-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaInteraction between the tRNA-Binding and C-Terminal Domains of Yeast Gcn2 Regulates Kinase Activity In Vivo
The survival of all living organisms depends on their capacity to adapt their gene expression program to variations in the environment. When subjected to various stresses, eukaryotic cells modulate general and gene-specific protein synthesis by phosphorylating the α-subunit of eukaryotic translation initiation factor 2 (eIF2α). The yeast Saccharomyces cerevisiae has a single eIF2α kinase, Gcn2, activated by uncharged tRNAs that accumulate in amino acid starved cells, which bind to a regulatory domain homologous to histidyl-tRNA synthetase (HisRS). Gcn2 also contains a C-terminal domain implicated in autoinhibition of Gcn2. Our findings identify a direct interaction between the CTD and a novel regulatory surface in the HisRS domain that is required for inhibition of Gcn2 function in non-starved cells, which is down-regulated by uncharged tRNA. The results further suggest that tRNA binding to the pseudo-active site in the HisRS domain remodels its proximal CTD-binding surface to weaken HisRS/CTD interaction and thereby release the autoinhibitory function of the CTD to activate kinase function. This study provides new molecular insights into how tRNA binding can modulate regulatory interactions among the HisRS, CTD, and kinase domains of Gcn2 to elicit kinase activation.
Published in the journal: . PLoS Genet 11(2): e32767. doi:10.1371/journal.pgen.1004991
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004991Summary
The survival of all living organisms depends on their capacity to adapt their gene expression program to variations in the environment. When subjected to various stresses, eukaryotic cells modulate general and gene-specific protein synthesis by phosphorylating the α-subunit of eukaryotic translation initiation factor 2 (eIF2α). The yeast Saccharomyces cerevisiae has a single eIF2α kinase, Gcn2, activated by uncharged tRNAs that accumulate in amino acid starved cells, which bind to a regulatory domain homologous to histidyl-tRNA synthetase (HisRS). Gcn2 also contains a C-terminal domain implicated in autoinhibition of Gcn2. Our findings identify a direct interaction between the CTD and a novel regulatory surface in the HisRS domain that is required for inhibition of Gcn2 function in non-starved cells, which is down-regulated by uncharged tRNA. The results further suggest that tRNA binding to the pseudo-active site in the HisRS domain remodels its proximal CTD-binding surface to weaken HisRS/CTD interaction and thereby release the autoinhibitory function of the CTD to activate kinase function. This study provides new molecular insights into how tRNA binding can modulate regulatory interactions among the HisRS, CTD, and kinase domains of Gcn2 to elicit kinase activation.
Introduction
Eukaryotic cells harbor stress-activated protein kinases that allow cells to reduce bulk protein synthesis while simultaneously activating the transcription of genes encoding stress management proteins. The target of these kinases is Ser-51 of the α-subunit of translation initiation factor 2 (eIF2α). Phosphorylation of eIF2α reduces the function of eIF2 in recruiting methionyl initiator tRNA to the 40S ribosomal subunit by impairing the recycling of eIF2-GDP to eIF2-GTP by guanine exchange factor eIF2B and thereby reducing the cellular concentration of eIF2·GTP·Met-tRNAiMet ternary complexes. The inhibition of ternary complex assembly diminishes the rate of general translation but enables translation preinitiation complexes to bypass multiple “decoy” AUG start codons in the mRNA leader of GCN4 mRNA and translate the coding sequences for Gcn4, a transcriptional activator of amino acid and vitamin biosynthetic genes in budding yeast (reviewed in [1]). A similar mechanism up-regulates translation of mammalian ATF4 and ATF5 mRNAs when eIF2α is phosphorylated by Gcn2 or one of the other mammalian eIF2α kinases PKR, PERK and HRI [2] [3]. PKR is a key component of the innate immune response, PERK is crucial for responding to ER stress, and HRI couples globin synthesis to heme availability in reticulocytes [4]. Interestingly, rodent Gcn2 mediates the animal’s aversion to amino acid-deficient diets [5], dampens protein synthesis in muscle during leucine starvation [6], and functions in lipid homeostasis [7] and in learning and memory formation [8]. Mammalian Gcn2 has also been implicated in tumor cell survival, innate and T-cell mediated immune responses, and DNA repair (reviewed in [9]); and recently mutations in human Gcn2 were linked to pulmonary hypertension [10]. Hence, elucidating the molecular mechanism of Gcn2 regulation is of importance to multiple aspects of human development and physiology.
Because eIF2α kinases act by inhibiting translation, their functions must be tightly regulated to limit eIF2α phosphorylation to the appropriate stress conditions. The Gcn2 kinase domain (KD) is intrinsically inert and depends on interactions with four other domains within Gcn2 to achieve an active conformation (Fig. 1A) [11]. Latency of Gcn2 KD activity depends on a rigid hinge connecting the N - and C-lobes of the KD, promoting a partially closed active site cleft and occluded ATP-binding pocket, and also on a non-productive orientation of helix αC in the N-lobe that blocks the proper positioning of ATP phosphates for catalysis (Fig. 1B) [12,13]. Binding of uncharged tRNA to the region immediately C-terminal to the KD, related in sequence to histidyl-tRNA synthetase (HisRS) is required to activate Gcn2 in amino acid-starved cells (Fig. 1A) [14,15,16,17]. Authentic HisRS is the enzyme that aminoacylates tRNAHis for protein synthesis. Consistent with the fact that Gcn2 is activated by starvation for any amino acid [15], the Gcn2 HisRS-related domain (henceforth, just HisRS domain) is not specific for binding histidyl tRNA [17]. An N-terminal segment in the HisRS domain that interacts with a portion of the KD containing the hinge is required for kinase activation [18], suggesting that tRNA binding might alter the HisRS-KD interface to evoke an active conformation of the KD. A pseudokinase domain (YKD), incapable of binding ATP or Mg+2 in vitro [19], is located just N-terminal to the KD and is also required for kinase activation (Fig. 1A) [16,20]. Our recent work established that the YKD must interact directly with the KD for kinase activation and identified likely KD-YKD contact sites that can be altered to either impair or constitutively activate Gcn2 kinase function in vivo [21].
Fig. 1. Summary of domain interactions in Gcn2 that couple binding of uncharged tRNA to activation of kinase function in starved cells. 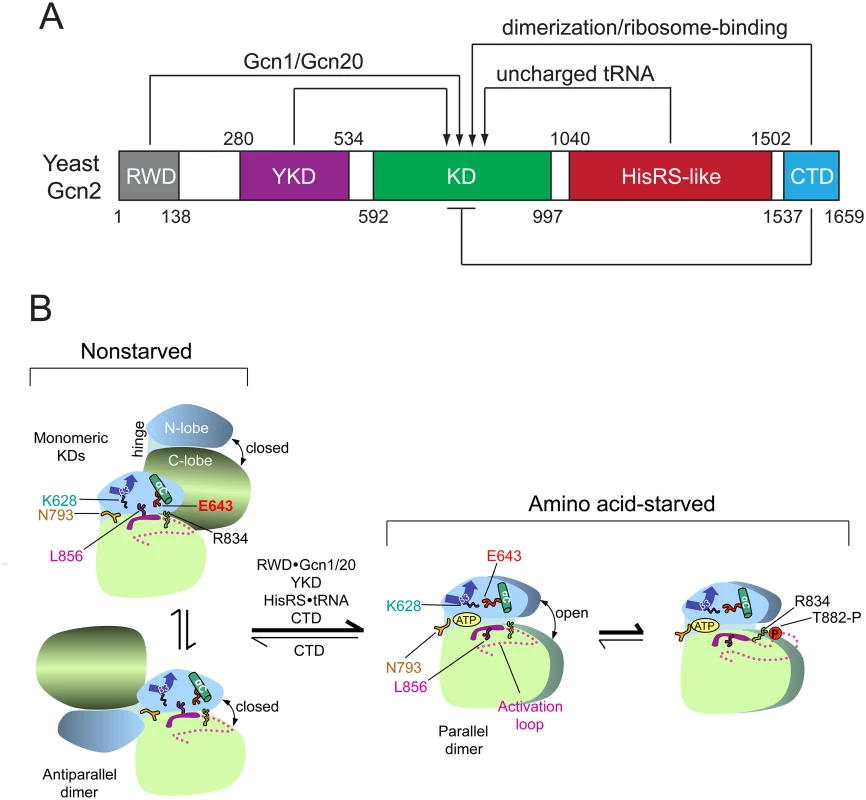
(A) Schematic diagram of the domain structure of yeast Gcn2 and interdomain interactions controlling kinase activity. The amino acid coordinates of the RWD/GI domain (pdb 2yz0), pseudokinase domain (YKD) [21], kinase domain (KD) [12], HisRS-like domain [14], and CTD [26] are derived from crystal structures (RWD, KD, CTD) or multiple sequence alignments (YKD and HisRS-like) of the respective domains. Stimulatory interactions are depicted with arrows, and involve domain interactions with Gcn1/Gcn20 (RWD) or uncharged tRNA (HisRS-like); or dimerization and ribosome-binding activities (CTD). The CTD also mediates autoinhibition of kinase activity, depicted with a bar. (B) Schematic summary of conformational rearrangements in the KD evoked by uncharged tRNA binding to the HisRS domain in amino acid-starved cells. The mode of dimerization and disposition of helix αC, β strand-3 (β3), and the activation loop, as well as key residues in the KD controlling ATP binding and catalysis (Lys-628, Asn-793, Glu-643, Arg-834, and Leu-856) are depicted for the inactive and active states of the Gcn2 KD that prevail in nonstarved or amino acid-starved cells, respectively. It is assumed that, in both states, Gcn2 dimerizes through self-interaction of the CTD and the schematic depicts only the disposition of the KDs within the full-length dimer. At low levels of uncharged tRNA in nonstarved cells (left), the KD exists in an equilibrium between two inactive conformations, with monomeric KDs (upper) or the KDs in an antiparallel dimer (lower). In both, ATP binding is hindered by a closed conformation of the N- and C-lobes and by Gln-793 (N793), which forms a flap over the ATP-binding pocket; and catalysis is blocked by hinge rigidity and rotation of αC to a nonproductive orientation that is stabilized by L856 and the inhibitory E643-R834 salt bridge. CTD/KD domain interaction also promotes this inactive conformation by an unknown mechanism. In amino acid-starved cells, binding of uncharged tRNA to the HisRS domain, along with stimulatory contributions of the RWD domain (engaged with Gcn1/Gcn20), the YKD domain, and the CTD, evokes a parallel mode of KD dimerization with αC properly oriented to form the stimulatory K628-E643 salt-bridge, and with ATP binding enhanced by a reduction in hinge rigidity that facilitates an open conformation of the N- and C-lobes and displaces the inhibitory N793 flap. Autophosphorylation of the activation loop ensues to produce a fully functional kinase locked into the active conformation. (See text for additional details; modified from Garriz et al [13].) The extreme C-terminal domain (CTD) of Gcn2 plays multiple roles in kinase regulation, both positive and negative, including dimerization, ribosome binding, and autoinhibition of the KD (Fig. 1A) [1]. Activation of Gcn2 is dependent on KD dimerization [22] in a back-to-back, parallel orientation (Fig. 1B), as described for the active dimer of PKR [23]. While the KD, HisRS region, and CTD are all capable of self-interaction as isolated domains, only the CTD is essential for dimerization and attendant activation of full-length Gcn2 [24,25]. Gcn2 likely dimerizes constitutively through CTD self-interaction [24], and it is possible that the mode of KD dimerization switches from the antiparallel orientation seen in the crystal structure of the inactive conformation of the Gcn2 KD [12] to the parallel, PKR-like mode of dimerization required for kinase function (Fig. 1B) [22] [23]. Recent work elucidated the three-dimensional structure of the CTD dimer, which is evident with some differences in both yeast and mammalian Gcn2 [26].
In addition to dimerization, the CTD mediates ribosome association of Gcn2 (Fig. 1A) [27], which depends on conserved basic residues that mediate RNA binding by the CTD in vitro and are crucial for activation of Gcn2 in vivo [28]. Gcn2 activation also requires trans-acting factors Gcn1 and Gcn20, which form a complex that must interact with both the N-terminal “RWD” domain of Gcn2 and translating ribosomes for Gcn2 activation in starved yeast cells [29,30,31,32,33]. These and other findings [34], support the model that Gcn2 is activated by uncharged tRNA that binds first to the decoding center of a translating ribosome and is subsequently transferred to the HisRS domain in Gcn2, with Gcn1/Gcn20 enhancing one or both of these reactions involving uncharged tRNA [33]. However, stable association of mammalian Gcn2 with ribosomes was not observed [26], and it was proposed that the RNA binding activity of the CTD supports tRNA binding by mammalian Gcn2, in the manner described previously for yeast Gcn2 [17].
