-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaTranscriptional Control of an Essential Ribozyme in Reveals an Ancient Evolutionary Divide in Animals
The processing of the 5′ end of nascent tRNAs is catalyzed by ribonuclease P (RNase P), an essential enzyme. In the ribonucleoprotein form of this enzyme, the RNase P RNA (RPR) functions as a ribozyme aided by protein cofactors. All previously examined eukaryotic RPR genes are transcribed from their own promoters by RNA pol III. In contrast, the Drosophila RPR gene is embedded in an intron of a recipient gene. We have shown that the embedded sequence, the only copy of RPR in the genome, is transcribed by pol II from the promoter of its recipient gene and encodes the functional RPR. Analysis of other animal genomes revealed that an embedded RPR is also present in the genomes of other insects and crustaceans. This feature provides evidence that the mode of transcription of RPR changed as the result of insertion into a recipient gene approximately 500 million years ago. This new, inserted type of RPR must first have appeared in the arthropod lineage in a common ancestor of insects and crustaceans.
Published in the journal: . PLoS Genet 11(1): e32767. doi:10.1371/journal.pgen.1004893
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004893Summary
The processing of the 5′ end of nascent tRNAs is catalyzed by ribonuclease P (RNase P), an essential enzyme. In the ribonucleoprotein form of this enzyme, the RNase P RNA (RPR) functions as a ribozyme aided by protein cofactors. All previously examined eukaryotic RPR genes are transcribed from their own promoters by RNA pol III. In contrast, the Drosophila RPR gene is embedded in an intron of a recipient gene. We have shown that the embedded sequence, the only copy of RPR in the genome, is transcribed by pol II from the promoter of its recipient gene and encodes the functional RPR. Analysis of other animal genomes revealed that an embedded RPR is also present in the genomes of other insects and crustaceans. This feature provides evidence that the mode of transcription of RPR changed as the result of insertion into a recipient gene approximately 500 million years ago. This new, inserted type of RPR must first have appeared in the arthropod lineage in a common ancestor of insects and crustaceans.
Introduction
RNase P catalyzes the essential removal of the 5′ leader sequence from precursor tRNAs (pre-tRNAs) [1]–[5]. With the exception of some protein-only variants in eukaryotes [6], [7], RNase P is a ribonucleoprotein (RNP) complex that consists of a catalytic RNA (RNase P RNA, RPR) and as many as ten protein cofactors (RNase P proteins, RPPs) in eukaryotes, up to five protein cofactors in archaea, and just one in bacteria [1], [2]. Conserved sequences and structural elements (including the active site) in all RPRs are suggestive of a shared evolutionary ancestry. By contrast, homology among RPPs is restricted to those of archaea and eukaryotes.
Biochemical characterization of bacterial RNase P has provided insights into how a single protein cofactor aids RNA catalysis by enhancing affinity for metal ions and substrate recognition [8], [9]. Comparisons of bacterial RNase P to its multi-subunit archaeal and eukaryotic counterparts provide an opportunity to examine whether structural and functional attributes of the RPR have been appropriated by additional protein cofactors. Of additional interest is understanding the role of these RPPs in regulating the function of RNase P during development and in response to environmental cues. In our efforts to develop Drosophila RNase P as a multicellular eukaryotic experimental model, we examined the transcription of RPR, and our work has unexpectedly shed some light on the evolution of this ancient ribozyme.
Eukaryotic RPRs that have been analyzed to date, ranging from yeast to human, are transcribed by pol III [2], [10]–[13]. The RPR gene in all Drosophila species examined [14]–[16] has been annotated in the last intron of ATPsynC/CG1746 [17], the pol II-transcribed gene that encodes subunit C of the F0 complex that is part of the mitochondrial ATP synthase [15]. We showed that the RPR locus within this gene does indeed produce, in a splicing-independent fashion, a functional RPR. In subsequent analysis of genomic databases, we found that such embedding of the RPR gene within a pol II-transcribed gene is also a characteristic in all insects and crustaceans examined. This common feature within a major group of arthropods suggests that the change from pol III to pol II transcription of RPR occurred approximately 500 million years ago [18].
Results
An intronic RPR is conserved and expressed in Drosophila species
Each of the twelve species of Drosophila for which genome sequence is available has a single copy of the RPR gene. In all cases, the RPR gene is inserted in the last intron of a recipient gene, ATPsynC/CG1746, with both genes arranged in the same 5′ to 3′ orientation (Fig. 1A). The RPR sequence is conserved, as are the ATPsynC exons and UTRs, but the other introns of the gene are not conserved (Fig. 1B). In keeping with a functional role, RPR-derived RNAs accumulate at higher levels (3 - to 5-fold in polyA+ samples and 20-fold in total RNA samples) than those corresponding to the preceding intron (Fig. 1C). Like its recipient gene, RPR is expressed throughout development in D. melanogaster and in multiple tissues (Fig. 1C and S1 Fig.).
Fig. 1. Drosophila RPR is embedded in an intron and ubiquitously expressed. 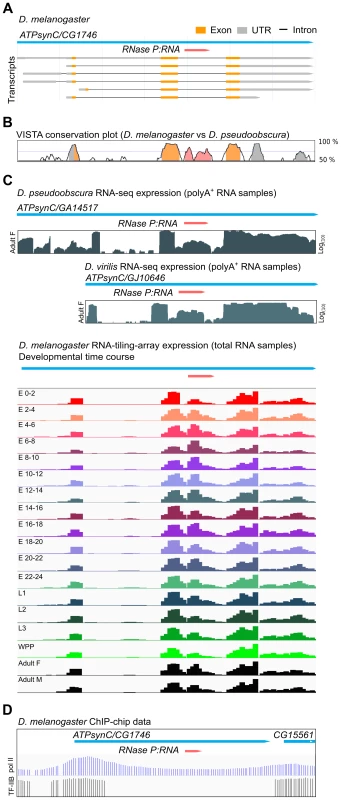
A. The RPR gene (pink) in D. melanogaster is present in the last intron of ATPsynC/CG1746. B. This arrangement is conserved in D. pseudoobscura (and other members of the genus). The exons of ATPsynC (orange peaks) are highly conserved between these species, as is the region within the intron that contains RPR (pink). The preceding intron is not conserved. Untranslated regions of ATPsynC are shown in grey. Peaks showing 75% or greater conservation are colored. C. Analysis of polyA-selected RNA [52] from D. pseudoobscura and D. virilis, and of total RNA from different developmental stages of D. melanogaster show that the region corresponding to RPR is expressed at higher levels than the preceding intron [52], [53]. Presence of RPR in polyA+ RNA is likely due to carryover (see also Materials and Methods). D. ChIP on chip data (D. melanogaster embryos) showing binding sites of pol II [63] and transcription factor IIB (TF-IIB) [64] in the 5′ region of ATPsynC/CG1746. E, embryonic stage in hours after egg laying; L, larval instar; WPP, white pre-pupae; F, female; M, male. Although the expression data suggest that RPR-derived RNAs are expressed, we could not identify an RPR promoter by sequence analysis. The flanking sequences required for transcription by pol III, which are found in known eukaryotic RPR genes [10]–[12], [19], are absent in the vicinity of the Drosophila ATPsynC-RPR genes (S2A Fig.). The Drosophila RPR genes also lack internal pol III recognition sequences that are characteristic of tRNA genes (S2A Fig.) [16], [20]. Analysis of data from genome-wide chromatin immunoprecipitation (ChIP) assays in D. melanogaster shows binding of pol II in the 5′ region of the ATPsynC-RPR locus (Fig. 1D) [20], but ChIP studies mapping pol III binding in Drosophila do not identify a pol III target in the vicinity of the RPR genes [16], [20]. Together, these findings show that Drosophila RPR is expressed, but sequence analysis did not identify a pol III promoter that could drive its independent expression.
Drosophila RPR expression requires a pol II promoter
The insertion of the RPR gene, which apparently lacks an independent promoter, into the last intron of ATPsynC in Drosophila suggests that RPR may be transcribed from the recipient gene promoter. To test this idea experimentally, we generated a reporter gene with the RPR-coding intron from D. virilis inserted between two red fluorescent protein (RFP) exons (Fig. 2A). The reporter gene was tested in D. melanogaster S2 cells in which D. virilis RPR can be distinguished from the endogenous D. melanogaster RPR by size and sequence differences. Transfected S2 cells expressed RFP from the reporter gene when driven by an Actin 5C (Act5C) promoter (pol II) [21]. RFP expression indicated that all the cis-elements required for correct splicing of the intron were present in the construct.
