-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaThe Tumor Suppressor BCL7B Functions in the Wnt Signaling Pathway
BCL7B, a member of the human BCL7 gene family, is deleted in patients with Williams-Beuren syndrome. Although several clinical studies have suggested that malignant diseases occurring in patients with Williams-Beuren syndrome are associated with aberrations in BCL7B, little is known regarding the physiological function of this gene. Here, we show that bcl-7, the only homolog of BCL7 gene family in Caenorhabditis elegans, regulates asymmetric cell differentiation in somatic “stem-like” seam cells through at least the Wnt pathway and promotes the apoptotic pathway. In addition, bcl-7 deletion mutants show enlarged nuclei in epidermis and germ cells. Furthermore, in KATOIII human gastric cancer cells, BCL7B knockdown induces nuclear enlargement, as observed in Caenorhabditis elegans, and promotes the multinucleated phenotype, both of which are reminiscent of malignant diseases. BCL7B also negatively regulates the Wnt-signaling pathway and positively regulates the apoptotic pathway, similar to Caenorhabditis elegans. Altogether, this study may open the door for understanding the function of BCL7 family in cell differentiation and malignancies.
Published in the journal: . PLoS Genet 11(1): e32767. doi:10.1371/journal.pgen.1004921
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004921Summary
BCL7B, a member of the human BCL7 gene family, is deleted in patients with Williams-Beuren syndrome. Although several clinical studies have suggested that malignant diseases occurring in patients with Williams-Beuren syndrome are associated with aberrations in BCL7B, little is known regarding the physiological function of this gene. Here, we show that bcl-7, the only homolog of BCL7 gene family in Caenorhabditis elegans, regulates asymmetric cell differentiation in somatic “stem-like” seam cells through at least the Wnt pathway and promotes the apoptotic pathway. In addition, bcl-7 deletion mutants show enlarged nuclei in epidermis and germ cells. Furthermore, in KATOIII human gastric cancer cells, BCL7B knockdown induces nuclear enlargement, as observed in Caenorhabditis elegans, and promotes the multinucleated phenotype, both of which are reminiscent of malignant diseases. BCL7B also negatively regulates the Wnt-signaling pathway and positively regulates the apoptotic pathway, similar to Caenorhabditis elegans. Altogether, this study may open the door for understanding the function of BCL7 family in cell differentiation and malignancies.
Introduction
Cytogenetic abnormalities of chromosome 7 occur frequently in patients with cancer. Patients with certain types of malignant transformation, such as acute lymphoblastic leukemia, myelodysplastic syndrome, or juvenile myelomonocytic leukemia, frequently have deletions or abnormalities in chromosome 7 [1]–[4]. Some of the genes located on chromosome 7 are thought to act as tumor-related genes, with roles in cancer initiation and/or progression. However, few studies have investigated the molecular mechanisms controlled by specific genes located on chromosome 7.
One of the most well-known diseases related to chromosome 7 microdeletions is Williams-Beuren syndrome (WBS), a contiguous gene syndrome with a dominant autosomal inheritance pattern. WBS patients show a variety of phenotypes, including elfin face, mental retardation, reduced spatial reasoning capacity, supravalvular aortic stenosis, and peripheral pulmonic stenosis. In the past three decades, several reports have described the occurrence of malignant diseases in WBS patients [5]–[9]. These reports have shown that patients with WBS are at an increased risk of malignant transformation due to aberrations in candidate genes, such as BCL7B.
BCL7B is a member of the BCL7 gene family; members of this gene family, including BCL7A and BCL7C, located on chromosomes 12 and 16, respectively, have a conserved amino-terminal region as their functional domain [10]. A number of studies have found that BCL7 family members are involved in cancer initiation, progression, and development. For example, decreased expression of BCL7A may be a risk factor for astrocytoma [11], Burkitt lymphoma [12], non-Hodgkin's lymphoma [13], mycosis fungoides [14], and cutaneous T cell lymphoma [15]. Although the BCL7 gene family is thought to have tumor-associated functions, little is known regarding the specific functional roles of BCL7 genes; this may be attributed to the functional redundancy among BCL7 family members, which makes it difficult to analyze the individual roles of BCL7 genes.
In this study, the functional significance of C28H8.1 (designated here as bcl-7), which shares 41% homology with the amino-terminal region of human BCL7 gene family and is the only homolog in Caenorhabditis elegans (S1A Fig.), was analyzed in the Wnt-signaling pathway and apoptotic pathway. In addition, we also analyzed the function of the BCL7B gene in both pathways in KATOIII cells, a human gastric cancer cell line [16].
Results
bcl-7 is required for normal seam cell development in C. elegans
First, we knocked down bcl-7 expression using the feeding RNA interference (RNAi) technique with a bcl-7-specific RNAi clone and observed the phenotypes associated with bcl-7 downregulation in wild-type C. elegans hermaphrodites. Downregulation of bcl-7 in wild-type worms resulted in the egg-laying defective (Egl) phenotype (S1B and S1E Fig.), the protruding vulva (Pvl) phenotype (S1C Fig.), and the burst phenotype (S1D Fig.), reminiscent of defects in epidermal barrier formation [17]. Therefore, we hypothesized that bcl-7 is involved in the development of the epidermis.
Next, to examine the phenotypes produced by bcl-7 knockout, we generated a bcl-7 deletion mutant, tm5268, containing a deletion of 0.7 kbp (Fig. 1A). Because the deletion covered almost all bcl-7 regions, including the amino-terminal domain, which is conserved and considered to be the functional domain [10], tm5268 is practically a null mutant. The tm5268 mutant showed a variety of phenotypes, including Pvl (the rate was 61.4%; S1F–I Fig.), the alae morphological variant (Fig. 1B–D, and 1L), and sterility (Ste), which suggest the phenotypes in deletion mutants not only reproduced the RNAi experiments but also indicated additional phenotypes. While the bcl-7 mutants had a normal number of vulval precursor cells at the larval stages, they showed Pvl phenotypes after young adult stages (S1G–H Fig.). In addition, the Pvl phenotype was also observed in bcl-7 heterozygotes at a rate of 14.3% (S1I Fig.). This result suggests that the phenotype of bcl-7 deletion mutants is semi-dominant similar to the phenotype of BCL7B deletion in human disease, such as Williams-Beuren syndrome. Furthermore, alae, the cuticle structures considered a hallmark of normal seam cell differentiation, were “incomplete” (alae with only one or two ridges) or absent in tm5268 worms in contrast to wild-type worms (Fig. 1B and 1C). The Pvl phenotype and alae malformation are caused by defects in epidermal cells, particularly epidermal stem-like seam cells [17], [18]. The presence of these phenotypes in bcl-7 deletion mutants suggests that BCL-7 influences the development of seam cells, which have both self-renewal potential and differentiation capability, in C. elegans.
Fig. 1. Knockout of bcl-7 inhibits normal seam cell development in Caenorhabditis elegans. 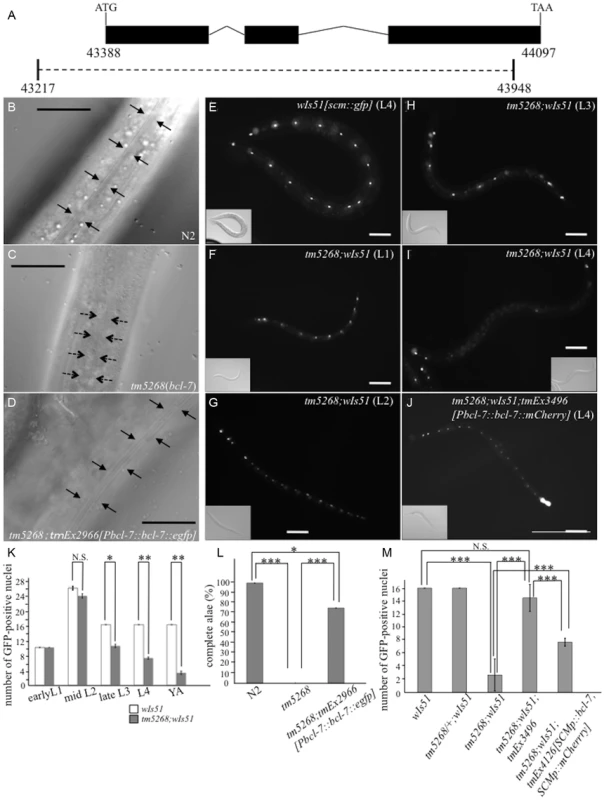
A: Structure of bcl-7. Black boxes, exons; bent lines, introns. The region deleted in the strain tm5268 is shown as a dotted line, below. The numbers indicate the location in the cosmid C28H8. B–D: Nomarski images of adult hermaphrodites. A wild-type hermaphrodite had complete alae (the region between arrows) (B); Alae were incomplete in bcl-7 (tm5268) hermaphrodites. A region between dotted arrows indicates partial alae with only two ridges. The area without arrows indicates regions without alae (C); a transgenic rescue line carrying Pbcl-7::bcl-7::egfp reporters (tm5268;tmEx2966) with complete alae (three ridges) (D). E–J: Examples of SCM::GFP localization in wild-type, tm5268, and tm5268;tmEx3496 hermaphrodites with wIs51 (SCM::GFP). tm5268;tmEx3496 is a transgenic rescue line carrying Pbcl-7::bcl-7::mCherry reporters. A wild-type L4 hermaphrodite expressing SCM::GFP in 16 seam cell nuclei (E), tm5268 hermaphrodites carrying scm::gfp reporters (H–G), and a tm5268;tmEx3496 hermaphrodite carrying scm::gfp reporters (J). Inserts show the Nomarski images of the same animals of the fluorescence images. K: Bar chart showing the average number of seam cells (about the worms at each larval stage, except for the middle L2 stage, and the young adult stage) or daughter cells (about the worms at middle L2 stage) expressing GFP in wild-type and tm5268 hermaphrodites (n = 21–46). L: Percentages of worms with complete alae in wild-type, tm5268, and tm5268;tmEx2966 adult hermaphrodites (n = 20–30). M: Bar chart showing the average number of seam cells expressing GFP in wild-type, tm5268, tm5268;tmEx3496, and tm5268;tmEx4126[SCMp::bcl-7, SCMp::mCherry] adult hermaphrodites with wIs51 (SCM::GFP) (n = 15–23). Error bars indicate the standard error of the mean (SEM). Asterisks indicate statistical significance compared with each other. *p<0.05. **p<0.005. ***p<0.001. N.S.: no significance. Scale bar = 25 µm. Then, we investigated whether the number of seam cells was altered in bcl-7 deletion mutants by analyzing transgenic worms carrying the scm::gfp transgene [19] as a marker of seam cell nuclei. In a wild-type L4-stage hermaphrodite, there were 16 seam cells on each side (Fig. 1E). By contrast, the number of seam cells was significantly reduced in the mutant worms (Fig. 1I), and scm::gfp-negative cells were observed more often in the V cell lineage than in the H and T cell lineages (Fig. 1I). Differences between wild-type worms and tm5268 worms were found in most larval stages, except for the early L1 stage (Fig. 1F–I and 1K). The expression pattern of another seam cell marker, cdh-3::gfp [20], which is localized to the cytoplasm of seam cells, also revealed that the number of GFP-positive cells was lower in bcl-7 mutants (S2A–D Fig.). Both the defect of alae and the decreased seam cell number were rescued by the introduction of bcl-7 genomic DNA, Pbcl-7::bcl-7::egfp (tm5268;tmEx2966) or Pbcl-7::bcl-7::mCherry (tm5268;tmEx3496) (Fig. 1D and 1J–M). The expression of the rescue constructs was ubiquitous, including in the hypodermis, from the embryonic stage to the adult stage, and BCL-7 was localized to the nuclei (S3A–H Fig.). Therefore, we hypothesized that BCL-7 functions cell-autonomously in seam cells. To test this hypothesis, we analyzed whether expressing a seam cell-specific construct rescues decreased seam cell number. The number of seam cells was significantly increased by the introduction of bcl-7 genomic DNA under a seam cell-specific promoter (tmEx4126[scmp::bcl-7, scmp::mCherry]) (Fig. 1M). These results suggest that BCL-7 is involved in the normal development of seam cells and functions cell-autonomously in seam cells.
Next, we addressed whether the observed decrease in seam cells in tm5268 worms resulted from the hyperactivation of apoptosis. To examine this, we analyzed whether the apoptotic pathway was hyperactivated in bcl-7 deletion mutant worms using worms with a mutation in the ced-3 gene, which encodes a member of the caspase family required for the execution of apoptosis in C. elegans [21]. However, bcl-7(III);ced-3(IV) double mutants did not exhibit increased numbers of seam cells compared with bcl-7 single mutants (the average seam cell number in adult, double-mutant hermaphrodites was 3.6 (n = 12)), suggesting that the decrease in the number of seam cells in bcl-7-deficient worms is not caused by hyperactivation of apoptosis.
In wild-type C. elegans, seam cells divide asymmetrically during each larval stage. The anterior daughter cell loses its seam cell properties and differentiates into a hyp7 cell, whereas the posterior cell keeps its self-renewal potential, remaining a seam cell (S4S Fig.) [22], [23]. To examine whether the reduction in seam cell numbers in tm5268 is followed by an increase in the hyp7 cell number, we used transgenic worms that expressed an adult-specific hypodermal marker, col-19::gfp [24], [25]. In wild-type animals, col-19::gfp was expressed in the hypodermal syncytial hyp7 cells and 16 seam cells (Fig. 2A, 2B and 2E). The number of GFP-positive hyp7 cells in tm5268 was not increased but tended to decrease compared with the number in the wild-type worms. Additionally, the number of GFP-positive seam cells in tm5268 was significantly decreased compared with the number in the wild-type worms (Fig. 2C–E). Interestingly, the nuclei of hyp7 cells from bcl-7 mutants were significantly enlarged and had an irregular shape compared to those from wild-type animals (7.66±0.10 µm and 6.77±0.14 µm, p<0.001; S5A–D Fig.).
