-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaEye Selector Logic for a Coordinated Cell Cycle Exit
Organs develop from groups of undifferentiated cells that proliferate and differentiate into specific cell types. During development, the coupling between proliferation and differentiation programs ensures that enough cells of the different cell types are generated. This is critical for proper organ formation and function. Here, we use the developing Drosophila eye to examine how the coupling between these two programs is achieved. During eye development, progenitors are amplified before they exit the cell cycle and enter the differentiation program. This amplification step depends on an expression burst of the mitotic trigger string/cdc25, which, by forcing cells into mitosis, synchronizes cells in G1 just before differentiation onset. Thus string regulation acts as a hub where differentiation and proliferation programs are integrated. We identify a DNA element that controls the burst of string expression prior to differentiation, and show that it is regulated by the same gene network that triggers eye development. The transcription factor Pax6/Eyeless is a key regulator in this network. Eyeless acts cooperatively with Sine oculis and Eyes absent to regulate string, through a positive feed-forward loop. This loop is negatively modulated by the progenitor-specific transcription factor Homothorax/Meis1. This work shows that transcription factors that instruct cells to acquire an eye fate also control their proliferation regime, thus guaranteeing the coupling between proliferation and differentiation.
Published in the journal: . PLoS Genet 11(2): e32767. doi:10.1371/journal.pgen.1004981
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004981Summary
Organs develop from groups of undifferentiated cells that proliferate and differentiate into specific cell types. During development, the coupling between proliferation and differentiation programs ensures that enough cells of the different cell types are generated. This is critical for proper organ formation and function. Here, we use the developing Drosophila eye to examine how the coupling between these two programs is achieved. During eye development, progenitors are amplified before they exit the cell cycle and enter the differentiation program. This amplification step depends on an expression burst of the mitotic trigger string/cdc25, which, by forcing cells into mitosis, synchronizes cells in G1 just before differentiation onset. Thus string regulation acts as a hub where differentiation and proliferation programs are integrated. We identify a DNA element that controls the burst of string expression prior to differentiation, and show that it is regulated by the same gene network that triggers eye development. The transcription factor Pax6/Eyeless is a key regulator in this network. Eyeless acts cooperatively with Sine oculis and Eyes absent to regulate string, through a positive feed-forward loop. This loop is negatively modulated by the progenitor-specific transcription factor Homothorax/Meis1. This work shows that transcription factors that instruct cells to acquire an eye fate also control their proliferation regime, thus guaranteeing the coupling between proliferation and differentiation.
Introduction
Selector genes are transcription factors that instruct the development of organs. The processes under the control of selector genes include the assignation of cell fates and their organ-specific responses to extracellular signals [1]. But organ development also requires the faithful execution of proliferation programs to ensure the expansion of progenitor cells and their coordinated exit from the cell cycle prior to the onset of differentiation. This coordinated cell cycle exit is critical to regulate organ size during development and to ensure tissue homeostasis during adult life. The power of selector genes to control the differentiation state of cells and their proliferation regimes explains why abnormal expression of these transcription factors is often associated to cancer [reviewed in 2,3]. However, how selector genes carry out the coordination between proliferation and differentiation programs is still unclear.
Structures of the nervous system, such as the retina, in which complex arrays of different cell types need to be assembled from multipotent proliferative progenitors, are especially sensitive to impairments of proliferation control mechanisms [reviewed in 4,5]. It is therefore likely that selector genes coordinate cell cycle exit with the processes of differentiation and patterning by co-regulating the transcription of cell cycle and patterning genes. However, this control may be direct or mediated by intermediate transcription factors.
The eye selector function is exerted by a network of transcription factors and signaling pathways, with many of the network genes shared by invertebrates and vertebrates. The Pax6 selector genes are on top of the retinal determination (RD) gene network in both animal groups [6]. Pax6 mutations are responsible for aniridia [7,8], while Pax6 overexpression is associated with retinoblastoma cancer progression through promotion of proliferation and cell survival [9–11].
In Drosophila, the RD gene network comprises a number of transcription factors and nuclear proteins, that includes members of conserved gene families: The Pax6 paralogues eyeless (ey) and twin of eyeless (toy); the Six family genes Optix (Six3) and sine-oculis (so; Six1,2); So’s partner, eyes absent (eya); dachshund (dac); and the Meis1 homologue homothorax (hth). These genes are not only connected through transcriptional cross-regulation, but also have been found to engage in protein complexes [reviewed in 12,13].
Research during the past years is yielding an increasingly clearer picture of how the process of eye specification and retinal patterning in Drosophila is controlled [reviewed in 13,14]. The eye primordium (also called “eye disc”) derives from the So-expressing embryonic cephalic neuroectoderm [15]. Within this domain, toy activates ey expression during late embryogenesis, which results in the specification of the eye-progenitor cells [16]. During larval life, ey-expressing progenitors are maintained proliferative and multipotent as long as they express hth [17–19]. Repression of hth starts during the third and last larval stage (L3), mediated by Decapentaplegic (Dpp a BMP2/4-like molecule) and Hedgehog (Hh) signals produced at a moving signaling center, called “morphogenetic furrow” (MF). hth repression is key, as it allows the upregulation of so, eya and dac [17,19]. Coinciding with hth repression, the expression of string (stg)-the Drosophila cdc25 phosphatase homologue [20–22] - is upregulated and this drives cells through a few consecutive mitotic rounds (the first mitotic wave, FMW), resulting in G1-synchronized ey-so-eya-dac-expressing cells (retinal precursors) [17,19,20]. The expression of ey, so and eya turns on the expression of the bHLH gene atonal (ato), the fly homologue of ath5/atoh7, which is necessary for the differentiation of precursor cells into photoreceptors, lens and pigment cells of the retina [23–26].
The information processing devices in networks such as the RD gene network are cis-regulatory elements (CREs), DNA sequences that allow binding of specific combinations of transcription factors, which in turn regulate transcription of the CRE target genes [27]. Therefore, CREs are key to understand the logic that drives the developmental processes directed by a gene network. In the Drosophila RD gene network, CREs from ey [28,29], so [30,31], eya [32], dac [33]; optix [34] and ato [24–26,35] have been isolated and studied in molecular detail. Not surprisingly, all rely on direct Pax6 input and at least so, dac and ato CREs also integrate direct regulation by the So:Eya complex [24–26,31,33]. But all of these genes are transcription factors, not effector genes. Is the logic acting upon transcription factors the same as that controlling specific outputs of the network’s function—such as cell cycle control?
In this paper we have addressed this issue by investigating the direct regulatory logic acting upon the eye-specific stg CRE. During Drosophila retina development, a transcriptional burst of stg is associated to the transition from proliferative progenitors to cell cycle quiescent precursors [19,20,22]. This peak of stg drives progenitors, which are mostly in the G2 phase of their cell cycle, through the FMW, leading to their G1 synchronization [19,36]. This synchronicity is essential: In stghwy mutants, which lack specifically this peak of stg expression, precursors are specified but do not become G1-synchronized. As a result, the patterning of the retina is aberrant [22]. Therefore, the study of stg transcriptional regulation in the eye offers an ideal model to understand how organ specific cell cycle and patterning programs are coupled during development.
We identified a distal 5′stg CRE, which we named stg-FMW (First Mitotic Wave) enhancer. When stg expression was driven by stg-FMW enhancer, it rescued the eye defects of stghwy mutants, indicating that stg-FMW contains most, if not all the regulatory information required for the accurate spatial-temporal expression of stg at the progenitor-precursor transition. Within this element, we characterized two positive inputs: one from both Pax6 proteins, Ey and Toy, and one from So:Eya. Interaction with these transcription factors occurs through two binding sites. We also identified one negative input: Hth. In agreement, assays in vivo suggested that Hth hampers Ey activation of stg-FMW. This fact could explain mechanistically the negative action of Hth on stg transcription. The picture that emerges is of a coherent feed-forward loop in which Ey and Toy play partially redundant activating roles, together with So:Eya, on stg transcription. Moreover, this activation is modulated by the negative input of the meis1 gene, hth.
Results
Identification of eye-specific stg CREs
As an entry point into the molecular mechanisms by which selector transcription factors activate organ-specific programs of cell division, we searched for an eye specific regulatory element of stg. Lehman and co-workers had scanned 38 Kb of the stg locus (from-35 to +3 relative to the transcription start site) and uncovered several CREs [21 and Fig. 1A]. These included CREs active in embryos and imaginal discs, but none of the fragments studied recapitulated the strong stripe of stg expression anterior to the MF (Fig. 1B). We re-analyzed a similar interval of 38.7 Kb (from the stg transcription start site [Chr3R: 25.081.410] to CG14506 [Chr3R: 25.120.100], the gene located immediately upstream of stg) by generating a new set of tiled reporter transgenes with an average fragments length of 5 Kb. Contiguous fragments overlap each other an average length of 1,5 Kb (Fig. 1A). This approach was selected to avoid splitting blocks of conserved sequence, as sequence conservation is often a landmark of CREs [37]. Again, none of the fragments from this interval revealed an expression pattern reminiscent of stg in the eye disc.
