-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaRRP6L1 and RRP6L2 Function in Silencing Regulation of Antisense RNA Synthesis
Arabidopsis FLOWERING LOCUS C (FLC) delays flowering; therefore, repressing expression of FLC provides a critical mechanism to regulate flowering. This mechanism involves multiple levels of regulation, including genetic regulation by transcription factors, and epigenetic regulation by modifications of genomic DNA and histones at the FLC locus. This work examines the role of non-coding RNAs in the epigenetic regulation of FLC, finding that the different RNAs produced from the FLC locus may have different functions in altering the epigenetic landscape at the FLC locus, and revealing that AtRRP6L1 and AtRRP6L2, two components of the exosome, an RNA-processing complex, play roles in regulating these non-coding RNAs. Therefore, this work explores the complex ties between RNA processing, non-coding RNAs, and epigenetic regulation of FLC, a key repressor of flowering.
Published in the journal: . PLoS Genet 10(9): e32767. doi:10.1371/journal.pgen.1004612
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004612Summary
Arabidopsis FLOWERING LOCUS C (FLC) delays flowering; therefore, repressing expression of FLC provides a critical mechanism to regulate flowering. This mechanism involves multiple levels of regulation, including genetic regulation by transcription factors, and epigenetic regulation by modifications of genomic DNA and histones at the FLC locus. This work examines the role of non-coding RNAs in the epigenetic regulation of FLC, finding that the different RNAs produced from the FLC locus may have different functions in altering the epigenetic landscape at the FLC locus, and revealing that AtRRP6L1 and AtRRP6L2, two components of the exosome, an RNA-processing complex, play roles in regulating these non-coding RNAs. Therefore, this work explores the complex ties between RNA processing, non-coding RNAs, and epigenetic regulation of FLC, a key repressor of flowering.
Introduction
The regulation of gene silencing occurs at multiple levels and non-coding RNAs (ncRNAs) have emerged as important regulators of genome silencing at the transcriptional and posttranscriptional levels [1]. At the chromatin structure level, chromatin remodeling factors and DNA - and histone-modifying enzymes alter chromatin to control its structure and compaction, thus affecting silencing [2]. ncRNAs significantly contribute to the regulation of chromatin structure and play important roles in eukaryotic genomes by affecting the epigenetic architecture, including both establishment and maintenance of epigenetic marks. ncRNAs can initiate heterochromatin formation through the RNA interference (RNAi) pathway, or independently of RNAi, and through the RNA processing machinery [3], [4].
The exosome is an evolutionarily conserved complex of RNase-like and RNA binding proteins involved in 3′ to 5′ decay and processing of various RNA substrates [5]–[10]. The exosome complex plays an important role in regulating both coding and ncRNAs [11]. The nuclear and cytoplasmic forms of the eukaryotic exosome complex share ten common subunits [12]. In most organisms, all nine subunits of the exosome core complex are inactive, and enzymatic activities are provided by the tenth catalytic RRP44 subunit, a 3′-5′ hydrolytic exoribinuclease, which also has endonucleolytic activity [13].
The nuclear form of eukaryotic exosome also associates with the second catalytic RRP6 subunit, a substoichiometric, nuclear-specific, 3′ to 5′ exoribonuclease [14]–[16]. The RRP6 subunit has a number of unique functions in the exosome [14], and also additional functions not associated with the exosome core [17]–[19]. Arabidopsis has three possible functional homologs of RRP6: the nuclear proteins AtRRP6L1 and AtRRP6L2, and the cytoplasmic protein AtRRP6L3 [20]. When we purified the exosome complex from Arabidopsis, AtRRP6L proteins were not detected in our experiments, which is likely due to the fact that, as a substoichiometric subunit restricted to a nuclear form, it was underrepresented in our preparations [8]. Therefore, whether these Arabidopsis RRP6Ls physically associate with the exosome core remains to be tested.
The exosome complex broadly affects epigenetic silencing of heterochromatic and euchromatic loci by regulating a variety of ncRNAs [11], [21]–[24]. In fission yeast (Schizosaccharomyces pombe), the exosome acts in several different small RNA (smRNA) pathways to affect constitutive and facultative heterochromatin silencing, in either RNAi-dependent or RNAi-independent manners [4], [25]–[27]. The exosome also acts in gene silencing through RNA quantity and quality surveillance, and in collaboration with the 3′ termination machinery [21], [22], [28]–[30].
Our previous genome-wide survey revealed that many exosome targets in Arabidopsis correspond to ncRNAs, many originating from heterochromatic loci, suggesting that the exosome participates in various silencing pathways in Arabidopsis [8]. Our recent analysis of exosome functions in smRNA-mediated silencing of genes in Arabidopsis showed that the exosome has little effect on the smRNAs that function in the main silencing mechanisms, siRNA-dependent methylation of DNA (RdDM). Rather, we showed that the exosome associates physically with long, polyadenylated RNAs transcribed from the scaffold regions of several heterochromatic loci, and exosome defects affected the level of histone H3K9me2, an epigenetic mark that alters chromatin structure [31]. We also found that the Arabidopsis rrp6 homologues AtRRP6L1 and AtRRP6L2 participate in these epigenetic mechanisms and may function redundantly [31]. With the exception of AtCSL4, the genes encoding the Arabidopsis exosome core complex subunits are essential for viability [8]. By contrast, unlike the core exosome subunits, the exosome nuclear catalytic subunit RRP6 and the Arabidopsis RRP6-Like proteins are not essential for viability [14], [20]; thus, the rrp6 and rrp6l mutants provide tools to study the role of the exosome during development.
Epigenetic regulation by long non-coding RNAs (lncRNAs) and histone modifications plays a key role in controlling the expression of Arabidopsis FLC (FLOWERING LOCUS C), which encodes a MADS-box transcription factor that suppresses flowering [32]–[35].
FLC regulation through chromatin modifications has been well studied [36], [37]. FLC expression requires H3K4 methylation, a permissive chromatin modification, on FLC chromatin, and H3K4 demethylation leads to FLC repression [37]–[39]. FLC silencing requires Polycomb complex 2 (PRC2) -deposited H3K27me3, a repressive chromatin modification [36], [40], [41]. Repressing the expression of FLC provides a central mechanism for both the vernalization pathway, which regulates flowering time in response to periods of prolonged cold, and the autonomous pathway, which regulates flowering independently of environmental signals [37].
The processing and metabolism of lncRNAs play a crucial role in FLC silencing, and different lncRNAs produced from the FLC locus may have distinct functions. For example, lncRNAs COOLAIR and COLDAIR participate in the epigenetic silencing of the FLC locus [40], [42]. The COOLAIR antisense transcript is produced from the FLC locus as two alternatively polyadenylated isoforms, AS I and AS II, and it was shown that the processing of AS I and II function in FLC epigenetic silencing by affecting H3K4 demethylation [37], [38], [42]. COOLAIR transcription does not appear to be required for vernalization, but it has been implicated in FLC repression early during cold treatment, possibly mediated by direct effects on the FLC promoter [37]. By contrast, the COLDAIR sense transcript is produced from within FLC intron 1, and plays a role in FLC silencing via recruitment of Polycomb repressing complex 2 (PRC2) to FLC during vernalization [40]. The establishment of H3K27 trimethylation to silence FLC in vernalization requires COLDAIR [40], but not AS I and AS II [41], [43]. Also, in the autonomous pathway, the proteins involved in 3′-end RNA processing act as the main factors in FLC silencing [38], [39], [44]–[46].
Here, we report that mutations of AtRRP6L1 and AtRRP6L2 result in delayed flowering in early-flowering Arabidopsis ecotypes that do not require vernalization for flowering. We found that AtRRP6L1 and AtRRP6L2 epigenetically regulate FLC silencing by regulating different antisense transcripts and modulating H3K4me3 and H3K27me3 histone modifications at the FLC locus. Moreover, we discovered a novel antisense transcript, termed Antisense Long (ASL), which originates from the FLC locus in wild type plants and is regulated by AtRRP6L1 and AtRRP6L2. Our study demonstrates that Arabidopsis RRP6L proteins play an important role in the regulation of genes expressed in specific developmental phases via participating in lncRNA-mediated epigenetic silencing.
Results
Mutations in AtRRP6L1 and AtRRP6L2 result in late flowering, caused by de-repression of FLC expression
We previously examined T-DNA mutations in Arabidopsis RRP6L1 and RRP6L2 and found that these mutations lead to de-repression of known heterochromatic loci and that RRP6L1 and 2 likely function redundantly in this process [31]. The T-DNA insertion allele of AtRRP6L1 was isolated from the Wisconsin population of T-DNA mutants [31], in the Wasilevskaya (Ws) ecotype, and the T-DNA insertion allele of AtRRP6L2 comes from the SALK collection, in the Columbia (Col-0) ecotype [31]. We previously used RT-PCR analysis to demonstrate that rrp6l1-3 mutant is a null allele and rrp6l2-3 is nearly null [31].
To control for the ecotype, we examined the phenotypes of rrp6l1-3, rrp6l2-3, and rrp6l1-3 rrp6l2-3 double mutants (hereafter rrp6l1/2) and compared them to wild-type plants of Col-0 and Ws ecotypes. When we examined the phenotype of different rrp6l1 and rrp6l2 alleles, we found that these single mutants did not show significant phenotypic alterations, although they exhibited a mild delay in flowering, as measured by leaf number at bolting (Fig. 1A). By contrast, the late-flowering phenotype becomes pronounced in rrp6l1/2 double mutants grown under long day conditions (Fig. 1A) and becomes very severe in plants grown under short day conditions (Fig. 1B). These findings indicate that AtRRP6L1 and AtRRP6L2 likely have redundant functions in the pathways activating flowering. The flowering defects in plants grown under short and long day conditions suggest that AtRRP6L1 and AtRRP6L2 function in the regulation of flowering through a pathway different from the photoperiod pathway, which promotes flowering in response to day length. The vernalization and autonomous pathways promote flowering through repression of FLC expression. During vernalization, cold temperatures repress FLC expression, and FRIGIDA (FRI) activates FLC expression [35]. Ecotypes that lack functional alleles of FRI, such as the early-flowering accessions Col-0 and Ws ecotypes used in this study, do not require vernalization for flowering. Thus, our data suggest that AtRRP6L1and AtRRP6L2 could be involved in regulation of FLC expression through the autonomous flowering pathway.
