-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaK-homology Nuclear Ribonucleoproteins Regulate Floral Organ Identity and Determinacy in Arabidopsis
Unlike animals, angiosperms (flowering plants) lack a germline that is set-aside early in embryo development. Contrariwise, reproductive success relies on the formation of flowers during adult life, which provide the germ cells and the means for fertilization. Therefore, timing of flowering and flower organ morphogenesis are critical developmental operations that must be finely regulated and coordinated to complete reproduction. Arabidopsis thaliana FLOWERING LOCUS WITH KH DOMAINS (FLK) and PEPPER (PEP) encode two KH-domain RNA-binding proteins phylogenetically related to human proteins characterized by their high developmental versatility. FLK and PEP modulate the mRNA expression of the MADS-box gene FLOWERING LOCUS C, key in flowering control. In this work we have found that FLK and PEP also play a pivotal role in flower organogenesis by post-transcriptionally regulating the MADS-box floral organ identity gene AGAMOUS (AG). Interestingly, FLK and PEP physically interact with proteins involved in AG pre-mRNA processing to secure correct AG function in the floral meristem and flower. Taken together, our results reveal the existence of a post-transcriptional regulatory activity controlling key master genes for floral timing and flower morphogenesis, which might be instrumental for coordinating both developmental phases.
Published in the journal: . PLoS Genet 11(2): e32767. doi:10.1371/journal.pgen.1004983
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004983Summary
Unlike animals, angiosperms (flowering plants) lack a germline that is set-aside early in embryo development. Contrariwise, reproductive success relies on the formation of flowers during adult life, which provide the germ cells and the means for fertilization. Therefore, timing of flowering and flower organ morphogenesis are critical developmental operations that must be finely regulated and coordinated to complete reproduction. Arabidopsis thaliana FLOWERING LOCUS WITH KH DOMAINS (FLK) and PEPPER (PEP) encode two KH-domain RNA-binding proteins phylogenetically related to human proteins characterized by their high developmental versatility. FLK and PEP modulate the mRNA expression of the MADS-box gene FLOWERING LOCUS C, key in flowering control. In this work we have found that FLK and PEP also play a pivotal role in flower organogenesis by post-transcriptionally regulating the MADS-box floral organ identity gene AGAMOUS (AG). Interestingly, FLK and PEP physically interact with proteins involved in AG pre-mRNA processing to secure correct AG function in the floral meristem and flower. Taken together, our results reveal the existence of a post-transcriptional regulatory activity controlling key master genes for floral timing and flower morphogenesis, which might be instrumental for coordinating both developmental phases.
Introduction
Development of multicellular organisms relies on exquisitely controlled transcriptional and post-transcriptional regulatory actions to govern gene expression and accurately respond to endogenous and environmental fluctuations. As exemplified in the reference plant Arabidopsis thaliana (Arabidopsis hereafter), reproductive success in angiosperms largely depends on two developmental events that initiate the reproductive phase: floral timing and flower morphogenesis. Upon flowering, the shoot apical meristem (SAM) transforms into an inflorescence meristem (IM) which will give rise to floral meristems (FMs) [1]. FM identity genes, such as LEAFY (LFY) [2] and APETALA1 (AP1) [3], are crucial in activating the floral homeotic genes that specify identity of concentric whorls of organs in the Arabidopsis flower [1]. According to the ABC(E) model [4–6], the class A genes AP1 and AP2 specify sepals and, together with the B function genes PISTILLATA (PI) and AP3, contribute to petal identity. Co-expression of B-genes and the C-function gene AGAMOUS (AG) confer male stamen identity, while AG alone specifies female carpels, defining the pistil or gynoecium situated in the innermost whorl. The model also establishes mutual antagonism between A and C activities and requirement of the E activity, represented by the redundant SEPALLATA function [4–9]. With the exception of AP2 (an AP2/EREBP) [10,11], all floral homeotic genes encode type II MADS-box transcription factors, a lineage comprising central regulators in most aspects of plant development [9,12,13].
In addition to floral organ identity, AG plays a crucial role in FM determinacy by repressing the homeobox stem-cell-identity gene WUSCHEL (WUS) [14,15]. WUS and LFY activate AG, which in turn, represses WUS both directly and through the activation of the transcriptional repressor KNUCKLES (KNU) [16], resulting in consumption of the stem cell niche [16–21]. Otherwise, continuing cell proliferation leads to an indeterminate pattern of alternating whorls of sepals and petals, as described in strong ag mutants [22].
Whereas transcriptional control of gene expression is key to development, it is nowadays widely accepted that post-transcriptional operations are crucial to secure proper gene regulation. For example, mounting evidence indicates that mRNA processing steps, such as splicing and polyadenylation, usually proceed co-transcriptionally in a tightly coordinated manner to ensure correct gene activity [23–25]. RNA-binding proteins from multifunctional ribonucleoprotein (RNP) complexes coat nascent transcripts to regulate different aspects of mRNA synthesis, affecting thus, the final levels of gene expression [26, 27].
It has been shown that, in addition to its transcriptional control, post-transcriptional regulation is essential to secure correct AG function during flower development, in particular AG intron 2 processing [28]. So far three Arabidopsis RNA-binding proteins (RNPs) were found to facilitate this process: HUA1, a nuclear CCCH-type zinc-finger protein [29], the RPR-domain (Regulation of nuclear pre-mRNA) protein HUA2 [30], and HUA ENHANCER 4 (HEN4), containing 5 K-homology (KH) domains and one of the few KH proteins functionally characterized in Arabidopsis [31,28]. Interestingly, hua1 hua2 hen4 triple mutants displayed stamen and carpel homeotic transformations, and loss of flower determinacy as a result of the reduced levels of mature AG mRNA. The fact that HUA1 binds to the AG pre-mRNA and physically associates with HEN4, suggests that both proteins belong to the same RNP regulatory complex [28].
Named after the human heterogeneous nuclear ribonucleoprotein K (hnRNP K) [32], the KH domain is an ancient RNA-binding module present in proteins whose disruption causes important developmental alterations in animals, including human syndromes as fragile-X [33,34], metastasis and cancer progression [35]. The hnRNP K is also representative of the remarkably versatile poly(C)-binding proteins (PCBP), characterized by a stereotypical triple-KH-domain configuration. PCBPs play roles in multiple developmental processes in animal systems, from erythropoiesis to neuronal differentiation [36–40]. The KH domain also provides a structural basis for protein-protein interactions, which most likely contributes to the multifunctionality of PCBPs [36,41].
In contrast, very little is known about plant PCBP-type hnRNPs and their relevance to plant development or morphogenesis is largely unexplored. So far, only two canonical PCBP-type hnRNP encoding genes, FLOWERING LOCUS WITH KH DOMAINS (FLK) [42,43] and PEPPER (PEP) [44], have been characterized in Arabidopsis to some extent. FLK promotes flowering in the autonomous pathway by negatively regulating the MADS-box floral repressor FLOWERING LOCUS C (FLC) [42,43,45]. PEP was originally described to interact with element(s) of the WUS pathway [44] and more recently we found that PEP is a positive regulator of FLC activity, hence antagonizing with FLK [46]. In line with this, the late flowering phenotype of flk plants (due to elevated levels of FLC) is rescued in the flk pep background [46]. However, in spite of the fact that PEP is expressed in FM and developing flowers, pep, flk or pep flk double mutants lack conspicuous floral defects, probably reflecting the compensation by overlapping activities [43,44,46].
In this work, we have functionally investigated the magnitude of PEP and FLK roles in flower patterning. Our genetic and molecular analyses place PEP as a positive regulator of the floral C-function by facilitating AG pre-mRNA processing and preventing premature polyadenylation in the large second intron. Here we also show that FLK also contributes to maintain the C-function. Furthermore, we provide evidence that PEP and FLK interact with the previously identified AG mRNA processing factors HUA1 and HEN4, strongly suggesting that all these proteins likely work together as components of a common post-transcriptional regulatory activity. Identifying PEP and FLK as new regulators of AG, broadens the scope of the developmental functions played by plant PCBPs, as they impinge upon the control of master regulatory genes, in this case AG, central during reproductive development.
Results
Genetic interactions with HEN4, HUA1 and HUA2 uncover the contribution of PEP to maintain the floral C-function
As mentioned above, PEP is expressed in FM and developing flowers, but pep flowers are largely normal. Thus, to test whether the role of PEP in floral patterning was masked by redundant gene activities, we combined the null pep-4 allele [44] (pep hereafter) with mutations in HEN4, HUA1 and HUA2, genes that encode post-transcriptional regulatory proteins [28].
HEN4 is a KH paralog relatively distant to PEP [31]. Unlike hen4-2 (hen4 hereafter) and pep single mutants (S1A Fig.) [28,44], ∼10% of hen4 pep flowers exhibited petaloid stamens (Fig. 1A, 1B, and S2B-S2D Fig.). Similarly, hua1-1 mutants (hua1 hereafter) appeared normal (S1B Fig.) [30], but hua1 pep double mutants displayed abundant petaloid transformations in the third whorl (40% of the flowers examined; Fig. 1C). We could not obtain hen4 hua1 pep triple homozygous mutants implying that PEP becomes essential in the hen4 hua1 background. This was noteworthy since hen4 hua1 double mutants flowers look wild-type [28]. Strikingly, introducing only one pep allele into hen4 hua1 plants (hen4 hua1 pep/+) led to conspicuous floral alterations including petaloid stamens in all flowers (Fig. 1D).