Fig. 2. RPR is processed from a recipient intron. 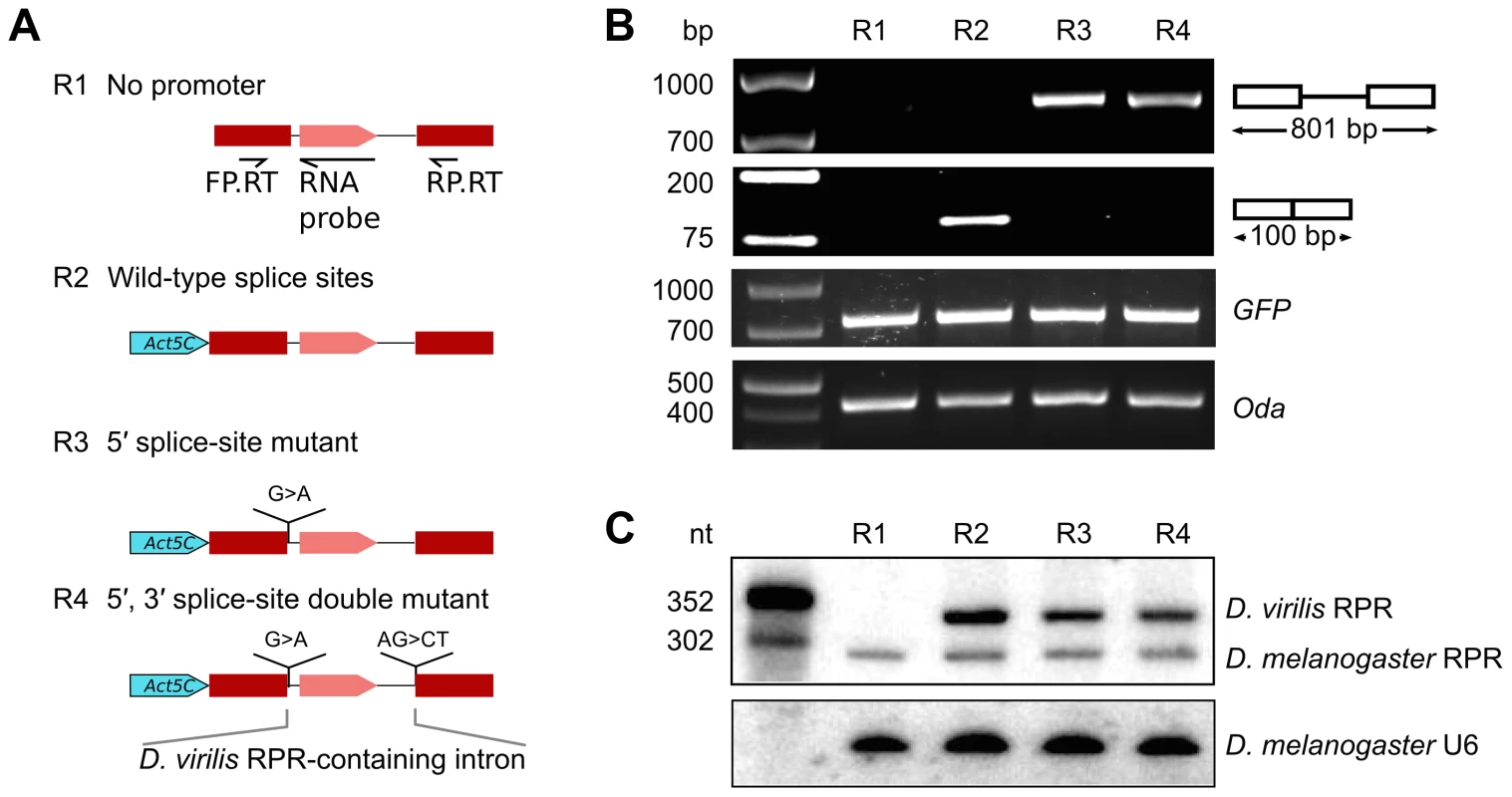
Various RFP reporter genes harboring the RPR-coding D. virilis intron were expressed in D. melanogaster S2 cultured cells to define sequences required for biogenesis of RPR. A. Schematic showing the reporter genes tested. B. RFP pre-mRNA and mRNA were analyzed by RT-PCR (using the primers FP.RT and RP.RT indicated in A). The presence of the protein was determined by fluorescent microscopy. C. RPR was detected by northern analysis. The antisense D. virilis RNA probe also detected the native D. melanogaster RPR because of sequence conservation. Controls used: GFP, expressed from a co-transfected plasmid to serve as a control for transfection efficiency; Oda (Ornithine decarboxylase antizyme) is a housekeeping transcript used to normalize input RNA for the RT-PCR experiment; U6 RNA was used as loading control the northern analysis. We analyzed RNA products from the reporter gene using RT-PCR and northern analysis. As expected from RFP expression, the mature RFP mRNA was expressed (R2 in Fig. 2B). D. virilis RPR was also expressed, demonstrating that the intron contained all the sequences necessary for production of mature RPR (R2 in Fig. 2C). The D. virilis RPR co-purified with RNase P activity (S3 Fig.), indicating that it assembled with endogenous RPPs to form a functional holoenzyme. D. virilis RPR was also expressed from a reporter gene with a UAS-Hsp70 promoter [22], showing that the production of RPR is not dependent on the identity of the pol II promoter (S4 Fig.). In a reporter gene lacking a promoter sequence (Fig. 2A), no RPR was detected by northern analysis (R1 in Fig. 2C). This finding ruled out the possibility that the RPR gene was transcribed by a cryptic promoter in the intron that we could not identify by sequence analysis. Importantly, the failure to produce RPR also showed that transcription of RPR was dependent on the pol II promoter of the recipient gene.
Splicing is not required for accumulation of RPR
To assess if splicing is required for processing of RPR from the intron, we designed splicing-deficient reporter genes and analyzed the RNA products using RT-PCR and northern analysis. We tested two reporter genes, one with a 5′ splice-site mutation and another with both 5′ and 3′ splice-site mutations (R3 and R4 in Fig. 2A). These mutations effectively blocked splicing as only the pre-mRNA for RFP was detected in the cells and no RFP expression was observed (Fig. 2B). In contrast, mature RPR accumulated (Fig. 2C), indicating that splicing is not required to process RPR from the primary transcript.
The embedded Drosophila RPR gene encodes the ribozyme required for RNase P activity
The embedded Drosophila RPR is the only RPR copy in the genome suggesting that it fulfills the essential function as the ribozyme component of RNase P. We examined the association of the RPR with Drosophila RNase P to verify its functional role in the enzyme. The holoenzyme was partially purified from D. melanogaster S2 tissue-culture cells using sequential, ion-exchange chromatography on DEAE-Sepharose (anionic) followed by SP-Sepharose (cationic). The presence of RNase P activity in fractions from each matrix was detected using a pre-tRNA processing assay (Fig. 3A) [23]. Peak activity from both matrices was found in fractions eluted with 300 to 500 mM NaCl. D. melanogaster RNase P cleaved pre-tRNAGly to yield two products identical in size to those generated by the Escherichia coli enzyme, which was used as a reference standard (Fig. 3A). The mature tRNA resulting from cleavage by D. melanogaster RNase P had a 5′ phosphate on G+1, an end group expected from RNase P catalysis (Fig. 3B). This inference was based on finding guanosine-3′,5′-bisphosphate (pGp) in a thin-layer chromatogram of the products from RNase T2 digestion of D. melanogaster RNase P-generated mature tRNAGly.
Fig. 3. D. melanogaster RPR co-purifies with RNase P and is required for its activity. 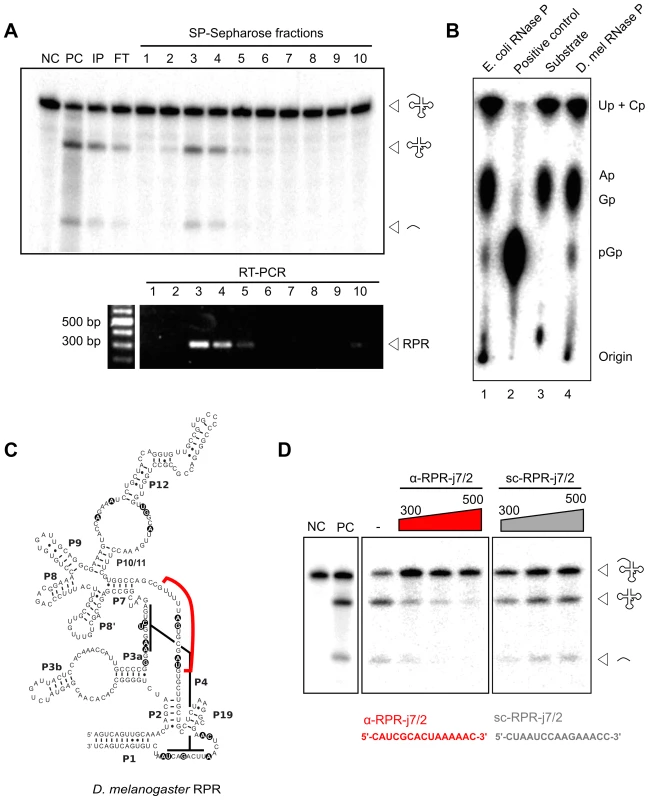
A. RNase P activity was partially purified from D. melanogaster S2 cells using sequential DEAE- and SP-Sepharose (above). Pre-tRNA processing assays established that the peak of activity eluted in 300–500 mM NaCl (fractions 3–5). RNA isolated from all fractions was subjected to RT-PCR using RPR-specific primers. Amplicons corresponding to the expected RPR size were detected in fractions 3–5 that showed maximal RNase P activity. B. Thin-layer chromatographic analysis of RNase T2-cleaved tRNAGly containing a 5′-pGp; the tRNAGly was first generated from cleavage of internally [α-32P]-GTP-labeled pre-tRNAGly by in vitro reconstituted E. coli RNase P or partially-purified D. melanogaster RNase P (lanes 1 and 4, respectively). The negative control (lane 3) shows RNase T2-cleaved internally labeled pre-tRNAGly that lacks a 5′-pGp, and the positive control (lane 2) shows RNase T2-cleaved 5′-labeled pre-tRNAGly that has a 5′-pGp. C. The predicted secondary structure of D. melanogaster RPR contains universally-conserved and functionally-important nucleotides (indicated by black circles). An antisense RNA oligonucleotide (red line; α-RPR-j7/2, complementary to a predicted single-stranded region between paired regions P7 and P2) was designed to inhibit RNase P activity. D. Partially-purified RNase P was inactivated with increasing concentrations of α-RPR-j7/2, but not with a scrambled oligo (sc-RPR-j7/2) that has the same nucleotide composition as α-RPR-j7/2. NC, negative control with no enzyme added; PC, positive control with in vitro reconstituted E. coli RNase P; IP, input; FT, flow-through. RPR present in the SP-Sepharose fractions was then detected using reverse-transcription and PCR (RT-PCR). The enrichment of RPR in fractions that also showed RNase P activity is consistent with its co-purification with the holoenzyme (Fig. 3A). To test if this co-purified RPR is required for RNase P activity, we designed an antisense RNA oligonucleotide (α-RPR-j7/2) that is complementary to a predicted single-stranded region that is part of the RPR active site (Fig. 3C). Incubation with α-RPR-j7/2 inhibited RNase P activity in a concentration-dependent fashion (Fig. 3D). In contrast, another oligonucleotide with the same nucleotide composition as α-RPR-j7/2 but a scrambled sequence (sc-RPR-j7/2) was ineffective at inhibiting activity even at the highest concentration tested. Together, these results confirm that the intronic RPR encodes the RNA component of D. melanogaster RNase P and is required for its activity.
