-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaA Dependent Pool of Phosphatidylinositol 4,5 Bisphosphate (PIP) Is Required for G-Protein Coupled Signal Transduction in Photoreceptors
PIP2 has been implicated in multiple functions at the plasma membrane. Some of these require its hydrolysis by receptor-activated phospholipase C, whereas others, such as membrane transport and cytoskeletal function, involve the interaction of the intact lipid with cellular proteins. The mechanistic basis underlying the segregation of these two classes of PIP2 dependent functions is unknown; it has been postulated that this might involve unique pools of PIP2 generated by distinct phosphoinsoitide kinases. We have studied this question in Drosophila photoreceptors, a model system where sensory transduction requires robust phospholipase C mediated PIP2 hydrolysis. We find that the activity of phosphatidylinositol-4-phosphate 5 kinase encoded by dPIP5K is required to support normal sensory transduction and PIP2 dynamics in photoreceptors. Remarkably, non-PLC dependent functions of PIP2, such as vesicular transport and the actin cytoskeleton, were unaffected in dPIP5K mutants. Thus, dPIP5K supports a pool of PIP2 that is readily available to PLC, but has no role in sustaining other non-PLC mediated PIP2 dependent processes. These findings support the existence of at least two non-overlapping pools of PIP2 at the plasma membrane, and provide a platform for future studies of PIP2 regulation at the plasma membrane.
Published in the journal: . PLoS Genet 11(1): e32767. doi:10.1371/journal.pgen.1004948
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004948Summary
PIP2 has been implicated in multiple functions at the plasma membrane. Some of these require its hydrolysis by receptor-activated phospholipase C, whereas others, such as membrane transport and cytoskeletal function, involve the interaction of the intact lipid with cellular proteins. The mechanistic basis underlying the segregation of these two classes of PIP2 dependent functions is unknown; it has been postulated that this might involve unique pools of PIP2 generated by distinct phosphoinsoitide kinases. We have studied this question in Drosophila photoreceptors, a model system where sensory transduction requires robust phospholipase C mediated PIP2 hydrolysis. We find that the activity of phosphatidylinositol-4-phosphate 5 kinase encoded by dPIP5K is required to support normal sensory transduction and PIP2 dynamics in photoreceptors. Remarkably, non-PLC dependent functions of PIP2, such as vesicular transport and the actin cytoskeleton, were unaffected in dPIP5K mutants. Thus, dPIP5K supports a pool of PIP2 that is readily available to PLC, but has no role in sustaining other non-PLC mediated PIP2 dependent processes. These findings support the existence of at least two non-overlapping pools of PIP2 at the plasma membrane, and provide a platform for future studies of PIP2 regulation at the plasma membrane.
Introduction
The detection and conversion of external stimuli into physiological outputs is a fundamental property of neurons and depends on intracellular signal transduction pathways. Phosphoinositides, the seven phosphorylated derivatives of phosphatidylinositol are key signalling molecules and of these the most abundant PIP2 has multiple roles in neurons. Several neuronal receptors (such as the metabotropic glutamate, growth factor and sensory receptors) transduce stimuli into cellular information using the hydrolysis of PIP2 by phospholipase C enzymes. Additionally, within the context of neuronal cell biology PIP2 has several roles including cytoskeletal function [1] [2] and several ion channels and transporters (eg: Kir, TRP and Na+/Ca2+ exchanger ) require PIP2 for their activity [3]. At the pre-synaptic terminal, a regulated cycle of PIP2 turnover is essential to regulate synaptic vesicle cycling. Thus PIP2 plays multiple roles at the plasma membrane of neurons; hence not surprisingly, changes in phosphoinositide metabolism have been linked to several inherited diseases of the human nervous system [reviewed in [4]]. Finally, one of the molecular targets of lithium, used in the treatment of bipolar disorders, is inositol monophosphatase a key regulator of PIP2 turnover in neurons [5].
Given the multiple functions of PIP2 at the plasma membrane, it is unclear if a common pool of PIP2 supports all these functions. Alternatively, if there are distinct pools, it is unclear how these are generated and sequestered on the nanoscale structure of the membrane. In principle, PIP2 can be generated by the activity of two classes of phosphatidylinositol phosphate kinase (PIPK) enzymes, designated PIP5K and PIP4K; PIP5K phosphorylates PI4-P at position 5 of the inositol ring, whereas PIP4K phosphorylates PI5-P at position 4 [[6]]. Although PIP4K and PIP5K synthesize the same end product, they are not functionally redundant [7] and studies of the mammalian enzymes has defined the molecular basis of substrate specificity [8]. Genes encoding PIP5K are present in all sequenced eukaryotes; however PIP4K appears to be a feature of metazoans; mammalian genomes contain three distinct genes for each of these two activities. However, the functional importance of these two classes of enzymes in generating plasma membrane PIP2 has remained unclear.
Drosophila photoreceptors are a well-established model for analyzing phosophoinositide signaling in-vivo [9]. In these cells, the absorption of photons is transduced into neuronal activity by G-protein coupled, phospholipase Cβ (PLCβ) mediated PIP2 hydrolysis [10]. Thus, during phototransduction, PIP2 needs to be resynthesized to match consumption by ongoing PLCβ activity. PIP2 turnover is tightly regulated in photoreceptors; mutants in molecules that regulate PIP2 turnover show defects in phototransduction [11]. However the role of PIPK enzymes in regulating PIP2 synthesis during phototransduction is unknown. In this study we have analyzed each of the three PIPK encoded in the Drosophila genome that could generate PIP2 in the context of phototransduction. Our analysis defines three pools of PIP2 supporting distinct molecular processes in photoreceptors.
Results
Multiple PIP kinases are expressed in the Drosophila eye
In silico analysis of the Drosophila genome sequence revealed that there are four distinct genes that encode open reading frames that include the Interpro domain IPR00002498 which is the “PIP kinase catalytic domain”. These include CG6355, CG3682, CG9985 and CG17471. Of these CG6355 encodes a FYVE domain containing protein that is the single ortholog of yeast Fab1, a protein with 1-phosphatidylinositol 3—phosphate 5 kinase activity [12][13]. CG17471 (dPIP4K) has recently been shown to encode a PIP4K activity that can generate PIP2 by 4 kinase activity using PI5P as a substrate [14]. The remaining two genes namely CG9985 (sktl) and CG3682 could encode putative PIP5K activity. sktl has been proposed to encode a Drosophila PIP5K [15][16]. CG3682 is an independent gene that also encodes a putative PIP5K activity. Previous studies have shown that the activation loop region of PIPKs contains specific residues that are conserved among PIP5K and are distinct from PIP4K enzymes [8]. A multiple alignment of PIP5K and PIP4K proteins from mammals and Drosophila reveals that sktl and CG3682 have activation loop residues that are highly diagnostic of those seen in mammalian PIP5K enzymes (Fig. 1A). Both SKTL and dPIP5K show high level of sequence similarity with all the isoforms of mammalian PIP5K. In catalytic domain the identity is more than 80%, whereas the overall sequence homology is from 55–65% with different mammalian isoforms. SKTL is ubiquitously expressed in all organs (Fig. 1B) suggesting its function in many/all cell types. In mammals this kind of expression pattern is evident for α and β isoforms of the PIP5K [17]. By contrast, the γ isoform of PIP5K is mostly expressed in neuronal tissues [18,19] an expression pattern recapitulated by dPIP5K. In addition dPIP5K has multiple splice variants with a conserved catalytic domain and variable C-terminal extensions [19]. The splicing pattern and protein isoforms of dPIP5K so generated as well as its expression pattern (enriched in the adult head Fig. 1B) recapitulates that seen for mammalian PIP5Kγ. The functional significance of the splice variants of dPIP5K remains to be established. In summary it is very likely that the Drosophila genome contains two genes that encode PIP5K activity namely sktl and CG3682. We have named CG3682 as dPIP5K. Thus, collectively there are three phosphoinositide kinases (PIPK), sktl, dPIP5K and dPIP4K all of which could generate PIP2.
Fig. 1. PIP kinase genes in Drosophila genome. 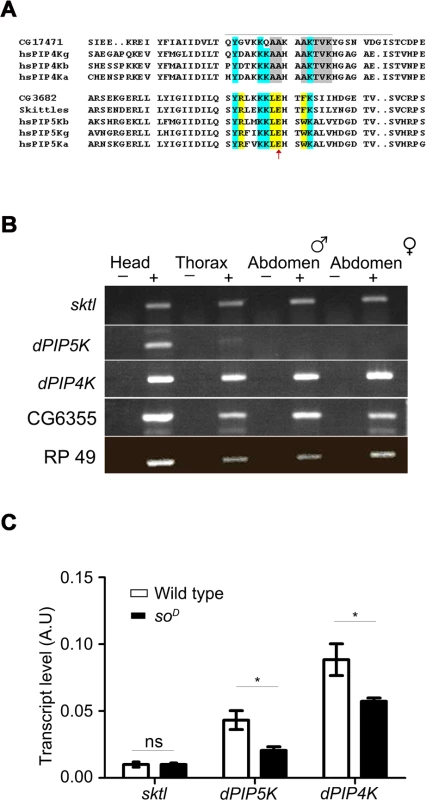
A) Multiple alignment of the protein sequences of PIP4K and PIP5K genes. The sequence around the activation loop is presented with the region indicated by a grey line. Amino acid residues common between PIP5K and PIP4K proteins are marked in blue; residues unique to PIP5K are marked in yellow and unique residues for PIP4K in grey. The red arrow indicates the single residue described as responsible for the unique substrate specificity of PIP4K and PIP5K. Notations used for gene names are; hs-Homo sapiens; a-alpha; b-beta; g-gamma; CG17471-Drosophila PIP4K. B) RNA expression pattern of PIP kinases in various fly tissues: Qualitative RT (reverse transcription) PCR analysis with RNA extracted from various fly tissues. The tissue sources are labeled above the lanes. ‘+’ denotes +RT and ‘−’ denotes −RT. The corresponding gene names are indicated on the left side of the agarose gel picture. C) Comparative real time PCR analysis showing eye enrichment of dPIP5K and dPIP4K; the X-axis indicates gene names and the Y-axis represents transcript level expression in arbitrary units (A.U). White bars represent expression levels from cDNA samples of wild type fly heads and black bars represent samples from heads of soD (mutants that lack eyes). Values shown are the means ± S.D of three independent samples. p values between wild type and soD samples were determined using an unpaired t-test. The stars represent level of significance (***p< 0.001; **p< 0.01; *p< 0.05) In order to identify the PIPK that would generate PIP2 in adult Drosophila photoreceptors, we studied the expression of all three genes. We found that while sktl and dPIP4K were ubiquitously expressed in adult Drosophila, dPIP5K expression was mainly restricted to the head (Fig. 1B). All three genes were expressed in the Drosophila retina; while sktl RNA was present at very low levels and showed no eye-enrichment, dPIP5K and dPIP4K, showed some degree of enrichment in the eye (Fig. 1C).
