-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaEssential Role for Endogenous siRNAs during Meiosis in Mouse Oocytes
In animals, the three main classes of small RNAs are microRNAs, short interfering RNAs, and PIWI-interacting RNAs. All three RNA species silence gene expression post-transcriptionally through interaction with the ARGONAUTE family of proteins. In mammals in particular, microRNAs are ubiquitously expressed, are essential for development, and perform numerous functions in a variety of cells and tissues. piRNAs are expressed almost exclusively in the germline, and are essential for male fertility and defense against transposons. Endogenous siRNAs are only expressed in germ cells and embryonic stem cells and have not been ascribed a functional role. By engineering a mouse that expresses a modified ARGONAUTE protein, we disrupt the function of endo-siRNAs exclusively in oocytes and find that females are infertile. Oocytes with an impaired siRNA pathway fail to complete meiosis I, and display severe spindle formation and chromosome alignment defects. Their transcriptome is widely perturbed and expression of the most abundant transposon is increased. These findings indicate that endo-siRNAs are essential for female fertility in mouse, are required for spindle formation, chromosome congression, and defense against transposons. This study unequivocally demonstrates an essential function for siRNAs in mammals, mediated through endonucleolytic cleavage of targets, and provides an explanation for the selective pressure that one AGO protein retains catalytic activity.
Published in the journal: . PLoS Genet 11(2): e32767. doi:10.1371/journal.pgen.1005013
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005013Summary
In animals, the three main classes of small RNAs are microRNAs, short interfering RNAs, and PIWI-interacting RNAs. All three RNA species silence gene expression post-transcriptionally through interaction with the ARGONAUTE family of proteins. In mammals in particular, microRNAs are ubiquitously expressed, are essential for development, and perform numerous functions in a variety of cells and tissues. piRNAs are expressed almost exclusively in the germline, and are essential for male fertility and defense against transposons. Endogenous siRNAs are only expressed in germ cells and embryonic stem cells and have not been ascribed a functional role. By engineering a mouse that expresses a modified ARGONAUTE protein, we disrupt the function of endo-siRNAs exclusively in oocytes and find that females are infertile. Oocytes with an impaired siRNA pathway fail to complete meiosis I, and display severe spindle formation and chromosome alignment defects. Their transcriptome is widely perturbed and expression of the most abundant transposon is increased. These findings indicate that endo-siRNAs are essential for female fertility in mouse, are required for spindle formation, chromosome congression, and defense against transposons. This study unequivocally demonstrates an essential function for siRNAs in mammals, mediated through endonucleolytic cleavage of targets, and provides an explanation for the selective pressure that one AGO protein retains catalytic activity.
Introduction
The RNase III enzyme DICER is responsible for biosynthesis of short-interfering RNAs (siRNAs) and microRNAs (miRNAs). DICER processes long double-stranded RNA (dsRNA) precursors into 21–23 bp-long duplexes known as siRNAs [1]. miRNAs are encoded by specific genomic loci and are processed from endogenous hairpin-shaped transcripts that are initially cleaved in the nucleus to a 70-bp miRNA precursor (pre-miRNA) by the Microprocessor complex, which is composed of the RNase III enzyme DROSHA and its partner, DiGeorge syndrome critical region 8 (DGCR8). The pre-miRNA is exported to the cytoplasm, where DICER cleaves the loop region of the molecule to generate the mature miRNA duplex [2].
Although both siRNAs and miRNAs are synthesized as duplexes, only one of the two strands, the ‘guide’ strand, is incorporated into the multi-protein complex RNA-induced silencing complex (RISC); the other strand (‘passenger’ strand) is discarded [3]. The guide strand recognizes a target mRNA by Watson-Crick base pairing and, based on the degree of sequence complementarity between the small RNA and target mRNA, either endonucleolytic cleavage or translational repression of the target mRNA follows [4]. In animals, siRNAs are perfectly complementary to their targets, and hence trigger mRNA cleavage, whereas miRNAs are usually only partially complementary and silence gene expression by translational repression and mRNA decay. Although it was initially postulated that mRNA levels did not change substantially in response to animal miRNAs, it was later shown that mRNA destabilization, prompted by deadenylation and decapping by the mRNA degradation machinery, is the main mode of regulation by mammalian miRNAs [5]. ARGONAUTE (AGO) proteins are at the core of RISC. In mammals, there are four AGO proteins (AGO1–4). All four can bind small RNAs and trigger translational repression, but only AGO2 possesses endonucleolytic activity and is the catalytic component of RISC [6].
We previously demonstrated a role for small RNAs during meiosis in mouse oocytes. Mice with an oocyte-specific deletion of Dicer are infertile due to defects during meiosis I [7,8]. Dicer-deficient females have morphologically normal ovaries and oocytes, produce normal numbers of oocytes, and ovulate similar numbers of eggs. However, Dicer null oocytes display meiotic catastrophe, with multiple disorganized meiotic spindles and severe chromosome congression defects. Expression of a subset of transposable elements is increased and the transcriptome is widely perturbed in Dicer null oocytes, with 18.4% of transcripts mis-regulated [7].
Deep sequencing of small RNAs demonstrated the presence of DICER-dependent miRNAs and endogenous siRNAs (endo-siRNAs), as well as DICER-independent PIWI interacting RNAs (piRNAs) in mouse oocytes [9,10]. Two populations of endo-siRNAs were found, one that corresponds to transposon-rich loci and another that maps to protein-coding genes. Interestingly, we found that some siRNAs are processed from dsRNAs formed by hybridization of transcripts from protein-coding genes to antisense transcripts from homologous pseudogenes and that these endo-siRNAs regulate the expression of endogenous genes. Therefore, the phenotype of Dicer null oocytes could be due to the absence of miRNAs or endo-siRNAs, or both. Using mRNA reporters, we assayed the ability of miRNAs to silence gene expression, looking at both translational repression and transcript levels. We found that miRNA activity decreases during oocyte growth and is suppressed in the full-grown oocyte. Furthermore, the very modest translational repression observed is not accompanied by message degradation [11]. Similarly, Suh et al. generated an oocyte-specific deletion of Dgcr8 and found that Dgcr8 null oocytes, which lack mature miRNAs, have a normal transcriptome and undergo normal meiotic maturation, fertilization, and embryonic development; consistent with these findings, Dgcr8 null mice have no discernable phenotype [12]. These two studies suggest that most likely endo-siRNAs, and not miRNAs, have an essential role during female meiosis.
It has recently been reported that mouse oocytes express a truncated DICER isoform, DICERO, which lacks the N-terminal DExD helicase domain, and which processes long dsRNAs much more efficiently than the somatic DICER isoform (DICERS), which is also expressed, albeit at lower levels [13]. The phenotype of DicerO null mice is virtually identical to the phenotype of mice with an oocyte-specific deletion of Dicer (which lack both DICERS and DICERO). Although DICERO can produce both miRNAs and endo-siRNAs when ectopically expressed in embryonic stem (ES) cells, miRNA levels appear slightly increased in DicerO null oocytes, suggesting that likely siRNAs are responsible for the observed phenotype. Whether this role of endo-siRNAs is mediated by endonucleolytic cleavage of mRNA targets remains unknown.
To test directly the role of endo-siRNAs through endonucleolytic cleavage in mouse oocytes, we expressed a catalytically inactive knock-in allele of Ago2 specifically in oocytes to disrupt the function of endo-siRNAs. We find that female mice expressing a catalytically inactive AGO2 (but not active AGO2) in their oocytes are infertile due to meiosis I defects. The phenotype is virtually identical to that in Dicer null females—female sterility, defects in spindle formation and chromosome congression, increase in abundance of transposable elements, and widespread changes in the transcriptome—and using live cell imaging, we characterize in more detail the meiotic defects. This study demonstrates a functional role for endogenous siRNAs through endonucleolytic cleavage in mammals and adds support to the evolutionary pressure to conserve ARGONAUTE endonucleolytic activity in animals.
Results and Discussion
Generation and characterization of an oocyte-specific catalytically inactive Ago2 allele
To eliminate the function of siRNAs we generated mice carrying a catalytically inactive form of AGO2 in their oocytes using a knock-in allele of Ago2 in which the catalytic DDH motif was mutated to ADH (Ago2ADH) [14]. This mutation inhibits endonucleolytic cleavage without affecting small RNA binding [6]. However, because mice carrying this allele die shortly after birth, we utilized a breeding scheme using Ago2ADH mice, Ago2fl/fl mice, and mice expressing Cre recombinase driven by the oocyte-specific Zp3 promoter to produce Ago2fl/ADH; Cre/+ females, referred to as Ago2ADH (S1 Fig.). These crosses also generated Ago2 null mice.