Fig. 1. PEP regulates flower reproductive organ identity and determinacy. 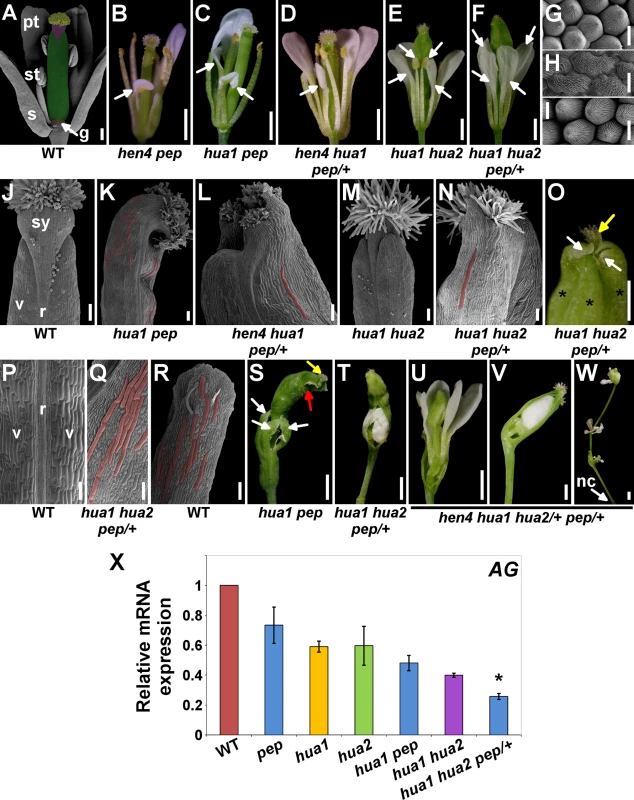
A) Scanning Electron Microscopy micrograph (SEM) of a post-anthesis wild-type flower. The different parts of the pistil have been artificially colored: stigma (yellow), style (purple), ovary (green), gynophore (g, orange). pt, petal; st, stamen; s, sepal. B-F) Post-anthesis flowers of different mutant backgrounds. Some outer organs were removed to better show petaloid stamens in the third whorl (arrows). G-I) SEM micrographs. Petal (G) and anther (I) adaxial surface in wild-type. Adaxial side in third whorl organs in hua1 hua2 pep/+ flowers (I). J) SEM apical portion of wild-type pistil. K-N) Top portion (SEM micrographs) of gynoecia/fruits from different mutant combinations. O) hua1 hua2 pep/+ pistils developed extra valves (asterisks) topped by white tissue that resembled that of sepal tips (white arrows). No style and very rudimentary stigmatic tissue were observed (yellow arrow). P) Medial view (SEM micrograph) of stage 17 wild-type fruit (according to [106]). Q) Abaxial surface of hua1 hua2 pep/+ gynoecia. R) Abaxial side of a wild-type sepal. S) A hua1 pep flower after outer organ abscission. Sepaloid carpels (white arrows) formed the fourth whorl that developed on a long gynophore. Similar structures were seen developing inside the sepaloid gynoecia which also contained rudiments of stigmatic tissue (yellow arrow) and further additional floral organs (red arrow). T) hua1 hua2 pep/+ fourth whorl. U-W) hen4 hua1 hua2/+ pep/+ flowers. Fourth whorl organs were removed to observe inner flowers (U, V). X) mRNA expression levels of AG in the wild type (WT) and diverse hua-pep mutant backgrounds, monitored by quantitative RT-PCR (qPCR). Error bars, standard deviation (SD). Asterisks indicate statistically significant differences from hua1 hua2 plants (*P < 0.05). On panels (K, L, N, Q) and (R) some giant cells appear false-colored. Scale bars: 1 mm (B, C, D, E, F, S, T, U, V, W), 500 μm (O), 200 μm (A), 100 μm (J, K, L, M, N, P, Q, R) and 10 μm (G, H, I). nc, nectaries; r, replum; sy, style; v, valve. Loss of HUA2 does not cause any obvious floral phenotype [30] and, although HUA2 and PEP interact during floral timing, hua2-4 pep flowers are normal [46]. However, this might not be surprising as our data indicate that the hua2-4 allele is leaky (S3 Fig.). We therefore used the null hua2-7 allele (hua2 hereafter, unless it is specified otherwise). Double mutants hua1 hua2 showed a variety of flower defects, including stamen-to-petal transformations (Fig. 1E and S1 Table), as reported for hua1 hua2-1 [30]. Unexpectedly, we were unable to isolate hua2 pep or hua1 hua2 pep individuals and only hua1 hua2 pep/+ plants were identified among the progeny. This background was sterile and showed a significant enhancement of the hua1 hua2 floral phenotype, including stronger petaloid transformations (Fig. 1E-I and S1 Table).
Dramatic alterations of fruit morphogenesis in pep, hua and hen mutant combinations
The fruit derives from the fertilized gynoecium carpels, whose formation, in turn, almost entirely depends on C-function [7,47]. We therefore decided to use carpel and fruit development as readout of how pep, hua, and hen mutant combinations affect C-function.
Although fruits in some of the mutant backgrounds were slightly shorter but normal looking (100% in hen4 pep, 20–60% in hua1 pep), we detected pistils with very distorted development, such as unfused carpels, and reduced style and stigma (S1D Fig.). In certain combinations, the apical portion of carpels was pointed with areas of white or pale green tissue conformed by smaller fringe cells as those in the apex of wild-type sepals (Fig. 1K, 1L, 1N, 1O and S2F, S2G Fig.).
The hua1 hua2 double mutant presented shorter pistils broadened at the tip [30] (Fig. 1M and S1D Fig.). However, hua1 hua2 pep/+ pistils were on average much shorter and crumpled (S1D Fig.). Indeed, close inspection of severely affected gynoecia in hua1 hua2 pep/+ by scanning electron microscopy (SEM) revealed that the carpel epidermis, rather than the wild-type characteristic vertical files of smooth cells (Fig. 1P), showed a wide range of epidermal cell sizes with epicuticular wax crenulations, including sepal-like giant cells [48–50] (Fig. 1N, 1Q, 1R). These alterations are typical of carpel-to-sepal transformation and were also seen in additional pep mutant combinations (Fig. 1K, 1L and S2H-S2K Fig.).
We detected that a significant percentage of hua1 pep pistils (40%) developed supernumerary valves (S1 Fig. and S2L Fig.). This trait is typical of loss of meristem determinacy and it was further enhanced in hua1 hua2 pep/+ (Fig. 1O and S1 Table). Terminal hua1 pep flowers, and at least a quarter of the hua1 hua2 pep/+ flowers exhibited conspicuously long gynophores and gynoecia that, strikingly, contained additional flowers inside. These basically consisted of petals and sepaloid gynoecia recapitulating the sepaloid features seen in the fourth whorl (Fig. 1S, 1T and S2M, S2O Fig. and see below). This phenotype, never observed in hua1 hua2 flowers (S1 Table), was reminiscent of that of ag mutants and also resembled the loss of HEN4 in the hua1 hua2 background [28]. In hen4 hua1 pep/+ a significant fraction of flowers (25%) contained supernumerary sepaloid valves (S2N Fig.), reflecting certain loss of determinacy in this genotype.
Overall, these results indicate that PEP, in collaboration with HUA and HEN genes, act as a positive regulator of the floral C-activity to, therefore, secure the downstream developmental programs depending on this function, such as fruit development.
The mutant combinations described above exhibit very similar developmental defects. Moreover, gene dosage effects in hua1 hen4 pep/+ and hua1 hua2 pep/+ plants illustrate the sensitivity of such backgrounds to PEP activity. These findings strongly suggest that PEP shares redundant developmental functions with HUA1, HUA2 and HEN4 despite their protein structural disparity. Accordingly, hen4 hua1 hua2/+ pep/+ plants showed very dramatic floral alterations (Fig. 1U-W and S2P-S2R Fig.). Hence, these factors were tentatively included in a common gene activity abbreviated as HUA-PEP along this work.
The lack of HUA-PEP activity causes sepaloid transformations in ful gynoecia
The MADS-box regulatory gene FRUITFULL (FUL) [51] is crucial for valve formation during ovary patterning, and it does so, in part, by preventing valves from adopting valve margin identity through the negative regulation of valve margin identity genes [52–56]. Upon fertilization, ful lignified valve cells remain small, arresting stomata development and silique growth. However, replum cells develop normally leading to a characteristic zig-zag configuration of this tissue in ful fruits [51] (Fig. 2A, 2B and S4A Fig.). Additionally, ful siliques show elongated styles [57] (Fig. 2A and S4A, S4K Fig.).
Fig. 2. The loss of HUA-PEP activity is epistatic over the ful phenotype. 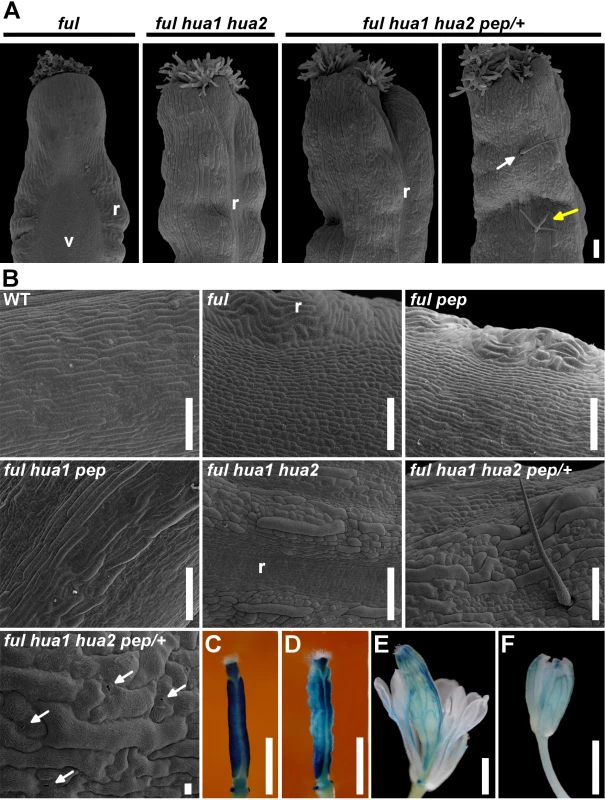
A) SEM images of the top portion of a ful fruit. The typical long style and wide zig-zag replum were suppressed in ful hua1 hua2 and ful hua1 hua2 pep/+ pistils, and sepaloid giant cells were observed on the valve surface. Simple or branched trichomes (white and yellow arrows, respectively) were occasionally observed on the surface of ful hua1 hua2 pep/+ pistils. B) SEM images of the abaxial ovary surface in wild-type (WT) and different mutant backgrounds. Observe interspersed stomata (arrows) in a ful hua1 hua2 pep/+ panel. C-F) GUS reporter whole-mount staining (ful-1) in ful (C), ful hua1 hua2 (D), ful hua1 hua2 pep/+ (E) pistils and wild-type sepal (F). Observe long gynophores and full petaloid conversion of stamens in (E). Scale bars: 100 μm (A, B), except 10 μm in the last B panel (ful hua1 hua2 pep/+ genotype), and 1 mm (C-F). r, replum; v, valve. To get more insights into the role of HUA-PEP activity during pistil development, we decided to characterize the behavior of hua-pep activity mutants in the ful background.
The ful-1 hua1 fruit was virtually identical to that of ful-1 plants [51] (ful hereafter; S4A Fig.). In contrast, ful pep, ful hua2 and ful hua1 pep siliques were progressively longer and showed shorter styles (S4A Fig.). In such backgrounds, valve epidermal cells were elongated and streaked, along with interspersed stomata. These phenotypes indicated that valves took onto sepaloid identity (Fig. 2B and S4B Fig.). In ful hua1 hua2, gynoecia were smaller and replum cells remained small as in wild-type unpollinated pistils [58], abolishing the characteristic zig-zag shape (Fig. 2A, 2B and S4A Fig.). Fertility in ful hua1 hua2 plants was severely reduced.
ful hua1 hua2 pep/+ plants were phenotypically identical to hua1 hua2 pep/+. In such combinations, we also found new floral organs developing inside swollen gynoecia that were often seating on long gynophores (Fig. 2E and S4A, S4E, S4I, S4L, S4M Fig.).
The glucuronidase (GUS) reporter harbored by the ful-1 transposon reflects the native expression pattern of FUL [51]. Pistils of ful or wild-type-looking heterozygous ful/+ plants displayed characteristic GUS activity in the valves, style and nectaries [51] (Fig. 2C and S4D, S4G Fig.). In ful hua1 hua2, strong GUS signal was detected in nectaries and apical territory preserving style identity, whereas valves presented a more irregular pattern (Fig. 2D and S4H Fig.). The GUS-staining pattern of ful hua1 hua2 pep/+ in the fourth whorl organs had little resemblance to that of a gynoecium, except in nectaries and style vestiges, notably evoking FUL expression in the sepal vasculature [51] (Fig. 2E, 2F and S4E, S4F, S4I Fig.).
Next, we treated flowers with the lignin-specific dye phloroglucinol. Mature wild-type fruits showed preferential staining in the valve margin, whereas in ful mutants valves were ectopically lignified [52,53] (S4J, S4K Fig.). Nonetheless, in equivalent flowers from ful hua1 hua2 pep/+ plants, lignification in the presumptive gynoecium was nearly restricted to branched red lines with striking resemblance to lignified sepal vasculature (S4L-S4N Fig.). Altogether, genetic and histochemical analyses indicate that the HUA-PEP gene activity is required to prevent gynoecium tissues from adopting sepaloid fate independently of their original identity (valve, valve margin), highlighting the role played by PEP to preserve carpel identity.
PEP is a positive regulator of AG functional mRNA levels
To determine whether PEP impinges upon AG regulation and therefore C-function, we measured mRNA levels from wild-type and mutant flower buds by quantitative PCR (qPCR). In consonance with the phenotypes described above, relative expression of AG decreased significantly in hua1 pep and hua1 hua2 double mutants, and reduced even further in hua1 hua2 pep/+ plants (Fig. 1X).