RPR genes in insects and crustaceans lack signals for pol III transcription
To determine if the insertion of RPR in a recipient gene is unique to the Drosophila genus or more widespread in the animal kingdom, we analyzed RPR genes in the genomes of additional animals. All newly identified genes were verified to encode RPRs by their resemblance to typical eukaryotic RPRs in secondary structures and location of conserved nucleotides, including those essential for catalysis (S5 Fig.) [13]. Strikingly, we found a divide that classifies animals into two groups—(i) insects and crustaceans that have embedded RPR genes lacking signals for pol III transcription (Fig. 4, Fig. 5, S2A Fig. and S6A Fig.), and (ii) other animals that have typical signals for pol III-dependent transcription (Fig. 4, Fig. 5, and S2B Fig.). We draw these conclusions from an examination of species in the four subphyla of extant arthropods [Hexapoda (Insecta and Entognatha), Crustacea, Myriapoda, Chelicerata] and some non-arthropods that had not been previously examined.
Fig. 4. Insects and crustaceans have RPR genes embedded in pol II recipient genes, while other animals have independent pol III genes. 
The location of RPR and the neighboring genes in representative species of insects, crustaceans, and other animals are shown. In insects and crustaceans (light grey), RPR genes lack pol III signals and are in an intron (see also S2 Fig.). The P. humanus gene lacks pol III signals and is currently annotated between two genes. Each recipient gene is color-coded (as in Fig. 5; homologous genes have the same color). In other sub-phyla of Arthropoda (Myriapoda and Chelicerata) and other phyla (Deuterostomia and Porifera) (dark grey), RPR is an independent pol III-regulated gene (see also S2 Fig.). RPRs without pol III signals, pink; RPRs with pol III signals, blue; proximal sequence element (PSE), blue star; TATA box 21–27 nucleotides upstream of RPR, green oval; 3′ poly-T stretch of 4–5 nucleotides, red triangle. Wavy lines indicate regions where either poor sequence quality or weak homology prevents accurate prediction of the exons. Scale bar is 1 kb. Fig. 5. RPR genes lacking pol III signals are only present in the arthropod clade that includes insects and crustaceans. 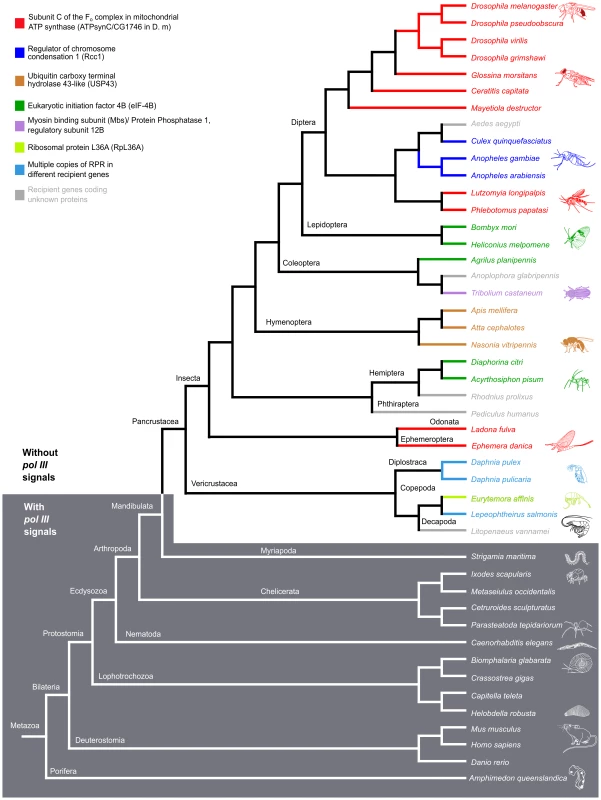
Phylogenetic relationship of animals showing two groups, those with RPR genes lacking pol III signals (light grey) and others with typical motifs found in type 3 pol III genes (dark grey). (See S2 Fig. for sequence motifs). The divide occurs in Arthropoda—species of Insecta and Vericrustacea (true crustaceans, including branchiopods and copepods) have RPR genes that lack pol III signals, whereas species of Myriapoda and Chelicerata have RPR genes with typical pol III signals. The RPR genes are associated with a variety of different recipient genes, indicated by different colored bars and named in the key (the same scheme is used in Fig. 4). In crustaceans (cyan), where there are multiple RPR genes in a single species, none is inserted in the ortholog of a gene identified as a recipient gene in insects. Within the Hexapoda and Crustacea, we examined species in eight orders of insects and three orders of crustaceans. All these RPR genes lack signals required for pol III transcription (S2A Fig.). In 26 out of 27 insect species, the RPR gene is present in an annotated pol II-dependent recipient gene and oriented in the same 5′ to 3′ direction (Fig. 4). The one exception is Pediculus humanus (human body louse) where RPR is in a poorly annotated region. Nevertheless, it is likely that the P. humanus RPR is part of a recipient gene because it lacks signals for pol III transcription. In the case of Tribolium castaneum (red flour beetle) and Heliconius melpomene (Postman butterfly), there are two copies of RPR in the same recipient gene (Fig. 4 and S7B Fig.). The two RPR copies are present in tandem within the same intron in T. castaneum, while they are present in two different introns of the same gene in H. melpomene. We were unable to examine species in Entognatha, the other Hexapod class, because there is no genomic sequence available. In the five crustaceans that we examined, there are two or more RPR genes in a given species and all lack signals for pol III transcription (Fig. 4 and S6 Fig.). For example, there are ten RPR-like genes in Daphnia pulex, which is consistent with the extensive gene duplications that have occurred in its genome (S6A Fig.) [24]. At least one D. pulex RPR gene is expressed [24] and may be a functional gene (Fig. 4). Finding an inserted type of RPR gene in insects and crustaceans is consistent with their close evolutionary relationship [25]–[28] (Fig. 5).
Within the Myriapoda and Chelicerata, we examined one myriapod (centipede) and four chelicerates (spider, tick, scorpion, and mite). All species have an RPR gene with typical signals for pol III-dependent transcription (S2B Fig.). The same was found for five non-arthropod animal species we examined [two molluscs (snail and oyster), two annelids (polychaete worm and leech), and a sponge] in which RPR had not been previously analyzed (S2B Fig.). RPR genes in all these non-insect and non-crustacean species are present in intergenic regions, except for the centipede Strigamia maritima, where the gene is found in an intron in the opposite orientation to the recipient gene. These genes have typical signals for pol III transcription (Fig. 4 and S2B Fig.). These arthropods (Myriapoda and Chelicerata) and all other animals examined to date have what has been considered a typical RPR that is transcribed by pol III (Fig. 4 and Fig. 5).
RPR resides in various recipient genes indicating a dynamic evolutionary history
Although the initial insertion of RPR into a recipient gene in the arthropod lineage appears to have been a single event, RPR moved again multiple times after this event as shown by its association with several different recipient genes (Fig. 5). In the eight orders of insects we examined, five different recipient genes were identified (Fig. 5). RPR recipient genes were also different within an order; for example, RPR is present in Regulator of chromosome condensation 1 (Rcc1) in mosquitoes, but it is in ATPsynC in the other species of Diptera.