Fig. 1. Identification of eye-specific stg CREs. 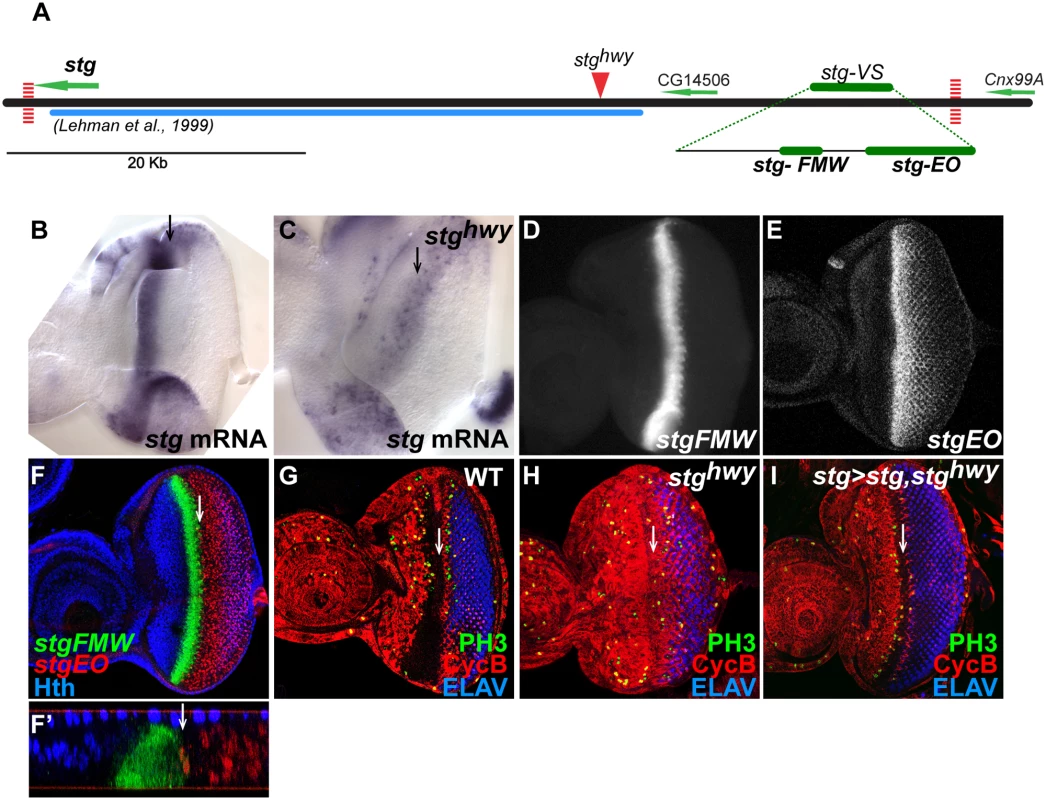
(A) Map of the stg genomic locus, covering a region of ~62 Kb, which includes CG14506. Green arrows delimit the coding regions and indicate direction of transcription. Vertical dashed red lines map the position of CTCF binding sites downstream of stg and Cnx99A. Solid blue line delimits the region previous screened by Lehman et al. [21]. Solid red arrowhead indicates the insertion point of the gypsy transposon in stghwy allele. Solid green lines indicate the position of the CREs that showed expression in the visual system, stg-VS, stg-FMW and stg-EO. (B, C) Expression of stg mRNA in wild type and stghwy eye imaginal discs. (B) Expression can be detected in the precursor cells domain, anterior to the MF and ocellar domain. (C) In stghwy homozygous eye imaginal discs expression can only be detected posterior to the MF. (D, E) Expression of dGFP driven by stg-FMW and stg-EO enhancer fragments. (F) Eye imaginal disc stained for stg-FMW (green), stg-EO (red), and Hth (blue). stg-FMW and stg-EO are expressed in non-overlapping domains. stg-FMW drives expression in precursor cells and abuts the domain of Hth expression (blue). stg-EO drives expression in cells posterior to the MF. (F’) Cross-section from an inset of F, showing that stg-FMW and stg-EO patterns do not overlap. (G-H) Wild-type (G) and stghwy homozygous (H) L3 eye imaginal discs stained for the G2 marker Cyclin B (red), the mitotic marker phospho-Histone H3 (PH3; green) and the photoreceptor marker ELAV (blue). (I) stghwy homozygous disc ectopically expressing Stg, driven by stgFMW-Gal4, and stained for cyclin B (red), PH3 (green) and ELAV (blue). Ectopic expression of Stg in the stg-FMW expression domain restores the pattern of Cyclin B (red) and mitosis (green) to wild type. In all images anterior is to the left. White arrow indicates the MF. Together with the Lehman study, our results suggested that the eye-specific CREs should be located further upstream [21]. To try to define the expected limit of the stg regulatory landscape we used several landmarks. First, the analysis of an extended genomic region revealed the existence of two class I insulator binding sites [38,39], one immediately downstream of the stg transcript (Chr3R: 25077239) and another downstream of Cnx99A (Chr3R: 25138877), delimiting a region of 61,6 Kb (Fig. 1A). Binding of class I insulators helps to establish chromatin boundaries between genes [39,40]. Therefore, we considered that this region might comprise the stg regulatory landscape and should include unidentified stg CREs. This interval includes CG14506 as well. However, this transcript is not conserved in all Drosophila species sequenced, although the adjacent sequences are highly conserved, suggesting that CG14506 is a bystander gene within the stg locus. Second, a regulatory mutation in the stg gene, highway (stghwy), had been shown to be associated to an insertion of an uncharacterized DNA sequence at around 30 Kb upstream of the stg transcription start site. The stghwy is a viable allele that results in slightly reduced, roughened eyes [22]. In stghwy mutant eye discs the peak of stg expression at the progenitor-precursor transition is lost (Fig. 1B, C). As a consequence, cells fail to undergo G1 arrest, and accumulate in G2, with high levels of mitotic cyclins, such as cyclin B [Fig. 1G, H and 22]. Since stghwy is an eye-specific regulatory allele of stg, we reasoned that the stghwy insertion might be affecting the CRE we were looking after, perhaps having landed in its vicinity.
A primer walking strategy was next employed to identify the nature of the DNA element and the exact insertion point in stghwy. Molecularly, we defined the mutation associated with stghwy as an insertion of a gypsy transposable element between positions Chr3R: 25115094 and Chr3R: 25115097 (Fig. 1A and S1A Fig.). Gypsy transposable elements are known to block enhancer-promoter interactions when located in between them [reviewed in 41]. This finding suggested that the insertion in stghwy was likely impairing the contacts between the eye-specific CRE and the stg promoter. Further, it predicted that the eye CREs should lie between the genes CG14506 and Cnx99A. When we extended our reporter transgene study to this region, we identified a fragment of 4.8 Kb, located distal to CG14506 and 52 Kb away from the stg promoter. This fragment was sufficient to drive expression of the reporter gene (destabilized Green Fluorescent Protein, (dGFP)) in eye discs, both in a stripe anterior to the MF as well as in cells posterior to it (Fig. 1A and S1B Fig.). In addition, this fragment showed enhancer activity in the dorsal anterior region of the eye disc, where the prospective ocellar region resides, and in the lamina region of the optic lobes. We named it stg-VisualSystem (stg-VS).
We next subdivided stg-VS into smaller overlapping fragments. This allowed the identification of two enhancer elements, of 539bp and 690bp respectively, that drive expression in different cell populations of the eye disc (Fig. 1A, D, E). The remaining sub-fragments of stg-VS failed to drive expression in the visual system or elsewhere. The 539bp enhancer drives strong dGFP expression in the FMW domain and precursor cells and recapitulates stg expression in the eye field after differentiation onset (S2 Fig.). Accordingly, the 539bp enhancer was called stg-First Mitotic Wave (stg-FMW) (Fig. 1D). The 690bp element drives expression in the ocellar domain, and in a subset of cells posterior to the MF. This fragment was named stg-EyeOcelli (stg-EO) (Fig. 1E). stg-FMW and stg-EO are expressed in adjacent, non-overlapping domains (Fig. 1F, F’) and together reconstitute the eye disc-specific pattern of stg transcription. Anteriorly, expression driven by stg-FMW abuts the Hth expression domain (Fig. 1F, F’), as was previously shown for stg mRNA [19]. Expression of stg-EO in the eye field overlaps the so-called second mitotic wave [SMW, 42]. To test that stg-FMW is a functional stg enhancer, we attempted to rescue the stghwy phenotype, by driving stg expression using a stg-FMW-GAL4 driver in stghwy homozygous individuals. stg-FMW-GAL4>UAS-stg rescued the adult eye phenotype and the pattern of cyclin B accumulation in L3 eye discs of stghwy mutants (Fig. 1G-I). This result supports the idea that stg-FMW is a functional, eye-specific stg CRE and, together with the data on enhancer activity throughout the stg locus, suggests that it may be the sole CRE responsible for stg expression at the FMW.
Ey and Toy are redundantly required to activate stg-FMW
The stg-FMW sequence shows a high degree of conservation (Fig. 2A). Using JASPAR and TRANSFAC models [43,44] we predicted the existence of putative transcription factor binding sites for components of the RD network. For the identification of Ey binding sites we generated our own position weight matrix from a set of published binding sites [24,30,34,45] (Fig. 2B and S3 Fig.). Evolutionarily conserved Ey binding sites in the genome of 12 Drosophila species were filtered using the CBS platform [46]. Two highly conserved regions were identified, which we refer to as Binding Site 1 (BS1) and BS2 (Fig. 2A, C). BS1 contains partially overlapping putative binding sites for Ey/Pax6 and So (Fig. 2C). BS2 contains one Ey/Pax6 conserved binding site, and a highly conserved consensus site for Hth lies adjacent to it (Fig. 2A, C).
Fig. 2. Ey and Toy are redundantly required to activate stg-FMW. 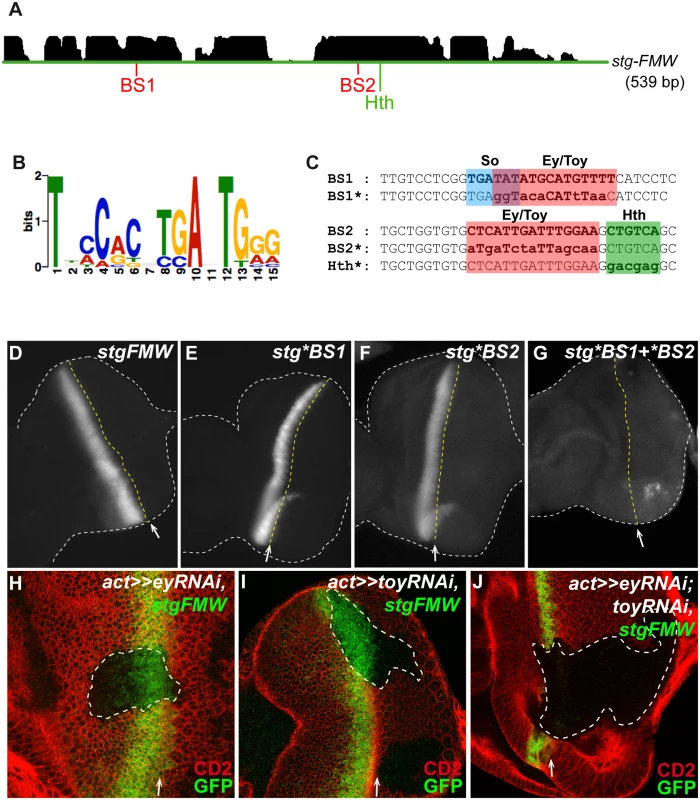
(A) The pattern of conservation of stg-FMW enhancer sequence in Drosophila species as displayed by the UCSC genome browser (http://genome.ucsc.edu) is shown at the top. Two highly conserved regions harboring binding sites for Pax6, BS1 and BS2, and Hth are shown below. (B) Logo of the optimized PAX6 Position Weight Matrix (PWM) used in this study. Nucleotide preference is represented by the height of the letter. (C) Partial sequence of BS1 and BS2. BS1 contains binding sites for So and Pax6 genes (Ey/Toy) highlighted in blue and red, respectively. BS1*: Mutated version of BS1. Ey/Pax6 mutated bases are shown in lowercase. BS2 contains adjacent binding sites for Pax6 (Ey/Toy) and Hth, highlighted in red and green respectively. Mutated core bases for Pax6 (Ey/Toy) and Hth are shown in lowercase in BS2*. (D—G) Analysis of the expression of stg-FMW upon mutation of BS1 or BS2. Representative L3 eye imaginal discs carrying the wild type stg-FMW (D), and the mutated versions stg*BS1 (E) and stg*BS2 (F) of the enhancer. (G) Expression of GFP is not affected upon mutation of BS1 or BS2 in stg-FMW. Simultaneous mutation of BS1 and BS2 (stg*BS1+*BS2) abolishes GFP expression in the eye field; small GFP dots can be detected but only at the dorsal and ventral margins of the eye disc. Discs are outlined by the white dashed lines. The position of the MF is indicated by the white arrow and yellow dashed line. (H—J) Clones of ey (H) or toy (I) RNAi, in the eye imaginal disc. Clones are outlined and marked by the absence of CD2 (red). Expression of stg-FMW (green) is not affected by downregulation of Ey or Toy. (J) Eye disc containing ey and toy double mutant clones, stained for CD2 (red) and stg-FMW (green). In clones of cells double mutant for ey and toy the expression of stg-FMW is lost. White arrow indicates the position of the MF. In all images anterior is to the left. To test the in vivo relevance of BS1 and BS2, we mutated the bases fitting the Ey/Pax6 consensus at each site. Transgenic lines carrying mutant versions of the stg-FMW enhancer harboring mutations in BS1 (stg*BS1), in BS2 (stg*BS2) or in both sites (stg *BS1+*BS2) were analyzed (Fig. 2D-G). Neither the stg*BS1 nor the stg*BS2 single mutants showed altered temporal or spatial expression (Fig. 2E, F), suggesting that the remaining site suffices for enhancer activity during eye development. However, when the two sites were simultaneously mutated (stg*BS1+*BS2), enhancer activity was lost (Fig. 2G). This shows that both sites are redundant for enhancer activity in vivo.
