-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaPooled Sequencing of 531 Genes in Inflammatory Bowel Disease Identifies an Associated Rare Variant in and Implicates Other Immune Related Genes
Crohn’s disease and ulcerative colitis are two forms of inflammatory bowel disease which cause chronic inflammation of the gastrointestinal tract. Common genetic variants in more than 160 regions of the human genome have been associated with an altered risk of these disorders, but leave much of the estimated genetic contribution to disease risk unexplained. We sought to establish whether rare genetic variants which alter the structure or function of the proteins encoded by genes also contribute to disease susceptibility. We used high throughput DNA sequencing to screen over 500 genes for such variants in nearly 500 patients and controls, and validated interesting variants in about 10,000 patients and 7,000 controls. We detected association of a limited number of rare variants from coding regions with disease, suggesting that they do not account for a large proportion of genetic susceptibility. However, they highlight the involvement of genes of potential importance in the development of inflammatory bowel disease, including those involved in the activation of immune cells, the regulation of immune response genes, and the degradation of proteins in cells.
Published in the journal: . PLoS Genet 11(2): e32767. doi:10.1371/journal.pgen.1004955
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004955Summary
Crohn’s disease and ulcerative colitis are two forms of inflammatory bowel disease which cause chronic inflammation of the gastrointestinal tract. Common genetic variants in more than 160 regions of the human genome have been associated with an altered risk of these disorders, but leave much of the estimated genetic contribution to disease risk unexplained. We sought to establish whether rare genetic variants which alter the structure or function of the proteins encoded by genes also contribute to disease susceptibility. We used high throughput DNA sequencing to screen over 500 genes for such variants in nearly 500 patients and controls, and validated interesting variants in about 10,000 patients and 7,000 controls. We detected association of a limited number of rare variants from coding regions with disease, suggesting that they do not account for a large proportion of genetic susceptibility. However, they highlight the involvement of genes of potential importance in the development of inflammatory bowel disease, including those involved in the activation of immune cells, the regulation of immune response genes, and the degradation of proteins in cells.
Introduction
The inflammatory bowel diseases (IBD), Crohn’s disease (CD) and ulcerative colitis (UC) are chronic inflammatory disorders of the gastrointestinal tract that can cause diarrhoea, abdominal pain, bleeding and weight loss. Collectively they affect approximately 827 per 100,000 individuals in European populations and their incidence is rising [1]. CD may affect any part of the gut with discontinuous penetrating lesions, whereas in UC the disease is limited to the colon and rectum and the lesions are continuous but superficial [2]. Both diseases are multi-factorial, with a complex aetiology that involves a combination of an underlying genetic predisposition and environmental triggers. A variety of factors have been proposed to contribute to the pathogenesis including changes within the intestinal microbiota, a defective mucosal barrier, and / or dysregulation of the immune response [3].
A meta-analysis of genome-wide association studies (GWAS) in CD and UC by the International IBD Genetics Consortium (IIBDGC), followed by extensive confirmation of association signals in more than 75,000 individuals has increased the number of IBD-associated loci to 163 [4]. The majority of these loci are associated with both CD and UC, which suggests that there is extensive overlap in the biological mechanisms involved in their pathogenesis. However, although our understanding of the aetiology of IBD has been substantially advanced by GWAS-based approaches, only a modest proportion of total disease variance can be explained by current genetic findings (<15%) [4]. It has been proposed that rare coding sequence variants may make a substantial contribution to disease variance, and confer disease risks large enough to warrant use in preventative screening [5]. Such variants would not be detectable by a conventional GWAS approach because they are not well tagged by the common SNPs on which GWAS panels are based [6].
New high throughput DNA sequencing technologies have made it feasible to investigate the contribution of rare variants to complex disease. In CD, it has long been known that low frequency coding variants in NOD2 make a substantial contribution to disease risk [7–9], and more recent high-throughput sequencing strategies have discovered several independent IBD associated rare variants in NOD2 and other genes from GWAS loci including IL23R, CARD9, IL18RAP, CUL2, C1orf106, PTPN22, RNF186 and MUC19 [10–12]. However, a recent large-scale sequencing study of the coding regions of 25 autoimmune candidate genes in more than 40,000 individuals yielded little evidence that rare variants drive the associations observed at susceptibility loci for common immune disorders, including CD [13]. Thus the exact contribution of rare coding variants to IBD and other immune disorders remains unknown.
Here we describe a targeted high throughput sequencing approach in pooled DNA samples from 474 CD patients and 480 population controls to screen all exons, splice sites, and proximal promoter regions in 531 positional and functional candidate genes. We sequenced CD patients with early-onset disease and/or strong family history to enrich for functional causal variants with stronger effects, and we looked beyond common loci using functionally-derived bioinformatics data such as pathway and protein network analysis to identify additional candidate genes involved in key processes such as the immune-response and autophagy. Potential functional variants and those with evidence of association with CD underwent validation genotyping in a follow up study including 6507 IBD cases and 3064 controls with replication of the top hits in an additional 3662 IBD cases and 3639 controls giving a total of over 10,000 IBD cases and over 7,000 controls for the final combined analysis. We discovered significant novel association of a rare coding variant in BTNL2 and suggestive associations of additional variants in potentially novel IBD genes.
Results
Sequencing of CD cases and controls
An overview of our strategy for the discovery of rare variants associated with CD is shown in Fig. 1. We selected 531 candidate genes for sequencing in phase I based on 5 selection criteria (Table 1 and described in Materials and Methods). A total of 6,249 exons, together with associated splice sites and proximal promoter regions, were sequenced in 474 CD cases and 480 population controls. Samples were sequenced in case-only or control-only pools of 12, 18 or 24 individuals using the Illumina Genome Analyzer II platform. An average of 98 million sequence reads were generated per pool, of which 87% could be aligned to the reference genome and 64% passed subsequent quality control steps (Materials and Methods). Of these, an average of 25.7 million reads mapped to the targeted genomic regions, which corresponded to a capture efficiency of 40.5%. We observed a mean read depth of >1000x per pool across the 1.57 million bases captured. Taking into consideration the number of individuals per pool, on average 90% of all bases had coverage greater than 4x per haploid genome (S1 Fig.).
Fig. 1. Summary of strategy for detecting rare variants associated with IBD. 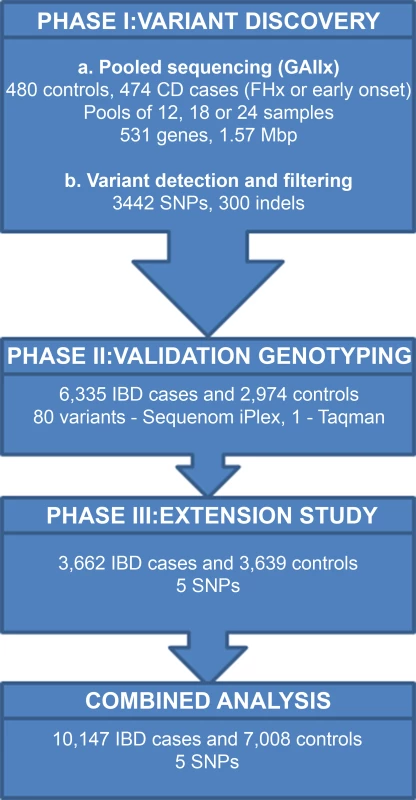
Overview of our rare variant screening strategy in IBD using DNA pools. We detected 3442 high quality variants in phase I based on stringent filtering criteria. We were able to validate 1252 of these variants using a) previously generated genotyping data for 153 SNPs in 634 of the individuals who were sequenced; b) case-control association p values for 1099 SNPs from CD Immunochip study [4]. We then performed validation genotyping of 80 variants in phase II in 6335 IBD cases and 2974 controls and extended the analysis of the top 5 SNVs to a further 3662 IBD cases and 3639 controls (phase III) to allow a final combined analysis of 10,147 IBD cases and 7,008 controls. In order to reduce false positives calls due to sequencing errors, we applied a stringent filtering procedure (Materials and Methods), after which the number of variants was approximately constant across all pools for all types of variants (S2 Fig.).