Using the recipient gene as an indicator, ATPsynC appears to be the oldest recipient gene for RPR in the insects, as it is the common recipient gene in species belonging to the most divergent orders—the highly derived Diptera and the basal Ephemeroptera and Odonata (Fig. 4 and Fig. 5). Moreover, in D. melanogaster, Ephemera danica (mayfly) and Ladona fulva (dragonfly), RPR resides in the same intron providing further support for ATPsynC being the original recipient site in insects (Fig. 4, Fig. 5, and S7A Fig.).
Another common recipient gene for RPR is eukaryotic initiation factor 4B (eIF-4B). RPR is present in eIF-4B in seven species belonging to three orders—Lepidoptera (moths and butterflies), Coleoptera (beetles) and Hemiptera (true bugs, including aphids) (Fig. 4, Fig. 5, and S7B Fig.). In five of the seven species, the insertion of RPR is in the same intron in eIF-4B. Although there is no significant conservation of its sequence, the intron can be identified based on the conserved amino acid sequence of the flanking exons (S7B Fig.). Presence of the RPR in the same intron is consistent with a common ancestor for these orders, but this is not supported by a well-established insect phylogeny [25]. An alternative explanation is that these were independent events and examples of recipient-site convergence. This idea is supported by the case of the Asian citrus psyllid (Diaphorina citri) and the bull-headed dung beetle (Onthophagus taurus) where RPR is in different eIF-4B introns (S7B Fig.), reflecting independent insertions of RPR into eIF-4B likely due to a bias for this recipient gene.
In D. melanogaster, the homologs of recipient genes in other insects and crustaceans are all expressed throughout development and in multiple tissues, with ATPsynC being one of the most highly expressed genes (S1 Fig.). This observation supports the idea that the expression pattern and level of expression may constrain possible recipient genes, so that only those genes with ubiquitous and high expression are suitable sites for insertion of RPR (S1 Fig.). In Tribolium castaneum, there are two RPR genes embedded in tandem in the myosin binding subunit/protein phosphatase 1 regulatory subunit 12B-like gene (Mbs/PPP1R12B). Although Mbs shows a low level of expression relative to the other recipient genes, the two copies of RPR may compensate for this (Fig. 4 and S1 Fig.). Analyzing more insect genomes and transcriptomes will provide information about genomic contexts suitable for functional insertion of RPR and may reveal common features of recipient sites.
RNase MRP RNA, a sister RNA to RPR, is regulated by pol III
RNase MRP has roles in mitochondrial DNA replication, nucleolar rRNA processing, and mRNA turnover, and is present only in eukaryotes. It is an RNP that shares eight protein subunits with RNase P [29]. Furthermore, the RNA subunit of RNase MRP (MRP RNA) resembles RPR and appears to have derived from a common ancestor by a gene duplication event early in eukaryotic evolution [17], [30]. The two RNAs, albeit similar in secondary structure, have distinctive features that enable their unambiguous identification.
Given our unexpected findings of a transcriptional switch for the RPR in insects and crustaceans, we conducted a survey of MRP RNA genes in 26 insect species (in addition to Drosophila [14], [31]). These newly identified genes encode bona fide MRP RNAs, as judged by secondary structures and the location of various previously established signature motifs; for example, a five-nucleotide “GARAR” consensus in L8 (the terminal loop which caps the P8 helix; [17]) is present in all of them. In all 26 cases, we found signals for pol III transcription (S8 Fig.) [14], [31]. Therefore, MRP RNA genes, in contrast to RPR genes, appear to have maintained pol III regulation throughout the animal kingdom, including insects and crustaceans.
Discussion
Eukaryotic RPR genes have been widely held as independent genes transcribed by RNA pol III. Contrary to this generalization, we found that crustaceans and insects have RPR genes that lack signals for pol III transcription and are embedded in a recipient gene. In Drosophila, we demonstrated that the embedded RPR is dependent on the pol II promoter of a recipient gene for expression and that the encoded RNA copurifies with and is required for RNase P activity. Our findings change the long-held view of RPR as a prototype pol III-dependent gene [12], [32], and have implications for the biogenesis and evolutionary genetics of RPR.
The biogenesis of RPR
In Drosophila species, the RPR gene is embedded in the last intron of the ATPsynC gene. We found splicing was not required to produce mature RPR using an experimental reporter system. In the native context, RPR could either be generated from the spliced-out intron or from the primary transcript, with additional processing required to trim sequences beyond the mature RPR termini. Certain classes of micro RNAs (miRNAs) [33] and intron-derived small nucleolar RNAs (snoRNAs) [34], [35] also require processing to generate their mature 5′ and 3′ termini. The intronic miRNAs, which also do not require splicing when assessed using reporters [33], are processed to their mature lengths by Drosha and Pasha/DGCR8 [36]. It is unlikely these endonucleases trim Drosophila RPR, because their recognition sequences are absent in the regions flanking the mature RPR. In the case of intron-derived snoRNAs, examples of both splicing-dependent and splicing-independent processing are found, wherein nucleolytic trimming guides the maturation of the snoRNA termini following the assembly of snoRNP proteins [34], [35]. Like snoRNP proteins aiding the processing of the snoRNAs, RPPs could play a role in the maturation of the intronic RPR, but details of the assembly of the RPPs on the intronic RPR remain to be investigated. To further understand the biogenesis of the intronic RPR, it will be important to identify the nucleases that act on the RPR ends to produce the mature form. We presume these enzymes were already present in the founder animal for processing other non-coding RNAs, and were co-opted to generate the mature RPR from the recipient gene transcript. If so, identifying the enzymes acting on RPR may also provide general information on the biogenesis of some other non-coding RNAs.
It has been reported that some other non-coding RNAs show differences in their transcriptional control and are transcribed by pol II in some organisms and pol III in others [37], [38]. The significance, if any, for the different mechanisms is unclear. One of the possible effects of a change in the transcriptional control of RPR is altered RNA activity (for example, from differences in modification). Testing this idea using RPR produced in vivo by pol II or pol III in a pre-tRNA processing assay will provide a tractable experimental model for determining whether the transcriptional shift between pol II and pol III has functional consequences for a non-coding RNA.
Evolutionary genetics of RPR
Based on sequence analysis, it has been hypothesized that RPR gene gave rise to the MRP RNA gene in eukaryotes, presumably through gene duplication followed by neofunctionalization of the new gene copy [17], [30] (Fig. 6A). While MRP RNA is under pol III regulation in all animals that we and others have examined, RPR has undergone a second genetic event that inserted it into a recipient gene in crustaceans and insects (Fig. 6A). Current data indicate that this genetic change, which caused embedding of RPR within the arthropod lineage, occurred approximately 500 million years ago in an ancestor of the insects and crustaceans, an estimate that is placed prior to the emergence of the insects at approximately 479 million years ago [18]. The species of crustaceans we examined are examples of the so-called true crustaceans (Vericrustacea) [26], which are closely related to the insects (Hexapoda); both are members of the epic Pancrustacea clade (Fig. 6B) [26]–[28]. The other major group of Pancrustaceans is the Oligostraca, that includes the seed shrimp, oar-feet, fish lice, and tongue worms, for which there is currently no genomic sequence. If Oligostraca species have an embedded RPR, this would support an earlier origin—in an ancestor of all pancrustaceans (Fig. 6B). As more genomes become available, we will be able to refine when a pol II-regulated RPR first occurred and test whether it was indeed a single event in arthropod evolution.