Fig. 1. The rrp6l1-3 and rrp6l2-3 mutants affect flowering time and gene expression. 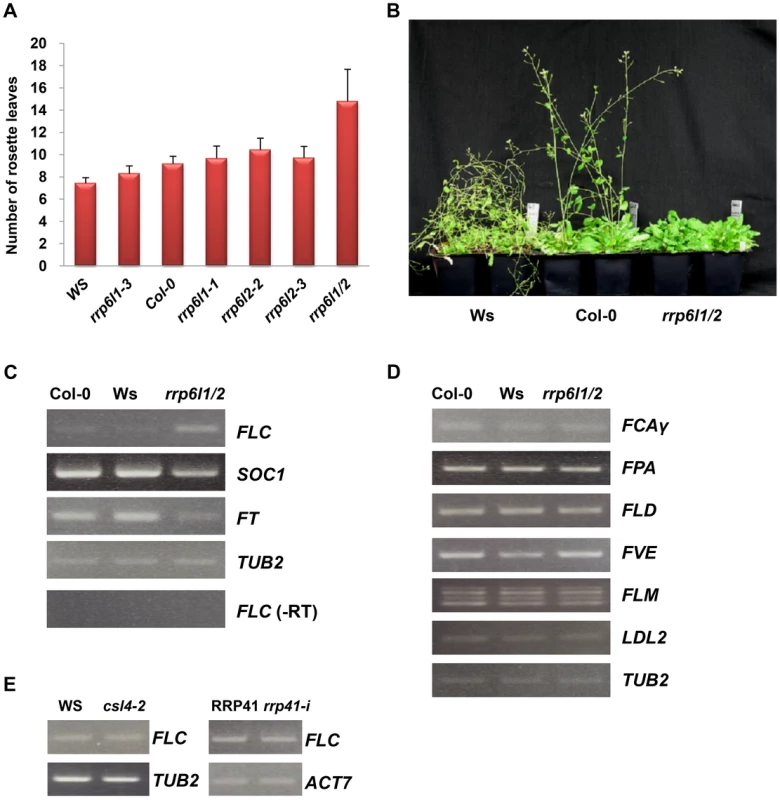
(A) The late-flowering phenotype of rrp6l1 and rrp6l2 mutants grown under long day conditions. Flowering time was measured as rosette leaf number at bolting. To control for effects of ecotype, the phenotype of mutants was compared to wild-type plants of Col-0 and Ws ecotypes. Error bars represent standard deviation (SD). (B) The late-flowering phenotype of rrp6l1-3 rrp6l2-3 mutants grown under short day conditions. 66-day-old plants are shown. (C) Effect of rrp6l1-3 and rrp6l2-3 mutations on the expression of the FLC, SOC1 and FT. RT-PCR showed that the rrp6l1-3 rrp6l2-3 double mutant (rrp6l1/2), has increased expression of FLC and decreased expression of SOC1 and FT, which act downstream of FLC. (-RT) is the no reverse transcriptase control. (D) The expression of FCA, FPA, FLD, FVE, FLM, and LDL2, genes involved in regulation of flowering time in the autonomous flowering pathway, is not affected in rrp6l1/2 mutants. (E) FLC expression is not affected in AtCSL4-2 T-DNA mutant and RRP41 iRNAi line. RRP41 corresponds to the iRNAi line grown without estradiol, and rrp41-i corresponds to line grown on estradiol-containing medium, to induce the RNAi-mediated knockdown of RRP41. TUBULIN 2 and ACTIN 7 were used as loading controls. To examine the roles of AtRRP6L1and AtRRP6L2 in regulation of FLC expression, we examined whether mutations of AtRRP6Ls affect the expression of FLC. We observed no change in FLC expression in the rrp6l1-3 and rrp6l2-3 single mutants, consistent with the degree of the observed phenotypic alterations, and we confirmed this observation with additional AtRRP6L1 and AtRRP6L2 T-DNA alleles (Fig. S1B). By contrast, we observed an increase in the levels of FLC transcript in the rrp6l1/2 double mutant relative to wild-type plants (Fig. 1C). These results suggest that AtRRP6L1 and AtRRP6L2 affect FLC expression and function redundantly in this process. To make sure that this phenotype does not result from transgressive segregation, we constructed AtRRP6Ls mutants using different AtRRP6L alleles isolated from the same Col-0 background and confirmed that this mutant combination causes a similar delay in flowering and derepression of FLC (Fig. S1A).
FLC acts as a dosage-dependent floral repressor [47]. The exosome complex and RRP6 subunits affect the processing and turnover of various RNAs, and thus regulate RNA quality and quantity [5], [8], [11]. To find out whether the FLC transcript, which increased in abundance in rrp6l1/2 mutants, corresponds to a functional transcript rather than nonfunctional byproduct, we examined the expression of the flowering genes SUPPRESSOR OF CONSTANS OVEREXPRESSION 1 (SOC1) and FLOWERING LOCUS T (FT), which act downstream of FLC. We found that the rrp6l1/2 mutants showed lower levels of both SOC1 and FT transcripts (Fig. 1C), indicating that the increased level of FLC transcript in rrp6l1/2 mutants corresponds to a functional FLC transcript, and the increase in FLC expression enhances the repression of the downstream genes. In addition to increased levels of FLC transcript, we also detected increased amounts of the unspliced FLC RNA in rrp6l1/2 mutants (Fig. 2B).
Fig. 2. The effect of the rrp6l1-3 and rrp6l2-3 mutations on expression of FLC sense and antisense transcripts, and on H3K4 methylation. 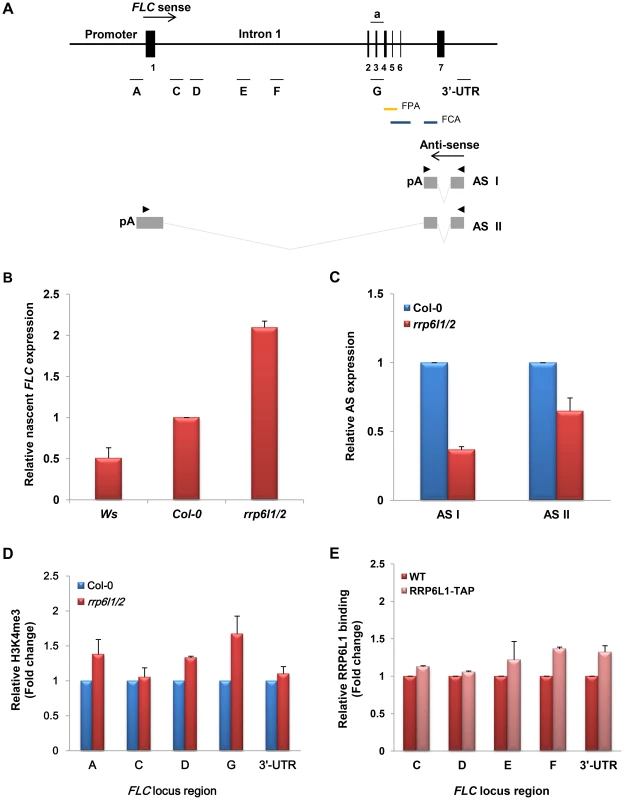
(A) The diagram of the FLC gene based on analysis of the transcription unit [38] Vertical bars and numbers denote the exons of the FLC sense transcript. The transcription start site of the FLC sense mRNA is indicated by an arrow. Two antisense transcripts, AS I (proximal) and AS II (distal), are depicted below the FLC diagram. Antisense transcripts are alternatively polyadenylated, with a proximal poly(A) site in sense intron 6 and a distal poly(A) site in the sense promoter region. Grey boxes correspond to AS I and II exons, and grey lines correspond to the spliced regions of the antisense RNAs. Horizontal bars (letters a, A to G and 3′-untranslated region, 3′UTR), correspond to the FLC regions used in qRT-PCR and ChIP. Arrowheads correspond to position of primers used for RT-PCR amplification of AS I and II. Blue bars correspond FCA binding region, yellow bar correspond to the FPA binding region. (B) Nascent FLC expression significantly increased in rrp6l1/2 mutants. Expression of FLC was measured by qRT-PCR and is shown relative to FLC expression in Col-0 and Ws wild type plants; (C) Expression of AS I and AS II transcripts in rrp6l1/2 mutants. Expression of AS I and AS II in rrp6l1/2 was compared to their expression in Col-0 wild type plants by qRT-PCR. The antisense transcript levels were normalized by total antisense transcript as described previously [38]. Error bars represent standard deviation (SD). (D) Effect of rrp6l1-3 and rrp6l2-3 mutations on the level of H3K4me3 examined by ChIP using antibodies against H3K4me3 in the various regions of FLC. The level of H3K4me3 in rrp6l1/2 mutants was normalized to the level of H3K4me3 in wild type Col-0 plants. (E) AtRRP6L1 protein physically interacts with the FLC locus. ChIP assays were done using the rrp6l1-3 mutant complemented by a functional AtRRP6L1-TAP transgene. AtRRP6L1-TAP recruitment was normalized to wild type Ws, the background of the rrp6l1-3 mutant. The error bars in ChIP experiments represent the standard error of the mean and correspond to the difference between 2 biological replicates. The autonomous pathway constitutively represses FLC, independent of environmental inputs [35]. To find out whether AtRRP6L1 and AtRRP6L2 affect FLC expression directly or by regulating the expression of the upstream genes that silence FLC in the autonomous pathway, we examined the expression of upstream genes in the AtRRP6L1 and AtRRP6L2 mutants. We found that the rrp6l1/2 plants showed no changes in expression of the genes that act upstream of FLC (Fig. 1D). These data imply that the de-repression of FLC observed in AtRRP6L mutants results from a direct effect of AtRRP6Ls on the expression of FLC, not from regulation of the genes acting upstream of FLC.
To test whether the effect of rrp6l1/2 on FLC expression requires the core exosome complex, we next examined AtCSL4-2 and AtRRP41, which encode core exosome complex subunits. We did not observe de-repression of FLC transcription in an AtRRP41 inducible RNAi line or in AtCSL4-2 T-DNA insertion mutants (Fig. 1E), suggesting that AtRRP6L1 and AtRRP6L2 likely function independently of the exosome in the regulation of FLC expression.
Mutations of AtRRP6Ls affect expression of the antisense transcripts that regulate expression of FLC
Antisense transcripts regulate FLC silencing and antisense expression appears to independently intersect with both the vernalization and autonomous pathways to repress FLC expression [37]. During the vegetative phase, the FLC locus produces two alternatively spliced, polyadenylated regulatory antisense (AS) transcripts, AS I and AS II [37] (Fig. 2A). Targeted 3′ end processing of these antisense transcripts affects the recruitment of histone chromatin remodelers to the locus, which results in reduced FLC transcription [38], [39], [46]. Therefore, we investigated whether the rrp6l1/2 mutants showed changes in the ratio of 3′ end processing and polyadenylation of these antisense transcripts. We found that, compared to wild type plants, the rrp6l1 or rrp6l2 single mutants, and the rrp6l1/2 double mutants had lower levels of processed AS I and II transcripts (Fig. 2C and Fig. S1C); also, the rrp6l1/2 plants showed reduced levels of AS I and II, consistent with the stronger phenotype of the rrp6l1/2 mutants (Fig. 1A and S1C). Interestingly, the pattern of down-regulation of AS I and II transcripts in rrp6l1/2 mutants was similar to the pattern observed in the mutants of cleavage stimulation factors CstF64 and CstF77, components of the cleavage polyadenylation machinery required for the 3′-end processing of AS I and II transcripts [38]. RRP6 plays an important role in formation of the 3′ ends of many RNAs in yeast and humans [14], [48], [49], and in budding yeast, also participates in the regulation of antisense transcripts derived from the PHO84 locus [21], [50]. Therefore, it is possible that AtRRP6L1 and AtRRP6L2 proteins, along with CstF, could participate in the 3′ end processing of both antisense transcripts. To further investigate the relationship between CstF64 and AtRRP6Ls, we attempted to construct a triple rrp6l1-3 rrp6l2-3 cstf64-2 mutant; however, since cstf64-2 mutants are sterile, we could not obtain the triple homozygous mutant. Taken together, our data suggest that AtRRP6L proteins could negatively regulate FLC expression by affecting the expression of the regulatory antisense transcripts.