The yeast Gcn2 CTD also appears to interact with the KD in a manner that impedes kinase activation (Fig. 1A) [17,18], as a mutation that constitutively activates Gcn2 kinase function, GCN2c-E803V (substitutes Glu-803 for Val in the KD) weakens interaction between the isolated KD and CTD and also increases tRNA binding by Gcn2 in vitro [17,18]. Consistent with an autoinhibitory function, eliminating the CTD from mouse Gcn2 activates eIF2α phosphorylation and abrogates stimulation by uncharged tRNA in vitro [35]. The finding that tRNA competed for interaction between the isolated KD and a HisRS-CTD segment of yeast Gcn2 in vitro [17] led to the proposal that the HisRS and CTD domains both dissociate from the KD on tRNA binding. However, complete dissociation of the HisRS domain seems incompatible with the subsequent finding that an N-terminal segment of the HisRS region interacts with the KD and is crucial for Gcn2 activation at a step following tRNA binding, suggesting that this portion of the HisRS domain remains engaged with the KD in the activated state [18]. In addition to the autoinhibitory CTD-KD interaction, the CTD mediates an inhibitory interaction with translation elongation factor eEF1A that can be overcome by uncharged tRNA [36].
While it is clear that tRNA binding to the HisRS domain is required for activation, and a stimulatory interaction of the HisRS-N region with the KD seems likely, it was unclear how tRNA binding might antagonize the autoinhibitory KD-CTD interaction and simultaneously promote stimulatory association of the YKD with the KD. In an effort to answer this question, we identified substitutions in the HisRS region that restore kinase activation by the gcn2-m2 variant, which harbors substitutions in conserved residues of the predicted HisRS active site cleft that impair tRNA binding in vitro and kinase activation in vivo [15,16,17]. We reasoned that such m2 suppressors could alter a regulatory surface in the HisRS whose interactions with another domain are modulated by tRNA binding in a manner mimicking the tRNA-bound state of WT Gcn2. Interestingly, the locations of these suppressors led us to identify a regulatory patch predicted to be surface-exposed and proximal to the region in the HisRS domain corresponding to the active site of authentic HisRS, below dubbed the “pseudo-active site”, which can be altered to either activate or impede Gcn2 function. Our finding that certain of the (Gcn-) inactivating substitutions strengthen HisRS-CTD interaction without affecting tRNA binding in vitro implies that one stimulatory consequence of tRNA binding is to weaken HisRS-CTD association. This inference leads to an appealing model for how tRNA binding releases the autoinhibitory KD-CTD interaction, promotes YKD-KD association, and thereby activates Gcn2.
Results
Identification of Gcd- substitutions in the HisRS domain as suppressors of the m2 mutation
In an effort to identify residues in the Gcn2 HisRS domain involved in regulation of kinase function by uncharged tRNA, we randomly mutagenized the coding sequence for the HisRS domain in a plasmid-borne gcn2-m2 allele and selected for suppressors of the sensitivity to 3-aminotriazole (3-AT) conferred by this allele in yeast cells. The m2 mutation substitutes two residues in highly conserved motif 2 in the pseudo-active site of the HisRS domain, impairing tRNA binding by Gcn2 in vitro and abolishing activation of Gcn2 kinase function in vivo [15,17]. Defective activation of Gcn2 confers sensitivity to 3-AT, an inhibitor of histidine biosynthesis, by preventing Gcn2-dependent induction of GCN4 translation and attendant derepression of histidine biosynthetic enzymes under Gcn4 control. Thus, transformants of a gcn2∆ strain harboring WT GCN2 or GCN2c-M788V, whose product is constitutively activated [37], grow well on 3-AT medium, whereas gcn2-m2 transformants do not (Fig. 2A, rows 1–3).
Fig. 2. Substitutions in the HisRS domain suppress the m2 mutation and constitutively activate Gcn2 in vivo. 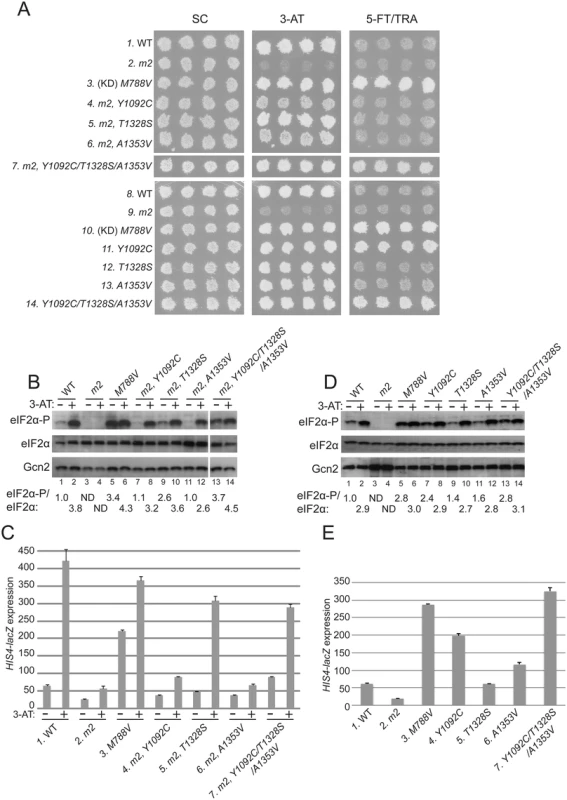
(A) Transformants of gcn2Δ strain H1149 containing derivatives of low-copy plasmid p722 with wild-type GCN2, gcn2-m2, GCN2c-M788V, or the indicated mutations affecting residues in the HisRS domain either in combination with m2 (rows 4–7) or separated from m2 (rows 11–14) were replica-plated to synthetic complete medium lacking uracil (SC-Ura), SC-Ura plus 30 mM 3-AT, or minimal synthetic medium (SD) supplemented with 0.5 mM 5-FT and 0.125 mM TRA (5FT/TRA) and incubated for 3 d at 30°C. For rows 3 & 10, (KD) signifies a kinase domain substitution by M788V. Images were cropped from results obtained from different plates examined in parallel in the same experiment. (B and D) Cultures of strains from panel A were grown in liquid SC medium lacking uracil and histidine (SC-Ura-His) to saturation, diluted into fresh SC-Ura-His at A600 of ≈0.2, and grown for 6 h at 30°C. 3-AT was added at 10 mM to one culture for 1 h before harvesting (even-numbered lanes). WCEs were resolved by SDS-PAGE and subjected to Western analysis using the indicated specific antibodies and enhanced chemiluminescence to detect immune complexes. (Note that strains examined in panel B harboring suppressor mutations also contain m2; whereas m2 is absent in the strains analyzed in panel D.) Western signals on the upper panel (eIF2α-P) were quantified by scanning densitometry of exposed films using ImageJ software, normalized for the corresponding signals in the middle panel (total eIF2α), and the mean ratios of the two signals (eIF2α-P/eIF2α) calculated from replicate measurements are indicated below the corresponding lanes. Standard errors are less than 6.5% of the mean values shown. The results in lanes 13–14 were cropped from the same Western blot containing lanes 1–12. (C and E) Strains from (A) were cultured in both nonstarvation and starvation conditions (C), or only in nonstarvation conditions (E), as described in Materials and Methods, and WCEs were prepared and assayed for β-galactosidase activities. Results are the means and S.E.M.s calculated from three transformants, with activity expressed as nanomoles of o-nitrophenyl-β-D-galactopyronoside hydrolyzed per minute per milligram of protein. (Note that strains examined in panel C harboring suppressor mutations also contain m2; whereas m2 is absent in the strains analyzed in panel E.) Interestingly, we identified 3 mutations in the HisRS domain that suppress the 3-ATS phenotype of the m2 mutation, with the strongest growth on 3-AT displayed by the m2,T1328S mutant (Fig. 2A, 3-AT, rows 4–6 vs. 2). As expected, a mutant allele combining all three suppressors with m2 also confers a strong 3-ATR phenotype (Fig. 2, row 7). Comparison of the single and triple suppressor alleles at elevated temperature (37°), which exacerbates sensitivity to 3-AT, reveals that combining the suppressor mutations in one allele confers greater resistance to 3-AT than that given by any of the single suppressors (S1 Fig.).
The allele combining all three suppressors with m2 additionally conferred resistance to a combination of tryptophan analog 5-fluorotryptophan and histidine analog triazolealanine (Fig. 2A, 5-FT/TRA, row 7). The 5FTR/TRAR phenotype signifies constitutive, Gcn4-mediated derepression of tryptophan and histidine biosynthetic enzymes, known as the Gcd- phenotype [37]. Accordingly, the GCN2c-M788V allele confers growth on 5-FT/TRA medium, whereas GCN2+ cells, and Gcn- strains like gcn2-m2, are sensitive to these analogs (Fig. 2A, rows 1–3). GCN2c-M788V alters the ATP binding pocket of the KD to elevate kinase activity at low levels of uncharged tRNA [12,37]. Thus, it appears that combining all three m2 suppressors confers constitutive activation of Gcn2, even in the presence of the m2 mutation.
In accordance with their suppression of the 3-ATS phenotype of m2, all three suppressor mutations also restored Gcn2 kinase function under starvation conditions. Western blot analysis of whole cell extracts (WCEs) revealed that 3-AT evokes the expected increase in eIF2α phosphorylated on Ser-51 (eIF2α-P) relative to total eIF2α in GCN2 cells, whereas m2 cells have no detectable eIF2α-P; and M788V cells display high-level eIF2α with and without 3-AT treatment (Fig. 2B, lanes 1–6). Importantly, each of the suppressor alleles restored 3AT induction of eIF2α-P in m2 cells, without increasing Gcn2 abundance (Fig. 2B, lanes 7–14). In agreement with its 5FTR/TRAR phenotype, the m2 mutant harboring all three suppressors also displayed a greater than WT level of eIF2α-P in nonstarvation conditions (Fig. 2B, lane 13 vs. 1), indicating constitutive activation of Gcn2. Consistent with these findings, the m2 suppressors increase expression of a Gcn4-dependent HIS4-lacZ reporter [37]. HIS4-lacZ expression in 3-AT-starved cells is ~8-fold lower in m2 versus WT cells, and each of the mutants containing one or more suppressor mutations displays substantially higher reporter expression in 3-AT treated cells compared to that seen in the m2 single mutant, although only a slight increase was observed for the A1353V suppressor (Fig. 2C). The particularly large increases in HIS4-lacZ expression observed for the m2 strains harboring T1328S or the triple suppressor mutation are consistent with their marked 3-AT-resistant phenotypes (Fig. 2A).
It is noteworthy that all of the suppressor strains display an induction of eIF2α-P in response to 3-AT treatment (Fig. 2B, lanes 7–14, 3-AT + vs.–), implying that their Gcn2 variants can be activated by uncharged tRNA accumulating in histidine-deprived cells. As demonstrated below, the m2 mutation reduces, but does not abolish tRNA binding by Gcn2 in vitro. It was possible, therefore, that the suppressor mutations overcome the activation defect of m2 simply by restoring robust tRNA binding by the HisRS domain. Alternatively, they could increase the ability of low-levels of tRNA bound by the m2 variant of the HisRS domain to activate Gcn2. If the latter was true, we reasoned that the suppressor mutations should elevate eIF2α phosphorylation when separated from the m2 mutation in nonstarvation conditions by enabling Gcn2 activation by the low, basal level of uncharged tRNA present in amino acid-replete cells. Consistent with this last prediction, when separated from m2, the Y1092C and triple suppressor mutations each evoked strong resistance to 5-FT/TRA (Fig. 2A, rows 10,11,14). They also conferred marked increases in eIF2α-P (Fig. 2D, lanes 5,7,13) and HIS4-lacZ expression (Fig. 2E, lanes 3,4,7) under nonstarvation conditions, comparable in degree to that given by GCN2c-M788V. The A1353V single mutation conferred smaller increases in eIF2α-P accumulation and derepression of HIS4-lacZ (Fig. 2D, lane 1 vs. 11; Fig. 2E, column 1 vs. 6). Thus, it appears that the triple suppressor mutation, Y1092C, and to a lesser extent A1353V increase the ability of low-level uncharged tRNA present in nonstarved cells to stimulate Gcn2 kinase function. The triple mutant was chosen as the exemplar Gcd- variant for subsequent biochemical studies described below.
Surprisingly, despite being the most effective suppressor of m2, the T1328S single mutation produced only a slight increase in eIF2α-P (Fig. 2D) and no increase in resistance to 5-FT/TRA or HIS4-lacZ expression (Fig. 2A & E). Thus, although T1328S restores robust activation of the m2 variant in starved cells, it does not appreciably activate otherwise WT Gcn2 in nonstarvation conditions.
m2 suppressors and GCN2c substitutions alter residues in the HisRS domain pseudo-active site
To evaluate the locations of the m2 suppressors in the predicted structure of the HisRS domain, we constructed a multiple sequence alignment of Gcn2 HisRS domain sequences from various fungal species (S2 Fig.), and also an alignment of a subset of these HisRS domain sequences with authentic HisRS enzymes from diverse eukaryotic species (S3 Fig.). The latter reveals regions of considerable sequence similarity between authentic HisRS and the Gcn2 HisRS domains spanning the region extending from motifs 1 and 2, conserved in all class II aminoacyl tRNA synthetases, portions of the insertion domain between motifs 2 and 3 and the HisA and HisB motifs unique to HisRS enzymes, and the N-terminal half of class II motif 3. Interestingly, the three m2 suppressors Y1092C, T1328S, and A1353V alter residues in the vicinity of motif 2, HisB, and within motif 3, respectively (Fig. 3A and S3 Fig.). Conserved residues of these motifs include active site residues that directly contact different moieties of the intermediate HAM formed in the first step of tRNA aminoacylation (Fig. 3A and S3 Fig., residues labeled with H (histidyl), P (phosphate), S (sugar), or A (adenine)). These critical residues are color-coded in the “ribbons” depiction of the crystal structure of the T. cruzi HisRS-HAM complex shown in Fig. 3C (salmon (H), cyan (P), orange (S), or dark gray (A)). Interestingly, six GCN2c mutations described previously [37] also alter residues located in or nearby the conserved HisRS motifs in the primary sequence, including F1134L and D1138N (motif 2), A1197G (insertion domain), N1295D and H1308Y (near HisA), and G1338D (motif 3) (Fig. 3A and S3 Fig.).
