes1, while treatment with 0.1 nM rapamycin for 24 hours led to decreases in these parameters in primary calvarial cells. (E) Rapamycin dose-dependently reduced the expression of Jagged1, NICD and Hes1 in MC3T3-E1 cells. Differentiating MC3T3-E1 (F, G) and primary cavarial cells from control and ΔTsc1 mice (H, I) were treated with 50 μM DAPT (F, H) or Notch1 siRNA (G, I) and then underwent immunoblotting to detect Runx2. V: vehicle; D: DAPT; N: negative control; S: si-Notch1. In consideration of the same negative role of Notch signaling in osteoblast differentiation [32,33,34,35] as mTORC1 signaling and the positive correlation between Notch and mTORC1[28,29,30], we next investigated the potential role of the Notch pathway in the mechanism underlying the impaired differentiation potential of mTORC1-activated cells. Jagged1 (a Notch ligand) protein expression was significantly enhanced in primary ΔTsc1 calvarial cells, and rapamycin reduced this expression to normal (Fig 6D) and dose-dependently decreased its expression in MC3T3-E1 cells (Fig 6E). In addition, a similar pattern of expression of the Notch transactivator NICD domain and Hes1 (a direct target of Notch) was also observed in these cells (Fig 6D and 6E). These results demonstrated that mTORC1 activated Notch signaling in preosteoblasts.
As the Notch pathway has been reported to reduce Runx2 activity [32], we next determined whether Notch signaling impaired differentiation of preosteoblasts upstream of Runx2. We found increased Runx2 expression in differentiating MC3T3-E1 and primary cavarial cells following suppression of the Notch pathway using the Notch inhibitor N-[N-(3, 5-difluorophenacetyl-L-alanyl)]-S-phenylglycinet-butylester (DAPT) (Fig 6F and 6H) and Notch siRNA (Fig 6G and 6I). We conclude that mTORC1 inhibited Runx2 expression by activating Notch signaling in preosteoblasts.
mTORC1 activates Notch pathway in preosteoblasts through the STAT3/p63/Jagged cascade
To define how activated mTORC1 influenced the Jagged1/Notch/Hes1 pathway, we determined the expression of STAT3 and p63, as STAT3 is a transcriptional activator of p63 [36] as well as the downstream target of mTOR [37], and p63 is a positive regulator of Jagged expression and Notch activity [38,39,40] which is induced by PI3K [41]. Cavarial cells with disruption of Tsc1 showed increased phosphorylation of STAT3 on Ser727 and expression of TP63 (total p63) (Fig 7A), and rapamycin reduced these parameters to normal levels. Knockdown of STAT3 by siRNA led to reduction in p63, Jagged1 and Hes1 without affecting the activity of mTORC1 (Fig 7B), and downregulation of p63 resulted in reduced Jagged1 and Hes1 but no changes of STAT3 phosphorylation and mTORC1 activity (Fig 7C). These findings suggested that the STAT3/p63 cascade is positively regulated by mTORC1 and serves as a conjunction between mTORC1 and the Notch pathway.
Fig. 7. mTORC1 impairs osteoblast differentiation through the STAT3/p63/Jagged/Notch cascade upstream of Runx2. 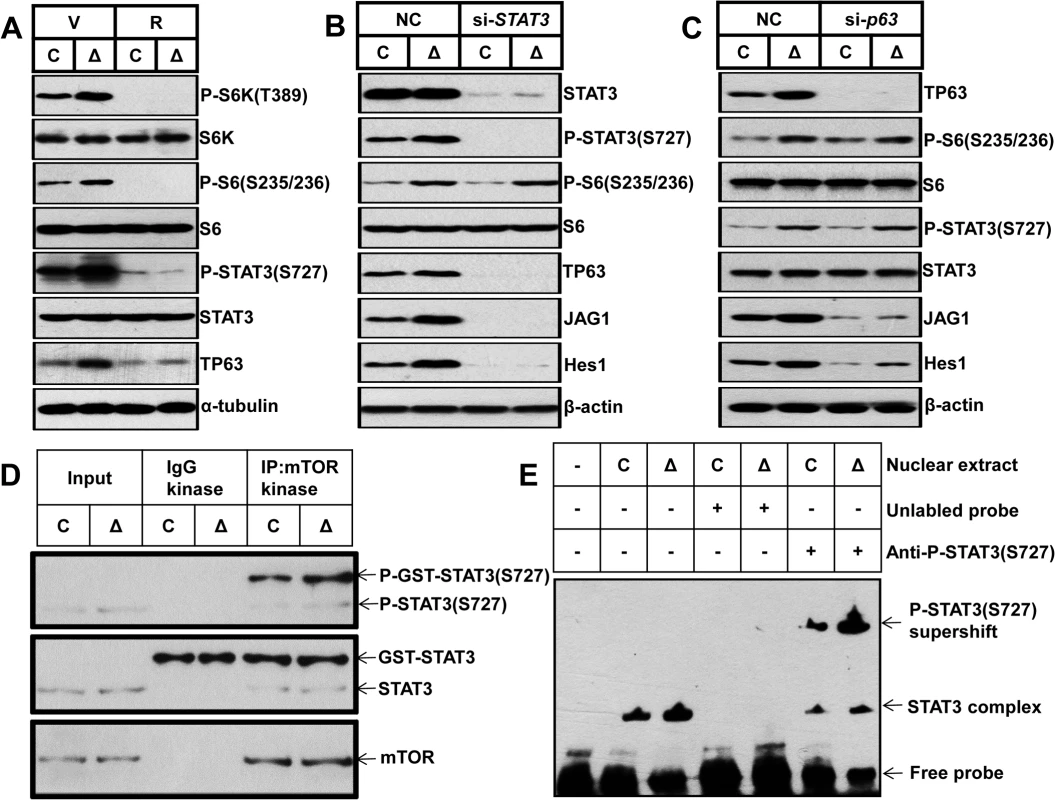
(A) Control and ΔTsc1 primary calvarial cells were treated with vehicle (V) or 0.1 nM rapamycin (R) for 24 hours and then underwent immunoblotting to detect phosphorylation of STAT3 (Ser727) and TP63 (total p63). Control and ΔTsc1 primary calvarial cells were treated with STAT3 (B) and p63 (C) siRNA and negative control (NC) for 48 hours and then underwent immunoblotting to detect mTORC1 activity, phosphorylation of STAT3(Ser727), TP63 (total p63) and activity of Notch pathway (JAG1, Hes1). (D) Cultured primary calvarial cells were immunoprecipitated with anti-mTOR antibody and the precipitated mTOR was assayed for kinase activity against recombinant GST-tagged full-length STAT3. (E) Nuclear extract from control and ΔTsc1 primary calvarial cells were analyzed using EMSA. Binding of STAT3 to biotin-labeled DNA probes is shown as “STAT3 complex”. To compete with the binding, an unlabeled STAT3 binding site DNA probe was added to the reaction in 200 times molar excess. Adding anti-pSTAT3 (S727) antibody to the reactions caused reduction of STAT3-DNA binding and bands of supershift. We next sought to define how mTORC1 regulates STAT3/p63 cascade in osteoblasts. As has been reported that mTORC1 directly phosphorylates STAT3 at Ser727 [37], we firstly examined whether mTORC1 could phosphorylate STAT3 at Ser727 in osteoblasts. We immunoprecipitated mTOR complex from control and ΔTsc1 calvarial cells and subjected the immunoprecipitates to in vitro kinase assays using a recombinant GST-tagged full-length STAT3 peptide. As shown in Fig 7D, mTOR immunoprecipitates could phosphorylate STAT3 at Ser727 in vitro and constitutive activated mTORC1 from ΔTsc1 calvarial cells enhanced the phosphorylation. Thus the elevated phosphorylation of STAT3 (S727) in ΔTsc1 osteoblasts is due to increased mTOR kinase activity. Next, we examined the role of phosphorylated STAT3 (S727) in the regulation of the p63 gene transcription by electrophoretic mobility shift assay (EMSA). Nuclear protein of osteoblasts bound specifically to a double-strand probe containing a consensus STAT3-specific binding sequence in the promoter region of ΔNp63 [36], and anti-pSTAT3 (S727) antibody significantly reduced the binding of STAT3 to p63 promoter (Fig 7E). Accordingly, nuclear protein from ΔTsc1 osteoblasts showed an increased binding of pSTAT3 (S727) to the probe (Fig 7E). Taken together, these results suggest that phosphorylated STAT3 (S727) may bind to p63 promoter to regulate its transcription in osteoblast, and mTORC1 activates the Notch pathway through STAT3/p63/Jagged cascade.