Tab. 1. Candidate gene selection strategies. 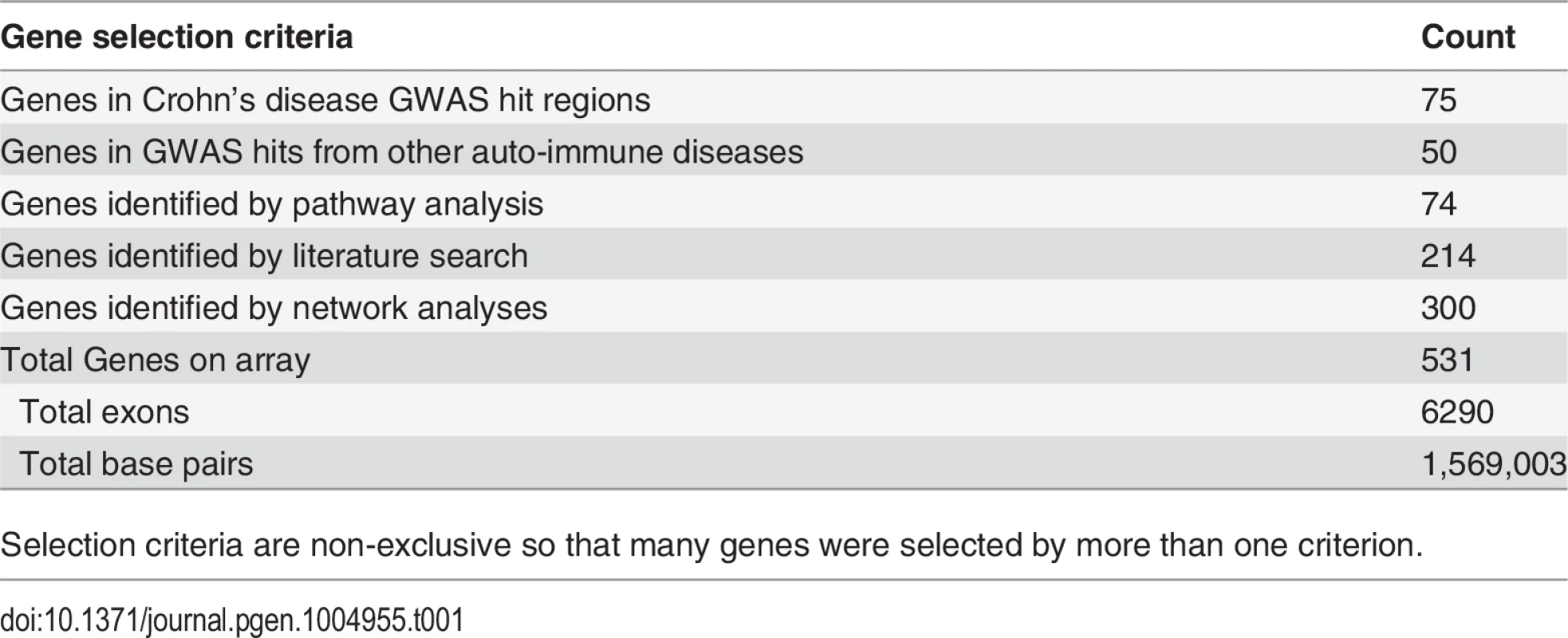
Selection criteria are non-exclusive so that many genes were selected by more than one criterion. Next, variant allele frequencies in each pool were estimated from base-call counts. We assessed the accuracy of this approach by comparing these estimates to minor allele frequencies (MAFs) derived from genotyping data generated by the Wellcome Trust Case Control Consortium (WTCCC); genotypes were available for 153 SNPs located in the captured genomic regions in 66.5% (388 controls, 246 cases) of the individuals sequenced in this study [14]. We observed a very strong correlation (Spearman Rank Correlation r = 0.977) between MAFs for the WTCCC genotypes and the pooled sequencing data (Fig. 2).
Fig. 2. Minor allele frequencies WTCCC vs pooled NGS (24 pools combined). 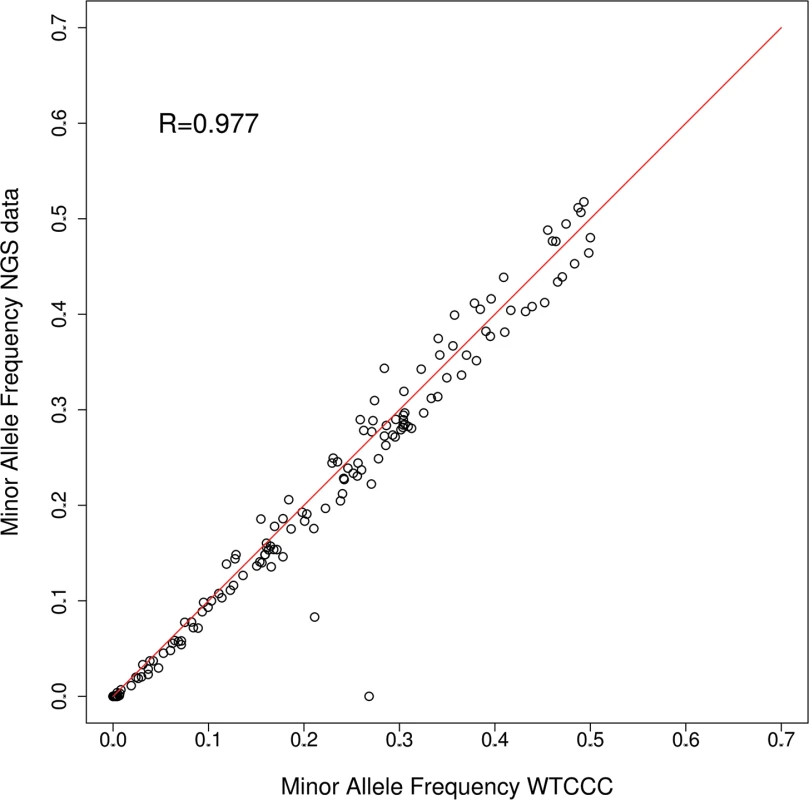
MAFs for 153 SNPs were compared between allele frequency estimates based on pooled NGS and genotyping data from the WTCCC [47] for 634 individuals. MAFs are strongly correlated between both datasets (Spearman rank correlation coefficient R = 0.976), with only two SNPs showing substantial differences. After filtering, 3,749 single nucleotide variants (SNVs, here used to refer to any single nucleotide variation regardless of minor allele frequency) were retained, of which over half were low frequency (<5%, S1 Table). Just over half of the SNVs were located in exons (51.1%; 1914 SNVs), with the remainder located in introns, untranslated regions (UTRs), putative splice sites and intergenic regions. We considered 106 of the SNVs (3%) to be novel because they were not present in dbSNP138 (http://www.ncbi.nlm.nih.gov/SNP/). Analysis of all SNVs yielded a transition/transversion ratio (Ti/Tv) of 2.41, which is expected given the bias toward coding sequences in our target regions and is in agreement with previous studies [11]. In addition to SNVs we identified 183 deletions and 117 insertions. Only 14 of these insertion/deletions (indels) were located in an exon (S1 Table). A high rate of true positives in our sequencing data was corroborated by the presence of 97% of our variants in dbSNP138, and the strong correlation between MAFs for the pooled sequencing data and the WTCCC genotype data. Regarding sensitivity of variant detection, the regions captured in our sequencing contain 1,599 variants with a MAF >5% in the phase I release of the 1000 Genomes project, 1,291 of which (80.7%) were detected in our pooled sequencing data.
Analysis of bias
Our strategy relied on the necessity of sequencing individuals in case-only or control-only DNA pools which could potentially inflate any biases that would arise due to sequencing batch effects. We therefore used principal component analysis to control for this and identify any outlier pools. Examination of PC axes 1 and 2 revealed pools 7 and 8 to be outliers. Both were case pools, although each represented a single lane of flow-cell data from two different runs of the GAII sequencer. Once these pools were removed the data showed reasonable separation of points, but there was a clear tendency for case and control pools to be separated along PC axis 1 (S3 Fig.), which led to an overall genomic inflation of 1.3 (Fig. 3). The extent of the systematic bias in the data meant that PC axes could not be used as covariates in a logistic regression to correct for it, as previously noted [15], nor could we apply methods designed to correct for overdispersion but not bias [16]. We therefore applied a genomic control method for downstream association analysis plus additional QC measure for removal of SNVs with strong over dispersion among pools (Materials and Methods). We note that it is possible that the high systematic bias reflects genuine causal influences given the candidature of all the genes sequenced, but equally we cannot exclude the possibility of experimental sources of bias.