To investigate whether somehow PEP (and HUA) control A and B function, we measured the transcript levels of the homeotic A - and B-class genes AP1 and PI, respectively. Results were inconclusive because, although expression of both genes declined moderately in some mutant strains (S5 Fig.), no morphological evidence of altered A - or B-floral functions was observed in any of the hua-pep mutant combinations examined. Therefore, our molecular and genetic data suggest that, in contrast to the C-function, it appears that HUA-PEP gene activity has little or no role in regulating A - and B-functions.
AG triggers several reproductive developmental programs in part by activating additional regulators that perform different subsets of its functions. For example, SPOROCYTELESS (SPL) stimulates stamen development, including organ identity [59–61], whereas the zinc-finger gene KNU cooperates with AG to repress WUS [19,21]. Consistently, SPL and KNU expression decreased markedly in hua1 pep and hua1 hua2 double mutants, and hua1 hua2 pep/+ plants (S6A, S6B Fig.). Accordingly, KNU gene expression monitored by a GUS-reporter construct was found to be less intense in hua1 pep developing flower organs, as compared to the wild type (S6C-S6H Fig.).
Interestingly, we found that the mutant phenotype of hua1 pep plants was completely rescued by increasing the dosage of AG gene with a genomic construct able to complement ag mutants [62] (S7 Fig.), thus reinforcing our hypothesis that AG functions depend on HUA-PEP activity genes. Collectively, these results might explain the organ identity and determinacy defects seen in pep hen hua combos and further support PEP as a positive regulator of AG.
PEP prevents AP1 expansion to inner whorls
One of the functions of AG is to prevent AP1 expression in the two inner whorls of organs where stamens and carpels normally form [8]. To examine the expression of AP1 we used the genomic GFP (green fluorescent protein)-based reporter gAP1::AP1-GFP, that largely mirrors endogenous AP1 expression [62]. As expected, in the wild type AP1-GFP signal was detected in sepals but absent in pistils (S8 Fig.). However, a number of hua1 pep pistils showed AP1-GFP fluorescence (Fig. 3A-D). These results are coherent with earlier work showing AP1 mRNA ectopic expression in inner whorls of hua1 hua2 and hua1 hua2 hen4 [28,30], and underscore the importance of PEP as a regulator of the C-function during flower organogenesis.
Fig. 3. Detection of the AP1-GFP protein in hua1 pep gynoecia. 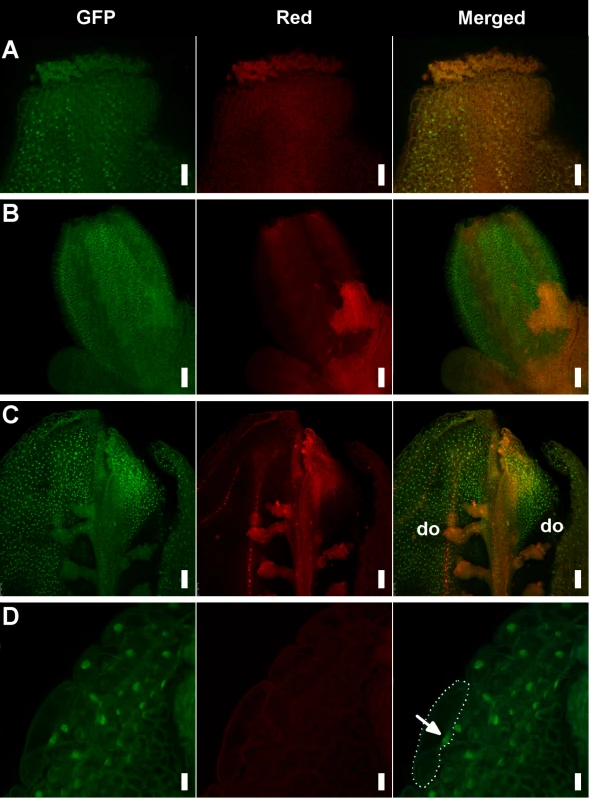
A) Apical region of a mildly affected gynoecium with recognizable pistil morphology. Specific AP1-GFP signal is detected in some style cells. B) Fourth whorl organs of a pre-anthesis flower displaying a severe sepaloid phenotype. C) Adaxial (inner) view of a manually open pistil with severe sepaloid transformations, but containing some developing ovules (do). D) Detail of a fourth whorl organ from panel D showing nuclear-localized AP1-GFP. A cell has been outlined with a dotted line and the nucleus marked with an arrow. Scale bars: 25 μm (A), 50 μm (B and C) and 10 μm (D). PEP overexpression impairs flower morphogenesis
Our loss-of-function genetic analyses show that components of the HUA-PEP function are redundantly required for the floral C-function. So we asked whether PEP alone could compensate for the deficiency in members of this activity. To test this idea, a 35S::PEP overexpressing construct [46] was introduced into the hua1 hua2 background. Strikingly, PEP overexpression, instead of rescuing, dramatically enhanced the hua1 hua2 mutant phenotypes. Homozygous hua1 hua2 35S::PEP flowers were sterile, and exhibited much stronger stamen-to-petal and carpel-to-sepal transformations than in hua1 hua2, as well as frequent severe indeterminacy defects, a trait never observed in hua1 hua2 plants (Fig. 4A-D, 4F-H, S9A-S9E Fig. and S1 Table). In line with the strong phenotypes observed, the levels of AG, KNU and SPL mRNAs in hua1 hua2 35S::PEP plants were significantly lower than those of hua1 hua2 mutants (Fig. 4I and S9K, S9L Fig.).
Fig. 4. PEP overexpression impairs flower morphogenesis and AG pre-mRNA processing. 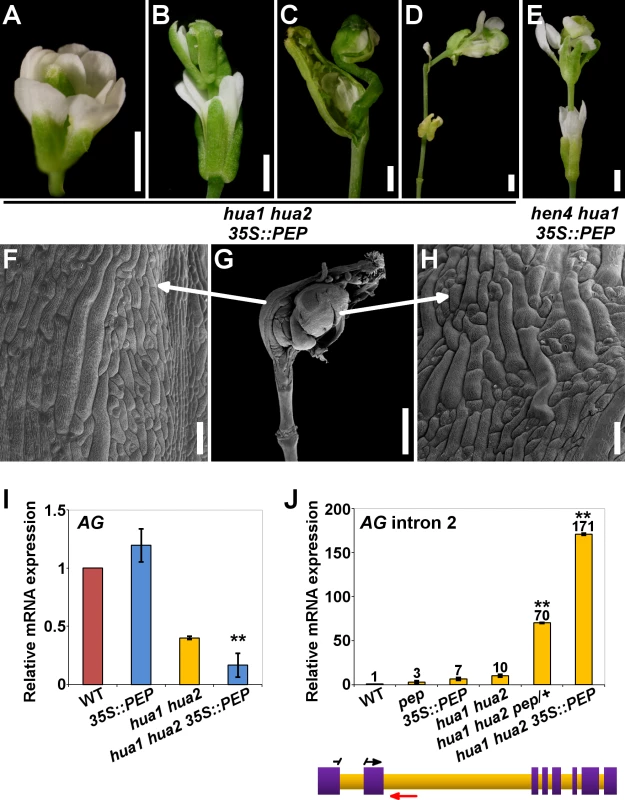
A-D) hua1 hua2 35S::PEP flowers. E) hen4 hua1 35S::PEP flower. In both genotypes, loss of determinacy was frequent. All flowers displayed severely transformed petaloid stamens and sepaloid carpels. F-H) SEM micrograph of a hua1 hua2 35S::PEP flower (G), and close-up views of the fourth whorl organ abaxial surface (F) and inner additional whorl organ (H), respectively. Sepaloid traits were found in these gynoecia. I, J) Relative expression levels of AG mRNA (I) and AG transcripts including intron 2 sequences (J), in wild-type plants (WT) and diverse hua-pep mutant backgrounds, monitored by qPCR. In (J), a diagram of the AG gene is shown below. Purple boxes denote exons whereas intronic regions are colored in orange. Relative positions of forward (black arrow) and reverse (red arrow) primers are indicated. To increase annealing specificity, the forward primer sequence was split between exons 1 and 2. Error bars, SD. Asterisks indicate statistically significant differences from hua1 hua2 plants (**P < 0.01). Scale bars are 1 mm (A-E, G) and 100 μm (F, H). We ruled out any RNA silencing effect as hua1 hua2 35S::PEP plants showed much higher PEP mRNA levels than wild-type individuals (S10D Fig.). Rather, PEP protein overproduction might exceed a certain critical threshold, leading to the strong phenotypes observed. Consistent with this idea, hemizygous hua1 hua2 35S::PEP/+ plants produced PEP mRNA levels higher than those of the wild type, yet much lower than in homozygous hua1 hua2 35S::PEP plants (S10D Fig.), and did not show the severe floral phenotypes of the latter, being indistinguishable from hua1 hua2 individuals (S10E-J Fig. and S1 Table).
Although, PEP overexpression in hen4, hua1 and hua2 single mutant backgrounds did not result in noticeable morphological alterations (S10A-C Fig.), we speculated whether excess of PEP was critically detrimental in more compromised conditions. In line with this interpretation, PEP overexpression in the wild-type looking hen4 hua1 plants [28] led to the same developmental abnormalities previously described for the strong deficient hua-pep backgrounds. A significant number of hen4 hua1 35S::PEP flowers (∼65%) displayed severe indeterminacy, closely resembling ag flowers (Fig. 4E and S9F-J Fig.). It is worth mentioning that this phenotype never occurred in hen4 hua1 pep/+, indicating that PEP gain-of-function has a stronger impact on floral determinacy in hen4 hua1 than reducing PEP activity, similarly as described for hua1 hua2 background (S1 Table).
PEP secures correct AG function by facilitating pre-mRNA processing
Mutations in HEN4, HUA1 and HUA2 led to a gradual decrease of AG mRNA levels concomitant with the accumulation of aberrant transcripts incorrectly terminated at the large second intron [28]. To test whether PEP impacts on this process, we carried out qPCR assays using intronic primers situated near the exon2/intron2 junction (Fig. 4J and S2 Table). The relative abundance of a PCR product increased progressively in various hua-pep mutant strains, notably in hua1 hua2 pep/+ and hua1 hua2 35S::PEP individuals, whereas it was barely detectable in the wild type (Fig. 4J). These values negatively correlated with the levels of correctly spliced AG transcript in the mutant backgrounds under study, and unambiguously indicated that altering levels of PEP has an important impact on the accumulation of these transcript species.
To examine transcript structure, polyadenylated RNA from hua1 hua2 pep/+ and hua1 hua2 35S::PEP plants was subjected to 3’ RACE (Rapid Amplification of cDNA Ends). Several products were obtained corresponding to transcripts comprising correctly spliced exons 1 and 2 followed by a variable stretch of nucleotides of intron two (105–368 nt), after which premature cleavage and polyadenylation events took place (S11A, S11B Fig.). These transcripts miss the last 6 exons, lacking the ability to encode a functional AG polypeptide. In plants, three polyadenylation signals define the site of processing: the far upstream element (FUE), the near upstream element (NUE), and the cleavage element (CE) [63]. Inspection of such RACE products revealed the presence of FUE, NUE and CE elements properly situated, strongly suggesting their implication in the premature termination event [63] (S11A, S11B Fig.).
FLK: an additional component of the HUA-PEP activity
FLK is expressed in all major organs, yet its loss of function did not cause any visible defect [42,43]. FLK interacts with PEP and HUA2 during flowering time regulation [46] but its possible role in flower morphogenesis has not yet been studied.