Fig. 2. BCL-7 is involved in asymmetric cell differentiation of the epidermis in Caenorhabditis elegans. 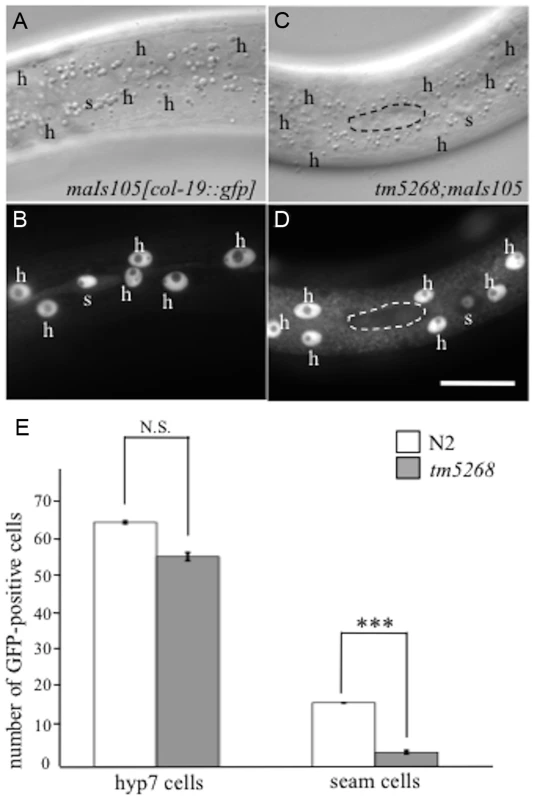
A–D: Examples of col-19p::gfp localization in wild-type and tm5268 adult hermaphrodites. Nomarski (A, C) and GFP images (B, D) of wild-type (A, B) and tm5268 (C, D) adult hermaphrodites carrying col-19::gfp reporters. A wild-type hermaphrodite expressing col-19p::gfp in both hyp7 cells (‘h’) and seam cells (‘s’), (B) and a tm5268 hermaphrodite expressing col-19p::gfp in hyp7 cells but not in seam cells (surrounded with a dotted oval) (D). E: Bar chart of the cell numbers expressing col-19::gfp in wild-type and tm5268 adult hermaphrodites (n = 15–20) for hyp7 and seam cells. Error bars indicate SEM. Asterisks indicate statistical significance compared with each other. ***p<0.001. N.S.: no significance. Scale bar = 25 µm. To determine whether the somatic stem-like cells acquire another fate in the cell lineage, we analyzed neural cells, including sensory PVD neurons, PDE neurons, and phasmids, derived from V - and T-cell lineages. PVD and PDE neurons originate from an asymmetric division of the V5 cell (S4S Fig.) [23], whereas phasmid cells are generated by the asymmetric T-cell division when the posterior daughter cell maintains the seam cell phenotype and the anterior daughter cell commits to the neural fate, giving rise to the phasmid (S4T Fig.) [23]. We analyzed the cell fates of PVD and PDE in transgenic worms carrying the PVD and PDE markers des-2::gfp and dat-1p::gfp, respectively [26], [27]. No extra PVD or PDE cells were found in any of the bcl-7 mutants (n = 10–15, S4A–J Fig.). In wild-type worms, phasmids can be detected by the uptake of fluorescent dye [28]. Similarly, in all bcl-7 mutants, both socket cells in the phasmid, but not other cells, absorbed the fluorescent dye in dye-filling assays (n = 10, S4K–N Fig.). These results suggest that stem-like cells fail to acquire a terminal differentiation fate in bcl-7 mutants.
Next, we hypothesized that more undifferentiated cells are present in bcl-7 deletion mutants because we observed an increase in the number of cells with enlarged nuclei, as well as the loss of differentiated-cell markers. To test this hypothesis, we analyzed the expression patterns of an undifferentiated state marker, egl-27/Mta [29], in wild type and tm5268 worms carrying the egl-27p::his-24::mCherry transgene. The expression pattern of egl-27 was different between wild type and bcl-7 mutant worms. In wild type worms, egl-27 was strongly expressed in the nuclei of intestinal cells (S6A–B Fig.) and weakly expressed in the nuclei of epidermis (S6C–D Fig.) during the L4 stage (n = 10). By contrast, egl-27 was strongly expressed ubiquitously particularly in the epidermal cells including both seam cells and hyp7 cells in bcl-7 mutant worms (n = 10, S6E–H Fig.). Furthermore, we analyzed the expression levels of undifferentiated cell markers using a quantitative real-time polymerase chain reaction (qRT-PCR) analysis. Both egl-27 and ceh-6, markers of the undifferentiated state in C. elegans, were significantly increased in the tm5268 mutants compared with the wild type worms (S6I–J Fig.). These results suggest that stem-like cells fail to acquire a terminal differentiation fate in bcl-7 mutants.
Loss of BCL-7 function affects gonadal size and germ cell differentiation
The Ste phenotype was observed in bcl-7 deletion mutants (Fig. 3M); specifically, no oocytes were detected in the homozygous mutants, and their gonads were shortened (Fig. 3D). In addition, the brood size of bcl-7 heterozygotes was significantly reduced compared with that of wild-type worms (Fig. 3M), which indicates that the genetic trait of bcl-7 mutation in C. elegans is haploinsufficiency, similar to that of human diseases. These results revealed that the loss of BCL-7 function affects not only seam cells but also the development of somatic gonads and/or germ cells. To further investigate the gonadal and germ cell phenotypes in tm5268 worms, we performed diamidinophenylindol (DAPI)-staining and fluorescence immunostaining with an anti-phospho-histone H3 (PH3) antibody as a mitotic marker [30]. In wild-type C. elegans hermaphrodites, the mitotic region covered approximately 10–15 cell diameters as previously reported (Fig. 3A) [31]. In bcl-7 mutant worms, PH3-positive cells were found, but tended to decrease compared with the wild type worms (Fig. 3D). In addition, they were occasionally observed farther from the distal tip cells (DTCs) than in wild type worms. These results suggest that the shortened gonad observed in tm5268 mutants was not because of the absence of mitosis but rather because of the decrease of mitosis and may be due to the defects of cell differentiation after mitosis. In addition, the average length of the major axis of germ cell nuclei was 3.17±0.03 µm in wild type worms (Fig. 3A–C, S7A Fig.). By contrast, bcl-7 mutants exhibited irregularly shaped and significantly larger germ cell nuclei measuring 4.42±0.08 µm (Fig. 3D–F, S7B Fig.). Furthermore, the number of germ cells in tm5268 worms was decreased compared with that in wild-type worms (Fig. 3D–F). Thus, BCL-7 is necessary for gonadal development, particularly for gonadal arm elongation, and for germ cell entry into meiosis. The Ste phenotype, shortened gonads, and enlarged germ cell nuclei in tm5268 worms were rescued by the introduction of bcl-7 genomic DNA under the bcl-7 promoter (Fig. 3G–I, 3M and S7C). The rescue construct was strongly expressed in the nuclei of somatic DTCs and weakly expressed in the nuclei of germ cells and gonadal sheath cells (S3I–L Fig.). These results suggest that the expression of BCL-7 in DTCs, germ cells, and/or gonadal sheath cells is necessary for its function.
Fig. 3. Knockout of bcl-7 affects gonadal development and germ cell proliferation in Caenorhabditis elegans. 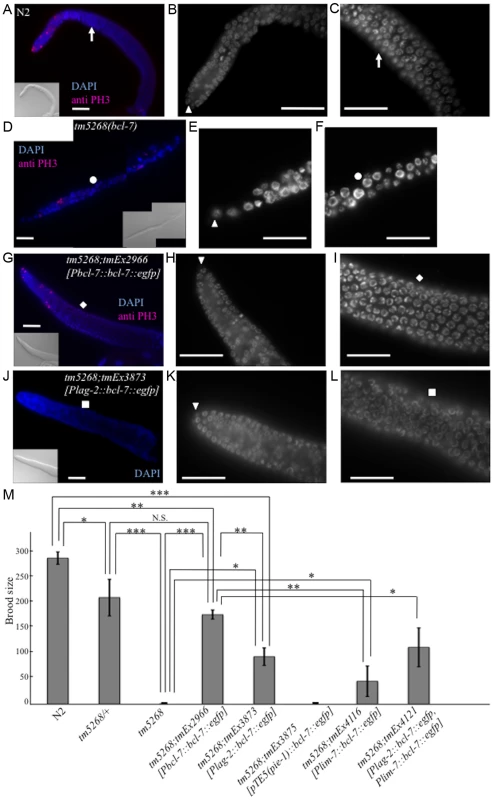
A–C: A dissected adult germline of a wild-type adult hermaphrodite. A dissected gonad stained with DAPI (blue) and an anti-PH3 antibody (pink) as a mitotic cell-specific marker (A), and the regions of mitosis (B) and meiosis (C) of the gonad stained with DAPI. The arrowhead indicates a distal tip cell (DTC). The white arrows indicate the corresponding position of the gonad. D–F: A dissected germline of a tm5268 adult hermaphrodite. A dissected gonad stained with DAPI (blue) and an anti-PH3 antibody (pink) as a mitotic cell-specific marker (D), and the regions of mitosis (E) and meiosis (F) of the gonad stained with DAPI. The arrowhead indicates a DTC. The white circles indicate the corresponding position of the gonad. G–I: A dissected germline of a tm5268;tmEx2966 adult hermaphrodite. A dissected gonad stained with DAPI (blue) and an anti-PH3 antibody (pink) as a mitotic cell-specific marker (G), and the regions of mitosis (H) and meiosis (I) of the gonad stained with DAPI. The arrowhead indicates a DTC. The white rhomboid indicates the corresponding position of the gonad. J–L: A dissected germline of a tm5268;tmEx3873 adult hermaphrodite carrying Plag-2::bcl-7::egfp as a DTC-specific rescue construct. A dissected gonad stained with DAPI (blue) (J), and the regions of mitosis (K) and meiosis (L) of the gonad stained with DAPI. The arrowhead indicates a DTC. The white squares indicate the corresponding position of the gonad. M: Bar chart indicating the brood size in wild-type, heterozygous and homozygous bcl-7 mutants, and transgenic rescue lines. The extrachromosomal array tmEx3875 contains pTE5::bcl-7::egfp as a germ cell-specific rescue construct, tmEx4116 contains lim-7::bcl-7::egfp as a gonadal sheath cell-specific rescue construct, and tmEx4121 contains both Plag-2::bcl-7::egfp and lim-7::bcl-7::egfp. Error bars indicate SEM. Asterisks indicate statistical significance. *p<0.05, **p<0.005, ***p<0.001. N.S.: no significance. Scale bar = 25 µm. To further examine the observed phenotypes, we introduced bcl-7 genomic DNA under the DTC-specific lag-2 promoter (Plag-2::bcl-7::egfp). The Ste phenotype of tm5268 worms was partially rescued by the expression of the bcl-7 gene in DTCs (Fig. 3M). In addition, thin gonads and irregularly shaped nuclei of germ cells were also partially rescued by the expression of this construct (Fig. 3J–L and S7D). These results suggest that BCL-7 expression in DTCs is necessary but insufficient for normal gonadal development. Therefore, we hypothesized that restoring BCL-7 expression in germ cells could rescue the Ste phenotype in bcl-7 deletion mutants (tm5268). To test this hypothesis, we expressed bcl-7 genomic DNA fused with the promoter, first intron, and 3′-untranslated region (UTR) of the pie-1 gene, which is expressed exclusively in germ cells (pTE-5(pie-1)::bcl-7) [30]. However, the Ste phenotype was not rescued by the introduction of pTE5(pie-1)::bcl-7 (tmEx3875) (Fig. 3M). Our study showed that BCL-7 was also expressed in gonadal sheath cells (S3I–L Fig., arrows indicate a part of gonadal sheath cells). Gonadal sheath cells play an important role in embryonic germline amplification and larval gonadal elongation [32]. Therefore, we examined whether the expression of BCL-7 in gonadal sheath cells is sufficient for normal gonadal development by introducing bcl-7 genomic DNA under a sheath cell-specific sequence (lim-7 promoter and first intron; tmEx4116[Plim-7::bcl-7::egfp]) [33], [34]. The Ste phenotype of tm5268 worms was partially rescued (Fig. 3M). In addition, we introduced both the DTC-specific rescue construct and the sheath cell-specific rescue construct (tmEx4121[Plag-2::bcl-7::egfp, Plim-7::bcl-7::egfp]) and found that the brood size was larger than with either the DTC-specific rescue or the gonadal sheath cell-specific rescue alone (Fig. 3M). These results suggest that BCL-7 expression in somatic DTCs and gonadal sheath cells is more important than its expression in germ cells for normal gonadal development.
One of the factors that regulates the size of the gonads and the timing of the entry into meiosis is LAG-2, a homolog of the Notch receptor ligand that is secreted by DTCs [35], [36]. We therefore hypothesized that DTC functions, including LAG-2 secretion, may be impaired in bcl-7 mutants. We analyzed DTCs using worms carrying the lag-2p::gfp transgene (qIs56) [37]. Two GFP-positive DTCs were detected in 100% (17/17) of wild-type worms (S8A and B Fig.), whereas only 19.5% (25/128) of bcl-7 mutant worms were positive for GFP in both DTCs. The remaining 80.5% (103/128) of bcl-7 mutant worms were positive for GFP in only one DTC. Furthermore, 12.5% (16/128) of bcl-7 mutants showed mispositioning of the DTCs with the expression of lag-2p::gfp (S8E–H Fig. and S1 Table). Interestingly, heterozygous bcl-7 mutants exhibited the same phenotypes; i.e., 30% (6/20) of heterozygous bcl-7 mutants had expression of lag-2p::gfp on only one DTC and 50% (10/20) showed mispositioning of the DTC with GFP expression (S1 Table, middle row). These differences between homozygous and heterozygous mutants further demonstrate that the phenotype is semi-dominant. These results suggest that BCL-7 partially controls the differentiation of DTCs.