Loss-of-function mutants in dPIP5K
In order to reveal the function of dPIP5K in vivo, a loss-of-function mutant was generated using ends-out homologous recombination [20]. This results in the insertion of a dominant selection marker (Pw+) flanked by multiple stop codons within the gene such that the kinase domain of dPIP5K was disrupted and the mutant allele should produce no protein. A total of eight independent knock-out alleles were isolated by following phenotypic markers, genetic mapping and molecular screening. Two of these namely dPIP5K18 and dPIP5K30 were studied in detail and are described in this study. All eight alleles were semi-lethal; very few homozygous mutant flies emerged as viable adults. Using a polyclonal antibody generated against a relatively unique C-terminal region of dPIP5K, we found that dPIP5K18 and dPIP5K30 were protein null alleles of dPIP5K (Fig. 2A).
Fig. 2. dPIP5K controls the light response in Drosophila photoreceptors. 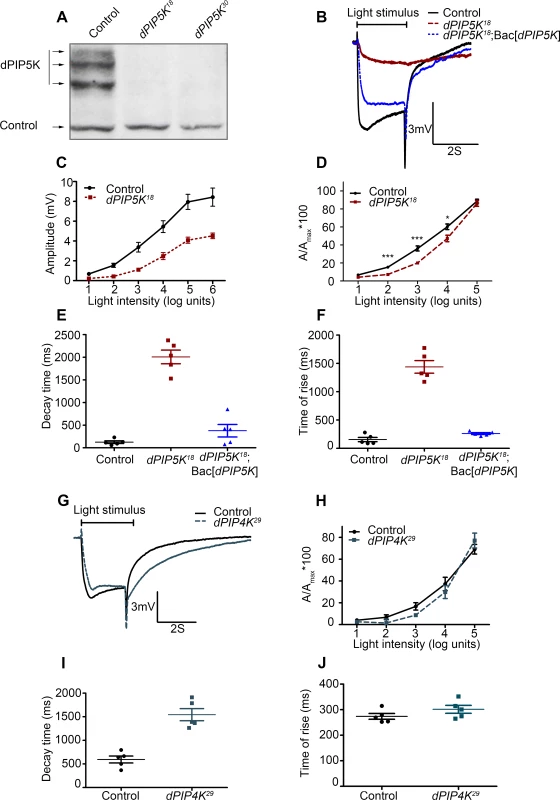
(A) Western blot of fly head extracts probed with dPIP5K specific antibody. The genotypes of the flies are labeled above each lane. The arrows indicate three bands corresponding to three different forms of dPIP5K detected with this antibody in wild type flies. All three bands are missing in both the knockout lines labeled as dPIP5K18 and dPIP5K30. A protein loading control is shown labeled with a black arrow. (B) Representative ERG traces depicting the response of control and dPIP5K18 photoreceptors to single 2s flash of green light of intensity 3 (cf. X-axis of Fig. 2D). The genotypes corresponding to each trace are indicated on the right side. Scale bar at the bottom shows the axis; X-axis represents time in seconds and Y-axis represents amplitude of the response in mV. The duration of the light pulse is indicated. Genotypes: Control-wild type; Mutant- dPIP5K18; Bac[dPIP5K] represents BAC clone containing the dPIP5K gene. (C) Graphical representation comparing the light response between control and dPIP5K18. The X-axis represents increasing light intensity in log units. The Y-axis represents peak amplitude of each response in mV. Error bars: Mean +/− S.D. (D) Intensity response function of the light response in control and dPIP5K18 flies. Wild type and dPIP5K18 flies with matched eye color are shown. The X-axis represents increasing light intensity in log units and Y-axis the peak response amplitude at each intensity normalized to the response at the maximum intensity. p values were determined using an unpaired t-test. The stars represent level of significance (***p< 0.001; **p< 0.01; *p< 0.05). Quantification of the decay time (time taken for the amplitude of the ERG response to reach 50% of its peak amplitude) (E) and the rise time of the ERG response (F) in control, dPIP5K18 and dPIP5K18; Bac[dPIP5K]. (G) Representative light responses from control and dPIP4K29 flies to single 2s flashes of green light. Scale bar at the bottom shows the axes; X-axis represents time in seconds and Y-axis represents amplitude of the response in mV. The duration of the light pulse is indicated. (H) Quantification of the intensity- response to light function from control and dPIP4K29 flies. The X-axis represents light intensity in log units and Y-axis represents the peak amplitude of the response at a given intensity normalized to the response at the maximum intensity. (I) Quantification of the decay time (time taken for the amplitude of the ERG response to reach 50% of its peak amplitude) (J) and the rise time of the ERG response in controls and dPIP4K29. In this study we also used a protein null allele of dPIP4K (dPIP4K29) that has already been described [14]. Homozygous deletions in sktl (eg: sktlΔ20) are larval lethal [15] and analysis using mitotic clones revealed that loss of sktl is also cell lethal in the developing eye. Thus for the analysis of sktl we have used an allelic combination sktlΔ20 / + and over expression of a kinase dead version of SKTL. These are not protein null alleles but represent the most severe alleles of sktl that give viable eyes.
Abnormal light response in dPIP5K18/30 photoreceptors
Since phototransduction in Drosophila involves rapid G-protein coupled PIP2 hydrolysis, it might be predicted that loss-of-function mutants in a PIPK that generates the PIP2 (which is the substrate for photoreceptor PLCβ) might also show a defective electrical response to light. We studied the response to light of mutants in all three genes encoding PIP kinases that can in principle generate PIP2, namely dPIP5K, sktl and dPIP4K. A widely accepted way to study the electrical response to light is the electroretinogram (ERG), an extracellular recording of light-induced electrical changes in the eye. Using ERG, we examined the electrophysiological responses of dPIP5K18 photoreceptors to light. Since very few homozygous mutant adults were obtained, FLP/FRT mediated mitotic recombination was used to obtain mosaic animals in whom the whole eye was homozygous mutant for dPIP5K18[21,22]. ERGs were performed on day 0 (< 24hrs post-eclosion) flies. Characteristically, wild type photoreceptors respond with a large depolarization associated with on and off-transients. By contrast, photoreceptors from dPIP5K18 produced a much smaller receptor potential in response to a stimulus of equivalent intensity associated with a very slow activation kinetics and response termination. Sample traces of voltage changes in response to a 2s stimulus of green light from wild type and mutants are shown (Fig. 2B). This phenotype was seen in all eight knock-out alleles of dPIP5K that we isolated. Responses in dPIP5K18 displayed abnormal kinetics: the time of rise to the peak of the response was substantially prolonged (Fig. 2F) and the time for decay back to baseline following the end of the light stimulus was also increased (Fig. 2E). dPIP5K18 photoreceptors did not display any on or off transients that represent synaptic activity at the first synapse between photoreceptors and the brain. An intensity response function analysis using flies of matched eye colour showed that dPIP5K18 photoreceptors have a reduction in response sensitivity when measured over several log units of light intensity (Fig. 2C,D). Introduction of a genomic rescue transgene in dPIP5K18 flies was able to largely correct the peak amplitude, response termination as well as restore both “on” and “off” transients completely (Fig. 2B, E, F). These results demonstrate that dPIP5K is required to support a normal electrical response to light in Drosophila photoreceptors.
By contrast light responses were unaffected in dPIP4K29 [protein null mutant of dPIP4K [14]] photoreceptors (Fig. 2G,H). Although the amplitude of ERG responses from dPIP4K29 look smaller than controls, this is because these flies are smaller than controls due to a growth defect during larval development [14]. However the kinetics of the light response were only marginally different in dPIP4K29. Finally we studied the most severe allele of sktl that gives rise to adult photoreceptors, namely sktlΔ20/+ (S1A Fig.); these flies gave normal response to light. Further overexpression of a kinase dead version of sktl had no effect on the electrical response to light as measured by ERGs (S1B Fig.). Together these results suggest that dPIP4K and sktl are most likely dispensable for a normal electrical response to light in Drosophila photoreceptors.
dPIP5K18 photoreceptors show no major defects in ultrastructure or levels of transduction proteins
A number of mechanisms could account for the abnormal light response in dPIP5K18 photoreceptors. PIP2 is known to be an allosteric regulator of a number of proteins involved in both vesicular transport as well as the cytoskeleton. Thus, loss of dPIP5K function might impact photoreceptor structure through defects in these processes and the abnormal light response may be a consequence of abnormal ultrastructure as seen in the case of mutants such as rdgA and rdgB [23].
To test this hypothesis, we studied photoreceptor ultrastructure using transmission electron microscopy (TEM). This revealed that photoreceptors R1-R7 from 0 day old flies were normal in dPIP5K18(Fig. 3A). Microvilli were completely intact and showed no vesiculation or blebbing and only minimal defects were seen at the base of the microvilli; however these changes did not increase with age or illumination and rhabdomere structure remained completely intact (Fig. 3B).