To explore FLK activity during flower development and to determine whether FLK participates in the HUA-PEP function, the null flk-2 mutant [43] (flk hereafter) was crossed to different hua-pep mutant combinations. flk hen4 double mutant flowers were wild-type in appearance (S12A, S12B Fig.). Unlike hua1 pep (Fig. 1 and S1 and S2 Fig.), flk hua1 and flk pep double mutant flowers also looked essentially normal (S12C Fig.) [46]. In contrast, flk pep hua1/+ plants showed some aberrant gynoecia, and petaloid stamens (S12D-F Fig.). Interestingly, stamen identity in the flk pep background, therefore, is sensitive to HUA1 gene-dosage since this trait is never observed in pep hua1/+, nor in flk pep flowers.
Next, the flk mutant was crossed to hua1 hua2 plants, a sensitized background repeatedly used to uncover gene activities involved in flower organ identity and determinacy [28,64–66, this work]. The resulting flk hua1 hua2 triple mutants were easily identified because of their conspicuous flower defects. flk hua1 hua2 flowers had two sets of petals and were “stamenless” (Fig. 5A and S12G Fig.), thus lacking fertilization and fruit set. Besides, flk hua1 hua2 gynoecium development was severely distorted with obvious sepaloid attributes (Fig. 5B, 5C). Nevertheless, the most defining feature was again the occurrence of indeterminate flowers (>50%) (Fig. 5B, 5D, 5E and S12H Fig.). As indicated above, hua1 hua2 flowers never show this severe developmental alteration, underscoring the contribution of the flk mutation to debilitate the floral C-function.
Fig. 5. Loss of FLK dramatically enhances the floral phenotypes of hua1 hua2 plants. 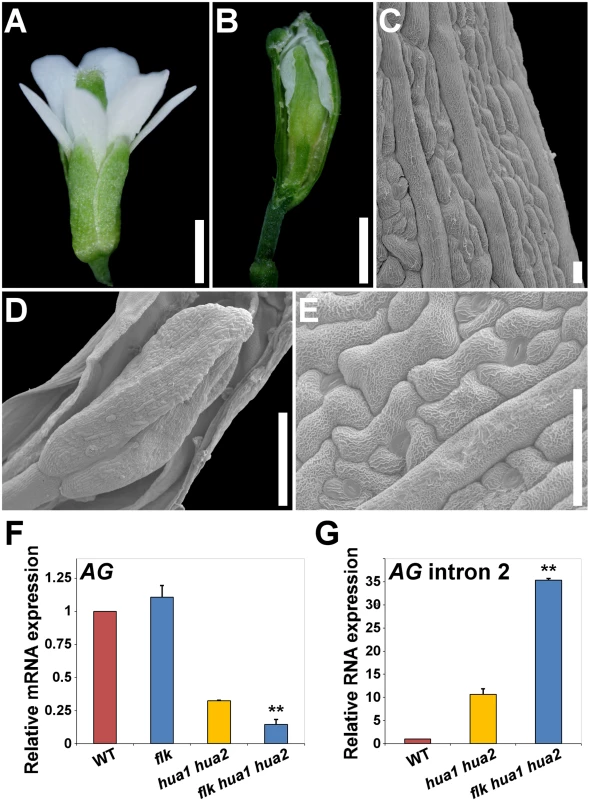
A) flk hua1 hua2 flower with all stamens converted into petals. B) Gynoecium with a long gynophore. A sepaloid valve was manually removed to better observe a new flower developing inside. C) SEM image of a sepaloid carpel with giant cells and epicuticular wax ridges. D) SEM magnification of the inner flower shown in (B). E) Close-up view of the sepaloid organ shown in (D). F, G) qPCR relative expression levels of AG mRNA (F), and AG transcripts including intron 2 sequences (G) in wild type (WT) and mutant backgrounds. Error bars, SD. Asterisks indicate statistically significant differences from hua1 hua2 plants (**P < 0.01). Scale bars: 1 mm (A, B), 20 μm (C) 500 μm (D) and 50 μm (E). Our qPCR gene expression data backed up the hypothesis of FLK as part of the HUA-PEP activity. In flk the expression levels of AG, KNU and SPL remained unaltered when compared to those of the wild type, whereas in flk hua1 hua2 significantly dropped, being even lower than in hua1 hua2 individuals (Fig. 5F and S12I Fig.). This result substantiates the floral defects detected. Conversely, levels of AG transcripts containing intron 2 sequences increased in flk hua1 hua2 (Fig. 5G), suggesting an influence of FLK on AG post-transcriptional regulation. Indeed, we performed 3’ RACE assays for RNA from flk hua1 hua2 and identified new aberrant transcripts indicating premature cleavage and polyadenylation within the large intron 2. As described above, polyadenylation signals were found around the presumptive maturation site (S11C Fig.). Altogether, these results strongly support FLK as an additional component of the HUA-PEP activity.
The components of the HUA-PEP activity physically associate
As mentioned in the introduction, RNA binding proteins participate in multimeric RNP complexes to perform their regulatory functions [36,41]. Our genetic and expression analyses indicated that genes of the HUA-PEP activity act in concert during floral organogenesis, which makes reasonable their interplay at the protein level. Nuclear localization of their products has been demonstrated [28,42,43,46,67] and, importantly, physical interaction between HEN4 and HUA1 has been already established [28]. Moreover, HEN4 was also computationally predicted to interact with PEP and FLK [68]. We therefore, conducted in vivo bimolecular fluorescence complementation (BiFC) assays in tobacco leaves using PEP, FLK, HEN4 and HUA1. Reconstituted yellow fluorescent protein (YFP) was detected in leaf cell nuclei when FLK-PEP, HEN4-PEP and HUA1-PEP interactions were assayed, respectively (Fig. 6 and S13 Fig.). Similarly, robust nuclear interaction was seen when FLK was tested against HUA1 and HEN4 (Fig. 6 and S13 Fig.). The HUA1-HEN4 BiFC interaction was used as a positive control (S13 Fig.). All associations were tested in both directions, thus endorsing specificity of the interactions (Fig. 6 and S13 Fig.).
Fig. 6. The hnRNPs PEP and FLK physically interact with HUA1 and HEN4. 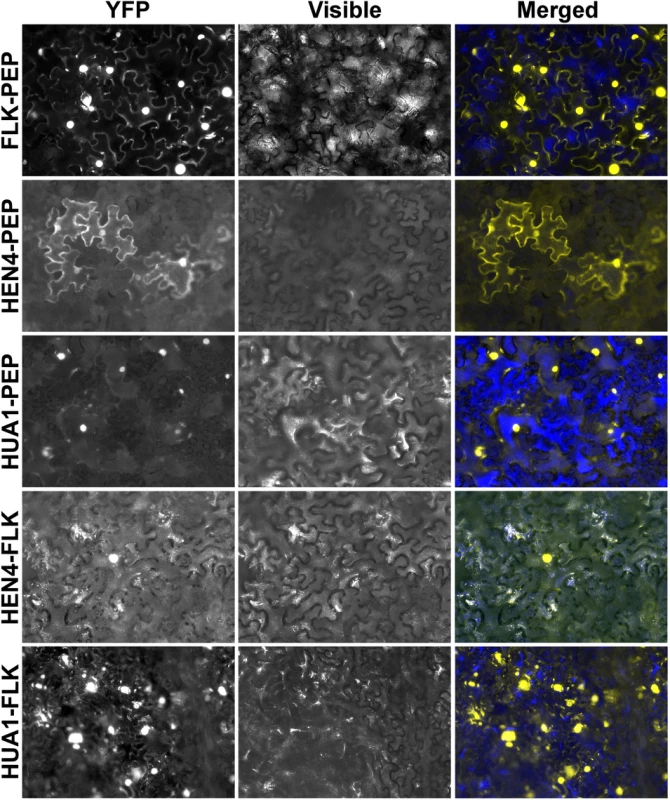
BiFC visualization of protein dimerization (yellow fluorescence) in Nicotiana benthamiana leaf cells agroinfiltrated with plasmids encoding fusion proteins. In each test, the first protein was fused to the C-terminal fragment of the YFP (YFPct), and the second protein to the N-terminal portion (YFPnt), respectively (see Materials and Methods section). We were also able to confirm in vivo protein homodimerization of PEP and FLK in our assays, corroborating the publicly available in silico data [68] (S13 Fig.). Homodimer formation was also seen in HUA1 BiFC experiments (S13 Fig.). These associations were further verified in yeast-two-hybrid assays (Y2H; S14 Fig.).
In a subset of our BiFC assays we detected, in addition to clear signal in the nuclei, specific cytoplasmic fluorescence (Fig. 6 and S13 Fig.). KH-domain containing proteins, particularly PCBPs, are known to participate in numerous RNA processing events in the nucleus and in the cytoplasm (RNA transport, stability, translation) [36–38]. Therefore, this extranuclear signal might reflect additional regulatory roles for PEP and FLK in this cell compartment.
Taken together, these results strongly suggest that PEP, FLK, HUA1 and HEN4 proteins physically associate likely reflecting their participation in common multimeric complexes involved in pre-mRNA processing. Additionally, these data further reinforce the assumption of FLK as a new partner of the HUA-PEP activity.
Discussion
PEP and FLK were previously identified to control flowering time through regulation of the FLC gene [46,42]. Now, our analyses show that PEP and FLK also play a key role in the specification of flower organ identity as components of the post-transcriptional machinery that ensures normal processing of the AG pre-mRNA. Genetic, functional and molecular interactions with additional RNA-binding proteins previously established as AG regulators [28] led us to define HUA-PEP as a common gene activity comprising HUA1, HUA2, HEN4, PEP and FLK.
PEP is a positive regulator of C-function activity during flower morphogenesis
We have demonstrated that PEP is functionally linked to the AG pre-mRNA processing pathway. Whereas hua1, hua2 and hen4 single mutants are phenotypically wild-type [28, this work], when these same mutants were combined with pep, we observed developmental abnormalities consistent with reduced C-function activity. Moreover, hua1 hua2 double mutant flower defects [30] were dramatically enhanced when combined by plants that were heterozygous for a mutation in PEP (hua1 hua2 pep/+), illustrating dosage-effects among HUA-PEP genes as previously reported for HUA1, HUA2 and HEN4 [28]. The intensity of these floral phenotypes correlated with a reduction in AG mRNA levels. As a result, the A-function gene AP1, which is normally expressed in whorls 1 and 2, was ectopically expressed in the inner whorls of hua1 pep flowers, consistent with a compromised C-function and the A-C antagonism [4,8]. The sepaloid transformations seen in gynoecium tissues when HUA-PEP genes were mutated in the ful background provided further evidence for the critical contribution of PEP to carpel identity.
Loss of PEP contributed to reduce the floral C-function activity. Surprisingly, PEP overexpression in hua1 hua2 and hua1 hen4 also caused a dramatic enhancement of flower mutant phenotypes. Although this might seem unexpected, there are many examples in which loss - and gain-of-function result in the same phenotypical alterations. Loss and overexpression of bancal, encoding a Drosophila homologue of vertebrate hnRNP K, generates appendage developmental defects [69]. In Xenopus embryos, both reduction and overexpression of the KH gene Mex3b, involved in neural plate formation, led to downregulation of target genes [70]. In Arabidopsis, increasing or reducing the expression of kinase-encoding genes FAB1A/B elicits the same pleiotropic alterations, which are attributed to perturbations in the protein complexes in which they participate [71].