mTORC1 impairs preosteoblast differentiation through the Notch pathway upstream of Runx2
As mTOR is a positive regulator of the Jagged1/Notch/Hes1 pathway, we next determined whether hyperactive Notch signaling was responsible for the impaired differentiation of preosteoblasts by mTORC1 activation. Firstly, we examined the correlation between the level of Notch activity and differentiation of preosteoblasts. Low activity of Notch signaling is required during differentiation of preosteoblasts, as the expression of Jagged1, NICD and Hes1 was decreased in parallel with an increase in the markers of osteoblast differentiation (Fig 8A), and inhibition of the Notch pathway by DAPT or si-Notch1 promoted osteocalcin expression (S7A Fig and Fig 8B) and mineralized nodules formation in MC3T3-E1 cells (S7B Fig and Fig 8C). Importantly, a reduction in Notch by DAPT (S7C and S7D Fig) or siRNA (Fig 8D and 8E) potentiated the differentiation of ΔTsc1 preosteoblasts.
Fig. 8. mTORC1 impairs osteoblast differentiation through the Notch pathway upstream of Runx2. 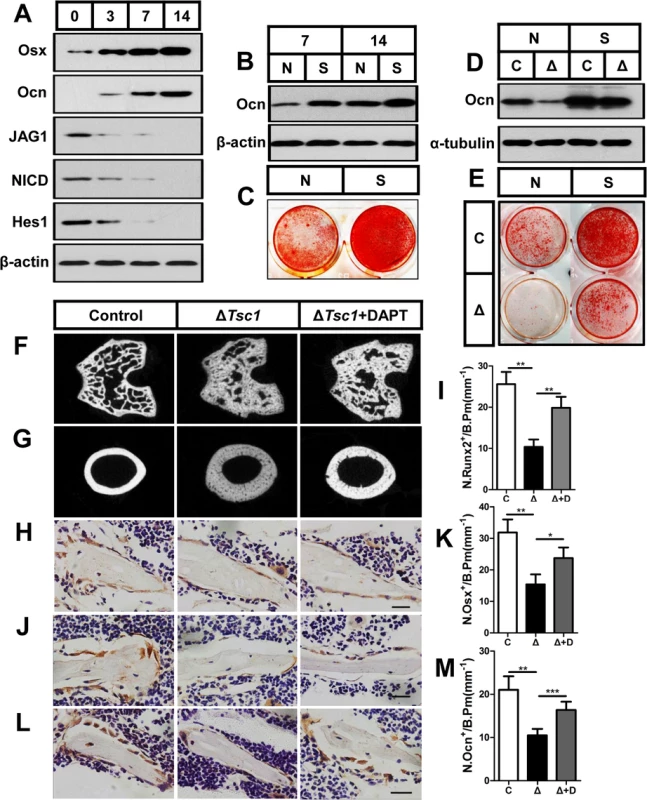
(A) Western blot analysis of the expression of Jagged1, NICD and Hes1 during differentiation of MC3T3-E1 cells. Differentiating MC3T3-E1 cells were treated with Notch1 siRNA (S) and then underwent immunoblotting to detect osteocalcin (B) on the 7th and 14th day and alizarin red staining (C) on the 14th day. Differentiating control (C) and ΔTsc1 (Δ) primary calvarial cells were treated with negative control (N) or si-Notch1 (S) and then underwent immunoblotting to detect osteocalcin (D) and alizarin red staining (E) on the 14th day. 6-week old ΔTsc1 mice were administered with DAPT (10mg/kg per day) or equivalent volume of DMSO for 4 weeks, micro-CT images of trabecular bone in secondary ossification center (F) and cortical bone of midshaft (G) of femur were shown. Immunohistochemistry staining for Runx2 (H), osterix (Osx) (J), and osteocalcin (Ocn) (L) in distal femur. Numbers of Runx2 (N.Runx2+) (I), osterix (N.Osx+) (K) and osteocalcin (N.Ocn+) (M) positive cells on the bone surface were measured as cells per millimeter of perimeter in sections (/B.Pm). All data are mean ± SD (n = 5 mice), all scale bars represent 50 μm. *P < 0.05, ** P < 0.01, *** P < 0.001 by t test. To test these results in vivo, we administered 6-week old mice with DAPT or equivalent volume of DMSO for 4 weeks. Micro-CT analysis of the distal femur showed a marked normalizing of bone architecture in DAPT-treated ΔTsc1 mice (Fig 8F and 8G and Table 4) when compared to their DMSO-treated control. Inhibition of Notch pathway by DAPT elevated expression of Runx2 and subsequent osterix and osteocalcin in osteoblasts (Fig 8H–8M), and significantly reversed the bone phenotypes of ΔTsc1 mice (Table 4). Therefore, Notch acts as an inhibitor of osteoblast differentiation, while deregulated mTORC1 activation impairs preosteoblast differentiation through activation of the Notch signaling pathway.
Tab. 4. Micro CT analysis of DAPT-treated ΔTsc1 mice at 10 weeks of age. 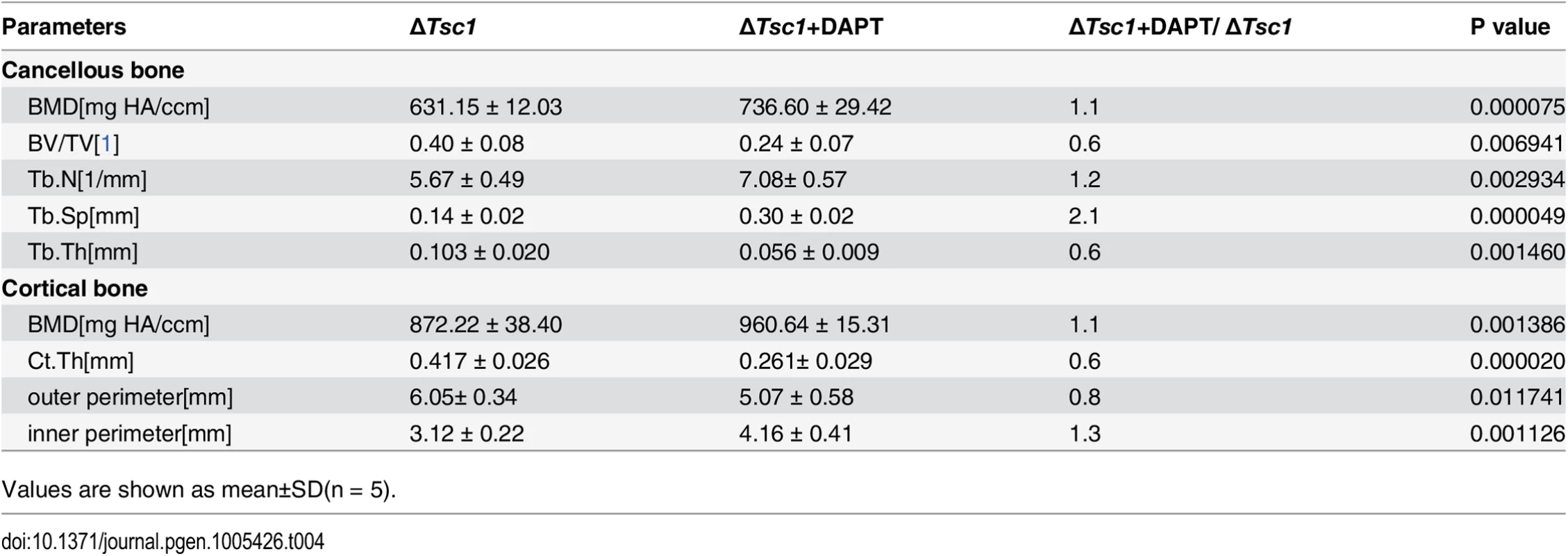
Values are shown as mean±SD(n = 5). Together, these in vitro and in vivo loss - and gain-of-function studies lend support for a central role of mTORC1 signaling in regulating both proliferation and differentiation of preosteoblast during bone homeostasis (S8 Fig).