Ovarian morphology in Ago2ADH females was normal, with follicles at different stages of development, as well as corpora lutea, indicating that ovulation had occurred (Fig. 1A). After hormone stimulation, Ago2ADH females yielded similar numbers of full-grown oocytes as their wild-type (Ago2fl/fl) or heterozygous (Ago2fl/ADH) counterparts; similar numbers were also present in Ago2 null females (Fig. 1B). This result indicated that siRNA function is not required for oocyte development or response to hormones. However, Ago2ADH females were unable to produce any offspring during a 6-month mating trial with several wild-type males, indicating female sterility.
Fig. 1. AGO2 catalytic activity is not required for oocyte growth and hormonal response. 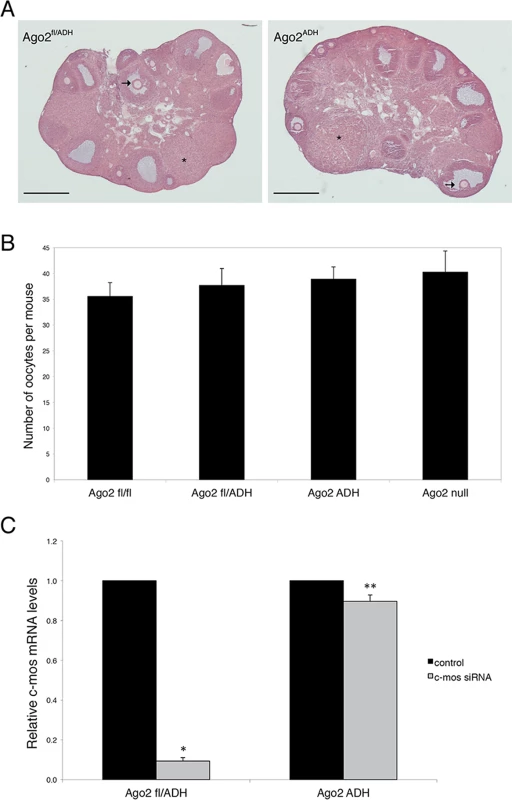
A) Histological sections of ovaries derived from Ago2fl/ADH (left panel) and Ago2ADH (right panel) females. Hematoxylin and eosin staining was performed as described in Materials and Methods. There were no obvious differences in ovary size, number of follicles, or follicular stages present between the two groups. The arrows indicate antral follicles, whereas the asterisks denote corpora lutea. Scale bar: 500 μm. B) Number of full-grown oocytes recovered from Ago2fl/fl, Ago2fl/ADH, Ago2ADH, and Ago2 null females. Oocyte collection after equine chorionic gonadotropin (eCG) priming was performed as described in Materials and Methods. The data are presented as the mean ± SEM; 29 Ago2fl/fl, 26 Ago2fl/ADH, 54 Ago2ADH, and 19 Ago2 null females were utilized. One-way ANOVA was used to compare the different groups and no statistical differences were found. C) Major reduction in AGO2 catalytic activity in oocytes from Ago2ADH mice. Full-grown oocytes were microinjected with c-Mos siRNA and c-Mos transcript levels were assayed by qRT-PCR 40 h later. The experiment was performed 3 times and statistical analysis was done using one-way ANOVA, followed by Bonferroni post-test. *p<0.001; **p< 0.05. To ascertain if oocytes carrying an Ago2ADH allele are incapable of endonucleolytic cleavage of small RNA targets, an RNAi assay was performed with Ago2ADH females. Full-grown oocytes were microinjected with c-Mos siRNA and 40 h later c-Mos mRNA levels were quantified by qRT-PCR. Whereas oocytes derived from Ago2fl/fl or Ago2fl/ADH females exhibited ~90% decrease in c-Mos transcript levels in c-Mos siRNA-treated oocytes compared to control oocytes, oocytes obtained from Ago2ADH females only showed a mild reduction (~10%) in c-Mos levels (Fig. 1C). These results demonstrated that oocytes from Ago2ADH females had extremely reduced AGO2 catalytic activity. This residual endonucleolytic activity may be due to persistent wild-type AGO2 levels that were present prior to Cre excision, because both mRNAs and proteins are often stable in oocytes.
AGO2 catalytic activity is required for completion of meiosis in mouse oocytes
To assess if AGO2 catalytic activity was required for meiotic maturation, full-grown oocytes were in vitro matured and spindle morphology was determined by immunofluorescence. Oocytes derived from Ago2fl/fl (Fig. 2A) or Ago2fl/ADH (Fig. 2B) females matured normally to metaphase II, as evidenced by the barrel-shaped meiotic spindle and extrusion of the first polar body. However, oocytes collected from Ago2ADH (Fig. 2C) or Ago2 null (Fig. 2D) females exhibited abnormal, disorganized spindles, with unaligned chromosomes. Some oocytes derived from Ago2ADH females extruded a polar body; nevertheless, upon closer examination it became clear that meiotic maturation was also abnormal in these oocytes, because partitioning of chromosomes between egg and polar body had not faithfully occurred (Fig. 2E, F).
Fig. 2. Abnormal meiotic spindles in Ago2ADH oocytes. 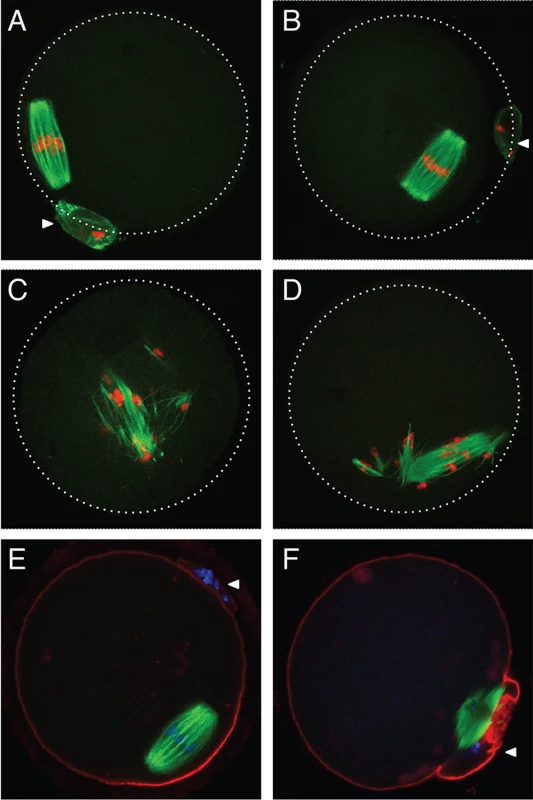
Oocytes from Ago2fl/fl, Ago2fl/ADH, Ago2ADH, and Ago2 null females were in vitro matured for 16 h. Immunofluorescence was performed as described in Materials and Methods. Arrowheads indicate the first polar body. A-D) Microtubules were stained with an antibody against (-tubulin (green) and DNA was counterstained with TO-PRO3 (red). A) Ago2fl/fl oocyte, B) Ago2fl/ADH oocyte, C) Ago2ADH oocyte, D) Ago2 null oocyte. E, F) Oocytes from Ago2fl/ADH (E) and Ago2ADH (F) females were stained with an antibody against (-tubulin (green), F-actin was labeled with Alexa 633-conjugated phaloidin (red), and DNA was counterstained with DAPI (blue). To characterize better the meiotic defects, oocytes were microinjected with cRNAs encoding Aurora kinase A (AURKA) fused to EGFP (to label spindle poles) and histone H2B fused to mCherry (to label chromosomes) and live imaging was performed during meiotic maturation (S1–S3 Movies). In Ago2fl/fl or Ago2fl/ADH oocytes (S1 Movie, Fig. 3A-B), the chromosomes remained centrally located and formed a sphere right after germinal vesicle breakdown (GVBD). In contrast, in Ago2ADH oocytes (S2 Movie, Fig. 3G-H), the chromosomes did not congress and instead scattered, covering a large area of the oocyte. Ago2fl/ADH oocytes proceeded to form a barrel-shaped metaphase I spindle, with chromosomes tightly aligned at the metaphase plate (Fig. 3C). Homologous chromosomes then separated at anaphase I (Fig. 3D), and migrated to opposite poles at telophase I (Fig. 3E), followed by cytokinesis, resulting in extrusion of the first polar body, completion of meiosis I and arrest at the metaphase stage of meiosis II (Fig. 3F). In contrast, in most Ago2ADH oocytes, the chromosomes remained dispersed and never aligned, and oocytes failed to enter anaphase I (Fig. 3G-L, S2 Movie). In a few Ago2ADH oocytes, after an initial dispersion of the chromosomes at GVBD, most chromosomes managed to align and form a metaphase I spindle, but there were always a few misaligned chromosomes, which resulted in a failure to enter anaphase and dispersion of chromosomes (S3 Movie).