Next, we attempted to determine the time course of BCL-7 function using a chromophore-assisted light inactivation (CALI) assay. We created a light-inactivatable BCL-7::KillerRed fusion protein by substituting egfp of the rescue construct with KillerRed. We generated transgenic worms (tmEx3878) that expressed the bcl-7 promoter-driven bcl-7::KillerRed construct (Pbcl-7::bcl-7::KillerRed) in the bcl-7-deficient background (i.e., tm5268). In the absence of green-light illumination, this transgene rescued the Ste phenotype (similar results were observed independently in three transgenic lines). When the animals were illuminated with a green light, the BCL-7 protein fused to KillerRed was inactivated [38], [39], allowing inactivation of BCL-7 function. We exposed tm5268;tmEx3878 to a green light starting from the comma stage of the embryo, the early larval L1 stage, or the L2 stage to the young adult stage and analyzed the Ste phenotype. All worms exposed to the green light at the L2 stage were fertile; however, approximately 90% (70/79) of worms illuminated at the comma stage and 54.5% (18/33) of worms illuminated at the early L1 stage retained the Ste phenotype (S7E Fig.). In addition, we performed a pulse experiment to identify the most critical stage in development for the function of BCL-7. We exposed the same transgenic worms to a green light during the comma and early L1 stages and found that only 20% of the worms were fertile (S7E Fig.). These results imply that the expression and function of BCL-7 in DTCs begin during the early L1 stage, which corresponds to the timing of asymmetric cell division in somatic DTCs.
RNAi-based screening for suppressors of the bcl-7 mutant phenotypes
Loss of BCL-7 function resulted in defects in both seam cells and the gonads, suggesting that BCL-7 controls the asymmetric division of cells in C. elegans. More than one genetic pathway is involved in the asymmetric division and differentiation of these cells. Therefore, we next sought to determine the role of BCL-7 in these pathways by screening for genes that could suppress the phenotypes of the bcl-7 mutant (tm5268). Using bcl-7 mutants, with wild-type worms as the control, we carried out a feeding RNAi screen of 96 genes involved in the development of seam cells and/or gonads, including genes in the WNT/ß-catenin and Notch pathways, heterochronic genes, and other genes that regulate the division and differentiation of seam cells and/or somatic gonadal precursors (SGPs) (functionally classified in S2 Table). We found that downregulation of either wrm-1 or lsy-22 suppressed the phenotypes of the bcl-7 mutant tm5268 (Fig. 4A–I and S3 Table). Downregulation of WRM-1, a homolog of ß-catenin [40]–[42], suppressed both seam cell reduction and Ste phenotypes. Although LSY-22 is considered a homolog of Groucho-like protein [43] and is also a member of the noncanonical Wnt pathway, lsy-22-specific RNAi only partially suppressed the Ste phenotype and did not affect the number of seam cells. Subsequently, we focused on WRM-1, which exhibited potent effects in both somatic cells and germ cells and was expressed in both seam cells and SGPs. Interestingly, the suppressor effect of wrm-1 RNAi appeared to be dependent on the strength of the RNAi clone (S3 Table). The effect of the wrm-1c RNAi clone (constructed with a cDNA generated from wild-type worms) on N2 was stronger than that of wrm-1 RNAi (from the Ahringer library) and comparable with the phenotypes of the wrm-1 mutant strains (WormBase; http://www.wormbase.org/). The effect of diluted wrm-1c RNAi seemed comparable to RNAi by a genomic clone on suppressing the bcl-7 mutant's phenotypes (Fig. 4E, S3 Table).
Fig. 4. Downregulation of wrm-1 or lsy-22 suppresses the phenotypes of bcl-7 mutants. 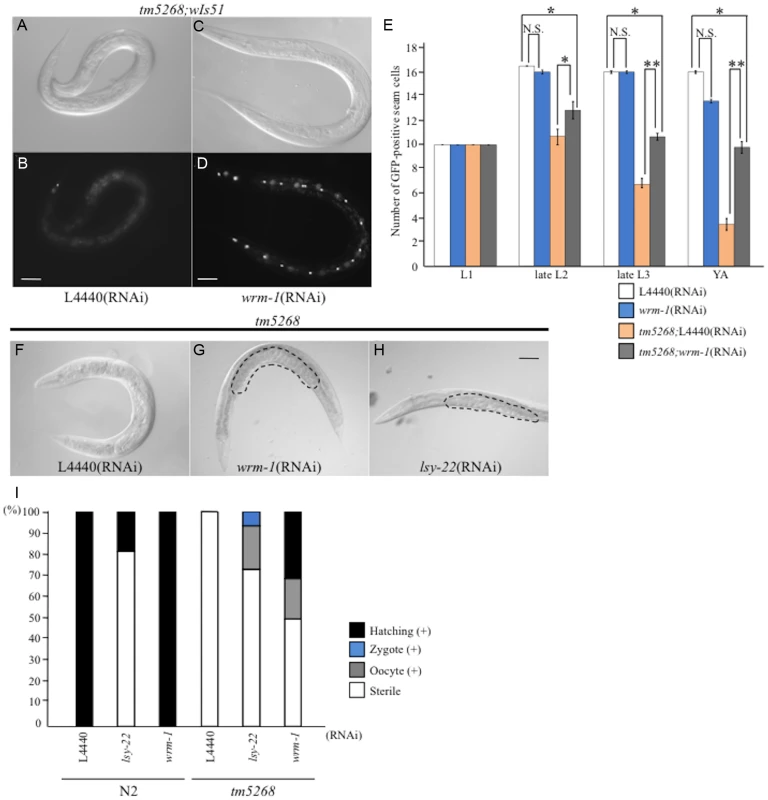
A–D: Nomarski (A, C) and GFP images (B, D) of tm5268 adult hermaphrodites carrying an scm::gfp reporter treated with empty vector (L4440) (A, B) or wrm-1-specific RNA (C, D). E: Bar chart representing the average number of seam cells in wild-type, tm5268, tm5268;L4440 (RNAi), or tm5268;wrm-1 (RNAi) hermaphrodites at the L1, late L2, late L3, or young adult stages. F–H: Nomarski images of tm5268 treated with L4440 (RNAi), wrm-1 (RNAi), or lsy-22 (RNAi). Eggs in the uterus are outlined with dotted lines (G, H). I: Percentages of the Ste phenotypes in wild-type and tm5268 adult hermaphrodites treated with L4440 (RNAi), wrm-1 (RNAi), or lsy-22 (RNAi). Numbers of examined animals were more than 60 for all strains.Error bars indicate SEM. Asterisks indicate statistical significance. *p<0.05. **p<0.005. N.S.: no significance. Scale bar = 50 µm. Based on the results of the suppressor screening, we hypothesized that BCL-7 negatively regulates WRM-1 expression. To test this hypothesis, we analyzed the expression levels of wrm-1 mRNA using qRT-PCR. Additionally, because WRM-1 is one of the three ß-catenin homologs in C. elegans, we also analyzed the expression levels of the other ß-catenin homologs, BAR-1 and SYS-1. Compared with wild type worms, bar-1 and sys-1 mRNAs were significantly increased in bcl-7 deletion mutants (S9A Fig.). There was also a trend for increased wrm-1 in the mutants (S9A Fig.). These results suggest that BCL-7 functions as a negative regulator of the expression of three ß-catenin homologs in C. elegans.
The phenotypes of the bcl-7 deletion mutants were different from the pop-1 deletion mutants and wrm-1(gf) mutants, which are associated with hyperactivation of the Wnt pathway. Therefore, we hypothesized that BCL-7 not only functions as a negative regulator but also affects the Wnt pathway in a different mechanism. Because WRM-1/ß-catenin regulates the levels of POP-1 in the nuclei of target cells during asymmetric cell division, we assumed that the cellular distribution of WRM-1 or POP-1 may be altered in bcl-7 mutants, and therefore we analyzed the localization of WRM-1 and POP-1 using WRM-1::GFP [44] and GFP::POP-1 [45] fusion reporter proteins. In wild-type worms, the cellular localization of WRM-1::GFP in mother seam cells at the L2 stage was observed near the cell cortex in the anterior half of the cells, as shown by previous reports (Fig. 5A–B) [44]. Furthermore, after division, the fluorescence intensity of WRM-1::GFP was stronger in posterior daughter cells than in anterior cells (Fig. 5E–F). In bcl-7 mutants, the cellular localization of WRM-1 before cell division was similar to that in wild-type worms (Fig. 5C–D). However, in daughter cells, the fluorescence intensity in the anterior cells was equal to or stronger than that in the posterior cells in approximately 50% of tm5268 mutant worms (Fig. 5G–I). The asymmetric localization pattern of GFP::POP-1 in tm5268 worms was also different from that in wild-type worms (Fig. 5J–N), as expected due to the change in WRM-1 localization.
Fig. 5. BCL-7 is involved in the asymmetric localization of Wnt components in Caenorhabditis elegans. 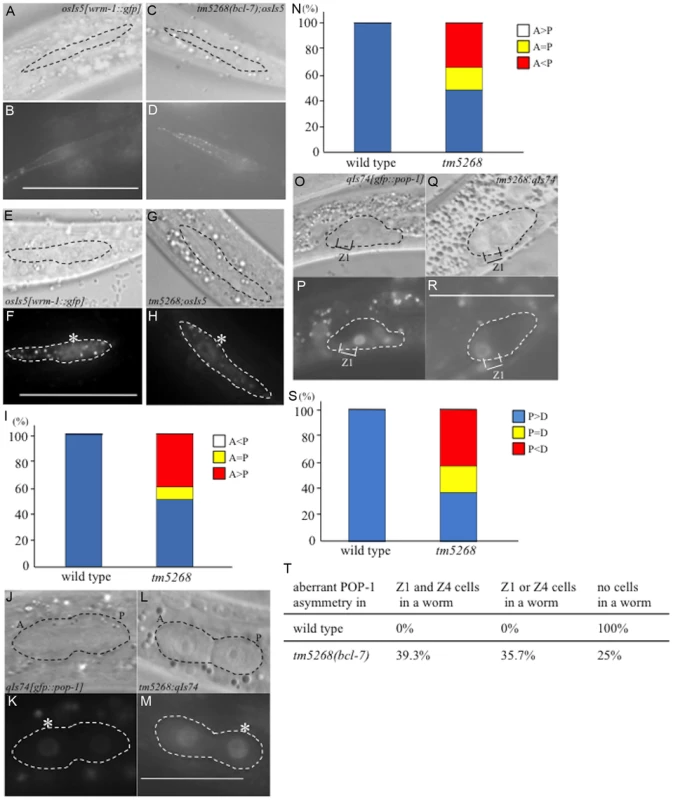
A–D: Nomarski (A, C) and GFP (B, D) images of wild type (A, B) and tm5268 (C, D) L2-stage hermaphrodites carrying a wrm-1::gfp reporter (osIs5) at pre-division states. Seam cells are traced with dotted ovals (A, C). E–H: Nomarski (E, G) and GFP (F, H) images showing the localization of WRM-1::GFP in two daughter cells of wild-type or tm5268 L2 hermaphrodites at post-divisions. The pairs of daughter cells are outlined with dotted ovals. I: Frequency of the localization patterns of WRM-1::GFP. The equal and unequal signs indicate the relative intensities of nuclear WRM-1::GFP between two daughter cells. J–M: Nomarski (J, L) and GFP (K, M) images showing the localization of GFP::POP-1 (qIs74) in two daughter cells of wild-type (J, K) or tm5268 (L, M) L1 hermaphrodites in post-division states. The pairs of daughter cells are outlined with dotted ovals. Asterisks indicate nuclei with stronger expression of GFP. N: The frequency of the localization patterns of GFP::POP-1. The equal and unequal signs indicate the relative intensities of nuclear GFP::POP-1 between two daughter cells. O–R: Nomarski (O, Q) and GFP (P, R) images showing the localization of GFP::POP-1 (qIs74) in SGPs of wild-type (O, P) or tm5268 (Q, R) early L1 hermaphrodites. SGPs are outlined with dotted ovals. S: The frequency of the localization patterns of GFP::POP-1. The equal and unequal signs indicate the relative intensities of nuclear GFP::POP-1 between two daughter cells. Anterior is oriented toward the left, and ventral is oriented toward the bottom. T: Percentages of worms with the aberrant POP-1 asymmetry in both Z1 and Z4 cells, either Z1 or Z4 cells, and no cells in one worm between wild type and bcl-7 mutant worms. Numbers of examined samples were more than 30 for all analyses. Scale bar = 25 µm. We also analyzed the localization of POP-1 in somatic gonadal precursor Z1 and Z4 cells and their daughter cells using gfp::pop-1 reporter transgenes. In wild-type worms, GFP expression was stronger in the nuclei of Z1.p and Z4.a cells than in Z1.a and Z4.p cells (Fig. 5O–P). By contrast, approximately 60% of tm5268 mutants showed aberrant POP-1 localization (Fig. 5Q–S). In addition, 35.7% of the tm5268 mutants showed both wild type and aberrant POP-1 asymmetry within the same gonad, as shown in Fig. 5T.
Next, we determined whether the impaired nuclear localization of POP-1 disturbs the asymmetric localization of gene products downstream of POP-1 in DTCs. HLH-2 is a downstream factor in the POP-1/TCF pathway [46], [47] and an important transcription factor regulating LAG-2; therefore, it is a determining factor in DTC development. We analyzed the localization of HLH-2 using an hlh-2p::gfp::hlh-2 reporter construct (qyIs174) and found that only 28.2% (13/46) of bcl-7 mutants showed hlh-2p::gfp::hlh-2 expression in two DTCs, whereas 100% (10/10) of wild type worms were positive for GFP in two DTCs (S8I–J Fig., and S4 Table). The remaining mutants showed decreased hlh-2p::gfp::hlh-2 expression, with either no expression observed or expression observed in only one DTC. In addition, 46% (21/46) of the bcl-7 mutants showed mispositioning of the DTCs (S8K–N Fig. and S4 Table). This result is consistent with the expression pattern of lag-2p::gfp in the mutant worms (S8C–H Fig.). HLH-2 is an important transcription factor that regulates LAG-2 and is thus a determining factor for DTCs; therefore, this result may also partially explain the defects in gonadal development in the bcl-7 mutant (tm5268). Taken together, our data suggest that BCL-7 regulates POP-1 distribution in the Z-cell lineage by controlling WRM-1 activity and therefore affects the expression pattern of HLH-2.