Since mutation of both Ey/Pax6 consensus-binding sites abolished enhancer activity, we next assayed whether Ey was required for stg-FMW activity. The expression of stg-FMW remains unaffected in clones where ey expression has been knocked-down using RNAi (Fig. 2H). As toy, a second Pax6 gene, is expressed coextensively with ey in the eye primordium [16], we tested if Toy was required for regulation of stg-FMW. As observed with ey, toy downregulation through RNAi did not affect enhancer activity (Fig. 2I).
Since Ey and Toy have similar expression patterns and binding site preferences [16,31,47], we next tested a potential redundant function of Ey and Toy in stg-FMW regulation. For this, we generated clones of cells in which both genes were simultaneously knocked-down by co-expression of ey-RNAi and toy-RNAi. In these cells the activity of stg-FMW was abolished in a cell-autonomous way (Fig. 2J). This result shows that both Ey and Toy redundantly activate the stg-FMW enhancer. A redundant function between Ey and Toy was further supported by the finding that in ey2 homozygous imaginal discs, where toy expression is maintained [16], the pattern and levels of expression of stg mRNA or stg-FMW were not affected (S4A-G Fig.).
To explore a potential “division of labor” between the two sites, with each of them specializing in only one of the two Pax6, we generated clones of ey-RNAi and toy-RNAi in the presence of stg-FMW mutated versions (stg*BS1 and stg*BS2) (S4H-K Fig.) Upon mutation of BS1 or BS2, downregulation of Ey did not abolish enhancer activity (S4F,G Fig.). Downregulation of Toy did not impact on the activity of the single-mutant versions of stg-FMW either (S4J, K Fig.). These results show that one Ey/Pax6 binding site suffices for enhancer activity, and that Ey and Toy do not have preferential binding in vivo.
Eya/So cooperate with Ey/Toy and act as positive regulators of stg-FMW
Region BS1 also contains a putative binding site for So (Fig. 2C). So is known to physically interact with the transcriptional co-activator Eya to regulate downstream genes [48]. We next tested the role of So and its transcriptional co-activator Eya in the regulation of stg-FMW. Both genes lay downstream of Ey in the RD gene network [reviewed in 14,49] and stg has been previously identified as a transcriptional target of the Eya:So complex [50]. Loss of function of Eya (S5A Fig.) or its downregulation by means of RNAi (Fig. 3A) in cell clones resulted in a cell-autonomous loss of enhancer activity in the precursor domain. A similar result was obtained when So expression was knocked down using RNAi (S5B Fig.). This loss of enhancer activity coincided with the maintenance of high levels of Hth expression (Fig. 3A” and S5B Fig.).
Fig. 3. So/Eya are positive regulators of stg-FMW. 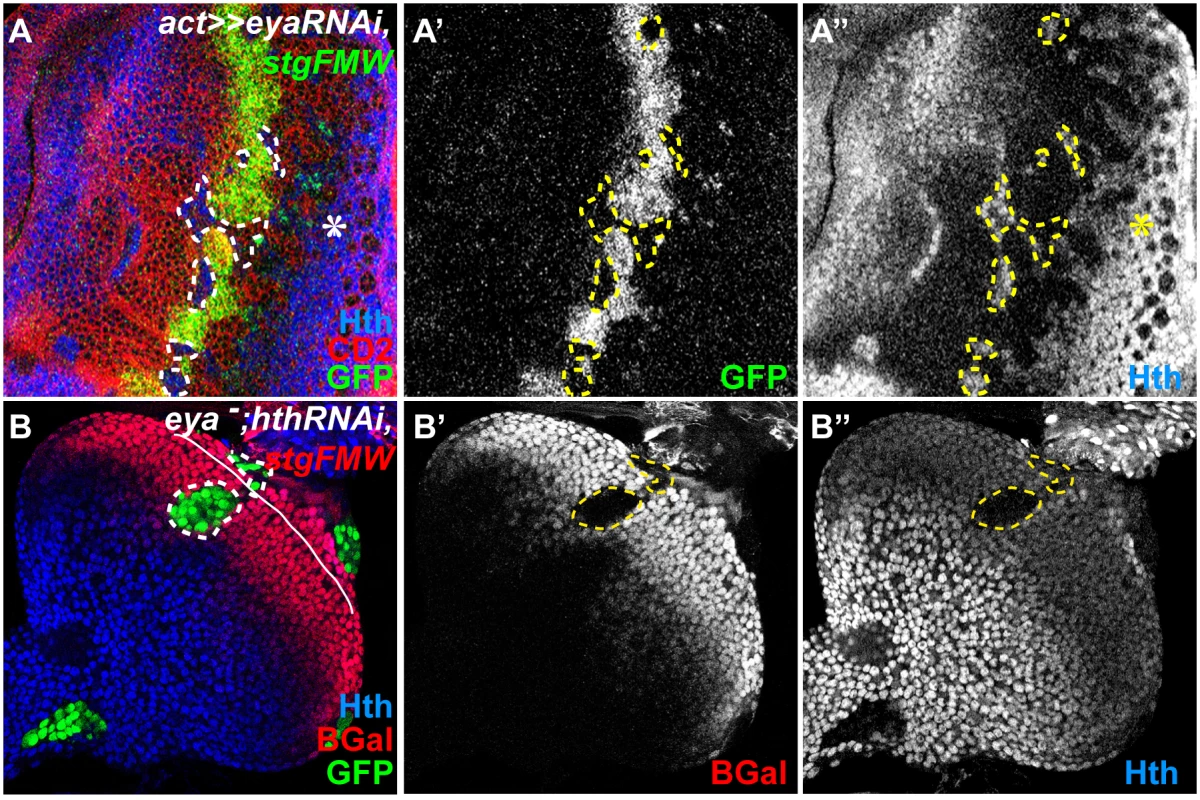
(A) Eye imaginal disc with clones of cells mutant for eya, labeled by the absence of CD2 (red). (A’) Within the clone cells, stg-FMW expression (green) is not activated. (A”) Hth expression (blue) is maintained in eya RNAi cells. (B) MARCM eya-hth- clones (GFP) stained for stg-FMW (lacZ- red) (B’) and Hth (B”)(blue), showing that eya is required for stg-FMW expression. Clones are outlined. The asterisk (*) in A indicates the normal expression pattern of Hth, during L3, in pigment progenitor cells posterior to the MF [17]. In all images anterior is to the left. We had previously shown that Hth could act as a repressor of stg transcription [19]. Additionally, Eya:So are negative regulators of Hth expression during eye development [17]. Therefore, the observed loss of stg-FMW enhancer activity could result from either the loss of Hth repression, or alternatively reflect a positive requirement of Eya:So for stg-FMW activation. To discriminate between these two hypotheses, we first checked whether ectopic expression of Hth could repress stg-FMW. In Hth-expressing clones stg-FMW activation was delayed, but not repressed (Fig. 4A). These findings were qualitatively different from the ones obtained upon RNAi-mediated eya knock-down, where loss of enhancer activity was always observed, irrespective of the position of the clones within the precursor cell domain. This suggested that indeed Eya:So acted as stg-FMW activators. To test this issue avoiding any interference by Hth, we generated clones of cells simultaneously mutant for eya (eya null) and hth (hth RNAi), using the MARCM system (Fig. 3B) [51]. In eya - hth- cells, stg-FMW activity was always lost in a cell-autonomous manner. However, in these eya - hth- double mutant cells expression of Ey was maintained (S5C Fig.). Therefore, these results show that Eya:So are required as stg-FMW activators independently of their role as hth transcriptional repressors.
Fig. 4. Hth defines the anterior domain of stg-FMW expression. 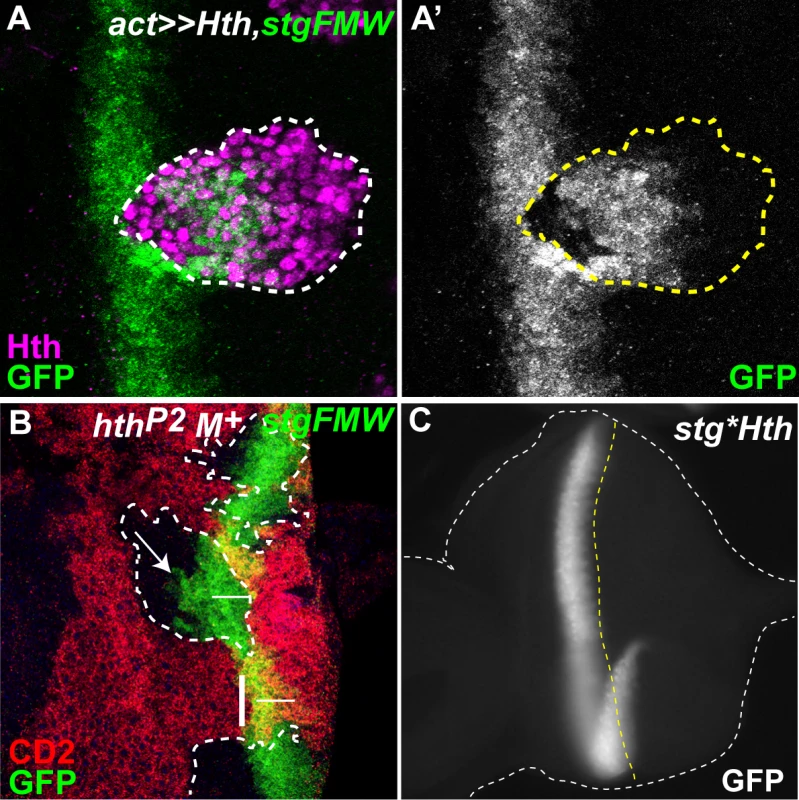
(A) Clone of cells ectopically expressing Hth-HA, labeled in magenta, and stained for stg-FMW (green). (A’) Expression of stg-FMW is only repressed within the anterior domain of the clone. (B) Eye disc containing clones of a null allele of hth (hthP2), stained for CD2 (red) and stg-FMW (green). Clone cells are labeled by the absence of CD2. Within the clone stg-FMW expression occurs in a few cell rows anterior to its normal domain of expression. Arrow indicates premature expression of GFP. The vertical dashed line limits the most anterior expression of stg-FMW in wild-type tissue. The horizontal line represents the width of stg-FWW expression in wild-type tissue. (C) Mutation of the Hth BS (stg*Hth) does not affect the spatial or temporal expression of stg-FMW. Since the RD nuclear protein Dac has also been found as part of the Eya/So complex [52] we tested if Dac also played a role in stg transcriptional regulation. In clones of a dac-null allele (dac3) the expression of stg-FMW remained unaltered (S5D Fig.), indicating that Dac is not a partner of Eya:So in the regulation of stg-FMW enhancer. This finding further indicates that different Eya:So targets may rely on the formation of different protein complexes.