Fig. 3. Abnormal chromosome segregation and spindle assembly in Ago2ADH oocytes. 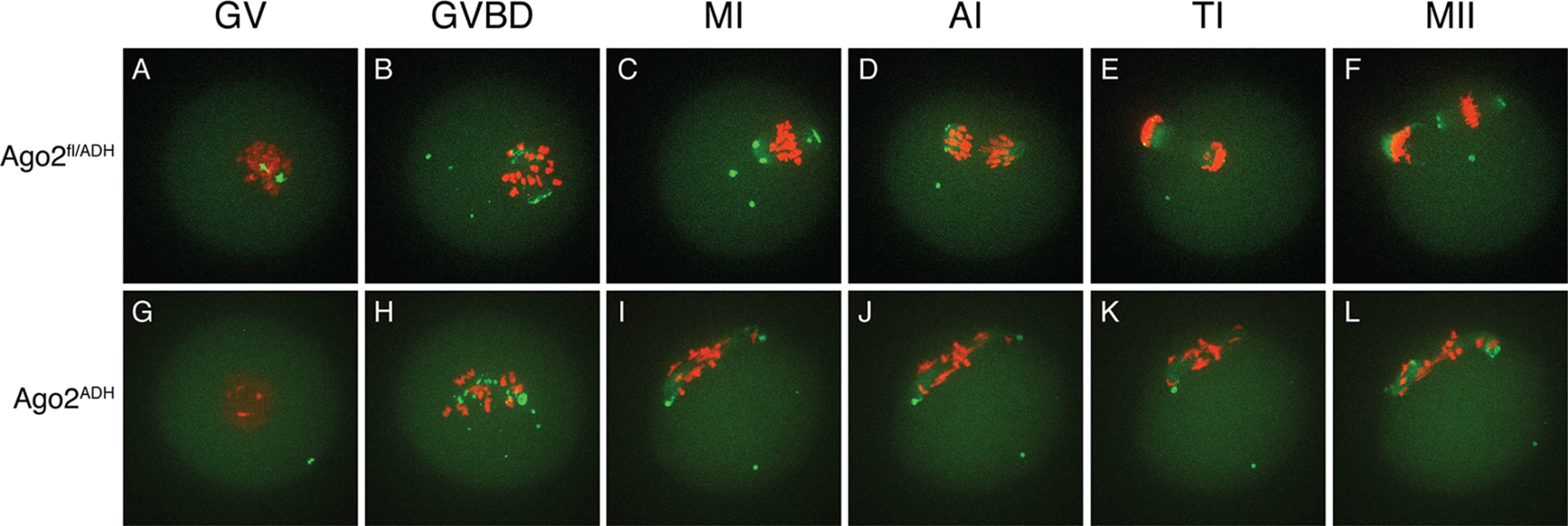
Chromosome and spindle dynamics in oocytes expressing AURKA-EGFP (green) and H2B-mCherry (red) were observed by time-lapse live confocal microscopy. Frames at the indicated stages of meiotic maturation were selected from the original time series (S1–S2 Movies), in which images were acquired every 18 min for 16 h. All images are maximal intensity projections of a confocal z series. A-F: Ago2fl/ADH oocytes; G-L: Ago2ADH oocytes. GV: germinal vesicle intact (A, G), GVBD: germinal vesicle breakdown (B, H), MI: metaphase I (C, I), AI: anaphase I (D, J), TI: telophase I (E, K), MII: metaphase II (F, L). The experiment was performed 3 times using at least 10 oocytes per group. Representative images are shown. The severe spindle defects observed in Dicer null oocytes have also been described in Ago2 null oocytes [15]. Although in the latter study the defect was attributed to reduced levels of miRNAs, it was later demonstrated that oocytes devoid of miRNAs have normal meiotic spindles [12]. By utilizing an allele of Ago2 that can bind small RNAs, but is catalytically inactive, we show that spindle formation and chromosome congression depend on the action of endo-siRNAs. Live imaging technology revealed that the defects start during GVBD, when chromosomes and microtubule organizing centers (MTOCs) scatter instead of forming a sphere [16], resulting in a long, abnormal spindle with unaligned chromosomes that fail to progress to anaphase I. The mechanism that links siRNAs with the spindle defects remains unknown. Given that the transcriptome of Ago2ADH oocytes is widely perturbed (see below), it is unlikely that a single protein is responsible for this phenotype.
Increase in MT retrotransposon levels in Ago2ADH oocytes
Because the levels of a subset of transposons are increased in Dicer-deficient oocytes [7,12], we investigated if this was also the case in the absence of AGO2 catalytic activity. Quantitative RT-PCR of the most abundant transposons in mouse oocytes revealed a significant increase in the levels of mouse transcript (MT), a member of the MaLR family of non-autonomous retrotransposons, in Ago2ADH and Ago2 null oocytes (Fig. 4). No significant differences were observed for the short interspersed repetitive elements (SINEs), long interspersed repetitive element 1 (LINE1 or L1), or intracisternal A-particle (IAP). This result differs somewhat from what we had previously described in Dicer null oocytes, where not only MT, but also Sine B1 and B2 elements were increased. This difference is likely due to differences in genetic background. We found that after re-deriving the Dicer null line, only MT levels were increased in oocytes (S2 Fig.), in agreement with a previous study [12], with DicerO null mice [13], and with Ago2ADH oocytes.
Fig. 4. Increased abundance of mouse transcript (MT) retrotransposon in Ago2ADH oocytes. 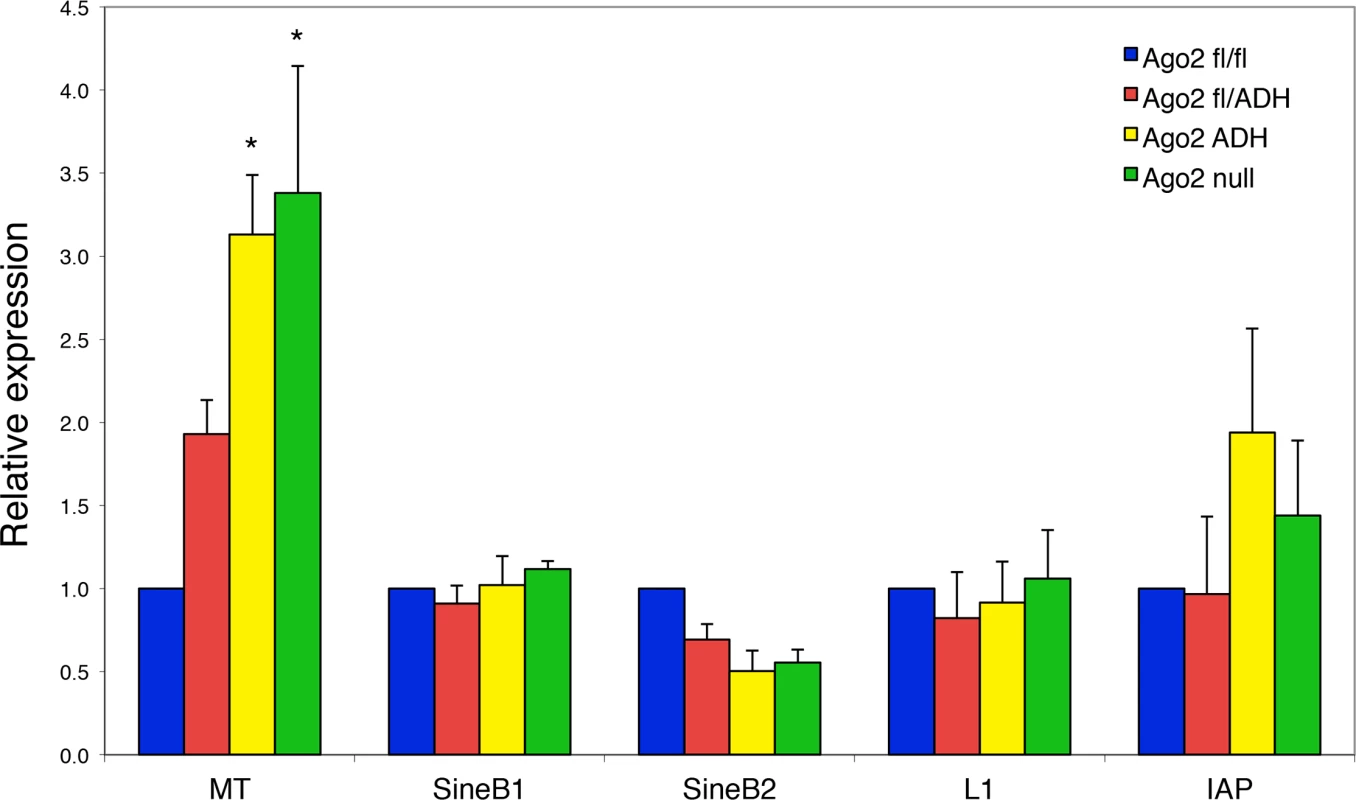
The levels of various transposons were determined by qRT-PCR in oocytes from different Ago2 genotypes, as described in Materials and Methods. Transposon levels in Ago2fl/fl oocytes were set as 1. Data are expressed as the mean ± SEM of four experiments. *p< 0.05 vs. Ago2fl/fl; two-way ANOVA, followed by Bonferroni post-test. PIWI family mutants are male sterile, but female fertile in mouse, indicating that the piRNA system is not essential during oogenesis [17]. This female fertility is not the case in flies and fish, where mutants that disrupt the piRNA system are female sterile [17]. The presence of endo-siRNAs that map to transposons in mouse oocytes likely explains why piRNAs are not essential in females, because both piRNAs and endo-siRNAs repress transposable elements in mouse oocytes. Because MT transcripts account for ~13% of all transcripts in the oocyte [18], a 3-fold increase in abundance is substantial and emphasizes the importance of siRNA action through endonucleolytic cleavage in transposon control.