BCL-7 affects the apoptotic pathway in C. elegans
BCL-7 has several functions in C. elegans. Therefore, we examined whether BCL-7 regulates the apoptotic pathway by analyzing the number of PLM neurons using transgenic worms carrying the Pmec-4::gfp transgene (bzIs8) [48] as a marker of PLM nuclei. The number of PLM neurons should either decrease or increase if the apoptotic pathway is activated or suppressed, respectively, in tm5268 mutant worms, because one cell in the PLM lineage undergoes to apoptosis. In wild type worms, two PLM neurons were observed in each worm as shown in S4O–P Fig. (n = 10). Similarly, in bcl-7 deletion mutants, two PLM neurons were exhibited in the tail of each worm (n = 12, S4Q–R Fig.). This result showed that extra neurons, which are caused by the suppression of the apoptotic pathway, were not found in bcl-7 mutants. Next, we analyzed whether the levels of apoptosis-related factors were increased in tm5268 mutant worms. The expression of the anti-apoptotic factor, ced-9, was significantly increased in tm5268 mutant worms compared with wild type worms (S9B Fig.). These results suggest that the apoptotic pathway is suppressed in tm5268 mutant worms.
BCL7B regulates the size of nuclei in human gastric cancer cells
As described above, BCL-7 likely affects the morphology of nuclei and functions in the Wnt-signaling pathway in the development of C. elegans. Because bcl-7 is a homolog of the human BCL7B gene, we wondered whether BCL7B has similar roles in humans. To examine this, we used KATOIII cells, which are derived from gastric signet-ring cell cancer and express only BCL7B of the BCL7 family members. First, we examined whether siRNA-mediated BCL7B knockdown results in nuclear enlargement, as was observed in C. elegans bcl-7 mutants. As expected, KATOIII cells transfected with BCL7B siRNA showed enlarged nuclei compared with control cells transfected with nontargeting siRNA (170.8 ±7.8 µm2 and 98.0±4.5 µm2; Fig. 6A–D, and S10A–B Fig.). In addition, the occurrence of multinucleated cells (containing two or more nuclei) was increased in BCL7B-knockdown cells compared with control cells (the rates of multinucleated cells were 18.8% and 5.7%, respectively; Fig. 6E–H). The average number of nuclei per cell was 1.13 in control cells and 1.41 in BCL7B-knockdown cells (P<0.05, Student's t-test). In general, cell nuclear enlargement and multinucleated cells are observed in undifferentiated cells, such as cancer cells. Therefore, we hypothesized that the downregulation of BCL7B is involved in cell differentiation. To test this hypothesis, we analyzed the expression levels of undifferentiated markers of human cells using qRT-PCR. We found that the stem cell markers, Nanog, Oct3/4, and Sox2, were increased in BCL7B-knockdown KATOIII cells (S11A Fig.). This result suggests that BCL7B affects cell differentiation in KATOIII cells, similar to its role in C. elegans. Because enlarged nuclei are generally a result of uncontrolled DNA synthesis [49], we next tested whether BCL7B-knockdown cells exhibited aneuploidy or cell cycle defects. According to the results of a cell cycle assay, BCL7B knockdown did not induce aneuploidy (S10C–D Fig.) but did result in a significant accumulation of cells in the G0/G1 phase and a decrease in cells in the S phase (Fig. 6J and S10C–E Fig.). Because the observed nuclear enlargement was not induced by an alteration in DNA synthesis, we hypothesized that this phenomenon was caused by increased RNA levels. To test this hypothesis, we analyzed the RNA content of the nucleus by determining the fluorescence intensity of ethidium bromide-stained cells with or without RNase. We found that the fluorescence intensity of ethidium bromide in cells transfected with BCL7B siRNA was significantly stronger than in control cells. Furthermore, the addition of RNase eliminated the difference between the control cells and BCL7B-knockdown cells (S12A–I Fig.). The expression of nuclear paraspeckle assembly transcript 1 (NEAT1), a noncoding RNA and the core molecule of nuclear paraspeckle [50], was increased in BCL7B-knockdown cells compared with control cells (S12J Fig.). These results suggest that BCL7B plays a role in cell cycle progression and in the maintenance of the nuclear structure.
Fig. 6. BCL7B functions as a tumor suppressor via multiple pathways. 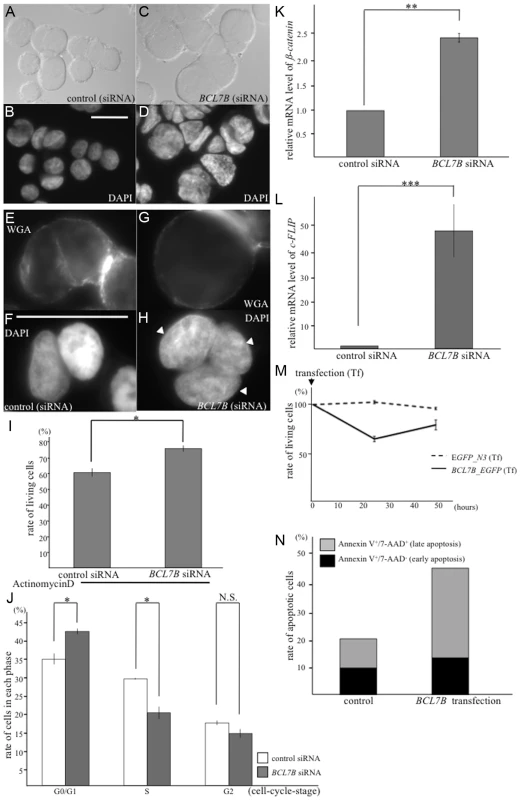
A–D: Nomarski (A, C) and DAPI (B, D) images of KATOIII cells transfected with control siRNA (A, B) and BCL7B-specific siRNA (C, D). E–H: Fluorescence images of KATOIII cells stained with WGA-Alexa Fluor 488 (E, G) and DAPI (F, H). These cells were transfected with control siRNA (E, F) or BCL7B-specific siRNA (G, H). The arrowheads indicate the presence of three nuclei in one cell. I: The percentages of the surviving KATOIII cells after transfection with control siRNA or BCL7B-specific siRNA and addition of actinomycin D. The number of living cells was counted 5 h after the addition of actinomycin D using trypan blue dye exclusion; this number was then divided by the total cell number. J: Percentages of KATOIII cells transfected with control siRNA or BCL7B-specific siRNA in the G0/G1, S, and G2 phases. More than twenty thousand cells were counted for each analysis. K, L: The expression levels of ß-catenin (K) and c-FLIP (L) by the qRT-PCR analysis. M: Survival rates of KATOIII cells transfected with EGFP, as a control, or BCL7B_EGFP. The number of living cells was counted using trypan blue dye exclusion, and this number was then divided by the total cell number. N: The rates of apoptotic KATOIII cells transfected with EGFP, as a control, or BCL7B_EGFP. Apoptotic cells were stained for Annexin-V and/or 7-AAD and monitored by flow cytometric analysis. More than ten thousand cells were counted. Each experiment was performed three times independently. Error bars indicate SEM. The asterisks indicate the statistical significance of differences between the two groups. *p<0.05, **p<0.005, ***p<0.001. N.S.: no significance. Scale bar = 25 µm. BCL7B regulates several signaling pathways including Wnt and apoptosis
Next, we examined whether BCL7B is involved in the Wnt signaling pathway in human cells, as observed with BCL-7 in C. elegans. We analyzed the expression of ß-catenin, an important member of the Wnt signaling pathway, and high-mobility group A1 (HMGA1), one of the target genes of the Wnt pathway [51]. According to a qRT-PCR analysis, the expression levels of ß-catenin and HMGA1 were significantly increased in BCL7B-knockdown cells (Fig. 6K, S10F and S10G Fig.). These results suggest that BCL7B functions as a negative regulator of the Wnt pathway in KATOIII cells.
We then analyzed the effects of BCL7B overexpression by transfecting KATOIII cells with a BCL7B_EGFP fusion construct or EGFP alone. Our experiments demonstrated that BCL7B overexpression modulated cell proliferation (S13A Fig.). Specifically, 25.5% of BCL7B-overexpressing cells died two days after transfection with BCL7B_EGFP, compared with 12.3% of control EGFP cells (Fig. 6M). Therefore, we hypothesized that overexpression of BCL7B may promote apoptosis. To test this hypothesis, we analyzed the number of apoptotic cells using flow cytometry. Indeed, overexpression of BCL7B resulted in increased apoptosis compared with the control (Fig. 6N, and S13B–C Fig.). To determine whether the apoptotic pathway was conversely inhibited in BCL7B-knockdown cells, we analyzed the survival of BCL7B-knockdown cells and control cells following treatment with actinomycin D. The rate of surviving cells was increased in BCL7B-knockdown cells compared with control cells (Fig. 6I). We next analyzed the expression of apoptosis inhibitors. qRT-PCR analysis showed that the expression of cellular FLICE-like inhibitory protein (c-FLIP), which inhibits apoptosis by antagonizing caspase-8 and caspase-10 [52], [53], was significantly upregulated in BCL7B-knockdown cells (Fig. 6L). Another apoptosis inhibitor, Bcl2, was also significantly increased but the apoptotic factor, PTEN, was not increased in BCL7B-knockdown KATOIII cells. Interestingly, Bax, which binds Bcl2 and functions as a positive regulator of apoptosis, was increased in BCL7B-knockdown KATOIII cells (S11B Fig.). However, the degree of increase in Bcl2 expression was much larger than the increase in Bax2 expression; therefore, the Bcl2/Bax ratio was increased in BCL7B-knockdown KATOIII cells, similar to what has been observed in other apoptosis-resistant cells [54]. Collectively, these results suggest that BCL7B is a positive regulator of apoptosis in KATOIII cells and support the hypothesis that BCL7B contributes to the apoptotic pathway.
Discussion
Malignant diseases are caused by defects in cell differentiation, cell cycle progression, DNA repair mechanisms, or apoptotic pathways [55]–[57]. Although previous studies have revealed the genetic backgrounds of certain types of cancer—for example, the MSH2 gene in familial nonpolyposis colon cancer [58], the BRCA1 gene in familial breast cancer [59]–[61], the APC gene in hereditary adenomatous polyposis [62], [63], and the RB gene in retinoblastoma [64], [65]—the functional mechanisms leading to cancer initiation, progression and development remain to be elucidated. BCL7B, located on chromosome 7, is a member of the BCL7 family of genes and is thought to be a tumor-associated gene [7]–[9]. In this study, we investigated the functional roles of the bcl-7 and BCL7B genes in the Wnt signaling pathway and apoptosis in C. elegans and in human gastric cancer cells, respectively. Our results revealed that BCL-7 regulates terminal cell differentiation in somatic “stem-like” seam cells and SGPs via negative regulation of the Wnt pathway in C. elegans. BCL-7 also functions as a positive regulator of apoptosis by inhibiting the expression of an anti-apoptotic factor. Additionally, similar to its role in C. elegans, human BCL7B functions as a negative regulator of Wnt signaling, presumably upstream of ß-catenin, and induces apoptosis in gastric cancer cells. Furthermore, this study revealed that BCL-7 and BCL7B are also involved in the mechanisms of nuclear enlargement, which is an important signature of malignancy. Collectively, our data suggest that the members of the BCL7 family and their homolog proteins may function as tumor suppressors by affecting multiple pathways (S14 Fig.). Therefore, patients with WBS who have a heterozygous deletion in BCL7B may be at risk of malignancy. This study may also be clinically significant in terms of the long-term medical care of WBS patients.
BCL-7 and BCL7B are involved in the Wnt signaling pathway
Our data demonstrated that BCL7B interacts with the Wnt signaling pathway, similar to its C. elegans homolog bcl-7. The knockdown of bcl-7 increased the expression of three ß-catenin homologs, bar-1, sys-1, and wrm-1. Therefore, BCL-7 may function as a negative regulator upstream of ß-catenin in the Wnt/ß-catenin pathway in C. elegans. In humans, BCL7B is a component of the SWI/SNF complex [66], which has multiple functions, including regulation of the Wnt pathway through transcriptional control of related signaling molecules [67]. In C. elegans, the SWI/SNF complex regulates asymmetric cell division and may be associated with Wnt signaling [68], [69]. Although there is currently no evidence of an association between BCL-7 and the SWI/SNF complex in C. elegans, our data are consistent with a recent report [68]. Therefore, the SWI/SNF complex may represent the link between BCL-7 and the Wnt pathway.
The Wnt pathway is known to regulate the asymmetric division of most somatic cells in C. elegans through the asymmetric localization of Wnt components [70]. Our results showed that bcl-7 knockout also induced defects in the localization of WRM-1 and POP-1 in the target cell nuclei. Specifically, bcl-7 deletion caused a reversal of cell polarity and occasionally a loss of cell polarity in seam cells and SGPs. Yamamoto et al. (2011) [71] found that the knockout of multiple Wnt ligands resulted in the randomization, but rarely the loss, of cell polarity. By contrast, knockout of apr-1/APC resulted in the symmetrical localization of WRM-1 to both the anterior and posterior nuclei by inhibiting the export of WRM-1 from the anterior nuclei [72], [73]. Thus, the functions of BCL-7 and multiple Wnt ligands or APR-1 may be similar. BCL-7 may affect the translocation of WRM-1 from the cortex to the nucleus or the export of WRM-1 from the nucleus. In addition, multiple molecules, including WRM-1 itself, regulate the asymmetric localization of WRM-1. Therefore, BCL-7 may also interact, either directly or indirectly, with WRM-1 itself to regulate its asymmetric localization in the nuclei of target cells and maintain both nuclear WRM-1 and nuclear POP-1 at appropriate levels in C. elegans.