Hth sharpens the onset of stg-FMW activity
Previous results suggested that Hth was a transcriptional repressor of stg [19]. However, as we described before, ectopic expression of Hth delayed the onset of stg-FMW activation, but did not block it (Fig. 4A), suggesting that hth could be involved in the precise timing of stg-FMW expression rather than in repressing it. To test this idea, we generated hth-mutant clones of a strong allele (hthP2) [53]. Since hth-clones grow poorly [18,19,54], we gave them a growth advantage by using the Minute technique [55]. In hth - M+ clones the anterior border of stg-FMW expression was shifted anteriorly (Fig. 4B). Therefore hth is required for the precise spatio-temporal activation of stg-FMW, delaying its initiation. Hth is a transcription factor and its action could be mediated through direct interaction with the stg-FMW enhancer. In fact, we identified a potential Hth BS in the stg-FMW sequence (Fig. 2C). However, mutation of this site (stg*hth) did not result in changes in stg-FMW expression (Fig. 4C and S6C Fig.). Although this result does not rule out a direct Hth-DNA interaction through a non-canonical site on the stg-FMW enhancer, it points to an indirect effect. In fact, it has been previously shown that Hth and Ey can form a protein complex in vivo [17]. The possibility that Hth affects stg-FMW through Ey is explored below.
So/Eya and Ey/Toy act preferentially through different binding sites in vivo
Our results show that during eye development Ey/ Toy and So plus Eya are all necessary to activate stg-FMW, although in the eye neither the Pax6 genes Ey/Toy or Eya/So are sufficient to do so. Molecularly, mutational analysis of the Pax6 binding sites suggested that Ey and Toy could exert their function through direct binding to BS1 and BS2. To test this hypothesis directly and grasp the molecular interactions underlying stg-FMW activity, we performed chromatin immunoprecipitation followed by quantitative real-time PCR (ChIP-qPCR) experiments. We used ectopic gene expression in wing discs as they can be used as a “blank slate” where to assess the functional consequences of expressing RD genes, including Ey. In addition, since in the wing disc Hth expression is restricted to the hinge, we bypass the potential repressor effect of Hth on Ey activity in most of the disc. To drive gene expression we used the dpp-GAL4 line, which is expressed in a stripe that bisects the wing disc along its anterior-posterior (A/P) axis (Fig. 5A). dpp-GAL4-driven Ey expression (dpp>ey) was sufficient to promote activation of the enhancer throughout the Dpp expression domain (Fig. 5B and S7 Fig.), in agreement with the potent eye-inducing ability of Ey [56,57]. In contrast, Toy was only able to induce expression from stg-FMW in a small subset of cells in the ventral hinge region (Fig. 5C). This suggests that although in the eye imaginal disc both Ey and Toy have the ability to promote enhancer activity, Ey is a stronger regulator of stg-FMW than Toy. We next analyzed the in vivo binding of Ey to stg-FMW by ChIP-qPCR in dpp>ey wing discs. We designed primers so that we could detect binding to region 1 (stg-BS1) or region 2 (stg-BS2). As positive control we used a region in the ato-3′ enhancer known to be bound by Ey [24] (Fig. 5I). As expected, we detected a high enrichment of Ey at ato-3’ relative to our negative control (Fig. 5I). ChIP-qPCR analysis showed that Ey binds to both BS1 and BS2, reinforcing the results described above showing that both sites are used in vivo. We consistently recovered higher amounts of chromatin from BS2 than from BS1, suggesting that Ey’s binding affinity towards BS2 region is higher (Fig. 5I), and that this site might be preferentially used by Ey in vivo.
Our previous experiments showed a requirement for the Eya:So complex in stg-FMW activation, and identified a putative So binding site on region BS1. Ectopic assays in the wing showed that co-expression of Eya and So (dpp>Eya,So) was able to activate the enhancer in a subset of hinge cells located along the A/P boundary (Fig. 5D and S7 Fig.). However, ectopic expression of So, alone or together with Dac, was not sufficient to activate stg-FMW. This observation supports the existence of an Eya:So complex within the precursor domain, whose targets are distinct and independent of the Dac:Eya:So complex. Eya and So can act as transcriptional regulators of Ey [29,49] and it could be argued that the observed stg-FMW activity might be indirect and due to Ey up-regulation. To test this point, we checked if ectopic expression of Eya:So in the Dpp domain induced Ey expression. Although ectopic Ey expression was easily detected in the antennal imaginal disc, we systematically failed to detect Ey expression in the wing or leg imaginal discs of dpp>Eya,So larvae (S7D,G Fig.). These results are in agreement with previous observations [16,48,58]. Nevertheless, and to rule out the possibility that undetectable levels of Ey might contribute to the activation of stg-FMW upon ectopic Eya:So expression (Fig. 5D), we used an RNAi to knock ey expression down when co-expressing Eya and So (dpp>eyRNAi,Eya,So; Fig. 5E). In these conditions, ectopic stg-FMW was induced in the same subset of cells as when induced by Eya and So alone (Fig. 5D, E). This shows that, in ectopic assays, the Eya:So complex has the capacity to promote transcription from the enhancer independently of Ey. This finding allowed us to test if BS1, which contains a putative So binding site, was indeed required for the activation of stg-FMW by Eya+So. In case this hypothesis were true, mutation of BS1 should preclude stg-FMW activation. To test this point, we checked Eya:So’s ability to activate the enhancer upon mutation of BS1 or BS2, when Ey expression was simultaneously attenuated (dpp>eyRNAi,Eya,So). In this background, mutation of BS1 (stg*BS1) prevented Eya+So from activating the enhancer (Fig. 5F). In contrast, when stg*BS2 was used, the pattern and expression levels of the reporter gene upon Eya+So expression were similar to those of wild-type stg-FMW (Fig. 5G). These results suggest that Eya:So complex most likely regulates stg-FMW activity through binding to stg-BS1. To test this hypothesis we performed ChIP-qPCR experiments using an HA-tagged Eya protein (Eya:HA). As Eya lacks a DNA binding domain, its association with DNA would only occur if forming a complex with its DNA-binding partner So [59]. Thus, Eya ChIP can be used as a read-out of Eya:So target DNA binding. In dpp>Eya:HA wing discs, anti-HA ChIP-qPCR showed enrichment of stg-BS1 and stg-BS2, although only that for BS2 was statistically significant. ato-3’, which was again used as positive control, showed also a significant enrichment, as so did the banA enhancer (also included as control; see below), although to a lower extent (S7H Fig.). Taken together, our results show that the Eya:So complex is able to bind BS2 (and likely also BS1). However, Eya:So regulation of stg-FMW relies mostly on BS1.
In addition to the positive regulators Toy, Ey, Eya and So, our experiments indicate that Hth contributes to the precision of the onset of stg-FMW expression, delaying its activation. The fact that a mutation that eliminates the single canonical Hth binding site does not affect the enhancer’s expression suggested that Hth’s action could be indirect, perhaps mediated through its known interaction with Ey [17]. To test this point, we evaluated the ability of Ey to activate stg-FMW in the presence of ectopic Hth (Fig. 5H). In the wing imaginal disc, ectopic expression of Hth strongly reduces the ability of Ey to activate transcription of the reporter gene, which can only be detected in spots in hinge cells (compare Fig. 5H with 5B). To address if Hth counteracts Ey positive role on stg transcription by preventing Ey’s binding to chromatin, we performed ChIP-qPCR assays in wing discs upon simultaneous expression of an HA-tagged Ey plus Hth (dpp>HA:Ey,Hth) (Fig. 5I, red bars). The amount of Ey bound to chromatin regions stg-BS1 and stg-BS2 in dpp>HA:Ey,Hth was slightly reduced compared to dpp>HA:Ey (Fig. 5I, blue bars). A stronger reduction was observed for ato-3′, the activity of which is known to be directly regulated by Ey binding [24,25]. These results show that Hth only moderately hampers Ey binding to its target DNA sites, something that could be happening through a direct Hth:Ey interaction. Additionally, we noted that banA, a CRE from the bantam gene, a known direct Hth target in the eye is also bound by Ey (Fig. 5I) [18]. In contrast to stg and ato sequences, we observe a 2-fold enrichment of the banA sequence upon ectopic co-expression of Hth and Ey. This is in agreement with the known binding of Hth to banA and likely reflects the previously described ability of Hth to interact with Ey [17,18].