Chromatin structure of the FLC locus is altered in rrp6l1/2 plants
Methylated H3K4 marks active chromatin states and the antisense transcripts synthesized from FLC may function in FLC silencing by recruiting chromatin remodeling factors that drive H3K4 demethylation [38], [39]. To investigate whether the decrease in the antisense transcripts in the rrp6l1/2 mutants leads to changes in histone modifications at the FLC locus, we used chromatin immunoprecipitation (ChIP) to analyze the levels of H3K4me3 at various regions of the FLC locus (Fig. 2A). We found that the rrp6l1/2 mutant had significantly increased levels of H3K4me3 along the entire length of FLC, compared with wild type (Fig. 2D). These data suggest that the decrease in the level of antisense transcripts in rrp6l1/2 mutants might lead to the decreased recruitment of chromatin remodeling factors required for H3K4 demethylation, thereby regulating the accessibility of the transcription machinery to the locus, and in turn leading to FLC de-silencing, similar to previously reported observations [38].
AtRRP6L1 directly interacts with the FLC locus
The expression of FLC sense and antisense transcripts together with the increased level of H3K4 trimethylation in rrp6l1/2 mutants suggested that AtRRP6s could participate in FLC transcriptional silencing by affecting the chromatin structure at the FLC locus. Therefore, we asked whether AtRRP6L proteins can directly interact with the FLC locus to participate in the silencing pathway. To address this question, we constructed transgenic rrp6l1-3 lines that were complemented by a wild type copy of AtRRP6L1 fused with the TAP-tag for affinity purification, AtRRP6L1-TAP (see Methods).
We then used ChIP on these lines to examine the association of AtRRP6L1 protein with several regions of FLC (Fig. 2A). ChIP showed a modest enrichment of AtRRP6L1 protein in the regions corresponding to the 3′-UTR and intron 1 of FLC (Fig. 2E). The AtRRP6L1 binding region within intron 1 appears to be further downstream of the 3′-end of the AS I transcript (with respect to the direction of AS I and II transcription), implying that AtRRP6L1 may bind in this region to process a longer antisense precursor transcript. The AtRRP6L1 binding regions do not overlap with the regions reported to be bound by FPA and FCA proteins, RNA-binding 3′-end processing factors required for the processing of the AS I transcript [39], [46]. However, FPA binds to the region between exon 4 and 5 (Fig. 2A), which is also downstream of the 3′-end region of AS I [46]; thus, AtRRP6L1 and FPA have somewhat similar, but not identical, binding patterns (Fig. 2A and E). The association of AtRRP6L1 protein with a larger region of the FLC locus (downstream of the 3′end of AS I) also suggests that AtRRP6L proteins might participate in the processing of different types of antisense RNAs, in addition to the known antisense transcripts derived from the FLC locus. Alternatively, AtRRP6L could also participate in co-transcriptional regulation of antisense transcription by binding to nascent antisense transcripts. Also, the level of AtRRP6L1 enrichment at the FLC locus was relatively modest (Fig. 2E). Thus, we cannot rule out the possibility that AtRRP6L proteins could associate with the locus by binding other protein and RNA complexes that physically interact with the locus.
A novel antisense transcript in the FLC locus
When we were examining the pattern of known antisense transcripts produced from the FLC locus, we observed the presence of a different antisense transcript in wild type plants, but not in rrp6l1/2 plants. Therefore, we set out to investigate the features of this antisense transcript by tiling RT-PCR using a set of primers that cover the entire FLC locus (Fig. S2A). We found that the transcript is a novel antisense RNA of over 2000 nucleotides in length. The sequence of this antisense RNA, which we termed ASL (Antisense Long), corresponds to intron 1 and the 3′-UTR region of the sense FLC transcript (Fig. 3A). Interestingly, 5′ region of the ASL transcript overlaps with the 5′ region of the AS I and II transcripts. Sequencing of ASL revealed that it has two different isoforms, 2,236 nt and 2,536 nt (ASLa and b, respectively), produced by alternative splicing (Fig. 3A). Moreover, ASL spans intron 1, an important region for maintenance of FLC silencing [51]. Notably, ASL also overlaps with the COLDAIR lncRNA, which is transcribed within intron 1 in the sense direction during vernalization [40] (Fig. 3A).
Fig. 3. Characterization of the ASL transcript. 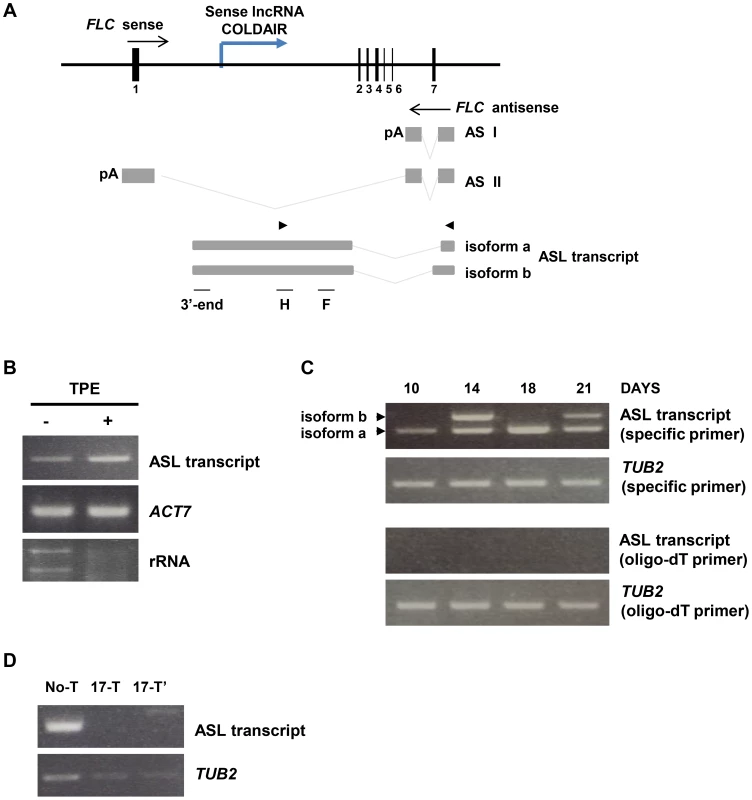
(A) The diagram of the FLC gene and ASL RNA produced from the FLC locus in wild type plants. Gray boxes depict antisense transcript exons and gray lines indicate spliced regions. Alternatively spliced antisense transcript isoforms found by tiling RT-PCR. Arrowheads indicate position of primers used for RT-PCR amplification of ASL. F, H and 3′-end indicate that the regions used for RT-PCR amplification after RNA-IP (in Fig. 5A and B). The blue line depicts the COLDAIR transcript. (B) The ASL transcript is capped at the 5′end. RNA samples of Col-0 plants were treated with Terminator 5′-Phosphate-Dependent Exonuclease (TPE), which degrades uncapped RNA. (−) corresponds to the RNA sample before TPE treatment and (+) corresponds to the TPE treated sample. ACTIN 7 and rRNA were used as capped and un-capped controls for TPE treatment, respectively. (C) Expression of ASL in Col-0 plants at 10, 14, 18 and 21 days after germination of vegetative phase. Arrowheads indicate the two alternatively spliced isoforms of ASL. cDNA was synthesized using either antisense RNA specific primers or oligo-dT primers. TUBULIN 2 was used as an internal control and was amplified from the sample reverse transcribed with either tubulin 2-specific primers (second row) or the oligo-dT primers (bottom row). (D) α-amanitin treatment of Col-0 (11 days seedlings). No-T corresponds to non-treated plants, and 17-T corresponds to plants treated with α-amanitin for 17 hours. 17-T and 17-T′ indicate independent biological replicates. To determine whether the ASL RNA has a 5′ cap, we used Terminator 5′-Phosphate-Dependent Exonuclease (TPE), which degrades uncapped RNA. We found that TPE treatment did not affect ASL levels, indicating that ASL has a 5′ cap (Fig. 3B). Next, we examined the 3′ end of ASL by performing cDNA synthesis primed by either sequence specific primer or oligo-dT primers. To our surprise, we found that ASL is not polyadenylated, as we detected ASL only from the cDNA primed by specific primers, not by oligo-dT (Fig. 3C).
In plants, the RNA polymerases RNA Pol IV and Pol V [52] participate in gene silencing through smRNA-mediated mechanisms [53]–[55]. To investigate which RNA polymerase synthesizes ASL, we treated plants with α-amanitin, an inhibitor of RNA Pol II, and then used RT-PCR to examine ASL levels. We did not detect ASL in plants treated with α-amanitin (Fig. 3D), implying that RNA Pol II synthesizes ASL. To confirm this, we also examined the presence of ASL in nrpd1 (Pol IV) and nrpe1 (Pol V) mutants; we detected ASL in these mutants, indicating that Pol IV and V do not affect ASL, although nrpd1 mutants showed a minor change in ASL levels (Fig. S2B).
Taken together, our data indicate that ASL is capped, synthesized by RNA Pol II and non-polyadenylated, and also suggest that it is distinct from the known antisense transcripts originating from the FLC locus. The tiling RT-PCR analysis indicates that the same promoter produces ASL, AS I, and AS II. Thus, it is possible that the ASL transcript could function differently from AS I and II in the silencing of FLC. Indeed, different antisense RNAs transcribed from same promoter of the human pseudogene PTENpg1 have different functions in transcriptional and post-transcriptional silencing of the tumor suppressor gene PTEN [56].
AtRRP6L1 and AtRRP6L2 regulate ASL levels
Next, we investigated how the AtRRP6Ls participate in the regulation of ASL expression. We found that the level of ASL transcript decreased in rrp6l1-3 and rrp6l2-3 single mutants (Fig. 4A and Fig. S2C). Moreover, we detected little or no ASL transcript in rrp6l1/2 double mutants (Fig. 4A), indicating that AtRRP6L proteins function as the main factors regulating the levels of the ASL transcript. Consistent with AtRRP6L functions in regulation of ASL expression, we observed that the level of the ASL transcript recovered to wild type levels in rrp6l1-3 mutant complemented by the wild type copy of AtRRP6L1-TAP (Fig. 4B). These data suggest that both AtRRP6L1 and AtRRP6L2 directly regulate the expression of ASL and are the main factors in this process.