Fig. 3. dPIP5K18 photoreceptors have normal ultrastructure and unaltered levels of transduction proteins. 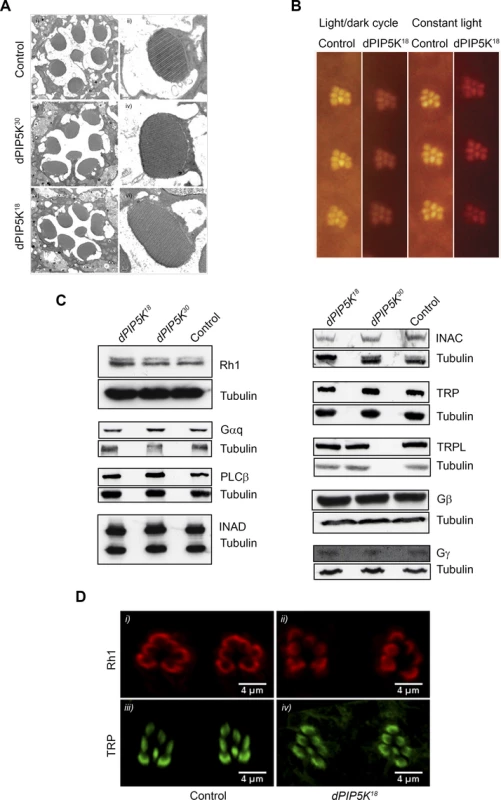
(A) TEM images showing the ultrastructure of control (i-ii), dPIP5K30 (iii-iv) and dPIP5K18 (v-vi). The cross sectional view of a single ommatidium (i, iii, v) and a high magnification view of a single rhabdomere (ii, iv, vi) are shown for each genotype. (B) Optical neutralization images of dPIP5K18 retinae showing normal rhabdomere ultrastructure in flies grown in a 12h L/D cycle as well as in 24 hrs constant light. Images shown are from flies aged nine days post-eclosion. (C) Western blot analysis of head extracts from wild type, dPIP5K18, dPIP5K30 probed with antibodies to each of the major phototransduction proteins. The antibodies used are indicated at the right side of each panel. Tubulin is used as loading control for each set of blots. (D) Single optical transverse sections of a control and dPIP5K18 retina probed with antibodies to Rhodopsin (Rh1) (i, ii) and TRP (iii, iv). A reduced light response could also result from reduction in the levels of key proteins required to support the phototransduction cascade. Thus the abnormal light response in dPIP5K18 photoreceptors could be due to altered levels of any one of the key proteins required for phototransduction such as Gq and Rh1. Gαq1, a severe hypomorph of dGq expressing less than 1% of the wild type protein levels shows more than 1000-fold reduction in sensitivity to light [24]and ninaE mutants characterized by large decrease in the level of Rh1 also show reduced sensitivity to light. Alternatively, the abnormal ERG could also be the consequence of reductions in the level of proteins like NORPA, TRP, INAD which act downstream of photon absorption to generate a normal electrical response to light. This led us to check the protein level of all the key components of the phototransduction cascade in dPIP5K18. Western blot analysis (Fig. 3C) showed that neither the levels of Rh1, Gαq, Gβ and Gγ, nor the levels of NORPA, TRP, TRPL, INAD and INAC were appreciably reduced compared to controls. This result suggests that the abnormal light response observed in dPIP5K18 is not due to reduced levels of any of the key protein required for phototransduction. Further the subcellular localization of Rh1 and TRP were also studied and found to be not different between control and dPIP5K18 (Fig. 3D). Together, these findings suggest that ultrastructural defects or changes in the levels and localization of the major transduction proteins cannot explain the abnormal electrical response in dPIP5K18 photoreceptors.
dPIP5K regulates light induced PIP2 dynamics in photoreceptors
dPIP5K is a PIP kinase that is predicted to convert PI4P into PIP2. To test its requirement in regulating the dynamics of light induced PIP2 turnover at the rhabdomeral membrane we used a live fly preparation in which a fluorescent biosensor consisting of the PH domain of PLCδ fused to GFP (hereafter called PIP2 biosensor) is expressed in photoreceptors. When eyes are illuminated with bright light (λmax 488 nm) high rates of PLC activation result in the hydrolysis of PIP2 and as a consequence the fluorescent biosensor is detached from the membrane and diffuses out of the microvillar cytoplasm resulting in the loss of the fluorescent pseudopupil signal. Under red light illumination that converts metarhodopsin to rhodopsin thus terminating PLCβ activity, PIP2 levels recover as a consequence of ongoing PIP2 resynthesis and the PIP2 biosensor signal in the pseudopupil recovers (Fig. 4A,B). The kinetics of the fluorescent pseudopupil are blocked in norpAP24 which lacks appreciable PLCβ activity (Fig. 4B). Using this approach we studied the kinetics of light induced PIP2 turnover in dPIP5K18 and compared it to wild type controls. This analysis revealed a clear delay in the kinetics of PIP2 resynthesis in dPIP5K18 compared to wild type controls and suggests that dPIP5K activity is required to support normal PIP2 resynthesis following phototransduction (Fig. 4B).
Fig. 4. dPIP5K controls PIP2 dynamics in Drosophila photoreceptors. 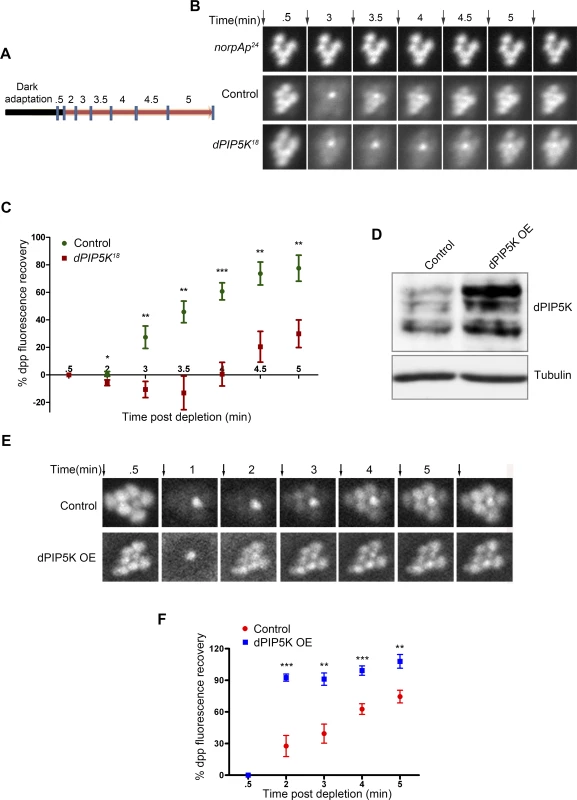
(A) Diagrammatic representation of the experimental protocol used to study PIP2 dynamics in the intact eye. The blue symbols indicate the time of image acquisition. The color of the bar indicates the light condition at which the fly was kept during the experiment. Black color indicates total dark and red indicates in red light illumination. The time points are labeled above the bar in minute. The detailed experimental procedure is discussed in material and methods section. (B) Fluorescent deep pseudopupil (dpp) imaging to study PIP2 dynamics using flies expressing the PIP2 biosensor (see main text for details). The time scale of the imaging is indicated on the top of each panel. Arrows indicate the timing of a 90 ms flash of blue light used for imaging the dpp. Images were acquired from control, dPIP5K18 and norpAP24. The genotypes used for the image acquisition are labeled at the left of the image panel. norpAP24, which is a protein null mutant of PLCβ, is used to show the dependence of dpp dynamics on PLCβ activity. (C) Quantitative representation of PIP2 dynamics. X-axis represents time in minutes between the depleting flash of blue light and the next image acquired. During this period eyes were illuminated in red light. Y-axis represents the level of fluorescence represented as a % of the value in the initial image. Error bars represents mean +/− S.D from five flies. p values were calculated using an unpaired t-test. The stars represent level of significance (***p< 0.001; **p< 0.01; *p< 0.05) (D) Western blot from head extracts depicting the level of dPIP5K protein expression in wild type flies and those overexpressing dPIP5K. The blot was probed with antibody to dPIP5K. Tubulin was used as loading control. (E) Representative images of dpp imaging in control flies and those overexpressing dPIP5K. (F) Quantification of PIP2 dynamics in flies overexpressing dPIP5K compared to controls. X-axis represents time in minutes and Y-axis represents the level of fluorescence represented as a % of the value in the initial image. Error bars represents mean +/− S.D from five flies. p values were determined using an unpaired t-test. The stars represent level of significance (***p< 0.001; **p< 0.01; *p< 0.05). We also tested the effects of overexpressing dPIP5K in adult photoreceptors; the level of protein overexpression was established by Western blots of retinal extracts using an antibody to dPIP5K (Fig. 4D). Overexpression of dPIP5K in photoreceptors resulted in a marked acceleration in the recovery of the PIP2 biosensor following stimulation with light (Fig. 4E, F). This result implies that dPIP5K is able to regulate the rate of PIP2 resynthesis following illumination in photoreceptors.
dPIP5K does not support cytoskeletal function at the microvillar membrane
PIP5K has been shown to have a role in actin remodeling in both yeast and mammalian systems [25]. In dPIP5K18, photoreceptor ultrastructure was largely normal by TEM analysis and growing flies under conditions of bright light illumination did not result in disruption of microvillar ultrastructure. This suggests that the actin cytoskeleton is unaffected by the absence of dPIP5K activity. Further, phalloidin staining suggested that the actin cytoskeleton was largely unaffected in dPIP5K18 (Fig. 5A). The Ezrin/Radixin/Moesin (ERM) family of proteins are regulated by PIP2 and act as cross-linkers between cortical actin and plasma membrane thus playing a key role in maintaining membrane projections, such as microvilli and filopodia [26][27]. ERM proteins are regulated by PIP2 and in the presence of PIP2 the active, phosphorylated form of the protein is attached to the membrane; on PIP2 hydrolysis the proteins are dephosphorylated and the inactive form is released to cytosol [28]. Drosophila has only a single ERM protein, dMoesin, which is required for morphogenesis and maintenance of microvillar structure in photoreceptors. Moesin localizes to the base of the rhabdomere in wild-type flies in the dark [29], but is dephosphorylated and translocates to the cytosol under bright illumination. Using immunolabelling we studied the distribution of p-Moesin in photoreceptors and found this to be no different between controls and dPIP5K18(Fig. 5B). Collectively these findings suggest that dPIP5K activity is not required to support cytoskeletal function in adult Drosophila photoreceptors.
