-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaAn Intracellular Transcriptomic Atlas of the Giant Coenocyte
Plants include both the green algae and land plants. Multiple times, root, stem, and leaf-like structures arose independently in plant lineages. In some instances, such as the siphonous algae, these structures arose in the absence of multicellularity. It has been argued by some that the morphology of multicellular land plant organs similarly arises independently of cell division patterns. Here, we explore the partitioning of gene transcripts within what is debatably the largest single-celled organism in the world, the siphonous alga Caulerpa taxifolia. We find that within this giant cell specific transcripts localize within pseudo-organs (morphological structures that are not comprised of cells or tissue). The overall pattern of transcript accumulation follows an apical-basal pattern within the cell. Moreover, transcripts related to different cellular processes, such as transcription and translation, localize to specific regions. Analyzing the signatures of transcript accumulation in land plant organs and the pseudo-organs of Caulerpa, we find that groups of transcripts accumulate together in morphological structures across evolution at rates higher than expected by chance. Together, our results demonstrate a relationship between transcript partitioning and organism morphology, independent from multicellularity, throughout diverse plant lineages.
Published in the journal: . PLoS Genet 11(1): e32767. doi:10.1371/journal.pgen.1004900
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004900Summary
Plants include both the green algae and land plants. Multiple times, root, stem, and leaf-like structures arose independently in plant lineages. In some instances, such as the siphonous algae, these structures arose in the absence of multicellularity. It has been argued by some that the morphology of multicellular land plant organs similarly arises independently of cell division patterns. Here, we explore the partitioning of gene transcripts within what is debatably the largest single-celled organism in the world, the siphonous alga Caulerpa taxifolia. We find that within this giant cell specific transcripts localize within pseudo-organs (morphological structures that are not comprised of cells or tissue). The overall pattern of transcript accumulation follows an apical-basal pattern within the cell. Moreover, transcripts related to different cellular processes, such as transcription and translation, localize to specific regions. Analyzing the signatures of transcript accumulation in land plant organs and the pseudo-organs of Caulerpa, we find that groups of transcripts accumulate together in morphological structures across evolution at rates higher than expected by chance. Together, our results demonstrate a relationship between transcript partitioning and organism morphology, independent from multicellularity, throughout diverse plant lineages.
Introduction
Convergent morphologies have arisen multiple times in plants (Viridiplantae). Diverse cellular architectures underlie these moprhologies, with varying relationships between the number of nuclei per cell and the number of cells within an organism. Within the Chlorophyta, Acetabularia possesses an anchoring rhizoid, supporting stalk, and photosynthetic cap, but is, during most of its life cycle, a unicellular organism reaching heights of up to 10 cm with a single nucleus [1]–[3]. Another green alga, Caulerpa, is one of the largest known single-celled organisms, with stolons (up to meters in length) producing fronds and holdfasts [4]–[9] (Fig. 1A–C). Unlike Acetabularia, which is a single-celled organism, Caulerpa is coenocytic, with numerous nuclei. Siphonocladous chlorophytes have a chambered body plan compartmentalizing variable numbers of nuclei, as in Cladophora. Land plants (Embryophyta) are multicellular organisms, in which organs are composed of tissues and distinct cell types. Developmental biology in land plants was historically influenced by cell theory and studies in animals, in which organismal level morphology is an emergent property of cell division and histogenesis [10], [11]. Animal development is a poor example for land plants, in which morphogenesis is dissociated from histogenesis because cellular lineages and division patterns are largely independent from organ morphology.
Fig. 1. The evolutionary and developmental context of siphonous morphology in Caulerpa taxifolia. 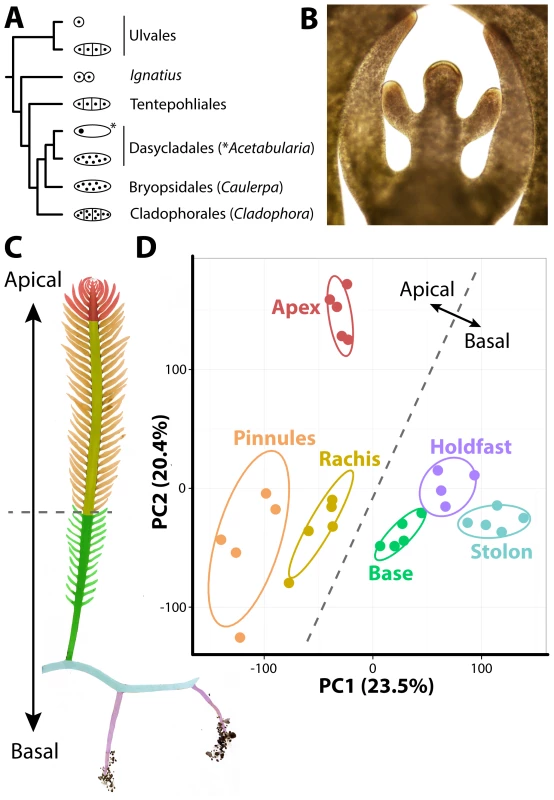
A) Evolutionary relationships among green algae with unicellular, siphonous, siphonocladous, and uninucleate multicellular lifestyles. Diagrams indicate body plan, redrawn from Cocquyt et al. [40]. B) The growing frond apex of Caulerpa taxifolia, producing young pinnules. C) Diagram of sampled regions. D) Principal Component Analysis (PCA) performed on organ RNA-Seq replicates based on transcript accumulation levels. 95% confidence ellipses are indicated for each sampled region. For convenience, a dotted line is provided separating apical from basal pseudo-organ types to relate data back to morphology. Colors indicate different sampled regions (as opposed to nodes in subsequent figures). Red, apex; orange, pinnules; yellow, rachis; green, frond base; blue, stolon; purple, holdfast. For the above reasons, it has been argued [11] that cell theory is not as applicable to plants as in animals with respect to explaining how complex morphologies arise. In its place, Kaplan and Hagemann [11] argued for organismal theory, which they define as “[interpreting] the living protoplasmic mass as a whole, rather than considering its constituent cells as the basic unit.” In other words, the morphology in plants arises at the organismal level rather than as an emergent cellular property. Kaplan and Hagemann assert that “higher plants are also siphonous, but at a subtler, microscopic level.” Some of the siphonous features they argue land plants possess include: 1) cell division through a phragmoplast, 2) plasmodesmata, 3) the symplasm, 4) a multinucleate endosperm and megagametophytes, 5) distinct cytohistological zonations of the shoot apical meristem throughout the Embryophyta, 6) that cell lineage is often independent of morphology (e.g., in leaves), 7) and convergent morphology in multicellular red algae and land plants with different cell lineage patterns.