Fig. 4. The level of ASL transcripts is decreased in rrp6l mutants. 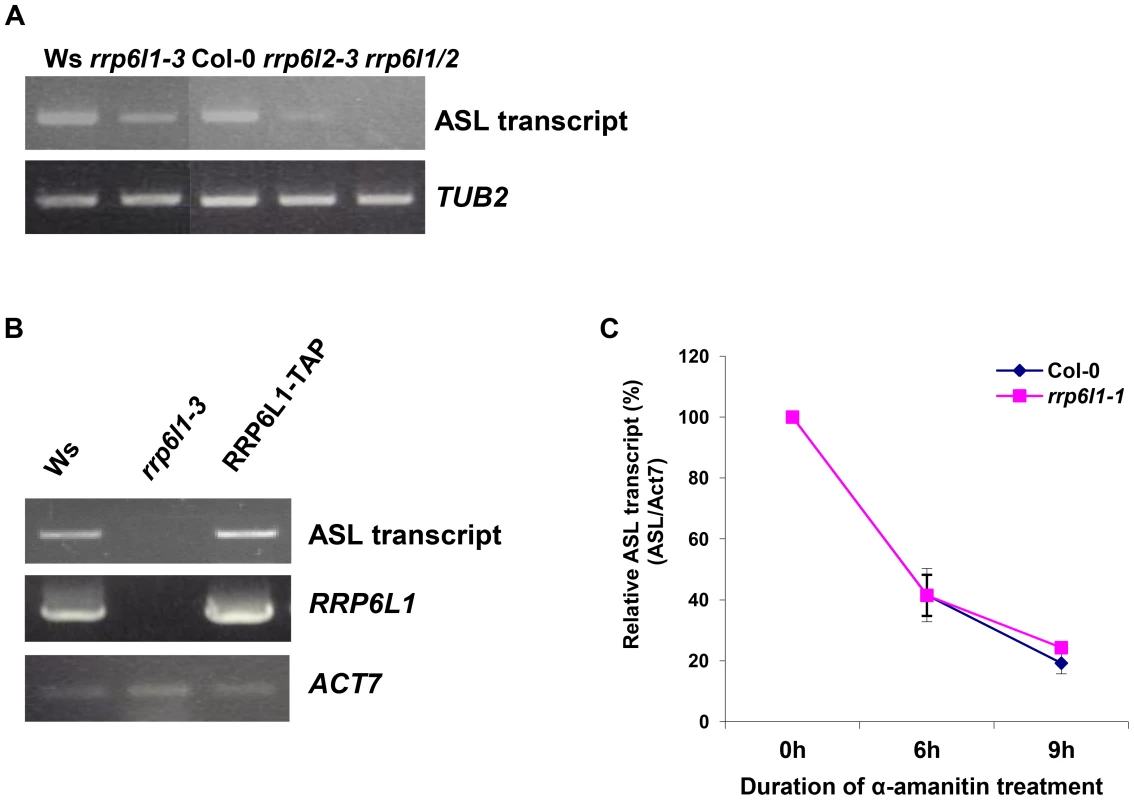
(A) Expression of ASL RNA in rrp6l1-3, rrp6l2-3 and rrp6l1/2 double mutants. (B) The level of ASL transcript expression is restored in the rrp6l1-3 mutant complemented by functional AtRRP6L1-TAP transgene. ACTIN 7 was used as a loading control. Transgenic plants are in Col-0 ecotype. (C) Decay rate of ASL transcript in rrp6l1-1 mutants. α-amanitin treatment was performed for 0, 6 and 9 hours. The expression of antisense transcript was normalized to ACTIN 7. RRP6 is a 3′ - 5′ exoribonuclease and RRP6 defects usually result in abnormal accumulation of various RNAs due to failure to degrade or process them [21], [49], [50], [57]. We next asked how AtRRP6Ls could regulate ASL levels. The rrp6l1/2 mutants showed nearly undetectably low levels of ASL. We reasoned that, if AtRRP6Ls regulate the stability of the ASL transcript, then we would observe a difference in ASL decay rate in AtRRP6L single mutants. Therefore, we used the rrp6l1-1 single mutant and conducted an α-amanitin chase to compare the rates of ASL transcript decay in rrp6l1-1 and wild type plants. We found that the rrp6l1-1 mutant and wild type had similar rates of ASL transcript decay (Fig. 4C), suggesting that AtRRP6L proteins do not directly participate in the degradation of the ASL transcript but rather affect its production or biogenesis.
AtRRP6L1 interacts with the ASL transcript, which physically associates with H3K27me3 regions of FLC
To find out whether AtRRP6L1 could play a direct role in the expression of ASL, we examined if AtRRP6L1 protein physically associates with the ASL transcript. To this end, we conducted RNA immunoprecipitation (RNA-IP) in wild type plants using antibodies against AtRRP6L1 protein (Fig. 5A). The RNA-IP showed that AtRRP6L1 protein physically associates with the ASL transcript (Fig. 5A). We also obtained identical results from RNA-IP in the rrp6l1-3 mutant complemented with AtRRP6L1-TAP (Fig. 5B). Together these results indicate that AtRRP6L1 protein likely participates directly in the regulation of ASL transcript levels.
Fig. 5. ASL transcript associates with AtRRP6L1 protein and H3K27 trimethylated regions and rrp6l1/2 mutants have decreased H3K27me3 and nucleosome density. 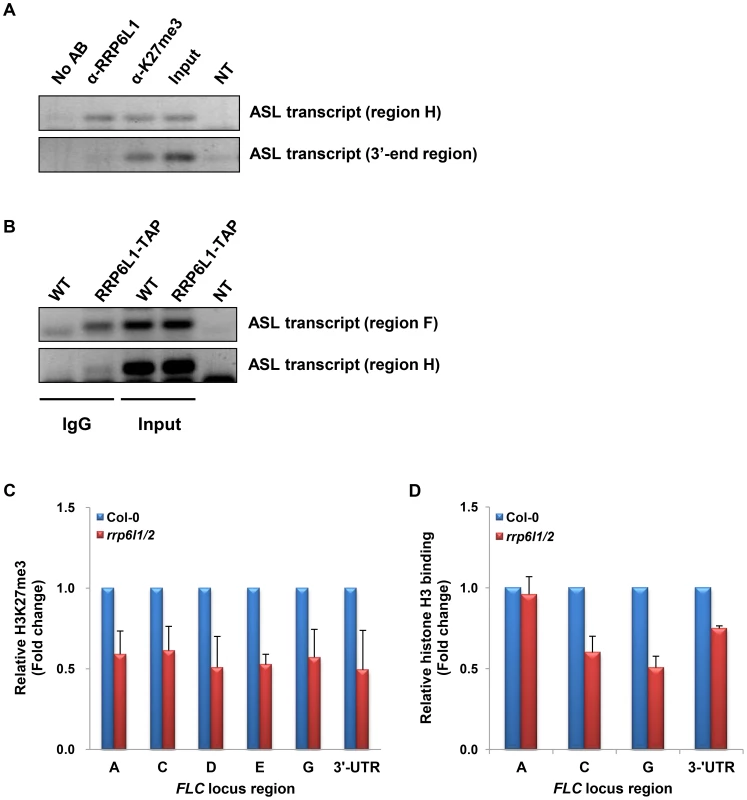
(A) ASL transcript directly associates with AtRRP6L1 protein and H3K27me3 regions of FLC. RNA-IP was performed using anti-AtRRP6L1 and anti-H3K27me3 antibodies to precipitate ASL RNA from wild type Col-0 plants. The regions used in RT-PCR (region H and the 3′end of ASL) are shown on Fig. 3A. No AB indicates no antibodies and is the negative control for RNA-IP. NT indicates no template and is the negative control for RT-PCR. (B) ASL transcript directly associates with AtRRP6L1 in rrp6l1-3 mutants complemented by functional AtRRP6L1-TAP transgene. Transgenic plants are in the Ws ecotype and Ws was used as negative control for RNA-IP. RNA-IP was performed using IgG antibodies to co-precipitate ASL RNA with RP6L1-TAP from wild type plants complemented with a transgene expressing RRP6L1-TAP. NT indicates no template and is the negative control for RT-PCR. (C) The level of H3K27me3 is decreased in rrp6l1/2 mutants. ChIP assays were performed using H3K27me3 antibodies. The level of H3K27me3 in rrp6l1/2 mutants was plotted relative to the level of H3K27me3 in Col-0 plants. The error bars in ChIP experiments represent the standard error of the mean and correspond to the difference between 2 biological replicates. (D) Nucleosome density is decreased in rrp6l1/2 mutants. Mnase-ChIP assays were performed using histone H3 antibodies. Nucleosomal density in rrp6l1/2 mutants was plotted relative to the level of nucleosomal density in Col-0 plants. The error bars in ChIP experiments represent the standard error of the mean and correspond to the difference between 2 biological replicates. We then hypothesized that the ASL RNA may play a role distinct from that of AS I and II. Histone remodeling factors affect FLC silencing and several lncRNAs affect H3K4 demethylation and H3K27 trimethylation [41]. The sense lncRNA COLDAIR participates in recruiting PRC2 and is necessary for the establishment of H3K27 trimethylation during vernalization [40], but this does not require AS I and II [41], [43]. However, H3K4 demethylation in the autonomous pathway does require AS I and II [37]–[39]. COLDAIR may also contribute to the maintenance of H3K27me3 during vegetative growth. In addition, the H3K27me3-binding protein LHP1 functions in the maintenance of H3K27me3 during the vegetative phase in actively dividing cells, suggesting that H3K27me3 maintenance could be important for FLC silencing during the vegetative phase after H3K27me3 has been established [58].
To examine whether ASL has a role distinct from that of AS I and II, we examined the level of the repressive histone mark, H3K27me3 at the FLC locus in rrp6l1/2 mutants. To our surprise, we observed that the rrp6l1/2 mutants showed significantly decreased levels of H3K27me3 along the entire FLC locus (Fig. 5C). The rrp6l1-3 single mutant also showed a mild decrease in the level of H3K27me3 (Fig. S1D). This observation is consistent with the very mild phenotype and the decreased level of the ASL transcript observed in rrp6l1 single mutants (Fig. 1A, 4A and S2C). Together, our data indicate that the knock-out of both AtRRP6L1 and AtRRP6L2 affected the levels of both H3K4me3 and H3K27me3 in the FLC locus (Fig. 3B). This is in contrast to AS I and II, which affect only the levels of H3K4me3 at FLC, at least in vernalization pathway [37], [41], [43].
The level of H3K27me3 correlates with the nucleosomal density [59], [60]. To examine whether the reduction in the level of H3K27me3 in rrp6l/2 mutants affects nucleosome positioning, we performed Micrococcal Nuclease (MNase)-ChIP assays using anti-H3 antibodies. MNase degrades nucleosome-free regions, allowing the estimation of nucleosome density. We found a lower nucleosomal density at the FLC locus in rrp6l1/2 mutants (Fig. 5D). These data indicate that the reduction of H3K27me3 levels observed in the rrp6l1/2 mutants could also result in relaxation of the chromatin state, which then allows factors involved in FLC transcription to gain easier access to the locus. Taken together, our results suggest that the regulation of the ASL transcript by AtRRP6L proteins may contribute to the maintenance of H3K27me3 at the FLC locus, which in turn contributes to the compact chromatin structure of the locus.
The rrp6l1/2 mutations lead to a decrease of H3K27me3 levels and also affect the nucleosome density at the FLC locus (Fig. 5C and D). Therefore, we asked whether the ASL transcript could be directly involved in H3K27 trimethylation, a role similar to that played by the COLDAIR lncRNA. To answer this question, we performed RNA-IP using antibodies against H3K27me3. We found that the ASL transcript physically associates with H3K27me3 regions (Fig. 5A). Taken together, our data suggest that the ASL transcript could function in the maintenance of H3K27 trimethylation during the vegetative phase.
Discussion
Here, we report that AtRRP6L1 and AtRRP6L2, the homologues of rrp6 subunits of the exosome in other organisms, negatively regulate the expression of FLC to promote flowering in Arabidopsis, and act redundantly in this process. This finding provides the first evidence that plant RRP6 proteins participate in the regulation of a specific developmental process. Our data indicate that AtRRP6L proteins play a role in the regulation of antisense RNA production and the chromatin landscape of the FLC locus. Moreover, we identified a novel antisense transcript produced from the FLC locus in wild type plants and found that AtRRP6L proteins regulate the expression of this transcript. ASL appears to be distinct from the previously described FLC antisense transcripts, AS I and II, and our data suggest that it could function in the regulation of levels of H3K27me3 at the FLC locus.