Fig. 5. Ey and So regulate stg-FMW through different binding sites in vivo. 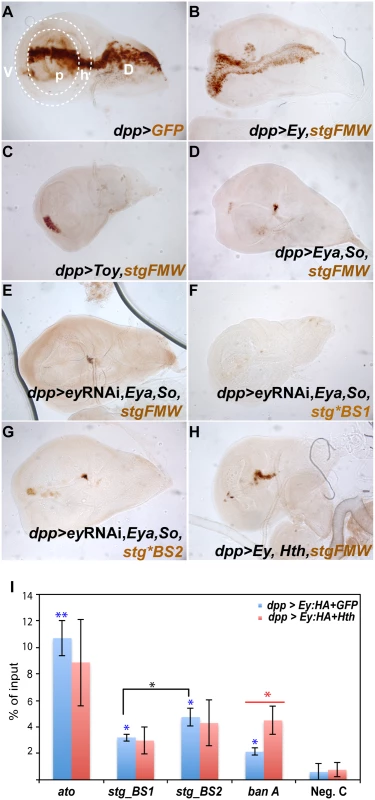
(A) L3 wing imaginal disc expressing GFP (brown) in the dpp-Gal4 domain, along the anterior-posterior axis. The wing pouch (p) and hinge (h) domains are delimited by the dashed white line. P: pouch, H: hinge, D: dorsal, V: ventral. (B- D) Expression of GFP (brown) driven by stg-FMW upon ectopic expression of Ey (B), Toy (C) and Eya/So (D) in the dpp-Gal4 domain. (B) Ey is sufficient to promote enhancer activity throughout the Dpp expression domain. (C) Toy is only sufficient to activate stg-FMW expression in the ventral hinge region. (D) Ectopic expression of the Eya/So complex can induce enhancer activity in the ventral and dorsal hinge cells of the DPP domain. (E) Activity of stg-FMW in discs ectopically expressing Eya/So and ey RNAi. Downregulation of Ey, does not affect the ability of Eya/SO to activate the enhancer. (F) Upon mutation of BS1 Eya/So are no longer able to activate stg-FMW enhancer. (G) Mutation of BS2 does not affect the ability of Eya/So to activate the stg-FMW enhancer even upon downregulation of Ey. (H) Ectopic expression of Hth and Ey in the Dpp domain. The activity of stg-FMW is severely compromised when Ey and Hth are co-expressed. (I) ChIP-qPCR experiments show that Ey-HA binds to BS1 and BS2 regions in vivo (blue bars). Anti-HA antibodies pulled down stg-BS2 more efficiently than stg-BS1. Sequences from ato-3’ were used as positive control. In the presence of Hth (red bars) the binding affinity of Ey to stg-BS1, stg-BS2, and ato-3’ is reduced (red bars). Each column shows the averages and standard error of the mean for three independent IPs and real-time PCRs. C: negative control region. Student’s t-test was used for statistical analysis. Blue asterisk: Ey-ChIP recovered chromatin from each region, in dpp>ey wing discs, compared to the negative control sample. Black asterisk: Comparison between stg-BS1 and stg-BS2 regions after Ey ChIP in dpp>ey wing discs. Red asterisk: Ey-ChIP recovered chromatin from dpp>ey compared to dpp>ey, hth wing discs. * p ≤ 0,05; ** p ≤ 0,005; Discussion
Selector genes lie atop organ-specific gene regulatory networks (GRNs), but it is still unclear what is the depth of their connectivity—i.e. whether selectors regulate a first layer of transcription factors that then relay their information, through consecutive layers down onto specific effector genes (those that determine the actual properties of the cells), or if they regulate the expression at all levels of those GRNs, connecting both to transcription factors and effector genes. This control, in any case, is established by their binding to specific CREs and still, in most organogenetic processes, our knowledge of the molecular logic used by selector genes in GRNs to control gene expression is fragmentary.
The Drosophila RD gene network is a good example of this. Despite the vast knowledge of its main components and their contribution to eye development, there is not much evidence about the molecular mechanisms that underlie their function. In particular, how interactions among the different RD gene network components take place and contribute to retina development, by acting not only on other components of the network (all transcription factors or nuclear co-factors) but also on effector genes. In this study we have addressed this question by investigating the mechanisms that regulate stg transcription in the developing eye.
stg codes for the universal phosphatase that triggers the G2-M transition [60]. Upregulation of stg expression during L3 is essential for the synchronous exit from mitosis of retinal progenitors, while simultaneously ensures their amplification at the FMW in order to produce a sufficient number of retinal precursors. It therefore works as an effector gene during the progenitor-precursor cell state transition.
We identified the eye-specific stg CRE and showed that it contains two conserved Pax6 binding sites. Drosophila has two Pax6 paralogues, toy and ey [16]. The expression of toy starts during early embryogenesis and is required for the activation of ey transcription in the eye primordium during late embryogenesis. During larval development, both toy and ey are coexpressed in the undifferentiated cells of the developing eye primordium [16]. However, while loss of ey function during larval stages results in smaller or absent eyes [61,62], no function in the eye had been attributed to the larval expression of toy. Here we show that both ey and toy act as positive regulators of the stg-FMW CRE, and that in the absence of ey, toy suffices to maintain stg-FMW CRE activity. However, ectopic experiments in the wing show that their activating capacity differs, with Ey proving to be a more efficient activator of stg CRE than Toy. This is consistent with a less powerful eye-inducing ability of Toy compared to Ey [16,56,57]. The discrepancy between the functional equivalence of Ey and Toy in the eye and their different eye-inducing ability in ectopic assays could be explained if Ey expression could facilitate the accessibility of Toy to (at least some) Ey targets. This would happen in the eye (where toy activates ey very early in the development of the eye primordium) but not in the wing, where none of the two Pax6 genes are normally expressed.
Our work shows that Ey is able to bind both BS1 and BS2, but shows higher affinity towards BS2 in vivo (Fig. 5I), suggesting that this site might play a key role in the enhancer activity. In agreement, we found that mutation of BS2, although not affecting stg-FMW pattern, causes a significant reduction in its expression levels (S6 Fig.). On the other hand, the transcriptional complex Eya:So is able to bind to BS1 and BS2 with similar affinities, but genetic analysis suggests that interaction with BS1 is critical for stg-FMW activity (Fig. 5E-G). However, while this analysis derives from wing disc assays, the mechanism of action in the eye disc might be more complex. While in the wing disc Ey shows a superior stg-FMW induction, in the eye removing Eya:So results in loss of enhancer’s activity, despite the fact that Ey and Toy expression remain. This suggests a model in which Toy/Ey and Eya:So cooperate to activate stg-FMW enhancer. A similar cooperation between Ey and So has been recently described for the activation of ato CREs [24,25], which are also active anterior to the MF with a pattern similar to that of stg-FMW [24–26].
The picture that emerges is that of a feed-forward loop, in which Pax6 genes activate Eya and So expression and then Pax6 and Eya:So control stg-FMW through direct binding. The engagement of Ey/Pax6 and Eya:So in a positive feed-forward loop has also been reported for the activation of dac, even though in this case two separate enhancer elements are involved [33]. Therefore, a similar gene regulatory motif involving Ey and Eya:So operates to control the expression of transcription factors and stg, this latter an effector gene. This may be a general feature of the gene networks where Pax6 proteins participate. For example, during the development of the vertebrate eye lens, Pax6 and c-Maf are similarly engaged in a positive feed-forward loop to activate the expression of crystallin genes [63]. This suggests that synergistic interactions among transcription factors within the same GRN determine the specificity of their recruitment to cell type-specific CREs.
A key step towards the activation of stg-FMW is the repression of Hth, which is mediated by Dpp and Hh [19]. Hth interferes with the coherent feed-forward loop formed by Ey and Eya:So (Fig. 6) at two points: First, Hth moderately hampers Ey binding to stg-CRE, something that could contribute to the temporal shifts that this enhancer suffers upon manipulating hth function (this work). This could happen through a direct Hth-Ey physical interaction [17]. And second, Hth also acts as a transcriptional repressor of Eya [17]. The resulting GRN allows integration of extracellular signals with tissue specification resulting in a short pulse of stg transcription as soon as MF-produced Dpp represses Hth. This pulse is thus made coincidental with the transition from progenitors into precursors (Fig. 6). The need of both Ey and Eya/So inputs for the enhancer’s activation acts as a molecular coincidence detector that ensures that the enhancer will only be active when the regulatory state of the cell is “correct”, avoiding spurious stg activation.
Fig. 6. The logic of the eye GRN ensures the pulse activation of the stg-FMW CRE in a narrow stripe. 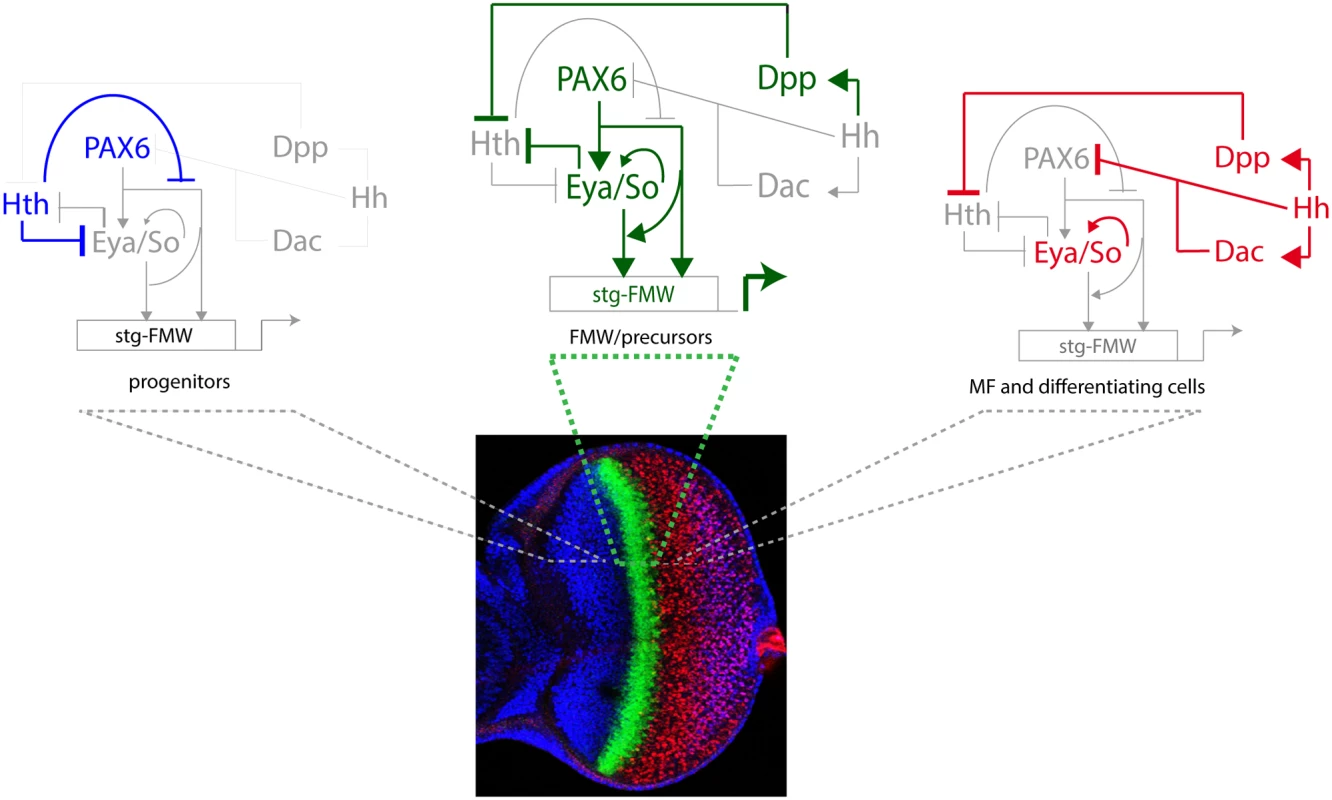
Activation requires the contribution of Ey, Toy (collectively referred to as Pax6 in the figure) and the Eya/So complex. In progenitors, Hth expression prevents the activation of stg-FMW by repressing Eya/So expression [17] and by reducing the binding and transcriptional activity of Ey (this work). Dpp, acting at long range from its site of production at the MF, represses Hth, thereby alleviating its double repression and allowing the sharp activation of stg coincidentally with the upregulation of Eya and So, which engage in an autoregulatory loop [78] in precursor cells. This loop makes their expression independent of Ey. In the eye, both Ey and Eya/So are required for stg CRE activation. This is represented by an arrow from Ey to Eya/So that would permit this latter genes to act as transcriptional activators together if Ey is present. At the MF and posterior to it, shorter range repression of Ey by Hh [79] results in the turning off of stg-FMW. In addition, after the MF So and Eya are required to keep the enhancer off. A similar concept, the switch of So from an activator to a repressor role at and after the MF, has been recently reported [29] and is in agreement with work showing that So plays essential roles during eye development acting as either transcriptional activator or transcriptional repressor [80]. Our results also point to a role for hth regulation as a precision mechanism, acting to guarantee a sudden, rather than gradual, activation of stg. It is through hth regulation that the system integrates the extracellular cues with the activity of the selector genes. This mechanism ensures coupling of growth with tissue specification. That hth and its vertebrate homologues may play a similar role in Ey/Pax6-regulated processes than the one we have described for stg CRE is a tantalizing hypothesis that needs to be investigated. Interestingly, loss of function of Hth does not suffice for enhancer activation in all cells of the anterior domain (Fig. 4). This seems to indicate that additional factors or signaling inputs contribute to stg-FMW activation. Dpp and Hh signaling are the obvious candidates. However, ectopic activation of either pathway does not change stg-FMW activity in progenitor cells (S8 Fig.) Altogether our data suggests the existence of still unknown anterior factors/signaling inputs that contribute to the regulation of stg-FMW expression onset.