Fig. 3. Quantile-quantile plot of chi-squared statistic. 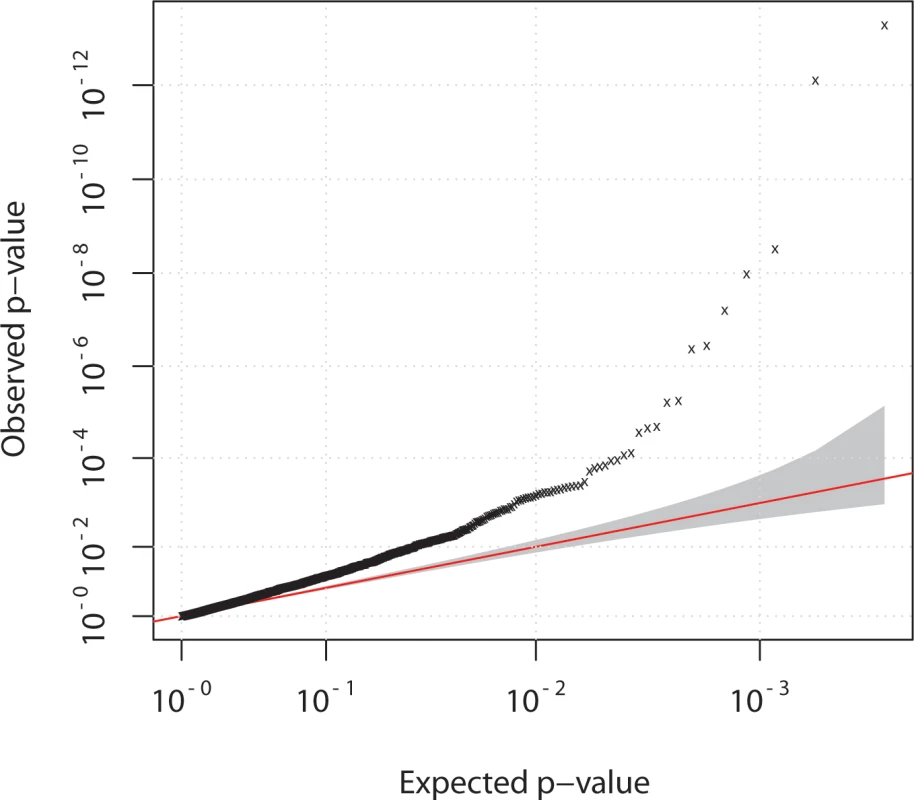
Data for case-control comparison of allele frequencies of 3442 variants, detected in pooled sequencing experiment for 42 case and 40 control pools. Overall genomic inflation (lambda) of 1.3 was observed.A genomic control correction was therefore applied for downstream association analysis (Materials and Methods). Association analysis
Variant level association with case-control status of pools was performed using logistic regression on 3,442 SNVs after exclusion of 307 SNVs that were too rare (had zero count in case or controls), or only had allele counts in excluded pools (Materials and Methods) (Fig. 3). Encouragingly, several known common and low frequency CD susceptibility variants were detected including variants in ATG16L1, IRGM, IL23R, CARD9 and NOD2, and rare variants in IL23R and NOD2 [7, 8, 10, 11, 17], all of which showed the expected CD odds ratios and allele frequencies in both cases and controls (S2 Table).
We noticed that 1,099 of 3,442 SNVs tested for association in our sequencing data were either included in the IBD Immunochip project directly (803) or by a suitable tagging SNP (r2≥0.8, n = 296) [4]. The IBD Immunochip dataset was therefore considered as an independent replication study for these 1099 variants. We found that 43 of the 141 variants (30.5%) that were at least nominally associated in our sequencing data (p<0.05) were also associated in the CD Immunochip data (p<5x10−8), resulting in a significant correlation between the two datasets (r = 0.446, p = 4.46x10−32).
The majority of variants identified by our study were rare, resulting in modest statistical power for the SNV-wise tests of association. We therefore applied gene-level association tests to investigate whether the burden of predicted functional variants non-synonymous and stop-gain variants) was different in cases compared to controls (Materials and Methods). In our discovery sequencing we identified 341 genes containing one or more functional variants. Thus the gene-burden test provided >90% power to detect a gene-level association where the cumulative MAF is 5% and the cumulative risk (OR) is 2.5 at an alpha level of 0.00015 (allowing for Bonferroni correction based on 341 genes/tests). We identified significant gene-level associations for BTNL2 (no. of variants = 18, p = 8.15x10−5) and NOD2 (no. of variants = 10, p = 9.03x10−6) (S3 Table). Since both genes contained substantially more functional variants than other genes that were tested we controlled for LD by permutation analysis (Materials and Methods), which resulted in loss of significance for BTNL2 (p = 0.022), whilst NOD2 remained significant (p<0.001). Repeating the analysis to include all intragenic variants (functional and non-functional) gave a similar outcome, although neither gene survived permutation testing (p>0.001).
Validation genotyping and extension study
In Phase II we selected 85 variants for validation of disease association by Sequenom (84 SNVs) or Taqman (1 SNV) genotyping in 6,335 IBD cases from the UK IBD Genetics Consortium (3,715 CD and 2,619 UC) and 2,974 controls (Materials and Methods). UC cases were included in the validation because of the extensive overlap in known associated loci for these two related phenotypes [4]. SNVs were selected based on at least nominal evidence of association in the pooled sequencing experiment (p < 0.05), and we prioritised those predicted to be functionally relevant (S1 Text). SNVs already genotyped as part of the IBD Immunochip experiment [4] were excluded. Post-genotyping quality control revealed that two SNVs failed to genotype, two were non-polymorphic and one was not in Hardy Weinberg equilibrium (p < 1x10−6 in controls) leaving a total of 80 SNVs (S4 Table). The genotyping call rate for all remaining SNVs was >90%. To allow validation of our variant calling analysis pipeline we genotyped an additional subset of 368 individuals previously included in our sequencing experiment and were able to show strong correlation between predicted and actual allele frequencies for all 80 SNVs (r = 0.94, p = 2.42x10−38) and low frequency SNVs (MAF<5%, r = 0.86, p = 1.69x10−24). In addition, allele frequencies derived from the pooled sequencing experiment were compared to those derived from all individuals in the phase II genotype data and revealed a highly significant correlation (r = 0.971, p < 6.58x10−48), further supporting the validity of the pooled sequencing approach. We followed up 3 insertion deletion polymorphisms by Taqman genotyping in 2,532 IBD cases and 3,545 controls (rs58682836/COBL frameshift delTTC, rs71297581/TYK2 upstream insC, and rs3833864/PIK3C upstream insC). The indel rs71297581 failed genotyping quality control, producing poor genotype clusters, and neither rs58682836 nor rs3833864 were associated with IBD (p > 0.5).
There was some evidence of association (p < 0.05) for 16 SNVs across 12 genes, CHTOP, ARIH2, NICN1, PLSCR1, IL12B, BTNL2, QRSL1, CALML5, GLT1D1, RTEL1, ATG4B and TBX21 (S5 Table). These were associated with either CD (11 variants), UC (5 variants) or IBD (12 variants), with 4 of these variants located in BTNL2. BTNL2 and IL12B map to established UC and IBD risk loci and have previously been implicated in UC and IBD respectively (6p21/HLA class II/UC and 5q31/IBD respectively), whilst ARIH2 and NICN1are within the same previously described IBD locus (3p21.3/IBD) but the genes themselves have not been implicated. Association of the other 10 genes and their respective variants with IBD has not been reported previously.
Since BTNL2 is within the MHC region and close to the common IBD associated locus in the HLA class 2 region we investigated the extent of LD across the 4 variants and their independence from the known risk locus using haplotype and conditional analysis within a set of cases and controls previously genotyped in both the Immunochip study and our follow up genotyping study (Materials and Methods). The analysis showed that the rare BTNL2 variants p.G454C and p.D336N (rs28362675 and rs41441651) were in almost complete LD with each other (r2 = 0.99) and remained associated with IBD even when the effect at the common SNPs was accounted for (p < 0.049), as did BTNL2 c.-118G>T (rs28362684, p = 0.039) but not the missense variant (p.S334L). Regarding association of the 80 variants with IBD, only the two highly correlated variants in BTNL2 (p.454C and p.D336N) surpassed the Bonferroni threshold for multiple testing (p < 0.0006 for 79 independent SNVs tested). However there was significant enrichment for association signals among the 79 variants, with nearly 3 times the number of significant results than would be expected by chance, with 14% of p-values for association with IBD (i.e. 11/79) being less than 0.05 (p = 0.00189).
