-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaPhosphorylation of Elp1 by Hrr25 Is Required for Elongator-Dependent tRNA Modification in Yeast
tRNA molecules function as adapters in protein synthesis, bringing amino acids to the ribosome and reading the genetic code through codon-anticodon base pairing. When the tRNA contains a uridine residue in the “wobble position” of its anticodon, which base-pairs with purine residues in the third position of a cognate codon, it is almost always chemically modified and modification is required for efficient decoding. In eukaryotic cells, these wobble uridine modifications require a conserved protein complex called Elongator. Our work shows that Elp1, Elongator's largest subunit, is phosphorylated on several sites. By blocking phosphorylation at these positions using mutations, we identified four phosphorylation sites that are important for Elongator's role in tRNA modification. We have also shown that Hrr25 protein kinase, a member of the casein kinase I (CKI) family, is responsible for modification of two of the sites that are important for Elongator function. Phosphorylation appears to affect interaction of the Elongator complex both with its kinase (Hrr25) and with Kti12, an accessory protein previously implicated in Elongator function. Our studies imply that Elp1 phosphorylation plays a positive role in Elongator-mediated tRNA modification and raise the possibility that wobble uridine modification may be regulated, representing a potential translational control mechanism.
Published in the journal: . PLoS Genet 11(1): e32767. doi:10.1371/journal.pgen.1004931
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004931Summary
tRNA molecules function as adapters in protein synthesis, bringing amino acids to the ribosome and reading the genetic code through codon-anticodon base pairing. When the tRNA contains a uridine residue in the “wobble position” of its anticodon, which base-pairs with purine residues in the third position of a cognate codon, it is almost always chemically modified and modification is required for efficient decoding. In eukaryotic cells, these wobble uridine modifications require a conserved protein complex called Elongator. Our work shows that Elp1, Elongator's largest subunit, is phosphorylated on several sites. By blocking phosphorylation at these positions using mutations, we identified four phosphorylation sites that are important for Elongator's role in tRNA modification. We have also shown that Hrr25 protein kinase, a member of the casein kinase I (CKI) family, is responsible for modification of two of the sites that are important for Elongator function. Phosphorylation appears to affect interaction of the Elongator complex both with its kinase (Hrr25) and with Kti12, an accessory protein previously implicated in Elongator function. Our studies imply that Elp1 phosphorylation plays a positive role in Elongator-mediated tRNA modification and raise the possibility that wobble uridine modification may be regulated, representing a potential translational control mechanism.
Introduction
Elongator is a conserved, multi-subunit protein complex containing six different polypeptides (Elp1-Elp6), first discovered in yeast in association with the elongating form of RNA polymerase II and initially proposed to play a role in transcriptional elongation [1], [2]. Although Elongator is non-essential in yeast, knockout of the mouse IKBKAP gene encoding Elongator's largest subunit leads to embryonic lethality and the protein is crucial for vascular and neural development [3]. The hereditary neuropathy Familial Dysautonomia results from human IKBKAP mutations, while mutations in other Elongator subunits have been associated with Amyotrophic Lateral Sclerosis [4] and Rolandic Epilepsy [5]. Elongator in Caenorhabditis elegans is also involved in neuronal function and development [6], [7], while in plants it plays a role in proliferation during organ growth [8].
While it is therefore clear that Elongator is important for neural function in higher organisms [9], [10], it has been proposed to have a bewildering range of seemingly unrelated functions. The Elp3 subunit of Elongator has a histone acetyltransferase (HAT) domain that can acetylate histones in vitro [11] and yeast Elongator mutants show changes in histone acetylation in vivo [12]. Elongator has also been proposed to acetylate α-tubulin and the neuronal protein Bruchpilot [7], [13], [14] and has been implicated in paternal DNA demethylation in mouse zygotes [15]. In yeast, Elongator mutants adapt slowly to changing growth conditions, show sensitivity to high temperature, rapamycin, caffeine, hydroxyurea and various other chemical stressors, and are resistant to zymocin [16], [17], a protein toxin secreted by the yeast Kluyveromyces lactis that kills other yeasts including Saccharomyces cerevisiae [18]. Yeast Elongator has also been implicated in transcriptional silencing, replication-coupled nucleosome assembly [17] and polarized secretion [19].
However, while these apparently diverse roles could imply that Elongator is multifunctional, work in yeast [20], C. elegans [6], plants [21] and mammals [22] has demonstrated that many if not all of these proposed functions reflect a primary role for Elongator in tRNA modification, specifically in the addition of mcm5 (5-methoxycarbonylmethyl) and ncm5 (5-carbamoylmethyl) groups to uridine when present at the ‘wobble’ position (U34) in tRNA anticodons. Eleven out of 13 such tRNAs in yeast contain either mcm5U, ncm5U or 5-methoxycarbonymethyl-2-thiouridine (mcm5s2U) in the wobble position and addition of the mcm5 and ncm5 moieties requires Elongator [20]. This role in tRNA modification explains the zymocin-resistant phenotype of Elongator mutants: zymocin is a tRNA anticodon nuclease that inactivates tRNAGlu(UUC) by cleaving the anticodon on the 3′ side of U34, but the mcm5 modification is necessary for tRNA recognition and cleavage [23], [24].
Wobble uridine-containing tRNAs read codons ending with a purine, and the mcm5/ncm5 modifications are needed to confer full decoding competence on these tRNAs [6], [20], [25]-[27]. Wobble uridine modification is most likely the primary role of Elongator, at least in yeast, because elevated expression of just two Elongator-dependent tRNAs, tRNALys(UUU) and tRNAGln(UUG), can suppress all the phenotypes associated with loss of Elongator function apart from zymocin resistance, which remains unaffected because elevated levels of the two tRNAs do not restore the tRNAGlu(UUC) modification required for cleavage by zymocin [28], [29]. Suppression of Elongator mutant phenotypes by elevated tRNA levels without restoration of wobble uridine modification strongly suggests that these phenotypes are caused by translational defects resulting from hypomodified tRNAs, a notion supported by findings that U34 modification promotes binding of tRNALys(UUU), tRNAGln(UUG) and tRNAGlu(UUC) to the ribosomal A-site [30]. Recent structural work indicates that Elp4, Elp5 and Elp6 are RecA-fold proteins that form a heterohexamer containing two copies of each polypeptide, which interacts with two copies of an Elp1-Elp2-Elp3 sub-complex [31]. The recombinant heterohexamer binds and hydrolyses NTPs and shows tRNA binding that is reduced when the NTP can be hydrolyzed [31], while a separate tRNA-binding motif in the C-terminal domain of Elp1 may also mediate tRNA interaction with the Elp1-Elp2-Elp3 sub-complex [32]. Thus while the existence of additional substrates cannot be excluded, it is now clear that Elongator plays a conserved role in wobble uridine modification [6], [21], [22] and that this role, through effects at the level of translation, is likely to underpin the majority of phenotypes resulting from Elongator deficiency, at least in yeast. Within Elongator, Elp3 is highly likely to catalyze the tRNA modification as it contains a ‘radical SAM’ domain [33] that in other proteins can mediate RNA modification reactions [34], and both its radical SAM and histone acetyltransferase (HAT) domains are required for wobble uridine modification [20], [29]. This is supported by the recent finding that recombinant archaeal Elp3 can catalyze modification of tRNA wobble uridines in an in vitro reaction containing SAM, acetyl-CoA, tRNA and Na2S2O4 [35].
Previously, we reported that the largest subunit of Elongator (Elp1) is a phosphoprotein and identified mutations in either HRR25 (encoding a casein kinase I) or SIT4 (encoding a protein phosphatase) that conferred zymocin resistance [36]-[38]. Elp1 was present as a hypophosphorylated isoform in hrr25 mutants and as a hyperphosphorylated isoform in sit4 mutants, whereas wild-type cells contained similar amounts of both isoforms [37], [39]. We therefore sought to investigate the potential functional significance of Elp1 phosphorylation by locating the phosphorylation sites on Elp1, identifying several such sites that are critical for Elongator-dependent tRNA modification. Our findings therefore raise the possibility that Elongator activity (and hence tRNA modification) could be regulated, potentially constituting a novel mechanism for translational control.
Results
Identification of in vivo phosphorylation sites in Elp1 and other Elongator subunits
To identify sites of phosphorylation in Elongator we affinity isolated the complex from yeast cells expressing a TAP-tagged version of Elp1. Tryptic digests of the affinity-purified material were subjected to phosphopeptide enrichment involving a two-step procedure using Hypersep SCX and TiO2, followed by tandem mass spectrometry to locate sites of phosphorylation. To maximize the chance of detecting phosphopeptides, in addition to isolating Elongator from wild-type yeast cells we also prepared and analyzed the complex from a sit4Δ strain, in which lack of Sit4 phosphatase leads to Elp1 hyperphosphorylation [37], [39]. We also analyzed Elongator prepared from a kti12Δ mutant in which Elongator is hypophosphorylated [37], [39] in case additional phosphorylation sites could also be detected under these circumstances. In this way we identified eight phosphorylation sites in Elp1, one each in Elp2 and Elp4 and two in Elp5 (Table 1). With the exception of Ser-222 in Elp4 [40], all of these sites are novel and were not identified in any of the recent proteome-wide phosphoproteomics studies.
Tab. 1. Summary of phosphorylation sites identified in Elp1 and other Elongator subunits. 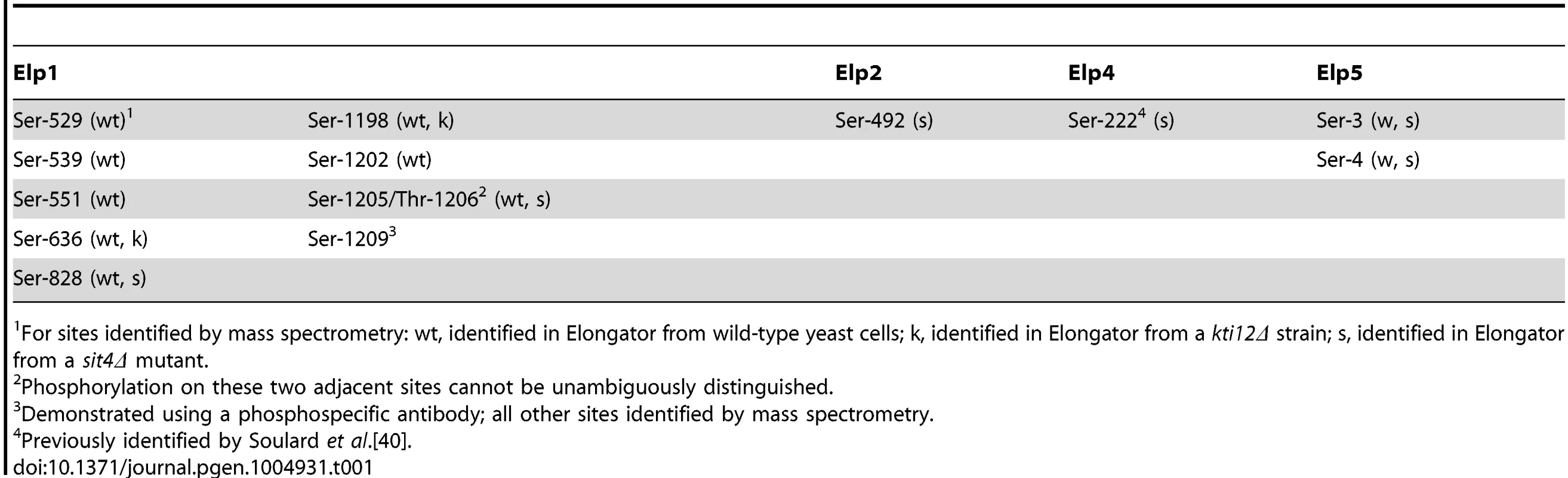
For sites identified by mass spectrometry: wt, identified in Elongator from wild-type yeast cells; k, identified in Elongator from a kti12Δ strain; s, identified in Elongator from a sit4Δ mutant. Since we previously showed that Elp1 phosphorylation state changes are associated with altered Elongator function [37], [39], we focused on the phosphorylation sites we identified in Elp1. S1 Fig. shows representative MS/MS spectra providing evidence for the eight sites identified in Elp1, which with one exception were identified within monophosphorylated peptides. Apart from Ser-1205/Thr-1206, where phosphorylation of the two sites cannot be unambiguously distinguished from the mass spectra, all phosphorylation sites can be identified with high confidence. All Elp1 sites identified in Elongator isolated from either the sit4Δ mutant or the kti12Δ mutant were also found in Elp1 from the wild-type strain (Table 1).
Since analysis of our mass spectrometry data also provided additional weak evidence for phosphorylation on Elp1 Ser-1209, we raised a phosphospecific antibody against a synthetic peptide carrying phosphate on this residue to examine whether it was a genuine phosphorylation site. As shown in Fig. 1B, when used to probe Elp1 by Western blotting, this phosphospecific antibody gave a strong signal that was lost when Ser-1209 was mutated to alanine, thereby demonstrating its phosphospecificity. Thus Elp1 Ser-1209 represents a ninth Elp1 phosphorylation site (Table 1). Fig. 1A shows that five of the nine sites are located centrally within Elp1. Four of these five sites map to the Elp1 amino-terminal domain, which is strongly predicted to form a β-propeller structure that may mediate interactions with other Elongator subunits or accessory proteins. The four remaining phosphorylation sites are tightly clustered close to the carboxy-terminus of Elp1, in a region that is located adjacent to its tRNA binding region [32] and predicted to be disordered (Fig. 1A). Phosphorylation sites are generally found to be enriched in disordered regions [41], in particular those sites that show dynamic variation in phosphorylation state [42]. Beyond Ser-1209, it is striking that every fourth residue between Thr-1212 and Thr-1230 is either threonine or serine (Fig. 1E). However, despite this intriguing pattern of phosphorylatable residues we were unable to detect phosphate groups in this region.