The role of Pax6 genes in cancer development appears to be linked with their function during organ development. They act as oncogenes in organs where their expression correlates with the maintenance of the progenitor state, as is the case of the retina and pancreas [reviewed in 2]. In both organs, the maintenance of Pax6 expression during adult stages associates with a failure to undergo differentiation and to tumor development. In contrast, cdc25 is commonly up-regulated in tumors, as expected from a mitotic gene, but this up-regulation is not tumor type-specific [60]. Our results raise the possibility that Pax6 genes may regulate cell cycle genes in collaboration with Eya/Six proteins also during vertebrate organogenesis, something that might be linked with their oncogenic potential.
Materials and Methods
Genotypes and genetic manipulations
The following fly stocks were used: w1118, stghwy [22], w; FRT82BhthP2/TM6B [64], w; dac3 FRT40A/ CyO [65], w; eyaE8 FRT40 [66,67], UAS-Toy [16], UAS-eya, UAS-so [24], UAS-so, UAS-dac (kindly provided by F. Pignoni), UAS-soRNAi (VDRC 8950); UAS-eyaRNAi (VDRC 43911); UAS-hthRNAi (VDRC 12764); UAS-eyRNAi (VDRC 42845) and UAS-toyRNAi (VDRC 15919), ey2 (Bloomington Stock Center) [61]. Standard genetic techniques were used to introduce stg-FMW reporters in the different genetic backgrounds. All crosses were kept at 25°C unless otherwise stated. Cells mutant for hthP2 were recovered using the Minute technique [55]. The fly strain yw,hsFLP; FRT82BhsCD2, y+M/TM2 was used. Mutant tissue was identified by the absence of CD2 staining. Clones were induced between 24 and 48 h or 48 and 72 h after egg laying (AEL) by a 45′ heat-shock (hs) at 37°C. The Flip-Out method [68] was used to induce gain of function clones. The line yw,hsFLP, act>hsCD2>Gal4 was used. Clones were generated at 36–60 h AEL, by a 20′ hs at 35.5°C. Flies were kept at 25°C, except when UAS-RNAi lines were used, in which case they were transferred to 29°C after hs. Dpp-Gal4/TM6B (FBti0002123) and Dpp-Gal4,UAS-GFP/ MKRS (kindly provided by M. Dominguez) were used to ectopically express RD genes in the wing imaginal disc.
Reporter assays and binding site identification
The FlyC31 system was used to generate all transgenic lines used in this study. Transgenes were inserted in either 2L (22A) or 3R(86FA) attP sites [69]. Insertions on either landing site yielded similar results. Two reporter vectors were used for assaying enhancer activity: pRVV54 that uses nuclear lacZ as reporter [70], and pBPUwdGFP which uses destabilized GFP as reporter [71]. stg-FMW was cloned into pBPGUw [72] vector to generate stgFMW-Gal4 line. Overlapping DNA fragments, covering >60 Kb of stg locus were amplified by PCR and introduced into either pBPUwdGFP or pRVV54 using the Gateway System. Mutant versions of stg-FMW were cloned into pBPUwdGFP and inserted in 2L (22A) and 3R(86A) sites, as wild-type versions of the enhancer. The megaprimer method was used to generate mutations on putative Ey and Hth binding sites [73]. Primers delimiting stg-FMW enhancer sequence and used for enhancer mutagenesis are listed in S1 Table.
Immunohistochemistry
Imaginal discs were dissected and fixed according to standard protocols. Primary antibodies used were guinea-pig anti-Hth [74], rabbit anti-PH3 (Sigma), rabbit anti β-galactosidase (Cappel), mouse anti β-galactosidase (Sigma), mouse anti-CD2 (Serotec), rabbit anti-GFP (Molecular Probes), mouse anti-Ey (Clements et al., 2008) and rabbit anti-cyclin B [75]. Mouse anti-Eya, rat anti-ELAV (7E8A10), and mouse anti-cyclin B were from Developmental Studies Hybridoma Bank (Iowa University). Fluorescently labeled secondary antibodies were from Molecular Probes. Anti-mouse-HRP (Sigma) was used for immunoperoxidase staining. Digoxigenin labelled stg RNA probe was produced from cDNA clone LD47579 (BDGP). ImageJ was used to quantify pixel intensities (http://imagej.nih.gov/ij/).
Mapping of stghwy mutation
A PCR based approach was used to map and characterize, at the molecular level, the nature of stghwy allele. Several primer combinations spanning the genomic region Chr3R: 25.081.410 to Chr3R: 25.141.369 (Drosophila Genome Release 5) were used to amplify fragments of approximately 5 Kb of DNA from control (w1118) and stghwy flies. An insertion was detected between genomic coordinates Chr3R: 25114731 and ChR3R: 25115810. Primers flanking and within this region were employed to amplify and sequence stghwy DNA.
ChIP q-PCR
Wing imaginal discs from wandering L3 larvae of the following genotypes, dpp>Ey-HA, GFP; dpp> Ey-HA, Hth-GFP and dpp> Eya:HA, GFP were used for Chip-qPCR analysis. Chromatin was prepared essentially as described in Estella et al. [76]. 30 μg of soluble chromatin, with a size average of 200 bp, was incubated with 3 μg of rabbit anti-HA antibody (AbCam). Ey:HA and Eya:HA bound chromatin complexes were pulled down with protein G magnetic beads (Invitrogen) according to Sandmann et al. [77]. Chromatin was eluted with 100 mM NaHCO3. To reverse crosslinking, samples were incubated overnight at 65°C after the addition of 160 mM NaCl. DNA was purified via phenol-chloroform extraction and ethanol precipitation. The PCRs were performed on 1 : 50 dilutions of the ChIP and input samples. Primers were designed to specifically amplify regions BS1 and BS2, which are 100 bps apart. Primers on ato-3’ enhancer were used as positive control [24]. Primers used are described in S1 Table.
Supporting Information
Zdroje
1. Mann RS, Carroll SB (2002) Molecular mechanisms of selector gene function and evolution. Curr Opin Genet Dev 12 : 592–600. 12200165
2. Li CG, Eccles MR (2012) PAX Genes in Cancer; Friends or Foes? Front Genet 3 : 6. doi: 10.3389/fgene.2012.00006 22303411
3. Blake JA, Ziman MR (2014) Pax genes: regulators of lineage specification and progenitor cell maintenance. Development 141 : 737–751. doi: 10.1242/dev.091785 24496612
4. Dyer MA, Cepko CL (2001) Regulating proliferation during retinal development. Nat Rev Neurosci 2 : 333–342. 11331917
5. Bassett EA, Wallace VA (2012) Cell fate determination in the vertebrate retina. Trends Neurosci 35 : 565–573. doi: 10.1016/j.tins.2012.05.004 22704732
6. Sinn R, Wittbrodt J (2013) An eye on eye development. Mech Dev 130 : 347–358. doi: 10.1016/j.mod.2013.05.001 23684892
7. Glaser T, Jepeal L, Edwards JG, Young SR, Favor J, et al. (1994) PAX6 gene dosage effect in a family with congenital cataracts, aniridia, anophthalmia and central nervous system defects. Nat Genet 7 : 463–471. 7951315
8. Sisodiya SM, Free SL, Williamson KA, Mitchell TN, Willis C, et al. (2001) PAX6 haploinsufficiency causes cerebral malformation and olfactory dysfunction in humans. Nat Genet 28 : 214–216. 11431688
9. Bai SW, Li B, Zhang H, Jonas JB, Zhao BW, et al. (2011) Pax6 regulates proliferation and apoptosis of human retinoblastoma cells. Invest Ophthalmol Vis Sci 52 : 4560–4570. doi: 10.1167/iovs.10-5487 21169528
10. Li L, Li B, Zhang H, Bai S, Wang Y, et al. (2011) Lentiviral vector-mediated PAX6 overexpression promotes growth and inhibits apoptosis of human retinoblastoma cells. Invest Ophthalmol Vis Sci 52 : 8393–8400. doi: 10.1167/iovs.11-8139 21948554
11. Wang J, Wang X, Wu G, Hou D, Hu Q (2013) MiR-365b-3p, down-regulated in retinoblastoma, regulates cell cycle progression and apoptosis of human retinoblastoma cells by targeting PAX6. FEBS Lett 587 : 1779–1786. doi: 10.1016/j.febslet.2013.04.029 23660406
12. Kumar JP (2011) My what big eyes you have: how the Drosophila retina grows. Dev Neurobiol 71 : 1133–1152. doi: 10.1002/dneu.20921 21604387
13. Treisman JE (2013) Retinal differentiation in Drosophila. Wiley Interdiscip Rev Dev Biol 2 : 545–557. doi: 10.1002/wdev.100 24014422
14. Amore G, Casares F (2010) Size matters: the contribution of cell proliferation to the progression of the specification Drosophila eye gene regulatory network. Dev Biol 344 : 569–577. doi: 10.1016/j.ydbio.2010.06.015 20599903
15. Cheyette BN, Green PJ, Martin K, Garren H, Hartenstein V, et al. (1994) The Drosophila sine oculis locus encodes a homeodomain-containing protein required for the development of the entire visual system. Neuron 12 : 977–996. 7910468
16. Czerny T, Halder G, Kloter U, Souabni A, Gehring WJ, et al. (1999) twin of eyeless, a second Pax-6 gene of Drosophila, acts upstream of eyeless in the control of eye development. Mol Cell 3 : 297–307. 10198632
17. Bessa J, Gebelein B, Pichaud F, Casares F, Mann RS (2002) Combinatorial control of Drosophila eye development by eyeless, homothorax, and teashirt. Genes Dev 16 : 2415–2427. 12231630
18. Peng HW, Slattery M, Mann RS (2009) Transcription factor choice in the Hippo signaling pathway: homothorax and yorkie regulation of the microRNA bantam in the progenitor domain of the Drosophila eye imaginal disc. Genes Dev 23 : 2307–2319. doi: 10.1101/gad.1820009 19762509
19. Lopes CS, Casares F (2010) hth maintains the pool of eye progenitors and its downregulation by Dpp and Hh couples retinal fate acquisition with cell cycle exit. Dev Biol 339 : 78–88. doi: 10.1016/j.ydbio.2009.12.020 20036228
20. Thomas BJ, Gunning DA, Cho J, Zipursky L (1994) Cell cycle progression in the developing Drosophila eye: roughex encodes a novel protein required for the establishment of G1. Cell 77 : 1003–1014. 8020091
21. Lehman DA, Patterson B, Johnston LA, Balzer T, Britton JS, et al. (1999) Cis-regulatory elements of the mitotic regulator, string/Cdc25. Development 126 : 1793–1803. 10101114