Recognising the relatively low power of the validation panel to detect significant association of rare variants with disease, we next carried out extended genotyping (Phase III) of the 5 top SNVs that had a p < 0.01 (and in the case of BTNL2 were independent of each other and the known common risk variants) in an additional panel of 3,662 IBD cases and 3,639 controls (Materials and Methods), and then performed a combined case-control analysis of all 10,147 IBD cases and 7,008 controls that were either sequenced or genotyped (Table 2). We confirmed a genome-wide significant association with BTNL2 p.G454C and increased risk of IBD at (p = 9.65x10−10, OR = 2.3 [95%CI = 1.75–3.04]). We detected association for 3 other variants of the 5 tested in phase III (p < 0.005). Notably, in the combined analysis the direction of the effect for each of the 5 SNPs is consistent with the effect in the validation panel (p < 0.031). However, the 3 additional associations do not meet correction for 79 independent tests (P<0.00063) and are therefore suggestive. They include two low frequency missense variants IL12B p.V298F and NICN1 p.H191R associated with a reduced risk for IBD and one noncoding variant ARIH2 c.338-6C>T which was associated with an increased risk (Table 2). Two of the 3 missense variants associated with IBD (IL12B p.V298F and BTNL2 p.G454C) were predicted to be damaging or non-tolerated by Polyphen2 [18] and/or SIFT (sorts intolerant from tolerant) or Provean [19]. IL1B encodes the p40 subunit common to both the interleukin-12 and interleukin-23 heterodimeric cytokines. The p.V298F variant is not in LD with the common risk variant at this locus (r2 = 0.001, D’ = 0.079), and is predicted to disrupt the structure of the p40 protein by the mCSM structure prediction tool [20], with a predicted stability change ΔΔG of −0.917. We also used the available structure of the IL12B (p40) and IL23A (p19) proteins to model the effect of the V298F mutation in IL12B (S4 Fig.). This indicated an altered conformational state of a region of p40 which is important for binding to its partner proteins IL23A (p19) and IL12A (p35) [21].
Tab. 2. Combined case-control association analysis of 5 sequence variants from the phase III extension study in 10,147 IBD cases and 7,008 controls from phases I–III. 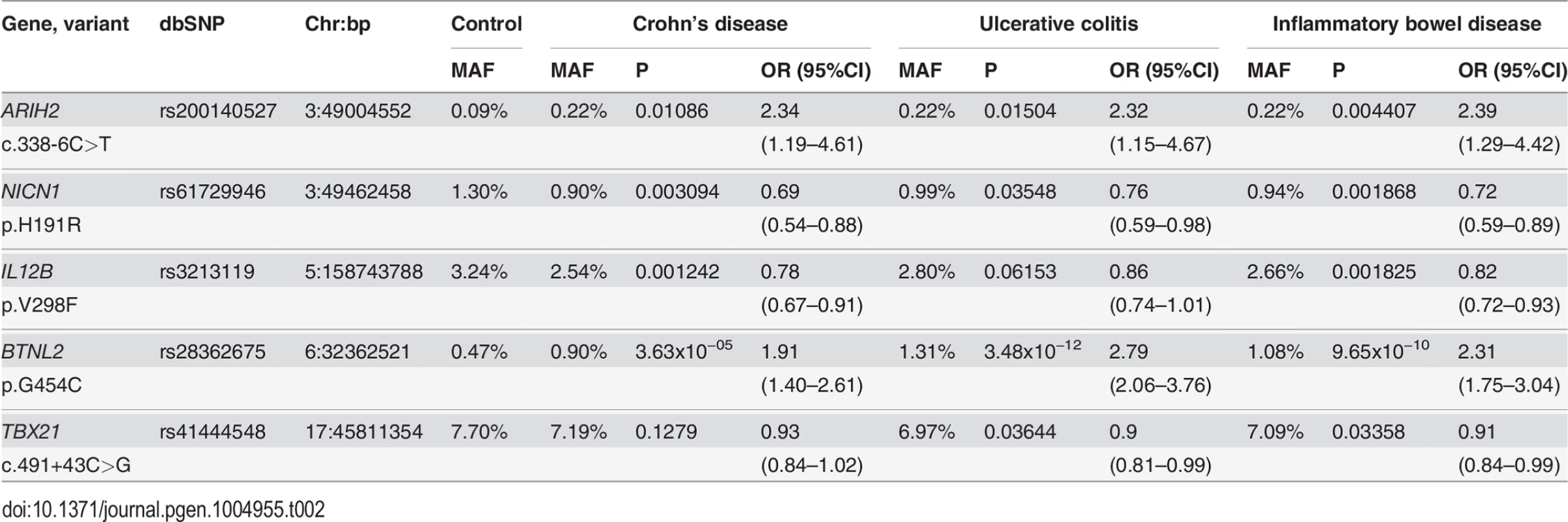
BTNL2
BTNL2 is located on chromosome 6p21.3, which contains two common and independent risk loci for IBD. The closest (approximately 200Kb proximal to BTNL2) is within the HLA class II region and is associated with UC (rs477515, p = 5x10−133). The other locus is much further away (approximately 1.1Mb distal of BTNL2) within the HLA-class I region, and associated with CD (rs9264942, p = 5x10−28) [4]. We observed that BTNL2 p.G454C was associated very strongly with UC (p = 3.5x10−12, Table 2) and also associated with CD but to a lesser extent (p = 3.6x10−5, Table 2). In view of the extended LD in this region, it is possible that these associations could be due to LD with the known common risk variants in the HLA class I or class II regions. We investigated this by further conditional logistic regression analysis using 1,638 IBD cases and 1,243 controls genotyped in both the Immunochip study and both our genotyping studies. We confirmed that BTNL2 p.G454C was not in LD with either of the two common IBD risk variants (r2 < 0.001, D’ < 0.7). Conditional analysis showed that BTNL2 p.G454C remained significantly associated with IBD when the effect at the common UC associated SNP (rs477515) was accounted for (p = 0.0045, S6 Table), or the common CD associated SNP (rs9264942) was accounted for (p = 4.83x10−5, S6 Table). Haplotype analysis showed that the risk “A” allele for the rare variant occurred on haplotypes containing either the non-risk or the risk allele for both of the common variants, further suggesting their independence. Given the strength of the effect of p.G454C in UC individuals in particular (Table 2) we carried out specific haplotype analysis using this and the common UC GWAS SNP in the class II HLA region and showed that haplotype A-A containing the risk allele at the rare variant (p.G454C) and the non-risk “A” allele at the common UC GWAS SNP (rs477515) respectively, although very rare, was increased in frequency in cases, (0.2%) compared to controls (0.07%) (S7 Table), and the haplotype G-A containing the risk allele at both the common and the rare variant had a much higher risk for disease (OR = 6.51 [95%CI = 1.87–22.72]) than the haplotype G-C that only had the risk allele at the common SNP and lacked the rare risk allele (OR = 1.38 [95%CI = 1.20–1.57]).
Discussion
In this study we investigated the contribution of rare variants to susceptibility to inflammatory bowel disease in a large set of candidate genes. Use of targeted next generation sequencing in combination with a DNA pooling strategy allowed us to screen over 500 genes for variants in more than 900 individuals, which is ten-fold more than were investigated in previous studies of IBD [10–12]. The results demonstrate that this is a cost-effective strategy for identifying low frequency variants that may be associated with disease. We were able to validate our approach by accurate estimation of the minor allele frequencies of 153 SNPs previously genotyped in individual case and control samples by the Affymetrix 500K SNP array, and by successfully reproducing the effect sizes (odds ratios) and allele frequencies of multiple common and low-frequency variants previously associated with IBD. We also demonstrated highly significant overlap of association for 1,099 SNPs that were common to our study and the recent GWAS/Immunochip meta-analysis for IBD [4], and showed a strong correlation between the allele frequencies and odds ratios of 80 SNVs that were genotyped by both pooled DNA sequencing and genotyping in our follow up study. Strong correlations between allele frequency estimates from pooled sequencing and genotyping have also been reported in previous studies of Crohn’s disease [10, 11], although read counts tended to underestimate actual frequencies for rare variants in one study [10]. However this approach could prove useful when supported by stringent quality control and validation measures.