Widespread changes in the oocyte transcriptome in the absence of AGO2 catalytic activity
Dicer-deficient oocytes exhibit dramatic changes in their transcriptome, as assayed by microarray analysis, with thousands of transcripts up - and down-regulated compared to wild-type oocytes [7,12]. To determine if the same molecular phenotype exists in the absence of AGO2 catalytic activity, we performed high-throughout RNA sequencing (RNA-seq) in full-grown Ago2fl/fl, Ago2ADH, and Ago2 null oocytes, as well as Dicer wild-type (WT) and knockout (KO) oocytes. We found extensive changes in transcript levels in Ago2ADH and Ago2 null oocytes. Using a false discovery rate (FDR) of 1%, 6441 transcripts were mis-regulated in Ago2ADH vs. Ago2fl/fl oocytes (3199 up-regulated and 3242 down-regulated) and 6142 transcripts were mis-regulated in Ago2 null vs. Ago2fl/fl oocytes (3050 up-regulated, 3092 down-regulated). Similarly, 6767 transcripts were mis-regulated in Dicer KO vs. WT oocytes (3195 up-regulated, 3572 down-regulated). Interestingly, although similar numbers of transcripts were down-regulated and up-regulated, as we had described for Dicer null oocytes, when the dataset was filtered by fold-change, a different picture surfaced. Of those transcripts whose abundance changed at least two-fold, the percentages that were up-regulated vs. down-regulated were 69%/31% in Ago2ADH vs. Ago2fl/fl oocytes, 68%/32% in Ago2 null vs. Ago2fl/fl, and 62%/38% in Dicer KO vs. WT oocytes. This finding indicates that the magnitude of change is greater in those transcripts that are up-regulated. This is indeed the case, as shown in S3 Fig., where the absolute values of fold-changes for the different comparisons were plotted for up-regulated and down-regulated transcripts. Because Cre-mediated recombination to excise the floxed allele of Ago2 and impair endo-siRNA function occurs in small, growing oocytes, and we utilized full-grown oocytes in our study, most likely the changes that we observe in the transcriptome are not only primary to disruption of siRNA function, but represent a complex array of downstream effects. Interrogating the transcriptome in growing oocytes should provide a better picture of the direct targets of endo-siRNAs.
As expected, there was an excellent correlation between the Ago2ADH and Dicer datasets (Fig. 5A). Of the 3242 transcripts that were down-regulated in Ago2ADH vs. Ago2fl/fl oocytes, 2385 (74%) were also down-regulated in Dicer KO vs. WT oocytes. Similarly, of the 3199 transcripts that were up-regulated in Ago2ADH vs. Ago2fl/fl oocytes, 2165 (68%) were also up-regulated in Dicer KO vs. WT oocytes. Comparable numbers were obtained when Ago2 null and Dicer datasets were compared (S4 Fig.). Also as expected, the transcriptome of Ago2ADH and Ago2 null oocytes was very similar, with only 33 transcripts (24 genes) whose abundance differs between these two groups, one of them being Ago2 itself (S5 Fig., S1 Table). Accordingly, the overlap between genes up-regulated compared to Ago2fl/fl oocytes in both groups or down-regulated in both groups is quite high (79–84%, S6 Fig.).
Fig. 5. Extensive transcriptome changes, and high correlation with Dicer KO oocytes, in Ago2ADH oocytes. 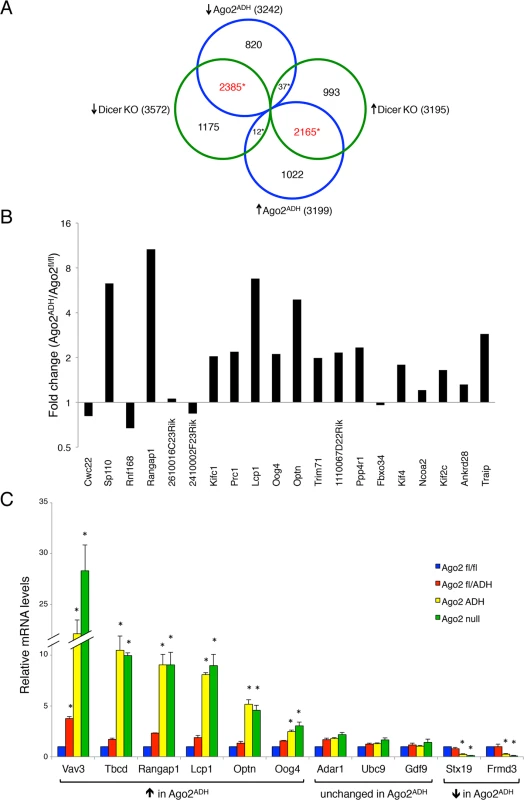
Oocytes from Ago2fl/fl, Ago2ADH, Ago2 null, Dicer WT, and Dicer KO females were subjected to RNA-seq. A) Comparison of transcripts up-regulated (↑) or down-regulated (↓) in Ago2ADH vs. Ago2fl/fl oocytes (blue circles) with those up-regulated (↑) or down-regulated (↓) in Dicer KO vs. Dicer WT oocytes (green circles). Mis-regulated transcripts were identified using an FDR of 0.01. The overlapping transcripts are shown in red. *p< 2.2e-16, Chi-square test. B) The majority of genes that produce endo-siRNAs in oocytes are up-regulated in the absence of AGO2 catalytic activity. Transcript levels in our RNA-seq dataset were compared in Ago2ADH vs. Ago2fl/fl oocytes for the 20 genes that produce the largest number of endo-siRNAs [9] and fold-changes were calculated. C) Validation of RNA-seq data by qRT-PCR. The relative abundance of 11 selected transcripts [6 up-regulated (↑), 3 unchanged, and 2 down-regulated (↓)] in our RNA-seq dataset when comparing Ago2ADH vs. Ago2fl/fl oocytes) was determined in oocytes of the different Ago2 genotypes by qRT-PCR. Transcript levels in Ago2fl/fl oocytes were set as 1. Data are expressed as the mean ± SEM of 3 experiments. *p< 0.05 vs. Ago2fl/fl; two-way ANOVA, followed by Bonferroni post-test. Although the overlap between genes mis-regulated in Ago2ADH and Dicer KO oocytes is quite high, there are many genes that are regulated differently in both groups. One possible explanation for these differences is that endo-siRNAs could have additional functions not mediated through AGO2-dependent endonucleolytic cleavage of target mRNAs. Also, AGO2 could cleave other, yet uncharacterized, DICER-independent small RNAs.