Fig. 6. Model for the evolutionary history of RPR. 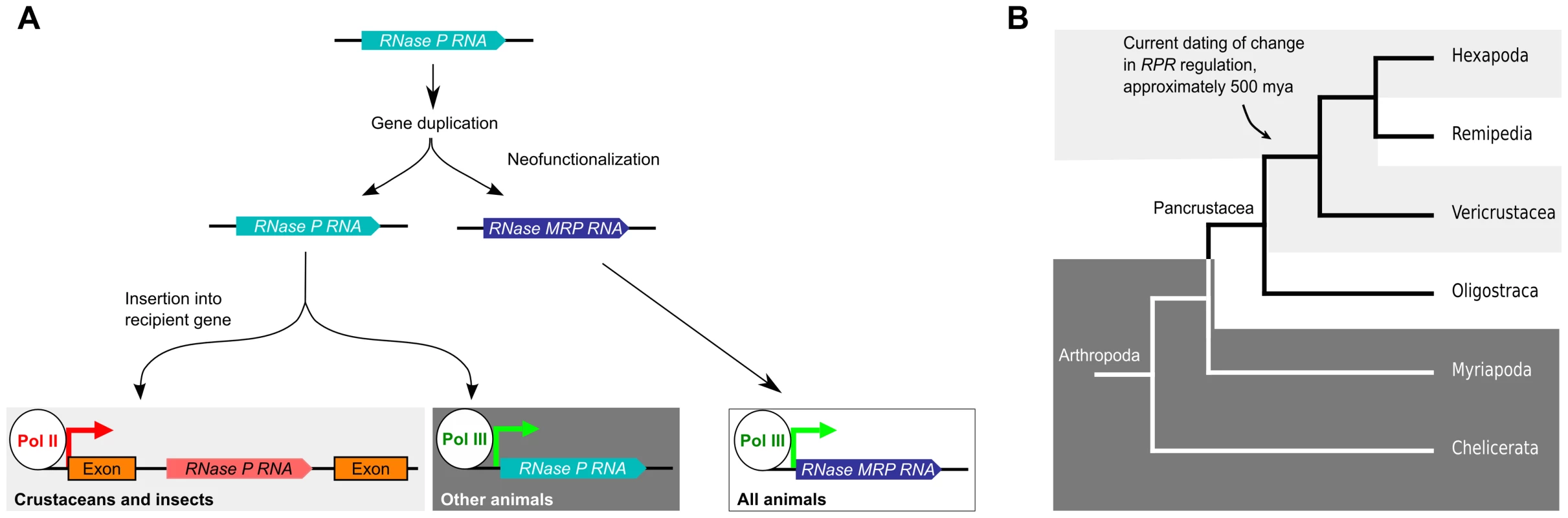
A. An ancestral RPR gene is thought to have undergone gene duplication and one of the daughter genes assumed the new functions of MRP RNA (neofunctionalization). MRP RNA is transcribed by pol III in all animals, as is RPR in animals previously characterized. We found that the RPR gene in crustaceans and insects has undergone another genetic event that inserted it, devoid of pol III signals, into a pol II-transcribed gene. B. A cladogram showing arthropod evolution (based on, [26], [27], [65]). In Hexapods and true crustaceans (Vericrustacea) (light grey), RPR is embedded in a pol II-regulated gene. In contrast, in Myriapoda and Chelicerata, RPR is a pol III gene (dark grey). The RPR genes in Remipedia and Oligostraca have not been characterized due to lack of genomic sequences (unshaded). The arrow indicates a node that connects branches where RPR is found in a recipient gene. These groups are thought to have diverged 500 million years ago [18], [66]. We predict that Remipedia RPR is also embedded in a recipient gene similar to the sister group Hexapoda. An analysis of RPR in Oligostraca will enable us to determine if embedding of RPR occurred earlier in arthropod evolution in an ancestor of all pancrustaceans. Generating an embedded RPR could have involved DNA - or RNA-mediated duplication and subsequent loss of any associated signals for pol III transcription [39]. It is also possible, given the catalytic function of RPR, that the insertion resulted from reverse splicing, similar to at least one route hypothesized for the spread of self-splicing group I and group II introns [40], although this activity has yet to be demonstrated for RPR. Regardless of the mechanism, the insertion caused a change in the regulation of RPR so that it became dependent on pol II transcription. This is not the case for MRP RNA (S8 Fig.), which shows the switch in transcriptional regulation occurred uniquely to RPR. As a first step towards determining any consequence of the change in RPR transcription, genome editing could be used to engineer, for example, D. melanogaster to have only a pol III-dependent RPR gene. Such a strategy would allow determination of the phenotypic consequences of reverting to the ancestral regulation of RPR.
Following the initial event that caused embedding in a recipient gene, RPR moved again multiple times into different recipient genes (Fig. 4, Fig. 5, and S7B Fig.). Insertion does not appear to be random because RPR inserted independently into the same gene more than once. In cases where RPR is present in two copies, such as T. castaneum (Fig. 4) and H. melpomene (S7B Fig.), both are present in the same recipient gene either in the same or two different introns, which is suggestive of local duplications. The crustaceans that we examined all had multiple RPR copies, however, these were associated with different recipient genes (S6A Fig.). While we have not identified a ‘signature’ of an insertion site, it appears that in all instances a pol II-regulated RPR has been retained and no case of a pol III-regulated RPR was found.
Our studies have shed some light on the evolution of RPR, a legacy of the RNA world and the first true trans-acting ribozyme discovered, and suggest that RPR transcription and subsequent processing entails the use of a different mechanism in a large group of animals. Although it is not known if this mode of biogenesis has functional consequences, our findings add to the variations in RNase P, an essential housekeeping enzyme, already noted for the diversity in its subunit composition [3], [5].
Materials and Methods
Cell culture and RNA isolation
D. melanogaster S2 cells [41] were grown in Schneider insect medium (Sigma) with 10% (v/v) fetal bovine serum. DNA transfections were performed using Effectene (Qiagen). Cells were harvested 30 h post transfection and total RNA was isolated using Trizol (Invitrogen).
Construction of RFP-RPR reporter genes
The intronic region containing RPR was amplified from D. virilis genomic DNA using PCR and the following primers: forward primer virilis intron, 5′-CTGCTTCATCTACAAGGTTCGTATTGGTTACC-3′ and reverse primer virilis intron, 5′-CCGATGAACTTCACCTGTTGTATTGGTTGTC-3′. A DsRed ORF (from pP(RedH-Stinger) [42] was used as a template to generate two exons, separated at nucleotide 323, which creates a match to the consensus Drosophila splice junction [43]. The exons were generated using PCR and the following primer pairs: Exon I forward primer RFP, 5′-TCCGATATCATGGCCTCCTCC-3′ and Exon I reverse primer RFP, 5′-GGTAACCAATACGAACCTTGTAGATGAAGCAG-3′; Exon II forward primer RFP, 5′-GACAACCAATACAACAGGTGAAGTTCATCGG-3′ and Exon II reverse primer RFP, 5′ -ACCTCTAGACTACAGGAACAGGTGGTG -3′. The intron and the two exons were combined using overlapping PCR [44] and cloned into pPACPL, which contains the Act5C promoter [21]. DsRed exons were also generated with splice mutations. Splice mutations were created by site-directed mutagenesis with the following primer pairs: 5′ splice mutant forward, 5′-CTGCTTCATCTACAAGATTCGTATTGGTTACC-3′ and 5′ splice mutant reverse, 5′-GGTAACCAATACGAATCTTGTAGATGAAGCAG-3′; 3′ splice mutant forward, 5′-CAATGACAACCAATACAACCTGTGAAGTTCATCGGCGTGAACT-3′ and 3′ splice mutant reverse, 5′-AGTTCACGCCGATGAACTTCACAGGTTGTATTGGTTGTCATTG-3′. In addition, the fragment containing the DsRed exons with the D. virilis intron was cloned into pUAST [22] to generate a reporter gene under control of the UAS promoter. This reporter was expressed using a Gal4 gene (pGaTB) [22] cloned into the pPACPL vector using the following primers, Gal4 forward, 5′-TCCGATATCATGAAGCTACTGTCTTCTATC-3′; Gal4 reverse, 5′-AAATCTAGATTACTCTTTTTTTGGGTTTGGTGGGGTATCTTC-3′.
Reverse transcription and PCR
cDNAs were prepared using an oligo dT primer (for mRNAs) or gene specific primers (for RPRs) by reverse transcription using an Omniscript RT kit (Qiagen). cDNAs were amplified with Taq DNA polymerase (NEB) using the recommended conditions and the following primer pairs: Forward DmelRPR, 5′-AGTCAGTTGCAAACTAGCATC-3′ and Reverse Dmel RPR, 5′ - AGTCAGTCACAGATTAGTCTGAATTG-3′; Forward GFP, 5′-TAAGATATCATGGTGAGCAAGGG-3′ and Reverse GFP, 5′ - ACCTCTAGATTACTTGTACAGCTCGTCC-3′; Forward Oda, 5′-GTCCTTCGGTAGAGCGACAT-3′ and Reverse Oda, 5′ - GCACCATCTCGACTTCGTCT-3′.
Northern blot analysis
D. melanogaster and D. virilis RPRs were detected using full-length anti-sense RNA probes labeled with [α-32P]-ATP in an in vitro transcription reaction. The DNA templates were generated from PCR-mediated amplification of the genomic DNA using the following primers (for both species): Forward primer-genomic, 5′-AGTCAGTTGCAAACTAGCATCTG-3′ and Reverse primer-genomic, 5′-TCACTATAGGAGTCAGTCACAGATTAGTCTG-3′. A T7 RNA polymerase promoter was introduced to the above PCR product using a second round of PCR using the same forward primer and the following reverse primer: 5′-GAGAATTCTAATACGACTCACTATAGGAGTCAGTCACAG-3′. D. virilis RPR was also detected using the following DNA oligo, 5′ - CCGCGACACACAATCACCTCTCGGCTTTTGTATGTTGTTACAGCAAC-3′. U6 RNA was detected using the DNA oligo, 5′ - GCAGGGGCCATGCTAATCTTCTCTGTATCG-3′. Both DNA oligos were 5′-labeled using [γ-32P]-ATP and T4 polynucleotide kinase. Eight micrograms of total RNA isolated from transfected cells was separated on a 7.5% (w/v) polyacrylamide gel containing 8 M urea, transferred to a nylon membrane (Hybond N+, GE Healthcare) and analyzed by northern hybridization. After pre-hybridization in the same hybridization buffer, RNA probes were hybridized in hybridization buffer (5X SSC, 1% (w/v) SDS, 5X Denhardt's solution, 200 µg/ml of sheared salmon sperm DNA) for 16 h at 65°C. DNA oligo probes were hybridized in QuikHyb buffer (GE Healthcare) for 16 h at 55°C. Membranes were washed with 2X SSC with 0.1% (w/v) SDS at 10°C below hybridization temperature. The binding of labeled probes to their complementary target RNAs was detected using phosphorimaging.