Discussion
In this study, we defined the essential role of mTORC1 signaling in osteoblastgenesis. We observed a decline in mTORC1 activity during differentiation of preosteoblasts and enhancement of osteoblastic differentiation following inhibition of mTORC1 in vitro and in vivo. Using conditional knockout cell and mouse models, we further revealed that activation of mTORC1 prevented preosteoblast differentiation through activation of the Notch signaling pathway.
mTORC1 is ubiquitously expressed in all types of cells to regulate growth and metabolism. Proliferation in many types of cells is promoted by mTORC1, thus it is not surprising that the activity of mTORC1 positively correlated with the proliferative rate of preosteoblasts. However, it is interesting that mTORC1 activity resulted in a decline in the differentiation of preosteoblasts. We speculate that the decline in mTORC1 activity is due to differentiated osteoblasts. As osteoblasts differentiate, calcification of the extracellular matrix gradually increases, which generates hyperosmolarity and cellular stress. Coincidentally, this type of stimulus has been shown to inhibit mTORC1 [42].
The role of mTORC1 in bone formation was originally identified in various cell lines and animals treated with rapamycin, however, the results were controversial. Depending on the cell type, rapamycin either stimulates or inhibits osteoblastic differentiation. In rat osteosarcoma (ROS 17/2.8) cells, rapamycin inhibited proliferation, but promoted osteogenic differentiation [16]. In C2C12 cells, rapamycin potentiated the effect of BMP-2 inducing late markers of osteoblast differentiation [17]. In Human Embryonic Stem Cells, rapamycin induced the up-regulation of early osteogenic markers and further promoted the expression of late osteoblastic marker mRNA and/or proteins and mineralized bone nodule formation following induction for 2–3 weeks [43]. On the other hand, rapamycin has been shown to block osteogenic protein-1 induction of alkaline phosphatase activity in fetal rat calvarial cells [19] and reduce alkaline phosphatase activity, osteocalcin expression and the calcium content in mesenchymal stem cells [20]. In MC3T3-E1 subclone 4 (MC-4) cells and primary mouse bone marrow stromal cells (BMSCs), rapamycin inhibited osteoblast differentiation by targeting osteoblast proliferation and the early stage of osteoblast differentiation [21]. Although the reasons for the discrepant results are not readily apparent, one possible explanation is that the cells used in these reports were in various stages of osteoblastic differentiation and mTORC1 activity is stage-specifically required during the differentiation process. The results of the present study demonstrated that inhibition of mTORC1 is essential for preosteoblast differentiation. Another possible explanation for these contradictory results could be related to the treatment conditions including concentration and duration. Rapamycin at a concentration as low as 0.1 nM is effective in significantly suppressing mTORC1 activity [21], while higher concentrations and long-term treatment may produce a decrease in cell viability and growth, and inhibition of mTORC2. Thus, we used a low dose of rapamycin (0.1 nM) as a precaution against non-specific effects, and obtained reliable results that low mTORC1 activity is crucial for osteogenesis.
Previous results in animals administered rapamycin have also been inconsistent. Administration of rapamycin for 14 days resulted in no change in cancellous bone volume in rats [23], and after a longer duration of treatment (28 days), rats still lacked an osteopenic phenotype despite increased bone turnover [24]. However, mice treated with a similar concentration and duration of treatment showed a phenotype with less new bone formation and lower trabecular bone mass [25], due to a reduced number of osterix and osteocalcin positive cells coupled with unaffected osteoclasts. In the present study, rapamycin-treated mice had a similar phenotype of bone loss as well as decreased numbers of osterix and osteocalcin positive cells, except that the mean density of these cells was enhanced and the number of osteoclasts was reduced by rapamycin. Although we could not rule out that the systemic effect of rapamycin acted on other cell types to affect bone mass indirectly, the results of rapamycin administration in vivo are in agreement with those from rapamycin treated cells in vitro, which consolidates our conclusions that hyperactive mTORC1 is not required for differentiation of preosteoblasts and rapamycin promotes preosteoblast differentiation by blocking mTORC1.
Clinically, rapamycin was initially identified as a macrocyclic antifungal agent [44] and is used for immunosuppression in transplantation. The experimental and clinical trials have showed that rapamycin reduced brain, kidney, and skin lesions of tuberous sclerosis complex (TSC) [45, 46]. Some TSC children are being started on rapamycin at a young age (< 5 years), and it is sometimes continued for many (>5) years [47, 48]. In this situation, bone densitometry and morphological measurements must be advised to monitor the possible side effects of rapamycin on bone. Rapamycin is likely to impair bone microarchitecture and quality of those children as indicated by the current study, although it presented no effect on growth in weight and height of them [48].
Recent mouse genetic studies also highlighted the critical role of mTORC1 in skeletal development, whereas the function of mTORC1 in osteoblast formation is uncertain and the underlying mechanisms are not defined. Mice with Pten [49] and Tsc2 [50] disrupted by OC (osteocalcin)-cre recombinase (ΔTsc2) in mature osteoblasts shared an elevated mTORC1 activity, and exhibited different phenotypes. Although both mouse models had uniform high bone mass, osteoblasts with deletion of Pten showed increased osteoblastic differentiation, while Tsc2 disruption led to impaired differentiation of osteoblasts and mineralized nodule formation. Because inhibition of mTORC1 by rapamycin restored the differentiation defects in osteoblasts due to disruption of Tsc2, we reason that impairing osteoblastic differentiation is the function of hyperactive mTORC1 in mature osteoblasts, while an mTORC1-independent mechanism may account for the distinct phenotype in mice with disruption of Pten. Akt, which has been reported to promote osteoblast differentiation and bone development [51], is highly activated following Pten deletion and significantly inhibited following Tsc2 disruption. Acceleration of osteoblast differentiation by Akt may exceed the inhibition due to mTORC1 activation and cause increased differentiation in osteoblasts with Pten ablation. ΔTsc2 mice presented a similar phenotype and same differentiation defect in osteoblasts as ΔTsc1 mice in the current study. However, in contrast to ΔTsc1 mice, ΔTsc2 mice exhibited a decreased proliferation rate of osteoblast, which indicates that mTORC1 may play different role in regulating proliferation of immature and mature osteoblasts, since OC (osteocalcin) is expressed exclusively in mature osteoblasts. Moreover, though mTORC1 activation resulted in a same defect of differentiation in mature osteoblasts and preosteoblasts, the underlying mechanisms may be different. In this sense, our work revealed the role of mTORC1 in preosteoblast differentiation under physiological and pathological conditions and explored a different mechanism (STAT3/p63/Jagged/Notch pathway) responsible. More recently, another similar phenotype of immature woven bone as that of ΔTsc1 mice has been reported in mice with disruption of Lkb1 in preosteoblasts (ΔLkb1) [52], but the increased trabecular bone mass was more severe in ΔLkb1 mice than in ΔTsc1 mice, as BV/TV increased 7-fold versus 2.4-fold. In contrast, cortical thickness, which showed a 2.3-fold increase in ΔTsc1 mice, was decreased 3-fold in ΔLkb1 mice and osteoclasts were increased inversely in ΔLkb1 mice when compared with ΔTsc1 mice. These discrepancies may also be attributed to the activation of mTORC1-independent pathways in ΔLkb1 mice, as AMP kinase (AMPK), the target of Lkb1, regulates many signaling pathways besides mTORC1. In addition, there is no evidence to show that rapamycin restores bone phenotypes in ΔLkb1 mice. Unlike Lkb1, mTORC1 is a well-established target of TSC-TBC, and the phenotype of immature woven bone in ΔTsc1 mice was reversed by rapamycin, which further confirmed that bone changes in ΔTsc1 mice were the result of mTORC1 activation. Together, these findings demonstrate that differentiation of both preosteoblasts and osteoblasts is attenuated by mTORC1 activation.