That land plants are truly siphonous is false: cell walls are a prominent features of land plants upon which morphology is dependent and land plants are multicellular organisms. However, it is useful to think about development in land plants from this unique perspective. That organ growth and morphogenesis are separate from cell division reduces the importance of cell-type specific transcript accumulation in these organisms. Transcriptomics and phylogenetics provide a mechanism to test hypotheses of cell versus organismal theory in siphonous green algae and land plants. Do the accumulation patterns of transcripts differ between single-celled and multicellular organisms with convergent morphology? Are groups of transcripts recurrently recruited to organs across large evolutionary distances regardless of whether an organism is multicellular?
Here, we provide a transcriptome of the giant coenocyte Caulerpa taxifolia. We detect a strong apical-basal gradient of transcript accumulation within the cell. Groups of transcripts with distinct functionalities accumulate in relevant pseudo-organs (morphological structures equivalent to a multicellular organ but not comprised of tissues or cells). Cell compartmentalization is partitioned in Caulerpa, despite its polynucleate condition, and transcripts are patterned according to the flow of genetic information, from transcription-to-translation in a basal-to-apical fashion. An analysis of the intersection of the transcriptomic atlases of a land plant (tomato, Solanum lycopersicum) and Caulerpa demonstrates a limited, recurrent recruitment of genes with similar functions to morphological structures. Our results provide a broad, evolutionary context for the relationship between the cell and organismal morphology at a molecular level within plants, confirming and expanding upon the organismal theory originally proposed by Kaplan and Hagemann [11].
Results/Discussion
Intracellular accumulation of transcripts
To develop a resource to address how organismal morphology can arise in the absence of multicellularity, we sequenced transcripts from multiple pseudo-organs and de novo assembled the intracellular transcriptome of Caulerpa taxifolia (see sequence submission information and S1–S4 Datasets). Caulerpa taxifolia stolons, upwards of 1 m in length, bear fronds (typically 15–30 cm long at maturity) with pinnately-arranged, tapered pinnules. The pinnules arise from active growth at the frond apex, which superficially resembles, in form and function, the apical cells and meristems of embryophytes (Fig. 1B). Caulerpa taxifolia is anchored by holdfasts, which take up phosphorus, nitrogen, and carbon from the substrate, and harbor both ecto - and endosymbiont bacteria that aid this process [9]. We sampled five replicates each of 1) the frond apex, 2) rachis, 3) pinnules, 4) the lower third of the frond base, 5) stolon, and 6) holdfast regions (Fig. 1C). One holdfast sample was lost when thawing for library preparation, reducing holdfast sampling to four replicates. The sample we sequenced was clonal in origin, having proceeded through numerous rounds of asexual reproduction. In its vegetative phase, Caulerpa taxifolia is a haplophasic diploid. Caulerpa taxifolia has one of the smallest genome sizes in its genus (∼100 Mbp, approximately the size of the Arabidopsis thaliana genome) and unlike other Caulerpa species does not exhibit extensive endopolyploidy [12], [13].
The transcriptome of Caulerpa taxifolia is dominated by patterning along the apical-basal axis. Throughout this manuscript, we use the terms “accumulation” and “abundance” in a relative sense to describe transcript accumulation patterns. Transcript accumulation in replicates derived from basal regions (holdfast, stolon, frond base) is highly similar and distinct from apical regions (frond apex, rachis, pinnules), as shown in a Principal Component Analysis (PCA) performed on replicates (Fig. 1D; S5–S20 Datasets). The growing frond apex in particular exhibits a unique transcriptomic signature, perhaps indicative of the “meristemplasm” found in this region, as previously described [5], [14].
A Self-Organizing Map (SOM) was used to partition transcripts into six clusters (nodes), each with a distinct accumulation pattern (Figs. 2, S1; S21 Dataset). These nodes explain prominent densities of transcripts with similar accumulation patterns across organs, as visualized using a PCA (Fig. 2A–B). The nodes are roughly organized along the apical-basal axis (Fig. 2C). For example, Node 3 transcripts exhibit high frond apex accumulation, and progressing basally to Node 5 transcripts which accumulate at high levels in the holdfast, nodes with intermediate accumulation patterns along the apical-basal axis are observed. The overall patterns of transcript accumulation, visualized using the combination of SOMs and PCA, can be explored for a random subset of genes in an interactive graphic we have prepared (http://danchitwood.github.io/CaulerpaGeneExpression/, for use with a Google Chrome web browser). Such strong apical-basal, intracellular partitioning of transcript accumulation is not surprising considering the influence of gravitropism on regeneration and anchoring [6], circadian movements of chloroplasts into and out of the apex, and cytoplasmic streaming along the frond and pinnule lengths [5].
Fig. 2. Intracellular accumulation of transcripts in a giant, single-celled organism. 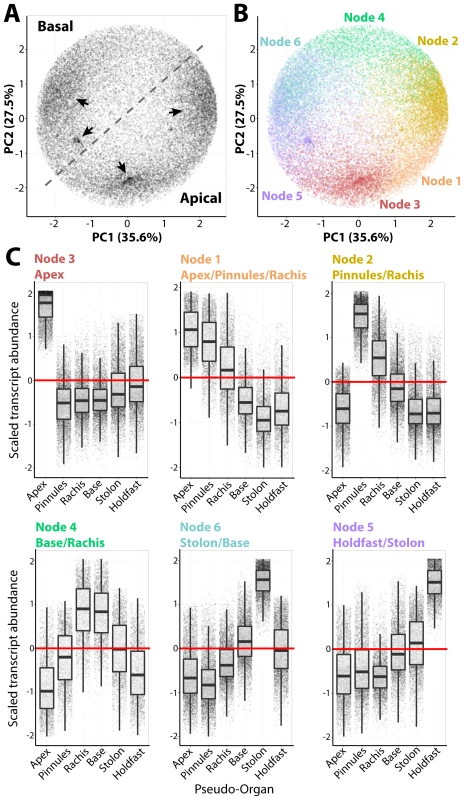
A) Principal Component Analysis (PCA) performed on transcript accumulation across sampled regions (the inverse of the PCA presented in Fig. 1D). Four major densities in the transcript accumulation variance structure are indicated by arrow. B) PCA was performed to visualize results of clustering by transcripts using Self-Organizing Maps (SOMs), visualized as different colors corresponding to nodes. C) Transcript accumulation profiles of genes belonging to different nodes, arranged with increasing abundance in an apical-to-basal direction. Scaled transcript abundance is such that the average abundance level across pseudo-organs for each transcript is 0 and variance is equal to 1. Scaled transcript abundance is shown as a boxplot and individual genes as jittered points (randomly displaced along the x-axis) to visualize transcript abundance distributions. Text for each node indicating those regions with scaled transcript abundance >0 is indicated. Transcripts belonging to each node are highly enriched for associated Gene Ontology (GO) terms, often specific to cellular functions and organelles (S22–S28 Datasets). For example, Node 2 transcripts, which accumulate at high levels in the pinnules and rachis, are enriched for photosynthetic GOs, but additionally those associated with mitochondria, respiration, the electron transport chain and ATP synthesis, as well as the production of secondary metabolites. Node 3 transcripts with high abundances in the frond apex are enriched for COPI/II vesicle coat proteins and kinases. Most surprising is the overwhelming concentration of transcripts associated with nuclear functions—DNA replication and damage, chromatin, RNAi, and even the subunits of DNA polymerase II—in the frond base, stolon, and holdfast.