As described above, the suppressor screening supported the idea that BCL-7 functions mostly as a negative regulator of the Wnt pathway in C. elegans. A weak downregulation of WRM-1 had little effect on wild type worms but was sufficient to partially suppress the phenotype induced by bcl-7 deficiency (S3 Table). In general, activating the expression of Wnt-related molecules, such as wrm-1(gf) mutants and pop-1 mutants, results in an increased number of seam cells, as indicated by downregulation of terminal differentiation markers and upregulation of stem cell markers (Fig. 2, S2 Fig., S6 Fig., S11 Fig.) [74], [75]. However, knockout of BCL-7, which is thought to result in the activation of the Wnt pathway, results in a decreased number of seam cells. This discrepancy may be caused by an additional function of BCL-7 other than its role in the Wnt pathway. For example, BCL-7 can affect seam cell development not only by suppressing the Wnt pathway but also by regulating the terminal cell differentiation of seam cells. The phenotype of fewer seam cells observed in tm5268 mutants is caused by the disturbance of these two mechanisms.
In this study, we show that BCL-7 suppresses the expression of the ß-catenin homologs, bar-1, sys-1, and wrm-1. However, RNAi-based screening showed that only wrm-1 was a suppressor of the tm5268 mutant phenotype. This discrepancy may be the result of differences in the degree of BCL-7′s effect on the mRNA expression level of these genes in tm5268 mutants. The expression of both bar-1 and sys-1 is markedly increased in tm5268 mutant worms (S9A Fig.); therefore, the moderate downregulation of these genes caused by feeding RNAi may not be enough to affect the phenotypes in bcl-7 deletion mutants. By contrast, the mRNA level of wrm-1 is only approximately doubled in tm5268 mutants, therefore, even weak downregulation of wrm-1 may be sufficient to partially suppress the phenotypes of tm5268 mutants.
Interestingly, our findings showed that knockdown of lsy-22 also partially suppressed the Ste phenotype in bcl-7-knockout mutants. LSY-22 is a homolog of the Groucho-like protein and is thought to promote expression of the Groucho homolog UNC-37 [43], thereby repressing the transcription of target genes in C. elegans. In our experiments, knockdown of lsy-22 by RNAi partially suppressed the phenotype of the bcl-7 mutant (tm5268). This result is inconsistent with a previous study [43] and suggests that the downregulation of lsy-22 inhibits the Wnt pathway.
BCL-7 may affect the differentiation of DTCs and the development of gonads
LAG-2/Notch is secreted from differentiated DTCs and regulates gonadal development. The Wnt signaling pathway is an important regulator of DTC differentiation. In this study, the DTC-specific expression of BCL-7 rescued both the Ste phenotype and the defects in gonadal development observed in bcl-7 mutant worms. Although lag-2 temperature-sensitive (ts) mutants or glp-1(ts) mutants exhibited defects in gonadal development, these mutants had only meiotic cells (no mitotic cells) in their gonads and regular germ cell sizes, unlike the bcl-7 mutant (tm5268) [76]–[78]. This result suggests that BCL-7 is involved in the normal cell differentiation of SGPs and subsequent gonadal development in bcl-7 mutants is affected by the impairment of normal LAG-2 secretion from differentiated DTCs.
Our study found that BCL-7 was also expressed in gonadal sheath cells, as shown in S3I–L Fig. Differentiated gonadal sheath cells are crucial for gonadal elongation, meiotic maturation of germ cells, and embryogenesis [32]. In this study, gonadal sheath cell-specific expression of BCL-7 partially rescued the Ste phenotype of the bcl-7 deletion mutant. This result suggests that BCL-7 functions not only in DTCs but also in gonadal sheath cells as a regulator of terminal cell differentiation. Furthermore, BCL-7 was also expressed in germ cells, although we did not demonstrate any cell-autonomous functions of BCL-7 in germ cells (Fig. 3M).
Taken together, these data suggest that BCL-7 functions cell-autonomously at least in DTCs and gonadal sheath cells, similar to in seam cells, and its functions are necessary for terminal cell differentiation and normal gonadal development.
bcl-7/BCL7B shares characteristics with other tumor-suppressor genes
In this study, we show that BCL-7 and BCL7B positively regulate the apoptotic pathway and negatively regulate the Wnt signaling pathway. These characteristics are common with certain tumor-suppressor genes. For example, p53, one of the most well-studied tumor suppressors, inhibits cancer initiation and progression through the induction of apoptosis in abnormal cells [79]–[83]. The results of our qRT-PCR analysis revealed that BCL-7 may function as a positive regulator of the apoptotic pathway in C. elegans (S9B Fig.). However, there was a discrepancy between the results of qRT-PCR and PLM number analysis, possibly because suppression of apoptosis may occur in the limited cell population or because the remaining cells that are inhibited apoptosis may not undergo terminal differentiation. BCL7B also functions as a positive regulator of apoptosis by repressing the anti-apoptotic factor Bcl2, much stronger than its repression of the pro-apoptotic factor Bax (Fig. 6L and S11A Fig.). The function of BCL-7 in the apoptotic pathway is in some ways similar to the function of p53, which activates the apoptotic pathway of target cells [54], [79]–[83]. In addition, BCL-7 and BCL7B also negatively regulate the Wnt signaling pathway through suppressing the expression of ß-catenin. Downregulation of apc, which promotes the degradation of ß-catenin (thereby affecting Wnt signaling), induces hyperactivation of the Wnt pathway and is involved in the development of colorectal cancer [84]. The role of BCL7B in the Wnt pathway is similar to APC. However, there are some differences between BCL7B and other tumor suppressors. For example, the Rb gene, which encodes the retinoblastoma (RB) protein, primarily functions to modulate the G1/S-phase cell cycle checkpoint [64], [65], [85], whereas BCL7B knockdown increased the rate of G1 arrest. This dissimilarity is further reflected by the finding that BCL7B knockdown does not induce hyperproliferation. Furthermore, the accumulation of cells in the G0/G1 phase observed in BCL7B-knockdown KATOIII cells is similar to the specific profile of quiescent cancer stem cells, which also accumulate in the G0/G1 phase [86]. Collectively, these findings suggest that BCL7B is a novel tumor suppressor gene and is required for the terminal cell differentiation.
Downregulation of BCL7B in KATOIII cells induced nuclear enlargement, which is considered a hallmark of undifferentiated cells, such as cancer cells, and is associated with the grade of malignancies in neoplastic diseases [49]. This phenotype was similar to the phenotype of the bcl-7 mutant (tm5268) in C. elegans. Because the mechanisms mediating nuclear enlargement are not clearly understood, it is difficult to compare the function of BCL7 with the functions of other tumor-related genes. However, we demonstrated that the mRNA expression of specific genes, e.g., NEAT1, is significantly increased in BCL7B-knockdown cells. Although a direct interaction between nuclear enlargement and an increase in RNA has not been clearly established, a variety of noncoding RNAs have been shown to play various roles in many types of diseases, including cancer. Additionally, the increase in mRNA expression may indicate that transcriptional hyperactivity occurs in BCL7B-knockdown cells, potentially through the loosening of chromatin structure [87]–[90]. Although our experiments did not evaluate the chromatin structure of BCL7B-knockdown cells, changes in the chromatin structure may explain the observed nuclear alterations in these cells. Furthermore, the mRNA expression levels of undifferentiated markers were significantly increased in both bcl-7 knockout worms and BCL7B-knockdown cells, similar to observations made in some types of malignant cancer cells [91]. Thus, understanding the roles of BCL7B may provide insights into malignant alterations in nuclei.
It should be noted that nuclear changes are not the only alterations that occur in malignant diseases. Poor differentiation, autonomous growth, unlimited proliferation, immortalization, metastatic ability, angiogenic ability, and other phenotypes also contribute significantly to the development of malignancies. Our study demonstrated that BCL7 is involved in mediating nuclear defects, an indicator of poor differentiation and immortalization, but our study did not provide evidence of an association between BCL7 and other malignant phenotypes. Furthermore, BCL7B-knockdown cells did not show hyperproliferation compared with control cells, but they did demonstrate phenotypes similar to cancer stem cells [86]. These results suggest that BCL7B activity contributes only to some malignant phenotypes and that cancer initiation may be caused by a combination of aberrations of BCL7 and abnormalities in other tumor-related genes. Therefore, further investigation of the role of BCL7 in malignant transformation and cancer progression and of molecules that associate with BCL7 family proteins is necessary.
Materials and Methods
Strains
All strains of C. elegans were seeded with Escherichia coli OP50. All experiments were performed at 20°C using standard techniques [92]. The wild-type strain Bristol N2 and some transgenic animals (wIs51, maIs105, osIs5, qIs74, arIs51, hmIs4, baIs4, bzIs8, stIs10165, qIs56 and qyIs174) were obtained from the Caenorhabditis Genetics Center (CGC, Minneapolis, MN). Strains carrying the following mutations were obtained from the trimethylpsoralen/ultraviolet-mutagenized library, as described previously [93]. Mutated strains were identified by polymerase chain reaction (PCR) amplification with primers spanning the deletion regions: bcl-7 (tm5268) III and ced-3 (tm1196) IV [48]. Strain tm5268 was backcrossed twice with N2 and balanced with hT2 [bli-4 (e937) let-? (q782) qIs48] (I;III). The primers used in this study for nested PCR screening were as follows: tm5268_1st round; 5′-TCC GGA TGA GTT GGA TTG TC-3′, 5′-TGT CAT TTC AGC GTC GCG CA-3′; 2nd round; 5′-GCT CCG TCA GAC TCG TAG AT-3′, 5′-AGT GGC TCC ACC TTG ATA GT-3′.
Constructs and transgenic worms
To determine the expression pattern of the rescue plasmid in the wild-type hermaphrodite, the bcl-7 genomic sequence, comprising a 0.3-kbp sequence upstream from the ATG initiation codon of bcl-7, as well as the full-length bcl-7 (0.7 kbp), was PCR amplified from N2 genomic DNA using the following primers: bcl-7_sense, 5′-GGT TCC GCG TGG ATC CCA TTT TGA CGC AAG ATT TGA GAG-3′; bcl-7_antisense, 5′-GCT CAC CAT GCG GCC GCA TGG TTG TTT TGA TGT CAT TTC A-3′. The pFX_egfp or the pFX_mCherry expression vector was digested with BamHI and NotI, and the bcl-7 fragment (Pbcl-7::bcl-7) was cloned into the distinct vectors to generate Pbcl-7::bcl-7::egfp and Pbcl-7::bcl-7::mCherry, respectively [94].
For the seam cell rescue experiment, full-length bcl-7 (0.7 kbp) was amplified from N2 genomic DNA; the pPD95.77_Pscm_mCherry expression vector was digested with SmaI and NotI, and the full-length bcl-7 sequence was cloned into the vector to generate Pscm::bcl-7.
For the gonadal sheath cell rescue experiment, the first intron of lim-7 and the full-length bcl-7 (0.7 kbp) were amplified from N2 genomic DNA using the following primers: lim-7(1st intron)_sense, 5′-TTC TGG TTC CGC GTG GAT CCG TGA GTG TTT TTT TTT TAA TTT G-3′ and lim-7(1st intron)_antisense, 5′-ATT TGC TGA GTA CAT ACG TTC TGA AAA ATG AAA GCT CGA-3′; bcl-7_sense, 5′-TCA TTT TTC AGA ACG TAT GTA CTC AGC AAA TAG ATC TCA-3′ and bcl-7_antisense, 5′-GCT CAC CAT GCG GCC GCT GGT TGT TTT GAT GTC ATT TCA G-3′. The pFX_egfp expression vector was digested with BamHI and BglII, and the lim-7(1st intron)::bcl-7 fragment was cloned into the vector to generate lim-7(1st intron)::bcl-7::egfp.
For the somatic DTC rescue experiment, the 3-kbp sequence upstream from the ATG initiation codon of lag-2 and the full-length bcl-7 (0.7 kbp) were amplified using the following primers: Plag-2_sense, 5′-GGT TCC GCG TGG ATC CTC TTA CAG GTT ATA TTA AAT TCT C-3′ and Plag-2_antisense, 5′-GCT GAG TAC ATA AGG CAA ATT TG-3′; bcl-7_sense, 5′-TGC CTT ATG TAC TCA GCA AAT AG-3′, and bcl-7_antisense, 5′-TCA AAA ATA GAG ATC TTG GTT GTT TTG ATG TCA TTT CAG-3′. The pFX_egfp expression vector was digested with BamHI and BglII, and the Plag-2::bcl-7 fragment was cloned into the vector to generate Plag-2::bcl-7::egfp.
For the germ cell rescue experiment, full-length bcl-7 (0.7 kbp) was amplified from N2 genomic DNA; the pTE5_egfp expression vector was digested with BamHI, and the full-length bcl-7 sequence was cloned into the vector to generate pTE5::bcl-7::egfp.
For the chromophore-assisted light inactivation (CALI) of BCL-7, we prepared Pbcl-7::bcl-7::KillerRed. To generate this construct, a bcl-7 genomic sequence, composed of a 0.3-kbp sequence upstream from the ATG initiation codon of bcl-7 and the full-length bcl-7 (0.7 kbp), was PCR amplified from N2 genomic DNA using the following primers: bcl-7-KR_sense, 5′-GGT TCC GCG TGG ATC CCA TTT TGA CGC AAG ATT TGA GAG-3′, and bcl-7-KR_antisense, 5′-GAA CAG GGC GGG GCC GCC CTC CAT TTA TGG TTG-3′. The KillerRed coding sequence was amplified from the commercially available pKillerRed-C vector (Evrogen, Moscow, Russia) using the following primers: KillerRed_sense, 5′-TAA ATG GAG GGC GGC CCC GC-3′, and KillerRed_antisense, 5′-GCT CAC CAT GCG GCC GCT CCT CGT CGC TAC CGA TGG CGC-3′. Pbcl-7::bcl-7 and KillerRed were cloned into the pFX vector at the BamHI and NotI sites to produce Pbcl-7::bcl-7::KillerRed.