Sequencing of coding and potential regulatory regions of 531 genes in a discovery set of 954 individuals, followed by genotyping in 17,131 individuals has allowed us to identify a novel disease associated genetic variant within a gene that maps to a region previously associated with IBD, and suggestive associations of other variants in a known IBD susceptibility gene and in other genes not previously implicated in IBD. The association of the rare variant p.G454C in BTNL2 reached genome-wide significance, and was independent of the known common risk variants for IBD in the HLA region in both a conditional and haplotype analysis. However, this is a complex region of the genome with extensive allelic variation and linkage disequilibrium, and additional as yet unknown IBD risk variants at this locus may exist that are independent of the two main HLA signals previously described but correlated with our rare variant. The glycine residue is highly conserved across all mammals and the cysteine substitution is predicted to be damaging by SIFT (score = 0.01) and probably damaging by PolyPhen2 (score = 0.997). This variant was in almost complete LD with another missense variant D336N which is not predicted to be damaging. BTNL2 codes for the butyrophilin like protein 2, which is a member of butyrophilin family that shares sequence homology with the B7 co-stimulatory molecules. The butyrophilins are implicated in T cell inhibition and the modulation of epithelial cell-T cell interactions [22]. BTNL2 negatively regulates T-cell activation independently of CD28 and CTLA-4, is predominantly expressed in gastrointestinal tissues including human terminal ileum (www.gtexportal.org), and is overexpressed in mouse models of colitis [23]. Recently it has been shown that BTNL2 promotes the expression of Foxp3, which is a transcription factor required for regulatory T cell development and function [24]. In view of its important role in immune modulation and homeostasis and an expression pattern restricted to intestinal epithelial and immune cells, mutations in BTNL2 may affect its ability to regulate T cell activation in response to mucosal inflammation. Common variants at the BTNL2 locus, have been previously shown to be associated with ulcerative colitis whilst being independent of the nearby known HLA susceptibility alleles [25]. Additional coding and loss-of-function variants in BTNL2, have been associated with susceptibility to other immune related disorders including adult-onset sarcoidosis [26, 27] and rheumatoid arthritis [28].
Although no variants other than the two rare and highly correlated missense mutations in BTNL2 surpassed the Bonferroni threshold for testing the 79 independent variants for association with IBD, there was significant enrichment for association signals among these 79 variants, and our extension study and combined analysis showed that the direction of the effect for all 5 SNVs tested was consistent with the initial finding. This suggests that there are likely to be additional true positives within phase II and III of our study that have not met the stringent Bonferroni threshold. This emphasises the difficulty in obtaining statistically robust evidence for association of rare variants even with a combined sample of 17,000 tested here and a relatively large effect size such as, for example, ARIH2 c.338-6C>T, OR = 2.39.
The association of common variants at the IL12B locus with both CD and UC is well established [4], although no obvious causal variant has yet been found. The association of the low frequency IL12B variant V298F with IBD which was detected in our sequencing experiment was retained in the combined analysis of 10,146 IBD and 7,008 controls, (p = 0.00183, OR = 0.82 [95%CI = 0.72–0.93]). IL12B encodes the IL12p40 subunit common to both IL12 and IL23, both of which are produced by activated dendritic cells and macrophages and lead to activation of distinct subsets of T-cells. We found that the minor allele of V298F is associated with a reduced risk of both CD and UC and is independent of the common risk variants at this locus. The variant is predicted to have a damaging or destabilizing effect on protein function or structure, and modeling of the effect of the mutation on the structure of the p40 subunit predicted an altered conformational state which could affect binding to its partner proteins. Thus the rare (Phe) allele may reduce the risk of IBD by attenuating the activation of T cell populations by IL12 and IL23.
We found two additional suggestive associations in ARIH2 and NICN1. Ariadne homolog 2 (ARIH2) is a member of an unusual family of E3 ubiquitin-protein ligases. Loss of ARIH2 has been shown to cause degradation of IκBβ in dendritic cells leading to dysregulated activation of NFκB. The SNP rs200140527 is associated with IBD, and is 6bp upstream of the splice acceptor site for exon 9 of ARIH2, although the C>T change is not predicted to affect the strength of the splice site [29]. Nicolin 1 (NICN1) is a nuclear protein and part of the neuronal tubulin polyglutamate complex [30] although very little else is known about its function. It is expressed in multiple tissues including the human terminal ileum and transverse colon (www.gtexportal.org). The nonsynonymous SNP p.H191R is associated with a protective effect for CD and UC in this study. NICN1 is on chromosome 3 at 49.46Mb, i.e. approximately 460kb proximal to ARIH2 on 3p21 and within a 2Mb locus previously associated with IBD that contains multiple independent genome-wide significant SNPs [4].
Previous sequencing studies have reported that rare coding variants make a limited contribution to the genetics of immune disorders and hypertriglyceridaemia, explaining 1–2% of their genetic variance [10–13, 31, 32]. However, these studies have generally sequenced a limited number of genes located in regions derived from the association of common variants with the disease. Our study highlights the challenges in identifying rare variant association for a polygenic complex trait like IBD. In sequencing more than 500 genes from both GWAS and pathway or network analysis combined with follow up genotyping in over 17,000 individuals we found genome-wide significant association of a rare variant in one gene and suggestive association of 3 SNVs in 3 other genes. However, our follow up studies were powered to detect associations of rare variants with relatively strong effects. For example, our phase II validation panel had 57% power to detect association of a low frequency variant with an allele frequency of 2.5% and OR = 1.3 at alpha level of 0.01 (to flag candidate associations), and 75% power to detect a rare variant with an allele frequency of 1% and OR of 1.6. In the combined analysis of 10,147 cases and 7,008 controls, we had 69% power to confirm association of a variant with a MAF of 0.025 and OR of 1.3 at alpha level of 0.0006 (correction for 79 SNV tests), but 89% power to confirm association for a variant with MAF 0.01 and an OR of 1.6. It is therefore likely that some rare variants with effect sizes of less than 1.6 remain undiscovered in these genes. It is also possible that a proportion of variants that are recognised as being suggestive of association in this study may turn out to be false positives, so further replication and subsequent functional studies will be required to prove causality.
If our 4 newly discovered associations were added to the 26 low frequency SNVs identified in 13 other genes from previously published studies of IBD [7, 10–12, 17, 33] this would total 30 IBD associations with low frequency SNVs in 17 of 548 sequenced genes. However, these screens have predominantly interrogated the coding regions of less than 3% of all known genes. Our study has targeted <25% (198) of all the known genes that map to the 163 IBD associated regions identified by the most recent mapping efforts of the International IBD Consortium [4]. A comprehensive evaluation of the true extent of the contribution of rare coding variants to IBD will have to await whole exome sequencing of very large numbers of case and controls [34], and whole genome sequencing to capture rare regulatory variants in non-coding regions.
The value of studies of rare variants in IBD lies not only in the discovery of additional risk variants which may aid future genetic profiling in at risk populations, but also in their potential to discover further genes and pathways involved in IBD. Our study provides additional evidence of the importance of the regulation of T cell activation and mucosal T cell responses involving BTNL2, and the potential role of proteosomal degradation in the pathogenesis of IBD.
Materials and Methods
Selection of candidate genes and design of the capture array
A total of 531 candidate genes were selected based on: (a) Crohn’s disease GWAS hits; (b) GWAS hits from other immune disorders; (c) Pathway analysis based on Gene-set enrichment analysis; (d) IBD related literature; and (e) Network Analysis (Table 1). Details of these selection criteria are provided in S1 Text. Exon coordinates from RefSeq [35] and Ensembl [36] were combined to include all potentially coding regions. Proximal promoters were included by selection of genomic regions from 200 bp upstream to 50 bp downstream of the transcription start site. Putative splice sites were included by addition of five bp each side of coding exons. In total 6,290 genomic intervals were successfully synthesized for the Agilent SureSelect DNA Capture Array. Capture probes (120 bp; 60bp tiling) corresponding to 1,569,003 bp of target sequence.