Cell compartmentalization and morphology
Multicellular land plants possess an inherent constraint at the cellular level. Generally, every cell must have a nucleus, plastids, mitochondria, and cytoskeletal components to carry out basic metabolism, cell division, and differentiation, although numerous exceptions exist. But how is cellular compartmentalization distributed over similar morphology in Caulerpa? One hypothesis is that because morphogenesis is decoupled from multicellularity in Caulerpa, the distribution of different cell compartment identities might consolidate within distinct organs. That is, each cell is subcompartmentalized in a multicellular land plant, whereas the siphonous body plan of Caulerpa may maintain compartmentalization in pseudo-organs. Indeed, GO enrichment analysis reveals that transcript identity loosely follows the flow of genetic information progressing in a basal to apical direction in Caulerpa (Fig. 2, S22–S28 Datasets). Transcripts associated with transcriptional gene regulation accumulate at high levels in the holdfasts, stolon, and frond base, whereas those associated with translation are more abundant in the pinnules. Vesicular trafficking and kinase activity, associated with the cytoplasm and plasma membrane, are enriched within the frond apex.
To explore the fundamental relationship between cell compartmentalization and organism morphology, we selected all transcripts belonging to significantly enriched GO terms related to transcriptional gene regulation, translation, and other important organellar and cell biological functions (Fig. 3, S29 Dataset). Transcripts encoding RNA polymerase II subunits are highly abundant in the holdfast. Those encoding numerous chromatin, epigenetic, DNA recombination, repair, and replication, and RNAi machinery components are highly abundant in the stolon. Transcripts related to translation accumulate at high levels in the photosynthetic tissues, mostly in the pinnules and somewhat in the rachis. Proteolysis transcripts are found in these regions too, but additionally in the frond apex where translational components accumulate at lower levels. COPI/II coat proteins and numerous kinases are highly abundant in the frond apex. The association of COPI/II trafficking with the apex, an active growth region containing white “meristemplasm,” is consistent with the previously reported enrichment of rough endoplasmic reticulum and Golgi bodies in this region [4], [5], [14].
Fig. 3. The relationship between cell compartmentalization and morphology. 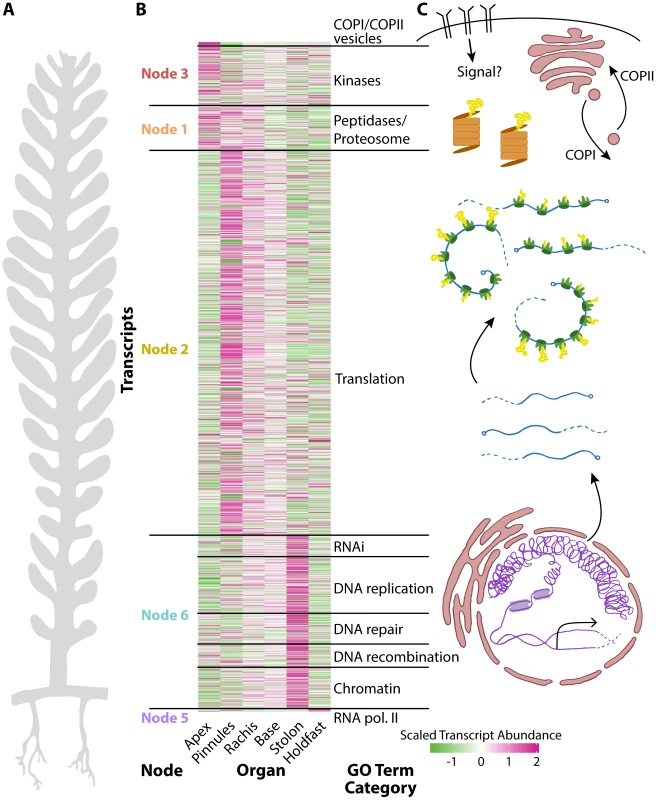
Panels within this figure correspond to each other, indicating a relationship between morphology, transcript accumulation, and cellular compartments. A) A diagram of Caulerpa morphology. Pseudo-organs roughly correspond to the apical-basal pattern of transcript accumulation shown in neighboring panel B) and the location of transcripts related to cellular compartments as shown in C). B) Heat map for genes belonging to select GO categories showing (left to right) node the parent GO term belongs to, transcript accumulation across pseudo-organs, and the general GO term category. Color indicates scaled transcript abundance, in which average transcript abundance is equal to 0 and variance equal to 1 for each transcript's abundance level across pseudo-organs. Green indicates low and magenta high scaled transcript abundance. C) Diagram of cellular compartments and the flow of genetic information from transcription to translation. The overall transcriptomic signature in Caulerpa—a single cell—is striking. From the holdfast to the frond apex, transcript accumulation loosely follows a nuclear-to-cytoplasm and transcriptional-to-translational pattern of identity (Fig. 3). The Caulerpa body plan is compartmentalized as if a single land plant cell, and different cellular compartments in Caulerpa are associated with different types of morphogenesis.
Recurrent signatures of transcript accumulation underlying plant morphology
Land plant morphology, and the numerous and diverse morphologies of various chlorophytes, are derived from the monophyletic inheritance of a core gene set. In some instances, as between land plants and Caulerpa, convergent structures with related functions (for example, leaves and fronds, and roots and holdfasts) have evolved using these genes. If land plant morphology is viewed from the perspective of organismal theory proposed by Kaplan and Hagemann [11], and land plants are even considered to be siphonous and cellularization patterns arbitrary, then the accumulation of transcripts throughout the organism can be compared to detect molecular homology.
To what degree have similar transcript accumulation profiles been recruited to morphological structures in land plants and Caulerpa? To answer this question, we analyzed the intersection of the Caulerpa transcriptomic atlas (Figs. 1–3) with a transcriptomic atlas from a land plant (tomato, Solanum lycopersicum cv. M82) that was derived using similar molecular and bioinformatics methods as presented here [15]–[17]. Putative homologous transcripts from tomato (see Materials and Methods) were used to assign Caulepra transcripts to a corresponding tomato self-organizing map node [17] (Fig. 4A). The distribution of genes from each Caulepra node across tomato nodes was then compared to the expected distribution using a χ2 test. A higher than expected enrichment of genes assigned to a particular tomato node indicates that a group of Caulerpa genes with similar accumulation patterns are associated with a specific accumulation profile in tomato (Fig. 4B).