Our data suggest that the impaired differentiation of preosteoblasts following mTORC1 activation is due to over-activated Notch signaling downstream. Notch signaling mediates communication between neighboring cells and decides their fate [32,33,34,35,53]. Specifically, gain of Notch function in preosteoblasts under the control of the type I collagen (Col1a1) promoter results in a phenotype similar to ΔTsc1 mice in the present study [32]. By inhibiting mTORC1 and the Notch pathway via rapamycin and DAPT/Si-Notch1, respectively, we showed that Notch acts downstream of mTORC1 and mediates the attenuation of osteoblastic differentiation by mTORC1. These findings are supported by a report which revealed that mTOR positively regulates Notch signaling in mouse and human cells through induction of the STAT3/p63/Jagged signaling cascade [28]. More recently, Wang et al. reported that mTORC1 activates Notch3 to accelerate the development of hypoxia-induced pulmonary hypertension [30] and Karbowniczek M et al. found that mTORC1 activates Notch in tuberous sclerosis complex and Drosophila external sensory organ development [54]. In contrast, Notch has also been shown to promote mTORC1 signaling by increasing Raptor protein expression in rat hepatoma cells and primary mouse hepatocytes [55]. We speculate that the interaction between mTORC1 and Notch may depend on cell type. Our data also confirmed that Runx2 is negatively regulated by mTORC1 and identified the Notch pathway as the responsible mechanism. Notch was identified as a major mediator of mTORC1 signaling in the impairment of preosteoblast differentiation in the present study.
In summary, this study clarified the potential role of mTORC1 signaling in the regulation of preosteoblast proliferation and differentiation and identified Notch signaling and Runx2 as critical downstream mediators. Pharmaceutical coordination of the pathways and agents in preosteoblasts may be beneficial in bone formation.
Materials and Methods
Cells
The preosteoblast cell line, MC3T3-E1, was maintained in alpha-MEM (Gibco) supplemented with 10% FBS (Gibco), 100 U/ml penicillin, and 100 mg/ml streptomycin sulfate, at 37°C with 5% CO2. For growth curve analysis, MC3T3-E1 cells were plated in six-well plates at a density of 8×104 cells/well and cultured until confluent (8th day). Growth rate was assessed by cell counting. Primary osteoblastic cells were prepared from the calvaria of 21-day-old fetal rats or newborn mice as described previously [56, 57, 58] and cultured using the same method as for MC3T3-E1 cells.
For osteogenic induction, 100 μg/ ml ascorbic acid (Sigma-Aldrich) and 10 mM β-glycerol phosphate (Sigma-Aldrich) were added to confluent cells. Rapamycin (Sigma-Aldrich) was added as stated in the Results section. Alizarin red staining was carried out according to standard techniques.
Mice
The Southern Medical University Animal Care and Use Committee approved all procedures involving mice. Mice importing, transporting, housing and breeding were conducted according to the recommendations of "The use of non-human primates in research." Newborn C57BL/6 mice were purchased from the Laboratory Animal Centre of Southern Medical University. Tsc1flox/flox [59] and Osx-GFP::Cre [60] mice were both purchased from The Jackson Laboratory. The background of Tsc1flox/flox mice is 129S4/SvJae, and Osx-GFP::Cre mice were backcrossed on to a 129S4/SvJae background for 8 generations prior to use. We performed genotyping using genomic DNA isolated from tail biopsies, and the primers used are shown in S1 Table. The specificity of recombination was examined by PCR using primers flanking the floxed allele (S1 Table). Mice were sacrificed by cervical dislocation to ameliorate suffering.
Preparation of decalcified sections, histochemistry and immunohistochemistry (IHC) and histomorphometric analyses
Femur tissues dissected from the mice were fixed using 4% paraformaldehyde in PBS at 4°C for 24 hours and then decalcified in 15% EDTA (pH 7.4) at 4°C for 14 days. The tissues were embedded in paraffin or optimal cutting temperature (OCT) compound (Sakura Finetek), and 2–5 μm sagittal-oriented sections were prepared for histological analyses. H&E and Toluidine blue staining was performed as previously described [61]. Tartrate-resistant acid phosphatase (TRAP) or alkaline phosphatase (ALP) staining was performed using a standard protocol (Sigma-Aldrich). For IHC, we incubated primary antibodies which recognized mouse phospho-S6 ribosomal protein (Ser235/236) (Cell Signaling, 1 : 100, #2211), proliferating cell nuclear antigen (PCNA) (Cell Signaling 1 : 200, #13110), osterix (Abcam, 1 : 500, ab22552), osteocalcin (Abcam, 1 : 500, ab93876), and Runx2 (Cell Signaling, 1 : 100) overnight at 4°C. All sections were observed and photographed on Olympus BX51 microscope. Immunohistochemical staining was evaluated by cell number counting and computerized optical density (OD) measurements. In proliferation analysis, osteoblast proliferation fraction was calculated as BrdU+ osteoblasts per total osteoblasts on bone surface. Osteoblasts on bone surface were discerned by morphology and calculated by two independent observers blinded to the groups. In immunohistochemistry assays, cells per bone perimeter (B.Pm) was used to calculate the number of positive cells, and integrated optical density per area of positive cells (IOD/area, mean density) was used to quantify the staining intensity by detecting in 6 different images taken at 100x magnification with Image Pro Plus 6.0 software (Media Cybernetics, MD,USA)[62]. Briefly, positively stained regions of the image were selected by HSI (hue, saturation and lightness) with S from 0–255, I from 0–210 and H from 0–25, and then the brown color was converted into grayscale signal. The grayscale signal was measured as mean optical intensity of staining (mean density) within the tissue masks. At least three mice per group were examined. Three equidistant sections spaced at 200 μm apart throughout the midsagittal section of femur were evaluated.