Fig. 1. Identification of functionally important sites of phosphorylation in Elp1. 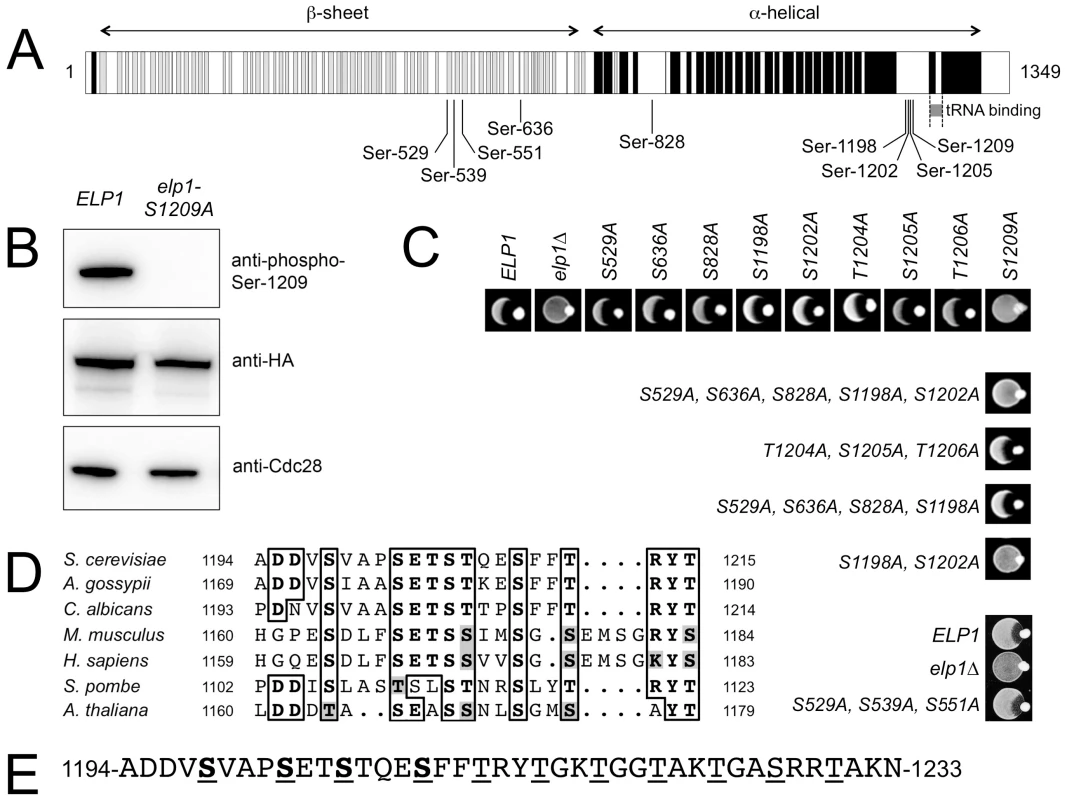
(A) Cartoon showing Elp1 indicating mapped sites of phosphorylation and structural elements predicted using PSIPRED [82], with predicted α-helix shown in black and β-sheet in grey. (B) Western blot of cell lysate prepared from cells expressing ELP1-6HA (ELP1) or elp1-S1209A-6HA showing recognition of Elp1 by a phosphospecific antibody raised against the phosphopeptide TSTQE(pS)FFTRY that recognizes Elp1 phosphorylated on Ser-1209; blots with anti-HA and anti-Cdc28 antibodies control for levels of Elp1 and total protein loading respectively. (C) Eclipse assays showing the zymocin phenotype of wild-type and a range of single or multiple elp1 phosphorylation site mutants. Lack of growth inhibition around the K. lactis colony placed at the edge of each patch of cells being tested indicates zymocin resistance and hence loss of Elongator function. (D) Conservation of the phosphorylation sites identified in the Elp1 C-terminal domain. The full sequences were first aligned using T-Coffee [83] but only the relevant detail from the alignment is shown. NCBI database accessions for the aligned sequences are Saccharomyces cerevisiae (NP_013488), Ashbya gossypii (NP_984907), Candida albicans (XP_710040), Mus musculus (NP_080355), Homo sapiens (NP_003631) Schizosaccharomyces pombe (NP_595335), Arabidopsis thaliana (NP_196872). (E) C-terminal phosphorylated region of S. cerevisiae Elp1 showing the downstream region with a repeating pattern of serine and threonine residues. Mapped phosphorylation sites are underlined and shown in bold, potential phosphorylation sites downstream of the mapped sites are underlined. To determine whether any of the Elp1 phosphorylation sites we had identified were likely to be functionally significant, we mutated each in turn to non-phosphorylatable alanine. These mutants were assayed for zymocin sensitivity by eclipse assay, in which growth inhibition of an S. cerevisiae strain around a colony of zymocin-producing K. lactis indicates loss of Elongator-dependent mcm5 modification of tRNAGlu(UUC) [16], [24]. Fig. 1C shows that with the exception of the S1209A mutant, no alanine substitution at a single phosphorylation site conferred detectable resistance to zymocin and hence loss of Elongator function. In contrast, the S1209A substitution conferred complete zymocin resistance in this assay. Combinations of alanine substitutions at multiple sites were therefore also generated and tested (Fig. 1C and S1 Table). This analysis indicated that any mutants in which both Ser-1198 and Ser-1202 had been replaced by alanine were also zymocin-resistant. Thus Ser-1198, Ser-1202 and Ser-1209 define three phosphorylated residues in Elp1 that are required for Elongator function, but with Ser-1198 and Ser-1202 apparently showing some redundancy. Furthermore, these residues are highly conserved in Elp1 in both lower and higher eukaryotes (Fig. 1D). Since Elp1 becomes hyperphosphorylated in cells lacking Sit4 phosphatase that are defective for Elongator function [37], [39], we also examined whether any alanine substitutions could reverse the zymocin resistance shown by a sit4Δ mutant. However, none of the mutations tested altered the zymocin-resistance phenotype of the sit4Δ strain (S1 Table). Similarly, we also tried mimicking constitutive phosphorylation at many of the sites by making glutamate or aspartate substitutions alone or in combination (S1 Table), but failed to find any substitution(s) that conferred a significant loss of function phenotype.
Phenotype of Elp1 C-terminal domain phosphorylation site mutants
To investigate the Elp1 C-terminal phosphorylated region, additional combinations of alanine, aspartate or glutamate substitutions at the phosphorylation sites were generated. In addition to using the eclipse assay, we monitored tolerance to intracellular expression of the zymocin tRNase (γ) subunit from the galactose-inducible GAL1 promoter as a more quantitative measure of zymocin resistance [43]. Since zymocin sensitivity provides a readout largely for the mcm5 modification state of just tRNAGlu(UUC), we also monitored Elongator function via efficiency of ochre suppression mediated by SUP4, a tRNATyr(UUA) that requires Elongator-dependent wobble uridine modification for efficient ochre (UAA) codon readthrough [20], [24]. This involved single copy integration of a plasmid that carried both SUP4 and a ura3 ochre allele [32], such that suppression (and hence Elongator's tRNA modification function) could be monitored by growth in the absence of uracil.
Fig. 2 shows that the S1209A substitution conferred complete resistance to intracellular expression of the zymocin γ subunit and greatly reduced SUP4-dependent ura3oc suppression, in each case conferring a phenotype comparable to that observed upon complete loss of Elongator function (elp1Δ). In contrast, an S1209D substitution showed considerable restoration of function, while the equivalent glutamate substitution (potentially a poorer mimic of phosphoserine due to its longer side chain) was less effective. The regain of functionality caused by the phosphomimic aspartate substitution therefore provides evidence that phosphorylation of Ser-1209 acts positively for Elongator function. Although the double S1198A S1202A substitution mutant conferred strong zymocin resistance it showed considerable residual Elongator function in the SUP4 assay, allowing for growth in the absence of uracil similar to that shown by the ELP1 wild-type control. However, concurrent glutamate substitutions at both positions largely restored zymocin sensitivity, supporting the notion that phosphorylation of these two sites acts positively for Elongator function. S1198A S1202A in combination with T1204A S1205A T1206A, a triple alanine substitution that on its own had essentially no effect on Elongator function in any of the assays, conferred stronger zymocin resistance than S1198A S1202A alone and dramatically reduced SUP4-dependent suppression efficiency, indicative of an additive Elongator defect in the quintuple mutant. The triple T1204A S1205A T1206A mutation was used here because phosphorylation at Ser-1205/Thr-1206 could not be unambiguously distinguished (see above). Mutants where acidic residues substituted combinations of these five positions, either alone or in combination with alanine substitutions, indicated that Elongator functionality was not greatly affected by acidic substitutions, although the T1204E S1205D T1206E triple substitution improved Elongator function when combined with the S1198A S1202A double mutant (seen most clearly by the eclipse assay). While the difference in the severity of phenotype observed between the zymocin - and SUP4-based assays for some elp1 mutants might reflect differential effects on tRNAGlu(UUC) and tRNATyr(UUA), we consider it more likely to reflect different loss of modification thresholds required to score positive in these assays; while as little as ∼40% reduction in modification may generate sufficient uncleavable tRNAGlu(UUC) to confer zymocin resistance [44], a much larger reduction in modification may be required before there is insufficient functional tRNATyr(UUA) to support effective ochre suppression. Our results therefore indicate that blocking phosphorylation at all four sites identified in this region of Elp1 leads to reduced Elongator function, with the S1209A mutant showing the greatest defect followed by S1198 S1202A and T1204A S1205A T1206A in decreasing order of severity, but with additivity between S1198 S1202A and T1204A S1205A T1206A leading to a defect as severe as that of S1209A. That acidic substitutions mimicking phosphorylation at each of the four sites conferred considerable Elongator function is consistent with the notion that phosphorylation at these sites functions positively for Elongator.
Fig. 2. Effect on Elongator function of phosphorylation site mutations in the C-terminal domain of Elp1. 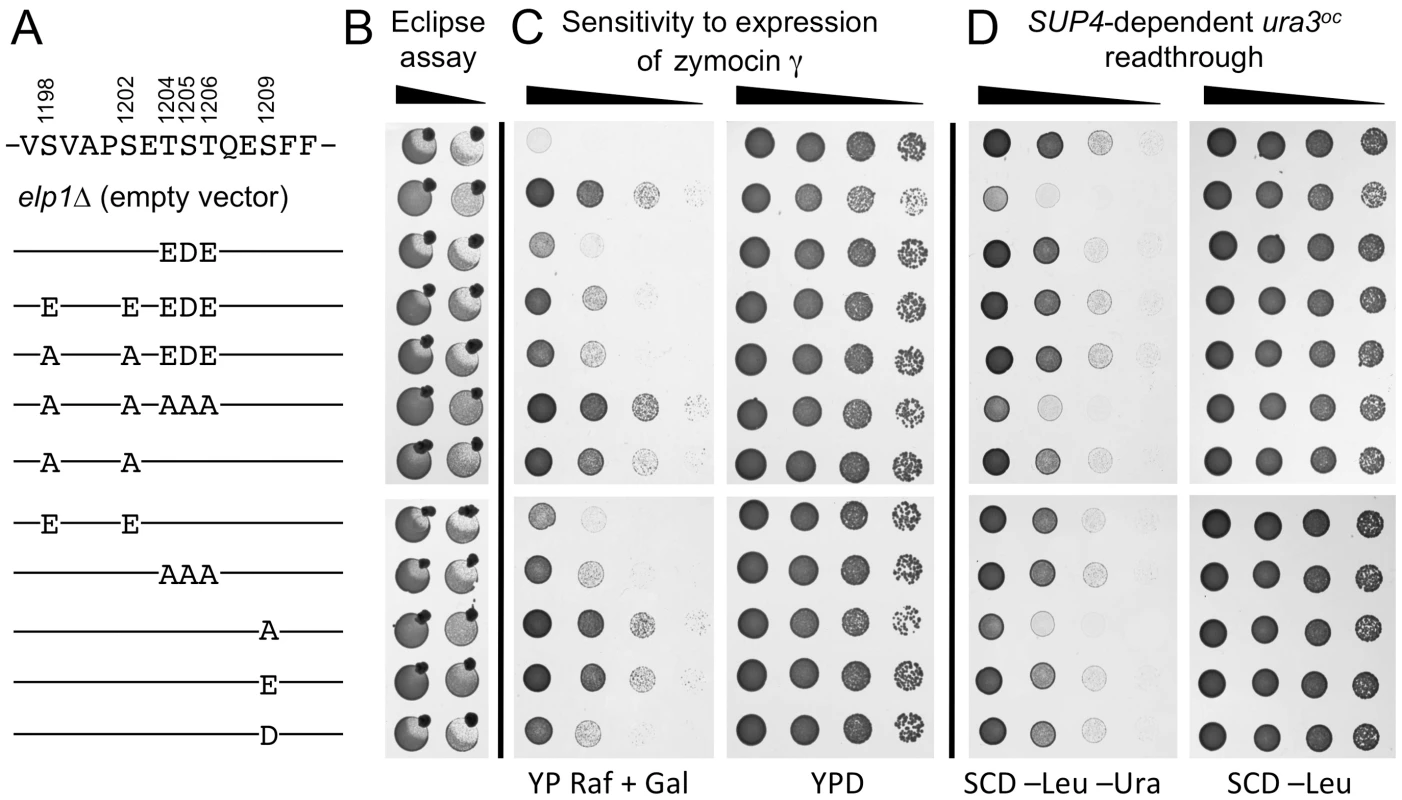
All strains were based on WAY034 (elp1Δ; panel B), YRDS119 (elp1Δ [pAE1]; panel C) or SBY138 (elp1Δ his3Δ1::pSB3; panel D) transformed with YCplac111, YCplac111-ELP1-6HA (wild-type) or mutant derivatives carrying mutations in ELP1 as shown. (A) Key to substitutions present in the different alleles tested (wild-type sequence at the top). (B) Zymocin sensitivity measured by eclipse assay (see Fig. 1 legend), with strains being tested at either 1.0 or 0.1 OD600/ml following growth in SCD-Leu medium. (C) Zymocin sensitivity monitored by intracellular expression of the zymocin γ subunit. Cells were grown on SCD-Leu-Ura to select for both plasmids, diluted to 1.0 OD600/ml and then equivalent 10-fold dilutions plated on YPD agar (to control for cell viability) and YP agar containing 2% raffinose and 2% galactose to induce expression of zymocin γ-toxin from pAE1. (D) Cells were grown in SCD-Leu (to select for YCplac111 and its derivatives), diluted to 1.0 OD600/ml and then equivalent 10-fold dilutions plated on SCD-Leu (to control for cell viability) and SCD-Leu-Ura (to assess efficiency of SUP4-dependent ura3oc22 suppression. To look directly at the tRNA wobble uridine modifications, we prepared tRNA from selected elp1 mutant strains and used LC-MS analysis to quantitate the levels of modified U34 nucleosides. Fig. 3 shows that in the elp1Δ control strain, mcm5U and ncm5U were absent from tRNA as expected. The S1209A and quintuple S1198 S1202A T1204A S1205A T1206A strains showed almost no mcm5U or ncm5U, consistent with the major defect in Elongator-dependent wobble uridine modification indicated by the phenotypic assays (Fig. 2). The S1198A S1202A double mutant showed reduced levels of mcm5U and ncm5U consistent with a less severe Elongator defect, while strains with the S1198E S1202E double or S1209D single phosphomimic alleles, or with the triple T1204A S1205A T1206A allele, had levels of mcm5U and ncm5U intermediate between those of the S1198A S1202A mutant and ELP1 wild-type. Taken together, the profiles for the ncm5 and mcm5 modification nucleosides are consistent with our phenotypic analyses (Fig. 2) and our data therefore support the notion that phosphorylation of Ser-1209 plays a major, positive role in Elongator-dependent tRNA modification and that phosphorylation at Ser-1198, Ser-1202 and Ser-1205/Thr-1206 makes a similar but partly redundant contribution to Elongator activity.