Fig. 3. Gcd- substitutions in the HisRS domain cluster near the predicted pseudo-active site. 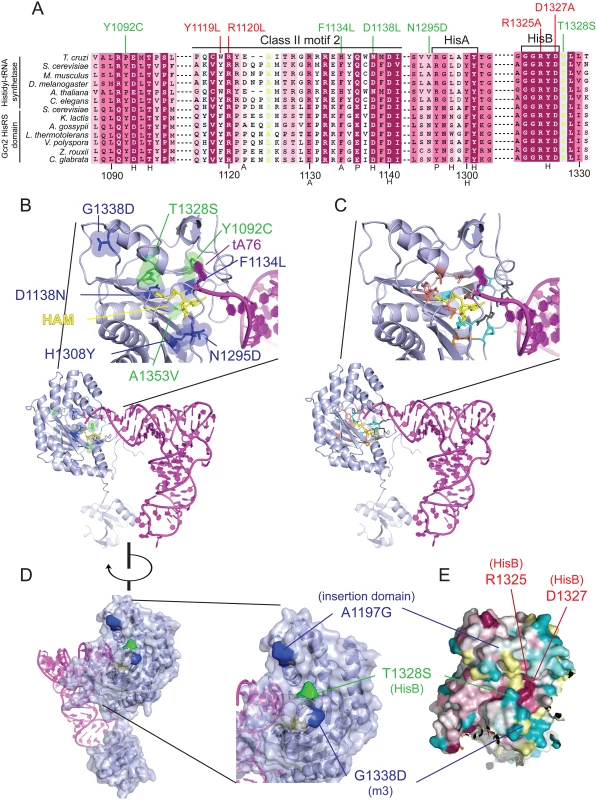
(A) Excerpt of the multiple sequence alignment shown in S2 Fig. of authentic HisRSs and HisRS domains from fungal Gcn2 sequences, identified on the left by abbreviations of their species of origin, built using the MUSCLE program. Residues are colored according to evolutionary sequence variation as analyzed with the CONSURF on-line server, with dark magenta corresponding to the most conserved residues. Yellow color indicates residues for which the data were insufficient to calculate a reliable conservation grade. Numbering corresponds to residue positions in full-length S. cerevisiae Gcn2 (residues 1088–1331). Regions of predicted conserved motifs are denoted above the T. cruzi HisRS sequence. Substitutions conferring Gcn- or Gcd- phenotypes are shown in red and green, respectively. Residues interacting directly with HAM in T. cruzi HisRS are indicated by black letters below the sequence, with H/P/A signifying interaction with histidyl/phosphate/adenine moieties, respectively. (B) Localization of Gcd− substitutions in the predicted 3-D structure of a monomer of the Gcn2 HisRS domain in complex with an uncharged tRNA, based on the crystal structure of T. cruzi authentic HisRS (PDB: 3HRK) and co-crystal structure of S. cerevisiae AspRS-tRNAAsp (Materials and Methods). Gcd- substitutions identified here are in green, while previously identified GCN2c substitutions are in blue. (C) The locations of T. cruzi HisRS (PDB: 3HRK) residues interacting directly with the histidyl/phosphate/sugar/adenine moieties of HAM (in yellow) are indicated in salmon/cyan/orange/dark gray, respectively. (D) Localization of surface-exposed Gcd- substitutions on the predicted 3-D structure of the HisRS domain, colored as in (B). (E) The degree of evolutionary conservation of Gcn2 HisRS domain residues shown in S2 Fig. was projected onto the 3-D structure of T. cruzi authentic HisRS monomer using the CONSURF program, with magenta/dark cyan corresponding to the most/least conserved residues, respectively, and yellow residues corresponding to indeterminate conservation. Because the structure of the Gcn2 HisRS domain is unknown, we used the sequence alignment between Gcn2 and authentic HisRSs (S3 Fig.) and the crystal structure of T. cruzi authentic HisRS to predict the locations of m2 suppressors and GCN2c substitutions in the three-dimensional structure of the Gcn2 HisRS domain (Fig. 3B). It is striking that all three m2 suppressors and 5 of the 6 previously identified GCN2c mutations alter residues within, or in proximity to, the pseudo-active site of the HisRS domain (green residues: m2 suppressors; blue residues previously known GCN2c mutations). In fact, several mutations alter residues corresponding to amino acids in HisRS that make direct contacts with the adenine (F1134L), histidyl (D1138N), or ribose (A1353V) moiety, while others are located only one or two residues away in the polypeptide chain from amino acids contacting the histidyl (Y1092C and T1328S) or phosphate (N1295) moiety of HAM (Fig. 3A, S3 Fig.; cf. Fig. 3B-C). In the cases of T1328S, A1197V and G1338D, these residues are predicted to be surface-exposed and (at least for T1328S and G1338D) in proximity to one another (Fig. 3D-E) at the “top” of the predicted pseudo-active site cleft of the Gcn2 HisRS domain (Fig. 3B).
The predicted locations of these last three substitutions led us to consider a model in which this surface of the HisRS domain interacts with another region in Gcn2 to regulate kinase function in a manner that is modulated by binding of uncharged tRNA to the pseudo-active site. In this view, the putative regulatory interaction involving this patch of the HisRS domain would be altered by the Gcd- m2 suppressors and GCN2c substitutions mapping in the HisRS domain in a way that mimics the effect of uncharged tRNA binding to the pseudo-active site of the WT Gcn2 HisRS domain.
Identification of Gcn- substitutions in the predicted HisRS pseudo-active site
We reasoned that if the foregoing hypothesis is correct, then it should be possible to isolate Gcn- substitutions affecting the same exposed surface of the HisRS domain altered by the m2 suppressor T1328S and Gcd- substitution G1338D, but with the opposite effect on its putative regulatory interactions with other Gcn2 domains. To test this idea, we first determined the degree of sequence conservation of residues on this face of the HisRS domain by projecting the sequence conservation scores obtained from the alignment of fungal Gcn2 HisRS domains (S2 Fig.) onto a surface representation of the crystal structure of T. cruzi HisRS (Fig. 3E). We then determined the phenotypes conferred by substituting two highly conserved residues, Arg-1325 and Asp-1327, which are surface exposed and located in proximity both to one another and the residues altered by T1328S and G1338D (Fig. 3D-E).
Strikingly, substitutions of Arg-1325 with Ala or Glu (R1325A, R1325E) and substitutions of Asp-1327 with Ala or Lys (D1327A, D1327K) completely abrogate Gcn2 function. Thus, all four substitutions confer strong sensitivity to 3-AT (Fig. 4A), eliminate detectable eIF2α-P in both nonstarvation and starvation conditions (Fig. 4B), and evoke low basal expression of the HIS4-lacZ reporter at levels comparable to, or even below, that given by the m2 mutation (Fig. 4C); and all of these Gcn- phenotypes occur without any reduction in the level of Gcn2 itself (Fig. 4B). These findings are consistent with the possibility that highly conserved residues Arg-1325 and Asp-1327 are critical constituents of a regulatory patch exposed on the surface of the HisRS domain near the pseudo-active site cleft (Fig. 3E).
Fig. 4. Mutations of conserved surface-exposed residues of the Gcn2 HisRS domain that impair activation of Gcn2 in vivo. 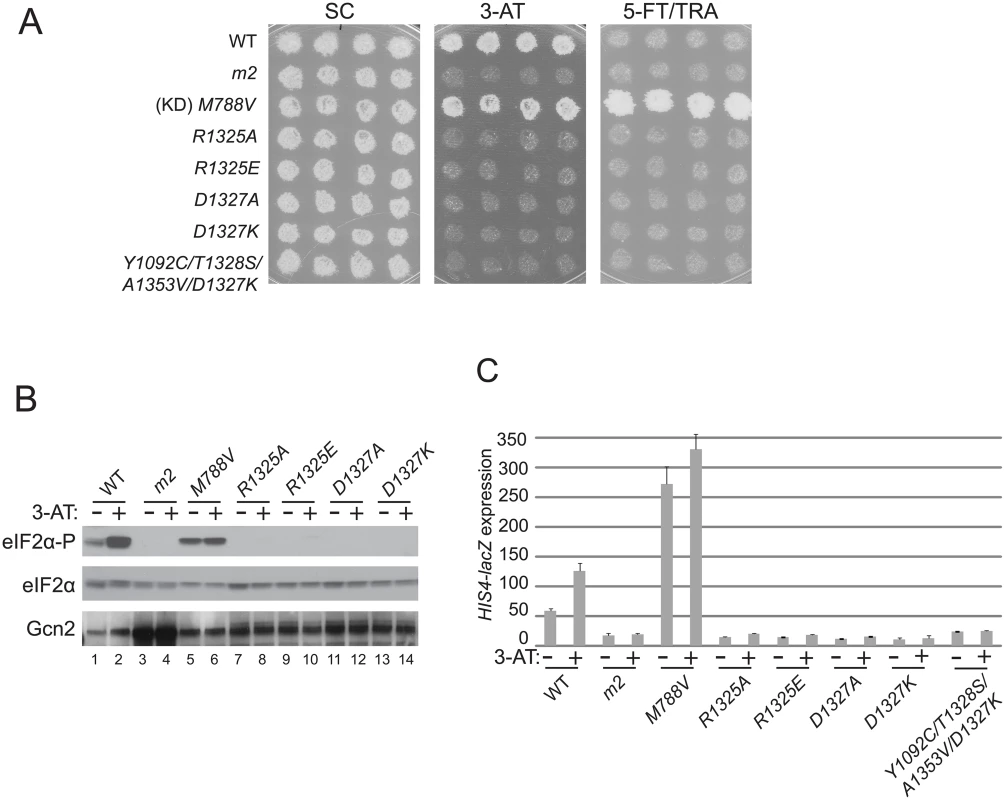
(A) Growth phenotypes of transformants of gcn2Δ strain H1149 containing the indicated plasmid-borne GCN2 alleles were analyzed as in Fig. 2A. (B) Cultures of strains from panel A were analyzed for levels of eIF2α-P as in Fig. 2B. (C) HIS4-lacZ expression was analyzed in strains from (A) as in Fig. 2C. Interestingly, a mutant combining the strong Gcn- mutation D1327K with the Gcd- triple m2 suppressor Y1092C/T1328S/A1353V described above exhibits a 3-AT-sensitive phenotype (Fig. 4A) and a defect in derepression of HIS4-lacZ expression (Fig. 4C) nearly indistinguishable from those seen for the D1327K single mutant, indicating that the strong activation defect conferred by D1327K is epistatic to the constitutively activating phenotype of the Gcd- suppressor substitutions.
Gcd- m2 suppressors and Gcn- substitutions of Asp-1327 in the HisRS domain do not alter tRNA binding by Gcn2
We proposed above that the Gcd- substitutions identified as m2 suppressors restore Gcn2 kinase function to the m2 variant by altering a regulatory interaction of the HisRS domain in a way that mimics the effect of uncharged tRNA and allows for Gcn2 activation at low levels of bound tRNA. To bolster this view and eliminate the alternative possibility that they simply overcome the effect of m2 of impairing tRNA binding, we purified the gcn2-m2 product and the Gcn2 variant harboring the m2 substitutions in combination with all three suppressor substitutions, and compared them to WT Gcn2 for binding [32P]-labeled total tRNA using a gel mobility shift assay to detect Gcn2-tRNA complexes. In accordance with previous results [15,17], the m2 product displayed an obvious defect in tRNA binding compared to WT Gcn2; however, unlike the results of our previous studies, it retained appreciable tRNA binding activity (Fig. 5A). (This disparity in results might be attributable to the fact that, unlike the gcn2-m2 protein examined here, this variant was unstable and subject to degradation when purified from a different yeast strain used in previous studies [17].) The fact that m2 does not abolish tRNA binding in vitro but completely impairs activation of Gcn2 in vivo might indicate that the m2 substitutions in the pseudo-active site cleft impair a regulatory interaction of the HisRS domain with another region in Gcn2 in addition to reducing tRNA binding. Importantly, the presence of all three suppressors in a quadruple mutant harboring the m2 substitutions did not increase the tRNA binding activity compared to that measured for gcn2-m2 (Fig. 5A). These findings are consistent with our conclusion that the suppressor substitutions restore eIF2α-P formation by enhancing kinase function at the low tRNA occupancy permitted by the m2 substitutions, rather than restoring high-level tRNA binding to the HisRS domain.