However, PEP overexpression in wild-type or single hua-pep mutant backgrounds rendered normal flowers, suggesting certain buffering capacity against PEP excess. Nevertheless, simultaneous inactivation of various HUA-PEP components (hua1 hua2 or hen4 hua1) when PEP is overexpressed might aggravate a detrimental excess of PEP, by likely disrupting protein stoichiometric equilibria [72]. In line with this hypothesis is the fact that hemizygous hua1 hua2 35S::PEP/+ plants, expressing higher levels of PEP than the wild type but much less than homozygous hua1 hua2 35S::PEP plants, do not differ from hua1 hua2 double mutants.
FLK is a member of the HUA-PEP gene activity
Our analyses have also uncovered a role for FLK in plant morphogenesis. FLK participates in the HUA-PEP activity during C-function maintenance. The genetic interaction between flk, pep, hua1 and hua2, the phenotypic similarities between flk hua1 hua2 (Fig. 5) and hen4 hua1 hua2 [28], the gene expression analyses, as well as FLK physical associations, firmly support this conclusion.
FLK represses FLC and thus promotes flowering whereas PEP and HUA2 are FLC activators [42,43,46,73,74]. During flower morphogenesis, however, FLK and PEP promote flower morphogenesis through the positive regulation of AG (this work). Taking into consideration the promiscuity of RNA-binding proteins, it is very plausible that components of the HUA-PEP activity might be participating in functionally distinct complexes. This is not unprecedented. For example Arabidopsis SR (serine/arginine rich) factors and the hnRNP AtGRP8 exhibit antagonistic and cooperative effects during circadian regulation [75]. Also, closely related MADS-box genes AGAMOUS-LIKE 24 (AGL24) and SHORT VEGETATIVE PHASE (SVP) accelerate and delay flowering, respectively. Later, AGL24 and SVP cooperate with AP1 to downregulate AG during first stages of floral development [76–78]. Similarly, FUL-SVP replaces FLC-SVP heterodimers counteracting the repressive effect of the latter on flowering time [79]. Moreover, AG and AP3/PI participate in the same protein complexes to specify stamen anlagen. However, many genes promoting carpel development that are induced by AG are, on the contrary, repressed by AP3/PI [80]. Functional versatility of the HUA-PEP activity, in turn, might be very advantageous to provide regulatory flexibility to modulate the highly dynamic and complex networks governing reproductive development.
Association of HUA-PEP proteins
PEP and FLK physically associate, as well as with HUA1 and HEN4, indicating that, probably, they all participate in common regulatory complexes. HUA2, however, might affect AG independently since no physical interaction between HUA2 and any other HUA-PEP component described here could be detected in a recent Y2H screen [81]. Formally, HUA2 molecular interactions might be mediated through HUA-PEP factors yet to be identified. We observed stronger phenotypes in hua-pep backgrounds when HUA2 was mutated. These results might be explained with the existence of two complementary subactivities: one incorporating the HUA2 function and another one comprising the remaining identified HUA-PEP factors. Simultaneous disruption of both complexes might account for more profound phenotypic defects. Lethality in hua2 pep mutants substantiates this notion.
PEP and FLK secure AG expression by mediating correct RNA processing
Our molecular analyses of hua-pep mutants are coincident with previous work showing accumulation of transcripts retaining intronic sequences at the expense of the functional AG mRNA [28]. A large intron where important regulatory motifs reside is a feature shared by AG, FLC and other MADS-box genes, that is conserved across species [82–87]. However, nascent transcripts are vulnerable to premature processing and large introns might increase the risk of cryptic signals recognizable by the splicing and/or polyadenylation machineries [88,89].
Transcript maturation mainly proceeds co-transcriptionally, increasing the fidelity of the process [24,90,91]. Altering PEP and FLK expression in the hua1 hua2 background had a profound effect on the accumulation of AG intron-retaining transcripts. Remarkably, FLC intron-containing transcripts also increased in pep plants [46]. We propose that the HUA-PEP proteins assist transcription elongation by “hiding” cryptic signals in the nascent RNA (Fig. 7A-C). Otherwise, these sites could be accessible to the corresponding processing machinery, giving rise to non-functional or prematurely terminated transcripts (Fig. 7D). Our hypothesis is consistent with the recent characterization of mammal PCBPs as global regulators of alternative polyadenylation. Knock down of PCBPs actually favors usage of cryptic intronic sites [40]. Interestingly, hnRNP K suppresses usage of a premature polyadenylation site for NEAT1, a long non coding RNA (lncRNA) operating in nuclear paraspeckles (ribonucleoprotein bodies) formation, thus increasing the ratio of the long effective transcript [92].
Fig. 7. The HUA-PEP activity facilitates pre-mRNA processing of target genes. 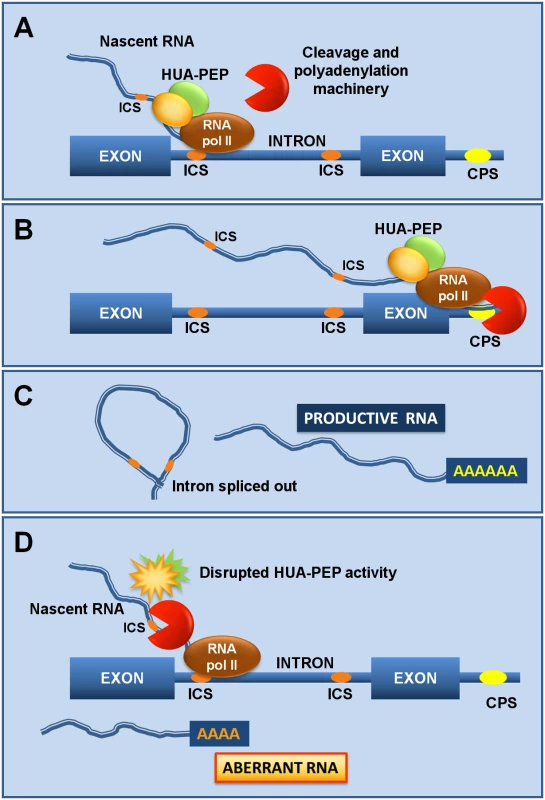
A) As the RNA polymerase (RNA pol II) activity progresses, the HUA-PEP hnRNP complex coats the nascent transcript, still chromatin-associated, thus sequestering intronic cryptic sites (ICS) from cleavage and polyadenylation. B) The elongation complex reaches the distal terminal cleavage and polyadenylation site (CPS), where correct termination occurs. C) Adequate intron excision and 3’ maturation take place. D) Conversely, an altered HUA-PEP activity does not prevent the RNA 3’ processing machinery to access cryptic motifs in the elongating transcript, producing thereby a prematurely terminated and ineffective RNA. By sequestering intronic polyadenylation motifs, PEP (and the remaining HUA-PEP factors) may also facilitate correct splicing, as documented for other PCBPs [36,41]. The U1 snRNP (U1), in addition to its splicing role, protects pre-mRNAs from premature termination at intronic polyadenylation sites [88,89], raising the attractive possibility of a connection with the HUA-PEP gene activity. The carboxyl-terminal (CTD) domain of eukaryotic RNA polymerase II coordinates transcription and transcript maturation [93]. The Arabidopsis KH protein SHINY1 (SHI1) interacts with a phosphatase that dephosphorylates particular residues in CTD, downregulating transcription of abiotic stress-related genes by preventing 5’ capping [94,95]. Uncovering new functional and molecular relationships among distinct HUA-PEP components will certainly provide a better understanding of the developmental programs regulated by this activity (floral timing; flower patterning) and the importance, at the regulatory level, of multifunctional plant PCBP-type hnRNPs.
Materials and Methods
Plant material
This work was carried out with the Arabidopsis thaliana Columbia (Col-0) accession as the wild type. Strains previously obtained in other accessions were backcrossed at least five times into Col-0 before any further experiment. Plant materials used in this study were pep-4 [44], flk-2 [43], hua2-4 [73]; hua2-7 [74], 35S::PEP [46], and ful-1 [51]. gAG::AG-GFP and gAP1::AP1-GFP [62] were provided by Gerco Angenent and Richard Immink (Wageningen University, The Netherlands). hen4-2 [28], hua1-1 and hua2-1 [30] were provided by Xuemei Chen (UC Riverside, USA). KNU::GUS [16] was provided by Anna M. Koltunow (CSIRO, Adelaide, Australia). Information about all primers used in this work and molecular genotyping can be found in S2 Table. Plants were grown in MS plates or soil as previously described [44].
Microscopy and histology
Phloroglucinol lignin staining [96,97] and GUS assays were performed essentially as described [44,96,97]. All GUS analyses, except in the case of ful-1/+, were performed in homozygous lines. Whole-mount pictures were taken under a Nikon SMZ1500 stereomicroscope. Histological sections (8 μm) were photographed under bright-field or dark-field illumination using a Nikon E800 microscope. In both cases Nikon Digital Camera DXM1200F was used operated by the ACT-1 2.70 program. Scanning electron microscopy (SEM) was according to [44]. For confocal laser scanning microscopy, all analyses were performed in homozygous lines. Samples were pre-treated with methanol/acetone (1 : 1 v/v) solution for 30 minutes at -20°C, and subsequently rinsed in PBS buffer (1.94 mM K2PO4; 8.06 mM Na2PO4; 2.7 mM KCl, 0.137 mM NaCl, pH 7.4) to be observed under a Leica TCS SPE confocal microscope. Pictures were taken with the LAS AF program.
Quantitative RT-PCR and RACE
For quantitative RT-PCR (qPCR), 5 μg of total RNA was extracted from young flower buds until stage 9, treated with DNase I, and used for cDNA synthesis with an oligo(dT) primer and RevertAid Premium Reverse Transcriptase (Thermo Scientific) following the manufacturer’s instructions. Subsequently, for each qPCR reaction, 0.5 μl of the cDNA was used as template. Relative changes in gene expression levels were determined using the LyghtCycler 1.5 system with the LightCycler FastStart DNA amplification kit according to the manufacturer (Roche Diagnostics). RNA levels were normalized to constitutively expressed genes OTC (ORNITINE TRANSCARBAMILASE) [98] and ACT2 [99], and the corresponding wild-type levels, as previously reported [46,100]. Each experiment was undertaken using three biological replicates with three technical replicates each. The standard deviation was calculated in Microsoft Excel. Statistical significance was estimated by the Student’s t-test according to [101] (*P < 0.05, **P < 0.01).
For 3’ rapid amplification of cDNA ends (3’ RACE), 5 μg of young flower bud total RNA was reverse transcribed using Maxima Reverse Transcriptase and the adaptor oligo d(T)-anchor (kit 5’/3’ RACE, Roche Diagnostics) as a primer. Then, AG cDNAs were amplified with High Fidelity PCR Enzyme Mix (Thermo Scientific) using forward primers situated in the exon 2 (S2 Table) and the PCR anchor (Roche Diagnostics) as a reverse primer hybridizing with the adaptor sequence, thus ensuring that only polyA-containing sequences were amplified. Amplified products were cloned into pSC-A plasmids and sequenced with M13F and M13R primers. Sequences were analyzed using CLUSTAL-W aligning [102].
Protein interactions
For bimolecular fluorescence complementation (BiFC), coding sequences of all genes under study were amplified from their respective cDNAs using Phusion Taq-polymerase (NEB). The corresponding primer sequences (S2 Table) were designed for cloning the resulting PCR amplicons via Gibson DNA assembly method [103], and cloned into both the pBJ36-SPYNE and pBJ36-SPYCE plasmids, containing N-terminal (nt) and C-terminal (ct) halves of the yellow fluorescent protein (YFP), respectively (YFPnt and YFPct) [104]. The 35S::SPYNE and 35S::SPYCE cassettes were then cloned via NotI into the binary vectors pGreen0229 and pGreen0179 [105], respectively. Transformed AGL-0 Agrobacterium tumefaciens cells were used to infect Nicotiana benthamiana leaves. YFP reconstituted fluorescence was visualized 72 h after inoculation under a Nikon Eclipse TE2000-U epifluorescence microscope. The reciprocal BiFC assays were also performed obtaining the same results as shown in Fig. 6 and S13 Fig. As negative controls, Nicotiana leaves were co-infiltrated with the corresponding recombinant YFPct construct and the empty YFPnt version, yielding no signal in any case.