Study participants and sample preparation
Crohn’s disease patients for the sequencing experiment (n = 474) were recruited from specialist IBD clinics in London and Newcastle [37] after informed consent and ethical review (REC 05/Q0502/127). Population controls for sequencing (n = 480) were obtained from the 1958 British Birth Cohort [38]. All individuals were of European ancestry. The chances of detecting rare variants with large effects in the sequencing stage was increased by selection of Crohn’s disease (CD) patients with an early age of onset <20 years (n = 204), or with a family history of IBD (n = 174) or both early onset and family history (n = 96). Additionally, 178 (86%) of those individuals with a family history also had at least one affected first degree relative. DNA samples were quantified in triplicate (Qubit, Life technologies) prior to pooling in equimolar amounts to a total of 3 μg of DNA. Pools of 24 CD case DNA samples or 24 control DNA samples were made with a total of 44 pools, 474 cases and 480 controls (including 9 pilot/test pools of 12 and one test pool of 18 CD cases; S1 Text) and libraries were prepared following standard protocols. The validation panel for phase II, consisted of 3,799 unrelated CD and 2,708 unrelated UC, patients recruited by the UK IBD Genetics Consortium [4] and the replication panel consisted of an additional 1644 CD cases and 2018 UC cases recruited from London and Newcastle (as described above). Additional population controls (n[validation] = 3,064; n[replication] = 3622) were from the 1958 British Birth cohort and the National Blood Donor Service [14]. All cases and controls analysed in the replication phase III were independent and unrelated to those sequenced in the phase I and phase II discovery cohort.
Read alignment and read quality control
Sequencing reads were aligned to the hg18 (NCBI 36) reference genome using Novoalign (version 2.07.09, Novocraft Technologies). We performed quality control using SAM tools [39] and removed PCR duplicates using Picard tools [40]. SNVs and indels were called using SAM-tools and filtered based on the following criteria: i) Phred base quality score ≥ 20, ii) any allele to have at least two base calls on each strand, iii) minimum base call count for any allele to be the equivalent to at least one expected chromosome count (N allele-specific base calls / N total base calls * 2 * N individuals in pool), with at least 0.3 expected chromosome counts attributable to each strand, iiii) criteria to be met in at least three different pools from at least two different batches. These parameters were optimized to reduce biases across all 44 pools (S2 Fig.). After filtering, base call counts were normalised to allele frequencies for each pool based on the total number of base calls that passed the filtering criteria. Variants were annotated using ANNOVAR [41]. Further details of read alignment, quality control and variant calling are provided in S1 Text.
Validation genotyping and extension study
After excluding variants previously implicated with IBD and variants analysed in the IBD Immunochip project [4], we selected 96 SNVs for follow up in phase II using the Sequenom iplex genotyping platform. We chose variants that a) surpassed multiple testing in the pooled sequencing based case-control comparison (p < 10−5), b) were modestly significant in the pooled sequencing based case-control comparison (p < 0.05) and had a low allele frequency (MAF < 5%), c) had functional consequence (within 20bp of a splice acceptor or donor site or non-synonymous variant), and were novel or low frequency (< 1%), d) were absent from one group (either controls or cases) and had a functional consequence (within 20bp of a splice acceptor or donor site or non-synonymous variant). In total 84 SNVs passed design and were genotyped via Sequenom iplex in 2,974 controls, 3,715 Crohn’s disease and 2,620 ulcerative colitis cases. Individuals for which more than 20% of SNVs could not be called were excluded from further analysis. One additional SNV (rs138274580/ATG4B) and 3 indels (rs58682836/COBL frameshift delTTC, rs71297581/TYK2 upstream insC, rs3833864/PIK3C upstream insC), that failed iplex design, were genotyped using the TaqMan chemistry (Life Technologies); SNP since they were ranked as high priority in all categories of our variant selection criteria (S1 Text). Finally we selected 5 SNVs with p<0.01 in any one phenotype (CD, UC or IBD) and, in the case of multiple SNVs in BTNL2, were indicated by LD and conditional regression analysis to be independent of each other and the known common risk variants, for replication genotyping via KASPTM chemistry at LGC Genomics (Hoddesdon, Herts, UK) in 3666 additional IBD cases and 3622 additional controls. In order to validate previous phases we also included a further 858 individuals who had been sequenced and/or undergone sequenom iplex genotyping. To investigate LD and independence of the BTNL2 variants from the known IBD GWAS hits within the MHC we used Immunochip data supplied by the UKIBD Genetics Consortium that was available for 1,638 of our genotyped IBD cases and 1,243 of genotyped controls.
Statistical analyses
Allele frequencies for each SNV in each pool were standardized and subjected to principal components analysis (PCA) to identify outlier pools and investigate systematic bias between cases and controls. PCA revealed considerable bias, such that cases and control pools could be largely separated by PC axis 1 alone. Various statistical methods for dealing with pooled SNV data have been proposed [16]. In light of the PCA results, we adopted a genomic control approach because it can correct for both overdispersion (additional variance that is distributed equally among pools) and bias (a consistent tendency for allele frequencies in case pools to be different from controls pools). For each SNV, a logistic regression across pools was performed using expected chromosome counts for the two most common alleles to form the dependent variable, and case-control status as the independent variable. The reversal of the conventional functional from allows for different pool sizes to be readily accounted for, and also appropriately reflects the study design (pool status is fixed by the experimenter, not pool allele frequencies). Genomic control was performed by dividing the chi-square statistic for association by the median chi-square statistic across all SNVs. We used evidence for especially strong SNV-specific overdispersion among pools (via a test of residual deviance from the logistic regression for association, p < 1.5x10−5) as an additional QC measure for removal of suspect SNVs.
Burden tests for significant association of a group of SNVs (e.g. all SNVs in a gene) were also performed taking in account both the pooled design and the presence of case-control bias. For a given set of n SNVs, genomic-control-corrected z2 values were summed and tested against the chi-squared distribution with n degrees of freedom. Significant sum-statistics were further tested via permutation of case-control status among pools, to correct for false positives that could be caused from linkage disequilibrium distributing the same signal among multiple SNVs. Note that our burden test allows SNV groups containing a mixture of both risk and protective variants to be tested appropriately.
Statistical analyses of pooled sequencing data was performed using R project for statistical computing (http://www.r-project.org/). Cases-control analysis of validation and replication genotyping data was performed with PLINK version 1.07 [42] using Armitage Trend Test. Additional conditional regression, linkage disequilibrium and haplotype analysis at known common IBD loci was performed using UNPHASED v3.0.12 [43].
Structural analysis of IL12B
The effect of the mutation Val298Phe on IL12B (p40) protein stability was examined using the tool mCSM, which predicts the effect of mutations in proteins using graph-based signatures [20]. The structure of the complex of human IL12B (p40) and IL23A (p19) is available in the RCSB Protein Data Bank [44] (PDB entry 4GRW), and was used as the template to model the structure of mutant IL12BV298F. The modelling procedure first generated the sequence alignment between the target (IL12BV298F) and the template structure (4GRW chain B) by running the tool T-Coffee [45]. The aligned sequences were then used as an input to the structure modelling package Modeller 9v8 [46]to generate 200 structures of IL12BV298F. Among these, only the one with the best Discrete Optimized Protein Energy score was selected for inspection of the mutation Val298Phe. The structure representation tool PyMol (Version 1.5.0.4, Schrödinger, LLC)was used for visual inspection and structural analysis. The interaction between IL12BV298F and IL23A was modelled by superimposing the IL12BV298F structure onto the human wild-type IL12B.