Given that a population of endo-siRNAs in oocytes derives from protein-coding genes, it was postulated that these small RNAs regulate expression of their precursor mRNAs [9,10]. To test this hypothesis, we analyzed our RNA-seq data for the transcript levels of the 20 genes that produce the largest number of siRNAs in oocytes [9]. The vast majority (15/20) are up-regulated in the absence of AGO2 catalytic activity (Fig. 5B), demonstrating a functional role for endo-siRNAs in the regulation of endogenous transcripts through endonucleolytic cleavage. The RNA-seq data were validated by performing qRT-PCR on several transcripts for which expression was either significantly increased, decreased, or unchanged in Ago2ADH oocytes, obtaining very similar results (Fig. 5C). We had previously demonstrated that the transcripts levels of genes that make siRNAs were increased in Dicer null oocytes [9], indicating a gene regulatory role for these small RNAs. Nevertheless, it was not clear if transcript regulation was due to endonucleolytic cleavage or if the mere production of siRNAs was diminishing the relative abundance of the transcript. Our results demonstrate that the action of siRNAs is through endonucleolytic cleavage of target mRNAs.
To gain insight into specific pathways that could be affected in Ago2ADH oocytes, gene ontology analysis of mis-regulated transcripts was performed using the database for annotation, visualization and integrated discovery (DAVID). For genes that are up-regulated in Ago2ADH oocytes, cell cycle, cell division, and regulation of translation, as well as microtubules and ribosomes were enriched (S7 Fig.); very similar categories were over-represented among genes up-regulated in Dicer KO oocytes (S8 Fig.). Many more categories were enriched among the genes that are down-regulated in Ago2ADH oocytes (S9 Fig.); these include RNA binding, nucleotide binding, cell cycle, chromosome, and transcription. And there was also an excellent correlation with those categories enriched for genes that are down-regulated in Dicer KO oocytes (S10 Fig.).
Although the miRNA pathway is dispensable in mouse oocytes, we were interested in determining if miRNA levels were normal in Ago2ADH oocytes, because Ago2 null oocytes have reduced miRNA levels [15]. The concentration of 5 abundant miRNAs was assayed in oocytes of different Ago2 genotypes. Mature miRNA levels were significantly decreased in both Ago2ADH and Ago2 null oocytes (S11A Fig.). Consistent with this finding, the modest miRNA-mediated translational repression, as assayed using luciferase reporters, was also reduced (S11B Fig.). Although AGO proteins stabilize mature miRNAs (and hence AGO loss leads to miRNA turnover), the catalytic activity of AGO2 is not required for this effect [19–21]. There are at least two possible explanations for the discrepancy with our results. First, Ago2ADH oocytes contain only one allele of Ago2 and hence the amount of protein is likely half the amount present in wild-type oocytes. Although Ago3 is the most abundant Ago transcript in mouse oocytes (S12A Fig.), Ago2 levels are substantial and a decrease in available AGO protein concentration may affect miRNA stability. Also, the levels of the other Ago transcripts are unchanged in both Ago2ADH and Ago2 null oocytes (S12B Fig.; only Ago2 and Ago3 transcript levels are shown because Ago1 and Ago4 mRNA levels are extremely low, undetectable in many samples, but are not up-regulated in Ago2ADH oocytes). Second, the aforementioned studies were performed in somatic cells, which lack endo-siRNAs. Because the catalytic activity of AGO2 is required for passenger strand cleavage and siRNA unwinding [22–25], in Ago2ADH oocytes siRNA duplexes would remain associated with AGO2, preventing miRNA binding and thus leading to more rapid miRNA turnover. The Zp3-driven Cre recombinase utilized to delete the floxed allele of Ago2 is active very early during oocyte growth [26], which takes ~ 3 weeks during which time transcription starts to decrease around mid-growth such that the full grown oocyte is transcriptionally inactive. Therefore, a small difference in miRNA stability can result over time in a highly significant decrease in miRNA levels. However, because mice whose oocytes are depleted of miRNAs show no discernable phenotype [12], the phenotype of Ago2ADH mice cannot be attributed to differences in oocyte miRNA levels.
In mammals, endo-siRNAs have only been described in mouse oocytes, ES cells, and male germ cells [9,10,27,28]. A physiological role for endo-siRNAs, however, has not been demonstrated in mammals. Mouse oocytes and ES cells lack the interferon response, an anti-viral defense mechanism against long dsRNA [29,30], and germ cells in the testis have also been suggested to be insensitive to interferon and hence tolerate dsRNA precursors that could generate endo-siRNAs [28]. In the mouse testis, ablation of Dicer or Drosha in germ cells leads to abnormal spermatogenesis, but male mice with a germ cell-specific ablation of Ago2 show no phenotype [31,32], suggesting that miRNAs are essential for spermatogenesis, but endo-siRNAs are dispensable in the male germline. In contrast, we find that endo-siRNAs are essential in the female germline in mouse. The presence in oocytes of DICERO that efficiently generates siRNAs from long dsRNA precursors, coupled with the absence of an interferon response, makes the mouse oocyte a privileged environment for siRNA action and may explain why this highly specialized cell relies on the siRNA pathway to regulate gene expression and protect genomic integrity. Given that DICERO is only expressed in mouse and rat oocytes, but not other rodent or non-rodent species [13], this essential role of siRNAs in oocytes may be restricted to the Muridae family.
Because most animal miRNAs silence their targets by translational repression, often linked to mRNA decay, but not by endonucleolytic cleavage, it has been puzzling that one mammalian AGO protein (AGO2) has retained catalytic activity. The finding that the catalytic activity of AGO2 is required for biosynthesis of one miRNA, miR-451 [14], and that this small RNA is essential for erythropoiesis [33] provided an answer to this conundrum. Our findings of an essential role of siRNAs through endonucleolytic cleavage during female meiosis strengthen the idea of evolutionary pressure that at least one AGO retain catalytic activity.
Materials and Methods
Animals
Ago2fl/+ animals [20] were crossed to Ago2ADH/+ mice [14]. The resulting Ago2fl/ADH females were crossed to Zp3-Cre males (Jackson Laboratories) and their progeny were intercrossed to produce Ago2fl/ADH; Cre/+ (Ago2ADH) mice (S1 Fig.). These crosses also generated Ago2 null mice. To determine fertility, two Ago2ADH and two Ago2fl/ADH female mice were bred with several males of proven fertility for a period of 6 months. Oocyte-specific Dicer null mice have been described [7]. All animal experiments were approved by the Institutional Animal Use and Care Committee of the University of Pennsylvania (protocol number 803551) and were consistent with National Institutes of Health guidelines.
Oocyte collection, meiotic maturation, and culture
Four - to six-week-old female mice were primed by intraperitoneal injection of 5 IU of equine chorionic gonadotropin (eCG) 48 h before oocyte collection. Full-grown, germinal vesicle (GV)-intact cumulus-enclosed oocytes were collected as previously described [34]. The collection medium was bicarbonate-free minimal essential medium (Earle’s salt) supplemented with polyvinylpyrrolidone (3 mg/mL) and 25 mM HEPES, pH 7.3 (MEM/PVP). Germinal vesicle breakdown was inhibited by including 2.5 μM milrinone [35]. The oocytes were transferred to CZB medium [36] containing 2.5 μM milrinone and cultured in an atmosphere of 5% CO2 in air at 37°C until microinjection was performed. In experiments in which oocyte maturation was assessed, after collection the oocytes were transferred to milrinone-free CZB medium and cultured for 16h in an atmosphere of 5% CO2 in air at 37°C.
Oocyte microinjection
GV oocytes were microinjected with approximately 5 pL of either siRNAs or cRNAs in MEM/PVP containing 2.5 μM milrinone as previously described [37]. c-Mos siRNA (CTGAACATTGCAAGACTAC; Dharmacon) was microinjected at 50 μM. For live imaging experiments, oocytes were microinjected with Aurka-Gfp cRNA (590 ng/μl) and H2b-mCherry cRNA (1035 ng/μL). miRNA reporters and firefly luciferase cRNAs were microinjected at 0.05 μg/μl.