Fig. 5. dPIP5K is not required to support cytoskeleton function and dynamin mediated endocytosis in adult photoreceptors. 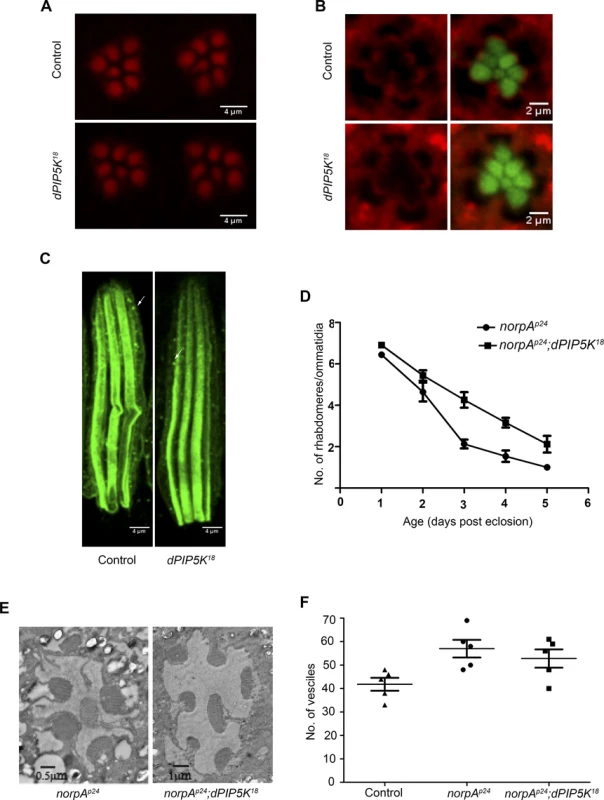
(A) Confocal images of phalloidin stained retinae from control and dPIP5K18 photoreceptors showing normal staining of the rhabdomeres. (B) Confocal images of retinae stained with a p-moesin specific antibody from control and dPIP5K18. Red pseudo color represents p-moesin staining and green marks the rhabdomeric region stained with phalloidin. (C) Confocal images showing longitudinal sections from retinae stained with an antibody to Rh1. The arrows indicated Rhodopsin Loaded Vesicle (RLV) involved in Rh1 endocytosis and recycling. (D) Rate of photoreceptor degeneration in norpAP24 and norpAP24; dPIP5K18 monitored using optical neutralization. The flies were reared in continuous light at 2300 lux. The X-axis represents age of the flies and the Y-axis represents number of rhabdomere visualized in each ommatidium. Error bars represent mean +/− S.D from 50 ommatidia taken from at least five flies. (E) Representative transmission electron micrographs of retinae from norpAP24 and norpAP24; dPIP5K18 flies showing the degree of preservation of ultrastructure. Images shown are from flies that are three days old gown under the same illumination conditions as for panel D. (F) Quantitative representation of Rhodopsin Loaded Vesicles (RLVs) in control, norpAp24 and norpAp24;dPIP5K18. The genotype of the fly is shown in the X-axis and Y-axis represents the count of RLVs. dPIP5K is not required to support rhodopsin turnover at the microvillar plasma membrane
In Drosophila photoreceptors during illumination rhodopsin, the G-protein coupled receptor for light is endocytosed into a vesicular compartment called rhodopsin loaded vesicles (RLV) [30], a key compartment in the turnover of rhodopsin. PIP2 is an important regulator of multiple steps in the endocytic cycle [31]. To test if dPIP5K supports the synthesis of the PIP2 pool regulating endocytosis, we observed the number of RLV in photoreceptors from dPIP5K18 photoreceptors; these were very similar to those in wild type (Fig. 5C). We also tested the effect of dPIP5K18 on the retinal degeneration phenotype of norpA this has previously been shown to depend on endocytosis. norpA mutants undergo light dependent retinal degeneration due to the accumulation of excessive amounts of metarhodopsin-arrestin2 (Arr2-Rh) complexes stabilised at the microvillar membrane [32,33]. This process has been shown to depend on clathrin-mediated endocytosis, a process requiring a number of PIP2 dependent steps. To test if this was affected in dPIP5K18, we generated double mutants of norpAP24;dPIP5K18 and studied the time-course of light dependent retinal degeneration in comparison with norpAP24 alone. If the PIP2 pool produced by dPIP5K was required to mediate endocytosis then one might expect reduced endocytosis of Arr2-Rh complexes in dPIP5K18 photoreceptors thus suppressing the retinal degeneration phenotype of norpAP24. Such an effect is seen when dynamin function is removed using the shibire mutant that results in suppression of degeneration in norpA [32]. However we found that light dependent retinal degeneration was not suppressed in norpAP24; dPIP5K18, although the time course of degeneration was marginally slower (Fig. 5D, E). Additionally the number of RLVs in norpAP24 and norpAP24; dPIP5K18 were not significantly different (Fig. 5F). These results suggest that dPIP5K function is not required to support the endocytic turnover of rhodopsin.
Subcellular localization of dPIP5K
Given our prediction that dPIP5K generates PIP2 that is used as a substrate for light induced PLCβ activity, it is likely that the enzyme is localized to the microvillar plasma membrane where PLCβ is localized. Initial fractionation experiments showed that almost all of the dPIP5K is localized to a membrane fraction (Fig. 6A) where it co-fractionates with key phototransduction proteins such as INAD. We attempted to establish the localization of dPIP5K expressed at endogenous levels using immunocytochemistry; under these conditions the dPIP5K antibody was able to detect the protein localized at the microvillar membrane (Fig. 6B); this was abolished in retinae from dPIP5K18 photoreceptors that are protein null alleles for this gene. Double labeling experiments showed that dPIP5K co-localizes with Rh1 at the microvillar membrane (Fig. 6C). We exploited a genetic tool [34] that allowed us to elevate the expression level of untagged endogenous dPIP5K in photoreceptors, expressed from the endogenous gene locus. Under these conditions too, we found dPIP5K clearly localized to the microvillar membrane (Fig. 6D). By contrast a dPIP4K::GFP transgene was excluded from the microvillar plasma membrane (Fig. 6E).
Fig. 6. Subcellular localization of different PIPKs in adult photoreceptors. 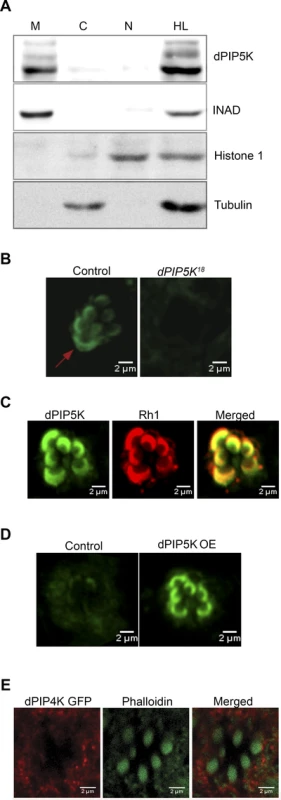
(A) Western blot showing localization of dPIP5K in different sub-cellular fractions prepared from adult Drosophila heads. Fractions shown are: HL-total head lysate; C-cytoplasm; N, nuclear; M-microsomal/membrane. An antibody to INAD is used as a membrane marker. Histone 1 has been used as a nuclear marker and Tubulin as a marker for cytosol. (B) Confocal image showing the distribution of endogenous dPIP5K as detected by a polyclonal antibody from wild type and dPIP5K18 photoreceptors. The enrichment of dPIP5K staining at the rhabdomere membrane (arrow) in wild type is missing in dPIP5K18. (C) Double staining experiments on wild type retinae showing the co-localization of dPIP5K (green) with Rh1 (red). (D) Localization of dPIP5K overexpressed from its endogenous genomic locus. Confocal image from retinae of control flies and those overexpressing dPIP5K using Rh1-GAL4. The protein is shown localized to the rhabdomere membrane. (E) Confocal image showing localization of overexpressed dPIP4K in adult Drosophila photoreceptors. Phalloidin staining marking the rhabdomeres shown in green and dPIP4K localization detected by antibody labeling shown in red. Merged image shows that dPIP4K is excluded from the rhabdomeres. Interaction of dPIP5K with rdgB
Photoreceptors of the Drosophila rdgB mutant show defects in the electrical response to light as well as light dependent degeneration. rdgB encodes a large multi domain protein including an N-terminal phosphatidylinositol transfer protein (PITP) domain [reviewed in [35]]. In vitro the PITP domain can bind and transfer phosphatidylinositol (PI) between two membrane bound compartments and it is presumed, though not demonstrated that the PI delivered to the acceptor compartment is the substrate for phosphorylation by PIPKs that generate phosphorylated versions of PI. In the case of PIP2 this would include the sequential phosphorylation of PI and PI4P by PI4K and PIP5K respectively. Although the precise molecular function of RDGB in photoreceptors in unknown, it has previously been shown that rdgB mutant photoreceptors have a defect in restoring the level of microvillar PIP2 following transduction triggered by a bright flash of light [36]. Thus rdgB mutants represent an opportunity to test the importance of a potential PIP5K that might generate microvillar PIP2 required for phototransduction. To test the relevance of dPIP5K in generating PIP2 required for G-protein coupled PLCβ activity, we generated photoreceptors that are double mutant rdgB9; dPIP5K18; importantly we used the rdgB9 allele that is a strong hypomorph and expresses a small amount of this protein and therefore has a residual response to light. We compared the light response of rdgB9 photoreceptors with those of rdgB9; dPIP5K18 (Fig. 7A). Under similar conditions, while rdgB9 photoreceptors have peak ERG amplitudes of ca. 1.5 mV (Fig. 7A), rdgB9; dPIP5K18 photoreceptors respond with a amplitude of only 0.4 mV (Fig. 7B). This observation suggests that dPIP5K function is required to support the residual light response in rdgB9 photoreceptors. We also studied a second phenotype of rdgB9 namely light dependent retinal degeneration and found that, rdgB9; dPIP5K18 photoreceptors degenerated faster than rdgB9 alone (Fig. 7C,D). By contrast loss of dPIP4K or sktl did not exacerbate the electrical response to light or the retinal degeneration phenotype of rdgB9.