Supporting Information
Zdroje
1. Molodecky NA, Soon IS, Rabi DM, Ghali WA, Ferris M, et al, (2012) Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology 142 : 46–54. doi: 10.1053/j.gastro.2011.10.001 22001864
2. Podolsky DK, (2002) Inflammatory bowel disease. N Engl J Med 347 : 417–29. 12167685
3. Khor B, Gardet A, and Xavier RJ, (2011) Genetics and pathogenesis of inflammatory bowel disease. Nature 474 : 307–17. doi: 10.1038/nature10209 21677747
4. Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, et al, (2012) Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491 : 119–24. doi: 10.1038/nature11582 23128233
5. Bodmer W and Bonilla C, (2008) Common and rare variants in multifactorial susceptibility to common diseases. Nat Genet 40 : 695–701. doi: 10.1038/ng.f.136 18509313
6. Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, et al, (2009) Finding the missing heritability of complex diseases. Nature 461 : 747–53. doi: 10.1038/nature08494 19812666
7. Hugot JP, Chamaillard M, Zouali H, Lesage S, Cezard JP, et al, (2001) Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature 411 : 599–603. 11385576
8. Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, et al, (2001) A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature 411 : 603–6. 11385577
9. Hampe J, Cuthbert A, Croucher PJ, Mirza MM, Mascheretti S, et al, (2001) Association between insertion mutation in NOD2 gene and Crohn’s disease in German and British populations. Lancet 357 : 1925–8. 11425413
10. Momozawa Y, Mni M, Nakamura K, Coppieters W, Almer S, et al, (2011) Resequencing of positional candidates identifies low frequency IL23R coding variants protecting against inflammatory bowel disease. Nat Genet 43 : 43–7. doi: 10.1038/ng.733 21151126
11. Rivas MA, Beaudoin M, Gardet A, Stevens C, Sharma Y, et al, (2011) Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nat Genet 43 : 1066–73. doi: 10.1038/ng.952 21983784
12. Beaudoin M, Goyette P, Boucher G, Lo KS, Rivas MA, et al, (2013) Deep resequencing of GWAS loci identifies rare variants in CARD9, IL23R and RNF186 that are associated with ulcerative colitis. PLoS Genet doi: 10.1371/journal.pgen.1003723 24415959
13. Hunt KA, Mistry V, Bockett NA, Ahmad T, Ban M, et al, (2013) Negligible impact of rare autoimmune-locus coding-region variants on missing heritability. Nature 498 : 232–5. doi: 10.1038/nature12170 23698362
14. Consortium TWTCC, (2007) Genome-wide Association Studies of 14,000 cases of Seven Common Human Diseases and 3,000 shared controls. Nature 447 : 661. 17554300
15. Dadd T, Weale ME, and Lewis CM, (2009) A critical evaluation of genomic control methods for genetic association studies. Genet Epidemiol 33 : 290–8. doi: 10.1002/gepi.20379 19051284
16. Yang X, Todd JA, Clayton D, and Wallace C, (2012) Extra-binomial variation approach for analysis of pooled DNA sequencing data. Bioinformatics 28 : 2898–904. doi: 10.1093/bioinformatics/bts553 22976083
17. King K, Sheikh MF, Cuthbert AP, Fisher SA, Onnie CM, et al, (2006) Mutation, selection, and evolution of the Crohn disease susceptibility gene CARD15. Hum Mutat 27 : 44–54. 16278823
18. Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, et al, (2010) A method and server for predicting damaging missense mutations. Nat Methods 7 : 248–9. doi: 10.1038/nmeth0410-248 20354512
19. Kumar P, Henikoff S, and Ng PC, (2009) Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 4 : 1073–81. doi: 10.1038/nprot.2009.86 19561590
20. Pires DE, Ascher DB, and Blundell TL, (2014) mCSM: predicting the effects of mutations in proteins using graph-based signatures. Bioinformatics 30 : 335–42. doi: 10.1093/bioinformatics/btt691 24281696
21. Yoon C, Johnston SC, Tang J, Stahl M, Tobin JF, et al, (2000) Charged residues dominate a unique interlocking topography in the heterodimeric cytokine interleukin-12. EMBO J 19 : 3530–41. 10899108
22. Abeler-Dorner L, Swamy M, Williams G, Hayday AC, and Bas A, (2012) Butyrophilins: an emerging family of immune regulators. Trends Immunol 33 : 34–41. doi: 10.1016/j.it.2011.09.007 22030238
23. Arnett HA, Escobar SS, Gonzalez-Suarez E, Budelsky AL, Steffen LA, et al, (2007) BTNL2, a butyrophilin/B7-like molecule, is a negative costimulatory molecule modulated in intestinal inflammation. J Immunol 178 : 1523–33. 17237401
24. Swanson RM, Gavin MA, Escobar SS, Rottman JB, Lipsky BP, et al, (2013) Butyrophilin-like 2 modulates B7 costimulation to induce Foxp3 expression and regulatory T cell development in mature T cells. J Immunol 190 : 2027–35. doi: 10.4049/jimmunol.1201760 23359506
25. Pathan S, Gowdy RE, Cooney R, Beckly JB, Hancock L, et al, (2009) Confirmation of the novel association at the BTNL2 locus with ulcerative colitis. Tissue Antigens 74 : 322–9. doi: 10.1111/j.1399-0039.2009.01314.x 19659809
26. Adrianto I, Lin CP, Hale JJ, Levin AM, Datta I, et al, (2012) Genome-wide association study of African and European Americans implicates multiple shared and ethnic specific loci in sarcoidosis susceptibility. PLoS One 7: e43907. doi: 10.1371/journal.pone.0043907 22952805
27. Valentonyte R, Hampe J, Huse K, Rosenstiel P, Albrecht M, et al, (2005) Sarcoidosis is associated with a truncating splice site mutation in BTNL2. Nat Genet 37 : 357–64. 15735647
28. Mitsunaga S, Hosomichi K, Okudaira Y, Nakaoka H, Kunii N, et al, (2013) Exome sequencing identifies novel rheumatoid arthritis-susceptible variants in the BTNL2. J Hum Genet 58 : 210–5. doi: 10.1038/jhg.2013.2 23364395
29. Reese MG, Eeckman FH, Kulp D, and Haussler D, (1997) Improved splice site detection in Genie. J Comput Biol 4 : 311–23. 9278062
30. Backofen B, Jacob R, Serth K, Gossler A, Naim HY, et al, (2002) Cloning and characterization of the mammalian-specific nicolin 1 gene (NICN1) encoding a nuclear 24 kDa protein. Eur J Biochem 269 : 5240–5. 12392556
31. Nejentsev S, Walker N, Riches D, Egholm M, and Todd JA, (2009) Rare variants of IFIH1, a gene implicated in antiviral responses, protect against type 1 diabetes. Science 324 : 387–9. doi: 10.1126/science.1167728 19264985
32. Johansen CT, Wang J, Lanktree MB, Cao H, McIntyre AD, et al, (2010) Excess of rare variants in genes identified by genome-wide association study of hypertriglyceridemia. Nat Genet 42 : 684–7. doi: 10.1038/ng.628 20657596
33. Ogura Y, Saab L, Chen FF, Benito A, Inohara N, et al, (2003) Genetic variation and activity of mouse Nod2, a susceptibility gene for Crohn’s disease. Genomics 81 : 369–77. 12676561
34. Kiezun A, Garimella K, Do R, Stitziel NO, Neale BM, et al, (2012) Exome sequencing and the genetic basis of complex traits. Nat Genet 44 : 623–30. doi: 10.1038/ng.2303 22641211
35. Pruitt KD, Tatusova T, Klimke W, and Maglott DR, (2009) NCBI Reference Sequences: current status, policy and new initiatives. Nucleic Acids Res 37: D32–6. doi: 10.1093/nar/gkn721 18927115
36. Flicek P, Amode MR, Barrell D, Beal K, Brent S, et al, (2011) Ensembl 2011. Nucleic Acids Res 39: D800–6. doi: 10.1093/nar/gkq1064 21045057
37. Prescott NJ, Fisher SA, Franke A, Hampe J, Onnie CM, et al, (2007) A Nonsynonymous SNP in ATG16L1 Predisposes to Ileal Crohn’s Disease and Is Independent of CARD15 and IBD5. Gastroenterology 132 : 1665–1671. 17484864
38. Power C and Elliott J, (2006) Cohort profile: 1958 British birth cohort (National Child Development Study). Int J Epidemiol 35 : 34–41. 16155052
39. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, et al, (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25 : 2078–9. doi: 10.1093/bioinformatics/btp352 19505943