Fig. 5. A subset of Gcn- substitutions of conserved surface-exposed residues in the HisRS domain impair kinase activity but not tRNA binding by purified Gcn2 in vitro. 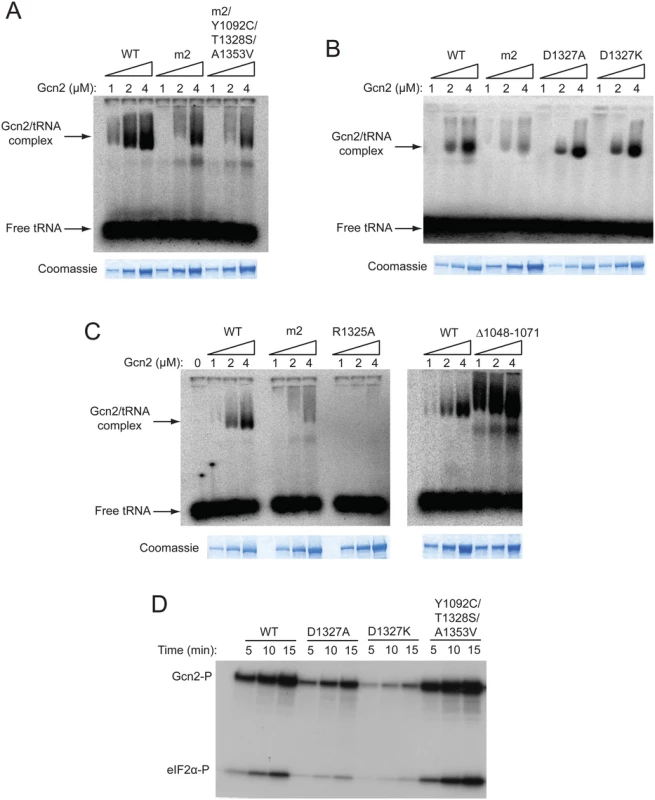
(A, B and C) The indicated purified Gcn2 proteins were incubated with [32P]-labeled total yeast tRNA in 20 μL of GMSA buffer. The Gcn2-tRNA complexes were resolved by electrophoresis through a 1% agarose gel in 1×MOPS buffer (1.5h, 100 V), transferred to a nitrocellulose membrane and visualized by autoradiography. Unbound [32P]-tRNA, which has a higher mobility, was present at essentially identical amounts in each lane at levels ~15-fold higher than the WT Gcn2/tRNA complexes formed at 4μM. The purified Gcn2 proteins used in the assays were visualized by staining with Coomassie brilliant blue following separation by SDS-PAGE (images labelled Coomassie). (D) The indicated Gcn2 proteins (ca. 0.25 μg) were incubated with 3 μCi of [γ-32P]ATP (6000 Ci/mmol, Amersham), 1 μg of recombinant eIF2α−ΔC purified from E. coli, and 0.5 μg of bovine serum albumin in 20 μL of kinase assay buffer at 30°C for the indicated times. Samples were resolved by 8%–16% SDS—PAGE and subjected to autoradiography. Positions of autophosphorylated Gcn2 (Gcn2-P) and phosphorylated eIF2α−ΔC (eIF2α-P) are indicated. We also examined whether Gcn- substitutions affecting the conserved, surface-exposed residues Arg-1325 and Asp-1327 proximal to the pseudo-active site affect tRNA binding. Importantly, we saw little or no effect on tRNA binding by the Gcn- substitutions D1327A and D1327K (Fig. 5B), implying that they impair activation of Gcn2 by disrupting the ability of bound tRNA to trigger activation of kinase function rather than reducing the amount of bound tRNA. By contrast, the Gcn- substitution of Arg-1325, R1325A, abolished tRNA binding by Gcn2 (Fig. 5C), making it likely that substitutions of this residue impair Gcn2 activation by reducing the level of bound tRNA, although they could also disrupt the proposed regulatory interactions involving the HisRS pseudo-active site. In accordance with previous findings, deletion of HisRS residues 1048–1071 evokes a greater than WT level of tRNA binding by Gcn2 (Fig. 5C), supporting our previous conclusion that removing this N-terminal segment of the HisRS domain impairs Gcn2 activation by disrupting a stimulatory interaction of the tRNA-bound HisRS domain with the KD rather than impairing tRNA binding by Gcn2 [18]. Based on its reduced electrophoretic mobility, the gcn2-∆1048–1071 variant might also exhibit a less compact conformation compared to WT Gcn2.
We wished to confirm that the key regulatory mutations of interest, the Gcn- substitutions D1327A and D1327K, and the Gcd- triple substitution Y1092C/T1328S/A1353V, alter Gcn2 kinase activity in vitro in the manner predicted by their phenotypes in vivo. To this end, we conducted in vitro kinase assays with the relevant purified Gcn2 proteins using [γ-32P]-ATP and a truncated form of recombinant eIF2α as substrates, and employed SDS-PAGE/autoradiography to detect the reaction products. It was shown previously that WT Gcn2 displays similar kinase activity whether purified from starved or nonstarved cells, but that the m2 mutation reduces kinase activity in vitro, indicating that WT Gcn2 becomes activated in vitro by deacylated tRNA in cell lysates prior to purification [16]. Thus, although yeast Gcn2 cannot be activated further by adding tRNA to kinase assays, the activity levels of Gcn2 variants with HisRS domain substitutions should reflect their abilities to be activated by tRNA during purification. Consistent with their Gcn- phenotypes, the D1327A and D1327K variants also exhibit substantially reduced autophosphorylation and eIF2α substrate phosphorylation activities in vitro (Fig. 5D). Moreover, the Gcd- variant Y1092C/T1328S/A1353V exhibits an obvious increase in kinase activity relative to WT Gcn2 (Fig. 5D).
Gcn- substitutions of Asp-1327 strengthen HisRS interaction with the CTD
Our finding that Gcn- variants D1327A/D1327K are completely defective for Gcn2 activation (Fig. 4A) but retain robust tRNA binding activity (Fig. 5B) made them good candidates for mutations that alter a regulatory interaction of the HisRS region that mediates allosteric activation of kinase function by uncharged tRNA. Previously, we demonstrated that distinct segments of the isolated HisRS domain interact with the isolated KD or CTD of Gcn2 [18]. As noted above, the N-terminal HisRS segment (HisRS-N) interacts with the KD and the Δ1048–1071 deletion in this region impairs Gcn2 activation without reducing tRNA binding, thus identifying a stimulatory HisRS-N/KD interaction [18]. Moreover, the C-terminal HisRS segment (HisRS-C) was shown to interact with the CTD [18], and as it encompasses Asp-1327, we hypothesized that the Gcn- D1327A/D1327K substitutions impair Gcn2 function by altering the HisRS-CTD interaction.
We obtained evidence supporting this hypothesis using the yeast two-hybrid assay. In agreement with previous results [24], a LexA-fusion to the WT Gcn2 HisRS domain shows little interaction with a fusion of the B42 activation domain to the CTD. Remarkably, introducing Gcn- substitutions D1327A or D1327K into the lexA-HisRS fusion greatly enhanced this two-hybrid interaction (Fig. 6A). By contrast, the Gcd- triple substitution Y1092C/T1328S/A1353V had no significant effect on the HisRS/CTD interaction when introduced into otherwise WT lexA-HisRS. Interestingly, however, these Gcd- substitutions diminished the enhanced two-hybrid interaction conferred by the D1327K Gcn- substitution (Fig. 6A).
Fig. 6. Gcn- substitutions D1327A/D1327K strengthen HisRS/CTD domain interaction in a manner antagonized by uncharged tRNA. 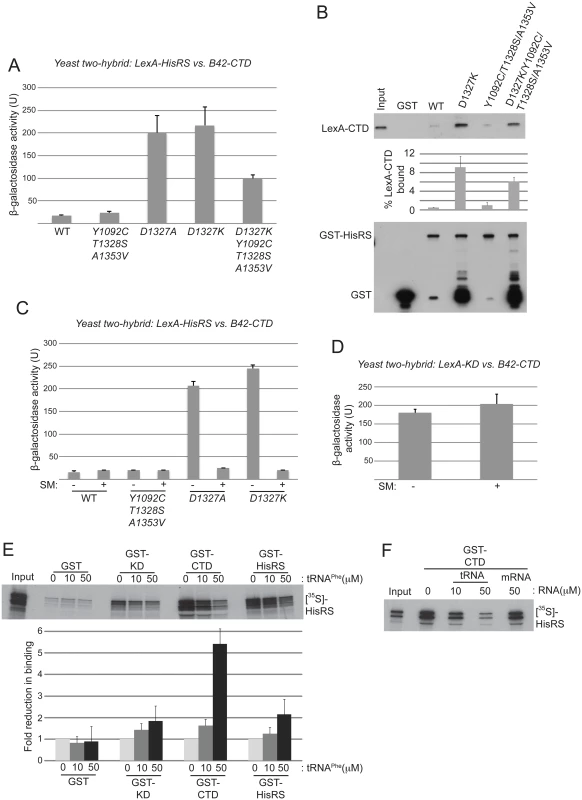
(A) Yeast two-hybrid analysis of domain interactions. Strain EGY48 carrying the lexAop-lacZ reporter plasmid was co-transformed with plasmids encoding lexA-HisRS (Gcn2 residues 970–1497) (with or without the indicated substitutions) and B42-CTD (Gcn2 residues 1497–1659). Strains were cultured and β-galactosidase activities were measured in WCEs. (B) In vitro binding of LexA-CTD (Gcn2 residues 1498–1659), expressed in yeast, to GST fusion proteins containing the Gcn2 HisRS domain (residues 970–1497), either WT or containing the indicating mutations. Aliquots of nuclease-treated whole-cell extracts (500 μg of total protein) from transformants of yeast gcn2Δ strain HQY132 carrying plasmids containing the LexA-CTD fusion protein were incubated with approximately equivalent amounts of GST or GST-HisRS (970–1497) fusion proteins immobilized on glutathione-Sepharose beads. After extensive washing, LexA fusion proteins bound to the GST proteins were resolved by SDS-PAGE and detected by immunoblot analysis with anti-LexA (Top) and anti-GST (bottom) antibodies. Results from quantification of LexA blots from replicate experiments are summarized in the histogram below as mean percentages of input amounts recovered in the pull-downs. (C) Yeast two-hybrid analysis as shown in (A). Samples for β-galactosidase assay were collected from cells under starvation (+) or nonstarvation (-) conditions as described in Materials and Methods. The results shown are averages of activities from three or more individual transformants. (D) Yeast two-hybrid analysis of CTD and KD interaction domains in Gcn2. Plasmids pHQ311 and pHQ428 encoding respectively the lexA-CTD(1498–1659) and B42-KD(720–999) segments were co-transformed into strain EGY48 carrying the lexAop-lacZ reporter plasmid. Samples for β-galactosidase assay were collected from cells under starvation (+) or nonstarvation (-) conditions. The results shown are averages of activities from three or more individual transformants. (E and F) In vitro binding of Gcn2 [35S]-HisRS(970–1497) segment translated in vitro to GST fusion proteins containing the Gcn2 KD (568–998), CTD (1498–1659) or HisRS (970–1497) domains. RNA (tRNAPhe or mRNA) was added to the reaction to the indicated final concentration. The mean fold-reductions in pull-downs of [35S]-HisRS(970–1497) on tRNAPhe addition were calculated from replicate experiments and presented in the histogram below the gel. In an effort to confirm the two-hybrid findings, we examined in vitro interaction of a LexA-CTD fusion expressed in yeast cells with immobilized GST fusions containing mutant or WT HisRS segments purified in yeast. As both the HisRS and CTD segments have RNA binding activity [17,28], the reactants were treated with micrococcal nuclease to eliminate indirect association between these segments bridged by RNA. Paralleling the two-hybrid results, the D1327K substitution greatly increased binding of LexA-CTD to GST-HisRS compared to the low-level binding observed for both WT GST-HisRS and the variant harboring the Gcd- triple substitution Y1092C/T1328S/A1353V; and introducing the Gcd- triple substitution reduced binding by the D1327K variant (Fig. 6B). It could be argued that the truncated species of the GST-HisRS-D1327K fusion that are not observed for WT GST-HisRS (lower blot, lane 3 vs. 2) mediate the relatively greater binding of LexA-CTD by GST-HisRS-D1327K; however, this interpretation is inconsistent with the fact that the GST-HisRS fusion harboring substitutions D1327K/Y1092C/T1328S/A1353V contains even greater levels of the truncated species but binds relatively smaller amounts of LexA-CTD compared to GST-HisRS-D1327K (Fig. 6B, lane 5 vs. 3). Our finding that Y1092C/T1328S/A1353V does not reduce the HisRS/CTD interaction when introduced into the otherwise WT HisRS segment might be explained by proposing that a physiologically relevant interaction between the WT HisRS and CTD domains cannot be captured outside of the context of full-length Gcn2 in two-hybrid or pull-down assays unless the HisRS segment contains the Gcn- substitutions D1327A/D1327K that stabilize HisRS/CTD association. Together, the findings in Fig. 6A-B suggest that the D1327A/D1327K substitutions impair activation of Gcn2 by strengthening the HisRS-CTD domain interaction, while the Gcd- substitution Y1092C/T1328S/A1353V activates Gcn2 by weakening HisRS/CTD association.