The regulation of lncRNAs could link AtRRP6L functions in development to epigenetic mechanisms
For most exosome complex subunits, mutations cause a lethal phenotype, which indicates that, for most organisms, development requires exosome-mediated regulation of diverse RNAs [8], [61]. Unlike exosome core subunits, RRP6 and AtRRP6-Like proteins are not essential for viability [14], [20]. Most studies of exosome-dependent and exosome-independent RRP6 functions in developmental processes have been performed in fission and budding yeast [18], [21], [24], [49], [57], [62]. In these systems, RRP6 participates in facultative gene silencing and also regulates the transition from mitosis to meiosis through RNA-mediated epigenetic mechanisms [24], [57]. Similar to these findings, we observed that AtRRP6L1 and AtRRP6L2 function in regulation of flowering time by repressing FLC expression. The defect in the AtRRP6L proteins results in mis-regulation of antisense RNA production from the FLC locus and affects the level of histone modification of the locus.
AS I and II transcripts function in FLC silencing in both vernalization and autonomous flowering pathways [37], possibly by affecting the level of H3K4 demethylation [38], [39], although the exact mechanism remains unknown. A decrease in the levels of processed AS I and II in mutants of CstF64 and CstF77, proteins involved in 3′-end processing, leads to increased levels of H3K4 trimethylation and subsequent de-repression of FLC [38]. Thus, AtRRP6Ls may contribute to 3′-end processing of the antisense RNAs similarly to the CstFs; indeed, 3′-end processing of various RNAs is one of the well-known functions of the exosome complex. The exosome processes a number of structural RNAs including rRNA, snRNA and snoRNA via trimming the 3′-ends of their precursors [63], [64]. Furthermore, RRP6 acts together with the Nrd1-Nab3 termination complex in budding yeast in non-canonical 3′-end processing and termination of the antisense RNA derived from PHO84, as well as processing of several other mRNAs [49], [50]. Disruption of the Nrd1-exosome pathway leads to de-repression of reporter genes integrated into heterochromatic regions and results in alteration of chromatin structure at specific loci and heterochromatic regions [11], [21], [22]. The exosome function in processing of mRNA and antisense lncRNA is likely to be conserved in plants, suggesting that AtRRP6L proteins may participate in regulating synthesis of the antisense RNAs derived from the FLC locus and this regulation could be important to maintain a repressive chromatin state for silencing of FLC.
Surprisingly, we found that the defect of AtRRP6Ls caused a reduction of the level of H3K27 trimethylation, which has not been reported in studies of AS I and II in the vernalization and autonomous flowering pathways [38], [43]. A different intronic sense lncRNA, COLDAIR, derived from intron 1 of FLC, physically associates with the PHD-PRC2 complex to establish H3K27 trimethylation during vernalization [40]. Moreover, the AS I and II transcripts are not required for PcG-mediated silencing via regulation of H3K27me3 trimethylation in the vernalization pathway [41], [43]. Together, these data suggest that other lncRNAs, not AS I and II, function in regulating H3K27 trimethylation.
AtRRP6Ls might affect the level of H3K27me3 indirectly, by regulating H3K4 demethylation through regulating either AS I and II, or ASL transcripts, via a mechanism similar to the interplay between Trithorax and Polycomb groups, which antagonistically regulate the levels of H3K4me3 and H3K27me3 at the FLC locus during vernalization [40], [41]. Alternatively, AtRRP6Ls may regulate the level of H3K27me3 directly via an unknown mechanism that functions independently of the silencing pathway involving AS I and II. Indeed, ARABIDOPSIS TRITHORAX 1 (ATX1), an Arabidopsis homolog of Trithorax 1, dynamically regulates activation of FLC through trimethylation but not dimethylation of H3K4 and atx1 mutations led to the loss of H3K4me3 and gain of H3K27me2 during the vegetative phase, but did not affect H3K4me2 and H3K27me3 [65]. This indicates that the regulation of the levels of H3K4me3 and H3K27me3 at the FLC locus could be independent of each other, at least during the vegetative phase. Together, the previous reports and our data suggest that the decrease of H3K27me3 in rrp6l1/2 mutants could be caused by the decrease in ASL RNA expression, and AtRRP6Ls may participate in the respective pathways for FLC silencing through regulating the expression of AS I, AS II, and ASL RNAs.
We identified ASL, a novel, long antisense RNA that is distinct from the previously-described antisense RNAs. We observed that the ASL transcript physically associates with H3K27me3, suggesting that it could play a role in H3K27 trimethylation and function differently than AS I and II RNAs. The PRC2 complex binds ncRNAs with high affinity but does not recognize specific sequences, while its binding affinity correlates with the length of the RNA [66], [67]. Thus, it is possible that ASL transcript could also participate in recruitment of the PRC2 complex to the FLC locus, which leads to maintenance of H3K27 trimethylation during vegetative growth. Taken together, the previous reports and our data suggest that the regulation of chromatin structure via various lncRNAs is a central mechanism in FLC silencing, and different lncRNAs may function in different chromatin modification pathways. The AtRRP6L proteins may play a role in silencing pathways by regulating antisense transcription.
ASL expression is regulated mostly by AtRRP6L1 and AtRRP6L2
ASL has several features in common with AS I and AS II. First, ASL is transcribed by RNA Pol II; second, it is alternatively spliced, existing in 2 isoforms; third, its transcription is driven by the same promoter that drives AS I and AS II; fourth, the 5′ part of ASL overlaps with the 5′ region of AS I and II. However, in contrast to AS I and II, the ASL transcript is long (over 2,000 nucleotides long), is non-polyadenylated, and extends into intron 1 of FLC. These differences suggest that ASL may have functions distinct from the functions of AS I and II in FLC silencing, and the mechanism of FLC silencing could be more complicated than previously thought.
RNA Pol II transcribes ASL. Various non-polyadenylated Pol II RNAs, such as snRNA, snoRNAs, and some mRNAs, are processed by the exosome, which is recruited by the Nrd1-Nab3-Sen1 termination complex in the noncanonical 3′ end-processing pathway [22], [49], [50], [63], [68]. Thus, the noncanonical 3′ end-processing pathway may also participate in processing of ASL, if this pathway is conserved in plants. However, Arabidopsis homologs of Nrd1, Nab3 and Sen1 have not yet been characterized.
The rrp6l1-3 and rrp6l2-3 single mutants showed decreased levels of ASL, and ASL was almost undetectable in the rrp6l1/2 double mutant. Based on the results of the α-amanitin chase experiments, the rrp6l1-1 mutation does not affect the stability of ASL, suggesting that AtRRP6L1 and AtRRP6L2 could be the main regulators of ASL synthesis. This finding is very intriguing, since defects in the exosome and RRP6 usually lead to abnormal accumulation of various RNAs due to failures of RNA degradation or processing [21], [22], [49], [50], [57]. This result may be caused by an unknown function of AtRRP6L proteins, which participate in either the synthesis or biogenesis of ASL, rather than in its degradation. Indeed, we previously reported that the expression of a number of loci decreased in AtRRP4 and AtRRP41 inducible RNAi plants and the AtCSL4-2 mutant [8], and many of these loci are located within euchromatic regions as well as in regions harboring H3K27me3 (unpublished data). Therefore, the exosome and AtRRP6Ls may function in regulation of RNA synthesis, different from their conventional functions in RNA degradation. Similarly, inactivation of the human homologue of RRP6 leads to dramatically reduced levels of Xist ncRNA involved in X-chromosome inactivation, although it remains to be seen whether this effect is direct [69].
In our study, we found that AtRRP6L1 protein physically associates with the ASL transcript, suggesting that AtRRP6L1 plays a direct role in regulation of ASL levels, likely through ASL synthesis rather than degradation. In addition, recent work demonstrated that another 5′-3′ exoribonuclease, Xrn1, also directly contributes to RNA synthesis of several mRNAs in budding yeast, by physically associating with chromatin and contributing to transcription elongation [70]. Alternatively, the decrease in ASL levels in the AtRRP6L mutants may indicate that different RNA decay proteins participate in degradation of these RNAs.
We found that the level of FLC transcript was unaffected in the exosome core subunit mutants, AtRRP4 and AtRRP41 inducible RNAi lines and AtCSL4-2 T-DNA mutants, suggesting that the function of AtRRP6L1 and AtRRP6L2 in regulation of FLC expression could be independent of the exosome complex. This could also indicate that the exosome core complex is not necessary for metabolism of RNAs produced from the FLC locus and different RNA decay factors could participate in their degradation. For example, in yeast, the XRN family of 5′ to 3′ exoribonucleases works in both the nucleus and cytoplasm, and has diverse functions in RNA metabolism [71], including in the degradation of XRN1-sensitive unstable antisense RNAs [72].
Possible mechanism of AtRRP6L protein function in FLC silencing
Silencing of FLC is regulated mainly through histone modifications rather than DNA methylation [73]. We previously reported that the exosome complex and AtRRP6L proteins function in DNA methylation-independent silencing and affect the histone modification pathway in some heterochromatic loci in Arabidopsis [31]. Along with our previous findings, regulation of FLC silencing mainly by histone modifications suggested that the FLC locus could be one of the targets of the AtRRP6L proteins. In accord, we observed that defects in AtRRP6L proteins caused a decrease in antisense RNAs, resulting in the alteration of histone modifications and de-repression of FLC.
It is intriguing to speculate that AtRRP6L proteins may have dual functions in FLC silencing via regulation of antisense transcription, which means that AtRRP6L proteins could participate in 2 different pathways, one involved in H3K4 demethylation and the other involved in H3K27 trimethylation. First, AtRRP6Ls could participate in the H3K4 demethylation pathway via regulating synthesis of AS I and AS II. Second, AtRRP6L proteins could function in the H3K27 trimethylation pathway via regulating the synthesis of ASL. However, more work will be needed to untangle the interrelationships of the different lncRNAs and the roles they play in the epigenetic architecture at FLC. How AtRRP6L proteins and the ncRNAs controlled by them help recruit chromatin modifiers to modulate silencing by affecting histone modifications that repress transcription remains an intriguing topic for future work.
Materials and Methods
Plant materials and growth conditions
The atrrp6l1-3 allele was isolated from the BASTA population from the University of Wisconsin [31]; the atrrp6l1-1, atrrp6l2-2, atrrp6l2-3, atrrp6l2-4 and atrrp6l3-1 alleles correspond to SALK_004432, SALK_113786, SALK_011429, and SALK_149898, and SALK_122492, respectively. iRNAi lines of RRP41, csl4-2, RNA Pol IV (SALK_128428, nrpd1a-3, nrpd1-3), RNA Pol V (SALK_029919, nrpd1b-11, nrpe1-11) mutants were described previously [8], [74], [75]. All Salk alleles are in the Col-0 ecotype and the University of Wisconsin alleles are in the Ws ecotype. The RNAi-mediated knockdown of RRP41 was induced by germinating and growing seedlings on ½× MS plates containing 8 mM 17β-estradiol, following a previously published method [8].
Long day and short day conditions for plant growth were 16 hours light/8 hours dark and 8 hours light/16 hours dark, respectively. Flowering time was measured by counting rosette leaf number at the time of flowering [76].