Fig. 7. dPIP5K is required to support rdgB dependent function in photoreceptors. 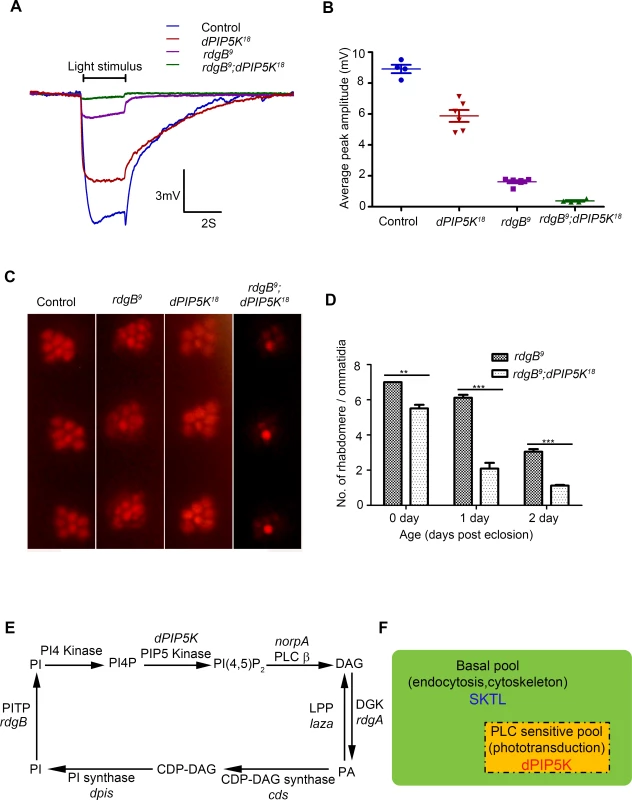
(A) Representative ERG traces depicting the response of control, dPIP5K18, rdgB9 and rdgB9; dPIP5K18. Responses to single 2s flash of green light (intensity 5 cf. X-axis of Fig. 2D) are depicted. The genotypes corresponding to each trace are indicated on the top of the graph. Scale bar at the bottom shows the axis; X-axis represents time in seconds and Y-axis represents amplitude of the response in mV. The duration of the light pulse is indicated. (B) Comparison of maximum amplitude of the light response among control, dPIP5K18, rdgB9 and rdgB9; dPIP5K18. Y-axis represents mean +/− S.D of the peak amplitude from five flies. (C) Representative optical neutralization images from control, rdgB9, dPIP5K18 and rdgB9; dPIP5K18 showing the exacerbation of degeneration in rdgB9; dPIP5K18 compared to rdgB9, dPIP5K18 alone does not show any degeneration. The representative images shown are collected from one-day-old flies maintained in L/D cycle (900 lux). (D) Quantification of the effect of dPIP5K18 loss of function on photoreceptor structure in rdgB9. (E) Schematic diagram showing the light induced PIP2 cycle in Drosophila photoreceptors. Genes encoding a given enzyme activity where identified are marked in italics. Notations used: PI(4,5)P2-phosphatidylinositol 4,5 bisphosphate, norpA- no receptor potential A, PLCβ-phospholipase C beta, DAG- diacylglycerol, DGK- diacylglycerol kinase, rdgA- retinal degeneration A, laza-lipid phosphate phosphohydrolase (PA phosphatase), PA-phosphatidic acid, CDP-DAG- Cytidine diphosphate diacylglycerol; CDS CDP-DAG synthase, PI- phosphatidylinositol, rdgB- retinal degeneration B, PITP- phosphatidylinositol transfer protein, PI(4)P- phosphatidylinositol 4-phosphate (F) Schematic representation of the pools of PIP2 in Drosophila photoreceptor membranes. Representations are only semi-quantitative. The total pool of PIP2 in the plasma membrane is shown bounded by the solid black line. The PLC sensitive PIP2 pool sensitive to light induced PLC activity is shown in the rectangle bounded by the broken/dashed line indicating that PLC will likely also use a non-dPIP5K dependent pool. The basal PIP2 pool is indicated in green. The major function of each pool is indicated. Enzymes responsible for the synthesis of each pool are marked. Discussion
The hydrolysis of PIP2 by PLC in response to receptor activation is a widespread mechanism of signalling at the plasma membrane. In some cells such as neurons, activation of cell surface receptors by neurotransmitter ligands (e.g glutamate, Ach) or sensory stimuli triggers high rates of PLC activation and rapid consumption of PIP2. Under these conditions, it is essential that levels of PIP2, the substrate for PLC are maintained as failure to do so would likely result in desensitization. In mammalian cells, multiple classes of PIPK, the enzymes that resynthesize PIP2 have been described; yet the contribution of these enzymes to PIP2 resynthesis following PLC activation during cell signalling in vivo remains unclear. Broadly two classes of PIPK can synthesize PIP2 have been described; PIP5K that phosphorylates PI4P at position 5 [37] or PIP4K that can phosphorylate PI5P at position 4 [38]. In this study we have analyzed the consequence of loss of each of these two classes of PIPK to resynthesis following PLC mediated PIP2 depletion during Drosophila phototransduction. Loss of dPIP5K function results in profound defects in the light activated electrical response as well as slower recovery of plasma membrane PIP2 levels. Conversely overexpression of dPIP5K was able to substantially accelerate the recovery of PIP2 levels following stimulation with a bright flash of light. We found that dPIP5K is localized to the microvillar plasma membrane, the site at which PIP2 needs to be produced to support ongoing light induced PLC activity. Finally, we found that loss of dPIP5K enhances the ERG defect in a hypomorphic allele of rdgB, a gene with a well-established defect in the response to light. Collectively these observations strongly suggest that dPIP5K activity underlies the conversion of PI4P to PIP2 at the microvillar membrane where it is then available as a substrate for light induced PLCβ activity (Fig. 7E). By contrast loss of the only PIP4K enzyme in the Drosophila genome has minimal effects on phototransduction and this enzyme is not targeted to the microvillar plasma membrane. Our findings also imply that dPIP4K activity (and hence the conversion of PI5P into PIP2) is dispensable for maintaining PIP2 levels during Drosophila phototransduction. This is consistent with a previous study which found no reduction in the levels of PIP2 in flies lacking dPIP4K function [14]. Our observations validate the conclusion from biochemical studies in mammalian cells that the levels of PI5P are substantially lower than those of PIP2 and hence it is unlikely to be the source of the majority of PIP2 in cells [38]. The identity of the PI4K isoform that generates the substrate, PI4P used by dPIP5K remains unknown although a recent study in mammalian systems suggests that PI4KIIIα is likely to be the relevant isoform [39].
Although the ERG response is severely affected in dPIP5K18, it is not abolished as seen in null mutants of PLCβ (norpA) that are not able to hydrolyse PIP2. Additionally, the resting levels of PIP2 as detected by the PIP2 biosensor are comparable to wild type and following a bright flash of light that depletes PIP2, its levels do recover albeit at a slower rate than in wild type photoreceptors. Given that dPIP5K18 is a protein null allele, these observations imply that there must be a second pool of PIP2 in dPIP5K18 cells that is able to support phototransduction and microvillar PIP2 re-synthesis albeit with lower efficiency (Fig. 7F). This second pool of PIP2 is likely available with low efficiency for PLC activity in the absence of the dPIP5K dependent pool thus accounting for the residual light response and observed PIP2 dynamics in dPIP5K18 photoreceptors.
We found that the ultrastructure of dPIP5K18 photoreceptors was essentially normal. This was particularly surprising given that in addition to phototransduction, PIP2 at the microvillar membrane is also expected to regulate multiple processes required to maintain normal microvillar structure including dynamin dependent endocytosis [40][32] as well as cytoskeletal function [41]. However, using multiple readouts we found that molecular readouts of endocytosis and cytoskeletal function were unaffected in dPIP5K18 photoreceptors (Fig. 5). These observations imply that the PIP2 required for these processes is not dependent on dPIP5K activity; rather PIP2 generated by a separate PIPK supports these processes. Thus far, dPIP4K has not been detected on the microvillar plasma membrane, dPIP4K29 photoreceptors show normal ultrastructure on eclosion and do not undergo light dependent microvillar degeneration; thus dPIP4K is unlikely to be the critical enzyme that generates the PIP2 required to support dynamin dependent endocytosis, p-Moesin localization or phototransduction. The Drosophila genome encodes an additional PIP5K activity, sktl that is expressed at low levels in the adult retina but is localized to both the microvillar and basolateral membrane and hence could synthesize PIP2 at both these locations. Complete loss of sktl function is cell lethal and overexpression of sktl in developing photoreceptors results in a severe block in rhabdomere biogenesis [42] whereas overexpression of sktl results in light dependent retinal degeneration in post-development photoreceptors. These findings presumably reflect an essential and non-redundant role for SKTL in supporting fundamental PIP2 dependent cellular processes such as endocytosis and cytoskeletal function that are not dependent on PIP2 hydrolysis by PLC. This model is consistent with the cell-lethal phenotype of photoreceptors that are null for sktl and previous studies showing a role for sktl in supporting cytoskeletal function and endocytosis in other Drosophila tissues and processes such as spermiogenesis [43] and oogenesis [44].
Collectively, our observations imply that there are at least two pools of PIP2 in photoreceptors; one generated by dPIP5K that is required to support a normal electrical response to light but is dispensable for non-PLC dependent functions of PIP2 in photoreceptors and another that is generated by enzymes other than dPIP5K (most likely SKTL) that is also capable of supporting PIP2 synthesis during the light response albeit with reduced efficiency. In summary the PIP2 pool synthesized by dPIP5K is unique in that it is required for a normal light response and apparently dispensable for other PIP2 dependent functions/processes. It also reflects the existence of distinct/segregated pools of PIP2 on the same microvillar plasma membrane that are maintained by distinct kinases.
A number of previous studies have shown that in multiple eukaryotic cell types, plasma membrane PIP2 levels are remarkably stable, undergoing transient fluctuations despite ongoing PLC mediated PIP2 hydrolysis [36,45–47]. However the reasons for this remarkable finding have remained unclear although pharmacological studies have suggested the importance of PIP2 resynthesis in this process [47,48]. One potential explanation for this idea is the existence at the plasma membrane of two pools of PIP2, a larger but less dynamic pool of that is not normally accessed by PLC and supporting non-PLC dependent functions of this lipid and a second, quantitatively smaller but more dynamic pool that is the substrate for PLC activity. What underpins such pools of PIP2? The existence of separate enzymes that generate unique pools of PIP2 has been previously suggested but there have been limited experimental studies to support this model. In murine platelets where thrombin induced PIP2 hydrolysis appears to be dependent on PIP5K1β but not PIP5Kγ [49]; since both these enzymes are expressed in platelets this implies the existence of two pools of PIP2 in these cells of which the PIP5K1β dependent pool is available for thrombin dependent PIP2 turnover. This finding together with our study in Drosophila photoreceptors implies that the plasma membrane in general may contain a specific pool of PIP2 dedicated for the use of receptor dependent PLC signalling and synthesized by a specific PIPK. It is possible that given the high rates of PLC activated PIP2 turnover at the plasma membrane (such as the microvillar membrane in photoreceptors) eukaryotic cells have evolved a mechanism to generate distinct PIP2 pool for this purpose so that other PIP2 dependent functions at the plasma membrane remain unaffected by ongoing receptor activated PIP2 hydrolysis. It is likely that dPIP5K and mammalian PIP5K1β represent PIP5K enzymatic activities required to support such a pool of PIP2 at the plasma membrane. It is presently unclear what properties might make dPIP5K more suitable for generating PIP2 in the context of receptor triggered PLC activity. One possibility is that the kinetic properties of the enzyme encoded by dPIP5K is distinct from that encoded by sktl allowing it to function in the context of high rates of PIP2 turnover. Alternatively (or additionally) within the nanoscale organization of the microvillar plasma membrane, it is possible that dPIP5K is segregated such that PIP2 generated by this enzyme is available within molecular distances of the phototransduction machinery. Interestingly, Drosophila photoreceptors contain within their microvillar membrane a macromolecular signalling complex organized by the PDZ domain protein INAD. It is presently not known if dPIP5K is part of a similar complex but the existence of such mechanisms has been previously shown for mammalian PIP5K1γ in the context of focal adhesion function [50,51]. Interestingly, it has been reported that the INAD protein complex that includes PLCβ is recruited to detergent resistant membranes during light stimulation [52] which themselves have been previously implicated in the formation of PIP2 microdomains and receptor activated PIP2 turnover [53][54]. It is possible that the two PIPKs, SKTL and dPIP5K show differential localization to such domains thus generating and segregating such pools of PIP2 and further studies in this direction are likely to provide insight into this issue. Nevertheless our study provided evidence for the concept of distinct PIPK enzymes as the basis for functionally distinct pools of PIP2 at the plasma membrane. Further analysis in this system is likely to reveal the molecular basis for the organization of PIP2 pools at cellular membranes.