X-ray and micro-CT analysis
The narcotized mice were analyzed using X-ray radiography. Quantitative analysis was performed in mice femora at 12 μm resolution on a micro-CT Scanner (Viva CT40; Scanco Medical AG, Bassersdorf, Switzerland) [63]. Briefly, scanning was performed at the lower growth plate in the femora and extended proximally for 300 slices. We started morphometric analysis with the first slice in which the femoral condyles were fully merged and extended for 100 slices proximally. Using a contouring tool, we segmented the trabecular bone from the cortical shell manually on key slices, and morphed the contours automatically to segment the trabecular bone on all slices. The three-dimensional structure and morphometry were constructed and analyzed for BV/TV (%), BMD (mg HA/mm3), Tb.N. (mm–1), Tb.Th. (mm) and Tb.Sp (mm). We also performed micro CT imaging in the mid-diaphysis of the femur and performed mid-shaft evaluation of 100 slices to quantify the cortical thickness, bone mineral density and outer/inner perimeter of the mid-shaft.
Western blot
Cells and tissues were lysated by 2% sodium dodecyl sulfate with 2 M urea, 10% glycerol, 10 mM Tris-HCl (pH 6.8), 10 mM dithiothreitol and 1 mM phenylmethylsulfonyl fluoride. The lysates were centrifuged and the supernatants were separated by SDS-PAGE and blotted onto a nitrocellulose (NC) membrane (Bio-Rad Laboratories). The membrane was then analyzed using specific antibodies and visualized by enhanced chemiluminescence (ECL Kit, Amersham Biosciences).
Preparation of undecalcified histology sections
To label the mineralization fronts, 10-week-old mice were subcutaneously injected with calcin (Sigma, 15 mg/kg body weight) in 2% sodium bicarbonate solution 10 days and 3 days before death [25]. After dissection, the femurs were fixed in 4% paraformaldehyde for 24 hours. They were then dehydrated through a graded series of ethanol (70–100%) and xylene before being embedded in methylmethacrylate (MMA) without prior decalcification [64]. 5 μm-thick sections were prepared for Goldner’s-Masson trichrome [65], and 10 μm-thick sections were prepared for double-labeling fluorescent analysis.
Bromodeoxyuridine incorporation
After initial culture for 48 hours, primary calvarial cells were replated and expanded for an additional 24 hours. The cells were then treated with BrdU labeling reagent (Invitrogen) for 6 hours according to the manufacturer’s instructions and washed with PBS. The cells were fixed with 70% ethanol for 25 min at 4°C, and then stained for immunocytochemical analysis. Three to five areas for each group (n = 3 slides) were counted by two independent observers blinded to the groups. BrdU-positive cells were scored visually. For in vivo proliferation analysis, mice were injected with BrdU (1ml/100g body weight) 2 hours before sacrifice.
Scanning electron microscopy (SEM)
The surface of the MMA embedded femurs were polished and acid-etched with 37% phosphoric acid for 2–10s. After washing for 5 min with 5% sodium hypochlorite they were coated with gold and palladium before examining with SEM (S-3700N, Hitachi, Japan).
Doxycycline treatment
To prevent the Osx promoter from driving Cre expression, pregnant mice were exposed to 200 μg/ml doxycycline (Sigma-Aldrich) in drinking water until their progeny had been processed for calvarial cell isolation. Doxycycline 100 μg/ml was added sequentially to the isolated cells until they reached confluence.
siRNA knockdown
We transiently transfected cells with siRNA using Lipofectamine RNAi MAX (Invitrogen, Carlsbad, CA, USA) in Opti-MEM medium (Invitrogen), according to the manufacturer’s instructions. The efficiency of transfection was measured by western blot. The sequences of siRNA used in this study were as follows: Notch1: sense: 5’-CCAAGAAGUUCCGGUUUGATT-3’, and anti-sense: 5’-UCAAACCGGAACUUCUUGGTT-3’; STAT3: 5’-CTGGATAACTTCATTAGCA-3’; p63 (all isoforms): 5’-CACAGACCACGCACAGAAUdTdT-3’, 5’-UCCAGAUGACUUCCAUCAAdTdT-3’ (1 : 1 ratio mixture). Non-specific siRNA sequences were used as negative controls: sense: 5’-UUCUCCGAACGUGUCACGUTT-3’, and anti-sense: 5’-ACGUGACACGUUCGGAGAATT-3’. (GenePharma, Shanghai, China).
In vitro kinase assay for mTORC1
Primary calvarial cells were lysed in ice-cold buffer (40mM HEPES (pH 7.4), 2mM EDTA, 10mM pyrophosphate, 10mM glycerophosphate, 0.3% CHAPS, and one tablet of EDTA-free protease inhibitors (Roche, Basel, Switzerland) per 25 ml). Supernatants were incubated with anti-mTOR antibody for 2h at 4°C, followed by addition of 30 μl of 50% slurry of protein G Sepharose beads for another 1h. Beads were then washed four times with lysis buffer and once kinase buffer (25mM HEPES (pH7.4), 50mM KCl, 10mM MgCl2, 250 μM ATP). 0.4μg of recombinant GST-tagged full-length STAT3 peptide (Creative BioMart, #1496H) was added to 30μl kinase buffer. Kinase assays were performed for 30 min at 30°C, and terminated by the addition of the 2×SDS sample buffer followed by boiling for 5 min.
EMSA
A total of 2μg of nuclear protein extracted from calvarial cells was incubated with biotin-labeled STAT3 binding-site DNA probe in binding buffer (EMSA kit; Thermo scientific) for 30 minutes at room temperature. The probe used for the reaction contains the STAT3 binding site of the ΔNp63 promoter with a sequence of 5'-GGATTCCTATTTCCCGTACATAATATGGAT-3'. After incubation, the samples were separated on a 6% polyacrylamide gel in Tris-borate ethylenediaminetetraacetic acid, transferred onto a nylon membrane, and fixed on the membrane by ultraviolet cross-linking. The biotin-labeled probe was detected with streptavidin-horseradish peroxidase (EMSA kit; Thermo scientific). A probe lacking nuclear extracts was used as a negative control. The specificity of the identified STAT3-DNA binding activity was confirmed by using a 200-fold excess of unlabeled probe containing a same sequence. For supershift analysis, 1 μg monoclonal anti-phosphorylated STAT3 (S727) (Cell Signaling Technology) was incubated with nuclear extracts for 30 minutes before the addition of the biotin-labeled DNA probe.
Statistics
All results are presented as the mean ± S.D. Curve analysis was determined using Prism (GraphPad). The data in each group were analyzed using unpaired, two-tailed Student’s t-test. The level of significance was set at P < 0.05.
Supporting Information
Zdroje
1. Erlebacher A, Filvaroff EH, Gitelman SE, Derynck R Toward a molecular understanding of skeletal development. Cell.1995; 80 : 371–378. 7859279
2. Manolagas SC Birth and death of bone cells: basic regulatory mechanisms and implications for the pathogenesis and treatment of osteoporosis. Endocr Rev.2000; 21 : 115–137. 10782361
3. Sandhu SK, Hampson G The pathogenesis, diagnosis, investigation and management of osteoporosis. J Clin Pathol.2011; 64 : 1042–1050. doi: 10.1136/jcp.2010.077842 21896577
4. Ducy P, Schinke T, Karsenty G The osteoblast: a sophisticated fibroblast under central surveillance. Science.2000; 289 : 1501–1504. 10968779
5. Long F Building strong bones: molecular regulation of the osteoblast lineage. Nat Rev Mol Cell Biol.2012; 13 : 27–38.
6. Laplante M, Sabatini DM mTOR signaling at a glance. Journal of Cell Science.2009; 122 : 3589–3594. doi: 10.1242/jcs.051011 19812304
7. Yang Q, Guan K Expanding mTOR signaling. Cell Research.2007; 17 : 666–681. 17680028
8. Dibble CC, Elis W, Menon S, Qin W, Klekota J, Asara JM, et al. TBC1D7 is a third subunit of the TSC1-TSC2 complex upstream of mTORC1. Mol Cell.2012; 47 : 535–546. doi: 10.1016/j.molcel.2012.06.009 22795129