Fig. 3. Quantitation of modified U34 nucleosides in tRNA from selected elp1 mutants. 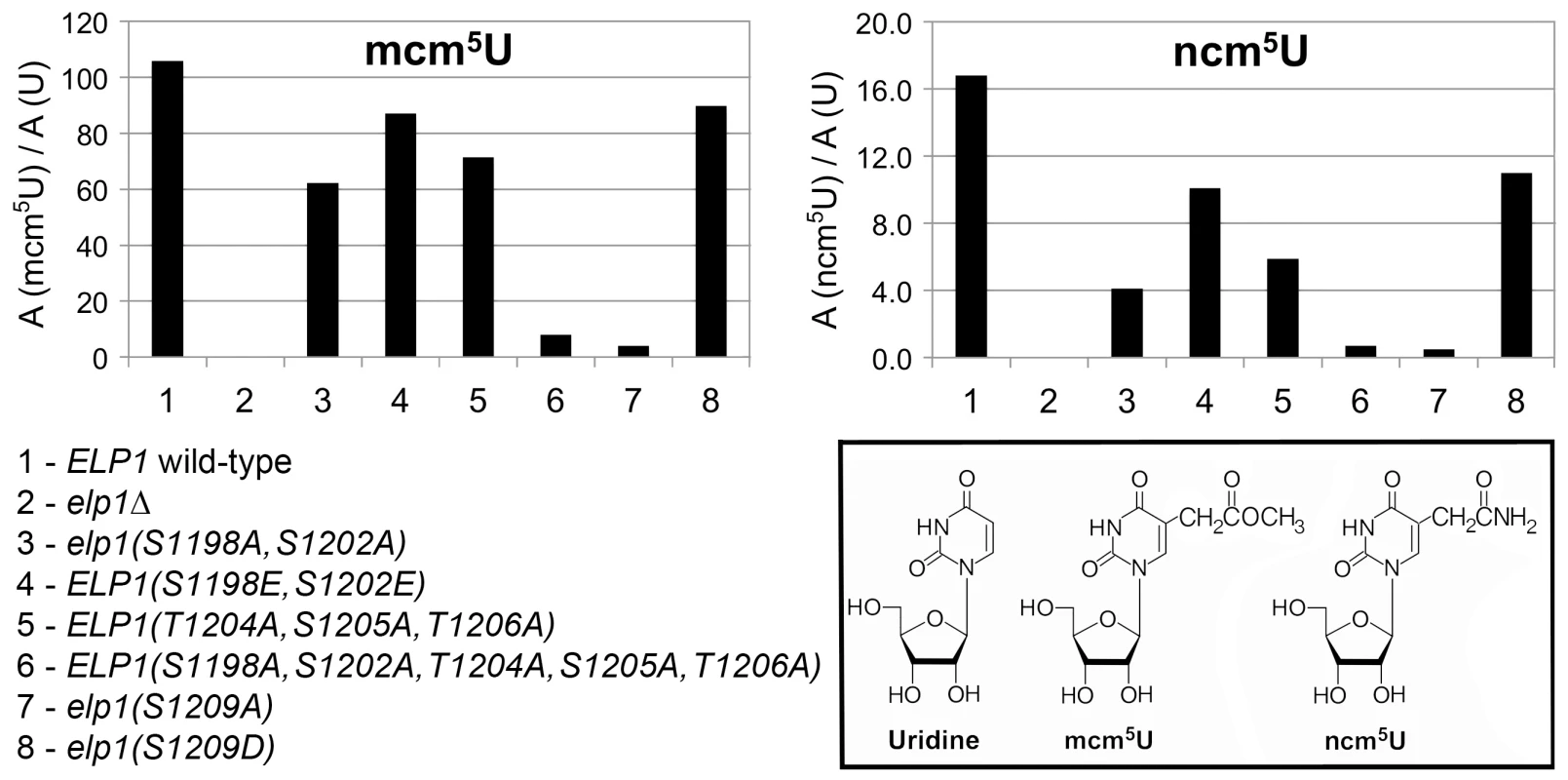
Content of ncm5U and mcm5U in small RNA samples from the indicated yeast strains. In each case, A (modified U)/A (U) indicates that the modified nucleoside signal was normalized using the total uridine content to allow comparison of the different samples. Inset: chemical structures of uridine and its Elongator-dependent mcm5U and ncm5U derivatives. Phosphorylation site mutations in Elp1 do not prevent Elongator assembly but lead to changes in association with Kti12 and with Hrr25 kinase
Phosphorylation site mutations that alter Elongator functionality could in principle do so by affecting assembly of the holo-Elongator complex, either because they interfere with assembly or because phosphorylation of these sites could regulate assembly of the Elongator complex. When myc-tagged Elp2 was used to immunoprecipitate Elongator from cell extracts, elp1 phosphorylation site mutations at positions 1198, 1202, 1205, 1206 and 1209, alone or in combination, did not affect the recovery of Elp1, Elp3 and Elp5 in the Elp2 immunoprecipitates in comparison with strains where Elp1 was wild-type (Fig. 4). In particular, there was no evidence for any changes in the assembly of the complex when Elp1 carried the S1209A substitution, which is essentially null for Elongator function as discussed above. Thus similar co-immune precipitation of all four proteins was observed, regardless of whether these mutations affected Elongator functionality. Although we have not tested every mutant elp1 allele constructed in this study, assembly of the Elp1-Elp2-Elp3 subcomplex and its association with the Elp4-Elp5-Elp6 subcomplex appear essentially normal, irrespective of the consequences of the mutations for Elongator functionality.
Fig. 4. Elongator assembly is unaffected by a range of phosphorylation site mutations in Elp1. 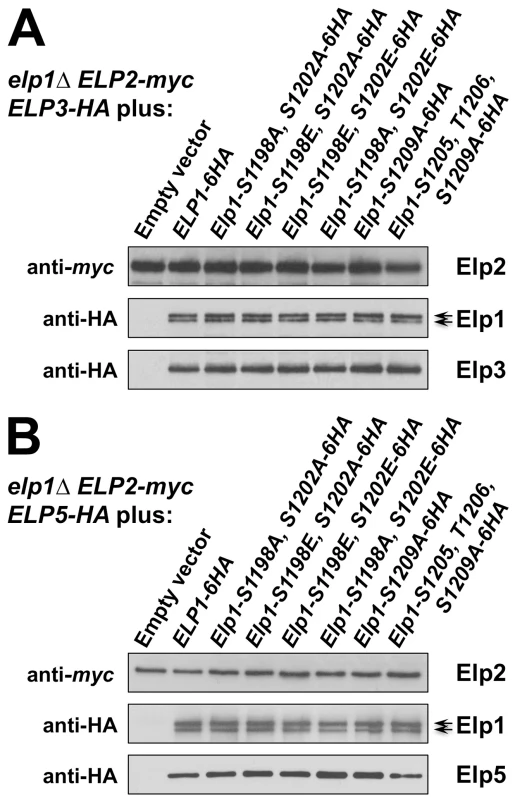
elp1Δ yeast strains expressing myc-tagged ELP2 and either HA-tagged ELP3 (A) or ELP5 (B) were transformed with either empty vector or plasmids expressing the indicated ELP1 alleles (Table S2). Elp2-myc was immunoprecipitated from extracts and immunoprecipitates were examined by Western blotting with anti-myc and anti-HA antibodies to detect co-immunoprecipitated Elp1, Elp3 and Elp5 as indicated. Note that Elp1 runs as a doublet as observed previously [84]. Since Elongator interacts with both its accessory factor Kti12 and with Hrr25 kinase and because Elongator function requires both Kti12 and Hrr25 kinase activity [16], [37], [45], [46], we next used co-immunoprecipitation to examine the effect of selected phosphorylation site mutations on Elongator's association with Kti12 and Hrr25. Elongator complex in which Elp1 carries the double S1198A S1202A mutation showed reduced interaction with Hrr25 that was not seen with the corresponding phosphomimic allele (S1198E S1202E), but combining S1198A S1202A with T1204A S1205A T1206A did not further reduce interaction with Hrr25 (Fig. 5) despite the greater loss of Elongator function in the quintuple mutant. Conversely, mutation of Elp1 Ser-1209 to alanine led to enhanced interaction between Elongator and Hrr25 (Fig. 5). In all ELP1 mutants tested, Elongator retained its ability to interact with Kti12, but the elp1-S1209A allele also led to enhanced Kti12 interaction (Fig. 6A). We examined the effect of the Elp1 S1209A substitution on Kti12 association in more detail by tagging the genomic copy of ELP1 with GFP and carefully quantitating the recovery of HA-tagged Kti12 in strains expressing either the wild-type or mutant Elp1-GFP fusions. This confirmed that the S1209A mutation leads to enhanced Kti12 association: in comparison with wild-type Elp1, Elp1-S1209A was reproducibly associated with approximately twice as much Kti12 (Fig. 6B, C). Thus the S1198A S1202A and S1209A mutations have opposite effects on association with Hrr25 and the 1209A mutation enhances association with Kti12, suggesting that phosphorylation at these sites affects the interaction between Elongator and key proteins required for its functionality.