Fig. 4. Recurrent recruitment of transcript accumulation to morphological structures in a land plant and Caulerpa. 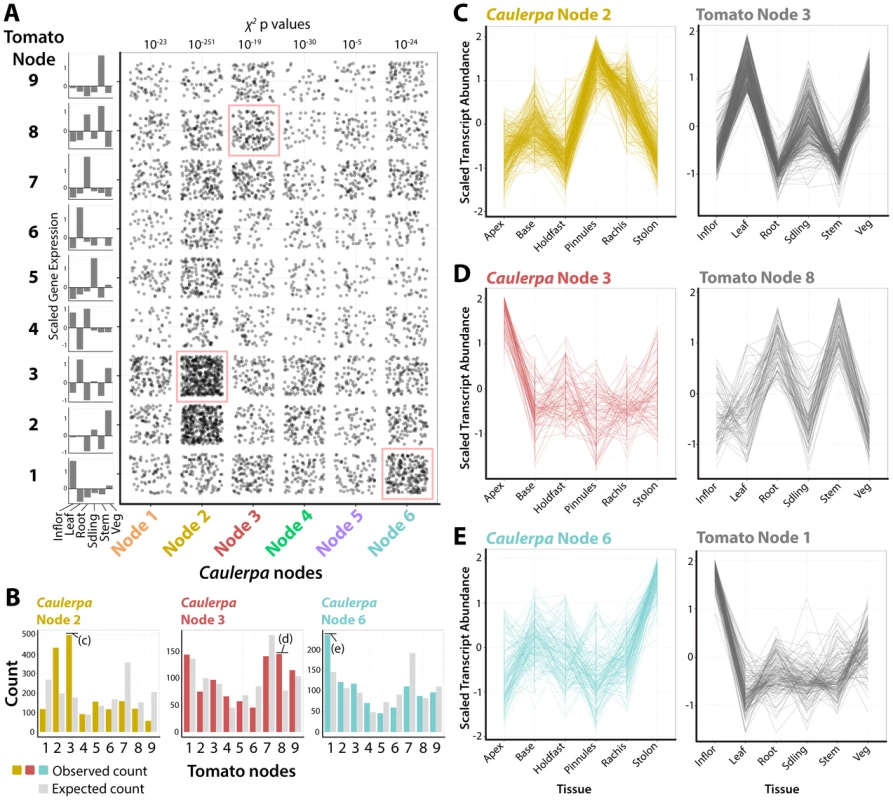
A) Intersection of transcript accumulation profiles by their Caulerpa node membership (x-axis) and corresponding accumulation pattern in tomato (Solanum lycopersicum). Each point corresponds to a Caulerpa transcript and its corresponding best BLAST-hit tomato homolog. χ2 p values indicate probability of Caulerpa transcript membership among tomato nodes differing from expected null distribution. Along the side of the graph is indicated averaged scaled transcript abundance across sampled regions in tomato. Intersections of transcript accumulation detailed in C–E are indicated with red boxes. B) Bar graphs showing expected (gray) and observed (yellow, Node 2; red, Node 4; blue, Node 6) distributions of Caulerpa BLAST hits in tomato against tomato nodes. Intersections of transcript accumulation detailed in C–E are indicated. C–E) Line graphs of select intersections of transcripts with similar accumulation profiles in Caulerpa, the homologs of which are enriched for specific transcript accumulation patterns in tomato. Details of gene identities are discussed in the text. Inflor. = inflorescence, Leaf = leaf, Root = root, Sdling = seedling, Stem = stem, Veg = vegetative apex. For example, Caulerpa Node 2 genes, which are highly abundant in the photosynthetically active pinnules and rachis (Fig. 2), are associated with tomato genes belonging to tomato Node 3, which are highly abundant in leaf, seedling, and vegetative apex samples (note: tomato and Caulerpa nodes are distinct and should not be confused with each other) (Fig. 4B, C). The intersection of Caulerpa Node 2 with tomato Node 3 is predominately photosynthetic genes (S30 Dataset). Although the molecular correspondence between photosynthetic structures is expected, other associations are less so. Caulepra Node 3 genes are highly abundant in the frond apex and are associated with tomato Node 8 genes that accumulate at high levels in the root and stem (Fig. 4D). Consistent with enriched GO terms in both Caulerpa and tomato, these genes are associated with vesicular trafficking (particularly COP cotamers) and vacuolar transporters (S30 Dataset). Interestingly, the Caulerpa Node 6 genes with high stolon abundance are associated with tomato Node 1 genes with high abundance in the inflorescence and relatively high abundance in the vegetative apex, both meristematic organs (Fig. 4E). The genes intersecting both nodes (S30 Dataset) are members of RNAi, chromatin, and DNA recombination, repair, and replication pathways, suggesting a molecular association between the stolon and meristems of land plants. The ability of the stolon to repetitively produce pseudo-organs (both fronds and holdfasts) and the enrichment of nuclear replication-associated transcripts indicates meristem-like identity at the molecular level.
The association between transcript accumulation profiles in Caulerpa and a land plant suggests, to a limited extent, molecular homology underlying morphology. Kaplan and Hagemann [11] argued that land plants, like Caulerpa, are siphonous. While the statement is extreme and not technically correct, reevaluating land plant development from this perspective is insightful, with respect to the role cells play in determining morphology. Morphogenesis and key patterning events in land plants rely on non-cell autonomous, symplastic movement of transcription factors and small RNAs, that transcend cell division patterns [18]–[21]. Spatially restricted transcripts in a siphonous organism, and their correspondence with land plant morphology, demonstrate that the plant form is achievable without cells and questions the centrality of cell division patterns in determining plant morphology.
Materials and Methods
RNA-seq library preparation, sequencing and preprocessing of reads
RNA-seq libraries were prepared from at least four replicates of the frond apex, rachis, pinnules, the frond base, stolon, and holdfast, sampling a prolifically growing Caulerpa taxifolia strain obtained from an aquarium in St. Louis, MO. The sampled strain was entrained to a circadian cycle using aquarium lighting roughly synchronized with the outside light-dark cycle. Sampling occurred mid-afternoon, at a time when chloroplasts were enriched in the frond apex (an important consideration, given the nightly retreat of chloroplasts into the frond base and stolon) [5], [22]. Large, intact fragments consisting of fronds, stolons, and holdfasts were removed from the marine aquarium and cleaned in synthetically prepared seawater for approximately 5 seconds to help reduce levels of outside contamination. Different samples corresponded to separately growing clones in the same aquarium. Samples were immediately immersed in liquid nitrogen after cleaning. The samples were then removed from the liquid nitrogen, dissected before they thawed, and placed into microcentrifuge tubes that were immersed again in liquid nitrogen. Samples were then stored at −80°C until library preparation. During thawing before library preparation, one holdfast microcentrifuge sample exploded and was removed from analysis, reducing the holdfast sample number to four. All five samples from other pseudo-organs were successfully processed.