Evidence that tRNA antagonizes HisRS/CTD interaction
A corollary of this last conclusion is that the HisRS-CTD domain interaction in WT Gcn2 stabilizes the inactive conformation of Gcn2, which would persist constitutively in the Gcn- mutants D1327A/D1327K with attendant impairment of Gcn2 activation. If so, then binding of uncharged tRNA to the HisRS region might be expected to weaken the HisRS-CTD domain interaction as one means of activating Gcn2. Supporting this possibility, we found that the enhanced HisRS-CTD two-hybrid interactions conferred by D1327A or D1327K in vivo were abolished by starving the cells for isoleucine/valine by treatment with sulfometuron methyl (SM) (Fig. 6C), an inhibitor of the ILV2-encoded biosynthetic enzyme, which is known to increase the level of uncharged tRNAIle and tRNAVal and activate Gcn2 in vivo [15,38]. By contrast, the previously demonstrated two-hybrid interaction between LexA-KD and B42-CTD fusion proteins was unaffected by SM treatment (Fig. 6D), as was expression of the two-hybrid reporter conferred by the LexA-B42 activator. These findings are consistent with the idea that accumulation of uncharged tRNAIle and tRNAVal and their attendant increased binding to the LexA-HisRS-D1327A and LexA-HisRS-D1327K fusions weakens the ability of these LexA-HisRS proteins to form complexes with the B42-CTD fusion in vivo. They also support the idea that the gcn2-D1327A and gcn2-D1327K variants are defective for a regulatory interaction with the CTD that is normally disrupted by uncharged tRNA binding to the HisRS domain.
To provide additional evidence that tRNA binding to the HisRS domain reduces its ability to interact with the CTD, we examined the effect of tRNA on this interaction in vitro. Consistent with previous results [18,24], [35S]-methionine labeled HisRS fragment can be pulled down with immobilized GST fusions containing the Gcn2 KD, CTD or HisRS region itself, with the last interaction reflecting dimerization of the HisRS domain [18] (Fig. 6E). Addition of increasing amounts of purified yeast tRNAPhe reduced interaction of the [35S]-HisRS fragment with all three GST fusions; however, the magnitude of the reduction was larger for GST-CTD (~5.4-fold) compared to GST-KD (~1.8-fold) or GST-HisRS (~2.1-fold). Moreover, interaction of [35S]-HisRS with GST-CTD was unaffected by an equivalent concentration of an unstructured model mRNA [39] (Fig. 6F), suggesting specificity for tRNA in weakening the HisRS-CTD domain interaction. Together, these findings provide evidence that a tight interaction between the HisRS and CTD domains favors the inactive conformation of Gcn2 and that tRNA binding to the HisRS domain activates Gcn2 at least partly by weakening the HisRS-CTD interaction. As discussed below, based on previous findings indicating that direct interaction of the CTD with the KD contributes to the latency of Gcn2 kinase function [18], we propose that the HisRS-CTD interaction helps to stabilize this inhibitory CTD-KD interaction in a manner that is diminished by uncharged tRNA binding to the HisRS domain in amino acid-starved cells.
Gcd- triple substitution increases protease sensitivity of full-length Gcn2
The model alluded to above envisions that the non-activated state of Gcn2 is characterized by domain interactions between the HisRS-C and CTD, and between the CTD and KD, which are destabilized by tRNA binding to the HisRS domain to evoke the activated state. We reasoned that the Y1092C/T1328S/A1353V Gcd- substitutions, which destabilize the HisRS/CTD interaction and confer constitutive activation of Gcn2, would evoke a conformational change in full-length Gcn2 that mimics the activated, tRNA-bound state of WT Gcn2. Supporting this possibility, we found that the Gcd- triple substitution increases the sensitivity of full-length purified Gcn2 to digestion by elastase, reducing the amount of full-length protein remaining after a fixed time of incubation compared to WT Gcn2 or the Gcn- variant D1327K (S4 Fig.). Trypsin digestion also reduced the amounts of the largest intermediates in addition to the full-length protein for the Y1092C/T1328S/A1353V variant compared to the WT and D1327 proteins (S4 Fig.). Judging by the amount of full-length Gcn2 remaining after partial digestion, the Gcn- variant D1327K appears to be somewhat less sensitive than WT Gcn2 to protease digestion, consistent with the tighter HisRS/CTD domain interaction conferred by D1327K (Fig. 6A-C); although this difference is less pronounced than that between WT and the Y1092C/T1328S/A1353V variant (S4 Fig.). These results support the idea that activation of Gcn2 by the Y1092C/T1328S/A1353V substitutions involves the elimination of inhibitory domain interactions, which favors a less compact conformation of Gcn2. It should be noted that in separate experiments we observed a decrease in protease sensitivity of WT Gcn2 on addition of excess tRNAPhe. While this result is ostensibly at odds with the notion that tRNA binding evokes a more extended, protease-sensitive conformation of Gcn2, it seems possible that contacts between tRNA and the HisRS or CTD domains would reduce protease access to these Gcn2 segments and compensate for loss of protein-domain interactions in the tRNA-free state of Gcn2.
Discussion
In this study, we used a genetic approach to identify a novel regulatory surface in the HisRS domain of Gcn2, juxtaposed to the pseudo-active site cleft where tRNA binds, which participates in the activation of kinase function in amino acid starved cells through its association with the Gcn2 CTD. One of the residues belonging to this regulatory surface, Thr-1328, was identified by isolating suppressors of the m2 lesion in motif 2 of the HisRS domain, a mutation that reduces tRNA binding to Gcn2 and abolishes activation of kinase function in starved cells. Two other m2 suppressors alter residues Tyr-1092 and Ala-1353 located within the pseudo-active site cleft. The suppressor substitution Y1092C, as well as the combination of all three suppressor substitutions in the same protein, confer constitutive activation of Gcn2 function in the absence of the m2 substitutions—the Gcd- phenotype—and we showed that the triple substitution does not suppress the tRNA binding defect evoked by m2. Hence, rather than influencing the level of tRNA binding, we propose that these mutations evoke a conformational change in the HisRS domain that mimics the consequences of tRNA binding to the WT HisRS region, which then mediates activation of the adjacent KD. In this view, the m2 suppressors allow rearrangement of Gcn2 to the active conformation at a lower occupancy of tRNA in the HisRS pseudo-active site. This alteration would compensate for the reduced affinity for tRNA of the m2 variant, and in the cases of Y1092C and A1353V allow for activation of otherwise WT Gcn2 by the basal level of uncharged tRNA in non-starved cells to produce the Gcd- phenotype. Our conclusion above that T1328S corrects the activation defect conferred by m2 but does not appreciably activate otherwise WT Gcn2, ie. T1328S is not Gcd-, indicates that replacing Thr with Ser at this position promotes tRNA binding only in the context of the m2 alterations of the HisRS pseudo-active site. This restricted efficacy of T1328S is consistent with the fact that Thr-1328 is not evolutionarily conserved in Gcn2 HisRS domains, and is even substituted with Ser in some species (S2 Fig.). Given that the m2 lesion abolishes Gcn2 activation in vivo but only reduces tRNA binding in vitro, the m2 substitutions might also impair regulatory interactions of the HisRS domain that can be compensated by the m2 suppressors.
A second line of evidence supporting this model is that all 6 previously identified GCN2c mutations affecting the HisRS domain [37] involve substitutions mapping within, or proximal to, the pseudo-active site cleft. These mutations were identified by screening randomly mutagenized GCN2 alleles for the Gcd- phenotype, rather than selecting for m2 suppressors. The striking clustering of these 6 GCN2c substitutions in the predicted structure of the HisRS domain suggests that the pseudo-active site cleft is the key regulatory hub in this domain. The GCN2c mutations D1138N and F1134L alter residues in proximity to those substituted by the m2 suppressors Y1092C and A1353V within the pseudo-active site (Fig. 3B) and thus, according to our model, would evoke a rearrangement of the active site that mimics the effect of tRNA binding. The GCN2c mutations G1338D and A1197G introduce substitutions proximal to the active site, but located on a distinct surface, with Gly-1338 nearly adjacent on that surface to Thr-1328 (altered by the m2 suppressor T1328S). We envision that this surface patch in the WT HisRS domain communicates with the pseudo-active site and is remodeled by tRNA binding in a manner mimicked by the Gcd- substitutions G1338D, A1197G, and m2 suppressor T1328S.
A third line of genetic evidence supporting this model came from making targeted alanine substitutions of two HisRS domain residues that are invariant among Gcn2 homologs and exposed on the putative regulatory surface that circumscribes the Gcd- substitutions G1338D, A1197G, and m2 suppressor T1328S. Ala or Lys substitutions of the highly conserved residue D1327 completely abolish Gcn2 function in vivo while retaining robust tRNA-binding activity in vitro. It is remarkable that Gcn- and Gcd- substitutions of nearby residues belonging to this patch of the HisRS surface have opposite effects on Gcn2 activation. We envision that the Gcn- substitutions D1327K/D1327A either impede the proposed conformational remodeling of this surface patch induced by tRNA binding or alter the affinity of the remodeled surface for its binding partner within Gcn2.
The latter possibility is supported by our finding that Gcn- substitutions D1327K/D1327A enhance interaction between the HisRS and CTD domains, whereas the Gcd- triple substitution Y1092C/T1328S/A1353V partially reverses this effect in the quadruple mutant also containing D1327K. These findings imply that tight binding between the HisRS regulatory patch identified here and the CTD stabilizes the inactive conformation of Gcn2. Consistent with this, the increased yeast two hybrid interactions between the HisRS and CTD domains evoked by Gcn- substitutions D1327K/D1327A are eliminated under conditions of isoleucine/valine starvation, in which the uncharged cognate tRNAs accumulate and Gcn2 is activated. Furthermore, interaction between the WT HisRS and CTD domains was antagonized in vitro by tRNA, but not by an equal concentration of unstructured mRNA. These findings support the idea that one aspect of Gcn2 activation by uncharged tRNA involves the ability of tRNA bound to the HisRS domain to weaken HisRS/CTD interaction.
As noted above, we previously identified an autoinhibitory CTD/KD interaction that appears to be disrupted by tRNA binding to the HisRS domain [17,18]. More recently, we obtained strong evidence that the YKD domain stimulates Gcn2 activity by directly interacting with the KD, and proposed that the inhibitory CTD/KD interaction would compete with this stimulatory YKD/KD interaction, and that tRNA binding to the HisRS domain would shift the balance towards the stimulatory YKD/KD interaction [21]. Integrating our current findings with these previous results suggests the attractive possibility that tRNA binding to the HisRS domain antagonizes the HisRS/CTD interaction to promote a more open conformation of Gcn2 in which the CTD is less tightly bound to the KD. This would allow the YKD to compete more effectively with the CTD for binding to the KD, thereby eliminating autoinhibition by the CTD and correcting structural impediments to kinase activity inherent in the Gcn2 KD (Fig. 7A). Thus, the ability of tRNA binding to weaken the HisRS/CTD interaction would provide a mechanism that serves to replace the inhibitory KD/CTD interaction with the stimulatory YKD/KD interaction. Consistent with the idea that activation of Gcn2 involves rearrangement to a more open conformation lacking domain interactions between the CTD and both the HisRS-C and KD, we found that the activating Gcd- substitution Y1092C/T1328S/A1353V increases the sensitivity of purified WT Gcn2 to digestion by elastase and trypsin. However, high-resolution structural analyses of full-length Gcn2 in the presence and absence of tRNA are clearly required for a rigorous test our model in Fig. 7A.