To generate the transgenic lines, constructs were injected into worms at 20–100 ng/µL along with Pmyo-2::DsRed, Plin-44::gfp, or scmp::mCherry as an injection marker (100 ng/µL). The transgenic strains constructed for this study were tmEx2966 [Pbcl-7::bcl-7::egfp, Pmyo-2::DsRed], tmEx3496 [Pbcl-7::bcl-7::mCherry, Plin-44::gfp], tmEx3873 [Plag-2::bcl-7::egfp, Pmyo-2::DsRed], tmEx3875 [pTE5::bcl-7::egfp, Pmyo-2::DsRed], tmEx3878 [Pbcl-7::bcl-7::KillerRed, Plin-44::gfp], tmEx4126 [scmp::bcl-7, scmp::mCherry], tmEx4116 [Plim-7::bcl-7::egfp, Pmyo-2::DsRed], and tmEx4121[Plag-2::bcl-7::egfp, Plim-7::bcl-7::egfp, Pmyo-2::DsRed].
The integrated arrays wIs51, containing scm::gfp, maIs105, containing col-19::gfp, and arIs51, containing cdh-3::gfp, were used to assay the numbers of seam cells and hyp7 cells as well as the differentiation of seam cells. The integrated arrays qIs74, containing pop-1::gfp, and osIs5, containing scm::wrm-1::gfp, were used to assay the distribution of POP-1 and WRM-1, respectively, in the seam cells. The integrated arrays hmIs5, containing Pdes-2::gfp, and baIs4, containing Pdat-1::gfp, were used to assay the formation of PVD and PDE sensory neurons, respectively. The integrated array bzIs8, containing Pmec-4::gfp, was used to assay the number of PLM neurons. The integrated array stIs10165, containing egl-27p::his-24::mCherry, was used to analyze the expression pattern of egl-27 as an undifferentiated marker. The integrated array qIs56, containing Plag-2::gfp, and qyIs174, containing Phlh-2::gfp::hlh-2, was used to assay development of DTCs. These transgenic strains were obtained through CGC.
Microscopy
Differential interference contrast and fluorescence images were obtained using a BX51 microscope equipped with a DP30BW CCD camera (Olympus Optical Co., Ltd, Tokyo, Japan).
Analysis of seam cells
To characterize seam cell phenotypes, we scored the number of seam cell nuclei and observed alae formation. The numbers of GFP-positive seam cell nuclei were counted at each larval stage (early L1, middle L2, late L3, and L4) and the young adult stage, and staging was assessed by vulval shapes and gonadal morphologies. Alae formation was observed through a Nomarski microscope at the young adult stage.
Dye-filling assay in C. elegans
We performed a dye-filling assay to observe the phenotype of the phasmid in bcl-7 mutants as described in a previous report [95]. The assay was performed by incubating the wild-type and bcl-7 (tm5268) hermaphrodites in the DiI solution (10 µg/mL DiI in M9 buffer) for 2 h at room temperature. Thereafter, these specimens were observed under a fluorescence microscope as above.
Self-brood size of C. elegans
To determine the average number of progeny produced by each strain, as well as by the transgenic worms, L4 worms were placed on individual NGM plates. Worms were transferred daily until egg laying ceased, and the total number of produced live progeny was then counted.
DAPI staining and immunostaining in C. elegans
Immunostaining of the gonads was performed as described previously [96]. Transgenic or wild-type animals were placed on a subbed slide in 5.0 µL of M9 buffer containing 1 mM levamisole. The heads or tails of these worms were cut off using a 27-gauge needle to extrude their gonads. The dissected gonads were fixed with 1.0% paraformaldehyde in phosphate-buffered saline (PBS) for 10 min and permeabilized with PBS containing 0.1% Triton X-100 for 3 min. Samples were blocked in PBS containing 2.5% bovine serum albumin (BSA) for at least 30 min. Fixed worms were incubated with an anti-PH3 antibody overnight at 4°C. Samples were washed three times with PBS containing 0.5% BSA, incubated with a secondary antibody conjugated to Alexa 594 (Invitrogen, San Diego, CA) for 1 h, and washed at least twice. Then, samples were suspended in 0.1 µg/mL Prolong Gold containing DAPI (Invitrogen) for more than 15 min and observed under a fluorescence microscope.
Chromophore-Assisted Light Inactivation (CALI) of BCL-7
For CALI of BCL-7, bcl-7 (tm5268) mutants expressing Pbcl-7::bcl-7::KillerRed were exposed to a light-emitting diode array (572 nm) from the comma-stage, early L1 stage, and L2 stage. At the adult stage, we observed the worms through a microscope and calculated the rates of worms exhibiting the Ste phenotype. In addition, to determine when bcl-7 function is most critical during development, we also performed the pulse experiments using the CALI method. bcl-7 (tm5268) mutants expressing Pbcl-7::bcl-7::KillerRed were exposed to a light-emitting diode array (572 nm) only during the comma and early L1 stages (the exposure period was 6 hrs). The mutants grew in a dark room at all other stages until they became adults. At the adult stage, the percent of worms with Ste phenotypes was calculated.
RNA interference
RNA interference analyses (RNAi) were performed by feeding animals with dsRNA-producing bacteria as described previously [97]. Briefly, the RNAi clones were transformed into E. coli HT115(DE3), and then, approximately 10–20 P0 animals at the early L1 stage were transferred to plates containing RNAi-bacteria grown on 100 µg/mL ampicillin and 1 mmol/L isopropyl-beta-D-thiogalactopyranoside (IPTG).
For the analysis of the phenotypes of bcl-7-knockdown worms, the Egl-grade was scored in the F1 generation at the young adult larval stage. More specifically, we observed the worms under a microscope and categorized them as follows: stage 1 : 1 - to 8-cell stage embryos present in the uterus; stage 2 : 16-cell stage to comma-stage embryos present in the uterus; or stage 3: postcomma-stage embryos present in the uterus [98]. We compared the frequencies of bcl-7-knockdown and control worms in each category.
For suppressor screening, the numbers of seam cells and of eggs were counted in the F1 generation at the L4 larval stage. We compared bcl-7 mutants to wild-type worms, and if there was any remission of the mutant phenotype, we identified the corresponding gene as a candidate suppressor gene.
All RNAi clones, except for sys-1, glp-1, cki-1, bro-1, lin-17, mig-14, and cwn-1, were taken from the Ahringer RNAi library. The sys-1, glp-1, cki-1, bro-1, lin-17, mig-14, cwn-1, and wrm-1c RNAi clones were constructed with the cDNAs generated from wild-type worms using the primers listed in S5 Table. These cDNA fragments were cloned into the L4440 (pPD129.36) vector.
Cell culture and transfection
KATOIII cells obtained through ATCC were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 20% fetal bovine serum at 37°C in a humidified 5% CO2 incubator. Transfection of 100 nM siRNA (ON-TARGETplus SMARTpool L-017228-00 or ON-TARGET plus siCONTROL non-targeting siRNA; Dharmacon RNAi Technologies, Lafayette, CO) into cells was performed using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's protocol.
Nuclear size assays
KATOIII cells were grown on 2-well chamber slides (Lab-Tek, Campbel, CA) at 1×105 cells/well for approximately 24 h prior to transfection. Forty-eight hours after transfection with control-siRNA or BCL7B-siRNA, the cells were fixed with 4% paraformaldehyde for 10 min at room temperature and permeabilized with 0.1% Triton X-100 for 3 min. Then, the cells were washed and incubated in 0.1 µg/mL DAPI stain (Life Technology, Carlsbad, CA) overnight at room temperature in the dark. Samples were observed under a fluorescence microscope (with a UV filter), and acquired images were digitally analyzed with ImageJ software (National Institutes of Health, Bethesda, MA).
Analysis of the number of the nuclei
KATOIII cells were grown on 2 well chamber slides (Lab-Tek) at 1×105 cells/well for approximately 24 h prior to transfection. Forty-eight hours after transfection of control-siRNA or BCL7B-siRNA, 1 mg/l wheat germ agglutinin (WGA)-Alexa Fluor 488 (Invitrogen) was added to the cells to stain the plasma membrane, which were then incubated for 10 min at 37°C in the dark. Subsequently, the cells were washed twice, fixed with 4% paraformaldehyde for 10 min at room temperature, and permeabilized with 0.1% Triton X-100 for 3 min. Then, the cells were washed and suspended in 0.1 µg/mL DAPI overnight at room temperature in the dark. The samples were then observed under a fluorescence microscope, and the numbers of the cells and the nuclei were counted; the number of nuclei per cell was also calculated for each sample.
RNA detection assays in KATOIII cells
KATOIII cells were grown on 2 well chamber slides (Lab-Tek) at 1×105 cells/well for approximately 24 h prior to transfection. Forty-eight hours after transfection with control-siRNA (slide-a and slide-b) or BCL7B-siRNA (slide-a and slide-b), the cells were fixed with 4% paraformaldehyde for 10 min at room temperature and permeabilized with 0.1% Triton X-100 for 3 min. Thereafter, 0.3 mg/mL RNase (Qiagen, Valencia, CA) was added to the cells on slide-a for all samples, while the cells on slide-b were not treated with RNase; all cells were incubated overnight at 4°C. Then, the cells were washed, incubated with the ethidium bromide (Life Technologies, Gaithersburg, MD) for more than 1 h, washed twice, and suspended in 0.1 µg/mL DAPI overnight at room temperature in the dark. Samples were observed under a fluorescence microscope (DAPI: UV filter; ethidium bromide: 594 nm filter). To adjust the focus of the DAPI image, a 25% neutral density (ND) filter was used, and the exposure time was 300 msec at each data acquisition point; for the EtBr image, a 25% ND filter was also used, and the exposure time was 100 msec at each data. Thereafter, acquired images were digitally analyzed with ImageJ software (National Institutes of Health).
Cell viability assays
To analyze the effects of BCL7B knockdown on cell viability, KATOIII cells were transfected with BCL7B-siRNA or control-siRNA (day 0) and incubated until day 2 at 37°C. Then, 0.1% actinomycin D was added to the medium, and the cells were incubated for more than 5 h at 37°C. Cells were then collected, supplemented with 0.2% trypan blue, transferred to a plastic disposable counting chamber, and counted with an automated cell counter (TC20, Bio-Rad laboratories, Hercules, CA).
qRT-PCR analysis
For worms, total RNA was isolated from young adult animals using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. For KATOIII cells, total RNA was extracted using TRIzol (Invitrogen). Total RNA was used for reverse transcription with the Superscript III reverse transcriptase (Invitrogen) using an oligo(dT) primer (for worms) or random primers (for KATOIII cells), according to the manufacturer's instructions, and was subsequently diluted with nuclease-free water (Sigma-Aldrich, St. Louis, MO). qRT-PCR amplification mixtures (25 µL) contained 25 ng of template cDNA, 12.5 µL of 2× SYBR Green I Master Mix buffer (Applied Biosystems, Framingham, MA), and 300 nM of each forward and reverse primers. Reactions were performed on an ABI PRISM 7500 Sequence Detector (Applied Biosystems). The cycling conditions comprised 10 min polymerase activation at 95°C. All data was normalized to ama-1 gene (for the worms) or GAPDH gene (for KATOIII cells). The primer pairs used in this study are listed in S6 Table and S7.
Cell cycle analysis
After 48 h of BCBL7B-siRNA or control-siRNA transfection, cells were applied to a CycleTEST PLUS DNA Reagent Kit (Becton Dickinson Biosciences, San Diego, CA) according to the manufacturer's instructions. Thereafter, cells were subjected to flow cytometry with a Cell Lab Quanta SC flow cytometer (Beckman-Coulter, Fullerton, CA). For the cell cycle analysis, unstained non-treated samples were used to set the EV gain (set at 0.25) and the FL1 photomultiplier tube (PMT) voltages (set at 3.26). These experiments were repeated three times independently.
Analysis of apoptotic cells
After 48h of BCBL7B-siRNA or control-siRNA transfection, cells were applied to an Annexin V/PE/7-AAD kit (BD Biosciences) according to the manufacturer's instructions. Thereafter, cells were subjected to flow cytometry. FL1 was used to measure the Annexin V fluorescence, and 7-AAD fluorescence was detected using FL3. Annexin V-positive and 7-AAD-positive cells were considered to be late apoptotic cells, and Annexin V-positive and 7-AAD-negative cells were considered to be early apoptotic cells [99], [100]. The gating lines were drawn based on the viable cells as the negative control, which were Annexin V-negative and 7-AAD-negative cells. Unstained and single-stained samples were used to set the EV gain (at 0.25), FL1 and FL3 PMT-voltages (at 4.08 and 4.50, respectively), and to compensate for Annexin V spillover into the 7-AAD channel. These experiments were repeated three times independently.
Statistical analyses
All the data were compared using Student's t-test.
Supporting Information
Zdroje
1. HasleH, OlsenJH, HansenJ, FriedrichU, TommerupN (1998) Occurrence of cancer in a cohort of 183 persons with constitutional chromosome 7 abnormalities. Cancer Genet Cytogenet 105 : 39–42.
2. HillierLW, FultonRS, FultonLA, GravesTA, PepinKH, et al. (2003) The DNA sequence of human chromosome 7. Nature 424 : 157–164.
3. Le BeauMM, EspinosaR, DavisEM, EisenbartJD, LarsonRA, et al. (1996) Cytogenetic and molecular delineation of a region of chromosome 7 commonly deleted in malignant myeloid diseases. Blood 88 : 1930–1935.