Libraries were created using a custom high-throughput method for Illumina RNA-seq with a poly-A enrichment step [15], and sequenced in 100 bp paired-end format at the UC Berkeley Genomics Sequencing Laboratory on two lanes of HiSeq 2000 platform (Illumina Inc. San Diego, CA, USA). Library making was undertaken exactly as published in Kumar et al. [15] without modification of the protocol. We believe that the freezing step during sample preparation is important to bypass the Caulerpa wounding response for successful RNA isolation.
Reads were preprocessed using custom perl scripts that involved removal of low quality reads with average Phred quality score <20, trimming of low-quality bases with Phred score <20 from the 3′ ends of the reads, and removal of adapter/primer contamination. In addition identical reads, which originated during the PCR enrichment step of the library preparation, were collapsed into a single read using a custom perl script in order to reduce the computational resources required for transcriptome assembly. However, PCR-duplicated reads were retained for downstream quantification of transcript abundances. The pre-processed reads were sorted into individual samples based on barcodes using fastx_barcode_splitter and barcodes were trimmed using fastx_trimmer from Fastx_toolkit (http://hannonlab.cshl.edu/fastx_toolkit/). A total of 420 million reads (210 million paired-end 100×100), obtained after preprocessing, were used for transcriptome assembly.
De novo transcriptome assembly and refinement
De novo transcriptome assembly was carried in a similar fashion as Ranjan et al. [23], but is described here again in detail. The Trinity software package (version r2013-02-16) was used to assemble, de novo, a Caulerpa taxifolia transcriptome using preprocessed RNA-seq reads [24]. The assembly was performed, using 24 large-memory computing nodes, at The Lonestar Linux Cluster at Texas Advance Computing Center (TACC, University of Texas). “Trinity.pl —seqType fq —JM 1000G —left reads-1.fq —right reads-2.fq —min_contig_length 200 —CPU 24 —bflyHeapSpaceMax 7G” was the command line used for assembly. Subsequently, assembly statistics and downstream analysis were performed in the iPlant atmosphere and Discovery computing atmosphere [25].
A total of 77,285 contigs with N50 (N50 is defined as the largest contig length such that using equal or longer contigs produces half the bases of the transcriptome) of 1243 bp, mean length of 807 bp and median of 433 bp, were assembled. In order to remove redundant and/or highly similar contigs, the contigs were then clustered using the CD-HIT-EST program from the CD-HIT suite at a sequence similarity threshold value (-c) of 0.95 and word-length (n) of 8, leaving other parameters at default [26]. This resulted in the final Caulerpa transcriptome assembly with 57,118 contigs and N50 of 813 bp, mean length of 632 bp and median of 381 bp (see sequence submission information). The prediction of likely coding sequences from 57,118 clustered contigs, using TransDecoder (http://transdecoder.sourceforge.net/), resulted in 35,827 putative open reading frames (ORFs)/coding sequences (CDS) (see sequence submission information).
Functional annotation of transcriptome
The contigs from the final Caulerpa transcriptome assembly were compared to the NCBI nr (nonredundant) database (ftp://ftp.ncbi.nlm.nih.gov/blast/db/FASTA/nr.gz), Arabidopsis protein database (ftp://ftp.arabidopsis.org/home/tair/Sequences/blast_datasets/TAIR10_blastsets/TAIR10_pep_20110103_representative_gene_model_updated) and tomato (Solanum lycopersicum) ITAG2.3 protein database (ftp://ftp.solgenomics.net/tomato_genome/annotation/ITAG2.3_release/ITAG2.3_proteins.fasta) using BLASTX with an e-value threshold of 1e-5 (S1 Dataset) [27]. BLAST searches against the nr database resulted in annotation of 24,589 contigs (43% of clustered contigs) of which 20,146 contigs had reasonably stringent e-value of less than 1−e10. 17427 (13698 with e-value <1e−10) and 17392 (13274 with e-value <1e−10) contigs were annotated against Arabidopsis and tomato protein databases. When BLASTX comparison was performed against the nr database, more than 49% of annotated C. taxifolia contigs found top BLAST hits against the sequences from members of Cholorophyta, such as Volvox carteri, Chlamydomonas reinhardtii, Chlorella variabilis, and Coccomyxa subellipsoidea. Almost half of the top BLAST hits to cholorophytes confirm high sequence similarity between C. taxifolia and chlorophytes. The Caulerpa contigs were, further, compared explicitly against protein sequences of the chlorophytes C. reinhardtii and V. carteri using BLASTx, and vice-versa using tBLASTn with an e-value threshold of 1e−5 [27]. Chlamydomonas reinhardtii and V. carteri protein sequences were downloaded from Phytozome v10 (http://phytozome.jgi.doe.gov/pz/portal.html). BLAST searches against V. carteri and C. reinhardtii sequences found a hit for 19048 (33%) and 20377(36%) of assembled Caulerpa contigs, respectively (S2 Dataset). Reciprocal BLAST searches of V. carteri and C. reinhardtii sequences against C. taxifolia contigs found homologs for 42% of sequences of each species.
The BLASTX output generated against the NCBI nr database, with maximum twenty hits for each contig, was used for Blast2GO analysis to annotate the contigs with GO terms describing biological processes, molecular functions, and cellular components [28]. Blast2GO performs GO annotation by applying an annotation rule (AR) on the found ontology terms from the BLAST-hits, which is based on annotation score. The default e-Value hit filter (1e−6) and annotation cut-off (55) was used to calculate the annotation score. Our gene ontology annotations are, by necessity, based in part on the inclusion of hits using a relatively low e-value threshold. It will be critical in the future to validate these assignments using functional analysis. After the Blast2GO mapping process, proper GO terms were generated followed by use of ANNEX and GO Slim, which are integrated in the Blast2GO software, to enrich the annotation (S3, S4 Datasets). Sequence descriptions were also generated from Blast2GO, which are arbitrary nomenclature based upon degrees of similarities identified in the nr database according to e-value and identity to blasted genes (S1, S2 Datasets). BLASTX against the nr database resulted in annotation of 24,589 contigs (43% of clustered contigs) of which only 14,206 (25% of clustered contigs) were assigned GO-terms. Given the problems associated with the de novo transcriptome assembly algorithms and lack of functional tools in Caulerpa, BLASTX annotation of 43% of clustered contigs and GO annotation of only 25% of clustered contigs is not surprising. Similar functional annotation for only a fraction of assembled contigs has been noted for other de novo assembled plant transcriptomes [23], [29], [30]. These non-annotated contigs likely correspond to 3′ or 5′ untranslated regions, non-coding RNAs, or short sequences not containing a known protein domain, some of which might represent potential Caulerpa-specific genes.