Immunohistochemistry, immunofluorescence and live cell imaging
For immunohistochemistry, whole ovaries were fixed for 16h in Bouin’s fixative, embedded in paraffin, sliced to 10-μm sections, and stained with hematoxylin and eosin.
Immunofluorescence was performed as previously described [38]. The meiotic spindle was stained with a mouse anti-(-tubulin monoclonal antibody conjugated to AlexaFluor 488 (1 : 100; Life Technologies), the cortical actin cap was visualized with Alexa Fluor 633-conjugated phalloidin (1 : 500; Life Technologies). DAPI (Sigma) and TO-PRO3 (Life Technologies), both at 1.5 μg/mL, were used to label DNA and were added to the mounting medium (Vectashield, Vector Laboratories).
cRNAs encoding AURKA-GFP and H2B-mCherry were synthesized as described [39]. Oocytes were microinjected with Aurka-Gfp and H2b-mCherry cRNAs, cultured for 5 h in CZB + milrinone, and then transferred to individual drops of milrinone-free CZB medium, where meiotic maturation was assessed through live imaging, as described [39]. Images of individual cells were acquired every 18 min during 16 h and processed using NIH ImageJ software.
mRNA quantitative RT—PCR
Total RNA was extracted from 20 full-grown oocytes using Trizol (Life Technologies), according to the manufacturer’s protocol, except that 2 ng of Egfp RNA was added to the Trizol at the beginning of RNA isolation to serve as an exogenous normalization gene. cDNA was prepared by reverse transcription of total RNA with Superscript II and random hexamer primers. One oocyte equivalent of the resulting cDNA was amplified using TaqMan probes and the ABI Prism Sequence Detection System 7000 (Applied Biosystems). Two replicates were run for each real-time PCR reaction; a minus template served as control. Quantification was normalized to Egfp within the log-linear phase of the amplification curve obtained for each probe/primer using the comparative CT method (ABI PRISM 7700 Sequence Detection System, User Bulletin 2, Applied Biosystems, 1997). The TaqMan gene expression assays used were: Mm00445082_m1 (Vav3), Mm00551650_m1 (Tbcd), Mm00441071_m1 (Rangap1), Mm00620601_m1 (Oog4), Mm00786153_s1 (Lcp1), Mm00725286_m1 (Optn), Mm00433565_m1 (Gdf9), Mm00508001_m1 (Adar1), Mm00459008_m1 (Stx19), Mm00556276_m1 (Frmd3), Mm00462977_m1 (Ago1), Mm03053414_g1 (Ago2), Mm01188534_m1 (Ago3), and Mm00462659_m1 (Ago4). For Ubc9, Egfp, and c-Mos, custom TaqMan Gene Expression Assays were used that had the following primers and probes: Ubc9 forward primer 5′-CAGGTGAGAGCCAAGGACAAA-3′, Ubc9 reverse primer 5′-GGCCCACTGTACAGCTAACA-3′, Ubc9 probe 5′-CTGGCCTGCATTGATC-3′; Egfp forward primer: 5′-GCTACCCCGACCACATGAAG-3′, Egfp reverse primer: 5′-CGGGCATGGCGGACTT-3′, Egfp probe: 5′-CAGCACGACTTCTTC-3′; c-Mos forward primer: 5′-GGGAACAGGTATGTCTGATGCA-3′, c-Mos reverse primer: 5′-CACCGTGGTAAGTGGCTTTATACA-3′, c-Mos probe: 5′-CCGAGCCAAACCCTC-3′.
Transposon quantitative RT—PCR
RNA isolation and reverse transcription were performed as above. Real-time PCR was done using one oocyte equivalent per reaction and SYBR Green master mix. β-actin served as an internal control for normalization. Primer sequences were: MT.fwd: 5’-TGTTAAGAGCTCTGTCGGATGTTG-3’; MT.rev: 5’-ACTGATTCTT CAGTCCCAGCTAAC-3’; SineB1.fwd: 5’-GTGGCGCACGCCTTTAATC-3’; SineB1.rev: 5’-GACAGGGTTTCTCTGTGTAG-3’; SineB2.fwd: 5’-GAGATGGCTCAGTGGTTAAG-3’; SineB2.rev: 5’-CTGTCTTCAGACACTCCAG-3’; Line L1 ORF2.fwd: 5’-TTTGGGACACAATGAAAGCA-3’; Line L1 ORF2.rev: 5’-CTGCCGTCTACTCCTCTTGG-3’; IAP LTR.fwd: 5’-TTGATAGTTGTGTTTTAAGTGGTAAATAAA-3’; IAP LTR.rev: 5’-AAAACACCACAAACCAAAATCTTCTAC-3’; actin.fwd: 5’ - CGGTTCCGATGCCCTGAGGCTCTT-3’; actin.rev: 5’-CGTCACACTTCATGATGGAATTGA-3’.
miRNA quantitative RT—PCR
miRNA levels were assayed using the TaqMan MicroRNA Cells-to-CT kit (Life Technologies), following the manufacturers’ instructions, with slight modifications. Briefly, 9.1 μl of lysis solution was added to a tube containing 50 previously frozen full-grown oocytes. The samples were incubated for 8 min at room temperature and then 0.9 μl of stop solution was added, followed by a 2 min incubation at room temperature. Reverse transcription was performed using MultiScribe reverse transcriptase and following a multiplex protocol where the different miRNA-specific primers are mixed at a final concentration of 250 nM each. The resulting cDNA was diluted 10 times and real-time PCR was performed as described for mRNAs, using snoRNA202 as normalizing control. The following small RNA TaqMan assays were used: 000391 (mmu-miR-16–5p), 000580 (mmu-miR-20a-5p), 000602 (mmu-miR-30b-5p), 002459 (mmu-miR-106a-5p), 002406 (mmu-let-7e-5p), and 001232 (snoRNA202).
RNA sequencing
Twenty oocytes were lysed in 5 μL of NuGen lysis buffer. Each tube contained oocytes derived from 3 or 4 different animals of the same genotype and collection was performed three times to obtain 3 replicates per group. The groups were: Ago2fl/fl, Ago2ADH, Ago2 null, Dicer WT and Dicer KO. The lysate was used for cDNA synthesis using the Ovation RNA-Seq System V2 (Nugen) according to the manufacturer’s protocol. The resulting cDNA was fragmented into 200bp using Covaris shearing, and the Ovation Ultralow DR Multiplex System (Nugen) was used for library construction. The size and concentration of the resulting libraries were checked on Bioanalyzer, quantified by qPCR and sequenced on Illumina HiSeq 2000 with PE50. Sequencing reads were mapped to the mm10 refGene transcriptome and genome using TopHat v2.0.3 [40] with options ‘--read-mismatches 1 --read-gap-length 1 --read-edit-dist 1 --max-multihits 100 --no-discordant --b2-very-sensitive --transcriptome-max-hits 100 --library-type fr-unstranded --no-coverage-search --no-novel-juncs’ for 36bp reads and ‘--read-mismatches 3 --read-edit-dist 3—max-multihits 100 --b2-very-sensitive --transcriptome-max-hits 100 --library-type fr-unstranded --no-coverage-search --no-novel-juncs’ for 50bp reads. Read counts were computed using htseq-count (http://dx.doi.org/10.1101/002824) with options ‘--stranded = no -mode = intersection-strict’. Differential expression analysis was performed using the DESeq R package (version 1.10.1) [41]. Gene ontology (GO) analysis was performed using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) online resource [42,43] and using only the molecular function, cellular component, and biological process terms in the gene ontology database. The RNA-seq data have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE57514 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE57514).
Luciferase assays
Oocytes were microinjected with reporters that contain four bulged miR-30c sites (RL-4xB) downstream of the Renilla luciferase coding sequence. As a control, a reporter where the four miR-30c sites were mutated (RL-4xM) was used [11,44]. For normalization, firefly luciferase cRNA was coinjected with the Renilla luciferase reporters. The experiments were performed as previously described [11].
Statistical analysis
All experiments were replicated at least three times, except for luciferase assays, which were performed twice. Data were analyzed by ANOVA, followed by Bonferroni post-test. RNA-seq data were analyzed using a Chi-square test. A p< 0.05 was considered statistically significant.