RNase P purification
D. melanogaster S2 cells were collected by centrifugation and washed once with phosphate-buffered saline. Packed cells (100 µL) were lysed in 400 µL of lysis buffer [15 mM HEPES (pH 7.9), 3 mM MgCl2, 50 mM NaCl, 1 mM dithiothreitol, 0.2 mM phenylmethylsulfonyl fluoride, 0.1% (v/v) Tween-20, 10% (v/v) glycerol, 0.2 U/μL of Ribolock RNase Inhibitor (Thermo Scientific)]. Cells were homogenized using a type A glass Dounce homogenizer (Wheaton) on ice and debris was removed by centrifugation at 2,500 g for 10 min. The crude lysate was mixed with 100 µL of diethylaminoethyl (DEAE)-Sepharose resin (GE Healthcare), which had been pre-equilibrated with lysis buffer at 4°C for 30 min. The resin was collected by centrifugation (2,500 g for 5 min) and washed twice, each with 1 mL of lysis buffer to remove weakly bound constituents. Fractions were eluted stepwise with increasing NaCl concentration (from 50 mM to 1 M) in lysis buffer, and tested for RNase P activity (as described below). Fractions with detectable activity were pooled and dialyzed twice, each with 500 volumes of lysis buffer (without NaCl and RNase inhibitor) for 2 h at 4°C. The dialysate was then mixed with 100 µL sulfopropyl (SP)-Sepharose resin (GE Healthcare), washed with 1 ml of lysis buffer, and bound constituents eluted with increasing NaCl concentration (as described above for the DEAE-Sepharose purification).
RNase P activity (pre-tRNA processing) assay and inhibition studies
Four μL of partially purified Drosophila RNase P fractions were assayed in a 20-μL reaction containing 10 mM HEPES (pH 7.9), 10 mM magnesium acetate, 200 mM ammonium acetate, 0.1% (v/v) Nonidet P-40 and 250 nM of in vitro transcribed pre-tRNAGly (tobacco chloroplast; without 3′-CCA), a trace amount of which had been internally labeled with [α-32P]-GTP (28). The reactions were incubated at 28°C for 10 min, and then terminated with 10 µL of 2X urea loading dye [8 M urea, 15 mM EDTA, 0.025% (w/v) xylene cyanol, 0.025% (w/v) bromophenol blue, 20% (v/v) phenol]. The products were separated on an 8% (w/v) polyacrylamide (19∶1) gel containing 8 M urea, and detected using phosphorimaging. Oligo-inhibition assays were performed as described earlier for bacterial and archaeal RNase P [23], [45], [46]. For these experiments, the RNA oligo (final concentration 300, 400 or 500 µM) was pre-incubated with 4 µL partially purified RNase P in assay buffer for 5 min at 28°C. After addition of substrate (pre-tRNAGly) in assay buffer, the reaction was incubated for 15 min at 28°C, and then terminated and characterized as described above.
Bioinformatics analysis
We performed sequence analysis using genomic data from multiple sources—i5K Pilot Project (Baylor College of Medicine, Human Genome Sequencing Center), NCBI Genome, Ensembl Genomes, VectorBase, Penaeus Genome Database (PAGE), BeeBase, wFleaBase, FlyBase, DOE Joint Genome Institute (Table S1), using the Infernal package (release 1.1) [47]. The secondary structures for newly discovered RPRs were drawn using a ClustalW2-aided multiple sequence alignment of the RNA sequences and Mfold [48]. The results of our search, which yielded 32 new RPRs, were independently validated by the fact that 9 of these 32 RPRs were identified using a different bioinformatics approach [49]. These putative RPRs were manually analyzed for size, the presence of conserved nucleotides (identity and location) and the length of P1 helix (which help define the 5′ and 3′ termini). The RPRs we identified diverged in the length and sequence from prototypes and were not identified using previous methods. Incorporating this information into covariance model-based searches will improve future RPR searches. Our results should also serve as a cautionary note for excluding putative RPR genes because they lack pol III promoters and terminators. MRP RNAs were identified and analyzed using an approach similar to that employed for RPRs.
The cladograms in Fig. 5 were generated using the NCBI taxonomy browser, Dendroscope [50], and current literature on arthropod phylogeny [25], [26]. The VISTA conservation graphs in Fig. 1B and S7A Fig. were generated using mVISTA [51]. For analysis of RPR expression during D. melanogaster development and in various tissues, data derived from the analysis of total RNA by tiling arrays were examined [52]. These data were obtained from the modENCODE consortium [53] as wig files and viewed using the Integrative Genomics Viewer [54] (S2 Table). For D. virilis and D. pseudoobscura, data from polyA-selected samples analyzed by RNA-seq were used (sam files). The SAMtools program was used to index and sort the RNA-seq reads [55] and the Integrative Genomics Viewer was used to visualize the reads. In both the total RNA and polyA-selected samples, reads corresponding to the RPR-containing intron are higher than those corresponding to the preceding intron in ATPsynC. The presence of RPR in the polyA+ sample may have resulted from incomplete removal of highly expressed RNAs, as has been observed in RNA-seq analyses for other non-polyA+ RNAs [56]. The reads for the polyA+ samples are as follows; D. virilis: RPR, 2435 reads/353 bp and preceding intron, 978 reads/683 bp; D. pseudoobscura, RPR 2478 reads/322bp and preceding intron 1980 reads/804 bp. For T. castaneum, RNA-seq data as raw reads (SRR1048514 and SRR1161702 in SRA format; refer to Table S1 for details) were downloaded from NCBI GEO, and ERR161589 as FASTA was downloaded from EMBL ENA. The SRR files were converted to FASTQ format using SRAtoolkit [57]. These reads were mapped to T. castaneum genome (Genome version Tcas3, from EMBL indexed using Bowtie2 [58]) and TopHat [59]. The mapped reads were then analyzed using Cufflinks [60] with annotated transcripts (Tcas3.22.gtf). The locus of T. castaneum recipient and housekeeping genes were identified in the version of the genome mentioned above, using TBLASTN (NCBI-BLAST+) (refer to Table S3 for loci) [61]. SAMtools was also used to extract genomic sequences flanking the identified RPRs and these sequences were aligned using ClustalW2 [62]. For each species, a consensus proximal sequence element (PSE) and TATA box was generated using an alignment of the U6 and 7SK RNA promoters. This consensus was used to search for candidate pol III promoter elements proximal to a given newly identified RPR sequence.
Supporting Information
Zdroje
1. Liu F, Altman S (2009) Ribonuclease P: Springer, New York.
2. ChamberlainJR, LeeY, LaneWS, EngelkeDR (1998) Purification and characterization of the nuclear RNase P holoenzyme complex reveals extensive subunit overlap with RNase MRP. Genes Dev 12 : 1678–1690.
3. RossmanithW (2011) Of P and Z: mitochondrial tRNA processing enzymes. Biochim Biophys Acta 1819 : 1017–1026.
4. HowardMJ, LiuX, LimWH, KlemmBP, FierkeCA, et al. (2013) RNase P enzymes: divergent scaffolds for a conserved biological reaction. RNA Biol 10 : 909–914.
5. LaiLB, VioqueA, KirsebomLA, GopalanV (2010) Unexpected diversity of RNase P, an ancient tRNA processing enzyme: challenges and prospects. FEBS Lett 584 : 287–296.
6. HolzmannJ, FrankP, LofflerE, BennettKL, GernerC, et al. (2008) RNase P without RNA: identification and functional reconstitution of the human mitochondrial tRNA processing enzyme. Cell 135 : 462–474.
7. GutmannB, GobertA, GiegeP (2012) PRORP proteins support RNase P activity in both organelles and the nucleus in Arabidopsis. Genes Dev 26 : 1022–1027.
8. GuentherUP, YandekLE, NilandCN, CampbellFE, AndersonD, et al. (2013) Hidden specificity in an apparently nonspecific RNA-binding protein. Nature 502 : 385–388.
9. SunL, HarrisME (2007) Evidence that binding of C5 protein to P RNA enhances ribozyme catalysis by influencing active site metal ion affinity. RNA 13 : 1505–1515.
10. MyslinskiE, AmeJC, KrolA, CarbonP (2001) An unusually compact external promoter for RNA polymerase III transcription of the human H1RNA gene. Nucleic Acids Res 29 : 2502–2509.
11. EderPS, SrinivasanA, FishmanMC, AltmanS (1996) The RNA subunit of ribonuclease P from the zebrafish, Danio rerio. J Biol Chem 271 : 21031–21036.
12. LeeJY, EvansCF, EngelkeDR (1991) Expression of RNase P RNA in Saccharomyces cerevisiae is controlled by an unusual RNA polymerase III promoter. Proc Natl Acad Sci U S A 88 : 6986–6990.
13. MarquezSM, ChenJL, EvansD, PaceNR (2006) Structure and function of eukaryotic Ribonuclease P RNA. Mol Cell 24 : 445–456.
14. PiccinelliP, RosenbladMA, SamuelssonT (2005) Identification and analysis of ribonuclease P and MRP RNA in a broad range of eukaryotes. Nucleic Acids Res 33 : 4485–4495.
15. TripoliG, D'EliaD, BarsantiP, CaggeseC (2005) Comparison of the oxidative phosphorylation (OXPHOS) nuclear genes in the genomes of Drosophila melanogaster, Drosophila pseudoobscura and Anopheles gambiae. Genome Biol 6: R11.