Mapping reads to contigs and normalized count data
RSEM (RNAseq by expectation maximization), which allows for an assessment of transcript abundances based on the mapping of RNA-seq reads to the assembled transcriptome, was used for transcript abundance estimation of the de novo assembled transcripts [31]. Due to read mapping ambiguity among de novo assembled transcripts, it is common to have the same read mapped to multiple contigs. RSEM models the reads mapped at multiple contigs taking into account length of target contigs, number of mismatches, sequencing errors, etc., and generates an estimated read count for each contig. Single end reads, retaining the PCR-duplicated reads, from individual libraries of each Caulerpa sample were mapped to clustered contigs using the perl script run_RSEM_align_n_estimate.pl that employs RSEM, followed by joining RSEM-estimated abundance values for each sample using merge_RSEM_frag_counts_single_table.pl, generating raw estimated counts for each contig from each Caulerpa sample (S5 Dataset). Subsequently, differential expression analysis for each organ pair was carried out using run_DE_analysis.pl, which involves the Bioconductor package EdgeR in the R statistical environment [32]. Contigs that had RSEM-estimated counts ≥30 for all samples combined were used for transcript abundance estimates. Normalization factors were calculated using the trimmed mean of M-values method to obtain normalized read count per million for each contig of a sample. This normalized reads per million was then used for the pair-wise differential expression analysis for each organ pair using EdgeR. The lists of significant differentially expressed contigs (FDR<0.05) for each organ-pair comparison are presented in S6–S20 Datasets. All the Perl scripts used for read mapping, generating read counts and differential expression analysis are documented with Trinity software suite [33].
Principal Component Analysis (PCA), Self Organizing Maps (SOM) clustering, and other statistical analyses
Those transcripts differentially expressed between at least one organ pair (S6–S20 Datasets) were subsequently used to find clusters of genes with similar transcript accumulation patterns using Self Organizing Maps (SOMs) [34]. Differentially expressed transcripts were averaged across replicates for each pseudo-organ sample. Averaged transcript abundance values were then scaled across pseudo-organs to arrive at scaled transcript accumulation patterns which were used to assign cluster membership. Scaling was performed using the scale() function in the base package using default settings, such that the average transcript abundance value across pseudo-organs was 0 and the variance equal to 1. To cluster transcripts across organs, a 3×2 hexagonal SOM was implemented, using the Kohonen package in R [35], [36]. 100 training iterations were used during clustering, over which the α-learning rate decreased from 0.05 to 0.01. Mean distance of transcript accumulation patterns to their closest unit stabilized after approximately 15 iterations of training.
A decision to use six clusters was arrived at by first analyzing the results of a Principal Component Analysis (PCA) on scaled transcript accumulation across tissues, using the prcomp() function in R with default settings. Average and scaled transcript accumulation levels across organs, as well as SOM cluster membership and PC values are provided (S21 Dataset). Four main densities in the variance attributable to accumulation patterns were discernable (arrows, Fig. 2A), and the results of a 4 cluster SOM largely overlap with the densities. Variance in accumulation among transcripts across organs was large, however, and the decision to specify 6 SOM clusters not only produced clusters with unique accumulation patterns and lower variance in abundance (Fig. 2C), but also yielded clusters with more interpretable GO enrichment categories (that is, significant GO enrichment consistent with known biology, such as photosynthetic GOs enriched in nodes with high pinnule transcript abundance). To verify that 6 GOs was indeed the maximum cluster number specifying unique transcript accumulation profiles without redundancy, we undertook partitioning of the PCA space into a variable number of SOM clusters over 100 iterations for each node number with random seeds. Linear Discriminant Analysis (LDA) was performed on genes maximizing separation of cluster identity using PCs 1–5 (PC6 explained negligible amounts of variance and could not be incorporated into the LDA) using the lda function from the MASS package [37]. The predict function (stats package) and table function (base package) were used to reallocate genes to predicted clusters (within MASS) using the linear discriminants. A high fraction of a node's originally assigned transcripts by SOM being reassigned correctly indicates little redundancy in node transcript accumulation patterns. Starting with 6 nodes, reassignment using LDA begins to drop before reaching a plateau of low reassignment rates, indicating that choosing 6 nodes maximizes the number of unique accumulation profiles represented by clusters without redundancy (see S1 Fig. for results).
Analysis of intersection between tomato and Caulerpa transcriptomic atlases was undertaken using data from Chitwood et al. [17]. Best BLASTX hits of Caulerpa transcripts to tomato (Solanum lycopersicum) (see “Functional annotation of transcriptome” above and S1, S2 Datasets) were used to assign tomato transcript accumulation patterns, across a number of organs, to Caulerpa transcript accumulation patterns. The distribution of tomato transcripts assigned to tomato SOM nodes was taken as the null distribution and compared to the number of Caulerpa transcripts assigned to each tomato node. p values, indicating the degree of significant difference between the two distributions, were obtained from χ2 values using the chisq.test function (stats package).
Clusters of transcripts were analyzed for GO enrichment terms at a 0.05 FDR cut-off using the “goseq” package in Bioconductor (S22–S28 Datasets) [38]. Unless otherwise specified, all statistical analyses on transcript accumulation were performed using R [36] and data visualization using the ggplot2 package [39].
Sequence submission
The quality filtered, barcode-sorted and trimmed short read dataset, which was used for transcriptome assembly, was deposited to the NCBI Short Read Archive under accessions SRR1228213–SRR1228223, SRR1228225–SRR1228232, SRR1228234–SRR1228238 and. SRR1228240–SRR1228244. All assembled contigs have been deposited at DDBJ/EMBL/GenBank under the accession GBCY00000000. The version described in this paper is the first version, GBCY01000000.
Sequences of all contigs of Caulerpa_final_transcriptome, obtained after clustering of transcripts, can be downloaded as a FASTA file at http://de.iplantcollaborative.org/dl/d/80CF0D47-5A80-4CE7-B6DF-F4A7ED803493/Caulerpa_final_transcriptome.fasta. The contigs were named as Ctaxi_contig plus a serial number with the Trinity identifiers.
Sequences of all predicted ORFs from the Caulerpa transcriptome assembly can be downloaded as a FASTA file at http://de.iplantcollaborative.org/dl/d/40273882-35DD-4930-9DBD-6D60CEAA7890/Caulerpa_predicted_ORFs.fasta. The ORFs were named as Ctaxi_predicted_CDS plus a serial number.
Supporting Information
Zdroje
1. HämmerlingJ (1953) Nucleo-cytoplasmic relationship in the development of Acetabularia. J Intern Rev Cytol 2 : 475–498.
2. MandoliDF (1992) Vegetative growth of Acetabularia acetabulum (Chlorophyta): Structural evidence for juvenile and adult phases in development. Journal of Phycology 5 : 669–677.
3. MandoliDF (1998) Elaboration of body plan and phase change during development of Acetabularia: How is the complex architecture of a giant unicell built? Annual review of plant biology 49 : 173–198.
4. DawesCJ, RhamstineE (1967) An ultra-structural study of the giant algal coenocyte, Caulerpa prolifera. J Phycol 3 : 177–126.
5. DawesCJ, BarilottiDC (1969) Cytoplasmic organization and rhythmic streaming in growing blades of Caulerpa prolifera. Amer J Bot 56 : 8–15.