Fig. 5. Phosphorylation site mutations in Elp1 lead to changes in Hrr25 association with Elongator. 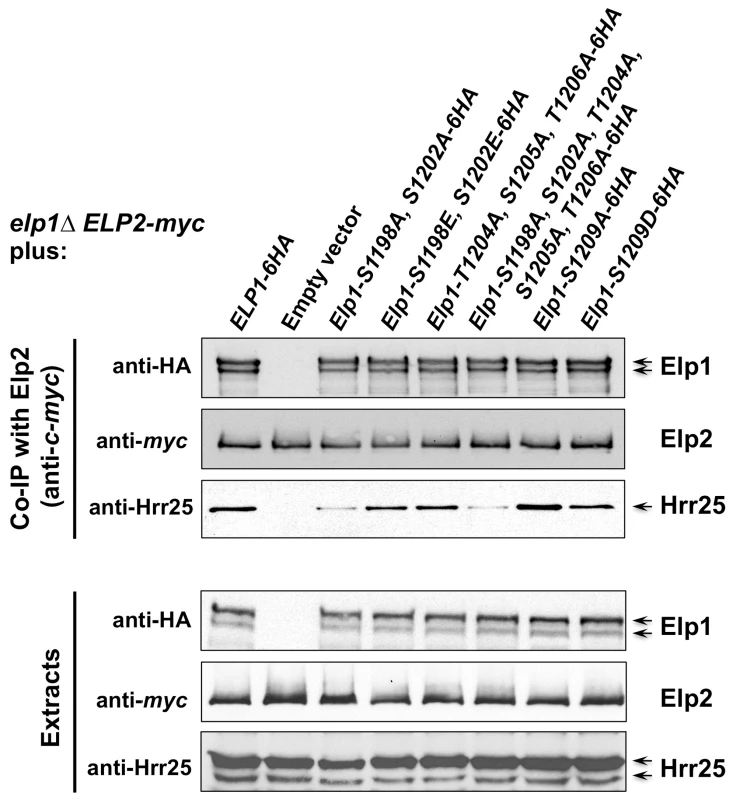
elp1Δ ELP2-myc3 KTI12-HA6 yeast strains were transformed with either empty vector or plasmids expressing the indicated HA6-tagged ELP1 alleles (Table S2). Elp2-myc was immunoprecipitated from extracts and immunoprecipitates were examined by Western blotting with anti-myc (to detect Elp2), anti-HA antibodies (to detect co-immunoprecipitated Elp1) and anti-Hrr25 (to detect co-immunoprecipitated Hrr25) as indicated. Western blots of the extracts before immunoprecipitation (lower panels) confirm similar levels of each protein in the different strains. Fig. 6. Phosphorylation site mutations in Elp1 lead to changes in Kti12 association with Elongator. 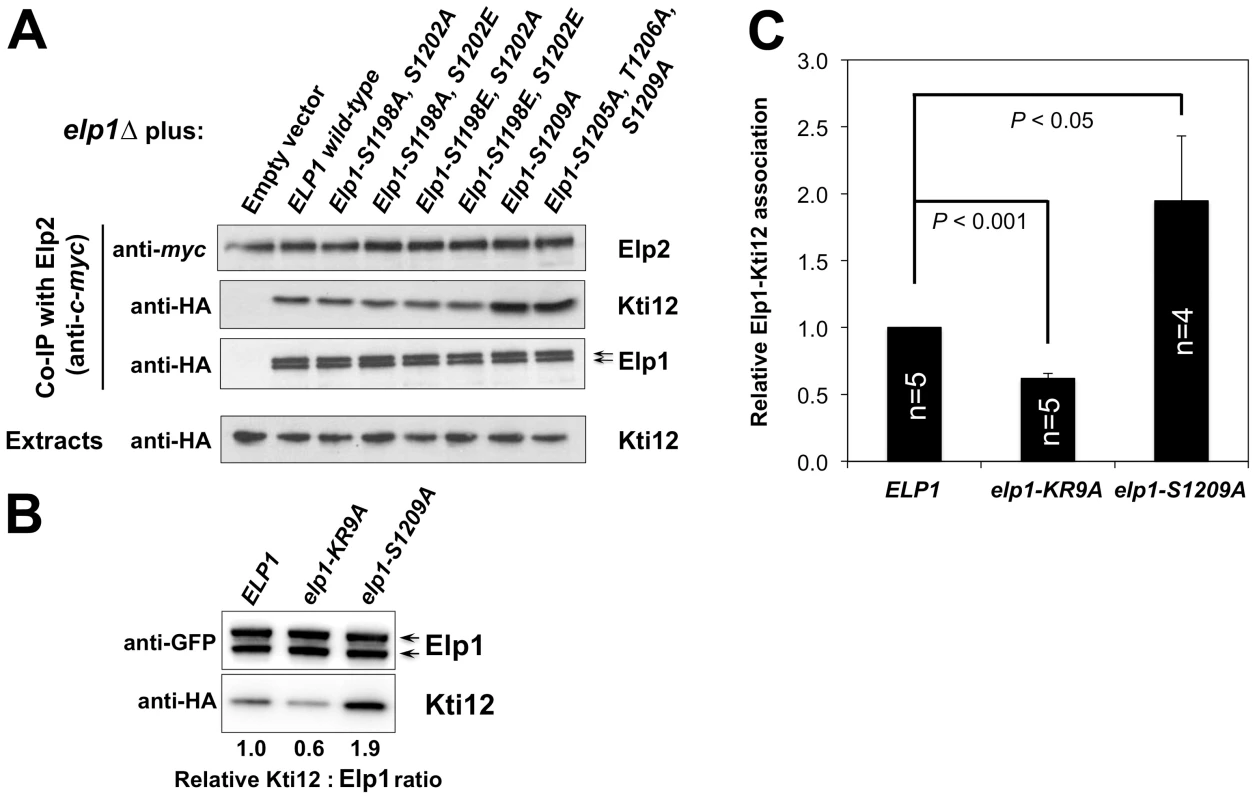
(A) elp1Δ ELP2-myc3 KTI12-HA6 yeast strains were transformed with either empty vector or plasmids expressing the indicated HA6-tagged ELP1 alleles (Table S2). Elp2-myc was immunoprecipitated from extracts and immunoprecipitates were examined by Western blotting with anti-myc (to detect Elp2) and anti-HA antibodies (to detect co-immunoprecipitated Elp1 and Kti12) as indicated. Western blots of the extracts before immunoprecipitation (lower panels) confirm similar levels of each protein in the different strains. (B) Co-immune precipitation of HA-tagged Kti12 with GFP-tagged Elp1. Extracts from strains expressing GFP-tagged wild-type or mutant Elp1 from its genomic locus together with HA-tagged Kti12 were subjected to immune precipitation using GFP-trap and then the immune precipitates examined by Western blotting with anti-GFP (to detect Elp1) and anti-HA antibodies (to detect co-immunoprecipitated Kti12) as indicated. elp1-KR9A, a tRNA binding domain mutation that reduces Kti12 association [32], is shown as a control. (C) Quantification of co-immunoprecipitation efficiency shown in (B). Immunoprecipitation of HA-tagged Kti12 was quantified by densitometry of the HA tag signals and normalized using the Elp1–GFP signals across the indicated number of replicates (n), setting the value for the wild-type strain in each case to 1.0. Error bars represent the standard deviation of the mean and the significance of the differences was analyzed using a one-way ANOVA, showing a statistically significant, two-fold increase in the association of Kti12 with Elp1-S1209A. Hrr25 directly phosphorylates Elp1 on functionally important sites
We next wished to identify the protein kinase(s) that mediate modification of the phosphorylation sites in the Elp1 carboxy-terminal domain that are important for Elongator function. To take an unbiased approach, we screened a library of 119 GST-protein kinase fusions [47] for their ability to phosphorylate Elp1 in vitro using TAP-purified Elongator as a substrate. Purified Elongator showed significant background phosphorylation specifically on Elp1 when incubated with [γ-32P]ATP in the absence of added kinase. However, five protein kinases were identified that could clearly phosphorylate Elp1 in vitro (Hrr25, Yck1, Yck2, Yck3 and Hal5: S2A Fig.). Given that Hrr25 interacts with Elongator [37], [48], [49] as seen in Fig. 5 and because Elp1 is already known to become hypophosphorylated in a hrr25 mutant [37], it is an excellent candidate for an in vivo Elp1 kinase. Hrr25, Yck1, Yck2 and Yck3 all belong to the casein kinase I (CKI) family of protein kinases [50], but while hrr25 mutants show clear defects in Elongator function [37], [38], Yck1, Yck2 and Yck3 are membrane associated via lipid modification [51], [52] and do not confer detectable zymocin resistance when the corresponding genes are deleted [38]. Thus we consider it unlikely that Yck1-3 mediate functionally important phosphorylation events on Elp1 in vivo and that they were identified in our in vitro screen due to shared substrate specificity with the related Hrr25 CKI isozyme. Preliminary analysis also failed to generate data supporting a role for Hal5 in Elongator function (S2B-S2C Fig.).
In further support of a role for Hrr25 as a direct Elp1 kinase, we next showed that the Elp1 kinase activity present in affinity-isolated Elongator preparations was due to Hrr25. This made use of a yeast strain dependent on an ‘analog-sensitive’ HRR25 allele (hrr25-I82G) in which the mutant Hrr25 kinase has acquired the capacity to be inhibited specifically by addition of the ATP analogs 1NM-PP1 or 3MB-PP1 [53]. When Elongator was isolated from the hrr25-I82G strain, the Elp1 phosphorylation observed upon incubation of the isolated complex with [γ-32P]ATP was blocked by addition of either 1NM-PP1 or 3MB-PP1 (Fig. 7A). This was in contrast to Elongator isolated from a control strain expressing wild-type HRR25, phosphorylation of which was refractory to these inhibitors. In fact the hrr25-I82G strain became zymocin resistant when grown in the presence of 1NM-PP1, emphasizing the positive role of Hrr25 kinase in Elongator's tRNA modification in vivo (Fig. 7B) and consistent with Elongator-minus phenotypes of hrr25 mutants [37], [38].
Fig. 7. Purified Elongator shows Hrr25-dependendent phosphorylation that requires Ser-1198 and Ser-1202. 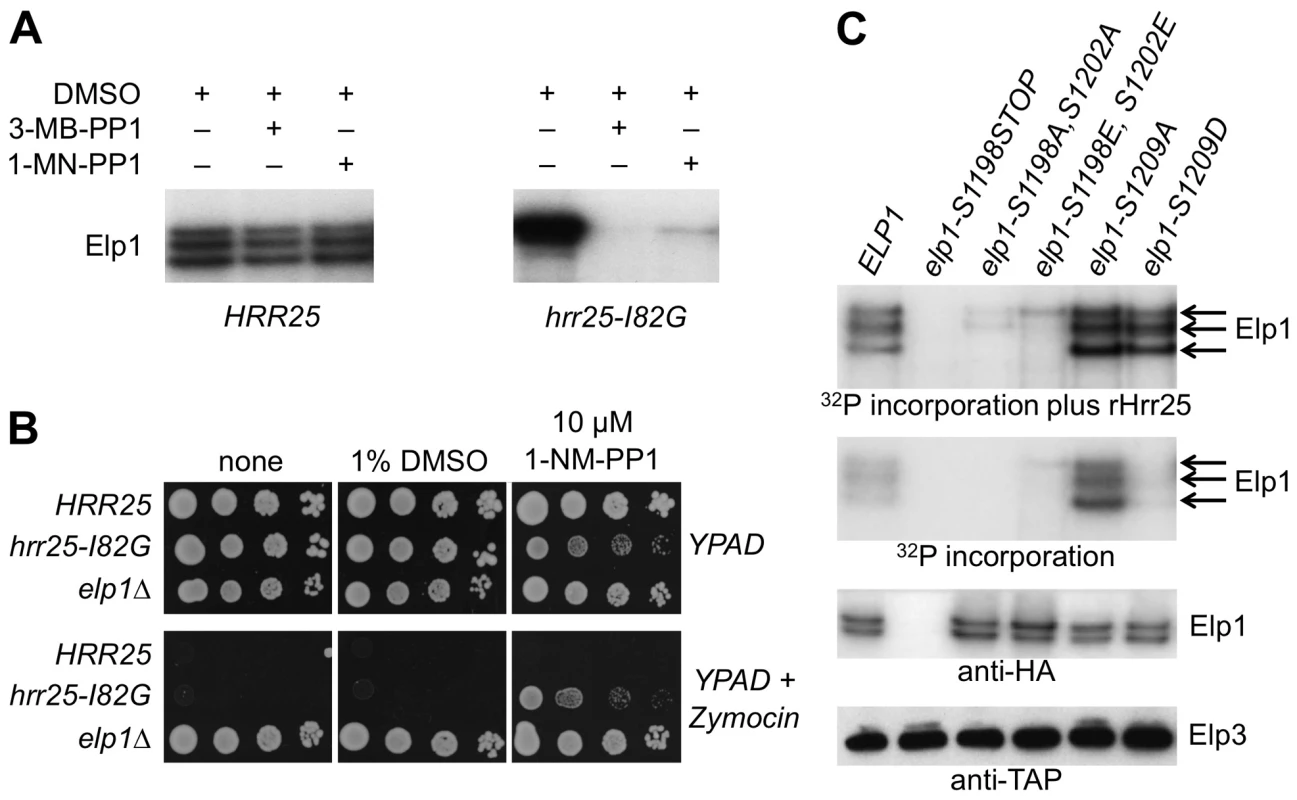
(A) Elongator was purified using TAP-tagged Elp3 from either a HRR25 wild-type or a hrr25-I82G analog-sensitive mutant strain and supplemented with [γ-32P]ATP to examine endogenous Elp1 phosphorylation in the presence of DMSO (drug vehicle), 1NM-PP1 or 3MB-PP1. Phosphorylated Elp1 was visualized by autoradiography following SDS-PAGE. (B) Equivalent 10-fold serial dilutions of HRR25 ELP1, hrr25-I82G ELP1 and HRR25 elp1Δ yeast strains were plated out on YPAD agar with or without 1% (v/v) zymocin in the presence or absence of 10 µM 1NM-PP1 or an equivalent volume of DMSO (drug vehicle) as indicated and photographed after 2 days' growth at 30°C. (C) Elongator was prepared as in (A) from elp1Δ ELP3-TAP strains carrying plasmids encoding the indicated ELP1 alleles and incubated with [γ-32P]ATP in the presence or absence of added recombinant Hrr25 (100 ng). Phosphorylated Elp1 was visualized by autoradiography following SDS-PAGE. Western blotting to detect Elp3-TAP and Elp1-6HA demonstrates equivalent recovery of Elongator from each strain. To determine whether Hrr25 phosphorylation of Elp1 occurred at any of the functionally relevant sites that we had identified, Elongator containing wild-type or mutant forms of Elp1 was purified and tested for incorporation of phosphate from [γ-32P]ATP both in the absence and presence of recombinant Hrr25. Fig. 7C shows that even when reactions were supplemented with Hrr25, Elp1 in which the C-terminal region had been deleted was not phosphorylated. Elongator in which both Elp1 Ser-1198 and Ser-1202 had been replaced by either alanine or glutamate showed negligible incorporation of radiolabel in comparison with the Elp1 in wild-type Elongator. In contrast, Elongator in which Elp1 Ser-1209 was substituted with either alanine or aspartate showed high levels of 32P incorporation into Elp1 in the presence of recombinant Hrr25. Since HRR25 is an essential gene we were unable to examine Ser-1209 phosphorylation with the anti-phophoserine-1209 antibody in the complete absence of Hrr25. However, in the kinase-dead hrr25-3 mutant, which shows loss of Elongator function and absence of the phosphorylated isoform detected in wild-type cells by gel shift assay [37], a similar level of Ser-1209 phosphorylation to that seen in a wild-type strain was observed (S3 Fig.). Although more complex interpretations are possible, these observations are therefore most simply explained if Ser-1198 and Ser-1202 are major sites of Hrr25 phosphorylation and Ser-1209 is not a major site for modification by the kinase. In the absence of added Hrr25, Elongator complex containing Elp1-S1209A still showed high levels of Elp1 phosphorylation that were not seen with the corresponding aspartate substitution or when Elp1 was wild-type, although after addition of recombinant Hrr25 the level of Elp1 phosphorylation of the Elp1-S1209A and Elp1-S1209D complexes was similar as noted above (Fig. 7C). These data are consistent with the enhanced interaction of Elp1-S1209A-containing Elongator with Hrr25 that was seen by co-immune precipitation (Fig. 5).
To complement these experiments, the ability of recombinant Hrr25 to phosphorylate synthetic peptides corresponding to the C-terminal phosphorylated region of Elp1 was tested, using both mass spectrometry to identify phosphorylated residues in reactions with unlabeled ATP and by monitoring incorporation of 32P-phosphate in reactions containing [γ-32P]ATP. When a peptide containing Elp1 residues 1193-1213 was phosphorylated by recombinant Hrr25 in vitro and analyzed by mass spectrometry, three types of phosphopeptide were detected: monophosphorylated peptide modified on either Ser-1198 (S4A Fig.) or Ser-1202 (S4B Fig.), and diphosphorylated peptide modified on both these residues (S4C Fig.). These data therefore confirm the identity of Hrr25 as a protein kinase that can directly phosphorylate Elp1 on serine residues that are important for Elongator function, and are consistent with a model in which phosphorylation at one of these two sites may then favor phosphorylation of the second site. When all phosphorylatable residues apart from Ser-1198 and Ser-1202 were changed to alanines, dual phosphorylation on Ser-1198 and Ser-1202 was still seen, indicating that phosphorylation of these two positions was not dependent on phosphorylation of Ser-1205 or any other downstream residues.