Fig. 7. Hypothetical model for the role of the CTD/HisRS domain interaction in stabilizing the inhibitory CTD/KD interaction. 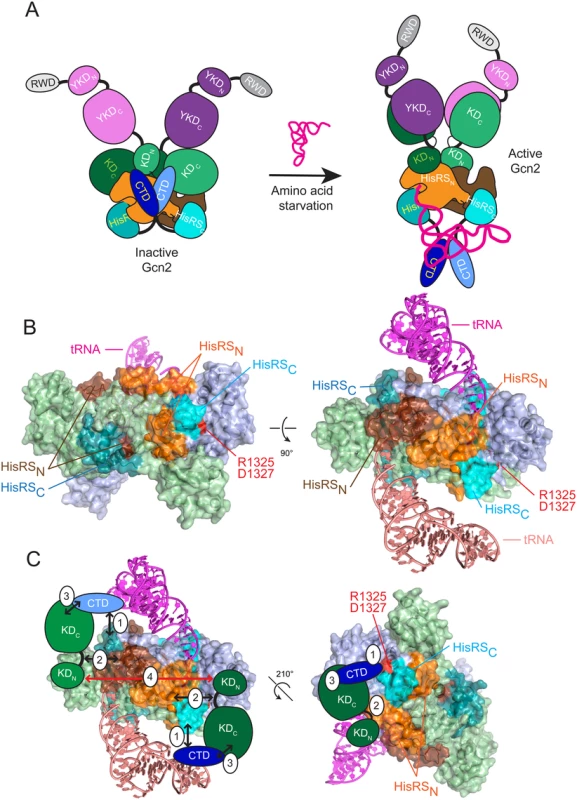
(A) (left) In nonstarved cells, Gcn2 exists as an inactive dimer with interactions between the CTD (light and dark blue), HisRS domain (light and dark orange for HisRSN and cyan for HisRSC) and KD (light and dark green) of each protomer. The KDs assume the anti-parallel mode of dimerization seen in the crystal structure of the inactive KD [12], and the HisRS domains dimerize as observed in crystal structures of authentic HisRS, as shown for T. cruzi HisRS in panel B. The YKD domain (light and dark purple) is not engaged with the KD owing to the inhibitory KD/CTD interaction, which is stabilized by CTD interaction with the HisRS domain. YKD/CTD interactions might also stabilize this conformation but were omitted for simplicity. (right) In amino acid-starved cells, uncharged tRNA binds to the HisRS domains and possibly also the CTD [17] (tRNA binding to only one protomer is depicted for simplicity), evoking a conformational change in the CTD-binding surface of the HisRS domain that triggers dissociation of the CTD/HisRS interaction, which in turn weakens the CTD/KD interaction to allow the stimulatory YKD/KD interaction to prevail. The KDs dimerize in the active back-to-back conformation observed in active PKR dimers [23]. (B) Two views of the crystal structure of the T. cruzi HisRS dimer complexed with HAM (PDB: 1KMM) [51] with modeled uncharged tRNA. HisRS residues Cys-67 to Arg-156, corresponding to the HisRS-N segment of Gcn2 (residues 1028–1120), are colored orange or brown in each protomer; and residues Lys-332 to Iso-380, corresponding to the Gcn2 HisRS-C segment (residues 1315–1383), colored light or dark cyan in each protomer. Arg-341 and Asp-343, corresponding to Gcn2 regulatory residues Arg-1325 and Asp-1327, are colored red. Modeled tRNAs are colored salmon (omitted for clarity in left panel) and magenta. (C) Two views of the proposed interaction of the Gcn2 CTD with the HisRS-C segment (1) and interaction of the hinge region of the KD with the HisRS-N segment (2), enabling cooperative binding between the KD and CTD (3), which is important for autoinhibition of kinase function in non-starvation conditions. Dimerization of the KDs (4) that occurs in the antiparallel arrangement observed in the crystal structure of the inactive conformation of the Gcn2 KD is indicated by a bidirectional red arrowhead [12]. A distinctive feature of authentic HisRS enzymes is that substrate binding involves an induced-fit mechanism in which histidine binding evokes movement of the “insertion domain” and HisA loop in a way that properly orients a key catalytic arginine residue (Arg-259/Arg-314 of E. coli/T. cruzi HisRS) for the formation of histidyl adenylate (HAM). Binding of ATP evokes additional motion of the m2 loop, which moves yet again on ejection of pyrophosphate following HAM formation [40] [41]. The presence of a bound HAM analogue increased the affinity of E. coli HisRS for tRNAHis [42], which might indicate that conformational changes induced by HAM binding also evoke a rearrangement of the active site that optimizes contacts with the acceptor stem of tRNAHis. Interestingly, it appears that the propensity of HisRS for histidine-induced rearrangement of the active site has been exploited to enable another HisRS-related protein, HisZ, to regulate the catalytic subunit of the octameric subfamily of ATP-phosphoribosyltransferase, HisG, an enzyme of histidine biosynthesis. HisZ contains the allosteric binding site for feedback-inhibition of HisG by histidine, and it is thought that conformational changes in the HisZ pseudo-active site evoked by histidine-binding evoke a remodeling of the HisG dimer interface to stabilize the inactive conformation [43]. Thus, in contrast to Gcn2, the binding of histidine rather than tRNA to the HisRS subunit (HisZ) allosterically regulates the catalytic activity of the binding partner (HisG), and the allosteric molecule (histidine) inhibits rather than stimulates the associated enzyme activity. Nevertheless, it seems plausible to propose that the pseudo-active site in the Gcn2 HisRS domain has evolved to evoke a conformational rearrangement of proximal, surface-exposed residues in response to binding of tRNA (rather than histidine) in the manner envisioned by our model. It is intriguing that the HisA loop, highly conserved among Gcn2 homologs (S2C Fig.), is predicted to be juxtaposed between the 3’ end of tRNA and the HisRS regulatory surface identified here (S5 Fig.), and thus could provide a path for transducing the aminoacylation status of the 3’ end of bound tRNA to the HisRS-CTD regulatory interface.
As noted above, we previously identified a positive regulatory interaction between the HisRS-N segment and the KD and mapped the KD-interacting region between residues 1028–1120 [18], which encompasses the N-terminal dimerization determinant we identified in the Gcn2 HisRS domain [18] (Fig. 7B, see orange and brown surfaces on the two protomers of the T. cruzi HisRS dimer). Interestingly, this region is contiguous with that corresponding to the portion of the HisRS-C segment that interacts with the CTD [18] (Fig. 7B, light and dark cyan surfaces that harbors the surface-exposed residues altered by the regulatory substitutions D1327A/D1327K and T1328S identified here (red residues in Fig. 7B). It is tempting to propose that the contiguity of the HisRS-N and HisR-C segments will juxtapose their respective interaction partners, the KD and CTD, and enable cooperativity in KD/CTD interaction (Fig. 7B-C). This model also seems compatible with the antiparallel mode of KD dimerization observed in the crystal structure of the inactive state of the Gcn2 KD [12] (Fig. 7C, red arrow connecting KDs in the two protomers). Eliminating the HisRS-C/CTD interaction on tRNA binding, as we proposed above, would eliminate the proposed cooperativity and destabilize CTD binding to the KD, allowing the YKD access to the KD instead (Fig. 7A). Release of the inhibitory HisRS-C/CTD interaction could also facilitate isomerization of the KDs to the parallel mode of dimerization required for their activation, and this alternative mode of dimerization could be further stabilized by the stimulatory YKD-KD interaction (Fig. 7A).
Materials and Methods
Sequence and structural alignments
Multiple sequence alignments were generated using MUSCLE at http://www.ebi.ac.uk/Tools/msa/muscle/. ConSurf [44] and PyMOL [45] were used to obtain sequence conservation scores and project the surface representation of sequence conservation on the crystal structure of the Trypanosoma cruzi authentic HisRS (PDB:3HRK, Fig. 3E). To obtain a hypothetical model of the Gcn2 HisRS-uncharged tRNA complex, the co-crystal structure of S. cerevisiae AspRS-tRNAAsp complex (PDB: 1ASZ[Ref: PubMed: 8313877]) was aligned with T. cruzi HisRS (PDB: 3HRK) by superimposing the highly conserved catalytic core domain (327 residues) using Dali pairwise comparison with default parameters [Ref: PubMed: 19481444]. This alignment produced a robust Z score of 12.9, a RMSD of 3.0 Å, and minimal clashes between tRNAAsp and HisRS. A similar alignment procedure was previously used to model HisRS interaction with tRNAHis. [Ref: PubMed 7556055; PubMed 11329259] The locations of Gcn2 residues involved in this study were then projected onto the T. cruzi HisRS crystal structure based on the sequence alignment between Gcn2 and authentic HisRSs (S3 Fig.).
Plasmids and strains
Plasmids employed are listed in Table 1. For Gcd- mutations identified by random mutagenesis, p2201 was subjected to error-prone PCR mutagenesis using the GeneMorph II kit (Stratagene) by using primer pairs PS-3 (5’-TCTATTTGATAACTCAGTTCCAAC-3’) and PS-4 (5’ - TCAGGAATATGTATAAGAAAGGTGAC-3’). The KpnI-NheI 1.8-kb GCN2 fragment encoding the HisRS-CTD was isolated from plasmid DNA prepared from a pool of E. coli transformants harboring mutagenized plasmids and subcloned into p2201. Plasmid DNA prepared from a pool of the resulting E. coli transformants was introduced into yeast strain H1149 and transformants were selected on SC-Ura medium containing 15 mM 3-AT. Resident plasmids were isolated from colony-purified transformants and subjected to DNA sequence analysis to identify the relevant mutations. As multiple mutations generally occurred, QuikChange® site-directed mutagenesis (Stratagene) was used to produce plasmids pSL501, pSL502 and pSL503, containing only single mutations in GCN2. Site-directed mutagenesis was also used to generate the novel derivatives (listed in parenthesis) of the following previously constructed plasmids: p2201 (pSL501-pSL507), p722 (pSL508-pSL525), pHQ430 (pSL535-pSL538), pHQ601 (pSL539-pSL541). Plasmids pSL526, pSL527, pSL529, pSL530 and pSL542 were generated by replacing the 3.0-kb BspEI-NheI fragment in pSL101 or pSL102 with the corresponding fragment from p722 derivatives harboring the appropriate GCN2 mutations.
Tab. 1. Plasmids used in this study. 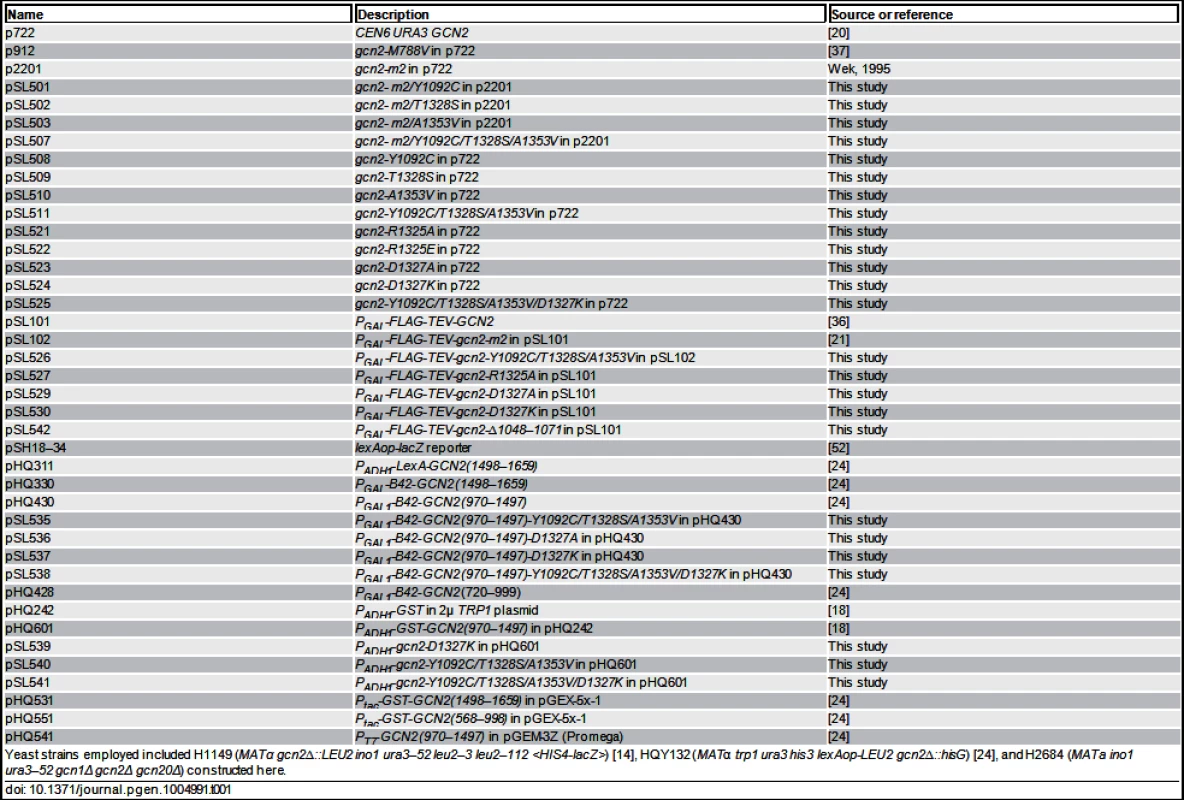
Yeast strains employed included H1149 (MATα gcn2∆::LEU2 ino1 ura3–52 leu2–3 leu2–112 <HIS4-lacZ>) [14], HQY132 (MATα trp1 ura3 his3 lexAop-LEU2 gcn2∆::hisG) [24], and H2684 (MATa ino1 ura3–52 gcn1Δ gcn2Δ gcn20Δ) constructed here. Protein purification
Transformants of H2684 bearing plasmids pSL101, pSL102, pSL526, pSL527, pSL529, pSL530 and pSL542 were grown to saturation in SC-Ura medium, diluted to A600 = 0.2 in SC-Ura containing 10% galactose as carbon source and grown to A600 of ∼2.5. Cells were harvested (∼25 g), washed with cold distilled water containing EDTA-free protease inhibitor cocktail (PIC) (Boehringer Mannheim) and 0.5 mM PMSF, resuspended in ice-cold binding buffer (BB) (100 mM sodium phosphate [pH 7.4], 500 mM NaCl, 0.1% Triton X-100, EDTA-free PIC, 1 μg/ml leupeptin, and 1 mM PMSF) and disrupted using SPEX freezer mill (model 6870). Lysates were clarified by centrifugation at 39,000 × g for 2 h at 4°C and mixed with 1 ml of M2-FLAG affinity resin (Sigma) overnight at 4°C. The resin was washed three times with 10 vol of BB and Gcn2 was eluted with 100 units of AcTEV protease in 500 μl of 1X TEV buffer (50mM Tris-HCl [pH 8.0], 0.5 mM EDTA, 1mM DTT). The eluates were concentrated with an Amicon Centricon filter (exclusion limit of Mr 10,000) and dialyzed against 10 mM Tris-HCl [pH 7.4], 50 mM NaCl, 20% glycerol and stored at −800 C. The eIF2α−ΔC protein was purified from E. coli as previously described [16]. Preparation of GST and GST fusion proteins of GCN2 were carried out as described previously [24].