6. JacobsWP, OlsonJ (1980) Developmental changes in the algal coenocyte Caulerpa prolifera (Siphonales) after inversion with respect to gravity. Amer J Bot 67 : 141–146.
7. MenzelD, GrantBR (1981) Fine structure study on the development of trabeculae in the siphonous green alga Caulerpa simpliciuscula C. Ag. Protoplasma 107 : 47–61.
8. MatilskyMB, JacobsWP (1983) Regeneration in the coenocytic marine alga, Caulerpa, with respect to gravity. American Journal of Botany 70 : 635–638.
9. ChisholmJR, DaugaC, AgeronE, GrimontPA, JaubertJM (1996) ‘Roots’ in mixotrophic algae. Nature 381 : 382.
10. Schwann T, Schleiden MJ (1847) Microscopical researches into the accordance in the structure and growth of animals and plants. London: Printed for the Sydenham Society.
11. Kaplan DR, Hagemann W (1991) The relationship of cell and organism in vascular plants. Bioscience 693–703
12. KapraunDR (2005) Nuclear DNA content estimates in multicellular green, red, and brown algae: phylogenetic considerations. Annals of Botany 95 : 7–44.
13. Varela-ÁlvarezE, Gómez GarretaA, Rull LluchJ, Salvador SolerN, SerraoEA, et al. (2012) Mediterranean species of Caulerpa are polyploidy with smaller genomes in the invasive ones. PLOS ONE 7: e47728.
14. JanseJM (1910) Über Organveränderung bei Caulerpa prolifera. Jahrb. Wiss Bot 48 : 73–110.
15. KumarR, IchihashiY, KimuraS, ChitwoodDH, HeadlandLR, et al. (2012) A High-Throughput Method for Illumina RNA-Seq Library Preparation. Front Plant Sci 3 : 202.
16. KoenigD, Jiménez-GómezJM, KimuraS, FulopD, ChitwoodDH, et al. (2013) Comparative transcriptomics reveals patterns of selection in domesticated and wild tomato. Proc Natl Acad Sci USA 110: E2655–62.
17. ChitwoodDH, MaloofJN, SinhaNR (2013) Dynamic transcriptomic profiles between tomato and a wild relative reflect distinct revelopmental architectures. Plant Physiol 162 : 537–52.
18. SessionsA, YanofskyMF, WeigelD (2000) Cell-cell signaling and movement by the floral transcription factors LEAFY and APETALA1. Science 289 : 779–82.
19. NakajimaK, SenaG, NawyT, BenfeyPN (2001) Intercellular movement of the putative transcription factor SHR in root patterning. Nature 413 : 307–11.
20. ChitwoodDH, NogueiraFT, HowellMD, MontgomeryTA, CarringtonJC, et al. (2009) Pattern formation via small RNA mobility. Genes Dev 23 : 549–54.
21. CarlsbeckerA, LeeJY, RobertsCJ, DettmerJ, LehesrantaS, et al. (2010) Cell signaling by microRNA 165/6 directs gene dose-dependent root cell fate. Nature 465 : 316–21.
22. MenzelD, Elsner-MenzelC (1989) Actin-based chloroplast rearrangements in the cortex of the giant coenocytic green alga Caulerpa. Protoplasma 150 : 1–8.
23. RanjanA, IchihashiY, FarhiM, ZumsteinK, TownsleyB, et al. (2014) De novo assembly and characterization of the transcriptome of the parasitic weed Cuscuta pentagona identifies genes associated with plant parasitism. Plant Physiol 166 : 1186–99.
24. GrabherrMG, HaasBJ, YassourM, LevinJZ, ThompsonDA, et al. (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29 : 644–652.
25. GoffSA, VaughnM, McKayS, LyonsE, StapletonAE, et al. (2011) The iPlant Collaborative: Cyberinfrastructure for Plant Biology. Front Plant Sci 2 : 34.
26. HuangY, NiuB, GaoY, FuL, LiW (2010) CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics 26 : 680–682.
27. AltschulSF, MaddenTL, SchafferAA, ZhangJ, ZhangZ, et al. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25 : 3389–3402.
28. GotzS, Garcia-GomezJM, TerolJ, WilliamsTD, NagarajSH, et al. (2008) High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res 36 : 3420–3435.
29. LiuS, LiW, WuY, ChenC, LeiJ (2013) De novo transcriptome assembly in chili pepper (Capsicum frutescens) to identify genes involved in the biosynthesis of capsaicinoids. PLOS ONE 8: e48156.
30. NakasugiK, CrowhurstRN, BallyJ, WoodCC, HellensRP, et al. (2013) De novo transcriptome sequence assembly and analysis of RNA silencing genes of Nicotiana benthamiana. PLOS ONE 8: e59534.
31. LiB, DeweyCN (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12 : 323.
32. RobinsonMD, OshlackA (2010) A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 11: R25.
33. HaasBJ, PapanicolaouA, YassourM, GrabherrM, BloodPD, et al. (2013) De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc 8 : 1494–1512.
34. Kohonen T (1997) Self-Organizing Maps. Springer, New York.
35. WehrensR, BuydensLMC (2007) Self - and super-organizing maps in R: the Kohonen package. J Stat Softw 21 : 1–19.
36. R Development Core Team (2014) R: A language and environment for statistical computing. R Foundation for Statistical Computing. Vienna, Austria.
37. Venables WN, Ripley BD (2002) Modern Applied Statistics with S. Fourth Edition. New York: Springer.
38. YoungMD, WakefieldMJ, SmythGK, OshlackA (2010) Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 11: R14.
39. Wickham H (2009) ggplot2: elegant graphics for data analysis. Springer, New York.
40. CocquytE, VerbruggenH, LeliaertF, De ClerckO (2010) Evolution and cytological diversification of the green seweeds (Ulvophyceae). Mol Biol Evol 27 : 2052–61.
Štítky
Genetika Reprodukční medicína
Článek Phosphorylation of Elp1 by Hrr25 Is Required for Elongator-Dependent tRNA Modification in YeastČlánek Naturally Occurring Differences in CENH3 Affect Chromosome Segregation in Zygotic Mitosis of HybridsČlánek Insight in Genome-Wide Association of Metabolite Quantitative Traits by Exome Sequence AnalysesČlánek ALIX and ESCRT-III Coordinately Control Cytokinetic Abscission during Germline Stem Cell DivisionČlánek Deciphering the Genetic Programme Triggering Timely and Spatially-Regulated Chitin Deposition
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 1
-
Všechny články tohoto čísla
- The Combination of Random Mutagenesis and Sequencing Highlight the Role of Unexpected Genes in an Intractable Organism
- Ataxin-3, DNA Damage Repair, and SCA3 Cerebellar Degeneration: On the Path to Parsimony?