When the 1193-1213 peptide was incubated with Hrr25 together with [γ-32P]ATP, it readily incorporated radiolabelled phosphate (Fig. 8B and C; peptide 89). Although the related peptide in which all serine and threonine residues apart from Ser-1198 and Ser-1202 had been substituted by alanines was still an excellent Hrr25 substrate (Fig. 8C; peptide 92), incorporation of 32P at later times was reduced in comparison (Fig. 8B), consistent with the possibility of additional phosphorylation to the right of Ser-1202. Additional single alanine substitutions at either Ser-1198 or Ser-1202 in peptide 92 greatly reduced but did not completely prevent phosphorylation (Fig. 8C, peptides 1001, 1002). However, replacement of both Ser-1198 and Ser-1202 by either alanines or glutamates completely blocked phosphorylation of the peptide despite the presence of the five serine and threonine residues downstream (peptides 90 and 91). Another peptide encompassing Elp1 residues 1207-1229 was not phosphorylated at all following incubation with Hrr25 (Fig. 8D, peptide 95), supporting the notion that neither Ser-1209 nor any of the repeating threonine and serine shown in Fig. 1E can be directly phosphorylated by Hrr25 kinase. These data are therefore consistent with interdependence of Ser-1198 and Ser-1202 phosphorylation by Hrr25 and absence of Hrr25 phosphorylation on the downstream serines and threonines.
Fig. 8. In vitro phosphorylation of peptides corresponding to the Elp1 C-terminal region by Hrr25. 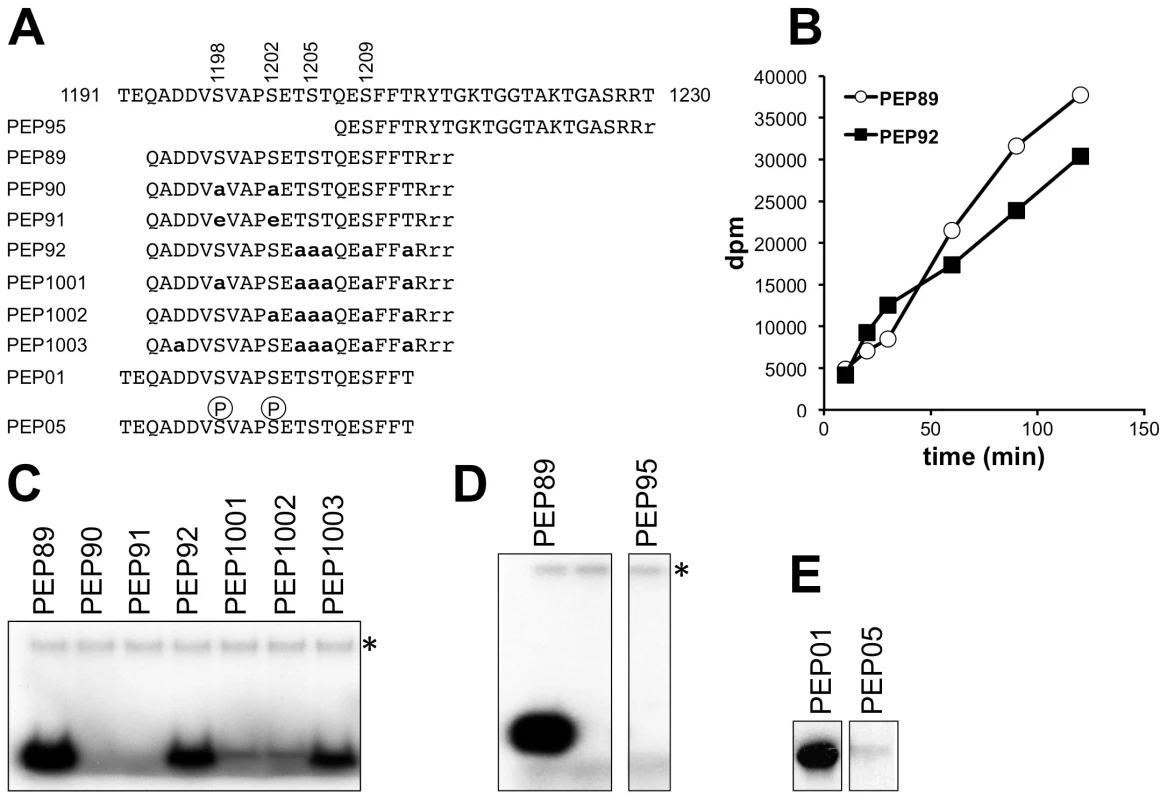
(A) Sequences of synthetic peptides used as substrates shown in alignment with Elp1 residues 1191-1230. Lower-case bold letters represent alanine and/or glutamate substitutions relative to the native Elp1 sequence. (B) Time course of Hrr25-dependent incorporation of 32P from [γ-32P]ATP into Peptides 89 and 92. (C-E) Incorporation of 32P from [γ-32P]ATP into peptides as assayed by SDS-PAGE and autoradiography. Note that in (D) and (E) two panels are shown in each case because some unnecessary intervening lanes were removed when preparing the figure. * indicates autophosphorylated recombinant Hrr25. Since phosphorylation of specific Ser or Thr residues by CKIs such as Hrr25 is often primed by phosphorylation at another Ser or Thr residue 2-4 positions upstream [54], it was possible that Hrr25 might phosphorylate Ser-1205 once Ser-1202 had been modified, and that phosphorylation at Ser-1205 might then promote modification of Ser-1209. However, phosphomimic glutamate substitutions at 1198 and 1202, which supported essentially normal Elongator functionality in vivo (Fig. 2), also prevented phosphorylation of the 1193-1213 peptide on any other downstream site. Furthermore, phosphorylation of a related peptide (Elp1 1192-1212; Fig. 8E), which was also a good in vitro substrate for Hrr25, was also completely blocked for Hrr25 phosphorylation when synthesized with phosphoserine at the two positions corresponding to Elp1 Ser-1198 and Ser-1202. This again supports the model that Ser-1198 and Ser-1202 are the only major sites of Hrr25 phosphorylation and also indicates that it is unlikely that efficient priming of phosphorylation by Hrr25 on downstream residues, such as Ser-1205 and Ser-1209, occurs as a result of the upstream phosphorylation events at Ser-1198 and Ser-1202. Thus although these experiments do not as yet fully explain all the intricacies of Hrr25 phosphorylation in this region, taken together they nonetheless demonstrate that Hrr25 phosphorylates Elp1 directly on Ser-1198 and Ser-1202, two serine residues that we identified as functionally relevant in vivo phosphorylation sites.
Discussion
Identification of functionally relevant phosphorylation sites in Elp1
We have identified nine phosphorylation sites on the Elp1 subunit of yeast Elongator complex and based on the phenotypes of non-phosphorylatable and phosphomimic mutations, provide evidence that phosphorylation on four sites near the Elp1 C-terminus (Ser-1198, Ser-1202, Ser-1205/Thr-1206 and Ser-1209) plays a positive role in Elongator function. Previously, we showed that a sit4 phosphatase mutant trapped Elp1 in a slower-migrating, hyperphosphorylated form whereas hrr25 kinase mutations led to presence of just a faster-migrating, hypophosphorylated Elp1 isoform [37], [39]. Both types of mutation cause loss of Elongator function [37], [39], suggesting that dynamic phosphorylation and dephosphorylation of Elp1 is needed in functional Elongator and predicting that mimicking constitutive phosphorylation on at least some of the sites we have identified should be inhibitory. It is therefore surprising that all of the phosphomimic alleles we tested conferred significant Elongator function. Possibly the acidic substitutions do not fully mimic constitutive phosphorylation and thereby allow for substantial residual function, even though constitutive phosphorylation might inhibit Elongator. Thus while our alanine substitution mutants lend support to the idea that phosphorylation at the mapped sites functions positively for Elongator activity, we cannot rule out a requirement for dynamic phosphorylation/dephosphorylation at the sites we have identified. It is also possible that additional, inhibitory phosphorylation sites exist that were not found in our study. For example, although we could not detect phosphorylation of Thr-1212 in vivo or in vitro, both alanine and glutamate substitutions at this site caused severe loss of Elongator function (S5 Fig.), consistent with the notion that dynamic phosphorylation and dephosphorylation of this site could be important if it is phosphorylated in vivo.
Hrr25 directly phosphorylates Elp1 on sites important for Elongator function
Despite conducting an unbiased, ‘kinome-wide’ screen to identify kinases responsible for Elp1 C-terminal domain phosphorylation, Hrr25, the yeast CKI that associates with Elongator and has already been implicated in Elongator function [37], [48], [49] was the sole convincing candidate to be identified. Through several lines of evidence, we have now established that Hrr25 directly phosphorylates Elp1 on two in vivo phosphorylation sites that we have mapped and shown to be relevant for Elongator function: Ser-1198 and Ser-1202. Thus purified Elongator shows Hrr25-dependent phosphorylation that requires the presence of these two residues, while peptides derived from the Elp1 C-terminal region show direct, interdependent phosphorylation by recombinant Hrr25 on Ser-1198 and Ser-1202. Conversely, we found no evidence for direct phosphorylation of Ser-1209 by Hrr25 using three different synthetic peptides containing Ser-1209, or using purified Elongator complex in which Ser-1198 and Ser-1202 were mutated but Ser-1209 was intact.
Two types of consensus phosphorylation site have been proposed for CKIs: pS [X]1-3 [ST] and [DE]2-4 [X]0-2 [ST], where pS indicates an upstream phosphoserine residue that is needed to prime phosphorylation at the downstream Ser or Thr (shown in bold and underlined) but which can be substituted by an acidic patch in the second class of motif [54]. Ser-1198 conforms to the latter type of motif and is closely related to other mapped Hrr25 phosphorylation sites [55]-[57] in yeast proteins (S6 Fig.). In contrast, Ser-1202 matches the former motif, suggesting that priming-independent phosphorylation at Ser-1198 by Hrr25 might then prime phosphorylation by Hrr25 at Ser-1202 and predicting that Ser-1198 should still be efficiently phosphorylated when Ser-1202 is replaced by alanine. However, the latter mutation largely blocked phosphorylation at Ser-1198 in our in vitro peptide kinase assays and instead we observed interdependent phosphorylation at these two residues. Furthermore, replacement of one of the two upstream aspartate residues with alanine did not obviously interfere with phosphorylation of Ser-1198 and Ser-1202. Thus although the sites are direct targets of Hrr25 and match the accepted consensus for CKI phosphorylation, dependency of Ser-1202 phosphorylation on prior Ser-1198 modification remains to be demonstrated. Similarly, Ser-1205 and Ser-1209 both fit the pS [X]1-3 [ST] consensus and might be modified once Ser-1202 phosphorylation has occurred, and yet we could not demonstrate Hrr25-dependent incorporation of phosphate at these sites even using a peptide where the upstream sites were already phosphorylated. Given the known substrate specificity of CKIs, it is therefore surprising that we can find no evidence for such priming of phosphorylation at Ser-1205 and Ser-1209 following modification of Ser1202. Furthermore, since the repeating pattern of phosphorylatable residues shown in Fig. 1E also fits the pS [X]1-3 [ST] motif we are even more surprised that we failed to detect phosphorylation of these sites either in vivo or in vitro.
In spite of the strong evidence we have provided that Hrr25 is a direct Elp1 kinase, inability to detect direct phosphorylation of either the key residue Ser-1209 or of Ser-1205/Thr-1206 by Hrr25 implies that at least one additional Elp1 kinase is involved in Elp1 phosphorylation. While these sites might become better substrates for Hrr25 in the context of fully assembled Elongator complex rather than within synthetic peptides, the fact that we detect normal levels of Ser-1209 phosphorylation in a hrr25 mutant that is defective for Elongator function strongly supports the involvement of a different kinase at Ser-1209.
Elp1 phosphorylation may regulate Elongator's interaction with Kti12 and Hrr25
Although all phosphorylation site mutants examined showed essentially normal assembly of the Elongator complex and retained the ability to interact with the accessory protein Kti12, the S1209A mutation enhanced association of Kti12 with Elongator in comparison with cells expressing either the wild-type protein or the S1198A S1202A mutant. Kti12 stoichiometry is important – either too much or too little interferes with Elongator function [39], [43], [45] – and thus Ser-1209 phosphorylation may regulate Elongator's interaction with its accessory protein. Furthermore, the S1209A mutation caused increased Hrr25 association with Elongator whereas the double S1198A 1202A mutation led to reduced Hrr25 association. Thus Hrr25 not only directly phosphorylates Elp1 but may also regulate its own interaction with Elongator through doing so. Although the S1209A single and S1198A S1202A T1204A S1205S T1206A quintuple mutants both conferred strong loss of Elongator function, their differing effects on Hrr25 association implies that they are defective for different reasons – phosphorylation at S1198A S1202A may stabilize Hrr25 binding whereas phosphorylation at Ser-1209 may be required to displace the kinase. Since the interaction between Hrr25 and Elongator is dependent on Kti12 [37] it is also possible that the enhanced interaction of both proteins with Elongator seen when Elp1 cannot be phosphorylated on Ser-1209 are directly related.
Intriguingly, enhanced interaction of both Kti12 and Hrr25 with the Elongator complex is also seen in hrr25 mutants that cause Elp1 phosphorylation defects and zymocin resistance [37 and S7 Fig.]. However, these properties of hrr25 mutants are more similar to those of the elp1-S1209A mutant, which is mutated at a site apparently not directly phosphorylated by Hrr25, rather than mirroring the properties of the elp1-S1198A, S1202A mutant that removes the only sites in Elp1 that we have shown to be direct Hrr25 targets. This suggests an as yet undiscovered connection between phosphorylation at Ser-1209 and Hrr25 kinase. Regardless of this, our data nonetheless indicate that Elp1 phosphorylation by Hrr25 and other kinases could modulate the interaction between Elongator and Kti12.
What type of role does Elp1 phosphorylation play in Elongator function?
Although there is clearly more to learn about the role of phosphorylation in Elongator function, two types of model can be proposed. On the one hand, phosphorylation might regulate Elongator, turning its wobble uridine modification activity up or down in response to growth conditions and the demand for protein synthesis, or perhaps in response to cellular stresses. Given that the translation of some mRNAs is particularly dependent on wobble base modification and Elongator function [58]-[60] and that tRNA modifications (including Elongator-dependent ones) oscillate during the cell cycle and in response to stress signals [61], [62], this raises the interesting possibility that Elongator may be part of a translational control mechanism functioning through tRNA modification. Our proposal that Elp1 phosphorylation operates in a largely positive sense for Elongator, based on the properties of the phosphorylation site mutants that we have examined, is consistent with such a regulatory role. The Elp1 kinase Hrr25 is needed for a wide range of cellular functions [49], [56], [57], [63]-[65] that do not appear to provide a clear insight into the signals that might regulate Elp1 phosphorylation. However, the Hrr25 requirement for full functionality of two different components of the translation machinery - wobble uridine-containing tRNAs [37, this work] and ribosomes [49], [57] - might reflect a role in regulation of the cell's capacity for protein synthesis. In addition, both hrr25 mutants [66] and Elongator-deficient yeast [17] are sensitive to hydroxyurea and methyl methanesulfonate. Since efficient translation of the ribonucleotide reductase gene RNR1 requires mcm5-modified tRNAs [62], [67], Elp1 phosphorylation may also be linked to the known role of Hrr25 in expression of genes such as RNR2 and RNR3 in response to DNA damage [66].