22. Mozer BA, Easwarachandran K (1999) Pattern formation in the absence of cell proliferation: tissue-specific regulation of cell cycle progression by string (stg) during Drosophila eye development. Dev Biol 213 : 54–69. 10452846
23. Jarman AP, Grell EH, Ackerman L, Jan LY, Jan YN (1994) Atonal is the proneural gene for Drosophila photoreceptors. Nature 369 : 398–400. 8196767
24. Zhang T, Ranade S, Cai CQ, Clouser C, Pignoni F (2006) Direct control of neurogenesis by selector factors in the fly eye: regulation of atonal by Ey and So. Development 133 : 4881–4889. 17108002
25. Zhou Q, Zhang T, Jemc JC, Chen Y, Chen R, et al. (2014) Onset of atonal expression in Drosophila retinal progenitors involves redundant and synergistic contributions of Ey/Pax6 and So binding sites within two distant enhancers. Dev Biol 386 : 152–164. doi: 10.1016/j.ydbio.2013.11.012 24247006
26. Tanaka-Matakatsu M, Du W (2008) Direct control of the proneural gene atonal by retinal determination factors during Drosophila eye development. Dev Biol 313 : 787–801. 18083159
27. Peter IS, Davidson EH (2011) Evolution of gene regulatory networks controlling body plan development. Cell 144 : 970–985. doi: 10.1016/j.cell.2011.02.017 21414487
28. Hauck B, Gehring WJ, Walldorf U (1999) Functional analysis of an eye specific enhancer of the eyeless gene in Drosophila. Proc Natl Acad Sci U S A 96 : 564–569. 9892673
29. Atkins M, Jiang Y, Sansores-Garcia L, Jusiak B, Halder G, et al. (2013) Dynamic rewiring of the Drosophila retinal determination network switches its function from selector to differentiation. PLoS Genet 9: e1003731. doi: 10.1371/journal.pgen.1003731 24009524
30. Niimi T, Seimiya M, Kloter U, Flister S, Gehring WJ (1999) Direct regulatory interaction of the eyeless protein with an eye-specific enhancer in the sine oculis gene during eye induction in Drosophila. Development 126 : 2253–2260. 10207149
31. Punzo C, Seimiya M, Flister S, Gehring WJ, Plaza S (2002) Differential interactions of eyeless and twin of eyeless with the sine oculis enhancer. Development 129 : 625–634. 11830564
32. Bui QT, Zimmerman JE, Liu H, Gray-Board GL, Bonini NM (2000) Functional analysis of an eye enhancer of the Drosophila eyes absent gene: differential regulation by eye specification genes. Dev Biol 221 : 355–364. 10790331
33. Pappu KS, Ostrin EJ, Middlebrooks BW, Sili BT, Chen R, et al. (2005) Dual regulation and redundant function of two eye-specific enhancers of the Drosophila retinal determination gene dachshund. Development 132 : 2895–2905. 15930118
34. Ostrin EJ, Li Y, Hoffman K, Liu J, Wang K, et al. (2006) Genome-wide identification of direct targets of the Drosophila retinal determination protein Eyeless. Genome Res 16 : 466–476. 16533912
35. Sun Y, Jan LY, Jan YN (1998) Transcriptional regulation of atonal during development of the Drosophila peripheral nervous system. Development 125 : 3731–3740. 9716538
36. Escudero LM, Freeman M (2007) Mechanism of G1 arrest in the Drosophila eye imaginal disc. BMC Dev Biol 7 : 13. 17335573
37. Narlikar L, Ovcharenko I (2009) Identifying regulatory elements in eukaryotic genomes. Brief Funct Genomic Proteomic 8 : 215–230. doi: 10.1093/bfgp/elp014 19498043
38. Bushey AM, Ramos E, Corces VG (2009) Three subclasses of a Drosophila insulator show distinct and cell type-specific genomic distributions. Genes Dev 23 : 1338–1350. doi: 10.1101/gad.1798209 19443682
39. Gurudatta BV, Corces VG (2009) Chromatin insulators: lessons from the fly. Brief Funct Genomic Proteomic 8 : 276–282. doi: 10.1093/bfgp/elp032 19752045
40. Negre N, Brown CD, Ma L, Bristow CA, Miller SW, et al. (2011) A cis-regulatory map of the Drosophila genome. Nature 471 : 527–531. doi: 10.1038/nature09990 21430782
41. Wallace JA, Felsenfeld G (2007) We gather together: insulators and genome organization. Curr Opin Genet Dev 17 : 400–407. 17913488
42. Baonza A, Murawsky CM, Travers AA, Freeman M (2002) Pointed and Tramtrack69 establish an EGFR-dependent transcriptional switch to regulate mitosis. Nat Cell Biol 4 : 976–980. 12447387
43. Sandelin A, Alkema W, Engstrom P, Wasserman WW, Lenhard B (2004) JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res 32: D91–94. 14681366
44. Wingender E (2008) The TRANSFAC project as an example of framework technology that supports the analysis of genomic regulation. Brief Bioinform 9 : 326–332. doi: 10.1093/bib/bbn016 18436575
45. Bailey TL, Elkan C (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2 : 28–36. 7584402
46. Blanco E, Corominas M (2012) CBS: an open platform that integrates predictive methods and epigenetics information to characterize conserved regulatory features in multiple Drosophila genomes. BMC Genomics 13 : 688. doi: 10.1186/1471-2164-13-688 23228284
47. Jacobsson L, Kronhamn J, Rasmuson-Lestander A (2009) The Drosophila Pax6 paralogs have different functions in head development but can partially substitute for each other. Mol Genet Genomics 282 : 217–231. doi: 10.1007/s00438-009-0458-2 19484263
48. Pignoni F, Hu B, Zavitz KH, Xiao J, Garrity PA, et al. (1997) The eye-specification proteins So and Eya form a complex and regulate multiple steps in Drosophila eye development. Cell 91 : 881–891. 9428512
49. Kumar JP (2009) The molecular circuitry governing retinal determination. Biochim Biophys Acta 1789 : 306–314. doi: 10.1016/j.bbagrm.2008.10.001 19013263
50. Jemc J, Rebay I (2007) Identification of transcriptional targets of the dual-function transcription factor/phosphatase eyes absent. Dev Biol 310 : 416–429. 17714699
51. Lee T, Luo L (1999) Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron 22 : 451–461. 10197526
52. Chen R, Amoui M, Zhang Z, Mardon G (1997) Dachshund and eyes absent proteins form a complex and function synergistically to induce ectopic eye development in Drosophila. Cell 91 : 893–903. 9428513
53. Rieckhof GE, Casares F, Ryoo HD, Abu-Shaar M, Mann RS (1997) Nuclear translocation of extradenticle requires homothorax, which encodes an extradenticle-related homeodomain protein. Cell 91 : 171–183. 9346235
54. Pichaud F, Casares F (2000) homothorax and iroquois-C genes are required for the establishment of territories within the developing eye disc. Mech Dev 96 : 15–25. 10940621
55. Morata G, Ripoll P (1975) Minutes: mutants of drosophila autonomously affecting cell division rate. Dev Biol 42 : 211–221. 1116643
56. Halder G, Callaerts P, Gehring WJ (1995) Induction of ectopic eyes by targeted expression of the eyeless gene in Drosophila. Science 267 : 1788–1792. 7892602
57. Salzer CL, Kumar JP (2010) Identification of retinal transformation hot spots in developing Drosophila epithelia. PLoS One 5: e8510. doi: 10.1371/journal.pone.0008510 20062803
58. Bonini NM, Bui QT, Gray-Board GL, Warrick JM (1997) The Drosophila eyes absent gene directs ectopic eye formation in a pathway conserved between flies and vertebrates. Development 124 : 4819–4826. 9428418
59. Tadjuidje E, Hegde RS (2013) The Eyes Absent proteins in development and disease. Cell Mol Life Sci 70 : 1897–1913. doi: 10.1007/s00018-012-1144-9 22971774
60. Boutros R, Lobjois V, Ducommun B (2007) CDC25 phosphatases in cancer cells: key players? Good targets? Nat Rev Cancer 7 : 495–507. 17568790
61. Quiring R, Walldorf U, Kloter U, Gehring WJ (1994) Homology of the eyeless gene of Drosophila to the Small eye gene in mice and Aniridia in humans. Science 265 : 785–789. 7914031
62. Clements J, Hens K, Merugu S, Dichtl B, de Couet HG, et al. (2009) Mutational analysis of the eyeless gene and phenotypic rescue reveal that an intact Eyeless protein is necessary for normal eye and brain development in Drosophila. Dev Biol 334 : 503–512. doi: 10.1016/j.ydbio.2009.08.003 19666017
63. Xie Q, Cvekl A (2011) The orchestration of mammalian tissue morphogenesis through a series of coherent feed-forward loops. J Biol Chem 286 : 43259–43271. doi: 10.1074/jbc.M111.264580 21998302
64. Kurant E, Pai CY, Sharf R, Halachmi N, Sun YH, et al. (1998) Dorsotonals/homothorax, the Drosophila homologue of meis1, interacts with extradenticle in patterning of the embryonic PNS. Development 125 : 1037–1048. 9463350
65. Mardon G, Solomon NM, Rubin GM (1994) dachshund encodes a nuclear protein required for normal eye and leg development in Drosophila. Development 120 : 3473–3486. 7821215
66. Bonini NM, Leiserson WM, Benzer S (1993) The eyes absent gene: genetic control of cell survival and differentiation in the developing Drosophila eye. Cell 72 : 379–395. 8431945
67. Hazelett DJ, Bourouis M, Walldorf U, Treisman JE (1998) decapentaplegic and wingless are regulated by eyes absent and eyegone and interact to direct the pattern of retinal differentiation in the eye disc. Development 125 : 3741–3751. 9716539
68. Struhl G, Basler K (1993) Organizing activity of wingless protein in Drosophila. Cell 72 : 527–540. 8440019
69. Bischof J, Maeda RK, Hediger M, Karch F, Basler K (2007) An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc Natl Acad Sci U S A 104 : 3312–3317. 17360644
70. Slattery M, Voutev R, Ma L, Negre N, White KP, et al. (2013) Divergent transcriptional regulatory logic at the intersection of tissue growth and developmental patterning. PLoS Genet 9: e1003753. doi: 10.1371/journal.pgen.1003753 24039600
71. Royo JL, Maeso I, Irimia M, Gao F, Peter IS, et al. (2011) Transphyletic conservation of developmental regulatory state in animal evolution. Proc Natl Acad Sci U S A 108 : 14186–14191. doi: 10.1073/pnas.1109037108 21844364
72. Pfeiffer BD, Jenett A, Hammonds AS, Ngo TT, Misra S, et al. (2008) Tools for neuroanatomy and neurogenetics in Drosophila. Proc Natl Acad Sci U S A 105 : 9715–9720. doi: 10.1073/pnas.0803697105 18621688