Reporter assays and Western blot analysis in whole cell extracts (WCEs)
β-galactosidase assays of HIS4-lacZ expression were conducted on WCEs prepared from cultures grown in SD medium containing only the required supplements. For non-starvation conditions, saturated cultures were diluted 1 : 50 and harvested in mid-logarithmic phase after 6 h of growth. For starvation conditions, cultures were grown for 2 h under repressing conditions and then for 6 h after the addition of 3-AT to 10 mM or sulfometuron methyl (SM) to 0.5 μg/ml. β-galactosidase activity was assayed as described previously [46] and expressed as nanomoles of o-nitrophenyl-β-D-galactopyranoside hydrolyzed per min per mg of protein.
For Western analysis, WCEs were prepared by trichloroacetic acid extraction, as described previously [47], and immunoblot analysis was conducted as described [24] using phosphospecific antibodies against eIF2α-P (Biosource International) and polyclonal antibodies against eIF2α [48] or Gcn2 [49].
Kinase and tRNA-binding assays with purified Gcn2
Assays of Gcn2 autophosphorylation were conducted as described previously [11]. Binding of tRNA by Gcn2 was measured using a gel mobility shift assay as described previously [21].
GST pull-down assays
Pull-downs of LexA-CTD in yeast WCEs with GST-HisRS fusion proteins were conducted as follows. Immobilization of GST fusion proteins on glutathione-Sepharose 4B beads was carried out by incubating the purified fusion proteins at 0.5 μg/μL of beads (bed volume) in buffer A (20mM Tris/HCl pH7.5, 100mM NaCl, 0.2mM EDTA, 1mM DTT) containing 0.1% Triton X-100 at room temperature for 30 min with rocking. The beads were washed and resuspended in the same buffer. Five hundred μg of WCE prepared from pHQ311 transformants of HQY132 was treated with 12,000 units of micrococcal nuclease in the presence of 2mM CaCl2 for 10 min at 37°C. Nuclease-treated WCE was then added to beads (10-μL bed volume) containing 5 μg of bound GST fusion proteins and the volume was increased to 200 μL with buffer A. The mixtures were incubated at 4°C for 2 h with rocking. The beads were collected by brief centrifugation in a microcentrifuge, washed three times with 500 μL of buffer A, resuspended in 40 μL of Tris-Glycine SDS Sample Buffer (Novex), and fractionated by SDS-PAGE, transferred to nitrocellulose membranes, and probed with antibodies against GST or LexA. The immune complexes were visualized by enhanced chemiluminescence (ECL; GE Healthcare Life Science) according to the vendor’s instructions.
Pull-downs of [35S]-HisRS domain fragments were conducted as follows. In vitro transcription/translation with [35S]-methionine was conducted using the TNT T7 Coupled Reticulocyte Lysate System (Promega) according to the vendor’s instructions. The resulting [35S]-HisRS domain fragments were partially purified by ammonium sulfate precipitation as described previously [50] and resuspended in 50 μL of buffer A (described above) containing 12.5% glycerol. Immobilization of GST fusion proteins on glutathione-Sepharose 4B beads was carried out by incubating the purified fusion proteins at 0.5 μg/μL of beads (bed volume) in buffer A containing 0.1% Triton X-100 at room temperature for 30 min with rocking. The beads were washed and resuspended in the same buffer. Five microliters of [35S]-HisRS domain fragments were added to beads (10-μL bed volume) containing 5 μg of bound GST fusion proteins along with the indicated amount of tRNAPhe (Sigma-Aldrich, # R4018), or synthetic mRNA (GGAAUCUCUCUCUCUCUCUCUGCUCUCUCUCUCUCUCUCUCUC) synthesized by T7 polymerase as described in [39], and the volume was increased to 200 μL with buffer A. The mixtures were incubated at 4°C for 2 h with rocking. The beads were collected by brief centrifugation in a microcentrifuge, washed three times with 500 μL of buffer A, resuspended in 40 μL of SDS sample buffer, and fractionated by SDS-PAGE. For detecting the [35S]-HisRS domain fragments, the gels were fixed with a solution of isopropanol:water:acetic acid (25 : 65 : 10), treated with Amplify (GE Healthcare Life Science), dried, and subjected to fluorography at −80°C.
Yeast two-hybrid analysis
Plasmids encoding the appropriate LexA - and B42-Gcn2 fusions were cotransformed into yeast strain HQY132. The transformants were selected on synthetic complete medium lacking uracil, histidine, and tryptophan (SC−Ura−His−Trp). Two-hybrid interactions were indicated by β-galactosidase activities in cell extracts of three or more independent transformants. For these assays, cells were grown for 38 h to saturation in SC−Ura−His−Trp and were diluted 1 : 50 into the same medium containing galactose (2%) and raffinose (1%) as carbon sources (SC/Gal/Raf−Ura−His−Trp). When indicated, sulfometuron was added to the medium at a final concentration of 0.5μg/mL. Cells were harvested in the mid-logarithmic phase after 6 h of growth. β-Galactosidase assays were carried out as described above.
Protease digestion of Gcn2
Aliquots of 8 μg of purified Gcn2 were incubated with 0.001 units of elastase (Sigma-Aldrich) or 2 pg of trypsin (Sigma-Aldrich) in 10 mM Tris-HCl [pH 7.4], 50 mM NaCl, 20% glycerol for 5 min at room temperature and reactions were quenched by adding SDS sample buffer to a final concentration of 1X followed by heat inactivation at 95°C for 5 min. Digested samples were separated by SDS/PAGE and stained with Coomassie brilliant blue.
Supporting Information
Zdroje
1. Hinnebusch AG (2005) Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol 59 : 407–450. 16153175
2. Lu PD, Harding HP, Ron D (2004) Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J Cell Biol 167 : 27–33. 15479734
3. Vattem KM, Wek RC (2004) Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc Natl Acad Sci U S A 101 : 11269–11274. 15277680
4. Ron D, Harding HP (2007) eIF2α Phosphorylation in Cellular Stress Responses and Disease. In: Mathews M, Sonenberg N, Hershey JWB, editors. Translational Control in Biology and Medicine. Cold Spring Harbor: Cold Spring Harbor Laboratory Press. pp. 345–368.
5. Hao S, Sharp JW, Ross-Inta CM, McDaniel BJ, Anthony TG, et al. (2005) Uncharged tRNA and sensing of amino acid deficiency in mammalian piriform cortex. Science 307 : 1776–1778. 15774759
6. Anthony TG, McDaniel BJ, Byerley RL, McGrath BC, Cavener DR, et al. (2004) Preservation of liver protein synthesis during dietary leucine deprivation occurs at the expense of skeletal muscle mass in mice deleted for eIF2 kinase GCN2. J Biol Chem 279 : 36553–36561. 15213227
7. Guo F, Cavener DR (2007) The GCN2 eIF2alpha kinase regulates fatty-acid homeostasis in the liver during deprivation of an essential amino acid. Cell Metab 5 : 103–114. 17276353
8. Costa-Mattioli M, Gobert D, Harding H, Herdy B, Azzi M, et al. (2005) Translational control of hippocampal synaptic plasticity and memory by the eIF2alpha kinase GCN2. Nature 436 : 1166–1173. 16121183
9. Murguia JR, Serrano R (2012) New functions of protein kinase Gcn2 in yeast and mammals. IUBMB Life 64 : 971–974. doi: 10.1002/iub.1090 23129244
10. Eyries M, Montani D, Girerd B, Perret C, Leroy A, et al. (2014) EIF2AK4 mutations cause pulmonary veno-occlusive disease, a recessive form of pulmonary hypertension. Nat Genet 46 : 65–69. doi: 10.1038/ng.2844 24292273
11. Qiu H, Hu C, Dong J, Hinnebusch AG (2002) Mutations that bypass tRNA binding activate the intrinsically defective kinase domain in GCN2. Genes Dev 16 : 1271–1280. 12023305
12. Padyana AK, Qiu H, Roll-Mecak A, Hinnebusch AG, Burley SK (2005) Structural basis for autoinhibition and mutational activation of eukaryotic initiation factor 2alpha protein kinase GCN2. J Biol Chem 280 : 29289–29299. 15964839
13. Garriz A, Qiu H, Dey M, Seo EJ, Dever TE, et al. (2008) A network of hydrophobic residues impeding helix {alpha}C rotation maintains latency of eIF2{alpha} kinase Gcn2. Mol Cell Biol. doi: 10.1128/MCB.00782-08 19114562
14. Wek RC, Jackson BM, Hinnebusch AG (1989) Juxtaposition of domains homologous to protein kinases and histidyl-tRNA synthetases in GCN2 protein suggests a mechanism for coupling GCN4 expression to amino acid availability. Proc Natl Acad Sci USA 86 : 4579–4583. 2660141
15. Wek SA, Zhu S, Wek RC (1995) The histidyl-tRNA synthetase-related sequence in the eIF-2α protein kinase GCN2 interacts with tRNA and is required for activation in response to starvation for different amino acids. Mol Cell Biol 15 : 4497–4506. 7623840
16. Zhu S, Sobolev AY, Wek RC (1996) Histidyl-tRNA synthetase-related sequences in GCN2 protein kinase regulate in vitro phosphorylation of eIF-2. J Biol Chem 271 : 24989–24994. 8798780
17. Dong J, Qiu H, Garcia-Barrio M, Anderson J, Hinnebusch AG (2000) Uncharged tRNA activates GCN2 by displacing the protein kinase moiety from a bipartite tRNA-binding domain. Mol Cell 6 : 269–279. 10983975
18. Qiu H, Dong J, Hu C, Francklyn CS, Hinnebusch AG (2001) The tRNA-binding moiety in GCN2 contains a dimerization domain that interacts with the kinase domain and is required for tRNA binding and kinase activation. EMBO J 20 : 1425–1438. 11250908
19. Murphy JM, Zhang Q, Young SN, Reese ML, Bailey FP, et al. (2014) A robust methodology to subclassify pseudokinases based on their nucleotide-binding properties. Biochem J 457 : 323–334. doi: 10.1042/BJ20131174 24107129
20. Wek RC, Ramirez M, Jackson BM, Hinnebusch AG (1990) Identification of positive-acting domains in GCN2 protein kinase required for translational activation of GCN4 expression. Mol Cell Biol 10 : 2820–2831. 2188100
21. Lageix S, Rothenburg S, Dever TE, Hinnebusch AG (2014) Enhanced interaction between pseudokinase and kinase domains in Gcn2 stimulates eIF2alpha phosphorylation in starved cells. PLoS Genet 10: e1004326. doi: 10.1371/journal.pgen.1004326 24811037
22. Dey M, Cao C, Sicheri F, Dever TE (2007) Conserved intermolecular salt-bridge required for activation of protein kinases PKR, GCN2 and PERK. J Biol Chem 282 : 6650–6660.
23. Dar AC, Dever TE, Sicheri F (2005) Higher-order substrate recognition of eIF2alpha by the RNA-dependent protein kinase PKR. Cell 122 : 887–900. 16179258
24. Qiu H, Garcia-Barrio MT, Hinnebusch AG (1998) Dimerization by translation initiation factor 2 kinase GCN2 is mediated by interactions in the C-terminal ribosome-binding region and the protein kinase domain. Mol Cell Biol 18 : 2697–2711. 9566889
25. Narasimhan J, Staschke KA, Wek RC (2004) Dimerization is required for activation of eIF2 kinase Gcn2 in response to diverse environmental stress conditions. J Biol Chem 279 : 22820–22832. 15010461
26. He H, Singh I, Wek SA, Dey S, Baird TD, et al. (2014) Crystal structures of GCN2 protein kinase C-terminal domains suggest regulatory differences in yeast and mammals. J Biol Chem 289 : 15023–15034. doi: 10.1074/jbc.M114.560789 24719324
27. Ramirez M, Wek RC, Hinnebusch AG (1991) Ribosome-association of GCN2 protein kinase, a translational activator of the GCN4 gene of Saccharomyces cerevisae. Mol Cell Biol 11 : 3027–3036. 2038314
28. Zhu S, Wek RC (1998) Ribosome-binding domain of eukaryotic initiation factor-2 kinase GCN2 facilitates translation control. J Biol Chem 273 : 1808–1814. 9430731
29. Marton MJ, Vasquez de Aldana CR, Qiu H, Charkraburtty K, Hinnebusch AG (1997) Evidence that GCN1 and GCN20, translational regulators of GCN4, function on enlongating ribosomes in activation of the eIF2α kinase GCN2. Mol Cell Biol 17 : 4474–4489. 9234705
30. Vazquez de Aldana CR, Marton MJ, Hinnebusch AG (1995) GCN20, a novel ATP binding cassette protein, and GCN1 reside in a complex that mediates activation of the eIF-2α kinase GCN2 in amino acid-starved cells. EMBO J 14 : 3184–3199. 7621831
31. Garcia-Barrio M, Dong J, Ufano S, Hinnebusch AG (2000) Association of GCN1/GCN20 regulatory complex with the conserved N-terminal domain of eIF2α kinase GCN2 is required for GCN2 activation in vivo. EMBO J 19 : 1887–1899. 10775272
32. Sattlegger E, Hinnebusch AG (2005) Polyribosome binding by GCN1 is required for full activation of eukaryotic translation initiation factor 2{alpha} kinase GCN2 during amino acid starvation. J Biol Chem 280 : 16514–16521. 15722345
33. Sattlegger E, Hinnebusch AG (2000) Separate domains in GCN1 for binding protein kinase GCN2 and ribosomes are required for GCN2 activation in amino acid-starved cells. EMBO J 19 : 6622–6633. 11101534
34. Visweswaraiah J, Lageix S, Castilho BA, Izotova L, Kinzy TG, et al. (2011) Evidence that eukaryotic translation elongation factor 1A (eEF1A) binds the Gcn2 protein C terminus and inhibits Gcn2 activity. J Biol Chem 286 : 36568–36579. doi: 10.1074/jbc.M111.248898 21849502
35. Jimenez-Diaz A, Remacha M, Ballesta JP, Berlanga JJ (2013) Phosphorylation of initiation factor eIF2 in response to stress conditions is mediated by acidic ribosomal P1/P2 proteins in Saccharomyces cerevisiae. PLoS One 8: e84219. doi: 10.1371/journal.pone.0084219 24391917
36. Visweswaraiah J, Lee SJ, Hinnebusch AG, Sattlegger E (2012) Overexpression of eukaryotic translation elongation factor 3 impairs Gcn2 protein activation. J Biol Chem 287 : 37757–37768. doi: 10.1074/jbc.M112.368266 22888004
37. Ramirez M, Wek RC, Vazquez de Aldana CR, Jackson BM, Freeman B, et al. (1992) Mutations activating the yeast eIF-2α kinase GCN2: Isolation of alleles altering the domain related to histidyl-tRNA synthetases. Mol Cell Biol 12 : 5801–5815. 1448107
38. Jia MH, Larossa RA, Lee JM, Rafalski A, Derose E, et al. (2000) Global expression profiling of yeast treated with an inhibitor of amino acid biosynthesis, sulfometuron methyl. Physiol Genomics 3 : 83–92. 11015603
39. Acker MG, Kolitz SE, Mitchell SF, Nanda JS, Lorsch JR (2007) Reconstitution of yeast translation initiation. Methods Enzymol 430 : 111–145. 17913637
40. Francklyn C, Arnez J (2005) Histidyl-tRNA Synthetases. In: Ibba M, Francklyn C, Cusack S, editors. Aminoacyl-tRNA Synthetases. Austin, TX: Landes Publishing. pp. 135–148.