Alternatively, Elp1 phosphorylation might be dynamic, with sequential phosphorylation and dephosphorylation of specific residues driving the biochemical mechanism through which Elongator carries out the tRNA modification reactions. Such a dynamic role might operate through modulation of Elongator's interaction with factors such as Kti12 or with tRNA. It is intriguing in this context that the C-terminal phosphorylated region in Elp1 is immediately adjacent to a basic region that binds specifically to tRNA and that mutation of the tRNA binding domain leads to reduced interaction with Kti12 [Fig. 6C and ref. 32]. Thus phosphorylation of Elp1 could potentially influence how Elp1 interacts with tRNA, perhaps through interaction between the acidic phosphate groups and the basic residues present in the tRNA binding domain.
In conclusion, while there is still much to learn about Elp1 phosphorylation and its involvement in Elongator function, our work clearly demonstrates the importance of at least four in vivo phosphorylation sites in the C-terminal domain of Elp1 for Elongator-dependent tRNA wobble uridine modification, shows that Hrr25 kinase directly modifies two of these sites and provides evidence that phosphorylation regulates the association between Elongator and both its accessory protein Kti12 and its kinase Hrr25.
Materials and Methods
Yeast strains and plasmids
Basic yeast methods, growth media, and routine recombinant DNA methodology were performed as previously described [68], [69]. All plasmids used in this study are listed in Table S2 and yeast strains are listed in Table S3. To generate yeast strains dependent on wild-type or mutant forms of ELP1, ELP1 was first deleted from BY4741 and WAY008 (ELP3-TAP) using pFA6a-KanMX6 and pCORE-UH deletion cassettes to obtain the elp1Δ knockout mutant strains WAP034 and WAY037, respectively. Wild-type or mutant ELP1 variants carried on the low-copy plasmid YCplac111-ELP1-6HA (Table S2) were then introduced into these elp1Δ strains as the sole source of Elp1. The mutant versions were made by either site directed mutagenesis (Qiagen QuikChange) using YCplac111-ELP1-6HA as template, or by replacing the relevant region using standard cloning procedures and synthetic DNA carrying the desired mutations. The majority of plasmids made using the latter approach were produced by DNA2.0, Inc (Menlo Park, CA, USA). All elp1 mutants made by site-directed mutagenesis were verified by DNA sequencing of the entire ELP1 ORF plus approximately 200 base pairs upstream and downstream. When mutations were introduced by cloning, the region that had been replaced was sequenced to exclude the possibility of gene synthesis errors. The wild-type and mutant versions of YCplac111-ELP1-6HA were transformed into WAY034, WAY037, or elp1Δ::KanMX6 ELP2-myc3 strains carrying additional epitope-tagged components (Table S3) for phenotypic screening, Elongator-complex purification or Western blot analysis, respectively. pFA6a-CTAP4-HIS3MX6 was made by ligating the 1322 bp BglII-PmeI fragment from pFA6a-3HA-HIS3MX6 to the 3603 bp BglII-PmeI fragment from pFA6a-CTAP4-NatMX6 [70] such that the NatMX marker was replaced by HIS3MX. Using this template, a C-terminal TAP tag was added to ELP1 in RL-343-F0 and RL-343-F1 (Table S3) using standard one-step tagging methodology [71]. sit4Δ::LEU2 and kti12Δ::LEU2 knockouts were made as previously described [39], [43], as was introduction of elp1Δ::KanMX6 in strains used for co-immune precipitation with myc-tagged Elp2 [16]. Construction of ELP1-HA6::KlTRP1 utilized the pYM3 tagging plasmid and S2/S3 primer set described by Knop et al. [72].
Purification of TAP-tagged Elongator
For phosphorylation site mapping, Elongator complexes were purified from WAY009, WAY010, WAY011, WAY-H-P1T and WAY-Has-P1T (Table S3) by two-step tandem affinity purification of Elp1-TAP from 2-12 liter cultures in YPAD medium grown to OD600 1.0-1.5, as described previously [73]. All purifications were done in the presence of PhosSTOP and Complete protease inhibitor cocktail (Roche). Elongator complexes in which Elp1 contained phosphorylation site mutations were similarly purified from WAY037 (elp1Δ::pCORE-UH ELP3-TAP::HIS3MX: Table S3) harboring YCplac111-ELP1-6HA or its mutant derivatives (Table S2), growing cells as above but in SCD-Leu to select for retention of the plasmid.
Phosphorylation site mapping
Elongator protein preparations were digested in solution with Trypsin (Trypsin Gold, mass spectrometry grade; Promega V5280). The resulting peptides were cleaned over Hypersep C18 columns (Thermo Scientific) to remove buffer contaminants, eluting the peptides in 70% acetonitrile, 0.1% trifluoroacetic acid (TFA). Phosphopeptide enrichment was done in a two-step procedure using a Hypersep SCX column (Thermo Scientific) followed by TiO2 enrichment of mono-phosphorylated peptides from the SCX flow-through. SCX binding and washing buffer contained 10 mM KH2PO4, 25% acetonitrile (pH 3.0) and the elution buffer contained 10 mM KH2PO4, 25% acetonitrile, 350 mM KCl (pH 3.0). The flow-through was reduced in volume to ∼100 µl, supplemented with 100 µl 80% acetonitrile/2% TFA containing 200 mg/ml 2,5-dihydroxybenzoic acid (DHB), pH 2.0, then the sample applied to 5 micron Titansphere TiO2 beads (GL Sciences) and rotated gently at room temperature for 1 h. The beads were washed twice with 80% acetonitrile/2% TFA, 200 mg/ml DHB (pH 2.0), then 3 times with 80% acetonitrile/2%TFA (pH 2.0), before eluting with 60 µl 400 mM NH4OH (pH 11.0) and then supplementing with 2 µl of 100% formic acid. All phosphopeptide fractions were cleaned over C18 and submitted for mass spectrometry in 0.1% TFA.
Phosphopeptide fractions were analyzed by LC-MS/MS using a Dionex U300 system (Dionex California) with a PepMap C18 column coupled to either an Orbitrap XL or Orbitrap Velos (Thermo Fisher Scientific). Peptides were eluted using a 45 min 5%-90% acetonitrile gradient, sequencing the top 5 most intense ions with the following settings: CID, FTMS 335-1800 Da, 60,000 resolution, MS/MS charge state 1+ rejected,>2+ accepted. Peak picking, recalibration and peptide mass fingerprinting was done using MaxQuant software [74], [75], searching masses against the Saccharomyces Genome Database orf_trans_all database (January 5th 2010 release) with 10 ppm MS error, ≤ 2 missed cleavages, Trypsin/P enzyme, variable modifications set to Acetyl (protein N-term), Oxidation (M), Phospho (ST) and Phospho (Y) and a fixed modification of Carbomidomethyl (C). The MS/MS tolerance was set to 0.5 Da and the false discovery rate for site, protein and peptide identification was set to 0.01. All phosphorylation sites identified in this way were verified by hand annotating the spectra. The analysis was done on four biological replicates of the finally optimized SCX-TiO2 protocol.
Anti-Elp1 PhosphoSer-1209 antibody
An anti-Elp1 phosphoSer-1209 antibody was raised in a rabbit by BioGenes GmbH, using a peptide antigen (H2N-CTSTQE-pS-FFTRY-CO.NH2, where pS indicates phosphoserine) and their standard procedures (see http://www.biogenes-antibodies.com). The phosphospecific fraction of the final bleed was purified by serum depletion using immobilized non-phosphorylated peptide (H2N-CTSTQESFFTRY-CO.NH2), then phosphospecific antibodies were isolated from the depleted serum using immobilized phosphopeptide. The purified phosphospecific antibody fraction was tested for phosphospecificity by Western blotting, showing loss of signal when protein extracts were prepared from an elp1-1209A mutant and following competition with the phosphopeptide. The antibody was stored at -20°C in 50% (v/v) glycerol and used at a 1 in 3000 dilution. A small excess of the non-phosphorylated peptide was added to the primary dilution before use to prevent any cross-reaction with the non-phosphorylated epitope.
Phenotypic assays
To test the effect of extracellular zymocin on wild-type and mutant strains, killer-eclipse assays were performed as described previously [76] using the K. lactis zymocin producer strain NCYC1368 and YPD plates prepared using Kobe I agar (Roth 5210). To test the effect of intracellular expression of zymocin's γ subunit on growth, wild-type and mutant elp1 strains were transformed with pAE1, which expresses the γ subunit from the galactose-inducible GAL1 promoter [43]. The response to zymocin γ-toxin induction was monitored on galactose plates after 3-4 days at 30°C. SUP4 suppression efficiency was measured following integration of pSB3 in single copy at the his3Δ0 locus and then monitoring growth on SCD-Leu-Ura after 3 days at 30°C as described previously [32]. To test the effect of Hrr25 inhibition on Elongator function, strains dependent on w.t. or analog-sensitive Hrr25 were grown overnight in YPAD medium, diluted to 1.0 A600/ml and then 10-fold dilutions were spotted onto YPAD agar with or without 1%(v/v) zymocin and containing 10 µM 1NM-PP1 or an equivalent volume of DMSO as drug vehicle control. Growth was documented after 2 days growth at 30°C. Zymocin was prepared using cell-free culture medium from K. lactis NCYC1368 which had been grown at 30°C for 2 days in YPAD medium, by 50-fold concentration using Amicon Ultra-15 Centrifugal Filter Units (Millipore) followed by sterilization by filtration.
Relative quantitation of tRNA wobble uridine modifications by HPLC-DAD-MS/MS analysis
elp1Δ yeast strains containing YCplac111-ELP1-HA6 or its mutant derivatives were grown in SCD-Leu to select for the plasmid and then total tRNA was prepared by RNA extraction and LiCl precipitation as described previously [32]. Prior to LC-MS/MS analysis, 5 µg of each tRNA sample were digested into nucleosides by incubation at 37°C for 2 h in the presence of 1/10 volume of 10× nuclease P1 buffer (0.2 M NH4OAc pH 5.0, ZnCl2 0.2 mM), 0.3 U nuclease P1 (Sigma Aldrich, Munich, Germany) and 0.1 U snake venom phosphodiesterase (Worthington, Lakewood, USA). Next, 1/10 volume of 10× fast alkaline phosphatase buffer and 1 U fast alkaline phosphatase (Fermentas, St. Leon-Roth, Germany) were added, and samples were incubated for additional 60 min at 37°C. The digested tRNA samples were analyzed on an Agilent 1260 HPLC series equipped with a diode array detector (DAD) and a triple quadrupol mass spectrometer (Agilent 6460). A Synergy Fusion RP column (4 µm particle size, 80 Å pore size, 250 mm length, 2 mm inner diameter) from Phenomenex (Aschaffenburg, Germany) was used at 35°C column temperature. The solvents consisted of 5 mM ammonium acetate buffer adjusted to pH 5.3 using acetic acid (solvent A) and pure acetonitrile (solvent B). The elution was performed at a flow rate of 0.35 ml/min using a linear gradient from 0% to 8% solvent B at 10 min, 40% solvent B at 20 min and 0% solvent B at 23 min. For additional 7 min, the column was rinsed with 100% solvent A to restore the initial conditions. Prior to entering the mass spectrometer, the effluent from the column was measured photometrically at 254 nm by the DAD for detection of the 4 canonical nucleosides. The triple quadruple mass spectrometer, equipped with an electrospray ion source (Agilent Jet Stream), was run at the following ESI parameters: gas (N2) temperature 350°C, gas (N2) flow 8 L/min, nebulizer pressure 50 psi, sheath gas (N2) temperature 350°C, sheath gas (N2) flow 12 L/min and capillary voltage 3000 V. The MS was operated in positive ion mode using Agilent MassHunter software and modified nucleosides were monitored by multiple reaction monitoring (dynamic MRM mode). Identification of ncm5U and mcm5U peaks was performed as described previously [77]. Peak areas were determined employing Agilent MassHunter Qualitative Analysis Software. In the case of the major nucleosides, peak areas were extracted from the recorded UV chromatograms in order to avoid saturation of the mass signals. For inter-sample comparability of the detected modifications, the peak areas of the modified nucleosides were normalized to the UV peak area of uridine.
Western blotting and co-immune precipitation
Detection of tagged proteins used anti-TAP (Thermo Scientific, CAB1001), anti-myc and anti-HA antibodies (Roche) and was performed as previously described [16], [45]. Protein concentrations were determined using Quick StartTM Bradford Protein Assay (BioRAD) [78] and checked with anti-Pfk1 antibodies recognizing yeast Pfk1 (1 : 50,000, kindly provided by Dr. J. Heinisch) or anti-Cdc19 serum (1 : 10,000, kindly provided by Dr. J. Thorner) so as to ensure equivalent loadings. For detection of the Hrr25 kinase in total yeast extracts and in immune precipitates, a generic anti-Hrr25 antibody [64] was used (1 : 10,000 dilution). Antibody cross-linking to Dynabeads M-270 Epoxy (Invitogen), preparation of protein extracts and immune precipitation were performed according to the manufacturer's instruction and as described previously [16], [79]. In general, 1 µg of antibody coupled to Dynabeads was used per 1 mg total cell extract in B60 buffer.
Identification of protein kinases that phosphorylate Elp1
All 119 GST-protein kinases constructs in the library described by Zhu et al. [47] were expressed in yeast and purified as described previously [80]. Yeast Elongator complex was isolated as described in the main paper and used as a substrate for the GST-kinases, which were initially assayed in 23 pools of 5 kinases under similar conditions to those described below. Reactions were separated by SDS-PAGE and radiolabelled Elp1 detected by autoradiography. Eight out of the 23 pools showed phosphorylation occurred above the background level observed due to co-purification of Hrr25 with Elongator (see Results). From these eight pools, 40 individual kinases were individually screened for their ability to phosphorylate Elp1. Reactions in which Elongator was omitted controlled for any radiolabeled bands that co-migrated with Elp1 and were therefore due to kinase autophosphorylation or co-purified kinase substrates.