4. ZenklusenJC, ContiCJ (1996) Cytogenetic, molecular and functional evidence for novel tumor suppressor genes on the long arm of human chromosome 7. Mol Carcinog 15 : 167–175.
5. AmentaS, MoschoviM, SofocleousC, KostaridouS, MavrouA, et al. (2004) Non-Hodgkin lymphoma in a child with Williams syndrome. Cancer Genet Cytogenet 154 : 86–88.
6. CulicV, CulicS, ArmandaV, ResicB, LasanR, et al. (2002) Single signal of the Williams syndrome chromosome region 1 gene in hyperploidic bone marrow cells of acute lymphoblastic leukemia in a Williams syndrome patient. Med Pediatr Oncol 38 : 205–207.
7. OnimoeGI, KahwashS, TermuhlenA, GrossTG, VargaE, et al. (2011) Bilateral burkitt lymphoma of the ovaries: a report of a case in a child with williams syndrome. Case Rep Med 2011 : 327263.
8. ThornburgCD, RoulstonD, CastleVP (2005) Burkitt lymphoma and Williams syndrome: a model for children with a multisystem disorder and malignancy. J Pediatr Hematol Oncol 27 : 109–111.
9. ZhukovaN, NaqviA (2013) Williams-Beuren Syndrome and Burkitt Leukemia. J Pediatr Hematol Oncol 35: e30–32.
10. JadayelDM, OsborneLR, CoignetLJ, ZaniVJ, TsuiLC, et al. (1998) The BCL7 gene family: deletion of BCL7B in Williams syndrome. Gene 224 : 35–44.
11. PotterN, KarakoulaA, PhippsKP, HarknessW, HaywardR, et al. (2008) Genomic deletions correlate with underexpression of novel candidate genes at six loci in pediatric pilocytic astrocytoma. Neoplasia 10 : 757–772.
12. ZaniVJ, AsouN, JadayelD, HewardJM, ShipleyJ, et al. (1996) Molecular cloning of complex chromosomal translocation t(8;14;12)(q24.1;q32.3;q24.1) in a Burkitt lymphoma cell line defines a new gene (BCL7A) with homology to caldesmon. Blood 87 : 3124–3134.
13. MortonLM, PurdueMP, ZhengT, WangSS, ArmstrongB, et al. (2009) Risk of non-Hodgkin lymphoma associated with germline variation in genes that regulate the cell cycle, apoptosis, and lymphocyte development. Cancer Epidemiol Biomarkers Prev 18 : 1259–1270.
14. CarboneA, BernardiniL, ValenzanoF, BottilloI, De SimoneC, et al. (2008) Array-based comparative genomic hybridization in early-stage mycosis fungoides: recurrent deletion of tumor suppressor genes BCL7A, SMAC/DIABLO, and RHOF. Genes Chromosomes Cancer 47 : 1067–1075.
15. van DoornR, ZoutmanWH, DijkmanR, de MenezesRX, CommandeurS, et al. (2005) Epigenetic profiling of cutaneous T-cell lymphoma: promoter hypermethylation of multiple tumor suppressor genes including BCL7a, PTPRG, and p73. J Clin Oncol 23 : 3886–3896.
16. SekiguchiM, SakakibaraK, FujiiG (1978) Establishment of cultured cell lines derived from a human gastric carcinoma. Jpn J Exp Med 48 : 61–68.
17. AmbrosV, HorvitzHR (1984) Heterochronic mutants of the nematode Caenorhabditis elegans. Science 226 : 409–416.
18. KohK, RothmanJH (2001) ELT-5 and ELT-6 are required continuously to regulate epidermal seam cell differentiation and cell fusion in C. elegans. Development 128 : 2867–2880.
19. ClucasC, CabelloJ, BüssingI, SchnabelR, JohnstoneIL (2002) Oncogenic potential of a C.elegans cdc25 gene is demonstrated by a gain-of-function allele. EMBO J 21 : 665–674.
20. KarpX, GreenwaldI (2004) Multiple roles for the E/Daughterless ortholog HLH-2 during C. elegans gonadogenesis. Dev Biol 272 : 460–469.
21. YuanJ, ShahamS, LedouxS, EllisHM, HorvitzHR (1993) The C. elegans cell death gene ced-3 encodes a protein similar to mammalian interleukin-1 beta-converting enzyme. Cell 75 : 641–652.
22. MossEG (2007) Heterochronic genes and the nature of developmental time. Curr Biol 17: R425–434.
23. SulstonJE, HorvitzHR (1977) Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol 56 : 110–156.
24. AbrahanteJE, MillerEA, RougvieAE (1998) Identification of heterochronic mutants in Caenorhabditis elegans. Temporal misexpression of a collagen::green fluorescent protein fusion gene. Genetics 149 : 1335–1351.
25. LiuZ, KirchS, AmbrosV (1995) The Caenorhabditis elegans heterochronic gene pathway controls stage-specific transcription of collagen genes. Development 121 : 2471–2478.
26. SmithCJ, O'BrienT, ChatzigeorgiouM, SpencerWC, Feingold-LinkE, et al. (2013) Sensory neuron fates are distinguished by a transcriptional switch that regulates dendrite branch stabilization. Neuron 79 : 266–280.
27. TreininM, GilloB, LiebmanL, ChalfieM (1998) Two functionally dependent acetylcholine subunits are encoded in a single Caenorhabditis elegans operon. Proc Natl Acad Sci U S A 95 : 15492–15495.
28. HermanMA, HorvitzHR (1994) The Caenorhabditis elegans gene lin-44 controls the polarity of asymmetric cell divisions. Development 120 : 1035–1047.
29. KagiasK, AhierA, FischerN, JarriaultS (2012) Members of the NODE (Nanog and Oct4-associated deacetylase) complex and SOX-2 promote the initiation of a natural cellular reprogramming event in vivo. Proc Natl Acad Sci U S A 109 : 6596–6601.
30. TenenhausC, SubramaniamK, DunnMA, SeydouxG (2001) PIE-1 is a bifunctional protein that regulates maternal and zygotic gene expression in the embryonic germ line of Caenorhabditis elegans. Genes Dev 15 : 1031–1040.
31. HsuJY, SunZW, LiX, ReubenM, TatchellK, et al. (2000) Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell 102 : 279–291.
32. KillianDJ, HubbardEJ (2005) Caenorhabditis elegans germline patterning requires coordinated development of the somatic gonadal sheath and the germ line. Dev Biol 279 : 322–335.
33. HallDH, WinfreyVP, BlaeuerG, HoffmanLH, FurutaT, et al. (1999) Ultrastructural features of the adult hermaphrodite gonad of Caenorhabditis elegans: relations between the germ line and soma. Dev Biol 212 : 101–123.
34. VoutevR, KeatingR, HubbardEJ, VallierLG (2009) Characterization of the Caenorhabditis elegans Islet LIM-homeodomain ortholog, lim-7. FEBS Lett 583 : 456–464.
35. HendersonST, GaoD, LambieEJ, KimbleJ (1994) lag-2 may encode a signaling ligand for the GLP-1 and LIN-12 receptors of C. elegans. Development 120 : 2913–2924.
36. HendersonST, GaoD, ChristensenS, KimbleJ (1997) Functional domains of LAG-2, a putative signaling ligand for LIN-12 and GLP-1 receptors in Caenorhabditis elegans. Mol Biol Cell 8 : 1751–1762.
37. BlellochR, Anna-ArriolaSS, GaoD, LiY, HodgkinJ, et al. (1999) The gon-1 gene is required for gonadal morphogenesis in Caenorhabditis elegans. Dev Biol 216 : 382–393.
38. BulinaME, LukyanovKA, BritanovaOV, OnichtchoukD, LukyanovS, et al. (2006) Chromophore-assisted light inactivation (CALI) using the phototoxic fluorescent protein KillerRed. Nat Protoc 1 : 947–953.
39. YoshinaS, SakakiK, Yonezumi-HayashiA, Gengyo-AndoK, InoueH, et al. (2012) Identification of a novel ADAMTS9/GON-1 function for protein transport from the ER to the Golgi. Mol Biol Cell 23 : 1728–1741.
40. EisenmannDM, MaloofJN, SimskeJS, KenyonC, KimSK (1998) The beta-catenin homolog BAR-1 and LET-60 Ras coordinately regulate the Hox gene lin-39 during Caenorhabditis elegans vulval development. Development 125 : 3667–3680.
41. MeneghiniMD, IshitaniT, CarterJC, HisamotoN, Ninomiya-TsujiJ, et al. (1999) MAP kinase and Wnt pathways converge to downregulate an HMG-domain repressor in Caenorhabditis elegans. Nature 399 : 793–797.
42. NatarajanL, WitwerNE, EisenmannDM (2001) The divergent Caenorhabditis elegans beta-catenin proteins BAR-1, WRM-1 and HMP-2 make distinct protein interactions but retain functional redundancy in vivo. Genetics 159 : 159–172.
43. FlowersEB, PooleRJ, TursunB, BashllariE, Pe'erI, et al. (2010) The Groucho ortholog UNC-37 interacts with the short Groucho-like protein LSY-22 to control developmental decisions in C. elegans. Development 137 : 1799–1805.
44. TakeshitaH, SawaH (2005) Asymmetric cortical and nuclear localizations of WRM-1/beta-catenin during asymmetric cell division in C. elegans. Genes Dev 19 : 1743–1748.
45. MathiesLD, SchvarzsteinM, MorphyKM, BlellochR, SpenceAM, et al. (2004) TRA-1/GLI controls development of somatic gonadal precursors in C. elegans. Development 131 : 4333–4343.
46. ChesneyMA, LamN, MorganDE, PhillipsBT, KimbleJ (2009) C. elegans HLH-2/E/Daughterless controls key regulatory cells during gonadogenesis. Dev Biol 331 : 14–25.
47. KarpX, GreenwaldI (2003) Post-transcriptional regulation of the E/Daughterless ortholog HLH-2, negative feedback, and birth order bias during the AC/VU decision in C. elegans. Genes Dev 17 : 3100–3111.
48. NawaM, Kage-NakadaiE, AisoS, OkamotoK, MitaniS, et al. (2012) Reduced expression of BTBD10, an Akt activator, leads to motor neuron death. Cell Death Differ 19 : 1398–1407.
49. Thomas SP, Vinay K (2005) Neoplasia. In: Vinay K, Abul AK, Nelson F, Richard MN, editors. Pathologic Basis of Disease, Philadelphia: Elsevier Saunders. pp. 270–342.
50. ClemsonCM, HutchinsonJN, SaraSA, EnsmingerAW, FoxAH, et al. (2009) An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell 33 : 717–726.
51. AkaboshiS, WatanabeS, HinoY, SekitaY, XiY, et al. (2009) HMGA1 is induced by Wnt/beta-catenin pathway and maintains cell proliferation in gastric cancer. Am J Pathol 175 : 1675–1685.
52. SafaAR (2012) c-FLIP, a master anti-apoptotic regulator. Exp Oncol 34 : 176–184.
53. SafaAR, PollokKE (2011) Targeting the Anti-Apoptotic Protein c-FLIP for Cancer Therapy. Cancers (Basel) 3 : 1639–1671.
54. González de AguilarJL, GordonJW, RenéF, de TapiaM, Lutz-BucherB, et al. (2000) Alteration of the Bcl-x/Bax ratio in a transgenic mouse model of amyotrophic lateral sclerosis: evidence for the implication of the p53 signaling pathway. Neurobiol Dis 7 : 406–415.
55. FuchsY, StellerH (2011) Programmed cell death in animal development and disease. Cell 147 : 742–758.
56. JacksonSP, BartekJ (2009) The DNA-damage response in human biology and disease. Nature 461 : 1071–1078.
57. NegriniS, GorgoulisVG, HalazonetisTD (2010) Genomic instability—an evolving hallmark of cancer. Nat Rev Mol Cell Biol 11 : 220–228.
58. FishelR, LescoeMK, RaoMR, CopelandNG, JenkinsNA, et al. (1993) The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell 75 : 1027–1038.
59. FordD, EastonDF, BishopDT, NarodSA, GoldgarDE (1994) Risks of cancer in BRCA1-mutation carriers. Breast Cancer Linkage Consortium. Lancet 343 : 692–695.
60. FordD, EastonDF, StrattonM, NarodS, GoldgarD, et al. (1998) Genetic heterogeneity and penetrance analysis of the BRCA1 and BRCA2 genes in breast cancer families. The Breast Cancer Linkage Consortium. Am J Hum Genet 62 : 676–689.
61. MikiY, SwensenJ, Shattuck-EidensD, FutrealPA, HarshmanK, et al. (1994) A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 266 : 66–71.
62. MorinPJ, SparksAB, KorinekV, BarkerN, CleversH, et al. (1997) Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science 275 : 1787–1790.
63. SuLK, VogelsteinB, KinzlerKW (1993) Association of the APC tumor suppressor protein with catenins. Science 262 : 1734–1737.
64. HunterT, PinesJ (1994) Cyclins and cancer. II: Cyclin D and CDK inhibitors come of age. Cell 79 : 573–582.
65. SherrCJ, RobertsJM (1995) Inhibitors of mammalian G1 cyclin-dependent kinases. Genes Dev 9 : 1149–1163.
66. KadochC, HargreavesDC, HodgesC, EliasL, HoL, et al. (2013) Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet 45 : 592–601.
67. ParkJI, VenteicherAS, HongJY, ChoiJ, JunS, et al. (2009) Telomerase modulates Wnt signalling by association with target gene chromatin. Nature 460 : 66–72.
68. SawaH, KouikeH, OkanoH (2000) Components of the SWI/SNF complex are required for asymmetric cell division in C. elegans. Mol Cell 6 : 617–624.
69. ShibataY, UchidaM, TakeshitaH, NishiwakiK, SawaH (2012) Multiple functions of PBRM-1/Polybromo - and LET-526/Osa-containing chromatin remodeling complexes in C. elegans development. Dev Biol 361 : 349–357.