Transgenic lines and vector construction
For chromatin immunoprecipitation (see below), we used the Tandem Affinity Purification (TAP) affinity tag to selectively precipitate RRP6L1 by expressing a RRP6L1 - TAP fusion protein. For construction of plant lines with affinity-tagged RRP6L1 for RNA-IP and ChIP, we complemented atrrp6l1-3 with the RRP6L1-TAP transgene. For RRP6L1-TAP complementation, the entire genomic region of RRP6L1 including 1.5 kb upstream from the ATG codon was amplified by PCR using LA taq polymerase (Takara) and cloned into TAP-tag carrying destination vector pDB1008 [8]. For complementation with the TAP tagged transgene, the homozygous atrrp6l1-3 mutant was transformed using Agrobacterium-mediated transformation and the progeny plants containing both the T-DNA insertion allele and the transgene were identified by PCR.
RNA analysis
Trizol reagent (Invitrogen) was used to isolate total RNA from seedlings. 10-, 14-, 18-, and 21-day-old seedlings were used for examining the expression of ASL. 11-day-old seedlings were used for investigating expression of genes and antisense RNAs examined in our study. For RT-qPCR, 2–4 µg of total RNA was digested with DNase I (Fermentas) and reverse transcribed for one hour at 42°C (oligo-dT primers) or at 50°C (gene-specific primers), with 100 units of PrimeScript reverse transcriptase (Takara). RT-qPCR (MyiQ-iCycler; Bio-Rad) was used to quantify transcripts using the comparative threshold cycle method (ΔΔCt, Table S1 shows primer sequences), with ACTIN 7 (At5g09810) as an internal reference.
For tiling RT-PCR, sets of serial primers were designed in intervals of 100–200 nt. After obtaining the full-length ASL transcript, another set of overlapping primers was designed to make sure the 3′ and 5′-ends of the RNA have been isolated. The PCR products amplified by tiling RT-PCR were cloned into the pBluescript KS vector and sequenced using T7 and T3 primers.
Chromatin Immunoprecipitation (ChIP) assays
ChIP was conducted following a previously-described method [77]. Each experiment used 1.5 grams of tissue from 11-day-old seedlings. All ChIP experiments used at least two biological replicates and at least two technical replicates. Anti-H3K4me3 (07-473) and anti-H3K27me3 (ab6002) were purchased from Millipore and Abcam, respectively. IgG Sepharose 6 Fast Flow (GE Healthcare) was used for ChIP using RRP6L1-TAP tagged line. The mock antibody control used an equal amount of chromatin that was not treated with antibody. The ChIPed DNA was purified using PCR purification kit (Fermentas) and qPCR was performed. Supplemental Table S1 lists the primers used for PCR.
Micrococcal nuclease (MNase) ChIP
MNase-ChIP was performed following a previously-described method [78]. Two grams of tissue from 11-day-old seedlings was fixed using 1% formaldehyde solution for 10 min and washed with distilled water several times. The fixed samples were homogenized with HONDA buffer (20 mM HEPES-KOH pH 7.4, 0.44 M sucrose, 1.25% Ficoll, 2.5% Dextran T40, 10 mM MgCl2, 0.5% Triton X-100, 5 mM DTT, 1 mM PMSF, 1% plant protease inhibitors) and then filtered through miracloth. After isolation of the nucleus-containing fraction by centrifugation, the fraction was treated with MNase (NEB) at 37°C for 10 min. Anti-histone H3 (ab1791) was used for the ChIP. The purification of ChIPed DNA and qPCR was performed as described in ChIP assay.
RNA Immunoprecipitation (RNA-IP)
RNA-IP assays were performed as described previously [31], [79]. Two grams of tissue from seedlings at 11-days-old was fixed with 1% formaldehyde. For purification of RRP6L1-TAP or RRP6L1 RNA complexes, the chromatin was incubated with prewashed IgG Sepharose 6 Fast Flow (GE Healthcare) or with polyclonal anti-AtRRP6L1 antibodies, respectively, at 4°C overnight. H3K27me3-RNA complex purification was performed using anti-H3K27me3 (ab6002) overnight following by incubation with protein A agarose beads. Immunoprecipitated RNA purification used phenol∶chloroform and PrimeScript reverse transcriptase (Takara) and sequence specific primers were used for cDNA synthesis. Supplemental Table S1 lists the primers used for PCR.
Alpha-amanitin treatment
Eleven-day-old seedlings were treated with 5 µM α-amanitin (Sigma) for 0, 6 and 9 h or 17 h. After RNA extraction, cDNA was synthesized using ASL-specific and ACTIN 7 primers, followed by qRT-PCR. The level of ASL transcript was normalized relative to the level of ACTIN 7 transcript.
Terminator 5′-Phosphate-Dependent Exonuclease (TPE) treatment
Total RNA extracted from 11-day-old seedlings was treated with TPE (Epicentre) at 42°C, and purified with phenol∶chloroform. Complementary cDNA was synthesized using RNA sequence specific primers followed by RT-qPCR.
Supporting Information
Zdroje
1. MarcheseFP, HuarteM (2014) Long non-coding RNAs and chromatin modifiers: Their place in the epigenetic code. Epigenetics 9 : 21–26 doi:10.4161/epi.27472
2. CedarH, BergmanY (2009) Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet 10 : 295–304 doi:10.1038/nrg2540
3. MatzkeMA, BirchlerJA (2005) RNAi-mediated pathways in the nucleus. Nat Rev Genet 6 : 24–35 doi:10.1038/nrg1500
4. BühlerM, HaasW, GygiSP, MoazedD (2007) RNAi-Dependent and -Independent RNA Turnover Mechanisms Contribute to Heterochromatic Gene Silencing. Cell 129 : 707–721 doi:10.1016/j.cell.2007.03.038
5. MitchellP, EP, ShevchenkoA, MannM, TollerveyD (1997) The Exosome: A Conserved Eukaryotic RNA Processing Complex Containing Multiple 3″→5″ Exoribonucleases. Cell 91 : 457–466.
6. BurkardKTD, ButlerJS (2000) A Nuclear 3″-5″ Exonuclease Involved in mRNA Degradation Interacts with Poly(A) Polymerase and the hnRNA Protein Npl3p. Molecular and Cellular Biology 20 : 604–616 doi:10.1128/MCB.20.2.604-616.2000
7. ChekanovaJA, ShawRJ, WillsMA, BelostotskyDA (2000) Poly(A) Tail-dependent Exonuclease AtRrp41p from Arabidopsis thaliana Rescues 5.8 S rRNA Processing and mRNA Decay Defects of the Yeast ski6 Mutant and Is Found in an Exosome-sized Complex in Plant and Yeast Cells. Journal of Biological Chemistry 275 : 33158–33166 doi:10.1074/jbc.M005493200
8. ChekanovaJA, GregoryBD, ReverdattoSV, ChenH, KumarR, et al. (2007) Genome-Wide High-Resolution Mapping of Exosome Substrates Reveals Hidden Features in the Arabidopsis Transcriptome. Cell 131 : 1340–1353 doi:10.1016/j.cell.2007.10.056
9. BelostotskyDA, SieburthLE (2009) Kill the messenger: mRNA decay and plant development. Current Opinion in Plant Biology 12 : 96–102 doi:10.1016/j.pbi.2008.09.003
10. LorentzenE, WalterP, FribourgS, Evguenieva-HackenbergE, KlugG, et al. (2005) The archaeal exosome core is a hexameric ring structure with three catalytic subunits. Nat Struct Mol Biol 12 : 575–581 doi:10.1038/nsmb952
11. BelostotskyD (2009) Exosome complex and pervasive transcription in eukaryotic genomes. Current Opinion in Cell Biology 21 : 352–358 doi:10.1016/j.ceb.2009.04.011
12. JanuszykK, LimaCD (2011) STRUCTURAL COMPONENTS AND ARCHITECTURES OF RNA EXOSOMES. Adv Exp Med Biol 702 : 9–28.
13. ChlebowskiA, TomeckiR, Gas LópezME, SéraphinB, DziembowskiA (2011) CATALYTIC PROPERTIES OF THE EUKARYOTIC EXOSOME. Adv Exp Med Biol 702 : 63–78.
14. BriggsMW, BurkardKTD, ButlerJS (1998) Rrp6p, the Yeast Homologue of the Human PM-Scl 100-kDa Autoantigen, Is Essential for Efficient 5.8 S rRNA 3′ End Formation. Journal of Biological Chemistry 273 : 13255–13263.
15. AllmangC, KufelJ, ChanfreauG, MitchellP, EP, et al. (1999) Functions of the exosome in rRNA, snoRNA and snRNA synthesis. The EMBO Journal 18 : 15399–54102.
16. ButlerJS, MitchellP (2011) Rrp6, Rrp47 AND COFACTORS OF THE NUCLEAR EXOSOME. Adv Exp Med Biol 702 : 91–104.
17. CallahanKP, ButlerJS (2008) Evidence for core exosome independent function of the nuclear exoribonuclease Rrp6p. Nucleic Acids Research 36 : 6645–6655 doi:10.1093/nar/gkn743
18. GrahamAC, KissDL, AndrulisED (2009) Core Exosome-independent Roles for Rrp6 in Cell Cycle Progression. Molecular Biology of the Cell 20 : 2242–2253 doi:10.1091/mbc.E08
19. KissDL, AndrulisED (2010) Genome-wide analysis reveals distinct substrate specificities of Rrp6, Dis3, and core exosome subunits. RNA 16 : 781–791 doi:10.1261/rna.1906710
20. LangeH, HolecS, CognatV, PieuchotL, Le RetM, et al. (2008) Degradation of a Polyadenylated rRNA Maturation By-Product Involves One of the Three RRP6-Like Proteins in Arabidopsis thaliana. Molecular and Cellular Biology 28 : 3038–3044 doi:10.1128/MCB.02064-07
21. CamblongJ, IglesiasN, FickentscherC, DieppoisG, StutzF (2007) Antisense RNA Stabilization Induces Transcriptional Gene Silencing via Histone Deacetylation in S. cerevisiae. Cell 131 : 706–717 doi:10.1016/j.cell.2007.09.014
22. VasiljevaL, KimM, TerziN, SoaresLM, BuratowskiS (2008) Transcription Termination and RNA Degradation Contribute to Silencing of RNA Polymerase II Transcription within Heterochromatin. Molecular Cell 29 : 313–323 doi:10.1016/j.molcel.2008.01.011
23. ZofallM, FischerT, ZhangK, ZhouM, CuiB, et al. (2009) Histone H2A.Z cooperates with RNAi and heterochromatin factors to suppress antisense RNAs. Nature 461 : 419–422 doi:10.1038/nature08321
24. ZofallM, YamanakaS, Reyes-TurcuFE, ZhangK, RubinC, et al. (2012) RNA Elimination Machinery Targeting Meiotic mRNAs Promotes Facultative Heterochromatin Formation. Science 335 : 96–100 doi:10.1126/science.1211651
25. BühlerM, SpiesN, BartelDP, MoazedD (2008) TRAMP-mediated RNA surveillance prevents spurious entry of RNAs into the Schizosaccharomyces pombe siRNA pathway. Nat Struct Mol Biol 15 : 1015–1023 doi:10.1038/nsmb.1481
26. BernardP, DrogatJ, DheurS, GenierS, JaverzatJP (2010) Splicing Factor Spf30 Assists Exosome-Mediated Gene Silencing in Fission Yeast. Molecular and Cellular Biology 30 : 1145–1157 doi:10.1128/MCB.01317-09
27. Reyes-TurcuFE, ZhangK, ZofallM, ChenE, GrewalSIS (2011) Defects in RNA quality control factors reveal RNAi-independent nucleation of heterochromatin. Nat Struct Mol Biol 18 : 1132–1138 doi:10.1038/nsmb.2122
28. UhlerJP, HertelC, SvejstrupJQ (2007) A role for noncoding transcription in activation of the yeast PHO5 gene. PNAS 104 : 8011–8016.