In vitro protein kinase assays
E. coli BL21 (DE3) pLysS (Novagen) was transformed with pTrcHis-HRR25 for overexpression and purification of His6-Hrr25 using HisPur cobalt resin (Thermo Scientific) according to the manufacturer's instructions. To analyze Elp1 phosphorylation in vitro, purified wild-type or mutant Elongator complex (1 µg) was incubated for 30 min at 30°C in 20 µl P-buffer (50 mM HEPES-KOH (pH 8.0), 5 mM MgCl2, 50 mM KCl) containing 100 µM ATP and 0.5 µCi of radiolabelled [γ-32P]ATP (3000 Ci/mmol), in the absence or presence of purified recombinant His6-Hrr25 (1 µg). Phosphorylation reactions were stopped by the addition of NuPAGE 4× LDS Sample Buffer (Life Technologies) and heated for 5 min at 95°C. Samples were run on a NuPAGE (4-12%) polyacrylamide/Bis-Tris SDS gel (Life Technologies) at 200 V for 1 h. Gels were stained with Bio-Safe Coomassie (BioRad), dried and subjected to autoradiography. To examine the effect of chemical inactivation of yeast Hrr25 on Elp1 phosphorylation in purified Elongator complex, Elongator was purified as above from strains WAY-H-P1T (wild-type HRR25) and WAY-Has-P1T (allele-sensitive hrr25 I82G) and assayed as described above in the presence or absence of 10 µM 3-MB-PP1 or 1-NM-PP1.
Time course phosphorylation reactions of synthetic peptides were carried out in 150 µl P-buffer containing 500 pmol synthetic peptide (Biomatik), 400 pmol recombinant His6-Hrr25, 1 mM ATP and 10 µCi [γ-32P]ATP (6000 Ci/mmol) and incubated at 30°C. Samples (5 µl) were collected at time intervals, spotted on Whatman Grade P81 Ion Exchange Cellulose Chromatography Paper, washed 3 times in 1% phosphoric acid, dried and quantitated by liquid scintillation counting [81]. To visualize phosphorylated peptides, 10 µl reactions were assembled at the above described stoichiometry, incubated for 30 min at 30°C and then terminated by addition of 4× SDS loading dye and heating at 95°C for 5 min. After separation by SDS-PAGE at 200 V using a NuPAGE 12% polyacrylamide/Bis-Tris gel (Life Technologies) with MES running buffer (50 mM MES, 50 mM Tris-base, 0.1% SDS, 1 mM EDTA, pH 7.3), phosphorylated peptides were visualized by autoradiography. For mass spectrometric analysis of phosphorylated peptides, 150 µl reactions were conducted as above but omitting the radiolabelled ATP.
Supporting Information
Zdroje
1. KroganNJ, KimM, AhnSH, ZhongG, KoborMS, et al. (2002) RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol Cell Biol 22 : 6979–6992.
2. OteroG, FellowsJ, LiY, de BizemontT, DiracAM, et al. (1999) Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol Cell 3 : 109–118.
3. ChenYT, HimsMM, ShettyRS, MullJ, LiuL, et al. (2009) Loss of mouse Ikbkap, a subunit of elongator, leads to transcriptional deficits and embryonic lethality that can be rescued by human IKBKAP. Mol Cell Biol 29 : 736–744.
4. SimpsonCL, LemmensR, MiskiewiczK, BroomWJ, HansenVK, et al. (2009) Variants of the elongator protein 3 (ELP3) gene are associated with motor neuron degeneration. Hum Mol Genet 18 : 472–481.
5. StrugLJ, ClarkeT, ChiangT, ChienM, BaskurtZ, et al. (2009) Centrotemporal sharp wave EEG trait in rolandic epilepsy maps to Elongator Protein Complex 4 (ELP4). Eur J Hum Genet 17 : 1171–1181.
6. ChenC, TuckS, BystromAS (2009) Defects in tRNA modification associated with neurological and developmental dysfunctions in Caenorhabditis elegans elongator mutants. PLoS Genet 5: e1000561.
7. SolingerJA, PaolinelliR, KlössH, ScorzaFB, MarchesiS, et al. (2010) The Caenorhabditis elegans Elongator complex regulates neuronal alpha-tubulin acetylation. PLoS Genet 6: e1000820.
8. NelissenH, FleuryD, BrunoL, RoblesP, De VeylderL, et al. (2005) The elongata mutants identify a functional Elongator complex in plants with a role in cell proliferation during organ growth. Proc Natl Acad Sci USA 102 : 7754–7759.
9. NguyenL, HumbertS, SaudouF, ChariotA (2010) Elongator - an emerging role in neurological disorders. Trends Mol Med 16 : 1–6.
10. TorresAG, BatlleE, Ribas de PouplanaL (2014) Role of tRNA modifications in human diseases. Trends Mol Med 20 : 306–314.
11. WittschiebenBO, OteroG, de BizemontT, FellowsJ, Erdjument-BromageH, et al. (1999) A novel histone acetyltransferase is an integral subunit of elongating RNA polymerase II holoenzyme. Mol Cell 4 : 123–128.
12. WinklerGS, KristjuhanA, Erdjument-BromageH, TempstP, SvejstrupJQ (2002) Elongator is a histone H3 and H4 acetyltransferase important for normal histone acetylation levels in vivo. Proc Natl Acad Sci USA 99 : 3517–3522.
13. CreppeC, MalinouskayaL, VolvertML, GillardM, CloseP, et al. (2009) Elongator controls the migration and differentiation of cortical neurons through acetylation of alpha-Tubulin. Cell 136 : 551–564.
14. MiskiewiczK, JoseLE, Bento-AbreuA, FislageM, TaesI, et al. (2011) ELP3 controls active zone morphology by acetylating the ELKS family member Bruchpilot. Neuron 72 : 776–788.
15. OkadaY, YamagataK, HongK, WakayamaT, ZhangY (2010) A role for the elongator complex in zygotic paternal genome demethylation. Nature 463 : 554–558.
16. FrohloffF, FichtnerL, JablonowskiD, BreunigKD, SchaffrathR (2001) Saccharomyces cerevisiae Elongator mutations confer resistance to the Kluyveromyces lactis zymocin. EMBO J 20 : 1993–2003.
17. LiQ, FazlyAM, ZhouH, HuangS, ZhangZ, et al. (2009) The Elongator complex interacts with PCNA and modulates transcriptional silencing and sensitivity to DNA damage agents. PLoS Genet 5: e1000684.
18. JablonowskiD, SchaffrathR (2007) Zymocin, a composite chitinase and tRNase killer toxin from yeast. Biochem Soc Trans 35 : 1533–1537.
19. RahlPB, ChenCZ, CollinsRN (2005) Elp1p, the yeast homolog of the FD disease syndrome protein, negatively regulates exocytosis independently of transcriptional elongation. Mol Cell 17 : 841–853.
20. HuangB, JohanssonMJ, BystromAS (2005) An early step in wobble uridine tRNA modification requires the Elongator complex. RNA 11 : 424–436.
21. MehlgartenC, JablonowskiD, WrackmeyerU, TschitschmannS, SondermannD, et al. (2010) Elongator function in tRNA wobble uridine modification is conserved between yeast and plants. Mol Microbiol 76 : 1082–1094.
22. LinF-J, ShenL, JangC-W, FalnesPØ, ZhangY (2013) Ikbkap/Elp1 Deficiency Causes Male Infertility by Disrupting Meiotic Progression. PLoS Genet 9: e1003516.
23. LuJ, HuangB, EsbergA, JohanssonMJ, BystromAS (2005) The Kluyveromyces lactis gamma-toxin targets tRNA anticodons. RNA 11 : 1648–1654.
24. JablonowskiD, ZinkS, MehlgartenC, DaumG, SchaffrathR (2006) tRNAGlu wobble uridine methylation by Trm9 identifies Elongator's key role for zymocin-induced cell death in yeast. Mol Microbiol 59 : 677–688.
25. BjorkGR, HuangB, PerssonOP, BystromAS (2007) A conserved modified wobble nucleoside (mcm5s2U) in lysyl-tRNA is required for viability in yeast. RNA 13 : 1245–1255.
26. AgrisPF (2008) Bringing order to translation: the contributions of transfer RNA anticodon-domain modifications. EMBO Rep 9 : 629–635.
27. JohanssonMJ, EsbergA, HuangB, BjorkGR, BystromAS (2008) Eukaryotic wobble uridine modifications promote a functionally redundant decoding system. Mol Cell Biol 28 : 3301–3312.
28. EsbergA, HuangB, JohanssonMJ, BystromAS (2006) Elevated levels of two tRNA species bypass the requirement for elongator complex in transcription and exocytosis. Mol Cell 24 : 139–148.
29. ChenC, HuangB, EliassonM, RydenP, BystromAS (2011) Elongator Complex Influences Telomeric Gene Silencing and DNA Damage Response by Its Role in Wobble Uridine tRNA Modification. PLoS Genet 7: e1002258.
30. RezguiVA, TyagiK, RanjanN, KonevegaAL, MittelstaetJ, et al. (2013) tRNA tKUUU, tQUUG, and tEUUC wobble position modifications fine-tune protein translation by promoting ribosome A-site binding. Proc Natl Acad Sci USA 110 : 12289–12294.
31. GlattS, LetoquartJ, FauxC, TaylorNM, SeraphinB, et al. (2012) The Elongator subcomplex Elp456 is a hexameric RecA-like ATPase. Nat Struct Mol Biol 19 : 314–320.
32. Di SantoR, BandauS, StarkMJR (2014) A conserved and essential basic region mediates tRNA binding to the Elp1 subunit of the Saccharomyces cerevisiae Elongator complex. Mol Microbiol 92 : 1227–1242.
33. ParaskevopoulouC, FairhurstSA, LoweDJ, BrickP, OnestiS (2006) The Elongator subunit Elp3 contains a Fe4S4 cluster and binds S-adenosylmethionine. Mol Microbiol 59 : 795–806.
34. GroveTL, RadleMI, KrebsC, BookerSJ (2011) Cfr and RlmN contain a single [4Fe-4S] cluster, which directs two distinct reactivities for S-adenosylmethionine: methyl transfer by SN2 displacement and radical generation. J Am Chem Soc 133 : 19586–19589.
35. SelvaduraiK, WangP, SeimetzJ, HuangRH (2014) Archaeal Elp3 catalyzes tRNA wobble uridine modification at C5 via a radical mechanism. Nat Chem Biol 10 : 810–812.
36. JablonowskiD, ButlerAR, FichtnerL, GardinerD, SchaffrathR, et al. (2001) Sit4p protein phosphatase is required for sensitivity of Saccharomyces cerevisiae to Kluyveromyces lactis zymocin. Genetics 159 : 1479–1489.
37. MehlgartenC, JablonowskiD, BreunigKD, StarkMJR, SchaffrathR (2009) Elongator function depends on antagonistic regulation by casein kinase Hrr25 and protein phosphatase Sit4. Mol Microbiol 73 : 869–881.
38. MehlgartenC, SchaffrathR (2003) Mutant casein kinase I (Hrr25p/Kti14p) abrogates the G1 cell cycle arrest induced by Kluyveromyces lactis zymocin in budding yeast. Mol Genet Genomics 269 : 188–196.
39. JablonowskiD, FichtnerL, StarkMJR, SchaffrathR (2004) The yeast elongator histone acetylase requires Sit4-dependent dephosphorylation for toxin-target capacity. Mol Biol Cell 15 : 1459–1469.
40. SoulardA, CremonesiA, MoesS, SchutzF, JenoP, et al. (2010) The rapamycin-sensitive phosphoproteome reveals that TOR controls protein kinase A toward some but not all substrates. Mol Biol Cell 21 : 3475–3486.
41. FukuchiS, HosodaK, HommaK, GojoboriT, NishikawaK (2011) Binary classification of protein molecules into intrinsically disordered and ordered segments. BMC Struc Biol 11 : 29.
42. TyanovaS, CoxJ, OlsenJ, MannM, FrishmanD (2013) Phosphorylation variation during the cell cycle scales with structural propensities of proteins. PLoS Comput Biol 9: e1002842.
43. ButlerAR, WhiteJH, FolawiyoY, EdlinA, GardinerD, et al. (1994) Two Saccharomyces cerevisiae genes which control sensitivity to G1 arrest induced by Kluyveromyces lactis toxin. Mol Cell Biol 14 : 6306–6316.
44. HuangB, LuJ, BystromAS (2008) A genome-wide screen identifies genes required for formation of the wobble nucleoside 5-methoxycarbonylmethyl-2-thiouridine in Saccharomyces cerevisiae. RNA 14 : 2183–2194.
45. FichtnerL, FrohloffF, BurknerK, LarsenM, BreunigKD, et al. (2002) Molecular analysis of KTI12/TOT4, a Saccharomyces cerevisiae gene required for Kluyveromyces lactis zymocin action. Mol Microbiol 43 : 783–791.
46. PetrakisTG, SogaardTM, Erdjument-BromageH, TempstP, SvejstrupJQ (2005) Physical and functional interaction between Elongator and the chromatin-associated Kti12 protein. J Biol Chem 280 : 19454–19460.
47. ZhuH, KlemicJF, ChangS, BertoneP, CasamayorA, et al. (2000) Analysis of yeast protein kinases using protein chips. Nat Genet 26 : 283–289.
48. GavinAC, BoscheM, KrauseR, GrandiP, MarziochM, et al. (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415 : 141–147.
49. SchäferT, MacoB, PetfalskiE, TollerveyD, BottcherB, et al. (2006) Hrr25-dependent phosphorylation state regulates organization of the pre-40S subunit. Nature 441 : 651–655.
50. WangX, HoekstraMF, DeMaggioAJ, DhillonN, VancuraA, et al. (1996) Prenylated isoforms of yeast casein kinase I, including the novel Yck3p, suppress the gcs1 blockage of cell proliferation from stationary phase. Mol Cell Biol 16 : 5375–5385.
51. BabuP, DeschenesRJ, RobinsonLC (2004) Akr1p-dependent palmitoylation of Yck2p yeast casein kinase 1 is necessary and sufficient for plasma membrane targeting. J Biol Chem 279 : 27138–27147.