70. SawaH (2012) Control of cell polarity and asymmetric division in C. elegans. Current Topics in Developmental Biology 101 : 55–76..
71. YamamotoY, TakeshitaH, SawaH (2011) Multiple Wnts redundantly control polarity orientation in Caenorhabditis elegans epithelial stem cells. PLoS Genet 7: e1002308.
72. MizumotoK, SawaH (2007) Cortical beta-catenin and APC regulate asymmetric nuclear beta-catenin localization during asymmetric cell division in C. elegans. Dev Cell 12 : 287–299.
73. MizumotoK, SawaH (2007) Two betas or not two betas: regulation of asymmetric division by beta-catenin. Trends Cell Biol 17 : 465–473.
74. GleasonJE, EisenmannDM (2010) Wnt signaling controls the stem cell-like asymmetric division of the epithelial seam cells during C. elegans larval development. Dev Biol 348 : 58–66.
75. RenH, ZhangH (2010) Wnt signaling controls temporal identities of seam cells in Caenorhabditis elegans. Dev Biol 345 : 144–155.
76. GaoD, KimbleJ (1995) APX-1 can substitute for its homolog LAG-2 to direct cell interactions throughout Caenorhabditis elegans development. Proc Natl Acad Sci U S A 92 : 9839–9842.
77. PepperAS, KillianDJ, HubbardEJ (2003) Genetic analysis of Caenorhabditis elegans glp-1 mutants suggests receptor interaction or competition. Genetics 163 : 115–132.
78. PepperAS, LoTW, KillianDJ, HallDH, HubbardEJ (2003) The establishment of Caenorhabditis elegans germline pattern is controlled by overlapping proximal and distal somatic gonad signals. Dev Biol 259 : 336–350.
79. GreenblattMS, BennettWP, HollsteinM, HarrisCC (1994) Mutations in the p53 tumor suppressor gene: clues to cancer etiology and molecular pathogenesis. Cancer Res 54 : 4855–4878.
80. LiD, MarchenkoND, SchulzR, FischerV, Velasco-HernandezT, et al. (2011) Functional inactivation of endogenous MDM2 and CHIP by HSP90 causes aberrant stabilization of mutant p53 in human cancer cells. Mol Cancer Res 9 : 577–588.
81. LiuDP, SongH, XuY (2010) A common gain of function of p53 cancer mutants in inducing genetic instability. Oncogene 29 : 949–956.
82. MiyashitaT, HarigaiM, HanadaM, ReedJC (1994) Identification of a p53-dependent negative response element in the bcl-2 gene. Cancer Res 54 : 3131–3135.
83. MiyashitaT, KrajewskiS, KrajewskaM, WangHG, LinHK, et al. (1994) Tumor suppressor p53 is a regulator of bcl-2 and bax gene expression in vitro and in vivo. Oncogene 9 : 1799–1805.
84. SuLK, VogelsteinB, KinzlerKW (1993) Association of the APC tumor suppressor protein with catenins. Science 262 : 1734–1737.
85. CuriaMC, De IureS, De LellisL, VeschiS, MammarellaS, et al. (2012) Increased variance in germline allele-specific expression of APC associates with colorectal cancer. Gastroenterology 142 : 71–77.e71.
86. Al-HajjM, WichaMS, Benito-HernandezA, MorrisonSJ, ClarkeMF (2003) Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A 100 : 3983–3988.
87. KouzaridesT (2007) Chromatin modifications and their function. Cell 128 : 693–705.
88. LiB, CareyM, WorkmanJL (2007) The role of chromatin during transcription. Cell 128 : 707–719.
89. MurataniM, TanseyWP (2003) How the ubiquitin-proteasome system controls transcription. Nat Rev Mol Cell Biol 4 : 192–201.
90. ZagerRA, JohnsonAC (2009) Renal ischemia-reperfusion injury upregulates histone-modifying enzyme systems and alters histone expression at proinflammatory/profibrotic genes. Am J Physiol Renal Physiol 296: F1032–1041.
91. JeterCR, LiuB, LiuX, ChenX, LiuC, et al. (2011) NANOG promotes cancer stem cell characteristics and prostate cancer resistance to androgen deprivation. Onocogene 30 : 3833–3845.
92. BrennerS (1974) The genetics of Caenorhabditis elegans. Genetics 77 : 71–94.
93. Gengyo-AndoK, MitaniS (2000) Characterization of mutations induced by ethyl methanesulfonate, UV, and trimethylpsoralen in the nematode Caenorhabditis elegans. Biochem Biophys Res Commun 269 : 64–69.
94. Gengyo-AndoK, YoshinaS, InoueH, MitaniS (2006) An efficient transgenic system by TA cloning vectors and RNAi for C. elegans. Biochem Biophys Res Commun 349 : 1345–1350.
95. StarichTA, HermanRK, KariCK, YehWH, SchackwitzWS, SchuylerMW, ColletJ, ThomasJH, RiddleDL (1995) Mutations affecting the chemosensory neurons of Caenorhabditis elegans. Genetics 139 : 171–188.
96. MaruyamaR, EndoS, SugimotoA, YamamotoM (2005) Caenorhabditis elegans DAZ-1 is expressed in proliferating germ cells and directs proper nuclear organization and cytoplasmic core formation during oogenesis. Dev Biol 277 : 142–154.
97. KamathRS, Martinez-CamposM, ZipperlenP, FraserAG, AhringerJ (2001) Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol 2: RESEARCH0002.
98. KoelleMR, HorvitzHR (1996) EGL-10 regulates G protein signaling in the C. elegans nervous system and shares a conserved domain with many mammalian proteins. Cell 84 : 115–125.
99. KoopmanG, ReutelingspergerCP, KuijtenGA, KeehnenRM, PalsST, et al. (1994) Annexin V for flow cytometric detection of phosphatidylserine expression on B cells undergoing apoptosis. Blood 84 : 1415–1420.
100. LecoeurH, LedruE, PrévostMC, GougeonML (1997) Strategies for phenotyping apoptotic peripheral human lymphocytes comparing ISNT, annexin-V and 7-AAD cytofluorometric staining methods. J Immunol Methods 209 : 111–123.
Štítky
Genetika Reprodukční medicína
Článek Phosphorylation of Elp1 by Hrr25 Is Required for Elongator-Dependent tRNA Modification in YeastČlánek Naturally Occurring Differences in CENH3 Affect Chromosome Segregation in Zygotic Mitosis of HybridsČlánek Insight in Genome-Wide Association of Metabolite Quantitative Traits by Exome Sequence AnalysesČlánek ALIX and ESCRT-III Coordinately Control Cytokinetic Abscission during Germline Stem Cell DivisionČlánek Deciphering the Genetic Programme Triggering Timely and Spatially-Regulated Chitin Deposition
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 1
-
Všechny články tohoto čísla
- The Combination of Random Mutagenesis and Sequencing Highlight the Role of Unexpected Genes in an Intractable Organism
- Ataxin-3, DNA Damage Repair, and SCA3 Cerebellar Degeneration: On the Path to Parsimony?
- α-Actinin-3: Why Gene Loss Is an Evolutionary Gain
- Origins of Context-Dependent Gene Repression by Capicua
- Transposable Elements Contribute to Activation of Maize Genes in Response to Abiotic Stress
- No Evidence for Association of Autism with Rare Heterozygous Point Mutations in Contactin-Associated Protein-Like 2 (), or in Other Contactin-Associated Proteins or Contactins
- Nur1 Dephosphorylation Confers Positive Feedback to Mitotic Exit Phosphatase Activation in Budding Yeast
- A Regulatory Hierarchy Controls the Dynamic Transcriptional Response to Extreme Oxidative Stress in Archaea
- Genetic Variants Modulating CRIPTO Serum Levels Identified by Genome-Wide Association Study in Cilento Isolates
- Small RNA Sequences Support a Host Genome Origin of Satellite RNA
- Phosphorylation of Elp1 by Hrr25 Is Required for Elongator-Dependent tRNA Modification in Yeast
- Genetic Mapping of MAPK-Mediated Complex Traits Across
- An AP Endonuclease Functions in Active DNA Demethylation and Gene Imprinting in
- Developmental Regulation of the Origin Recognition Complex
- End of the Beginning: Elongation and Termination Features of Alternative Modes of Chromosomal Replication Initiation in Bacteria
- Naturally Occurring Differences in CENH3 Affect Chromosome Segregation in Zygotic Mitosis of Hybrids
- Imputation of the Rare G84E Mutation and Cancer Risk in a Large Population-Based Cohort
- Polycomb Protein SCML2 Associates with USP7 and Counteracts Histone H2A Ubiquitination in the XY Chromatin during Male Meiosis
- A Genetic Strategy for Probing the Functional Diversity of Magnetosome Formation
- Interactions of Chromatin Context, Binding Site Sequence Content, and Sequence Evolution in Stress-Induced p53 Occupancy and Transactivation
- The Yeast La Related Protein Slf1p Is a Key Activator of Translation during the Oxidative Stress Response
- Integrative Analysis of DNA Methylation and Gene Expression Data Identifies as a Key Regulator of COPD
- Proteasomes, Sir2, and Hxk2 Form an Interconnected Aging Network That Impinges on the AMPK/Snf1-Regulated Transcriptional Repressor Mig1
- Functional Interplay between the 53BP1-Ortholog Rad9 and the Mre11 Complex Regulates Resection, End-Tethering and Repair of a Double-Strand Break
- Estrogenic Exposure Alters the Spermatogonial Stem Cells in the Developing Testis, Permanently Reducing Crossover Levels in the Adult
- Protein Poly(ADP-ribosyl)ation Regulates Immune Gene Expression and Defense Responses
- Sumoylation Influences DNA Break Repair Partly by Increasing the Solubility of a Conserved End Resection Protein
- A Discrete Transition Zone Organizes the Topological and Regulatory Autonomy of the Adjacent and Genes
- Elevated Mutation Rate during Meiosis in
- The Intersection of the Extrinsic Hedgehog and WNT/Wingless Signals with the Intrinsic Hox Code Underpins Branching Pattern and Tube Shape Diversity in the Airways
- MiR-24 Is Required for Hematopoietic Differentiation of Mouse Embryonic Stem Cells
- Tissue-Specific Effects of Genetic and Epigenetic Variation on Gene Regulation and Splicing
- Heterologous Aggregates Promote Prion Appearance via More than One Mechanism
- The Tumor Suppressor BCL7B Functions in the Wnt Signaling Pathway
- , A -Acting Locus that Controls Chromosome-Wide Replication Timing and Stability of Human Chromosome 15
- Regulating Maf1 Expression and Its Expanding Biological Functions
- A Polyubiquitin Chain Reaction: Parkin Recruitment to Damaged Mitochondria
- RecFOR Is Not Required for Pneumococcal Transformation but Together with XerS for Resolution of Chromosome Dimers Frequently Formed in the Process
- An Intracellular Transcriptomic Atlas of the Giant Coenocyte
- Insight in Genome-Wide Association of Metabolite Quantitative Traits by Exome Sequence Analyses
- The Role of the Mammalian DNA End-processing Enzyme Polynucleotide Kinase 3’-Phosphatase in Spinocerebellar Ataxia Type 3 Pathogenesis
- The Global Regulatory Architecture of Transcription during the Cell Cycle
- Identification and Functional Characterization of Coding Variants Influencing Glycemic Traits Define an Effector Transcript at the Locus
- Altered Ca Kinetics Associated with α-Actinin-3 Deficiency May Explain Positive Selection for Null Allele in Human Evolution
- Genetic Variation in the Nuclear and Organellar Genomes Modulates Stochastic Variation in the Metabolome, Growth, and Defense
- PRDM9 Drives Evolutionary Erosion of Hotspots in through Haplotype-Specific Initiation of Meiotic Recombination
- Transcriptional Control of an Essential Ribozyme in Reveals an Ancient Evolutionary Divide in Animals
- ALIX and ESCRT-III Coordinately Control Cytokinetic Abscission during Germline Stem Cell Division
- Century-scale Methylome Stability in a Recently Diverged Lineage
- A Re-examination of the Selection of the Sensory Organ Precursor of the Bristle Sensilla of
- Antagonistic Cross-Regulation between Sox9 and Sox10 Controls an Anti-tumorigenic Program in Melanoma
- A Dependent Pool of Phosphatidylinositol 4,5 Bisphosphate (PIP) Is Required for G-Protein Coupled Signal Transduction in Photoreceptors
- Deciphering the Genetic Programme Triggering Timely and Spatially-Regulated Chitin Deposition
- Aberrant Gene Expression in Humans
- Fascin1-Dependent Filopodia are Required for Directional Migration of a Subset of Neural Crest Cells
- The SWI2/SNF2 Chromatin Remodeler BRAHMA Regulates Polycomb Function during Vegetative Development and Directly Activates the Flowering Repressor Gene
- Evolutionary Constraint and Disease Associations of Post-Translational Modification Sites in Human Genomes
- A Truncated NLR Protein, TIR-NBS2, Is Required for Activated Defense Responses in the Mutant
- The Genetic and Mechanistic Basis for Variation in Gene Regulation
- Inactivation of PNKP by Mutant ATXN3 Triggers Apoptosis by Activating the DNA Damage-Response Pathway in SCA3
- DNA Damage Response Factors from Diverse Pathways, Including DNA Crosslink Repair, Mediate Alternative End Joining
- hnRNP K Coordinates Transcriptional Silencing by SETDB1 in Embryonic Stem Cells
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The Global Regulatory Architecture of Transcription during the Cell Cycle
- A Truncated NLR Protein, TIR-NBS2, Is Required for Activated Defense Responses in the Mutant
- Proteasomes, Sir2, and Hxk2 Form an Interconnected Aging Network That Impinges on the AMPK/Snf1-Regulated Transcriptional Repressor Mig1
- The SWI2/SNF2 Chromatin Remodeler BRAHMA Regulates Polycomb Function during Vegetative Development and Directly Activates the Flowering Repressor Gene
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