Materials and Methods
Fly culture and genetics
Flies (Drosophila melanogaster) were reared on medium containing corn flour, sugar, yeast powder, and agar along with antibacterial and antifungal agents. Flies were maintained at 25°C and 50% relative humidity. There was no internal illumination within the incubator and the flies were subjected to light pulses of short duration only when the incubator door was opened. When required, flies were grown in an incubator with timed illumination from a white light source (Intensity mentioned in the figure legends of each experiment).
The wild-type strain was Red Oregon-R. The following fly alleles and insertions were obtained for the experiments described here: soD, norpAp24 (Bloomington Stock Center), sktlΔ20 (Hugo Bellen), rdgB9 (R. C. Hardie, Cambridge University), dPIP5K overexpression line - GS200386 (DGRC-Kyoto).
Generation of a dPIP5K antibody
In order to generate an antibody to dPIP5K, the antigenic fragment (an ∼ 250 amino acid unique sequence at the C-terminus of dPIP5K) was expressed as a recombinant protein and purified by affinity chromatography. Polyclonal antibodies were generated in rats using standard immunization protocols.
Generation of dPIP5K knockout by homologous recombination
A knockout of dPIP5K was generated using ‘ends-out’ homologous recombination [20]. A 5.4 kb sequence of dPIP5K genomic sequence was used to generate the donor construct. It consisted of two pieces of genomic dPIP5K cloned as insert 1 (3.17 kb) and insert 2 (2.3 kb) separated by a marker gene white (Pw+) which was flanked by stop codons. These targeting sequences were cloned into the vector pW25 [55]. Transgenic flies carrying this construct were generated and used to perform homologous recombination as previously described [20]. Potential recombinants, which were mapped onto chromosome II, were subjected to molecular analyses using a PCR-based method. Finally, eight individual mutant alleles of dPIP5K (termed as PC4, PC5, PC8, PC18, PC30, PC33, PC60, and PC62) were confirmed and one of these dPIP5K18 was characterized in detail and used in all experiments described in this study. Since homozygous mutants in dPIP5K are semi-lethal during pupal development we recombined dPIP5K18 onto a chromosome with an FRT site at 42B. This allele was used to generate mosaic animals in which only adult retinae were homozygous mutant [21,22].
dPIP5K genomic rescue fly
A BAC clone encompassing dPIP5K,CH321-03B05 (in attB-Pacman-CmR vector) was obtained from p[acman] resource [56]. This BAC clone was 57.178 Kb long and included the dPIP5K gene with extended 5’ and 3’ regions having the promoter and most of the regulatory regions of the gene. The presence of dPIP5K in the clone was verified by PCR using specific primers. This clone was microinjected into embryos and inserted via ΦC31 integration into the VK22attP docking site in the fly genome to generate the wild-type dPIP5K genomic transgene {Bac[dPIP5K]}. Classical genetic crosses were used to move Bac[dPIP5K] into the dPIP5K18 mutant background. Protein expression from Bac[dPIP5K] was verified using Western blotting using a dPIP5K specific antibody.
Western immunoblotting
Heads from 1 day old flies were homogenized in 2× SDS-PAGE sample buffer followed by boiling at 95°C for 5 min. Samples were separated using SDS-PAGE and electro blotted onto nitrocellulose membrane (Hybond-C Extra; GE Healthcare) using semidry transfer assembly (Bio-Rad). Following blocking with 5% Blotto (Santa Cruz Biotechnology, CA), blots were incubated overnight at 4°C in appropriate dilutions of primary antibodies [anti-α-tubulin (1 : 5000 dilution; E7 DSHB), anti-Gαq (1 : 1000 dilution), anti-TRP (1 : 5000 dilution), anti-Rh1 (1 : 200,4C5 DSHB), anti-INAD (1 : 2000) and anti-NORPA (1 : 1000)]. Protein immunoreacted with the primary antibody was visualized after incubation in 1 : 10,000 dilution of appropriate secondary antibody coupled to horseradish peroxidase (Jackson Immuno Research Laboratories) for 2 h at room temperature. Blots were developed with ECL (GE Healthcare) and imaged using a LAS 4000 instrument (GE Healthcare).
Optical neutralization and scoring retinal degeneration
Flies were immobilized on ice, decapitated using a blade and fixed on a glass slide using a drop of colorless nail varnish. It was imaged using 40× oil objective of Olympus BX43 microscope. Quantitation of degeneration was done as previously described [57]
Isolation of pure retinal tissue
Pure preparations of retinal tissue were collected using previously described methods [58]. Briefly, 0 - to 12-hr-old flies were snap-frozen in liquid nitrogen and dehydrated in acetone at −20°C for 48 hr. The acetone was then drained off and the retinae dried at room temperature. They were cleanly separated from the head at the level of the basement membrane using a scalpel blade.
RNA extraction and QPCR
RNA was extracted from Drosophila head using TRIzol reagent (Invitrogen). Purified RNA was treated with amplification grade DNase I (Invitrogen). cDNA conversion was done using SuperScript II RNase H–Reverse Transcriptase (Invitrogen) and random hexamers (Applied Biosystems). Quantitative PCR (QPCR) was performed with the Applied Biosystem 7500 Fast Real Time PCR instrument. Primers were designed at the exon-exon junction following the parameters recommended for QPCR. Transcript levels of the ribosomal protein 49 (RP49) were used for normalization across samples. Three separate samples were collected from each genotype, and duplicate measures of each sample were conducted to ensure the consistency of the data. The primers used for QPCR were as follows:
RP49 fwd: CGGATCGATATGCTAAGCTGT;RP49 rev: GCGCTTGTTCGATCCGTA;
dPIP4K fwd: CATCCGTACGTTGTGGAGAG; dPIP4K rev: AGATCCACATCGTTGCTCAG;
sktl fwd: CTCATGTCCATGTGTGCGTC; sktl rev: TTAATGGTGCTCATCAGTG;
dPIP5K fwd: AGCAGAGAAAACCGCTTAGG; dPIP5K rev: GGCGATTCACTGACTTATTCC
Subcellular fractionation
Fractionation was performed as described in [52]; in short frozen fly heads were homogenized in lysis buffer (20 mM Hepes, 30 mM NaCl, 5 mM EDTA) with protease inhibitors (Roche) at 4°C. Homogenate was centrifuged at 600×g for 3 min to remove chitonous material. The supernatant was spun at 55,000 rpm (ca. 100K g) for 30 minutes at 4°C (Beckman Ultracentriguge, Optima LE-80K ultracentrifuge, using a SW 50.1 rotor) to separate membrane fraction from cytosol. Equal volume of each fraction were subjected to SDS PAGE and analyzed by Western immunoblotting.
Immunohistochemistry
For immunofluorescence, retinae from flies (0–12 hour post eclosion) were dissected under low red light in phosphate buffered saline (PBS) and then fixed in 4% paraformaldehyde in PBS with 1mg/ml saponin in fixing solution for 30 min at room temperature. Fixed eyes were washed 3 times in PBST (PBS with 0.3% TritonX-100) for 10 minutes. The tissues were then incubated in blocking solution [5% fetal bovine serum (FBS) in PBST] for 2 hours at room temperature, after which the tissues were incubated with primary antibodies diluted in blocking solution [anti-p-Moesin-1 : 200[29]], anti-Rh1–1 : 50 (4C5, Developmental Studies Hybridoma Bank), anti-TRP-1 : 250,anti-dPIP5K-1 : 100, anti-GFP-1 : 200 (Abcam)] overnight at 4°C. Appropriate secondary antibodies conjugated with a fluorophore were used at 1 : 300 dilutions [Alexa Fluor 633/568 IgG, (Molecular Probes)] and incubated for 2 hours at room temperature. Wherever required, during the incubation with secondary antibody, Alexafluor 568–phalloidin (Invitrogen) was also added to the tissues to stain the F-actin. After three washes in PBST, the tissues were washed in PBS for 10 min, mounted in mounting medium (70% glycerol in PBS). The whole-mounted preparations were viewed under Olympus FV1000 laser scanning confocal microscope.
Electroretinogram recordings
Flies were anesthetized and immobilized at the end of a disposable pipette tip using a drop of low melt wax. Recordings were done using glass microelectrodes filled with 0.8% w/v NaCl solution. Voltage changes were recorded between the surface of the eye and an electrode placed on the thorax. Following fixing and positioning, flies were dark adapted for 6 min. ERG was recorded with 2 second flashes of green light stimulus. Stimulating light was delivered from a LED light source to within 5 mm of the fly’s eye through a fiber optic guide. Calibrated neutral density filters were used to vary the intensity of the light source. Voltage changes were amplified using a DAM50 amplifier (WPI) and recorded using pCLAMP 10.2. Analysis of traces was performed using Clampfit (Axon Laboratories).
PIP2 dynamics
To monitor PIP2 dynamics in live flies, transgenic flies expressing PH-PLCδ::GFP (PIP2 biosensor) were anesthetized and immobilized at the end of a pipette tip using a drop of low melt wax and fixed by clay on the stage of an Olympus IX71 microscope. The fluorescent deep pseudopupil (dpp, a virtual image that sums rhabdomere fluorescence from ∼20–40 adjacent ommatidia) was focused and imaged using a 10× objective. Time-lapse images were taken by exciting GFP using a 90ms flash of blue light and collecting emitted fluorescence. The program used for this purpose was created in Micromanager. Following preparation, flies were dark adapted for seven minutes after which the eye was stimulated with a 90 ms flash of blue light. The blue light used to excite GFP was also the stimulus to rapidly convert the majority of rhodopsin (R) to metarhodopsin (M) thus activating the phototransduction cascade and triggering depletion of rhabdomeric PIP2. Between the blue light stimulations, photoreceptors were exposed to long wavelength (red) light that reconverts M to R. The resurgence in dpp fluorescence with time indicates translocation of the probe from cytoplasm to rhabdomere membrane upon PIP2 re-synthesis. The dpp intensity was measured using ImageJ from NIH (Bethesda, MD, USA). Cross sectional areas of rhabdomeres of R1-R6 photoreceptors were measured and the mean intensity values per unit area were calculated.
Electron microscopy
Samples for TEM were prepared as mentioned in Ref.44. Briefly eyes were bisected in ice-cold fixative (2.5% glutaraldehyde in 0.1 M PIPES buffer [pH 7.4]). After 10hrs of fixation at 4°C, eyes were washed with 0.1M PIPES, post-fixed in 1% OsO4 (30min), and stained en bloc in 2% uranyl acetate (1 hr). Eyes were dehydrated in ethanol series and embedded in epoxy. Ultrathin sections (50 nm) were cut and viewed on a Tecnai G2 Spirit Bio-TWIN electron microscope.