29. HouseleyJ, KotovicK, HageAE, DT (2007) Trf4 targets ncRNAs from telomeric and rDNA spacer regions and functions in rDNA copy number control. The EMBO Journal 26 : 4996–5006.
30. CamblongJ, BeyrouthyN, GuffantiE, SchlaepferG, SteinmetzLM, et al. (2009) Trans-acting antisense RNAs mediate transcriptional gene cosuppression in S. cerevisiae. Genes & Development 23 : 1534–1545 doi:10.1101/gad.522509
31. ShinJ-H, WangH-LV, LeeJ, DinwiddieBL, BelostotskyDA, et al. (2013) The Role of the Arabidopsis Exosome in siRNA–Independent Silencing of Heterochromatic Loci. PLoS Genet 9: e1003411 doi:10.1371/journal.pgen.1003411.s007
32. MichaelsSD, AmazinoRM (1999) FLOWERING LOCUS C Encodes a Novel MADS Domain Protein That Acts as a Repressor of Flowering. The Plant Cell 11 : 949–956.
33. SheldonCC, RouseDT, FinneganEJ, PeacockWJ, DennisES (2000) The molecular basis of vernalization: The central role of FLOWERING LOCUS C (FLC). PNAS 97 : 3753–3758.
34. ShindoC, AranzanaMJ, ListerC, BaxterC, NichollsC, et al. (2005) Role of FRIGIDA and FLOWERING LOCUS C in Determining Variation in Flowering Time of Arabidopsis. PLANT PHYSIOLOGY 138 : 1163–1173 doi:10.1104/pp.105.061309
35. AmasinoRM, MichaelsSD (2010) The Timing of Flowering. PLANT PHYSIOLOGY 154 : 516–520 doi:10.1104/pp.110.161653
36. HeY (2012) Chromatin regulation of flowering. Trends in Plant Science 17 : 556–562 doi:10.1016/j.tplants.2012.05.001
37. IetswaartR, WuZ, DeanC (2012) Flowering time control: another window to the connection between antisense RNA and chromatin. Trends in Genetics 28 : 445–453 doi:10.1016/j.tig.2012.06.002
38. LiuF, MarquardtS, ListerC, SwiezewskiS, DeanC (2010) Targeted 3′ Processing of Antisense Transcripts Triggers Arabidopsis FLC Chromatin Silencing. Science 327 : 94–97 doi:10.1126/science.1180278
39. LiuF, QuesadaV, CrevillénP, BäurleI, SwiezewskiS, et al. (2007) The Arabidopsis RNA-Binding Protein FCA Requires a Lysine-Specific Demethylase 1 Homolog to Downregulate FLC. Molecular Cell 28 : 398–407 doi:10.1016/j.molcel.2007.10.018
40. HeoJB, SungS (2011) Vernalization-Mediated Epigenetic Silencing by a Long Intronic Noncoding RNA. Science 331 : 76–79 doi:10.1126/science.1197349
41. KimD-H, SungS (2012) Environmentally coordinated epigenetic silencing of FLC by protein and long noncoding RNA components. Current Opinion in Plant Biology 15 : 51–56 doi:10.1016/j.pbi.2011.10.004
42. SwiezewskiS, LiuF, MagusinA, DeanC (2009) Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature 462 : 799–802 doi:10.1038/nature08618
43. HelliwellCA, RobertsonM, FinneganEJ, BuzasDM, DennisES (2011) Vernalization-Repression of Arabidopsis FLC Requires Promoter Sequences but Not Antisense Transcripts. PLoS ONE 6: e21513 doi:10.1371/journal.pone.0021513.s003
44. HeY, MichaeliSD, AmasinoRM (2003) Regulation of Flowering Time by Histone Acetylation in Arabidopsis. Science 302 : 1751–1754 doi:10.1126/science.1091109
45. SimpsonGG, PPD, QuesadaV, HendersonI, DeanC (2003) FY Is an RNA 3′ End-Processing Factor that Interacts with FCA to Control the Arabidopsis Floral Transition. Cell 113 : 777–787.
46. HornyikC, TerziLC, SimpsonGG (2010) The Spen Family Protein FPA Controls Alternative Cleavage and Polyadenylation of RNA. DEVCEL 18 : 203–213 doi:10.1016/j.devcel.2009.12.009
47. LeeI, MichaeliSD, MassherdtAS, AmazinoRM (1994) The late-flowering phenotype of FRIGIDA and mutations in LUMINIDEPENDENS is suppressed in the Ladsberg erecta strain of Arabidopsis. The Plant Journal 6 : 903–909.
48. KuaiL, FangF, ButlerJS, ShermanF (2004) Polyadenylation of rRNA in Saccharomyces cerevisiae. PNAS 101 : 8581–8586.
49. VasiljevaL, BuratowskiS (2006) Nrd1 Interacts with the Nuclear Exosome for 3′ Processing of RNA Polymerase II Transcripts. Molecular Cell 21 : 239–248 doi:10.1016/j.molcel.2005.11.028
50. CastelnuovoM, RahmanS, GuffantiE, InfantinoV, StutzF, et al. (2013) Bimodal expression of PHO84 is modulated by early termination of antisense transcription. Nat Struct Mol Biol 20 : 851–858 doi:10.1038/nsmb.2598
51. SheldonCC, ConnAB, ESD, PeacockWJ (2002) Different Regulatory Regions Are Required for the Vernalization-Induced Repression of FLOWERING LOCUS C and for the Epigenetic Maintenance of Repression. THE PLANT CELL ONLINE 14 : 2527–2537 doi:10.1105/tpc.004564
52. KannoT, HuettelB, MetteMF, AufsatzW, JaligotE, et al. (2005) Atypical RNA polymerase subunits required for RNA-directed DNA methylation. Nat Genet 37 : 761–765 doi:10.1038/ng1580
53. WierzbickiAT, HaagJR, PikaardCS (2008) Noncoding Transcription by RNA Polymerase Pol IVb/Pol V Mediates Transcriptional Silencing of Overlapping and Adjacent Genes. Cell 135 : 635–648 doi:10.1016/j.cell.2008.09.035
54. MatzkeM, KannoT, DaxingerL, HuettelB, MatzkeAJ (2009) RNA-mediated chromatin-based silencing in plants. Current Opinion in Cell Biology 21 : 367–376 Available: message:%3CA233D938F69F5A49AF9FECB3A02B40AF5DBA82C5@UM-MBX-T01.um.umsystem.edu%3E.
55. HaagJR, PikaardCS (2011) Multisubunit RNA polymerases IV and V: purveyors of non-coding RNA for plant gene silencing. Nature Publishing Group 12 : 483–492 doi:10.1038/nrm3152
56. JohnssonP, AckleyA, VidarsdottirL, LuiW-O, CorcoranM, et al. (2013) A pseudogene long-noncoding-RNA network regulates PTEN transcription and translation in human cells. Nat Struct Mol Biol 20 : 440–446 doi:10.1038/nsmb.2516
57. LardenoisA, LiuY, WaltherT, ChalmelF, EvrardB, et al. (2011) Execution of the meiotic noncoding RNA expressionprogram and the onset of gametogenesis in yeastrequire the conserved exosome subunit Rrp6. PNAS 108 : 1058–1063 doi:10.1073/pnas.1016459108/-/DCSupplemental/pnas.201016459SI.pdf
58. DerkachevaM, SteinbachY, WildhaberT, aacuteIM, MahrezW, et al. (2013) Arabidopsis MSI1 connects LHP1 to PRC2 complexes. The EMBO Journal 32 : 2073–2085 doi:10.1038/emboj.2013.145
59. FrancisNJ (2004) Chromatin Compaction by a Polycomb Group Protein Complex. Science 306 : 1574–1577 doi:10.1126/science.1100576
60. YuanW, WuT, FuH, DaiC, WuH, et al. (2012) Dense Chromatin Activates Polycomb Repressive Complex 2 to Regulate H3 Lysine 27 Methylation. Science 337 : 971–975 doi:10.1126/science.1225237
61. AllmangC, EP, PodtelejnikovA, MannM, TollerveyD, et al. (1999) The yeast exosome and human PM–Scl are related complexes of 3′→5′ exonucleases. Genes & Development 13 : 2148–2158.
62. SugiyamaT, Sugioka-SugiyamaR (2011) Red1 promotes the elimination of meiosis-specific mRNAs in vegetatively growing fission yeast. The EMBO Journal 30 : 1027–1039 doi:10.1038/emboj.2011.32
63. GrzechnikP, KufelJ (2008) Polyadenylation Linked to Transcription Termination Directs the Processingof snoRNA Precursors in Yeast. Molecular Cell 32 : 247–258 doi:10.1016/j.molcel.2008.10.003
64. SchmidM, JensenTH (2008) The exosome: a multipurpose RNA-decay machine. Trends in Biochemical Sciences 33 : 501–510 doi:10.1016/j.tibs.2008.07.003
65. PienS, FleuryD, MylneJS, CrevillenP, InzeD, et al. (2008) ARABIDOPSIS TRITHORAX1 Dynamically Regulates FLOWERING LOCUS C Activation via Histone 3 Lysine 4 Trimethylation. The Plant Cell 20 : 580–588 doi:10.1105/tpc.108.058172
66. DavidovichC, ZhengL, GoodrichKJ, CechTR (2013) Promiscuous RNA binding by Polycomb repressive complex 2. Nat Struct Mol Biol 20 : 1250–1257 doi:10.1038/nsmb.2679
67. KanekoS, SonJ, ShenSS, ReinbergD, BonasioR (2013) PRC2 binds active promoters and contacts nascent RNAs in embryonic stem cells. Nat Struct Mol Biol 20 : 1258–1264 doi:10.1038/nsmb.2700
68. SteinmetzEJ, ConradNK, BrowDA, CordenJL (2001) RNA-binding protein Nrd1 directs poly(A)-independent 3′-end formation of RNA polymerase II transcripts. Nature 413 : 327–331 doi:10.1038/35095090
69. CiaudoC, BourdetA, Cohen-TannoudjiM, DietzHC, RougeulleC, et al. (2006) Nuclear mRNA Degradation Pathway(s) Are Implicated in Xist Regulation and X Chromosome Inactivation. PLoS Genet 2: e94 doi:10.1371/journal.pgen.0020094.sg001
70. HaimovichG, MedinaDA, CausseSZ, GarberM, Millán-ZambranoG, et al. (2013) Gene Expression Is Circular: Factors for mRNA Degradation Also Foster mRNA Synthesis. Cell 153 : 1000–1011 doi:10.1016/j.cell.2013.05.012
71. NagarajanVK, JonesCI, NewburySF, GreenPJ (2013) XRN 5′→3′ exoribonucleases: Structure, mechanisms and functions. Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms 1829 : 590–603 doi:10.1016/j.bbagrm.2013.03.005
72. van DijkEL, ChenCL, d'Aubenton-CarafaY, GourvennecS, KwapiszM, et al. (2011) XUTs are a class of Xrn1-sensitive antisense regulatory non-coding RNA in yeast. Nature 475 : 114–117 doi:10.1038/nature10118
73. BastowR, MylneJS, ListerC, LippmanZ, MartienssenRA, et al. (2004) Vernalization requires epigeneticsilencing of FLC by histonemethylation. Nature 427 : 164–167.