For yeast two-hybrid assays, the cDNAs for PEP, FLK, HEN4 and HUA1 genes were amplified with the proof-reading Phusion Taq-polymerase (New England Biolabs, Inc.) using the corresponding primers (S2 Table). The resulting products were cloned into the pB42AD (+Trp) and pGilda (+His) vectors via Gibson DNA assembly procedure [103]. The integrity of constructs was checked by sequencing. The yeast strain EGY48 (-Ura) was cotransformed with the corresponding combinations of pGilda and pB42AD constructs. Empty vectors were used as negative controls. Positive colonies were selected on solid media (-Ura, -His, -Trp +glucose). Induction for testing protein-protein association was assayed growing the resulting yeast strains on plates or liquid in the presence of galactose and raffinose (DB Falcon). X-gal was used for colorimetric assays on plates, and ONPG (2-Nitrophenyl β-D-galactopyranoside, SIGMA) for β-galactosidase liquid experiments. The Clontech protocol book was followed for all these procedures.
Supporting Information
Zdroje
1. Ó’Maoiléidigh DS, Graciet E, Wellmer F (2014) Gene networks controlling Arabidopsis thaliana flower development. New Phytol 201 : 16–30. doi: 10.1111/nph.12444 23952532
2. Weigel D, Alvarez J, Smyth DR, Yanofsky MF, Meyerowitz EM (1992) LEAFY controls floral meristem identity in Arabidopsis. Cell 69 : 843–859. 1350515
3. Mandel MA, Gustafson-Brown C, Savidge B, Yanofsky MF (1992) Molecular characterization of the Arabidopsis floral homeotic gene apetala1. Nature 360 : 273–277. doi: 10.1038/360273a0 1359429
4. Coen ES, Meyerowitz EM (1991) The war of the whorls: genetic interactions controlling flower development. Nature 353 : 31–37. doi: 10.1038/353031a0 1715520
5. Pelaz S, Ditta GS, Baumann E, Wisman E, Yanofsky MF (2000) B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405 : 200–203. doi: 10.1038/35012103 10821278
6. Ditta G, Pinyopich A, Robles P, Pelaz S, Yanofsky MF (2004) The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr Biol 14 : 1935–1940. doi: 10.1016/j.cub.2004.10.028 15530395
7. Yanofsky MF, Ma H, Bowman JL, Drews GN, Feldmann KA, et al. (1990) The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 346 : 35–39. doi: 10.1038/346035a0 1973265
8. Gustafson-Brown C, Savidge B, Yanofsky MF (1994) Regulation of the arabidopsis floral homeotic gene APETALA1. Cell 76 : 131–143. 7506995
9. Causier B, Schwarz-Sommer Z, Davies B (2010) Floral organ identity: 20 years of ABCs. Semin Cell Dev Biol 21 : 73–79. doi: 10.1016/j.semcdb.2009.10.005 19883777
10. Jofuku KD, den Boer BG, Van Montagu M, Okamuro JK (1994) Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 6 : 1211–1225. 7919989
11. Weigel D (1995) The APETALA2 domain is related to a novel type of DNA binding domain. Plant Cell 7 : 388–389. doi: 10.1105/tpc.7.4.388 7773013
12. Parenicová L, de Folter S, Kieffer M, Horner DS, Favalli C, et al. (2003) Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: new openings to the MADS world. Plant Cell 15 : 1538–1551. 12837945
13. Smaczniak C, Immink RGH, Angenent GC, Kaufmann K (2012) Developmental and evolutionary diversity of plant MADS-domain factors: insights from recent studies. Development 139 : 3081–3098. doi: 10.1242/dev.074674 22872082
14. Laux T, Mayer KF, Berger J, Jürgens G (1996) The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122 : 87–96. 8565856
15. Mayer KF, Schoof H, Haecker A, Lenhard M, Jürgens G, et al. (1998) Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95 : 805–815. 9865698
16. Payne T, Johnson SD, Koltunow AM (2004) KNUCKLES (KNU) encodes a C2H2 zinc-finger protein that regulates development of basal pattern elements of the Arabidopsis gynoecium. Development 131 : 3737–3749. doi: 10.1242/dev.01216 15240552
17. Lenhard M, Bohnert A, Jürgens G, Laux T (2001) Termination of stem cell maintenance in Arabidopsis floral meristems by interactions between WUSCHEL and AGAMOUS. Cell 105 : 805–814. 11440722
18. Lohmann JU, Hong RL, Hobe M, Busch MA, Parcy F, et al. (2001) A molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell 105 : 793–803. 11440721
19. Sun B, Xu Y, Ng K-H, Ito T (2009) A timing mechanism for stem cell maintenance and differentiation in the Arabidopsis floral meristem. Genes Dev 23 : 1791–1804. doi: 10.1101/gad.1800409 19651987
20. Liu X, Kim YJ, Müller R, Yumul RE, Liu C, et al. (2011) AGAMOUS terminates floral stem cell maintenance in Arabidopsis by directly repressing WUSCHEL through recruitment of Polycomb Group proteins. Plant Cell 23 : 3654–3670. doi: 10.1105/tpc.111.091538 22028461
21. Sun B, Looi L-S, Guo S, He Z, Gan E-S, et al. (2014) Timing mechanism dependent on cell division is invoked by Polycomb eviction in plant stem cells. Science 343 : 1248559. doi: 10.1126/science.1248559 24482483
22. Bowman JL, Smyth DR, Meyerowitz EM (1989) Genes directing flower development in Arabidopsis. Plant Cell 1 : 37–52. doi: 10.1105/tpc.1.1.37 2535466
23. Pandya-Jones A, Black DL (2009) Co-transcriptional splicing of constitutive and alternative exons. RNA 15 : 1896–1908. doi: 10.1261/rna.1714509 19656867
24. Tian B, Manley JL (2013) Alternative cleavage and polyadenylation: the long and short of it. Trends Biochem Sci 38 : 312–320. doi: 10.1016/j.tibs.2013.03.005 23632313
25. Bentley DL (2014) Coupling mRNA processing with transcription in time and space. Nat Rev Genet 15 : 163–175. doi: 10.1038/nrg3662 24514444
26. Ankö M-L, Neugebauer KM (2012) RNA-protein interactions in vivo: global gets specific. Trends Biochem Sci 37 : 255–262. doi: 10.1016/j.tibs.2012.02.005 22425269
27. Müller-McNicoll M, Neugebauer KM (2013) How cells get the message: dynamic assembly and function of mRNA-protein complexes. Nat Rev Genet 14 : 275–287. doi: 10.1038/nrg3434 23478349
28. Cheng Y, Kato N, Wang W, Li J, Chen X (2003) Two RNA binding proteins, HEN4 and HUA1, act in the processing of AGAMOUS pre-mRNA in Arabidopsis thaliana. Dev Cell 4 : 53–66. 12530963
29. Li J, Jia D, Chen X (2001) HUA1, a regulator of stamen and carpel identities in Arabidopsis, codes for a nuclear RNA binding protein. Plant Cell 13 : 2269–2281. 11595801
30. Chen X, Meyerowitz EM (1999) HUA1 and HUA2 are two members of the floral homeotic AGAMOUS pathway. Mol Cell 3 : 349–360. 10198637
31. Lorković ZJ, Barta A (2002) Genome analysis: RNA recognition motif (RRM) and K homology (KH) domain RNA-binding proteins from the flowering plant Arabidopsis thaliana. Nucleic Acids Res 30 : 623–635. 11809873
32. Siomi H, Matunis MJ, Michael WM, Dreyfuss G (1993) The pre-mRNA binding K protein contains a novel evolutionarily conserved motif. Nucleic Acids Res 21 : 1193–1198. 8464704
33. De Boulle K, Verkerk AJ, Reyniers E, Vits L, Hendrickx J, et al. (1993) A point mutation in the FMR-1 gene associated with fragile X mental retardation. Nat Genet 3 : 31–35. doi: 10.1038/ng0193-31 8490650
34. Siomi H, Choi M, Siomi MC, Nussbaum RL, Dreyfuss G (1994) Essential role for KH domains in RNA binding: impaired RNA binding by a mutation in the KH domain of FMR1 that causes fragile X syndrome. Cell 77 : 33–39. 8156595
35. Gao R, Yu Y, Inoue A, Widodo N, Kaul SC, et al. (2013) Heterogeneous nuclear ribonucleoprotein K (hnRNP-K) promotes tumor metastasis by induction of genes involved in extracellular matrix, cell movement, and angiogenesis. J Biol Chem 288 : 15046–15056. doi: 10.1074/jbc.M113.466136 23564449
36. Makeyev AV, Liebhaber SA (2002) The poly(C)-binding proteins: a multiplicity of functions and a search for mechanisms. RNA 8 : 265–278. 12003487
37. Choi HS, Hwang CK, Song KY, Law P-Y, Wei L-N, et al. (2009) Poly(C)-binding proteins as transcriptional regulators of gene expression. Biochem Biophys Res Commun 380 : 431–436. doi: 10.1016/j.bbrc.2009.01.136 19284986
38. Chaudhury A, Chander P, Howe PH (2010) Heterogeneous nuclear ribonucleoproteins (hnRNPs) in cellular processes: Focus on hnRNP E1’s multifunctional regulatory roles. RNA 16 : 1449–1462. doi: 10.1261/rna.2254110 20584894
39. Cao W, Razanau A, Feng D, Lobo VG, Xie J (2012) Control of alternative splicing by forskolin through hnRNP K during neuronal differentiation. Nucleic Acids Res 40 : 8059–8071. doi: 10.1093/nar/gks504 22684629
40. Ji X, Wan J, Vishnu M, Xing Y, Liebhaber SA (2013) αCP Poly(C) binding proteins act as global regulators of alternative polyadenylation. Mol Cell Biol 33 : 2560–2573. doi: 10.1128/MCB.01380-12 23629627
41. Mikula M, Bomsztyk K (2011) Direct recruitment of ERK cascade components to inducible genes is regulated by heterogeneous nuclear ribonucleoprotein (hnRNP) K. J Biol Chem 286 : 9763–9775. doi: 10.1074/jbc.M110.213330 21233203
42. Lim M-H, Kim J, Kim Y-S, Chung K-S, Seo Y-H, et al. (2004) A new Arabidopsis gene, FLK, encodes an RNA binding protein with K homology motifs and regulates flowering time via FLOWERING LOCUS C. Plant Cell 16 : 731–740. doi: 10.1105/tpc.019331 14973162
43. Mockler TC, Yu X, Shalitin D, Parikh D, Michael TP, et al. (2004) Regulation of flowering time in Arabidopsis by K homology domain proteins. Proc Natl Acad Sci USA 101 : 12759–12764. doi: 10.1073/pnas.0404552101 15310842
44. Ripoll JJ, Ferrándiz C, Martínez-Laborda A, Vera A (2006) PEPPER, a novel K-homology domain gene, regulates vegetative and gynoecium development in Arabidopsis. Dev Biol 289 : 346–359. doi: 10.1016/j.ydbio.2005.10.037 16356489
45. Michaels SD, Amasino RM (1999) FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11 : 949–956. 10330478
46. Ripoll JJ, Rodríguez-Cazorla E, González-Reig S, Andújar A, Alonso-Cantabrana H, et al. (2009) Antagonistic interactions between Arabidopsis K-homology domain genes uncover PEPPER as a positive regulator of the central floral repressor FLOWERING LOCUS C. Dev Biol 333 : 251–262. doi: 10.1016/j.ydbio.2009.06.035 19576878
47. Ferrándiz C, Pelaz S, Yanofsky MF (1999) Control of carpel and fruit development in Arabidopsis. Annu Rev Biochem 68 : 321–354. doi: 10.1146/annurev.biochem.68.1.321 10872453
48. Bowman JL (1993) Arabidopsis: An Atlas of Morphology and Development. Berlin & New York: Springer-Verlag. 450 p. 25590127
49. Roeder AHK, Chickarmane V, Cunha A, Obara B, Manjunath BS, et al. (2010) Variability in the control of cell division underlies sepal epidermal patterning in Arabidopsis thaliana. PLoS Biol 8: e1000367. doi: 10.1371/journal.pbio.1000367 20485493
50. Roeder AHK, Cunha A, Ohno CK, Meyerowitz EM (2012) Cell cycle regulates cell type in the Arabidopsis sepal. Development 139 : 4416–4427. doi: 10.1242/dev.082925 23095885
51. Gu Q, Ferrándiz C, Yanofsky MF, Martienssen R (1998) The FRUITFULL MADS-box gene mediates cell differentiation during Arabidopsis fruit development. Development 125 : 1509–1517. 9502732
52. Ferrándiz C, Liljegren SJ, Yanofsky MF (2000) Negative regulation of the SHATTERPROOF genes by FRUITFULL during Arabidopsis fruit development. Science 289 : 436–438. 10903201
53. Liljegren SJ, Roeder AHK, Kempin SA, Gremski K, Østergaard L, et al. (2004) Control of fruit patterning in Arabidopsis by INDEHISCENT. Cell 116 : 843–853. 15035986
54. Dinneny JR, Weigel D, Yanofsky MF (2005) A genetic framework for fruit patterning in Arabidopsis thaliana. Development 132 : 4687–4696. doi: 10.1242/dev.02062 16192305
55. Martínez-Laborda A, Vera A (2009) Arabidopsis fruit development. In: Østergaard L, editor. Annual Plant Reviews Volume 38: Fruit Development and Seed Dispersal. Oxford: Wiley-Blackwell. pp. 172–203.