16. HernandezGJr, ValafarF, StumphWE (2007) Insect small nuclear RNA gene promoters evolve rapidly yet retain conserved features involved in determining promoter activity and RNA polymerase specificity. Nucleic Acids Res 35 : 21–34.
17. Davila LopezM, RosenbladMA, SamuelssonT (2009) Conserved and variable domains of RNase MRP RNA. RNA Biol 6 : 208–220.
18. MisofB, LiuS, MeusemannK, PetersRS, DonathA, et al. (2014) Phylogenomics resolves the timing and pattern of insect evolution. Science 346 : 763–767.
19. AmeJC, SchreiberV, FraulobV, DolleP, de MurciaG, et al. (2001) A bidirectional promoter connects the poly(ADP-ribose) polymerase 2 (PARP-2) gene to the gene for RNase P RNA. structure and expression of the mouse PARP-2 gene. J Biol Chem 276 : 11092–11099.
20. IsogaiY, TakadaS, TjianR, KelesS (2007) Novel TRF1/BRF target genes revealed by genome-wide analysis of Drosophila Pol III transcription. EMBO J 26 : 79–89.
21. KrasnowMA, SaffmanEE, KornfeldK, HognessDS (1989) Transcriptional activation and repression by Ultrabithorax proteins in cultured Drosophila cells. Cell 57 : 1031–1043.
22. BrandAH, PerrimonN (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118 : 401–415.
23. LaiLB, ChanPP, CozenAE, BernickDL, BrownJW, et al. (2010) Discovery of a minimal form of RNase P in Pyrobaculum. Proc Natl Acad Sci U S A 107 : 22493–22498.
24. ColbourneJK, PfrenderME, GilbertD, ThomasWK, TuckerA, et al. (2011) The ecoresponsive genome of Daphnia pulex. Science 331 : 555–561.
25. TrautweinMD, WiegmannBM, BeutelR, KjerKM, YeatesDK (2012) Advances in insect phylogeny at the dawn of the postgenomic era. Annu Rev Entomol 57 : 449–468.
26. RegierJC, ShultzJW, ZwickA, HusseyA, BallB, et al. (2010) Arthropod relationships revealed by phylogenomic analysis of nuclear protein-coding sequences. Nature 463 : 1079–1083.
27. OakleyTH, WolfeJM, LindgrenAR, ZaharoffAK (2013) Phylotranscriptomics to bring the understudied into the fold: monophyletic ostracoda, fossil placement, and pancrustacean phylogeny. Mol Biol Evol 30 : 215–233.
28. von ReumontBM, JennerRA, WillsMA, Dell'ampioE, PassG, et al. (2012) Pancrustacean phylogeny in the light of new phylogenomic data: support for Remipedia as the possible sister group of Hexapoda. Mol Biol Evol 29 : 1031–1045.
29. EsakovaO, KrasilnikovAS (2010) Of proteins and RNA: the RNase P/MRP family. RNA 16 : 1725–1747.
30. ZhuY, StribinskisV, RamosKS, LiY (2006) Sequence analysis of RNase MRP RNA reveals its origination from eukaryotic RNase P RNA. Rna 12 : 699–706.
31. SchneiderMD, BainsAK, RajendraTK, DominskiZ, MateraAG, et al. (2010) Functional characterization of the Drosophila MRP (mitochondrial RNA processing) RNA gene. Rna 16 : 2120–2130.
32. BaerM, NilsenTW, CostiganC, AltmanS (1990) Structure and transcription of a human gene for H1 RNA, the RNA component of human RNase P. Nucleic Acids Res. 18 : 97–103.
33. KimYK, KimVN (2007) Processing of intronic microRNAs. EMBO J 26 : 775–783.
34. HiroseT, ShuMD, SteitzJA (2003) Splicing-dependent and -independent modes of assembly for intron-encoded box C/D snoRNPs in mammalian cells. Mol Cell 12 : 113–123.
35. RichardP, KissAM, DarzacqX, KissT (2006) Cotranscriptional recognition of human intronic box H/ACA snoRNAs occurs in a splicing-independent manner. Mol Cell Biol 26 : 2540–2549.
36. HanJ, LeeY, YeomKH, KimYK, JinH, et al. (2004) The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev 18 : 3016–3027.
37. AebyE, UlluE, YepiskoposyanH, SchimanskiB, RoditiI, et al. (2010) tRNASec is transcribed by RNA polymerase II in Trypanosoma brucei but not in humans. Nucleic Acids Res 38 : 5833–5843.
38. DieciG, PretiM, MontaniniB (2009) Eukaryotic snoRNAs: a paradigm for gene expression flexibility. Genomics 94 : 83–88.
39. KatjuV (2012) In with the old, in with the new: the promiscuity of the duplication process engenders diverse pathways for novel gene creation. Int J Evol Biol 2012 : 341932.
40. LambowitzAM, BelfortM (1993) Introns as mobile genetic elements. Annu Rev Biochem 62 : 587–622.
41. SchneiderI (1972) Cell lines derived from late embryonic stages of Drosophila melanogaster. J Embryol Exp Morphol 27 : 353–365.
42. PaululatA, HeinischJJ (2012) New yeast/E. coli/Drosophila triple shuttle vectors for efficient generation of Drosophila P element transformation constructs. Gene 511 : 300–305.
43. ReeseMG, EeckmanFH, KulpD, HausslerD (1997) Improved splice site detection in Genie. J Comput Biol 4 : 311–323.
44. BonanoVI, OlteanS, Garcia-BlancoMA (2007) A protocol for imaging alternative splicing regulation in vivo using fluorescence reporters in transgenic mice. Nat Protoc 2 : 2166–2181.
45. ChildsJL, PooleAW, TurnerDH (2003) Inhibition of Escherichia coli RNase P by oligonucleotide directed misfolding of RNA. Rna 9 : 1437–1445.
46. GruegelsiepeH, WillkommDK, GoudinakisO, HartmannRK (2003) Antisense inhibition of Escherichia coli RNase P RNA: mechanistic aspects. Chembiochem 4 : 1049–1056.
47. NawrockiEP, EddySR (2013) Infernal 1.1 : 100-fold faster RNA homology searches. Bioinformatics 29 : 2933–2935.
48. ZukerM (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31 : 3406–3415.
49. YusufD, MarzM, StadlerPF, HofackerIL (2010) Bcheck: a wrapper tool for detecting RNase P RNA genes. BMC Genomics 11 : 432.
50. HusonDH, RichterDC, RauschC, DezulianT, FranzM, et al. (2007) Dendroscope: An interactive viewer for large phylogenetic trees. BMC Bioinformatics 8 : 460.
51. FrazerKA, PachterL, PoliakovA, RubinEM, DubchakI (2004) VISTA: computational tools for comparative genomics. Nucleic Acids Res 32: W273–279.
52. GraveleyBR, BrooksAN, CarlsonJW, DuffMO, LandolinJM, et al. (2011) The developmental transcriptome of Drosophila melanogaster. Nature 471 : 473–479.
53. RoyS, ErnstJ, KharchenkoPV, KheradpourP, NegreN, et al. (2010) Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science 330 : 1787–1797.
54. RobinsonJT, ThorvaldsdottirH, WincklerW, GuttmanM, LanderES, et al. (2011) Integrative genomics viewer. Nat Biotechnol 29 : 24–26.
55. LiH, HandsakerB, WysokerA, FennellT, RuanJ, et al. (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25 : 2078–2079.
56. TarazonaS, Garcia-AlcaldeF, DopazoJ, FerrerA, ConesaA (2011) Differential expression in RNA-seq: a matter of depth. Genome Res 21 : 2213–2223.
57. LeinonenR, SugawaraH, ShumwayM (2011) International Nucleotide Sequence Database C (2011) The sequence read archive. Nucleic Acids Res 39: D19–21.
58. LangmeadB, SalzbergSL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9 : 357–359.
59. TrapnellC, PachterL, SalzbergSL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25 : 1105–1111.
60. TrapnellC, WilliamsBA, PerteaG, MortazaviA, KwanG, et al. (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28 : 511–515.
61. CamachoC, CoulourisG, AvagyanV, MaN, PapadopoulosJ, et al. (2009) BLAST+: architecture and applications. BMC Bioinformatics 10 : 421.
62. LarkinMA, BlackshieldsG, BrownNP, ChennaR, McGettiganPA, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23 : 2947–2948.
63. LiXY, MacArthurS, BourgonR, NixD, PollardDA, et al. (2008) Transcription factors bind thousands of active and inactive regions in the Drosophila blastoderm. PLoS Biol 6: e27.
64. MacArthurS, LiXY, LiJ, BrownJB, ChuHC, et al. (2009) Developmental roles of 21 Drosophila transcription factors are determined by quantitative differences in binding to an overlapping set of thousands of genomic regions. Genome Biol 10: R80.
65. GiribetG, EdgecombeGD (2012) Reevaluating the arthropod tree of life. Annu Rev Entomol 57 : 167–186.
66. GlennerH, ThomsenPF, HebsgaardMB, SorensenMV, WillerslevE (2006) Evolution. The origin of insects. Science 314 : 1883–1884.