73. Lai R, Bekessy A, Chen CC, Walsh T, Barnard R (2003) Megaprimer mutagenesis using very long primers. Biotechniques 34 : 52–54, 56. 12545538
74. Casares F, Mann RS (1998) Control of antennal versus leg development in Drosophila. Nature 392 : 723–726. 9565034
75. Jacobs H, Richter D, Venkatesh T, Lehner C (2002) Completion of mitosis requires neither fzr/rap nor fzr2, a male germline-specific Drosophila Cdh1 homolog. Curr Biol 12 : 1435–1441. 12194827
76. Estella C, McKay DJ, Mann RS (2008) Molecular integration of wingless, decapentaplegic, and autoregulatory inputs into Distalless during Drosophila leg development. Dev Cell 14 : 86–96. doi: 10.1016/j.devcel.2007.11.002 18194655
77. Sandmann T, Jakobsen JS, Furlong EE (2006) ChIP-on-chip protocol for genome-wide analysis of transcription factor binding in Drosophila melanogaster embryos. Nat Protoc 1 : 2839–2855. 17406543
78. Pauli T, Seimiya M, Blanco J, Gehring WJ (2005) Identification of functional sine oculis motifs in the autoregulatory element of its own gene, in the eyeless enhancer and in the signalling gene hedgehog. Development 132 : 2771–2782. 15901665
79. Firth LC, Baker NE (2009) Retinal determination genes as targets and possible effectors of extracellular signals. Dev Biol 327 : 366–375. doi: 10.1016/j.ydbio.2008.12.021 19135045
80. Anderson AM, Weasner BM, Weasner BP, Kumar JP (2012) Dual transcriptional activities of SIX proteins define their roles in normal and ectopic eye development. Development 139 : 991–1000. doi: 10.1242/dev.077255 22318629
Štítky
Genetika Reprodukční medicína
Článek 2014 Reviewer Thank YouČlánek Closing the Gap between Knowledge and Clinical Application: Challenges for Genomic TranslationČlánek Discovery of CTCF-Sensitive Cis-Spliced Fusion RNAs between Adjacent Genes in Human Prostate CellsČlánek K-homology Nuclear Ribonucleoproteins Regulate Floral Organ Identity and Determinacy in ArabidopsisČlánek A Nitric Oxide Regulated Small RNA Controls Expression of Genes Involved in Redox Homeostasis inČlánek Contribution of the Two Genes Encoding Histone Variant H3.3 to Viability and Fertility in MiceČlánek The Genetic Architecture of the Genome-Wide Transcriptional Response to ER Stress in the Mouse
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 2
-
Všechny články tohoto čísla
- 2014 Reviewer Thank You
- Systematic Cell-Based Phenotyping of Missense Alleles Empowers Rare Variant Association Studies: A Case for and Myocardial Infarction
- African Glucose-6-Phosphate Dehydrogenase Alleles Associated with Protection from Severe Malaria in Heterozygous Females in Tanzania
- Genomics of Divergence along a Continuum of Parapatric Population Differentiation
- microRNAs Regulate Cell-to-Cell Variability of Endogenous Target Gene Expression in Developing Mouse Thymocytes
- A Rolling Circle Replication Mechanism Produces Multimeric Lariats of Mitochondrial DNA in
- Closing the Gap between Knowledge and Clinical Application: Challenges for Genomic Translation
- Partially Redundant Enhancers Cooperatively Maintain Mammalian Expression Above a Critical Functional Threshold
- Discovery of Transcription Factors and Regulatory Regions Driving Tumor Development by ATAC-seq and FAIRE-seq Open Chromatin Profiling
- Mutations in Result in Ocular Coloboma, Microcornea and Cataracts
- A Genome-Wide Hybrid Incompatibility Landscape between and
- Recurrent Evolution of Melanism in South American Felids
- Discovery of CTCF-Sensitive Cis-Spliced Fusion RNAs between Adjacent Genes in Human Prostate Cells
- Tissue Expression Pattern of PMK-2 p38 MAPK Is Established by the miR-58 Family in
- Essential Role for Endogenous siRNAs during Meiosis in Mouse Oocytes
- Matrix Metalloproteinase 2 Is Required for Ovulation and Corpus Luteum Formation in
- Evolutionary Signatures amongst Disease Genes Permit Novel Methods for Gene Prioritization and Construction of Informative Gene-Based Networks
- RR-1 Cuticular Protein TcCPR4 Is Required for Formation of Pore Canals in Rigid Cuticle
- GC-Content Evolution in Bacterial Genomes: The Biased Gene Conversion Hypothesis Expands
- Proteotoxic Stress Induces Phosphorylation of p62/SQSTM1 by ULK1 to Regulate Selective Autophagic Clearance of Protein Aggregates
- K-homology Nuclear Ribonucleoproteins Regulate Floral Organ Identity and Determinacy in Arabidopsis
- A Nitric Oxide Regulated Small RNA Controls Expression of Genes Involved in Redox Homeostasis in
- HYPER RECOMBINATION1 of the THO/TREX Complex Plays a Role in Controlling Transcription of the Gene in Arabidopsis
- Mitochondrial and Cytoplasmic ROS Have Opposing Effects on Lifespan
- Structured Observations Reveal Slow HIV-1 CTL Escape
- An Integrative Multi-scale Analysis of the Dynamic DNA Methylation Landscape in Aging
- Combining Natural Sequence Variation with High Throughput Mutational Data to Reveal Protein Interaction Sites
- Transhydrogenase Promotes the Robustness and Evolvability of Deficient in NADPH Production
- Regulators of Autophagosome Formation in Muscles
- Genomic Selection and Association Mapping in Rice (): Effect of Trait Genetic Architecture, Training Population Composition, Marker Number and Statistical Model on Accuracy of Rice Genomic Selection in Elite, Tropical Rice Breeding Lines
- Eye Selector Logic for a Coordinated Cell Cycle Exit
- Inflammation-Induced Cell Proliferation Potentiates DNA Damage-Induced Mutations
- The DNA Polymerase δ Has a Role in the Deposition of Transcriptionally Active Epigenetic Marks, Development and Flowering
- Contribution of the Two Genes Encoding Histone Variant H3.3 to Viability and Fertility in Mice
- Membrane Recognition and Dynamics of the RNA Degradosome
- P-TEFb, the Super Elongation Complex and Mediator Regulate a Subset of Non-paused Genes during Early Embryo Development
- is a Long Non-coding RNA in JNK Signaling in Epithelial Shape Changes during Drosophila Dorsal Closure
- A Pleiotropy-Informed Bayesian False Discovery Rate Adapted to a Shared Control Design Finds New Disease Associations From GWAS Summary Statistics
- Genome-wide Association Study Identifies Shared Risk Loci Common to Two Malignancies in Golden Retrievers
- and Hyperdrive Mechanisms (in Mouse Meiosis)
- Elevated In Vivo Levels of a Single Transcription Factor Directly Convert Satellite Glia into Oligodendrocyte-like Cells
- Systemic Delivery of MicroRNA-101 Potently Inhibits Hepatocellular Carcinoma by Repressing Multiple Targets
- Pooled Sequencing of 531 Genes in Inflammatory Bowel Disease Identifies an Associated Rare Variant in and Implicates Other Immune Related Genes
- Abscission Is Regulated by the ESCRT-III Protein Shrub in Germline Stem Cells
- Temperature Stress Mediates Decanalization and Dominance of Gene Expression in
- Transcriptome Wide Annotation of Eukaryotic RNase III Reactivity and Degradation Signals
- The Exosome Component Rrp6 Is Required for RNA Polymerase II Termination at Specific Targets of the Nrd1-Nab3 Pathway
- Sex-specific -regulatory Variation on the X Chromosome
- Regulation of Toll-like Receptor Signaling by the SF3a mRNA Splicing Complex
- Modeling of the Human Alveolar Rhabdomyosarcoma Chromosome Translocation in Mouse Myoblasts Using CRISPR-Cas9 Nuclease
- Asymmetry of the Budding Yeast Tem1 GTPase at Spindle Poles Is Required for Spindle Positioning But Not for Mitotic Exit
- TIM Binds Importin α1, and Acts as an Adapter to Transport PER to the Nucleus
- Antagonistic Roles for KNOX1 and KNOX2 Genes in Patterning the Land Plant Body Plan Following an Ancient Gene Duplication
- The Genetic Architecture of the Genome-Wide Transcriptional Response to ER Stress in the Mouse
- Fatty Acid Synthase Cooperates with Glyoxalase 1 to Protect against Sugar Toxicity
- Region-Specific Activation of mRNA Translation by Inhibition of Bruno-Mediated Repression
- An Essential Role of the Arginine Vasotocin System in Mate-Guarding Behaviors in Triadic Relationships of Medaka Fish ()
- Interaction between the tRNA-Binding and C-Terminal Domains of Yeast Gcn2 Regulates Kinase Activity In Vivo
- Hyper-Acetylation of Histone H3K56 Limits Break-Induced Replication by Inhibiting Extensive Repair Synthesis
- Prodomain Removal Enables Neto to Stabilize Glutamate Receptors at the Neuromuscular Junction
- Recent Selective Sweeps in North American Show Signatures of Soft Sweeps
- Identification and Functional Analysis of Healing Regulators in
- A Multi-Megabase Copy Number Gain Causes Maternal Transmission Ratio Distortion on Mouse Chromosome 2
- Drosophila Casein Kinase I Alpha Regulates Homolog Pairing and Genome Organization by Modulating Condensin II Subunit Cap-H2 Levels
- The Hippo Pathway Regulates Homeostatic Growth of Stem Cell Niche Precursors in the Ovary
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Genomic Selection and Association Mapping in Rice (): Effect of Trait Genetic Architecture, Training Population Composition, Marker Number and Statistical Model on Accuracy of Rice Genomic Selection in Elite, Tropical Rice Breeding Lines
- Discovery of Transcription Factors and Regulatory Regions Driving Tumor Development by ATAC-seq and FAIRE-seq Open Chromatin Profiling
- Evolutionary Signatures amongst Disease Genes Permit Novel Methods for Gene Prioritization and Construction of Informative Gene-Based Networks
- Proteotoxic Stress Induces Phosphorylation of p62/SQSTM1 by ULK1 to Regulate Selective Autophagic Clearance of Protein Aggregates
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