Supporting Information
Zdroje
1. Klopfenstein DR, Tomishige M, Stuurman N, Vale RD (2002) Role of phosphatidylinositol(4,5)bisphosphate organization in membrane transport by the Unc104 kinesin motor. Cell 109 : 347–358. 12015984
2. Moss SE (2012) How actin gets the PIP. Sci Signal 5: pe7. doi: 10.1126/scisignal.2002839 22375053
3. Hilgemann DW, Feng S, Nasuhoglu C (2001) The Complex and Intriguing Lives of PIP2 with Ion Channels and Transporters. Sci STKE 2001: RE19. 11734659
4. Mccrea HJ, De Camilli P (2009) Mutations in Phosphoinositide Metabolizing Enzymes and Human Disease Mutations in Phosphoinositide Metabolizing Enzymes and Human Disease. Physiol 24 : 8–16.
5. Ackermann KE, Gish BG, Honchar MP, Sherman WR (1987) Evidence that inositol 1-phosphate in brain of lithium-treated rats results mainly from phosphatidylinositol metabolism. Biochem J 242 : 517–524. 3036092
6. Hinchliffe KA, Ciruela A, Irvine RF (1998) PIPkins1, their substrates and their products: new functions for old enzymes. Biochim Biophys Acta 1436 : 87–104. 9838059
7. Kunz J, Wilson MP, Kisseleva M, Hurley JH, Majerus PW, et al. (2000) The activation loop of phosphatidylinositol phosphate kinases determines signaling specificity. Mol Cell 5 : 1–11. 10678164
8. Kunz J, Fuelling A, Kolbe L, Anderson RA (2002) Stereo-specific substrate recognition by phosphatidylinositol phosphate kinases is swapped by changing a single amino acid residue. J Biol Chem 277 : 5611–9. 11733501
9. Hardie RC, Raghu P (2001) Visual transduction in Drosophila. Nature 413 : 186–93.
10. Raghu P, Hardie RC (2009) Regulation of Drosophila TRPC channels by lipid messengers. Cell Calcium 45 : 566–573. 19362736
11. Raghu P, Yadav S, Mallampati NBN (2012) Lipid signaling in Drosophila photoreceptors. Biochim Biophys Acta 1821 : 1154–1165. 22487656
12. Cooke FT, Dove SK, McEwen RK, Painter G, Holmes AB, et al. (1998) The stress-activated phosphatidylinositol 3-phosphate 5-kinase Fab1p is essential for vacuole function in S. cerevisiae. Curr Biol 8 : 1219–1222. 9811604
13. Rusten TE, Rodahl LM, Pattni K, Englund C, Samakovlis C, et al. (2006) Fab1 phosphatidylinositol 3-phosphate 5-kinase controls trafficking but not silencing of endocytosed receptors. Mol Biol Cell 17 : 3989–4001. 16837550
14. Gupta A, Toscano S, Trivedi D, Jones DR, Mathre S, et al. (2013) Phosphatidylinositol 5-phosphate 4-kinase (PIP4K) regulates TOR signaling and cell growth during Drosophila development. Proc Natl Acad Sci U S A 110 : 5963–5968. doi: 10.1073/pnas.1219333110 23530222
15. Hassan BA, Prokopenko SN, Breuer S, Zhang B, Paululat A, et al. (1998) skittles, a Drosophila phosphatidylinositol 4-phosphate 5-kinase, is required for cell viability, germline development and bristle morphology, but not for neurotransmitter release. Genetics 150 : 1527–1537. 9832529
16. Gervais L, Claret S, Januschke J, Roth S, Guichet A (2008) PIP5K-dependent production of PIP2 sustains microtubule organization to establish polarized transport in the Drosophila oocyte. Development 135 : 3829–3838. doi: 10.1242/dev.029009 18948416
17. Loijens JC, Anderson RA (1996) Type I phosphatidylinositol-4-phosphate 5-kinases are distinct members of this novel lipid kinase family. 271 : 32937–32943. 8955136
18. Wenk MR, Pellegrini L, Klenchin VA, Di Paolo G, Chang S, et al. (2001) PIP kinase Igamma is the major PI(4,5)P(2) synthesizing enzyme at the synapse. Neuron 32 : 79–88. 11604140
19. Giudici M-L, Emson PC, Irvine RF (2004) A novel neuronal-specific splice variant of Type I phosphatidylinositol 4-phosphate 5-kinase isoform gamma. Biochem J 379 : 489–496. 14741049
20. Gong WJ, Golic KG (2003) Ends-out, or replacement, gene targeting in Drosophila. Proc Natl Acad Sci U S A 100 : 2556–2561. 12589026
21. Xu T, Rubin GM (1993) Analysis Of Genetic Mosaics In Developing and Adult Drosophila Tissues. Development 117 : 1223–1237. 8404527
22. Stowers RS, Schwarz TL (1999) A genetic method for generating Drosophila eyes composed exclusively of mitotic clones of a single genotype. Genetics 152 : 1631–1639. 10430588
23. Harris WA, Stark WS (1977) Hereditary retinal degeneration in Drosophila melanogaster. A mutant defect associated with the phototransduction process. JGenPhysiol 69 : 261–91. 139462
24. Scott K, Becker A, Sun Y, Hardy R, Zuker C (1995) Gq alpha protein function in vivo: genetic dissection of its role in photoreceptor cell physiology. Neuron 15 : 919–927. 7576640
25. Coppolino MG, Dierckman R, Loijens J, Collins RF, Pouladi M, et al. (2002) Inhibition of phosphatidylinositol-4-phosphate 5-kinase Ialpha impairs localized actin remodeling and suppresses phagocytosis. J Biol Chem 277 : 43849–43857. 12223494
26. Bretscher A (1999) Regulation of cortical structure by the ezrin-radixin-moesin protein family. Curr Opin Cell Biol 11 : 109–116. 10047517
27. Fiévet B, Louvard D, Arpin M (2007) ERM proteins in epithelial cell organization and functions. Biochim Biophys Acta 1773 : 653–660.
28. Yonemura S, Matsui T, Tsukita S, Tsukita S (2002) Rho-dependent and -independent activation mechanisms of ezrin / radixin / moesin proteins: an essential role for polyphosphoinositides in vivo. J Cell Sci 115 : 2569–2580. 12045227
29. Karagiosis SA, Ready DF (2004) Moesin contributes an essential structural role in Drosophila photoreceptor morphogenesis. Development 131 : 725–732. 14724125
30. Satoh AK, O’Tousa JE, Ozaki K, Ready DF (2005) Rab11 mediates post-Golgi trafficking of rhodopsin to the photosensitive apical membrane of Drosophila photoreceptors. JCellSci 132 : 1487–1497. 15728675
31. Zoncu R, Perera RM, Sebastian R, Nakatsu F, Chen H, et al. (2007) Loss of endocytic clathrin-coated pits upon acute depletion of phosphatidylinositol 4,5-bisphosphate. Proc Natl Acad Sci U S A 104 : 3793–3798. 17360432
32. Alloway PG, Howard L, Dolph PJ (2000) The formation of stable rhodopsin-arrestin complexes induces apoptosis and photoreceptor cell degeneration. Neuron 28 : 129–38. 11086989
33. Kiselev A, Socolich M, Vinos J, Hardy RW, Zuker CS, et al. (2000) A molecular pathway for light-dependent photorecptor apoptosis in Drosophila. Neuron 28 : 139–152. 11086990
34. Toba G, Ohsako T, Miyata N, Ohtsuka T, Seong K-H, et al. (1999) The Gene Search System: A Method for Efficient Detection and Rapid Molecular Identification of Genes in Drosophila melanogaster. Genet 151 : 725–737. 9927464
35. Trivedi D, Padinjat R (2007) RdgB proteins: Functions in lipid homeostasis and signal transduction. Biochim Biophys Acta 1771 : 692–699. 17543578
36. Hardie RC, Raghu P, Moore S, Juusola M, Baines RA, et al. (2001) Calcium influx via TRP channels is required to maintain PIP2 levels in Drosophila photoreceptors. Neuron 30 : 149–59. 11343651
37. King CE, Stephens LR, Hawkins PT, Guy GR, Michell RH (1987) Multiple metabolic pools of phosphoinositides and phosphatidate in human erythrocytes incubated in a medium that permits rapid transmembrane exchange of phosphate. Biochem J 244 : 209–217. 2821998
38. Rameh LE, Tolias KF, Duckworth BC, Cantley LC (1997) A new pathway for synthesis of phosphatidylinositol-4,5-bisphosphate. Nature 390 : 192–196. 9367159
39. Nakatsu F, Baskin JM, Chung J, Tanner LB, Shui G, et al. (2012) PtdIns4P synthesis by PI4KIIIα at the plasma membrane and its impact on plasma membrane identity. J Cell Biol 199 : 1003–1016. doi: 10.1083/jcb.201206095 23229899
40. Orem NR, Dolph PJ (2002) Loss of the phospholipase C gene product induces massive endocytosis of rhodopsin and arrestin in Drosophila photoreceptors. Vision Res 42 : 497–505. 11853766
41. Chang HY, Ready DF (2000) Rescue of photoreceptor degeneration in rhodopsin-null Drosophila mutants by activated Rac1. Science (80-) 290 : 1978–80. 11110667
42. Raghu P, Coessens E, Manifava M, Georgiev P, Pettitt T, et al. (2009) Rhabdomere biogenesis in Drosophila photoreceptors is acutely sensitive to phosphatidic acid levels. J Cell Biol 185 : 129–145. doi: 10.1083/jcb.200807027 19349583
43. Fabian L, Wei HC, Rollins J, Noguchi T, Blankenship JT, et al. (2010) Phosphatidylinositol 4,5-bisphosphate directs spermatid cell polarity and exocyst localization in Drosophila. Mol Biol Cell 21 : 1546–1555. doi: 10.1091/mbc.E09-07-0582 20237161
44. Gervais L, Claret S, Januschke J, Roth S, Guichet A (2008) PIP5K-dependent production of PIP2 sustains microtubule organization to establish polarized transport in the Drosophila oocyte. Development 135 : 3829–3838. doi: 10.1242/dev.029009 18948416
45. Willars GB, Nahorski SR, Challiss RA (1998) Differential regulation of muscarinic acetylcholine receptor-sensitive polyphosphoinositide pools and consequences for signaling in human neuroblastoma cells. J Biol Chem 273 : 5037–5046. 9478953
46. Várnai P, Balla T (1998) Visualization of phosphoinositides that bind pleckstrin homology domains: calcium - and agonist-induced dynamic changes and relationship to myo-[3H]inositol-labeled phosphoinositide pools. J Cell Biol 143 : 501–510.