74. HerrAJ, JensenMB, DalmayT, BaulcombeDC (2005) RNA Polymerase IV Directs Silencing of Endogenous DNA. Science 308 : 118–120 doi:10.1126/science.1106910
75. PontierD, YahubyanG, VegaD, BulskiA, Saez-VasquezJ, et al. (2005) Reinforcement of silencing at transposons and highly repeated sequences requires the concerted action of two distinct RNA polymerases IV in Arabidopsis. Genes & Development 19 : 2030–2040 doi:10.1101/gad.348405
76. KoornneefM, HanhartCJ, van der VeenJH (1991) A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol Gen Genet 229 : 57–66.
77. MorohashiK, XieZ, GrotewoldE (2009) Gene-specific and genome-wide ChIP approaches to study plant transcriptional networks. Methods Mol Biol 553 : 3–12 doi:_10.1007/978-1-60327-563-7_1
78. ZhuY, RowleyMJ, BöhmdorferG, WierzbickiAT (2013) A SWI/SNF Chromatin-Remodeling Complex Actsin Noncoding RNA-Mediated Transcriptional Silencing. Molecular Cell 49 : 298–309 doi:10.1016/j.molcel.2012.11.011
79. TerziLC, SimpsonGG (2009) Arabidopsis RNA immunoprecipitation. The Plant Journal 59 : 163–168 doi:10.1111/j.1365-313X.2009.03859.x
Štítky
Genetika Reprodukční medicína
Článek An Evolutionarily Conserved Role for the Aryl Hydrocarbon Receptor in the Regulation of MovementČlánek Requirement for Drosophila SNMP1 for Rapid Activation and Termination of Pheromone-Induced ActivityČlánek Co-regulated Transcripts Associated to Cooperating eSNPs Define Bi-fan Motifs in Human Gene NetworksČlánek Identification of a Regulatory Variant That Binds FOXA1 and FOXA2 at the Type 2 Diabetes GWAS LocusČlánek tRNA Modifying Enzymes, NSUN2 and METTL1, Determine Sensitivity to 5-Fluorouracil in HeLa CellsČlánek Derlin-1 Regulates Mutant VCP-Linked Pathogenesis and Endoplasmic Reticulum Stress-Induced ApoptosisČlánek A Genetic Assay for Transcription Errors Reveals Multilayer Control of RNA Polymerase II FidelityČlánek The Proprotein Convertase KPC-1/Furin Controls Branching and Self-avoidance of Sensory Dendrites inČlánek Regulation of p53 and Rb Links the Alternative NF-κB Pathway to EZH2 Expression and Cell SenescenceČlánek BMPs Regulate Gene Expression in the Dorsal Neuroectoderm of and Vertebrates by Distinct MechanismsČlánek Unkempt Is Negatively Regulated by mTOR and Uncouples Neuronal Differentiation from Growth Control
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 9
-
Všechny články tohoto čísla
- Translational Regulation of the Post-Translational Circadian Mechanism
- An Evolutionarily Conserved Role for the Aryl Hydrocarbon Receptor in the Regulation of Movement
- Eliminating Both Canonical and Short-Patch Mismatch Repair in Suggests a New Meiotic Recombination Model
- Requirement for Drosophila SNMP1 for Rapid Activation and Termination of Pheromone-Induced Activity
- Co-regulated Transcripts Associated to Cooperating eSNPs Define Bi-fan Motifs in Human Gene Networks
- Targeted H3R26 Deimination Specifically Facilitates Estrogen Receptor Binding by Modifying Nucleosome Structure
- Role for Circadian Clock Genes in Seasonal Timing: Testing the Bünning Hypothesis
- The Tandem Repeats Enabling Reversible Switching between the Two Phases of β-Lactamase Substrate Spectrum
- The Association of the Vanin-1 N131S Variant with Blood Pressure Is Mediated by Endoplasmic Reticulum-Associated Degradation and Loss of Function
- Identification of a Regulatory Variant That Binds FOXA1 and FOXA2 at the Type 2 Diabetes GWAS Locus
- Regulation of Flowering by the Histone Mark Readers MRG1/2 via Interaction with CONSTANS to Modulate Expression
- The Actomyosin Machinery Is Required for Retinal Lumen Formation
- Plays a Conserved Role in Assembly of the Ciliary Motile Apparatus
- Hidden Diversity in Honey Bee Gut Symbionts Detected by Single-Cell Genomics
- Ribosome Rescue and Translation Termination at Non-Standard Stop Codons by ICT1 in Mammalian Mitochondria
- tRNA Modifying Enzymes, NSUN2 and METTL1, Determine Sensitivity to 5-Fluorouracil in HeLa Cells
- Causal Variation in Yeast Sporulation Tends to Reside in a Pathway Bottleneck
- Tissue-Specific RNA Expression Marks Distant-Acting Developmental Enhancers
- WC-1 Recruits SWI/SNF to Remodel and Initiate a Circadian Cycle
- Clonal Expansion of Early to Mid-Life Mitochondrial DNA Point Mutations Drives Mitochondrial Dysfunction during Human Ageing
- Methylation QTLs Are Associated with Coordinated Changes in Transcription Factor Binding, Histone Modifications, and Gene Expression Levels
- Differential Management of the Replication Terminus Regions of the Two Chromosomes during Cell Division
- Obesity-Linked Homologues and Establish Meal Frequency in
- Derlin-1 Regulates Mutant VCP-Linked Pathogenesis and Endoplasmic Reticulum Stress-Induced Apoptosis
- Stress-Induced Nuclear RNA Degradation Pathways Regulate Yeast Bromodomain Factor 2 to Promote Cell Survival
- The MAPK p38c Regulates Oxidative Stress and Lipid Homeostasis in the Intestine
- Widespread Genome Reorganization of an Obligate Virus Mutualist
- Trans-kingdom Cross-Talk: Small RNAs on the Move
- The Vip1 Inositol Polyphosphate Kinase Family Regulates Polarized Growth and Modulates the Microtubule Cytoskeleton in Fungi
- Myosin Vb Mediated Plasma Membrane Homeostasis Regulates Peridermal Cell Size and Maintains Tissue Homeostasis in the Zebrafish Epidermis
- GLD-4-Mediated Translational Activation Regulates the Size of the Proliferative Germ Cell Pool in the Adult Germ Line
- Genome Wide Association Studies Using a New Nonparametric Model Reveal the Genetic Architecture of 17 Agronomic Traits in an Enlarged Maize Association Panel
- Translational Regulation of the DOUBLETIME/CKIδ/ε Kinase by LARK Contributes to Circadian Period Modulation
- Positive Selection and Multiple Losses of the LINE-1-Derived Gene in Mammals Suggest a Dual Role in Genome Defense and Pluripotency
- Out of Balance: R-loops in Human Disease
- A Genetic Assay for Transcription Errors Reveals Multilayer Control of RNA Polymerase II Fidelity
- Altered Behavioral Performance and Live Imaging of Circuit-Specific Neural Deficiencies in a Zebrafish Model for Psychomotor Retardation
- Nipbl and Mediator Cooperatively Regulate Gene Expression to Control Limb Development
- Meta-analysis of Mutations in Autism Spectrum Disorders: A Gradient of Severity in Cognitive Impairments
- The Proprotein Convertase KPC-1/Furin Controls Branching and Self-avoidance of Sensory Dendrites in
- Hydroxymethylated Cytosines Are Associated with Elevated C to G Transversion Rates
- Memory and Fitness Optimization of Bacteria under Fluctuating Environments
- Regulation of p53 and Rb Links the Alternative NF-κB Pathway to EZH2 Expression and Cell Senescence
- Interspecific Tests of Allelism Reveal the Evolutionary Timing and Pattern of Accumulation of Reproductive Isolation Mutations
- PRO40 Is a Scaffold Protein of the Cell Wall Integrity Pathway, Linking the MAP Kinase Module to the Upstream Activator Protein Kinase C
- Low Levels of p53 Protein and Chromatin Silencing of p53 Target Genes Repress Apoptosis in Endocycling Cells
- SPDEF Inhibits Prostate Carcinogenesis by Disrupting a Positive Feedback Loop in Regulation of the Foxm1 Oncogene
- RRP6L1 and RRP6L2 Function in Silencing Regulation of Antisense RNA Synthesis
- BMPs Regulate Gene Expression in the Dorsal Neuroectoderm of and Vertebrates by Distinct Mechanisms
- Unkempt Is Negatively Regulated by mTOR and Uncouples Neuronal Differentiation from Growth Control
- Atkinesin-13A Modulates Cell-Wall Synthesis and Cell Expansion in via the THESEUS1 Pathway
- Dopamine Signaling Leads to Loss of Polycomb Repression and Aberrant Gene Activation in Experimental Parkinsonism
- Histone Methyltransferase MMSET/NSD2 Alters EZH2 Binding and Reprograms the Myeloma Epigenome through Global and Focal Changes in H3K36 and H3K27 Methylation
- Bipartite Recognition of DNA by TCF/Pangolin Is Remarkably Flexible and Contributes to Transcriptional Responsiveness and Tissue Specificity of Wingless Signaling
- The Olfactory Transcriptomes of Mice
- Muscular Dystrophy-Associated and Variants Disrupt Nuclear-Cytoskeletal Connections and Myonuclear Organization
- Interplay of dFOXO and Two ETS-Family Transcription Factors Determines Lifespan in
- Evidence for Widespread Positive and Negative Selection in Coding and Conserved Noncoding Regions of
- Genome-Wide Association Meta-analysis of Neuropathologic Features of Alzheimer's Disease and Related Dementias
- Rejuvenation of Meiotic Cohesion in Oocytes during Prophase I Is Required for Chiasma Maintenance and Accurate Chromosome Segregation
- Admixture in Latin America: Geographic Structure, Phenotypic Diversity and Self-Perception of Ancestry Based on 7,342 Individuals
- Local Effect of Enhancer of Zeste-Like Reveals Cooperation of Epigenetic and -Acting Determinants for Zygotic Genome Rearrangements
- Differential Responses to Wnt and PCP Disruption Predict Expression and Developmental Function of Conserved and Novel Genes in a Cnidarian
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Admixture in Latin America: Geographic Structure, Phenotypic Diversity and Self-Perception of Ancestry Based on 7,342 Individuals
- Nipbl and Mediator Cooperatively Regulate Gene Expression to Control Limb Development
- Genome Wide Association Studies Using a New Nonparametric Model Reveal the Genetic Architecture of 17 Agronomic Traits in an Enlarged Maize Association Panel
- Histone Methyltransferase MMSET/NSD2 Alters EZH2 Binding and Reprograms the Myeloma Epigenome through Global and Focal Changes in H3K36 and H3K27 Methylation
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