9. Hay N Upstream and downstream of mTOR. Genes & Development.2004; 18 : 1926–1945.
10. Barron RP, Kainulainen VT, Forrest CR, Krafchik B, Mock D, Sandor GK Tuberous sclerosis: clinicopathologic features and review of the literature. J Craniomaxillofac Surg.2002; 30 : 361–366. 12425991
11. Bernauer TA, Mirowski GW, Caldemeyer KS Tuberous sclerosis. Part II. Musculoskeletal and visceral findings. J Am Acad Dermatol.2001; 45 : 450–452. 11511845
12. Pui MH, Kong HL, Choo HF Bone changes in tuberous sclerosis mimicking metastases. Australas Radiol.1996; 40 : 77–79. 8838896
13. DICKERSON WW Nature of certain osseous lesions in tuberous sclerosis. AMA Arch Neurol Psychiatry.1955; 73 : 525–529. 14360867
14. HOLT JF, DICKERSON WW The osseous lesions of tuberous sclerosis. Radiology.1952; 58 : 1–8. 14883368
15. Wood AR, Esko T, Yang J, Vedantam S, Pers TH, Gustafsson S, et al. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat Genet.2014 : 46–1173.
16. Ogawa T, Tokuda M, Tomizawa K, Matsui H, Itano T, Konishi R, et al. Osteoblastic differentiation is enhanced by rapamycin in rat osteoblast-like osteosarcoma (ROS 17/2.8) cells. Biochem Biophys Res Commun.1998; 249 : 226–230. 9705862
17. Vinals F, Lopez-Rovira T, Rosa JL, Ventura F Inhibition of PI3K/p70 S6K and p38 MAPK cascades increases osteoblastic differentiation induced by BMP-2. FEBS Lett.2002; 510 : 99–104. 11755539
18. Martin SK, Fitter S, Bong LF, Drew JJ, Gronthos S, Shepherd PR, et al. NVP-BEZ235, a dual pan class I PI3 kinase and mTOR inhibitor, promotes osteogenic differentiation in human mesenchymal stromal cells. J Bone Miner Res.2010; 25 : 2126–2137. doi: 10.1002/jbmr.114 20499346
19. Shoba LN, Lee JC Inhibition of phosphatidylinositol 3-kinase and p70S6 kinase blocks osteogenic protein-1 induction of alkaline phosphatase activity in fetal rat calvaria cells. J Cell Biochem.2003; 88 : 1247–1255. 12647306
20. Isomoto S, Hattori K, Ohgushi H, Nakajima H, Tanaka Y, Takakura Y Rapamycin as an inhibitor of osteogenic differentiation in bone marrow-derived mesenchymal stem cells. J Orthop Sci.2007; 12 : 83–88. 17260122
21. Singha UK, Jiang Y, Yu S, Luo M, Lu Y, Zhang J, et al. Rapamycin inhibits osteoblast proliferation and differentiation in MC3T3-E1 cells and primary mouse bone marrow stromal cells. J Cell Biochem.2008; 103 : 434–446. 17516572
22. Yeh LC, Ma X, Ford JJ, Adamo ML, Lee JC Rapamycin inhibits BMP-7-induced osteogenic and lipogenic marker expressions in fetal rat calvarial cells. J Cell Biochem.2013; 114 : 1760–1771. doi: 10.1002/jcb.24519 23444145
23. Joffe I, Katz I, Sehgal S, Bex F, Kharode Y, Tamasi J, et al. Lack of change of cancellous bone volume with short-term use of the new immunosuppressant rapamycin in rats. Calcif Tissue Int.1993; 53 : 45–52. 8348384
24. Romero DF, Buchinsky FJ, Rucinski B, Cvetkovic M, Bryer HP, Liang XG, et al. Rapamycin: a bone sparing immunosuppressant? J Bone Miner Res.1995; 10 : 760–768. 7543725
25. Xian L, Wu X, Pang L, Lou M, Rosen CJ, Qiu T, et al. Matrix IGF-1 maintains bone mass by activation of mTOR in mesenchymal stem cells. Nat Med.2012; 18 : 1095–1101. doi: 10.1038/nm.2793 22729283
26. Artavanis-Tsakonas S, Rand MD, Lake RJ Notch signaling: cell fate control and signal integration in development. Science.1999; 284 : 770–776. 10221902
27. Chiba S Notch signaling in stem cell systems. Stem Cells.2006; 24 : 2437–2447. 16888285
28. Ma J, Meng Y, Kwiatkowski DJ, Chen X, Peng H, Sun Q, et al. Mammalian target of rapamycin regulates murine and human cell differentiation through STAT3/p63/Jagged/Notch cascade. J Clin Invest.2010; 120 : 103–114. doi: 10.1172/JCI37964 20038814
29. Karbowniczek M, Zitserman D, Khabibullin D, Hartman T, Yu J, Morrison T, et al. The evolutionarily conserved TSC/Rheb pathway activates Notch in tuberous sclerosis complex and Drosophila external sensory organ development. J Clin Invest.2010; 120 : 93–102. doi: 10.1172/JCI40221 20038815
30. Wang W, Liu J, Ma A, Miao R, Jin Y, Zhang H, et al. mTORC1 Is Involved in Hypoxia-Induced Pulmonary Hypertension Through the Activation of Notch3. J Cell Physiol.2014; 229 : 2117–2125. doi: 10.1002/jcp.24670 24825564
31. Wang D, Christensen K, Chawla K, Xiao G, Krebsbach PH, Franceschi RT Isolation and characterization of MC3T3-E1 preosteoblast subclones with distinct in vitro and in vivo differentiation/mineralization potential. J Bone Miner Res.1999; 14 : 893–903. 10352097
32. Engin F, Yao Z, Yang T, Zhou G, Bertin T, Jiang MM, et al. Dimorphic effects of Notch signaling in bone homeostasis. Nat Med.2008; 14 : 299–305. doi: 10.1038/nm1712 18297084
33. Deregowski V, Gazzerro E, Priest L, Rydziel S, Canalis E Notch 1 overexpression inhibits osteoblastogenesis by suppressing Wnt/beta-catenin but not bone morphogenetic protein signaling. J Biol Chem.2006; 281 : 6203–6210. 16407293
34. Hilton MJ, Tu X, Wu X, Bai S, Zhao H, Kobayashi T, et al. Notch signaling maintains bone marrow mesenchymal progenitors by suppressing osteoblast differentiation. Nat Med.2008; 14 : 306–314. doi: 10.1038/nm1716 18297083
35. Zanotti S, Smerdel-Ramoya A, Stadmeyer L, Durant D, Radtke F, Canalis E Notch inhibits osteoblast differentiation and causes osteopenia. Endocrinology.2008; 149 : 3890–3899. doi: 10.1210/en.2008-0140 18420737
36. Chu WK, Dai PM, Li HL, Chen JK Transcriptional activity of the DeltaNp63 promoter is regulated by STAT3. J Biol Chem.2008; 283 : 7328–7337. doi: 10.1074/jbc.M800183200 18198175
37. Yokogami K, Wakisaka S, Avruch J, Reeves SA Serine phosphorylation and maximal activation of STAT3 during CNTF signaling is mediated by the rapamycin target mTOR. Curr Biol.2000; 10 : 47–50. 10660304
38. Sasaki Y, Ishida S, Morimoto I, Yamashita T, Kojima T, Kihara C, et al. The p53 family member genes are involved in the Notch signal pathway. J Biol Chem.2002; 277 : 719–724. 11641404
39. Laurikkala J, Mikkola ML, James M, Tummers M, Mills AA, Thesleff I p63 regulates multiple signalling pathways required for ectodermal organogenesis and differentiation. Development.2006; 133 : 1553–1563. 16524929
40. Murata K, Ota S, Niki T, Goto A, Li CP, Ruriko UM, et al. p63—Key molecule in the early phase of epithelial abnormality in idiopathic pulmonary fibrosis. Exp Mol Pathol.2007; 83 : 367–376. 17498688
41. Barbieri CE, Barton CE, Pietenpol JA Delta Np63 alpha expression is regulated by the phosphoinositide 3-kinase pathway. J Biol Chem.2003; 278 : 51408–51414. 14555649
42. Parrott LA, Templeton DJ Osmotic stress inhibits p70/85 S6 kinase through activation of a protein phosphatase. J Biol Chem.1999; 274 : 24731–24736. 10455142
43. Lee K, Yook J, Son M, Kim M, Koo D, Han Y, et al. Rapamycin Promotes the Osteoblastic Differentiation of Human Embryonic Stem Cells by Blocking the mTOR Pathway and Stimulating the BMP/Smad Pathway. Stem Cells and Development.2010; 19 : 557–568. doi: 10.1089/scd.2009.0147 19642865
44. Vezina C, Kudelski A, Sehgal SN Rapamycin (AY-22,989), a new antifungal antibiotic. I. Taxonomy of the producing streptomycete and isolation of the active principle. J Antibiot (Tokyo).1975; 28 : 721–726.