Supporting Information
Zdroje
1. Bernstein E, Caudy AA, Hammond SM, Hannon GJ (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409 : 363–366. 11201747
2. Kim VN, Han J, Siomi MC (2009) Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 10 : 126–139. doi: 10.1038/nrm2632 19165215
3. Czech B, Hannon GJ (2011) Small RNA sorting: matchmaking for Argonautes. Nat Rev Genet 12 : 19–31. doi: 10.1038/nrg2916 21116305
4. Carthew RW, Sontheimer EJ (2009) Origins and Mechanisms of miRNAs and siRNAs. Cell 136 : 642–655. doi: 10.1016/j.cell.2009.01.035 19239886
5. Guo H, Ingolia NT, Weissman JS, Bartel DP (2010) Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466 : 835–840. doi: 10.1038/nature09267 20703300
6. Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, et al. (2004) Argonaute2 is the catalytic engine of mammalian RNAi. Science 305 : 1437–1441. 15284456
7. Murchison EP, Stein P, Xuan Z, Pan H, Zhang MQ, et al. (2007) Critical roles for Dicer in the female germline. Genes Dev 21 : 682–693. 17369401
8. Tang F, Kaneda M, O’Carroll D, Hajkova P, Barton SC, et al. (2007) Maternal microRNAs are essential for mouse zygotic development. Genes Dev 21 : 644–648. 17369397
9. Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, et al. (2008) Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature 453 : 534–538. doi: 10.1038/nature06904 18404147
10. Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, et al. (2008) Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature 453 : 539–543. doi: 10.1038/nature06908 18404146
11. Ma J, Flemr M, Stein P, Berninger P, Malik R, et al. (2010) MicroRNA activity is suppressed in mouse oocytes. Curr Biol 20 : 265–270. doi: 10.1016/j.cub.2009.12.042 20116252
12. Suh N, Baehner L, Moltzahn F, Melton C, Shenoy A, et al. (2010) MicroRNA function is globally suppressed in mouse oocytes and early embryos. Curr Biol 20 : 271–277. doi: 10.1016/j.cub.2009.12.044 20116247
13. Flemr M, Malik R, Franke V, Nejepinska J, Sedlacek R, et al. (2013) A retrotransposon-driven dicer isoform directs endogenous small interfering RNA production in mouse oocytes. Cell 155 : 807–816. doi: 10.1016/j.cell.2013.10.001 24209619
14. Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ (2010) A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature 465 : 584–589. doi: 10.1038/nature09092 20424607
15. Kaneda M, Tang F, O’Carroll D, Lao K, Surani MA (2009) Essential role for Argonaute2 protein in mouse oogenesis. Epigenetics Chromatin 2 : 9. doi: 10.1186/1756-8935-2-9 19664249
16. Schuh M, Ellenberg J (2007) Self-organization of MTOCs replaces centrosome function during acentrosomal spindle assembly in live mouse oocytes. Cell 130 : 484–498. 17693257
17. Cook MS, Blelloch R (2013) Small RNAs in germline development. Curr Top Dev Biol 102 : 159–205. doi: 10.1016/B978-0-12-416024-8.00006-4 23287033
18. Peaston AE, Evsikov AV, Graber JH, de Vries WN, Holbrook AE, et al. (2004) Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev Cell 7 : 597–606. 15469847
19. Winter J, Diederichs S (2011) Argonaute proteins regulate microRNA stability: Increased microRNA abundance by Argonaute proteins is due to microRNA stabilization. RNA Biol 8 : 1149–1157. doi: 10.4161/rna.8.6.17665 21941127
20. O’Carroll D, Mecklenbrauker I, Das PP, Santana A, Koenig U, et al. (2007) A Slicer-independent role for Argonaute 2 in hematopoiesis and the microRNA pathway. Genes Dev 21 : 1999–2004.
21. Zamudio JR, Kelly TJ, Sharp PA (2014) Argonaute-Bound Small RNAs from Promoter-Proximal RNA Polymerase II. Cell 156 : 920–934. doi: 10.1016/j.cell.2014.01.041 24581493
22. Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD (2005) Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell 123 : 607–620. 16271386
23. Leuschner PJF, Ameres SL, Kueng S, Martinez J (2006) Cleavage of the siRNA passenger strand during RISC assembly in human cells. EMBO Rep 7 : 314–320. 16439995
24. Rand TA, Petersen S, Du F, Wang X (2005) Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell 123 : 621–629.
25. Miyoshi K, Tsukumo H, Nagami T, Siomi H, Siomi MC (2005) Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes Dev 19 : 2837–2848. 16287716
26. Lira SA, Kinloch RA, Mortillo S, Wassarman PM (1990) An upstream region of the mouse ZP3 gene directs expression of firefly luciferase specifically to growing oocytes in transgenic mice. Proc Natl Acad Sci USA 87 : 7215–7219. 2402504
27. Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R (2008) Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev 22 : 2773–2785. doi: 10.1101/gad.1705308 18923076
28. Song R, Hennig GW, Wu Q, Jose C, Zheng H, et al. (2011) Male germ cells express abundant endogenous siRNAs. Proc Natl Acad Sci USA 108 : 13159–13164. doi: 10.1073/pnas.1108567108 21788498
29. Stein P, Zeng F, Pan H, Schultz RM (2005) Absence of non-specific effects of RNA interference triggered by long double-stranded RNA in mouse oocytes. Dev Biol 286 : 464–471. 16154556
30. Yang S, Tutton S, Pierce E, Yoon K (2001) Specific double-stranded RNA interference in undifferentiated mouse embryonic stem cells. Mol Cell Biol 21 : 7807–7816. 11604515
31. Hayashi K, de Sousa Chuva Lopes SM, Kaneda M, Tang F, Hajkova P, et al. (2008) MicroRNA biogenesis is required for mouse primordial germ cell development and spermatogenesis. PLoS One 3: e1738. doi: 10.1371/journal.pone.0001738 18320056
32. Wu Q, Song R, Ortogero N, Zheng H, Evanoff R, et al. (2012) The RNase III enzyme DROSHA is essential for microRNA production and spermatogenesis. J Biol Chem 287 : 25173–25190. doi: 10.1074/jbc.M112.362053 22665486
33. Papapetrou EP, Korkola JE, Sadelain M (2010) A genetic strategy for single and combinatorial analysis of miRNA function in mammalian hematopoietic stem cells. Stem Cells 28 : 287–296. doi: 10.1002/stem.257 19911427
34. Schultz RM, Montgomery RR, Belanoff JR (1983) Regulation of mouse oocyte meiotic maturation: implication of a decrease in oocyte cAMP and protein dephosphorylation in commitment to resume meiosis. Dev Biol 97 : 264–273. 6189752
35. Wiersma A, Hirsch B, Tsafriri A, Hanssen RG, Van de Kant M, et al. (1998) Phosphodiesterase 3 inhibitors suppress oocyte maturation and consequent pregnancy without affecting ovulation and cyclicity in rodents. J Clin Invest 102 : 532–537. 9691090
36. Chatot CL, Ziomek CA, Bavister BD, Lewis JL, Torres I (1989) An improved culture medium supports development of random-bred 1-cell mouse embryos in vitro. J Reprod Fertil 86 : 679–688. 2760894
37. Kurasawa S, Schultz RM, Kopf GS (1989) Egg-induced modifications of the zona pellucida of mouse eggs: effects of microinjected inositol 1,4,5-trisphosphate. Dev Biol 133 : 295–304. 2785065
38. Romanova LG, Anger M, Zatsepina OV, Schultz RM (2006) Implication of nucleolar protein SURF6 in ribosome biogenesis and preimplantation mouse development. Biol Reprod 75 : 690–696. 16855206
39. Balboula AZ, Stein P, Schultz RM, Schindler K (2014) Knockdown of RBBP7 unveils a requirement of histone deacetylation for CPC function in mouse oocytes. Cell Cycle 13 : 0–11.
40. Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, et al. (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36.
41. Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11: R106.