67. LeuJH, ChenSH, WangYB, ChenYC, SuSY, et al. (2011) A review of the major penaeid shrimp EST studies and the construction of a shrimp transcriptome database based on the ESTs from four penaeid shrimp. Mar Biotechnol (NY) 13 : 608–621.
68. EddySR (2002) A memory-efficient dynamic programming algorithm for optimal alignment of a sequence to an RNA secondary structure. BMC Bioinformatics 3 : 18.
69. AltschulSF, MaddenTL, SchafferAA, ZhangJ, ZhangZ, et al. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25 : 3389–3402.
70. BurgeCB, KarlinS (1998) Finding the genes in genomic DNA. Curr Opin Struct Biol 8 : 346–354.
71. Graveley BR, May G, Brooks AN, Carlson JW, Cherbas L, et al. (2011) The D. melanogaster transcriptome: modENCODE RNA-Seq data for dissected tissues.
72. BehrensS, PeussR, MilutinovicB, EggertH, EsserD, et al. (2014) Infection routes matter in population-specific responses of the red flour beetle to the entomopathogen Bacillus thuringiensis. BMC Genomics 15 : 445.
73. HepatR, SongJJ, LeeD, KimY (2013) A viral histone h4 joins to eukaryotic nucleosomes and alters host gene expression. J Virol 87 : 11223–11230.
Štítky
Genetika Reprodukční medicína
Článek Phosphorylation of Elp1 by Hrr25 Is Required for Elongator-Dependent tRNA Modification in YeastČlánek Naturally Occurring Differences in CENH3 Affect Chromosome Segregation in Zygotic Mitosis of HybridsČlánek Insight in Genome-Wide Association of Metabolite Quantitative Traits by Exome Sequence AnalysesČlánek ALIX and ESCRT-III Coordinately Control Cytokinetic Abscission during Germline Stem Cell DivisionČlánek Deciphering the Genetic Programme Triggering Timely and Spatially-Regulated Chitin Deposition
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 1
-
Všechny články tohoto čísla
- The Combination of Random Mutagenesis and Sequencing Highlight the Role of Unexpected Genes in an Intractable Organism
- Ataxin-3, DNA Damage Repair, and SCA3 Cerebellar Degeneration: On the Path to Parsimony?
- α-Actinin-3: Why Gene Loss Is an Evolutionary Gain
- Origins of Context-Dependent Gene Repression by Capicua
- Transposable Elements Contribute to Activation of Maize Genes in Response to Abiotic Stress
- No Evidence for Association of Autism with Rare Heterozygous Point Mutations in Contactin-Associated Protein-Like 2 (), or in Other Contactin-Associated Proteins or Contactins
- Nur1 Dephosphorylation Confers Positive Feedback to Mitotic Exit Phosphatase Activation in Budding Yeast
- A Regulatory Hierarchy Controls the Dynamic Transcriptional Response to Extreme Oxidative Stress in Archaea
- Genetic Variants Modulating CRIPTO Serum Levels Identified by Genome-Wide Association Study in Cilento Isolates
- Small RNA Sequences Support a Host Genome Origin of Satellite RNA
- Phosphorylation of Elp1 by Hrr25 Is Required for Elongator-Dependent tRNA Modification in Yeast
- Genetic Mapping of MAPK-Mediated Complex Traits Across
- An AP Endonuclease Functions in Active DNA Demethylation and Gene Imprinting in
- Developmental Regulation of the Origin Recognition Complex
- End of the Beginning: Elongation and Termination Features of Alternative Modes of Chromosomal Replication Initiation in Bacteria
- Naturally Occurring Differences in CENH3 Affect Chromosome Segregation in Zygotic Mitosis of Hybrids
- Imputation of the Rare G84E Mutation and Cancer Risk in a Large Population-Based Cohort
- Polycomb Protein SCML2 Associates with USP7 and Counteracts Histone H2A Ubiquitination in the XY Chromatin during Male Meiosis
- A Genetic Strategy for Probing the Functional Diversity of Magnetosome Formation
- Interactions of Chromatin Context, Binding Site Sequence Content, and Sequence Evolution in Stress-Induced p53 Occupancy and Transactivation
- The Yeast La Related Protein Slf1p Is a Key Activator of Translation during the Oxidative Stress Response
- Integrative Analysis of DNA Methylation and Gene Expression Data Identifies as a Key Regulator of COPD
- Proteasomes, Sir2, and Hxk2 Form an Interconnected Aging Network That Impinges on the AMPK/Snf1-Regulated Transcriptional Repressor Mig1
- Functional Interplay between the 53BP1-Ortholog Rad9 and the Mre11 Complex Regulates Resection, End-Tethering and Repair of a Double-Strand Break
- Estrogenic Exposure Alters the Spermatogonial Stem Cells in the Developing Testis, Permanently Reducing Crossover Levels in the Adult
- Protein Poly(ADP-ribosyl)ation Regulates Immune Gene Expression and Defense Responses
- Sumoylation Influences DNA Break Repair Partly by Increasing the Solubility of a Conserved End Resection Protein
- A Discrete Transition Zone Organizes the Topological and Regulatory Autonomy of the Adjacent and Genes
- Elevated Mutation Rate during Meiosis in
- The Intersection of the Extrinsic Hedgehog and WNT/Wingless Signals with the Intrinsic Hox Code Underpins Branching Pattern and Tube Shape Diversity in the Airways
- MiR-24 Is Required for Hematopoietic Differentiation of Mouse Embryonic Stem Cells
- Tissue-Specific Effects of Genetic and Epigenetic Variation on Gene Regulation and Splicing
- Heterologous Aggregates Promote Prion Appearance via More than One Mechanism
- The Tumor Suppressor BCL7B Functions in the Wnt Signaling Pathway
- , A -Acting Locus that Controls Chromosome-Wide Replication Timing and Stability of Human Chromosome 15
- Regulating Maf1 Expression and Its Expanding Biological Functions
- A Polyubiquitin Chain Reaction: Parkin Recruitment to Damaged Mitochondria
- RecFOR Is Not Required for Pneumococcal Transformation but Together with XerS for Resolution of Chromosome Dimers Frequently Formed in the Process
- An Intracellular Transcriptomic Atlas of the Giant Coenocyte
- Insight in Genome-Wide Association of Metabolite Quantitative Traits by Exome Sequence Analyses
- The Role of the Mammalian DNA End-processing Enzyme Polynucleotide Kinase 3’-Phosphatase in Spinocerebellar Ataxia Type 3 Pathogenesis
- The Global Regulatory Architecture of Transcription during the Cell Cycle
- Identification and Functional Characterization of Coding Variants Influencing Glycemic Traits Define an Effector Transcript at the Locus
- Altered Ca Kinetics Associated with α-Actinin-3 Deficiency May Explain Positive Selection for Null Allele in Human Evolution
- Genetic Variation in the Nuclear and Organellar Genomes Modulates Stochastic Variation in the Metabolome, Growth, and Defense
- PRDM9 Drives Evolutionary Erosion of Hotspots in through Haplotype-Specific Initiation of Meiotic Recombination
- Transcriptional Control of an Essential Ribozyme in Reveals an Ancient Evolutionary Divide in Animals
- ALIX and ESCRT-III Coordinately Control Cytokinetic Abscission during Germline Stem Cell Division
- Century-scale Methylome Stability in a Recently Diverged Lineage
- A Re-examination of the Selection of the Sensory Organ Precursor of the Bristle Sensilla of
- Antagonistic Cross-Regulation between Sox9 and Sox10 Controls an Anti-tumorigenic Program in Melanoma
- A Dependent Pool of Phosphatidylinositol 4,5 Bisphosphate (PIP) Is Required for G-Protein Coupled Signal Transduction in Photoreceptors
- Deciphering the Genetic Programme Triggering Timely and Spatially-Regulated Chitin Deposition
- Aberrant Gene Expression in Humans
- Fascin1-Dependent Filopodia are Required for Directional Migration of a Subset of Neural Crest Cells
- The SWI2/SNF2 Chromatin Remodeler BRAHMA Regulates Polycomb Function during Vegetative Development and Directly Activates the Flowering Repressor Gene
- Evolutionary Constraint and Disease Associations of Post-Translational Modification Sites in Human Genomes
- A Truncated NLR Protein, TIR-NBS2, Is Required for Activated Defense Responses in the Mutant
- The Genetic and Mechanistic Basis for Variation in Gene Regulation
- Inactivation of PNKP by Mutant ATXN3 Triggers Apoptosis by Activating the DNA Damage-Response Pathway in SCA3
- DNA Damage Response Factors from Diverse Pathways, Including DNA Crosslink Repair, Mediate Alternative End Joining
- hnRNP K Coordinates Transcriptional Silencing by SETDB1 in Embryonic Stem Cells
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The Global Regulatory Architecture of Transcription during the Cell Cycle
- A Truncated NLR Protein, TIR-NBS2, Is Required for Activated Defense Responses in the Mutant
- Proteasomes, Sir2, and Hxk2 Form an Interconnected Aging Network That Impinges on the AMPK/Snf1-Regulated Transcriptional Repressor Mig1
- The SWI2/SNF2 Chromatin Remodeler BRAHMA Regulates Polycomb Function during Vegetative Development and Directly Activates the Flowering Repressor Gene
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