47. Balla A, Kim YJ, Varnai P, Szentpetery Z, Knight Z, et al. (2008) Maintenance of hormone-sensitive phosphoinositide pools in the plasma membrane requires phosphatidylinositol 4-kinase IIIalpha. Mol Biol Cell 19 : 711–721. 18077555
48. Kennedy ED, Challiss RA, Ragan CI, Nahorski SR (1990) Reduced inositol polyphosphate accumulation and inositol supply induced by lithium in stimulated cerebral cortex slices. 267 : 781–786. 2339988
49. Wang Y, Chen X, Lian L, Tang T, Stalker TJ, et al. (2008) Loss of PIP5KIbeta demonstrates that PIP5KI isoform-specific PIP2 synthesis is required for IP3 formation. Proc Natl Acad Sci U S A 105 : 14064–14069. doi: 10.1073/pnas.0804139105 18772378
50. Di Paolo G, Pellegrini L, Letinic K, Cestra G, Zoncu R, et al. (2002) Recruitment and regulation of phosphatidylinositol phosphate kinase type 1 gamma by the FERM domain of talin. Nature 420 : 85–89. 12422219
51. Ling K, Doughman RL, Firestone AJ, Bunce MW, Anderson RA (2002) Type I gamma phosphatidylinositol phosphate kinase targets and regulates focal adhesions. Nature 420 : 89–93. 12422220
52. Sanxaridis PD, Cronin MA, Rawat SS, Waro G, Acharya U, et al. (2007) Light-induced recruitment of INAD-signaling complexes to detergent-resistant lipid rafts in Drosophila photoreceptors. Mol Cell Neurosci 36 : 36–46. 17689976
53. Pike LJ, Miller JM (1998) Cholesterol depletion delocalizes phosphatidylinositol bisphosphate and inhibits hormone-stimulated phosphatidylinositol turnover. J Biol Chem 273 : 22298–22304. 9712847
54. Morris JB, Huynh H, Vasilevski O, Woodcock EA (2006) Alpha1-adrenergic receptor signaling is localized to caveolae in neonatal rat cardiomyocytes. J Mol Cell Cardiol 41 : 17–25. 16730745
55. Rong YS, Titen SW, Xie HB, Golic MM, Bastiani M, et al. (2002) Targeted mutagenesis by homologous recombination in D. melanogaster. Genes Dev 16 : 1568–1581. 12080094
56. Venken KJT, He Y, Hoskins RA, Bellen HJ (2006) P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science 314 : 1747–1751. 17138868
57. Georgiev P, Garcia-Murillas I, Ulahannan D, Hardie RC, Raghu P (2005) Functional INAD complexes are required to mediate degeneration in photoreceptors of the Drosophila rdgA mutant. J Cell Sci 118 : 1373–1384. 15755798
58. Fujita SC, Inoue H, Yoshioka T, Hotta Y (1987) Quantitative tissue isolation from Drosophila freeze-dried in acetone. Biochem J 243 : 97–104. 3111462
Štítky
Genetika Reprodukční medicína
Článek Phosphorylation of Elp1 by Hrr25 Is Required for Elongator-Dependent tRNA Modification in YeastČlánek Naturally Occurring Differences in CENH3 Affect Chromosome Segregation in Zygotic Mitosis of HybridsČlánek Insight in Genome-Wide Association of Metabolite Quantitative Traits by Exome Sequence AnalysesČlánek ALIX and ESCRT-III Coordinately Control Cytokinetic Abscission during Germline Stem Cell DivisionČlánek Deciphering the Genetic Programme Triggering Timely and Spatially-Regulated Chitin Deposition
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 1
-
Všechny články tohoto čísla
- The Combination of Random Mutagenesis and Sequencing Highlight the Role of Unexpected Genes in an Intractable Organism
- Ataxin-3, DNA Damage Repair, and SCA3 Cerebellar Degeneration: On the Path to Parsimony?
- α-Actinin-3: Why Gene Loss Is an Evolutionary Gain
- Origins of Context-Dependent Gene Repression by Capicua
- Transposable Elements Contribute to Activation of Maize Genes in Response to Abiotic Stress
- No Evidence for Association of Autism with Rare Heterozygous Point Mutations in Contactin-Associated Protein-Like 2 (), or in Other Contactin-Associated Proteins or Contactins
- Nur1 Dephosphorylation Confers Positive Feedback to Mitotic Exit Phosphatase Activation in Budding Yeast
- A Regulatory Hierarchy Controls the Dynamic Transcriptional Response to Extreme Oxidative Stress in Archaea
- Genetic Variants Modulating CRIPTO Serum Levels Identified by Genome-Wide Association Study in Cilento Isolates
- Small RNA Sequences Support a Host Genome Origin of Satellite RNA
- Phosphorylation of Elp1 by Hrr25 Is Required for Elongator-Dependent tRNA Modification in Yeast
- Genetic Mapping of MAPK-Mediated Complex Traits Across
- An AP Endonuclease Functions in Active DNA Demethylation and Gene Imprinting in
- Developmental Regulation of the Origin Recognition Complex
- End of the Beginning: Elongation and Termination Features of Alternative Modes of Chromosomal Replication Initiation in Bacteria
- Naturally Occurring Differences in CENH3 Affect Chromosome Segregation in Zygotic Mitosis of Hybrids
- Imputation of the Rare G84E Mutation and Cancer Risk in a Large Population-Based Cohort
- Polycomb Protein SCML2 Associates with USP7 and Counteracts Histone H2A Ubiquitination in the XY Chromatin during Male Meiosis
- A Genetic Strategy for Probing the Functional Diversity of Magnetosome Formation
- Interactions of Chromatin Context, Binding Site Sequence Content, and Sequence Evolution in Stress-Induced p53 Occupancy and Transactivation
- The Yeast La Related Protein Slf1p Is a Key Activator of Translation during the Oxidative Stress Response
- Integrative Analysis of DNA Methylation and Gene Expression Data Identifies as a Key Regulator of COPD
- Proteasomes, Sir2, and Hxk2 Form an Interconnected Aging Network That Impinges on the AMPK/Snf1-Regulated Transcriptional Repressor Mig1
- Functional Interplay between the 53BP1-Ortholog Rad9 and the Mre11 Complex Regulates Resection, End-Tethering and Repair of a Double-Strand Break
- Estrogenic Exposure Alters the Spermatogonial Stem Cells in the Developing Testis, Permanently Reducing Crossover Levels in the Adult
- Protein Poly(ADP-ribosyl)ation Regulates Immune Gene Expression and Defense Responses
- Sumoylation Influences DNA Break Repair Partly by Increasing the Solubility of a Conserved End Resection Protein
- A Discrete Transition Zone Organizes the Topological and Regulatory Autonomy of the Adjacent and Genes
- Elevated Mutation Rate during Meiosis in
- The Intersection of the Extrinsic Hedgehog and WNT/Wingless Signals with the Intrinsic Hox Code Underpins Branching Pattern and Tube Shape Diversity in the Airways
- MiR-24 Is Required for Hematopoietic Differentiation of Mouse Embryonic Stem Cells
- Tissue-Specific Effects of Genetic and Epigenetic Variation on Gene Regulation and Splicing
- Heterologous Aggregates Promote Prion Appearance via More than One Mechanism
- The Tumor Suppressor BCL7B Functions in the Wnt Signaling Pathway
- , A -Acting Locus that Controls Chromosome-Wide Replication Timing and Stability of Human Chromosome 15
- Regulating Maf1 Expression and Its Expanding Biological Functions
- A Polyubiquitin Chain Reaction: Parkin Recruitment to Damaged Mitochondria
- RecFOR Is Not Required for Pneumococcal Transformation but Together with XerS for Resolution of Chromosome Dimers Frequently Formed in the Process
- An Intracellular Transcriptomic Atlas of the Giant Coenocyte
- Insight in Genome-Wide Association of Metabolite Quantitative Traits by Exome Sequence Analyses
- The Role of the Mammalian DNA End-processing Enzyme Polynucleotide Kinase 3’-Phosphatase in Spinocerebellar Ataxia Type 3 Pathogenesis
- The Global Regulatory Architecture of Transcription during the Cell Cycle
- Identification and Functional Characterization of Coding Variants Influencing Glycemic Traits Define an Effector Transcript at the Locus
- Altered Ca Kinetics Associated with α-Actinin-3 Deficiency May Explain Positive Selection for Null Allele in Human Evolution
- Genetic Variation in the Nuclear and Organellar Genomes Modulates Stochastic Variation in the Metabolome, Growth, and Defense
- PRDM9 Drives Evolutionary Erosion of Hotspots in through Haplotype-Specific Initiation of Meiotic Recombination
- Transcriptional Control of an Essential Ribozyme in Reveals an Ancient Evolutionary Divide in Animals
- ALIX and ESCRT-III Coordinately Control Cytokinetic Abscission during Germline Stem Cell Division
- Century-scale Methylome Stability in a Recently Diverged Lineage
- A Re-examination of the Selection of the Sensory Organ Precursor of the Bristle Sensilla of
- Antagonistic Cross-Regulation between Sox9 and Sox10 Controls an Anti-tumorigenic Program in Melanoma
- A Dependent Pool of Phosphatidylinositol 4,5 Bisphosphate (PIP) Is Required for G-Protein Coupled Signal Transduction in Photoreceptors
- Deciphering the Genetic Programme Triggering Timely and Spatially-Regulated Chitin Deposition
- Aberrant Gene Expression in Humans
- Fascin1-Dependent Filopodia are Required for Directional Migration of a Subset of Neural Crest Cells
- The SWI2/SNF2 Chromatin Remodeler BRAHMA Regulates Polycomb Function during Vegetative Development and Directly Activates the Flowering Repressor Gene
- Evolutionary Constraint and Disease Associations of Post-Translational Modification Sites in Human Genomes
- A Truncated NLR Protein, TIR-NBS2, Is Required for Activated Defense Responses in the Mutant
- The Genetic and Mechanistic Basis for Variation in Gene Regulation
- Inactivation of PNKP by Mutant ATXN3 Triggers Apoptosis by Activating the DNA Damage-Response Pathway in SCA3
- DNA Damage Response Factors from Diverse Pathways, Including DNA Crosslink Repair, Mediate Alternative End Joining
- hnRNP K Coordinates Transcriptional Silencing by SETDB1 in Embryonic Stem Cells
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The Global Regulatory Architecture of Transcription during the Cell Cycle
- A Truncated NLR Protein, TIR-NBS2, Is Required for Activated Defense Responses in the Mutant
- Proteasomes, Sir2, and Hxk2 Form an Interconnected Aging Network That Impinges on the AMPK/Snf1-Regulated Transcriptional Repressor Mig1
- The SWI2/SNF2 Chromatin Remodeler BRAHMA Regulates Polycomb Function during Vegetative Development and Directly Activates the Flowering Repressor Gene
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