56. Ripoll JJ, Roeder AHK, Ditta GS, Yanofsky MF (2011) A novel role for the floral homeotic gene APETALA2 during Arabidopsis fruit development. Development 138 : 5167–5176. doi: 10.1242/dev.073031 22031547
57. Ferrándiz C, Gu Q, Martienssen R, Yanofsky MF (2000) Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER. Development 127 : 725–734. 10648231
58. Bowman JL, Baum SF, Eshed Y, Putterill J, Alvarez J (1999) Molecular genetics of gynoecium development in Arabidopsis. Curr Top Dev Biol 45 : 155–205. 10332605
59. Ito T, Wellmer F, Yu H, Das P, Ito N, et al. (2004) The homeotic protein AGAMOUS controls microsporogenesis by regulation of SPOROCYTELESS. Nature 430 : 356–360. doi: 10.1038/nature02733 15254538
60. Ito T, Ng K-H, Lim T-S, Yu H, Meyerowitz EM (2007) The homeotic protein AGAMOUS controls late stamen development by regulating a jasmonate biosynthetic gene in Arabidopsis. Plant Cell 19 : 3516–3529. doi: 10.1105/tpc.107.055467 17981996
61. Liu X, Huang J, Parameswaran S, Ito T, Seubert B, et al. (2009) The SPOROCYTELESS/NOZZLE gene is involved in controlling stamen identity in Arabidopsis. Plant Physiol 151 : 1401–1411. doi: 10.1104/pp.109.145896 19726570
62. Urbanus SL, de Folter S, Shchennikova AV, Kaufmann K, Immink RGH, et al. (2009) In planta localisation patterns of MADS domain proteins during floral development in Arabidopsis thaliana. BMC Plant Biol 9 : 5. doi: 10.1186/1471-2229-9-5 19138429
63. Loke JC, Stahlberg EA, Strenski DG, Haas BJ, Wood PC, et al. (2005) Compilation of mRNA polyadenylation signals in Arabidopsis revealed a new signal element and potential secondary structures. Plant Physiol 138 : 1457–1468. doi: 10.1104/pp.105.060541 15965016
64. Li J, Chen X (2003) PAUSED, a putative exportin-t, acts pleiotropically in Arabidopsis development but is dispensable for viability. Plant Physiol 132 : 1913–1924. 12913148
65. Wang W, Chen X (2004) HUA ENHANCER3 reveals a role for a cyclin-dependent protein kinase in the specification of floral organ identity in Arabidopsis. Development 131 : 3147–3156. doi: 10.1242/dev.01187 15175247
66. Ji L, Liu X, Yan J, Wang W, Yumul RE, et al. (2011) ARGONAUTE10 and ARGONAUTE1 regulate the termination of floral stem cells through two microRNAs in Arabidopsis. PLoS Genet 7: e1001358. doi: 10.1371/journal.pgen.1001358 21483759
67. Jali SS, Rosloski SM, Janakirama P, Steffen JG, Zhurov V, et al. (2014) A plant-specific HUA2-LIKE (HULK) gene family in Arabidopsis thaliana is essential for development. Plant J. doi: 10.1111/tpj.12629.
68. Lin M, Shen X, Chen X (2011) PAIR: the predicted Arabidopsis interactome resource. Nucleic Acids Res 39: D1134–D1140. Database: PAIR. http://www.cls.zju.edu.cn/pair/home.pair. Accessed 10 September 2014. doi: 10.1093/nar/gkq938 20952401
69. Charroux B, Angelats C, Fasano L, Kerridge S, Vola C (1999) The levels of the bancal product, a Drosophila homologue of vertebrate hnRNP K protein, affect cell proliferation and apoptosis in imaginal disc cells. Mol Cell Biol 19 : 7846–7856. 10523673
70. Takada H, Kawana T, Ito Y, Kikuno RF, Mamada H, et al. (2009) The RNA-binding protein Mex3b has a fine-tuning system for mRNA regulation in early Xenopus development. Development 136 : 2413–2422. doi: 10.1242/dev.029165 19542354
71. Hirano T, Matsuzawa T, Takegawa K, Sato MH (2011) Loss-of-function and gain-of-function mutations in FAB1A/B impair endomembrane homeostasis, conferring pleiotropic developmental abnormalities in Arabidopsis. Plant Physiol 155 : 797–807. doi: 10.1104/pp.110.167981 21173023
72. Veitia RA, Bottani S, Birchler JA (2013) Gene dosage effects: nonlinearities, genetic interactions, and dosage compensation. Trends Genet 29 : 385–393. doi: 10.1016/j.tig.2013.04.004 23684842
73. Doyle MR, Bizzell CM, Keller MR, Michaels SD, Song J, et al. (2005) HUA2 is required for the expression of floral repressors in Arabidopsis thaliana. Plant J 41 : 376–385. doi: 10.1111/j.1365-313X.2004.02300.x 15659097
74. Wang Q, Sajja U, Rosloski S, Humphrey T, Kim MC, et al. (2007) HUA2 caused natural variation in shoot morphology of A. thaliana. Curr Biol 17 : 1513–1519. doi: 10.1016/j.cub.2007.07.059 17764945
75. Streitner C, Köster T, Simpson CG, Shaw P, Danisman S, et al. (2012) An hnRNP-like RNA-binding protein affects alternative splicing by in vivo interaction with transcripts in Arabidopsis thaliana. Nucleic Acids Res 40 : 11240–11255. doi: 10.1093/nar/gks873 23042250
76. Gregis V, Sessa A, Colombo L, Kater MM (2006) AGL24, SHORT VEGETATIVE PHASE, and APETALA1 redundantly control AGAMOUS during early stages of flower development in Arabidopsis. Plant Cell 18 : 1373–1382. doi: 10.1105/tpc.106.041798 16679456
77. Liu C, Zhou J, Bracha-Drori K, Yalovsky S, Ito T, et al. (2007) Specification of Arabidopsis floral meristem identity by repression of flowering time genes. Development 134 : 1901–1910. doi: 10.1242/dev.003103 17428825
78. Liu C, Chen H, Er HL, Soo HM, Kumar PP, et al. (2008) Direct interaction of AGL24 and SOC1 integrates flowering signals in Arabidopsis. Development 135 : 1481–1491. doi: 10.1242/dev.020255 18339670
79. Balanzà V, Martínez-Fernández I, Ferrándiz C (2014) Sequential action of FRUITFULL as a modulator of the activity of the floral regulators SVP and SOC1. J Exp Bot 65 : 1193–1203. doi: 10.1093/jxb/ert482 24465009
80. ÓMaoiléidigh DS, Wuest SE, Rae L, Raganelli A, Ryan PT, et al. (2013) Control of reproductive floral organ identity specification in Arabidopsis by the C function regulator AGAMOUS. Plant Cell 25 : 2482–2503. doi: 10.1105/tpc.113.113209 23821642
81. Janakirama P (2013) Functional characterization of the HUA2 gene family in Arabidopsis thaliana. PhD Thesis, University of Western Ontario. Available: http://ir.lib.uwo.ca/etd/1109. Accessed 15 September 2014.