- α-Actinin-3: Why Gene Loss Is an Evolutionary Gain
- Origins of Context-Dependent Gene Repression by Capicua
- Transposable Elements Contribute to Activation of Maize Genes in Response to Abiotic Stress
- No Evidence for Association of Autism with Rare Heterozygous Point Mutations in Contactin-Associated Protein-Like 2 (), or in Other Contactin-Associated Proteins or Contactins
- Nur1 Dephosphorylation Confers Positive Feedback to Mitotic Exit Phosphatase Activation in Budding Yeast
- A Regulatory Hierarchy Controls the Dynamic Transcriptional Response to Extreme Oxidative Stress in Archaea
- Genetic Variants Modulating CRIPTO Serum Levels Identified by Genome-Wide Association Study in Cilento Isolates
- Small RNA Sequences Support a Host Genome Origin of Satellite RNA
- Phosphorylation of Elp1 by Hrr25 Is Required for Elongator-Dependent tRNA Modification in Yeast
- Genetic Mapping of MAPK-Mediated Complex Traits Across
- An AP Endonuclease Functions in Active DNA Demethylation and Gene Imprinting in
- Developmental Regulation of the Origin Recognition Complex
- End of the Beginning: Elongation and Termination Features of Alternative Modes of Chromosomal Replication Initiation in Bacteria
- Naturally Occurring Differences in CENH3 Affect Chromosome Segregation in Zygotic Mitosis of Hybrids
- Imputation of the Rare G84E Mutation and Cancer Risk in a Large Population-Based Cohort
- Polycomb Protein SCML2 Associates with USP7 and Counteracts Histone H2A Ubiquitination in the XY Chromatin during Male Meiosis
- A Genetic Strategy for Probing the Functional Diversity of Magnetosome Formation
- Interactions of Chromatin Context, Binding Site Sequence Content, and Sequence Evolution in Stress-Induced p53 Occupancy and Transactivation
- The Yeast La Related Protein Slf1p Is a Key Activator of Translation during the Oxidative Stress Response
- Integrative Analysis of DNA Methylation and Gene Expression Data Identifies as a Key Regulator of COPD
- Proteasomes, Sir2, and Hxk2 Form an Interconnected Aging Network That Impinges on the AMPK/Snf1-Regulated Transcriptional Repressor Mig1
- Functional Interplay between the 53BP1-Ortholog Rad9 and the Mre11 Complex Regulates Resection, End-Tethering and Repair of a Double-Strand Break
- Estrogenic Exposure Alters the Spermatogonial Stem Cells in the Developing Testis, Permanently Reducing Crossover Levels in the Adult
- Protein Poly(ADP-ribosyl)ation Regulates Immune Gene Expression and Defense Responses
- Sumoylation Influences DNA Break Repair Partly by Increasing the Solubility of a Conserved End Resection Protein
- A Discrete Transition Zone Organizes the Topological and Regulatory Autonomy of the Adjacent and Genes
- Elevated Mutation Rate during Meiosis in
- The Intersection of the Extrinsic Hedgehog and WNT/Wingless Signals with the Intrinsic Hox Code Underpins Branching Pattern and Tube Shape Diversity in the Airways
- MiR-24 Is Required for Hematopoietic Differentiation of Mouse Embryonic Stem Cells
- Tissue-Specific Effects of Genetic and Epigenetic Variation on Gene Regulation and Splicing
- Heterologous Aggregates Promote Prion Appearance via More than One Mechanism
- The Tumor Suppressor BCL7B Functions in the Wnt Signaling Pathway
- , A -Acting Locus that Controls Chromosome-Wide Replication Timing and Stability of Human Chromosome 15
- Regulating Maf1 Expression and Its Expanding Biological Functions
- A Polyubiquitin Chain Reaction: Parkin Recruitment to Damaged Mitochondria
- RecFOR Is Not Required for Pneumococcal Transformation but Together with XerS for Resolution of Chromosome Dimers Frequently Formed in the Process
- An Intracellular Transcriptomic Atlas of the Giant Coenocyte
- Insight in Genome-Wide Association of Metabolite Quantitative Traits by Exome Sequence Analyses
- The Role of the Mammalian DNA End-processing Enzyme Polynucleotide Kinase 3’-Phosphatase in Spinocerebellar Ataxia Type 3 Pathogenesis
- The Global Regulatory Architecture of Transcription during the Cell Cycle
- Identification and Functional Characterization of Coding Variants Influencing Glycemic Traits Define an Effector Transcript at the Locus
- Altered Ca Kinetics Associated with α-Actinin-3 Deficiency May Explain Positive Selection for Null Allele in Human Evolution
- Genetic Variation in the Nuclear and Organellar Genomes Modulates Stochastic Variation in the Metabolome, Growth, and Defense
- PRDM9 Drives Evolutionary Erosion of Hotspots in through Haplotype-Specific Initiation of Meiotic Recombination
- Transcriptional Control of an Essential Ribozyme in Reveals an Ancient Evolutionary Divide in Animals
- ALIX and ESCRT-III Coordinately Control Cytokinetic Abscission during Germline Stem Cell Division
- Century-scale Methylome Stability in a Recently Diverged Lineage
- A Re-examination of the Selection of the Sensory Organ Precursor of the Bristle Sensilla of
- Antagonistic Cross-Regulation between Sox9 and Sox10 Controls an Anti-tumorigenic Program in Melanoma
- A Dependent Pool of Phosphatidylinositol 4,5 Bisphosphate (PIP) Is Required for G-Protein Coupled Signal Transduction in Photoreceptors
- Deciphering the Genetic Programme Triggering Timely and Spatially-Regulated Chitin Deposition
- Aberrant Gene Expression in Humans
- Fascin1-Dependent Filopodia are Required for Directional Migration of a Subset of Neural Crest Cells
- The SWI2/SNF2 Chromatin Remodeler BRAHMA Regulates Polycomb Function during Vegetative Development and Directly Activates the Flowering Repressor Gene
- Evolutionary Constraint and Disease Associations of Post-Translational Modification Sites in Human Genomes
- A Truncated NLR Protein, TIR-NBS2, Is Required for Activated Defense Responses in the Mutant
- The Genetic and Mechanistic Basis for Variation in Gene Regulation
- Inactivation of PNKP by Mutant ATXN3 Triggers Apoptosis by Activating the DNA Damage-Response Pathway in SCA3
- DNA Damage Response Factors from Diverse Pathways, Including DNA Crosslink Repair, Mediate Alternative End Joining
- hnRNP K Coordinates Transcriptional Silencing by SETDB1 in Embryonic Stem Cells
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The Global Regulatory Architecture of Transcription during the Cell Cycle
- A Truncated NLR Protein, TIR-NBS2, Is Required for Activated Defense Responses in the Mutant
- Proteasomes, Sir2, and Hxk2 Form an Interconnected Aging Network That Impinges on the AMPK/Snf1-Regulated Transcriptional Repressor Mig1
- The SWI2/SNF2 Chromatin Remodeler BRAHMA Regulates Polycomb Function during Vegetative Development and Directly Activates the Flowering Repressor Gene
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