45. Herry I, Neukirch C, Debray MP, Mignon F, Crestani B Dramatic effect of sirolimus on renal angiomyolipomas in a patient with tuberous sclerosis complex. Eur J Intern Med.2007; 18 : 76–77. 17223050
46. Micozkadioglu H, Koc Z, Ozelsancak R, Yildiz I Rapamycin therapy for renal, brain, and skin lesions in a tuberous sclerosis patient. Ren Fail.2010; 32 : 1233–1236. doi: 10.3109/0886022X.2010.517345 20954988
47. Canpolat M, Per H, Gumus H, Yikilmaz A, Unal E, Patiroglu T, et al. Rapamycin has a beneficial effect on controlling epilepsy in children with tuberous sclerosis complex: results of 7 children from a cohort of 86. Childs Nerv Syst.2014; 30 : 227–240. doi: 10.1007/s00381-013-2185-6 23743820
48. Kotulska K, Chmielewski D, Borkowska J, Jurkiewicz E, Kuczynski D, Kmiec T, et al. Long-term effect of everolimus on epilepsy and growth in children under 3 years of age treated for subependymal giant cell astrocytoma associated with tuberous sclerosis complex. Eur J Paediatr Neurol.2013; 17 : 479–485. doi: 10.1016/j.ejpn.2013.03.002 23567018
49. Liu X, Bruxvoort KJ, Zylstra CR, Liu J, Cichowski R, Faugere MC, et al. Lifelong accumulation of bone in mice lacking Pten in osteoblasts. Proc Natl Acad Sci U S A.2007; 104 : 2259–2264. 17287359
50. Riddle RC, Frey JL, Tomlinson RE, Ferron M, Li Y, DiGirolamo DJ, et al. Tsc2 is a molecular checkpoint controlling osteoblast development and glucose homeostasis. Mol Cell Biol.2014; 34 : 1850–1862. doi: 10.1128/MCB.00075-14 24591652
51. Mukherjee A, Rotwein P Akt promotes BMP2-mediated osteoblast differentiation and bone development. J Cell Sci.2009; 122 : 716–726. doi: 10.1242/jcs.042770 19208758
52. Lai LP, Lotinun S, Bouxsein ML, Baron R, McMahon AP Stk11 (Lkb1) deletion in the osteoblast lineage leads to high bone turnover, increased trabecular bone density and cortical porosity. Bone.2014.
53. Canalis E, Parker K, Feng JQ, Zanotti S Osteoblast lineage-specific effects of notch activation in the skeleton. Endocrinology.2013; 154 : 623–634. doi: 10.1210/en.2012-1732 23275471
54. Karbowniczek M, Zitserman D, Khabibullin D, Hartman T, Yu J, Morrison T, et al. The evolutionarily conserved TSC/Rheb pathway activates Notch in tuberous sclerosis complex and Drosophila external sensory organ development. J Clin Invest.2010; 120 : 93–102. doi: 10.1172/JCI40221 20038815
55. Shimobayashi M, Hall MN Making new contacts: the mTOR network in metabolism and signalling crosstalk. Nat Rev Mol Cell Biol.2014; 15 : 155–162. doi: 10.1038/nrm3757 24556838
56. Rao LG, Ng B, Brunette DM, Heersche JN Parathyroid hormone - and prostaglandin E1-response in a selected population of bone cells after repeated subculture and storage at -80C. Endocrinology.1977; 100 : 1233–1241. 191237
57. Bhargava U, Bar-Lev M, Bellows CG, Aubin JE Ultrastructural analysis of bone nodules formed in vitro by isolated fetal rat calvaria cells. Bone.1988; 9 : 155–163. 3166832
58. Bellows CG, Aubin JE Determination of numbers of osteoprogenitors present in isolated fetal rat calvaria cells in vitro. Dev Biol.1989; 133 : 8–13. 2707489
59. Kwiatkowski DJ, Zhang H, Bandura JL, Heiberger KM, Glogauer M, El-Hashemite N, et al. A mouse model of TSC1 reveals sex-dependent lethality from liver hemangiomas, and up-regulation of p70S6 kinase activity in Tsc1 null cells. Hum Mol Genet.2002; 11 : 525–534. 11875047
60. Rodda SJ, McMahon AP Distinct roles for Hedgehog and canonical Wnt signaling in specification, differentiation and maintenance of osteoblast progenitors. Development.2006; 133 : 3231–3244. 16854976
61. Ye L, Mishina Y, Chen D, Huang H, Dallas SL, Dallas MR, et al. Dmp1-deficient mice display severe defects in cartilage formation responsible for a chondrodysplasia-like phenotype. J Biol Chem.2005; 280 : 6197–6203. 15590631
62. Iakovlev VV, Gabril M, Dubinski W, Scorilas A, Youssef YM, Faragalla H, et al. Microvascular density as an independent predictor of clinical outcome in renal cell carcinoma: an automated image analysis study. Lab Invest.2012; 92 : 46–56. doi: 10.1038/labinvest.2011.153 22042086
63. Hildebrand T, Laib A, Muller R, Dequeker J, Ruegsegger P Direct three-dimensional morphometric analysis of human cancellous bone: microstructural data from spine, femur, iliac crest, and calcaneus. J Bone Miner Res.1999; 14 : 1167–1174. 10404017
64. Sun Y, Lu Y, Chen L, Gao T, D'Souza R, Feng JQ, et al. DMP1 processing is essential to dentin and jaw formation. J Dent Res.2011; 90 : 619–624. doi: 10.1177/0022034510397839 21297011
65. Zhou X, Zhang Z, Feng JQ, Dusevich VM, Sinha K, Zhang H, et al. Multiple functions of Osterix are required for bone growth and homeostasis in postnatal mice. Proc Natl Acad Sci U S A.2010; 107 : 12919–12924. doi: 10.1073/pnas.0912855107 20615976
Štítky
Genetika Reprodukční medicína
Článek Loss and Gain of Natural Killer Cell Receptor Function in an African Hunter-Gatherer PopulationČlánek Let-7 Represses Carcinogenesis and a Stem Cell Phenotype in the Intestine via Regulation of Hmga2Článek Binding of Multiple Rap1 Proteins Stimulates Chromosome Breakage Induction during DNA ReplicationČlánek SLIRP Regulates the Rate of Mitochondrial Protein Synthesis and Protects LRPPRC from DegradationČlánek Protein Composition of Infectious Spores Reveals Novel Sexual Development and Germination Factors inČlánek The Formin Diaphanous Regulates Myoblast Fusion through Actin Polymerization and Arp2/3 RegulationČlánek Runx1 Transcription Factor Is Required for Myoblasts Proliferation during Muscle Regeneration
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 8
-
Všechny články tohoto čísla
- Putting the Brakes on Huntington Disease in a Mouse Experimental Model
- Identification of Driving Fusion Genes and Genomic Landscape of Medullary Thyroid Cancer
- Evidence for Retromutagenesis as a Mechanism for Adaptive Mutation in
- TSPO, a Mitochondrial Outer Membrane Protein, Controls Ethanol-Related Behaviors in
- Evidence for Lysosome Depletion and Impaired Autophagic Clearance in Hereditary Spastic Paraplegia Type SPG11
- Loss and Gain of Natural Killer Cell Receptor Function in an African Hunter-Gatherer Population