42. Huang DW, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4 : 44–57. doi: 10.1038/nprot.2008.211 19131956
43. Huang DW, Sherman BT, Lempicki RA (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37 : 1–13.
44. Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, et al. (2005) Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science 309 : 1573–1576. 16081698
Štítky
Genetika Reprodukční medicína
Článek 2014 Reviewer Thank YouČlánek Closing the Gap between Knowledge and Clinical Application: Challenges for Genomic TranslationČlánek Discovery of CTCF-Sensitive Cis-Spliced Fusion RNAs between Adjacent Genes in Human Prostate CellsČlánek K-homology Nuclear Ribonucleoproteins Regulate Floral Organ Identity and Determinacy in ArabidopsisČlánek A Nitric Oxide Regulated Small RNA Controls Expression of Genes Involved in Redox Homeostasis inČlánek Contribution of the Two Genes Encoding Histone Variant H3.3 to Viability and Fertility in MiceČlánek The Genetic Architecture of the Genome-Wide Transcriptional Response to ER Stress in the Mouse
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 2
-
Všechny články tohoto čísla
- 2014 Reviewer Thank You
- Systematic Cell-Based Phenotyping of Missense Alleles Empowers Rare Variant Association Studies: A Case for and Myocardial Infarction
- African Glucose-6-Phosphate Dehydrogenase Alleles Associated with Protection from Severe Malaria in Heterozygous Females in Tanzania
- Genomics of Divergence along a Continuum of Parapatric Population Differentiation
- microRNAs Regulate Cell-to-Cell Variability of Endogenous Target Gene Expression in Developing Mouse Thymocytes
- A Rolling Circle Replication Mechanism Produces Multimeric Lariats of Mitochondrial DNA in
- Closing the Gap between Knowledge and Clinical Application: Challenges for Genomic Translation
- Partially Redundant Enhancers Cooperatively Maintain Mammalian Expression Above a Critical Functional Threshold
- Discovery of Transcription Factors and Regulatory Regions Driving Tumor Development by ATAC-seq and FAIRE-seq Open Chromatin Profiling
- Mutations in Result in Ocular Coloboma, Microcornea and Cataracts
- A Genome-Wide Hybrid Incompatibility Landscape between and
- Recurrent Evolution of Melanism in South American Felids
- Discovery of CTCF-Sensitive Cis-Spliced Fusion RNAs between Adjacent Genes in Human Prostate Cells
- Tissue Expression Pattern of PMK-2 p38 MAPK Is Established by the miR-58 Family in
- Essential Role for Endogenous siRNAs during Meiosis in Mouse Oocytes
- Matrix Metalloproteinase 2 Is Required for Ovulation and Corpus Luteum Formation in
- Evolutionary Signatures amongst Disease Genes Permit Novel Methods for Gene Prioritization and Construction of Informative Gene-Based Networks
- RR-1 Cuticular Protein TcCPR4 Is Required for Formation of Pore Canals in Rigid Cuticle
- GC-Content Evolution in Bacterial Genomes: The Biased Gene Conversion Hypothesis Expands
- Proteotoxic Stress Induces Phosphorylation of p62/SQSTM1 by ULK1 to Regulate Selective Autophagic Clearance of Protein Aggregates
- K-homology Nuclear Ribonucleoproteins Regulate Floral Organ Identity and Determinacy in Arabidopsis
- A Nitric Oxide Regulated Small RNA Controls Expression of Genes Involved in Redox Homeostasis in
- HYPER RECOMBINATION1 of the THO/TREX Complex Plays a Role in Controlling Transcription of the Gene in Arabidopsis
- Mitochondrial and Cytoplasmic ROS Have Opposing Effects on Lifespan
- Structured Observations Reveal Slow HIV-1 CTL Escape
- An Integrative Multi-scale Analysis of the Dynamic DNA Methylation Landscape in Aging
- Combining Natural Sequence Variation with High Throughput Mutational Data to Reveal Protein Interaction Sites
- Transhydrogenase Promotes the Robustness and Evolvability of Deficient in NADPH Production
- Regulators of Autophagosome Formation in Muscles
- Genomic Selection and Association Mapping in Rice (): Effect of Trait Genetic Architecture, Training Population Composition, Marker Number and Statistical Model on Accuracy of Rice Genomic Selection in Elite, Tropical Rice Breeding Lines
- Eye Selector Logic for a Coordinated Cell Cycle Exit
- Inflammation-Induced Cell Proliferation Potentiates DNA Damage-Induced Mutations
- The DNA Polymerase δ Has a Role in the Deposition of Transcriptionally Active Epigenetic Marks, Development and Flowering
- Contribution of the Two Genes Encoding Histone Variant H3.3 to Viability and Fertility in Mice
- Membrane Recognition and Dynamics of the RNA Degradosome
- P-TEFb, the Super Elongation Complex and Mediator Regulate a Subset of Non-paused Genes during Early Embryo Development
- is a Long Non-coding RNA in JNK Signaling in Epithelial Shape Changes during Drosophila Dorsal Closure
- A Pleiotropy-Informed Bayesian False Discovery Rate Adapted to a Shared Control Design Finds New Disease Associations From GWAS Summary Statistics
- Genome-wide Association Study Identifies Shared Risk Loci Common to Two Malignancies in Golden Retrievers
- and Hyperdrive Mechanisms (in Mouse Meiosis)
- Elevated In Vivo Levels of a Single Transcription Factor Directly Convert Satellite Glia into Oligodendrocyte-like Cells
- Systemic Delivery of MicroRNA-101 Potently Inhibits Hepatocellular Carcinoma by Repressing Multiple Targets
- Pooled Sequencing of 531 Genes in Inflammatory Bowel Disease Identifies an Associated Rare Variant in and Implicates Other Immune Related Genes
- Abscission Is Regulated by the ESCRT-III Protein Shrub in Germline Stem Cells
- Temperature Stress Mediates Decanalization and Dominance of Gene Expression in
- Transcriptome Wide Annotation of Eukaryotic RNase III Reactivity and Degradation Signals
- The Exosome Component Rrp6 Is Required for RNA Polymerase II Termination at Specific Targets of the Nrd1-Nab3 Pathway
- Sex-specific -regulatory Variation on the X Chromosome
- Regulation of Toll-like Receptor Signaling by the SF3a mRNA Splicing Complex
- Modeling of the Human Alveolar Rhabdomyosarcoma Chromosome Translocation in Mouse Myoblasts Using CRISPR-Cas9 Nuclease
- Asymmetry of the Budding Yeast Tem1 GTPase at Spindle Poles Is Required for Spindle Positioning But Not for Mitotic Exit
- TIM Binds Importin α1, and Acts as an Adapter to Transport PER to the Nucleus
- Antagonistic Roles for KNOX1 and KNOX2 Genes in Patterning the Land Plant Body Plan Following an Ancient Gene Duplication
- The Genetic Architecture of the Genome-Wide Transcriptional Response to ER Stress in the Mouse
- Fatty Acid Synthase Cooperates with Glyoxalase 1 to Protect against Sugar Toxicity
- Region-Specific Activation of mRNA Translation by Inhibition of Bruno-Mediated Repression
- An Essential Role of the Arginine Vasotocin System in Mate-Guarding Behaviors in Triadic Relationships of Medaka Fish ()
- Interaction between the tRNA-Binding and C-Terminal Domains of Yeast Gcn2 Regulates Kinase Activity In Vivo
- Hyper-Acetylation of Histone H3K56 Limits Break-Induced Replication by Inhibiting Extensive Repair Synthesis
- Prodomain Removal Enables Neto to Stabilize Glutamate Receptors at the Neuromuscular Junction
- Recent Selective Sweeps in North American Show Signatures of Soft Sweeps
- Identification and Functional Analysis of Healing Regulators in
- A Multi-Megabase Copy Number Gain Causes Maternal Transmission Ratio Distortion on Mouse Chromosome 2
- Drosophila Casein Kinase I Alpha Regulates Homolog Pairing and Genome Organization by Modulating Condensin II Subunit Cap-H2 Levels
- The Hippo Pathway Regulates Homeostatic Growth of Stem Cell Niche Precursors in the Ovary
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Genomic Selection and Association Mapping in Rice (): Effect of Trait Genetic Architecture, Training Population Composition, Marker Number and Statistical Model on Accuracy of Rice Genomic Selection in Elite, Tropical Rice Breeding Lines
- Discovery of Transcription Factors and Regulatory Regions Driving Tumor Development by ATAC-seq and FAIRE-seq Open Chromatin Profiling
- Evolutionary Signatures amongst Disease Genes Permit Novel Methods for Gene Prioritization and Construction of Informative Gene-Based Networks
- Proteotoxic Stress Induces Phosphorylation of p62/SQSTM1 by ULK1 to Regulate Selective Autophagic Clearance of Protein Aggregates
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