41. Merritt EA, Arakaki TL, Gillespie JR, Larson ET, Kelley A, et al. (2010) Crystal structures of trypanosomal histidyl-tRNA synthetase illuminate differences between eukaryotic and prokaryotic homologs. J Mol Biol 397 : 481–494. doi: 10.1016/j.jmb.2010.01.051 20132829
42. Bovee ML, Yan W, Sproat BS, Francklyn CS (1999) tRNA discrimination at the binding step by a class II aminoacyl-tRNA synthetase. Biochemistry 38 : 13725–13735. 10521280
43. Champagne KS, Sissler M, Larrabee Y, Doublie S, Francklyn CS (2005) Activation of the hetero-octameric ATP phosphoribosyl transferase through subunit interface rearrangement by a tRNA synthetase paralog. J Biol Chem 280 : 34096–34104. 16051603
44. Landau M, Mayrose I, Rosenberg Y, Glaser F, Martz E, et al. (2005) ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res 33: W299–302. 15980475
45. DeLano WL (2002) The PyMOL Molecular Graphics System. Palo Alto, CA.
46. Moehle CM, Hinnebusch AG (1991) Association of RAP1 binding sites with stringent control of ribosomal protein gene transcription in Saccharomyces cerevisiae. Mol Cell Biol 11 : 2723–2735. 2017175
47. Reid GA, Schatz G (1982) Import of proteins into mitochondria. Yeast cells grown in the presence of carbonyl cyanide m-chlorophenylhydrazone accumulate massive amounts of some mitochondrial precursor polypeptides. J Biol Chem 257 : 13056–13061. 6290491
48. Cigan AM, Pabich EK, Feng L, Donahue TF (1989) Yeast translation initiation suppressor sui2 encodes the α subunit of eukaryotic initiation factor 2 and shares identity with the human α subunit. Proc Natl Acad Sci USA 86 : 2784–2788. 2649894
49. Romano PR, Garcia-Barrio MT, Zhang X, Wang Q, Taylor DR, et al. (1998) Autophosphorylation in the activation loop is required for full kinase activity in vivo of human and yeast eukaryotic initiation factor 2α kinases PKR and GCN2. Mol Cell Biol 18 : 2282–2297. 9528799
50. Bardwell L, Cooper AJ, Friedberg EC (1992) Stable and specific association between the yeast recombination and DNA repair proteins RAD1 and RAD10 in vitro. Mol Cell Biol 12 : 3041–3049. 1620114
51. Arnez JG, Augustine JG, Moras D, Francklyn CS (1997) The first step of aminoacylation at the atomic level in histidyl-tRNA synthetase. Proc Natl Acad Sci U S A 94 : 7144–7149. 9207058
52. Golemis EA, Gyuris J, Brent R (1996) Interaction trap/two-hybrid system to identify interacting proteins. In: Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG et al., editors. Current Protocols in Molecular Biology. New York: John Wiley. pp. 20.21.21–20.21.28.
Štítky
Genetika Reprodukční medicína
Článek 2014 Reviewer Thank YouČlánek Closing the Gap between Knowledge and Clinical Application: Challenges for Genomic TranslationČlánek Discovery of CTCF-Sensitive Cis-Spliced Fusion RNAs between Adjacent Genes in Human Prostate CellsČlánek K-homology Nuclear Ribonucleoproteins Regulate Floral Organ Identity and Determinacy in ArabidopsisČlánek A Nitric Oxide Regulated Small RNA Controls Expression of Genes Involved in Redox Homeostasis inČlánek Contribution of the Two Genes Encoding Histone Variant H3.3 to Viability and Fertility in MiceČlánek The Genetic Architecture of the Genome-Wide Transcriptional Response to ER Stress in the Mouse
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 2
-
Všechny články tohoto čísla
- 2014 Reviewer Thank You
- Systematic Cell-Based Phenotyping of Missense Alleles Empowers Rare Variant Association Studies: A Case for and Myocardial Infarction
- African Glucose-6-Phosphate Dehydrogenase Alleles Associated with Protection from Severe Malaria in Heterozygous Females in Tanzania
- Genomics of Divergence along a Continuum of Parapatric Population Differentiation
- microRNAs Regulate Cell-to-Cell Variability of Endogenous Target Gene Expression in Developing Mouse Thymocytes
- A Rolling Circle Replication Mechanism Produces Multimeric Lariats of Mitochondrial DNA in
- Closing the Gap between Knowledge and Clinical Application: Challenges for Genomic Translation
- Partially Redundant Enhancers Cooperatively Maintain Mammalian Expression Above a Critical Functional Threshold
- Discovery of Transcription Factors and Regulatory Regions Driving Tumor Development by ATAC-seq and FAIRE-seq Open Chromatin Profiling
- Mutations in Result in Ocular Coloboma, Microcornea and Cataracts
- A Genome-Wide Hybrid Incompatibility Landscape between and
- Recurrent Evolution of Melanism in South American Felids
- Discovery of CTCF-Sensitive Cis-Spliced Fusion RNAs between Adjacent Genes in Human Prostate Cells
- Tissue Expression Pattern of PMK-2 p38 MAPK Is Established by the miR-58 Family in
- Essential Role for Endogenous siRNAs during Meiosis in Mouse Oocytes
- Matrix Metalloproteinase 2 Is Required for Ovulation and Corpus Luteum Formation in
- Evolutionary Signatures amongst Disease Genes Permit Novel Methods for Gene Prioritization and Construction of Informative Gene-Based Networks
- RR-1 Cuticular Protein TcCPR4 Is Required for Formation of Pore Canals in Rigid Cuticle
- GC-Content Evolution in Bacterial Genomes: The Biased Gene Conversion Hypothesis Expands
- Proteotoxic Stress Induces Phosphorylation of p62/SQSTM1 by ULK1 to Regulate Selective Autophagic Clearance of Protein Aggregates
- K-homology Nuclear Ribonucleoproteins Regulate Floral Organ Identity and Determinacy in Arabidopsis
- A Nitric Oxide Regulated Small RNA Controls Expression of Genes Involved in Redox Homeostasis in
- HYPER RECOMBINATION1 of the THO/TREX Complex Plays a Role in Controlling Transcription of the Gene in Arabidopsis
- Mitochondrial and Cytoplasmic ROS Have Opposing Effects on Lifespan
- Structured Observations Reveal Slow HIV-1 CTL Escape
- An Integrative Multi-scale Analysis of the Dynamic DNA Methylation Landscape in Aging
- Combining Natural Sequence Variation with High Throughput Mutational Data to Reveal Protein Interaction Sites
- Transhydrogenase Promotes the Robustness and Evolvability of Deficient in NADPH Production
- Regulators of Autophagosome Formation in Muscles
- Genomic Selection and Association Mapping in Rice (): Effect of Trait Genetic Architecture, Training Population Composition, Marker Number and Statistical Model on Accuracy of Rice Genomic Selection in Elite, Tropical Rice Breeding Lines
- Eye Selector Logic for a Coordinated Cell Cycle Exit
- Inflammation-Induced Cell Proliferation Potentiates DNA Damage-Induced Mutations
- The DNA Polymerase δ Has a Role in the Deposition of Transcriptionally Active Epigenetic Marks, Development and Flowering
- Contribution of the Two Genes Encoding Histone Variant H3.3 to Viability and Fertility in Mice
- Membrane Recognition and Dynamics of the RNA Degradosome
- P-TEFb, the Super Elongation Complex and Mediator Regulate a Subset of Non-paused Genes during Early Embryo Development
- is a Long Non-coding RNA in JNK Signaling in Epithelial Shape Changes during Drosophila Dorsal Closure
- A Pleiotropy-Informed Bayesian False Discovery Rate Adapted to a Shared Control Design Finds New Disease Associations From GWAS Summary Statistics
- Genome-wide Association Study Identifies Shared Risk Loci Common to Two Malignancies in Golden Retrievers
- and Hyperdrive Mechanisms (in Mouse Meiosis)
- Elevated In Vivo Levels of a Single Transcription Factor Directly Convert Satellite Glia into Oligodendrocyte-like Cells
- Systemic Delivery of MicroRNA-101 Potently Inhibits Hepatocellular Carcinoma by Repressing Multiple Targets
- Pooled Sequencing of 531 Genes in Inflammatory Bowel Disease Identifies an Associated Rare Variant in and Implicates Other Immune Related Genes
- Abscission Is Regulated by the ESCRT-III Protein Shrub in Germline Stem Cells
- Temperature Stress Mediates Decanalization and Dominance of Gene Expression in
- Transcriptome Wide Annotation of Eukaryotic RNase III Reactivity and Degradation Signals
- The Exosome Component Rrp6 Is Required for RNA Polymerase II Termination at Specific Targets of the Nrd1-Nab3 Pathway
- Sex-specific -regulatory Variation on the X Chromosome
- Regulation of Toll-like Receptor Signaling by the SF3a mRNA Splicing Complex
- Modeling of the Human Alveolar Rhabdomyosarcoma Chromosome Translocation in Mouse Myoblasts Using CRISPR-Cas9 Nuclease
- Asymmetry of the Budding Yeast Tem1 GTPase at Spindle Poles Is Required for Spindle Positioning But Not for Mitotic Exit
- TIM Binds Importin α1, and Acts as an Adapter to Transport PER to the Nucleus
- Antagonistic Roles for KNOX1 and KNOX2 Genes in Patterning the Land Plant Body Plan Following an Ancient Gene Duplication
- The Genetic Architecture of the Genome-Wide Transcriptional Response to ER Stress in the Mouse
- Fatty Acid Synthase Cooperates with Glyoxalase 1 to Protect against Sugar Toxicity
- Region-Specific Activation of mRNA Translation by Inhibition of Bruno-Mediated Repression
- An Essential Role of the Arginine Vasotocin System in Mate-Guarding Behaviors in Triadic Relationships of Medaka Fish ()
- Interaction between the tRNA-Binding and C-Terminal Domains of Yeast Gcn2 Regulates Kinase Activity In Vivo
- Hyper-Acetylation of Histone H3K56 Limits Break-Induced Replication by Inhibiting Extensive Repair Synthesis
- Prodomain Removal Enables Neto to Stabilize Glutamate Receptors at the Neuromuscular Junction
- Recent Selective Sweeps in North American Show Signatures of Soft Sweeps
- Identification and Functional Analysis of Healing Regulators in
- A Multi-Megabase Copy Number Gain Causes Maternal Transmission Ratio Distortion on Mouse Chromosome 2
- Drosophila Casein Kinase I Alpha Regulates Homolog Pairing and Genome Organization by Modulating Condensin II Subunit Cap-H2 Levels
- The Hippo Pathway Regulates Homeostatic Growth of Stem Cell Niche Precursors in the Ovary
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Genomic Selection and Association Mapping in Rice (): Effect of Trait Genetic Architecture, Training Population Composition, Marker Number and Statistical Model on Accuracy of Rice Genomic Selection in Elite, Tropical Rice Breeding Lines
- Discovery of Transcription Factors and Regulatory Regions Driving Tumor Development by ATAC-seq and FAIRE-seq Open Chromatin Profiling
- Evolutionary Signatures amongst Disease Genes Permit Novel Methods for Gene Prioritization and Construction of Informative Gene-Based Networks
- Proteotoxic Stress Induces Phosphorylation of p62/SQSTM1 by ULK1 to Regulate Selective Autophagic Clearance of Protein Aggregates
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