- Trans-Reactivation: A New Epigenetic Phenomenon Underlying Transcriptional Reactivation of Silenced Genes
- Early Developmental and Evolutionary Origins of Gene Body DNA Methylation Patterns in Mammalian Placentas
- Strong Selective Sweeps on the X Chromosome in the Human-Chimpanzee Ancestor Explain Its Low Divergence
- Dominance of Deleterious Alleles Controls the Response to a Population Bottleneck
- Transient 1a Induction Defines the Wound Epidermis during Zebrafish Fin Regeneration
- Systems Genetics Reveals the Functional Context of PCOS Loci and Identifies Genetic and Molecular Mechanisms of Disease Heterogeneity
- A Genome Scale Screen for Mutants with Delayed Exit from Mitosis: Ire1-Independent Induction of Autophagy Integrates ER Homeostasis into Mitotic Lifespan
- Non-synonymous FGD3 Variant as Positional Candidate for Disproportional Tall Stature Accounting for a Carcass Weight QTL () and Skeletal Dysplasia in Japanese Black Cattle
- The Relationship between Gene Network Structure and Expression Variation among Individuals and Species
- Calmodulin Methyltransferase Is Required for Growth, Muscle Strength, Somatosensory Development and Brain Function
- The Wnt Frizzled Receptor MOM-5 Regulates the UNC-5 Netrin Receptor through Small GTPase-Dependent Signaling to Determine the Polarity of Migrating Cells
- Nbs1 ChIP-Seq Identifies Off-Target DNA Double-Strand Breaks Induced by AID in Activated Splenic B Cells
- CCNYL1, but Not CCNY, Cooperates with CDK16 to Regulate Spermatogenesis in Mouse
- Evidence for a Common Origin of Blacksmiths and Cultivators in the Ethiopian Ari within the Last 4500 Years: Lessons for Clustering-Based Inference
- Of Fighting Flies, Mice, and Men: Are Some of the Molecular and Neuronal Mechanisms of Aggression Universal in the Animal Kingdom?
- Hypoxia and Temperature Regulated Morphogenesis in
- The Homeodomain Iroquois Proteins Control Cell Cycle Progression and Regulate the Size of Developmental Fields
- Evolution and Design Governing Signal Precision and Amplification in a Bacterial Chemosensory Pathway
- Rac1 Regulates Endometrial Secretory Function to Control Placental Development
- Let-7 Represses Carcinogenesis and a Stem Cell Phenotype in the Intestine via Regulation of Hmga2
- Functions as a Positive Regulator of Growth and Metabolism in
- The Nucleosome Acidic Patch Regulates the H2B K123 Monoubiquitylation Cascade and Transcription Elongation in
- Rhoptry Proteins ROP5 and ROP18 Are Major Murine Virulence Factors in Genetically Divergent South American Strains of
- Exon 7 Contributes to the Stable Localization of Xist RNA on the Inactive X-Chromosome
- Regulates Refractive Error and Myopia Development in Mice and Humans
- mTORC1 Prevents Preosteoblast Differentiation through the Notch Signaling Pathway
- Regulation of Gene Expression Patterns in Mosquito Reproduction
- Molecular Basis of Gene-Gene Interaction: Cyclic Cross-Regulation of Gene Expression and Post-GWAS Gene-Gene Interaction Involved in Atrial Fibrillation
- The Spalt Transcription Factors Generate the Transcriptional Landscape of the Wing Pouch Central Region
- Binding of Multiple Rap1 Proteins Stimulates Chromosome Breakage Induction during DNA Replication
- Functional Divergence in the Role of N-Linked Glycosylation in Smoothened Signaling
- YAP1 Exerts Its Transcriptional Control via TEAD-Mediated Activation of Enhancers
- Coordinated Evolution of Influenza A Surface Proteins
- The Evolutionary Potential of Phenotypic Mutations
- Genome-Wide Association and Trans-ethnic Meta-Analysis for Advanced Diabetic Kidney Disease: Family Investigation of Nephropathy and Diabetes (FIND)
- New Routes to Phylogeography: A Bayesian Structured Coalescent Approximation
- SLIRP Regulates the Rate of Mitochondrial Protein Synthesis and Protects LRPPRC from Degradation
- Satellite DNA Modulates Gene Expression in the Beetle after Heat Stress
- SHOEBOX Modulates Root Meristem Size in Rice through Dose-Dependent Effects of Gibberellins on Cell Elongation and Proliferation
- Reduced Crossover Interference and Increased ZMM-Independent Recombination in the Absence of Tel1/ATM
- Suppression of Somatic Expansion Delays the Onset of Pathophysiology in a Mouse Model of Huntington’s Disease
- Protein Composition of Infectious Spores Reveals Novel Sexual Development and Germination Factors in
- The Evolutionarily Conserved LIM Homeodomain Protein LIM-4/LHX6 Specifies the Terminal Identity of a Cholinergic and Peptidergic . Sensory/Inter/Motor Neuron-Type
- SmD1 Modulates the miRNA Pathway Independently of Its Pre-mRNA Splicing Function
- piRNAs Are Associated with Diverse Transgenerational Effects on Gene and Transposon Expression in a Hybrid Dysgenic Syndrome of .
- Retinoic Acid Signaling Regulates Differential Expression of the Tandemly-Duplicated Long Wavelength-Sensitive Cone Opsin Genes in Zebrafish
- The Formin Diaphanous Regulates Myoblast Fusion through Actin Polymerization and Arp2/3 Regulation
- Genome-Wide Analysis of PAPS1-Dependent Polyadenylation Identifies Novel Roles for Functionally Specialized Poly(A) Polymerases in
- Runx1 Transcription Factor Is Required for Myoblasts Proliferation during Muscle Regeneration
- Regulation of Mutagenic DNA Polymerase V Activation in Space and Time
- Variability of Gene Expression Identifies Transcriptional Regulators of Early Human Embryonic Development
- The Drosophila Gene Interacts Genetically with and Shows Female-Specific Effects of Divergence
- Functional Activation of the Flagellar Type III Secretion Export Apparatus
- Retrohoming of a Mobile Group II Intron in Human Cells Suggests How Eukaryotes Limit Group II Intron Proliferation
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Exon 7 Contributes to the Stable Localization of Xist RNA on the Inactive X-Chromosome
- YAP1 Exerts Its Transcriptional Control via TEAD-Mediated Activation of Enhancers
- SmD1 Modulates the miRNA Pathway Independently of Its Pre-mRNA Splicing Function
- Molecular Basis of Gene-Gene Interaction: Cyclic Cross-Regulation of Gene Expression and Post-GWAS Gene-Gene Interaction Involved in Atrial Fibrillation
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