82. Sieburth LE, Meyerowitz EM (1997) Molecular dissection of the AGAMOUS control region shows that cis elements for spatial regulation are located intragenically. Plant Cell 9 : 355–365. doi: 10.1105/tpc.9.3.355 9090880
83. Sheldon CC, Conn AB, Dennis ES, Peacock WJ (2002) Different regulatory regions are required for the vernalization-induced repression of FLOWERING LOCUS C and for the epigenetic maintenance of repression. Plant Cell 14 : 2527–2537. 12368502
84. Hong RL, Hamaguchi L, Busch MA, Weigel D (2003) Regulatory elements of the floral homeotic gene AGAMOUS identified by phylogenetic footprinting and shadowing. Plant Cell 15 : 1296–1309. 12782724
85. Kooiker M, Airoldi CA, Losa A, Manzotti PS, Finzi L, et al. (2005) BASIC PENTACYSTEINE1, a GA binding protein that induces conformational changes in the regulatory region of the homeotic Arabidopsis gene SEEDSTICK. Plant Cell 17 : 722–729. doi: 10.1105/tpc.104.030130 15722463
86. Causier B, Bradley D, Cook H, Davies B (2009) Conserved intragenic elements were critical for the evolution of the floral C-function. Plant J 58 : 41–52. doi: 10.1111/j.1365-313X.2008.03759.x 19054363
87. Schauer SE, Schlüter PM, Baskar R, Gheyselinck J, Bolaños A, et al. (2009) Intronic regulatory elements determine the divergent expression patterns of AGAMOUS-LIKE6 subfamily members in Arabidopsis. Plant J 59 : 987–1000. doi: 10.1111/j.1365-313X.2009.03928.x 19473325
88. Kaida D, Berg MG, Younis I, Kasim M, Singh LN, et al. (2010) U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature 468 : 664–668. doi: 10.1038/nature09479 20881964
89. Berg MG, Singh LN, Younis I, Liu Q, Pinto AM, et al. (2012) U1 snRNP determines mRNA length and regulates isoform expression. Cell 150 : 53–64. doi: 10.1016/j.cell.2012.05.029 22770214
90. Dahan O, Gingold H, Pilpel Y (2011) Regulatory mechanisms and networks couple the different phases of gene expression. Trends Genet 27 : 316–322. doi: 10.1016/j.tig.2011.05.008 21763027
91. Elkon R, Ugalde AP, Agami R (2013) Alternative cleavage and polyadenylation: extent, regulation and function. Nat Rev Genet 14 : 496–506. doi: 10.1038/nrg3482 23774734
92. Naganuma T, Nakagawa S, Tanigawa A, Sasaki YF, Goshima N, et al. (2012) Alternative 3’-end processing of long noncoding RNA initiates construction of nuclear paraspeckles. EMBO J 31 : 4020–4034. doi: 10.1038/emboj.2012.251 22960638
93. Hsin J-P, Manley JL (2012) The RNA polymerase II CTD coordinates transcription and RNA processing. Genes Dev 26 : 2119–2137. doi: 10.1101/gad.200303.112 23028141
94. Chen T, Cui P, Chen H, Ali S, Zhang S, et al. (2013) A KH-domain RNA-binding protein interacts with FIERY2/CTD phosphatase-like 1 and splicing factors and is important for pre-mRNA splicing in Arabidopsis. PLoS Genet 9: e1003875. doi: 10.1371/journal.pgen.1003875 24146632
95. Jiang J, Wang B, Shen Y, Wang H, Feng Q, et al. (2013) The arabidopsis RNA binding protein with K homology motifs, SHINY1, interacts with the C-terminal domain phosphatase-like 1 (CPL1) to repress stress-inducible gene expression. PLoS Genet 9: e1003625. doi: 10.1371/journal.pgen.1003625 23874224
96. Liljegren SJ, Ditta GS, Eshed Y, Savidge B, Bowman JL, et al. (2000) SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature 404 : 766–770. doi: 10.1038/35008089 10783890
97. Alonso-Cantabrana H, Ripoll JJ, Ochando I, Vera A, Ferrándiz C, et al. (2007) Common regulatory networks in leaf and fruit patterning revealed by mutations in the Arabidopsis ASYMMETRIC LEAVES1 gene. Development 134 : 2663–2671. doi: 10.1242/dev.02864 17592013
98. Pérez-Pérez JM, Ponce MR, Micol JL (2004) The ULTRACURVATA2 gene of Arabidopsis encodes an FK506-binding protein involved in auxin and brassinosteroid signaling. Plant Physiol 134 : 101–117. doi: 10.1104/pp.103.032524 14730066
99. An YQ, McDowell JM, Huang S, McKinney EC, Chambliss S, et al. (1996) Strong, constitutive expression of the Arabidopsis ACT2/ACT8 actin subclass in vegetative tissues. Plant J 10 : 107–121. 8758981
100. González-Reig S, Ripoll JJ, Vera A, Yanofsky MF, Martínez-Laborda A (2012) Antagonistic gene activities determine the formation of pattern elements along the mediolateral axis of the Arabidopsis fruit. PLoS Genet 8: e1003020. doi: 10.1371/journal.pgen.1003020 23133401
101. Pfaffl MW, Horgan GW, Dempfle L (2002) Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res 30: e36. 11972351
102. Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22 : 4673–4680. 7984417
103. Gibson DG (2011) Enzymatic assembly of overlapping DNA fragments. Meth Enzymol 498 : 349–361. doi: 10.1016/B978-0-12-385120-8.00015-2 21601685
104. Walter M, Chaban C, Schütze K, Batistic O, Weckermann K, et al. (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40 : 428–438. doi: 10.1111/j.1365-313X.2004.02219.x 15469500
105. Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM (2000) pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol 42 : 819–832. 10890530
106. Smyth DR, Bowman JL, Meyerowitz EM (1990) Early flower development in Arabidopsis. Plant Cell 2 : 755–767. 2152125
Štítky
Genetika Reprodukční medicína
Článek 2014 Reviewer Thank YouČlánek Closing the Gap between Knowledge and Clinical Application: Challenges for Genomic TranslationČlánek Discovery of CTCF-Sensitive Cis-Spliced Fusion RNAs between Adjacent Genes in Human Prostate CellsČlánek A Nitric Oxide Regulated Small RNA Controls Expression of Genes Involved in Redox Homeostasis inČlánek Contribution of the Two Genes Encoding Histone Variant H3.3 to Viability and Fertility in MiceČlánek The Genetic Architecture of the Genome-Wide Transcriptional Response to ER Stress in the Mouse
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 2
-
Všechny články tohoto čísla
- 2014 Reviewer Thank You
- Systematic Cell-Based Phenotyping of Missense Alleles Empowers Rare Variant Association Studies: A Case for and Myocardial Infarction
- African Glucose-6-Phosphate Dehydrogenase Alleles Associated with Protection from Severe Malaria in Heterozygous Females in Tanzania
- Genomics of Divergence along a Continuum of Parapatric Population Differentiation
- microRNAs Regulate Cell-to-Cell Variability of Endogenous Target Gene Expression in Developing Mouse Thymocytes
- A Rolling Circle Replication Mechanism Produces Multimeric Lariats of Mitochondrial DNA in
- Closing the Gap between Knowledge and Clinical Application: Challenges for Genomic Translation
- Partially Redundant Enhancers Cooperatively Maintain Mammalian Expression Above a Critical Functional Threshold
- Discovery of Transcription Factors and Regulatory Regions Driving Tumor Development by ATAC-seq and FAIRE-seq Open Chromatin Profiling
- Mutations in Result in Ocular Coloboma, Microcornea and Cataracts
- A Genome-Wide Hybrid Incompatibility Landscape between and
- Recurrent Evolution of Melanism in South American Felids
- Discovery of CTCF-Sensitive Cis-Spliced Fusion RNAs between Adjacent Genes in Human Prostate Cells
- Tissue Expression Pattern of PMK-2 p38 MAPK Is Established by the miR-58 Family in
- Essential Role for Endogenous siRNAs during Meiosis in Mouse Oocytes
- Matrix Metalloproteinase 2 Is Required for Ovulation and Corpus Luteum Formation in
- Evolutionary Signatures amongst Disease Genes Permit Novel Methods for Gene Prioritization and Construction of Informative Gene-Based Networks
- RR-1 Cuticular Protein TcCPR4 Is Required for Formation of Pore Canals in Rigid Cuticle
- GC-Content Evolution in Bacterial Genomes: The Biased Gene Conversion Hypothesis Expands
- Proteotoxic Stress Induces Phosphorylation of p62/SQSTM1 by ULK1 to Regulate Selective Autophagic Clearance of Protein Aggregates
- K-homology Nuclear Ribonucleoproteins Regulate Floral Organ Identity and Determinacy in Arabidopsis
- A Nitric Oxide Regulated Small RNA Controls Expression of Genes Involved in Redox Homeostasis in
- HYPER RECOMBINATION1 of the THO/TREX Complex Plays a Role in Controlling Transcription of the Gene in Arabidopsis
- Mitochondrial and Cytoplasmic ROS Have Opposing Effects on Lifespan
- Structured Observations Reveal Slow HIV-1 CTL Escape
- An Integrative Multi-scale Analysis of the Dynamic DNA Methylation Landscape in Aging
- Combining Natural Sequence Variation with High Throughput Mutational Data to Reveal Protein Interaction Sites
- Transhydrogenase Promotes the Robustness and Evolvability of Deficient in NADPH Production
- Regulators of Autophagosome Formation in Muscles
- Genomic Selection and Association Mapping in Rice (): Effect of Trait Genetic Architecture, Training Population Composition, Marker Number and Statistical Model on Accuracy of Rice Genomic Selection in Elite, Tropical Rice Breeding Lines
- Eye Selector Logic for a Coordinated Cell Cycle Exit
- Inflammation-Induced Cell Proliferation Potentiates DNA Damage-Induced Mutations
- The DNA Polymerase δ Has a Role in the Deposition of Transcriptionally Active Epigenetic Marks, Development and Flowering
- Contribution of the Two Genes Encoding Histone Variant H3.3 to Viability and Fertility in Mice
- Membrane Recognition and Dynamics of the RNA Degradosome
- P-TEFb, the Super Elongation Complex and Mediator Regulate a Subset of Non-paused Genes during Early Embryo Development
- is a Long Non-coding RNA in JNK Signaling in Epithelial Shape Changes during Drosophila Dorsal Closure
- A Pleiotropy-Informed Bayesian False Discovery Rate Adapted to a Shared Control Design Finds New Disease Associations From GWAS Summary Statistics
- Genome-wide Association Study Identifies Shared Risk Loci Common to Two Malignancies in Golden Retrievers
- and Hyperdrive Mechanisms (in Mouse Meiosis)
- Elevated In Vivo Levels of a Single Transcription Factor Directly Convert Satellite Glia into Oligodendrocyte-like Cells
- Systemic Delivery of MicroRNA-101 Potently Inhibits Hepatocellular Carcinoma by Repressing Multiple Targets
- Pooled Sequencing of 531 Genes in Inflammatory Bowel Disease Identifies an Associated Rare Variant in and Implicates Other Immune Related Genes
- Abscission Is Regulated by the ESCRT-III Protein Shrub in Germline Stem Cells
- Temperature Stress Mediates Decanalization and Dominance of Gene Expression in
- Transcriptome Wide Annotation of Eukaryotic RNase III Reactivity and Degradation Signals
- The Exosome Component Rrp6 Is Required for RNA Polymerase II Termination at Specific Targets of the Nrd1-Nab3 Pathway
- Sex-specific -regulatory Variation on the X Chromosome
- Regulation of Toll-like Receptor Signaling by the SF3a mRNA Splicing Complex
- Modeling of the Human Alveolar Rhabdomyosarcoma Chromosome Translocation in Mouse Myoblasts Using CRISPR-Cas9 Nuclease
- Asymmetry of the Budding Yeast Tem1 GTPase at Spindle Poles Is Required for Spindle Positioning But Not for Mitotic Exit
- TIM Binds Importin α1, and Acts as an Adapter to Transport PER to the Nucleus
- Antagonistic Roles for KNOX1 and KNOX2 Genes in Patterning the Land Plant Body Plan Following an Ancient Gene Duplication
- The Genetic Architecture of the Genome-Wide Transcriptional Response to ER Stress in the Mouse
- Fatty Acid Synthase Cooperates with Glyoxalase 1 to Protect against Sugar Toxicity
- Region-Specific Activation of mRNA Translation by Inhibition of Bruno-Mediated Repression
- An Essential Role of the Arginine Vasotocin System in Mate-Guarding Behaviors in Triadic Relationships of Medaka Fish ()
- Interaction between the tRNA-Binding and C-Terminal Domains of Yeast Gcn2 Regulates Kinase Activity In Vivo
- Hyper-Acetylation of Histone H3K56 Limits Break-Induced Replication by Inhibiting Extensive Repair Synthesis
- Prodomain Removal Enables Neto to Stabilize Glutamate Receptors at the Neuromuscular Junction
- Recent Selective Sweeps in North American Show Signatures of Soft Sweeps
- Identification and Functional Analysis of Healing Regulators in
- A Multi-Megabase Copy Number Gain Causes Maternal Transmission Ratio Distortion on Mouse Chromosome 2
- Drosophila Casein Kinase I Alpha Regulates Homolog Pairing and Genome Organization by Modulating Condensin II Subunit Cap-H2 Levels
- The Hippo Pathway Regulates Homeostatic Growth of Stem Cell Niche Precursors in the Ovary
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Genomic Selection and Association Mapping in Rice (): Effect of Trait Genetic Architecture, Training Population Composition, Marker Number and Statistical Model on Accuracy of Rice Genomic Selection in Elite, Tropical Rice Breeding Lines
- Discovery of Transcription Factors and Regulatory Regions Driving Tumor Development by ATAC-seq and FAIRE-seq Open Chromatin Profiling
- Evolutionary Signatures amongst Disease Genes Permit Novel Methods for Gene Prioritization and Construction of Informative Gene-Based Networks
- Proteotoxic Stress Induces Phosphorylation of p62/SQSTM1 by ULK1 to Regulate Selective Autophagic Clearance of Protein Aggregates
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