40. Wysoker A, Tibbetts K, Fennell T, and Weisburd B, Picard Tools, 2009.
41. Wang K, Li M, and Hakonarson H, (2010) ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38: e164. doi: 10.1093/nar/gkq603 20601685
42. Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, et al, (2007) PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am J Hum Genet 81 : 559–575. 17701901
43. Dudbridge F, (2003) Pedigree disequilibrium tests for multilocus haplotypes. Genet Epidemiol 25 : 115–21. 12916020
44. Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, et al, (2000) The Protein Data Bank. Nucleic Acids Res 28 : 235–42. 10592235
45. Notredame C, Higgins DG, and Heringa J, (2000) T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol 302 : 205–17. 10964570
46. Eswar N, Eramian D, Webb B, Shen MY, and Sali A, (2008) Protein structure modeling with MODELLER. Methods Mol Biol 426 : 145–59. doi: 10.1007/978-1-60327-058-8_8 18542861
47. Wellcome Trust Case Control Consortium, (2007) Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447 : 661–678. 17554300
Štítky
Genetika Reprodukční medicína
Článek 2014 Reviewer Thank YouČlánek Closing the Gap between Knowledge and Clinical Application: Challenges for Genomic TranslationČlánek Discovery of CTCF-Sensitive Cis-Spliced Fusion RNAs between Adjacent Genes in Human Prostate CellsČlánek K-homology Nuclear Ribonucleoproteins Regulate Floral Organ Identity and Determinacy in ArabidopsisČlánek A Nitric Oxide Regulated Small RNA Controls Expression of Genes Involved in Redox Homeostasis inČlánek Contribution of the Two Genes Encoding Histone Variant H3.3 to Viability and Fertility in MiceČlánek The Genetic Architecture of the Genome-Wide Transcriptional Response to ER Stress in the Mouse
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 2
-
Všechny články tohoto čísla
- 2014 Reviewer Thank You
- Systematic Cell-Based Phenotyping of Missense Alleles Empowers Rare Variant Association Studies: A Case for and Myocardial Infarction
- African Glucose-6-Phosphate Dehydrogenase Alleles Associated with Protection from Severe Malaria in Heterozygous Females in Tanzania
- Genomics of Divergence along a Continuum of Parapatric Population Differentiation
- microRNAs Regulate Cell-to-Cell Variability of Endogenous Target Gene Expression in Developing Mouse Thymocytes
- A Rolling Circle Replication Mechanism Produces Multimeric Lariats of Mitochondrial DNA in
- Closing the Gap between Knowledge and Clinical Application: Challenges for Genomic Translation
- Partially Redundant Enhancers Cooperatively Maintain Mammalian Expression Above a Critical Functional Threshold
- Discovery of Transcription Factors and Regulatory Regions Driving Tumor Development by ATAC-seq and FAIRE-seq Open Chromatin Profiling
- Mutations in Result in Ocular Coloboma, Microcornea and Cataracts
- A Genome-Wide Hybrid Incompatibility Landscape between and
- Recurrent Evolution of Melanism in South American Felids
- Discovery of CTCF-Sensitive Cis-Spliced Fusion RNAs between Adjacent Genes in Human Prostate Cells
- Tissue Expression Pattern of PMK-2 p38 MAPK Is Established by the miR-58 Family in
- Essential Role for Endogenous siRNAs during Meiosis in Mouse Oocytes
- Matrix Metalloproteinase 2 Is Required for Ovulation and Corpus Luteum Formation in
- Evolutionary Signatures amongst Disease Genes Permit Novel Methods for Gene Prioritization and Construction of Informative Gene-Based Networks
- RR-1 Cuticular Protein TcCPR4 Is Required for Formation of Pore Canals in Rigid Cuticle
- GC-Content Evolution in Bacterial Genomes: The Biased Gene Conversion Hypothesis Expands
- Proteotoxic Stress Induces Phosphorylation of p62/SQSTM1 by ULK1 to Regulate Selective Autophagic Clearance of Protein Aggregates
- K-homology Nuclear Ribonucleoproteins Regulate Floral Organ Identity and Determinacy in Arabidopsis
- A Nitric Oxide Regulated Small RNA Controls Expression of Genes Involved in Redox Homeostasis in
- HYPER RECOMBINATION1 of the THO/TREX Complex Plays a Role in Controlling Transcription of the Gene in Arabidopsis
- Mitochondrial and Cytoplasmic ROS Have Opposing Effects on Lifespan
- Structured Observations Reveal Slow HIV-1 CTL Escape
- An Integrative Multi-scale Analysis of the Dynamic DNA Methylation Landscape in Aging
- Combining Natural Sequence Variation with High Throughput Mutational Data to Reveal Protein Interaction Sites
- Transhydrogenase Promotes the Robustness and Evolvability of Deficient in NADPH Production
- Regulators of Autophagosome Formation in Muscles
- Genomic Selection and Association Mapping in Rice (): Effect of Trait Genetic Architecture, Training Population Composition, Marker Number and Statistical Model on Accuracy of Rice Genomic Selection in Elite, Tropical Rice Breeding Lines
- Eye Selector Logic for a Coordinated Cell Cycle Exit
- Inflammation-Induced Cell Proliferation Potentiates DNA Damage-Induced Mutations
- The DNA Polymerase δ Has a Role in the Deposition of Transcriptionally Active Epigenetic Marks, Development and Flowering
- Contribution of the Two Genes Encoding Histone Variant H3.3 to Viability and Fertility in Mice
- Membrane Recognition and Dynamics of the RNA Degradosome
- P-TEFb, the Super Elongation Complex and Mediator Regulate a Subset of Non-paused Genes during Early Embryo Development
- is a Long Non-coding RNA in JNK Signaling in Epithelial Shape Changes during Drosophila Dorsal Closure
- A Pleiotropy-Informed Bayesian False Discovery Rate Adapted to a Shared Control Design Finds New Disease Associations From GWAS Summary Statistics
- Genome-wide Association Study Identifies Shared Risk Loci Common to Two Malignancies in Golden Retrievers
- and Hyperdrive Mechanisms (in Mouse Meiosis)
- Elevated In Vivo Levels of a Single Transcription Factor Directly Convert Satellite Glia into Oligodendrocyte-like Cells
- Systemic Delivery of MicroRNA-101 Potently Inhibits Hepatocellular Carcinoma by Repressing Multiple Targets
- Pooled Sequencing of 531 Genes in Inflammatory Bowel Disease Identifies an Associated Rare Variant in and Implicates Other Immune Related Genes
- Abscission Is Regulated by the ESCRT-III Protein Shrub in Germline Stem Cells
- Temperature Stress Mediates Decanalization and Dominance of Gene Expression in
- Transcriptome Wide Annotation of Eukaryotic RNase III Reactivity and Degradation Signals
- The Exosome Component Rrp6 Is Required for RNA Polymerase II Termination at Specific Targets of the Nrd1-Nab3 Pathway
- Sex-specific -regulatory Variation on the X Chromosome
- Regulation of Toll-like Receptor Signaling by the SF3a mRNA Splicing Complex
- Modeling of the Human Alveolar Rhabdomyosarcoma Chromosome Translocation in Mouse Myoblasts Using CRISPR-Cas9 Nuclease
- Asymmetry of the Budding Yeast Tem1 GTPase at Spindle Poles Is Required for Spindle Positioning But Not for Mitotic Exit
- TIM Binds Importin α1, and Acts as an Adapter to Transport PER to the Nucleus
- Antagonistic Roles for KNOX1 and KNOX2 Genes in Patterning the Land Plant Body Plan Following an Ancient Gene Duplication
- The Genetic Architecture of the Genome-Wide Transcriptional Response to ER Stress in the Mouse
- Fatty Acid Synthase Cooperates with Glyoxalase 1 to Protect against Sugar Toxicity
- Region-Specific Activation of mRNA Translation by Inhibition of Bruno-Mediated Repression
- An Essential Role of the Arginine Vasotocin System in Mate-Guarding Behaviors in Triadic Relationships of Medaka Fish ()
- Interaction between the tRNA-Binding and C-Terminal Domains of Yeast Gcn2 Regulates Kinase Activity In Vivo
- Hyper-Acetylation of Histone H3K56 Limits Break-Induced Replication by Inhibiting Extensive Repair Synthesis
- Prodomain Removal Enables Neto to Stabilize Glutamate Receptors at the Neuromuscular Junction
- Recent Selective Sweeps in North American Show Signatures of Soft Sweeps
- Identification and Functional Analysis of Healing Regulators in
- A Multi-Megabase Copy Number Gain Causes Maternal Transmission Ratio Distortion on Mouse Chromosome 2
- Drosophila Casein Kinase I Alpha Regulates Homolog Pairing and Genome Organization by Modulating Condensin II Subunit Cap-H2 Levels
- The Hippo Pathway Regulates Homeostatic Growth of Stem Cell Niche Precursors in the Ovary
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Genomic Selection and Association Mapping in Rice (): Effect of Trait Genetic Architecture, Training Population Composition, Marker Number and Statistical Model on Accuracy of Rice Genomic Selection in Elite, Tropical Rice Breeding Lines
- Discovery of Transcription Factors and Regulatory Regions Driving Tumor Development by ATAC-seq and FAIRE-seq Open Chromatin Profiling
- Evolutionary Signatures amongst Disease Genes Permit Novel Methods for Gene Prioritization and Construction of Informative Gene-Based Networks
- Proteotoxic Stress Induces Phosphorylation of p62/SQSTM1 by ULK1 to Regulate Selective Autophagic Clearance of Protein Aggregates
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