52. RobinsonLC, MenoldMM, GarrettS, CulbertsonMR (1993) Casein kinase I-like protein kinases encoded by YCK1 and YCK2 are required for yeast morphogenesis. Mol Cell Biol 13 : 2870–2881.
53. BodenmillerB, WankaS, KraftC, UrbanJ, CampbellD, et al. (2010) Phosphoproteomic analysis reveals interconnected system-wide responses to perturbations of kinases and phosphatases in yeast. Sci Signal 3: rs4.
54. KennellyPJ, KrebsEG (1991) Consensus sequences as substrate specificity determinants for protein kinases and protein phosphatases. J Biol Chem 266 : 15555–15558.
55. CorbettKD, HarrisonSC (2012) Molecular architecture of the yeast monopolin complex. Cell Rep 1 : 583–589.
56. PfaffenwimmerT, ReiterW, BrachT, NogellovaV, PapinskiD, et al. (2014) Hrr25 kinase promotes selective autophagy by phosphorylating the cargo receptor Atg19. EMBO Rep 15 : 862–870.
57. RayP, BasuU, RayA, MajumdarR, DengH, et al. (2008) The Saccharomyces cerevisiae 60S ribosome biogenesis factor Tif6p is regulated by Hrr25p-mediated phosphorylation. J Biol Chem 283 : 9681–9691.
58. BauerF, MatsuyamaM, CandiracciJ, DieuM, ScheligaJ, et al. (2012) Translational control of cell division by elongator. Cell Rep 1 : 424–433.
59. ZinshteynB, GilbertWV (2013) Loss of a Conserved tRNA Anticodon Modification Perturbs Cellular Signaling. PLoS Genet 9: e1003675.
60. DedonPC, BegleyTJ (2014) A system of RNA modifications and biased codon use controls cellular stress response at the level of translation. Chem Res Toxicol 27 : 330–337.
61. ChanCT, DyavaiahM, DeMottMS, TaghizadehK, DedonPC, et al. (2010) A quantitative systems approach reveals dynamic control of tRNA modifications during cellular stress. PLoS Genet 6: e1001247.
62. PatilA, DyavaiahM, JosephF, RooneyJP, ChanCT, et al. (2012) Increased tRNA modification and gene-specific codon usage regulate cell cycle progression during the DNA damage response. Cell Cycle 11 : 3656–3665.
63. CyertMS (2003) Calcineurin signaling in Saccharomyces cerevisiae: how yeast go crazy in response to stress. Biochem Biophys Res Commun 311 : 1143–1150.
64. KatisVL, LippJJ, ImreR, BogdanovaA, OkazE, et al. (2010) Rec8 phosphorylation by casein kinase 1 and Cdc7-Dbf4 kinase regulates cohesin cleavage by separase during meiosis. Devel Cell 18 : 397–409.
65. TanakaC, TanLJ, MochidaK, KirisakoH, KoizumiM, et al. (2014) Hrr25 triggers selective autophagy-related pathways by phosphorylating receptor proteins. J Cell Biol 207 : 91–105.
66. HoY, MasonS, KobayashiR, HoekstraM, AndrewsB (1997) Role of the casein kinase I isoform, Hrr25, and the cell cycle-regulatory transcription factor, SBF, in the transcriptional response to DNA damage in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 94 : 581–586.
67. BegleyU, DyavaiahM, PatilA, RooneyJP, DiRenzoD, et al. (2007) Trm9-Catalyzed tRNA Modifications link translation to the DNA damage response. Mol Cell 28 : 860–870.
68. Amberg DC, Burke D, Strathern JN (2005) Methods in yeast genetics: a Cold Spring Harbor Laboratory course manual. New York: Cold Spring Harbor Laboratory Press.
69. Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual: Cold Spring Harbor Laboratory Press.
70. Van DriesscheB, TafforeauL, HentgesP, CarrAM, VandenhauteJ (2005) Additional vectors for PCR-based gene tagging in Saccharomyces cerevisiae and Schizosaccharomyces pombe using nourseothricin resistance. Yeast 22 : 1061–1068.
71. LongtineMS, McKenzieA3rd, DemariniDJ, ShahNG, WachA, et al. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14 : 953–961.
72. KnopM, SiegersK, PereiraG, ZachariaeW, WinsorB, et al. (1999) Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15 : 963–972.
73. Séraphin B, Puig O, Bouveret E, Rutz B, Caspary F (2002) Tandem affinity purification to enhance interacting protein identification. In: Golemis E, editor. Protein-protein interactions: a molecular cloning manual. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press. pp.313–328.
74. GietzRD, SuginoA (1988) New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74 : 527–534.
75. BerbenG, DumontJ, GilliquetV, BollePA, HilgerF (1991) The YDp plasmids: a uniform set of vectors bearing versatile gene disruption cassettes for Saccharomyces cerevisiae. Yeast 7 : 475–477.
76. KishidaM, TokunagaM, KatayoseY, YajimaH, Kawamura-WatabeA, et al. (1996) Isolation and genetic characterization of pGKL killer-insensitive mutants (iki) from Saccharomyces cerevisiae. Biosci Biotechnol Biochem 60 : 798–801.
77. KellnerS, NeumannJ, RosenkranzD, LebedevaS, KettingRF, et al. (2014) Profiling of RNA modifications by multiplexed stable isotope labelling. Chem Commun 50 : 3516–3518.
78. BradfordMM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72 : 248–254.
79. ZachariaeW, ShinTH, GalovaM, ObermaierB, NasmythK (1996) Identification of subunits of the anaphase-promoting complex of Saccharomyces cerevisiae. Science 274 : 1201–1204.
80. SutherlandCM, HawleySA, McCartneyRR, LeechA, StarkMJR, et al. (2003) Elm1p is one of three upstream kinases for the Saccharomyces cerevisiae SNF1 complex. Curr Biol 13 : 1299–1305.
81. Hardie DG, Haystead TAJ, Salt IP, Davies SP (1999) Assay and purification of protein–serine/threonine kinases. In: Hardie DG, editor. Protein Phosphorylation: a Practical Approach. 2nd edition. Oxford: Oxford University Press. pp.201–220.
82. BuchanDW, MinneciF, NugentTC, BrysonK, JonesDT (2013) Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res 41: W349–W357.
83. NotredameC, HigginsDG, HeringaJ (2000) T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol 302 : 205–217.
84. FichtnerL, JablonowskiD, SchierhornA, KitamotoHK, StarkMJR, et al. (2003) Elongator's toxin-target (TOT) function is nuclear localization sequence dependent and suppressed by post-translational modification. Mol Microbiol 49 : 1297–1307.
Štítky
Genetika Reprodukční medicína
Článek Naturally Occurring Differences in CENH3 Affect Chromosome Segregation in Zygotic Mitosis of HybridsČlánek Insight in Genome-Wide Association of Metabolite Quantitative Traits by Exome Sequence AnalysesČlánek ALIX and ESCRT-III Coordinately Control Cytokinetic Abscission during Germline Stem Cell DivisionČlánek Deciphering the Genetic Programme Triggering Timely and Spatially-Regulated Chitin Deposition
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 1
-
Všechny články tohoto čísla
- The Combination of Random Mutagenesis and Sequencing Highlight the Role of Unexpected Genes in an Intractable Organism
- Ataxin-3, DNA Damage Repair, and SCA3 Cerebellar Degeneration: On the Path to Parsimony?
- α-Actinin-3: Why Gene Loss Is an Evolutionary Gain
- Origins of Context-Dependent Gene Repression by Capicua
- Transposable Elements Contribute to Activation of Maize Genes in Response to Abiotic Stress
- No Evidence for Association of Autism with Rare Heterozygous Point Mutations in Contactin-Associated Protein-Like 2 (), or in Other Contactin-Associated Proteins or Contactins
- Nur1 Dephosphorylation Confers Positive Feedback to Mitotic Exit Phosphatase Activation in Budding Yeast
- A Regulatory Hierarchy Controls the Dynamic Transcriptional Response to Extreme Oxidative Stress in Archaea
- Genetic Variants Modulating CRIPTO Serum Levels Identified by Genome-Wide Association Study in Cilento Isolates
- Small RNA Sequences Support a Host Genome Origin of Satellite RNA
- Phosphorylation of Elp1 by Hrr25 Is Required for Elongator-Dependent tRNA Modification in Yeast
- Genetic Mapping of MAPK-Mediated Complex Traits Across
- An AP Endonuclease Functions in Active DNA Demethylation and Gene Imprinting in
- Developmental Regulation of the Origin Recognition Complex
- End of the Beginning: Elongation and Termination Features of Alternative Modes of Chromosomal Replication Initiation in Bacteria
- Naturally Occurring Differences in CENH3 Affect Chromosome Segregation in Zygotic Mitosis of Hybrids
- Imputation of the Rare G84E Mutation and Cancer Risk in a Large Population-Based Cohort
- Polycomb Protein SCML2 Associates with USP7 and Counteracts Histone H2A Ubiquitination in the XY Chromatin during Male Meiosis
- A Genetic Strategy for Probing the Functional Diversity of Magnetosome Formation
- Interactions of Chromatin Context, Binding Site Sequence Content, and Sequence Evolution in Stress-Induced p53 Occupancy and Transactivation
- The Yeast La Related Protein Slf1p Is a Key Activator of Translation during the Oxidative Stress Response
- Integrative Analysis of DNA Methylation and Gene Expression Data Identifies as a Key Regulator of COPD
- Proteasomes, Sir2, and Hxk2 Form an Interconnected Aging Network That Impinges on the AMPK/Snf1-Regulated Transcriptional Repressor Mig1
- Functional Interplay between the 53BP1-Ortholog Rad9 and the Mre11 Complex Regulates Resection, End-Tethering and Repair of a Double-Strand Break
- Estrogenic Exposure Alters the Spermatogonial Stem Cells in the Developing Testis, Permanently Reducing Crossover Levels in the Adult
- Protein Poly(ADP-ribosyl)ation Regulates Immune Gene Expression and Defense Responses
- Sumoylation Influences DNA Break Repair Partly by Increasing the Solubility of a Conserved End Resection Protein
- A Discrete Transition Zone Organizes the Topological and Regulatory Autonomy of the Adjacent and Genes
- Elevated Mutation Rate during Meiosis in
- The Intersection of the Extrinsic Hedgehog and WNT/Wingless Signals with the Intrinsic Hox Code Underpins Branching Pattern and Tube Shape Diversity in the Airways
- MiR-24 Is Required for Hematopoietic Differentiation of Mouse Embryonic Stem Cells
- Tissue-Specific Effects of Genetic and Epigenetic Variation on Gene Regulation and Splicing
- Heterologous Aggregates Promote Prion Appearance via More than One Mechanism
- The Tumor Suppressor BCL7B Functions in the Wnt Signaling Pathway
- , A -Acting Locus that Controls Chromosome-Wide Replication Timing and Stability of Human Chromosome 15
- Regulating Maf1 Expression and Its Expanding Biological Functions
- A Polyubiquitin Chain Reaction: Parkin Recruitment to Damaged Mitochondria
- RecFOR Is Not Required for Pneumococcal Transformation but Together with XerS for Resolution of Chromosome Dimers Frequently Formed in the Process
- An Intracellular Transcriptomic Atlas of the Giant Coenocyte
- Insight in Genome-Wide Association of Metabolite Quantitative Traits by Exome Sequence Analyses
- The Role of the Mammalian DNA End-processing Enzyme Polynucleotide Kinase 3’-Phosphatase in Spinocerebellar Ataxia Type 3 Pathogenesis
- The Global Regulatory Architecture of Transcription during the Cell Cycle
- Identification and Functional Characterization of Coding Variants Influencing Glycemic Traits Define an Effector Transcript at the Locus
- Altered Ca Kinetics Associated with α-Actinin-3 Deficiency May Explain Positive Selection for Null Allele in Human Evolution
- Genetic Variation in the Nuclear and Organellar Genomes Modulates Stochastic Variation in the Metabolome, Growth, and Defense
- PRDM9 Drives Evolutionary Erosion of Hotspots in through Haplotype-Specific Initiation of Meiotic Recombination
- Transcriptional Control of an Essential Ribozyme in Reveals an Ancient Evolutionary Divide in Animals
- ALIX and ESCRT-III Coordinately Control Cytokinetic Abscission during Germline Stem Cell Division
- Century-scale Methylome Stability in a Recently Diverged Lineage
- A Re-examination of the Selection of the Sensory Organ Precursor of the Bristle Sensilla of
- Antagonistic Cross-Regulation between Sox9 and Sox10 Controls an Anti-tumorigenic Program in Melanoma
- A Dependent Pool of Phosphatidylinositol 4,5 Bisphosphate (PIP) Is Required for G-Protein Coupled Signal Transduction in Photoreceptors
- Deciphering the Genetic Programme Triggering Timely and Spatially-Regulated Chitin Deposition
- Aberrant Gene Expression in Humans
- Fascin1-Dependent Filopodia are Required for Directional Migration of a Subset of Neural Crest Cells
- The SWI2/SNF2 Chromatin Remodeler BRAHMA Regulates Polycomb Function during Vegetative Development and Directly Activates the Flowering Repressor Gene
- Evolutionary Constraint and Disease Associations of Post-Translational Modification Sites in Human Genomes
- A Truncated NLR Protein, TIR-NBS2, Is Required for Activated Defense Responses in the Mutant
- The Genetic and Mechanistic Basis for Variation in Gene Regulation
- Inactivation of PNKP by Mutant ATXN3 Triggers Apoptosis by Activating the DNA Damage-Response Pathway in SCA3
- DNA Damage Response Factors from Diverse Pathways, Including DNA Crosslink Repair, Mediate Alternative End Joining
- hnRNP K Coordinates Transcriptional Silencing by SETDB1 in Embryonic Stem Cells
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The Global Regulatory Architecture of Transcription during the Cell Cycle
- A Truncated NLR Protein, TIR-NBS2, Is Required for Activated Defense Responses in the Mutant
- Proteasomes, Sir2, and Hxk2 Form an Interconnected Aging Network That Impinges on the AMPK/Snf1-Regulated Transcriptional Repressor Mig1
- The SWI2/SNF2 Chromatin Remodeler BRAHMA Regulates Polycomb Function during Vegetative Development and Directly Activates the Flowering Repressor Gene
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



